Abstract
Deceptive pollination often involves volatile organic compound emissions that mislead insects into performing nonrewarding pollination. Among deceptively pollinated plants, Arum maculatum is particularly well-known for its potent dung-like volatile organic compound emissions and specialized floral chamber, which traps pollinators—mainly Psychoda phalaenoides and Psychoda grisescens—overnight. However, little is known about the genes underlying the production of many Arum maculatum volatile organic compounds, and their influence on variation in pollinator attraction rates. Therefore, we performed de novo transcriptome sequencing of Arum maculatum appendix and male floret tissue collected during anthesis and postanthesis, from 10 natural populations across Europe. These RNA-seq data were paired with gas chromatography–mass spectrometry analyses of floral scent composition and pollinator data collected from the same inflorescences. Differential expression analyses revealed candidate transcripts in appendix tissue linked to malodourous volatile organic compounds including indole, p-cresol, and 2-heptanone. In addition, we found that terpene synthase expression in male floret tissue during anthesis significantly covaried with sex- and species-specific attraction of Psychoda phalaenoides and Psychoda grisescens. Taken together, our results provide the first insights into molecular mechanisms underlying pollinator attraction patterns in Arum maculatum and highlight floral chamber sesquiterpene (e.g. bicyclogermacrene) synthases as interesting candidate genes for further study.
Keywords: Chemical ecology, GC–MS, molecular ecology, plant-pollinator interactions, RNA-seq, sesquiterpenes
Introduction
Angiosperms have evolved to produce a wide array of specialized metabolites to mediate their interactions with other organisms and their environment, including volatile organic compounds (VOCs) emitted from flowers, leaves, fruits, and roots. While VOCs represent a relatively small proportion of all plant metabolomic diversity (Knudsen and Gershenzon 2020), they play important functional roles in defence against predators, pathogens, and abiotic stresses (Pichersky and Gershenzon 2002; Holopainen 2004; Irwin et al. 2004) and as cues for pollinator attraction (Pellmyr and Thien 1986; Knudsen and Tollsten 1993; Whitehead and Peakall 2009; Junker and Parachnowitsch 2015). Pollination, particularly by insects, appears to be a major driver of angiosperm diversification (Van der Niet and Johnson 2012; Schiestl and Johnson 2013; Hernández-Hernández and Wiens 2020). Recent research has hypothesized and partially demonstrated that floral trait diversity is the result of complex interactions between plant genomic diversity among populations, pollinator network composition, and environmental conditions—all of which vary through space and time (Thompson et al. 2013, 2017; Friberg et al. 2019). Large molecular and ecological datasets, with wide spatial and temporal coverage, are therefore still needed in order to further our understanding of the evolution of key traits such as floral scent.
Pollinator-mediated selection is known to influence floral morphology (Bröderbauer et al. 2013; Gervasi and Schiestl 2017) and colour (Schemske and Bradshaw 1999; Newman et al. 2012; Trunschke et al. 2021), and recent studies have also identified strong evidence for variation in VOC emissions driven by pollinator preferences. Notably, comparative community ecology data and gas chromatography–mass spectrometry (GC–MS) have been combined to identify convergent VOC bouquets in unrelated plant species with similar pollinators (Fenster et al. 2004; Schiestl and Johnson 2013; Junker and Parachnowitsch 2015), and divergent VOC bouquets in related species with different pollinators (Dobson et al. 1997; Urru et al. 2010; Byers et al. 2014; Friberg et al. 2019). Moreover, the discovery of biochemical pathways underlying floral VOCs has greatly accelerated by next-generation sequencing technologies such as mRNA sequencing (Dhandapani et al. 2017; Wong, Pichersky, et al. 2017; Xiao et al. 2019). Here, we aimed to combine these techniques, in order to understand how pollinator-mediated selection influences transcriptomic variation underlying floral scent divergence among populations of deceptively pollinated Arum maculatum L. (Araceae).
A. maculatum is a common European woodland flower with a long history of study, due to its deceptive pollination strategy which involves olfactory deception through brood-site mimicry (Schmucker 1925; Knoll 1926; Vogel 1965; Lack and Diaz 1991). Specifically, A. maculatum emit VOCs (e.g. 2-heptanone, indole, and p-cresol) during anthesis which are also present in cow manure (Kite 1995) and decomposing organic material (Gfrerer et al. 2021, 2022): the breeding substrates of Psychoda phalaenoides L. and Psychoda (Psycha) grisescens (Tonnoir) (Espíndola et al. 2011; Szenteczki et al. 2021). These 2 dipteran (Psychodidae) moth flies are trapped at varying frequencies across the geographic distribution of A. maculatum (Espíndola et al. 2011; Szenteczki et al. 2021; Gfrerer et al. 2021). Geographic variation in A. maculatum floral scent has also been observed, first in England (Kite et al. 1998; Diaz and Kite 2002) and France (Chartier et al. 2011, 2013), and recently, across most of the species distribution range (Gfrerer et al. 2021; Szenteczki et al. 2021), and may be linked to variation in pollinator attraction (Gfrerer et al. 2021; Szenteczki et al. 2021).
A. maculatum VOC emissions during anthesis are highly variable, and also include several sesquiterpenes and other aliphatic, aromatic, monoterpene, and nitrogen-containing minor compounds which may also contribute to pollinator attraction (Kite 1995; Gfrerer et al. 2021; Szenteczki et al. 2021). Intra- and interpopulation VOC variation mainly centers around variation in the proportion of indole to terpenes (Chartier et al. 2013; Szenteczki et al. 2021), which are respectively produced by aromatic amino acid (AAA; i.e. phenylalanine, tyrosine, and tryptophan) metabolism and terpene synthases (TPSs) via the mevalonate or methylerythritol 4-phosphate (MEP) pathways. Experiments in England with scented bait traps have shown that indole, 2-heptanone, and p-cresol are attractive to P. phalaenoides (Kite et al. 1998). Large-scale field studies have further demonstrated that sesquiterpenes may also play a role in differential attraction of P. phalaenoides and P. grisescens (Szenteczki et al. 2021) and influence fruit set size (Gfrerer et al. 2021).
Another recognizable feature of A. maculatum is its inflorescence morphology. Like other monoecious Araceae, A. maculatum produce densely clustered, unisexual male and female florets arranged along a central spike (the spadix), which is surrounded by a leaf-like bract (the spathe). The spathe surrounds the fertile flowers, creating a basal trap chamber with only a narrow opening at the top of the chamber, which is surrounded by a ring of hair-like sterile flowers. The scent of the floral chamber during anthesis appears to be dominated by 1 sesquiterpene, bicyclogermacrene (Kite et al. 1998), which was previously misidentified as germacrene B (Kite 1995). The purple colour of the appendix (the sterile apex of the spadix) and stamens is due to anthocyanin pigments (Williams et al. 1981). Purple pigmentation in the appendix of Araceae appears to be attractive to Sphaeroceridae pollinators (Roháček et al. 1990; Angioy et al. 2004), but it is not currently known whether it is also attractive to Psychodidae, the main pollinators of A. maculatum. Tradeoffs between pigmentation and VOCs could also exist; however these would be limited to benzenoids (e.g. p-cresol and skatole) and not terpenes, which form a proportionally larger part of the floral scent bouquet in A. maculatum (Chartier et al. 2016).
The A. maculatum reproductive cycle takes place over 2 days (reviewed in Gibernau 2004): on the evening of the first day, the appendix is thermogenic, heating up to 15°C above ambient air temperature. Pollinators are attracted by VOCs emitted by the appendix, and fall into the trap chamber, where they crawl over the male and female florets. No rewards are provided to pollinators within the chamber. A small amount of stigmatic secretions have been observed, but the low (9–12.5%) sucrose content suggests that this fluid is not nectar (Lack and Diaz 1991), and more likely is produced to collect pollen grains (Paiva et al. 2021). The following morning, after the female florets are no longer receptive, pollen is released onto the trapped pollinators. Finally, pollinators are able to escape later in the day, as the sterile hairs at the opening of the trap chamber begin to wither.
In the closely related Arum italicum, sesquiterpene biosynthesis appears to begin in the male florets several days prior to anthesis, but the full range of characteristic floral VOCs such as p-cresol and skatole are only detected on the day of anthesis, in appendix tissue (Leguet et al. 2014). To date, the biosynthetic pathways underlying the production of p-cresol and skatole have only been characterized in bacteria (Selmer and Andrei 2001; Liu et al. 2018), and it is unclear whether A. maculatum and other angiosperms use similar AAA fermentation pathways to produce these VOCs. Furthermore, tissue-specific RNA-seq of Arum concinnatum identified more diverse transcript expression in male floret tissue on the day of anthesis rather than in the appendix (Onda et al. 2015), even though A. concinnatum also emits dung-like VOCs from its appendix (Urru et al. 2010). Consequently, many questions remain regarding the specific biosynthetic pathways underlying A. maculatum VOCs, the localization of their expression, and whether mRNA expression related to VOC production varies with geographic distance or pollinator community composition.
