Abstract
Leaf angle is an important agronomic trait determining maize (Zea mays) planting density and light penetration into the canopy and contributes to the yield gain in modern maize hybrids. However, little is known about the molecular mechanisms underlying leaf angle beyond the ZmLG1 (liguleless1) and ZmLG2 (Liguleless2) genes. In this study, we found that the transcription factor (TF) ZmBEH1 (BZR1/BES1 homolog gene 1) is targeted by ZmLG2 and regulates leaf angle formation by influencing sclerenchyma cell layers on the adaxial side. ZmBEH1 interacted with the TF ZmBZR1 (Brassinazole Resistant 1), whose gene expression was also directly activated by ZmLG2. Both ZmBEH1 and ZmBZR1 are bound to the promoter of ZmSCL28 (SCARECROW-LIKE 28), a third TF that influences leaf angle. Our study demonstrates regulatory modules controlling leaf angle and provides gene editing targets for creating optimal maize architecture suitable for dense planting.
Various regulatory modules control leaf angle in maize.
Introduction
Maize yields have increased dramatically since the late 1930s due in large part to the increased planting densities (Duvick, 2005). The selection of upright leaf architecture enables plants to be grown at higher density while minimizing the shading of neighboring plants and increasing the efficiency of light capture. Leaf angle, the trait defined as the inclination between the leaf blade midrib and the vertical culm, contributes to upright leaf architecture. Recently, the identification of two quantitative trait loci, Upright Plant Architecture1 (UPA1) and Upright Plant Architecture2 (UPA2), which conferred upright plant architecture by reducing the leaf angle and then dramatically improved planting density and enhanced grain yield (Tian et al., 2019), further supported the critical role of leaf angle alteration in maize breeding.
The establishment of the leaf angle in maize is largely determined by the ligular region, which encompasses the ligule and auricle and serves as a hinge at the blade-sheath boundary, allowing the leaf blade to project away from the stem. Two classical genes, ZmLG1 (Liguleless1), a Squamosa Promoter Binding Protein domain-containing TF (Moreno et al., 1997), and ZmLG2 (Liguleless2) ZmLG2, a basic leucine zipper TF family member (Harper and Freeling, 1996; Walsh et al., 1998), play key roles in ligular region formation that alters leaf angle. In the lg1 mutants, ligules and auricles are not formed, and the plants exhibit excessively erect leaves (Moreno et al., 1997). In lg2, ligules and auricles are often absent around the midrib, but developed at the leaf margins and positioned incorrectly, also causing upright plant architecture with extremely small leaf angles (Lambert and Johnson, 1978; Walsh et al., 1998; Mantilla-Perez and Salas Fernandez, 2017). Introgression of lg2 mutant alleles into maize hybrid lines resulted in increased grain yield (Pendleton et al., 1968; Lambert and Johnson, 1978); however, the extremely erect leaf limits the commercial use of this allele. Even though, ZmLG1 and ZmLG2 could provide genetic clues to optimize leaf angle for dense plantings, such as identifying their downstream targets. The genetic basis by which genes affect leaf angle in maize needs further exploration.
Several genes in maize have been identified to control leaf angle (Ku et al., 2011; Zhang et al., 2014; Tian et al., 2019; Cao et al., 2020; Ren et al., 2020), and most of them are related to the phytohormone brassinosteroids (BRs), e.g. BRD1 (brassinosteroid C-6 oxidase1) and BRI1 (brassinosteroid insensitive 1). The overexpression of BRD1, which encodes brassinosteroid C-6 oxidase1 to catalyze the last step of brassinosteroid synthesis, increased the leaf angle by enlarging the auricle and decreasing the number of sclerenchyma cells on the adaxial side, and knockout of the function of BRI1, which encodes a Leu-rich repeat receptor kinase that is responsible for BR binding to start BR signal transduction, resulted in upright leaves with decreased auricle formation (Makarevitch et al., 2012; Tian et al., 2019).
BR signaling is sent from the BRI1 receptor kinase at the cell surface to the BZR1/BES1 (brassinazole resistant 1/BRI1-EMS-suppressor 1) TFs, such as BZR1 in Arabidopsis (Arabidopsis thaliana), which directly binds to the BR response element (BRRE, CGTGT/CG) to regulate the expression of downstream BR-responsive genes (Li and Jin, 2007). BZR1 family members act not only as major TFs in the BR signaling pathway but also as mediators to participate in plant development and abiotic stress (Wang et al., 2014; Gruszka, 2020). In Arabidopsis, BZR1 promotes phloem and xylem differentiation (Saito et al., 2018) and interacts with GATA2 (a GATA-type TF), a TF in the light signaling pathway, to regulate hypocotyl elongation of seedlings (Luo et al., 2010). A recent study showed that anther locule development was also regulated by BZR1 (Chen et al., 2019). In addition, BZR1 serves as a regulator in abiotic stress. For instance, BZR1 positively modulates plant freezing tolerance through C-repeat binding factor (CBF)-dependent and CBF-independent pathways (Li et al., 2017). In rice (Oryza sativa), OsBZR1 was identified by its homology to Arabidopsis BZR1, and the loss-of-function of OsBZR1 reduced BR sensitivity, presenting a dwarfism phenotype with erect leaves, indicating the critical roles of OsBZR1 in the BR signaling pathway (Bai et al., 2007). To date, very few studies have focused on the maize BZR TF family, and our understanding of the role of BZRs in maize is still limited.
In this study, we screened the downstream target of ZmLG2 and identified a BZR1/BES1 homolog gene named ZmBEH1. The loss-of-function of ZmBEH1 results in upright plant architecture. Interestingly, maize ZmBZR1, which also influences leaf angle, also acts downstream of ZmLG2 and interacts with ZmBEH1 to coregulate the expression of ZmSCL28, a rice Dwarf and Low Tillering (DLT; Tong et al., 2009) homolog that alters leaf angle in maize. These findings reveal that the ZmLG2-ZmBEH1/BZR1-ZmSCL28 cascades primarily regulate the leaf angle in maize, which will deepen our understanding of the BR response in maize and help to optimize maize plant architecture for dense planting.
Results
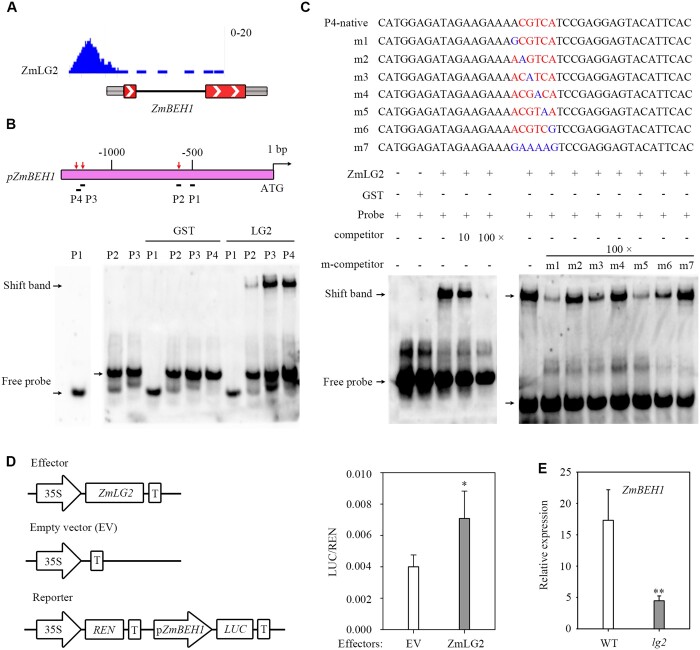
ZmLG2 binds to the promoter of ZmBEH1
ZmLG2, which encodes a bZIP TF, is a classical regulator controlling maize ligule development (Harper and Freeling, 1996; Walsh, et al., 1998; Bolduc et al., 2012). To study the regulatory mechanism mediated by ZmLG2, we analyzed its ChIP-seq results from the maize 104 TF ChIP-Seq data collection (Tu et al., 2020). Interestingly, the results indicated that genes participating in BR signaling might be the downstream targets of ZmLG2, since genes, e.g. ZmBEH1 (BZR1/BES1 homolog gene 1), ZmBEH2 (BZR1/BES1 homolog gene 2), ZmBIR1 (BAK1-interacting receptor-like kinase 1), ZmBRH (Brassinosteroid-responsive RING-H2), ZmBRL3 (BRI1 suppressor 1-like 3), and ZmBSK1 (BR-signaling kinase 1), all had a strong binding peak of ZmLG2 in front of its transcription start site (TSS; Figure 1A;Supplemental Figure S1A). To test the regulatory effect of ZmLG2 on BR signaling, we searched the maize stock center (https://maizecoop.cropsci.uiuc.edu/request/) and identified a mutant with a transposon inserted into ZmBEH1, a homologous gene of rice BZR3 (Supplemental Figure S2). Then, we first focused our research on identifying the regulatory cascade between ZmLG2 and ZmBEH1.
Figure 1.
