ABSTRACT
Lake Cadagno, a permanently stratified high-alpine lake with a persistent microbial bloom in its chemocline, has long been considered a model for the low-oxygen, high-sulfide Proterozoic ocean. Although the lake has been studied for over 25 years, the absence of concerted study of the bacteria, phytoplankton, and viruses, together with primary and secondary production, has hindered a comprehensive understanding of its microbial food web. Here, the identities, abundances, and productivity of microbes were evaluated in the context of Lake Cadagno biogeochemistry. Photosynthetic pigments together with 16S rRNA gene phylogenies suggest the prominence of eukaryotic phytoplankton chloroplasts, primarily chlorophytes. Chloroplasts closely related to those of high-alpine-adapted Ankyra judayi persisted with oxygen in the mixolimnion, where photosynthetic efficiency was high, while chloroplasts of Closteriopsis-related chlorophytes peaked in the chemocline and monimolimnion. The anoxygenic phototrophic sulfur bacterium Chromatium dominated the chemocline along with Lentimicrobium, a genus of known fermenters. Secondary production peaked in the chemocline, which suggested that anoxygenic primary producers depended on heterotrophic nutrient remineralization. The virus-to-microbe ratio peaked with phytoplankton abundances in the mixolimnion and were at a minimum where Chromatium abundance was highest, trends that suggest that viruses may play a role in the modulation of primary production. Through the combined analysis of bacterial, eukaryotic, viral, and biogeochemical spatial dynamics, we provide a comprehensive synthesis of the Lake Cadagno microbial loop. This study offers a new ecological perspective on how biological and geochemical connections may have occurred in the chemocline of the Proterozoic ocean, where eukaryotic microbial life is thought to have evolved.
KEYWORDS: Lake Cadagno, Proterozoic, ancient ocean, meromictic, microbial loop, viral ecology
INTRODUCTION
Roughly 2.3 billion years (Gyr) ago, the oceans were sulfide rich (1) and anoxygenic photosynthesis was a dominant mode of microbial primary production (2). Following the great oxidation event 2.4 to 2.1 Gyr ago (3), anoxygenic photosynthesis continued to sustain microbial life in the euxinic environments below oxygenated surface waters (1, 2, 4). Habitats in which anoxygenic photosynthesis significantly contributes to primary production continue to persist, such as in sulfur-rich meromictic lakes (5) like Lake Cadagno (6). These permanently stratified lakes are resistant to seasonal mixing events that would otherwise redistribute oxygen throughout the water column (7). In these systems, biogeochemical processes provide a link between the contrasting dominant modes of primary production, thereby directly connecting the food webs of the oxygenated and anoxic habitats. By resolving these biogeochemical connections, we may obtain insights into how oxygenic and anoxygenic photosynthesis may have evolved in the Proterozoic ocean’s chemocline.
Meromictic Lake Cadagno is a well-established ancient ocean model (8–12) owing to its permanently stratified oxygenated surface and euxinic bottom waters, its shallow chemocline within the photic zone, and sulfate and molybdenum concentrations lower than those of modern oceans. The microbial community composition of meromictic Lake Cadagno typifies the reconstructed signatures of life in the Proterozoic ocean (8, 10, 12). Purple and green sulfur bacteria (PSB and GSB, respectively) are among the hallmark bloom-forming microbes that are frequently observed in the chemocline of Lake Cadagno (13, 14). Within the chemocline, carbon, nitrogen, sulfur, and iron cycling are integrated through metabolic activities of the anoxygenic photosynthetic purple sulfur and green sulfur bacteria (15–18). Two PSB, Thiodictyon syntrophicum and Chromatium okenii, have been identified as the greatest contributors to primary production in Lake Cadagno (14, 16, 19). However, recent measurements of primary production have been limited to the chemocline (14, 20), and measurements of secondary production do not exist. Phytoplankton (including modern eukaryotes and cyanobacteria) likely coexisted with anoxygenic phototrophic sulfur bacteria in the Proterozoic ocean (21, 22). High phytoplankton abundances and productivity were expected in the oxic zone, because phytoplankton produces organic matter via oxygenic photosynthesis. In order to better understand microbial loop linkages across the stratified mixolimnion-chemocline transition in Lake Cadagno, paired measurements of productivity, microbial community composition, and diversity are essential next steps.
Lake Cadagno studies to date have focused on the phytoplankton and prokaryotic members of the microbial community and have yet to include viruses. Viruses are known to impact both phytoplankton and heterotrophic microbial populations through lysis and lysogeny and may hold a central position in the marine food webs and biogeochemical cycling (23, 24). Through the lysis of their microbial hosts, viruses play a role in the conversion of particulate organic carbon to dissolved organic carbon, which is predicted to be capable of sustaining the microbial loop (25, 26). Viruses have been identified in meromictic lakes (27–29), but their abundances across the vertical water column of Lake Cadagno are unknown.
To better understand the role the microbial loop plays in sustaining the food web across the stratified layers of Lake Cadagno, microbial community members (phytoplankton, prokaryotes, and viruses) were studied in the context of biogeochemical and primary and secondary production measurements. We hypothesized the mixolimnion to have low primary production relative to the chemocline, because the chemocline is known to be inhabited by a persistent microbial bloom of the primary producers phototrophic sulfur bacteria (14). We also expected high secondary production and viral abundance to be an indicator of effective organic matter recycling within the Lake Cadagno water column, because photoautotrophs, both phototrophic sulfur bacteria and phytoplankton, rely on heterotrophs and viruses for the remineralization of organic matter and nutrient cycling (30, 31). Through a combination of physical, chemical, and biological analyses, this work provides new evidence on how transitions between permanently stratified lake habitats and assemblages may sustain microbial food webs, informing our understanding of the aquatic ecosystems of early Earth.
RESULTS
Positions of the mixolimnion, chemocline, and monimolimnion.
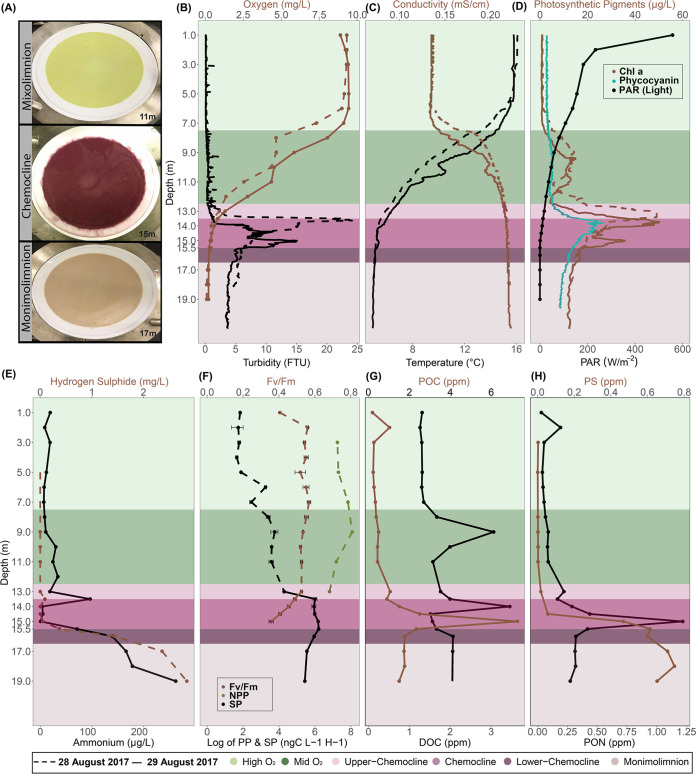
Transitions between the major stratified layers sampled from Lake Cadagno were visually apparent by the color of pigmented biomass captured on 0.22-μm filters (Fig. 1A). From the surface to a depth of 12.5 m, the oxygen levels fell from a maximum of 9.24 mg/L to 2.79 mg/L (Fig. 1B, light green, day 2). As hypoxia is defined as 2.00 mg O2/L (32), 12.5 m marks the boundary of the oxic mixolimnion zone and the upper chemocline (Fig. 1, top light purple). The chemocline was at a depth of 13.0 to 15.5 m, as determined based on near-zero oxygen and light levels (day 2), peaked turbidity, a slight decrease in temperature, and a simultaneous rise in conductivity (Fig. 1B to D, dark purple). At 14.5 m, oxygen levels were near zero and <1% of surface light penetrated (Fig. 1B and D, day 2). Within the chemocline, the deep chlorophyll maximum (DCM) and peak turbidity shifted overnight between days 1 and 2, but in both cases, peak turbidity was observed 0.5 to 1.5 m below the DCM (Fig. 1B and D, day 2). Peak phycocyanin concentrations (26.53 μg/L) and basal fluorescence (Fb = 66.0) were observed at the DCM (day 2, 13.5 to 14 m) and above a secondary chlorophyll peak at 15 m (Fig. 1D; see Fig. S2 in the supplemental material). The monimolimnion was defined as the zone between 15.5 m and the benthos. Gradual increases in particulate sulfur (0.74 ppm), H2S (2.84 mg/L), and ammonia (262.4 μg/L) concentrations were observed in the monimolimnion, as well as a decline in turbidity and photosynthetic pigments (Fig. 1B, E, and H).
FIG 1.
Depth profiles of biogeochemical parameters of Lake Cadagno. Dashed and solid lines represent 28 and 29 August 2017, respectively. Standard deviations for each sample are indicated by horizontal bars. (A) Photographs of biomass collected on 0.22-μm (142-mm-diameter) filters from the Lake Cadagno mixolimnion, chemocline, and monimolimnion strata. (B to F) Depth profiles of oxygen and turbidity (B), conductivity and temperature (C), Chl a, phycocyanin, and photosynthetically active radiation (PAR) (D), hydrogen sulfide and ammonium concentrations (E), net primary production (NPP) and secondary production (SP) rates (F), particulate organic carbon (POC) and dissolved organic carbon (DOC) concentrations (G), and particulate sulfur (PS) and particulate organic nitrogen (PON) concentrations (H). In panels B to H, the high O2 mixolimnion, mid O2 mixolimnion, low O2 mixolimnion-chemocline transition zone, chemocline, lower-chemocline, and anoxic monimolimnion layers are indicated by light green, dark green, light lilac, lilac, dark lilac, and light brown backgrounds, respectively.
Line plot depicting the results of the experiment to monitor the health of photosynthetic cells. A fast repetition rate fluorometer was used to measure basal fluorescence (Fb; blue line) and maximum fluorescence (Fm; black line), which were then used to calculate the maximum photosynthetic quantum yield (Fv/Fm; red line), a proxy for the health of photosynthetic cells. Download FIG S2, PDF file, 0.1 MB (127.4KB, pdf) .
Copyright © 2022 Saini et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Microbial production at the oxic-anoxic interface.
Between 7.0 and 11.0 m in the lower mixolimnion, there was a slight rise (up to 13.12 μg/L on day 1 and 11.80 μg/L on day 2) in chlorophyll a (Chl a) (Fig. 1D). The Chl a peak in the mixolimnion at 9.0 m corresponded with a peak in net primary production (NPP; 3,210 ng C L−1 h−1) (Fig. 1D and F). The NPP peak declined in the lower mixolimnion (11.0 m) and upper chemocline (13.0 m). Though NPP was not measured below 13.0 m, basal and maximum fluorescence measured through the chemocline gave an indication of phytoplankton photosynthetic activity and efficiency (Fig. S2). In the oxic-anoxic boundary, a maximum quantum yield (Fv/Fm, where Fv is variable fluorescence and Fm is maximal fluorescence) (an indicator of photosynthetic efficiency) of 0.49 was observed between 13.0 and 13.5 m and declined to 0.36 at 15.0 m (Fig. 1F), where the light was absent. Overlapping with this oxygenic photosynthesis, the highest rates of secondary productivity (SP) were observed through the chemocline (Fig. 1F). SP was significantly positively associated with some indicators of both photosynthetic microbes (Pearson correlation coefficient: Chl a, R = 0.96, P value = 1.7e−05; phycocyanin, R = 0.80, P value = 0.005) and total biomass (Pearson correlation coefficient: turbidity, R = 0.78, P value = 0.007; total cell counts, R = 0.65, P value = 0.04) (Fig. S3). The peak of dissolved organic carbon (DOC) (14.0 m) followed the peaks of particulate organic carbon (POC) and particulate organic nitrogen (PON) (15.0 m) (Fig. 1G and H). Below the chemocline, some organic and inorganic compounds continued to rise (particulate sulfur, H2S, NH4+), while PON and POC dropped (Fig. 1).
Grid and heat map depicting the Pearson correlation R coefficients and P values determined between data types; strong positive correlations are indicated by blue hues, strong negative correlations are indicated by red, and no correlation is indicated by yellow. Prefix of parameter name indicates the parameter type category. A, abundance: 16S rRNA amplicon-based relative abundances, flow cytometry-generated PLP and VLP counts, and phycobilisome-containing cell counts. D, diversity: alpha and beta diversity calculated from 16S-based genotypic and flow cytometry-based phenotypic Bray-Curtis distance measures. FCS, flow cytometry-derived data. N, nutrients, including DOC, POC, PON, PS, ammonium, and sulfate. P, physicochemical parameters from CTD, including light, conductivity, temperature, oxygen, Chl a, phycocyanin, turbidity. Secondary production is included with no prefix. Download FIG S3, PDF file, 0.6 MB (657.4KB, pdf) .
