Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is evolving with mutations in the spike protein, especially in the receptor-binding domain (RBD). The failure of public health measures in some countries to contain the spread of the disease has given rise to novel viral variants with increased transmissibility. However, key questions about how quickly the variants can spread remain unclear. Herein, we performed a structural investigation using molecular dynamics simulations and determined dissociation constant (KD) values using surface plasmon resonance assays of three fast-spreading SARS-CoV-2 variants, alpha, beta, and gamma, as well as genetic factors in host cells that may be related to the viral infection. Our results suggest that the SARS-CoV-2 variants facilitate their entry into the host cell by moderately increased binding affinities to the human ACE2 receptor, different torsions in hACE2 mediated by RBD variants, and an increased spike exposure time to proteolytic enzymes. We also found that other host cell aspects, such as gene and isoform expression of key genes for the infection (ACE2, FURIN, and TMPRSS2), may have few contributions to the SARS-CoV-2 variant infectivity. In summary, we concluded that a combination of viral and host cell factors allows SARS-CoV-2 variants to increase their abilities to spread faster than the wild type.
Introduction
We are facing an unprecedented pandemic situation caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2),1,2 responsible for the death of almost 6.3 million people and infected more than 524 million people worldwide, as on May 20, 2022.3 The first outbreak of SARS-CoV-2 was reported in December 2019 at Wuhan, China, causing a severe respiratory syndrome known as Coronavirus Disease 2019 (COVID-19).3 The virus surface glycoprotein spike (S) is composed of two subunits, S1 and S2. S1 has the receptor-binding domain (RBD) that interacts with the human angiotensin-converting enzyme 2 (hACE2),4 while S2 mediates the viral and host cellular membrane fusion.5,6 The process of virus invasion occurs only after the cleavage of two proteolytic sites in the S protein, S1/S2, processed by a furin-like protein convertase (FURIN), and S2′, processed by a transmembrane protease surface serine 2 (TMPRSS2).7−10 A third protease, lysosomal cysteine protease cathepsin L (CTSL), also enhances virus entry.10 These proteolytic processes cleave the S1 subunit and induce drastic conformational changes in the S2 subunit, allowing the membrane fusion and release of virus genetic information inside the host cell.
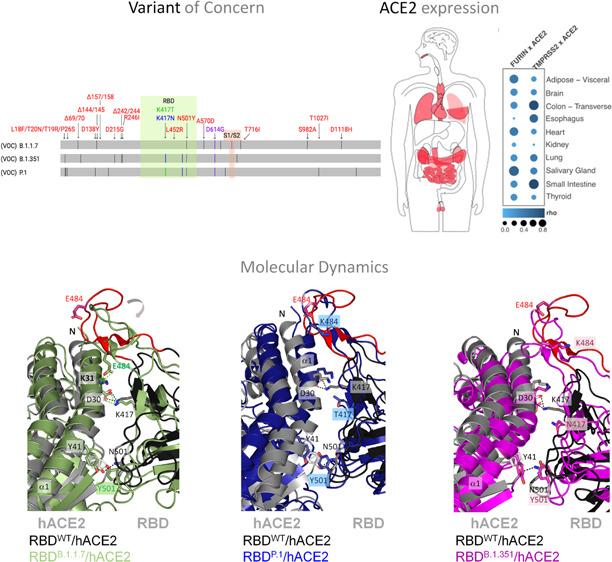
Actually, the world has already gone through three waves, and others are plausible with the emergence of novel SARS-CoV-2 variants. According to World Health Organization criteria, a variant of concern (VOC) is defined mainly based on its increased transmissibility, higher virulence, or altered clinical characteristics of the disease.11 The first VOC was reported in the United Kingdom, also called B.1.1.7 (alpha variant; September 2020), which quickly became prevalent in the British population.12 Now, there are three more worldwide spread variants: (i) B.1.351, which originated in South Africa (beta variant; May 2020); (ii) P.1, which originated in Brazil (gamma variant; November 2020); (iii) and B.1.617.2, which originated in India (delta variant; October 2020).13 It is evident that these variants have already affected the trajectory of the pandemic in countries where they originated. The SARS-CoV-2 population seems to be still adapting to its human host because an accumulation of multiple mutations in the S protein is being observed in new variants (Figure 1a). Some virus variants have been reported to increase transmissibility and lethality and to be associated with decreased effectiveness of neutralizing antibodies (Tables S1 and S2).14,15 Herein, we performed computational and experimental assays to investigate the changes in the binding affinity of three fast-spreading SARS-CoV-2 variants, the alpha, beta and gamma ones, and their cellular entrance, the hACE2. We thus discuss the possible implications of our molecular interaction results for the higher spread and infectivity of the variants.
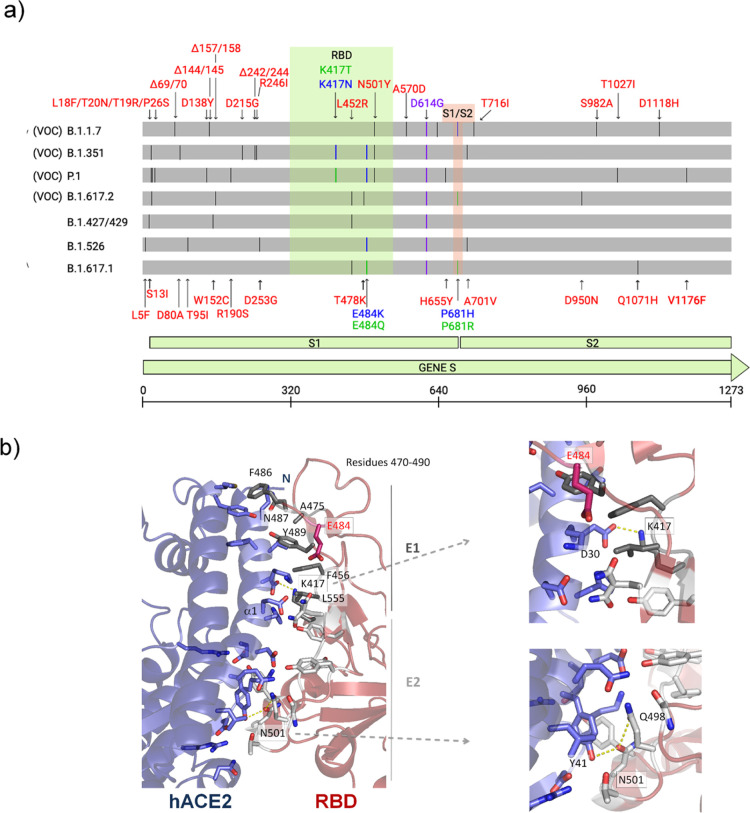
Figure 1.
