Abstract
Hypoxic tumor microenvironment (TME) plays critical roles in induction of cancer stem cell-like phenotype in breast cancer and contribute to chemoresistance. However, the mechanism underlying stemness reprogramming of breast cancer cells (BCs) by hypoxic TME remains largely unknown. In the present study, we illustrated that HIF-2α, but not HIF-1α, induces stemness in BCs under hypoxia through SOD2-mtROS-PDI/GRP78-UPRER pathway, linking mitochondrial metabolic state to endoplasmic reticulum (ER) response via mitochondrial reactive oxygen species (mtROS) level. HIF-2α activates endoplasmic reticulum unfolded protein response (UPRER) in drug-sensitive MCF7 and T47D cells to induce drug-resistant stem-like phenotype. Genetic depletion or pharmacological inhibition (YQ-0629) of HIF-2α abolished hypoxia-induced stem-like phenotype in vitro and in vivo. Mechanistically, HIF-2α activates transcription of superoxide dismutase 2 (SOD2) under hypoxia and thereby decreases mtROS level. With less mtROS transported to endoplasmic reticulum, the expression and activity of protein disulfide isomerase (PDI) is suppressed, allowing glucose-regulated protein 78 (GRP78) to dissociate from receptor proteins of UPRER and bind misfolded protein to activate UPRER, which eventually confer chemoresistance and stem-like properties to BCs. Moreover, the increase in mtROS and PDI levels caused by HIF-2α knockdown and the subsequent UPRER inhibition could be substantially rescued by mitoTEMPOL (a mtROS scavenger), 16F16 (a PDI inhibitor), or GRP78 overexpression. Overall, we reported the critical roles of HIF-2α-SOD2-mtROS-PDI/GRP78-UPRER axis in mediating hypoxia-induced stemness in BCs, highlighting the interaction between organelles and providing evidence for further development of targeted HIF-2α inhibitor as a promising therapeutic strategy for chemoresistant breast cancer.
Subject terms: Cancer microenvironment, Cancer stem cells, Protein aggregation
Introduction
Solid tumor microenvironment (TME) is often characterized by hypoxia, which promotes the transformation of breast cancer cells (BCs) to breast cancer stem cells (BCSCs) [1]. Such transformation confers therapeutic resistance to breast cancer and limits clinical treatment benefits [2–5]. Hypoxia-induced transcription factors (HIFs) including HIF-2α (also known as EPAS1) play important roles in regulating cancer cell stemness by activating multiple transcriptional programs under hypoxia [6–8]. Our previous study showed that HIF-2α promotes the chemoresistant stem-like phenotype in BCs via activation of Wnt and Notch pathways [9]. These results highlight the plasticity of cancer cells and indicate the complicated composition of cancer stem cells (CSCs), supporting the function of HIF-2α in CSC phenotype conversion and chemoresistance acquisition.
HIF-2α is involved in stemness reprogramming and acquisition of chemoresistance through various pathways, and is often related to poor clinical outcomes [10–12]. For example, HIF-2α enhances ALKBH5-mediated m6A-demethylation of NANOG mRNA to induce the BCSC phenotype [6]. HIF-2α also regulates Wnt/β-catenin signaling to maintain pancreatic cancer stemness [13]. Previous studies of the stemness conversion have mainly focused on mitochondria [14, 15], whereas multiple organelles are actually involved in the stemness conversion process and the underlying mechanism remains unclear.
Hypoxia in TME alters protein synthesis by endoplasmic reticulum unfolded protein response (UPRER) [16–19]. Emerging evidence has revealed that HIF-2α regulates UPRER to promote tumorigenesis and cancer cell survival, and suppresses apoptosis in leukemic cells under hypoxia [20]. Recent work has unveiled a critical role of UPRER in cell identity remodeling and acquisition of pluripotency, through the regulation of mitochondrial reactive oxygen species (mtROS) generation and protein-folding environment [21–23]. In this study, we found that UPRER was activated under hypoxia to induce the stemness remodeling via a HIF-2α/mtROS-dependent signaling pathway, which eventually contributes to the chemoresistance of BCs. Mechanistically, HIF-2α decreased mtROS levels by transcriptionally up-regulating SOD2, causing less mtROS to be transported from mitochondria to ER which, in turn, suppressed PDI expression and activity in ER. PDI competes with GRP78 for binding to misfolded protein to allow GRP78 bind with UPRER sensors and act as an “off” switch for UPRER. The inhibition of PDI enhanced binding of GRP78 to misfolded protein, causing dissociation of GRP78 from UPRER sensors and ultimately activation of UPRER. Moreover, we found that a novel HIF-2α targeted inhibitor, YQ-0629, sensitized tumor cells to paclitaxel in mouse PDX model, implying the clinical relevance of the reported HIF-2α-SOD2-mtROS-PDI/GRP78-UPRER axis. This work provides crucial insights into hypoxia-dependent stemness reprogramming and identifies a novel mechanism involving crosstalk between mitochondria and ER, encouraging the development of HIF-2α-targeting strategies for breast cancer patients, especially those who are resistant to chemotherapy.
Results
HIF-2α mediates hypoxia-induced breast cancer cell stemness remodeling and chemotherapy resistance
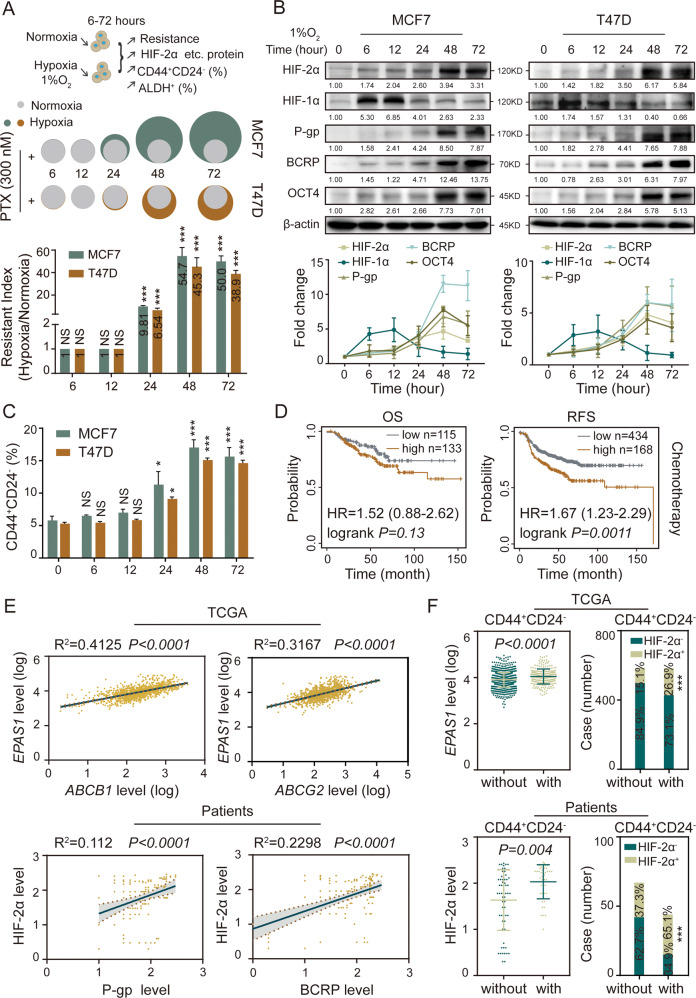
Hypoxia induces stemness in BCs [24]. To explore whether hypoxia is responsible for chemoresistance acquisition in BCs, we exposed MCF7 and T47D cells to paclitaxel (PTX) under hypoxia (1% O2) or normoxia (20% O2). MCF7 and T47D showed no significant change in IC50 values and resistance index (RI) after being cultured under hypoxia for up to 12 h. However, MCF7 and T47D displayed higher IC50 values and RI after being cultured under hypoxia for 24, 48, and 72 h (Figs. 1A and S1A, B). Furthermore, hypoxia also induced significant multi-chemoresistance of MCF7, BT474, and HCC1937 cells to adriamycin (ADR), mitoxantrone (MX) and cisplatin (DDP) (Fig. S1C, data of BT474 and HCC1937 not shown), confirming that chronic hypoxia induced chemoresistance in BCs.
Fig. 1. Hypoxia induces stemness reprogramming of BCs by activating HIF-2α.
A The cell viability rate was detected in MCF7 and T47D cells cultured with different concentrations of paclitaxel (PTX) under hypoxia for 0–72 h by MTT assay. The gray color represents normoxia (20% O2), green and brown color represents hypoxia (1% O2); the area indicates the cell viability rate. IC50 fold change and resistance index (RI) values were compared relative to normoxia. h, hour. B The expression levels of HIF-2α, HIF-1α, P-gp, BCRP and OCT4 in MCF7 and T47D cells were detected by western bolt under hypoxia for 0–72 h. C The proportion of CD44+CD24− subpopulation in MCF7 and T47D cells were detected by flow cytometry under hypoxia for 0–72 h. D The survival was analyzed between HIF-2α high expression and low expression cases received chemotherapy in overall survival (OS) and relapse-free survival (RFS) by Kaplan–Meier plot analysis (http://kmplot.com/analysis/index.php?p=service). E The correlations were analyzed among HIF-2α, P-gp, and BCRP in mRNA and protein levels from TCGA database (n = 1169) and our sample’s bank (n = 110). F The mRNA and protein levels of HIF-2α (EPAS1) was compared in CD44+CD24− and non-CD44+CD24− patients from TCGA dataset (n = 1169) and our sample’s bank (n = 110) (left panel). The right panel displays correlations between HIF-2α+ and CD44+CD24− phenotype in mRNA and protein level. NS non-significant; *P < 0.05, ***P < 0.001, compared to 0 h/normoxia; Student’s t test, One-way ANOVA test, Pearson correlation analysis, Mann–Whitney U analysis, Pearson χ2 test. Error bars, mean ± SD (n = 3).
HIF family members play important roles in stemness remodeling and chemoresistance acquisition, but the specific functions of HIF-1α and HIF-2α remains unclear. We observed that the expression of HIF-2α, but not HIF-1α, and resistance-related proteins P-gp (also known as ABCB1) and BCRP (also known as ABCG2) increased over time in MCF7 and T47D under hypoxia (Fig. 1B). Meanwhile, the expression of CSC marker OCT4 was elevated, and the percentage of CD44+CD24− and ALDH1+ BCSC populations were increased (Figs. 1B, C and S1D). These results indicated that hypoxia promoted chemoresistance of BCs in a time-dependent manner, and induced their transformation into BCSCs, with a simultaneous and specific increase in HIF-2α expression. Higher HIF-2α expression also predicted shorter overall survival (OS) and relapse-free survival (RFS) in breast cancer patients receiving chemotherapy (Fig. 1D). The Pearson’s correlation analysis of the TCGA dataset containing 1169 breast cancer patients and our in-house dataset containing 110 breast cancer tissues confirmed that HIF-2α, but not HIF-1α, was positively correlated with P-gp and BCRP (Figs. 1E and S1E). Consistently, breast cancer tissues that were CD44+CD24− had higher levels of HIF-2α (not HIF-1α), suggesting that patients with higher level of HIF-2α are likely to have a higher percentage of stem-like cells in their cancer tissues (Figs. 1F and S1F). Together, these results indicate that long-term hypoxia drives stemness remodeling and induces chemoresistance in BCs by up-regulating HIF-2α.
HIF-2α regulates the GRP78-UPRER pathway to remodel stemness and drive chemoresistance in BCs
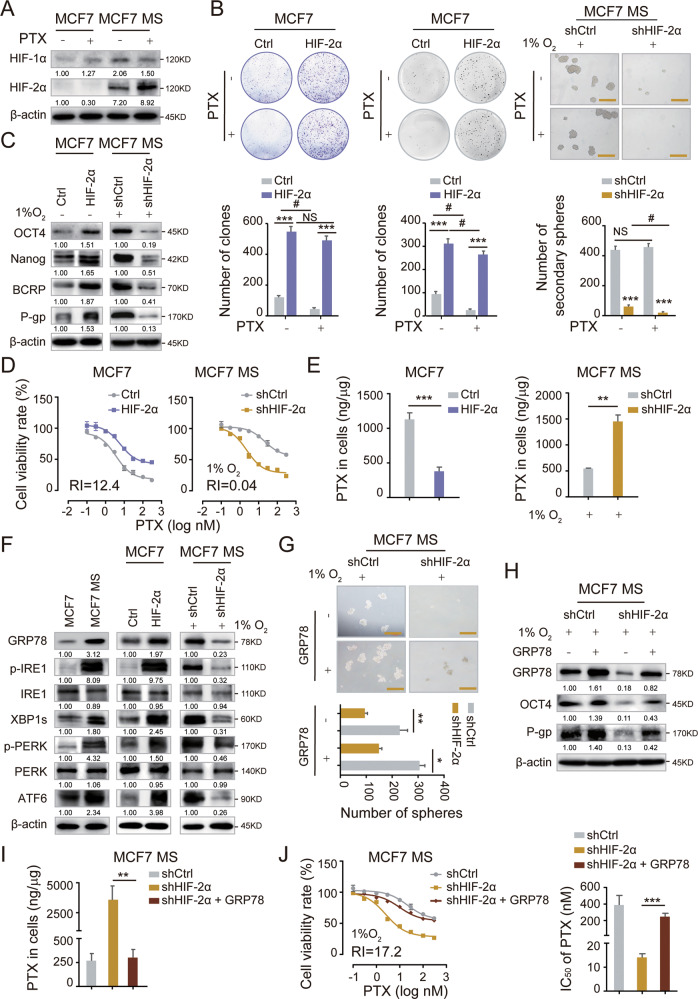
We first generated MCF7 and T47D mammospheres (MCF7 MS and T47D MS), confirmed their BCSC-like properties and chemoresistance to PTX (Fig S2A–F, [25]). We observed that HIF-2α was highly expressed in MCF7 MS and T47D MS, PTX treatment further increased the expression of HIF-2α, but not HIF-1α (Figs. 2A and S3A). To further determine the function of HIF-2α, we overexpressed HIF-2α (HIF-2α OE) in MCF7 and T47D, as well as knocked down HIF-2α (HIF-2α KD) in MCF7 MS and T47D MS by lentiviral infection and compared the knockdown efficiency (Fig. S3E–G). HIF-2α overexpression significantly enhanced colony formation capacity of MCF7. In contrast, HIF-2α knockdown dramatically reduced the formation of primary and secondary mammospheres from MCF7 MS (Fig. 2B; data of primary mammosphere formation not shown), indicating that HIF-2α maintains the long-term self-renewal capacity of BCSCs. Similar results were achieved with T47D and T47D MS (Fig. S3H). Importantly, HIF-2α overexpression increased the expression of pluripotent transcription factors OCT4 and Nanog in MCF7 and T47D, and HIF-2α knockdown significantly decreased these expression in MCF7 MS and T47D MS (Figs. 2C and S3I). Moreover, we noticed that HIF-2α overexpression induced PTX resistance in MCF7, whereas HIF-2α knockdown remarkably increased the sensitivity of MCF7 MS to PTX treatment (Fig. 2D). We also achieved similar results T47D with and T47D MS and validated this finding (Fig. S3J). Consistently, HIF-2α overexpression increased the expression of drug resistance-related proteins BCRP and P-gp in MCF7 and T47D, and HIF-2α silencing in MCF7 MS and T47D MS reduced the expression of these proteins (Figs. 2C and S3I). Additionally, HIF-2α overexpression decreased the intracellular accumulation of PTX, which is transported by P-gp, and vice versa (Fig. 2E). Collectively, these data further confirmed that hypoxia-induced stemness remodeling and chemoresistant properties in BSCSs is mainly mediated by HIF-2α.
Fig. 2. HIF-2α is required for the self-renewal maintenance of BCSCs by activating UPRER.
