Abstract
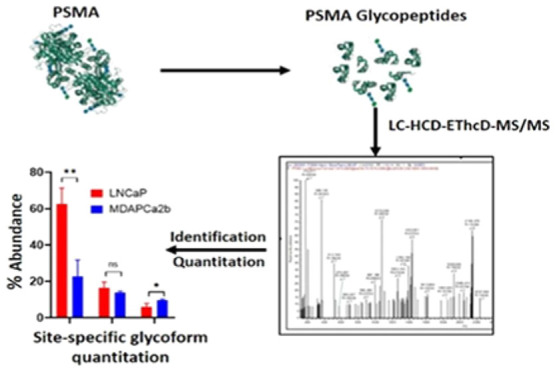
The composition of N-linked glycans that are conjugated to the prostate-specific membrane antigen (PSMA) and their functional significance in prostate cancer progression have not been fully characterized. PSMA was isolated from two metastatic prostate cancer cell lines, LNCaP and MDAPCa2b, which have different tissue tropism and localization. Isolated PSMA was trypsin-digested, and intact glycopeptides were subjected to LC-HCD-EThcD-MS/MS analysis on a Tribrid Orbitrap Fusion Lumos mass spectrometer. Differential qualitative and quantitative analysis of site-specific N-glycopeptides was performed using Byonic and Byologic software. Comparative quantitative analysis demonstrates that multiple glycopeptides at asparagine residues 51, 76, 121, 195, 336, 459, 476, and 638 were in significantly different abundance in the two cell lines (p < 0.05). Biochemical analysis using endoglycosidase treatment and lectin capture confirm the MS and site occupancy data. The data demonstrate the effectiveness of the strategy for comprehensive analysis of PSMA glycopeptides. This approach will form the basis of ongoing experiments to identify site-specific glycan changes in PSMA isolated from disease-stratified clinical samples to uncover targets that may be associated with disease progression and metastatic phenotypes.
1. Introduction
Prostate cancer (PCa) remains a significant public health problem in the United States and worldwide. In the USA, it is the third most common form of cancer. In 2022, it is estimated that there will be more than 268,490 new cases and 34,500 deaths from the disease in the country.1,2 Globally, it is responsible for 3.8% of all deaths caused by cancer in men.3 The clinical diversity of PCa is dramatic, ranging from asymptomatic indolent disease to aggressive metastatic and lethal forms.4,6 There is a limitation in the utility of currently used clinical risk stratification strategies, as they do not accurately discriminate aggressive from indolent disease.5 The consequence is the systemic over- and under-treatment that continues to occur in the clinical management of the disease. Therefore, there is a critical need for novel and effective biomarkers with high sensitivity and specificity for PCa diagnosis and prognosis.7,8 These markers will be vital in the emerging area of precision medicine for improved diagnosis, prognosis, and treatment options in patients. In addition to the assessment of genomic and proteomic biomarkers using biochemical approaches, there has been a significant increase in the application of magnetic resonance imaging for PCa diagnosis.9 This has been augmented with the availability of novel positron emission tomography (PET) tracers that target surface proteins and provide enhanced sensitivity to the process.9 There are ongoing efforts focusing on the development of novel markers for PCa diagnosis and treatment. The advent of precision medicine has spurred an increase in the development of theranostics, molecules with dual functions for diagnosis and treatment based on the conjugated radionuclide.10 Most of these proteins/molecules are cell surface proteins or small molecules that bind to their corresponding cell surface receptors that are translocated into targeted cells. These molecules are at the pinnacle of precision medicine, where cancer cells are uniquely targeted by imaging and therapeutics. The ultimate objective is to target cancer cells with the right therapeutic treatment based on their unique molecular characteristics, at the appropriate time and with the optimal dose.
The prostate-specific membrane antigen (PSMA), also known as folate hydrolase 1 (FOLH1) or glutamate carboxypeptidase 2, was previously studied as a PCa biomarker in tissues and body fluids using conventional biochemical methods with mixed results.11 The applied methodology has been significantly improved, and the molecule is a promising PCa theranostic for image-guided approaches.12,14 PSMA is not exclusively expressed in prostatic tumors; the protein is expressed in low abundance in prostate epithelium, in the neo-vasculature of exclusive solid tumor types, and in some healthy tissues including proximal renal tubes, duodenum, and ganglia of the nervous system.15 In PCa, PSMA abundance increases with disease severity, and up to 100-fold higher abundance has been observed in advanced aggressive forms of the disease compared to normal tissue.16,17 The PSMA receptor plays a critical role in PCa cell growth and proliferation.18 There are ongoing efforts and clinical trials to evaluate and develop novel PSMA-based applications for PCa diagnosis, prognosis, and treatment. Gallium 68 PSMA-11 (Ga 68 PSMA-11) was recently approved by the US FDA for PET imaging of PSMA positive PCa lesions in men.19,22
PSMA is a type II transmembrane glycoprotein that is expressed abundantly on the plasma membrane of PCa cells.13,20,21 Glycoproteins mediate and modulate multiple critical cellular processes. These include modulating cell adhesion, cell signaling, protein structure stabilization, and trafficking, in addition to processes and mechanisms that promote tumorigenesis. Aberrant protein glycosylation occurs in many cancers, where these changes impact progression, severity, and ultimately disease outcome.23−25 Carbohydrate domains comprise approximately 30% of the molecular mass of PSMA, and it is plausible that the high levels of N-linked glycosylation of the protein affects its structure and functions. In fact, Ghosh and Heston demonstrated that PSMA protein displays differences in overall glycosylation across cell lines and that deglycosylation enzymatically or by mutagenesis completely abolishes PSMA enzyme activity.26 Although there are 10 predicted N-linked glycosylation sites at asparagine residues at positions 51, 76, 121, 140, 153, 195, 336, 459, 476, and 638 of its primary amino acid sequence, the composition of N-linked glycans that are conjugated to PSMA has not been fully characterized. In addition, the potential role of glycoforms in disease progression and in the ongoing development of PSMA-based prognostic and diagnostic markers and therapeutics against PCa is unknown. We hypothesize that differential glycosylation of PSMA correlates with disease severity and that qualitative and quantitative changes in PSMA glycoforms occur as PCa progress and metastasize to other organs and sites. Workflows that enable site-specific identification and quantitation of PSMA glycans in disease-stratified PCa cells and clinical samples are needed to allow for discovery of glyco-markers that correlate with disease severity.
Here, we have utilized a liquid chromatography (LC)/mass spectrometry (MS)/MS strategy incorporating different, but complementary fragmentation methods [higher energy collision dissociation (HCD) and electron transfer dissociation higher energy collision dissociation (EThcD)] on a Tribrid Orbitrap Fusion Lumos instrument for site-specific N-linked glycan characterization of PSMA in two PCa cell lines with different metastatic-site localizations. We have taken the advantage of recent MS technological advancements27−37 and the development of integrated post-acquisition data processing algorithms, combined peptide, glycan search engines, and algorithms that allow for the detection and characterization of non-redundant intact N-glycopeptides.38−55 We hereby report the site-specific glycosylation profiles at eight out of ten predicted N-linked glycosylation sites of PSMA. We identified N-linked glycans conjugated to asparagine residues 51, 76, 121, 195, 336, 459, 476, and 638 of the eight glycopeptides with site-specific microheterogeneity in the glycoforms that are amenable to a global analysis strategy. PSMA from these PCa cells contain high-mannose, hybrid, and complex types of N-linked glycans. Some of the glycopeptides show significant qualitative and quantitative changes between the lymph node metastatic LNCaP cells and the bone metastatic MDAPCa2b cells. These glycoforms may form the basis for the development of prognostic markers for PCa disease stratification.
2. Experimental Procedures
2.1. Materials
Reagents were obtained from Thermo Fisher Scientific, unless otherwise stated. Trypsin was purchased from Promega (Madison, WI). Anti-human PSMA antibodies waere purchased from Cell Signaling Technologies Inc. [Danvers, MA (Catalogue #D4S1F, cs-12702)]. PNGase-F was from N-Zyme Scientifics (Doylestown, PA). Endo-H glycosidase and the lectins, Aleuria aurantia lectin (AAL), concanavalin A (ConA), wheat germ agglutinin (WGA), and Ulex europaeus agglutinin (UEA), were from Vector Laboratories Inc. (Burlingame, CA). Maackia amurensis lectin (MAA/MAL II) was from bioWorld (Dublin, OH).
2.2. PCa Cell Lines
Two PCa cell lines, LNCaP cells, clone FGC (CRL-1740) and MDAPCa2b (ATCC CRL-2422) were obtained from ATCC and used in the study. These two cell lines were selected because they expressed substantial amounts of PSMA whereas the other commonly used PCa cell lines, for example, PC3, DU145, and RWPE1, do not express the protein. In addition, other cell lines that express the proteins such as C4-2 and C4–2B were produced after orthotopic injection into castrated male mice and could possess phenotypic changes associated with their development in mice. LNCaP and MDAPCa2b cells are both androgen-dependent and undergo metastasis to different niches. LNCaP was isolated from a supraclavicular lymph node of a 50-year patient with metastatic PCa.56 MDAPCa2b was isolated from a bone metastasis site of a 63 year old man with androgen-independent PCa.57 The molecular characterization, expression, and mutation profiles of oncogenes and PCa progression markers of these cell lines have been previously comprehensively described.58 The cells were authenticated and confirmed to be devoid of mycoplasma contamination prior to culturing. The cells have similar doubling time (∼34 h), and they are routinely used in studying many aspects of prostate tumorigenesis. Cells with passage numbers below 20 were used in all the experiments. The origins and malignant tumorigenic characteristics of the lines that have been used in the study are summarized in Table S1. For each cell line, three independent sets of cell cultures were grown under standard conditions as we have previously described.59
2.3. PSMA Purification
Cultured cells were scrapped and lysed in RIPA buffer fortified with protease and phosphatase inhibitors. The concentrations of the lysates were determined using bicinchoninic acid assay. The lysates were stored at −80 °C until required for PSMA immunoprecipitation. To minimize non-specific binding to protein A/G beads and IgG, 500 μg of protein lysates were precleared by incubation with 35 μL of packed protein A/G agarose beads (Thermo Scientific) mixed with 2 μg of normal rabbit IgG for 12 h at 4 °C with gentle mixing. Protein A/G and antibody complex non-specific binding proteins were removed after pelleting by centrifugation. The soluble supernatant fractions were collected and used for PSMA immunoprecipitation. A total of 6 μg of anti-PSMA antibodies (cs-12702, Cell Signaling Technologies, Inc) was added per 500 μg of precleared protein lysates before incubation for a minimum of 12 h at 4 °C with gentle agitation. A total of 2 μg of non-specific normal rabbit IgG was mixed with 500 μg of lysates and incubated under similar conditions for negative control experiments. After 12 h, each sample was incubated with 35 μL of packed protein A/G agarose beads for 12 h on a rotatory platform at 4 °C. The beads were washed using RIPA buffer before eluting by heating the beads at 95 °C in 140 μL of Laemmli sample buffer for 10 min. We also used the same set of antibodies conjugated to cyanogen bromide sepharose beads and conformational epitope-specific antibody-affinity chromatography.60 The eluted proteins were fractionated in sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) gels that were used for western blot, colloidal Coomassie, and silver staining to visualize the proteins. These experiments were performed using lysates from three independent biological experiments as described in Section 2.2.
