Summary
Objective
Fibroblast Growth Factor 12 (FGF12) may represent an important modulator of neuronal network activity and has been associated with developmental and epileptic encephalopathy (DEE). We sought to identify the underlying pathomechanism of FGF12-related disorders.
Methods
Patients with pathogenic variants in FGF12 were identified through published case reports, GeneMatcher and whole exome sequencing of own case collections. The functional consequences of two missense and two copy number variants (CNVs) were studied by co-expression of wildtype and mutant FGF12 in neuronal-like cells (ND7/23) with the sodium channels NaV1.2 or NaV1.6, including their beta-1 and beta-2 sodium channel subunits (SCN1B and SCN2B).
Results
Four variants in FGF12 were identified for functional analysis: one novel FGF12 variant in a patient with autism spectrum disorder and three variants from previously published patients affected by DEE. We demonstrate the differential regulating effects of wildtype and mutant FGF12 on NaV1.2 and NaV1.6 channels. Here, FGF12 variants lead to a complex kinetic influence on NaV1.2 and NaV1.6, including loss- as well as gain-of function changes in fast and slow inactivation.
Interpretation
We could demonstrate the detailed regulating effect of FGF12 on NaV1.2 and NaV1.6 and confirmed the complex effect of FGF12 on neuronal network activity. Our findings expand the phenotypic spectrum related to FGF12 variants and elucidate the underlying pathomechanism. Specific variants in FGF12-associated disorders may be amenable to precision treatment with sodium channel blockers.
Funding
DFG, BMBF, Hartwell Foundation, National Institute for Neurological Disorders and Stroke, IDDRC, ENGIN, NIH, ITMAT, ILAE, RES and GRIN
Keywords: FGF12, Sodium channels, Epilepsy, SCN2A, SCN8A, Autism spectrum disorder
Research in context.
Evidence before this study
Fibroblast growth factors homologous factors are known to modulate voltage gated sodium channels. FGF12 modulates the subtypes NaV1.2, NaV1.5 and NaV1.6 but not NaV1.1. Recently one recurrent variant was shown to cause developmental and epileptic encephalopathy by shifting the voltage dependence of fast inactivation of NaV1.6 to more depolarized potentials.
Added value of this study
We could demonstrate the detailed regulating effect of FGF12 on NaV1.2 and NaV1.6 and confirmed the complex effect of wildtype FGF12 on neuronal network activity. Our findings expand the phenotypic spectrum related to FGF12 variants and elucidate the underlying pathomechanism. Specific variants in FGF12-associated disorders may be amenable to precision treatment with sodium channel blockers.
Implications of all the available evidence
Our study suggests a more complex mechanism underlaying FGF12 dependent voltage gated sodium channel regulation as known until now. Additionally, we were able to elucidate the complex gain- and loss-of function mechanisms implicated in FGF12-related disorders. Future experiments using primary neuronal cultures expressing the different FGF12 variants will help to gain a better understanding of the regulatory effects on action potential initiation and propagation as well as on the firing behaviour of neurons.
Alt-text: Unlabelled box
Introduction
Voltage-gated sodium channels (NaVs) play an important role in the initiation and propagation of action potentials in neurons. NaVs are expressed in the central and the peripheral nervous system, the skeletal muscle and the heart.1 A total of nine different alpha subunits are known, of which variants in five of these subunits (NaV1.1 [SCN1A], NaV1.2 [SCN2A], NaV1.3 [SCN3A], NaV1.6 [SCN8A] and NaV1.7 [SCN9A]) have been related to epilepsy or intellectual disability.2, 3, 4 Sodium channels are modulated by different proteins. Of these, four members of the FGF family also called FGF11 subfamily, namely FGF11, FGF12, FGF13 and FGF14, play a major role in modulating sodium channels fast inactivation kinetics.5 All members of the FGF11 subfamily have two or more transcripts that differ in their N-terminal length, suggesting that they are involved in different molecular mechanisms (Figure 1a).6 Accordingly, they exert a broad spectrum of modulating effects on various channels. FGF14 differentially modulates the fast inactivation of NaV1.2 and NaV1.6, and slows the recovery from inactivation of NaV1.6.7 FGF13 is able to modulate NaV1.5 by influencing fast inactivation as well as the recovery from fast inactivation.8 FGF12 is known to interact with NaV1.2, NaV1.5 and NaV1.6, while it seems not to bind to or modulate NaV1.1.9, 10, 11, 12
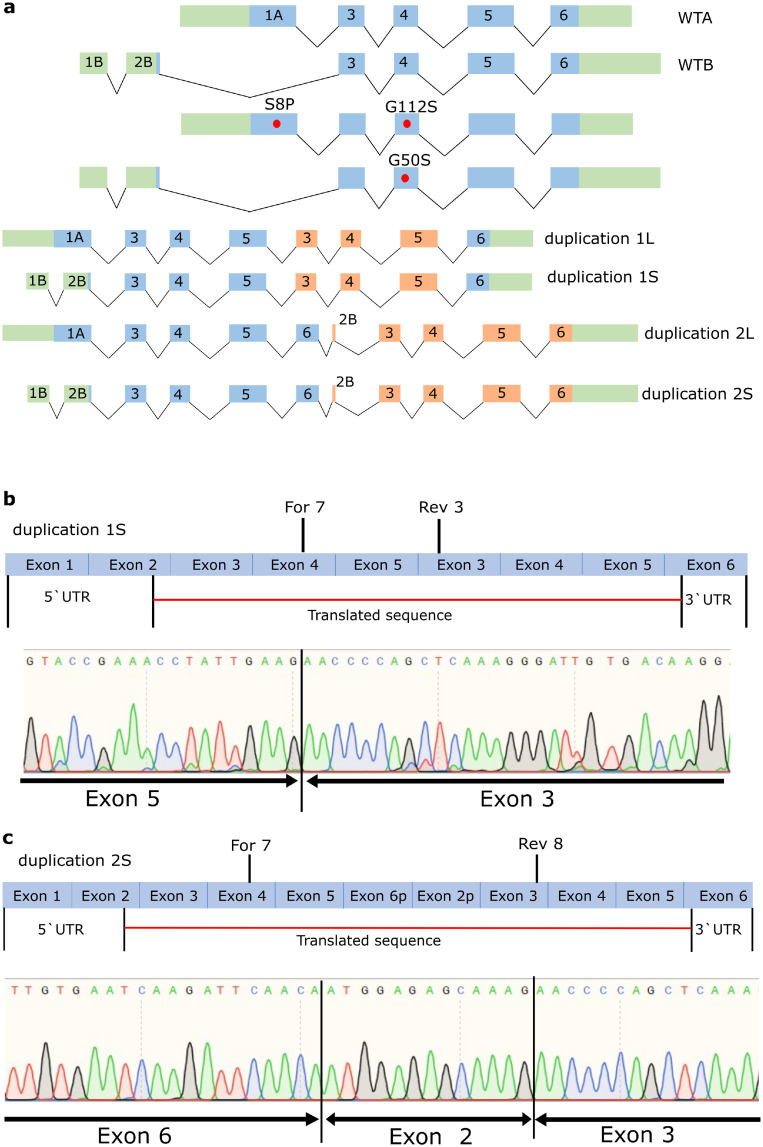
Figure 1.
Characterisation of FGF12 CNVs. a. Schematic drawings of the different FGF12 WTs as well as the different missense variants and CNVs. Green: UTR sites, blue: translated exons, orange: duplicated translated exons, red dots: missense variants. b. Characterisation of duplication 1 using RT-PCR from cDNA extracted from venous blood of the two patients described in Willemsen et al., 2020. Primer For 7 and Rev 3 were used for the amplification of the CNV, and the sequencing result of this PCR product showed a duplication encompassing Exon 3 to Exon 5. c. Characterisation of duplication 2 using RT-PCR from RNA extracted from Fibroblasts of the patient described in Willemsen et al., 2020. Primer For 7 and Rev 8 were used for the amplification of the CNV, and the sequencing result of this PCR product showed a duplication encompassing the whole gene of FGF12 (Exon 2 to Exon 6).
Variants in genes encoding members of this subfamily have been associated with generalized epilepsy with febrile seizures plus (GEFS+; FGF13), spinocerebellar ataxia (FGF14) or developmental and epileptic encephalopathy (DEE; FGF12 and FGF13).13, 14, 15, 16 However, the detailed pathophysiological mechanism behind FGF12-associated DEE is still unclear.9
Here we investigated the modifying effects of FGF12 on NaV1.2 and NaV1.6 in the wildtype state. Additionally, we carried out functional analyses of four FGF12 variants, including a novel variant of FGF12 causing autism spectrum disorder (ASD).
Methods
Patients and variants collection
The variants in this narrative case series were collected through GeneMatcher reports,17 literature review and whole exome sequencing in two novel cases identified by the Institute of Human genetics UKE Hamburg-Eppendorf and the Clinic for Neuropediatric UKSH Kiel. Further pathogenic variants were not detected based on the classification criteria of the American College of Medical Genetics.18 Segregation analysis was performed if applicable.
Ethics
For the novel cases, written informed consent was obtained from all participants or their legal representatives. The current study was approved by each local ethics committee the University of Leipzig Germany (224/16-ek, 402/16-ek), the ethics committee of the Hamburg Medical Association (PV7038) and the medical ethical committee of the Radboudumc Nijmegen the Netherlands (CMO-Nr.:2014-1254).
Sample Size
We collected two new patients with FGF12 variants for clinical analysis. Furthermore, we analysed in total four different FGF12 variants (three already published variants and one newly identified variant). All variants were analysed using ND7/23 cells and a minimum of ten cells per protocol and variant was recorded each.
Fibroblast generation
The fibroblasts were generated from a skin biopsy with a size of 3-4 mm. First, the subcutaneous adipose tissue was removed from the biopsy and washed twice with PBS + Penicillin/Streptomycin. Afterwards, the biopsy was cut in 1-millimetre pieces and transferred to a 15 mL tube with 2 mL collagenase solution (collagenase diluted in 2 mL DMEM) and incubated for 3 hours at 37°C. After the incubation, 6 mL DMEM + 20% FCS was added to dilute the collagenase. Once spun down, the medium was exchanged with 6 mL fresh DMEM-medium, supplemented with 20% FCS. The sample was split into 4 T25 flasks and incubated for 7 days at 37°C in 5% CO2. The medium was refreshed every 3-4 days, as cells started to grow.
Transcript identification
For two duplications, we had to determine the structure of the resulting transcript. The duplication 3q28q29(chr3:191.860.089-192.451.114), which was found in one individual, and the duplication 3q28q29 (chr3:191876978-192454675), which was identified in two related individuals, have been published previously within a clinical report.19 To analyse the transcript of the first duplication, we performed RNA purification from fibroblasts and retrotranscription to cDNA, using the Isolation II RNA Mini kit (Bioline) and the First strand cDNA synthesis kit for RT-PCR (Roche), respectively, according to manufacturer protocols. For the CNV-specific PCR, the following primers were used: FGF12For7: CTC TCT TCA ATC TAA TTC CCG TGG and FGF12Rev8: GTA GTC GCT GTT TTC GTC CTT GGT C. For the analysis of the second duplication, RNA from venous blood of both patients was isolated and retro-transcribed in cDNA followed by a CNV-specific PCR using the following primers: FGF12For7: CTC TCT TCA ATC TAA TTC CCG TGG and FGF12Rev3: CCT TGT CAC AAT CCC TTT GAG CTG G. All three PCR products were sent for sequencing to LGC Genomics GmbH (Berlin, Germany).
