Abstract
Cellular proliferation depends on the accurate and timely replication of the genome. Several genetic diseases are caused by mutations in key DNA replication genes; however, it remains unclear whether these genes influence the normal program of DNA replication timing. Similarly, the factors that regulate DNA replication dynamics are poorly understood. To systematically identify trans-acting modulators of replication timing, we profiled replication in 184 cell lines from three cell types, encompassing 60 different gene knockouts or genetic diseases. Through a rigorous approach that considers the background variability of replication timing, we concluded that most samples displayed normal replication timing. However, mutations in two genes showed consistently abnormal replication timing. The first gene was RIF1, a known modulator of replication timing. The second was MCM10, a highly conserved member of the pre-replication complex. Cells from a single patient carrying MCM10 mutations demonstrated replication timing variability comprising 46% of the genome and at different locations than RIF1 knockouts. Replication timing alterations in the mutated MCM10 cells were predominantly comprised of replication delays and initiation site gains and losses. Taken together, this study demonstrates the remarkable robustness of the human replication timing program and reveals MCM10 as a novel candidate modulator of DNA replication timing.
Introduction
Cell proliferation is one of the most fundamental aspects of development and becomes mis-regulated in many genetic diseases, in cancer, and during aging and tissue degeneration. A central part of cell proliferation is the replication of DNA, which occurs during S phase and spans roughly a third of the cell cycle in actively dividing cells. Accordingly, delays in DNA replication and S phase completion have been implicated in several developmental diseases that are characterized by growth and developmental defects of various tissues. In addition, some disease-associated gene mutations disrupt the temporal progression of DNA replication, causing certain genomic loci to replicate earlier or later than they normally would (reviewed in (1–3)). Such alterations to replication dynamics have been described in diseases caused by mutations in central DNA replication initiation factors, such as CDC45 that is part of large deletion occurring in DiGeorge/Veleocardiofacial (VCF) syndrome (4,5), RECQ4 (RECQL4) in Rothmund–Thomson syndrome (RTS) (6–9) and components of the ORC and MCM complexes and associated genes in Meier–Gorlin syndrome (10–14). Aberrant replication initiation and progression have also been reported to result from LMNA mutations affecting nuclear Lamin A and C in Hutchinson–Gilford progeria (HGPS) (9), FANCD2 deficiency in a subtype of Fanconi Anemia (FA) (15), DNMT3B mutations affecting DNA methylation in ICF1 syndrome (16,17) and loss-of-function mutations in BLM in Bloom syndrome (BLM) (18). These studies used various approaches for assaying replication dynamics, most of which were underpowered to comprehensively characterize the genomic effects of these disease mutations on DNA replication timing. In addition, previous studies have not fully considered natural polymorphism in DNA replication timing (19,20) when interpreting replication timing alterations in disease. Thus, it remains largely unknown to what extent alterations in DNA replication dynamics associate with human developmental diseases. Deciphering these links is important for understanding the etiology of these diseases and for bridging genetic alterations and disease phenotypes via intermediary molecular phenotypes.
Similar to replication timing alterations in developmental diseases, very little is known about the regulatory factors that determine the temporal order of DNA replication progression in mammalian cells. A single well-described modulator of the replication timing program is RIF1, which has been shown to regulate global replication timing in yeasts, flies, mice and human cells (21–26). Mutations in RIF1 cause widespread delays and advances in replication timing across numerous regions in the genome, many spanning several megabases of DNA (27–29) and in some cases have even been suggested to define the entire replication timing program (30). Apart from RIF1, several studies have described more modest replication timing alterations following knock-out of DNA polymerase theta (Pol θ) (31,32) or PREP1 (33). Nonetheless, systematic studies of the effects of trans-acting regulators on DNA replication timing are lacking. More generally, the dearth of well-described regulators of DNA replication timing is surprising and warrants further investigation. It could be due to lack of comprehensive assays for testing the effects of trans-acting mutations on DNA replication timing, or to a fundamental essentiality of the replication timing program that would preclude the identification of such factors due to cell lethality.
Here, we set to comprehensively test for replication timing alterations in relevant human developmental diseases and in knockouts (KOs) of DNA replication-related genes. We analyzed a total of 184 patient or mutant cell lines and compared them to 167 normal cell lines. Our results point to the rarity of replication timing alterations, suggesting that replication dynamics represent an essential and rigid cellular program. We stress the importance of methodological aspects for the rigorous identification of replication timing alterations and rule out several previously suggested regulators and diseases impacting replication timing. In particular, through simulations we identify a set of genomic regions with greater tendency for inter-individual variation, and show that this tendency occurs in normal samples and is independent of genic mutations in trans. Last, we report substantial replication timing alterations in RIF1 KO cells, and—newly discovered here—in four induced pluripotent stem cell lines from a single patient (and from one reprogramming event) carrying mutations in MCM10. MCM10 (minichromosomal maintenance complex member 10) is a conserved and essential gene that is critical for the proper function of the CDC45: MCM2-7: GINS (CMG) DNA replicative helicase, which binds to origins of replication and is required for replication initiation. Specifically, MCM10 promotes reconfiguration of CMG complexes to enable bidirectional unwinding of DNA at replication origins. In addition, there is growing evidence that MCM10 is required for the processivity of the progressing replisome, although the specific mechanisms remain unclear (34–36). Our results point to the presence of still-elusive factors that regulate human replication timing.
Results
Abundant replication timing variation is observed a priori in disease cell lines and gene knockouts
To identify potential modulators of DNA replication timing in human cells, we generated replication timing profiles for 184 cell lines from individuals with genetic diseases or with introduced gene KOs (hereafter, ‘mutant/affected’) in three cell types, compared with 167 healthy or wild-type (WT) samples (hereafter, ‘WT/unaffected’) (Tables 1 and 2; Supplementary Material, Table S1). The analyzed cell types included lymphoblastoid cell lines (LCLs), which are EBV-transformed lymphoid cells widely available from many individuals; induced pluripotent stem cells (iPSCs), which are not transformed but not as commonly available across specific patient cohorts; and HAP1 cells, which are nearly-haploid human cell lines derived from a chronic myeloid leukemia patient and readily amenable to CRISPR/Cas9-mediated gene knockout (37,38). The selected mutant/affected cell lines included all those (to our knowledge) with previous evidence of replication timing alterations, strong links to DNA replication or related pathways such as nucleotide metabolism, DNA repair, chromatin structure (20), and cell lines with alterations in chromosome structure (e.g. disease-associated aneuploidies or repeat expansions). In total, 60 genes or genetic diseases were analyzed across the cell types. Of note, samples of the same disease or KO may contain different mutations (Supplementary Material, Table S1). DNA from proliferating cell cultures was subjected to whole genome sequencing (WGS) and replication timing was inferred for each sample based on DNA copy number fluctuations along chromosomes, as previously described (19,39) (see Materials and Methods).
Table 1.
Summary of cell types analyzed in this study. For LCL and iPSC samples, ‘unique individuals’ excludes repeated clones or sequencing of the same individual. In HAP1, all cell lines were derived from a single individual therefore ‘unique mutant samples’ signifies different KO types. HAP1 also includes 26 KO lines involving 22 genes with three genes having multiple KO clones. Among the 26 KO lines, some were sequenced before and after diploidization and are considered the same unique mutant sample. See Supplementary Material, Table S1 for more details
| Cell type | WT/unaffected samples | Unique WT/unaffected individuals | Mutant/affected samples | Unique mutant/affected samples | Unique genetic diseases or genes affected |
|---|---|---|---|---|---|
| LCL | 137 | 124 | 134 | 117 | 32 |
| HAP1 | 6 | 1 | 32 | 26 | 21 |
| iPSC | 24 | 24 | 18 | 11 | 7 |
Table 2.
Abbreviations of diseases used in this study
| Disease name | Abbreviation | Number of unique mutant/affected samples | Number of total mutant/affected samples |
|---|---|---|---|
| Ataxia-Oculomotor Apraxia 2 | AOA2 | 4 | 5 |
| Ataxia-Telangiectasia | AT | 3 | 4 |
| Bloom syndrome | BLM | 4 | 9 |
| Breast Cancer, Type 1 | BRCA1 | 3 | 3 |
| Beckwith-Wiedemann Syndrome | BWS | 2 | 2 |
| Cornelia de Lange Syndrome 1 | CDLS1 | 2 | 2 |
| Chronic myeloid leukemia | CML | 1 | 1 |
| DiGeorge Syndrome | DGS | 3 | 3 |
| Myotonic dystrophy type I | DM1 | 14 | 14 |
| Fanconi anemia | FA | 5 | 5 |
| Friedreich’s ataxia | FRDA | 13 | 13 |
| Fragile-X syndrome | FXS | 14 | 20 |
| Huntington’s disease | HD | 13 | 13 |
| Hutchinson-Gilford Progeria Syndrome | HGPS | 5 | 6 |
| Immunodeficiency-centromeric instability-facial anomalies syndrome 1 | ICF1 | 1 | 1 |
| Lesch–Nyhan Syndrome | LNS | 3 | 4 |
| Mental retardation, autosomal dominant 1 caused by a deletion in MBD5 | MRD1 | 1 | 1 |
| Rothmund-Thomson Syndrome | RTS | 3 | 4 |
| Rett Syndrome | RTT | 5 | 9 |
| Rett Syndrome, congenital variant | RTTC | 2 | 2 |
| Spinal and bulbar muscular atrophy | SBMA | 1 | 1 |
| Spinocerebellar ataxia type 1 | SCA1 | 3 | 3 |
| Seckel Syndrome | SCKL1 | 1 | 1 |
| Sotos Syndrome 1 | SOTOS1 | 1 | 1 |
| Down Syndrome | TRI21 | 2 | 2 |
| Williams-Beuren Syndrome | WBS | 2 | 2 |
| Wolf-Hirschhorn Syndrome | WHS | 1 | 1 |
| Werner Syndrome | WRN | 3 | 3 |
| Turner Syndrome | XO | 2 | 2 |
| Translocation of the X chromosome | XTRANS | 2 | 2 |
| 4 X chromosomes | XXXX/XXXXY | 2 | 3 |
| Klinefelter Syndrome | XXY | 2 | 2 |
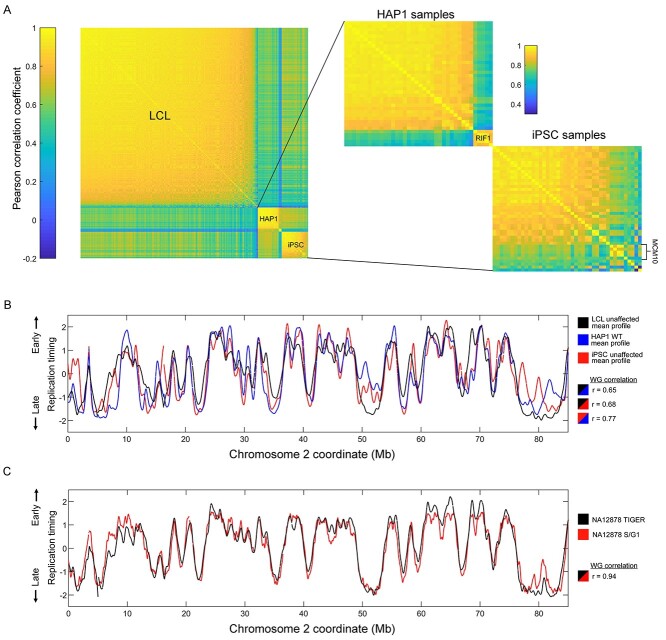
Replication timing profiles had a median Pearson’s correlation coefficient of 0.86 (0.31–0.90) among all LCL samples, 0.86 (0.71–0.90) among LCL unaffected samples and 0.90 (0.73–0.95) between repeats of unaffected LCL samples (Fig. 1A and B, Supplementary Material, Fig. S1). There was also a high correlation (r = 0.94) of the unaffected LCL sample NA12878 to its replication profile generated by sequencing S and G1 phase DNA (40), further demonstrating the high quality of replication timing profiles generated in this study (Fig. 1C). Heterozygous carriers of disease mutations were classified as affected if there was evidence of haploinsufficiency for the implicated genes (e.g. BRCA1, HGPS) and otherwise classified as unaffected (e.g. the recessive diseases RTS, LNS, AT and FRDA; Supplementary Material, Table S1); the replication timing correlation to non-carrier, unaffected samples (Supplementary Material, Fig. S2) also supported this classification.
