Abstract
Historically, the primary focus of cancer research has been molecular and clinical studies of a few essential pathways and genes. Recent years have seen the rapid accumulation of large-scale cancer omics data catalysed by breakthroughs in high-throughput technologies. This fast data growth has given rise to an evolving concept of ‘big data’ in cancer, whose analysis demands large computational resources and can potentially bring novel insights into essential questions. Indeed, the combination of big data, bioinformatics and artificial intelligence has led to notable advances in our basic understanding of cancer biology and to translational advancements. Further advances will require a concerted effort among data scientists, clinicians, biologists and policymakers. Here, we review the current state of the art and future challenges for harnessing big data to advance cancer research and treatment.
Subject terms: Computational biology and bioinformatics, Cancer genomics, Cancer epigenetics, Cancer therapy
The increasing size of cancer datasets requires new ways of thinking for analysing and integrating these data. In this Review, Jiang et al. discuss considerations and strategies for wielding ‘big data’ ― large, information-rich datasets ― in basic research and for translational applications such as identifying biomarkers, informing clinical trials and developing new assays and treatments.
Introduction
Cancer is a complex process, and its progression involves diverse processes in the patient’s body1. Consequently, the cancer research community generates massive amounts of molecular and phenotypic data to study cancer hallmarks as comprehensively as possible. The rapid accumulation of omics data catalysed by breakthroughs in high-throughput technologies has given rise to the notion of ‘big data’ in cancer, which we define as a dataset with two basic properties; first, it contains abundant information that can give novel insights into essential questions, and second, its analysis demands a large computer infrastructure beyond equipment available to an individual researcher — an evolving concept as computational resources evolve exponentially following Moore’s law. A model example of such big data is the dataset collected by The Cancer Genome Atlas (TCGA)2. TCGA contains 2.5 petabytes of raw data — an amount 2,500 times greater than modern laptop storage in 2022 — and requires specialized computers for storage and analysis. Further, between its initial release in 2008 to March 2022, at least 10,242 articles and 11,054 NIH grants cited TCGA according to a PubMed search, demonstrating its transformative value as a community resource that has markedly driven cancer research forward.
Big data are not unique to the cancer field, and play an essential role in many scientific disciplines, notably cosmology, weather forecasting and image recognition. However, datasets in the cancer field differ from those in other fields in several key aspects. First, the size of cancer datasets is typically markedly smaller. For example, in March 2022, the US National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database3 — the largest genomics data repository to our knowledge — contained approximately 1.1 million samples with ‘cancer’ as a keyword. However, ImageNet, the largest public repository for computer vision, contains 15 million images4. Second, cancer research data are typically heterogeneous and may contain many dimensions measuring distinct aspects of cellular systems and biological processes. Modern multi-omics workflows may generate genome-wide mRNA expression, chromatin accessibility and protein expression data on single cells5, together with a spatial molecular readout6. The comparatively limited data size in each modality and the high heterogeneity among them necessitate the development of innovative computational approaches for integrating data from different dimensions and cohorts.
The subject of big data in cancer is of immense scope, and it is impossible to cover everything in one review. We therefore focus on key big-data analyses that led to conceptual advances in our understanding of cancer biology and impacted disease diagnosis and treatment decisions. Further, we detail reviews in the pertaining sections to direct interested readers to relevant resources. We acknowledge that our limited selection of topics and examples may omit important work, for which we sincerely apologize.
In this Review, we begin by describing major data sources. Next, we review and discuss data analysis approaches designed to leverage big datasets for cancer discoveries. We then introduce ongoing efforts to harness big data in clinically oriented, translational studies, the primary focus of this Review. Finally, we discuss current challenges and future steps to push forward big data use in cancer.
Common data types
There are five basic data types in cancer research: molecular omics data, perturbation phenotypic data, molecular interaction data, imaging data, and textual data. Molecular omics data describe the abundance or status of molecules in cellular systems and tissue samples. Such data are the most abundant type generated in cancer research from patient or preclinical samples, and include information on DNA mutations (genomics), chromatin or DNA states (epigenomics), protein abundance (proteomics), transcript abundance (transcriptomics) and metabolite abundance (metabolomics) (Table 1). Early studies relied on data from bulk samples to provide insights into cancer progressions, tumour heterogeneity and tumour evolution, by using well-designed computational approaches7–10. Following the development of single-cell technologies and decreases in sequencing costs, current molecular data can be generated at multisample and single-cell levels11,12 and reveal tumour heterogeneity and evolution at a much higher resolution. Furthermore, genomic and transcriptomic readouts can include spatial information13, revealing cancer clonal evolutions within distinct regions and gene expression changes associated with clone-specific aberrations. Although more limited in resolution, conventional bulk analyses are still useful for analysing large patient cohorts as the generation of single-cell and spatial data is costly and often feasible for only a few tumours per study.
Table 1.
Common molecular omics data types in cancer research
| Data type | Technology | Description |
|---|---|---|
| DNA mutations | Whole-exome/whole-genome sequencing | Reveals DNA nucleotide mutations, such as single-nucleotide missense mutations, frameshift insertions or deletions, nonsense mutations149,150, copy number alterations151, DNA non-coding variations in regulatory regions that may impact the gene regulatory network152 and large structural variants, such as genome rearrangements and chromothripsis152. Whole-exome sequencing can provide focused readouts if only protein-coding alterations are needed. Single-cell genome sequencing is possible on a few cells153 |
| Chromatin accessibility | ATAC-seq or DNase I-seq | Reveals accessible chromatin regions in bulk cells, a hallmark of active DNA regulatory elements154. Coupled with cell barcoding techniques, ATAC-seq technologies can reveal chromatin accessibility at the single-cell level155 |
| Histone modification | ChIP–seq | Identifies the genome-wide location of DNA-binding proteins or histones with diverse modifications156. Single-cell ChIP–seq can reveal chromatin states for hundreds of cells157 |
| DNA methylation | Bisulfite sequencing and BeadChip | Bisulfite conversion of unmethylated cytosine to uracil, coupled with sequencing or BeadChip, enables genome-wide profiling of DNA methylation patterns158. Single-cell bisulfite sequencing can provide methylation readout at up to 50% of CpG dinucleotides on the genome scale159 |
| Transcriptomics | Microarrays | Reveal gene expression level or transcript isoforms from diverse patient sample types160 |
| RNA-seq |
Reveals gene expression level, transcript isoforms or fusions from diverse patient sample types160 Droplet-based161,162, plate-based163 or MicroWell164 technologies can assign DNA barcodes to individual cells, enabling transcriptomics profiling in single cells |
|
| Spatial transcriptomic techniques165 | Generate gene expression data with spatial location information based on positional barcoding, such as spatial transcriptomics166 and Slide-seq167, or in situ sequencing, such as FISSEQ168. Certain technologies, such as spatial transcriptomics, cannot achieve single-cell spatial resolution because the detection spot diameter covers multiple cells | |
| Proteomics | Mass spectrometry | Profiles protein expression and phosphorylation in bulk samples on the genome scale169 |
| Protein array | Profiles protein expression and phosphorylation on a few targets with antibodies available170 | |
| CITE-seq | Based on antibodies tagged with DNA barcodes171, single-cell sequencing can generate transcriptomics readouts and levels on a few cell-surface targets | |
| Flow cytometry | Based on antibodies tagged with fluorophores, sorting technologies can profile protein levels on the single-cell level focused on a few targets | |
| Mass cytometry |
Based on antibodies tagged with metal isotopes172, mass spectrometry technologies can profile protein levels on the single-cell level focused on several targets A few technologies, such as imaging mass cytometry173, and multiplexed ion beam imaging174, can profile more than 30 protein antibody intensities in a tissue slice with spatial and single-cell resolution |
|
| CODEX | Can profile more than 30 protein antibody intensities in a tissue slice with spatial and single-cell resolution using antibodies with nucleotide imaging tags175 | |
| Metabolomics | NMR spectroscopy | Can reveal metabolites from patient samples on the basis of resonance frequencies of atoms and their immediate chemical environment in the magnetic field176 |
| Mass spectrometry | Reveals metabolites from samples on the basis of mass-to-charge ratios and comparisons in a database of known metabolites177 (unlike NMR spectroscopy, which can be used to determine structures of unknown molecules) |
By default, most technologies work on bulk samples. When applicable, single-cell or spatial solutions are discussed in the description. ATAC-seq, assay for transposase-accessible chromatin using sequencing; ChIP–seq, chromatin immunoprecipitation followed by sequencing; CODEX, co-detection by indexing; DNase I-seq, DNase I hypersensitive site sequencing; FISSEQ, fluorescent in situ sequencing; RNA-seq, RNA sequencing.
Perturbation phenotypic data describe how cell phenotypes, such as cell proliferation or the abundance of marker proteins, are altered following the suppression or amplification of gene levels14 or drug treatments15,16. Common phenotyping experiments include perturbation screens using CRISPR knockout17, interference or activation18; RNA interference19; overexpression of open reading frames20; or treatment with a library of drugs15,16. As a limitation, the generation of perturbation phenotypic data from clinical samples is still challenging due to the requirement of genetically manipulable live cells.
Molecular interaction data describe the potential function of molecules through their interacting with diverse partners. Common molecular interaction data types include data on protein–DNA interactions21, protein–RNA interactions22, protein–protein intercations23 and 3D chromosomal interactions24. Similar to perturbation phenotypic data, molecular interaction datasets are typically generated using cell lines as their generation requires a large quantity of material that often exceeds that available from clinical samples.
Clinical data such as health records25, histopathology images26 and radiology images27,28 can also be of considerable value. The boundary between molecular omics and image data is not absolute as both can include information of the other type, for example in datasets that contain imaging scans and information on protein expression from a tumour sample (Table 1).
Data repositories and analytic platforms
We provide an overview of key data resources for cancer research organized in three categories. The first category comprises resources from projects that systematically generate data (Table 2); for example, TCGA generated transcriptomic, proteomic, genomic and epigenomic data for more than 10,000 cancer genomes and matched normal samples, spanning 33 cancer types. The second category describes repositories presenting processed data from the aforementioned projects (Table 3), such as the Genomic Data Commons, which hosts TCGA data for downloading. The third category includes Web applications that systematically integrate data across diverse projects and provide interactive analysis modules (Table 4). For example, the TIDE framework systematically collected public data from immuno-oncology studies and provided interactive modules to study pathways and regulation mechanisms underlying tumour immune evasion and immunotherapy response29.
Table 2.
