Abstract
Traditional Chinese medicines (TCMs) have been widely used for treating ischemic heart disease (IHD), and secondary metabolites are generally regarded as their pharmacologically active components. However, the effects of nucleic acids in TCMs remain unclear. We reported for the first time that a 22-mer double-strand RNA consisting of HC83 (a tRNA-derived fragment [tRF] from the 3′ end of tRNAGln(UUG) of ginseng) and its complementary sequence significantly promoted H9c2 cell survival after hypoxia/reoxygenation (H/R) in vitro. HC83_mimic could also significantly improve cardiac function by maintaining both cytoskeleton integrity and mitochondrial function of cardiomyocytes. Further in vivo investigations revealed that HC83_mimic is more potent than metoprolol by >500-fold against myocardial ischemia/reperfusion (MI/R) injury. In-depth studies revealed that HC83 directly downregulated a lncRNA known as myocardial infarction-associated transcript (MIAT) that led to a subsequent upregulation of VEGFA expression. These findings provided the first evidence that TCM-derived tRFs can exert miRNA-like functions in mammalian systems, therefore supporting the idea that TCM-derived tRFs are promising RNA drug candidates shown to have extraordinarily potent effects. In summary, this study provides a novel strategy not only for discovering pharmacologically active tRFs from TCMs but also for efficiently exploring new therapeutic targets for various diseases.
Keywords: MT: oligonucleotides, therapies and applications, ginseng, tRF, ischemic heart disease, lncRNA, MIAT/VEGFA pathway
Graphical abstract
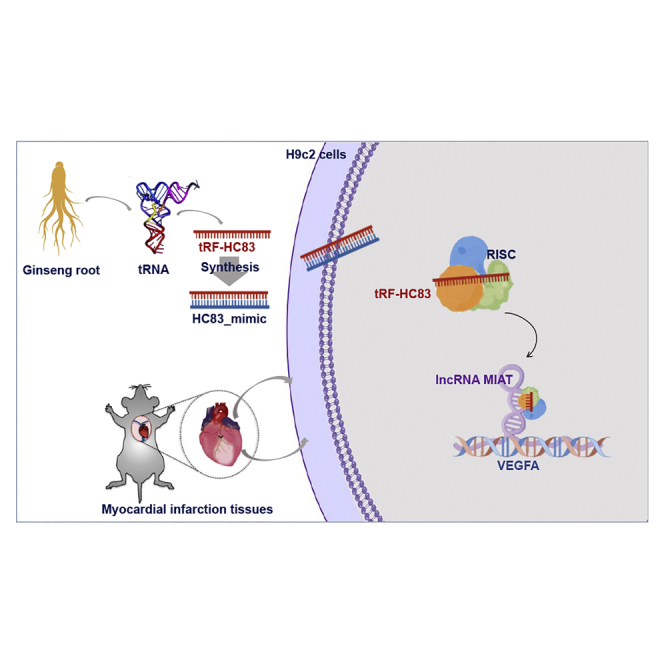
This research reported for the first time that a 22-mer tRF derived from ginseng exhibited cardioprotective effects against ischemia/reperfusion injury by targeting MIAT/VEGFA in a miRNA-like pathway. This raised a new perspective that TCM-derived tRFs could serve as a huge sequence library for the development of miRNA therapeutics.
Introduction
Ischemic heart disease (IHD), also known as coronary heart disease, is a leading cause of morbidity and mortality globally. According to the World Health Organization, IHD and stroke accounted for a combined 15.2 million deaths in 2016.1 Traditional Chinese medicines (TCMs) have been widely used for the treatment of IHD. Most of the effective treatments are Chinese medicine preparations, such as Du Shen Tang, Compound Danshen Dripping Pills, and Tongxinluo capsules, which are extensively applied to clinic.2,3 In the past few decades, major efforts in the active compounds of TCMs have been made on the secondary metabolites, such as saponins, alkaloids, and flavonoids.4,5 However, intensive evidence showed that the effects of using these single compounds alone were inferior to total TCMs extract.6,7 This indicates that TCMs extract contains other effective constituents such as macromolecular nucleic acids, which may be responsible for the overall pharmacological activities of TCMs.
Current evidence indicates that small RNAs with a length of <200 nt derived from plants, including those used in TCMs, are able to regulate gene expression in a cross-kingdom manner in mammalian systems.8, 9, 10, 11 The suppressive effects of honeysuckle-derived microRNA (miRNA) on viral infection demonstrated that miRNAs are a class of important active compounds in TCMs.10 In addition to miRNA, a small RNA from Chinese herb medicine, hongjingtian, has been shown to possess significant antipulmonary fibrotic effects.11 Recent studies proved that other small RNA species more than miRNA derived from corn inhibited the proliferation of HeLa cells, indicating the promise of exploring the function of other small RNA species.12 By detecting small RNA sequences from TCMs in human blood cells and tissues, a large amount of TCMs-derived small RNAs were demonstrated to enter mammalian cells and tissues and regulate mammalian gene expression in a sequence-dependent manner, which promotes the possibility of developing TCMs-derived small RNAs as molecular drugs.13
Transfer RNA (tRNA), the most abundant species of small RNA, can undergo endonucleolytic cleavage by specific ribonucleases into tRNA-derived fragments (tRFs). Recent studies have revealed that endogenous tRFs are associated with cellular functions and disease pathologies by mediating post-transcriptional gene expression through targeting mRNA in the pathway analogous to miRNA and small interfering RNA (siRNA).14, 15, 16 Recently, our studies demonstrated that a tRF derived from the medicinal plant Chinese yew produced effects comparable to those of Taxol by targeting TRPA1 on ovarian cancer.17 These findings, therefore, prompted us to speculate that tRFs in TCMs may be an important group of active compounds and hold great potential as RNAi-based therapeutics.
Ginseng, reputed to be the most valuable and famous TCM, known as the “king” of TCM, is extensively used as the effective therapeutic interventions for various human diseases, including cardiovascular disease.18 In particular, ginsenosides, which are regarded as the main active components of ginseng, have been found to play an important role in cardioprotection.19 Therefore, we undertook to investigate whether tRFs derived from ginseng possess any protective effects against myocardial ischemia/reperfusion injury. We document here our findings on one such tRF derived from ginseng, HC83, which exhibited significant cardioprotective effects with high potency against ischemia/reperfusion injury both in vitro and in vivo. The mechanism of action appears to involve the direct downregulation of a long non-coding RNA (lncRNA) called MIAT (myocardial infarction-associated transcript) that leads to a subsequent upregulation of vascular endothelial growth factor A (VEGFA) expression.
Results
Ginseng tRNAs protected cardiomyocytes
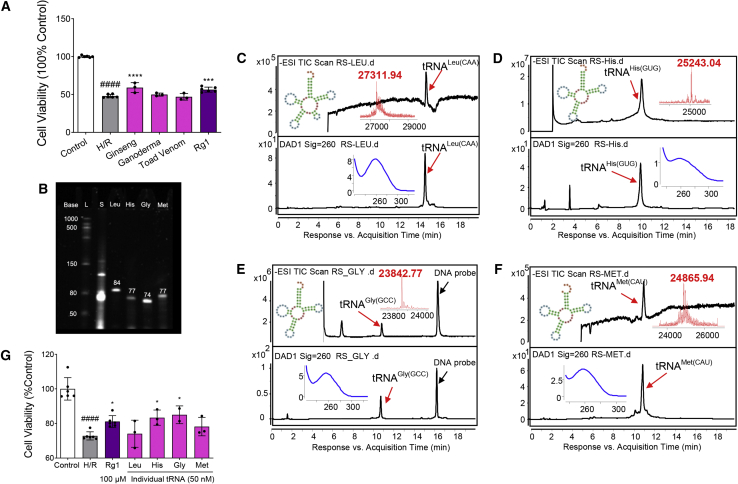
tRNA-enriched fractions (TEFs) containing a complete set of tRNA species of ginseng were prepared to investigate the cardioprotective effects of ginseng tRNAs. As shown in Figure 1A, hypoxia/reoxygenation (H/R) injury significantly decreased the cell viability of H9c2 cells to 48% of the control group. TEFs, at 100 nM, increase cell viability to approximately 60%. In contrast, no significant effects were observed with TEF similarly prepared from other TCMs, such as ganoderma and toad venom, which served as negative controls.
Figure 1.
Ginseng tRNAs promoted cell survival after H/R injury
(A) Effects of tRNA-enriched fractions (TEFs) derived from ginseng, ganoderma, and toad venom on H9c2 cell viability at 100 nM after exposure to hypoxia/reoxygenation (H/R). Ginsenoside Rg1 at 100 μM was used as a positive control. TEF derived from ginseng exhibited a significant cardioprotective effect. Data are shown as the means ± SDs of 3 independent experiments. ####p < 0.0001 versus control; ∗∗∗p < 0.001 versus H/R; ∗∗∗∗p < 0.0001 versus H/R. p values were calculated using 1-way ANOVA. (B) Urea-PAGE analysis of small RNAs (lane 2) and 4 individual tRNAs purified from ginseng, namely tRNALeu(CAA), tRNAHis(GUG), tRNAGly(GCC), and tRNAMet(CAU) (lanes 3–6, respectively). Unique bands were observed in the predicted positions at 84, 77, 74, and 77 mer, respectively. A low-range single-stranded RNA (ssRNA) ladder (lane 1) was used as the marker. (C–F) Representative mass spectra (top) and UHPLC-MS (bottom) chromatograms of tRNALeu(CAA), tRNAHis(GUG), tRNAGly(GCC), and tRNAMet(CAU), respectively. Red, deconvolution mass spectra. Blue, UV absorption spectra. (G) Effects of the 4 individual tRNAs at a concentration of 50 nM on cell survival after H/R determined by MTT. tRNAHis(GUG) and tRNAGly(GCC) showed significant cardioprotective effects. Ginsenoside Rg1 at 100 μM was used as a positive control. Data are shown as the means ± SDs of 3 independent experiments. ####p < 0.0001 versus control; ∗p < 0.05 versus H/R. p values were calculated using 1-way ANOVA.
tRNA sequences and their relative abundance in ginseng roots have been identified by using next-generation sequencing (NGS) technique in our previous study.20 The top 10 were specifically isolated from TEFs and analyzed by using urea-denatured polyacrylamide gel electrophoresis (urea-PAGE) and ultra-high-performance liquid chromatography-quadrupole time-of-flight mass spectrometry (UHPLC-QTOF/MS). Four tRNA species, including tRNALeu(CAA), tRNAHis(GUG), tRNAGly(GCC), and tRNAMet(CAU), demonstrate top content and high quality. As shown in Figure 1B and Table 1, their unique bands were observed in their predicted positions at 84, 77, 74, and 77 mer, respectively. UHPLC-QTOF/MS analysis showed that all four tRNAs of ginseng gave baseline resolved peaks in UV spectra at 260 nm (Figures 1C–1F). The deconvoluted mass spectrum (see inset) showed that the accurate molecular weights of these tRNAs were higher than those theoretically calculated from the gene sequences that were reported in National Center for Biotechnology Information (NCBI): KF431956.1. The added mass may be a result of chemical modifications on these native tRNAs (Table 1). These results confirmed that highly purified tRNAs with post-transcriptional modifications from ginseng were obtained.