A The protein expression of HIF-1α and HIF-2α were measured in MCF7 and MCF7 MS cells cultured with or without PTX (3 nM) for 48 h by western blot. B The self-renewal ability was detected in HIF-2α-overexpressed (HIF-2α OE) MCF7 cells and HIF-2α-silencing (HIF-2α KD) MCF7 MS cells cultured with or without PTX (3 nM) for 48 h. Scale bar, 250 μm. C The protein expression of OCT4, NANOG, BCRP and P-gp were measured in HIF-2α OE MCF7 cells and HIF-2α KD MCF7 MS cells. D The cell viability rate was detected in HIF-2α OE MCF7 cells and HIF-2α KD MCF7 MS cells cultured with different concentrations of PTX for 48 h. Resistance index (RI) value was calculated. E The intracellular accumulation of PTX was detected in HIF-2α OE MCF7 cells and HIF-2α KD MCF7 MS cells by HPLC-MS. F The expression levels of GRP78, IRE1, XBP1s, PERK, and ATF6; phosphorylation levels of p-IRE1 and p-PERK were measured in MCF7 cells, MCF7 MS cells, HIF-2α OE MCF7 cells and HIF-2α KD MCF7 MS cells. G The self-renewal ability rescued in GRP78-overexpressing and HIF-2α-silencing (GRP78 OE + HIF-2α KD) MCF7 MS cells, compared with HIF-2α KD MCF7 MS cells. Scale bar, 250 μm. H The expression levels of GRP78, OCT4 and P-gp were measured in GRP78 OE + HIF-2α KD MCF7 MS cells. I The intracellular accumulation of PTX was detected in GRP78 OE + HIF-2α KD MCF7 MS cells by HPLC-MS. J The cells viability rate was detected in GRP78 OE + HIF-2α KD MCF7 MS cells cultured with different concentrations of PTX (left panel), the related IC50 of PTX were calculated (right panel). NS, non-significant; #P < 0.05, ##P < 0.01, ###P < 0.001, compared to treatment without PTX, *P < 0.05, **P < 0.01, ***P < 0.001, compared to Ctrl/shCtrl/shHIF-2α; Student’s t test, two-way ANOVA test. Error bars, mean ± SD (n = 3).
Cells undergo endoplasmic reticulum stress (ERS) when their ER homeostasis is disturbed by hypoxia [26], and they respond to ERS by activating UPRER. UPRER activation has been reported to involve in pluripotency acquisition and chemotherapy resistance [22, 27]. Therefore, we examined the activation status of UPRER in MCF7 and MCF7 MS, as well as in HIF-2α-overexpressing MCF7 and HIF-2α-knockdown MCF7 MS. The expression levels of GRP78, XBP1s, and ATF6 as well as the phosphorylation levels of p-IRE1 and p-PERK were higher in MCF7 MS compared to MCF7 (Fig. 2F), suggesting that UPRER was activated in BCSCs. Consistently, HIF-2α-overexpression increased the expression and the phosphorylation levels of the aforementioned proteins in MCF7, whereas HIF-2α knockdown achieved opposite phenomenon in MCF7 MS (Fig. 2F), confirming the regulation of UPRER by HIF-2α. To determine whether HIF-2α maintains stemness and chemoresistance through the UPRER, we tested if forced expression of GRP78 could restore stemness and chemoresistance in HIF-2α-knockdown MCF7 MS. Interestingly, forced expression of GRP78 rescued the oncosphere formation ability in HIF-2α depleted MCF7 MS (Fig. 2G). Additionally, forced expression of GRP78 also abolished HIF-2α-knockdown-induced inhibition of OCT4 and P-gp (Fig. 2H), increasing the intracellular accumulation of PTX and reducing chemoresistance (Fig. 2I, J). Collectively, these data indicate that HIF-2α drives stemness remodeling in BCs and induces chemoresistance by activating the GRP78-UPRER.
HIF-2α modulates SOD2 and mtROS level to activate UPRER
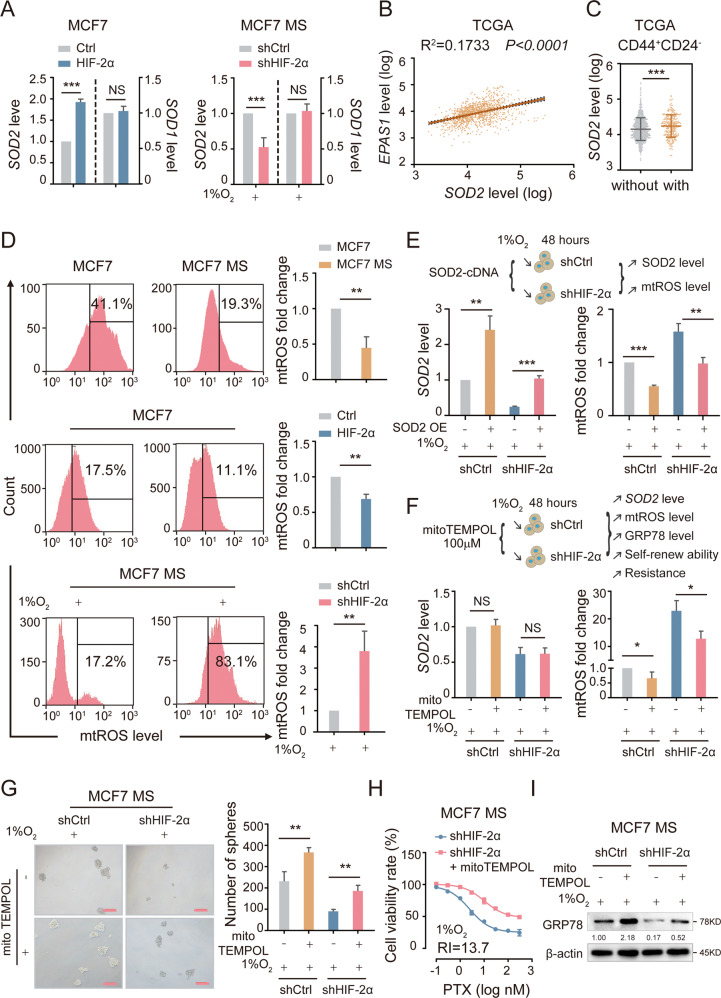
Next, we sought to investigate the detailed mechanism for the regulation of stemness and GRP78-UPRER by HIF-2α. The antioxidant gene superoxide dismutase 2 (SOD2) has been found to interact with HIF-2α [10, 28, 29]. SOD2 catalyzes the reduction of mtROS in the mitochondrial matrix, thereby protecting cells from oxidative damage [30–32]. To determine whether HIF-2α affects the SOD2-mtROS level to regulate the GRP78-UPRER, we examined the mRNA expression of SOD2 in HIF-2α-overexpressing MCF7 and HIF-2α-knockdown MCF7 MS. Results showed that HIF-2α overexpression increased the mRNA level of SOD2, whereas HIF-2α knockdown reduced the expression of SOD2 (Fig. 3A). As a control, the mRNA level of SOD1 was not affected by either overexpression or knockdown of HIF-2α (Fig. 3A). Furthermore, we analyzed the TCGA dataset and found that HIF-2α expression was positively correlated with SOD2 expression, and patients with the CD44+CD24− phenotype expressed SOD2 at a higher level (Fig. 3B, C). Together with the previous finding that patients with the CD44+CD24− phenotype had a higher expression of HIF-2α (Fig. 1F), our results confirmed that HIF-2α activates SOD2 in BCSCs and this axis is clinically relevant. mtROS level was also lower in MCF7 MS than in MCF7 cells, indicating that the intracellular mtROS level of BCSCs was lower than that of BCs. Consistently, the overexpression or knockdown of HIF-2α decreased or increased the level of mtROS respectively (Fig. 3D). These data suggest that HIF-2α is an upstream regulator of SOD2 and mtROS.
Fig. 3. HIF-2α activates UPRER via regulating SOD2-mtROS axis.
A The mRNA level of SOD2 and SOD1 were detected in HIF-2α OE MCF7 cells and HIF-2α KD MCF7 MS cells. B The correlation between HIF-2α and SOD2 was detected in mRNA level from TCGA database (n = 1169). C The mRNA expression of SOD2 was compared in CD44+CD24− and non CD44+CD24− patients from TCGA database (n = 1169). D The levels of mtROS were detected in MCF7 and MCF7 MS cells, HIF-2α OE MCF7 cells and HIF-2α KD MCF7 MS cells. E The mRNA level of SOD2 and mtROS level were measured in SOD2-overexpressing and HIF-2α-silencing (SOD2 OE + HIF-2α KD) MCF7 MS cells, under 1% O2. F The mRNA level of SOD2 and mtROS level were measured in HIF-2α KD MCF7 MS cells cultured with mitoTEMPOL (100 µM) for 48 h, under 1% O2. G The self-renewal ability rescued in HIF-2α KD MCF7 MS cells cultured mitoTEMPOL (100 µM) for 48 h, compared with HIF-2α KD MCF7 MS cells, under 1% O2. Scale bar, 250 μm. H The cells viability rate was detected in HIF-2α KD MCF7 MS cells cultured with mitoTEMPOL (100 µM) for 48 h, under 1% O2. I The expression level of GRP78 was detected in HIF-2α KD MCF7 MS cells cultured with mitoTEMPOL (100 µM) for 48 h, under 1% O2. NS, non-significant; *P < 0.05, **P < 0.01, ***P < 0.001, compared to MCF7 cells/Ctrl/shCtrl/shHIF-2α; Student’s t test, two-way ANOVA test, Mann–Whitney U analysis, Pearson correlation analysis. Error bars, mean ± SD (n = 3).
To further validate whether HIF-2α regulates mtROS level in BCSCs via SOD2, we overexpressed SOD2 in HIF-2α-knockdown MCF7 MS. As expected, SOD2 overexpression abrogated the HIF-2α-knockdown-induced increase in mtROS level (Figs. 3E and S4A). We also incubated HIF-2α-knockdown MCF7 MS with mitoTEMPOL, a mtROS-specific scavenger, to further verify if mtROS mediate the downstream effects of HIF-2α. MitoTEMPOL treatment reduced the level of mtROS without affecting SOD2 expression (Figs. 3F and S4B), confirming the suitability of mitoTEMPOL for testing the role of mtROS in the function of HIF-2α. We then observed that mitoTEMPOL rescued the oncosphere formation capacity in HIF-2α-knockdown MCF7 MS (Fig. 3G). Moreover, mitoTEMPOL restored the chemoresistance of HIF-2α-knockdown MCF7 MS (Figs. 3H and S4C). Additionally, to explore whether UPRER is involved, we examined the protein level of GRP78 in the same groups. Our results showed that mitoTEMPOL rescued the protein expression of GRP78 in HIF-2α-knockdown MCF7 MS (Figs. 3I and S4D). In sum, HIF-2α upregulates SOD2 to reduce mtROS level, leading to the downstream biological effects including GRP78-UPRER activation, stemness remodeling, and chemoresistance acquisition.
PDI competitively inhibits the binding of GRP78 to misfolded proteins to prevent UPRER activation
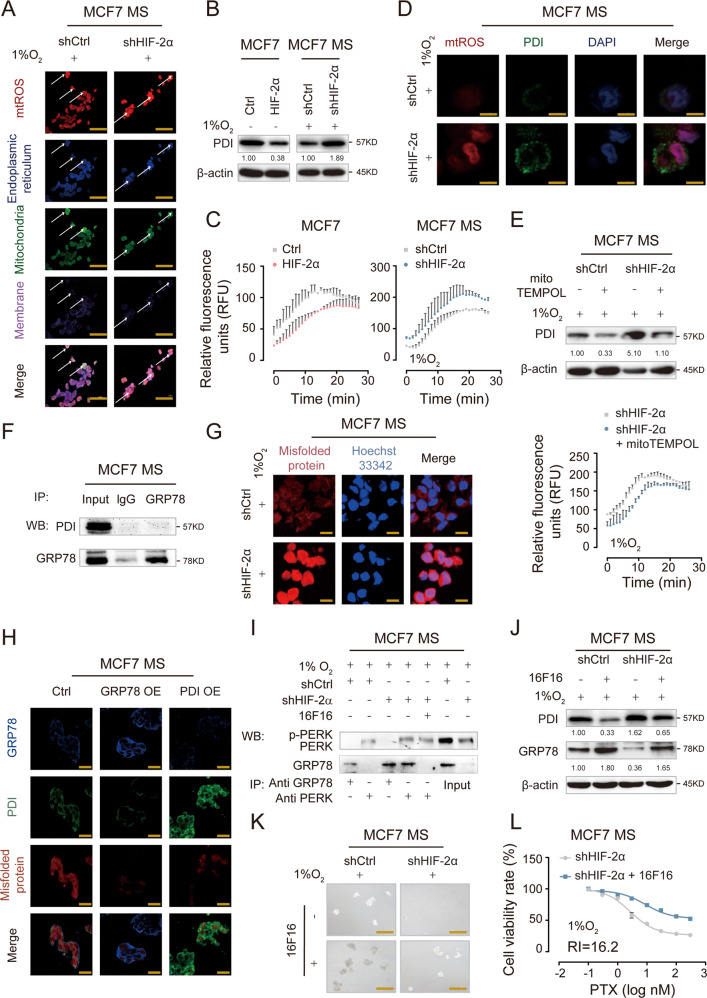
MtROS are mainly generated during the process of oxidative phosphorylation at the mitochondrial electron transport chain and are transported from mitochondria to multiple intracellular organelles as signaling molecules [31, 32]. In particular, mtROS are transported to ER and are involved in the regulation of ER-related pathways [33]. To elucidate how the modulation of SOD2-mtROS level by HIF-2α is transduced intracellularly to activate UPRER, we detected the subcellular location of mtROS. We observed that mtROS was mainly located in mitochondria in control cells, while the increased mtROS primarily accumulated in ER in HIF-2α-knockdown MCF7 MS (Fig. 4A), indicating that excess mtROS induced by HIF-2α knockdown were mainly transported from mitochondria to ER. Protein disulfide isomerase (PDI) is a critical protein in ER that has two major functions: one is to act as an oxidoreductase, and the other is to function as a chaperone protein to bind misfolded protein for subsequent degradation. The enzyme activity of PDI can be increased when the ER environment becomes more oxidative [34, 35]. Therefore, we hypothesized that HIF-2α might regulate PDI activity in ER through mtROS transport. To test our hypothesis, we first detected the expression and enzymatic activity of PDI after HIF-2α overexpression and knockdown. We found that the expression and enzymatic activity of PDI was reduced by HIF-2α overexpression in MCF7, whereas the opposite was observed upon HIF-2α knockdown in MCF7 MS (Fig. 4B, C). Not only that, HIF-2α silencing increased the levels of mtROS and PDI in MCF7 MS, with their subcellular location overlapping (Fig. 4D), supporting our hypothesis that mtROS might be transported to ER to regulate PDI. We then exposed HIF-2α-knockdown MCF7 MS to mitoTEMPOL and detected the expression and enzymatic activity of PDI (Fig S4E). We found that mitoTEMPOL significantly decreased PDI expression and enzymatic activity (Fig. 4E). These results suggest that HIF-2α regulates the expression and catalytic activity of PDI in BCSCs through the signaling molecules - mtROS.
Fig. 4. PDI competitively binding to misfolded proteins with GRP78 to activate UPRER.
A The location of mtROS in mitochondria and ER was observed by confocal microscope. Scale bar, 10 μm. B The protein expression of PDI was detected in HIF-2α OE MCF7 cells and HIF-2α KD MCF7 MS cells, under 1% O2. C The enzyme activities of PDI in HIF-2α OE MCF7 cells and HIF-2α KD MCF7 MS cells were measured, under 1% O2. D The level and location of mtROS and PDI were measured in HIF-2α KD MCF7 MS cells were detected by immunofluorescence microscopy, under 1% O2. Scale bar, 5 μm. E The protein expression and enzyme activity of PDI were measured in HIF-2α KD MCF7 MS cells cultured with mitoTEMPOL (100 µM), under 1% O2. F The direct interaction of GRP78 and PDI in MCF7 MS cells was determined by co-immunoprecipitation (co-IP). G The level of misfolded protein was detected in HIF-2α KD MCF7 MS cells by confocal microscope. Scale bar, 10 μm. H The expression levels of GRP78, PDI and misfolded proteins were detected in the GRP78-overexpressing (GRP78 OE), PDI-overexpressing (PDI OE) MCF7 MS cells by immunofluorescence microscopy, under 1% O2. Scale bar, 20 μm. I The combination and dissociation of GRP78 and PERK was confirmed by co-immunoprecipitation (co-IP) in HIF-2α KD MCF7 MS cells cultured with 16F16 (100 µM), under 1% O2. J–L The protein expressions of PDI and GRP78, cell viability rate and self-renewal ability were detected in HIF-2α KD MCF7 MS cells cultured with 16F16 (100 µM), under 1% O2. Scale bar, 250 μm.
