Abstract
Background
The incidence of cutaneous melanoma is increasing in Italy, in parallel with the implementation of gene panels. Therefore, a revision of national genetic assessment criteria for hereditary melanoma may be needed. The aim of this study was to identify predictors of susceptibility variants in the largest prospective cohort of Italian high-risk melanoma cases studied to date.
Materials and methods
From 25 Italian centers, we recruited 1044 family members and germline sequenced 940 cutaneous melanoma index cases through a shared gene panel, which included the following genes: CDKN2A, CDK4, BAP1, POT1, ACD, TERF2IP, MITF and ATM. We assessed detection rate according to familial status, region of origin, number of melanomas and presence and type of non-melanoma tumors.
Results
The overall detection rate was 9.47% (5.53% analyzing CDKN2A alone), ranging from 5.14% in sporadic multiple melanoma cases (spoMPM) with two cutaneous melanomas to 13.9% in familial cases with at least three affected members. Three or more cutaneous melanomas in spoMPM cases, pancreatic cancer and region of origin predicted germline status [odds ratio (OR) = 3.23, 3.15, 2.43, P < 0.05]. Conversely, age > 60 years was a negative independent predictor (OR = 0.13, P = 0.008), and was the age category with the lowest detection rate, especially for CDKN2A. Detection rate was 19% when cutaneous melanoma and pancreatic cancer clustered together.
Conclusions
Gene panel doubled the detection rate given by CDKN2A alone. National genetic testing criteria may need a revision, especially regarding age cut-off (60) in the absence of strong family history, pancreatic cancer and/or a high number of cutaneous melanomas.
Key words: melanoma, germline, predictors, gene panel, CDKN2A, susceptibility
Highlights
-
•
Gene panel testing doubles germline variants detection rate compared to CDKN2A-only testing.
-
•
If only two melanoma events at > 60 years of age are reported, eligibility to genetic testing requires careful consideration.
-
•
Pancreatic cancer is a strong predictor of germline status.
Introduction
The incidence of cutaneous melanoma (CM) is growing worldwide,1,2 including in former low-CM-incidence countries such as Italy. At the same time, the use of gene panels for hereditary melanoma testing has been increasingly implemented in clinical practice.
According to AIRTUM (Italian Association of Cancer Registries), the average annual CM incidence in Italy was 20.4/100 000 per year from 2008 until 2016, with regional differences, as northern Italian incidence was twice that of southern Italy. A 20% increase in this rate was projected for 2020, with CM representing 4% of all new cancer diagnoses.3
Five to twelve percent of all CM cases belong to either melanoma-prone families or families with melanoma-related multicancer clustering.4 A subset of these families fall in the definition of hereditary melanoma, i.e. families in which the risk is attributable to a specific germline risk allele. Indeed, melanoma can be part of the spectrum of multi-tumor hereditary cancer syndromes caused by high and intermediate penetrance alleles in known predisposition genes such as CDKN2A, CDK4, POT1, BAP1, TERT, ACD, TERF2IP, MITF and ATM.4, 5, 6, 7, 8, 9, 10, 11, 12, 13 Additional genes, such as GOLM1, EBF3, POLE and NEK11, have been recently linked to melanoma susceptibility, but are not yet included in clinical genetic testing due to the lack of risk estimates.14, 15, 16, 17, 18
A 2009 Italian Melanoma Intergroup (IMI) study established that the prevalence of CDKN2A pathogenic/likely pathogenic variant (PV/LPV) carriers was 33% in all recruited families and 25% in families with only two CM cases, concluding that clinical genetic testing in those latter families was justified in terms of the likelihood of identifying a PV.19 In another 2016 IMI study involving multiple primary melanoma (MPM) cases, 19% of cases carried CDKN2A or, rarely, MITF variants. CDKN2A detection rate varied from 36.6% to 58.8% in familial MPM cases and from 8.2% to 17.6% in sporadic MPM (spoMPM) cases.20 Based on these findings, coupled with the fact that Italy was a low-incidence melanoma country, genetic assessment criteria for hereditary melanoma were less conservative compared to those adopted in high-incidence countries, where the frequency of phenocopies is greater. In our recent retrospective study on an Italian cohort of 273 CDKN2A-negative CM cases categorized as high risk according to our current genetic assessment criteria,12 and analyzed through a multi-gene panel shared within the IMI, we showed that 9% of cases carried PV/LPV in high- or intermediate-penetrance genes such as BAP1, POT1, ACD, MITF or ATM. ATM emerged as a moderate-risk gene both in the aforementioned study and in a subsequent international survey conducted within the GenoMEL and Melanostrum consortia.13 Another study, involving familial and spoMPM cases enrolled from 2009 to 201721 from central and southern Italy, reported a low CDKN2A detection rate, in contrast with previous studies on northern Italian cohorts, but consistent with other studies on central Italian cohorts conducted during the last 5 years.22, 23, 24
As stated earlier, genetic assessment is currently based on a clinical score that takes into account both CM incidence in a specific area and the presence of cancers associated with CM within the so-called melanoma-dominant or melanoma-subordinate syndromes, in order to guide eligibility to genetic testing and the choice of the most appropriate genetic panel.4 However, based on the aforementioned changing incidence, it may be time for a revision of such criteria.
