Abstract
The ykzB and ykoL genes encode two peptides, of 51 and 60 amino acids, the functions of which are unknown. The ykzB and tnrA genes are contiguous and transcribed divergently. Expression of ykzB and ykoL is induced by glutamate and is under the control of the TnrA global regulator of nitrogen utilization. TnrA regulated its own synthesis in glutamate minimal medium. Two DNA sequences (TnrAB1 and TnrAB2) homologous to the TnrA binding site are present in the region between tnrA and ykzB. Deletion mapping indicated that the TnrAB2 binding site was involved in activation of the ykzB promoter. In addition, transcription of tnrA depends on the presence of the TnrAB1 binding site. The ykzB and ykoL genes are probably in the same transcriptional unit. A single promoter involved in transcription in the presence of glutamate was mapped by primer extension. ykoL expression was induced by phosphate limitation and depended on the PhoP-PhoR two-component regulatory system. Its promoter was mapped to the region between ykoL and ykzB. Four boxes similar to the PhoP binding site are present upstream from the ykoL promoter. These boxes are probably recognized by PhoP∼P during the activation of transcription in phosphate limitation conditions.
In Bacillus subtilis, the expression of several genes, such as glnA (glutamine synthetase), ansA (asparaginase), ansB (aspartase), ureABC (urease), gabP (γ-aminobutyrate permease), nasABCDEF (nitrate assimilatory enzymes), and nrgAB (ammonium permease and nitrogen-regulated PII-like protein), that are involved in the transport and catabolism of nitrogen-containing compounds is controlled by nitrogen availability (2, 8, 9, 19, 20, 28). Three regulatory proteins, GlnR, TnrA, and CodY, and a sigma factor called SigL are involved in the control of nitrogen metabolism in response to nitrogen supply. GlnR and TnrA are involved in the regulation of genes with nitrogen source-dependent expression. GlnR and TnrA are very similar and belong to the MerR family of regulators. GlnR is a negative regulator of glnRA operon expression and represses several TnrA-regulated genes (24). It has been shown in vitro that GlnR binds simultaneously to two operators in the glnRA promoter region. TnrA activates transcription of the gabP, ureABC, nrgAB, and nas genes in response to nitrogen limitation. TnrA is also a negative regulator of the ammonium assimilatory enzymes, glutamine synthetase and glutamate synthase, during nitrogen-limited growth (29). The TnrA protein is active only during nitrogen limitation, whereas GlnR is active in conditions of nitrogen excess. All of the promoters positively regulated by TnrA contain a GlnR/TnrA binding site (TGT-NAN7-TNACA) located upstream from the −35 region (9). The CodY protein controls several genes in nitrogen metabolism, acetate metabolism, and competence. CodY repression occurs in cells growing in a medium rich in amino acids. CodY is thought to bind to a DNA structure containing AT-rich DNA sequences (25). The sigL gene in B. subtilis encodes a sigma factor equivalent to the sigma 54 of gram-negative bacteria (4). SigL is required for the transcription of several operons involved in the catabolism of arginine, ornithine (roc operons), isoleucine, and valine (bkd operon) (5). Specific transcription factors controlling these operons have been identified (3, 5). However, there is no global nitrogen regulatory system equivalent to the NtrB/NtrC two-component system of gram-negative bacteria.
Under phosphate starvation conditions, several genes in B. subtilis, including phoA and phoB (alkaline phosphatases), pstS (inorganic phosphate transport system), tuaA (teichuronic acid biosynthesis), are activated by the two-component (PhoP-PhoR) regulatory system response regulator and histidine kinase. All pho-regulated promoters require a minimum of four consensus PhoP binding sequence repeats (TT(A/T/C)A(C/T)A) for activation (7). During the B. subtilis function analysis program, 1,200 mutants were constructed using the pMUTIN4 vector to study the function and regulation of the unknown “y” genes (27). This plasmid can be used to construct lacZ fusions. The capacity of various amino acids to act as nitrogen sources or inducers of gene expression and induction by phosphate starvation have been studied. Upon screening of this collection of mutants, we found that the expression of 30 y genes was increased by the presence of glutamate in the growth medium. In one of these mutant strains, lacZ expression was strongly induced by glutamate as well as by phosphate starvation. This strain contains pMUTIN4 inserted into the ykoL gene. In this work, we describe the regulation of the ykzB-ykoL operon and of the divergent gene tnrA. We found that this divergon was positively controlled by TnrA and by the two-component system, PhoP-PhoR.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The B. subtilis strains used in this work are described in Table 1. Escherichia coli DH5α [supE44 Δlac U169 (φ80 lacZΔM15) hsdR17 secA1 endA1 gyrA96 thi-1 relA1] was grown at 37°C in Luria-Bertani (LB) medium and used for cloning experiments. B. subtilis was grown in LB medium and transformed with plasmid or chromosomal DNA as previously described (12). Transformants were selected on SP plates (4) supplemented with erythromycin (1 μg/ml), lincomycin (25 μg/ml), chloramphenicol (5 μg/ml), or kanamycin (5 μg/ml). B. subtilis 168 derivative strains were grown in minimal medium [11.4 mM K2SO4, 62 mM K2HPO4 · 5H2O, 44 mM KH2PO4, 3.4 mM sodium citrate · 7H2O, 0.8 mM MgSO4 · 7H2O, 0.4% glucose, 0.005% l-tryptophan, 4 mg of FeCl3/liter, 0.2 mg of MnSO4/liter, 5.5 mg of CaCl2/liter 1.7 mg of ZnCl2/liter, 0.43 mg of CuCl2 · 2H2O/liter, 0.6 mg of CaCl2 · 6H2O/liter 0.6 mg of Na2MoO4 · 2H2O/liter, 10 mM (NH4)2SO4 or potassium glutamate adjusted to pH 7 with 10 N NaOH] or in low-phosphate defined medium (LPDM) or high-phosphate defined medium (HPDM) (11), each supplemented with 50 μg of l-tryptophan/ml. B. subtilis tnrA mutants were constructed using tnrA::Tn917 chromosomal DNA (strain SF706T provided by S. H. Fischer) transferred by transformation with selection for transposon-encoded erythromycin resistance. B. subtilis phoP-phoR mutants were constructed using chromosomal DNA from the strain MH5913 (provided by F. M. Hulett) and selected for tetracycline resistance.
