Summary
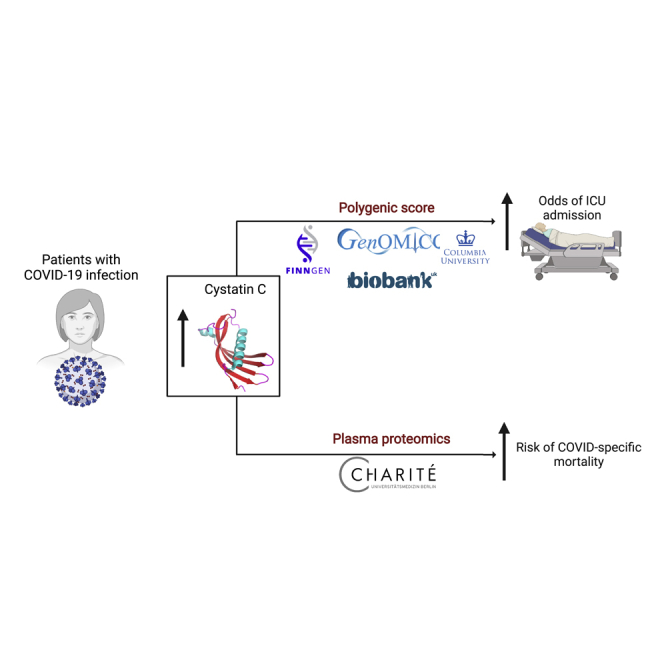
COVID-19 has highly variable clinical courses. The search for prognostic host factors for COVID-19 outcome is a priority. We performed logistic regression for ICU admission against a polygenic score (PGS) for Cystatin C (CyC) production in patients with COVID-19. We analyzed the predictive value of longitudinal plasma CyC levels in an independent cohort of patients hospitalized with COVID-19. In four cohorts spanning European and African ancestry populations, we identified a significant association between CyC-production PGS and odds of critical illness (n cases=2,319), with the strongest association captured in the UKB cohort (OR 2.13, 95% CI 1.58-2.87, p=7.12e-7). Plasma proteomics from an independent cohort of hospitalized COVID-19 patients (n cases = 131) demonstrated that CyC production was associated with COVID-specific mortality (p=0.0007). Our findings suggest that CyC may be useful for stratification of patients and it has functional role in the host response to COVID-19.
Subject areas: Virology, human metabolism
Graphical abstract
Highlights
-
•
Cystatin C varies independently of renal function in COVID-19
-
•
A polygenic score for cystatin C production predicts critical COVID-19 illness
-
•
Elevated serum cystatin C is associated with COVID-19 mortality
Virology; Human metabolism
Introduction
COVID-19 is caused by a single pathogen, severe acute respiratory coronavirus 2 (SARS-CoV-2), and remains the main burden to global health with >500M cases and ∼6M deaths to date (Dong et al., 2020). The clinical course of COVID-19 ranges from asymptomatic infection or mild viral pneumonia to acute respiratory distress syndrome (ARDS) necessitating hospitalization and respiratory support (Wiersinga et al., 2020), potentially with fatal outcomes. Although variants with differing virulence are emerging (Wolter et al., 2022), there is clear evidence that host factors are the most important determinant of outcome. Acquired characteristics, such as increasing age, comorbidities (Zhou et al., 2020), and high body mass index (BMI) (Gao et al., 2021) as well as inherited germline predisposition (Initiative, 2021; Pairo-Castineira et al., 2021) are known to impact on risk for poor outcome.
Mild COVID-19 is associated with sustained upregulated type 1 IFN signaling, which correlates with symptom severity (Brennan et al., 2022). In contrast, inappropriately attenuated early IFN signaling is a hallmark of severe COVID-19 (Hadjadj et al., 2020; van der Wijst et al., 2021), potentially driven by germline loss-of-function mutations in pathway components (Zhang et al., 2020b) or pre-existing anti-type 1 IFN autoantibodies (van der Wijst et al., 2021). Incomplete innate antiviral responses cause prolonged viraemia (Fajnzylber et al., 2020) and hyperactivated inflammatory response (Hadjadj et al., 2020; Zhang et al., 2020a), termed cytokine release syndrome (CRS), which is thought to be a common pathway of critical illness in COVID-19 (Moore and June, 2020). Glucocorticoids, such as dexamethasone, have highly pleiotropic immune suppressive effects including the inhibition of cytokine production and type 1 IFN signaling, potentially explaining the benefits (RECOVERY Collaborative Group et al., 2021) and potential harms of glucocorticoid treatment (Brenner et al., 2020), respectively.
We have recently reported that the production of cystatin C (CyC), a secreted protein routinely used as a marker of renal function, is induced by glucocorticoid signaling (Kleeman et al., 2021). Using CyC and renal function data in the prospectively collected UK Biobank (UKB) cohort, we determined the genetic basis of the unmeasured (latent) trait CyC-production and validated a polygenic score (PGS) for CyC-production (Kleeman et al., 2021). This CyC-production PGS was predictive and prognostic in a range of inflammatory conditions, including cancer, potentially capturing inter-individual variation in endogenous glucocorticoid production (Kleeman et al., 2021). As a result, we hypothesized that CyC-production PGS might be associated with adverse COVID-19 outcomes such as hospitalization, critical illness (such as ICU admission), and death.