Then, we explored the underlying mechanism by which PDI regulates the UPRER. GRP78 functions as an “off” switch for UPRER, hence we first mocked docking of PDI to GRP78, and found the two proteins did not match in spatial structure (Fig. S4F). We further confirmed that there was no direct interaction between PDI and GRP78 in MCF7 MS (Figs. 4F and S4G). Therefore, we presumed that a potential indirect mechanism mediates the regulation of UPRER by PDI. Given that the activation of UPRER depends on the binding of GRP78 to misfolded protein and the subsequent dissociation of GRP78 from the UPRER transmembrane protein sensors [36, 37], we detected the intracellular level of misfolded protein in HIF-2α-knockdown MCF7 MS. We found that the level of misfolded protein significantly increased upon HIF-2α knockdown (Figs. 4G and S4H). To search for potential interaction among PDI, GRP78, and misfolded proteins, we constructed stably GRP78 or PDI overexpressed MCF7 MS. We found the protein level of GRP78 was decreased in PDI-overexpressing MCF7 MS, and vice versa. Additionally, the level of misfolded protein was remarkably reduced in both GRP78-overexpressing and PDI-overexpressing MCF7 MS (Fig. 4H and S4I). 16F16 is a specific PDI inhibitor. Strikingly, 16F16 treatment enhanced the binding of GRP78 to misfolded proteins in HIF-2α-knockdown MCF7 MS, allowing the dissociation of GRP78 from PERK. This result confirmed that PDI competes with GRP78 for binding to misfolded proteins to activate UPRER (Fig. 4I). We also detected the expression of GRP78 to further validate if PDI regulates GRP78 to affect UPRER. We found that 16F16 released the HIF-2α-knockdown-induced inhibition on GRP78 expression, as well as suppressed the expression and activity of PDI in HIF-2α-knockdown MCF7 MS (Figs. 4J and S4K). Consistently, 16F16 restored mammosphere forming ability and chemoresistance in HIF-2α-knockdown MCF7 MS (Figs. 4K, L and S4L, M). These results indicate that PDI competitively inhibits the binding of GRP78 to misfolded proteins to prevent the activation of UPRER and HIF-2α activates UPRER by suppressing PDI expression and activity.
HIF-2α silencing reduces stemness and reverses chemoresistance via SOD2-mtROS-UPRER pathway in vivo
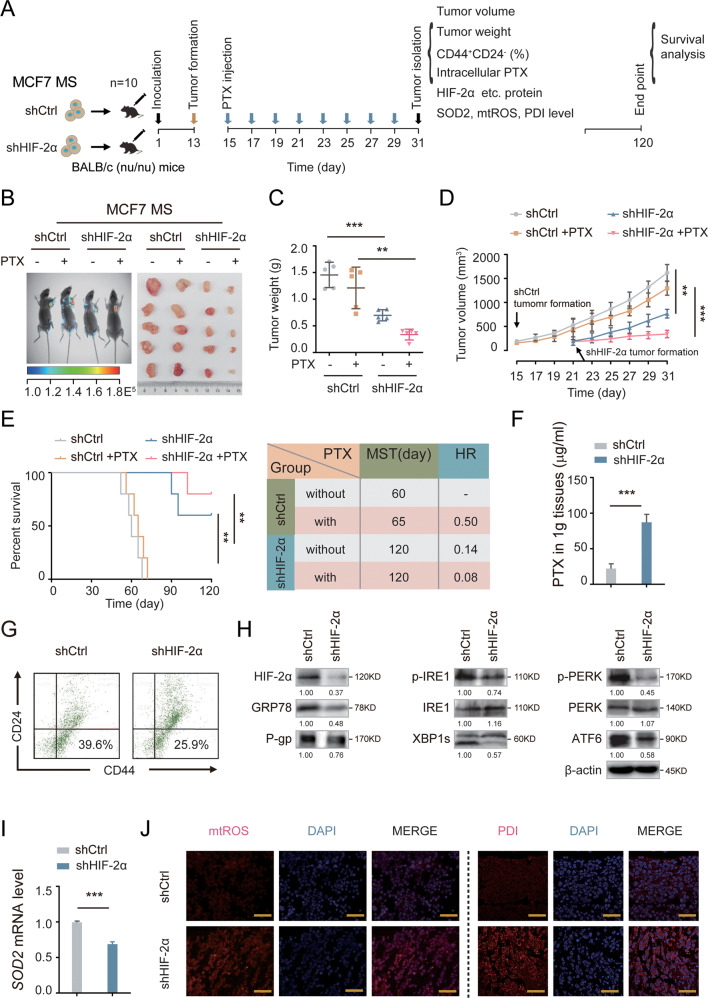
We next confirmed the role of HIF-2α in tumorigenesis in vivo. We injected BALB/c (nu/nu) mice subcutaneously with control or HIF-2α-knockdown MCF7 MS, and monitored tumor growth as well as measured the percentage of BCSC population and related pathway indicators (Fig. 5A). HIF-2α depletion remarkably suppressed the growth of xenograft tumors, and the inhibitory effect was even stronger when treated in combination with PTX (Fig. 5B, C). CSCs are responsible for early tumorigenesis [24]. We defined the formation of a detectable tumor as when a tumor reaches the of size 125 mm3 and observed xenograft tumor formation at about 1 month post injection. HIF-2α depletion resulted in a significant delay in tumor formation (at day 21) compared to the control group (at day 15), suggesting that HIF-2α knockdown reduces the tumor initiating ability of MCF7 MS (Fig. 5D). Additionally, we observed that HIF-2α knockdown significantly improved survival in the xenograft mouse model, especially when treated in combination with PTX (Fig. 5E). The accumulation of PTX in HIF-2α-knockdown xenograft tissues was much higher than that in control tissues (Fig. 5F). Furthermore, we found that the percentage of CD44+CD24− cells was also lower in HIF-2α-knockdown xenograft tissues than in control tissues (Fig. 5G). These findings imply that HIF-2α depletion decreases self-renewal capacity and reverses chemoresistance in BCSCs in vivo. Consistently, HIF-2α depletion dramatically reduced the expression of P-gp, GRP78, and other UPRER-related proteins in vivo (Fig. 5H). Similar results were obtained for the mRNA level of SOD2 (Fig. 5I). The mtROS level and PDI expression were also increased in HIF-2α-knockdown xenograft tissues, compared to control tissues (Fig. 5J). Overall, these data indicate that HIF-2α silencing impairs stemness and chemoresistance of BCSCs through SOD2-mtROS-PDI/GRP78-UPRER pathway in vivo.
Fig. 5. Silencing HIF-2α suppresses the early tumorigenesis and increases the sensitivities of PTX via SOD2-mtROS-PDI/GRP78-UPRER in vivo.
A The diagram showed the time of tumor formation in BALB/c (nu/nu) mice transplanted with control or HIF-2α KD MCF7 MS cells (1 × 105). 15 days after the inoculation, the mice were intraperitoneally injected with or without PTX (5 mg/kg) once every other day till to the 31th day (n = 5). The other mice (n = 5) were observed survival till the 120th day. B Small animal imaging showed the expression of green fluorescent protein (GFP) in the xenografted mice of each group. n = 5. C The tumor weights were measured in each group after sacrifice of xenograft mice at the 31th days. n = 5. Two-way ANOVA test. D The growth curves of tumor volumes were measured in xenograft mice were measured every other day. n = 5. Two-way ANOVA test. E The survival of the mice in each group (left) were analyzed by Kaplan Meier-plotter curve. Median survival times (MST). Hazard ratio (HR). n = 5. Two-way ANOVA test. F The PTX accumulation were measured in the HIF-2α KD xenografted tissue by HPLC-MS. n = 3. Student’s t test. G The proportion of CD44+CD24− cell in the HIF-2α KD xenografted tumor were detected by flow cytometry. H The protein expressions of HIF-2α, GRP78, P-IRE1, IRE1, XBP1s, p-PERK, PERK, ATF6 and P-gp were detected in the HIF-2α KD xenografted tissue. I The mRNA expression of SOD2 was detected in the HIF-2α KD xenografted tissue. n = 3. Student’s t test. J The level of mtROS and PDI were detected in the HIF-2α KD xenografted tissue. Scale bar, 100 μm. *P < 0.05, **P < 0.01, ***P < 0.001, compared to shCtrl.
A specific HIF-2α inhibitor YQ-0629 synergizes with PTX in vivo
To further explore the clinical relevance of HIF-2α and the reported downstream pathway in breast cancer, we aimed to find a specific HIF-2α inhibitor. The HIF-2α protein contains three domains with known structures: a b-Helix-loop-helix (bHLH) segment, Per-Arnt-Sim (PAS) domains, and terminal transactivation domains (TADs). PAS-B domain is the potential active site for ligand binding [38]. The amino acid sequence targeted by the HIF-2α shRNA we used was located in PAS-B domain (Fig. S5A). We found the crystal structure of the PAS-B region from Protein Data Bank (PDB ID: 4XT2), and it has already been proved that active small molecule compounds binding this active site can be successfully screened out [39]. First, we used two libraries of small molecular compounds containing a total of 1 million compounds for docking into this site. 81 compounds (Table 1) with S values less than −9 were screened out. 17 compounds with high drug-forming property were further screened out based on ADME evaluation. Three compounds including YQ-0629 were demonstrated strong inhibitory effects on cell viability. We further performed a Biacore assay to verify the binding of the three compounds to HIF-2α active sites in vitro (Fig. S5B). Only YQ-0629 showed a strong affinity with HIF-2α PAS-B domain (dissociation constant [Kd] 5.75E−5 M) and the potential binding site is predicted to be the residue Cys339 (Fig. 6A). To investigate whether YQ-0629 inhibits HIF-2α in vivo, we detected the protein expression and intracellular location of HIF-2α after MCF7 MS culturing with YQ-0629 (10 µM). We observed that YQ-0629 could inhibit the expression of HIF-2α with or without the presence of PTX (3 nM) (Fig. 6B, C). We also confirmed that YQ-0629 did not bind to HIF-1α or affect the expression of HIF-1α (Fig. S5C, D). These data indicate that YQ-0629 is a specific inhibitor of HIF-2α.
Table 1.
The reault of docking by MOE.
| mol | rseq | mseq | S | rmsd_refine | FP:PLIF | PLIF_ligidx | E_conf | E_place | E_score1 | E_refine | E_score2 | a_acc | a_don | logP(o/w) | logS | SlogP | TPSA | Weight |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fc1cc(C)c(NC( = O)CN2C( = O)C(N(C( = O)CC)C) = Nc3c2cccc3)cc1 | 1 | 194 | −10.938254 | 1.7831233 | 1 2 6 13 | [[10 10 10 10 10] [10 10 10 10 10] 21 21] | 73.83429 | −65.439285 | −9.6604252 | −36.246143 | −10.938254 | 4 | 1 | 2.046 | −4.86901 | 3.0178199 | 82.080002 | 396.422 |
| O = C(N(C)C = 1 C( = O)N(CC( = O)Nc2ccc(OC)cc2)c2c(N = 1)cccc2)CC | 1 | 195 | −10.912217 | 1.6352755 | 4 | 14 | 53.837105 | −64.515892 | −9.6641216 | −30.77556 | −10.912217 | 5 | 1 | 1.516 | −4.46394 | 2.5789001 | 91.309998 | 394.431 |
| Fc1cc(C)c(NC( = O)CN2C( = O)C(N(C( = O)CC)C) = Nc3c2cccc3)cc1 | 1 | 194 | −10.709622 | 1.3912883 | 1 2 6 13 | [[10 10 10 10 10] [10 10 10 10 10] 21 21] | 66.496262 | −86.975899 | −9.3012972 | −32.216003 | −10.709622 | 4 | 1 | 2.046 | −4.86901 | 3.0178199 | 82.080002 | 396.422 |
| Fc1c(NC( = O)CN2C( = O)C(N(C( = O)CC)C) = Nc3c2cccc3)ccc(F)c1 | 1 | 191 | −10.616098 | 1.5885445 | 1 2 66 6 13 | [[10 10 10 10 10] [10 10 10 10 10] 26 21 21] | 87.036522 | −74.541992 | −9.252636 | −36.326653 | −10.616098 | 4 | 1 | 1.901 | −5.00352 | 2.8485 | 82.080002 | 400.385 |
| Fc1cc(C)c(NC( = O)CN2C( = O)C(N(C( = O)CC)C) = Nc3c2cccc3)cc1 | 1 | 194 | −10.552382 | 1.3751938 | 1 2 6 | [[10 10 10 10 10] [10 10 10 10 10] 21] | 80.821815 | −90.90596 | −9.8919563 | −31.125942 | −10.552382 | 4 | 1 | 2.046 | −4.86901 | 3.0178199 | 82.080002 | 396.422 |
| Clc1ccc(NC( = O)CN2C( = O)C(N(C( = O)CC)C) = Nc3c2cccc3)cc1 | 1 | 192 | −10.400867 | 1.2550629 | 6 13 | [17 17] | 81.382957 | −66.724831 | −9.3333635 | −29.644501 | −10.400867 | 4 | 1 | 2.152 | −5.14785 | 3.2237 | 82.080002 | 398.85 |