Here, we aim to address each of the aforementioned gaps in our knowledge. Our specific objectives were to: (1) investigate variation in A. maculatum transcript expression between appendix and male floret tissue during anthesis, (2) use these results to identify differentially expressed transcripts putatively related to VOC biosynthesis, and (3) to characterize and compare differential expression in these transcripts across Europe, with a focus on populations with divergent pollinator attraction patterns. We predicted that during anthesis, AAA metabolism would be highly expressed in appendix tissue, while sesquiterpene synthases would be highly expressed in male floret tissue. We further expected to observe differential expression of transcripts associated with AAA metabolism and TPSs in inflorescences with dung-like (i.e. indole, 2-heptanone, and p-cresol) vs. sesquiterpene-dominated floral bouquets, respectively. Finally, given that the A. maculatum population in Forêt du Gâvre, France, appears to attract almost exclusively P. grisescens (Espíndola et al. 2011; Szenteczki et al. 2021), we predicted that putative locally adapted transcripts associated with particular VOCs may exist within this population. To test these predictions, we surveyed VOC variation, pollinator attraction, and mRNA expression in both appendix and male floret tissue, across most of the A. maculatum species distribution range.
Materials and methods
Sampling design
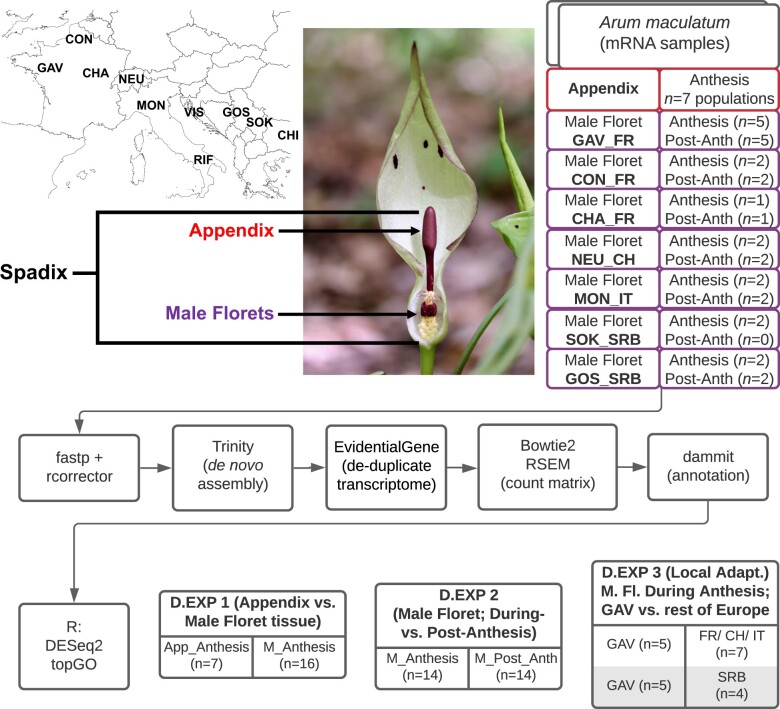
We sampled 10 natural populations of A. maculatum between April and May 2019 (Fig. 1): 3 in France (Forêt du Gâvre, Conteville, and Chaumont), 1 in Switzerland (Neuchâtel), 2 in Italy (Montese and Rifreddo), 1 in Croatia (Visuć), 2 in Serbia (Gostilje and Sokobanja), and 1 in Bulgaria (Chiflik). Full details on sample data and population coordinates are given in Supplementary Table 1.
Fig. 1.
Overview of the sampling design, bioinformatic pipeline, and differential expression analyses in this study. Inset photo: A. maculatum inflorescence, with the lower spathe chamber dissected to reveal the male and female florets. Inset map: population codes, placed over their approximate locations.
Before collecting tissue samples, we noninvasively sampled the dynamic headspace VOCs emitted by each A. maculatum inflorescence, following the methods detailed in (Szenteczki et al. 2021). Briefly, this involved placing polydimethylsiloxane coated Twister stir bars (Gerstel) in glass tubes near the appendices of A. maculatum inflorescences on the evening of the day of anthesis (18:00–19:00), and pumping air over them for 30 min at an air flow rate of 200 ml/min. All samples were analyzed using a gas chromatography (Agilent 7890a) mass spectrometry (Agilent 5975c) system. Desorption of the VOCs was carried out in a thermal desorption unit (TDU, Gerstel) with an initial temperature of 50°C, followed by a ramp of 60°C/min until 250°C (followed by a 3.5-min hold time). Before injection, the VOCs were cryo-focused (CIS, Gerstel) at −80°C, then released with a ramp of 12°C/min until 260°C (hold time 6 min). PTV solvent vent at 14 PSI and 50 ml/min flow was used for injection into a HP5-MS column in constant flow mode (0.9 ml/min) with helium as the carrier gas. The Mass Selective Detector (MSD) transfer line was set at 280°C. Scan mode was used for the MS acquisition with a solvent delay (1 min), over a mass range of 33–300 m/z; MS source and quadrupole temperatures were set at 230 and 150°C, respectively. Electron ionization (70 eV) was used. Major ions were recorded for each integrated peak using Agilent Chemstation, and compound identifications were derived from NIST 2.3 (library v.17) and published Kovats retention indices; all names used in our analyses should therefore be considered hypotheses. Appendix temperatures were recorded at the time of VOC sampling using FLIR thermographic imaging, and ambient air temperature was recorded using a thermistor.
Immediately following VOC sampling (i.e. during anthesis), we used sterile scalpel blades to collect 1-mm thick slices of appendix and/or male floret tissue. As a control, male floret tissue samples were also collected from inflorescences the following morning, after pollinator attraction had ended. All samples were individually preserved in RNAlater (Thermo Fischer Scientific) and stored at −80°C until extraction. After collecting each during-anthesis male floret tissue sample, we closed and sealed the small window we cut in floral chamber, so that pollinator trapping was not affected. During the morning following anthesis, we collected all insects trapped within inflorescences, preserved them in 70% ethanol and identified them to at least the suborder level. Psychodidae were further identified to species level (including sex) following taxonomic descriptions and illustrations (Ježek 1990).
RNA-seq library preparation
All samples and tissue types were extracted using identical methods; approximately 100 mg of tissue was rinsed in sterile RNase-free water, flash frozen in liquid nitrogen, powdered in a QIAGEN TissueLyser II, and immediately extracted using a QIAGEN RNeasy Plant Mini Kit, following the manufacturer’s protocol. Extraction quality and concentration were verified using an Agilent Fragment Analyzer. Three Illumina TruSeq Stranded mRNA polyA libraries were prepared from these samples, following the manufacturer’s recommended protocol. The first library contained the appendix RNA samples, and the second and third each contained a mix of during anthesis and postanthesis male floret tissue samples from all populations. Each library was then sequenced on its own lane (i.e. 3 lanes in total) using an Illumina HiSeq 4000 (150-bp paired-end sequencing) at the Lausanne Genomic Technologies Facility (Switzerland).
Preprocessing raw read data
We performed an initial quality filtering of all raw read files using fastp (Chen et al. 2018) to remove adapter sequences and polyA tails, trim reads with phred quality values below 30, and remove reads with >1 “N” bases. Then, we used rCorrector (Song and Florea 2015) to remove reads containing erroneous k-mers (25-mers). Finally, we discarded unfixable read pairs following quality filtering with FilterUncorrectablePEfastq.py (https://github.com/harvardinformatics/TranscriptomeAssemblyTools).
De novo transcriptome assembly, annotation, and expression quantification
We merged all of the quality filtered reads and assembled a single de novo reference A. maculatum transcriptome (i.e. from all samples, tissue types, and populations) using Trinity v2.11.0 (Grabherr et al. 2011) with in silico normalization, a k-mer size of 25, minimum contig size of 200 bp, and strand-specific assembly enabled. Then, we tested 2 methods to filter out redundant transcripts with poor coding potential (i.e. minor isoforms resulting from incomplete assembly, sequencing errors, and/or heterozygosity introduced by our wide geographic sampling range): (1) CD-HIT (Li and Godzik 2006; Fu et al. 2012) clustering at 95 % identity with a word size of 5 and (2) the tr2aacds v4 pipeline from EvidentialGene (Gilbert 2019), which classifies sequences into primary, alternate, and redundant sets based on coding sequence length and similarity. When using EvidentialGene, we kept only the primary transcripts, removing the alternate and redundant isoforms. We then calculated assembly scores and assessed transcriptome completeness of both filtering methods with BUSCO v5.0.0, using the Embryophyta_odb10 dataset (Seppey et al. 2019).
Next, we used dammit v1.2 (Scott et al. 2019) to annotate our deduplicated assembly. This pipeline uses Transdecoder (https://github.com/TransDecoder/TransDecoder) to identify candidate coding regions and searches the OrthoDB, Pfam-A, Rfam, and uniref90 protein databases for transcript annotations, with an E-value threshold of 1 × 10−5. Unannotated transcripts were not removed from the final A. maculatum reference transcriptome. We also uploaded the EvidentialGene output to the Kyoto Encyclopedia of Genes and Genomes (KEGG) Automatic Annotation Server (Moriya et al. 2007) to generate KEGG Orthology (KO) annotations and pathway maps. Finally, we mapped the cleaned raw reads of each sample to our annotated reference transcriptome using Bowtie2 v2.3.5.1 (Langmead and Salzberg 2012) and quantified transcript expression using RSEM v1.3.2 (Li and Dewey 2011).