Figure 1.
Overview of replication timing data. (A) Correlation matrices of replication profiles for all samples. (B) Replication profiles and whole genome correlations of the WT/unaffected mean profiles of the different cell types. (C) Replication timing profile comparison and whole genome correlation for the sample NA12878 generated with TIGER (see Materials and Methods) or S/G1 sequencing.
For iPSCs and HAP1 cells, the median between-sample correlations were 0.95 (0.66–0.96) and 0.90 (0.66–0.93), respectively. The correlations of samples within a given cell type were somewhat lower than expected, which we attribute to several low-correlating mutant/affected samples (to be further discussed below), a less stringent approach to filtering in anticipation of replication timing variation (see Materials and Methods) and the variability of sample source and WGS method and sequencing depth. There was no observable difference between isogenic haploid and diploid HAP1 samples (Supplementary Material, Fig. S3).
To analyze replication timing variation between mutant/affected and WT/unaffected cell lines, we performed an analysis of variance (ANOVA) in sliding windows across the autosomes (sex chromosomes were not considered since the samples included both male and female individuals). ANOVA was applied to raw data (before smoothing) in windows of 185 kb of uniquely alignable sequence (76 bins of 2500 bp; Methods) with a step of a quarter window. We studied both individual samples as well as samples grouped by mutated gene or genetic disease and compared each to the control samples of the same cell type. This was performed to capture potential outlier samples with unique phenotypes that could be explained by mutation type or variable penetrance. Individual windows with a Bonferroni-corrected P-value < 0.05 were considered to have variant replication timing, and overlapping variant windows were subsequently merged into continuous variant regions. Given the variable numbers of mutant/affected and WT/unaffected samples in the grouped analyses, we permissively allowed samples to show an inconsistent direction of replication timing difference compared with other samples, as long as the group ANOVA was significant.
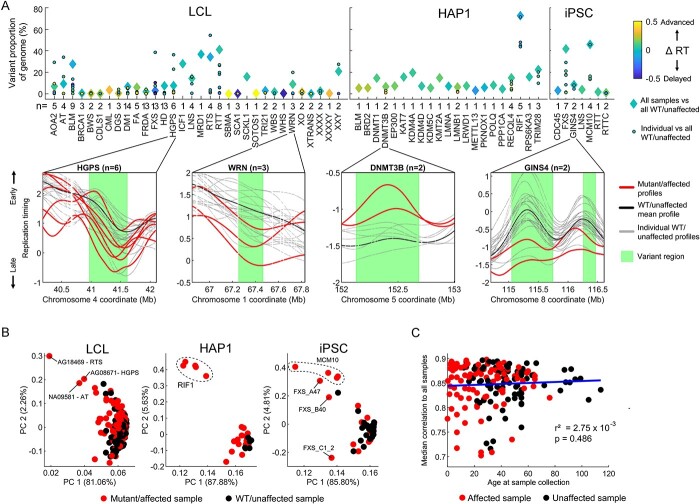
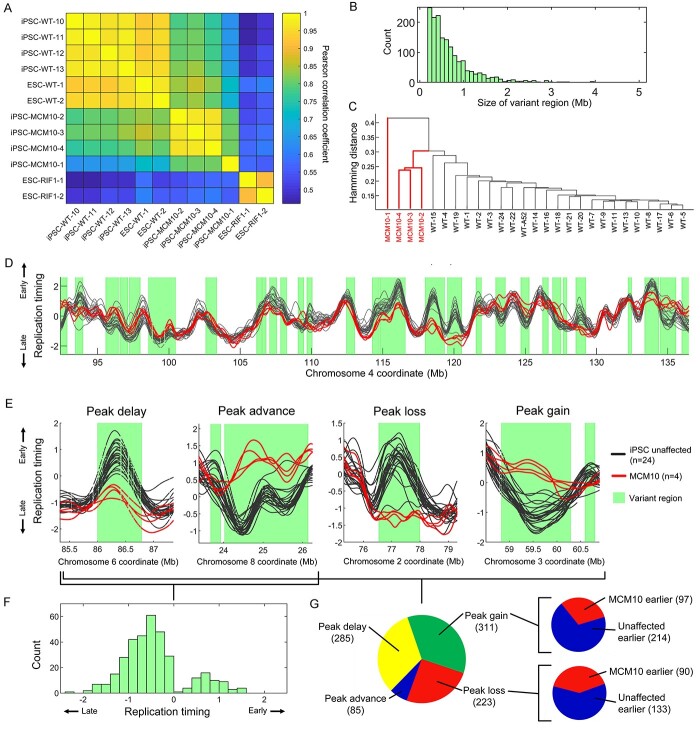
All samples grouped by mutated gene, and nearly all individual mutant/affected samples (177/184), contained at least one genomic region with a priori replication timing variation compared with the WT/unaffected samples (Fig. 2A, Supplementary Material, Fig. S4A). Across all cell types, individual mutant/affected samples showed a median variant replication timing covering 1.33% of the autosomes. Samples grouped by mutated gene showed a median replication timing variation covering 6.67% of the autosomes. RIF1 KOs in HAP1 cells contained the highest proportion of variant replication timing at 72.65%, with the five individual samples ranging from 43.79 to 71.47% genome variation compared with WT/unaffected (Supplementary Material, Fig. S4B). Several other gene mutations were also associated with higher-than-average genome variation (e.g. RTT, RTS, MCM10, FXS in iPSCs).
Figure 2.
Analysis of variance detects significant replication timing variation in all mutated gene groups and most individual samples. (A) Top: Proportion of the autosomal genome with variant replication timing detected via ANOVA with a Bonferroni-corrected P-value of <0.05. For each mutated gene, both grouped and individual samples were evaluated against WT/unaffected samples for each cell type. Colors represent the extent of bias toward replication timing advances or delays. The total number of mutant/affected samples is noted for each gene. Bottom: four example gene mutations are shown, with variant regions depicted based on the grouped analysis. For LCL samples, only 20 unaffected samples are shown. (B) PCA of the entire autosomal replication timing profiles. Clustering by mutation is observed for the RIF1 KO in HAP1 cells and for samples with mutated MCM10 in iPSCs. (C) Relationship of individual donor age at sample collection to median correlation of autosomal replication timing among LCL samples. Only samples with a mean correlation >0.7 to other LCL samples are included (see Materials and Methods and Supplementary Material, Fig. S5B for all samples).
In the 99 850 variant regions called across all mutated gene groups and individuals, the median absolute difference in replication timing from the mean WT/unaffected value was 0.50 units of standard deviation. In virtually all of the 30 strongest cases (with variation spanning ≥20% of the genome), replication timing variations included both advances and delays in roughly equal proportions; the most directionally biased sample was a HAP1 RIF1 KO sample with a mean delay across variant regions of 0.14 standard deviations (Fig. 2A).
Despite many mutated gene groups showing a substantial genomic proportion of variant replication timing, closer inspection ruled out most as candidates for gene-related dysregulation of replication timing. For example, replication timing in variant regions for AOA2, RTS, BLM, HGPS and FXS (in LCLs, across autosomes) was largely driven by one or two outlier samples, while the rest strongly resembled the unaffected profile (Supplementary Material, Fig. S4C). Accordingly, principal components analysis (PCA) of replication timing profiles did not reveal clustering by mutated gene except for the five RIF1 KOs in HAP1 cells and the four iPSC samples derived from a single reprogramming event from a patient with mutated MCM10 (Fig. 2B). Singular outlier samples with abnormal replication timing will be further investigated below.
Apart from disease state, we also assessed other factors that could influence local or global replication timing variation in our samples. Sequencing cohort or batch effects seemed to be minimal based on PC analysis, with the exception of one cohort of samples, which had the lowest-coverage sequencing (Supplementary Material, Fig. S5A). We also ruled out that replication timing is significantly influenced by a person’s age in our sample set. First, we compared the correlations of replication timing profiles in 186 LCLs from donors of known ages (ranging from 0 to 114 years old) at sample collection (Fig. 2C, Supplementary Material, Fig. S5B). If replication timing changes with age, we would expect deviation from the average replication profile in older (or younger) samples. However, we found no change in replication timing correlation to other samples as a function of a sample’s age. Secondly, PCA of unaffected replication timing profiles did not show stratification by age (Supplementary Material, Fig. S5C). Although the unknown number of cell culture passages that each cell line underwent may confound the analysis of ‘age’, the large number of samples analyzed here effectively rules out a strong influence of aging on replication timing, at least in LCLs. Also consistent with the minimal or no effect of age on LCL replication timing, we did not observe any notable replication timing variability in diseases associated with accelerated aging (HGPS, WRN, BLM) (Fig. 2A, see further below).
Taken together, variability in replication timing is detected in most mutant/affected individuals and mutated gene groups. However, variability is not necessarily the result of a gene mutation-related modulation of replication timing but may instead be driven by a subset of outlier samples or by background technical or biological variation, as further explored below.
Recalibration of false discovery rates using simulations of replication timing variation
Identifying genuine variability in replication timing as a result of a diseased state or gene mutation may be confounded by background replication timing variability, which may arise due to technical factors or be related to common polymorphisms that influence local replication timing (19,20). For example, as shown above, a subset of outlier samples led to the identification of variability in replication timing that is not shared with other samples of the same disease or gene mutation. Variant detection can be made stricter by adjusting the significance threshold or by requiring that all samples within a group follow the same trend. These remedies are expected to be heavily influenced by the number of samples compared in each mutated gene and WT/unaffected group and were therefore not implemented in initial analyses.