Copyright © 2022 Saini et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
PLP and VLP abundances and trends.
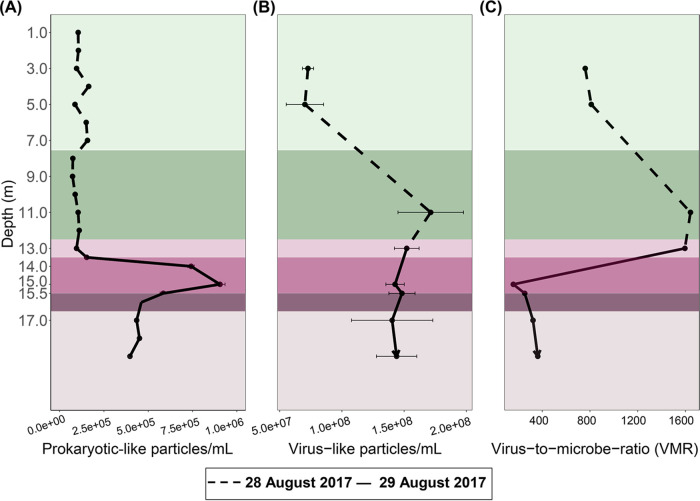
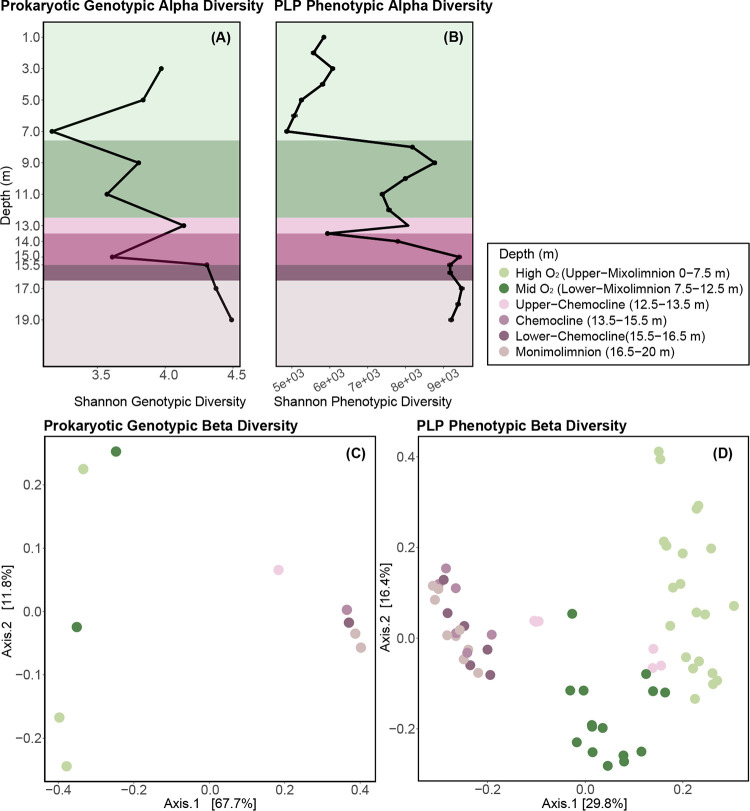
The concentration of prokaryote-like particles (PLPs; prokaryotic cell counts inferred from flow cytometry [FCM] data) was highest in the chemocline at 15.0 m, roughly half that in the monimolimnion, and lowest in the mixolimnion (Fig. 2A). The flow cytometry PLP counts were in line with the fluorescent in situ hybridization (FISH) counts of the previous year (July 2016), which reported up to 106 cells/mL in chemocline (33), and also concurred with the turbidity (Fig. 1D), which is used as a proxy for cell density in Lake Cadagno (12, 34). In contrast, free virus-like particles (VLPs; viral counts inferred from flow cytometry data) were highest at the bottom of the mixolimnion, lowest in the upper mixolimnion, and relatively invariable through the rest of the water column (Fig. 2B). As a result, the virus-to-microbe ratio (VMR) peaked at the bottom of the mixolimnion (1,642 at 11 m), but decreased drastically (158 at 15 m) when prokaryote-like particles rose in the chemocline (Fig. 2C).
FIG 2.
Dashed and solid lines represent 28 and 29 August 2017, respectively. Standard deviations for each sample are indicated by horizontal bars. (A to C) Abundances of prokaryote-like particles (A) and free virus-like particles (B) from flow cytometry analyses and the virus-to-microbe ratio as VLP/PLP (C).
Bacterial community composition through the mixolimnion, chemocline, and monimolimnion.
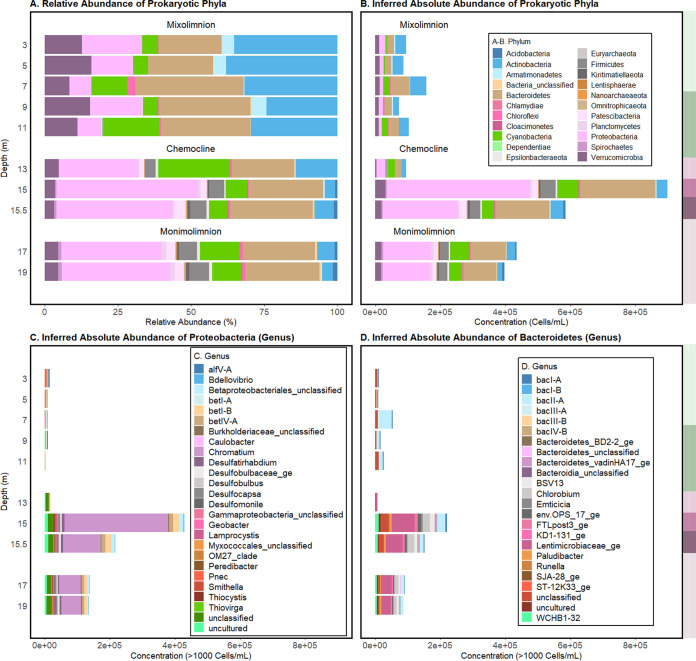
Both relative (operational taxonomic unit [OTU] count scaled by total OTUs) and inferred absolute (OTU count scaled by total FCM PLP counts [35]) abundances were considered to assess the abundances of microbial taxa through Lake Cadagno (Fig. 3A and B). Inferred absolute abundances of prokaryote-classified cells (“Bacteria” and “Archaea” OTU count scaled by total FCM PLP counts [35]) were at a minimum in the oxic mixolimnion (day 1, 0 to 12.5 m) (Fig. 3B; Table S1). On average, the mixolimnion community consisted primarily of the phyla Actinobacteria (35.37% of total OTUs; 33,841 cells/mL), Bacteroidetes (21.49% of total OTUs; 20,557 cells/mL), Proteobacteria (20.46% of total OTUs; 19,579 cells/mL), and Cyanobacteria (5.46% of total OTUs; 5,229 cells/mL) (Fig. 3A and B; Table S1).
FIG 3.
(A and B) Relative (OTU count scaled by total OTUs) (A) and inferred absolute (OTU count scaled by total FCM PLP counts [35]) (B) abundances of prokaryotic phyla at the sampled depths in Lake Cadagno. (C) Genera in the Proteobacteria phylum. (D) Genera in the Bacteroidetes phylum. To the right of each panel, the high O2 mixolimnion, mid O2 mixolimnion, low O2 mixolimnion-chemocline transition zone, chemocline, lower-chemocline, and anoxic monimolimnion layers are indicated by light green, dark green, light lilac, lilac, dark lilac, and light brown backgrounds, respectively, and correspond with the depths reported on the left-most y axis.
Relative abundances (OTU read count/total sample read count) and inferred abundances (relative abundances × FCM cell counts) of prokaryotic OTUs and chloroplasts. Download Table S1, XLSX file, 0.2 MB (179KB, xlsx) .
Copyright © 2022 Saini et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In the chemocline, the inferred absolute abundance of prokaryote-classified cells peaked at 15 m (Fig. 3A, day 2), which corresponded with peak turbidity, POC, and PON (Fig. 1). The major phyla represented at this depth were Proteobacteria (48.52%; 439,195 cells/mL) and Bacteroidetes (25.40%; 229,927 cells/mL). The genus that dominated these phyla at 15 m was the purple sulfur bacterium (PSB) Chromatium (35.25%; 319,051 cells/mL). Chromatium (Proteobacteria) was significantly positively correlated with turbidity (R = 0.93; P value = 7.25e−05) and total cell counts (R = 0.99; P value = 8.66e−09), suggesting its significant contribution to the chemocline biomass (Fig. S3). The inferred absolute abundances of an unclassified Lentimicrobiaceae OTU, the second most abundant OTU in the chemocline at 15 m, was significantly positively correlated with secondary production (R = 0.8, P value = 5.09e−03), turbidity (R = 0.91, P value = 2.28e−04), and PLPs (R = 0.85, P value = 1.82e−03) yet negatively correlated with light (R = −0.78, P value = 7.33e−03) and oxygen concentration (R = −0.87; P value = 1.10e−03) (Fig. S3). Considering that Lentimicrobium species are known anaerobic fermenters (36), their strong correlation with secondary production and microbial biomass suggests their unknown contribution toward recycling of the chemocline biomass. Furthermore, though Firmicutes was not a dominant phylum itself, an unclassified genus of the family Erysipelotrichaceae (Firmicutes) (3.91%; 35,435 cells/mL), was among the abundant genera at 15 m (Fig. S4; Table S1). A green sulfur bacterium (GSB), Chlorobium (2.52% 20,849 cells/mL), was the fifth most abundant genus of the chemocline. The phototrophic sulfur bacteria (PSB) Thiocystis, Thiodictyon, and Lamprocystis and sulfate-reducing Desulfocapsa and Desulfobulbus, previously observed in Lake Cadagno (14, 17, 37), were present at less than 0.01% of the total OTUs in the chemocline (Table S1).
Inferred absolute abundances of bacterial genera through the water column of meromictic Lake Cadagno. Absolute abundances are inferred by multiplying the relative abundances (OTU read count/total sample read count) by FCM cell counts. Download FIG S4, PDF file, 0.01 MB (6.9KB, pdf) .
Copyright © 2022 Saini et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
On average, the total PLP counts in the monimolimnion (17.0 to 19.0 m) were less than half that of the peak chemocline counts and more than twice the average mixolimnion counts (Fig. 2A). The average relative and inferred (Fig. 3A and B) abundances of the dominant populations reflected those of the chemocline at 15 m, which included Proteobacteria (34.05% of total OTUs; 148,019 cells/mL), Bacteroidetes (24.65%; 107,165 cells/mL), Cyanobacteria (13.51% of total OTUs; 58,726 cells/mL), and Actinobacteria (5.84%; 25,425 cells/mL). In addition to bacteria, archaeal OTUs of the Methanoregula and Woesearchaeia genera were identified in the monimolimnion but in low abundances (<1,000 cells/mL) (Table S1), which may be partially attributed to primer bias against archaea in the 16S rRNA gene V4 amplification performed (38, 39).
Phytoplankton in the mixolimnion, chemocline, and monimolimnion.
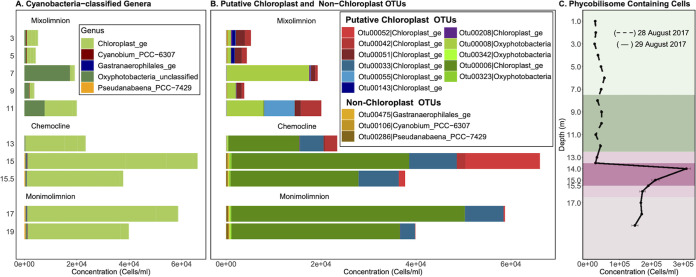
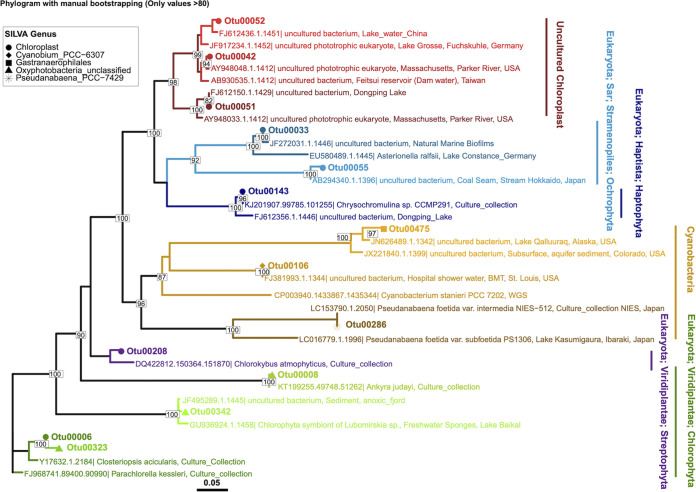
Cyanobacteria were consistently among the abundant phyla at each depth, but at no depth did they dominate (Fig. 3A). Within this phylum, Oxyphotobacteria and chloroplast-classified OTUs dominated, representing 94 to 100% of the total Cyanobacteria-classified OTUs (Fig. 4A). Of the low-abundance cyanobacterial genera, Cyanobium was found throughout the water column, while Pseudanabaena and Gastranaerophilales were predominantly found at 15 m and deeper (Fig. 4A and B). To better understand the relationship between the microbial guilds of Lake Cadagno, we sought phylogenetic evidence to support the origin of these putative chloroplast OTUs, as described in previous literature (40, 41). The phylogeny of all Cyanobacteria-classified OTUs and their two nearest neighbors in SILVA (42) indicated that the chloroplast and Oxyphotobacteria OTUs clustered in clades with chloroplast 16S rRNA genes of cultured eukaryotic algae (Fig. 4B): Chlorophyta (Otu00008, Otu000342, Otu00006, Otu00323), Ochrophyta (Otu00033), Streptophyta (Otu00208), Haptophyta (Otu00143), and uncultured chloroplasts (Otu00042, Otu00051, Otu00052, Otu00055).
