Abstract
The global COVID-19 pandemic remains a critical public health threat, as existing vaccines and drugs appear insufficient to halt the rapid transmission. During an outbreak from May to August 2021 in Taiwan, patients with severe COVID-19 were administered NRICM102, which was a traditional Chinese medicine (TCM) formula developed based on its predecessor NRICM101 approved for treating mild cases. This study aimed to explore the mechanism of NRICM102 in ameliorating severe COVID-19-related embolic and fibrotic pulmonary injury. NRICM102 was found to disrupt spike protein/ACE2 interaction, 3CL protease activity, reduce activation of neutrophils, monocytes and expression of cytokines (TNF-α, IL-1β, IL-6, IL-8), chemokines (MCP-1, MIP-1, RANTES) and proinflammatory receptor (TLR4). NRICM102 also inhibited the spread of virus and progression to embolic and fibrotic pulmonary injury through reducing prothrombotic (vWF, PAI-1, NET) and fibrotic (c-Kit, SCF) factors, and reducing alveolar type I (AT1) and type II (AT2) cell apoptosis. NRICM102 may exhibit its protective capability via regulation of TLRs, JAK/STAT, PI3K/AKT, and NET signaling pathways. The study demonstrates the ability of NRICM102 to ameliorate severe COVID-19-related embolic and fibrotic pulmonary injury in vitro and in vivo and elucidates the underlying mechanisms.
Chemical compounds studied in this article: 5-caffeoylquinic acid (PubChem CID: 5280633), 3-caffeoylquinic acid (PubChem CID: 1794427), 4-caffeoylquinic acid (PubChem CID: 9798666), Quercetin 3-galactoside (PubChem CID: 5281643), Quercetin 3-glucoside (PubChem CID: 5280804), Quercetin 3-rhamnoside (PubChem CID: 5280459), Baicalin (PubChem CID: 64982), Norwogonin 7-glucuronide (PubChem CID: 44258552), Oroxyloside (PubChem CID: 14655552), Glycyrrhizic acid (PubChem CID: 14982)
Keywords: COVID-19, NRICM102, Pulmonary embolism, Pulmonary fibrosis, Traditional Chinese medicine
Graphical Abstract
1. Introduction
Multiple vaccines and antiviral agents have received emergency use authorizations in the two years after the onset of the COVID-19 outbreak, yet ending the global pandemic remains an elusive goal. Emergence of highly transmissible variants with immune-evasive properties as a result of ongoing SARS-CoV-2 evolution continues to cause numerous infections, overwhelming healthcare systems worldwide. In this context, a pan-sarbecovirus therapeutic that can treat infections caused by all variants of concern is urgently needed.
Lung involvement is of the greatest concern in COVID-19. When the virus replicates and spreads in the lungs, it stimulates severe immune responses. In the stage of systemic hyperinflammation, uncontrolled cytokine release may trigger a cytokine storm and lead to lung injury, venous thromboembolic diseases (pulmonary embolism or deep vein thrombosis), acute respiratory distress syndrome (ARDS), and even fatal complications such as multi-organ failure [1]. Since the onset of the pandemic, when substantial numbers of severe and critical cases were reported, the disease progression has often been accompanied by a high prevalence of venous thromboembolic syndrome in hospitalized patients; for those in the ICU, the rate can be as high as 27% [2], [3], [4], [5], [6], [7], [8].