In this study, conducted on an Italian cohort prospectively recruited partly using a centralized telecounseling service, we aimed to evaluate the impact of a panel that includes all major established and candidate susceptibility genes on the overall mutation yield. Moreover, we sought to identify the best predictors for CM susceptibility, in order to lay the groundwork for a possible revision of national genetic testing assessment criteria.
Methods
Study cohort
Since 2016, we carried out genetic testing using a germline panel that included CDKN2A, CDK4, BAP1, POT1, ACD, TERF2IP, MITF and ATM, on a consecutive series of high-risk melanoma cases and their affected and unaffected family members enrolled from 25 different IMI centers until the end of 2021. Out of 940 probands (i.e. the index cases), 798 were recruited and tested at the IRCCS Ospedale Policlinico San Martino, Genoa, 92 at the IRCCS Veneto Institute of Oncology, Padua and 50 at the Institute of Biomolecular Chemistry, National Research Council, Sassari.
Inclusion criteria were: CM family history with at least another first/second-degree affected relative (familial cases); personal history of MPMs in the absence of CM family history (spoMPM cases); and presence, in a single CM proband and/or in first/second-degree relatives, of other known CM-associated cancers, i.e. pancreatic cancer, kidney cancer, mesothelioma or uveal melanoma, for a total of at least two cancer events (syndromic cases).
Of the 798 samples analyzed in Genoa, 401 underwent genetic counseling at the local clinic, 211 were referred from other centers for molecular analysis only and 186 were recruited through the genetic telecounseling service developed within IMI. All participants signed a local IRB-approved informed consent (IRB approval status: reviewed and approved by CER Liguria; approval #534/2020). Next-generation sequencing workflow and statistical analysis are described in Supplementary Methods, available at https://doi.org/10.1016/j.esmoop.2022.100525.
Results
Gene panel testing doubles CDKN2A detection rate
Our cohort consisted of 1044 family members, including 940 probands from northern (705), central (119) and southern (116) Italy, covering the majority of Italian regions (Supplementary Figure S1, available at https://doi.org/10.1016/j.esmoop.2022.100525). Of these, 474, 375 and 91 were familial, spoMPM and syndromic cases, respectively (Table 1).
Table 1.
Detection rate according to personal and family history
| Category | Gene panel |
CDKN2A |
Non-CDKN2A |
CDK4 |
BAP1 |
ATM |
POT1 |
MITF |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mut | % | Mut | % | Mut | % | Mut | % | Mut | % | Mut | % | Mut | % | Mut | % | |
| Fam 2 CM cases (N = 373)a | 34 | 9.12 | 21 | 5.63 | 14 | 3.75 | 0 | 0 | 1 | 0.27 | 6 | 1.61 | 3 | 0.8 | 4 | 1.07 |
| Fam 3 or more CM cases (N = 101) | 14 | 13.9 | 10 | 9.9 | 4 | 3.96 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 3.96 |
| spoMPM 2 CM events (N = 253) | 13 | 5.14 | 8 | 3.16 | 5 | 1.98 | 0 | 0 | 0 | 0 | 1 | 0.4 | 0 | 0 | 4 | 1.58 |
| spoMPM 3 or more CM events(N = 122) | 16 | 13.1 | 9 | 7.38 | 7 | 5.74 | 1 | 0.82 | 1 | 0.82 | 0 | 0 | 1 | 0.82 | 4 | 3.28 |
| Syndromic (N = 91) | 12 | 13.2 | 4 | 4.4 | 8 | 8.79 | 0 | 0 | 3 | 3.3 | 3 | 3.3 | 0 | 0 | 2 | 2.2 |
| Total (N = 940) | 89 | 9.47 | 52 | 5.53 | 38 | 4.04 | 1 | 0.11 | 5 | 0.53 | 10 | 1.06 | 4 | 0.43 | 18 | 1.91 |
| P value | 0.016 | 0.11 | 0.052 | |||||||||||||
The table shows the rate of PVs/LPVs in each gene depending on personal and family history, and the corresponding Fisher’s exact test derived P value.
