ABSTRACT
The cross-kingdom interactions between Candida albicans and Actinomyces viscosus play critical roles in root caries. However, the key pathway by which C. albicans regulates its interactions with A. viscosus is unclear. Here, we first employed 39 volunteers with root caries and 37 caries-free volunteers, and found that the abundances of C. albicans and A. viscosus were significantly increased in the individuals with root caries and showed a strong positive correlation. Their dual-species combination synergistically promoted biofilm formation and root caries in rats. The arginine biosynthesis pathway of C. albicans was significantly upregulated in dual-species biofilms and dental plaques from another 10 root caries volunteers compared with the 10 caries-free volunteers. The exogenous addition of arginine increased the cariogenicity of the dual-species biofilm. The C. albicans ARG4, a key gene from the arginine biosynthesis pathway, null mutant failed to promote dual-species biofilm formation and root caries in rats; however, the addition of arginine restored its synergistic actions with A. viscosus. Our results identified the critical roles of the C. albicans arginine biosynthesis pathway in its cross-kingdom interactions with A. viscosus for the first time and indicated that targeting this pathway was a practical way to treat root caries caused by multiple species.
IMPORTANCE Root caries is a critical problem that threatens the oral health of the elderly population. Our results identified the essential roles of the C. albicans arginine biosynthesis pathway in its cross-kingdom interactions with A. viscosus in root caries for the first time and indicated that targeting this pathway was a practical way to treat root caries caused by multiple species.
KEYWORDS: root caries, multispecies infection, Candida albicans, Actinomyces viscosus, arginine biosynthesis pathway
INTRODUCTION
Oral diseases are a major health burden for many countries and affect individuals throughout their lifetimes, causing pain, discomfort, disfigurement, and even death (https://www.who.int/news-room/fact-sheets/detail/oral-health). Oral diseases affect approximately 3.5 billion people worldwide, of which 2.3 billion people suffer from permanent tooth caries, the most common oral problem (1). Root caries is among the important reasons for tooth loosening in the elderly population, with prevalence ranging from 25% to 100% (2–4). Facilitating the prevention and treatment of root caries is among the important missions necessary to achieve the plan “8020 better oral health for older people” proposed by the WHO (5).
Oral microorganisms have been indicated to play crucial roles in the development of root caries. Actinomyces viscosus is an early colonizing microorganism of the root surface and a key pathogenic agent for root caries. A. viscosus was found to be the dominant bacterium in all plaque samples from root surface caries (6) and accounted for 100% of the isolation frequencies (7). The cariogenic factors of A. viscosus include the strong ability of cell adhesion (8) and the capability to metabolize several kinds of carbohydrates, such as starch, sucrose, glucose, and fructose, which results in the production of large amounts of acids and the rapid demineralization of the infected teeth (9). Acid production can also reduce the growth of other oral bacteria in root caries plaque because A. viscosus has a tolerance to acid (9).
Candida albicans is a common symbiotic fungus in the oral cavity, respiratory and digestive tracts, and urogenital system. C. albicans is highly associated with oral candidiasis (10) and dental caries in the oral cavity, especially in Early Childhood Caries (ECC) and root caries (11, 12). The isolation frequency of C. albicans in root caries lesions was found to be approximately 40% (13), while both the isolation frequency and detection abundance of C. albicans from root caries lesions were found to be much higher than those from sound root surfaces (12). C. albicans can penetrate into the dentin tubules and bind to collagen, and then secrete hydrolases to degrade collagen under acidic conditions to promote the caries process (14, 15).
The cross-kingdom interactions between C. albicans and many oral bacteria, such as Streptococcus, Actinomyces, Fusobacterium, and Helicobacter species, contribute to the development of different oral diseases (16–19). C. albicans and Actinomyces could coaggregate tightly, especially when C. albicans was in the hyphal state (20, 21). C. albicans and streptococci had a synergistic partnership, as streptococci promoted C. albicans to invade the oral mucosae, while C. albicans promoted streptococci to form biofilms on abiotic surfaces and in the oral cavity (18, 22, 23). Interactions between Streptococcus mutans and C. albicans could result in the formation of a more complicated biofilm with the elevation of extracellular polysaccharide production by S. mutans and hyphal formation of C. albicans, respectively, to promote dental caries development (16, 24–26). We also found that C. albicans could affect the interactions between S. mutans and Streptococcus sanguinis to promote the development of dental caries (12). Streptococcus gordonii could also promote C. albicans biofilm formation and hyphal development (17). C. albicans and Staphylococcus aureus synergistically interacted to promote pathogenic potential, increase resistance to antibiotics and help Candida circumvent the host immune system (27, 28). However, the key pathways by which C. albicans regulates its interactions with oral bacteria are still unclear.
We previously found that C. albicans increased the cariogenic abilities of A. viscosus in vitro, while voriconazole inhibited their cross-kingdom interactions (29, 30). However, the key pathway by which C. albicans regulates its cross-kingdom interactions with A. viscosus and the effects of their coinfection in root caries are still unclear. Thus, in this study, we aimed to identify the C. albicans pathway that is critical for its interactions with A. viscosus in clinical root caries samples, dual-species biofilms, and root caries rat models.
RESULTS
Increased detection rates and abundances of C. albicans and A. viscosus in root caries.
Seventy-six volunteers, including 39 patients with root caries in the root caries (RC) group and 37 healthy people in the healthy control (HC) group, were recruited to compare the detection rates and abundances of C. albicans and A. viscosus in supragingival dental plaque. The rate of C. albicans detection was 82.05% in the RC group and was significantly higher than that in the HC group (51.35%) (Fig. 1A; P < 0.05). The rate of A. viscosus detection was 82.05% in the RC group and was also significantly higher than that in the HC group (70.27%) (Fig. 1B; P < 0.05). The abundances of C. albicans and A. viscosus were also significantly enriched in the RC group (19.75 ± 12.92 copies/ng and 41.84 ± 23 copies/ng, respectively) compared with those in the HC group (6.399 ± 6.669 copies/ng and 18.57 ± 11.33 copies/ng, respectively) (Fig. 1C and D; P < 0.05). Notably, the abundances of C. albicans and A. viscosus were positive (Fig. 1E), suggesting that there is a strong correlation between C. albicans and A. viscosus in root caries.
FIG 1.
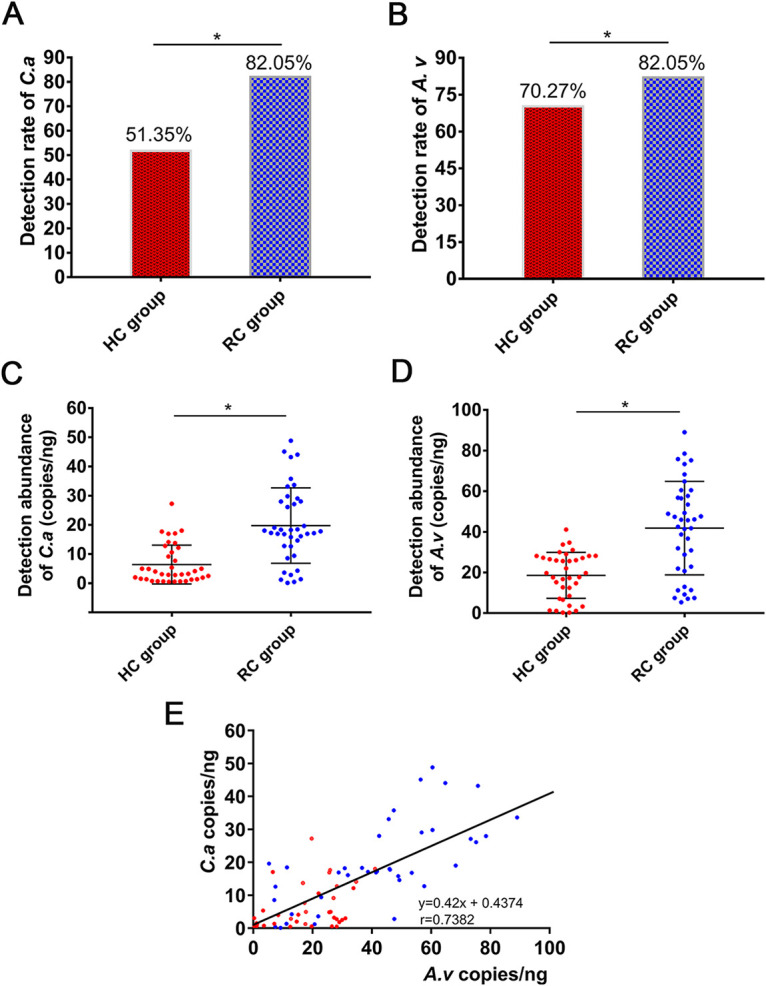
Increased amounts of C. albicans and A. viscosus in root carious lesions. (A, B) Detection rates of C. albicans and A. viscosus by PCR in the RC and HC groups. (A) C. albicans; (B) A. viscosus. (*, P < 0.05). (C, D) The abundances of C. albicans and A. viscosus determined by qPCR in the RC and HC groups. (C) C. albicans; (D) A. viscosus (*, P < 0.05). (E) Correlation and linear regression analysis between the abundances of C. albicans and A. viscosus among the plaque samples of all recruited subjects. The red dots represented samples from the HC group and the blue dots represented samples from the RC group. The Pearson correlation coefficient r value is 0.7382 (r = 0.7382, r2 = 0.5449, P ≤ 0.05).