| Clc1ccc(NC( = O)CN2C( = O)C(N(C( = O)CC)C) = Nc3c2cccc3)cc1 | 1 | 192 | −10.360132 | 1.2362972 | 1 6 | [[9 9 9 9 9] 17] | 83.137192 | −61.30851 | −9.4128256 | −29.487608 | −10.360132 | 4 | 1 | 2.152 | −5.14785 | 3.2237 | 82.080002 | 398.85 |
| Fc1c(NC( = O)CN2C( = O)C(N(C( = O)CC)C) = Nc3c2cccc3)ccc(F)c1 | 1 | 191 | −10.352764 | 2.0071845 | 1 2 66 6 13 | [[10 10 10 10 10] [10 10 10 10 10] 26 21 21] | 79.611908 | −48.871712 | −9.5374498 | −31.998219 | −10.352764 | 4 | 1 | 1.901 | −5.00352 | 2.8485 | 82.080002 | 400.385 |
| O = C(N(CCOC)Cc1nc(COc2cc(C)ccc2)sc1)CC | 1 | 206 | −10.315837 | 1.373821 | [] | 53.685596 | −63.378716 | −10.607458 | −36.705509 | −10.315837 | 4 | 0 | 2.608 | −2.92336 | 3.9483199 | 51.66 | 348.467 | |
| O = C(N(CCOC)Cc1nc(COc2c(C)c(C)cc(C)c2)sc1)CC | 1 | 202 | −10.18891 | 1.1696758 | [] | 49.33707 | −72.034058 | −11.159831 | −23.807611 | −10.18891 | 4 | 0 | 3.237 | −3.55775 | 4.5651598 | 51.66 | 376.521 | |
| O = C(N(CCOC)Cc1nc(COc2c(C)c(C)cc(C)c2)sc1)CC | 1 | 202 | −10.165962 | 1.6515702 | [] | 44.253685 | −54.726841 | −9.7377605 | −20.87141 | −10.165962 | 4 | 0 | 3.237 | −3.55775 | 4.5651598 | 51.66 | 376.521 | |
| O = C(N(CCOC)Cc1nc(COc2cc(C)ccc2)sc1)CC | 1 | 206 | −10.154015 | 1.8054231 | [] | 47.665268 | −73.104424 | −9.5320988 | −32.237152 | −10.154015 | 4 | 0 | 2.608 | −2.92336 | 3.9483199 | 51.66 | 348.467 | |
| O = C(N(CCOC)Cc1nc(COc2cc(C)ccc2)sc1)CC | 1 | 206 | −10.148293 | 1.1232054 | 10 | 6 | 41.12479 | −74.544441 | −10.236934 | −33.357601 | −10.148293 | 4 | 0 | 2.608 | −2.92336 | 3.9483199 | 51.66 | 348.467 |
| O = C(N(CCOC)Cc1nc(COc2c(C)c(C)cc(C)c2)sc1)CC | 1 | 202 | −10.147232 | 1.6236573 | 37 26 29 3 | [22 6 6 22] | 46.595928 | −68.276367 | −9.9389849 | −22.807604 | −10.147232 | 4 | 0 | 3.237 | −3.55775 | 4.5651598 | 51.66 | 376.521 |
| Fc1cc(C)c(NC( = O)CN2C( = O)C(N(C( = O)CC)C) = Nc3c2cccc3)cc1 | 1 | 194 | −10.083389 | 1.8171558 | 45 | 21 | 82.07254 | −79.647301 | −9.9758043 | −26.444155 | −10.083389 | 4 | 1 | 2.046 | −4.86901 | 3.0178199 | 82.080002 | 396.422 |
| O = C(N(CCOC)Cc1nc(COc2cc(C)c(C)cc2)sc1)CC | 1 | 205 | −10.046181 | 1.2799406 | [] | 46.098122 | −87.489403 | −9.7152643 | −27.573259 | −10.046181 | 4 | 0 | 2.904 | −3.39728 | 4.2567401 | 51.66 | 362.494 | |
| O = C(N(CCOC)Cc1nc(COc2c(C)c(C)cc(C)c2)sc1)CC | 1 | 202 | −10.021337 | 1.0244079 | [] | 47.566261 | −94.499832 | −11.015378 | −21.993591 | −10.021337 | 4 | 0 | 3.237 | −3.55775 | 4.5651598 | 51.66 | 376.521 | |
| O = C(N(CCOC)Cc1nc(COc2c(C)c(C)cc(C)c2)sc1)CC | 1 | 202 | −10.013167 | 2.2391009 | 51 | 6 | 47.740715 | −43.478626 | −9.8501482 | -17.340368 | −10.013167 | 4 | 0 | 3.237 | −3.55775 | 4.5651598 | 51.66 | 376.521 |
| O = C(N(CCOC)Cc1nc(COc2cc(C)c(C)cc2)sc1)CC | 1 | 205 | −9.8725662 | 1.3905082 | 10 | 6 | 47.896034 | −45.029846 | −10.007674 | −24.523638 | −9.8725662 | 4 | 0 | 2.904 | −3.39728 | 4.2567401 | 51.66 | 362.494 |
| Fc1ccc(NC( = O)CN2C( = O)C(N(C( = O)CC)C) = Nc3c2cccc3)cc1 | 1 | 200 | −9.81569 | 1.0334991 | [] | 83.840248 | −78.816048 | −9.863452 | −26.088251 | −9.81569 | 4 | 1 | 1.713 | −4.70854 | 2.7093999 | 82.080002 | 382.395 | |
| Fc1ccc(NC( = O)CN2C( = O)C(N(C( = O)CC)C) = Nc3c2cccc3)cc1 | 1 | 200 | −9.8077593 | 1.3528427 | [] | 95.229691 | −60.510231 | −9.8809338 | −26.250235 | −9.8077593 | 4 | 1 | 1.713 | −4.70854 | 2.7093999 | 82.080002 | 382.395 | |
| Fc1ccc(NC( = O)CN2C( = O)C(N(C( = O)CC)C) = Nc3c2cccc3)cc1 | 1 | 200 | −9.7154455 | 1.441017 | 1 2 6 | [[9 9 9 9 9] [9 9 9 9 9] 17] | 72.269409 | −82.974785 | −10.168523 | −25.800505 | −9.7154455 | 4 | 1 | 1.713 | −4.70854 | 2.7093999 | 82.080002 | 382.395 |
| Brc1ccc(NC( = O)CN2C( = O)C(N(C( = O)CC)C) = Nc3c2cccc3)cc1 | 1 | 150 | −9.6700525 | 1.3923471 | 6 | 17 | 90.105148 | −40.248928 | −9.6419649 | −18.469656 | −9.6700525 | 4 | 1 | 2.358 | −5.50395 | 3.3327999 | 82.080002 | 443.301 |
| O = C(N(CCOC)Cc1nc(COc2cc(C)ccc2)sc1)CC | 1 | 206 | −9.5716019 | 1.651099 | 39 | 22 | 51.783993 | −68.938942 | −10.407656 | −27.115789 | −9.5716019 | 4 | 0 | 2.608 | −2.92336 | 3.9483199 | 51.66 | 348.467 |
| Fc1cc(C)c(NC( = O)CN2C( = O)C(N(C( = O)CC)C) = Nc3c2cccc3)cc1 | 1 | 194 | −9.5423374 | 1.8602611 | [] | 90.812431 | −76.622543 | −10.010983 | −16.224051 | −9.5423374 | 4 | 1 | 2.046 | −4.86901 | 3.0178199 | 82.080002 | 396.422 | |
| O = C(N(C)C = 1 C( = O)N(CC( = O)Nc2ccc(C)cc2)c2c(N = 1)cccc2)CC | 1 | 201 | −9.5378523 | 1.4987394 | 1 6 | [[9 9 9 9 9] 17] | 83.412437 | −68.649963 | −8.6353951 | −17.296104 | −9.5378523 | 4 | 1 | 1.858 | −4.88748 | 2.87872 | 82.080002 | 378.432 |
| Fc1ccc(NC( = O)CN2C( = O)C(N(C( = O)CC)C) = Nc3c2cccc3)cc1 | 1 | 200 | −9.5346565 | 1.4158621 | 1 6 | [[9 9 9 9 9] 17] | 85.169342 | −68.123856 | −10.437311 | −23.037125 | −9.5346565 | 4 | 1 | 1.713 | −4.70854 | 2.7093999 | 82.080002 | 382.395 |
| O = C(N(C)C = 1 C( = O)N(CC( = O)Nc2cc(C)c(C)cc2)c2c(N = 1)cccc2)CC | 1 | 198 | −9.5189009 | 1.6912936 | [] | 62.66856 | −33.40675 | −8.6570959 | −12.775396 | −9.5189009 | 4 | 1 | 2.191 | −5.3614 | 3.18714 | 82.080002 | 392.459 | |
| Clc1ccc(NC( = O)CN2C( = O)C(N(C( = O)CC)C) = Nc3c2cccc3)cc1 | 1 | 192 | −9.4825964 | 1.6797754 | 6 | 17 | 84.100311 | −80.925598 | −9.1933918 | −22.722916 | −9.4825964 | 4 | 1 | 2.152 | −5.14785 | 3.2237 | 82.080002 | 398.85 |
| O = C(N(CCOC)Cc1nc(COc2cc(C)ccc2)sc1)CC | 1 | 206 | −9.4407234 | 1.1775275 | [] | 46.461788 | −60.646614 | −9.5731077 | −24.327597 | −9.4407234 | 4 | 0 | 2.608 | −2.92336 | 3.9483199 | 51.66 | 348.467 | |
| O = C(N(C)C = 1 C( = O)N(CC( = O)Nc2ccc(OC)cc2)c2c(N = 1)cccc2)CC | 1 | 195 | −9.4121208 | 1.8469723 | 6 | 17 | 104.67824 | −77.250793 | −9.8469458 | −17.24333 | −9.4121208 | 5 | 1 | 1.516 | −4.46394 | 2.5789001 | 91.309998 | 394.431 |
| Brc1ccc(NC( = O)CN2C( = O)C(N(C( = O)CC)C) = Nc3c2cccc3)cc1 | 1 | 150 | −9.3616257 | 1.5611087 | 1 2 6 13 | [[9 9 9 9 9] [9 9 9 9 9] 17 17] | 85.092247 | −91.566315 | −10.05342 | −16.262087 | −9.3616257 | 4 | 1 | 2.358 | −5.50395 | 3.3327999 | 82.080002 | 443.301 |
| O = C(N(CCOC)Cc1nc(COc2cc(C)c(C)cc2)sc1)CC | 1 | 205 | −9.3223333 | 1.5094248 | 28 60 | [[22 44 43 42 23] [22 44 43 42 23]] | 74.316414 | −51.635536 | −12.669622 | −20.374947 | −9.3223333 | 4 | 0 | 2.904 | −3.39728 | 4.2567401 | 51.66 | 362.494 |
| Brc1ccc(NC( = O)CN2C( = O)C(N(C( = O)CC)C) = Nc3c2cccc3)cc1 | 1 | 150 | −9.2992764 | 1.781745 | 1 2 6 | [[9 9 9 9 9] [9 9 9 9 9] 17] | 82.239342 | −42.763515 | −9.2397318 | −16.375626 | −9.2992764 | 4 | 1 | 2.358 | −5.50395 | 3.3327999 | 82.080002 | 443.301 |
| O = C(N(C)C = 1 C( = O)N(CC( = O)Nc2ccc(C)cc2)c2c(N = 1)cccc2)CC | 1 | 201 | −9.2312002 | 1.4929682 | [] | 118.03013 | −46.209911 | −8.457756 | −16.017235 | −9.2312002 | 4 | 1 | 1.858 | −4.88748 | 2.87872 | 82.080002 | 378.432 | |
| O = C(N(C)C = 1 C( = O)N(CC( = O)Nc2c(C)cc(C)cc2)c2c(N = 1)cccc2)CC | 1 | 197 | −9.200736 | 1.8307776 | [] | 68.142166 | −68.19297 | −9.8695173 | −11.049742 | −9.200736 | 4 | 1 | 2.191 | −5.04795 | 3.18714 | 82.080002 | 392.459 | |
| O = C(N(CCOC)Cc1nc(COc2cc(C)c(C)cc2)sc1)CC | 1 | 205 | −9.1963415 | 1.5497842 | 39 | 22 | 59.502342 | −64.809776 | −12.160249 | −20.262173 | −9.1963415 | 4 | 0 | 2.904 | −3.39728 | 4.2567401 | 51.66 | 362.494 |
| O = C(N(C)C = 1 C( = O)N(CC( = O)Nc2ccc(OC)cc2)c2c(N = 1)cccc2)CC | 1 | 195 | −9.1187716 | 1.6638592 | 3 14 | [14 14] | 84.711357 | −53.852581 | −8.5630455 | −14.621253 | −9.1187716 | 5 | 1 | 1.516 | −4.46394 | 2.5789001 | 91.309998 | 394.431 |
| Fc1c(NC( = O)CN2C( = O)C(N(C( = O)CC)C) = Nc3c2cccc3)ccc(F)c1 | 1 | 191 | −9.1121922 | 1.4418428 | 1 2 | [[10 10 10 10 10] [10 10 10 10 10]] | 92.921974 | −67.421486 | −10.055465 | −19.784554 | −9.1121922 | 4 | 1 | 1.901 | −5.00352 | 2.8485 | 82.080002 | 400.385 |
| FC(F)(F)c1c(OC)ccc(C( = O)Nc2cc(C(F)(F)F)cc(-c3onc4c3CCCC4)c2)c1 | 1 | 104 | −9.1094694 | 1.8124239 | 16 | [[5 9 8 7 6]] | 141.37689 | −59.811256 | −7.6863132 | −1.4543294 | −9.1094694 | 2 | 1 | 6.00752 | −7.64869 | 7.14184 | 64.360001 | 484.396 |
| Clc1c(OC)ccc(NC( = O)CN2C( = O)C(N(C( = O)CC)C) = Nc3c2cccc3)c1 | 1 | 167 | −9.0432253 | 1.5727638 | 1 6 | [[33 33 33 33 33] 22] | 111.14684 | −94.654655 | −10.549702 | −10.234877 | −9.0432253 | 5 | 1 | 2.143 | −5.19823 | 3.2323 | 91.309998 | 428.876 |
| Fc1ccc(NC( = O)CN2C( = O)C(N(C( = O)CC)C) = Nc3c2cccc3)cc1 | 1 | 200 | −9.0261774 | 1.1828657 | 6 13 | [17 17] | 97.467995 | −78.931458 | −10.31346 | −18.311417 | −9.0261774 | 4 | 1 | 1.713 | −4.70854 | 2.7093999 | 82.080002 | 382.395 |
| O = C(NCc1occc1)CCCc1sc(C( = O)Nc2ccc(C)cc2)nn1 | 1 | 204 | −10.460769 | 1.9126331 | 4 16 | [12 12] | 32.185101 | −54.236809 | −8.3161516 | −33.550163 | −10.460769 | 4 | 2 | 2.356 | −4.63966 | 3.59729 | 97.120003 | 384.46 |
| O = C1N2C( = NC(CN(Cc3occc3)Cc3ccccc3)=C1)C = C(C)C = C2 | 1 | 45 | −10.189792 | 0.8146282 | 12 13 1 14 | [4 4 [34 41 43 47 45 39] 13] | 62.182995 | −86.637306 | −10.502284 | −29.542992 | −10.189792 | 3 | 0 | 2.653 | −5.3023 | 4.4127998 | 49.049999 | 359.429 |
| Clc1ccc(CSC = 2n3nc(-c4sccc4)cc3C(=O)NN = 2)cc1 | 1 | 127 | −10.149225 | 0.8502005 | 4 | 15 | 122.89852 | −85.182693 | −11.545432 | −41.840046 | −10.149225 | 3 | 1 | 4.503 | −6.32458 | 4.3273001 | 59.279999 | 374.876 |
| S(Cc1ccc(C)cc1)C = 1n2nc(-c3sccc3)cc2C(=O)NN = 1 | 1 | 31 | −10.102728 | 1.0717729 | 4 | 15 | 118.09198 | −76.843628 | −12.053961 | −38.377987 | −10.102728 | 3 | 1 | 4.209 | −6.06421 | 3.9823201 | 59.279999 | 354.458 |
| Clc1ccc(CSC = 2n3nc(-c4occc4)cc3C(=O)NN = 2)cc1 | 1 | 43 | −9.9872217 | 0.9513763 | 3 4 | [[12 16 20 18 14] 15] | 142.22092 | −78.559776 | −11.87368 | −43.721352 | −9.9872217 | 3 | 1 | 3.688 | −6.26948 | 3.8587999 | 72.419998 | 358.809 |