2.4. Trypsin Digestion
Coomassie-stained PSMA bands were processed for trypsin digestion as previously described.61 Protein bands were excised into small pieces <1 mm2. Gel pieces were destained in 50 mM ammonium bicarbonate in 50% acetonitrile before dehydration of the gel pieces in 100% acetonitrile. Reduction was performed by incubating the gels in 10 mM dithiothreitol in 50 mM ammonium bicarbonate at 56 °C for 60 min. The proteins were alkylated using 55 mM iodoacetamide in 50 mM ammonium bicarbonate for 45 min in the dark at room temperature. The reduced and alkylated protein were digested with trypsin at 37 °C for 18 h. Trypsin digestion generates eight glycopeptides with single N-linked glycan sites and a single peptide that contains two N-linked glycosylation sites. Peptides extraction was performed as described previously.58 The eluted peptides were dried using SpeedVac before enrichment of glycopeptides by affinity purification with hydrophilic interaction liquid chromatography (HILIC) TopTips (Glygen, Columbia, MD). Briefly, tip activation was achieved by washing three times with water. Activated tips were equilibrated using the binding buffer comprising 15 mM ammonium acetate, pH 3.5 in 85% acetonitrile. The peptide mixtures were dissolved in binding buffer, applied, and reloaded onto the tips three times to maximize binding. The bound peptides were washed three times with the binding buffer to remove non-glycopeptides. Elution of bound glycopeptides was carried out with 10 μL of water twice before drying them under vacuum. The dried peptides were resuspended in normalized volumes of buffer A comprising 5% acetonitrile and 0.1% formic acid (FA). The peptide concentrations were determined by spectrophotometry on a NanoDrop system before storage at −80 °C until ready for further analysis.
2.5. LC–MS/MS Analysis
PSMA glycopeptides were resuspended in 0.1% FA at a normalized concentration of 0.5 μg/μL and analyzed on an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher) connected to an EASY-nLC 1200 system. Peptides were injected into an Easy-Spray PepMap C18 pre-column (cat. # 164946 Thermo Fisher Scientific). The LC was coupled to a Thermo Scientific PepMap C18 analytic column (cat. # 164570 Thermo Fisher Scientific). Two micrograms of each sample was injected, and separation of peptides was performed using a binary-gradient solvent system consisting of buffer A [0.1% (v/v) FA in water] and buffer B [0.1% (v/v) FA in 80% acetonitrile]. The LC running conditions were as follows: flow rate 250 nL/min, isocratic 8% solvent B over 8 min, 8–12% solvent B over 5 min, 12–30% solvent B over 100 min, 30–60% solvent B over 20 min, 60–98% solvent B over 5 min, and isocratic 98% solvent B over 10 min. Every sample injection was followed by two column washes in the sequence runs. For intact glycopeptide analysis, we have utilized a high-resolution, high-sensitivity glycopeptide-specific method that incorporates the two scanning fragmentation methods HCD and EThcD.31−36 The instrument methods and scan parameters are summarized in Figure S1. In this method, the oxonium ions such as HexNAc, HexHexNAc, NeuAc, NeuAc-H2O, and HexNAcHexNeuAc with m/z 204.09, 366.13, 292.10 274.09, and 657.23, respectively) trigger the acquisition of ETD spectra. This retains the N-linked glycan on the asparagine peptide backbone whereas HCD provides peptide backbone sequence data. Differences between observed and theoretical precursors and product-ion masses for Orbitrap Fusion Lumos analyses were generally better than 2 ppm. The MS instrument was run automatically in an automatic data-dependent acquisition (DDA) acquisition mode switching between MS1 and MS2 scans. MS1 acquisitions with the m/z range 375–2000 were measured in the Orbitrap under the following conditions: resolution, 60,000; 4 × 105 automatic gain control (AGC); and 75 ms injection time. This was followed by oxonium-ion triggered MS2 EThcD MS/MS scans of glycopeptide precursor ions prioritized by the highest-charge states and intensity. The peptides were detected in the Orbitrap under the following conditions: 30,000 resolution, 3 × 105 AGC, and 200 ms injection time. EThcD scans were acquired under optimized conditions as reported previously.37 The three biological samples from the two cells were analyzed in triplicate technical replicate injections, generating a sum nine spectral data files for each cell line. The files have been deposited in the ProteomeXchange Consortium through the PRIDE repository (http://www.ebi.ac.uk/pride/archive/) with the identifier PXD033028.
2.6. LC–MS/MS Data Analysis
The MS raw data were used for integrated post-acquisition data processing using algorithms that combine peptide and glycan search engines for glycopeptide identification and characterization.38−46 The searches were performed against the UniProt human FOLH1 (PSMA) database (Q04609) using Byonic (Protein Metrics, San Carlos, CA). The human 132 N-glycan database from the Byonic human N-glycan database was included in the search. The searches were performed using the following parameters: (i) enzyme, trypsin with a maximum of two missed cleavages, (ii) static modification, carbamidomethylation of cysteine (C), and (iii) dynamic modifications, methionine (M) oxidation, asparagine deamidation of (N) and glutamine (Q), and N-linked glycan residues were used as variable modifications of asparagine (N) residues. The mass tolerance for the searches was set at 20 ppm for precursors and 20 ppm for product ions. The precursor peak picking was offset by 1 or 2 to allow for incorrect precursor monoisotopic selection in the analyses. The “normal” multicore option was enabled for the Byonic searches. The database search results were filtered at 1% FDR and confidence thresholds of the Byonic score >100. The spectra were inspected manually to confirm retention time consistency and for the presence of oxonium ions, for HexNAc, NeuAc, NeuAc-H2O, HexHexNAc, HexHexNAcFuc, and HexNAcHexNeuAc with m/z of 204.09, 292.10, 274.09, 366.13, 512.20, and 657.23, respectively. In the case of fucosylated and sialylated glycopeptides, monoisotopic peaks and corresponding diagnostic oxonium ions were closely inspected in the case of 2Fuc-1.02 = 1NeuAc to eliminate the false assignment of a sialic acid to the presence of two fucoses (NeuAc to 2Fuc). The peptides that contain the 10 PSMA N-linked glycosylation sites including eight that are within the m/z range for detection under these experimental conditions are shown in Table 1 below.
Table 1. Predicted PSMA N-Linked Glycopeptides after Complete Trypsin Digestiona.
| mass (MH2+) | position | peptide sequence | ASN-position |
|---|---|---|---|
| 581.2909 | 45–55 | SSNEATN*ITPK | 51 |
| 1290.6659 | 73–94 | FLYN*FTQIPHLAGTEQNFQLAK | 76 |
| 1205.1022 | 102–122 | EFGLDSVELAHYDVLLSYPN*K | 121 |
| 3278.0575 | 123–181 | THPNYISIINEDGNEIFN*TSLFEPPPPGYEN*VSDIVPPFSAFSPQGMPEGDLVYVNYAR | 140, 153 |
| 339.6656 | 194–199 | IN*C#SGK | 195 |
| 906.9494 | 325–341 | VPYNVGPGFTGN*FSTQK | 336 |
| 971.9789 | 446–463 | GVAYINADSSIEGN*YTLR | 459 |
| 945.9762 | 464–479 | VDC#TPLMYSLVHN*LTK | 476 |
| 455.2374 | 638–645 | N*FTEIASK | 638 |
The N-linked glycosylated asparagine residues are in bold and marked with asterisks (*) and carbamidomethylated cysteines with a number sign (#).
Comparative analysis to determine quantitative differences in expression of PSMA glycopeptides in the two PCa cells was performed using MS1 raw data and Byonic search results using Byologic (Protein Metrics Inc). All the MS raw files and their corresponding Byonic search results from the three independent cultures for the two cell lines and their triplicate technical experiments were uploaded into one project for comparative analysis. In these analyses, the peak area of the extracted ion chromatogram (XIC) of any glycopeptide was integrated and normalized against the sum of XICs of all the PSMA glycopeptides that are identified in each MS run. Site-specific abundance of glycoforms was determined by obtaining the sum of all glycopeptides containing the same glycan at the asparagine residue, where the peptide sequences may be different due to trypsin miscleavage or variable modifications. The relative quantitation for each of the eight site-specific glycopeptides was determined at the total glycopeptide and glycosite levels. Each sample was analyzed with triplicate injections, and the average normalized abundance of a given N-glycopeptide from the three replicates was used to determine its relative abundance between the two cell lines.
2.7. Lectin Affinity Purification and PSMA Detection
Two hundred micrograms of protein lysates was solubilized in lectin-binding buffer [20 mM TBS (pH 7.0), 1 mM CaCl2 chloride, 1 mM MgCl2, and 1 mM MnCl2] and incubated for 18 h at 4 °C with 100 μL of agarose beads conjugated with the lectins: AAL, ConA, WGA, MAA/MAL II, and UEA as we have described previously.62 The agarose beads were pelleted and washed three times with lectin-binding buffer. The bound proteins were released by solubilization in 100 μL of Laemmli buffer boiled for 10 min at 95 °C. The released proteins were fractionated in 4–12% Tris/bis SDS–PAGE gels followed by western blot detection of membranes with anti-PSMA rabbit monoclonal antibodies (D4S1F, Cell Signaling Technologies). PSMA bands were visualized using donkey anti-rabbit secondary IR-dye conjugated antibodies using an Odyssey CLx scanner (LI-COR Biosciences).