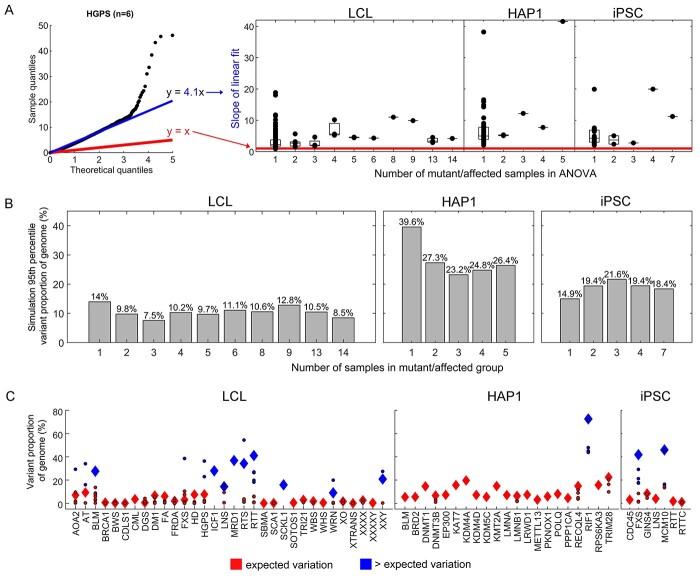
When inspecting the ANOVA variant results using quantile-quantile (QQ) plots, we observed widespread inflation (and in some cases, deflation) in the obtained P-values (Fig. 3A, Supplementary Material, Fig. S6A). This inflation is likely related to the continuous nature of the DNA replication profiles from which the data are sampled. It did not result from the sliding window method, as it was still observed in an ANOVA variant search with non-overlapping windows (Supplementary Material, Fig. S6B). Importantly, the extent of P-value inflation was different for each mutated gene group and individual, which makes it challenging to determine an appropriate threshold for rejecting the null hypothesis. Strict multiple testing correction did not mitigate this challenge, as the P-values were inflated beyond Bonferroni-corrected significance thresholds. An alternative method for multiple-test correction that could be considered in this case is q-value transformation, where P-values are adjusted based on false-discovery rate (FDR). However, in the ANOVA tests for replication timing variation, such FDR would be independently calculated based on the P-values for each mutated gene group or individual. This creates a different FDR value for each analysis even when the number of mutant/affected samples being analyzed is equal and the WT/unaffected samples remain the same (Supplementary Material, Fig. S6C).
Figure 3.
Variability in replication timing. (A) P-value inflation quantified with QQ plots for the different mutant/affected-WT/unaffected ANOVA tests. Theoretical quantiles (the uniform distribution of P-values) and ANOVA test quantiles should be linearly related with a slope of 1 (red) if they are generated from the same distribution. The linear fit of the ANOVA quantiles to theoretical values (blue) quantifies the deviation as inflation or deflation of P-values. The boxplot demonstrates the P-value inflation statistic in the different ANOVA tests for mutated/affected gene groups and individuals. (B) The 95th percentile of variant proportions of the genome from the 1000 simulations for different mutant/affected group sizes. The number of WT/unaffected samples matched that available for each cell type (137 in LCL, 6 in HAP1 and 24 in iPSCs). (C) Same as Figure 1A, highlighting (blue) the mutant/affected groups and individuals exceeding the 95th percentile cutoff.
To overcome these statistical challenges of analyzing sample groups with different sizes, we sought to determine a universal FDR by empirically calculating the expected variability in our replication timing data for any relevant number of compared samples. To achieve this, we permuted the samples, randomly assigning them into mutant/affected and WT/unaffected groups and repeating the ANOVA variant search. Given that the majority of mutant/affected samples fell within the correlation distribution of WT/unaffected samples (Fig. 1A), we used all samples (WT/unaffected and mutant/affected) with a correlation >0.7 to other samples. This cutoff removed genuine outlier samples such as RIF1 and others, while the inclusion of mutant/affected samples in the ‘WT/unaffected’ permutations maximized the number of samples available for this analysis. The latter enabled us to test permuted WT/unaffected groupings up to the actual number of WT/unaffected samples available for a given cell type. We thus tested WT/unaffected and mutant/affected groups with varying numbers of samples in each, performing 1000 sample permutations for each pair of groups sizes. In this analysis, FDR is akin to the expected variability of replication timing based on the number of samples in each compared group.
In these ‘simulations’ of background variability, the proportion of the genome found to be variant decreased with increasing numbers of WT/unaffected samples, for all cell types (Supplementary Material, Fig. S7). This is expected, since including more controls effectively rules out many false positive outlier observations. Importantly, there was a substantial dispersion around the median variation across the simulations (Supplementary Material, Fig. S8). For example, in simulations of LCLs with one affected sample and 137 unaffected samples (i.e. all available), the median genome variation was 0.18% but the 5th and 95th percentile (representing typical limits of low and high variation) were 0 and 13.97%, respectively. Therefore, we would argue that 13.97% genome variation (the 95th percentile) represents an upper limit for expected variation in analyses with one mutant/affected and 137 WT/unaffected samples. To further illustrate the application of a 95th percentile cutoff for expected variation, consider simulations for LCLs with three mutant/affected and 137 WT/unaffected samples—the same number of actual samples available for BRCA1, DGS, SCA1 and WRN. In this case, the median proportion of the genome with replication timing variation was 0.28% and the 95th percentile was 7.54%. If we use the 95th percentile as the upper cutoff for expected variation, then among the LCL-affected groups that had three affected samples we would rule out BRCA1 (0.58%), DGS (0.51%) and SCA1 (0.15%) as having an extent of variation within the expected range (Fig. 3B). In contrast, the group containing the three WRN samples does have an extent of variation (8.93%) exceeding this 95th percentile cutoff and would therefore remain as a candidate for later analyses of replication timing variation.
We applied the 95th percentile cutoff to all samples and groups (Fig. 3B), which resulted in 155 of 184 individual mutant/affected samples, and 52 of 60 mutated gene groups being classified as within the expected range of replication timing variability (Fig. 3C). Therefore, these gene mutations (at least in the analyzed cell type) were concluded to likely not influence replication timing. The several mutant/affected samples that did exhibit variation above the expected range of replication timing variability will be analyzed further below. Background heterogeneity in replication timing data still emerges as a critical factor requiring rigorous consideration in any search for replication timing differences between samples or groups.
Replication timing variation is non-randomly distributed across the genome
Based on the above simulations, variability in replication timing can be expected in a substantial percentage of the genome, depending on cell type. We asked if this variability (in both simulations and true analyses) is uniformly distributed across the genome or clustered in specific regions and/or specific replication times. For the true mutant/affected individuals and gene groups, the regions of variant replication timing were bimodally distributed to late- and early-replicating regions of the genome (Supplementary Material, Fig. S9). Furthermore, variant regions across all individual mutant/affected analyses overlapped more than would be expected by chance. The median proportion of variant region coordinates shared between two mutant/affected samples of the same cell type (excluding samples without variant regions) was 2.43% (Supplementary Material, Fig. S10A). Comparatively, variant regions only covered a median of 1.33% of the autosomes in individual mutant/affected analyses so therefore we would roughly expect only 0.0177% of coordinates to overlap by chance (2.43% × 2.43%). Surprisingly, variant regions across different cell types also showed high overlap (Supplementary Material, Fig. S10B). Using a Fisher’s exact test, the average P-value for variant region overlap across all mutant/affected samples (including analyses where mutant/affected sample pairs did not overlap at all) was 3.77 × 10−5 (Supplementary Material, Fig. S10C; 7.47 × 10−8 among mutant/affected sample pairs with non-zero overlap). Taken together, we conclude that replication timing variability tends to localize to particular regions of the genome.
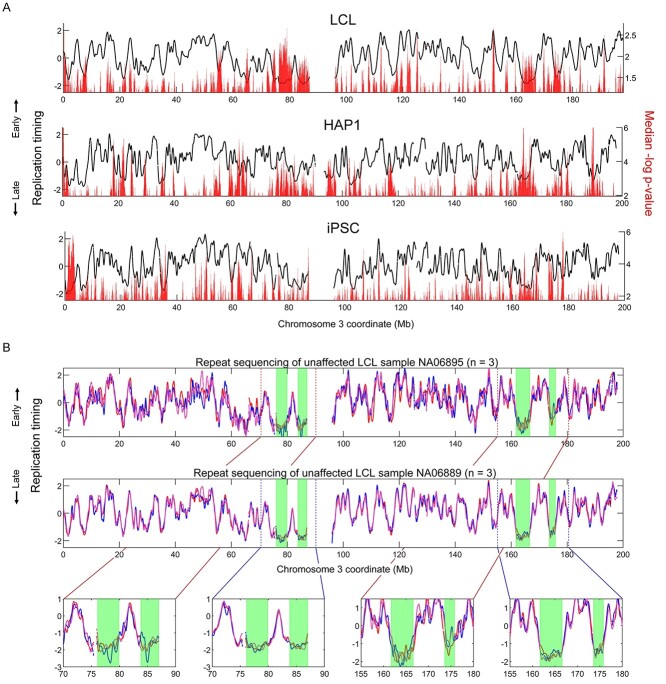
To analyze where replication timing variation tends to occur in the genome in each cell type, we used the randomized sample grouping simulations to calculate the median variation P-value in each sliding window across the genome. Replication timing variation was disproportionately more common in early- and late-replicating parts of the genome (Supplementary Material, Fig. S11A) in a similar bimodal distribution to where true mutant/affected variant regions fell (Supplementary Material, Fig. S9). This observed concentration of variating in early- or late-replicating regions occurs despite greater statistical power to detect variation in mid-replicating regions (Supplementary Material, Fig. S11B). Variant regions in simulations were not overwhelmingly biased toward either early- or late-replicating areas. Across all simulations, the average replication timing of variant regions was −0.036 (±standard deviation of 1.00) in LCL, −0.038 (±0.99) in HAP1 and −0.038 (±0.98) in iPSC. However, variation appeared to more statistically significant in the earliest replicating regions of the genome, suggesting that zones of DNA replication initiation tend to be more variable between samples. Indeed, greater replication timing variation was often observed at peaks, as well as valleys, in the replication timing profiles, which represent regions that contain sites of DNA replication initiation and termination, respectively (Fig. 4A). Notably, different peaks and valleys showed different intensities of variation. The genomically variable regions detected in the simulations were consistent with the variable regions in the analyses of mutant/affected samples (Supplementary Material, Fig. S12).
Figure 4.
Localized variability in replication timing. (A) The median P-value in each sliding window across 1000 simulations using all mutant/affected group sizes tested against the total number of WT/unaffected samples available for each cell type (137 for LCL, 6 for HAP1 and 24 for iPSCs). Only P-values below the median are shown. (B) Replication timing of the LCL unaffected samples NA06895 and NA06889 (each of which includes three repetitions). Late-replicating regions with notable variation are highlighted in green.
