Summary
The immense regenerative potential of the liver is attributed to the ability of its two key cell types – hepatocytes and cholangiocytes – to trans-differentiate to one another either directly or through intermediate progenitor states. However, the dynamic features of decision-making between these cell-fates during liver development and regeneration remains elusive. Here, we identify a core gene regulatory network comprising c/EBPα, TGFBR2, and SOX9 which is multistable in nature, enabling three distinct cell states – hepatocytes, cholangiocytes, and liver progenitor cells (hepatoblasts/oval cells) – and stochastic switching among them. Predicted expression signature for these three states are validated through multiple bulk and single-cell transcriptomic datasets collected across developmental stages and injury-induced liver repair. This network can also explain the experimentally observed spatial organization of phenotypes in liver parenchyma and predict strategies for efficient cellular reprogramming. Our analysis elucidates how the emergent dynamics of underlying regulatory networks drive diverse cell-fate decisions in liver development and regeneration.
Subject areas: Cell biology, In silico biology, Systems biology
Graphical abstract
Highlights
-
•
Identified minimal regulatory network to model liver development and regeneration
-
•
Changes in phenotypic landscapes by in-silico perturbations of regulatory networks
-
•
Ability to explain physiological spatial patterning of liver cell types
-
•
Decoded strategies for efficient reprogramming among liver cell phenotypes
Cell biology; In silico biology; Systems biology
Introduction
The liver, the largest internal organ in the body, performs key physiological functions. It possesses remarkable regenerative ability and is capable of restoring its mass, architecture, and function completely after injury (Gadd et al., 2020; Kopp et al., 2016). Both the major cell types seen in liver parenchyma – hepatocytes and cholangiocytes (biliary epithelial cells) – have been shown to be capable of dividing extensively (Kopp et al., 2016) and trans-differentiate into one another, thus contributing to liver regeneration (Deng et al., 2018; Schaub et al., 2018; Yanger et al., 2013).
In the liver, hepatocytes and cholangiocytes perform very different functions. Although the former executes most metabolic functions, including bile secretion; the latter are biliary epithelial cells that line the bile duct tubules and serve to transport bile from liver to the small intestine. Developmentally, hepatocytes and cholangiocytes are both formed from a common progenitor cell type known as hepatoblasts. Hepatoblasts can co-express markers for both hepatocytes (HNF4α, CK18) and cholangiocytes (CK19) (Gordillo et al., 2015), a characteristic trait of such bipotent cells witnessed during embryonic development (Zhou and Huang, 2011). Cellular plasticity seen in vivo between cholangiocytes and hepatocytes suggests that although they may be terminally differentiated, they retain the ability to trans-differentiate into one another during injury-induced repair (Deng et al., 2018; Schaub et al., 2018). These observations suggest that these differentiated cells in liver can carry permissive chromatin from their progenitors, which may be important for their developmental and reprogramming competence (Li et al., 2020). However, the intracellular and tissue-level dynamics of cell-state transitions between these cell types in a liver remains unclear.
Phenotypic plasticity is a hallmark of injury response too. For instance, during chronic liver injury, a subset of resident liver cells are capable of undergoing epithelial-mesenchymal transition (EMT) and later regenerate hepatocytes through mesenchymal-epithelial transition (MET) (Choi and Diehl, 2009). EMT/MET has been well-investigated in such pathophysiological conditions (Nieto et al., 2016). However, in another manifestation of plasticity, the cholangiocytes can transition to hepatocytes via an intermediate biphenotypic state which expresses both hepatocytic and biliary markers (Deng et al., 2018; Manco et al., 2019), as identified via lineage tracing. Importantly, these reprogrammed cells are more proliferative and less apoptotic than the native hepatocytes (Manco et al., 2019). The biphenotypic state seen is reminiscent of earlier observations about the existence of oval cells – bipotent cells that expressed hepatoblast marker AFP and could differentiate to both lineages (Kopp et al., 2016). Similarly, SOX9+ hepatocytes have also been implicated to behave as bipotent progenitor cells after liver injury (Han et al., 2019; Tsuchiya and Yu, 2019). In liver homeostasis, liver progenitor cells (LPCs) have been identified that can differentiate into both hepatocytes and cholangiocytes in culture and can repopulate a liver after transplantation (Li et al., 2020). Thus, although molecular and functional similarities and differences in these different types of bipotent ‘hybrid’ cells identified are still being accrued, it is intriguing to note that across the contexts of embryonic development, homeostasis, liver injury and regeneration, one or more bipotent cell types have been identified.
Across these different contexts of liver development, injury repair and reprogramming, a variety of ‘master regulators’ have been identified that are capable of either inducing a hepatocyte cell-fate over cholangiocytes or vice versa or driving trans-differentiation of cholangiocytes to hepatocytes or vice versa. However, there are surprisingly few studies that attempt to uncover the fundamental traits of underlying gene regulatory networks formed by these ‘master regulators’ that govern cell fate transitions in liver development and regeneration.
Here, we first identify a core gene regulatory network involved in both developmental and regenerative cell-state transitions between hepatocytes and cholangiocytes and elucidate the dynamics of this network through a mechanism-based mathematical model. Our simulations suggest that this network is capable of enabling three states that can be mapped to hepatocytes, cholangiocytes and a progenitor-like phenotype. These model predictions are supported by analysis of diverse gene expression datasets collected during development and reprogramming scenarios. Further, these states were shown to be capable of transitioning to one another, under the influence of biological noise and/or external perturbations, deciphering possible mechanistic basis for observed trans-differentiation. We also decipher how the emergent properties of this regulatory network can give rise to spatial patterning of these liver cell types. Our mathematical model unravels design principles of the underlying liver cell-fate decision network as well as lays a predictive framework to investigate the dynamics of cell commitment and reprogramming in liver.
Results
Multistability in gene regulatory network underlying hepatocyte-cholangiocyte cell-fate commitment and plasticity
First, we identified a core gene regulatory network that is integral to the observed plasticity among hepatocytes, hepatoblasts and cholangiocytes. Various molecular players have been implicated in remarkable cellular plasticity of liver cells both in the context of liver development (Gérard et al., 2017; Lau et al., 2018; Zong and Stanger, 2011) and regeneration (Li et al., 2020). Among these, TGFβ signaling is of key importance in the differentiation of cholangiocytes from the hepatoblasts. High levels of TGFβ signaling is required near the portal vein for differentiation of biliary cells in vivo (Clotman et al., 2005). Consistently, TGFβ can mediate hepatocyte to cholangiocyte trans-differentiation in vivo (Schaub et al., 2018), and can suppress the transcription and activity of HNF4α, a well-known hepatocyte inducer (Cozzolino et al., 2013; Li et al., 2000; Lucas et al., 2004). Overexpression of TGFBR2 in hepatoblasts in vitro can drive them into a cholangiocyte fate (Takayama et al., 2014). Thus, TGFβ signaling can be thought of as a ‘master regulator’ of cholangiocyte cell-fate. Similarly, a ‘master regulator’ of hepatocyte cell fate is c/EBPα, whose overexpression transcriptionally inhibits TGFBR2 and can drive hepatocyte differentiation in hepatoblasts (Takayama et al., 2014). Furthermore, suppression of c/EBPα can stimulate biliary cell differentiation via increased Hnf6 and Hnf1b expression in periportal hepatoblasts (Yamasaki et al., 2006). In these hepatoblasts, overexpression of TGFBR2 inhibits c/EBPα, thus forming a ‘toggle switch’ or mutually inhibitory feedback loop between TGFBR2 and c/EBPα. Such ‘toggle switches’ are hallmarks of cellular decision-making between various ‘sibling’ cell fates (Zhou and Huang, 2011).
Another important player in biliary development is SOX9, which can repress the expression of both c/EBPα and TGFBR2 in mature biliary cells (Antoniou et al., 2009; O’Neill et al., 2014). SOX9, together with SOX4, can coordinate development of the bile duct (Poncy et al., 2015), during which it is upregulated by TGFβ signaling through the Jagged1-Notch axis (Wang et al., 2018). SOX9, similar to c/EBPα and TGFβ/TGFBR2 signaling, has been proposed to self-activate either directly or indirectly (Du et al., 2018; Duan and Derynck, 2019; Friedman, 2015). Put together, these interactions constitute a gene regulatory network comprising SOX9, c/EBPα and TGFBR2 (Figure 1A) which is involved in hepatocyte-cholangiocyte cell-fate decisions in the liver. Our goal here is not to identify a comprehensive network, but a minimal network motif that can potentially explain diverse instances of cell differentiation and reprogramming seen in liver development and injury repair. Thus, the nodes represented here – SOX9, c/EBPα and TGFBR2 – can be viewed as proxies for their corresponding co-factors and their regulons implicated in cell-fate commitment.
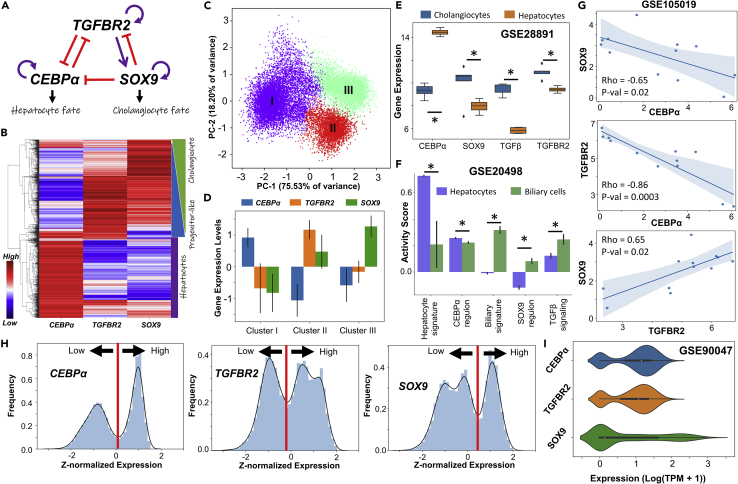
Figure 1.
Multiple phenotypes enabled by a regulatory network driving cell-fate decisions in liver
(A) Gene regulatory network (GRN) underlying plasticity in the hepatocyte-cholangiocyte cell-fate decision. Purple arrows represent activation links; red hammers represent inhibitory links.
(B) Heatmap showing hierarchical clustering in ensemble of steady state solutions obtained from the GRN. Red represents higher expression levels while blue denotes lower expression levels, plotted as z-scores.
(C) Principal Component Analysis (PCA) showing three major clusters in steady state solutions obtained.
(D) Quantification of steady state levels from the three clusters identified by PCA. Error bars represent the SD over n = 3 replicates.
