Abstract
Early diagnosis and treatment of patients with aggressive prostate cancer (PCa) remains a clinically unmet need. We aimed to determine the levels of small extracellular vesicle (sEV)-associated microRNAs (miRs); miR-4737, miR-6068, and miR-6076 in a large panel of PCa cells and delineate the biological significance of miR-6068 in promoting PCa cells. sEVs were isolated from the conditioned medium of PCa cells, followed by RNA extraction and quantitative Real-Time PCR analysis. Functional assays were performed, and the protein expression of hypermethylated in cancer 2 (HIC2), as a potential miR-6068 target gene, was evaluated in PCa tissues by immunohistochemistry. sEV-associated miR-6068, miR-4737, and miR-6076 levels displayed large and significant differences compared to normal cells. miR-6068 was explicitly upregulated in sEV of PC-3 and CWR-R1ca cells (P<0.010). Suppression of miR-6068 in CWR-R1ca cells decreased cell proliferation, colony formation, and cell migration. In contrast, upregulation of miR-6068 in RC77T/E cells decreased HIC2 levels and increased cell aggressive phenotypes. The overexpression of HIC2 in PCa tissues was primarily observed in the cytoplasm compared to benign prostatic hyperplasia (BPH) and normal tissues (P<0.0001). This study confirms the differential packaging of miR-4737, miR-6068, and miR-6076 in sEVs of PCa cells. MiR-6068 promotes PCa cells to acquire aggressive phenotypes by inhibiting the HIC2/Sirtuin 1 (SIRT1) axis.
Keywords: Prostate cancer, small extracellular vesicles, miR-6068, functional assays, HIC2
Introduction
Prostate cancer (PCa) is the second most common leading cause of cancer-related deaths among elderly men in the United States. According to a recent report by the American Cancer Society, it is estimated that about 191,930 new cases and 33,330 deaths will be diagnosed with PCa [1]. African-American (AA) men are highly susceptible to and develop more aggressive PCa when compared to Caucasian-American (CA) men, with 158.3 new cases diagnosed per 100,000 men [1]. Early diagnosis and treatment of aggressive PCa remains a current clinical challenge. Although prostate-specific antigen (PSA) is the gold standard screening biomarker for diagnosis of PCa, it is not associated with the mortality rate of PCa patients [2]. In some cases, the negative consequences of using PSA in screening PCa patients can lead to overdiagnosis, overtreatment, and other treatment complications [3,4]. Nevertheless, most newly used PCa detection, and diagnosis tools have less sensitivity and specificity, making it more challenging to treat PCa patients successfully (5). PCa is an androgen-driven disease where tumor cells at early stages of PCa are sensitive to androgen [5]. Under androgen depletion therapy (ADT), the standard goal of treatment is to achieve circulating testosterone level of less than 50 ng/dL [6]. Although ADT is effective, treatment of metastatic PCa and molecular mechanisms underlying the development of androgen-sensitive tumors to castration-resistant PCa (CRPC) is still an unmet medical need. To address this challenge, it is necessary to identify new molecular biomarkers that can stratify patients according to their aggressive phenotypes.
As mean of cell communication, small extracellular vesicles (sEVs) are best described as small extracellular vesicles (30-150 nm in diameter) released by most cells as a result of the fusion of an intermediate endocytic compartment with the plasma membrane [7]. sEVs carry different biomolecules differentially loaded into their cargo, including proteins, mRNAs, microRNAs (miRs), DNAs, and lipids. The contents of sEVs cargo often vary under various pathologic conditions that reflect the original host cell [8]. microRNAs are small noncoding RNAs that suppress gene expression at the posttranscriptional level [9]. Accordingly, a number of circulating sEV-associated miRs are associated with tumor progression and metastasis [10]. Hypermethylated in cancer 2 (HIC2) is a transcriptional factor associated with HIC1 [11]. However, its biological function in PCa aggressiveness has not been investigated.
This study investigated the expression levels of three miRs in sEVs isolated from the conditioned medium of a large panel of PCa cell lines. We also elucidated the mechanism by which the sEV-associated miR-6068 promoted the aggressive behavior of PCa cells and identified HIC2 as a potential target of miR-6068. HIC2 protein expression was examined in eighty PCa and non-cancerous tissue specimens and was correlated to the available clinical outcomes of PCa patients.
Materials and methods
Cell culture
Human PCa DU-145, C4-2B, LNCaP, CWR22-RV1, PC-3 cells, and normal prostatic epithelial RWPE-1 cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). CWR-R1ca was purchased from Millipore Sigma (Burlington, MA, USA). CWR-R1ca are aggressive castration-recurrent cells derived from CWR-R1 cells depleted from fibroblasts [12]. RC77T/E and RC77N/E cell lines were kindly provided by Dr. J.S. Rhim (Uniformed Services University) and maintained as reported [12]. RC77T/E cells derived from 63-year-old African American PCa patients at Gleason score 7 [13]. DU-145, C4-2B, LNCaP, CWR22-RV1, and PC-3 cells were cultured in Dulbecco’s Modification of Eagle’s Medium (DMEM) supplemented with 10% FBS (Corning, Manassas, VA) and 1% penicillin/streptomycin (Life Technologies Corp., Grand Island, NY). CWR-R1ca cell line was cultured in RPMI-1640 media supplemented with 10% FBS and 1% antibiotic mixture. RWPE-1, RC77 N/E, and RC77 T/E cells were cultured in keratinocyte-SFM media kit (Gibco Laboratories, Carlsbad, CA) supplemented with EGF (5 ng/mL) and bovine pituitary extract (0.05 mg/mL). Cells were maintained at 37°C in 5% CO2 and humidified incubator as previously described [14]. All cells tested negative for mycoplasma and kept in the culture for short time period while doing the experiments.
sEVs collection, isolation, and characterization
PCa and normal prostate epithelial cells were grown up to 70% confluency [14]. Cells were washed with PBS and maintained in a complete medium that contained 10% sEVs-free FBS and 1% antibiotics. Utilization of sEVs-free FBS will reduce cell stress due to serum starvation. sEVs were collected from conditioned medium after 36 hours of incubation using Capturem EV isolation kits (Takara Bio USA Inc., Mountain View, CA). Briefly, sEVs containing media were collected in 50 mL tubes, centrifuged at 3000xg for 10 min. Next, the supernatant was transferred into another 50 mL filtration unit and centrifuged at 1000xg for 3 min, followed by sEVs elution, aliquoted, and stored at -80°C for further use. After isolation, the size of sEVs was measured by ZetaPals zeta potential analyzer (Brookhaven Instruments Corp., Holtsville, NY) and qNano method (Izon Science Ltd, Cambridge, MA) and as we previously described [15].
Quantitative RT-PCR (qPCR)
The dysregulated sEV-associated miRs isolated and PCa cells were evaluated by qPCR analysis using Bio-Rad CFX69 Touch thermal cycler (Bio-Rad, Hercules, CA). Total RNA was then extracted from the sEVs and their corresponding cells using TRIzol reagent following the standard protocol (ThermoFisher Scientific, Waltham, MA). Finally, cDNA specific to microRNA was synthesized using microRNA cDNA Poly (A) polymerase tailing kit (Applied Biological Materials Inc., Richmond, BC, Canada). Bright green-No dye SYBR mixture and hsa-miR-4737, hsa-miR-6068, has-miR-6076, U6, and 5S rRNA primers (Applied Biological Materials Inc., Richmond, BC, Canada) were used in the study. The fold change of miRs expression was calculated compared to the reference genes U6 and 5S rRNA by comparative Ct method as described [16]. The expression of hic2 in miR-6068-transfected and non-transfected cells was evaluated by qPCR using the following set of primers (forward: 5’-CATTGATGCACCCCCAGGAA-3’; backward: 5’-ATGACGTCACACAGGAAGCC-3’) and β-actin as an internal control (forward: 5’-TGAGACCTTCAACACCCCAGCCATG-3’; backward: GTAGATGGGCACAGTGTGGGTG).
