Abstract
Lineage plasticity is implicated in treatment resistance in multiple cancers. In lung adenocarcinomas (LUADs) amenable to targeted therapy, transformation to small cell lung cancer (SCLC) is a recognized resistance mechanism. Defining molecular mechanisms of neuroendocrine (NE) transformation in lung cancer has been limited by a paucity of pre-/post-transformation clinical samples. Detailed genomic, epigenomic, transcriptomic, and protein characterization of combined LUAD/SCLC tumors, as well as pre-/post-transformation samples, support that NE transformation is primarily driven by transcriptional reprogramming rather than mutational events. We identify genomic contexts in which NE transformation is favored, including frequent loss of the 3p chromosome arm. We observed enhanced expression of genes involved in PRC2 complex and PI3K/AKT and NOTCH pathways. Pharmacological inhibition of the PI3K/AKT pathway delayed tumor growth and NE transformation in an EGFR-mutant patient-derived xenograft model. Our findings define a novel landscape of potential drivers and therapeutic vulnerabilities of neuroendocrine transformation in lung cancer.
INTRODUCTION
Lineage plasticity describes the capacity of cells to transition from one committed identity to that of a distinct developmental lineage. This phenotypic flexibility can promote survival of cancer cells under unfavorable conditions, such as hypoxia or selective pressure from oncogenic driver-targeted therapy(1,2),(3). The histological transformation of lung adenocarcinoma (LUAD) to an aggressive neuroendocrine (NE) derivative resembling small cell lung cancer (SCLC) is a signature example of lineage plasticity in cancer. Initially described in the prostate setting as a mechanism of resistance to androgen suppression(4–6), a similar process of NE transformation was identified in LUADs harboring EGFR mutations(7), and subsequently found to occur more broadly in lung cancers(8). Transformed SCLC (T-SCLC) is associated with a notably poor prognosis, similar or worse than that of de novo SCLC(3). The increased practice of tumor re-biopsy upon disease progression has improved the ability to identify histologic transformation, which in EGFR-mutant LUAD may comprise up to 14% of cases of acquired resistance to osimertinib(9,10).
Identification of the molecular mechanisms promoting lineage plasticity in clinical samples is key to identifying patients at high risk of transformation and may define strategies to prevent or treat this phenomenon. Little is known about the molecular alterations occurring during NE transformation in human tumors, including in lung cancer. Transcriptomic analyses of prostate cancer undergoing NE histologic transformation have been performed, but only on relapsed and post-transformation samples(11,12). A paucity of well-annotated paired pre- and post-transformation clinical samples has been a major hurdle in defining mechanisms of lineage plasticity in lung cancer. Previous genomic studies in small numbers of cases, have suggested that concomitant inactivation of TP53 and RB1 is necessary but not sufficient, and have identified few other recurrent genomic alterations(3,9,13).
On rare occasions, pathologic examination of resected cancers reveals more than one histology in single tumors. We hypothesized that such cases might represent lineage plasticity captured in temporal and spatial proximity to the occurrence of a histologic shift. Detailed molecular characterization of such cases could provide novel insight into key drivers of histologic transformation. Here we report the first comprehensive characterization of NE transformation, including genomic, transcriptomic, epigenomic and protein analyses, in a cohort of mixed histology LUAD/SCLC samples. In addition to our primary analysis of mixed histology tumors with discrete areas of LUAD and SCLC, we include analyses in matched pre- and post-transformation cases, with reference to control “pure” LUAD and SCLC. Our strategy provides novel insights into molecular drivers and potential therapeutic vulnerabilities of NE transformation in lung cancer.
RESULTS
Genomic landscape defines potential novel predictors of NE transformation
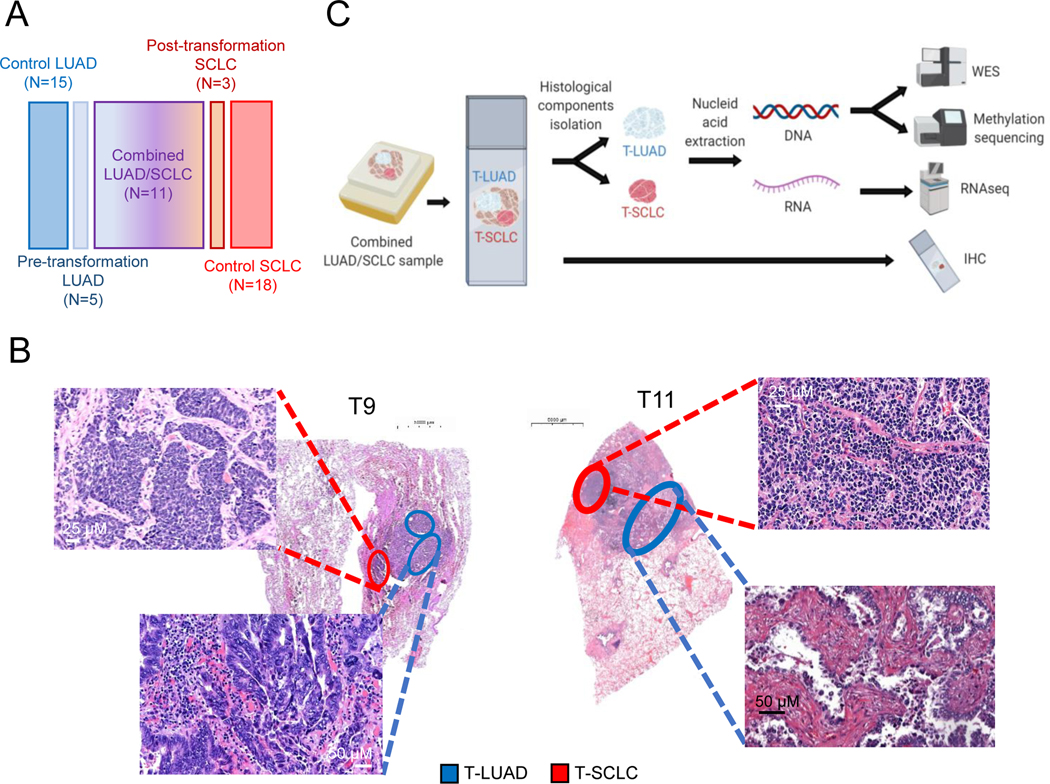
For in-depth characterization of NE transformation, we analyzed clinical specimens consisting of combined LUAD/SCLC histology exhibiting clear spatial separation (n=11); pre-transformation LUADs (n=5) and post-transformation SCLCs (n=3), including one matched case; never-transformed LUADs (n=15); and de novo SCLCs (n=18) (Figure 1A and Supplementary Tables S1–S4). Microdissection was performed for independent genomic, epigenomic and transcriptomic analyses (Figure 1B, 1C and Supplementary Figure S1).
Figure 1. Multilayer molecular characterization of SCLC transformation.
Related to Supplementary Figure S1. (A) Schematic composition of the cohort under study. (B) Illustrative H&E images of two of our combined histology samples, showing spatial separation of both independently isolated histologic components. (C) Schema of processing of combined histology samples for molecular analyses.
Our selection of combined histology samples for this analysis was predicated on the assumption that the LUAD and SCLC components were clonally related. Alternatively, it was possible that these represented “collision tumors” derived from two independent oncogenic events. To distinguish between these alternatives, we performed whole exome sequencing (WES) of all LUAD and SCLC samples from combined histology specimens and the matched pre- and post-transformation pair (T12) (Supplementary Tables S5,6). Sequencing revealed multiple shared mutations in all cases, confirming that matched LUAD and SCLC components were clonally related (Figure 2A). For one case lacking WES data, we were able to confirm genetic relatedness in the RNAseq data (Figure S2A). We therefore refer to these hereafter as T-LUAD and T-SCLC with the T referring to histologic transformation, without presumption of directionality. Higher tumor purity in the T-SCLC component was observed, consistent with the low stromal content of SCLC relative to LUAD(14,15) (Supplementary Figure S2B). Suggestive of temporal proximity, we did not observe consistent differences in tumor ploidy, tumor mutation burden, or predicted neoantigen burden between T-LUAD and T-SCLC components (Supplementary Figures S2C–E).
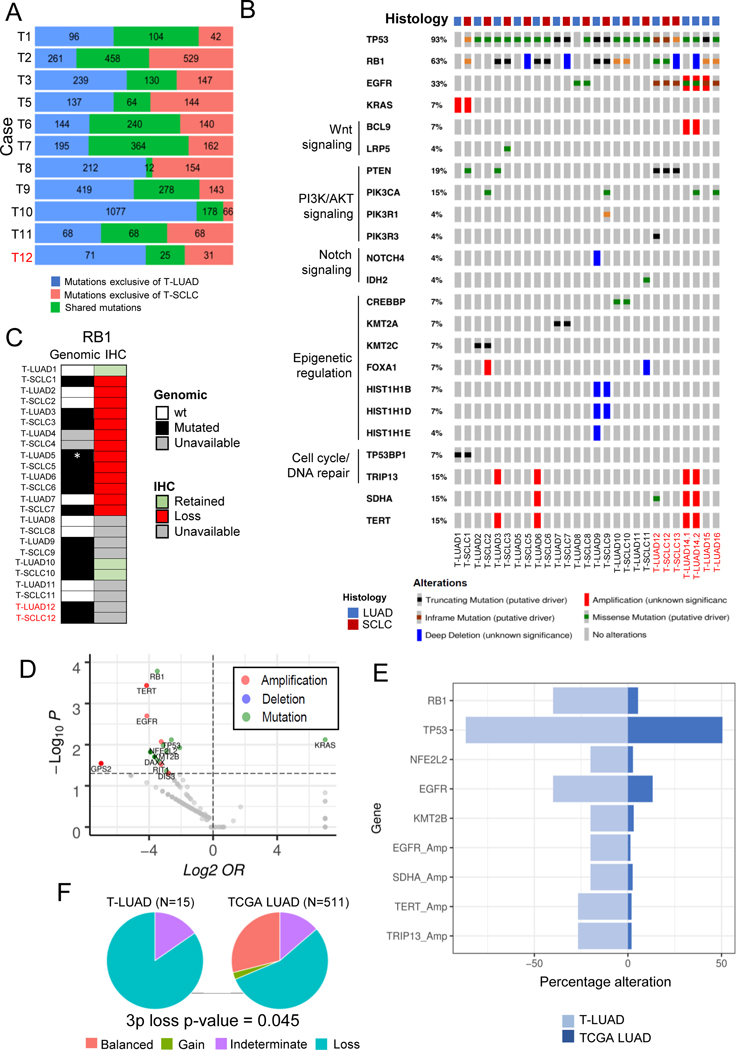
Figure 2. Genomic characterization of SCLC transformation.
Related to Supplementary Figures S2–3. (A) Bar plot showing number of exonic mutations occurring specifically in the T-LUAD and T-SCLC components, and of mutations shared between these. (B) Oncoprint showing the most prevalent likely driver/non-VUS mutations and CNAs in the transformation samples, grouped by recurrent pathways. (C) Heatmap showing complementary genomic and immunohistochemical characterization of RB1 alterations. (D) Volcano plot showing enrichment of mutations/CNAs in T-LUAD versus TCGA LUAD cohort. (E) Bar plot showing prevalence (%) of mutations/CNA enriched in T-LUAD versus TCGA LUAD with over 25% prevalence in our cohort. (F) Pie charts showing the abundance of 3p chromosome arm lost in our T-LUAD cases versus TCGA LUAD. p-value for enrichment in 3p loss was calculated using the Fisher’s exact test for count data. Samples IDs in black and red indicate that they come from a combined histology specimen or a pre-/post-transformation specimen, respectively. Cohort sizes for these analyses were N=15 for T-LUAD and N=515 for LUAD TCGA (mutations) or N=511 for LUAD TCGA (CNAs).
We next sought to define mutational processes that might contribute to lineage plasticity and histologic transformation through mutational signature analysis. Smoking signature was dominant in 7 out of 11 cases but did not differ consistently between T-LUAD and T-SCLC (Supplementary Figure S2F). APOBEC signature, previously proposed to be a predictor of SCLC-transformation in triple EGFR/TP53/RB1 mutant tumors(9), was prominent in 5 out of 11 of the T-LUAD samples (Supplementary Figure S2F).
