Abstract
Organisms such as Saccharomyces capable of utilizing several different sugars selectively ferment glucose when less desirable carbon sources are also available. This is achieved by several mechanisms. Glucose down-regulates the transcription of genes involved in utilization of these alternate carbon sources. Additionally, it causes posttranslational modifications of enzymes and transporters, leading to their inactivation and/or degradation. Two glucose sensing and signaling pathways stimulate glucose-induced inactivation of maltose permease. Pathway 1 uses Rgt2p as a sensor of extracellular glucose and causes degradation of maltose permease protein. Pathway 2 is dependent on glucose transport and stimulates degradation of permease protein and very rapid inactivation of maltose transport activity, more rapid than can be explained by loss of protein alone. In this report, we characterize signal generation through pathway 2 using the rapid inactivation of maltose transport activity as an assay of signaling activity. We find that pathway 2 is dependent on HXK2 and to a lesser extent HXK1. The correlation between pathway 2 signaling and glucose repression suggests that these pathways share common upstream components. We demonstrate that glucose transport via galactose permease is able to stimulate pathway 2. Moreover, rapid transport and fermentation of a number of fermentable sugars (including galactose and maltose, not just glucose) are sufficient to generate a pathway 2 signal. These results indicate that pathway 2 responds to a high rate of sugar fermentation and monitors an intracellular metabolic signal. Production of this signal is not specific to glucose, glucose catabolism, glucose transport by the Hxt transporters, or glucose phosphorylation by hexokinase 1 or 2. Similarities between this yeast glucose sensing pathway and glucose sensing mechanisms in mammalian cells are discussed.
Glucose is a global metabolic regulator in Saccharomyces that controls the expression of many genes involved in carbohydrate utilization, gluconeogenesis, mitochondrial biogenesis, and cell cycle regulation (reviewed in references 15, 24, and 25). In part, this regulation is achieved at the level of transcription, causing induction or repression of different sets of genes via multigene transcription regulators such as Mig1p and Rgt1p. Glucose also acts at the posttranslational level by decreasing the activity and stability of certain target proteins. The overall effect of these glucose-regulated processes is to speed the transition from utilization of alternate carbon sources, such as maltose, galactose, sucrose, and ethanol, to the fermentation of glucose. We are interested in identifying the mechanisms by which glucose regulates MAL gene expression. One such mechanism is glucose-induced inactivation of maltose permease (23, 31, 39).
Maltose permease is required for maltose transport and MAL gene induction (6, 7). Maltose permease is subject to glucose-induced inactivation. Addition of glucose to maltose-fermenting cells causes an initial very rapid loss of maltose transport activity that is faster than can be explained by loss in maltose permease protein alone (31). This is followed by a slower decrease in maltose transport activity that correlates with the proteolysis of maltose permease protein (31, 39). Proteolysis of maltose permease requires ubiquitination and endocytosis of the permease and degradation by the vacuolar proteases (31, 32, 39). The mechanism of rapid inactivation is as yet unknown, but it is distinct from the mechanism of proteolysis, since it does not require ubiquitination or endocytosis and occurs in end3 and doa4 mutant strains (31, 32).
Several glucose sensing and signaling pathways have been identified in Saccharomyces (reviewed in references 3, 24, 25, 29, 38, and 49). Snf3p and Rgt2p are integral membrane proteins that act as glucose receptors responding to low and high extracellular glucose concentrations, respectively, to regulate induction of HXT genes encoding glucose transporters (34, 35, 36). Downstream effectors of the Snf3p- and Rgt2p-dependent signaling pathway are largely unidentified, except for Grr1p, an F-box protein (27, 35). Snf1 protein kinase is required for the derepression of genes for the utilization of alternate energy sources such as maltose, galactose, sucrose, and nonfermentable carbon sources. Snf1p is the catalytic subunit of a large multiprotein complex whose activity is down-regulated in response to high rates of glucose fermentation (reviewed in references 5 and 24). It is the central regulatory component in the glucose repression pathway. HXK2, protein phosphatase type 1 (encoded by GLC7 and REG1), and other upstream components of the glucose repression pathway are responsible for the inactivation of Snf1 kinase in high glucose (24, 30, 44). The most recently identified glucose sensing and signaling pathway regulates pseudohyphal differentiation. Gpr1p is a G-protein-coupled receptor that is suggested to respond to extracellular glucose levels and signal via Gpa2p, a Gα subunit, and protein kinase A to stimulate pseudohyphal differentiation (29, 38, 49).
We demonstrated that two glucose sensing and signaling pathways stimulate glucose-induced inactivation of maltose permease (23). Rgt2p was found to function as the glucose sensor in pathway 1, the glucose transport-independent pathway. Pathway 1 stimulates proteolysis of maltose permease but not the very rapid inactivation of transport activity (23). Pathway 2 was shown to be dependent on high rates of glucose transport and thus is similar to the glucose repression pathway. Pathway 2 stimulates proteolysis and the rapid inactivation of transport activity noted above. In this report, we explore glucose signal generation in pathway 2 and relate this to the glucose repression pathway. We show that HXK2 is an upstream regulator of pathway 2. Our results provide evidence that the initial steps of sugar fermentation, including transport and phosphorylation, function as metabolic gatekeepers and generate an intracellular signal that stimulates pathway 2. Moreover, in addition to glucose, other fermentable sugars, even maltose, are able to trigger the inactivation of maltose permease and repression. Thus, pathway 2 and the glucose repression pathway share upstream components. Moreover, the intracellular metabolic signal that stimulates both pathways is not specific to glucose. It can be generated by the rapid transport and fermentation of any one of a number of sugars, even those utilizing transporters and/or catabolic pathways different than those used by glucose.
MATERIALS AND METHODS
Strains and plasmids.