Transfection of PCa cells with miR-6068 mimic and inhibitor
About 70% confluent CWR-R1ca cells were transfected with miR-6068 inhibitor and highly validated non-silencing siRNA allStars as a negative control using HiPerFect transfection reagent (Qiagen, Germantown, MD). The concentration of miR inhibitor was optimized to 50 nM in optimum medium (Gibco Laboratories, Carlsbad, CA). On the other hand, RC-77 T/E and PC-3 cell lines were transfected with 25 nM miR-6068 mimic. This process was followed by total RNA and protein extractions after 24 h and 72 h of transfection, respectively. Micro-cDNA and cDNA were synthesized, and qPCR was performed to examine the transfection efficiency and evaluate the miR and its target genes. The fold change of miR-6068 expression was calculated relative to negative control using U6 and 5S rRNA as internal controls, as we previously reported [14].
Cell proliferation assay
About 2 × 103 transfected and control cells were seeded in a 96-well plate and maintained at 37°C in 5% CO2 atmosphere for 72 h as previously described [17]. Cell proliferation was assessed using cell counting kit-8 (Dojindo Molecular Tech. Inc., Rockville, MD) following the standard protocol. After 4 h of incubation, the developed color was measured by microplate reader (AccuSkan FC plate reader, ThermoFisher Scientific, Waltham, MA) at 450 nm.
Clonogenic assay
Four hundred PCa cells transfected with either miR inhibitor or mimic or all-star negative control were seeded in 6-well plate. Cell culture media were changed every other day, and cells were maintained for 2-3 weeks until the colonies developed as described [17]. Next, the developed colonies were washed with PBS, fixed with 4% paraformaldehyde for 5 min, and stained with 0.5% crystal-violet (Hardy Diagnostics, Santa Maria, CA) for 30 min. The plates were then rinsed with tap water and left overnight to dry out before imaging and counting.
Migration assay
According to our prior study, transwell migration assay was conducted in 24-well chambers [17]. Briefly, transfected cells were resuspended in serum-free media and added to the upper chamber, whereas the lower chamber included 10% FBS complete medium. Cells were incubated for 24 h, and the migrated cells were then fixed and stained by crystal-violet dye (Hardy Diagnostics, Santa Maria, CA, USA). Finally, stained migrated cells are counted under light microscope (Leica Microsystem Inc., Buffalo Grove, IL).
miRNA target prediction
TargetScan, Diana-tools, and miRWalk bioinformatic programs searched for miR-6068 target genes. The three bioinformatic tools identified the predicted and reproduced target genes selected and then validated in transfected cells with either miR mimic or inhibitor.
Western blot analysis
Western blot analysis was performed as previously described [17]. Briefly, protein lysates were collected, and their concentrations were measured using Bradford method (ThermoFisher Scientific, Waltham, MA). About 30 µg protein lysate was uploaded onto a precast 4-20% SDS-PAGE gel, and the resolved proteins were then transferred onto a nitrocellulose membrane (Bio-Rad, Hercules, CA, USA). This process was followed by incubating the membranes in a blocking buffer containing 5% BSA for 1 h at room temperature. The blocked membranes were incubated overnight at 4°C with anti-HIC2 (Cat.#22788-1-AP, dilution 1:1000), anti-SIRT1 (Cat.#13161-1-AP, dilution 1:1000) from Proteintech (Rosemont, IL, USA), and anti-GAPDH (Cat.#sc-365604; Santa Cruz Biotechnology, Dallas, TX, USA) primary antibodies. After extensive washing, the membranes were incubated with the appropriate secondary antibodies for 1 h at room temperature. The signal was then developed by Clarity Western ECL substrate (Bio-Rad, Hercules, CA) using C-Digit scanner (Li-COR Biosciences, Lincoln, NE).
Immunohistochemical (IHC) analysis
The protocol of this study was approved by the Institutional Review Board (IRB) of Edward Via College of Osteopathic Medicine (VCOM), Virginia (IRB#2020-036). Prior to initiating this study, written informed consents were obtained from all patients. Tissue microarray (TMA) slide comprised 80 human cases comprising 50 PCa tissue specimens, and 30 normal and hyperplastic prostate tissues were purchased (US Biomax, Inc., Derwood, MD). IHC analysis was performed as we previously described [17]. Briefly, PCa tissue sections were de-waxed in xylene and rehydrated in a descending series of ethyl alcohol. The tissue slides were then heated in EDTA buffer at pH 8.0 for 20 min. The tissue sections were then incubated in 3% H2O2 to block the action of tissue endogenous peroxidases. The TMA slide was incubated with anti-HIC2 antibody overnight at 4°C. The developed antigen-antibody complex was detected by VECTASTAIN Elite ABC HRP Kit (Vector Laboratories, Burlingame, CA). The tissue slides were counterstained with hematoxylin, and images were acquired using a Nikon light microscope (Eclipse 80i Nikon Ins., Melville, NY). The histochemical score was calculated as previously described [18].
Statistical analysis
Data are presented as mean ± standard error of the mean (SEM). Relative fold change is calculated using the comparative ct method (2^ΔΔCT) using reference genes as internal controls [15]. For comparisons between experimental and control group, we used an unpaired student t-test and one-way analysis of variance (ANOVA) followed by post-hoc Tukey’s test for multiple groups. Graphs were generated by GraphPad Prism Version 9.0 (GraphPad Software, Inc., La Jolla, CA). Data were considered significant at p-value less than 0.05.
Results
Characterization of small extracellular vesicles (sEVs) isolated from PCa cells
The study design and characterization of PCa-associated sEVs are summarized in Figure 1A. After isolating sEVs from the conditioned media of PCa and normal cells, the characterization of sEVs was performed by measuring the sEVs particle size using qNano method and ZetaPals zeta potential analyzer. In addition, surface protein markers of sEVs were previously validated by Western blot analysis according to our published studies [15,17]. As shown in Figure 1B, 1C, the size of identified sEVs derived from blood and conditioned medium of PCa cells ranged between 30-165 nm in diameter, which follows the guideline of the International Society for Extracellular Vesicles [19].
Figure 1.
