Abstract
Intestinal microecology is the main component of human microecology. Intestinal microecology consists of intestinal microbiota, intestinal epithelial cells, and intestinal mucosal immune system. These components are interdependent and establish a complex interaction network that restricts each other. According to the impact on the human body, there are three categories of symbiotic bacteria, opportunistic pathogens, and pathogenic bacteria. The intestinal microecology participates in digestion and absorption, and material metabolism, and inhibits the growth of pathogenic microorganisms. It also acts as the body's natural immune barrier, regulates the innate immunity of the intestine, controls the mucosal barrier function, and also participates in the intestinal epithelial cells' physiological activities such as hyperplasia or apoptosis. When the steady‐state balance of the intestinal microecology is disturbed, the existing core intestinal microbiota network changes and leads to obesity, diabetes, and many other diseases, especially irritable bowel syndrome, inflammatory bowel disease (IBD), and colorectal malignancy. Intestinal diseases, including tumors, are particularly closely related to intestinal microecology. This article systematically discusses the research progress on the relationship between IBD and intestinal microecology from the pathogenesis, treatment methods of IBD, and the changes in intestinal microbiota.
Keywords: inflammatory bowel disease, intestinal epithelial barrier, intestinal microbiota, intestinal mucosal barrier
The article systematically discussed the research progress of the relationship between inflammatory bowel disease and intestinal microecology from the pathogenesis and treatment methods of inflammatory bowel disease and the changes in intestinal flora.
1. INTRODUCTION
It is currently known that as many as 1014 microorganisms aggregate in the nutrient‐rich gut, which together constitutes the microbiota with very diverse characteristics. 1 These microbes perform many functions in the gut, such as preventing pathogen infection, aiding digestion, providing nutrients, and forming a mucosal barrier to the immune system. 2 Some researchers believe that the microbial diversity of the human gut is the result of the co‐evolution of the microbial community and its host. 3 Human Microecology Project and the Human Intestinal System Genome Project (MetaHIT) have focused on the function and structure of gut microbes. 4 , 5 Studies between obese and non‐obese individuals in twins have shown that obese individuals have reduced diversity in their gut microbiota and altered characterization of metabolic pathways. 6 , 7 Between the host epithelial cells and immune cells and specific gut microbes there are pattern receptors can mutual recognition, microbes through Pregnane X receptor (PXR) and Toll‐like receptors (TLR) control the mucosal barrier function modulates gut innate immunity and affects gut community structure. 8 After the microbiota and metabolites bind to receptors, they activate signaling pathways and interact with related key proteins, thereby participating in the regulation of macrophage polarization by host immune cells and the destruction and regeneration of intestinal epithelial cells. 9
Pathogen recognition by immunoglobulins (Ig) is an important aspect in immune adaptation, and microbial symbionts increase the frequency of antimicrobial IgM + IgD + and B cells in the gut. This enrichment affects follicular B cells, and microbial symbionts influence host immunity by enriching antibacterial‐specific frequencies in the pre‐immune B cell pool, thereby affecting the mucosal immune system. 10 Bifidobacterium, Faecacterium, Ruminococcus, and Prevotella were negatively associated with markers of chronic inflammation such as hsCRP and interleukin (IL)‐6. Gut microbiota may control chronic inflammatory response and thus participate in the pathogenesis of atherosclerotic cardiovascular disease (ACVD). 11 Studies show that the progression of myocarditis to fatal heart disease depends on cardiac myosin‐specific TH17 cells tagged in the gut by commensal Bacteroidetes peptidomimetics, involved in immune responses to exacerbate myocardial inflammation. 12 Studies in obese patients have shown reduced diversity of gut microbiota and abundance of lactobac in obese individuals, and although the causal role of altered gut microbiome and obesity is controversial, animal and human studies have shown that gut metabolites (such as butyrate) can be used to prevent and treat obesity and its complications. 13 A metagenomic study demonstrated a significant reduction in gut bacterial diversity in patients with autoimmune liver disease (AILD), in whom functional analyses revealed altered metabolic pathways in the gut microbiome of AILD patients. 14 A study of pregnant women with gestational diabetes found that the microbes in pregnant women and newborns changed significantly at almost the same time, mainly Firmicutes and Proteobacteria. 15 Studies have shown that the gut microbiota is involved in the early stages of Rheumatoid Arthritis (RA). The results of the analysis found that, compared with healthy controls, the diversity of RA patients led to dysbiosis, and especially genes related to Bacteroidetes and synthetic lipopolysaccharide were enriched in RA. 16
In this review, the intestinal epithelial‐mucosal barrier is considered to be spatiotemporally controlled and organized by commensal bacteria, and intestinal epithelial cells and effector molecules. 17 To avoid the attack of pathogenic bacteria, viruses, and sometimes opportunistic pathogenic bacteria, the immune barrier in the intestinal tract is composed of epithelial cell layer and mucus layer, and active and passive immunity with symbiotic bacteria together forms the intestinal barrier system.
1.1. The relationship between gut bacteria and the mucus layer
The intestinal mucus layer is divided into inner and outer mucus layers. The outer mucus layer has a loose structure, a thin and thick texture, and is in direct contact with the microbiota. it is distributed in the small intestine and colon, but the mucus layer on the surface of the small intestine does not completely cover the epithelial cells of the small intestine. he mucus layer in the colon is more complex than that in the small intestine, one of the main locations the immune response occurs where the mechanical barrier of mucus layer in the colon while providing energy to maintain microbial symbiosis. 18 How the host controls mucus barrier integrity and maintains symbiosis is unclear. A major function of the intestinal mucus layer is the production of mucin secreted by intercrypt goblet cells (icGCS) on the colon surface. 19 Dense O‐glycosylated mucin 2 (MUC2) is the core molecule in the mucus layer. 20 Deficiency of MUC2 results in a thinner mucus layer in the colon, which is less likely to colonize commensal bacteria. 21
If the microbiota is imbalanced, many bacteria and metabolites can disrupt the mucus layer and affect its recovery of the mucus layer. A study has shown that the toxins produced by Akkermansia municiphila and Bacteroides fragilis release dissolved proteolytic enzymes of the mucus layer. 22 Sulfate‐reducing bacteria can also degrade mucin proteins, and the mucus layer also provides energy for the activity of the microbiota. 23 Goblet cells secrete a large amount of mucin. When pathogenic bacteria invade, Toll receptors receive a signal to instruct MUC2 protein to encapsulate pathogenic bacteria and opportunistic bacteria and metabolites out of the inner mucus layer and then secrete new mucin to form renewal. 24 , 25 When the intestinal mucosal barrier is destroyed, the Paneth cells in the small intestinal crypts undergo gene mutations or contain abnormal transcription factors, and the Nod2 gene mutation in the antibacterial peptide subgroup will reduce the secretion of mucin and antibacterial substances after being stimulated by the microbiota. 26 , 27 However, symbiotic bacteria also have a protective effect on the mucus layer. Studies shows that intestinal microbiota metabolites produce short‐chain fatty acids (SCFA) and butyric acid, and these products can upregulate the secretion of MUC2. 28 Mucin also participates in the anti‐pathogen invasion and protects the crypt stem cells at the base of Paneth cells in the crypts of the small intestine and colon to normally maintain the renewal of intestinal epithelial cells. 29 , 30 WFDC2 is an anti‐protease molecule expressed by goblet cells and inhibits bacterial growth. In vivo, WFDC2 maintains the integrity of tight junctions (TJ) between epithelial cells and prevents the invasion of commensal bacteria and mucosal inflammation, a role in mucosal barrier homeostasis. 31
1.2. The relationship between gut bacteria and intestinal epithelial cells
Intestinal epithelial cells are the second immune barrier of the intestine and complete a renewal cycle every 3–6 days. To maintain intestinal homeostasis, colonic epithelial cells are covered by a thick inner and outer mucus layer, which contains some symbiotic bacteria (eg. Bacteroidetes. Fermicutes, etc). The inner mucus layer does not contain commensal bacteria. 19 , 32 Although the mucus layer serves as a physical barrier to the large microbiota present in the large intestine, the mechanism that separates bacteria from the colonic epithelium has not been fully elucidated. Secretory fragments and IgA of intestinal epithelial cells assemble into sIgA; the sIgA found on the surface of the intestinal mucosa can recognize the intestinal microbiota, especially gram‐negative facultative anaerobic bacilli, encapsulate the bacteria, avoid bacterial colonization in the intestinal epithelium, block the junction between bacteria and mucus, and activate autophagy to clear bacteria and metabolites. 33 The mechanism of maintaining intestinal epithelial barrier homeostasis has not yet been elucidated. Some studies have found that if Lypd8 gene expression is blocked, bacteria can directly bind to receptors on the cell membrane. Lypd8−/− mice show spontaneous inflammatory response than normal mice or high‐fat diet (HFD) mice. 34 Lypd8 gene is selectively expressed in the uppermost epithelial cells of the colorectal glands, possibly. It is in the Ly6/PLAUR domain containing eight (Lypd8) proteins, which can bind to the flagella of gram‐negative bacilli or Proteus mirabilis to inhibit the movement of the flagella, thereby preventing bacteria from entering the inner mucus layer, inhibiting the inflammatory response of epithelial cells, and separating bacteria and epithelial cell layer. 35 , 36
