Background:
Atherosclerotic cardiovascular disease is the main cause of mortality worldwide and is strongly influenced by circulating low-density lipoprotein (LDL) cholesterol levels. Only a few genes causally related to plasma LDL cholesterol levels have been identified so far, and only 1 gene, ANGPTL3, has been causally related to combined hypocholesterolemia. Here, our aim was to elucidate the genetic origin of an unexplained combined hypocholesterolemia inherited in 4 generations of a French family.
Methods:
Using next-generation sequencing, we identified a novel dominant rare variant in the LIPC gene, encoding for hepatic lipase, which cosegregates with the phenotype. We characterized the impact of this LIPC-E97G variant on circulating lipid and lipoprotein levels in family members using nuclear magnetic resonance–based lipoprotein profiling and lipidomics. To uncover the mechanisms underlying the combined hypocholesterolemia, we used protein homology modeling, measured triglyceride lipase and phospholipase activities in cell culture, and studied the phenotype of APOE*3.Leiden.CETP mice after LIPC-E97G overexpression.
Results:
Family members carrying the LIPC-E97G variant had very low circulating levels of LDL cholesterol and high-density lipoprotein cholesterol, LDL particle numbers, and phospholipids. The lysophospholipids/phospholipids ratio was increased in plasma of LIPC-E97G carriers, suggestive of an increased lipolytic activity on phospholipids. In vitro and in vivo studies confirmed that the LIPC-E97G variant specifically increases the phospholipase activity of hepatic lipase through modification of an evolutionarily conserved motif that determines substrate access to the hepatic lipase catalytic site. Mice overexpressing human LIPC-E97G recapitulated the combined hypocholesterolemic phenotype of the family and demonstrated that the increased phospholipase activity promotes catabolism of triglyceride-rich lipoproteins by different extrahepatic tissues but not the liver.
Conclusions:
We identified and characterized a novel rare variant in the LIPC gene in a family who presents with dominant familial combined hypocholesterolemia. This gain-of-function variant makes LIPC the second identified gene, after ANGPTL3, causally involved in familial combined hypocholesterolemia. Our mechanistic data highlight the critical role of hepatic lipase phospholipase activity in LDL cholesterol homeostasis and suggest a new LDL clearance mechanism.
Keywords: cholesterol, HDL; cholesterol, LDL; LIPC protein, human; Lipc protein, mouse; phospholipases
Clinical Perspective.
What Is New?
The first gain-of-function variant in the LIPC gene (LIPC-E97G) encoding hepatic lipase in a family with combined hypocholesterolemia (low low-density lipoprotein and low high-density lipoprotein cholesterol) was identified.
This unique LIPC-E97G variant specifically increases the phospholipase activity of hepatic lipase without affecting triglyceride lipase activity.
The hypocholesterolemic phenotype related to LIPC-E97G variant is attributable to an increased clearance of cholesterol within triglyceride-rich lipoprotein remnants predominantly by extrahepatic tissues.
What Are the Clinical Implications?
This novel gain-of-function variant in LIPC potentially represents the second cause of familial combined hypocholesterolemia, after loss-of-function variants in ANGPTL3.
This study highlights an unexpected and critical role of the phospholipase activity of hepatic lipase (encoded by LIPC) in low-density lipoprotein cholesterol metabolism and identifies it as a potential novel drug target.
Additional data are warranted to clarify the impact of LIPC-E97G–related combined hypocholesterolemia on atherosclerosis and atherosclerotic cardiovascular disease‚ due to the occurrence of documented coronary stenosis and evolutive carotid atherosclerosis in the index case.
Editorial, see p 740
The development of atherosclerotic cardiovascular disease (ASCVD), the main cause of mortality worldwide, is strongly influenced by circulating lipoprotein levels.1,2 Elevated low-density lipoprotein (LDL) cholesterol (LDL-C) is a well-established causal risk factor for ASCVD, whereas the role of high-density lipoprotein cholesterol (HDL-C) in ASCVD development remains to be further characterized.1,2 Plasma LDL-C concentrations are strongly affected by genetics, with heritability explaining ≈40% to 60% of plasma levels.3 Genetically modulated LDL-C concentrations typically have a polygenic origin but can also be caused by a codominant disease attributable to biallelic or monoallelic pathogenic variants.4,5 So far, however, only a few genes with a strong causal relationship to plasma LDL-C levels have been identified.
Even fewer monogenic causes of low LDL-C, familial hypobetalipoproteinemia (FHBL), have been identified. These monogenic disorders were recently classified into 2 classes based on molecular mechanisms: FHBL attributable to lipoprotein secretion defects (FHBL-SD1, -SD2, and -SD3 for MTTP, APOB, and SAR1B-related FHBL) or to enhanced catabolism of apolipoprotein B (apoB)–containing lipoprotein (FHBL-EC1 and -EC2 for ANGPTL3- and PCSK9-related FHBL).6 Furthermore, pathogenic variants in the ANGPTL3 gene have been identified in cases of familial combined hypolipidemia, a condition characterized by very low concentrations of circulating very-low-density lipoprotein (VLDL), LDL, and HDL.7 Despite the suspected polygenic origins of unexplained cases of extremely elevated or reduced LDL-C concentrations, polygenic risk scores have so far yielded lower effect sizes compared with the effect of monogenic variants.8,9 This suggests that additional monogenic variants that cause very low plasma LDL-C concentrations or combined hypolipidemia remain to be characterized.
Knowledge of the molecular mechanisms and genes implicated in extreme lipoprotein phenotypes is of direct clinical relevance. Accordingly, many of the recent therapeutics developed to lower plasma LDL-C levels and that provided breakthroughs in ASCVD risk reduction involved the products of genes implicated in FHBL and familial combined hypolipidemia such as PCSK9 and ANGPTL3.1 Despite the effectiveness of these new therapies in lowering LDL-C, a need to identify additional mechanisms to effectively lower plasma LDL-C remains.
To identify new causative genes in LDL-C metabolism, we have built up large cohorts of patients with extreme LDL-C phenotypes as part of the CHOPIN (Cholesterol Personalized Innovation) French national research program. In the present study, we identified and characterized a novel rare variant in the LIPC gene, encoding for hepatic lipase (HL), in a family who presents a dominant familial combined hypocholesterolemia. This first gain-of-function variant identifies LIPC as the second gene, after ANGPTL3, causally involved in familial combined hypocholesterolemia.
Methods
Extended methods are available in the online Supplemental Material. The data, methods, and study materials used to conduct the research are available from the corresponding author on reasonable request.
Family Studies
The proband and his relatives were recruited in Lyon to investigate the genetic cause of a familial combined hypolipidemia (GENELIP study [From Known to New Genes in Dyslipidemia]; ClinicalTrials.com registration No. NCT03939039). GENELIP aims to decipher the mechanisms involved in the occurrence or modulation of dyslipidemia in patients referred for primary dyslipidemia. After the identification of 1 potential candidate gene and a confirmative cosegregation analysis, the members of the pedigree were recruited to the HYPOCHOL study (A Genetically-Based Strategy to Identify New Targets in Cholesterol Metabolism; ClinicalTrials.com registration No. NCT02354079) of the CHOPIN program for functional studies. Written informed consent was obtained from all participants in accordance with the principles of the Declaration of Helsinki and the French bioethics law. GENELIP and HYPOCHOL protocols were approved by the University Hospital Center of Lyon (France) and Nantes (CHU Nantes, France), respectively. The GENELIP study also obtained the agreement of the ethical committee of the Commission Nationale de l’Informatique et des Libertés (No. 920434).
Targeted Next-Generation Sequencing
After genomic DNA extraction, DNA from the proband and patient IV.2 was sequenced and analyzed as previously detailed with the DysliSEQ custom design.10 This panel includes coding exons and intron/exon junctions of 311 genes selected from published literature: (1) genes identified in monogenic dyslipidemia, (2) genes identified in genome-wide association studies in lipid metabolism through direct or indirect effects, (3) genes associated with dyslipidemia in mice, and (4) single-nucleotide polymorphisms already described in familial hypercholesterolemia genetic risk scores and in genome-wide association studies (with P>5.10-8). Among these genes, a first intention panel was defined for FHBL (APOB, PCSK9, ANGPTL3), abetalipoproteinemia (ABL, OMIM 200100; MTTP), and chylomicron retention disease (CMRD, OMIM No. 246700; SAR1B). In the absence of a variant in the first intention panel, relevant variants were selected as previously described10 (Table S1.3). The only relevant variant common to the proband and his granddaughter and that cosegregated with the combined hypocholesterolemia observed in the family after Sanger sequencing was the E97G variant in the LIPC gene. In addition, the patients exhibited a combined hypolipidemia and a decrease of phospholipids concentration, as observed in mice after LIPC overexpression.11
Mouse Studies
All mouse studies were performed with APOE*3.Leiden.CETP mice that have a humanized plasma lipid profile12 and that were bred at Leiden University Medical Centre, Leiden, the Netherlands. All mice experiments were approved by the ethics committee of Pays de la Loire (France, 006) and the Ministère de l’enseignement supérieur de la recherche et de l’innovation (France; APAFIS 26862) or the Central Committee on Animal Experimentation of the Netherlands (AVD11600202010187) and Animal Welfare Body of the Leiden University Medical Center and conducted in accordance with institutional guidelines. At ≈10 weeks of age, male mice were placed on a diet containing 0.5% cholesterol and 15% cocoa butter (Ssniff, No. S8854-E035 EF 4021-04T) for 3 to 5 weeks. Mice were injected intravenously with 3×1010 or 3×1011 genome copies of adeno-associated viruses (AAV8) under the thyroxin-binding globulin (TBG) promoter and containing enhanced green fluorescent protein (eGFP), wild-type lipase C, hepatic type (LIPC), or LIPC-E97G (Vector Biolabs). Mice were monitored weekly for changes in plasma cholesterol and triglyceride levels and body weight. Four weeks after injection, experiments to phenotypically characterize the mice were started as described in the Supplemental Material, each with at least a 1-week interval.
Statistical Analyses
For mice studies, a 1-way ANOVA with Tukey correction for multiple comparisons was used, with a 2-sided P value cutoff set at P<0.05. For cell culture studies, a nonparametric Mann-Whitney test was used with a 2-sided P value cutoff set at P<0.05.