Analyses of the most prevalent mutations and copy number alterations (CNAs), including variants of both known and unknown significance, revealed almost universal TP53 loss in both T-LUAD and T-SCLC (93%, Figure 2B), with only two T-LUADs (T-LUAD1 and T-LUAD8) showing wild type TP53. RB1 mutations/deletions were less frequently detected (63% of samples), identified in 8 out of 14 T-LUADs, and in 8 out of 11 T-SCLCs (Figures 2B, C). However, IHC in samples for which tissue was available showed that Rb protein expression was lost in all but one T-LUAD with wild type RB1 (T-LUAD1) and in all T-SCLC samples (Figure 2C). These results show that loss of RB1 function can be independent of apparent genomic alterations, highlighting the importance of complementary genomic and IHC profiling for confirmation of RB1 activity. Additionally, to confirm whether the mutations in TP53 and RB1 occurring only in one of the two matched histological components for a given case were truly private (T1 and T8), we performed targeted sequencing (Supplementary Figure S2G) that confirmed TP53 and RB1 mutations in T1 are exclusive of the T-SCLC component. However, targeted sequencing showed that the TP53 mutation found in T-SCLC8 was not truly private, as it was detected at near subclonal levels also in T-LUAD8 (VAF=0.06). Oncogenic EGFR mutations were present in 33% of T-LUAD samples (Figure 2B), further illustrating that NE transformation can occur outside the EGFR mutant setting(8). Within matched pairs, we observed common mutations of both known and unknown significance highlighting genetic relatedness. There were no recurrent mutational distinctions between paired T-LUAD and T-SCLC seen in more than two cases in this dataset, suggesting that while a preexisting genetic context may facilitate plasticity, NE transformation itself may not be mutationally driven. Mutation profile of the T-SCLC samples was similar to that described for de novo SCLC (Supplementary Figure S2H).
To better define the context that permits lineage plasticity, we focused on the most commonly altered genes in this sample set, present in both the T-LUAD and T-SCLC components (Figure 2B). Notably, these include factors involved in WNT signaling (BCL9, LRP5); PI3K/AKT signaling (PTEN, PIK3CA, PIK3R1, etc.); Notch signaling (NOTCH4, IDH2); epigenetic regulation (KMT2A/C, CREBBP, FOXA1, etc); and cell cycle/DNA repair (TRIP13 and TP53BP). The presence of these pathway alterations in T-LUAD samples implies that they may occur early in the NE transformation process and prime LUAD for lineage transition.
Next, we compared the frequency of mutations/copy number alteration (CNAs) identified in the T-LUADs in our cohort to those of the cancer genome atlas (TCGA) LUADs (Figures 2D,E, Supplementary Figure S3A and Supplementary Table S7). We focused on the differentially mutated genes showing alterations in ≥ 20% of T-LUAD samples, to filter for those more likely to have a role in transformation promotion. As expected, we found enrichment of TP53 (p=0.008) and RB1 (p<0.001) alterations in T-LUAD(3,9). Consistent with previous reports of NE transformation in EGFR-mutant LUAD, we found enrichment in EGFR alterations (p=0.012) in the T-LUAD cohort(9,13). We noted decreased frequency of KRAS mutations in our T-LUAD (p=0.008) (Supplementary Figure S3A); this may suggest that KRAS-mutant LUADs are less likely to undergo NE transformation, or, alternatively, may be attributable to the historical lack of potent targeted inhibitors of KRAS.
Novel observations in this analysis included mutations on NFE2L2 (p=0.010), a transcription factor involved in response to oxidative stress(16); KMT2B (p=0.014), an epigenetic regulator; and amplifications in EGFR (p=0.002), TERT (p<0.001) and TRIP13 (p<0.001), a gene involved in cell division. These genes were rarely altered (<5%) in the TGCA LUADs (Figure 2E and Supplementary Figure S3A). Of note, none of these enrichments were significant when performing multiple hypothesis testing, most likely due to the small size of our cohort. Validation in a larger cohort would be required to support a role for these alterations as predictors of susceptibility to SCLC transformation. Interestingly, we observed recurrent loss of the 3p chromosome arm in 85% of our pre-transformation LUAD cases, a significantly higher rate than observed in TCGA LUADs (p=0.045, Figure 2F).
Genomic evolution of SCLC transformation
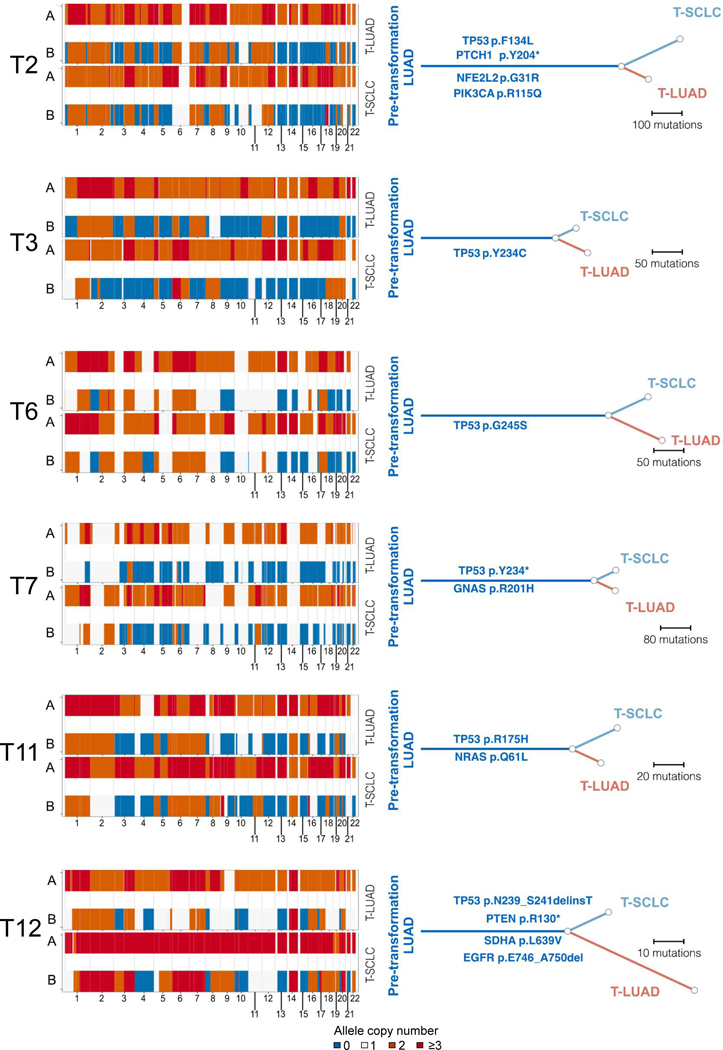
The paired nature of the combined histology tumors provided an opportunity to explore serial events in branched evolution of the distinct histologic lineages. WES data was of sufficient quality to allow reconstruction of the clonal history for 5 of the cases under study (Figure 3). For each of these, we identified exclusive or enriched mutations in the T-SCLC components, but no common driver mutations across cases were observed. The integer copy number segmentation profile (Figure 3, left) was suggestive of genomic instability consistent with TP53 inactivation, which was identified as an early event in most samples. Within these segments, at a gene level, we observed several recurrent focal non-VUS copy number changes (Supplementary Table S8). Many of the oncogenic driver mutations were shared within each matched pair (Figure 3, right), suggesting that they occurred before the histologic divergence. Interestingly, both pre- and post-transformation samples in all of these cases were genome doubled, implying that WGD occurred before transformation. (Figure 3, right).
Figure 3. Genomic mutation evolution of SCLC transformation.
Chromosomal gain/losses (at a segment level) in both alleles for matched LUAD and SCLC components for each case (left) and reconstruction of clonal evolution (right) in 4 combined histology and 1 pair of pre- and post-transformation cases. All oncogenic and hotspot mutations are annotated along their respective branch. Samples IDs in black and red indicate that they come from a combined histology specimen or a pre-/post-transformation specimen, respectively.
T-SCLC spans all SCLC subtypes
Work from our group and others has highlighted the inter-tumoral heterogeneity of SCLCs(15). De novo SCLCs can be divided into discrete molecularly defined subtypes based on dominant expression of one of four transcriptional regulators: ASCL1 (SCLC-A), NEUROD1 (SCLC-N), POU2F3 (SCLC-P), and YAP1 (SCLC-Y). However, little is known about the molecular subtyping of T-SCLC tumors, or whether these tumors consistently align with one of these four defined subtypes.
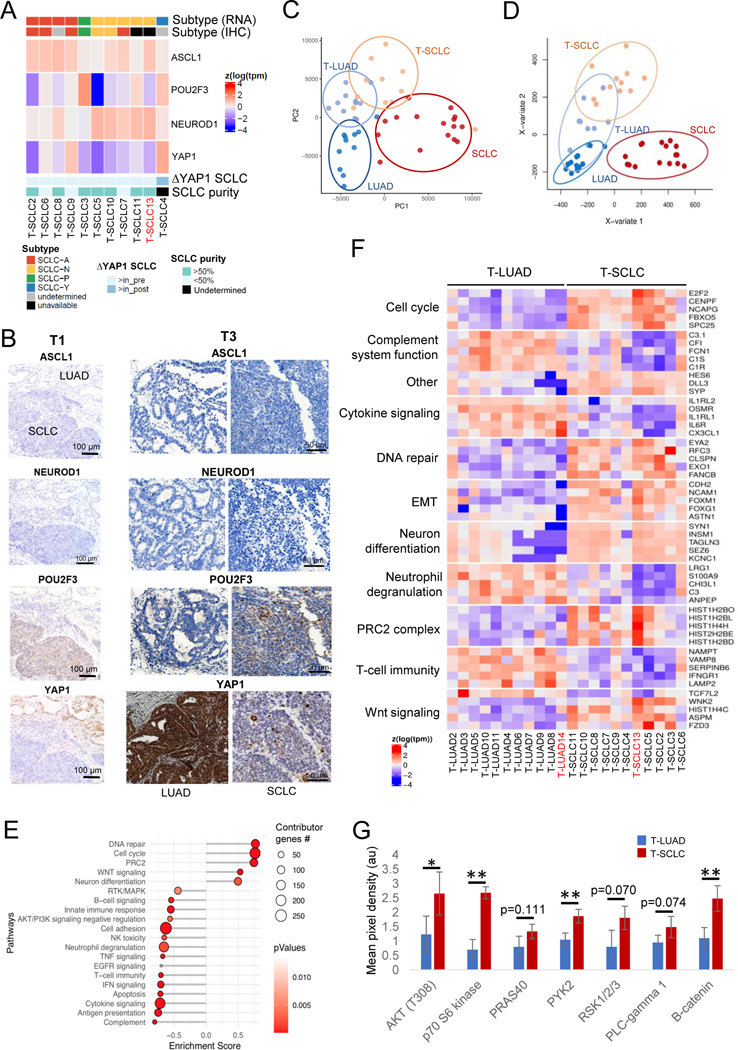
To study if T-SCLCs were enriched in any subtype, we analyzed relative expression of these four transcriptional regulators (mRNA and protein by IHC), together with canonical NE markers (mRNA), in the T-LUADs and T-SCLC samples (Figures 4A, S3B and Supplementary Table S9). As expected, we observed increased expression of NE markers in T-SCLC versus T-LUAD consistent with NE transformation, which was further accentuated in de novo SCLC (Figure S3B). Expression of three transcription factors (ASCL1, NEUROD1 and POU2F3) was consistently low in the T-LUADs. However, expression of YAP1 was higher in all but one (T4) T-LUADs than in their matched T-SCLCs (Figure 4A). YAP1 expression was higher in never-transformed LUADs than in T-LUAD (Supplementary Figure S3C), consistent with the oncogenic role of this Hippo pathway effector in LUAD(17,18) and with its incompatibility with NE features in lung cancer(19). We observed good concordance between IHC and RNA data. Where discrepant, we assigned the subtype based on relative RNA expression, following current consensus(15).
Figure 4. Transcriptomic, epigenomic and protein characterization of SCLC transformation.
