Abstract
Zebrafish (Danio rerio) have attracted much attention over the past decade as a reliable model for gut microbiome research. Owing to their low cost, strong genetic and development coherence, efficient preparation of germ‐free (GF) larvae, availability in high‐throughput chemical screening, and fitness for intravital imaging in vivo, zebrafish have been extensively used to investigate microbiome‐host interactions and evaluate the toxicity of environmental pollutants. In this review, the advantages and disadvantages of zebrafish for studying the role of the gut microbiome compared with warm‐blooded animal models are first summarized. Then, the roles of zebrafish gut microbiome on host development, metabolic pathways, gut‐brain axis, and immune disorders and responses are addressed. Furthermore, their applications for the toxicological assessment of aquatic environmental pollutants and exploration of the molecular mechanism of pathogen infections are reviewed. We highlight the great potential of the zebrafish model for developing probiotics for xenobiotic detoxification, resistance against bacterial infection, and disease prevention and cure. Overall, the zebrafish model promises a brighter future for gut microbiome research.
Keywords: gut microbiome, host physiology, probiotic treatment, toxicological assessment, zebrafish
Owing to their unique attributes, zebrafish have been harnessed as a powerful model for investigating host‐microbiome interactions using diverse techniques. In this review, the roles of zebrafish gut microbiome on their host development, gut‐brain axis, immune responses and metabolic pathways are summarized. Their applications on the toxicological assessment of environmental pollutants, the exploration of the pathogenic infection process, and the applications of probiotics are also reviewed.
1. INTRODUCTION
The gut microbiome is widely acknowledged to coexist with the animal host, consisting of symbiotic and pathogenic bacteria in a dynamic balance state that produce a wide variety of signaling molecules. These bacteria colonize the intestine and perform functions the host itself cannot accomplish; in turn, they rely on the habitat provided by the host. A healthy gut microbiome plays a significant and irreplaceable role in determining the host’s overall health.
Zebrafish is an omnivorous freshwater fish that belongs to the small carp family. It has been widely used for research purposes on embryology and tissue regeneration, molecular genetics, reproductive biology, and toxicology given its numerous advantages, including high fecundity, short lifespan, highly annotated genome, optical clarity of embryo and larvae, and suitability for high‐throughput screening in vivo. 1 , 2 , 3 So far, mammalian host models have played a predominant role in evaluating microbial functions and the influence of exterior substances on host health. 4 Using the vertebrate zebrafish model to study the gut microbiome brings many advantages. First of all, zebrafish shares homology with the human genome 5 and is similar to the intestine of mammals in terms of structure and mode of action. 6 Moreover, owing to its transparency, it is feasible to apply in situ real‐time imaging technology to the whole organism. 7 Furthermore, given that in zebrafish the innate immune system arises first, and adaptive immunity develops after 2–3 weeks, it is possible to examine the relationship between the innate immune system and gut microbiome. 8 , 9 Last but not least, germ‐free (GF) zebrafish provide a robust system for dissecting or manipulating microbial signals owing to its cost‐effectiveness and the convenience of the techniques for constructing sterile zebrafish. 10 , 11 Accordingly, it is possible to directly determine causality between the gut microbiome and disease‐associated alterations in functional and mechanistic studies. 12 Studies on gut microbiome using the zebrafish model have been considered a pioneering and vital field of research in recent years.
This review focuses on the application of a series of GF zebrafish‐derived models that unveil how the gut microbiome affects host development, metabolism, and immunity. Moreover, the roles of the gut microbiome in microbiota homeostasis and vertebrate microbiome‐host interactions relevant to human health are elucidated, providing a theoretical foundation and support for further application in disease treatment. A flowchart of our review is shown in Figure 1.
FIGURE 1.
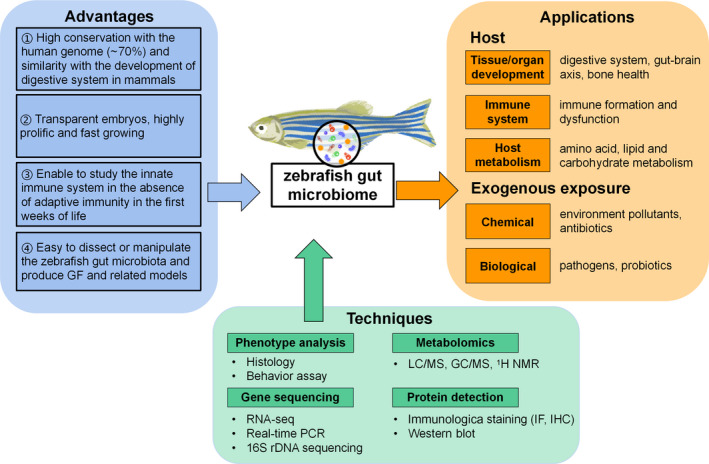
Currently available applications and techniques for research on gut microbiome‐host interactions with zebrafish models
2. COMPARISON BETWEEN ZEBRAFISH AND STANDARD WARM‐BLOODED ANIMAL MODELS (MICE AND RATS)
The advantages and limitations of the zebrafish model used for host homeostasis and gut microbiome studies compared with murine models are comprehensively summarized in Table 1. Given these unique attributes, the vertebrate animal zebrafish has become an ideal model for studying the gut microbiome. Though the gut microbiome structure of zebrafish may differ significantly from humans, its complexity and diversity can also provide valuable information and reference in comparative studies of the gut microbiome. 13
TABLE 1.
The merits and limitations of common animal models (mice, rats, and zebrafish) in gut microbiome research
| Animal model | Advantages | Limitations | Ref. |
|---|---|---|---|
| Mice and rats |
|
|
14, 15, 16, 17 |
| Zebrafish |
|
|
5, 7, 9, 11, 18, 19, 20, 21 |
3. THE ROLES OF THE GUT MICROBIOME IN TISSUE DEVELOPMENT AND PHYSIOLOGICAL FUNCTION
3.1. Digestive system
Zebrafish provide effective models to research the functions of the gut microbiome for host intestinal tract development, including gene expression, cell proliferation, tissue differentiation, and related functions. The embryos initially develop in an essentially axenic chorion and first encounter microorganisms in the external environment after hatching (approximately 48 or 96 h post‐fertilization). Most larval organs interact with the microbiota during hatching. The zebrafish gut microbiome has been found to aggregate into different communities during development, and these communities gradually become different from the external environment and from each other. 21 The first comparison of gene expression between the digestive tract of GF zebrafish and conventional zebrafish was conducted by Rawls et al. in 2004. Two hundred genes were found to be regulated by the gut microbiome, among which the expression of 59 genes was conserved in the mouse intestine. The expression levels of these microbiota‐related genes were mainly correlated with epithelial cell turnover, nutrient uptake, xenobiotic metabolism, and immune response. 22 It has been established that the spatial distribution of the gut microbiome is related to both its host and itself, impacting the overall growth kinetics. 23 Yossa et al. first documented inhibited growth and increased mortality in a bacteria‐dysbiosis zebrafish model induced by antibiotics. 1 Furthermore, the proliferation, differentiation, morphology, and related functions of intestinal cells of zebrafish are reportedly affected by the lack or the variation of gut microbiome. 22 , 24 Hill et al. found that, during early development, the growth and division of pancreatic β cells require the participation of gut microbiome and certain bacteria, which secrete β‐cell expansion factor A (BefA) proteins to induce the proliferation of β cells. 25 In addition, next‐generation sequencing showed that the hypoglycemic effect of BefA was highly correlated with an increase in beneficial bacteria (such as Oscillospria, Lactobacillus, and Bifidobacterium) and a decrease in opportunistic pathogens (Acinetobacter). 26
3.2. The gut‐brain axis
The gut microbiome has been recognized to profoundly affect the neurochemistry and central nervous system in zebrafish. Importantly, microbial colonization is required for the normal development and physiological function of the nervous system in zebrafish. In this regard, it has been found that sterile or antibiotic‐treated zebrafish exhibited increased locomotor behavior or hyperactivity; colonization with different strains of Vibrio cholerae or Aeromonas veronii could hinder locomotor hyperactivity. However, interference with heat‐killed bacteria or microbiome‐associated molecular patterns could not inhibit this abnormal phenotype in GF larvae. 27 Besides, treatment with Lactobacillus plantarum strain alleviated anxiety and depressive‐like behavior and alleviated the stress response in zebrafish with an intestinal disorder. 28 Manipulating the gut microbiome composition in zebrafish may also affect the nervous system. By co‐culturing GF zebrafish with six selected bacteria, either single strain or mixed strains, Weitekamp et al. showed that different bacterial species had different effects on their host's behavior, which might be correlated with colonization success in the host's intestine. 29 Borrelli et al. found variations in the gut microbial composition in the probiotic Lactobacillus rhamnosus treatment group, with a significant increase in Firmicutes and decrease in Proteobacteria, including potential pathogens (such as Plesiomonas and Vibrio). In this respect, zebrafish’s social and explorative behavior could be significantly altered; the expression levels of endogenous neuroactive molecules, brain‐derived neurotrophic factor, and serotonin were modulated to a certain extent by feeding with probiotic L. rhamnosus. 30 A GF zebrafish study revealed a potential mode of action where melatonin could regulate disorders of neurotransmitter secretion induced by caffeine via the gut‐microbiome‐brain axis. 31 Additionally, Cuomo et al. documented that the administration of L. rhamnosus in larvae led to DNA methylation code of the Tph1A and BDNF promoter gene reconstruction in the gut and the brain of zebrafish. Accordingly, alterations in the gut microbiome may influence the host epigenetic landscape, resulting in long‐term consequences for specific gene regions. 32