Table 1.
Information of individual tRNAs purified from ginseng TEF
| tRNA | Sequence (5′ > 3′)a | Length (mer) | Molecular weight (Da) |
|
|---|---|---|---|---|
| Calculated | Actual | |||
| tRNALeu(CAA) | 5′-GCCUUGGUGGUGAAAUG GUAGACACGCGAGACUCAA AAUCUCGUGCUAAAGAGCG UGGAGGUUCGAGUCCUCU UCAAGGCACCA-3′ |
84 | 27,200.38 | 27,311.94 |
| tRNAHis(GUG) | 5′-GCGGAUGUAGCCAAGUG GAUCAAGGCAGUGGAUUG UGAAUCCACCAUGCGCGG GUUCAAUUCCCGUCGUU CGCCCCA-3′ |
77 | 24,838.9 | 25,243.04 |
| tRNAGly(GCC) | 5′-GCGGAUAUAGUCGAAUG GUAAAAUUUCUCUUUGCC AAGGAGAAGACGCGGGUU CGAUUCCCGCUAUCCGCCCCA-3′ |
74 | 23,812.29 | 23,842.77 |
| tRNAMet(CAU) | 5′-CGCGGAGUAGAGCAGUU UGGUAGCUCGCAAGGCUC AUAACCUUGAGGUCACGG GUUCAAAUCCUGUCUCC GCAACCA-3′ |
77 | 24,806.90 | 24,865.94 |
Sequences were acquired by next-generation sequencing of ginseng TEF.
Figure 1G shows that only two of the four tRNAs—tRNAGly(GCC) and tRNAHis(GUG)—showed significant cardioprotective effects, increasing cell viability by approximately 10% at a concentration of 50 nM, which is comparable to the effects of the positive control ginsenoside Rg1 at 100 μM.19 This may translate to a 2,000-fold higher potency exhibited by the ginseng tRNAs in comparison to ginsenosides. Thus, these tRNAs may contribute more significantly to the overall cardioprotective effects of ginseng than that of ginsenosides.
Ginseng tRFs protected cardiomyocytes
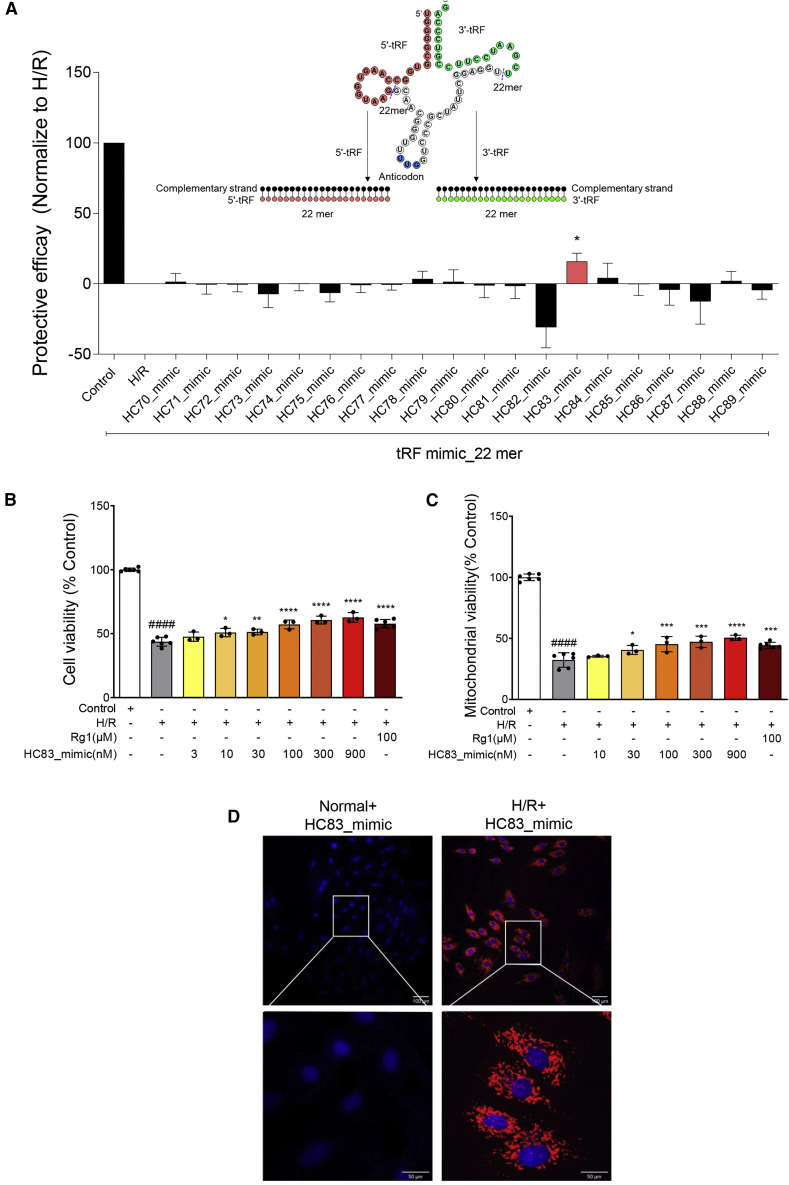
Twenty 22-mer tRF_mimics consisting of tRFs of ginseng roots and their complementary sequence were synthesized and subsequently screened at 300 nM for cardioprotective activities using the cell viability assay. As shown in Figure 2A, one of the tRF_mimics, namely HC83_mimic, increased cell viability by 15% (p < 0.05 versus H/R), thus exhibiting significant protective effects against H/R injury. The HC83_mimic, which is a double-strand tRF_mimic consisting of HC83 (a 22 mer 3′ tRF derived from tRNAGln(UUG) of ginseng roots) and its complementary sequence, was then studied in more detail to confirm its efficacy and potency for cardioprotection.
Figure 2.
Ginseng tRNA-derived fragments (tRFs) increased cell viability and mitochondrial viability
(A) Screening of protective activity 20 tRF_mimics derived from ginseng tRNA at 300 nM on H9c2 cells exposed to H/R (n = 6 per group). Diagrams of 5′ tRFs (red) and 3′ tRFs (green) are shown as insets. Red nucleotides represent the sequence of 5′ tRF, and green nucleotides represent the sequence of 3′ tRF. Both of them are derived from the tRNA of ginseng, which acts as the guide strand of tRF_mimic. HC83_mimic consisting of HC83 (a 22-mer 3′ tRF derived from tRNAGln(UUG) of ginseng roots) and its complementary sequence increased cell viability significantly. (B) Dose-dependent investigation of HC83_mimic by cell viability assay (n = 3 per group). (C) Dose-dependent investigation of HC83_mimic by mitochondrial viability assay (n = 3 per group). HC83_mimic increased cell viability and mitochondrial viability of H9c2 cells following H/R treatment in a dose-dependent manner. Ginsenoside Rg1 at 100 μM was used as a positive control. Data are shown as the means ± SDs. ####p < 0.0001 versus control; ∗p < 0.05 versus H/R; ∗∗p < 0.01 versus H/R; ∗∗∗p < 0.001 versus H/R; and ∗∗∗∗p < 0.0001 versus H/R. p values were calculated using 1-way ANOVA. (D) Fluorescence imaging of normal (a, b) and H/R-injured (c, d) H9c2 cells, which were treated with Cy5-labeled HC83_mimic at a concentration of 300 nM. Images were acquired by using a confocal laser microscope. Nuclei were stained with Hoechst 33342 (blue). HC83_mimics can efficiently enter into H/R-injured cardiomyocytes. a and c were observed under 20× objective, scale bars, 100 μm; b and d were magnified views of marked areas, scale bars, 50 μm.
HC83_mimic increased the cell viability of H9c2 cells following H/R treatment in a dose-dependent manner (Figure 2B). The maximum effect of 20% was achieved at 900 nM (H/R: 43% survival versus HC83_mimic: 63% survival). In addition, HC83_mimic exhibited a protective effect (57% survival) at 100 nM that was comparable to that of ginsenoside Rg1 at 100 μM. The data suggest that HC83_mimic is 1,000-fold more potent than ginsenoside Rg1. Similar findings were obtained when mitochondrial viability was evaluated. HC83_mimic was demonstrated to dose dependently enhance the mitochondrial viability by 8% to 18% as compared to the H/R group (p < 0.05–0.0001) (Figure 2C).
HC83_mimic was fluorescently labeled with Cy5 and visualized by confocal laser microscopy. Interestingly, a large amount of Cy5-labeled HC83_mimic (red fluorescence in Figure 2D) was observed in H/R-treated H9c2 cells, while no red fluorescence was observed in normal cells, implying that naked tRF_mimics can efficiently enter into H/R-injured cardiomyocytes but not the normal cells. For further confirmation, we also analyzed the effects of HC83_mimic transfected with lipofectamine RNAiMAX reagents in normal and H/R-injured H9c2 cells, respectively. The results showed that HC83_mimic has little effect and low cytotoxicity in normal H9c2 cells, but shows some cytotoxicity in H/R-injured H9c2 cells when transfected with transfection reagents (Figure S1). Therefore, we proposed that it is an advantage to take naked-HC83_mimic rather than transfecting with transfection reagents.
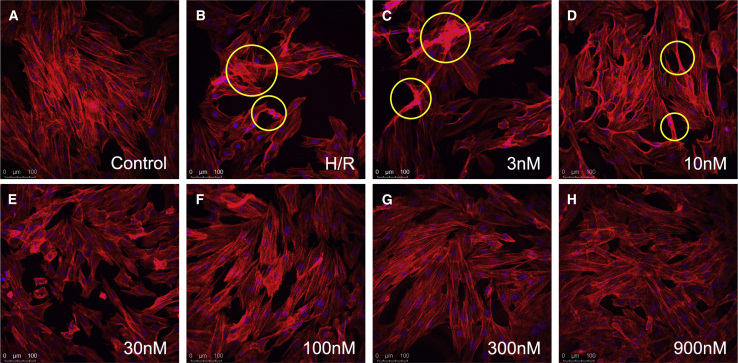
HC83_mimic relieved H/R-induced cytoskeleton destruction
Figure 3A shows the filamentous actin (F-actin) stained in red in control H9c2 cells. These long, fusiform fibers appeared in close contact with one another and were uniformly distributed along the long axis of the cells. After H/R treatment, the H9c2 cells exhibited irregular cell shapes with obvious cell shrinkage (Figure 3B). The F-actin (red) was ruffled around the nuclei (blue) and became much less intense or even absent (those marked with yellow circles in Figure 3). When treated with HC83_mimic (≥100 nM), the H9c2 cells showed minimal cytoskeletal damage, as shown in Figures 3F–3H. It may be concluded that HC83_mimic can remarkably relieve cytoskeleton destruction caused by H/R stress in a dose-dependent manner, which is consistent with the cell viability findings.
