Abstract
Extracellular secretion of proteins via the type II or general secretion pathway in gram-negative bacteria requires the assistance of at least 12 gene products that are thought to form a complex apparatus through which secreted proteins are translocated. Although this apparatus is specifically required only for the outer membrane translocation step during transport across the bacterial cell envelope, it is believed to span both membranes. The EpsE, EpsL, and EpsM proteins of the type II apparatus in Vibrio cholerae are thought to form a trimolecular complex that is required to either control the opening and closing of the secretion pore or to transduce energy to the site of outer membrane translocation. EpsL is likely to play an important role in this relay by interacting with both the cytoplasmic EpsE protein and the cytoplasmic membrane protein EpsM, which is predominantly exposed on the periplasmic side of the membrane. We have now extended this model and mapped the separate regions within EpsL that contain the EpsE and EpsM binding domains. By taking advantage of the species specificity of the type II pathway, we have used chimeric proteins composed of EpsL and its homologue, ExeL, from Aeromonas hydrophila together with either EpsE or its Aeromonas homologue, ExeE, to complement the secretion defect in both epsL and exeL mutant strains. These studies have mapped the species-specific EpsE binding site to the N-terminal cytoplasmic region between residues 57 and 216 of EpsL. In addition, the species-specific EpsM binding site was mapped to the C-terminal half of EpsL by coimmunoprecipitation of EpsM with different EpsL-ExeL chimeras. This site is present in the region between amino acids 216 and 296, which contains the predicted membrane-spanning segment of EpsL.
Extracellular secretion in gram-negative bacteria requires complex systems that transport the secreted proteins across the cell envelope. Although a number of pathways have evolved for this purpose, the general secretion pathway (GSP) appears to be the most widely distributed and has been discovered in both animal and plant pathogens, including Vibrio cholerae, Aeromonas hydrophila, Pseudomonas aeruginosa, and Erwinia chrysanthemi (10, 13, 14, 36, 42). This pathway is required only for the outer membrane translocation step, since translocation across the cytoplasmic membrane appears to be assisted by the Sec system, and mutations in GSP genes result in the accumulation of folded proteins in the periplasm (for a review, see reference 29). The secretion apparatus must be very specific, since a limited number of proteins are secreted and resident periplasmic proteins, that are smaller than some of the secreted proteins, do not escape into the extracellular environment. In addition, secretion also appears to be host specific in that most species cannot secrete proteins from a closely related species and cross-complementation of mutations by genes from another species is very limited (6, 10, 17, 21, 34). Protein-protein interactions are therefore likely to play a major role during the secretion process. There must be very precise and highly stoichiometric interactions not only between the different components of the apparatus but also between these components and the proteins to be secreted. The secreted proteins must interact with one or more components of this apparatus during their transport, and these interactions are likely responsible for the molecular recognition of a specific, as yet unidentified, secretion signal on the folded proteins destined for export (7, 33).
Twelve to 16 GSP proteins are required for outer membrane translocation (30). The majority of these components are not synthesized with classical N-terminal signal sequences and appear to be inner membrane proteins, despite the fact that they are specifically involved in transport of secreted proteins from the periplasm to the extracellular environment (3, 32, 35, 41). The GspG-K proteins resemble prepilin subunits of the type IV class and are processed by prepilin peptidase, which is itself encoded by the gsp operon in a number of bacteria (24–26). The GspD protein does contain an N-terminal signal sequence and has been shown to be an outer membrane protein (9). Studies have shown that the GspD homolog of P. aeruginosa, XcpQ, the pIV protein of fI phage, the YscC protein of the Yersinia enterocolitica type III pathway, and the PilQ protein of P. aeruginosa all form large, multimeric complexes which are thought to form the secretion port through which the substrates of these various pathways pass (2, 16, 18). These conclusions were strengthened by the recent results from the laboratories of Pugsley and Russel showing that PulD and the phage pIV protein form ion-conducting channels in planar lipid bilayers (19, 23).
In V. cholerae, extracellular secretion is assisted by the eps genes and the prepilin peptidase gene vcpD (20, 27, 36, 37, 38). The eps genes encode components of the Eps secretion apparatus that spans both membranes. The vcpD gene product is thought to be required for the formation of this apparatus, since it has been predicted to process the prepilin-like protein EpsG (36) and shown to process the prepilin-like protein EpsI (20). Mutations in the vcpD and eps genes result in defects in outer membrane translocation of several proteins, including the principal virulence factor cholera toxin and protease. In addition, the biogenesis of the outer membrane protein OmpU is affected. Inactivation of the vcpD gene also results in decreased ability to colonize infant mice (20).
Initial characterization of the Eps components revealed that EpsE is a cytoplasmic protein containing an ATP-binding motif that is required for its activity (34). Subcellular fractionation and immunoblot analysis have shown that EpsE interacts with the integral cytoplasmic membrane protein EpsL, which results in stabilization and membrane association of EpsE (34). Additional support for interaction between proteins E and L came from studies of these proteins in P. aeruginosa and E. crysanthemi, were it was shown that overproduction of the L protein inhibits secretion and concomitant overproduction of the E protein relieves this inhibition (1, 31). Coimmunoprecipitation experiments have shown that EpsL, in turn, interacts with another cytoplasmic membrane protein, EpsM (35). This latter interaction appears to stabilize EpsL and increase the amount of EpsE in the membrane, suggesting that EpsM may induce a conformational change within EpsL that results in increased affinity for EpsE. Purified EpsE demonstrates autophosphorylation activity, suggesting that it is a protein kinase that may regulate the extracellular secretion process (34). The interaction of the EpsE-EpsL-EpsM trimolecular complex with the rest of the secretion apparatus may result in transmembrane signaling that opens the secretory channel or pore, permitting extracellular proteins to cross the outer membrane. If, on the other hand, EpsE is an ATPase and the demonstrated in vitro autophosphorylation activity of EpsE is only an intermediate step in the ATPase activity, EpsL and EpsM could possibly transduce energy from EpsE and the cytoplasmic membrane to the site of outer membrane translocation (35).