Another notable category of replication timing variants were large (>1 Mb), very late-replicating regions void of clearly defined peaks (e.g. Fig. 4A and B). These structures were most prominently present in LCLs. For example, chromosome 3 in LCLs contained four of these late-replicating regions that together covered 14 Mb (Fig. 4B). Within these regions, even replication timing of repeat samples from the same individual varied considerably. For example, in three repetitions each of the unaffected LCL samples NA06895 and NA06889, the correlation of replication timing within these late-replicating zones was on average 0.69 and 0.65, respectively, markedly lower than the correlations of 0.89 and 0.97 for the rest of the chromosome. Taken together, some genomic regions are relatively enriched for background variation and could be expected to show up in any sample comparison, whether mutant/affected compared with WT/unaffected or in control comparisons.
Most samples with high variability in replication timing are false-positives
By defining the range of background replication timing variability, 29 individual mutant/affected samples and nine mutated gene groups were identified as candidates for representing trans-acting modulators of replication timing (Fig. 3C). Of those, three candidate LCL mutated gene groups—ICF1, MRD1 and SCKL1—contained only one sample each. Despite having greater than expected variation compared with unaffected samples, MRD1 and SCKL1 had an overall high correlation to the mean LCL unaffected replication timing profile (r = 0.85 and 0.92, respectively). We suspect that these samples may show a high degree of variation due to low frequency subclonal deletions or duplications that might have escaped filtering and were ultimately amplified during data processing (specifically, normalization of the genome to a copy number of two before the ANOVA variant search; see Materials and Methods). Therefore, we do not consider individual samples with both high replication timing variability and high correlation to unaffected samples as strong candidates for having altered replication timing. In contrast, ICF1 showed both high variability and low correlation (r = 0.67) to the mean LCL unaffected replication timing profile. Since we only analyzed a single sample with this disease in LCLs, we cannot rule out other explanations for the high level of variation in this sample (e.g. a secondary somatic mutation, or technical factors). Furthermore, we analyzed two repeats of an HAP1 KO of the DNA methyltransferase DNMT3B gene, mutated in ICF1 syndrome, but did not find elevated replication timing variability. We thus conclude that ICF1/DNMT3B is not a strong candidate for altered replication timing.
Following a similar rationale, we eliminated additional individual mutant/affected samples with high replication timing variability yet high correlation to the mean WT/unaffected profile. In LCLs, 15 individual affected samples exhibited replication timing variation above expectation (Fig. 3C), of which seven demonstrated high correlation (r > 0.8) to the mean LCL unaffected profile (Supplementary Material, Fig. S13). These included sole individual outliers among WRN and XXY samples, effectively removing this gene mutation and aneuploidy state, respectively, as candidate regulators of replication timing. Among the eight LCL individual affected samples with low (r ≤ 0.8) correlation, six were from the cohort with the lowest-coverage sequencing. Given this overrepresentation of low-coverage samples, we regarded these samples as possible false positives; this further eliminated the RTS sample group as an a priori candidate for regulating replication timing, as well as the gene mutations in AT, HGPS, LNS and RTT—all of which contained outlier samples that belonged to either this low-coverage or to the high-correlation samples. Consistent with apparent replication timing variation (Fig. 2) being false-positives, we found variation of similar magnitude, and in some instances, the same locations, in simulated data (e.g. Supplementary Material, Fig. S14 for HGPS and WRN). Following the filtering of outliers due to low-coverage sequencing, two remaining individual affected outliers remained in AOA2 and in FXS. However, re-sequencing these individual samples did not reproduce the high replication timing variability, ruling out the corresponding gene mutations as likely causes of the initially observed variation. No gene mutations were eliminated in HAP1 cells nor iPSCs based on individual mutant/affected sample correlation to the mean WT/unaffected profile.
After the above elimination, four mutated gene groups remained. The mutated gene group of Fragile-X syndrome (FXS) in iPSCs (although not in LCLs), and four of its seven individual affected samples, had replication timing variability above the expected background threshold. Of the FXS iPSC individual affected samples, three showed low correlation to the unaffected mean profile (Supplementary Material, Fig. S15). However, the abnormal replication timing was not shared among the clones or re-sequenced samples available for two of the three FXS affected individuals. Therefore, based on the available samples we conclude that the FMR1 gene, mutated in FXS, is unlikely to be a trans-acting regulator of replication timing as the associated variation is not consistently observed among genetically identical samples.
There were nine affected samples from four individuals with Bloom syndrome (BLM) involving two types of mutations in the BLM (RECQL3) gene (Supplementary Material, Table S1). The BLM samples showed variant replication timing covering 0.14–13.6% of the genome and 27.6% when analyzed as a group (Fig. 2A). Of those, two individuals (NA04408 and NA09960) as well as their re-sequenced samples showed high correlation to the mean LCL unaffected replication timing profile and relatively invariant replication timing (Supplementary Material, Fig. S16). The remaining two individuals (NA03403 and GM16375) as well as their re-sequenced samples had significant replication timing differences from unaffected samples, typically encompassing novel or lost peaks (Supplementary Material, Fig. S16). These peak gains and losses as well as the overall replication profiles were not fully consistent among the three experimental repetitions of sample NA03403, suggesting possible technical or biological noise in this particular individual. Additionally, replication timing variation was not associated with the two types of BLM mutations observed in BLM samples (Supplementary Material, Table S1). While we hypothesize that true replication timing variation is present in these two BLM samples, we refrain from ascribing them to the BLM gene mutation directly given the lack of consistency across all BLM samples. Moreover, a BLM KO in HAP1 failed to show altered replication timing, further suggesting that the BLM gene is not directly involved in global replication timing regulation. It is possible that the replication timing variability observed in only half of the LCL BLM individuals may occur due to a secondary somatic mutation in another, potentially unknown regulator of DNA replication timing, especially considering that loss-of-function mutations in the BLM RecQ helicase results in increased somatic crossing-over and spontaneous mutation rate (41).
MCM10 is a novel candidate regulator of DNA replication timing
After a systematic analysis of 60 mutated genes or genetic diseases, only mutations in MCM10 and RIF1 demonstrated consistent variability in replication timing, low correlation to WT/unaffected replication timing, and clustering of replication timing in PCA all consistently related to the mutated gene (Fig. 2A and B, Supplementary Material, Fig. S17). RIF1, a known modulator of replication timing, showed variant replication timing covering 72.65% of the genome. The locations of variation were shared among the individual HAP1 RIF1 mutant samples, which also had highly correlated replication timing profiles at a similar level to the correlation of WT HAP1 samples (Supplementary Material, Fig. S17).
Samples with an MCM10 mutation demonstrated high deviation in replication timing from the corresponding unaffected iPSC profile, with variation covering 46.0% of the genome. We verified that the mutated MCM10 iPSCs were not spontaneously differentiated (and therefore demonstrating the replication timing of another cell type) by comparing them to various repli-seq profiles of differentiated cells (42) (Supplementary Material, Fig. S18). We also confirmed that the abnormal replication timing in mutated MCM10 samples was not the result of copy number alterations that escaped filtering (Supplementary Material, Fig. S19). To further rule out technical artifacts (e.g. incomplete reprogramming) as underlying the observed replication timing variation, we compared the affected MCM10 samples to another 300 unaffected iPSC samples. The 300 iPSC samples were all similar to each other as well as to the 24 control iPSCs directly used in this study, and none resembled the MCM10-affected samples (Supplementary Material, Fig. S20).
Given the established and unique role of RIF1 in DNA replication timing, it is possible that the MCM10 mutations operate in the same pathway or indirectly (e.g. by means of a secondary mutation) impinge on RIF1 function. To compare the mutated MCM10 profiles to RIF1 profiles in a similar cell type, we generated two RIF1 KO clones in haploid ESCs (43,44) using CRISRP/Cas9. The WT/unaffected ESC and iPSC replication profiles were similar (Fig. 5A), allowing for direct comparison of the RIF1 KO in ESCs to the mutated MCM10 samples in iPSCs. RIF1 ESC KOs were consistent among themselves (r = 0.97) yet significantly differed from WT controls (r = 0.56) (Fig. 5A), showing variation across 56.6% of the genome in grouped analysis (39.5 and 37.6% individually), similar to HAP1 RIF1 KOs. In contrast to previous reports (30), the replication profiles in ESC RIF1 KOs (as well as HAP1 RIF1 KO) did not appear to be ‘lost’ and resemble a flat profile, but rather showed a consistent, fluctuating profile that differed from WT at well-defined sites (Supplementary Material, Figs S17 and S21A). Importantly, MCM10 and RIF1 showed different alterations in DNA replication timing (Fig. 5A). When variant regions in these two gene mutations were merged, 94.2% of the genome demonstrated variant replication timing (Supplementary Material, Fig. S22). Thus, the mutated MCM10 iPSCs we analyzed appear to harbor a previously undescribed alteration of the DNA replication timing program.
Figure 5.
Mutations in MCM10 are associated with extensive replication timing variation. (A) Whole genome correlation of the replication timing profiles of several unaffected (denoted as WT) iPSCs, mutated MCM10 samples, ESC RIF1 KO mutants and normal (denoted as WT) ESC samples. (B) The distribution of the sizes of all MCM10 variant regions. (C) Clustering of mutated MCM10 and unaffected (denoted as WT) iPSC samples by peak presence. Sample MCM10-1 is an outlier, as it was in its genome-wide correlation values (Supplementary Material, Fig. S17), indicating lower data quality; samples MCM10-3 and MCM10-4 are repetitions of the MCM10–1 cell line. (D) Example of replication timing variants in MCM10. (E) Examples of peak alterations in mutated MCM10 samples. (F) The distribution of the change in replication timing at peaks within variant regions in mutated MCM10 samples relative to unaffected iPSCs. (G) The number of peak delays, advances, losses and gains in mutated MCM10 samples compared with unaffected iPSCs at MCM10-variant regions. Insets indicate the relative changes in replication timing at peak gain and loss sites.
These cell lines represent experimental repetitions and different iPSC clones derived from reprogramming of a sample from a single patient. This male individual was found to carry compound heterozygous mutations in MCM10, including a missense variant allele inherited from the father and a nonsense variant allele inherited from the mother (34). The patient presented at age 16 months with profoundly reduced natural killer cell numbers and succumbed to cytomegalovirus infection at age 24 months. These MCM10 mutations were previously shown to prevent its nuclear localization, causing de-stabilization of the replisome, reduced origin firing, genome instability and reduced cell proliferation (34,35). Given this established role of MCM10 in DNA replication dynamics, and the otherwise robustness of the replication timing program and scarcity of genetic alterations that modify it (above), we conclude that MCM10 is an attractive new candidate regulator of the replication timing program.
MCM10 replication profiles differed from the unaffected iPSC profiles across 1613 variant regions, spanning 46.0% of the genome (13.4–45.8% in individual samples; Supplementary Material, Figs S21B and S23). Variant regions spanned between 196 Kb (a single sliding window in ANOVA) to a maximum of 4.9 Mb, with a median of 532 Kb (mean 713 Kb; Fig. 5B). Interestingly, we noticed that much of the variation in mutated MCM10 samples localized to replication timing peaks—proxies for replication initiation sites (Fig. 5D, Supplementary Material, Fig S23). Indeed, MCM10 samples clustered separately from unaffected samples at peak locations (Fig. 5C), indicating that a fundamental difference in replication between MCM10 and unaffected cells resides at replication initiation sites.