Functional investigations
Mutagenesis
The human FGF12 wildtype (WT) and the variants were purchased from Genescript as g-blocks and cloned with the HiFi DNA Assembly Kit (purchased from NEB) into the pCMV6 Vector (purchased from Origene) together with the two human β1- and β2-subunits of voltage-gated Na+ channels as well as the antibiotic puromycin as selection marker. The four proteins were separated by the P2A, E2A and T2A sequences, respectively, to separate the proteins during translation. Except for the stop codon, the sequence of FGF12A corresponded to the Ensembl transcript ENST00000454309.6 and Consensus Coding Sequence Database transcript CCDS3301 and FGF12B corresponded to the Ensembl transcript ENST00000445105.6 and Consensus Coding Sequence Database transcript CCDS46983.
The human NaV1.6 channel construct was purchased from Origene and modified to introduce TTX resistance by a known point mutation (c.1112A > G, p.Y371C).20 The WT open reading frame included the canonical SCN8A coding sequence, with a C-terminal P2A sequence combined with an eGFP. The SCN8A coding sequence (5940 bp) contained the splice isoform 5 N of exon 5. Apart from the TTX resistance change and the stop codon, it was identical to the coding sequences of Ensembl transcript ENST00000354534.11 and Consensus Coding Sequence Database transcript CCDS44891. To engineer the TTX-resistance into the human NaV1.6 channel, site-directed mutagenesis was performed using PCR with Pfu polymerase (Promega; mutagenic primers are available upon request). To record currents from NaV1.2 channels, we exchanged the NaV1.6 cDNA with the NaV1.2 cDNA and introduced TTX resistance by a known point mutation (c.1154 T>C, p.F385S).21 Except for the stop codon and TTX resistance change, the cDNA was identical to the coding sequence of Ensembl transcript ENST00000636071.2 and Consensus Coding Sequence Database transcript CCDS33313. To engineer the TTX-resistance into the human NaV1.2 channel, site-directed mutagenesis was performed using PCR with Phusion HF polymerase (NEB; mutagenic primers are available upon request). Further mutations in the constructs were excluded by sequencing the whole open reading frame prior using the clones for physiological experiments.
Transfection of ND7/23 cells
ND7/23 is a hybrid cell line derived from neonatal rat dorsal root ganglia neurons fused with mouse neuroblastoma cells.22 It was purchased from Sigma Aldrich and cultured in Dulbecco's modified Eagle nutrient medium (Invitrogen) supplemented with 10% fetal calf serum (PAN-Biothech) and 1% L-glutamine 200 mM (Biochrom) at 37°C, with 5% CO2 humidified atmosphere. ND7/23 cells were plated in 35 mm petri dishes following the standard protocol for Lipofectamine™ 3000 (Invitrogen) transfections.
Transfections of WT or mutant human FGF12 cDNAs were then performed together with the human SCN8A cDNA, encoding the NaV1.6 channel α-subunit with an engineered TTX resistance or the human SCN2A cDNA, encoding the NaV1.2 channel α-subunit with an engineered TTX resistance. For co-expression of FGF12 with both β-subunits and the α subunit, 3·4 µg of cDNA were used (3 µg of the α-subunit and 0·4 µg of the FGF12 with both β-subunits) as described previously.3 After 24 h, cells were selected by adding 1·5 µg/mL puromycin (InvivoGen) to the medium. Electrophysiological recordings were performed 72 h after transfection only from cells expressing all four proteins, which were recognized by (i) a green fluorescence (α-subunit) and (ii) a puromycin resistance (FGF12 and both beta subunits).
Electrophysiology
For recordings of transfected ND7/23 cells, 500 nM TTX was added to the bath solution to block all endogenous Na+ currents. Standard whole-cell voltage clamp recordings were performed using an Axopatch 200B amplifier, a Digidata 1440 A digitizer and Clampex 10.2 data acquisition software (Molecular Devices) as described previously.3 Leakage and capacitive currents were automatically subtracted using a prepulse protocol (−P/4). Cells were held at −100 mV. Currents were filtered at 10 kHz and digitized at 20 kHz. Cells were visualized using an inverted microscope (DM IL LED, Leica). All recordings of transfected ND7/23 cells were performed 10 min after establishing the whole-cell configuration at room temperature to avoid larger shifts in voltage dependence. Borosilicate glass pipettes had a final tip resistance of 1.5–3.5 MΩ when filled with internal recording solution (see below). We carefully checked that the maximal voltage error due to residual series resistance after up to 90% compensation was always <5 mV. The pipette solution contained (in mM): 10 NaCl, 1 EGTA, 10 HEPES, 140 CsF (pH was adjusted to 7·3 with CsOH, osmolarity was adjusted to 310 mOsm/kg with mannitol). The bath solution contained (in mM): 140 NaCl, 3 KCl, 1 MgCl2, 1 CaCl2, 10 HEPES, 20 TEACl (tetraethylammonium chloride), 5 CsCl and 0·1 CdCl2 (pH was adjusted to 7·3 with CsOH, osmolarity was adjusted to 320 mOsm/kg with mannitol).
Statistics
For voltage-clamp recordings in ND7/23 cells, the biophysical parameters of human WT NaV1.6 and human WT NaV1.2 channels were obtained as described previously3using a test pulse of -10 mV or -20 mV in all protocols for the recordings of NaV1.6 and a test pulse to 0 mV for the recordings of NaV1.2. A detailed description is provided in the Supplementary material. All data were analysed using Clampfit software of pClamp 10.6 (Axon Instruments) with respect to the voltage dependent activation, fast inactivation as well as the slow inactivation and recovery from inactivation. In addition, Clampfit software was used to fit the single exponential curve for the time constant of fast inactivation. Microsoft Excel (Microsoft Corporation, Redmond, WA, USA) was used for rearrangement and transformation of the data received from the Clampfit software according to the described analysis method in the Supplementary Material. Gnuplot 5·2 (Freeware, T. Williams & C.Kelly) was used to fit the Boltzmann-function to the data points. Statistics were performed using Graphpad software (Graphpad prism, San Diego, CA, USA). All data were tested for normal distribution. For comparison of multiple groups, one-way ANOVA with Holm-Sidak´s post hoc test was used for normally distributed data and ANOVA on ranks with Dunn's post hoc test was used for not normally distributed data.
Role of funding source
The funding sources had no involvement in the study design, the analysis and interpretation of the data, the writing of this manuscript or in the decision to submit this manuscript for publication.
Results
Patients
We identified two novel patients through our own patient collection and GeneMatcher, both harbouring a de novo variant in FGF12 and suffering from DEE or ASD, respectively.
Patient 1 (male) is the first child of healthy unrelated parents. He was born at term with normal birth measures (3190 g, 49 cm, OFC 34 cm) and normal postnatal adaptation. He had a first generalized seizure at the age of 3.5 months. An initial EEG showed multifocal bi-hemispheric epileptic activity. Consequently, his development was moderately delayed, and he was diagnosed with ASD at the age of 1,5 years. At the current age of 11.5 years, he still has a marked speech delay, intellectual disability with autistic behaviour and ongoing absence-like episodes treated with oxcarbacepine and valproate. Whole exome sequencing revealed a de novo variant in FGF12 (NM_021032.4: c.334G>A, p.G112S), with a known association to DEE (ClinVar accession number: VCV000522854.4).
Patient 2 (male) was a 4·5-year-old boy. At 1·5 years of age, he was diagnosed with ASD. He showed developmental delay with speech delay, feeding difficulties and intellectual disability. ECG, EEG and MRI were normal. Whole exome sequencing revealed a de novo variant in the FGF12 gene (NM_021032.4: c.23C>T, p.S8P), which has not been previously described.
Variants
Through our literature review, we identified three further copy number variants (CNVs) (3q28q29(191,876,978-192,454,675) X3; 3q28q29(191,876,968-192,454,685) X3; 3q28q29(191,860,089-192,451,114) X3) which have been published clinically before.19,23 The first duplication was already characterized on a molecular level23 while the other two duplications were not further characterized. None of them had been functionally analysed. All of these missense variants and copy number variants were associated with DEE.
Characterisation of the duplications
To functionally investigate the duplications, it was first necessary to determine the correct resulting transcript form. The first duplication (3q28q29(191.876.978-192.454.675) X3 had already been analysed and published before.23 This duplication encompasses exons 3 to 5 (NM_004113) and is referred to as duplication 1 for the following functional characterization (Figure 1b, supplementary file 1). The second duplication (3q28q29(191,876,968-192,454,685) X3) is very similar to the first duplication but spans 10 bp more in each direction. To evaluate if this expansion leads to a different transcript, we used reverse transcription PCR from isolated RNA from venous blood of the patient. The 332 bp encompassing PCR product demonstrated exon 4, 5 followed by exon 3 (NM_004113) (Figure 1b). Thus, this duplication encompasses the same exons as duplication 1. The third duplication (3q28q29(191,860,089-192,451,114) X3) was analysed using reverse transcription PCR from isolated RNA of the patient's fibroblasts. The 542 bp encompassing PCR product showed exon 4, 5, and 6, which was followed by a part of exon 2 and exon 3 (NM_004113) (Figure 1c). Thus, this duplication (referred here as duplication 2) encompasses the whole gene except for exon 1, partially exon 2 and partially exon 6, since these parts include just the UTR sites (Supplementary file 1).