2.8. Endoglycosidase Treatment of PSMA
To validate site occupancy of PSMA N-linked glycans, immunoaffinity purified PSMA was digested with trypsin as described in Section 2.4 to generate peptides. Twenty micrograms of peptides was incubated with 2 μL of PNGase-F [N-Zyme Scientifics, concentration 106 Units/mL (2.0 mg/mL)] in the presence of 18O heavy water (H218O) and incubated for 18 h at 37 °C. This endoglycosidase cleaves the amide bond between the innermost N-acetylglucosamine (GlcNAc) and asparagine residues of all N-linked glycans including high-mannose, hybrid, and complex-type glycan glycoproteins and glycopeptides. The cleavage products are a deamidated protein or peptide and a free glycan. This reaction converts asparagines into aspartates. Since 18O heavy water is included, deamidation increases the mass of aspartic acid by 2.98 Da. Specific deamidation events and mass changes can be identified by high-resolution MS. In additional experiments, PSMA tryptic peptides were treated with endoglycosidase H (Endo H). Endo H cleaves the bond between the two N-acetylglucosamine subunits in N-linked glycans directly proximal to the asparagine residue of high-mannose and hybrid glycan chains. This results in the formation of a truncated sugar molecule with one N-acetylglucosamine residue being retained on the asparagine residue of the glycopeptide. The mass of the remaining N-acetylglucosamine residue (m/z 204.1) can be used for the identification of the modified asparagine residue in the glycopeptide.62,63
3. Results and Discussion
We have performed differential qualitative and quantitative characterization of intact N-glycopeptides of PSMA isolated from prostate cell lines with different distant metastatic localization sites. The objective was to identify differences in site-specific glycoforms of PSMA between LNCaP cells that were originally isolated from a supraclavicular lymph node metastatic site and MDAPCa2b cell lines that are bone-metastatic. The experimental workflow used to characterize PSMA glycoforms in this study is summarized in Figure 1.The origins and malignant characteristics of the two cell lines are summarized in Table S1. As demonstrated by western blot, the expression of PSMA varies across PCa cell lines representing different malignancy phenotypes (Figure 2). GAPDH was used as controls for the experiments. Our data demonstrate that some of the cell lines that are commonly used in PCa research including RWPE-1 and PC3 do not express significant amounts of PSMA and were therefore not included in the PSMA glycosylation characterization experiments. Our results reveal variability in glycosylation abundance that highlights the potential for glycoform expression differences between cell types.
Figure 1.
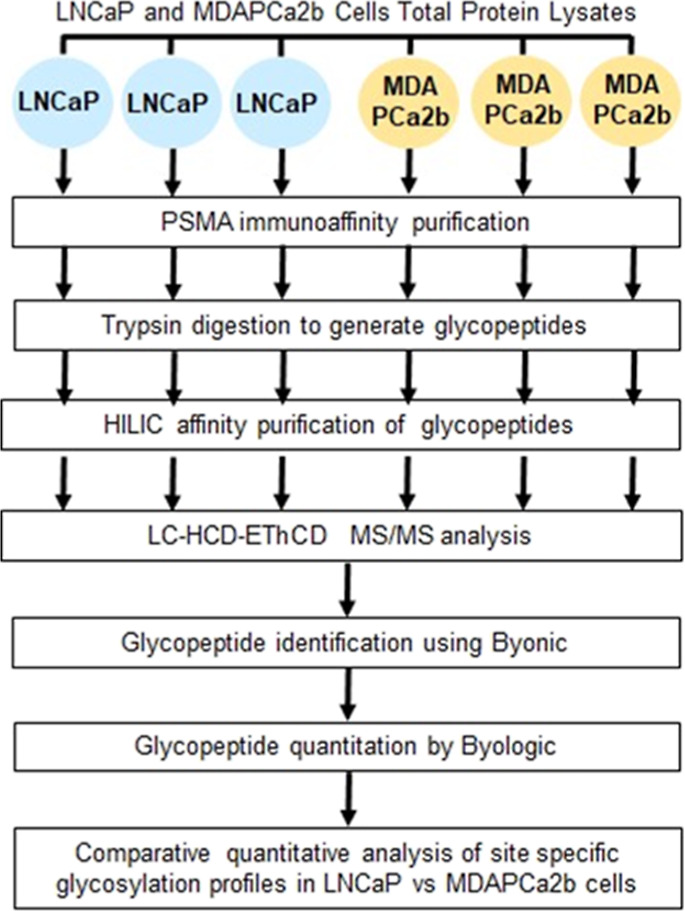
Schematic diagram of the experimental workflow. Summary of the workflow for qualitative and quantitative analysis of intact PSMA N-linked glycopeptides in PCa cells with different malignant phenotypes.
Figure 2.
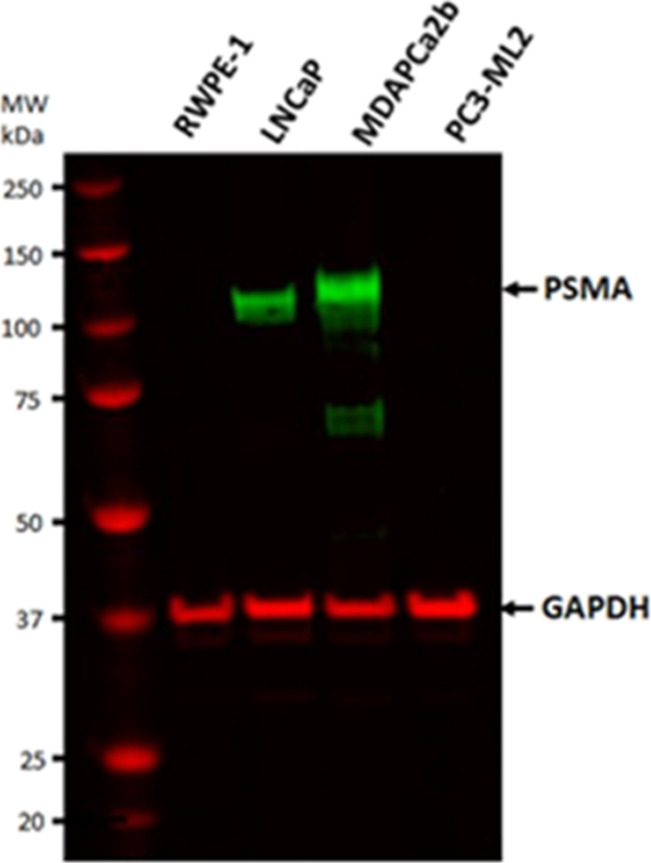
Western blot detection of PSMA expression in four PCa cell lines with different malignancy phenotypes. (A) PSMA was detected in LNCaP and MDAPCa2b but not in the RWPE-1 control lines and PC3-ML2 cell lines.
PSMA has 10 potential N-glycosylation sites at asparagine residues at positions 51, 76, 121, 140, 153, 195, 336, 459, 476, and 638 of its primary amino acid sequence. Digestion with trypsin generates nine PSMA glycopeptides. The identification and characterization of the peptides and glycan occupancy of PSMA has been previously carried out by using mutation analysis of transfected systems or using insect cells that allow for glycan analysis.63,64 The m/z and sequences are shown in Table 1. Eight peptides possess single glycosites; however, two sites ASN-140 and ASN-153 are contained in a single peptide with sequence THPNYISIINEDGNEIFN140TS LFEPPPPGYEN153VSDIVPPFSAFSPQGMPEGDLVYVNYAR. The mass of the peptide should be significantly higher with carbohydrates being conjugated to the two sites compromising the interpretation of the glycan composition in the chosen workflow. The option was to use chymotrypsin to generate peptides which are amenable to MS/MS glycopeptide characterization using standard conditions. This approach would allow for the determination of glycans at all the 10 sites independently. Chymotrypsin digestion should result in the generation of a short peptide with the sequence NTSLF (position 140–144) with ASN-140 with an MH2+ m/z of 291.1501 (without conjugating glycans) and the peptide containing ASN-153 with the sequence ENVSDIVPPF (position 152–161) and a MH2+ m/z of 558.7822 (without conjugating glycans). However, these peptides are not detected in our analyses using intact glycopeptides with and without HILIC purification and even after PNGase-F cleavage. In the case of the NTSLF peptide, the lack of detection could be due to its low m/z, which is below the scan range for the acquisition method (375–2000). In the case of the ENVSDIVPPF peptide, the presence of two prolines at y2 and y3 positions may have an effect on its fragmentation efficiency. Cleavage at proline residues frequently occurs from the N terminal to the residue; this results in the proline effect, with the generation of abundant b or y fragment ions from cleavage at the bond, with low-intensity ions at the rest of the residues. In additional experiments using Glu-C protease in bicarbonate buffer, theoretically, a short peptide with sequence IFNTSLFE (position 138–145) should be generated. This peptide was not detected in our analyses. Instead, the peptide with sequence LAHYDVLLSYPNKTHPNYISIINED GNEIFNTSLFE with two missed cleavage sites at Glu 133 and Glu 138 is detected at a very low frequency and with very poor fragmentation and identification scores. The ASN-140-containing site IFNTSLFE peptide is not identified in these analyses. The peptide with sequence PPPPGYENVSDIVPPFSAFSPQGMPE with the ASN-153 glycosylation site is also not identified in our analysis using intact glycopeptides with and without HILIC purification. These peptides are not detected even after PNGase-F cleavage to release their N-linked glycans. Therefore, the characterization of these two sites is not included in the current study.
3.1. PSMA N-Glycosylation Site Occupancy
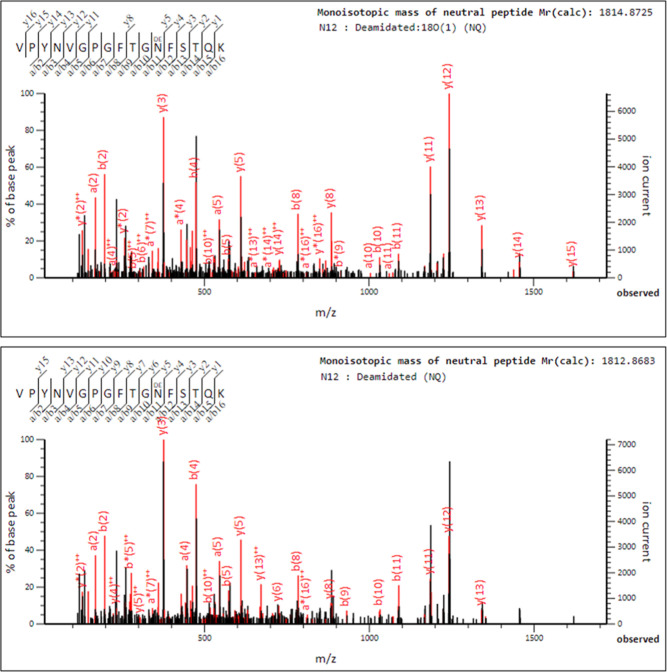
The site occupancy of PSMA N-linked glycans was confirmed after proteolytic digestion of affinity purified PSMA and treatment with PNGase-F in the presence of 18O heavy water (H218O). In this reaction, asparagines are deamidated into aspartic acid with an increase in mass of the aspartic acid by 2.98 Da. The mass shift is specific, reliable, and not subject to false positive misidentification due to overlap with artefactual deamidation events leading to a 0.98 Da mass shift that is commonly observed in asparagine residues in peptides due to hydrolysis during routine proteomics sample processing. Figure 3 shows a representative MS/MS fragmentation spectrum and fragmentation tables for the ASN-336 PSMA glycopeptide VPYNVGPGFTGN336FSTQK deamidation in normal 16O water (lower panel) and 18O water, confirming the N-linked glycosylation status of the asparagine residue (upper panel). Similar data were obtained for the other seven glycosylation sites.
Figure 3.