Results
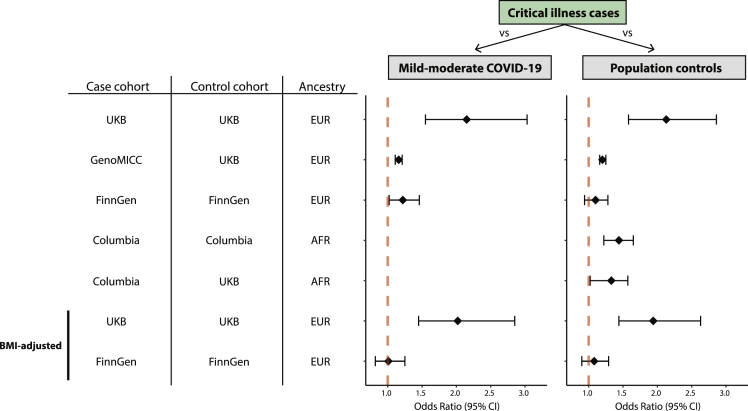
In four cohorts (UKB validation set, GenOMICC, FinnGen, Columbia) spanning European and African ancestry populations, we identified a significant association between Z-scored CyC-production PGS and odds of critical illness (Figure 1, p < 0.05, OR>1.20, Table S1), with the strongest association captured in the UKB cohort versus population controls (OR 2.13, 95% CI 1.58–2.87, p = 7.12 × 10−7), reflecting approximately a 2-fold increased odds of critical illness per one SD of CyC-production PGS. In UKB, we identified an additional significant association between CyC-production and odds of hospitalization (OR 1.24, 95% CI 1.09–1.41, p = 0.00077). In the Columbia cohort of African ancestry participants, CyC-production PGS was associated with significantly increased odds of hospitalization (OR 1.20, 95% CI 1.05–1.34, p = 0.0175) and critical illness (OR 1.44, 95% CI 1.22–1.65, p = 0.000956). Age data were unavailable for the Columbia population controls (STAR Methods) and, in order to perform an age-adjusted analysis, we incorporated ancestry-matched UKB controls (n = 2334) to independently test the significant association between CyC-production PGS and ICU admission (Figure 1, OR 1.31, 95% CI 1.02–1.57, p = 0.053, Table S1). All other analyses for non-critical illness phenotypes showed a consistent direction of effect (odds ratio >1), but did not reach statistical significance (V).
Figure 1.
Forest plot logistic regression analyses for odds of critical illness as a function of Z-scored CyC-production polygenic score (PGS)
All analyses were adjusted for age (except Columbia-vs-Columbia comparison), ancestry principal components, and sex. Control populations comprised either mild-moderate COVID-19-positive patients or ancestry-matched population controls. A mild-moderate COVID-19 control population was not available for the Columbia cohort. Critical illness refers to ICU admission for all analyses except for the Columbia cohort where it reflects a composite outcome of death or invasive respiratory support.
We and others have reported that serum CyC is correlated with increased body mass index and obesity (Kleeman et al., 2021; Muntner et al., 2008), which are risk factors for adverse COVID-19 outcomes (Gao et al., 2021; Initiative, 2021). To investigate whether the association between CyC-production PGS and COVID-19 morbidity is partly explained by BMI we repeated our analyses with BMI as an additional covariate, where BMI data were available (UKB, FinnGen). In UKB the effect size for each clinical outcome was only mildly attenuated by adjustment for BMI (Figure 1 and Table S1), and associations with hospitalization and ICU admission remained significant. Unexpectedly, BMI adjustment abrogated the association between CyC-production and ICU admission in the FinnGen cohort (OR 1.01, 95% CI 0.82–1.25, p = 0.163). Next, we investigated potential reasons for this unexpected result. In FinnGen, BMI is recorded for approximately 70% of participants, and so we hypothesized that there could be systematic differences between patients with BMI recorded versus those without. Although there was no evidence of population stratification between these groups (Figure S1), a genome-wide association study (GWAS) for BMI recording (yes/no) identified a large number of highly significant loci (Figure S2), with the top signal in the HLA locus (rs3957146). This spurious heritability is likely to be explained by a significant participant bias between groups (Pirastu et al., 2021), and implies that the association between CyC-production and ICU admission is enriched in a specific subset of the population, potentially capturing a gene-environment interaction in the Finnish population.