Illustration of mutations in the S protein gene in different variants of SARS-CoV-2. (a) The RBD (residues 333–526) and the furin cleavage site (S1/S2) (residues 682–685) are colored in light green and salmon boxes, respectively. All mutations are represented by black lines and typed in red. Mutations that are found to be mutated for different residues are colored in blue and green. The green line shows different mutations in the same location. All SARS-CoV-2 variants shown in the figure have D614G, an advantage mutation for SARS-CoV-2. D614G has enhanced spike stability and transmission but does not significantly increase the binding affinity for hACE2 at 37 °C.37−39 VOC: variant of concern. (b) Cartoon representation of the RBD/hACE2 interface complex (PDB ID: 6M0J4). RDB presents two different regions, E1 (residues 417, 455–456, and 470–490) and E2 (444–454 and 493–505). The residues mutated in RBD of SARS-CoV-2 variants are K417, E484 (does not make part of the interface), and N501. RBDB.1.1.7 (alpha variant) has the mutation N501Y; RBDB.1.351 (beta variant) has K417N, E484K, and N501Y mutations; and RBDP.1 (gamma variant) has K417T, E484K, and N501Y mutations.
Materials and Methods
The initial coordinates for the RBD/hACE2 complex were obtained from the Protein Data Bank (PDB ID 6M0J), and RBD mutations were obtained from a site-directed mutation in wild-type RBD. From the ACE2/RBD complex structures, protonation states of ionizable residues were computed in an implicitly aqueous environment at pH 7.0 (more details are in the Supporting Information). The systems were explicitly solvated with TIP3P water models in a triclinic box and neutralized keeping the NaCl concentration at 150 mM. The systems were consecutively equilibrated in isothermal–isochoric (NVT) ensemble, relaxed during a 5000 ps annealing, and equilibrated in isothermal–isobaric (1 bar; NpT) ensembles at 310 K for 1000 ps. MD simulations in the NpT ensemble were carried out for 100 ns in a periodic box, in which the Zn2+ position was restricted throughout the simulations. In order to confirm the convergence of the simulation between hACE2 and RBD of the alpha (B.1.1.7) and gamma (P.1) variants and the wild type (WT), we simulated 300 ns of these complexes, using the same conditions used in the simulations of 100 ns. All details about computational analyses and the binding free energy calculation methods are provided in the Supporting Information. Experimental equilibrium dissociation constants (KD’s) calculated for RBD and its variants in the complex with dimeric hACE2 protein were obtained by surface plasmon resonance (SPR) assays using a Biacore 8K system (GE Healthcare), as described in more detail in the Supporting Information. To investigate the expression profiles of ACE2, FURIN, and TMPRSS2, we gathered transcripts per million (TPM) normalized gene expression data of 16,705 samples from 32 healthy tissues directly at the GTEx (genotype-tissue expression) portal (https://gtexportal.org/), where gender and age information were also collected. We downloaded RNA sequencing data sets to investigate isoform-based expression levels of ACE2 at ENA projects; more information about the genes and splicing isoforms expression analyses are provided in the Supporting Information.
Results and Discussion
In order to evaluate the potential impact of fast-spreading variants B.1.1.7 (RBDB.1.1.7), B.1.351 (RBDB.1.351), and P.1 (RBDP.1) on their interaction with hACE2, we performed molecular dynamic (MD) simulations for 100 ns using RBD (residues 333–526) and hACE2 (residues 19–615) (Figures 2 and S1). SARS-CoV-2 RDB binds to hACE2 through two different regions, E1 (residues 417, 455–456, and 470–490) and E2 (residues 444–454 and 493–505) (Figure 1b).16 While E1 mediates more hydrophobic interactions, E2 mediates mostly polar interactions with hACE2. RBDP.1 has mutations in K417T, E484K, and N501Y; RBDB.1.351 has mutations in K417N, E484K, and N501Y; and RBDB.1.1.7 has only the N501Y mutation. K417 and N501 are in the protein complex interface, whereas E484 is located in a mobile loop distant from the interface (Figure 1b). From ref (17), we verified that mutations in the protein interface cause a hydrophobicity gain (N501Y of +2.2 kcal/mol, K417T of +4.6 kcal/mol, and K417N of +0.4 kcal/mol) that may result in an enhancement of complex stability.
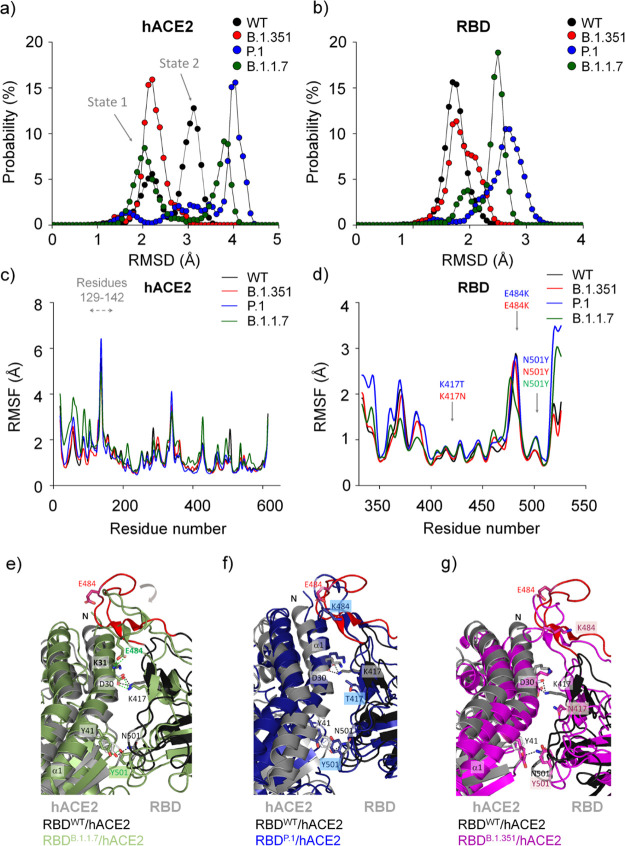
Figure 2.