MS/MS Fragmentation spectrum for ASN-336 PSMA glycopeptides after PNGase-F cleavage of the glycans in the presence of light and heavy water. Spectrum showing b and y ions from the PSMA glycopeptide with sequence VPYNVGPGFTGN*FSTQK in the presence of 18O heavy water; the deamidation results in an increase in mass of the aspartic acid by 2.98 Da in the upper panel. A similar spectrum showing b and y ions after ASN-336 deamidation in the presence of normal 16O water with a mass increase of 0.98 Da of the aspartic acid is shown in the lower panel.
3.2. Identification of Intact N-Glycopeptides
Affinity purified site-specific PSMA N-linked glycopeptides of PSMA from the two cell lines were analyzed HCD and HCD-triggered EThcD MS/MS scans with triplicate sample injections. Examples of XICs MS/MS spectra with the HexNAc oxonium ion (HexNAc+) with the m/z range 204.08–204.09 and HexHexNAc with the m/z range 366.139–366.143 for MDAPCa2b are shown in Figure S2 for an ASN-336 glycopeptide with sequence VPYNVGPGFTGNFSTQK conjugated with the glycan FA4G4S2 (m/z 1224.7557 4+). To identify N-glycopeptides, Byonic and Byologic software (Versions 3.5; Protein Metrics Inc.) were used to extract tandem MS fragmentation data from HCD and EThCD scans from the raw files.64,65 The following search parameters were used for the identification and assignment of glycopeptides: precursor and product ions mass tolerance 20 ppm, with an FDR <1% and Byonic scores >100. The N-glycopeptide identifications were verified as true positives by manual inspection of the spectra to verify retention times and confirm the presence of the correct tandem mass spectrum ions (b/y and c/z) fragment ions, together with oxonium ions corresponding to the identified glycan including those for fucose and sialic acid to differentiate the assignments between NeuAc and 2Fuc.54,64,65 Positive glycan identifications were only included for further analysis upon identification in at least two of the three biological culture replicates. A recent study reported that glycopeptide assignments from Byonic includes spurious and more false positives than true positives.66 Most of the reported deficiencies were attributed to searches using MS data containing only CID spectra. In this study, all the MS data have been derived from HCD and EThcD fragmentation, and the filtering criteria that were used for Byonic searches that are described above include recommendations from Go et al.,66 and we are confident with the assignments and identifications that have been used for qualitative and quantitative analyses.
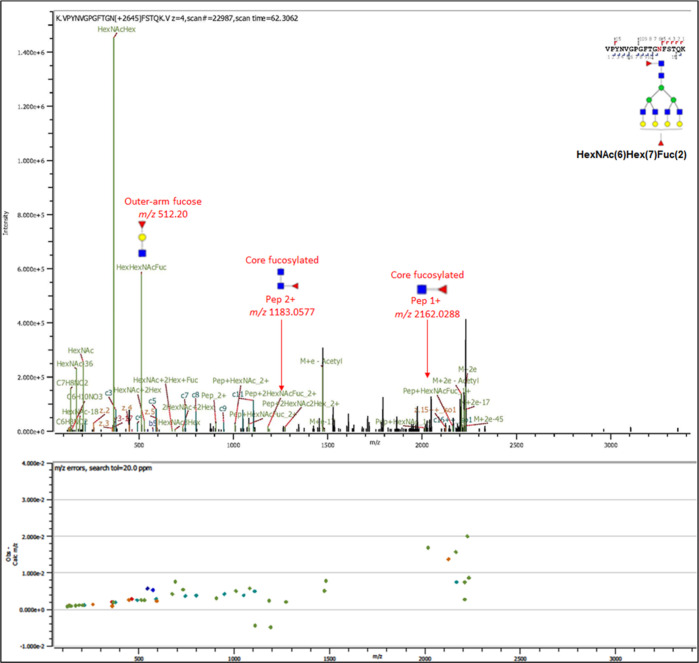
We utilized multiple fragmentation techniques including HCD and EThcD-MS/MS to generate composite glycan and peptide fragment ions in the same MS/MS spectra. The spectra comprise oxonium ions derived from the glycans, b/y and c/z ions from amino acid peptide backbones, and glycosidic fragments that include sequential monosaccharides residue losses from the intact precursor glycopeptide. HexNAc, HexHexNAc, NeuAc, NeuAc-H2O, and HexNAcHexNeuAc diagnostic oxonium ions with m/z 204.09, 366.13, m/z 292.10, m/z 274.09, and m/z 657.23, respectively, are identified at high intensities in the spectra. In addition, peptide backbone-derived b and y fragments are identified albeit at low intensities. For some of the glycopeptides, the glycosidic fragments conjugated with the peptide are also identified and annotated in the spectra (Figure 4). Taken together, these fragments were sufficient to identify the glycopeptides with detailed spectral characterization and annotation for both glycans and peptide. Further information is useful in the characterization of glycosidic linkages including core- and outer-arm fucosylation. Specific fragment ions were used to confirm the presence and type of fucosylation in some of the identified PSMA glycans. Outer-arm fucose was confirmed by the presence of the HexHexNAcFuc (m/z 512.20) oxonium fragment ion. The presence of core fucosylated residue was confirmed by the presence of the diagnostic ions of the 2 + charged peptide+2HexNAc + Fuc (m/z 1183.0577) and 1 + charged peptide + HexNAc + Fuc (m/z 2162.0288) for the ASN-336 peptide VPYNVGPGFTGN(2HexNAcFuc)FSTQK. Figure 4 shows an MS–MS spectrum for the 4 + charged glycopeptide from LNCaP cells with m/z 1115.2188 conjugated with a complex glycan with the following composition FA4G4F depicted as HexNAc(6)Hex(7)Fuc(2). The glycan contains both outer-arm (terminal) fucose and core fucose, and these ions are annotated in the spectrum. The high concordance in the accuracy of the experimental and reference masses showed by error plots of the spectra confirmed the accuracy and robustness of the assignments.
Figure 4.
MS/MS spectra of an intact PSMA N-glycopeptide with sequence VPNYVGPGFTGN*FSTQK and conjugated with the glycan FA4G4F [depicted as HexNAc(6)Hex(7)Fuc(2)] at ASN-336. Both b and y peptide fragment ions and oxonium ions include outer-arm terminal fucose fragment ions HexNAc(1)Hex(1)Fuc(1) with m/z 512.20 and composite fragment ions comprise the intact peptide and core fucosylated [HexNAc(2)Fuc(1) sugar moiety with m/z 1183.0577 (+2) and peptide + HexNAc + Fuc m/z 2162.0288 (+1)].
3.3. Site-Specific N-Glycosylation Microheterogeneity and Intact N-Glycopeptide Relative Quantitation
Relative abundance of specific PSMA glycopeptides in the two cell lines was quantitated using Byologic (Protein Metrics). In this automated process, the XIC peak area of a unique glycopeptide was integrated and normalized against the sum of abundance of all the identified PSMA glycopeptides to provide relative quantitation of the glycoform within each sample. The relative abundance of each glycan at each site was obtained by comparing XIC peak areas for the different glycans for peptides with identical amino acid sequences. For specific glycopeptides detected with multiple charge states, the XIC areas of these different glycans were summed according to the detected glycan composition. This information was used to calculate the relative abundance of each glycopeptide at each of the eight glycosites and the total glycopeptides at the respective sites.
The mean relative abundance of unique N-glycopeptides at a specific glycosylation site in the two cell lines was determined by normalizing the abundance of the glycopeptide against the sum of all glycopeptides of PSMA at the same site. The top five most abundant glycopeptides across the two cell lines were used for comparative analysis. The top five glycopeptides of each cell line were grouped to make a profile list for each glycosite. For these specific comparisons, the glycans that are used for comparisons were identified in at least two of the three biological replicates. For most of the glycosites, the sum of the relative abundances of the top five identified glycans comprise >90% of the total abundance of all the glycans that are identified for the site. These results are summarized for each of the targeted eight N-glycosylation sites, and individual comparisons for ASN-76 are shown in Figure 5. Similar data for the other seven N-linked glycosylation sites are shown in Figures S3A–G. These glycans and their structures are listed in Table S2. The compositions have been simplified using a four-abbreviation nomenclature to represent them in the following order HexNAc-Hex-Fuc-NeuAc (HexNAc representing N-acetylhexosamine, Hex for hexose, Fuc for fucose, and NeuAc for sialic acid) in column 2 of the table. The Oxford nomenclature for the glycans is provided in the third column, and the consortium for functional glycomics styled structures is provided in column 4. In the text, glycans are described as AxGxFxSx according to the Oxford glycan nomenclature, where x represents the number of the following structures that are present in the glycan, that is, A, antenna; G, galactose on antenna; F, fucose; S, sialic acid; and N,N-acetylglucosamine and with FA corresponding to core fucosylation. For example, FA3G3S2 represents a core-fucosylated triantennary trigalactosylated bisialylated glycan.
Figure 5.
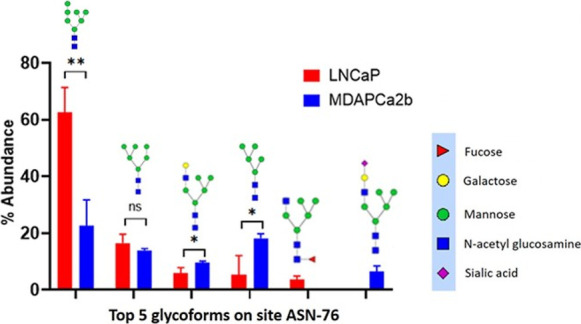
Site-specific comparative quantitative glycan analysis for the relative abundance of the five most abundant glycopeptides at the ASN-76 glycosylation site in LNCaP (red bars) and MAPCa2b cells (blue bars). Error bars show the standard deviations of the means across triplicate analyses from each cell line. Multiple unpaired t-tests were used to determine differences in abundance. There are glycans that are differentially expressed between the different cell lines. Man7: LNCaP versus MDAPCa2b, P = 0.0053**. M5G1: LNCaP versus MDAPCa2b, P = 0.034593*; Man5: LNCaP versus MDAPCa2b, P = 0.032044*; FM5G1 was exclusively detected in LNCaP cells whereas M5G1S1 was exclusively detected in MDAPCa2b cells.
For ASN-51, there was a significant reduction in glycosylation levels in both cell lines. This is consistent with the PNGase-F cleavage experiments that show the presence of both deamidated and non-deamidated asparagine residues for ASN-51. This could also provide the explanation for the low Byonic score that is observed for the site. In contrast, only deamidated asparagine residues are detected for the other seven sites. MS/MS spectra are shown for ASN-51, ASN-76, and ASN-336 in Figure S4A–D.