The intestinal mucosal barrier is capable of secreting antimicrobial proteins, some of which are activated only when the barrier is attacked. An example is SPRR2A protein that can work in low salt concentration and acidic environment. SPRR2A binds with negatively charged lipids and can destroy liposomes of negatively charged lipids. After intestinal microbiota colonization or pathogen infection, the expression of small protein 2A is significantly upregulated through the activation of the TLR‐MyD88 signaling pathway. 37 The intestinal microbiota of SPRR2A knockout mice was altered, and 16sRNA gene sequence analysis showed an increase in the abundance of gram‐positive bacteria in the intestine, indicating that SPRR2A knockout increased the susceptibility of gram‐positive bacteria in the intestine. The researchers observed that the SPRR2A protein is induced through STAT6. Intraperitoneal injection of IL‐13 into the wild‐type mice also increased SPRR2A expression. STAT6 acts downstream of IL‐4 and IL‐13 to induce type 2 immunity. This indicated that SPRR2A could be upregulated through the IL‐4/13‐STAT6 pathway, thereby eliminating pathogenic bacteria and protecting the integrity of the epithelial barrier. 38
Clostridium difficile toxin A (TcdA) is a major exotoxin that contributes to the destruction of the colonic epithelium during C. difficile infection and is thought to be a major contributor to colonic inflammation. A study used untransformed NCM460 human colon cells, in which C. difficile toxin A activated the PGE2 pathway through EP1 receptor agonists to activate FasL transcription in enterocytes, and FasL perforated the mitochondrial outer membrane, leading to apoptosis, destroying epithelial cells. 39 In addition, TcdA contains a carbohydrate‐binding combinatorial repeat oligopeptide (CROPs) domain that mediates its attachment to the cell surface. TcdA binds with sulfated groups in sulfated glycosaminoglycans (sGAG) by recognizing low‐density lipoproteins receptor (LDLR) and enters epithelial cells; while cells lacking LDLR show reduced sensitivity to TcdA. 40
Parikh et al. discovered a new absorptive cell, BEST4+/OTOP2+ cells, which help goblet cells repair barrier damage by regulating the GC‐C signaling pathway. 31 Guillermin et al. found that Wnt/β‐catenin signaling is an important pathway for epithelial cell proliferation in intestinal crypts. 41 TcdA destroys intestinal epithelial cells by inhibiting Wnt/β‐catenin signaling. 42 Wnt signaling is responsible for proliferation of intestinal crypt stem cell and cooperates with Scr‐YAP signaling to inhibit inflammatory damage and promote intestinal epithelial regeneration. 41 , 43 Metabolites are produced by intestinal microbiota through fermentation. Among them, SCFA families, including acetate, propionate, butyrate, play a role in protecting the intestinal mucosal barrier in the intestine. SCFAs can be directly taken up by epithelial cells into the lamina propria, and sometimes can also pass through the lamina propria. As transporter, SCFAs have a stronger affinity with SMCT‐1 (sodium‐coupled monocarboxylate transporter 1) than MCT‐1 (monocarboxylate transporter 1). 44 , 45 SCFAs can also activate inflammasomes to produce a large number of protective cytokines by regulating the expression of TLRs, and improving the self‐repair ability of epithelial cells. 46 , 47 Shao et al. found that the transplantation of B. fragilis into germ‐free mice could restore cell oxidative phosphorylation and increase ATP levels, and maintain epithelial cell integrity through increased colon epithelial cell metabolism. 48
1.3. The relationship between intestinal microbiota and TJs
TJs are multiprotein complexes composed of regulatory molecules, including kinases, transmembrane proteins, and peripheral membrane proteins, which protect the intestinal epithelial barrier and regulate intestinal permeability. 49 , 50 The TJs family mainly includes claudins, occludins, and JAM, ZO‐1, ZO‐2, and zonulin. 50 Enteropathogenic bacteria can release toxic proteins through their secretion system or direct secretion, transport them to the transport system in the host cell, and deliver them into the intestinal epithelial cells, affecting the expression and localization of TJ proteins and destructing intestinal epithelial cells. At the same time, dendritic cells release dendrites through epithelial cells to directly capture bacteria, identify invasive bacteria in advance, and strengthen the expression of TJ proteins to protect barrier integrity. 51 It is known that enteropathogenic Escherichia coli (EPEC) causes enteritis to destroy the TJs of the intestinal epithelium. The experiment found that after infection with EPEC, it will accumulate near the TJ proteins, encoding specific type III secreted effector proteins through the effector EspG1 to induce consumption of tricellulin. 52 Shigella flexneri also affects the expression and distribution of ZO‐1, Cldn1, and occludin, disrupting the epithelial barrier. 53 Intestinal bacteria can also directly secrete some enzymes or toxic proteins into the extracellular space, such as zonula occluden toxin (ZOT), which can activate intracellular signaling pathways leading to mislocalization of TJ proteins and damage to the intestinal mucosal barrier. 54 In contrast, intestinal epithelial cells contain protective antimicrobial proteins, and in a mouse model of inflammatory bowel disease (IBD), it was found that REG3A and Resistin‐like molecule‐beta can bind with negatively charged lipids to form pore membranes and lead to the destruction of liposomes to control inflammation. 55 , 56 In vivo, in the absence of the antibacterial protein WFDC2 secreted by goblet cells, epithelial cells separate and pathogenic bacteria cross the barrier, and normal expression of WFDC2 maintains the integrity of TJs between epithelial cells and prevents the invasion of commensal bacteria and mucosal inflammation. 31 In addition, the quinone oxidoreductase 1 (NQO1) protein involved in the formation of TJs is itself resistant to the acute inflammatory response induced by C. difficile toxin A, and Wang et al. found that NQO1 knockout mice spontaneously showed weak intestinal inflammation, marked loss of colonic epithelial TJ proteins, mucosal barrier disruption, and higher levels of epithelial cell apoptosis. 57 Studies have found that bacterial metabolite SCFAs can also regulate TJ proteins to protect the integrity of the barrier and have shown that higher levels of SCFAs in the gut can regulate NFκB and AMPK signaling pathway phosphorylation, upregulate the expression of ZO‐1 and claudin TJ protein, and enhance epithelial cell transmembrane resistance (TER). 58 Interestingly, some intestinal epithelial TJs are receptors for bacterial toxins. Saitoh et al. have shown that Claudin3 and Claudin4 were the earliest discovered Clostridium perfringens enterotoxin (CPE) receptors, and the C‐terminal region can bind to Claudin3/4, acting as cytotoxin and perforin, and break down TJ structures and increase the permeability of paraepithelial pathways, trigger epithelial cell apoptosis, and affect the physiological activities of intestinal epithelial cells. 59 C. difficile gets attached to the epithelium in the intestine through the action of C. difficile transferase (CDT). 60 There is evidence that a lipolysis stimulated lipoprotein receptor (LSR), which exists between consecutive epithelial cells, mediates the entry of CDT into target cells and is considered to be the host recognition receptor for CDT. 42 , 61 Tube cell binding promotes Clostridial adhesion and colonization, ultimately resulting in cell death that disrupts intestinal mucosal barrier integrity. 62 , 63
In conclusion, the relationship between gut microflora and gut barrier is a complex and intimate one. Although the aforementioned studies have demonstrated the interregulation between intestinal flora and intestinal barrier under physiological conditions, there are some potential mechanisms of action that need to be elucidated by more research evidence.
2. INTESTINAL MICROECOLOGY AND IBD
IBD is usually divided into two categories: ulcerative colitis (UC) and Crohn's disease (CD). IBD is considered to be caused by genetic susceptibility, diet, environmental and microbial factors that drive and trigger immune responses in genetically susceptible individuals. 64 , 65
2.1. IBD and microbial homeostasis
The current study puts the core of the pathogenesis of IBD seems to be all point to the interaction between the gut microbiota and the gut epithelium. Increased epithelial permeability in the colon is one of the indicators of the severity of IBD. Goblet cells in gastrointestinal epithelial cells regulate that defects in the mucus secretion can lead to changes in the expression of TJ proteins, resulting in the destruction of TJs in epithelial cells and in the imbalance of intestinal homeostasis and the termination of symbiosis (Figure 1).
FIGURE 1.
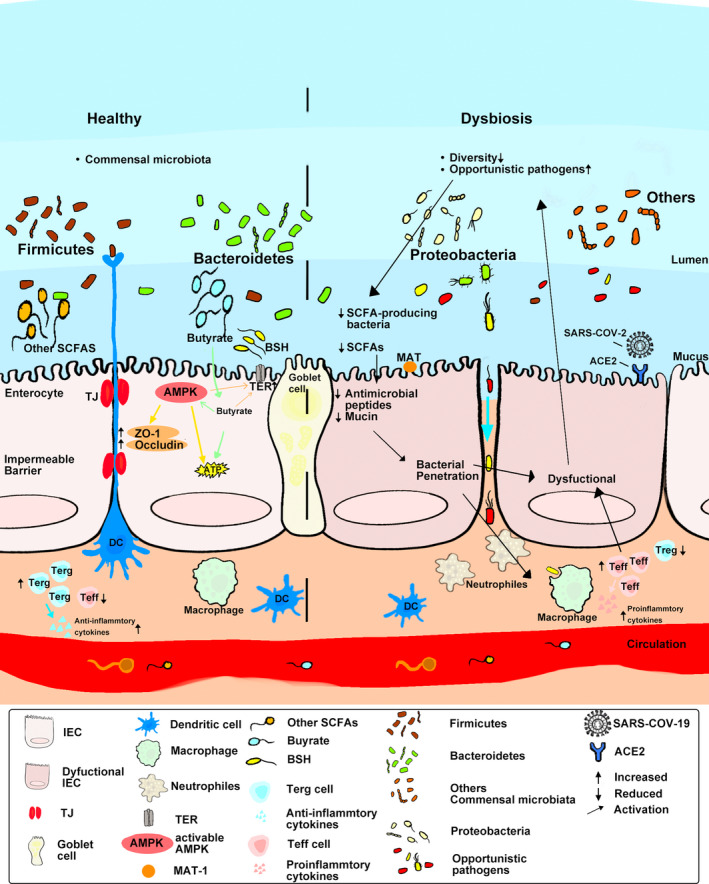
Factors influencing dysbiosis and intestinal inflammation in intestinal bowel disease.
On the one hand, the Integrative Human Microbiome Project (HMP2 or iHMP) tracks patients and analyzes the relationship between gut microbial activity and host disease activity through a multi‐omics approach. On the other hand, substantial evidence has demonstrated that microbial taxa enriched in IBD patients have similar pro‐inflammatory roles, such as Bacteroidetes, Firmicutes, and Proteobacteria. Furthermore, disturbances in the microbiota‐metabolic network with different subtypes of bacteria group‐characterized in patients interaction networks behave differently, including a balance of Lachnospira and Ruminococcus gnavus. 66 The study found that some gut‐derived microbiota differed in classification and function between enteritis‐susceptible samples and healthy samples. Enterococcus faecium, as the most different species, was positively correlated with the disease severity of patients. Transplant feces Enterococcus strains, compared with healthy people, the strains transplanted from UC patients can cause the increased expression of inflammatory factors related to UC in IL‐10 knockout mice. 67 .