Synergistic interactions between C. albicans and A. viscosus promoted biofilm formation and cariogenicity.
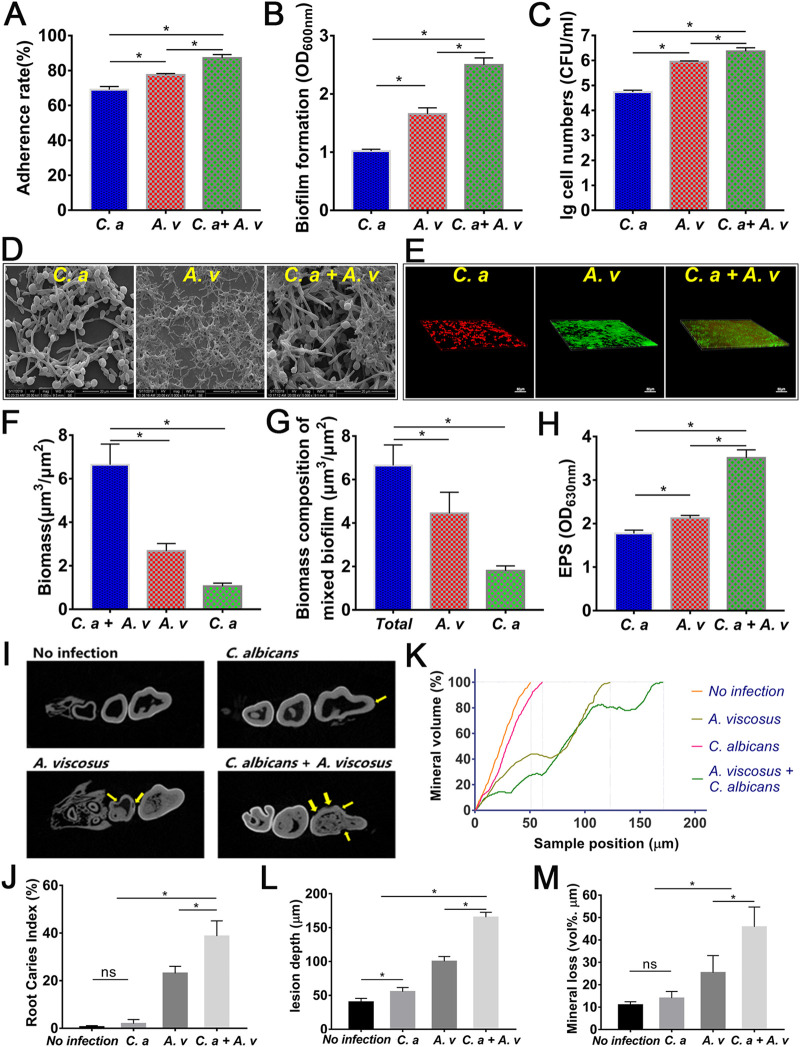
We then investigated the cariogenicity of the C. albicans and A. viscosus dual-species combinations due to their positive correlation in clinical samples. During biofilm formation, the adherence rate in the dual-species group (87.17%) was higher than that in the single-species groups (69.06% for C. albicans, 77.51% for A. viscosus) (Fig. 2A; P < 0.05). The biofilm biomass and viable cells of the dual-species group were significantly elevated compared with those of the single-species groups (Fig. 2B and C; P < 0.05). These results indicated that the combination of C. albicans and A. viscosus enhanced cell adherence, cell growth, and biofilm formation. The dual-species combination formed thicker and denser biofilms (Fig. 2D and E). A higher proportion of hyphal forms of C. albicans was observed in the dual-species biofilms than in the C. albicans mono-species biofilms (Fig. 2D and E). The numbers of both C. albicans and A. viscosus cells were elevated in the dual-species group (Fig. 2F and G). To test whether viable cell-cell contact is necessary for the enhanced biofilm formation, we combined viable cells, heat-killed cells, and cell supernatants of C. albicans and A. viscosus, respectively, and found that only the combination of viable cells significantly enhanced biofilm formation, indicating that cell-cell contact was essential for the interactions between C. albicans and A. viscosus (Fig. S1).
FIG 2.
C. albicans synergistically interacted with A. viscosus to promote the cariogenicity and root caries. (A to C) The adherence rates (A), biofilm formations (B), and CFU (C) from the three groups: C. albicans single-species; A. viscosus single species; C. albicans + A. viscosus dual-species (*, P < 0.05). (D, E) Structural observations of biofilms formed by C. albicans, A. viscosus, C. albicans, and A. viscosus through SEM (D) and FISH (E) analysis. C. albicans was stained with red color while A. viscosus was stained with green color. (F, G) Biomasses of the three kinds of biofilms from FISH observation result, quantitatively calculated by COMSTAT. (F) Biomasses of C. albicans single-species, A. viscosus single-species, and C. albicans + A. viscosus dual-species FISH-visualized biofilms, respectively, quantified by COMSTAT. (G) Biomass of C. albicans + A. viscosus FISH-visualized biofilm and the respective biomass compositions of C. albicans and A. viscosus in this dual-species biofilm (*, P < 0.05). (H) Water insoluble EPS productions of three groups: C. albicans single-species, A. viscosus single species, C. albicans + A. viscosus dual-species (*, P < 0.05). (I) Representative micro-CT images of rat jaws from uninfected rats and rats infected with C. albicans, A. viscosus, or C. albicans + A. viscosus, respectively. Yellow arrows indicated root carious lesions. (J) Root caries index scores according to Doff’s system (*, P < 0.05; ns, not significant). (K) The mineral volume curves of teeth from uninfected rats and rats infected with C. albicans, A. viscosus, or C. albicans + A. viscosus, respectively. (L) The lesion depths curves of teeth from uninfected rats and rats infected with C. albicans, A. viscosus, or C. albicans + A. viscosus, respectively (*, P < 0.05). (M) The mineral losses of teeth from uninfected rats and rats infected with C. albicans, A. viscosus, or C. albicans + A. viscosus, respectively (*, P < 0.05; ns, not significant).
Moreover, the dual-species biofilm produced more water insoluble extracellular polysaccharides (EPS), the key cariogenic virulence factor, compared with that of C. albicans or A. viscosus single-species biofilms (Fig. 2H; P < 0.05), indicating that the cross-kingdom interactions between C. albicans and A. viscosus enhanced cariogenicity.
C. albicans synergized with A. viscosus to promote root caries in rats.
We further evaluated whether the cross-kingdom interactions of C. albicans and A. viscosus could promote the development of root caries in rat model. Rats infected with C. albicans alone formed very little root caries, while the rats infected with A. viscosus formed typical root caries, indicating the strong cariogenic ability of A. viscosus (Fig. 2I and J). Coinfection with C. albicans and A. viscosus synergistically increased the formation and severity of root caries compared to those of C. albicans or A. viscosus single-species infection (Fig. 2I and J). The rats coinfected with C. albicans and A. viscosus had the highest root caries score, lowest mineral volume, and largest lesion depth and mineral loss of the jaw (Fig. 2J to M; P < 0.05). These results demonstrated that C. albicans could synergize with A. viscosus to promote root caries in vivo.
The highly activated C. albicans arginine biosynthesis pathway in dual-species biofilms.
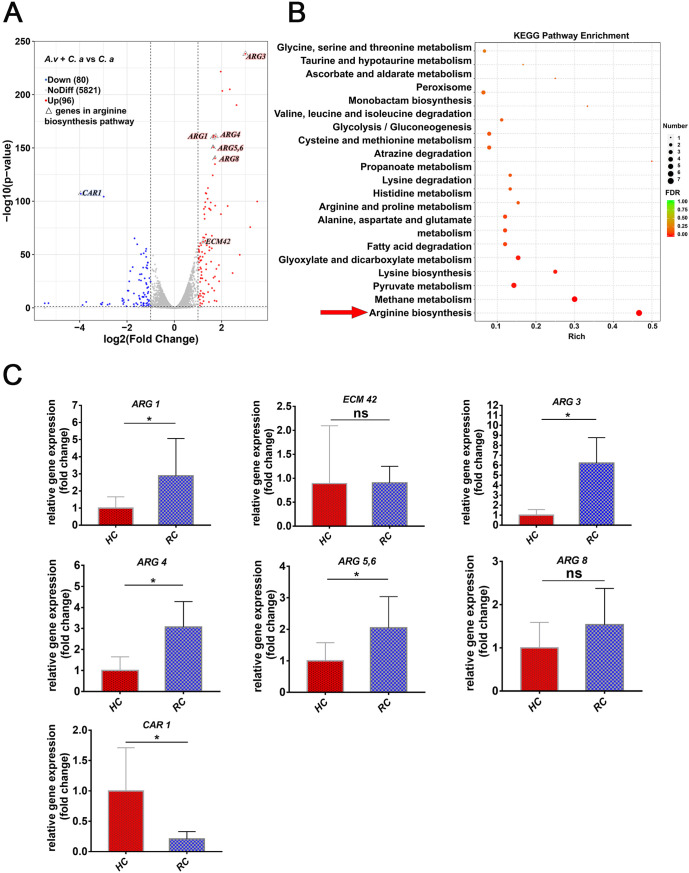
To further identify the key pathway by which C. albicans regulates its synergistic interaction with A. viscosus, we analyzed the transcriptome of C. albicans from the dual-species biofilm compared with the C. albicans single-species biofilm (Fig. S2). There were 176 differentially expressed genes (DEGs) between the two groups (FDR < 5%, |log2FoldChange| >1). 96 genes were upregulated and 80 genes were downregulated in the dual-species biofilm (Fig. 3A; Fig. S2C). The expressions of genes related to arginine biosynthesis of C. albicans were significantly increased, while the expression of arginine degradation associated gene CAR1 was significantly decreased (Fig. 3A). Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis further confirmed that the DEGs were most enriched in the arginine biosynthesis pathway of C. albicans (Fig. 3B). Gene expressions in the arginine biosynthesis pathway, including that of ARG1, ECM42, ARG3, ARG4, ARG5,6, ARG8, and CAR1, were then confirmed by quantitative PCR (qPCR) analysis. The results confirmed that the expression levels of ARG1, ECM42, ARG3, ARG4, and ARG5,6 were significantly upregulated and that the expression of CAR1 was significantly downregulated in the dual-species biofilm compared with those in the C. albicans single-species biofilm (Fig. S3); this result was consistent with the transcriptome analysis (Fig. 3A), indicating the key roles of the arginine biosynthesis pathway in dual-species biofilm.