ZmLG2 directly regulates the expression of ZmBEH1. A, ChIP-Seq shows that ZmLG2 binds to the ZmBEH1 promoter. B, EMSA of ZmLG2 binding to the promoter of ZmBEH1. Top, diagram of the promoter fragments of ZmBEH1. The locations of four Biotin-labeled probes named P1-P4 are indicated by black lines. Red arrows indicate the probes that can be bound by ZmLG2. Bottom, ZmLG2 directly binds to P2-P4 probes with different affinities in EMSA. Black arrows indicate the positions of the bound complex and free probe. GST is the control of ZmLG2-GST. C, Top, diagram of the P4 native and mutated (m1 to m7) fragments acting as probes in EMSA. The ZmLG2 binding motif is labeled in red, and the nucleotide substitutions in the mutated fragments (m1 to m7) are labeled in blue. Bottom, ZmLG2-GST, but not GST by itself, specifically binds to the Biotin-labeled ZmBEH1 P4 probe. Ten- and 100-fold excess amounts of the unlabeled probe were used to compete for the binding of the ZmLG2 protein to the labeled probe. m1 to m7 unlabeled mutated probes were used as competitors (m-competitor) at 100-fold in EMSA. “−” indicates the absence of this component. “+” indicates the presence. Black arrows indicate the positions of the bound complex and free probe. D, Dual-luciferase assay shows relative transactivation of ZmLG2 to the ZmBEH1 promoter. The coding sequence of ZmLG2 driven by the 35S promoter was used as an effector, and the empty vector (EV) was used as an effector control in the transient luciferase assay performed in maize protoplasts. The vector with the REN gene driven by a 35S promoter and firefly luciferase (LUC) gene driven by the promoter sequence from ZmBEH1 was used as the reporter. The LUC/REN ratio represents the relative activity of the promoters. n = 5. E, RT-qPCR analyses show the relative expression of ZmBEH1 in the ligular region of WT and lg2 mutants. Total RNA was isolated from the ligular region of V3 leaves when the seedlings were grown for 18 d. n = 7. Error bars indicate SD. Statistical significance was determined by a two-sided t test: *P < 0.05, **P < 0.01.
Based on ChIP-Seq results, the predicted binding motif of ZmLG2 was CGTCA (Supplemental Figure S1B). To test the reliability of ZmLG2 binding to this motif in the ZmBEH1 promoter, four short genomic regions within 1.3 kb of the ZmBEH1 promoter with the CGTCA motif inside were synthesized as probes (named P1 to P4) to perform an electrophoretic mobility shift assay (EMSA) with purified recombinant ZmLG2-GST fusion protein. As shown in Figure 1B, the ZmLG2-GST fusion protein has high affinity with the P3 (−1,212 to −1,171 bp) and P4 (−1,239 to −1,198 bp) probes, low affinity with the P2 (−610 to −569 bp) probe and no affinity with the P1 (−522 to −492 bp) probe, indicating that the bases near the motif may play important roles in the affinity of the ZmLG2 protein to DNA probes. Next, we performed a binding site selection experiment by mutating the individual residues within and beyond the CGTCA motif. The mutation of A (m1), one base in front of the motif, and mutation of C (m5), the 4th base within the motif, caused strong competition between unlabeled mutated competitor and labeled probe, indicating that these two sites were not key in binding; the mutation of G (m3) and A (m6) in the motif markedly reduced the competition between unlabeled mutated competitor and labeled probe, indicating that they played some roles in ZmLG2 binding; moreover, the mutation on C (m2), T (m4), and entire motif (m7) lost the competition, indicating the importance of the 1st (C) and 3rd (T) base in the motif on TF binding (Figure 1C). In addition, a dual-luciferase (LUC) transient transcriptional activity assay was performed in maize leaf protoplasts with ZmLG2 driven by the 35S promoter as an effector and luciferase driven by the ZmBEH1 promoter as the reporter. The results confirmed that the ZmBEH1 promoter could be not only bound but also activated by ZmLG2 in vivo (Figure 1D).
We further checked the expression profiles of ZmBEH1 in the lg2 mutant through reverse transcription-quantitative PCR (RT-qPCR). RNA was isolated from the ligular region of 18-d-old V3 leaves. At this stage, the auricle started expanding but did not mature (Supplemental Figure S3). The expression of ZmBEH1 in lg2 mutants was significantly reduced, which was only approximately one-fourth of that in the wild type (WT; Figure 1E). All these results indicate that ZmLG2 positively regulates the expression of ZmBEH1 and that ZmBEH1 is the direct downstream target of ZmLG2.
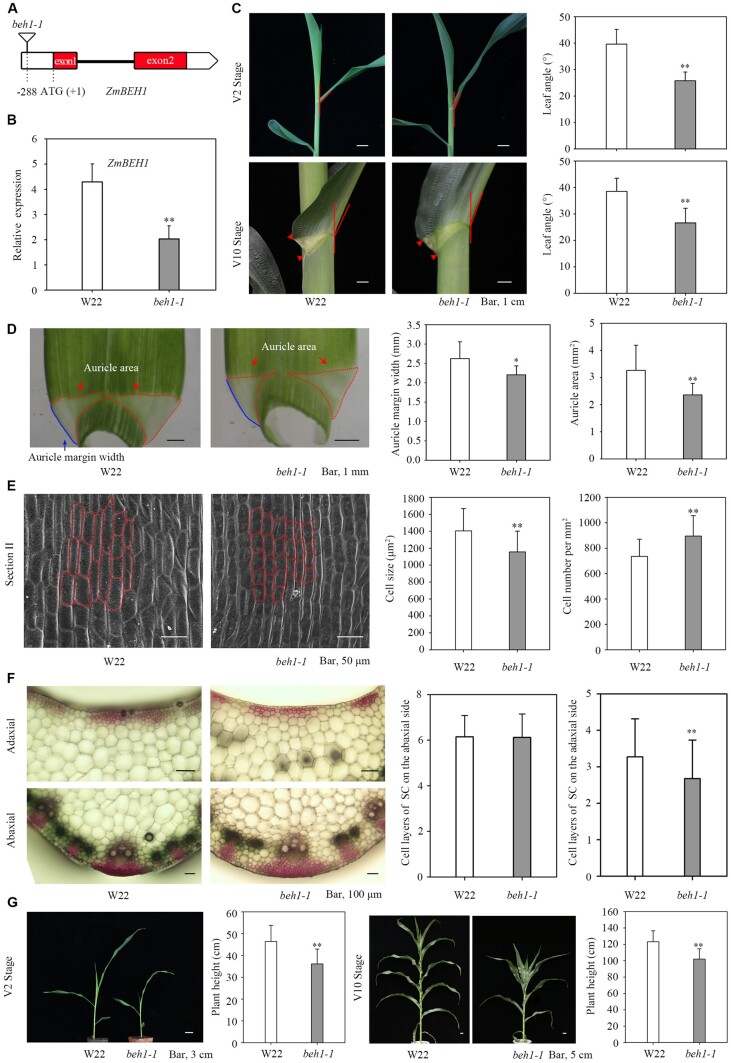
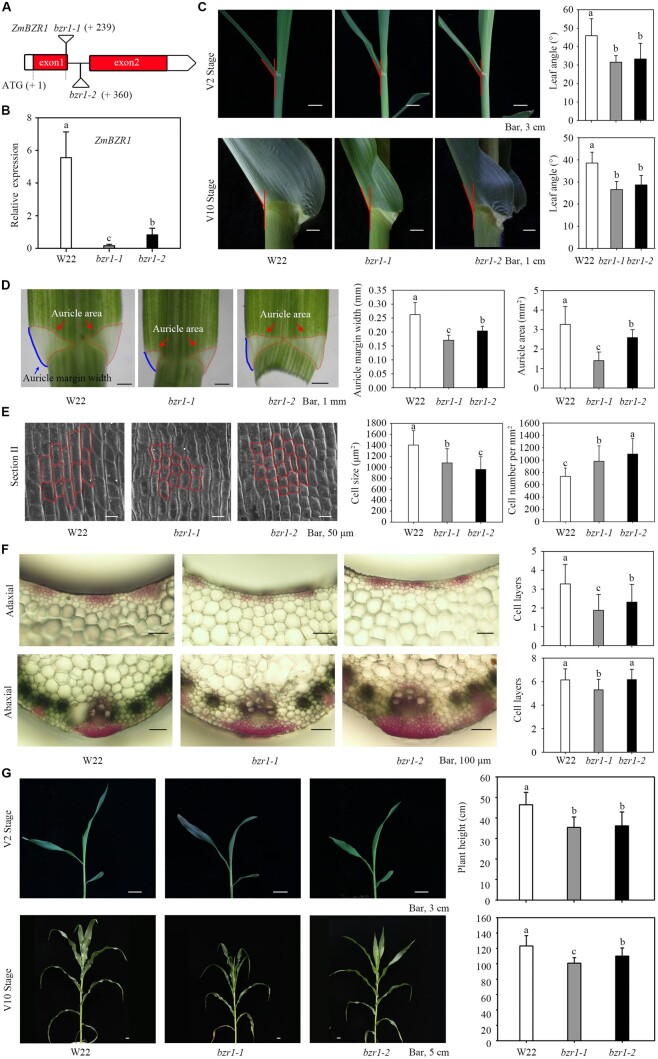
ZmBEH1 Mu-insertion mutant and CRISPR lines showed reduced leaf angle
As a direct downstream target of ZmLG2, we investigated whether ZmBEH1 is also involved in ligule development. To perform this study, we obtained a uniform Mu insertion mutant of ZmBEH1 (named beh1-1) from the maize stock center. Genomic DNA and RT-qPCR results showed that the Mu transposon was inserted into the 5′-UTR of ZmBEH1 and dramatically decreased its expression in the mutant (Figure 2, A and B; Supplemental Figure S4). The decreased expression of ZmBEH1 indeed caused erect leaves, with the leaf angle of the mutants decreasing 34.9% in the V2 stage and 30.9% in the V10 stage compared to the WT (Figure 2C). We noticed a smaller area and margin width of the auricle in beh1-1 compared to the area and margin width of the auricle in the WT, which might result in decreased auricle size and leaf angle at V2 and V10 stages (Figure 2D). We then checked the cell sizes and numbers using scanning microscopy by dividing the auricle into three sections (I, close to the midrib; II, middle distance to the midrib; and III, the margin of the auricle, Supplemental Figure S5B). Overall, the cell size in all three sections was decreased in the beh1-1 mutant, and the most obvious difference between WT and beh1-1 was observed in section II. In addition, beh1-1 had more cells when counting in the same area as the area counted in the WT in all three sections (Figure 2E;Supplemental Figure S6A), indicating that ZmBEH1 may control the cell size and cell number in the auricle region to influence the leaf angle. Accordingly, cross-sections of the ligular region revealed a reduced number of sclerenchyma cell layers at the adaxial blade-sheath junction in the mutant at the midrib (Figure 2F;Supplemental Figure S5A), suggesting that in maize, leaf angle is mediated at least in part through the regulation of adaxial sclerenchyma development.