TABLE 1.
B. subtilis strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| 168 | trpC2 | Laboratory stock |
| FA5 | trpC2 amyE::(ykoL600′-′lacZ cat) | This study |
| FA6 | trpC2 amyE::(ykoL600′-′lacZ cat) | This study |
| tnrA::Tn917 | ||
| FA18 | trpC2 amyE::(ykoL600′-′lacZ cat) | This study |
| phoPR::tet | ||
| FA19 | trpC2 amyE::(ykoL600′-′lacZ cat) | This study |
| tnrA::Tn917 phoPR::tet | ||
| FA20 | trpC2 amyE::(ykoL200′-′lacZ cat) | This study |
| FA21 | trpC2 amyE::(ykzB′-′lacZ cat) | This study |
| FA22 | trpC2 amyE::(tnrA′-′lacZ cat) | This study |
| FA23 | trpC2 amyE::(ykoL200′-′lacZ cat) tnrA::Tn917 | This study |
| FA24 | trpC2 amyE::(ykzB′-′lacZ cat) tnrA::Tn917 | This study |
| FA25 | trpC2 amyE::(tnrA′-′lacZ cat) tnrA::Tn917 | This study |
| FA26 | trpC2 amyE::(ykoL200′-′lacZ cat) phoPR::tet | This study |
| FA27 | trpC2 amyE::(ykzB′-′lacZ cat) phoPR::tet | This study |
| FA28 | trpC2 amyE::(tnrA′-′lacZ cat) phoPR::tet | This study |
| FA29 | trpC2 amyE::(tnrA′-′lacZ cat) ΔtnrAB2 | This study |
| FA30 | trpC2 amyE::(ykzB′-′lacZ cat) ΔtnrAB1 | This study |
| FA31 | trpC2 amyE::(tnrA′-′lacZ cat) ΔtnrAB1B2 | This study |
| FA32 | trpC2 amyE::(ykzB′-′lacZ cat) ΔtnrAB1B2 | This study |
| FA33 | pMUTIN4MCS::ykoM erm | This study |
| SF706T | tnrA::Tn917 amyE::neo φ(gabP-lacZ)706 trpC2 | S. H. Fisher |
| MH5913 | pheA1 trpC2 phoPR::tet | F. M. Hulett |
DNA manipulations.
Standard procedures were used to extract plasmids from E. coli (1, 22). Restriction enzymes were used as recommended by the manufacturers. DNA fragments were amplified by PCR (18, 21) using the Taq DNA polymerase (Stratagene). DNA sequences were determined by the dideoxy-chain termination method (23) using the modified T7 DNA polymerase (26) (Pharmacia).
β-Galactosidase assays.
β-Galactosidase activity was estimated on plates by 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) hydrolysis. β-Galactosidase specific activities were determined as described previously (16, 17) and expressed as nanomoles of o-nitrophenol produced per minute per milligram of protein. Induction by glutamate was measured in samples taken in exponential growth phase, while induction by phosphate starvation was measured at the beginning of the stationary phase.
Plasmid constructs.
pMUTIN4MCS (27) was used for gene disruption and for the construction of transcriptional fusions with the E. coli lacZ gene. The BFS1843 (ykoL) mutant was constructed in K. Devine's laboratory by insertion of pMUTIN4MCS between bases 1397629 and 1397747 of the B. subtilis genome (13). The ykoM′-lacZ transcriptional fusion was constructed using a HindIII/BamHI DNA fragment corresponding to the ykoM central region generated by PCR with oligonucleotides YKOMH (5′-GCCGAAGCTTCCGTGATAGCAAAGAGCACGGTTT-3′) and YKOMB (5′-CGCGGATCCGCTCCCTTACTAAAAACCCGTTTC-3′). This fragment was inserted between the HindIII and BamHI sites of pMUTIN4MCS, and the recombinant plasmid was used to transform B. subtilis 168. Integration of the construct into the ykoM gene was selected by erythromycin resistance, giving strain FA33.