Figure 3.
HC83_mimic relieved H/R-induced cytoskeleton destruction
(A) The fluorescence images of F-actin (red) and nuclei (blue) of normal H9c2 cells. (B) The fluorescence images of H/R-injured H9c2 cells. (C–H) The images of H/R-injured cells that have been rescued with varying concentrations of HC83_mimic (3, 10, 30, 100, 300, and 900 nM). The yellow circles denote the ruffled F-actin. When treated with HC83_mimic (≥100 nM), the H9c2 cells showed minimal cytoskeletal damage. All of the images were acquired by using a confocal laser microscope with a 20× objective lens. Scale bars, 100 μm.
HC83_mimic attenuated H/R-induced mitochondrial dysfunction
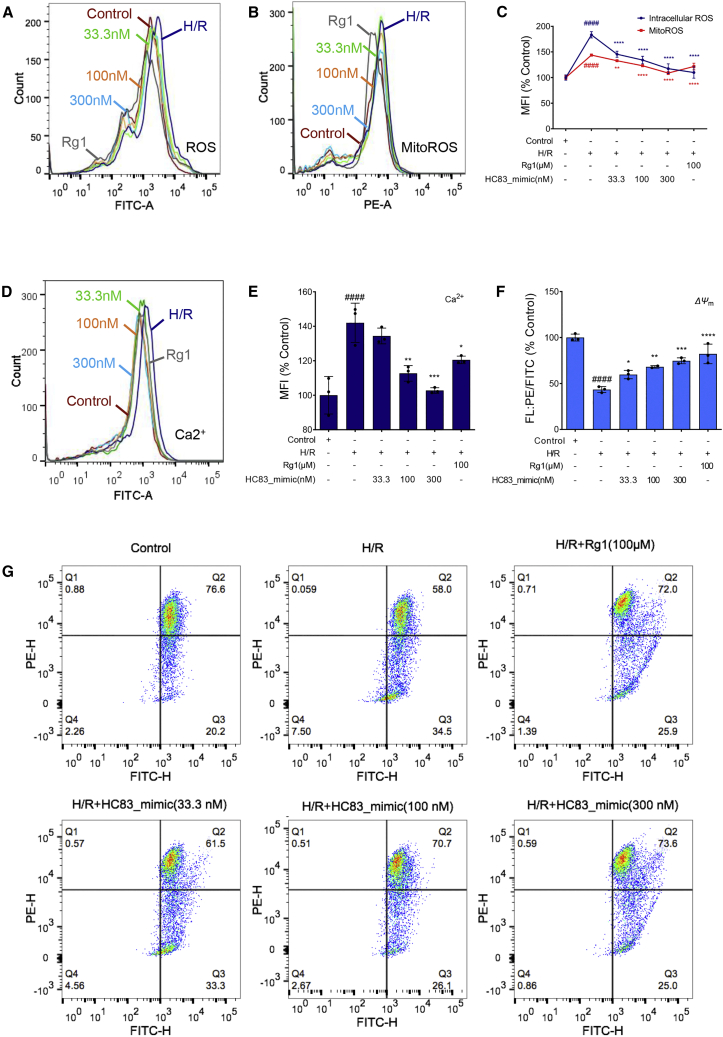
H/R induced significant increases in intracellular reactive oxygen species (ROS) (Figure 4A) and mitochondrial ROS (mitoROS) (Figure 4B) to 184% and 143%, respectively, when compared to their respective controls (p < 0.0001). HC83_mimic dose dependently reversed such effects, reaching 121% (p < 0.0001 versus H/R) and 108% (p < 0.01∼0.0001 versus H/R), respectively, at a concentration of 300 nM (Figure 4C). These results demonstrated that HC83_mimic could significantly suppress both intracellular and mitoROS generation induced by H/R.
Figure 4.
HC83_mimic attenuated H/R-induced mitochondrial dysfunction
HC83_mimic decreased the fluorescence intensity of (A) intracellular ROS and (B) mitochondrial ROS. (C) Quantification of the intracellular ROS and mitochondrial ROS level by MFI (median fluorescence intensity). (D) HC83_mimic inhibited the fluorescence intensity of intracellular Ca2+ overproduced by H/R. (E) Quantification of the intracellular Ca2+ level by MFI. (F) Quantification of the mitochondrial membrane potential (ΔΨm) was expressed as the ratio of red to green fluorescence intensity. (G) HC83_mimic stabilized ΔΨm in H/R-injured H9c2 cells. Disruption of ΔΨm due to mitochondria depolarization was presented as the shift from red to green fluorescence emission. All of the fluorescence intensities in H/R-treated H9c2 cells were detected by flow cytometry after H/R-injured H9c2 cells were treated with HC83_mimic (33.3, 100, and 300 nM). Quantification was calculated with MFI. Ginsenoside Rg1 at 100 μM was used as a positive control. Data are shown as the means ± SDs of 3 independent experiments. ####p < 0.0001 versus control; ∗p < 0.05 versus H/R; ∗∗p < 0.01 versus H/R; ∗∗∗p < 0.001 versus H/R; and ∗∗∗∗p < 0.0001 versus H/R. p values were calculated using 1-way ANOVA.
In addition, intracellular Ca2+ levels were remarkably increased by H/R (142% of control, p < 0.0001), which were dose dependently reversed by HC83_mimic and totally abolished at 300 nM (Figures 4D and 4E, p < 0.01 ∼ 0.001 versus H/R). These results indicated that HC83_mimic suppressed the accumulation of intracellular Ca2+, which would result in Ca2+ overload and consequently cell damage/death.21
Consistently, Figure 4G showed that H/R caused remarkable disruption of mitochondrial membrane potential (ΔΨm), a key indicator of mitochondrial respiratory efficiency,22 presenting as the shift from red (phycoerythrin [PE]) to green (fluorescein isothiocyanate [FITC]) fluorescence emission. The ratio of red to green fluorescence intensity following H/R was significantly reduced to 43% of the control group (100%) (p < 0.0001; Figure 4F). Again, HC83_mimic treatment dose dependently attenuated the loss of mitochondrial membrane potential.
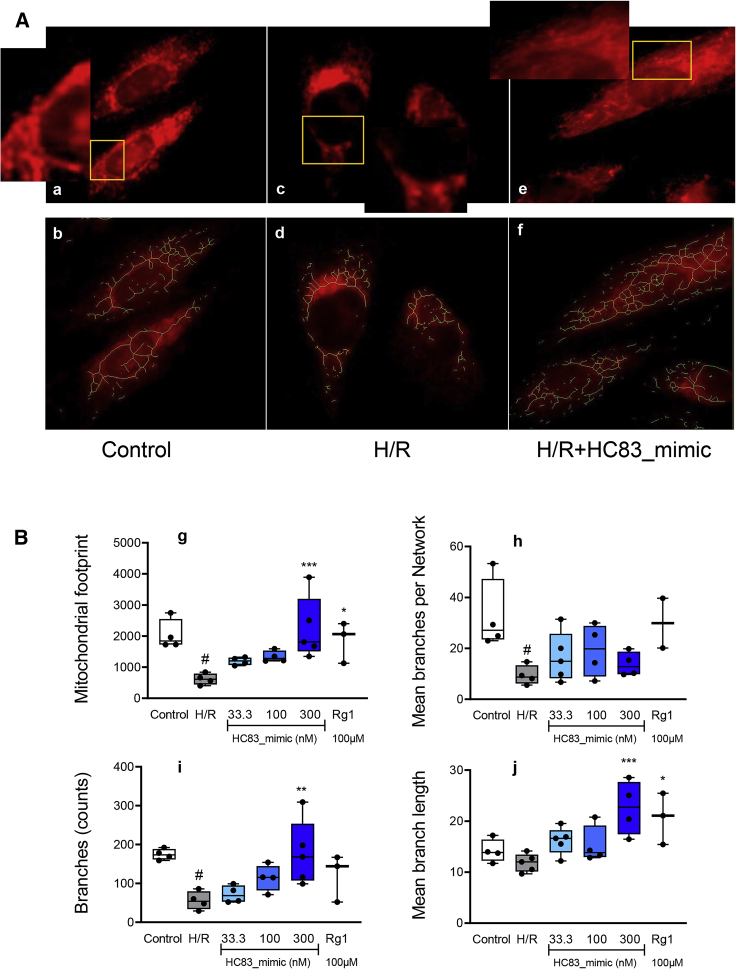
Mitochondrial function is strictly related to the morphology and network. Therefore, we analyzed the mitochondrial morphology and quantified the mitochondrial network after staining mitochondria in H9c2 cells by using Mito-Tracker Red CMXRos (C1035, Beyotime Biotechnology).23 The mitochondrial images were captured from a Delta Vision Elite Imaging System (GE Healthcare Life Sciences) and analyzed by using an Image J (NIH) macro tool, MiNA.24,25 The mitochondrial footprint, branches, mean branches per network, and mean branch length were obtained. As shown in Figure 5A, long and highly interconnected mitochondria were observed in normal H9c2 cells (a), and a branched mitochondrial network was spread evenly throughout the cells (b). When induced by H/R, the percentage of cross-linked mitochondria and the length of the network branches were significantly decreased (c, d), whereas they were restored by treating them with HC83_mimic (e, f). These results were also quantified by using GraphPad Prism 9.0 and displayed in Figure 5B. H/R induced a significant reduction in the number of mitochondria (g) as well as in the length of mitochondrial branches (h). Both the branches and the mean number of branches per network were also decreased (i, j). In comparison, treating with various concentrations of HC83_mimic (33.3, 100, and 300 nM) resulted in a significant improvement in all four mitochondrial metrics (g–j). Taken together, it may be concluded from these results that HC83_mimic can prevent mitochondrial dysfunction by preserving mitochondrial integrity.
Figure 5.
Visualization and quantification of the mitochondrial network in H9c2 cells by using a MiNA ImageJ macro tool
(A) Visualization of representative cells for mitochondrial morphology analysis by using a Delta Vision Elite Imaging System. All of the mitochondria in cells were immunofluorescence stained with Mito-Tracker Red. a, c, and e: Representative cells; the high magnification images are labeled with yellow frames; b, d, and f: Skeletonization of mitochondrial shapes and structures (green). (B) The original images were processed with the MiNA ImageJ macro tool by using “unsharp mask,” “CLAHE,” “median,” “binarize,” “skeletonize,” and the MiNA plugin for quantifying the mitochondrial footprint (g), mean branches per network (h), branches (i), and mean branch length (j). Ginsenoside Rg1 (100 μM) was used as a positive control. Data are shown as the means ± SDs. #p <0.01 versus control; ∗p < 0.05 versus H/R; ∗∗p < 0.001 versus H/R; ∗∗∗p < 0.001 versus H/R. p values were calculated using 1-way ANOVA.