FIG 3.
Activation of the C. albicans arginine biosynthesis pathway in dual-species biofilm and clinical root caries plaque samples. (A) Volcano plot of centered and scaled FPKM values of DEGs indicating significant expression changes in the arginine biosynthesis pathway. The ARG1, ECM42, ARG3, ARG4, ARG5,6, ARG8, and CAR1 genes in the arginine biosynthesis pathway were marked. (B) KEGG pathway enrichment analysis indicating that the arginine biosynthesis pathway was the most DEG-enriched pathway. The yellow arrow showed the arginine biosynthesis pathway. (C) The differential expression of the arginine biosynthesis-associated genes: ARG1, ECM42, ARG3, ARG4, ARG5,6, ARG8, and the arginine degradation-associated gene: CAR1 were confirmed from the root caries plaques (RC groups) compared with the HC groups by qPCR (*, P < 0.05; ns, not significant).
Enhanced activation of the C. albicans arginine biosynthesis pathway in clinical root caries.
To further confirm that the arginine biosynthesis pathway of C. albicans was also upregulated in clinical root caries, another 20 volunteers, including 10 patients with root caries (RC group) and 10 caries-free individuals (HC group), were recruited. The expression levels of the genes associated with arginine biosynthesis (ARG1, ARG3, ARG4, and ARG5,6) were significantly elevated while CAR1 expression was decreased in the root caries plaques in the RC group compared with those in the sound root surface plaques in the HC group (Fig. 3C), which was in line with the transcriptome analysis in the dual-species biofilms and indicated that the enrichment of the C. albicans arginine biosynthesis pathway played key roles in the development of root caries.
The addition of arginine increased the growth and biofilm formation of C. albicans, A. viscosus, and their dual-species combination.
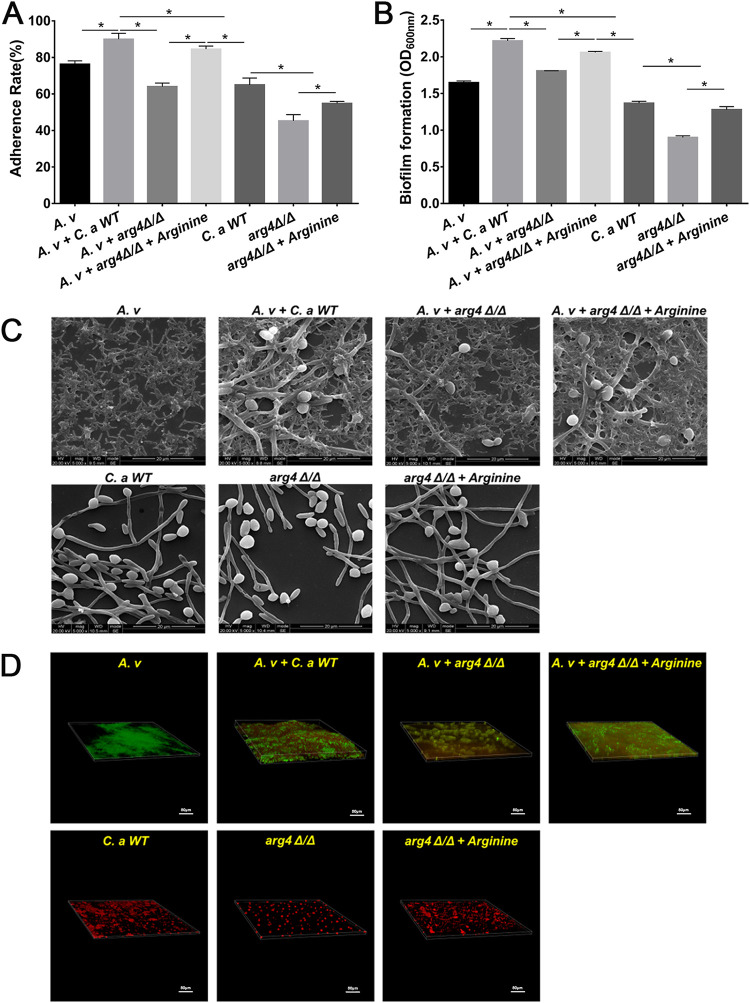
We further evaluated the effect of the corresponding product of the arginine biosynthesis pathway (arginine) on the growth of C. albicans and A. viscosus. As shown in Fig. 4, the addition of arginine promoted C. albicans biofilm formation, similar to the dual-species combination, compared with that in the group without arginine (Fig. 4A; P < 0.05). More hyphal formation in the C. albicans single-species biofilm with the addition of arginine and the dual-species biofilm was observed (Fig. 4B). C. albicans formed denser and more compact biofilm with the addition of arginine (Fig. 4C), with higher biomass than that of C. albicans single-species biofilm without arginine (Fig. 4C and D; P < 0.05). The addition of arginine also promoted the formation of A. viscosus biofilms. A. viscosus formed denser and more compact biofilms with a greater total biomass with the addition of arginine (Fig. 4E and F). Moreover, the addition of arginine also enhanced the formation of dual-species biofilm. The total biomass of the dual-species biofilm with the addition of arginine was elevated significantly (Fig. 4G; P < 0.05), suggesting that arginine increased the growth and biofilm formation of the C. albicans and A. viscosus combination.
FIG 4.
The addition of arginine promoted the growth of C. albicans, A. viscosus, and dual-species biofilms. (A) Effect of arginine on C. albicans biofilm formation: Total biomasses quantified with CV assay of three kinds of biofilms formed by C. albicans, A. viscosus, C. albicans + A. viscosus, respectively (*, P < 0.05). (B, C) Effect of arginine on A. viscosus biofilm structure: Three structures of biofilms from C. albicans, A. viscosus, C. albicans + A. viscosus, respectively, observed with SEM and FISH. C. albicans was stained with red color while A. viscosus was stained with green color. (D) Biomasses of the biofilms according to FISH observation quantitatively calculated by COMSTAT (*, P < 0.05; ns, not significant). (E) Effect of arginine on the A. viscosus biofilm formation: Total biomasses of A. viscosus biofilm formed with or without arginine addition quantified by CV assay (*, P < 0.05). (F) Effect of arginine on A. viscosus biofilm structure: Structures of A. viscosus biofilm formed with or without arginine addition observed by SEM. (G) Effect of arginine on A. viscosus and C. albicans dual-species biofilm formation: Total biomasses of dual-species biofilm formed with or without arginine addition quantified by CV assay (*, P < 0.05).
The C. albicans arginine biosynthesis pathway regulated its cross-kingdom interaction with A. viscosus in dual-species biofilms.
To confirm the essential role of the C. albicans arginine biosynthesis pathway in the regulation of its cross-kingdom interactions with A. viscosus, the ARG4 null mutant (arg4Δ/Δ) was employed (Table S1). The arg4Δ/Δ mutant failed to exhibit enhanced cell adhesion in both the dual-species and single-species groups compared with that of the wild-type (WT) strain, while the addition of arginine recovered the promotion (Fig. 5A). The arg4Δ/Δ mutant exhibited reduced biofilm formation in both the dual-species and single-species biofilms compared to that of the WT strain, and the addition of arginine also reversed the reduction (Fig. 5B to D). In C. albicans WT and A. viscosus dual-species biofilm, the colonization of both C. albicans and A. viscosus were increased, compared with that in the single-species biofilm (Fig. 5D). However, the colonization of C. albicans in the arg4Δ/Δ and A. viscosus dual-species biofilm was not obviously increased compared with that in the arg4Δ/Δ single-biofilm (Fig. 5D). The addition of arginine restored the C. albicans colonization in the dual-species biofilm (Fig. 5D). These results indicated the essential roles of the arginine biosynthesis pathway of C. albicans in its cross-kingdom interactions with A. viscosus and suggested that targeting the arginine biosynthesis pathway could block their interaction in dual-species biofilm.
FIG 5.
C. albicans arginine biosynthesis pathway regulated cross-kingdom interactions in dual-species biofilms. (A, B) Determination of the effects of the arg4Δ/Δ mutant and arginine supplementation on C. albicans growth through adherence rate (A) and biofilm formation (B) (*, P < 0.05). (C, D) The biofilm structures of the arg4Δ/Δ mutant and with arginine supplementation determined via SEM (C) and FISH observations (D). C. albicans or arg4Δ/Δ mutant was stained with red color while A. viscosus was stained with green color.
The arginine biosynthesis pathway of C. albicans promoted root caries.