Based on hydrophobicity plots and the positive-inside rule, it is likely that the transmembrane domain of EpsL spans residues 254 to 271 with the N terminus of EpsL facing the cytoplasm and the C terminus exposed to the periplasmic space (43). This is in good agreement with the topologies of the two EpsL homologues OutL and XcpY, whose transmembrane domains have been mapped by fusion protein analysis to the corresponding hydrophobic regions in these two proteins (3, 12). It is likely that the cytoplasmic N terminus of EpsL interacts with EpsE, since EpsL is responsible for membrane association of EpsE, which is otherwise a soluble cytoplasmic protein (34). Preliminary results suggest that protein E binds within the first 248 residues of protein L, since an N-terminal OutL fragment spanning residues 1 to 248 inhibits secretion when overexpressed in wild-type (wt) E. crysanthemi and this inhibition can be suppressed by the concomitant overexpression of OutE (31). Binding to EpsM most likely occurs via the transmembrane and/or periplasmic domain of EpsL, since hydrophobicity plots of EpsM suggest that this protein is predominantly exposed on the periplasmic face of the cytoplasmic membrane and the transmembrane domain spans residues 26 to 42 (unpublished data). To test these hypotheses, regions were exchanged between EpsL and the homologous ExeL protein from A. hydrophila and EpsL-ExeL chimeras with altered specificity were constructed. These chimeras were used to identify and map both the EpsE and EpsM binding domains to specific regions of EpsL by complementation analysis in V. cholerae and A. hydrophila and by coimmunoprecipitation experiments with anti-EpsL and anti-EpsM antibodies.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| E. coli strains | ||
| MC1061 | F−lac | 4 |
| BL21(DE3) | F−ompT (rB− mB−) prophage λ carrying T7 RNA polymerase gene | 39 |
| V. cholerae strains | ||
| TRH7000 | El Tor; Δ(ctxAB), wild type with respect to extracellular protein secretion | 11 |
| Mut8 | TRH7000 with Kmr insertion in epsL gene | 34 |
| A. hydrophila strains | ||
| Ah65 | Wild type | 4 |
| 89A | Kmr insertion in exeL gene | This study |
| Plasmids | ||
| pMMB207, pMMB208 | Broad-host-range cloning vectors; Cmr | 8, 22 |
| pMMB532 | epsM in pT7-5 | This study |
| pMS49 | epsL in pMMB208 | This study |
| pMS55 | exeL154epsL in pMMB208 | This study |
| pMS56 | epsL154exeL in pMMB208 | This study |
| pMS57 | exeL216epsL in pMMB208 | This study |
| pMS59 | exeL57epsL in pMMB208 | This study |
| pMS60 | epsL296exeL in pMMB208 | This study |
| pMS73 | epsL57exeL in pMMB208 | This study |
| pMS74 | exeL296epsL in pMMB208 | This study |
| pMS77 | epsE epsL in pMMB208 | This study |
| pMS79 | epsE exeL296epsL in pMMB208 | This study |
| pMS92 | epsL216exeL in pMMB208 | This study |
| pMS96 | exeE exeL154epsL in pMMB208 | This study |
| pMS97 | exeE exeL216epsL in pMMB208 | This study |
| pMS98 | epsE epsL154exeL in pMMB208 | This study |
| pMS99 | epsE epsL216exeL in pMMB208 | This study |
| pMS100 | epsE epsL296exeL in pMMB208 | This study |
| pMS101 | exeE exeL296epsL in pMMB208 | This study |
| pRJ84.1 | exeL in pMMB208 | This study |
| pT7-5 | T7 RNA polymerase promoter φ10, Apr | 40 |
| pWD600 | etxA etxB (E. coli heat-labile enterotoxin A and B subunit genes), Tcr | 5 |
Construction of fusion proteins.
PCR fragments containing different portions of epsL or exeL were introduced into the appropriate sites of exeL in pRJ84.1 or epsL in pMS49, respectively, in order to make in-frame translational fusions between EpsL and ExeL. The resulting plasmids were conjugated from appropriate E. coli strains into wt V. cholerae TRH7000, V. cholerae epsL mutant Mut 8, wt A. hydrophila Ah65, and A. hydrophila exeL mutant 89A and assayed for complementation or inhibition of secretion as previously described (34).
Immunoprecipitation.
Triton X-100-soluble extracts of E. coli strain BL21(DE3) expressing epsM from pMMB532 and wt epsL or different chimeras under noninducing conditions were subjected to immunoprecipitation with anti-EpsL or anti-EpsM antibodies as previously described (35). The precipitated material was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting using biotinylated anti-EpsL immunoglobulin G (IgG) and horseradish peroxidase-conjugated streptavidin. Peroxidase activity was visualized with the Supersignal chemiluminescent substrate (Pierce).
Toxin assays.
The amount of toxin in culture supernatants and cell lysates of V. cholerae cells was determined by GM1 enzyme-linked immunosorbent assay as described previously (34). Aerolysin activity was assayed in cell lysates and culture supernatants of A. hydrophila cells as previously described (15, 34).