HC83_mimic reversed the upregulation of lncRNA MIAT and downregulation of VEGFA induced by H/R
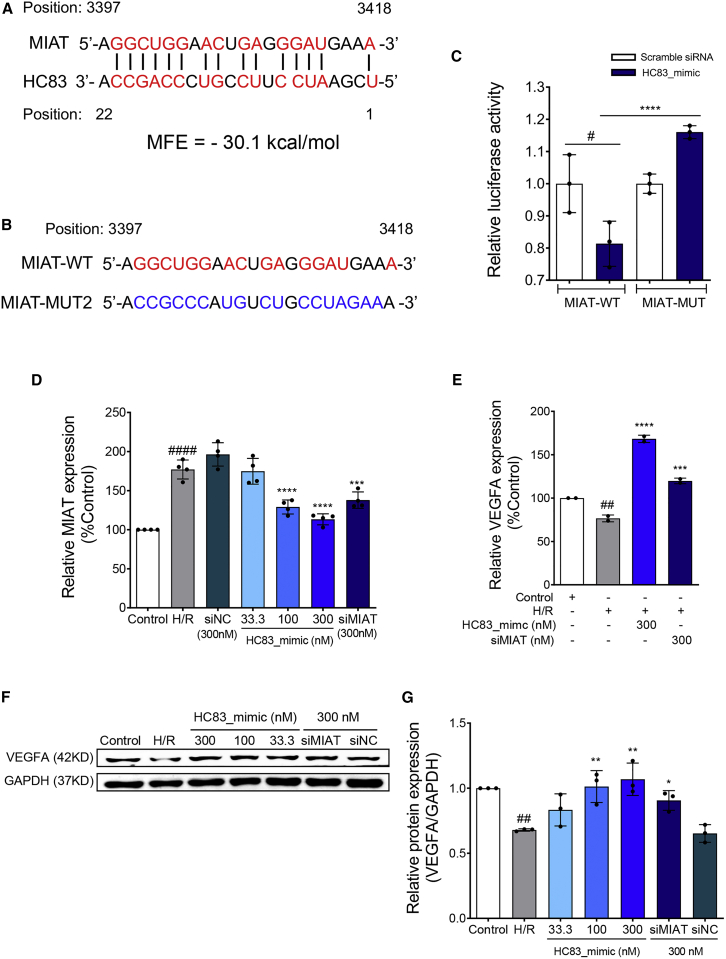
The target RNAs of HC83 were bioinformatically predicted with a well-proven analysis pipeline.26,27 Briefly, the sequence of HC83 was successively loaded into TargetScan and miRanda to predict its potential target against an in-house constructed 3′ UTR database of rat mRNA and lncRNA. The program parameters were set as follows: structure: ≥140, energy: <−10, context + score: 0. The top 10 targets were prioritized and filtered by literature. Table S1 shows the top 10 candidate targets with the highest structure scores with more negative energy.26,27 The results showed that MIAT, a long non-coding RNA, was identified as the most likely target for HC83 because it is the most implicated in the pathogenesis of IHD.28 MIAT contains a putative HC83 binding site in the nucleotide 3397–3418 region with minimum free energy (MFE) of −30.1 kcal/mol (Figure 6A). To confirm the binding site in MIAT, the wild-type (WT) or mutated HC83 binding site in MIAT (Figure 6B) was cloned into the pmiR-RB-Report vector and transfected into 293T cells. Luciferase reporter assay revealed that HC83_mimic significantly reduced the luciferase activity of MIAT-WT, indicating that HC83 binds at this site. However, the effect of HC83_mimic was not seen when the sequence of this site in MIAT was mutated (MIAT-MUT) (Figure 6C).29
Figure 6.
HC83 protected H/R-injured H9c2 cells by directly targeting lncRNA MIAT/VEGFA pathway
(A) Prediction of the target of HC83. The position and minimal free energy (MFE) of the HC83-binding site located within the lncRNA MIAT is schematically presented. The aligned nucleotides were presented as red. MIAT contains a putative HC83 binding site in the nucleotide 3397–3418 region with MFE of −30.1 kcal/mol. (B) The sequence of the wild-type (MIAT-WT) and mutated MIAT (MIAT-MUT) in the HC83-binding site. Red: The complementary sequence alignment between HC83 and MIAT. Blue: The mutated nucleotide sequence in MIAT. (C) Luciferase activity determined by luciferase reporter assay co-transfected with MIAT-WT or MIAT-MUT and HC83_mimic or scramble siRNA. HC83_mimic significantly reduced the luciferase activity of MIAT-WT. Data are shown as the means ± SDs of 3 independent experiments. #p < 0.05 versus MIAT-WT + scramble siRNA; ∗∗∗∗p < 0.0001 versus MIAT-WT + HC83_mimic. p values were calculated using 1-way ANOVA. (D) The RNA expression level of MIAT in H/R-injured H9c2 cells treated with HC83_mimic (33.3, 100, and 300 nM), siMIAT (positive control, 300 nM) and siNC (negative control, 300 nM) by using quantitative real-time PCR analysis. HC83_mimic reversed the upregulation of MIAT expression induced by H/R injury. (E) The mRNA expression level of VEGFA in H/R-injured H9c2 cells treated with HC83_mimic (300 nM) or siMIAT (300 nM) by using quantitative real-time PCR analysis. (F and G) The protein expression level of VEGFA in H/R-injured H9c2 cells treated with HC83_mimic (33.3, 100, and 300 nM) by using western blot analysis. siMIAT and siNC were used at 300 nM. Band intensity was normalized to those of GAPDH. HC83_mimic reversed the downregulation of VEGFA expression induced by H/R injury. The data are shown as the means ± SDs of 3 independent experiments. ##p < 0.01 versus control; ####p < 0.0001 versus control, ∗p < 0.05 versus H/R; ∗∗p < 0.01 versus H/R, ∗∗∗p < 0.001 versus H/R, ∗∗∗∗p < 0.0001 versus H/R. p values were calculated using 1-way ANOVA.
Quantitative real-time PCR experiments showed that the expression of MIAT was remarkably increased by 77% in H9c2 cells upon H/R treatment (p < 0.0001 versus control). More important, such upregulation of MIAT expression was significantly reversed in a dose-dependent manner by HC83_mimic and nearly abolished at 300 nM (p < 0.0001 versus H/R) (Figure 6D). At this concentration, HC83_mimic exerted a stronger suppressive effect than that of the siMIAT, a positive-control siRNA against MIAT.30 In contrast, the scrambled negative sequence (siNC) has no effect on the expression of MIAT (Figure 6D). We also analyzed the effects of HC83_mimic in the MIAT knockdown H9c2 cells; when H 9c2 cells were treated with siMIAT, the effects of HC83_mimic were abolished (Figure S2). All of these data strongly suggest a direct binding of HC83 in MIAT.
Simultaneous to the upregulation of MIAT following H/R, VEGFA, which is known to modulate angiogenesis in IHD,31,32 was observed to be downregulated at the mRNA level. Interestingly, treatment with HC83_mimic not only reversed this downregulation but also caused an overexpression of mRNA to approximately 168% of control (Figure 6E). Corroborating with the mRNA expression, VEGFA expression at the protein level was similarly reduced in H/R-injured cells, which was abolished by HC83_mimic (Figures 6F and 6G). These results may suggest that HC83_mimic protects cardiomyocytes against H/R injuries by directly binding to MIAT, thus preventing the upregulation of MIAT and downregulation of VEGFA.
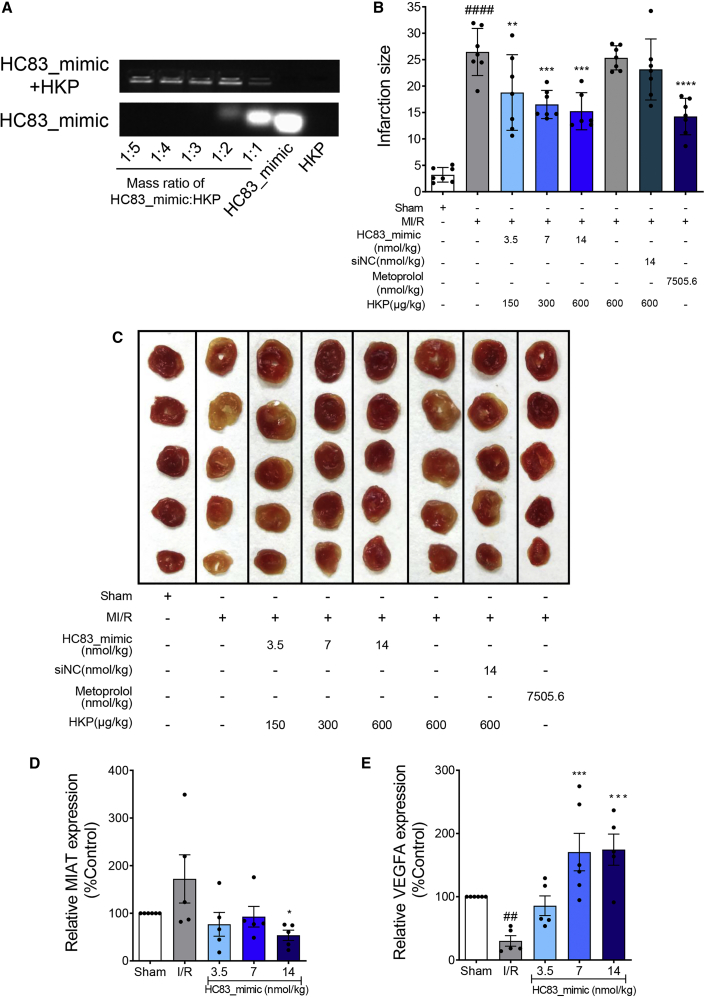
HC83_mimic ameliorated ischemic/reperfusion (I/R)-induced myocardial infarction in vivo
HC83_mimic was encapsulated in histidine-lysine polymer (HKP) nanoparticles for in vivo administration by intravenous injection. As shown in Figure 7A, when HKP was used at a mass ratio of ≥3 relative to the tRF_mimic, HC83_mimic was completely encapsulated. Figures 7B and 7C showed that the infarctions in the myocardium induced by ischemia/reperfusion injury were significantly and dose dependently reduced by treating with HC83_mimic. At the highest dose of 14 nmol/kg, the mean infarction size was significantly decreased by 42.4% in comparison to the myocardial ischemia/reperfusion (MI/R) group (15.2% ± 3.5% versus 26.4% ± 4.4%, p < 0.05). Comparable cardioprotective effects were obtained with positive control metoprolol but at a dose approximately 500-fold higher than that of HC83_mimic. As expected, both HKP (vehicle) and siNC did not exhibit any effects (Figure 7B). The quantitative real-time-PCR results showed that HC83_mimic can significantly reverse both the upregulation of MIAT and the downregulation of VEGFA induced by myocardial infarction in vivo (Figures 7D and 7E). Taken together, these findings strongly support the conclusion that HC83_mimic can effectively protect the heart against I/R injury both in vitro and in vivo by targeting the MIAT/VEGFA pathway.