However, currently no evidence directly demonstrates a causal relationship between gut microbe‐host symbiosis and IBD. Qian et al. observed that HLA‐B27 transgenic rats housed in a non‐sterile environment developed spontaneous immune‐mediated colonic inflammation. 68 Im et al. also found that in a mouse model deficient in IL‐2 and IL‐10 genes, mice with defective cell receptor genes in a non‐sterile environment developed symptoms of colitis. 69 , 70 Therefore, the changes in intestinal microbiota in the intestinal mucosa or the organ cavity may be secondary to intestinal inflammation. The intestinal environment with IBD may alter the metabolic activities of the strains, making it difficult for the strains to colonize, and then stimulate the intestinal mucosa to destroy the intestinal immune barrier. To reconstruct the gut microbial environment in IBD, the intestinal commensal microbiota forms a unique dominant microbiota. In addition, it was found that the dynamically changing microbiota in healthy people highly overlapped with the gut microbiota that was reduced in CD patients, and it was also observed that alleles of CD in patients were highly correlated with changes in colonic mucosal microbial composition. 71 In addition, a systematic review found that the amount of Faecalibacterium prausnitzii was significantly reduced in both CD patients and UC patients, indicating that the expression level of F. prausnitzii was negatively correlated with the degree of IBD. 72 The immune mechanism of the intestinal mucosal barrier in IBD is still unclear. Studies have found that if there is an inflammatory damage to the intestinal mucosa, the stimulated intestinal epithelial barrier can adapt to a certain degree of inflammation without causing an immune cascade. 73
The production of indopoacrylic acid (IA) by the Peptostreptococcus species can regulate intestinal immune responses through the xenobiotic sensor pregnane X receptor(PXR). 74 Metagenomic studies have found reduced biosynthetic gene clusters of IA in IBD patients, resulting in an inability to mitigate the inflammatory response to restore the intestinal epithelial barrier. 75 Statistics show that the level of bile salt hydrolases (BSH) in the gut microbiota of IBD patients is significantly lower than that in healthy individuals, especially in Firmicutes in CD patients. Lower BSH level fails to inhibit NF‐B signaling by activating farnesoid‐activated X receptor (FXR). 76
Colonic inflammation activates the immune response, produces IFNγ, and then generates reactive oxygen species (ROS) through phagocytic innate immune cells. These free radicals eventually provide a suitable environment for anaerobic microorganisms, reduce microbial diversity, reduce the abundance of inhibiting inflammatory microbiota, and fail to activate inhibiting inflammatory pathways to relieve inflammation, disrupted the balance of microbial homeostasis in IBD patients. 77In particular, in the colonization of germ‐free mice with microbiota from IBD patients, research shows that microbial adhesion to epithelial cells stimulates the production of Th17 cells and reduces the number of RORγt+ Tregs to break the regulatory process of inflammation, providing evidence for disease mechanisms in the gut microbiota of IBD (Table 1).
TABLE 1.
Major dysbiosis of gut microbiome during intestinal bowel disease
| Microorganism | Type | Dysbiosis | Model | References |
|---|---|---|---|---|
| Bifidobacteria, Firmicutes, Firmicutes prausnitzii | CD/UC | Decrease | Humans | 72 |
| Firmicutes | 2‐, 4‐ and 6‐ Trinitrobenzenesul fonic acid (TNBS) colitis | Decrease | Animal (rats and mice) | 66 |
| Bacteroidetes, Bacteroides, Enterobacteriaceae | Increase | 91 | ||
| Helicobacteraceae, Mucispirillum, Desulfovibrio | Experimental colitis | Increase | T‐bet(−/−), Rag2(−/−) mice | 117 |
| Enterobacteriaceae and adherent/invasive E. coli | Experimental colitis | Increase | IL‐10(−/−)mice | 69, 86 |
| Akkermansia muciniphila, Bacteroides distasonis, Enterobacteriaceae, Clostridium ramnosum | DSS‐colitis | Increase | F1Mice | 88 |
| Porphyromonas genera, Bacteroides | Experimental colitis | Increase | F11r(−/−), Rag1(−/−)mice | 73 |
| Lachnospira and Ruminococcus | DSS‐colitis/TNBS colitis | Decrease | Animal (rats and mice) | 66 |
| Enterococcus faecium | Experimental colitis | Increase | IL‐10(−/−)mice | 67 |
| Fusobacterium nucleatum | Experimental colitis | Increase | Humans | 90 |
| Faecalibacterium, Roseburia | CD | Decrease | Humans | 82 |
Abbreviations: CD, Crohn's disease; DSS, dextran sodium sulfate; UC, ulcerative colitis.
In addition to bacterial microbes, the microbiome includes both fungi and viruses. In 1988, an antibody to the cell wall component of Saccharomyces cerevisiae was linked to CD. 78 Studies have demonstrated that S. cerevisiae colonization can promote purine metabolism in mice, leading to elevated uric acid levels to enhance the inflammatory response. 79
2.2. Intestinal microecological imbalance and IBD
At present, a large number of studies have indicated that metabolites regulate the interaction between the gut microbiota and the host. Among these metabolites, bile acids, SCFAs, and tryptophan metabolites are closely related to the pathogenesis of IBD. 80 CARD9 is one of the IBD susceptibility genes that integrate signals downstream of the pattern recognition receptor. When Card9−/− mice fail to normally mediate p38‐JNK signaling and Toll‐like receptor signaling, then the microbiota of mice is unable to normally metabolize tryptophan to a ligand for the aromatic hydrocarbon receptor and fails to activate the IL‐22 pathway, leading to an increase in the number of proinflammatory microorganisms and aggravating the inflammatory response. 81 Roseburia hominis and F. prausnitzii are currently recognized acetic acid‐butyric acid converters, which are enriched in the mucus layer and play an important role in producing anti‐inflammatory effects. In some studies R. hominis and other SCFA‐producing bacteria decreased, and the levels of SCFA and unconjugated bile acids in feces decreased. 82 , 83 However, there are a large number of butyric acid metabolites in the human intestine, such as Clostridium and methanogens. Butyrate promotes Treg development and specifically regulates MUC expression in goblet cells, and the increased secretion of mucus enhances the mucosal barrier; the reduction in butyric acid synthesis leads to the slowdown of reconstitution of intestinal mucosal barrier. 84
Pathogens and viruses and some opportunistic bacteria in the gut microbiota may promote inflammatory damage, but commensal bacteria also play a protective role in the gut against inflammatory damage. Different microbiota plays different roles in IBD. Studies have shown that when the number of anaerobic bacteria in the intestine is high, the inflammatory damage is the most serious, the adhesion is strong, and the exudation and infiltration of mononuclear cells are the main factors. 85 Adherent/invasive E. coli (AIEC) isolated from IBD patients and non‐IBD patients express different virulence factors, and the virulence factors of non‐IBD patient strains are absent in AIEC. 86 AIEC is a typical opportunistic pathogen. AIEC avoids the immune system, invasion into epithelial cells and macrophages. In addition to the autophagy machinery that impairs the IBD host, it can avoid macrophage phagocytosis. Persistent replication of macrophages leads to increased TNFα levels and excessive colonization of AIEC, leading to a sustained immune response. 87 Some studies have also suggested that Mycobacterium paratuberculosis, Helicobacter pylori, Myxobacterium, Desulfovibrio, and Porphyromonas may play a role in promoting the pathogenesis of IBD, but there is still a lack of sufficient experimental evidence. 88 , 89
Fusobacterium nucleatum targeted by NOD2 in colonic epithelial cells' upregulation of CARD3 activates the IL‐17F/NF‐κB signaling pathway, promotes intestinal inflammation, and causes relapse in patients. 90 On the contrary, Lactobacillus can reduce the receptors of pro‐inflammatory factors, and reduce the expression of IL‐6, TNF‐α,and Bcl‐2 family proteins, thereby reducing the inflammatory response. 91 Recent studies have indicated that AIEC bacteria can invade epithelial cells and induce TNF‐α to activate downstream signals of the T‐like receptor transduction pathway, thereby activating NF‐κB to initiate abnormal immune responses, through the adhesion of CEACAM‐6 (carcinoembryonican‐tigen associated cell adhesion molecule‐6) on the mucosa. 92 This abnormal immune response leads to changes in the integrity of the intestinal mucus‐epithelial barrier, which in turn enhances IBD susceptibility. Intestinal microbiota imbalance reduces mucus secretion and exposes the intestinal epithelium to damage by intestinal toxic metabolites. However, animal experiments have shown that germ‐free mice have reduced plasma cells, lymphoid follicles, T cells, and Paneth cells, resulting in mucosal damage. IgA was also reduced, and IgG‐producing lymphocytes in the intestinal mucosa of IBD patients significantly increased compared with controls. 93 It indicates that symbiotic microbiota can also increase anti‐inflammatory factors, and symbiotic microbiota can promote 5‐fluorouracil‐induced mucus secretion in rat goblet cells, constituting an important part of the mucus‐epithelial barrier. 94 The occurrence of abnormal immune responses and the destruction of the mucus‐epithelial barrier are the two core links of the injury mechanism.
Recently, SARS‐CoV‐2 is known to initially affect the respiratory system, but studies have found that it may lead to multiorgan dysfunction syndrome (MODS); a study found after a patient was infected with the virus, he developed symptoms of IBD. 95 The binding of SARS‐CoV‐2 to angiotensin‐converting enzyme receptors 2 (ACE2) hinders the absorption of nutrients, disrupts the intestinal barrier, leads to an imbalance in the microbiota of the gut microbiome, and causes symptoms of intestinal inflammation. COVID‐19‐related changes were found in the gut microbiota. Wei et al. observed that viral RNA was detected in feces in 69% of patients with diarrhea infected with COVID‐19, although only 17% of patients with diarrhea developed respiratory symptoms. 96 At present, studies on mouse colon showed that the Bacteroidetes species can regulate ACE2 expression, and that changes in Firmicutes species can also cause variable ACE2 expression. It has been found that after respiratory symptoms have subsided for a period of time, the virus can still be detected in the feces of COVID‐19 patients. It is speculated that the gastrointestinal tract may be one of the sites for virus replication.
The researchers found that nearly half of the feces of COVID‐19 infected patients were positive, and positive patients contained more high abundance of opportunistic bacteria, including Collinsella aerofaciens, Collinsella tanakaei, Streptococcus infantis, and Morganella morganii. Colonization of Coprobacillus, Clostridium ramosum, and Clostridium hathewayi is positively associated with COVID‐19 patient severity. 97 The increase in opportunistic bacteria is more common in critically ill patients and is associated with poor prognosis. 98 In vulnerable populations, the immune cascade generated in the short term by COVID‐19, “cytokine storm syndrome” (CRS), that mechanism possible involves that butyrate increases mucin production by goblet cells, thereby activating regulatory T cells, producing cytokines, resulting in increased ACE2 expression, enhancing SARS‐CoV‐2 in the intestine, and results in replication in the gut leading to intestinal dysbiosis and further fecal‐oral transmission. 99 Viral infection may reflect a potentially dysregulated microbial homeostasis,as discussed earlier, colitis symptoms in COVID‐19 patients may be caused by the destruction of the intestinal mucosal layer and thus the intestinal microecological imbalance, and the reduced microbial diversity aggravates the inflammatory response.