The Saccharomyces strains used in this study are listed in Table 1. Strain CMY1001 was derived from strain 100-1A (MATa mal11Δ::URA3 MAL12 MAL13 leu2-3,112 ura3-52) (6) by two-step gene replacement of the mal11Δ::URA3 with the hemagglutinin (HA)-tagged MAL61/HA maltose permease gene as described in Jiang et al. (23) and Medintz et al. (31). Strains CMY1001 (wild type), CMY1005 (grr1Δ), and CMY1006 (hxk2Δ) are isogenic and were described in detail in previous publications (23, 31).
TABLE 1.
Effect of various fermentable carbon sources on expression of maltose transport and maltasea
| Strain | Carbon source | Maltose transport activity (nmol/mg [dry wt]/min) | Maltase activity (nmol of PNPG/mg of protein/min) |
|---|---|---|---|
| CMY1001 | Maltose | 6.38 | 603 |
| CMY1001 | Glucose | 0.04 | <1 |
| CMY1001 | Galactose | 0.07 | 8 |
| CMY1001 | Fructose | 0.02 | <1 |
| CMY1001 | Mannose | 0.09 | 1 |
| CMY1001 | Maltose plus glucose | 0.04 | 3 |
| CMY1001 | Maltose plus fructose | 0.08 | 20 |
| CMY1001 | Maltose plus mannose | 0.41 | 69 |
| CMY1001 | Maltose plus galactose | 5.14 | 588 |
| CMY1001(pADH1-GAL2) | Maltose | 2.36 | 313 |
| CMY1001(pADH1-GAL2) | Maltose plus galactose | 0.75 | 146 |
Strain CMY1001 was grown in rich medium supplemented with 2% of various fermentable carbon sources (as indicated in the table). When two different carbon sources were used in combination, the concentration of each one was 2%. CMY1001 transformed with plasmid pADH1-GAL2 was grown in selective medium lacking uracil and containing the indicated carbon sources at concentrations of 2%. Cells were grown to early log phase (OD600 of about 0.3), harvested, and the steady-state maltose transport and maltase activities were determined as described in Materials and Methods. PNPG, p-nitrophenyl-β-d-glucoside.
Strains CMY1014 and CMY1015 are derived from CMY1006. In strain CMY1014 the open reading frame of GLK1 was replaced by HIS3, and in strain CMY1015 the open reading frame of HXK1 was replaced by TRP1 by PCR-based gene disruption. The primers for disrupting HXK1 are primer 1, 5′TAAGAAA CAATTGTGGCTTGCAATACTCAATTAGAATTCTTTTCTTTTAATCAA GCAGATTGTACTGAGAGTGC3′, and primer 2, 5′CGTAATTGGATCTTT GCTTGCGTCACCAGTCCATCCATAGATATCTCTCAATGCCGATTTCG GCCTATTGG3′. The primers for disrupting GLK1 are primer 3, 5′CCGCTA TCAACAGAACCCCAACCCCCCCATCAGTGCCAACTCAGCTTCCCTTG GTGAGCGCTAGGAG3′, and primer 4, 5′TCAGCTCAACGCCACAGGCG GCACCACTCCGGAACCATCCTGGCCACACCGCATAGATCCGTCG3′. Strain CMY1013 is derived from strain CMY1001. HXK1 was disrupted with primers 1 and 2, and GLK1 was disrupted with primers 3 and 4. Each gene disruption was verified by Southern analysis. Strain 100-1B is isogenic to strain 100-1A, except at the MAL1 locus, and has the genotype MAL11 mal12Δ::LEU2 MAL13 (6).
Plasmids pADH1-GAL2 and pHXT1-LacZ were obtained from Sabire Ozcan and Mark Johnston. Plasmid pADH1-GAL2 is a URA3 2μm plasmid containing the GAL2 coding region under the control of the constitutive ADH1 promoter (S. Ozcan, personal communication). Plasmids pRS405-MAL61/HA and pMAL61/HA carry the HA-tagged MAL61/HA maltose permease gene (described in reference 31) in plasmid vectors pRS405 and pRS416, respectively (42).
Inactivation assay protocol.
The maltose permease inactivation assay protocol was described in detail by Medintz et al. (31). Strains were grown at 30°C to very early log phase (optical density at 600 nm [OD600] of 0.3) in either rich medium or selective medium lacking the appropriate nutrient for plasmid selection, supplemented with 2% maltose or 2% maltose plus 2% galactose (as indicated below), with the following exception. Strain 100-1B(pMAL61/HA) does not grow on maltose as the sole carbon source. Thus, strains 100-1A(pMAL61/HA) and 100-1B(pMAL61/HA) were pregrown on selective medium containing 2% galactose to very early log phase, at which time 2% maltose was added to the medium in order to induce the expression of the MAL genes.
Cells were harvested and transferred to nitrogen-starvation medium (yeast nitrogen base without amino acids and ammonium sulfate) plus 2% glucose (or another sugar as indicated) (32). Cell samples were taken at the times indicated below over a 3-h period, and for each sample, maltose transport rates were determined and total cell extracts were prepared for Western analysis. Growth dilution at any given time was calculated as the OD600 at time zero divided by the OD600 at the given time X).
Western blot analysis.
Western blot analysis was carried out as described by Medintz et al. (31). The protein concentration of total cell extract was determined using the protein assay kit from Sigma. The Mal61/HA protein in the extracts was detected using anti-HA specific antibody and the ECL Western blotting kit (Amersham). The relative amount of each band on the ECL-Hyperfilm was measured by densitometric comparison to the zero-time sample. Western analysis was done in duplicate from duplicate cell cultures.
Sugar transport assays.