Figure 2.
Phenotypes of beh1-1 plants. A, Uniform Mu-mediated insertion of ZmBEH1. A maize transposon-insertion line beh1-1 was obtained from the maize Uniform Mu resource, which carried a Mu insertion (UFMu-13557) in the 5′-UTR of ZmBEH1. B, Expression of the ZmBEH1 gene in WT inbred line W22 and beh1-1 plants at the V2 stage. C, Leaf angle changes between W22 and beh1-1 plants at the V2 and V10 stages. The leaf angles of the second and sixth leaves were measured at the V2 and V10 stages, respectively. n = 20. D, Quantitative measurement of auricles in W22 and beh1-1 plants at the V2 stage. Twenty replicates were used for auricle margin width and area. E, Scanning electron microscopic observation of auricles in W22 and beh1-1 plants at the V2 stage. The auricles were divided into three parts (I–III), representing the midrib, 1/2 and the margin of the auricle (Supplemental Figure S5B). Part II of the auricle is presented here, and partial cellular outlines are labeled. Four replicates were used for SEM. F, Cross-sections of the ligular region from W22 and beh1-1 plants at the V2 stage. The sclerenchyma cell (SC) layers were stained red with safranin. The number of SC cell files on the adaxial and abaxial sides was calculated from 10 replicates. G, Plant height of W22 and beh1-1 plants at the V2 and V10 stages. n = 20. Error bars are SD. *P < 0.05, **P < 0.01 determined by Student’s t test.
The decreased expression of ZmBEH1 also influenced the growth rate in the mutant, since the total heights were much shorter in the mutant than in the WT in both the 15-d-old seedling stage (V2 stage) and 54-d-old seedling stage (V10 stage; Figure 2G). Interestingly, when the WT height stopped increasing after silking, the beh1-1 mutant could catch up.
To confirm the function of ZmBEH1, we used the CRISPR/Cas9 system to edit ZmBEH1 specifically. The two obtained CRISPR mutants that segregated CAS9 were named beh1-2 and beh1-3, which have a 76 bp deletion and one base pair change, respectively (Supplemental Figure S7A). Consistent with the phenotype of beh1-1, beh1-2, and beh1-3 exhibited erect leaves, fewer layers of sclerenchyma cells on the adaxial side and shortened plant height at the V2 stage (Supplemental Figure S7, B and C). However, the plant heights were consistently shorter than the plant heights in the WT throughout the whole life cycle, which was different from the results of beh1-1, which reached the WT height after flowering. Collectively, the evidence from these lines suggests that ZmBEH1 plays a role in leaf angle formation in maize.
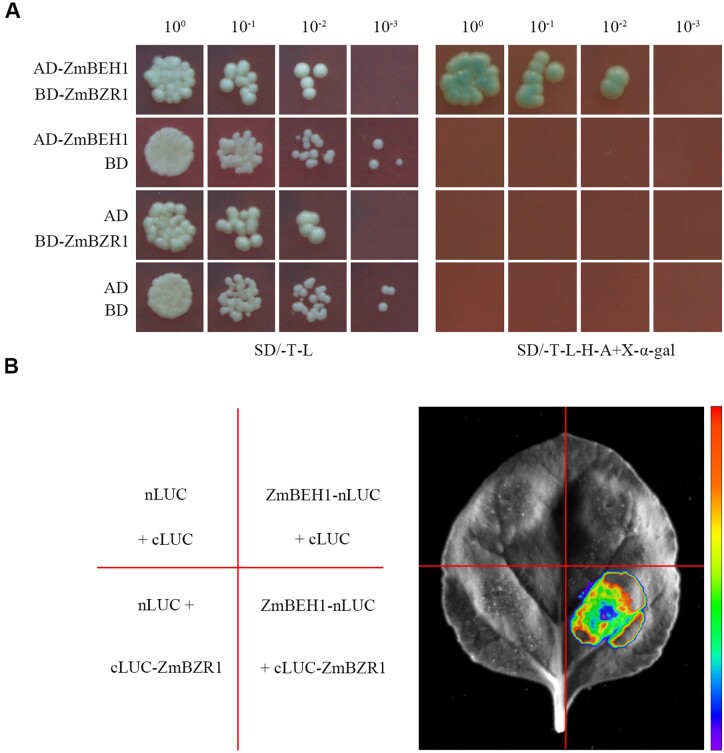
Yeast two-hybrid and split luciferase assays showed that ZmBEH1 interacts with ZmBZR1
To identify the partners that interact with ZmBEH1, we screened the B73 leaf cDNA yeast two-hybrid library using ZmBEH1 as bait. A protein that shares a close phylogenetic relationship with OsBZR1 and AtBZR1/BES1 (Supplemental Figure S2) was identified to have a strong interaction with ZmBEH1 (Figure 3A), which we named ZmBZR1.
Figure 3.
ZmBEH1 interacts with ZmBZR1. A, Interaction between ZmBEH1 and ZmBZR1 in yeast two-hybrid assays. Different concentrations of cotransformed yeast cells were spotted on synthetic dropout (SD) medium without tryptophan and leucine (SD/-T-L) or without tryptophan, leucine, histidine, and adenine plus 20 mg/mL X-α-gal (SD/-T-L-H-A + X-α-gal). B, Interaction between ZmBEH1 and ZmBZR1 in the split luciferase assay.
To further confirm the interaction between ZmBZR1 and ZmBEH1 in vivo, we then performed a split luciferase assay with ZmBEH1 fused to the N-terminus of LUC (ZmBEH1-nLUC) and ZmBZR1 fused to the C-terminus of LUC (cLUC-ZmBZR1). When the Agrobacterium tumefaciens strain GV3101 containing both the ZmBEH1-nLUC and cLUC-ZmBZR1 constructs was injected into the tobacco (Nicotiana tabacum) leaf, a strong luciferase signal was detected, suggesting that the interaction between ZmBEH1 and ZmBZR1 also occurred in vivo (Figure 3B).