To better understand how the MCM10 mutations influence replication initiation, we characterized four categories of peak change within MCM10 variant regions: peak advance or delay, and peak gain or loss relative to unaffected profiles (Fig. 5E). Of 627 peaks shared between MCM10 and unaffected, 285 showed replication timing delay while only 85 were advanced. This demonstrated substantial DNA replication initiation defects in mutated MCM10 cells (Fig. 5C, F and G). The median absolute change in replication timing in peak advances and delays was 0.65 units of standard deviation. Replication advances were more common in (but not exclusive to) later replicating parts of the genome, with the median peak advance having an unaffected (normal state) replication timing value of 0.42 units of standard deviation below the mean. Replication delays, on the other hand, were more common in very early-replicating parts of the genome with the median delayed peak having an unaffected replication value of 1.29 units of standard deviation above the mean. Furthermore, MCM10 demonstrated 311 peak gains and 223 peak losses relative to unaffected cells. At a majority of sites of either peak gain and loss, unaffected samples remained earlier replicating than MCM10 (Fig. 5G). Additionally, both peak gains and losses occurred more frequently at earlier replicating parts of the genome (Supplementary Material, Fig. S24). The median peak gain site had an unaffected replication timing value of 0.77 and an MCM10 replication value of 0.44 units of standard deviation above the mean. For the median peak loss, the replication timing values were 0.26 and 0.47, respectively.
We further characterized the chromatin states in the variant regions in mutated MCM10 samples. Using established 18-state chromHMM chromatin models from four reference iPSC cell lines (see Materials and Methods) (45,46), we assessed the proportion of each state in MCM10 variant regions and separately across the autosomal genome. Across all the four MCM10 samples and four iPSC chromatin reference lines, we found that variant regions were depleted of active transcriptional states with a mean fold-change of 0.68× for active transcription start sites and 0.63× for strong transcription (Supplementary Material, Fig. S25). Variant regions were also depleted of repressive states with a mean fold-change of 0.69× for bivalent transcription start sites and 0.71× for repressed Polycomb (Supplementary Material, Fig. S25). Variant regions in MCM10 samples were enriched for active enhancer, heterochromatin, and quiescent states with a mean fold-change of 1.17×, 1.12× and 1.05×, respectively (Supplementary Material, Fig. S25). We next compared MCM10 chromatin state enrichments to the two ESC e KO clones. To reflect the different cell type, chromatin states in e KO variant regions were calculated using four ESC reference lines (see Materials and Methods). As in MCM10 samples, RIF1 KO variant regions were depleted of repressive states with a mean fold-change of 0.71× for bivalent transcription start sites and 0.69× for repressed Polycomb compared with the full genome (Supplementary Material, Fig. S25). Contrariwise, RIF1 variant regions were enriched for active transcription states with a mean fold-change of 1.21× for active transcription start sites and 1.36× for strong transcription (Supplementary Material, Fig. S25). This is consistent with findings that RIF1 KO cells showed broad depletion of active transcription marks (30) and further supports the conclusion that Rif1 and MCM10 influence the replication timing of different genomic regions.
In conclusion, impairment of MCM10 appears to exert a global influence on genomic replication timing, in particular perturbing normal replication initiation in regions depleted of active transcription chromatin states. Replication timing variants included both replication delays at sites of shared initiation between unaffected and MCM10 cells and gains and losses of replication initiation sites, predominantly at early-replicating genomic regions.
Discussion
Identifying genetic alterations that lead to reprogramming of DNA replication timing can illuminate the molecular mechanisms of DNA replication control. However, almost no such factors have been identified to date despite intensive efforts. Here, we took, to our knowledge, the most comprehensive characterization of replication timing alterations in gene knockouts and genetic diseases. Apart from the previously described role of RIF1 in replication timing control, we propose a novel role for MCM10 and a possible albeit potentially indirect involvement of the BLM helicase in DNA replication timing. MCM10 is a conserved and essential component of the DNA replication initiation machinery (36) and we show here that disease-associated mutations in the MCM10 gene in a single patient lead to extensive perturbation of origin firing genome-wide.
Notably, we did not observe similar replication timing aberrations in cells mutated for other central components of the DNA replication initiation machinery, such as GINS4 and RECQL4. The replication timing phenotype of mutated MCM10 cells therefore appears to be highly specific, consistent with the diverse roles of MCM10 in stabilizing the CDC45:MCM2-7:GINS helicase as well as the proceeding replisome (35). Further research will be required to validate the mutated MCM10 phenotype, better understand the mechanisms by which MCM10 may control replication initiation timing, and link MCM10 mutations to genome stability and cellular and disease phenotypes. In particular, although we identify a consistent replication timing phenotype across different experimental repetitions and patient-derived iPSC clones of the mutated MCM10 cells, they are all derived from the same individual and reprogramming event. Identification of additional individuals with MCM10 mutations and complementary studies using engineered cell lines (35) will further establish the role of MCM10 in replication timing. However, naturally occurring mutations in MCM10, such as the compound heterozygote analyzed in this study, are extraordinarily rare.
Although we ultimately ruled out most tested genes including those with previously suggested roles in replication timing (with the exception of RIF1), the majority of tests initially resulted in the positive identification of changes in DNA replication timing. We show that this is expected given the genome-wide nature of variant search, which is especially pronounced in the case of replication timing data due to its chromosomal continuity. We thus emphasize the need for rigorous consideration of multiple testing in any genome-wide search for replication timing alterations in any biological system. In contrast, many previous studies used arbitrary thresholds for identifying variants and determining whether a gene mutation influences DNA replication timing. By applying an empirical false-discovery correction, we were able to rule out many candidate replication timing regulators, including some that were proposed by previous studies.
A main limitation of our study is that we focused on three specific cell types, among which LCLs and HAP1 cells are both transformed or derived from tumor cells, while iPSCs are pluripotent stem cells. It is conceivable that alterations in replication timing would only be observed in certain normal cell lineages. This is consistent with the specific symptoms and affected tissues in patients with replication gene mutations. Thus, it remains of interest to study replication timing alterations in different genetic backgrounds in a variety of differentiated cell types. Furthermore, the effects of human disease-associated variants may differ depending on the relative pathogenicity of a variant, different variants and genes that could be implicated in a given disease, and the heterogeneous nature of patient-derived samples. In addition, although our analysis controls for sample number and is powerful enough to detect subtle changes, the limited patient availability for many of the studied diseases (Fig. 2A, Supplementary Material, Table S1) leaves open the possibility that different or additional variation is present in other patients. Furthermore, heterozygosity, haploinsufficiency, individual or cell-type-specific penetrance may produce variable effects on replication timing in different patients or cell samples. For example, alternate mutations elsewhere in a gene may also affect replication timing while those observed in this study do not. Variation may also exist at smaller scales that are undetectable at the resolution of this study’s ANOVA analyses (185 kb) or replication timing data (2500 bp), although any such alterations would necessarily be limited in their extent due to the continues nature of replication timing. Thus, we cannot rule out any of the tested genes and diseases as having an impact on replication dynamics.
Another limitation of our study is the use of bulk cell samples for analysis. Newer single cell approaches (47) have the potential to reveal stochastic events that are specific to individual cells rather than being shared across a population of cells. For instance, a genetic mutation may affect the activity of different replication origins or replication forks in each cell, thus evading detection in bulk analysis yet still impacting tissue physiology in an affected human. Other assays, such as fiber-based techniques to measure replication initiation and progression (48), also hold the potential of revealing replication alterations for which bulk replication profiling is agnostic to; it should be noted, however, that similar false-positive identification of replication perturbation is possible with these assays, especially when they only sample a small fraction of the genome. It is also interesting to note that some of the studied gene mutations have an effect on cell cycle progression and/or cell proliferation (12,13,17,49,50) yet did not show any replication timing phenotypes in our assay. This has been noted before as ‘scaling’ of the replication timing program (51,52) and has an unclear molecular basis that may also require more detailed assays such as single cell profiling in order to fully understand at the molecular level.
Notwithstanding these caveats, we find it remarkable that the replication timing program is so robust to a wide array of genetic perturbation in trans. While cis-acting polymorphisms can affect local replication timing in different cell types and across many genomic loci (19,20), it appears that global changes in replication timing may not be compatible with cell or human health. It is intriguing to consider the possible reasons for this rigidness of the replication timing program. In particular, interactions of replication timing with gene regulation and with genome stability—and even the intersection of the two (53)—may define DNA replication timing as an essential cellular program.
Materials and Methods
Cell lines
LCL
We analyzed 271 LCL samples from 245 individuals. These samples covered 32 genetic diseases across 117 individuals (134 total samples) and 124 individuals (137 samples) of presumed healthy status (Table 1). One hundred thirty-eight of the LCL samples were obtained from the Coriell Institute for Medical Research (Camden, NJ) as either DNA samples or cell cultures. Five FA affected cell lines were obtained from the International Fanconi Anemia Tissue and Cell Repository housed at the Rockefeller University. LCLs were cultured in Roswell Park Memorial Institute 1640 medium (Corning Life Sciences, Tewksbury, MA, USA), supplemented with 15% fetal bovine serum (FBS; Corning). Culture was maintained at 37°C with 5% CO2 in a humidified atmosphere. Sample identification numbers and genotypes are available in Supplementary Material, Table S1.
Among the remaining samples, the repeat expansion cohort provided 55 affected LCL samples from six diseases and 48 presumed healthy samples (54). The Illumina platinum family provided 17 presumed healthy samples (55).
HAP1
HAP1 WT and KO cell lines were obtained from Horizon (https://horizondiscovery.com). Information regarding specific KO strains is available on the provider’s website. All CRISPR KOs were validated using Sanger sequencing. The cell lines were cultured following the provider’s recommendations, in IMDM medium supplemented with 10% FBS and 1% Penicillin/Streptomycin. Culture was maintained at 37°C with 5% CO2 in a humidified atmosphere. Cells were passaged every 2–3 days.
