Key Points
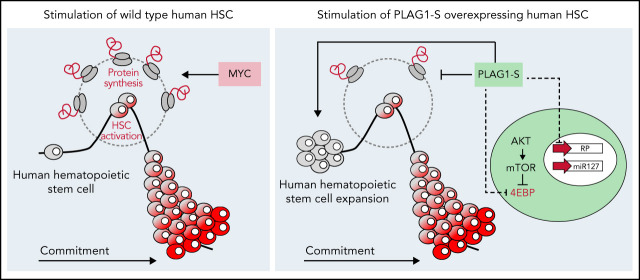
The PLAG1 transcription factor promotes human HSC self-renewal and dormancy.
PLAG1 enforces stemness by dampening expression of translation machinery activated in HSC-stimulatory conditions.
Visual Abstract
Abstract
Hematopoietic stem cell (HSC) dormancy is understood as supportive of HSC function and its long-term integrity. Although regulation of stress responses incurred as a result of HSC activation is recognized as important in maintaining stem cell function, little is understood of the preventive machinery present in human HSCs that may serve to resist their activation and promote HSC self-renewal. We demonstrate that the transcription factor PLAG1 is essential for long-term HSC function and, when overexpressed, endows a 15.6-fold enhancement in the frequency of functional HSCs in stimulatory conditions. Genome-wide measures of chromatin occupancy and PLAG1-directed gene expression changes combined with functional measures reveal that PLAG1 dampens protein synthesis, restrains cell growth and division, and enhances survival, with the primitive cell advantages it imparts being attenuated by addition of the potent translation activator, c-MYC. We find PLAG1 capitalizes on multiple regulatory factors to ensure protective diminished protein synthesis including 4EBP1 and translation-targeting miR-127 and does so independently of stress response signaling. Overall, our study identifies PLAG1 as an enforcer of human HSC dormancy and self-renewal through its highly context-specific regulation of protein biosynthesis and classifies PLAG1 among a rare set of bona fide regulators of messenger RNA translation in these cells. Our findings showcase the importance of regulated translation control underlying human HSC physiology, its dysregulation under activating demands, and the potential if its targeting for therapeutic benefit.
Introduction
Hematopoietic stem cells (HSCs) ensure long-term multilineage blood regeneration through their enduring self-renewal capacity.1,2 As such, transplantation of hematopoietic stem and progenitor cells (HSPCs) sourced from bone marrow (BM) or umbilical cord blood (CB), can be life-saving for patients with myriad malignant and nonmalignant disorders.3,4 Widespread application of HSPC therapies remains limited by disease-causing mutations in autologous HSPCs and difficulties sourcing HLA-matched allogenic BM.3,5 Efforts to bridge these gaps include attempts to achieve therapeutic HSPC genome editing and amplification of less plentiful but immunologically superior CB HSPCs.6-13 However, these goals remain challenged by our incomplete understanding of the fundamental biology underlying human HSC identity and fate-decisions, especially in stimulatory regenerative conditions where HSCs are predisposed to depletion.6,14-19
Despite their extensive regenerative capacity, the most potent HSCs live principally in a state of quiescence and dormancy, wherein cells exhibit reduced anabolic activity that preserves the integrity of the HSC pool.20-23 Indeed, conditions that cause HSPCs to exit dormancy, such as the regenerative stress of ex vivo manipulation or transplantation, compromises the pool of long-term (LT) repopulation-competent HSCs.24-26 Interventions that restrain mitochondrial metabolism or cell cycle to limit HSC activation in these settings can aid in preservation of mouse and human HSCs.21,27-31 Importantly, however, the process of protein biosynthesis, which in many contexts is co-regulated with metabolism and can fuel cellular growth and division, has not been well explored for its role in human HSC dormancy and transplantation paradigms.
In their native environment murine HSCs, like several other model stem cell types,32-35 require tightly controlled, low levels of protein synthesis,36,37 understood to limit exhaustive proliferation and safeguard the proteome to evade attrition or death from proteotoxicity.37-40 In response to stimulatory conditions, the activation of stress effectors in murine HSCs serves to rebalance proteostasis by favoring diminished translation rates.41,42 These studies have begun to elucidate the multifactorial contributions to HSC dormancy and fate; however, they raise a number of key questions: Under activating conditions, what are the dynamics of translation in stem vs mature human hematopoietic cells? Can translation modulation be supportive of human HSCs? Can translation regulation be decoupled from stress response? And what factor(s) control translation in human HSCs?
PLAG1 is a zinc finger transcription factor (TF), first discovered as being rearranged in pleomorphic adenomas of the salivary gland and plays essential regulatory roles in mammalian fetal growth and development.43,44 We now identify PLAG1 as an essential regulator of human HSC dormancy and self-renewal by acting as a novel negative regulator of protein synthesis independently of triggering intrinsic stress-responsive effectors. Our study supports an emerging paradigm that dysregulation of protein synthesis is a key clinical demand on human HSCs and its modulation could be leveraged for therapeutic benefit.
Methods
CB Lin−CD34+ culture and analysis
Lin−CD34+ CB cells were isolated, transduced with lentiviral vectors (supplemental Methods available on the Blood web site), and cultured in StemSpan Serum Free Expansion Medium with 20 ng/mL thrombopoietin and interleukin-6 and 100 ng/mL SCF and FLT3 ligand at 37°C 5% CO2. Culture-derived cells were analyzed by xenotransplantation, extreme limiting dilution analysis (ELDA),45 colony-forming unit (CFU) assay, flow cytometry, immunofluorescence microscopy, quantitative polymerase chain reaction, CUT&RUN,46 or RNA-sequencing (RNA-seq) (supplemental Methods; supplemental Tables 7-10).
Mouse xenotransplantations
Mouse work was done in pathogen-free facilities in compliance with ethical regulations approved by McMaster University’s Animal Research Ethics Board. Age- and sex-matched NSG mice (Jackson Laboratories) were sublethally irradiated (315 cGy) 1 day before intrafemoral injection of HSPCs, and endpoint tissues were processed by passage through 40-μM cell strainer as previously described.47
Informed consent was received for all of the human cord blood samples used.
Results
PLAG1 is enriched and essential in human HSCs
We previously showed that when co-overexpressed, short isoforms of PLAG1 (-S and -B) (supplemental Figure 1A) and the USF2 TFs can cooperatively transactivate the pro-self-renewal gene MSI2.48 However, although USF2 expression is stable across the human hematopoietic hierarchy, PLAG1 is specifically elevated in HSCs from human CB48-51 and murine52 and human53 BM (supplemental Figure 1B-E), altogether suggesting that PLAG1 may have important unexplored functions in the most primitive hematopoietic cells.
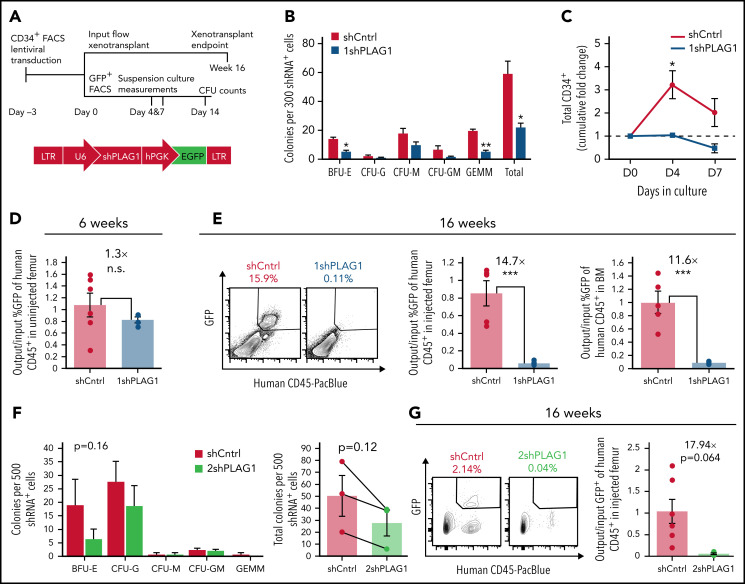
To evaluate this, we queried the in vitro and in vivo potential of CB-derived Lin−CD34+ HSPCs expressing PLAG1-targeting short hairpin RNAs (Figure 1A; supplemental Figure 1F). PLAG1-depleted HSPCs generated fewer colonies, due mainly to reduced burst forming erythroid (BFU-E) and primitive granulocyte-erythroid-megakaryocyte-monocyte (CFU-GEMM) colonies (Figure 1B). This was mirrored in suspension culture where shPLAG1 significantly reduced total nucleated cell and CD34+ outputs over 7 days (Figure 1C; supplemental Figure 1G). Six weeks following NSG mouse xenotransplantation, when engraftment is largely contributed by progenitors, there was modest but nonsignificant reduction in the representation of shPLAG1-expressing GFP+ cells (Figure 1D). However, 16 weeks after transplant when the graft is sustained by bona fide HSCs, engraftment by shPLAG1-expressing cells was significantly impaired 11- to 14-fold (Figure 1E). Patterns in reduced CFU output, and impairment of long-term BM reconstitution were replicated by a second independent PLAG1-targeting hairpin (Figure 1F-G; supplemental Figure 1H).
Figure 1.