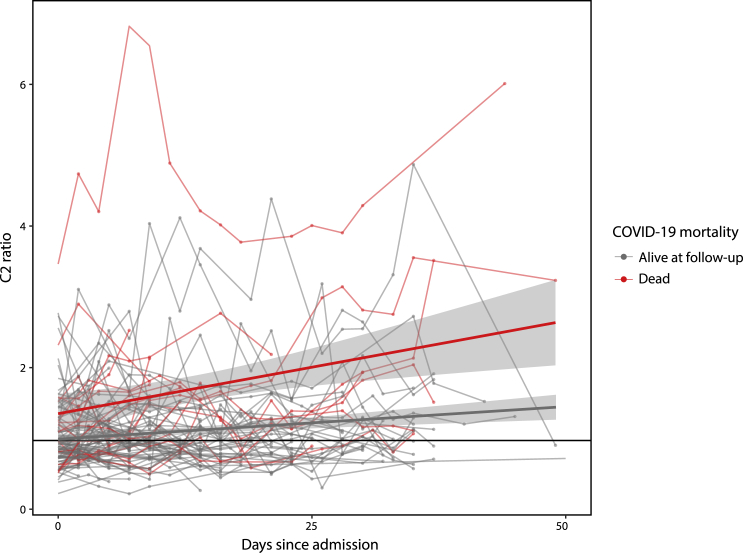
While it has proven challenging to implement polygenic scores into routine clinical practice (Chatterjee et al., 2016), CyC is a secreted protein that can be readily detected with validated clinical assays. We hypothesized that the ratio of estimated glomerular filtration rate (eGFR) calculated from creatinine (Inker et al., 2012), an alternative marker of renal function, to eGFR calculated from CyC (termed C2 ratio, a surrogate for CyC-production as eGFR is inversely related to serum CyC/creatinine) would be associated with adverse COVID-19 outcomes. We analyzed longitudinal plasma proteomics data, including the quantification of CyC, from a recently reported (Demichev et al., 2021) cohort of 309 patients admitted to the Charité University Hospital, Berlin, Germany, of which 131 patients had PCR-confirmed SARS-CoV-2 infection, clinical follow-up and paired serum creatinine measurements. These patients were treated prior to the RECOVERY dexamethasone trial (RECOVERY Collaborative Group et al., 2021) and, therefore, did not receive steroids as part of their COVID-19 management. Analysis of longitudinal patient data demonstrated that COVID-specific mortality was associated with progressive elevations in the C2 ratio (Figure 2, p = 0.0007, t-test on linear regression coefficients). A C2 ratio >1 was associated with a significantly increased risk of COVID-specific mortality in univariate (HR 3.01, 95% CI 2.28–3.74, p = 0.0030) and multivariate (HR 2.81, 95% CI 2.19–3.45, p = 0.0013) Cox regression, incorporating repeated measures for each patient (if applicable) and accounting for their interdependence. Furthermore, a multivariate analysis incorporating only the earliest timepoint (n = 126) for each patient replicated a significant association between C2 ratio >1 and COVID-19 mortality (HR 2.74, 95% CI 1.14–6.58, p = 0.023).
Figure 2.
Dynamic longitudinal changes in creatinine-CyC (C2 ratio) during the first 50 days following admission with PCR-confirmed SARS-CoV-2
Each line signifies repeated measures from a single patient, with the color signifying whether the patient was alive (gray) or dead (red) at follow-up cut-off (December 2020). Data points are annotated with linear regression lines and 95% confidence intervals.
Discussion
In this study, we demonstrated through two entirely independent approaches that CyC is a relevant predictor of outcome in diverse populations of patients with COVID-19. We found that a PGS for CyC predicts the risk of ICU admission in three independent cohorts and, in a fourth independent cohort, that increased renal function normalized plasma levels of CyC are predictive of COVID-specific mortality.
CyC is used routinely as a passive marker of renal function, but varies in a renal function-independent manner in the context of COVID-19. As such, eGFR estimates from CyC alone may be a confounded marker of renal function in patients with COVID-19, consistent with recent reports (Liu et al., 2021). The biological explanation of these observations remains incomplete but is likely to include CyC’s molecular function as a potent inhibitor of cysteine proteases (Kopitar-Jerala, 2006), such as components of antigen presentation pathways, and its role as a glucocorticoid response gene (Kleeman et al., 2021). Altogether, we propose that CyC is both a predictive biomarker that could be readily implemented to risk-stratify patients with COVID-19, but also a driver in the pathophysiology of COVID-19 and other inflammatory diseases.
Limitations of the study
This study has limitations. Our conclusions are based on a relatively small number of COVID-19 cases, necessitating future large prospective studies to interrogate the generalizability of both CyC-production PGS and the C2 ratio as prognostic markers for COVID-19 and other diseases. Analyses incorporating population controls have the potential to increase power but implicitly assume that the overwhelming majority of the control group would not experience an adverse outcome if exposed to COVID-19. However, this assumption does not appear to confound genetic associations (Initiative, 2021). Although proteomics-based measurements of CyC are sufficiently accurate and precise our findings should be replicated using standard clinical CyC assays.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| UK Biobank | https://www.ukbiobank.ac.uk/ | |
| FinnGen | https://www.finngen.fi/en/ | |
| GenOMICC | https://genomicc.org/ | |
| Columbia COVID-19 Biobank | https://www.vagelos.columbia.edu/research/researchers/core-and-shared-facilities/new-instruments-and-facilities/columbia-university-biobank | |
| Software and algorithms | ||
| PLINK | https://www.cog-genomics.org/plink/ | |
| Hail | https://hail.is/ | |
| R | https://www.r-project.org/ | |
| KING | https://www.kingrelatedness.com/ | |
| TOPMED | https://imputation.biodatacatalyst.nhlbi.nih.gov/# | |
| MAMA | https://github.com/JonJala/mama/ | |
Resource availability
Lead contact
Further information and requests for resources should be directed to the Lead Contact, Tobias Janowitz (janowitz@cshl.edu).
Materials availability
The study did not generate any new reagents or materials.