Large-scale projects generating cancer genomic datasets
| Project | Samples | Data type | Size | Description |
|---|---|---|---|---|
| TCGA | Primary cancers, matched normal samples, some metastatic samples | Gene expression, DNA mutations, DNA methylation, chromatin accessibility, CNA, protein expression, histopathology images | 11,315 cancer genomes from 33 cancer types | Joint effort between the US National Cancer Institute and the US National Human Genome Research Institute |
| ICGC | Primary cancers, matched normal samples, some metastatic samples | Gene expression, DNA mutations, DNA methylation, CNA, protein expression | 25,000 cancer genomes from 22 cancer types | A global cancer genomics effort for documenting somatic mutations that drive common tumour types |
| PCAWG | Samples from TCGA and ICGC | DNA variations from whole-genome sequencing | 2,658 cancer genomes from 38 tumour types | Revealed 288,457 structural variations across topologically associated domains152 |
| LINCS | Human cell lines | Differential expression upon treatment or genetic perturbations | 1.4 million gene expression profiles in 50 cell types, focused on approximately 1,000 landmark genes | Probes how cell models respond to chemical or genetic perturbations through use of microarrays focused on approximately 1,000 genes that are most representative of variations in the transcriptome16 |
| CCLE | Human cancer cell lines | Gene expression, DNA mutations, promoter methylation, CNA, metabolomics, drug sensitivity, CRISPR/RNAi genome-wide screens, protein expression for a few targets | 1,072 cell lines | Provides a data encyclopedia of human cancer cell lines178 |
| CPTAC | Human cancers and normal tissue | Protein expression and post-translational modifications | Almost 4,000 samples from 14 tumour sites | A national effort to understand the molecular basis of cancer through large-scale proteome genomics |
| Human Protein Atlas | Human cancers, normal tissues, cell models | IHC images, gene expression | 3.1 million annotated IHC tissue images for most protein-coding genes, spanning 17 cancer types | Aims to map all human proteins in tumours and tissues using IHC179 |
| GENIE | Human cancers | Exome mutations focused on common cancer-related genes | 136,096 cases from 110 cancer sites | A registry assembled through 19 cancer centres worldwide, aggregating sequencing data obtained during routine medical practice from patients with cancer |
| CAMELYON | Sentinel lymph nodes of patients with metastatic breast cancer | H&E-stained slides | 1,399 whole-slide images with pathology annotations of metastases regions | A challenge to evaluate new and existing algorithms for automated detection and classification of breast cancer metastases in whole-slide images of lymph nodes110 |
| TARGET | Paediatric cancers | Gene expression, DNA mutation (whole-genome and whole-exome sequencing), DNA methylation | 6,196 cancer genomes spanning 9 cancer types | Applies a comprehensive genomic approach to determine molecular changes that drive childhood cancers |
CCLE, Cancer Cell Line Encyclopedia; CNA, copy number alteration; CPTAC, Clinical Proteomic Tumour Analysis Consortium; H&E, haematoxylin and eosin; ICGC, International Cancer Genome Consortium; IHC, immunohistochemistry; PCAWG, Pan-Cancer Analysis of Whole Genomes; TARGET, Therapeutically Applicable Research to Generate Effective Treatments; TCGA, The Cancer Genome Atlas.
Table 3.
Data repositories hosting cancer genomics data
| Repository | Datasets included | Sample size | Description |
|---|---|---|---|
| GDC | 20 data-generation programmes, including TCGA, TARGET, GENIE and CPTAC | 85,552 cases from 67 primary cancer sites | Provides the cancer research community with a unified repository that enables data sharing across genomic studies |
| IDC | 115 data collections, including cohorts from TCGA, CPTAC and other projects | 61,134 cases from 21 primary cancer sites | Connects researchers with publicly available cancer imaging data and provides a cloud computing environment integrated with other cancer research data commons180 |
| TCIA | 169 data collections, including cohorts from TCGA, CPTAC and other projects | 65,508 cases from 69 disease types, including cancer and non-cancer types (for example, COVID-19) | De-identifies and hosts cancer medical images for public download, but not cloud computing use like IDC. Parts of its data are included in IDC. Also includes some private data collections |
| GEO | 177,063 data series; 53,740 contain ‘cancer’ as a keyword | 5,102,810 samples; 1,118,082 samples contain ‘cancer’ as a keyword in metadata | Host data submissions from various studies. It contains many individual biology studies that may support knowledge rediscovery |
| Array Express | 16,345 experiments; 3,293 contain ‘cancer’ as a keyword | 894,309 samples; 236,935 of them contain ‘cancer’ as a keyword in their metadata | A popular genomics data repository |
| FDC | 81,883 human datasets deposited in GEO and ArrayExpress | 3,707,349 samples in total, not restricted to cancer | Helps researchers annotate metadata in GEO and ArrayExpress to enable automatic algorithmic analysis and knowledge rediscovery34 |
CPTAC, Clinical Proteomic Tumour Analysis Consortium; FDC, Framework for Data Curation; GDC, Genomic Data Commons; GEO, Gene Expression Omnibus; IDC, Imaging Data Commons; TARGET, Therapeutically Applicable Research to Generate Effective Treatments; TCGA, The Cancer Genome Atlas; TCIA, The Cancer Imaging Archive.
Table 4.
Web applications that enable interactive analysis of cancer datasets
| Web application | Data sources integrated | Functions |
|---|---|---|
| cBioportal | 344 cancer omics data cohorts from large-scale projects, such as TCGA and GENIE, and many homogenized datasets from individual studies181 | Interactive analysis and visualization modules to find associations among different data types and clinical outcomes |
| UCSC Xena | 139 omics data cohorts from large-scale projects, such as TCGA, ICGC and GTEX, and many homogenized datasets from individual studies182 | |
| TIDE | Approximately 33,000 samples in 188 tumour cohorts from public databases, repurposed through computational models to study tumour immune evasion; 998 tumours from 12 immunotherapy clinical studies; 8 CRISPR screens in immunological models | Interactive data analysis and visualization modules to identify cancer immune evasion regulators, predict immune checkpoint blockade response from pretreatment transcriptomic profiles and evaluate new immunotherapy biomarkers in public cohorts29 |
| PRECOG | 166 gene expression datasets, collected from GEO and ArrayExpress | Query associations between gene expression and survival outcomes31 |
| RABIT | 686 ChIP–seq profiles representing 150 transcription factors with 7,484 TCGA tumour profiles in 18 cancer types | Presents transcription factors and RBPs shaping gene expression patterns in diverse cancer types by integrating ChIP–seq data from diverse cell models, with information on transcription factor and RBP motifs and tumour gene expression profiles183 |
| TISCH | 79 public single-cell RNA-seq datasets, including 2,045,746 cells | Shows gene expression levels in diverse cell populations in tumours184 |
| DepMap | Genome-wide CRISPR screen data from 1,086 cell lines and RNAi screen data from 710 cell lines, paired with omics profiles and drug sensitivities of cell models | Queries the effects of perturbing genes on cell line fitness. Also presents a cell line’s gene expression, copy number alterations and DNA mutations |
| Tres | 36 single-cell RNA-seq datasets from 168 tumours spanning 19 cancer types, 8 T cell transcriptomics datasets from immunotherapy response studies and 8 genome-wide genetic screens in T cells | Uses single-cell transcriptomic data from solid tumours to identify signatures of T cells that are resilient to immunosuppressive signals54. Users can query whether a gene is a positive or a negative marker of tumour-resilient T cells, or input gene expression profiles of T cells or T cell-enriched samples to predict the clinical efficacies of T cells in immune checkpoint blockade and adoptive cell transfer |
ChIP–seq, chromatin immunoprecipitation followed by sequencing; GEO, Gene Expression Omnibus; GTEX, Genotype-Tissue Expression Project; ICGC, International Cancer Genome Consortium; RBP, RNA-binding protein; RNA-seq, RNA sequencing; TCGA, The Cancer Genome Atlas.
In addition to cancer-focused large-scale projects enumerated in Table 2, many individual groups have deposited genomic datasets that are useful for cancer research in general databases such as GEO3 and ArrayExpress30. Curation of these datasets could lead to new resources for cancer biology studies. For example, the PRECOG database contains 166 transcriptomic studies collected from GEO and ArrayExpress with patient survival information for querying the association between gene expression and prognostic outcome31.
Integrative analysis
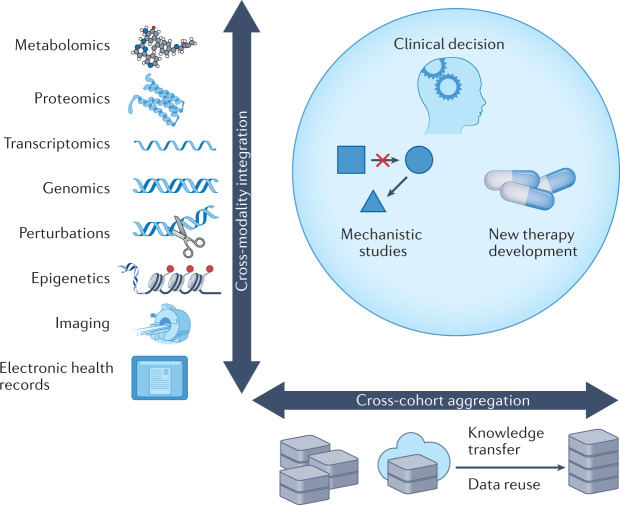
Although data-intensive studies may generate omics data on hundreds of patients, the data scale in cancer research is still far behind that in other fields, such as computer vision. Cross-cohort aggregation and cross-modality integration can markedly enhance the robustness and depth of big data analysis (Fig. 1). We discuss these strategies in the following subsections.
Fig. 1. Considerations for using big data in translational applications and basic research.
Clinical decisions, basic research and the development of new therapies should consider two orthogonal dimensions when leveraging big-data resources; integrating data across many data modalities and integrating data from different cohorts, which may include the transfer of knowledge from pre-existing datasets.
Cross-cohort data aggregation
Integration of datasets from multiple centres or studies can achieve more robust results and potentially new findings, especially where individual datasets are noisy, incomplete or biased with certain artefacts. A landmark of cross-cohort data aggregation is the discovery of the TMPRSS2–ERG fusion and a less frequent TMPRSS2–ETV1 fusion as oncogenic drivers in prostate cancer. A compendium analysis across 132 gene-expression datasets representing 10,486 microarray experiments first identified ERG and ETV1 as highly expressed genes in six independent prostate cancer cohorts32, further studies identified their fusions with TMPRSS2 as the cause of ERG and ETV1 overexpression. Another example is an integrative study of tumour immune evasion across many clinical datasets that revealed that SERPINB9 expression consistently correlates with intratumoural T cell dysfunction and resistance to immune checkpoint blockade29. Further studies found SERPINB9 activation to be an immune checkpoint blockade resistance mechanism in cancer cells29 and immunosuppressive cells33.
