Abstract
Background & Aims:
The CD47-signal regulatory protein alpha (SIRPα) signaling pathway plays important roles in immune homeostasis and tissue inflammatory response. Activation of the Hedgehog/SMO/Gli1 pathway regulates cell growth, differentiation, and immune function. However, it remains unknown whether and how the CD47-SIRPα interaction may regulate Hedgehog/SMO/Gli1 signaling in mesenchymal stem cell (MSC)-mediated immune regulation during sterile inflammatory liver injury.
Approach & Results:
In a mouse model of ischemia/reperfusion (IR)-induced sterile inflammatory liver injury, we found that adoptive transfer of MSCs increased CD47 expression and ameliorated liver IRI. However, deletion of CD47 in MSCs exacerbated IR-induced liver damage, with increased serum ALT levels, macrophage/neutrophil infiltration, and pro-inflammatory mediators. MSC treatment augmented SIRPα, Hedgehog/SMO/Gli1, and Notch1 intracellular domain (NICD), whereas CD47-deficient MSC treatment reduced these gene expressions in IR-stressed livers. Moreover, disruption of myeloid SMO or Notch1 increased IR-triggered liver inflammation with diminished Gli1 and NICD but enhanced NEK7 and NLRP3 activation in MSC-transferred mice. Using a MSC/macrophage co-culture system, we found that MSC CD47 and macrophage SIRPα expression were increased after LPS stimulation. The CD47-SIRPα interaction increased macrophage Gli1 and NICD nuclear translocation, whereby NICD interacted with Gli1 and regulated its target gene Dvl2, which in turn inhibited NEK7/NLRP3 activity.
Conclusions:
The CD47-SIRPα signaling activates the Hedgehog/SMO/Gli1 pathway, which controls NEK7/NLRP3 activity through a direct interaction between Gli1 and NICD. NICD is a novel coactivator of Gli1, and the target gene Dvl2 regulated by the NICD-Gli1 complex is crucial for the modulation of NLRP3-driven inflammatory response in MSC-mediated immune regulation. Our findings provide novel potential therapeutic targets in MSC-mediated immunotherapy of sterile inflammatory liver injury.
Keywords: SIRPα, Notch, NICD, Dvl2, NLRP3
Mesenchymal stem cells (MSCs) are multipotent cells capable of differentiation into mesenchymal lineages. They have become the most frequently used cell type for tissue repair and regeneration because of their remarkable immunoregulatory properties. MSCs exert extensive immunomodulation through interaction with immune cells in innate and adaptive immune systems (1). Although MSC-based therapy has been successfully applied in various immune-mediated diseases in humans (2), some critical questions remain elusive as the low immunosuppressive efficacy of engrafted cells could not meet the primary endpoints in many clinical trials of MSC immunotherapy (3). Thus, understanding immunoregulatory mechanisms of MSCs to develop novel approaches has emerged as one of the key challenges of MSC therapy.
CD47 (Cluster of Differentiation 47), which is also known as integrin-associated protein (IAP), is a cell surface protein of the immunoglobulin (Ig) superfamily that is expressed by virtually all cells in the body (4). CD47 expression is upregulated in hematopoietic stem cells (HSCs) after mobilization or induced inflammation (5). Ligation of CD47 can induce intracellular signaling resulting in cell activation and differentiation in response to stress (6). By binding to signal regulatory protein alpha (SIRPα), an inhibitory transmembrane receptor present on myeloid cells, CD47 can regulate cell functions in the monocyte/macrophage lineage (7). Indeed, SIRPα is abundantly expressed in macrophages and has been implicated in regulating innate immunity during inflammatory responses (8), indicating that SIRPα is an essential endogenous regulator of tissue inflammation.
The Hedgehog signaling pathway has been shown to regulate cell differentiation, tissue development, homeostasis, and regeneration (9, 10). Activation of the Hedgehog pathway involves two essential proteins, the G-protein-coupled receptor smoothened (SMO) and the twelve-pass transmembrane protein patched 1 (PTCH1). The Hedgehog signaling is activated upon binding the ligand to PTCH1, which activates SMO, leading to the promotion of the downstream targeting gene, the transcriptional activator Gli1 (11). Indeed, increasing Gli1 activity inhibits pro-inflammatory mediators and tissue inflammation, whereas deletion of Gli1 promotes immune cell activation and inflammatory response (12, 13). Moreover, the Notch1 pathway has been indicated to control the homeostasis of innate cell populations and regulate immune cell development and function (14). Activation of Notch1 and its ligand Jagged-1 increases cell growth and differentiation during liver regeneration (15). Disruption of the transcription factor RBP-J increases cell apoptosis/necrosis and inflammatory response, leading to aggravated liver injury (16). Our recent study has shown that activation of Notch1 signaling inhibits innate immune responses during liver inflammatory injury (17, 18). Although both the Hedgehog signaling and Notch1 pathway are essential for the modulation of tissue inflammatory responses (19), it remains largely unknown as to whether and how the CD47-SIRPα signaling may regulate the Hedgehog/SMO/Gli1 and Notch1 pathways to control innate immune response in MSC-mediated immune modulation during liver inflammatory injury.
Here, we identify a novel functional role and regulatory mechanism of the CD47-mediated Hedgehog/SMO/Gli1 signaling in MSC-mediated immune regulation. We demonstrate that interaction between MSC CD47 and macrophage SIRPα activates Hedgehog/SMO/Gli1 and Notch1 pathways. The CD47-mediated Hedgehog/SMO/Gli1 signaling controls NEK7/NLRP3 activation through a direct interaction between NICD and Gli1, which in turn regulates the target gene Dvl2 leading to inhibited NEK7/NLRP3 function in IR-triggered liver inflammation.
Experimental Procedures
Animals.
The floxed SMO (SMOFL/FL) mice (The Jackson Laboratory, Bar Harbor, ME) and the mice expressing Cre recombinase under the control of the Lysozyme 2 (Lyz2) promoter (LysM-Cre; The Jackson Laboratory) were used to generate myeloid-specific SMO knockout (SMOM-KO) mice (Supplementary Fig. 1). The generation of myeloid-specific Notch1 knockout (Notch1M-KO) mice, as described (17). This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health. Animal protocols were approved by the Institutional Animal Care and Use Committee of The University of California at Los Angeles. See Supplementary Materials.
Mouse liver IRI model.
We used an established mouse model of warm hepatic ischemia (90min) followed by reperfusion (6h), as described (20). Some animals were injected via tail vein with bone marrow-derived MSCs, genetically modified MSCs (1x106cells in PBS/mouse), or MSCs pre-labeled with 5-chloromethylfluorescein diacetate (CMFDA) green fluorescent dye (Invitrogen) 24h prior to ischemia. See Supplementary Materials.
Hepatocellular function assay.
Serum alanine aminotransferase (sALT) levels, an indicator of hepatocellular injury, were measured by IDEXX Laboratories (Westbrook, ME).
Histology, immunohistochemistry, and immunofluorescence staining.
Liver sections (5-μm) were stained with hematoxylin and eosin (H&E). The severity of IRI was analyzed by Suzuki's histological criteria (21). Histological changes were scored from 0 to 4 based on the degree of sinusoidal congestion, cytoplasmic vacuolization, and necrosis of parenchymal cells (Supplementary Table 1). 0 = no sinusoidal congestion, cytoplasmic vacuolization, and necrosis of parenchymal cells, 1 = minimal sinusoidal congestion/cytoplasmic vacuolization and single-cell necrosis of parenchymal cells, 2 = mild sinusoidal congestion/cytoplasmic vacuolization and up to 30% necrosis of parenchymal cells, 3 = moderate sinusoidal congestion/cytoplasmic vacuolization and up to 60% necrosis of parenchymal cells, and 4 = severe sinusoidal congestion/cytoplasmic vacuolization and more than 60% necrosis of parenchymal cells. Liver macrophages and neutrophils were detected using CD11b, CD68, and Ly6G rat mAbs. The primary mouse SMO, Gli1, NICD, Dvl2, NLRP3 mAbs were used for immunofluorescence and immunohistochemistry staining. Images for immunofluorescence staining were captured using a fluorescence microscope (Keyence BZ-X810, Osaka, Japan). See Supplementary Materials.
Quantitative RT-PCR analysis.
Quantitative real-time PCR was performed as described (22). Quantitative real-time PCR was carried out using the QuantStudio 3 (Applied Biosystems). Amplification conditions were: 50°C (2min), 95°C (5min), followed by 40 cycles of 95°C (15sec) and 60°C (30sec). Primer sequences used to amplify IL-1β, TNF-α, IL-6, CXCL2, CXCL10, NICD, Gli1, and β-actin are shown in Supplementary Table 2. See Supplementary Materials.
Western blot analysis.