PLAG1 is enriched and essential in human HSCs. (A) Schematic of Lin−CD34+ CB HSPC in vitro and in vivo functional assay timelines and lentivectors used for PLAG1 knockdown. (B) Primary CFU output by Lin−CD34+ HSPCs expressing control or (1sh) PLAG1-targeting hairpins. (C) Cumulative in vitro CD34+ cell fold change of cultured of Lin−CD34+ HSPCs expressing 1shPLAG1 or control hairpins. (D) GFP+ engraftment in the uninjected femur of primary NSG mice 6 weeks after xenotransplantation of Lin−CD34+ cells expressing either 1shPLAG1 (n = 4) or control (n = 6) hairpins normalized to input % GFP+ levels. (E) GFP+ engraftment in the injected femur and bone marrow of primary NSG mice 16 weeks after xenotransplantation of Lin−CD34+ cells expressing either 1shPLAG1 (n = 4) or control (n = 5) hairpins normalized to input % GFP+ levels. (F) Primary CFU output by Lin−CD34+ HSPCs expressing control or a second (2sh) PLAG1-targeting hairpin. (G) GFP+ engraftment in the injected femur and bone marrow of primary NSG mice 16 weeks after xenotransplantation of Lin−CD34+ cells expressing either 2shPLAG1 (n = 3) or control (n = 6) hairpins normalized to input % GFP+ levels. Data are presented as average ± SEM unless otherwise indicated. Each point represents one mouse or an individual CB unit. ***P < .005, **P < .01, *P < .05. n.s., not significant. See also supplemental Figure 1.
PLAG1-S is a positive regulator of human HSPC fitness
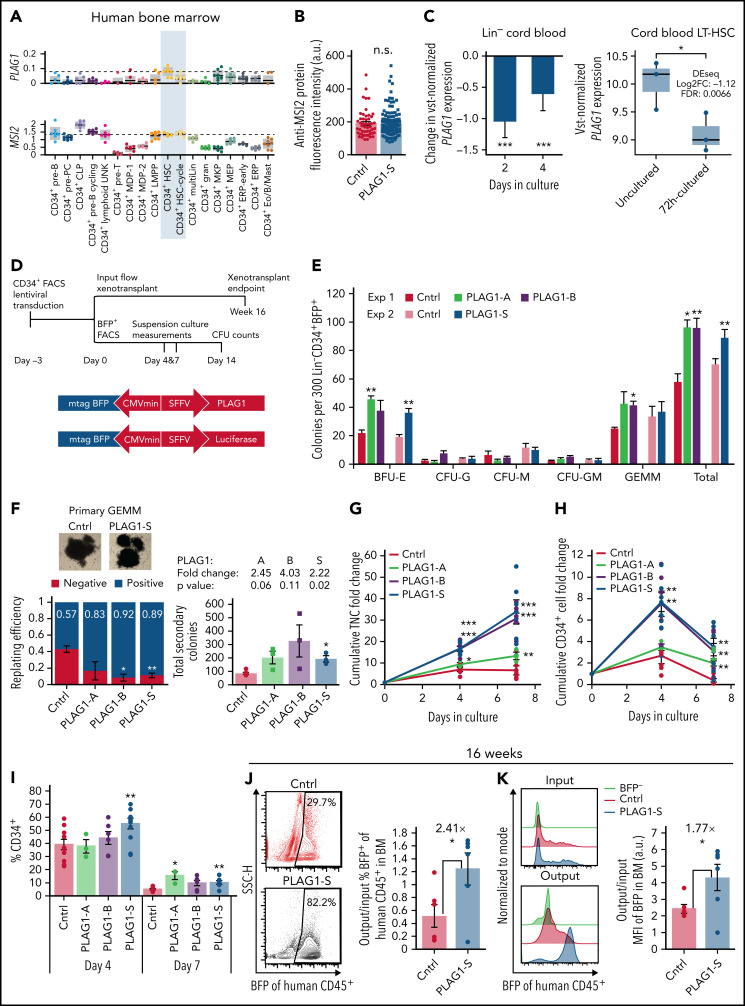
Mining expression profile data, we next uncovered intriguing differences in the expression profiles of PLAG1 and MSI2 signifying that the role of PLAG1 in human HSC physiology may transcend the PLAG1/USF2-MSI2 regulatory axis. First, in the human hierarchy, PLAG1 expression is highly restricted to noncycling HSCs, whereas MSI2 is strongly expressed in both noncycling and cycling HSCs and CD34+ progenitors (Figure 2A).54 Second, upon 5-fluorouracil stress induction of mouse HSC cycling, Msi2 is elevated, whereas Plag1 is repressed (supplemental Figure 2A).55 Most importantly and unexpectedly, PLAG1-S overexpression without USF2 in Lin−CD34+ cells is insufficient to enhance MSI2 protein expression (Figure 2B). Moreover, PLAG1 levels are reduced in human HSPCs activated by transplantation (supplemental Figure 2B)28 or culture (Figure 2C; supplemental Figure 2C).39,56,57 Therefore, to explore the potential for an independent function of PLAG1 in modulating human HSPC fate decisions in these contexts, we assayed Lin−CD34+ CB cells in vitro and in vivo upon gain of PLAG1 (Figure 2D; supplemental Figure 2D).
Figure 2.
PLAG1-S is a positive regulator of human HSPC fitness. (A) PLAG1 and MSI2 transcript expression in human bone marrow cell populations determined by single-cell RNA-seq.54 (B) MSI2 protein expression measured by immunofluorescence microscopy in PLAG1-S overexpressing Lin−CD34+ cells. (C) Change in variance-stabilizing transformed (vst) PLAG1 transcript expression in Lin− cord blood cells cultured for 2 or 4 days showing the P value from 1-tailed Student t-test39 and in 72-hour cultured long-term (Lin−CD34+CD38−CD45RA−CD90+CD49f+) CB HSCs showing the P value from 1-tailed Student t-test and differential expression from DEseq analysis.56 (D) Schematic of Lin−CD34+ CB HSPC in vitro and in vivo functional assay timelines and lentivectors used for overexpression of PLAG1 protein isoforms. (E) Primary CFU output by BFP+ Lin−CD34+ cells overexpressing PLAG1-A, B, or S, or Luciferase control (n = 3 per experiment). (F) Secondary CFU replating efficiency (for each condition, 12 GEMMs from 3 distinct CB units were replated into new wells. Negative indicates no secondary colonies were derived from the primary GEMM, Positive indicates at least 1 secondary colony was derived from the primary GEMM) and the total number of secondary colonies on positive plates with images of representative primary GEMM colonies used. (Square data points are from experiment 1 and circle data points are from experiment 2, n=3 per experiment.) (G) Cumulative in vitro total nucleated cell (TNC) and (H) CD34+ cell fold change of cultured of Lin−CD34+ cells overexpressing PLAG1-A (n = 3), B, or S, or Luciferase control (n = 6). (I) Frequency of CD34 positivity in PLAG1-A (n = 3), B, or S, or Luciferase control (n = 6) overexpressing cultures after 4 and 7 days ex vivo. (J) Representative flow plots and quantification relative to input proportions of BFP representation in CD45+ human grafts in bone marrow of primary NSG mice 16 weeks after receiving Lin−CD34+ cells overexpressing either PLAG1-S or Luciferase control (n = 6). (K) Representative flow plots of input and output BFP fluorescence intensity and quantification of output/input BFP median fluorescence intensity in bone marrow of primary NSG mice 16 weeks after receiving Lin−CD34+ cells overexpressing either PLAG1-S or Luciferase control (n = 6). Data are presented as average ± SEM unless otherwise indicated. Each point represents 1 mouse or an individual CB unit. ***P < .005, **P < .01, *P < .05. n.s., not significant. See also supplemental Figure 2.
Overexpression of each of the 3 PLAG1 isoforms (supplemental Figure 1A)43,44,48 increased BFU-E output with only the shorter isoforms (-S and -B) enhancing GEMM replating efficiency (Figure 2E-F) and CD34+ cell expansion in culture over 7 days (peak advantage at day 4) (Figure 2G-I; supplemental Figure 2E). In PLAG1-S overexpressing (PLAG1-SOE) cultures enhanced CD34 maintenance occurs concurrently with a reduction of committed CD33+ cells and an elevated frequency of CD34+CD71+ (BFU-E) progenitors (supplemental Figure 1F,G). Altogether, these findings point to the short isoforms of PLAG1 as important positive regulators of human HSPCs and contextualize our past observation that these isoforms are preferentially expressed in the HSC-enriched compartment of human CB.48
Given the strong and comparable phenotypes between PLAG1-S and PLAG1-B, we prioritized the shortest form, PLAG1-S, for assessment in competitive repopulation assays (Figure 2D). Following a 4- or 6-week repopulation period in NSG mice, PLAG1-SOE and control short-term progenitors are similarly capable of contributing to engraftment (supplemental Figure 2H). After 16 weeks, the proportion and intensity of the BFP transduction marker relative to input levels was however significantly higher in the BM of PLAG1-SOE recipients compared with control (Figure 2J-K; supplemental Figure 2I). Because BFP intensity from this bidirectional promoter vector provides a surrogate measure for transgene expression40,58 this indicates that cotransplanted cells overexpressing PLAG1-S to higher levels outcompete those expressing lower levels. Importantly, the enhanced fitness of PLAG1-SOE HSCs was neither associated with splenomegaly (supplemental Figure 2J) nor elevation of the CD34+ compartment; and despite erythroid bias in vitro, in the in vivo niche PLAG1-SOE grafts displayed balanced multilineage output (supplemental Figure 2K; supplemental Table 1).