Maltose transport was measured as the uptake of 1 mM 14C-maltose (described in references 9 and 32). Similar methods were used to measure the uptake of 14C-glucose, with the exception that the substrate concentration was varied from 0.2 mM to 10 mM in order to determine the Km of glucose transport for the maltose- or maltose-plus-galactose-grown cells. Assays were done in duplicate cultures.
Maltase assays.
Maltase activity was determined as described by Dubin et al. (10). The values reported are the average of duplicate assays obtained by using extracts from at least duplicate cultures. Standard errors were less than 20%.
RESULTS
HXK2 is an important regulator of pathway 2.
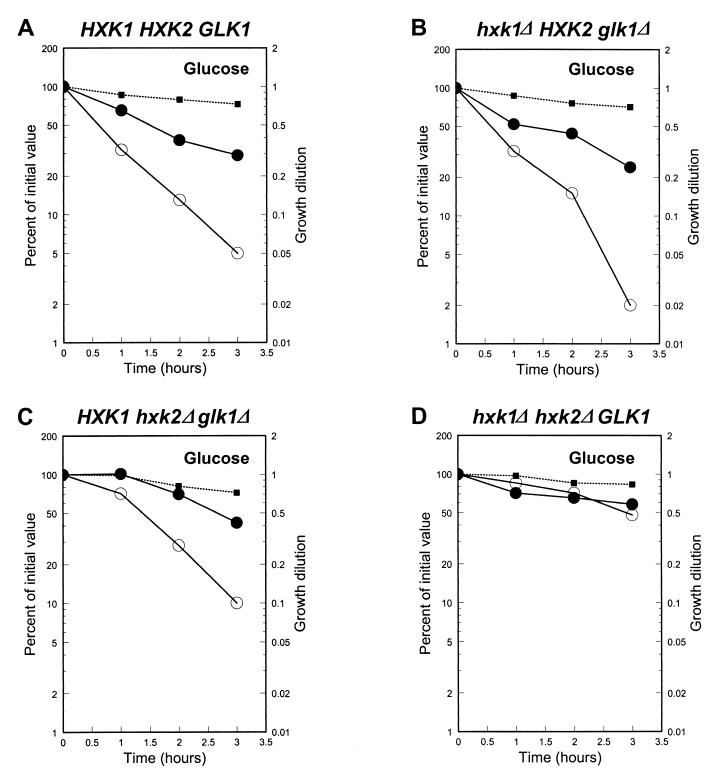
HXK1, HXK2, and GLK1 encode Saccharomyces cerevisiae hexokinase 1, hexokinase 2, and glucokinase, respectively (reviewed in reference 25). These enzymes catalyze the first step in glycolysis and reportedly are involved in high-affinity glucose transport (1, 2, 43). HXK2 is an upstream negative regulator of the glucose repression pathway (reviewed in reference 25). To test if HXK2 also is a regulator of pathway 2, we constructed a series of deletion mutations by PCR-based methods, creating a series of isogenic strains, each expressing only one of these three kinase genes, and assayed glucose-induced inactivation of maltose permease. The results are shown in Fig. 1. It is important to note that pathway 1, the Rgt2p-dependent pathway, is active in this strain series, but this did not interfere with our analysis. Pathway 1 causes only degradation of maltose permease and not rapid inactivation of maltose transport activity (23). Thus, activation of pathway 2 can be determined by comparing the rate of loss of transport activity to the loss in permease protein. If the two are coincident, signaling through pathway 2 is inactive.
FIG. 1.
Effect of HXK1, HXK2, and GLK1 mutations on glucose-induced inactivation of maltose permease. Strains CMY1001 (HXK1 HXK2 GLK1) (A), CMY1013 (hxk1Δ HXK2 glk1Δ) (B), CMY1014 (HXK1 hxk2Δ glk1Δ) (C), and CMY1015 (hxk1Δ hxk2Δ GLK1) (D) were grown in rich medium supplemented with 2% maltose plus 2% galactose. Standard inactivation assays were carried out as described in Materials and Methods (31) on early-log-phase cells. The relative levels of Mal61/HAp protein (●), maltose transport (○) and growth dilution (■) are plotted in the same panel. The relative protein level and transport activity at time X are compared to the corresponding values at time zero. Growth dilution was calculated as the OD600 at time zero divided by the OD600 at time X. Maltose transport activities (in nmol/mg [dry weight]/min) at time zero for the strains and growth conditions of this experiment were as follows: for CMY1001, 5.43; for CMY1013, 6.31; for CMY1014, 5.56; and for CMY1015, 6.47.
Glk1p is not sufficient to induce inhibition of maltose permease (Fig. 1D). The hxk1 hxk2 GLK1 strain exhibits no rapid inactivation of maltose transport activity. Instead, the slow loss in transport activity is paralleled by the loss in maltose permease protein. In contrast, glucose-induced inactivation of maltose permease in the strain expressing only Hxk2p is comparable to that observed in the HXK1 HXK2 GLK1 strain. In particular, the rate of inactivation of maltose transport activity is as rapid as or possibly more rapid than that in the wild-type strain. Thus, HXK2 expression alone is sufficient to produce full levels of signaling through pathway 2. Taken together, these results indicate that HXK2 is an important regulator of pathway 2 signal generation.
In the absence of HXK2, HXK1 is able to stimulate rapid inactivation of maltose transport but not to the same extent as HXK2. Similar results were reported regarding the effects of hxk2Δ and hxk1Δ mutations on glucose repression of maltase gene expression (21). Loss of Hxk2p significantly relieves glucose repression of maltase, while loss of Hxk1p alone has little effect. Residual glucose repression is observed in the hxk2Δ mutant, but this is relieved by deletion of HXK1. This finding is consistent with reports on HXK1 and HXK2 expression patterns that demonstrate increased HXK1 expression in hxk2 mutants (9, 20).