(E and F) (E) Gene expression levels of SOX9, c/EBPα, TGFβ and TGFBR2 in hepatocytes and cholangiocytes (GSE28891).
(F ) Activity levels of the gene expression signatures quantified through single sample Gene Set Enrichment Analysis (ssGSEA) in hepatocytes and biliary cells (GSE20498): c/EBPαregulon, SOX9 regulon and TGFβ signaling. In E, F panels; ∗ represents a statistically significant difference in the activity/expression levels (Students’ two tailed t test; pvalue<= 0.05). (G)Scatterplots showing pairwise correlations in human liver progenitor-like population in culture (GSE105019). Spearman correlation coefficient (Rho) and pvalue (p-val) are given.
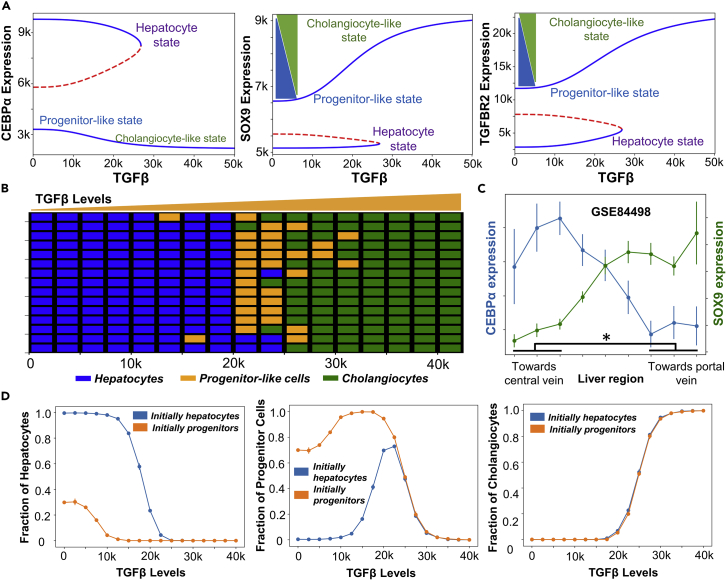
(H) Simulation data indicating the multimodal nature of steady state values in the expression of the various nodes in the network.
(I) Single-cell RNA-seq data (GSE90047) for a population composed of hepatocytes, hepatoblasts and cholangiocytes: distributions of gene expression levels of SOX9, c/EBPα, and TGFBR2.
To understand the distinct phenotypes enabled by the gene regulatory network involving c/EBPα, TGFBR2 and SOX9 (Figure 1A), we performed dynamical simulations on this regulatory network, using an ensemble of kinetic parameter sets for a set of ordinary differential equations (ODEs) representing the regulatory interactions within a network. The parameter sets sampled randomly from a biologically plausible regime capture the inherent variability in a cell population. For each parameter set, various possible steady-state (phenotype) combinations are collated to identify the robust dynamical features emerging from this network topology (see STAR methods).
For the ensemble of steady-state solutions thus obtained, hierarchical clustering suggested the evidence of three distinct cell-states, through a heatmap Figure 1B). A c/EBPα-high state can be attributed to a mature hepatocyte (Akai et al., 2014), whereas a SOX9-high state can be mapped on to a cholangiocyte profile (Dianat et al., 2014). Thus, progenitor-like (hepatoblasts or oval cells) state is likely to have low levels of both c/EBPα and SOX9, with possible enrichment of TGFBR2 (Takayama et al., 2014).Further, we observed a continuum between the progenitor-like and cholangiocyte phenotypes, pointing toward a gradient of cholangiocyte fate determination and maturation with possible intermediate states, as experimentally reported (Yang et al., 2017). Further credence for three states was obtained via principal component analysis (PCA), through which three distinct clusters of points were visually discernible (Figure 1C). The c/EBPα-high state (cluster I) had relatively low levels of SOX9 and TGFBR2. The TGFBR2-high state (cluster II) had moderate to low levels of SOX9 and low levels of c/EBPα. The SOX9-high state (cluster III) had low levels of both TGFBR2 and c/EBPα (Figure 1D). These observations were corroborated by experimental data from transcriptomic analysis of hepatocytes and cholangiocytes (GSE28891) (Shin et al., 2011) (Figure 1E). As predicted by our model, levels of c/EBPα were significantly higher in hepatocytes than in cholangiocytes, but those of SOX9, TGFβ and TGFBR2 were significantly higher in cholangiocytes than in hepatocytes (Figure 1E). In another dataset where the adult hepatocyte and cholangiocyte signatures were found to be significantly enriched in hepatocytes and cholangiocytes respectively, the c/EBPα regulon activity was higher in hepatocytes than in the biliary cell population whereas the SOX9 regulon and overall TGFβ signaling followed the opposite trend (GSE20498; Figure 1F) (Cullen et al., 2010). Finally, we show that in yet another dataset consisting of hepatocytes and cholangiocytes (Raven et al., 2017), c/EBPα levels were significantly higher in hepatocytes than in cholangiocytes whereas TGBFR2 expression, TGFB1 expression, and SOX9 regulon activity were higher in biliary cells compared to hepatocytes (GSE98034; Figure S1A). This analysis suggests that multistable dynamics of this gene regulatory network investigated here can allow for three distinct phenotypes – hepatocytes, cholangiocytes and bipotent progenitors (hepatoblasts/oval cells) – whose molecular footprints as predicted here are corroborated by diverse high-throughput transcriptomic datasets.
Next, we quantified pairwise correlation between these master regulators, and noted that while c/EBPα is negatively correlated with SOX9 and TGFBR2, SOX9 and TGFBR2 correlate positively (Figure S1B) in our simulations. This trend is consistent with observations in a population of primary hepatocytes and hepatocyte progenitor cells (Figure 1G; GSE105019) (Fu et al., 2019) and with the evidence during the hepatocyte-ductal trans-differentiation (O’Neill et al., 2014). Further, we plotted histograms for gene expression values of c/EBPα, SOX9 and TGFBR2 obtained from simulations, revealing their bimodality (Figure 1H). Such bimodality was validated from single cell RNA-seq performed specifically on a population consisting of hepatocytes, hepatoblasts and cholangiocytes (Figure 1I; GSE90047) (Yang et al., 2017).
The above-mentioned observations are largely also seen in simulations of an expanded gene regulatory network that includes a separate node showing the TGFβ ligand itself (Figures S1C–S1E). Besides allowing for the three above-mentioned phenotypes, this extended network also enabled another one with high levels of c/EBPα and SOX9 (Figure S1E). This phenotype may correspond to bipotent SOX9+ hepatocytes as has been observed in various context of liver injury and regeneration (Han et al., 2019; Tanimizu et al., 2014). Put together, we demonstrate that the network motif involving c/EBPα, SOX9 and TGFBR2 can recapitulate various cell phenotypes observed in liver development and regeneration: hepatocytes, cholangiocytes and their bipotent progenitors.
Consistent role of c/EBPα, SOX9, TGFβ signaling during cell fate decisions in liver development, regeneration and reprogramming
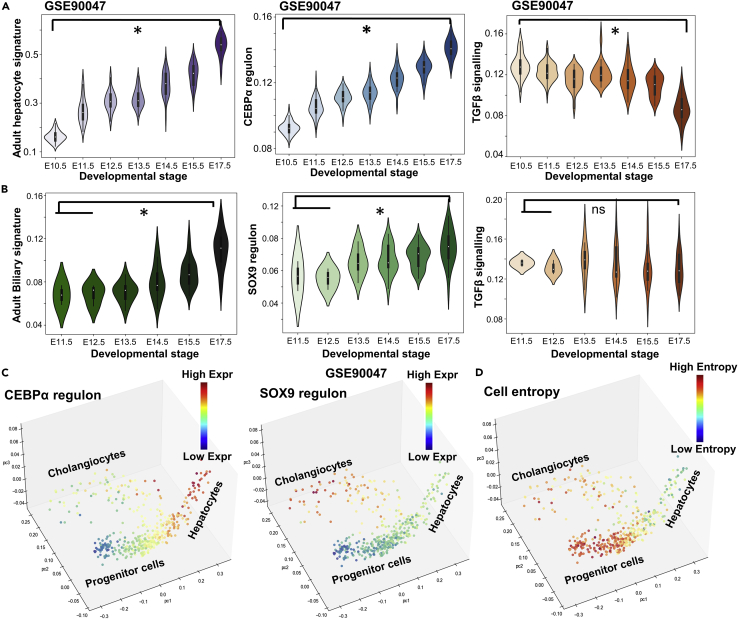
To gain further confidence in the network curated and our model predictions, we quantified the activity of various metrics relevant to our curated gene regulatory network in a single-cell RNA-seq dataset (GSE90047) for sorted hepatoblasts, hepatocytes and cholangiocytes from E10.5-E17.5 mouse fetal livers (Yang et al., 2017). As cells differentiated from hepatoblasts to hepatocytes, the adult hepatocyte signature and the c/EBPα regulon activity increased, whereas TGFβ signaling levels consistently decreased (Figure 2A). Similarly, as cells differentiated into cholangiocytes, activity for adult biliary signature and SOX9 regulon increase (Figure 2B). TGFβ signaling did not show an expected decrease; instead its variance increased. In the extended network, nonetheless, mature cholangiocytes had visibly lower levels of TGFβ than the bulk of developing cholangiocytes (Figures S1C and S1E), as observed in matured cholangiocytes (Antoniou et al., 2009). These observations were further corroborated by plotted the cell-fate trajectories at an individual cell level on a 3D PCA space, clearly showing the increase of c/EBPα levels in the hepatocyte branch and that of SOX9 higher in cholangiocyte branch respectively (Figure 2C). However, TGFβ signaling was active mostly in the cholangiocyte branch with intermediate levels of expression in the progenitor branch (Figure S2A), further underscoring the role of TGFβ signaling in commitment to the cholangiocyte fate. Similar expression patterns to those seen in liver development were observed in adult liver stem cells (GSE64292). Mature hepatocytes have very low levels of TGFβ signaling and SOX9 regulon activities (Figure S2C). Conversely, a subset of adult stem cells had high TGFβ signaling and SOX9 regulon activities, indicating a possible commitment toward the cholangiocyte fate (Figure S2C).
Figure 2.
Activity of c/EBPα, SOX9, and TGFβ signaling during development of hepatocytes and cholangiocytes from hepatoblasts
(A) Experimentally observed adult hepatocyte signature, c/EBPα regulon activity and TGFβ signaling activity in a population of single cells of hepatocytes/hepatoblasts at different developmental times (GSE90047). ∗ represents a statistically significant difference in activity levels (Students’ two-tailed t test; pvalue<0.05) while comparing E10.5 and E17.5.