The ykzB, ykoL, and tnrA promoter regions were obtained by PCR amplification using B. subtilis chromosomal DNA as the template. The corresponding PCR-amplified DNA fragments were introduced into the EcoRI and BamHI sites of pAC5 (15) to create lacZ translational fusions (Fig. 1). These constructs were used to transform B. subtilis. Recombinant strains were selected using chloramphenicol resistance. In these strains, the lacZ fusions were integrated as single copies at the amyE locus. The synthetic primers used for PCR amplification were as follows: YKOL1 (5′-CGGAATTCCCTGATCTGTCTTACGG-3′) or YKOLE (5′-CGGAATTCGATCGGTTCAAAACGGAC-3′) and YKOL2 (5′-CGGGATCCCGAAGCTCAGAAATCGT-3′); YKZBE (5′-CGGAATTCGAATGATCTTCTGTGGTC-3′), YKZBE2 (5′-CGGAATTCCAAATAGAAGATTTTTGAAAAAATAC-3′), or YKZBE3 (5′-CGGAATTCATAATGCGTAACACCCAA-3′) and YKZBB (5′-CGGGATCCGCCAGTGAGACTGCTTT-3′); TNRAE (5′-CGGAATTCGCCAGTGAGACTGCTTT-3′), TNRAE2 (5′-CGGAATTCCAAAAATCTTCTATTTGATGTTAG-3′), or TNRAE3 (5′-CGGAATTCGCGTCAGGTTTTTTCTCT-3′) and TNRAB (5′-CGGGATCCGAATGATCTTCTGTGGTC-3′).
FIG. 1.
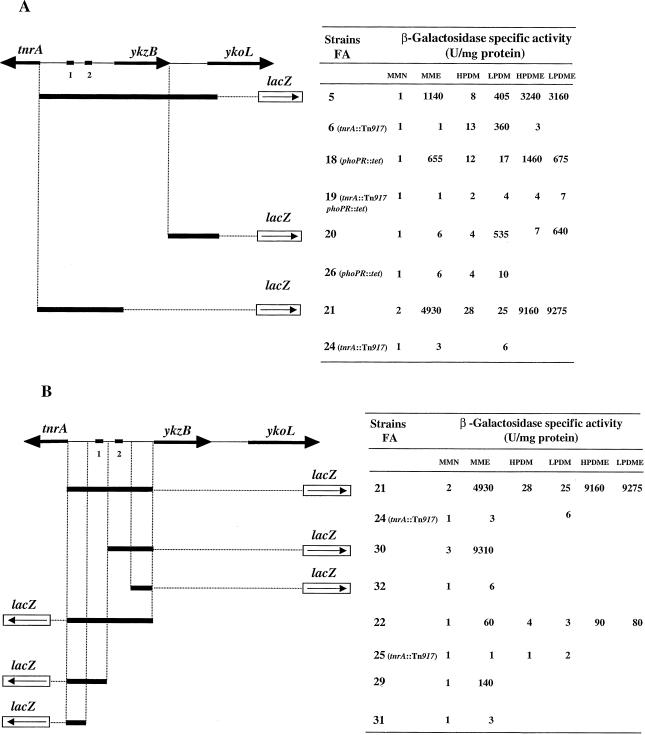
Schematic representation of the DNA regions studied and lacZ fusions. Black arrows show the orientation of transcription. Black lines in bold type (on thick black lines) represents the PCR-amplified DNA fragments fused to the lacZ gene. β-Galactosidase assays were performed using cells harvested at the end of the exponential growth phase. MMN is minimal medium with 10 mM ammonium sulfate; MME is minimal medium with 10 mM potassium glutamate; LPDME and HPDME correspond to LPDM and HPDM in which ammonium sulfate has been replaced by 10 mM potassium glutamate. Black boxes 1 and 2 indicate positions of the TnrAB1 and TnrAB2 sites, respectively. The values presented at the right are the means of two or more assays.
RNA extraction and primer extension.
RNA extraction, primer extension, and oligonucleotide labeling reactions were performed as described previously (6). B. subtilis 168 was grown in minimal medium with ammonium sulfate or potassium glutamate as the sole nitrogen source to activate glutamate-inducible promoters and in LPDM or HPDM to activate phosphate starvation-inducible promoters. RNA was extracted, and primer extension was performed using the YKZBPE primer (5′-AACGTCTCCAGCATTTCATCACGC-3′) for the ykzB transcript and the YKOLPE primer (5′-GGCGTCGTTCTTTTTCTAAAGCGG-3′) for the ykoL transcript.