Lung injury occurs when SARS-CoV-2 infects endothelial cells, including alveolar type I (AT1) and alveolar type II (AT2) cells, and triggers capillary permeability, adhesion and extravasation of neutrophils and monocytes into the alveolar interstitial space by the release of cytokines [9]. SARS-CoV-2-activated neutrophils, monocytes and macrophages thereby secrete a multitude of cytokines, procoagulants, and complements via pathogen-associated and damage-associated molecular patterns (PAMPs and DAMPs), which not only promotes viral attack and clearance but also induces further vascular injury, enhancing the risk for thrombosis. COVID-19 patients’ blood neutrophils form large amounts of neutrophil extracellular traps (NETs), as evidenced by increased concentrations of myeloperoxidase (MPO)-DNA and citrullinated histone H3, to contain infection [10], [11], but NETs can also increase coagulation by enhancing thrombin generation and platelet adhesion [11]. There are other factors that contribute to the prothrombotic environment. Downregulation of angiotensin-converting enzyme 2 (ACE2) by SARS-CoV-2 infection causes microvascular thrombosis through upregulation of angiotensin II and consequently elevates platelet activator inhibitor-1 (PAI-1) levels [10], [11], [12]. Moreover, hospitalized patients with COVID-19 show elevated levels of fibrinogen and vWF, which promote microvascular thrombosis [13]. On the other hand, infected circulating monocytes participate in aberrant activities that may induce inflammatory responses, cause cytokine storm and contribute to lung injury [9], [14]. Furthermore, AT2 cell dysfunction or loss (apoptosis or necrosis) induced by SARS-CoV-2 is harmful to the injured lung because decreases in the amounts of surfactant lipids increase the risk for alveolar collapse and atelectasis, and reductions in the numbers of AT2 cells impair AT1 cell regeneration, affecting alveolar repair and stimulating fibrosis.
Therefore, in addition to developing antiviral agents that target spike protein (Dalbavancin [15] and Ceftazidime [16]), the 3CL protease (Paxlovid) [17], and RNA-dependent RNA polymerase inhibitors (Remdesivir [18] and Molnupiravir [19]), halting the progression of lung injury and embolism in those diagnosed with COVID-19 by regulating the immune response is more challenging yet equally important.
Understanding the value of a traditional Chinese medicine (TCM) integrated approach in the treatment of COVID-19, the World Health Organization (WHO) encouraged those engaged in clinical service and research and evidence-based evaluation to effectively enhance the contribution of TCM in global pandemics [20]. In line with the strategy, novel TCM formulae NRICM101 and NRICM102 have been developed and applied with satisfactory results in Taiwan since 2020.
NRICM101 is composed of herbs with antiviral and immunomodulatory properties, and has shown effectiveness in interrupting the progression of COVID-19. Focal ground-glass opacities dissipate in chest X-ray images after NRICM101 is administered to COVID-19 patients. Clinical observations have shown that intervention with NRICM101 relieves fever, stabilizes cardiopulmonary function, and shortens the length of hospital stay, without reports of adverse effects. In pharmacological assays, the TCM formula has demonstrated abilities to neutralize SARS-CoV-2, block virus infection and 3CL protease activity, inhibit virus growth, and suppress the expression of the cytokines IL-6 and TNF-α [21].
Given the potential of this multiple-target agent to halt the pandemic, an enhanced version NRICM102 has been developed and administered in Taiwan for patients with severe COVID-19. Clinical outcome analyses have demonstrated reduced mortality of severe-to-critical patients with COVID-19 [22]. This study aims to provide scientific evidence that NRICM102 ameliorates pulmonary embolism and fibrosis, and respiratory functions via regulation of specific immune responses. In particular, RNA-sequencing analysis was carried out to illustrate the gene regulation and signaling pathways related to NRICM102 activity.
2. Material and methods
2.1. Chemicals and reagents
The human bronchial epithelial cells (BEAS-2B cells) were purchased from the Bioresource Collection and Research Center (BCRC, Taiwan). Recombinant SARS-CoV-2 spike protein subunit 1 (S1) was purchased from GeneTex International Corporation, UK (cat no.: GTX135817-pro). LPS (Escherichia coli, O55:B5) and bleomycin were purchased from Sigma (MO, USA).