Protein was extracted from liver tissue or cell cultures as described (22). Protein was extracted from liver tissue or cell cultures and subjected to 4-20% SDS-polyacrylamide gel electrophoresis (PAGE) and then transferred to nitrocellulose membrane (Bio-Rad). The nuclear and cytosolic fractions were prepared with NE-PER Nuclear and Cytoplasmic Extraction Reagents (ThermoFisher Scientific). The SIRPα, p-GSK3β, GSK3β, NICD, β-catenin, XBP1, ASC, cleaved caspase-1, Lamin B2, β-actin (Cell Signaling Technology), CD47, SMO, Dvl2, NRX (Santa Cruz Biotechnology), Gli1 (Invitrogen), NEK7, and NLRP3 (Abcam) mAbs were used. The membranes were incubated with Abs, and then added Western ECL substrate mixture (Bio-Rad) for imaging with the iBright FL1000 (ThermoFisher Scientific). Relative quantities of protein were determined by comparing the β-actin expression using iBright image analysis software (ThermoFisher Scientific). See Supplementary Materials.
Isolation of primary hepatocytes, Kupffer cells, BMMs, and bone marrow-derived MSCs.
Primary hepatocytes and Kupffer cells from the SMOFL/FL, SMOM-KO, or wild-type (WT) mice were isolated as described (22). In brief, the livers were perfused in situ with warmed (37°C) HBSS solution, followed by a collagenase-buffer (collagenase type IV, Sigma-Aldrich). Perfused livers were dissected and teased through 70-μm nylon mesh cell strainers (BD Biosciences). Nonparenchymal cells (NPCs) were separated from hepatocytes by centrifuging at 50 × g for 2min three times. NPCs were suspended in HBSS and layered onto a 50%/25% two-step Percoll gradient (Sigma) in a 50-ml conical centrifuge tube and centrifuged at 1800 × g at 4°C for 15min. Kupffer cells in the middle layer were collected and allowed to attach onto cell culture plates in DMEM for 15min at 37°C. Bone marrow-derived MSCs were isolated, as described (23). In brief, the bone marrow cells were cultured with α-MEM. After 24h, the culture medium was refreshed to remove non-adherent cells. MSCs were used in the experiments only after 2 to 3 expansion passages to ensure depletion of monocytes/macrophages. Murine bone-derived macrophages (BMMs) were generated as described (22). See Supplementary Materials.
In vitro transfection.
MSCs (1x106) were transfected with CRISPR/Cas9 CD47 knockout (KO), and BMMs (1x106) were transfected with CRISPR/Cas9 Notch1 KO, SIRPα KO, Dvl2 KO, NRX KO, CRISPR Dvl2 or SIRPα activation, and CRISPR control vector (Santa Cruz Biotechnology) by using Lipofectamine™ 3000 according to the manufacturer's instructions (Invitrogen). After 24-48h, cells were supplemented with 100 ng/ml of lipopolysaccharide (LPS) for an additional 6h.
Co-culture of MSCs and macrophages.
MSCs were co-cultured with macrophages as previously described (24). In brief, BMMs (1x106/well) were plated on 0.1% gelatin-coated six-well plates for 24h and transfected with CRISPR/Cas9 Notch1 KO, SIRPα KO, Dvl2 KO, NRX KO, CRISPR Dvl2 activation or CRISPR control vector. After 48h, MSCs (2x105/well) were co-cultured with macrophages followed by LPS (100 ng/mL) stimulation for 6h. Co-culture was washed extensively to remove MSCs. The attached macrophages were collected for FACS analysis (Supplementary Fig. 2) and used for further experiments.
ELISA assay.
Murine serum and cell culture supernatants were harvested for cytokine analysis. ELISA kit (ThermoFisher Scientific) was used to measure levels of IL-1β.
Immunoprecipitation analysis.
BMMs from co-culture were lysed in NP-40 lysis buffer (ThermoFisher Scientific) containing protease inhibitors. The lysates were incubated with Gli1 antibody (Santa Cruz Biotechnology), NICD antibody (Cell Signaling Technology), or control IgG and protein A/G beads at 4 °C overnight. After immunoprecipitation, the immunocomplexes were washed with lysis buffer, and analyzed by standard immunoblot procedures.
Chromatin immunoprecipitation (ChIP).
The ChIP analysis was carried out using ChIP Assay Kit according to the manufacturer's instructions (Abcam). For sequential ChIP, sheared chromatin was first immunoprecipitated with NICD antibody (Cell Signaling Technology) and then eluted with a second immunoprecipitation using Gli1 antibody (Novus Biologicals). DNA from each immunoprecipitation reaction was examined by PCR. The primer for the Gli1-responsive region of Dvl2 promoter: forward: 5’- GATGGGAAGTTTAGGGGCCAA -3’, reverse: 5’- AACTATAAGAGGGGCGGGGAT -3’. See Supplementary Materials.
ChIP-sequencing (ChIP-seq).
ChIP-DNA was amplified to generate a library for sequencing with an Illumina HiSeq3000 (Illumina) at the Technology Center for Genomics & Bioinformatics (TCGB) at UCLA. See Supplementary Materials.
RNA in situ hybridization.
RNA in situ hybridization (ISH) was carried out using the RNAscope 2.5 HD Assay-RED KIT (324510, Advanced Cell Diagnostics, CA) according to the manufacturer's instructions. Mouse Dvl-2 (Mm-Dvl-2-C1, 1038481-C1), negative and positive control probes were purchased from Advanced Cell Diagnostics. See Supplementary Materials.
Caspase-1 enzymatic activity assay.
Caspase-1 enzymatic activity was determined by a colorimetric assay kit (R&D System), as described (18). Briefly, after co-cultured with MSC, A 50μl of cell lysate from BMMs was added to 50μl of caspase-1 reaction buffer in a 96-well flat bottom microplate. Each sample was then added to 200mM caspase-1 substrate, WEHD-pNA, followed by 2h of incubation at 37°C. The enzymatic activity of caspase-1 was measured on an ELISA reader at 405nm wavelength.
Statistical analysis.
Data are expressed as mean±SD and analyzed by Permutation t-test and Pearson correlation. Per comparison, two-sided p values less than 0.05 were considered statistically significant. Multiple group comparisons were made using one-way ANOVA followed by Bonferroni's post hoc test. When groups showed unequal variances, we applied Welch's ANOVA to make multiple group comparisons. All analyses were used by SAS/STAT software, version 9.4.
Results
Disruption of CD47 in MSCs exacerbates IR-induced liver injury and augments proinflammatory mediators.
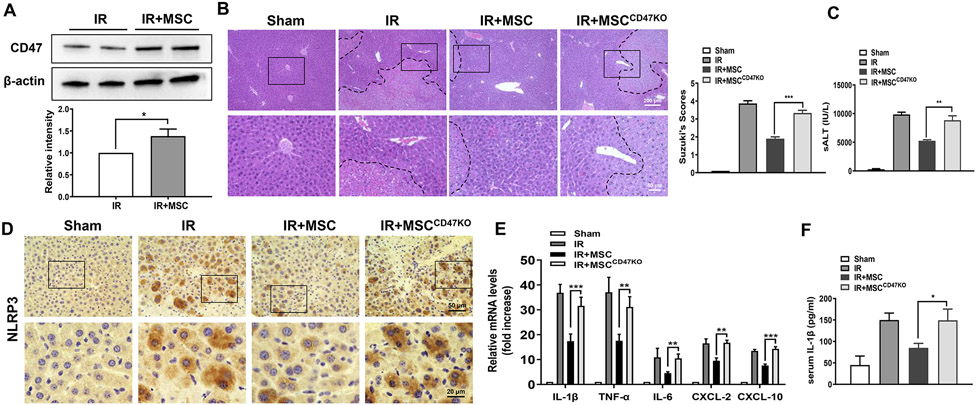
In a mouse model of hepatic IRI, we found that adoptive transfer of MSCs increased CD47 expression in IR-stressed livers (Fig. 1A). Unlike in MSC control-treated livers, which showed well-preserved hepatic architecture, with minimal sinusoidal congestion and without edema, vacuolization or necrosis (Fig. 1B), disruption of CD47 in MSCs by transfecting CRISPR/Cas9-CD47 KO vector showed moderate or severe sinusoidal congestion, cytoplasmic vacuolization, and hepatocellular necrosis (Fig. 1B). The serum ALT (sALT) levels were significantly increased after the adoptive transfer of CD47-deficient MSCs compared to the MSC control-treated mice (Fig 1C). We further confirmed that they have similar amounts of MSCs in mice after the adoptive transfer of MSCs or CD47-deficient MSCs pre-labeled with CMFDA green fluorescent dye. Immunofluorescence staining revealed that disruption of CD47 in MSCs did not affect MSC homing in ischemic livers (Supplementary Fig. 3). As our previous studies have demonstrated that NLRP3 is crucial for triggering liver inflammatory response in liver IRI (18, 22, 25), we then highlight the NLRP3-mediated liver inflammation in liver IRI. Indeed, immunohistochemistry staining revealed that CD47-deficient MSC treatment augmented NLRP3 expression (Fig. 1D) in ischemic livers accompanied by increased expression of pro-inflammatory IL-1β, TNF-α, IL-6, CXCL2, CXCL-10 (Fig. 1E), and IL-1β release (Fig. 1F), compared to the MSC control-treated groups.