Cells were harvested at 70–80% confluence with 0.05% trypsin at 37°C for 5 min. Dissociation of cells was checked using microscopy. Cells were split into two samples containing approximately 2 × 106 cells each, one of which was used for FACS analysis and the other for DNA extraction. Cells collected for FACS analysis were pelleted at 1000 rpm at 4°C for 5 min, washed once in 500 mL of ice-cold PBS and resuspended in 250 μL of ice-cold PBS. Cells were fixed in 750 μL of ice-cold (−20°C) ethanol with constant gentle vortexing and stored at 4°C. Fixed cells were washed with 400 μL of ice-cold PBS and centrifuged at 1000 rpm at 4°C for 5 min. Cells were resuspended in 400 μL of PBS, 400 μL of Accutase and incubated at room temperature for 20 min. Cells were then pelleted and resuspended in 400 μL of PBS. RNase A (10 mg/mL) treatment was done at 37°C for 30 min. Propidium iodide at a concentration of 1 mg/mL was added and staining was done at room temperature in the dark for 30 min. Cells were passed through a polystyrene cell-strainer-capped tube before flow cytometry analysis. Analysis of PI-stained cells was done with a FACSAria Fusion Sorter. Cells of known ploidy (haploid/diploid) were used as controls for each run. All HAP1 KO samples were sequenced as either diploidized samples, or both as haploid and diploidized samples. In those cases where both haploid and diploid samples were analyzed from the same KO clone, no significant differences were observed in replication timing as a function of ploidy, and the two samples were considered experimental repetitions of the same KO clone.
iPSC
The iPSCs from MCM10-, CDC45- and the GINS4-deficient individuals were cultured in feeder-free condition with Stemflex (Gibco, A3349401). iPSC lines for RTT, RTTC, LNS and matched unaffected controls were obtained from the Coriell Institute (see Supplementary Material, Table S1). iPSC lines GM27622 and GM27629 were grown in mTeSR Plus Basal medium and iPSCs for FXS and matched controls (56,57) were grown in mTeSR1 medium (STEMCELL Technologies). Cell lines GM27437, GM26077, GM27730 and GM260105 were grown on a layer of Mouse Embryonic Fibroblast feeder cells (Gibco) coated with D-MEM/F-12 supplemented with 20% of KnockOut Serum replacement (Gibco) and 10 μg/mL of Basic Fibroblast Growth Factor (bFGF) (Gibco, PHG0264). Feeder-dependent cells were transferred and adapted to Matrigel conditions following the recommendations of STEMCELL Technologies. All cell lines were grown at 37°C, 5% CO2, and passaged by dissociating to single cells with Accutase (Sigma, A6964) and plating at a density of 1 × 106 cells/well in Matrigel-coated six well plates. For the first 24 h after passaging, cells were cultured with 10 μM ROCK inhibitor (Y-27632; STEMCELL Technologies).
ESC
The WT and RIF1 KO human stem cells were cultured in StemFlex media (Thermo Fisher A3349401) on Geltrex (Thermo Fisher A1413302). Upon reaching 70% confluency, cultures were passaged at a ratio of 1:10, or cryopreserved in a solution of freezing media containing 40% FBS (Gemini Bio-Products 900-108) and 10% DMSO (Sigma Aldrich D2650). Passaging was performed by TrypLE (Life Technologies 12605036) dissociation to small clusters of cells, and plated in media containing 10 μM Rock inhibitor Y-27632 (Selleckchem S1049) was added to media and removed within 24–48 h. Cells beyond passage 10 were no longer supplemented with Rock inhibitor. All embryo and ESC research was reviewed and approved by the Columbia University Embryonic Stem Cell Committee and the Institutional Review Board.
In preparation for CRISPR, guide RNAs were designed targeting relevant genes using software from cripsr.mit.edu or Benchling.com. Guides were chosen with the highest index score in a region closest to the DNA region of interest. Nucleofection was performed using Amaxa Cell Line Nucleofector Kit II, program A-23 with a Cas9-GFP plasmid (Addgene, 44 719) and guide RNA. Cells were plated, cultured for 2 additional days, stained with 10 μg/mL Hoechst 33342, and subsequently sorted via FACS for cells that had both haploid DNA content and GFP positivity. Single colonies were propagated, and duplicates were made for cryopreservation and for DNA isolated for PCR and Sanger sequencing.
Fluorescence-activated cell sorting (FACS) was performed using the FACS-Aria machine at the Columbia University Stem Cell Initiative flow cytometry core. Populations were gated first for cells, followed by gating for single cells. Cells were suspended in media containing 10% FBS in PBS (Life Technologies 14190-250) throughout the analysis. Live cells were kept on ice during transportation and analysis. Analysis was performed using FloJo software (BD Biosciences).
Generation of replication timing profiles
DNA was extracted using the MasterPure™ Complete DNA and RNA Purification Kit (Lucigen) following the manufacturer’s instructions. PCR-free whole genome sequencing was performed using paired-end reads (GeneWiz, South Plainfield, NJ, see Supplementary Material, Table S1). Sequencing reads were converted into non-mapped bam files and marked for Illumina adaptors and duplicate reads with Picard Tools (v1.138) (http://broadinstitute.github.io/picard/) commands ‘FastqToSam’, ‘MarkIlluminaAdapters’ and ‘MarkDuplicates’. Bam files were aligned to hg19 with BWA mem (58) (v0.7.17). GC-corrected read depth data for each sample were then generated via TIGER (39) using a read length of 36 bp for alignability filtering and a bin size of 2500 bp. All other parameters were TIGER defaults.
The raw post-GC-corrected data were then filtered for copy number alterations using permissive parameters such as to retain replication timing information and any potential disease-related replication timing alterations. To remove clonal or sub-clonal aneuploidies, individual autosomes were first removed if they had a copy number >2.2 or <1.8. This step removed whole chromosomes from diseased samples with known aneuploidies (e.g. TRI21) along with healthy samples with sub-clonal aneuploidies. Notably, chromosome 1q is removed in analyses of all MCM10 and HAP1 due to high copy number (all mutated MCM10 samples and 50% of HAP1 WT samples were affected). Next, regions of large (>1 Mb) duplications or deletion were manually removed by visually comparing the distribution of raw data across all samples. To filter outlier and smaller CNVs, windows 4 standard deviations above or below the mean copy number per chromosome were removed. Each sample was then filtered via the TIGER command ‘TIGER_segment_filt’ (using the MATLAB function ‘segment’, R2 = 0.04, standard deviation threshold = 1.5). These steps optimally corrected samples across all cell types and data qualities.
Replication timing values were generated by smoothing the filtered GC-corrected data with a cubic smoothing spline (MATLAB command ‘csaps’, smoothing parameter = 1 × 10−17, roughly the equivalent of smoothing across 250 kb windows). Only regions of >20 continuous 2500 bp windows were included and smoothing was not performed over data gaps >100 kbp or reference genome gaps >50 kb. The smoothed profiles were then normalized to an autosomal mean of zero and a standard deviation of one. Importantly, smoothed profiles were only used for visualization and correlation but not for detecting replication timing variant regions.
Detection of replication timing variant regions via ANOVA
Analysis of variance (ANOVA) was performed on autosomes to detect regions of replication timing variation among samples. We performed the variant analysis both for individual mutant/affected samples against all WT/unaffected samples of the same cell type and again with samples grouped by mutated gene. For the RIF1 KO of ESCs, unaffected iPSCs were used. To avoid regions with different numbers of analyzed samples due to sample-specific filtered regions or chromosomes, we substituted individual windows of WT/unaffected samples where less than five samples have missing data with the average filtered unsmoothed data of the WT/unaffected samples. All filtered unsmoothed data were then mean-shifted to an autosomal genome copy number of 2.
We performed one-way ANOVA in a sliding window of 76 × 2500 bp bins (185 kb window) with a quarter step of 19 bins (47.5 kb) on the filtered GC-corrected unsmoothed data (43). The corresponding P-value for each window was calculated with the MATLAB function ‘anova1’. ANOVA was not performed in windows with complete missing data for one or more mutant/affected samples to avoid a local sample number disparity. We called variant regions as windows with a P-value less than the Bonferroni-corrected threshold based on the number of ANOVA tests performed for each individual mutant/affected sample or mutated gene group. Adjacent significant windows were merged, and the P-value was recalculated over the merged region (in later analyses for FDR calculation, only the non-merged windows were used). The proportion of the genome with variant replication timing was then calculated from the total length of regions assigned as variant divided by the length of the genome analyzed.
Comparison of replication timing profiles, PC analysis and age analysis
All replication timing profile correlation was calculated as Pearson’s correlation coefficients. In comparing the TIGER and S/G1 replication timing profiles of sample NA12878, S/G1 coordinates were lifted from hg38 to hg19 with vcf-liftover (https://github.com/hmgu-itg/VCF-liftover) and interpolated to TIGER window coordinates with the MATLAB function ‘interp1’.
PC analysis was performed on the replication timing profiles of all autosomes with the MATLAB function ‘pca’. To determine if age influenced DNA replication timing, PC analysis was only performed on 83 WT/unaffected individuals with known age. In calculating the relationship between age and replication timing correlation, the median correlation of a sample to all other samples was used. A linear model was fit using the MATLAB function ‘fitlm’. The linear model excluded samples with a correlation ≤0.7 to all other LCL samples. However, when including all samples in the analysis, the linear correlation was still insignificant (Supplementary Material, Fig. S5B). Although including all samples produced a marginally higher correlation (r2 = 2.75 × 10−3 to r2 = 0.189), this was driven by a few samples from young individuals with abnormal replication profiles.
In comparing repli-seq profiles of ESC and differentiated cells to MCM10, coordinates were interpolated to TIGER window coordinates with the MATLAB function ‘interp1’.
P-value inflation analysis and ANOVA simulations
For each mutant/affected-WT/unaffected ANOVA test, the slope of the QQ-plot relationship of theoretical vs sample quantiles was extracted with the MATLAB function ‘fitlm’. To determine FDR for each analysis, q-values were calculated by the MATLAB function ‘mafdr’ using the Benjamini-Hochberg method (59). FDR for each ANOVA test was determined as the proportion of q-values less than the original Bonferroni-corrected P-value threshold compared with its original P-value (i.e. the proportion of windows identified as false positive) (Supplementary Material, Fig. S6).
ANOVA simulations used all samples with a >0.7 correlation to all other samples regardless of mutant/affected or WT/unaffected status. The simulations were performed for different combinations of sampled mutant/affected and WT/unaffected samples. In 1000 iterations for each combination, samples were randomly assigned into the WT/unaffected or mutant/affected groups and an ANOVA test was performed identically to the true mutant/affected-WT/unaffected tests. For each iteration, the proportion of the genome with variant replication timing and the P-values for each window were analyzed.
Overlap in regions of replication timing variability
Overlap percentage was calculated in pairs of all individual mutant/affected samples (184 × 184 tests) where the variant regions of one query sample were compared with the variant regions in one subject sample. For each analysis, the number of overlapping nucleotides in the variant regions of the query and subject sample were calculated with BEDtools intersect (60). The overlapping percentage was determined as the number of overlapping nucleotides divided by the total length of variant replication timing regions for the query sample. Therefore, the overlap percentage can differ for two samples depending on which is the query or subject sample. For the significance of overlap, a Fisher’s exact test was performed on pairs of individual mutant/affected variant regions using BEDtools fisher. The P-value from the two-tailed t-test was used.
For determining the proportion of the genome with variant replication timing covered by both MCM10 and RIF1 KO in ESCs, variant regions were merged with BEDtools merge. The sum of the merged variant regions was divided by the length of the genome available for analysis in MCM10 (which was shorter than RIF1 by 7.12 Mb).
Identifying and clustering replication timing peaks
Chromosome 1p was removed for all peak analyses as it was filtered out in MCM10. For identifying peaks in MCM10 and iPSC unaffected samples, the pairwise distances of local maxima in the individual affected samples were calculated with the MATLAB function ‘pdist’. Hierarchical clustering was then performed on the pairwise distance matrix using the MATLAB function ‘linkage’ using the average method and the default metric of Euclidean distance. Peak clusters and ranges were next calculated with the MATLAB function ‘cluster’ using a cutoff of 20 000 bp as the distance criterion for forming clusters. For determining peak overlap, only peaks present in at least 75% of MCM10 or unaffected samples were compared. A peak was considered shared if the range of an MCM10 peak and unaffected peak overlapped as calculated by BEDtools intersect.