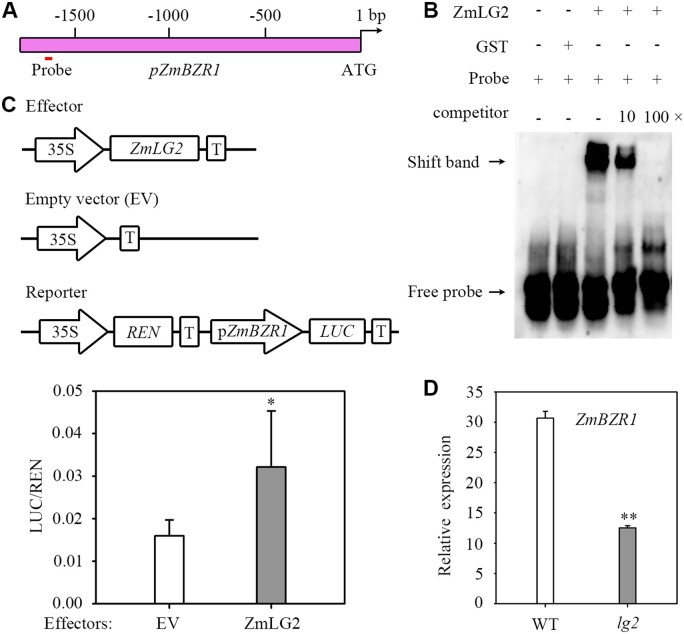
ZmLG2 also regulates the expression of ZmBZR1
Since ZmLG2 directly regulates the expression of ZmBEH1, which is homologous to ZmBZR1, we wondered whether ZmLG2 could also regulate ZmBZR1. Although ZmBZR1 was not among the set of predicted ZmLG2 downstream BR signaling genes from the ChIP-seq results, a weak binding peak existed in the G-browser, which did not pass the statistical selection (Supplemental Figure S8), and there was a ZmLG2 binding motif at the ZmBZR1 promoter 1,650 bp upstream of the initiation codon (Figure 4A). We then tested whether ZmLG2 could directly bind to the ZmBZR1 promoter using an EMSA. As shown in Figure 4B, ZmLG2 showed strong binding affinity to the probe, and binding was inhibited with unlabeled DNA probes at increasing concentrations, confirming the direct binding of ZmLG2 to the ZmBZR1 promoter. Next, a dual-LUC transient assay performed in maize protoplasts confirmed that ZmLG2 could activate the expression of ZmBZR1, as the ZmLG2 protein significantly induced the expression of LUC driven by the ZmBEH1 promoter (Figure 4C). Furthermore, we conducted RT-qPCR on the ZmBZR1 gene in the ligular region of V3 leaves in lg2 and WT seedlings as we did on ZmBEH1 and found that the expression of ZmBZR1 was significantly reduced (more than 2-fold) in the ligular region in the lg2 mutant compared with the expression of ZmBZR1 in the WT, further confirming the positive regulation of ZmLG2 on the expression of ZmBZR1 (Figure 4D).
Figure 4.
ZmLG2 directly activates the expression of ZmBZR1. A, Diagram of the promoter fragments of ZmBZR1. The location of the binding site is indicated. B, ZmLG2 directly binds to the ZmBZR1 promoter in EMSA. “−” indicates the absence of this component. “+” indicates the presence. The arrows indicate the positions of the bound complex and free probe. GST is the control of ZmLG2-GST. C, ZmLG2 activated the expression of ZmBZR1 in a dual-luciferase assay. The coding sequence of ZmLG2 driven by the 35S promoter was used as an effector, and the empty vector (EV) was used as an effector control in the transient luciferase assay performed in maize protoplasts. The vector with the REN gene driven by a 35S promoter and firefly luciferase (LUC) gene driven by the promoter sequence from ZmBZR1 was used as the reporter. The LUC/REN ratio represents the relative activity of the promoters. n = 5. D, RT-qPCR analyses showing the relative expression of ZmBZR1 in the ligular region of WT and lg2 mutants. n = 7. Error bars indicate SD. *P < 0.05, **P < 0.01 determined by Student’s t test.
ZmBZR1 Mu-insertion mutants and CRISPR lines showed reduced leaf angle
Since ZmBZR1 was positively regulated by ZmLG2, similar to ZmBEH1, we suspect that it also participates in leaf angle formation. To test this hypothesis, we obtained two potential loss-of-function mutants of ZmBZR1: one had a uniform Mu inserted into the first exon of ZmBZR1, and the other had a Mu inserted into the first intron, which was referred to as bzr1-1 and bzr1-2, respectively (Figure 5A;Supplemental Figure S4). We found that the expression of ZmBZR1 in both mutant alleles was significantly decreased (Figure 5B), and the plants exhibited the erect leaf phenotype similar to beh1-1. Compared with the WT, the leaf angle of bzr1-1 and bzr1-2 decreased 31.3% and 27.5% at the V2 stage, respectively. Moreover, in the V10 stage, bzr1-1 and bzr1-2 decreased 33.2% and 31.1%, respectively (Figure 5C). Correspondingly, the area and margin width of the auricle region decreased in both the bzr1-1 and bzr1-2 mutants (Figure 5D). We noticed that the auricle in both mutants showed a smaller cell size than the cell size in the WT, suggesting that decreased cell size in mutants may confer a reduced auricle area (Figure 5E;Supplemental Figure S6B). Similar to ZmBEH1, bzr1-1 and bzr1-2 also showed fewer sclerenchyma layers on the adaxial side than WT (Figure 5F).
Figure 5.
Characterization of bzr1-1 and bzr1-2 mutants. A, Gene model shows the inserted position of uniform Mu (UFMu-13537 and UFMu-03258) in the 1st exon (bzr1-1) and 1st intron (bzr1-2) of the ZmBZR1 gene. B, Expression analysis of ZmBZR1 in W22, bzr1-1 and bzr1-2 plants at the V2 stage. C, Leaf angle changes between W22 and bzr1-1 and bzr1-2 plants in the V2 and V10 stages, n = 20. D, Quantitative measurement of auricles in W22, bzr1-1 and bzr1-2 plants at the V2 and V10 stages. Twenty replicates were used for auricle margin width and area. E, Scanning electron microscopic observation of auricles in W22, bzr1-1 and bzr1-2 plants at the V2 stage. The auricles were divided into three parts (I–III), representing the midrib, 1/2 and the margin of the auricle (Supplemental Figure S5B). Part II of the auricle is presented here, and partial cellular outlines are labeled. Four replicates were used for SEM. F, Cross-sections of the ligular region from W22 and bzr1-1 and bzr1-2 plants at the V2 stage. The sclerenchyma cell (SC) layers stained red with safranin. The number of SC cell files on the adaxial (top) and abaxial sides (bottom) was calculated from 10 replicates. G, Plant height of W22 and bzr1-1 and bzr1-2 plants in the V2 and V10 stages. n = 20. Error bars are SD. Different letters above the columns indicate statistically significant differences between groups (Student’s t test).
At the V10 stage, bzr1-1 reached approximately 81.8%, and bzr1-2 reached approximately 89.3% of the WT height (Figure 5G). However, when comparing another nine important agronomic traits, including ear height, leaf length, leaf width, and tassel branch number, between bzr1 and the WT plants at the blister stage (Supplemental Figure S9), the difference observed between bzr1-1 and WT was much more dramatic than the difference in bzr1-2, indicating that the Mu inserted in the first intron of ZmBZR1 caused a weak allele.
In addition, another two mutant alleles of ZmBZR1 (bzr1-3 and bzr1-4), which presented 42 bp and 72 bp deletions in the first exon, were generated through the CRISPR/Cas9 system (Supplemental Figure S10A). The reduced leaf angle and shortened plant height were consistent with the phenotype of bzr1-1 (Supplemental Figure S10B).
ZmBEH1 and ZmBZR1 act synergistically to regulate leaf angle
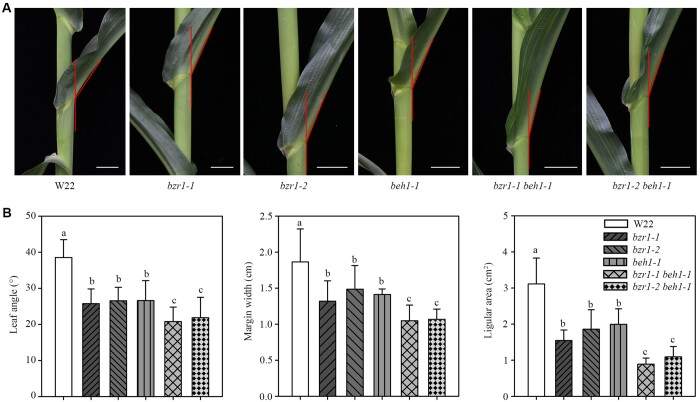
We suspected that ZmBZR1 and ZmBEH1 work redundantly in regulating maize leaf angle. To test this hypothesis, we created double mutants of bzr1-1 beh1-1 and bzr1-2 beh1-1 and measured the leaf angle at the V10 stage. Compared with the single mutant plants, the leaf angle in the double mutants decreased by approximately 6° and 5° in bzr1-1 beh1-1 and bzr1-2 beh1-1, respectively (Figure 6A). Consistently, double mutant plants had upright leaf angles, narrower margin widths, and smaller areas of auricles, resulting in reduced auricle size at the V10 stage (Figure 6B). Except for these differences, the double mutants were almost identical to the single mutants in terms of other agronomic traits, e.g. leaf length and width (Supplemental Figure S11, A and B). Together with the observation that ZmBZR1 and ZmBEH1 physically interact, we propose that they might cooperate with each other to regulate leaf angle.
Figure 6.
Leaf angle observation from single and double mutants of bzr1 and beh1. A, Leaf angle morphology of W22, single mutants of bzr1-1, bzr1-2, and beh1-1, double mutants of bzr1-1 beh1-1 and bzr1-2 beh1-1. Bar, 3 cm. B, Quantitative measurements of leaf angle (n = 20), auricle margin width (n = 10), and auricle area (n = 10) in W22 single and double mutants. Error bars are SD. Different letters above the columns indicate statistically significant differences between groups (Student’s t test).