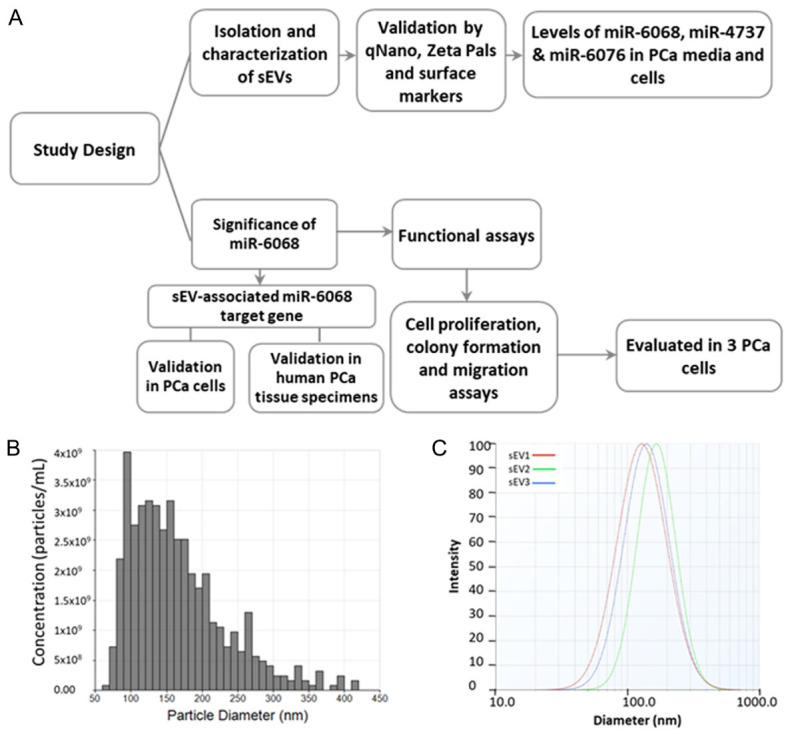
Study design and characterization of PCa-associated small extracellular vesicles. (A) Flowchart shows the study design. (B, C) Particle size of sEVs in triplicates (sEV-1 to 3) was measured by qNano method (B) and ZataPals Zeta potential analyzer (C).
Differential expression of sEV-associated miRs in PCa cells
Our previous study conducted miR profiling for sEVs derived from the blood of PCa patients and normal volunteers [15]. To examine their functional significance, we selected three dysregulated miRs (miR-4737, miR-6068, and miR-6076) to be validated in PCa cells and their respected sEVs isolated from culture media using qPCR analysis. Selection criteria based on the differential enrichment of these miRs in the tumor as compared to normal subjects (miR-6068), race as a risk factor of PCa patients (miR-6076), and Gleason score (miR-4737). The panel includes nine cell lines; two of them were normal epithelial prostate cells; RC77 N/E, which was established from African American (AA) patient, and RWPE-1 from Caucasian American (CA) man; one AA PCa cells (RC77T/E), and 6 CA PCa cells (CWR22-RV1, LNCaP, C4-2B, DU-145, PC-3, and CWR-R1ca). As illustrated in Figure 2, there was a differential loading of miRs in sEVs isolated from different PCa cells compared to the endogenous cellular expression of miRs. For example, miR-6068 was significantly upregulated (P<0.001) on the cellular level of CWR-R1ca but downregulated (P<0.01) in C4-2B and PC-3 cells when compared to normal RWPE1 and RC77 N/E cells. On the sEVs level, this miR was upregulated (P<0.001) in PC-3 and CWR-R1ca compared to sEVs derived from other PCa and normal cells. It is evident that miR-4737 was upregulated (P<0.01) in 22RV1 and PC-3 cells and downregulated (P<0.001) in C4-2B and RC77 T/E cells. The sEV-associated miR-4737 was upregulated (P<0.01) in PC-3 cells, whereas downregulated (P<0.001) in sEVs derived from CWR-R1ca and RC77 T/E cells. Concerning miR-6076 level, it was upregulated in DU145 cells (P<0.001) and downregulated (P<0.05) in LNCaP, C4-2B, PC-3, and RC77 T/E cells. sEV-associated miR-6076 was downregulated (P<0.05) in most of PCa cells except in CWR-R1ca cells, where it was upregulated (P<0.01).
Figure 2.
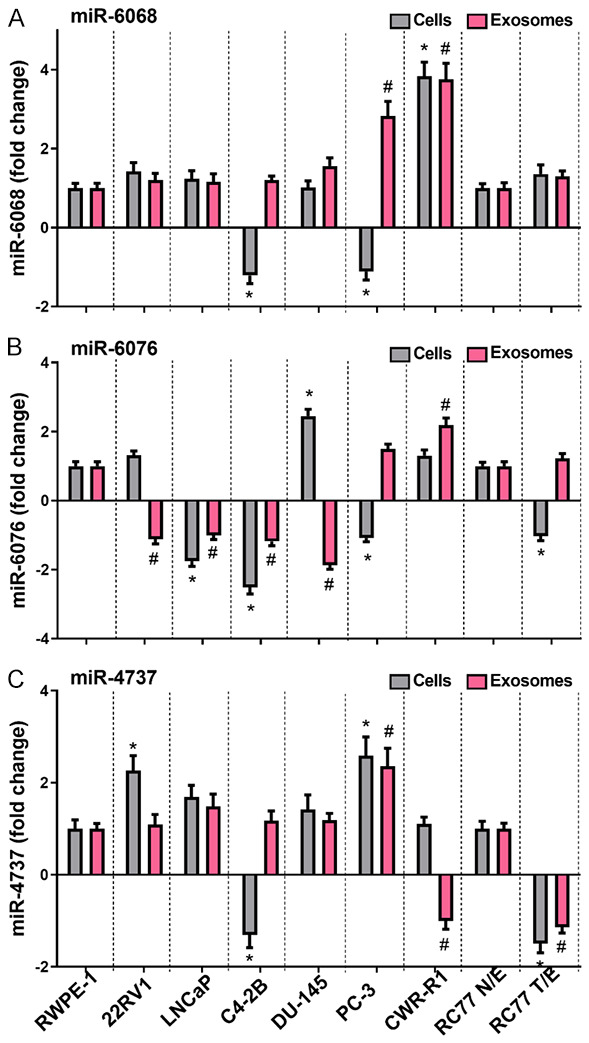
Expression of sEV-associated miR-6068, miR-6076 and miR-4737 in PCa cells. Total RNA was extracted from PCa and normal prostate cells and their respected sEVs. Micro-cDNA was synthesized, and qPCR was performed for miR-6068 (A), miR-6076 (B) and miR-4737 (C). U6 and 5srRNA were used as internal controls. Expression level was calculated as fold change relative to normal cell lines. Experiments were conducted in triplicates. Data were considered significant at P<0.05 compared to normal cells (*) or their respected sEVs (#).
Suppression of endogenous miR-6068 expression decreases cell proliferation, colony formation, and migration in CWR-R1ca cells
We selected CWR-R1ca cells to be transfected with miR-6068 inhibitor and RC77 T/E and PC-3 cells as a model for mimic transfection based on qPCR results. After transfection, we determined the functional significance of sEV-associated miR-6068 in these cells. After 24 h of transfection, cell proliferation, clonogenic, and migration assays were performed to determine the potential role of this miR in PCa aggressiveness. As expected, suppression of endogenous miR-6068 inhibited cell proliferation (P<0.001), colony formation (P=0.015), and migration (P=0.008) of CWR-R1ca cells (Figure 3). These results suggest the oncogenic properties of sEV-associated miR-6068.
Figure 3.