(B) Experimentally observed adult biliary signature, SOX9 regulon activity and TGFβ signaling activity in a single-cell RNA-seq dataset containing mature/immature cholangiocytes with developmental time (GSE90047). ∗ represents a statistically significant difference in activity/expression levels (Students’ two tailed t-test; pvalue<= 0.05); ns indicated non-significant.
(C) Trajectory analysis of hepatocytes and cholangiocytes as they form from progenitor cells (colored by c/EBPαand SOX9 regulons showing their specificity to hepatocyte and cholangiocyte fate respectively).
(D) Cell entropy values seen to be the maximum in progenitor cells and decreases as cells enter more mature states.
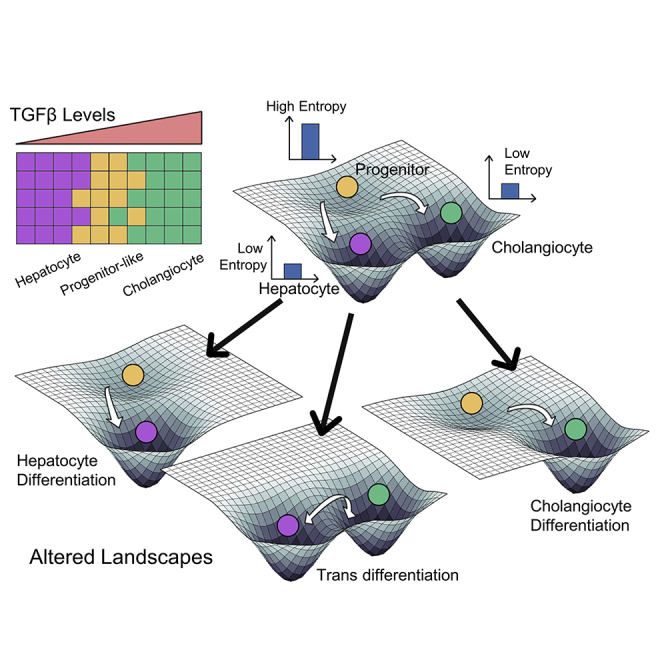
It was been earlier reported that progenitor cells generally have a larger transcriptional diversity quantified at a single-cell level (Gulati et al., 2020). To assess the transcriptional diversity in the cells, we computed the Shannon entropy values for individual cells (Teschendorff and Enver, 2017). Intriguingly, we found out that entropy is the highest in the context of the progenitor cell and significantly lower in the hepatocytes indicative of a more differentiated state (Figure 2D).
After investigating developmental datasets, we investigated samples from alcoholic steatosis liver, where we observed that the steatosis liver samples showed a strong enrichment of the adult hepatocyte signature along with high c/EBPα expression (GSE102683; Figure 3A). This observation is expected as hepatocytes form a majority of the liver mass. EpCAM has been reported to be expressed in both mouse normal cholangiocytes and oval cells. TROP2, is expressed exclusively in oval cells (Okabe et al., 2009). However, TROP2 high oval cells have been also reported to be poised toward developing into cholangiocytes (Seow et al., 2021). We found that EPCAM+ and TROP2+ populations were enriched for an adult biliary signature with a concurrent high expression of SOX9 (GSE102683; Figure 3A) (Ceulemans et al., 2017). This co-expression could indicate toward a possible enrichment of cholangiocytes in these samples, given that both TROP2 and EPCAM expression has been observed in cholangiocytes (Seow et al., 2021). Finally, progenitor cell samples isolated via side population techniques and progenitor cell niches isolated via laser microdissection showed relatively lower levels of c/EBPα, SOX9, adult hepatocyte and biliary signatures (GSE102683; Figure 3A) (Ceulemans et al., 2017). These cells instead showed an enrichment for TGFBR2 indicative of enrichment of progenitor cell populations in these samples (GSE102683; Figure 3A). Furthermore, when all these samples were analyzed together, the pairwise correlations between c/EBPα, SOX9, TGFBR2 and adult liver cell signatures showed consistent trends with what our model predicted (GSE102683; Figure S2B). Together, these observations suggest that the emergent dynamics of mechanistic model comprising c/EBPα, SOX9, and TGFβ signaling can explain how these pathways act in concert to allow for different cell fates during various liver developmental and liver pathological scenarios.
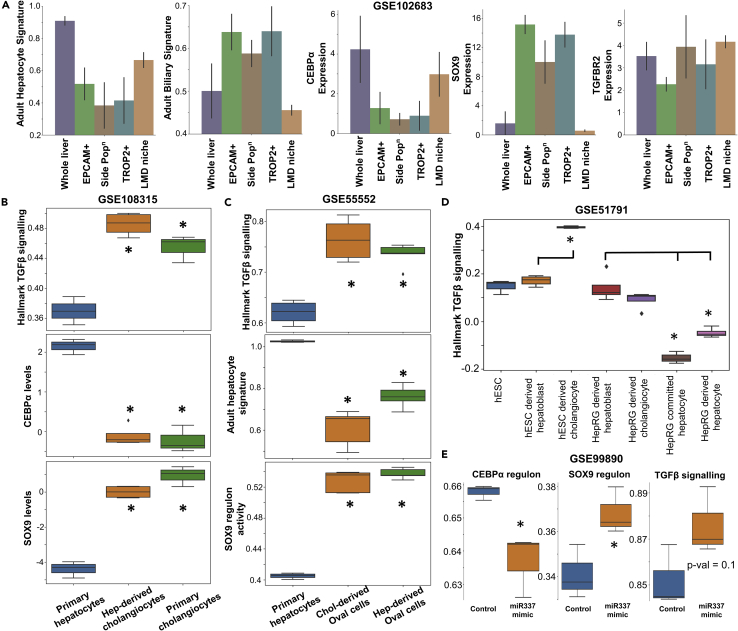
Figure 3.
Prevalence of c/EBPα, SOX9 and TGFβ signaling in the adult liver and in reprogramming experiments
(A) Quantification of c/EBPα, SOX9, TGFBR2 expression levels along with activity of adult hepatocyte and adult biliary signatures in whole liver, EPCAM+, TROP2+, side cell populations and laser micro-dissected niche of adult liver stem cells via bulk RNA sequencing (GSE102683).
(B) Experimentally observed gene expression levels of c/EBPα, SOX9 and activity of TGFβ signaling in a population of hepatocytes, cholangiocytes and cholangiocytes derived from hepatocytes (GSE108315).
(C) Experimentally observed activity levels of TGFβ signaling, hepatocyte signature and SOX9 regulon activity in hepatocytes and oval cells derived from either hepatocytes or cholangiocytes (GSE55552).
(D) Experimentally observed activity levels of TGFβ signaling in several reprogrammed cells (GSE51791).
(E) c/EBPα regulon, SOX9 regulon and TGFβ signaling activity in hepatoblasts treated with miR-337 mimic showing a development toward cholangiocyte fate (GSE99890). ∗ represents a statistically significant difference in activity/expression levels (Students’ two-tailed t test; pvalue<= 0.05).
Next, we assessed if gene expression changes observed in hepatic development and steatosis are seen in the context of liver reprogramming or during liver injury induced trans-differentiation. TGFβ treatment can convert hepatocytes to cholangiocytes via trans-differentiation. Thus, we compared the TGFβ signaling levels in a peripheral bile duct RNA-seq dataset from mouse livers (GSE108315) comprising cholangiocytes, hepatocytes, and hepatocyte-derived cholangiocytes. TGFβ signaling was lower in primary hepatocytes, but enriched in primary cholangiocytes and those derived (trans-differentiated) from hepatocytes (Schaub et al., 2018). Consistent trends are noted for SOX9, while c/EBPα levels show the opposite trends, as expected (Figure 3B). In another instance of trans-differentiation, in mouse oval cells (progenitor-like cells during liver regeneration) either derived from cholangiocytes or hepatocytes through chronic injury (Tarlow et al., 2014), the levels of SOX9 regulon and TGFβ signaling were higher than in primary hepatocytes, but the adult hepatocyte signature was lower (GSE55552; Figure 3C). Finally, during reprogramming from human embryonic stem cells (hESCs) and/or hepatic stem cell line (HepaRG) (Dianat et al., 2014), hepatocytes – both mature or immature – exhibit low levels of TGFβ signaling activity (compare fourth column with the sixth and seventh columns in Figure 3D), whereas cholangiocyte had higher or comparable levels of TGFβ signaling activity as compared to hepatoblast cell state (compare second column with third column in Figure 3D), indicative of different maturation levels of cholangiocytes (GSE51791).
Previously, miR-337 has been implicated to promote a cholangiocyte fate during liver development (Demarez et al., 2018). Thus, we examined the differential gene expression programs induced by miR-337 mimic in in vitro cultures of immortalized hepatoblasts from E12.5 wild-type mice liver. c/EBPα regulon activity decreased significantly, but SOX9 regulon and TGFβ signaling increased (GSE99890; Figure 3E), indicating the functional role of these players in committing to a cholangiocyte cell-fate. As another case study, we investigated hepatoblasts expressing adult stem cell marker LGR5 that have been shown to be capable of establishing both the hepatocyte and cholangiocyte colonies (Prior et al., 2019). We found a bimodal distribution of SOX9 regulon and TGFβ signaling activity in the entire hepatoblast population as the developmental time progressed; however, no such shift in the levels was observed specifically within LGR5+ hepatoblasts (Figure S2D), suggesting that a switch in TGFβ signaling and SOX9 levels are required to commit the hepatoblast cells toward a cholangiocyte or a hepatocyte cell fate. Therefore, the analysis of these single cell and bulk RNA-seq datasets highlight how the emergent dynamics of c/EBPα, TGFBR2 and SOX9 as predicted by a minimal model signaling is capable of recapitulating the trajectories of cell-fate decisions as observed experimentally during hepatic development, regeneration and liver cell reprogramming.