Moreover, the zebrafish model revealed the roles of the gut microbiome in neuroendocrine response. The intestine, vital for controlling food intake and maintaining energy balance, represents one of the most important endocrine systems in vivo. 33 The gut microbiome is capable of promoting enteroendocrine cells (EECs) to secrete gut hormones (e.g., gut peptide YY, cholecystokinin, oxyntomodulin, and glucagon‐like peptide‐1). These hormones act on the central nervous system through blood circulation via vagal afferent fibers and work mainly on the hypothalamus. Furthermore, current evidence suggests that the gut microbiome can influence the sensing ability and modulation of EECs. Ye et al. revealed that a high‐fat (HF) diet altered EECs morphology and converted them into a state insensitive to nutrients, termed “EEC silencing.” It has also been shown that a high‐fat diet could alter the gut microbiome composition, especially with the proliferation of Acinetobacter. They further identified a strain of Acinetobacter that can induce EEC silencing. 24 Likewise, EECs also transmit signals from the gut microbiome to regulate intestinal and vagal pathways. Researchers found that the gut microbiome could produce tryptophan catabolite to activate the transient receptor potential ankyrin A1 channels on EECs and then cause rapid activation of cells in the intestine and vagus nerve. 34
3.3. Bone health
The gut microbiome has been established as a primary regulator of zebrafish bone metabolism. The relationship between the microbiota (or probiotics) and bone homeostasis and development has been explored in recent years; direct evidence of how the gut microbiome communicates with its host to regulate bone mineral density has been obtained. 35 Nevertheless, the effects of the microbiome on zebrafish bone metabolism have also been studied. It was found that supplementation of L. rhamnosus to conventional zebrafish microbiome led to faster backbone calcification and correlated with stimulation of the insulin‐like growth factor system. 36 Moreover, L. rhamnosus feeding could regulate genes involved in osteocyte formation and suppress bone formation inhibitors in zebrafish. 37 It is well established that inflammatory bowel disease (IBD) is correlated with a higher risk of low bone density. Zebrafish share similarities with humans in terms of bone development given that their scales represent a good readout model to assess bone metabolism. Accordingly, zebrafish represent an excellent model to verify the relationships between intestinal inflammation and bone metabolism. 38 After supplementation of defatted soybean meal to cause intestinal inflammation in adult zebrafish, Carnovali et al. found that intense acute intestinal inflammation was related to temporary osteoporosis‐like phenotype at the edge of the scales. Besides, the chronic inflammatory state with continuous IL‐8 expression was highly correlated with the resorption of lacunae at the center of the scale. 39 However, it remains unknown whether intestinal inflammation causes changes in microbial structure or its secreted metabolites, nor is it clear how the gut microbiome regulates bone metabolism. Further studies are required to reveal the mechanism of the action of the gut microbiome in bone signaling pathways after dietary intervention or probiotic treatment. The above findings also emphasize the need to explore new strategies to further improve bone disease treatment by regulating the composition and abundance of the targeted gut microbiome.
3.4. Immunity system
3.4.1. Immune system development
An increasing body of evidence suggests that the gut microbiome is involved in the normal development of the zebrafish immune system, with the ability to mount immune responses to different stresses such as injuries and infections, especially neutrophils. 40 , 41 A study by Masud et al. comprehensively described how innate immune cells were produced in early development and how the gut microbiome impacted immune cell production, differentiation, and function using a zebrafish model. 42 Interestingly, Bates et al. showed that the internal microbiome accounted for the normal neutrophil levels in the zebrafish gut by modulating intestinal tumor necrosis factor receptor, Myd88, and alkaline phosphatase. 43 Koch et al. consistently found that a normal gut microbiome or single commensal bacterial species (from phylum Bacteroidetes or Firmicutes) could induce changes in intestinal leukocytes and host gene expression; these changes were dependent on innate immune adaptor gene Myd88. 44 Besides, Brugman et al. revealed that adaptive immune deficiency was associated with excessive growth of Vibrio species at larval stages, and overgrowth could be inhibited with the development of adaptive immunity. It was further demonstrated that adaptive immune processes could control the proliferation of Vibrio species in mutants with the loss of adaptive immunity. It was found that the adoptive transfer of T lymphocytes to Rag1‐deficient recipients effectively suppressed the expansion of the Vibrio species in vitro. 45
The gut microbiome has been reported to modulate the activity of gut neutrophils and other leukocytes. Kanther et al. found that the number of systemic neutrophils and the expression of myeloperoxidase were increased, and the location and migration of neutrophils was changed in GF zebrafish colonized with gut microbiome. 46 With live imaging of larvae, Wiles et al. discovered that the expression of gut‐related macrophages and proinflammatory cytokine tumor necrosis factor‐α (TNF‐α) in the liver was induced by a Vibrio symbiont derived from zebrafish intestine through its swimming motility and chemotaxis. 47 In addition, serum amyloid A (SAA), a host factor secreted by intestinal epithelium cells, was potently upregulated in the gut after microbial colonization of zebrafish and mediated the migratory behavior of tissue‐specific neutrophils caused by microbial stimuli. 48 , 49 Murdoch et al. showed that SAA secreted by the gut in reaction to microbiome changes, acting as a systemic signal to determine the fate of neutrophils. Importantly, SAA could reduce the inflammatory response and bacterial killing ability while improving the capacity of neutrophils to migrate to the wound. The intestinal SAA could also restore neutrophils to normal levels in GF zebrafish. 50
Intestinal microbial metabolites also play a vital role in determining neutrophil levels. Cholan et al. discovered that butyrate isolated and synthesized by gut microbiome in adult zebrafish could significantly reduce the number of neutrophils recruited after embryonic trauma. 51 In addition, GF zebrafish transplanted with hybrid sturgeon gut microbiota, treated with a para‐probiotic and postbiotic supplement diet, showed that the gene TGF‐β and the levels of non‐specific immune‐associated genes (lysozyme, Defbl‐1, C3a) were significantly upregulated. In contrast, the levels of the proinflammatory gene IL‐1β significantly decreased. 52 These findings highlight the need to maintain stability and homeostasis of the intestinal microecology to protect host health and prevent chronic inflammation.
3.4.2. Immune dysfunction
Invasion of pathogenic bacteria can disturb the homeostasis of the intestinal microbiome and result in perturbations of the intestinal immune system. Subsequently, the innate immune system is activated to mediate pathogen clearance and inflammation. Yang et al. demonstrated that, in response to invasion of pathogenic bacteria, the intestinal microbial structure was susceptible to changes with increased abundance of pathogens and decreased abundance of beneficial bacteria. Rolig et al. found that zebrafish lacking an enteric nervous system exhibited microbiome‐dependent inflammation; increased inflammation levels were associated with an excess of proinflammatory bacterial lineages and a lack of anti‐inflammatory ones. 53 Furthermore, transgenic lines expressing fluorescent proteins were subjected to pathogen infection and provided readout models for immune system activation and in vivo visualization of immune responses to pathogens. Additionally, it has been found that antimicrobial peptide genes, including defensin1, lectin, and hepcidin, increased at the mRNA level in the intestine after pathogenic infection. 54
The intestinal microbiome is a central factor associated with IBD with dysfunction or the loss of integrity of the intestinal barrier. Using zebrafish and mouse models, Kaya et al. substantiated that the expression of gut G‐protein‐coupled receptor 35 was dependent on the gut microbiome, and it increased when inflammation was triggered. 55 Interestingly, a close relationship was found between gene GPR35 single‐nucleotide polymorphism and increased risk of IBD. 56 In a project exploring susceptibility genes for IBD, mutations in the ubiquitin‐like protein with PHD and RING finger domains 1, a highly conserved gene of methylation, were identified. Besides, current evidence shows that dysfunctional ubiquitin‐like protein with PHD and RING finger domains 1 could induce hypomethylation of the TNF‐α promoter, releasing transcriptional repression of the promoter and resulting in TNF‐α upregulation within the intestinal epithelium. 57 Notably, the upregulated expression of TNF‐α contributed to the occurrence of microbiota‐dependent chronic inflammation, such as the shedding and apoptosis of epithelial cells, recruitment of immune cells, and impairment of intestinal barrier. By establishing a Shigella‐zebrafish infection model, Willis et al. found that Shigella‐mediated stem cell‐driven granulopoiesis could activate the innate immune system and protect against superinfection. 58
3.5. The host metabolism
Given the substantial number of metabolism‐associated genes shared with humans, 59 the zebrafish model has been extensively employed to investigate the relationship between different metabolic patterns and the gut microbiome. Substantial evidence suggests that changes in the gut microbiome and their metabolites are closely related to glucose metabolism, insulin resistance, and recovery of pancreatic function in type 2 diabetes mellitus (T2DM). 60 T2DM zebrafish represents a promising model to study host‐microbial interactions in human obesity, metabolic syndrome, and related diseases. Intriguingly, it was found that some strains of the genera Aeromonas and Shewanella could release BefA proteins to induce upregulation of host pancreatic β cells, thereby increasing insulin levels and regulating blood glucose levels. 25 Furthermore, the intestinal microbial metabolites (endotoxin, short‐chain fatty acids [SCFAs], secondary bile acids, and indole) are reportedly involved in glucose regulation by participating in glucagon‐like peptide‐1 secretion. 61 In addition, the α‐diversity (the Chao1 index) was decreased in T2DM adult zebrafish compared with the healthy controls. 62 Furthermore, Bootorab et al. found that blood glucose levels were decreased after probiotic L. rhamnosus administration via downregulation of proinflammatory cytokines (such as TNF‐α and IL‐1β) involved in T2DM therapeutic signaling pathways. 63 Importantly, it has been found that Escherichia coli could utilize glucose and produce acidic byproducts of glucose metabolism in the zebrafish gut. These acidic products significantly reduced the colonization rate of the classical and EI Tor biotypes of V. cholerae to prevent or treat cholera infection. 64
