Abstract
MYC drives the tumorigenesis of human cancers, including prostate cancer (PrCa), thus deubiquitinase (DUB) that maintains high level of c-Myc oncoprotein is a rational therapeutic target. Several ubiquitin-specific protease (USP) family members of DUB have been reported to deubiquitinate c-Myc, but none of them is the physiological DUB for c-Myc in PrCa. By screening all the DUBs, here we reveal that OTUD6A is exclusively amplified and overexpressed in PrCa but not in other cancers, eliciting a prostatic-specific oncogenic role through deubiquitinating and stabilizing c-Myc oncoprotein. Moreover, genetic ablation of OTUD6A efficiently represses prostatic tumorigenesis of both human PrCa cells and the Hi-Myc transgenic PrCa mice, via reversing the metabolic remodeling caused by c-Myc overexpression in PrCa. These results indicate that OTUD6A is a physiological DUB for c-Myc in PrCa setting and specifically promotes prostatic tumorigenesis through stabilizing c-Myc oncoprotein, suggesting that OTUD6A could be a unique therapeutic target for Myc-driven PrCa.
Subject terms: Deubiquitylating enzymes, Oncogenes
Introduction
MYC is an important driver oncogene for prostate cancer (PrCa) [1], the second most frequent cancer in men worldwide [2]. Amplification of MYC locus, one of the most common chromosome alternation in PrCa, occurs in over 8% of primary PrCa and 20% of metastatic PrCa [3–8], and MYC amplification is a prognostic factor for poor survival of PrCa patients [9–11]. Moreover, overexpression of c-Myc mRNA and protein has been detected in PrCa patients, including 80–90% of all primary PrCa [5, 12–14]. Compared with loss of PTEN and p53, activation of c-Myc occurs at the earliest prostatic intraepithelial neoplasia (PIN) phase of PrCa [15]. Notably, overexpression of c-Myc alone is enough to immortalize human prostate epithelial cells [16, 17], while genetically engineered mouse model (GEMM) with prostate-specific overexpression of c-Myc leads to PIN and invasive adenocarcinoma, with a penetrance of 100% [18–20], recapturing the phenotype of human PrCa. To date, several Myc-targeted therapies have been developed, including MYC inhibitor MYCi361/MYCi975 [21] and 10074-G5 [22], as well as PIN1 inhibitor [23] and PP2A activator [24, 25], all of which eventually destabilize c-Myc protein, suggesting that perturbing c-Myc protein stability could be promising for PrCa therapy.
c-Myc is an unstable protein that undergoes quick ubiquitination and degradation mediated by E3 ligases, such as FBXW7 (F-Box and WD Repeat Domain Containing 7) [26, 27] and SPOP (Speckle Type BTB/POZ Protein), the ubiquitin adapter of the Cullin 3-based E3 ligase complex [28]. The deubiquitination of c-Myc has been reported to be mediated by deubiquitinases (DUBs), such as USP22 [29], USP28 [30], USP36 [31] and USP37 [32], in breast, colon and lung cancers. However, it remains elusive whether these DUBs deubiquitinate and stabilize c-Myc in PrCa setting and could be used as therapeutic target for Myc-driven PrCa. In this study, we screened and identified that OTUD6A, an OTU (ovarian tumor) family member of DUB, is a physiological DUB for c-Myc in PrCa, and depletion of endogenous OTUD6A in human PrCa cell lines and in the Hi-Myc transgenic PrCa mice significantly repressed prostatic tumorigenesis through reversing Myc-driven metabolic remodeling.
Materials and methods
General cloning
Flag-OTUB1, Flag-OTUB2, Flag-OTUD3, Flag-OTUD5, Flag-OTUD6A, Flag-OTUD6B and Flag-OTUD7B were generated as previous described [33]. Expression vectors Flag-OTUD6A-N (aa 1-140) and Flag-OTUD6A-C (aa 141-288) were constructed by cloning the corresponding cDNAs into pcDNA3-HA vector. Flag-OTUD6A-C152A and HA-c-Myc-T58AS62A mutants were constructed using the Site-Directed Mutagenesis Kit (Agilent, Santa Clara, CA, USA) following the manufacturer’s instruction. The full-length GST-OTUD6A-WT and GST-OTUD6A-C152A constructs were constructed by cloning OTUD6A cDNAs into pGEX-4T1 vector. The lentivirus HA-OTUD6A construct was constructed by cloning OTUD6A cDNAs into pLenti-hygromycin-GFP vector. The shRNAs for OTUD6A were purchased from Thermo Scientific Open Biosystem.
Antibodies
Rabbit polyclonal anti-OTUD6A antibody (# PA5-62772) and mouse monoclonal anti-c-Myc antibody (# MA1-980) were purchased from Thermo Fisher (Waltham, MA, USA) for IHC staining. Mouse anti-c-Myc antibody (sc-40), mouse anti-USP22 (sc-390585), mouse monoclonal anti-α Tubulin (sc-8035) antibody and normal mouse IgG (sc-2025) were purchased from Santa Cruz (Dallas, TX, USA). Rabbit polyclonal anti-USP28 antibody (17707-1-AP) and rabbit polyclonal anti-USP36 antibody (14783-1-AP) were purchased from Proteintech (Rosemont, IL, USA). Mouse monoclonal anti-HA antibody (901513) was purchased from BioLegend (San Diego, CA, USA). Rabbit polyclonal anti-Flag antibody (F7425), mouse monoclonal anti-Flag antibody (F3165), rabbit polyclonal anti-HA antibody (H6908), mouse monoclonal anti-Vinculin antibody (V9131), mouse monoclonal anti-FLAG® M2 affinity agarose gel (A2220), mouse monoclonal anti-HA-agarose (A2095), anti-mouse IgG (whole molecule)-peroxidase antibody (A4416) and anti-rabbit IgG (whole molecule)-peroxidase antibody (A4914) were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Cell culture and transfection
Human embryonic kidney 293T (HEK293T) were gifts kindly from Dr. William G. Kaelin, Jr in Harvard Medical School. PC3, DU145, 22Rv1, C4-2, MCF7, NCI-H1299 and HeLa cells were purchased from ATCC. HEK293T and HeLa were maintained in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum (FBS), 100 Units of penicillin and 100 mg/ml streptomycin. PC3, DU145, 22Rv1, C4-2, MCF7 and NCI-H1299 cells were cultured in RPMI-1640 containing 10% FBS, 100 Units of penicillin and 100 mg/ml streptomycin. Cell culture transfection, lentiviral virus packaging and subsequent infection of various cell lines were performed according to the protocol described previously [34].
Generation of knockout cell lines using the CRISPR/Cas9 technology
The sgRNA was designed and cloned into Lenti-CRISPRv2 vector according to the standard protocol [35]. The lentivirus particle was produced in HEK293T cells and used to infect PrCa cell lines. The OTUD6A-KO cells were generated using CRISPR/Cas9 with a guide sequence of 5'-CCTGGCCGGCTTCAAGCGCG-3'. Briefly, 22Rv1 and C4-2 cancer cells were infected with lentivirus, purified with puromycin, followed by picking for single clones 3 to 4 weeks later. The OTUD6A knockout single clone cells were identified by immunoblotting analysis. For further validation of knockout by Sanger sequencing, the genomic DNA was extracted and subjected to PCR with forward primer (5′-TCGAAGACCTGGCCAAGATG-3′) and reverse primer (5′-CGGGATCGCTTTCATCTCCA-3′). The PCR product was then sequenced with both the forward and reverse primers. For other DUB knockout using CRISPR/Cas9, two guiding RNAs were designed and mixed for depletion of individual gene, and the guiding RNA were 5′-TCTACTCTTGACACTAGCGT-3′ and 5′-GTGTATTCGAAAGTTGAAGG-3′ for USP28; 5′-GTCCCGCAGAAGTGGCGTGT-3′ and 5′-CCCACCTATGGTGCAGTTCG-3′ for USP22; 5′-GGAGGGTATCTCAGATCACG-3′ and 5′-CACTCCAAAACGCCGCCAAG-3′ for USP36; 5′-GGATCGATTAAGACTGTAGC-3′ and 5′-CTGATGAGCCGGTACGAATG-3′ for USP37; 5′-AGGACCGGGCGGATATCATG-3′ and 5′-GGACCATTGGCAAGTATCGG-3′ for USP21; 5′-GGGGCTTACCGACTTGGAGC-3′ and 5′-CAGTCCGTGGTAGGTCAGCT-3′ for MINDY1; 5′-TTAGTGAACTGACCGTAGAA-3′ and 5′-CTTGTACTACTACGATGGCC-3′ for MINDY4; 5′-AGTGTACCGAGTGCGGCCAG-3′ and 5′-GTGTAGCACTACGTCCGGGT-3′ for VCPIP.
Generation of OTUD6A knockout mice
The female heterozygous Otud6a+/− mice in a C57BL/6N background (B6/N-Otud6atm1/Nju) were generated in Model Animal Research Center of Nanjing University (Nanjing, China), and then crossed with the Hi-Myc mice in a FVB background gift kindly from Dr. Yong Yang (China Pharmaceutical University, Nanjing, China). The Hi-Myc mice specifically expressed human c-Myc oncoprotein in prostate, which was driven by the rat Probasin promoter with two androgen response elements, i.e., ARR2/Probasin [18]. The genetic locus of Otud6a was on chromosome X. In order to get the male mice with or without Otud6a, the male Hi-Myc mice were crossed with female heterzygous Otud6a+/− mice. Then the male littermates, including four genotypes, i.e., Otud6a+/y, Otud6a−/y, Hi-Myc; Otud6a+/y or Hi-Myc; Otud6a−/y were used for evaluating the effect on prostatic tumorigenesis.
Xenograft mouse model
For xenograft experiments, the 6-week-old, athymic male nude mice in a BALB/C background were used and randomly assigned for injection with different stable cell lines. OTUD6A-depleted 22Rv1 and control cells were mixed with matrigel (v/v, 1:1) and inoculated into the flank of nude mice (2 × 106 cells per injection, six mice for each group). Alternatively, 22Rv1 cells stably expressing HA-c-Myc-WT or HA-Myc-T58AS62A with/without depletion of OTUD6A were injected subcutaneously into nude mice (2 × 106 cells per mice and six mice for each cell line). For measuring the mRNA levels of Otud6a, different mouse tumors/tissues were collected from C57BL/6N mice. All animal experiments were approved by the Animal Use and Care Committee of the School of Life Science and Technology, Xi’an Jiaotong University, based on the Guide for the Care and Use of Laboratory Animals: eighth edition (ISBN10: 0-309-15396-4).