| S(Cc1ccc(C)cc1)C = 1n2nc(-c3occc3)cc2C(=O)NN = 1 | 1 | 7 | −9.9405518 | 0.6945121 | 3 4 | [[12 16 20 18 14] 15] | 137.4003 | −98.104568 | −12.835557 | −39.99435 | −9.9405518 | 3 | 1 | 3.394 | −6.00911 | 3.5138199 | 72.419998 | 338.391 |
| Fc1ccc(C(NC( = O)c2cc(OC)ccc2)c2oc(-c3ccccc3)nn2)cc1 | 1 | 385 | −9.8510714 | 2.1505692 | 12 13 10 | [14 14 9] | 76.445915 | −26.643658 | −8.9426355 | −23.171175 | −9.8510714 | 4 | 1 | 4.972 | −7.21102 | 4.4991999 | 77.25 | 403.413 |
| O = C(Nc1cc(C( = O)C)ccc1)c1c2c(nc(-c3cnccc3)c1)cccc2 | 1 | 94 | −9.8059988 | 1.1290568 | 12 13 10 | [14 14 9] | 3.94593 | −62.999607 | −12.244148 | −26.206341 | −9.8059988 | 4 | 1 | 3.674 | −5.39272 | 4.7516999 | 71.949997 | 367.408 |
| O = C(Nc1cc(OC)c(OC)c(OC)c1)c1c2c(nc(-c3cnccc3)c1)cccc2 | 1 | 552 | −9.8008347 | 1.0850925 | 12 3 10 | [20 [19 31 47 33 21 25] 8] | 58.65123 | −60.098019 | −11.729462 | −14.375378 | −9.8008347 | 6 | 1 | 3.22548 | −5.23159 | 4.5749002 | 82.57 | 415.449 |
| Clc1ccc(CSC2 = NC = Cn3nc(-c4sccc4)cc23)cc1 | 1 | 41 | −9.7324276 | 0.6792789 | [] | 9.6543798 | −81.243019 | −11.77093 | −37.725327 | −9.7324276 | 2 | 0 | 3.792 | −6.02345 | 5.6532001 | 30.18 | 357.889 | |
| S(Cc1ccc(OC)cc1)C1 = NC = Cn2nc(-c3ccccc3)cc12 | 1 | 23 | −9.5824881 | 1.1068726 | 2 | 13 | 7.6172404 | −61.247097 | −10.116899 | −27.129368 | −9.5824881 | 3 | 0 | 3.626 | −5.71699 | 4.9468999 | 39.41 | 347.442 |
| O(CCn1c(Cc2ccccc2)nc2c1cccc2)c1cc(C)ccc1 | 1 | 10 | −9.5771179 | 0.8715386 | [] | 39.945278 | −66.531174 | −11.243786 | −22.246656 | −9.5771179 | 2 | 0 | 5.667 | −5.65198 | 5.28089 | 27.049999 | 342.442 | |
| Clc1ccc(CSC2 = NC = Cn3nc(-c4ccccc4)cc23)cc1 | 1 | 29 | −9.5222225 | 0.9016182 | 2 | 13 | 3.7078457 | −74.063538 | −11.286135 | −32.970608 | −9.5222225 | 2 | 0 | 4.262 | −6.4009 | 5.5917001 | 30.18 | 351.861 |
| S(Cc1ccc(C)cc1)C1 = NC = Cn2nc(-c3sccc3)cc12 | 1 | 6 | −9.5210714 | 0.7866034 | [] | 4.9888377 | −83.027557 | −10.850096 | −33.233433 | −9.5210714 | 2 | 0 | 3.498 | −5.76308 | 5.3082199 | 30.18 | 337.471 | |
| S(Cc1ccc(C)cc1)C1 = NC = Cn2nc(-c3ccccc3)cc12 | 1 | 3 | −9.4389629 | 1.1168524 | 2 | 13 | −0.938683 | −84.930847 | −10.968632 | −29.40621 | −9.4389629 | 2 | 0 | 3.968 | −6.14053 | 5.2467198 | 30.18 | 331.443 |
| O = C(Nc1cc2N(C( = O)c3occc3)CCCc2cc1)c1cc(OC)ccc1 | 1 | 139 | −9.4326887 | 1.2540237 | 10 11 | [17 17] | 90.964813 | −49.745277 | −11.406919 | −24.559771 | −9.4326887 | 3 | 1 | 3.102 | −5.40646 | 4.1334701 | 71.779999 | 376.412 |
| Brc1ccc(CSC2 = NC = Cn3nc(-c4ccccc4)cc23)cc1 | 1 | 302 | −9.3804808 | 1.0461311 | 2 | 13 | 7.9948578 | −61.308563 | −11.300909 | −25.608639 | −9.3804808 | 2 | 0 | 4.468 | −6.757 | 5.7007999 | 30.18 | 396.312 |
| S(Cc1ccc(C)cc1)C1 = NC = Cn2nc(-c3c(C)ccc(C)c3)cc12 | 1 | 55 | −9.3762856 | 1.2313714 | [] | 9.3400259 | −71.844727 | −10.716241 | −16.636683 | −9.3762856 | 2 | 0 | 4.599 | −7.08837 | 5.8635602 | 30.18 | 359.497 | |
| S(Cc1ccc(OC)cc1)C1 = NC = Cn2nc(-c3ccc(F)cc3)cc12 | 1 | 75 | −9.3619413 | 1.3067884 | 2 | 13 | 18.820042 | −57.730362 | −9.95998 | −25.407757 | −9.3619413 | 3 | 0 | 3.779 | −6.01197 | 5.086 | 39.41 | 365.432 |
| Clc1ccc(CSC2 = NC = Cn3nc(-c4c(C)cccc4)cc23)cc1 | 1 | 88 | −9.3481798 | 0.9533808 | 1 | [[11 30 38 40 36 15]] | 5.2840195 | −74.416542 | −10.528507 | −27.199154 | −9.3481798 | 2 | 0 | 4.558 | −6.87482 | 5.9001198 | 30.18 | 365.888 |
| N(Cc1ccc(C)cc1)C = 1n2nc(-c3occc3)nc2N = C(C)C = 1 | 1 | 1 | −9.3377171 | 1.1166085 | [] | 124.29549 | −63.576096 | −11.017919 | −31.1987 | −9.3377171 | 3 | 1 | 1.921 | −5.98017 | 3.8071201 | 68.239998 | 319.368 | |
| S( = O)( = O)(N(C)c1ccc(C)cc1)c1sc(NC( = O)/C = C/c2ccccc2)nn1 | 1 | 542 | −9.3339987 | 2.4967248 | 10 4 16 | [[3 11] 18 18] | 30.843954 | −44.116062 | −9.7825909 | −13.754408 | −9.3339987 | 5 | 1 | 3.646 | −6.61198 | 3.3235199 | 92.260002 | 414.51 |
| O = C(Nc1cc(OC)c(OC)cc1)c1c2c(nc(-c3cnccc3)c1)cccc2 | 1 | 210 | −9.3253307 | 1.1258233 | 12 3 | [16 [14 31 43 33 18 24]] | 17.510517 | −74.765373 | −13.496837 | −13.778906 | −9.3253307 | 5 | 1 | 3.48274 | −5.18121 | 4.5662999 | 73.339996 | 385.423 |
| O(C)c1ccc(CNC = 2n3nc(-c4cnccc4)nc3N = C(C)C = 2)cc1 | 1 | 21 | −9.2783651 | 1.013528 | 1 | [[11 30 43 37 14 18]] | 95.112495 | −80.941689 | −12.355953 | −23.961651 | −9.2783651 | 5 | 1 | 1.631 | −4.73104 | 3.3092999 | 77.220001 | 346.394 |
| Fc1c(C( = O)Nc2cc(CCc3n(C)c4c(n3)cccc4)ccc2)cccc1 | 1 | 115 | −9.2757092 | 1.3320879 | [] | 34.493172 | −59.985386 | −10.428094 | −17.55064 | −9.2757092 | 2 | 1 | 4.989 | −5.65978 | 5.1090398 | 46.919998 | 373.431 | |
| S(CC( = O)N(C)c1ccccc1)C1 = NC(c2ccc(OC)cc2)=NC = 2N1NC( = O)C = 2 | 1 | 643 | −9.2612324 | 1.3560218 | 10 11 4 | [[9 12] 9 19] | 36.53323 | −63.482971 | −11.788753 | −13.550608 | −9.2612324 | 5 | 1 | 3.668 | −6.20003 | 2.3958001 | 86.599998 | 421.481 |
| O = C(NCc1occc1)c1c(C)nc(COc2ccc(C)cc2)s1 | 1 | 9 | −9.2602644 | 1.686085 | 5 6 | [[1 4 5 3 2] 4] | 27.167377 | −58.323898 | −12.913135 | −27.493792 | −9.2602644 | 3 | 1 | 2.534 | −4.51044 | 4.3947401 | 64.360001 | 342.419 |
| O(C)c1ccc(CNC = 2n3nc(-c4cnccc4)nc3N = C(CCC)C = 2)cc1 | 1 | 125 | −9.2581339 | 1.5960585 | 20 21 1 | [[8 8 8 8 8] [8 8 8 8 8] [11 30 43 34 14 18]] | 168.3857 | −49.01144 | −10.639342 | −14.098823 | −9.2581339 | 5 | 1 | 2.548 | −5.44803 | 4.0895 | 77.220001 | 374.448 |
| Clc1c(CNC = 2n3c(nnc3)N = C(c3ccc(OC)cc3)C = 2)cccc1 | 1 | 81 | −9.2345181 | 1.6097397 | 15 1 | [8 [15 31 39 41 33 19]] | 84.841797 | −67.491676 | −12.882171 | −27.361973 | −9.2345181 | 4 | 1 | 4.317 | −5.79835 | 3.9291 | 64.330002 | 365.824 |
| S(Cc1ccc(C)cc1)C1 = NC = Cn2nc(-c3c(C)cccc3)cc12 | 1 | 19 | −9.2313328 | 1.0720456 | 1 2 | [[11 29 41 43 35 15] 14] | 0.95992428 | −80.215492 | −11.268885 | −24.245464 | −9.2313328 | 2 | 0 | 4.264 | −6.61445 | 5.55514 | 30.18 | 345.47 |
| Clc1ccc(CSC2 = NC = Cn3nc(-c4ccc(F)cc4)cc23)cc1 | 1 | 97 | −9.2252741 | 1.1121234 | 2 | 13 | 13.107337 | −69.086746 | −10.447957 | −28.599001 | −9.2252741 | 2 | 0 | 4.415 | −6.69588 | 5.7308002 | 30.18 | 369.851 |
| O = C(Nc1ccc(C)cc1)c1sc(C2N(C( = O)c3ccccc3)CCC2)nn1 | 1 | 281 | −9.1642857 | 1.8345664 | 5 24 | [[1 5 6 2 3] [1 5 6 2 3]] | 64.915665 | −66.42865 | −10.25529 | −14.549685 | −9.1642857 | 4 | 1 | 3.873 | −5.18932 | 4.1716199 | 75.190002 | 392.483 |
| O = C(Nc1ccccc1)CC1N(Cc2occc2)C( = O)N(c2cc(OC)ccc2)C1 = O | 1 | 607 | −9.1215267 | 1.2033887 | 10 11 | [19 19] | −31.805082 | −33.053543 | −10.671512 | −5.9882545 | −9.1215267 | 4 | 1 | 2.22 | −5.20056 | 3.9208 | 92.089996 | 419.437 |
| O = C(Nc1c(OC)cc(OC)cc1)c1c2c(nc(-c3cnccc3)c1)cccc2 | 1 | 211 | −9.0648451 | 1.6063622 | 3 | [[15 31 43 33 17 24]] | 25.543804 | −70.667473 | −11.988891 | −9.2061634 | −9.0648451 | 5 | 1 | 3.731 | −5.18121 | 4.5662999 | 73.339996 | 385.423 |
| Clc1ccc(CSC = 2n3nc(-c4ccc(C)cc4)cc3C(=O)NN = 2)cc1 | 1 | 194 | −9.0527763 | 1.4399766 | 15 1 2 4 | [14 [13 17 24 28 26 15] 12 14] | 138.23361 | −91.044739 | −10.411779 | −18.228752 | −9.0527763 | 3 | 1 | 5.271 | −7.17595 | 4.5742202 | 59.279999 | 382.875 |
| O = C(Nc1cc2N(C( = O)c3occc3)CCCc2cc1)c1cc(OC)c(OC)cc1 | 1 | 430 | −9.039814 | 1.4730424 | 10 11 | [21 21] | 112.04241 | −70.091591 | −11.564325 | −11.970048 | −9.039814 | 4 | 1 | 2.80774 | −5.45684 | 4.1420698 | 81.010002 | 406.438 |
| Clc1ccc(CN(C)c2c(-c3onc(-c4c(C)cccc4)n3)cccn2)cc1 | 1 | 273 | −9.0395203 | 1.1121174 | 19 | 19 | 78.985283 | −76.667534 | −11.215223 | −10.100689 | −9.0395203 | 3 | 0 | 5.036 | −7.99421 | 5.6632199 | 55.049999 | 390.874 |
| O = C(Nc1c(CCc2n(C)c3c(n2)cccc3)cccc1)Cc1cc(OC)ccc1 | 1 | 337 | −9.0243988 | 1.3242083 | 8 26 | [7 7] | 7.9607992 | −86.649765 | −12.920856 | −5.0245686 | −9.0243988 | 3 | 1 | 4.88 | −5.47665 | 4.9074101 | 56.150002 | 399.494 |
| S(Cc1ccc(C = C)cc1)C1 = NC = Cn2nc(-c3c(C)cccc3)cc12 | 1 | 40 | −9.0214014 | 0.8013189 | 1 2 | [[11 34 42 44 40 15] 14] | 15.069262 | −78.46283 | −10.606567 | −16.755856 | −9.0214014 | 2 | 0 | 4.8 | −7.23722 | 5.88972 | 30.18 | 357.481 |
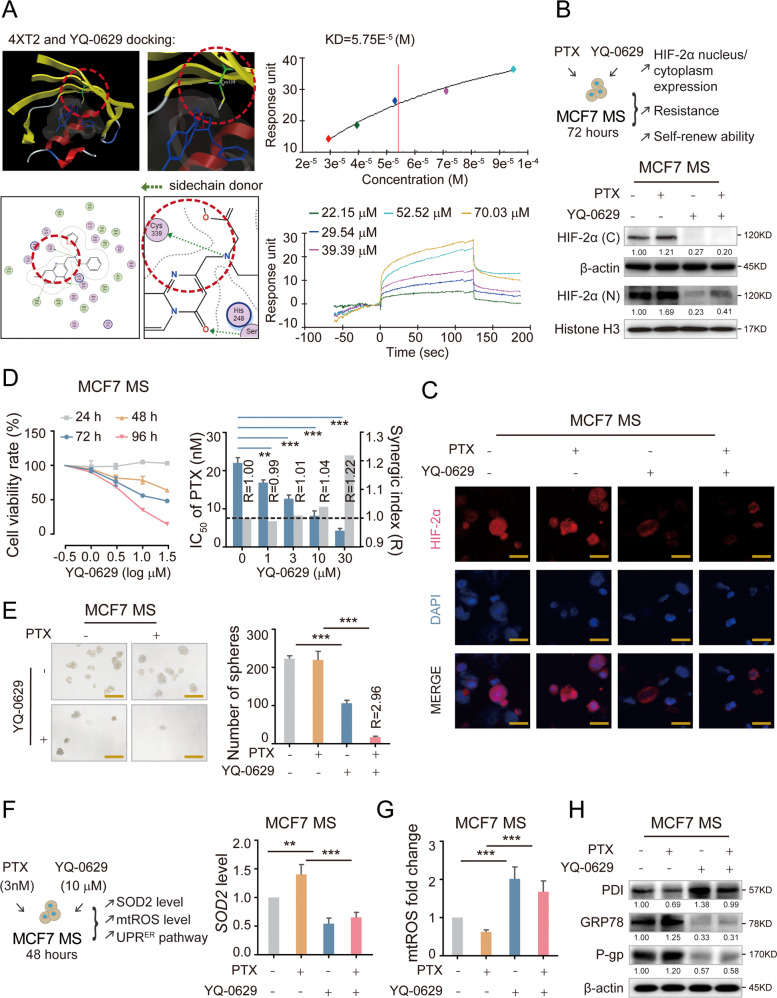
Fig. 6. YQ-0629 targets HIF-2α to suppress stem trait of BCSCs and synergy the sensitization to PTX in vitro.
A The chemical structure of YQ-0629 and docking conformation showed the interaction of the YQ-0629 with the active site of HIF-2α through MOE software (left). YQ-0629 and HIF-2α PAS-B domain was confirmed direct binding by surface plasmon resonance (SPR)-based Biacore assay (right). B The expression level of HIF-2α was detected in the nucleus (N) and cytoplasm (C) of MCF7 MS cells cultured with YQ-0629 (10 µM) alone, PTX (3 nM) alone, or YQ-0629 (10 µM) combined with PTX (3 nM) for 72 h. C The expression and location of HIF-2α were detected in MCF7 MS cells cultured with YQ-0629 (10 µM) alone, PTX (3 nM) alone, or YQ-0629 (10 µM) combined with PTX (3 nM) for 72 h by immunofluorescence staining. Scale bar, 10 μm. D The cell viability rate of MCF7 MS cells cultured with different concentrations of YQ-0629 for 24–96 hours were determined by CCK-8 assay (left). The IC50 values of PTX and synergic index (R) were calculated in MCF7 MS cells cultured with indicated dose of YQ-0629 for 72 h (right). n = 3. Student’s t test. E The self-renewal ability was detected in MCF7 MS cells cultured with YQ-0629 (10 µM) alone, PTX (3 nM) alone, or YQ-0629 (10 µM) combined with PTX (3 nM) for 72 h. n = 3. Synergic index (R). Two-way ANOVA test. Scale bar, 250 μm. F–H The mRNA expression of SOD2 and mtROS level, the expressions level of PDI, GRP78, and P-gp were detected in the MCF7 MS cells cultured with indicated dose of PTX and YQ-0629 for 72 h. n = 3. Two-way ANOVA test. **P < 0.01, **P < 0.01, ***P < 0.001, compared to MCF7 MS cells/MCF7 MS cells + PTX treatment.