For ASN 76, the predominant glycopeptides that are identified in the two cell lines contain high-mannose structures. In the case of LNCaP cells, 84% of the top five identified peptides possess high-mannose glycans. In MDAPCa2b cells, 54% of the top five identified glycopeptides are conjugated with high-mannose structures. For the two cell lines, Man7, Man6, and Man5 structures are predominantly identified, and tandem mass spectra for ASN-76 glycopeptides are provided in Figure S5A–C. The MS/MS spectra for the ASN-76 sialylated M5G1S1 glycopeptide is shown in Figure S6. There are significantly different expression abundance levels at some of the sites between the two cells, and these are provided in the figure captions. ASN-121 contains a mixture of complex carbohydrate structures with sialylated and fucosylated glycans in addition to Man8 structures. The ASN-195 site is predominantly composed of fucosylated and sialylated structures. Importantly, among the most abundant glycans, fucosylated structures are exclusively detected in LNCaP cells, whereas in MDAPCa2b cells, the complex glycans possess both fucosylated and sialylated glycans. The ASN-336 site has the most significant heterogeneity and diversity in detected glycans. The detected glycans are mainly complex structures, and high mannoses are detected at extremely low abundance. The top five glycoforms comprising about 75% of the glycoforms at the site for LNCaP cells are composed of four main glycans that are found predominantly in the cell line. These include FA4G4F, FA4G4, FA2G4F2, and FA4G4F2S1. Core fucosylation was detected among the top five in both cell lines whereas the following sialylated complex glycans are the most abundant in MDAPCa2b cells: FA4G4S2, FA4G4S3, FA4G4F2S1, and FA4G4F2S2.
For ASN 459, the predominant glycopeptides that are identified in the two cell lines contain high-mannose structures. In the case of LNCaP cells, 83% of the top five identified peptides possess high-mannose glycans. In MDAPCa2b cells, 81% of the top five identified glycopeptides are conjugated with high-mannose structures. For the two cell lines, Man8, Man7, Man6, and Man5 structures are the predominant glycoforms. A similar situation is observed for ASN-476, where 96% of the top five identified peptides possess high-mannose glycans. In MDAPCa2b cells, 88% of the top five identified glycopeptides are conjugated with high-mannose structures that include Man9, Man8, Man7, and Man6. Finally, for ASN-638, the top five identified glycopeptides in LNCaP and MDAPCa2b cells exclusively possess high-mannose structures including Man8, Man7, Man6, Man5, and Man4.
A summary of the five most abundant glycans that are identified in the eight PSMA N-linked glycosylation sites that we have characterized is provided in Table S2. Among these glycans, in total 24 unique glycans are identified in LNCaP cells and 29 are identified in MDAPCa2b cells. Thirteen glycans are shared between the two cells, whereas 11 glycans are identified exclusively in LNCaP cells and 16 are identified exclusively in MDAPCa2b cells (Figure S7). In LNCaP cells, 10 out of the 11 glycans (91%) are fucosylated and only 2 (18%) possess sialic acid. In contrast, in MDAPCa2b cells, 11 out of 16 glycans (69%) are fucosylated and 13 out of 16 glycans (81%) possess sialic acid structures.
Previous studies to characterize the glycosylation pathways and identify PSMA glycans in PCa cell lines and in human-derived tissue samples have produced inconsistent results. Using exoglycosidase and endoglycosidase treatments, Holmes et al. concluded that in vivo-derived PSMA from tumor tissue or serum was primarily the N-linked complex-type, whereas, only high-mannose-type N-linked glycans are present on the PSMA from LNCaP cells.67 In subsequent studies, the recombinant baculovirus/insect cell system-expressed PSMA was determined to possess primarily N-linked high-mannose-type glycans.68 Barinka et al. confirmed that all predicted N-glycosylation sites are occupied with an oligosaccharide moiety, but the authors did not perform extensive characterization of the glycans even though the studies performed using S2 cells transfected with PSMA cDNA showed the presence of both complex and high-mannose-type glycans.69 In other studies using endoglycosidases with different specificities, PSMA was shown to be N-glycosylated with a mixture of high-mannose, hybrid, and complex types of oligosaccharides in LNCaP cells and a HEK293 human cell line transfected with a plasmid encoding full-length PSMA.70 Walczak and Danishefsky have demonstrated the synthesis of a fully sialylated triantennary N-linked glycan that is relevant to PSMA.71 Our current study provides the first comprehensive qualitative and quantitative characterization of N-linked glycosylation of PSMA in two PCa cell lines and provides greater detail of diverse glycan structural modification. It is important to note that the data that we report for PMSA glycosylation in LNCaP cells are similar but more comprehensive when compared to those in a recent article by Yuan et al.,72 where PSMA glycosylation in these cells was compared against insect S2 Schneiders’ cells and human HEK293T17 cells. The site-specific glycoforms that are found in LNCaP cells PSMA are generally concordant between our study and this study. Briefly, high-mannose glycans are highly abundant and are the dominant glycoforms at ASN-459, ASN-476, and ASN-638. The same situation is observed in MDAPCa2b cells. These asparagine residues are located on the PSMA surface facing the plasma membrane in X-ray structures of the physiological PSMA homodimer.73 In our studies, ASN-76 possesses mainly high-mannose and hybrid glycans but no complex structures. We also detected glycoforms conjugated to ASN-51 but in significantly low levels compared to the other seven glycosites. The other three sites ASN-121, ASN-195, and ASN-336 are predominantly conjugated with complex glycans with biantennary, triantennary, and tetraantennary structures. These asparagine residues are located in the exposed apical domain of the protein that faces away from the plasma membrane.73 These results are consistent with the recently published data.72 However, our study demonstrates significant differences in the fucosylation and sialylation levels of PSMA glycans in the two cell lines at these three sites. The bone metastatic MDAPCa2b cells possess significantly higher levels of sialylation compared to lymph node metastatic LNCaP cells, where sialylation is observed on a single glycan. This is consistent with previous reports of extremely low or lack of sialylation of PSA secreted by LNCaP cells.74,75 and also the recent observations of lack of PSMA sialylation in LNCaP cells.72 The upregulation of sialylated glycans on the plasma membrane cancer cells contribute to increased aggressiveness, metastasis, and resistance to cell death.76−80 Increased sialylation of PSA, another molecule that is used in PCa screening and prognosis, has been recently demonstrated to be associated with high-grade PCa.81,82
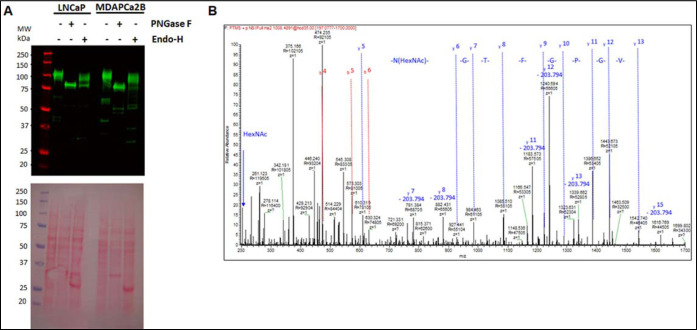
3.4. Qualitative Analysis of Endoglycosidase-Treated PSMA Glycopeptides
In complementary experiments, cell lysates from the two cell lines were treated with the glucosidases, PNGase-F and Endo H. Figure 6A shows western blot detection of the PSMA and GAPDH using total protein lysates from LNCaP cells and MDAPCa2b cells without glycosidase treatment, with PNGase-F cleavage and with Endo-H cleavage. In the case of PNGase-F treatment, a single band is detected at the expected molecular mass of completely deglycosylated PSMA. In the case of Endo-H, two bands were detected at higher mass compared to the PNGase-F-generated band. Since Endo-H cleaves the bond in the diacetylchitobiose core of the oligosaccharide between the two GlcNAcs directly proximal to the ASN residues of high-mannose and hybrid glycan chains, the western blot results are consistent with PSMA possessing both high-mannose, hybrid, and complex N-linked glycans. Figure 6
Figure 6.
(A,B) Detection of the PSMA total protein lysates from LNCaP and MDAPCa2b cells treated with endoglycosidases. (A). Western blot analysis of PSMA in LNCaP and MDAPCa2b cells after treatment with PNGase-F and Endo-H. (B). MS/MS analysis of PSMA glycopeptides after cleavage with Endo-H glycosidase identifies HexNAc-modified ASN-336 in the VPYNVGPGFTGN*FSTQK glycopeptide.
These results confirm our observations in the preceding sections that PSMA is modified by complex glycans in addition to high-mannose and hybrid structures. To validate the biochemical data, immunoaffinity purified PSMA from the two PCa cell lines were trypsin-digested before treatment with PNGase-F and Endo-H, respectively, before LC/MS/MS analyses. Figure 6B shows MS/MS spectra with the remaining N-acetylglucosamine residue (m/z 204.1) after Endo-H glycosidase treatment (neutral loss of HexNAc is annotated in the spectrum).
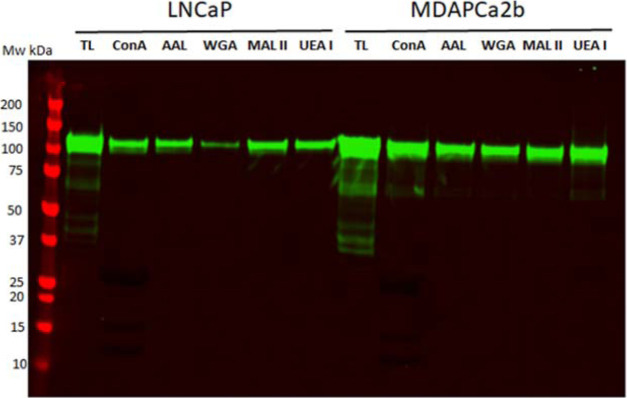
3.5. Differential Lectin Capture of PSMA
Total protein lysates from the two cell lines were used for lectin affinity capture and profiling for five lectins, ConA (D-mannosyl general specificity, branched N-linked hexasaccharide), AAL (preferentially binds to Fucα1-6/3GlcNAc), WGA (preferentially binds to (GlcNAcβ1-4)2–5, Neu5Ac Manβ1-4GlcNAcβ1-4GlacNAc), MAA/MAL II (Neu5Ac/Gcα2-3Galβ1-4GlcNAc), and UEA 1 (Fucα1-2Galβ).83 Following lectin binding, bound glycosylated proteins were eluted and separated by SDS–PAGE before western blot and detection of PSMA. As shown in Figure 7, the five lectins affinity purify PSMA from the two cells with different abundance, and the results are consistent with our MS data. These results support the MS detection of the different categories of N-linked glycans in PSMA in LNCaP and MDAPca2b cells. These include high mannosylated, fucosylated, and sialylated carbohydrates in addition to H-type and Lewis antigen structures.
Figure 7.
PSMA detection after lectin affinity purification. Total protein lysates from the two cell lines were incubated with agarose beads conjugated with ConA, AAL, WGA, MAA/MAL II, and UAE1 lectins overnight. Proteins bound to the lectins were eluted and separated by SDS-gel and used for western blot to detect PSMA.