Immunoblots (IB) and immunoprecipitation (IP)
Cells were lysed in EBC buffer (50 mM Tris pH 7.5, 120 mM NaCl, 0.5% NP-40) supplemented with protease inhibitors (cOmplete Mini, Roche, Grenzacherstrasse, CH, Switzerland) and phosphatase inhibitors (phosphatase inhibitor cocktail set I and II, Calbiochem, Burlington, MA, USA). The lysates were then resolved by SDS-PAGE and immunoblotted with indicated antibodies. For immunoprecipitation, 0.5 to 1 mg lysates were incubated with the appropriate beads for 4 h at 4 °C. Immuno-complexes were washed four times with NETN buffer (20 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA and 0.5% NP-40) before being resolved by SDS-PAGE and immunoblotted for indicated proteins. The primary antibodies were diluted in 5% BSA in TBST and secondary antibodies were diluted in 5% non-fat milk for immunoblotting analysis.
In vivo ubiquitination assays
Denatured in vivo ubiquitination assays were performed as previous described [34]. Briefly, HEK293T cells were transfected with His-ubiquitin and indicated plasmids. Forty-eight hours after transfection, 30 μM MG132 (BML-PI102, Enzo Life Science, Farmingale, NY, USA) was added to block proteasome degradation for 6 h and cells were harvested in denatured buffer A (6 M guanidine-HCl, pH 8.0, 0.1 M Na2HPO4/NaH2PO4, 10 mM imidazole). After sonication, the ubiquitinated proteins were purified by incubation with Ni-NTA resin (88221, Thermo Fisher, Waltham, MA, USA) for 3 h at room temperature. The pull-down products were washed sequentially twice in buffer A, twice in buffer A/TI mixture (v/v, 1:3) and once in buffer TI (25 mM Tris-HCl, pH 6.8, 20 mM imidazole). The poly-ubiquitinated proteins were separated by SDS-PAGE for immunoblot analyses.
Protein half-life cycloheximide (CHX) chasing assay
To measure the half-life of c-Myc protein, a cycloheximide (CHX)-based assay was performed following our previously described experimental procedures [34]. Briefly, cells were transfected with indicated constructs in 10-cm dish, and the cells were split into 6 of 6-cm dish 24 h after transfection. Another 24 h later, cells were treated with 200 µg/ml CHX (239764, Sigma-Aldrich, St. Louis, MO, USA) for indicated time before harvest for immunoblot analysis of indicated proteins.
Purification of GST-OTUD6A protein
The full-length GST-OTUD6A wild type and C152A mutant proteins were purified from BL21 E. Coli as previous described [34]. Briefly, the pGEX-4T1-GST-OTUD6A-WT and C152A constructs were transfected into BL21 E. Coli, and the proteins were induced by 100 μM of isopropyl β-D-1-thiogalactopyranoside (IPTG) at 18 °C overnight. Then, the bacteria were collected and lysed in PBS with proteinase inhibitor (cOmplete Mini, Roche, Grenzacherstrasse, CH, Switzerland) under sonication for 15 min at 4 °C. The lysis was centrifuged at 10, 000 RPM for 15 min to remove bacterial debris, and the supernatant was incubated with 500 µl Glutathione Sepharose (17-5279-01, GE, Amersham, UK) for 3 h. After washed with PBS for four times, the GST-OTUD6A proteins were further eluted with PBS within 10 mM fresh-made reduced glutathione (3541, Sigma, St. Louis, MO, USA).
In vitro deubiquitination assay
The in vitro deubiquitination assay of c-Myc was performed with HA-c-Myc proteins derived from HEK293T cells that transfected with HA-c-Myc plasmids and pretreated with 10 µM MG132 overnight. Briefly, the HA-c-Myc protein was pulled down using HA-agarose, and the protein-loaded HA-agarose with 10 µg c-Myc protein was further incubated with 5 µg bacterial purified GST-OTUD6A-WT or C152A mutant proteins under vortex at 30 °C using Eppendorf Thermomixer for 1 h. Finally, the reaction was stopped by adding 3X SDS loading buffer, boiled at 98 °C for 10 min, and analyzed by SDS-PAGE.
Quantitative polymerase chain reaction (qPCR)
Cells were harvested in TRIsol (Thermo Fisher, Waltham, MA, USA), and mRNA was extracted according to standard protocol. The mRNA was then reverse transcripted into cDNA and subjected to qPCR for HK2 and LDHA. For the mRNA levels of OTU DUB family members in different tissues or organs, mRNA samples were extracted from tissues or organs dissected from C57B/6N mice and the mRNA levels of individual OTU family member were measured. The primers used for qPCR are as below: hHK2-f: GAGCCACCACTCACCCTACT; hHK2-r: CCAGGCATTCGGCAATGTG; hLDHA-f: ATGGCAACTCTAAAGGATCAGC; hLDHA-r: CCAACCCCAACAACTGTAATCT; msOtud1-f: AGAGGCAGGACAAGTACCTGA; USP37-f: CCAGTGGAGCGAAACAAAGC; USP37-r: CCTCTGCATCCTTACTTGGTACT; msOtud1-r: CCCGTACACAGTCTTGCTGAC; msOtud2-f: GCCAAATCGCCGCTATCAC; msOtud2-r: ATGTCCCGGTCGCTGAGAT; msOtud3-f: CTTCGTGGAAGATGACATTCCC; msOtud3-r: AAAGGGGCATTAAGCTGATGG; msOtud4-f: AAGTTGGAGATAAATGCCACCAG; msOtud4-r: GACACCGTGTTCCAGCTTTCT; msOtud5-f: CAGTGAAGACGAGTATGAAGCTG; msOtud5-r: AGCCCGAAATAGACAGGCAC; msOtud6a-f: ACAGCAGTGTGGATTCGGTT; msOtud6a-r: GAATGGCAGCGACCTTTTCC; msOtud6b-f: AGCTCACGGAAGATGTTGCTA; msOtud6b-r: TTTTGTGCTTTTGAAATCCGAGG; msActin-f: GGCTGTATTCCCCTCCATCG; and msActin-r: CCAGTTGGTAACAATGCCATGT.
Colony formation assay
PrCa cells were cultured in 10% FBS containing RPMI-1640 media before being plated into 6-well plate at 1000 cells per well. Two to three weeks later, cells were fixed with 10% acetic acid/10% methanol for 10 min, stained with 0.4% crystal violet/20% ethanol solution, followed by counting the colony numbers under bright-field microscope.
Seahorse XF24 extracellular bioenergetics analysis
The extracellular acidification rate of PrCa cells was measured using Seahorse XF24 analyzer (Agilent, Santa Clara, CA, USA) as previous described [34]. The cell numbers and FCCP concentration were optimized based on previous reports and titration experiments. Seahorse XF basal media (103576-100, Agilent, Santa Clara, CA, USA) containing no glucose, no pyruvate and 2 mM glutamine was used. The final concentrations of glucose, oligomycin (75351, Sigma-Aldrich, St. Louis, MO, USA) and 2-DG (D8375, Sigma-Aldrich, St. Louis, MO, USA) were 10 mM, 1 μM and 50 mM, respectively, unless indicated otherwise.
Immunohistochemistry (IHC) assay
Human prostate tissue arrays (PR807a) were purchased from US Biomax, Inc. (Rockville, MD, USA) and subjected to IHC staining with either OTUD6A (PA5-62722, 1:500, Thermo Fisher, Waltham, MA, USA) or c-Myc (MA1-980, 1:100, Thermo Fisher, Waltham, MA, USA). This array included 66 PrCa tissues, 6 normal prostate tissues, 7 hyperplasia and 1 paracancerous tissues. Mouse prostate tissues were dissected and fixed in 4% paraformaldehyde. Fixed prostate samples were embedded in paraffin according to standard procedures, and 5 µm sections were subjected to H&E staining or IHC staining. The stained slides were scanned with Pannoramic DESK (3D HISTECH) with the Pannoramic Scanner software.
Mouse genotyping
Mouse tail tips were collected at 7-day-old, boiled in lysis buffer A (25 mM NaOH, 0.2 mM EDTA) for 15 min, and then neutralized with buffer B (40 mM Tris-HCl, pH 8.0). Then, 1 µl of DNA solution was used for further PCR with specific primers: lazRev: GTCTGTCCTAGCTTCCTCACTG; Otud6a-SU: CATGCTACGGGGACCCAAC; Otud6a-wt2R: TCTCGAGATTCATCTTTTCGAGGTC; Hi-Myc-F: AAACATGATGACTACCAAGCTTGGC; Hi-Myc-R: ATGATAGCATCTTGTTCTTAGTCTTTTTCTTAATAGGG.
RNA-Seq
Total RNA was extracted from prostate tissue or tumor of Otud6a+/y, or Otud6a−/y, or Hi-Myc; Otud6a+/y or Hi-Myc mice. The extraction, quantification, qualification, library preparation and subsequent RNA-sequencing of RNA samples were conducted by Novogene Co., Ltd (Beijing, China). RNA-seq was performed by using Illumina NovaSeq 6000 platform. The RNA-sequencing data generated in this study have been deposited into the Gene Expression Omnibus Database under GSE189716.
Statistical analysis
The quantitative data from multiple repeat experiments were analyzed by a two-tailed unpaired Student’s t test or one-way ANOVA, and presented as MEAN ± SD. The IHC staining score of human prostate tissue array was analyzed by chi-square test. When p < 0.05, the data were considered as statistically significant.