Differential expression analyses
RSEM outputs were imported into R v.4.1.0 (R Core Team 2021) using tximport 1.20 (Soneson et al. 2015). Transcripts expressed below 0.75 counts per million (CPM) in at least 7 samples (i.e. the smallest sample size for a group in our analyses) were then filtered out prior to subsequent analyses in in DESeq2 1.3.2 (Love et al. 2014), using the apeglm log2fold change shrinkage estimation algorithm (Zhu et al. 2019). Transcripts with False Discovery Ratio (FDR)-corrected P-values <0.05 and log2fold changes greater than 1.0 or less than −1.0 were considered as significantly differentially expressed among groups. We further identified significantly enriched gene sets in each tissue type using Gene Ontology (GO) term analysis with the R package TopGO (Alexa and Rahnenfuhrer 2010) and visualized the result using the R package rrvgo 1.4 (Sayols 2020), an implementation of the semantic similarity-based GO summary tool REViGO (Supek et al. 2011).
We performed 3 sets of differential expression analyses, each addressing one of the main aims of our study. Our first comparison was between appendix (n = 7) and male floret (n = 16) tissue collected during anthesis (i.e. at the same phenological time point). Second, we compared male floret tissue collected during anthesis (n = 14) against their paired control samples from the morning following anthesis (n = 14). Paired control samples were not collected for 2 inflorescences from Sokobanja, Serbia; these individuals were excluded from this analysis. Finally, we compared transcript expression in during-anthesis male floret tissue sampled in the Forêt du Gâvre population against all other populations, to identify transcripts putatively linked to this population’s exclusive attraction of P. grisescens. To account for some of the effects of isolation by distance, we further split this analysis along the 2 main neutral genetic clusters (Espíndola and Alvarez 2011): specifically, we compared during-anthesis male floret tissue from Forêt du Gâvre (n = 5) against during-anthesis male floret tissue from (1) France, Switzerland, and Italy (n = 7) and (2) Serbia (n = 4).
Identifying transcripts underlying VOC biosynthesis
Building on the results of our differential expression analyses, we characterized and compared the expression of metabolic pathways underlying key A. maculatum VOCs known to be involved in the attraction of Psychodidae pollinators (Kite et al. 1998): indole, p-cresol, 2-heptanone, and sesquiterpenes. First, we reconstructed entire metabolic pathways using automated annotations, GO terms, and KEGG pathway maps generated from (1) our complete reference transcriptome and (2) sets of significantly differentially expressed genes. Since the genes responsible for the production of 2-heptanone and p-cresol in A. maculatum are unclear and unknown respectively, we also used homology searches to identify additional candidate genes. Here, we performed BLASTp searches of genes known involved in the biosynthesis of the aforementioned VOCs in other plants and bacteria, against a database of all of our Transdecoder predicted peptide sequences using, with an e-value cutoff of 1 × 10−5. After all candidate transcripts were identified, we visualized variation in their average expression across all tissue types and populations using the R package pheatmap (Kolde 2019).
Coinertia analysis—TPSs correlated with VOCs and pollinator attraction
We extracted the coding sequences and DESeq2 normalized expression of all TPSs in our transcriptome (i.e. using uniref annotations, GO/KO terms, and PFAM domains), performed a multiple sequence alignment of these sequences using Clustal Omega (Sievers et al. 2011), created a Maximum Likelihood phylogeny using automatic substitution model selection and default parameters in IQ-TREE (Nguyen et al. 2015), and visualized the result using iTOL v5 (Letunic and Bork 2021). Then, we investigated whether TPS expression was correlated with (1) total terpene emissions during anthesis or (2) the composition (i.e. sex and/or species) of trapped pollinators within inflorescences. Specifically, we performed coinertia analysis using the R packages vegan (Oksanen et al. 2020), RVAideMemoire (Hervé 2021), and ade4 (Thioulouse et al. 2018), comparing the TPS expression matrix produced above to corresponding matrices of (1) proportional emissions of terpene VOCs and (2) proportions of Psychodidae species trapped by the same A. maculatum inflorescences. Prior to this analysis, the gene-level TPS expression matrix was log-transformed and scaled, while the VOC and pollinator matrices were centered log ratio transformed and scaled.
Results
Assembly and annotation quality
Between 27,933,352 and 66,825,898 (median: 40,354,964) paired-end reads from individual tissue samples passed all quality filtering steps (Supplementary Table 1). These were subsequently assembled into 273,346 Trinity genes, comprising 593,392 transcript isoforms. Trinity quality statistics (e.g. N50 of 1,699 bp) are given in Supplementary Table 2. EvidentialGene filtered out a larger number of transcripts from this raw assembly (Supplementary Table 3) while maintaining a higher BUSCO scores than CD-HIT clustering (e.g. 95.6% complete vs. 92.2% complete; full result in Supplementary Table 4). The EvidentialGene output was therefore annotated and used for all subsequent analyses. A total of 18,411 unique annotations were generated. A total of 17,435 annotations mapped to Eukaryota (94.7%); 12,676 of these annotations further mapped to Mesangiospermae (68.9%); a relatively small number of annotations mapped to bacterial genes (692 results; 3.8%).
A total of 49,779 transcripts remained in our expression matrix following prefiltering in DESeq2 (i.e. >0.75 CPM in at least 7 samples). Principal components analysis of filtered transcript expression (Supplementary Fig. 1) revealed highly divergent transcript expression among male floret and appendix tissue during anthesis along the first PCA axis. Along the second PCA axis, samples were further divided between 2 main geographic regions—namely, northern populations (i.e. France, Switzerland, and northern Italy) and Balkan populations (i.e. Croatia, Serbia, and Bulgaria). This regional split is consistent with the 2 main genetic clusters identified using neutral (AFLP) markers (Espíndola and Alvarez 2011).
Differential transcript expression between appendix and male floret tissue
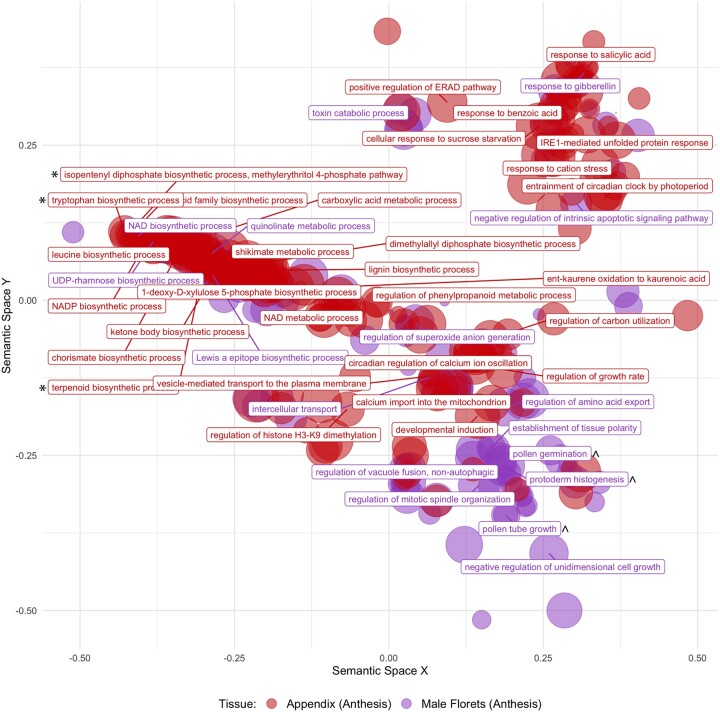
Differential expression analysis comparing appendix and male floret tissue (both collected during anthesis) revealed 8,683 transcripts with significantly greater expression in appendix tissue and 6,581 transcripts with significantly greater expression in male floret tissue (FDR-corrected P-values <0.05; log2fold change ±1), after controlling for population effects. Furthermore, we found that VOC biosynthetic activity is significantly elevated in A. maculatum appendix tissue during anthesis. Many major biosynthetic pathways (e.g. tryptophan/indole synthesis, terpene synthesis, and phenylpropanoid synthesis) were significantly enriched in the appendix tissue during anthesis, whereas transcripts related to pollen production were significantly enriched in male floret tissue (Fig. 2).
Fig. 2.