Figure 7.
HC83_mimic decreased I/R-induced myocardial infarction size in vivo
(A) Agarose gel electrophoresis analysis of HC83_mimic-HKP nanoparticle to optimize the mass ratio. HC83_mimic was encapsulated with HKP at a mass ratio of 1:5 to 1:1. Naked HC83_mimic and HKP were used as control. When the mass ratio is above 1:3, all of the HC83_mimics were encapsulated. (B) Quantification of myocardial infarction size. (C) Representative pictures of myocardial infarction tissues stained with TTC. The white area represented infarcted myocardium. HC83_mimic significantly reduced the myocardial infarction size, at the dose of 14 nmol/kg, the mean infarction size was significantly decreased by 42.4% in comparison to the MI/R group. All of the HC83_mimics and siNC were encapsulated in HKP nanoparticles for in vivo administration by intravenous injection. HKP was used as a vehicle. Data are shown as the means ± SEMs (n = 6–7 per group). ####p < 0.0001 versus control; ∗∗p < 0.01 versus MI/R; ∗∗∗p < 0.001 versus MI/R; ∗∗∗∗p < 0.0001 versus MI/R. p values were calculated using 1-way ANOVA. (D) The RNA expression levels of MIAT. The results showed that HC83_mimic at 14 nmol/kg can significantly reverse the upregulation of MIAT induced by myocardial infarction in vivo (p < 0.05). (E) The RNA expression levels of VEGFA. The results showed that HC83_mimic at 7 and 14 nmol/kg can significantly reverse the downregulation of VEGFA induced by myocardial infarction in vivo (p < 0.001). Data are shown as the means ± SEMs, n = 5–6. ##p < 0.01 versus control; ∗p < 0.05 versus H/R; ∗∗∗p < 0.001 versus H/R. p values were calculated using 1-way ANOVA.
Discussion
Small non-coding RNAs (sncRNAs) are powerful regulators of gene expression, and they have been implicated in a variety of pathological processes such as cardiovascular disease and cancer.33,34 As the most abundant species of sncRNA, tRNA and tRFs are extensively reported to be involved in various diseases and exert multiple biological functions.35,36 Indeed, it has been proven that tRFs derived from Chinese yew exhibited efficient antitumor activities, which is the first report on the pharmacological activities of plant-derived tRFs.17 Together with the findings that TCM-derived small RNAs could be the active compounds responsible for their pharmacological activities,10,11 the present study expanded the active compounds in TCMs to tRFs. HC83, a 22 mer 3′ tRF derived from tRNAGln(UUG) of ginseng roots, exhibited extraordinarily potent effects on protecting cardiomyocytes from I/R injury both in vitro and in vivo. In addition, HC83 could improve cardiac function by maintaining both cytoskeleton integrity and mitochondrial function of cardiomyocytes, which are always damaged during H/R stress. Undoubtedly, a normal cytoskeleton is essential to keep the morphology and physical structure of cells to provide mechanical integrity for cellular progression. However, the myocardial cytoskeleton is highly sensitive to H/R stress, and the pathological remodeling of the cytoskeleton not only changes the metabolism and gene expression but also will lead to cell death.37,38 Mitochondrial dysfunction is also a major consequence of H/R as the indispensable role of mitochondria in energy generation. Signal pathways, including overproduction of intracellular and mitoROS, the overload of Ca2+, or collapse of mitochondrial membrane potential (ΔΨm), will impair mitochondrial functions and ultimately lead to cell apoptosis.39, 40, 41 To date, a variety of therapeutic approaches have been explored to relieve the cytoskeleton destruction or repair mitochondrial functions. Either total ginseng extract or individual ginsenoside has been proven to have effects on such pathologic conditions related to IHD.42,43 However, most of these molecules are required to execute functions at a high dosage of up to 1 μmol, which may cause severe side effects. Our study demonstrated that as low a dosage as 1 nmol of ginseng’s tRF_mimics is required to exert pharmacological effects, indicating an extraordinarily potent effect with a remarkable decrease of side effects linked to tRF therapy. Moreover, these tRFs derived from ginseng are also proposed to be developed as a novel type of drug candidate for various diseases induced by either cytoskeleton destruction or mitochondrial dysfunction, such as cancer and neurodegenerative diseases.44
There is increasing evidence that has led to a conceptual consensus that tRFs act in a way that is analogous to that of siRNAs or miRNAs.15,16 Consistent with the recognition, a lncRNA called MIAT was identified both bioinformatically and experimentally as the target of HC83. The results shown in Figures 6A–6D verified that HC83 can dramatically downregulate the MIAT expression by binding to its nucleotide 3397–3418 region. However, HC83 shows poor complementary matching with the seed region of MIAT (Figure 6A), which may reduce both the activity and gene silence efficiency. The MIAT is a well-proven risk factor for IHD; it was found to be aberrantly expressed in IHD patients participating in fundamental biological processes and pathophysiological developments in cardiovascular diseases.28,45 Previous studies have reported that MIAT is upregulated in the I/R-injured hearts of mice, and repressing it significantly improves cardiac functions.46,47 In this study, similar results were observed in H/R-injured cardiomyocytes. Therefore, HC83 may act in a more “potent” manner than small molecules, most likely RNA interference (RNAi), by which the harmful gene, MIAT, can be “silenced” directly. If so, then the HC83 could act catalytically during the RNAi process, and thus only a few molecules of HC83 per cell are required to silence MIAT molecules.48 This also explains why the tRFs of ginseng exhibited 3–4 orders of magnitude higher potency than that of small molecular drugs.
lncRNAs exert their functions in part via participating in gene expression-regulating networks; thus, downstream mRNA changes occurred concomitantly with the changes in lncRNA levels.49 VEGFA is one of the most potent cytokines in inducing pathological angiogenesis, which is essential for the salvage of ischemic myocardium at the early stage of MI.50 Intramyocardial injection of VEGFA, the most direct way to elevate the content of VEGFA in cardiomyocytes, was proposed as a therapeutic strategy to enhance collateral vessel formation in myocardial tissues.51,52 It has been examined in detail that VEGFA expression can be regulated by MIAT through sponging and sequestering the gene-repressing miRNA in endothelial cells and cancer cells in a competing endogenous RNA (ceRNA) regulatory manner.53,54 However, it remains unclear in damaged myocardial cells. In this study, a reciprocal negative regulation was identified between the MIAT level and the expression of VEGFA in H/R-injured H9c2 cells, where the knockdown of MIAT by HC83 caused a significant increase in both mRNA and protein levels of VEGFA. This relationship indicated a correlation of lncRNA with mRNA—that MIAT can directly bind VEGFA transcripts and cause steric hindrance to block translation, which may undergo other mechanisms different from ceRNA regulations.55 Although the more detailed mechanism underlying the MIAT-VEGFA interaction remains a further elaboration, we put forward a notion that the MIAT/VEGFA pathway was directly responsible for tRF-mediated post-transcriptional silencing in IHD.
This study substantiated that tRFs are the potent active components in TCMs responsible for their pharmacological activities, thus improving the promise of developing them as novel RNA drugs for RNAi-based therapeutics. tRF naturally presents in plants as a single strand, while in this study a double-stranded HC83_mimic, which contains not only the tRF single strand but also a completely complementary sequence, was applied to pharmacological studies. This construction strategy was adopted because (1) double-stranded tRF_mimic is substantially more chemically and biologically stable than single stranded;56 (2) double-stranded structure can facilitate the efficient loading of the tRF into the RNA-induced silencing complex (RISC), thereby enhancing the silencing effects;57 (3) double-stranded tRF_mimic shows more ability to cross cellular boundaries, thus enhancing the delivery efficiency.58 Moreover, we have previously demonstrated that tRF can exert functions similar to miRNA, which usually is in the form of synthetic miRNA_mimic to mimic the function of the endogenous miRNAs.17 As a result, double-stranded tRF_mimic has a considerable of advantages over single stranded as a therapeutic strategy. Unlike the conventional method of RNA drug development, which is based on the sequence of well-defined targeted genes relevant to diseases, our study presented a strategy to discover unknown or overlooked targets of diseases based on the sequence of tRF. As revealed in this study, the target of the tRF of ginseng was unambiguously identified and experimentally validated. The aberrantly expressed lncRNA linked to diseases was demonstrated as the direct target of tRF. Although extensive efforts have been made to develop siRNA drugs that target mRNAs,4,59,60 the low abundance, which is at least one order of magnitude lower than mRNAs, and the tissue-specific expression of lncRNA make it a positive advantage to be the target of siRNA drugs, resulting in a low-dose administration with high efficiency.61,62 Meanwhile, as we all know, the RNAi approach has been successful in the downregulation of gene expression; nevertheless, it remains a particular challenge to upregulate gene expression. lncRNAs as therapeutic targets could easily achieve the upregulation of desired genes.55,63,64 Therefore, therapeutic by targeting lncRNA presents a medical significance, emphasizing the importance of our strategy to discover new targets of diseases.
In summary, we provide the first evidence that tRF derived from ginseng exhibited extraordinarily potent protective effects against MI/R injury both in vitro and in vivo. This is also the first study demonstrating that TCM-derived tRFs exhibit potent pharmacological activities and exert function with a miRNA-like pathway. This indicates that TCM-derived tRFs could be an enormous benefit in providing a new sequence library for the development of miRNA therapeutics. We also present an innovative strategy bypassing the traditional approaches of siRNA drug exploitation, which leads to the promotion of the efficiency and feasibility of exploring new miRNA therapeutics as well as new targets. By using this new strategy, we have illustrated that RNA derived from TCMs plays regulatory roles by directly targeting lncRNA, which is different from siRNA downregulating gene expression by directly targeting mRNA. This indicates that the targets of diseases based on RNAi therapy could be extended to lncRNA, realizing the full potential targets of miRNA therapeutics. In a word, our studies provide a novel strategy not only for discovering pharmacologically active tRFs from TCMs but also for efficiently exploring new therapeutic targets for various diseases.