Experimental model and subject details
Ethical approval
All patient-based research in this analysis was performed in accordance to international and institutional guidelines. UK Biobank obtained ethics approval from the North West Multi-Centre Research Ethics Committee which covers the UK (approval number: 11/NW/0382) and obtained informed consent from all participants. UKB data were accessed as part of this analysis under applications 58510, 19655 and 41849. The FinnGen study protocol was approved by the Ethical Review Board of the Hospital District of Helsinki and Uusimaa (Nr HUS/990/2017). The FinnGen study is approved by the Finnish Institute for Health and Welfare (THL), approval number THL/2031/6.02.00/2017, amendments THL/1101/5.05.00/2017, THL/341/6.02.00/2018, THL/2222/6.02.00/2018, THL/283/6.02.00/2019, THL/1721/5.05.00/2019, Digital and population data service agency VRK43431/2017-3, VRK/6909/2018-3, VRK/4415/2019-3, the 5Social Insurance Institution (KELA) KELA 58/522/2017, KELA 131/522/2018, KELA 70/522/2019, KELA 98/522/2019, and Statistics Finland TK-53-1041-17. Access to FinnGen study data were obtained through a research proposal with Dr. Andrea Ganna, a consortium partner of FinnGen and a co-author of the work, which granted only sharing of results but not individual-level data sharing. The collection of samples to the Columbia University COVID-19 Biobank was approved by the Institutional Review Board (IRB) of Columbia University (IRB protocol number AAAS7370), while the genetic analyses were approved under Columbia University IRB protocol number AAAS7948. The GenOMICC study was approved by the appropriate research ethics committees in Scotland (15/SS/0110) and England, Wales and Northern Ireland (19/WM/0247).
COVID-19 phenotype extraction
For UKB, GenOMICC and FinnGen cohorts, the critical illness phenotype refers to patients with a documented admission to ICU. As this data does not exist for the Columbia cohort, we defined critical illness as death from COVID-19 or treatment with invasive respiratory support.
UK biobank (UKB)
COVID-19 phenotype data were accessed as part of application 58510, which was registered for access to COVID-19-specific data fields. COVID-19 test results (for England, Wales and Scotland), COVID-19 Hospital Episode Statistics (HES) for admissions, discharge destination and critical care admissions and COVID-19 death certificate data were downloaded from the UK Biobank data portal on July 23rd 2021. These data were processed to identify patients admitted to hospital with COVID-19 as the primary diagnosis (hospitalized outcome), patients admitted to ICU with COVID-19 (ICD10 code U07.1) as the primary diagnosis (ICU outcome) and patients that died from COVID-19 (death outcome). The latter outcome included patients admitted with a primary diagnosis of COVID-19 who died during their admission (discharge destination code 11001) or where COVID-19 was listed as the primary cause of death. The mild-moderate control cohort comprised patients with a positive COVID-19 test but without recorded hospital/ICU admission or death. The population control cohort comprised patients without recorded positive COVID-19 test, COVID-19-related admission or death.
GenOMICC
GenOMICC and ISARIC4C patients were recruited based on their admission to high-dependency or intensive care units for continuous cardiorespiratory monitoring after testing positive for COVID-19. Presence of SARS-CoV-2 particles was confirmed by local clinical testing. Phenotypic information with matched genetic data, age, sex, confirmed COVID-19 diagnosis and first part of the participant postcode was available for 3,650 (74.4%) of the 4.906 unrelated European-ancestry individuals in the GenOMICC cohort. After exclusion of participants in postcode-ascribed local authority regions with fewer than 50 individuals, 2,188 patients remained and were included in the analysis (see Supplementary Note).
Columbia COVID-19 biobank
The Columbia University COVID-19 Biobank was established in response to the New York City infection surge in March 2020. The Biobank recruited COVID-19 cases (n = 1,166) of diverse ancestry among all patients who were treated at Columbia University Irving Medical Center between March and May 2020. All cases were diagnosed by positive SARS-CoV-2 PCR test based on nasopharyngeal samples. Hospital record data were utilized to identify hospitalized cases (n = 1,067). We extracted a composite endpoint capturing critical illness that comprised patients who died from COVID-19 or had severe respiratory failure defined by intubation and requirement for invasive respiratory support (n = 496). Population controls are derived from the same general patient population as the cases, and were genotyped as healthy controls for other studies. Age data were unavailable for these population controls as these participants were fully anonymized.
FinnGen
The FinnGen study combines electronic health records data from Finnish national health registries. These include the infectious disease registry and data on all SARS-CoV-2 positive individuals included in FinnGen Release 7 (n = 3,496). The data were collected from the register on May 27th 2021. ICU admitted COVID-19 cases were defined based on information on ICU admission or invasive ventilation (n = 165). Hospitalized cases were defined based on hospitalization information available in the register (n = 319). The COVID-positive control population comprised all the individuals not included in the previous two categories (n = 2835) while population controls included all the individuals in FinnGen, excluding participants with a history of COVID-19 infection (n = 305,996).
Quantification and statistical analysis
Cohort genomic data quality control (QC)
UK biobank (UKB)
UKB-provided measured genotype, imputed genotype (GRCh37, imputed data release 3) and phenotype data73 was accessed as part of application 58510. For sample QC we excluded subjects with sex chromosome aneuploidy (field 22019), discordant genetic sex (fields 31 and 22,001), excess heterozygosity and missing rate (field 22,027). Genetic ancestry was classified as described previously (Kleeman et al., 2021). For all analyses using imputed data, we filtered to variants with INFO score >0.8 and MAF >1% across whole cohort.
GenOMICC
GenOMICC and ISARIC4C individuals (data release 7) were genotyped using the Illumina Global Screening Array v.3.0 and had their relatedness inferred using KING 2.1, as previously described (Pärn et al., 2018). For brevity, the combined cohort is referred to as GenOMICC. Sample QC was performed by excluding subjects with sex chromosome aneuploidy, discordant genetic sex, and SNP call rates <97%. Genotyped array variants were filtered to have call rates of >99%, MAF >1%, and Hardy–Weinberg equilibrium (HWE) p ≥ 10−6. Genome-wide SNPs were imputed using the TOPMed reference panel (Taliun et al., 2021). Excluding all non-European ancestry samples, removing all but one of each set of related individuals (up to third degree), and further excluding individuals who had withdrawn from the study, a total of 4,906 individuals had complete genetic information in GenOMICC.