Significantly enriched GO terms when comparing A. maculatum appendix (n = 7) and male floret (n = 16) transcript expression during anthesis. Nonredundant GO terms are visualized in semantic similarity space (allowed similarity = 0.8); the full list of GO terms represented above is available in Supplementary Appendix 2. [*] indicates parent groups linked to the biosynthesis of volatile compounds and [^] indicates parent groups linked to pollen production
Differential transcript expression in male florets during anthesis vs. postanthesis
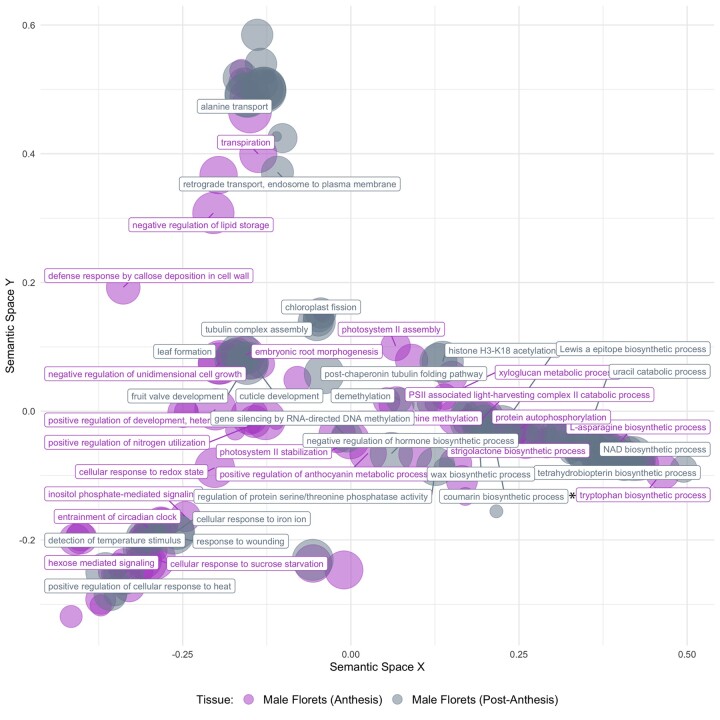
When comparing during-anthesis and postanthesis male floret tissue samples in a differential expression analysis that incorporated the paired nature of these samples, we identified 3,847 transcripts with significantly greater expression during anthesis and 2,920 transcripts with significantly greater expression postanthesis (FDR-corrected P-values <0.05; log2fold change ±1). While tryptophan (i.e. indole) biosynthesis was elevated in male florets during anthesis, we did not identify increased expression of other putative VOC biosynthetic pathways (Fig. 3). The full list of significantly enriched GO terms from both our first and second set of differential expression analyses are available in Supplementary Appendix 2.
Fig. 3.
Significantly enriched GO terms when comparing A. maculatum transcript expression in male floret tissue during anthesis (n = 14), vs. approximately 18 h after anthesis (n = 14). Nonredundant GO terms are visualized in semantic similarity space (allowed similarity = 0.8); the full list of GO terms represented above is available in Supplementary Appendix 2. [*] indicates parent groups linked to the biosynthesis of volatile compounds
Differential transcript expression associated with exclusive attraction of P. grisescens
Our third and final set of differential expression analyses, comparing male floret tissue during anthesis among populations, revealed that (1) 84 and 9 transcripts were, respectively, differentially expressed in Forêt du Gâvre vs. all other French, Swiss, and northern Italian populations (FDR-corrected P-value <0.05; log2fold change ±1) and (2) 327 and 175 transcripts were, respectively, differentially expressed in Forêt du Gâvre vs. Serbian populations (FDR-corrected P-value <0.05; log2fold change ±1). However, no transcripts putatively linked to VOC biosynthesis were identified among these transcripts.
Candidate genes underlying A. maculatum floral VOCs
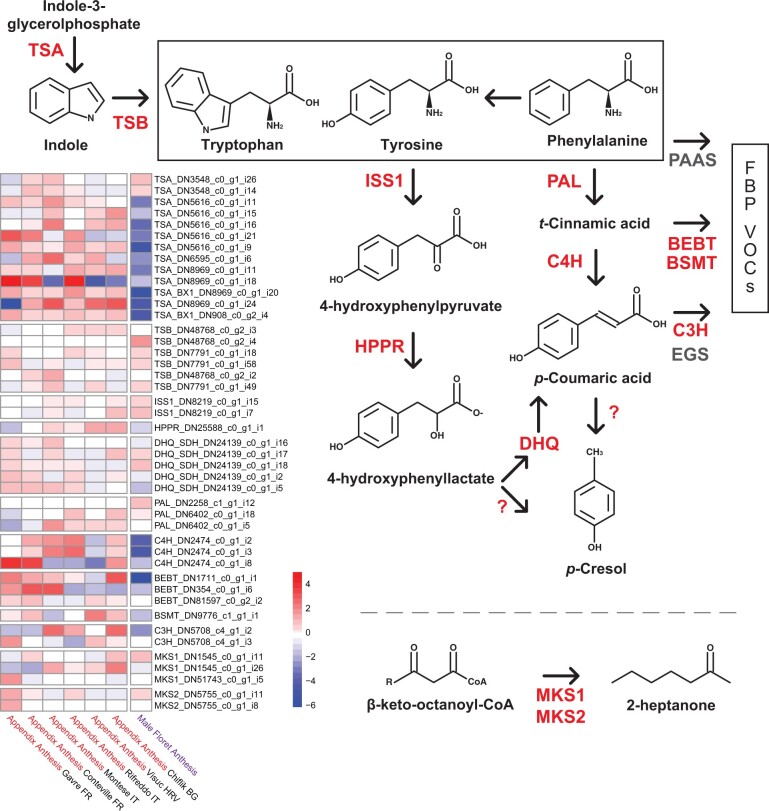
Consistent with recent large-scale surveys of A. maculatum floral scent (Gfrerer et al. 2021; Szenteczki et al. 2021), we observed considerable within-population variation in VOC bouquet composition among our samples (Supplementary Fig. 2 and Supplementary Table 5). We further identified candidate genes putatively linked to the biosynthesis of several dung-mimicking A. maculatum VOCs. First, we identified transcripts in both appendix and male floret tissue homologous with tryptophan synthase alpha (TSA) subunit, which catalyzes the conversion of indole-3-glycerolphosphate to indole.
Second, we identified homologs of 2 proteins, which catalyze the degradation of tyrosine to 4-hydroxyphenyllactate: aromatic aminotransferase (ISS1) and hydroxyphenylpyruvate reductase (HPPR). Pearson correlation tests further identified several transcripts correlated with HPPR expression in appendix tissue during anthesis (Supplementary Table 6); notably, this included a putative dehydratase/shikimate dehydrogenase (Pearson r = 0.988, P < 0.001). This enzyme may further catalyze the conversion of 4-hydroxyphenyllactate to p-coumaric acid. However, we did not identify any homologs of bacterial proteins linked to the production of 4-hydroxyphenylacetate and/or p-cresol (Saito et al. 2018), such as hydroxyphenylacetate decarboxylase.
Third, while phenylalanine-derived precursor molecules for several common floral benzenoid/phenylpropanoid (FBP) compounds—particularly p-coumaric acid—appear to be produced by A. maculatum, transcripts responsible for the production of benzenoid and phenylpropanoid VOCs generally appeared to be absent or weakly expressed in A. maculatum. Finally, we identified putative homologs of methylketone synthases (MKS1 and MKS2), which are known to be involved in 2-heptanone biosynthesis (Ben-Israel et al. 2009; Khuat et al. 2019). Heatmaps of the expression patterns for all of the aforementioned candidate transcripts are shown in Fig. 4.
Fig. 4.
A simplified overview of key proteins involved in producing aromatic amino acid-derived A. maculatum VOCs, and a heatmap of their vst-transformed expression during anthesis. Color scale represents whether expression in a given group (i.e. tissue type and population) is above or below the transcript’s mean expression across all samples. Genes in gray are absent from the A. maculatum transcriptome. Gene names: BEBT = benzyl alcohol O-benzoyltransferase; BSMT, benzoic acid/salicylic acid carboxyl methyltransferase; DHQ, 3-dehydroquinate dehydratase/shikimate dehydrogenase; EGS, eugenol synthase; MKS, methylketone synthase; PAAS, phenylacetaldehyde synthase; TSA/B, tryptophan synthase alpha/beta subunit; ?, unknown gene(s).
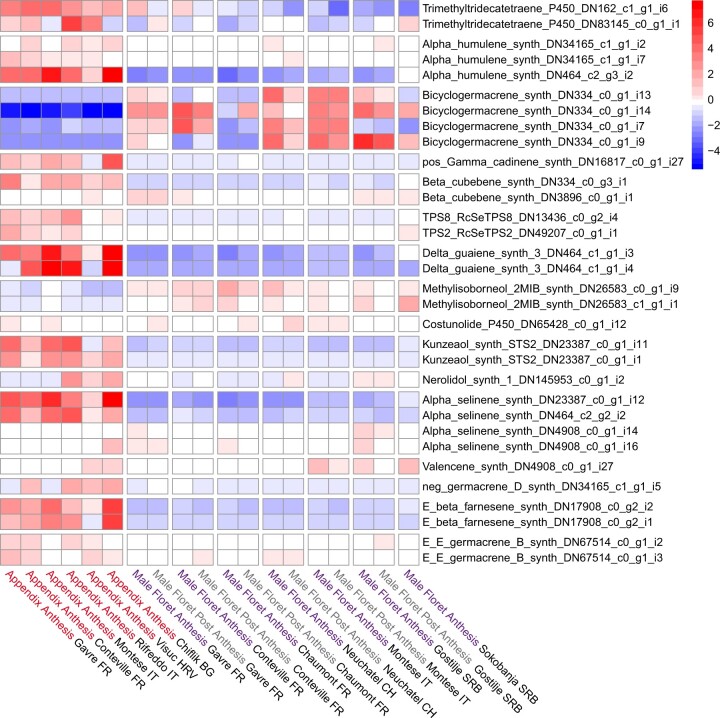
We identified A. maculatum transcripts putatively encoding all proteins in both the mevalonate and MEP pathways. During anthesis, the MEP pathway appears to be more highly expressed than the mevalonate pathway, particularly in appendix tissue (Supplementary Fig. 3). The first enzyme of the MEP pathway, 1-deoxy-d-xylulose-5-phosphate synthase (DXS), which was highly expressed in appendix tissue, also featured the greatest isoform diversity among all terpene backbone synthesis genes. Furthermore, we identified 108 putative TPS transcripts expressed in appendix and male floret tissue during anthesis. A phylogeny of all putative A. maculatum TPSs is given in Supplementary Fig. 4, and expression patterns of TPS transcripts that passed our initial CPM filtering threshold are visualized in Fig. 5.