The main weakness of our analysis is the comparison of two metastatic PCa cell lines and the analysis of clinical disease stratification will be required for further validation of the observations. These studies will be necessary to determine whether unique glycoforms have the ability to discriminate different stages of PCa and disease progression and their potential utility in diagnosis and prognosis. We were also not able to identify the glycoforms that are conjugated to the ASN-140 and ASN-153 glycosylation sites of the protein (Figure S8). In addition, complementary orthogonal experiments that evaluate the expression profiles of specific glycosyltransferases that may be associated with differential fucosylation, and sialylation of PSMA glycans that is observed in the two cell lines would uncover the molecular mechanisms and targets that mediate them. In our study, we have combined two complementary MS fragmentation methods HCD and EThcD for intact N-linked glycan analyses using DDA methods. In a recent study, a comprehensive comparison of EThcD, stepped collision energy HCD (sceHCD), EThcD-sceHCD, higher-energy collisional dissociation product-dependent electron-transfer dissociation (HCD-pd-ETD), and sceHCD-pd-ETD of intact N-glycopeptide analyses of clinical samples concluded that EThcD-sceHCD is more suitable for analyzing clinical samples compared to the other methods.84 In addition, efforts toward using data-independent acquisition (DIA) methods for intact N-glycopeptide analysis are ongoing, and HCD MS/MS is the method of choice for fragmentation since ETD MS/MS has not been directly integrated for DIA.85 The disadvantage of HCD fragmentation in intact glycopeptide analyses is the loss of conjugated glycans and poor yield of peptide backbone fragments. On the other hand, ETD fragmentation technique is capable of unambiguous mapping of glycosites. Further development of the technology could lead to promising applications and utility of DIA in analyzing intact N-glycopeptide in the future. However, overall this study provides novel comparative glycosylation data for PSMA in PCa cell lines with different tissue tropisms and metastatic niches.
4. Conclusions
In this study, we performed comparative qualitative and quantitative analysis of site-specific intact N-glycopeptides between two phenotype stratified PCa cell lines. We employed complementary MS-based fragmentation methods including HCD and EThcD and qualitative and quantitative glycoproteomics data analysis to characterize site-specific N-glycopeptides of PSMA. Our workflow is enabled by the multiscan capabilities of Tribrid Orbitrap Lumos Fusion MS to yield a robust glycopeptide and glycan structural elucidation. This approach allows simultaneous identification of glycans and glycopeptides in the same acquisition/analysis scheme using multiple algorithms provided in commercially available software (Byonic and Byologic).
We have mapped and characterized the N-linked glycosite occupancy and provided the glycosylation profile of PSMA directly from PCa cells using stable isotope labeling after glycosidase treatment for identification and validation of N-linked glycosylation sites. Our results demonstrate significant differences in the expression of several glycans in two cell lines LNCaP and MDAPCa2b, which have different phenotypes. There are significantly more glycans identified in MDAPCa2b compared to LNCaP cells at each of the eight N-linked glycosylation sites that were studied. We demonstrate the expression profiles and distribution of specific glycan groups at different asparagine residues in PSMA from different cell lines based on the quaternary structure of the protein. We further demonstrate differential expression of specific classes of glycans that possess glycans with different levels of fucosylation and sialylation and are potentially associated with specific disease phenotypes.
Since these cell lines represent distinct disease phenotypes, their differential glycan PSMA expression profiles may be associated with modulating progression and tissue- or organ-specific metastasis in PCa. A major clinical challenge is the inability of current diagnostic tests to distinguish between aggressive and indolent forms of PCa. Uncovering potential prognostic biomarkers in the form of assayable glycan modification in functionally relevant proteins such as PSMA may provide opportunity to develop tools to assist in precision treatment efforts. As PSMA protein is shed at detectable amounts in post-digital rectal examination (DRE) urines,86,87 our next focus is to characterize the glycan expression profiles of PSMA in this prostate proximal fluid that is obtained from men by minimally invasive methods. The objective these experiments is to identify site-specific glycan changes in PSMA from post-DRE urines from disease-stratified patients that correlate with disease severity as demonstrated in this study using PCa cell lines. These studies will form the basis of developing site-specific PSMA glycoform-based prognostic markers for PCa disease stratification in the future.
Acknowledgments
This work was supported by funds from the DOD/CDMRP through the Prostate Cancer Research Program (PCRP) grant to W81XWH-18-1-0228 PC170843 to JON. Opinions, interpretations, conclusions, and recommendations of the author are not necessarily endorsed by the Department of Defense (USAMRAA).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c02265.
Instrument methods and scan parameters that were used for LC/MS/MS analysis, representative XICs of tandem mass spectra of the HexNAc oxonium ion (HexNAc+) with the m/z range 204.08–204.09, top five most abundant glycoforms at eight N-linked glycosylation sites in PSMA, MS/MS spectra for trypsin-derived and PNGase-F-treated PSMA peptides corresponding to three N-linked glycosylation sites, tandem mass spectra for ASN-76 glycopeptides with oligomannose structures including Man7, Man6, and Man5 from LNCaP and MDAPCa2b cells, MS/MS spectra for an ASN-76 sialylated glycopeptide, Venn diagram indicating the number of top five most abundant glycans detected in LNCaP and MDAPCa2b cells, non-identification of ASN-140- and ASN-153-containing peptides after chymotrypsin and Glu-C digestion, origins and malignant characteristics of the PCa cells used in the study, and summary of the top five most abundant glycans identified at eight N-linked glycosylation sites in eight PSMA glycopeptides from LNCaP and MDAPCa2b cells (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- https://seer.cancer.gov/statfacts/html/prost.html#content (accessed on March 10, 2022).
- Siegel R. L.; Miller K. D.; Fuchs H. E.; Jemal A. Cancer statistics, 2022. Ca-Cancer J. Clin. 2022, 72, 7–33. 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- Bray F.; Ferlay J.; Soerjomataram I.; Siegel R. L.; Torre L. A.; Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca—Cancer J. Clin. 2018, 68, 394–424. 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- Berman D. M.; Epstein J. I. When is prostate cancer really cancer?. Urol. Clin. 2014, 41, 339. 10.1016/j.ucl.2014.01.006. [DOI] [PubMed] [Google Scholar]
- Schröder F. H.; Hugosson J.; Roobol M. J.; Tammela F. H.; Zappa J.; Nelen M. J.; Kwiatkowski T. L.; Lujan M.; Määttänen V.; Lilja M.; et al. Screening and prostate cancer mortality: results of the European Randomised Study of Screening for Prostate Cancer (ERSPC) at 13 years of follow-up. Lancet 2014, 384, 2027. 10.1016/s0140-6736(14)60525-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilt T. J.; Brawer M. K.; Jones K. M.; Barry M. J.; Aronson W. J.; Fox S.; Gingrich J. R.; Wei J. T.; Gilhooly P.; Grob B. M.; et al. Radical prostatectomy versus observation for localized prostate cancer. N. Engl. J. Med. 2012, 367, 203. 10.1056/nejmoa1113162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blute M. L. Jr; Abel E. J.; Downs T. M.; Kelcz F.; Jarrard D. F. Addressing the need for repeat prostate biopsy: new technology and approaches. Nat. Rev. Urol. 2015, 12, 435. 10.1038/nrurol.2015.159. [DOI] [PubMed] [Google Scholar]
- Lomas D. J.; Ahmed H. U. All change in the prostate cancer diagnostic pathway. Nat. Rev. Clin. Oncol. 2020, 17, 372. 10.1038/s41571-020-0332-z. [DOI] [PubMed] [Google Scholar]
- O’Dwyer E.; Bodei L.; Morris M. J. The Role of Theranostics in Prostate Cancer. Semin. Radiat. Oncol. 2021, 31, 71–82. 10.1016/j.semradonc.2020.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solnes L. B.; Shokeen M.; Pandit-Taskar N. Novel Agents and Future Perspectives on Theranostics. Semin. Radiat. Oncol. 2021, 31, 83–92. 10.1016/j.semradonc.2020.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horoszewicz J. S.; Kawinski E.; Murphy G. P. Monoclonal antibodies to a new antigenic marker in epithelial prostatic cells and serum of prostatic cancer patients. Anticancer Res. 1987, 7, 927. [PubMed] [Google Scholar]
- Beckett M. L.; Cazares L. H.; Vlahou A.; Schellhammer P. F.; Wright G. L. Jr. Prostate-specific membrane antigen levels in sera from healthy men and patients with benign prostate hyperplasia or prostate cancer. Clin. Cancer Res. 1999, 5, 4034. [PubMed] [Google Scholar]
- Sokoloff R. L.; Norton K. C.; Gasior C. L.; Marker K. M.; Grauer L. S. A dual-monoclonal sandwich assay for prostate-specific membrane antigen: Levels in tissues, seminal fluid and urine. Prostate 2000, 43, 150–157. . [DOI] [PubMed] [Google Scholar]
- Xiao Z.; Adam B. L.; Cazares L. H.; Clements M. A.; Davis J. W.; Schellhammer P. F.; Dalmasso E. A.; Wright G. L. Jr. Quantitation of serum prostate-specific membrane antigen by a novel protein biochip immunoassay discriminates benign from malignant prostate disease. Cancer Res. 2001, 61, 6029. [PubMed] [Google Scholar]
- Van de Wiele C.; Sathekge M.; de Spiegeleer B.; De Jonghe P. J.; Debruyne P. R.; Borms M.; Beels L.; Maes A. PSMA expression on neovasculature of solid tumors. Histol. Histopathol. 2020, 35, 919–927. 10.14670/HH-18-215. [DOI] [PubMed] [Google Scholar]
