Abstract
MicroRNAs (miRNAs) play various roles in the implantation and pregnancy process. Abnormal regulation of miRNAs leads to reproductive disorders such as repeated implantation failure (RIF). During the window of implantation, different miRNAs are released from the endometrium, which can potentially reflect the status of the endometrium for in vitro fertilization (IVF). The focus of this review is to determine whether endometrial miRNAs may be utilized as noninvasive biomarkers to predict the ability of endometrium to implant and provide live birth during IVF cycles. The levels of certain miRNAs in the endometrium have been linked to implantation potential and pregnancy outcomes in previous studies. Endometrial miRNAs could be employed as non-invasive biomarkers in the assisted reproductive technology (ART) cycle to determine the optimal time for implantation. Few human studies have evaluated the association between ART outcomes and endometrial miRNAs in RIF patients. This review may pave the way for more miRNA transcriptomic studies on human endometrium and introduce a specific miRNA profile as a multivariable prediction model for choosing the optimal time in the IVF cycle.
Keywords: miRNAs, art, transcriptome, pregnancy, endometrial receptivity
1 Introduction
Implantation is a process in which the blastocyst attaches to and attacks the mother’s endometrium within the time frame of the implantation window (Achache and Revel, 2006). The receptive endometrium plays an active role in the implantation process (Kliman and Frankfurter, 2019). In response to steroid hormones, the endometrial tissue undergoes morphological, cellular, and molecular changes in cycles (Davidson and Coward, 2016; Kliman and Frankfurter, 2019). Reproductive specialists currently use methods such as transvaginal ultrasound and hormonal analysis of serum to predict endometrial receptivity for embryo transfer (ET). However, these methods do not provide beneficial predictions for the outcome of in vitro fertilization (IVF) (Craciunas et al., 2019; Horcajadas et al., 2008; Quinn and Casper, 2009). Taking into account the molecular changes of the endometrium during the implantation window provides us with crucial information regarding endometrial receptivity, which is of great importance (Craciunas et al., 2019).
Thus far, a range of single molecules, including miRNAs, have been examined as biomarkers of uterine receptivity (Edgell et al., 2013). Reproductive disorders such as polycystic ovary syndrome (PCOS), repeated implantation failure (RIF), and endometriosis are linked to abnormal miRNA regulation (Liang et al., 2017; Shokrzadeh et al., 2018). About 15%–20% of infertile couples who undergo IVF-ET suffer from Repeated implantation failure (RIF) (Simon and Laufer, 2012). About 15%–20% of infertile couples who undergo IVF-ET suffer from RIF. MiRNAs might have the capacity to predict RIF. The present review may help to identify various biomarkers through miRNA detection.
2 Repeated implantation failure
The inability of an embryo to implant into the uterine wall after multiple transfers during IVF treatment is referred to as RIF. However, due to the lack of a unified definition, various definitions of RIF are proposed in IVF centers. RIF is defined by some sources as the non-implantation of embryos in three consecutive cycles with the transfer of up to three high-quality embryos in each cycle, taking into account the number of embryos transferred in each cycle and the IVF success rate (Shufaro and Schenker, 2011). In some centers, the absence of a sac approximately 45 days (week 5 onwards) after the transfer of at least three embryos or the transfer of more than 10 embryos in multiple transfers is considered RIF (Salehpour et al., 2016). The incidence and prevalence of RIF are rarely reported due to the various definitions of this condition (Table 1) (Maesawa et al., 2015). Various types of RIF are classified into three broad categories (Timeva et al., 2014), including endometrial RIF, idiopathic RIF, and multifactorial RIF (Figure 1). The main causes for this complication are fetal defects, decreased uterine receptivity, abnormal anatomy of the uterus, and the medical condition of the mother (Margalioth et al., 2006). Other factors influencing RIF include chromosomal and uterine abnormalities, hormonal and placental disorders, smoking, certain medications, maternal heart and kidney disease, and the quality of the transferred embryo (Coughlan et al., 2014).
TABLE 1.
Definition of RIF.
| References | Definition |
|---|---|
| Choi et al. (2016) | Failure to conceive in at least three previous IVF cycles with good quality embryos |
| Revel et al. (2011) | At least four ART cycles with embryo transfer failure |
| Shi et al. (2017) | Three embryo transfer failures in which at least four morphologically high-grade embryos were transferred |
| Rekker et al. (2018) | Three unsuccessful IVF cycles with embryo transfer |
| Xu et al. (2019) | Three transplantation failures with at least four good-quality embryos |
| Tan et al. (2005) | Non-pregnancy after transfer of more than 10 high-quality embryos, after 2–6 IVF cycles |
FIGURE 1.

Types of RIF and their causes. The failure of an embryo to implant into the uterine wall after multiple transfers during IVF treatment is referred to as RIF or recurrent implantation failure. RIF types are divided into three categories: Endometrial RIF, which occurs due to the low thickness (≤6 mm) of the endometrium; idiopathic RIF, which is unexplained failure to achieve pregnancy after the transfer of good quality embryos; multifactorial RIF, which is caused by a wide variety of reasons (male-related factors, genetic abnormalities, infections, immunological factors, psychological factors, lifestyle, and other similar variables).
Thus far, RIF has been managed in a variety of ways, but no consensus has emerged on the most effective method. Some of the RIF-management methods are blastocyst transfer, assisted hatching, co-culture system, sequential transfer, hysteroscopy, endometrial scratching, salpingectomy for tubal disease, extra number embryo transfer, natural cycle, oocyte donation, intra-tubal ET, immune therapy, and endometrial receptivity array (ERA) (Choi et al., 2016; Katzorke et al., 2016).
3 Relationship between hormone balance and microRNAs in repeated implantation failure
The human endometrium undergoes cyclical changes due to sex steroid hormones (Klinge, 2012). Previous studies have examined the relationship between sex steroid hormones and miRNAs at different stages of the menstrual cycle. According to these studies, some endometrial miRNAs regulate the expression of estrogen and progesterone, and on the other hand, estrogen and progesterone are involved in regulating the expression of some endometrial miRNAs. The effect of sex steroid hormones on the expression of endometrial miRNAs in animal models such as zebrafish (Cohen et al., 2008) as well as humans (Klinge, 2012) has been investigated. Reed et al. (2018) reported increased expression of miR-181b and let-7e, and decreased expression of mi-R27b in cultured human endometrial stromal cells exposed to estradiol. Similarly, the induction of miR-125b and miR-133a expression has been reported in the cell culture of human endometrial epithelial cells (Chen et al., 2016; Pan et al., 2017). At the time of ovulation, women with high blood progesterone levels under the ovulation stimulation protocol had a higher endometrial expression of miR-30b, miR-125b, miR-424, and miR-451 than women with low blood progesterone levels (Li et al., 2011). Another study compared the expression levels of several miRNAs in the mid-secretory and late proliferative phases of human endometrial epithelial cells. This study indicated that miR-29b, miR-29c, miR-30b, miR-30d, miR-31, miR-193a-3p, miR-200c, miR-203, miR-204, miR-210, miR-345, and miR-582-5p levels were higher in the mid-secretory phase compared to the late proliferative phase. On the contrary, the expression of miR-105, miR-127, miR-134, miR-214, miR-222, miR-369-5p, miR-370, miR-376a, miR-382, miR450, miR-503, and miR-542-3p was lower in the mid-secretory phase compared to the late proliferative phase (Kuokkanen et al., 2010).
The actions of estrogen and progesterone are related to the altered expression of their receptors in the human endometrium. There is not much information about the regulation of estrogen receptors by miRNAs in the human endometrium. However, one study reported that endometrial cancer cells transfected with a miR-107 mimic had lower estrogen receptor expression (Bao W et al., 2019). In addition, miR-22-5p transfection in endometrial stromal cells of female endometriosis in the culture medium altered the estrogen receptor expression (Xiao et al., 2020). Moreover, miR-194-3p transfection in cultured endometrial stromal cells resulted in a significant reduction in progesterone receptor protein levels (PR-A and PR-B) (Pei et al., 2018). Zhou et al. (2016) reported a decrease in protein levels of PR-A and PR-B in endometrial stromal cells transfected with miR-196a (Zhou et al., 2016). They also identified PRs as targets for miR-196a, miR-297, miR-575, miR-628-3p, miR-635, miR-921, miR-938, and miR-1184. It should be noted that miR-92a transfection in the endometrial stromal cell line resulted in progesterone resistance and increased cell proliferation (Zhou et al., 2016).
4 The role of endometrial microRNAs in repeated implantation failure
According to the literature, the expression of miRNAs varies in different phases and pathological conditions of the endometrium (Shariati et al., 2019; Shokrzadeh et al., 2019) It has been established that the upregulation of certain miRNAs promotes implantation (pro-implantation miRNAs) while the upregulation of others causes implantation failure (anti-implantation miRNAs) (Reza et al., 2019). MiRNAs that are involved in implantation can also be classified based on their roles into categories such as proliferation, decidualization, angiogenesis, and apoptosis, among others (Table 2) (Figures 2A,B).