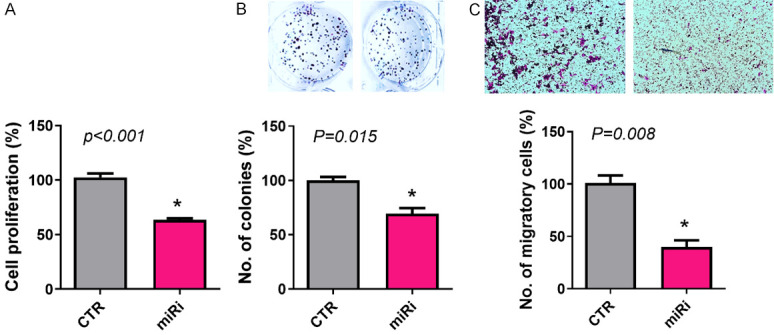
Suppression of endogenous expression of miR-6068 decreases cell viability, colony formation, and migration. CWR-R1a cells were transfected with miR-6068 inhibitor and non-specific miR as a negative control. After 24 h of transfection, the cells bearing miR-6068 were trypsinized and seeded for performing cell proliferation (A), colony formation (B), and migration (C) assays. Representative inserts showing cell colonies and migration of results generated from three independent experiments. *Depicts significance at P<0.05.
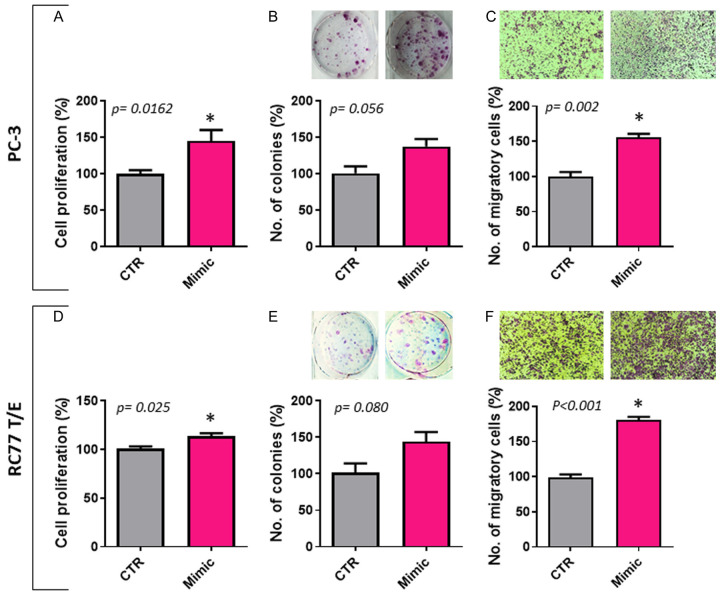
Ectopic expression of miR-6068 increases cell proliferation, colony formation, and migration in PC-3 and RC77 T/E cells
Selection of PC-3 and RC77 T/E cells was based on relatively lower endogenous expression of miR-6068 in these cells. PC-3 cells represent metastatic castration-resistant PCa (mCRPC), and RC77 T/E cells represent immortalized primary African American PCa at Gleason score 7. As shown in Figure 4, transfection of these two cells with miR-6068 mimic increased cell proliferation (P<0.05) and migration (P<0.01). There was a trend of increased colony formation, but it did not reach the significance level in PC-3 and RC77 T/E cells (P=0.056 and P=0.080, respectively).
Figure 4.
Ectopic expression of sEV-associated miR-6068 increases cell viability, colony formation, and migration. PC-3 and RC77T/E cells were transfected with miR-6068 mimic and non-specific miR as a negative control. After 24h of transfection, transfected cells were trypsinized and seeded for conducting cell proliferation (A, D), colony formation (B, E), and migration (C, F) assays. Representative inserts showing cell colonies and migration of results generated from three independent experiments. *Significance was calculated at P<0.05.
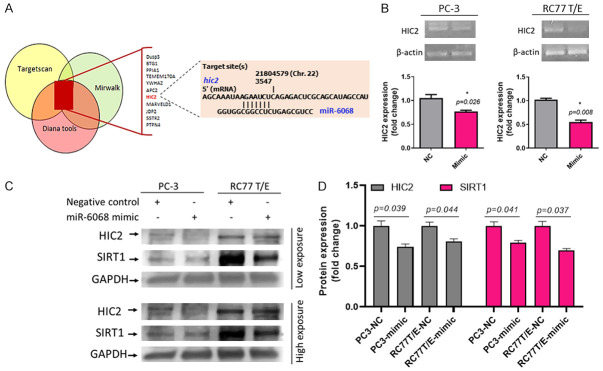
miR-6068 as a possible target for HIC2
To determine whether miR-6068 has an oncogenic role in PCa cells, we first identified the 11 common target genes of miR-6068 based on three bioinformatics tools; TargetScan, Diana-Tools, and miRWalk (Figure 5A). We selected the most relevant genes, and according to the results of qPCR and immunoblotting, we identified that miR-6068 inhibits HIC2 expression on RNA and protein levels. Transfection of PC-3 and RC77 T/E cells with miR-6068 mimic downregulates HIC2 gene (P=0.026 and P=0.008, respectively) on RNA level and its protein level (P<0.05) in the two cells (Figure 5B-D) suggesting that miR-6068 may regulate the expression of HIC2. The original immunoblots of HIC2 and Sirtuin 1 (SIRT1) were provided in Supplementary Figure 1. A further step was taken to identify the downstream gene of HIC2. Interestingly, targeting the HIC2 expression by miR-6068 mimic leads to inhibition of SIRT1 expression in RC77T/E and PC-3 cells.
Figure 5.
Hic2 is a possible gene target of miR-6068 in miR-mimic-transfected PCa cells. A: Selected top listed target genes of miR-6068 using three different prediction bioinformatic algorithms explaining the binding site of miR-6068 on the 3’-UTR of hic2 gene. B: Transient transfection of PCa cells with miR-6068 downregulates the hic2 gene. C: Western blot analysis (Left) shows a decrease in HIC2 and SIRT1 after transfection of PCa cells with miR-6068. D: The quantification of HIC2 and SIRT1 protein expression was performed. *Significance was calculated at P<0.05. The experiments were repeated at least twice.
HIC2 expression in human PCa tissue specimens
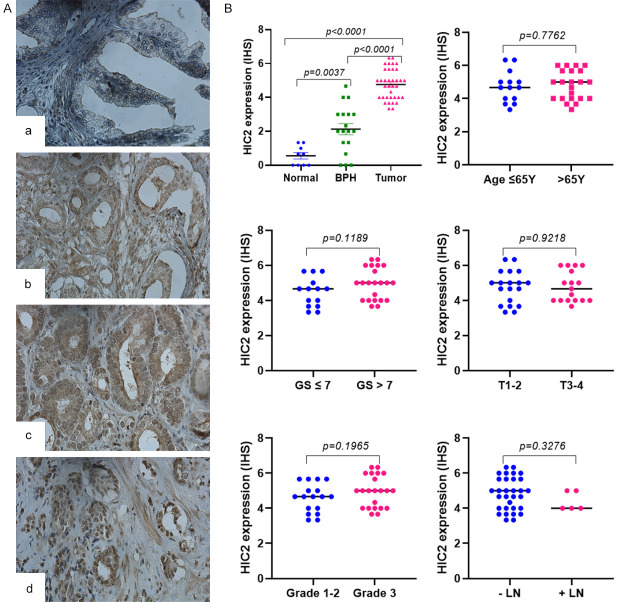
To validate the in vitro data of HIC2 as a potential target of miR-6068 in human tissues, eighty tissues cores comprising fifty PCa and thirty normal and hyperplastic tissues were stained with anti-HIC2 antibodies. The available clinical information of PCa patients and their age-matched healthy individuals is provided in Supplementary Table 1. As shown in Figure 6, immunohistochemical staining of HIC2 reveals unequivocal cytoplasmic and nuclear protein overexpression in PCa tissue (P<0.0001) compared to BPH and normal tissues. Of note, BPH had more expression of HIC2 (P=0.004) than normal prostatic tissues. When PCa tissues were considered, there were no significant differences between HIC protein expression and age at diagnosis (P=0.776), Gleason score (P=0.119), tumor stages (P=0.922), tumor grades (P=0.197) and lymph node involvement (P=0.328).