Results
A Novel Variant in LIPC Cosegregates With Familial Combined Hypocholesterolemia
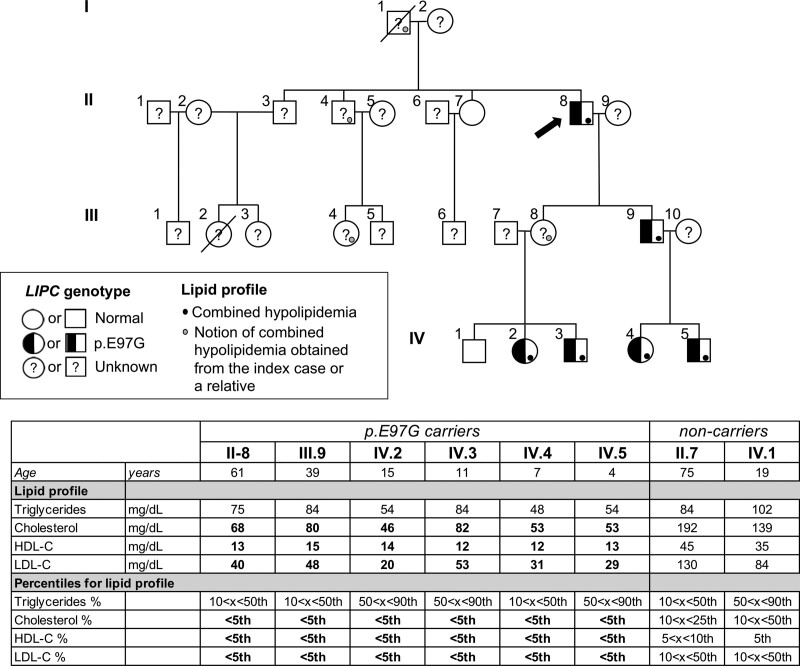
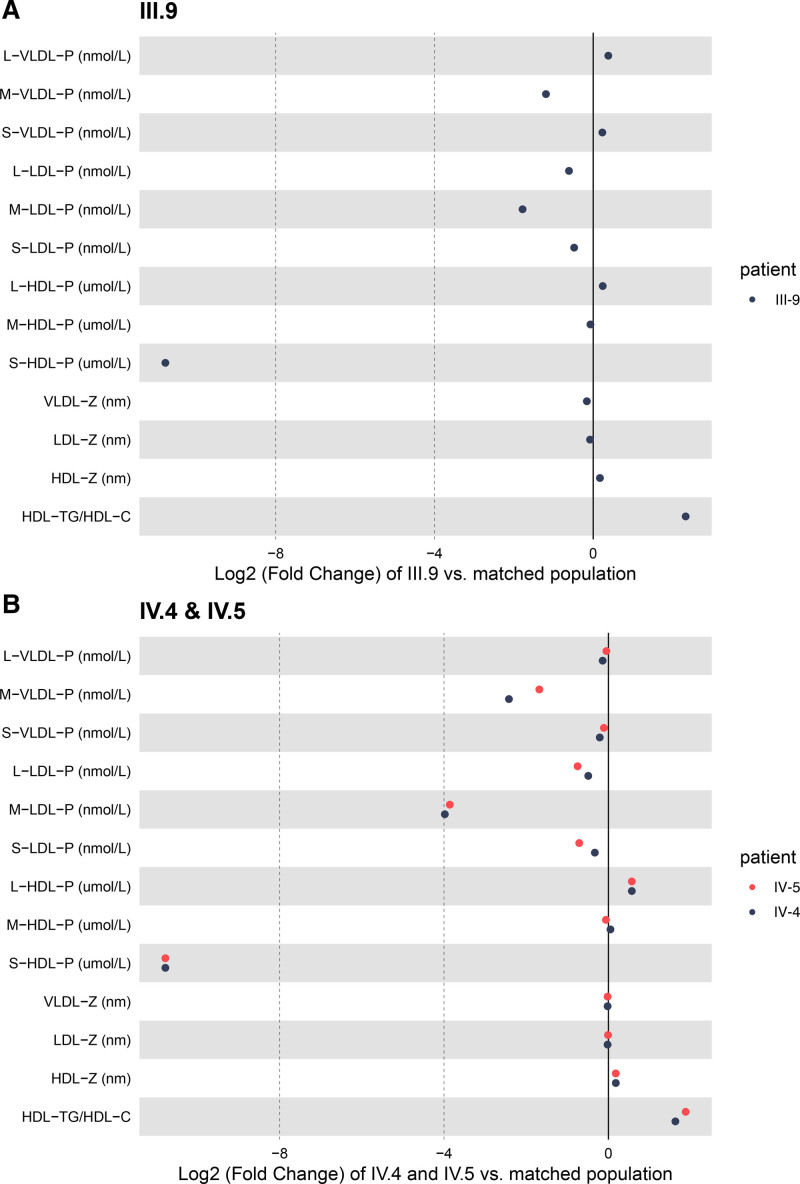
A 61-year-old patient was admitted to the lipid clinic of the University Hospital in Lyon (France) to explore a combined hypocholesterolemia inherited as a dominant phenotype over 4 generations. He was referred for low LDL-C before statin treatment while presenting with coronary artery disease requiring a first coronary stenting. He needed subsequent iterative coronary stentings at 62 and 73 years of age. In addition, he had evolving silent carotid artery plaques, which progressed from 7% (NASCET [North American Symptomatic Carotid Endarterectomy Trial]) with an increased intima-media thickness (1.23 mm) when he was 61 years of age to 75% when he was 73 years of age. He was a former heavy smoker between 18 and 36 years of age and had well-controlled type 2 diabetes since 53 years of age. He had a body mass index of 25.3 kg/m2 and android adiposity. A phenotypic characterization of the patient (patient II.8, Figure 1) revealed extremely low values of circulating cholesterol, LDL-C, HDL-C, phospholipids, and apolipoprotein A1 levels (below the fifth percentile for age and sex; Figure 1 and Table S1.1). In contrast, circulating triglycerides and apoB levels were within the low but normal range. Liver transaminases were mildly increased (aspartate aminotransferase, 54 UI/L; alanine aminotransferase, 54 UI/L when he was 71 years of age), but the fibrosis-4 indexes (a liver fibrosis score) were within the normal interval range (fibrosis-4 index, 1.5 and 1.0 when he was 62 and 74 years of age, respectively), suggesting the absence of liver fibrosis. To clarify the origin of the hypocholesterolemic phenotype, family members of the patient were recruited for plasma and DNA collection. Plasma lipid analyses revealed that a combined hypocholesterolemia below the fifth percentile for age and sex was present in multiple family members and followed an autosomal dominant mode of inheritance (Figure 1 and Table S1.1). More detailed nuclear magnetic resonance lipoprotein profiling of 3 affected family members (III.9, IV.4, IV.5) revealed profound changes in lipoprotein composition and sizes compared with age- and sex-matched individuals (Figure 2 and Table S1.2). Whereas VLDL particle numbers of affected family members were within the normal range, VLDL particle size was reduced compared with control subjects (Figure 2). In contrast, LDL particle size was normal, but LDL particle numbers were low in affected family members, concordant with circulating apoB levels, which were within the lower range in these individuals (Figure 2 and Table S1.1). Furthermore, we found an elevated number of large HDL particles and a near absence of small and medium HDL particles in affected family members, resulting in an elevated average HDL particle size (Figure 2). These large HDL particles are relatively triglyceride enriched, as indicated by the low HDL-C in these individuals and the elevated HDL-triglycerides/HDL-C ratio (Figure 2). In terms of ASCVD, only the proband (II.8) had ASCVD, with no knowledge of clinical ASCVD in the other family members.
Figure 1.
A novel variant in LIPC cosegregates with familial combined hypocholesterolemia. Pedigree of family with familial combined hypocholesterolemia. Squares indicate male family members; circles, female family members. Slashes indicate deceased individuals. Roman numerals to the left of the pedigree indicate the generation; numerals to the upper left of each symbol indicate the individual family member. Basic lipid parameters of the recruited family members are indicated in the table below the pedigree. Values of total cholesterol, triglyceride, and low-density lipoprotein cholesterol (LDL-C), and high-density lipoprotein cholesterol (HDL-C) levels below the fifth percentile for age and sex are in bold.
Figure 2.
Lipoprotein particle quantification by nuclear magnetic resonance. Log2-fold changes in plasma lipoprotein particle quantities and diameters as measured by nuclear magnetic resonance and compared with an age-matched control population for family members III.9 (A, adults) and IV.4 and IV.5 (B, children). HDL indicates high-density lipoprotein; HDL-C, high-density lipoprotein cholesterol; L, large; LDL, low-density lipoprotein; M, medium; P, particles; S, small; TG, triglycerides; VLDL, very-low-density lipoprotein; and Z, diameter.
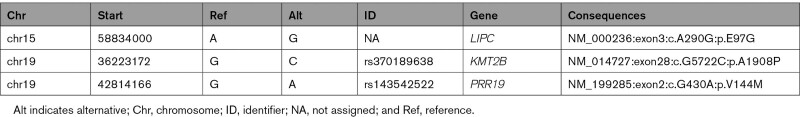
To elucidate the genetic origin of the hypocholesterolemia in the family, DNA from the proband was sequenced with the DysliSEQ custom next-generation sequencing panel design based on ontology of lipid disorders.10 No rare single nucleotide or copy number variation was found in coding exons of genes involved in FHBL (ANGPTL3, APOB, MTTP, PCSK9, SAR1B), and the polygenic risk score (0.91, 49th percentile) did not support a polygenic hypobetalipoproteinemia. Our rare variant filtering criteria (Table S1.3) enabled the detection of a rare missense variant, P.(Glu97Gly) or E97G, in the LIPC gene. The LIPC gene encodes for HL, a protein known to be involved in lipid metabolism. The LIPC-E97G variant is not reported in any large genetic data set (gnomAD and >300 000– individuals of the UK Biobank database) and cosegregated with the combined hypocholesterolemia in the family (Figure 1). To further confirm the absence of another candidate gene, we performed a whole-genome sequencing analysis of all recruited family members, leading to the identification of 3 variants that cosegregate with the hypocholesterolemic phenotype (Table 1). Of these variants, the LIPC-E97G variant was predicted to be the most conserved (Genomic Evolutionary Rate Profiling score, 4.6) and the most deleterious by 9 prediction algorithms (Table S1.4). This result confirmed that the combined hypocholesterolemia might be related to the LIPC-E97G variant.