Stochastic state switching and cellular reprogramming strategies among liver cell phenotypes based on gene regulatory network dynamics
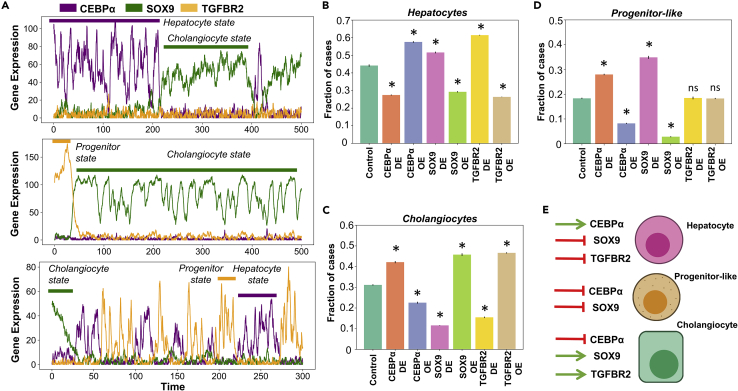
After observing distinct cellular phenotypes enabled by this regulatory network, we examined if these phenotypes could stochastically switch among one other under the influence of noise in gene expression. Previous experimental and computational studies have demonstrated the importance of intrinsic and extrinsic gene expression noise in cell fate switching in development and disease (Balázsi et al., 2011), including pathological conditions in liver (Sahoo et al., 2020a). We conducted stochastic simulations for the core gene regulatory network (Figure 1A) (Kohar and Lu, 2018) and observed switching among cell fates for various parametric combinations. We observed that hepatocytes (high levels of c/EBPα) and cholangiocytes (high levels of SOX9) could switch between one another (Figure 4A– top panel) indicative of trans-differentiation as observed experimentally (Deng et al., 2018; Schaub et al., 2018). Similarly, we also observed that hepatoblasts (characterized by high levels of TGFBR2 and low levels of both SOX9 and c/EBPα) could switch to a cholangiocyte cell fate (characterized by high level of SOX9) and stably exist for relatively longer periods of time (Figure 4A– middle panel). Finally, we also observed certain parameter sets showed dynamic transitions between all three cell states – hepatocytes, cholangiocytes and hepatoblasts (Figure 4A– bottom panel), transitions which could not be seen in absence of noise (Figure S4). These results indicate that multistable features of this gene regulatory network can confer cellular plasticity and consequent cell-fate switching in liver, which may be of paramount importance during injury repair and reprogramming.
Figure 4.
Stochastic cell state switching and predicted cellular reprogramming strategies
(A) Noise induced stochastic switching simulations of the gene regulatory network (GRN) under different representative parameter sets. Biological states have been defined based on the expression levels of the individual genes. (B-D) Simulation results quantifying the levels of (B) hepatocytes (C) cholangiocytes and (D) progenitor-like cells under various perturbations to the gene regulatory network.
(E) Schematic showing the list of perturbations that are likely to be the most effective in enriching for a given cell type. ∗ represents a statistically significant difference in the proportion of cases resulting in a particular phenotype compared to control case (Students’ two-tailed t test; pvalue<0.05); ns denotes a non-significant difference.
After elucidating the dynamics of a core regulatory network for cellular decision-making during liver development and homeostasis, we predicted the cellular reprogramming efficiencies of different perturbations on this network and the final proportions of cell fates obtained. We performed either overexpression (OE) or down-regulation (DE) of c/EBPα, SOX9 and TGFBR2 individually and calculated the ratio of hepatocytes, cholangiocytes, progenitor-like cells and SOX9+ hepatocytes (see STAR methods for how the cell types have been defined for this analysis). Overall, we found that hepatocytes can be best enriched by either performing a c/EBPα OE or TGFBR2 DE (Figure 4B). On the other hand, cholangiocytes can be best enriched for by SOX9 OE or TGFBR2 OE (Figure 4C). For the progenitor-like state (high levels of TGFBR2; low levels of both c/EBPα and SOX9), one might expect that overexpression of TGFBR2 levels would lead to an enrichment of this state. However, we find out that TGFBR2 OE has a weaker impact in enriching progenitor cell population as compared to c/EBPα DE or SOX9 DE (Figure 4D). This observation lends support to the hypothesis that to maintain a progenitor cell state population, both differentiation programs (cholangiocyte-inducing and hepatocyte-inducing) need to be inactive (Figure 4E). This observation is likely to be more generic, given that various underlying gene regulatory networks for cell-fate decisions have similar topology as the one shown here (Duddu et al., 2020; Zhou and Huang, 2011). All above-mentioned trends remain qualitatively unchanged when TGFβ was perturbed too. Further, SOX9+ hepatocytes, as expected, were most enriched for by SOX9 OE instead of by TGFBR2 OE, although SOX9 is activated by TGFβ signaling (Figures S3A and S3B), implying that enriching SOX9+ hepatocytes may need a more specific agonist facilitating SOX9 transcription, instead of a generic activation of TGFβ signaling pathway. These simulations offer testable predictions for executing effective reprogramming strategies when liver cells of one phenotype switch to another sibling fate.
Overall, these observations endorse that concerted changes in levels of these ‘master regulators’ can orchestrate corresponding phenotypic plasticity and heterogeneity, as seen in vitro and in vivo.
Bifurcation analysis reveals spatial pattern formation and other dynamical properties associated with gene regulatory network in liver cells
After investigating different possible phenotypes enabled by the network comprising c/EBPα, SOX9 and TGFBR2, we investigated the role of TGFβ in controlling the transitions between hepatocyte and cholangiocyte cell states. For a set of kinetic parameters estimated from literature, we mapped different cell-states observed at various levels of TGFβ through a bifurcation (dose-response curve) diagram (see STAR methods). At lower TGFβ levels, we found c/EBPα can exist at two levels – high and low – mapping onto a hepatocyte and a progenitor/cholangiocyte fate respectively. Correspondingly, when c/EBPα levels were high, TGFBR2 and SOX9 levels were both low. On the other hand, when TGFBR2 and SOX9 were high, the levels of c/EBPα were lower, thus indicating bistability (Figure 5A). As levels of TGFβ increase, the levels of both TGFBR2 and SOX9 increase with a concurrent disappearance of the hepatocyte cell state (Figure 5A), which can be interpreted as the maturation of progenitor cells into a distinct cholangiocyte-like cell state and subsequent destabilization of the hepatocyte cell fate at higher levels of TGFβ. In developmental contexts, it has been observed that the levels of TGFBR2 increase along with those of SOX9 as cholangiocytes develop/mature from a hepatoblast stage (Takayama et al., 2014), thus corroborating the bifurcation diagram seen here. This trend is also consistent with a graded mode in which cholangiocytes develop, i.e., not an abrupt change to a mature state (Yang et al., 2017). Furthermore, from the bifurcation diagram, we see that the progenitor cell state has higher level of SOX9 compared to hepatocytes, but lower than cholangiocytes (Figure 5A). This observation can explain in vivo scenarios where progenitors having high levels of SOX9 can give rise to both biliary cells and hepatocytes in the context of liver development (Furuyama et al., 2011).
Figure 5.
Bifurcation analysis reveals spatial patterns and hysteresis as emergent properties of the gene regulatory network
(A) Bifurcation analysis showing the stable (solid blue lines) and unstable steady state (dashed red lines) solutions in c/EBPα, SOX9, and TGFBR2 levels, as a function of increasing TGFβ levels.
(B) Spatial patterning emergent from the dynamics of the gene regulatory network as a function of a gradient of TGFβ signaling under the influence of gene expression noise.
(C) Experimentally observed expression levels of c/EBPα and SOX9 via RNA-seq data (GSE84498) in different regions of liver (toward central vein or portal vein). ∗ represents a statistically significant difference in SOX9 and c/EBPα expression levels (Students’ two-tailed t test; pvalue<= 0.05).
(D) Effects of different initial conditions on the stochastic dynamics of GRN, showing hysteresis in the system – fraction of hepatocytes, progenitor cells and cholangiocytes. To test for hysteresis, two distinct initial conditions used for the system is either a pure hepatocyte or a progenitor-like population, and the fraction of the two cell populations are plotted. Error bars represent SD values (n = 3).
After examining the temporal dynamics of cell-fate commitment in the liver, we focused on spatial patterning of these phenotypes. Cholangiocytes are more abundantly found near the portal vein. On the contrary, hepatocytes are found much more abundantly on the parenchymal side (Clotman et al., 2005). Hepatic progenitor cells (similar to hepatoblasts/oval cells in our model) have been proposed to be sandwiched between the cholangiocytes and hepatocytes in the adult liver (Ko et al., 2020). Hepatoblasts in periportal region are likely to adopt a cholangiocyte fate and eventually form the bile ducts, whereas hepatoblasts in other regions of the lobule form hepatocytes (Raynaud et al., 2011). Such spatial segregation has been proposed to be a result of TGFβ gradient (Ayabe et al., 2018; Clotman et al., 2005) and/or by mechanical cues induced Notch signaling variations in the periportal region (Kaylan et al., 2018). We probed whether we could reproduce this spatial patterning of hepatocytes/hepatoblast-like cells/cholangiocytes by simulations capturing a simple gradient of TGFβ influence the gene regulatory network. Our simulations could reproduce the experimentally observed spatial pattern of phenotypes even without considering the local interactions between the neighboring cells in the simulation (i.e., the dynamics of each cell is independent of the neighboring cells). We demonstrated that under high levels of TGFβ, a cholangiocyte cell state are more prevalent, whereas at intermediate and low levels of TGFβ, hepatocytes and progenitor-like cells are seen to be abundant with the hepatocyte abundance increasing as the levels of TGFβ drop (Figure 5B). The analysis shows that spatial pattering is emergent from the gene regulatory network itself without necessarily invoking additional spatial interactions between neighbors. Our simulation observations are further corroborated by analysis of spatial gene expression profiles in liver tissue (GSE84498) (Halpern et al., 2017). We observed that the expression levels of c/EBPα drop toward the portal vein region with a concurrent increase in the levels of the SOX9 (Figure 5C). These observations provide insights into the spatial organization of distinct cell types seen in liver development and homeostasis as a function of spatiotemporal dynamics of underlying gene regulatory networks.
A hallmark feature of multistable regulatory networks is hysteresis. To characterize the hysteretic behavior for this network, we first considered a “pure population” of hepatocytes and monitored, as a function of TGFβ levels, the proportion of cells that switched to the progenitor/cholangiocyte state and the proportion of cells that did not switch states. Similarly, we started from a population of “pure progenitors” and tracked the relative proportion of cells in the three states (hepatocytes, cholangiocytes, and progenitor cells) as a function of TGFβ levels. For a non-hysteretic system in nature, the proportions of cells in each of these three states would have been the same from the two starting populations (only hepatocytes or only progenitor cells) for a given level of TGFβ. However, we observed distinctly different profiles for the different starting populations (Figure 5D). For instance, at TGFβ = 0, and starting from a population of only hepatocytes, the cells show minimal switching, instead maintain hepatocyte fate (Figure 5D– left panel). But if one starts from a population of only progenitor cells, at TGFβ = 0, only ∼30% switch to a hepatocyte fate and the remaining 70% retain their progenitor cell fate. Similarly, at TGFβ = 15,000 molecules, if we start from a population of only progenitor cells, almost none of them switch to a hepatocyte fate but if we start from a population of only hepatocytes ∼20% of the cells will switch to a progenitor cell fate (Figure 5D– middle panel). We also observed that the proportion of cholangiocytes obtained (when starting from a pure population of either hepatocytes or oval cells) has little effect on its starting population (Figure 5D– right panel), which is expected, given that the cholangiocyte state is stable mostly beyond the bistable region where the hysteretic pattern is less pronounced. Although hysteresis in liver cell phenotypes requires experimental validation, it has been experimentally witnessed for other multistable biological systems such as epithelial-mesenchymal plasticity and lactose metabolism (Celià-Terrassa et al., 2018; Ozbudak et al., 2004).