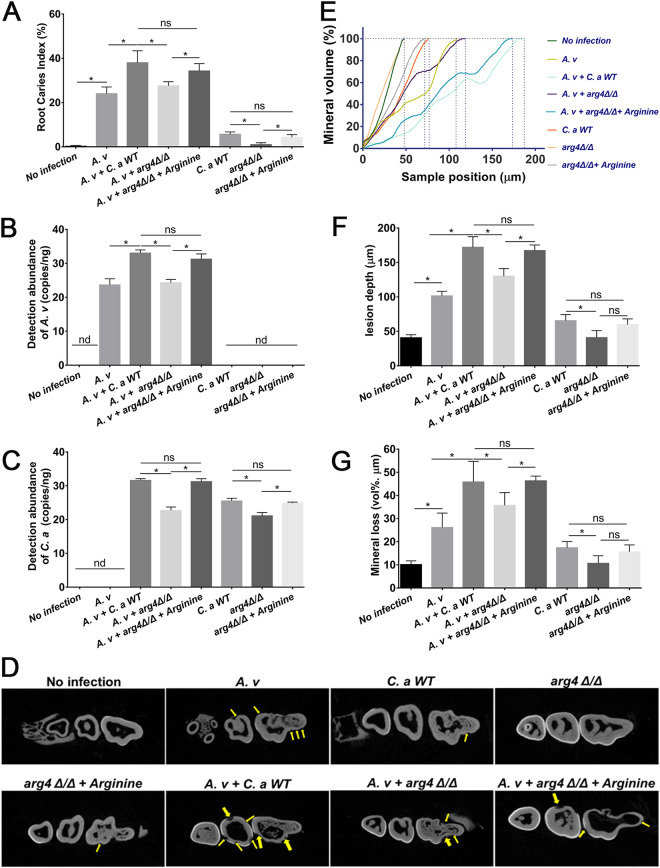
We then investigated the contribution of the arginine biosynthesis pathway of C. albicans to the development of root caries in vivo. In the rat root caries model, coinfection with C. albicans WT and A. viscosus caused the most remarkable root caries lesions (Fig. 6A, D; P < 0.05), with elevated colonization of C. albicans and A. viscosus on the root surfaces (Fig. 6B, C; P < 0.05). A reduced colonization of arg4Δ/Δ was observed in the teeth (Fig. 6C; P < 0.05). Combination of arg4Δ/Δ and A. viscosus also failed to promote the colonization of A. viscosus (Fig. 6B; P < 0.05) and the development of root caries (Fig. 6A, D; P < 0.05), as the rats showed similar hard-tissue destruction, mineral volume, lesion depth and mineral loss compared to those of the rats infected by the A. viscosus single-species (Fig. 6D, G; P < 0.05). However, the addition of arginine increased the colonization of arg4Δ/Δ and A. viscosus in their combination (Fig. 6B, C; P < 0.05). The addition of arginine enhanced the development of root caries in rats coinfected with arg4Δ/Δ and A. viscosus (Fig. 6A, D; P < 0.05), and increased hard-tissue destruction, lesion depth and mineral loss and decreased mineral volumes (Fig. 6D, G; P < 0.05), indicating that the C. albicans arginine biosynthesis pathway is essential for the development of root caries.
FIG 6.
C. albicans arginine biosynthesis pathway regulated the development of root caries and affected the demineralization of the teeth in root caries. (A) Root caries index scores according to Doff’s system (*, P < 0.05; ns, not significant). (B, C) Levels of C. albicans (B) and A. viscosus (C) from the root-caries rats quantified via qPCR (*, P < 0.05; ns, not significant; nd, not detectable). (D) Representative micro-CT images of jaws from uninfected rats and rats infected with A. viscosus, A. viscosus + C. albicans, A. viscosus+ arg4 Δ/Δ, A. viscosus+ arg4 Δ/Δ + Arginine, C. albicans, arg4 Δ/Δ, or arg4 Δ/Δ + Arginine, respectively. Yellow arrows indicated root carious lesions. (E to G) The mineral volume curves (E), lesion depths (F), and mineral losses (G) of teeth from uninfected rats and rats infected with A. viscosus, A. viscosus+ C. albicans, A. viscosus+ arg4 Δ/Δ, A. viscosus+ arg4 Δ/Δ + Arginine, C. albicans/arg4 Δ/Δ, or arg4 Δ/Δ + Arginine, respectively (*, P < 0.05; ns, not significant).
DISCUSSION
The incidence of root caries has increased in recent years and it has become a major oral problem in the elderly population, but the treatment of root caries is challenging (2, 31). The prevention and treatment of root caries has become one of the major issues to improve oral health. C. albicans and A. viscosus are two resident symbiotic opportunistic microorganisms in the oral cavity (32, 33), and their cross-kingdom interactions play important roles in the development of root caries. We found that the isolation frequencies and abundances of both species were significantly higher in the root caries plaque samples than in the sound root surface plaque samples (Fig. 1A to D). The abundances of the two species showed a positive correlation (Fig. 1E).
The hyphal state of C. albicans is the main virulent form, and it can efficiently mediate microbial adhesion and biofilm formation process. In the dual-species biofilm, we found that A. viscosus could promote the hyphal formation of C. albicans, thus promoting its virulence. Morse et al. (34) suggested that the coculture of C. albicans and A. viscosus could significantly increase the hyphal content in the biofilm, and the expression levels of C. albicans virulence genes, such as ALS3, EPA1, PLD1, SAP4, and SAP6, in the coculture biofilm were significantly increased. Our previous work also showed that cariogenic virulence such as biofilm proliferation, acid production, acid resistance, sugar production, and biofilm formation were significantly enhanced under C. albicans and A. viscosus coculture conditions (30). However, the specific pathway by which C. albicans regulates the interactions between C. albicans and A. viscosus is still unclear. In our study, KEGG enrichment analysis of the RNA-Seq-identified DEGs suggested that the C. albicans arginine biosynthesis pathway, including six upregulated genes (ARG1, ECM42, ARG3, ARG4, ARG5,6, ARG8) and one downregulated gene (CAR1), was remarkably changed in the dual-species biofilm and clinical root caries plaques (Fig. 3; Fig. S3). KEGG analysis also indicated that the alanine, aspartate, and glutamate metabolism pathway, and pantothenate and CoA biosynthesis pathway were upregulated in the dual-species group (Fig. 3). Glutamate is the essential substrate of arginine biosynthesis, while Acetyl-CoA is one of the important enzymes that metabolize glutamate to produce CoA. The produced glutamate and COA then activate the arginine biosynthesis procedure (35). KEGG analysis also showed that the lysine biosynthesis pathway was significantly enriched, and the expression levels of genes related to lysine biosynthesis (such as HOM1, LYR22, LYR4, etc.) were significantly upregulated in the dual-species biofilm, while an increase in intracellular lysine can promote arginine secretion (36). These upregulation pathways in the dual-species biofilms indicated that A. viscosus might enhance glutamate metabolism and CoA biosynthesis of C. albicans and then effectively promote arginine biosynthesis. In addition, A. viscosus might also increase the secretion of synthesized arginine through the upregulation of lysine biosynthesis of C. albicans. The mechanism of amino acid biosynthesis and metabolism in regulating the interaction between different species is complex. The cross talk between the different amino acid biosynthesis and metabolism from C. albicans and A. viscosus interactions require further evaluation.
In this study, we evaluated the important role of the arginine biosynthesis pathway of C. albicans in its cross-kingdom interaction with A. viscosus. The arginine biosynthesis process in the mitochondria and cytoplasm was summarized in Fig. S4A (35, 37). The deletion of ARG4, the key gene from the arginine biosynthesis pathway, eliminated synergistic interactions with A. viscosus, while the addition of arginine complemented the virulence deficiencies of the arg4Δ/Δ mutant in both the single- and dual-species groups (Fig. 5). The addition of arginine could also promote the biofilm formation of C. albicans, A. viscosus, dual-species, and hyphal formation of C. albicans (Fig. 4 and 5), while the neutral amino acids tyrosine and the acidic amino acid glutamate could not promote the biofilm formation of C. albicans and A. viscosus (unpublished data), indicating the critical role of the arginine biosynthesis pathway of C. albicans in its cross-kingdom interaction with A. viscosus (Fig. S4B), and targeting this pathway is a practical strategy to reduce the development of root caries. Further investigations are still needed to reveal the mechanisms by which the arginine biosynthesis pathway regulates the growth and virulence of A. viscosus.