Significant P values are marked in bold.
CM, cutaneous melanoma; Fam, familial cases; LPV, likely pathogenic variant; N, number of cases; Mut, number of cases with a PV/LPV; PV, pathogenic variant; spoMPM, apparently sporadic multiple primary melanoma cases.
One case had both an MITF PV and an ATM LPV.
CDKN2A detection rate was 5.53% (52 cases), while adding the other genes in the panel increased the detection rate up to 9.47% (89 cases), almost doubled compared to CDKN2A alone.
The most frequent PV was the CDKN2A p.Gly101Trp founder, predominantly found in Ligurian cases.25, 26, 27 Other known or candidate CDKN2A founders or recurring PVs were also present, such as the p.Arg24Pro28 distributed throughout the Italian peninsula and the p. Glu27Ter only found in Ligurian cases.29 Another candidate founder, the p.Pro48Thr PV, typically found in the northeastern part of the country, was identified in a case from Veneto.30 The remaining variants were all found in one or two cases each, without specific regional clusterings (Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2022.100525). Concerning the p. Ala127Pro variant of unknown significance (VUS), even though a deleterious effect on the protein function has been recently reported,31 the variant did not co-segregate with melanoma in the family in which we found it and, therefore, we did not reclassify it as LPV. Overall, CDKN2A PVs represented 57.3% of all identified LPV/PVs.
In the POT1 gene, we detected a novel variant, c.258_259insT, in a familial case who developed three melanomas (age 43, 44 and 47) and an atypical Spitz nevus. Of note, the known POT1 founder p.Ser270Asn was identified in a patient from Emilia-Romagna.7
We also found two novel BAP1 variants in two syndromic patients fulfilling the BAP1-tumor predisposition syndrome diagnostic criteria.8 The first one, c.677delT, was found in a case diagnosed with ocular melanoma at age 29 and chromophobe renal cell carcinoma at age 44, who also had a family history of mesothelioma. The other BAP1 variant, p.Trp202Ter, was found in a patient with prostate cancer (age 60), CM (age 68), and family history of pancreatic cancer, breast cancer and mesothelioma.
The variants more frequently found in non-CDKN2A genes were those conferring intermediate risk, such as the MITF p.Glu318Lys (20.2% of all variants) and ATM LPV/PVs (11.23%). Indeed, the frequency of PVs/LPVs in the intermediate- to low-penetrance genes MITF and ATM (N = 23/940, 2.98%) was more than twice as that of PVs/LPVs in non-CDKN2A high-penetrance genes (N = 10/940, 1.06%), as expected, considering that lower-penetrant alleles are generally found at a higher allele frequency.
The complete list of all PV, LPV and VUS found in our study cohort is reported in Supplementary Tables S1 and S2, available at https://doi.org/10.1016/j.esmoop.2022.100525. Total detection rate in Liguria was twice the detection rate observed in cases from other regions (13.37% versus 6.72%, OR = 42.93, P < 0.01). When considering CDKN2A alone, this difference was even higher (8.48% versus 3.45%, OR = 4.23, P < 0.01), whereas distribution of variants in non-CDKN2A genes did not significantly vary across regions.
Detection rate by personal and family history of melanoma: sporadic MPM with only two CM and first melanoma diagnosis over 60 show the lowest rate
Gene panel detection rate varied significantly according to personal and family history, ranging from 5.14% in spoMPM with only two CMs to 13.86% in melanoma families with three or more affected members (P = 0.02) (Table 1). The greatest difference was seen between spoMPM with only two CMs and spoMPM with three or more CMs, familial cases with three CMs and syndromic cases. However, no pairwise comparison was significant after false discovery rate (FDR) correction. Similar differences, although not significant, were seen for CDKN2A PV/LPVs but not for non-CDKN2A PV/LPVs grouped together (Table 1).