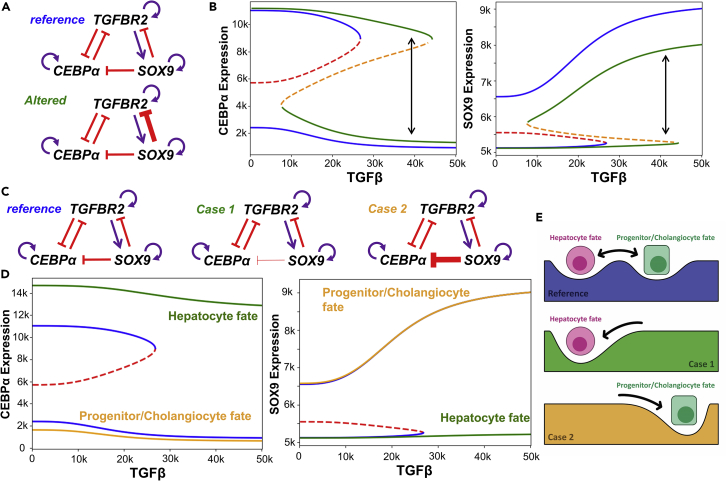
Finally, we also investigated possible trajectories of cellular reprogramming enabled by modulating the strength of individual edges in the network. We hypothesize that such alteration of strength of induvial edges can, in principle, happen via epigenetic regulations, but there is no current evidence for the same. Nonetheless, it is important to understand how such alterations might affect the dynamics of the gene regulatory network in question that can be later investigated experimentally. Given that TGFBR2 DE was shown to enrich for hepatocytes, we probed the impact of a stronger inhibition of TGFBR2 by SOX9 (Figure 6A). For the control case, the bifurcation diagram showed that at TGFβ =0, the system is bistable (Figure 6A; blue curve in Figure 6B), i.e., in the absence of TGFβ signaling, a certain fraction of progenitor-like cells will be present in the population, thus blocking a transition to all-hepatocyte population, when starting from a population of only cholangiocytes. However, on increasing the strength of suppression of TGFBR2 by SOX9, bifurcation diagram changed, now enabling a scenario of only hepatocyte cells at TGFβ =0, with modest changes in steady state expression values of c/EBPα and SOX9 (green curve in Figure 6B). Further, cholangiocytes and hepatocytes can co-exist and likely stochastically switch even at higher values of TGFβ signaling, without necessarily requiring transitioning via a hepatoblast state. Thus, a stronger inhibition of TGFBR2 by SOX9 provides a permissive situation for experimentally observed direct trans-differentiation among the two mature liver cell types (Deng et al., 2018; Schaub et al., 2018).
Figure 6.
Impact of altering edge strengths on cell reprogramming landscape
(A) Schematic showing the reference and the altered gene regulatory network in which the link strength of SOX9 inhibiting TGFBR2 has been increased.
(B) Changes in bifurcation diagram on altering the circuit as shown in A. Blue solid lines show stable steady states whereas the red dashed line shows the unstable states for the reference bifurcation diagram. Green solid lines and orange dashed lines are the stable and unstable states for the altered GRN in A. Bidirectional arrow is indicative of direct transition of hepatocytes to cholangiocytes at higher levels of TGFβ ligand.
(C) Schematic showing the reference and the two cases of the gene regulatory network in which the link strength of SOX9 inhibiting c/EBPα has been decreased (Case 1) or increased (Case 2).
(D) Reference bifurcation diagram is shown in blue solid lines and red dashed lines. Bifurcation diagram for case 1 is shown in green solid line (hepatocyte state only) while the orange solid curve refers to case 2 (progenitor/cholangiocyte state only). Note that there are no unstable states in the green and the orange profiles indicating that only one fate is enabled by the parameter set.
(E) Schematic showing the changes in the landscape brought about by changing the strength of SOX9 suppression of c/EBPα in which for case 1 and case 2 only one stable state is allowed while for the reference case 2 stable states are possible.
Next, we probed the impact of altering the link strength from SOX9 to c/EBPα. When this inhibition is weaker (case 1), the progenitor-like and cholangiocyte states are not observed irrespective of the value of TGFβ (green curve in Figure 6D). In contrast, in case of a stronger inhibition (case 2), hepatocyte state disappears (orange curve in Figure 6D). Thus, by changing the link strength from SOX9 to c/EBPα, landscape of cellular plasticity can be transformed such that only one cell state (‘attractor’) exists (Figure 6E), compromising on the ability to switch cell-fates in the liver. These observations suggest potent mechanisms for cells to lock into specific cell-fates as well as pinpoint how a delicate balance between different links in underlying regulatory network allow multistability and consequent phenotypic plasticity – a fundamental tenet of development and regeneration across organs and organisms.
Discussion
Phenotypic plasticity is a ubiquitous phenomenon via which cells can exhibit diverse phenotypic states which can interconvert among themselves. It is thought to be a ‘bet-hedging’ strategy that can be implemented by cell populations to survive through various bottlenecks, such as drug-tolerant persisters seen ranging from microbial populations to cancer cells (Sahoo et al., 2020b; van Boxtel et al., 2017). Understanding the first principles behind cell-fate decision making in developmental systems can be paramount to elucidating mechanistic underpinnings of phenotypic plasticity in biological systems in general. Furthermore, such an understanding can propel rational strategies for reprogramming of cell types in vitro (Qian et al., 2018). The hepatocyte-cholangiocyte decision-making offers an ideal system, given their common progenitor (hepatoblasts) that can differentiate into these divergent cell-fate trajectories. Furthermore, once differentiated, these cell still retain the capacity to trans-differentiate to one another during liver injury (Gadd et al., 2020).
Here, we identified and analyzed a minimalistic gene regulatory circuit that can, in principle, explain various cellular phenotypes observed in the liver and their cell-fate decision under the influence of specific signaling cues. This network comprising SOX9, TGFBR2 and c/EBPα is multistable in nature; enabling multiple co-existing phenotypes that can switch in presence of biological noise. The experimentally observed spatial patterning of these cell types in the liver can also be explained as an emergent property of this gene regulatory network without explicitly considering spatial interactions between the neighboring cells in our minimalistic model. Finally, we explore possible perturbations in the gene regulatory network that can enrich for various cell phenotypes, driving reprogramming.
The in-silico model constructed here, like all model systems, has limitations. First, the hepatocyte and cholangiocyte phenotypes are proxied by individual ‘master regulator’ transcription factors, which makes it tricky to identify various heterogeneous ‘micro-states’ such as Axin+ hepatocytes, TertHigh hepatocytes, hybrid periportal hepatocytes and Lgr5+ hepatoblasts (Li et al., 2020; Prior et al., 2019). Second, we have excluded several potentially important key regulators. For instance, we have not considered SOX4 as a distinct node in the gene regulatory network as its expression of SOX4 is largely correlated with that of SOX9 in multiple datasets we analyzed (Figure S5). Including SOX4 in subsequent models can help identify ‘micro-states’ during development and disease. Similarly, we excluded c/EBPβ, whose overexpression has been shown to promote cholangiocyte differentiation by enhancing TGFBR2 expression (Takayama et al., 2014). This observation, along with antagonistic trends in expression levels and/or regulation of c/EBPβ and c/EBPα (Jakobsen et al., 2013; Takayama et al., 2014), suggests that c/EBPβ can also be included in subsequent models to elucidate its role in development of cholangiocytes. Expanding the regulatory network to include these other nodes may alleviate this limitation; for instance, on including TGFβ, our simulations showed the emergence of SOX9+ hepatocytes. These cells are reported to behave as bipotent progenitors after liver injury (Han et al., 2019; Tanimizu et al., 2017). Third, we only investigate the transcriptional control of these cell-fates; they can be affected by other layers of regulation such as epigenetics (Aloia, 2021). Future efforts to interrogate the impact of chromatin changes on cell-fate decisions (Jia et al., 2019) will be important. Fourth, our model cannot rigorously distinguish biological differences that might exist between developmental bipotent progenitors (hepatoblasts) and adult bipotent progenitors (oval cells) or between these progenitors and cells undergoing hepatocyte-to-cholangiocyte conversion (intermediate, and possibly metastable, states). Within the scope of this study, these diverse biological entities are considered to be similar, but they may be different in in vivo conditions. For instance, excessive activation of TGFβ signaling in the liver is associated with appearance of “hepatobiliary” features; i.e. “hybrid” cells co-expressing both hepatocyte and cholangiocyte markers (Raynaud et al., 2011). Another manifestation of “hybrid” cells may be those co-expressing SOX9 with HNF4α and HNF1α (Akai et al., 2014). A systematic approach to map the regulatory networks involved in cell decision making during liver development and injury repair can characterize various cell-fates observed experimentally.
Our analysis opens up a wide range of questions for which experiments can be designed to test out novel hypothesis. Based on the bifurcation analysis presented here and earlier literature, TGFβ treatment is known to promote SOX9 levels by increasing activity of Notch/TGFβ signaling (Wang et al., 2018), but it still remains to be seen if the prolonged treatment of TGFβ antagonists can push the mature/immature cholangiocytes into a progenitor-like/hepatocyte cell fate. Furthermore, even if the program is reversible in the initial stages, is there a point of no return that prevents cholangiocytes from reverting back to hepatocytes/progenitor-like states, similar to those observed in other instances of phenotypic plasticity? (Tripathi et al., 2020) If yes, what mechanisms underlie a distinction between cell decision-making (reversible) and cell-fate ‘locking’ (irreversible). Juxtaposing stability versus plasticity (switching ability) in a multistable system can offer new insights into the operating principles invoked during canalization as well as organ injury repair. TGFβ is a well-known inducer of a mesenchymal state in liver development and regeneration (Choi and Diehl, 2009; Karkampouna et al., 2012) as well as in cancer. However, cholangiocytes are known to be epithelial, even though TGFβ appears to be crucial in their development. How the cholangiocytes maintain an “epithelial phenotype” under the influence of TGFβ signaling remains to be answered. Potentially, interplay of TGFβ with other signaling pathways such as Notch signaling can help resolve these context-specific discrepancies.
