Abstract
Current anti–hepatitis B virus (HBV) therapies have little effect on covalently closed circular DNA (cccDNA) and fail to eliminate HBV. The clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 system has been reported to directly target cccDNA and exert antiviral effects. In this study, we hypothesized that the inhibition of the DNA repair machinery, which is important for the repair of CRISPR‐induced double‐strand breaks, may enhance the effect of CRISPR targeting cccDNA, and we investigated the antiviral effect of potential combination therapy. The antiviral effect of CRISPR targeting cccDNA (HBV‐CRISPR) was evaluated in HBV‐susceptible HepG2‐hNTCP‐C4 cells expressing Cas9 (HepG2‐hNTCP‐C4‐iCas9) or primary human hepatocytes (PHHs) expressing Cas9. Following HBV infection, HBV‐CRISPR reduced cccDNA levels, accompanied by decreases in pregenomic RNA (pgRNA) levels and supernatant HBV DNA, hepatitis B surface antigen and hepatitis B e antigen levels in HepG2‐hNTCP‐C4‐iCas9 cells, and PHHs. HBV‐CRISPR induced indel formation in cccDNA and up‐regulated poly(adenosine diphosphate ribose) polymerase (PARP) activity in HBV‐infected HepG2‐hNTCP‐C4‐iCas9 cells. The suppression of PARP2‐Histone PARylation factor 1 (HPF1) (involved in the initial step of DNA repair) with small interfering RNA (siRNA) targeting either PARP2 or HPF1 increased the reduction in pgRNA and cccDNA by HBV‐CRISPR in HBV‐infected HepG2‐hNTCP‐C4‐iCas9 cells. The suppression of DNA Ligase 4 (LIG4) (essential for nonhomologous end joining [NHEJ]) but not breast cancer susceptibility gene (BRCA) (essential for homologous recombination) enhanced the antiviral effect of HBV‐CRISPR in HBV‐infected HepG2‐hNTCP‐C4‐iCas9 cells. Finally, the clinically available PARP inhibitor olaparib increased the reductions in pgRNA and cccDNA levels induced by HBV‐CRISPR in HBV‐infected HepG2‐hNTCP‐C4‐iCas9 cells and PHHs. Conclusion: The suppression of the NHEJ‐mediated DNA repair machinery enhances the effect of CRISPR targeting cccDNA. The combination of CRISPR and olaparib may represent a therapy for HBV elimination.
The suppression of the NHEJ‐mediated DNA repair machinery enhances the effect of CRISPR targeting cccDNA. The combination of CRISPR and olaparib may represent a novel therapy for HBV elimination.
INTRODUCTION
Hepatitis B virus (HBV) is a 3.2‐kb incomplete double‐stranded DNA virus of the genus Hepadnaviridae.[ 1 ] It is estimated that there are approximately 250 million patients with chronic hepatitis B (CHB) patients worldwide.[ 2 ] Within 5 years, 8%–20% of patients with untreated CHB develop cirrhosis, and 2%–8% of patients with cirrhosis develop hepatocellular carcinoma every year.[ 3 , 4 ] HBV is a significant public health problem.
HBV forms covalently closed circular DNA (cccDNA), which serves as a template for HBV messenger RNA (mRNA), in infected hepatocytes. cccDNA is stable in the infected hepatocyte nucleus and imposes a therapeutic barrier. The current treatment options for CHB are either interferons (IFNs) or nucleos(t)ide analogs (NUCs). IFN exerts anti‐HBV effects through direct antiviral and indirect immunomodulatory effects. NUCs suppress HBV replication by inhibiting HBV polymerase.[ 3 , 5 ] IFN exerts some effect on cccDNA, but its effect is limited, and IFN treatment has strong side effects. NUCs can strongly suppress HBV replication, but they have little effect on cccDNA and present the risk of causing resistant viruses to emerge. Furthermore, when NUC treatment is discontinued, HBV reactivation may occur from existing cccDNA in hepatocytes.[ 6 ] Thus, it is difficult to eradicate cccDNA from infected hepatocytes with currently available antiviral therapies. Several potential new therapies, including capsid assembly modulators,[ 7 , 8 , 9 , 10 ] nucleic acid–based polymers, and antisense oligonucleotides tested in clinical trials, also do not directly target cccDNA. Therefore, further development of therapeutics targeting cccDNA is awaited.
Clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 technology introduces double‐strand breaks (DSBs) in arbitrary regions of the genome and edits genes. In addition to gene editing, the application of CRISPR/Cas9 to treat diseases such as hemophilia B[ 11 ] and hereditary tyrosinemia type 1[ 12 ] has recently been investigated. Gene therapy using CRISPR/Cas9 for the treatment of β‐thalassemia is already in clinical trials.[ 13 ] In HBV, it has been reported that the CRISPR/Cas9 system is capable of directly targeting cccDNA and exerting antiviral effects in HBV‐susceptible cells[ 14 , 15 , 16 ] and humanized liver chimeric mice.[ 17 ] However, its effects have been limited in previous studies and need to be increased for clinical application. When CRISPR/Cas9 introduces DSBs into double‐stranded DNA, the cleaved DNA is repaired by the DNA repair machinery via homologous recombination (HR) and nonhomologous end joining (NHEJ).[ 18 ] Therefore, we hypothesized that the antiviral effect of CRISPR/Cas9 targeting cccDNA would be amplified by the inhibition of the CRISPR‐induced DSB repair pathway.
Here, we showed that the suppression of the NHEJ‐mediated DNA repair machinery, but not the HR‐mediated machinery, increased the antiviral effect of CRISPR/Cas9 targeting cccDNA. We further demonstrated that the antiviral effect of CRISPR/Cas9 was increased by the inhibition of poly(adenosine diphosphate ribose) polymerase (PARP), which is important for the initial step of DNA repair, using the clinically applicable PARP inhibitor olaparib. Our study therefore provides a potential combination therapy targeting cccDNA to cure patients with CHB.
METHODS
Cell culture
Primary human hepatocytes (PHHs) were collected from humanized liver chimeric NOG‐TKm30 mice[ 19 ] using the two‐step collagenase–pronase liver perfusion method as previously reported.[ 20 ] The collected PHHs were seeded in 12‐well or 24‐well collagen type I‐coated microplates (AGC Techno Glass Co., Ltd.). The PHH culture medium was Dulbecco's modified Eagle's medium (DMEM) (Nacalai Tesque, Inc.) supplemented with 5 μg/ml L‐proline (Sigma‐Aldrich), 32.2 μg/ml 2‐phospho‐L‐ascorbic acid trisodium (Sigma‐Aldrich), 0.02 μg/ml dexamethasone (Sigma‐Aldrich), 0.005 μg/ml recombinant human epidermal growth factor (Sigma‐Aldrich), 25 μl/L human insulin solution (Sigma‐Aldrich), 20 mM 4‐(2‐hydroxyethyl)‐1‐piperazine ethanesulfonic acid (Nacalai Tesque, Inc.), 0.37% NaHCO3 (Nacalai Tesque, Inc.), 2% dimethyl sulfoxide (DMSO) (Nacalai Tesque, Inc.), 10% fetal bovine serum (FBS; Life Technologies), 100 units/ml penicillin, 100 μg/ml streptomycin, and 0.25 μg/ml amphotericin B (Life Technologies). The culture medium was changed on the indicated days. HepG2‐hNTCP‐C4‐iCas9 cells were maintained in DMEM/F‐12 (Life Technologies) containing G418 Disulfate Aqueous Solution and 10% FBS at 37°C under 5% CO2. Doxycycline (DOX) was administered at 1 μg/ml to induce Cas9.
HBV infection
The HBV genome is integrated in the genome of Hep38.7‐tet cells, and these cells release HBV genotype D in the culture supernatant.[ 21 , 22 ] Here, the culture supernatant of Hep38.7‐tet cells was collected and filtered through a 0.45‐μm filter (Merck Millipore). The collected culture supernatant was concentrated 30‐fold and used as the in vitro HBV inoculum. HepG2‐hNTCP‐C4‐iCas9 cells and PHHs were incubated with culture medium containing the HBV inoculum, 2% DMSO, and 4% polyethylene glycol 8000 (Promega) for 24 h. After incubation with the HBV inoculum, PHHs and HepG2‐hNTCP‐C4‐iCas9 cells were washed with phosphate‐buffered saline (PBS) containing 2% DMSO, and the culture medium was changed.