Overall, median age of diagnosis (AOD) was 48 years [interquartile range (IQR) = 39-59 years]. The median AOD of PV/LPV carriers was 46 years (IQR = 38-54 years), both for CDKN2A and the whole panel.
The detection rate was 7.69%, 5.48% and 1.19% in the age groups ≤40, 41-60 and ≥60 years, respectively, when counting only CDKN2A PV/LPVs, but it increased to 11.54%, 9.76% and 4.76%, respectively, when the whole panel was considered, with a significant difference observed between ≤40 and >60-year-old cases (FDR P = 0.04).
The association of age with detection rate was even more significant when only CDKN2A was considered for the analysis (P < 0.01). In the pairwise comparisons, detection rate in cases aged ≤40 years and 41-60 years significantly differed from that of >60-year-old cases (FDR P < 0.01 and FDR P = 0.033, respectively).
Indeed, the detection rate was at its lowest in cases where the earliest AOD was over 60 years old, especially in familial melanoma cases, in whom no variants were found, and in sporadic MPM cases with only two CMs (0% CDKN2A and 2.82% panel, see Supplementary Table S3, available at https://doi.org/10.1016/j.esmoop.2022.100525).
Detection rate by cancer events: pancreatic cancer as one of the strongest predictors
To assess the impact of type and number of cancer events on the detection rate, we divided cases into categories based on the presence, in the case or in the family, of: two CM events, three or more CM events, CM and pancreatic cancer, CM and other selected cancers except pancreatic cancer (uveal melanoma, mesothelioma, kidney cancer), and co-occurrence of CM, pancreatic cancer and at least one other selected cancer.
As shown in Table 2, the detection rate differed among categories, ranging from 4.06% (only 2 CM events) to 6.43% (≥3 CM events) for CDKN2A PV/LPVs, and from 5.84% to 10.61%, respectively, when including the whole panel. In the pairwise comparison, overall detection rate differences across categories remained significant when comparing two CM events with the co-occurrence of CM and pancreatic cancer (FDR P < 0.001).
Table 2.
Detection rate according to cancer events
| Category | Gene panel |
CDKN2A |
Non-CDKN2A |
CDK4 |
BAP1 |
ATM |
POT1 |
MITF |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mut | % | Mut | % | Mut | % | Mut | % | Mut | % | Mut | % | Mut | % | Mut | % | |
| CM only—2 events (N = 394) | 23 | 5.84 | 16 | 4.06 | 8 | 0.02 | 0 | 0 | 0 | 0 | 3 | 0.76 | 0 | 0 | 5 | 1.27 |
| CM only—≥ 3 events (N = 311) | 33 | 10.61 | 20 | 6.43 | 13 | 0.04 | 1 | 0.32 | 0 | 0 | 2 | 0.64 | 3 | 0.96 | 7 | 2.25 |
| CM + PC (N = 121) | 23 | 19.01 | 15 | 12.4 | 8 | 0.07 | 0 | 0 | 0 | 0 | 4 | 3.31 | 0 | 0 | 4 | 3.31 |
| CM + other cancers (N = 103) | 9 | 8.74 | 1 | 0.97 | 8 | 0.08 | 0 | 0 | 4 | 3.88 | 1 | 0.97 | 1 | 0.97 | 2 | 1.94 |
| CM + PC + other cancers (N = 11) | 1 | 10 | 0 | 0 | 1 | 0.1 | 0 | 0 | 1 | 10 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total (N = 940) | 89 | 9.47 | 52 | 5.53 | 38 | 4.04 | 1 | 0.11 | 5 | 0.53 | 10 | 1.06 | 4 | 0.43 | 18 | 1.91 |
| P value | <0.01 | <0.01 | <0.01 | |||||||||||||
The table shows the detection rate according to the type and number of cancer events, and the corresponding Fisher’s exact test derived P value.
Genes in which no PVs/LPVs were detected are not reported.
Significant P values are marked in bold.
CM, cutaneous melanoma; LPV, likely pathogenic variant; Mut, number of cases with a PV/LPV; PC, pancreatic cancer; PV, pathogenic variant.