RESULTS
Characterization of glutamate-induced genes.
Within the framework of the European systematic function analysis of B. subtilis genes, 1,200 mutants were constructed. Genes to be interrupted by the pMUTIN4MCS (27) vector were selected from the 3,000 unknown open reading frames identified during the complete B. subtilis genome sequencing project (13). The corresponding proteins show no significant similarities to known proteins in databases. The unknown genes were interrupted by single or double crossover, leading to (i) inactivation of the target gene and (ii) transcriptional fusions between the promoters and the reporter gene, lacZ. We were interested in nitrogen metabolism and amino acid utilization. To investigate the potential function of the unknown genes, we tested the growth of the various mutant strains on minimal medium plates supplemented with X-Gal and containing a single amino acid (proline, alanine, aspartate, glutamine, or glutamate) or ammonium sulfate as the sole nitrogen source. The induction or repression of target gene expression on the different plates was estimated by β-galactosidase activity. One of the mutants tested, BFA1843, which showed a strong activity on plates was chosen for further study. In this strain, the expression of the lacZ fusion was strongly induced by glutamate, resulting in blue colonies on plates supplemented with this amino acid. β-Galactosidase specific activities were determined to be about 1 U/mg of protein in ammonium sulfate minimal medium and 2,100 U/mg in glutamate minimal medium. It was therefore 2,000 times higher with glutamate (10 mM) than with ammonium sulfate (10 mM) as the sole nitrogen source. The integration of pMUTIN4MCS into BFA1843 interrupts the ykoL gene. The results obtained show that ykoL expression is induced by glutamate.
Analysis of the ykoL promoter.
To identify the promoter of the ykoL gene, PCR-amplified DNA fragments containing sequences located upstream from ykoL were fused to lacZ, creating translational fusions. These fusions were integrated into the B. subtilis chromosome at the amyE locus of strain 168. β-Galactosidase activity was determined in minimal medium containing 10 mM ammonium sulfate or 10 mM glutamate (Fig. 1A). The fusion present in strain FA5, extending from the tnrA NH2-terminal region to the ykoL NH2-terminal region, gave a high level of β-galactosidase activity in glutamate medium. lacZ expression levels were 1,000 times higher with glutamate than with ammonia for this strain. The results also show that the ykoL promoter is located between tnrA and ykoL. Strain FA20, which contains a lacZ fusion with a DNA fragment located between the ykzB COOH-terminal region and the ykoL NH2-terminal region, expressed β-galactosidase at a low level in ammonium sulfate and glutamate minimal media. These results suggest that the glutamate-inducible promoter is not present upstream from the ykoL gene but probably resides in the tnrA-ykzB intergenic region. To test this hypothesis, strain FA21, containing a DNA fragment extending from the tnrA NH2-terminal region to the ykzB NH2-terminal region, was constructed and integrated by a double crossover event at the amyE locus of strain 168. β-Galactosidase activity in FA21 cells grown in minimal medium containing 10 mM glutamate was 2,500 times higher than that in cells grown in ammonium sulfate. These results indicate that the glutamate-dependent promoter is located upstream from the ykzB gene and that the ykzB and ykoL genes probably form an operon.
Determination of the ykzB transcriptional start site.
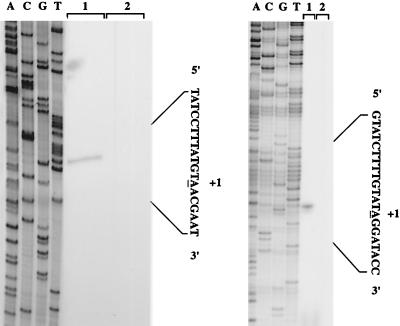
Primer extension experiments were performed to determine the ykzB transcriptional initiation start site. Total RNA was extracted from strain 168 grown in minimal medium containing either 10 mM glutamate or 10 mM ammonium sulfate as the nitrogen source. The YKZBPE and YKOLPE primers, which are complementary to the ykzB and ykoL coding sequences, respectively, were used for the extension reactions. One signal was detected using the YKZBPE primer and RNA extracted from a culture in glutamate medium, whereas no cDNA was detected with cells grown in ammonium sulfate (Fig. 2). The transcription initiation start site was located 29 bp upstream from the translation initiation codon of ykzB. Examination of the upstream nucleotide sequence revealed a putative −10 region similar to the Pribnow box (Fig. 3). However, the −35 region differs from the consensus sequence for ςA-dependent promoters. The same transcription initiation site was detected with the YKOLPE primer (data not shown). These results confirm that ykzB expression is induced by glutamate and that ykzB and ykoL are located in the same transcriptional unit.
FIG. 2.