FIG 4.
Abundances and phylogenies of Lake Cadagno phytoplankton based on 16S rRNA gene amplicon sequencing and flow cytometry. Mixolimnion samples (0 to 11.0 m) were collected on day 1, and chemocline and monimolimnion samples (12.0 to 19.0 m) were collected on day 2. Abundances in panels B and C are acquired by scaling 16S rRNA gene read counts by FCM cell counts. (A and B) Inferred concentrations (cells/mL) of OTUs were assigned to the phylum Cyanobacteria, and of those, the inferred concentrations (cells/mL) of OTUs were classified as putative chloroplasts and non-chloroplasts at the genus level (B). (C) Depth profile of phycobilin-containing cell counts based on flow cytometry with a 640-nm laser. The high O2 mixolimnion, mid O2 mixolimnion, low O2 mixolimnion-chemocline transition zone, chemocline, lower-chemocline, and anoxic monimolimnion layers are indicated by light green, dark green, light lilac, lilac, dark lilac, and light brown backgrounds, respectively.
In the lower mixolimnion, between 7.0 and 11.0 m, where high oxygen, Chl a, and NPP persisted and peak Fv/Fm was observed (Fig. 1D and F), Otu00008, which is most closely related to the chloroplast of the cultured chlorophyte Ankyra judayi, dominated the subset of Cyanobacteria-classified OTUs, where they represented up to 89.97% of the total cyanobacterial OTUs of the mixolimnion (Fig. 4B and D). At 11.0 m, the next most abundant OTUs were Otu00042 and Otu00055, which belonged to a clade of chloroplasts from uncultured organisms, representing 21.53% and 31.63% of the total cyanobacterial OTUs, respectively.
A primary chlorophyll peak was identified in the chemocline between 13.0 and 14.0 m, where FCM-based counts of phycobilin-containing cells (309,333 cells/mL) (Fig. 4C), phycocyanin pigments (26 μg/L, day 2) (Fig. 1D), and Chl a pigments (45 μg/L, day 2) rose sharply (Fig. 1D, day 2). While Cyanobacteria-classified OTUs represented nearly a quarter of all OTUs at the primary chemocline chlorophyll peak, they represented only 7.30% of the total OTUs at the secondary chemocline chlorophyll peak (15 m) (Table S1). The most abundant chloroplast OTU throughout the chemocline, Otu00006, was most closely related to the chloroplasts of the cultured chlorophytes Parachlorella kessleri and Closteriopsis acicularis, the latter genus (43) observed in meromictic Lake Tanganyika (Fig. 5). The next most dominant chloroplast-like OTUs, Otu00033 and Otu00052, clustered with cultured Ochrophyta chloroplasts and uncultured chloroplasts, respectively (Fig. 5). In the monimolimnion, Otu00006 and Otu00033 dominated the Cyanobacteria-classified OTUs (Fig. 5), which were most closely related to Chlorophyta and Ochrophyta chloroplasts, respectively.
FIG 5.
Phylogenetic tree of the representative 16S rRNA gene amplicon sequences of putative chloroplast and nonchloroplast OTUs shown in Fig. 4B assigned as Cyanobacteria, along with their two nearest neighbor sequences from the SILVA database. Clades are color coded consistently with the coloring of OTUs shown in the bar plot of Fig. 4B.
Prokaryotic genotypic and phenotypic diversity trends.
Bacterial genotypic diversity (16S rRNA gene) and PLP phenotypic diversity (based on individual cell features distinguished by FCM [44]) both dropped at 7.0 m and rose in the lower mixolimnion, where oxygenic primary production peaked (Fig. 6A and B; Fig. 1F). Overall, genotypic alpha diversity (Shannon) trends were nearly uniform, except at the peak of turbidity (15.0 m), where PLP phenotypic alpha diversity peaked and moderate genotypic alpha diversity was observed (Fig. 6A and B). Below the chemocline, consistently high genotypic alpha diversity and phenotypic alpha diversity were observed in the anoxic monimolimnion (Fig. 6). The increases in genotypic and phenotypic alpha diversity in the lower chemocline and monimolimnion were consistent with previous 16S rRNA gene-based studies of Lake Cadagno (33) and other permanently stratified systems, including Powell Lake (45), Ursu Lake (46), and Fara Fund Lake (46). Given the evidence that in microbial ecology “diversity begets diversity” (47), the observed increase in substrate diversity in the lower chemocline and monimolimnion (Fig. 1) likely facilitated greater microbial functional diversity, such as the anaerobic methane oxidation and phosphorus recycling described at those depths (8, 48). In addition, the increased particulate matter (Fig. 1) may have created greater habitat heterogeneity, which is also likely to lead to greater community diversity. Finally, it is also possible that the high diversity in the monimolimnion may have been partially attributed to the accumulation of sinking cells from the mixolimnion and chemocline, which may or may not still be living but contribute to community diversity measures.
FIG 6.
Trends in microbial community genotypic (16S rRNA gene amplicon sequencing, including prokaryotic and chloroplast-related OTUs) and phenotypic (PLP features based on flow cytometry [44]) alpha and beta diversity in Lake Cadagno. (A) Variation in genotypic alpha diversity (Shannon) with depth. (B) Variation in PLP phenotypic alpha diversity (Shannon). Mixolimnion samples (0 to 11.0 m) were collected on day 1, and chemocline and monimolimnion samples (12.0 to 19.0 m) were collected on day 2. Colors of data points represent the depth strata sampled. (C) PCoA represents the genotypic beta diversity dissimilarity (Bray-Curtis distance) of microbial communities. (D) PCoA represents the phenotypic beta diversity dissimilarity (Bray-Curtis distance) of microbial communities.
In the principal-coordinate analysis (PCoA) ordinations of bacterial community dissimilarity (Bray-Curtis), the first two components combined accounted for 76.9% and 46.2% of all observed variation in genotypic and phenotypic beta diversity, respectively (Fig. 6C and D). When both genotypic diversity and phenotypic diversity were considered, the oxic mixolimnion and the coclustering anoxic chemocline and monimolimnion samples separated along the first axis (Fig. 6C and D). In the phenotypic diversity ordination (Fig. 6D), the mixolimnion samples separated along the second axis by whether they originated from the high-oxygen (1.0 to 7.0 m, light green) or the mid-oxygen (8.0 to 11.0 m, dark green) zone (Fig. 6C and D). As with phenotypic alpha diversity, oxygen and light were negatively correlated with phenotypic and genotypic beta diversity (Fig. S3).
DISCUSSION
In this study, the investigation of viral, microbial, and biogeochemical spatial dynamics through Lake Cadagno’s water column improves our understanding of how biological and geochemical connections between the spatially segregated food webs in the Proterozoic ocean model may have manifested (2, 8, 10, 12).
Eukaryotic algae associated with oxygenic phototrophy in the mixolimnion.
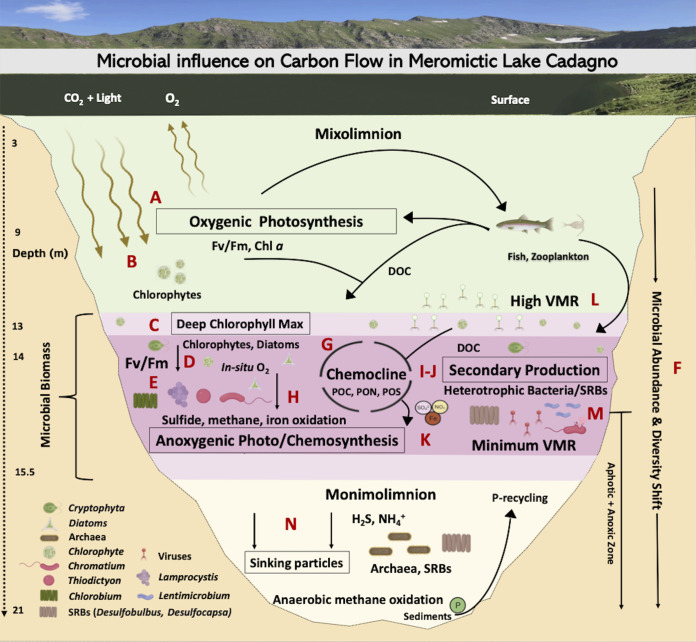
With the major transition to an oxygenated ocean 800 to 700 millions of years ago, the ever-reducing levels of toxic sulfide and relief of nitrogen limitations likely facilitated the evolution of modern eukaryote precursors, as proposed in the Proterozoic ocean model (2, 49). In the oxic mixolimnion of Lake Cadagno, we observed low levels of sulfur and ammonia in combination with high abundances of eukaryotic algal chloroplast OTUs and photosynthetic activity simultaneously with the low abundances of small (<40-μm) prokaryote-like particles. These patterns reflected the conditions predicted during the early period of eukaryotic evolution in the Proterozoic ocean (Fig. 7A). The chloroplast sequences identified in the mixolimnion were dominated by close relatives of the freshwater chlorophyte Ankyra judayi (OTU00008) (Fig. 7B), which has not yet been described in Lake Cadagno. This first report of Ankyra judayi in Lake Cadagno may reflect the dearth of studies focusing on the eukaryotic phytoplankton composition of the Lake Cadagno mixolimnion, especially those using modern molecular techniques. Likely due to its ability to endure high UV radiation, Ankyra judayi has been observed to outcompete other phytoplankton and to grow to high densities in a high-alpine Andean lake (50). These traits may explain the evidence for its presence in high-alpine Lake Cadagno in the sun-lit mixolimnion during the month of August, when UV intensity is known to peak (51), and may reflect traits of early photosynthesizing eukaryotes. Our findings are consistent with prior microscopy-based studies reporting that the majority of the algal biomass was attributed to eukaryotic algae in the Lake Cadagno mixolimnion (20). Moreover, eukaryotes also persisted in the anoxic zone (15 to 19 m), however, Fv/Fm data suggested that they were not photosynthetically active and their presence may be explained by settling from photic zones. These observations of the Lake Cadagno mixolimnion provide one possible model for the period of early evolution of eukaryotes in the oxic strata of the Proterozoic ocean, where, though spatially segregated, they may have cooccurred with abundant and productive chemocline and monimolimnion bacteria.
FIG 7.
Microbial loop of meromictic Lake Cadagno. Microbiology icons were created with BioRender.
Revisiting the microbial linkages between in situ oxygen production and anoxygenic processes in the chemocline.
The processes of the major microbial oxygenic and anoxygenic guilds have been reported to contribute almost equally to organic matter production in the Lake Cadagno chemocline (20), as they may have in ancient ocean chemoclines. We observed the coexistence of oxygenic (chlorophytes, diatoms, cyanobacteria) and anoxygenic (PSB and GSB) phototrophs in the Lake Cadagno chemocline (Fig. 7C to E); however, unlike previous conclusions (33, 51), cyanobacterial 16S amplicon reads were rare, while those of chloroplasts of chlorophytes and stramenopiles were abundant in this study. The presence of chlorophytes and stramenopiles in the Lake Cadagno chemocline were consistent with previous studies that indicated in situ oxygen production by photosynthetic algae and diatoms (52, 53). Given that eukaryotes also contain phycobilisomes (54) and the abundance of chloroplast 16S rRNA reads of eukaryotic algae observed here, we propose that previous conclusions that attributed phycobilin-based cell counts to cyanobacterial blooms (33, 51) be revisited. Future studies should investigate the relative contribution of cyanobacteria and eukaryotic phytoplankton to oxygenic photosynthesis in the chemocline.