The highest CDKN2A detection rate was seen when pancreatic cancer was present either in the proband or in the family (12.4%), whereas the contribution of other cancers to the detection rate was minimal (0.97%). In the pairwise comparison, CDKN2A detection rate differences remained significant between two CM events and the co-occurrence of CM and pancreatic cancer (FDR P < 0.01) and between the presence of pancreatic cancer and the presence of other associated cancers in the absence of pancreatic cancer (FDR P < 0.01).
Conversely, for non-CDKN2A genes, the contribution of other cancers to the detection rate (7.77%) was similar to that of pancreatic cancer (6.61%).
Pancreatic cancer was apparently more frequent in cases/families with an ATM PV/LPV (4/10, 40%) than in those with wild-type germline ATM status (128/930, 13.76%, OR = 4.1, P = 0.039). Similarly, pancreatic cancer diagnoses were more frequent in cases/families with the MITF p.Glu318Lys variant (4/18, 22.2%) than in those without the variant (128/922, 13.9%). However, in the latter case, the difference was not significant (P > 0.05).
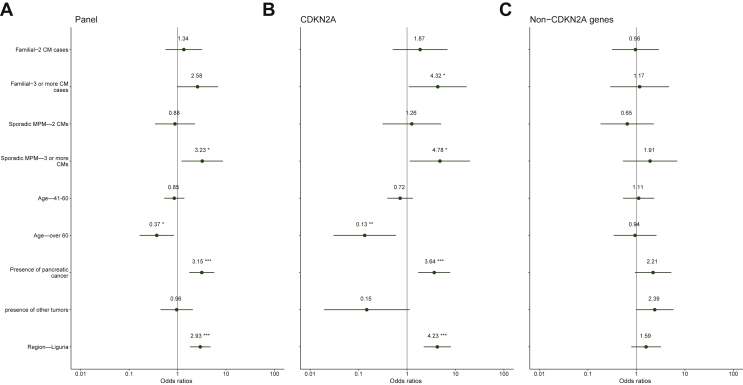
Overall, in the multiple logistic regression model, the main independent predictors of detecting a germline PV/LPV were: three or more CM events in spoMPM (OR = 3.23, P = 0.020), presence of pancreatic cancer in the case or in the family (OR = 3.15, P < 0.001) and Liguria as the region of origin (OR = 2.93, P < 0.001), whereas age > 60 was a negative predictor (OR = 0.37, P = 0.017), as shown in Figure 1A. When looking at CDKN2A, these associations were even stronger (Figure 1B).
Figure 1.
Forest plot presenting the results of multiple logistic regression analysis for detection rate of all genes.
(A) Panel genes, (B) CDKN2A and (C) non-CDKN2A genes. Significant values are marked with asterisks. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
CM, cutaneous melanoma; MPM, multiple primary melanoma.
No variables were significantly associated with the presence of non-CDKN2A PV/LPVs (Figure 1C).
Discussion
Given the steady increase of CM incidence in Italy, we investigated whether the current melanoma genetic testing criteria are still appropriate, by evaluating the association of the detection rate obtained by a multi-gene panel with personal and familial characteristics currently used to assess genetic testing eligibility. This work is part of a broader effort to adjust genetic testing criteria in the context of Italy’s current epidemiological picture, similarly to what is done by other countries. Indeed, international scores present issues preventing their applicability worldwide. For instance, some scores have been validated in countries with a CM incidence not comparable with that of southern Europe. Moreover, other scores are either CDKN2A-centered, or exclude melanoma-associated cancers other than pancreatic cancer.32, 33, 34, 35
In our cohort, selected with less conservative criteria compared to high-incidence countries, single-gene (CDKN2A) testing leads to a low rate of identified PVs (<5% in selected categories). Nevertheless, CDKN2A PVs still represent the majority of identified variants, due to the presence of founder or recurring PVs, especially in northwestern Italy.25, 26, 27,29
Criteria such as young age at diagnosis, presence of pancreatic cancer, number of family members and melanomas are mainly relevant for CDKN2A, which still accounts for slightly more than half of germline PV/LPVs found, and all of them were independent predictors of CDKN2A germline status.
In contrast, even though none of the included variables emerged as an independent predictor of non-CDKN2A genes germline status, an association with non-melanoma tumors was observed for non-CDKN2A PV/LPVs carriers. However, non-CDKN2A genes were grouped together due to the small individual detection rate, although their cancer spectrums and possibly other characteristics do not necessarily overlap, which may have hampered the identification of one or more predictors. Moreover, given the low overall detection rate of non-CDKN2A genes, our cohort might have been underpowered to see associations with these genes.