Fig. 1. Disruption of CD47 in MSCs exacerbates IR-induced liver injury and augments proinflammatory mediators.
WT mice were adoptive transferred MSCs or CD47-deficient MSCs (1x106) 24h prior to liver ischemic insult. (A) The CD47 expression was detected by Western blot assay in ischemic livers. The representative of three experiments. (B) Representative histological staining (H&E) of ischemic liver tissue (n=4-6 mice/group) and Suzuki's histological score. Scale bars, 200μm and 50μm. (C) Liver function was evaluated by serum ALT levels (IU/L) (n=4-6 samples/group). (D) Immunohistochemistry staining of NLRP3 in ischemic livers (n=4-6 mice/group). Scale bars, 50μm and 20μm. (E) qRT-PCR analysis of IL-1β, TNF-α, IL-6, CXCL-2, and CXCL-10 in ischemic livers (n=3-4 samples/group). (F) ELISA analysis of serum IL-1β levels (n=3-4 samples/group). All data represent the mean±SD. *p<0.05, **p<0.01, ***p<0.001.
The CD47-SIRPα interaction activates Hedgehog/SMO/Gli1 pathway, Notch1 signaling, and inhibits NEK7/NLRP3 activation in IR-stressed livers.
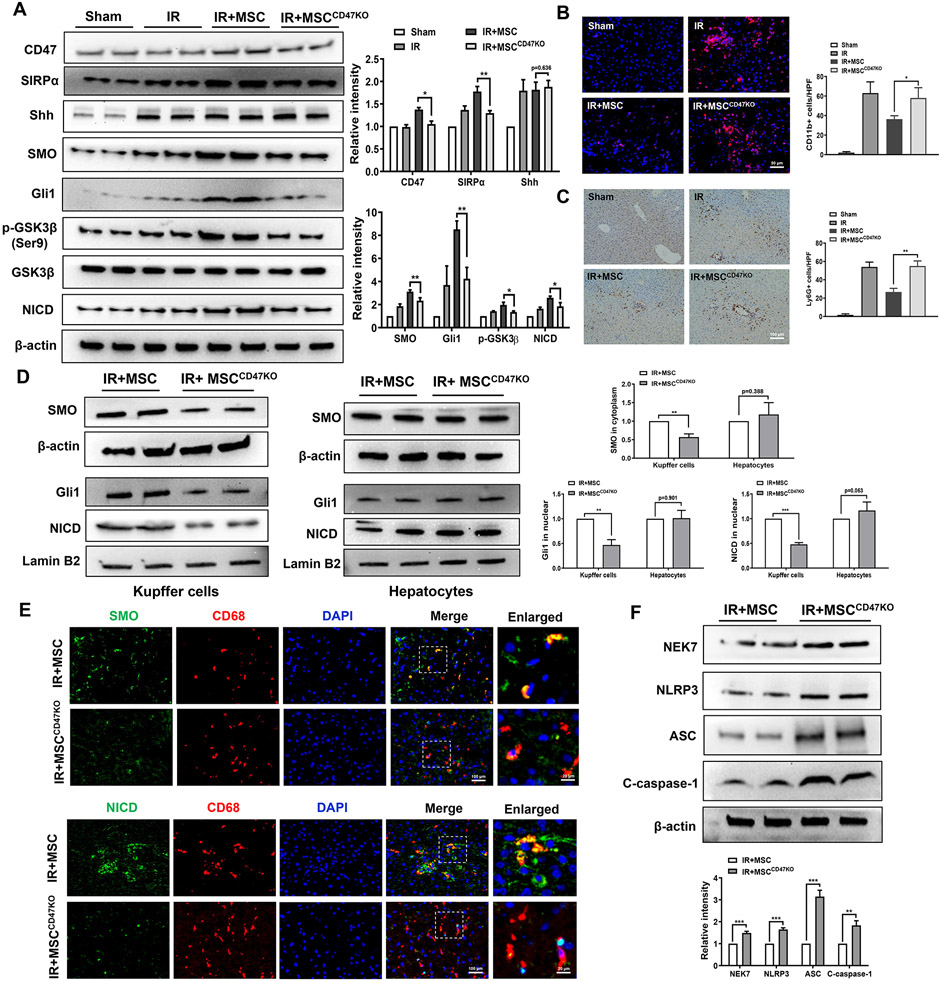
We then analyzed whether CD47 may influence the Hedgehog/SMO/Gli1 pathway and Notch1 signaling in MSC-treated livers after IR stress. Indeed, adoptive transfer of MSCs increased CD47 and SIRPα expression in IR stressed livers (Fig. 2A). MSC treatment augmented SMO, Gli1, p-GSK3β, and Notch1 intracellular domain (NICD) expression (Fig. 2A). However, CD47-deficient MSC treatment reduced SIRPα, SMO, Gli1, p-GSK3β, and NICD expression in ischemic livers (Fig. 2A). Unlike MSC-treated livers, CD47-deficient MSC treatment augmented CD11b+ macrophage and Ly6G+ neutrophil infiltration (Fig. 2B and 2C). Interestingly, reduced expression of SMO, nuclear Gli1, and NICD was observed in Kupffer cells but not in hepatocytes from WT mice treated with CD47-deficient MSCs, compared to the MSC control-treated groups (Fig. 2D). Immunofluorescence staining revealed that CD47-deficient MSC treatment diminished SMO and NICD expression in ischemic livers (Fig. 2E). The expression of NEK7, NLRP3, ASC, and cleaved caspase-1 was significantly decreased in MSC-treated livers (Fig. 2F). However, CD47-deficient MSC treatment markedly increased NEK7, NLRP3, ASC, and cleaved caspase-1 in ischemic livers (Fig. 2F).
Fig. 2. The CD47-SIRPα interaction activates Hedgehog/SMO/Gli1 pathway, Notch1 signaling, and inhibits NEK7/NLRP3 activation in IR-stressed livers.
WT mice were adoptive transferred MSCs or CD47-deficient MSCs (1x106) 24h prior to liver ischemic insult. (A) Western blot analysis and relative density ratio of CD47, SIRPα, Shh, SMO, Gli1, p-GSK3β, GSK3β, and NICD in ischemic livers. (B) Immunofluorescence staining of CD11b+ macrophages in ischemic livers (n=4-6 mice/group). Quantification of CD11b+ macrophages, Scale bars, 50μm. (C) Immunohistochemistry staining of Ly6G+ neutrophils in ischemic livers (n=4-6 mice/group). Quantification of Ly6G+ neutrophils, Scale bars, 100μm. (D) Western blot analysis and relative density ratio of SMO, Gli1, and NICD in Kupffer cells and hepatocytes. (E) Immunofluorescence staining for SMO or NICD expression in macrophages from the WT liver tissues (n=3-4 samples/group). DAPI was used to visualize nuclei. Scale bars, 100μm and 20μm. (F) Western blot analysis and relative density ratio of NEK7, NLRP3, ASC, and cleaved caspase-1 in ischemic livers. All Western blots represent three experiments, and the data represent the mean±SD. *p<0.05, **p<0.01, ***p<0.001.
Myeloid SMO deficiency in MSC-treated livers aggravates IR-induced hepatocellular damage and promotes NLRP3 inflammasome activation in IR-stressed livers.