A general approach for cross-cohort aggregation is to obtain public datasets that are related to a new research topic or have similar study designs to a new dataset. However, use of public data for a new analysis is challenging because the experimental design behind each published dataset is unique, requiring labour-intensive expert interpretation and manual standardization. A recent framework for data curation provides natural language processing and semi-automatic functions to unify datasets with heterogeneous meta-information into a format usable for algorithmic analysis34 (Framework for Data Curation in Table 3).
Although data aggregation may generate robust hypotheses, batch effects caused by differences in laboratories, individual researcher’s techniques or platforms or other non-biological factors may mask or reduce the strength of signals uncovered35, and correcting for these effects is therefore a critical step in cross-cohort aggregations36,37. Popular batch effect correction approaches include the ComBat package, which uses empirical Bayes estimators to compute corrected data36, and the Seurat package, which creates integrated single-cell clusters anchored on similar cells between batches38. Despite the availability of batch correction methods, analysis of both original and corrected data is essential to draw reliable conclusions as batch correction can introduce false discoveries39.
Cross-modality data integration
Cross-modality integration of different data types is a promising and productive approach for maximizing the information gained from data as the information embedded in each data type is often complementary and synergistic40. Cross-modality data integration is exemplified by projects such as TCGA, which provides genomic, transcriptomic, epigenomic and proteomic data on the same set of tumours (Table 2). Cross-modality integration has led to many novel insights regarding factors associated with cancer progression. For example, the phosphorylation status of proteins in the EGFR signalling pathway — an indicator of EGFR signalling activity — is highly correlated with the expression of genes encoding EGFR ligands in head and neck cancers but not receptor expression, copy number alterations, protein levels or phosphorylations41, suggesting that patients should be stratified to receive anti-EGFR therapies on the basis of ligand abundance instead of receptor status.
A recent example of cross-modality data integration used single-cell multi-omics technologies that allowed genome-wide transcriptomics and chromatin accessibility data to be measured together with a handful of proteins of interest42. The advantages of using cross-modality data were clear as during cell lineage clustering, CD8+ T cell and CD4+ T cell populations could be clearly separated in the protein data but were blended when the transcriptome was analysed42. Conversely, dendritic cells formed distinct clusters when assessed on the basis of transcriptomic data, whereas they mixed with other cell types when assessed on the basis of cell-surface protein levels. Chromatin accessibility measured by assay for transposase-accessible chromatin using sequencing (ATAC-seq) further revealed T cell sublineages by capturing lineage-specific regulatory regions. For each cell, the study first identified neighbouring cells through similarities in each data modality. Then, the study defined the weights of the different data modalities in the lineage classification as their accuracy for predicting molecular profiles of the target cell from the profiles of neighbouring cells. The resulting cell clustering, using the weighted distance averaged across single-cell RNA, protein and chromatin accessibility data, was then shown to improve cell lineage separation42.
Another common type of multimodal data analysis involves integrating molecular omics data and data on physical interaction networks (typically those involving protein–protein or protein–DNA interactions) to understand how individual genes interact with each other to drive oncogenesis and metastasis43–46. For example, an integrative pan-cancer analysis of TCGA detected 407 master regulators organized into 24 modules, partly shared across cancer types, that appear to canalize heterogeneous sets of mutations47. In another study, an analysis of 2,583 whole-tumour genomes across 27 cancers by the Pan-Cancer Analysis of Whole Genomes Consortium revealed rare mutations in the promoters of genes with many interactions (such as TP53, TLE4 and TCF4), and these mutations correlated with low downstream gene expression45. These examples of integrating networks and genomics data demonstrate a promising way to identify rare somatic mutations with a causal role in oncogenesis.
Knowledge transfer through data reuse
Existing data can be leveraged to make new discoveries. For example, cell-fraction deconvolution techniques can infer the composition of individual cell types in bulk-tumour transcriptomics profiles48. Such methods typically assemble gene expression profiles of diverse cell types from many existing datasets and perform regression or signature-enrichment analysis to deconvolve cell fractions49 or lineage-specific expression50,51 in a bulk-tumour expression profile.
Other data reuse examples come from single-cell transcriptomics data analysis. As single-cell RNA sequencing (scRNA-seq) has a high number of zero counts (dropout)52, analyses based on a limited number of genes may lead to unreliable results53, and genome-wide signatures from bulk data can therefore complement such analyses. For example, the transcriptomic data atlas collected from cytokine treatments in bulk cell cultures has enabled the reliable inference of signalling activities in scRNA-seq data34. Further, single-cell signalling activities inferred through bulk data have been used to reveal therapeutic targets, such as FIBP, to potentiate cellular therapies in solid tumours and molecular programmes of T cells that are resilient to immunosuppression in cancer54. In another example, the analysis of more than 50,000 scRNA-seq profiles from 35 pancreatic adenocarcinomas and control samples revealed edge cells among non-neoplastic acinar cells, whose transcriptomes have drifted towards malignant pancreatic adenocarcinoma cells55; TCGA bulk pancreatic adenocarcinoma data were then used to validate the edge-cell signatures inferred from the single-cell data.
Data reuse can assist the development of new experimental tests. For example, existing tumour whole-exome sequencing data were used to optimize a circulating tumour DNA assay by maximizing the number of alterations detected per patient, while minimizing gene and region selection size56. The resulting circulating tumour DNA assay can provide a comprehensive view of therapy resistance and cancer relapse and metastasis by detecting alterations in DNA released from multiple tumour regions or different tumour sites57.
Although the data scale in cancer research is typically much smaller than in other fields, the number of input features, such as genes or imaging pixels, can be extremely high. Training a machine learning model with a high number of input dimensions (a large number of features) and small data size (a small number of training samples) is likely to lead to overfitting, in which the model learns noise from training data and cannot generalize on new data58. Transfer learning approaches are a promising way of addressing this disparity related to data reuse. These approaches involve training a neural network model on a large, related dataset, and then fine-tuning the model on the smaller, target dataset. For example, most cancer histopathology artificial intelligence (AI) frameworks start from pretrained architectures from ImageNet — an image database containing 15 million images with detailed hierarchical annotations4 — and then fine-tune the framework on new imaging datasets of smaller sizes. As a further example of this approach, a few-shot learning framework enabled the prediction of drug response using data from only several patient-derived samples and a model pretrained using in vitro data from cell lines59. Despite these successful applications, transfer learning should be used with caution as it may produce mostly false predictions when data properties are markedly different between the pretraining set and the new dataset. Training a lightweight model60 or augmenting the new dataset61 are alternative solutions.
Data-rich translational studies
Many clinical diagnoses and decisions, such as histopathology interpretations, are inherently subjective and rely on interpreters’ experience or the availability of standardized diagnostic nomenclature and taxonomy. Such subjective factors may bring interpretive error62–64 and diagnostic discrepancies, for example when senior stature can have an undue influence on diagnostic decisions — the so-called big-dog effect65. Big-data approaches can provide complementary options that are systematic and objective to guide diagnosis and clinical decisions.
Diagnostic biomarkers trained from data cohorts
A major focus of translational big-data studies in cancer has been the development of genomics tests for predicting disease risk, some of which have already been approved by the US Food and Drug Administration (FDA) and commercialized for clinical use66. Distinct from biomarker discoveries through biological mechanisms and empirical observations, big data-derived tests analyse genome-scale genomics data from many patients and cohorts to generate a gene signature for clinical assays67. Such predictors mainly help clinicians determine the minimal therapy aggressiveness needed to minimize unnecessary treatment and side effects. The success of such tests depends on their high negative predictive value — the proportion of negative tests that reflect true negative results — so as not to miss patients who need aggressive therapy options66.
Some early examples of diagnostic biomarker tests trained from big data include prognosis assays for patients with oestrogen receptor (ER)- or progesterone receptor (PR)-positive breast cancer, such as Oncotype DX68,69, MammaPrint67,70, EndoPredict71 and Prosigna72. These tests are particularly useful as adjuvant endocrine therapy alone can bring sufficient clinical benefit to ER/PR-positive, HER2-negative patients with early-stage breast cancer73. Thus, patients stratified as being at low risk can avoid unnecessary additional chemotherapy. Predictors for other cancer types include Oncotype DX biomarkers for colon cancer74 and prostate cancer75 and Pervenio for early-stage lung cancer76.
In the early applications discussed above, large-scale data from genome-scale experiments served in the biomarker discovery stage but not in their clinical implementation. Owing to the high cost of genome-wide experiments and patent issues, the biomarker tests themselves still need to be performed through quantitative PCR or NanoString gene panels. However, the rapid decline of DNA sequencing costs in recent years could allow therapy decisions to be informed directly by genomics data and bring notable advantages over conventional approaches77. Gene alterations relevant to therapy decisions could involve diverse forms, including single-nucleotide mutations, DNA insertions, DNA deletions, copy number alterations, gene rearrangements, microsatellite instability and tumour mutational burden78–80. These alterations can be detected by combining hybridization-based capture and high-throughput sequencing. The MSK-IMPACT81 and FoundationOne CDx82 tests profile 300–500 genes and can use DNA from formalin-fixed, paraffin-embedded tumour specimens to detect oncogenic alterations and identify patients who may benefit from various therapies.
Variant interpretation in clinical decisions is still challenging as the oncogenic impact of each mutation depends on its clonality83, zygosity84 and co-occurrences with other mutations85. Sequencing data can uncover tumorigenic processes (such as DNA repair defects, exogenous mutagen exposure and prior therapy histories81) by identifying underlying mutational signatures, such as DNA substitution classes and sequence contexts86. Future computational frameworks for therapy decisions should therefore consider many dimensions of variants and inferred biological processes, together with other clinical data, such as histopathology data, radiology images and health records.
Data-rich assays that complement precision therapies currently focus on specific genomic aberrations. However, epigenetic therapies, such as inhibitors that target histone deacetylases87, have a genome-wide effect and are typically combined with other treatments, and therefore current genomics assays may not readily evaluate their therapeutic efficacy. We could not find any clinical datasets of histone deacetylase inhibitors deposited in the NCBI GEO database when writing this Review, indicating there are many unexplored territories of data-driven predictions for this broad category of anticancer therapies.