TABLE 2.
Endometrial miRNAs and implantation.
| miRNA | Anti/pro implantation | Target gene | Role | Species | References |
|---|---|---|---|---|---|
| miR-17–92 | Pro-implantation | E2Fs, TGFβ | Decidualization | - | Mogilyansky and Rigoutsos (2013) |
| miR-21 | Pro-implantation | RECK, MMP9, PTEN | Proliferation | Mice | Hu et al. (2008) |
| miR-21 | Pro-implantation | KLF12 | Decidualization | Human | Jiang et al. (2013) |
| miR-29a | Pro-implantation | Bak1, Bmf, Bcl-w | Apoptosis | Rat | Xia et al. (2014a) |
| miR-101a | Pro-implantation | Cox-2 | Decidualization | Mice | Chakrabarty et al. (2007) |
| miR-199a | Pro-implantation | Cox-2 | Decidualization | Mice | Chakrabarty et al. (2007) |
| miR-199a | Pro-implantation | Muc1 | Decidualization | Mice | Inyawilert et al. (2014) |
| miR-199a | Pro-implantation | Grb10 | Proliferation and apoptosis | Rat | Xia et al. (2014b) |
| miR-22 | Anti-implantation | Tiam1, Rac1 | Cell migration-motility | Mice | Ma et al. (2015) |
| Let-7 | Pro-implantation | Muc-1 | Inhibits proliferation-promotes differentiation | Mice | Inyawilert et al. (2015) |
| miR-200 | Anti-implantation | Zeb1, Zeb2, PTEN | Proliferation and apoptosis | Mice | Jimenez et al. (2016) |
| miR-30d | Pro-implantation | H19, NNMT | Proliferation, hormonal responses, methylation status | Human | Moreno-Moya et al. (2014) |
| miR-98 | Anti-implantation | Bcl-xL | Promotes proliferation and inhibits apoptosis | Rat | Xia et al. (2014c) |
| miR-141 | Anti-implantation | PTEN | Proliferation, apoptosis | Mice | Liu et al. (2013) |
| miR-143 | Pro-implantation | LIFR | Proliferation, invasion, decidualization | Rat | Tian et al. (2015) |
| miR-181a | Pro-implantation | KLF19 | Decidualization, differentiation | Human | Zhang et al. (2015) |
| miR-193 | Pro-implantation | GRB7 | Migration | Mice | Li et al. (2014) |
| miR-429 | Anti-implantation | Pcdh8 | Invasion | Mice | Li et al. (2015a) |
| miR-451 | Pro-implantation | Ankrd46 | Angiogenesis, invasion, and proliferation | Mice | Li et al. (2015b) |
| miR-222 | Anti-implantation | CDKN1C, p57kip2 | Differentiation cell cycle, decidualization | Human | Qian et al. (2009) |
| miR-424 | Anti-implantation | - | - | Human | Li et al. (2011) |
| miR-30b | Pro implantation | P4HA2 | - | Human | Li et al. (2011) |
| miR-125b | Anti-implantation | MMP26 | Migration and invasion | Mice | Li et al. (2011) |
FIGURE 2.
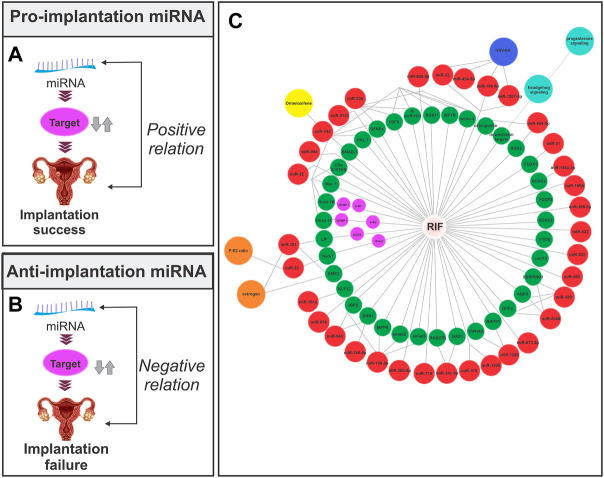
Relationship between the endometrial miRNA expression and implantation. (A) Pro-implantation miRNA. The expression of the miRNA has a positive association with the implantation outcome. (B) Anti-implantation miRNA. The expression of the miRNA has a negative association with the implantation outcome. (C) Endometrial miRNAs lead to implantation failure through their effect on target mRNA in the endometrial tissue of RIF patients. The red circle represents miRNA, the green and purple circles represent mRNA, the orange circle represents hormones, the yellow circle represents the drug, the blue circle represents the signaling pathway, and the navy blue circle represents infection. The miRNA-mRNA network is based on Section 4in this article.
The first study to investigate the different expressions of miRNAs in the secretory phase endometrium of RIF patients was published in 2011, and it found 13 miRNAs that could be used to diagnose and treat RIF (Revel et al., 2011). Since then, many human studies have found different expressions of several miRNAs in the endometrial tissue or peripheral blood of women with RIF (Figure 2C). These studies have shown that the profile of miRNAs in the pre-receptive and receptive endometrium of RIF patients is different from that of normal individuals, indicating the role of miRNAs in the implantation failure in these patients. The following is an overview of some RIF-related miRNAs (Table 3).
TABLE 3.
Endometrial miRNAs and repeated implantation failure.
| Year | Sample | Group compare | miRNA measurement | miR | Expression pattern | Targets/regulators | References |
|---|---|---|---|---|---|---|---|
| 2011 | EB | Receptive endometrium in RIF patients vs. fertile patients | TaqMan | miR-23b, miR-145, miR-99a, miR-27b, miR 652, miR-139-5p, miR-195, miR-342-3p, miR-150, miR-374b and miR-32, miR-628-5p, miR-874 |
Upregulated in RIF, and Downregulated in RIF | N-cadherin, H2AFX, Netrin-4, SFRP4 | Revel et al. (2011) |
| 2015 | EB | Infertile patients with RIF, vs. control group | Real-Time PCR | miR-22 | Upregulated in RIF | Tiam1/Rac1 | Ma et al. (2015) |
| 2015 | EB | Women with a normal menstrual cycle | - | miR-181a | - | KLF12 | Zhang et al. (2015) |
| 2016 | EB | Healthy volunteers and RIF | miRNA microarrays | miR-138-1-3p, miR-29b-1-5p, miR-363-3p, miR-34b-3p, miR-146a-5p, miR-363-3p | Upregulated in RIF | S100P, CXCL13, SIX1 | Choi et al. (2016) |
| 2017 | EB | Receptive endometrium in RIF patients vs. infertile patients | miRNA Complete labeling | miR-1207-5p, miR-4306, miR-572, miR-5739, miR-30b, miR-6088 and miR-374a-5p, miR-145-5p, miR-196b-5p, miR-199a-5p, miR-449a, miR-424-5p, miR-125b-5p,miR-21-5p | Downregulated in RIF and Upregulated in RIF | ERα, RTKN | Shi et al. (2017) |
| 2018 | Blood, EB | Fertile and RIF patients | Small RNA sequencing | miR-30b-5p, miR-30d-3p, miR-30d-5p, miR-30a-5p | Dysregulation | CDK5, STAT3 | Rekker et al. (2018) |
| 2019 | Blood | RIF in women with or without metabolic syndrome | Real‐Time PCR | miR‐21, miR‐223, miR-146a | Increased in RIF‐MS patients, declined in RIF‐MS patients | - | Sheikhansari et al. (2019) |
| 2019 | EB | RIF and healthy female controls | - | miR-489, miR-199A, miR-522, miR-369-3p, miR-422 | Considered as the key regulatory factors during RIF | UBE2I, PLK4, XPO1, AURKB, NUP107, E2F4, SIN3A | Wang and Liu (2020) |
| 2020 | EB | Normal fertile women and RIF | Real‐Time PCR | miR-31 | Increase in RIF | FOXP3 | Dehghan and Salehi (2020) |
| 2020 | EB | RIF | RT-qPCR | miR-152-3p, miR-155-5p, miR-455-3p, miR-4423-3p | Overexpression | - | Drissennek et al. (2020) |
| 2020 | EB | Women with and without RIF | Microarray, RT-qPCR | miR-148a-3p | Upregulated in RIF | HOXA8 | Zhang et al. (2020) |
| 2020 | Human endometrial HEC-1A cell | - | Microarray | miR-15, miR-22, miR-196-5p and miR-1207-5p | Upregulated in RIF, and Downregulated in RIF | - | Bortolotti et al. (2020) |
| 2020 | EB | RIF, control group | RT-qPCR | miR-145 | Upregulated in RIF | PAI-1 | Liu et al. (2020b) |
4.1 MiR-145
MiR-145 was first discovered in 2001 on chromosome 18 in mice, and then in 2003, on chromosome 5 in humans (Yuan et al., 2019). It is abundant in mesoderm-derived tissues, such as the uterus, ovaries, testes, prostate, and heart, and plays a key role in endometrial differentiation. (Chang et al., 2017). This miRNA inhibits the SMAD-1 pathway, angiogenesis, and stromal cell differentiation while also regulating decidua cell proliferation (Sirohi et al., 2019). MiR-145 shows a threefold increase in RIF patients compared to fertile individuals (Liu et al., 2020a). Targets of this miRNA include insulin-like growth factor 1 receptor (IGF1R), rhotekin (RTKN), estrogen receptor alpha (Era), octamer-binding transcription factor 4 (OCT4), SRY-related HMG-box 11 (SOX11), mucin 1 (Muc1), PAL-1, homeobox A10 (HOXA10), and homeobox A11 (HOXA11). The attachment of the mouse embryo to the endometrium is inhibited during an increase in miR-145 levels or a decrease in IGF1R levels in endometrial epithelial cells (Kang et al., 2015). The expression of miR-145 in the endometrium of mice treated with ormeloxifene, which is a non-steroidal oral medication used to prevent endometrial receptivity on the first day of pregnancy, is increased (Sirohi et al., 2018). Moreover, miR-145 affects receptivity and implantation by targeting PAL-1 and reducing its expression in the endometrium of RIF patients (Liu et al., 2020a). This miRNA inhibits SOX11 in endometrial cancer and suppresses the proliferation, migration, and invasion of HCC-1 cell lines while increasing the induction of apoptosis. In addition, by inhibiting OCT4, it also prevents the growth of endometrial cancer cells (Chang et al., 2017). Elevated miR-145 expression inhibits HOXA10 and HOXA11. Inhibition of these genes (HOXA10 and HOXA11) by acting on IGFBP-1 and avB3 leads to infertility (Nazarian et al., 2022).