Arginine is one of the most versatile amino acids in eukaryotic cells and contributes to protein synthesis, cell growth, sexual reproduction, hormone metabolism, signal transduction, osmotic pressure homeostasis, metabolic energy production, nitrogen metabolism, and urea biosynthesis (38, 39). Arginine at the appropriate concentration was essential for the growth and pathogenicity of various microorganisms. Novick et al. (40) isolated an arginine auxotroph Escherichia coli mutant and found that it grew slowly in the absence of arginine but grew at a normal rate in the presence of arginine. Hartmann et al. (41) found that arginine could help Halobacteria grow in the anaerobic state. Tonon et al. observed that arginine increased the growth of wine lactic acid bacteria (42). Senouci-Rezkallah et al. suggested that arginine stimulated the growth of Bacillus cereus under low-acid conditions (43). Vrancken et al. found that arginine enhanced the resistance of Lactobacillus fermentum to environmental stresses, such as acid, temperature, salt stress, and osmotic pressure factors (44). Huang’s results showed that arginine stimulated the growth of Streptococcus thermophilus T1C2 by enhancing resistance to a low intracellular pH under high extracellular osmotic pressure (45). Zhang et al. (38) indicated that three ARG genes involved in arginine biosynthesis were essential for growth, conidiogenesis, sexual reproduction, hyphal growth, and pathogenicity in Magnaporthe oryzae. The M. oryzae argΔ/Δ mutants exhibited significantly delayed conidial germination and decreased pathogenicity, while exogenous arginine could partially restore the infection defects in invasive hyphal growth and pathogenicity (38). Similarly, the addition of arginine could restore the pathogenicity of Fusarium oxysporum f. sp. melonis arginine auxotroph mutants (46). Our results suggested that the C. albicans arginine biosynthesis pathway was significantly activated in the dual-species interactions, while hyphal and biofilm formation were also increased with the significant upregulation of hyphae-associated genes under anaerobic conditions, including RAS1, NCE103, GPR1, PDE2, TPK1, UME6, MEP2, NTH1, TOP1, PTP3, etc. (unpublished data). Arginine biosynthesis was reported to be associated with oxidative stress (47), while the transition of C. albicans yeast to hyphae also generated ROS as a by-product of oxidative phosphorylation in mitochondria (47, 48). C. albicans can also eliminate oxidative stress through the antioxidant pathway and enzymes (49–51). Our transcriptome analysis indicated that a series of antioxidant genes were significantly upregulated in the dual-species group, including HSP78, HSP21, CIP1, SOD2, SOD5, FRE9, FRE10, CFL1, CFL2, CFL4, CFL11, CTR2, ZRT1, and ZRT2, etc., indicating the increased capacity of C. albicans to eliminate oxidative stress. In addition, the low concentration of additional of arginine can directly enhance the growth, biofilm formation, and cariogenic capability of A. viscosus, indicating that the upregulated arginine was the key factor in promoting the cross-kingdom interaction. However, the specific mechanisms by which arginine regulates the effects on A. viscosus and hyphal growth of C. albicans in dual-species combination still need further investigation.
Currently, the relevance of arginine in the oral cavity is still unclear. Arginine could be catabolized by local arginase secreted from host or bacterial cells, such as macrophages and Porphyromonas gingivalis, to produce urea and ornithine, thus, increasing the production of polyamines and promote the growth of some bacteria to aggravate the inflammation and tissue destruction (52). Many studies have shown that the arginase activity was positively related to the degree of periodontal inflammation (53–55), indicating the importance of arginine in the development of oral diseases. It is worth noting that arginine could also be an effective therapeutic agent against caries, especially when combined with high-concentration fluoride (56–61), by inhibiting the growth of some cariogenic bacteria, such as Streptococcus mutans and Streptococcus sobrinus (57), and promoting the remineralization of enamel (60). However, the arginine concentrations used in these studies were high (1.5% mass fraction, or even higher at 8% to 10% mass fraction) (56–61). In our study, arginine was added at a concentration of 0.2%, which was much lower than the potential anti-caries concentrations. The concentration from our study was similar to those that promoted the growth of different microorganisms. Zhang et al. (38) indicated that exogenous arginine supplementation could partially recover the aerial hyphal growth and pathogenicity of M. oryzae arg Δ/Δ mutants, but the recovery effect was dependent on the concentration of arginine: the concentration with better recovery effect was 2.5 mM (approximately 0.05% mass fraction), while higher concentration led to the decreased recovery effect. Huang et al. (45) also found that 1.2g/L (approximately 0.12% mass fraction) arginine exhibited the best promotion effect on S. thermophilus T1C2 growth and that growth was inhibited when the initial arginine concentration exceeded 1.2g/L (approximately 0.12% mass fraction). Similar results were observed by Mira et al. in Oenococcus oeni (62). Arginine inhibited S. mutans and S. sobrinus growth only when the concentration was over 0.4%, while arginine at a concentration ≤0.4% did not affect the growth of S. mutans or S. sobrinus (57). Our results indicated that arginine at low concentration could enhance the pathogenicity of A. viscosus and C. albicans, and in the root caries caused by C. albicans and A. viscosus coinfection. Therefore, the recommendation of arginine-containing caries prevention products and the arginine concentration are worth further careful consideration.
In summary, we identified for the first time that the arginine biosynthesis pathway of C. albicans was critical for the regulation of its cross-kingdom interactions with A. viscosus and for promoting the occurrence and development of root caries, while targeting this pathway can be a new practical strategy to reduce root caries.
MATERIALS AND METHODS
Strains and culture conditions.
A. viscosus ATCC 19246 and C. albicans WT (SC 5314, ATCC MYA-2876) were obtained from the State Key Laboratory of Oral Diseases. The ARG4 null mutant of C. albicans was also employed to confirm the role of the arginine biosynthesis pathway (Table S1). Briefly, C. albicans BWP17 was knocked out in three genes, including ARG4/URA3/HIS1. Therefore, as a compromise for the deletion of URA3 and HIS1 genes in BWP17 and to obtain the null mutant of ARG4 (C. albicans arg4Δ/Δ), BWP17 was cultured in media with additional uracil and histidine as described previously (63). A. viscosus was grown on BHY medium (brain heat infusion medium containing 5 g/l yeast extract) anaerobically (85% N2, 10% H2, and 5% CO2) at 37°C (30) and C. albicans WT was grown on yeast extract peptone dextrose (YPD) medium aerobically at 37°C. The coculture mixture (OD600nm of each microorganism = 0.1) was grown on yeast nitrogen base (YNB) supplemented with Na2HPO4-NaH2PO4, N-acetylglucosamine, casamino acids, and sucrose medium anaerobically (85% N2, 10% H2, and 5% CO2) at 37°C (30). The medium of C. albicans arg4Δ/Δ (BWP 17) was supplemented with 0.2% histidine and 0.2% uracil.
Microbial detection in clinical samples.
In total, 76 volunteers were recruited in our study. Thirty-nine subjects (aged 45 to 75 years old) were clinically diagnosed with root caries by radiography and clinical probing and were divided into RC group, while the other 37 caries-free subjects (aged 45 to 75 years old) acted as the HC group. All participants were in good general health. Ethical approval for the study was granted by the Institutional Review Board of the West China Hospital of Stomatology, Sichuan University (WCHSIRB-D-2020-072). Written informed consent was obtained from each participant that was recruited in this research. The sampling standards were designed as described previously (12). Briefly, in the RC group, after drying and isolating the tooth with sterile cotton rolls, the decayed plaque of root caries was collected with a dental spoon excavator. In the HC group, after drying and isolating the chosen sampling tooth, the supragingival plaque on the root surface was collected with a dental spoon excavator. Each sample was suspended in 1 mL TE buffer and stored at −80°C.
Total DNA of each sample was extracted with the DNeasy PowerSoil Kit (Qiagen, Valencia, CA, USA). Concentration and quality (A260 nm and A280 nm) measurements of the extracted DNA were performed with a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
The detection rates of C. albicans and A. viscosus were determined by PCR (12). The detection abundances of C. albicans and A. viscosus were quantified by qPCR (12, 64) according to the standard curves of A. viscosus and C. albicans (65). Correlation analysis and linear regression models were constructed to observe the change trends of the abundances of the two microorganisms. The primers used in this part were listed in Tables S2 and S3.
Adherence assay.
The adhesion assay was performed as described previously (66). Biofilms were formed in 48-well plates (single species groups: 500 μL C. albicans, arg4Δ/Δ or A. viscosus, respectively, for each well; dual species groups: 250 μL C. albicans or arg4Δ/Δ + 250 μL A. viscosus for each well) under stationary conditions after 24-h incubation in YNBB medium anaerobically (85% N2, 10% H2, and 5% CO2) at 37°C. The total cells including the cells from the suspension and the cells that were adhered to the well bottom were thoroughly mixed, and then OD600nm was recorded to quantify the total bacteria (OD600nm of total cells). Adherent cells were obtained by removing the suspension and resuspending the remaining cells in equal volume of medium (OD600nm of adherent cells). The adhesion rate was calculated with the formula: adhesion rate = OD600nm of adherent cells/OD600nm of total cells (including the adherent cells and the suspension cells) × 100%.
Crystal violet assay and CFU counts.
In 96-well plates, 24-h biofilms were produced (single species groups: 200 μL C. albicans, arg4Δ/Δ or 200 μL A. viscosus, respectively, for each well; dual species groups: 100 μL C. albicans/arg4Δ/Δ + 100 μL A. viscosus for each well) under stationary conditions in YNBB medium anaerobically (85% N2, 10% H2, and 5% CO2) at 37°C. The total biomass of each biofilm was quantified by crystal violet assay as previously described (30). The biofilms were sequentially fixed with methanol and stained with 0.1% (wt/vol) crystal violet for 15 to 30 min. The suspension was removed, and the cells were resolubilized with 33% (vol/vol) glacial acetic acid. Total biomass was determined with OD600nm of the suspension. To quantify the viable cells in biofilms, CFU counts were performed as described previously (30). Briefly, the biofilms were scraped off and resuspended with equal volume of medium. Then, the suspensions were 1:10 serially diluted and viable biofilm cells were quantified by CFU counts after plating the proper dilutions on YPD agar and incubating for 24 h.
Scanning electron microscopy observation and fluorescence in situ hybridization observation.