Preparation and composition of NRICM102. The NRICM102 decoction was prepared by the TCM pharmacies in Taichung Veterans General Hospital. It consisted of 10 traditional Chinese medicines (TCMs), including Scutellaria Root (Scutellaria baicalensis), Heartleaf Houttuynia Herb (Houttuynia cordata), Prepared Monkshood Daughter Root (Aconitum carmichaelii), Indian Bread (Poria cocos), Mongolian Snakegourd Fruit (Trichosanthes kirilowii), Fragrant Solomonseal Rhizome (Polygonatum odoratum), Baked Licorice Root and Rhizome (Glycyrrhiza glabra), Magnolia Bark (Magnolia officinalis), Oriental Wormwood Herb (Artemisia scoparia), and Pinellia tuber (Pinellia ternata). The NRICM102 formulation in 1200 mL of water was boiled down to 300 mL. The decoction was further concentrated by freeze-drying to obtain a test sample of 17.7 ± 0.5 g per 300 mL. The single TCMs (5 g) of NRICM102 were respectively extracted by H2O (100 mL) refluxing for 1 hr. After filtering, the extracts were concentrated by freeze-drying to obtain the single TCM extracts.
2.2. HPLC analysis of NRICM102
HPLC was performed on a Shimadzi UPLC Nexera X3 series instrument (Shimadzu, Kyoto, Japan) including an LC-40D X3 (pump), CBM-40 (system controller), DGU-405 (degassing unit), Sil‐40C X3 (autosampler), and CTO-40S (column oven) and equipped with an ODS COSMOSIL 5C18-AR-II column (4.6 mm × 250 mm). In HPLC fingerprint analysis, the mobile phase consisted of double-distilled water with 0.3% phosphoric acid and acetonitrile (ACN) using a gradient condition. The mobile phase flow rate and the injection volume were 1.0 mL/min and 10 μL, respectively. The column oven set at 40 ℃. The concentration of the tested sample was 10 mg/mL in ddH2O.
2.3. Identification of the components of NRICM102
The NRICM102 decoction (2000 mL) was separated on a Diaion HP-20 open column (EtOH/H2O) and a C18 flash column and further purified by preparative HPLC, yielding the components. The chemical structures of all isolated components were determined by NMR analyses, including 1H, 13C, DEPT, HSQC, HMBC, and NOESY analyses, and high-resolution electrospray ionization mass spectroscopy was performed. The findings were compared with literature data.
2.4. Bio-Layer Interferometry (BLI)
Bio-Layer Interferometry binding events were detected and monitored in real time using a FortéBio Octet Red 96e Biolayer Interferometer (Molecular Devices). A recombinant SARS-CoV-2 variant RBD protein (Sino Biological) was immobilized on HIS1K sensor tips for 600 s at 100 µg/mL in PBS. The sensor tips were then blocked with 1% BSA for 5 min. NRICM102 was resuspended in kinetic buffer (PBST, NaCl adjusted to 350 mM). Sample testing was done sequentially using baseline, association, and dissociation steps for 60, 300, and 300 s, respectively. The data were aligned using association signals and curves fitted with a 1:1 best-fit model in FortéBio’s data analysis software. The sensor tips created atypical binding events to the immobilized protein through nonspecific binding effects. To reduce signals associated with the atypical binding events, we employed reference sensor subtraction. A separate set of blank sensors without loaded proteins were exposed to predefined conditions.
2.5. ACE2-spike protein inhibition enzyme-linked immunosorbent assay (ELISA)
ELISA was carried out to determine the ability of NRICM102 to disrupt the interaction between the recombinant SARS-CoV-2 variant RBD protein and human ACE2 (Sino Biological). Briefly, microplate wells were coated with the recombinant SARS-CoV-2 variant RBD protein (0.1–2 μg/well). After blocking with 1% BSA for 1 hr at 37 ℃, serial dilutions of NRICM102 (1/10X, 1/50X, 1/100X, 1/150X, 1/300X, 1/600X, 1/900X, 1/1200, 1/1500, 1/2000X, 1/3000X, and 1/6000X) were added to the wells to react with the recombinant SARS-CoV-2 variant RBD protein at 37 ℃. When the reaction was completed, the human ACE2 recombinant protein (0.2 μg/mL, Sino Biological) was added to each well, and the plate was incubated for 40 min at 37 ℃. After that, the plates were incubated with rabbit anti-human IgG-HRP (Immunology consultants laboratories, Inc.) for 40 min. The HRP substrate 3,3',5,5' tetramethylbenzidine (TMB, SeraCare) was added to each well for color development. The reaction was stopped with 1N HCl, and signal intensity was quantified at OD 450 nm (SPECTROstar Nano, BMG LABTECH).