Figure 6.
Protein expression of HIC2 in PCa tissue specimens. HIC2 expression was evaluated by immunohistochemistry in tissue microarray slide comprising 50 tissue cores of PCa, 20 BPH, and ten normal prostate tissues. A: Immunohistochemical staining of HIC2 in tissues collected from normal individuals (a) and PCa tissue cores at different Gleason scores (GSs): b (GS<7), c (GS=7) and d (GS>7). B: Quantification of HIC2 expression in PCa tissues considering age, Gleason score, pathological stage, tumor grades, and lymph node involvement. Significance of the data was calculated at P<0.05. Magnification is 400X.
Discussion
This study evaluated the expression level of sEV-associated miR-4737, miR-6068, and miR-6076 in a large panel of PCa and normal cells. It was reported that these miRs are differentially expressed in different malignancies. For example, miR-4737 was identified as a new classifier in breast cancer tissue [20]. The high expression level of miR-6068 was found to be associated with the overall survival of patients with colorectal cancer [21]. It was also upregulated in tissues and plasma of lung squamous cell carcinoma patients [22]. Additionally, miR-6076 was used in combination with other miRs to screen patients with ovarian cancer [23] and discriminate PCa from other types of cancers [24]. Accumulated evidence shows that current sEV-associated miR-4737, miR-6068, and miR-6076 as promising novel biomarkers for diagnosis and/or prognosis of PCa patients. First, miR-4737 and miR-6068 are upregulated in several PCa cells and their respected sEVs. However, miR-6076 was downregulated in most PCa cell lines and associated sEVs. The present study reports that miR-6068 was upregulated in sEVs derived from conditioned media collected from PC-3 and CWR-R1ca cells compared to RC77T/E and normal cells. We also determined the biological significance of miR-6068 by suppressing its endogenous level in CWR-R1ca cells using a miR-6068 inhibitor. Suppression of miR-6068 resulted in a significant decrease in cell proliferation and migration. Irrefutable evidence shows that ectopic upregulation of miR-6068 in RC77 T/E and PC-3 cells promoted cell proliferation and migration. Consistent with our findings, miR-6068 was listed among five miRs associated with positive lymph node status in endometrial cancer [25].
A growing body of evidence shows that sEV-associated cargo bioactive molecules are associated with tumor progression, angiogenesis, drug resistance, and metastasis [8,26,27]. In this context, a number of studies have utilized sEVs-associated miRs in the stratification of PCa patients according to the clinical outcomes. For example, Huang et al. reported high expression of sEV-associated miR-1290 and miR-375 as prognostic markers associated with poor survival in plasma of CRPC patients [28]. In addition, miRs isolated from serum-derived sEVs of PCa patients showed a direct association of miR-141 and miR-375 with tumor metastasis [29]. HIC1 is involved in many cellular processes such as cell survival, growth, and motility. It is epigenetically silenced in solid cancers, including PCa [30,31]. Remarkably, hypermethylation of HIC1 promoter is not only found in solid tumors but also in normal tissues, including breast ductal [32], prostate epithelial [33], and brain tissues [34]. These findings suggest another uncharacterized regulatory mechanism than hypermethylation involved in HIC1 repression. In PCa cells, HIC1 silencing increases cell migration by promoting epithelial-mesenchymal transition (EMT) [35]. Several studies reported a number of HIC1 downstream target genes, including CXCR7, LCN2, SIRT1, ATOH1, CCND1, and P57KIP2, which are involved in the regulation of cell cycle, angiogenesis, and metastasis [30,31,36-38]. However, the potential role of HIC2 in regulating PCa progression and metastasis is still largely unknown. We report HIC2 as a target gene of miR-6068 and its downstream gene SIRT1, which adds another line of evidence to the role of miR-6068 in PCa. Sirtuins are differentially expressed in cancer types by exerting either oncogenic or tumor-suppressive activities based on the cellular context and experimental conditions [39]. For example, pharmacological inhibition of SIRT1 induces cell apoptosis and reduces tumor growth and chemoresistance in PCa cells [39]. On the contrary, knockout of SIRT1 in mouse-model increased tumor survival [40]. The link between HIC2 and SIRT1 was reported by Song et al., where HIC2 acts as a transcriptional activator of SIRT1 [41], and this finding may explain the expression pattern of HIC2/SIRT1-axis in PCa cells. By examining the protein expression of HIC2 in PCa tissue specimens, the protein was overexpressed in tumors compared to normal tissues. However, there was no difference in the protein expression at different stages of the disease, suggesting that HIC2 is involved in PCa initiation but not at late stages of the disease. The biological interaction between miRs and protein-coding gene(s) is a complex process where a single miR can control multiple target genes [42,43]. Some miRs are enriched in sEVs by RNA-binding proteins of tumor cells which is a tissue- and disease-specific process [44,45]. These enriched miRs in sEVs regulate tumor progression and metastasis and can be used as diagnostic/prognostic classifiers and as therapeutic targets [46,47]. It has been reported that tumor suppressor proteins such as p53 are overexpressed in tumor tissues and their mutational forms were corelated with poor clinical outcomes [48]. RB transcriptional corepressor 1 (RB1) loss of function was reported in PCa aggressive phenotypes. Approximately, 20% of 156 PCa tissues expressed RB1 and this expression was correlated with the copy number of the gene locus [49]. This could explain the expression pattern of HIC2 in PCa tissues used in our study. However, further studies are needed to assess the level of miR-6068 in a large number of tissue specimens and circulating blood collected from primary and metastatic PCa patients in addition to benign prostatic hyperplasia. Although we used a large panel of cell lines representing a wide variety of PCa heterogeneity, human cell lines still have limitations [50]. However, the goal of this study was to elucidate the molecular functions of these miRs. Therefore, we first examined their expression in different PCa cells to identify their endogenous expression. We used miR mimic and inhibitor to determine their role in PCa cell proliferation and migration based on the expression pattern. Finally, we examined the most relevant miR-target gene expression using IHC analysis in real PCa tissue specimens. Taken together, we report a differential packaging of sEV-associated miR-4737, miR-6068, and miR-6076 in PCa and normal cells. In PCa cells, two miRs are upregulated (miR-4737 and miR-6068) and one is downregulated (miR-6076). Knockdown of miR-6068 in CWR-R1ca cells suppresses cell proliferation, colony formation and migration, while ectopic expression of the miR in RC77 T/E cells promotes PCa aggressive phenotype. Thus, we anticipate that miR-6068 promotes the aggressive phenotypes of PCa cells by targeting HIC2/SIRT1 axis.