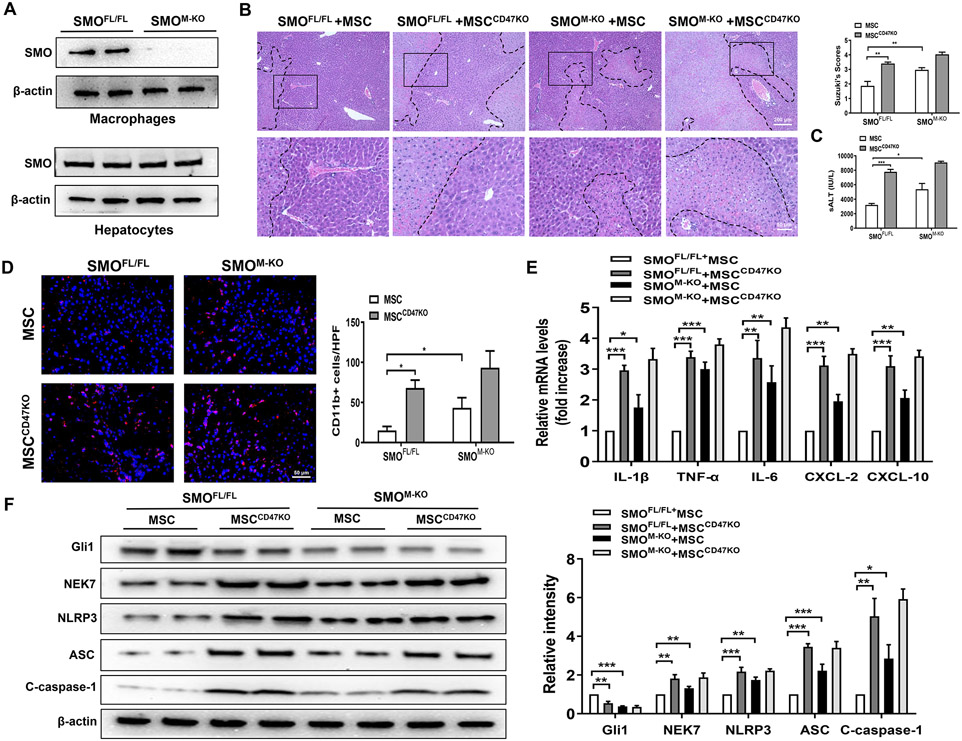
As increased SMO/Gli1 expression was found in hepatic Kupffer cells after MSC treatment, we next determined whether myeloid SMO/Gli1 may play a role in regulating the NLRP3 activation in IR-stressed livers after MSC treatment. We isolated both hepatocytes and liver macrophages (Kupffer cells) from the SMOFL/FL and SMOM-KO ischemic livers. SMOM-KO did not change hepatocyte SMO expression. However, the SMO expression was lacking in liver macrophages from the SMOM-KO but not the SMOFL/FL livers (Fig. 3A). Unlike livers in the MSC-treated SMOFL/FL controls, MSC-treated SMOM-KO or CD47-deficient MSC treatment augmented IR-induced liver damage as evidenced by the increased histological Suzuki's scores (Fig. 3B) and sALT levels (Fig. 3C). MSC-treated SMOM-KO or CD47-deficient MSC treatment increased CD11b+ macrophage infiltration (Fig. 3D) and proinflammatory IL-1β, TNF-α, IL-6, CXCL-2, and CXCL-10 expression (Fig. 3E). Strikingly, MSC-treated SMOM-KO or CD47-deficient MSC treatment markedly reduced Gli1 but augmented NEK7, NLRP3, ASC, and cleaved caspase-1 protein expression in ischemic livers (Fig. 3F), compared to the MSC-treated SMOFL/FL mice.
Fig. 3. Myeloid SMO deficiency in MSC-treated livers aggravates IR-induced hepatocellular damage and promotes NLRP3 inflammasome activation in IR-stressed livers.
The SMOFL/FL and SMOM-KO mice were adoptive transferred MSCs or CD47-deficient MSCs (1x106) 24h prior to liver ischemic insult. (A) Western blot analysis and relative density ratio of SMO in ischemic livers. (B) Representative histological staining (H&E) of ischemic liver tissue (n=4-6 mice/group) and Suzuki's histological score. Scale bars, 200μm and 50μm. (C) Serum ALT levels (IU/L) (n=4-6 samples/group). (D) Immunofluorescence staining of CD11b+ macrophages in ischemic livers (n=4-6 mice/group). Quantification of CD11b+ macrophages, Scale bars, 50μm. (E) qRT-PCR analysis of IL-1β, TNF-α, and IL-6, CXCL-2, and CXCL-10 (n=3-4 samples/group). (F) Western blot analysis and relative density ratio of Gli1, NEK7, NLRP3, ASC, and cleaved caspase-1 in ischemic livers. All Western blots represent three experiments, and the data represent the mean±SD. *p<0.05, **p<0.01, ***p<0.001.
Disruption of myeloid Notch1 in MSC-treated livers enhances NEK7/NLRP3 function in IR-stressed livers.
As MSCs promoted Notch1 signaling, we then determined the role of Notch1 signaling in MSC-mediated immune regulation. Unlike in MSC-treated Notch1FL/FL controls, MSC-treated Notch1M-KO or CD47-deficient MSC treatment increased IR-induced liver damage (Fig. 4A), sALT levels (Fig. 4B), CD11b+ macrophage (Fig. 4C) and Ly6G+ neutrophil (Fig. 4D) accumulation. Moreover, MSC-treated Notch1M-KO or CD47-deficient MSC treatment inhibited NICD but enhanced NEK7/NLRP3, ASC, and caspase-1 activation (Fig. 4E). The expression of IL-1β, TNF-α, IL-6, CXCL-2, and CXCL-10 was significantly increased in the MSC-treated Notch1M-KO or CD47-deficient MSC-treated livers, compared to the MSC-treated Notch1FL/FL controls (Fig. 4F).
Fig. 4. Disruption of myeloid Notch1 in MSC-treated livers enhances NEK7/NLRP3 function in IR-stressed livers.
The Notch1FL/FL and Notch1M-KO mice were adoptive transferred MSCs or CD47-deficient MSCs (1x106) 24h prior to liver ischemic insult. (A) Representative histological staining (H&E) of ischemic liver tissue (n=4-6 mice/group) and Suzuki's histological score. Scale bars, 200μm and 50μm. (B) Serum ALT levels (IU/L) (n=4-6 samples/group). (C) Immunofluorescence staining of CD11b+ macrophages in ischemic livers (n=4-6 mice/group). Quantification of CD11b+ macrophages, Scale bars, 50μm. (D) Immunohistochemistry staining of Ly6G+ neutrophils in ischemic livers (n=4-6 mice/group). Quantification of Ly6G+ neutrophils, Scale bars, 50μm. (E) Western blot analysis and relative density ratio of NICD, NEK7, NLRP3, ASC, and cleaved caspase-1 ischemic livers. (F) qRT-PCR analysis of IL-1β, TNF-α, IL-6, CXCL-2, and CXCL-10 (n=3-4 samples/group). All Western blots represent three experiments, and the data represent the mean±SD. *p<0.05, **p<0.01, ***p<0.001.
The CD47-SIRPα signaling regulates the interaction between macrophage Gli1 and NICD in MSC-mediated immune regulation.
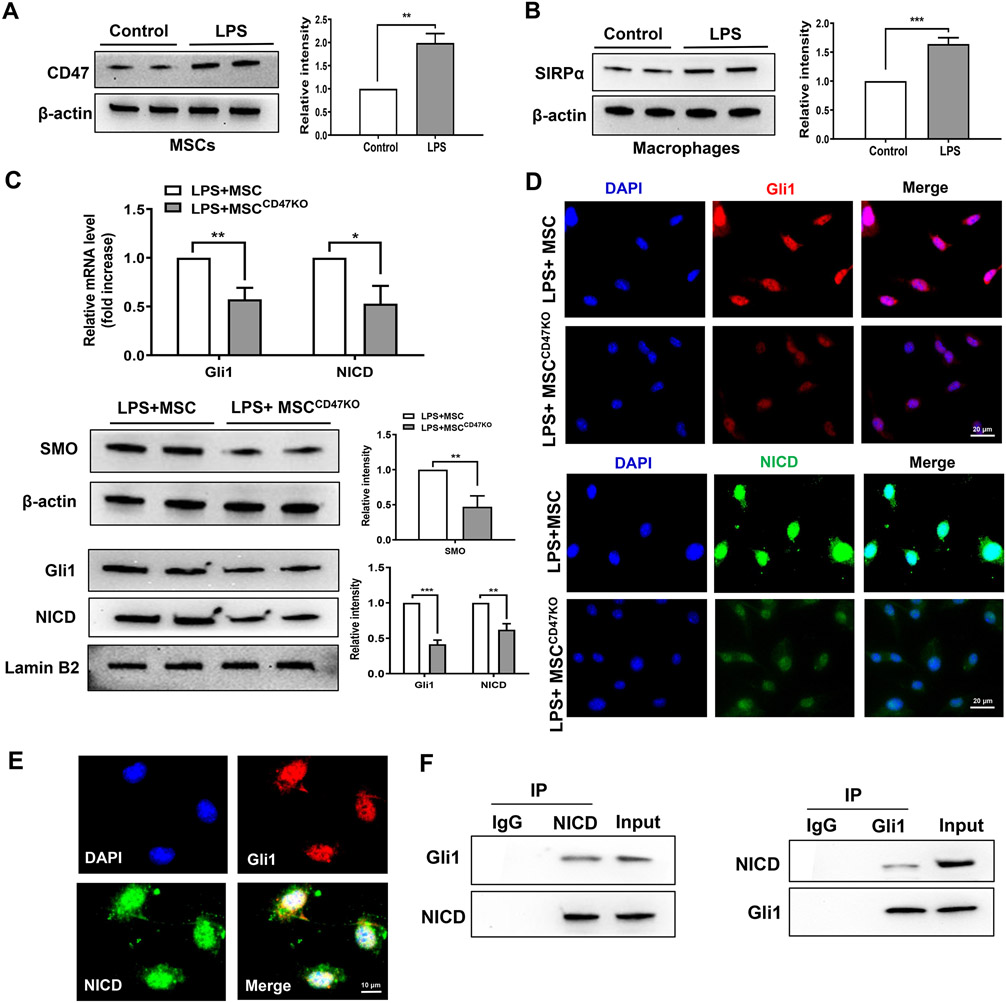
As the CD47-SIRPα interaction activates both Hedgehog/SMO/Gli1 and Notch1 pathways in the modulation of NLRP3 function after adoptive transfer of MSCs in vivo, we next tested whether there is crosstalk between the Hedgehog/SMO/Gli1 pathway and Notch1 signaling in MSC-mediated immune regulation. Using MSC/macrophage co-culture, we found increased CD47 expression in MSCs (Fig. 5A) and SIRPα expression in macrophages (Fig. 5B) after LPS stimulation. Moreover, increased mRNA and protein expression of Gli1 and NICD were observed in LPS-stimulated macrophages after co-culture with MSCs but not CD47-deficient MSCs (Fig. 5C). This was further confirmed by immunofluorescence staining, which showed_increased nuclear Gli1 and NICD expression in macrophages after co-culture with MSCs (Fig. 5D), whereas ablation of CD47 in MSCs diminished nuclear Gli1 and NICD expression in macrophages after co-culture (Fig. 5D). Interestingly, Gli1 and NICD were co-localized in the nucleus (Fig. 5E). Co-immunoprecipitation assays revealed that NICD bound to endogenous Gli1 in macrophages after co-culture with MSCs (Fig. 5F). Furthermore, disruption of SIRPα by CRISPR/Cas9-SIRPα KO reduced mRNA levels of Gli1 and NICD in LPS-stimulated macrophages after co-culture with MSCs (Supplementary Fig. 4A). Western blot assay showed that SIRPα deletion reduced nuclear Gli1 and NICD protein levels (Supplementary Fig. 4B). Immunofluorescence staining revealed that Gli1 and NICD expression were decreased (Supplementary Fig. 4C) in macrophages after co-culture with MSCs compared to the control vector-transfected cells. In contrast, induction of SIRPα by CRISPR-SIRPα activation increased Gli1 and NICD mRNA and protein expression (Supplementary Fig. 5A and 5B). Immunofluorescence staining showed that induction of SIRPα increased Gli1 and NICD expression (Supplementary Fig. 5C) in LPS-stimulated macrophages after transfection with CRISPR-SIRPα activation vector.