Fig. 5.
Heatmap of vst-transformed expression of putative terpene synthases in A. maculatum. Colour scale represents whether expression in a given group (i.e. tissue type, stage of anthesis, and population) is above or below the transcript’s mean expression across all samples.
Some of the putative TPSs we identified are homologs of proteins known to produce common A. maculatum VOCs (e.g. humulenes and germacrenes); others, such as 2-methylisoborneol (2-MIB) synthase, catalyze the production of terpenes not previously described in A. maculatum. However, in the case of the latter, the transcript we identified likely encodes a novel TPS, given that we identified an uncharacterized Colocasia esculenta (Araceae) gene, which shares 92.5% identity with our transcript, as opposed to approximately 22.8% identity with the bacterial 2-MIB synthase (Supplementary Fig. 5). Transcripts annotated as trimethyltridecatetraene synthase (Cyt P450 92C6) may represent another such novel TPS. This gene was the only putative VOC synthase we identified that was significantly overexpressed (FDR-corrected P-values <0.05; log2fold change >1) in appendix tissue samples emitting high quantities of an unnamed sesquiterpene with a nonpolar Kovats Retention Index of 1681, which is known to be a strong predictor of P. grisescens attraction (Szenteczki et al. 2021).
Male floret TPS transcript expression is correlated with pollinator attraction
Coinertia analyses did not identify covariation between putative TPS expression in male floret tissue, and proportional emissions of terpenes in VOC bouquets (P = 0.964, Monte-Carlo test, 999 replicates). However, we did identify significant concordance between putative TPS expression in male floret tissue and Psychodidae pollinator composition trapped within inflorescences (P = 0.047, Monte-Carlo test, 999 replicates); the first coinertia axis split TPS expression based on species (i.e. P. grisescens vs. P. phalaenoides), while the second coinertia axis split TPS expression based on sex (Supplementary Fig. 6). Consequently, while our coinertia analyses could not disentangle covariation in TPS expression and specific terpene VOC emissions, it appears that TPS expression in male florets is correlated with sex- and/or species-specific attraction of Psychodidae.
Discussion
High-throughput transcriptome sequencing is rapidly advancing our understanding of the biosynthesis of floral volatile compounds. In this study, we were able to overcome some of the challenges associated with studying the genes underlying VOC biosynthesis in nonmodel systems (Wong, Pichersky, et al. 2017), by surveying transcript expression variation in multiple VOC-emitting floral tissues, across the Europe-wide distribution range of A. maculatum (Fig. 1). This allowed us to identify (1) elevated VOC biosynthetic activity in the appendix of A. maculatum during anthesis (Fig. 2), (2) candidate transcripts for the production of several VOCs such as p-cresol, 2-heptanone, and sesquiterpenes (Figs. 4 and 5), and (3) covariation between male floret TPS expression and the relative proportions of P. phalaenoides and P. grisescens trapped within inflorescences (Supplementary Fig. 6).
Differential expression analyses
During anthesis, diverse VOC metabolic processes were significantly enriched in A. maculatum appendix tissue when compared against male floret tissue. This result is consistent with previous research, suggesting that the thermogenic appendix plays a central role in diffusing the potent VOC bouquets of Araceae (Meeuse 1975; Angioy et al. 2004; Barthlott et al. 2009). Although we expected to observe elevated TPS activity in male florets during anthesis compared to the morning following anthesis, it appears that many TPSs continued to be expressed at approximately equal levels in paired control samples collected the following morning; several factors may explain this pattern. First, given that sesquiterpenes are known to be synthesized and stored in A. italicum male floret tissue during the days leading up to anthesis (Leguet et al. 2014), mevalonate pathway and TPS expression may have peaked prior to our during-anthesis tissue sampling. However, Araceae are known to emit particularly concentrated VOC blends during anthesis (Skubatz et al. 1995, 1996), which should require continuous biosynthesis and emission (Widhalm et al. 2015). It is therefore likely that the large number of TPSs we identified in the A. maculatum transcriptome include those which are most relevant to pollinator attraction.
The timing of our postanthesis tissue sampling may have also influenced the above result. Postanthesis sampling occurred within 1 h after we collected and preserved all pollinators within the floral chamber; it is therefore likely that most pollinators normally would have still been trapped at this time. Both early (Dormer 1960; Prime 1960; Lack and Diaz 1991) and more recent studies (Bröderbauer et al. 2013) of A. maculatum have highlighted morphological features related to pollinator retention, namely the sterile flowers partially blocking the exit of the trap, and downward-pointing papillate cells on the inner wall of the spathe. However, continued expression of some TPS transcripts during the morning following anthesis may also highlight sesquiterpenes putatively involved in pollinator retention within the trap chamber. Consequently, our paired “postanthesis” samples may not have acted as a negative control for comparison to during-anthesis samples, as we initially intended. Further research is needed to confirm whether the end of male floret VOC emissions may also serve as a cue for pollinator release, which occurs during the afternoon on the second day of the pollination cycle (Prime 1960; Lack and Diaz 1991; Chartier et al. 2013).
After comparing male floret transcript expression among populations during anthesis, we did not identify any putative VOC synthases which were unique to Forêt du Gâvre, France. High inter-individual variation in VOC emissions has been widely observed in A. maculatum (Gfrerer et al. 2021; Szenteczki et al. 2021) and this variability is also evident in our transcriptomic dataset. Although our differential expression analyses were unable to identify unique VOC synthases in the Forêt du Gâvre population, targeted investigations into candidate genes underlying key compound classes did provide further insights into species-specific pollinator attraction in A. maculatum.
Candidate transcripts linked to A. maculatum VOCs
Most inter-individual variation in A. maculatum floral scent centers around variation in the ratio of AAA-derived VOCs to sesquiterpenes (Szenteczki et al. 2021). Correspondingly, we identified abundant expression of transcripts putatively involved in the biosynthesis of these 2 compound classes. As predicted, indole synthesis via the TSA subunit was highly expressed in appendix tissue in all studied populations. Some of the TSA transcripts we identified also appear to be homologs of BX1, which more efficiently cleaves indole-3-glycerol phosphate without any interaction with the beta subunit (Frey et al. 1997; Kriechbaumer et al. 2008). Conversely, TSB was more abundantly expressed in male floret tissue, which would limit indole emissions within the floral trap chamber.
Skatole (3-methylindole) has also been reported in A. maculatum VOC emissions (Gfrerer et al. 2021; Szenteczki et al. 2021), but we did not identify any homologs of indoleacetate decarboxylase in the A. maculatum transcriptome, which would produce this compound (Whitehead et al. 2008). This may be due to low emissions of skatole by inflorescences sampled in this study; ultimately, the mechanism by which A. maculatum produces skatole remains unclear. Prior to this study, it was also unclear whether A. maculatum produced p-cresol (4-methylphenol) via a pathway homologous to tyrosine degradation in bacteria (Selmer and Andrei 2001; Saito et al. 2018), or using 1 or more novel proteins. We were able to identify candidate transcripts (i.e. ISS1 and HPPR homologs) which may catalyze the conversion of tyrosine to p-coumaric acid, but were ultimately unable to identify transcripts which could further produce p-cresol from p-coumaric acid.
Homologs of phenylalanine ammonia-lyase (PAL) and cinnamate 4-hydroxylase (C4H), which produce p-coumaric acid from phenylalanine, where also abundantly expressed in the appendix during anthesis. However, downstream proteins producing common FBP VOCs (e.g. eugenol, isoeugenol, benzyl benzoate, benzaldehyde, and phenylacetaldehyde; Muhlemann et al. 2014) were largely absent or lowly expressed in both our VOC and transcriptomic datasets. Typically, p-coumarate-3-hydroxylase (C3H) would further modify p-coumaric acid into precursors for the aforementioned FBP VOCs, but this gene was expressed at relatively low levels in appendix tissue. Interestingly, experimental knock-outs of C3H in Petunia × hybrida have demonstrated that downregulation of this gene also leads to unexpected production of p-cresol (Kim et al. 2019). Given that we observed similar patterns in A. maculatum, it appears that flowering plants may have a novel mechanism for producing p-cresol involving p-coumaric acid, which is distinct from known bacterial proteins such as hydroxyphenylacetate decarboxylase.