Functional characterization of the effect of different FGF12 WT isoforms on NaV1.6 and NaV1.2 channels
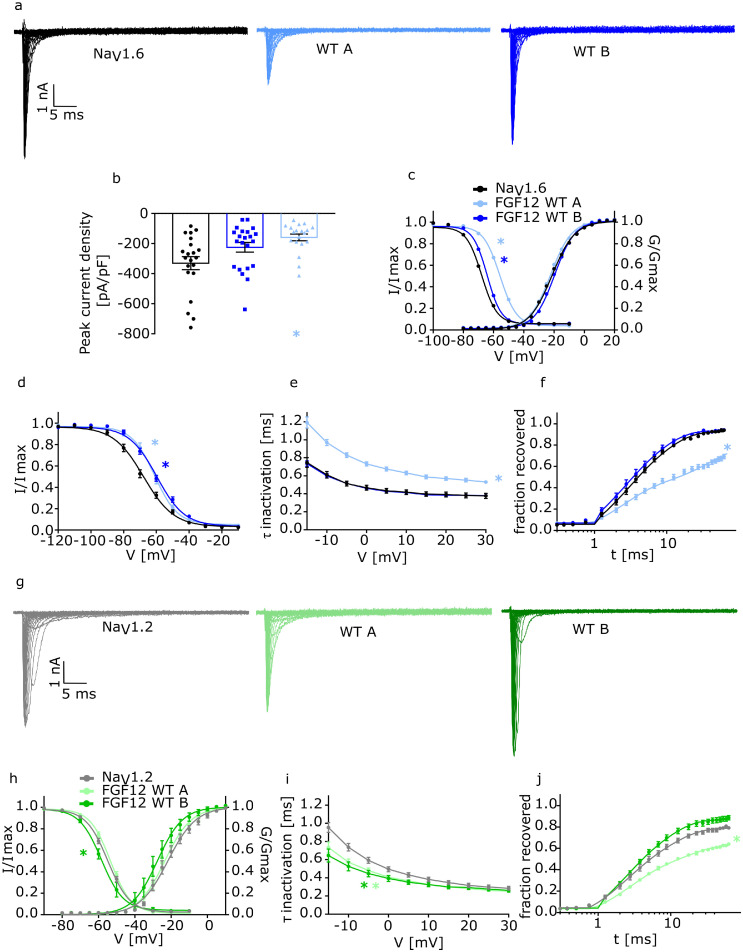
FGF12 has two different homologs which result from alternative splicing and are only different in their N-terminal part (FGF12A, long N-terminal part: WTA; FGF12B, short N-terminal part: WTB) (Figure 1a). Since both homologs include the core structure which is necessary for the interaction of FGF12 with NaVs, we characterized the effects of both WTs on NaV1.2 and NaV1.6. The effect on NaV1.5 was not investigated since NaV1.5 is not expressed in the brain. Expression of FGF12A together with NaV1.6 showed a reduction in Na+ current density (NaV1.6: -330 [-420·0 – -241·5] pA/pF; NaV1.6+ WTA: -158·90 [-206·5 – -111·3] pA/pF; p=0·0022)(Figure 2a and b, Table 1), and altered voltage-dependent gating kinetics including a depolarizing shift of fast inactivation (NaV1.6: -67·8 [-69·6 – -66·1] mV; NaV1.6+WTA: -55·8 [-57·4 – -54·2] mV; p=0·0001) (Figure 2c, Table 1) and slow inactivation (NaV1.6: -68·0 [-71·4 – -64·5] mV; NaV1.6+WTA: -61·3 [-64·4 – -58·1] mV; p=0·0072) (Figure 2d, Table 1) in comparison to NaV1.6 alone. Additionally, fast inactivation (NaV1.6: 0·61 [0·54 – 0·69] ms; NaV1.6+WTA:0·97 [0·90 – 1·04] ms; p=<0·0001) and the slow time constant (τ2) of recovery from fast inactivation were slowed (NaV1.6: 8·12 [6·93 – 9·32] ms; NaV1.6+WTA: 31·88 [21·86 – 41·91] ms; p=<0·0001; Figure 2e and f, Table 1). Interestingly the channels activation kinetics were not changed. Taken together, FGF12A resulted in a mixed gain- and loss-of overall ion channel function (GOF/LOF) in NaV1.6. Conversely, FGF12B led to a depolarizing shift of fast inactivation (NaV1.6: -67·8 [-69·6 – -66·1] mV, NaV1.6+WTB: -64·37 [-65·6 – -63·2] mV; p=0·002) as well as to a depolarizing shift of slow inactivation (NaV1.6: -68·0 [-71·4 – -64·5] mV; NaV1.6+WTB: -59·9 [-62·8 – -57·0] mV; p=0·0005), resulting in a gain-of-function (GOF) effect on NaV1.6 (Figure 2c and d, Table 1). Similarly, as for WTA the activation kinetic of the channel was not affected, too. In contrast to WTA, neither the time constant of the fast inactivation nor the recovery from fast inactivation were affected.
Figure 2.
Electrophysiological analysis of the two different FGF12 WT isoforms (WTA and WTB). Functional analysis of the effect of the two different FGF12 WTs on NaV1.2 and NaV1.6 compared to the channels without FGF12. Statistically significant effects for each condition are indicated by coloured asterisks.
a. Representative traces of NaV1.6 currents in ND7/23 cells expressing NaV1.6 without FGF12, with FGF12 WTA or FGF12 WTB in response to voltage steps from -80 to +35 mV in 5 mV steps.
b. Mean Current amplitudes of analysed ND7/23 cells injected with NaV1.6 without FGF12 (n=21; 10 transfections), with FGF12 WTA (n=19; 8 transfections; p = 0·002) or FGF12 WTB (n=22; 16 transfections; p=0·1). For the statistical analysis ANOVA on ranks with Dunn´s post hoc test was used. Expression of NaV1.6 together with FGF12 WTA showed a statistically significantly reduced current density compared to NaV1.6 alone.
c. Mean voltage-dependent activation and fast inactivation of NaV1.6 without FGF12 (n=21; 10 transfections), with FGF12 WTA (n=19/16; 8/6 transfections) or FGF12 WTB (n=22; 16 transfections). Lines illustrate Boltzmann Function fit to the data points. All fast inactivation curves (FGF12 WTA: p (V1/2) = 0·0001; FGF12 WTB: p (V1/2) = 0·002) showed a statistically significant shift to more depolarized potentials in comparison to NaV1.6 without FGF12. For the statistical analysis of the fast inactivation one-way ANOVA with Holm-Sidak´s post hoc test was used. All data are shown as means ± SEM.
d. Mean voltage-dependent slow inactivation of NaV1.6 without FGF12 (n=15; 8 transfections), with FGF12 WTA (n=11; 8 transfections) or FGF12 WTB (n=15; 12 transfections). Lines illustrate Boltzmann Function fit to the data points. All slow inactivation curves showed a statistically significant shift to more depolarized potentials (WTA: p (V1/2) = 0·007; WTB: p (V1/2) = 0·0005) in comparison to NaV1.6 without FGF12. For the statistical analysis of the slow inactivation one-way ANOVA with Holm-Sidak´s post hoc test was used. All data are shown as means ± SEM.
e. Mean voltage-dependent fast inactivation time constant of NaV1.6 without FGF12 (n=21; 10 transfections), with FGF12 WTA (n=19; 8 transfections; p = <0·0001) or FGF12 WTB (n=22; 16 transfections; p = >0·999). For the statistical analysis ANOVA on ranks with Dunn´s post hoc test was used. All data are shown as means ± SEM.
f. Time course of recovery from fast inactivation of NaV1.6 without FGF12 (n=10; 6 transfections), with FGF12 WTA (n=10; 8 transfections; p (τ2 = 0·0001) or FGF12 WTB (n=11; 8 transfections; p (τ2) = 0·9) determined at -100 mV. Lines represent fits of biexponential functions yielding the time constants τ1 and τ2. A1 was set to 0·3. For the statistical analysis one-way ANOVA with Holm-Sidak´s post hoc test was used. All data are shown as means ± SEM. All values of electrophysiological results, numbers and p-values are listed in Table 1 as means with the 95% confidence interval.
g. Representative traces of NaV1.2 currents in ND7/23 cells expressing NaV1.2 without FGF12, with FGF12 WTA or FGF12 WTB in response to voltage steps from -80 to +35 mV in 5 mV steps.
h. Mean voltage-dependent activation and fast inactivation of NaV1.2 without FGF12 (n=14/13; 6/5 transfections), with FGF12 WTA (n=13; 5 transfections) or FGF12 WTB (n=11; 3 transfections). The fast inactivation of WTB (p (V1/2) = 0·0328) was statistically significantly shifted to more hyperpolarized potentials than NaV1.2 without FGF12. For the statistical analysis ANOVA on ranks with Dunn´s post hoc test was used. Lines illustrate Boltzmann Function fit to the data points. All data are shown as means ± SEM.
i. Mean voltage-dependent fast inactivation time constant of NaV1.2 without FGF12 (n=14; 6 transfections), with FGF12 WTA (n=13; 5 transfections; p = 0·01) or FGF12 WTB (n=11; 3 transfections; p = 0·003). All deactivation curves showed a statistically significantly faster deactivation in comparison to channels only containing NaV1.2 subunit. For the statistical analysis one-way ANOVA with Holm-Sidak´s post hoc test was used. All data are shown as means ± SEM.
j. Time course of recovery from fast inactivation of NaV1.2 without FGF12 (n=12; 5 transfections), with FGF12 WTA (n=11; 5 transfections; p (τ2) = 0·001) or FGF12 WTB (n=10; 3 transfections; p (τ2) = 0·9) determined at -100 mV. Lines represent fits of biexponential functions yielding the time constants τ1 and τ2. A1 was set to 0·3. For the statistical analysis ANOVA on ranks with Dunn´s post hoc test was used. All data are shown as means ± SEM. All values of electrophysiological results, numbers and p-values are listed in Table 2 as means with the 95% confidence interval.
Table 1.
Biophysical parameters of NaV1.6 with FGF12 variants.