Besides oxygenic and anoxygenic primary production, aerobic respiration has also been proposed to be coupled with biogeochemical processes of the ancient ocean chemocline (2). However, evidence for which microbes are associated with such activity in Proterozoic analogs is sparse. Though typically thought of for its role in photoautotrophy, Chromatium, the dominant chemocline microbe in this study, is also known to perform light-independent chemolithoautotrophy and has been reported to contribute up to 40% of total dark carbon fixation through aerobic sulfide oxidation (53) (Fig. 7L). In that study, aerobic respiration by Chromatium okenii cooccurred with in situ production of oxygen by photosynthetic algae (53), methane oxidation by methanotrophs. and iron oxidation likely by Chlorobium and Rhodobacter, where iron oxidation accounted for 10% of total primary production in the chemocline (52, 55) (Fig. 7H). Through their oxygen production, the oxygenic phototrophs (chlorophytes, diatoms, cyanobacteria) observed in the chemocline may play a critical role by enabling these oxidative metabolic processes central to Lake Cadagno’s biogeochemical cycling. These observations indicate that the ancient purple sulfur bacteria may have likely adapted the aerobic metabolism with the rise in oxygenic phototrophs following a great oxygenation event and support the proposal of a mixed community of oxygenic and anoxygenic primary producers in the chemocline of the Proterozoic ocean model (2). Future work targeting the ecological and metabolic interactions between chlorophytes, diatoms, cyanobacteria, and chemocline bacteria at the oxic-anoxic interface of Lake Cadagno will improve our understanding of the evolution of life in ancient oceans.
Potential for nitrogen metabolism by phototrophic sulfur bacteria and photosynthetic microbial eukaryotes in Proterozoic oceans.
The nitrogen cycle likely mediated by N2-fixing cyanobacteria and purple and green sulfur bacteria has been proposed to provide inorganic nitrogen essential for ancient ocean photoautotrophy (2, 12). The shift of oxygenic to anoxygenic photosynthesis observed in this study was coincident with a decreasing efficiency of oxygenic photosynthetic microbes (Fv/Fm), increasing concentrations of hydrogen sulfide, and a shift in bacterial abundance and beta diversity (Fig. 7F). Among the anoxygenic phototrophic sulfur bacteria, Chromatium okenii has been found to perform >80% of the average bulk N2 fixation (12) and 40% of the ammonia uptake (19) in the chemocline of Lake Cadagno. The highest abundance of Chromatium at the peak of PON and turbidity observed here suggested the potential for ongoing N fixation and nitrogen assimilation in the Lake Cadagno chemocline. In addition to Chromatium, other phototrophic purple sulfur bacteria (Lamprocystis and Thiodictyon) and green sulfur bacteria (Chlorobium) are also capable of nitrogen fixation and assimilation (12, 15, 19). These genera are also observed in this study but in low abundances compared to Chromatium. While N2 fixation is important, ammonia assimilation has been considered to be the major N source, capable of sustaining ~80% of autotrophic carbon fixation in the Lake Cadagno chemocline (12). The eukaryotic phytoplankton observed in this study may also be capable of nitrogen assimilation (56); however, direct evidence for this in Lake Cadagno is lacking. Overall, the diazotrophic purple (Chromatium, Lamprocystis, and Thiodictyon) and green (Chlorobium) sulfur bacteria, along with the microbial eukaryotes (chlorophytes), may have assimilated the upwardly diffusing ammonia and facilitated the nitrogen needed for organic matter production (POC, PON, PS) in the Lake Cadagno chemocline (Fig. 7G). Such relief from nitrogen limitation may have supported the persistence of PSB, GSB, and chlorophytes in the permanently stratified Proterozoic ocean (57, 58).
Nutrient remineralization through secondary production contributes to chemocline biomass.
While it is broadly recognized that the PSB and GSB that dominated the Lake Cadagno chemocline in this study are largely responsible for anoxygenic primary production in this stratum (Fig. 7E) (14, 59), we found that as with primary production, secondary production rates, reported here for the first time, were the highest in the chemocline, exceeding secondary production in the mixolimnion and monimolimnion by 42.4- and 1.9-fold, respectively (Fig. 7I). Here, we estimated that the N generated by secondary production can contribute on average 8.05% ± 1.06% (standard deviation [SD]) of the autotrophic N demand in the chemocline, while N2 fixation has been reported to support up to 7.3% (12). In summary, the observations of peak secondary production in the zone of greatest biomass suggested that remineralization is important for supplying nutrients to the active chemocline microbial assemblage.
By pairing these secondary production rates with a description of heterotrophic populations present, we shed light on the clades that may be involved in nutrient remineralization in the chemocline. Lentimicrobium was the second most abundant genus in this study and has been previously implicated in fermentation under limited light and oxygen (36). Sulfate-reducing bacteria (SRBs) (Fig. 7J), such as the observed Desulfocapsa and Desulfobulbus, are known to form aggregates with phototrophic PSB (e.g., Thiodictyon syntrophicum, Lamprocystis purpurea), and both cooccurred in the chemocline (37, 60, 61). Through secondary production, heterotrophic microbes are likely involved in the decomposition of particulate organic matter (POC, PON) and particulate sulfur (PS) and the supply of inorganic nutrients necessary to carry out primary production and sustaining populations of oxygenic and anoxygenic photoautotrophs in the chemocline (Fig. 7K). Trends in putative autotrophic and heterotrophic population abundances combined with the secondary productivity rates provide evidence for microbial potential toward recycling of organic matter proposed in the Proterozoic ocean model (2, 8, 10, 12).
Potential for the role of viruses in modulating microbial activity and nutrient cycling.
Viruses serve as top-down controls on host populations and reprogram host metabolism during infection (23, 24, 62). Evidence for viral activity has been observed in ancient cyanobacterial mat analogues, including their potential for resource scavenging through the degradation of host phycobilisomes, a pigment central to photosynthesis (63). The observed colocalization of high VMR and photosynthetically active phytoplankton in the lower mixolimnion and mixolimnion-chemocline transition may be explained by ongoing lytic viral infections of phytoplankton. Viral lysis products may contribute to the DOC peak and support the high secondary production near the DCM (Fig. 7L). Such a scenario was observed in the phytoplankton Ostreococcus virus-host system, where host densities were maintained concurrently with continual lysis due to phase switching between resistant and susceptible hosts (64). If this scenario is occurring, the sustained lysis of phytoplankton has the potential to release DOC in a viral shunt near the mixolimnion-chemocline transition. A DOC peak was indeed observed slightly below this depth in Lake Cadagno. However, given that the VMR is a feature that emerges from a number of underlying virus-microbe interactions, viral life history traits, and environmental cofactors (65), the prediction of sustained phytoplankton viral predation at the mixolimnion-chemocline transition requires empirical confirmation.
Previous studies have identified genomic evidence for viral infection of the presumed major carbon assimilators in the chemocline, Thiodictyon syntrophicum and Chromatium okenii (14). Genomes of these organisms sequenced from Lake Cadagno contain CRISPR elements (66, 67), which derive from a type of acquired immunity against invading viral and plasmid DNA. This suggests that viruses may play a role in the modulation of carbon and sulfur cycles, as has been proposed in the hydrothermal vent (68) and wetland (69) microbial communities, where viruses have been found to carry genes central to methanogenesis (mcrA) and sulfur reduction (dsrA, dsrD). Active viral lysis also provides a new possible mechanism to explain the observed low abundances of Chromatium okenii, which have previously been attributed to microbial predation (19). Despite genomic evidence suggesting active infection of these populations (66, 67), VLP counts did not rise with PLP counts in the chemocline. Yet, our observation of a minimum VMR (Fig. 7M) and peak microbial abundances in the Lake Cadagno chemocline supported an emerging trend in stratified meromictic lakes, as the same observation was made in meromictic Ace Lake (Vestfold Hills of East Antarctica) (27). Such coincident low VMR and high microbial abundances have been recognized as a hallmark of the power-law relationship proposed to describe the VMR dynamics in aquatic systems (65). We propose viruses as an important component of the Proterozoic ocean microbial community. Population-specific studies of viral infection are needed in Lake Cadagno to understand the impact of viruses on the evolution and function of microbes that underlie its biogeochemistry and to shed light on the roles viruses may have played in the ancient ocean.
Conclusion.
This work highlights how biogeochemical exchange within microbial guilds may be driving biomass accumulation to support the food web of the permanently stratified Lake Cadagno. Ultimately, the organic matter generated through eukaryotic and bacterial activity and viral remineralization in the chemocline can be made available to zooplankton and fish. The collective microbial biomass, including viruses, will eventually sink to the sediments contributing to the organic matter burial (Fig. 7N) (17, 70). The observed trends suggest a high degree of interconnectedness and ecological importance of microbial guilds in stratified ancient oceans.
This study inspires future research directions toward a better ecological and biogeochemical understanding of Lake Cadagno and similar meromictic ancient ocean analogues. Our community composition and secondary production data suggested that the ecological role and metabolic potential of heterotrophic bacteria that recycle chemocline biomass in the presence of sulfur and ammonia, such as the second most abundant bacterium, Lentimicrobium, and their potential toward recycling chemocline biomass were intriguing unknowns. Furthermore, eukaryotic phytoplankton has been known to contribute to the deep chlorophyll maximum of Lake Cadagno’s chemocline for 2 decades (20), yet insights into the cellular machinery that allows them to photosynthesize under limited light remain unknown. Further, while primary producers (phytoplankton, phototrophic sulfur bacteria) and heterotrophic bacteria are central to the realized function of the lake biogeochemistry and ecology, viruses may shuffle the repertoire of genes controlling both microbial guilds, hence modulating biogeochemical (carbon, nitrogen and sulfur) cycles of the lake. Our empirical observations provide a better understanding of the role that microbes of the chemocline (oxic-anoxic boundary) may have played in the Proterozoic transition from the primarily anoxygenic to oxygenic metabolisms that dominate nutrient biogeochemical cycling in modern oceans (2, 71, 72).
MATERIALS AND METHODS
Water sample collection and physicochemical profiling.
This study was conducted in Lake Cadagno (21 m deep), a high alpine meromictic lake situated at 1,921 m above sea level in the Southern Alps of Switzerland (73). Due to the extensive number of samples collected and experiments performed for process measurements, sampling was done over two subsequent days. The oxic mixolimnion (0 to 10 m) was sampled on 28 August 2017 (day 1), and the chemocline (11 to 16 m) and monimolimnion (17 to 19 m) were sampled on 29 August 2017 (day 2). Internal waves are known to affect the position of Lake Cadagno’s stratified water masses, though their frequency is unknown (34). To control for this anticipated variability, a number of parameters are collected on both days to identify the depths of the zones, guiding the sampling of the mixolimnion on day 1 and the chemocline and monimolimnion on day 2. Water sampling and characterization of physical (turbidity, temperature, oxygen, conductivity), biological (Chl a, phycocyanin), and chemical (H2S, NH4+) parameters of the water column were performed as previously described (18). Water was sampled using a double-diaphragm Teflon pump (Almatec PSG Germany GmbH) connected to an acid-washed low-density polyethylene (LDPE) line deployed from a platform that was anchored above the deepest part of Lake Cadagno (46.55087°N, 8.71152°E).
Vertical lake profiles were determined using an autonomous conductivity-temperature-depth sensor (CTD; Ocean Seven 316 Plus CTD; Idronaut S.R.L.). The CTD recorded pressure (dbar), temperature (°C), and conductivity (mS/cm); dissolved oxygen (mg/L) was measured with a pressure-compensated polarographic sensor. The error in oxygen profiles for day 2 was manually adjusted by referring to day 1 measurements, which indicated approximately zero oxygen within and below the chemocline. CTD profiles from the whole water column (0 to 20 m, days 1 and 2) were used to identify the distribution of the mixolimnion, chemocline, and monimolimnion and guided water sampling strategy. The CTD was equipped with an LI-192 underwater quantum sensor (Li-Cor Biosciences, NE, USA) that continuously recorded photosynthetically active radiation (PAR-W/m2) and a TriLux multiparameter algal sensor (Chelsea Technologies, Ltd., Surrey, UK) that measured in vivo Chl a (μg/L), an indicator of phytoplankton (including eukaryotic autotrophs and cyanobacteria) biomass (74), and phycocyanin (μg/L), a pigment characteristic of cyanobacteria (75).
Chemical parameters. (i) Dissolved compounds.
Samples for the analysis of dissolved compounds (DOC) were filtered through an AcroPak filter cartridge (Pall) with a 0.8-μm prefilter and a 0.2-μm final filter. Filters were collected in 30 mL acid-washed and pyrolyzed Pyrex tubes filled to the top and supplemented with 100 μL 2 M HCl. Tubes were stored in the dark at 4°C until processed (November 2017; University of the Geneva) using a Shimadzu TOC-LCPH analysis system. Blanks consisting of Milli-Q water were made, and calibration was done using the standard from 0.2 to 5 ppm. Analyses of dissolved ammonium (NH4+) and hydrogen sulfide (H2S) were done on-site at the Alpine Biology Centre (CBA, Piora, Switzerland) with freshly collected water samples using a UV-visible spectrophotometer (DR 3800; Hach) (76).
(ii) Particulate compounds.