In determining peak advances or delays, only peaks overlapping MCM10 variant regions were considered. The relative change in replication timing was determined as the change in mean replication timing of MCM10 samples to unaffected samples within the shared peak range. For peak gains or losses, only peaks present in at least 75% of MCM10 or unaffected samples were compared. Peak gains were defined as peaks present in MCM10 but not unaffected and peak losses were defined as peaks present in unaffected but not MCM10. Replication timing changes in peak gains and losses were calculated within the range of the peak cluster using either the mean unaffected value or mean MCM10 value, as applicable.
For clustering MCM10 and unaffected samples by peak use, all 6234 peaks present in any of the samples were included. The binary presence or absence of the 6234 peaks in MCM10 and unaffected samples was clustered with the MATLAB function ‘linkage’ using the average method and the metric of Hamming distance (the percentage of coordinates that differ).
Chromatin state analysis
Chromatin state enrichment in MCM10 and RIF1 variant regions were assessed using established 18-state ChromHMM chromatin tracks (45,46). For the four iPSC MCM10 samples, we used the reference tracks iPS-18 (E019), iPS-20b (E020), iPS DF 6.9 (E021) and iPS DF 19.11 (E022). For the two ESC RIF1 KO samples, we used the ESC reference tracks H9 (E008), HUES48 (E014), HUES6 (E015) and HUES64 (E016). For each reference track in the matched cell type, we first evaluated the proportion of each chromatin state (% of bases) across all autosomes (limiting the analysis to all coordinates that had a replication timing value) of MCM10 and RIF1 KO samples using BEDtools intersect. Next, we evaluated chromatin state proportions only within variant regions. We calculated enrichment as the fold-change in state proportions between the variant regions and autosomes.
Supplementary Material
Acknowledgements
MCM10 and GINS4 iPSCs were a gift from Dr Jordan Orange and generated through the support of NIH-NIAID R01AI120989. Fanconi Anemia cell lines were obtained from the International Fanconi Anemia Tissue and Cell Repository housed at Rockefeller University.
Conflict of Interest statement. The authors declare no conflicts of interest.
Contributor Information
Madison Caballero, Department of Molecular Biology and Genetics, Cornell University, Ithaca, NY 14853, USA.
Tiffany Ge, Department of Molecular Biology and Genetics, Cornell University, Ithaca, NY 14853, USA.
Ana Rita Rebelo, Department of Molecular Biology and Genetics, Cornell University, Ithaca, NY 14853, USA.
Seungmae Seo, Vagelos College of Physicians and Surgeons, Columbia University, New York, NY 10032, USA.
Sean Kim, Department of Molecular Biology and Genetics, Cornell University, Ithaca, NY 14853, USA.
Kayla Brooks, Department of Molecular Biology and Genetics, Cornell University, Ithaca, NY 14853, USA.
Michael Zuccaro, Department of Pediatrics and Naomi Berrie Diabetes Center, Columbia University, New York, NY 10032, USA; Columbia University Stem Cell Initiative, New York, NY 10032, USA.
Radhakrishnan Kanagaraj, The Francis Crick Institute, London NW1 1AT, UK.
Dan Vershkov, The Azrieli Center for Stem Cells and Genetic Research, Department of Genetics, Silberman Institute of Life Sciences, The Hebrew University, Jerusalem 91904, Israel.
Dongsung Kim, Department of Molecular Biology and Genetics, Cornell University, Ithaca, NY 14853, USA; Weill Institute for Cell and Molecular Biology, Cornell University, Ithaca, NY, USA.
Agata Smogorzewska, Laboratory of Genome Maintenance, The Rockefeller University, New York, NY, USA.
Marcus Smolka, Department of Molecular Biology and Genetics, Cornell University, Ithaca, NY 14853, USA; Weill Institute for Cell and Molecular Biology, Cornell University, Ithaca, NY, USA.
Nissim Benvenisty, The Azrieli Center for Stem Cells and Genetic Research, Department of Genetics, Silberman Institute of Life Sciences, The Hebrew University, Jerusalem 91904, Israel.
Stephen C West, The Francis Crick Institute, London NW1 1AT, UK.
Dieter Egli, Department of Pediatrics and Naomi Berrie Diabetes Center, Columbia University, New York, NY 10032, USA; Columbia University Stem Cell Initiative, New York, NY 10032, USA.
Emily M Mace, Vagelos College of Physicians and Surgeons, Columbia University, New York, NY 10032, USA.
Amnon Koren, Department of Molecular Biology and Genetics, Cornell University, Ithaca, NY 14853, USA.
Data availability
Raw sequence data are available on SRA under the bioproject PRJNA754107 (HAP1 samples and iPSC and LCL samples approved for non-restricted data access) and on dbGaP with accession numbers phs001957 (ESCs) and phs002597 (iPSCs and LCLs).
Funding
National Institutes of Health grants (1DP2GM123495 to A.K., R01GM123018 to M.B.S.); NYSTEM IDEA award (#C32564GG to D.E.); NIH-NIAID (R01AI137275 to E.M.M.).
References
- 1. Bellelli, R. and Boulton, S.J. (2021) Spotlight on the replisome: aetiology of DNA replication-associated genetic diseases. Trends Genet., 37, 317–336. [DOI] [PubMed] [Google Scholar]
- 2. O’Driscoll, M. (2017) The pathological consequences of impaired genome integrity in humans; disorders of the DNA replication machinery. J. Pathol., 241, 192–207. [DOI] [PubMed] [Google Scholar]
- 3. Schmit, M. and Bielinsky, A.-K. (2021) Congenital diseases of DNA replication: clinical phenotypes and molecular mechanisms. Int. J. Mol. Sci., 22, E911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. D’Antoni, S., Mattina, T., Di Mare, P., Federico, C., Motta, S. and Saccone, S. (2004) Altered replication timing of the HIRA/Tuple1 locus in the DiGeorge and Velocardiofacial syndromes. Gene, 333, 111–119. [DOI] [PubMed] [Google Scholar]
- 5. Yeshaya, J., Amir, I., Rimon, A., Freedman, J., Shohat, M. and Avivi, L. (2009) Microdeletion syndromes disclose replication timing alterations of genes unrelated to the missing DNA. Mol. Cytogenet., 2, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sangrithi, M.N., Bernal, J.A., Madine, M., Philpott, A., Lee, J., Dunphy, W.G. and Venkitaraman, A.R. (2005) Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell, 121, 887–898. [DOI] [PubMed] [Google Scholar]
- 7. Im, J.-S., Park, S.-Y., Cho, W.-H., Bae, S.-H., Hurwitz, J. and Lee, J.-K. (2015) Rec QL4 is required for the association of Mcm 10 and Ctf 4 with replication origins in human cells. Cell Cycle, 14, 1001–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thangavel, S., Mendoza-Maldonado, R., Tissino, E., Sidorova, J.M., Yin, J., Wang, W., Monnat, R.J., Falaschi, A. and Vindigni, A. (2010) Human RECQ1 and RECQ4 helicases play distinct roles in DNA replication initiation. Mol. Cell. Biol., 30, 1382–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rivera-Mulia, J.C., Desprat, R., Trevilla-Garcia, C., Cornacchia, D., Schwerer, H., Sasaki, T., Sima, J., Fells, T., Studer, L., Lemaitre, J.-M.et al. (2017) DNA replication timing alterations identify common markers between distinct progeroid diseases. Proc. Natl. Acad. Sci. U. S. A., 114, E10972–E10980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fenwick, A.L., Kliszczak, M., Cooper, F., Murray, J., Sanchez-Pulido, L., Twigg, S.R.F., Goriely, A., McGowan, S.J., Miller, K.A., Taylor, I.B.et al. (2016) Mutations in CDC45, encoding an essential component of the pre-initiation complex, cause Meier-Gorlin syndrome and craniosynostosis. Am. J. Hum. Genet., 99, 125–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bicknell, L.S., Bongers, E.M.H.F., Leitch, A., Brown, S., Schoots, J., Harley, M.E., Aftimos, S., Al-Aama, J.Y., Bober, M., Brown, P.A.J.et al. (2011) Mutations in the pre-replication complex cause Meier-Gorlin syndrome. Nat. Genet., 43, 356–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bicknell, L.S., Walker, S., Klingseisen, A., Stiff, T., Leitch, A., Kerzendorfer, C., Martin, C.-A., Yeyati, P., Al Sanna, N., Bober, M.et al. (2011) Mutations in ORC1, encoding the largest subunit of the origin recognition complex, cause microcephalic primordial dwarfism resembling Meier-Gorlin syndrome. Nat. Genet., 43, 350–355. [DOI] [PubMed] [Google Scholar]
- 13. Guernsey, D.L., Matsuoka, M., Jiang, H., Evans, S., Macgillivray, C., Nightingale, M., Perry, S., Ferguson, M., LeBlanc, M., Paquette, J.et al. (2011) Mutations in origin recognition complex gene ORC4 cause Meier-Gorlin syndrome. Nat. Genet., 43, 360–364. [DOI] [PubMed] [Google Scholar]
- 14. Burrage, L.C., Charng, W.-L., Eldomery, M.K., Willer, J.R., Davis, E.E., Lugtenberg, D., Zhu, W., Leduc, M.S., Akdemir, Z.C., Azamian, M.et al. (2015) De Novo GMNN mutations cause autosomal-dominant primordial dwarfism associated with Meier-Gorlin syndrome. Am. J. Hum. Genet., 97, 904–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Madireddy, A., Kosiyatrakul, S.T., Gerhardt, J., Boisvert, R.A., Vuono, E.A., Moyano, E.H., Garcia Rubio, M.L., Owen, N., Yan, Z., Olson, S.et al. (2016) FANCD2 facilitates replication through common fragile sites. Mol. Cell, 64, 388–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hansen, R.S., Stöger, R., Wijmenga, C., Stanek, A.M., Canfield, T.K., Luo, P., Matarazzo, M.R., D’Esposito, M., Feil, R., Gimelli, G.et al. (2000) Escape from gene silencing in ICF syndrome: evidence for advanced replication time as a major determinant. Hum. Mol. Genet., 9, 2575–2587. [DOI] [PubMed] [Google Scholar]
- 17. Lana, E., Mégarbané, A., Tourrière, H., Sarda, P., Lefranc, G., Claustres, M. and De Sario, A. (2012) DNA replication is altered in immunodeficiency centromeric instability facial anomalies (ICF) cells carrying DNMT3B mutations. Eur. J. Hum. Genet., 20, 1044–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hand, R. and German, J. (1977) Bloom’s syndrome: DNA replication in cultured fibroblasts and lymphocytes. Hum. Genet., 38, 297–306. [DOI] [PubMed] [Google Scholar]