| Beta | WTA | WTB | S8P | G50S | G112S | duplication 1S | duplication 1L | duplication 2S | duplication 2L | |
|---|---|---|---|---|---|---|---|---|---|---|
| Current density, pA/pF | -330·8 [-420·0 – -241·5] |
-158·9 [-206·5 - -111·3] (p=0·002) |
-224·9 [-292·0 - -157·7] |
-155·9 [-196·0 - -115·9] |
-338·3 [-447·3 - -229·2] |
-373·1 [-474·6 - -271·6] (p=0·0004) |
-150·7 [-185·6 - -115·7] |
-158·2 [-203·2 - -113·2] |
-214·4 [-306·7 - -122·1] |
-120·8 [-158·5 - -83·0] |
| c | 21 | 19 | 22 | 15 | 25 | 13 | 19 | 15 | 15 | 15 |
| Activation | ||||||||||
| V0·5act, mV | -21·1 [-24·0 - -18·1] |
-22·0 [-24·6 - -19·4] |
-19·5 [-20·9 - -18·1] |
-22·6 [-24·7 - -20·4] |
-20·9 [-22·9 - -18·8] |
-25·4 [-28·74- -22·1] |
-18·9 [-21·3 - -16·5] |
-19·1 [-21·1 - -17·1] |
-18·9 [-20·8 - -16·9] |
-19·9 [-22·0 - -17·9] |
| kact, mV | -6·24 [-6·82 - -5·65] |
-6·16 [-6·82 - -5·50] |
-6·45 [-6·79 - -6·11] |
-5·79 [-6·36 - -5·22] |
-5·78 [-6·42 - -5·14] |
-4·56 [-5·61 - -3·50] (p=0·04) |
-7·45 [-8·07 - -6·84] (p=0·01) |
-7·44 [-8·04 - -6·84] |
-6·86 [-7·36 - -6·36] |
-6·89 [-7·65 - -6·14] |
| C | 21 | 19 | 22 | 15 | 25 | 13 | 19 | 15 | 15 | 15 |
| Fast Inactivation | ||||||||||
| V0·5inact, mV | -67·8 [-69·6 - -66·1] |
-55·8 [-57·4 - -54·2] (p=0·0001) |
-64·4 [-65·6 - -63·2] (p=0·002) |
-56·6 [–57·4 - -55·8] |
-59·8 [-61·0 - -58·7] (p=0·0001) |
-51·1 [-52·7 - -49·4] (p=0·04) |
-66·8 [-68·2 - -65·4] (p=0·02) |
-68·1 [-69·1 - -67·1] (p=<0·0001) |
-64·6 [-66·1 - -63·2] |
-58·7 [-60·5 - -56·9] |
| kinact, mV | 4·58 [4·31 – 4·85] |
5·15 [4·87 – 5·43] (p=0·004) |
4·40 [4·19 – 4·61] |
5·27 [5·11 – 5·43] |
4·78 [4·64 – 4·93] |
5·47 [4·87 – 6·07] |
5·44 [5·16 – 5·72] (p=<0·0001) |
5·63 [5·19 – 6·06] |
4·51 [4·25 – 4·78] |
5·93 [5·70 – 6·17] (p=0·0003) |
| c | 21 | 16 | 22 | 15 | 25 | 10 | 19 | 12 | 15 | 11 |
| Slow Inactivation | ||||||||||
| V0·5inact, mV | -68·0 [-71·4 - -64·5] |
-61·3 [-64·4 - -58·1] (p=0·007) |
-59·9 [-62·8 - -57·0] (p=0·0005) |
-63·7 [-66·1 - -61·4] |
-58·6 [-61·0 - -56·3] |
-60·2 [-62·8 - -57·5] |
-60·7 [-63·1 - -58·2] |
-67·2 [-68·5 - -66·0] (p=0·002) |
-59·3 [-61·9 - -56·6] |
-62·1 [-65·0 - -59·1] |
| kinact, mV | 8·76 [8·08 – 9·44] |
6·69 [5·65 – 7·74] (p=0·0002) |
8·27 [7·82 – 8·73] |
6·16 [5·50 – 6·82] |
7·55 [7·14 – 7·95] |
6·74 [5·42 – 8·06] |
8·08 [7·26 – 8·91] |
9·76 [8·10 – 11·41] (p=0·005) |
8·76 [7·95 – 9·57] |
6·91 [6·26 – 7·55] |
| c | 15 | 11 | 15 | 10 | 16 | 10 | 10 | 10 | 10 | 10 |
| Time course of fast inactivation | ||||||||||
| τ at -10 mV, ms | 0·61 [0·54 – 0·69] |
0·97 [0·90 – 1·04] (p=<0·0001) |
0·60 [0·55 – 0·65] |
1·30 [1·22 – 1·38] (p=0·001) |
0·68 [0·64 – 0·73] (p=0·02) |
1·1 [0·99 – 1·22] |
0·59 [0·54 – 0·64] |
0·65 [0·58 – 0·72] (p=0·002) |
0·57 [0·53 – 0·60] |
0·95 [0·86 – 1·05] |
| c | 21 | 19 | 22 | 15 | 25 | 13 | 19 | 15 | 15 | 15 |
| Time course of recovery at100 mV | ||||||||||
| τ1, ms | 2·24 [1·71 – 2·78] |
2·71 [1·89 – 3·54] |
2·28 [1·53 – 3·03] |
2·29 [1·89 – 2·68] |
2·77 [2·31 – 3·23] |
3·24 [2·63 – 3·85] |
1·81 [1·33 – 2·30] |
3·33 [2·63 – 4·03] |
2·26 [1·74 – 2·79] |
2·52 [1·83 – 3·21] |
| τ2, ms | 8·12 [6·93 – 9·32] |
31·88 [21·86 – 41·91] (p=<0·0001) |
6·84 [5·15 – 8·54] |
8·05 [7·10 – 9·00] (p=<0·0001) |
7·64 [6·67 – 8·62] |
71·95 [31·50 – 112·40] |
6·88 [5·77 – 7·99] |
18·09 [16·73 – 19·44] |
5·94 [5·26 – 6·61] |
23·56 [12·94 – 34·19] |
| A2 | 0·57 [0·56 – 0·58] |
0·37 [0·33 – 0·41] (p=<0·0001) |
0·56 [0·55 – 0·57] |
0·41 [0·40 – 0·43] |
0·57 [0·56 – 0·59] |
0·53 [0·40 – 0·66] |
0·56 [0·54 – 0·58] |
0·40 [0·38 – 0·41] |
0·55 [0·53 – 0·57] |
0·35 [0·32 – 0·38] |
| c | 10 | 10 | 11 | 11 | 12 | 11 | 12 | 12 | 12 | 12 |
Data are presented as mean [CI], k: slope factor, c: cell number. The recovery of fast inactivation was fitted to a biexponential function and the amplitude of the first exponential function (A1) was set to 0·3. statistically significant differences between the two WTs and the corresponding channel alone or the variants and the corresponding WTs are shown in brackets.
The effect of FGF12A on NaV1.2 was markedly different from the effect on NaV1.6. Here, we did not find any change in current density, the activation or fast inactivation (Figure 2g and h, Table 2). Similar to NaV1.6 channels, FGF12A, when co-expressed with NaV1.2, showed an accelerated fast inactivation (NaV1.2:0·74 [0·65 – 0·83] ms; NaV1.2+FGF12A: 0·57 [0·49 – 0·65] ms; p=0·02) (Figure 2i, Table 2) and a slowing of the slow time constant (τ2) of the recovery from fast inactivation (NaV1.2: 9·34 [7·52 – 11·16] ms; NaV1.2+FGF12A: 17·68 [13·00 – 22·36] ms; p=0·0013) (Figure 2j, Table 2). For co-expression of FGF12B and NaV1.2, we found a hyperpolarizing shift of fast inactivation (NaV1.2:-54·9 [-56·4 – -53·3] mV; NaV1.2+FGF12B: -57·6 [-62·1 – -53·1] mV; p=0·0328) (Figure 2h, Table 2) as well as an accelerated fast inactivation (NaV1.2:0·74 [0·65 – 0·83] ms; NaV1.2+ FGF12B:0·53 [0·41 – 0·64] ms; p=0·003) (Figure 2i, Table 2), resulting in an overall loss-of-function (LOF). All other kinetic values were not statistically significantly altered.
Table 2.
Biophysical parameters of NaV1.2 with FGF12 variants.
| Beta | WTA | WTB | S8P | G50S | G112S | duplication 1S | duplication 1L | duplication 2S | duplication 2L | |
|---|---|---|---|---|---|---|---|---|---|---|
| Current density, pA/pF | -327·6 [-445·3 - -209·8] |
-394·1 [ [-521·6 - -266·5] |
-349·7 [-478·9 - -220·5] |
-239·9 [-333·7 - -146·1] |
-416·2 [-585·0 – 247·3] |
-257·2 [-369·2 – 145·3] |
-516·7 [-712·4 - -320·9] |
-310·1 [-503·4 - -116·8] |
-496·3 [-790·4 - -202·2] |
-278·7 [-437·1 - -120·3] |
| c | 13 | 13 | 11 | 11 | 12 | 11 | 12 | 11 | 12 | 12 |
| Activation | ||||||||||
| V0·5act, mV | -21·3 [-25·0 - -17·5] |
-23·0 [-26·8 - -19·2] |
-26·5 [-31·8 - -21·1] |
-23·0 [-26·1 - -19·9] |
-24·9 [-28·1 - -21·7] |
-21·4 [-24·9 - -17·9] |
-21·3 [-25·1 - -17·5] |
-21·1 [-28·0 - -14·2] |
-23·7 [-29·2 - -18·1] |
-20·5 [-24·9 - -16·1] |
| kact, mV | -6·8 [-7·9 - -5·7] |
-6·2 [-7·3 - -5·1] |
-5·1 [-6·7 - -3·5] |
-6·24 [-7·50 - -4·98] |
-6·05 [-7·21 - -4·90] |
-5·93 [-6·97 - -4·89] |
-6·16 [-7·06 - -5·27] |
-6·24 [-8·11 - -4·38] |
-5·34 [-7·03 - -3·64] |
-6·70 [-7·79 - -5·60] |
| c | 13 | 13 | 11 | 11 | 12 | 11 | 12 | 11 | 12 | 12 |
| Fast Inactivation | ||||||||||
| V0·5inact, mV | -54·9 [-56·4 - -53·3] |
-53·4 [-55·0 - -51·7] |
-57·6 [-62·1 - -53·1] (p=0·03) |
-51·7 [-53·1 - -50·3] |
-57·6 [-60·1 - -55·2] |
-48·3 [-50·5 - -46·0] (p=0·01) |
-58·2 [-59·7 - -56·6] |
-60·4 [-62·3 - -58·4] (p=0·009) |
-58·0 [-60·6 - -55·3] |
-53·3 [-54·9 – 51·8] |
| kinact, mV | 5·77 [5·48 – 6·07] |
5·43 [5·26 – 5·60] |
5·20 [4·92 – 5·47] (p=0·003) |
5·54 [5·23 – 5·86] |
5·74 [5·50 – 5·97] (p=0·03) |
5·51 [5·15 – 5·86] |
6·79 [6·29 – 7·29] (p=<0·0001) |
6·60 [6·01 – 7·18] (p=0·0001) |
5·24 [5·00 – 5·47] |
6·15 [5·87 – 6·43] (p=0·006) |
| c | 13 | 13 | 11 | 10 | 13 | 11 | 11 | 11 | 12 | 12 |
| Slow Inactivation | ||||||||||
| V0·5inact, mV | -61·6 [-66·2 - -56·9] |
-60·8 [-63·1 - -58·5] |
-59·4 [-64·3 - -54·6] |
-60·39 [-63·3 - -57·4] |
-61·7 [-64·8 - -58·6] |
-57·0 [-59·7 - -54·3] |
-57·5 [-59·6 - -55·3] |
-61·2 [-64·5 – 57·9] |
-57·5 [-59·9 - -55·1] |
-56·7 [-59·1 - -54·2] |
| kinact, mV | 6·29 [4·99 – 7·59] |
5·40 [4·64 – 6·17] |
6·04 [4·72 – 7·37] |
5·61 [4·70 – 6·52] |
5·81 [4·93 – 6·70] |
5·17 [4·63 – 5·71] |
6·65 [5·78 – 7·51] |
6·55 [6·03 – 7·07] (p=0·04) |
6·55 [5·87 – 7·23] |
5·40 [4·90 – 5·90] |
| c | 10 | 10 | 10 | 11 | 12 | 11 | 10 | 10 | 10 | 10 |
| Time course of fast inactivation | ||||||||||
| τ at -10 mV, ms | 0·74 [0·65 – 0·83] |
0·57 [0·49 – 0·65] (p=0·02) |
0·53 [0·41 – 0·64] (p=0·003) |
0·72 [0·61 – 0·82] |
0·61 [0·54 – 0·67] |
1·01 [0·79 – 1·24] (p=0·009) |
0·64 [0·54 – 0·74] |
0·61 [0·54 – 0·68] |
0·53 [0·45 – 0·61] |
0·68 [0·59 – 0·78] |
| c | 13 | 13 | 11 | 11 | 12 | 11 | 12 | 11 | 12 | 12 |
| Time course of recovery at 100 mV | ||||||||||
| τ1, ms | 2·33 [1·66 – 3·00] |
2·00 [1·33 – 2·60] |
1·90 [1·48 – 2·32] |
2·22 [1·68 – 2·76] |
2·73 [2·21 – 3·24] |
2·81 [2·25 – 3·36] |
2·39 [1·74 – 3·03] |
2·79 [2·27 – 3·32] |
2·16 [1·47 – 2·86] |
2·60 [2·03 – 3·16] |
| τ2, ms | 9·34 [7·52 – 11·16] |
17·68 [13·00 - 22·36] (p=0·001) |
8·06 [6·46 – 9·66] |
10·40 [7·87 – 12·93] (p=0·001) |
10·07 [8·57 – 11·58] |
22·80 [13·63 – 31·96] |
9·22 [7·55 – 10·90] |
13·86 [10·98 – 16·74] |
8·51 [6·84 – 10·19] |
14·02 [11·23 – 16·81] |
| A2 | 0·44 [0·41 – 0·47] |
0·30 [0·28 – 0·33] (p=<0·0001) |
0·53 [0·48 – 0·58] (p=0·003) |
0·39 [0·35 – 0·43] (p=0·003) |
0·52 [0·49 – 0·54] |
0·28 [0·26 – 0·31] |
0·53 [0·51 – 0·54] |
0·40 [0·37 – 0·44] (p=0·001) |
0·53 [0·51 – 0·56] |
0·34 [0·30 – 0·38] |
| c | 12 | 11 | 10 | 10 | 12 | 10 | 11 | 10 | 11 | 10 |
Data are presented as mean [CI], k: slope factor, c: cell number. The recovery of fast inactivation was fitted to a biexponential function and the amplitude of the first exponential function (A1) to 0·3. Statistically significant differences between the two WTs and the corresponding channel alone or the variants and the corresponding WTs are shown in brackets.