Mounting evidence suggests that a disturbed gut microbiome can disrupt energy homeostasis and lipid metabolism. The gut microbiome can contribute to increased lipid accumulation in the intestinal epithelium. Most differentially expressed genes between conventional and GF zebrafish larvae are reportedly involved in lipid metabolism. 65 After being colonized with gut microbiota from donors disrupted by 12% palmitic acid (PA), the recipient GF zebrafish exhibited endoplasmic reticulum stress and liver injury. The transplantation of the PA‐altered microbiome in recipients boosted the continuous absorption of PA in vivo, resulting in increased PA, which entered the liver and exacerbated liver toxicity. 66 Qiao et al. screened a panel of bacteria associated with host lipid deposition by generating a diet‐induced fat accumulation zebrafish model. 67 Then, gut microbiome samples were collected from the adult zebrafish of the control and the HF diet groups and transplanted into GF zebrafish. It was found that the intestinal microbiome from the donors fed the HF diet induced more lipid deposition in the recipient GF zebrafish. Nonetheless, to the best of our knowledge, few studies have sought to clarify the critical role of probiotics in modulating the gut microbiome and their potential effects in the treatment of lipid turbulence. For instance, Falcinelli et al. found that L. rhamnosus supplementation led to increased Firmicutes and decreased Actinobacteria levels. These variations induced the downregulation of genes related to triglyceride and cholesterol metabolism. Moreover, they regulated lipid processing, lowered lipid content, and increased fatty acid levels in the host, 68 finally attenuating metabolic disorders caused by the HF diet in zebrafish. 69
The gut microbiome can metabolize amino acids and is conversely influenced by the amino acids. 51 , 70 , 71 By observing the evolution of Aeromonas in gnotobiotic zebrafish experimentally, researchers found that Aeromonas could sense host‐derived amino acid signals to modulate its motility via a process called chemokinesis, and these bacteria subsequently enter the intestine. 72 Wang et al. found that the abundance of Hyphomicrobium, Paracoccus, and Plesiomonas was significantly correlated with leucine metabolism in zebrafish after treatment with 300 μg/L sodium ρ‐perfluorous nonenoxybenzene sulfonate. 73 Another study demonstrated that the gut microbiome was significantly changed in T2DM adult zebrafish with downregulation of the metabolic pathways of arginine, proline, and phenylalanine, suggesting that the gut microbiome of T2DM zebrafish may adversely affect host health by inhibiting the metabolism of these amino acids. 62 The gut microbial community of zebrafish supplemented with a gluten formulated diet displayed activated metabolic KEGG pathways related to threonine, serine, and glycine metabolism. 74 An increasing body of evidence suggests that upregulation of these metabolic pathways is associated with oncogenesis. 75 , 76 In addition, gut microbiota can ferment dietary fibers into SCFAs in the gut, among which butyric acid is a profoundly essential SCFA with anti‐inflammatory properties. It was reported that both the levels of butyrate and the abundance of butyrate‐producing bacteria were generally low in inflammatory diseases associated with intestinal dysbiosis in mammals. 77 , 78 To validate these findings, Cholan et al. first isolated 3 main SCFAs (butyrate, acetate, and propionate) from ferments of gut microbiome using a zebrafish model in vitro. Then, these 3 SCFAs were supplemented into the tanks of zebrafish that underwent tail wound injury. Importantly, butyrate could reduce the recruitment of neutrophils and M1‐type proinflammatory macrophages to the wound and enhance the anti‐inflammatory ability of zebrafish. 51 Furthermore, butyrate sensitivity was dependent on the maturity of the intestine. Moreover, a strain of Pediococcus pentosaceus isolated from the gut microbiome in zebrafish was used to demonstrate that the host’s resistance to Aeromonas hydrophila could be enhanced by increasing the abundance of SCFA‐produced bacteria, butyrate levels, and the expression of IL‐1β. 79 These findings provide compelling evidence that the gut microbiome can metabolize and produce SCFAs, and play a conservative role in the immunity of zebrafish. In return, SCFAs can regulate the composition of the gut microbiome. Li et al. found that dietary supplementation with SCFAs could induce inhibition of pathogens and enrichment of beneficial bacteria while improving innate immunity, enhancing antioxidative capacity, and increasing the host’s disease resistance (by protecting zebrafish against A. hydrophila). 80
4. EXOGENOUS EXPOSURE
4.1. Toxicity of environmental pollutants
Nowadays, environmental pollutants and their residuals are frequently detected in water environments. They can accumulate in organisms, seriously harming the ecological environment and human health. It has been established that the gut microbiome is extremely sensitive to those xenobiotics found in the environment, including drugs, diet, and pollutants. The gut microbiome plays a pivotal role in the fate of xenobiotics, influencing host xenobiotic metabolism and preventing systemic toxin absorption. 81 In recent years, the zebrafish model has been widely used for toxicological assessment from environmental pollutants to the intestinal microbiome by monitoring microbial richness, diversity, structure, and ecological behavior. Many studies have indicated that persistent exposure to hazards results in morphological alterations or pathological changes in the intestinal tract and disturbs the abundance and structural composition of the intestinal microbiome in zebrafish. These disturbances led to nutrient uptake, energy metabolism, and immune function disorders in the host. The applications of the zebrafish model for the study of the effects of diverse environmental pollutants on intestinal microbiome are summarized in Table 2.
TABLE 2.
The application of the zebrafish model for the study of the relationship between exogenous substances and intestinal bacteria
| Category | Environment pollutants | Age | Exposure time | Exposure dose | Methods | Ref. |
|---|---|---|---|---|---|---|
| Antibiotic | Streptomycin | Larvae | 10 days | 0.1, 1.0, 10.0 μg/ml | 16S rDNA sequencing using DADA2 | 87 |
| Tetracycline | Juvenile | 30 days | Low (1 μg/L) and high (100 μg/L) environmental concentrations | Histopathological analysis, Real‐time PCR for genes in the liver, Metabolite profiling, identification, and pathway analysis, 16S rDNA sequencing, PICRUSt for functional prediction | 88 | |
| Sulfamethoxazole (SMX) or oxytetracycline (OTC) | Adult | 6 weeks | SMX (100 mg/kg body weight), OTC (80 mg/kg body weight) | 16S rDNA sequencing, Biochemical assay, Real‐time PCR for genes related to nutrient transportation | 89 | |
| OTC | Juvenile | 30 days | Low (1 μg/L) and high (100 μg/L) environmental concentrations | Real‐time PCR for genes of thyroid hormones in the brain, 16S rDNA sequencing | 90 | |
| SMX or OTC | Adult | 6 weeks | SMX (260 ng/L), OTC (420 ng/L) environmental concentrations | Biochemical assay, Gut morphology, Real‐time PCR for genes related to inflammation, 16S rDNA sequencing | 91 | |
| OTC | Adult | 2 months | Low (0.1 and 10 μg/L) represents environmental concentrations, high (10 000 μg/L) elucidates the mode of action | 16S rDNA sequencing, Behavior assay, Biochemical analysis, LC/MS nontargeted metabolomic analysis, Correlation analysis of changed bacteria and metabolites, Energetic reserves analysis, in silico metagenome analysis of functional profile inference | 92, 93, 94 | |
| Microplastic | Polystyrene microplastics (PS‐MPs) | Embryo | 7 days | 100 and 1000 μg/L of 2 sizes (5 and 50 μm) | 16S rDNA sequencing, GC/MS for metabolite analysis, Real‐time PCR for genes related to glycolysis and lipid metabolism, Measurements of oxidative stress | 95 |
| PS‐MPs | Embryo | 7 days | 10, 100, and 1000 μg/L | Biochemical indicator analysis, Real‐time PCR for glycolipid‐ and phospholipid‐related genes, 16S rDNA sequencing, LC/MS nontargeted metabolomic analysis, Correlation analysis of altered bacteria and metabolites | 96 | |
| PS‐MPs | Adult | 14 days | 100 and 1000 μg/L of 2 sizes (5 and 50 μm) | Histopathological analysis, 16S rDNA sequencing, Real‐time PCR for genes related to inflammation | 97 | |
| PS‐MPs | Adult | 21 days | 50 and 500 μg/L | Histological analysis, Biochemical analysis, 16S rDNA sequencing, 1H NMR for metabolomic analysis | 98 | |
| PS‐MPs | Adult | 21 days | Three sizes (100 nm, 5 μm, and 200 μm) | Histopathological analysis, Cytokine analysis for TNF‐α and TLR2, Single‐cell RNA sequencing for transcriptome heterogeneity of intestinal cells, 16S rDNA sequencing | 99 | |
| PS‐MPs and PS‐NPs | Adult | 21 days | 10 μg/L and 1 mg/L of MPs (8 μm) and NPs (80 nm) | 16S rDNA sequencing, Real‐time PCR for genes related to inflammation pathways in the intestine | 100 | |
| Environmental endocrine‐disrupting chemical | Methylparaben | Adult and larvae | Adult (96 h), Larvae (168 h) | Environmental concentration (30 μg/L) or non‐effect concentration (50 mg/L) | Evaluation of the utilization of the carbon sources by microbiota, Calculation of the Shannon diversity and Shannon evenness | 101 |
| Atrazine, estradiol, polychlorinated biphenyl [PCB]126, and PCB153 | Adult | 7 days | 1 μg/L nominal concentrations | Metagenomic sequencing, MEGAN analysis of the functional profile of gut microbiota, Measurements of oxidative stress, Biochemical analysis for intestinal and hepatic status | 102 | |