Columbia COVID-19 biobank
DNA of whole blood samples was extracted using standard procedures and genotyping was performed using the Illumina Global Diversity Array (GDA) chip. The controls were genotyped using the Illumina Multiethnic Global Ancestry (MEGA) chip. The analysis of intensity clusters and genotype calls were performed in Illumina Genome Studio software; all SNPs were called on forward DNA strand and standard quality control (QC) filters were applied, including per-SNP genotyping rate >95%, per-individual genotyping rate >90%, minor allele frequency (MAF) > 0.01, and Hardy–Weinberg equilibrium (HWE) test p value > 10-08 in controls. The duplicates and cryptic relatedness in the given cohort were determined and excluded based on the estimated pairwise KING kinship coefficients >0.0884. After QC, the dataset consisted of 6,757 individuals (1,029 cases and 5,728 controls) genotyped for 1,096,321 SNPs with overall genotyping rate of 99.9%. The imputation analysis was performed using TOPMed imputation server. A total of 13,439,413 common markers imputed at high quality (r2 > 0.8 and MAF >0.01) were used in downstream analyses. Genetic ancestry was then classified as described previously (Kleeman et al., 2021), followed by joint PCA analysis to eliminate ancestry outliers, and to select controls closely matched to the cases in the PCA space. The largest ancestry populations in the COVID-19 case cohort were Admixed American (n = 542) and African (n = 220). We opted to focus on the African ancestry population as there were insufficient Admixed American participants (n = 707) in UKB to derive a modified ancestry-specific CyC-production PGS (described below). The final case-control cohort comprised 220 African ancestry COVID-19 patients and 2,341 controls. Genetic analyses were approved under Columbia University IRB protocol number AAAS7948.
FinnGen
Chip genotype data processing and QC Samples were genotyped with Illumina (Illumina Inc., San Diego, CA, USA) and Affymetrix arrays (Thermo Fisher Scientific, Santa Clara, CA, USA).
Genotype calls were made with GenCall and zCall algorithms for Illumina and AxiomGT1 algorithm for Affymetrix data. Chip genotyping data produced with previous chip platforms and reference genome builds were lifted over to build version 38 (GRCh38/hg38) as previously described (Pärn et al., 2018). In sample-wise quality control, individuals with ambiguous gender, high genotype missingness (>5%), excess heterozygosity (+-4SD) and non-Finnish ancestry were excluded. In variant-wise quality control variants with high missingness (>2%), low HWE p-value (<1 × 10−6) and minor allele count, MAC<3 were excluded.
UKB matched controls
GenoMICC
UKB genotype data were accessed as part of application 19,655. We included only White British individuals (field 22,006), again removing all but one of each set of related individuals (up to third degree, field 22,021), and further excluding individuals who had withdrawn from the study. A total of 336,015 individuals had complete genetic information in UKB. Individuals with history of COVID-19 diagnosis (defined as above) or from local authority districts with fewer than 50 participants were excluded, leaving a total of 330,227 individuals (see Supplementary Note). Differences in genotyping and imputation between GenOMICC cases and UKB control requires stringent QC to avoid confounding. For each cohort, we removed SNPs absent from the CyC PGS as well as SNPs with HWE p < 10−6, MAF <1%, INFO <0.8, or call rates <99% using QCTOOL v2. Taking forward the SNPs passing QC filters in both studies, we next excluded any SNPs with differences in MAF larger than 10% between the two cohorts. The final dataset used for PGS calculation included 980,733 SNPs (95.08% of the original 1,031,528 PGS SNPs). Between GenOMICC and UKB, there were 75 individuals present in both cohorts. These were used to confirm there were no systematic differences in PGS between cohorts, and then were then excluded from the UKB controls (see Supplementary Note).
Columbia COVID-19 biobank
For age-adjusted sensitivity analysis incorporating UKB population controls, UKB genotype data were accessed as part of application 41,849. Genetic ancestry was assigned as described previously (Kleeman et al., 2021) and related subjects (>3rd degree using UKB-provided KING kinship estimates) were removed, altogether identifying 8,152 unrelated African ancestry controls. We merged the genotyping data from the UKB controls with the Columbia COVID-19 cases, keeping only the overlapping markers for downstream analyses. We then performed joint PCA analysis to eliminate ancestry outliers, and to select controls closely matched to the cases in the PCA space, leaving 217 cases and 2,334 ancestry-matched UKB controls. The merged UKB-Columbia cohort was imputed using the TOPMed imputation server (Taliun et al., 2021). After standard QC, a total of 10,704,006 common markers imputed at high quality (r2 > 0.8 and MAF >0.01) were used in downstream logistic regression analyses.