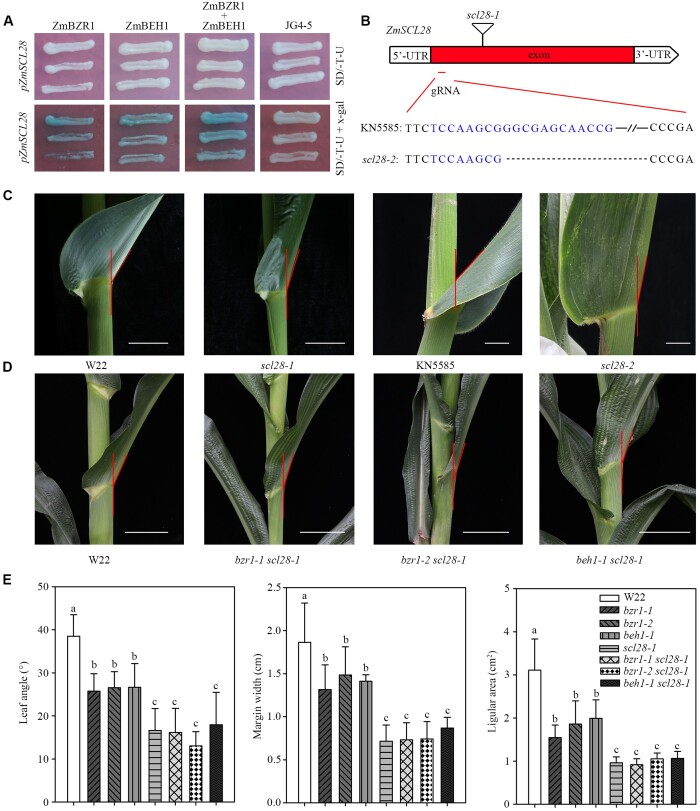
Yeast one-hybrid analysis identified ZmSCL28 as a downstream target of ZmBZR1 and ZmBEH1
To study how these TFs regulate leaf angle, we used yeast one-hybrid assays to identify their potential downstream targets. Y1H assays showed that the ZmSCL28 promoter can be targeted by both ZmBZR1 and ZmBEH1 (Figure 7A). ZmSCL28 encodes a GRAS domain TF that shared 77.8% sequence similarity with rice DLT (Supplemental Figure S12). In rice, OsBZR1 binds to the CGTGCG element (named BRRE motif) in the promoter of the DLT gene to suppress its expression, and a similar regulation has also been reported in Arabidopsis (Li and Jin, 2007; Tong et al., 2009).
Figure 7.
ZmSCL28 works downstream of ZmBZR1 and ZmBEH1. A, Y1H assays showed that ZmBZR1 and ZmBEH1 bound to the promoter of ZmSCL28. B, Gene model presents the positions of two mutant alleles in ZmSCL28. The triangle shows the position of a Mu insertion (UFMu-09491) from line scl28-1 in the W22 genetic background. The position of a single-guide RNA (sgRNA) designed to mutate ZmSCL28 using CRISPR/Cas9 technology is labled in exon. The sequence of sgRNA is highlighted, and the symbols “…” indicate the deletion caused by CRISPR/Cas9-induced mutations in line scl28-2 in the KN5585 genetic background. C, Leaf angle morphology in W22 and scl28-1, KN5585 and scl28-2 at the V10 stage. Bar, 5 cm. D, Leaf angle morphology in the WT W22 and double mutants of bzr1-1 scl28-1, bzr1-2 scl28-1, and beh1-1 scl28-1 at the V10 stage. Bar, 5 cm. E, Quantitative measurements of leaf angle (n = 20), auricle margin width (n = 10), and auricle area (n = 10) in single and double mutants. Error bars are SD. Different letters above the columns indicate statistically significant differences between groups (Student’s t test).
Next, we obtained a ZmSCL28 uniform Mu insertion line (scl28-1) and confirmed the insertion in the first exon by PCR (Figure 7B;Supplemental Figure S4). The plant architecture of scl28-1 is similar to the plant architecture of bzr1-1, bzr1-2, and beh1-1, conferring erect leaves, short auricle margin width, small auricle size and decreased plant height, but the leaf angle is even smaller (Figure 7, C and E; Supplemental Figure S13, A and C). Accordingly, the cross-sections of the ligular region exhibited a similar variation as the mutants of ZmBZR1 and ZmBEH1, with the scl-28 mutant showing fewer sclerenchyma layers on the adaxial side than WT (Supplemental Figure S14).
To further confirm this observation, we generated a second loss-of-function mutant of SCL28 by CRISPR/Cas9 (Figure 7B). This scl28-2 line has a 49 bp deletion located 50 bp downstream of the start codon. The phenotype of scl28-2 is similar to the phenotype of scl28-1, e.g. small leaf angle, upright leaf, and short plant height (Figure 7, C and E; Supplemental Figure S13, A and C), confirming that ZmSCL28 also plays important roles in controlling plant architecture.
To genetically test the relationship among ZmSCL28, ZmBZR1, and ZmBEH1, double mutants of bzr1-1 scl28-1, bzr1-2 scl28-1, and beh1-1 scl28-1 were generated. The double mutants had significantly reduced leaf angle, margin width, the area of the auricle region, and plant height compared with the single mutants of bzr1-1, bzr1-2, and beh1-1, which had no significant difference with the mutant of scl28-1. (Figure 7, D and E; Supplemental Figure S13, B and C). All these results support the conclusion that ZmSCL28 is the direct target of ZmBZR1 and ZmBEH1 to regulate leaf angle in maize.
Discussion
Erect leaves improve light interception and increase planting density, thus enhancing the yield per unit of land (Duvick, 2005; Tian et al., 2011). Therefore, the selection of compact maize architecture with a small leaf angle is a long-term goal in maize breeding ZmLG2 is one of the most important loci controlling leaf angle formation in maize; however, mutations in this gene abolish normal ligules and cause extremely erect leaf angles, which limit its use in commercial crop production. Therefore, we believe it is important to explore the downstream targets of ZmLG2 to achieve both small leaf angle and normal ligule development.
Interestingly, our study based on ChIP-seq provides evidence that ZmLG2 may affect leaf angle development by regulating the expression of genes involved in BR signaling (Supplemental Figure S1A). Specifically, we found that ZmLG2 directly binds and activates the expression of ZmBEH1 (Figure 1), a BZR1/BES1 homolog gene. This binding depends on the specific motif of CGTCA, and the first (C) and third (T) bases within the motif played important roles in ZmLG2 binding, followed by G and A, the second and fifth bases (Figure 1C). The binding affinity of ZmLG2 might also be influenced by the adjuvant environment of the motif, since only three out of four motifs within 1.3 Kb of ZmBEH1 could be bound by ZmLG2, and the tandem duplication of CGTCA within 36 bp (−1,221 to −1,185) might help trap ZmLG2 on the motif (Figure 1C). Despite the weak binding of ZmBZR1 (Supplemental Figure S8) in ChIP-seq, EMSA proved that ZmLG2 directly binds to the ZmBZR1 promoter and activates its expression (Figure 4). The discovery of the physical interaction between ZmBEH1 and ZmBZR1 further proved a close relationship between these two homologous genes as direct downstream targets of ZmLG2 (Figure 3).
The function of BZR1 is well documented in Arabidopsis. In the presence of BR, BIN2 is inactivated, BZR1 is dephosphorylated by PP2A, moves into the nucleus to promote the expression of BR-dependent genes, and then influences various developmental processes and stress responses (He et al., 2005; Yin et al., 2005; Sun et al., 2010; Tang et al., 2011; Wang et al., 2012). However, our current knowledge about BZR1 and its homologous genes is still rather limited in monocots, and there are no reports about ZmBZR1 and ZmBEH1 mutants in maize. The overexpression of maize BZR1 in Arabidopsis enlarged organ size and produced larger seeds and larger cotyledons and leaves, which reveals its function in regulating cell size (Zhang et al., 2020). Recent report on the ZmBZR1 homolog gene ZmBES1/BZR1-5 also supports the role of BZR family numbers in controlling maize kernel size development (Sun et al., 2020). However, ZmBZR1 could not bind all downstream target genes identified from AtBZR1, indicating that some differences existed in BZR1 between dicot and monocot plants (Zhang et al., 2020). The phenotypes of the ZmBZR1 and ZmBEH1 maize mutants were quite similar; both were semidwarf and presented decreased leaf angles (Figures 2 and 5), indicating that the decreased levels of ZmBZR1 and ZmBEH1 might affect the genes related to cell extension to regulate plant architecture. Indeed, we observed reduced cell size in the auricle region and reduced numbers of sclerenchyma cell layers at the adaxial site of the ligular region (Figure 2D–F). However, how ZmBEH1 and ZmBZR1 regulate cell size and sclerenchyma cell development at adaxial/abaxial sites needs further exploration.