The pathogenic role of fungi in inflammation is not well understood; a study of adult IBD patients revealed clear differences in the relative abundance of specific fungi between IBD patients and healthy individuals; especially in CD patients, fungal expansion is more conducive than bacterial expansion, but the causality is not yet clarified, and further research is needed to clarify the reasons. 62
The intestinal microbiota plays both pro‐inflammatory and anti‐inflammatory roles in the pathogenesis of IBD, and there is no sufficient evidence for a causal relationship between intestinal homeostasis and IBD. In conclusion, the gut microbial homeostasis or imbalanced state plays an important role in the pathophysiological role of IBD, mainly by protecting or attacking the mucus‐epithelial barrier and affecting the immune system through commensal bacteria and metabolites.
3. REGULATION OF THE INTESTINAL MICROBIOTA TO TREAT IBD
The interaction between the intestinal microbiota and the gastrointestinal tract of the host, as well as its role in the occurrence and development of gastrointestinal diseases, provides potential therapeutic targets for the prevention and treatment of gastrointestinal diseases. 100 Clinically commonly used drugs from different directions to treat IBD. Developed in 1940, sulfasalazine is decomposed by the bacterial nitrate reductase in the colon and is used for the treatment of IBD and rheumatoid arthritis. 101 5‐Aminosalicylic acid (ASA) is the active part of sulfasalazine's therapeutic action as it enhances the expression of mucosal proteins, but 5‐ASA is considered a topical treatment drug, which is more effective in treating mild to moderate UC than other disease sites and ulcerseverity. 102 The most common side effects in ASA treatment are gastrointestinal symptoms, including epigastric pain, nausea, and diarrhea. 103 Azithromycin and metronidazole are cell‐permeable and potent activators of T‐cell apoptosis, treating IBD primarily through immunomodulatory effects. 104 Rifaximin is a non‐systemic bactericidal antibiotic that currently relieves symptoms in CD patients, but is ineffective against translocated bacteria. 105 Its current mechanism of action still needs to be further explored, and the side effects are obvious. In addition to the aforementioned therapeutic drugs, the following ways can be used to treat IBD.
3.1. Probiotics
Probiotics and prebiotics are substances that can be selectively utilized by the host's microbiota and converted into substances beneficial to the host's health. 106 , 107 The American Gastroenterological Association recommends that probiotics be used to improve functional gastrointestinal symptoms in IBD. 108 Multiple randomized, placebo‐controlled clinical trials provide partial evidence for the role of probiotics in relieving clinical symptoms in patients with IBD. 109 , 110 , 111 Szajewska et al. performed randomized controlled trials (RCT, N = 4208) in children with diarrhea with secondary to acute gastroenteritis, that those children received rhamnosus infection compared to placebo or no treatment,the clinical level of diarrhea score did not decrease, but the duration of diarrhea was significantly shorter. 112 Bifidobacterium dentium regulates the glycosylation of the intestinal mucus layer, which has a unique role in regulating mucin production and can act as a mucinogenic agent. B. dentium can increase the expression of mucin genes and MUC2 protein, and its secreted metabolite acetate is also able to increase MUC2 protein expression in vitro. 113
In a study prebiotics and synbiotics significantly reduced disease activity index in patients with active UC, and probiotic supplementation could increase the quantity of Bifidobacteria in the IBD patients. 114 A controlled trial from Cochrane, including 22 eligible RCTs, found that probiotics were effective in preventing recurrence of quiescent UC. 115 Probiotic effector molecules, including fimbriae, lipoteichoic acid, exopolysaccharides and various surface proteins, alter the gastrointestinal microenvironment by reducing pH, and competing with pathogenic bacteria for nutrients and binding sites. 116 Wang et al. found a gene in Lactocaccus lactis I‐1631 that is specifically involved in aerobic respiration in fermented dairy products, and in the T‐bet(−/−)Rag2(−/−) model, in L. lactis I‐1631 the sodA gene can be made not to express. 117
3.2. Fecal microbiota transplantation
Fecal microbiota transplantation (FMT) involves transferring a fecal sample from a healthy donor to a recipient, either by various transplant modalities such as endoscopy, rectal enema, nasogastric/nasoenteric tube (into the upper stomach/small intestine) or by mouth. Ingestion of capsules containing fecal matter restores the microbial environment. 118 American Gastroenterologinal Association (AGA) suggest that FMT mainly improves mucosal inflammation rather than functional gastrointestinal symptoms. 119 , 120
FMT is especially good at treating refractory CDI, and studies have shown that the efficacy of FMT in a single treatment of CDI can be up to 75%–90%; the efficiency rate is higher after multiple FMT treatments. 121 , 122 Clinical and experimental evidence also shows that the FMT treatment relieves symptoms in IBD patients. The results of double‐blind trials show that after the FMT treatment, the diversity of gut microbes in UC patients is increased, and the gut of patients is enriched with Eubacterium and Roseburia inulivorans. 123 , 124 The meta‐analysis found that for patients with CDI and IBD, the cure rate was 81%, and the overall cure rate was 89% (95% CI = 83%–93%). 125 FMT performed through gastroscope to the distal duodenum can significantly improve abdominal distension symptoms in patients with IBS. 126
The cure rate of FMT is lower than that of other methods of surgery in randomized trials. 127 , 128 However, Lima et al. believed that the choice of FMT transplantation method has a direct impact on the cure rate and recurrence rate. Colonoscopy or nasojejunal tube to transplant fresh or frozen feces is better than fecal capsules and enemas. The researchers used UC patient‐derived strains colonized germ‐free or genetically engineered mice, and metagenomic analysis found that the transferable strains could promote metabolism and participate in mucosal immunity. The induction of mucosal regulatory T cells and the strains of Odoribacter splanchnicus colonization resulted in an increase in Foxp3/RORγt tregs induction of IL‐10, and production of SCFAs, repairing of the host mucosal and epithelial barrier integrity, found in the gut of FMT mice. 129
Yet there is substantial evidence that FMT is effective as adjunctive therapy. 130 However, some randomized controlled studies have not found FMT as significantly effective as it lacks a treatment‐related mechanism. 131 , 132
3.3. Diet intervention
Whether diet intervention can affect the structure and abundance of host intestinal microbiota species is unknown, but studies have shown that symptoms in IBD patients are triggered by some specific dietary conditions. For example, polyglycans have been observed to exacerbate inflammatory symptoms in inactive IBD patients without abdominal pain, diarrhea. 133 , 134 Dietary fiber is metabolized in the gut to SCFAs, resulting in the subsequent activation of GPCR and Treg, and resulting in a reduced inflammatory response and increased mucosal tolerance, thereby protecting CD patients. 135 , 136 In addition, the interaction between gut microbiota and dietary fiber and protein concentration alleviates inflammation and is also a factor that improves intestinal permeability in mice. 137
Multiple randomized controlled trials have shown that a low‐FODMAP dietary intervention can adequately relieve symptoms in patients with IBS, accompanied by decreased abundance of Bifidobacterium adolescentis, Bifidobacterium longum, F. prausnitzii, and decreased fecal butyrate levels. 138 , 139 In a crossover trial of patients with remission or mild UC, researchers found that a targeted intake of either a low‐fat diet or a high‐fiber diet for 4 weeks significantly improved quality of life, a low‐fat diet significantly improved dysregulated gut microbiota. 140 Studies based on animal models of IBD have found that hydrolyzed protein diets regulate bacterial abundance and metabolism, and their mechanism of action reduces the abundance of pathogenic bacteria (e.g., E. coli, C. perfringens), and the abundance of secondary bile acid–producing bacteria Clostridium hiranonis, increased associated with elevated levels of secondary bile acids (lithocholic acid, deoxycholic acid). 141 Based on individual differences in the composition and function of the baseline microbiota, therefore, individualized dietary interventions of patients should be considered. 142
3.4. Traditional Chinese medicine
The active ingredients of traditional Chinese medicine (TCM) have a two‐way regulating effect on intestinal homeostasis. Some active ingredients contribute to the homeostasis of intestinal microecology, while others inhibit the growth of probiotics and destroy the homeostasis of intestinal microorganisms. Liu et al. found that the main components of Kuijieyuan decoction (KD) include emodin, and KD intervention increased the proportion of Alloprevotella, Treponema, Prevotellaceae, and Prevotella. In the experimental UC model, KD, through regulating the intestinal microbiota and reducing the level of TLR‐dependent phosphorylated PI3K/AKT/NF‐κB, protected the colonic mucosa. 143 Emodin from Polygonum multiflorum can also protect the colonic mucosal barrier through increases in the mRNA and protein expression of the VDR and its downstream molecules, Nrf2 and HO‐1. 144 Although these experiments demonstrate the protective effect of emodin, Cheng et al. have found that its long‐term administration produces toxic metabolites, because colon microorganisms convert rhubarb anthraquinones to rhein, which accumulates over time and causes melanosis coli to increase the risk of colon cancer. 145 Shao et al. indicated that Sophora flavescens Aiton EtOAc extract (SFE) inhibits oxidative stress and immune inflammation in UC mice, mainly manifested as linoleic acid, arachidonic acid, and fatty acid metabolism recovery, and reduces the expression of related inflammatory markers. SFEs can construct a bacterial metabolite co‐expression network to reduce mucosal damage, suppress immune inflammation, and reduce oxidative stress in UC mice by enriching Lactobacillus, Roseburia, norank_f_Muribaculaceae, Anaerotruncus, Candidatus Saccharimona, and Parasutterella. 146 Purple sweet potato anthocyanin extract (PSPAE) is also able to maintain intestinal homeostasis and reduce bacterial intestinal inflammation by modulating gut microbiota in mice with dextran sodium sulfate (DSS)‐induced chronic colitis. 147 A randomized controlled clinical study demonstrated that after Ulmus macrocarpa Hance was administered, the number of Eubacterium ventriosum, Blautia faecis, and Ruminococcus gnavus in feces increased, and the ability of patients to synthesize bile acids improved. Maintaining bile acid homeostasis can protect the microbial structure and improve immunity. 148
Experiments have confirmed that the intestinal microbiota is also involved in the mechanism of Chinese herbal extracts to keep the intestinal barrier intact. Four flavonoid extracts and three astragalosides from Bunge‐Salisb (ACE) astragalus mongolica inhibit tumor growth and metastasis by modulating intestinal microbiota, inhibiting the overgrowth of opportunistic pathogens, and protecting the colonization of probiotics such as Prevotellacea. In particular, ACE by mediating the intestinal SDF‐1/CXCR4 signaling pathway through the microbiota‐metabolic network it can futher repair the integrity of the intestinal barrier and by reduce the expression of Cyclin D1 and C‐myc, increasing the level of propionic acid and butyric acid. 149 Extract leaves (EL), 150 Bilobalide (BI), 151 Rhodiola crenulata extract (RCE), 152 ginger extract, 153 ethanol extract of Ganoderma lucidum (GL95), 154 flavonoid extracts of Smilax glabra Roxb one of the functions of these extracts is to significantly increase the diversity and richness of the gut microbiota and reverse gut dysbiosis. 155 Gallic acid can regulate the ratio of Firmicutes and Proteobacteria in the gut microbiota of DSS‐UC mice to induce metabolic changes; the main focus is on increasing carbohydrate metabolism (glucose‐related metabolism) and bile acid (BA) metabolism and reducing amino acid metabolism, thereby restoring intestinal microbial homeostasis. High‐fat diet (HFD)‐induced gut dysbiosis is thought to alter gene expression by improving Firmicutes/Bacteroides abundance using the flavonoid extract from Smilax glabra Roxb. 156 Researchers also observed that sugar molecules and ginsenosides in the water extract of ginseng (WEG) can act as energy substrates for specific gut bacteria. Thus beneficially modulating the gut microbiota, remodeled gut microbial ecosystem subsequently triggers multiple molecular and cellular signaling pathways (such as butyrate or TGR5 signaling). 157 Danggui Shaoyao powder is a TCM formula, which has been used for the treatment of Alzheimer's disease. The relative abundance of beneficial bacteria Akkermansia was upregulated, and the pathogenic bacteria Lactobacillus and Erysipelotrichaceae were downregulated in rat feces after the intervention. 158
In conclusion, due to the incomplete and sufficient evidence, the efficacy and adverse effects of probiotics, FMT, dietary intervention, and TCM treatment are still controversial. Although probiotics do not significantly improve the severity of symptoms or prevent diarrhea recurrence, they can help the host to increase the number of beneficial bacteria, and their effectors can participate in the recovery of intestinal mucosal barrier integrity. Dietary intervention is particularly suitable for IBD caused by bad diet habits, such as high‐fat diet (HFB), which can directly alter gut microbes and metabolites. TCM extracts and compounds can repair the intestinal epithelial‐mucosal barrier and restore the intestinal microbial balance by activating multiple immune‐related signaling pathways. In the future, more rigorous randomized or double‐blind clinical experiments should be conducted to reduce individual differences. Although the above evidence shows that these therapies can be used to alleviate and treat the symptoms of IBD, there are still potential mechanisms of action that need further studies to elucidate.