Derivation and application of CyC-production polygenic scores
CyC-production polygenic score (PGS) was derived and validated as described previously (Kleeman et al., 2021), using summary statistics from the UKB European (EUR) population, implemented in LDpred2 (Privé et al., 2020). For African ancestry populations, we used a modified version of the CyC-production PGS to capture ancestry-specific linkage disequilibrium structure. To develop this modified score, we performed multi-ancestry meta-analysis implemented in the ‘mama’ package for R, according to the package tutorial (https://github.com/JonJala/mama/tree/mainline/tutorial). For this analysis, we provided eGFR-CyC and eGFR-Cr summary statistics for EUR, Central and South Asian (CSA) and AFR populations in UKB, with GWAS performed as described previously (Kleeman et al., 2021), implemented in BOLT-LMM (Loh et al., 2018). To generate AFR-specific summary statistics for the CyC-production latent trait, we performed structural equation modeling using GenomicSEM (Grotzinger et al., 2019) as described previously (Kleeman et al., 2021). To compute CyC-production PGS on a per-patient level, we utilized the PLINK2 linear scoring function (--score), avoiding the exclusion of duplicate dbSNP IDs. The sample-level PGS was normalized by Z-scoring in each cohort. For the primary UKB case-control analysis, we included subjects in a predefined held-out validation cohort comprising 50,000 randomly selected unrelated (>3rd degree using UKB-provided KING kinship estimates) European participants (Kleeman et al., 2021), which was not used for polygenic score derivation.
Logistic regression for adverse COVID-19 outcomes against polygenic score
Where data were available, we examined three relevant clinical outcomes (hospitalization, ICU admission and death) using two ancestry-matched control populations (COVID-positive patients with mild-moderate disease, or population controls), generating up to 6 analysis per cohort where data were available. We modeled each binary clinical outcome as a function of Z-scored CyC-production PGS and relevant covariates (summarized in Table S1). The coefficients of the regression model were extracted to calculate the odds ratio and 95% confidence interval for each outcome per standard deviation of CyC-production PGS.
GWAS for BMI recorded/not recorded in FinnGen
We performed a GWAS of having (n = 245,936) vs not having (n = 96,563) BMI recorded for the whole FinnGen cohort. We ran association tests with SAIGE for each variant with a minimum allele count of 5 from the imputation pipeline. We filtered the results to include variants with an imputation INFO >0.6 and MAF >1%.
Quantitative proteomics analysis of plasma cystatin C
The Charité Hospital patient cohort, recruited as part of the Pa-COVID-19 study, has been described previously (Demichev et al., 2021). Briefly, patients with a PCR-confirmed diagnosis of SARS-CoV-2 infection were eligible for inclusion in the study. Plasma sampling for plasma proteomics by mass spectrometry was performed three times per week subsequent to inclusion. Sample processing, mass spectrometry and data analysis were performed as described previously (Demichev et al., 2021), allowing for quantification of plasma CyC levels in 309 patients. Out of these patients, 131 had available paired serum creatinine for at least one timepoint, as well as clinical outcome data (COVID-specific mortality). For patients with at least one creatinine measurement, missing data were imputed with the most recent value. Plasma CyC levels were scaled by a factor of 300, so that the cohort mean was comparable to the mean serum CyC recorded in the UKB cohort (field 30720, units mg/L). For each patient, a creatinine-CyC (C2) ratio was calculated at each timepoint, using CKD-EPI eGFR equations with the race term set to 0. We performed Cox regression for in-hospital mortality against C2 ratio, with age, sex, BMI, C-reactive protein and Charlson comorbidity index as model covariates. To account for patient-specific factors where repeated measures were included, the robust standard error term was included. The time interval corresponded to days from blood sampling to death or follow-up cut-off date (December 2020).
Acknowledgments
We thank all patients and their families who have volunteered to participate in clinical research, without whom this study would not have been possible. This work was conducted using the UK Biobank resource under application numbers 58510, 19655, and 41849. The FinnGen project is funded by two grants from Business Finland (HUS 4685/31/2016 and UH 4386/31/2016) and the following industry partners: AbbVie Inc., AstraZeneca UK Ltd, Biogen MA Inc., Bristol Myers Squibb (and Celgene Corporation & Celgene International II Sàrl), Genentech Inc., Merck Sharp & Dohme Corp, Pfizer Inc., GlaxoSmithKline Intellectual Property Development Ltd., Sanofi US Services Inc., Maze Therapeutics Inc., Janssen Biotech Inc, Novartis AG, and Boehringer Ingelheim. Following biobanks are acknowledged for delivering biobank samples to FinnGen: Auria Biobank (www.auria.fi/biopankki), THL Biobank (www.thl.fi/biobank), Helsinki Biobank (www.helsinginbiopankki.fi), Biobank Borealis of Northern Finland (https://www.ppshp.fi/Tutkimus-ja-opetus/Biopankki/Pages/Biobank-Borealis-briefly-in-English.aspx), Finnish Clinical Biobank Tampere (www.tays.fi/en-US/Research_and_development/Finnish_Clinical_Biobank_Tampere), Biobank of Eastern Finland (www.ita-suomenbiopankki.fi/en), Central Finland Biobank (www.ksshp.fi/fi-FI/Potilaalle/Biopankki), Finnish Red Cross Blood Service Biobank (www.veripalvelu.fi/verenluovutus/biopankkitoiminta) and Terveystalo Biobank (www.terveystalo.com/fi/Yritystietoa/Terveystalo-Biopankki/Biopankki/). All Finnish Biobanks are members of BBMRI.fi infrastructure (www.bbmri.fi). Finnish Biobank Cooperative -FINBB (https://finbb.fi/) is the coordinator of BBMRI-ERIC operations in Finland. The Finnish biobank data can be accessed through the Fingenious services (https://site.fingenious.fi/en/) managed by FINBB. The Columbia University Biobank was supported by the Vagelos College of Physicians & Surgeons as well as the Precision Medicine Resource and Biomedical Informatics Resource of Irving Institute for Clinical and Translational Research, home of the Columbia University’s Clinical and Translational Science Award (CTSA), funded by the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant Number UL1TR001873. The genotyping was made possible by the Columbia University Biobank and its COVID-19 Genomics Workgroup members, including Andrea Califano, Wendy Chung, Christine K. Garcia, David B. Goldstein, Iuliana Ionita-Laza, Krzysztof Kiryluk, Richard Mayeux, Sheila M. O’Byrne, Danielle Pendrick, Muredach P. Reilly, Soumitra Sengupta, Peter Sims, and Anne-Catrin Uhlemann. The work was further supported by the German Ministry of Education and Research (BMBF), as part of the National Research Node “Mass spectrometry in Systems Medicine (MSCoreSys),” under grant agreements 031L0220 (to M.R.) and 161L0221 (to V.D.). A.K. was supported by grant no. K25 (K25DK128563) from the NIH/National Institute of Diabetes and Digestive and Kidney Diseases. TJ funding CSHL, Northwell Health, Pershing Square Foundation, and the Dr. Lee MacCormick Edwards Charitable Foundation.HVM receives funding from the Simons Center for Quantitative Biology at Cold Spring Harbor Laboratory. S.K. is supported by the Starr Centennial Scholarship at the Cold Spring Harbor Laboratory School of Biological Sciences. P.R.H.J.T. and J.F.W. acknowledge funding from the UK Medical Research Council Human Genetics Unit (grant no. MC_UU_00007/10). Computational analyses were performed with assistance from the US National Institutes of Health Grant S10OD028632-01.