Related to Supplementary Figures S4–5. (A) Heatmap showing mRNA expression of the SCLC subtype-determining TFs, tumor purity, highest TF expressed by IHC in the T-SCLC component and YAP1 mRNA expression in the T-SCLC component relative to their matched T-LUAD component, in the transformation samples. (B) IHC images for subtype-determining TFs in the SCLC-P T-SCLC cases (ch1 and ch3). (C) PCA analysis on the transcriptomes of our pre- and post-transformation samples, and of our control LUAD and de novo SCLC samples. (D) PLSDA analyses on the methylome of our T-LUAD and T-SCLC samples, and of our control LUAD and SCLC samples. (E) Pathway enrichment analyses on the DEGs of the T-LUAD versus T-SCLC comparison. (F) Heatmap highlighting DEGs of interest, grouped by recurrent pathways. (G) Bar plot showing differential phosphorylation of genes involved in the AKT/Wnt signaling pathways, and differential expression of β-catenin, as determined by an antibody array on pre- and post-transformation clinical and PDX samples. Samples IDs in black and red indicate that they come from a combined histology specimen or a pre-/post-transformation specimen, respectively. p-values legend: * p<0.05, **p<0.01.
Notably, we were able to detect all four SCLC subtypes among the T-SCLC samples, suggesting that lineage plasticity in LUAD can give rise to any of the four SCLC subtypes. Interestingly, two of the samples (T1 and T3) were categorized as SCLC-P, with high POU2F3 levels exclusive to the T-SCLC component, and no expression of any of the other transcription factors by IHC (Figures 4A,B and Supplementary Table S9). Tuft cells, a rare population of lung cells, have been previously hypothesized to be the cell of origin for SCLC-P, based on a very similar POU2F3-dependent gene expression program exclusive in this cell type of the normal lung(20). However, no POU2F3 protein expression was observed in the matched T-LUAD components of these samples (Figures 4A,B and Supplementary Table S9). Furthermore, mRNA levels of other tuft cell markers(20) were also not elevated in these T-LUADs relative to the rest of pre-transformation or control LUADs (Supplementary Figure S3D). This suggests that a tuft cell-like gene expression program is induced in this T-SCLC subtype, independent of the cell of origin. These data demonstrate that T-SCLCs conform to all major subtypes of de novo SCLCs, and suggest a tuft cell-independent origin of SCLC-P.
Gene expression and methylation analyses identify pathways involved in NE transformation
We performed transcriptomic (RNAseq, Supplementary Table S10) and methylation (EPIC) analyses of T-LUADs, T-SCLCs, control LUADs and de novo SCLCs (Figure 1C and Supplementary Tables S1–3,11). Principal component analyses (PCA) of RNAseq data showed dissimilar expression patterns for control LUAD and de novo SCLC, as expected (Figure 4C). T-LUADs clustered together in adjacency to control LUADs, and T-SCLCs in proximity to de novo SCLCs. T-LUAD and T-SCLC did appear to represent intermediate phenotypes, and demonstrated substantial overlap in expression profile (Figure 4C). This suggests that T-LUADs might be distinctly primed to transform, relative to other LUAD, and that T-SCLC retains some transcriptomic features of T-LUAD. PCA analysis of methylation profiling by EPIC revealed that T-SCLCs exhibit distinct methylation profiles to those of de novo SCLCs, and show proximity to the methylome of T- and control LUADs (Figure 4D). This implies that tumors undergoing NE transformation retain broad scale epigenomic features of the LUAD from which they derived.
To further analyze the transcriptional changes occurring during NE transformation, we performed differential gene expression and pathway enrichment analyses (GSEA) of T-LUAD and T-SCLC samples (Figure 4E). As expected, EGFR expression was downregulated upon transformation (Supplementary Figure S4A). T-SCLC demonstrated increased expression of NE markers such as SYP, SYN1 and INSM1; and genes associated with Notch signaling inhibition, including DLL3 and HES6(15). Pathway enrichment analyses performed on differentially expressed genes (DEG) in T-LUAD vs. T-SCLC samples (Figures 4E,F) showed T-SCLC-specific upregulation of genes involved in (1) neural differentiation (including SEZ6, TAGLN3 and KCNC1); (2) cell cycle progression (including E2F2, CENPF and FBXO5); (3) DNA repair (including FANCB, EYA2 and RFC3); (4) chromatin remodeling (including HDAC2); and (5) PRC2 complex (including HIST1H2BO, HIST1H2BL and HISH1H4H) (Figures 4E,F). We further confirmed a consistent increase in the mRNA expression of EZH2, enconding the enzymatic subunit of the PRC2 complex (Supplementary Figure S4B), previously strongly implicated in lineage plasticity and neuroendocrine transformation in prostate cancer(4). GSEA analyses also showed a gene expression signature of induced WNT signaling in T-SCLC, with downregulation of the negative regulator of WNT signaling TCF7L2 and overexpression of WNT pathway activators such as WNK2, ASPM and FZD3 (Figures 4E–F). This was further supported by protein analyses results of T-LUAD and T-SCLC samples and patient-derived xenografts (PDXs) (Figures 4G and S4C,D and Supplementary Table S12). We observed increased expression of the major WNT signaling effector, β-catenin, and increased phosphorylation of PYK2, a protein involved in WNT signaling activation(21) in T-SCLC (Figures 4G and S4C and Supplementary Tables S11,S12). Among other changes, NE transformation was also associated with global downregulation of receptor tyrosine kinase signaling, inhibition of apoptotic induction, suppression of anti-tumor immune activation, and induction of PI3K/AKT signaling (Figures 4E,F), which was also confirmed at the protein level in protein array assays (Figures 4G and S4C) and western blot (Figure S4D).
Integration of gene expression and DNA methylation data
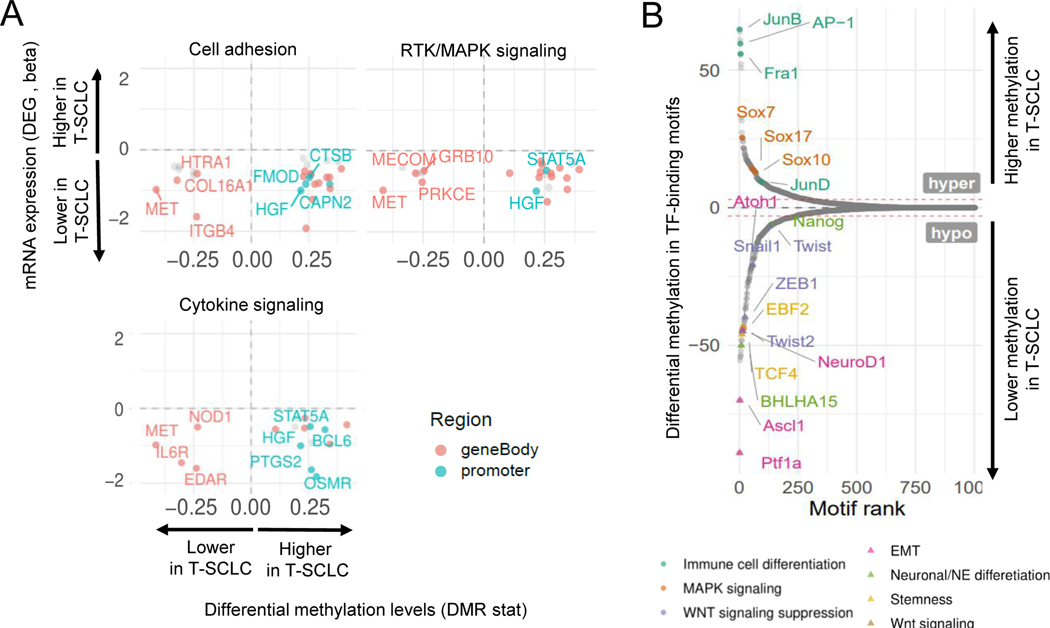
Integrative analyses of transcriptomic and epigenomic data showed that a substantial number of differentially expressed genes were also differentially methylated in T-SCLC relative to T-LUAD, consistent with epigenomic reprogramming upon NE transformation in lung. We observed cell adhesion, neuron differentiation, cytokine signaling and neutrophil degranulation pathways to be among the top pathways differentially affected by methylation (Figure 5A and Supplementary Figure S4E).
Figure 5. Integrative RNA and methylation analyses of SCLC transformation.
Related to Supplementary Figure S4. (A) Scatter plots showing DEGs exhibiting differential methylation levels in T-LUAD versus control LUAD comparison, grouped by pathways of interest. Significantly differentially expressed (q value < 0.05 and beta >= log2(1.5)) and methylated (FDR < 0.5 and differential methylation level greater than 0.1) sites are highlighted. Those genes where increased gene body or promoter methylation is correlated to expression positively and negatively, respectively, are labeled. (B) Plot exhibiting differentially methylated transcription factor binding domains in T-SCLC versus T-LUAD. Interested TFs in this study are highlighted and labeled.
Methylation occurring in TF-binding motifs can inhibit TF engagement and affect regulation of target gene expression(22). Analysis of differential methylation of TF-binding motifs revealed hypomethylation of binding motifs for genes involved in (1) neuronal and NE differentiation (including ASCL1 and NEUROD1); (2) WNT signaling activators (TCF4, EBF2); and (3) stemness (NANOG, BHLHA15); and (4) EMT (SNAI1, TWIST1/2, ZEB1, among others) in T-SCLC relative to T-LUAD (Figure 5B). We also found T-SCLC-specific hypermethylation of binding motifs for TFs involved in MAPK signaling (JUNB/D, AP-1, FOSL1/2); and WNT signaling suppression (SOX7/10/17) (Figure 5B). These data suggest that epigenomic reprogramming upon transformation leads to altered methylation of key TF-binding motifs, driving expression phenotypes observed during histological transition (Figures 4E–F).
Notably, three TFs, FOXN4 (β=3.38, q-value=0.031), ONECUT2 (β=3.10, q-value=0.014) and POU3F2 (β=2.02, q-value=0.083), were among the top differentially expressed genes upregulated in T-SCLCs (Supplementary Figure S5A). ONECUT2 and POU3F2 have been previously implicated in acquisition and maintenance of the neuroendocrine phenotype in prostate cancer(23,24). FOXN4 has been previously shown to interact with ASCL1 to modulate Notch signaling(25). To assess the role of these TFs as drivers of NE transformation, we overexpressed FOXN4, ONECUT2, and POU3F2 each independently in two EGFR-mutant LUAD cell lines (PC9 and HCC827, Supplementary Figures S5B–C). Ectopic overexpression of these factors downregulated EGFR expression in both lines (Supplementary Figure S5B), as also observed after ASCL1 or NEUROD1 overexpression (Supplementary Figure S5D). ONECUT2 and POU3F2 overexpression increased osimertinib resistance in one of the cell lines under study (Supplementary Figure S5E), and osimertinib exposure increased FOXN4 and ONECUT2 expression expression in both cell lines (Supplementary Figures S5F,G). These results suggest that although these transcription factors might not individually induce NE transformation, they may contribute to transformation through downregulation of EGFR expression, a commonly observed phenotype in EGFR-mutant T-SCLC3,13. Additionally, these results highlight a link between anti-EGFR therapy and upregulation of TFs implicated in transformation.
Taken together, these data highlight that while epigenetic reprogramming in NE transformation results in induction of transcriptional changes affecting several key signaling pathways, some epigenomic features are maintained during NE transformation, differentiating these tumors from de novo SCLC. Transformation to a neuroendocrine phenotype may be promoted by the PRC2 complex and other epigenetic modifiers, and appears to be characterized by activation of PI3K and WNT signaling pathways, acquisition of a mesenchymal phenotype, and suppression of anti-tumor immune response pathways.