While not derived from amino acids, 2-heptanone is another abundant A. maculatum VOC with a “fermented” scent. In this study, we were able to identify several methylketone synthase homologs (MKS1 and MKS2), which produce 2-heptanone and related compounds in tomato plants (Ben-Israel et al. 2009; Khuat et al. 2019). 2-heptanone is known to be attractive to fruit flies (Prokopy et al. 1998) and beetles (Torto et al. 2007); however, it has not yet been linked with the attraction of specific pollinators of A. maculatum (Espíndola et al. 2011; Szenteczki et al. 2021). While this compound is infrequent in VOC bouquets, it can be a proportionally large component of VOC emissions when it is produced (Szenteczki et al. 2021); further research is therefore needed to determine whether this compound is also under selection (e.g. by Drosophilidae).
We also found that DXS activity was elevated in A. maculatum appendix tissue during anthesis. An identical expression pattern was also identified in A. concinnatum, where it was hypothesized that DXS is the rate-limiting step in monoterpene synthesis (Onda et al. 2015). If DXS is also a rate-limiting enzyme in A. maculatum, then this may explain the proportionally smaller quantities of monoterpenes in its floral blend, in comparison to other VOC chemical families.
Finally, we identified many putative TPSs in the A. maculatum transcriptome, the majority of which appear to produce sesquiterpene VOCs. These include cytochrome P450 enzymes (P450s), which can further modify volatile terpenes through oxidation, methylation, or acylation (Hamberger and Bak 2013). P450s were the first proteins identified in Arum appendices (Yáhiel et al. 1974), and our transcriptomic data further confirm that a diverse suite of P450s are expressed in A. maculatum appendix and male floret tissue during anthesis. Notably, we found that a trimethyltridecatetraene synthase (Cyt P450 92C6) homolog appears to be correlated with the production of at least 1 unnamed A. maculatum sesquiterpene (Kovats RI 1681). This unnamed sesquiterpene was the single strongest predictor of P. grisescens attraction in a large-scale survey of A. maculatum pollinators (Szenteczki et al. 2021). Until now, it has not been possible to experimentally test whether this compound alone is attractive to Psychodidae. However, the candidate gene we identified could be useful in future research aiming to produce this compound through heterologous expression in yeasts (Dhandapani et al. 2017; Kutyna and Borneman 2018).
Our results also confirm that bicyclogermacrene synthase (Crocoll et al. 2010) is consistently and almost exclusively expressed in male floret tissue during anthesis, in accordance with the dominance of this sesquiterpene in A. maculatum floral chamber scent (Kite et al. 1998). Interestingly, previous studies have demonstrated that 9-methyl germacrene B is a sex pheromone produced by male Lutzomyia longipalpis (Psychodidae) to attract females (Hamilton et al. 1996; Hamilton et al. 1999). Consequently, A. maculatum may emit bicyclogermacrene, or a closely related sesquiterpene compound as a as part of their deceptive pollination strategy (i.e. as a pheromone mimic). Such terpene emissions in the floral chamber could stimulate the movement of trapped Psychodidae over the male and female florets, aiding in pollination and pollen dispersal; however, further experiments are needed to test these hypotheses. A recent gas chromatography–electroantennography (GC–EAD; Schiestl and Marion-Poll 2002) study demonstrated that P. phalaenoides antennae responded most strongly to p-cresol, germacrene D, and several other unnamed mono- and sesquiterpenes (Gfrerer et al. 2022); these findings are consistent with our transcriptomic results. However, P. grisescens antennal responses were not tested as part of this study. Consequently, the results of our coinertia analyses could be used to formulate hypotheses for future GC–EAD research.
Male floret-specific TPSs and their influence on pollinator attraction
There is growing evidence to suggest that tissue-specific transcript expression of VOC synthases is a common characteristic of deceptive pollination systems (Wong, Amarasinghe, et al. 2017, and references therein), and our results confirm similar patterns in A. maculatum. Specifically, our coinertia analyses identified significant covariation between male floret TPS expression, and the communities of Psychodidae pollinators trapped by inflorescences. While the floral scent of A. maculatum is often described as dung-like due to abundant emissions of amino acid-derived VOCs, our results suggest that terpene VOCs may be subject to pollinator-mediated selection as well. Recent large-scale ecological studies also support this hypothesis, with sesquiterpene compounds, rather than so-called “dung-mimicking” compounds, being a better predictor of variation in pollinator attraction patterns (Szenteczki et al. 2021) and fruit set size (Gfrerer et al. 2021) in A. maculatum.
Some of the main compounds related to sex- and species-specific attraction in our coinertia analysis included humulene (covarying with female P. phalaenoides), and the aforementioned trimethyltridecatetraene synthase homolog, which may produce an unnamed sesquiterpene with a Kovats Retention Index of 1681 (covarying with P. grisescens). Notably, these same compounds were highlighted as key predictors of pollinator species trapped by A. maculatum inflorescences in a random forest analysis with a larger sample size (Szenteczki et al. 2021). Further research is therefore needed in order to assess the importance of male floret VOCs for pollinator attraction in A. maculatum and to confirm the true identities of the compounds produced by the candidate TPSs we identified.
Conclusion
Our data provide a deeper understanding of the relationships between transcript expression, floral scent, and pollinator attraction in A. maculatum, and may lead to the identification of new VOC biosynthetic genes in the future. It appears that A. maculatum inflorescences employ a combination of highly diverse appendix VOCs including 2-heptanone, indole, and p-cresol to attract a broad range of coprophilous dipterans to their inflorescences, and specialized sesquiterpene emissions to further lure specific Psychodidae into the floral trap chamber. Male floret-specific VOCs, particularly bicyclogermacrene, may also play a role in retaining pollinators until the pollination cycle is complete. While Kite and colleagues (Kite 1995; Kite et al. 1998) separately characterized A. maculatum appendix and trap chamber VOC emissions, all subsequent studies analyzed total headspace profiles (i.e. a blend of appendix and trap chamber VOCs). Our results highlight a need for further focused study of VOCs within the trap chamber, given that specific sesquiterpenes appear to be an important aspect of this highly specialized lure-and-trap pollination system.
Data availability
Supplementary Appendices 1 and 2 contain supporting figures and tables referenced in the main text. Raw Illumina RNA-seq reads have been deposited in the NCBI SRA database under the BioProject Accession PRJNA856436 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA856436). Additional data, including the files and R code needed to reproduce all analyses and figures, have been archived in the Zenodo repository 6806366 (https://doi.org/10.5281/zenodo.6806365).
Supplemental material is available at G3 online.
Supplementary Material
Acknowledgments
We thank Monica Fleisher, Andrea Galmán, and Alberto Garcia Jimenez for their dedicated assistance during our field sampling and Laurent Oppliger and colleagues for allowing us to sample VOCs and tissue from the native population of A. maculatum at the Jardin Botanique de Neuchâtel. We also thank Dessislava Savova Bianchi for her support during RNA extractions, Johann Weber for his advice on our RNA-seq library preparation, and Eran Pichersky for his advice on the biosynthesis of some floral VOCs.
Funding
This project was funded by the Swiss National Science Foundation through grant P1NEP3_191659 awarded to MAS and grant 31003A_163334 awarded to NA and SR.
Author contributions
MAS, ALG, MG, SR, and NA designed the experiments. MAS and ALG conducted the field sampling. MAS prepared tissue samples for RNA-Seq library preparation, and ALG and MG analyzed the headspace VOC samples and identified pollinators. MAS and JG designed and performed the bioinformatic analyses, and MG contributed to the interpretation of the results. MAS wrote the manuscript. All coauthors contributed to the revision of the manuscript, and have approved it for publication.
Conflicts of interest
None declared.
Contributor Information
Mark A Szenteczki, Université de Neuchâtel, Institut de Biologie, 2000 Neuchâtel, Switzerland.
Adrienne L Godschalx, Université de Neuchâtel, Institut de Biologie, 2000 Neuchâtel, Switzerland.
Jérémy Gauthier, Geneva Natural History Museum, 1208 Genève, Switzerland.
Marc Gibernau, CNRS—University of Corsica, Laboratory Sciences for the Environment (SPE—UMR 6134), Natural Resources Project, 20000 Ajaccio, France.
Sergio Rasmann, Université de Neuchâtel, Institut de Biologie, 2000 Neuchâtel, Switzerland.
Nadir Alvarez, Geneva Natural History Museum, 1208 Genève, Switzerland; Department of Genetics and Evolution, University of Geneva, 1211 Geneva, Switzerland.
Literature cited
- Alexa A, Rahnenfuhrer J. topGO: Enrichment Analysis for Gene Ontology. R Package Version 2.0; 2010.