Primer extension analysis of ykzB and ykoL mRNAs. (Left) Total RNA was isolated from B. subtilis 168 grown in minimal medium containing 10 mM glutamate (lane 1) or 10 mM ammonium sulfate (lane 2) as the sole nitrogen source. (Right): Total RNA was isolated from B. subtilis 168 grown in LPDM (lane 1) or HPDM (lane 2). The RNA preparations were used as templates for reverse transcriptase. The oligonucleotide primers used for reverse transcription were also used to prime dideoxy sequencing reactions from the corresponding plasmids (lanes A, C, G, and T). Positions of transcription start sites (+1) are underlined.
FIG. 3.
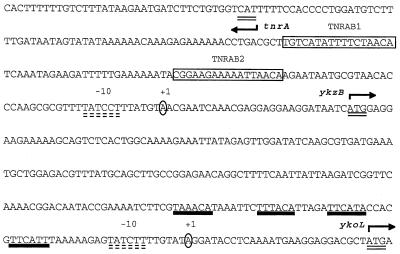
Sequence of the promoter region located between tnrA and the ykzB-ykoL operon. Potential −10 sequences are indicated by double dashed lines, and the transcriptional start site (+1) is circled. Translational initiation codons are double underlined, and bent arrows indicate the direction of transcription. The TrnA binding sites are boxed, and the binding sites for PhoP are marked by thick black lines.
ykoL expression is induced by phosphate starvation.
During the systematic function analysis program, it was found that the ykoL gene is expressed under phosphate limitation conditions. We analyzed the expression of ykoL (FA20) in LPDM and in HPDM (Fig. 1A). As expected, β-galactosidase activity was 100 times higher in cells grown in low-phosphate medium than in cells grown in high-phosphate medium. In parallel, we measured the activity in strain FA21 in low- and high-phosphate media. ykzB was not expressed under phosphate limitation, suggesting the existence in the ykzB-ykoL operon of an internal promoter allowing ykoL expression during phosphate starvation.
Determination of the ykoL transcriptional start site under phosphate limitation conditions.
To determine the transcriptional start site of the ykoL gene, RNA was extracted from strain 168 grown under low- or high-phosphate conditions. Primer extension experiments were performed using the YKOLPE primer. ykoL mRNA was absent in high-phosphate medium but was detectable in low-phosphate conditions (Fig. 2). The transcription initiation site was located 28 bp upstream from the translation initiation codon of ykoL. The nucleotide sequence of the region preceding position +1 contained a putative −10 region similar to the consensus sequence of ςA-dependent promoters. In contrast, the potential −35 region diverged greatly from the consensus sequence of the −35 region of promoters recognized by the ςA-containing RNA polymerase. These results confirm that ykoL has its own promoter, activated during phosphate starvation.
Regulation of ykoL expression by the two-component PhoP-PhoR system.
As the PhoP-PhoR system activates the transcription of genes required for phosphate uptake and utilization during phosphate limitation, we expected that ykoL, which is induced in low-phosphate conditions, would be regulated by the PhoP-PhoR system. Evidence in favor of this hypothesis was provided by the presence of four putative PhoP binding sites TT(A/T/C)A(C/T)A with a spacing of 4 to 7 bp between repeats (Fig. 3) (7) and essential for promoter activity as demonstrated for the tuaA system (14). We tested whether ykoL was indeed regulated by the PhoP-PhoR system using strain FA20, in which the phoP-phoR genes were deleted. β-Galactosidase specific activity in the resulting FA26 strain was determined in cells grown in LPDM and HPDM (Fig. 1A). We found that the fusion was not induced by phosphate limitation in FA26, whereas it was induced in the parental strain FA20. β-Galactosidase activity in FA20 was 100 times higher in LPDM than in HPDM, whereas it was low in both media for the phoP-phoR mutant. Therefore, the expression of ykoL is controlled by the two-component PhoP-PhoR system in phosphate starvation conditions.
Regulation of ykzB and ykoL expression by TnrA.
During nitrogen-limited growth in B. subtilis, TnrA induces the expression of several genes, including ureABC, nasA, and gabP. As TnrA is involved in the regulation of nitrogen metabolism genes, we thought it likely that the expression of ykzB would be regulated by this transcriptional factor. To test this, we introduced a null mutation of the tnrA structural gene into strain FA21. β-Galactosidase specific activity was measured in the resulting strain, FA24, grown in minimal medium with ammonia or glutamate (Fig. 1A). Activity was much lower in the tnrA-defective strain grown in glutamate than in the wild-type strain (1,500 times higher), indicating that ykzB expression was positively regulated by TnrA. The tnrA mutation was also introduced by transformation into strain FA5, to give strain FA6. β-Galactosidase specific activity was assayed in cultures of FA6 in minimal medium containing 10 mM ammonia or 10 mM glutamate as the sole nitrogen source. The product of tnrA was also involved in the induction of ykoL expression (Fig. 1A). According to Wray et al. (30), all TnrA-regulated promoters contain a common inverted repeat sequence centered 49 to 51 bp upstream from the transcriptional start site. Two such putative TnrA sites, TnrAB1 and TnrAB2, were found centered 93 and 50 bp upstream from the ykzB +1 position (Fig. 3). The first is identical to the proposed TnrA binding site, whereas the second has 15 of a possible 17 matches with the consensus sequence. The role of each site in the regulation of ykzB was then assessed. Expression of the fusion in the FA21 strain, covering both TnrA sites, was fully induced by glutamate. Using the same lacZ fusion, we sequentially deleted TnrAB1 and TnrAB2 sites to give strains FA30 and FA32, respectively. In strain FA30 (ΔtnrAB1), β-galactosidase specific activity was 3,000 times higher in cells grown in the presence of glutamate than in cells grown in ammonia (Fig. 1B). In contrast, β-galactosidase specific activity was very low in strain FA32 (ΔtnrAB1 ΔtnrAB2). These results indicate that the expression of ykzB is induced by the transcriptional nitrogen regulatory factor TnrA and that activation occurs through the TnrA site located 50 bp upstream from the transcriptional start site (TnrAB2).