Clinical trials guided by molecular data
Genome-wide and multimodal data have begun to play a role in matching patients in prospective multi-arm clinical trials, particularly those investigating precision therapies. For example, the WINTHER trial prospectively matched patients with advanced cancer to therapy on the basis of DNA sequencing (arm A, through Foundation One assays) or RNA expression (arm B, comparing tumour tissue with normal tissue through Agilent oligonucleotide arrays) data from solid tumour biopsies88. Such therapy matches by omics data typically lead to off-label drug use. The WINTHER study concluded that both data types were of value for improving therapy recommendations and patient outcomes. Furthermore, there were no significant differences between DNA sequencing and RNA expression with regard to providing therapies with clinical benefits88, which was corroborated by a later study89.
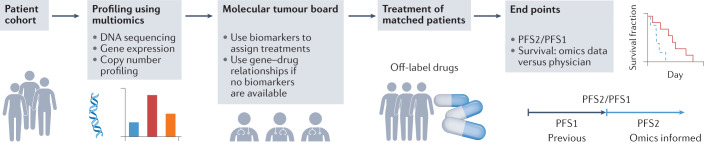
Other, similar trials have demonstrated the utility of matching patients for off-label use of targeted therapies on the basis of genome-wide genomics or transcriptomics data89–92 (Fig. 2). In these studies, the fraction of enrolled patients who had therapies matched by omics data ranged from 19% to 37% (WINTHER, 35%88; POG, 37%89; MASTER, 31.8%92; MOSCATO 01, 19.2%90; CoPPO, 20%91). Among these matched patients, about one third demonstrated clinical benefits (WINTHER, 25%88; POG, 46%89; MASTER, 35.7%92; MOSCATO 01, 33%90; CoPPO, 32%91). Except for the POG study, all studies used the end point defined by the Von Hoff model, which compares progression-free survival (PFS) for the trial (PFS2) with the PFS recorded for the therapy preceding enrolment (PFS1) and defines clinical benefit as a PFS2/PFS1 ratio of more than 1.3 (ref.93).
Fig. 2. Prospective clinical studies guided by omics data to use off-label drugs.
Recent umbrella clinical trials88–92 have focused on multi-omics profiling of the tumours of enrolled patients by generating and analysing genome-wide data — including data from DNA sequencing, gene expression profiling, and copy number profiling — to prioritize treatments. After multi-omics profiling, a multidisciplinary molecular tumour board led by clinicians selects the best therapies on the basis of the current known relationships between drugs, genes and tumour vulnerabilities. For each therapy, the relevant altered vulnerabilities could include direct drug targets, genes in the same pathway, indirect drug targets upregulated or downregulated by drug treatment, or other genes interacting with the drug targets through physical or genetic interactions. This process then results in patients being treated with off-label targeted therapies. The end points for evaluating clinical efficacy include the ratio of the progression-free survival (PFS) associated with omics data-guided therapies (PFS2) and the PFS associated with previous therapy (PFS1), or differences in survival between patients treated with omics data-guided therapies and patients treated with therapies guided by physician’s choice alone.
A recent study demonstrated the feasibility and value of an N-of-one strategy that collected multimodal data, including immunohistochemistry data for multiple protein markers, RNA levels and genomics alterations in cell-free DNA from liquid biopsies94 (Fig. 2). A broad multidisciplinary molecular tumour board (MTB) then made personalized decisions using these multimodal omics data. Overall, patients who received MTB-recommended treatments had significantly longer PFS and overall survival than those treated by independent physician choice. Similarly, another study also demonstrated overall survival benefits brought by MTB recommendations95.
With these initial successes, emerging clinical studies aim to collect additional data beyond bulk-sample sequencings — such as tumour cell death response following various drug treatments96 or scRNA-seq data collected on longitudinal patient samples — to study therapy response and resistance mechanisms97. Besides omics data generated from tumour samples, cross-modality data integration is a potential strategy to improve therapy recommendations. One such promising direction involves the study and application of synthetic lethal interactions98–104, which, once integrated with tumour transcriptomic profiles, can accurately score drug target importance and predict clinical outcomes for many anticancer treatments, including targeted therapies and immunotherapies98. We foresee that new data modalities and assays will provide additional ways to design clinical trials.
Artificial intelligence for data-driven cancer diagnosis
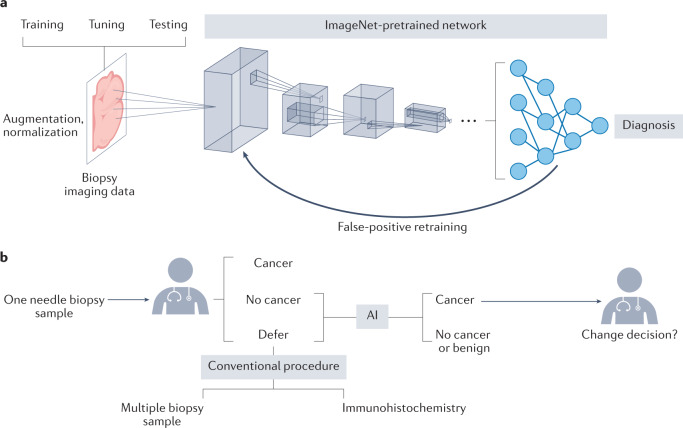
Genomics datasets, such as gene expression levels or mutation status, can typically be aligned to each other on gene dimensions. However, data types in clinical diagnoses, such as imaging data or text reports, may not directly align across samples in any obvious way. AI approaches based on deep neural networks (Fig. 3a) are an emerging method for integrating these data types for clinical applications105.
Fig. 3. Data-driven artificial intelligence to support cancer diagnosis.
a | A common artificial intelligence (AI) framework in cancer detection uses a convolutional neural network (CNN) to detect the presence of cancer cells from a diagnostic image. CNNs use convolution (weighted sum of a region patch) and pooling (summarize values in a region to one value) to encode image regions into low-dimensional numerical vectors that can be analysed by machine learning models. The CNN architecture is typically pretrained with ImageNet data, which is much larger than any cancer biology imaging dataset. To increase the reliability of the AI framework, the input data can be augmented through rotation or blurring of tissue images to increase data size. The data are separated into non-overlapping training, tuning and test sets to train the AI model, tune hyperparameters and estimate the prediction accuracy on new inputs, respectively. False-positive predictions are typically essential data points for retraining the AI model. b | An example of the application of AI in informing clinical decisions, as per the US Food and Drug Administration-approved AI test Paige Prostate. From one needle biopsy sample, the pathologist can decide whether cancer cells are present. If the results are negative (‘no cancer’) or if the physician cannot make a firm diagnosis (‘defer’), the Paige Prostrate AI can analyse the image and prompt the pathologist with regard to potential cancer locations if any are detected. The alternative procedure involves evaluating multiple biopsy samples and performing immunohistochemistry tests on prostate cancer markers, independently from the AI test185.
The most popular application of AI for analysing imaging data involves clinical outcome prediction and tumour detection and grading from tissue stained with haematoxylin and eosin (H&E)26. In September 2021, the FDA approved the use of the AI software Paige Prostate106 to assist pathologists in detecting cancer regions from prostate needle biopsy samples107 (Fig. 3b). This approval reflects the accelerating momentum of AI applications on histopathology images108 to complement conventional pathologist practices and increase analysis throughput, particularly for less experienced pathologists. The CAMELYON challenge for identifying tumour regions provided 1,399 manually annotated whole-slide H&E-stained tissue images of sentinel lymph nodes from patients with breast cancer for training AI algorithms109. The top performers in the challenge used deep learning approaches, which achieved similar performance in detecting lymph node metastasis as expert pathologists110. Other studies have trained deep neural networks to predict patient survival outcomes111, gene mutations112 or genomic alterations113, on the basis of analysing a large body of H&E-stained tissue images with clinical outcome labels or genomics profiles.
Besides histopathology, radiology is another application of AI imaging analysis. Deep convolutional neural networks that use 3D computed tomography volumes have been shown to predict the risk of lung cancer with an accuracy comparable to that of predictions by experienced radiologists114. Similarly, convolutional neural networks can use computed tomography data to stratify the survival duration of patients with lung cancer and highlight the importance of tumour-surrounding tissues in risk stratification115.
AI frameworks have started to play an important role in analysing electronic health records. A recent study evaluating the effect of different eligibility criteria on cancer trial outcomes using electronic health records of more than 60,000 patients with non-small-cell lung cancer revealed that many patient exclusion criteria commonly used in clinical trials had a minimal effect on trial hazard ratios25. Dropping these exclusion criteria would only marginally decrease the overall survival and result in more inclusive trials without compromising patient safety and overall trial success rates25. Besides images and health records, AI trained on other data types also has broad clinical applications, such as early cancer detection through liquid biopsies capturing cell-free DNA116,117 or T cell receptor sequences118, or genomics-based cancer risk predictions119,120. Additional examples of AI applications in cancer are available in other reviews40,121.
New AI approaches have started to play a role in biological knowledge discovery. The saliency map122 and class activation map123 can highlight essential portions of input images that drive predicted outcomes. Also, in a multisample cohort, clustering data slices on the basis of deep learning-embedded similarities can reveal human-interpretable features associated with a clinical outcome. For example, clustering similar image patches related to colorectal cancer survival prediction revealed that high-risk survival predictions are associated with a tumour–adipose feature, characterized by poorly differentiated tumour cells adjacent to adipose tissue124. Although the molecular mechanisms underlying this association are unclear, this study provided an example of finding imaging features that could help cancer biologists pinpoint new disease mechanisms.
Despite the promising results described above, few AI-based algorithms have reached clinical deployment due to several limitations26. First, the performance of most AI predictors deteriorates when they are applied to test data generated in a setting different from that in which their training data are generated. For example, the performance of top algorithms from the CAMELYON challenge dropped by about 20% when they were evaluated on the basis of data from other centres108. Such a gap may arise from differences in image scanners (if imaging data are being evaluated), sample collection protocols or study design, emphasizing the need for reliable data homogenization. Second, supervised AI training requires a large amount of annotated data, and acquiring sufficient human-annotated data can be challenging. In imaging data, if a feature for a particular diagnosis is present in only a fraction of image regions, an algorithm will need many samples to learn the task. Furthermore, if features are not present in the training data, the AI will not make meaningful predictions; for example, the AI framework of AlphaFold2 can predict wild type protein structures with high accuracy, but it cannot predict the impact of cancer missense mutations on protein structures because the training data for AlphaFold2 do not contain altered structures of these mutated proteins125.
Many studies of AI applications that claim improvements lack comparisons with conventional clinical procedures. For example, the performance study of Paige Prostate evaluated cancer detection using an H&E-stained tissue image from one needle biopsy sample126. However, the pathologist may make decisions on the basis of multiple needle biopsy samples and immunohistochemistry stains for suspicious samples instead of relying on one H&E-stained tissue image (Fig. 3b). Therefore, rigorous comparison with conventional clinical workflows is necessary for each application before the advantage of any AI framework is claimed.