Overexpression of GAL2 suppresses the resistance to glucose-induced inactivation of grr1Δ.
We wished to test whether the HXT-encoded transporters are essential for signaling via pathway 2 or if rapid glucose entry mediated by another transporter is sufficient. grr1Δ mutant strains are completely resistant to glucose-induced inactivation of maltose permease because both glucose sensing and signaling pathways 1 and 2 are absent (23) (Fig. 2A). Loss of Grr1p blocks the Rgt2p-dependent signaling pathway 1 and causes a dramatic decrease in HXT gene expression and glucose transport (23, 34). GAL2 encodes the S. cerevisiae galactose transporter which recent studies demonstrate is able to transport glucose, albeit with lower affinity (28, 33). Plasmid pADH1-GAL2, a high-copy plasmid expressing GAL2 from the constitutive ADH1 promoter, was introduced into a grr1Δ mutant strain to determine if glucose transport via Gal2p is able to stimulate pathway 2.
FIG. 2.
Effect of overexpression of GAL2 on glucose-induced inactivation of maltose permease in maltose-grown grr1Δ strain. Strain CMY1005 (grr1Δ) was transformed with either the URA3 vector (pRS416) (A) or the GAL2 overexpression plasmid pADH1-GAL2 (B). Strains were grown in selective medium lacking uracil and containing 2% maltose, and the standard inactivation assay protocol was carried out (as discussed in the legend to Fig. 1). Maltose transport activities (in nmol/mg [dry weight]/min) at time zero were 2.31 for strain CMY1005(pRS416) and 1.90 for strain CMY1005(pADH1-GAL2). ●, Mal61/HAp protein; ○, maltose transport; ■, growth dilution.
Overexpression of GAL2 partially restores glucose transport in the grr1Δ strain (from a Vmax of 2.9 to 7.7 nmol/mg [dry weight]/min) but not to levels observed in the GRR1 strain (19.2 nmol/mg [dry weight]/min). This Gal2p-mediated glucose transport also partially restores glucose-induced proteolysis of maltose permease, and rapid inactivation of maltose transport activity is observed (Fig. 2B). Since pathway 1 is completely blocked in this grr1Δ strain, signaling is entirely via pathway 2. Thus, glucose transport by an HXT hexose transporter is not essential for stimulating pathway 2-dependent inactivation.
Galactose is capable of stimulating inactivation of maltose permease and repression of maltase expression.
Galactose is generally considered to be a nonrepressing sugar. We wished to test its ability to stimulate inactivation of maltose permease. Our parental strain CMY1001 grows slowly on galactose because galactose transport rates are low (data not shown), and, like many laboratory strains, it carries a gal2 mutant allele. When pregrown in medium containing only maltose or maltose plus galactose, galactose is not able to stimulate inactivation of maltose permease (data not shown).
Plasmid pADH1-GAL2 was introduced into CMY1001 to allow high-level expression of galactose permease even in the absence of galactose. Strain CMY1001(pADH1-GAL2) was grown in medium containing either maltose alone or maltose plus galactose to induce expression of the GAL structural genes (GAL1, GAL7, and GAL10) (reviewed in reference 25). In maltose-grown CMY1001(pADH1-GAL2) cells, galactose does not stimulate inactivation of maltose permease (Fig. 3A). However, when strain CMY1001(pADH1-GAL2) is grown on galactose plus maltose, galactose is capable of stimulating very rapid inactivation of maltose transport and maltose permease proteolysis (Fig. 3B). Additionally, in strain CMY1001(pADH1-GAL2) galactose represses maltose-induced expression of maltose transport activity and maltase (Table 1) but only in maltose-plus-galactose-grown cells and only when GAL2 is overexpressed. Thus, under conditions where galactose is both rapidly transported and metabolized, galactose is a repressing sugar and stimulates inactivation of maltose permease.
FIG. 3.
Induction of the galactose utilization genes is required for galactose to stimulate inactivation of maltose permease. Strain CMY1001 was transformed with the GAL2 overexpression plasmid pADH1-GAL2. Transformants were grown in selective medium lacking uracil and supplemented with 2% maltose (A) or 2% maltose plus 2% galactose (B). Inactivation assays performed as described in Methods and Materials (Fig. 1) except 2% galactose was used instead of glucose to induce inactivation. Maltose transport activities (in nmol/mg [dry weight]/min) at time zero for strain CMY1001(pADH1-GAL2) were 1.90 (maltose grown) and 0.61 (maltose plus galactose grown). ●, Mal61/HAp protein; ○, maltose transport; ■, growth dilution.
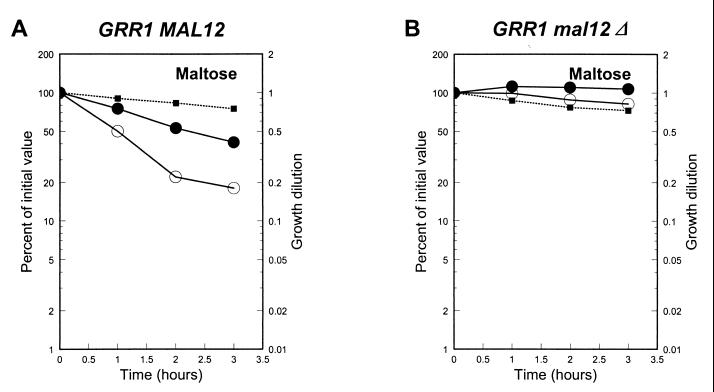
Galactose-induced inactivation of maltose permease was assayed in grr1Δ and grr1Δ(pADH1-GAL2) strains. In maltose-plus-galactose-grown grr1Δ(pADH1-GAL2), galactose-induced inactivation of maltose permease was comparable to that seen in the GRR1 strain carrying plasmid pADH1-GAL2 and grown under the same conditions (data not shown). Since Hxt expression is very low in grr1Δ strains and pathway 1 is blocked (23), this signal must entirely result from rapid galactose transport by Gal2p and rapid catabolism of galactose by the galactose fermentation enzymes. Taken together, these results indicate that rapid fermentation of galactose is sufficient to produce a potent signal for maltose permease inactivation, but that galactose transport in the absence of catabolism is not.