Plasmids
We selected a guide RNA (gRNA) targeting the HBV genome (gRNA21) with reference to a previous report[ 15 ] and designed control gRNAs targeting neither HBV nor the human genome. The gRNAs were cloned into a lentiviral gRNA/Cas9 expression vector (lentiCRISPR v2; addgene#52961) or a lentiviral gRNA expression vector (lentiGuide‐Puro; addgene#52963). HepG2‐hNTCP‐C4‐iCas9 cells were established through the transduction of a lentiviral vector expressing DOX‐inducible Cas9 (Lenti‐iCas9‐neo; Addgene #85400) into HepG2‐hNTCP‐C4 cells,[ 23 ] followed by green fluorescent protein selection with fluorescence‐assisted cell sorting.
Measurement of HBV DNA, hepatitis B surface antigen, and hepatitis B e antigen levels
HBV DNA levels were determined with a COBAS TaqMan HBV Test (Roche Diagnostics). Hepatitis B surface (HBs) and hepatitis B e( HBe) antigen levels were determined with a chemiluminescence enzyme immunoassay (CLEIA System; Fujirebio). The samples were diluted 10‐fold for measurement.
RNA isolation and real‐time polymerase chain reaction
RNA was isolated with the RNeasy Mini Kit (Qiagen). To analyze pregenomic RNA (pgRNA), DNase (RNase‐Free DNase Set; Qiagen) was used to remove the genomic DNA from the isolated RNA. The isolated RNA was reverse‐transcribed using ReverTra Ace qPCR RT Master Mix (Toyobo) to synthesize complementary DNA. The following primers were used in TaqMan gene expression assays for real‐time polymerase chain reaction (RT‐PCR): human PARP2 (Hs00193931_m1), human Histone PARylation factor 1 (HPF1) (Hs04188909_g1), human DNA Ligase 4 (LIG4) (Hs01866071_u1), human breast cancer susceptibility gene (BRCA) 1 (Hs01556193_m1), human BRCA2 (Hs00609073_m1), and human β‐actin (Hs99999903_m1). All primers used in the TaqMan gene‐expression assays were purchased from Life Technologies. The following forward and reverse primer set was prepared to analyze pgRNA: 5′‐TGTCCTACTGTTCAAGCCTCCAA‐3′ (forward) and 5′‐GAGAGTAACTCCACAGTAGCTCCAA‐3′ (reverse). The expression levels of all messenger RNAs (mRNAs) were normalized to the levels of the internal control mRNA (human β‐actin).
DNA isolation and real‐time PCR analysis of HBV cccDNA
DNA was extracted with the QIAamp DNA Mini Kit (Qiagen). The following primer set and probe were used to detect genotype D cccDNA: primers, 5′‐TCCCCGTCTGTGCCTTCTC‐3′ (1420–1438) and 5′‐GCACAGCTTGGAGGCTTGA‐3′ (1738–1756); probe, 5′‐FAM‐CCGTGTGCACTTCG‐3′ (1449–1462). The PCR protocol was previously described.[ 20 ] HBV cccDNA levels were normalized to the expression levels of human RNase P using the TaqMan RNase P Control Reagents Kit (#4316844; Life Technologies). DNA was treated with T5 exonuclease before PCR according to the manufacturer's instruction in some experiments, as specified in the Results.
Western blotting
The western blotting procedure has been previously described.[ 24 ] The cell protein elution solution was prepared using lysis buffer with a protease inhibitor (Nacalai Tesque, Inc.) and a phosphatase inhibitor (Nacalai Tesque, Inc.). We used the following antibodies for immunodetection: anti‐Cas9 (#14697; Cell Signaling Technology) and anti‐β‐actin (A5316; Sigma–Aldrich).
Transfection of small interfering RNA
Six small interfering RNAs (siRNAs) targeting human PARP2 (s19502), HPF1 (s276774), LIG4 (s8179), BRCA1 (s457), BRCA2 (s2083), and corresponding negative controls (#4390843) were purchased from Life Technologies. Transfections were performed using Lipofectamine RNAiMAX (Life Technologies).
Immunofluorescent staining
HepG2‐hNTCP‐iCas9 cells were seeded in chambered cover glasses (Matsunami Glass Ind., Ltd.). The cells were fixed with ice‐cold acetone‐methanol. The cells were blocked with PBS/0.2% bovine serum albumin. Hepatitis B core (HBc) antigen monoclonal antibodies were produced by immunizing BALB/c mice with recombinant HBc protein (clone 7B2, culture supernatant of the hybridoma) as previously described.[ 25 ] As a secondary antibody, a goat anti‐mouse antibody labeled with Alexa Fluor 594 (Cell Signaling Technology) was used. Cell nuclei were stained using 4′,6‐diamidino‐2‐phenylindole. Immunofluorescent staining was performed as previously described.[ 20 ] Stained cells were analyzed using a fluorescence microscope (Invitrogen EVOS FL Auto 2 Imaging System; Life Technologies).
Analysis of CRISPR‐induced mutations at the targeted locus with next‐generation sequencing
HBV‐infected HepG2‐hNTCP‐C4‐iCas9 cells were lentivirally transduced with gRNA targeting HBV or nontargeting gRNA. Then, 9 days after treatment with DOX to induce Cas9 expression, DNA was extracted. Then, 250 ng of total DNA was digested overnight at 37°C with 10 U of Plasmid‐Safe ATP‐Dependent DNase (Lucigen Corp.) as previously reported.[ 26 ] DNA sequences around the gRNA target locus were amplified by PCR using KOD FX (Toyobo) and the primers listed in Table S1. The amplicon was sequenced on an Illumina NovaSeq 6000, and the data were analyzed using CRISPRpic to detect CRISPR‐induced indels at the targeted locus as previously reported.[ 27 ] The indel ratio was defined as the sum of the insertion ratio, deletion ratio, and complex deletion ratio calculated by CRISPRpic.
PARP activity
HBV‐infected HepG2‐hNTCP‐C4‐iCas9 cells were lentivirally transduced with gRNA targeting HBV. Then, 9 days after treatment with DOX for Cas9 expression, PARP activity was measured with a Poly (ADP‐Ribose) Polymerase Assay Kit (4677‐096‐K; Bio‐Techne) according to the manufacturer's protocol.
Statistical analysis
The data are presented as the means ± SEM. Comparisons between two groups were performed by an unpaired two‐sided t test. Analysis of variance was performed to detect overall differences among multiple groups, followed by the Tukey–Kramer test. In the experiment evaluating the concentration‐dependent inhibitory effect of olaparib on cccDNA, comparisons between the control group (0 μM) and treated groups (1 μM or 10 μM) were performed with Dunnett's test. A value of p < 0.05 indicated statistical significance.
RESULTS
CRISPR/Cas9 targets cccDNA and exerts an antiviral effect, with a significant reduction in cccDNA, in HBV‐infected hepatoma cells
First, to evaluate the antiviral effect of the CRISPR/Cas9 system targeting cccDNA (HBV‐CRISPR) in HBV‐infected cells, we lentivirally introduced a DOX‐inducible Cas9 expression vector into HepG2‐hNTCP‐C4 cells and generated a stable HBV‐susceptible cell line with inducible Cas9 expression (HepG2‐hNTCP‐C4‐iCas9). We confirmed Cas9 expression in a DOX dose‐dependent manner in these cells without apparent leaky expression in the absence of DOX (Figure 1A). After HBV inoculation, HBc‐positive cells were detected (Figure 1B), confirming that HepG2‐hNTCP‐C4‐iCas9 cells retain HBV susceptibility. We then lentivirally transduced the HBV‐gRNA or NC‐gRNA vector into HepG2‐hNTCP‐C4‐iCas9 cells (Figure 1C). After HBV infection, DOX administration in HepG2‐hNTCP‐C4‐iCas9 cells transduced with HBV gRNA significantly reduced intracellular cccDNA levels relative to those in cells transduced with NC gRNA (Figure 1C,D). The reduction of cccDNA levels was confirmed with DNA treated with T5 exonuclease, to eliminate relaxed circular DNA (rcDNA) potentially contaminating the purified DNA (Figure S1). To examine the direct effect of HBV‐CRISPR on cccDNA, we sequenced cccDNA in these cells and identified a significantly higher ratio of indel formation at the locus targeted by HBV gRNA in HBV gRNA–transduced cells than in NC gRNA–transduced cells (8.63 ± 2.65% vs. 0.006 ± 0.0003%) (Figure 1E). DOX administration also significantly reduced HBV DNA, HBs antigen, and HBe antigen levels in the supernatant and intracellular pgRNA levels in HBV gRNA–transduced cells relative to NC gRNA–transduced cells (Figure 1F). These data suggested that HBV‐CRISPR directly cleaved cccDNA and exerted an antiviral effect accompanied by a reduction in cccDNA levels in HBV‐infected cells.