Author contributions
S.O.K. and T.J. conceived and designed the study. S.O.K. performed a statistical and computational analysis in the UK Biobank and Charité Hospital cohorts. M.C., P.R.H.J.T., and A.K. performed replication statistical analyses under the supervision of J.F.W., K.K, A.G. and K.B. H.V.M. provided input for the interpretation of results and design of trans-ancestry analyses. P.R.H.J.T. and K.B. provided direction for BMI-adjusted analyses. P.T., F.K., V.D., and M.R. assisted with the analysis of the Charité Hospital cohort. S.O.K. and T.J. wrote the article with input from all co-authors, and both had access to all summary statistics from all cohorts. All co-authors approved the final version of the article, and take full responsibility for the accuracy of data and statistical analyses.
Declaration of interests
P.R.H.J.T. is a salaried employee of BioAge Labs Inc. The remaining authors declare no competing interests.
Inclusion and diversity
We worked to ensure ethnic or other types of diversity in the recruitment of human subjects. The author list of this paper includes contributors from the location where the research was conducted who participated in the data collection, design, analysis, and/or interpretation of the work.
Published: October 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105040.
Supplemental information
Data and code availability
-
•
Due to the data use agreements for the datasets analyzed in this manuscript, we are unable to directly share or distribute any patient-level data. All summary statistics are published alongside the study, and polygenic scores will be reposited on PGS Catalog on the acceptance of the peer-reviewed manuscript. UK Biobank data can be requested through the application process detailed at https://www.ukbiobank.ac.uk/.
-
•
All code has been reposited on Github at https://github.com/Janowitz-Lab/cyc_covid.
-
•
Where data use agreements allow, additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Brennan C.M., Nadella S., Zhao X., Dima R.J., Jordan-Martin N., Demestichas B.R., Kleeman S.O., Ferrer M., Gablenz E.C., von Mourikis N., et al. Oral famotidine versus placebo in non-hospitalised patients with COVID-19: a randomised, double-blind, data-intense, phase 2 clinical trial. Gut. 2022:326952. doi: 10.1136/gutjnl-2022-326952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner E.J., Ungaro R.C., Gearry R.B., Kaplan G.G., Kissous-Hunt M., Lewis J.D., Ng S.C., Rahier J.-F., Reinisch W., Ruemmele F.M., et al. Corticosteroids, but not TNF antagonists, are associated with adverse COVID-19 outcomes in patients with inflammatory bowel diseases: results from an international registry. Gastroenterology. 2020;159:481–491.e3. doi: 10.1053/j.gastro.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee N., Shi J., García-Closas M. Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat. Rev. Genet. 2016;17:392–406. doi: 10.1038/nrg.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demichev V., Tober-Lau P., Lemke O., Nazarenko T., Thibeault C., Whitwell H., Röhl A., Freiwald A., Szyrwiel L., Ludwig D., et al. A time-resolved proteomic and prognostic map of COVID-19. Cell Syst. 2021;12:780–794.e7. doi: 10.1016/j.cels.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/s1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajnzylber J., Regan J., Coxen K., Corry H., Wong C., Rosenthal A., Worrall D., Giguel F., Piechocka-Trocha A., Atyeo C., et al. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat. Commun. 2020;11:5493. doi: 10.1038/s41467-020-19057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Piernas C., Astbury N.M., Hippisley-Cox J., O’Rahilly S., Aveyard P., Jebb S.A. Associations between body-mass index and COVID-19 severity in 6·9 million people in England: a prospective, community-based, cohort study. Lancet Diabetes Endocrinol. 2021;9:350–359. doi: 10.1016/s2213-8587(21)00089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotzinger A.D., Rhemtulla M., de Vlaming R., Ritchie S.J., Mallard T.T., Hill W.D., Ip H.F., Marioni R.E., McIntosh A.M., Deary I.J., et al. Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nat. Hum. Behav. 2019;3:513–525. doi: 10.1038/s41562-019-0566-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Initiative C.-19H.G. Mapping the human genetic architecture of COVID-19. Nature. 2021;600:472–477. doi: 10.1038/s41586-021-03767-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inker L.A., Schmid C.H., Tighiouart H., Eckfeldt J.H., Feldman H.I., Greene T., Kusek J.W., Manzi J., Van Lente F., Zhang Y.L., et al. Estimating glomerular filtration rate from serum creatinine and cystatin C. N. Engl. J. Med. 2012;367:20–29. doi: 10.1056/nejmoa1114248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleeman S.O., Ferrer M., Demestichas B., Bankier S., Lee H., Heywood T., Ruusalepp A., Bjorkegren J.L.M., Walker B.R., Meyer H.V., Janowitz T. Cystatin C is a glucocorticoid response gene predictive of cancer immunotherapy failure. medRxiv. 2021 doi: 10.1101/2021.08.17.21261668. Preprint at. [DOI] [Google Scholar]