To observe the biofilm structure, scanning electron microscopy (SEM) observation and fluorescence in situ hybridization (FISH) observations were performed. Then, 24-h biofilm samples were produced on sterile glass slides at the bottom of each well of 24-well plates (single species groups: 1,000 μL C. albicans, arg4Δ/Δ or A. viscosus, respectively, for each well; dual species groups: 500 μL C. albicans/arg4Δ/Δ + 500 μL A. viscosus for each well) under stationary conditions in YNBB medium anaerobically (85% N2, 10% H2, and 5% CO2) at 37°C. SEM analysis was carried out as previously described (30). Each sample was observed by SEM imaging (FEI, Hillsboro, USA) at 5,000× magnification. The FISH procedure was performed as previously described (30) and observed with an Eclipse FV1000 inverted confocal laser scanning microscope (Olympus Corporation, Japan). The sequences of oligonucleotide probes (30) were listed in Table S4. The probes were synthesized by Sangon Biotech (Shanghai, China).
Anthrone-sulfuric acid assay.
The water-insoluble EPS production ability of 24-h biofilms were analyzed by anthrone-sulfuric acid assay. In 96-well plates, 24-h biofilms were produced (single species groups: 200 μL C. albicans, arg4Δ/Δ or A. viscosus, respectively, for each well; dual species groups: 100 μL C. albicans/arg4Δ/Δ + 100 μL A. viscosus for each well) under stationary conditions in YNBB medium anaerobically (85% N2, 10% H2, and 5% CO2) at 37°C. The biofilms were resuspended, and the precipitates were obtained and washed with sterile water to remove the water-soluble EPS. Then, each water-insoluble EPS sample was extracted with 0.4M NaOH under agitation for 2 h. Three milliliters of 0.2% anthrone-sulfuric acid reagent was mixed into each supernatant sample and then incubated in a water bath at 95°C for 6 min. The water-soluble EPS production ability was determined by OD625nm.
RNA sequencing and data analysis.
Total RNA in each sample was extracted with TRIzol reagent (Invitrogen, Carlsbad, USA). RNA sequencing (RNA-Seq) was performed by Illumina NovaSeq (Shanghai Personal Biotechnology Co., Ltd., China) as described elsewhere (67).
A total of 6,030 genes were analyzed. Differential gene expression analysis was performed using fragments per kilobase per million (FPKM) values. The Pearson correlation coefficient was estimated to analyze the correlation of gene expression levels between samples and principal-component analysis (PCA) was used to cluster samples in each group. Differentially expressed genes (DEGs) were defined with the criteria of absolute log2-fold change (FC) > 1 and adjusted P value < 0.05. DEGs were regarded as upregulated if their expression levels in dual-species biofilm samples were higher than those in the Candida albicans single-species biofilm, and vice versa. The expression of DEGs in each treatment was visualized as a volcano plot and heatmap. DEGs were submitted for functional enrichment analyses to Gene Orthology (GO) and KEGG annotations.
Analysis of the gene expression levels in biofilms.
The biofilms were collected and quantitative PCR (qPCR) was performed to evaluate the expression levels of arginine biosynthesis-associated DEGs (ARG1, ARG3, ARG4, etc.). RNA isolation with TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) and purification procedures were conducted as previously described (68). To synthesize first-strand cDNAs, RNA reverse transcription was performed with a PrimeScript RT reagent kit with gDNA Eraser (TaKaRa Biotechnology, Japan). Specific primers for the tested genes were designed using Primer3Plus (http://www.primer3plus.com/cgi-bin/dev/primer3plus.cgi) and were listed in Table S5. The qPCR mixture and procedure were carried out as previously described (68). Relative fold changes in the expression of associated genes were evaluated with the 2-ΔΔCt method (69), and the 18S rRNA gene expression level was used to normalize the expression level of different genes.
Gene expression analysis in clinical samples.
In total, another 20 volunteers were recruited. Ten subjects (aged 45 to 75 years) were clinically diagnosed with root caries by radiography and clinical probing and were divided into the RC group, while the other 10 caries-free subjects (aged 45 to 75 years) acted as the HC group. All participants were in good general health. Ethical approval for the study was granted by the Institutional Review Board of the West China Hospital of Stomatology, Sichuan University (WCHSIRB-D-2020-072). Clinical plaque samples were collected as described above. RNA from each sample was extracted with TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) and qPCR was performed to compare the expression levels of genes in the arginine biosynthesis pathway in the RC group and HC group.
Root caries rat model.
The rat model was established to investigate the promotion ability of C. albicans and A. viscosus interactions on root caries and the corresponding pathogenesis in vivo. The experiment was started after approval was obtained from the animal research committee of West China School of Stomatology, Sichuan University (WCHSIRB-D-2020-127). Male 17-day-old specific pathogen free (SPF) Sprague-Dawley (SD) rats purchased from Dashuo Inc. (Chengdu, China) were used for the in vivo experiment (five rats in each group). The root caries model was established as described in a previous study (12). Briefly, the rats were fed with 5% (wt/vol) sucrose-containing water and caries-promoting diet (Diet 2000) every day. The rats were infected daily for 3 consecutive days with A. viscosus, C. albicans WT, arg4Δ/Δ mutant, and C. albicans-A.viscosus combinations according to the designated groups (109 CFU/mL, 200 mL each rat). Ten days after the initial infection, the rats were anesthetized and underwent the gingivectomy surgery. On days 38 to 40, the rats were reinoculated with microbes. On day 66, the rats were sacrificed and the jaws were removed aseptically. The dental plaque of each jaw was collected to detect the abundances of microorganisms through qPCR as described above. Each jaw was stained with mercurochrome for 18 h to record the root caries score according to Doff’s criterion (70). Then, the jaws were subjected to the micro–computed tomography (μCT 50, SCANCO Medical AG, Brüttisellen, Switzerland) analysis (71). They were scanned at a medium resolution, with parameters of 70 kVp and 200 μA. Each sample was rotated 360° within 14.3 min. SCANCO evaluation software version 1.1.11.0 (SCANCO Medical AG) was used to acquire and analyze Micro-CT images. A line in the selected sectional view of each jaw was chosen as the region of interest (ROI) to be quantitatively analyzed. The mineral volume, lesion depth, and mineral loss of the ROI were measured by SCANCO evaluation software to evaluate the degree of root caries.
Statistical analysis.
For the clinical sample detections, differences between the two groups were compared with t test or Kruskal-Wallis analysis after a homogeneity test of variance with Levene’s test. For the other experiments, differences among multiple groups were compared using one-way ANOVA and post hoc Tukey’s multiple comparisons after a homogeneity test of variance with Levene’s test, and two independent groups were analyzed with t test after the homogeneity of variance test. Statistical analysis was performed using SPSS software (Version 20.0; IBM Corp., Armonk, USA) with a significance level of 0.05, and then all figures were generated with GraphPad Prism7 software (version 7.00 for Windows; GraphPad Prism, Inc, La Jolla, USA).
Data availability.
RNA sequencing data have been deposited in the public database Sequence Read Archive with accession no. (PRJNA753272). All data sets generated and/or analyzed in the current study are available from the corresponding author on reasonable request.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (82071111, 81870778, 81600858, 81991500, 81991501), Key Research and Development Projects of Science and Technology Department of Sichuan Province (2020YFSY0019, 2021YFQ0064), and Applied Basic Research Programs of Sichuan Province (2020YJ0227).
We declare no conflicts of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Ling Zou, Email: zouling@scu.edu.cn.
Biao Ren, Email: renbiao@scu.edu.cn.