FIGURE 1.
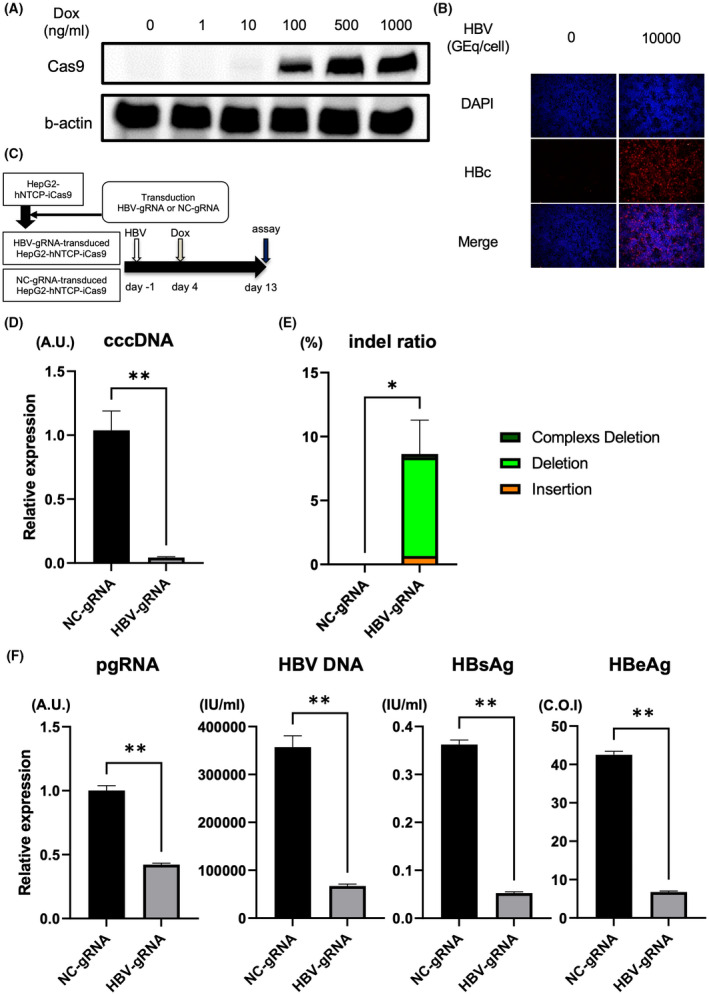
Clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 targets covalently closed circular DNA (cccDNA) and exerts an antiviral effect accompanied by a significant reduction in cccDNA in HBV‐infected hepatoma cells. (A) Western blot of Cas9 and b‐actin in HepG2‐hNTCP‐C4‐iCas9 cells 1 day after doxycycline (DOX) treatment. (B) Representative images of immunofluorescent staining of hepatitis B core (HBc) 13 days after hepatitis B virus (HBV) inoculation. (C–F) HepG2‐hNTCP‐C4‐iCas9 cells lentivirally transduced with the HBV guide RNA (gRNA) or NC gRNA vector were inoculated with HBV (10,000 GEq/cell). (C) Experimental protocol. (D) Intracellular cccDNA levels in HepG2‐hNTCP‐C4‐iCas9 cells 13 days after HBV inoculation (10,000 GEq/cell) (n = 4, **p < 0.01). (E) Indel ratio in the cccDNA of HepG2‐hNTCP‐C4‐iCas9 cells 13 days after HBV inoculation (n = 4, *p < 0.05). (F) Intracellular pregenomic RNA (pgRNA) levels and supernatant HBV DNA, hepatitis B surface antigen (HBsAg) and hepatitis B e antigen (HBeAg) levels in HepG2‐hNTCP‐C4‐iCas9 cells 13 days after HBV inoculation (10,000 GEq/cell) (n = 4, **p < 0.01). DAPI, 4′,6‐diamidino‐2‐phenylindole. NC, negative control.
HBV‐CRISPR exerts an antiviral effect, with a significant reduction in cccDNA, in HBV‐infected PHHs
We next evaluated the antiviral effect of HBV‐CRISPR in HBV‐infected primary hepatocytes. PHHs isolated from humanized liver chimeric mice were lentivirally transduced with the Cas9 gRNA and HBV gRNA (or NC gRNA)–expressing vectors after HBV infection (Figure 2A). HBV‐gRNA/Cas9 transduction significantly reduced intracellular cccDNA levels, which was accompanied by decreases in supernatant HBV DNA, HBs antigen and HBe antigen levels, and intracellular pgRNA levels (Figure 2B). These data suggested that HBV‐CRISPR exerts an antiviral effect accompanied by a significant reduction in cccDNA levels in HBV‐infected hepatocytes.
FIGURE 2.
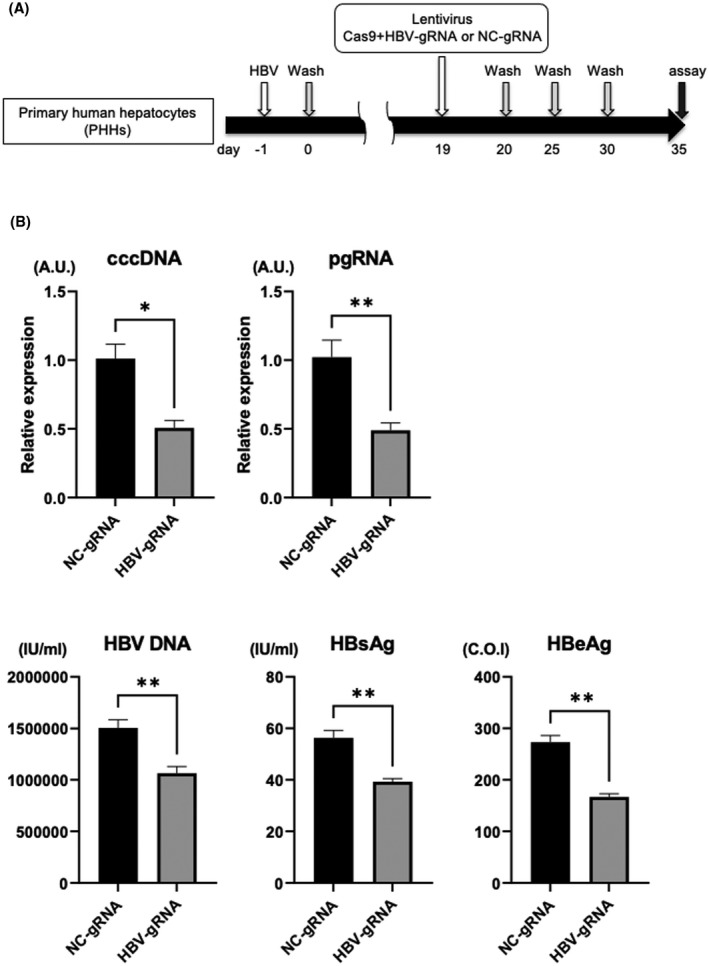
HBV‐CRISPR exerts an antiviral effect accompanied by a significant reduction in cccDNA in HBV‐infected primary human hepatocytes (PHHs). PHHs isolated from humanized liver chimeric mice were lentivirally transduced with tandem Cas9 gRNA and HBV gRNA (or NC gRNA)–expressing vectors (10 multiplicity of infection [MOI]) 19 days after HBV inoculation (500 GEq/cell). (A) Experimental protocol. (B) cccDNA levels, intracellular pgRNA levels, and supernatant HBV DNA, HBs antigen, and HBe antigen levels in PHHs 35 days after HBV inoculation (n = 4; *p < 0.05, **p < 0.01).