New therapy development aided by big-data analysis
Developing a new drug is costly, is time-intensive and suffers from a high failure rate127. The development of new therapies is a promising direction for big-data applications. To our knowledge, no FDA-approved cancer drugs have been developed primarily through big-data approaches; however, some big data-driven preclinical studies have attracted the attention of the pharmaceutical industry for further development and may soon make impactful contributions to clinics128.
Big data have been used to aid the repurposing of existing drugs to treat new diseases129,130 and the design of synergistic combinations131–134. By creating a network of 1.2 billion edges among diseases, tissues, genes, pathways and drugs by mining more than 40 million documents, one study revealed that the combination of vandetanib and everolimus could inhibit ACVR1, a drug efflux transporter, as a potential therapy for diffuse intrinsic pontine glioma135.
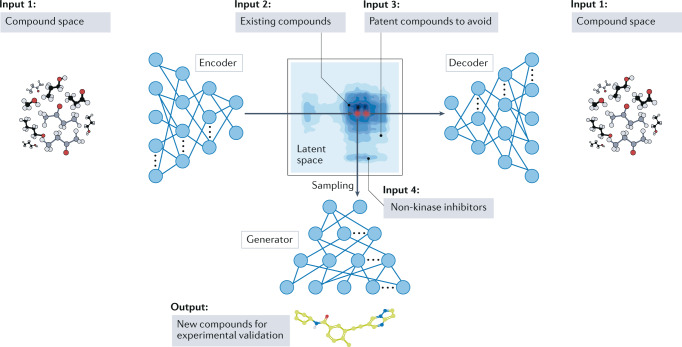
Recent studies have combined pharmacological data and AI to design new drugs (Fig. 4). A deep generative model was used to design new small molecules inhibiting the receptor tyrosine kinase DDR1 on the basis of information on existing DDR1 inhibitors and compound libraries, with the lead candidate demonstrating favourable pharmacokinetics in mice136. Deep generative models are neural networks with many layers that learn complex characteristics of specific datasets (such as high-dimensional probability distributions) and can use them to generate new data similar to the training data137. For each specific drug design application, such a framework can encode distinct data into the neural network parameters and thus naturally incorporate many data types. A network aiming to find novel kinase inhibitors, for example, may include data on the structure of existing kinase inhibitors, non-kinase inhibitors and patent-protected molecules that are to be avoided136.
Fig. 4. Design of new kinase inhibitors using a generative artificial intelligence model.
The variational autoencoder, trained with the structures of many compounds, can encode a molecular structure into a latent space of numerical vectors and decode this latent space back into the compound structure. For each target, such as the receptor tyrosine kinase DDR1, the variational autoencoder can create embeddings of compound categories, such as existing kinase inhibitors, patented compounds and non-kinase inhibitors. Sampling the latent space for compounds that are similar to existing on-target inhibitors and not patented compounds or non-kinase inhibitors can generate new candidate kinase inhibitors for downstream experimental validation. Adapted from ref.136, Springer Nature Limited.
AI can also be used for the virtual screening of bioactive ligands on target protein structures. Under the assumption that biochemical interactions are local among chemical groups, convolutional neural networks can comprehensively integrate training data from previous virtual screening studies to outperform previous docking methods based on minimizing empirical scores138. Similarly, a systematic evaluation revealed that deep neural networks trained using large and diverse datasets composed of molecular descriptors and drug biological activities could predict the activity of test-set molecules better than other approaches139.
Big data in front of narrow therapeutic bottlenecks
During dynamic tumour evolution, cancers generally become more heterogeneous and harbour a more diverse population of cells with different treatment sensitivities. Drug resistance can eventually evolve from a narrow bottleneck of a few cells140. Furthermore, the difference between a treatment dose with antitumour effects and toxicity leading to either clinical trial failure or treatment cessation is small66. These two challenges are common reasons for anticancer therapy failures as increasing drug combinations to target rare cancer cells will quickly lead to unacceptable toxic effects. An essential question is whether big data can bring solutions to overcome heterogeneous tumour evolution towards drug resistance while avoiding intolerable toxic effects.
Ideally, well-designed drug combinations should target various subsets of drug-tolerant cells in tumours and induce robust responses. Computational methods have been developed to design synergistic drug pairs131,141; however, drug synergy may not be predictable for certain combinations even with comprehensive training data. A recent community effort assessed drug synergy prediction methods trained on AstraZeneca’s large drug combination dataset, consisting of 11,576 experiments from 910 combinations across 85 molecularly characterized cancer cell lines134. The results showed that none of the methods evaluated could make reliable predictions for approximately 20% of the drug pairs whose targets independently regulate downstream pathways.
There could be a theoretical limitation of the power of drug combinations in killing heterogeneous tumour cells while avoiding toxic effects on normal tissues. A recent study mining 15 single-cell transcriptomics datasets revealed that inhibition of four cell-surface targets is necessary to kill at least 80% of tumour cells while sparing at least 90% of normal cells in tumours142. However, a feasible drug-target combination may not exist to kill a higher fraction of tumour cells while sparing normal cells.
An important challenge accompanying therapy design efforts is the identification of genomic biomarkers that could predict toxicity. A community evaluation demonstrated that computational methods could predict the cytotoxicity of environmental chemicals on the basis of the genotype data of lymphoblastoid cell lines143. Further, a computational framework has been used to predict drug toxicity by integrating information on drug-target expression in tissues, gene network connectivity, chemical structures and toxicity annotations from clinical trials144. However, these studies were not explicitly designed for anticancer drugs, which are challenging with regard to toxicity prediction due to their extended cytotoxicity profiles.
Challenges and future perspectives
While many big-data advancements are encouraging and impressive, considerable challenges remain regarding big-data applications in cancer research and the clinic. Omics data often suffer from measurement inconsistencies between cohorts, marked batch effects and dependencies on specific experimental platforms. Such a lack of consistency is a major hurdle towards clinical translation. Consensus on the measurement, alignment and normalization of tumour omics data will be critical for each data type35. Besides these technical challenges, structural and societal challenges also exist and may impede the progress of the entire cancer data science field. We discuss these in the following subsections.
Less-than-desirable data availability
A key challenge of cancer data science is the insufficient availability of data and code. A recent study found that machine learning-based studies in the biomedical domain compare poorly with those in other areas regarding public data and source code availability145. Sometimes, the clinical information accompanying published cancer genomics data is not provided or complete, even when security and privacy issues are resolved. One possible reason for this bottleneck is related to data release policies and data stewardship costs. Although many journals require the public release of data, such requirements are often met by deposition of data into repositories that require author and institutional approval-of-access requests due to intellectual property and various other considerations. Furthermore, deposited data may be missing critical information, such as missing cell barcodes for single-cell sequencing data or low-resolution images in the case of histopathology data.
In our opinion, the mitigation of these issues will require the enforcement of policies regarding public data availability by funding agencies and additional community efforts to examine the fulfilment of open data access. For example, a funding agency may suspend a project if the community readers report any violations of data release agreements upon publication of articles. The allocation of budgets in grants for patient de-identification upon manuscript submission and financial incentives for checking data through independent data stewardship services upon paper acceptance could markedly help facilitate data and code availability. One notable advance in data availability through industry–academia alliances has come in the form of data-sharing initiatives; specifically, making large repositories of patient tumour sequencing and clinical data available for online queries to researchers in partner institutions146. Such initiatives typically involve query-only access (that is, without allowing downloads), but are an encouraging way to expand the collaborative network between academia and industry entities that generate massive amounts of data.
Data-scale gaps
As mentioned earlier, the datasets available for cancer therapeutics are substantially smaller than those available in other fields. One reason for such a gap is that the generation of medical data depends on professionally trained scientists. To close the data-scale gap, more investments will be required to automate the generation of at least some types of annotated medical data and patient omics data. Rare cancers especially suffer from a lack of preclinical models, clinical samples and dedicated funding147. Moreover, the usability of biomedical data is typically constrained by the genetic background of the population. For example, the frequency of actionable mutations may differ among East Asian, European and American populations148.
A further reason for the data-scale gap is a lack of data generation standards in cancer clinical and biology studies. For example, most clinical trials do not yet collect omics data from patients. With the exponential decrease in sequencing cost, collection of omics data in clinical trials should, in our opinion, be markedly expanded, and possibly be made mandatory as a standard requirement. Further, current data repositories, such as ClinicalTrials.gov and NCBI GEO, do not have common metalanguage standards, whose incorporation would markedly improve the development of algorithms applied to their analysis. Although semi-automated frameworks are becoming available to homogenize metadata34, the foundational solution should be establishing common vocabularies and systematic meta-information standards in critical fields.
Conclusion
Data science and AI are transforming our world through applications as diverse as self-driving cars, facial recognition and language translation, and in the medical world, the interpretation of images in radiology and pathology. We already have available tumour data to facilitate biomedical breakthroughs in cancer through cross-modality integration, cross-cohort aggregation and data reuse, and extraordinary advancements are being made in generating and analysing such data. However, the state of big data in the field is complex, and in our view, we should acknowledge that ‘big data’ in cancer are not yet so big. Future investments from the global research community to expand cancer datasets will be critical to allow better computational models to drive basic research, cancer diagnostics and the development of new therapies.
Acknowledgements
The authors are supported by the intramural research budget of the US National Cancer Institute.
Glossary
- Few-shot learning
A machine learning method that classifies new data using only a few training samples by transferring knowledge from large, related datasets.
- Saliency map
A map of important image locations that support machine learning outputs.
- Class activation map
A coarse-resolution map of important image regions for predicting a specific class using activations and gradients in the final convolutional layer.
Author contributions
P.J. and E.R. designed the scope and structure of the Review, assembled write-up components and finalized the manuscript. C.S. wrote the text on tumour evolution and heterogeneity. S.H. wrote the text on transcriptional dysregulation. P.J. wrote the sections related to spatial genomics and artificial intelligence. P.J., E.R. and K.A. wrote the section on cancer diagnosis and treatment decisions. S.S. and P.J. prepared Tables 1–4.