Particulate organic carbon (POC) and nitrogen (PON) and particulate organic sulfur (POS) were collected by filtering 150 to 500 mL of lake water (volume necessary to begin to clog the filter) on precombusted 47-mm GF/F filters (5 h at 550°C) (catalog no. WH1825-047; Whatman, WI, USA), which were then acidified (500 μL 1 M HCL, added twice at a 1-h interval) and dried in an oven (65°C) overnight in acid-washed Pyrex petri dishes. Samples were sealed using Parafilm and aluminum foil until analysis (November 2017, University of Geneva) using an elemental analyzer (2400 series II CHNS/O elemental analyzer; PerkinElmer). Procedural blanks using Milli-Q water were performed (24 to 25 November 2017) in duplicate to measure the background signal, which was used to correct the concentrations of samples. POC, PON, and POS were expressed in mg/L (ppm). Because purple sulfur bacteria also contain inorganic sulfur globules inside their cell (61, 67), particulate organic sulfur (POS) is referred to as particulate sulfur (PS).
Biological parameters. (i) Primary production.
Primary productivity was determined using incubation with NaH14CO3 (catalog no. NEC086H005MC; Perkin Elmer, Waltham, MA, USA) (5 mCi, 1 mL in glass ampoule), which was freshly diluted into 4 mL Milli-Q water at pH 9.6 (adjusted using NaOH; Sigma) and added at a dose of 1 mCi/L of lake water, as described previously (77). Incubation was carried out along an incubation chain deployed at six depths for 23 h on 28 to 29 August 2017. Each depth chain consisted of a metallic ring fixed to a rope at the desired depth (3, 5, 7, 9, 11, and 13 m). The ring was surrounded by six arms on a horizontal plane, each of which ended with a bottle holder. Seventy-five-milliliter acid-washed glass bottles were filled to the top with lake water (ca. 100 mL) at the corresponding depth and spiked with 14C prior to being deployed on each level of the incubation chain. At each depth, three transparent bottles were incubated for activity measurements at in situ natural light intensities, and three amber bottles were incubated for dark community respiration measurements. At the end of the incubation, each bottle was filtered onto 47-mm GF/F filters (catalog no. WH1825-047; Whatman, WI, USA), which were then stored in a plastic petri dish. Petri dishes were placed in a sealed box containing calcium hydroxide (slaked lime) powder, and after the addition of HCl (1 M, 500 μL) onto the filters, inorganic carbon was degassed overnight. The calcium hydroxide captures the degassed 14C, which is further treated as solid radioactive waste. Filters were then collected into 20-mL scintillation vials, supplemented with 10 mL of Ultima Gold AB cocktail, and shaken manually. Primary productivity was expressed in micromoles of net carbon fixation per hour either per liter of lake water or per Chl a, considering a dissolved inorganic carbon concentration of 1.8 mM C (16).
(ii) Maximum quantum yield.
The maximum quantum yield (Fv/Fm) informs the photosynthetic health and biological activity of algae based on photophysiological characteristics of the chlorophyll photosystem II. Maximum quantum yield was determined using a fast repetition rate fluorometer (FRRF; FastOcean PTX coupled to a FastAct base unit; Chelsea Technologies). Water for maximum quantum yield measurements was preconcentrated 10-fold by gently resuspending the cells as the sample passed through a 0.22-μm 47-mm polycarbonate filter (catalog no. GVWP04700; Millipore, Darmstadt, Germany) using a hand pump (pressure below 15 mbar) and a 47-mm filtration unit (catalog no. 300-4000; Nalgene). This step was done to ensure a high enough method sensitivity and has been shown to alter neither Chl a nor cell integrity of samples from Lake Geneva (78). The FRRF was used in single turnover mode to record, in a 45-min dark-adapted natural sample, the basal (Fb) and maximal (Fm) fluorescence following exposure to intense light. The sample was then gently filtered on a 0.22-μm, 47-mm filter (catalog no. GVWP04700; Millipore, Darmstadt, Germany) to record the residual fluorescence (Fr). For each sample, Fb − Fr represented the initial dark-adapted fluorescence, F0, associated with intact cells. Fv/Fm was then calculated using the ratio (Fm − F0)/Fm.
(iii) FCM.
Total cell counts from fresh unpreserved samples collected on-site, as well as 0.22-μm-filtered cell-free Milli-Q water controls, were generated in triplicate on a BD Accuri C6 cytometer (Becton, Dickinson, San José, CA, USA) using two lasers (blue, 488 nm; red, 640 nm) and four fluorescence detectors (lasers, 488 nm: FL1 = 533/30, FL2 = 585/40, FL3 = 670; laser, 640 nm: FL4 = 675/25). Standard deviations of the triplicates were calculated to assess variations and to evaluate the potential for counting biases obtained from the flow cytometry (FCM) cell counts. FL4 detectors were specifically used to detect emission from phycobilin, a pigment proxy for cyanobacteria. Cells were stained with SYBR green I (catalog no. S7563; Molecular Probes, Eugene, OR) at a ratio of 1:10,000 (vol/vol), following incubation for 13 min at 37°C in the dark (13). A histogram of event counts versus green fluorescence (FL1 > 1,100) allowed quantification of total cells.
VLPs were counted using flow cytometry (79) after prefiltration of lake water through 55-μm mesh to remove higher organisms such as zooplankton (20), and the lake water was subsequently passed through 0.22-μm filters to remove large particles and cells. To discern viral particles from cell-sized particles or instrument background, 0.22-μm- and 0.02-μm-filtered Milli-Q water samples were used as cell- and virus-free controls, respectively. In triplicate, a 1-mL sample from the filtrate was fixed by adding 10 μL of glutaraldehyde (25% stock solution) in sample cryovials. The samples were fixed for 15 min at room temperature, subsequently stored in liquid nitrogen for shipping, and then stored at –80°C until they were processed in February 2018. Free virus-like particle counts were obtained using a FACSCalibur flow cytometer with a 15-mW 488-nm laser (Becton, Dickinson Biosciences, Grenoble, France). Samples were thawed at 37°C and then diluted in autoclaved and 0.02-μm-filter-sterilized Tris-EDTA (0.1 mM Tris-HCL and 1 mM EDTA, pH 8). Samples were then stained for 10 min at 75°C using SYBR green I (Molecular Probes) at a 1:10,000 final concentration (80). Standard deviations were calculated to assess the potential bias resulting during VLP counting.
(iv) Phenotypic alpha diversity of PLPs.
FCM-based phenotypic fingerprints have been proven to be a rapid, inexpensive, and robust method to spatially and temporally assess microbial community dynamics (44) and have been considered effective in assessing microbial diversity in aquatic systems (81, 82). Raw flow cytometry files were used to estimate phenotypic traits (morphology and nucleic acid content) and were transformed and normalized using transform and flowBasis functions in the Phenoflow package (44) (https://github.com/rprops/Phenoflow_package) in R (v4.1.0) (83) and RStudio (v1.4.1106) (84). Briefly, the height values of the two fluorescence parameters (FL1, FL3) and two scatter parameters (side scatter [SSC], forward scatter [FSC]) recorded for each cell were treated with a hyperbolic arcsine transformation. These phenotypic intensity values were then scaled to a 0 to 1 range. Each parameter was then normalized based on the average maximum FL1-H intensity value over the data set. The cytometry fingerprint represents community structure in terms of the phenotypic attributes of the cells that comprise the community; essentially, any feature that influences the way the laser interacts with passing particles, such as morphology, size, and nucleic acid content, is captured in the phenotypic fingerprint. Using this approach, prokaryotic phenotypic diversity metrics have been shown to be highly correlated with community taxonomic diversity (44). To calculate phenotypic alpha diversity, Phenoflow calculates Hill number diversity indices (Dq) from order 0 to 2, where 0 represents the richness and q > 1 represents richness and evenness, also referred to as D0 (richness), D1 (Shannon), and D2 (inverse Simpson) (85). Phenotypic beta diversity was estimated from the phenotypic feature vectors using the Bray-Curtis distance metric, and the beta diversity ordination was then calculated using nonmetric multidimensional scaling. Fluorescence detectors (lasers, 488 nm: FL1-H = 533/30, FL3-H = 670) were used to target PLP diversity. Polygon gating was applied to filter out the noise observed in control samples (see Fig. S1 in the supplemental material). Clockwise, the polygon gate coordinates (x, y) were as follows: (7, 6), (15, 6), (15, 17), and (7, 10). Kernel density estimates were calculated by applying a binning grid (128 × 128) to FSC-H (forward scatter height), SSC-H (side scatter height), FL1-H, and FL3-H. The obtained kernel densities values were assigned into one-dimensional vectors, termed “phenotypic fingerprints.”
Scatter plot of flow cytometry events visualized with a BD6 flow cytometer interface (A and B) and RStudio Phenoflow package (A to D) for one replicate of the 1-m sample. FCM events representing prokaryote-like particles (PLPs) using automatic gating via a four-quadrant layout (A) and manual gating (B and C). (D) FCM events in 0.2-μm-filtered, cell-free Milli-Q water (control). The red outline represents the gating strategy for cell counts and phenotypic diversity analysis. This gate was applied to all samples. Download FIG S1, PDF file, 1.4 MB (1.5MB, pdf) .
Copyright © 2022 Saini et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(v) Secondary production.
Heterotrophic bacterial production, also termed secondary production, was estimated by the microcentrifuge [3H]leucine bacterial production method (86). A working solution of 5 μCi mL−1 leucine was made from a manufacturer’s stock solution of radioactive leucine, l-[3,4,5-3H(N)]-leucine (catalog no. NET460A005MC; PerkinElmer, MA, USA). To reduce the potential for autodegradation of the leucine radiolabel, stock [3H]leucine material was stored in the dark at 2 to 4°C (never frozen) and used for the incorporation experiment within 5 days. Two replicates and one trichloroacetic acid (TCA)-killed control (5% [vol/vol] final concentration) (catalog no. 91228; Sigma) were used for each depth. Briefly, 1.5 mL of lake water samples was incubated with a mixture of radioactive and nonradioactive leucine at final concentrations of 20 nM. Using the [3H]leucine working solution, enough “hot” leucine was added to each tube to create a sufficient signal (50% or 85% of hot leucine was used in the mixture for samples collected above and below 13-m depths, respectively). Incubations were conducted for 2 h in a dark incubator at in situ temperatures that corresponded to their sample depths. The saturation of leucine incorporation over this period was tested with 20, 30, and 40 nM total leucine. At the end of the incubation, 200 μL of 50% TCA was added to all but the control tubes to terminate leucine incorporation. To facilitate the precipitation of proteins, bovine serum albumin (BSA; Sigma) (100 mgL−1 final concentration) was added, and then samples were centrifuged at 16,000 × g for 10 min (87). The supernatant was discarded, and the resultant precipitated proteins were washed with 1.5 mL of 5% TCA by vigorous vortexing and again centrifuged (16,000 × g for 10 min). The supernatant was discarded. Subsequently, 1.5 mL of UltimaGold LLT (part no. 6013681; PerkinElmer, MA, USA) was added to each vial, mixed, and allowed to sit for >24 h at room temperature before the radioactivity was determined using a liquid scintillation counter (Beckman LS6500). To calculate biomass production, the following conversion factors were applied: molecular weight of leucine of 131.2 g/mol, the fraction of leucine per protein of 0.073, the ratio of cellular carbon to the protein of 0.86, and 2.22 × 106 dpm/μCi to convert disintegrations per minute (the measure of radioactivity) to μCi. A factor of 1.55 kg C mol leucine−1 was used to convert the incorporation of leucine to carbon equivalents, assuming no isotope dilution (88).
The contribution of remineralization toward autotrophic N demand was calculated using the following equations:
| (1) |
where BR is bacterial respiration proxy for remineralization, BP is bacterial production, BGE is bacterial growth efficiency (=0.26) (89), and bacterial respiration is expressed in mol C L−1 day−1.
| (2) |
where BR is bacterial respiration proxy for remineralization, C:N ratio is the ratio of particulate organic carbon (POC) to particulate organic nitrogen (PON), and N generated by heterotrophic bacteria is expressed in μmol N L−1 day−1.
| (3) |
where the average C fixation rate between a depth of 13.7 to 15.5 m is 31.1 μmol C L−1 day−1 (12), the C:N ratio is the ratio of particulate organic carbon (POC) to particulate organic nitrogen (PON), and the autotrophic N demand is expressed in μmol N L−1 day−1.
| (4) |
Statistical analyses.
The magnitude and significance of relationships between physical and biological parameters were determined with linear regressions and visualized with scatterplots using R (v4.1.0) (83) in RStudio (1.4.1106) (84) using ggplot2 (3.3.3) (90) and ggpubr (0.4.0) (91) packages (92). An R of >0.70 was considered a strong positive correlation, and a P value of <0.05 was considered statistically significant.
Illumina MiSeq 16S rRNA gene amplicon sequence analyses. (i) 16S rRNA gene amplicon sequencing.