Table 1.
Genetic Variants Identified With Whole-Genome Sequencing That Cosegregate With the Hypocholesterolemic Phenotype of our Family
LIPC-E97G Modifies the Structural Conformation of HL
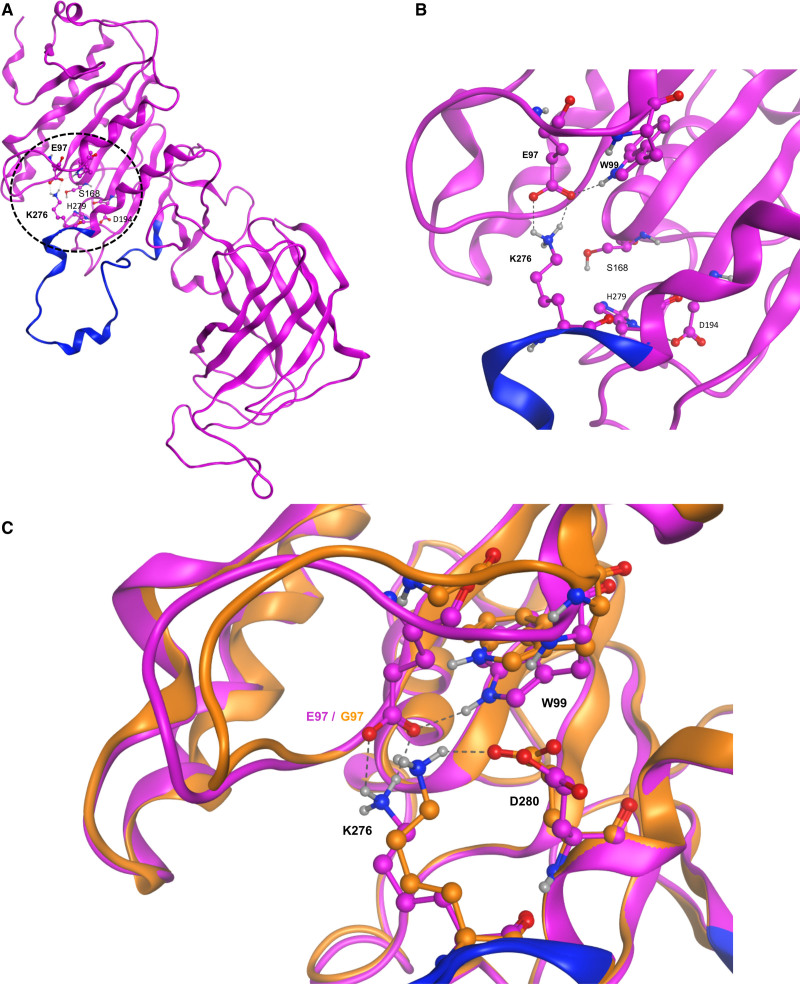
HL is part of a family of glycerol-sn-1-fatty acid hydrolases that includes lipoprotein lipase (LPL) and endothelial lipase (EL).13 HL has intermediate triglyceride lipase and phospholipase activities and hydrolyses triglycerides and phospholipids on circulating lipoproteins such as HDL, intermediate-density lipoprotein, and chylomicron remnants.14 Besides its lipolytic actions, HL also functions as a ligand to facilitate the uptake of lipoproteins by heparan sulfate proteoglycans or other cell surface receptors.15 To determine how the E97G variant might affect HL functionality, we performed an in silico analysis using homology modeling. A homology model for HL was created from a recently established crystal structure of LPL (6OB0,16 Figure S1). Using this model, we observed that E97 is not part of the catalytic triad (S168, D194, and H279) but lies in relatively close proximity to the triad owing to protein folding (Figure 3A). Furthermore, we observed the existence of a salt bridge between E97 and K276, an amino acid present in the lid region of HL (residues 244–277; Figure 3B). This lid region has been shown to determine substrate access to the HL catalytic site.17 The conformation of the lateral chain of K276 was found to be similar to the corresponding lateral chain in LPL (Lys265, lid region from residues 233–266), indicating a conservation of this particular motif between the 2 lipases (Figure S1). The importance of this structural motif is underlined by the observation that K265 is hydrogen bonded to the inhibitor present in the catalytic site of an experimental structure of LPL (Figure S1).
Figure 3.
E97G modifies the structural conformation of HL. A, Global structure of the homology model of hepatic lipase (HL). The N and C termini are on the top left and right, respectively. Dashed circle indicates the zone of particular interest in this study, with the location of the E97 and K276 residues and of the amino acids of the catalytic triad (S168, D194, and H279). B, Closer view of the structural motif (salt bridge and hydrogen bond) involving E97, W99, and K276. Residues of the lid domain are shown in blue. C, Detailed view of the surroundings of the K276 residue in the E97G mutant model (orange) and the HL homology model (purple) The name of some residues is indicated for clarity. Dashed lines represent the interactions (salt bridge, hydrogen bonds) between the various residues. Residues of the lid domain are shown in blue.
We next virtually mutated E97G in the HL homology model and optimized the geometry/energy of the resulting structure. The E97G variant significantly changes the conformation of K276 (Figure 3C). Instead of with E97, K276 is now involved in a salt bridge with the carboxylate group of D280 [d(Hz3…OE1)=1.80 Å] in the mutant model. These results show that E97 and K276 might be important for structural features of the catalytic site. By significantly affecting the conformation of K276, E97G alters the HL lid region and might consequently modify the substrate access to the HL catalytic site.
LIPC-E97G Alters HL Substrate Specificity
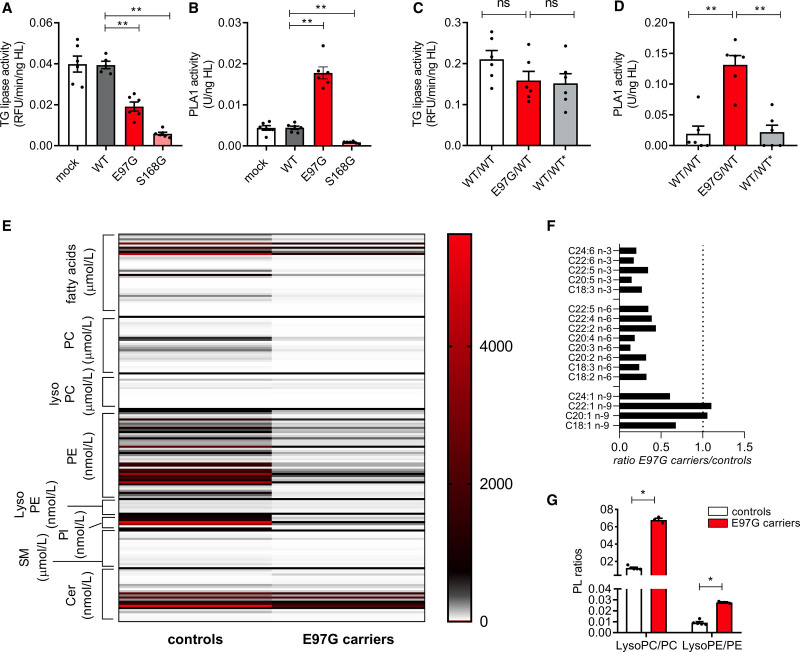
We next set out to determine the impact of the E97G variant on the HL triglyceride lipase and phospholipase activities. We overexpressed wild-type human LIPC (LIPC-WT), LIPC-E97G, or LIPC-S168G, a catalytically inactive mutant, in immortalized human hepatocytes and determined triglyceride lipase and phospholipase activities in the medium after treatment with heparin, which releases HL from cell surface heparan sulfate proteoglycan binding sites (Figure S2A). Triglyceride lipase and phospholipase activities of the released HL were identical between mock-treated and LIPC-WT–overexpressing cells when corrected for the amount of released HL (Figure 4A and 4B) but were nearly absent in the LIPC-S168G–overexpressing cells. A striking finding was that cells overexpressing LIPC-E97G had a moderately reduced triglyceride lipase activity level but a 4-fold increased phospholipase activity level (P<0.01; Figure 4A and 4B). To investigate whether the heterozygous presence of E97G, as found in our family, similarly affects triglyceride lipase and phospholipase activities, we introduced the E97G variant into immortalized human hepatocytes using CRISPR-Cas9 and recorrected the variant to the wild-type allele (Figure S2B). After heparin treatment, the heterozygous presence of E97G did not affect triglyceride lipase activity in the medium compared with wild-type cells (Figure 4C and 4D). However, we observed a 7-fold increase in HL phospholipase activity (P<0.01) with heterozygous E97G presence (Figure 4C and 4D). This effect was completely reversed when E97G was corrected to the wild-type allele (Figure 4C and 4D). These data suggest that the E97G variant specifically increases HL phospholipase activity without increasing triglyceride lipase activity.
Figure 4.
E97G alters hepatic lipase substrate specificity. A and B, Triglyceride (TG) lipase activity and phospholipase A1 (PLA1) activity in medium of heparin-treated immortalized human hepatocytes with overexpression of wild-type LIPC (LIPC-WT), LIPC-E97G, or LIPC-S168G. C and D, Triglyceride lipase activity and PLA1 activity in medium of heparin-treated immortalized human hepatocytes with a wild-type allele, a heterozygous presence of the E97G variant, or a corrected wild-type allele. Each enzymatic activity was corrected for the amount of released hepatic lipase (HL). E, Lipidomics data of plasma of control individuals (n=5) or E97G carriers (n=3). Values are depicted in nanomoles per liter (phosphatidylethanolamine [PE], lysophosphatidylethanolamine [LysoPE], phosphatidylinositol [PI], ceramides [Cer]) or micromoles per liter (fatty acids [FAs], phosphatidylcholine [PC], lysophosphatidylcholine [LysoPC], sphingomyelin [SM]). F, Ratios of plasma FA levels between control individuals (n=5) and E97G carriers (n=3). G, Phospholipid (PL) ratios of lysophospholipids/phospholipids in control individuals (n=5) or family members (n=3). Cell culture data are of 3 independent experiments with a technical duplicate. Statistical significance determined by Mann-Whitney tests. ns Indicates not significant; and RFU, relative fluorescence units. *P<0.05; **P<0.01; ***P<0.001.