PLAG1-S overexpression promotes self-renewal of human LT-HSCs
To evaluate the capacity of PLAG1-S to promote LT-HSC self-renewal, we performed serial gold-standard primary xenotransplantation in limiting dilution to quantify HSC frequencies in the culture-derived progeny of Lin−CD34+ cells immediately after induction of ectopic PLAG1-S (Figure 3A). Fourteen weeks following primary xenotransplantation, only 33% of mice transplanted with control cells, compared with 66% of mice transplanted with PLAG1-SOE cells met multilineage BM engraftment criteria (Figure 3B,D). ELDA45 determined HSC frequencies of 1 in 715 PLAG1-SOE vs 1 in 11 157 control cells, representing 15.6-fold enhancement of functionally validated HSCs (Figure 3C-D). A similar analysis of splenic grafts revealed 75-fold enhancement in primitive cells capable of repopulating this environment (supplemental Figure 3A-B). Notably, HSC renewal achieved by ectopic MSI2 does not occur at this early timepoint posttransduction,47 highlighting a unique functional capacity of PLAG1-S enacted via MSI2-independent means.
Figure 3.
PLAG1-S overexpression promotes self-renewal of long-term human HSCs. (A) Schematic of primary and secondary xenotransplantation in limiting dilution format. (B) Representative flow plots of human CD45+BFP+ multilineage (CD33+, CD19+) engraftment of primary recipient mice in injected femur. Percent human CD45+BFP+ engraftment in injected femur of primary recipient mice across multiple cell input doses. Dashed line indicates cutoff for calling engraftment, which was >0.5% human chimerism including both myeloid (CD45+BFP+CD33+) and lymphoid (CD45+BFP+CD19+) lineages. (C-D) Quantification of HSC frequency by ELDA45 of injected femur of primary recipient mice. Shaded area under the curve represents 95% confidence interval of HSC frequency. (E) Percent human CD45+BFP+ engraftment in injected femur of secondary recipient mice across multiple cell input doses. Dashed line indicates cutoff for calling engraftment, which was the same as for primary mice. (F-G) Quantification of HSC frequency by ELDA of injected femur or uninjected bone marrow of secondary recipient mice and of in vivo expansion. Shaded area under the curve represents 95% confidence interval of HSC frequency. Total BFP+ cells within whole-body BM of primary mice were extrapolated, as previously,47 based on femur and hind limb counts and proportional accounting from Colvin et al,107 and in vivo expansion is measured as the fold difference of total BFP+ HSCs in donor mice relative to total day 0 HSCs initially transplanted into the 6 donor mice. Data are presented as average ± SEM unless otherwise indicated. Each point represents 1 mouse. See also supplemental Figure 3.
At the secondary transplant endpoint, we confirmed that recipients of PLAG1-SOE cells exhibited enduring multilineage reconstitution and heightened engraftment because of a 4.8-fold higher HSC frequency relative to control (Figure 3E-G; supplemental Figure 3C). Accounting for initial HSC input into primary recipients, we find that enhanced engraftment in secondary recipients is not the result of enhanced renewal in vivo (Figure 3G) and is primarily attributed to the initial ex vivo promotion of the HSC compartment. These results quantitatively demonstrate that PLAG1-SOE does not impart excessive HSC self-renewal characteristic of clonal hematopoiesis or premalignancy, and that the potent promotion of HSCs in a stimulatory setting is not associated with detrimental exhaustion of the LT-HSC compartment.
To test for the effect of PLAG1-S on HSC renewal over longer periods in vitro, cells cultured for 7 additional days were subjected to limiting dilution xenotransplantation (supplemental Figure 3D). Relative to immediately postinduction of ectopic PLAG1-S, the number of functional HSCs continue to increase 1.6-fold and sustain 4.3-fold higher frequencies relative to control (supplemental Figure 3E-G). Thus, the potent stem cell advantage endowed by PLAG1-S, though sustained in culture, can be maximally achieved shortly after PLAG1-S induction, underscoring PLAG1-S as an early actor in promoting HSC function to improve hematopoietic repopulation.
PLAG1-S enforces a pro-HSC transcriptional state
Genomic binding and transcriptomic profiles were next used to uncover molecular targets underpinning the positive regulation of HSCs by PLAG1-S. CUT&RUN performed in PLAG1-SOE Lin−CD34+ cells identified 9788 reproducible genomic binding sites46,59 (supplemental Figure 4A; supplemental Table 2). Consistent with our published chromatin immunoprecipitation sequencing in K562 cells48 and its known role as a TF, PLAG1-S sites are principally located in promoter regions (58.4%) (Figure 4A). De novo motif discovery revealed that PLAG1-S is predominantly (35%) bound to G-rich core consensus sequences expected for PLAG family members, likewise supporting the specificity of the CUT&RUN profile generated (Figure 4B). To a lesser extent, PLAG1-S is also bound to non-canonical motifs, including those associated with other zinc finger, GATA, or RUNX TFs (Figure 4B; supplemental Table 3).
Figure 4.
PLAG1-S enforces a pro-HSC transcriptional state. (A) Loci annotations and distribution of PLAG1-S binding sites in the Lin-CD34+ genome identified by CUT&RUN. (B) Enriched motifs among PLAG1-S genomic binding sites determined by HOMER indicating the % of PLAG1-S targets bound to the consensus and P value of the enrichment relative to genome-wide background occurrence of the consensus. (C) Volcano plot of differential gene expression in PLAG1-S overexpressing Lin−CD34+ cells. Red- or blue-colored genes are significantly changed by adjusted P value < .05 and green- and purple-colored genes are directly bound by PLAG1-S. (D) PLAG1-S overexpression and shPLAG1 transcriptomic alignment to DMAP signatures of hematopoietic compartments.49 Numbers above or below the bars indicate the empirical P value determined based on the percentage of times for which the observed value (set of up- or downregulated genes) was as large or larger in that population than random values (equal number of randomly selected genes) based on 1000 trials. (E) Enrichment map of significantly enriched gene sets (FDR < 0.1) in PLAG1-SOE Lin−CD34+ cells compared with control. Genes bound by PLAG1-S in Lin-CD34+ cells (CUT&RUN q-value cutoff of 0.05) are intersected to gene sets by Mann-Whitney U test (P < .05) and the width of green edges correlates with increasing statistical significance of the overlap. Node size reflects the number of genes in the gene set. (F) Forty-one of 46 gene sets from the “Establishment Protein Localization Translation” cluster that are overrepresented among PLAG1-S genomic binding sites (g:Profiler FDR < 0.1) and the list of bound leading-edge genes driving negative enrichments in this cluster. See also supplemental Figure 4. FDR, false discovery rate.
RNA-seq of HSPCs directly following up- or down-modulation of PLAG1 levels (supplemental Tables 4 and 5) corroborates the respective immunophenotypic and functional outcomes both through the expression of surface markers (supplemental Figure 4B) and by global alignment to transcriptional states of 20 human hematopoietic cell subpopulations,49 which show positive associations to primitive and erythroid signatures and negative associations to myeloid signatures correlated with high PLAG1 levels in vitro (Figure 4D).
Ectopic PLAG1-S significantly altered the expression of 543 genes (supplemental Table 4; Figure 4C). Consistent with its understood function as a transcription activator, 60% of the 291 upregulated genes are proximally bound by PLAG1-S. Thirty percent of downregulated genes are also directly bound, which similarly to other recent publications,60 suggests an underappreciated role for PLAG1-S in negative regulation of gene expression. Gene set enrichment analysis (GSEA) revealed that coordinated pathway-level changes are dominated by negative enrichments (supplemental Table 6). The top-most negatively enriched gene sets coalesce in the largest cluster of altered signatures and point to a synchronized attenuation of messenger RNA (mRNA) translation machinery in PLAG1-SOE HSPCs (Figure 4E; supplemental Figure 4C). These negative enrichments are driven largely by reduced expression of genes encoding ribosomal proteins (RPs) (supplemental Figure 4D). Moreover, there is a significant overrepresentation and overlap of genes belonging to the cluster of protein synthesis gene sets in the repertoire of genes directly bound by PLAG1-S (Figure 4E [green edges] and 4F; supplemental Table 7). Altogether, this speaks to an unexpected ability of PLAG1-S to intersect with the regulation of protein synthesis machinery to regulate fate decisions in human HSPCs.