In line with previous data,36 pancreatic cancer was positively associated with the presence of ATM PV/LPVs, whereas the association with MITF was not significant.37 Most non-pancreatic cancers occurred in cases with non-CDKN2A PV/LPVs.
Non-pancreatic cancers were more frequent in CM cases carrying BAP1 PVs, consistent with the BAP1-tumor predisposition syndrome (BAP1-TPDS) cancer spectrum.8 However, CM is only one of the cancers included in BAP1-TPDS and is not the most prevalent, hence this cohort, selected with a focus on melanoma, underestimates the actual BAP1 detection rate found when BAP1-TPDS criteria are fulfilled.
The study has some limitations. As mentioned before, due to their low individual detection rate, all non-CDKN2A genes were analyzed as one group, despite their heterogeneity in terms of tumor spectrum, penetrance and possibly other characteristics. Moreover, the limited availability of co-segregation data may have affected the classification of VUS, with a possible underestimation of the detection rate.
While the detection rate obtained by gene panel almost doubled versus CDKN2A testing alone, our data suggest that, when the earliest melanoma occurs over 60 years of age, particularly in spoMPMs with only two CMs and in families with only two affected members, the probability of an underlying PV/LPV in a known melanoma-risk gene is extremely low. Hence, in those cases, before genetic referral, clinicians should consider more common risk factors, e.g. phototype, UV exposure and signs of actinic damage. Conversely, the presence of strong predictors such as at least three melanoma diagnoses or personal/familial presence of pancreatic cancer increased the detection rate up to nearly 20%.
The strength of our study lies in the prospective and consistent recruitment throughout the country, partly due to centralized telecounseling and genetic testing, resulting in the largest Italian cohort of high-risk melanoma cases sequenced by a shared gene panel at the germline level. We updated the number and type of germline mutation status predictors in Italian CM cases, laying the bases for a national revision of genetic assessment criteria.
Acknowledgements
We wish to thank the participating patients and families, whose generosity and cooperation have made this study possible. We acknowledge the contribution of Roberta La Starza (University of Perugia, Italy) and Elena Maccaroni (AOU Ospedali Riuniti di Ancona, Italy) to this study.
Funding
This work was supported by the Italian Melanoma Intergroup (IMI), Emme Rouge (M.B. funded by IMI/Emme Rouge grants), Italian Ministry of Health [grant number RF-2016-02362288 to P.G.] and Ricerca Corrente/5x1000 to IRCCS Ospedale Policlinico San Martino to P.G., Lega Italiana per la Lotta contro I Tumori (LILT) 2019 to P.G. Part of the work was funded by the Fondazione AIRC ‘Programma di ricerca 5 per Mille 2018-ID#21073’ to EPigenetic Immune-oncology Consortium Airc (EPICA) investigators (G.P., P.G.) and institutional funding of IOV-IRCCS (5x1000) [under grant number BIGID219MENI (L.E., C.M.)].
Disclosure
The authors have declared no conflicts of interest.
Supplementary data
References
- 1.Leiter U., Keim U., Garbe C. In: Sunlight, Vitamin D and Skin Cancer. Reichrath J., editor. Vol. 1268. Springer International Publishing; 2020. Epidemiology of Skin Cancer: Update 2019; pp. 123–139. (Advances in Experimental Medicine and Biology). [DOI] [PubMed] [Google Scholar]
- 2.GLOBOCAN 2018. https://gco.iarc.fr/ Available at.
- 3.AIRTUM i numeri del cancro in Italia. https://www.aiom.it/i-numeri-del-cancro-in-italia/ Available at.