The mild decreases in plant height and leaf angle of ZmBZR1 and ZmBEH1 mutants are quite different from other BR signaling- and biosynthesis-related mutants reported in maize. zmbri1, to our knowledge, was the earliest reported maize mutant participating in the BR signaling pathway. The zmbri1-RNAi plants showed strongly decreased plant height and upright leaves (Kir et al., 2015). Other BR biosynthetic maize mutants, e.g. na1, na2, and brd1, all exhibited severe dwarf stature, altered leaf morphology, and suppressed floral organ growth, similar to the BR signaling mutants of zmbri1 (Hartwig et al., 2011; Makarevitch et al., 2012; Best et al., 2016). In contrast, the loss of function of ZmBZR1 and ZmBEH1 had only a mild influence on maize height and leaf angle. Despite the similar phenotypes between ZmBZR1 and ZmBEH1 mutants, the two genes were not redundant, at least in regulating leaf angle formation, since the bzr1-1 beh1-1 and bzr1-2 beh1-1 double mutants displayed smaller leaf angles than any of the single mutants (Figure 6), indicating that as a gene family with multiple numbers, other genes belonging to the BZR/BES1 family, such as ZmBEH2 (Zm00001d039439), which was also bound by ZmLG2 according to the ChIP-seq results (Supplemental Figure S1A), may also play roles in regulating maize growth and leaf angle formation downstream of ZmLG2. The upright leaf and semidwarf phenotypes are ideal maize plant architectures pursued by breeders. Therefore, the BZR1 and BEH1 genes and their family numbers may provide ideal alleles for maize molecular breeding. A recent GWAS on leaf angle across the canopy in sorghum reported that the expression of BZR1/BES1 and Dw3 (a gene involved in auxin transport) was highly and positively associated with the distribution of leaf angles at different layers, and a gradient of expression existed in the collar tissue from bottom to the top of the canopy (Mantilla-Perez et al., 2020), further emphasizing the value of manipulation of the BZR1 family in creating the ideal plant architecture. However, studies on allele effects on the BZR1 and BEH1 gene families are still missing in maize and other monocot plants, and we need to discover the best haplotypes of these genes for breeding. As upstream members, the ZmBZR1 and ZmBEH1 genes regulate other genes to transduce BR signaling and control maize development. Interestingly, our Y1H assay proved that ZmSCL28 was a direct downstream target of both ZmBZR1 and ZmBEH1. ZmSCL28 is the homolog gene of DLT in rice, and the mutation of ZmSCL28 shared similar phenotypes with dlt, e.g. dwarf and erect leaves, indicating the conserved function of these TFs in the different species. In rice, DLT was bound by OsBZR1 in the promoter through the BR-response element, and the loss-of-function mutant dlt was similar to BR-insensitive mutants (Tong et al., 2009). In maize, not only ZmBZR1 but also ZmBEH1 could bind to the BR-response element in ZmSCL28. We also genetically proved that ZmSCL28 is the downstream target of ZmBZR1 and ZmBEH1, since the double mutants of bzr1-1 scl28-1, bzr1-2 scl28-1, and beh1-1 scl28-1 have phenotypes almost identical to the scl28-1 single mutant. For example, the leaf angle was 16.6° in the scl28-1 mutant and 16.1° in the bzr1-1 scl28-1 double mutant; however, the angle size was much larger than the angle size of single mutants of bzr1-1 (25.7°), bzr1-2 (26.5°), and beh-1 (26.6°; Figure 7). The DLT in rice promotes tillering, which needs to be tightly regulated to pursue ideal tillering numbers (Tong et al., 2009; Liu et al., 2021). The phenotypes in maize scl28 mutants that are stronger than bzr1-1 and beh-1 also indicate its importance as a downstream target in regulating maize development and architecture.
Taken together, we propose a ZmLG2-BEH1/BZR1-SCL28 regulatory cascade that affects leaf angle formation in maize (Figure 8). In this model, we showed that ZmBEH1 is directly activated by ZmLG2 and that ZmBEH1 can interact with ZmBZR1. ZmBZR1 is also directly activated by ZmLG2. Furthermore, we found that both ZmBEH1 and ZmBZR1 bind to the ZmSCL28 promoter to regulate leaf angle. As the downstream targets of ZmLG2, the loss-of-function mutants of ZmBEH1, ZmBZR1, and ZmSCL28 had normal development of leaf ligules, as expected; however, the size of the leaf angle was dramatically reduced. Therefore, we propose that these three target loci, ZmBZR1, ZmBEH1, and ZmSCL28, are ideal targets to manipulate leaf angle to generate upright and semidwarf plant architecture, the two important traits pursued by breeders to increase planting density and yields.
Figure 8.
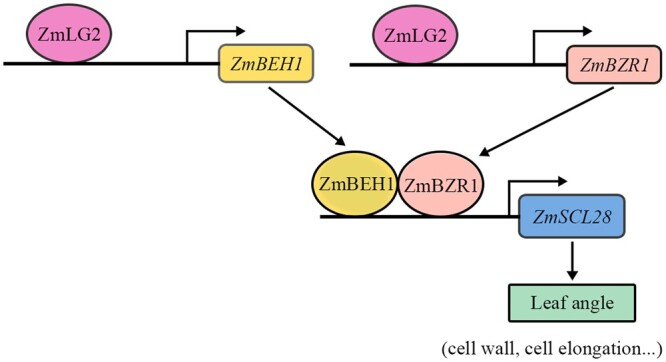
A proposed ZmLG2-ZmBEH1/BZR1-ZmSCL28 module regulates leaf angle in maize. ZmLG2 binds to the promoter of ZmBEH1 to activate its expression. ZmBEH1 interacts with ZmBZR1, which is also a direct target of ZmLG2, to regulate the expression of a downstream target gene, ZmSCL28. These proposed regulatory cascades may control the leaf angle size in maize.
Materials and methods
Plant materials and growth conditions
The maize Uniform Mu mutants with stock numbers UFMu-13537, UFMu-03258, and UFMu-13557 were requested from the maize genetic cooperation stock center. After two generations of backcrossing to maize inbred line W22, the BC2F1 population that segregated the WT and mutant phenotypes was obtained. The stable single mutants from each gene were crossed to each other to obtain the double mutant.
To observe the phenotypes at the seedling stage, seeds were planted in soil and grown in a growth room at 28°C with a 12-h light/22°C, 12-h dark cycle, and 60% relative humidity.
Yeast one-hybrid
To test the binding of ZmBZR1 and ZmBEH1 to the ZmSCL28 promoter, the full-length CDSs of ZmBZR1 and ZmBEH1 were amplified and cloned into the pJG4-5 vector at the EcoRI and XhoI restriction sites, and the fusion plasmids were cotransformed with the EcoRI- and KpnI-digested LacZ reporter vector (LacZi2μ) driven by the ZmSCL28 promoter. The empty vector pJG4-5 and the LacZ reporter driven by the ZmSCL28 promoter served as negative controls.
The transformed yeast (Saccharomycetes) strain EGY48 was plated on SD/-Trp-Ura medium and grown at 30°C for 3 d. Then, the transformants were tested on an SD screening medium containing X-gal (5-bromo-4-chloro-3-indolyl-b-D-galactopyranoside) at 30°C for 4 d. The sequences of primers used in Y1H are listed in Supplemental Table S1.
Transient expression assay in maize protoplast
For the dual-luciferase transient expression assay, ZmBZR1 and ZmBEH1 promoter segments were amplified from inbred line B73 and recombined into the pGreenII 0800-LUC vector at the HindIII and BamHI restriction sites to generate the pZmBZR1/BEH1::LUC plasmids as reporters. The Renilla luciferase (REN) gene driven by the 35S promoter in the pGreenII 0800-LUC vector was used as the internal control. The full-length CDS of ZmLG2 was amplified and recombined into the pHW-3xAvi vector via Gateway LR Clonase II Enzyme mix driven by the 35S promoter, forming the effectors. The empty pHW-3 × Avi vector was used as control. Transient dual-luciferase assays were performed in maize protoplasts collected from the leaves of 12-d-old etiolated seedlings of inbred line B73. The luciferase signal was detected using dual-luciferase assay reagents following the manufacturer’s instructions. Relative LUC activity was calculated by normalizing LUC activity to REN activity. The sequences of primers used in the transient expression assay are listed in Supplemental Table S1.
Electrophoretic Mobility Shift Assay
EMSA was performed as described previously (Song et al., 2020). The protein fused with GST was purified first. Full-length ZmLG2 cDNA was amplified with gene-specific primers and cloned into the expression vector Pcold-GST. The construct was transformed into Escherichia coli BL21 cells grown at 37°C. At OD600 = 0.4–0.5, the culture solution was placed at 16°C and allowed to stand for 30 min. Isopropylthio-β-galactoside was added at a final concentration of 1 mM, and the culture was incubated at 16°C for 24 h. The Pcold-GST-LG2-expressing GST-LG2 fusion protein was purified with BeaverBeads GSH (Beaver, Suzhou, China) and used for EMSA. The probes with biotin were synthesized according to the manufacturer’s recommendation (Supplemental Table S1). The affinity bands labeled with biotin probes were detected by an EMSA kit (Beyotime Biotechnology), and approximately 10 nm of probe was used for each binding assay. For the competition assay, an unlabeled probe was added to the reactions to detect the binding specificity.