Overall, we demonstrate how a minimalistic gene regulatory network can provide insights in elucidating the dynamical properties of cell-fate decisions during liver development and regeneration. Our approach highlights common “first principles” that appear to be followed in varied biological contexts corresponding to healthy and diseased liver. Finally, our study provides a conceptual framework to guide cellular reprogramming strategies in pre-clinical and/or clinical setups by a better predictive understanding of emergent dynamics of networks driving liver cell-fate decisions.
Limitations of the study
Our minimalistic model explains how different cell types in the liver can switch among one another, but does not have the granularity to distinguish among certain biological micro-states or phenotypes seen in liver during development, or injury repair. Including other relevant master regulators to the network considered here would be critical in explaining such phenotypes. Also, other layers of biological regulation such as epigenetic, metabolic, and translational are not yet integrated into our framework to describe their role in liver cell plasticity. Finally, some of our model predictions and hypotheses would require further experimental validation in a context-specific manner.
STAR★Methods
Key resources table
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Mohit Kumar Jolly (mkjolly@iisc.ac.in).
Materials availability
This study did not generate new unique reagents.
Method details
RACIPE simulations
Random Circuit Perturbation (RACIPE) is a computational framework that allows an extensive exploration of the dynamical properties of a gene regulatory network (Huang et al., 2017). Only the network topology is provided as an input to simulation framework, which is then modeled as a set of x ODEs (where x is the number of nodes in the gene regulatory network). The change in concentration of each node in the network depends on production rate of the node, the effect of regulatory links incident on the node (modeled as a shifted Hill’s function (Lu et al., 2013)) and the degradation rate of the node. Each parameter in the set of unknown parameters for ODEs is randomly sampled from a biologically relevant range. The sampling of parameters is done so as to ensure that it generates a representative ensemble of models for a specific circuit topology. The range for production rate varies from 1 to 100 whereas the range for the degradation rates varies from 0.1 to 1 (arbitrary units). The fold change parameter associated with each link is assumed to be in the range of 1- to 100-fold. The Hills coefficients sampled are assumed to vary from 1 to 6. Furthermore, the threshold parameter is carefully chosen based on the gene network topology features such as inward regulations to ensure that each link in the network has a roughly equal chance of being functional or not functional. This ensures that steady state solutions obtained sample most if not all of the possible states allowed by the gene regulatory network without biasing the system towards a particular set of solutions.
After such sampling, the set of parameterized ODEs is solved to get different possible steady state solutions. The set of ODEs can be multistable, i.e., multiple sets of steady state concentrations satisfy the set of ODEs. The program samples 10000 different sets of parameters. For each parameter set, RACIPE chooses a random set of initial conditions (n = 100) for each node in the network and solves, using Euler’s method, with the set of coupled ODEs that represent interactions among the different nodes in a network. For each given parameter set and initial conditions, RACIPE reports the steady-state values for each of the nodes in the network. The steady state values were then Z-normalized where the z-normalized expression value (zi) is given by the term:
where Emean and Estd is the mean and standard deviation of the expression levels of a given node across all its steady state solutions.
The above procedure was followed to generate RACIPE results for the network topology given in Figure 1. The Z-normalized steady state values were then plotted as a clustermap as in Figure 1B. PCA was performed with the major clusters being labeled via hierarchical clustering setting n = 3 as in Figure 1C. Kernel density maps overlaid on top of histograms for steady state expression for each node was plotted as in Figure 1H. Scatterplots for steady state solutions for each pair of nodes was plotted as presented in Figure S1B. The variant circuit as shown in Figure S1C was also simulated and processed using the methods described above to generate Figures S1D and S1E.
The perturbation analysis in Figures 4B–4D and Figure S3A were done by performing RACIPE analysis on the system by either over expressing (OE) or down expressing (DE) the given node by 10-fold. The Z score normalization of these perturbation data was done with respect to the control case. Similarly, perturbation analysis was done on variant circuit shown in Figure S1C (shown in Figure S3B).
Bifurcation analysis
We simulated a system of coupled ODEs using PyDSTool (Clewley, 2012) to create the bifurcations diagrams in the manuscript. The following set of ODEs were simulated:
whereGI is the the production rate of the gene i, is the degradation rate of the gene i, Hs is the shifted hill’s function that is given by the term:
Each edge in the network has an associated shifted hill’s function which captures the effect of pair of nodes, i and j (j affecting i). is the fold change from the basal synthesis rate of i because of j. Therefore, λ> 1 for activatory links and λ< 1 for inhibitory links.is the threshold value for the interaction and is the hills coefficient. The initial conditions for c/EBPα, SOX9 and TGFBR2 were set to be 12000, 120 and 120 respectively. TGFβ levels was varied from 0 to 50000 units to create the bifurcation diagram. The parameters for the reference bifurcation diagram have been listed in Table S1. Simulation of this system resulted in Figure 5A. For creating the bifurcations in Figure 6B, the parameter was changed from 0.5 (reference circuit) to 0.35 (altered circuit). Similarly, for the bifurcations in Figure 6D, the parameter was varied from 0.5 to 0.65 (Case 1) and to 0.35 (Case 2).
Stochastic simulations (including noise) – Hysteresis and spatial dynamics
For the stochastic simulations in Figure 4A we used the webserver facility of Gene Circuit Explorer (GeneEx) to simulate stochastic dynamics of gene regulatory circuit as shown in Figure 1A—https://geneex.jax.org/. To account for stochastic gene expression effects (because of cell-to-cell variation and low copy numbers in individual cells, etc), the webserver of GeneEx includes a noise term based on a Wiener process (Wt) with a fixed variance for all genes. We have now reported these values along with the parameter sets and the initial conditions in Table S2.
The noise parameters for Table S2 were chosen heuristically to show the various scenarios that can exist as a result of introduction of the noise. Noise values were chosen so that it is high enough that clear switching between states can be seen while not too high for the clearly distinguishable cell states to be lost because of widely ranging fluctuations in the gene expression levels. So far, we have not used a numerical relationship to identify the exact ratio between the steady state value and the noise, but operate within an appropriate range of values to observe the reported behavior.
For simulating the hysteresis in the system we simulated the above-described set of ODEs and with the parameter sets listed in Table S1 in the presence of noise at different values of TGFβ (0–40000 in steps of 2500) which was incorporated as follows:
-
1.
We used Wiener process to simulate the noise because it is a continuous Gaussian white noise.
-
2.Wiener process is a continuous noise with the properties that
-
1.W(t = 0) = 0
-
2.The increments in W are Gaussian and independent
-
3.For our model we have used a diagonal noise
-
1.
Here represents the deterministic equation and is the coefficient of the noise, and represents the Wiener process. For our case is a diagonal matrix, so each of the differential equations for c/EBPα, TGFBR2, and SOX9 has their individual noise terms. For the simulation we used the noise values () of 1000, 1000, 10 respectively. This assumes that the noise level should be proportional to the difference in the absolute values between the steady states for a given gene. For example, for the parameter set we have considered SOX9 has a smaller absolute difference between the steady state values compared to TGFBR2 and CEBPα, hence the variance in noise is also lower compared to the other two scenarios. By imposing the said assumption, we make sure that the noise levels are not too high to cause an absolute destabilization of any given steady state.
The system was started from either a hepatocyte state (11000, 2500, 5100) or a progenitor-like state (2100, 12500, 6500) where the values are in the order of c/EBPα, TGFBR2, and SOX9 respectively. A cell was deemed to be hepatocyte if the c/EBPα level was above 6200 while it was deemed a progenitor-like cell if the level of TGFBR2 was between 10,000 and 17,500 and values above that were labeled to be cholangiocytes. This analysis was carried out for 3 sets of replicates each with 500 instances to estimate the mean and standard deviation for the fraction of cases for a given cell type.
For spatial simulations, we started the system from an all-hepatocyte state and simulated 15 cells at each level of TGFβ (0 to 40000 in steps of 2500). Same classification of biological phenotypes as above was used to assign cell types on the spatial pattern.
Data analysis
All bulk microarray and RNA-Seq pre-processed datasets were obtained from publicly available GEO datasets. Gene signatures for adult hepatocyte program, adult biliary program and adult hepatobiliary program were obtained from (Segal et al., 2019). The gene sets specifying c/EBPα regulon and SOX9 regulon were obtained from (Møller and Natarajan, 2020). Hallmark TGFβ signaling pathway was obtained from MSigDB (Liberzon et al., 2011). Gene set activity were estimated via ssGSEA for all bulk samples and via AUCell for all single cell datasets (Aibar et al., 2017; Subramanian et al., 2005). For trajectory analysis the coordinates of each cell was obtained by performing a 3D PCA on the gene sets provided in (Segal et al., 2019).
We have used Shannon entropy as a proxy for the transcriptional diversity of the single cells in the dataset GSE90047. We calculate the Shannon entropy for the distribution of all genes’ expression (in TPM) as a single quantity for a given cell.
For TGFβ signaling, we included multiple metrics: expression levels of TGFB1, expression levels of TGFBR2, TGFβ signaling activity ssGSEA scores. Representative cases for individual datasets are shown in corresponding main figures; all pairwise comparisons are given in Table S3. Most entries across datasets show the expected trend of TGFβ signaling levels at a reduced level in the hepatocytes (fold-change levels <1 when compared to cholangiocytes and/or oval cells).
Quantification and statistical analysis
We computed the Spearman correlation coefficients and used corresponding pvalues to gauge the strength of correlations. For statistical comparison between groups, we used a two-tailed Student’s t-test under the assumption of unequal variances and computed significance. Details of statistical analysis, definitions for significance and abbreviations can be found in figure legends.
Acknowledgments
MKJ was supported by Ramanujan Fellowship awarded by Science and Engineering Research Board (SERB), Department of Science and Technology (DST),Government of India (SB/S2/RJN-049/2018), and by Infosys Foundation, Bangalore. SS and AM are supported in part by KVPY fellowship (DST). Atchuta Srinivas Duddu is acknowledged for the design of the graphical abstract.
Author contributions
M.K.J. conceived and supervised research; S.S. and A.M. performed research; S.S. and A.M.D. analyzed data; S.S. prepared the first draft of the manuscript; all authors edited and wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: September 16, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104955.
Supplemental information
Data and code availability
-
•
This paper analyses existing, publicly available data. The accession numbers for the datasets are listed in the key resources table.