4.2 MiR-22
MiR-22 is an anti-implantation miRNA whose expression is increased during the normal cycle window of implantation in RIF patients. MiR-22 leads to the dysregulation of decidualization in endometrial stromal cells by targeting Tiam1. Tiam1 with the help of Race1 is involved in stromal cell decidualization, uterine receptivity regulation, migration, and implantation (Grewal et al., 2008). An increased expression of miR-22 leads to a decreased expression of Tiam1. As a result, the reduction of Tiam1/Race1 signaling will lead to implantation failure in RIF patients (Ma et al., 2015). The abnormal expression of miR-22 and Tiam1/Race1 has been linked to a decrease in the progesterone/estradiol (P/E2) ratio in RIF patients. Transfection of miR-22 in cultured stromal cells isolated from the endometrium of female endometriosis leads to changes in estrogen receptor (ER) expression. MiR-22 suppresses the estrogen signaling pathway by targeting estrogen receptor 1 (ESR1), which is essential for the formation of the male glands (Xiao et al., 2020; Li et al., 2021; Shekibi et al., 2022).
4.3 MiR-181
The expression of miR-181 is reduced in RIF patients (Chu et al., 2015). Leukemia inhibitory factor (LIF) and Kruppel-like factor 12 (KLF12) are the targets of miR-181. Estrogen has been shown to regulate empty spiracles homeobox 2 (EMX2) levels, which in turn control miR-181 expression. In fact, estrogen reduces the expression of EMX2 and EMX2 suppresses the expression of miR-181 (Troy et al., 2003). Subsequently, the reduction of miR-181 expression leads to a rise in LIF levels, resulting in implantation success. LIF is a proinflammatory cytokine from the interleukin 6 (IL-6) family that plays an important role in preparing the uterus for embryo implantation (Aghajanova et al., 2009). Mariee et al. (2012) reported decreased LIF expression in RIF patients. MiR-181a is a member of the miR-181 family whose expression is suppressed by estrogen. MiR-181a also inhibits the expression of KLF12, which is required for endometrial receptivity. It has been reported that KLF12 expression is increased in the endometrium of RIF and endometriosis patients (Maillot et al., 2009; Zhang et al., 2015).
4.4 MiR-424-5p
MiR-424-5p can be a useful marker in assessing endometrial receptivity. The expression of this miRNA increases in the endometrium of RIF patients (Rekker et al., 2018). Moreover, decreased miR-424-5p expression has been reported in the mid-secretory endometrium of fertile women (Rekker et al., 2018). The targets of miR-424-5p include the secreted phosphoprotein 1 (SPP1), serum/glucocorticoid regulated kinase 2 (SGK2), and angiogenin (Ang) genes. Osteopontin is a glycoprotein encoded by SPP1 and its expression in the endometrium is associated with infertility. MiR-424-5p targets osteopontin and thus regulates adhesion and cell migration during implantation (Johnson et al., 2014; Kang et al., 2014). Progesterone also regulates osteopontin expression during the endometrial menstrual cycle (Casals et al., 2010). SGK2 is a protein kinase, that is, involved in cell proliferation as well as the regulation of endometrial receptivity by acting on ion channels (Gamper et al., 2002). In addition, Ang is a gene, that is, regulated by miR-424-5p and encodes the vascular endothelial growth factor (VEGF) protein. MiR-424-5p expression is reduced in tissues with high progesterone levels compared to those with normal progesterone levels. The role of miR-424-5p in cancer has also been reported, which, given the miR-424-5p targets listed above, could introduce a common molecular pathway between implantation and cancer (Kolanska et al., 2021).
4.5 MiR-155-5p
MiR-155-5p expression is increased in RIF patients (Chen P et al., 2021; Drissennek et al., 2022). MiR-155-5p is involved in implantation by targeting genes such as membrane palmitoylated protein 5 (MPP5), insulin-like growth factor 2 (IGF2), and transforming growth factor beta (TGFβ). TGFβ is involved in leukocyte extravasation signaling, which has been reported to play a role in implantation. Alteration in miR-155-5p expression contributes to implantation failure because it leads to the inhibition of smad2/3 as well as the suppression of essential processes in implantation (cell proliferation, migration, apoptosis, and invasion) (Lin et al., 2018; Chen et al., 2019; Luo et al., 2020). Because smad2/3 is one of the most important genes in TGFβ signaling, miR-155-5p may be involved in implantation failure by altering this pathway (Drissennek et al., 2022). MiR-155-5p affects the function of the MPP5 protein in the menstrual cycle of RIF patients. MPP5 expression in the normal menstrual cycle gradually decreases from the beginning of the proliferation stage to the end of the secretory stage (Li et al., 2017). IGF2 is another protein whose expression is greatly increased during the implantation window and miR-155-5p suppresses the expression of this protein (Whitby et al., 2018).
4.6 MiR-31
MiR-31 is a candidate for endometrial receptivity whose expression is increased in RIF patients compared to healthy women (Shi et al., 2017). MiR-31 expression is increased in the secretory phase of fertile women compared to the proliferative phase (Azhari et al., 2022). Moreover, miR-31 expression is increased in the serum of fertile patients in the secretory phase. MiR-31 expression is also reduced in the endometrial secretory phase of RIF patients compared to the proliferative phase. In addition, miR-31 expression is increased in the early secretory stage compared to the midluteal stage in the endometrium of women with regular menstrual cycles. Considering the above, miR-31 plays an important role in implantation (Ghafouri-Fard et al., 2021; Kresowik et al., 2014). MiR-31 target genes include forkhead box P3 (FOXP3) and C-X-C motif chemokine 12 (CXCL12), which are suppressed by miR-31 in the secretory phase of fertile women (Azhari et al., 2022). Increased expression of miR-31 leads to decreased expression of FOXP3 (immune suppressor), which can be considered the reason for implantation failure and recurrent miscarriage in RIF patients (Azarpoor et al., 2020). Another target of this miRNA is MMPs, which are involved in decidualization during implantation, defense mechanisms, and immune responses (Azhari et al., 2022).
4.7 MiR-34c-5p
Another miRNA of interest in RIF is miR-34c-5p (Tan et al., 2020). This miRNA is involved in endometrial receptivity and inflammation (Cai et al., 2018; Gao et al., 2019). An increase in the levels of this miRNA leads to a decrease in GAS1 during implantation, which results in implantation failure. Thus, miR-34c-5p is negatively associated with implantation. Tan et al. found an increase in miR-34c-5p in exosomes, indicating that endometrial receptivity-associated miRNAs are present in the small extracellular vesicles (sEVs) of uterine fluid. During endometrial implantation, miR-34c-5p is increased in these vesicles to suppress RAB27B and it is simultaneously decreased in the endometrium; therefore, miR-34c-5p levels in sEVs can be used as a marker to assess the physiological condition of the uterus and confirm the most appropriate time for implantation (Tan et al., 2020).