RESULTS
We have previously demonstrated that EpsL is present in the cytoplasmic membrane of V. cholerae and that it is involved in the membrane association and stabilization of EpsE (34, 35). In addition, EpsL was shown to interact with another cytoplasmic membrane protein, EpsM, and this interaction appears to stabilize both EpsL and EpsM and protect them from proteolytic degradation (34, 35). In order to identify the EpsE and EpsM binding domains within EpsL, a protein hybrid approach was used in which regions of EpsL and ExeL from A. hydrophila were exchanged. The chimeras were then analyzed for the ability to interact with the EpsE and EpsM proteins.
Species-specific complementation of secretion-defective mutants.
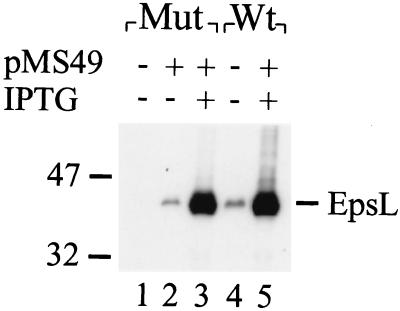
Prior to the construction of different EpsL-ExeL hybrids, the abilities of EpsL and ExeL to functionally replace each other were tested. epsL and exeL mutants of V. cholerae and A. hydrophila, respectively, were analyzed for extracellular secretion of their respective toxins in the absence and presence of plasmid-encoded epsL and exeL (Table 2). The amounts of cholera toxin and aerolysin present in the growth medium and periplasm were determined, and secretion was calculated as a percentage of the total toxin produced. Only EpsL could restore secretion in the epsL mutant, and only ExeL could complement the secretion defect in the exeL mutant. These results indicated that the function of protein L is species specific. In these experiments, complementation of the secretion defect was obtained in both species by the basal low-level expression of the L genes from the plasmid without the addition of isopropyl-β-d-thiogalactopyranoside (IPTG). However, when the L proteins were overproduced by the addition of IPTG, the ability of the L proteins to complement the mutants was prevented (data not shown). In addition, overexpression of plasmid-encoded epsL (using IPTG at 0.02 mM or a higher concentration) in the wt V. cholerae strain also resulted in inhibition of secretion, suggesting that the function of the secretion apparatus is sensitive to unbalanced levels of its components (Table 3). For comparison, the EpsL level in the epsL mutant expressing plasmid-encoded epsL in the absence or presence of IPTG and wt V. cholerae expressing endogenous and plasmid-encoded epsL after IPTG addition is shown in Fig. 1. Just as in P. aeruginosa and E. chrysanthemi (1, 31), overexpression of epsE alone rescued the secretion defect (data not shown). Since the GspE proteins alone can relieve the inhibitory effect of overproduced GspL proteins, these data suggest that the dominant-negative effect of GspL overproduction results primarily from the titration of GspE.
TABLE 2.
Species-specific complementation by EpsL and ExeL
| Plasmid-encoded protein | Secretion of toxin (% of total)a
|
|
|---|---|---|
| V. cholerae epsL mutant | A. hydrophila exeL mutant | |
| None | 7 | 2 |
| EpsL | 72 | 2 |
| ExeL | 11 | 88 |
Secretion of enterotoxin in V. cholerae and aerolysin in A. hydrophila was monitored in the absence or presence of plasmid-encoded epsL or exeL without IPTG induction.
TABLE 3.
Effect of EpsL chimera overexpression on secretion in wt V. cholerae
| Plasmid-encoded proteina | Secretion of toxin in wt V. cholerae (% of total) |
|---|---|
| None | 85 |
| EpsL | 5 |
| ExeL57EpsL | 30 |
| ExeL154EpsL | 6 |
| ExeL216EpsL | 26 |
| ExeL296EpsL | 80 |
| EpsL296ExeL | 10 |
| EpsL216ExeL | 18 |
| EpsL154ExeL | 86 |
| EpsL57ExeL | 85 |
| ExeL | 80 |
Induced with 0.1 mM IPTG.
FIG. 1.
Levels of EpsL production in mutant and wt strains of V. cholerae containing a plasmid expressing epsL. Triton X-100 extracts of epsL mutant (lanes 1 to 3) and wt V. cholerae (lanes 4 and 5) expressing plasmid-encoded epsL were subjected to SDS-PAGE and immunoblot analysis with anti-EpsL antibodies. Lanes: 1, Mut8 (epsL mutant); 2, Mut8/pMS49; 3, Mut8/pMS49 grown in the presence of 0.1 mM IPTG; 4, TRH7000 (wt); 5, TRH7000/pMS49 grown in the presence of 0.1 mM IPTG. The values on the left show the positions of molecular size markers (in kilodaltons).
Identification of species-specific regions within the L protein.