-
•
All codes used in the manuscript are available at: https://github.com/sarthak-sahoo-0710/2021_liver_cell_plasticity.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Aibar S., González-Blas C.B., Moerman T., Huynh-Thu V.A., Imrichova H., Hulselmans G., Rambow F., Marine J.C., Geurts P., Aerts J., et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akai Y., Oitate T., Koike T., Shiojiri N. Impaired hepatocyte maturation, abnormal expression of biliary transcription factors and liver fibrosis in C/EBPα (Cebpa)-knockout mice. Histol. Histopathol. 2014;29:107–125. doi: 10.14670/HH-29.107. [DOI] [PubMed] [Google Scholar]
- Aloia L. Epigenetic regulation of cell-fate changes that determine adult liver regeneration after injury. Front. Cell Dev. Biol. 2021;9:643055. doi: 10.3389/fcell.2021.643055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniou A., Raynaud P., Cordi S., Zong Y., Tronche F., Stanger B.Z., Jacquemin P., Pierreux C.E., Clotman F., Lemaigre F.P. Intrahepatic bile ducts develop according to a newmode of tubulogenesis regulated by the transcription factor SOX9. Gastroenterology. 2009;136:2325–2333. doi: 10.1053/j.gastro.2009.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayabe H., Anada T., Kamoya T., Sato T., Kimura M., Yoshizawa E., Kikuchi S., Ueno Y., Sekine K., Camp J.G., et al. Optimal hypoxia regulates human iPSC-derived liver bud differentiation through intercellular TGFB signaling. Stem Cell Rep. 2018;11:306–316. doi: 10.1016/j.stemcr.2018.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balázsi G., Van Oudenaarden A., Collins J.J. Cellular decision making and biological noise: from microbes to mammals. Cell. 2011;144:910–925. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celià-Terrassa T., Bastian C., Liu D.D., Ell B., Aiello N.M., Wei Y., Zamalloa J., Blanco A.M., Hang X., Kunisky D., et al. Hysteresis control of epithelial-mesenchymal transition dynamics conveys a distinct program with enhanced metastatic ability. Nat. Commun. 2018;9:5005. doi: 10.1038/s41467-018-07538-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceulemans A., Verhulst S., Van Haele M., Govaere O., Ventura J.J., Van Grunsven L.A., Roskams T. RNA-sequencing-based comparative analysis of human hepatic progenitor cells and their niche from alcoholic steatohepatitis livers. Cell Death Dis. 2017;8:e3164. doi: 10.1038/cddis.2017.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S.S., Diehl A.M. Epithelial-to-mesenchymal transitions in the liver. Hepatology. 2009;50:2007–2013. doi: 10.1002/hep.23196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewley R. Hybrid models and biological model reduction with PyDSTool. PLoS Comput. Biol. 2012;8:e1002628. doi: 10.1371/journal.pcbi.1002628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clotman F., Jacquemin P., Plumb-Rudewiez N., Pierreux C.E., Van Der Smissen P., Dietz H.C., Courtoy P.J., Rousseau G.G., Lemaigre F.P. Control of liver cell fate decision by a gradient of TGFβ signaling modulated by Onecut transcription factors. Genes Dev. 2005;19:1849–1854. doi: 10.1101/gad.340305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzolino A.M., Alonzi T., Santangelo L., Mancone C., Conti B., Steindler C., Musone M., Cicchini C., Tripodi M., Marchetti A. TGFβ overrides HNF4α tumor suppressing activity through GSK3β inactivation: implication for hepatocellular carcinoma gene therapy. J. Hepatol. 2013;58:65–72. doi: 10.1016/j.jhep.2012.08.023. [DOI] [PubMed] [Google Scholar]
- Cullen J.M., Falls J.G., Brown H.R., Yoon L.W., Cariello N.F., Faiola B., Kimbrough C.L., Jordan H.L., Miller R.T. Time course gene expression using laser capture microscopy-extracted bile ducts, but not hepatic parenchyma, reveals acute alpha-naphthylisothiocyanate toxicity. Toxicol. Pathol. 2010;38:715–729. doi: 10.1177/0192623310373774. [DOI] [PubMed] [Google Scholar]
- Demarez C., Gérard C., Cordi S., Poncy A., Achouri Y., Dauguet N., Rosa D.A., Gunning P.T., Manfroid I., Lemaigre F.P. MicroRNA-337-3p controls hepatobiliary gene expression and transcriptional dynamics during hepatic cell differentiation. Hepatology. 2018;67:313–327. doi: 10.1002/hep.29475. [DOI] [PubMed] [Google Scholar]
- Deng X., Zhang X., Li W., Feng R.X., Li L., Yi G.R., Zhang X.N., Yin C., Yu H.Y., Zhang J.P., et al. Chronic liver injury induces conversion of biliary epithelial cells into hepatocytes. Cell Stem Cell. 2018;23:114–122.e3. doi: 10.1016/j.stem.2018.05.022. [DOI] [PubMed] [Google Scholar]
- Dianat N., Dubois-Pot-Schneider H., Steichen C., Desterke C., Leclerc P., Raveux A., Combettes L., Weber A., Corlu A., Dubart-Kupperschmitt A. Generation of functional cholangiocyte-like cells from human pluripotent stem cells and HepaRG cells. Hepatology. 2014;60:700–714. doi: 10.1002/hep.27165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X., Pan Z., Li Q., Liu H., Li Q. SMAD4 feedback regulates the canonical TGF-β signaling pathway to control granulosa cell apoptosis. Cell Death Dis. 2018;9:151. doi: 10.1038/s41419-017-0205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan D., Derynck R. Transforming growth factor-β (TGF-β)-induced up-regulation of TGF-β receptors at the cell surface amplifies the TGF-β response. J. Biol. Chem. 2019;294:8490–8504. doi: 10.1074/jbc.RA118.005763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duddu A.S., Sahoo S., Hati S., Jhunjhunwala S., Jolly M.K. Multi-stability in cellular differentiation enabled by a network of three mutually repressing master regulators. J. R. Soc. Interface. 2020;17:20200631. doi: 10.1098/rsif.2020.0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman A.D. C/EBPα in normal and malignant myelopoiesis. Int. J. Hematol. 2015;101:330–341. doi: 10.1007/s12185-015-1764-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu G.B., Huang W.J., Zeng M., Zhou X., Wu H.P., Liu C.C., Wu H., Weng J., Zhang H.D., Cai Y.C., et al. Expansion and differentiation of human hepatocyte-derived liver progenitor-like cells and their use for the study of hepatotropic pathogens. Cell Res. 2019;29:8–22. doi: 10.1038/s41422-018-0103-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama K., Kawaguchi Y., Akiyama H., Horiguchi M., Kodama S., Kuhara T., Hosokawa S., Elbahrawy A., Soeda T., Koizumi M., et al. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat. Genet. 2011;43:34–41. doi: 10.1038/ng.722. [DOI] [PubMed] [Google Scholar]
- Gadd V.L., Aleksieva N., Forbes S.J. Epithelial plasticity during liver injury and regeneration. Cell Stem Cell. 2020;27:557–573. doi: 10.1016/j.stem.2020.08.016. [DOI] [PubMed] [Google Scholar]
- Gérard C., Tys J., Lemaigre F.P. Gene regulatory networks in differentiation and direct reprogramming of hepatic cells. Semin. Cell Dev. Biol. 2017;66:43–50. doi: 10.1016/j.semcdb.2016.12.003. [DOI] [PubMed] [Google Scholar]
- Gordillo M., Evans T., Gouon-Evans V. Orchestrating liver development. Development. 2015;142:2094–2108. doi: 10.1242/dev.114215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati G.S., Sikandar S.S., Wesche D.J., Manjunath A., Bharadwaj A., Berger M.J., Ilagan F., Kuo A.H., Hsieh R.W., Cai S., et al. Single-cell transcriptional diversity is a hallmark of developmental potential. Science. 2020;367:405–411. doi: 10.1126/science.aax0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern K.B., Shenhav R., Matcovitch-Natan O., Tóth B., Lemze D., Golan M., Massasa E.E., Baydatch S., Landen S., Moor A.E., et al. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature. 2017;542:352–356. doi: 10.1038/nature21065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Wang Y., Pu W., Huang X., Qiu L., Li Y., Yu W., Zhao H., Liu X., He L., et al. Lineage tracing reveals the bipotency of SOX9 + hepatocytes during liver regeneration. Stem Cell Reports. 2019;12:624–638. doi: 10.1016/j.stemcr.2019.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Lu M., Jia D., Ben-Jacob E., Levine H., Onuchic J.N. Interrogating the topological robustness of generegulatory circuits. PLoS Comput. Biol. 2017;13:e1005456. doi: 10.1101/084962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen J.S., Waage J., Rapin N., Bisgaard H.C., Larsen F.S., Porse B.T. Temporal mapping of CEBPA and CEBPB binding during liver regeneration reveals dynamic occupancy and specific regulatory codes for homeostatic and cell cycle gene batteries. Genome Res. 2013;23:592–603. doi: 10.1101/gr.146399.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia W., Deshmukh A., Mani S.A., Jolly M.K., Levine H. A possible role for epigenetic feedback regulation in the dynamics of the Epithelial-Mesenchymal Transition (EMT) Phys. Biol. 2019;16:066004. doi: 10.1101/651620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkampouna S., ten Dijke P., Dooley S., Kruithof-de Julio M. TGFβ signaling in liver regeneration. Curr. Pharm. Des. 2012;18:4103–4113. doi: 10.2174/138161212802430521. [DOI] [PubMed] [Google Scholar]
- Kaylan K.B., Berg I.C., Biehl M.J., Brougham-Cook A., Jain I., Jamil S.M., Sargeant L.H., Cornell N.J., Raetzman L.T., Underhill G.H. Spatial patterning of liver progenitor cell differentiation mediated by cellular contractility and Notch signaling. Elife. 2018;7:38536. doi: 10.7554/eLife.38536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko S., Russell J.O., Molina L.M., Monga S.P. Liver progenitors and adult cell plasticity in hepatic injury and repair: knowns and unknowns. Annu. Rev. Pathol. Mech. Dis. 2020;15:23–50. doi: 10.1146/annurev-pathmechdis-012419-032824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohar V., Lu M. Role of noise and parametric variation in the dynamics of gene regulatory circuits. NPJ Syst. Biol. Appl. 2018;4:40. doi: 10.1038/s41540-018-0076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp J.L., Grompe M., Sander M. Stem cells versus plasticity in liver and pancreas regeneration. Nat. Cell Biol. 2016;18:238–245. doi: 10.1038/ncb3309. [DOI] [PubMed] [Google Scholar]