Water was collected from 3.0, 5.0, 7.0, 9.0, 11.0, 13.0, 15.0, 15.5, 17.0, and 19.0 m (20 L per depth). One sample from each depth was collected for 16S sequencing, representing five from the mixolimnion, three from the chemocline, and two from the monimolimnion. As described in prior studies of Lake Cadagno (20), samples were prefiltered with a 55-μm filter to remove large particles and organisms, such as zooplankton. Using a peristaltic pump, the subsequent biomass was captured on 0.22-μm filters (catalog no. GPWP14250; 142-mm Express Plus filter; Millipore, Darmstadt, Germany) as previously performed for bacterial studies in Lake Cadagno (33, 37, 93, 94). Filters were flash-frozen in liquid nitrogen and stored at –80°C until extraction of DNA by use of a DNeasy blood and tissue kit (catalog no. 69504; Qiagen, Germantown, MD, USA) combined with a QIAshredder (catalog no. 79654; Qiagen, CA, USA), as described previously (95). The first elutions of DNA (50 μL each) were stored (4°C) until sequencing. Dual indexed primers were used, and the V4 region of the 16S rRNA gene amplicon was targeted and amplified, as described previously (96). Sequencing libraries were prepared using the Illumina Nextera Flex kit (Illumina, CA, USA) and sequenced using the MiSeq platform (500 cycles; Illumina) at the University of Michigan Microbiome Core facility.
The sequenced reads were quality controlled (mothur, minlength = 240, maxhomop = 8, maxlength = 275), assembled, trimmed, aligned using SILVA v132 16S rRNA reference sequences (mothur, reference position start = 1968, end = 11550). Following the removal of chimers, sequences were clustered at 97% identity into representative operational taxonomic units (OTU) using mothur (97). Samples that resulted in fewer than 5,000 reads were resequenced to ensure that the full profile was captured with at least two replicates per stratified zone (i.e., mixolimnion, chemocline, and monimolimnion). The OTUs were taxonomically classified using SILVA 132 (98) and TaxAss freshwater (version 1) (99) 16S rRNA gene databases.
(ii) Microbial community analysis.
Downstream community analysis was carried out by the phyloseq package (100) using R (4.1.0) (83) in RStudio (1.4.1106) (84) to analyze the genotypic alpha and beta diversity of prokaryotic communities. Bacterial and archaeal taxa were merged using merge_phyloseq. To minimize the impact of rare taxa, taxa not observed at least three times in 20% of the samples were removed using the filter_taxa function in phyloseq. OTU counts were transformed to relative abundance (0 to 1 range) by transform_sample_counts. OTUs were not rarefied to preserve rare OTUs for diversity analyses (101). For genotypic beta diversity PCoA (16S amplicon data), OTU counts were Hellinger transformed with the microbiome R package (1.15.3) (102).
To resolve the challenges associated with compositional relative abundance data (103, 104), “inferred absolute abundances” were calculated by multiplying relative abundances by flow cytometry (FCM)-generated cell counts (35) for each sample. Briefly, the relative abundances of prokaryotic OTUs of a given taxon (taxon read count/total read count in sample) were multiplied by the PLP concentrations (cells/mL) for that sample, as determined by flow cytometry. Scaling the 16S amplicon read counts by flow cytometry cell counts provided a more accurate assessment of changes in microbial populations through the water column. This is most strongly demonstrated in the differences observed between Fig. 3A and B. When the total cell counts increase in the chemocline, the magnitude of the changes in populations can be represented only when reads are scaled by cell counts (Fig. 3B).
(iii) Chloroplast 16S rRNA gene phylogeny.
The Alignment, Classification, and Tree (ACT) functions of SILVA were used to generate the phylogenetic tree of the representative 16S rRNA gene amplicon sequences of putative chloroplast and nonchloroplast OTUs that had been assigned as Cyanobacteria (105). Representative sequences of chloroplast OTUs were aligned to the SILVA 132 sequences (SINA 1.2.11). Two neighbors per query sequence (95% minimum sequence identity) were added to the tree with the chloroplast OTU sequences. The advanced variability profile was selected as “auto,” and sequences above 90% identity were kept in order to consider only the highest-confidence, most-homologous sequence. The phylogenetic tree of the OTUs was bootstrapped with RAxML (version 8) (106) with 20 maximum likelihood (ML) and 100 bootstrapped searches. The tree was annotated using the R package ggtree (2.2.4) (107) and Adobe Illustrator (25.2.1).
Data availability.
Raw sequences were deposited in the NCBI Short Read Archive (SRA) and are available under BioProject accession no. PRJNA717659. The code used for running the mothur analysis, statistical analyses, and figure generation is available in the project GitHub repository at https://github.com/DuhaimeLab/Lake_Cadagno_microbial_loop_Saini_et_al_2021.
ACKNOWLEDGMENTS
This project was funded by Swiss National Science Foundation grant no. PP00P2-138955 and through financial support to J.S.S. from the Swiss Confederation and the Ernst and Lucy Schmidheiny Foundation, which graciously enabled the development of this paper.
We thank the Duhaime Lab at the University of Michigan for supporting a learning environment to carry out the sequencing, flow cytometry, and ecological analysis and for productive feedback during the manuscript development. We thank Sandro Peduzzi and the Alpine Biology Center Foundation (Switzerland) for providing accommodations during Lake Cadagno sampling and for offering comments on the manuscript. The University of Michigan Microbiome Core facility aided in the generation of Lake Cadagno 16S rRNA gene data.
Contributor Information
Melissa B. Duhaime, Email: duhaimem@umich.edu.
María Mercedes Zambrano, Corporación CorpoGen.
REFERENCES
- 1.Canfield DE. 1998. A new model for Proterozoic ocean chemistry. Nature 396:450–453. doi: 10.1038/24839. [DOI] [Google Scholar]
- 2.Johnston DT, Wolfe-Simon F, Pearson A, Knoll AH. 2009. Anoxygenic photosynthesis modulated Proterozoic oxygen and sustained Earth’s middle age. Proc Natl Acad Sci USA 106:16925–16929. doi: 10.1073/pnas.0909248106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lyons TW, Reinhard CT, Planavsky NJ. 2014. The rise of oxygen in Earth’s early ocean and atmosphere. Nature 506:307–315. doi: 10.1038/nature13068. [DOI] [PubMed] [Google Scholar]
- 4.Shen Y, Buick R, Canfield DE. 2001. Isotopic evidence for microbial sulphate reduction in the early Archaean era. Nature 410:77–81. doi: 10.1038/35065071. [DOI] [PubMed] [Google Scholar]
- 5.Gulati RD, Zadereev ES, Degermendzhi AG (ed). 2017. Ecology of meromictic lakes. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 6.Tonolla M, et al. 2017. Lake Cadagno: microbial life in crenogenic meromixis, p 155–186. In Gulati RD, Zadereev ES, Degermendzhi AG (ed), Ecology of meromictic lakes. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 7.Stewart KM, Walker KF, Likens GE. 2009. Meromictic lakes, p 589–602. In Likens GE (ed), Encyclopedia of inland waters. Academic Press, New York, NY. [Google Scholar]
- 8.Xiong Y, Guilbaud R, Peacock CL, Cox RP, Canfield DE, Krom MD, Poulton SW. 2019. Phosphorus cycling in Lake Cadagno, Switzerland: a low sulfate euxinic ocean analogue. Geochim Cosmochim Acta 251:116–135. doi: 10.1016/j.gca.2019.02.011. [DOI] [Google Scholar]
- 9.Canfield DE, Farquhar J, Zerkle AL. 2010. High isotope fractionations during sulfate reduction in a low-sulfate euxinic ocean analog. Geology 38:415–418. doi: 10.1130/G30723.1. [DOI] [Google Scholar]
- 10.Dahl TW, Anbar AD, Gordon GW, Rosing MT, Frei R, Canfield DE. 2010. The behavior of molybdenum and its isotopes across the chemocline and in the sediments of sulfidic Lake Cadagno, Switzerland. Geochim Cosmochim Acta 74:144–163. doi: 10.1016/j.gca.2009.09.018. [DOI] [Google Scholar]
- 11.Wirth SB, Gilli A, Niemann H, Dahl TW, Ravasi D, Sax N, Hamann Y, Peduzzi R, Peduzzi S, Tonolla M, Lehmann MF, Anselmetti FS. 2013. Combining sedimentological, trace metal (Mn, Mo) and molecular evidence for reconstructing past water-column redox conditions: the example of meromictic Lake Cadagno (Swiss Alps). Geochim Cosmochim Acta 120:220–238. doi: 10.1016/j.gca.2013.06.017. [DOI] [Google Scholar]
- 12.Philippi M, Kitzinger K, Berg JS, Tschitschko B, Kidane AT, Littmann S, Marchant HK, Storelli N, Winkel LHE, Schubert CJ, Mohr W, Kuypers MMM. 2021. Purple sulfur bacteria fix N2 via molybdenum-nitrogenase in a low molybdenum Proterozoic ocean analogue. Nat Commun 12:4774. doi: 10.1038/s41467-021-25000-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danza F, Storelli N, Roman S, Lüdin S, Tonolla M. 2017. Dynamic cellular complexity of anoxygenic phototrophic sulfur bacteria in the chemocline of meromictic Lake Cadagno. PLoS One 12:e0189510. doi: 10.1371/journal.pone.0189510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Nezio F, Beney C, Roman S, Danza F, Buetti-Dinh A, Tonolla M, Storelli N. 2021. Anoxygenic photo- and chemo-synthesis of phototrophic sulfur bacteria from an alpine meromictic lake. FEMS Microbiol Ecol 97:fiab010. doi: 10.1093/femsec/fiab010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halm H, Musat N, Lam P, Langlois R, Musat F, Peduzzi S, Lavik G, Schubert CJ, Singha B, LaRoche J, Kuypers MMM. 2009. Co-occurrence of denitrification and nitrogen fixation in a meromictic lake, Lake Cadagno (Switzerland). Environ Microbiol 11:1945–1958. doi: 10.1111/j.1462-2920.2009.01917.x. [DOI] [PubMed] [Google Scholar]
- 16.Storelli N, Peduzzi S, Saad MM, Frigaard N-U, Perret X, Tonolla M. 2013. CO2 assimilation in the chemocline of Lake Cadagno is dominated by a few types of phototrophic purple sulfur bacteria. FEMS Microbiol Ecol 84:421–432. doi: 10.1111/1574-6941.12074. [DOI] [PubMed] [Google Scholar]
- 17.Posth NR, Bristow LA, Cox RP, Habicht KS, Danza F, Tonolla M, Frigaard N-U, Canfield DE. 2017. Carbon isotope fractionation by anoxygenic phototrophic bacteria in euxinic Lake Cadagno. Geobiology 15:798–816. doi: 10.1111/gbi.12254. [DOI] [PubMed] [Google Scholar]
- 18.Ellwood MJ, Hassler C, Moisset S, Pascal L, Danza F, Peduzzi S, Tonolla M, Vance D. 2019. Iron isotope transformations in the meromictic Lake Cadagno. Geochim Cosmochim Acta 255:205–221. doi: 10.1016/j.gca.2019.04.007. [DOI] [Google Scholar]
- 19.Musat N, Halm H, Winterholler B, Hoppe P, Peduzzi S, Hillion F, Horreard F, Amann R, Jørgensen BB, Kuypers MMM. 2008. A single-cell view on the ecophysiology of anaerobic phototrophic bacteria. Proc Natl Acad Sci USA 105:17861–17866. doi: 10.1073/pnas.0809329105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Camacho A, Erez J, Chicote A, Florin M, Squires MM, Lehmann C, Backofen R. 2001. Microbial microstratification, inorganic carbon photoassimilation and dark carbon fixation at the chemocline of the meromictic Lake Cadagno (Switzerland) and its relevance to the food web. Aquat Sci 63:91–106. doi: 10.1007/PL00001346. [DOI] [Google Scholar]
- 21.Falkowski PG, Katz ME, Knoll AH, Quigg A, Raven JA, Schofield O, Taylor FJR. 2004. The evolution of modern eukaryotic phytoplankton. Science 305:354–360. doi: 10.1126/science.1095964. [DOI] [PubMed] [Google Scholar]
- 22.Hamilton TL, Bryant DA, Macalady JL. 2016. The role of biology in planetary evolution: cyanobacterial primary production in low-oxygen Proterozoic oceans. Environ Microbiol 18:325–340. doi: 10.1111/1462-2920.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Breitbart M, Bonnain C, Malki K, Sawaya NA. 2018. Phage puppet masters of the marine microbial realm. Nat Microbiol 3:754–766. doi: 10.1038/s41564-018-0166-y. [DOI] [PubMed] [Google Scholar]
- 24.Zimmerman AE, Howard-Varona C, Needham DM, John SG, Worden AZ, Sullivan MB, Waldbauer JR, Coleman ML. 2020. Metabolic and biogeochemical consequences of viral infection in aquatic ecosystems. Nat Rev Microbiol 18:21–34. doi: 10.1038/s41579-019-0270-x. [DOI] [PubMed] [Google Scholar]