Transcriptomic and epigenomic analyses of T-LUADs reveal early molecular alterations in NE transformation
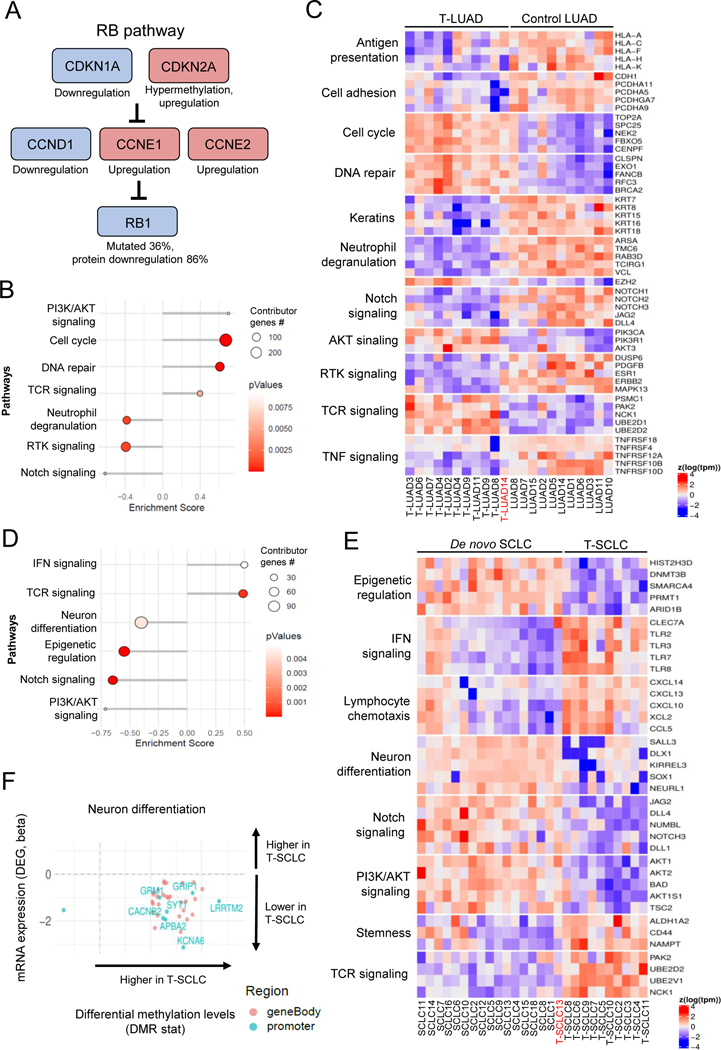
To identify transcriptional changes that may predispose to NE transformation, we next compared the transcriptomic and methylomic profiles of T-LUAD and control (never-transformed) LUADs (Figures 6A–C). In the T-LUAD samples, we observed relative downregulation of a variety of keratin genes (KRT7, KRT8 and KRT15, among others, Figure 6C), consistent with a potential partial loss of LUAD phenotype(26). As expected, we also observed multiple alterations in the RB pathway (Figure 6A). RB1 mutations and Rb protein loss were found in 36% (4/11) and in 86% (6/7), respectively, of T-LUADs. We also observed differential expression of members upstream RB1 (possibly in compensation for RB1 functional deficiency(27,28)) including upregulation of CDKN2A associated with an increases in gene body methylation (Figures 6A and Supplementary Figure S6A); downregulation of CCND1 (Cyclin D1) and upregulation of CCNE1/2 (Cyclin E1/2). These results are consistent with prior observations that RB1 loss of function precedes NE transformation(3,9).
Figure 6. Integrative RNA and methylation analyses of T-LUAD and T-LUSC versus their control counterparts.
Related to Supplementary Figure S6 (A) Alterations in the RB pathway identified in T-LUAD. (B) Pathway enrichment analyses on the DEGs of the T-LUAD versus control LUAD comparison. (C) Heatmap highlighting DEGs of interest, grouped by recurrent pathways, of the T-LUAD versus control LUAD comparison. (D) Pathway enrichment analyses on the DEGs of T-SCLC versus de novo SCLC comparison. (E) Heatmap highlighting DEGs of interest, grouped by recurrent pathways, of T-SCLC versus de novo SCLC comparison. (F) Scatter plots showing DEGs exhibiting differential methylation levels in T-SCLC versus de novo SCLC comparison, grouped by pathways of interest. Significantly differentially expressed (q value < 0.05 and beta >= log2(1.5)) and methylated (FDR < 0.5 and differential methylation level greater than 0.1) sites are highlighted. Those genes where increased gene body or promoter methylation is correlated to expression positively and negatively, respectively, are labeled. Samples IDs in black and red indicate that they come from a combined histology specimen or a pre-/post-transformation specimen, respectively.
We also identified DEGs representative of some of the same pathways identified when comparing T-LUAD and T-SCLC samples, suggesting progressive differential regulation of these pathways in NE transformation (Figures 6B–C). These included up-regulation of genes enriched in cell cycle progression (TOP2A, CENPF, FBXO5), DNA repair pathways (CLSPN, EXO1, FANCB), and PI3K/AKT signaling (PIK3CA, PIK3R1, AKT3); as well as downregulation of RTK signaling (DUSP6, ERBB2, and MAPK13), cell adhesion (CDH1 (E-cadherin), PCDHA11, PCDHA9) and anti-tumor immune response (multiple genes involved in neutrophil degranulation, TNF signaling and antigen presentation). Consistent with the known role of Notch signaling in suppressing NE tumor growth(29), these analyses revealed early downregulation of genes involved in Notch signaling, including Notch receptors NOTCH1/2/3, and ligands JAG2 and DLL4 (Figures 6B–C). Consistent with an overall retention of genome methylation patterns of LUAD, integrative analyses with transcriptomic and methylation data revealed that none of these pathways was likely being differentially regulated by gene-specific methylation (Supplementary Figure S6B).
These results suggest that an intermediate phenotype is captured in T-LUAD specimens, which is further accentuated upon NE transformation to T-SCLC. This phenotype is characterized by partial loss of LUAD features and of dependence on RTK signaling, and by the upregulation of gene programs promoting AKT signaling, cell cycle progression and DNA repair, as well as downregulation of genes related to immune response and Notch signaling.
Molecular comparison of de novo and T-SCLCs reveals differential signaling and immune pathways regulation
Finally, we sought to explore molecular differences between transformed and de novo SCLCs. Comparison of the transcriptome of T-SCLCs to that of our control de novo SCLCs revealed lower expression of genes involved in neuron differentiation (SALL3, DLX1, and NEURL1); Notch signaling (JAG2, DLL1/4, and NOTCH3); PI3K/AKT pathway (AKT1/2, BAD, and TSC2); and epigenetic regulators (HIST2H3D, SMARCA4 and ARID1B) (Figures 6D–E). We also observed higher expression of genes involved in stemness (such as CD44, NAMPT or the aldehyde dehydrogenase ALDH1A2); IFN signaling (TLR2/3/7/8, CLEC7A), lymphocyte chemotaxis (CXCL10/13/14, XCL and, CCL5) and TCR signaling (PAK2, UBE2D2, and NCK1) in T-SCLCs relative to de novo SCLCs. Integrative transcriptome/methylome analyses (Figure 6F and Supplementary Figure S6C) indicated that the suppressed neuronal phenotype in T-SCLCs was associated with a high number of differentially methylated genes in that pathway, suggesting epigenetic reprogramming (Figure 6F). These results suggest that T-SCLC is characterized by decreased neuronal features, an accentuated stem-like/plastic phenotype, and increased ability to induce an anti-tumor immune response relative to de novo SCLC. These data further support that inhibition of Notch signaling may be particularly key for SCLC transformation and persists after histological transition.
AKT inhibition delays tumor growth and augments the anti-tumor effect of osimertinib in an EGFR mutant PDX model of NE transformation
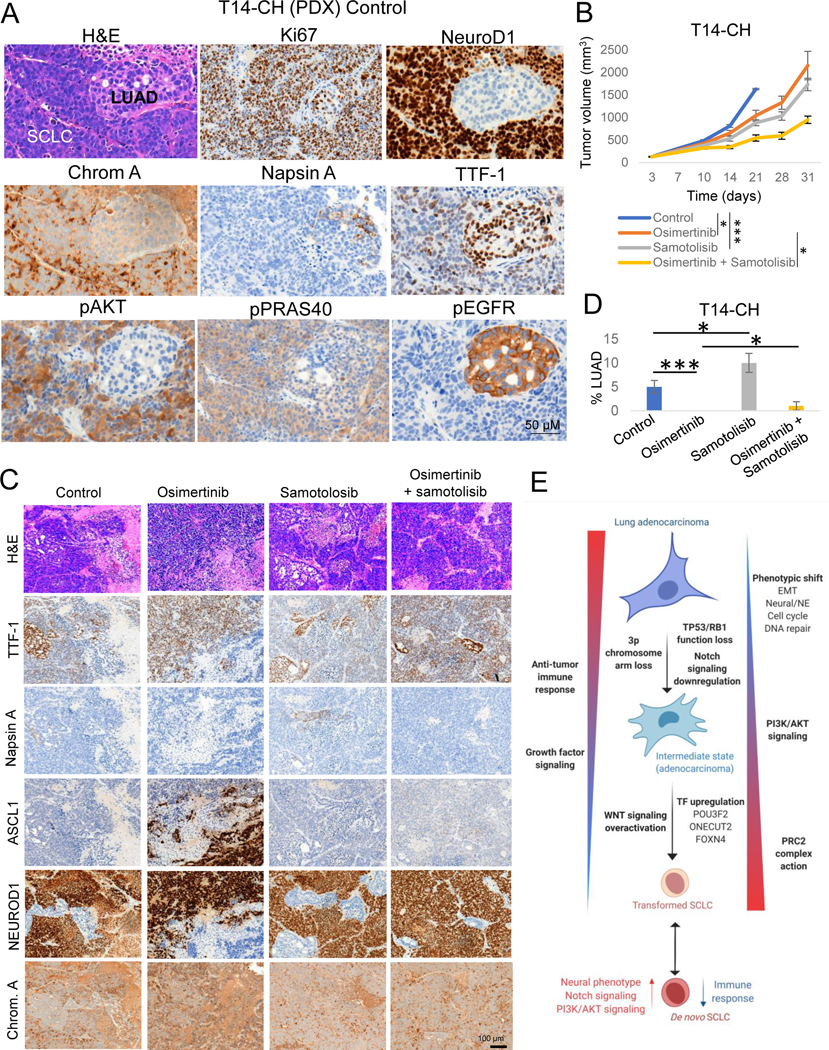
The identification of novel therapeutic approaches to treat or prevent emergence of T-SCLC is a major clinical need. Toward this end we tested different targeted therapies on an EGFR mutant PDX model (Figures 7A–D and Supplementary Figures S7A–D) exhibiting combined LUAD and NE histology with small cell features (T14-CH, Figures 7A,C). This model was derived from a patient with LUAD whose tumor subsequently underwent SCLC transformation (T14, Supplementary Table S2). The resulting PDX from the original LUAD exhibited combined histology after serial passaging. Targeted sequencing confirmed the presence of the same EGFR mutation as in the pre-transformation LUAD clinical sample (EGFR S768_D770dup).
Figure 7. Potential therapeutic approaches for SCLC transformation.
Related to Supplementary Figure S7. (A) H&E and IHC markers of interest images showing combined LUAD and SCLC histology in the T14-CH PDX. (B) In vivo tumor growth of the combined LUAD/NE EGFR-mutant PDX model T14-CH with the EGFR inhibitor Osimertinib, the AKT inhibitor Samotolisib, or their combination. Group mean tumor size ± SEM is shown. Statistical differences in tumor sizes were assessed by a two-tailed Studentś t-test, using the tumor sizes for day 21 (control group endpoint) for those comparisons involving the control group, and on day 31 (experiment endpoint) for those comparisons involving the Osimertinib-treated group. (C) Representative H&E and IHC stains for the LUAD markers TTF-1 and Napsin A and the NE markers ASCL1, NEUROD1 and Chromogranin A, of tumors in each treatment arm. (D) Percentages of LUAD component per treatment group, showing the median ± standard deviation per group. Statistical differences were assessed by a two-tailed Studentś t-test. Diagnosis of each histological component was performed by a pathologist using morphological criteria and differential staining of LUAD (TTF-1, Napsin A), NE (ASCL1, NEUROD1, Chromogranin A) and other supporting (Ki67, pEGFR) markers. (E) Schematic of molecular and phenotype changes on the different steps of SCLC transformation. Our data suggest that transformation from LUAD to SCLC may be a progressive process involving multiple signaling pathways and phenotypic changes. This process may be initiated by the loss of TP53 and RB1, decreased dependence on RTK signaling and Notch signaling downregulation, and involve progressive activation of AKT and WNT signaling pathways, epigenomic regulation by the PRC2 complex and a number of additional epigenetic enzymes, acquisition of a neuronal and EMT phenotype, and downregulation of genes involved in multiple immune response pathways. Created with BioRender.com.p-values legend: * p<0.05, **p<0.01, ***<0.001.