Structural aspects obtained by molecular dynamics simulations of different SARS-CoV-2 RBD variants while interacting with hACE2. Backbone RMSD probability distributions are shown for (a) hACE2 and (b) RBD. Backbone RMSF are shown for (c) hACE2 and (d) RBD. The Panels (e–g) show different conformational changes of each RBDvariant/hACE2 complex in relation to the RBDWT/hACE2 complex. Note that the main structural differences are localized in the loop 474–488 caused by the N501Y mutation in RBDB.1.1.7 (e) that is partially recovered in RBDP.1 (f) and RBDB.1.351 (g). Residues 469−490 are colored in red in the RBDWT/hACE2 complex. The structure models of RBDWT/hACE2, RBDB.1.1.7/hACE2, RBDP.1/hACE2 and RBDB.1.351/hACE2 were taken from a snapshot at 40 ns, 19 ns, 61 ns, and 42 ns, respectively, from each MD simulation trajectory. We observed in another independent calculation simulated for 300 ns for hACE2 in complex with RBDWT, RBDB.1.1.7, and RBDP.1, showing the same structural behavior (Figures S15–S17).
MD simulation analysis showed that hACE2 presented different backbone root-mean-square deviation (RMSD) and backbone root-mean-square fluctuation (RMSF) profiles in the presence of different RBD variants: RBDB.1.1.7, RBDB.1.351, and RBDP.1 (Figures 2 and S2). Consistent with our previous study,16 hACE2 sampled in two major conformational states (state 1 of RMSD of ∼2.4 Å and state 2 of ∼3.2 Å RMSD, Figure 2a), characterized by a significant motion of residues 129–142 located distantly from the interface (Figure 2c). In its interaction with RBDWT and RBDB.1.1.7, hACE2 presented RMSDWT values of ∼2.2 and ∼3.1 Å and RMSDB.1.1.7 values of ∼2.0 and ∼3.8 Å, both sampling two conformational states. We verified that this conformational change is also related to the closing of the hACE2 enzymatic cavity caused by RBD. In contrast, in the interaction with RBDP.1, hACE2 sampled three states, presenting RMSD values of ∼1.8, ∼2.8, and ∼4.0 Å (Figure 2a). While interacting with RBDB.1.351, hACE2 visits one major state (closed enzymatic cavity) (Figure 2a). Probability distributions of the backbone RMSD along the MD trajectory showed that RBD mutants sampled a broader conformational ensemble in comparison to RBDWT (Figure 2b). Interestingly, hACE2 presents significant changes in the solvent-accessible surface area (SASA) in complex with different RBD variants, while RBDs share similar values (Figure S4). This observation suggests that RBDs induce different conformational states of hACE2.
Curiously, the RMSF profile is slightly different for the RBDB.1.1.7/hACE2 complex in relation to the others: while RBDB.1.1.7 decreases its RMSF values for residues 350–400 (distance of the interface), hACE2 increases its RMSF values for residues 19–140, which are located in the interface (Figure 2c−d). The K417 mutation does not significantly affect its RMSF values, yet mutation in N501Y slightly increases its flexibility in RBDP.1 and RBDB.1.1.7, and it is restored in RBDB.1.351. Intriguingly, the N501Y mutation in RBDB.1.1.7 causes a loss of flexibility in residues 480–486, which is restored by the E484K mutation in RBDP.1 and RBDB.1.351 (Figure 2d–g). Based on the analysis of the snapshots of RBD/hACE2 complexes, it is evident that the mutation of N501Y in RBDB.1.1.7 causes a significant folding in the loop containing residues 474–488 (the more mobile region of the RBD), favoring the formation of a hydrogen bonding between E484RBD and K31hACE2 (Figure 2e and Table S3). In agreement, a similar folding is also observed in the cryo-electron microscopy of the trimeric spike structure of the B.1.1.7 variant (Figure S3).18 Our analyses of hydrogen bonding occupancies revealed the absence of hydrogen-bonding interaction between E484RBD and K31hACE2 in the other complexes (Figure 2f,g; Table S3). Additionally, RBDP.1 and RBDB.1.351 have an E484K mutation, causing a charge-repelling impact with K31hACE2. These variants also present the mutations K417T and K417N, respectively. K417 and E484 mutations seem to reverse the effect of the N501Y mutation in the loop of residues 474–488, restoring a more original conformational state (Figures 2c–g and S2a,b).
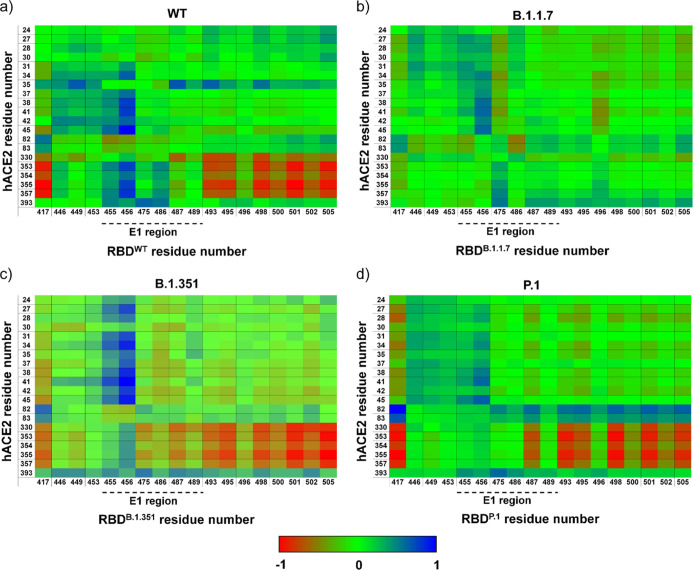
We also calculated the correlation matrix of the spatial displacements of Cα atoms to study internal motions of the hACE2/RBDWT complex and compared them with different SARS-CoV-2 RBD mutants (Figures 3 and S5; Tables S4 and S5). The sets of intermolecular and intramolecular correlation pairs changed between different SARS-CoV-2 RBD mutants and RBDWT (Tables S4 and S5). Both correlated and anticorrelated pairs were affected in E1 and E2 regions of RBD variants, mostly in RBDB.1.1.7, whereas the other two mutants tend to show similar patterns to RBDWT (Figure 3). We also calculated the principal components using MD trajectories to provide the main structural insights into the RBD/hACE2 complexes from eigenvalues and orthogonal eigenvectors (Figures 4 and S6). These orthogonal eigenvectors are associated with the maximum correlation between residue pairs. Thus, we observed 100 snapshots corresponding to MD trajectories obtained by the first eigenvalue of the RBDWT/hACE2 and RBDvariants/hACE2 complexes (Figure 4). The movement of the RBD and hACE is different in each analyzed complex, demonstrating that RBD induces different torsions in hACE2 (Figure 4 and Supporting Information, Movies S1 and S2). In this regard, we hypothesized that observed torsions in hACE2 may be important for efficiently mediating the structural transition of the spike protein from the prefusion to the intermediate fusion state,19 facilitating the virus entry into the host cell. Therefore, the different conformational changes in RBDvariants interacting with hACE2 could facilitate the transition process from prefusion to postfusion state.