Results
High c-Myc expression in prostate cancer is maintained at posttranslational level
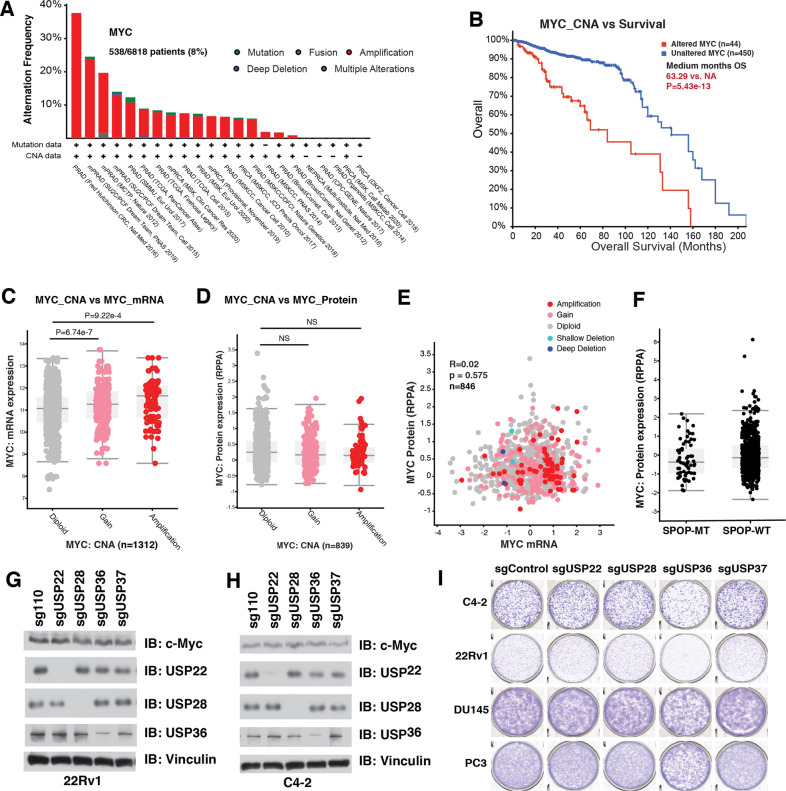
Oncogenic MYC is amplified in about 8% of all PrCa patients in the Cancer Genome Atlas (TCGA) database [36, 37], and amplified MYC indicates worse prognosis of PrCa patients (Fig. 1A, B). In keeping with that, the mRNA levels of c-Myc in MYC-amplified PrCa patients were significantly higher than those in patients with diploid MYC (Fig. 1C and Supplementary Table S1A). However, there was no difference in c-Myc protein levels between the PrCa patients with either amplified or diploid MYC (Fig. 1D and Supplementary Table S1B). Moreover, there was no correlation between the mRNA and protein levels of c-Myc in PrCa patients (Fig. 1E), suggesting that at least in PrCa setting, c-Myc protein is subjected to both transcriptional and posttranslational regulation. SPOP is the most frequent somatic mutation in primary PrCa, accounting for ~10% of all primary cases (Supplementary Fig. S1A) [38, 39], and SPOP has been previously reported as an ubiquitin adapter of Cullin 3 E3 ubiquitin ligase for c-Myc [28]. However, SPOP mutation status was not associated with increased c-Myc protein levels in PrCa patients (Fig. 1F). FBXW7, another E3 ligase of c-Myc [26, 27], was not frequently mutated in PrCa patients, and FBWX7 expression was neither a prognosis marker for PrCa patients (Supplementary Fig. S1B, C). These analyses indicate that high c-Myc expression in PrCa patients might be more likely due to the deubiquitination process.
Fig. 1. Overexpression of c-Myc in prostate cancer is due to both transcriptional and posttranslational regulation.
A MYC is amplified in PrCa patients in TCGA database. B MYC amplification indicates worse prognosis of PrCa patients. C MYC amplification indicates higher mRNA levels of c-Myc oncoprotein. D MYC amplification does not indicate higher protein levels of c-Myc oncoprotein. E c-Myc protein abundance was not correlated with its mRNA levels in human PrCa samples in TCGA database. R = 0.02, p = 0.575. F SPOP mutation did not correlated with high c-Myc expression in human PrCa patients in TCGA. G–H Depletion of USP22, USP28, USP36 or USP37 did not change the protein level of c-Myc in 22Rv1 and C4-2 PrCa cells. 22Rv1 (G) and C4-2 (H) cells were infected lentivirus to deplete endogenous USP22, USP28, USP36 or USP37, and selected with puromycin for 72 h, and collected for immunoblot analysis. I Depletion of USP36, but not USP22, USP28 or USP37, compromised the tumorigenesis ability of PrCa cell lines. The PrCa cells (22Rv1, C4-2, DU145 and PC3) treated as in (G) and (H) were subjected to colony formation assay, and stained with crystal violet. The relevant raw data are provided in Supplementary Materials.
The ubiquitination process of c-Myc can be antagonized by DUBs, including USP22 [29], USP28 [30], USP36 [31] and USP37 [32], in several cancer types but not in PrCa setting. However, all of these USPs for c-Myc were not frequently amplified in PrCa patients (Supplementary Fig. S2A), and the genetic alternations of these DUBs were not prognosis markers for PrCa patients (Supplementary Fig. S2B). To further examine whether these DUBs play essential roles in prostatic tumorigenesis and whether they are suitable therapeutic targets for PrCa with high c-Myc expression, we depleted these USPs in a panel of human PrCa cells, and found that depletion of these USPs did not reduce c-Myc levels (Fig. 1G, H and Supplementary Fig. S2C–E), while only depletion of USP36 reduced the proliferation of these PrCa cells assessed in a clonogenic assay (Fig. 1I). In keeping with previous report, deletion of USP28 [30] and USP36 [31] reduced c-Myc protein expression in breast cancer cells MCF7 and lung cancer cells H1299, while depletion of USP37 had a preferential effect in H1299 cells (Supplementary Fig. S2F, G) [32]. Although knockout of USP36 also repressed the proliferation of PrCa cells, it had limit effect on c-Myc level, suggesting that other substrate other than c-Myc mediates the oncogenic role of USP36. These findings suggest that, at least in PrCa, c-Myc protein levels might be regulated by other DUBs other than USPs, stimulating us to further screen the bona fide DUB for c-Myc that could be a potential therapeutic target in PrCa setting.
Identification of OTUD6A as the physiological DUB for c-Myc in PrCa
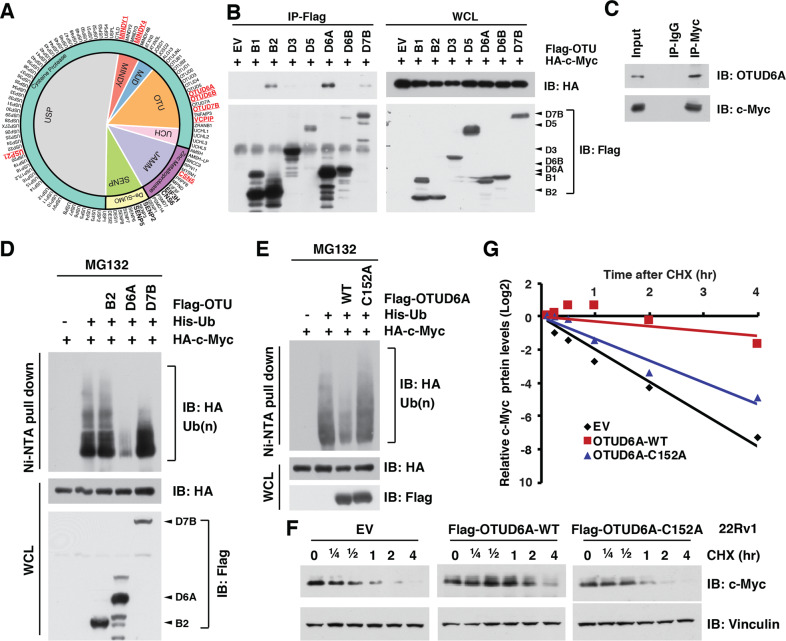
We first screened and found that 14 DUBs were frequently (>2% of total cases) mutated in PrCa patients (Supplementary Fig. S3A), while 10 in the 14 DUBs predicted worse prognosis (Supplementary Fig. S3B–E), among which 8 DUBs have enzymatic activity (Supplementary Fig. S3F). Among these 8 DUBs, 4 candidate DUBs belong to the OTU family of DUB (Fig. 2A), the second largest DUB family [40], thus we mainly focused on OTU family members for their potential role in regulating c-Myc protein stability in PrCa setting. We found that c-Myc interacted with only OTUB2, OTUD6A and OTUD7B (Fig. 2B, C), among which only OTUD6A dramatically removed the poly-ubiquitin chain from c-Myc (Fig. 2D and Supplementary Fig. S4A–C). In contrast, depletion of MINDY1/4, USP21 and VCPIP did not reduce c-Myc protein level (Supplementary Fig. S4D). Moreover, we generated a catalytic-dead mutant version of OTUD6A (namely OTUD6A-C152A), which was deprived of its DUB enzymatic activity (Supplementary Fig. S4E). Compared with the wild type OTUD6A (OTUD6A-WT), OTUD6A-C152A mutant was incapable of deubiquitinating c-Myc (Fig. 2E), supporting the notion that OTUD6A is likely a bona fide DUB for c-Myc oncoprotein and deubiquitinates c-Myc in a catalytic activity-dependent manner.
Fig. 2. OTUD6A deubiquitinates and stabilizes c-Myc.
A A schematic diagram to show the screening of potential DUB candidates that might play oncogenic role in PrCa. Amplified genes were marked in bold, and potential oncogenic DUB candidates in red-bold. B OTUB2, OTUD6A, and OTUD7B, but not other OTU family members interacted with c-Myc. Immunoblot of Flag-immunoprecipitants and whole-cell lysis (WCL) derived from HEK293T cells transfected with haemagglutinin (HA)-c-Myc and Flag-tagged OTU DUB family members. C Endogenous OTUD6A binds with c-Myc protein. Immunoblot of c-Myc-immunoprecipitants derived from 22Rv1 cells. D OTUD6A, but not OTUB2 or OTUD7B, deubiquitinated c-Myc. Immunoblot of WCL and nickel-nitrilotriacetic acid (Ni-NTA) pull-downs from HEK293T cells transfected with HA-c-Myc, His-ubiquitin (His-Ub) and Flag-tagged constructs. Cells were pretreated with 10 µM of MG132 for 6 h before harvesting. E Catalytic-dead mutant of OTUD6A (OTUD6A-C152A) failed to deubiquitinate c-Myc oncoprotein. Immunoblot of WCL and Ni-NTA pull-downs from HEK293T cells transfected with HA-c-Myc, His-Ub and Flag-tagged OTUD6A or the catalytic-dead mutant OTUD6A-C152A. Cells were pretreated with 10 µM of MG132 for 6 h before harvesting. F, G Ectopic WT, but not the C152A mutant form of OTUD6A, extended the protein half-life of c-Myc in 22Rv1 cells. Immunoblot (F) and quantification (G) of cell lysis derived from 22Rv1 cells transient transfected with either empty vector (EV), OTUD6A-WT or OTUD6A-C152A, and treated with 200 µg/ml of cycloheximide (CHX) for indicated time. The protein abundances were in G. The relevant raw data are provided in Supplementary Materials.