PLAG1-S dampens protein synthesis and promotes dormancy in stimulated human HSPCs
The state of attenuated translation machinery in PLAG1-SOE HSPCs is intriguing given that tightly controlled protein synthesis is a hallmark of stem cell biology.32-35,37 Although others have demonstrated that culturing human HSPCs, like their murine counterparts, promotes exit from dormancy, loss of quiescence, and differentiation,24,25,28,39 little is known of their translation dynamics within immediate and prolonged timeframes, and in comparison with more committed cells.42 To gain these insights and contextualize the HSPC-specific PLAG1-S regulon, we measured O-propargyl-puromycin (OP-Puro) incorporation in CB cells upon culture-induced stimulation.37,61,62 As early as 4 hours after being placed in culture, Lin−CD34+ cells activate protein synthesis, and these levels progressively increase, peaking at 9.5-fold after 48 hours. Subsequently, translation rates drastically decline between 48 and 72 hours and plateau between 3 and 10 days, but do not return to baseline levels (Figure 5A; supplemental Figure 5A). In contrast, more mature Lin−CD34− cells experience only a modest elevation of translation after 24 hours that diminishes by 48 hours (Figure 5B), indicating that human HSPCs selectively undergo an immediate but transient pro-translation response when placed into culture. Evaluating PLAG1-SOE HSPCs we observed that transcriptional reprogramming of protein biosynthetic processes indeed supports a 13% reduction in global translation rates in Lin−CD34+ cells ex vivo (Figure 5C; supplemental Figure 5B-C). This appears selective to primitive hematopoietic cells because comparably handled PLAG1-SOE K562 and Lin−CD34− cells display unchanged and heighted translation levels, respectively (supplemental Figure 5D).
Figure 5.
PLAG1-S dampens protein synthesis and promotes dormancy in stimulated human HSPCs. (A) OP-Puro incorporation dynamics measured as median fluorescence intensity (MFI) in cultured Lin−CD34+ cells with representative flow cytometry plots (n = 5 for 0 and 24 hours; n = 3 for 4, 48, and 72 hours). Red and blue asterisks denote statistical significance relative to previous timepoint or T0, respectively. (B) Fold difference of OP-Puro MFI relative to T0 in cultured Lin−CD34+ compared with Lin−CD34− CB fractions (n = 4 for 24 hours, n = 2 for 48 hours). Blue statistics are relative to 1× levels at T0 and red statistics are between cell types at matched time points. (C) OP-Puro incorporation by PLAG1-SOE and control Lin−CD34+ cells on day 4 of ex vivo culture (n = 8). Data from 3 experiments normalized to the average MFI in control cells per experiment. (D) Reduced size of PLAG1-SOE Lin−CD34+ cells on day 4 of ex vivo culture determined by flow cytometric MFI of FSC-H profiles (n = 7, left, each point is from a culture of an individual CB unit) and immunofluorescence microscopy (right, each point is a single cell; scale bar = 25 μm). (E) Cell-cycle analysis by Hoechst and Ki67 staining of PLAG1-SOE and control Lin−CD34+ cells on days 4 and 7 of ex vivo culture (n = 3). (F) Representative flow plots for PLAG1-SOE and control Lin−CD34+ cells stained for 7-AAD and Annexin V with apoptosis measurements of surface positivity of Annexin V on day 4 (n = 5) and day 7 (n = 4) of ex vivo culture. (G) Heatmap of log2FC of transcripts coding EIF2 subunits (bottom) and intracellular flow cytometric measures of EIF2S1 protein expression (n = 4) in PLAG1-SOE relative to control Lin−CD34+ cells on day 4 of ex vivo culture (top). (H) GSEA of the PLAG1-SOE transcriptome to curated targets of ATF4 generated by Han et al74 and used by van Galen et al73 and FPKM heatmap of ATF4 targets differentially expressed in PLAG1-SOE HSPCs (p.adj < .1). (I) Negative enrichment of gene sets related to unfolded protein response (P < .05). Data are presented as average ± SEM unless otherwise indicated. Each point represents an individual CB unit otherwise indicated. ***P < .005, **P < .01, *P < .05. See also supplemental Figure 5. FSC-H, Forward Scatter Height; n.s., not significant; p.adj, adjusted P value.
Anabolic processes such as protein biosynthesis are generally correlated with cellular enlargement and division,63-67 both of which can predict HSC exhaustion.24,68 To this point, the diameter of untransduced cultured HSPCs is significantly enlarged coincident with the peak in translation rates, which notably precedes when the first cellular division is expected28 (supplemental Figure 5A). Likewise, we find that depressed translation in PLAG1-SOE HSPCs is associated with restraint in size (Figure 5D). GSEA did not uncover consensus control over cell-cycle progression by PLAG1-S (supplemental Table 6; Figure 4E). However, Hoechst/Ki-67 staining indicated that PLAG1-SOE HSPCs are slightly restrained in cell-cycle progression (Figure 5E), paralleling the correlation between PLAG1 expression and dormant HSCs (Figure 2A-C). Although this may likely be secondary to translation regulation, we also noticed that expression of CDKN1C, an essential regulator of murine HSC quiescence and renewal,69,70 appears directly activated 2.5-fold by PLAG1-S in HSPCs (supplemental Tables 2 and 4), possibly contributing to the cell-cycle profile of these cells.
In vitro survival, which could contribute to amplifying a stem cell pool experiencing infrequent and/or slow division dynamics, is enhanced in PLAG1-SOE cells, as measured by Annexin V (Figure 5F). Given that translation dynamics are highly interconnected with stress signaling that can dictate survival decisions, we profiled stress effectors in PLAG1-SOE HSPCs. Firstly, pro-apoptotic p53 targets, which can be induced by imbalances in ribosome components, are not activated within the PLAG1-SOE transcriptome (supplemental Figure 5E).71,72 Low levels of EIF2 complex restrains global translation while dichotomously promoting translation of the integrated stress response effector ATF4 in CB HSPCs73 and muscle stem cells to support regeneration.35 However, neither EIF2 subunits nor ATF4 targets73,74 are differentially regulated in PLAG1-SOE HSPCs, suggesting this pathway is not significantly at play (Figure 5G-H). Lastly, CB HSPCs are selectively sensitive to stress associated with misfolded proteins40 and low translation rates in murine HSCs is a mechanism that protects their proteostatic integrity.38 To this point, reduced translation in PLAG1-SOE HSPCs is associated with depressed expression of unfolded protein response signatures (Figure 5I). Altogether, this suggests that dampened translation rates imparted by PLAG1-S is protective and forestalls stress responses.
In sum, we show that human HSPCs selectively experience an immediate and transient pro-translation response when exposed to stimulatory conditions and, through transcriptional programming, PLAG1-S limits translation in human HSPCs to mitigate the impact of culture-induced protein stress and HSC activation. This manifests as PLAG1-S-induced reductions in cell enlargement, division, differentiation, and death in human HSPCs, altogether significantly enhancing HSC fitness and output in vivo.
PLAG1-S activates imprinted loci to support human HSPCs
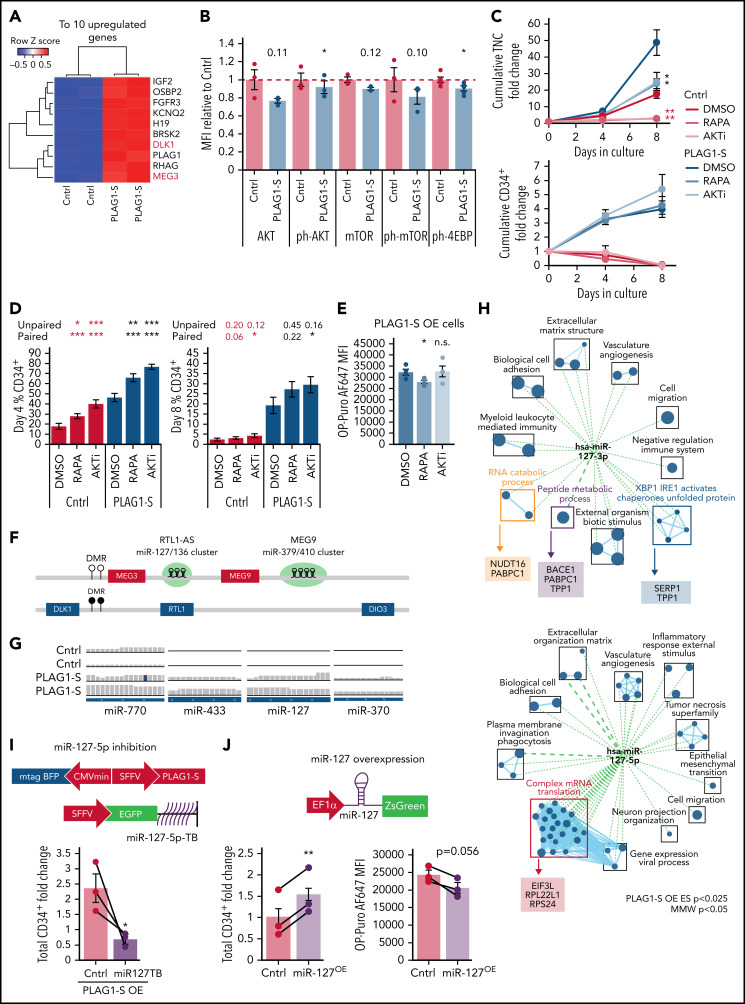
A notable finding of the PLAG1-S regulatory network is its direct binding and activation of DLK1/MEG3 and IGF2/H19 (Figures 4C and 6A; supplemental Figure 6A), affirming that as in mouse tissues,75 these imprinted loci are targets of PLAG1-S in primitive human hematopoietic cells. PLAG1-induced activation of IGF2 can stimulate mitogenic and PI3K-AKT-mTOR signaling to promote tumorigenic growth and division.75-77 However, H19 and MEG3 act in opposition to PI3K-AKT-mTOR signaling in support of fetal murine HSC quiescence and function.78,79 When activated, this pathway can stimulate protein synthesis dually through phospho-dependent activation of RPS6K and inhibition of translation initiation-regulating 4EBPs (supplemental Figure 6B). At the transcript level, PI3K-AKT-mTOR signaling signatures are both up- and downregulated in PLAG1-SOE HSPCs (supplemental Figure 6C). Definitive characterization of the pathway flux by intracellular flow cytometry (IFC) reveals subtle reductions in AKT and 4EBP1 phosphorylation, while RPS6 phospho-status was unchanged (Figure 6B; supplemental Figure 6D), suggesting selective repression of 4EBP1-regulated translation initiation could partially contribute to reduced protein synthesis in PLAG1-SOE HSPCs. Human HSPC fitness can be enhanced by pharmacological inhibition of AKT (AKTi), which promotes quiescence80 or addition of the mTOR inhibitor, rapamycin.81 Given the pleiotropism of these signaling factors, we investigated whether combining PLAG1-SOE with these inhibitors could produce combinatorial effects on human HSPC output. As demonstrated previously in human Lin−CD34+ cells treated with AKTi80 and murine Lin−Sca+ cells treated with rapamycin,82 either inhibitor reduced total cells in both control and PLAG1-SOE cultures (Figure 6C) while enhancing the proportion of primitive CD34+ cells (Figure 6D), resulting in maintenance of similar total CD34+ cell outputs (Figure 6C). Only with rapamycin were these growth dynamics associated with a reduction in translation rates in PLAG1-SOE cells (Figure 6E), whereas neither treatment altered apoptosis in control or PLAG1-SOE cultures (supplemental Figure 6E).