To confirm a change in HL substrate specificity in carriers of the E97G variant, we carried out detailed lipidomics on plasma of family members (III.9, IV.4, IV.5) and compared these data with data from normolipidemic individuals (Table S1.5). HL has phospholipase A1 activity and hydrolyzes phospholipids at the SN-1 position to form a fatty acid and a lysophospholipid. In agreement with increased HL phospholipase activity in carriers of the E97G variant, plasma lipidomics data of the E97G carriers showed a clear reduction in overall fatty acids compared with control subjects, with a moderate reduction in fatty acid/apoB ratios (Figure 4E and Table S1.5). Polyunsaturated fatty acids were significantly more reduced than monounsaturated fatty acids, in agreement with the literature18 on in vitro HL substrate preference (Figure 4F). A more striking finding was that phospholipid concentrations were significantly reduced in plasma of E97G carriers, and a concomitant 3- to 5-fold increase in lysophospholipids/phospholipids ratios was observed (Figure 4E and 4G). Furthermore, an increased ratio of lysophospholipids and a decreased ratio of phospholipids versus circulating apoB levels were found in E97G carriers compared with control subjects, suggesting significant changes in the composition of circulating apoB-containing lipoprotein particles (Figure S2C). Together, these data suggest that the E97G variant alters substrate specificity by significantly increasing HL phospholipase activity levels.
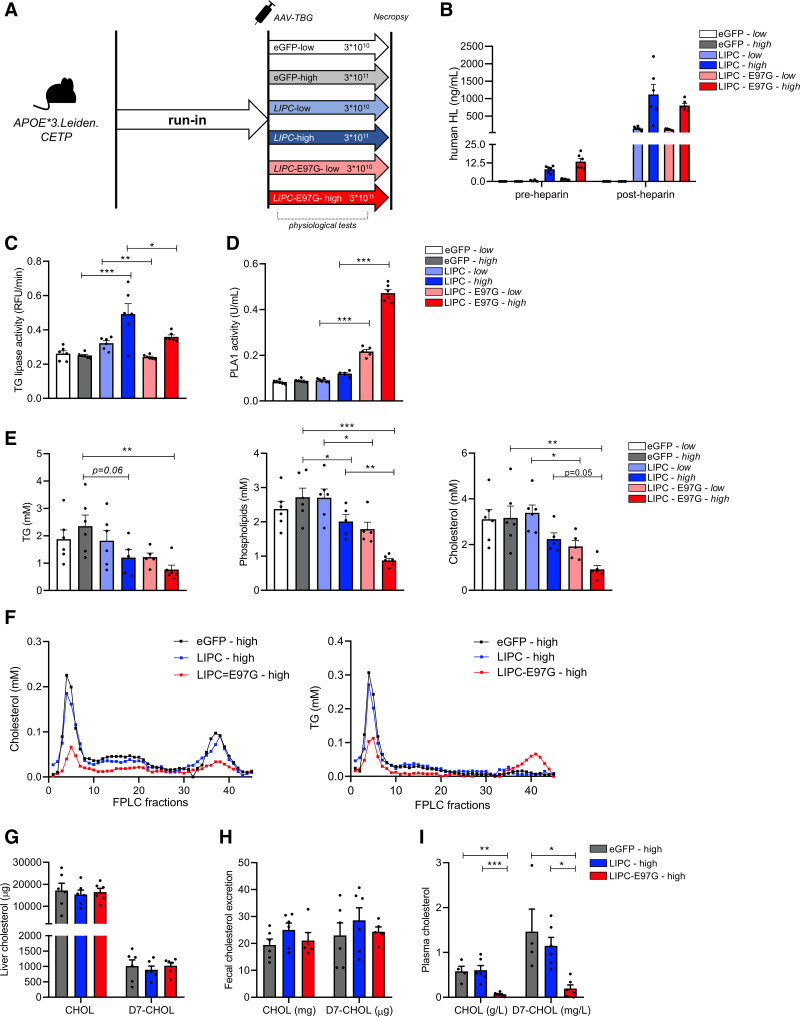
Overexpression of the Human LIPC-E97G Variant Markedly Lowers LDL-C and HDL-C in APOE*3.Leiden.CETP Mice
To determine how the increased phospholipase activity of the E97G variant causes combined hypocholesterolemia, we overexpressed LIPC-WT, LIPC-E97G, and an eGFP control in hepatocytes of the humanized APOE*3.Leiden.CETP mouse model. To do so, we used 2 different doses of adeno-associated virus (AAV8; 3×1010 genome copies [low] and 3×1011 genome copies [high]; Figure 5A). On a Western diet, APOE*3.Leiden.CETP mice have a humanized lipoprotein profile, with cholesterol being carried principally on VLDL and LDL.12 To confirm the successful dose-dependent overexpression of human HL, we performed ELISA on preheparin and postheparin plasma and found similar levels of HL in mice overexpressing LIPC-WT and LIPC-E97G (Figure 5B). The large increase in circulating HL after heparin injection also confirms the localization of human HL on endothelial heparan sulfate proteoglycan (Figure 5B). Confirming our cell culture and lipidomics studies, the overexpression of E97G in mice strongly increased plasma phospholipase activity, but not triglyceride lipase activity levels, in postheparin plasma compared with eGFP or LIPC-WT overexpression (Figure 5C and 5D).
Figure 5.
Overexpression of the LIPC-E97G variant markedly lowers LDL-C and HDL-C in APOE*3.Leiden.CETP mice. A, Experimental setup of mice study overexpressing AAV-TBG-eGFP, AAV-TBG-LIPC, and AAV-TBG-LIPC-E97G in APOE*3.Leiden.CETP mice. B, Human hepatic lipase (HL) levels as determined by ELISA in preheparin and postheparin plasma of mice overexpressing low or high doses of AAV-TBG-eGFP, AAV-TBG-LIPC, and AAV-TBG-LIPC-E97G. C and D, Triglyceride (TG) lipase activity and phospholipase A1 (PLA1) activity in plasma of APOE*3.Leiden.CETP mice overexpressing low or high doses of AAV-TBG-eGFP, AAV-TBG-LIPC, and AAV-TBG-LIPC-E97G and after heparin injection. E, Triglycerides (TG), phospholipids, and cholesterol concentrations in plasma of APOE*3.Leiden.CETP mice overexpressing low or high doses of AAV-TBG-eGFP, AAV-TBG-LIPC, and AAV-TBG-LIPC-E97G. F, Cholesterol and triglyceride (TG) concentrations in fast protein liquid chromatography (FPLC)–separated pooled plasma of APOE*3.Leiden.CETP mice overexpressing high doses of AAV-TBG-eGFP, AAV-TBG-LIPC, and AAV-TBG-LIPC-E97G. G through I, Cholesterol (CHOL) and D7-cholesterol (D7-CHOL) levels extracted from liver (G), feces (H), and plasma (I) of APOE*3.Leiden.CETP mice overexpressing high doses of AAV-TBG-eGFP, AAV-TBG-LIPC, and AAV-TBG-LIPC-E97G and injected with D7-cholesterol 3 days before death. n=5 or 6 per group. A 1-way ANOVA with Tukey correction for multiple comparisons was used for statistical analysis, with a P value cutoff at P<0.05. eGFP indicates enhanced green fluorescent protein; and LIPC, lipase C, hepatic type. *P<0.05; **P<0.01; ***P<0.001.
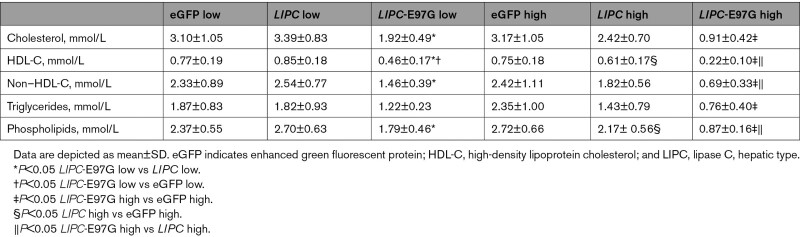
Overexpression of LIPC-WT had only a modest impact on plasma lipid levels (Table 2 and Figure 5E). In contrast, LIPC-E97G overexpression did strongly affect plasma lipoprotein metabolism, with significant dose-dependent reductions in plasma cholesterol, HDL-C, non–HDL-C, phospholipid, and triglyceride levels (Table 2 and Figure 5E). In line with the phenotype observed in familial carriers of the LIPC-E97G, cholesterol levels in VLDL, LDL, and HDL fractions were markedly lower in mice overexpressing LIPC-E97G compared with LIPC- and eGFP-overexpressing mice (Figure 5F). In contrast to family data, triglyceride levels in VLDL and LDL fractions were also markedly lower in mice overexpressing LIPC-E97G (Figure 5F). Furthermore, a mild enrichment of triglycerides in HDL was observed in mice overexpressing LIPC-E97G, as was seen in our familial LIPC-E97G carriers (Figure 5F, Figure S3A, and Table S2.1). We confirmed the profound lowering of plasma lipid levels on overexpression of LIPC-E97G during different metabolic challenges, including postprandial lipemia and fasting/refeeding experiments (Figure S3B and S3C). Overall, these data show that overexpression of LIPC-E97G in mice results in increased hepatic phospholipase activity and leads to a profound reduction in circulating plasma lipid levels.
Table 2.
Plasma Parameters APOE*3.Leiden.CETP Mice
Functional Consequences of the Overexpression of the LIPC-E97G Variant
We first hypothesized that the reduced LDL-C and HDL-C levels associated with the LIPC-E97G variant were caused by an increased hepatic cholesterol clearance such as an increased hepatic cholesteryl ester uptake or increased hepatic receptor–mediated endocytosis of circulating lipoproteins as a result of hydrolysis of phospholipids on the lipoprotein surface.19,20 A moderate reduction in the hepatic expression of 3-hydroxy-3-methylglutaryl-CoA reductase Hmgcr, the rate-limiting enzyme in cholesterol synthesis (Figure S4), was found. However, no significant enrichment of either hepatic triglycerides or free or esterified cholesterol was observed in the livers of mice overexpressing the E97G variant (Figure S4 and Table S2.2). Consistently, after an intravenous injection of D7-labeled cholesterol 3 days before death, we found no increase in hepatic D7-cholesterol and fecal D7-cholesterol excretion in LIPC-E97G-high–overexpressing mice (Figure 5G and 5H). Given that D7-cholesterol labels were cleared more rapidly from the blood of LIPC-E97G-high–overexpressing mice, this suggests that hepatic catabolism and reverse cholesterol transport are unlikely to be the main cause of the reduced plasma cholesterol levels (Figure 5I). In addition, no impact of the overexpression of LIPC-E97G on VLDL secretion rates compared with overexpression of LIPC-WT or eGFP was found (Figure S4C).