Figure 3.
Correlation matrix obtained by MD trajectories of RBD and its variants interacting with hACE2. Painels (a–d) show heat maps representing the correlation matrix of the interface residues. Anti-correlated and correlated pairs are colored in red and blue, respectively. Note that correlated and anti-correlated pairs are affected in E1 and E2 regions of RBD variants, mostly in RBDB.1.1.7. Heat maps of RBDB.1.351 and RBDP.1 have similar patterns to that of RBDWT.
Figure 4.
MD trajectories obtained by principal component analysis. Principal component analysis using MD trajectories of hACE2 in complex with (a) RBDWT, (b) RBDB.1.1.7, (c) RBDB.1.351, and (d) RBDP.1. Each panel has 100 snapshots corresponding to MD trajectories obtained from the first component. The movements of the RBD variants induce different torsions in the hACE2 (Supporting Information, Movies S1 and S2).
We then investigated the contributions of RBDvariants in the hACE2 complex formation by calculating the binding free energy using molecular mechanics combined with Poisson-Boltzmann surface area (MM-PBSA). The binding free energy ratio between RBDWT/hACE2 and RBDvariants/hACE2 complexes presented values ranging from ∼1.0-fold to ∼1.4-fold (Table S6 and Figure S6). Most contributions for slight increase in the binding affinity for RBDvariants/hACE2 complexes are localized on the interaction of interface residues (Figure S7). We also performed SPR assays to measure the dissociation constant (KD) values using hACE2 and different constructions of trimeric spike and RBD variants (Tables 1 and S7; Figures S8 and S9). RBDvariants bind hACE2 with KD values ranging from 1.8 ± 0.5 to 4.8 ± 1.4 nM, showing that they have higher binding affinity for hACE2 than RBDWT (KD = 15.0 ± 4.1 nM). Trimeric spike variants also increased their binding affinities for hACE2, ranging from 1.4 ± 0.3 to 2.1 ± 0.3 nM, when compared to trimeric spikeWT (KD = 6.9 ± 1.3 nM). Our data are consistent with those of other studies showing that variants of the SARS-CoV-2 spike protein have greater affinity for hACE2 than wild type (Table S8).20−24 Although there are only slight differences in the KD values, significant differences in ka and kd rates point to altered binding kinetics (Tables 1 and S7). Decreases in the kd rates of hACE2 binding by these variants indicate a longer permanence of the spike/hACE2 complex when compared to the wild type, which is necessary for the successful cleavage of the spike protein and cell invasion by the virus. Decreased kd rates agree with an enhanced goodness of fit for virus–ACE2 protein interactions.
Table 1. Kinetic Parameters Obtained from SPR Assaysa.
| variant | ka (105 M–1 s–1) | kd (10–4 s–1) | KD (nM)b |
|---|---|---|---|
| RBDWT | 6.6 ± 2.0 | 93.4 ± 1.7 | 15.0 ± 4.1 |
| RBDB.1.1.7 (alpha) | 9.0 ± 3.0 | 15.5 ± 0.1 | 1.8 ± 0.5 |
| RBDB.1.351 (beta) | 9.0 ± 2.7 | 40.9 ± 1.3 | 4.8 ± 1.4 |
| RBDP.1 (gamma) | 9.0 ± 3.4 | 27.8 ± 0.9 | 3.3 ± 1.0 |
| spike trimerWT | 0.7 ± 0.2 | 4.8 ± 0.8 | 6.7 ± 1.3 |
| spike trimerB.1.1.7 (alpha) | 0.8 ± 0.2 | 1.5 ± 0.1 | 1.9 ± 0.2 |
| spike trimerB.1.351 (beta) | 1.6 ± 0.9 | 2.1 ± 0.7 | 1.4 ± 0.3 |
| spike trimerP.1 (gamma) | 1.2 ± 0.5 | 2.4 ± 0.5 | 2.1 ± 0.3 |
Equilibrium dissociation constants (KD’s) calculated for RBD and its variants in complex with the dimeric hACE2 protein. Experimental KD values were also measured using a trimeric spike protein and its variants for interacting with the dimeric form of hACE2 (residues 18–740) fused with a human IgG1 Fc tag at the C-terminus. The mutations in each spike constructions are shown in parentheses: RBDB.1.1.7 (N501Y); RBDB.1.351 (K417N, E484K, and N501Y); RBDP.1 (K417T, E484K, and N501Y); spike trimerB.1.1.7 (H69-V70del, Y144del, N501Y, A570D, D614G, P681H, T716I, S982A, and D1118H); spike trimerB.1.351 (L18F, D80A, D215G, L242-A243-L244del, R246I, K417N, E484K, N501Y, D614G, and A701V); spike trimerP.1 (L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I, and V1176F). In spike trimer constructions, the proline substitutions (F817P, A892P, A899P, A942P, K986P, and V987P) were introduced to stabilize the trimeric prefusion state of the SARS-CoV-2 spike protein, and alanine substitutions (R683A and R685A) were introduced to abolish the furin cleavage site. More details are shown in Table S7. The SPR assays were performed in three biological replicates.
KD = kd/ka.