We further found that the OTU domain, but not the N-terminal domain of OTUD6A, was essential for its interaction with c-Myc (Supplementary Fig. S4F, G). Notably, overexpression of OTUD6A-WT extended the half-life of endogenous c-Myc oncoprotein in both 22Rv1 PrCa cells (Fig. 2F, G) and HEK293T cells (Supplementary Fig. S4H, I), while the catalytic-dead OTUD6A-C152A mutant did not (Fig. 2F, G and Supplementary Fig. S4H, I). These results suggest that OTUD6A regulates the ubiquitination process and protein stability of c-Myc oncoprotein in PrCa setting (Supplementary Fig. S4J).
OTUD6A is exclusively amplified in PrCa and OTUD6A protein level positively correlates with c-Myc abundance in PrCa patients
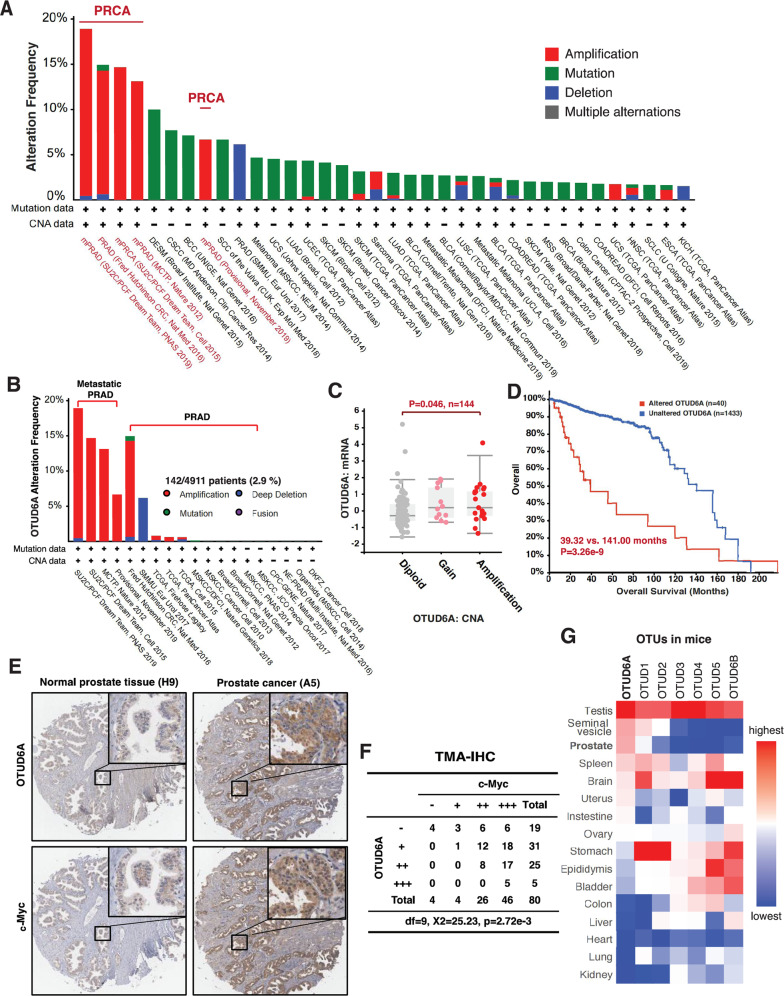
To further determine the possible oncogenic role of OTUD6A in PrCa, we systemically analyzed OTUD6A genetic variations in all cancer types in TCGA, and found that OTUD6A was exclusively amplified in human PrCa but not other cancer types (Fig. 3A and Supplementary Fig. S5A–C). In addition, amplification of OTUD6A occurred more frequently in metastatic PrCa patients than non-metastatic PrCa (Fig. 3B). OTUD6A amplification led to a higher level of OTUD6A mRNA (Fig. 3C), and indicated worse prognosis of PrCa patients (Fig. 3D), indicating that OTUD6A could be a biomarker and a potential therapeutic target for PrCa.
Fig. 3. OTUD6A is amplified and overexpressed in human PrCa, and positively correlated with c-Myc oncoprotein level.
A OTUD6A was exclusively amplified in PrCa in TCGA. The genetic alternation of OTUD6A gene was analyzed among all cancer studies in TCGA database and classified by cancer study. These cancer studies with low frequency of OTUD6A alternation were not shown. B OTUD6A amplification preferentially occurred in metastatic PrCa in TCGA. C OTUD6A amplification led to increase in OTUD6A mRNA expression. N = 144, p = 0.046 in OTUD6A amplification group vs. diploid group. D OTUD6A gene alteration indicated worse prognosis of human PrCa. The overall survival of PrCa patients in TCGA with vs. without OTUD6A genetic alternation were compared (Median Months Survival: 39.32 vs. 120.00, p = 2.143 × 10−3, altered group: 40 cases; unaltered group: 2854 cases). E Representative immunohistochemistry (IHC) staining images of OTUD6A and c-Myc in normal prostate tissue and PrCa samples. F The expressions of OTUD6A were positively correlated with c-Myc staining scores in human PrCa samples. Chi-squared test, df = 9, χ2 = 24.709, p = 2.72 × 10−3. G Otud6a was highly expressed in sexual tissues/organs, including testis and prostate, in mice. Otud6a mRNA levels in different tissues/organs derived from C57 mice were analyzed by qPCR. The relevant raw data are provided in Supplementary Materials.
To further determine the protein abundance of OTUD6A in human PrCa samples, we immuno-stained OTUD6A in a human PrCa tissue chip, and found that near 70% (56 in 80) of samples had mild or moderate OTUD6A staining, mainly in the cytoplasm (Fig. 3E, F, Supplementary Fig. S5D, and Supplementary Table S2). Those PrCa cases with moderate OTUD6A staining had moderate or strong c-Myc staining, and 5 cases with strong OTUD6A staining were all with strong c-Myc staining (Fig. 3E, F, Supplementary Fig. S5E, and Supplementary Table S2). More importantly, the OTUD6A protein levels were positively correlated with c-Myc expression levels in these human PrCa samples (Fig. 3F).
To date, little is known about the distribution and function of OTUD6A, thus we measured the mRNA expression of Otud6a and other OTU family members of DUB in different tissues of mice (Fig. 3G). Notably, Otud6a was highly expressed in sexual organs/tissues, such as testis, seminal vesicle and prostate in mice (Fig. 3G), consisting with its expression pattern in human (Supplementary Fig. S5F). Interesting, AR and FOXA1 are predicted transcription factor for OTUD6A based on the Cistrome [41], and both AR and FOXA1 bind with the promoter of OTUD6A (Supplementary Fig. S5G–I), which partially explain the unique overexpression manner of OTUD6A in PrCa. Unlike Otud6a, other OTUD family members had no or very low expression in prostate and distinct expression patterns in other tissues, such as high OTUD1 and OTUD2 expressions in stomach, as well as high OTUD5 and OTUD6B expressions in brain (Fig. 3G). These results further supporting the unique role of OTUD6A in prostatic tumorigenesis.
Depletion of OTUD6A represses prostatic tumorigenesis in vitro and in xenograft mice
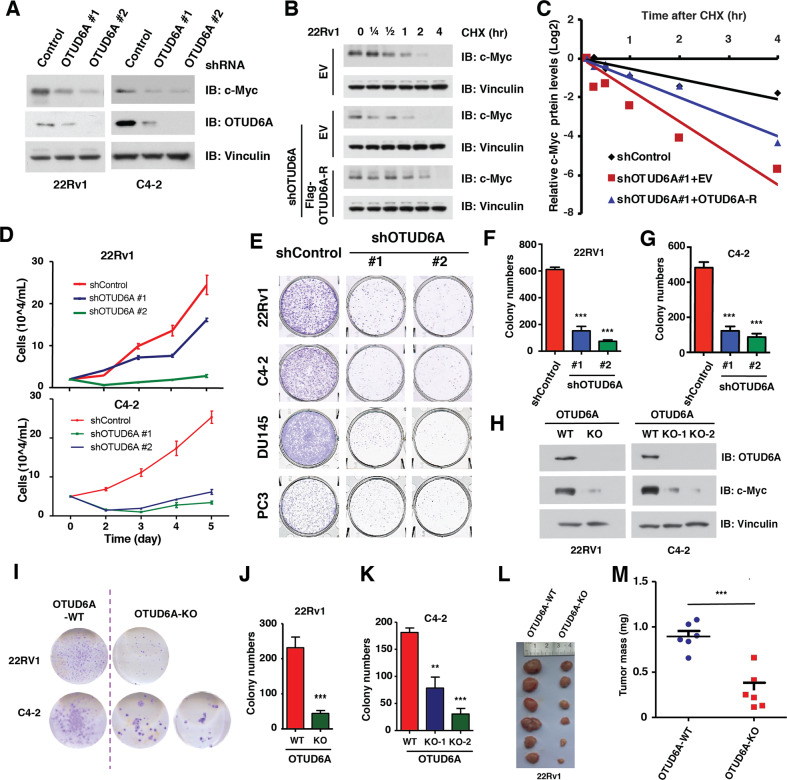
To further explore whether OTUD6A is a driving oncogene that promotes prostatic tumorigenesis, we depleted OTUD6A with shRNA in a panel of PrCa cell lines, and found that OTUD6A depletion led to reductions of c-Myc abundance (Fig. 4A and Supplementary Fig. S6A, B) and shortening of the protein half-life of c-Myc, which could be rescued by transient transfection with shRNA-resistant OTUD6A constructs (OTUD6A-R) in both 22Rv1 (Fig. 4B, C) and C4-2 PrCa cells (Supplementary Fig. S6C, D), indicating that OTUD6A regulates the protein stability of c-Myc in PrCa cells in vitro.