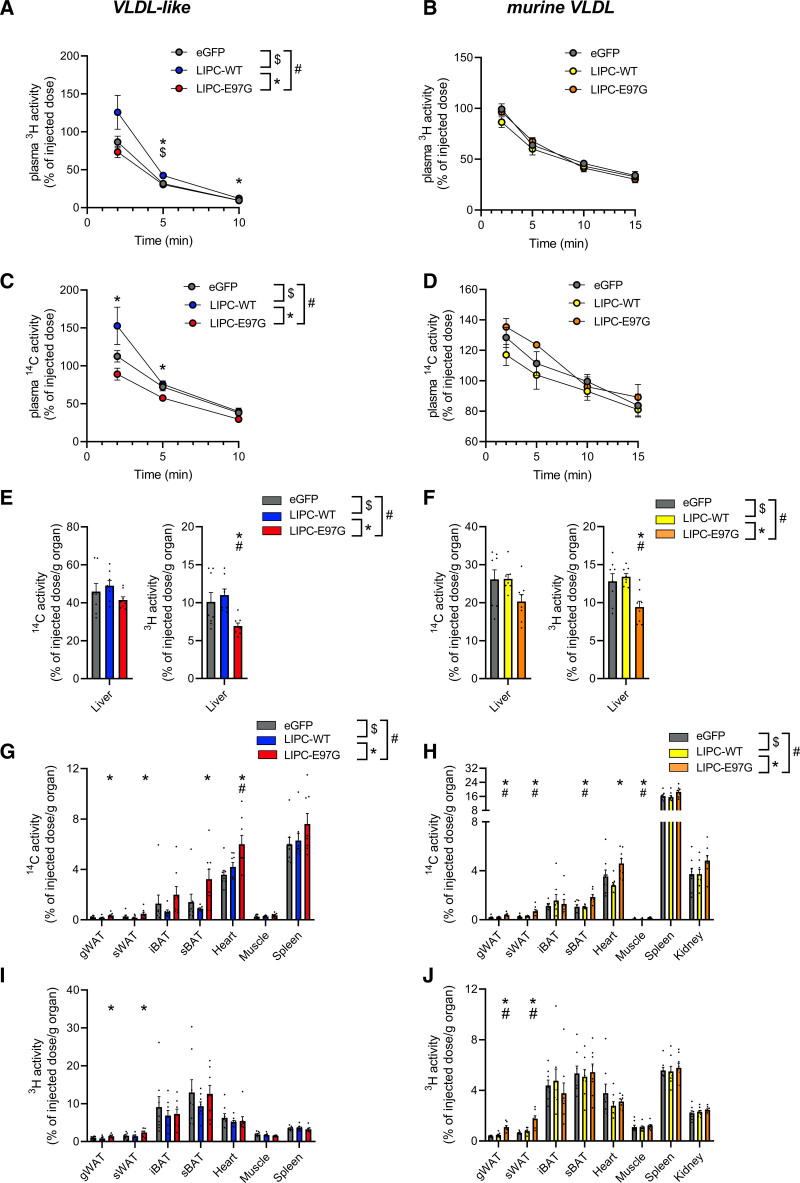
To more directly investigate the impact of LIPC-E97G on lipoprotein kinetics, we overexpressed high doses of eGFP, LIPC-WT, or LIPC-E97G in APOE*3.Leiden.CETP mice and injected these mice with VLDL-like particles or murine VLDL particles labeled with hydrolysable glycerol tri[3H]oleate ([3H]triolein) and [14C]cholesteryl oleate (CO), which is not hydrolysable by HL and is a tracer of triglyceride-rich lipoprotein (TGRL) core remnant particle uptake. Plasma decay of radiolabeled VLDL-like particles was followed, and mice were killed 10 minutes (VLDL-like particles) or 15 minutes (murine VLDL) after injection to determine tissue distribution of the radiolabels (Figure 6A and 6B). No major impact of LIPC-WT overexpression was found on [3H]triolein and [14C]CO decay, whereas [3H]triolein and [14C]CO plasma levels were cleared moderately faster in LIPC-E97G– compared with LIPC-WT–overexpressing mice when injected with VLDL-like particles but not when injected with murine VLDL (Figure 6A–6D). More remarkable differences were observed with a detailed analysis of tissue distributions. These data indicated that [3H]triolein uptake was significantly and [14C]CO uptake was moderately reduced in the liver after clearance of both types of particles (Figure 6E and 6F). Instead, [3H]triolein and [14C]CO labels were increasingly taken up by different adipose tissue depots and oxidative tissues (Figure 6G–6I). These data suggest that the LIPC-E97G variant is responsible for the combined hypocholesterolemic phenotype observed in our family by promoting the uptake of cholesterol-containing remnants by extrahepatic tissues. In support, a nonsignificant increase in cholesterol levels was observed in the muscle and gonadal white adipose tissue of mice overexpressing LIPC-E97G (Figure S5). It should be noted that these effects are likely not mediated through an apoB-mediated pathway because no significant differences in the tissue clearance profile were observed between the clearance of VLDL-like particles (without apoB) and endogenous murine VLDL (with apoB).
Figure 6.
Overexpression of the LIPC-E97G variant promotes peripheral cholesterol uptake. A and C, Decay of plasma 3H activity (glycerol tri[3H]oleate, hydrolysable; A) and plasma 14C activity ([14C]cholesteryl oleate, nonhydrolysable) levels (C) in APOE*3.Leiden.CETP mice overexpressing AAV-TBG-eGFP, AAV-TBG-LIPC, and AAV-TBG-LIPC-E97G injected with very-low-density lipoprotein (VLDL)–like particles. B and D, Decay of plasma 3H activity (glycerol tri[3H]oleate, hydrolysable; B) and plasma 14C activity ([14C]cholesteryl oleate, nonhydrolysable) levels (D) in APOE*3.Leiden.CETP mice14 overexpressing AAV-TBG-eGFP, AAV-TBG-LIPC, and AAV3-TBG-LIPC-E97G injected with radiolabeled murine VLDL. E, Liver 14C activity (cholesteryl ester, nonhydrolysable; left) and liver 3H activity (triolein, hydrolysable) levels (right) in APOE*3.Leiden.CETP mice overexpressing AAV-TBG-eGFP, AAV-TBG-LIPC, and AAV-TBG-LIPC-E97G injected with VLDL-like particles. F, Liver 14C activity (cholesteryl ester, nonhydrolysable; left) and liver 3H activity (triolein, hydrolysable levels (right) in APOE*3.Leiden.CETP mice overexpressing AAV-TBG-eGFP, AAV-TBG-LIPC, and AAV-TBG-LIPC-E97G injected with radiolabeled murine VLDL. G and I, Tissue 14C activity (cholesteryl ester, nonhydrolysable; G) and 3H activity (triolein, hydrolysable; I) levels in APOE*3.Leiden.CETP mice overexpressing AAV-TBG-eGFP, AAV-TBG-LIPC, and AAV-TBG-LIPC-E97G injected with VLDL-like particles. H and J, Tissue 14C activity (cholesteryl ester, nonhydrolysable; H) and 3H activity (triolein, hydrolysable; J) levels in APOE*3.Leiden.CETP mice overexpressing AAV-TBG-eGFP, AAV-TBG-LIPC, and AAV-TBG-LIPC-E97G injected with radiolabeled murine VLDL. n=8 per group. A 1-way ANOVA with Tukey correction for multiple comparisons was used for statistical analysis, with a P value cutoff at P<0.05. gWAT indicates gonadal white adipose tissue; iBAT, interscapular brown adipose tissue; sBAT, subscapular brown adipose tissue; and sWAT, subcutaneous white adipose tissue; and TGRL‚ triglyceride-rich lipoprotein. *P<0.05, human wild-type (LIPC-WT) vs LIPC-E97G. $P<0.05, enhanced green fluorescent protein (eGFP) vs LIPC-WT. #P<0.05, eGFP vs LIPC-E97G.
Discussion
In the present study, we identified a rare gain-of-function variant in the LIPC gene, encoding HL, as a novel cause of monogenic dominant familial combined hypocholesterolemia. We show that this variant, E97G, modifies an evolutionarily conserved motif that determines substrate access to the HL catalytic site. This modification specifically increases HL phospholipase activity and enhances lipoprotein catabolism. Mice overexpressing LIPC-E97G recapitulate the combined hypocholesterolemic phenotype and demonstrate that the increased HL phospholipase activity caused by the variant promotes TGRL catabolism by extrahepatic tissues. Together, our data uncover a novel type of hypocholesterolemia that is caused by an increased peripheral lipoprotein clearance as a result of increased HL phospholipase activity. Whether this drastic reduction in circulating lipids translates into decreased atherosclerosis remains to be established. Indeed, whereas no linkage to ASCVD was found in the family, the proband had evolving documented coronary artery stenosis and carotid atherosclerosis.
Our data highlight, for the first time, the physiological importance of the phospholipase activity of HL in controlling lipoprotein metabolism. In our family, the increased phospholipase activity profoundly changes lipoprotein characteristics, with an increased presence of large triglyceride-rich HDL and low HDL-C, low quantities of LDL particles, and a small VLDL size. These changes are largely what would be expected from an LIPC gain-of-function variant based on literature, with the exception of the triglyceride enrichment of HDL. Indeed, individuals with an LIPC gain of function are characterized by elevated HDL-triglycerides, HDL-phospholipids, and intermediate-density lipoprotein levels.21 Our data suggest that the HL phospholipase activity is a pivotal factor controlling lipoprotein catabolism. With its increased preference for phospholipids, the E97G variant makes HL more closely resemble its family member EL in terms of substrate preference.22 EL has mainly phospholipase activity and has historically been linked to the modulation of plasma HDL levels.22,23 Recent studies have, however, suggested that an increased EL activity and subsequent accelerated LDL catabolism are involved in the LDL-C reduction induced by angiopoietin-like 3 inhibition.24,25 Furthermore, a study on individuals with rare EL variants causing hyperalphalipoproteinemia (ie, high HDL-C levels) indicated that many of these patients also had elevated LDL-C levels, and the inhibition of EL with a monoclonal antibody increased both HDL-C and LDL-C levels in nonhuman primates and humans.26,27 Together, these studies highlight the important role of intravascular phospholipases in VLDL and LDL metabolism, on top of their major role in HDL metabolism.