| Bisphenol A (BPA) and BPA alternatives | Embryo | 10 days | Semi‐log spaced concentration ranges | Behavior testing, 16S rDNA sequencing | 103 | |
| Estradiol (E2) | Embryo | 10 days | Non‐teratogenic concentrations, ranging from 0.34 to 3.5 μM E2 | Behavior testing, 16S rDNA sequencing, LC–MS/MS for targeted chemistry analysis, Nontargeted mass spectrometric analysis for metabolites identification | 104 | |
| BPA and E2 | Adult | 5 weeks | BPA (2000 μg/L), E2 (2000 ng/L) | TG content test in the liver, Real‐time PCR for VTG gene expression in muscle, 16S rDNA sequencing | 105 | |
| BPA | Adult | 3 months | 2 and 20 μg/L | 16S rDNA sequencing, Physiological analysis of the intestine | 106 | |
| Antimicrobial agent/fungicide | Triclosan | Adult | 7 days | 100 μg/g fish | 16S rDNA sequencing, Microbial correlation network analysis | 107 |
| Triclosan | Embryo | 10 days | 0.16–0.30 μM | 16S rDNA sequencing, LC–MS for targeted and nontargeted chemistry analysis | 3 | |
| Imazalil | Adult | 1, 7, 21 days | 100 and 1000 μg/L | Gut histological analysis, 16S rDNA sequencing, GC/MS‐based metabolomic analysis, Real‐time PCR for genes related to glycolysis and lipid metabolism | 108 | |
| Carbendazim | Adult | 21 days | 30 and 100 μg/L | Determination of hepatic biochemical parameters, 16S rDNA sequencing, Real‐time PCR for genes related to glycolysis and lipid metabolism, Hepatic RNA‐seq analysis | 109 | |
| Triclosan | Adult and larvae | 120 days (larvae), 7 days (adult) | 0.03, 0.3, 3, 30, 100, and 300 ng/ml | 16S rDNA sequencing | 110 | |
| Pesticide | Dieldrin | Adult | 4 months | 16 and 163.5 ng/g dry weight | Histopathology analysis, 16S rDNA sequencing, Predicted relative metabolomic turnover to predict how the microbial alteration affects the exchange of metabolites | 111 |
| Difenoconazole | Adult | 21 days | 0.4 mg/L | Histopathological analysis of liver, Biochemical analysis, RNA‐seq for differentially expressed genes in the liver and real‐time PCR for confirmation, 16S rDNA sequencing | 112 | |
| Propamocarb | Adult | 7 days | 100 and 1000 μg/L | Histopathological analysis of liver, Biochemical analysis, Real‐time PCR for genes related to glycolysis and lipid metabolism in the liver, GC/MS‐based hepatic metabolomic analysis, 16S rDNA sequencing | 113 | |
| Imidacloprid | Adult | 21 days | 100 and 1000 μg/L | Gut histology analysis, Enzyme activity and ELISA detection in the gut, Real‐time PCR for oxidative stress‐related genes and inflammatory‐related genes, 16S rDNA sequencing | 114 | |
| Chlorpyrifos | Adult | 21 days | 30, 100, and 300 μg/L | Gut histology analysis, 16S rDNA sequencing, Antioxidant enzyme analysis, Real‐time PCR for genes related to glycolysis and lipid metabolism, GC/MS‐based hepatic metabolomic analysis | 115 | |
| Persistent organic pollutant | Benzo[a]pyrene | Adult | 15 days | 100 μg/L | 16S rDNA sequencing, Real‐time PCR for genes related to inflammatory pathways in the intestine | 116 |
| Polybrominated diphenyl ethers | Adult | 7 days | Environmentally realistic concentration (5.0 ng/L) | Metagenomic sequencing, Biochemical analysis in the gut and liver | 117 | |
| Sodium ρ‐perfluorous nonenoxybenzene sulfonate | Adult | 7 and 21 days | 3, 30 and 300 μg/L | LC–MS/MS for hepatic metabolites, 16S rDNA sequencing, Real‐time PCR for genes related to glycolipid metabolism in the liver, Gut microbiota‐differential metabolites correlation analysis | 73 | |
| Heavy metal | Lead | Adult | 7 days | 10 and 30 μg/L | Gut histopathological analysis, Real‐time PCR for genes related to glucose and lipid metabolism, 16S rDNA sequencing, GC/MS‐based hepatic metabolomic analysis | 118 |
| Lead | Adult | 14 days | 0.8 g/kg of food | Histological analysis, 16S rDNA sequencing, Phylogenetic analysis | 119 | |
| Cadmium | Embryo | 7 days | Ecologically relevant concentrations (0, 1.25, 2.5, and 5 μg/L) | Locomotion analysis, 16S rDNA sequencing, Real‐time PCR for neuronal gene expression | 18 | |
| Engineered nanoparticle | nTiO2, nZnO, nSe | Embryo | 3 months | 100 μg/L | Histological analysis, 16S rDNA sequencing, Ecological process analysis, Network analysis for gut microbial interactions | 120 |
| Metalloid | Arsenic | Embryo | 20 days | Low (10 ppb), medium (50 ppb), and high (100 ppb) found in contaminated water | 16S rDNA sequencing using DADA and QIIME | 121 |
Bacterial transplantation or probiotic treatment has been applied to modulate gut microbiota dysbiosis and lipid metabolism disorders induced by xenobiotics exposure. Zang et al. discovered that the abundance and species of zebrafish gut microbiome were disrupted by triclosan, which could be restored by administration of L. plantarum ST‐III, with alleviation of lipid metabolism disorder and a decreased number of inflammatory cells. Besides, intestinal metabolic syndrome and neurodegenerative diseases caused by triclosan exposure were also attenuated by probiotic treatment. 82 Chen et al. showed that perfluorobutane sulfonate (PFBS) exposure caused dysregulation of the gut microbial community, and maternal transfer of PFBS to offspring increased the risks to aquatic populations. 83 , 84 However, L. rhamnosus administration inhibited the disorders caused by PFBS and regulated the metabolic activities of the host indirectly. It was found that β‐oxidation and fatty acid synthesis were increased, and blood cholesterol levels were reduced. 85 Furthermore, probiotic feeding can prevent PFBS‐induced intestinal disturbances and ferroptosis. 86
4.2. Pathogenic infections
Many pathogens have been investigated using the zebrafish model in recent years, including Aeromonas, Salmonella, Mycobacterium, Vibrio, etc. (Table 3). The zebrafish model, as a natural host model, provides a complete picture of the infection period from exposure to colonization since it allows high‐clarity in vivo imaging combined with genetic manipulation. Accordingly, a more comprehensive understanding of the pathogen infection process and cellular response can be obtained in its entirety. 122 Indeed, in recent years, with the increasing use of the zebrafish infection model, mechanisms that underlie how the host cells first recognize microbiome and the initial communication among the various cell types (including non‐immune cells) during bacterial infection have been gradually unveiled.
TABLE 3.
Summary of the studies on zebrafish model for pathogenic bacteria infection
| Relevant diseases | Infectious agent | Age | Route of administration | Ref. |
|---|---|---|---|---|
| Tuberculosis | Mycobacterium marinum | Larvae | Injection | 123 |
| Embryos and adult | Injection | 124 | ||
| Larvae | Injection | 125 | ||
| Salmonellosis | Salmonella | Adult | Immersion | 126 |
| Fish motile aeromonad septicemia | Aeromonas veronii and Aeromonas hydrophila | Larvae and adult | Immersion for larvae, immersion and injection for adult zebrafish | 127 |
| Aeromonas sp. and Vibrio sp. | Larvae (5 dpf) | Immersion | 128 | |
| Cholera diarrhea | Vibrio cholerae | Larvae | Immersion | 129 |
| Adult | Immersion | 130 | ||
| Diarrheal illness, hemolytic uremic syndrome | Enterohemorrhagic Escherichia coli | Larvae (4 dpf) | Immersion | 131 |
| Streptococcosis, meningitis, sepsis | Streptococcus agalactiae | Adult | Injection | 132 |
In 2016, Caruffo et al. conducted research using in vitro co‐aggregation assays and in vivo infection experiments on larval zebrafish. It was suggested that the protective effect of yeast against Vibrio anguillarum was correlated with antipathogen effects and immune regulation in vivo, rather than modulation of the gut microbiome. 133 Nevertheless, contrasting studies have suggested that probiotic treatment probably plays a pivotal role by modulating gut microbial composition to protect the host from infectious diseases. It has been demonstrated that gut bacteria themselves (endogenous bacteria) or exogenous addition of protective bacteria in larvae could prevent or decrease the chance of pathogenic infection and improve the survival rate of fish. Vargas et al. showed that V. anguillarum changed the gut microbial β‐diversity, and probiotic yeasts could inhibit the enrichment of Vogesella and Ensifer, which were identified as a negative predictive factor of survival rate in larvae. 134 Besides, probiotics of selected bacteria with high surface glycotope Galα1‐3Galβ1‐(3)4GlcNAc‐R (α‐Gal) content were effective and safe for Mycobacterium marinum. It was found that probiotics with high α‐Gal content activated gut microbial structure modification, B‐cell maturation, and anti‐α‐Gal antibody‐mediated control of Mycobacteria. Meanwhile, they stimulated innate immune responses and reduced oxidative stress. 135 Given the complexity of the gut microbiome, it was difficult to identify in situ endogenous bacteria that provided this protective effect. However, this problem has gradually been overcome with the rapid development of gnotobiotic zebrafish technology. He et al. discovered a significant correlation between the spatial position of Lactobacillus in the intestine and their protective activity in zebrafish infection, and they divided them into 3 types: mucus type (>70% in mucus), mucosa type (>70% in mucosa), and hybrid types (others). 136 Interestingly, the hybrid types were more efficient in protecting zebrafish against pathogenic infections. Besides, Stressmann et al. explored whether native microbial communities could protect their host using GF, conventional, and reconventionalized zebrafish infection models. Two independent protection strategies, individual‐level protection by bacterium Chryseobacterium massiliae and community‐level resistance to infection, were identified against the same pathogen. 137 On the basis of these studies, López‐Igual et al. engineered toxins split by inteins and delivered them by conjugation into a mixture of bacteria. They found that the engineered toxin could specifically conjugate and kill antibiotic‐resistant V. cholerae in the bacteria mixture in vitro. Furthermore, the in vivo study showed that their split toxin‐intein could also target specific strains of Vibrio species in zebrafish larvae that are well recognized natural hosts for the pathogens. 138 Importantly, these studies provide the basis for developing targeted probiotic strains or engineered toxins to protect the host against specific pathogenic infections.