Taken together, even a small gain in the binding affinity of the mutants may enhance infection, facilitating hACE2/spike complex formation, necessary for membrane fusion and virus entry.25 It is also possible that actions of allosteric domains in the spike protein, priming spike/hACE2 binding,26,27 benefit from the mutations in the SARS-CoV-2 variants. It is relevant to mention that spike/hACE2 interactions and the binding affinity for complex formation also depend on the microenvironment. For instance, the presence of heparin or heparan sulfate,28,29 among others, so far not well-studied, positively or negatively acting allosteric factors that might provide an explanation for the differences between KD values from our experimental kinetic data and other studies. In addition to any favoring of the spike/hACE2 complex formation, mutants may facilitate FURIN binding and promote cleavage, as shown for the SARS-CoV-2 spike mutant protein D614G.30 Further biological factors are probably associated with the increased infectivity of SARS-CoV-2 variants. It has been reported that D614G mutation can increase the density of spike on the surface of the virion due to a decrease in the premature loss of the S1 subunit.31 It is known that P.1, B.1.1.7, and B.1.351 variants have lower affinity for neutralizing antibodies, which may be a way for the virus to escape the immune system of an infected individual.21,32−34 Additionally, factors related to the expression of key genes and their variants for SARS-CoV-2 infections may be critical.
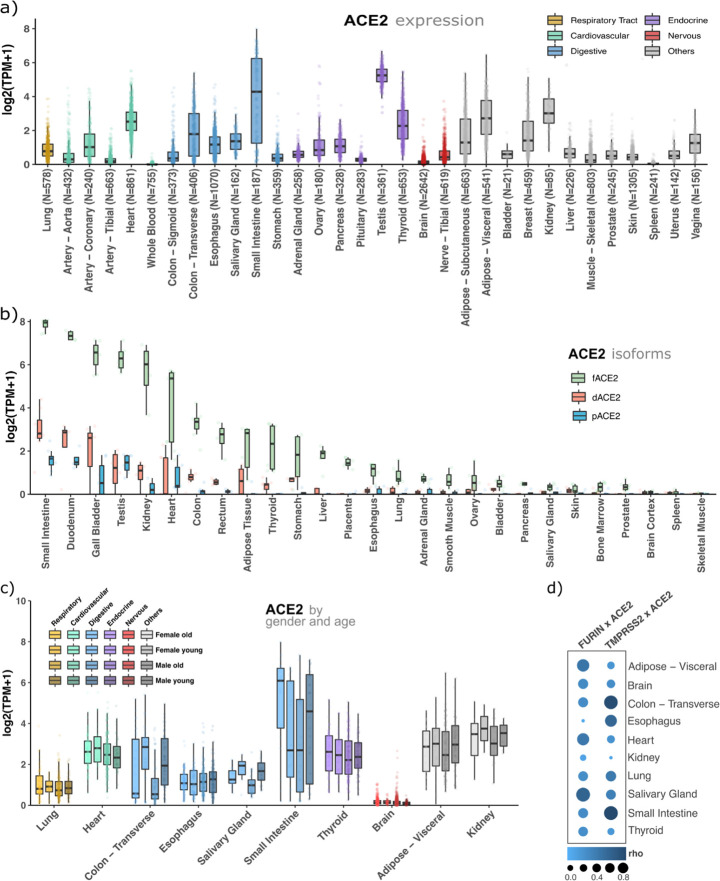
We investigated whether expression profiles of three genes essential for the viral entrance into the human cells, ACE2, TMPRSS2, and FURIN, may be determinative for infection rates. We downloaded and analyzed approximately 23,000 samples (∼1,000 individuals) from 32 healthy tissues from the genotype-tissue expression database (GTEx), and to gain more insights into their expression patterns, we also sectioned them in gender, donors’ age (young < 40; old > 60 years old), and ACE2 splicing isoforms (Figure 5). ACE2 has a high or middle expression in organs of the respiratory tract (lung), cardiovascular (heart), digestive (colon and small intestine), endocrine (testis, thyroid), adipose tissue, breasts, and kidneys (Figure 5a). TMPRSS2 has a similar expression pattern. Notably, both genes are low expressed in the brain (Figures 5a and S10). Conversely, FURIN is highly expressed in almost all human tissues (Figure S11). Altogether, some of these tissues presenting a high expression of ACE2 plus TMPRSS2 or FURIN are already known to be critical organs for the disease (e.g., lung and heart). Additionally, a new ACE2 splicing isoform was recently reported,35,36 which lacks the S1 RBD interaction region, is induced by interferons, but has no metallopeptidase activity. Then, we investigated the expression of this isoform (dACE2) as well as the full-length (fACE2) and other ACE2 alternative isoforms (pACE2) (Figure S12). Figure 5b shows that the full-length isoform, which contains receptor regions that interact with RBD, is majorly expressed in all human tissues. Next, we also investigated the expression of these three genes by gender and age (young < 40; old > 60 years old). We did not find any significant differential expression by age or gender for ACE2 (Figure 5c), TMPRSS2 (Figure S13), or FURIN (Figure S14). Finally, we found a high expression correlation between ACE2 and the proteases, FURIN and TMPRSS2, suggesting a functional synergy in organs usually affected by the SARS-CoV-2 (Figure 5d). In summary, we confirmed that ACE2 and either proteases, FURIN or TMPRSS2, are co-expressed in organs already described as affected by SARS-CoV-2, such as the respiratory, cardiovascular, digestive, and some endocrine organs. Using these expression data (Materials and Methods in the Supporting Information), no significant expression differences were found for these genes in terms of gender, age, and ACE2 splicing isoforms. These results were not expected because the severity of the disease is associated with age and gender. Therefore, these results support the hypothesis that only the presence of ACE2 and proteases do not explain the relationships between infectivity and lethality with age and gender.
Figure 5.
ACE2 is expressed in critical tissues for the disease, its splicing isoforms are less expressed than its full-length sequence, and there is no significant expression difference between gender or age for this gene. (a) ACE2 gene expression profile in 32 human healthy tissues. (b) Expression profile of ACE2 splicing isoforms. (c) ACE2 expression profile segmented by age (young < 40 years of age; old > 60 years of age) and gender. (d) Expression correlation (Rho = Spearman’s correlation coefficient) between ACE2 and FURIN or ACE2 and TMPRSS2.
Overall, our computational and experimental data show that the SARS-CoV-2 variants moderately increase their binding affinities to hACE2, which is not enough to explain the increase of transmission and lethality caused by the virus. We therefore suggest that the decrease of kd rate values could positively influence the trimeric spike cleavage, by increasing the exposure time of its cleavage site to proteolytic enzymes. Furthermore, SARS-CoV-2 variants modulate different torsions in hACE2, which may efficiently mediate the structural transition of the spike protein from the pre-fusion to the intermediate fusion state19, facilitating the virus entry into the host cell. The individual genetic background, such as ACE2, FURIN, and TMPRSS2 polymorphisms and expression patterns, among other factors, together with circulating and upcoming SARS-CoV-2 variants will determine the course and duration of the pandemic. Therefore, combining all these components allows SARS-CoV-2 variants to increase their abilities to spread faster than the WT.