Inhibition of PARP2‐HPF1 enhances the antiviral effect of HBV‐CRISPR in HBV‐infected hepatoma cells
It has been reported that CRISPR targeting cccDNA decreases the cccDNA pool but also generates mutated cccDNAs that escape repetitive cleavage by CRISPR.[ 28 , 29 ] HBV uses the host cellular DNA repair machinery for cccDNA biosynthesis,[ 30 ] so we hypothesized that cccDNA cleaved by CRISPR may be repaired by the host DNA repair pathway and that its inhibition may enhance the antiviral effect of HBV‐CRISPR. It has been recently reported that the initial step in the repair of DNA DSBs is the bridging of two nucleosomes and the alignment of the cleaved DNA in a position suitable for ligation, mediated by PARP2‐HPF1.[ 31 ] In HBV‐infected HepG2‐hNTCP‐C4‐iCas9 cells transduced with HBV‐gRNA, although there was no difference in the expression levels of PARP2 and HPF1 between the DOX‐treated and nontreated groups (Figure 3A,B), PARP activity was significantly increased in the DOX‐treated group (Figure 3C). Importantly, in HBV‐infected HepG2‐hNTCP‐C4‐iCas9 cells transduced with HBV‐gRNA supplemented with DOX, the knockdown of PARP2 and/or HPF1 with siRNA significantly reduced the expression levels of cccDNA and intracellular pgRNA (Figure 3D–G). These data suggested that the PARP2‐HPF1 complex may be involved in the repair of cccDNA cleaved by CRISPR. Moreover, the co‐suppression of PAPR2 and HPF1 did not exert an additional antiviral effect relative to either HPF1 or PARP2 suppression (Figure 3D–G), suggesting that both proteins may be indispensable for this process.
FIGURE 3.
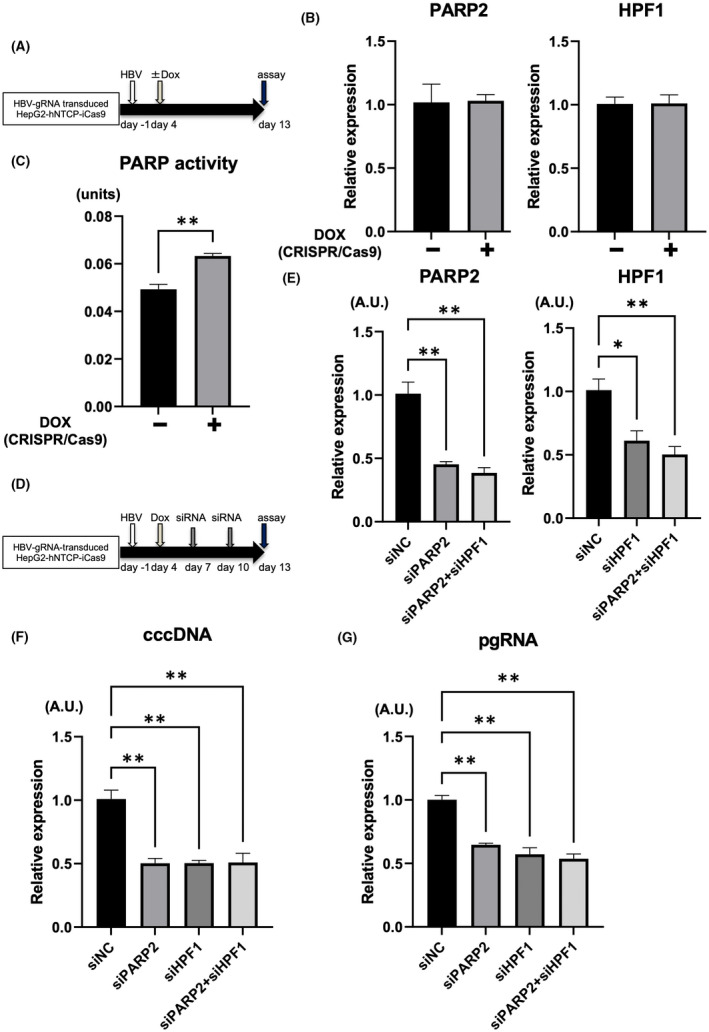
Inhibition of poly(adenosine diphosphate ribose) polymerase 2 (PARP2)–HPF1 enhances the antiviral effect of HBV‐CRISPR in HBV‐infected hepatoma cells. (A–C) HepG2‐hNTCP‐C4‐iCas9 cells transduced with HBV gRNA were treated with or without DOX 4 days after HBV inoculation (10,000 GEq/cell). (A) Experimental protocol. (B) Messenger RNA (mRNA) levels of PARP2 and HPF1 in HepG2‐hNTCP‐C4‐iCas9 cells transduced with HBV gRNA 13 days after HBV inoculation (n = 4). (C) PARP activity in HepG2‐hNTCP‐C4‐iCas9 cells transduced with HBV gRNA 13 days after HBV inoculation (n = 4, **p < 0.01). (D–G) HepG2‐hNTCP‐C4‐iCas9 cells transduced with HBV gRNA were treated with DOX 4 days after HBV inoculation (10,000 GEq/cell) and were then treated with small interfering RNA (siRNA) 7 and 10 days after HBV inoculation. (D) Experimental protocol. (E) mRNA levels of PARP2 and HPF1 in HBV gRNA–transduced HepG2‐hNTCP‐C4‐iCas9 cells 13 days after HBV inoculation (n = 4; *p < 0.05, **p < 0.01). (F) Expression levels of cccDNA in HBV gRNA–transduced HepG2‐hNTCP‐C4‐iCas9 cells 13 days after HBV inoculation (n = 4; **p < 0.01). (G) Expression levels of pgRNA in HBV gRNA–transduced HepG2‐hNTCP‐C4‐iCas9 cells 13 days after HBV inoculation (n = 4; **p < 0.01). HPF1, Histone PARylation factor 1.
Inhibition of the NHEJ pathway enhances the antiviral effect of HBV‐CRISPR in HBV‐infected hepatoma cells
After the initial step, DNA DSBs are repaired via either the NHEJ or HR pathway. We therefore investigated the effect of inhibiting the NHEJ‐mediated or HR‐mediated DNA repair pathway on the antiviral effect of HBV‐CRISPR by using siRNA targeting LIG4 or BRCA (genes essential for NHEJ or HR, respectively) (Figure 4A).[ 32 , 33 ] In HBV‐infected HepG2‐hNTCP‐C4‐iCas9 cells transduced with HBV gRNA supplemented with DOX, the inhibition of LIG4 significantly reduced the expression levels of cccDNA and intracellular pgRNA (Figure 4B), while the inhibition of BRCA1 or BRCA2 increased the expression levels of cccDNA (Figure 4C). These data suggested the involvement of the NHEJ pathway in the repair of cccDNA cleaved by CRISPR. In addition, neither the inhibition of LIG4 nor the inhibition of PARP2 affected cccDNA levels in the absence of CRISPR, indicating that the DNA repair machinery may be important for the repair of CRISPR‐induced DSBs in cccDNA but is not involved in cccDNA biosynthesis (Figure S2A,B). Interestingly, LIG4 inhibition significantly decreased pgRNA levels without altering cccDNA levels in the absence of CRISPR (Figure S2B,C), suggesting that LIG4 is involved in not only DSB repair of cccDNA but also the translation of cccDNA to pgRNA.
FIGURE 4.