5. FUTURE RESEARCH PROSPECTS ON GUT MICROBIOME BY USING THE ZEBRAFISH MODEL
Given that the longest survival time of GF zebrafish is 30 days, 22 it remains difficult to obtain adult GF zebrafish. Accordingly, more emphasis should be placed on improving our current understanding of the nutrient ratio required to feed sterile zebrafish to prolong their lifespan. Among the studies on the role of intestinal microbes in host tissue development and physiological function, few have assessed their influence on vascular development and hematopoiesis. Importantly, during early development stages, the zebrafish embryo is transparent and has simple vasculature. These features can be harnessed to establish transgenic zebrafish lines with fluorescent vascular or hematopoietic stem cells. Then, through manipulating the intestinal microbiome and in vivo imaging, the microbial influence on vascular development and hematopoiesis can be investigated. Likewise, in gut‐brain axis studies, it is necessary to develop new behavioral models of neurobehavioral diseases based on the intestinal microbiome to explore the relationship among microbiota, intestinal disease, and brain function, delve into the gut‐microbiome‐brain axis, and enhance human cognition in neurobehavioral diseases.
Furthermore, the zebrafish model can be helpful to explore disease causes; for instance, metagenomic sequencing can be used to compare the composition and abundance of bacteria in disease and healthy states. It is expected that analysis of the heterogeneity in intestinal microbial composition can be used to establish a pathological classification to assist clinicians in disease diagnosis. Importantly, unveiling the molecular communications between animals and their resident gut microbiome in pathogenic states could yield further insights into the mechanisms of the influences of the gut microbiome on human health and the etiology of diseases. Moreover, it is urgent to determine which groups of bacteria residing in the gut microbiome are responsible for the pathological changes in the host state and evaluate beneficial groups of bacteria. The use of GF zebrafish model to study the effects of intestinal bacterial colonization is still subject to many limitations that need to be overcome for future research on probiotics and prebiotic‐driven studies.
Interestingly, Schlomann et al. found significant differences in spatial distribution and cohesion of bacterial strains in zebrafish via bacterial colonization and in vivo imaging experiments. Importantly, this research provided a framework for precise microbial engineering. 139 Theoretically, it is possible to selectively displace bacterial communities in some intestinal regions or remove them altogether by controlling cohesion. Indeed, this study inspires us to explore the relationships between spatial structure, cohesion, and flow that may help to clarify diseases caused by microbial imbalance. Disorders caused by changes in community composition can be treated by targeted changes in bacterial aggregation, providing new targets and horizons for human disease prevention and control.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest.
AUTHOR CONTRIBUTIONS
Xiaoting Zhong: information collection, writing—draft and modification. Jinglin Li: information collection, visualization. Furong Lu: information collection. Jingjing Zhang & Lianxian Guo: supervision, conceptualization, writing—review and editing.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (31970777), Discipline Construction Project of Guangdong Medical University (4SG21014G, 4SG21003G), and the Natural Science Foundation of Guangdong Province (2020A151501457).
Zhong X, Li J, Lu F, Zhang J, Guo L. Application of zebrafish in the study of the gut microbiome. Anim Models Exp Med. 2022;5:323‐336. doi: 10.1002/ame2.12227
Funding information
The National Natural Science Foundation of China (31970777); Discipline construction project of Guangdong Medical University (4SG21014G, 4SG21003G); The Natural Science Foundation of Guangdong Province (2020A151501457)
Contributor Information
Jingjing Zhang, Email: jingjing.zhang@live.com.
Lianxian Guo, Email: glx525@gdmu.edu.cn.
REFERENCES
- 1. Yossa R, Sarker PK, Vandenberg GW. Preliminary evidence of the contribution of the intestinal microflora to biotin supply in zebrafish Danio rerio (Hamilton‐Buchanan). Zebrafish. 2011;8(4):221‐227. [DOI] [PubMed] [Google Scholar]
- 2. Gonzales JM, Law SHW. Feed and feeding regime affect growth rate and gonadosomatic index of adult zebrafish (Danio rerio). Zebrafish. 2013;10(4):532‐540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Weitekamp CA, Phelps D, Swank A, et al. Triclosan‐selected host‐associated microbiota perform xenobiotic biotransformations in larval zebrafish. Toxicol Sci. 2019;172(1):109‐122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sathkumara HD, Eaton JL, Field MA, Govan BL, Ketheesan N, Kupz A. A murine model of tuberculosis/type 2 diabetes comorbidity for investigating the microbiome, metabolome and associated immune parameters. Animal Model Exp Med. 2021;4(2):181‐188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Howe K, Clark MD, Torroja CF, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013;496(7446):498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lickwar CR, Camp JG, Weiser M, et al. Genomic dissection of conserved transcriptional regulation in intestinal epithelial cells. PLoS Biol. 2017;15(8):e2002054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Meijer AH, van der Vaart M, Spaink HP. Real‐time imaging and genetic dissection of host‐microbe interactions in zebrafish. Cell Microbiol. 2014;16(1):39‐49. [DOI] [PubMed] [Google Scholar]
- 8. Trede NS, Langenau DM, Traver D, Look AT, Zon LI. The use of zebrafish to understand immunity. Immunity. 2004;20(4):367‐379. [DOI] [PubMed] [Google Scholar]
- 9. Page DM, Wittamer V, Bertrand JY, et al. An evolutionarily conserved program of B‐cell development and activation in zebrafish. Blood. 2013;122(8):e1‐e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pham LN, Kanther M, Semova I, Rawls JF. Methods for generating and colonizing gnotobiotic zebrafish. Nat Protoc. 2008;3(12):1862‐1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Melancon E, Gomez De La Torre S, Canny S, et al. Best practices for germ‐free derivation and gnotobiotic zebrafish husbandry. Methods Cell Biol. 2017;138:61‐100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee J‐G, Lee S, Jeon J, et al. Host tp53 mutation induces gut dysbiosis eliciting inflammation through disturbed sialic acid metabolism. Microbiome. 2022;10(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nag D, Farr DA, Walton MG, Withey JH. Zebrafish models for pathogenic vibrios. J Bacteriol. 2020;202(24):e00165‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hildebrand F, Nguyen TLA, Brinkman B, et al. Inflammation‐associated enterotypes, host genotype, cage and inter‐individual effects drive gut microbiota variation in common laboratory mice. Genome Biol. 2013;14(1):R4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Barc MC, Bourlioux F, Rigottier‐Gois L, et al. Effect of amoxicillin‐clavulanic acid on human fecal flora in a gnotobiotic mouse model assessed with fluorescence hybridization using group‐specific 16S rRNA probes in combination with flow cytometry. Antimicrob Agents Chemother. 2004;48(4):1365‐1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McCoy KD, Geuking MB, Ronchi F. Gut microbiome standardization in control and experimental mice. Curr Protoc Immunol. 2017;117:23.1.1‐23.1.13. [DOI] [PubMed] [Google Scholar]
- 17. Kamareddine L, Najjar H, Sohail MU, Abdulkader H, al‐Asmakh M. The microbiota and gut‐related disorders: insights from animal models. Cell. 2020;9:2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xia Y, Zhu J, Xu Y, Zhang H, Zou F, Meng X. Effects of ecologically relevant concentrations of cadmium on locomotor activity and microbiota in zebrafish. Chemosphere. 2020;257:127220. [DOI] [PubMed] [Google Scholar]
- 19. Wallace KN, Akhter S, Smith EM, Lorent K, Pack M. Intestinal growth and differentiation in zebrafish. Mech Dev. 2005;122(2):157‐173. [DOI] [PubMed] [Google Scholar]