Fig. 5. The CD47-SIRPα signaling regulates the interaction between macrophage Gli1 and NICD in MSC-mediated immune regulation.
Bone marrow-derived macrophages (BMMs, 1x106) were co-cultured with MSCs or CD47-deficient MSCs (2x105) for 6h followed by LPS (100 ng/ml) stimulation. (A) (B) Western blot analysis and relative density ratio of CD47 and SIRPα in LPS-stimulated macrophages. (C) Analysis of mRNA levels and protein expression of Gli1 and NICD in macrophages after co-culture with MSCs or CD47-deficient MSCs by qRT-PCR and Western blot assay. (D) Immunofluorescence staining of nuclear Gli1 (red) and NICD (green) in macrophages after co-culture with MSCs or CD47-deficient MSCs. DAPI was used to visualize nuclei (blue). Scale bars, 20μm. (E) Immunofluorescence staining for macrophage Gli1 (red) and NICD (green) co-localization in the nucleus after co-culture with MSCs. DAPI was used to visualize nuclei (blue). Scale bars, 10μm. (F) Immunoprecipitation analysis of NICD and Gli1 in macrophages after co-culture with MSCs. All Western blots represent three experiments, and the data represent the mean±SD. *p<0.05, **p<0.01, ***p<0.001.
The Gli1-NICD interaction targets Dvl2 and modulates NEK7/NLRP3 function in MSC-mediated immune regulation.
To explore the potential mechanism of the Gli1-NICD interaction in the modulation of NEK7/NLRP3 activation in MSC-mediated immune regulation, we performed Gli1 ChIP coupled to massively parallel sequencing (ChIP-Seq) (Fig. 6A). Clearly, Gli1 ChIP-seq peaks were identified within the Dvl2 gene. One was located in the promoter region, and the others were located within the intron or exon (Fig. 6B). To validate the ChIP-seq peak located in the Dvl2 promoter region, ChIP-PCR was performed using Gli1 and NICD antibodies in MSC-treated BMMs. After ChIP with NICD or Gli1 antibody, the primer was designed to detect the Gli1 DNA-binding site in Dvl2 promoter by PCR analysis. To confirm that NICD is co-localized with Gli1 on the promoter of Dvl2, sequential ChIPs were performed. The first ChIP was performed with Gli1 antibody, and the second ChIP was carried out with either NICD or Gli1 antibody using the chromatin eluted from the first ChIP. Following the second ChIP, both NICD and Gli1 were still bound to the Gli1-binding motif in the Gli1-chromatin complex (Fig. 6C), confirming that NICD and Gli1 are present at the same promoter region of Dvl2. Hence, Dvl2 is a target gene regulated by the NICD-Gli1 complex. Moreover, RNA in situ hybridization assay revealed that the target gene Dvl2 transcript expression was upregulated in LPS-stimulated macrophages after co-culture with MSC (Fig. 6D). In contrast, disruption of CD47 in MSCs downregulated macrophage Dvl2 transcripts after co-culture (Fig. 6D). Increased Dvl2 and β-catenin and reduced NRX protein expression were observed in LPS-stimulated macrophages after co-culture with MSCs but not CD47-deficient MSCs (Fig. 6E). Disruption of CD47 in MSCs augmented macrophage NEK7, NLRP3, ASC, and cleaved caspase-1 expression (Fig. 6E) as well as increased proinflammatory IL-1β, TNF-α, IL-6, CXCL-2, and CXCL-10 after co-culture (Fig. 6F).
Fig. 6. The Gli1-NICD interaction targets Dvl2 and modulates NEK7/NLRP3 function in MSC-mediated immune regulation.
(A) Experimental design of Gli1 ChIP-seq analysis. BMMs were collected and fixed after co-culture with MSCs. Following chromatin shearing and Gli1 antibody selection, the precipitated DNA fragments bound by Gli1-containing protein complexes were used for sequencing. (B) Localization of Gli1-binding sites on the mouse Dvl2 gene. The 15 exons, 14 introns, 3' untranslated region (UTR), 5’ UTR, and transcription start sites (TSS) of the mouse Dvl2 gene on chromosome 11 are shown. (C) ChIP-PCR analysis of Gli1 and NICD binding to the Dvl2 promoter. Protein-bound chromatin was prepared from BMMs and immunoprecipitated with Gli1 or NICD antibodies. For sequential ChIP, the protein-bound chromatin was first immunoprecipitated with the Gli1 antibody followed by elution with a second immunoprecipitation using NICD antibody, and then the immunoprecipitated DNA was analyzed by PCR. The normal IgG was used as a negative control. (D) RNA in situ hybridization for Dvl2 transcripts in LPS-stimulated macrophages after co-culture with MSCs or CD47-deficient MSCs (n=3-4 samples/group). (E) Western blot analysis and relative density ratio of SMO, Gli1, NICD, Dvl2, NRX, NEK7, NLRP3, ASC, and cleaved caspase-1 in macrophages after co-culture with MSCs or CD47-deficient MSCs. (F) qRT-PCR analysis of IL-1β, TNF-α, IL-6, CXCL-2, and CXCL-10 in LPS-stimulated macrophages (n=3-4 samples/group). All Western blots represent three experiments, and the data represent the mean±SD. *p<0.05, **p<0.01, ***p<0.001.
Notch1 signaling is required for the Hedgehog/SMO/Gli1 pathway-mediated Dvl2 activation and NEK7/NLRP3 inhibition in MSC-mediated immune regulation.
To elucidate the mechanistic role of the Notch1 signaling in MSC-mediated immune regulation, we used macrophage/MSC co-culture followed by LPS stimulation. Immunofluorescence staining revealed that macrophage Gli1 expression was increased in SMO-proficient (SMOFL/FL) after co-culture with MSCs but not CD47-deficient MSCs (Fig. 7A). The protein expression of macrophage Dvl2 and β-catenin was augmented, while NRX and NEK7/NLRP3 were reduced after co-culture with MSCs (Fig. 7B). However, disruption of CD47 in MSCs reduced Dvl2 and β-catenin but augmented NRX and NEK7/NLRP3 expression in LPS-stimulated SMOFL/FL macrophages after co-culture (Fig. 7B). Interestingly, unlike in control vector-treated groups, immunofluorescence staining showed that CRISPR/Cas9-mediated Notch1 KO inhibited Dvl2 expression in macrophages after co-culture with MSCs (Fig. 7C). Moreover, Notch1 deficiency diminished Dvl2 and β-catenin but augmented NRX and NEK7/NLRP3 protein expression (Fig. 7D) and increased caspase-1 activity (Fig. 7E) and mRNA levels coding for IL-1β, TNF-α, IL-6, CXCL-2, and CXCL-10 (Fig. 7F) in macrophages after co-culture.