Our study suggests that the increased phospholipase activity of HL promotes peripheral catabolism of VLDL and LDL. It is well established that the phospholipase activity of HL promotes hepatic phospholipids uptake from circulating lipoproteins,28 and we initially hypothesized that an increased hepatic clearance of lipoproteins was responsible for the observed hypocholesterolemia. However, the reduced uptake of radiolabeled VLDL-like particles and murine VLDL by the liver and the absence of changes in D7-cholesterol in feces argue against this hypothesis. Instead, the LIPC-E97G variant seems to promote the increase in core remnant uptake in peripheral tissues such as adipose tissue and muscle. Peripheral clearance of TGRL has previously been demonstrated in the context of cold exposure: Local LPL-mediated remodeling of TGRL promoted whole-particle TGRL uptake through CD36 in brown adipose tissue and white adipose tissue for further lipolytic processing of lipoprotein by lysosomal acid lipase.29,30 Besides CD36, endothelial scavenger receptor B1 might also be involved in peripheral whole particle uptake, as was elegantly shown in the context of LDL transcytosis and atherosclerosis.31 In this respect, it is of interest to note that both CD36 and scavenger receptor B1 have a great affinity for anionic phospholipids such as phosphatidylserine, which have been previously shown to accumulate on the surface of HL-remodeled lipoproteins.32–35 Further studies are warranted to further decipher the molecular mechanisms between phospholipase-induced remodeling and peripheral lipoprotein uptake.
The present study is not without limitations. Although we used the humanized APOE*3.Leiden.CETP mouse model, the moderate overexpression of LIPC-E97G in these mice is not fully comparable to the heterozygous presence of the E97G variant in our family. The hypocholesterolemic phenotype is overall recapitulated, but there are clear differences with respect to triglyceride levels and HDL size and composition. Indeed, the presence of large, triglyceride-enriched HDL particles in our family is peculiar, and we do not have a clear explanation for this phenotype. Previous studies have shown that HL-deficient individuals also have triglyceride-enriched HDL and that the inhibition of EL with a monoclonal antibody significantly increased HDL particle size in humans.36 Unfortunately, the phenotypic differences in HDL size and composition between mice and humans precluded more extensive studies in mice, and further studies could not be performed in family members for personal reasons. It should also be noted that we did not observe a major impact of a moderate LIPC-WT overexpression on plasma lipid levels or on plasma lipoprotein clearance, in contrast to previously published studies.11,15,37 The presence of endogenous mouse HL and the moderate level of overexpression might play a role here. We do feel that the strong and consistent phenotype on LIPC-E97G overexpression further highlights the major impact that this variant has on lipoprotein metabolism.
From a clinical perspective, the impact of our LIPC-E97G variant on ASCVD development remains unclear, which is a clear limitation of our study. Whereas the low LDL-C levels in our family would suggest a beneficial impact on ASCVD risk, the strong decrease in HDL-C, the near absence of small HDL particles, and the fact that our proband was initially referred after an acute coronary syndrome might suggest otherwise. It should be noted, however, that even the role of wild-type HL, but also EL, in ASCVD development remains a matter of debate because HL and EL have both proatherogenic and antiatherogenic properties.26,38 Furthermore, human genetic studies are not consistent, although overall data suggest that a reduction in HL activity is associated with a modestly increased ASCVD risk.39,40 Because low HDL-C does not seem to be causally linked to ASCVD,41 it could be that our complex clinical phenotype is an exception to the statement that any decrease in LDL-C translates into an ASCVD reduction. To confirm or refute such a hypothesis and to accurately study the impact of the LIPC-E97G variant on ASCVD risk would require a heterozygous transgenic model with a physiological expression profile, a human-like lipoprotein profile, and no endogenous Lipc, which was beyond the scope of the present study. In addition, the fact that the LIPC-E97G variant was associated with an increased catabolism of TGRLs in extrahepatic tissues suggests that this new pathway can promote the lowering of circulating lipids without increasing the risk of hepatic steatosis, an undesirable side effect that has halted the development of some new lipid-lowering drugs.42
Conclusions
We identify a rare variant in the LIPC gene as a second cause of familial combined hypocholesterolemia by increased clearance. This study highlights the importance of the phospholipase activity of HL in the regulation of lipoprotein metabolism, as well as the need for an increased understanding of the link between lipase specificity (triglycerides/phospholipids) and lipoprotein catabolism (TGRL remnants/HDL). Future studies that combine the study of specific LIPC variants and corresponding molecular modeling investigations might shed further light on these aspects. Our observations are of direct clinical relevance because they highlight that an increased hepatic phospholipase activity can drastically lower plasma LDL-C levels. Because the proband developed ASCVD despite low LDL-C plasma levels, additional models and studies will be required to specify the impact of the E97G variant on atherosclerosis progression and to decipher whether this approach could be a promising area of research to lower ASCVD risk.
Article Information
Acknowledgments
The authors warmly thank the members of the family and Taissia Lelekov Boissard, PhD (RCA), who proactively participated in the segregation study. They are most grateful to the Bioinformatics Core Facility of Nantes BiRD, member of Biogenouest, Institut Français de Bioinformatique (ANR-11-INBS-0013), for the use of its resources and for its technical support. Part of this research was conducted using the UK Biobank resource under application 49823.
Sources of Funding
This work was supported by the French national project CHOPIN funded by the Agence Nationale de la Recherche (ANR-16-RHUS-0007) and coordinated by the CHU of Nantes, the INSTINCTIVE research program funded by the Fondation pour la Recherche Médicale (FRM: EQU201903007846) and a grant from the Fondation de France (Engt No. 00047967) and the foundation GENAVIE. Dr Rimbert is supported by a postdoctoral fellowship grant from the Institut de France–Fondation Lefoulon-Delalande. This work was further supported by the Netherlands Cardiovascular Research Initiative, an initiative with support of the Dutch Heart Foundation (CVON-GENIUS-2 to Dr Rensen).
Disclosures
Dr Cariou reports personal fees from Abbott and Akcea, as well as grants and personal fees from Amgen, AstraZeneca, Bristol Myers Squibb, Gilead, Eli Lilly, Novartis, Novo Nordisk, Sanofi, and Regeneron, outside the submitted work. Dr Moulin reports grants, personal fees, and nonfinancial support from Akcea, Amgen, Boehringer, Ionis, and Novo, outside the submitted work. Dr Rensen reports grants from Eli Lilly, Lipigon, and Retrotope, outside the submitted work. Dr Amigo is a stock owner of Biosfer Teslab and has a patent on the lipoprotein profiling described in this article. The other authors report no conflicts.
Supplemental Material
Figures S1–S5
Tables S1 and S2
Expanded Materials & Methods
Supplementary Material
Nonstandard Abbreviations and Acronyms
- apoB
- apolipoprotein B
- ASCVD
- atherosclerotic cardiovascular disease
- CHOPIN
- Cholesterol Personalized Innovation
- CO
- cholesteryl oleate
- eGFP
- enhanced green fluorescent protein
- EL
- endothelial lipase
- FHBL
- familial hypobetalipoproteinemia
- GENELIP
- From Known to New Genes in Dyslipidemia
- HYPOCHOL
- A Genetically-Based Strategy to Identify New Targets in Cholesterol Metabolism
- HDL
- high-density lipoprotein
- HDL-C
- high-density lipoprotein cholesterol
- HL
- hepatic lipase
- LDL
- low-density lipoprotein
- LDL-C
- low-density lipoprotein cholesterol
- LIPC
- lipase C, hepatic type (hepatic lipase)
- LIPC-WT
- wild-type human LIPC
- LPL
- lipoprotein lipase
- TBG
- thyroxin-binding globulin
- TGRL
- triglyceride-rich lipoprotein
- VLDL
- very-low-density lipoprotein
W. Dijk and M. Di Filippo contributed equally.
C. Le May, P. Moulin, and B. Cariou contributed equally.
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/circulationaha.121.057978.
Circulation is available at www.ahajournals.org/journal/circ
For Sources of Funding and Disclosures, see page 737.
Contributor Information
Wieneke Dijk, Email: wieneke.dijk@inrae.fr.
Mathilde Di Filippo, Email: mathilde.di-filippo@chu-lyon.fr.
Sander Kooijman, Email: s.kooijman@lumc.nl.
Robin van Eenige, Email: r.van_eenige@lumc.nl.
Antoine Rimbert, Email: antoine.rimbert@univ-nantes.fr.
Amandine Caillaud, Email: amandine.caillaud@univ-nantes.fr.
Aurélie Thedrez, Email: aureliethedrez@gmail.com.
Damien Garçon, Email: Damien.Garcon@ircm.qc.ca.
Thibaud Sotin, Email: thibaud.sotin@etu.univ-nantes.fr.
Pierre Lindenbaum, Email: pierre.lindenbaum@univ-nantes.fr.
Jean-Paul Pais de Barros, Email: jppais@u-bourgogne.fr.
Laurence Duvillard, Email: laurence.duvillard@chu-dijon.fr.
Karim Si-Tayeb, Email: karimsitayeb@gmail.com.
Nuria Amigo, Email: namigo@biosferteslab.com.
Jean-Yves Le Questel, Email: jean-yves.le-questel@univ-nantes.fr.
Patrick C.N. Rensen, Email: p.c.n.rensen@lumc.nl.
Cédric Le May, Email: cedric.lemay@univ-nantes.fr.
Philippe Moulin, Email: philippe.moulin@chu-lyon.fr.