- Ghosh A.; Heston W. D. Tumor target prostate specific membrane antigen (PSMA) and its regulation in prostate cancer. J. Cell. Biochem. 2004, 91, 528–539. 10.1002/jcb.10661. [DOI] [PubMed] [Google Scholar]
- Sweat S. D.; Pacelli A.; Murphy G. P.; Bostwick D. G. Prostate-specific membrane antigen expression is greatest in prostate adenocarcinoma and lymph node metastases. Urology 1998, 52, 637–640. 10.1016/s0090-4295(98)00278-7. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Guo Z.; Du T.; Chen J.; Wang W.; Xu K.; Lin T.; Huang H. Prostate specific membrane antigen (PSMA): A novel modulator of p38 for proliferation, migration, and survival in prostate cancer cells. Prostate 2012, 73, 835–841. 10.1002/pros.22627. [DOI] [PubMed] [Google Scholar]
- Rahbar K.; Afshar-Oromieh A.; Jadvar H.; Ahmadzadehfar H. PSMA Theranostics: Current Status and Future Directions. Mol. Imag. 2018, 17, 153601211877606. 10.1177/1536012118776068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor N.; Sathianathen N. J.; Buteau J.; Koschel S.; Juanilla M. A.; Kapoor J.; Azad A.; Hofman M. S.; Murphy D. G. Prostate-specific membrane antigen theranostics in advanced prostate cancer: an evolving option. BJU Int. 2020, 126, 525–535. 10.1111/bju.15143. [DOI] [PubMed] [Google Scholar]
- Hadaschik B. A.; Giesel F. L.; Chi K. Prostate-specific Membrane Antigen Imaging and Theranostics Impact Patient Outcomes. Eur. Urol. Focus 2021, 7, 229–230. 10.1016/j.euf.2020.09.005. [DOI] [PubMed] [Google Scholar]
- Kumar A. S. R.; Lawrentschuk N.; Hofman M. S. Prostate-specific membrane antigen PET/computed tomography for staging prostate cancer. Curr. Opin. Urol. 2020, 30, 628–634. 10.1097/MOU.0000000000000799. [DOI] [PubMed] [Google Scholar]
- Munkley J.; Elliott D. J. Hallmarks of glycosylation in cancer. Oncotarget 2016, 7, 35478–35489. 10.18632/oncotarget.8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamczyk B.; Tharmalingam T.; Rudd P. M. Glycans as cancer biomarkers. Biochim. Biophys. Acta 2012, 1820, 1347–1353. 10.1016/j.bbagen.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Gilgunn S.; Conroy P. J.; Saldova R.; Rudd P. M.; O’Kennedy R. J. Aberrant PSA glycosylation--a sweet predictor of prostate cancer. Nat. Rev. Urol. 2013, 10, 99–107. 10.1038/nrurol.2012.258. [DOI] [PubMed] [Google Scholar]
- Ghosh A.; Heston W. D. Effect of carbohydrate moieties on the folate hydrolysis activity of the prostate specific membrane antigen. Prostate 2003, 57, 140–151. 10.1002/pros.10289. [DOI] [PubMed] [Google Scholar]
- Chandler K. B.; Costello C. E. Glycomics and glycoproteomics of membrane proteins and cell-surface receptors: Present trends and future opportunities. Electrophoresis 2016, 37, 1407–1419. 10.1002/elps.201500552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaysen-Andersen M.; Packer N. H. Advances in LC-MS/MS-based glycoproteomics: getting closer to system-wide site-specific mapping of the N- and O-glycoproteome. Biochim. Biophys. Acta 2014, 1844, 1437–1452. 10.1016/j.bbapap.2014.05.002. [DOI] [PubMed] [Google Scholar]
- Thaysen-Andersen M.; Packer N. H.; Schulz B. L. Maturing Glycoproteomics Technologies Provide Unique Structural Insights into the N-glycoproteome and Its Regulation in Health and Disease. Mol. Cell. Proteomics 2016, 15, 1773–1790. 10.1074/mcp.o115.057638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott N. E.; Cordwell S. J. Enrichment and identification of bacterial glycopeptides by mass spectrometry. Methods Mol. Biol. 2015, 1295, 355–368. 10.1007/978-1-4939-2550-6_25. [DOI] [PubMed] [Google Scholar]
- Scott N. E.; Parker B. L.; Connolly A. M.; Paulech J.; Edwards A. V.; Crossett B.; Falconer L.; Kolarich D.; Djordjevic S. P.; Højrup P.; et al. Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, higher energy collisional dissociation, and electron transfer dissociation MS applied to the N-linked glycoproteome of Campylobacter jejuni. Mol. Cell. Proteomics 2011, 10, PS1–S18. 10.1074/mcp.m000031-mcp201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford K. L.; Zeng W.; Heazlewood J. L.; Bacic A. Characterization of protein N-glycosylation by tandem mass spectrometry using complementary fragmentation techniques. Front. Plant Sci. 2015, 6, 674. 10.3389/fpls.2015.00674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q.; Zhao X.; Zhao Q.; Lv X.; Ma C.; Li X.; Zhao Y.; Peng B.; Ying W.; Qian X. Strategy integrating stepped fragmentation and glycan diagnostic ion-based spectrum refinement for the identification of core fucosylated glycoproteome using mass spectrometry. Anal. Chem. 2014, 86, 6804–6811. 10.1021/ac501154a. [DOI] [PubMed] [Google Scholar]
- Aboufazeli F.; Kolli V.; Dodds E. D. A comparison of energy-resolved vibrational activation/dissociation characteristics of protonated and sodiated high mannose N-glycopeptides. J. Am. Soc. Mass Spectrom. 2015, 26, 587–595. 10.1007/s13361-014-1070-1. [DOI] [PubMed] [Google Scholar]
- Marino F.; Bern M.; Mommen G. P. M.; Leney A. C.; van Gaans-van den Brink J. A. M.; Bonvin A. M. J. J.; Becker C.; van Els C. A. C. M.; Heck A. J. R. Extended O-GlcNAc on HLA Class-I-Bound Peptides. J. Am. Chem. Soc. 2015, 137, 10922–10925. 10.1021/jacs.5b06586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S. W.; Pu T. H.; Viner R.; Khoo K. H. Novel LC-MS2 product dependent parallel data acquisition function and data analysis workflow for sequencing and identification of intact glycopeptides. Anal. Chem. 2014, 86, 5478–5486. 10.1021/ac500945m. [DOI] [PubMed] [Google Scholar]
- Yu Q.; Wang B.; Chen Z.; Urabe G.; Glover M. S.; Shi X.; Guo L. W.; Kent K. C.; Li L. Electron-Transfer/Higher-Energy Collision Dissociation (EThcD)-Enabled Intact Glycopeptide/Glyco-proteome Characterization. J. Am. Soc. Mass Spectrom. 2017, 28, 1751–1764. 10.1007/s13361-017-1701-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin M.; Bern Q.; Xing J.; Ho R.; Viner M.; Mayr M. Glycoproteomic analysis of the secretome of human endothelial cells. Mol. Cell. Proteomics 2013, 12, 956. 10.1074/mcp.M112.024018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F.; Glinskii O. V.; Glinsky V. V. Glycobioinformatics: current strategies and tools for data mining in MS-based glycoproteomics. Proteomics 2013, 13, 341–354. 10.1002/pmic.201200149. [DOI] [PubMed] [Google Scholar]
- Woodin C. L.; Maxon M.; Desaire H. Software for automated interpretation of mass spectrometry data from glycans and glycopeptides. Analyst 2013, 138, 2793–2803. 10.1039/c2an36042j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L.; Xin L.; Shan B.; Lajoie G. A.; Ma B. GlycoMaster DB: software to assist the automated identification of N-linked glycopeptides by tandem mass spectrometry. J. Proteome Res. 2014, 13, 3881–3895. 10.1021/pr401115y. [DOI] [PubMed] [Google Scholar]
- Eshghi S. T.; Shah P.; Yang W.; Li X.; Zhang H. GPQuest: A Spectral Library Matching Algorithm for Site-Specific Assignment of Tandem Mass Spectra to Intact N-glycopeptides. Anal. Chem. 2015, 87, 5181–5188. 10.1021/acs.analchem.5b00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S. W.; Liang S. Y.; Pu T. H.; Chang F. Y.; Khoo K. H. Sweet-Heart - an integrated suite of enabling computational tools for automated MS2/MS3 sequencing and identification of glycopeptides. J. Proteonomics 2013, 84, 1–16. 10.1016/j.jprot.2013.03.026. [DOI] [PubMed] [Google Scholar]
- Mayampurath A.; Yu C. Y.; Song E.; Balan J.; Mechref Y.; Tang H. Computational framework for identification of intact glycopeptides in complex samples. Anal. Chem. 2014, 86, 453–463. 10.1021/ac402338u. [DOI] [PubMed] [Google Scholar]
- Lynn K. S.; Chen C. C.; Lih T. M.; Cheng C. W.; Su W. C.; Chang C. H.; Cheng C. Y.; Hsu W. L.; Chen Y. J.; Sung T. Y. MAGIC: an automated N-linked glycoprotein identification tool using a Y1-ion pattern matching algorithm and in silico MS2 approach. Anal. Chem. 2015, 87, 2466–2473. 10.1021/ac5044829. [DOI] [PubMed] [Google Scholar]
- Zhu Z.; Su X.; Go E. P.; Desaire H. New glycoproteomics software, GlycoPep Evaluator, generates decoy glycopeptides de novo and enables accurate false discovery rate analysis for small data sets. Anal. Chem. 2014, 86, 9212–9219. 10.1021/ac502176n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park G. W.; Kim J. Y.; Hwang H.; Lee J. Y.; Ahn Y. H.; Lee H. K.; Ji E. S.; Kim K. H.; Jeong H. K.; Yun K. N.; Kim Y. S.; Ko J. H.; An H. J.; Kim J. H.; Paik Y. K.; Yoo J. S. Integrated GlycoProteome Analyzer (I-GPA) for Automated Identification and Quantitation of Site-Specific N-Glycosylation. Sci. Rep. 2016, 6, 21175. 10.1038/srep21175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaysen-Andersen M.; Venkatakrishnan V.; Loke I.; Laurini C.; Diestel S.; Parker B. L.; Packer N. H. Human neutrophils secrete bioactive paucimannosidic proteins from azurophilic granules into pathogen-infected sputum. J. Biol. Chem. 2015, 290, 8789–8802. 10.1074/jbc.m114.631622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker B. L.; Thaysen-Andersen M.; Solis N.; Scott N. E.; Larsen M. R.; Graham M. E.; Packer N. H.; Cordwell S. J. Site-specific glycan-peptide analysis for determination of N-glycoproteome heterogeneity. J. Proteome Res. 2013, 12, 5791–5800. 10.1021/pr400783j. [DOI] [PubMed] [Google Scholar]
- Medzihradszky K. F.; Kaasik K.; Chalkley R. J. Tissue-Specific Glycosylation at the Glycopeptide Level. Mol. Cell. Proteomics 2015, 14, 2103–2110. 10.1074/mcp.m115.050393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker B. L.; Thaysen-Andersen M.; Fazakerley D. J.; Holliday M.; Packer N. H.; James D. E. Terminal Galactosylation and Sialylation Switching on Membrane Glycoproteins upon TNF-Alpha-Induced Insulin Resistance in Adipocytes. Mol. Cell. Proteomics 2016, 15, 141–153. 10.1074/mcp.m115.054221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.; Szeto S. S.; Kong R. P.; Law C. H.; Li G.; Quan Q.; Zhang Z.; Wang Y.; Chu I. K. Online two-dimensional porous graphitic carbon/reversed phase liquid chromatography platform applied to shotgun proteomics and glycoproteomics. Anal. Chem. 2014, 86, 12172–12179. 10.1021/ac503254t. [DOI] [PubMed] [Google Scholar]