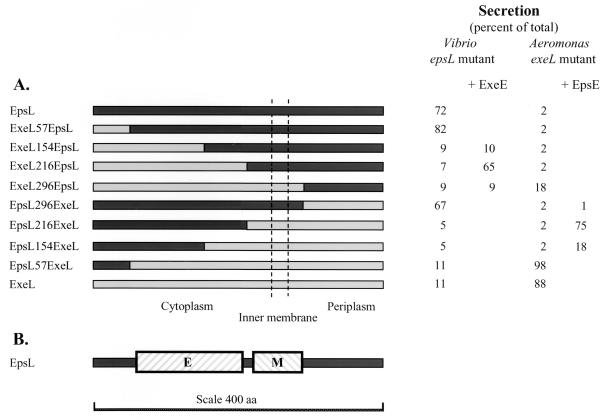
The recognition that EpsL and ExeL could only support secretion in their original hosts gave us the opportunity to map the region(s) involved in species specificity by segment swapping. Chimeric genes were constructed by PCR and expressed in the epsL and exeL mutants of V. cholerae and A. hydrophila (Fig. 2A). Replacement of the N-terminal 57 amino acids or the C-terminal 107 residues of EpsL with ExeL residues to create ExeL57EpsL and EpsL296ExeL did not diminish the activity of EpsL (Fig. 2A). These results suggest that neither the extreme N terminus nor the C terminus of EpsL is involved in species-specific protein-protein interactions in V. cholerae. However, replacement of the N-terminal 154 or the C-terminal 187 residues of EpsL in ExeL154EpsL and EpsL216ExeL produced proteins that could no longer complement the secretion defect in the epsL mutant strain (Fig. 2A). It appears, therefore, that an internal portion spanning residues 57 through 296 of EpsL determines species specificity and may contain the regions that interact with the rest of the secretion machinery, including the EpsE and EpsM proteins. The results obtained with the same chimeras in the A. hydrophila exeL mutant complemented those obtained with V. cholerae. In the exeL mutant, the ExeL protein with its first 57 residues replaced with EpsL residues was functional and could restore aerolysin secretion (Fig. 2A). The ExeL296EpsL fusion could support a low level of secretion but was not able to restore secretion to wt levels, as was the case with the reverse chimera EpsL296ExeL in V. cholerae.
FIG. 2.
Effects of various EpsL-ExeL chimeras on secretion. (A) Schematic representation of EpsL, ExeL, and EpsL-ExeL chimeras and their effects on secretion in the V. cholerae epsL mutant in the absence and presence of exeE and the A. hydrophila exeL mutant in the absence and presence of epsE. Secretion is presented as a percentage of the total toxin produced in cultures grown without IPTG induction. (B) The wt EpsL protein, in which the species-specific EpsE and EpsM binding domains are shown as cross-hatched boxes. aa, amino acids.
In order to map the region within EpsL involved in species-specific protein-protein interactions more precisely, we analyzed the different EpsL-ExeL chimeras for the ability to inhibit secretion when overproduced (Table 3). First of all, we observed that the chimeras ExeL57EpsL and EpsL296ExeL, which are functional in V. cholerae, behaved similarly to wt EpsL, in that they also exhibited a dominant negative effect when overproduced in wt V. cholerae. Likewise, the nonfunctional chimeras ExeL296EpsL, EpsL57ExeL, and EpsL154ExeL did not interfere with secretion, suggesting that they have lost the capability to functionally interact with both the EpsE and EpsM proteins (Table 3). In contrast, overexpression of the nonfunctional chimeras ExeL154EpsL, ExeL216EpsL, and EpsL216ExeL did inhibit secretion in the wt strain, suggesting that these chimeras retain the ability to interact with either EpsE or EpsM but not the ability to interact with both. Therefore, while these chimeras cannot complement the secretion defect, they can act as dominant negative inhibitors of secretion by titrating out one of the components of the secretion apparatus. For example, if the chimeras have an intact EpsE binding site, then the inhibition might be similar to that observed with the wt EpsL protein and when overproduced, these chimeras would remove EpsE from the secretion apparatus, thus rendering it nonfunctional. Alternatively, if the EpsE binding region is not intact but the chimeras are still able to interact with EpsM, the insertion of these hybrids into the secretion apparatus would prevent the formation of a functional EpsE-EpsL complex by forming an EpsL-EpsM complex to which EpsE could not bind. If this interpretation is correct, then these latter chimeras should provide excellent reagents for the identification of the two potential sites of interaction for EpsE and EpsM within the EpsL segment between residues 57 and 296. Based on the results obtained by overexpression of the chimeras, it is possible that one site is present between residues 57 and 216 and the other is present between amino acids 216 and 296. Finally, consistent with the data obtained with V. cholerae, analysis of the chimeras in wt A. hydrophila gave very similar results (data not shown), with the exception of ExeL216EpsL, which did not inhibit secretion.
Mapping of the EpsE binding domain within EpsL.
Hydropathy plots suggest that EpsL is a cytoplasmic membrane protein which has one membrane-spanning region its C-terminal half (residues 254 to 271). This segment is preceded by positively charged amino acids, which, according to the positive-inside rule (43), suggests that the N-terminal two-thirds of this protein is located on the cytoplasmic side of the inner membrane whereas the C terminus is periplasmic. This has been confirmed by membrane topology analysis of fusion proteins composed of the EpsL homologues OutL and XcpY and either β-lactamase or alkaline phosphatase (3, 12). If EpsE associates with the cytoplasmic membrane via interaction with EpsL only on the cytoplasmic face of the membrane, it suggests that the N-terminal two-thirds of EpsL contains the region that interacts with EpsE. On the other hand, if portions of EpsE are embedded in the cytoplasmic membrane, then other regions of EpsL could be involved in the membrane association of EpsE. To identify the region within EpsL that interacts with the EpsE protein, the nonfunctional ExeL154EpsL, ExeL216EpsL, and ExeL296EpsL proteins were analyzed for the ability to restore secretion in the presence of ExeE in the V. cholerae epsL mutant (Fig. 2A). The addition of ExeE could not restore secretion in the presence of ExeL154EpsL or ExeL296EpsL, but secretion could be obtained in the presence of ExeL216EpsL. This suggests that ExeL216EpsL both contains an intact ExeE binding domain and is able to interact functionally with the rest of the Eps apparatus. Thus, the reason the ExeL216EpsL chimera cannot complement the secretion defect in the epsL mutant in the absence of ExeE is that it has lost its EpsE binding domain. Since ExeL57EpsL can replace wt EpsL and support secretion in the presence of EpsE and since ExeL216EpsL can restore secretion in the epsL mutant in the presence of ExeE, this strongly suggests that the species-specific protein E binding site is located between cytoplasmic residues 57 and 216 of EpsL. The fact that ExeL154EpsL did not function in the presence of either EpsE or ExeE suggests that the site of fusion in this chimera is likely to be within the EpsE protein binding domain and, thus, disrupts the association of either protein EpsE or ExeE with this hybrid. Furthermore, these data also suggest that the reason ExeL296EpsL was not functional with either protein EpsE or ExeE is likely the inability of this chimera to interact with another component of the apparatus, possibly the EpsM protein (see below). Also, consistent with this is the inability of ExeL296EpsL to inhibit secretion in the wt V. cholerae strain, since our data suggest that this chimera has lost its binding sites for both EpsE and EpsM. The results obtained with the reverse chimeras in Aeromonas in the presence of epsE were entirely consistent with the Vibrio results (Fig. 2A). In this strain background, the chimera EpsL216ExeL was functional in the presence of EpsE while EpsL296ExeL could not support secretion. Finally, a small increase in secretion was obtained in the presence of EpsE and EpsL154ExeL, supporting the suggestion that the protein E binding site is close to position 154.