Peer review
Peer review information
Nature Reviews Cancer thanks Itai Yanai, Anjali Rao and the other, anonymous, reviewers for their contribution to the peer review of this work.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Array Express: https://www.ebi.ac.uk/arrayexpress/
CAMELYON: https://camelyon17.grand-challenge.org/
cBioportal: https://www.cbioportal.org/
CCLE: https://depmap.org/portal/ccle/
CPTAC: https://proteomics.cancer.gov/data-portal
CytoSig: https://cytosig.ccr.cancer.gov/
DepMap: https://depmap.org/portal
DNA sequencing costs: https://www.genome.gov/about-genomics/fact-sheets/DNA-Sequencing-Costs-Data
DrugCombDB: http://drugcombdb.denglab.org/
FDC: https://curate.ccr.cancer.gov/
GENIE: https://www.aacr.org/professionals/research/aacr-project-genie
GEO: https://www.ncbi.nlm.nih.gov/geo
Human Protein Atlas: https://www.proteinatlas.org/humanproteome/pathology
ICGC: https://dcc.icgc.org/
IDC: https://datacommons.cancer.gov/repository/imaging-data-commons
LINCS: https://clue.io/
PCAWG: https://dcc.icgc.org/pcawg
PRECOG: https://precog.stanford.edu/
RABIT: http://rabit.dfci.harvard.edu/
TARGET: https://ocg.cancer.gov/programs/target/data-matrix
TCIA: https://www.cancerimagingarchive.net/
TCGA: https://gdc.cancer.gov/
TIDE: http://tide.dfci.harvard.edu/
TISCH: http://tisch.comp-genomics.org/
Tres: https://resilience.ccr.cancer.gov/
UCSC Xena: https://xena.ucsc.edu/
Contributor Information
Peng Jiang, Email: peng.jiang@nih.gov.
Eytan Ruppin, Email: eytan.ruppin@nih.gov.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Weinstein JN, et al. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 2013;45:1113–110. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng J, et al. ImageNet: a large-scale hierarchical image database. 2009 IEEE Conf. Computer Vis. Pattern Recognit. 2009 doi: 10.1109/cvprw.2009.5206848. [DOI] [Google Scholar]
- 5.Stuart T, Satija R. Integrative single-cell analysis. Nat. Rev. Genet. 2019;20:257–272. doi: 10.1038/s41576-019-0093-7. [DOI] [PubMed] [Google Scholar]
- 6.Ji AL, et al. Multimodal analysis of composition and spatial architecture in human squamous cell carcinoma. Cell. 2020;182:1661–1662. doi: 10.1016/j.cell.2020.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deshwar AG, et al. PhyloWGS: reconstructing subclonal composition and evolution from whole-genome sequencing of tumors. Genome Biol. 2015;16:35. doi: 10.1186/s13059-015-0602-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roth A, et al. PyClone: statistical inference of clonal population structure in cancer. Nat. Methods. 2014;11:396–398. doi: 10.1038/nmeth.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller CA, et al. SciClone: inferring clonal architecture and tracking the spatial and temporal patterns of tumor evolution. PLoS Comput. Biol. 2014;10:e1003665. doi: 10.1371/journal.pcbi.1003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carter SL, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Minussi DC, et al. Breast tumours maintain a reservoir of subclonal diversity during expansion. Nature. 2021;592:302–308. doi: 10.1038/s41586-021-03357-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laks E, et al. Clonal decomposition and DNA replication states defined by scaled single-cell genome sequencing. Cell. 2019;179:1207–1221.e22. doi: 10.1016/j.cell.2019.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao T, et al. Spatial genomics enables multi-modal study of clonal heterogeneity in tissues. Nature. 2022;601:85–91. doi: 10.1038/s41586-021-04217-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Przybyla L, Gilbert LA. A new era in functional genomics screens. Nat. Rev. Genet. 2022;23:89–103. doi: 10.1038/s41576-021-00409-w. [DOI] [PubMed] [Google Scholar]
- 15.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subramanian A, et al. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell. 2017;171:1437–1452.e17. doi: 10.1016/j.cell.2017.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat. Rev. Genet. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilbert LA, et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsherniak A, et al. Defining a cancer dependency map. Cell. 2017;170:564–576.e16. doi: 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johannessen CM, et al. A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature. 2013;504:138–142. doi: 10.1038/nature12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robertson G, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 22.Hafner M, et al. CLIP and complementary methods. Nat. Rev. Methods Prim. 2021;1:20. doi: 10.1038/s43586-021-00018-1. [DOI] [Google Scholar]
- 23.Vidal M, Cusick ME, Barabási A-L. Interactome networks and human disease. Cell. 2011;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kempfer R, Pombo A. Methods for mapping 3D chromosome architecture. Nat. Rev. Genet. 2020;21:207–226. doi: 10.1038/s41576-019-0195-2. [DOI] [PubMed] [Google Scholar]
- 25.Liu R, et al. Evaluating eligibility criteria of oncology trials using real-world data and AI. Nature. 2021;592:629–633. doi: 10.1038/s41586-021-03430-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Laak J, Litjens G, Ciompi F. Deep learning in histopathology: the path to the clinic. Nat. Med. 2021;27:775–784. doi: 10.1038/s41591-021-01343-4. [DOI] [PubMed] [Google Scholar]
- 27.Hosny A, Parmar C, Quackenbush J, Schwartz LH, Hjwl A. Artificial intelligence in radiology. Nat. Rev. Cancer. 2018;18:500–510. doi: 10.1038/s41568-018-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang P, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018;24:1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parkinson H, et al. ArrayExpress — a public database of microarray experiments and gene expression profiles. Nucleic Acids Res. 2007;35:D747–D750. doi: 10.1093/nar/gkl995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gentles AJ, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 2015;21:938–945. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 33.Jiang L, et al. Direct tumor killing and immunotherapy through anti-serpinB9 therapy. Cell. 2020;183:1219–1233.e18. doi: 10.1016/j.cell.2020.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang P, et al. Systematic investigation of cytokine signaling activity at the tissue and single-cell levels. Nat. Methods. 2021;18:1181–1191. doi: 10.1038/s41592-021-01274-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leek JT, et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat. Rev. Genet. 2010;11:733–739. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 37.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stuart T, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nygaard V, Rødland EA, Hovig E. Methods that remove batch effects while retaining group differences may lead to exaggerated confidence in downstream analyses. Biostatistics. 2016;17:29–39. doi: 10.1093/biostatistics/kxv027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boehm KM, Khosravi P, Vanguri R, Gao J, Shah SP. Harnessing multimodal data integration to advance precision oncology. Nat. Rev. Cancer. 2022;22:114–126. doi: 10.1038/s41568-021-00408-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang C, et al. Proteogenomic insights into the biology and treatment of HPV-negative head and neck squamous cell carcinoma. Cancer Cell. 2021;39:361–379.e16. doi: 10.1016/j.ccell.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hao Y, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klein MI, et al. Identifying modules of cooperating cancer drivers. Mol. Syst. Biol. 2021;17:e9810. doi: 10.15252/msb.20209810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hofree M, Shen JP, Carter H, Gross A, Ideker T. Network-based stratification of tumor mutations. Nat. Methods. 2013;10:1108–1115. doi: 10.1038/nmeth.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reyna MA, et al. Pathway and network analysis of more than 2500 whole cancer genomes. Nat. Commun. 2020;11:729. doi: 10.1038/s41467-020-14367-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng F, et al. Interpretation of cancer mutations using a multiscale map of protein systems. Science. 2021;374:eabf3067. doi: 10.1126/science.abf3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paull EO, et al. A modular master regulator landscape controls cancer transcriptional identity. Cell. 2021;184:334–351. doi: 10.1016/j.cell.2020.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Avila Cobos F, Alquicira-Hernandez J, Powell JE, Mestdagh P, De Preter K. Benchmarking of cell type deconvolution pipelines for transcriptomics data. Nat. Commun. 2020;11:5650. doi: 10.1038/s41467-020-19015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Newman AM, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Newman AM, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019;37:773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang K, et al. Deconvolving clinically relevant cellular immune cross-talk from bulk gene expression using CODEFACS and LIRICS stratifies patients with melanoma to anti-PD-1 therapy. Cancer Discov. 2022;12:1088–1105. doi: 10.1158/2159-8290.CD-21-0887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kharchenko PV, Silberstein L, Scadden DT. Bayesian approach to single-cell differential expression analysis. Nat. Methods. 2014;11:740–742. doi: 10.1038/nmeth.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suvà ML, Tirosh I. Single-cell RNA sequencing in cancer: lessons learned and emerging challenges. Mol. Cell. 2019;75:7–12. doi: 10.1016/j.molcel.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y, et al. A T cell resilience model associated with response to immunotherapy in multiple tumor types. Nat. Med. 2022 doi: 10.1038/s41591-022-01799-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gopalan V, et al. A transcriptionally distinct subpopulation of healthy acinar cells exhibit features of pancreatic progenitors and PDAC. Cancer Res. 2021;81:3958–3970. doi: 10.1158/0008-5472.CAN-21-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Newman AM, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014;20:548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heitzer E, Haque IS, Roberts CES, Speicher MR. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 2019;20:71–88. doi: 10.1038/s41576-018-0071-5. [DOI] [PubMed] [Google Scholar]
- 58.Hastie, T., Friedman, J. & Tibshirani, R. The Elements of Statistical Learning (Springer, 2001).