Functional characterization of FGF12 variants on NaV1.6 and NaV1.2 channels
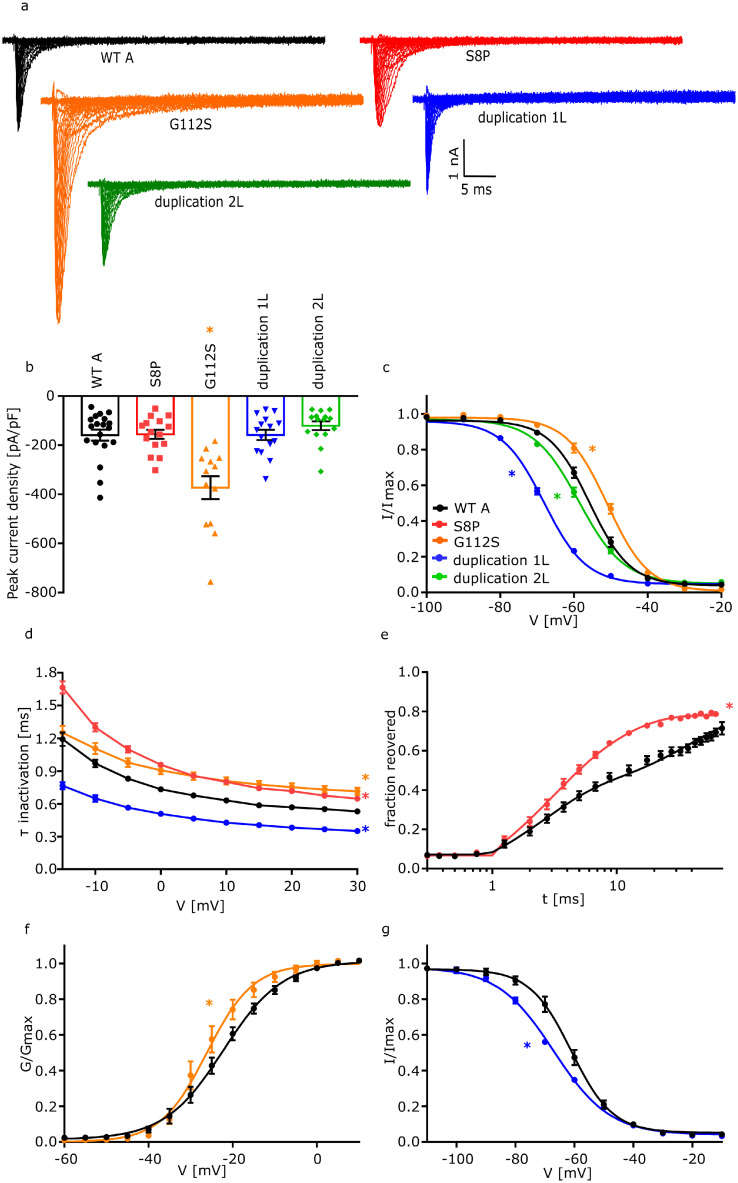
Here, we describe the functional effects caused by FGF12 variants by co-expression of the different variants with either NaV1.2 or NaV1.6 channels, comparing them to the corresponding WT recordings. The missense variant p.(Ser8Pro) (S8P) is located at the N-terminal part of FGF12A. Since the only difference between FGF12 WTs is the different length of the N-terminal part, the variant S8P can only be found in FGF12 WTA (Figure 1a). The co-expression of S8P with NaV1.6 showed no effect on current density, activation, fast inactivation or slow inactivation (Figure 3a and b, Table 1), but slowed fast inactivation (NaV1.6+WTA: 0·97 [0·90 – 1·04] ms; NaV1.6+S8P: 1·30 [1·22 – 1·38] ms; p=0·001) and accelerated the slow component (τ2) of the recovery from fast inactivation (NaV1.6+WTA: 31·88 [21·86 – 41·91] ms; NaV1.6+S8P: 8·05 [7·10 – 9·00] ms; p=<0·0001) (Figure 3d and e, Table 1) compared to the coexpression of NaV1.6 with FGF12A. Expression of S8P with NaV1.2 demonstrated a similar effect as in NaV1.6. Here, no changes in current density, activation, fast inactivation or slow inactivation were observed, whereas recovery from fast inactivation was accelerated (NaV1.2+WTA: 17·68 [13·00 – 22·36] ms; NaV1.2+S8P: 10·40 [7·87 – 12·93] ms; p=0·0135) (Figure 4 a, b, c and d, Table 2). Thus, S8P showed a GOF effect on both NaV1.6 and NaV1.2 channels.
Figure 3.
Electrophysiological analysis of FGF12 variants on NaV1.6. Functional analysis of the effect of two FGF12 missense variants (S8P and G112S) and two FGF12 CNVs (duplication 1L and duplication 2L) on NaV1.6 compared to WTA. Statistically significant effects for each condition are indicated by coloured asterisks.
a. Representative traces of NaV1.6 currents in ND7/23 cells expressing NaV1.6 with FGF12 WTA or the FGF12 variants, respectively, in response to voltage steps from -80 to +35 mV in 5 mV steps.
b. Mean Current amplitudes of analysed ND7/23 cells injected with NaV1.6 with WTA (n=19; 8 transfections), S8P (n=15; 4 transfections; p = >0·999), G112S (n=13; 5 transfections; p = 0·0004), duplication 1L (n=15; 7 transfections; p = >0·999) or duplication 2L (n=15; 6 transfections; p = 0·999). Expression of NaV1.6 together with G112S showed a statistically significantly elevated current density compared to NaV1.6 with WTA. For the statistical analysis ANOVA on ranks with Dunn´s post hoc test was used.
c. Mean voltage-dependent fast inactivation of NaV1.6 with FGF12 WTA (n=16; 6 transfections), G112S (n=10; 3 transfections; p (V1/2) = 0·04), duplication 1L (n=12; 5 transfections; p (V1/2) = <0·0001) or duplication 2L (n=11; 5 transfections; p (k) = 0·0003). Lines illustrate Boltzmann Function fit to the data points. G112S showed a statistically significant shift to more depolarized potentials in comparison to WTA while both Duplications show a hyperpolarizing shift in comparison to WTA. For the statistical analysis ANOVA on ranks with Dunn´s post hoc test was used. All data are shown as means ± SEM.
d. Mean voltage-dependent fast inactivation time constant of NaV1.6 FGF12 WTA (n=19; 8 transfections), S8P (n=15; 4 transfections; p = 0·001), G112S (n=13; 5 transfections; p = 0·05) or duplication 1L (n=15; 7 transfections; p = 0·002). S8P and G112S show a statistically significantly slowed fast inactivation in comparison to WTA while duplication 1L shows an accelerated fast inactivation. For the statistical analysis ANOVA on ranks with Dunn´s post hoc test was used. All data are shown as means ± SEM.
e. Time course of recovery from fast inactivation of NaV1.6 with FGF12 WTA (n=10; 8 transfections) or FGF12 S8P (n=11; 4 transfections; p (τ2) = <0·0001) determined at -100 mV. The variant S8P leads to a statistically significantly accelerated recovery of fast inactivation in comparison to WTA. Lines represent fits of biexponential functions yielding the time constants τ1 and τ2. A1 was set to 0·3. For the statistical analysis ANOVA on ranks with Dunn´s post hoc test was used.
f. Mean voltage-dependent activation of NaV1.6 with FGF12 WTA (n=19; 8 transfections) or G112S (n=13; 5 transfections; p (k) = 0·04). G112S shows a hyperpolarizing shift in comparison to WTA. Lines illustrate Boltzmann Function fit to the data points. For the statistical analysis ANOVA on ranks with Dunn´s post hoc test was used.
g. Mean voltage-dependent slow inactivation of NaV1.6 with FGF12 WTA (n=11; 8 transfections) or FGF12 duplication 1L (n=10; 5 transfections; p (V1/2) = 0·002). Lines illustrate Boltzmann Function fit to the data points. FGF12 duplication 1L shows a shift to more hyperpolarized potentials in comparison to NaV1.6 with FGF12 WTA. For the statistical analysis of the slow inactivation one-way ANOVA with Holm-Sidak´s post hoc test was used. All data are shown as means ± SEM. All values of electrophysiological results, numbers and p-values are listed in Table 1 and are shown as means with the 95% confidence interval.
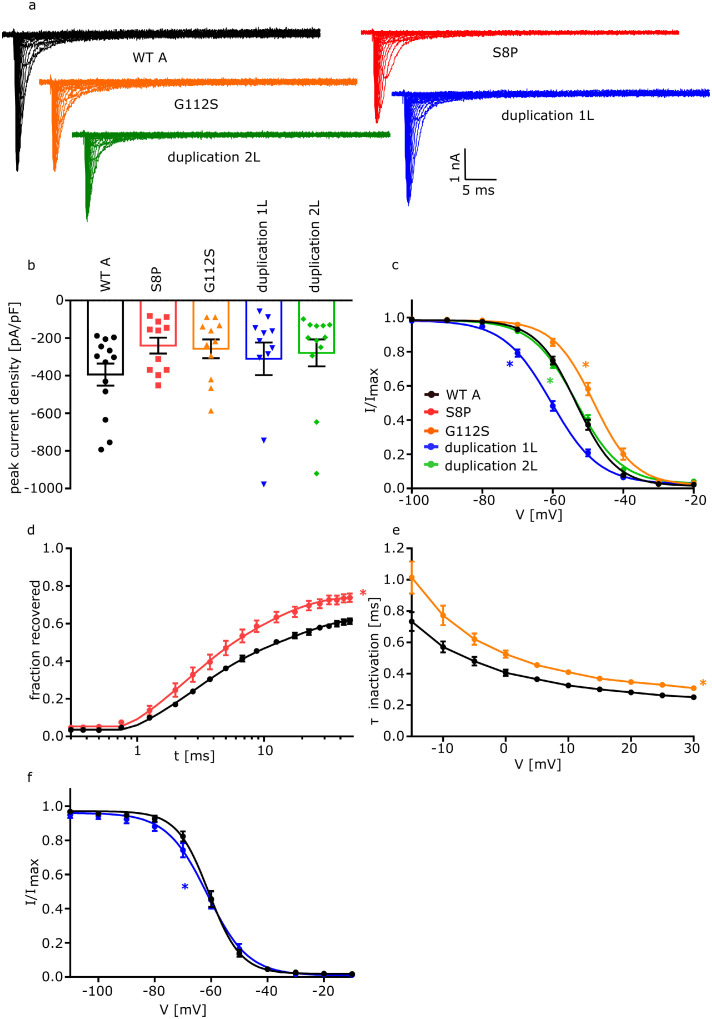
Figure 4.