Acknowledgements
We thank Ms. Jennifer Arnold from VCOM’s Grant Development Office for her editing and proofreading of the manuscript. This work was partially supported by NIH/NCI (R21CA194750) and VCOM/Delta Collaboration Research Program, VCOM/REAP Funding (ZYA) and by the Egyptian Higher Education through PhD-Joint Supervision Program (MSAG).
The protocol of this study was approved by the Institutional Review Board (IRB) of Edward Via College of Osteopathic Medicine (VCOM), Virginia (IRB#2020-036).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Saini S. PSA and beyond: alternative prostate cancer biomarkers. Cell Oncol (Dordr) 2016;39:97–106. doi: 10.1007/s13402-016-0268-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prensner JR, Rubin MA, Wei JT, Chinnaiyan AM. Beyond PSA: the next generation of prostate cancer biomarkers. Sci Transl Med. 2012;4:127rv3. doi: 10.1126/scitranslmed.3003180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caram ME, Skolarus TA, Cooney KA. Limitations of prostate-specific antigen testing after a prostate cancer diagnosis. Eur Urol. 2016;70:209–210. doi: 10.1016/j.eururo.2015.12.045. [DOI] [PubMed] [Google Scholar]
- 5.Fang D, Zhou L. Androgen deprivation therapy in nonmetastatic prostate cancer patients: Indications, treatment effects, and new predictive biomarkers. Asia Pac J Clin Oncol. 2019;15:108–120. doi: 10.1111/ajco.13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y, Sawyers CL, Scher HI. Targeting the androgen receptor pathway in prostate cancer. Curr Opin Pharmacol. 2008;8:440–448. doi: 10.1016/j.coph.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang X, Liang M, Dittmar R, Wang L. Extracellular microRNAs in urologic malignancies: chances and challenges. Int J Mol Sci. 2013;14:14785–14799. doi: 10.3390/ijms140714785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saber SH, Ali HEA, Gaballa R, Gaballah M, Ali HI, Zerfaoui M, Abd Elmageed ZY. Exosomes are the driving force in preparing the soil for the metastatic seeds: lessons from the prostate cancer. Cells. 2020;9:564. doi: 10.3390/cells9030564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shukla GC, Singh J, Barik S. MicroRNAs: processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol. 2011;3:83–92. [PMC free article] [PubMed] [Google Scholar]
- 10.Akoto T, Saini S. Role of exosomes in prostate cancer metastasis. Int J Mol Sci. 2021;22:3528. doi: 10.3390/ijms22073528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deltour S, Pinte S, Guerardel C, Leprince D. Characterization of HRG22, a human homologue of the putative tumor suppressor gene HIC1. Biochem Biophys Res Commun. 2001;287:427–434. doi: 10.1006/bbrc.2001.5624. [DOI] [PubMed] [Google Scholar]
- 12.Shourideh M, DePriest A, Mohler JL, Wilson EM, Koochekpour S. Characterization of fibroblast-free CWR-R1ca castration-recurrent prostate cancer cell line. Prostate. 2016;76:1067–1077. doi: 10.1002/pros.23190. [DOI] [PubMed] [Google Scholar]
- 13.Theodore S, Sharp S, Zhou J, Turner T, Li H, Miki J, Ji Y, Patel V, Yates C, Rhim JS. Establishment and characterization of a pair of non-malignant and malignant tumor derived cell lines from an African American prostate cancer patient. Int J Oncol. 2010;37:1477–1482. doi: 10.3892/ijo_00000800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ali HEA, Lung PY, Sholl AB, Gad SA, Bustamante JJ, Ali HI, Rhim JS, Deep G, Zhang J, Abd Elmageed ZY. Dysregulated gene expression predicts tumor aggressiveness in African-American prostate cancer patients. Sci Rep. 2018;8:16335. doi: 10.1038/s41598-018-34637-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ali HEA, Gaballah MSA, Gaballa R, Mahgoub S, Hassan ZA, Toraih EA, Drake BF, Abd Elmageed ZY. Small extracellular vesicle-derived microRNAs stratify prostate cancer patients according to gleason score, race and associate with survival of African American and Caucasian men. Cancers (Basel) 2021;13:5236. doi: 10.3390/cancers13205236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 17.Gaballa R, Ali HEA, Mahmoud MO, Rhim JS, Ali HI, Salem HF, Saleem M, Kandeil MA, Ambs S, Abd Elmageed ZY. Exosomes-mediated transfer of Itga2 promotes migration and invasion of prostate cancer cells by inducing epithelial-mesenchymal transition. Cancers (Basel) 2020;12:2300. doi: 10.3390/cancers12082300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abd Elmageed ZY, Moroz K, Srivastav SK, Fang Z, Crawford BE, Moparty K, Thomas R, Abdel-Mageed AB. High circulating estrogens and selective expression of ERbeta in prostate tumors of Americans: implications for racial disparity of prostate cancer. Carcinogenesis. 2013;34:2017–2023. doi: 10.1093/carcin/bgt156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, Ayre DC, Bach JM, Bachurski D, Baharvand H, Balaj L, Baldacchino S, Bauer NN, Baxter AA, Bebawy M, Beckham C, Bedina Zavec A, Benmoussa A, Berardi AC, Bergese P, Bielska E, Blenkiron C, Bobis-Wozowicz S, Boilard E, Boireau W, Bongiovanni A, Borras FE, Bosch S, Boulanger CM, Breakefield X, Breglio AM, Brennan MA, Brigstock DR, Brisson A, Broekman ML, Bromberg JF, Bryl-Gorecka P, Buch S, Buck AH, Burger D, Busatto S, Buschmann D, Bussolati B, Buzas EI, Byrd JB, Camussi G, Carter DR, Caruso S, Chamley LW, Chang YT, Chen C, Chen S, Cheng L, Chin AR, Clayton A, Clerici SP, Cocks A, Cocucci E, Coffey RJ, Cordeiro-da-Silva A, Couch Y, Coumans FA, Coyle B, Crescitelli R, Criado MF, D’Souza-Schorey C, Das S, Datta Chaudhuri A, de Candia P, De Santana EF, De Wever O, Del Portillo HA, Demaret T, Deville S, Devitt A, Dhondt B, Di Vizio D, Dieterich LC, Dolo V, Dominguez Rubio AP, Dominici M, Dourado MR, Driedonks TA, Duarte FV, Duncan HM, Eichenberger RM, Ekstrom K, El Andaloussi S, Elie-Caille C, Erdbrugger U, Falcon-Perez JM, Fatima F, Fish JE, Flores-Bellver M, Forsonits A, Frelet-Barrand A, Fricke F, Fuhrmann G, Gabrielsson S, Gamez-Valero A, Gardiner C, Gartner K, Gaudin R, Gho YS, Giebel B, Gilbert C, Gimona M, Giusti I, Goberdhan DC, Gorgens A, Gorski SM, Greening