Teresa R. O'Meara, University of Michigan
REFERENCES
- 1.Anonymous. 2018. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 392:1789–1858. doi: 10.1016/S0140-6736(18)32279-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cai J, Palamara J, Manton DJ, Burrow MF. 2018. Status and progress of treatment methods for root caries in the last decade: a literature review. Aust Dent J 63:34–54. doi: 10.1111/adj.12550. [DOI] [PubMed] [Google Scholar]
- 3.Hayes M, Burke F, Allen PF. 2017. Incidence, prevalence and global distribution of root caries. Monogr Oral Sci 26:1–8. doi: 10.1159/000479301. [DOI] [PubMed] [Google Scholar]
- 4.Gao YB, Hu T, Zhou XD, Shao R, Cheng R, Wang GS, Yang YM, Li X, Yuan B, Xu T, Wang X, Feng XP, Tai BJ, Hu Y, Lin HC, Wang B, Si Y, Wang CX, Zheng SG, Liu XN, Rong WS, Wang WJ, Yin W. 2018. How root caries differs between middle-aged people and the elderly: findings from the 4th National Oral Health Survey of China. Chin J Dent Res 21:221–229. doi: 10.3290/j.cjdr.a41078. [DOI] [PubMed] [Google Scholar]
- 5.WHO Center for Health Development. 2006. 8020 better oral health for older people. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 6.Syed SA, Loesche WJ, Pape HL, Jr, Grenier E. 1975. Predominant cultivable flora isolated from human root surface caries plaque. Infect Immun 11:727–731. doi: 10.1128/iai.11.4.727-731.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellen RP, Banting DW, Fillery ED. 1985. Longitudinal microbiological investigation of a hospitalized population of older adults with a high root surface caries risk. J Dent Res 64:1377–1381. doi: 10.1177/00220345850640121001. [DOI] [PubMed] [Google Scholar]
- 8.Liu T, Gibbons RJ, Hay DI, Skobe Z. 1991. Binding of Actinomyces viscosus to collagen: association with the type 1 fimbrial adhesin. Oral Microbiol Immunol 6:1–5. doi: 10.1111/j.1399-302x.1991.tb00443.x. [DOI] [PubMed] [Google Scholar]
- 9.Komiyama K, Khandelwal RL, Heinrich SE. 1988. Glycogen synthetic and degradative activities by Actinomyces viscosus and Actinomyces naeslundii of root surface caries and noncaries sites. Caries Res 22:217–225. doi: 10.1159/000261109. [DOI] [PubMed] [Google Scholar]
- 10.Zhou Y, Cheng L, Liao B, Shi Y, Niu Y, Zhu C, Ye X, Zhou X, Ren B. 2021. Candida albicans CHK1 gene from two-component system is essential for its pathogenicity in oral candidiasis. Appl Microbiol Biotechnol 105:2485–2496. doi: 10.1007/s00253-021-11187-0. [DOI] [PubMed] [Google Scholar]
- 11.Bachtiar EW, Bachtiar BM. 2018. Relationship between Candida albicans and Streptococcus mutans in early childhood caries, evaluated by quantitative PCR. F1000Res 7:1645. doi: 10.12688/f1000research.16275.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du Q, Ren B, He J, Peng X, Guo Q, Zheng L, Li J, Dai H, Chen V, Zhang L, Zhou X, Xu X. 2021. Candida albicans promotes tooth decay by inducing oral microbial dysbiosis. ISME J 15:894–908. doi: 10.1038/s41396-020-00823-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beighton D, Ludford R, Clark DT, Brailsford SR, Pankhurst CL, Tinsley GF, Fiske J, Lewis D, Daly B, Khalifa N. 1995. Use of CHROMagar Candida medium for isolation of yeasts from dental samples. J Clin Microbiol 33:3025–3027. doi: 10.1128/jcm.33.11.3025-3027.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayer FL, Wilson D, Hube B. 2013. Candida albicans pathogenicity mechanisms. Virulence 4:119–128. doi: 10.4161/viru.22913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pereira D, Seneviratne CJ, Koga-Ito CY, Samaranayake LP. 2018. Is the oral fungal pathogen Candida albicans a cariogen? Oral Dis 24:518–526. doi: 10.1111/odi.12691. [DOI] [PubMed] [Google Scholar]
- 16.Lobo CIV, Rinaldi TB, Christiano CMS, De Sales Leite L, Barbugli PA, Klein MI. 2019. Dual-species biofilms of Streptococcus mutans and Candida albicans exhibit more biomass and are mutually beneficial compared with single-species biofilms. J Oral Microbiol 11:1581520. doi: 10.1080/20002297.2019.1581520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bamford CV, d’Mello A, Nobbs AH, Dutton LC, Vickerman MM, Jenkinson HF. 2009. Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect Immun 77:3696–3704. doi: 10.1128/IAI.00438-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diaz PI, Xie Z, Sobue T, Thompson A, Biyikoglu B, Ricker A, Ikonomou L, Dongari-Bagtzoglou A. 2012. Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect Immun 80:620–632. doi: 10.1128/IAI.05896-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, Zhou X, Liao B, Zhou Y, Cheng L, Ren B. 2021. The cross-kingdom interaction between Helicobacter pylori and Candida albicans. PLoS Pathog 17:e1009515. doi: 10.1371/journal.ppat.1009515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arzmi MH, Dashper S, Catmull D, Cirillo N, Reynolds EC, McCullough M. 2015. Coaggregation of Candida albicans, Actinomyces naeslundii and Streptococcus mutans is Candida albicans strain dependent. FEMS Yeast Res 15:fov038. doi: 10.1093/femsyr/fov038. [DOI] [PubMed] [Google Scholar]
- 21.Cavalcanti IM, Nobbs AH, Ricomini-Filho AP, Jenkinson HF, Del Bel Cury AA. 2016. Interkingdom cooperation between Candida albicans, Streptococcus oralis and Actinomyces oris modulates early biofilm development on denture material. Pathog Dis 74. [DOI] [PubMed] [Google Scholar]
- 22.Souza JGS, Bertolini M, Thompson A, Barão VAR, Dongari-Bagtzoglou A. 2020. Biofilm interactions of Candida albicans and mitis group streptococci in a titanium-mucosal interface model. Appl Environ Microbiol 86. doi: 10.1128/AEM.02950-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sobue T, Diaz P, Xu H, Bertolini M, Dongari-Bagtzoglou A. 2016. Experimental models of C. albicans-streptococcal co-infection. Methods Mol Biol 1356:137–152. doi: 10.1007/978-1-4939-3052-4_10. [DOI] [PubMed] [Google Scholar]
- 24.Cavalcanti YW, Wilson M, Lewis M, Del-Bel-Cury AA, da Silva WJ, Williams DW. 2016. Modulation of Candida albicans virulence by bacterial biofilms on titanium surfaces. Biofouling 32:123–134. doi: 10.1080/08927014.2015.1125472. [DOI] [PubMed] [Google Scholar]
- 25.Falsetta ML, Klein MI, Colonne PM, Scott-Anne K, Gregoire S, Pai CH, Gonzalez-Begne M, Watson G, Krysan DJ, Bowen WH, Koo H. 2014. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect Immun 82:1968–1981. doi: 10.1128/IAI.00087-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim D, Sengupta A, Niepa TH, Lee BH, Weljie A, Freitas-Blanco VS, Murata RM, Stebe KJ, Lee D, Koo H. 2017. Candida albicans stimulates Streptococcus mutans microcolony development via cross-kingdom biofilm-derived metabolites. Sci Rep 7:41332. doi: 10.1038/srep41332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fehrmann C, Jurk K, Bertling A, Seidel G, Fegeler W, Kehrel BE, Peters G, Becker K, Heilmann C. 2013. Role for the fibrinogen-binding proteins coagulase and Efb in the Staphylococcus aureus-Candida interaction. Int J Med Microbiol 303:230–238. doi: 10.1016/j.ijmm.2013.02.011. [DOI] [PubMed] [Google Scholar]
- 28.Hu Y, Niu Y, Ye X, Zhu C, Tong T, Zhou Y, Zhou X, Cheng L, Ren B. 2021. Staphylococcus aureus synergized with Candida albicans to increase the pathogenesis and drug resistance in cutaneous abscess and peritonitis murine models. Pathogens 10:1036. doi: 10.3390/pathogens10081036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng L, Zou L, Wu J, Liu H, Luo T, Zhou X, Li W, Ren B. 2019. Voriconazole inhibits cross-kingdom interactions between Candida albicans and Actinomyces viscosus through the ergosterol pathway. Int J Antimicrob Agents 53:805–813. doi: 10.1016/j.ijantimicag.2019.02.010. [DOI] [PubMed] [Google Scholar]
- 30.Deng L, Li W, He Y, Wu J, Ren B, Zou L. 2019. Cross-kingdom interaction of Candida albicans and Actinomyces viscosus elevated cariogenic virulence. Arch Oral Biol 100:106–112. doi: 10.1016/j.archoralbio.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 31.Wierichs RJ, Meyer-Lueckel H. 2015. Systematic review on noninvasive treatment of root caries lesions. J Dent Res 94:261–271. doi: 10.1177/0022034514557330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janus MM, Willems HM, Krom BP. 2016. Candida albicans in multispecies oral communities; a keystone commensal? Adv Exp Med Biol 931:13–20. doi: 10.1007/5584_2016_5. [DOI] [PubMed] [Google Scholar]
- 33.Sosroseno W, Herminajeng E, Bird P. 2015. The effect of immune status, age and genetic background on induction of oral tolerance to Actinomyces viscosus in mice. Biomed Pharmacother 70:294–298. doi: 10.1016/j.biopha.2014.12.039. [DOI] [PubMed] [Google Scholar]