Fig. 4. Depletion of OTUD6A leads to destabilization of c-Myc and inhibits prostatic tumorigenesis in vitro and in PrCa xenograft mice.
A Depletion of OTUD6A led to reduced protein levels of c-Myc in 22Rv1 and C4-2 PrCa cells. 22Rv1 and C4-2 cells were infected with shControl or shOTUD6A virus, and selected with puromycin for 72 h, followed by IB assay for indicated proteins. B, C Depletion of OTUD6A reduced the half-life of c-Myc protein in 22Rv1 cells. Cells were infected with shControl or shOTUD6A virus and selected with puromycin for 3 days, followed by transient transfection with EV or shRNA-resistant Flag-OTUD6A constructs (Flag-OTUD6A-R). These cells were then treated with 200 µg/ml of CHX for indicated time before harvest, followed by IB assay for indicated proteins (B) and quantification (C). D Depletion of OTUD6A suppressed cell proliferation of 22Rv1 and C4-2 PrCa cells. 22Rv1 and C4-2 cells as in Fig. 4A were subjected to growth curve analysis. E Representative images showed the colony growth of human PrCa cells after depletion of endogenous OTUD6A. F, G Depletion of OTUD6A suppressed the colony growth of 22Rv1 and C4-2 cells. The cells as in Fig. 4A were subjected to colony formation assay. ***p < 0.001. H Depletion of OTUD6A using CRISPR/Cas9 led to reduced protein levels of c-Myc in 22Rv1 and C4-2 cells. Immunoblot of WCL derived from 22Rv1 cells with OTUD6A knockout by CRISPR/Cas9. I–K Depletion of OTUD6A by CRIPSR/Cas9 suppressed the colony growth of 22Rv1 and C4-2 cells. OTUD6A knockout cells as in Fig. 4H were subjected to colony formation assay (I) and quantification (J, K). **p < 0.01, ***p < 0.001. L, M Depletion of OTUD6A suppressed the xenograft tumor growth of 22Rv1 cells. OTUD6A-WT and OTUD6A-KO cells (as in Fig. 4H) were subcutaneously injected into 6-week-old male null mice (N = 6). The tumor mass was imaged (L) and weighed (M) at end of the experiment. ***p < 0.001. The relevant raw data are provided in Supplementary Materials.
In keeping with that, depletion of OTUD6A retarded the proliferation of all PrCa cell lines we tested (Fig. 4D and Supplementary Fig. S6E, F). Moreover, deletion of endogenous OTUD6A reduced the tumorigenesis capacity of these PrCa cell lines in a clonogenic assay (Fig. 4E–G and Supplementary Fig. S6G, H). On the other hand, the 22Rv1 cells that stably expressed HA-OTUD6A also had relative higher level of c-Myc protein, resulting in increased cell proliferation and tumorigenesis in a clonogenic assay (Supplementary Fig. S6I–K). To further exclude the potential off-target effect caused by shRNA, we generated OTUD6A knockout PrCa cell lines by using CRISPR/Cas9 technology in both 22Rv1 and C4-2 cells (Supplementary Fig. S6L), and validated these KO cell lines with sequencing (Supplementary Fig. S6M, N). We found that knockout of OTUD6A led to lower abundance of c-Myc oncoprotein (Fig. 4H), as well as a compromised tumorigenesis capacity in both 22Rv1 and C4-2 PrCa cells (Fig. 4I–K), but not in MCF7 breast cancer cells or H1299 lung cancer cells (Supplementary Fig. S6O). Finally, the tumors derived from OTUD6A-null 22Rv1 cells had significant lower tumorigenesis ability in a xenograft mouse model than OTUD6A-WT 22Rv1 cells (Fig. 4L, M). Taken together, these data suggest that the OTUD6A promotes the prostatic tumorigenesis in vitro and in xenograft mice (Supplementary Fig. S6P).
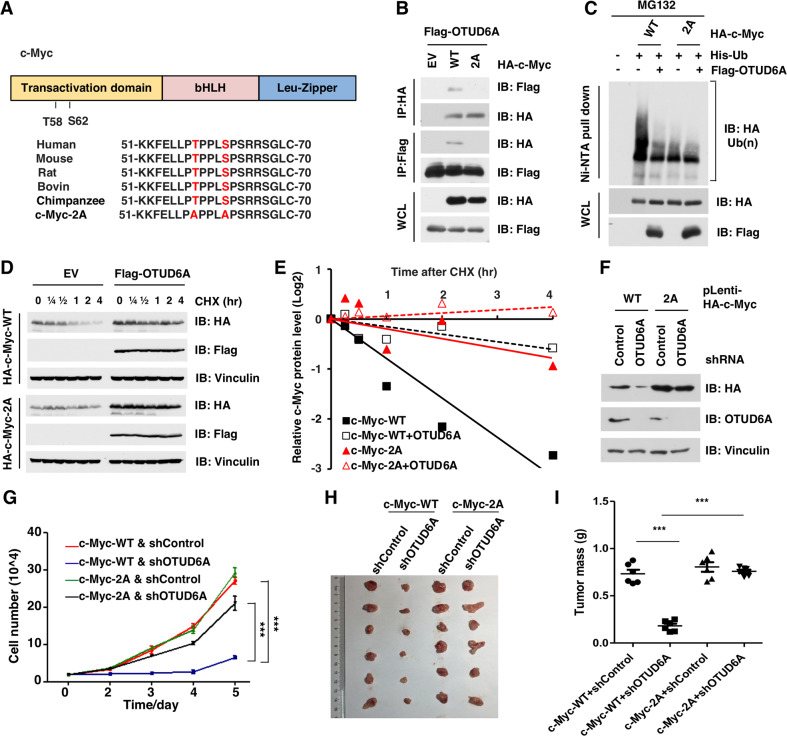
The phosphorylation of serine 62 (S62) and threonine 58 (T58) regulates the ubiquitination and degradation of c-Myc protein (Fig. 5A) [25–27, 42–44]. To determine whether the deubiquitination process of c-Myc by OTUD6A is related to the phosphorylation events on c-Myc, we further compared the effect of OTUD6A on both wild type c-Myc (c-Myc-WT) and non-phosphorylation mutant (T58AS62A, hereafter c-Myc-2A, Fig. 5A). Ectopic expression of OTUD6A bound with only c-Myc-WT but not c-Myc-2A (Fig. 5B). Consistently, OTUD6A removed the poly-ubiquitin chain from only wild type c-Myc, while exerted minimal effect on the non-phosphorylation mutant c-Myc-2A (Fig. 5C). Moreover, ectopic expression of OTUD6A only extended the protein half-life of wild type c-Myc, but not of the 2A mutant (Fig. 5D, E).
Fig. 5. OTUD6A promotes prostatic tumorigenesis by deubiquiting and stabilizing c-Myc oncoprotein.
A Alignment the conserved TXXXS consensus of c-Myc among species. B The non-degradable version of the c-Myc-T58AS62A (2A) mutant did not interact with OTUD6A. Immunoblot of Flag-immunoprecipitants, HA immunoprecipitants and WCL derived from HEK293T transfected with Flag-OTUD6A and HA-tagged constructs. C OTUD6A removed the poly-ubiquitin chain from c-Myc protein depending on the phosphorylation events. Immunoblot of Ni-NTA pull-downs and WCL derived from HEK293T cells transfected with His-Ub, Flag-OTUD6A and HA-tagged constructs. Cells were pretreated with 30 μM of MG132 for 6 h before harvest. D, E Ectopic expression of OTUD6A extended the half-life of WT c-Myc protein, but not of the 2A mutant. Immunoblot of WCL derived from HEK293T cells transfected with Flag-OTUD6A and HA-tagged constructs. The cells were treated with CHX (200 µg/ml) for indicated time before harvest. F Depletion of endogenous OTUD6A reduced the protein level of WT c-Myc, but not of the c-Myc 2A mutant. Immunoblot of WCL derived from 22Rv1 cells stably expressed with either HA-c-Myc-WT or HA-c-Myc-2A, with or without depletion of OTUD6A by shRNA. G Depletion of OTUD6A reduced the cell growth of 22Rv1 that stably expressed WT c-Myc, but not of the cell expressing 2A mutant. Cells as in Fig. 5F were subjected to growth curve assay. ***p < 0.001. H, I Depletion of OTUD6A reduced the tumorigenesis ability of 22Rv1 in a xenograft assay. Cells as in Fig. 5F were subcutaneously injected into 6-week-old male null mice (N = 6). The tumor mass was imaged (H) and weighed (I) at end of the experiment. ***p < 0.001. The relevant raw data are provided in Supplementary Materials.
Furthermore, depletion of OTUD6A significantly reduced the protein level of c-Myc-WT but not of c-Myc-2A mutant (Fig. 5F). In addition, depletion of OTUD6A inhibited the proliferation of c-Myc-WT-expressing cells but not the c-Myc-2A-expressing cells (Fig. 5G). In line with that, depletion of OTUD6A significantly restricted the growth of xenograft tumor derived from c-Myc-WT-expressing cells, but not of c-Myc-2A-expressing cells (Fig. 5H, I). These data together indicate that OTUD6A promotes prostatic tumorigenesis mainly by deubiquitinating and stabilizing c-Myc oncoprotein.
Depletion of Otud6a suppresses Myc-driven prostatic tumorigenesis in Hi-Myc transgenic mice
To assess whether loss of Otud6a inhibits Myc-driven prostate tumors in vivo, Otud6a knockout mice were generated and further crossed with the Hi-Myc PrCa mouse model. Human OTUD6A gene locates on chromosome X.p13 [40], and mouse Otud6a gene locates on mouse chromosome X.p6, indicating that female mouse has two alleles while male mouse has only one allele of Otud6a gene. To this end, Otud6a null sperm was used for the in vitro fertilization with wild type egg, and the female Otud6a+/− mice (P0, passage zero) were further crossed with male wild type mice, generating four genotypes of P1 litters, including female wild type Otud6a+/+, male wild type Otud6a+/y, a heterozygous female Otud6a+/− and a hemizygous male Otud6a−/y (Supplementary Fig. S7A, B). Next, the male Otud6a−/y mice (P1) were further crossed with female wild type to generating two genotypes of P2 litters, including wild type male Otud6a+/y and heterozygous female Otud6a+/− mice (Supplementary Fig. S7A, B).