- Angioy AM, Stensmyr MC, Urru I, Puliafito M, Collu I, et al. Function of the heater: the dead horse arum revisited. Proc Biol Sci. 2004;271:S13–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthlott W, Szarzynski J, Vlek P, Lobin W, Korotkova N.. A torch in the rain forest: thermogenesis of the Titan arum (Amorphophallus titanum). Plant Biol. (Stuttg). 2009;11(4):499–505. [DOI] [PubMed] [Google Scholar]
- Ben-Israel I, Yu G, Austin MB, Bhuiyan N, Auldridge M, Nguyen T, Schauvinhold I, Noel JP, Pichersky E, Fridman E, et al. Multiple biochemical and morphological factors underlie the production of methylketones in tomato trichomes. Plant Physiol. 2009;151(4):1952–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröderbauer D, Weber A, Diaz A.. The design of trapping devices in pollination traps of the genus Arum (Araceae) is related to insect type. Bot J Linn Soc. 2013;172(3):385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers KJRP, Bradshaw HD, Riffell JA.. Three floral volatiles contribute to differential pollinator attraction in monkeyflowers (Mimulus). J Exp Biol. 2014;217(Pt 4):614–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartier M, Liagre S, Weiss-Schneeweiss H, Kolano B, Bessière J-M, Schönenberger J, Gibernau M.. Floral traits and pollination ecology of European Arum hybrids. Oecologia. 2016;180(2):439–451. [DOI] [PubMed] [Google Scholar]
- Chartier M, Pélozuelo L, Buatois B, Bessière J-M, Gibernau M.. Geographical variations of odour and pollinators, and test for local adaptation by reciprocal transplant of two European Arum species. Funct Ecol. 2013;27(6):1367–1381. [Google Scholar]
- Chartier M, Pélozuelo L, Gibernau M.. Do floral odor profiles geographically vary with the degree of specificity for pollinators? Investigation in two sapromyophilous Arum species (Araceae). Ann. la Société Entomol. Fr. 2011;47(1–2):71–77. [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J.. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; 2021.
- Crocoll C, Asbach J, Novak J, Gershenzon J, Degenhardt J.. Terpene synthases of oregano (Origanum vulgare L.) and their roles in the pathway and regulation of terpene biosynthesis. Plant Mol Biol. 2010;73(6):587–603. [DOI] [PubMed] [Google Scholar]
- Dhandapani S, Jin J, Sridhar V, Sarojam R, Chua N-H, Jang I-C.. Integrated metabolome and transcriptome analysis of Magnolia champaca identifies biosynthetic pathways for floral volatile organic compounds. BMC Genomics. 2017;18(1):463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz A, Kite G.. A comparison of the pollination ecology of Arum maculatum and A. italicum in England. Watsonia. 2002;24:171–182. [Google Scholar]
- Dobson HEM, Arroyo J, Bergström G, Groth I.. Interspecific variation in floral fragrances within the genus Narcissus (Amaryllidaceae). Biochem Syst Ecol. 1997;25(8):685–706. [Google Scholar]
- Dormer KJ. The truth about pollination in Arum. New Phytol. 1960;59(3):298–301. [Google Scholar]
- Espíndola A, Alvarez N.. Comparative phylogeography in a specific and obligate pollination antagonism. PLoS One. 2011;6(12):e28662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espíndola A, Pellissier L, Alvarez N.. Variation in the proportion of flower visitors of Arum maculatum along its distributional range in relation with community-based climatic niche analyses. Oikos. 2011;120(5):728–734. [Google Scholar]
- Fenster CB, Armbruster WS, Wilson P, Dudash MR, Thomson JD.. Pollination syndromes and floral specialization. Annu Rev Ecol Evol Syst. 2004;35(1):375–403. [Google Scholar]
- Frey M, Chomet P, Glawischnig E, Stettner C, Grün S, Winklmair A, Eisenreich W, Bacher A, Meeley RB, Briggs SP, et al. Analysis of a chemical plant defense mechanism in grasses. Science. 1997;277(5326):696–699. [DOI] [PubMed] [Google Scholar]
- Friberg M, Schwind C, Guimarães PR, Raguso RA, Thompson JN.. Extreme diversification of floral volatiles within and among species of Lithophragma (Saxifragaceae). Proc Natl Acad Sci U S A. 2019;116(10):4406–4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W.. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28(23):3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gervasi DDL, Schiestl FP.. Real-time divergent evolution in plants driven by pollinators. Nat Commun. 2017;8(14691):14691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gfrerer E, Laina D, Gibernau M, Fuchs R, Happ M, Tolasch T, Trutschnig W, Hörger AC, Comes HP, Dötterl S, et al. Floral scents of a deceptive plant are hyperdiverse and under population-specific phenotypic selection. Front Plant Sci. 2021;12:719092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gfrerer E, Laina D, Wagner R, Gibernau M, Hörger AC, et al. Antennae of psychodid and sphaerocerid flies respond to a high variety of floral scent compounds of deceptive Arum maculatum L. Sci Rep. 2022;12:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibernau M. Pollination in the genus Arum—a review. Aroideana. 2004;27:148–166. [Google Scholar]
- Gilbert DG. Longest protein, longest transcript or most expression, for accurate gene reconstruction of transcriptomes? bioRxiv 829184, 2019.
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamberger B, Bak S.. Plant P450s as versatile drivers for evolution of species-specific chemical diversity. Philos Trans R Soc Lond B Biol Sci. 2013;368(1612):20120426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton JG, Dawson GW, Pickett JA.. 9-Methylgermacrene-B; proposed structure for novel homosesquiterpene from the sex pheromone glands of Lutzomyia longipalpis (Diptera: Psychodidae) from Lapinha, Brazil. J Chem Ecol. 1996;22(8):1477–1491. [DOI] [PubMed] [Google Scholar]
- Hamilton JGC, Ibbotson HC, Hooper AM, Pickett JA.. 9-Methylgermacrene-B is confirmed as the sex pheromone of the sandfly Lutzomyia longipalpis from Lapinha, Brazil, and the absolute stereochemistry defined as S. Chem Commun. 1999;(23):2335–2336. [Google Scholar]
- Hernández-Hernández T, Wiens JJ.. Why are there so many flowering plants? A multiscale analysis of plant diversification. Am Nat. 2020;195(6):948–963. [DOI] [PubMed] [Google Scholar]
- Hervé M. RVAideMemoire: Diverse Basic Statistical and Graphical Functions. R Package Version 0.9-81; 2021.
- Holopainen JK. Multiple functions of inducible plant volatiles. Trends Plant Sci. 2004;9(11):529–533. [DOI] [PubMed] [Google Scholar]
- Irwin RE, Adler LS, Brody AK.. The dual role of floral traits: pollinator attraction and plant defense. Ecology. 2004;85(6):1503–1511. [Google Scholar]
- Ježek J. Redescriptions of nine common palaearctic and holarctic species of Psychodini end. (Diptera: Psychodidae). Acta Entomol Musei Natl Pragae. 1990;43:33–83. [Google Scholar]
- Junker RR, Parachnowitsch AL.. Working towards a holistic view on flower traits—how floral scents mediate plant-animal interactions in concert with other floral characters. J Indian Inst Sci. 2015;95:43–67. [Google Scholar]
- Khuat VLU, Bui VTT, Tran HTD, Truong NX, Nguyen TC, et al. Characterization of Solanum melongena thioesterases related to tomato methylketone synthase 2. Genes. 2019;10(7):549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Swanson RT, Alvarez MI, Johnson TS, Cho KH, Clark DG, Colquhoun TA.. Down regulation of p-coumarate 3-hydroxylase in petunia uniquely alters the profile of emitted floral volatiles. Sci Rep. 2019;9(1):8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kite GC. The floral odour of Arum maculatum. Biochem Syst Ecol. 1995;23(4):343–354. [Google Scholar]
- Kite GC, Hetterscheid WLA, Lewis MJ, Boyce PC, Ollerton J, et al. Inflorescence odours and pollinators of Arum and Amorphophallus (Araceae). In Owens SJ, Rudall PJ, editors. Reproductive Biology. Royal Botanic Gardens Kew; London, UK; 1998. p. 295–315. [Google Scholar]
- Knoll F. Insekten und Blumen, die Arum—Blütenstände und ihre Besucher. Abhandlungen der Zool.-Botan. Gesellschaft in Wien. 1926;12:379–481. [Google Scholar]
- Knudsen JT, Gershenzon J.. The chemical diversity of floral scent. In Pichersky E, Dudareva N, editors. Biology of Plant Volatiles. CRC Press; 2020. p. 57–78. [Google Scholar]
- Knudsen JT, Tollsten L.. Trends in floral scent chemistry in pollination syndromes: floral scent composition in moth‐pollinated taxa. Bot J Linn Soc. 1993;113(3):263–284. [Google Scholar]
- Kolde R. pheatmap: Pretty Heatmaps. R Package Version 1.0.12; 2019.
- Kriechbaumer V, Weigang L, Fiesselmann A, Letzel T, Frey M, Gierl A, Glawischnig E.. Characterisation of the tryptophan synthase alpha subunit in maize. BMC Plant Biol. 2008;8:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutyna DR, Borneman AR.. Heterologous production of flavour and aroma compounds in Saccharomyces cerevisiae. Genes. 2018;9(7):326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lack AJ, Diaz A.. The pollination of Arum maculatum L.—a historical review and new observations. Watsonia. 1991;18:333–342. [Google Scholar]
- Langmead B, Salzberg SL.. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leguet A, Gibernau M, Shintu L, Caldarelli S, Moja S, Baudino S, Caissard J-C.. Evidence for early intracellular accumulation of volatile compounds during spadix development in Arum italicum L. and preliminary data on some tropical Aroids. Naturwissenschaften. 2014;101(8):623–635. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P.. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49(W1):W293–W296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN.. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A.. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22(13):1658–1659. [DOI] [PubMed] [Google Scholar]
- Liu D, Wei Y, Liu X, Zhou Y, Jiang L, Yin J, Wang F, Hu Y, Nanjaraj Urs AN, Liu Y, et al. Indoleacetate decarboxylase is a glycyl radical enzyme catalysing the formation of malodorant skatole. Nat Commun. 2018;9(1):4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeuse BJD. Thermogenic respiration in aroids. Annu Rev Plant Physiol. 1975;26(1):117–126. [Google Scholar]
- Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M.. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:182–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlemann JK, Klempien A, Dudareva N.. Floral volatiles: from biosynthesis to function. Plant Cell Environ. 2014;37(8):1936–1949. [DOI] [PubMed] [Google Scholar]
- Newman E, Anderson B, Johnson SD.. Flower colour adaptation in a mimetic orchid. Proc Biol Sci. 2012;279(1737):2309–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ.. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, et al. vegan: Community Ecology Package. R Package Version 2.5-7; 2020.