Acknowledgments
The authors acknowledge the National Council for Scientific and Technological Development (CNPq), the Coordination for the Improvement of Higher Education Personnel (CAPES, grant nos 88887.374931/2019-00 and 88887.620198/2021-00, Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Finance Code 01), Rede Virus MCTI (grant no FINEP 0459/20), and the São Paulo Research Foundation (FAPESP, grant nos 2019/00195-2, 2020/04680-0, 2016/09047-8, 2018/07366-4, 2018/15579-8, 2017/19541-2, 2017/18246-7, 2017/17636-6, 2020/14158-9, and 2020/06091-1), Brazil, for financial support.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c07240.
MD simulations; MM-PBSA; protein–protein binding assays using SPR; genes and splicing isoform expression analyses; SASA analysis; MM-PBSA studies; hydrogen bonding occupancy analysis; and cross-correlation analysis (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Andersen K. G.; Rambaut A.; Lipkin W. I.; Holmes E. C.; Garry R. F. The Proximal Origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhama K.; Patel S. K.; Sharun K.; Pathak M.; Tiwari R.; Yatoo M. I.; Malik Y. S.; Sah R.; Rabaan A. A.; Panwar P. K.; Singh K. P.; Michalak I.; Chaicumpa W.; Martinez-Pulgarin D. F.; Bonilla-Aldana D. K.; Rodriguez-Morales A. J. SARS-CoV-2 Jumping the Species Barrier: Zoonotic Lessons from SARS, MERS and Recent Advances to Combat This Pandemic Virus. Trav. Med. Infect. Dis. 2020, 37, 101830. 10.1016/j.tmaid.2020.101830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . Coronavirus (COVID-19) Dashboard. 2021, https://covid19.who.int (accessed Aug 24, 2021).
- Lan J.; Ge J.; Yu J.; Shan S.; Zhou H.; Fan S.; Zhang Q.; Shi X.; Wang Q.; Zhang L.; Wang X. Structure of the SARS-CoV-2 Spike Receptor-Binding Domain Bound to the ACE2 Receptor. Nature 2020, 581, 215–220. 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Li W.; Choe H.; Farzan M. Insights from the Association of SARS-CoV S-Protein with Its Receptor, ACE2. Adv. Exp. Med. Biol. 2006, 581, 209–218. 10.1007/978-0-387-33012-9_36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A. R.; Perlman S. Coronaviruses: An Overview of Their Replication and Pathogenesis. Methods Mol. Biol. 2015, 1282, 1–23. 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacka I.; Bertram S.; Müller M. A.; Allen P.; Soilleux E.; Pfefferle S.; Steffen I.; Tsegaye T. S.; He Y.; Gnirss K.; Niemeyer D.; Schneider H.; Drosten C.; Pöhlmann S. Evidence That TMPRSS2 Activates the Severe Acute Respiratory Syndrome Coronavirus Spike Protein for Membrane Fusion and Reduces Viral Control by the Humoral Immune Response. J. Virol. 2011, 85, 4122–4134. 10.1128/jvi.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G.; Gosalia D. N.; Rennekamp A. J.; Reeves J. D.; Diamond S. L.; Bates P. Inhibitors of Cathepsin L Prevent Severe Acute Respiratory Syndrome Coronavirus Entry. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 11876–11881. 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M.; Kleine-Weber H.; Schroeder S.; Krüger N.; Herrler T.; Erichsen S.; Schiergens T. S.; Herrler G.; Wu N.-H.; Nitsche A.; Müller M. A.; Drosten C.; Pöhlmann S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M.-M.; Yang W.-L.; Yang F.-Y.; Zhang L.; Huang W.-J.; Hou W.; Fan C.-F.; Jin R.-H.; Feng Y.-M.; Wang Y.-C.; Yang J.-K. Cathepsin L Plays a Key Role in SARS-CoV-2 Infection in Humans and Humanized Mice and Is a Promising Target for New Drug Development. Signal Transduct. Targeted Ther. 2021, 6, 134. 10.1038/s41392-021-00558-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . Proposed working definitions of SARS-CoV-2 Variants of Interest and Variants of Concern. 2021, https://www.who.int/docs/default-source/coronaviruse/situation-reports/20210225_weekly_epi_update_voc-special-edition.pdf?sfvrsn=1eacfa47_7&download=true (accessed May 8, 2021).
- PHE . Investigation of novel SARS-COV-2 variant. 2021, https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/959438/Technical_Briefing_VOC_SH_NJL2_SH2.pdf (accessed May 8, 2021).
- Tracking SARS-CoV-2 variants. 2021, https://www.who.int/activities/tracking-SARS-CoV-2-variants (accessed Jun 3, 2021).