- 25.Weinbauer MG, Bettarel Y, Cattaneo R, Luef B, Maier C, Motegi C, Peduzzi P, Mari X. 2009. Viral ecology of organic and inorganic particles in aquatic systems: avenues for further research. Aquat Microb Ecol 57:321–341. doi: 10.3354/ame01363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Z, Gonsior M, Schmitt-Kopplin P, Zhan Y, Zhang R, Jiao N, Chen F. 2019. Microbial transformation of virus-induced dissolved organic matter from picocyanobacteria: coupling of bacterial diversity and DOM chemodiversity. ISME J 13:2551–2565. doi: 10.1038/s41396-019-0449-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lauro FM, DeMaere MZ, Yau S, Brown MV, Ng C, Wilkins D, Raftery MJ, Gibson JAE, Andrews-Pfannkoch C, Lewis M, Hoffman JM, Thomas T, Cavicchioli R. 2011. An integrative study of a meromictic lake ecosystem in Antarctica. ISME J 5:879–895. doi: 10.1038/ismej.2010.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sime-Ngando T, Bettarel Y, Colombet J, Palesse S, Ram ASP, Charpin M, Amblard C. 2016. Lake Pavin: a pioneer site for ecological studies of freshwater viruses, p 229–244. In Sime-Ngando T, Boivin P, Chapron E, Jezequel D, Meybeck M (ed), Lake Pavin: history, geology, biogeochemistry, and sedimentology of a deep meromictic maar lake Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 29.Wu Y-T, Yang C-Y, Chiang P-W, Tseng C-H, Chiu H-H, Saeed I, Baatar B, Rogozin D, Halgamuge S, Degermendzhi A, Tang S-L. 2018. Comprehensive insights into composition, metabolic potentials, and interactions among archaeal, bacterial, and viral assemblages in meromictic Lake Shunet in Siberia. Front Microbiol 9:1763. doi: 10.3389/fmicb.2018.01763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christie-Oleza JA, Sousoni D, Lloyd M, Armengaud J, Scanlan DJ. 2017. Nutrient recycling facilitates long-term stability of marine microbial phototroph-heterotroph interactions. Nat Microbiol 2:17100. doi: 10.1038/nmicrobiol.2017.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coutinho FH, Silveira CB, Gregoracci GB, Thompson CC, Edwards RA, Brussaard CPD, Dutilh BE, Thompson FL. 2017. Marine viruses discovered via metagenomics shed light on viral strategies throughout the oceans. Nat Commun 8:15955. doi: 10.1038/ncomms15955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaquer-Sunyer R, Duarte CM. 2008. Thresholds of hypoxia for marine biodiversity. Proc Natl Acad Sci USA 105:15452–15457. doi: 10.1073/pnas.0803833105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Danza F, Ravasi D, Storelli N, Roman S, Lüdin S, Bueche M, Tonolla M. 2018. Bacterial diversity in the water column of meromictic Lake Cadagno and evidence for seasonal dynamics. PLoS One 13:e0209743. doi: 10.1371/journal.pone.0209743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Egli K, Wiggli M, Fritz M, Klug J, Gerss J, Bachofen R. 2004. Spatial and temporal dynamics of a plume of phototrophic microorganisms in a meromictic alpine lake using turbidity as a measure of cell density. Aquat Microb Ecol 35:105–113. doi: 10.3354/ame035105. [DOI] [Google Scholar]
- 35.Props R, Kerckhof F-M, Rubbens P, De Vrieze J, Hernandez Sanabria E, Waegeman W, Monsieurs P, Hammes F, Boon N. 2017. Absolute quantification of microbial taxon abundances. ISME J 11:584–587. doi: 10.1038/ismej.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun L, Toyonaga M, Ohashi A, Tourlousse DM, Matsuura N, Meng X-Y, Tamaki H, Hanada S, Cruz R, Yamaguchi T, Sekiguchi Y. 2016. Lentimicrobium saccharophilum gen. nov., sp. nov., a strictly anaerobic bacterium representing a new family in the phylum Bacteroidetes, and proposal of Lentimicrobiaceae fam. nov. Int J Syst Evol Microbiol 66:2635–2642. doi: 10.1099/ijsem.0.001103. [DOI] [PubMed] [Google Scholar]
- 37.Decristophoris P, Peduzzi S, Ruggeri-Bernardi N, Hahn D, Tonolla M. 2009. Fine scale analysis of shifts in bacterial community structure in the chemocline of meromictic Lake Cadagno, Switzerland. J Limnol 68:16–24. doi: 10.4081/jlimnol.2009.16. [DOI] [Google Scholar]
- 38.Fadeev E, Cardozo-Mino MG, Rapp JZ, Bienhold C, Salter I, Salman-Carvalho V, Molari M, Tegetmeyer HE, Buttigieg PL, Boetius A. 2021. Comparison of two 16S rRNA primers (V3-V4 and V4-V5) for studies of arctic microbial communities. Front Microbiol 12:637526. doi: 10.3389/fmicb.2021.637526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willis C, Desai D, LaRoche J. 2019. Influence of 16S rRNA variable region on perceived diversity of marine microbial communities of the Northern North Atlantic. FEMS Microbiol Lett 366:fnz152. doi: 10.1093/femsle/fnz152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burja AM, Tamagnini P, Bustard MT, Wright PC. 2001. Identification of the green alga, Chlorella vulgaris (SDC1) using cyanobacteria derived 16S rDNA primers: targeting the chloroplast. FEMS Microbiol Lett 202:195–203. doi: 10.1111/j.1574-6968.2001.tb10803.x. [DOI] [PubMed] [Google Scholar]
- 41.Hamilton M, Mascioni M, Hehenberger E, Bachy C, Yung C, Vernet M, Worden AZ. 2021. Spatiotemporal variations in Antarctic protistan communities highlight phytoplankton diversity and seasonal dominance by a novel cryptophyte lineage. mBio 12:e02973-21. doi: 10.1128/mBio.02973-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stoyneva MP, Gartner G, Cocquyt C, Vyverman W. 2005. Closteriopsis petkovii—a new green algal species from Lake Tanganyika (Africa). Phyton (Horn, Austria) 45:237–247. [Google Scholar]
- 44.Props R, Monsieurs P, Mysara M, Clement L, Boon N. 2016. Measuring the biodiversity of microbial communities by flow cytometry. Methods Ecol Evol 7:1376–1385. doi: 10.1111/2041-210X.12607. [DOI] [Google Scholar]
- 45.Haas S, Desai DK, LaRoche J, Pawlowicz R, Wallace DWR. 2019. Geomicrobiology of the carbon, nitrogen and sulphur cycles in Powell Lake: a permanently stratified water column containing ancient seawater. Environ Microbiol 21:3927–3952. doi: 10.1111/1462-2920.14743. [DOI] [PubMed] [Google Scholar]
- 46.Andrei A-Ş, Robeson MS, Baricz A, Coman C, Muntean V, Ionescu A, Etiope G, Alexe M, Sicora CI, Podar M, Banciu HL. 2015. Contrasting taxonomic stratification of microbial communities in two hypersaline meromictic lakes. ISME J 9:2642–2656. doi: 10.1038/ismej.2015.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanentzap AJ, Fitch A, Orland C, Emilson EJS, Yakimovich KM, Osterholz H, Dittmar T. 2019. Chemical and microbial diversity covary in fresh water to influence ecosystem functioning. Proc Natl Acad Sci USA 116:24689–24695. doi: 10.1073/pnas.1904896116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schubert CJ, Vazquez F, Lösekann-Behrens T, Knittel K, Tonolla M, Boetius A. 2011. Evidence for anaerobic oxidation of methane in sediments of a freshwater system (Lago di Cadagno). FEMS Microbiol Ecol 76:26–38. doi: 10.1111/j.1574-6941.2010.01036.x. [DOI] [PubMed] [Google Scholar]
- 49.Knoll AH, Bergmann KD, Strauss JV. 2016. Life: the first two billion years. Philos Trans R Soc Lond B Biol Sci 371:20150493. doi: 10.1098/rstb.2015.0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cabrera S, López M, Tartarotti B. 1997. Phytoplankton and zooplankton response to ultraviolet radiation in a high-altitude Andean lake: short- versus long-term effects. J Plankton Res 19:1565–1582. doi: 10.1093/plankt/19.11.1565. [DOI] [Google Scholar]
- 51.Luedin SM, Storelli N, Danza F, Roman S, Wittwer M, Pothier JF, Tonolla M. 2019. Mixotrophic growth under micro-oxic conditions in the purple sulfur bacterium “Thiodictyon syntrophicum.” Front Microbiol 10:384. doi: 10.3389/fmicb.2019.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Milucka J, Kirf M, Lu L, Krupke A, Lam P, Littmann S, Kuypers MMM, Schubert CJ. 2015. Methane oxidation coupled to oxygenic photosynthesis in anoxic waters. ISME J 9:1991–2002. doi: 10.1038/ismej.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berg JS, Pjevac P, Sommer T, Buckner CRT, Philippi M, Hach PF, Liebeke M, Holtappels M, Danza F, Tonolla M, Sengupta A, Schubert CJ, Milucka J, Kuypers MMM. 2019. Dark aerobic sulfide oxidation by anoxygenic phototrophs in anoxic waters. Environ Microbiol 21:1611–1626. doi: 10.1111/1462-2920.14543. [DOI] [PubMed] [Google Scholar]
- 54.Glazer AN, Wedemayer GJ. 1995. Cryptomonad biliproteins—an evolutionary perspective. Photosynth Res 46:93–105. doi: 10.1007/BF00020420. [DOI] [PubMed] [Google Scholar]
- 55.Berg JS, Michellod D, Pjevac P, Martinez-Perez C, Buckner CRT, Hach PF, Schubert CJ, Milucka J, Kuypers MMM. 2016. Intensive cryptic microbial iron cycling in the low iron water column of the meromictic Lake Cadagno. Environ Microbiol 18:5288–5302. doi: 10.1111/1462-2920.13587. [DOI] [PubMed] [Google Scholar]
- 56.Glibert PM, Wilkerson FP, Dugdale RC, Raven JA, Dupont CL, Leavitt PR, Parker AE, Burkholder JM, Kana TM. 2016. Pluses and minuses of ammonium and nitrate uptake and assimilation by phytoplankton and implications for productivity and community composition, with emphasis on nitrogen-enriched conditions. Limnol Oceanogr 61:165–197. doi: 10.1002/lno.10203. [DOI] [Google Scholar]
- 57.Brocks JJ, Love GD, Summons RE, Knoll AH, Logan GA, Bowden SA. 2005. Biomarker evidence for green and purple sulphur bacteria in a stratified Palaeoproterozoic sea. Nature 437:866–870. doi: 10.1038/nature04068. [DOI] [PubMed] [Google Scholar]
- 58.Moczydłowska M, Landing ED, Zang W, Palacios T. 2011. Proterozoic phytoplankton and timing of Chlorophyte algae origins. Palaeontology 54:721–733. doi: 10.1111/j.1475-4983.2011.01054.x. [DOI] [Google Scholar]
- 59.Peduzzi S, Storelli N, Welsh A, Peduzzi R, Hahn D, Perret X, Tonolla M. 2012. Candidatus “Thiodictyon syntrophicum”, sp. nov., a new purple sulfur bacterium isolated from the chemocline of Lake Cadagno forming aggregates and specific associations with Desulfocapsa sp. Syst Appl Microbiol 35:139–144. doi: 10.1016/j.syapm.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 60.Tonolla M, Demarta A, Peduzzi S, Hahn D, Peduzzi R. 2000. In situ analysis of sulfate-reducing bacteria related to Desulfocapsa thiozymogenes in the chemocline of meromictic Lake Cadagno (Switzerland). Appl Environ Microbiol 66:820–824. doi: 10.1128/AEM.66.2.820-824.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peduzzi S, Tonolla M, Hahn D. 2003. Isolation and characterization of aggregate-forming sulfate-reducing and purple sulfur bacteria from the chemocline of meromictic Lake Cadagno, Switzerland. FEMS Microbiol Ecol 45:29–37. doi: 10.1016/S0168-6496(03)00107-7. [DOI] [PubMed] [Google Scholar]
- 62.Howard-Varona C, Lindback MM, Bastien GE, Solonenko N, Zayed AA, Jang HB, Andreopoulos B, Brewer HM, Glavina Del Rio T, Adkins JN, Paul S, Sullivan MB, Duhaime MB. 2020. Phage-specific metabolic reprogramming of virocells. ISME J 14:881–895. doi: 10.1038/s41396-019-0580-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Voorhies AA, Eisenlord SD, Marcus DN, Duhaime MB, Biddanda BA, Cavalcoli JD, Dick GJ. 2016. Ecological and genetic interactions between cyanobacteria and viruses in a low-oxygen mat community inferred through metagenomics and metatranscriptomics. Environ Microbiol 18:358–371. doi: 10.1111/1462-2920.12756. [DOI] [PubMed] [Google Scholar]
- 64.Yau S, Krasovec M, Benites LF, Rombauts S, Groussin M, Vancaester E, Aury J-M, Derelle E, Desdevises Y, Escande M-L, Grimsley N, Guy J, Moreau H, Sanchez-Brosseau S, Van de Peer Y, Vandepoele K, Gourbiere S, Piganeau G. 2020. Virus-host coexistence in phytoplankton through the genomic lens. Sci Adv 6:eaay2587. doi: 10.1126/sciadv.aay2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wigington CH, Sonderegger D, Brussaard CPD, Buchan A, Finke JF, Fuhrman JA, Lennon JT, Middelboe M, Suttle CA, Stock C, Wilson WH, Wommack KE, Wilhelm SW, Weitz JS. 2016. Re-examination of the relationship between marine virus and microbial cell abundances. Nat Microbiol 1:15024. doi: 10.1038/nmicrobiol.2015.24. [DOI] [PubMed] [Google Scholar]