Maltose weakly signals inactivation of maltose permease.
Inactivation assays are carried out in nitrogen-starvation medium that blocks protein synthesis, including synthesis of maltose permease. Thus, we were able to test the possibility that maltose itself may stimulate inactivation of maltose permease. Strain 100-1A contains the MAL1 locus and no other MAL genes (6). This strain was transformed with a plasmid carrying the HA epitope-tagged maltose permease gene MAL61/HA to enable us to follow permease protein levels. The results shown in Fig. 4A demonstrate that maltose stimulates proteolysis of maltose permease and inactivation of maltose transport. Deletion of GRR1 has little effect on this maltose-stimulated inactivation of maltose permease (data not shown), indicating that neither Rgt2p-dependent signaling (pathway 1) nor glucose transport-dependent signaling (pathway 2) is required for maltose-induced inactivation of maltose permease.
FIG. 4.
Maltase is required for maltose-stimulated inactivation of maltose permease. Strains 100-1A(pRS405/MAL61/HA) (MAL12) (A) and 100-1B(pMAL61/HA) (mal12Δ) (B) were grown in selective medium lacking uracil and containing 2% galactose to early log phase (OD600, 0.1), and then 2% maltose was added to the culture to induce MAL gene expression for another 6 h. Cells were harvested and transferred to nitrogen-starvation medium containing 2% maltose to stimulate inactivation of maltose permease. Maltose transport activities (in nmol/mg [dry weight]/min) at time zero for the strains and growth conditions of this experiment were 4.15 for strain 100-1A(pRS405/MAL61/HA) and 0.55 for 100-1B(pMAL61/HA). ●, Mal61/HAp protein; ○, maltose transport; ■, growth dilution.
Maltase, encoded by MAL12 in strains carrying the MAL1 locus, catalyzes the first step of maltose metabolism, that is, hydrolysis of maltose into two molecules of glucose. To determine if intracellular glucose production is required for maltose-stimulated inactivation of maltose permease, we used strain 100-1B, which is isogenic to strain 100-1A except that it contains a deletion of MAL12. We found that loss of maltase results in complete insensitivity to maltose-induced inactivation of maltose permease (Fig. 4B) without affecting glucose-induced inactivation (data not shown). Thus, the ability of maltose to induce inactivation of maltose permease is dependent on the intracellular production of glucose by maltose hydrolysis. Maltose transport per se is not the inactivation signal.
Effects of other carbon sources on maltase expression and maltose permease inactivation.
Fructose and mannose inhibit maltose induction of MAL structural gene expression, but the effectiveness of these hexoses is significantly less than that of glucose (Table 1). Both hexoses are transported by the HXT-encoded transporters and phosphorylated by the HXK-encoded hexokinases but with lower affinity than glucose (reviewed in reference 3). In addition, we found that fructose and mannose also stimulate rapid inhibition of maltose transport activity but to different extents (data not shown). Fructose is as potent an inducer as glucose, but mannose is relatively ineffective. Neither fructose nor mannose stimulates significant proteolysis of maltose permease. Their potency as inducers of inactivation compared to glucose correlates with their decreased affinity for the hexose transporters and phosphorylation enzymes.
Our previous results showed that ethanol is not able to stimulate inactivation of maltose permease (31). Several other nonfermentable carbon sources, including glycerol, lactate, and acetate, were also tested. None were found to stimulate inactivation of maltose permease (data not shown).
DISCUSSION
Our results indicate that glucose sensing and signaling pathway 2 overlaps upstream with the glucose repression pathway, a key component of which is Snf1 protein kinase (reviewed in references 5 and 24). Both pathways are dependent on high rates of glucose transport (23, 24). We show here that HXK2 is an important regulator of pathway 2 (Fig. 1). Moreover, comparison of the results presented in Table 1 and Fig. 2 and 3 demonstrates a clear correlation between the ability of various sugars to repress MAL gene expression and the ability to induce maltose permease inactivation. In results to be reported elsewhere, members of our group demonstrate that REG1 is a negative regulator of pathway 2 (H. Jiang, S. Liu, K. Tatchell, and C. A. Michels, submitted for publication). Reg1p binds to Glc7p, the catalytic subunit of protein phosphatase type 1, and directly controls Snf1p kinase activity in response to glucose (5, 24). Thus, REG1 is an important negative regulator of both glucose repression and pathway 2. Recently, Sherwood and Carlson (40, 41), described GSF2, a negative regulator of glucose repression that acts upstream of Snf1 kinase. A gsf2Δ strain (constructed by P. Sherwood in strain CMY1001) exhibited no rapid glucose-induced inactivation of maltose transport activity, suggesting that Gsf2p also is a negative regulator of pathway 2 (data not shown). In summary, our studies indicate that the upstream sensing and signaling components of pathway 2 are shared with the glucose repression pathway.
Several lines of evidence presented here indicate that rapid sugar fermentation is required to generate the pathway 2 signal stimulating inactivation of maltose permease. This is also true of the glucose repression pathway. We found that in grr1Δ mutant strains, which are deficient in high-glucose signal production through both pathways 1 and 2, overproduction of galactose permease partially restores glucose transport and glucose-induced inactivation of maltose permease as well. Thus, glucose transport via the HXT-encoded high-affinity glucose transporters is not essential, and other transport proteins can substitute so long as the Vmax of glucose transport is sufficiently high. We found that inactivation of maltose permease can be stimulated by galactose, usually considered a nonrepressing sugar, but that galactose-induced inactivation requires not only overexpression of the galactose transporter but also induction of the other GAL genes (Fig. 3). This suggests that galactose transport alone is not sufficient for signaling and that utilization is also required. It is important to note that, galactose utilization bypasses the hexokinases. Thus, Hxk1p and Hxk2p do not have special sensing and signaling functions beyond their enzymatic role in glycolysis.