- Wang Z.; Schmidt F.; Weisblum Y.; Muecksch F.; Barnes C. O.; Finkin S.; Schaefer-Babajew D.; Cipolla M.; Gaebler C.; Lieberman J. A.; Oliveira T. Y.; Yang Z.; Abernathy M. E.; Huey-Tubman K. E.; Hurley A.; Turroja M.; West K. A.; Gordon K.; Millard K. G.; Ramos V.; Da Silva J.; Xu J.; Colbert R. A.; Patel R.; Dizon J.; Unson-O’Brien C.; Shimeliovich I.; Gazumyan A.; Caskey M.; Bjorkman P. J.; Casellas R.; Hatziioannou T.; Bieniasz P. D.; Nussenzweig M. C. mRNA Vaccine-Elicited Antibodies to SARS-CoV-2 and Circulating Variants. Nature 2021, 592, 616–622. 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer C. K.; Ayres F.; Hermanus T.; Madzivhandila M.; Kgagudi P.; Oosthuysen B.; Lambson B. E.; de Oliveira T.; Vermeulen M.; van der Berg K.; Rossouw T.; Boswell M.; Ueckermann V.; Meiring S.; von Gottberg A.; Cohen C.; Morris L.; Bhiman J. N.; Moore P. L. SARS-CoV-2 501Y.V2 Escapes Neutralization by South African COVID-19 Donor Plasma. Nat. Med. 2021, 27, 622–625. 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- Silva de Souza A.; Rivera J. D.; Almeida V. M.; Ge P.; de Souza R. F.; Farah C. S.; Ulrich H.; Marana S. R.; Salinas R. K.; Guzzo C. R. Molecular Dynamics Reveals Complex Compensatory Effects of Ionic Strength on the Severe Acute Respiratory Syndrome Coronavirus 2 Spike/Human Angiotensin-Converting Enzyme 2 Interaction. J. Phys. Chem. Lett. 2020, 11, 10446–10453. 10.1021/acs.jpclett.0c02602. [DOI] [PubMed] [Google Scholar]
- Kyte J.; Doolittle R. F. A Simple Method for Displaying the Hydropathic Character of a Protein. J. Mol. Biol. 1982, 157, 105–132. 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Zhu X.; Mannar D.; Srivastava S. S.; Berezuk A. M.; Demers J.-P.; Saville J. W.; Leopold K.; Li W.; Dimitrov D. S.; Tuttle K. S.; Zhou S.; Chittori S.; Subramaniam S. Cryo-Electron Microscopy Structures of the N501Y SARS-CoV-2 Spike Protein in Complex with ACE2 and 2 Potent Neutralizing Antibodies. PLoS Biol. 2021, 19, e3001237 10.1371/journal.pbio.3001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y.; Zhang J.; Xiao T.; Peng H.; Sterling S. M.; Walsh R. M. Jr.; Rawson S.; Rits-Volloch S.; Chen B. Distinct Conformational States of SARS-CoV-2 Spike Protein. Science 2020, 369, 1586–1592. 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan M.; Ferguson I. D.; Miao W.; Khavari P. A. SARS-CoV-2 B.1.1.7 and B.1.351 Spike Variants Bind Human ACE2 with Increased Affinity. Lancet Infect. Dis. 2021, 21, 1070. 10.1016/s1473-3099(21)00262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier D. A.; De Marco A.; Ferreira I. A. T. M.; Meng B.; Datir R. P.; Walls A. C.; Kemp S. A.; Bassi J.; Pinto D.; Silacci-Fregni C.; Bianchi S.; Tortorici M. A.; Bowen J.; Culap K.; Jaconi S.; Cameroni E.; Snell G.; Pizzuto M. S.; Pellanda A. F.; Garzoni C.; Riva A.; Elmer A.; Kingston N.; Graves B.; McCoy L. E.; Smith K. G. C.; Bradley J. R.; Temperton N.; Ceron-Gutierrez L.; Barcenas-Morales G.; Harvey W.; Virgin H. W.; Lanzavecchia A.; Piccoli L.; Doffinger R.; Wills M.; Veesler D.; Corti D.; Gupta R. K.; Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA Vaccine-Elicited Antibodies. Nature 2021, 593, 136–141. 10.1038/s41586-021-03412-7. [DOI] [PubMed] [Google Scholar]
- Tian F.; Tong B.; Sun L.; Shi S.; Zheng B.; Wang Z.; Dong X.; Zheng P.. Mutation N501Y in RBD of Spike Protein Strengthens the Interaction between COVID-19 and Its Receptor ACE2. 2021, bioRxiv 2021.02.14.431117. [Google Scholar]
- Laffeber C.; de Koning K.; Kanaar R.; Lebbink J. H. G.. Experimental Evidence for Enhanced Receptor Binding by Rapidly Spreading SARS-CoV-2 Variants. 2021, bioRxiv 2021.02.22.432357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y.; Zhang J.; Xiao T.; Lavine C. L.; Rawson S.; Peng H.; Zhu H.; Anand K.; Tong P.; Gautam A.; Lu S.; Sterling S. M.; Walsh R. M.; Rits-Volloch S.; Lu J.; Wesemann D. R.; Yang W.; Seaman M. S.; Chen B.. Structural Basis for Enhanced Infectivity and Immune Evasion of SARS-CoV-2 Variants. 2021, bioRxiv 2021.04.13.439709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton D. J.; Wrobel A. G.; Xu P.; Roustan C.; Martin S. R.; Rosenthal P. B.; Skehel J. J.; Gamblin S. J. Receptor Binding and Priming of the Spike Protein of SARS-CoV-2 for Membrane Fusion. Nature 2020, 588, 327–330. 10.1038/s41586-020-2772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paola L.; Hadi-Alijanvand H.; Song X.; Hu G.; Giuliani A. The Discovery of a Putative Allosteric Site in the SARS-CoV-2 Spike Protein Using an Integrated Structural/Dynamic Approach. J. Proteome Res. 2020, 19, 4576–4586. 10.1021/acs.jproteome.0c00273. [DOI] [PubMed] [Google Scholar]