- 19. Koren, A., Handsaker, R.E., Kamitaki, N., Karlić, R., Ghosh, S., Polak, P., Eggan, K. and McCarroll, S.A. (2014) Genetic variation in human DNA replication timing. Cell, 159, 1015–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ding, Q., Edwards, M.M., Wang, N., Zhu, X., Bracci, A.N., Hulke, M.L., Hu, Y., Tong, Y., Hsiao, J., Charvet, C.J.et al. (2021) The genetic architecture of DNA replication timing in human pluripotent stem cells. Nat. Commun., 12, 6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Peace, J.M., Ter-Zakarian, A. and Aparicio, O.M. (2014) Rif 1 regulates initiation timing of late replication origins throughout the S. cerevisiae genome. PLoS One, 9, e98501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mattarocci, S., Shyian, M., Lemmens, L., Damay, P., Altintas, D.M., Shi, T., Bartholomew, C.R., Thomä, N.H., Hardy, C.F.J. and Shore, D. (2014) Rif 1 controls DNA replication timing in yeast through the PP 1 phosphatase Glc 7. Cell Rep., 7, 62–69. [DOI] [PubMed] [Google Scholar]
- 23. Kanoh, Y., Matsumoto, S., Fukatsu, R., Kakusho, N., Kono, N., Renard-Guillet, C., Masuda, K., Iida, K., Nagasawa, K., Shirahige, K.et al. (2015) Rif 1 binds to G quadruplexes and suppresses replication over long distances. Nat. Struct. Mol. Biol., 22, 889–897. [DOI] [PubMed] [Google Scholar]
- 24. Hayano, M., Kanoh, Y., Matsumoto, S., Renard-Guillet, C., Shirahige, K. and Masai, H. (2012) Rif 1 is a global regulator of timing of replication origin firing in fission yeast. Genes Dev., 26, 137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hiraga, S.-I., Alvino, G.M., Chang, F., Lian, H.-Y., Sridhar, A., Kubota, T., Brewer, B.J., Weinreich, M., Raghuraman, M.K. and Donaldson, A.D. (2014) Rif 1 controls DNA replication by directing Protein Phosphatase 1 to reverse Cdc 7-mediated phosphorylation of the MCM complex. Genes Dev., 28, 372–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Munden, A., Rong, Z., Sun, A., Gangula, R., Mallal, S. and Nordman, J.T. (2018) Rif 1 inhibits replication fork progression and controls DNA copy number in Drosophila. Elife, 7, e39140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yamazaki, S., Ishii, A., Kanoh, Y., Oda, M., Nishito, Y. and Masai, H. (2012) Rif 1 regulates the replication timing domains on the human genome. EMBO J., 31, 3667–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Foti, R., Gnan, S., Cornacchia, D., Dileep, V., Bulut-Karslioglu, A., Diehl, S., Buness, A., Klein, F.A., Huber, W., Johnstone, E.et al. (2016) Nuclear architecture organized by Rif 1 underpins the replication-timing program. Mol. Cell, 61, 260–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cornacchia, D., Dileep, V., Quivy, J.-P., Foti, R., Tili, F., Santarella-Mellwig, R., Antony, C., Almouzni, G., Gilbert, D.M. and Buonomo, S.B.C. (2012) Mouse Rif 1 is a key regulator of the replication-timing programme in mammalian cells. EMBO J., 31, 3678–3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Klein, K.N., Zhao, P.A., Lyu, X., Sasaki, T., Bartlett, D.A., Singh, A.M., Tasan, I., Zhang, M., Watts, L.P., Hiraga, S.-I.et al. (2021) Replication timing maintains the global epigenetic state in human cells. Science, 372, 371–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Baldacci, G., Hoffmann, J.-S. and Cadoret, J.-C. (2014) Impact of the DNA polymerase Theta on the DNA replication program. Genom. Data, 3, 90–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fernandez-Vidal, A., Guitton-Sert, L., Cadoret, J.-C., Drac, M., Schwob, E., Baldacci, G., Cazaux, C. and Hoffmann, J.-S. (2014) A role for DNA polymerase θ in the timing of DNA replication. Nat. Commun., 5, 4285. [DOI] [PubMed] [Google Scholar]
- 33. Palmigiano, A., Santaniello, F., Cerutti, A., Penkov, D., Purushothaman, D., Makhija, E., Luzi, L., diFagagna, F., Pelicci, P.G., Shivashankar, V.et al. (2018) PREP1 tumor suppressor protects the late-replicating DNA by controlling its replication timing and symmetry. Sci. Rep., 8, 3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mace, E.M., Paust, S., Conte, M.I., Baxley, R.M., Schmit, M.M., Patil, S.L., Guilz, N.C., Mukherjee, M., Pezzi, A.E., Chmielowiec, J.et al. (2020) Human NK cell deficiency as a result of biallelic mutations in MCM10. J. Clin. Invest., 130, 5272–5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Baxley, R.M., Leung, W., Schmit, M.M., Matson, J.P., Yin, L., Oram, M.K., Wang, L., Taylor, J., Hedberg, J., Rogers, C.B.et al. (2021) Bi-allelic MCM10 variants associated with immune dysfunction and cardiomyopathy cause telomere shortening. Nat. Commun., 12, 1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Baxley, R.M. and Bielinsky, A.K. (2017) Mcm 10: a dynamic scaffold at eukaryotic replication forks. Genes, 8, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Essletzbichler, P., Konopka, T., Santoro, F., Chen, D., Gapp, B.V., Kralovics, R., Brummelkamp, T.R., Nijman, S.M.B. and Bürckstümmer, T. (2014) Megabase-scale deletion using CRISPR/Cas 9 to generate a fully haploid human cell line. Genome Res., 24, 2059–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kotecki, M., Reddy, P.S. and Cochran, B.H. (1999) Isolation and characterization of a near-haploid human cell line. Exp. Cell Res., 252, 273–280. [DOI] [PubMed] [Google Scholar]
- 39. Koren, A., Massey, D.J. and Bracci, A.N. (2021) TIGER: inferring DNA replication timing from whole-genome sequence data. Bioinformatics, 37, 4001–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Massey, D.J., Kim, D., Brooks, K.E., Smolka, M.B. and Koren, A. (2019) Next-generation sequencing enables spatiotemporal resolution of human centromere replication timing. Genes, 10, E269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cunniff, C., Bassetti, J.A. and Ellis, N.A. (2017) Bloom’s syndrome: clinical spectrum, molecular pathogenesis, and cancer predisposition. Mol. Syndromol., 8, 4–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rivera-Mulia, J.C., Buckley, Q., Sasaki, T., Zimmerman, J., Didier, R.A., Nazor, K., Loring, J.F., Lian, Z., Weissman, S., Robins, A.J.et al. (2015) Dynamic changes in replication timing and gene expression during lineage specification of human pluripotent stem cells. Genome Res., 25, 1091–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Edwards, M.M., Zuccaro, M.V., Sagi, I., Ding, Q., Vershkov, D., Benvenisty, N., Egli, D. and Koren, A. (2021) Delayed DNA replication in haploid human embryonic stem cells. Genome Res., 31, 2155–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sagi, I., Chia, G., Golan-Lev, T., Peretz, M., Weissbein, U., Sui, L., Sauer, M.V., Yanuka, O., Egli, D. and Benvenisty, N. (2016) Derivation and differentiation of haploid human embryonic stem cells. Nature, 532, 107–111. [DOI] [PubMed] [Google Scholar]
- 45. Kundaje, A., Meuleman, W., Ernst, J., Bilenky, M., Yen, A., Heravi-Moussavi, A., Kheradpour, P., Zhang, Z., Wang, J., Ziller, M.J.et al. (2015) Integrative analysis of 111 reference human epigenomes. Nature, 518, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ernst, J. and Kellis, M. (2012) Chrom HMM: automating chromatin-state discovery and characterization. Nat. Methods, 9, 215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Massey, D.J. and Koren, A. (2022) High-throughput analysis of single human cells reveals the complex nature of DNA replication timing control. bio Rxiv, 2021.05.14.443897. 10.1101/2021.05.14.443897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hulke, M.L., Massey, D.J. and Koren, A. (2020) Genomic methods for measuring DNA replication dynamics. Chromosom. Res., 28, 49–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Faragher, R.G., Kill, I.R., Hunter, J.A., Pope, F.M., Tannock, C. and Shall, S. (1993) The gene responsible for Werner syndrome may be a cell division “counting” gene. Proc. Natl. Acad. Sci. U. S. A., 90, 12030–12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Outwin, E., Carpenter, G., Bi, W., Withers, M.A., Lupski, J.R. and O’Driscoll, M. (2011) Increased RPA1 gene dosage affects genomic stability potentially contributing to 17p13.3 duplication syndrome. PLoS Genet., 7, e1002247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Koren, A., Soifer, I. and Barkai, N. (2010) MRC1-dependent scaling of the budding yeast DNA replication timing program. Genome Res., 20, 781–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Alvino, G.M., Collingwood, D., Murphy, J.M., Delrow, J., Brewer, B.J. and Raghuraman, M.K. (2007) Replication in hydroxyurea: it’s a matter of time. Mol. Cell. Biol., 27, 6396–6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Koren, A. (2014) DNA replication timing: coordinating genome stability with genome regulation on the X chromosome and beyond. BioEssays, 36, 997–1004. [DOI] [PubMed] [Google Scholar]
- 54. Dolzhenko, E., vanVugt, J.J.F.A., Shaw, R.J., Bekritsky, M.A., vanBlitterswijk, M., Narzisi, G., Ajay, S.S., Rajan, V., Lajoie, B.R., Johnson, N.H.et al. (2017) Detection of long repeat expansions from PCR-free whole-genome sequence data. Genome Res., 27, 1895–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Eberle, M.A., Fritzilas, E., Krusche, P., Källberg, M., Moore, B.L., Bekritsky, M.A., Iqbal, Z., Chuang, H.-Y., Humphray, S.J., Halpern, A.L.et al. (2017) A reference data set of 5.4 million phased human variants validated by genetic inheritance from sequencing a three-generation 17-member pedigree. Genome Res., 27, 157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Johannesson, B., Sagi, I., Gore, A., Paull, D., Yamada, M., Golan-Lev, T., Li, Z., LeDuc, C., Shen, Y., Stern, S.et al. (2014) Comparable frequencies of coding mutations and loss of imprinting in human pluripotent cells derived by nuclear transfer and defined factors. Cell Stem Cell, 15, 634–642. [DOI] [PubMed] [Google Scholar]
- 57. Urbach, A., Bar-Nur, O., Daley, G.Q. and Benvenisty, N. (2010) Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell Stem Cell, 6, 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li, H. (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. ar Xiv, 1303.3997.
- 59. Benjamini, Y. and Hochberg, Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol), 57, 289–300. [Google Scholar]
- 60. Quinlan, A.R. and Hall, I.M. (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics, 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence data are available on SRA under the bioproject PRJNA754107 (HAP1 samples and iPSC and LCL samples approved for non-restricted data access) and on dbGaP with accession numbers phs001957 (ESCs) and phs002597 (iPSCs and LCLs).