Mapping of the EpsM binding domain within EpsL.
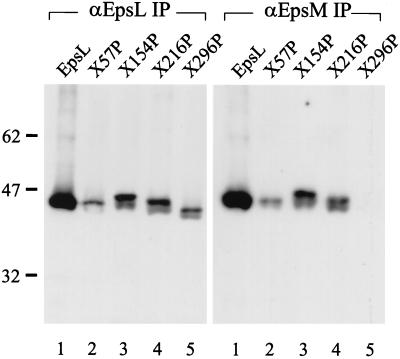
We next wanted to determine if the second region of EpsL involved in species-specific protein-protein interactions could contain the EpsM binding site. We had previously found that EpsL and EpsM form a stable complex that can be coimmunoprecipitated. We also observed that this complex formation does not require other Eps components and can occur in E. coli (35). Therefore, in order to analyze the EpsL-EpsM interaction only, the different chimeras were expressed in E. coli strain BL21(DE3) in the presence of epsM. The cells were subjected to Triton X-100 extraction, followed by immunoprecipitation with either anti-EpsL or anti-EpsM antibodies. The precipitated samples were then analyzed by SDS-PAGE and immunoblotting with biotinylated anti-EpsL IgG as described previously (35) (Fig. 3). Only chimeras containing EpsL sequences at the C terminus could be analyzed in this manner, since the anti-EpsL antibodies only recognize this region of EpsL. Hybrids containing portions of ExeL at the C terminus were thus not precipitated or detected. Nonetheless, we found that anti-EpsL antibodies could precipitate wt EpsL, as well as ExeL57EpsL, ExeL154EpsL, ExeL216EpsL, and ExeL296EpsL (Fig. 3). On the other hand, in the presence of EpsM, anti-EpsM could precipitate all of the chimeras except ExeL296EpsL, suggesting that this hybrid has lost its ability to interact with EpsM. Replacement of the region between residues 216 and 296 of EpsL appears to have resulted in loss of stable EpsM interaction. This is consistent with the inability of this chimera to inhibit secretion in the wt V. cholerae strain, while all of the other chimeras that contain N-terminal portions of ExeL are able to prevent secretion when overproduced. It is also consistent with the ability of hybrid ExeL216EpsL but not ExeL296EpsL to restore secretion in the epsL mutant in the presence of ExeE. These results indicate that chimera ExeL216EpsL must interact properly with EpsM. Although we could not determine if EpsL296ExeL can be coimmunoprecipitated with EpsM due to the specificity of the anti-EpsL antibody, this chimera must be able to functionally interact with EpsM, since it can restore secretion to wt levels in the epsL mutant. It appears, therefore, that residues N terminal to position 216 and C terminal to position 296 are not required for species-specific EpsM interactions.
FIG. 3.
Mapping of the EpsM binding domain. Triton X-100 extracts of E. coli BL21(DE3) producing EpsM and either wt EpsL or different EpsL-ExeL chimeras were immunoprecipitated with either anti-EpsL (αEpsL IP) or anti-EpsM (αEpsM IP) antibodies. Samples were subjected to SDS-PAGE and immunoblot analysis with biotinylated anti-EpsL IgG and horseradish peroxidase-coupled streptavidin. Lanes: 1, EpsL; 2, ExeL57EpsL; 3, ExeL154EpsL; 4, ExeL216EpsL; 5, ExeL296EpsL. The values on the left show the positions of molecular size markers (in kilodaltons).
DISCUSSION
In order to understand the mechanism of secretion and characterize the organization of the GSP apparatus in V. cholerae, we have continued to dissect the relevant interactions between the individual Eps components. These interactions are thought to be highly stoichiometric, since overproduction of some of the components interferes with the function of the secretion apparatus. For instance, overproduction of protein L results in inhibition of secretion and concomitant overproduction of protein E relieves this inhibitory effect (references 1 and 31 and our present study). This suggests that the dominant negative effect of protein L overproduction results primarily from the titration of protein E. Another example of a dominant negative effect on secretion comes from studies of Pugsley and colleagues, who found that when the prepilin-like protein PulG was overproduced, secretion was likewise inhibited (28).
In this study, we have extended the analysis of the V. cholerae Eps and A. hydrophila Exe apparatus and the EpsE-EpsL-EpsM trimolecular complex by mapping the regions within the EpsL protein that are involved in interactions with EpsE and EpsM. Because the EpsL protein from V. cholerae and ExeL from A. hydrophila cannot replace each other, we have been able to map protein-protein interacting domains within protein EpsL and ExeL by constructing and analyzing different EpsL-ExeL chimeras. This type of analysis has been fruitful previously when domains important for protein-protein interactions within EpsE were analyzed (34). In this study, segment swapping was useful for the analysis of protein-protein interacting domains between the EpsL protein and both the EpsE and EpsM proteins. However, it should be noted that in these experiments we have only mapped species-specific interactions and there may be additional sites of contact between these components. Other experiments are required to determine whether additional contacts exist between these proteins.