- 59.Ma J, et al. Few-shot learning creates predictive models of drug response that translate from high-throughput screens to individual patients. Nat. Cancer. 2021;2:233–244. doi: 10.1038/s43018-020-00169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raghu M, Zhang C, Kleinberg J, Bengio S. Transfusion: understanding transfer learning for medical imaging. Adv. Neural Inf. Process. Syst. 2019;33:3347–3357. [Google Scholar]
- 61.Zoph B, et al. Rethinking pre-training and self-training. Adv. Neural Inf. Process. Syst. 2020;34:3833–3845. [Google Scholar]
- 62.Meier FA, Varney RC, Zarbo RJ. Study of amended reports to evaluate and improve surgical pathology processes. Adv. Anat. Pathol. 2011;18:406–413. doi: 10.1097/PAP.0b013e318229bf20. [DOI] [PubMed] [Google Scholar]
- 63.Nakhleh RE. Error reduction in surgical pathology. Arch. Pathol. Lab. Med. 2006;130:630–632. doi: 10.5858/2006-130-630-ERISP. [DOI] [PubMed] [Google Scholar]
- 64.Nakhleh RE, et al. Interpretive diagnostic error reduction in surgical pathology and cytology: guideline from the College of American Pathologists Pathology and Laboratory Quality Center and the Association of Directors of Anatomic and Surgical Pathology. Arch. Pathol. Lab. Med. 2016;140:29–40. doi: 10.5858/arpa.2014-0511-SA. [DOI] [PubMed] [Google Scholar]
- 65.Raab SS, et al. The ‘Big Dog’ effect: variability assessing the causes of error in diagnoses of patients with lung cancer. J. Clin. Oncol. 2006;24:2808–2814. doi: 10.1200/JCO.2005.04.3661. [DOI] [PubMed] [Google Scholar]
- 66.Jiang P, Sellers WR, Liu XS. Big data approaches for modeling response and resistance to cancer drugs. Annu. Rev. Biomed. Data Sci. 2018;1:1–27. doi: 10.1146/annurev-biodatasci-080917-013350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van’t Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 68.Sparano JA, et al. Adjuvant chemotherapy guided by a 21-gene expression assay in breast cancer. N. Engl. J. Med. 2018;379:111–121. doi: 10.1056/NEJMoa1804710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kalinsky K, et al. 21-gene assay to inform chemotherapy benefit in node-positive breast cancer. N. Engl. J. Med. 2021;385:2336–2347. doi: 10.1056/NEJMoa2108873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cardoso F, et al. 70-gene signature as an aid to treatment decisions in early-stage breast cancer. N. Engl. J. Med. 2016;375:717–729. doi: 10.1056/NEJMoa1602253. [DOI] [PubMed] [Google Scholar]
- 71.Filipits M, et al. A new molecular predictor of distant recurrence in ER-positive, HER2-negative breast cancer adds independent information to conventional clinical risk factors. Clin. Cancer Res. 2011;17:6012–6020. doi: 10.1158/1078-0432.CCR-11-0926. [DOI] [PubMed] [Google Scholar]
- 72.Parker JS, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 74.You YN, Rustin RB, Sullivan JD. Oncotype DX® colon cancer assay for prediction of recurrence risk in patients with stage II and III colon cancer: a review of the evidence. Surg. Oncol. 2015;24:61–66. doi: 10.1016/j.suronc.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 75.Klein EA, et al. A 17-gene assay to predict prostate cancer aggressiveness in the context of Gleason grade heterogeneity, tumor multifocality, and biopsy undersampling. Eur. Urol. 2014;66:550–560. doi: 10.1016/j.eururo.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 76.Kratz JR, et al. A practical molecular assay to predict survival in resected non-squamous, non-small-cell lung cancer: development and international validation studies. Lancet. 2012;379:823–832. doi: 10.1016/S0140-6736(11)61941-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Beaubier N, et al. Integrated genomic profiling expands clinical options for patients with cancer. Nat. Biotechnol. 2019;37:1351–1360. doi: 10.1038/s41587-019-0259-z. [DOI] [PubMed] [Google Scholar]
- 78.Snyder A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Van Allen EM, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rizvi NA, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zehir A, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 2017;23:703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li M. Statistical methods for clinical validation of follow-on companion diagnostic devices via an external concordance study. Stat. Biopharm. Res. 2016;8:355–363. doi: 10.1080/19466315.2016.1202859. [DOI] [Google Scholar]
- 83.Litchfield K, et al. Meta-analysis of tumor- and T cell-intrinsic mechanisms of sensitization to checkpoint inhibition. Cell. 2021;184:596–614.e14. doi: 10.1016/j.cell.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bielski CM, et al. Widespread selection for oncogenic mutant allele imbalance in cancer. Cancer Cell. 2018;34:852–862.e4. doi: 10.1016/j.ccell.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.El Tekle G, et al. Co-occurrence and mutual exclusivity: what cross-cancer mutation patterns can tell us. Trends Cancer Res. 2021;7:823–836. doi: 10.1016/j.trecan.2021.04.009. [DOI] [PubMed] [Google Scholar]
- 86.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cheng Y, et al. Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal Transduct. Target. Ther. 2019;4:62. doi: 10.1038/s41392-019-0095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rodon J, et al. Genomic and transcriptomic profiling expands precision cancer medicine: the WINTHER trial. Nat. Med. 2019;25:751–758. doi: 10.1038/s41591-019-0424-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pleasance E, et al. Whole genome and transcriptome analysis enhances precision cancer treatment options. Ann. Oncol. 2022 doi: 10.1016/j.annonc.2022.05.522. [DOI] [PubMed] [Google Scholar]
- 90.Massard C, et al. High-throughput genomics and clinical outcome in hard-to-treat advanced cancers: results of the MOSCATO 01 trial. Cancer Discov. 2017;7:586–595. doi: 10.1158/2159-8290.CD-16-1396. [DOI] [PubMed] [Google Scholar]
- 91.Tuxen IV, et al. Copenhagen Prospective Personalized Oncology (CoPPO) — clinical utility of using molecular profiling to select patients to phase I trials. Clin. Cancer Res. 2019;25:1239–1247. doi: 10.1158/1078-0432.CCR-18-1780. [DOI] [PubMed] [Google Scholar]
- 92.Horak P, et al. Comprehensive genomic and transcriptomic analysis for guiding therapeutic decisions in patients with rare cancers. Cancer Discov. 2021;11:2780–2795. doi: 10.1158/2159-8290.CD-21-0126. [DOI] [PubMed] [Google Scholar]
- 93.Von Hoff DD, et al. Pilot study using molecular profiling of patients’ tumors to find potential targets and select treatments for their refractory cancers. J. Clin. Oncol. 2010;28:4877–4883. doi: 10.1200/JCO.2009.26.5983. [DOI] [PubMed] [Google Scholar]
- 94.Kato S, et al. Real-world data from a molecular tumor board demonstrates improved outcomes with a precision N-of-one strategy. Nat. Commun. 2020;11:4965. doi: 10.1038/s41467-020-18613-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hoefflin, R. et al. Personalized clinical decision making through implementation of a molecular tumor board: a German single-center experience. JCO Precis. Oncol. 1–16 10.1200/po.18.00105 (2018). [DOI] [PMC free article] [PubMed]
- 96.Irmisch A, et al. The Tumor Profiler Study: integrated, multi-omic, functional tumor profiling for clinical decision support. Cancer Cell. 2021;39:288–293. doi: 10.1016/j.ccell.2021.01.004. [DOI] [PubMed] [Google Scholar]
- 97.Cohen YC, et al. Identification of resistance pathways and therapeutic targets in relapsed multiple myeloma patients through single-cell sequencing. Nat. Med. 2021;27:491–503. doi: 10.1038/s41591-021-01232-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lee JS, et al. Synthetic lethality-mediated precision oncology via the tumor transcriptome. Cell. 2021;184:2487–2502.e13. doi: 10.1016/j.cell.2021.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang B, et al. The tumor therapy landscape of synthetic lethality. Nat. Commun. 2021;12:1275. doi: 10.1038/s41467-021-21544-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pathria G, et al. Translational reprogramming marks adaptation to asparagine restriction in cancer. Nat. Cell Biol. 2019;21:1590–1603. doi: 10.1038/s41556-019-0415-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Feng, X. et al. A platform of synthetic lethal gene interaction networks reveals that the GNAQ uveal melanoma oncogene controls the Hippo pathway through FAK. Cancer Cell35, (2019). [DOI] [PMC free article] [PubMed]
- 102.Lee JS, et al. Harnessing synthetic lethality to predict the response to cancer treatment. Nat. Commun. 2018;9:2546. doi: 10.1038/s41467-018-04647-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cheng K, Nair NU, Lee JS, Ruppin E. Synthetic lethality across normal tissues is strongly associated with cancer risk, onset, and tumor suppressor specificity. Sci. Adv. 2021;7:eabc2100. doi: 10.1126/sciadv.abc2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sahu AD, et al. Genome-wide prediction of synthetic rescue mediators of resistance to targeted and immunotherapy. Mol. Syst. Biol. 2019;15:e8323. doi: 10.15252/msb.20188323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Elemento O, Leslie C, Lundin J, Tourassi G. Artificial intelligence in cancer research, diagnosis and therapy. Nat. Rev. Cancer. 2021;21:747–752. doi: 10.1038/s41568-021-00399-1. [DOI] [PubMed] [Google Scholar]
- 106.Raciti P, et al. Novel artificial intelligence system increases the detection of prostate cancer in whole slide images of core needle biopsies. Mod. Pathol. 2020;33:2058–2066. doi: 10.1038/s41379-020-0551-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Office of the Commissioner. FDA authorizes software that can help identify prostate cancer. https://www.fda.gov/news-events/press-announcements/fda-authorizes-software-can-help-identify-prostate-cancer (2021).
- 108.Campanella G, et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 2019;25:1301–1309. doi: 10.1038/s41591-019-0508-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Litjens G, et al. 1399 H&E-stained sentinel lymph node sections of breast cancer patients: the CAMELYON dataset. GigaScience. 2018;7:giy065. doi: 10.1093/gigascience/giy065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ehteshami Bejnordi B, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318:2199–2210. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wulczyn E, et al. Deep learning-based survival prediction for multiple cancer types using histopathology images. PLoS ONE. 2020;15:e0233678. doi: 10.1371/journal.pone.0233678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Coudray N, et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med. 2018;24:1559–1567. doi: 10.1038/s41591-018-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kather JN, et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 2019;25:1054–1056. doi: 10.1038/s41591-019-0462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ardila D, et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat. Med. 2019;25:954–961. doi: 10.1038/s41591-019-0447-x. [DOI] [PubMed] [Google Scholar]
- 115.Hosny A, et al. Deep learning for lung cancer prognostication: a retrospective multi-cohort radiomics study. PLoS Med. 2018;15:e1002711. doi: 10.1371/journal.pmed.1002711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zviran A, et al. Genome-wide cell-free DNA mutational integration enables ultra-sensitive cancer monitoring. Nat. Med. 2020;26:1114–1124. doi: 10.1038/s41591-020-0915-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mathios D, et al. Detection and characterization of lung cancer using cell-free DNA fragmentomes. Nat. Commun. 2021;12:5060. doi: 10.1038/s41467-021-24994-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Beshnova D, et al. De novo prediction of cancer-associated T cell receptors for noninvasive cancer detection. Sci. Transl. Med. 2020;12:eaaz3738. doi: 10.1126/scitranslmed.aaz3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Katzman JL, et al. DeepSurv: personalized treatment recommender system using a Cox proportional hazards deep neural network. BMC Med. Res. Methodol. 2018;18:24. doi: 10.1186/s12874-018-0482-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ching T, Zhu X, Garmire LX. Cox-nnet: an artificial neural network method for prognosis prediction of high-throughput omics data. PLoS Comput. Biol. 2018;14:e1006076. doi: 10.1371/journal.pcbi.1006076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kann BH, Hosny A, Hjwl A. Artificial intelligence for clinical oncology. Cancer Cell. 2021;39:916–927. doi: 10.1016/j.ccell.2021.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kadir T, Brady M. Saliency, scale and image description. Int. J. Comput. Vis. 2001;45:83–105. doi: 10.1023/A:1012460413855. [DOI] [Google Scholar]
- 123.Zhou, B., Khosla, A., Lapedriza, A., Oliva, A. & Torralba, A. Learning deep features for discriminative localization. 2016 IEEE Conf. Comput. Vis. Pattern Recognit. (CVPR)10.1109/cvpr.2016.319https://www.computer.org/csdl/proceedings/cvpr/2016/12OmNqH9hnp (2016).