Figure 6.
PLAG1-S activates imprinted loci to support human HSPCs. (A) Heatmap of top 10 differentially expressed transcripts in the transcriptome of PLAG1-SOE Lin−CD34+ HSPCs. (B) Intracellular flow cytometry of components of the PI3K signaling pathway, including phospho-S473 AKT, phospho-S2448 mTOR, and phospho-Thr37/46 4EBP1, in PLAG1-SOE Lin−CD34+ cells on day 4 of culture. Numbers above PLAG1-SOE bars show the paired Student t-test P value relative to control (n = 3, ph-4EBP1 n = 5). (C) Total nucleated cell (top) and CD34+ cell (bottom) fold change in Lin−CD34+BFP+ cultures overexpressing either PLAG1-S or Luciferase control and treated with 50 nM rapamycin (RAPA), 1 μM AKT inhibitor (AKTi), or vehicle (DMSO) (n = 4). Student t-test P values in red are relative to Cntrl-DMSO and in black are relative to PLAG1-SOE-DMSO. (D) CD34 positivity in PLAG1-SOE or control HSPCs following 4 and 8 days of ex vivo culture with RAPA, AKTi, or vehicle (n = 4). Student t-test P values in red are relative to Cntrl-DMSO and in black are relative to PLAG1-SOE-DMSO. (E) OP-Puro incorporation by PLAG1-SOE HSPCs cultured in the presence of RAPA, AKTi, or vehicle on day 4 of culture (n = 4). (F) Schematic of the imprinted human DLK1/MEG3 locus, which encodes miRNA mega-clusters miR127/136 (7 miRNAs) and miR-379/410 (39 miRNAs). (G) RNA-seq read tracks for miRNA transcripts from this locus detected in PLAG1-SOE HSPCs. (H) Overlap of the PLAG1-S overexpression gene set enrichment map (P < .025) to signatures of miR-127-5p and miR-127-3p validated targets (Mann-Whitney U test, P < .05). (I-J) Schematic of lentivectors used for dual PLAG1-S overexpression and miR127-5p inhibition via a sponge consisting of multiple bulged 26-mer target sequences (miR127TB) or miR127 overexpression. (I) CD34+ cell fold change ex vivo when PLAG1-S and the miR127-5p inhibitor are coexpressed in Lin−CD34+ cells (n = 3). (J) CD34+ cell fold change ex vivo when miR127 is overexpressed in Lin−CD34+ cells (n = 3); and OP-Puro incorporation Lin−CD34+ cells overexpressing miR127 (n = 3). Data are presented as average ± SEM unless otherwise indicated. Each point represents an individual CB unit. ***P < .005, **P < .01, *P < .05. n.s., not significant. See also supplemental Figure 6.
The DLK1/MEG3 locus also encodes the largest microRNA (miRNA) mega-cluster in the mammalian genome, with possible roles transcending PI3K-AKT-mTOR regulation83,84 (Figure 6F). RNA-seq read counts identified 4 miRNAs from within this locus (miR-770, miR-433, miR-127, and miR-370) reproducibly overrepresented in PLAG1-SOE cells (Figure 6G). Comparison of experimentally supported miRNA targets to transcripts downregulated in PLAG1-SOE HSPCs found the highest overlap for miR-127 (supplemental Figure 6F),85,86 specifically including genes involved in complex cap-dependent translation and RNA and peptide metabolic processing (Figure 6H), providing impetus to test its role downstream of PLAG1-S. Simultaneous ectopic expression of PLAG1-S and an inhibitory miR-127-5p sponge87,88 netted significantly reduced CD34+ output (Figure 6I; supplemental Figure 6G,H), and in the 2 CBs in which sample amounts supported testing OP-Puro incorporation, this was associated with an increase in protein synthesis (supplemental Figure 6I). Finally, overexpression of miR-127 enhanced CD34+ output while imparting reduced levels of protein synthesis (Figure 6J; supplemental Figure 6J-L). Together, these results suggest that miR-127 partially contributes to the effects of PLAG1-S in promoting a specific translational state and primitive cell maintenance.
MYC-induced translation impairs PLAG1-S-mediated stemness in human HSPCs
MYC is a potent tissue nonspecific transcriptional activator of cytoplasmic translation and nuclear ribosome biogenesis.89 In the murine context, Myc deletion impairs hematopoietic differentiation,90-92 and Myc expression is concomitantly activated with translation machinery when murine HSCs exit dormancy.93 Recent findings in cultured human HSPCs also establish that MYC drives their ex vivo activation via promotion of anabolic programs.56 Therefore, we next investigated whether PLAG1-S acts through repression of MYC and whether MYC-mediated activation of translation could influence the ability of PLAG1-S to promote stemness.
Neither c-MYC protein levels, post-translational regulation, nor the expression of MYC target genes are reduced in PLAG1-SOE HSPCs (supplemental Results; supplemental Figure 7A-D). In fact, in contrast to diminished cytosolic ribosome expression, the expression of MYC-regulated nuclear ribosome assembly targets trend upward in PLAG1-SOE HSPCs (Figure 7A), altogether suggesting PLAG1-S acts autonomously of MYC repression. As such, modulation of MYC expression could serve as a tool to independently activate translation rates as a means to query the dependency of PLAG1-S-enforced stemness on its attenuation of protein synthesis. Indeed, modest c-MYC overexpression via the PGK promoter endowed a 25% increase in OP-Puro incorporation by Lin−CD34+ cells (supplemental Figure 7E-F). Next, BFP+GFP+ PLAG1-S and c-MYC co-overexpressing Lin−CD34+ cells and their control counterparts were assessed for primitive cell maintenance ex vivo (Figure 7B-C). Consistent with other reports,56,90-92 relative to control, ectopic c-MYC independently promotes hematopoietic differentiation, as measured by loss of CD34+ and gain of CD33+ cells (Figure 7D-E, purple vs red). Over culture c-MYCOE cells also become enlarged relative to control (Figure 7F, purple vs red) and display the highest rates of active translation (Figure 7G). We observed that PLAG1-S overexpression significantly reduced protein production rates in c-MYCOE HSPCs (Figure 7G), and concomitantly countered c-MYC-induced cellular enlargement and pro-differentiative phenotypes (Figure 7D-F top, purple vs green). Finally, after 7 days we observed that cells co-overexpressing PLAG1-S and MYC have significantly reduced primitive cell output relative to cells solely overexpressing PLAG1-S (Figure 7D-E, bottom, green vs blue). Together, these findings provide important proof of principle that dampened translation is key to the HSC-supportive programming imparted by PLAG1-S.
Figure 7.
MYC-induced translation impairs PLAG1-S-mediated stemness in human HSPCs. (A) Up- (red) or down- (blue) regulation of MYC ribosome biogenesis targets in PLAG1-SOE HSPCs. (B) Schematic of PLAG1-S and c-MYC overexpression lentivectors. (C) Representative sorting gates for dual-overexpression of PLAG1-S and c-MYC or controls in Lin−CD34+ cells. (D) CD34 (n = 4) and (E) CD33 (n = 3) positivity in BFP+GFP+ HSPCs over 4 and 7 days of ex vivo culture. (F) Cell size determined by flow cytometric MFI of FSC-H (n = 3-4) in BFP+GFP+ HSPC cultures on day 4 and 7. (G) OP-Puro incorporation by BFP+GFP+ HSPCs on day 4 of ex vivo culture (n = 4). Data are presented as average ± SEM unless otherwise indicated. Each point represents an individual CB unit. ***P < .005, **P < .01, *P < .05. n.s., not significant. See also supplemental Figure 7.