4.8 MiR-1290
MiR-1290 expression is increased in the endometrium of RIF patients (Liu et al., 2021). An elevated expression of this miRNA has also been observed in the endometrial extracellular vesicles of RIF patients (Ponsuksili et al., 2014). MiR-1290 has an inhibitory role in endometrial cell proliferation, and YWHAZ and RAP2C are the targets of this miRNA. MiR-1290 reduces YWHAZ in RIF patients. An increase in miR-375 and miR-1305 has also been reported in RIF (Luo et al., 2021).
4.9 MiR-148a-3p
The HOX genes, specifically the HOXA10 and HOXA11 genes, have an important role in implantation (Cakmak and Taylor, 2011). In addition, HOXC8 is involved in cell proliferation, differentiation, migration, adhesion, and tumorigenesis (Liu et al., 2018). HOXC8 is introduced as a target for miR-148a-3p (Zhang et al., 2020). With the increase of miR-148a-3p (miR-148/152 family) in RIF patients, HOXC8 is suppressed, which leads to implantation failure due to reduced decidualization. According to Chen et al. (2013), miR-148a-3p also suppresses tumors and is involved in various processes such as differentiation and development (Choi et al., 2016; Zhang et al., 2020).
4.10 MiR-21 and MiR‐146a
Given the increase in inflammatory miRNAs and cytokines in RIF, Sheikhansari et al. (2019) reported that the expression of miR-21 was increased in the RIF-metabolic syndrome (MS) group and the expression of miR-223 and miR-146a was reduced in this group. The reduction of miR‐146a leads to an increase in inflammatory factors and the inhibition of the IRAK1‐TRAF6‐NF‐κB pathway (Ghaebi et al., 2019). The function of miR‐21 can lead to inflammatory responses, suppression of the immune system, or stimulation of inflammation by inhibiting the TGFβ signaling pathway (Sheedy, 2015).
4.11 MiR-135b-5p
MiR-135b-5p is increased in the endometrium of RIF patients compared to healthy women (Shang et al., 2022). The targets of miR-135b-5p are podoplanin (PDPN) and angiotensinogen (AGT) (Shang et al., 2022). The reduction of miR-135b-5p expression plays a role in increasing the decidualization of endometrial stromal cells (Wang et al., 2021). Moreover, this miRNA is introduced as a biomarker in breast cancer due to its role in proliferation and migration (Bao C et al., 2019).
4.12 Other microRNAs
In addition to the miRNAs mentioned above, a number of miRNAs have been introduced in the literature, the expression of which is different in RIF patients at different stages or when compared to the control group. However, further validation and determination of specificity, sensitivity, and accuracy are required before these miRNAs can be studied in RIF patients. For this purpose, we merely covered basic information on these miRNAs in this section.
Shi et al. (2017) found a decrease in miR-4668-5p expression and an increase in miR-429, miR-5088, and miR-374 expressions in the RIF group (Shi et al., 2017). MiR-374 is involved in implantation by activating Wnt/β-catenin signaling. RIF patients also have lower levels of targets associated with miR-429 and miR-5088, which include dipeptidyl peptidase-4 (DPP4), SERPING1, and aquaporin 3 (AQP3). An increase in miR-30b expression is not associated with RIF since this miRNA is also overexpressed in the normal endometrium (Shi et al., 2017). MiR-152-3p is another RIF-related miRNA, which suppresses cell proliferation, migration, and angiogenesis (Haouzi et al., 2009; Drissennek et al., 2020).
Other miRNAs associated with RIF include miR-489, miR-199A, miR-369-3p, miR-422, and miR-522. The role of miR-489 has been established in many cancers, and the genes associated with this miRNA include RBBP6, NHS, ATRX, and XPO1. Genes associated with miR-199A, which is reduced in endometriosis, include CTDSPL2, HOXA9, LUC7L3, EML4, HYOU1, and PDS5B. Moreover, UBE2I, PLK4, XPO1, AURKB, and NUP107 are other genes involved in cell division and endometrial stromal cell differentiation, which have an elevated or reduced expression in RIF patients. Other associations that affect the function of miRNAs include miRNA-transcription factor (TF) interactions. According to the findings of Wang and Liu (2020), E2F4 and SIN3A are among the TFs effective in RIF, which are linked to genes related to several miRNAs involved in this condition (Wang and Liu, 2020). Furthermore, the HTR1A, NR3C1, and GABRA3 genes are essential in determining the medication treatment of RIF. Due to its identified features in RIF, NR3C1 has attracted the most attention (Tuckerman et al., 2010).
MiR-23a and miR-23b, in conjunction with the long non-coding RNA (lncRNA) PART1, decrease DUSP5 and ultimately inhibit the mitogen-activated protein kinase (MAPK) pathway in RIF patients. In addition, the association of miR-96-5p with the phosphatidylinositol 3-kinase (PI3K)-protein kinase B (Akt) pathway and the PTEN gene leads to endometrial abnormalities and affects endometrial receptivity (Chen C. H et al., 2021).
The expressions of miR-138-1-3p, miR-29b-1-5p, miR-363-3p, miR-34b-3p, miR-146a-5p, and miR-363-3p in RIF patients were different (Choi et al., 2016). Finally, it is worth noting that the study of polymorphisms in some miRNAs in RIF patients has gained much attention in recent years (Lee et al., 2020).
5 Are microRNAs potential biomarkers of repeated implantation failure detection in assisted reproductive technology?
Finding suitable strategies with acceptable specificity and sensitivity and minimal invasiveness is essential for determining the window of implantation.
The role of miRNAs in regulating biological cascades makes them potent diagnostic biomarkers. MiRNAs obtained from plasma, plasma exosomes, follicular fluid, uterine fluid, and endometrial tissue are presented as diagnostic biomarkers to estimate the window of implantation. Chen P et al. (2021) showed that endometrial-specific miRNAs can be considered diagnostic biomarkers for RIF prediction. They compared endometrial tissue biopsies from RIF and non-RIF patients using ERA technology and miRNA expedition. They identified three miRNAs, including miR-20b-5p, miR-155-5p, and miR-718, which can serve as biomarkers in RIF with 90% accuracy (specificity: 100.0%; sensitivity: 80.0%; positive prediction value: 100.0%; negative prediction value: 85.7%) (Chen P et al., 2021). In another study on miRNAs extracted from plasma and plasma exosomes, miR-150-5p, miR-150-3p, miR-149-5p, and miR-146b-3p were introduced as candidates for non-invasive biomarkers of RIF (Zeng et al., 2021). Moreover, a study on cumulus cells and follicular fluid of the RIF patients’ oocytes identified the overexpression of miR-34-5p and miR-26-5p as an indicator of a successful pregnancy (Habibi et al., 2022). In addition, von Grothusen et al. (2022) reported the decline of the expression of miR-486-5p and miR-92b-3p in the uterine fluid as a non-invasive biomarker of RIF conditions.
In this regard, targeting these miRNAs to either down or upregulate them may be an effective treatment for RIF. Therefore, manipulating miRNA-related signaling in favor of implantation by targeting these miRNAs can be considered a potential therapeutic approach (Figure 3) (Krützfeldt, 2016). Further studies are necessary to introduce miRNAs that can be used to determine the window of implantation time.
FIGURE 3.
MiRNAs as biomarkers. In this graph, mRNAs and miRNAs are compared as two biomarker candidates. MiRNAs can be used as therapeutic targets in a number of ways. Depending on whether the miRNA is upregulated or downregulated in the disease, there are generally two approaches: miRNA inhibition and miRNA replacement. If an increase in the miRNA expression leads to pathology (e.g., RIF), the use of miRNA antagomirs in therapeutic methods will prevent the binding of miRNAs to the target mRNA and will reduce the symptoms of the disease. On the other hand, if a reduction in miRNA expression leads to pathology, a miRNA delivery system (miRNA mimic) can be used. There are several techniques for detecting miRNAs (northern blotting, reverse transcription quantitative real-time PCR (RT-qPCR), microarray technology, nanomaterial-based methods, and nucleic acid amplification).
6 Conclusion and prospects
RIF is a growing problem in the field of reproductive medicine. Repetition of failed cycles in RIF patients is very costly for the patients and affects the physical and psychological health of those under treatment. Determining the expression of miRNAs and key genes in the endometrium can be beneficial in predicting the success rate of implantation in clinics. Since one type of miRNA can affect several target signaling pathways, and subsequently, change the fate of the cell, it is possible to use miRNAs to prevent implant failure or treat RIF patients. Nevertheless, research on the treatment of RIF using miRNA-targeting strategies is still lacking. In this review, we identified three miRNAs that are capable of acting as biomarkers in RIF, namely miR-20b-5p, miR-155-5p, and miR-718. In conclusion, specific endometrial miRNAs are suitable as diagnostic or therapeutic biomarkers in RIF.
Author contributions
SG, AA, and ME: literature review and manuscript drafting; KH and MK: literature review and manuscript editing; BN and SG: study design and critical revisions.