- 4.Leachman S.A., Lucero O.M., Sampson J.E., et al. Identification, genetic testing, and management of hereditary melanoma. Cancer Metastasis Rev. 2017;36(1):77–90. doi: 10.1007/s10555-017-9661-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldstein A.M., Chan M., Harland M., et al. High-risk melanoma susceptibility genes and pancreatic cancer, neural system tumors, and uveal melanoma across GenoMEL. Cancer Res. 2006;66(20):9818–9828. doi: 10.1158/0008-5472.CAN-06-0494. [DOI] [PubMed] [Google Scholar]
- 6.Puntervoll H.E., Yang X.R., Vetti H.H., et al. Melanoma prone families with CDK4 germline mutation: phenotypic profile and associations with MC1R variants. J Med Genet. 2013;50(4):264–270. doi: 10.1136/jmedgenet-2012-101455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi J., Yang X.R., Ballew B., et al. Rare missense variants in POT1 predispose to familial cutaneous malignant melanoma. Nat Genet. 2014;46(5):482–486. doi: 10.1038/ng.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walpole S., Pritchard A.L., Cebulla C.M., et al. Comprehensive study of the clinical phenotype of germline BAP1 variant-carrying families worldwide. J Natl Cancer Inst. 2018;110(12):1328–1341. doi: 10.1093/jnci/djy171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horn S., Figl A., Rachakonda P.S., et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339(6122):959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 10.Aoude L.G., Pritchard A.L., Robles-Espinoza C.D., et al. Nonsense mutations in the shelterin complex genes ACD and TERF2IP in familial melanoma. J Natl Cancer Inst. 2015;107(2) doi: 10.1093/jnci/dju408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bertolotto C., Lesueur F., Giuliano S., et al. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature. 2011;480(7375):94–98. doi: 10.1038/nature10539. [DOI] [PubMed] [Google Scholar]
- 12.Pastorino L., Andreotti V., Dalmasso B., et al. Insights into genetic susceptibility to melanoma by Gene panel testing: potential pathogenic variants in ACD, ATM, BAP1, and POT1. Cancers. 2020;12(4):1007. doi: 10.3390/cancers12041007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalmasso B., Pastorino L., Nathan V., et al. Germline ATM variants predispose to melanoma: a joint analysis across the GenoMEL and MelaNostrum consortia. Genet Med. 2021;23(11):2087–2095. doi: 10.1038/s41436-021-01240-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teerlink C.C., Huff C., Stevens J., et al. A nonsynonymous variant in the GOLM1 Gene in cutaneous malignant melanoma. J Natl Cancer Inst. 2018;110(12):1380–1385. doi: 10.1093/jnci/djy058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Artomov M., Stratigos A.J., Kim I., et al. Rare variant, Gene-based association study of hereditary melanoma using whole-exome sequencing. J Natl Cancer Inst. 2017;109(12):7–10. doi: 10.1093/jnci/djx083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aoude L.G., Heitzer E., Johansson P., et al. POLE mutations in families predisposed to cutaneous melanoma. Fam Cancer. 2015;14(4):621–628. doi: 10.1007/s10689-015-9826-8. [DOI] [PubMed] [Google Scholar]
- 17.Christodoulou E., van Doorn R., Visser M., et al. NEK11 as a candidate high-penetrance melanoma susceptibility gene. J Med Genet. 2019;57(3) doi: 10.1136/jmedgenet-2019-106134. [DOI] [PubMed] [Google Scholar]
- 18.Dalmasso B., Ghiorzo P. Evolution of approaches to identify melanoma missing heritability. Expert Rev Mol Diagn. 2020;20(5):523–531. doi: 10.1080/14737159.2020.1738221. [DOI] [PubMed] [Google Scholar]
- 19.Bruno W., Ghiorzo P., Battistuzzi L., et al. Clinical genetic testing for familial melanoma in Italy: a cooperative study. J Am Acad Dermatol. 2009;61(5):775–782. doi: 10.1016/j.jaad.2009.03.039. [DOI] [PubMed] [Google Scholar]