EpsE is a soluble cytoplasmic protein in the absence of EpsL. However, in the presence of EpsL, EpsE is associated with the membrane, and therefore it is likely that the region of EpsL that is located on the cytoplasmic side of the membrane is responsible for this function (34). Recent results obtained by Py and colleagues (31) support this hypothesis. When a truncated form of the EpsL homologue OutL containing cytoplasmic residues 1 to 248 was overexpressed, secretion was inhibited. The inhibition of secretion could be suppressed by the concomitant overexpression of OutE, suggesting that the E binding domain is most likely present within the cytoplasmic domain of the L protein. In this study, we have extended the analysis of the EpsE-EpsL interaction and mapped the species-specific EpsE binding region further to the N-terminal portion of EpsL, between residues 57 and 216. However, as discussed above, other sites within EpsL may also be involved in EpsE interactions. The result obtained with one of the chimeras, ExeL296EpsL in A. hydrophila, supports this latter hypothesis, since expression of this chimera in the A. hydrophila mutant only partially restores secretion (18% of the wt level) while the reverse chimera EpsL296ExeL appears to fully restore secretion in the V. cholerae epsL mutant. The reduced secretion level may be due to misfolding of ExeL296EpsL, or alternatively, the site of fusion in this chimera is close to a region that is involved in species-specific interactions. Interestingly, when wt EpsE is coproduced with this chimera in the A. hydrophila mutant, the level of secretion is increased to approximately 55% (data not shown), suggesting that there is a second site of interaction for protein E. ExeE appears to discriminate between this site in EpsL and ExeL, while EpsE may be more promiscuous and can interact with this site in both EpsL and ExeL. This observation is consistent with those made in an earlier study of EpsE (34). In that study, we showed that while ExeE could not function and replace EpsE in V. cholerae, EpsE could replace ExeE in A. hydrophila. These results suggest there is an additional site of interaction between proteins E and L that extends past residue 296. If this is the case, the EpsE protein must also interact with the periplasmic domain of EpsL and it is therefore possible that protein EpsE spans the membrane. Alternatively, EpsE may only transiently interact with the periplasmic domain of EpsL during its membrane insertion, where this region of EpsL may be involved in inserting EpsE into the membrane and then releasing it at some stage. Further analysis has to be done to confirm this observation.
EpsM appears to interact predominantly with the C-terminal domain of EpsL. It is likely that the species-specific region of interaction resides between residues 216 and 296. The basis for this conclusion is as follows. (i) ExeL216EpsL, but not ExeL296ExeL, could be coimmunoprecipitated with EpsM. (ii) ExeL216EpsL was functional and restored secretion in the V. cholerae epsL mutant in the presence of ExeE, suggesting that the EpsM binding site is intact in this chimera. (iii) EpsL296ExeL could replace EpsL in V. cholerae and restore secretion. Because the membrane-spanning region is thought to include residues 254 to 271, it is likely that EpsM and EpsL interact with each other in the cytoplasmic membrane. As discussed for the EpsE binding site, there may also be additional sites of interaction between EpsL and EpsM. An additional site of interaction could be present within the periplasmic domains of EpsL and EpsM, since both proteins contain periplasmic C-terminal tails that consist of more than 100 residues. However, it is also possible that the periplasmic domain of EpsL is not involved in the interaction with EpsM and might, instead, serve other functions. It may be required for the stability and/or oligomerization of protein L, as was suggested by Py and colleagues (31).
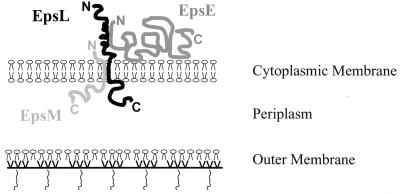
In conclusion, we have mapped the species-specific EpsE and EpsM binding domains to two separate sites within EpsL. At least one important species-specific interaction with EpsE occurs via the cytoplasmic region of EpsL. The species-specific EpsM binding site is localized to the C-terminal half of EpsL that contains the transmembrane segment (Fig. 2B and 4). Furthermore, the suggestion that EpsL connects EpsE with EpsM and the rest of the secretion apparatus has been strengthened (Fig. 4). Since the EpsL binding sites for EpsE and EpsM are close but do not overlap, there is a possibility that these proteins also interact with each other. Further analysis must be performed in order to determine whether there are any interactions between these two components.
FIG. 4.
Model of species-specific protein-protein interactions within the EpsE-EpsL-EpsM complex. EpsL spans the cytoplasmic membrane with the N terminus exposed on the cytoplasmic side and the C terminus present in the periplasm. EpsL is responsible for membrane association of EpsE, and the species-specific interaction between these components occurs via their N termini on the cytoplasmic side of the membrane. Another species-specific contact exists between the membrane-spanning regions of EpsL and EpsM. EpsM is predominantly exposed on the periplasmic side of the membrane.
ACKNOWLEDGMENTS
We thank Zain Dossani and Ravindra Jahagirdar for excellent technical assistance.
This work was supported by the American Red Cross (M.S.), the Medical Research Council of Canada (grant MT-10470 to S.P.H.), and the U.S. Department of Agriculture (grant 98-02052 to M.B.).