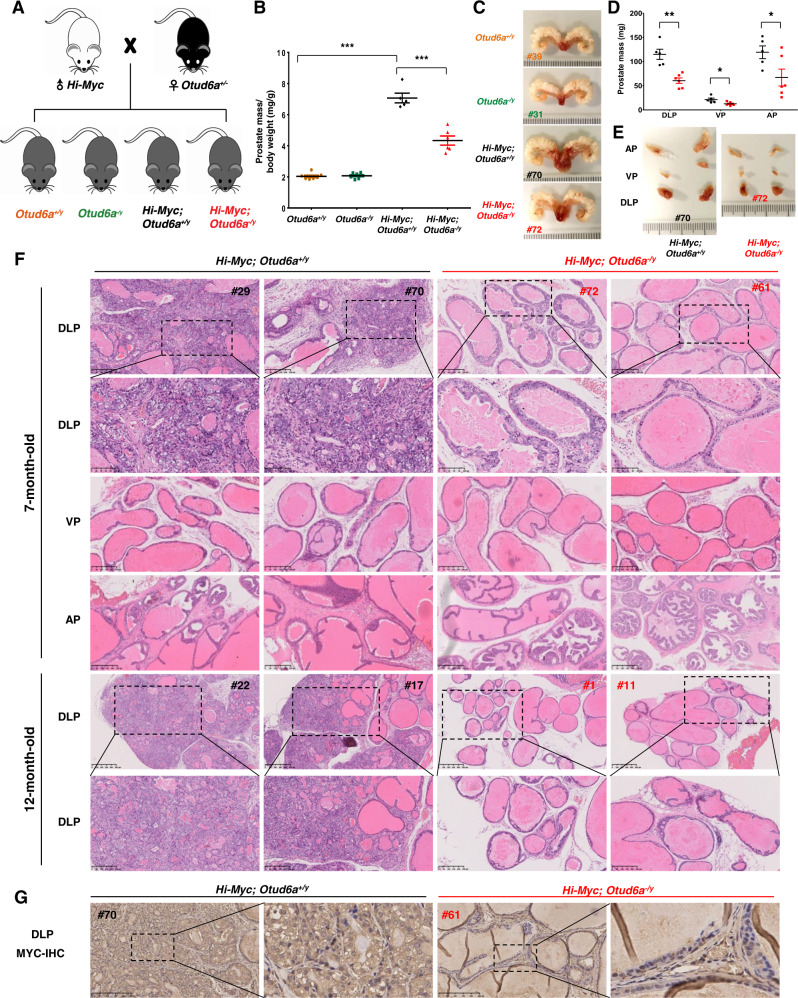
In order to obtain male mice with Hi-Myc; Otud6a+/y mice and Hi-Myc; Otud6a−/y genotypes to compare their tumor burden in prostate, the female heterozygous Otud6a+/− mice (P2) were further crossed with male Hi-Myc mice to obtain cohorts of Otud6a+/y and Otud6a−/y mice with or without MYC overexpression (Fig. 6A and Supplementary Fig. S7C). There was a total of four genotypes of male litters, including Otud6a+/y, Otud6a−/y, Hi-Myc; Otud6a+/y and Hi-Myc; Otud6a−/y (Fig. 6A). In consistent with previous reports [18], overexpression of c-Myc in the Hi-Myc; Otud6a+/Y mice led to severe prostate carcinoma phenotype (Fig. 6B, C and Supplementary Fig. S7D, E). Notably, Hi-Myc; Otud6a−/y mice had a dramatically reduction of invasive prostate carcinoma phenotype (Fig. 6B, C and Supplementary Fig. S7D, E). In Hi-Myc mice, the most dramatic hyperplasia phenotype was in the dorsolateral prostate (DLP), compared with the ventral prostate (VP) and anterior prostate (AP) lobes [18]. Thus, we further dissected the prostate tumor into different lobes and found that the weight of all lobes in Hi-Myc; Otud6a−/y mice were significantly lower than those in Hi-Myc; Otud6a+/y mice (Fig. 6D, E).
Fig. 6. Depletion of OTUD6A restricts prostatic tumorigenesis in Hi-Myc mice.
A A schematic diagram shows the animal experimental design for the crossing of Hi-Myc mice and OTUD6A null mice. B Depletion of Otud6a reduced the prostatic tumorigenesis of Hi-Myc PrCa mice. The prostate tumors/tissues were collected and weighed at 7-month of age. N = 7, 7, 5, and 6 for Otud6a+/y, Otud6a−/y, Hi-Myc;Otud6a+/y and Hi-Myc;Otud6a−/y group, respectively. ***p < 0.001. One-way ANOVA. C Representative images for the prostate tissue or tumor in Otud6a+/y, Otud6a−/y, Hi-Myc;Otud6a+/y and Hi-Myc;Otud6a−/y mice. D Depletion of Otud6a reduced the tumor weight in the different lobes of prostate in Hi-Myc mice. The DLP (dorsolateral prostate), VP (ventral prostate) and AP (anterior prostate) lobes of prostate tissue were collected and weighed for each mice at 7-month of age. N = 5 and 6 for Hi-Myc;Otud6a+/y and Hi-Myc;Otud6a−/y group, respectively. *p < 0.05, **p < 0.001. One-way ANOVA. E Representative images for the different lobes of prostate tumor in Hi-Myc;Otud6a+/y and Hi-Myc;Otud6a−/y mice. F Representative H&E staining images for the different lobes of prostate tumor in Hi-Myc;Otud6a+/y and Hi-Myc;Otud6a−/y mice at 7-month and 12-month of age. G Representative IHC staining images of c-Myc for the DLP lobe of prostate tumor in Hi-Myc;Otud6a+/y and Hi-Myc;Otud6a−/y mice.
The histological analysis indicated that Hi-Myc; Otud6a+/y mice had invasive carcinoma phenotypes in the whole DLP lobe and there was a lack of normal secretory lumens phenotype (Fig. 6F, top two panels). In contrast, the Hi-Myc; Otud6a−/y mice had substantially decreased invasive lesions and only mild hyperplasia phenotype (Fig. 6F, top two panels). Even at 12-month of age where the penetrance of Myc-driven PrCa phenotype reached to 100% [18], the Hi-Myc; Otud6a−/y mice did not develop invasive lesions as Hi-Myc; Otud6a+/y mice did (Fig. 6F, bottom two panels). Moreover, Hi-Myc; Otud6a+/y mice had only mild hyperplasia lesions in the VP and AP lobes, while the Hi-Myc; Otud6a−/y mice had few or even no PIN lesions (Fig. 6F, middle two panels). Further immunostaining assays indicated that c-Myc protein was highly expressed in both nuclear and cytoplasm of the epithelial cells in Hi-Myc; Otud6a+/y mice, while the basal and stromal cells had relative low c-Myc expression (Fig. 6G). More importantly, in Hi-Myc; Otud6a−/y mice, c-Myc staining signals were mainly in the nuclei of the epithelial cells and relatively weaker than those in Hi-Myc; Otud6a+/y mice (Fig. 6G), further supporting the role of OTUD6A in governing c-Myc protein stability and abundance in prostate. Taken together, the results in the Hi-Myc transgenic PrCa mice indicate that OTUD6A controls c-Myc stability and levels in prostate and that depletion of Otud6a suppresses the prostatic tumorigenesis driven by c-Myc in vivo.
Depletion of OTUD6A reverses Myc-induced metabolic remodeling in PrCa
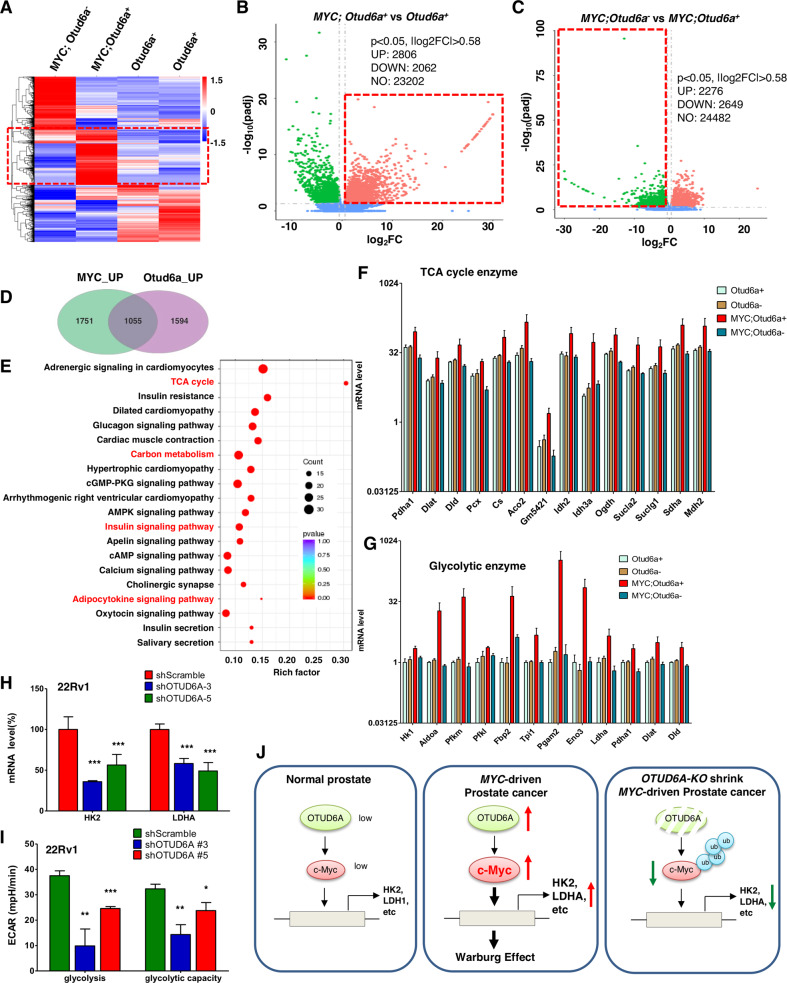
c-Myc is a key transcriptional factor for a series of genes, which provides mechanistic explanation for its oncogenic role in various cancers, including breast cancer and PrCa [45]. To further explore how OTUD6A/c-Myc axis regulates prostate tumorigenesis, the transcriptional profiles of prostate tissue/tumor samples from Otud6a+/y, Otud6a−/y, Hi-Myc; Otud6a+/y and Hi-Myc; Otud6a−/y mice were profiled with RNA-sequencing (Fig. 7A). First, Hi-Myc; Otud6a+/y mice had a distinct gene expression profile compared with the wild type Otud6a+/y mice, including reduced level of tumor suppressor gene Nkx3.1, in consistent with previous report [18]. Compared with Otud6a+/y mice, a total of 2806 genes were up-regulated in Hi-Myc; Otud6a+/y (Fig. 7B and Supplementary Table S3), and those genes were direct or indirect transcription targets of c-Myc transcriptional factor. More importantly, compared with Hi-Myc; Otud6a+/y mice, a total of 2649 genes were downregulated in Hi-Myc; Otud6a−/y mice (Fig. 7C and Supplementary Table S4), indicating Otud6a positively regulated genes. Further analysis of these differential expressed genes identified a total of 1055 co-regulated genes that were induced by c-Myc overexpression and further reduced after depletion of endogenous Otud6a (Fig. 7D, Supplementary Fig. S8A and Supplementary Table S5). The KEGG pathway analysis indicated that the 1055 genes were mainly enriched in metabolic function, including those in tricarboxylic acid cycle (TCA cycle or Krebs cycle) and glycolysis (Fig. 7E and Supplementary Table S6). Notably, genes in glucagon signaling, insulin signaling pathway and AMPK pathway were also significantly affected by c-Myc overexpression and further reversed by depletion of endogenous Otud6a in the transgenic mice (Fig. 7E). Further in-depth analysis showed that 14 TCA cycle genes (Fig. 7F) and 12 glycolysis genes (Fig. 7G) were up-regulated by c-Myc overexpression and further downregulated by depletion of endogenous Otud6a. These results indicate that c-Myc overexpression induces a transcriptional remodeling to promote prostatic tumorigenesis, which could be antagonized by depletion of endogenous OTUD6A.