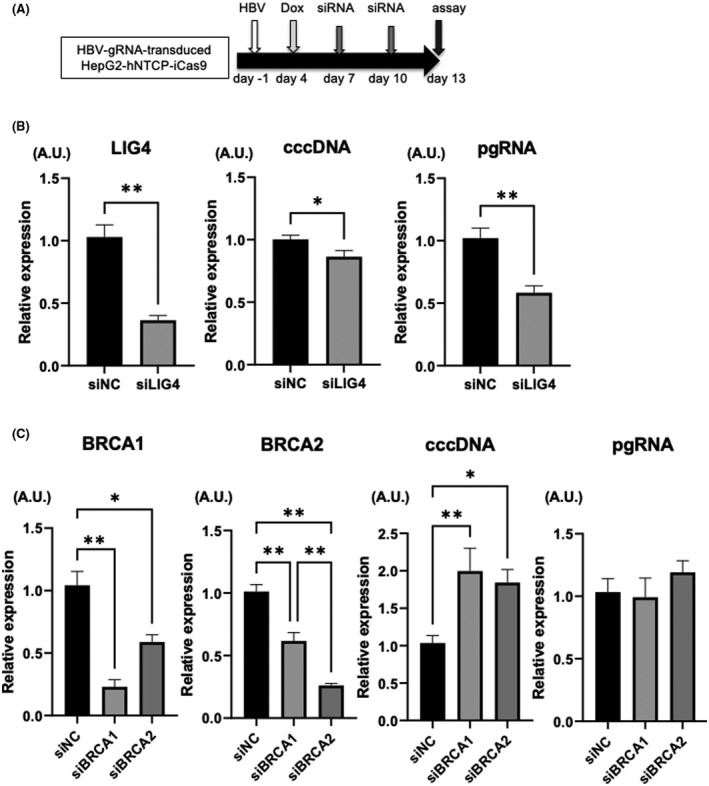
Inhibition of the nonhomologous end joining (NHEJ) pathway enhances the antiviral effect of HBV‐CRISPR in HBV‐infected hepatoma cells. (A–C) HepG2‐hNTCP‐C4‐iCas9 cells transduced with HBV gRNA were treated with DOX 4 days after HBV inoculation (10,000 GEq/cell) and then with siRNA 7 and 10 days after HBV inoculation. (A) Experimental protocol. (B) Expression levels of LIG4, cccDNA, and pgRNA in HBV gRNA–transduced HepG2‐hNTCP‐C4‐iCas9 cells 13 days after HBV inoculation (n = 4; *p < 0.05, **p < 0.01, siLIG4‐treated group vs. siNC‐treated group). (C) Expression levels of BRCA1, BRCA2, cccDNA, and pgRNA in HBV gRNA–transduced HepG2‐hNTCP‐C4‐iCas9 cells 13 days after HBV inoculation (n = 4; *p < 0.05, **p < 0.01, siBRCA1‐treated group or siBRCA2‐treated group vs. siNC‐treated group). BRCA, breast cancer susceptibility gene; LIG4, DNA Ligase 4.
Olaparib enhances the antiviral effect of HBV‐CRISPR in HBV‐infected hepatoma cells and PHHs
Next, we sought a clinically available drug to enhance the effect of HBV‐CRISPR. Because PARP activity increased after HBV‐CRISPR treatment and PARP2 inhibition increased the obtained antiviral effect, we investigated the effect of blocking PARP activity on the antiviral effect of HBV‐CRISPR using olaparib, a clinically available PARP inhibitor.[ 7 , 8 , 9 , 34 ] We confirmed that 1 μM olaparib modestly but significantly suppressed PARP activity (Figure S3A,B). Olaparib treatment did not cause a significant antiviral effect, as assessed by cccDNA and pgRNA levels in HBV‐infected HepG2‐hNTCP‐C4‐iCas9 cells transduced with HBV gRNA in the absence of DOX (Figure 5A–C). Under DOX supplementation, however, olaparib treatment significantly down‐regulated the expression levels of cccDNA and pgRNA relative to vehicle treatment in HBV‐infected HepG2‐hNTCP‐C4‐iCas9 cells transduced with HBV gRNA (Figure 5D–F). The inhibitory effect of olaparib on cccDNA was dose‐dependent (Figure 3C). The CRISPR‐induced indel rate was lower in cells treated with olaparib than in untreated cells, suggesting that olaparib suppresses NHEJ‐mediated DSB repair with indel formation (Figure 3D). Finally, we also examined the potential of combination therapy with HBV‐CRISPR and olaparib in HBV‐infected PHHs (Figure 5G). PHHs isolated from humanized liver chimeric mice were lentivirally transduced with Cas9‐expressing and HBV gRNA–expressing vectors after HBV infection (Figure 5G). Then, we further treated HBV‐infected PHHs with either olaparib or vehicle. Olaparib treatment significantly down‐regulated the expression levels of pgRNA and cccDNA relative to vehicle treatment (Figure 5H,I). Collectively, these findings suggested that treatment with HBV‐CRISPR together with olaparib may be a combinational anti‐HBV therapy targeting cccDNA (Figure 6).
FIGURE 5.
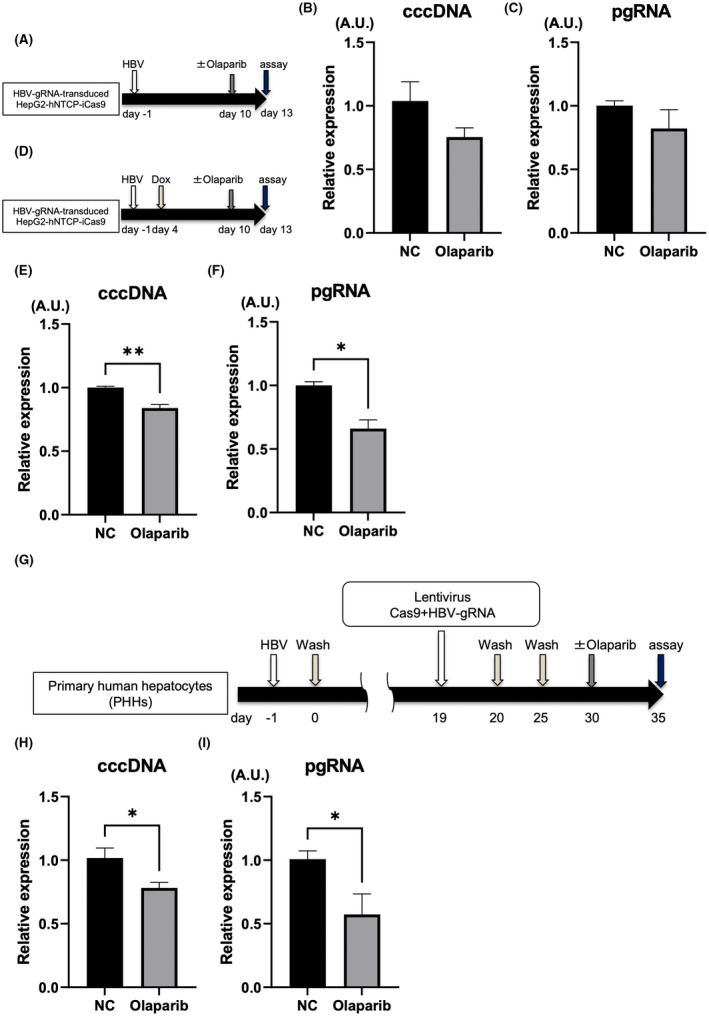
Olaparib enhances the antiviral effect of HBV‐CRISPR in HBV‐infected hepatoma cells and PHHs. (A–C) HepG2‐hNTCP‐C4‐iCas9 cells transduced with HBV gRNA were treated with or without olaparib (1 μM) 10 days after HBV inoculation (10,000 GEq/cell). (A) Experimental protocol. (B) Intracellular cccDNA levels in HBV gRNA–transduced HepG2‐hNTCP‐C4‐iCas9 cells 13 days after HBV inoculation. (C) pgRNA levels in HBV gRNA–transduced HepG2‐hNTCP‐C4‐iCas9 cells 13 days after HBV inoculation. (D–F) HepG2‐hNTCP‐C4‐iCas9 cells transduced with HBV gRNA were treated with DOX 4 days after HBV inoculation (10,000 GEq/cell) and then with olaparib (1 μM) or vehicle 10 days after HBV inoculation. (D) Experimental protocol. (E) cccDNA levels in HBV gRNA–transduced HepG2‐hNTCP‐C4‐iCas9 cells 13 days after HBV inoculation (n = 4; *p < 0.05). (F) Intracellular pgRNA levels in HBV gRNA–transduced HepG2‐hNTCP‐C4‐iCas9 cells 13 days after HBV inoculation (n = 4; **p < 0.01). (G–I) PHHs isolated from humanized liver chimeric mice were lentivirally transduced with tandem Cas9‐expressing and HBV gRNA–expressing vectors (10 MOI) 19 days after HBV inoculation (500 GEq/cell) and were then treated with olaparib (1 μM) or vehicle. (G) Experimental protocol. (H) cccDNA levels in PHHs 35 days after HBV inoculation (n = 4; *p < 0.05). (I) Intracellular pgRNA levels in PHHs 35 days after HBV inoculation (n = 4, *p < 0.05).