REFERENCES
- 1.Ball G, Chapon-Herve V, Bleves S, Michel G, Bally M. Assembly of XcpR in the cytoplasmic membrane is required for extracellular protein secretion in Pseudomonas aeruginosa. J Bacteriol. 1999;181:382–388. doi: 10.1128/jb.181.2.382-388.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bitter W, Koster M, Latijnhouwers M, De Cock H, Tommassen J. Formation of oligomeric rings by XcpQ and PilQ, which are involved in protein transport across the outer membrane of Pseudomonas aeruginosa. Mol Microbiol. 1998;27:209–219. doi: 10.1046/j.1365-2958.1998.00677.x. [DOI] [PubMed] [Google Scholar]
- 3.Bleves S, Lazdunski A, Filloux A. Membrane topology of three Xcp proteins involved in exoprotein transport by Pseudomonas aeruginosa. J Bacteriol. 1996;178:4297–4300. doi: 10.1128/jb.178.14.4297-4300.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casadaban M J, Cohen S N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 5.Dallas W S. Conformity between heat-labile toxin genes from human and porcine enterotoxigenic Escherichia coli. Infect Immun. 1983;40:647–652. doi: 10.1128/iai.40.2.647-652.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Groot A, Filloux A, Tommassen J. Conservation of xcp genes, involved in the two-step protein secretion process, in different Pseudomonas species and other gram-negative bacteria. Mol Gen Genet. 1991;229:278–284. doi: 10.1007/BF00272167. [DOI] [PubMed] [Google Scholar]
- 7.Filloux A, Michel G, Bally M. GSP-dependent protein secretion in gram-negative bacteria: the Xcp system of Pseudomonas aeruginosa. FEMS Microbiol Rev. 1998;22:177–198. doi: 10.1111/j.1574-6976.1998.tb00366.x. [DOI] [PubMed] [Google Scholar]
- 8.Furste J P, Pansegrau W, Frank R, Blocker H, Scholz P, Bagdasarian M, Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 9.Hardie K R, Lory S, Pugsley A P. Insertion of an outer membrane protein in Escherichia coli requires a chaperone-like protein. EMBO J. 1996;15:978–988. [PMC free article] [PubMed] [Google Scholar]
- 10.He S Y, Lindeberg M, Chatterjee A K, Collmer A. Cloned Erwinia chrysanthemi out genes enable Escherichia coli to selectively secrete a diverse family of heterologous proteins to its milieu. Proc Natl Acad Sci USA. 1991;88:1079–1083. doi: 10.1073/pnas.88.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirst T R, Sanchez J, Kaper J B, Hardy S J, Holmgren J. Mechanism of toxin secretion by Vibrio cholerae investigated in strains harboring plasmids that encode heat-labile enterotoxins of Escherichia coli. Proc Natl Acad Sci USA. 1984;81:7752–7756. doi: 10.1073/pnas.81.24.7752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Housby J N, Thomas J D, Wharam S D, Reeves P J, Salmond G P. Conditional mutations in OutE and OutL block exoenzyme secretion across the Erwinia carotovora outer membrane. FEMS Microbiol Lett. 1998;165:91–102. doi: 10.1111/j.1574-6968.1998.tb13132.x. [DOI] [PubMed] [Google Scholar]
- 13.Howard S P, Critch J, Bedi A. Isolation and analysis of eight exe genes and their involvement in extracellular protein secretion and outer membrane assembly in Aeromonas hydrophila. J Bacteriol. 1993;175:6695–6703. doi: 10.1128/jb.175.20.6695-6703.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jahagirdar R, Howard S P. Isolation and characterization of a second exe operon required for extracellular protein secretion in Aeromonas hydrophila. J Bacteriol. 1994;176:6819–6826. doi: 10.1128/jb.176.22.6819-6826.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang B, Howard S P. The Aeromonas hydrophila exeE gene, required both for protein secretion and normal outer membrane biogenesis, is a member of a general secretion pathway. Mol Microbiol. 1992;6:1351–1361. doi: 10.1111/j.1365-2958.1992.tb00856.x. [DOI] [PubMed] [Google Scholar]
- 16.Koster M, Bitter W, De Cock H, Allaoui A, Cornelis G R, Tommassen J. The outer membrane component, YscC, of the Yop secretion machinery of Yersinia enterocolitica forms a ring-shaped multimeric complex. Mol Microbiol. 1997;26:789–797. doi: 10.1046/j.1365-2958.1997.6141981.x. [DOI] [PubMed] [Google Scholar]
- 17.Lindeberg M, Boyd C M, Keen N T, Collmer A. External loops at the C terminus of Erwinia chrysanthemi pectate lyase C are required for species-specific secretion through the Out type II pathway. J Bacteriol. 1998;180:1431–1437. doi: 10.1128/jb.180.6.1431-1437.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Linderoth N A, Simon M N, Russel M. The filamentous phage pIV multimer visualized by scanning transmission electron microscopy. Science. 1997;278:1635–1638. doi: 10.1126/science.278.5343.1635. [DOI] [PubMed] [Google Scholar]
- 19.Marciano D K, Russel M, Simon S M. An aqueous channel for filamentous phage export. Science. 1999;284:1516–1519. doi: 10.1126/science.284.5419.1516. [DOI] [PubMed] [Google Scholar]
- 20.Marsh J W, Taylor R K. Identification of the Vibrio cholerae type 4 prepilin peptidase required for cholera toxin secretion and pilus formation. Mol Microbiol. 1998;29:1481–1492. doi: 10.1046/j.1365-2958.1998.01031.x. [DOI] [PubMed] [Google Scholar]