4. CONCLUSION
The relationship between intestinal microbiota and disease is constantly being studied, but it still faces uncharted territory. An immediate question is about the causal relationship between intestinal microbiota and disease, which requires repeated validation in germ‐free animals, disease models, and clinical evidence. Therapeutic strategies targeting the intestinal microbiota have shown great potential, although the best way to “precisely” modulate the intestinal microbiota is still in the research stage. There is no doubt that the modulation of gut microbes will be more widely used for overall health improvement and adjuvant therapy. With further research, the true meaning of gut dysbiosis with disease states can be better understood, and the intestinal microbiota homeostasis that promotes overall health will be better defined, which will facilitate patient recovery and have broadly positive effects on public health.
In healthy individuals, commensal microbes perform functions that are beneficial to host health (left), in which the gut microbiota produces short‐chain fatty acid (SCFA), mainly butyrate, that help to modulate the gut microbiome. By the intestinal epithelium, the dense and complete mucus layer composed of goblet cells and tight junction (TJ) proteins can effectively prevent bacteria from invading the epithelial layer. SCFAs can provide energy for epithelial cells and help maintain the overall intestinal barrier function. SCFAs can also regulate the differentiation of T cells to provide immune cells. In contrast, gut microbiota homeostasis is disrupted in IBD patients (right), infected with viruses such as SARA‐COV‐2, or microbiota dysbiosis due to changes in the dietary environment. Microbial diversity occurs and leads to decreased immunity, including the reduction in SCFA‐producing bacteria, and in turn leads to the reduction in SCFAs, which are required for the maintenance of intestinal barrier function. This also affects the differentiation of immune cells, causing the imbalance in Treg and Teff cells, producing more pro‐inflammatory cytokines, and aggravating the intestinal inflammatory damage. The mucosal layer is damaged, and therefore the intestinal barrier function is disrupted, resulting in intestinal inflammation.
AUTHOR CONTRIBUTIONS
JC and QHF wrote the manuscript. TY S and XQM contributed to revising the manuscript . All authors gave final approval for publication.
CONFLICT OF INTEREST
The authors declare that there are no conflicts of interest to disclose.
Supporting information
Appendix S1
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grant no. 81774449).
Fu Q, Song T, Ma X, Cui J. Research progress on the relationship between intestinal microecology and intestinal bowel disease. Anim Models Exp Med. 2022;5:297‐310. doi: 10.1002/ame2.12262
REFERENCES
- 1. Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci U S A. 1998;95:6578‐6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. O'Hara A, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688‐693. doi: 10.1038/sj.embor.7400731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837‐848. [DOI] [PubMed] [Google Scholar]
- 4. Lloréns‐Rico V, Raes J. Tracking humans and microbes. Nature. 2019;569:632‐633. [DOI] [PubMed] [Google Scholar]
- 5. Huttenhower C, Gevers D, Knight R, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207‐214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brave M, Lukin DJ, Mani S. Microbial control of intestinal innate immunity. Oncotarget. 2015;6:19962‐19963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Qin J, Li R, Raes J, Arumugam M. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59‐65. doi: 10.1038/nature08821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Watnick PI, Jugder BE. Microbial control of intestinal homeostasis via enteroendocrine cell innate immune signaling. Trends Microbiol. 2020;28(2):141‐149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhou X, Li W, Wang S, et al. YAP aggravates inflammatory bowel disease by regulating M1/M2 macrophage polarization and gut microbial homeostasis. Cell Rep. 2019;27:1176‐1189.e1175. [DOI] [PubMed] [Google Scholar]
- 10. Chen Y, Chaudhary N, Yang N, et al. Microbial symbionts regulate the primary Ig repertoire. J Exp Med. 2018;215:1397‐1415. doi: 10.1084/jem.20171761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van den Munckhof ICL, Kurilshikov A, ter Horst R, et al. Role of gut microbiota in chronic low‐grade inflammation as potential driver for atherosclerotic cardiovascular disease: a systematic review of human studies. Obes Rev. 2018;19:1719‐1734. doi: 10.1111/obr.12750 [DOI] [PubMed] [Google Scholar]
- 12. Gil‐Cruz C, Perez‐Shibayama C, de Martin A, et al. Microbiota‐derived peptide mimics drive lethal inflammatory cardiomyopathy. Science (New York, N.Y.). 2019;366:881‐886. doi: 10.1126/science.aav3487 [DOI] [PubMed] [Google Scholar]
- 13. Vallianou N, Stratigou T, Christodoulatos G, Dalamaga M. Understanding the role of the gut microbiome and microbial metabolites in obesity and obesity‐associated metabolic disorders: current evidence and perspectives. Curr Obes Rep. 2019;8:317‐332. doi: 10.1007/s13679-019-00352-2 [DOI] [PubMed] [Google Scholar]
- 14. Zheng Y, Ran Y, Zhang H, Wang B, Zhou L. The microbiome in autoimmune liver diseases: metagenomic and metabolomic changes. Front Physiol. 2021;12:715852. doi: 10.3389/fphys.2021.715852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen T, Qin Y, Chen M, et al. Gestational diabetes mellitus is associated with the neonatal gut microbiota and metabolome. BMC Med. 2021;19:120. doi: 10.1186/s12916-021-01991-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jeong Y, Kim JW, You HJ, Park SJ, Ji GE. Gut microbial composition and function are altered in patients with early rheumatoid arthritis. J Clin Med. 2019;8:693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goto Y, Kiyono H. Epithelial barrier: an interface for the cross‐communication between gut flora and immune system. Immunol Rev. 2015;245:147‐163. [DOI] [PubMed] [Google Scholar]
- 18. Birchenough GMH, Johansson MEV. Forming a mucus barrier along the colon. Science. 2020;370:402‐403. doi: 10.1126/science.abe7194 [DOI] [PubMed] [Google Scholar]
- 19. Nyström E, Martinez‐Abad B, Arike L, et al. An intercrypt subpopulation of goblet cells is essential for colonic mucus barrier function. Science (New York, N.Y.). 2021;372:eabb1590. doi: 10.1126/science.abb1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bergstrom K, Shan X, Casero D, et al. Proximal colon‐derived O‐glycosylated mucus encapsulates and modulates the microbiota. Science (New York, N.Y.). 2020;370:467‐472. doi: 10.1126/science.aay7367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Velcich A, Yang W, Heyer J, et al. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science. 2002;295:1726‐1729. [DOI] [PubMed] [Google Scholar]
- 22. Song ZM, Liu F, Chen YM, Liu YJ, Wang XD, du SY. CTGF‐mediated ERK signaling pathway influences the inflammatory factors and intestinal flora in ulcerative colitis. Biomed Pharmacother. 2019;111:1429‐1437. doi: 10.1016/j.biopha.2018.12.063 [DOI] [PubMed] [Google Scholar]
- 23. Rabizadeh S, Rhee K‐J, Wu S, et al. Enterotoxigenic bacteroides fragilis: a potential instigator of colitis. Inflamm Bowel Dis. 2007;13(12):1475‐1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Birchenough GM, Nyström EE, Johansson ME, Hansson GC. Innate immunity a sentinel goblet cell guards the colonic crypt by triggering Nlrp6‐dependent Muc2 secretion. Science. 2016;352(6293):1535‐1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Salzman NH, Ghosh D, Huttner KM, Paterson Y, Bevins CL. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature. 2003;422:522‐526. [DOI] [PubMed] [Google Scholar]
- 26. Kobayashi KS, Chamaillard M, Ogura Y, et al. Nod2‐dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731‐734. [DOI] [PubMed] [Google Scholar]
- 27. Kaser A, Lee AH, Franke A, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743‐756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Burger‐van Paassen N, Vincent A, Puiman PJ, et al. The regulation of intestinal mucin MUC2 expression by short‐chain fatty acids: implications for epithelial protection. Biochem J. 2009;420:211‐219. [DOI] [PubMed] [Google Scholar]
- 29. Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003‐1007. [DOI] [PubMed] [Google Scholar]
- 30. Johansson M, Hansson GC. Keeping bacteria at a distance. Science. 2011;334:182‐183. [DOI] [PubMed] [Google Scholar]
- 31. Parikh K, Antanaviciute A, Fawkner‐Corbett D, et al. Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature. 2019;567:49‐55. doi: 10.1038/s41586-019-0992-y [DOI] [PubMed] [Google Scholar]
- 32. Johansson ME, Hansson GC. Mucus and the goblet cell. Dig Dis. 2013;31:305‐309. doi: 10.1159/000354683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Corthésy B. Role of secretory immunoglobulin a and secretory component in the protection of mucosal surfaces. Future Microbiol. 2010;5:817‐829. doi: 10.2217/fmb.10.39 [DOI] [PubMed] [Google Scholar]
- 34. Hsu CC, Okumura R, Motooka D, et al. Alleviation of colonic inflammation by Lypd8 in a mouse model of inflammatory bowel disease. Int Immunol. 2021;33:359‐372. [DOI] [PubMed] [Google Scholar]
- 35. Okumura R, Kurakawa T, Nakano T, et al. Lypd8 promotes the segregation of flagellated microbiota and colonic epithelia. Nature. 2016;532:117‐121. [DOI] [PubMed] [Google Scholar]
- 36. Okumura R, Kodama T, Hsu CC, et al. Lypd8 inhibits attachment of pathogenic bacteria to colonic epithelia. Mucosal Immunol. 2020;13:75‐85. [DOI] [PubMed] [Google Scholar]