Materials and methods
RNA isolation and individual tRNA purification
Roots of Panax ginseng C. A. Mey were freshly collected from Fu-Song Town, Ji-Lin, China, and immediately stored in liquid nitrogen until use. Small RNAs (<200 mer) were extracted from ginseng by using a polysaccharase-aided RNA isolation (PARI) method, as previously described.20 Total tRNAs of ginseng (also referred to as TEFs) were further separated from small RNA samples by 6% urea-PAGE (Bio-Rad) and the bands corresponding to their predicted positions were recovered by electroelution in a 3-kD molecular weight cutoff dialysis tubing (Spectrum). Four individual tRNAs, namely tRNAHis(GUG), tRNAGly(GCC), tRNAMet(CAU), and tRNALeu(UAA), were purified from small RNA samples by specifically capturing the targeted tRNA molecules with biotinylated DNA probes.65 The probes were designed and synthesized based on the NGS data of ginseng tRNAs reported previously (Table S2). The integrity and purity of individual tRNAs were analyzed by 6% urea- PAGE (Bio-Rad). The average mass of each tRNA was determined by using an Agilent 6545 Q-TOF coupled to a 1290 Infinity UHPLC system as described previously.20
Design and synthesis of ginseng tRNA-derived fragments
Two tRF species, 5′ tRF and 3′ tRF, 22 mer in length, which are most prevalent in the plant kingdom, including TCM,66 were designed based on the gene sequences of 10 tRNA isoacceptors that are present in high abundance in ginseng roots (Table S3).20 The 5′ tRF series contains 5′ termini of mature tRNAs and are formed mainly by the cleavage in the D-domain of tRNAs. The 3′ tRFs contain 3′ CCA termini of mature tRNAs and are formed by the cleavage in the T-domain (see insets in Figure 2A). tRFs naturally present in ginseng root have been analyzed by using the UHPLC-QTOF-MS method,20 but were hardly detected due to both the extremely low abundance of tRFs and the insufficient sensitivity of mass spectrometers (Figure S3). As a result, all of the tRFs were chemically synthesized as guide strands and hybridized with their complementary strands (also referred to as passenger strands) to form tRF_mimics.67 The qualities of these tRF_mimics were analyzed by using an LC-MS method described in our previous studies.20
Cell culture and H/R treatments
Rat cardiomyocyte-derived cell line H9c2 was purchased from American Type Culture Collection (ATCC) and cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% v/v fetal bovine serum (FBS) and 1% v/v penicillin/streptomycin at humidified atmosphere containing 5% CO2 at 37°C. Hypoxic treatment was conducted by incubating cells in glucose-free DMEM under conditions of 94.9% N2/5% CO2/0.1% O2 for 12 h in a hypoxystation (Whitley H35 hypoxystation, Don Whitley Scientific), and then reoxygenation was carried out by incubating cells in growth medium for 6 h under normoxic conditions, as described above. All of the drugs were added into the medium before and retained in the culture medium over the entire period of reoxygenation.
Cell viability assay
H9c2 cells were plated in 96-well plates (Labware) at a density of 5,000 cells per well and allowed to adhere for 24 h before treatment. After H/R treatment, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution (0.5 mg/mL solution, 100 μL per well) was added and incubated at 37°C for 4 h. A volume of 150 μL dimethyl sulfoxide (DMSO) was then added and well shaken, and the resulting solutions were calorimetrically measured at 570 nm using a SpectraMax Paradigm multi-mode microplate reader (Molecular Devices,) to obtain the optical densities. Ginsenoside Rg1 (100 μM) was used as a positive control.
Mitochondrial viability assay
H9c2 cells were plated in 96-well black plates (Corning) at a density of 5,000 cells per well and allowed to adhere for 24 h before treatment. After H/R treatment, mitochondrial viability stain solution was added according to the manufacturer’s instructions. Fluorescence intensity was detected at 550 nM excitation and 590 nM emission using SpectraMax Paradigm multi-mode microplate reader (Molecular Devices). Ginsenoside Rg1 (100 μM) was used as a positive control.
Fluorescent detection of Cy-5 labeled HC83_mimic
H9c2 cells were cultured in μ-slide 8-well plates (Ibidi GmbH) at a density of 1 × 104 cells per well with a growth medium and allowed to adhere for 24 h before treatment. After exposure to H/R treatment with Cy-5 labeled HC83_mimic at 300 nM, the H9c2 cell nuclei were stained with Hoechst 33342 (Sigma) according to the manufacturer’s instruction. Images were then captured using a Leica TCS SP8 Confocal Laser Microscope with a 20× objective (TCS SP8).
Immunofluorescent imaging of F-actin
H9c2 cells were cultured in μ-slide 8-well plates (Ibidi GmbH) with a growth medium and allowed to adhere for 24 h before treatment. After exposure to H/R treatment with HC83_mimic (3–900 nM), the F-actin of cytoskeleton and cell nuclei were stained with rhodamine phalloidin (Cytoskeleton) and Hoechst 33342 (Sigma), respectively. Images were captured using a Leica TCS SP8 Confocal Laser Microscope with a 20× objective (TCS SP8).
Intracellular and mitoROS concentrations
H9c2 cells were cultured in 6-well plates (Ibidi GmbH) at a density of 10 × 104 cells per well and allowed to adhere for 24 h before treatment. After exposure to H/R treatment with HC83_mimic (33.3, 100, and 300 nM), all of the cells were collected and washed twice with pre-warmed PBS. Then, intracellular ROS levels were determined by using CM-H2DCFDA Oxidative Stress Indicator (Invitrogen), while mitoROS levels were measured by MitoSOX Red Mitochondrial Superoxide Indicator (Invitrogen). After being incubated with indicators, all of the cells were harvested by trypsinization and subjected to ROS concentration analysis by using a BD FACSAria III flow cytometry. ROS levels were finally evaluated by the median fluorescence intensity (MFI), which was calculated by FlowJo software.68 Ginsenoside Rg1 (100 μM) was used as a positive control.
Intracellular calcium concentrations
Intracellular calcium concentrations were measured using Fluo-4 AM Calcium Indicator (Invitrogen) according to the manufacturer’s instructions. The fluorescence intensity was determined by BD FACSAria III flow cytometry (Ex 488 nm/Em 530 nm), and the calcium concentration was evaluated by the MFI, which was calculated by FlowJo software.68 Ginsenoside Rg1 (100 μM) was used as a positive control.
Mitochondrial membrane potential
Mitochondrial membrane potential (ΔΨm) was measured using a MitoProbe JC-1 Assay Kit (Invitrogen). The fluorescence intensity was determined by BD FACSAria III flow cytometry with Ex 488 nm/Em 530nm and Ex 550 nm/Em 600 nm filters. ΔΨm was evaluated by calculating the ratio of red to green fluorescence. Ginsenoside Rg1 (100 μM) was used as a positive control.
Visualization and quantification of the mitochondrial network
H9c2 cells were cultured in μ-slide 8-well plates (Ibidi GmbH) with a growth medium and allowed to adhere for 24 h before treatment. Cells with non-treated or exposed to H/R treatment with HC83_mimic (33.3, 100, and 300 nM) were stained with Mito-Tracker Red CMXRos (C1035, Beyotime Biotechnology).23 After being fixed in 4% paraformaldehyde, images were captured using a Delta Vision Elite Imaging System with a 40× objective (GE Healthcare Life Sciences).24,25 The original images were processed with the MiNA Image J macro tool by using “unsharp mask,” “CLAHE,” “median,” “binarize,” “skeletonize,” and the MiNA plugin for analyzing the mitochondrial footprint, mean branches per network, branches, and mean branch length. The results were also quantified by using GraphPad Prism 9.0. Ginsenoside Rg1 (100 μM) was used as a positive control.
Prediction of the target of HC83
TargetScan and miRanda software were used to predict the target of HC83 in combination with an in-house constructed rat 3′ UTR mRNA and lncRNA database (KangChen Bio-tech).26,27 In TargetScan, the context plus score (context+) was set at 0, which is based on site type and site number to filter out low-efficacy targets.69 In miRanda, high-efficacy targets with a structure score >140 were retained. MFE was calculated by RNAhybrid software.70 The intersection of results from the two programs was used. The top 10 scored targets (including mRNA and lncRNA) with the highest structure scores from miRanda and more negative context+ score from TargetScan underwent a thorough literature search for functional annotation (Table S1).
Luciferase reporter assay
A WT fragment of rat MIAT containing the HC83 binding site (MIAT-WT) was PCR amplified and subcloned into the downstream of the luciferase gene in the pmiR-RB-Report vector (Ribobio). A mutant derivative devoid of the HC83 binding site was constructed to generate the MIAT-MUT vector. HEK293T cells were plated in 96-well plates at a density of 1 × 104 cells per well and allowed to adhere for 24 h. After co-transfected for 48 h with 50 nM HC83_mimic or scrambled siRNA (scrambled siRNA, sense: GGCGUGCGAAAUGGAGUUCGGA, antisense: UCCGAACUCCAUUUCGCAC GCC) and 50 ng of luciferase constructs by using Lipo6000 transfection reagent (Beyotime), firefly luciferase activity was measured with a Dual-Glo Luciferase Assay System (Promega) and normalized to the Renilla luciferase activity.
Quantitative real-time PCR analysis
Expression levels of lncRNA and mRNA were quantified by SYBR Green real-time PCR. Briefly, total RNA was extracted from H9c2 cells treated with HC83_mimic after exposure to H/R by using TRIzol reagent. After DNase I (Thermo Fisher Scientific) treatment, cDNA libraries were constructed by using RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). Then, quantitative real-time PCR reactions were conducted in the ViiA 7 Real-Time PCR System (Life Technologies) using GoTaq qPCR Master Mix (Promega) according to the manufacturer’s instructions. A siRNA against MIAT (siMIAT) reported previously (sense: CCAGGCUCCUUUAAACCAATT, antisense: UUGGUUUAAAGGAGCCUGGTT) was used as the positive control.30 Each experiment was carried out in triplicate. Relative expression differences were calculated by the 2−ΔΔCt method.71 Threshold cycle (Ct) values were normalized to the expression levels of β-actin. The PCR primers were designed using Primer-BLAST and subsequently synthesized by BGI (Table S4). The amplification efficiency for primers was also verified. Briefly, total RNA derived from H9c2 cells was subjected to several dilutions for the amplification efficiency of primers. The reverse transcription process was carried out to generate cDNA, which is used as a template for qPCR and amplified as described above. The calibration curves were constructed by plotting the Ct value against log concentration (1–10−4). Then, the amplification efficiency was calculated from the slope ().72 The primers were shown to have high specificity, with linear amplification efficiencies between 91.8% and 104.2% (Figure S4; Table S4).