- Kopitar-Jerala N. The role of cystatins in cells of the immune system. FEBS Lett. 2006;580:6295–6301. doi: 10.1016/j.febslet.2006.10.055. [DOI] [PubMed] [Google Scholar]
- Liu Y., Xia P., Cao W., Liu Z., Ma J., Zheng K., Chen L., Li X., Qin Y., Li X. Divergence between serum creatine and cystatin C in estimating glomerular filtration rate of critically ill COVID-19 patients. Ren. Fail. 2021;43:1104–1114. doi: 10.1080/0886022x.2021.1948428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh P.-R., Kichaev G., Gazal S., Schoech A.P., Price A.L. Mixed-model association for biobank-scale datasets. Nat. Genet. 2018;50:906–908. doi: 10.1038/s41588-018-0144-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J.B., June C.H. Cytokine release syndrome in severe COVID-19. Science. 2020;368:473–474. doi: 10.1126/science.abb8925. [DOI] [PubMed] [Google Scholar]
- Muntner P., Winston J., Uribarri J., Mann D., Fox C.S. Overweight, obesity, and elevated serum cystatin C levels in adults in the United States. Am. J. Med. 2008;121:341–348. doi: 10.1016/j.amjmed.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A.D., Rawlik K., Pasko D., Walker S., Parkinson N., Fourman M.H., Russell C.D., et al. Genetic mechanisms of critical illness in COVID-19. Nature. 2021;591:92–98. doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- Pärn K., Fontarnau J.N., Isokallio M.A., Sipilä T., Kilpelainen E., Palotie A., Ripatti S., Palta P. 2018. Genotyping chip data lift-over to reference genome build GRCh38/hg38. protocols.io. [Google Scholar]
- Pirastu N., Cordioli M., Nandakumar P., Mignogna G., Abdellaoui A., Hollis B., Kanai M., Rajagopal V.M., Parolo P.D.B., Baya N., et al. Genetic analyses identify widespread sex-differential participation bias. Nat. Genet. 2021;53:663–671. doi: 10.1038/s41588-021-00846-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privé F., Arbel J., Vilhjálmsson B.J. LDpred2: better, faster, stronger. Bioinformatics. 2020;36:5424–5431. doi: 10.1093/bioinformatics/btaa1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RECOVERY Collaborative Group. Horby P., Lim W.S., Emberson J.R., Mafham M., Bell J.L., Linsell L., Staplin N., Brightling C., Ustianowski A., Elmahi E., et al. Dexamethasone in hospitalized patients with Covid-19. N. Engl. J. Med. 2021;384:693–704. doi: 10.1056/nejmoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taliun D., Harris D.N., Kessler M.D., Carlson J., Szpiech Z.A., Torres R., Taliun S.A.G., Corvelo A., Gogarten S.M., Kang H.M., et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590:290–299. doi: 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiersinga W.J., Rhodes A., Cheng A.C., Peacock S.J., Prescott H.C. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19) JAMA. 2020;324:782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- van der Wijst M.G.P., Vazquez S.E., Hartoularos G.C., Bastard P., Grant T., Bueno R., Lee D.S., Greenland J.R., Sun Y., Perez R., et al. Type I interferon autoantibodies are associated with systemic immune alterations in patients with COVID-19. Sci. Transl. Med. 2021;13:eabh2624. doi: 10.1126/scitranslmed.abh2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolter N., Jassat W., Walaza S., Welch R., Moultrie H., Groome M., Amoako D.G., Everatt J., Bhiman J.N., Scheepers C., et al. Early assessment of the clinical severity of the SARS-CoV-2 omicron variant in South Africa: a data linkage study. Lancet. 2022;399:437–446. doi: 10.1016/s0140-6736(22)00017-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Bastard P., Bolze A., Jouanguy E., Zhang S.-Y., COVID Human Genetic Effort. Cobat A., Notarangelo L.D., Su H.C., Abel L., Casanova J.-L. Life-Threatening COVID-19: defective interferons unleash excessive inflammation. Med. 2020;1:14–20. doi: 10.1016/j.medj.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Bastard P., Liu Z., Pen J.L., Moncada-Velez M., Chen J., Ogishi M., Sabli I.K.D., Hodeib S., Korol C., et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370:eabd4570. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/s0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Due to the data use agreements for the datasets analyzed in this manuscript, we are unable to directly share or distribute any patient-level data. All summary statistics are published alongside the study, and polygenic scores will be reposited on PGS Catalog on the acceptance of the peer-reviewed manuscript. UK Biobank data can be requested through the application process detailed at https://www.ukbiobank.ac.uk/.
-
•
All code has been reposited on Github at https://github.com/Janowitz-Lab/cyc_covid.
-
•
Where data use agreements allow, additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.