YQ-0629 significantly inhibited cell viability of MCF7 MS in a time- and dose-dependent manner (Fig. 6D, left panel). We treated MCF7 MS with YQ-0629 and PTX to test if a synergistic effect exists between the two drugs. We observed that the IC50 values of PTX at 72 h gradually decreased with the increase of YQ-0629 dose. Importantly, the synergistic indices (R values) were all greater than 1, with a greatest R value of 1.22 when PTX is used in combination with 30 μM YQ-0629, implying the presence of a synergistic effect between YQ-0629 and PTX on tumor suppression (Fig. 6D, right panel). Notably, YQ-0629 treatment either used alone or in combination with PTX (R = 2.96), impaired the self-renewal ability of MCF7 MS (Fig. 6E). Additionally, the effect of YQ-0629 treatment in MCF7 MS was consistent with that of HIF-2α depletion on downstream pathway related parameters (Fig. 6F, G). Collectively, these data suggested that YQ-0629, as a HIF-2α targeted inhibitor, suppresses self-renewal capacity and synergizes with PTX to reverse the chemoresistance of MCF7 MS.
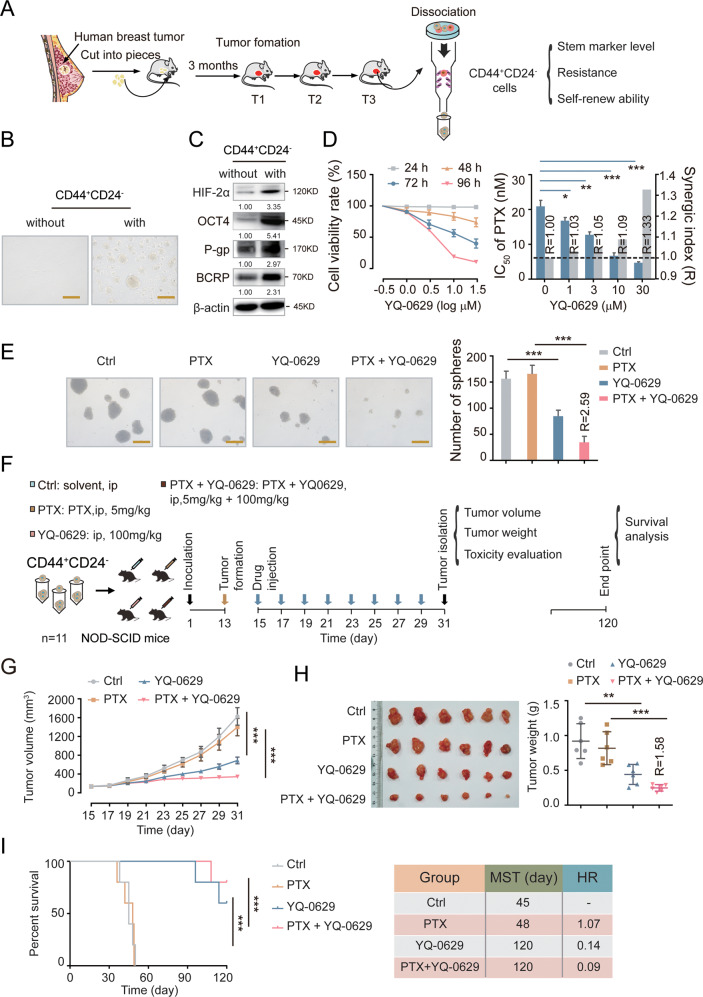
Next, we conducted a more stringent preclinical assessment on the efficacy of YQ-0629. First, we sorted out the CD44+CD24− BCSC population from the clinical samples of breast cancer patients and detected the expression levels of stemness-related, chemoresistance-related, and self-renewal-related markers (Fig. 7A). The CD44+CD24− cells displayed BCSC properties (Fig. 7B, C). Next, we treated the CD44+CD24− cells with YQ-0629 and found that YQ-0629 inhibited cell viability in a time- and dose-dependent manner (Fig. 7D, left panel). Similarly, a synergistic effect was observed between PTX and YQ-0629 (R = 1.33, Fig. 7D, right panel). Consistently, YQ-0629 alone or in combination with PTX (R = 2.59) decreased mammosphere forming ability (Fig. 7E). We further explored the effect of YQ-0629 in vivo (Fig. 7F). YQ-0629 remarkably suppressed xenograft tumor growth and prolonged survival in the transplanted mice, especially when combined with PTX (Fig. 7G, I), demonstrating that YQ-0629 synergizes with PTX not only for the inhibition of tumor growth (R = 1.58) but also for the prolongation of survival. Finally, we evaluated the toxicity of YQ-0629 in vital organs. No noticeable change to the bladder, heart, kidney, liver and lung was observed, indicating that YQ-0629 was safe at the dosage we used (Fig. S5E). Overall, our data demonstrated that YQ-0629 specifically targets HIF-2α to inhibit BCSC self-renewal capacity via the SOD2-mtROS-PDI/GRP78-UPRER pathway and synergizes with PTX to overcome chemoresistance in BCSCs.
Fig. 7. YQ-0629 inhibits growth of BCSCs derived from breast cancer patients and synergistically increases anti-BCSCs activity of PTX in vivo.
A The diagram showed the process of sorting out CD44+CD24− phenotype cells from primary human BCs by MACS. B The mammosphere’s morphology of CD44+CD24− cells were examined. Scale bar, 250 μm. C The expression of HIF-2α, OCT4, P-gp, and BCRP were detected in CD44+CD24− cells. D The cell viability rate of CD44+CD24− cells cultured with different concentrations of YQ-0629 for 24–72 h were determined by CCK-8 assay (left). The IC50 values of PTX and synergic index (R) were calculated in the MCF7 MS cells cultured with indicated dose of YQ-0629 for 72 h. n = 3. Student’s t test. E The self-renewal ability was measured in CD44+CD24− cells cultured with YQ-0629 (10 µM) alone, PTX (3 nM) alone, or YQ-0629 (10 µM) combined with PTX (3 nM) for 72 h. n = 3. Synergic index (R). Two-way ANOVA test. Scale bar, 250 μm. F The diagram showed the time of tumor formation in NOD-SCID mice transplanted with CD44+CD24− cells (1 × 105). 15 days after the inoculation, the mice were intraperitoneally injected with PTX (5 mg/kg), or YQ-0629 (100 mg/kg), or PTX (5 mg/kg) plus YQ-0629 (100 mg/kg), or PEG-35 castor oil as control, once every other day till to the 31th day for test (n = 6). And the other mice (n = 5) were observed survival till the 120th day. G The growth curves of tumor volumes were measured in xenograft mice of each group every other day (n = 6). Two-way ANOVA test. H The tumor weights in each group were measured (n = 6). Synergic index (R). Two-way ANOVA test. I The survival of the mice was analyzed in each group by Kaplan Meier-plotter curves. Median survival times (MST). Hazard ratio (HR). (n = 5). Two-way ANOVA test. *P < 0.05, **P < 0.01, ***P < 0.001, compared to non CD44+CD24− cells/Ctrl/Ctrl + PTX treatment.
Discussion
Here, we provided compelling in vitro and in vivo evidences that HIF-2α, induced by hypoxic TME, promotes the transition of chemosensitive BCs to chemoresistant BCSCs. Mechanistically, HIF-2α suppresses PDI, which competes with GRP78 for binding to misfolded proteins, to allow GRP78 dissociation from sensors of UPRER to turn on “the switch” of UPRER and thereby promote stemness in BCs. More specifically, HIF-2α decreased mtROS production and transportation from mitochondria to ER by transcriptionally upregulating SOD2, which consequently inhibited the expression and activity of PDI and promoted the binding of GRP78 with misfolded proteins. More importantly, we screened out a small molecule HIF-2α inhibitor that synergizes with PTX for inhibition of tumor growth in vitro and in vivo, and represents a promising candidate for targeted therapy of breast cancer.
In the adaptive response to hypoxia, HIF-2α acts as a crucial regulator of cancer stemness [13, 40–42]. Consistent with previous studies, we confirmed that HIF-2α was responsible for the elevation of OCT4 [43], P-gp [44], and BCRP [45] under hypoxia. Furthermore, we showed that HIF-2α, but not HIF-1α, was highly expressed in CD44+CD24− breast cancer tissues. Emerging evidence demonstrates that hypoxia induces cellular reprogramming to reshape cancer cell stemness [46]. However, current studies mainly focus on changes occurring inside mitochondria, such as impairment of oxidative phosphorylation, promotion of glycolytic activity, and increase in mtROS production [47]. Our study added another layer to the mechanism by which HIF-2α regulates the signal transduction between mitochondria and ER in breast cancer. We showed that HIF-2α promoted breast cancer stemness via its regulation of UPRER. These findings highlight the significance of UPRER activation in HIF-2α-induced stemness and chemoresistance acquisition.
UPRER is crucial for cancer stemness acquisition and maintenance. Although previous report revealed a potential interaction between HIF-2α and UPRER in hematopoietic stem and progenitor cells (HSPCs) [20], we found that HIF-2α regulates UPRER differently in BC. First, all three downstream pathways of UPRER (IRE1, PERK, and ATF6) were inhibited in HIF-2α-knockdown BCSCs. Second, in our study, although an increase in mtROS transportation from mitochondria to ER was also observed in HIF-2α-knockdown BCSCs, this event led to dysregulation of protein synthesis and impairment of stemness in BCSCs rather than induction of apoptosis. We speculate that the extent of hypoxia in TME and the HIF-2α expression level vary tremendously among different tissues and might contribute to such discrepancy, but the underlying reason need to be further investigated.
Aggregation of misfolded proteins in ER triggers ERS, and UPRER is involved in this event to control cell fate [36]. GRP78 is a sentinel marker of ERS and is widely used as a “switch” for UPRER activation [48]. In addition to the mechanism that GRP78 disassociates from ER transmembrane protein sensors and activates UPRER via binding to misfolded proteins, misfolded proteins are also regulated by GRP78 via other mechanisms [49]. The complete mechanism of this complicated process needs to be further clarified by more studies. Here we found that PDI [34], a multifunctional redox chaperone, was repressed for its activity and expression when the transportation of mtROS from mitochondria to ER was decreased. Like GRP78, PDI was also capable of binding to and disassociating from misfolded proteins. The inhibition of PDI allowed more misfolded protein to be bound with GRP78, which in turn caused GRP78 dissociation from UPRER sensors and eventually activated UPRER. This finding revealed the critical function of PDI in controlling the binding between GRP78 and misfolded proteins and in fine-tuning UPRER activation in response to ERS.
ROS level regulates cancer stemness in a wide variety of cancers [50, 51]. Although previous studies have shown that lower ROS level regulated by a branch of UPRER positive feedback loop played an essential role in stemness maintenance, the specific mechanism for crosstalk between ROS level and UPRER remains poorly understood [33, 52]. Our study revealed that mtROS was generated in mitochondria and then transported to ER to modulate PDI expression and activity. In the ER lumen, the homeostatic redox environment is required for proper protein folding and disulfide bond formation. PDI is an ER oxidoreductase that catalyzes disulfide bond formation and oxidative protein folding. The regulation of PDI by mtROS functions as a “bridge” linking mitochondrial oxidative phosphorylation to UPRER activation.
We also screened out a novel specific HIF-2α inhibitor YQ-0629. In hypoxic TME, HIF-2α proteins are stabilized and heterodimerize with ARNT to regulate related gene profile [38]. We found that YQ-0629 could interact with the PAS-B pocket of HIF-2α protein, preventing the dimerization of HIF-2α with ARNT and promoting its degradation. YQ-0629 is a highly specific inhibitor for HIF-2α which does not affect HIF-1α. Although several small molecules have been identified to selectively bind to the PAS-B domain, no study have shown the clinical value of HIF-2α inhibitor for breast cancer treatment [53–56]. In this study, we verified that YQ-0629 not only suppressed breast cancer stemness in vitro, but also synergized with PTX to inhibit tumor growth and prolong survival in vivo, highlighting the great potential of YQ-0629 for the targeted treatment of breast cancer.
In summary, we elucidated that HIF-2α remodeled stemness in breast cancer and conferred chemoresistance to BCs via SOD2-mtROS-PDI/GRP78-UPRER axis. We also developed a novel HIF-2α targeted inhibitor YQ-0629, which synergizes with PTX to suppress tumor growth in vitro and in vivo and represents a promising small molecule compound for breast cancer targeted therapy.
Material and methods
Reagents
Recombinant human SOD2 (Abcam, ab93946), Paclitaxel (Sigma, T7402), Doxorubicin, (Sigma, D1515), Mitoxantrone (Sigma, M2305000), Cisplatin (Sigma, P4394), Docetaxel (Sigma, 01885), Mito TEMPO (Sigma, SML0737), PDI inhibitor 16F16 (Sigma, SML0021), YQ-0629 (Topscience, chemdiv diversity discovery library), (3-Aminopropyl) triethoxysilane (sigma, A3648), Glutaraldehyde solution (sigma, G7651).
Cell culture and lentiviral transfection
Human breast cancer cell lines MCF7 and T47D were purchased from ATCC (MCF7: ATCC cat.HTB-22, RRID: CVCL_0031; T47D: ATCC cat.HTB-133, RRID: CVCL_0553). The two cell lines have been authenticated. MCF7, MCF7 MS and T47D, T47D MS cells were cultured as reported previously by He et al. [25, 57]. For hypoxia culture, cells were incubated in the hypoxia incubator (Thermo Fisher Scientific, HERACELL 150i) at 1% O2. Mammosphere cells derived from breast cancer patients were also cultured in DMEM-F12 medium supplemented with 2% B27, 10 μg/l b-FGF, and 20 μg/l EGF.
Lentiviral vectors were purchased from Shanghai Genechem Co., Ltd. HIF-2α-RNAi sequence is seen in Supplementary Table 1. HIF-2α cDNA (HIF-2α-OE) was cloned into a Ubi-MCS-3FLAG-SV40-EGFP-IRES-puromycin vector. GRP78, PDI and SOD2 cDNAs (GRP78-OE, PDI-OE, SOD2-OE) were cloned into a Ubi-MCS-SV40- puromycin. MCF7, T47D, MCF7 MS, and T47D MS cells were seeded into 6-well adhesion plates or ultra-low adhesion plates (Corning) at a density of 2 × 105 cells per well. The next day, lentiviral vectors and polybrene were mixed with medium (NC, MOI = 10; MOI = 20, HIF-2α-OE, GRP78-OE, PDI-OE, SOD2-OE; shCtrl, MOI = 20; shHIF-2α, MOI = 25). After transfection for 12 h, fresh culture medium was added, and culturing for another 48 h. Puromycin (2 μg/ml) was added to select stably transfected cells. This process was repeated 2 to 3 times until all cells expressed green fluorescent protein (GFP).
RNA isolation and real-time PCR
RNA isolation and real-time PCR were performed as described previously by Ma et al. [58]. Primer sequences are shown in Supplementary Table 2.
Immunofluorescence and misfolded protein detection
Paraffin-embedded and OCT-embedded samples were sectioned at 4 μm thickness. Antigen retrieval was performed by a pressure cooker for 10 min in 0.01 citrate buffer (pH 6.0) to remove aldehyde links formed during initial fixation of tissues (this step is just for paraffin-embedded samples, skip for OCT-embedded fresh tissue samples). Cells for immunofluorescence were fixed with 4% paraformaldehyde for 20 min at room temperature, washed with PBS and permeabilized with 0.2% Triton X-100 in PBS for 15 min. Thereafter, cells were blocked in PBS with the Normal Goat Serum (Solarbio) for 30 min at 37 °C. Then, samples were incubated with primary antibodies HIF-2α, HIF-1α, PDI, GRP78 (Seen in Supplementary Table 3) overnight at 4 °C. Incubation of Alexa Fluor-conjugated secondary antibodies (EarthOx Life Sciences) were carried out for 30 min at 37 °C protected from light. Misfolded protein was detected with the PROTEOSTAT® Aggresome Detection Kit (ENZ-51035; Enzo Life Sciences, USA) according to the manufacturer’s instructions. DAPI was used counterstaining the nuclei, ER-trackerTM was used for staining the ER (ThermoFisher, E12353), CellMaskTM was used for staining the plasma membranes (ThermoFisher, C10046), MitoGreen was used for staining the mitochondria (KeyGEN BioTECH, KGMP0072) and MitoSOXTM (Thermo Fisher Scientific, m36008) was used for staining mtROS in cells. Images were obtained by laser scanning confocal microscopy (Nikon ECLIPSE Ti).