We targeted the major signaling pathways upregulated upon NE transformation in our transcriptomic and protein analyses and prioritized targets with the highest potential for clinical translation. Inhibitors assessed samotolisib (AKT inhibitor)(30), DS3201-b (EZH1/2 inhibitor)(31) and G007-LK (WNT inhibitor)(32). The T14-CH model was treated with either vehicle or single agent samotolisib, DS3201-b, or G007-LK, without or with osimertinib. Osimertinib monotherapy significantly delayed tumor growth (Figure 7B, T/C=63.9%, p=0.011 at control group endpoint). Combinations of DS-3201b or G007-LK with osimertinib did not achieve greater tumor growth inhibition than osimertinib monotherapy alone (Supplementary Figure S7A).
Interestingly, the AKT inhibitor samotolisib caused significant delay in tumor growth as a single agent (T/C=54.2%, p<0.001) and the combination of samotolisib with osimertinib further inhibited tumor growth in this model, with a T/C value of 33.54% at control group endpoint (p<0.001) (Figure 7B). The combination group tumors showed an average 55.8% size reduction relative to the osimertinib-treated group at experiment endpoint. Western blots for pAKT and pPRAS40 in these tumors confirmed AKT signaling inhibition by samotolosib (Figure S7B). Mice body weight measurements suggested that the combination did not cause significantly greater toxicity than osimertinib monotherapy (Supplementary Figure S7C).
Tumors were collected and analyzed for histology and for IHC expression of the LUAD markers TTF-1 and Napsin A, and of the NE markers ASCL1, NEUROD1 and Chromogranin A (Figures 7A,C). Control tumors demonstrated mixed histology, with areas of LUAD showing high TTF-1 and mild Napsin A staining with no expression of any of the NE markers tested, and areas of SCLC-like cells with lower expression of TTF-1 and no expression of Napsin A (Figures 7A,C). The SCLC component showed high levels of NEUROD1 and Chromogranin A, but no ASCL1 staining (Figures 7A,C). Osimertinib monotherapy completely depleted the LUAD component of these tumors (Figure 7D) suggesting that after transformation, osimertinib exerts a selective pressure enriching for the NE component.
Osimertinib-treated tumors showed areas of mutually exclusive staining for ASCL1 or NEUROD1, all of them positive for Chromogranin A (Figure 7C). Samotolisib-treated tumors showed enrichment in the LUAD component as compared to control tumors (Figure 7C,D), suggesting that AKT inhibition may be exerting a selective pressure against the NE component in these tumors, which was consistent with increased pAKT and pRAS40 IHC staining In the NE component (Figures 7A and S7D). The addition of samotolisib to osimertinib tenuated the depletion of LUAD and completely suppressed the emergence of ASCL1 positive SCLC seen with osimertinib alone (Figures 7B and D).
These results further support a role of AKT signaling in NE transformation in the lung and suggest that combined AKT and EGFR inhibition may constrain NE relapse in EGFR-mutant tumors at risk of transformation.
DISCUSSION
Cancer cell promiscuity in lineage commitment is a reflection of the exceptional heterogeneity of tumors, and an important source of treatment failure. The advent of potent and specific targeted inhibitors for mutational drivers in LUAD, like the use of highly effective anti-androgenic agents in prostate cancer, has prompted increasing recognition of lineage plasticity as a primary barrier to successful management of cancer. While frequently considered in the context of acquired therapeutic resistance, lineage plasticity in cancer is also evident independent of drug selection. In this study, we took advantage of the long-standing recognition of mixed histology lung cancers to gain insight into the molecular phenotypic landscapes underlying histologic transformation between LUAD and SCLC lineages. Whole exome sequencing confirmed that the histologically distinct components of mixed tumors were clonally related, reflecting distinct lineage pathways derived from a shared tumorigenic founder. TMB was similar in the LUAD and SCLC components in these samples, reflecting their presumed temporal proximity. By focusing primarily on a cohort of biphenotypic tumors in which the distinct lineages are in temporal and spatial proximity, we have the opportunity to identify consistent molecular changes that characterize this transformation. In this study, we provide the first comprehensive multi-omic characterization of NE transformation in lung cancer, including genomic, transcriptomic, epigenomic and protein analyses of matched samples. Primary limitations of this study include the use of FFPE samples, with reduced sequencing quality relative to fresh samples, and the limited number of samples available for analysis. These factors should be taken into consideration in interpreting this dataset.
One conclusion that may be taken from our data concerns the degree to which activation of lineage plasticity can result in distinct cell fates. De novo SCLC has been classified into four distinct subtypes based on differential expression of master transcriptional regulators(3,15,33). Examining a cohort of mixed histology LUAD/SCLC tumors, we find that the T-SCLC derivatives do not consistently fall into one of these subtypes – rather we find all four subtypes clearly represented among just 11 cases. This observation underscores the degree to which plasticity in lung cancer can activate diverse transcriptional programs. Particularly surprising to us was the identification of mixed histology tumors in which the T-SCLC component expressed POU2F3, defining the subtype SCLC-P. LUAD is believed to derive from type II pneumocytes(34). Based on its expression profile, SCLC-P had been proposed to arise from transformation of tuft cells, a rare pulmonary cell that is the exclusive source of POU2F3 expression in lung(20). The identification of two independent cases of clonally linked T-LUAD and POU2F3-expressing T-SCLC calls into question the cell of origin of SCLC-P and highlights the capacity of lineage plasticity to allow cancer cells to transdifferentiate between clearly distinct biological lineages.
Several features of the analysis of mixed histology T-LUAD/T-SCLC tumors reflect prior observations made regarding NE transformation of LUAD, reinforcing the relevance of this approach. Consistent with previous publications(9,13), we observe inactivation of TP53 and RB1 in essentially all T-SCLC, and in nearly all of the paired T-LUAD (Figure 7E). Notably, inactivation of these two key tumor suppressors was not always evident in exome sequencing, highlighting the importance of determination of TP53 and RB1 protein expression as a complement to genetic testing. While NE transformation of LUAD was originally observed in EGFR-mutant LUAD under selective pressure of EGFR TKI treatment, we confirm here similar histologic transformation regardless of EGFR mutation, including in the treatment naïve setting(3,8). A novel finding here is the high frequency of 3p chromosome arm loss in T-LUADs (Figure 7E)(35). What genes resident on 3p singly or in combination could account for this observation is currently unclear, but 3p loss may represent a novel risk factor for NE transformation, which combined with the determination of the RB1/TP53 genomic and expression status, may increase the sensitivity to predict this histological transition.
The paucity of recurrent mutations across samples in our cohort suggests that NE transformation in lung is not dependent on a common mutational driver, but rather may be primarily dependent on epigenetic shifts in gene expression programs. Transcriptional analysis of the T-LUAD and T-SCLC components of our mixed histology tumor set, relative to control (non-transformed) LUAD and de novo SCLC, suggests that T-LUADs and T-SCLCs occupy intermediate, transitional states – states that overlap both with their apparent non-transforming histology and with each other. Our data point to a number of signaling pathways that appear to shift in consistent patterns from T-LUAD to T-SCLC (Figure 7E). These shifts include higher expression of genes in cell cycle and DNA repair, consistent with the highly proliferative capacity of SCLC tumors(36). Higher expression of neuroendocrine and mesenchymal features in T-SCLC agrees with previous reports suggesting that NE transformation may occur through an intermediate EMT stem-like state(37,38). Our data correlates this with putative methylation-induced repression of cell adhesion molecules, and induced expression of mesenchymal effectors such as CDH2 (N-cadherin) and NCAM1 associated with demethylation of binding motifs of key mediators of EMT, such as SNAI1 and TWIST1 in T-SCLC.
Our data implicates multiple pathways known to regulate stem and progenitor cell biology in lineage plasticity and NE transformation (Figure 7E), notably including upregulation of PRC2 complex activity, induction of WNT signaling and suppression of the Notch pathway. The induction of PRC2 activity is in keeping with its apparent role in NE transformation in prostate cancer(4,39). WNT signaling too has been previously implicated in lineage plasticity(40) and in the maintenance of a NE phenotype in the prostate(41,42). Given the previously defined role of Notch signaling in suppression of NE tumor growth, we believe that sustained inhibition of Notch signaling may be a prerequisite for NE transformation in lung.
We note that SCLCs are notoriously immune “cold” tumors relative to NSCLCs(14,15,43). Consistent with this, we see a progressive suppression in anti-tumor immune response pathways including cytokine signaling, T-cell immunity, and neutrophil degranulation from control LUAD to T-LUAD, from T-LUAD to T-SCLC, and from T-SCLC to de novo SCLC.
Finally, we also find consistent evidence of PI3K/AKT pathway activation in T-SCLC. Emerging data support a role for PI3K/AKT signaling in lineage plasticity and NE transformation(3,44). Overactivation of this pathway may predict higher risk of transformation, as mutations on AKT pathway members are enriched in LUAD at high risk of transformation(9), and we consistently found upregulation of genes involved in this pathway in T-LUAD as compared to LUAD. AKT also has been identified as a driver of NE phenotypic shift in non-tumoral prostate and lung cells(45). In line with these findings, our results suggest that T-SCLC may be sensitive to AKT inhibition, which delays NE relapse in an EGFR-mutant PDX model of NE transformation in combination with osimertinib. These results support a novel therapeutic approach to delay or revert NE transformation in lung.
NE transformation in lung cancer induces a highly lethal and recalcitrant tumor profile that currently lacks effective treatments. A better understanding of molecular drivers of NE transformation in lung cancer can nominate therapeutic targets to treat or prevent transformation. Through detailed analysis of transformation pairs, we provide a comprehensive molecular characterization of NE transformation in lung cancer, describing the signaling pathways and phenotypes altered during histologic transformation mediated by lineage plasticity, and defining a potential therapeutic approach to inhibit emergence of T-SCLC in targeted treatment of lung adenocarcinoma.
MATERIALS AND METHODS
Clinical samples
We identified 11 formalin-fixed paraffin-embedded (FFPE) tumors with combined LUAD and SCLC histology, from which independent isolation of both histological components was possible (N=11, Supplementary Tables S1–2, Supplementary Figure S1). As the components of these mixed histology tumors are not temporally ordered, we refer to the component parts of these mixed histology tumors as “T-LUAD” and “T-SCLC” with the T referring to histologic transformation. We identified an additional 5 pre-transformation LUAD and 3 post-transformation SCLC cases for which tissue material was available (Supplementary Tables S1, S3). As controls we included a group of never-transformed LUADs (N=15) and a set of de novo SCLC samples (N=18) (Supplementary Tables S1, S4). All study subjects had provided signed informed consent for biospecimen analyses under an Institutional Review Board-approved protocol.
Tissue isolation
For microdissection, hematoxylin and eosin (H&E)-stained FFPE tumor slides of tumors with combined LUAD/SCLC were independently evaluated by two pathologists. Where possible, multiple FFPE blocks of each tumor were reviewed, with the aim of selecting areas containing exclusively the LUAD or the SCLC component. Where individual slides with pure components were not available, slides containing both histologic components with complete physical separation were selected. Between 10 and 20 unstained sections (USS) at 10μm prepared on uncharged slides from corresponding FFPE blocks were used for microdissection of each case. Every 10 sections, an additional section was stained with H&E for confirmation of histology. The areas corresponding to each histological component on the initial H&E were dissected using a clean blade and the tissue collected in 0.5ml nuclease free tubes for nucleic acid extraction. Alternatively, 1.0–1.5mm core punches were made from LUAD and SCLC areas on the FFPE blocks and placed in 0.5ml nuclease free tubes for nucleic acid extraction, exclusively in cases where each histologic component was located in a different block, and where no histologic cross-contamination was confirmed by pathological review.