Fig. 7. Notch1 signaling is required for the Hedgehog/SMO/Gli1 pathway-mediated Dvl2 activation and NEK7/NLRP3 inhibition in MSC-mediated immune regulation.
(A) BMMs were isolated from SMOFL/FL mice and co-cultured with MSCs or CD47-deficient MSCs followed by LPS stimulation. Immunofluorescence staining for Gli1 expression in macrophages (n=3-4 samples/group). DAPI was used to visualize nuclei. Scale bars, 40μm and 20μm. (B) Western blot analysis and relative density ratio of Gli1, Dvl2, NRX, β-catenin, NEK7, and NLRP3 in LPS-stimulated macrophages. (C) BMMs were isolated from SMOFL/FL mice and transfected with CRISPR/Cas9-Notch1 KO or control vector, and then co-cultured with MSCs followed by LPS stimulation. Immunofluorescence staining for Dvl2 expression in macrophages (n=3-4 samples/group). DAPI was used to visualize nuclei. Scale bars, 100μm and 40μm. (D) Western blot analysis and relative density ratio of NICD, Dvl2, NRX, β-catenin, NEK7, and NLRP3 in LPS-stimulated macrophages. (E) Caspase-1 activity (U) in macrophages after co-culture (n=3-4 samples/group). (F) qRT-PCR analysis of TNF-α, IL-1β, IL-6, CXCL-2, and CXCL-10 (n=3-4 samples/group). All Western blots represent three experiments, and the data represent the mean±SD. *p<0.05, **p<0.01, ***p<0.001.
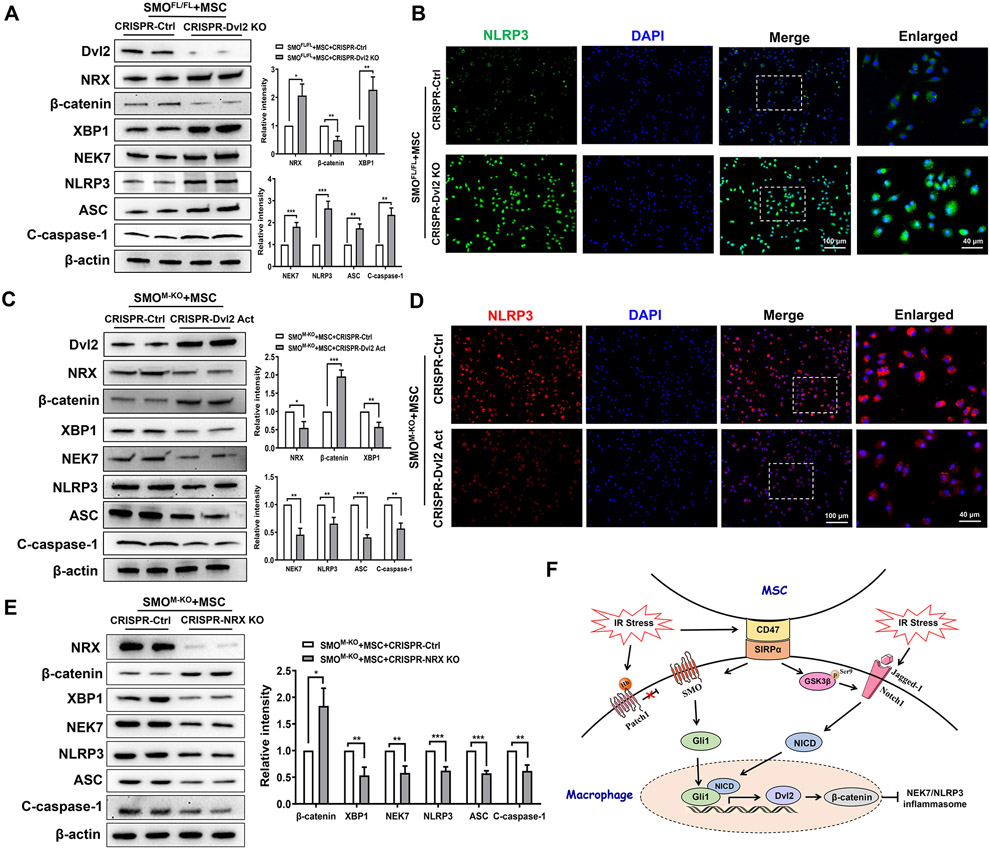
Dvl2 is crucial to regulate NLRP3-driven inflammatory response in MSC-mediated immune regulation.
To further test the functional role of Dvl2 in the regulation of NEK7/NLRP3 activation in MSC-mediated immune regulation, BMMs were isolated from the SMOFL/FL and SMOM-KO mice and co-cultured with MSCs. Indeed, CRISPR/Cas9-mediated Dvl2 KO diminished β-catenin while increasing NRX, XBP1, NEK7, NLRP3, and ASC cleaved caspase-1 expression in SMOFL/FL macrophages after co-culture with MSCs, compared to the control vector-treated controls (Fig. 8A). This result was confirmed by immunofluorescence staining, which showed that disruption of macrophage Dvl2 increased ROS production (Supplementary Fig. 6A) and NLRP3 expression (Fig. 8B) and pro-inflammatory IL-1β, TNF-α, IL-6, CXCL-2, and CXCL-10 expression (Supplementary Fig. 6B). Notably, CRISPR-mediated Dvl2 activation increased β-catenin yet diminished NRX, XBP1, NEK7, NLRP3, ASC, and cleaved caspase-1 expression in SMOM-KO macrophages after co-culture with MSCs (Fig. 8C). Activation of Dvl2 inhibited macrophage NLRP3 expression after co-culture with MSCs compared to the control vector-treated groups (Fig. 8D) by immunofluorescence staining. To determine the role of NRX in the regulation of NLRP3 activation in MSC-mediated immune regulation, BMMs from SMOM-KO mice were co-cultured with MSCs. Strikingly, CRISPR/Cas9-mediated NRX KO augmented β-catenin but diminished XBP1, NEK7, NLRP3, ASC, and cleaved caspase-1 expression in macrophages after co-culture with MSCs, compared to the control vector-treated controls (Fig. 8E).
Fig. 8. Dvl2 is crucial to regulate NLRP3-driven inflammatory response in MSC-mediated immune regulation.
BMMs were isolated from SMOFL/FL and SMOM-KO mice and transfected with the CRISPR/Cas9-Dvl2 KO, CRISPR-Dvl2 activation, CRISPR/Cas9-NRX KO or control vector, and then co-cultured with MSCs followed by LPS stimulation. (A) Western blots analysis and relative density ratio of Dvl2, NRX, β-catenin, XBP1, NEK7, NLRP3, ASC, and cleaved caspase-1 in LPS-stimulated macrophages after transfection of CRISPR/Cas9-Dvl2 KO or control vector. (B) Immunofluorescence staining for NLRP3 expression in macrophages (n=3-4 samples/group). DAPI was used to visualize nuclei. Scale bars, 100μm and 40μm. (C) Western blots analysis and relative density ratio of Dvl2, NRX, β-catenin, XBP1, NEK7, NLRP3, ASC, and cleaved caspase-1 in LPS-stimulated macrophages after transfection of CRISPR-Dvl2 activation or control vector. (D) Immunofluorescence staining for NLRP3 expression in macrophages (n=3-4 samples/group). DAPI was used to visualize nuclei. Scale bars, 100μm and 40μm. (E) Western blots analysis and relative density ratio of NRX, β-catenin, XBP1, NEK7, NLRP3, ASC, and cleaved caspase-1 in LPS-stimulated macrophages after transfection of CRISPR/Cas9-NRX KO or control vector. (F) The schematic figure depicts putative molecular mechanisms by which CD47-mediated Hedgehog/SMO/Gli1 signaling regulates NEK7/NLRP3 activation in MSC-mediated immune regulation. All Western blots represent three experiments, and the data represent the mean±SD. *p<0.05, **p<0.01, ***p<0.001.
Discussion
In this study, we reveal the mechanistic role of the CD47-mediated Hedgehog signaling in the modulation of NLRP3-driven inflammatory response in MSC-mediated immune regulation. We demonstrate that: i) the interaction between MSC CD47 and macrophage SIRPα activates Hedgehog/SMO/Gli1 pathway and Notch1 signaling in IR stressed livers; ii) NICD is co-localized and interacts with Gli1, which in turn regulates its target gene Dvl2 activation; iii) Dvl2 is crucial in the regulation of NEK7/NLRP3 activity in MSC-mediated immune regulation. Our results highlight the importance of the CD47-mediated Hedgehog/SMO/Gli1 signaling as a key regulator of the NLRP3 function in MSC-mediated immune regulation during IR stress-induced liver injury.