Funding
This project was financially supported by a grant from the Immunology Research Center of Tabriz University of Medical Sciences (Grant No. 65133) and this article has been extracted from the Ph.D. thesis of Sepide Gohari Taban.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.936173/full#supplementary-material
References
- Achache H., Revel A. (2006). Endometrial receptivity markers, the journey to successful embryo implantation. Hum. Reprod. Update 12 (6), 731–746. 10.1093/humupd/dml004 [DOI] [PubMed] [Google Scholar]
- Aghajanova L., Altmäe S., Bjuresten K., Hovatta O., Landgren B-M., Stavreus-Evers A., et al. (2009). Disturbances in the LIF pathway in the endometrium among women with unexplained infertility. Fertil. Steril. 91 (6), 2602–2610. 10.1016/j.fertnstert.2008.04.010 [DOI] [PubMed] [Google Scholar]
- Azarpoor A., Ardeshirylajimi A., Mohammadi Y. S., Dehghan Z., Salehi M. (2020). The expression of miR-31 and its target gene FOXP3 in recurrent implantation failure patients. Int. J. Women's Health Reprod. Sci. 8, 389–395. 10.15296/ijwhr.2020.62 [DOI] [Google Scholar]
- Azhari F., Pence S., Hosseini M. K., Balci B. K., Cevik N., Bastu E., et al. (2022). The role of the serum exosomal and endometrial microRNAs in recurrent implantation failure. J. Matern. Fetal. Neonatal Med. 35 (5), 815–825. 10.1080/14767058.2020.1849095 [DOI] [PubMed] [Google Scholar]
- Bao C., Lu Y., Chen J., Chen D., Lou W., Ding B., et al. (2019). Exploring specific prognostic biomarkers in triple-negative breast cancer. Cell Death Dis. 10 (11), 807. 10.1038/s41419-019-2043-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W., Zhang Y., Li S., Fan Q., Qiu M., Wang Y., et al. (2019). miR-107-5p promotes tumor proliferation and invasion by targeting estrogen receptor-α in endometrial carcinoma. Oncol. Rep. 41 (3), 1575–1585. 10.3892/or.2018.6936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolotti D., Soffritti I., D’Accolti M., Gentili V., Di Luca D., Rizzo R., et al. (2020). HHV-6A infection of endometrial epithelial cells affects miRNA expression and trophoblast cell attachment. Reprod. Sci. 27, 779–786. 10.1007/s43032-019-00102-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H., Zhu X-X., Li Z-F., Zhu Y-P., Lang J-H. (2018). MicroRNA dysregulation and steroid hormone receptor expression in uterine tissues of rats with endometriosis during the implantation window. Chin. Med. J. 131 (18), 2193–2204. 10.4103/0366-6999.240808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakmak H., Taylor H. S. (2011). Implantation failure: molecular mechanisms and clinical treatment. Hum. Reprod. Update 17 (2), 242–253. 10.1093/humupd/dmq037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casals G., Ordi J., Creus M., Fábregues F., Carmona F., Casamitjana R., et al. (2010). Osteopontin and alphavbeta3 integrin as markers of endometrial receptivity: the effect of different hormone therapies. Reprod. Biomed. Online 21 (3), 349–359. 10.1016/j.rbmo.2010.04.012 [DOI] [PubMed] [Google Scholar]
- Chen P., Guo Y., Li T., Jia L., Wang Y., Zhou Y., et al. (2021). Comprehensive analysis of micro RNAs in recurrent implantation failures patients and construction of prediction models based on circulating micro RNAs. Res. Sq. 10.21203/rs.3.rs-145125/v1 [DOI] [Google Scholar]
- Chakrabarty A., Tranguch S., Daikoku T., Jensen K., Furneaux H., Dey S. K., et al. (2007). MicroRNA regulation of cyclooxygenase-2 during embryo implantation. Proc. Natl. Acad. Sci. U. S. A. 104 (38), 15144–15149. 10.1073/pnas.0705917104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L., Yuan Z., Shi H., Bian Y., Guo R. (2017). miR-145 targets the SOX11 3’UTR to suppress endometrial cancer growth. Am. J. Cancer Res. 7 (11), 2305–2317. [PMC free article] [PubMed] [Google Scholar]
- Chen C. H., Lu F., Yang W. J., Yang P. E., Chen W. M., Kang S. T., et al. (2021). A novel platform for discovery of differentially expressed microRNAs in patients with repeated implantation failure. Fertil. Steril. 116 (1), 181–188. 10.1016/j.fertnstert.2021.01.055 [DOI] [PubMed] [Google Scholar]
- Chen C., Zhao Y., Yu Y., Li R., Qiao J. (2016). MiR-125b regulates endometrial receptivity by targeting MMP26 in women undergoing IVF-ET with elevated progesterone on HCG priming day. Sci. Rep. 6 (1), 25302. 10.1038/srep25302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Zheng S., Yang C., Ma B., Jiang D. (2019). MiR-155-5p inhibits the proliferation and migration of VSMCs and HUVECs in atherosclerosis by targeting AKT1. Eur. Rev. Med. Pharmacol. Sci. 23 (5), 2223–2233. 10.26355/eurrev_201903_17270 [DOI] [PubMed] [Google Scholar]
- Chen Y., Song Y-X., Wang Z-N. (2013). The microRNA-148/152 family: multi-faceted players. Mol. Cancer 12 (1), 43. 10.1186/1476-4598-12-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Kim H-R., Lim E. J., Park M., Yoon J. A., Kim Y. S., et al. (2016). Integrative analyses of uterine transcriptome and microRNAome reveal compromised LIF-STAT3 signaling and progesterone response in the endometrium of patients with recurrent/repeated implantation failure (RIF). PloS one 11 (6), e0157696. 10.1371/journal.pone.0157696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu B., Zhong L., Dou S., Wang J., Li J., Wang M., et al. (2015). miRNA-181 regulates embryo implantation in mice through targeting leukemia inhibitory factor. J. Mol. Cell Biol. 7 (1), 12–22. 10.1093/jmcb/mjv006 [DOI] [PubMed] [Google Scholar]
- Cohen A., Shmoish M., Levi L., Cheruti U., Levavi-Sivan B., Lubzens E., et al. (2008). Alterations in micro-ribonucleic acid expression profiles reveal a novel pathway for estrogen regulation. Endocrinology 149 (4), 1687–1696. 10.1210/en.2007-0969 [DOI] [PubMed] [Google Scholar]
- Coughlan C., Ledger W., Wang Q., Liu F., Demirol A., Gurgan T., et al. (2014). Recurrent implantation failure: definition and management. Reprod. Biomed. Online 28 (1), 14–38. 10.1016/j.rbmo.2013.08.011 [DOI] [PubMed] [Google Scholar]
- Craciunas L., Gallos I., Chu J., Bourne T., Quenby S., Brosens J. J., et al. (2019). Conventional and modern markers of endometrial receptivity: a systematic review and meta-analysis. Hum. Reprod. Update 25 (2), 202–223. 10.1093/humupd/dmy044 [DOI] [PubMed] [Google Scholar]
- Davidson L. M., Coward K. (2016). Molecular mechanisms of membrane interaction at implantation. Birth Defects Res. C Embryo Today 108 (1), 19–32. 10.1002/bdrc.21122 [DOI] [PubMed] [Google Scholar]
- Dehghan Z., Salehi M. (2020). The expression of miR-31 and its target gene FOXP3 in recurrent implantation failure patients. Int. J. Women’s Health Reproduction Sci. 8, 389–395. 10.15296/ijwhr.2020.62 [DOI] [Google Scholar]
- Drissennek L., Baron C., Brouillet S., Entezami F., Hamamah S., Haouzi D., et al. (2020). Endometrial miRNome profile according to the receptivity status and implantation failure. Hum. Fertil. 25, 356–368. 10.1080/14647273.2020.1807065 [DOI] [PubMed] [Google Scholar]
- Drissennek L., Baron C., Brouillet S., Entezami F., Hamamah S., Haouzi D., et al. (2022). Endometrial miRNome profile according to the receptivity status and implantation failure. Hum. Fertil. 25 (2), 356–368. 10.1080/14647273.2020.1807065 [DOI] [PubMed] [Google Scholar]
- Edgell T. A., Rombauts L. J., Salamonsen L. A. (2013). Assessing receptivity in the endometrium: the need for a rapid, non-invasive test. Reprod. Biomed. Online 27 (5), 486–496. 10.1016/j.rbmo.2013.05.014 [DOI] [PubMed] [Google Scholar]
- Gamper N., Fillon S., Feng Y., Friedrich B., Lang P., Henke G., et al. (2002). K+ channel activation by all three isoforms of serum-and glucocorticoid-dependent protein kinase SGK. Pflugers Arch. 445 (1), 60–66. 10.1007/s00424-002-0873-2 [DOI] [PubMed] [Google Scholar]
- Gao H-X., Su Y., Zhang A-L., Xu J-W., Fu Q., Yan L., et al. (2019). MiR-34c-5p plays a protective role in chronic obstructive pulmonary disease via targeting CCL22. Exp. Lung Res. 45 (1-2), 1–12. 10.1080/01902148.2018.1563925 [DOI] [PubMed] [Google Scholar]