DNA Extraction
FFPE tissue was deparaffinized using heat treatment (90°C for 10’ in 480μL PBS and 20μL 10% Tween 20), centrifugation (10,000xg for 15’), and ice chill. Paraffin and supernatant were removed, and the pellet was washed with 1mL 100% EtOH followed by an incubation overnight in 400μl 1M NaSCN for rehydration and impurity removal. Tissues were subsequently digested with 40μl Proteinase K (600 mAU/ml) in 360μl Buffer ATL at 55°C. DNA isolation proceeded with the DNeasy Blood & Tissue Kit (QIAGEN catalog # 69504) according to the manufacturer’s protocol modified by replacing AW2 buffer with 80% ethanol. DNA was eluted in 0.5X Buffer AE.
RNA/DNA dual extraction from FFPE tissue
FFPE sections were deparaffinized in mineral oil. Briefly, 800μL mineral oil (Fisher Scientific, #AC415080010) and 180μL Buffer PKD were mixed with the sections, Proteinase K was added for tissue digestion, and the sample was incubated at 56°C for 15 minutes. Phase separation was encouraged with centrifugation, and the aqueous phase was chilled 3 minutes to precipitate RNA. After centrifugation for 15 minutes at 20,000g, RNA-containing supernatant was removed for extraction, while DNA remained in the pellet. Nucleic acids were subsequently extracted using the AllPrep DNA/RNA Mini Kit (QIAGEN, #80204) according to the manufacturer’s instructions. RNA was eluted in nuclease-free water and DNA in 0.5X Buffer ATE.
RNA/DNA dual extraction from frozen tissue
Frozen tissues were weighed and homogenized in RLT and nucleic acids were extracted using the AllPrep DNA/RNA Mini Kit (QIAGEN, #80204) according to the manufacturer’s instructions. RNA was eluted in nuclease-free water and DNA in 0.5X Buffer EB.
Whole exome sequencing from DNA
After PicoGreen quantification and quality control by Agilent BioAnalyzer, 100–500 ng of DNA were used to prepare libraries using the KAPA Hyper Prep Kit (Kapa Biosystems KK8504) with 8 cycles of PCR. After sample barcoding, 100 ng of library were captured by hybridization using the xGen Exome Research Panel v1.0 (IDT) according to the manufacturer’s protocol. PCR amplification of the post-capture libraries was carried out for 12 cycles. Samples were run on a HiSeq 4000 in a 100bp/100bp paired end run, using the HiSeq 3000/4000 SBS Kit (Illumina).
Whole exome sequencing from previous DNA libraries
After PicoGreen quantification and quality control by Agilent BioAnalyzer, 100 ng of library transferred from the DMP were captured by hybridization using the xGen Exome Research Panel v1.0 (IDT) according to the manufacturer’s protocol. PCR amplification of the post-capture libraries was carried out for 8 cycles. Samples were run on a HiSeq 4000 in a 100bp/100bp paired end run, using the HiSeq 3000/4000 SBS Kit (Illumina).
WES sequencing quality
Tumor & normal specimens were sequenced to an average median coverage of 120x and 71x respectively. Median duplication rate was 39% and uniquely mapped reads were above 99% for both. In-depth sample level coverage and alignment metrics as computed using Picard tools http://broadinstitute.github.io/picard/ is provided in Supplementary Table S13. Tumor purity ranged from 25% to 89% tumor cell content as estimated from sequencing data, except for Case 10 where the algorithm was unable to predict purity, which is expected in low purity samples.
Whole Exome Analysis
We used a comprehensive in-house WES pipeline TEMPO - Time efficient mutational profiling in oncology (https://github.com/mskcc/tempo) that performs alignment using BWA-mem algorithm followed by mutation calling using Strekla2 and Mutect2 variant callers. The combined, annotated and filtered variant calls were used for downstream analysis. Details of the variant call processing are described at https://ccstempo.netlify.com/variant-annotation-and-filtering.html#somatic-snvs-and-indels and are previously described as well(46). Copy-number analysis was performed with FACETS (https://github.com/mskcc/facets), processed using facets-suite (https://github.com/mskcc/facets-suite), and manual reviewed and refitted using facets-preview (https://github.com/taylor-lab/facets-preview). To delineate mutational processes driving the acquisition of somatic alterations, mutational signatures were decomposed for all tumor samples that had a minimum of 5 single-nucleotide somatic mutations using the R package mutation-signatures (https://github.com/mskcc/mutation-signatures). Further, a given signature was considered to be ‘dominant’ if the proportion of mutations contributing to the signature was at least 20% of all mutations detected in the sample.
Purity, ploidy, tumor mutational burden (TMB), genome doubling, and cancer cell fractions for all mutations in all specimens were inferred from sequencing data. We estimated neoantigen load by taking the number of variants estimated to have strong class I MHC binding affinity by NetMHC 4.0(47) and normalizing it by the TMB. We summarized the top occurring somatic variants located on cancer genes in an oncoprint using the R package ComplexHeatmaps version 2.0.0 (https://github.com/jokergoo/ComplexHeatmap)(48). Cancer genes were genes defined as “OncoKB Annotated” on the Cancer Gene List downloaded on June 2020 (https://www.oncokb.org/cancerGenes). All other plots for this analysis were created using ggplot version 3.3.2 (https://github.com/tidyverse/ggplot2).
Comparison to TCGA
We compared somatic mutations and gene level calls (CNAs) in cancer genes in our T-LUAD samples to those in The Cancer Genome Atlas Lung Adenocarcinoma (TCGA-LUAD) cohort. The mutations for TCGA-LUAD50 cohort were extracted using the R package TCGAmutations (https://github.com/PoisonAlien/TCGAmutations) and selecting cancer type as “LUAD” by using an in-built R function “TCGAmutations::tcga_load(study = “LUAD”)”. For this set of TCGA samples and for our LUAD cohort, we predicted gene-level CNAs using the FACETS algorithm. A Fisher exact test was then performed via R function https://rdrr.io/bioc/maftools/man/mafCompare.html from maftools R package v.2.0.16 (https://github.com/PoisonAlien/maftools)51 to identify significantly altered mutations whereas that for gene-level CNAs (amplifications and deletions) was performed separately using custom R code. For both mutations and CNAs, genes with p<0.05 were considered significantly altered. The results were summarized in a volcano plot using the R packages ggplot and ‘EnhancedVolcano’ version 1.7.4 (https://github.com/kevinblighe/EnhancedVolcano). False Discovery Rates, q-values, were calculated for multiple hypothesis testing using the Benjamini-Hochberg method. All analysis was performed using the R environment for statistical computing.
Genomic Evolution
Evolutionary relationships among samples were inferred using the union of somatic mutations called in any of the tumors for a given case/patient. The genomic evolution trees were manually constructed based on the most parsimonious sequence of events using mutations and copy number events with the shared alterations represented by the trunk and the private alterations represented by the branches with the trunk and arm lengths being proportional to the relative number of shared and private alterations. Somatic variant sites included were those 1) covered at 20-fold or greater in all tumor and 10-fold or greater in matched normal specimens, 2) supported by greater than 3 reads in the tumor, 3) present in less than 3 reads in the matched normal, and 4) with a variant allele fraction in any affected tumor of greater than 2% for hotspots and 5% for non-hotspots. In order to ensure that private alterations are truly private, 1) we genotyped bam files from matched patient samples to find whether or not the said alteration is present, albeit at a sub-threshold level; if the alteration was indeed genotyped, it was not considered as truly private 2) we excluded mutation loci where the matched patient sample shows a copy number loss in the region.Samples with sufficient purity (greater than 30%) were selected for this analysis. Genotyping was performed using GetBaseCountsMultiSample v.1.2.2 (https://github.com/mskcc/GetBaseCountsMultiSample). Mutational cancer cell fraction (CCF) was inferred using annotate-maf-wrapper function from facets-suite (https://github.com/mskcc/facets-suite) which uses the variant allele fraction, locus-specific read coverage, and an analytical estimate of tumor purity, as previously described(46). WGD was also inferred using methods implemented in facets-suite utilizing FACETS(49) output and mutational timing analysis in the context of WGD was performed as previously described(50).
Methylation sequencing
After PicoGreen quantification (ThermoFisher, #P11496) and quality control by Agilent BioAnalyzer, 170–750 ng of genomic DNA were sheared using a LE220-plus Focused-ultrasonicator (Covaris, #500569). Samples were cleaned using Sample Purification Beads from the TruSeq Methyl Capture EPIC LT Library Prep Kit (Illumina, #FC-151–1002) according to the manufacturer’s instructions with modifications. Briefly, samples were incubated for 5 minutes after addition of SPB, 50 μL RSB were added for resuspension, and resuspended samples were incubated for 2 minutes. Sequencing libraries were prepared using the KAPA Hyper Prep Kit (Kapa Biosystems KK8504) without PCR amplification. Post-ligation cleanup proceeded according to Illumina’s instructions with 110 μL Sample Purification Mix. After purification, 3–4 samples were pooled equimolar and methylome regions were captured using EPIC oligos. Capture pools were bisulfite converted and amplified with 11–12 cycles of PCR. Pools were sequenced on a NovaSeq 6000 or HiSeq 4000 in a 150/150bp or 100bp/100bp paired end run, using the NovaSeq 6000 S4 Reagent Kit (300 Cycles) or HiSeq 3000/4000 SBS Kit (Illumina). The average number of read pairs per sample was 51 million.
DNA methyl capture EPIC data processing
The Bismark pipeline (51) was adopted to map bisulfite treated EPIC sequencing reads and determine cytosine methylation states. Trim Galore v0.6.4 was used to remove raw reads with low-quality (less than 20) and adapter sequences. The trimmed sequence reads were C(G) to T(A) converted and mapped to similarly converted reference human genome (hg19)(52) using default Bowtie 2(53) settings within Bismark. Uniquely aligned reads (52% - 83%) were retained. Duplicated reads (30% - 90%) were discarded. The remaining alignments were then used for cytosine methylation calling by Bismark methylation extractor. The average cytosine coverage ranges from 1.2X to 11X across 49 samples. The details of statistics on EPIC data can be found in Supplementary Table S14. Since CHG and CHH (H: one of A, T, C) shows minimal presence (<1%), we will focus on cytosines in CpG content. Number of CpGs were further filtered by C coverage >= 5 in all samples to yield 735,636 CpGs that will be used in the subsequent analyses.
Differential methylation analysis
Differentially methylated CpGs (DMCs) were identified using DSS R package(54,55) on the basis of dispersion shrinkage followed by Wald statistical test for beta-binomial distributions. Any CpGs with FDR < 0.05 and methylation percentage difference greater than 10% were considered significant DMCs. Differentially methylated regions (DMRs) were subsequently called based on the DMCs. The called DMRs were required to satisfy the minimum length of 50bps and minimum 3 CpGs in the region; two neighboring DMRs were merged if less than 50bps apart; and significant CpGs were those that occupy at least 50% of all CpGs population in the called DMRs as default in DSS package. Pairwise comparisons were conducted for pre-transformation LUAD vs control LUAD, post-transformation SCLC vs de novo SCLC, and post-transformation SCLC vs pre-transformation LUAD. The DMRs were mapped to gene regions at promoters and gene bodies, and differential methylation levels were subsequently associated with differential gene expression values in selected pathways. In addition to pairwise comparisons, principal component analysis (PCA) and partial least square discriminant analysis (PLSDA) were also performed to classify samples into groups and identify influential CpGs using mixOmics R package(55).
Motif enrichment analysis
Differential methylation may influence transcription factor (TF) binding. To identify overrepresented known TF motifs due to differential methylation for the post-transformation SCLC compared with pre-transformation LUAD, “findMotifsGenome.pl” from HOMER(56) was applied to DMCs (+/− 50bps) overlapping with gene promoter regions. DMCs regions with hyper- and hypo-methylation in SCLC were explored separately to show the effects from different methylation status. The significantly enriched TFs were defined as those with p value ≤ 0.05.