Although various studies have demonstrated that MSCs exert immunosuppressive effects via secreting multiple paracrine soluble factors (26), direct cell-cell interaction-dependent mechanisms may be crucial for MSC-mediated immune regulation. Indeed, MSCs possess broad immunoregulatory properties through interaction with immune cells in innate and adaptive immune systems (1). MSCs can induce differentiation of CD4+ T cells to regulatory T cells (Tregs) via secreting a multitude of immune-modulatory factors, cytokines, and growth factors (27). Our previous study showed that MSCs promoted the Hippo/YAP pathway and controlled macrophage-mediated inflammatory response through the secretion of PGE2 (25), suggesting that MSC-macrophage interaction exerts a distinct function in the modulation of innate immunity during inflammatory responses. The current studies revealed that adoptive transfer of MSCs promoted CD47 and SIRPα expression leading to alleviated IR-induced liver damage. However, CD47-deficient MSC treatment exacerbated IR-induced liver injury, indicating that MSC-mediated CD47 and its ligand receptor SIRPα are essential for MSC-mediated immune regulation in IR-stressed livers. Interestingly, MSC treatment promoted the Hedgehog/SMO/Gli1 pathway and activated Notch1 signaling whereas treatment with CD47-deficient MSCs inhibited Hedgehog downstream SMO/Gli1 and Notch1 intracellular domain (NICD) but enhanced NLRP3-inflammasome activation in IR-stressed livers. These results suggest that the CD47-SIRPα interaction plays a central role in the regulation of Hedgehog/SMO/Gli1 pathway and Notch1signaling in MSC-mediated immune regulation during liver IRI.
The Hedgehog/SMO/Gli1 pathway has been shown to regulate cell differentiation, tissue development, homeostasis, and regeneration (9). Hedgehog signaling is activated upon binding of the ligand to PTCH1, which leads to SMO activation of the transcriptional activator Gli1 (11). As increasing Gli1 activity inhibits pro-inflammatory mediators and deletion of Gli1 promotes immune cell activation and tissue inflammation (12), the Hedgehog/SMO/Gli1 pathway induced by CD47-SIRPα interaction could function as a negative regulator of NLRP3-driven inflammatory response during liver IRI. As expected, we found that myeloid SMO deficiency exacerbated IR-induced liver damage even with concomitant MSC treatment. Disruption of SMO activated NEK7, an essential mediator for the initiation of NLRP3 activation (28). Notably, MSC treatment augmented Gli1 and inhibited NEK7/NLRP3 activation in SMO-proficient mice. However, adoptive transfer of CD47-deficient MSCs into SMO-proficient mice depressed Gli1 but enhanced NLRP3 activation accompanied by increased proinflammatory mediators in IR-stressed livers, suggesting the CD47-SIRPα interaction activates the Hedgehog/SMO/Gli1 pathway, which in turn inhibits NEK7/NLRP3 activity in IR-triggered liver inflammation.
We also found that CD47-SIRPα interaction activated Notch1 signaling in IR-stressed livers after MSC treatment. Indeed, the Notch signaling pathway relies on a proteolytic cascade to release its transcriptionally active intracellular domain (NICD), which binds to the nuclear recombinant recognition sequence binding protein at the Jκ site (RBP-J) to activate Notch target genes (29). Disruption of the transcription factor RBP-J increases cell apoptosis/necrosis and aggravated liver inflammatory injury (16). Increasing Notch1 expression contributes to cell growth and survival during liver regeneration (15). Moreover, our recent studies have demonstrated that activation of Notch1 modulates liver inflammatory response by controlling TLR4 signaling to program innate and adaptive immunity (17), suggesting that Notch signaling regulates innate immune cell homeostasis and function. In line with these findings, our current results revealed that disruption of CD47 in MSCs reduced NICD while augmenting NEK7/NLRP3 activity and IR-induced inflammatory injury, implying that Notch1 could be one of the critical signaling molecules in MSC-mediated immune regulation.
As our current studies implicate that both Hedgehog/SMO/Gli1 pathway and Notch1 signaling are involved in the regulation of NLRP3 function in liver IRI after MSC intervention, the question arises as to what molecular mechanisms may confer the Hedgehog/SMO/Gli1 signaling with its ability to regulate NLRP3 function in MSC-mediated immune regulation. Using MSC/macrophage co-culture system, we found that the expression of CD47 in MSCs and SIRPα in macrophages was markedly increased after LPS stimulation. This indicates that CD47-SIRPα interaction is vital for cross-communication between MSCs and macrophages in MSC-mediated immune regulation. In line with our in vivo findings, which showed that MSC treatment augmented Gli1 and NICD, while CD47-deficient MSC treatment reduced Gli1 and NICD nuclear translocation in Kupffer cells from IR-stressed liver, our in vitro data revealed that disruption of SIRPα reduced mRNA and protein levels of Gli1 and NICD in LPS-stimulated macrophages after co-culture with MSCs (Supplementary Fig. 4). Moreover, activation of SIRPα augmented Gli1 and NICD activity in LPS-stimulated macrophages (Supplementary Fig. 5). These results suggest that CD47-SIRPα interaction is essential for modulating Gli1 and NICD activity in MSC-mediated immune regulation. Thus, we speculate that nuclear localization of endogenous Gli1 and NICD is necessary for transcriptional activity in MSC-mediated immune regulation during liver IRI. Our in vitro study provided further evidence, revealing that macrophage Gli1 and NICD co-localized in the nucleus and increased nuclear expression of Gli1 and NICD in response to LPS stimulation. Notably, NICD interacted with Gli1 via direct binding. Furthermore, the ChIP and ChIP-sequencing data revealed that NICD was co-localized with Gli1 on the promoter of Dvl2, suggesting that Dvl2 is a target gene of Gli1 regulated by the Gli1 and NICD complex. Moreover, disruption of Notch1 signaling reduced Dvl2 expression, indicating that NICD acts as a transcriptional coactivator of Gli1 in MSC-mediated immune regulation.
Another striking finding is that Dvl2 is key for controlling NLRP3 function in MSC-mediated immune regulation. Indeed, Dvl2 is an essential component of the Wnt/β-catenin pathway (30). The Wnt signal is propagated following the binding of the Wnt ligand to a heterodimeric complex consisting of a Frizzled receptor and an LDL-receptor-related protein (LRP) 5/6 protein, which recruits Dishevelled (Dvl) protein resulting in inhibition of β-catenin phosphorylation and thereby promoting β-catenin stabilization (31). Activation of Dvl2 stimulated β-catenin-dependent transcription of Wnt target genes (32), while disruption of Dvl2 resulted in constitutive β-catenin degradation and failure to β-catenin accumulation in response to the Wnt signal (33), suggesting that Dvl2 serves as a critical regulator of the β-catenin signaling. In line with these findings, we found that MSCs promoted Dvl2 transcript expression, whereas disruption of the SMO inhibited Dvl2 transcript expression (Supplementary Fig. 7A) accompanied by diminished protein levels of Gli1, Dvl2, and β-catenin but augmented NRX and NEK7/NLRP3 levels (Supplementary Fig. 7B) in LPS-stimulated macrophages after co-culture. MSC treatment activated macrophage Notch1 signaling while ablation of Notch1 diminished Dvl2 and augmented NRX expression after co-culture. These data indicate that both the Hedgehog/SMO/Gli1 and Notch1 pathways can positively regulate Dvl2 function in MSC-mediated immune regulation. NRX, a thioredoxin-related redox-regulating protein, is a negative regulator of β-catenin activation (34). Our previous study has demonstrated that activation of β-catenin inhibits IR-triggered liver inflammation via modulating its target gene XBP1 activation (20, 25). Thus, we speculate that Dvl2 controls NLRP3-driven liver inflammation through the regulation of NRX/β-catenin/XBP1 signaling. As expected, disruption of macrophage Dvl2 augmented NRX and XBP1 but reduced β-catenin expression, whereas activation of Dvl2 inhibited NRX and XBP1 but enhanced β-catenin signaling leading to reduced NEK7/NLRP3 activity after co-culture with MSCs. Consistent with this finding, disruption of NRX promoted β-catenin signaling and inhibited XBP1 and NLRP3 function in SMO-deficient macrophages after co-culture with MSCs. Taken together, these results reveal a novel role of Dvl2 in controlling dynamic crosstalk with the NLRP3 in MSC-mediated immune regulation.
Figure 8F depicts putative molecular mechanisms by which CD47-mediated Hedgehog/SMO/Gli1 signaling may regulate NEK7/NLRP3 activation in MSC-mediated immune regulation. The interaction between MSC CD47 and macrophage SIRPα activates the Hedgehog/SMO/Gli1 signaling and Notch1, leading to increased macrophage Gli1 and NICD nuclear translocation. Strikingly, macrophage Gli1 and NICD co-localized in the nucleus, whereby NICD interacted with Gli1 and regulated its target gene Dvl2, which in turn inhibited NEK7/NLRP3 activity in IR-triggered liver inflammation.
In conclusion, we identify a previously unrecognized role of CD47-mediated Hedgehog/SMO/Gli1 signaling in regulating NEK7/NLRP3 function in MSC-mediated immune regulation. Our findings demonstrate that Hedgehog/SMO/Gli1 signaling controls NLRP3-driven liver inflammation through a direct interaction between Gli1 and NICD. NICD is a novel coactivator of Gli1, and the target gene Dvl2 regulated by the NICD-Gli1 complex is crucial for the modulation of NEK7/NLRP3-driven inflammatory response in MSC-mediated immune regulation. Our findings provide novel potential therapeutic targets in MSC immunotherapy of liver sterile inflammatory injury.