- 34.Morse DJ, Wilson MJ, Wei X, Bradshaw DJ, Lewis MAO, Williams DW. 2019. Modulation of Candida albicans virulence in in vitro biofilms by oral bacteria. Lett Appl Microbiol 68:337–343. doi: 10.1111/lam.13145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turner SA, Ma Q, Ola M, Martinez de San Vicente K, Butler G. 2018. Dal81 regulates expression of arginine metabolism genes in Candida parapsilosis. mSphere 3. doi: 10.1128/mSphere.00028-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marbaniang CN, Gowrishankar J. 2012. Transcriptional cross-regulation between Gram-negative and Gram-positive bacteria, demonstrated using ArgP-argO of Escherichia coli and LysG-lysE of Corynebacterium glutamicum. J Bacteriol 194:5657–5666. doi: 10.1128/JB.00947-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gibbons GF, Howard DH. 1986. Arginine auxotrophs of Candida albicans deficient in argininosuccinate lyase. J Gen Microbiol 132:263–268. doi: 10.1099/00221287-132-2-263. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y, Shi H, Liang S, Ning G, Xu N, Lu J, Liu X, Lin F. 2015. MoARG1, MoARG5,6 and MoARG7 involved in arginine biosynthesis are essential for growth, conidiogenesis, sexual reproduction, and pathogenicity in Magnaporthe oryzae. Microbiol Res 180:11–22. doi: 10.1016/j.micres.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 39.Wu G, Bazer FW, Davis TA, Kim SW, Li P, Marc Rhoads J, Carey Satterfield M, Smith SB, Spencer TE, Yin Y. 2009. Arginine metabolism and nutrition in growth, health and disease. Amino Acids 37:153–168. doi: 10.1007/s00726-008-0210-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Novick RP, Maas WK. 1961. Control by endogenously synthesized arginine of the formation of ornithine transcarbamylase in Escherichia coli. J Bacteriol 81:236–240. doi: 10.1128/jb.81.2.236-240.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hartmann R, Sickinger HD, Oesterhelt D. 1980. Anaerobic growth of halobacteria. Proc Natl Acad Sci USA 77:3821–3825. doi: 10.1073/pnas.77.7.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tonon T, Lonvaud-Funel A. 2000. Metabolism of arginine and its positive effect on growth and revival of Oenococcus oeni. J Appl Microbiol 89:526–531. doi: 10.1046/j.1365-2672.2000.01142.x. [DOI] [PubMed] [Google Scholar]
- 43.Senouci-Rezkallah K, Schmitt P, Jobin MP. 2011. Amino acids improve acid tolerance and internal pH maintenance in Bacillus cereus ATCC14579 strain. Food Microbiol 28:364–372. doi: 10.1016/j.fm.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 44.Vrancken G, Rimaux T, Wouters D, Leroy F, De Vuyst L. 2009. The arginine deiminase pathway of Lactobacillus fermentum IMDO 130101 responds to growth under stress conditions of both temperature and salt. Food Microbiol 26:720–727. doi: 10.1016/j.fm.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 45.Huang S, Ai ZW, Sun XM, Liu GF, Zhai S, Zhang M, Chen H, Feng Z. 2016. Influence of arginine on the growth, arginine metabolism and amino acid consumption profiles of Streptococcus thermophilus T1C2 in controlled pH batch fermentations. J Appl Microbiol 121:746–756. doi: 10.1111/jam.13221. [DOI] [PubMed] [Google Scholar]
- 46.Namiki F, Matsunaga M, Okuda M, Inoue I, Nishi K, Fujita Y, Tsuge T. 2001. Mutation of an arginine biosynthesis gene causes reduced pathogenicity in Fusarium oxysporum f. sp. melonis. Mol Plant Microbe Interact 14:580–584. doi: 10.1094/MPMI.2001.14.4.580. [DOI] [PubMed] [Google Scholar]
- 47.Dutton LC, Paszkiewicz KH, Silverman RJ, Splatt PR, Shaw S, Nobbs AH, Lamont RJ, Jenkinson HF, Ramsdale M. 2016. Transcriptional landscape of trans-kingdom communication between Candida albicans and Streptococcus gordonii. Mol Oral Microbiol 31:136–161. doi: 10.1111/omi.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schröter C, Hipler UC, Wilmer A, Künkel W, Wollina U. 2000. Generation of reactive oxygen species by Candida albicans in relation to morphogenesis. Arch Dermatol Res 292:260–264. doi: 10.1007/s004030050484. [DOI] [PubMed] [Google Scholar]
- 49.Mayer FL, Wilson D, Jacobsen ID, Miramón P, Slesiona S, Bohovych IM, Brown AJ, Hube B. 2012. Small but crucial: the novel small heat shock protein Hsp21 mediates stress adaptation and virulence in Candida albicans. PLoS One 7:e38584. doi: 10.1371/journal.pone.0038584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dantas AdS, Day A, Ikeh M, Kos I, Achan B, Quinn J. 2015. Oxidative stress responses in the human fungal pathogen, Candida albicans. Biomolecules 5:142–165. doi: 10.3390/biom5010142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lorenz MC, Bender JA, Fink GR. 2004. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell 3:1076–1087. doi: 10.1128/EC.3.5.1076-1087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grandvaux N, Gaboriau F, Harris J, ten Oever BR, Lin R, Hiscott J. 2005. Regulation of arginase II by interferon regulatory factor 3 and the involvement of polyamines in the antiviral response. FEBS J 272:3120–3131. doi: 10.1111/j.1742-4658.2005.04726.x. [DOI] [PubMed] [Google Scholar]
- 53.Gheren LW, Cortelli JR, Rodrigues E, Holzhausen M, Saad WA. 2008. Periodontal therapy reduces arginase activity in saliva of patients with chronic periodontitis. Clin Oral Invest 12:67–72. doi: 10.1007/s00784-007-0146-8. [DOI] [PubMed] [Google Scholar]
- 54.Sun J, Wang D, Wang A, Wang Q. 2011. Study on the difference of arginase expression between normal and periodontitis gingival tissues (in Chinese). Contemporary Medicine 17. [Google Scholar]
- 55.Sun J, Wang A, Gao Y. 2012. Expression of arginase in periodontal tissues of experimental periodontitis in rats (in Chinese). Chin J Conserv Dent 22:445. [Google Scholar]
- 56.Kuriki N, Asahi Y, Sotozono M, Machi H, Noiri Y, Hayashi M, Ebisu S. 2021. Next-generation sequencing for determining the effect of arginine on human dental biofilms using an in situ model. Pharmacy (Basel) 9:18. doi: 10.3390/pharmacy9010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang X, Zhang K, Deng M, Exterkate RAM, Liu C, Zhou X, Cheng L, Ten Cate JM. 2017. Effect of arginine on the growth and biofilm formation of oral bacteria. Arch Oral Biol 82:256–262. doi: 10.1016/j.archoralbio.2017.06.026. [DOI] [PubMed] [Google Scholar]
- 58.Wolff MS, Schenkel AB. 2018. The anticaries efficacy of a 1.5% arginine and fluoride toothpaste. Adv Dent Res 29:93–97. doi: 10.1177/0022034517735298. [DOI] [PubMed] [Google Scholar]
- 59.Nascimento MM, Alvarez AJ, Huang X, Browngardt C, Jenkins R, Sinhoreti MC, Ribeiro APD, Dilbone DA, Richards VP, Garrett TJ, Burne RA. 2019. Metabolic profile of supragingival plaque exposed to arginine and fluoride. J Dent Res 98:1245–1252. doi: 10.1177/0022034519869906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bijle MNA, Ekambaram M, Lo EC, Yiu CKY. 2018. The combined enamel remineralization potential of arginine and fluoride toothpaste. J Dent 76:75–82. doi: 10.1016/j.jdent.2018.06.009. [DOI] [PubMed] [Google Scholar]
- 61.Agnello M, Cen L, Tran NC, Shi W, McLean JS, He X. 2017. Arginine improves pH homeostasis via metabolism and microbiome modulation. J Dent Res 96:924–930. doi: 10.1177/0022034517707512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mira De Orduña R, Patchett ML, Liu SQ, Pilone GJ. 2001. Growth and arginine metabolism of the wine lactic acid bacteria Lactobacillus buchneri and Oenococcus oeni at different pH values and arginine concentrations. Appl Environ Microbiol 67:1657–1662. doi: 10.1128/AEM.67.4.1657-1662.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ghosh S, Navarathna DH, Roberts DD, Cooper JT, Atkin AL, Petro TM, Nickerson KW. 2009. Arginine-induced germ tube formation in Candida albicans is essential for escape from murine macrophage line RAW 264.7. Infect Immun 77:1596–1605. doi: 10.1128/IAI.01452-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu B, Zhang W, Gou S, Huang H, Yao J, Yang Z, Liu H, Zhong C, Liu B, Ni J, Wang R. 2017. Intramolecular cyclization of the antimicrobial peptide Polybia-MPI with triazole stapling: influence on stability and bioactivity. J Pept Sci 23:824–832. doi: 10.1002/psc.3031. [DOI] [PubMed] [Google Scholar]
- 65.Lee C, Kim J, Shin SG, Hwang S. 2006. Absolute and relative QPCR quantification of plasmid copy number in Escherichia coli. J Biotechnol 123:273–280. doi: 10.1016/j.jbiotec.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 66.Hasan S, Danishuddin M, Khan AU. 2015. Inhibitory effect of zingiber officinale towards Streptococcus mutans virulence and caries development: in vitro and in vivo studies. BMC Microbiol 15:1. doi: 10.1186/s12866-014-0320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo J, Ma Z, Peng J, Mo J, Li Q, Guo J, Yang F. 2021. Transcriptomic analysis of Raphidocelis subcapitata exposed to erythromycin: the role of DNA replication in hormesis and growth inhibition. J Hazard Mater 402:123512. doi: 10.1016/j.jhazmat.2020.123512. [DOI] [PubMed] [Google Scholar]
- 68.Xu X, Zhou XD, Wu CD. 2011. The tea catechin epigallocatechin gallate suppresses cariogenic virulence factors of Streptococcus mutans. Antimicrob Agents Chemother 55:1229–1236. doi: 10.1128/AAC.01016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Doff RS, Rosen S, App G. 1977. Root surface caries in the molar teeth of Rice rats. I. A method for quantitative scoring. J Dent Res 56:1013–1016. doi: 10.1177/00220345770560080301. [DOI] [PubMed] [Google Scholar]
- 71.Wu T, Li B, Zhou X, Hu Y, Zhang H, Huang Y, Xu HHK, Guo Q, Li M, Feng M, Peng X, Weir MD, Cheng L, Ren B. 2018. Evaluation of novel anticaries adhesive in a secondary caries animal model. Caries Res 52:14–21. doi: 10.1159/000481832. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4; Tables S1 to S5. Download spectrum.00782-22-s0001.pdf, PDF file, 0.4 MB (489.3KB, pdf)
Data Availability Statement
RNA sequencing data have been deposited in the public database Sequence Read Archive with accession no. (PRJNA753272). All data sets generated and/or analyzed in the current study are available from the corresponding author on reasonable request.