- 66.Luedin SM, Pothier JF, Danza F, Storelli N, Frigaard N-U, Wittwer M, Tonolla M. 2018. Complete genome sequence of “Thiodictyon syntrophicum” sp. nov. strain Cad16T, a photolithoautotrophic purple sulfur bacterium isolated from the alpine meromictic Lake Cadagno. Stand Genomic Sci 13:14. doi: 10.1186/s40793-018-0317-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Luedin SM, Liechti N, Cox RP, Danza F, Frigaard N-U, Posth NR, Pothier JF, Roman S, Storelli N, Wittwer M, Tonolla M. 2019. Draft genome sequence of Chromatium okenii isolated from the stratified alpine Lake Cadagno. Sci Rep 9:1936. doi: 10.1038/s41598-018-38202-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anantharaman K, Duhaime MB, Breier JA, Wendt KA, Toner BM, Dick GJ. 2014. Sulfur oxidation genes in diverse deep-sea viruses. Science 344:757–760. doi: 10.1126/science.1252229. [DOI] [PubMed] [Google Scholar]
- 69.Dalcin Martins P, Danczak RE, Roux S, Frank J, Borton MA, Wolfe RA, Burris MN, Wilkins MJ. 2018. Viral and metabolic controls on high rates of microbial sulfur and carbon cycling in wetland ecosystems. Microbiome 6:138. doi: 10.1186/s40168-018-0522-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Berg JS, Lepine M, Laymand E, Han X, Vogel H, Morlock MA, Gajendra N, Gilli A, Bernasconi SM, Schubert CJ, Su G, Lever MA. 2022. Ancient and modern geochemical signatures in the 13,500-year sedimentary record of Lake Cadagno. Front Earth Sci 9:754888. doi: 10.3389/feart.2021.754888. [DOI] [Google Scholar]
- 71.Azam F, Malfatti F. 2007. Microbial structuring of marine ecosystems. Nat Rev Microbiol 5:782–791. doi: 10.1038/nrmicro1747. [DOI] [PubMed] [Google Scholar]
- 72.Zhang C, Dang H, Azam F, Benner R, Legendre L, Passow U, Polimene L, Robinson C, Suttle CA, Jiao N. 2018. Evolving paradigms in biological carbon cycling in the ocean. Natl Sci Rev 5:481–499. doi: 10.1093/nsr/nwy074. [DOI] [Google Scholar]
- 73.Del Don C, Hanselmann KW, Peduzzi R, Bachofen R. 2001. The meromictic alpine Lake Cadagno: orographical and biogeochemical description. Aquat Sci 63:70–90. doi: 10.1007/PL00001345. [DOI] [Google Scholar]
- 74.Martinez E, Gorgues T, Lengaigne M, Fontana C, Sauzède R, Menkes C, Uitz J, Di Lorenzo E, Fablet R. 2020. Reconstructing global chlorophyll-a variations using a non-linear statistical approach. Front Mar Sci 7:464. doi: 10.3389/fmars.2020.00464. [DOI] [Google Scholar]
- 75.Ogashawara I. 2020. Determination of phycocyanin from space—a bibliometric analysis. Remote Sensing 12:567. doi: 10.3390/rs12030567. [DOI] [Google Scholar]
- 76.Bridgewater L, American Public Health Association, American Water Works Association, Water Environment Federation . 2012. Standard methods for the examination of water and wastewater, 22nd ed. American Public Health Association, Washington, DC. [Google Scholar]
- 77.Hassler CS, Ridgway KR, Bowie AR, Butler ECV, Clementson LA, Doblin MA, Davies DM, Law C, Ralph PJ, van der Merwe P, Watson R, Ellwood MJ. 2014. Primary productivity induced by iron and nitrogen in the Tasman Sea: an overview of the PINTS expedition. Mar Freshw Res 65:517–537. doi: 10.1071/MF13137. [DOI] [Google Scholar]
- 78.Moisset SAM, Cabanes DJE, Blanco-Ameijeiras S, Hassler CS. 2019. Response of phytoplankton from the metalimnetic chlorophyll maximum to macro- and micro-nutrients amendments in Lake Geneva. J Great Lakes Res 45:290–299. doi: 10.1016/j.jglr.2019.02.001. [DOI] [Google Scholar]
- 79.Marie D, Brussaard CPD, Thyrhaug R, Bratbak G, Vaulot D. 1999. Enumeration of marine viruses in culture and natural samples by flow cytometry. Appl Environ Microbiol 65:45–52. doi: 10.1128/AEM.65.1.45-52.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jacquet S, Dorigo U, Personnic S. 2013. A few tests prior to flow cytometry and epifluorescence analyses of freshwater bacterio- and virioplankton communities. Flow Cytom 1:1–30. [Google Scholar]
- 81.Buysschaert B, Favere J, Vermijs L, Baetens V, Naka A, Boon N, De Gusseme B. 2019. Flow cytometric fingerprinting to assess the microbial community response to changing water quality and additives. Environ Sci: Water Res Technol 5:1672–1682. doi: 10.1039/C9EW00283A. [DOI] [Google Scholar]
- 82.Wanderley BMS, A Araújo DS, Quiroga MV, Amado AM, Neto ADD, Sarmento H, Metz SD, Unrein F. 2019. flowDiv: a new pipeline for analyzing flow cytometric diversity. BMC Bioinformatics 20:274. doi: 10.1186/s12859-019-2787-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.R Core Team. 2020. A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org. [Google Scholar]
- 84.RStudio Team. 2020. RStudio: integrated development for R. RStudio, PBC, Boston, MA. [Google Scholar]
- 85.Rubbens P, Props R, Kerckhof F-M, Boon N, Waegeman W. 2021. PhenoGMM: Gaussian mixture modeling of cytometry data quantifies changes in microbial community structure. mSphere 6:e00530-20. doi: 10.1128/mSphere.00530-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fourquez M, Bressac M, Deppeler SL, Ellwood M, Obernosterer I, Trull TW, Boyd PW. 2020. Microbial competition in the subpolar Southern Ocean: an Fe–C co-limitation experiment. Front Mar Sci 6:776. doi: 10.3389/fmars.2019.00776. [DOI] [Google Scholar]
- 87.Van Wambeke F, Heussner S, Diaz F, Raimbault P, Conan P. 2002. Small-scale variability in the coupling/uncoupling of bacteria, phytoplankton and organic carbon fluxes along the continental margin of the Gulf of Lions, Northwestern Mediterranean Sea. J Mar Syst 33-34:411–429. doi: 10.1016/S0924-7963(02)00069-6. [DOI] [Google Scholar]
- 88.Kirchman DL. 1993. Leucine incorporation as a measure of biomass production by heterotrophic bacteria. In Kemp PF, Sherr BF, Sherr EB, Cole JJ (ed), Handbook of methods in aquatic microbial ecology. CRC Press, Boca Raton, FL. [Google Scholar]
- 89.del Giorgio PA, Cole JJ. 1998. Bacterial growth efficiency in natural aquatic systems. Annu Rev Ecol Syst 29:503–541. doi: 10.1146/annurev.ecolsys.29.1.503. [DOI] [Google Scholar]
- 90.Wickham H. 2011. ggplot2. WIREs Comp Stat 3:180–185. doi: 10.1002/wics.147. [DOI] [Google Scholar]
- 91.Kassambara A. 2018. ggpubr: “ggplot2” based publication ready plots (version 0.1.7). https://CRAN.R-project.org/package=ggpubr.
- 92.Wei T, Simko V. 2017. R package “corrplot”: visualization of a correlation matrix (version 0.84). https://cran.r-project.org/web/packages/corrplot/.
- 93.Tonolla M, Peduzzi R, Hahn D. 2005. Long-term population dynamics of phototrophic sulfur bacteria in the chemocline of Lake Cadagno, Switzerland. Appl Environ Microbiol 71:3544–3550. doi: 10.1128/AEM.71.7.3544-3550.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bosshard PP, Santini Y, Grüter D, Stettler R, Bachofen R. 2000. Bacterial diversity and community composition in the chemocline of the meromictic alpine Lake Cadagno as revealed by 16S rDNA analysis. FEMS Microbiol Ecol 31:173–182. doi: 10.1111/j.1574-6941.2000.tb00682.x. [DOI] [PubMed] [Google Scholar]
- 95.Duhaime M, Cable R. 2017. DNA extraction from filters using QIAgen DNeasy and QIAshredder v1. protocolsio doi: 10.17504/protocols.io.jx5cpq6. [DOI] [Google Scholar]
- 96.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rohwer RR, Hamilton JJ, Newton RJ, McMahon KD. 2018. TaxAss: leveraging a custom freshwater database achieves fine-scale taxonomic resolution. mSphere 3:e00327-18. doi: 10.1128/mSphere.00327-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.McMurdie PJ, Holmes S. 2014. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 10:e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lahti L, Shetty S. 2018. Introduction to the microbiome R package. http://bioconductor.statistik.tu-dortmund.de/packages/3.6/bioc/vignettes/microbiome/inst/doc/vignette.html.
- 103.Lin H, Peddada SD. 2020. Analysis of compositions of microbiomes with bias correction. Nat Commun 11:3514. doi: 10.1038/s41467-020-17041-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Weiss S, Xu ZZ, Peddada S, Amir A, Bittinger K, Gonzalez A, Lozupone C, Zaneveld JR, Vázquez-Baeza Y, Birmingham A, Hyde ER, Knight R. 2017. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 5:27. doi: 10.1186/s40168-017-0237-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pruesse E, Peplies J, Glöckner FO. 2012. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pfeiffer W, Stamatakis A. 2010. Hybrid MPI/Pthreads parallelization of the RAxML phylogenetics code. In 2010 IEEE International Symposium on Parallel Distributed Processing, Workshops and PhD Forum (IPDPSW), Atlanta, GA.
- 107.Yu G. 2020. Using ggtree to visualize data on tree-like structures. Curr Protoc Bioinformatics 69:e96. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Line plot depicting the results of the experiment to monitor the health of photosynthetic cells. A fast repetition rate fluorometer was used to measure basal fluorescence (Fb; blue line) and maximum fluorescence (Fm; black line), which were then used to calculate the maximum photosynthetic quantum yield (Fv/Fm; red line), a proxy for the health of photosynthetic cells. Download FIG S2, PDF file, 0.1 MB (127.4KB, pdf) .
Copyright © 2022 Saini et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Grid and heat map depicting the Pearson correlation R coefficients and P values determined between data types; strong positive correlations are indicated by blue hues, strong negative correlations are indicated by red, and no correlation is indicated by yellow. Prefix of parameter name indicates the parameter type category. A, abundance: 16S rRNA amplicon-based relative abundances, flow cytometry-generated PLP and VLP counts, and phycobilisome-containing cell counts. D, diversity: alpha and beta diversity calculated from 16S-based genotypic and flow cytometry-based phenotypic Bray-Curtis distance measures. FCS, flow cytometry-derived data. N, nutrients, including DOC, POC, PON, PS, ammonium, and sulfate. P, physicochemical parameters from CTD, including light, conductivity, temperature, oxygen, Chl a, phycocyanin, turbidity. Secondary production is included with no prefix. Download FIG S3, PDF file, 0.6 MB (657.4KB, pdf) .
Copyright © 2022 Saini et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relative abundances (OTU read count/total sample read count) and inferred abundances (relative abundances × FCM cell counts) of prokaryotic OTUs and chloroplasts. Download Table S1, XLSX file, 0.2 MB (179KB, xlsx) .
Copyright © 2022 Saini et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Inferred absolute abundances of bacterial genera through the water column of meromictic Lake Cadagno. Absolute abundances are inferred by multiplying the relative abundances (OTU read count/total sample read count) by FCM cell counts. Download FIG S4, PDF file, 0.01 MB (6.9KB, pdf) .
Copyright © 2022 Saini et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Scatter plot of flow cytometry events visualized with a BD6 flow cytometer interface (A and B) and RStudio Phenoflow package (A to D) for one replicate of the 1-m sample. FCM events representing prokaryote-like particles (PLPs) using automatic gating via a four-quadrant layout (A) and manual gating (B and C). (D) FCM events in 0.2-μm-filtered, cell-free Milli-Q water (control). The red outline represents the gating strategy for cell counts and phenotypic diversity analysis. This gate was applied to all samples. Download FIG S1, PDF file, 1.4 MB (1.5MB, pdf) .
Copyright © 2022 Saini et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Raw sequences were deposited in the NCBI Short Read Archive (SRA) and are available under BioProject accession no. PRJNA717659. The code used for running the mothur analysis, statistical analyses, and figure generation is available in the project GitHub repository at https://github.com/DuhaimeLab/Lake_Cadagno_microbial_loop_Saini_et_al_2021.