- Onda Y, Mochida K, Yoshida T, Sakurai T, Seymour RS, Umekawa Y, Pirintsos SA, Shinozaki K, Ito K.. Transcriptome analysis of thermogenic Arum concinnatum reveals the molecular components of floral scent production. Sci Rep. 2015;5:8753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva ÉAS, Ballego-Campos I, Gibernau M.. True nectar or stigmatic secretion? Structural evidence elucidates an old controversy regarding nectaries in Anthurium. Am J Bot. 2021;108(1):37–50. [DOI] [PubMed] [Google Scholar]
- Pellmyr O, Thien LB.. Insect reproduction and floral fragrances: keys to the evolution of the angiosperms? Taxon. 1986;35(1):76–85. [Google Scholar]
- Pichersky E, Gershenzon J.. The formation and function of plant volatiles: perfumes for pollinator attraction and defense. Curr Opin Plant Biol. 2002;5(3):237–243. [DOI] [PubMed] [Google Scholar]
- Prime CT. 1960. Lords and Ladies. Collins: london. [Google Scholar]
- Prokopy RJ, Hu X, Jang EB, Vargas RI, Warthen JD.. Attraction of mature Ceratitis capitata females to 2-heptanone, a component of coffee fruit odor. J. Chem. Ecol. 1998;24(8):1293–1304. [Google Scholar]
- Roháček J, Beck-Haug I, Dobat K.. Sphaeroceridae associated with flowering Arum maculatum (Araceae) in the vicinity of Tübingen, SW-Germany. Senckenb Biol. 1990;71:259–268. [Google Scholar]
- Saito Y, Sato T, Nomoto K, Tsuji H.. Identification of phenol- and p-cresol-producing intestinal bacteria by using media supplemented with tyrosine and its metabolites. FEMS Microbiol Ecol. 2018;94:fiy125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayols S. rrvgo: A Bioconductor Package to Reduce and Visualize Gene Ontology Terms. R Package Version 1.4; 2020.
- Schemske DW, Bradshaw HD.. Pollinator preference and the evolution of floral traits in monkeyflowers (Mimulus). Proc Natl Acad Sci U S A. 1999;96(21):11910–11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl FP, Johnson SD.. Pollinator-mediated evolution of floral signals. Trends Ecol Evol. 2013;28(5):307–315. [DOI] [PubMed] [Google Scholar]
- Schiestl FP, Marion-Poll F.. Detection of physiologically active flower volatiles using gas chromatography coupled with electroantennography. In Jackson JF, Linskens HF (editors). Analysis of Taste and Aroma. Berlin, Heidelberg: Springer; 2002. p. 173–198. [Google Scholar]
- Schmucker T. Beiträge zur Biologie und Physiologie von Arum maculatum. Flora Oder Allg Bot Zeitung. 1925;118–119:460–475. [Google Scholar]
- Scott C, Pierce T, Johnson LK, Brown CT, Reid C, et al. dib-lab/dammit: dammit v1.2; 2019.
- Selmer T, Andrei PI.. p-Hydroxyphenylacetate decarboxylase from Clostridium difficile. A novel glycyl radical enzyme catalysing the formation of p-cresol. Eur J Biochem. 2001;268(5):1363–1372. [DOI] [PubMed] [Google Scholar]
- Seppey M, Manni M, Zdobnov EM.. BUSCO: assessing genome assembly and annotation completeness. Methods Mol Biol. 2019;1962:227–245. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skubatz H, Kunkel DD, Howald WN, Trenkle R, Mookherjee B.. The Sauromatum guttatum appendix as an osmophore: excretory pathways, composition of volatiles and attractiveness to insects. New Phytol. 1996;134(4):631–640. [DOI] [PubMed] [Google Scholar]
- Skubatz H, Kunkel DD, Patt JM, Howald WN, Hartman TG, Meeuse BJ.. Pathway of terpene excretion by the appendix of Sauromatum guttatum. Proc Natl Acad Sci U S A. 1995;92(22):10084–10088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C, Love MI, Robinson MD.. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research. 2015;4:1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Florea L.. Rcorrector: efficient and accurate error correction for Illumina RNA-seq reads. Gigascience. 2015;4:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T.. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6(7):e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szenteczki MA, Godschalx AL, Galmán A, Espíndola A, Gibernau M, Alvarez N, Rasmann S.. Spatial and temporal heterogeneity in pollinator communities maintains within-species floral odour variation. Oikos. 2021;130(9):1487–1499. [Google Scholar]
- Thioulouse J, Dray S, Dufour A-B, Siberchicot A, Jombart T, Pavoine S.. Multivariate Analysis of Ecological Data with ade4. New York, NY: Springer; 2018. [Google Scholar]
- Thompson JN, Schwind C, Friberg M.. Diversification of trait combinations in coevolving plant and insect lineages. Am Nat. 2017;190(2):171–184. [DOI] [PubMed] [Google Scholar]
- Thompson JN, Schwind C, Guimarães PR, Friberg M.. Diversification through multitrait evolution in a coevolving interaction. Proc Natl Acad Sci U S A. 2013;110(28):11487–11492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torto B, Boucias DG, Arbogast RT, Tumlinson JH, Teal PEA.. Multitrophic interaction facilitates parasite–host relationship between an invasive beetle and the honey bee. Proc Natl Acad Sci U S A. 2007;104(20):8374–8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trunschke J, Lunau K, Pyke GH, Ren Z-X, Wang H.. Flower color evolution and the evidence of pollinator-mediated selection. Front Plant Sci. 2021;12:617851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urru I, Stökl J, Linz J, Krügel T, Stensmyr MC, Hansson BS.. Pollination strategies in Cretan Arum lilies. Biol. J. Linn. Soc. 2010;101(4):991–1001. [Google Scholar]
- Van der Niet T, Johnson SD.. Phylogenetic evidence for pollinator-driven diversification of angiosperms. Trends Ecol Evol. 2012;27(6):353–361. [DOI] [PubMed] [Google Scholar]
- Vogel S. Kesselfallen–Blumen. Umschau. 1965;65:12–17. [Google Scholar]
- Whitehead MR, Peakall R.. Integrating floral scent, pollination ecology and population genetics. Funct. Ecol. 2009;23(5):863–874. [Google Scholar]
- Whitehead TR, Price NP, Drake HL, Cotta MA.. Catabolic pathway for the production of skatole and indoleacetic acid by the acetogen Clostridium drakei, Clostridium scatologenes, and Swine manure. Appl Environ Microbiol. 2008;74(6):1950–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widhalm JR, Jaini R, Morgan JA, Dudareva N.. Rethinking how volatiles are released from plant cells. Trends Plant Sci. 2015;20(9):545–550. [DOI] [PubMed] [Google Scholar]
- Williams CA, Harborne JB, Mayo SJ.. Anthocyanin pigments and leaf flavonoids in the family Araceae. Phytochemistry. 1981;20(2):217–234. [Google Scholar]
- Wong DCJ, Amarasinghe R, Rodriguez-Delgado C, Eyles R, Pichersky E, Peakall R.. Tissue-specific floral transcriptome analysis of the sexually deceptive orchid Chiloglottis trapeziformis provides insights into the biosynthesis and regulation of its unique UV-B dependent floral volatile, chiloglottone 1. Front Plant Sci. 2017;8:1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DCJ, Pichersky E, Peakall R.. The biosynthesis of unusual floral volatiles and blends involved in orchid pollination by deception: current progress and future prospects. Front Plant Sci. 2017;8:1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Zhang Y, Wang M.. Discovery and engineering of cytochrome P450s for terpenoid biosynthesis. Trends Biotechnol. 2019;37(6):618–631. [DOI] [PubMed] [Google Scholar]
- Yáhiel V, Cotte-Martinon MG, Ducet G.. Un cytochrome de type P450 dans le spadice d’arum. Phytochemistry. 1974;13(9):1649–1651. [Google Scholar]
- Zhu A, Ibrahim JG, Love MI.. Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics. 2019;35(12):2084–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supplementary Appendices 1 and 2 contain supporting figures and tables referenced in the main text. Raw Illumina RNA-seq reads have been deposited in the NCBI SRA database under the BioProject Accession PRJNA856436 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA856436). Additional data, including the files and R code needed to reproduce all analyses and figures, have been archived in the Zenodo repository 6806366 (https://doi.org/10.5281/zenodo.6806365).
Supplemental material is available at G3 online.