Electrophysiological analysis of FGF12 variants on NaV1.2. Functional analysis of the effect of 2 FGF12 missense variants (S8P and G112S) and two FGF12 CNVs (duplication 1L and duplication 2L) on NaV1.2 compared to WTA. Statistically significant effects for each condition are indicated by coloured asterisks.
a. Representative traces of NaV1.2 currents in ND7/23 cells expressing NaV1.2 with FGF12 WTA or the FGF12 variants, respectively, in response to voltage steps from -80 to +35 mV in 5 mV steps.
b. Mean Current amplitudes of analysed ND7/23 cells injected with NaV1.2 with WTA (n=13; 5 transfections), S8P (n=11; 6 transfections; p = 0·2), G112S (n=11; 4 transfections; p = 0·3), duplication 1L (n=11; 4 transfections; p = 0·4) or duplication 2L (n=12; 6 transfections; p = 0·1). None of the variants lead to a change in the current density of NaV1.2. For the statistical analysis ANOVA on ranks with Dunn´s post hoc test was used.
c. Mean voltage-dependent fast inactivation of NaV1.2 with FGF12 WTA (n=13; 5 transfections), G112S (n=11; 4 transfections; p (V1/2) = 0·01), duplication 1L (n=11; 4 transfections; p (V1/2) = 0·009) or duplication 2L (n=12; 6 transfections; p (k) = 0·006). Lines illustrate Boltzmann Function fit to the data points. G112S showed a statistically significant shift to more depolarized potentials in comparison to WTA while duplication 1L shows a hyperpolarizing shift in comparison to WTA and duplication 2L changes the slope of the fast inactivation. All data are shown as means ± SEM. For the statistical analysis ANOVA on ranks with Dunn´s post hoc test was used.
d. Time course of recovery from fast inactivation of NaV1.2 with FGF12 WTA (n=11; 5 transfections) or FGF12 S8P (n=10; 4 transfections; p (τ2) = 0·01) determined at -100 mV. The variant S8P leads to a statistically significantly accelerated recovery of fast inactivation in comparison to WTA. Lines represent fits of biexponential functions yielding the time constants τ1 and τ2. A1 was set to 0·3. For the statistical analysis ANOVA on ranks with Dunn´s post hoc test was used.
e. Mean voltage-dependent fast inactivation time constant of NaV1.2 FGF12 WTA (n=11; 5 transfections) or G112S (n=11; 4 transfections; p = 0·009). G112S show a statistically significantly slowed fast inactivation in comparison to WTA. All data are shown as means ± SEM. For the statistical analysis one-way ANOVA with Holm-Sidak´s post hoc test was used.
f. Mean voltage-dependent slow inactivation of NaV1.2 with FGF12 WTA (n=10; 5 transfections) or FGF12 duplication 1L (n=10; 4 transfections; p (k)= 0·04). Lines illustrate Boltzmann Function fit to the data points. FGF12 duplication 1L shows a change in the slope of the slow inactivation in comparison to NaV1.2 with FGF12 WTA. All data are shown as means ± SEM. All values of electrophysiological results, numbers and p-values are listed in Table 2 and are shown as means with the 95% confidence interval. For the statistical analysis of the slow inactivation one-way ANOVA with Holm-Sidak´s post hoc test was used.
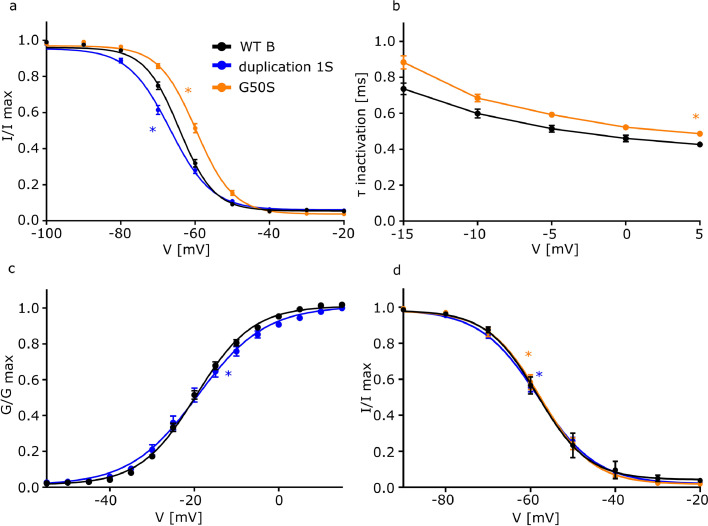
The second missense variant p.(Gly50Ser) (G50S) corresponds to WTB and is the same variant as p.(Gly112Ser) (G112S) which corresponds to WTA. This variant is located directly in the interaction interface between FGF12 and NaV1.6/NaV1.2 (Figure 1a). Co-expression of G112S with NaV1.6 led to an increased current density compared to WTA (NaV1.6+WTA: -158·9 [-206·5 – -111·3] pA/pF; NaV1.6+G112S: -373·1 [-474·6 – -271·6] pA/pF; p=0·0004) (Figure 3a and b, Table 1). Additionally, voltage-dependence of fast inactivation was shifted to more depolarized potentials (NaV1.6+WTA: -55·8 [-57·4 – -54·2] mV; NaV1.6+G112S: -51·1 [-52·7 – -49·4] mV; p=0·04), and fast inactivation was slowed (Figure 3c and d, Table 1). Furthermore, G112S showed a shift in the slope factor of the channel's voltage dependence of activation (NaV1.6+WTA: -6·16 [-6·82 – -5·50] mV; NaV1.6+G112S: -4·56 [-5·61 – -3·50] mV; p=0·04), indicating a GOF effect when co-expressed with NaV1.6 (Figure 3f, Table 1). Just the recovery from fast inactivation as well as the slow inactivation were not affected. Co-expression of G50S together with NaV1.6 however only showed changes in fast inactivation, with a depolarizing shift of the voltage-dependence (NaV1.6+WTB: -64·4 [-65·6 – -63·2] mV; NaV1.6+G50S: -59·8 [-61·0 – -58·7] mV; p=0·0001) and a slowing of fast inactivation (NaV1.6+WTB: 0·60 [0·55 – 0·65] ms; NaV1.6+G50S: 0·68 [0·64 – 0·73] ms; p=0·02) (Figure 5a and b, Table 1) compared to WTB. A similar trend could be observed for G112S when expressed with the NaV1.2 channel. Again, a depolarizing shift of the voltage-dependence of fast inactivation (NaV1.2+WTA: -53·4 [-55·0 – -51·7] mV; NaV1.2+G112S: -48·3 [-50·5 – -46·0] mV; p=0·01) and an additional slowing of fast inactivation (NaV1.2+WTA: 0·57 [0·49 – 0·65] ms; NaV1.2+G112S: 1·01 [0·79 – 1·24] ms; p=0·009) led to a GOF of NaV1.2, comparable to a co-expression with the NaV1.6 channel (Figure 4c and e, Table 2). We noted a minor effect for the co-expression of G50S together with NaV1.2, with a change in just the slope factor of fast inactivation (NaV1.2+WTB: 5·20 [4·92 – 5·47] mV; NaV1.2+G50S: 5·74 [5·50 – 5·97] mV; p=0·03) (Figure 5d, Table 2).
Figure 5.
Electrophysiological analysis of FGF12 variants (NM_004113) on NaV1.6 or NaV1.2. Functional analysis of the effect of one FGF12 missense variant (G50S) and one FGF12 CNV (duplication 1S) on NaV1.6 and NaV1.2 compared to WTB. Statistically significant effects for each condition are indicated by coloured asterisks.
a. Mean voltage-dependent fast inactivation of NaV1.6 with FGF12 WTB (n=22; 16 transfections), G50S (n=25; 13 transfections; p (V1/2) = 0·0001) or duplication 1S (n=19; 9 transfections; p (V1/2) = 0·02). Lines illustrate Boltzmann Function fit to the data points. G50S showed a statistically significant shift to more depolarized potentials in comparison to WTB while duplication 1S showed a hyperpolarizing shift in comparison to WTB. For the statistical analysis one-way ANOVA with Holm-Sidak´s post hoc test was used. All data are shown as means ± SEM.
b. Mean voltage-dependent fast inactivation time constant of NaV1.6 FGF12 WTB (n=22; 16 transfections) or G50S (n=25; 13 transfections; p = 0·02). G50S showed a statistically significantly slowed fast inactivation in comparison to WTB. For the statistical analysis one-way ANOVA with Holm-Sidak´s post hoc test was used. All data are shown as means ± SEM.
c. Mean voltage-dependent activation of NaV1.6 with FGF12 WTB (n=22; 16 transfections) or duplication 1S (n=19; 9 transfections; p (k) = 0·01). Duplication 1S shows a change in the slope of the activation curve in comparison to WTB. Lines illustrate Boltzmann Function fit to the data points. For the statistical analysis ANOVA on ranks with Dunn´s post hoc test was used.
d. Mean voltage-dependent fast inactivation of NaV1.2 with FGF12 WTB (n=11; 3 transfections), G50S (n=12; 6 transfections; p (k) = 0·03) or duplication 1S (n=11; 4 transfections; p (k) = <0·0001). Lines illustrate Boltzmann Function fit to the data points. Both variants show a statistically significant change in the slope of the fast inactivation in comparison to WTB. For the statistical analysis ANOVA on ranks with Dunn´s post hoc test was used. All data are shown as means ± SEM.
Interestingly, both duplication cases showed a completely different effect on NaV1.6 and NaV1.2 channels, compared to the effects of the missense variants detailed above. Since we used both transcript forms to analyse the different missense variants, we also used both forms to analyse the duplications. WTA without the last exon followed by exon 2, 3, 4 and 5 is subsequently referred to as duplication 1L, while WTB without the last exon followed by Exon 3, 4, 5 and 6 will be referred to as duplication 1S (Figure 1a and b). For duplication 2 we used WTA followed by WTB as duplication 2L, and WTB followed by WTB as duplication 2S (Figure 1a and c).