- 37. Zhang C, Hu Z, Lone AG, et al. Small proline‐rich proteins (SPRRs) are epidermally produced antimicrobial proteins that defend the cutaneous barrier by direct bacterial membrane disruption. eLife. 2022;11:e76729. doi: 10.7554/eLife.76729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hu Z, Zhang C, Sifuentes‐Dominguez L, et al. Small proline‐rich protein 2A is a gut bactericidal protein deployed during helminth infection. Science (New York, N.Y.). 2021;374:eabe6723. doi: 10.1126/science.abe6723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kim YH, Kim H. Clostridium difficiletoxin a upregulates Bak expression through PGE2 pathway in human colonocytes. J Microbiol Biotechnol. 2019;29:1675‐1681. [DOI] [PubMed] [Google Scholar]
- 40. Tao L, Tian S, Zhang J, Liu Z, Dong M. Sulfated glycosaminoglycans and low‐density lipoprotein receptor contribute to Clostridium difficile toxin a entry into cells. Nat Microbiol. 2019;4(1):1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Guillermin O, Angelis N, Sidor CM, Idgway RR, Thompson BJ. Wnt and Src signals converge on YAP‐TEAD to drive intestinal regeneration. EMBO J. 2021;40:e105770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Anderson DM, Sheedlo MJ, Jensen JL, Lacy DB. Structural insights into the transition of Clostridioides difficile binary toxin from prepore to pore. Nat Microbiol. 2020;5:102‐107. doi: 10.1038/s41564-019-0601-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lima BB, Fonseca BF, Amado N, Lima DM, Brito G. Clostridium difficiletoxin a attenuates Wnt/β‐catenin signaling in intestinal epithelial cells. Infect Immun. 2014;82:2680‐2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gill PA, Van Zelm MC, Muir JG, Gibson PR. Review article: short chain fatty acids as potential therapeutic agents in human gastrointestinal and inflammatory disorders. Aliment Pharmacol Ther. 2018;48:15‐34. [DOI] [PubMed] [Google Scholar]
- 45. Felmlee MA, Jones RS, Rodriguezcruz V, Follman KE, Morris ME. Monocarboxylate transporters (SLC16): function, regulation, and role in health and disease. Pharmacol Rev. 2020;72:466‐485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Eitel J, Suttorp N, Opitz B. Innate immune recognition and inflammasome activation in listeria monocytogenes infection. Front Microbiol. 2010;1:149. doi: 10.3389/fmicb.2010.00149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sturm A, Dignass A. Epithelial restitution and wound healing in inflammatory bowel disease. World J Gastroenterol. 2008;14:348‐353. doi: 10.3748/wjg.14.348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shao X, Sun S, Zhou Y, et al. Bacteroides fragilis restricts colitis‐associated cancer via negative regulation of the NLRP3 axis. Cancer Lett. 2021;523:170‐181. [DOI] [PubMed] [Google Scholar]
- 49. Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9:799‐809. [DOI] [PubMed] [Google Scholar]
- 50. Farquhar M, Palade G. Junctional complexes in various epithelia. J Cell Biol. 1963;17:375‐412. doi: 10.1083/jcb.17.2.375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rescigno M, Urbano M, Valzasina B, et al. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol. 2001;2:361‐367. doi: 10.1038/86373 [DOI] [PubMed] [Google Scholar]
- 52. Morampudi V, Graef FA, Stahl M, et al. Tricellular tight junction protein Tricellulin is targeted by the enteropathogenic Escherichia coli effector EspG1, leading to epithelial barrier disruption. Infect Immun. 2017;85:e00700‐16. doi: 10.1128/iai.00700-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Flamant M, Aubert P, Rolli‐Derkinderen M, et al. Enteric glia protect against shigella flexneri invasion in intestinal epithelial cells: a role for S‐nitrosoglutathione. Gut. 2011;60:473‐484. [DOI] [PubMed] [Google Scholar]
- 54. Fasano A, Fiorentini C, Donelli G, Uzzau S, Goldblum SE. Zonula occludens toxin modulates tight junctions through protein kinase C‐dependent Actin reorganization, in vitro. J Clin Investig. 1995;96:710‐720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Darnaud M, Dos Santos A, Gonzalez P, et al. Enteric delivery of regenerating family member 3 alpha alters the intestinal microbiota and controls inflammation in mice with colitis. Gastroenterology. 2017;154(4):1009‐1023.e14. [DOI] [PubMed] [Google Scholar]
- 56. Nam ST, Hwang JH, Kim DH, Lu LF, Kim H. NQO1‐knockout mice are highly sensitive to clostridium difficile toxin A‐induced enteritis. J Microbiol Biotechnol. 2016;26:1446‐1451. [DOI] [PubMed] [Google Scholar]
- 57. Wang J, Tian S, Yu H, et al. The response of colonic mucosa‐associated microbiota composition, mucosal immune homeostasis, and barrier function to early‐life galactooligosaccharides intervention in suckling piglets. J Agric Food Chem. 2018;67(2):578‐588. [DOI] [PubMed] [Google Scholar]
- 58. Shrestha A, Hendricks MR, Bomberger JM, Mcclane BA. Bystander host cell killing effects of Clostridium perfringens enterotoxin. mBio. 2016;7:e02015‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Saitoh Y, Suzuki H, Tani K, et al. Tight junctions. Structural insight into tight junction disassembly by Clostridium perfringens enterotoxin. Science. 2015;347:775‐778. [DOI] [PubMed] [Google Scholar]
- 60. Walther W, Petkov S, Kuvardina ON, Aumann J, Schlag PM. Novel Clostridium perfringens enterotoxin suicide gene therapy for selective treatment of claudin‐3‐ and −4‐overexpressing tumors. Gene Ther. 2012;19:494‐503. [DOI] [PubMed] [Google Scholar]
- 61. Papatheodorou P, Carette JE, Bell GW, et al. Lipolysis‐stimulated lipoprotein receptor (LSR) is the host receptor for the binary toxin Clostridium difficile transferase (CDT). Proc Natl Acad Sci U S A. 2011;108:16422‐16427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sokol H, Leducq V, Aschard H, et al. Fungal microbiota dysbiosis in IBD. Gut. 2017;66(6):1039‐1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Papatheodorou P, Hornuss D, Nölke T, et al. Clostridium difficile binary toxin CDT induces clustering of the lipolysis‐stimulated lipoprotein receptor into lipid rafts. mBio. 2013;4:e00244‐13. doi: 10.1128/mBio.00244-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Xavier R, Podolsky D. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427‐434. doi: 10.1038/nature06005 [DOI] [PubMed] [Google Scholar]
- 65. Lloyd‐Price J, Arze C, Ananthakrishnan AN, et al. Multi‐omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569:655‐662. doi: 10.1038/s41586-019-1237-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li D, Achkar JP, Haritunians T, et al. A pleiotropic missense variant in SLC39A8 is associated with Crohn's disease and human gut microbiome composition. Gastroenterology. 2016;151:724‐732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Seishima J, Iida N, Kitamura K, Yutani M, Kaneko S. Gut‐derived enterococcus faecium from ulcerative colitis patients promotes colitis in a genetically susceptible mouse host. Genome Biol. 2019;20:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Qian BF, Tonkonogy SL, Sartor RB. Aberrant innate immune responses in TLR‐ligand activated HLA‐B27 transgenic rat cells. Inflamm Bowel Dis. 2010;14:1358‐1365. [DOI] [PubMed] [Google Scholar]
- 69. Im E, Jung J, Pothoulakis C, Rhee S. Disruption of Pten speeds onset and increases severity of spontaneous colitis in Il10(−/−) mice. Gastroenterology. 2014;147:667‐679.e610. doi: 10.1053/j.gastro.2014.05.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sadlack B, Merz H, Schorle H, Schimpl A, Feller AC, Horak I. Ulcerative colitis‐like disease in mice with a disrupted interleukin‐2 gene. Cell. 1993;75:253‐261. [DOI] [PubMed] [Google Scholar]
- 71. Colman RJ, Tsai YT, Jackson K, et al. Achieving target infliximab drug concentrations improves blood and fecal neutrophil biomarkers in Crohn's disease. Inflamm Bowel Dis. 2021;27:1045‐1051. doi: 10.1093/ibd/izaa241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhao H, Xu H, Chen S, He J, Nie Y. Systematic review and meta||nalysis of the role of Faecalibacterium prausnitzii alteration in inflammatory bowel disease. J Gastroenterol Hepatol. 2020;36(2):320‐328. [DOI] [PubMed] [Google Scholar]
- 73. Khounlotham M, Kim W, Peatman E, et al. Compromised intestinal epithelial barrier induces adaptive immune compensation that protects from colitis. Immunity. 2012;37:563‐573. doi: 10.1016/j.immuni.2012.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Huang K, Mukherjee S, DesMarais V, et al. Targeting the PXR‐TLR4 signaling pathway to reduce intestinal inflammation in an experimental model of necrotizing enterocolitis. Pediatr Res. 2018;83:1031‐1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wlodarska M, Luo C, Kolde R, et al. Indoleacrylic acid produced by commensal Peptostreptococcus species suppresses inflammation. Cell Host Microbe. 2017;22:25‐37.e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ogilvie LA, Jones BV. Dysbiosis modulates capacity for bile acid modification in the gut microbiomes of patients with inflammatory bowel disease: a mechanism and marker of disease? Gut. 2012;61:1642‐1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Li J, Qiu H, Gong H, Tong W. Broad‐spectrum reactive oxygen species scavenging and activated macrophage‐targeting microparticles ameliorate inflammatory bowel disease. Biomacromolecules. 2021;22:3107‐3118. [DOI] [PubMed] [Google Scholar]
- 78. Main J, McKenzie H, Yeaman GR, et al. Antibody to Saccharomyces cerevisiae (bakers' yeast) in Crohn's disease. BMJ. 1988;297:1105‐1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Chiaro TR, Soto R, Zac Stephens W, et al. A member of the gut mycobiota modulates host purine metabolism exacerbating colitis in mice. Sci Transl Med. 2017;9:eaaf9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lavelle A, Sokol H. Gut microbiota‐derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2020;17:223‐237. doi: 10.1038/s41575-019-0258-z [DOI] [PubMed] [Google Scholar]