- 20. Roeselers G, Mittge EK, Stephens WZ, et al. Evidence for a core gut microbiota in the zebrafish. ISME J. 2011;5(10):1595‐1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stephens WZ, Burns AR, Stagaman K, et al. The composition of the zebrafish intestinal microbial community varies across development. ISME J. 2016;10(3):644‐654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rawls JF, Samuel BS, Gordon JI. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc Natl Acad Sci U S A. 2004;101:4596‐4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jemielita M, Taormina MJ, Burns AR, et al. Spatial and temporal features of the growth of a bacterial species colonizing the zebrafish gut. mBio. 2014;5:e01751‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ye L, Mueller O, Bagwell J, Bagnat M, Liddle RA, Rawls JF. High fat diet induces microbiota‐dependent silencing of enteroendocrine cells. eLife. 2019;8:e48479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hill JH, Franzosa EA, Huttenhower C, Guillemin K. A conserved bacterial protein induces pancreatic beta cell expansion during zebrafish development. eLife. 2016;5:e20145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Qin Q, Chen Y, Li Y, et al. Intestinal microbiota play an important role in the treatment of type I diabetes in mice with BefA protein. Front Cell Infect Microbiol. 2021;11:719542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Phelps D, Brinkman NE, Keely SP, et al. Microbial colonization is required for normal neurobehavioral development in zebrafish. Sci Rep. 2017;7(1):11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Davis DJ, Bryda EC, Gillespie CH, Ericsson AC. Microbial modulation of behavior and stress responses in zebrafish larvae. Behav Brain Res. 2016;311:219‐227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Weitekamp CA, Kvasnicka A, Keely SP, et al. Monoassociation with bacterial isolates reveals the role of colonization, community complexity and abundance on locomotor behavior in larval zebrafish. Anim Microbiome. 2021;3(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Borrelli L, Aceto S, Agnisola C, et al. Probiotic modulation of the microbiota‐gut‐brain axis and behaviour in zebrafish. Sci Rep. 2016;6:30046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang Z, Peng Q, Huo D, et al. Melatonin regulates the neurotransmitter secretion disorder induced by caffeine through the microbiota‐gut‐brain axis in zebrafish (Danio rerio). Front Cell Dev Biol. 2021;9:678190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cuomo M, Borrelli L, Della Monica R, et al. DNA methylation profiles of Tph1A and BDNF in gut and brain of L. Rhamnosus‐treated zebrafish. Biomolecules. 2021;11(2):142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bauer PV, Hamr SC, Duca FA. Regulation of energy balance by a gut‐brain axis and involvement of the gut microbiota. Cell Mol Life Sci. 2016;73(4):737‐755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ye L, Bae M, Cassilly CD, et al. Enteroendocrine cells sense bacterial tryptophan catabolites to activate enteric and vagal neuronal pathways. Cell Host Microbe. 2021;29(2):179‐196.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sjögren K, Engdahl C, Henning P, et al. The gut microbiota regulates bone mass in mice. J Bone Miner Res. 2012;27(6):1357‐1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Avella MA, Place A, Du S‐J, et al. Lactobacillus rhamnosus accelerates zebrafish backbone calcification and gonadal differentiation through effects on the GnRH and IGF systems. PLoS ONE. 2012;7(9):e45572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maradonna F, Gioacchini G, Falcinelli S, et al. Probiotic supplementation promotes calcification in Danio rerio larvae: a molecular study. PLoS ONE. 2013;8(12):e83155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mariotti M, Carnovali M, Banfi G. Danio rerio: the Janus of the bone from embryo to scale. Clin Cases Miner Bone Metab. 2015;12(2):188‐194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Carnovali M, Valli R, Banfi G, Porta G, Mariotti M. Soybean meal‐dependent intestinal inflammation induces different patterns of bone‐loss in adult zebrafish scale. Biomedicine. 2021;9(4):393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Murdoch CC, Rawls JF. Commensal microbiota regulate vertebrate innate immunity—insights from the zebrafish. Front Immunol. 2019;10:2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rolig AS, Parthasarathy R, Burns AR, Bohannan BJM, Guillemin K. Individual members of the microbiota disproportionately modulate host innate immune responses. Cell Host Microbe. 2015;18(5):613‐620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Masud S, Torraca V, Meijer AH. Modeling infectious diseases in the context of a developing immune system. Curr Top Dev Biol. 2017;124:277‐329. [DOI] [PubMed] [Google Scholar]
- 43. Bates JM, Akerlund J, Mittge E, Guillemin K. Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe. 2007;2(6):371‐382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Koch BEV, Yang S, Lamers G, Stougaard J, Spaink HP. Intestinal microbiome adjusts the innate immune setpoint during colonization through negative regulation of MyD88. Nat Commun. 2018;9(1):4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brugman S, Schneeberger K, Witte M, et al. T lymphocytes control microbial composition by regulating the abundance of vibrio in the zebrafish gut. Gut Microbes. 2014;5(6):737‐747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kanther M, Tomkovich S, Xiaolun S, et al. Commensal microbiota stimulate systemic neutrophil migration through induction of serum amyloid A. Cell Microbiol. 2014;16(7):1053‐1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wiles TJ, Schlomann BH, Wall ES, Betancourt R, Parthasarathy R, Guillemin K. Swimming motility of a gut bacterial symbiont promotes resistance to intestinal expulsion and enhances inflammation. PLoS Biol. 2020;18(3):e3000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Davison JM, Lickwar CR, Song L, Breton G, Crawford GE, Rawls JF. Microbiota regulate intestinal epithelial gene expression by suppressing the transcription factor hepatocyte nuclear factor 4 alpha. Genome Res. 2017;27(7):1195‐1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kanther M, Sun X, Mühlbauer M, et al. Microbial colonization induces dynamic temporal and spatial patterns of NF‐κB activation in the zebrafish digestive tract. Gastroenterology. 2011;141(1):197‐207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Murdoch CC, Espenschied ST, Matty MA, Mueller O, Tobin DM, Rawls JF. Intestinal serum amyloid a suppresses systemic neutrophil activation and bactericidal activity in response to microbiota colonization. PLoS Pathog. 2019;15(3):e1007381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cholan PM, Han A, Woodie BR, et al. Conserved anti‐inflammatory effects and sensing of butyrate in zebrafish. Gut Microbes. 2020;12(1):1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wu X, Teame T, Hao Q, et al. Use of a paraprobiotic and postbiotic feed supplement (HWF™) improves the growth performance, composition and function of gut microbiota in hybrid sturgeon (Acipenser baerii × Acipenser schrenckii). Fish Shellfish Immunol. 2020;104:36‐45. [DOI] [PubMed] [Google Scholar]
- 53. Rolig AS, Mittge EK, Ganz J, et al. The enteric nervous system promotes intestinal health by constraining microbiota composition. PLoS Biol. 2017;15(2):e2000689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yang H‐T, Zou S‐S, Zhai L‐J, et al. Pathogen invasion changes the intestinal microbiota composition and induces innate immune responses in the zebrafish intestine. Fish Shellfish Immunol. 2017;71:35‐42. [DOI] [PubMed] [Google Scholar]
- 55. Kaya B, Doñas C, Wuggenig P, et al. Lysophosphatidic acid‐mediated GPR35 signaling in CX3CR1 macrophages regulates intestinal homeostasis. Cell Rep. 2020;32(5):107979. [DOI] [PubMed] [Google Scholar]
- 56. Yansen Z, Lingang Z, Dali L, et al. Inflammatory bowel disease susceptible gene GPR35 promotes bowel inflammation in mice. Yi Chuan. 2021;43(2):169‐181. [DOI] [PubMed] [Google Scholar]
- 57. Marjoram L, Alvers A, Deerhake ME, et al. Epigenetic control of intestinal barrier function and inflammation in zebrafish. Proc Natl Acad Sci U S A. 2015;112(9):2770‐2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Willis AR, Torraca V, Gomes MC, et al. Shigella‐induced emergency granulopoiesis protects zebrafish larvae from secondary infection. mBio. 2018;9(3):e00933‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lakstygal AM, de Abreu MS, Lifanov DA, et al. Zebrafish models of diabetes‐related CNS pathogenesis. Prog Neuropsychopharmacol Biol Psychiatry. 2019;92:48‐58. [DOI] [PubMed] [Google Scholar]
- 60. Zhang H‐M, Chen S‐W, Xie C‐G, et al. Effects and mechanism of ShenQi compound recipe on inflammation maker in GK rats. Zhong Yao Cai. 2006;29(3):249‐253. [PubMed] [Google Scholar]