DW, Gross JC, Gualerzi A, Gupta GN, Gustafson D, Handberg A, Haraszti RA, Harrison P, Hegyesi H, Hendrix A, Hill AF, Hochberg FH, Hoffmann KF, Holder B, Holthofer H, Hosseinkhani B, Hu G, Huang Y, Huber V, Hunt S, Ibrahim AG, Ikezu T, Inal JM, Isin M, Ivanova A, Jackson HK, Jacobsen S, Jay SM, Jayachandran M, Jenster G, Jiang L, Johnson SM, Jones JC, Jong A, Jovanovic-Talisman T, Jung S, Kalluri R, Kano SI, Kaur S, Kawamura Y, Keller ET, Khamari D, Khomyakova E, Khvorova A, Kierulf P, Kim KP, Kislinger T, Klingeborn M, Klinke DJ 2nd, Kornek M, Kosanovic MM, Kovacs AF, Kramer-Albers EM, Krasemann S, Krause M, Kurochkin IV, Kusuma GD, Kuypers S, Laitinen S, Langevin SM, Languino LR, Lannigan J, Lasser C, Laurent LC, Lavieu G, Lazaro-Ibanez E, Le Lay S, Lee MS, Lee YXF, Lemos DS, Lenassi M, Leszczynska A, Li IT, Liao K, Libregts SF, Ligeti E, Lim R, Lim SK, Line A, Linnemannstons K, Llorente A, Lombard CA, Lorenowicz MJ, Lorincz AM, Lotvall J, Lovett J, Lowry MC, Loyer X, Lu Q, Lukomska B, Lunavat TR, Maas SL, Malhi H, Marcilla A, Mariani J, Mariscal J, Martens-Uzunova ES, Martin-Jaular L, Martinez MC, Martins VR, Mathieu M, Mathivanan S, Maugeri M, McGinnis LK, McVey MJ, Meckes DG Jr, Meehan KL, Mertens I, Minciacchi VR, Moller A, Moller Jorgensen M, Morales-Kastresana A, Morhayim J, Mullier F, Muraca M, Musante L, Mussack V, Muth DC, Myburgh KH, Najrana T, Nawaz M, Nazarenko I, Nejsum P, Neri C, Neri T, Nieuwland R, Nimrichter L, Nolan JP, Nolte-’t Hoen EN, Noren Hooten N, O’Driscoll L, O’Grady T, O’Loghlen A, Ochiya T, Olivier M, Ortiz A, Ortiz LA, Osteikoetxea X, Ostergaard O, Ostrowski M, Park J, Pegtel DM, Peinado H, Perut F, Pfaffl MW, Phinney DG, Pieters BC, Pink RC, Pisetsky DS, Pogge von Strandmann E, Polakovicova I, Poon IK, Powell BH, Prada I, Pulliam L, Quesenberry P, Radeghieri A, Raffai RL, Raimondo S, Rak J, Ramirez MI, Raposo G, Rayyan MS, Regev-Rudzki N, Ricklefs FL, Robbins PD, Roberts DD, Rodrigues SC, Rohde E, Rome S, Rouschop KM, Rughetti A, Russell AE, Saa P, Sahoo S, Salas-Huenuleo E, Sanchez C, Saugstad JA, Saul MJ, Schiffelers RM, Schneider R, Schoyen TH, Scott A, Shahaj E, Sharma S, Shatnyeva O, Shekari F, Shelke GV, Shetty AK, Shiba K, Siljander PR, Silva AM, Skowronek A, Snyder OL 2nd, Soares RP, Sodar BW, Soekmadji C, Sotillo J, Stahl PD, Stoorvogel W, Stott SL, Strasser EF, Swift S, Tahara H, Tewari M, Timms K, Tiwari S, Tixeira R, Tkach M, Toh WS, Tomasini R, Torrecilhas AC, Tosar JP, Toxavidis V, Urbanelli L, Vader P, van Balkom BW, van der Grein SG, Van Deun J, van Herwijnen MJ, Van Keuren-Jensen K, van Niel G, van Royen ME, van Wijnen AJ, Vasconcelos MH, Vechetti IJ Jr, Veit TD, Vella LJ, Velot E, Verweij FJ, Vestad B, Vinas JL, Visnovitz T, Vukman KV, Wahlgren J, Watson DC, Wauben MH, Weaver A, Webber JP, Weber V, Wehman AM, Weiss DJ, Welsh JA, Wendt S, Wheelock AM, Wiener Z, Witte L, Wolfram J, Xagorari A, Xander P, Xu J, Yan X, Yanez-Mo M, Yin H, Yuana Y, Zappulli V, Zarubova J, Zekas V, Zhang JY, Zhao Z, Zheng L, Zheutlin AR, Zickler AM, Zimmermann P, Zivkovic AM, Zocco D, Zuba-Surma EK. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Persson H, Kvist A, Rego N, Staaf J, Vallon-Christersson J, Luts L, Loman N, Jonsson G, Naya H, Hoglund M, Borg A, Rovira C. Identification of new microRNAs in paired normal and tumor breast tissue suggests a dual role for the ERBB2/Her2 gene. Cancer Res. 2011;71:78–86. doi: 10.1158/0008-5472.CAN-10-1869. [DOI] [PubMed] [Google Scholar]
- 21.Slattery ML, Herrick JS, Pellatt DF, Mullany LE, Stevens JR, Wolff E, Hoffman MD, Wolff RK, Samowitz W. Site-specific associations between miRNA expression and survival in colorectal cancer cases. Oncotarget. 2016;7:60193–60205. doi: 10.18632/oncotarget.11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pu Q, Huang Y, Lu Y, Peng Y, Zhang J, Feng G, Wang C, Liu L, Dai Y. Tissue-specific and plasma microRNA profiles could be promising biomarkers of histological classification and TNM stage in non-small cell lung cancer. Thorac Cancer. 2016;7:348–354. doi: 10.1111/1759-7714.12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yokoi A, Matsuzaki J, Yamamoto Y, Yoneoka Y, Takahashi K, Shimizu H, Uehara T, Ishikawa M, Ikeda SI, Sonoda T, Kawauchi J, Takizawa S, Aoki Y, Niida S, Sakamoto H, Kato K, Kato T, Ochiya T. Integrated extracellular microRNA profiling for ovarian cancer screening. Nat Commun. 2018;9:4319. doi: 10.1038/s41467-018-06434-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Urabe F, Matsuzaki J, Yamamoto Y, Kimura T, Hara T, Ichikawa M, Takizawa S, Aoki Y, Niida S, Sakamoto H, Kato K, Egawa S, Fujimoto H, Ochiya T. Large-scale circulating microRNA profiling for the liquid biopsy of prostate cancer. Clin Cancer Res. 2019;25:3016–3025. doi: 10.1158/1078-0432.CCR-18-2849. [DOI] [PubMed] [Google Scholar]
- 25.Canlorbe G, Wang Z, Laas E, Bendifallah S, Castela M, Lefevre M, Chabbert-Buffet N, Darai E, Aractingi S, Mehats C, Ballester M. Identification of microRNA expression profile related to lymph node status in women with early-stage grade 1-2 endometrial cancer. Mod Pathol. 2016;29:391–401. doi: 10.1038/modpathol.2016.30. [DOI] [PubMed] [Google Scholar]
- 26.Kawakami K, Fujita Y, Kato T, Mizutani K, Kameyama K, Tsumoto H, Miura Y, Deguchi T, Ito M. Integrin beta4 and vinculin contained in exosomes are potential markers for progression of prostate cancer associated with taxane-resistance. Int J Oncol. 2015;47:384–390. doi: 10.3892/ijo.2015.3011. [DOI] [PubMed] [Google Scholar]