Fig. 7. The OTUD6A/c-Myc axis remodels prostate cancer metabolism.
A Heatmap showed the differentially expressed genes (DEG) in prostate tissue and prostate tumors derived from Otud6a+/y (N = 5), Otud6a−/y (N = 5), Hi-Myc;Otud6a+/y (N = 4) and Hi-Myc;Otud6a−/y mice (N = 5). B Volcano plot showed the DEG between Otud6a+/y (N = 5) vs. Hi-Myc;Otud6a+/y mice (N = 4). p < 0.05, |log2FC | > 0.58. C Volcano plot showed the DEG between Hi-Myc;Otud6a−/y (N = 5) vs. Hi-Myc;Otud6a+/y mice (N = 4). p < 0.05, |log2FC | > 0.58. D Venn diagram showed the genes that were co-regulated by c-Myc and Otud6a in prostate tumors. E Functional enrichment KEGG assay for the genes that were co-regulated by c-Myc and Otud6a. F The mRNA expression of genes in tricarboxylic acid (TCA) cycle from prostate tissue and prostate tumors. G The mRNA expression of genes in glycolysis from prostate tissue and prostate tumors. H Depletion of OTUD6A led to reduced mRNA expressions of HK2 and LDHA in 22Rv1 cells. Cells were depleted of OTUD6A by shRNA, and the mRNA levels were measured by qPCR. ***p < 0.001. I Depletion of OTUD6A reduced the glycolysis level in 22Rv1 cells. Cells were depleted of OTUD6A by shRNA, and the cancer cell metabolism was measured by XF24 Seahorse extracellular flux analyzer. *p < 0.05, **p < 0.01, ***p < 0.001. J A schematic diagram shows that c-Myc induces metabolic remodeling and prostate tumorigenesis, which could be antagonized by depletion of Otud6a.
c-Myc is a driving transcription factor for controlling cellular metabolism, including glucose, glutamine and lipid metabolism [46], through the transcription of enzymes, such as HK2 [47] and LDHA [48]. The metabolic role of c-Myc has been determined in several cancer types, including colon, lung and breast cancer [46], but it remains largely elusive in PrCa. A recent study reported that c-Myc interferes metabolism of PrCa through up-regulating fatty acid synthesis genes, including ATP citrate lyase (ACLY), acetyl-CoA carboxylase 1 (ACC1) and fatty acid synthase (FASN) [49]. To this end, we found that depletion of Otud6a in mice reversed the transcriptional profile caused by overexpression of c-Myc, preferentially for those genes with metabolic function (Fig. 7E–G). In consistent with the RNA-seq results of prostate tumor derived from Hi-Myc; Otud6a+/y and Hi-Myc; Otud6a−/y mice, depletion of endogenous OTUD6A in a panel of human PrCa cell lines also led to decrease in the mRNA levels of HK2 and LDHA (Fig. 7H and Supplementary Fig. S8B, C). In keeping with this notion, in human PrCa samples, the mRNA levels of HK2 and LDHA were positively correlated with c-Myc expressions (Supplementary Fig. S8D–G). In addition, depletion of endogenous OTUD6A in PrCa cells led to decrease in the glycolysis capability measured by the XF24 Seahorse extracellular analyzer (Fig. 7I and Supplementary Fig. S9A, B). Taken together, these results indicate that metabolic abnormality is a significant phenotype in Myc-driven PrCa, and that OTUD6A might be a promising therapeutic target for combating PrCa with c-Myc overexpression and metabolic abnormality (Fig. 7J).
Discussion
The overexpression of c-Myc has been detected in most PrCa patients, and overexpressing c-Myc alone leads to transform of normal cells and prostatic tumorigenesis in vitro and in vivo, indicating c-Myc as a potent therapeutic for PrCa. Previously, antisense oligomers of MYC mRNA have been designed to target c-Myc for the treatment of PrCa [50, 51], and recent studies showed that several Myc-targeted therapeutic approaches eventually perturb c-Myc protein stability. For example, small molecule inhibitors of c-Myc, MYCi361/MYCi975, have been developed to efficiently repress prostatic tumorigenesis [21]. Notably, treatment with these inhibitors leads to the stabilization and degradation of c-Myc protein in PrCa cells [21]. Recently, BET inhibitions [52] and degraders [53] have been proven to be efficient in repressing the proliferation and tumorigenesis of PrCa cells in vitro and in xenograft mice, and these treatments also lead to reduction of c-Myc protein stability and abundance. Moreover, PP2A activator [24] and PIN1 inhibitors [23] have also been used to treat Myc-driven cancer through a mechanism of promoting proteasome-mediated degradation of c-Myc oncoprotein. These studies all together suggest that targeting c-Myc protein stability could be rational for designing therapeutic approach for Myc-driven cancers.
Although several USP DUBs [29–32] have been identified to be effective in deubiquitinating c-Myc in several types of cancer, but none of these USPs is responsible for deubiquitinating c-Myc in PrCa setting. We found that OTUD6A might be a prostate-specific DUB for c-Myc protein, and furthermore, depletion of OTUD6A retarded prostatic tumorigenesis in human PrCa cells-derived xenograft mouse model and in Hi-Myc GEMM, suggesting that OTUD6A might be a potential therapeutic target for PrCa. More importantly, c-Myc also plays a key role in the growth of normal cells, therefore a cancer specific upstream regulator of c-Myc, such as OTUD6A in PrCa setting, might be more suitable for therapeutic target. Given the limited expression spectrum in normal tissues, and the prostate-specific oncogenic role, OTUD6A could be a rational and promising therapeutic approach for the treatment of Myc-driven PrCa.
Supplementary information
Table S1. The CNA and mRNA of MYC in prostate cancer patients in TCGA.
Table S2. IHC staining score of OTUD6A and cMyc in human PrCa tissue array.
Table S3. List of upregulted 2086 genes in MYC;OTUD6A-WT mice compared with OTUD6A-WT mice.
Table S4. List of downregulated genes in MYC;Otud6a-null mice compared with MYC;Otud6a-wt mice.
Table S5. Raw RNA-seq reads of mouse prostate tissue/tumor.
Table S6. KEGG enrichment analysis of co-regulated genes by OTUD6A and c-Myc.
Acknowledgements
We thank Dr. Zhihui Feng, Dr. Jian Ding, and Dr. Yongping Shao for suggestions and comments on this work. We thank Dr. Yong Yang from China Pharmaceutical University for his kind help for providing Hi-Myc mice as gift.
Author contributions
The idea was conceived by WW, LZ, JKL, and JGL. YP and JL designed and performed most of the experiments with assistance from ZW, CC, TZ, PG, ZH, HL, YW, JG, and JZ. YP and JL wrote the manuscript. WW, LZ, JKL, and JGL supervised the study and edited the manuscript. All authors commented on the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China [81802787, 31700684, 31870848, 31770917 and 32171102], the National Basic Research Program of China [973 Program 2015CB856302 and 2015CB553602], the China Postdoctoral Science Foundation [2018M643673], and the National Key Research and Development Projects of China [2019YFD1002400].
Data availability
The data that support the findings of this study are available from the corresponding authors upon reasonable request. RNA-seq data discussed in this paper have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE189716.
Competing interests
The authors declare no competing interests.
Ethical approval
All animal experiments were approved by the Animal Use and Care Committee of the School of Life Science and Technology, Xi’an Jiaotong University, based on the Guide for the Care and Use of Laboratory Animals: eighth edition (ISBN10: 0-309-15396-4).
Footnotes
The original online version of this article was revised: Due to a figure error.
Edited by A Degterev
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yunhua Peng, Jing Liu, Zhen Wang, Chunping Cui.
Change history
4/19/2022
A Correction to this paper has been published: 10.1038/s41418-022-00978-1
Contributor Information
Lingqiang Zhang, Email: zhanglq@nic.bmi.ac.cn.
Jiankang Liu, Email: jkliu@uor.edu.cn.
Jiangang Long, Email: jglong@mail.xjtu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-022-00960-x.