- 81. Lamas B, Richard ML, Leducq V, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med. 2016;22:598‐605. doi: 10.1038/nm.4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Machiels K, Joossens M, Sabino J, et al. A decrease of the butyrate‐producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2014;63:1275‐1283. [DOI] [PubMed] [Google Scholar]
- 83. Desreumaux P, Neut C, Barnich N, et al. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn's disease. Gastroenterology. 1998;115:1405‐1413. [DOI] [PubMed] [Google Scholar]
- 84. Ye Z, Zhang N, Wu C, et al. A metagenomic study of the gut microbiome in Behcet's disease. Microbiome. 2018;6:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Mourelle M, Salas A, Guarner F, Crespo E. Stimulation of transforming growth factor beta1 by enteric bacteria in the pathogenesis of rat intestinal fibrosis. Gastroenterology. 1998;114:519‐526. [DOI] [PubMed] [Google Scholar]
- 86. Barrios‐Villa E, Pea C, Lozano‐Zaraín P, Cevallos M, Rocha‐Gracia R. Comparative genomics of a subset of adherent/invasive Escherichia coli strains isolated from individuals without inflammatory bowel disease. Genomics. 2019;112:1813‐1820. [DOI] [PubMed] [Google Scholar]
- 87. Palmela C, Chevarin C, Xu Z, et al. Adherent‐invasive Escherichia coli in inflammatory bowel disease. Gut. 2018;67:574‐587. [DOI] [PubMed] [Google Scholar]
- 88. Gobert AP, Sagrestani G, Delmas E, et al. The human intestinal microbiota of constipated‐predominant irritable bowel syndrome patients exhibits anti‐inflammatory properties. Sci Rep. 2016;6:1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Cook R, Fulcher JA, Tobin NH, et al. Effects of HIV viremia on the gastrointestinal microbiome of young MSM. AIDS (London, England). 2019;33:793‐804. doi: 10.1097/qad.0000000000002132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Chen Y, Chen Y, Cao P, Su W, Dong W. Fusobacterium nucleatum facilitates ulcerative colitis through activating IL‐17F signaling to NF‐κB via the upregulation of CARD3 expression. J Pathol. 2020;250:170‐182. [DOI] [PubMed] [Google Scholar]
- 91. Brandtzaeg P, Halstensen TS, Kett K, et al. Immunobiology and immunopathology of human gut mucosa: humoral immunity and intraepithelial lymphocytes. Gastroenterology. 1989;97:1562‐1584. [DOI] [PubMed] [Google Scholar]
- 92. Barnich N, Carvalho FA, Glasser AL, et al. CEACAM6 acts as a receptor for adherent‐invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J Clin Investig. 2007;117:1566‐1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Baldassano RN, Schreiber S, Johnston RB, Fu RD, Macdermott RP. Crohn's disease monocytes are primed for accentuated release of toxic oxygen metabolites. Gastroenterology. 1993;105:60‐66. [DOI] [PubMed] [Google Scholar]
- 94. Prisciandaro LD, Geier MS, Butler RN, Cummins AG, Howarth GS. Probiotic factors partially improve parameters of 5‐fluorouracil‐induced intestinal mucositis in rats. Cancer Biol Ther. 2011;11:671‐677. [DOI] [PubMed] [Google Scholar]
- 95. Ricciuto A, Lamb CA, Benchimol EI, et al. Inflammatory bowel disease clinical activity is associated with COVID‐19 severity especially in younger patients. J Crohns Colitis. 2021;16(4):591‐600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Wei XS, Wang X, Niu YR, Ye LL, Zhou Q. Diarrhea is associated with prolonged symptoms and viral carriage in COVID‐19. Clin Gastroenterol Hepatol. 2020;18:1753‐1759.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Zuo T, Zhang F, Lui G, Yun KY, Ng SC. Alterations in gut microbiota of patients with COVID‐19 during time of hospitalization. Gastroenterology. 2020;159:944‐955.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Tang L, Gu S, Gong Y, Li B, Li L. Clinical significance of the correlation between changes in the major intestinal bacteria species and COVID‐19 severity. Engineering. 2020;6:1178‐1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Oliveira G, Oliveira C, Pinzan CF, Salis L, Cardoso C. Microbiota modulation of the gut‐lung Axis in COVID‐19. Front Immunol. 2021;12:635471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Guarner F, Malagelada J. Gut flora in health and disease. Lancet (London, England). 2003;361:512‐519. doi: 10.1016/s0140-6736(03)12489-0 [DOI] [PubMed] [Google Scholar]
- 101. Zheng H, Chen M, Li Y, et al. Modulation of gut microbiome composition and function in experimental colitis treated with sulfasalazine. Front Microbiol. 2017;8:1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Cevallos SA, Lee J, Velazquez EM, Foegeding NJ, Bumler AJ. 5‐Aminosalicylic acid ameliorates colitis and checks dysbiotic Escherichia coli expansion by activating PPAR‐γ signaling in the intestinal epithelium. mBio. 2021;12:e03227‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Carter F , Alsayb M , Marshall JK , Yuan Y. Mesalamine (5‐ASA) for the prevention of recurrent diverticulitis. Cochrane Database Syst Rev . 2017; 10: Cd009839 . doi: 10.1002/14651858.CD009839.pub2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Levine A, Kori M, Kierkus J, et al. Azithromycin and metronidazole versus metronidazole‐based therapy for the induction of remission in mild to moderate paediatric Crohn's disease: a randomised controlled trial. Gut. 2018;68(2):239‐247. [DOI] [PubMed] [Google Scholar]
- 105. Sartor RB. Review article: the potential mechanisms of action of rifaximin in the management of inflammatory bowel diseases. Aliment Pharmacol Ther. 2016;43:27‐36. [DOI] [PubMed] [Google Scholar]
- 106. Salminen S, Collado MC, Endo A, Hill C, Vinderola G. Publisher correction: the International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat Rev Gastroenterol Hepatol. 2021;18:649‐667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Swanson KS, Gibson GR, Hutkins RW, Reimer RA, Sanders ME. The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of synbiotics. Nat Rev Gastroenterol Hepatol. 2020;17:1‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Colombel JF, Shin A, Gibson PR. AGA clinical practice update on functional gastrointestinal symptoms in patients with inflammatory bowel disease: expert review. Clin Gastroenterol Hepatol. 2019;17:380‐390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Goldenberg JZ, Mertz D, Johnston BC, et al. Probiotics to prevent Clostridium difficile infection in patients receiving antibiotics. JAMA. 2018;320:499‐500. [DOI] [PubMed] [Google Scholar]
- 110. Zhang XF, Guan XX, Tang YJ, Sun JF, Fan JM. Clinical effects and gut microbiota changes of using probiotics, prebiotics or synbiotics in inflammatory bowel disease: a systematic review and meta‐analysis. Eur J Nutr. 2021;60:2855‐2875. [DOI] [PubMed] [Google Scholar]
- 111. Rufino MN, da Costa AL, Jorge EN, et al. Synbiotics improve clinical indicators of ulcerative colitis: systematic review with meta‐analysis. Nutr Rev. 2021;80:157‐164. [DOI] [PubMed] [Google Scholar]
- 112. Szajewska H, Kołodziej M, Gieruszczak‐Białek D, Skórka A, Ruszczyński M, Shamir R. Systematic review with meta‐analysis: lactobacillus rhamnosus GG for treating acute gastroenteritis in children—a 2019 update. Aliment Pharmacol Ther. 2019;49:1376‐1384. doi: 10.1111/apt.15267 [DOI] [PubMed] [Google Scholar]
- 113. Engevik MA, Luk B, Chang‐Graham AL, et al. Bifidobacterium dentiumfortifies the intestinal mucus layer via autophagy and calcium signaling pathways. mBio. 2019;10:e01087‐19. doi: 10.1128/mBio.01087-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Chen M, Feng Y, Liu W. Efficacy and safety of probiotics in the induction and maintenance of inflammatory bowel disease remission: a systematic review and meta‐analysis. Ann Palliat Med. 2021;10:11821‐11829. doi: 10.21037/apm-21-2996 [DOI] [PubMed] [Google Scholar]
- 115. Rooks MG, Veiga P, Wardwell‐Scott LH, et al. Gut microbiome composition and function in experimental colitis during active disease and treatment‐induced remission. ISME J. 2014;8:1403‐1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Zhai Q, Zhang Q, Tian F, Zhao J, Zhang H, Chen W. The synergistic effect of lactobacillus plantarum CCFM242 and zinc on ulcerative colitis through modulating intestinal homeostasis. Food Funct. 2019;10:6147‐6156. [DOI] [PubMed] [Google Scholar]
- 117. Wang G, Liu Y, Lu Z, et al. The ameliorative effect of a Lactobacillus strain with good adhesion ability against dextran sulfate sodium‐induced murine colitis. Food Funct. 2019;10(1):397‐409. [DOI] [PubMed] [Google Scholar]
- 118. Mullish B, Quraishi MN, Segal JP, et al. Clostridium difficilethe use of faecal microbiota transplant as treatment for recurrent or refractory infection and other potential indications: joint British Society of Gastroenterology (BSG) and Healthcare Infection Society (HIS) guidelines. Gut. 2018;67:1920‐1941. doi: 10.1136/gutjnl-2018-316818 [DOI] [PubMed] [Google Scholar]
- 119. Smits HH, Engering A, van der Kleij D, et al. Selective probiotic bacteria induce IL‐10‐producing regulatory T cells in vitro by modulating dendritic cell function through dendritic cell‐specific intercellular adhesion molecule 3‐grabbing nonintegrin. J Allergy Clin Immunol. 2005;115:1260‐1267. [DOI] [PubMed] [Google Scholar]
- 120. He X, Gao J, Peng L, Hu T, Cao H. Bacterial O‐GlcNAcase genes abundance decreases in ulcerative colitis patients and its administration ameliorates colitis in mice. Gut. 2020;70:1872‐1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Rao K, Malani PN. Diagnosis and treatment of Clostridioides (clostridium) difficile infection in adults in 2020. JAMA. 2020;323:1403‐1404. [DOI] [PubMed] [Google Scholar]