FIGURE 6.
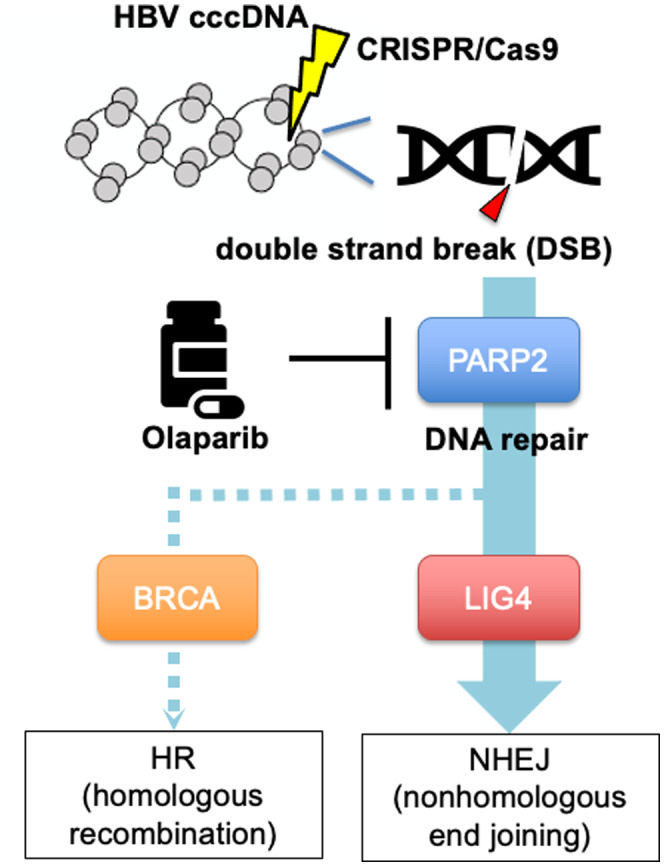
DNA repair pathway of CRISPR‐induced double‐strand breaks (DSBs) in HBV cccDNA and therapeutic targeting of the pathway by olaparib. HR, homologous recombination.
DISCUSSION
CRISPR/Cas9 gene‐editing technology introduces double‐strand breaks at arbitrary locations based on the design of a gRNA with a complementary sequence to the genome. It has been reported that CRISPR/Cas9 targeting HBV cccDNA decreases the expression level of cccDNA,[ 14 , 15 ] which may provide a route to a potential therapy for curing HBV. In the present study, we also showed that HBV‐CRISPR directly cut cccDNA and reduced intracellular cccDNA levels in HBV‐infected NTCP‐expressing hepatoma cells, supporting the potential of HBV‐CRISPR to achieve a complete cure. In previous studies, the antiviral effect of HBV‐CRISPR has been examined using HBV‐integrated cell lines or NTCP‐expressing hepatoma cell lines. We confirmed the therapeutic efficacy of HBV‐CRISPR on cccDNA in HBV‐infected primary hepatocytes for the first time in this study. A safe and highly efficient delivery method needs to be developed for clinical translation of this therapy. A recent report showed that the CRISPR system was successfully delivered into human hepatocytes from humanized liver chimeric mice using genetically modified AAV,[ 17 ] which could be a potential HBV‐CRISPR delivery option in patients. In particular, a genetically modified AAV vector (AAV.GT5) has been reported to have high transduction efficiency in human hepatocytes and low reactivity with neutralizing antibodies,[ 10 ] which would enable repeated administration to deliver CRISPR into all HBV‐infected hepatocytes in the liver. A potential concern about CRISPR treatment is that it may induce DSBs of the HBV DNA integrated in the host genome and cause genome instability and cytotoxicity. It was previously reported that HBV regions integrated into the host genome were enriched on the 3′ side of the hepatitis B x protein (HBx) gene.[ 35 ] Therefore, to avoid CRISPR therapy–induced damage to the host genome, one strategy is to design gRNA outside the open reading frame of HBx, as we did in this study. Nevertheless, the potential risk of host DNA damage needs to be considered for CRISPR therapy.
HBV‐CRISPR decreases the cccDNA pool but also generates mutated cccDNAs that escape repetitive cleavage by CRISPR due to limited mismatch tolerance of the CRISPR system.[ 28 , 29 ] Therefore, to enhance the effect of HBV‐CRISPR and eliminate cccDNA, we targeted the machinery important for the repair of cccDNA cleaved by CRISPR in the present study. The formation and maintenance of cccDNA are thought to be dependent on the host's DNA repair mechanism.[ 30 ] However, the detailed underlying mechanism and its kinetics following CRISPR‐mediated cleavage have not been clarified. We found that CRISPR‐mediated HBV cleavage induced PARP activity and that the suppression of either PARP2 or HPF1 significantly enhanced the CRISPR‐mediated decrease in cccDNA. These findings suggested that the PARP2‐HPF1 complex, which is important for the initial DNA alignment step during the repair of host DNA DSBs,[ 31 ] may also be important for the repair of cccDNA DSBs. Moreover, we showed that olaparib augmented the CRISPR‐mediated decrease in cccDNA. Although olaparib is known to block base excision repair of DNA single‐strand breaks through PARP1 inhibition, olaparib may enhance the CRISPR‐mediated anti‐cccDNA effect via inhibition of PARP2‐mediated DSB repair of cccDNA. We showed that the suppression of LIG4, an essential gene for NHEJ, enhanced the CRISPR‐mediated decrease in cccDNA, suggesting that NHEJ may play an important role in the repair of cccDNA DSBs. In contrast, the suppression of BRCA, an essential gene for HR, suppressed the CRISPR‐mediated decrease in cccDNA. It is widely accepted that the NHEJ and HR pathways are complementary and that the inhibition of HR components significantly increases NHEJ efficiency, which might explain this result.
Kostyushev et al. also recently evaluated the involvement of the NHEJ and HR pathways in the repair of CRISPR‐mediated HBV cleavage.[ 28 ] They showed that the inhibition of NHEJ with the DNA‐PKcs inhibitor NU7026 increased the anti‐HBV activity of CRISPR in HBV genome–integrated cells,[ 28 ] which is consistent with our findings. On the other hand, they showed that NU7026 treatment increased cytoplasmic cccDNA levels in cells harboring a CRISPR system targeting cccDNA and speculated that the mutation of cccDNA prevents its degradation via an unknown mechanism. This is inconsistent with our finding that the inhibition of the NHEJ pathway significantly increased the reduction in the cytoplasmic cccDNA pool mediated by HBV‐CRISPR. Although the precise reason for this discrepancy is unclear, it should be noted that Kostyushev et al. used two HBV genome‐integrated cell lines (HepG2‐1.1merHBV and HepG2‐1.5merHBV) that were different from our HBV‐infected cells and inhibited NHEJ with small molecules. In the case of the use of HBV genome‐integrated cells in particular, the effect of CRISPR on the integrated HBV genome cannot be ignored. Cleavage and repair associated with errors in the HBV genome integrated in host DNA may continuously produce mutated cccDNA and affect intracellular cccDNA pools. Therefore, HBV genome–integrated cells may not be ideal for studying cccDNA biology. In our present study using an HBV infection model, we consistently observed that NHEJ inhibition enhanced the CRISPR‐mediated cccDNA reduction in hepatoma cells and primary hepatocytes. Furthermore, we showed that the clinically available PARP inhibitor olaparib enhanced the CRISPR‐induced cccDNA reduction in these cells. In summary, our study demonstrates that the suppression of the NHEJ‐mediated DNA repair pathway is beneficial for enhancing the therapeutic activity of CRISPR targeting cccDNA.