Discussion
We identify PLAG1-S as a novel positive regulator of human HSC dormancy and self-renewal. We demonstrate that PLAG1-S enacts multifaceted and combinatorial programs to endow HSPCs with an in situ-like rate of protein production and simultaneously restrain growth, proliferation, differentiation, and death to ultimately enhance human HSC preservation and function in stimulatory culture and transplantation settings.
The role of PLAG1 in supporting healthy HSC self-renewal is strikingly at odds with its reported functions in oncogenic contexts. In a murine cbfβ-translocated acute myeloid leukemia model ectopic PLAG1 promotes proliferation and its elevation via lentiviral insertions was one of several co-occurring molecular abnormalities associated with the onset and progression of primate myelodysplastic clonal hematopoiesis,94,95 phenotypes notably absent upon PLAG1-S overexpression in the background of healthy human HSPCs. Additionally, effectors downstream of PLAG1-IGF2 reported to support solid tumors75-77,96 appear inert or repressed in PLAG1-SOE HSPCs, implicating context-specific counteracting mechanisms. It is particularly interesting that 4EBP1 displayed reduced activation in this setting, given that phospho-4EBP1/2 is currently the most predictive indicator of translation levels in murine HSCs.97 4EBP1 phosphorylation is unlikely, however, to fully account for the dampened translation observed in PLAG1-SOE HSPCs, where together with multiple functional nodes, including binding and modulation of RP genes and miR-127, PLAG1-S consolidates the restraint of translation to enhance HSC function. Interestingly, rapamycin treatment of PLAG1-SOE HSPCs has an additive effect on diminished protein production, possibly because of the preferential inhibition of mTOR-RPS6K1 over mTOR-4EBP1.98 However, the similar CD34+ cell growth dynamics upon AKT inhibition without further reduction of translation also suggests the possible contribution of other dormancy-promoting processes.99 Although neither AKTi nor rapamycin enhanced total CD34+ cell output with PLAG1-S overexpression, the elevated proportion of CD34+ cells from these cultures could suggest their functional nature differs from vehicle-treated cells. As such, an interesting question for future investigation is whether pharmacological inhibition of AKT, mTOR, or other pathways in PLAG1-SOE HSPCs could further enhance their in vivo repopulating fitness, as has been shown for untransduced CB.10,12,80,81
Insights garnered from murine models have demonstrated that HSCs strictly control their protein production.37 Hyperactivation of protein biosynthetic processes can lead to murine HSC impairment or depletion by driving their dormancy and quiescence exit, differentiation, and compromising their proteome integrity.38,42,93 We now address these phenomena from a human perspective, adding activation of protein synthesis as a selective and robust feature of the molecular summary of compromised human HSC function upon culture-induced stimulation that is commonly required for clinical applications. By preceding cell division kinetics, the proteostatic response likely also acts as an early determinant of cell fate100 and is thus an important but underappreciated checkpoint for therapeutic procedures. The intersection of translation, dormancy, and stemness being elucidated in model stem cells is mirrored in PLAG1-SOE human HSPCs, where concurrent with diminished translation we observe reduced differentiation, enlargement, division, death, and enhanced self-renewal, and by toggling c-MYC-driven protein production as a molecular tool, we provide proof of principle that diminished translation is an essential modality by which PLAG1-S enhances HSPC output. Together, our results forward the notion that the physiological importance of low translation rates in homeostatic niche-associated HSCs can be harnessed for therapeutic benefit. To this point, rebalancing proteostasis in murine satellite cells and HSCs by activating stress-responsive effectors can improve their long-term regenerative capacities.35,41,42 Importantly however, ectopic PLAG1-S does not enact the integrated stress response and depresses unfolded protein response signatures, suggesting that by directly tuning the translation machinery PLAG1-S preempts and averts pro-apoptotic branches of stress signaling.40 Others have also demonstrated that translation inhibitors can effectively eliminate primitive leukemic cells while sparing healthy HSC.101,102 Together with our findings, this highlights the possibility of achieving a stem cell advantage by optimizing the timing and dosage of such compounds. The recent success of transient mRNA delivery systems103 also provides an exciting opportunity through which the rapid pro-stem effect of PLAG1-S could be realized to its full advantage. Our identification of ectopic PLAG1-S as a highly context-selective regulator of protein synthesis also provides impetus for its future investigation in physiological contexts as a regulator of HSC translation, and as a modulator of translation and/or stemness in other primitive cell settings where its expression appears enriched.83,104-106 Finally, our findings underscore that addressing the current deficit in our understanding of translation dynamics and its regulators in human HSCs in vivo when subject to demands of disease or injury could substantively inform future HSC-focused regenerative therapies.
Altogether, our discovery and characterization of PLAG1-S as a novel regulator of human HSC dormancy and self-renewal has derived insights germane to the appreciation of translation control in determining human HSC fate and function and highlights the promise of exploiting regulators of this fundamental feature of stem cell physiology to enhance regenerative therapies.
Supplementary Material
The online version of this article contains a data supplement.
Acknowledgments
The authors acknowledge Minomi Subpanditha and Zoya Tabunshchyk for their help with fluorescence-activated cell sorting. The authors thank Sonam Bhatia, Anna Dvorkin-Gheva and Andy Zeng for bioinformatic assistance, Lina Liu for technical assistance, all members of the Hope laboratory for experimental support and advice, and Jonathan Draper, Brad Doble, Matthew Miller, Kerstin Kaufmann, and Stephanie Xie for critical assessment of this work.
This research was made possible through funding from the Ontario Institute for Cancer Research (IA-033) and the Canadian Institutes of Health Research (CIHR) (126030) to K.J.H., a Michael DeGroote Doctoral Fellowship and CIHR Frederick Banting and Charles Best Canada Graduate Scholarship to A.K.C. and to J.X., an NSERC Alexander Graham Bell Canada Graduate Scholarship to M.S.B., a CIHR Canada Graduate Scholarship-Masters award and Ontario Graduate Scholarships to H.T.T.C., and a Canadian Blood Services Graduate Fellowship to S.R.
Footnotes
CUT&RUN and RNA-seq data SuperSeries accession number is GSE181992 and can be accessed at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE181992.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.K.C. and M.S.B. designed and performed experiments and analyzed and interpreted results; J.X., H.T.T.C., and S.R. assisted with cord blood experiments; A.K.C., H.T.T.C., and V.V. under supervision of G.D.B., performed RNA-seq data analysis; G.K. under supervision of E.L. and J.E.D., assisted with CUT&RUN and microRNA experiments; S.A.M., under supervision of D.D.D.C., performed CUT&RUN data analysis; A.K.C. and K.J.H. wrote the manuscript; K.J.H. conceived and supervised the project and designed and interpreted results; and all authors reviewed the manuscript.
Conflict-of-interest disclosure: J.E.D. serves on the SAB for Graphite Bio, receives royalties from Trillium Therapeutics Inc/Pfizer and receives a commercial research grant from Celgene/BMS. The remaining authors declare no competing financial interests.
Correspondence: Kristin Hope, Princess Margaret Cancer Centre, 101 College St, Toronto, ON M5G 1L7, Canada; e-mail: kristin.hope@uhnresearch.ca.