- 124.Wulczyn E, et al. Interpretable survival prediction for colorectal cancer using deep learning. NPJ Digit. Med. 2020;4:71. doi: 10.1038/s41746-021-00427-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Buel GR, Walters KJ. Can AlphaFold2 predict the impact of missense mutations on structure? Nat. Struct. Mol. Biol. 2022;29:1–2. doi: 10.1038/s41594-021-00714-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.US Food and Drug Administration. Evaluation of automatic class III designation for Paige Prostate. https://www.accessdata.fda.gov/cdrh_docs/reviews/DEN200080.pdf (2021).
- 127.Calcoen D, Elias L, Yu X. What does it take to produce a breakthrough drug? Nat. Rev. Drug Discov. 2015;14:161–162. doi: 10.1038/nrd4570. [DOI] [PubMed] [Google Scholar]
- 128.Jayatunga MKP, Xie W, Ruder L, Schulze U, Meier C. AI in small-molecule drug discovery: a coming wave? Nat. Rev. Drug Discov. 2022;21:175–176. doi: 10.1038/d41573-022-00025-1. [DOI] [PubMed] [Google Scholar]
- 129.Pushpakom S, et al. Drug repurposing: progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019;18:41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 130.Jahchan NS, et al. A drug repositioning approach identifies tricyclic antidepressants as inhibitors of small cell lung cancer and other neuroendocrine tumors. Cancer Discov. 2013;3:1364–1377. doi: 10.1158/2159-8290.CD-13-0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kuenzi BM, et al. Predicting drug response and synergy using a deep learning model of human cancer cells. Cancer Cell. 2020;38:672–684.e6. doi: 10.1016/j.ccell.2020.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ling A, Huang RS. Computationally predicting clinical drug combination efficacy with cancer cell line screens and independent drug action. Nat. Commun. 2020;11:5848. doi: 10.1038/s41467-020-19563-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Aissa AF, et al. Single-cell transcriptional changes associated with drug tolerance and response to combination therapies in cancer. Nat. Commun. 2021;12:1628. doi: 10.1038/s41467-021-21884-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Menden MP, et al. Community assessment to advance computational prediction of cancer drug combinations in a pharmacogenomic screen. Nat. Commun. 2019;10:2674. doi: 10.1038/s41467-019-09799-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Carvalho DM, et al. Repurposing vandetanib plus everolimus for the treatment of ACVR1-mutant diffuse intrinsic pontine glioma. Cancer Discov. 2021 doi: 10.1158/2159-8290.CD-20-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhavoronkov A, et al. Deep learning enables rapid identification of potent DDR1 kinase inhibitors. Nat. Biotechnol. 2019;37:1038–1040. doi: 10.1038/s41587-019-0224-x. [DOI] [PubMed] [Google Scholar]
- 137.Ruthotto L, Haber E. An introduction to deep generative modeling. GAMM-Mitteilungen. 2021;44:e202100008. doi: 10.1002/gamm.202100008. [DOI] [Google Scholar]
- 138.Wallach, I., Dzamba, M. & Heifets, A. AtomNet: a deep convolutional neural network for bioactivity prediction in structure-based drug discovery. Preprint at https://arxiv.org/abs/1510.02855 (2015).
- 139.Ma J, Sheridan RP, Liaw A, Dahl GE, Svetnik V. Deep neural nets as a method for quantitative structure–activity relationships. J. Chem. Inf. Model. 2015;55:263–274. doi: 10.1021/ci500747n. [DOI] [PubMed] [Google Scholar]
- 140.Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018;15:81–94. doi: 10.1038/nrclinonc.2017.166. [DOI] [PubMed] [Google Scholar]
- 141.Bansal M, et al. A community computational challenge to predict the activity of pairs of compounds. Nat. Biotechnol. 2014;32:1213–1222. doi: 10.1038/nbt.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ahmadi S, et al. The landscape of receptor-mediated precision cancer combination therapy via a single-cell perspective. Nat. Commun. 2022;13:1613. doi: 10.1038/s41467-022-29154-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Eduati F, et al. Prediction of human population responses to toxic compounds by a collaborative competition. Nat. Biotechnol. 2015;33:933–940. doi: 10.1038/nbt.3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gayvert KM, Madhukar NS, Elemento O. A data-driven approach to predicting successes and failures of clinical trials. Cell Chem. Biol. 2016;23:1294–1301. doi: 10.1016/j.chembiol.2016.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.McDermott MBA, et al. Reproducibility in machine learning for health research: still a ways to go. Sci. Transl. Med. 2021;13:eabb1655. doi: 10.1126/scitranslmed.abb1655. [DOI] [PubMed] [Google Scholar]
- 146.AP News. Caris Precision Oncology Alliance partners with the National Cancer Institute, part of the National Institutes of Health, to expand collaborative clinical research efforts. Associated Presshttps://apnews.com/press-release/pr-newswire/technology-science-business-health-cancer-221e9238956a7a4835be75cb65832573 (2021).
- 147.Alvi MA, Wilson RH, Salto-Tellez M. Rare cancers: the greatest inequality in cancer research and oncology treatment. Br. J. Cancer. 2017;117:1255–1257. doi: 10.1038/bjc.2017.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Park KH, et al. Genomic landscape and clinical utility in Korean advanced pan-cancer patients from prospective clinical sequencing: K-MASTER program. Cancer Discov. 2022;12:938–948. doi: 10.1158/2159-8290.CD-21-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bailey MH, et al. Retrospective evaluation of whole exome and genome mutation calls in 746 cancer samples. Nat. Commun. 2020;11:4748. doi: 10.1038/s41467-020-18151-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ellrott K, et al. Scalable open science approach for mutation calling of tumor exomes using multiple genomic pipelines. Cell Syst. 2018;6:271–281.e7. doi: 10.1016/j.cels.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zare F, Dow M, Monteleone N, Hosny A, Nabavi S. An evaluation of copy number variation detection tools for cancer using whole exome sequencing data. BMC Bioinforma. 2017;18:286. doi: 10.1186/s12859-017-1705-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pan-cancer analysis of whole genomes. Nature578, 82–93 (2020). [DOI] [PMC free article] [PubMed]
- 153.Gawad C, Koh W, Quake SR. Single-cell genome sequencing: current state of the science. Nat. Rev. Genet. 2016;17:175–188. doi: 10.1038/nrg.2015.16. [DOI] [PubMed] [Google Scholar]
- 154.Corces MR, et al. The chromatin accessibility landscape of primary human cancers. Science. 2018;362:eaav1898. doi: 10.1126/science.aav1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Buenrostro JD, et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Furey TS. ChIP–seq and beyond: new and improved methodologies to detect and characterize protein–DNA interactions. Nat. Rev. Genet. 2012;13:840–852. doi: 10.1038/nrg3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Rotem A, et al. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat. Biotechnol. 2015;33:1165–1172. doi: 10.1038/nbt.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Papanicolau-Sengos A, Aldape K. DNA methylation profiling: an emerging paradigm for cancer diagnosis. Annu. Rev. Pathol. 2022;17:295–321. doi: 10.1146/annurev-pathol-042220-022304. [DOI] [PubMed] [Google Scholar]
- 159.Smallwood SA, et al. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat. Methods. 2014;11:817–820. doi: 10.1038/nmeth.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Cieślik M, Chinnaiyan AM. Cancer transcriptome profiling at the juncture of clinical translation. Nat. Rev. Genet. 2018;19:93–109. doi: 10.1038/nrg.2017.96. [DOI] [PubMed] [Google Scholar]
- 161.Macosko EZ, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zheng GXY, et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ramsköld D, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gierahn TM, et al. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat. Methods. 2017;14:395–398. doi: 10.1038/nmeth.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Rao A, Barkley D, França GS, Yanai I. Exploring tissue architecture using spatial transcriptomics. Nature. 2021;596:211–220. doi: 10.1038/s41586-021-03634-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ståhl PL, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353:78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- 167.Rodriques SG, et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science. 2019;363:1463–1467. doi: 10.1126/science.aaw1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lee JH, et al. Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat. Protoc. 2015;10:442–458. doi: 10.1038/nprot.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ellis MJ, et al. Connecting genomic alterations to cancer biology with proteomics: the NCI Clinical Proteomic Tumor Analysis Consortium. Cancer Discov. 2013;3:1108–1112. doi: 10.1158/2159-8290.CD-13-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Li J, et al. TCPA: a resource for cancer functional proteomics data. Nat. Methods. 2013;10:1046–1047. doi: 10.1038/nmeth.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Stoeckius M, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods. 2017;14:865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Bendall SC, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Jackson HW, et al. The single-cell pathology landscape of breast cancer. Nature. 2020;578:615–620. doi: 10.1038/s41586-019-1876-x. [DOI] [PubMed] [Google Scholar]
- 174.Keren L, et al. A structured tumor-immune microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging. Cell. 2018;174:1373–1387.e19. doi: 10.1016/j.cell.2018.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Schürch CM, et al. Coordinated cellular neighborhoods orchestrate antitumoral immunity at the colorectal cancer invasive front. Cell. 2020;183:838. doi: 10.1016/j.cell.2020.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Beckonert O, et al. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc. 2007;2:2692–2703. doi: 10.1038/nprot.2007.376. [DOI] [PubMed] [Google Scholar]
- 177.Jang C, Chen L, Rabinowitz JD. Metabolomics and isotope tracing. Cell. 2018;173:822–837. doi: 10.1016/j.cell.2018.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ghandi M, et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature. 2019;569:503–508. doi: 10.1038/s41586-019-1186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Uhlén M, et al. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 180.Fedorov A, et al. NCI Imaging Data Commons. Cancer Res. 2021;81:4188–4193. doi: 10.1158/0008-5472.CAN-21-0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Goldman MJ, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020;38:675–678. doi: 10.1038/s41587-020-0546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Jiang P, Freedman ML, Liu JS, Liu XS. Inference of transcriptional regulation in cancers. Proc. Natl Acad. Sci. USA. 2015;112:7731–7736. doi: 10.1073/pnas.1424272112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Sun D, et al. TISCH: a comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res. 2021;49:D1420–D1430. doi: 10.1093/nar/gkaa1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kristiansen G. Markers of clinical utility in the differential diagnosis and prognosis of prostate cancer. Mod. Pathol. 2018;31:S143–S155. doi: 10.1038/modpathol.2017.168. [DOI] [PubMed] [Google Scholar]