- Ghaebi M., Abdolmohammadi-Vahid S., Ahmadi M., Eghbal-Fard S., Dolati S., Nouri M., et al. (2019). T cell subsets in peripheral blood of women with recurrent implantation failure. J. Reprod. Immunol. 131, 21–29. 10.1016/j.jri.2018.11.002 [DOI] [PubMed] [Google Scholar]
- Ghafouri-Fard S., Abak A., Tondro Anamag F., Shoorei H., Majidpoor J., Taheri M. (2021). The emerging role of non-coding RNAs in the regulation of PI3K/AKT pathway in the carcinogenesis process. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie 137, 111279. 10.1016/j.biopha.2021.111279 [DOI] [PubMed] [Google Scholar]
- Grewal S., Carver J. G., Ridley A. J., Mardon H. J. (2008). Implantation of the human embryo requires Rac1-dependent endometrial stromal cell migration. Proc. Natl. Acad. Sci. U. S. A. 105 (42), 16189–16194. 10.1073/pnas.0806219105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habibi B., Novin M. G., Salehpour S., Novin M. G., Yeganeh S. M., Nazarian H. (2022). Expression analysis of genes and MicroRNAs involved in recurrent implantation failure: new noninvasive biomarkers of implantation. Biomed. Biotechnol. Res. J. (BBRJ) 6 (1), 145. 10.4103/bbrj.bbrj_246_21 [DOI] [Google Scholar]
- Haouzi D., Mahmoud K., Fourar M., Bendhaou K., Dechaud H., De Vos J., et al. (2009). Identification of new biomarkers of human endometrial receptivity in the natural cycle. Hum. Reprod. 24 (1), 198–205. 10.1093/humrep/den360 [DOI] [PubMed] [Google Scholar]
- Horcajadas J. A., Mínguez P., Dopazo J., Esteban F. J., Domínguez F., Giudice L. C., et al. (2008). Controlled ovarian stimulation induces a functional genomic delay of the endometrium with potential clinical implications. J. Clin. Endocrinol. Metab. 93 (11), 4500–4510. 10.1210/jc.2008-0588 [DOI] [PubMed] [Google Scholar]
- Hu S-J., Ren G., Liu J-L., Zhao Z-A., Yu Y-S., Su R-W., et al. (2008). MicroRNA expression and regulation in mouse uterus during embryo implantation. J. Biol. Chem. 283 (34), 23473–23484. 10.1074/jbc.M800406200 [DOI] [PubMed] [Google Scholar]
- Inyawilert W., Fu T-Y., Lin C-T., Tang P-C. (2015). Let-7-mediated suppression of mucin 1 expression in the mouse uterus during embryo implantation. J. Reproduction Dev. 61, 138–144. 10.1262/jrd.2014-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inyawilert W., Fu T-Y., Lin C-T., Tang P-C. (2014). MicroRNA-199a mediates mucin 1 expression in mouse uterus during implantation. Reprod. Fertil. Dev. 26 (5), 653–664. 10.1071/RD12097 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Shen X., Liu H., Zhen X., Sun H., Hu Y., et al. (2013). O-230 Mirna-21 is a novel regulator of NUR77 and KLF12 mediated decidual prolactin expression in human endometrial stromal cells. Hum. Reprod. 28, i93–i95. 10.1093/humrep/det192 [DOI] [Google Scholar]
- Jimenez P. T., Mainigi M. A., Word R. A., Kraus W. L., Mendelson C. R. (2016). miR-200 regulates endometrial development during early pregnancy. Mol. Endocrinol. 30 (9), 977–987. 10.1210/me.2016-1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson G. A., Burghardt R. C., Bazer F. W. (2014). Osteopontin: a leading candidate adhesion molecule for implantation in pigs and sheep. J. Anim. Sci. Biotechnol. 5 (1), 56. 10.1186/2049-1891-5-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y-J., Forbes K., Carver J., Aplin J. D. (2014). The role of the osteopontin–integrin αvβ3 interaction at implantation: functional analysis using three different in vitro models. Hum. Reprod. 29 (4), 739–749. 10.1093/humrep/det433 [DOI] [PubMed] [Google Scholar]
- Kang Y-J., Lees M., Matthews L. C., Kimber S. J., Forbes K., Aplin J. D., et al. (2015). miR-145 suppresses embryo–epithelial juxtacrine communication at implantation by modulating maternal IGF1R. J. Cell Sci. 128 (4), 804–814. 10.1242/jcs.164004 [DOI] [PubMed] [Google Scholar]
- Katzorke N., Vilella F., Ruiz M., Krüssel J-S., Simon C. (2016). Diagnosis of endometrial-factor infertility: current approaches and new avenues for research. Geburtshilfe Frauenheilkd. 76 (06), 699–703. 10.1055/s-0042-103752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliman H. J., Frankfurter D. (2019). Clinical approach to recurrent implantation failure: evidence-based evaluation of the endometrium. Fertil. Steril. 111 (4), 618–628. 10.1016/j.fertnstert.2019.02.011 [DOI] [PubMed] [Google Scholar]
- Klinge C. M. (2012). miRNAs and estrogen action. Trends Endocrinol. Metab. 23 (5), 223–233. 10.1016/j.tem.2012.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolanska K., Bendifallah S., Canlorbe G., Mekinian A., Touboul C., Aractingi S., et al. (2021). Role of miRNAs in normal endometrium and in endometrial disorders: comprehensive review. J. Clin. Med. 10 (16), 3457. 10.3390/jcm10163457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kresowik J. D., Devor E. J., Van Voorhis B. J., Leslie K. K. (2014). MicroRNA-31 is significantly elevated in both human endometrium and serum during the window of implantation: a potential biomarker for optimum receptivity. Biol. Reprod. 91 (117), 17. 10.1095/biolreprod.113.116590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krützfeldt J. (2016). Strategies to use microRNAs as therapeutic targets. Best. Pract. Res. Clin. Endocrinol. Metab. 30 (5), 551–561. 10.1016/j.beem.2016.07.004 [DOI] [PubMed] [Google Scholar]
- Kuokkanen S., Chen B., Ojalvo L., Benard L., Santoro N., Pollard J. W., et al. (2010). Genomic profiling of microRNAs and messenger RNAs reveals hormonal regulation in microRNA expression in human endometrium. Biol. Reprod. 82 (4), 791–801. 10.1095/biolreprod.109.081059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Ahn E. H., Ryu C. S., Kim J. O., An H. J., Cho S. H., et al. (2020). Association between microRNA machinery gene polymorphisms and recurrent implantation failure. Exp. Ther. Med. 19 (4), 3113–3123. 10.3892/etm.2020.8556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Liu D., Wang J., Deng H., Luo X., Shen X., et al. (2017). Meta-analysis identifies candidate key genes in endometrium as predictive biomarkers for clinical pregnancy in IVF. Oncotarget 8 (60), 102428–102436. 10.18632/oncotarget.22096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., He J., Chen X., Ding Y., Wang Y., Long C., et al. (2014). Mmu-miR-193 is involved in embryo implantation in mouse uterus by regulating GRB7 gene expression. Reprod. Sci. 21 (6), 733–742. 10.1177/1933719113512535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Qiao J., Wang L., Li L., Zhen X., Liu P., et al. (2011). MicroRNA array and microarray evaluation of endometrial receptivity in patients with high serum progesterone levels on the day of hCG administration. Reprod. Biol. Endocrinol. 9 (1), 29. 10.1186/1477-7827-9-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Hu R., Wang C., Guo F., Li X., Wang S., et al. (2021). miR-22 inhibits proliferation and invasion in estrogen receptor α-positive endometrial endometrioid carcinomas cells. Mol. Med. Rep. 24 (3), 2393–2399. 10.3892/mmr.2014.2123 [DOI] [PubMed] [Google Scholar]
- Li Z., Gou J., Jia J., Zhao X. (2015). MicroRNA-429 functions as a regulator of epithelial–mesenchymal transition by targeting Pcdh8 during murine embryo implantation. Hum. Reprod. 30 (3), 507–518. 10.1093/humrep/dev001 [DOI] [PubMed] [Google Scholar]
- Li Z., Jia J., Gou J., Zhao X., Yi T. (2015). MicroRNA-451 plays a role in murine embryo implantation through targeting Ankrd46, as implicated by a microarray-based analysis. Fertil. Steril. 103 (3), 834. 10.1016/j.fertnstert.2014.11.024 [DOI] [PubMed] [Google Scholar]