REFERENCES
- 1.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132(4):631-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doulatov S, Notta F, Laurenti E, Dick JE. Hematopoiesis: a human perspective. Cell Stem Cell. 2012;10(2):120-136. [DOI] [PubMed] [Google Scholar]
- 3.Chabannon C, Kuball J, Bondanza A, et al. Hematopoietic stem cell transplantation in its 60s: a platform for cellular therapies. Sci Transl Med. 2018;10(436):eaap9630. [DOI] [PubMed] [Google Scholar]
- 4.Smith AR, Wagner JE. Alternative haematopoietic stem cell sources for transplantation: place of umbilical cord blood. Br J Haematol. 2009;147(2): 246-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gragert L, Eapen M, Williams E, et al. HLA match likelihoods for hematopoietic stem-cell grafts in the U.S. registry. N Engl J Med. 2014;371(4):339-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emiloju OE, Potdar R, Jorge V, Gupta S, Varadi G. Clinical advancement and challenges of ex vivo expansion of human cord blood cells. Clin Hematol Int. 2019; 2(1):18-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta AO, Wagner JE. Umbilical cord blood transplants: current status and evolving therapies. Front Pediatr. 2020;8:570282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang X, Guo B, Capitano M, Broxmeyer HE. Past, present, and future efforts to enhance the efficacy of cord blood hematopoietic cell transplantation. F1000 Res. 2019;8:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kindwall-Keller TL, Ballen KK. Umbilical cord blood: the promise and the uncertainty. Stem Cells Transl Med. 2020; 9(10):1153-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boitano AE, Wang J, Romeo R, et al. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science. 2010;329(5997): 1345-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wagner JE Jr, Brunstein CG, Boitano AE, et al. Phase I/II trial of StemRegenin-1 expanded umbilical cord blood hematopoietic stem cells supports testing as a stand-alone graft. Cell Stem Cell. 2016;18(1):144-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fares I, Chagraoui J, Gareau Y, et al. Cord blood expansion. Pyrimidoindole derivatives are agonists of human hematopoietic stem cell self-renewal. Science. 2014;345(6203):1509-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bak RO, Dever DP, Porteus MH. CRISPR/Cas9 genome editing in human hematopoietic stem cells. Nat Protoc. 2018;13(2):358-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Lima M, McMannis J, Gee A, et al. Transplantation of ex vivo expanded cord blood cells using the copper chelator tetraethylenepentamine: a phase I/II clinical trial. Bone Marrow Transplant. 2008;41(9):771-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Lima M, McNiece I, Robinson SN, et al. Cord-blood engraftment with ex vivo mesenchymal-cell coculture. N Engl J Med. 2012;367(24):2305-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delaney C, Heimfeld S, Brashem-Stein C, Voorhies H, Manger RL, Bernstein ID. Notch-mediated expansion of human cord blood progenitor cells capable of rapid myeloid reconstitution. Nat Med. 2010; 16(2):232-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horwitz ME, Chao NJ, Rizzieri DA, et al. Umbilical cord blood expansion with nicotinamide provides long-term multilineage engraftment. J Clin Invest. 2014; 124(7):3121-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shpall EJ, Quinones R, Giller R, et al. Transplantation of ex vivo expanded cord blood. Biol Blood Marrow Transplant. 2002;8(7):368-376. [DOI] [PubMed] [Google Scholar]
- 19.Horwitz ME. Ex vivo expansion or manipulation of stem cells to improve outcome of umbilical cord blood transplantation. Curr Hematol Malig Rep. 2016;11(1):12-18. [DOI] [PubMed] [Google Scholar]
- 20.Nakamura-Ishizu A, Takizawa H, Suda T. The analysis, roles and regulation of quiescence in hematopoietic stem cells. Development. 2014;141(24):4656-4666. [DOI] [PubMed] [Google Scholar]
- 21.Liang R, Arif T, Kalmykova S, et al. Restraining lysosomal activity preserves hematopoietic stem cell quiescence and potency. Cell Stem Cell. 2020;26(3): 359-376.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu J, Gjini J, Arif T, Moore K, Lin M, Ghaffari S. Using mitochondrial activity to select for potent human hematopoietic stem cells. Blood Adv. 2021;5(6): 1605-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson A, Laurenti E, Oser G, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135(6):1118-1129. [DOI] [PubMed] [Google Scholar]
- 24.Glimm H, Oh IH, Eaves CJ. Human hematopoietic stem cells stimulated to proliferate in vitro lose engraftment potential during their S/G(2)/M transit and do not reenter G(0). Blood. 2000;96(13):4185-4193. [PubMed] [Google Scholar]
- 25.Passegué E, Wagers AJ, Giuriato S, Anderson WC, Weissman IL. Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. J Exp Med. 2005;202(11):1599-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szilvassy SJ, Meyerrose TE, Grimes B. Effects of cell cycle activation on the short-term engraftment properties of ex vivo expanded murine hematopoietic cells. Blood. 2000;95(9):2829-2837. [PubMed] [Google Scholar]
- 27.Kaufmann KB, Zeng AGX, Coyaud E, et al. A latent subset of human hematopoietic stem cells resists regenerative stress to preserve stemness. Nat Immunol. 2021; 22(6):723-734. [DOI] [PubMed] [Google Scholar]
- 28.Laurenti E, Frelin C, Xie S, et al. CDK6 levels regulate quiescence exit in human hematopoietic stem cells. Cell Stem Cell. 2015;16(3):302-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luchsinger LL, Strikoudis A, Danzl NM, et al. Harnessing hematopoietic stem cell low intracellular calcium improves their maintenance in vitro. Cell Stem Cell. 2019;25(2):225-240.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakada D, Saunders TL, Morrison SJ. Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature. 2010;468(7324):653-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oedekoven CA, Belmonte M, Bode D, et al. Hematopoietic stem cells retain functional potential and molecular identity in hibernation cultures. Stem Cell Reports. 2021;16(6):1614-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sampath P, Pritchard DK, Pabon L, et al. A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2008;2(5):448-460. [DOI] [PubMed] [Google Scholar]
- 33.Sanchez CG, Teixeira FK, Czech B, et al. Regulation of ribosome biogenesis and protein synthesis controls germline stem cell differentiation. Cell Stem Cell. 2016; 18(2):276-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blanco S, Bandiera R, Popis M, et al. Stem cell function and stress response are controlled by protein synthesis. Nature. 2016;534(7607):335-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zismanov V, Chichkov V, Colangelo V, et al. Phosphorylation of eIF2α is a translational control mechanism regulating muscle stem cell quiescence and self-renewal. Cell Stem Cell. 2016;18(1):79-90. [DOI] [PubMed] [Google Scholar]
- 36.Magee JA, Signer RAJ. Developmental stage-specific changes in protein synthesis differentially sensitize hematopoietic stem cells and erythroid progenitors to impaired ribosome biogenesis. Stem Cell Reports. 2021;16(1):20-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Signer RA, Magee JA, Salic A, Morrison SJ. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature. 2014;509(7498):49-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hidalgo San Jose L, Sunshine MJ, Dillingham CH, et al. Modest declines in proteome quality impair hematopoietic stem cell self-renewal. Cell Rep. 2020;30(1):69-80.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie SZ, Garcia-Prat L, Voisin V, et al. Sphingolipid modulation activates proteostasis programs to govern human hematopoietic stem cell self-renewal. Cell Stem Cell. 2019;25(5):639-653.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Galen P, Kreso A, Mbong N, et al. The unfolded protein response governs integrity of the haematopoietic stem-cell pool during stress. Nature. 2014;510(7504):268-272. [DOI] [PubMed] [Google Scholar]
- 41.Goncalves KA, Silberstein L, Li S, et al. Angiogenin promotes hematopoietic regeneration by dichotomously regulating quiescence of stem and progenitor cells. Cell. 2016;166(4):894-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kruta M, Sunshine MJ, Chua BA, et al. Hsf1 promotes hematopoietic stem cell fitness and proteostasis in response to ex vivo culture stress and aging. Cell Stem Cell. 2021;28(11):1950-1965.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Juma AR, Damdimopoulou PE, Grommen SV, Van de Ven WJ, De Groef B. Emerging role of PLAG1 as a regulator of growth and reproduction. J Endocrinol. 2016;228(2):R45-R56. [DOI] [PubMed] [Google Scholar]
- 44.Van Dyck F, Declercq J, Braem CV, Van de Ven WJ. PLAG1, the prototype of the PLAG gene family: versatility in tumour development (review). Int J Oncol. 2007;30(4):765-774. [PubMed] [Google Scholar]
- 45.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347(1-2):70-78. [DOI] [PubMed] [Google Scholar]
- 46.Skene PJ, Henikoff JG, Henikoff S. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat Protoc. 2018;13(5):1006-1019. [DOI] [PubMed] [Google Scholar]
- 47.Rentas S, Holzapfel N, Belew MS, et al. Musashi-2 attenuates AHR signalling to expand human haematopoietic stem cells. Nature. 2016;532(7600):508-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belew MS, Bhatia S, Keyvani Chahi A, Rentas S, Draper JS, Hope KJ. PLAG1 and USF2 co-regulate expression of Musashi-2 in human hematopoietic stem and progenitor cells. Stem Cell Reports. 2018;10(4):1384-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Novershtern N, Subramanian A, Lawton LN, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144(2):296-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amon S, Meier-Abt F, Gillet LC, et al. Sensitive quantitative proteomics of human hematopoietic stem and progenitor cells by data-independent acquisition mass spectrometry. Mol Cell Proteomics. 2019;18(7):1454-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Notta F, Zandi S, Takayama N, et al. Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science. 2016;351(6269):aab2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cabezas-Wallscheid N, Klimmeck D, Hansson J, et al. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell. 2014;15(4): 507-522. [DOI] [PubMed] [Google Scholar]
- 53.Pellin D, Loperfido M, Baricordi C, et al. A comprehensive single cell transcriptional landscape of human hematopoietic progenitors. Nat Commun. 2019;10(1):2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hay SB, Ferchen K, Chetal K, Grimes HL, Salomonis N. The Human Cell Atlas bone marrow single-cell interactive web portal. Exp Hematol. 2018;68:51-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Umemoto T, Hashimoto M, Matsumura T, Nakamura-Ishizu A, Suda T. Ca2+-mitochondria axis drives cell division in hematopoietic stem cells. J Exp Med. 2018;215(8):2097-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.García-Prat L, Kaufmann KB, Schneiter F, et al. Dichotomous regulation of lysosomes by MYC and TFEB controls hematopoietic stem cell fate. bioRxiv. 2021:2021.2002.2024.432720.