Yeast two-hybrid assays
The yeast two-hybrid assay was performed using the Matchmaker™ Gold Yeast Two-Hybrid System (Clontech, Dalian, China). The full-length coding region of ZmBZR1 was ligated into the EcoRI- and PstI-digested pGBKT7 plasmid as bait, and the full-length coding region of ZmBEH1 was fused into the pGADT7 vector as prey via Gateway LR Clonase II Enzyme mix. The bait and prey constructs were cotransformed into the Y2H Gold yeast strain and grown at 30°C for 3 d. Next, the transformants were tested on SD screening medium containing X-α-Gal (5-bromo-4-chloro-3-indoxyl-α-D-galactopyranoside) at 30°C for 4 d. Empty vectors were cotransformed as negative controls. The sequences of primers used in Y2H are listed in Supplemental Table S1.
The split luciferase complementation assay
The split luciferase complementation assay was performed to examine the interaction between ZmBEH1 and ZmBZR1 using the constructs nLUC and cLUC. The full-length CDS of ZmBEH1 without the stop codon and ZmBZR1 with the stop codon were amplified and cloned into BamHI- and SalI-digested nLUC and KpnI- and BamHI-digested cLUC, respectively, forming the nLUC-ZmBEH1 and ZmBZR1-cLUC constructs. Agrobacterium strain GV3101 cells carrying all constructs were transiently infiltrated into N.benthamiana leaves. After culturing for 3 d, luciferin (1 mM) was sprayed to activate luciferase, and the fluorescence signals were observed by the Chemiluminescent Imaging System (Tanon-5200). The primers used in the split luciferase complementation assay are listed in Supplemental Table S1.
RNA extraction and RT-qPCR analysis
Total RNA was extracted using TRIzol reagent (Ambion) from maize leaves. The RNA samples were treated with DNase I (Thermo) and the concentration was measured by DS-11. Subsequently, cDNA was prepared using M-MLV Reverse Transcriptase and qPCR was conducted using the SYBR Premix Ex Taq kit (Trans) on a Step One System (Applied Biosystems). The quantification method (2−ΔCt) was used and the gene expression level was estimated using three independent experiments. The maize Ubi2 (UniProtKB/TrEMBL, Q42415) gene was used as an internal control to normalize the data. The process of qPCR consisted of an initial denaturation step at 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 30 s. The primers for RT-qPCR are shown in Supplemental Table S1.
Phylogenetic analysis of ZmBZR1/BEH1 and ZmSCL28
The full-length amino acid sequences of ZmBZR1/BEH1 and ZmSCL28 were used to BLAST search in the protein databases of GRAMENE (http://www.gramene.org/), TAIR (http://www.arabidopsis.org), and RICEDATE (http://www.ricedata.cn/gene/) to identify the homologous genes in maize, Arabidopsis and rice. The amino acid sequences were aligned using ClustalW-2.0.10 software. A phylogenetic tree was constructed based on this alignment result using the neighbor-joining method in MEGA version 6 with the following parameters: Poisson model, pairwise deletion, uniform rates, and bootstrap method (1,000 replicates).
Microscopy observations and phenotypic analysis
For scanning electron microscopy (SEM) analysis, the auricle regions of W22 and Mu mutants at the V2 stage were sampled to examine the cell size and cell number. The samples were fixed with 2.5% (v/v) glutaraldehyde in phosphate buffer for more than 12 h at 4°C, washed with PBS every 20 min for a total of 5 times, and then soaked in osmic acid overnight. The next day, the samples were dehydrated by a graded series of ethanol from 45% to 100% for approximately 30 min at each step. Subsequently, the samples were strictly dried, coated with gold-palladium, and observed under SEM (JSM-6610LV). Cell size was determined as the average cell area of 20 cells per auricle region (n = 4). Cell numbers were counted at 1 mm2.
For the cross-section analysis, the ligular regions of WT and mutants at the V2 stage were dissected using an ultrathin blade. The ligular regions were placed in a 0.1% safranin solution for 5 min. After staining, the samples were destained in 75% ethanol for 5 min. After decolorization, the samples were soaked in distilled water. The cross-section of leaf ligular region was observed using a light microscope (NikonNi-U). The abaxial and adaxial sclerenchyma cell layers were counted in the recorded images.
Transformation
The CRISPR/Cas9 constructs for ZmBEH1, ZmBZR1, and ZmSCL28 were generated using the previously reported vector pBUE411 (Hui-Li Xing, 2014). The specific target sites and PAMs (the last three nucleotides) of ZmBEH1, ZmBZR1, and ZmSCL28 were designed and cloned into the pBUE411 vector. These constructs were introduced into the A.tumefaciens strain EHA105 and transformed into the maize inbred line KN5585 at WIMI Biotechnology Co., Ltd. The target regions of ZmBEH1, ZmBZR1, and ZmSCL28 were amplified from KN5585 and transgenic lines, and then the PCR products were sequenced to identify the mutations. For ZmBEH1, we obtained two independent homozygous knockout lines named beh1-2 and beh1-3. For ZmBZR1, we obtained two independent homozygous knockout lines named bzr1-3 and bzr1-4. For ZmSCL28, we obtained one homozygous knockout line named scl28-2. According to the selective marker gene, the glyphosate sensitivity of transgenic lines was detected, and knockout lines that were very sensitive to herbicide were selected for further research, so that the CAS9 and bar genes were removed from the plants. The T1 plant was self-crossed to generate T2 plants for further research. The phenotypes of these mutants were investigated under greenhouse or normal field planting conditions, together with their wild-type KN5585. The primers were shown in Supplemental Table S1.
Statistical analysis
To determine statistical significance, we employed Student’s t test (*P < 0.05, **P < 0.01).
Accession numbers
Sequence data from this article can be found in Maize GDB Database under the following accession numbers: ZmLG2 (Zm00001d042777), ZmBZR1 (Zm00001d021927), ZmBEH1 (Zm00001d046305), and ZmSCL28 (Zm00001d045507).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. ZmLG2 binds to the promoter of BR signaling-related genes.
Supplemental Figure S2 . Phylogenetic analysis of BZR1/BES1 family proteins in maize, rice and Arabidopsis.
Supplemental Figure S3. Tissues from the ligular region of V3 leaves were harvested for RT-qPCR assay.
Supplemental Figure S4. PCR identification of beh1-1, bzr1-1, bzr1-2, and scl28-1 mutants.
Supplemental Figure S5. Diagram of the cross-section and scanning electron microscopic observation of the auricle.
Supplemental Figure S6. Scanning electron microscopic observation of auricles in W22, beh1-1, bzr1-1, and bzr1-2.
Supplemental Figure S7. Phenotypes of beh1-2 and beh1-3 mutants.
Supplemental Figure S8. ZmLG2 ChIP-Seq binding profile for ZmBZR1.
Supplemental Figure S9. Agronomic characters of the bzr1-1 mutant.
Supplemental Figure S10. Characterization of the CRISPR/Cas9-mediated knock-out mutants of ZmBZR1.
Supplemental Figure S11. Phenotypes of the single and double mutants of bzr1-1, bzr1-2, and beh1-1.
Supplemental Figure S12. Phylogenetic analysis of SCL family.
Supplemental Figure S13. Phenotypes of scl28-1/scl28-2 and bzr1-1 scl28-1, bzr1-2 scl28-1, beh1-1 scl28-1 double mutant plants.
Supplemental Figure S14. Cross-sections of the ligular region from W22 and scl28-1 plants at the V2 stage.
Supplemental Table S1. The list of primers used in this paper.
Supplementary Material
Acknowledgments
We thank the maize genetics cooperation stock center for kindly providing uniform Mu mutants with the stock numbers UFMu-13537, UFMu-03258, and UFMu-13557. We thank Prof. Sarah Hake (University of California, Berkeley) for offering seeds of lg2 mutant. We would also like to thank Prof. Qijun Chen (China Agricultural University) for sharing the plasmid pBUE411.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31901552, 92035302, and 32100438), Hong Kong GRF 14109420, and China Postdoctoral Science Foundation (2020M672858 and 2021T140677).
Conflict of interest statement. The authors declare no competing financial interests.
Contributor Information
Xiaokun Wang, State Key Laboratory of Crop Biology, College of Agronomic Sciences, Shandong Agricultural University, Tai’an, Shandong 271018, China.
Xianglan Wang, State Key Laboratory of Crop Biology, College of Agronomic Sciences, Shandong Agricultural University, Tai’an, Shandong 271018, China.
Shilei Sun, State Key Laboratory of Crop Biology, College of Agronomic Sciences, Shandong Agricultural University, Tai’an, Shandong 271018, China.
Xiaoyu Tu, Joint Center for Single Cell Biology, School of Agriculture and Biology, Shanghai Jiao Tong University, Shanghai 200240, China.
Kande Lin, State Key Laboratory of Agrobiotechnology, School of Life Sciences, The Chinese University of Hong Kong, Hong Kong, China; The South China Botanical Garden, Chinese Academy of Sciences, Guangzhou, China.
Lei Qin, State Key Laboratory of Crop Biology, College of Agronomic Sciences, Shandong Agricultural University, Tai’an, Shandong 271018, China.
Xingyun Wang, State Key Laboratory of Crop Biology, College of Agronomic Sciences, Shandong Agricultural University, Tai’an, Shandong 271018, China.
Gang Li, State Key Laboratory of Crop Biology, College of Agronomic Sciences, Shandong Agricultural University, Tai’an, Shandong 271018, China.