- 122. Johnson S, Lavergne V, Skinner AM, et al. Clinical practice guideline by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA): 2021 focused update guidelines on management of Clostridioides difficile infection in adults. Clin Infect Dis. 2021;73:e1029‐e1044. [DOI] [PubMed] [Google Scholar]
- 123. Hvas CL, Dahl Jørgensen SM, Jørgensen SP, et al. Fecal microbiota transplantation is superior to fidaxomicin for treatment of recurrent Clostridium difficile infection. Gastroenterology. 2019;156:1324‐1332.e3. [DOI] [PubMed] [Google Scholar]
- 124. Leonardi I, Paramsothy S, Doron I, Semon A, Iliev ID. Fungal trans‐kingdom dynamics linked to responsiveness to fecal microbiota transplantation (FMT) therapy in ulcerative colitis. Cell Host Microbe. 2020;27:823‐829.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Chen T, Zhou Q, Zhang D, et al. Effect of Faecal microbiota transplantation for treatment of Clostridium difficile infection in patients with inflammatory bowel disease: a systematic review and meta‐analysis of cohort studies. J Crohns Colitis. 2018;12:710‐717. [DOI] [PubMed] [Google Scholar]
- 126. Holvoet T, Joossens M, Vázquez‐Castellanos JF, et al. Fecal microbiota transplantation reduces symptoms in some patients with irritable bowel syndrome with predominant abdominal bloating: short‐ and long‐term results from a placebo‐controlled randomized trial. Gastroenterology. 2021;160:145‐157. [DOI] [PubMed] [Google Scholar]
- 127. Paramsothy S, Nielsen S, Kamm MA, et al. Specific bacteria and metabolites associated with response to fecal microbiota transplantation in patients with ulcerative colitis‐ScienceDirect. Gastroenterology. 2019;156:1440‐1454. [DOI] [PubMed] [Google Scholar]
- 128. Tariq R, Pardi DS, Bartlett MG, Khanna S. Low cure rates in controlled trials of fecal microbiota transplantation for recurrent Clostridium difficile infection: a systematic review and meta‐analysis. Clin Infect Dis. 2019;68:1351‐1358. [DOI] [PubMed] [Google Scholar]
- 129. Lima SF, Gogokhia L, Viladomiu M, et al. Transferable IgA‐coated odoribacter splanchnicus in responders to fecal microbiota transplantation for ulcerative colitis limits colonic inflammation. Gastroenterology. 2021;162(1):166‐178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Koretz RL. Probiotics in gastroenterology: how pro is the evidence in adults? Am J Gastroenterol. 2018;113:1125‐1136. [DOI] [PubMed] [Google Scholar]
- 131. Ooijevaar RE, Terveer EM, Verspaget HW, Kuijper EJ, Keller JJ. Clinical application and potential of fecal microbiota transplantation. Annu Rev Med. 2019;70:335‐351. [DOI] [PubMed] [Google Scholar]
- 132. Sokol H, Landman C, Seksik P, et al. Fecal microbiota transplantation to maintain remission in Crohn's disease: a pilot randomized controlled study. Microbiome. 2020;8:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Ruemmele FM, Dan T. Differences in the management of pediatric and adult onset ulcerative colitis—lessons from the joint ECCO and ESPGHAN consensus guidelines for the management of pediatric ulcerative colitis. J Crohns Colitis. 2014;8(1):1‐4. [DOI] [PubMed] [Google Scholar]
- 134. Cox SR, Clarke H, O'Keeffe M, et al. Nutrient, fibre, and FODMAP intakes and food‐related quality of life in patients with inflammatory bowel disease, and their relationship with gastrointestinal symptoms of differing aetiologies. J Crohns Colitis. 2021;12:2041‐2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Kovatcheva‐Datchary P, Shoaie S, Lee S, et al. Simplified intestinal microbiota to study microbe‐diet‐host interactions in a mouse model. Cell Rep. 2019;26:3772‐3783.e3776. doi: 10.1016/j.celrep.2019.02.090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Albenberg LG, Wu GD. Diet and the intestinal microbiome: associations, functions, and implications for health and disease. Gastroenterology. 2014;146:1564‐1572. doi: 10.1053/j.gastro.2014.01.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Llewellyn SR, Britton GJ, Contijoch EJ, et al. Interactions between diet and the intestinal microbiota Alter intestinal permeability and colitis severity in mice. Gastroenterology. 2018;154:1037‐1046.e1032. doi: 10.1053/j.gastro.2017.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Wilson B, Rossi M, Kanno T, et al. β‐Galactooligosaccharide in conjunction with low FODMAP diet improves irritable bowel syndrome symptoms but reduces fecal Bifidobacteria. Am J Gastroenterol. 2020;115(6):906‐915. [DOI] [PubMed] [Google Scholar]
- 139. Cox SR, Lindsay JO, Fromentin S, et al. Effects of low FODMAP diet on symptoms, fecal microbiome, and markers of inflammation in patients with quiescent inflammatory bowel disease in a randomized trial. Gastroenterology. 2020;158:176‐188. [DOI] [PubMed] [Google Scholar]
- 140. Viennois E, Gewirtz AT, Chassaing B. Connecting the dots: dietary fat, microbiota dysbiosis, altered metabolome and colon cancer. Gastroenterology. 2021;162(1):38‐39. [DOI] [PubMed] [Google Scholar]
- 141. Wang S, Martins R, Sullivan MC, et al. Diet‐induced remission in chronic enteropathy is associated with altered microbial community structure and synthesis of secondary bile acids. Microbiome. 2019;7:1‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Gill SK, Rossi M, Bajka B, Whelan K. Dietary fibre in gastrointestinal health and disease. Nat Rev Gastroenterol Hepatol. 2020;18:1‐16. [DOI] [PubMed] [Google Scholar]
- 143. Liu B, Piao X, Niu W, et al. Kuijieyuan decoction improved intestinal barrier injury of ulcerative colitis by affecting TLR4‐dependent PI3K/AKT/NF‐κB oxidative and inflammatory signaling and gut microbiota. Front Pharmacol. 2020;11:1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Shang L, Liu Y, Li J, Pan G, Zhou F, Yang S. Emodin protects sepsis associated damage to the intestinal mucosal barrier through the VDR/ Nrf2 /HO‐1 pathway. Front Pharmacol. 2021;12:724511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Cheng Y, Zhang H, Qu L, et al. Identification of rhein as the metabolite responsible for toxicity of rhubarb anthraquinones. Food Chem. 2020;331:127363. [DOI] [PubMed] [Google Scholar]
- 146. Shao J, Li Z, Gao Y, et al. Construction of a “bacteria‐metabolites” co‐expression network to clarify the anti–ulcerative colitis effect of flavonoids of Sophora flavescens Aiton by regulating the “host–microbe” interaction. Front Pharmacol. 2021;12:710052. doi: 10.3389/fphar.2021.710052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Sun J, Chen H, Kan J, Gou Y, Jin C. Anti‐inflammatory properties and gut microbiota modulation of an alkali‐soluble polysaccharide from purple sweet potato in DSS‐induced colitis mice. Int J Biol Macromol. 2020;153:708‐722. [DOI] [PubMed] [Google Scholar]
- 148. Kim K, Veerappan K, Woo N, et al. Ulmus macrocarpa Hance extract modulates intestinal microbiota in healthy adults: a randomized, placebo‐controlled clinical trial. J Microbiol (Seoul, Korea). 2021;59:1150‐1156. doi: 10.1007/s12275-021-1329-8 [DOI] [PubMed] [Google Scholar]
- 149. Gu J, Sun R, Wang Q, Liu F, Tang D, Chang X. Astragalus Mongholicus Standardized Bunge‐ Salisb. Extract efficiently suppresses colon cancer progression through gut microbiota modification in CT26‐bearing mice. Front Pharmacol. 2021;12:714322. doi: 10.3389/fphar.2021.714322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Zhai Z, Niu K, Liu Y, Lin C, Wu X. Eucommia ulmoides the gut microbiota‐bile acids‐TGR5 Axis mediates leaf extract alleviation of injury to colonic epithelium integrity. Front Microbiol. 2021;12:727681. doi: 10.3389/fmicb.2021.727681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Zhang H, Wang Y, Su Y, Fang X, Guo W. The alleviating effect and mechanism of bilobalide on ulcerative colitis. Food Funct. 2021;12:6226‐6239. doi: 10.1039/d1fo01266e [DOI] [PubMed] [Google Scholar]
- 152. Wang Y, Tao H, Huang H, et al. The dietary supplement Rhodiola crenulata extract alleviates dextran sulfate sodium‐induced colitis in mice through anti‐inflammation, mediating gut barrier integrity and reshaping the gut microbiome. Food Funct. 2021;12:3142‐3158. doi: 10.1039/d0fo03061a [DOI] [PubMed] [Google Scholar]
- 153. Ma Z, Wang HJ, Ma XJ, et al. Modulation of gut microbiota and intestinal barrier function during alleviation of antibiotic‐associated diarrhea with Rhizoma Zingiber officinale (ginger) extract. Food Funct. 2020;11:10839‐10851. doi: 10.1039/d0fo01536a [DOI] [PubMed] [Google Scholar]
- 154. Guo W, Pan YY, Li L, Li TT, Liu B, Lv XC. Ethanol extract of Ganoderma lucidum ameliorates lipid metabolic disorders and modulates the gut microbiota composition in high‐fat diet fed rats. Food Funct. 2018;9:3419‐3431. doi: 10.1039/c8fo00836a [DOI] [PubMed] [Google Scholar]
- 155. Bai Y, Wang SW, Wang XX, et al. The flavonoid‐rich Quzhou fructus Aurantii extract modulates gut microbiota and prevents obesity in high‐fat diet‐fed mice. Nutr Diabetes. 2019;9:30. doi: 10.1038/s41387-019-0097-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Li Y, Xie Z, Gao T, et al. A holistic view of gallic acid‐induced attenuation in colitis based on microbiome‐metabolomics analysis. Food Funct. 2019;10:4046‐4061. doi: 10.1039/c9fo00213h [DOI] [PubMed] [Google Scholar]
- 157. Zhou S, Zhou J, Xu JD, et al. Ginseng ameliorates exercise‐induced fatigue potentially by regulating the gut microbiota. Food Funct. 2021;12:3954‐3964. doi: 10.1039/d0fo03384g [DOI] [PubMed] [Google Scholar]
- 158. Yin J, Lu J, Lei P, et al. Danggui‐Shaoyao‐san improves gut microbia dysbiosis and hepatic lipid homeostasis in fructose‐fed rats. Front Pharmacol. 2021;12:671708. doi: 10.3389/fphar.2021.671708 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1