Taken together, our findings suggest that the signal that stimulates pathway 2 results from a high rate of utilization of any of several fermentable sugars and does not specifically require Hxt-dependent transport or hexokinase-dependent phosphorylation. Rather, metabolic flux through the early steps of fermentation (transport and phosphorylation) appears to be the significant and controlling factor for all of the sugars tested. The hexokinases (and glucokinase) appear to function as glucose sensors in glucose-induced inactivation of maltose permease because of their pivotal role in controlling the metabolic flux through the initial steps of glycolysis. This control is achieved indirectly by regulating the rate of facilitated glucose transport as a result of regulating the rate of glucose phosphorylation (43).
The exact nature of the signal generated for pathway 2 activation appears to be similar to or the same as that for the glucose repression pathway. A working model for the mechanism of repression and derepression of genes in S. cerevisiae in response to the availability of glucose has been proposed in which the AMP-to-ATP ratio might act as a signal for glucose repression in the Snf1 protein kinase signal transduction pathway (48). Wilson et al. (48) suggest that high ATP levels (such as those which result from rapid fermentation of glucose, fructose, mannose, or galactose) could decrease AMP levels via an adenylate kinase reversible reaction generating two ADP molecules from AMP and ATP. Changes in the AMP-to-ATP ratio might be an alternate candidate for this second messenger. This same signal could also regulate pathway 2.
Sugar transport and sugar phosphorylation have been implicated in glucose sensing in mammalian pancreatic α- and β-cells (3, 4, 13, 16, and 19; reviewed in reference 24) as well as in the Saccharomyces glucose repression pathway, but again neither the signal molecule nor the sensor have been identified. It is interesting to note that yeast hexokinase 2 functions in the glucose-sensing pathway in pancreatic β-cells of transgenic mice (12, 47), suggesting some common mechanisms. Moreover, Snf1p and Snf4p exhibit homology to components of the AMP-activated kinase of mammalian cells, yet the yeast enzyme has not been shown to respond directly to AMP in vitro (18, 48). GLUT2 encodes a mammalian low-affinity glucose transporter and is thought to play a permissive role in controlling glucose metabolism. In pancreatic β-cells, underexpression of GLUT2 results in decreased insulin secretion in response to increased glucose concentrations (reviewed in references 3 and 45). AtT-20ins cells are derived from anterior pituitary cells that express high-affinity glucose transporter GLUT1. These cells can secrete insulin but not in response to glucose. When transfected with GLUT2 cDNA, AtT-20ins cells become glucose responsive (22). However, the role of GLUT2 in glucose sensing remains controversial. In normal β-cells, glucose transport capacity is in 100-fold excess compared to the capacity for glucose phosphorylation and catabolism, and phosphorylation, which is catalyzed predominantly by low-affinity glucokinase, appears to be the rate-limiting step of glycolysis (reviewed in reference 11). Accumulating evidence suggests an important role for pancreatic cell glucokinase in glucose sensing. There is a correlation between glucokinase levels and the ability to secrete insulin in response to glucose (reviewed in reference 3). Glucokinase inhibitors also inhibit glucose-stimulated insulin secretion (26), and mutations in glucokinase result in maturity onset diabetes of the young (8, 13, 14, 17, 46). In glucagon-producing pancreatic α-cells, glucokinase also may serve as a glucose sensor and mediate glucagon release in response to extracellular glucose concentration (18). Our results reported here suggest a homologous role for hexokinase in Saccharomyces, particularly hexokinase 2, in glucose sensing by pathway 2 in the inactivation of maltose permease.
ACKNOWLEDGMENTS
We thank Mark Johnston and Sabire Ozcan for providing strains and plasmids and for helpful discussions and critical reading of the manuscript. We thank Peter Sherwood for the construction of the GSF2Δ of strain CMY1001.
This work was supported by a grant from the National Institute of General Medical Sciences (GM49280) to C.A.M. and was carried out in partial fulfillment of the requirements for the Ph.D. degree from the Graduate School of the City University of New York (H.J. and I.M.).