- 27.Sanchez CA, Andahur EI, Valenzuela R, Castellon EA, Fulla JA, Ramos CG, Trivino JC. Exosomes from bulk and stem cells from human prostate cancer have a differential microRNA content that contributes cooperatively over local and pre-metastatic niche. Oncotarget. 2016;7:3993–4008. doi: 10.18632/oncotarget.6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang X, Yuan T, Liang M, Du M, Xia S, Dittmar R, Wang D, See W, Costello BA, Quevedo F, Tan W, Nandy D, Bevan GH, Longenbach S, Sun Z, Lu Y, Wang T, Thibodeau SN, Boardman L, Kohli M, Wang L. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur Urol. 2015;67:33–41. doi: 10.1016/j.eururo.2014.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zedan AH, Osther PJS, Assenholt J, Madsen JS, Hansen TF. Circulating miR-141 and miR-375 are associated with treatment outcome in metastatic castration resistant prostate cancer. Sci Rep. 2020;10:227. doi: 10.1038/s41598-019-57101-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 31.Cheng G, Sun X, Wang J, Xiao G, Wang X, Fan X, Zu L, Hao M, Qu Q, Mao Y, Xue Y, Wang J. HIC1 silencing in triple-negative breast cancer drives progression through misregulation of LCN2. Cancer Res. 2014;74:862–872. doi: 10.1158/0008-5472.CAN-13-2420. [DOI] [PubMed] [Google Scholar]
- 32.Zhang W, Zeng X, Briggs KJ, Beaty R, Simons B, Chiu Yen RW, Tyler MA, Tsai HC, Ye Y, Gesell GS, Herman JG, Baylin SB, Watkins DN. A potential tumor suppressor role for Hic1 in breast cancer through transcriptional repression of ephrin-A1. Oncogene. 2010;29:2467–2476. doi: 10.1038/onc.2010.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morton RA Jr, Watkins JJ, Bova GS, Wales MM, Baylin SB, Isaacs WB. Hypermethylation of chromosome 17P locus D17S5 in human prostate tissue. J Urol. 1996;156:512–516. doi: 10.1097/00005392-199608000-00073. [DOI] [PubMed] [Google Scholar]
- 34.Rood BR, Zhang H, Weitman DM, Cogen PH. Hypermethylation of HIC-1 and 17p allelic loss in medulloblastoma. Cancer Res. 2002;62:3794–3797. [PubMed] [Google Scholar]
- 35.Hao M, Li Y, Wang J, Qin J, Wang Y, Ding Y, Jiang M, Sun X, Zu L, Chang K, Lin G, Du J, Korinek V, Ye DW, Wang J. HIC1 loss promotes prostate cancer metastasis by triggering epithelial-mesenchymal transition. J Pathol. 2017;242:409–420. doi: 10.1002/path.4913. [DOI] [PubMed] [Google Scholar]
- 36.Van Rechem C, Rood BR, Touka M, Pinte S, Jenal M, Guerardel C, Ramsey K, Monte D, Begue A, Tschan MP, Stephan DA, Leprince D. Scavenger chemokine (CXC motif) receptor 7 (CXCR7) is a direct target gene of HIC1 (hypermethylated in cancer 1) J Biol Chem. 2009;284:20927–20935. doi: 10.1074/jbc.M109.022350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng J, Wang J, Sun X, Hao M, Ding T, Xiong D, Wang X, Zhu Y, Xiao G, Cheng G, Zhao M, Zhang J, Wang J. HIC1 modulates prostate cancer progression by epigenetic modification. Clin Cancer Res. 2013;19:1400–1410. doi: 10.1158/1078-0432.CCR-12-2888. [DOI] [PubMed] [Google Scholar]
- 38.Van Rechem C, Boulay G, Pinte S, Stankovic-Valentin N, Guerardel C, Leprince D. Differential regulation of HIC1 target genes by CtBP and NuRD, via an acetylation/SUMOylation switch, in quiescent versus proliferating cells. Mol Cell Biol. 2010;30:4045–4059. doi: 10.1128/MCB.00582-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carafa V, Altucci L, Nebbioso A. Dual tumor suppressor and tumor promoter action of sirtuins in determining malignant phenotype. Front Pharmacol. 2019;10:38. doi: 10.3389/fphar.2019.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Di Sante G, Pestell TG, Casimiro MC, Bisetto S, Powell MJ, Lisanti MP, Cordon-Cardo C, Castillo-Martin M, Bonal DM, Debattisti V, Chen K, Wang L, He X, McBurney MW, Pestell RG. Loss of Sirt1 promotes prostatic intraepithelial neoplasia, reduces mitophagy, and delays PARK2 translocation to mitochondria. Am J Pathol. 2015;185:266–279. doi: 10.1016/j.ajpath.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song JY, Lee SH, Kim MK, Jeon BN, Cho SY, Lee SH, Kim KS, Hur MW. HIC2, a new transcription activator of SIRT1. FEBS Lett. 2019;593:1763–1776. doi: 10.1002/1873-3468.13456. [DOI] [PubMed] [Google Scholar]
- 42.Xu P, Wu Q, Yu J, Rao Y, Kou Z, Fang G, Shi X, Liu W, Han H. A systematic way to infer the regulation relations of miRNAs on target genes and critical miRNAs in cancers. Front Genet. 2020;11:278. doi: 10.3389/fgene.2020.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu C, Zhang S, Wang Q, Zhang X. Tumor suppressor miR-1 inhibits tumor growth and metastasis by simultaneously targeting multiple genes. Oncotarget. 2017;8:42043–42060. doi: 10.18632/oncotarget.14927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teng Y, Ren Y, Hu X, Mu J, Samykutty A, Zhuang X, Deng Z, Kumar A, Zhang L, Merchant ML, Yan J, Miller DM, Zhang HG. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nat Commun. 2017;8:14448. doi: 10.1038/ncomms14448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu YF, Xu X, Gin A, Nshimiyimana JD, Mooers BHM, Caputi M, Hannafon BN, Ding WQ. SRSF1 regulates exosome microRNA enrichment in human cancer cells. Cell Commun Signal. 2020;18:130. doi: 10.1186/s12964-020-00615-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Z, Ma YY, Wang J, Zeng XF, Li R, Kang W, Hao XK. Exosomal microRNA-141 is upregulated in the serum of prostate cancer patients. Onco Targets Ther. 2016;9:139–148. doi: 10.2147/OTT.S95565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thind A, Wilson C. Exosomal miRNAs as cancer biomarkers and therapeutic targets. J Extracell Vesicles. 2016;5:31292. doi: 10.3402/jev.v5.31292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Incognito LS, Cazares LH, Schellhammer PF, Kuban DA, Van Dyk EO, Moriarty RP, Wright GL Jr, Somers KD. Overexpression of p53 in prostate carcinoma is associated with improved overall survival but not predictive of response to radiotherapy. Int J Oncol. 2000;17:761–769. doi: 10.3892/ijo.17.4.761. [DOI] [PubMed] [Google Scholar]
- 49.Sharma A, Yeow WS, Ertel A, Coleman I, Clegg N, Thangavel C, Morrissey C, Zhang X, Comstock CE, Witkiewicz AK, Gomella L, Knudsen ES, Nelson PS, Knudsen KE. The retinoblastoma tumor suppressor controls androgen signaling and human prostate cancer progression. J Clin Invest. 2010;120:4478–4492. doi: 10.1172/JCI44239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilding JL, Bodmer WF. Cancer cell lines for drug discovery and development. Cancer Res. 2014;74:2377–2384. doi: 10.1158/0008-5472.CAN-13-2971. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.