Silin Zhong, The South China Botanical Garden, Chinese Academy of Sciences, Guangzhou, China.
Pinghua Li, State Key Laboratory of Crop Biology, College of Agronomic Sciences, Shandong Agricultural University, Tai’an, Shandong 271018, China.
P.L. and XL.W. designed the research; XK.W., XL.W., S.S., X.T., K.L., L.Q., XY.W. performed the experiments; XK. W, XL.W., G.L., S.Z., and P. L. wrote the article.
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/General-Instructions) is: Pinghua Li (pinghuali@sdau.edu.cn).
References
- Bai MY, Zhang LY, Gampala SS, Zhu SW, Song WY, Chong K, Wang ZY (2007) Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc Natl Acad Sci USA 104:13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best NB, Hartwig T, Budka J, Fujioka S, Johal G, Schulz B, Dilkes BP (2016) Nana plant2 encodes a maize ortholog of the Arabidopsis brassinosteroid biosynthesis gene DWARF1, identifying developmental interactions between brassinosteroids and gibberellins. Plant Physiol 171:2633–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc N, O'Connor D, Moon J, Lewis M, Hake S (2012) How to pattern a leaf. Cold Spring Harb Symp Quant Biol 77:47–51 [DOI] [PubMed] [Google Scholar]
- Cao Y, Zeng H, Ku L, Ren Z, Han Y, Su H, Dou D, Liu H, Dong Y, Zhu F (2020) ZmIBH1-1 regulates plant architecture in maize. J Exp Bot 71:2943–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LG, Gao Z, Zhao Z, Liu X, Li Y, Zhang Y, Liu X, Sun Y, Tang W (2019) BZR1 family transcription factors function redundantly and indispensably in BR signaling but exhibit BRI1-independent function in regulating anther development in Arabidopsis. Mol Plant 12:1408–1415 [DOI] [PubMed] [Google Scholar]
- Duvick DN (2005) Genetic progress in yield of United States maize (Zea mays L.). Maydica 50 :193–202 [Google Scholar]
- Gruszka D (2020) Exploring the brassinosteroid signaling in monocots reveals novel components of the pathway and implications for plant breeding. Int J Mol Sci 21: 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper L, Freeling M (1996) Interactions of liguleless1 and liguleless2 function during ligule induction in maize. Genetics 144:1871–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig T, Chuck GS, Fujioka S, Klempien A, Weizbauer R, Potluri DP, Choe S, Johal GS, Schulz B (2011) Brassinosteroid control of sex determination in maize. Proc Natl Acad Sci USA 108:19814–19819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang ZY (2005) BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307: 1634–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kir G, Ye H, Nelissen H, Neelakandan AK, Kusnandar AS, Luo A, Inzé D, Sylvester AW, Yin Y, Becraft PW (2015) RNA interference knockdown of BRASSINOSTEROID INSENSITIVE1 in maize reveals novel functions for brassinosteroid signaling in controlling plant architecture. Plant Physiol 169:826–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku L, Wei X, Zhang S, Zhang J, Guo S, Chen Y (2011) Cloning and characterization of a putative TAC1 ortholog associated with leaf angle in maize (Zea mays L.). PLoS ONE 6:e20621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert RJ, Johnson RR (1978) Leaf angle, tassel morphology, and the performance of maize hybrids. Crop Sci 18:499–502 [Google Scholar]
- Li JM, Jin H (2007) Regulation of brassinosteroid signaling. Trends Plant Sci 12:37–41 [DOI] [PubMed] [Google Scholar]
- Li H, Ye K, Shi Y, Cheng J, Zhang X, Yang S (2017) BZR1 positively regulates freezing tolerance via CBF-dependent and CBF-independent pathways in Arabidopsis. Mol Plant 10:545–559 [DOI] [PubMed] [Google Scholar]
- Liu Y, Wang H, Jiang Z, Wang W, Xu R, Wang Q, Zhang Z, Li A, Liang Y, Ou S, et al. (2021) Genomic basis of geographical adaptation to soil nitrogen in rice. Nature 590:600–605 [DOI] [PubMed] [Google Scholar]
- Luo XM, Lin WH, Zhu S, Zhu JY, Sun Y, Fan XY, Cheng M, Hao Y, Oh E, Tian M, et al. (2010) Integration of light- and brassinosteroid-signaling pathways by a GATA transcription factor in Arabidopsis. Dev Cell 19:872–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevitch I, Thompson A, Muehlbauer GJ, Springer NM (2012) Brd1 gene in maize encodes a brassinosteroid C-6 oxidase. PLoS ONE 7:e30798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantilla-Perez MB, Bao Y, Tang L, Schnable PS, Salas-Fernandez MG (2020) Toward "smart canopy" sorghum: discovery of the genetic control of leaf angle across layers. Plant Physiol 184:1927–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantilla-Perez MB, Salas Fernandez MG (2017) Differential manipulation of leaf angle throughout the canopy: current status and prospects. J Exp Bot 68:5699–5717 [DOI] [PubMed] [Google Scholar]
- Moreno MA, Harper LC, Krueger RW, Dellaporta SL, Freeling M (1997) liguleless1 encodes a nuclear-localized protein required for induction of ligules and auricles during maize leaf organogenesis. Genes Dev 11:616–628 [DOI] [PubMed] [Google Scholar]
- Pendleton JW, Smith GE, Winter SR, Johnston TJ (1968) Field investigations of the relationships of leaf angle in corn (Zea mays L.) to grain yield and apparent photosynthesis. Agronomy Journal 60:422–424 [Google Scholar]
- Ren Z, Wu L, Ku L, Wang H, Zeng H, Su H, Wei L, Dou D, Liu H, Cao Y, et al. (2020) ZmILI1 regulates leaf angle by directly affecting liguleless1 expression in maize. Plant Biotechnol J 18:881–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M, Kondo Y, Fukuda H (2018) BES1 and BZR1 redundantly promote phloem and xylem differentiation. Plant Cell Physiol 59:590–600 [DOI] [PubMed] [Google Scholar]
- Song Z, Yan T, Liu J, Bian Y, Heng Y, Lin F, Jiang Y, Wang Deng X, Xu D (2020) BBX28/BBX29, HY5 and BBX30/31 form a feedback loop to fine-tune photomorphogenic development. Plant J 104:377–390 [DOI] [PubMed] [Google Scholar]
- Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, He JX, Bai MY, Zhu S, Oh E, et al. (2010) Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell 19:765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Yu H, Qu J, Cao Y, Ding L, Feng W, Khalid MHB, Li W, Fu F (2020) Maize ZmBES1/BZR1-5 decreases ABA sensitivity and confers tolerance to osmotic stress in transgenic Arabidopsis. Int J Mol Sci 21:996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Yuan M, Wang R, Yang Y, Wang C, Oses-Prieto JA, Kim TW, Zhou HW, Deng Z, Gampala SS, et al. (2011) PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat Cell Biol 13: 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F, Bradbury PJ, Brown PJ, Hung H, Sun Q, Flint-Garcia S, Rocheford TR, McMullen MD, Holland JB, Buckler ES (2011) Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat Genet 43:159–162 [DOI] [PubMed] [Google Scholar]
- Tian J, Wang C, Xia J, Wu L, Xu G, Wu W, Li D, Qin W, Han X, Chen Q, et al. (2019) Teosinte ligule allele narrows plant architecture and enhances high-density maize yields. Science 365:658–664 [DOI] [PubMed] [Google Scholar]
- Tong H, Jin Y, Liu W, Li F, Fang J, Yin Y, Qian Q, Zhu L, Chu C (2009) DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J 58:803–816 [DOI] [PubMed] [Google Scholar]
- Tu X, Mejía-Guerra MK, Valdes Franco JA, Tzeng D, Chu PY, Shen W, Wei Y, Dai X, Li P, Buckler ES, et al. (2020) Reconstructing the maize leaf regulatory network using ChIP-seq data of 104 transcription factors. Nat Commun 11:5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh J, Waters CA, Freeling M (1998) The maize gene liguleless2 encodesa basic leucine zipper protein involvedin the establishment of the leaf blade-sheath boundary. Genes Dev 12:208–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Bai MY, Oh E, Zhu JY (2012) Brassinosteroid signaling network and regulation of photomorphogenesis. Annu Rev Genet 46:701–724 [DOI] [PubMed] [Google Scholar]
- Wang W, Bai MY, Wang ZY (2014) The brassinosteroid signaling network-a paradigm of signal integration. Curr Opin Plant Biol 21:147–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14:327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J (2005) A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120: 249–259 [DOI] [PubMed] [Google Scholar]
- Zhang X, Guo W, Du D, Pu L, Zhang C (2020) Overexpression of a maize BR transcription factor ZmBZR1 in Arabidopsis enlarges organ and seed size of the transgenic plants. Plant Sci 292:110378. [DOI] [PubMed] [Google Scholar]
- Zhang J, Ku LX, Han ZP, Guo SL, Liu HJ, Zhang ZZ, Cao LR, Cui XJ, Chen YH (2014) The ZmCLA4 gene in the qLA4-1 QTL controls leaf angle in maize (Zea mays L.). J Exp Bot 65:5063–5076 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.