REFERENCES
- 1.Bisson L F, Fraenkel D G. Involvement of kinases in glucose and fructose uptake by Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1983;80:1730–1734. doi: 10.1073/pnas.80.6.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bisson L F, Fraenkel D G. Transport of 6-deoxyglucose in Saccharomyces cerevisiae. J Bacteriol. 1983;155:995–1000. doi: 10.1128/jb.155.3.995-1000.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bisson L F, Coons D M, Krucheberg A L, Lewis D A. Yeast sugar transporters. Crit Rev Biochem Mol Biol. 1993;28:259–308. doi: 10.3109/10409239309078437. [DOI] [PubMed] [Google Scholar]
- 4.Byrne M M, Sturis J, Fajans S S, Ortiz F J, Stoltz A, Stoffel M, Smith M J, Bell G I, Halter J B, Polonsky K S. Altered insulin secretory responses to glucose in subjects with a mutation in the MODY gene on chromosome 20. Diabetes. 1995;44:699–704. doi: 10.2337/diab.44.6.699. [DOI] [PubMed] [Google Scholar]
- 5.Carlson M. Glucose repression in yeast. Curr Opin Microbiol. 1999;2:202–207. doi: 10.1016/S1369-5274(99)80035-6. [DOI] [PubMed] [Google Scholar]
- 6.Charron M J, Dubin R A, Michels C A. Structural and functional analysis of the MAL1 locus of Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:3891–3899. doi: 10.1128/mcb.6.11.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng Q, Michels C A. MAL11 and MAL61 encode the inducible high-affinity maltose transporter of Saccharomyces cerevisiae. J Bacteriol. 1991;173:1812–1820. doi: 10.1128/jb.173.5.1817-1820.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devchand P R, Keller H, Peters J M, Vazquez M, Gonzalez F J, Wahli W. The PPARalpha-leukotriene B4 pathways to inflammation control. Nature. 1996;384:39–43. doi: 10.1038/384039a0. [DOI] [PubMed] [Google Scholar]
- 9.De Winde J H, Crauwels M, Hohmann S, Winderickx J. Differential requirement of the yeast sugar kinases for sugar sensing in establishing the catabolite-repressed state. Eur J Biochem. 1996;241:633–643. doi: 10.1111/j.1432-1033.1996.00633.x. [DOI] [PubMed] [Google Scholar]
- 10.Dubin R A, Needleman R B, Gossett D, Michels C A. Identification of the structural gene encoding maltase within the MAL6 locus of Saccharomyces carlbergensis. J Bacteriol. 1985;164:605–610. doi: 10.1128/jb.164.2.605-610.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Efrat S, Tal M, Lodish H F. The pancreatic β-cell glucose sensor. Trends Biochem Sci. 1994;19:535–538. doi: 10.1016/0968-0004(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 12.Epstein P N, Boschero A C, Atwater I, Cai X, Overbeek P A. Expression of yeast hexokinase in pancreatic β cells of transgenic mice reduces blood glucose, enhances insulin secretion, and decreases diabetes. Proc Natl Acad Sci USA. 1992;89:12038–12042. doi: 10.1073/pnas.89.24.12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Froguel P, Vaxillaire M, Sun F, Velho G, Zouali H, Butel M O, Lesage S, Vionnet N, Clement K, Fougerousse F, Tanizawa Y, Weissenbach J, Beckman J S, Passa G M, Permutt M A, Cohen D. Close linkage of glucokinase locus on chromosome 7p to early-onset non-insulin-dependent diabetes mellitus. Nature. 1992;356:162–164. doi: 10.1038/356162a0. [DOI] [PubMed] [Google Scholar]
- 14.Froguel P, Zouali H, Vionnet N, Velho G, Vaxillaire M, Sun F, Lesage S, Stoffel M, Takeda J, Passa P. Familial hyperglycemia due to mutations in glucokinase. Definition of a subtype of diabetes mellitus. N Engl J Med. 1993;328:697–702. doi: 10.1056/NEJM199303113281005. [DOI] [PubMed] [Google Scholar]
- 15.Gancedo J M. Yeast carbon catabolite repression. Microbiol Mol Biol Rev. 1998;62:334–361. doi: 10.1128/mmbr.62.2.334-361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Germen W H, Fajan S S, Ortiz F J, Smith M J, Sturis J, Bell G I, Polonsky K S, Halter J B. Abnormal insulin secretion, not insulin resistance, is the genetic or primary defect of MODY in the RW pedigree. Diabetes. 1994;43:40–46. doi: 10.2337/diab.43.1.40. [DOI] [PubMed] [Google Scholar]
- 17.Gidh-Jain M, Takeda J, Xu L Z, Lange A J, Vionnet N, Stoffel M, Froguel P, Velho G, Sun F, Cohen D, Patel P, Lo Y-M D, Hatterslley A T, Luthman H, Wedell A, St. Charles R, Harrison R W, Weber I T, Bell G I, Pilkis S J. Glucokinase mutations associated with non-insulin-dependent (type 2) diabetes mellitus have decreased enzymatic activity: implications for structure/function relationships. Proc Natl Acad Sci USA. 1993;90:1932–1936. doi: 10.1073/pnas.90.5.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardie D G, Carling D. The AMP-activated protein kinase. Fuel gauge of the mammalian cell? Eur J Biochem. 1997;246:259–273. doi: 10.1111/j.1432-1033.1997.00259.x. [DOI] [PubMed] [Google Scholar]
- 19.Heimberg H, De Vos A, Moens K, Quartier E, Bouwens L, Pipeleers D, Van Schaftingen E, Madsen O, Schuit F. The glucose sensor protein glucokinase is expressed in glucagon-producing α-cells. Proc Natl Acad Sci USA. 1996;93:7036–7041. doi: 10.1073/pnas.93.14.7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrero P, Galindez J, Ruiz N, Martinezcampa C, Mareno F. Transcriptional regulation of the Saccharomyces cerevisiae HXK1, HXK2, and GLK1 genes. Yeast. 1995;11:137–144. doi: 10.1002/yea.320110205. [DOI] [PubMed] [Google Scholar]
- 21.Hu, Z., Y. Yue, H. Jiang, B. Zhang, P. W. Sherwood, and C. A. Michels. Analysis of the mechanism by which glucose inhibits maltose induction of MAL gene expression in Saccharomyces. Genetics, in press. [DOI] [PMC free article] [PubMed]
- 22.Hughes S D, Johnson J H, Quaade C, Newgard C B. Engineering of glucose-stimulated insulin secretion and biosynthesis in non-islet cells. Proc Natl Acad Sci USA. 1992;89:688–692. doi: 10.1073/pnas.89.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang H, Medintz I, Michels C A. Two glucose sensing/signaling pathways stimulate glucose-induced inactivation of maltose permease in Saccharomyces. Mol Biol Cell. 1997;8:1293–1304. doi: 10.1091/mbc.8.7.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnston M. Feasting, fasting and fermenting: glucose sensing in yeast and other cells. Trends Genet. 1999;15:29–33. doi: 10.1016/s0168-9525(98)01637-0. [DOI] [PubMed] [Google Scholar]