Western blot analysis
Cell lysates were extracted using RIPA buffer (Cell Signaling Technology) supplemented with protease and phosphatase inhibitor cocktails (Roche). Protein concentrations were determined by bicinchoninic acid (BCA) protein assay (Bio-Rad). After denaturation at 95°C for 10 min, proteins were separated by 10% SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) membranes, and blocked with 5% bovine serum albumin (Thermo Fisher Scientific) for 1 h. Membranes containing target proteins were incubated overnight with primary antibody against VEGFA (1:1,000 dilution, Abcam) or glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (1:1,000 dilution, Cell Signaling Technology), followed by washing in 1× TBST (Tris-buffered saline with 0.1% Tween 20 detergent) and incubation with a fluorescent secondary antibody (Abcam) for detection. Band intensity was measured with Image J and normalized to those of GAPDH. A siRNA described above (siMIAT) and a siNC were used as positive and negative controls, respectively.
Preparation of HKP-HC83_mimic nanoparticles
HKP (2.4 mg) was dissolved in 10 mL RNase-free water and kept overnight at 4°C. An equal volume of HC83_mimic solutions (80 μg/mL) was then added to the HKP solution and incubated at room temperature for 30 min. The encapsulation efficiency of HC83_mimic in HKP nanoparticles was evaluated by gel retardation on 4% agarose gel electrophoresis (Bio-Rad).73
Animal care
C57BL/6 mice (male, 20 ± 2 g) were obtained from the Model Animal Research Center of Yangzhou University. All of the mice were housed in a controlled room with a 12-h light/dark cycle and access to food and water ad libitum. All of the experimental procedures were performed in accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals, and the protocols used were approved by the Animal Ethics Committee of China Pharmaceutical University.
MI/R model in mice
MI/R was performed as previously described.74 In brief, mice were anesthetized with 4% chloral hydrate injected intraperitoneally (10 mL/kg). The left descending coronary artery (LAD) was ligated with a 6–0 silk suture at approximately 3–4 mm from its origin. After 20 min, the occlusion was released to allow reperfusion. The sham group underwent the same procedure but without LAD ligation.
Mice were assigned randomly into 8 groups (n = 6–7/group) including sham, MI/R, MI/R + HC83_mimic at 3.5, 7, and 14 nmol/kg (encapsulated in HKP nanoparticles), MI/R + HKP at 600 μg/kg, MI/R + siNC at 14 nmol/kg (encapsulated in HKP nanoparticles), and MI/R + metoprolol at 7,505.6 nmol/kg (AstraZeneca), which is a clinical cardioprotectant used as a positive control. All RNA drugs and saline (for controls) were administered intravenously just after LAD ligation. Metoprolol was intragastrically administered just after LAD ligation. Mice were sacrificed 24 h after reperfusion, and hearts were collected for infarct size assessment.
Analysis of infarct size of cardiac necrosis
All hearts were sliced into 2-mm short-axis sections and incubated in 2% 2,3,5-triphenyltetrazolium chloride (TTC) at 37°C for 20 min in the dark, and then photographed digitally. The white infarcted areas were measured by Image J and expressed as a percentage of the total left ventricular (LV) area. Data were analyzed by GraphPad Prism 6.0 and represented as means ± SEMs.
Statistical analysis
GraphPad Prism 6.0 was used for statistical analysis. Data are expressed as means ± SDs. One-way ANOVA was used for multiple comparisons. For analysis of the infarction size of animal study, data are expressed as means ± SEMs. In all of the figures, significance was tested at ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
Acknowledgments
This work was financially funded by Macao Science and Technology Development Fund, Macau SAR (file no. 0082/2019A2 to Z.-H.J.). We are grateful to Dr. Yang Lu (Sirnaomics) for the gift of HKP for RNA delivery in vivo and his technical advice. We thank Dr. Pei Luo of Macau University of Science and Technology for technical advice in in vitro and in vivo studies. This work was partially supported by Ruina (Zhuhai Hengqin) Biothechnology.
Author contributions
Z.-H.J., T.-M.Y., and K.H. conceived the study. K.H., T.-M.Y., and J.-P.K. designed the experiments. K.H., T.-M.Y., F.L., X.-R.M., Q.L., J.-C.L., and Y.P. conducted the experiments. K.H. and F.L. analyzed the data. K.H., T.-M.Y., and Z.-H.J. drafted the manuscript. All of the authors have reviewed and approved the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.08.014.
Supplemental information
Data availability
The data that support the findings of this study are available within the article and its supplemental information.
References
- 1.Organization W.H. 2016. The Top 10 Causes of Death.https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death [Google Scholar]
- 2.Guo J., Yong Y., Aa J., Cao B., Sun R., Yu X., Huang J., Yang N., Yan L., Li X., et al. Compound danshen dripping pills modulate the perturbed energy metabolism in a rat model of acute myocardial ischemia. Sci. Rep. 2016;6:37919. doi: 10.1038/srep37919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai W.W., Xing Y.F., Wang B., Lu X.T., Wang Y.B., Sun Y.Y., Liu X.Q., Guo T., Zhao Y.X. Tongxinluo improves cardiac function and ameliorates ventricular remodeling in mice model of myocardial infarction through enhancing angiogenesis. Evid. Based. Complement. Alternat. Med. 2013;2013:813247. doi: 10.1155/2013/813247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balwani M., Sardh E., Ventura P., Peiró P.A., Rees D.C., Stölzel U., Bissell D.M., Bonkovsky H.L., Windyga J., Anderson K.E., et al. Phase 3 trial of RNAi therapeutic Givosiran for acute intermittent porphyria. N. Engl. J. Med. 2020;382:2289–2301. doi: 10.1056/NEJMoa1913147. [DOI] [PubMed] [Google Scholar]
- 5.Wang Z., Wu W., Guan X., Guo S., Li C., Niu R., Gao J., Jiang M., Bai L., Leung E.L., et al. 20(S)-Protopanaxatriol promotes the binding of P53 and DNA to regulate the antitumor network via multiomic analysis. Acta Pharm. Sin. B. 2020;10:1020–1035. doi: 10.1016/j.apsb.2020.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Irfan M., Kwak Y.S., Han C.K., Hyun S.H., Rhee M.H. Adaptogenic effects of Panax ginseng on modulation of cardiovascular functions. J. Ginseng Res. 2020;44:538–543. doi: 10.1016/j.jgr.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yarnell E. Synergy in herbal medicines: Part 1. J. Restor. Med. 2015;4:60–73. doi: 10.14200/jrm.2015.4.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L., Hou D., Chen X., Li D., Zhu L., Zhang Y., Li J., Bian Z., Liang X., Cai X., et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 2012;22:107–126. doi: 10.1038/cr.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chin A.R., Fong M.Y., Somlo G., Wu J., Swiderski P., Wu X., Wang S.E. Cross-kingdom inhibition of breast cancer growth by plant miR159. Cell Res. 2016;26:217–228. doi: 10.1038/cr.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Z., Li X., Liu J., Dong L., Chen Q., Liu J., Kong H., Zhang Q., Qi X., Hou D., et al. Honeysuckle-encoded atypical microRNA2911 directly targets influenza A viruses. Cell Res. 2015;25:39–49. doi: 10.1038/cr.2014.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du J., Liang Z., Xu J., Zhao Y., Li X., Zhang Y., Zhao D., Chen R., Liu Y., Joshi T., et al. Plant-derived phosphocholine facilitates cellular uptake of anti-pulmonary fibrotic HJT-sRNA-m7. Sci. China Life Sci. 2019;62:309–320. doi: 10.1007/s11427-017-9026-7. [DOI] [PubMed] [Google Scholar]
- 12.Shekhawat M., Jahagirdar D., Yadav S., Sharma N.K. Induction of apoptosis in HeLa by corn small RNAs. Nutr. Cancer. 2019;71:348–358. doi: 10.1080/01635581.2018.1526307. [DOI] [PubMed] [Google Scholar]
- 13.Huang F., Du J., Liang Z., Xu Z., Xu J., Zhao Y., Lin Y., Mei S., He Q., Zhu J., et al. Large-scale analysis of small RNAs derived from traditional Chinese herbs in human tissues. Sci. China Life Sci. 2019;62:321–332. doi: 10.1007/s11427-018-9323-5. [DOI] [PubMed] [Google Scholar]
- 14.Goodarzi H., Liu X., Nguyen H.C.B., Zhang S., Fish L., Tavazoie S.F. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 2015;161:790–802. doi: 10.1016/j.cell.2015.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar P., Anaya J., Mudunuri S.B., Dutta A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014;12:78–91. doi: 10.1186/s12915-014-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuscu C., Kumar P., Kiran M., Su Z., Malik A., Dutta A. tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA. 2018;24:1093–1105. doi: 10.1261/rna.066126.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao K.Y., Yan T.M., Zhang J.Z., Chan T.F., Li J., Li C., Lai-Han Leung E., Gao J., Zhang B.X., Jiang Z.H. A tRNA-derived fragment from Chinese yew suppresses ovarian cancer growth via targeting TRPA1. Mol. Ther. Nucleic Acids. 2022;27:718–732. doi: 10.1016/j.omtn.2021.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J.H. Cardiovascular diseases and Panax ginseng: a review on molecular mechanisms and medical applications. J. Ginseng Res. 2012;36:16–26. doi: 10.5142/jgr.2012.36.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong G., Chen T., Ren X., Zhang Z., Huang W., Liu L., Luo P., Zhou H. Rg1 prevents myocardial hypoxia/reoxygenation injury by regulating mitochondrial dynamics imbalance via modulation of glutamate dehydrogenase and mitofusin 2. Mitochondrion. 2016;26:7–18. doi: 10.1016/j.mito.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 20.Yan T., Hu K., Ren F., Jiang Z. LC-MS/MS profiling of post-transcriptional modifications in ginseng tRNA purified by a polysaccharase-aided extraction method. Biomolecules. 2020;10:621–636. doi: 10.3390/biom10040621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marsh J.D., Smith T.S. Calcium overload and ischemic myocardial Injury.pdf. Circulation. 1991;83:709–711. doi: 10.1161/01.cir.83.2.709. [DOI] [PubMed] [Google Scholar]
- 22.Petrosillo G., Colantuono G., Moro N., Ruggiero F.M., Tiravanti E., Di Venosa N., Fiore T., Paradies G. Melatonin protects against heart ischemia-reperfusion injury by inhibiting mitochondrial permeability transition pore opening. Am. J. Physiol. Heart Circ. Physiol. 2009;297:H1487–H1493. doi: 10.1152/ajpheart.00163.2009. [DOI] [PubMed] [Google Scholar]