References
- 1.Borén J, Chapman MJ, Krauss RM, Packard CJ, Bentzon JF, Binder CJ, Daemen MJ, Demer LL, Hegele RA, Nicholls SJ, et al. Low-density lipoproteins cause atherosclerotic cardiovascular disease: pathophysiological, genetic, and therapeutic insights: a consensus statement from the European Atherosclerosis Society Consensus Panel. Eur Heart J. 2020;41:2313–2330. doi: 10.1093/eurheartj/ehz962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dagenais GR, Leong DP, Rangarajan S, Lanas F, Lopez-Jaramillo P, Gupta R, Diaz R, Avezum A, Oliveira GBF, Wielgosz A, et al. Variations in common diseases, hospital admissions, and deaths in middle-aged adults in 21 countries from five continents (PURE): a prospective cohort study. Lancet. 2020;395:785–794. doi: 10.1016/S0140-6736(19)32007-0 [DOI] [PubMed] [Google Scholar]
- 3.Hagenbeek FA, Pool R, van Dongen J, Draisma HHM, Jan Hottenga J, Willemsen G, Abdellaoui A, Fedko IO, den Braber A, Visser PJ, et al. Heritability estimates for 361 blood metabolites across 40 genome-wide association studies. Nat Commun. 2020;11:39. doi: 10.1038/s41467-019-13770-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berberich AJ, Hegele RA. The complex molecular genetics of familial hypercholesterolaemia. Nat Rev Cardiol. 2019;16:9–20. doi: 10.1038/s41569-018-0052-6 [DOI] [PubMed] [Google Scholar]
- 5.Balder JW, Rimbert A, Zhang X, Viel M, Kanninga R, Van DIjk F, Lansberg P, Sinke R, Kuivenhoven JA. Genetics, lifestyle, and low-density lipoprotein cholesterol in young and apparently healthy women. Circulation. 2018;137:820–831. doi: 10.1161/CIRCULATIONAHA.117.032479 [DOI] [PubMed] [Google Scholar]
- 6.Bredefeld C, Peretti N, Hussain MM, Di Filippo M, Granot E, Cuerq C, Poinsot P, Moulin P, Charrière S, Brin M, et al. New classification and management of abetalipoproteinemia and related disorders. Gastroenterology. 2021;160:1912–1916. doi: 10.1053/j.gastro.2020.11.040 [DOI] [PubMed] [Google Scholar]
- 7.Musunuru K, Pirruccello JP, Do R, Peloso GM, Guiducci C, Sougnez C, Garimella KV, Fisher S, Abreu J, Barry AJ, et al. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N Engl J Med. 2010;363:2220–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rimbert A, Vanhoye X, Coulibaly D, Marrec M, Pichelin M, Charrière S, Peretti N, Valéro R, Wargny M, Carrié A, et al. Phenotypic differences between polygenic and monogenic hypobetalipoproteinemia. Arterioscler Thromb Vasc Biol. 2021;41:e63–e71. doi: 10.1161/ATVBAHA.120.315491 [DOI] [PubMed] [Google Scholar]
- 9.Oetjens MT, Kelly MA, Sturm AC, Martin CL, Ledbetter DH. Quantifying the polygenic contribution to variable expressivity in eleven rare genetic disorders. Nat Commun. 2019;10:4897. doi: 10.1038/s41467-019-12869-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marmontel O, Rollat-Farnier PA, Wozny A, Charrière S, Vanhoye X, Simonet T, Chatron N, Collin-Chavagnac D, Nony S, Dumont S, et al. Development of a new expanded next-generation sequencing panel for genetic diseases involved in dyslipidemia. Clin Genet. 2020;98:589–594. doi: 10.1111/cge.13832 [DOI] [PubMed] [Google Scholar]
- 11.Dichek HL, Brecht W, Fan J, Ji ZS, McCormick SPA, Akeefe H, Conzo L, Sanan DA, Weisgraber KH, Young SG, et al. Overexpression of hepatic lipase in transgenic mice decreases apolipoprotein B-containing and high density lipoproteins: evidence that hepatic lipase acts as a ligand for lipoprotein uptake. J Biol Chem. 1998;273:1896–1903. doi: 10.1074/jbc.273.4.1896 [DOI] [PubMed] [Google Scholar]
- 12.Westerterp M, Van Der Hoogt CC, De Haan W, Offerman EH, Dallinga-Thie GM, Jukema JW, Havekes LM, Rensen PCN. Cholesteryl ester transfer protein decreases high-density lipoprotein and severely aggravates atherosclerosis in APOE*3-Leiden mice. Arterioscler Thromb Vasc Biol. 2006;26:2552–2559. doi: 10.1161/01.ATV.0000243925.65265.3c [DOI] [PubMed] [Google Scholar]
- 13.Choi SY, Hirata KI, Ishida T, Quertermous T, Cooper AD. Endothelial lipase: a new lipase on the block. J Lipid Res. 2002;43:1763–1769. doi: 10.1194/jlr.r200011-jlr200 [DOI] [PubMed] [Google Scholar]
- 14.Santamarina-Fojo S, González-Navarro H, Freeman L, Wagner E, Nong Z. Hepatic lipase, lipoprotein metabolism, and atherogenesis. Arterioscler Thromb Vasc Biol. 2004;24:1750–1754. doi: 10.1161/01.ATV.0000140818.00570.2d [DOI] [PubMed] [Google Scholar]
- 15.Dugi KA, Amar MJA, Haudenschild CC, Shamburek RD, Bensadoun A, Hoyt RF, Fruchart-Najib J, Madj Z, Brewer HB, Santamarina-Fojo S. In vivo evidence for both lipolytic and nonlipolytic function of hepatic lipase in the metabolism of HDL. Arterioscler Thromb Vasc Biol. 2000;20:793–800. doi: 10.1161/01.atv.20.3.793 [DOI] [PubMed] [Google Scholar]
- 16.Arora R, Nimonkar AV, Baird D, Wang C, Chiu CH, Horton PA, Hanrahan S, Cubbon R, Weldon S, Tschantz WR, et al. Structure of lipoprotein lipase in complex with GPIHBP1. Proc Natl Acad Sci U S A. 2019;116:10360–10365. doi: 10.1073/pnas.1820171116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dugi KA, Dichek HL, Santamarina-Fojo S. Human hepatic and lipoprotein lipase: the loop covering the catalytic site mediates lipase substrate specificity. J Biol Chem. 1995;270:25396–25401. doi: 10.1074/jbc.270.43.25396 [DOI] [PubMed] [Google Scholar]
- 18.Yang P, Subbaiah PV. Regulation of hepatic lipase activity by sphingomyelin in plasma lipoproteins. Biochim Biophys Acta. 2015;1851:1327–1336. doi: 10.1016/j.bbalip.2015.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brasaemle DL, Cornely-Moss K, Bensadoun A. Hepatic lipase treatment of chylomicron remnants increases exposure of apolipoprotein E. J Lipid Res. 1993;34:455–465. [PubMed] [Google Scholar]
- 20.Lambert G, Chase MB, Dugi K, Bensadoun A, Brewer HB, Santamarina-Fojo S. Hepatic lipase promotes the selective uptake of high density lipoprotein-cholesteryl esters via the scavenger receptor B1. J Lipid Res. 1999;40:1294–1303. [PubMed] [Google Scholar]
- 21.Connelly PW. The role of hepatic lipase in lipoprotein metabolism. Clin Chim Acta. 1999;286:243–255. doi: 10.1016/s0009-8981(99)00105-9 [DOI] [PubMed] [Google Scholar]
- 22.McCoy MG, Sun G-S, Marchadier D, Maugeais C, Glick JM, Rader DJ. Characterization of the lipolytic activity of endothelial lipase. J Lipid Res. 2002;43:921–929. [PubMed] [Google Scholar]
- 23.Maugeais C, Tietge UJF, Broedl UC, Marchadier D, Cain W, McCoy MG, Lund-Katz S, Glick JM, Rader DJ. Dose-dependent acceleration of high-density lipoprotein catabolism by endothelial lipase. Circulation. 2003;108:2121–2126. doi: 10.1161/01.CIR.0000092889.24713.DC [DOI] [PubMed] [Google Scholar]
- 24.Adam RC, Mintah IJ, Alexa-Braun CA, Shihanian LM, Lee JS, Banerjee P, Hamon SC, Kim HI, Cohen JC, Hobbs HH, et al. Angiopoietin-like protein 3 governs LDL-cholesterol levels through endothelial lipase-dependent VLDL clearance. J Lipid Res. 2020;61:1271–1286. doi: 10.1194/jlr.RA120000888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu L, Soundarapandian MM, Castoreno AB, Millar JS, Rader DJ. LDL-cholesterol reduction by ANGPTL3 inhibition in mice is dependent on endothelial lipase. Circ Res. 2020;127:1112–1114. doi: 10.1161/CIRCRESAHA.120.317128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cole J, Blackhurst DM, Solomon GAE, Ratanjee BD, Benjamin R, Marais AD. Atherosclerotic cardiovascular disease in hyperalphalipoproteinemia due to LIPG variants. J Clin Lipidol. 2021;15:142–150.e2. doi: 10.1016/j.jacl.2020.12.007 [DOI] [PubMed] [Google Scholar]
- 27.Le Lay JE, Du Q, Mehta MB, Bhagroo N, Hummer BT, Falloon J, Carlson G, Rosenbaum AI, Jin CY, Kimko H, et al. Blocking endothelial lipase with monoclonal antibody MEDI5884 durably increases high density lipoprotein in nonhuman primates and in a phase 1 trial. Sci Transl Med. 2021;13:eabb0602. doi: 10.1126/scitranslmed.abb0602 [DOI] [PubMed] [Google Scholar]
- 28.Landin B, Nilsson A, Twu JS, Schotz MC. A role for hepatic lipase in chylomicron and high density lipoprotein phospholipid metabolism. J Lipid Res. 1984;25:559–563. [PubMed] [Google Scholar]
- 29.Fischer AW, Jaeckstein MY, Gottschling K, Heine M, Sass F, Mangels N, Schlein C, Worthmann A, Bruns OT, Yuan Y, et al. Lysosomal lipoprotein processing in endothelial cells stimulates adipose tissue thermogenic adaptation. Cell Metab. 2021;33:547–564.e7. doi: 10.1016/j.cmet.2020.12.001 [DOI] [PubMed] [Google Scholar]
- 30.Bartelt A, Bruns OT, Reimer R, Hohenberg H, Ittrich H, Peldschus K, Kaul MG, Tromsdorf UI, Weller H, Waurisch C, et al. Brown adipose tissue activity controls triglyceride clearance. Nat Med. 2011;17:200–205. doi: 10.1038/nm.2297 [DOI] [PubMed] [Google Scholar]
- 31.Huang L, Chambliss KL, Gao X, Yuhanna IS, Behling-Kelly E, Bergaya S, Ahmed M, Michaely P, Luby-Phelps K, Darehshouri A, et al. SR-B1 drives endothelial cell LDL transcytosis via DOCK4 to promote atherosclerosis. Nature. 2019;569:565–569. doi: 10.1038/s41586-019-1140-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mezdour H, Jones R, Dengremont C, Castro G, Maeda N. Hepatic lipase deficiency increases plasma cholesterol but reduces susceptibility to atherosclerosis in apolipoprotein E-deficient mice. J Biol Chem. 1997;272:13570–13575. doi: 10.1074/jbc.272.21.13570 [DOI] [PubMed] [Google Scholar]