2.6. Inhibition assay of 3CL protease
The assay was performed as previously described [21]. Briefly, recombinant SARS-CoV-2 3CL protease (Pharmtekx, Taipei, Taiwan) was incubated with NRICM102 in reaction buffer (25 mM Tris, 100 mM NaCl, 1 mM EDTA, 1 mM DTT, pH 7.3) on ice for 30 min, and a fluorogenic protease substrate peptide (Dabcyl-KTSAVLQSGFRKME(Edans)-OH, obtained from Kelowna International Scientific Inc., New Taipei City, Taiwan) was subsequently added to initiate the proteolytic reaction. The reaction was monitored at 538 nm with excitation at 355 nm at 37 °C by using a Cytation 5 cell imaging multi-mode reader (BioTek, Vermont, USA) for 1 hr. The inhibition was calculated and plotted with GraphPad Prism.
2.7. Animals
Six- to eight-week-old male C57 and ICR mice were purchased from the National Laboratory Animal Breeding and Research Center (Taipei, Taiwan). Fourteen- to sixteen-week-old male K18-hACE2 transgenic mice were purchased from the Jackson Laboratory and inbred in the Laboratory Animal Center of National Taiwan University College of Medicine (Taipei, Taiwan). All the animals were treated with standard environmental and food conditions, i.e. 22 ± 1 °C, 55 ± 5% humidity and 12 hr light/dark cycle with free food and water access. All the animals were randomly assigned to groups in a double-blind manner to reduce the experimental bias.
2.8. S1-induced pulmonary embolism in K18-hACE2 mice
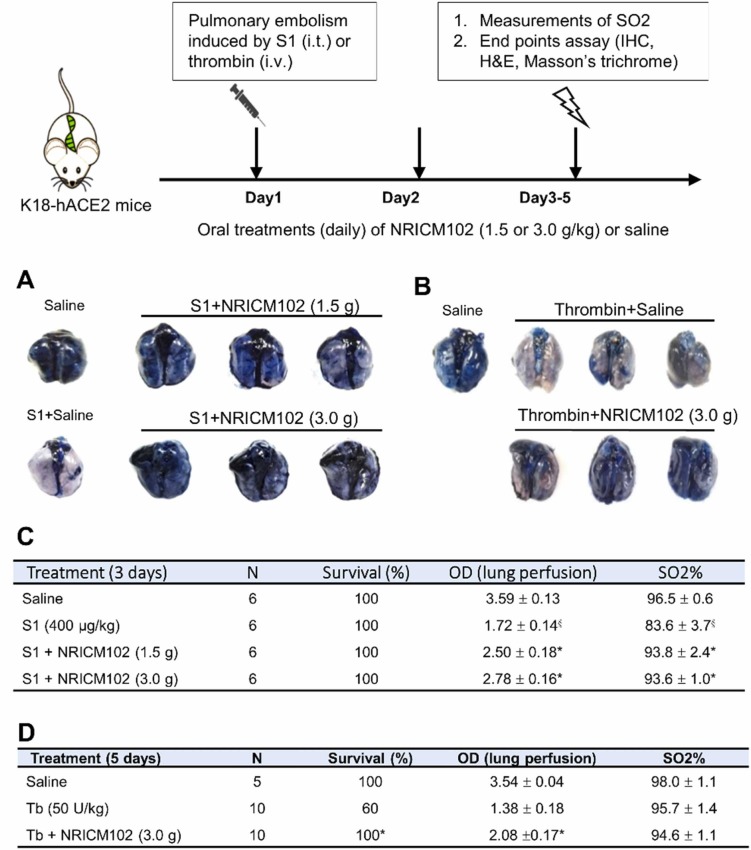
S1 was given intratracheally to K18-hACE2 mice by methods according to a previous report [23]. Briefly, mice were anesthetized with intraperitoneal injection of xylazine (6 mg/kg) and ketamine (60 mg/kg). A small skin incision was created on the neck of each mouse. S1 (400 μg/kg in 2 mL/kg) was dissolved in sterile normal saline and instilled into the tracheal lumen. The incision was closed after instillation to allow the mice to recover. For drug treatment, mice were treated with NRICM102 (1.5 or 3.0 g/kg, p.o.) or vehicle (saline) as a control (Ctrl) daily for 3 days after the treatment with S1. The animal groups were as follows: saline control (Ctrl), S1+saline, S1+NRICM102 (1.5 g) and S1+NRICM102 (3.0 g). The mice were sacrificed to collect their lungs after 3 days. For lung perfusion detection, mice were perfused with 0.5 mL of 1% Evans blue via the