- 61. Xiong R, Zhao C, Zhong M, Zhang X, Liu W. Effects of Shenqi compound on intestinal microbial metabolites in patients with type 2 diabetes: a protocol for systematic review and meta analysis. Medicine (Baltimore). 2020;99(48):e23017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Okazaki F, Zang L, Nakayama H, et al. Microbiome alteration in type 2 diabetes mellitus model of zebrafish. Sci Rep. 2019;9(1):867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bootorabi F, Saadat F, Falak R, et al. Gut micobiota alteration by lactobacillus rhamnosus reduces pro‐inflammatory cytokines and glucose level in the adult model of zebrafish. BMC Res Notes. 2021;14(1):302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nag D, Breen P, Raychaudhuri S, Withey JH. Glucose metabolism by Escherichia coli inhibits vibrio cholerae intestinal colonization of zebrafish. Infect Immun. 2018;86(12):e00486‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sheng Y, Ren H, Limbu SM, et al. The presence or absence of intestinal microbiota affects lipid deposition and related genes expression in zebrafish (Danio rerio). Front Microbiol. 2018;9:1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ding Q, Zhang Z, Ran C, et al. The hepatotoxicity of palmitic acid in zebrafish involves the intestinal microbiota. J Nutr. 2018;148(8):1217‐1228. [DOI] [PubMed] [Google Scholar]
- 67. Qiao F, Tan F, Li L‐Y, et al. Alteration and the function of intestinal microbiota in high‐fat‐diet‐ or genetics‐induced lipid accumulation. Front Microbiol. 2021;12:741616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Falcinelli S, Picchietti S, Rodiles A, et al. Lactobacillus rhamnosus lowers zebrafish lipid content by changing gut microbiota and host transcription of genes involved in lipid metabolism. Sci Rep. 2015;5:9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Falcinelli S, Rodiles A, Hatef A, et al. Dietary lipid content reorganizes gut microbiota and probiotic L. rhamnosus attenuates obesity and enhances catabolic hormonal milieu in zebrafish. Sci Rep. 2017;7(1):5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ma N, Ma X. Dietary amino acids and the gut‐microbiome‐immune axis: physiological metabolism and therapeutic prospects. Compr Rev Food Sci Food Saf. 2019;18(1):221‐242. [DOI] [PubMed] [Google Scholar]
- 71. Hoseinifar SH, Safari R, Dadar M. Dietary sodium propionate affects mucosal immune parameters, growth and appetite related genes expression: insights from zebrafish model. Gen Comp Endocrinol. 2017;243:78‐83. [DOI] [PubMed] [Google Scholar]
- 72. Robinson CD, Sweeney EG, Ngo J, et al. Host‐emitted amino acid cues regulate bacterial chemokinesis to enhance colonization. Cell Host Microbe. 2021;29(8):1221‐1234.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wang C, Zhao Y, Jin Y. The emerging PFOS alternative OBS exposure induced gut microbiota dysbiosis and hepatic metabolism disorder in adult zebrafish. Comp Biochem Physiol C Toxicol Pharmacol. 2020;230:108703. [DOI] [PubMed] [Google Scholar]
- 74. Koo H, Hakim JA, Powell ML, et al. Metagenomics approach to the study of the gut microbiome structure and function in zebrafish Danio rerio fed with gluten formulated diet. J Microbiol Methods. 2017;135:69‐76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Locasale JW. Serine, glycine and one‐carbon units: cancer metabolism in full circle. Nat Rev Cancer. 2013;13(8):572‐583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Amelio I, Cutruzzolá F, Antonov A, Agostini M, Melino G. Serine and glycine metabolism in cancer. Trends Biochem Sci. 2014;39(4):191‐198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Takahashi K, Nishida A, Fujimoto T, et al. Reduced abundance of butyrate‐producing bacteria species in the fecal microbial community in Crohn’s disease. Digestion. 2016;93(1):59‐65. [DOI] [PubMed] [Google Scholar]
- 78. Pascal V, Pozuelo M, Borruel N, et al. A microbial signature for Crohn’s disease. Gut. 2017;66(5):813‐822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Shan C, Li M, Liu Z, et al. Enhances host resistance against pathogen by increasing IL‐1β production: understanding probiotic effectiveness and administration duration. Front Immunol. 2021;12:766401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Li S, Heng X, Guo L, Lessing DJ, Chu W. SCFAs improve disease resistance via modulate gut microbiota, enhance immune response and increase antioxidative capacity in the host. Fish Shellfish Immunol. 2021;120:560‐568. [DOI] [PubMed] [Google Scholar]
- 81. Jin Y, Wu S, Zeng Z, Fu Z. Effects of environmental pollutants on gut microbiota. Environ Pollut. 2017;22(2):1‐9. [DOI] [PubMed] [Google Scholar]
- 82. Zang L, Ma Y, Huang W, et al. Dietary lactobacillus plantarum ST‐III alleviates the toxic effects of triclosan on zebrafish (Danio rerio) via gut microbiota modulation. Fish Shellfish Immunol. 2019;84:1157‐1169. [DOI] [PubMed] [Google Scholar]
- 83. Chen L, Lam JCW, Hu C, Tsui MMP, Lam PKS, Zhou B. Perfluorobutanesulfonate exposure skews sex ratio in fish and transgenerationally impairs reproduction. Environ Sci Technol. 2019;53(14):8389‐8397. [DOI] [PubMed] [Google Scholar]
- 84. Tang L, Song S, Hu C, et al. Parental exposure to perfluorobutane sulfonate disturbs the transfer of maternal transcripts and offspring embryonic development in zebrafish. Chemosphere. 2020;256:127169. [DOI] [PubMed] [Google Scholar]
- 85. Chen L, Lam JCW, Tang L, et al. Probiotic modulation of lipid metabolism disorders caused by perfluorobutanesulfonate pollution in zebrafish. Environ Sci Technol. 2020;54(12):7494‐7503. [DOI] [PubMed] [Google Scholar]
- 86. Hu C, Liu M, Tang L, Liu H, Sun B, Chen L. Probiotic intervention mitigates the metabolic disturbances of perfluorobutanesulfonate along the gut‐liver axis of zebrafish. Chemosphere. 2021;28(4):131374. [DOI] [PubMed] [Google Scholar]
- 87. Pindling S, Azulai D, Zheng B, Dahan D, Perron GG. Dysbiosis and early mortality in zebrafish larvae exposed to subclinical concentrations of streptomycin. FEMS Microbiol Lett. 2018;365(18):fny188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Keerthisinghe TP, Wang F, Wang M, et al. Long‐term exposure to TET increases body weight of juvenile zebrafish as indicated in host metabolism and gut microbiome. Environ Int. 2020;139:105705. [DOI] [PubMed] [Google Scholar]
- 89. Zhou L, Limbu SM, Qiao F, du ZY, Zhang M. Influence of long‐term feeding antibiotics on the gut health of zebrafish. Zebrafish. 2018;15(4):340‐348. [DOI] [PubMed] [Google Scholar]
- 90. Li J, Dong T, Keerthisinghe TP, et al. Long‐term oxytetracycline exposure potentially alters brain thyroid hormone and serotonin homeostasis in zebrafish. J Hazard Mater. 2020;399:123061. [DOI] [PubMed] [Google Scholar]
- 91. Zhou L, Limbu SM, Shen M, et al. Environmental concentrations of antibiotics impair zebrafish gut health. Environ Pollut. 2018;23(5):245‐254. [DOI] [PubMed] [Google Scholar]
- 92. Almeida AR, Tacão M, Machado AL, et al. Long‐term effects of oxytetracycline exposure in zebrafish: a multi‐level perspective. Chemosphere. 2019;222:333‐344. [DOI] [PubMed] [Google Scholar]
- 93. Almeida AR, Alves M, Domingues I, Henriques I. The impact of antibiotic exposure in water and zebrafish gut microbiomes: a 16S rRNA gene‐based metagenomic analysis. Ecotoxicol Environ Saf. 2019;186:109771. [DOI] [PubMed] [Google Scholar]
- 94. Almeida AR, Domingues I, Henriques I. Zebrafish and water microbiome recovery after oxytetracycline exposure. Environ Pollut. 2021;272:116371. [DOI] [PubMed] [Google Scholar]
- 95. Wan Z, Wang C, Zhou J, et al. Effects of polystyrene microplastics on the composition of the microbiome and metabolism in larval zebrafish. Chemosphere. 2019;217:646‐658. [DOI] [PubMed] [Google Scholar]
- 96. Zhao Y, Qin Z, Huang Z, Bao Z, Luo T, Jin Y. Effects of polyethylene microplastics on the microbiome and metabolism in larval zebrafish. Environ Pollut. 2021;282:117039. [DOI] [PubMed] [Google Scholar]
- 97. Jin Y, Xia J, Pan Z, Yang J, Wang W, Fu Z. Polystyrene microplastics induce microbiota dysbiosis and inflammation in the gut of adult zebrafish. Environ Pollut. 2018;235:322‐329. [DOI] [PubMed] [Google Scholar]
- 98. Qiao R, Sheng C, Lu Y, Zhang Y, Ren H, Lemos B. Microplastics induce intestinal inflammation, oxidative stress, and disorders of metabolome and microbiome in zebrafish. Sci Total Environ. 2019;662:246‐253. [DOI] [PubMed] [Google Scholar]
- 99. Gu W, Liu S, Chen L, et al. Single‐cell RNA sequencing reveals size‐dependent effects of polystyrene microplastics on immune and secretory cell populations from zebrafish intestines. Environ Sci Technol. 2020;546:3417‐3427. [DOI] [PubMed] [Google Scholar]
- 100. Xie S, Zhou A, Wei T, et al. Nanoplastics induce more serious microbiota dysbiosis and inflammation in the gut of adult zebrafish than microplastics. Bull Environ Contam Toxicol. 2021;107(4):640‐650. [DOI] [PubMed] [Google Scholar]
- 101. de Carvalho Penha LC, Coimbra Rola R, da Silva Junior FM, de Martinez Gaspar Martins C. Toxicity and sublethal effects of methylparaben on zebrafish (Danio rerio) larvae and adults. Environ Sci Pollut Res Int. 2021;28(33):45534‐45544. [DOI] [PubMed] [Google Scholar]