- Lau H.H., Ng N.H.J., Loo L.S.W., Jasmen J.B., Teo A.K.K. The molecular functions of hepatocyte nuclear factors – in and beyond the liver. J. Hepatol. 2018;68:1033–1048. doi: 10.1016/j.jhep.2017.11.026. [DOI] [PubMed] [Google Scholar]
- Li J., Ning G., Duncan S.A. Mammalian hepatocyte differentiation requires the transcription factor HNF-4α. Genes Dev. 2000;14:464–474. doi: 10.1101/gad.14.4.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Li L., Hui L. Cell plasticity in liver regeneration. Trends Cell Biol. 2020;30:329–338. doi: 10.1016/j.tcb.2020.01.007. [DOI] [PubMed] [Google Scholar]
- Liberzon A., Subramanian A., Pinchback R., Thorvaldsdóttir H., Tamayo P., Mesirov J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Jolly M.K., Levine H., Onuchic J.N., Ben-Jacob E. MicroRNA-based regulation of epithelial-hybrid-mesenchymal fate determination. Proc. Natl. Acad. Sci. USA. 2013;110:18144–18149. doi: 10.1073/pnas.1318192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas S.D., López-Alcorocho J.M., Bartolomé J., Carreño V. Nitric oxide and TGF-β1 inhibit HNF-4α function in HEPG2 cells. Biochem. Biophys. Res. Commun. 2004;321:688–694. doi: 10.1016/j.bbrc.2004.07.025. [DOI] [PubMed] [Google Scholar]
- Manco R., Clerbaux L.A., Verhulst S., Bou Nader M., Sempoux C., Ambroise J., Bearzatto B., Gala J.L., Horsmans Y., van Grunsven L., et al. Reactive cholangiocytes differentiate into proliferative hepatocytes with efficient DNA repair in mice with chronic liver injury. J. Hepatol. 2019;70:1180–1191. doi: 10.1016/j.jhep.2019.02.003. [DOI] [PubMed] [Google Scholar]
- Møller A.F., Natarajan K.N. Predicting gene regulatory networks from cell atlases. Life Sci. Alliance. 2020;3:e202000658. doi: 10.26508/LSA.202000658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto M.A., Huang R.Y.Y.J., Jackson R.A.A., Thiery J.P.P. EMT: 2016. Cell. 2016 doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- O’Neill K.E., Thowfeequ S., Li W.C., Eberhard D., Dutton J.R., Tosh D., Slack J.M.W. Hepatocyte-ductal transdifferentiation is mediated by reciprocal repression of SOX9 and C/EBPα. Cell. Reprogram. 2014;16:314–323. doi: 10.1089/cell.2014.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe M., Tsukahara Y., Tanaka M., Suzuki K., Saito S., Kamiya Y., Tsujimura T., Nakamura K., Miyajima A. Potential hepatic stem cells reside in EpCAM+ cells of normal and injured mouse liver. Development. 2009;136:1951–1960. doi: 10.1242/dev.031369. [DOI] [PubMed] [Google Scholar]
- Ozbudak E.M., Thattai M., Lim H.N., Shraiman B.I., van Oudenaarden A. Multistability in the lactose utilization network of Escherichia coli. Nature. 2004;427:737–740. doi: 10.1038/nature02298. [DOI] [PubMed] [Google Scholar]
- Poncy A., Antoniou A., Cordi S., Pierreux C.E., Jacquemin P., Lemaigre F.P. Transcription factors SOX4 and SOX9 cooperatively control development of bile ducts. Dev. Biol. 2015;404:136–148. doi: 10.1016/j.ydbio.2015.05.012. [DOI] [PubMed] [Google Scholar]
- Prior N., Hindley C.J., Rost F., Meléndez E., Lau W.W.Y., Göttgens B., Rulands S., Simons B.D., Huch M. Lgr5+ stem/progenitor cells reside at the apex of a heterogeneous embryonic hepatoblast pool. Development. 2019;146:dev174557. doi: 10.1242/dev.174557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y., McBride C., Del Vecchio D. Programming cells to work for us. Annu. Rev. Control. Robot. Auton. Syst. 2018;1:411–440. doi: 10.1146/annurev-control-060117-105052. [DOI] [Google Scholar]
- Raven A., Lu W.Y., Man T.Y., Ferreira-Gonzalez S., O’Duibhir E., Dwyer B.J., Thomson J.P., Meehan R.R., Bogorad R., Koteliansky V., et al. Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration. Nature. 2017;547:350–354. doi: 10.1038/nature23015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud P., Carpentier R., Antoniou A., Lemaigre F.P. Biliary differentiation and bile duct morphogenesis in development and disease. Int. J. Biochem. Cell Biol. 2011;43:245–256. doi: 10.1016/j.biocel.2009.07.020. [DOI] [PubMed] [Google Scholar]
- Sahoo S., Singh D., Chakraborty P., Jolly M.K. Emergent properties of the HNF4α-PPARγ network may drive consequent phenotypic plasticity in NAFLD. J. Clin. Med. 2020;9:870. doi: 10.3390/jcm9030870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo S., Subbalakshmi A.R., Jolly M.K. Phenotypic Switching. Elsevier; 2020. The fundamentals of phenotypic plasticity; pp. 1–21. [DOI] [Google Scholar]
- Schaub J.R., Huppert K.A., Kurial S.N.T., Hsu B.Y., Cast A.E., Donnelly B., Karns R.A., Chen F., Rezvani M., Luu H.Y., et al. De novo formation of the biliary system by TGFβ-mediated hepatocyte transdifferentiation. Nature. 2018;557:247–251. doi: 10.1038/s41586-018-0075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal J.M., Kent D., Wesche D.J., Ng S.S., Serra M., Oulès B., Kar G., Emerton G., Blackford S.J.I., Darmanis S., et al. Single cell analysis of human foetal liver captures the transcriptional profile of hepatobiliary hybrid progenitors. Nat. Commun. 2019;10:3350. doi: 10.1038/s41467-019-11266-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seow J.J.W., Pai R., Mishra A., Shepherdson E., Lim T.K.H., Goh B.K.P., Chan J.K.Y., Chow P.K.H., Ginhoux F., DasGupta R., Sharma A. Single-cell RNA-seq reveals angiotensin-converting enzyme 2 and transmembrane serine protease 2 expression in TROP2+ liver progenitor cells: implications in coronavirus disease 2019-associated liver dysfunction. Front. Med. 2021;8:603374. doi: 10.3389/fmed.2021.603374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S., Walton G., Aoki R., Brondell K., Schug J., Fox A., Smirnova O., Dorrell C., Erker L., Chu A.S., et al. Foxl1-Cre-marked adult hepatic progenitors have clonogenic and bilineage differentiation potential. Genes Dev. 2011;25:1185–1192. doi: 10.1101/gad.2027811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K., Kawabata K., Nagamoto Y., Inamura M., Ohashi K., Okuno H., Yamaguchi T., Tashiro K., Sakurai F., Hayakawa T., et al. CCAAT/enhancer binding protein-mediated regulation of TGFβ receptor 2 expression determines the hepatoblast fate decision. Development. 2014;141:91–100. doi: 10.1242/dev.103168. [DOI] [PubMed] [Google Scholar]
- Tanimizu N., Ichinohe N., Yamamoto M., Akiyama H., Nishikawa Y., Mitaka T. Progressive induction of hepatocyte progenitor cells in chronically injured liver. Sci. Rep. 2017;7:39990. doi: 10.1038/srep39990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimizu N., Nishikawa Y., Ichinohe N., Akiyama H., Mitaka T. Sry HMG box protein 9-positive (Sox9+) epithelial cell adhesion molecule-negative (EpCAM-) biphenotypic cells derived from hepatocytes are involved in mouse liver regeneration. J. Biol. Chem. 2014;289:7589–7598. doi: 10.1074/jbc.M113.517243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarlow B.D., Pelz C., Naugler W.E., Wakefield L., Wilson E.M., Finegold M.J., Grompe M. Bipotential adult liver progenitors are derived from chronically injured mature hepatocytes. Cell Stem Cell. 2014;15:605–618. doi: 10.1016/j.stem.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff A.E., Enver T. Single-cell entropy for accurate estimation of differentiation potency from a cell’s transcriptome. Nat. Commun. 2017;8:15599. doi: 10.1038/ncomms15599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi S., Levine H., Jolly M.K. Thephysics of cellular decision making during epithelial-mesenchymal transition. Annu. Rev. Biophys. 2020;49:1–18. doi: 10.1146/annurev-biophys-121219-081557. [DOI] [PubMed] [Google Scholar]
- Tsuchiya A., Lu W.Y. Liver stem cells: plasticity of the liver epithelium. World J. Gastroenterol. 2019;25:1037–1049. doi: 10.3748/wjg.v25.i9.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Boxtel C., van Heerden J.H., Nordholt N., Schmidt P., Bruggeman F.J. Taking chances and making mistakes : non-genetic phenotypic heterogeneity and its consequences for surviving in dynamic environments. J. R. Soc. Interface. 2017;14:20170141. doi: 10.1098/rsif.2017.0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Feng Y., Aimaiti Y., Jin X., Mao X., Li D. TGFβ signaling controls intrahepatic bile duct development may through regulating the Jagged1-Notch-Sox9 signaling axis. J. Cell. Physiol. 2018;233:5780–5791. doi: 10.1002/jcp.26304. [DOI] [PubMed] [Google Scholar]
- Yamasaki H., Sada A., Iwata T., Niwa T., Tomizawa M., Xanthopoulos K.G., Koike T., Shiojiri N. Suppression of C/EBPα expression in periportal hepatoblasts may stimulate biliary cell differentiation through increased Hnf6 and Hnf1b expression. Development. 2006;133:4233–4243. doi: 10.1242/dev.02591. [DOI] [PubMed] [Google Scholar]
- Yang L., Wang W.H., Qiu W.L., Guo Z., Bi E., Xu C.R. A single-cell transcriptomic analysis reveals precise pathways and regulatory mechanisms underlying hepatoblast differentiation. Hepatology. 2017;66:1387–1401. doi: 10.1002/hep.29353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanger K., Zong Y., Maggs L.R., Shapira S.N., Maddipati R., Aiello N.M., Thung S.N., Wells R.G., Greenbaum L.E., Stanger B.Z. Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes Dev. 2013;27:719–724. doi: 10.1101/gad.207803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.X., Huang S. Understanding gene circuits at cell-fate branch points for rational cell reprogramming. Trends Genet. 2011;27:55–62. doi: 10.1016/j.tig.2010.11.002. [DOI] [PubMed] [Google Scholar]
- Zong Y., Stanger B.Z. Molecular mechanisms of bile duct development. Int. J. Biochem. Cell Biol. 2011;43:257–264. doi: 10.1016/j.biocel.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
This paper analyses existing, publicly available data. The accession numbers for the datasets are listed in the key resources table.
-
•
All codes used in the manuscript are available at: https://github.com/sarthak-sahoo-0710/2021_liver_cell_plasticity.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.