This study is subject to the following limitations: (1) The lack of in vivo experiments, (2) the lack of confirmation of the quantitative PCR‐based cccDNA quantification assay by southern blotting, (3) potential rcDNA contamination in the cccDNA assay due to the absence of T5 exonuclease pretreatment, and (4) the use of hepatoma cells, in which the therapeutic effect of CRISPR monotherapy is very high, instead of PHH cells in most experiments.
In summary, we showed in this study that CRISPR targeting HBV cccDNA cleaves cccDNA and exerts an antiviral effect accompanied by a significant reduction in cccDNA in HBV‐infected cells. We also found that the inhibition of the NHEJ pathway enhances the efficacy of CRISPR in HBV‐infected cells. Finally, olaparib enhances the efficacy of CRISPR in HBV‐infected cells, providing clinically applicable experimental evidence of a combination therapy targeting cccDNA in patients with CHB.
AUTHOR CONTRIBUTIONS
Study concept: Kazuhiro Murai and T.K. Data acquisition: Kazuhiro Murai, Takahiro Kodama, Hayato Hikita, Akiyoshi Shimoda, Makoto Fukuoka, Keisuke Fukutomi, Satoshi Shigeno, Yuto Shiode, Daisuke Motooka, Yuki Tahata, Yuki Makino, Ryoko Yamada, Ryotaro Sakamori, Tomohide Tatsumi, Yuichiro Higuchi, Hiroshi Suemizu, Kei Miyakawa, and Akihide Ryo. Formal analysis: Kazuhiro Murai, Takahiro Kodama, Hayato Hikita, Akiyoshi Shimoda, Makoto Fukuoka, Keisuke Fukutomi, Satoshi Shigeno, Yuto Shiode, Daisuke Motooka, Yuki Tahata, Yuki Makino, Ryoko Yamada, Ryotaro Sakamori, Tomohide Tatsumi, Yuichiro Higuchi, Hiroshi Suemizu, Kei Miyakawa, and Akihide Ryo. Manuscript draft: Kazuhiro Murai, Takahiro Kodama, and T.Tak. Funding acquisition: Kazuhiro Murai, Takahiro Kodama, and Tetsuo Takehara. Resources: Yuichiro Higuchi, Hiroshi Suemizu, Kei Miyakawa, and Akihide Ryo. Project administration, supervision, review, and editing: Tetsuo Takehara. All authors read and approved the manuscript.
Funding information
Supported by the Japan Agency for Medical Research and Development (22fk0310512, 21fk0310106s0105, 22fk0210110h0001, and 22fk0310524s0101) and by a Grant‐in‐Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (20 K17020).
CONFLICT OF INTEREST
H.H. is on the speakers' bureau of Gilead.
Supporting information
Table S1 Next‐generation sequencing primer list
Figure S1 HepG2‐hNTCP‐C4‐iCas9 cells lentivirally transduced with the hepatitis B virus (HBV) guide RNA (gRNA) or NC gRNA vector were inoculated with HBV (10,000 GEq/cell). Extracted DNA was treated with T5 exonuclease according to the manufacturer’s protocol. Intracellular covalently closed circular DNA (cccDNA) levels were measured in HepG2‐hNTCP‐C4‐iCas9 cells 13 days after HBV inoculation (10,000 GEq/cell) (n = 4; *p < 0.05)
Figure S2 (A–C) HepG2‐hNTCP‐C4‐iCas9 cells transduced with HBV gRNA were treated with small interfering (siRNA) 7 and 10 days after HBV inoculation (10,000 GEq/cell). (A) Experimental protocol. (B) Expression levels of cccDNA in HBV gRNA–transduced HepG2‐hNTCP‐C4‐iCas9 cells 13 days after HBV inoculation (n = 4). (C) Expression levels of pgRNA in HBV gRNA–transduced HepG2‐hNTCP‐C4‐iCas9 cells 13 days after HBV inoculation (n = 4; *p < 0.05, siLIG4‐treated group vs. siNC‐treated group).
Figure S3 (A–C) HepG2‐hNTCP‐C4‐iCas9 cells transduced with HBV gRNA were treated with or without olaparib (0 μM, 1 μM, and 10 μM) 10 days after HBV inoculation (10,000 GEq/cell). (A) Experimental protocol. (B) Poly(adenosine diphosphate ribose) polymerase (PARP) activity in HepG2‐hNTCP‐C4‐iCas9 cells transduced with HBV gRNA 13 days after HBV inoculation (n = 4; **p < 0.01). (C) Intracellular cccDNA levels in HBV gRNA–transduced HepG2‐hNTCP‐C4‐iCas9 cells 13 days after HBV inoculation (n = 4; *p < 0.05). (D) Indel ratio in the cccDNA of HepG2‐hNTCP‐C4‐iCas9 cells 13 days after HBV inoculation (n = 4)
Murai K, Kodama T, Hikita H, Shimoda A, Fukuoka M, Fukutomi K, et al; Inhibition of nonhomologous end joining‐mediated DNA repair enhances anti‐HBV CRISPR therapy. Hepatol Commun. 2022;6:2474–2487. 10.1002/hep4.2014
REFERENCES
- 1. Rajoriya N, Combet C, Zoulim F, Janssen HLA. How viral genetic variants and genotypes influence disease and treatment outcome of chronic hepatitis B. Time for an individualised approach? J Hepatol. 2017;67:1281–97. [DOI] [PubMed] [Google Scholar]
- 2. Ward JW, Hinman AR. What is needed to eliminate hepatitis B virus and hepatitis C virus as Global Health threats. Gastroenterology. 2019;156:297–310. [DOI] [PubMed] [Google Scholar]
- 3. Terrault NA, Bzowej NH, Chang KM, Hwang JP, Jonas MM, Murad MH, et al. AASLD guidelines for treatment of chronic hepatitis B. Hepatology. 2016;63:261–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hung TH, Liang CM, Hsu CN, Tai WC, Tsai KL, Ku MK, et al. Association between complicated liver cirrhosis and the risk of hepatocellular carcinoma in Taiwan. PLoS One. 2017;12:e0181858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yang HC, Chen PJ. The potential and challenges of CRISPR‐Cas in eradication of hepatitis B virus covalently closed circular DNA. Virus Res. 2018;244:304–10. [DOI] [PubMed] [Google Scholar]
- 6. Di Bisceglie AM, Lok AS, Martin P, Terrault N, Perrillo RP, Hoofnagle JH. Recent US Food and Drug Administration warnings on hepatitis B reactivation with immune‐suppressing and anticancer drugs: just the tip of the iceberg? Hepatology. 2015;61:703–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Golan T, Hammel P, Reni M, Van Cutsem E, Macarulla T, Hall MJ, et al. Maintenance Olaparib for germline. N Engl J Med. 2019;381:317–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pujade‐Lauraine E, Ledermann JA, Selle F, Gebski V, Penson RT, Oza AM, et al. Olaparib tablets as maintenance therapy in patients with platinum‐sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT‐Ov21): a double‐blind, randomised, placebo‐controlled, phase 3 trial. Lancet Oncol. 2017;18:1274–84. [DOI] [PubMed] [Google Scholar]
- 9. Robson M, Im SA, Senkus E, Xu B, Domchek SM, Masuda N, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med. 2017;377:523–33. [DOI] [PubMed] [Google Scholar]
- 10. Ito M, Takino N, Nomura T, Kan A, Muramatsu SI. Engineered adeno‐associated virus 3 vector with reduced reactivity to serum antibodies. Sci Rep. 2021;11:9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ohmori T, Nagao Y, Mizukami H, Sakata A, Muramatsu SI, Ozawa K, et al. CRISPR/Cas9‐mediated genome editing via postnatal administration of AAV vector cures haemophilia B mice. Sci Rep. 2017;7:4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rossidis AC, Stratigis JD, Chadwick AC, Hartman HA, Ahn NJ, Li H, et al. In utero CRISPR‐mediated therapeutic editing of metabolic genes. Nat Med. 2018;24:1513–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Frangoul H, Altshuler D, Cappellini MD, Chen YS, Domm J, Eustace BK, et al. CRISPR‐Cas9 gene editing for sickle cell disease and β‐thalassemia. N Engl J Med. 2021;384:252–60. [DOI] [PubMed] [Google Scholar]