- Liang J., Wang S., Wang Z. (2017). Role of microRNAs in embryo implantation. Reprod. Biol. Endocrinol. 15 (1), 90. 10.1186/s12958-017-0309-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Chen Y., Liu L., Shen A., Zheng W. (2018). MicroRNA-155-5p suppresses the migration and invasion of lung adenocarcinoma A549 cells by targeting Smad2. Oncol. Lett. 16 (2), 2444–2452. 10.3892/ol.2018.8889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Wang M., Zhang H., Sui C. (2021). Altered microRNA profiles of extracellular vesicles secreted by endometrial cells from women with recurrent implantation failure. Reprod. Sci. 28 (7), 1945–1955. 10.1007/s43032-020-00440-y [DOI] [PubMed] [Google Scholar]
- Liu H., Zhang M., Xu S., Zhang J., Zou J., Yang C., et al. (2018). HOXC8 promotes proliferation and migration through transcriptional up-regulation of TGFβ1 in non-small cell lung cancer. Oncogenesis 7 (2), 1. 10.1038/s41389-017-0016-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Gao R., Chen X., Zhang H., Zheng A., Yang D., et al. (2013). Possible roles of mmu-miR-141 in the endometrium of mice in early pregnancy following embryo implantation. PLoS One 8 (6), e67382. 10.1371/journal.pone.0067382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Zhao H., Li W., Bao H., Qu Q., Ma D., et al. (2020). Up-regulation of miR-145 may contribute to repeated implantation failure after IVF–embryo transfer by targeting PAI-1. Reprod. Biomed. Online 40 (5), 627–636. 10.1016/j.rbmo.2020.01.018 [DOI] [PubMed] [Google Scholar]
- Liu X., Zhao H., Li W., Bao H., Qu Q., Ma D. (2020). Upregulation of miR-145 may contribute to repeated implantation failure in vitro fertilization by targeting PAI-1. Reprod. Biomed. Online 40, 627–636. 10.1016/j.rbmo.2020.01.018 [DOI] [PubMed] [Google Scholar]
- Luo J., Zhu L., Zhou N., Zhang Y., Zhang L., Zhang R., et al. (2021). Construction of circular RNA–MicroRNA–messenger RNA regulatory network of recurrent implantation failure to explore its potential pathogenesis. Front. Genet. 11, 627459. 10.3389/fgene.2020.627459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Dong J., He X., Shen L., Long C., Liu F., et al. (2020). MiR-155-5p exerts tumor-suppressing functions in Wilms tumor by targeting IGF2 via the PI3K signaling pathway. Biomed. Pharmacother. 125, 109880. 10.1016/j.biopha.2020.109880 [DOI] [PubMed] [Google Scholar]
- Ma H-L., Gong F., Tang Y., Li X., Li X., Yang X., et al. (2015). Inhibition of endometrial Tiam1/Rac1 signals induced by miR-22 up-regulation leads to the failure of embryo implantation during the implantation window in pregnant mice. Biol. Reprod. 92 (6152), 152. 10.1095/biolreprod.115.128603 [DOI] [PubMed] [Google Scholar]
- Maesawa Y., Yamada H., Deguchi M., Ebina Y. (2015). History of biochemical pregnancy was associated with the subsequent reproductive failure among women with recurrent spontaneous abortion. Gynecol. Endocrinol. 31 (4), 306–308. 10.3109/09513590.2014.994601 [DOI] [PubMed] [Google Scholar]
- Maillot G., Lacroix-Triki M., Pierredon S., Gratadou L., Schmidt S., Bénès V., et al. (2009). Widespread estrogen-dependent repression of micrornas involved in breast tumor cell growth. Cancer Res. 69 (21), 8332–8340. 10.1158/0008-5472.CAN-09-2206 [DOI] [PubMed] [Google Scholar]
- Margalioth E., Ben-Chetrit A., Gal M., Eldar-Geva T. (2006). Investigation and treatment of repeated implantation failure following IVF-ET. Hum. Reprod. 21 (12), 3036–3043. 10.1093/humrep/del305 [DOI] [PubMed] [Google Scholar]
- Mariee N., Li T., Laird S. (2012). Expression of leukaemia inhibitory factor and interleukin 15 in endometrium of women with recurrent implantation failure after IVF; correlation with the number of endometrial natural killer cells. Hum. Reprod. 27 (7), 1946–1954. 10.1093/humrep/des134 [DOI] [PubMed] [Google Scholar]
- Mogilyansky E., Rigoutsos I. (2013). The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 20 (12), 1603–1614. 10.1038/cdd.2013.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Moya J. M., Vilella F., Martínez S., Pellicer A., Simón C. (2014). The transcriptomic and proteomic effects of ectopic overexpression of miR-30d in human endometrial epithelial cells. Mol. Hum. Reprod. 20 (6), 550–566. 10.1093/molehr/gau010 [DOI] [PubMed] [Google Scholar]
- Nazarian H., Novin M. G., Khaleghi S., Habibi B. (2022). Small non-coding RNAs in embryonic pre-implantation. Curr. Mol. Med. 22 (4), 287–299. 10.2174/1566524021666210526162917 [DOI] [PubMed] [Google Scholar]
- Pan J. l., Yuan D. z., Zhao Y. b., Nie L., Lei Y., Liu M., et al. (2017). Progesterone-induced miR-133a inhibits the proliferation of endometrial epithelial cells. Acta Physiol. 219 (3), 683–692. 10.1111/apha.12762 [DOI] [PubMed] [Google Scholar]
- Pei T., Liu C., Liu T., Xiao L., Luo B., Tan J., et al. (2018). miR-194-3p represses the progesterone receptor and decidualization in eutopic endometrium from women with endometriosis. Endocrinology 159 (7), 2554–2562. 10.1210/en.2018-00374 [DOI] [PubMed] [Google Scholar]
- Ponsuksili S., Tesfaye D., Schellander K., Hoelker M., Hadlich F., Schwerin M., et al. (2014). Differential expression of miRNAs and their target mRNAs in endometria prior to maternal recognition of pregnancy associates with endometrial receptivity for in vivo-and in vitro-produced bovine embryos. Biol. Reprod. 91 (6135), 135. 10.1095/biolreprod.114.121392 [DOI] [PubMed] [Google Scholar]
- Qian K., Hu L., Chen H., Li H., Liu N., Li Y., et al. (2009). Hsa-miR-222 is involved in differentiation of endometrial stromal cells in vitro . Endocrinology 150 (10), 4734–4743. 10.1210/en.2008-1629 [DOI] [PubMed] [Google Scholar]
- Quinn C., Casper R. (2009). Pinopodes: a questionable role in endometrial receptivity. Hum. Reprod. Update 15 (2), 229–236. 10.1093/humupd/dmn052 [DOI] [PubMed] [Google Scholar]
- Reed B. G., Babayev S. N., Chen L. X., Carr B. R., Word R. A., Jimenez P. T., et al. (2018). Estrogen-regulated miRNA-27b is altered by bisphenol A in human endometrial stromal cells. Reproduction 156 (6), 559–567. 10.1530/REP-18-0041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekker K., Altmäe S., Suhorutshenko M., Peters M., Martinez-Blanch J. F., Codoñer F. M., et al. (2018). A two-cohort RNA-seq study reveals changes in endometrial and blood miRNome in fertile and infertile women. Genes 9 (12), 574. 10.3390/genes9120574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revel A., Achache H., Stevens J., Smith Y., Reich R. (2011). MicroRNAs are associated with human embryo implantation defects. Hum. Reprod. 26 (10), 2830–2840. 10.1093/humrep/der255 [DOI] [PubMed] [Google Scholar]
- Reza A. M. M. T., Choi Y. J., Han S. G., Song H., Park C., Hong K., et al. (2019). Roles of microRNAs in mammalian reproduction: from the commitment of germ cells to peri-implantation embryos. Biol. Rev. Camb. Philos. Soc. 94 (2), 415–438. 10.1111/brv.12459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehpour S., Zamaniyan M., Saharkhiz N., Zadeh Modares S., Hosieni S., Seif S., et al. (2016). Does intrauterine saline infusion by intrauterine insemination (IUI) catheter as endometrial injury during IVF cycles improve pregnancy outcomes among patients with recurrent implantation failure?: an RCT. Int. J. Reprod. Biomed. 14 (9), 583–588. 10.29252/ijrm.14.9.583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Cheng Y., Li M., Wang H., Zhang J., Guo X., et al. (2022). Identification of key endometrial microRNAs and its target genes associated with pathogenesis of recurrent implantation failure by integrated bioinformatics analysis. Front. Genet. 13, 919301. 10.3389/fgene.2022.919301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shariati M. B. H., Niknafs B., Seghinsara A. M., Shokrzadeh N., Alivand M. R. (2019). Administration of dexamethasone disrupts endometrial receptivity by alteration of expression of miRNA 223, 200a, LIF, Muc1, SGK1, and ENaC via the ERK1/2-mTOR pathway. J. Cell. Physiol. 234 (11), 19629–19639. 10.1002/jcp.28562 [DOI] [PubMed] [Google Scholar]