References
- 1.Koh CM, Bieberich CJ, Dang CV, Nelson WG, Yegnasubramanian S, De Marzo AM. MYC and prostate cancer. Genes Cancer. 2010;1:617–28. doi: 10.1177/1947601910379132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 3.Freedman ML, Haiman CA, Patterson N, McDonald GJ, Tandon A, Waliszewska A, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci USA. 2006;103:14068–73. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amundadottir LT, Sulem P, Gudmundsson J, Helgason A, Baker A, Agnarsson BA, et al. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–8. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 5.Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, et al. Integrative molecular concept modeling of prostate cancer progression. Nat Genet. 2007;39:41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- 6.Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–9. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 7.Zheng SL, Sun J, Wiklund F, Smith S, Stattin P, Li G, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358:910–9. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 8.Al Olama AA, Kote-Jarai Z, Giles GG, Guy M, Morrison J, Severi G, et al. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat Genet. 2009;41:1058–60. doi: 10.1038/ng.452. [DOI] [PubMed] [Google Scholar]
- 9.Ribeiro FR, Jeronimo C, Henrique R, Fonseca D, Oliveira J, Lothe RA, et al. 8q gain is an independent predictor of poor survival in diagnostic needle biopsies from prostate cancer suspects. Clin Cancer Res. 2006;12:3961–70. doi: 10.1158/1078-0432.CCR-05-1977. [DOI] [PubMed] [Google Scholar]
- 10.Ribeiro FR, Henrique R, Martins AT, Jeronimo C, Teixeira MR. Relative copy number gain of MYC in diagnostic needle biopsies is an independent prognostic factor for prostate cancer patients. Eur Urol. 2007;52:116–25. doi: 10.1016/j.eururo.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 11.Fromont G, Godet J, Peyret A, Irani J, Celhay O, Rozet F, et al. 8q24 amplification is associated with Myc expression and prostate cancer progression and is an independent predictor of recurrence after radical prostatectomy. Hum Pathol. 2013;44:1617–23. doi: 10.1016/j.humpath.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 12.Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, Montgomery K, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci USA. 2004;101:811–6. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–9. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 14.Dhanasekaran SM, Dash A, Yu J, Maine IP, Laxman B, Tomlins SA, et al. Molecular profiling of human prostate tissues: insights into gene expression patterns of prostate development during puberty. FASEB J. 2005;19:243–5. doi: 10.1096/fj.04-2415fje. [DOI] [PubMed] [Google Scholar]
- 15.Gurel B, Iwata T, Koh CM, Jenkins RB, Lan F, Van Dang C, et al. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Mod Pathol. 2008;21:1156–67. doi: 10.1038/modpathol.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gil J, Kerai P, Lleonart M, Bernard D, Cigudosa JC, Peters G, et al. Immortalization of primary human prostate epithelial cells by c-Myc. Cancer Res. 2005;65:2179–85. doi: 10.1158/0008-5472.CAN-03-4030. [DOI] [PubMed] [Google Scholar]
- 17.Williams K, Fernandez S, Stien X, Ishii K, Love HD, Lau YF, et al. Unopposed c-MYC expression in benign prostatic epithelium causes a cancer phenotype. Prostate. 2005;63:369–84. doi: 10.1002/pros.20200. [DOI] [PubMed] [Google Scholar]
- 18.Ellwood-Yen K, Graeber TG, Wongvipat J, Iruela-Arispe ML, Zhang J, Matusik R, et al. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell. 2003;4:223–38. doi: 10.1016/S1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]
- 19.Kim J, Eltoum I-EA, Roh M, Wang J, Abdulkadir SA. Interactions between cells with distinct mutations in c-MYC and Pten in prostate cancer. PLoS Genet. 2009;5:e1000542. doi: 10.1371/journal.pgen.1000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellis L, Ku S, Li Q, Azabdaftari G, Seliski J, Olson B, et al. Generation of a C57BL/6 MYC-driven mouse model and cell line of prostate cancer. Prostate. 2016;76:1192–202. doi: 10.1002/pros.23206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han H, Jain AD, Truica MI, Izquierdo-Ferrer J, Anker JF, Lysy B, et al. Small-molecule MYC inhibitors suppress tumor growth and enhance immunotherapy. Cancer cell. 2019;36:483–97 e415. doi: 10.1016/j.ccell.2019.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clausen DM, Guo J, Parise RA, Beumer JH, Egorin MJ, Lazo JS, et al. In vitro cytotoxicity and in vivo efficacy, pharmacokinetics, and metabolism of 10074-G5, a novel small-molecule inhibitor of c-Myc/Max dimerization. J Pharm Exp Ther. 2010;335:715–27. doi: 10.1124/jpet.110.170555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allen-Petersen BL, Sears RC. Mission possible: advances in MYC therapeutic targeting in cancer. BioDrugs. 2019;33:539–53. doi: 10.1007/s40259-019-00370-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leonard D, Huang W, Izadmehr S, O’Connor CM, Wiredja DD, Wang Z, et al. Selective PP2A enhancement through biased heterotrimer stabilization. Cell. 2020;181:688–701 e616. doi: 10.1016/j.cell.2020.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arnold HK, Sears RC. Protein phosphatase 2A regulatory subunit B56alpha associates with c-myc and negatively regulates c-myc accumulation. Mol Cell Biol. 2006;26:2832–44. doi: 10.1128/MCB.26.7.2832-2844.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23:2116–25. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN, et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci USA. 2004;101:9085–90. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geng C, Kaochar S, Li M, Rajapakshe K, Fiskus W, Dong J, et al. SPOP regulates prostate epithelial cell proliferation and promotes ubiquitination and turnover of c-MYC oncoprotein. Oncogene. 2017;36:4767–77. doi: 10.1038/onc.2017.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim D, Hong A, Park HI, Shin WH, Yoo L, Jeon SJ, et al. Deubiquitinating enzyme USP22 positively regulates c-Myc stability and tumorigenic activity in mammalian and breast cancer cells. J Cell Physiol. 2017;232:3664–76. doi: 10.1002/jcp.25841. [DOI] [PubMed] [Google Scholar]
- 30.Popov N, Wanzel M, Madiredjo M, Zhang D, Beijersbergen R, Bernards R, et al. The ubiquitin-specific protease USP28 is required for MYC stability. Nat Cell Biol. 2007;9:765–74. doi: 10.1038/ncb1601. [DOI] [PubMed] [Google Scholar]
- 31.Sun XX, He X, Yin L, Komada M, Sears RC, Dai MS. The nucleolar ubiquitin-specific protease USP36 deubiquitinates and stabilizes c-Myc. Proc Natl Acad Sci USA. 2015;112:3734–9. doi: 10.1073/pnas.1411713112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan J, Deng Q, Jiang C, Wang X, Niu T, Li H, et al. USP37 directly deubiquitinates and stabilizes c-Myc in lung cancer. Oncogene. 2015;34:3957–67. doi: 10.1038/onc.2014.327. [DOI] [PubMed] [Google Scholar]
- 33.Yuan L, Lv Y, Li H, Gao H, Song S, Zhang Y, et al. Deubiquitylase OTUD3 regulates PTEN stability and suppresses tumorigenesis. Nat Cell Biol. 2015;17:1169–81. doi: 10.1038/ncb3218. [DOI] [PubMed] [Google Scholar]
- 34.Liu J, Peng Y, Shi L, Wan L, Inuzuka H, Long J, et al. Skp2 dictates cell cycle-dependent metabolic oscillation between glycolysis and TCA cycle. Cell Res. 2021;31:80–93. doi: 10.1038/s41422-020-0372-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–4. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Disco. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z, Song Y, Ye M, Dai X, Zhu X, Wei W. The diverse roles of SPOP in prostate cancer and kidney cancer. Nat Rev Urol. 2020;17:339–50. doi: 10.1038/s41585-020-0314-z. [DOI] [PubMed] [Google Scholar]
- 39.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–9. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mevissen TE, Hospenthal MK, Geurink PP, Elliott PR, Akutsu M, Arnaudo N, et al. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell. 2013;154:169–84. doi: 10.1016/j.cell.2013.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu T, Ortiz JA, Taing L, Meyer CA, Lee B, Zhang Y, et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 2011;12:R83. doi: 10.1186/gb-2011-12-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeh E, Cunningham M, Arnold H, Chasse D, Monteith T, Ivaldi G, et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol. 2004;6:308–18. doi: 10.1038/ncb1110. [DOI] [PubMed] [Google Scholar]
- 43.Sears R, Leone G, DeGregori J, Nevins JR. Ras enhances Myc protein stability. Mol Cell. 1999;3:169–79. doi: 10.1016/S1097-2765(00)80308-1. [DOI] [PubMed] [Google Scholar]
- 44.Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–14. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, metabolism, and cancer. Cancer Disco. 2015;5:1024–39. doi: 10.1158/2159-8290.CD-15-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim JW, Gao P, Liu YC, Semenza GL, Dang CV. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol Cell Biol. 2007;27:7381–93. doi: 10.1128/MCB.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, et al. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci USA. 1997;94:6658–63. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh KB, Hahm ER, Kim SH, Wendell SG, Singh SV. A novel metabolic function of Myc in regulation of fatty acid synthesis in prostate cancer. Oncogene. 2021;40:592–602. doi: 10.1038/s41388-020-01553-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Devi GR, Beer TM, Corless CL, Arora V, Weller DL, Iversen PL. In vivo bioavailability and pharmacokinetics of a c-MYC antisense phosphorodiamidate morpholino oligomer, AVI-4126, in solid tumors. Clin Cancer Res. 2005;11:3930–8. doi: 10.1158/1078-0432.CCR-04-2091. [DOI] [PubMed] [Google Scholar]
- 51.Iversen PL, Arora V, Acker AJ, Mason DH, Devi GR. Efficacy of antisense morpholino oligomer targeted to c-myc in prostate cancer xenograft murine model and a Phase I safety study in humans. Clin Cancer Res. 2003;9:2510–9. [PubMed] [Google Scholar]
- 52.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raina K, Lu J, Qian Y, Altieri M, Gordon D, Rossi AM, et al. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc Natl Acad Sci USA. 2016;113:7124–9. doi: 10.1073/pnas.1521738113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. The CNA and mRNA of MYC in prostate cancer patients in TCGA.
Table S2. IHC staining score of OTUD6A and cMyc in human PrCa tissue array.
Table S3. List of upregulted 2086 genes in MYC;OTUD6A-WT mice compared with OTUD6A-WT mice.
Table S4. List of downregulated genes in MYC;Otud6a-null mice compared with MYC;Otud6a-wt mice.
Table S5. Raw RNA-seq reads of mouse prostate tissue/tumor.
Table S6. KEGG enrichment analysis of co-regulated genes by OTUD6A and c-Myc.
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon reasonable request. RNA-seq data discussed in this paper have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE189716.