- 21.Michel L O, Sandkvist M, Bagdasarian M. Specificity of the protein secretory apparatus: secretion of the heat-labile enterotoxin B subunit pentamers by different species of gram-bacteria. Gene. 1995;152:41–45. doi: 10.1016/0378-1119(94)00691-k. [DOI] [PubMed] [Google Scholar]
- 22.Morales V M, Backman A, Bagdasarian M. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene. 1991;97:39–47. doi: 10.1016/0378-1119(91)90007-x. [DOI] [PubMed] [Google Scholar]
- 23.Nouwen N, Ranson N, Saibil H, Wolpensinger B, Engel A, Ghazi A, Pugsley A P. Secretin PulD: association with pilot PulS, structure, and ion-conducting channel formation. Proc Natl Acad Sci USA. 1999;96:8173–8177. doi: 10.1073/pnas.96.14.8173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nunn D N, Lory S. Product of the Pseudomonas aeruginosa gene pilD is a prepilin leader peptidase. Proc Natl Acad Sci USA. 1991;88:3281–3285. doi: 10.1073/pnas.88.8.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nunn D N, Lory S. Components of the protein-excretion apparatus of Pseudomonas aeruginosa are processed by the type IV prepilin peptidase. Proc Natl Acad Sci USA. 1992;89:47–51. doi: 10.1073/pnas.89.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nunn D N, Lory S. Cleavage, methylation, and localization of the Pseudomonas aeruginosa export proteins XcpT, -U, -V, and -W. J Bacteriol. 1993;175:4375–4382. doi: 10.1128/jb.175.14.4375-4382.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Overbye L J, Sandkvist M, Bagdasarian M. Genes required for extracellular secretion of enterotoxin are clustered in Vibrio cholerae. Gene. 1993;132:101–106. doi: 10.1016/0378-1119(93)90520-d. [DOI] [PubMed] [Google Scholar]
- 28.Pugsley A P. Processing and methylation of PuIG, a pilin-like component of the general secretory pathway of Klebsiella oxytoca. Mol Microbiol. 1993;9:295–308. doi: 10.1111/j.1365-2958.1993.tb01691.x. [DOI] [PubMed] [Google Scholar]
- 29.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pugsley A P, Francetic O, Possot O M, Sauvonnet N, Hardie K R. Recent progress and future directions in studies of the main terminal branch of the general secretory pathway in Gram-negative bacteria—a review. Gene. 1997;192:13–19. doi: 10.1016/s0378-1119(96)00803-7. [DOI] [PubMed] [Google Scholar]
- 31.Py B, Loiseau L, Barras F. Assembly of the type II secretion machinery of Erwinia chrysanthemi: direct interaction and associated conformational change between OutE, the putative ATP-binding component and the membrane protein OutL. J Mol Biol. 1999;289:659–670. doi: 10.1006/jmbi.1999.2803. [DOI] [PubMed] [Google Scholar]
- 32.Reeves P J, Douglas P, Salmond G P. Beta-lactamase topology probe analysis of the OutO NMePhe peptidase, and six other Out protein components of the Erwinia carotovora general secretion pathway apparatus. Mol Microbiol. 1994;12:445–457. doi: 10.1111/j.1365-2958.1994.tb01033.x. [DOI] [PubMed] [Google Scholar]
- 33.Russel M. Macromolecular assembly and secretion across the bacterial cell envelope: type II protein secretion systems. J Mol Biol. 1998;279:485–499. doi: 10.1006/jmbi.1998.1791. [DOI] [PubMed] [Google Scholar]
- 34.Sandkvist M, Bagdasarian M, Howard S P, DiRita V J. Interaction between the autokinase EpsE and EpsL in the cytoplasmic membrane is required for extracellular secretion in Vibrio cholerae. EMBO J. 1995;14:1664–1673. doi: 10.1002/j.1460-2075.1995.tb07155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sandkvist M, Hough L P, Bagdasarian M M, Bagdasarian M. Direct interaction of the EpsL and EpsM proteins of the general secretion apparatus in Vibrio cholerae. J Bacteriol. 1999;181:3129–3135. doi: 10.1128/jb.181.10.3129-3135.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sandkvist M, Michel L O, Hough L P, Morales V M, Bagdasarian M, Koomey M, DiRita V J. General secretion pathway (eps) genes required for toxin secretion and outer membrane biogenesis in Vibrio cholerae. J Bacteriol. 1997;179:6994–7003. doi: 10.1128/jb.179.22.6994-7003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sandkvist M, Morales V, Bagdasarian M. A protein required for secretion of cholera toxin through the outer membrane of Vibrio cholerae. Gene. 1993;123:81–86. doi: 10.1016/0378-1119(93)90543-c. [DOI] [PubMed] [Google Scholar]
- 38.Sandkvist M, Overbye L J, Sixma T K, Hol W G J, Bagdasarian M. Assembly of Escherichia coli heat-labile enterotoxin and its secretion from Vibrio cholerae. In: Kado C, Crossa J, Sequeira L, editors. Molecular mechanisms of bacterial virulence. Dordrecht, The Netherlands: Academic Publishers; 1993. pp. 293–309. [Google Scholar]
- 39.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 40.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas J D, Reeves P J, Salmond G P. The general secretion pathway of Erwinia carotovora subsp. carotovora: analysis of the membrane topology of OutC and OutF. Microbiology. 1997;143:713–720. doi: 10.1099/00221287-143-3-713. [DOI] [PubMed] [Google Scholar]
- 42.Tommassen J, Filloux A, Bally M, Murgier M, Lazdunski A. Protein secretion in Pseudomonas aeruginosa. FEMS Microbiol Rev. 1992;9:73–90. doi: 10.1016/0378-1097(92)90336-m. [DOI] [PubMed] [Google Scholar]
- 43.von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]