- Olotu F. A.; Omolabi K. F.; Soliman M. E. S. Leaving No Stone Unturned: Allosteric Targeting of SARS-CoV-2 Spike Protein at Putative Druggable Sites Disrupts Human Angiotensin-Converting Enzyme Interactions at the Receptor Binding Domain. Inform. Med. Unlocked 2020, 21, 100451. 10.1016/j.imu.2020.100451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mycroft-West C. J.; Su D.; Pagani I.; Rudd T. R.; Elli S.; Gandhi N. S.; Guimond S. E.; Miller G. J.; Meneghetti M. C. Z.; Nader H. B.; Li Y.; Nunes Q. M.; Procter P.; Mancini N.; Clementi M.; Bisio A.; Forsyth N. R.; Ferro V.; Turnbull J. E.; Guerrini M.; Fernig D. G.; Vicenzi E.; Yates E. A.; Lima M. A.; Skidmore M. A. Heparin Inhibits Cellular Invasion by SARS-CoV-2: Structural Dependence of the Interaction of the Spike S1 Receptor-Binding Domain with Heparin. Thromb. Haemostasis 2020, 120, 1700–1715. 10.1055/s-0040-1721319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q.; Chen C. Z.; Swaroop M.; Xu M.; Wang L.; Lee J.; Wang A. Q.; Pradhan M.; Hagen N.; Chen L.; Shen M.; Luo Z.; Xu X.; Xu Y.; Huang W.; Zheng W.; Ye Y. Heparan Sulfate Assists SARS-CoV-2 in Cell Entry and Can Be Targeted by Approved Drugs in Vitro. Cell Discov. 2020, 6, 80. 10.1038/s41421-020-00222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad A.; Alshawaf E.; Marafie S. K.; Abu-Farha M.; Abubaker J.; Al-Mulla F. Higher Binding Affinity of Furin for SARS-CoV-2 Spike (S) Protein D614G Mutant Could Be Associated with Higher SARS-CoV-2 Infectivity. Int. J. Infect. Dis. 2021, 103, 611–616. 10.1016/j.ijid.2020.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; Cai Y.; Xiao T.; Lu J.; Peng H.; Sterling S. M.; Walsh R. M. Jr.; Rits-Volloch S.; Zhu H.; Woosley A. N.; Yang W.; Sliz P.; Chen B. Structural Impact on SARS-CoV-2 Spike Protein by D614G Substitution. Science 2021, 372, 525–530. 10.1126/science.abf2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobeil S. M.-C.; Janowska K.; McDowell S.; Mansouri K.; Parks R.; Stalls V.; Kopp M. F.; Manne K.; Saunders K.; Edwards R. J.; Haynes B. F.; Henderson R. C.; Acharya P.. Effect of Natural Mutations of SARS-CoV-2 on Spike Structure, Conformation and Antigenicity. 2021, bioRxiv 2021.03.11.435037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D.; Dejnirattisai W.; Supasa P.; Liu C.; Mentzer A. J.; Ginn H. M.; Zhao Y.; Duyvesteyn H. M. E.; Tuekprakhon A.; Nutalai R.; Wang B.; Paesen G. C.; Lopez-Camacho C.; Slon-Campos J.; Hallis B.; Coombes N.; Bewley K.; Charlton S.; Walter T. S.; Skelly D.; Lumley S. F.; Dold C.; Levin R.; Dong T.; Pollard A. J.; Knight J. C.; Crook D.; Lambe T.; Clutterbuck E.; Bibi S.; Flaxman A.; Bittaye M.; Belij-Rammerstorfer S.; Gilbert S.; James W.; Carroll M. W.; Klenerman P.; Barnes E.; Dunachie S. J.; Fry E. E.; Mongkolsapaya J.; Ren J.; Stuart D. I.; Screaton G. R. Evidence of Escape of SARS-CoV-2 Variant B.1.351 from Natural and Vaccine-Induced Sera. Cell 2021, 184, 2348–2361.e6. 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W.; Zhou D.; Supasa P.; Liu C.; Mentzer A. J.; Ginn H. M.; Zhao Y.; Duyvesteyn H. M. E.; Tuekprakhon A.; Nutalai R.; Wang B.; López-Camacho C.; Slon-Campos J.; Walter T. S.; Skelly D.; Costa Clemens S. A.; Naveca F. G.; Nascimento V.; Nascimento F.; Fernandes da Costa C.; Resende P. C.; Pauvolid-Correa A.; Siqueira M. M.; Dold C.; Levin R.; Dong T.; Pollard A. J.; Knight J. C.; Crook D.; Lambe T.; Clutterbuck E.; Bibi S.; Flaxman A.; Bittaye M.; Belij-Rammerstorfer S.; Gilbert S. C.; Carroll M. W.; Klenerman P.; Barnes E.; Dunachie S. J.; Paterson N. G.; Williams M. A.; Hall D. R.; Hulswit R. J. G.; Bowden T. A.; Fry E. E.; Mongkolsapaya J.; Ren J.; Stuart D. I.; Screaton G. R. Antibody Evasion by the P.1 Strain of SARS-CoV-2. Cell 2021, 184, 2939–2954.e9. 10.1016/j.cell.2021.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blume C.; Jackson C. L.; Spalluto C. M.; Legebeke J.; Nazlamova L.; Conforti F.; Perotin J.-M.; Frank M.; Butler J.; Crispin M.; Coles J.; Thompson J.; Ridley R. A.; Dean L. S. N.; Loxham M.; Reikine S.; Azim A.; Tariq K.; Johnston D. A.; Skipp P. J.; Djukanovic R.; Baralle D.; McCormick C. J.; Davies D. E.; Lucas J. S.; Wheway G.; Mennella V. A Novel ACE2 Isoform Is Expressed in Human Respiratory Epithelia and Is Upregulated in Response to Interferons and RNA Respiratory Virus Infection. Nat. Genet. 2021, 53, 205–214. 10.1038/s41588-020-00759-x. [DOI] [PubMed] [Google Scholar]
- Onabajo O. O.; Banday A. R.; Stanifer M. L.; Yan W.; Obajemu A.; Santer D. M.; Florez-Vargas O.; Piontkivska H.; Vargas J. M.; Ring T. J.; Kee C.; Doldan P.; Tyrrell D. L.; Mendoza J. L.; Boulant S.; Prokunina-Olsson L. Interferons and Viruses Induce a Novel Truncated ACE2 Isoform and Not the Full-Length SARS-CoV-2 Receptor. Nat. Genet. 2020, 52, 1283–1293. 10.1038/s41588-020-00731-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante J. A.; Liu Y.; Liu J.; Xia H.; Johnson B. A.; Lokugamage K. G.; Zhang X.; Muruato A. E.; Zou J.; Fontes-Garfias C. R.; Mirchandani D.; Scharton D.; Bilello J. P.; Ku Z.; An Z.; Kalveram B.; Freiberg A. N.; Menachery V. D.; Xie X.; Plante K. S.; Weaver S. C.; Shi P.-Y. Spike Mutation D614G Alters SARS-CoV-2 Fitness. Nature 2021, 592, 116–121. 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozono S.; Zhang Y.; Ode H.; Sano K.; Tan T. S.; Imai K.; Miyoshi K.; Kishigami S.; Ueno T.; Iwatani Y.; Suzuki T.; Tokunaga K. SARS-CoV-2 D614G Spike Mutation Increases Entry Efficiency with Enhanced ACE2-Binding Affinity. Nat. Commun. 2021, 12, 848. 10.1038/s41467-021-21118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.; Jackson C. B.; Mou H.; Ojha A.; Peng H.; Quinlan B. D.; Rangarajan E. S.; Pan A.; Vanderheiden A.; Suthar M. S.; Li W.; Izard T.; Rader C.; Farzan M.; Choe H. SARS-CoV-2 Spike-Protein D614G Mutation Increases Virion Spike Density and Infectivity. Nat. Commun. 2020, 11, 6013. 10.1038/s41467-020-19808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.