- 23.Chodkowski M., Serafińska I., Brzezicka J., Golke A., Słońska A., Krzyżowska M., Orłowski P., Bąska P., Bańbura M.W., Cymerys J. Human herpesvirus type 1 and type 2 disrupt mitochondrial dynamics in human keratinocytes. Arch. Virol. 2018;163:2663–2673. doi: 10.1007/s00705-018-3890-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vowinckel J., Hartl J., Butler R., Ralser M. MitoLoc: a method for the simultaneous quantification of mitochondrial network morphology and membrane potential in single cells. Mitochondrion. 2015;24:77–86. doi: 10.1016/j.mito.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valente A.J., Maddalena L.A., Robb E.L., Moradi F., Stuart J.A. A simple ImageJ macro tool for analyzing mitochondrial network morphology in mammalian cell culture. Acta Histochem. 2017;119:315–326. doi: 10.1016/j.acthis.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 26.Enright A.J., John B., Gaul U., Tuschl T., Sander C., Marks D.S. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia D.M., Baek D., Shin C., Bell G.W., Grimson A., Bartel D.P. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat. Struct. Mol. Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao J., He Q., Li M., Chen Y., Liu Y., Wang J. LncRNA MIAT: myocardial infarction associated and more. Gene. 2016;578:158–161. doi: 10.1016/j.gene.2015.12.032. [DOI] [PubMed] [Google Scholar]
- 29.Clement T., Salone V., Rederstorff M. Vol. 1296. Humana Press; 2015. Dual Luciferase Gene Reporter Assays to Study miRNA Function, Small Non-coding RNAs 2015/03/21; pp. 187–198. [DOI] [PubMed] [Google Scholar]
- 30.Qu X., Du Y., Shu Y., Gao M., Sun F., Luo S., Yang T., Zhan L., Yuan Y., Chu W., et al. MIAT is a pro-fibrotic long non-coding RNA governing cardiac fibrosis in post-infarct myocardium. Sci. Rep. 2017;7:42657. doi: 10.1038/srep42657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Z.M., Huang F., Huang W.Q. Angiogenic lncRNAs: a potential therapeutic target for ischaemic heart disease. Life Sci. 2018;211:157–171. doi: 10.1016/j.lfs.2018.09.022. [DOI] [PubMed] [Google Scholar]
- 32.Yu B., Wang S. Angio-LncRs: LncRNAs that regulate angiogenesis and vascular disease. Theranostics. 2018;8:3654–3675. doi: 10.7150/thno.26024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calin G.A., Croce C.M. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 34.van Rooij E., Olson E.N. MicroRNA therapeutics for cardiovascular disease: opportunities and obstacles. Nat. Rev. Drug Discov. 2012;11:860–872. doi: 10.1038/nrd3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma U., Conine C.C., Shea J.M., Boskovic A., Derr A.G., Bing X.Y., Belleannee C., Kucukural A., Serra R.W., Sun F., et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016;351:391–396. doi: 10.1126/science.aad6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen Y., Yu X., Zhu L., Li T., Yan Z., Guo J. Transfer RNA-derived fragments and tRNA halves: biogenesis, biological functions and their roles in diseases. J. Mol. Med. 2018;96:1167–1176. doi: 10.1007/s00109-018-1693-y. [DOI] [PubMed] [Google Scholar]
- 37.Sequeira V., Nijenkamp L.L.A.M., Regan J.A., van der Velden J. The physiological role of cardiac cytoskeleton and its alterations in heart failure. Biochim. Biophys. Acta. 2014;1838:700–722. doi: 10.1016/j.bbamem.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 38.Chen C.Y., Caporizzo M.A., Bedi K., Vite A., Bogush A.I., Robison P., Heffler J.G., Salomon A.K., Kelly N.A., Babu A., et al. Suppression of detyrosinated microtubules improves cardiomyocyte function in human heart failure. Nat. Med. 2018;24:1225–1233. doi: 10.1038/s41591-018-0046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murphy E., Steenbergen C. Mechanisms underlying acute protection from cardiac ischemia-reperfusion injury. Physiol. Rev. 2008;88:581–609. doi: 10.1152/physrev.00024.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takano H., Zou Y., Hasegawa H., Akazawa H., Nagai T., Komuro I. Oxidative stress-induced signal transduction pathways in cardiac myocytes: involvement of ROS in heart diseases. Antioxid. Redox Signal. 2003;5:789–794. doi: 10.1089/152308603770380098. [DOI] [PubMed] [Google Scholar]
- 41.Green D.R., Reed J.C. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 42.Cai B.X., Li X.Y., Chen J.H., Tang Y.B., Wang G.L., Zhou J.G., Qui Q.Y., Guan Y.Y. Ginsenoside-Rd, a new voltage-independent Ca2+ entry blocker, reverses basilar hypertrophic remodeling in stroke-prone renovascular hypertensive rats. Eur. J. Pharmacol. 2009;606:142–149. doi: 10.1016/j.ejphar.2009.01.033. [DOI] [PubMed] [Google Scholar]
- 43.Kiefer D., Pantuso T. Panax ginseng. Am. Fam. Phys. 2003;68:1539–1542. [PubMed] [Google Scholar]
- 44.Lin M.T., Beal M.F. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 45.Ishii N., Ozaki K., Sato H., Mizuno H., Saito S., Takahashi A., Miyamoto Y., Ikegawa S., Kamatani N., Hori M., et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J. Hum. Genet. 2006;51:1087–1099. doi: 10.1007/s10038-006-0070-9. [DOI] [PubMed] [Google Scholar]
- 46.Chen L., Zhang D., Yu L., Dong H. Targeting MIAT reduces apoptosis of cardiomyocytes after ischemia/reperfusion injury. Bioengineered. 2019;10:121–132. doi: 10.1080/21655979.2019.1605812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bai X., Yang C., Jiao L., Diao H., Meng Z., Wang L., Cui H., Sun L., Zhang Y., Yang B. LncRNA MIAT impairs cardiac contractile function by acting on mitochondrial translocator protein TSPO in a mouse model of myocardial infarction. Signal Transduct. Target. Ther. 2021;6:172. doi: 10.1038/s41392-021-00538-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hannon G.J. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 49.Batista P.J., Chang H.Y. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cochain C., Channon K.M., Silvestre J.S. Angiogenesis in the infarcted myocardium. Antioxid. Redox Signal. 2013;18:1100–1113. doi: 10.1089/ars.2012.4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shyu K.G., Wang M.T., Wang B.W., Chang C.C., Leu J.G., Kuan P., Chang H. Intramyocardial injection of naked DNA encoding HIF-1a:VP16 hybrid to enhance angiogenesis in an acute myocardial infarction model in the rat. Cardiovasc. Res. 2002;54:576–583. doi: 10.1016/s0008-6363(02)00259-6. [DOI] [PubMed] [Google Scholar]
- 52.Zhou L., Ma W., Yang Z., Zhang F., Lu L., Ding Z., Ding B., Ha T., Gao X., Li C. VEGF165 and angiopoietin-1 decreased myocardium infarct size through phosphatidylinositol-3 kinase and Bcl-2 pathways. Gene Ther. 2005;12:196–202. doi: 10.1038/sj.gt.3302416. [DOI] [PubMed] [Google Scholar]
- 53.Yan B., Yao J., Liu J.Y., Li X.M., Wang X.Q., Li Y.J., Tao Z.F., Song Y.C., Chen Q., Jiang Q. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ. Res. 2015;116:1143–1156. doi: 10.1161/CIRCRESAHA.116.305510. [DOI] [PubMed] [Google Scholar]
- 54.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fatemi R.P., Velmeshev D., Faghihi M.A. De-repressing lncRNA-targeted genes to upregulate gene expression: focus on small molecule therapeutics. Mol. Ther. Nucleic Acids. 2014;3:e196. doi: 10.1038/mtna.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Z. Springer; 2009. MicroRNA Interference Technologies, MicroRNA Interference Technologies; pp. 59–73. [Google Scholar]
- 57.Lam J.K.W., Chow M.Y.T., Zhang Y., Leung S.W.S. siRNA versus miRNA as therapeutics for gene silencing. Mol. Ther. Nucleic Acids. 2015;4:e252. doi: 10.1038/mtna.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 59.Garber K. Alnylam launches era of RNAi drugs. Nat. Biotechnol. 2018;36:777–778. doi: 10.1038/nbt0918-777. [DOI] [PubMed] [Google Scholar]
- 60.Kosmas C.E., Munoz Estrella A., Sourlas A., Silverio D., Hilario E., Montan P.D., Guzman E. Inclisiran: a new promising agent in the management of hypercholesterolemia. Diseases. 2018;6 doi: 10.3390/diseases6030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G., et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prensner J.R., Chinnaiyan A.M. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu G., Wong-Staal F., Li Q.X. Development of new RNAi therapeutics. Histol. Histopathol. 2007;22:211–217. doi: 10.14670/HH-22.211. [DOI] [PubMed] [Google Scholar]
- 64.Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat. Rev. Drug Discov. 2013;12:433–446. doi: 10.1038/nrd4018. [DOI] [PubMed] [Google Scholar]
- 65.Miyauchi K., Ohara T., Suzuki T. Automated parallel isolation of multiple species of non-coding RNAs by the reciprocal circulating chromatography method. Nucleic Acids Res. 2007;35:e24. doi: 10.1093/nar/gkl1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Loss-Morais G., Waterhouse P.M., Margis R. Description of plant tRNA-derived RNA fragments (tRFs) associated with argonaute and identification of their putative targets. Biol. Direct. 2013;8:6–10. doi: 10.1186/1745-6150-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Z. The guideline of the design and validation of miRNA mimics. Methods Mol. Biol. 2011;676:211–223. doi: 10.1007/978-1-60761-863-8_15. [DOI] [PubMed] [Google Scholar]
- 68.Mizrahi O., Ish Shalom E., Baniyash M., Klieger Y. Quantitative flow cytometry: concerns and recommendations in clinic and research. Cytometry B Clin. Cytom. 2018;94:211–218. doi: 10.1002/cyto.b.21515. [DOI] [PubMed] [Google Scholar]
- 69.Grimson A., Farh K.K.-H., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krüger J., Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 72.van Kasteren P.B., van der Veer B., van den Brink S., Wijsman L., de Jonge J., van den Brandt A., Molenkamp R., Reusken C.B.E.M., Meijer A. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J. Clin. Virol. 2020;128:104412. doi: 10.1016/j.jcv.2020.104412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chou S.T., Leng Q., Scaria P., Woodle M., Mixson A.J. Selective modification of HK peptides enhances siRNA silencing of tumor targets in vivo. Cancer Gene Ther. 2011;18:707–716. doi: 10.1038/cgt.2011.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li F., Fan X., Zhang Y., Pang L., Ma X., Song M., Kou J., Yu B. Cardioprotection by combination of three compounds from ShengMai preparations in mice with myocardial ischemia/reperfusion injury through AMPK activation-mediated mitochondrial fission. Sci. Rep. 2016;6:37114. doi: 10.1038/srep37114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available within the article and its supplemental information.