Supplementary Material
Financial support:
This work was supported by the NIH Grants (R01AI139552, R21AI146742, R21AI112722, R21AI115133 to B.KE), (P01AI120944, R01DK062357 to J.W.KW), and The Dumont Research Foundation.
Abbreviations:
- ASC
apoptosis-associated speck-like protein containing a caspase recruitment domain
- BMMs
bone marrow-derived macrophages
- CD47
cluster of differentiation 47
- ChIP
chromatin immunoprecipitation
- ChIP-seq
ChIP-sequencing
- CMFDA
5-chloromethylfluorescein diacetate
- CRISPR
clustered regularly interspaced short palindromic repeats
- Cas9
CRISPR associated protein 9
- Dvl2
dishevelled segment polarity protein 2
- Gli1
GLI family zinc finger 1
- GSK3β
glycogen synthase kinase 3β
- IRI
ischemia/reperfusion injury
- LPS
lipopolysaccharide
- MSCs
mesenchymal stem cells
- NICD
Notch1 intracellular domain
- NEK7
NIMA related kinase 7
- NLRP3
NLR family pyrin domain containing 3
- NRX
nucleoredoxin
- ROS
reactive oxygen species
- sALT
serum alanine aminotransferase
- Shh
sonic hedgehog
- SIRPα
signal regulatory protein alpha
- SMO
smoothened
- XBP1
X-box-binding protein 1
References
- 1.Contreras RA, Figueroa FE, Djouad F, Luz-Crawford P. Mesenchymal Stem Cells Regulate the Innate and Adaptive Immune Responses Dampening Arthritis Progression. Stem Cells Int 2016;2016:3162743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang LT, Ting CH, Yen ML, Liu KJ, Sytwu HK, Wu KK, et al. Human mesenchymal stem cells (MSCs) for treatment towards immune- and inflammation-mediated diseases: review of current clinical trials. J Biomed Sci 2016;23:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galipeau J, Sensebe L. Mesenchymal Stromal Cells: Clinical Challenges and Therapeutic Opportunities. Cell Stem Cell 2018;22:824–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindberg FP, Gresham HD, Schwarz E, Brown EJ. Molecular cloning of integrin-associated protein: an immunoglobulin family member with multiple membrane-spanning domains implicated in alpha v beta 3-dependent ligand binding. J Cell Biol 1993;123:485–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaiswal S, Jamieson CH, Pang WW, Park CY, Chao MP, Majeti R, et al. CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell 2009;138:271–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soto-Pantoja DR, Kaur S, Roberts DD. CD47 signaling pathways controlling cellular differentiation and responses to stress. Crit Rev Biochem Mol Biol 2015;50:212–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matozaki T, Murata Y, Okazawa H, Ohnishi H. Functions and molecular mechanisms of the CD47-SIRPalpha signalling pathway. Trends Cell Biol 2009;19:72–80. [DOI] [PubMed] [Google Scholar]
- 8.Kong XN, Yan HX, Chen L, Dong LW, Yang W, Liu Q, et al. LPS-induced down-regulation of signal regulatory protein {alpha} contributes to innate immune activation in macrophages. J Exp Med 2007;204:2719–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ingham PW, Nakano Y, Seger C. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat Rev Genet 2011;12:393–406. [DOI] [PubMed] [Google Scholar]
- 10.Petrova R, Joyner AL. Roles for Hedgehog signaling in adult organ homeostasis and repair. Development 2014;141:3445–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Briscoe J, Therond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol 2013;14:416–429. [DOI] [PubMed] [Google Scholar]
- 12.Mathew E, Collins MA, Fernandez-Barrena MG, Holtz AM, Yan W, Hogan JO, et al. The transcription factor GLI1 modulates the inflammatory response during pancreatic tissue remodeling. J Biol Chem 2014;289:27727–27743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JJ, Rothenberg ME, Seeley ES, Zimdahl B, Kawano S, Lu WJ, et al. Control of inflammation by stromal Hedgehog pathway activation restrains colitis. Proc Natl Acad Sci U S A 2016;113:E7545–E7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Radtke F, MacDonald HR, Tacchini-Cottier F. Regulation of innate and adaptive immunity by Notch. Nat Rev Immunol 2013;13:427–437. [DOI] [PubMed] [Google Scholar]
- 15.Kohler C, Bell AW, Bowen WC, Monga SP, Fleig W, Michalopoulos GK. Expression of Notch-1 and its ligand Jagged-1 in rat liver during liver regeneration. Hepatology 2004;39:1056–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu HC, Qin HY, He F, Wang L, Fu W, Liu D, et al. Canonical notch pathway protects hepatocytes from ischemia/reperfusion injury in mice by repressing reactive oxygen species production through JAK2/STAT3 signaling. Hepatology 2011;54:979–988. [DOI] [PubMed] [Google Scholar]
- 17.Lu L, Yue S, Jiang L, Li C, Zhu Q, Ke M, et al. Myeloid Notch1 deficiency activates the RhoA/ROCK pathway and aggravates hepatocellular damage in mouse ischemic livers. Hepatology 2018;67:1041–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin Y, Li C, Xu D, Zhu J, Wei S, Zhong A, et al. Jagged1-mediated myeloid Notch1 signaling activates HSF1/Snail and controls NLRP3 inflammasome activation in liver inflammatory injury. Cell Mol Immunol 2020;17:1245–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swiderska-Syn M, Syn WK, Xie G, Kruger L, Machado MV, Karaca G, et al. Myofibroblastic cells function as progenitors to regenerate murine livers after partial hepatectomy. Gut 2014;63:1333–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ke B, Shen XD, Kamo N, Ji H, Yue S, Gao F, et al. beta-catenin regulates innate and adaptive immunity in mouse liver ischemia-reperfusion injury. Hepatology 2013;57:1203–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki S, Toledo-Pereyra LH, Rodriguez FJ, Cejalvo D. Neutrophil infiltration as an important factor in liver ischemia and reperfusion injury. Modulating effects of FK506 and cyclosporine. Transplantation 1993;55:1265–1272. [DOI] [PubMed] [Google Scholar]
- 22.Yue S, Zhu J, Zhang M, Li C, Zhou X, Zhou M, et al. The myeloid heat shock transcription factor 1/beta-catenin axis regulates NLR family, pyrin domain-containing 3 inflammasome activation in mouse liver ischemia/reperfusion injury. Hepatology 2016;64:1683–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li C, Kong Y, Wang H, Wang S, Yu H, Liu X, et al. Homing of bone marrow mesenchymal stem cells mediated by sphingosine 1-phosphate contributes to liver fibrosis. J Hepatol 2009;50:1174–1183. [DOI] [PubMed] [Google Scholar]
- 24.Chakrabarti R, Celia-Terrassa T, Kumar S, Hang X, Wei Y, Choudhury A, et al. Notch ligand Dll1 mediates cross-talk between mammary stem cells and the macrophageal niche. Science 2018;360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li C, Jin Y, Wei S, Sun Y, Jiang L, Zhu Q, et al. Hippo Signaling Controls NLR Family Pyrin Domain Containing 3 Activation and Governs Immunoregulation of Mesenchymal Stem Cells in Mouse Liver Injury. Hepatology 2019;70:1714–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JW, Fang X, Krasnodembskaya A, Howard JP, Matthay MA. Concise review: Mesenchymal stem cells for acute lung injury: role of paracrine soluble factors. Stem Cells 2011;29:913–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Negi N, Griffin MD. Effects of mesenchymal stromal cells on regulatory T cells: Current understanding and clinical relevance. Stem Cells 2020;38:596–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He Y, Zeng MY, Yang D, Motro B, Nunez G. NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature 2016;530:354–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 2009;137:216–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boutros M, Paricio N, Strutt DI, Mlodzik M. Dishevelled activates JNK and discriminates between JNK pathways in planar polarity and wingless signaling. Cell 1998;94:109–118. [DOI] [PubMed] [Google Scholar]
- 31.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell 2009;17:9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang X, McGann JC, Liu BY, Hannoush RN, Lill JR, Pham V, et al. Phosphorylation of Dishevelled by protein kinase RIPK4 regulates Wnt signaling. Science 2013;339:1441–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gammons MV, Rutherford TJ, Steinhart Z, Angers S, Bienz M. Essential role of the Dishevelled DEP domain in a Wnt-dependent human-cell-based complementation assay. J Cell Sci 2016;129:3892–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Funato Y, Michiue T, Asashima M, Miki H. The thioredoxin-related redox-regulating protein nucleoredoxin inhibits Wnt-beta-catenin signalling through dishevelled. Nat Cell Biol 2006;8:501–508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.