Western blot analysis
Cells were harvested in RIPA lysis buffer (50 mM Tris-HCI; pH 7.5, 120 mM NaCl, 0,5% NP-40, 200 mM Na3VO4, 1 mM EDTA, 0.5% sodium deoxycholate, 1% SDS) containing 1 × protease inhibitor cocktail (Sigma) for 10 min on ice. An equal amount of protein from the cell lysates was resuspended in the gel sample buffer, resolved by SDS-PAGE, and transferred to PVDF membrane (Millipore). After blocking with 5% BSA, membranes were incubated with specific primary antibodies (Supplementary Table 3) overnight at 4 °C. After washing with TBST, corresponding secondary antibodies were incubated at room temperature for 1 h. The immunoreactivity of the signals was visualized by using ECL system (DNR MicroChemi system). The uncropped western blots can be seen in the Supplementary Material.
Cell viability assay
Cell viability assay was performed as reported previously [25] by first seeding MCF7, T47D cells (2000 cells/well), or MCF7 MS, T47D MS cells (5000 cells/well) into a 96-well cell culture plate for overnight. Indicated concentrations of Paclitaxel (PTX), adriamycin (ADR), mitoxantrone (MX) and cisplatin (DDP), YQ-0629 or vehicle (DMSO) alone were then added to the wells in 100 μl DMEM or DMEM-F12 medium. The calculation formula of synergistic index (R) is R = (S-exp)/(S-obs). S-exp referred to the product of cell viability rate after the two single drugs respectively acted on cells, and it’s expressed by the formula S-exp = (S-EA) × (S-EB). S-obs referred to the cell survival rate after the two drugs acted on cells in combination. R > 1.0 indicated that the two drugs had a synergistic effect, and R < 1.0 indicated that the two drugs had no synergistic effect.
Colony formation assay
For soft agar colony formation assay, 6-well plates were coated with a bottom layer of 1.2% SeaPlaque low melting temperature agarose (Lonza Rockland, ME USA) in phenol red-free medium supplemented with 20% FBS. 2000 cells were mixed in 0.6% agarose and the same medium, and applied as the top agarose layer. The top agarose layer was overlaid with 600 μl medium. Plates were incubated at 37 °C in 5% CO2 for 3 weeks until colonies formed. Colonies were stained with 100 μl MTT (5 mg/ml) in each well and incubated for 30 min at 37 °C. Colonies were calculated using the analysis software Quantity One (BioRad, Herculed, California, USA).
For plate colony formation assay, 4000 cells were plated into each well of six well plates and grown for 14 days. At the end of the experiment, cell colonies were fixed with 4% paraformaldehyde, and stained with 0.5% crystal violet (Sigma–Aldrich 46364). The number of colonies was counted under a Nikon eclipse TE2000-U microscope with pictures taken. The assay was performed three times in triplicate.
Sphere formation assay
After treated with indicated concentrations of PTX, Mito TEMPO, PDI inhibitor 16F16, or YQ-0629 for 48 h, MCF7 MS, T47D MS or Patient MS cells (2000 cells/ml) were cultured in ultra-low adhesion plates (Corning) in DMEM-F12 medium containing 2% B27, 10 μg/l b-FGF, 20 μg/l EGF. After culturing for 14 days, spheres with diameter >150 μm were counted. At least three independent experiments were conducted.
Flow cytometry
For cell surface markers analysis, cells were resuspended in PBS and stained with fluorescent-conjugated antibodies against CD44 (Biolegend, 103011) and CD24 (BD, 311106) for 30 min at 37 °C in darkness. For ALDH1+ detection, the treatment of cells was followed the manual protocol (STEMCELL, #01700). For the mtROS detection, resuspended cells were stained with 5 μM MitoSOXTM (Thermo Fisher Scientific, m36008) and reagent working solution at 37 °C for 30 min in darkness. Specimens were subsequently analyzed by MACSQuatTM Flow cytometer (Becton-Dickinson, USA).
Coimmunoprecipitation assay
Cells were harvested and lysed in the lysis buffer on ice for 30 min. After centrifugation at 4 °C at 12,000 × g for 10 min, GRP78 (Supplementary Table 3) antibodies were added to the supernatant with rolling at 4 °C overnight. Protein G or A agarose (Beyotime Biotechnology) was then added to samples, and samples were rolled at 4 °C for 2 h. After beads were washed three times with lysis buffer, pellets were dissolved into 2 × SDS loading buffer. After centrifugation and boiled at 100 °C for 10 min, proteins were analyzed by immunoblotting with the PDI, PERK and GRP78 antibodies (Supplementary Table 3).
Patients and tissue samples
Tumor samples were taken from 110 patients with breast carcinoma who underwent neither radiation nor chemotherapy prior to surgery at the First Affiliated Hospital of China Medical University, Liaoning Province, China between 2006 and 2008. According to the pathological staining, cancer was diagnosed. The patient age, menopausal status, tumor type, tumor size and lymph node metastasis were acquired from clinical records. All samples were collected from patients with informed consent, and all related procedures were performed with the approval of the internal review and ethics boards of the indicated hospitals.
Immunohistochemistry (IHC)
Immunohistochemistry was carried out as previously described [25] Paraffin-embedded sections (4 μm) from breast cancer were dewaxed and dehydrated, and antigen retrieval was performed by high pressure sections in Citrate Antigen Unmasking Solution (Beyotime, P0081) for 10 min. After blocked with the Normal Goat Serum (Solarbio), sections were incubated overnight at 4 °C using primary antibodies (shown in Supplementary Table 3). Following incubation with a biotinylated secondary antibody (anti-rabbit, 1:200; Vector Laboratories) for 30 min at 37 °C, antigens were revealed with 3,3’-diaminobenzidine (Solarbio).
Mouse xenografts
BALB/c (nu/nu) mice (Hua Fukang Biological Technologies Inc, Beijing) at 4–6 weeks of age were bred in pathogen-free conditions at the Animal Center of China Medical University. The mice were grouped according to their body weight to snake-shaped grouping method. The investigator was not blinded to the group.
For the study of tumorigenic abilities of MCF7 and MCF7 MS cells, different quantity of cells was suspended in PBS and Matrigel (200 μl, 1:1, BD Biosciences). Then the cell mixture was subcutaneously injected into the right flank of nude mice (n = 5/group). Tumor volume (V) was monitored with digital calipers using the following formula: Width2 × Length/2. After 31 days, the mice were euthanized, and the xenograft tumors were excised for the study.
For the study of effects of HIF-2α knockdown on sensitivities of xenograft tumors to PTX, equal numbers (1 × 105) of suspended stably sh-Ctrl-transfected MCF7 MS, or sh-HIF-2α-transfected MCF7 MS cells were injected subcutaneously into the mammary fat pad of nude mice. When tumor volumes reached over 125 mm3, xenograft mice were randomly divided into four groups: sh-Ctrl alone, sh-Ctrl + PTX, sh-HIF-2α alone, sh-HIF-2α + PTX groups (n = 10/group). The mice in sh-Ctrl + PTX and sh-HIF-2α + PTX groups were intraperitoneally injected with PTX (5 mg/kg), and mice in sh-Ctrl and sh-HIF-2α groups were intraperitoneally injected with PEG-35 castor oil as control once every other day. After 31 days inoculation, some of them (n = 5/group) were sacrificed and tumors were weighted and harvested for further test. Before sacrificed, mice were anesthetized with chloral hydrate then photographed using Living Image software (Perkin-Elmer). And the survival of other mice (n = 5/group) was observed till to the 120th day.
Patient derived xenogtaft (PDX) experiments
To establish patient-derived xenografts (PDX), primary tumor specimens were collected from breast cancer patients who underwent tumor resection at the First Affiliated Hospital of China Medical University (Shenyang, China) between 2016 and 2019. Clinical features of patients were provided in Supplementary Table 4. Eight-week-old NOD-SCID mice under pathogen-free conditions were used for PDX transplantation. Briefly, a small incision was made on the abdomen of anaesthetized NOD-SCID mice to reveal the mammary gland, and primary breast tumor samples were minced into 1–2 mm3 sized fragments and injected directly into the fourth pair of mammary fat pads of the mice. The incision was then closed with sutures. The time from cancer samples collection to mice implantation ranges from 1–3 h. The tumor formation was monitored in the next 3–4 months since implantation. After PDX transplant succeeded, xenografted tumors were minced into 1–2 mm3 sized fragments. Some fragments were transplanted again to generate secondary PDX, and some were digested with Collagenase/Hyaluronidase (Stem Cell Technology, 07912) overnight then added 3 ml pre-warmed 0.25% Trypsin-EDTA (Gibco, Thermo Fisher Scientific, 25200056) to digest fragments into single cell suspension. 1 × 107 cell suspension was incubated with CD44 Microbeads (Miltenyi Biotec, 130–095–194) and CD24 MicroBeads Kit (Miltenyi Biotec, 130–0950951) to separate CD44+CD24− BCs. Amplified CD44+CD24− BC cells in MammoCultTM medium (Stem Cell Technology, 05620) with 10% FBS (Stem Cell Technology, 05620).
For the study of effects of YQ-0629 on sensitivities of the PDX to PTX, equal numbers (1 × 105) of suspended patient MS cells were injected subcutaneously into the mammary fat pad of NOD-SCID mice. The mice were grouped according to their body weight to snake-shaped grouping method. The investigator was nor blinded to the group. When tumor volumes reached over 125 mm3, the PDX mice were randomly divided into four groups: Control, PTX, YQ-0629, PTX + YQ-0629 groups (n = 11/group). The mice in PTX group were intraperitoneally injected with PTX (5 mg/kg), the mice in YQ-0629 group were intraperitoneally injected with YQ-0629 (100 mg/kg), the mice in PTX + YQ-0629 group were intraperitoneally injected with PTX (5 mg/kg) plus YQ-0629 (100 mg/kg), and mice in Control group were intraperitoneally injected with PEG-35 castor oil once every other day. After 31 days inoculation, some of them (n = 6/group) were sacrificed and tumors were weighted and harvested for further test. And the survival of other mice (n = 5/group) was observed till to 120th day. All above animal studies were approved by Animal Research Committee at China Medical University.
High-performance liquid chromatography–tandem mass spectrometry (HPLC-MS)
The procedures were performed by our previous reports [45]. Cells and tissues were prepared by removing proteins through a liquid-liquid extraction method. Cells (1 × 107) or xenografts tissues (0.1–0.2 g) were added 500 μl methanol and fully homogenized. After centrifuging at 1000 rpm for 10 min, keeping the supernatant, 10 μl docetaxel (1 μg/ml, Sigma 1224562), as an internal standard (IS), 50 μl NaHCO3 saturated solution, and 1 ml tertbutyl methyl ether mixture was added and fully mixed. Then the mixture solution was centrifugated at 1 × 104 rpm for 10 min, and the upper organic phase was kept and evaporated to dryness at 4 °C. The dry residue was dissolved in 100 ul mobile phase with vortex-mixing for 1 min, then the reconstituted extract was taken and centrifugated at 13,000 rpm for 10 min at 4 °C, and 10 ul of supernatant fluid was kept and injected into the HPLC-MS/MS system for analysis. Controls and samples were analyzed on a 3500 MS/MS system from Applied AB Sciex (Ontario, Canada) coupled to an Agilent HPLC 1290 system (Agilent, Santa Clara, CA, USA). Separations were accomplished on an Agilent ZORBAX Eclipse Plus C 18 (2.1 mm × 100 mm, 1.8 µm) with an Agilent guard cartridge at temperature of 30 °C. The mobile phase consisted of methanol and 0.1% formic acid water (70:30) which was delivered at a flow rate of 0.3 ml/min. The injection volume was 5 µl. The mass spectrometer was operated in a positive ion mode with a TurboIonSpray source using ESI ionization in MRM mode.
Computation of protein–protein docking
The structural models of HIF-2α-PAS domain (4XT2), HIF-1α-PAS domain (4H6J), GRP78 polypeptide-binding pocket (6ASY), reduced PDI (4EKZ), oxidized PDI (4EL1) were downloaded from the Protein Data Bank (http://www.rcsb.org/pdb/home/home.do). The chemical library was from chemdiv diversity discovery library (Topscience Co., Ltd). The docking model between GRP78 and PDI was performed by MOE2016 software (Chemical Computing Group Inc).
PDI assay
PDI activity was assayed by measuring the PDI-catalyzed reduction of insulin in the presence of DTT, thus measuring the aggregation of reduced insulin B chains at 620 nm. Briefly, total protein extracts of different samples were treated according to present manual [59].
Biacore assay
HIF-2α was immobilized on an NTA Sensor Chip in Biacore T200 (GE Healthcare). Binding of HIF-2α to YQ-0629 was analyzed at 25 °C in PBS-P or PBS-P with 2% DMSO at pH7.4 buffer with a flow rate of 30 µg/ml min or 20 µg/ml min. The kinetics and dissociation constant (KD) were calculated with Biacore T200 Evaluation Software (GE Healthcare).
TCGA data processing
Gene’s expression analyses were based on the The Cancer Genome Altas (TCGA) Reasearch Nerwork: http://cancergenome.nih.gov/. In TCGA, total breast cancer patients were 1169 cases. The positive criteria of HIF-2α, HIF1α, P-gp, and BCRP, CD44+CD24- were based on the ROC curve.
Statistical analysis
Statistical analyses were conducted in GraphPad Prism 7 (RRID: SCR_002798). Results were p34resented as the mean ± standard deviation (SD) for at least three experiments. Student’s t test was used to compare differences between two groups. When the variance of each group is not uniform, Wilcoxon or Welch“s T-est can be used for analysis. One-way ANOVA or two-way ANOVA was used to compare differences among three or more groups. Mann–Whitney U analysis was used to compare mRNA expressions of HIF-2α or HIF1α in CD44+CD24− and non-CD44+CD24− breast cancer patients from TCGA database, and protein levels of HIF-2α or HIF-1α in CD44+CD24− and non- CD44+CD24− breast cancer patients from our tissue bank. Pearson χ2 test was used to analyze correlations between HIF-2α or HIF1α and CD44+CD24− mRNA level, or between HIF-2α or HIF-1α and CD44+CD24− protein level. A P < 0.05 was considered statistically significant.
Supplementary information
Acknowledgements
The authors would like to acknowledge the Key Laboratory of Precision Diagnosis and Treatment of Gastrointestinal Tumors, Ministry of Education (China Medical University, Shenyang, China) for providing the space and equipment for conducting the experiments.
Author contributions
YYY, MJW, FJ, TY and MH conceived the study design. YYY, YYZ, LH, BBW, DMY, XML, YW, WFY, HSZ, BC, ZNJ, JW, and YZ performed experiments and collected all data. YYY, MJW, TY, FJ, MH, LZ, and HZW collected and analyzed data and drafted the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (NSFC, No. 81373427, 81902708, 81972794, 81702616), NSFC-Liaoning joint fund key program (No. U20A20413), Key R&D Guidance Plan Projects in Liaoning Province (2019JH8/10300011), and Shenyang S&T Projects (19–109–4–09, 20–204–4–22), Program for Shenyang High Level Talent Innovation and Entrepreneurship Team (2019-SYRCCY-B-01), Major Special S&T Projects in Liaoning Province (2019JH1/10300005), Liaoning provincial department of education scientific research project (QN2019034), Science and Technology Program of Liaoning Province (2017225036).
Data availability
The authors declare that all data supporting the findings of this study are available within the paper in the main text or the Supplementary Materials.
Materials availability
All data on which the conclusions of the paper rely are available to readers.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All above animal studies were approved by Animal Research Committee at China Medical University. All samples were collected from patients with informed consent, and all related procedures were performed with the approval of the internal review and ethics boards of the indicated hospitals.
Consent for publication
All contributing authors agree to the publication of this article.
Footnotes
Edited by G. Del Sal
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yuanyuan Yan, Miao He.
Contributor Information
Tao Yu, Email: yutao@cancerhosp-ln-cmu.com.
Feng Jin, Email: jinfeng@cmu.edu.cn.
Minjie Wei, Email: weiminjiecmu@163.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-022-00963-8.