Induction of ykzB and ykoL by glutamate in phosphate starvation conditions.
In strain FA5, a high level of lacZ expression was observed in the presence of glutamate in both high- and low-phosphate conditions (Fig. 1A). In the phoPR strain FA18, lacZ expression was high in the presence of glutamate. As expected, this expression was completely abolished in the tnrA phoPR double mutant (FA19). In the FA20 strain containing the ykzB ykoL intergenic region, lacZ expression was induced in low-phosphate medium, confirming the existence of a PhoPR-dependent promoter. Conversely, in strain FA21, which contains the ykzB promoter, lacZ expression was induced only by glutamate.
Autoregulation of tnrA expression.
It has been reported that TnrA positively regulates its own synthesis (9). We investigated the role of the two putative TnrA binding sites in the autoregulation of TnrA, by constructing a tnrA′-′lacZ translational fusion using a DNA fragment extending from the ykzB NH2-terminal region to the tnrA NH2-terminal region. This fusion was integrated into the chromosome of strain 168 to give strain FA22 (Fig. 1B). β-Galactosidase specific activity was measured in cultures of cells grown in minimal medium containing 10 mM ammonia or 10 mM glutamate as the sole nitrogen source. Expression of the tnrA′-′lacZ fusion was about 60 times stronger in the presence of glutamate than in the presence of ammonia. A tnrA::Tn917 null mutation was introduced by transformation into strain FA22, to give strain FA25. In this strain, β-galactosidase specific activity was very weak, confirming that TnrA is involved in the activation of its own expression. No effect of phosphate limitation was observed on tnrA expression.
To study the function of the two putative TnrA binding sites, TnrAB1 and TnrAB2, two DNA fragments, one containing one site (TnrAB1) and the other containing neither, were synthesized and used to construct two translational lacZ fusions, giving strains FA29 and FA31, respectively. β-Galactosidase specific activity was assayed in cultures of the FA29 and FA31 strains (Fig. 1B). In cultures of the FA29 strain, β-galactosidase specific activity was strong in minimal medium containing glutamate whereas in strain FA31 (ΔtnrAB1 ΔtnrAB2), it was as weak as that in ammonia, indicating that the TnrAB1 site is essential for tnrA expression. Thus, TnrA regulated the induction of its own expression through the specific TnrA binding site TnrAB1.
DISCUSSION
The ykzB and ykoL genes encode two peptides of 51 and 60 amino acids, respectively. This study shows that the expression of ykzB and ykoL is induced by glutamate. The expression of ykoL is also induced by phosphate limitation.
We demonstrated that the product of tnrA was involved in the activation of transcription in minimal medium containing glutamate. The induction by glutamate is rather unexpected because the glutamate pool is very large, up to 0.1 M, according to Hu et al. (10). TnrA activity may be modulated by a metabolite produced by the product of glnA, as previously suggested (29). Alternatively, the uptake of glutamate may be involved in the induction process. The effect of glutamate on the expression of tnrA or the target genes is also not known. In particular, we cannot exclude a direct interaction of glutamate with TnrA. Like other regulatory genes, tnrA expression is positively autoregulated, leading to a significant increase in the level of the corresponding protein during induction. This may partly account for the very high level of induction observed for ykzB. Indeed, ykzB is one of the mostly highly expressed of the 1,200 y genes of B. subtilis studied during the European functional analysis program (unpublished results). The tnrA-regulated genes contain a common inverted repeat (TGT-NAN7-TNACA) located upstream from the transcriptional start site. This sequence has been shown to be the binding site for the TnrA protein (30). It has been shown that high-level activation of nrgAB expression occurs only if the TnrA site is centered 49 to 51 bp upstream from the transcriptional start site, indicating that the relative alignment of the −35 region of the promoter and the TnrA site is critical for activation (30). Two DNA sequences homologous to the TnrA binding site are present in the region between tnrA and ykzB. Deletion mapping showed that the promoter proximal box (TnrAB2) was involved in the activation of the ykzB promoter. The TnrAB2 site is centered 50 bp upstream from the transcriptional start site of the ykzB promoter, confirming that the distance between the putative TnrA binding site and the −35 sequence is crucial for promoter activation. The promoters of TnrA-regulated genes contain several mismatches within the −35 and −10 regions. We found that the −35 region of the ykzB promoter differed from the consensus sequence for promoters transcribed by the ςA form of RNA polymerase. Although the tnrA promoter was not mapped by primer extension, a similar result was observed for expression of the tnrA gene itself. One putative TnrA binding site, TnrAB1, is necessary for induction of the expression of tnrA by glutamate. Removal of the upstream TnrA binding site led to a slight increase in tnrA and ykzB expression. This may be due to the effects of competition between the proteins bound at the target sites.