right ventricle. The lungs were excised and photographed, and the degree of vascular occlusion was evaluated independently by detecting the optical density (OD, absorbance at 620 nm) of Evans blue by methods modified from those in a previous report [24]. Lung oxygen saturation (SO2%), the percentage of hemoglobin bound with oxygen, was measured using an iSTAT G3+ detection kit (Abbott Point of Care Canada Limited, Canada). The moving activity of mice was assessed by tracking their distance traveled in 3 min in a behavioral observation box (60 × 40 × 60 cm3) before sacrifice. The results were then analyzed using a video-tracking system (SMART v2.5.21, Panlab, Spain). The survival rates were calculated immediately (day 0) and at 72 hr (day 3) after administration of S1.
2.9. Thrombin-induced pulmonary embolism in ICR mice
Acute experimental pulmonary embolism was induced by injection of α-thrombin (50 U/kg, bovine, Sigma-Aldrich, St. Louis, USA) through the inferior vena cava in 100 μL of sterile saline by methods modified from those in previous reports [25], [26]. The animal groups were as follows: the saline control (Ctrl) group, the thrombin group and the thrombin+NRICM102 (3.0 g/kg/day, p.o. for 5 days) group. We repeated the abovementioned methods to perform lung perfusion and lung oxygen saturation analyses as well as movement observations. The survival rates were also calculated in the same manner.
2.10. Bleomycin-induced lung injury in C57BL/6 mice
C57BL/6 male mice were exposed to bleomycin (BLM) by intratracheal instillation with methods modified from those in previous reports [27], [28]. Briefly, mice were anesthetized with intraperitoneal injection of xylazine (6 mg/kg) and ketamine (60 mg/kg). A small skin incision was made on the neck of each mice. BLM (2 U/kg; Sigma) was dissolved in 40 μL of PBS and instilled into the tracheal lumen. After inoculation, the incision was closed, and the animal was allowed to recover. For drug treatment, the mice were treated with NRICM102 (1.5 and 3.0 g/kg, p.o.) or vehicle (saline) as a control (Ctrl) daily for 20 days before sacrifice. The body weights and survival rates were calculated immediately from day 0 to day 21 after administration of S1. Plethysmography was used to measure pulmonary function in vivo with a previously reported method [29].