References
- 1.Keith B, Simon MC. Hypoxia-inducible factors, stem cells, and cancer. Cell. 2007;129:465–72. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Samanta D, Gilkes DM, Chaturvedi P, Xiang L, Semenza GL. Hypoxia-inducible factors are required for chemotherapy resistance of breast cancer stem cells. Proc Natl Acad Sci USA. 2014;111:E5429–38. doi: 10.1073/pnas.1421438111. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Fekir K, Dubois-Pot-Schneider H, Desert R, Daniel Y, Glaise D, Rauch C, et al. Retrodifferentiation of human tumor hepatocytes to stem cells leads to metabolic reprogramming and chemoresistance. Cancer Res. 2019;79:1869–83.. doi: 10.1158/0008-5472.CAN-18-2110. [DOI] [PubMed] [Google Scholar]
- 4.Cazet AS, Hui MN, Elsworth BL, Wu SZ, Roden D, Chan CL, et al. Targeting stromal remodeling and cancer stem cell plasticity overcomes chemoresistance in triple negative breast cancer. Nat Commun. 2018;9:2897. doi: 10.1038/s41467-018-05220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng Y, Huang G, Chen F, Testroet ED, Li H, Li H, et al. Hypoxia enhances buffalo adipose-derived mesenchymal stem cells proliferation, stemness, and reprogramming into induced pluripotent stem cells. J Cell Physiol. 2019;234:17254–68. doi: 10.1002/jcp.28342. [DOI] [PubMed] [Google Scholar]
- 6.Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc Natl Acad Sci USA. 2016;113:E2047–56. doi: 10.1073/pnas.1521518113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gordan JD, Bertout JA, Hu CJ, Diehl JA, Simon MC. HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell. 2007;11:335–47. doi: 10.1016/j.ccr.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johansson E, Grassi ES, Pantazopoulou V, Tong B, Lindgren D, Berg TJ, et al. CD44 Interacts with HIF-2alpha to modulate the hypoxic phenotype of perinecrotic and perivascular glioma cells. Cell Rep. 2017;20:1641–53.. doi: 10.1016/j.celrep.2017.07.049. [DOI] [PubMed] [Google Scholar]
- 9.Yan Y, Liu F, Han L, Zhao L, Chen J, Olopade OI, et al. HIF-2alpha promotes conversion to a stem cell phenotype and induces chemoresistance in breast cancer cells by activating Wnt and Notch pathways. J Exp Clin Cancer Res. 2018;37:256. doi: 10.1186/s13046-018-0925-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He C, Danes JM, Hart PC, Zhu Y, Huang Y, de Abreu AL, et al. SOD2 acetylation on lysine 68 promotes stem cell reprogramming in breast cancer. Proc Natl Acad Sci USA. 2019;116:23534–41.. doi: 10.1073/pnas.1902308116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niu Y, Bao L, Chen Y, Wang C, Luo M, Zhang B, et al. HIF2-induced long noncoding RNA RAB11B-AS1 promotes hypoxia-mediated angiogenesis and breast cancer metastasis. Cancer Res. 2020;80:964–75.. doi: 10.1158/0008-5472.CAN-19-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bai J, Chen WB, Zhang XY, Kang XN, Jin LJ, Zhang H, et al. HIF-2alpha regulates CD44 to promote cancer stem cell activation in triple-negative breast cancer via PI3K/AKT/mTOR signaling. World J Stem Cells. 2020;12:87–99. doi: 10.4252/wjsc.v12.i1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Q, Lou Y, Zhang J, Fu Q, Wei T, Sun X, et al. Hypoxia-inducible factor-2alpha promotes tumor progression and has crosstalk with Wnt/beta-catenin signaling in pancreatic cancer. Mol Cancer. 2017;16:119. doi: 10.1186/s12943-017-0689-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoo HC, Park SJ, Nam M, Kang J, Kim K, Yeo JH, et al. A Variant of SLC1A5 is a mitochondrial glutamine transporter for metabolic reprogramming in cancer cells. Cell Metab. 2020;31:267–83 e12. doi: 10.1016/j.cmet.2019.11.020. [DOI] [PubMed] [Google Scholar]
- 15.Green NH, Galvan DL, Badal SS, Chang BH, LeBleu VS, Long J, et al. MTHFD2 links RNA methylation to metabolic reprogramming in renal cell carcinoma. Oncogene. 2019;38:6211–25.. doi: 10.1038/s41388-019-0869-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wobma HM, Tamargo MA, Goeta S, Brown LM, Duran-Struuck R, Vunjak-Novakovic G. The influence of hypoxia and IFN-gamma on the proteome and metabolome of therapeutic mesenchymal stem cells. Biomaterials. 2018;167:226–34.. doi: 10.1016/j.biomaterials.2018.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang K, Xu P, Sowers JL, Machuca DF, Mirfattah B, Herring J, et al. Proteome analysis of hypoxic glioblastoma cells reveals sequential metabolic adaptation of one-carbon metabolic pathways. Mol Cell Proteom. 2017;16:1906–21.. doi: 10.1074/mcp.RA117.000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miranda-Galvis M, Teng Y. Targeting hypoxia-driven metabolic reprogramming to constrain tumor progression and metastasis. Int J Mol Sci. 2020;21:5487. doi: 10.3390/ijms21155487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koditz J, Nesper J, Wottawa M, Stiehl DP, Camenisch G, Franke C, et al. Oxygen-dependent ATF-4 stability is mediated by the PHD3 oxygen sensor. Blood. 2007;110:3610–7. doi: 10.1182/blood-2007-06-094441. [DOI] [PubMed] [Google Scholar]
- 20.Rouault-Pierre K, Lopez-Onieva L, Foster K, Anjos-Afonso F, Lamrissi-Garcia I, Serrano-Sanchez M, et al. HIF-2alpha protects human hematopoietic stem/progenitors and acute myeloid leukemic cells from apoptosis induced by endoplasmic reticulum stress. Cell Stem Cell. 2013;13:549–63. doi: 10.1016/j.stem.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 21.Mogilenko DA, Haas JT, L’Homme L, Fleury S, Quemener S, Levavasseur M, et al. Metabolic and innate immune cues merge into a specific inflammatory response via the UPR. Cell. 2019;177:1201–16 e19. doi: 10.1016/j.cell.2019.03.018. [DOI] [PubMed] [Google Scholar]
- 22.Simic MS, Moehle EA, Schinzel RT, Lorbeer FK, Halloran JJ, Heydari K, et al. Transient activation of the UPR(ER) is an essential step in the acquisition of pluripotency during reprogramming. Sci Adv. 2019;5:eaaw0025. doi: 10.1126/sciadv.aaw0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Galen P, Kreso A, Mbong N, Kent DG, Fitzmaurice T, Chambers JE, et al. The unfolded protein response governs integrity of the haematopoietic stem-cell pool during stress. Nature. 2014;510:268–72. doi: 10.1038/nature13228. [DOI] [PubMed] [Google Scholar]
- 24.Prager BC, Xie Q, Bao S, Rich JN. Cancer stem cells: the architects of the tumor ecosystem. Cell Stem Cell. 2019;24:41–53. doi: 10.1016/j.stem.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He M, Fu YZ, Yan YY, Xiao QH, Wu HZ, Yao WF, et al. The Hedgehog signalling pathway mediates drug response of MCF-7 mammosphere cells in breast cancer patients. Clin Sci. 2015;129:809–22. doi: 10.1042/CS20140592. [DOI] [PubMed] [Google Scholar]
- 26.Koumenis C, Naczki C, Koritzinsky M, Rastani S, Diehl A, Sonenberg N, et al. Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2alpha. Mol Cell Biol. 2002;22:7405–16. doi: 10.1128/MCB.22.21.7405-7416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Obacz J, Avril T, Le Reste PJ, Urra H, Quillien V, Hetz C, et al. Endoplasmic reticulum proteostasis in glioblastoma-From molecular mechanisms to therapeutic perspectives. Sci Signal. 2017;10:eaal2323. doi: 10.1126/scisignal.aal2323. [DOI] [PubMed] [Google Scholar]
- 28.Scortegagna M, Ding K, Oktay Y, Gaur A, Thurmond F, Yan LJ, et al. Multiple organ pathology, metabolic abnormalities and impaired homeostasis of reactive oxygen species in Epas1−/− mice. Nat Genet. 2003;35:331–40. doi: 10.1038/ng1266. [DOI] [PubMed] [Google Scholar]
- 29.Oktay Y, Dioum E, Matsuzaki S, Ding K, Yan LJ, Haller RG, et al. Hypoxia-inducible factor 2alpha regulates expression of the mitochondrial aconitase chaperone protein frataxin. J Biol Chem. 2007;282:11750–6. doi: 10.1074/jbc.M611133200. [DOI] [PubMed] [Google Scholar]
- 30.Candas D, Li JJ. MnSOD in oxidative stress response-potential regulation via mitochondrial protein influx. Antioxid Redox Signal. 2014;20:1599–617. doi: 10.1089/ars.2013.5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Idelchik M, Begley U, Begley TJ, Melendez JA. Mitochondrial ROS control of cancer. Semin Cancer Biol. 2017;47:57–66. doi: 10.1016/j.semcancer.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sabharwal SS, Schumacker PT. Mitochondrial ROS in cancer: initiators, amplifiers or an Achilles’ heel? Nat Rev Cancer. 2014;14:709–21. doi: 10.1038/nrc3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eletto D, Chevet E, Argon Y, Appenzeller-Herzog C. Redox controls UPR to control redox. J Cell Sci. 2014;127:3649–58. doi: 10.1242/jcs.153643. [DOI] [PubMed] [Google Scholar]
- 34.Wang L, Wang X, Wang CC. Protein disulfide-isomerase, a folding catalyst and a redox-regulated chaperone. Free Radic Biol Med. 2015;83:305–13. doi: 10.1016/j.freeradbiomed.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 35.Bhandary B, Marahatta A, Kim HR, Chae HJ. An involvement of oxidative stress in endoplasmic reticulum stress and its associated diseases. Int J Mol Sci. 2012;14:434–56. doi: 10.3390/ijms14010434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 37.Cao SS, Kaufman RJ. Unfolded protein response. Curr Biol. 2012;22:R622–6. doi: 10.1016/j.cub.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 38.Wu D, Potluri N, Lu J, Kim Y, Rastinejad F. Structural integration in hypoxia-inducible factors. Nature. 2015;524:303–8. doi: 10.1038/nature14883. [DOI] [PubMed] [Google Scholar]
- 39.Scheuermann TH, Stroud D, Sleet CE, Bayeh L, Shokri C, Wang H, et al. Isoform-selective and stereoselective inhibition of hypoxia inducible factor-2. J Med Chem. 2015;58:5930–41. doi: 10.1021/acs.jmedchem.5b00529. [DOI] [PubMed] [Google Scholar]
- 40.Gerri C, Marass M, Rossi A, Stainier DYR. Hif-1alpha and Hif-2alpha regulate hemogenic endothelium and hematopoietic stem cell formation in zebrafish. Blood. 2018;131:963–73.. doi: 10.1182/blood-2017-07-797795. [DOI] [PubMed] [Google Scholar]
- 41.Wang X, Dong J, Jia L, Zhao T, Lang M, Li Z, et al. HIF-2-dependent expression of stem cell factor promotes metastasis in hepatocellular carcinoma. Cancer Lett. 2017;393:113–24.. doi: 10.1016/j.canlet.2017.01.032. [DOI] [PubMed] [Google Scholar]
- 42.Wigerup C, Pahlman S, Bexell D. Therapeutic targeting of hypoxia and hypoxia-inducible factors in cancer. Pharm Ther. 2016;164:152–69. doi: 10.1016/j.pharmthera.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 43.Covello KL, Kehler J, Yu H, Gordan JD, Arsham AM, Hu CJ, et al. HIF-2alpha regulates Oct-4: effects of hypoxia on stem cell function, embryonic development, and tumor growth. Genes Dev. 2006;20:557–70. doi: 10.1101/gad.1399906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao ZJ, Yuan WD, Yuan JQ, Yuan K, Wang Y. Downregulation of HIF-2alpha reverse the chemotherapy resistance of lung adenocarcinoma A549 cells to Cisplatin. Med Sci Monit. 2018;24:1104–11.. doi: 10.12659/MSM.906107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He M, Wu H, Jiang Q, Liu Y, Han L, Yan Y, et al. Hypoxia-inducible factor-2alpha directly promotes BCRP expression and mediates the resistance of ovarian cancer stem cells to adriamycin. Mol Oncol. 2019;13:403–21. doi: 10.1002/1878-0261.12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ullmann P, Nurmik M, Begaj R, Haan S, Letellier E. Hypoxia- and MicroRNA-Induced metabolic reprogramming of tumor-initiating cells. Cells. 2019;8:528. doi: 10.3390/cells8060528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshida GJ. Metabolic reprogramming: the emerging concept and associated therapeutic strategies. J Exp Clin Cancer Res. 2015;34:111. doi: 10.1186/s13046-015-0221-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Madhavan S, Nagarajan S. GRP78 and next generation cancer hallmarks: an underexplored molecular target in cancer chemoprevention research. Biochimie. 2020;175:69–76. doi: 10.1016/j.biochi.2020.05.005. [DOI] [PubMed] [Google Scholar]
- 49.Wang X, Bi X, Zhang G, Deng Y, Luo X, Xu L, et al. Glucose-regulated protein 78 is essential for cardiac myocyte survival. Cell Death Differ. 2018;25:2181–94.. doi: 10.1038/s41418-018-0109-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qian X, Nie X, Yao W, Klinghammer K, Sudhoff H, Kaufmann AM, et al. Reactive oxygen species in cancer stem cells of head and neck squamous cancer. Semin Cancer Biol. 2018;53:248–57.. doi: 10.1016/j.semcancer.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 51.Lee KM, Giltnane JM, Balko JM, Schwarz LJ, Guerrero-Zotano AL, Hutchinson KE, et al. MYC and MCL1 cooperatively promote chemotherapy-resistant breast cancer stem cells via regulation of mitochondrial oxidative phosphorylation. Cell Metab. 2017;26:633–47 e7. doi: 10.1016/j.cmet.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang CW, Chen YS, Tsay YG, Han CL, Chen YJ, Yang CC, et al. ROS-independent ER stress-mediated NRF2 activation promotes warburg effect to maintain stemness-associated properties of cancer-initiating cells. Cell Death Dis. 2018;9:194. doi: 10.1038/s41419-017-0250-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feng Z, Zou X, Chen Y, Wang H, Duan Y, Bruick RK. Modulation of HIF-2alpha PAS-B domain contributes to physiological responses. Proc Natl Acad Sci USA. 2018;115:13240–5. doi: 10.1073/pnas.1810897115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cho H, Du X, Rizzi JP, Liberzon E, Chakraborty AA, Gao W, et al. On-target efficacy of a HIF-2alpha antagonist in preclinical kidney cancer models. Nature. 2016;539:107–11.. doi: 10.1038/nature19795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen W, Hill H, Christie A, Kim MS, Holloman E, Pavia-Jimenez A, et al. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature. 2016;539:112–7. doi: 10.1038/nature19796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tella SH, Taieb D, Pacak K. HIF-2alpha: Achilles’ heel of pseudohypoxic subtype paraganglioma and other related conditions. Eur J Cancer. 2017;86:1–4. doi: 10.1016/j.ejca.2017.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Melzer C, von der Ohe J, Lehnert H, Ungefroren H, Hass R. Cancer stem cell niche models and contribution by mesenchymal stroma/stem cells. Mol Cancer. 2017;16:28. doi: 10.1186/s12943-017-0595-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu L, Zhao M, Jin X, Ney G, Yang KB, Peng F, et al. Adaptive endoplasmic reticulum stress signalling via IRE1alpha-XBP1 preserves self-renewal of haematopoietic and pre-leukaemic stem cells. Nat Cell Biol. 2019;21:328–37.. doi: 10.1038/s41556-019-0285-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaplan A, Gaschler MM, Dunn DE, Colligan R, Brown LM, Palmer AG, 3rd, et al. Small molecule-induced oxidation of protein disulfide isomerase is neuroprotective. Proc Natl Acad Sci USA. 2015;112:E2245–52. doi: 10.1073/pnas.1500439112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the paper in the main text or the Supplementary Materials.
All data on which the conclusions of the paper rely are available to readers.