When co-expressing duplication 1L with NaV1.6, the voltage-dependence of fast inactivation was shifted to more hyperpolarized potentials (NaV1.6+WTA: -55·8 [-57·4 – -54·2] mV; NaV1.6+duplication1L: -68·10 [-69·1 – -67·1] mV; p=<0·001), the time constant (τ) of fast inactivation was decreased (NaV1.6+WTA: 0·97 [0·90 – 1·04] ms; NaV1.6+duplication1L: 0·65 [0·58 – 0·72] ms; p=0·002) and the voltage-dependence of slow inactivation was shifted to more hyperpolarized potentials (NaV1.6+WTA: -61·3 [-64·4 – -58·1] mV; NaV1.6+duplication1L: -67·2 [-68·5 – -66·0] mV; p=0·002) when compared to WTA conditions (Figure 3c, d and g, Table 1), indicating a clear LOF. Neither the activation nor the recovery from inactivation were statistically significantly altered. Co-expression of duplication 1S with NaV1.6 showed a hyperpolarizing shift of the fast inactivation curve (NaV1.6+WTB: -64·4 [-65·6 – -63·2] mV; NaV1.6+duplication1S: -66·8 [-68·2 – -65·4] mV; p=0·02) and a change in the slope factor of the activation (NaV1.6+WTB: -6·45 [-6·79 – -6·11] mV; NaV1.6+duplication1S: -7·45 [-8·07 – -6·84] mV; p=0·01), leading to a LOF (Figure 5a, c, Table 1). The co-expression of duplication1L with NaV1.2 showed similar effects as with NaV1.6, only without a change of the τ of fast inactivation (Figure 4c and f, Table 2). A similar trend was observed for the co-expression of duplication 1S with NaV1.2, which leads to a change in the slope factor in the fast inactivation curve (NaV1.2+WTB: 5·20 [4·92 – 5·47] mV; NaV1.2+duplication1S: 6·79 [6·29 – 7·29] mV; p=<0·0001) (Figure 5d). In summary, the expression of duplication 1 consistently leads to a LOF in both NaV1.6 and NaV1.2 channels.
The last variant, duplication 2L, only showed a change of the slope factor of the voltage-dependence of fast inactivation (NaV1.6+WTA: -5·15 [4·87 – 5·43] mV; NaV1.6 + duplication2L: -5·93 [5·70 – 6·17] mV; p=0·0003) when co-expressed with the NaV1.6 channel (Figure 3c, Table 1), while the co-expression of duplication 2S with NaV1.6 channel did not result in any effect on ion channel function. A similar trend could be observed for co-expression with NaV1.2. Here, duplication 2L only caused a minor change in the slope of the fast inactivation curve (NaV1.2+WTA: 5·43 [5·26 – 5·60] mV; NaV1.2+duplication2L: 6·15 [5·87 – 6·43] mV; p=0·006), while duplication 2S had no effect (Figure 4c, Table 2). Thus, duplication 2 gives rise to a LOF effect on both NaV1.6 and NaV1.2.
Discussion
FGF12 had previously been suspected to be an important regulator of neuronal activity through modulation of different sodium channel subtypes.5 Here, we carried out a detailed analysis of the interaction of FGF12 with Nav1.2 and Nav1.6, which are known to be epilepsy-associated channels, and compared the physiological state with changes in ion channel function induced by missense and copy-number variants in FGF12.
Our functional analysis of WT FGF12 confirmed the gain-of-function (GOF) effect on NaV1.6 and the loss-of-function (LOF) effect on the voltage-dependent fast inactivation of NaV1.2, both of which have been described previously.9,12 Furthermore, we characterized the differential effects of WT FGF12 on both channels in detail, providing evidence that FGF12 not just affects the voltage-dependence, but also the time constants of the fast inactivation of both channels. For NaV1.6, an additional effect on the voltage dependence of slow inactivation was evident.
Physiologically, FGF12 is expressed in two different transcripts resulting from alternative splicing, which differ only in their N-terminal part. Our results demonstrated that the expression of WTA, with its longer N-terminal part, has a stronger effect on the fast inactivation of NaV1.6 than WTB with its shorter N-terminal part, which is in line with previous studies.9 Additionally, the long N-terminal part of FGF12 WTA seems to affect the time constants of the fast inactivation, since WTB did not alter the time constants of fast inactivation. A similar finding was already reported in a previous study.24 Furthermore, it is interesting that the recovery of fast inactivation is biphasic and just the slow component of the recovery of inactivation (τ2) was statistical significantly changed. Similar findings were already published for FGF13 WTA and FGF14 WTA, homologs of FGF12 WTA. 7,8,15 For both of them a much slower recovery from inactivation in comparison to the according WTB was shown. FGF13 WTA for example needs about 1000 ms to recover completely.15 Thus, WTB caused a clear GOF of NaV1.6 channels, while WTA resulted in mixed GOF and LOF changes. The effect of the FGF12 WTs on NaV1.2 was less pronounced than for NaV1.6, but still reduced channel function.
Previously, missense variants or copy number variants (CNVs) in FG12 have been solely identified in patients with DEE.25 Here we describe two additional patients, one with the recurrent variant G50S/G112S and a similar epileptic phenotype as described earlier,25 and one patient with the novel variant S8P showing a completely novel phenotype consisting of autism spectrum disorder and developmental delay without seizures.
For sodium channels, like NaV1.6 and NaV1.2, it has already been shown that disease-causing variants can be associated with intellectual disability with or without seizures, depending primarily on the resulting kinetic changes within the channel.3,26 LOF variants in NaV1.6 have mainly been associated with intellectual disability, while GOF variants lead to an epileptic phenotype. However, variants have also recently been described as LOF causing epileptic seizures.27 For NaV1.2, a similar genotype-phenotype correlation has been observed, with the difference that NaV1.2 LOF caused by missense variants can also cause severe late onset epilepsy phenotypes, while truncating variants (which by definition cause complete LOF) are associated with ASD.28 In general, the FGF12 phenotype described here is dominated by a severe, early onset DEE spectrum disease and isolated ASD. Since the ASD patient was only 4.5 years old at recruitment, we cannot exclude that the patient could get late onset seizures. Compared to other phenotypes associated to sodium channel genes like SCN2A28 or SCN8A29 the SCN2A-phenotype is far more variable including a very benign neonatal/infantile onset epilepsy in 15% of cases up to severe DEE including ASD and a very severe DEE in most SCN8A cases.
Interestingly, the functional analysis of variant S8P, which was found in a patient with ASD, caused GOF changes in NaV1.6 and NaV1.2 channels, mainly by affecting the time constant of fast inactivation components of both channels. These changes lead to a stronger deceleration of fast inactivation of NaV1.6 and reduced the regulating effect of WTA on the recovery from fast inactivation especially on the slow component (τ2). This implies that the mutation partially impairs the stability of the slow recovering component of the channel. Additionally, we could show that the effect on the recovery from fast inactivation of FGF12 WTA on NaV1.2 was reduced as well, which results in a GOF for both channels. This finding is similar to the recently identified effects for FGF13 variants also showing a faster recovery from inactivation. In contrast to our findings for the FGF12 variant S8P, the FGF13 variants additionally caused a depolarizing shift in the voltage dependent steady state inactivation.15 The phenotype of the patients harbouring a FGF13 variant showed a similar phenotype as the FGF12 patients with the missense variants G50S/G112S and R52H/R114H.15 In contrast the patient carrying the FGF12 variant S8P showed ASD without seizures. This is an unexpected finding, since ASD has so far primarily been associated with LOF of NaV1.6 or NaV1.2 channels but measurements in other cell systems may provide more electrophysiological details.3,28,30
For the second missense variant G50S/G112S (WTB/WTA variants), only two patients have been described before. Here, we identified an additional patient with this variant showing a similar age of onset at about three months of age, and phenotypic features including generalized and focal seizures as well as moderate intellectual disability. This variant seems to influence NaV1.6 in both forms in a similar way leading to a 5-mV depolarizing shift in the voltage-dependence of fast inactivation as well as to a decelerated fast inactivation. The same effect is visible for G112S on NaV1.2 while G50S shows just a small effect on the voltage-dependence of fast inactivation on NaV1.2 but does not influence the time constant of fast inactivation. In summary, the gain-of function effect of the variant G50S/G112S on NaV1.6 on the voltage-dependence of fast inactivation was similar to the effect of the already described variant R52H/R114H on NaV1.69, which is located just two amino acids apart in the interaction interface of FGF12 to the NaVs. The similar effect on the voltage-dependence of fast inactivation and the very close localization of these two variants suggests that both mutated FGF12 proteins may potentially interfere with channel function through a similar mechanism.
Analysis of the effect of CNVs on NaV1.6 and NaV1.2 revealed a key difference when compared to the effect of the missense variants R52H/R114H and G50S/G112S. Both CNVs primarily affected the channels’ fast inactivation, leading to a clear LOF of both channels. One possible explanation may be that the duplication 1L partially lost the ability to interact with NaV1.6, since the inactivation kinetics were similar to that of NaV1.6 expressed without FGF12. Only the effect on the peak current density and the recovery from fast inactivation were comparable to co-expressed WTA and NaV1.6. In contrast, duplication 1L seems to negatively regulate NaV1.2 since fast inactivation, which is not changed by WTA, showed a hyperpolarizing shift during co-expression with duplication 1L. This interaction also appeared to affect the slow inactivation, which was unchanged by wildtype co-expression. Further experiments are needed to analyse if the interaction between the CNVs and the channels is weakened or changed in another way, since some of the kinetic parameters which are changed by WTA are not changed anymore by the CNVs.
Interestingly, both duplications caused a similar phenotype regarding seizure types, age of onset and moderate to severe developmental delay/intellectual disability. This overlaps with the symptomatic spectrum associated with the missense variants R52H/R114H and G50S/G112S. The only difference between the point mutations and the CNVs was the age of onset, with later onset in the duplication cases at a median age of onset of 15·5 months (n = 4).19,23 This finding is in line with previous studies, where NaV1.2 LOF variants were shown to be associated with a later disease onset.28
In summary, the missense variant G50S/G112S led to a clear GOF effect on both channels, while both duplications were associated with LOF effects. The phenotype of the patients with these variants showed overlap to the established genotype-phenotype spectrum of GOF and LOF variants in SCN2A, but patients with LOF variants are known to show severe DEE with a later age of onset.28 Thus, no clear genotype-phenotype correlation was visible for these variants. But since the cohort is small further patients could be helpful to establish a genotype-phenotype correlation. Surprisingly, the variant S8P led to a GOF effect on both channels, which is a functional effect that has not been previously described for ASD associated with variants in sodium channel genes.
In a previous study, 6 out of 17 patients carrying one of the already published FGF12 variants showed a partial response to phenytoin or lamotrigine, sodium channel blockers that enhance the channels’ fast inactivation by shifting voltage-dependence towards more hyperpolarized potentials.25 However, not all patients responded well to phenytoin, suggesting that these variants may have an additional effect which could not be identified until now. This hypothesis is supported by the FGF12 mouse variant model fgf12R52H/+ which suffers from spontaneous seizures and dies of sudden unexplained death in epilepsy (SUDEP) at around 16 days of age.31 Veliskova and co-workers treated these mice with phenytoin, but the survival time was just marginally prolonged to about 19 days.31 Another previous study showed the modifying effect of a hypomorphic variant in GABRA2 which increased seizure severity in mouse models carrying human SCN8A variants. This suggests that GABRA2 could be a therapeutic target. 32
To the best of our knowledge, this is the first time a detailed biophysical and physiological modification of the NaV1.2 and NaV1.6 channel function by wildtype FGF12 has been investigated. We found a divergent effect on the sodium channel function, with a trend towards GOF in NaV1.6 and a trend towards LOF in NaV1.2. Secondly, we described the underlying pathomechanism of four FGF12 missense and copy-number variants leading to complex gain- or loss-of function changes in NaV1.6 and NaV1.2. Lastly, we identified two new patients - one with DEE and one with ASD - expanding the phenotypic spectrum of FGF12-associated disorders to include ASD, which has not been previously described. Thus, FGF12 seems to be an important modulator of neuronal activity driven by sodium channel subtypes and confirms its role as an epilepsy gene with an overlap to intellectual disability and autism spectrum disorder.