- 25.Johnston M, Carlson M. Regulation of carbon and phosphate utilization. In: Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces: gene expression. Vol. 2. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 193–281. [Google Scholar]
- 26.Lenzen S. Hexose recognition mechanisms in pancreatic β-cells. Biochem Soc Trans. 1990;18:105–107. doi: 10.1042/bst0180105. [DOI] [PubMed] [Google Scholar]
- 27.Li F, Johnston M. Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp1: coupling glucose sensing to gene expression and the cell cycle. EMBO J. 1997;16:101–110. doi: 10.1093/emboj/16.18.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang H, Gaber R F. A novel signal transduction pathway in Saccharomyces cerevisiae defined by Snf3-regulated expression of HXT6. Mol Biol Cell. 1996;7:1953–1966. doi: 10.1091/mbc.7.12.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorenz, M. C., X. Pan, T. Harashima, M. E. Cardenase, Y. Xue, J. Hirsch, and J. Heitman. The G protein-coupled receptor Gpr1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Genetics, in press. [DOI] [PMC free article] [PubMed]
- 30.Ludin K, Jiang R, Carlson M. Glucose-regulated interaction of a regulatory subunit of protein phosphatase 1 with the Snf1 protein kinase in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1998;95:6245–6250. doi: 10.1073/pnas.95.11.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Medintz I, Jiang H, Han E-K, Cui W, Michels C A. Characterization of the glucose-induced inactivation of maltose permease in Saccharomyces cerevisiae. J Bacteriol. 1996;178:2245–2254. doi: 10.1128/jb.178.8.2245-2254.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Medintz I, Jiang H, Michels C A. The role of ubiquitin conjugation in glucose-induced proteolysis of Saccharomyces maltose permease. J Biol Chem. 1998;273:34454–34462. doi: 10.1074/jbc.273.51.34454. [DOI] [PubMed] [Google Scholar]
- 33.Nishizawa K, Shimoda E, Kasahara M. Substrate recognition domain of the Gal2 galactose transporter in yeast Saccharomyces cerevisiae as revealed by chimeric galactose-glucose transporters. J Biol Chem. 1995;270:2423–2426. doi: 10.1074/jbc.270.6.2423. [DOI] [PubMed] [Google Scholar]
- 34.Ozcan S, Johnston M. Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol Cell Biol. 1995;15:1564–1572. doi: 10.1128/mcb.15.3.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ozcan S, Dover J, Rosenwald A G, Wolfl S, Johnston M. Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc Natl Acad Sci USA. 1996;93:12428–12432. doi: 10.1073/pnas.93.22.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ozcan S, Leong T, Johnston M. Rgt1p of Saccharomyces cerevisiae, a key regulator of glucose-induced genes, is both an activator and a repressor of transcription. Mol Cell Biol. 1996;16:6419–6426. doi: 10.1128/mcb.16.11.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ozcan S, Dover J, Johnston M. Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 1998;17:2566–2573. doi: 10.1093/emboj/17.9.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan X, Heitman J. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:4874–4887. doi: 10.1128/mcb.19.7.4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Riballo E, Herwijer M, Wolf D H, Lagunas R. Catabolite inactivation of the yeast maltose transporter occurs in the vacuole after internalization by endocytosis. J Bacteriol. 1995;177:5622–5627. doi: 10.1128/jb.177.19.5622-5627.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sherwood P W, Carlson M. Mutations in GSF1 and GSF2 alter glucose signaling in Saccharomyces cerevisiae. Genetics. 1997;147:557–566. doi: 10.1093/genetics/147.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sherwood P W, Carlson M. Efficient export of the glucose transporter Hxt1p from the endoplasmic reticulum requires Gsf2p. Proc Natl Acad Sci USA. 1999;96:7415–7420. doi: 10.1073/pnas.96.13.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sikorsky R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smits H-P, Smits G J, Postma P W, Walsh M C, Van Dam K. High-affinity glucose uptake in Saccharomyces cerevisiae is not dependent on the presence of glucose-phosphorylation enzymes. Yeast. 1996;12:439–447. doi: 10.1002/(SICI)1097-0061(199604)12:5%3C439::AID-YEA925%3E3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 44.Tu J, Carlson M. REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae. EMBO J. 1995;14:5939–5946. doi: 10.1002/j.1460-2075.1995.tb00282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Unger R H. Diabetic hyperglycemia: link to impaired glucose transport in pancreatic β cells. Science. 1991;251:1200–1205. doi: 10.1126/science.2006409. [DOI] [PubMed] [Google Scholar]
- 46.Vionnet N, Stoffel M, Takeda J, Yasuda K, Bell G I, Souali H, Lesage S, Velho G, Passa F, Froguel P, Cohen D. Nonsense mutation in the glucokinase gene causes early-onset non-insulin-dependent diabetes mellitus. Nature. 1992;356:721–722. doi: 10.1038/356721a0. [DOI] [PubMed] [Google Scholar]
- 47.Voss-McCowan M E, Xu B, Epstein P N. Insulin synthesis, secretory competence, and glucose utilization are sensitized by transgenic yeast hexokinase. J Biol Chem. 1994;26:15814–15818. [PubMed] [Google Scholar]
- 48.Wilson W A, Hawley S A, Hardie D G. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr Biol. 1996;6:1426–1434. doi: 10.1016/s0960-9822(96)00747-6. [DOI] [PubMed] [Google Scholar]
- 49.Xue Y, Batlle M, Hirsch J P. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Gα subunit and functions in a Ras-independent pathway. EMBO J. 1998;17:1996–2007. doi: 10.1093/emboj/17.7.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]