- 20.Bruno W., Pastorino L., Ghiorzo P., et al. Multiple primary melanomas (MPMs) and criteria for genetic assessment: MultiMEL, a multicenter study of the Italian Melanoma Intergroup. J Am Acad Dermatol. 2016;74(2):325–332. doi: 10.1016/j.jaad.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 21.Casula M., Paliogiannis P., Ayala F., et al. Germline and somatic mutations in patients with multiple primary melanomas: a next generation sequencing study. BMC Cancer. 2019;19(1):772. doi: 10.1186/s12885-019-5984-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pellegrini C., Maturo M.G., Martorelli C., et al. Characterization of melanoma susceptibility genes in high-risk patients from Central Italy. Melanoma Res. 2017;27(3):258–267. doi: 10.1097/CMR.0000000000000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Simone P., Bottillo I., Valiante M., et al. A Single center retrospective review of patients from central Italy tested for melanoma predisposition genes. Int J Mol Sci. 2020;21(24):9432. doi: 10.3390/ijms21249432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dika E., Patrizi A., Rossi C., et al. Clinical histopathological features and CDKN2A/CDK4/MITF mutational status of patients with multiple primary melanomas from Bologna: Italy is a fascinating but complex mosaic. Ital J Dermatol Venereol. 2021;156(5):599–605. doi: 10.23736/S2784-8671.20.06496-2. [DOI] [PubMed] [Google Scholar]
- 25.Mantelli M., Barile M., Ciotti P., et al. High prevalence of the G101W germline mutation in theCDKN2A (P16ink4a) gene in 62 Italian malignant melanoma families. Am J Med Genet. 2002;107(3):214–221. [PubMed] [Google Scholar]
- 26.Mantelli M., Pastorino L., Ghiorzo P., et al. Early onset may predict G101W CDKN2A founder mutation carrier status in Ligurian melanoma patients. Melanoma Res. 2004;14(6):443–448. doi: 10.1097/00008390-200412000-00002. [DOI] [PubMed] [Google Scholar]
- 27.Ghiorzo P., Pastorino L., Bonelli L., et al. INK4/ARF germline alterations in pancreatic cancer patients. Ann Oncol. 2004;15(1):70–78. doi: 10.1093/annonc/mdg498. [DOI] [PubMed] [Google Scholar]
- 28.Fargnoli M.C., Chimenti S., Peris K., et al. CDKN2a/p16INK4a mutations and lack of p19ARF involvement in familial melanoma kindreds. J Invest Dermatol. 1998;111(6):1202–1206. doi: 10.1046/j.1523-1747.1998.00412.x. [DOI] [PubMed] [Google Scholar]
- 29.Ghiorzo P., Gargiulo S., Pastorino L., et al. Impact of E27X, a novel CDKN2A germ line mutation, on p16 and p14ARF expression in Italian melanoma families displaying pancreatic cancer and neuroblastoma. Hum Mol Genet. 2006;15(18):2682–2689. doi: 10.1093/hmg/ddl199. [DOI] [PubMed] [Google Scholar]
- 30.Menin C., Vecchiato A., Scaini M.C., et al. Contribution of susceptibility gene variants to melanoma risk in families from the Veneto region of Italy [Letter to the Editor] Pigment Cell Melanoma Res. 2011;24(4):728–730. doi: 10.1111/j.1755-148X.2011.00876.x. [DOI] [PubMed] [Google Scholar]
- 31.Kimura H., Paranal R.M., Nanda N., et al. Functional CDKN2A assay identifies frequent deleterious alleles misclassified as variants of uncertain significance. eLife. 2022;11 doi: 10.7554/eLife.71137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pissa M., Helkkula T., Appelqvist F., et al. CDKN2A genetic testing in melanoma-prone families in Sweden in the years 2015-2020: implications for novel national recommendations. Acta Oncol. 2021;60(7):888–896. doi: 10.1080/0284186X.2021.1914346. [DOI] [PubMed] [Google Scholar]
- 33.Holland E.A., Lo S., Kelly B., et al. FRAMe: familial risk assessment of melanoma—a risk prediction tool to guide CDKN2A germline mutation testing in Australian familial melanoma. Fam Cancer. 2021;20(3):231–239. doi: 10.1007/s10689-020-00209-x. [DOI] [PubMed] [Google Scholar]
- 34.Taylor N.J., Mitra N., Qian L., et al. Estimating CDKN2A mutation carrier probability among global familial melanoma cases using GenoMELPREDICT. J Am Acad Dermatol. 2019;81(2):386–394. doi: 10.1016/j.jaad.2019.01.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Potjer T.P., Helgadottir H., Leenheer M., et al. CM-Score: a validated scoring system to predict CDKN2A germline mutations in melanoma families from Northern Europe. J Med Genet. 2018;55(10):661–668. doi: 10.1136/jmedgenet-2017-105205. [DOI] [PubMed] [Google Scholar]
- 36.Lesueur F., Easton D.F., Renault A.L., et al. First international workshop of the ATM and cancer risk group (4-5 December 2019) Fam Cancer. 2021;21(2):211–227. doi: 10.1007/s10689-021-00248-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ciccarese G., Dalmasso B., Bruno W., et al. Clinical, pathological and dermoscopic phenotype of MITF p.E318K carrier cutaneous melanoma patients. J Transl Med. 2020;18(1):78. doi: 10.1186/s12967-020-02253-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.