RNA sequencing
Approximately 500ng of FFPE RNA or 100ng of fresh frozen RNA per sample were used for RNA library construction using the KAPA RNA Hyper library prep kit (Roche, Switzerland) per the manufacturer’s instructions with minor modifications. Customized adapters with unique molecular indexes (UMI) (Integrated DNA Technologies, US) and Sample-specific dual-indexes primers (Integrated DNA Technologies, US) were added to each library. The quantity of libraries was measured with Qubit (Thermo Fisher Scientific, US) and quality measured by TapStation Genomic DNA Assay (Agilent Technologies, US). Equal amounts of each RNA library (around 500ng) were pooled for hybridization capture with IDT Whole Exome Panel V1 (Integrated DNA Technologies, US) using a customized capture protocol modified from NimbleGen SeqCap Target Enrichment system (Roche, Switzerland). The captured DNA libraries were then sequenced on an Illumina HiSeq4000 with paired end reads (2Å~100bp), at 50millions reads/sample.
RNASeq Analysis
In-line UMI sequences were trimmed from the sequencing reads with Marianas (https://github.com/mskcc/Marianas) and aligned to human GRCh37 genome using STAR 2.7.0 (https://github.com/alexdobin/STAR)(57) with Ensembl v75 gene annotation. Hybrid selection specific metrics and Alignment metrics were calculated for the BAM files using CalculateHsMetrics and CollectRnaSeqMetrics, respectively, from Picard Toolkit (https://github.com/broadinstitute/picard) to determine the quality of the capture.
We quantified RNA-seq reads with Kallisto v.0.45.0(58) to obtain transcript counts and abundances. Kallisto was run with 100 bootstrap samples, sequence based bias correction, and in strand specific mode, which processed only the fragments where the first read in a pair is pseudoaligned to the reverse strand of a transcript. Differential gene expression analysis, principle component analysis, and transcript per million (TPM) normalization by size factors, were done from Kallisto output files using Sleuth v0.30.0 run in gene mode(59). Differentially expressed genes were identified using the Wald test. Genes were marked significant if the False Discovery Rates, q, calculated using the Benjamini-Hochberg menthod, was less than 0.05, and beta(Sleuth-based estimation of log2 fold change)>1.25, which approximately correlated to a log2 fold change of 2 in our data. The log of the normalized TPM values for selected significant genes, were rescaled using a z-score transformation, and plotted in a heatmap using the ComplexHeatmap Library in R. RNAseq data QC is provided in Supplementary Table S15.
To confirm genetic relatedness for the case lacking WES data, known tumor mutations from MSK-IMPACT were genotyped in both RNA samples using GetBaseCountsMultiSample v.1.2.2 (https://github.com/mskcc/GetBaseCountsMultiSample). For locations where transcripts were expressed, mutations with at least 10 variant alleles were annotated as present.
Pathway enrichment
Gene set enrichment analysis (GSEA)(60) was performed on full sets of gene expression data across the previously mentioned three comparisons. Genes were ranked on p value scores computed as -log10(p value)*(sign of beta). Gene set annotations were taken from Molecular Signatures Database (MSigDB v7.0.1)(60,61). Gene sets tagged by KEGG(62,63) and REACTOME(64) pathways were retained for further analysis. The significance level of enrichment was evaluated using permutation test and the p value was adjusted by Benjamini-Hochberg procedure. Any enriched gene sets with adjusted p value ≤ 0.05 were regarded as significant. This analysis was conducted using ClusterProfiler R package(65). The enriched gene sets that are influenced by DMCs were selected and pathway annotations concatenated manually to remove redundancy and achieve high level generality. When the pathway terms were merged, median enrichment score was taken as the new group enrichment score, p values were aggregated using Fisher’s method from the Aggregation R package(66), and core enrichment of genes were collapsed.
Phospho-kinase array
Protein samples were quantified with the Bradford method (#5000205, Bio-Rad) and 200 ug aliquots were used in the phospho-kinase array (#ARYC003C, R&D-Biotechne), which was performed using the manufacturer’s instructions. Quantification of spots was performed using the Image Studio software (Version 3.1, Li-Cor). Technical replicates (2 per array) per sample were averaged. Two-tailed Student’s T-test was performed on these values, comparing the T-LUAD and T-LUSC groups.
Cell line transductions
PC9 cell line was purchased from Millipore Sigma (#90071810-VL) and HCC827 cell line was purchased from ATCC (#CRL-2868). Both cell lines were authenticated by the STR method and regularly tested (last tested April 2022) for Mycoplasma with the MycoAlert kit (#LT07–218, Lonza) and maintained in RPMI 1640 10% FBS. Both cell lines were used after little passages after purchase (5–10). Lentiviruses were produced as previously described(67) with FOXN4 (#EX-I2262-Lv151, GeneCopoeia), POU3F2 (#EX-A3238-Lv151, GeneCopoeia) and ONECUT2 (#EX-Z4476-Lv151, GeneCopoeia) overexpression lentiviral plasmids, with a EGFP overexpression plasmid as control plasmid (#EX-EGFP-Lv151, Genecopoeia). Cell lines were transduced at high MOI as previously described(67) with overnight virus incubation.
Immunoblotting
Protein extraction and western blot were performed as previously described(68). Antibodies for FOXN4 (#PA539174, ThermoFisher), ONECUT2 (#ab28466, Abcam), POU3F2 (#12137, Cell Signaling Technology), EGFR (#4267, Cell Signaling Technology), ASCL1 (#556604, BD), NEUROD1 (#ab109224, Abcam), pAKT (S473, #4060S, Cell Signaling Technology), pPRAS40 (T246, #13175, Cell Signaling Technology), Beta catenin (#8480, Cell Signaling Technology), Vinculin (#13901, Cell Signaling Technology) and actin (#3700, Cell Signaling Technology) were used.
RT-qPCR
RNA extraction, reverse transcription and quantitative PCR were performed as previously described(69). FOXN4 expression was normalized to that of GAPDH. Fluorescent probes against FOXN4 (#4351372, Applied Biosystems) and GAPDH (#4331182, Applied Biosystems) were used.
In vivo treatments
For treatments, 5–10 NOD.Cg-Prkdc<scid> Il2rg<tm1Wjl>/SzJ (NSG) mice were engrafted per treatment arm and incubated until they reached 100–150 mm3. At that point, mice were randomized into groups and treated with either vehicle, Osimertinib (25 mg/kg/day, by oral gavage), DS-3201b (50 mg/kg/day, by oral gavage), Samotolisib (10 mg/kg/day, by oral gavage), G007-LK (20 mg/kg/day, intraperitoneally), or combinations of Osimertinib with either of the other inhibitors using the same concentrations as in monotherapy, 5 days a week. For most treatment arms, mice were sacrificed when tumors reached ~1500 mm3 and fixed in formalin 10% O/N for paraffin embedding. For the Samotolisib and Samotolisib + Osimertinib groups, mice were sacrificed in parallel to the Osimertinib-treatment group, for direct comparison. Tumors and mice body weight were measured twice a week. T/C values were calculated by normalizing average tumor size of the treatment arm of interest with the average tumor size of the treatment group, both at control arm endpoint (day 21). FFPE tumors were stained as previously described(33) using the TTF-1 (#M3575, Dako), Napsin A (#NCL-NapsinA, Leica(Novocast)), ASCL1 (#556604, BD Pharmigen), NEUROD1 (#ab205300, Abcam) and Chromogranin A (#A0430, Dako) antibodies. All mice experiments. All animal experiments were approved by the Memorial Sloan Kettering Cancer Center (MSKCC) Animal Care and Use Committee.
Data reporting:
Whole exome sequencing of combined LUAD and SCLC tumor samples- SRA accession ID: PRJNA727969
RNAseq of combined LUAD and SCLC tumors to look at gene expression changes- Arrayexpress Accession ID: E-MTAB-10399
Methylation capture EPIC sequencing of combined LUAD and SCLC tumors to look at gene expression changes- ArrayExpress accession ID: E-MTAB-10617
Supplementary Material
STATEMENT OF SIGNIFICANCE
The difficulty in collection of transformation samples has precluded the performance of molecular analyses, and thus little is known about the lineage plasticity mechanisms leading to LUAD-to-SCLC transformation. Here, we describe biological pathways dysregulated upon transformation, and identify potential predictors and potential therapeutic vulnerabilities of neuroendocrine transformation in the lung.
ACKNOWLEDGEMENTS
Supported by NCI R01 CA197936 and U24 CA213274 (CMR), the Druckenmiller Center for Lung Cancer Research (CMR, TS, AQV), Parker Institute for Cancer Immunotherapy grant (TS); International Association for the Study of Lung Cancer grant (TS), NIH K08 CA-248723 (AC) and the Van Andel Institute - Stand Up to Cancer Epigenetics Dream Team grant (CMR). Stand Up to Cancer is a division of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the Scientific Partner of SU2C. We acknowledge the use of the Integrated Genomics Operation Core, funded by the NCI Cancer Center Support Grant (CCSG, P30 CA08748), Cycle for Survival, and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology. We also acknowledge Maria Corazon Mariana and Emily Lin from the PPBC Biobank for their invaluable help. The PPBC Biobank and Pathology Core Facility are supported by the NCI Cancer Center Support Grant P30-CA008748.
Footnotes
CONFLICT OF INTEREST
CAID receives research support from Bristol Myers Squibb.
CMR has consulted regarding oncology drug development with AbbVie, Amgen, Ascentage, Astra Zeneca, Bicycle, Celgene, Daiichi Sankyo, Genentech/Roche, Ipsen, Jazz, Lilly, Pfizer, PharmaMar, Syros, and Vavotek. CMR serves on the scientific advisory boards of Bridge Medicines, Earli, and Harpoon Therapeutics.