- 57.Dircio-Maldonado R, Flores-Guzman P, Corral-Navarro J, et al. Functional integrity and gene expression profiles of human cord blood-derived hematopoietic stem and progenitor cells generated in vitro. Stem Cells Transl Med. 2018;7(8):602-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Amendola M, Venneri MA, Biffi A, Vigna E, Naldini L. Coordinate dual-gene transgenesis by lentiviral vectors carrying synthetic bidirectional promoters. Nat Biotechnol. 2005;23(1):108-116. [DOI] [PubMed] [Google Scholar]
- 59.Skene PJ, Henikoff S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife. 2017;6:e21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wong J, Damdimopoulos A, Damdimopoulou P, et al. Transcriptome analysis of the epididymis from Plag1 deficient mice suggests dysregulation of sperm maturation and extracellular matrix genes. Dev Dyn. 2020;249(12):1500-1513. [DOI] [PubMed] [Google Scholar]
- 61.Hidalgo San Jose L, Signer RAJ. Cell-type-specific quantification of protein synthesis in vivo. Nat Protoc. 2019;14(2):441-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu J, Xu Y, Stoleru D, Salic A. Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc Natl Acad Sci USA. 2012;109(2):413-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cook M, Tyers M. Size control goes global. Curr Opin Biotechnol. 2007;18(4):341-350. [DOI] [PubMed] [Google Scholar]
- 64.Donati G, Montanaro L, Derenzini M. Ribosome biogenesis and control of cell proliferation: p53 is not alone. Cancer Res. 2012;72(7):1602-1607. [DOI] [PubMed] [Google Scholar]
- 65.Schmoller KM, Skotheim JM. The biosynthetic basis of cell size control. Trends Cell Biol. 2015;25(12):793-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Polymenis M, Aramayo R. Translate to divide: сontrol of the cell cycle by protein synthesis. Microb Cell. 2015;2(4):94-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamamoto K, Mak TW. Mechanistic aspects of mammalian cell size control. Dev Growth Differ. 2017;59(1):33-40. [DOI] [PubMed] [Google Scholar]
- 68.Lengefeld J, Cheng C-W, Maretich P, et al. Cell size is a determinant of stem cell potential during aging. bioRxiv. 2020:2020.2010.2027.355388. [DOI] [PMC free article] [PubMed]
- 69.Matsumoto A, Takeishi S, Kanie T, et al. p57 is required for quiescence and maintenance of adult hematopoietic stem cells. Cell Stem Cell. 2011;9(3):262-271. [DOI] [PubMed] [Google Scholar]
- 70.Zou P, Yoshihara H, Hosokawa K, et al. p57(Kip2) and p27(Kip1) cooperate to maintain hematopoietic stem cell quiescence through interactions with Hsc70. Cell Stem Cell. 2011;9(3):247-261. [DOI] [PubMed] [Google Scholar]
- 71.Golomb L, Volarevic S, Oren M. p53 and ribosome biogenesis stress: the essentials. FEBS Lett. 2014;588(16):2571-2579. [DOI] [PubMed] [Google Scholar]
- 72.Nii T, Marumoto T, Tani K. Roles of p53 in various biological aspects of hematopoietic stem cells. J Biomed Biotechnol. 2012;2012:903435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van Galen P, Mbong N, Kreso A, et al. Integrated stress response activity marks stem cells in normal hematopoiesis and leukemia. Cell Rep. 2018;25(5): 1109-1117.e5. [DOI] [PubMed] [Google Scholar]
- 74.Han J, Back SH, Hur J, et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol. 2013;15(5):481-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Declercq J, Van Dyck F, Braem CV, et al. Salivary gland tumors in transgenic mice with targeted PLAG1 proto-oncogene overexpression. Cancer Res. 2005;65(11): 4544-4553. [DOI] [PubMed] [Google Scholar]
- 76.Huang W, Li BR, Feng H. PLAG1 silencing promotes cell chemosensitivity in ovarian cancer via the IGF2 signaling pathway. Int J Mol Med. 2020;45(3):703-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Declercq J, Van Dyck F, Van Damme B, Van de Ven WJ. Upregulation of Igf and Wnt signalling associated genes in pleomorphic adenomas of the salivary glands in PLAG1 transgenic mice. Int J Oncol. 2008;32(5):1041-1047. [PubMed] [Google Scholar]
- 78.Qian P, He XC, Paulson A, et al. The Dlk1-Gtl2 locus preserves LT-HSC function by inhibiting the PI3K-mTOR pathway to restrict mitochondrial metabolism. Cell Stem Cell. 2016;18(2):214-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Venkatraman A, He XC, Thorvaldsen JL, et al. Maternal imprinting at the H19-Igf2 locus maintains adult haematopoietic stem cell quiescence. Nature. 2013;500(7462):345-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen S, Gao R, Kobayashi M, et al. Pharmacological inhibition of AKT activity in human CD34+ cells enhances their ability to engraft immunodeficient mice. Exp Hematol. 2017;45:74-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rohrabaugh SL, Campbell TB, Hangoc G, Broxmeyer HE. Ex vivo rapamycin treatment of human cord blood CD34+ cells enhances their engraftment of NSG mice. Blood Cells Mol Dis. 2011;46(4): 318-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Luo Y, Li L, Zou P, et al. Rapamycin enhances long-term hematopoietic reconstitution of ex vivo expanded mouse hematopoietic stem cells by inhibiting senescence. Transplantation. 2014;97(1):20-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Berg JS, Lin KK, Sonnet C, et al. Imprinted genes that regulate early mammalian growth are coexpressed in somatic stem cells. PLoS One. 2011;6(10):e26410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kircher M, Bock C, Paulsen M. Structural conservation versus functional divergence of maternally expressed microRNAs in the Dlk1/Gtl2 imprinting region. BMC Genomics. 2008;9(1):346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, et al. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 2018;46(D1): D239-D245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vlachos IS, Paraskevopoulou MD, Karagkouni D, et al. DIANA-TarBase v7.0: indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res. 2015; 43(Database issue):D153-D159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lechman ER, Gentner B, van Galen P, et al. Attenuation of miR-126 activity expands HSC in vivo without exhaustion. Cell Stem Cell. 2012;11(6):799-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gentner B, Schira G, Giustacchini A, et al. Stable knockdown of microRNA in vivo by lentiviral vectors. Nat Methods. 2009;6(1):63-66. [DOI] [PubMed] [Google Scholar]
- 89.van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010; 10(4):301-309. [DOI] [PubMed] [Google Scholar]
- 90.Laurenti E, Varnum-Finney B, Wilson A, et al. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell. 2008;3(6):611-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Poortinga G, Wall M, Sanij E, et al. c-MYC coordinately regulates ribosomal gene chromatin remodeling and Pol I availability during granulocyte differentiation. Nucleic Acids Res. 2011;39(8):3267-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wilson A, Murphy MJ, Oskarsson T, et al. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 2004;18(22):2747-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cabezas-Wallscheid N, Buettner F, Sommerkamp P, et al. Vitamin A-retinoic acid signaling regulates hematopoietic stem cell dormancy. Cell. 2017;169(5): 807-823.e19. [DOI] [PubMed] [Google Scholar]
- 94.Espinoza DA, Fan X, Yang D, et al. Aberrant clonal hematopoiesis following lentiviral vector transduction of HSPCs in a rhesus macaque. Mol Ther. 2019;27(6):1074-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Landrette SF, Kuo YH, Hensen K, et al. Plag1 and Plagl2 are oncogenes that induce acute myeloid leukemia in cooperation with Cbfb-MYH11. Blood. 2005;105(7):2900-2907. [DOI] [PubMed] [Google Scholar]
- 96.Abi Habib W, Brioude F, Edouard T, et al. Genetic disruption of the oncogenic HMGA2-PLAG1-IGF2 pathway causes fetal growth restriction. Genet Med. 2018;20(2):250-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Signer RA, Qi L, Zhao Z, et al. The rate of protein synthesis in hematopoietic stem cells is limited partly by 4E-BPs. Genes Dev. 2016;30(15):1698-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc Natl Acad Sci USA. 2008;105(45):17414-17419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wu F, Chen Z, Liu J, Hou Y. The Akt-mTOR network at the interface of hematopoietic stem cell homeostasis. Exp Hematol. 2021;103:15-23. [DOI] [PubMed] [Google Scholar]
- 100.Grinenko T, Eugster A, Thielecke L, et al. Hematopoietic stem cells can differentiate into restricted myeloid progenitors before cell division in mice. Nat Commun. 2018; 9(1):1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Callahan KP, Minhajuddin M, Corbett C, et al. Flavaglines target primitive leukemia cells and enhance anti-leukemia drug activity. Leukemia. 2014;28(10):1960-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Stevens BM, Khan N, D’Alessandro A, et al. Characterization and targeting of malignant stem cells in patients with advanced myelodysplastic syndromes. Nat Commun. 2018;9(1):3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chabanovska O, Galow AM, David R, Lemcke H. mRNA – a game changer in regenerative medicine, cell-based therapy and reprogramming strategies. Adv Drug Deliv Rev. 2021;179:114002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kim E, Kim M, Hwang SU, et al. Neural induction of porcine-induced pluripotent stem cells and further differentiation using glioblastoma-cultured medium. J Cell Mol Med. 2019;23(3):2052-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Juma AR, Grommen SVH, O’Bryan MK, et al. PLAG1 deficiency impairs spermatogenesis and sperm motility in mice. Sci Rep. 2017;7(1):5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Goto Y, Ibi M, Sato H, et al. PLAG1 enhances the stemness profiles of acinar cells in normal human salivary glands in a cell type-specific manner. J Oral Biosci. 2020;62(1):99-106. [DOI] [PubMed] [Google Scholar]
- 107.Colvin GA, Lambert JF, Abedi M, et al. Murine marrow cellularity and the concept of stem cell competition: geographic and quantitative determinants in stem cell biology. Leukemia. 2004;18(3):575-583. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.