- 33.Linton MF, Tao H, Linton EF, Yancey PG. SR-BI: a multifunctional receptor in cholesterol homeostasis and atherosclerosis. Trends Endocrinol Metab. 2017;28:461–472. doi: 10.1016/j.tem.2017.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crawford SE, Borensztajn J. Plasma clearance and liver uptake of chylomicron remnants generated by hepatic lipase lipolysis: evidence for a lactoferrin-sensitive and apolipoprotein E-independent pathway. J Lipid Res. 1999;40:797–805. [PubMed] [Google Scholar]
- 35.Tait JF, Smith C. Phosphatidylserine receptors: role of CD36 in binding of anionic phospholipid vesicles to monocytic cells. J Biol Chem. 1999;274:3048–3054. doi: 10.1074/jbc.274.5.3048 [DOI] [PubMed] [Google Scholar]
- 36.Le Lay JE, Du Q, Mehta MB, Bhagroo N, Hummer BT, Falloon J, Carlson G, Rosenbaum AI, Jin C, Kimko H, et al. Blocking endothelial lipase with monoclonal antibody MEDI5884 durably increases high density lipoprotein in nonhuman primates and in a phase 1 trial. Sci Transl Med. 2021;13:eabb0602. doi: 10.1126/scitranslmed.abb0602 [DOI] [PubMed] [Google Scholar]
- 37.Amar MJA, Dugi KA, Haudenschild CC, Shamburek RD, Foger B, Chase M, Bensadoun A, Hoyt RF, Brewer HB, Santamarina-Fojo S. Hepatic lipase facilitates the selective uptake of cholesteryl esters from remnant lipoproteins in apoE-deficient mice. J Lipid Res. 1998;39:2436–2442. [PubMed] [Google Scholar]
- 38.Jansen H, Verhoeven AJM, Sijbrands EJG. Hepatic lipase: a pro- or anti-atherogenic protein? J Lipid Res. 2002;43:1352–1362. doi: 10.1194/jlr.r200008-jlr200 [DOI] [PubMed] [Google Scholar]
- 39.Johannsen TH, Kamstrup PR, Andersen RV, Jensen GB, Sillesen H, Tybjaerg-Hansen A, Nordestgaard BG. Hepatic lipase, genetically elevated high-density lipoprotein, and risk of ischemic cardiovascular disease. J Clin Endocrinol Metab. 2009;94:1264–1273. doi: 10.1210/jc.2008-1342 [DOI] [PubMed] [Google Scholar]
- 40.Voight BF, Peloso GM, Orho-Melander M, Frikke-Schmidt R, Barbalic M, Jensen MK, Hindy G, Hólm H, Ding EL, Johnson T, et al. Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. Lancet. 2012;380:572–580. doi: 10.1016/S0140-6736(12)60312-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nordestgaard BG. Triglyceride-rich lipoproteins and atherosclerotic cardiovascular disease: new insights from epidemiology, genetics, and biology. Circ Res. 2016;118:547–563. doi: 10.1161/CIRCRESAHA.115.306249 [DOI] [PubMed] [Google Scholar]
- 42.Blom DJ, Raal FJ, Santos RD, Marais AD. Lomitapide and mipomersen—inhibiting microsomal triglyceride transfer protein (MTP) and apoB100 synthesis. Curr Atheroscler Rep. 2019;21:48. doi: 10.1007/s11883-019-0809-3 [DOI] [PubMed] [Google Scholar]
- 43.Balder JW, Lansberg PJ, Hof MH, Wiegman A, Hutten BA, Kuivenhoven JA. Pediatric lipid reference values in the general population: the Dutch Lifelines cohort study. J Clin Lipidol. 2018;12:1208–1216. doi: 10.1016/j.jacl.2018.05.011 [DOI] [PubMed] [Google Scholar]
- 44.Balder JW, de Vries JK, Nolte IM, Lansberg PJ, Kuivenhoven JA, Kamphuisen PW. Lipid and lipoprotein reference values from 133,450 Dutch Lifelines participants: age- and gender-specific baseline lipid values and percentiles. J Clin Lipidol. 2017;11:1055–1064. doi: 10.1016/j.jacl.2017.05.007 [DOI] [PubMed] [Google Scholar]
- 45.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinforma. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Q, Pierce-Hoffman E, Cummings BB, Alföldi J, Francioli LC, Gauthier LD, Hill AJ, O’Donnell-Luria AH, Genome Aggregation Database Production Team, Genome Aggregation Database Consortium, Karczewski KJ, MacArthur DG. Landscape of multi-nucleotide variants in 125,748 human exomes and 15,708 genomes. Nat. Commun. 2020;11:2539. doi: 10.1038/s41467-019-12438-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012;6:80–92. doi: 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, et al. Twelve years of SAMtools and BCFtools. GigaScience. 2021;10:giab008. doi: 10.1093/gigascience/giab008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lindenbaum P, Redon R. Bioalcidae, samjs and vcffilterjs: object-oriented formatters and filters for bioinformatics files. Bioinformatics. 2018;34:1224–1225. doi: 10.1093/bioinformatics/btx734 [DOI] [PubMed] [Google Scholar]
- 51.Yang H, Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc. 2015;10:1556–1566. doi: 10.1038/nprot.2015.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Talmud PJ, Shah S, Whittall R, Futema M, Howard P, Cooper JA, Harrison SC, Li K, Drenos F, Karpe F, et al. Use of low-density lipoprotein cholesterol gene score to distinguish patients with polygenic and monogenic familial hypercholesterolaemia: a case-control study. Lancet. 2013;381:1293–1301. doi: 10.1016/S0140-6736(12)62127-8 [DOI] [PubMed] [Google Scholar]
- 53.Mallol R, Amigó N, Rodríguez MA, Heras M, Vinaixa M, Plana N, Rock E, Ribalta J, Yanes O, Masana L, et al. Liposcale: a novel advanced lipoprotein test based on 2D diffusion-ordered 1H NMR spectroscopy. J Lipid Res. 2015;56:737–746. doi: 10.1194/jlr.D050120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reis A, Rudnitskaya A, Blackburn GJ, Mohd Fauzi N, Pitt AR, Spickett CM. A comparison of five lipid extraction solvent systems for lipidomic studies of human LDL. J Lipid Res. 2013;54:1812–1824. doi: 10.1194/jlr.M034330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mouillot T, Rizk M, Pais de Barros J-P, Gilloteau A, Busson A, Bernard-Chabert B, Thiefin G, Barraud H, Bronowicki J-P, Richou C, et al. Fatty acid composition of the erythrocyte membrane and risk of hepatocellular carcinoma in cirrhotic patients. Aliment Pharmacol Ther. 2020;52:1503–1515. doi: 10.1111/apt.16022 [DOI] [PubMed] [Google Scholar]
- 56.Bourgeois T, Jalil A, Thomas C, Magnani C, Le Guern N, Gautier T, Pais de Barros J-P, Bergas V, Choubley H, Mazzeo L, et al. Deletion of lysophosphatidylcholine acyltransferase 3 in myeloid cells worsens hepatic steatosis after a high-fat diet. J Lipid Res. 2021;62:100013. doi: 10.1194/jlr.RA120000737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tran TTT, Postal BG, Demignot S, Ribeiro A, Osinski C, Pais de Barros J-P, Blachnio-Zabielska A, Leturque A, Rousset M, Ferré P, et al. Short term palmitate supply impairs intestinal insulin signaling via ceramide production. J Biol Chem. 2016;291:16328–16338. doi: 10.1074/jbc.M115.709626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Holmes R, VandeBerg Cox. Vertebrate hepatic lipase genes and proteins: a review supported by bioinformatic studies. Open Access Bioinformatics. 2011;2011:85. doi: 10.2147/OAB.S18401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bellissent-Funel M-C, Hassanali A, Havenith M, Henchman R, Pohl P, Sterpone F, van der Spoel D, Xu Y, Garcia AE. Water determines the structure and dynamics of proteins. Chem Rev. 2016;116:7673–7697. doi: 10.1021/acs.chemrev.5b00664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 63.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, De Beer TAP, Rempfer C, Bordoli L, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Benkert P, Biasini M, Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27:343–350. doi: 10.1093/bioinformatics/btq662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schippers IJ, Moshage H, Roelofsen H, Mu M, Ruiters M, Kuipers F, Centre BT. Immortalized human hepatocytes as a tool for the study of hepatocytic. Cell Biol Toxicol. 1997;13:375–386. doi: 10.1023/a:1007404028681 [DOI] [PubMed] [Google Scholar]
- 66.Concordet JP, Haeussler M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018;46:W242–W245. doi: 10.1093/nar/gky354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Eenige R, Verhave PS, Koemans PJ, Tiebosch IACW, Rensen PCN, Kooijman S. RandoMice, a novel, user-friendly randomization tool in animal research. PLoS One. 2020;15:37096. doi: 10.1371/journal.pone.0237096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Löfgren L, Forsberg GB, Ståhlman M. The BUME method: a new rapid and simple chloroform-free method for total lipid extraction of animal tissue. Sci Rep. 2016;6:27688.doi: 10.1038/srep27688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barrilero R, Gil M, Amigó N, Dias CB, Wood LG, Garg ML, Ribalta J, Heras M, Vinaixa M, Correig X. LipSpin: a new bioinformatics tool for quantitative 1 H NMR lipid profiling. Anal Chem. 2018;90:2031–2040. doi: 10.1021/acs.analchem.7b04148 [DOI] [PubMed] [Google Scholar]
- 70.Rensen PC, van Dijk MC, Havenaar EC, Bijsterbosch MK, Kruijt JK, van Berkel TJ. Selective liver targeting of antivirals by recombinant chylomicrons--a new therapeutic approach to hepatitis B. Nat Med. 1995;1:221–225. doi: 10.1038/nm0395-221 [DOI] [PubMed] [Google Scholar]
- 71.Basu D, Manjur J, Jin W. Determination of lipoprotein lipase activity using a novel fluorescent lipase assay. J Lipid Res. 2011;52:826–832. doi: 10.1194/jlr.D010744 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.