- Sheedy F. J. (2015). Turning 21: induction of miR-21 as a key switch in the inflammatory response. Front. Immunol. 6, 19. 10.3389/fimmu.2015.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikhansari G., Soltani-Zangbar M. S., Pourmoghadam Z., Kamrani A., Azizi R., Aghebati-Maleki L., et al. (2019). Oxidative stress, inflammatory settings, and microRNA regulation in the recurrent implantation failure patients with metabolic syndrome. Am. J. Reprod. Immunol. 82 (4), e13170. 10.1111/aji.13170 [DOI] [PubMed] [Google Scholar]
- Shekibi M., Heng S., Nie G. (2022). MicroRNAs in the regulation of endometrial receptivity for embryo implantation. Int. J. Mol. Sci. 23 (11), 6210. 10.3390/ijms23116210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C., Shen H., Fan L-J., Guan J., Zheng X-B., Chen X., et al. (2017). Endometrial microRNA signature during the window of implantation changed in patients with repeated implantation failure. Chin. Med. J. 130 (05), 566–573. 10.4103/0366-6999.200550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokrzadeh N., Alivand M. R., Abedelahi A., Hessam Shariati M. B., Niknafs B. (2019). Calcitonin administration improves endometrial receptivity via regulation of LIF, Muc-1 and microRNA Let-7a in mice. J. Cell. Physiol. 234 (8), 12989–13000. 10.1002/jcp.27969 [DOI] [PubMed] [Google Scholar]
- Shokrzadeh N., Alivand M. R., Abedelahi A., Hessam Shariati M. B., Niknafs B. (2018). Upregulation of HB‐EGF, Msx. 1, and miRNA Let-7a by administration of calcitonin through mTOR and ERK1/2 pathways during a window of implantation in mice. Mol. Reprod. Dev. 85 (10), 790–801. 10.1002/mrd.23061 [DOI] [PubMed] [Google Scholar]
- Shufaro Y., Schenker J. G. (2011). Implantation failure, etiology, diagnosis and treatment. Int. J. Infertil. Fetal Med. 2 (1), 1–7. 10.5005/jp-journals-10016-1009 [DOI] [Google Scholar]
- Simon A., Laufer N. (2012). Assessment and treatment of repeated implantation failure (RIF). J. Assist. Reprod. Genet. 29 (11), 1227–1239. 10.1007/s10815-012-9861-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirohi V. K., Gupta K., Kapoor R., Dwivedi A. (2019). MicroRNA-145 targets Smad1 in endometrial stromal cells and regulates decidualization in rat. J. Mol. Med. 97 (4), 509–522. 10.1007/s00109-019-01744-6 [DOI] [PubMed] [Google Scholar]
- Sirohi V. K., Gupta K., Kumar R., Shukla V., Dwivedi A. (2018). Selective estrogen receptor modulator ormeloxifene suppresses embryo implantation via inducing miR-140 and targeting insulin-like growth factor 1 receptor in rat uterus. J. Steroid Biochem. Mol. Biol. 178, 272–282. 10.1016/j.jsbmb.2018.01.006 [DOI] [PubMed] [Google Scholar]
- Tan B. K., Vandekerckhove P., Kennedy R., Keay S. D. (2005). Investigation and current management of recurrent IVF treatment failure in the UK. BJOG Int. J. Obstetrics Gynaecol. 112 (6), 773–780. 10.1111/j.1471-0528.2005.00523.x [DOI] [PubMed] [Google Scholar]
- Tan Q., Shi S., Liang J., Zhang X., Cao D., Wang Z., et al. (2020). MicroRNAs in small extracellular vesicles indicate successful embryo implantation during early pregnancy. Cells 9 (3), 645. 10.3390/cells9030645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S., Su X., Qi L., Jin X-H., Hu Y., Wang C-L., et al. (2015). MiR-143 and rat embryo implantation. Biochim. Biophys. Acta 1850 (4), 708–721. 10.1016/j.bbagen.2014.11.023 [DOI] [PubMed] [Google Scholar]
- Timeva T., Shterev A., Kyurkchiev S. (2014). Recurrent implantation failure: the role of the endometrium. J. Reprod. Infertil. 15 (4), 173–183. [PMC free article] [PubMed] [Google Scholar]
- Troy P. J., Daftary G. S., Bagot C. N., Taylor H. S. (2003). Transcriptional repression of peri-implantation EMX2 expression in mammalian reproduction by HOXA10. Mol. Cell. Biol. 23 (1), 1–13. 10.1128/mcb.23.1.1-13.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuckerman E., Mariee N., Prakash A., Li T. C., Laird S. (2010). Uterine natural killer cells in peri-implantation endometrium from women with repeated implantation failure after IVF. J. Reprod. Immunol. 87 (1-2), 60–66. 10.1016/j.jri.2010.07.001 [DOI] [PubMed] [Google Scholar]
- von Grothusen C., Frisendahl C., Modhukur V., Lalitkumar P. G., Peters M., Faridani O. R., et al. (2022). Uterine fluid microRNAs are dysregulated in women with recurrent implantation failure. Hum. Reprod. 37 (4), 734–746. 10.1093/humrep/deac019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Liu Y. (2020). Identification of key genes, regulatory factors, and drug target genes of recurrent implantation failure (RIF). Gynecol. Endocrinol. 36 (5), 448–455. 10.1080/09513590.2019.1680622 [DOI] [PubMed] [Google Scholar]
- Wang Z., Liu Y., Liu J., Kong N., Jiang Y., Jiang R., et al. (2021). ATF3 deficiency impairs the proliferative–secretory phase transition and decidualization in RIF patients. Cell Death Dis. 12 (4), 387. 10.1038/s41419-021-03679-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby S., Salamonsen L. A., Evans J. (2018). The endometrial polarity paradox: differential regulation of polarity within secretory-phase human endometrium. Endocrinology 159 (1), 506–518. 10.1210/en.2016-1877 [DOI] [PubMed] [Google Scholar]
- Xia H. F., Jin X. H., Cao Z. F., Hu Y., Ma X. (2014). Micro RNA expression and regulation in the uterus during embryo implantation in rat. FEBS J. 281 (7), 1872–1891. 10.1111/febs.12751 [DOI] [PubMed] [Google Scholar]
- Xia H-F., Cao J-L., Jin X-H., Ma X. (2014). MiR199a is implicated in embryo implantation by regulating Grb10 in rat. Reproduction 147 (1), 91–99. 10.1530/REP-13-0290 [DOI] [PubMed] [Google Scholar]
- Xia H-F., Jin X-H., Cao Z-F., Shi T., Ma X. (2014). MiR-98 is involved in rat embryo implantation by targeting Bcl-xl. FEBS Lett. 588 (4), 574–583. 10.1016/j.febslet.2013.12.026 [DOI] [PubMed] [Google Scholar]
- Xiao L., Pei T., Huang W., Zhou M., Fu J., Tan J., et al. (2020). MicroRNA22-5p targets ten-eleven translocation and regulates estrogen receptor 2 expression in infertile women with minimal/mild endometriosis during implantation window. PloS one 15 (7), e0234086. 10.1371/journal.pone.0234086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Zhou M., Cao Y., Zhang D., Han M., Gao X., et al. (2019). Genome-wide analysis of long noncoding RNAs, microRNAs, and mRNAs forming a competing endogenous RNA network in repeated implantation failure. Gene 720, 144056. 10.1016/j.gene.2019.144056 [DOI] [PubMed] [Google Scholar]
- Yuan D-z., Lei Y., Zhao D., Pan J-l., Zhao Y-b., Nie L., et al. (2019). Progesterone-induced miR-145/miR-143 inhibits the proliferation of endometrial epithelial cells. Reprod. Sci. 26 (2), 233–243. 10.1177/1933719118768687 [DOI] [PubMed] [Google Scholar]
- Zeng H., Fu Y., Shen L., Quan S. (2021). MicroRNA signatures in plasma and plasma exosome during window of implantation for implantation failure following in-vitro fertilization and embryo transfer. Reprod. Biol. Endocrinol. 19 (1), 180. 10.1186/s12958-021-00855-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Ni T., Dang Y., Ding L., Jiang J., Li J., et al. (2020). MiR-148a-3p may contribute to flawed decidualization in recurrent implantation failure by modulating HOXC8. J. Assist. Reprod. Genet. 37 (10), 2535–2544. 10.1007/s10815-020-01900-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Zhang H., Jiang Y., Xue B., Diao Z., Ding L., et al. (2015). MicroRNA-181a is involved in the regulation of human endometrial stromal cell decidualization by inhibiting Krüppel-like factor 12. Reprod. Biol. Endocrinol. 13 (1), 23. 10.1186/s12958-015-0019-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Fu J., Xiao L., Yang S., Song Y., Zhang X., et al. (2016). miR-196a overexpression activates the MEK/ERK signal and represses the progesterone receptor and decidualization in eutopic endometrium from women with endometriosis. Hum. Reprod. 31 (11), 2598–2608. 10.1093/humrep/dew223 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.