- 14. Kennedy EM, Bassit LC, Mueller H, Kornepati AVR, Bogerd HP, Nie T, et al. Suppression of hepatitis B virus DNA accumulation in chronically infected cells using a bacterial CRISPR/Cas RNA‐guided DNA endonuclease. Virology. 2015;476:196–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ramanan V, Shlomai A, Cox DB, Schwartz RE, Michailidis E, Bhatta A, et al. CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus. Sci Rep. 2015;5:10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Scott T, Moyo B, Nicholson S, Maepa MB, Watashi K, Ely A, et al. ssAAVs containing cassettes encoding SaCas9 and guides targeting hepatitis B virus inactivate replication of the virus in cultured cells. Sci Rep. 2017;7:7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stone D, Long KR, Loprieno MA, De Silva Feelixge HS, Kenkel EJ, Liley RM, et al. CRISPR‐Cas9 gene editing of hepatitis B virus in chronically infected humanized mice. Mol Ther Methods Clin Dev. 2021;20:258–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jasin M, Haber JE. The democratization of gene editing: insights from site‐specific cleavage and double‐strand break repair. DNA Repair (Amst). 2016;44:6–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Uehara S, Higuchi Y, Yoneda N, Kawai K, Yamamoto M, Kamimura H, et al. An improved TK‐NOG mouse as a novel platform for humanized liver that overcomes limitations in both male and female animals. Drug Metab Pharmacokinet. 2022;42:100410. [DOI] [PubMed] [Google Scholar]
- 20. Murai K, Hikita H, Kai Y, Kondo Y, Fukuoka M, Fukutomi K, et al. Hepatitis C virus infection suppresses hepatitis B virus replication via the RIG‐I‐like helicase pathway. Sci Rep. 2020;10:941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ladner SK, Otto MJ, Barker CS, Zaifert K, Wang GH, Guo JT, et al. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrob Agents Chemother. 1997;41:1715–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ogura N, Watashi K, Noguchi T, Wakita T. Formation of covalently closed circular DNA in Hep38.7‐Tet cells, a tetracycline inducible hepatitis B virus expression cell line. Biochem Biophys Res Commun. 2014;452:315–21. [DOI] [PubMed] [Google Scholar]
- 23. Iwamoto M, Watashi K, Tsukuda S, Aly HH, Fukasawa M, Fujimoto A, et al. Evaluation and identification of hepatitis B virus entry inhibitors using HepG2 cells overexpressing a membrane transporter NTCP. Biochem Biophys Res Commun. 2014;443:808–13. [DOI] [PubMed] [Google Scholar]
- 24. Yamai T, Hikita H, Fukuoka M, Fukutomi K, Murai K, Nakabori T, et al. SIRT1 enhances hepatitis virus B transcription independent of hepatic autophagy. Biochem Biophys Res Commun. 2020;527:64–70. [DOI] [PubMed] [Google Scholar]
- 25. Matsunaga S, Kawakami S, Matsuo I, Okayama A, Tsukagoshi H, Kudoh A, et al. Wheat germ cell‐free system‐based production of hemagglutinin‐neuraminidase glycoprotein of human parainfluenza virus type 3 for generation and characterization of monoclonal antibody. Front Microbiol. 2014;5:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fukutomi K, Hikita H, Murai K, Nakabori T, Shimoda A, Fukuoka M, et al. Capsid allosteric modulators enhance the innate immune response in hepatitis B virus‐infected hepatocytes during interferon administration. Hepatol Commun. 2021;6:281–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee H, Chang HY, Cho SW, Ji HP. CRISPRpic: fast and precise analysis for CRISPR‐induced mutations via prefixed index counting. NAR Genom Bioinform. 2020;2:lqaa012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kostyushev D, Kostyusheva A, Brezgin S, Zarifyan D, Utkina A, Goptar I, et al. Suppressing the NHEJ pathway by DNA‐PKcs inhibitor NU7026 prevents degradation of HBV cccDNA cleaved by CRISPR/Cas9. Sci Rep. 2019;9:1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Anderson EM, Haupt A, Schiel JA, Chou E, Machado HB, Strezoska Z, et al. Systematic analysis of CRISPR‐Cas9 mismatch tolerance reveals low levels of off‐target activity. J Biotechnol. 2015;211:56–65. [DOI] [PubMed] [Google Scholar]
- 30. Tang L, Sheraz M, McGrane M, Chang J, Guo JT. DNA polymerase alpha is essential for intracellular amplification of hepatitis B virus covalently closed circular DNA. PLoS Pathog. 2019;15:e1007742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bilokapic S, Suskiewicz MJ, Ahel I, Halic M. Bridging of DNA breaks activates PARP2‐HPF1 to modify chromatin. Nature. 2020;585:609–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Neiger HE, Siegler EL, Shi Y. Breast cancer predisposition genes and synthetic lethality. Int J Mol Sci. 2021;22:5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. Increasing the efficiency of precise genome editing with CRISPR‐Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015;33:538–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hussain M, Mateo J, Fizazi K, Saad F, Shore N, Sandhu S, et al. Survival with Olaparib in metastatic castration‐resistant prostate cancer. N Engl J Med. 2020;383:2345–57. [DOI] [PubMed] [Google Scholar]
- 35. Furuta M, Tanaka H, Shiraishi Y, Unida T, Imamura M, Fujimoto A, et al. Characterization of HBV integration patterns and timing in liver cancer and HBV‐infected livers. Oncotarget. 2018;9:25075–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Next‐generation sequencing primer list
Figure S1 HepG2‐hNTCP‐C4‐iCas9 cells lentivirally transduced with the hepatitis B virus (HBV) guide RNA (gRNA) or NC gRNA vector were inoculated with HBV (10,000 GEq/cell). Extracted DNA was treated with T5 exonuclease according to the manufacturer’s protocol. Intracellular covalently closed circular DNA (cccDNA) levels were measured in HepG2‐hNTCP‐C4‐iCas9 cells 13 days after HBV inoculation (10,000 GEq/cell) (n = 4; *p < 0.05)
Figure S2 (A–C) HepG2‐hNTCP‐C4‐iCas9 cells transduced with HBV gRNA were treated with small interfering (siRNA) 7 and 10 days after HBV inoculation (10,000 GEq/cell). (A) Experimental protocol. (B) Expression levels of cccDNA in HBV gRNA–transduced HepG2‐hNTCP‐C4‐iCas9 cells 13 days after HBV inoculation (n = 4). (C) Expression levels of pgRNA in HBV gRNA–transduced HepG2‐hNTCP‐C4‐iCas9 cells 13 days after HBV inoculation (n = 4; *p < 0.05, siLIG4‐treated group vs. siNC‐treated group).
Figure S3 (A–C) HepG2‐hNTCP‐C4‐iCas9 cells transduced with HBV gRNA were treated with or without olaparib (0 μM, 1 μM, and 10 μM) 10 days after HBV inoculation (10,000 GEq/cell). (A) Experimental protocol. (B) Poly(adenosine diphosphate ribose) polymerase (PARP) activity in HepG2‐hNTCP‐C4‐iCas9 cells transduced with HBV gRNA 13 days after HBV inoculation (n = 4; **p < 0.01). (C) Intracellular cccDNA levels in HBV gRNA–transduced HepG2‐hNTCP‐C4‐iCas9 cells 13 days after HBV inoculation (n = 4; *p < 0.05). (D) Indel ratio in the cccDNA of HepG2‐hNTCP‐C4‐iCas9 cells 13 days after HBV inoculation (n = 4)