- 102. Chen L, Zhang W, Hua J, et al. Dysregulation of intestinal health by environmental pollutants: involvement of the estrogen receptor and aryl hydrocarbon receptor. Environ Sci Technol. 2018;524:2323‐2330. [DOI] [PubMed] [Google Scholar]
- 103. Catron TR, Keely SP, Brinkman NE, et al. Host developmental toxicity of BPA and BPA alternatives is inversely related to microbiota disruption in zebrafish. Toxicol Sci. 2019;167(2):468‐483. [DOI] [PubMed] [Google Scholar]
- 104. Catron TR, Swank A, Wehmas LC, et al. Microbiota alter metabolism and mediate neurodevelopmental toxicity of 17β‐estradiol. Sci Rep. 2019;91:7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Liu Y, Yao Y, Li H, et al. Influence of endogenous and exogenous estrogenic endocrine on intestinal microbiota in zebrafish. PLoS ONE. 2016;11(10):e0163895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Chen L, Guo Y, Hu C, Lam PKS, Lam JCW, Zhou B. Dysbiosis of gut microbiota by chronic coexposure to titanium dioxide nanoparticles and bisphenol a: implications for host health in zebrafish. Environ Pollut. 2018;234:307‐317. [DOI] [PubMed] [Google Scholar]
- 107. Gaulke CA, Barton CL, Proffitt S, et al. Triclosan exposure is associated with rapid restructuring of the microbiome in adult zebrafish. PLoS ONE. 2016;115:e0154632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Jin C, Luo T, Zhu Z, et al. Imazalil exposure induces gut microbiota dysbiosis and hepatic metabolism disorder in zebrafish. Comp Biochem Physiol C Toxicol Pharmacol. 2017;202:85‐93. [DOI] [PubMed] [Google Scholar]
- 109. Bao Z, Zhao Y, Wu A, et al. Sub‐chronic carbendazim exposure induces hepatic glycolipid metabolism disorder accompanied by gut microbiota dysbiosis in adult zebrafish (Daino rerio). Sci Total Environ. 2020;739:140081. [DOI] [PubMed] [Google Scholar]
- 110. Tang N, Fan P, Yu X, et al. Effects of long‐term triclosan exposure on microbiota in zebrafish. Front Microbiol. 2021;12:604313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Hua Q, Adamovsky O, Vespalcova H, et al. Microbiome analysis and predicted relative metabolomic turnover suggest bacterial heme and selenium metabolism are altered in the gastrointestinal system of zebrafish (Danio rerio) exposed to the organochlorine dieldrin. Environ Pollut. 2021;268(Pt B):115715. [DOI] [PubMed] [Google Scholar]
- 112. Jiang J, Chen L, Wu S, et al. Effects of difenoconazole on hepatotoxicity, lipid metabolism and gut microbiota in zebrafish (Danio rerio). Environ Pollut. 2020;265(Pt A):114844. [DOI] [PubMed] [Google Scholar]
- 113. Zhang R, Pan Z, Wang X, et al. Short‐term propamocarb exposure induces hepatic metabolism disorder associated with gut microbiota dysbiosis in adult male zebrafish. Acta Biochim Biophys Sin (Shanghai). 2019;511:88‐96. [DOI] [PubMed] [Google Scholar]
- 114. Luo T, Wang X, Jin Y. Low concentrations of imidacloprid exposure induced gut toxicity in adult zebrafish (Danio rerio). Comp Biochem Physiol C Toxicol Pharmacol. 2021;241:108972. [DOI] [PubMed] [Google Scholar]
- 115. Wang X, Shen M, Zhou J, Jin Y. Chlorpyrifos disturbs hepatic metabolism associated with oxidative stress and gut microbiota dysbiosis in adult zebrafish. Comp Biochem Physiol C Toxicol Pharmacol. 2019;216:19‐28. [DOI] [PubMed] [Google Scholar]
- 116. Xie S, Zhou A, Xu N, et al. Benzo[a]pyrene induces microbiome dysbiosis and inflammation in the intestinal tracts of western mosquitofish (Gambusia affinis) and zebrafish (Danio rerio). Fish Shellfish Immunol. 2020;105:24‐34. [DOI] [PubMed] [Google Scholar]
- 117. Chen L, Hu C, Lok‐Shun Lai N, et al. Acute exposure to PBDEs at an environmentally realistic concentration causes abrupt changes in the gut microbiota and host health of zebrafish. Environ Pollut. 2018;240:17‐26. [DOI] [PubMed] [Google Scholar]
- 118. Xia J, Lu L, Jin C, et al. Effects of short term lead exposure on gut microbiota and hepatic metabolism in adult zebrafish. Comp Biochem Physiol C Toxicol Pharmacol. 2018;209:1‐8. [DOI] [PubMed] [Google Scholar]
- 119. Patsiou D, Del Rio‐Cubilledo C, Catarino AI, et al. Exposure to pb‐halide perovskite nanoparticles can deliver bioavailable pb but does not alter endogenous gut microbiota in zebrafish. Sci Total Environ. 2020;715:136941. [DOI] [PubMed] [Google Scholar]
- 120. Chen P, Huang J, Rao L, et al. Environmental effects of nanoparticles on the ecological succession of gut microbiota across zebrafish development. Sci Total Environ. 2022;806(Pt 4):150963. [DOI] [PubMed] [Google Scholar]
- 121. Dahan D, Jude BA, Lamendella R, Keesing F, Perron GG. Exposure to arsenic alters the microbiome of larval zebrafish. Front Microbiol. 2018;9:1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Mitchell KC, Withey JH. Danio rerio as a native host model for understanding pathophysiology of vibrio cholerae . Methods Mol Biol. 2018;1839:97‐102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Volkman HE, Pozos TC, Zheng J, Davis JM, Rawls JF, Ramakrishnan L. Tuberculous granuloma induction via interaction of a bacterial secreted protein with host epithelium. Science. 2010;327(5964):466‐469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Johansen MD, Kasparian JA, Hortle E, Britton WJ, Purdie AC, Oehlers SH. Mycobacterium marinum infection drives foam cell differentiation in zebrafish infection models. Dev Comp Immunol. 2018;88:169‐172. [DOI] [PubMed] [Google Scholar]
- 125. Hu W, Yang S, Shimada Y, et al. Infection and RNA‐seq analysis of a zebrafish tlr2 mutant shows a broad function of this toll‐like receptor in transcriptional and metabolic control and defense to Mycobacterium marinum infection. BMC Genomics. 2019;201:878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Howlader DR, Sinha R, Nag D, et al. Zebrafish as a novel model for non‐typhoidal salmonella pathogenesis, transmission and vaccine efficacy. Vaccine. 2016;34(42):5099‐5106. [DOI] [PubMed] [Google Scholar]
- 127. Ran C, Qin C, Xie M, et al. Aeromonas veronii and aerolysin are important for the pathogenesis of motile aeromonad septicemia in cyprinid fish. Environ Microbiol. 2018;20(9):3442‐3456. [DOI] [PubMed] [Google Scholar]
- 128. Xin G‐Y, Li W‐G, Suman TY, Jia PP, Ma YB, Pei DS. Gut bacteria vibrio sp. and Aeromonas sp. trigger the expression levels of proinflammatory cytokine: first evidence from the germ‐free zebrafish. Fish Shellfish Immunol. 2020;106:518‐525. [DOI] [PubMed] [Google Scholar]
- 129. Logan SL, Thomas J, Yan J, et al. The type VI secretion system can modulate host intestinal mechanics to displace gut bacterial symbionts. Proc Natl Acad Sci U S A. 2018;115(16):E3779‐E3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Breen P, Winters AD, Theis KR, Withey JH. Vibrio cholerae infection induces strain‐specific modulation of the zebrafish intestinal microbiome. Infect Immun. 2021;89(9):e0015721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Stones DH, Fehr AGJ, Thompson L, et al. Zebrafish (Danio rerio) as a vertebrate model host to study colonization, pathogenesis, and transmission of foodborne O157. mSphere. 2017;2(5):e00365‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Zhang Q‐L, Li H‐W, Wu W, et al. The response of microbiota community to infection in zebrafish intestine. Front Microbiol. 2019;10:2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Caruffo M, Navarrete NC, Salgado OA, et al. Protective yeasts control pathogenicity and modulate the innate immune response of challenged zebrafish (Danio rerio) larvae. Front Cell Infect Microbiol. 2016;6:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Vargas O, Gutiérrez MS, Caruffo M, et al. Probiotic yeasts and infection modify the microbiome of zebrafish larvae. Front Microbiol. 2021;12:647977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Pacheco I, Díaz‐Sánchez S, Contreras M, et al. Probiotic bacteria with high alpha‐gal content protect zebrafish against mycobacteriosis. Pharmaceuticals (Basel). 2021;14(7):635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. He S, Ran C, Qin C, et al. Anti‐infective effect of adhesive probiotic lactobacillus in fish is correlated with their spatial distribution in the intestinal tissue. Sci Rep. 2017;7(1):13195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Stressmann FA, Bernal‐Bayard J, Perez‐Pascual D, et al. Mining zebrafish microbiota reveals key community‐level resistance against fish pathogen infection. ISME J. 2021;15(3):702‐719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. López‐Igual R, Bernal‐Bayard J, Rodríguez‐Patón A, Ghigo JM, Mazel D. Engineered toxin‐intein antimicrobials can selectively target and kill antibiotic‐resistant bacteria in mixed populations. Nat Biotechnol. 2019;37(7):755‐760. [DOI] [PubMed] [Google Scholar]
- 139. Schlomann BH, Wiles TJ, Wall ES, Guillemin K, Parthasarathy R. Bacterial cohesion predicts spatial distribution in the larval zebrafish intestine. Biophys J. 2018;115(11):2271‐2277. [DOI] [PMC free article] [PubMed] [Google Scholar]