As the expression of both ykzB and ykoL is induced by glutamate, these two genes probably belong to the same transcriptional unit. Moreover, only one promoter involved in transcription in the presence of glutamate was found by primer extension. The ykzB and ykoL genes are followed by ykoM on the chromosomal DNA of B. subtilis. However, we have shown that the expression of ykoM is not induced by glutamate and that this gene does not belong to the same transcriptional unit (data not shown). ykoL gene expression is also induced by phosphate starvation. Activation of ykoL transcription depends on the PhoP-PhoR regulatory system. The ykoL promoter, which is activated by phosphate limitation, was mapped to the region between ykoL and ykzB. Four boxes with similarity to the PhoP binding site are present upstream from the ykoL promoter. These boxes are probably recognized by PhoP during the activation of transcription in phosphate limitation conditions. Previous work has shown that the predominant binding region in promoters controlled by PhoP is located at approximately the same position (i.e., 21 to 60 bp upstream from the transcription start site). The binding region contained multiple TT(A/T)ACA repeats separated by approximately 5 bp. At least four of these repeats were present in each promoter (7). In the ΔtrnA ΔphoPR double-mutant strain, ykoL was not induced by glutamate or phosphate limitation, confirming that the expression of this gene is subject to double regulation.
The function of the YkzB and YkoL peptides is still unknown. However, we have shown that in a strain in which ykzB and ykoL were deleted and in strains with deletions of ykzB alone or ykoL alone, there was no difference in the ykzB activation of transcription by the tnrA gene product in the presence of glutamate (data not shown). Therefore, the products of these two genes are not involved in induction of the operon in the presence of glutamate. The link between glutamate induction and regulation by phosphate limitation is difficult to interpret at the molecular level and needs further investigation.
ACKNOWLEDGMENTS
We thank F. M. Hulett and S. H. Fisher for the gift of strains MH5913 and SF706T, respectively, and for helpful discussions. We also thank J. Bignon and S. Mazard for technical support, J. Knight for correcting the manuscript, and C. Dugast for secretarial assistance.
D. Robichon received a fellowship from the European Commission (contract BIO4-CT-95-0278). This work was supported by grants from the Institut Pasteur, Université Paris 7, and the Centre National de la Recherche Scientifique.
REFERENCES
- 1.Amory A, Kunst F, Aubert E, Klier A, Rapoport G. Characterization of the sacQ genes from Bacillus licheniformis and Bacillus subtilis. J Bacteriol. 1987;169:324–333. doi: 10.1128/jb.169.1.324-333.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkinson M R, Fisher S H. Identification of genes and gene products whose expression is activated during nitrogen-limited growth in Bacillus subtilis. J Bacteriol. 1991;173:23–27. doi: 10.1128/jb.173.1.23-27.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calogero S, Gardan R, Glaser P, Schweizer J, Rapoport G, Débarbouillé M. RocR, a novel regulatory protein controlling arginine utilization in Bacillus subtilis, belongs to the NtrC/NifA family of transcriptional activators. J Bacteriol. 1994;176:1234–1241. doi: 10.1128/jb.176.5.1234-1241.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Débarbouillé M, Martin-Verstraete I, Kunst F, Rapoport G. The Bacillus subtilis sigL gene encodes an equivalent of ς54 from Gram-negative bacteria. Proc Natl Acad Sci USA. 1991;88:9092–9096. doi: 10.1073/pnas.88.20.9092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Débarbouillé M, Gardan R, Arnaud M, Rapoport G. Role of BkdR, a transcriptional activator of the SigL-dependent isoleucine and valine degradation pathway in Bacillus subtilis. J Bacteriol. 1999;181:2059–2066. doi: 10.1128/jb.181.7.2059-2066.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derré I, Rapoport G, Devine K, Rose M, Msadek T. ClpE, a novel type of HSP100 ATPase, is part of the CtsR heat shock regulon of Bacillus subtilis. Mol Microbiol. 1999;32:581–593. doi: 10.1046/j.1365-2958.1999.01374.x. [DOI] [PubMed] [Google Scholar]
- 7.Eder S, Liu W, Hulett F M. Mutational analysis of the phoD promoter in Bacillus subtilis: implications for PhoP binding and promoter activation of Pho regulon promoters. J Bacteriol. 1999;181:2017–2025. doi: 10.1128/jb.181.7.2017-2025.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferson A E, Wray L V, Jr, Fisher S H. Expression of the Bacillus subtilis gabP gene is regulated independently in response to nitrogen and amino acid availability. Mol Microbiol. 1996;22:693–701. doi: 10.1046/j.1365-2958.1996.d01-1720.x. [DOI] [PubMed] [Google Scholar]