REFERENCES
- 1.Lawson DA, Kessenbrock K, Davis RT, Pervolarakis N, Werb Z. Tumour heterogeneity and metastasis at single-cell resolution. Nat Cell Biol [Internet]. Springer US; 2018;20:1349–60. Available from: 10.1038/s41556-018-0236-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501:328–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quintanal-Villalonga Á, Chan JM, Yu HA, Pe’er D, Sawyers CL, Sen T, et al. Lineage plasticity in cancer: a shared pathway of therapeutic resistance. Nat Rev Clin Oncol [Internet]. Springer US; 2020;17:360–71. Available from: 10.1038/s41571-020-0340-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, Zheng D, Zhou T, Song H, Hulsurkar M, Su N, et al. Androgen deprivation promotes neuroendocrine differentiation and angiogenesis through CREB-EZH2-TSP1 pathway in prostate cancers. Nat Commun [Internet]. Springer US; 2018;9. Available from: 10.1038/s41467-018-06177-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyoshi Y, Uemura H, Kitami K, Satomi Y, Kubota Y, Hosaka M. Neuroendocrine differentiated small cell carcinoma presenting as recurrent prostate cancer after androgen deprivation therapy. BJU Int. 2001;88:982–3. [DOI] [PubMed] [Google Scholar]
- 6.Wright ME, Tsai MJ, Aebersold R. Androgen receptor represses the neuroendocrine transdifferentiation process in prostate cancer cells. Mol Endocrinol. 2003;17:1726–37. [DOI] [PubMed] [Google Scholar]
- 7.Marcoux N, Gettinger SN, O’Kane G, Arbour KC, Neal JW, Husain H, et al. EGFR-mutant adenocarcinomas that transform to small-cell lung cancer and other neuroendocrine carcinomas: Clinical outcomes. J Clin Oncol. 2019;37:278–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujita S, Masago K, Katakami N, Yatabe Y. Transformation to SCLC after treatment with the ALK inhibitor alectinib. J Thorac Oncol [Internet]. International Association for the Study of Lung Cancer; 2016;11:e67–72. Available from: 10.1016/j.jtho.2015.12.105 [DOI] [PubMed] [Google Scholar]
- 9.Offin M, Chan JM, Tenet M, Rizvi HA, Shen R, Riely GJ, et al. Concurrent RB1 and TP53 Alterations Define a Subset of EGFR-Mutant Lung Cancers at risk for Histologic Transformation and Inferior Clinical Outcomes. J Thorac Oncol. 2019;14:1784–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sequist L V, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aggarwal R, Huang J, Alumkal JJ, Zhang L, Feng FY, Thomas GV, et al. Clinical and genomic characterization of treatment-emergent small-cell neuroendocrine prostate cancer: A multi-institutional prospective study. J Clin Oncol. 2018;36:2492–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aggarwal RR, Quigley DA, Huang J, Zhang L, Beer TM, Rettig MB, et al. Whole-genome and transcriptional analysis of treatment-emergent small-cell neuroendocrine prostate cancer demonstrates intraclass heterogeneity. Mol Cancer Res. 2019;17:1235–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JK, Lee J, Kim S, Kim S, Youk J, Park S, et al. ClonalHistory & genetic predictors of transformation into small-cell carcinomas from lung adenocarcinomas. J Clin Oncol. 2017;35:3065–74. [DOI] [PubMed] [Google Scholar]
- 14.Fischer JR, Schinde M, Stein N, Lahm H, Gallati H, Krammer PH, et al. Selective suppression of cytokine secretion in patients with small-cell lung cancer. Ann Oncol. 1995;6:921–6. [DOI] [PubMed] [Google Scholar]
- 15.Rudin CM, Poirier JT, Byers LA, Dive C, Dowlati A, George J, et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer [Internet]. 2019;19(5):289–297 Available from: 10.1038/s41568-019-0133-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryoo I geun, Kwak MK. Regulatory crosstalk between the oxidative stress-related transcription factor Nfe2l2/Nrf2 and mitochondria. Toxicol Appl Pharmacol [Internet]. Elsevier; 2018;359:24–33. Available from: 10.1016/j.taap.2018.09.014 [DOI] [PubMed] [Google Scholar]
- 17.Lin L, Sabnis AJ, Chan E, Olivas V, Cade L, Pazarentzos E, et al. The Hippo effector YAP promotes resistance to RAF- and MEK-targeted cancer therapies. Nat Genet. Nature Publishing Group; 2015;47:250–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sardo F Lo, Strano S, Blandino G. YAP and TAZ in lung cancer: Oncogenic role and clinical targeting. Cancers (Basel). 2018;10:1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito T, Matsubara D, Tanaka I, Makiya K, Tanei ZI, Kumagai Y, et al. Loss of YAP1 defines neuroendocrine differentiation of lung tumors. Cancer Sci. 2016;107:1527–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang YH, Klingbeil O, He XY, Wu XS, Arun G, Lu B, et al. POU2F3 is a master regulator of a tuft cell-like variant of small cell lung cancer. Genes Dev. 2018;32:915–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao C, Chen G, Kuan SF, Zhang DH, Schlaepfer DD, Hu J. FAK/PYK2 promotes the Wnt/β-catenin pathway and intestinal tumorigenesis by phosphorylating GSK3β. Elife. 2015;4:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Domcke S, Bardet AF, Adrian Ginno P, Hartl D, Burger L, Schübeler D. Competition between DNA methylation and transcription factors determines binding of NRF1. Nature. Nature Publishing Group; 2015;528:575–9. [DOI] [PubMed] [Google Scholar]
- 23.Guo H, Ci X, Ahmed M, Hua JT, Soares F, Lin D, et al. ONECUT2 is a driver of neuroendocrine prostate cancer. Nat Commun [Internet]. Springer US; 2019;10:1–13. Available from: 10.1038/s41467-018-08133-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bishop JL, Thaper D, Vahid S, Davies A, Ketola K, Kuruma H, et al. The master neural transcription factor BRN2 is an androgen receptor–suppressed driver of neuroendocrine differentiation in prostate cancer. Cancer Discov. 2017;7:54–71. [DOI] [PubMed] [Google Scholar]
- 25.Misra K, Luo H, Li S, Matise M, Xiang M. Asymmetric activation of Dll4-Notch signaling by Foxn4 and proneural factors activates BMP/TGFβ signaling to specify V2b interneurons in the spinal cord. Dev. 2014;141:187–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kummar S, Fogarasi M, Canova A, Mota A, Ciesielski T. Cytokeratin 7 and 20 staining for the diagnosis of lung and colorectal adenocarcinoma. Br J Cancer. 2002;86:1884–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Béguelin W, Rivas MA, Calvo Fernández MT, Teater M, Purwada A, Redmond D, et al. EZH2 enables germinal centre formation through epigenetic silencing of CDKN1A and an Rb-E2F1 feedback loop. Nat Commun [Internet]. Springer US; 2017;8:1–16. Available from: 10.1038/s41467-017-01029-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim N, Song M, Kim S, Seo Y, Kim Y, Yoon S. Differential regulation and synthetic lethality of exclusive RB1 and CDKN2A mutations in lung cancer. Int J Oncol. 2016;48:367–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sriuranpong V, Borges MW, Ravi RK, Arnold DR, Nelkin BD, Baylin SB, et al. Notch signaling induces cell cycle arrest in small cell lung cancer cells. Cancer Res. 2001;61:3200–5. [PubMed] [Google Scholar]
- 30.Bendell JC, Varghese AM, Hyman DM, Bauer TM, Pant S, Callies S, et al. A first-in-human phase 1 study of LY3023414, an Oral PI3K/mTOR dual inhibitor, in patients with advanced cancer. Clin Cancer Res. 2018;24:3253–62. [DOI] [PubMed] [Google Scholar]
- 31.Yamagishi M, Hori M, Fujikawa D, Ohsugi T, Honma D, Adachi N, et al. Targeting Excessive EZH1 and EZH2 Activities for Abnormal Histone Methylation and Transcription Network in Malignant Lymphomas. Cell Rep [Internet]. ElsevierCompany.; 2019;29:2321–2337.e7. Available from: 10.1016/j.celrep.2019.10.083 [DOI] [PubMed] [Google Scholar]
- 32.Leibold J, Ruscetti M, Cao Z, Ho YJ, Baslan T, Zou M, et al. Somatic Tissue Engineering in Mouse Models Reveals an Actionable Role for WNT Pathway Alterations in Prostate Cancer Metastasis. Cancer Discov. 2020;10:1038–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baine MK, Hsieh M-S, Lai WV, Egger JV, Jungbluth AA, Daneshbod Y, et al. SCLC Subtypes Defined by ASCL1, NEUROD1, POU2F3, and YAP1: A Comprehensive Immunohistochemical and Histopathologic Characterization. J Thorac Oncol [Internet]. Elsevier Inc; 2020;15:1823–35. Available from: 10.1016/j.jtho.2020.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferone G, Lee MC, Sage J, Berns A. Cells of origin of lung cancers: Lessons from mouse studies. Genes Dev. 2020;34:1017–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petersen I, Langreck H, Wolf G, Schwendel A, Psille R, Vogt P, et al. Small-cell lung cancer is characterized by a high incidence of deletions on chromosomes 3p, 4q, 5q, 10q, 13q and 17p. Br J Cancer. 1997;75:79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Travis WD. Update on small cell carcinoma and its differentiation from squamous cell carcinoma and other non-small cell carcinomas. Mod Pathol. Nature Publishing Group; 2012;25:18–30. [DOI] [PubMed] [Google Scholar]
- 37.Mu P, Zhang Z, Benelli M, Karthaus WR, Hoover E, Chen CC, et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53-and RB1-deficient prostate cancer. Science (80- ). 2017;355:84–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akamatsu S, Wyatt AW, Lin D, Lysakowski S, Zhang F, Kim S, et al. The placental gene PEG10 promotes progression of neuroendocrine prostate cancer. Cell Rep [Internet]. The Authors; 2015;12:922–36. Available from: 10.1016/j.celrep.2015.07.012 [DOI] [PubMed] [Google Scholar]
- 39.Dardenne E, Beltran H, Benelli M, Gayvert K, Berger A, Puca L, et al. N-Myc Induces an EZH2-Mediated Transcriptional Program Driving Neuroendocrine Prostate Cancer. Cancer Cell [Internet]. Elsevier Inc.; 2016;30:563–77. Available from: 10.1016/j.ccell.2016.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sánchez-Danés A, Larsimont JC, Liagre M, Muñoz-Couselo E, Lapouge G, Brisebarre A, et al. A slow-cycling LGR5 tumour population mediates basal cell carcinoma relapse after therapy. Nature. Springer US; 2018;562:434–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moparthi L, Pizzolato G, Koch S. Wnt activator FOXB2 drives the neuroendocrine differentiation of prostate cancer. Proc Natl Acad Sci U S A. 2019;116:22189–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin X, Spöttl G, Maurer J, Nölting S and Auernhammer CJ. Inhibition of Wnt / β -Catenin Signaling in Neuroendocrine Tumors In Vitro : Antitumoral Effects. Cancers 2020;12(2):345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang W, Hodkinson P, McLaren F, MacKinnon A, Wallace W, Howie S, et al. Small cell lung cancer tumour cells induce regulatory T lymphocytes, and patient survival correlates negatively with FOXP3+ cells in tumour infiltrate. Int J Cancer. 2012;131:928–37. [DOI] [PubMed] [Google Scholar]
- 44.Chen Y, Chi P, Rockowitz S, Iaquinta PJ, Shamu T, Shukla S, et al. ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss. Nat Med. 2013;19:1023–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park JW, Lee JK, Sheu KM, Wang L, Balanis NG, Nguyen K, et al. Reprogramming normal human epithelial tissues to a common, lethal neuroendocrine cancer lineage. Science (80- ). 2018;362:91–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jonsson P, Bandlamudi C, Cheng ML, Srinivasan P, Chavan SS, Friedman ND, et al. Tumour lineage shapes BRCA-mediated phenotypes. Nature. 2019;571:576–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jurtz V, Paul S, Andreatta M, Marcatili P, Peters B, Nielsen M. NetMHCpan-4.0: Improved Peptide–MHC Class I Interaction Predictions Integrating Eluted Ligand and Peptide Binding Affinity Data. J Immunol. 2017;199:3360–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–9. [DOI] [PubMed] [Google Scholar]
- 49.Shen R, Seshan VE. FACETS: Allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 2016;44:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bielski CM, Zehir A, Penson AV, Donoghue MTA, Chatila W, Armenia J, et al. Genome doubling shapes the evolution and prognosis of advanced cancers. Nat Genet. Springer US; 2018;50:1189–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krueger F, Andrews SR. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Erratum: Initial sequencing and analysis of the human genome: International Human Genome Sequencing Consortium (Nature (2001) 409 (860–921)). Nature. 2001;412:565–6. [DOI] [PubMed] [Google Scholar]
- 53.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park Y, Wu H. Differential methylation analysis for BS-seq data under general experimental design. Bioinformatics. 2016;32:1446–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rohart F, Gautier B, Singh A, Lê Cao KA. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput Biol. 2017;13:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol Cell [Internet]. Elsevier Inc.; 2010;38:576–89. Available from: 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–7. [DOI] [PubMed] [Google Scholar]
- 59.Pimentel H, Bray NL, Puente S, Melsted P, Pachter L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods. 2017;14:687–90. [DOI] [PubMed] [Google Scholar]
- 60.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. Oxford University Press; 2019;47:D590–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28:1947–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jassal B, Matthews L, Viteri G, Gong C, Lorente P, Fabregat A, et al. The reactome pathway knowledgebase. Nucleic Acids Res. Oxford University Press; 2020;48:D498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu G, Wang LG, Han Y, He QY. ClusterProfiler: An R package for comparing biological themes among gene clusters. Omi A J Integr Biol. 2012;16:284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yi L, Pimentel H, Bray NL, Pachter L. Gene-level differential analysis at transcript-level resolution. Genome Biol. Genome Biology; 2018;19:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hulton CH, Costa EA, Shah NS, Quintanal-Villalonga A, Heller G, de Stanchina E, et al. Direct genome editing of patient-derived xenografts using CRISPR-Cas9 enables rapid in vivo functional genomics. Nat Cancer [Internet]. Springer US; 2020;1:359–69. Available from: 10.1038/s43018-020-0040-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gardner EE, Lok BH, Schneeberger VE, Desmeules P, Miles LA, Arnold PK, et al. Chemosensitive Relapse in Small Cell Lung Cancer Proceeds through an EZH2-SLFN11 Axis. Cancer Cell [Internet]. Elsevier Inc.; 2017;31:286–99. Available from: 10.1016/j.ccell.2017.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Quintanal-Villalonga Á, Ferrer I, Guruceaga E, Cirauqui C, Marrugal Á, Ojeda L, et al. FGFR1 and FGFR4 oncogenicity depends on n-cadherin and their co-expression may predict FGFR-targeted therapy efficacy. EBioMedicine. 2020;53:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.