- Chen Z.; Huang J.; Li L. Recent advances in mass spectrometry (MS)-based glycoproteomics in complex biological samples. Trends Anal. Chem. 2019, 118, 880–892. 10.1016/j.trac.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P.; Wang X.; Yang W.; Toghi Eshghi S.; Sun S.; Hoti N.; Chen L.; Yang S.; Pasay J.; Rubin A.; Zhang H. Integrated Proteomic and Glycoproteomic Analyses of Prostate Cancer Cells Reveal Glycoprotein Alteration in Protein Abundance and Glycosylation. Mol. Cell. Proteomics 2015, 14, 2753–2763. 10.1074/mcp.m115.047928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L. Y.; Moh E. S.; Parker B. L.; Bern M.; Packer N. H.; Thaysen-Andersen M. Toward Automated N-Glycopeptide Identification in Glycoproteomics. J. Proteome Res. 2016, 15, 3904–3915. 10.1021/acs.jproteome.6b00438. [DOI] [PubMed] [Google Scholar]
- Horoszewicz J. S.; Leong S. S.; Kawinski E.; Karr J. P.; Rosenthal H.; Chu T. M.; Mirand E. A.; Murphy G. P. LNCaP model of human prostatic carcinoma. Cancer Res. 1983, 43, 1809–18. [PubMed] [Google Scholar]
- Navone N. M.; Olive M.; Ozen M.; Davis R.; Troncoso P.; Tu S. M.; Johnston D.; Pollack A.; Pathak S.; von Eschenbach A. C.; Logothetis C. J. Establishment of two human prostate cancer cell lines derived from a single bone metastasis. Clin. Cancer Res. 1997, 3, 2493. [PubMed] [Google Scholar]
- van Bokhoven A.; Varella-Garcia M.; Korch C.; Johannes W. U.; Smith E. E.; Miller H. L.; et al. Molecular characterization of human prostate carcinoma cell lines. Prostate 2003, 57, 205–225. 10.1002/pros.10290. [DOI] [PubMed] [Google Scholar]
- Burch T. C.; Isaac G.; Booher C. L.; Rhim J. S.; Rainville P.; Langridge J.; Baker A.; Nyalwidhe J. O. Comparative Metabolomic and Lipidomic Analysis of Phenotype Stratified Prostate Cells. PLoS One 2015, 10, e0134206 10.1371/journal.pone.0134206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T.; Toriyabe Y.; Berkman C. E. Purification of prostate-specific membrane antigen using conformational epitope-specific antibody-affinity chromatography. Protein Expr. Purif. 2006, 49, 251–255. 10.1016/j.pep.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Yang L.; Dutta S. M.; Troyer D. A.; Lin J. B.; Lance R. A.; Nyalwidhe J. O.; Drake R. R.; Semmes O. J. Dysregulated expression of cell surface glycoprotein CDCP1 in prostate cancer. Oncotarget 2015, 6, 43743–43758. 10.18632/oncotarget.6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyalwidhe J. O.; Betesh L. R.; Powers T. W.; Jones E. E.; White K. Y.; Burch T. C.; Brooks J.; Watson M. T.; Lance R. S.; Troyer D. A.; Semmes O. J.; Mehta A.; Drake R. R. Increased bisecting N-acetylglucosamine and decreased branched chain glycans of N-linked glycoproteins in expressed prostatic secretions associated with prostate cancer progression. Proteonomics Clin. Appl. 2013, 7, 677–689. 10.1002/prca.201200134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K. Y.; Rodemich L.; Nyalwidhe J. O.; Comunale M. A.; Clements M. A.; Lance R. S.; Schellhammer P. F.; Mehta A. S.; Semmes O. J.; Drake R. R. Glycomic characterization of prostate-specific antigen and prostatic acid phosphatase in prostate cancer and benign disease seminal plasma fluids. J. Proteome Res. 2009, 8, 620–630. 10.1021/pr8007545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bern M.; Kil Y. J.; Becker C. Byonic: advanced peptide and protein identification software. Curr. Protoc. Bioinf. 2012, 40, 13. 10.1002/0471250953.bi1320s40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.; Chen Z.; Zhang J.; An M.; Wu J.; Yu Q.; Skilton S. J.; Bern M.; Ilker Sen K.; Li L.; Lubman D. M. Differential Quantitative Determination of Site-Specific Intact N-Glycopeptides in Serum Haptoglobin between Hepatocellular Carcinoma and Cirrhosis Using LC-EThcD-MS/MS. J. Proteome Res. 2019, 18, 359–371. 10.1021/acs.jproteome.8b00654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go E. P.; Zhang S.; Ding H.; Kappes J. C.; Sodroski J.; Desaire H. The opportunity cost of automated glycopeptide analysis: case study profiling the SARS-CoV-2 S glycoprotein. Anal. Bioanal. Chem. 2021, 413, 7215–7227. 10.1007/s00216-021-03621-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E. H.; Greene T. G.; Tino W. T.; Boynton A. L.; Aldape H. C.; Misrock S. L.; Murphy G. P. Analysis of glycosylation of prostate-specific membrane antigen derived from LNCaP cells, prostatic carcinoma tumors, and serum from prostate cancer patients. Prostate Suppl. 1996, 29, 25–29. . [DOI] [PubMed] [Google Scholar]
- Xiao Z.; Jiang X.; Beckett M. L.; Wright G. L. Jr Generation of a baculovirus recombinant prostate-specific membrane antigen and its use in the development of a novel protein biochip quantitative immunoassay. Protein Expr. Purif. 2000, 19, 12–21. 10.1006/prep.2000.1222. [DOI] [PubMed] [Google Scholar]
- Barinka C.; Šácha P.; Sklenář J.; Man P.; Bezouška K.; Slusher B. S.; Konvalinka J. Identification of the N-glycosylation sites on glutamate carboxypeptidase II necessary for proteolytic activity. Protein Sci. 2004, 13, 1627–1635. 10.1110/ps.04622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak M. A.; Danishefsky S. J. Solving the convergence problem in the synthesis of triantennary N-glycan relevant to prostate-specific membrane antigen (PSMA). J. Am. Chem. Soc. 2012, 134, 16430–16433. 10.1021/ja307628w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlcochová P.; Barinka C.; Tykvart J.; Sácha P.; Konvalinka J. Prostate-specific membrane antigen and its truncated form PSM. Prostate 2009, 69, 471–479. 10.1002/pros.20894. [DOI] [PubMed] [Google Scholar]
- Yuan W.; Liu B.; Sanda M.; Wei R.; Benicky J.; Novakova Z.; Barinka C.; Goldman R. Glycoforms of human prostate-specific membrane antigen (PSMA) in human cells and prostate tissue. Prostate 2022, 82, 132–144. 10.1002/pros.24254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesters J. R.; Barinka C.; Li W.; Tsukamoto T.; Majer P.; Slusher B. S.; Konvalinka J.; Hilgenfeld R. Structure of glutamate carboxypeptidase II, a drug target in neuronal damage and prostate cancer. EMBO J. 2006, 25, 1375–1384. 10.1038/sj.emboj.7600969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peracaula R.; Tabarés G.; Royle L.; Harvey D. J.; Dwek R. A.; Rudd P. M.; de Llorens R. Altered glycosylation pattern allows the distinction between prostate-specific antigen (PSA) from normal and tumor origins. Glycobiology 2003, 13, 457–470. 10.1093/glycob/cwg041. [DOI] [PubMed] [Google Scholar]
- Ideo H.; Kondo J.; Nomura T.; Nonomura N.; Inoue M.; Amano J. Study of glycosylation of prostate-specific antigen secreted by cancer tissue-originated spheroids reveals new candidates for prostate cancer detection. Sci. Rep. 2020, 10, 2708. 10.1038/s41598-020-59622-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büll C.; den Brok M. H.; Adema G. J. Sweet escape: sialic acids in tumor immune evasion. Biochim. Biophys. Acta 2014, 1846, 238–246. 10.1016/j.bbcan.2014.07.005. [DOI] [PubMed] [Google Scholar]
- Büll C.; Stoel M. A.; den Brok M. H.; Adema G. J. Sialic acids sweeten a tumor’s life. Cancer Res. 2014b, 74, 3199–204. 10.1158/0008-5472.CAN-14-0728. [DOI] [PubMed] [Google Scholar]
- Dall’Olio F.; Malagolini N.; Trinchera M.; Chiricolo M. Sialosignaling: sialyltransferases as engines of self-fueling loops in cancer progression. Biochim. Biophys. Acta 2014, 1840, 2752. 10.1016/j.bbagen.2014.06.006. [DOI] [PubMed] [Google Scholar]
- Lu J.; Gu J. Significance of β-galactoside α2,6 sialyltranferase 1 in cancers. Molecules 2015, 20, 7509–7527. 10.3390/molecules20057509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce O. M.; Läubli H. Sialic acids in cancer biology and immunity. Glycobiology 2016, 26, 111–128. 10.1093/glycob/cwv097. [DOI] [PubMed] [Google Scholar]
- Hatano K.; Yoneyama T.; Hatakeyama S.; Tomiyama E.; Tsuchiya M.; Nishimoto M.; Yoshimura K.; Miyoshi E.; Uemura H.; Ohyama C.; Nonomura N.; Fujita K. Simultaneous analysis of serum α2,3-linked sialylation and core-type fucosylation of prostate-specific antigen for the detection of high-grade prostate cancer. Br. J. Cancer 2022, 126, 764–770. 10.1038/s41416-021-01637-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama T.; Yamamoto H.; Yoneyama M. S.; Tobisawa Y.; Hatakeyama S.; Narita T.; Kodama H.; Momota M.; Ito H.; Narita S.; et al. Characteristics of α2,3-sialyl N-glycosylated PSA as a biomarker for clinically significant prostate cancer in men with elevated PSA level. Prostate 2021, 81, 1411–1427. 10.1002/pros.24239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D.; Mao L. Cancer biomarker discovery: lectin-based strategies targeting glycoproteins. Dis. Markers 2012, 33, 1–10. 10.1155/2012/308738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W.; Zheng S.; Su T.; Cheng J.; Mao Y.; Zhong Y.; Liu Y.; Chen J.; Zhao W.; Lin T.; Liu F.; Li G.; Yang H.; Zhang Y. Comparative N-glycoproteomics analysis of clinical samples via different mass spectrometry dissociation methods. Front. Chem. 2022, 10, 839470. 10.3389/fchem.2022.839470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig C.; Gillet L.; Rosenberger G.; Amon S.; Collins B. C.; Aebersold R. Data-independent acquisition-based SWATH-MS for quantitative proteomics: a tutorial. Mol. Syst. Biol. 2018, 14, e8126 10.15252/msb.20178126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Principe S.; Kim Y.; Fontana S.; Ignatchenko V.; Nyalwidhe J. O.; Lance R. S.; Troyer D. A.; Alessandro R.; Semmes O. J.; Kislinger T.; Drake R. R.; Medin J. A. Identification of prostate-enriched proteins by in-depth proteomic analyses of expressed prostatic secretions in urine. J. Proteome Res. 2012, 11, 2386–2396. 10.1021/pr2011236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.; Ignatchenko V.; Yao C. Q.; Kalatskaya I.; Nyalwidhe J. O.; Lance R. S.; Gramolini A. O.; Troyer D. A.; Stein L. D.; Boutros P. C.; Medin J. A.; Semmes O. J.; Drake R. R.; Kislinger T. Identification of differentially expressed proteins in direct expressed prostatic secretions of men with organ-confined versus extracapsular prostate cancer. Mol. Cell. Proteomics 2012, 11, 1870–1884. 10.1074/mcp.m112.017889. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.