- 9.Fisher S H. Regulation of nitrogen metabolism in Bacillus subtilis: vive la différence! Mol Microbiol. 1999;32:223–232. doi: 10.1046/j.1365-2958.1999.01333.x. [DOI] [PubMed] [Google Scholar]
- 10.Hu P, Leighton T, Ishkhanova G, Kustu S. Sensing of nitrogen limitation by Bacillus subtilis: comparison to enteric bacteria. J Bacteriol. 1999;181:5042–5050. doi: 10.1128/jb.181.16.5042-5050.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hulett F M, Bookstein C, Jensen K. Evidence for two structural genes for alkaline phosphatase in Bacillus subtilis. J Bacteriol. 1990;172:735–740. doi: 10.1128/jb.172.2.735-740.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kunst F, Msadek T, Bignon J, Rapoport G. The DegS/DegU and ComP/ComA two-component systems are part of a network controlling degradative enzyme synthesis and competence in Bacillus subtilis. Res Microbiol. 1994;145:393–402. doi: 10.1016/0923-2508(94)90087-6. [DOI] [PubMed] [Google Scholar]
- 13.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 14.Liu W, Hulett F M. Comparison of PhoP binding to the tuaA promoter with PhoP binding to other Pho-regulon promoters establishes a Bacillus subtilis Pho core binding site. Microbiology. 1998;144:1443–1450. doi: 10.1099/00221287-144-5-1443. [DOI] [PubMed] [Google Scholar]
- 15.Martin-Verstraete I, Débarbouillé M, Klier A, Rapoport G. Mutagenesis of the Bacillus subtilis “−12, −24” promoter of the levanase operon and evidence for the existence of an upstream activating sequence. J Mol Biol. 1992;226:85–99. doi: 10.1016/0022-2836(92)90126-5. [DOI] [PubMed] [Google Scholar]
- 16.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 17.Msadek T, Dartois V, Kunst F, Herbaud M-L, Denizot F, Rapoport G. ClpP of Bacillus subtilis is required for competence development, motility, degradative enzyme synthesis, growth at high temperature and sporulation. Mol Microbiol. 1998;27:899–914. doi: 10.1046/j.1365-2958.1998.00735.x. [DOI] [PubMed] [Google Scholar]
- 18.Mullis K B, Faloona F A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 19.Nakano M M, Yang F, Hardin P, Zuber P. Nitrogen regulation of nasA and the nasB operon, which encode genes required for nitrate assimilation in Bacillus subtilis. J Bacteriol. 1995;177:573–579. doi: 10.1128/jb.177.3.573-579.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakano M M, Hoffman T, Zhu Y, Jahn D. Nitrogen and oxygen regulation of Bacillus subtilis nasDEF encoding NADH-dependent nitrite reductase by TnrA and ResDE. J Bacteriol. 1998;180:5344–5350. doi: 10.1128/jb.180.20.5344-5350.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, et al. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 23.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schreier H J, Brown S W, Hirschi K D, Nomellini J F, Sonenshein A L. Regulation of Bacillus subtilis glutamine synthetase gene expression by the product of the glnR gene. J Mol Biol. 1989;210:51–63. doi: 10.1016/0022-2836(89)90290-8. [DOI] [PubMed] [Google Scholar]
- 25.Serror P, Sonenshein A L. Interaction of CodY, a novel Bacillus subtilis DNA-binding protein, with the dpp promoter region. Mol Microbiol. 1996;20:843–852. doi: 10.1111/j.1365-2958.1996.tb02522.x. [DOI] [PubMed] [Google Scholar]
- 26.Tabor S, Richardson C C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci USA. 1987;84:4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vagner V, Dervyn E, Ehrlich S D. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology. 1998;144:3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- 28.Wray L V, Jr, Atkinson M R, Fisher S H. The nitrogen-regulated Bacillus subtilis nrgAB operon encodes a membrane protein and a protein highly similar to the Escherichia coli glnB-encoded PII protein. J Bacteriol. 1994;176:108–114. doi: 10.1128/jb.176.1.108-114.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wray L V, Jr, Ferson A E, Rohrer K, Fisher S H. TnrA, a transcription factor required for global nitrogen regulation in Bacillus subtilis. Proc Natl Acad Sci USA. 1996;93:8841–8845. doi: 10.1073/pnas.93.17.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wray L V, Jr, Zalieckas J M, Fisher S H. Mutational analysis of the TnrA-binding sites in the Bacillus subtilis nrgAB and gabP promoter regions. J Bacteriol. 1998;180:2943–2949. doi: 10.1128/jb.180.11.2943-2949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]