Contributors
SSe performed genetic and functional analysis and wrote the manuscript together with YW.
JRL, SSy, MP, TB and AvdV performed detailed clinical evaluation and further genetic analysis and edited the manuscript.
NS helped to conceptualize the used constructs for the electrophysiological measurements and edited the manuscript.
HG prepared cDNA samples from venous blood from the patients and edited the manuscript.
MW prepared fibroblasts from a skin biopsy of the patient and edited the manuscript.
UBSH re-analysed the functional data and edited the manuscript.
IH planned the study together with YW, supervised the analysis and edited the manuscript.
CMB reviewed the results and edited the manuscript.
YW planned and coordinated the study, supervised the analysis, wrote, and edited the manuscript together with SSe.
SSe and UBSH analysed and verified the data.
All authors have read and approved the manuscript.
Data sharing statement
Anonymized data not published within this article will be made available by request from any qualified investigator.
Declaration of interests
None of the authors has any competing interests.
Acknowledgements
Y.W. was supported by the DFG/FNR INTER Research Unit FOR2715 of the German Research Foundation (We4896/4-1 and WE4896/4-2) and by the BMBF Treat-ION grant (01GM1907).
I.H. received support through the German Research Foundation (HE5415/6-1) and the DFG/FNR INTER Research Unit FOR2715 (He5415/7-1 and He5415/7-2). I.H. was supported by The Hartwell Foundation through an Individual Biomedical Research Award. This work was also supported by the National Institute for Neurological Disorders and Stroke (K02 NS112600), including support through the Centre Without Walls on Ion channel function in epilepsy (“Channelopathy-associated Research Centre”, U54 NS108874), the Eunice Kennedy Shriver National Institute of Child Health and Human Development through the Intellectual and Developmental Disabilities Research Centre (IDDRC) at Children's Hospital of Philadelphia and the University of Pennsylvania (U54 HD086984), and by intramural funds of the Children's Hospital of Philadelphia through the Epilepsy NeuroGenetics Initiative (ENGIN). Research reported in this publication was also supported by the National Centre for Advancing Translational Sciences of the National Institutes of Health under Award Number UL1TR001878. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This project was also supported in part by the Institute for Translational Medicine and Therapeutics’ (ITMAT) Transdisciplinary Program in Translational Medicine and Therapeutics at the Perelman School of Medicine of the University of Pennsylvania. I.H. also received support through the International League Against Epilepsy (ILAE). The study also received support through the EuroEPINOMICS-Rare Epilepsy Syndrome (RES) Consortium and by the Genomics Research and Innovation Network (GRIN, grinnetwork.org).
UBSH received support from the DFG/FNR INTER Research Unit FOR2715 of the German Research Foundation (HE8155/1-1, HE8155/1-2) and by the BMBF Treat-Ion grant (01GM1907G).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104234.
Appendix. Supplementary materials
References
- 1.Catterall W.A. Voltage-gated sodium channels at 60: structure, function and pathophysiology: Voltage-gated sodium channels. J Physiol. 2012;590:2577–2589. doi: 10.1113/jphysiol.2011.224204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brunklaus A., Ellis R., Reavey E., Semsarian C., Zuberi S.M. Genotype phenotype associations across the voltage-gated sodium channel family. J Med Genet. 2014;51:650–658. doi: 10.1136/jmedgenet-2014-102608. [DOI] [PubMed] [Google Scholar]
- 3.Liu Y., Schubert J., Sonnenberg L., et al. Neuronal mechanisms of mutations in SCN8A causing epilepsy or intellectual disability. Brain J Neurol. 2019;142:376–390. doi: 10.1093/brain/awy326. [DOI] [PubMed] [Google Scholar]
- 4.Zhang S., Zhang Z., Shen Y., et al. SCN9A epileptic encephalopathy mutations display a gain-of-function phenotype and distinct sensitivity to oxcarbazepine. Neurosci Bull. 2020;36:11–24. doi: 10.1007/s12264-019-00413-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldfarb M., Schoorlemmer J., Williams A., et al. Fibroblast growth factor homologous factors control neuronal excitability through modulation of voltage-gated sodium channels. Neuron. 2007;55:449–463. doi: 10.1016/j.neuron.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smallwood P.M., Munoz-Sanjuan I., Tong P., et al. Fibroblast growth factor (FGF) homologous factors: new members of the FGF family implicated in nervous system development. Proc Natl Acad Sci. 1996;93:9850–9857. doi: 10.1073/pnas.93.18.9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laezza F., Lampert A., Kozel M.A., et al. FGF14 N-terminal splice variants differentially modulate Nav1.2 and Nav1.6-encoded sodium channels. Mol Cell Neurosci. 2009;42:90–101. doi: 10.1016/j.mcn.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang J., Wang Z., Sinden D.S., et al. FGF13 modulates the gating properties of the cardiac sodium channel Nav 1.5 in an isoform-specific manner. Channels. 2016;10:410–420. doi: 10.1080/19336950.2016.1190055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siekierska A., Isrie M., Liu Y., et al. Gain-of-function FHF1 mutation causes early-onset epileptic encephalopathy with cerebellar atrophy. Neurology. 2016;86:2162–2170. doi: 10.1212/WNL.0000000000002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wildburger N.C., Ali S.R., Hsu W.J., et al. Quantitative proteomics reveals protein–protein interactions with fibroblast growth factor 12 as a component of the voltage-gated sodium channel 1.2 (Nav1.2) macromolecular complex in mammalian brain*. Mol Cell Proteomics. 2015;14:1288–1300. doi: 10.1074/mcp.M114.040055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu C., Dib-Hajj S.D., Renganathan M., Cummins T.R., Waxman S.G. Modulation of the cardiac sodium channel Nav1.5 by fibroblast growth factor homologous factor 1B. J Biol Chem. 2003;278:1029–1036. doi: 10.1074/jbc.M207074200. [DOI] [PubMed] [Google Scholar]
- 12.Wang C., Wang C., Hoch E.G., Pitt G.S. Identification of novel interaction sites that determine specificity between fibroblast growth factor homologous factors and voltage-gated sodium channels. J Biol Chem. 2011;286:24253–24263. doi: 10.1074/jbc.M111.245803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puranam R.S., He X.P., Yao L., et al. Disruption of Fgf13 causes synaptic excitatory-inhibitory imbalance and genetic epilepsy and febrile seizures plus. J Neurosci. 2015;35:8866–8881. doi: 10.1523/JNEUROSCI.3470-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al-Mehmadi S., Splitt M., For DDD Study Group, et al. FHF1 (FGF12) epileptic encephalopathy: table. Neurol Genet. 2016;2:e115. doi: 10.1212/NXG.0000000000000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fry A.E., Marra C., Derrick A.V., et al. Missense variants in the N-terminal domain of the A isoform of FHF2/FGF13 cause an X-linked developmental and epileptic encephalopathy. Am J Hum Genet. 2021;108:176–185. doi: 10.1016/j.ajhg.2020.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalski A., Atici J., Kreuz F.R., Hellenbroich Y., Schwinger E., Zühlke C. Mutation analysis in the fibroblast growth factor 14 gene: frameshift mutation and polymorphisms in patients with inherited ataxias. Eur J Hum Genet. 2005;13:118–120. doi: 10.1038/sj.ejhg.5201286. [DOI] [PubMed] [Google Scholar]
- 17.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richards S., Aziz N., Bale S., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet Med. 2015;17:405–423. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Willemsen M.H., Goel H., Verhoeven J.S., et al. Epilepsy phenotype in individuals with chromosomal duplication encompassing FGF12. Epilepsia Open. 2020;5:301–306. doi: 10.1002/epi4.12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leffler A., Herzog R.I., Dib-Hajj S.D., Waxman S.G., Cummins T.R. Pharmacological properties of neuronal TTX-resistant sodium channels and the role of a critical serine pore residue. Pflüg Arch - Eur J Physiol. 2005;451:454–463. doi: 10.1007/s00424-005-1463-x. [DOI] [PubMed] [Google Scholar]
- 21.Rush A.M., Dib-Hajj S.D., Waxman S.G. Electrophysiological properties of two axonal sodium channels, Nav 1.2 and Nav 1.6, expressed in mouse spinal sensory neurones: Sodium channels in sensory neurones. J Physiol. 2005;564:803–815. doi: 10.1113/jphysiol.2005.083089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wood J.N., Bevan S.J., Coote P.R., et al. Novel cell lines display properties of nociceptive sensory neurons. Proc R Soc Lond B Biol Sci. 1990;241:187–194. doi: 10.1098/rspb.1990.0084. [DOI] [PubMed] [Google Scholar]
- 23.Shi R.-M., Kobayashi T., Kikuchi A., et al. Phenytoin-responsive epileptic encephalopathy with a tandem duplication involving FGF12. Neurol Genet. 2017;3:e133. doi: 10.1212/NXG.0000000000000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dover K., Solinas S., D'Angelo E., Goldfarb M. Long-term inactivation particle for voltage-gated sodium channels: Voltage-gated sodium channels. J Physiol. 2010;588:3695–3711. doi: 10.1113/jphysiol.2010.192559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trivisano M., Ferretti A., Bebin E., et al. Defining the phenotype of FHF1 developmental and epileptic encephalopathy. Epilepsia. 2020;61 doi: 10.1111/epi.16582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ben-Shalom R., Keeshen C.M., Berrios K.N., An J.Y., Sanders S.J., Bender K.J. Opposing effects on Na V 1.2 function underlie differences between SCN2A variants observed in individuals with autism spectrum disorder or infantile seizures. Biol Psychiatry. 2017;82:224–232. doi: 10.1016/j.biopsych.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johannesen K.M., Liu Y., Koko M., et al. Genotype-phenotype correlations in SCN8A -related disorders reveal prognostic and therapeutic implications. Brain. 2021 doi: 10.1093/brain/awab321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolff M., Johannesen K.M., Hedrich U.B.S., et al. Genetic and phenotypic heterogeneity suggest therapeutic implications in SCN2A-related disorders. Brain. 2017;140:1316–1336. doi: 10.1093/brain/awx054. [DOI] [PubMed] [Google Scholar]
- 29.Larsen J., Carvill G.L., Gardella E., et al. The phenotypic spectrum of SCN8A encephalopathy. Neurology. 2015;84:480–489. doi: 10.1212/WNL.0000000000001211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y., Koko M., Lerche H. A SCN8A variant associated with severe early onset epilepsy and developmental delay: loss- or gain-of-function? Epilepsy Res. 2021;178 doi: 10.1016/j.eplepsyres.2021.106824. [DOI] [PubMed] [Google Scholar]
- 31.Velíšková J., Marra C., Liu Y., et al. Early onset epilepsy and sudden unexpected death in epilepsy with cardiac arrhythmia in mice carrying the early infantile epileptic encephalopathy 47 gain-of-function FHF1(FGF12) missense mutation. Epilepsia. 2021;62:1546–1558. doi: 10.1111/epi.16916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu W., Hill S.F., Xenakis J.G., Pardo-Manuel de Villena F., Wagnon J.L., Meisler M.H. Gabra2 is a genetic modifier of Scn8a encephalopathy in the mouse*. Epilepsia. 2020;61:2847–2856. doi: 10.1111/epi.16741. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.