2.11. Histopathology and immunohistochemistry
For immunohistochemical (IHC) staining, we collected 15–20 consecutive sections (approximately 20–30 µm thick) of lung tissue at the same level from different groups for staining. All tissue sections were fixed, permeabilized, and blocked and were randomly selected for specific marker staining with primary antibodies (diluted in PBS containing 3% albumin at 4 °C overnight). Antibodies against S1 RBD (1:100), citrullinated histone H3 (CitH3, NET, 1:50); Ly6G (1:100), MPO (1:100), vWF (1:100), PAI-1 (1:100), PDPN (AT1, 1:100), SFTPC (AT2, 1:100), MIF (1:100) and TLR4 (1:100) were obtained from GeneTex (Irvine, CA, USA); antibodies against CD11b (1:50) and CD31 (also known as platelet endothelial cell adhesion molecule 1 [PECAM-1]) (1:50) were obtained from Abcam (Cambridge, UK); SCF (1:50) and cCasp3 (1:50) antibodies were obtained from Santa Cruz (Santa Cruz Biotechnology, Inc., CA, USA); a p-NFκB P65 antibody was obtained from BD Pharmingen (1:50, BD Pharmingen, San Diego, CA, USA); and a c-Kit antibody was obtained from Invitrogen (1:200, Invitrogen, Frederick, MD, USA). After washing, all the sections were stained with secondary antibodies conjugated with Alexa Fluor® 488, Alexa Fluor® 555, or Alexa Fluor® 647 (all from Cell Signaling Technology Inc., MA, USA). To counterstain the DNA in the nuclei, all sections on coverslips were mounted with medium containing 4',6-diamidino-2-phenylindole (DAPI). All the properly stained sections on coverslips were examined using a laser-scanning confocal microscope (Zeiss LSM780, Carl Zeiss, Jena, Germany). The distribution and number of immunopositive stained cells or the immunopositive areas were identified and quantified as the relative stained areas (%) using imaging software (Zen 2011, black edition, Carl Zeiss MicroImaging GmbH, 1997–2011) and AlphaEase FC (Alpha Innotech, San Leandro, CA, USA) across the entire image field of regions of interest sampled from each group under appropriate magnification (30×∼100×) in 3–5 independent experiments. For tissue fibrosis detection, a Masson's trichrome staining protocol was followed [30].
2.12. Monocyte isolation and cytokine array
Peripheral blood mononuclear cells (PBMCs) were isolated from blood samples of healthy donors. Human monocytes were isolated from PBMCs as described previously [31]. In brief, PBMCs were separated from whole blood by using Ficoll-Paque™ density gradient centrifugation, and monocytes (98% pure CD14+) were isolated from PBMCs by using a classical monocyte isolation kit (Miltenyi Biotec). The isolated monocytes were treated with S1 (100 μg/mL) and NRICM102 for 24 hr. The supernatant was prepared for cytokine determination. The cytokine expression was detected with a human XL cytokine array kit (Cytokine Array, R&D) that can detect 109 kinds of cytokines.
2.13. RNA sequencing and RNA-Seq data analysis
SimpliNano™ Biochrom Spectrophotometer (Biochrom, MA, USA) was used to check the purity and quantity of each RNA sample. RNA degradation and integrity were monitored with a Qsep 100 DNA/RNA Analyzer (BiOptic Inc., Taiwan). Sequencing libraries were generated from the total RNA of the samples by using a KAPA mRNA HyperPrep Kit (KAPA Biosystems, Roche, Basel, Switzerland). High-throughput sequencing (Illumina NovaSeq 6000 platform) was performed to obtain the raw data, and FastQC and MultiQC were used to assess quality [32]. High-quality raw paired-end read data were obtained by using Trimmomatic (v 0.38) for subsequent analysis [33]. Alignment of read pairs from each sample to a reference genome was performed with HISAT2 software (v 2.1.0) [34], [35], after which the reads mapped to individual genes were counted by using FeatureCounts (v 2.0.0). Differentially expressed gene (DEG) analysis of case and control samples was performed by using DEGseq (v 1.40.0) [36] or DESeq2 (v 1.26.0) [37], [38]. Gene Ontology (GO) functional annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses [39], [40] were performed by using clusterProfiler (v 3.14.3) [41]. A protein–protein interaction (PPI) network was constructed for the differentially expressed genes using STRINGdb (https://string-db.org/).
2.14. Epithelial–mesenchymal transition (EMT) and fibroblast to myofibroblast transformation (FMT) assays
For the EMT assay, BEAS-2B cells were cultured in dishes coated with bovine serum albumin (BSA, Bionovas), native fibronectin human protein (Gibco) and bovine collagen I (Gibco). The cells were grown at 37 °C under 5% CO2 in bronchial epithelial cell growth basal medium (BEGM, Lonza). BEAS-2B cells (6 × 103) were seeded in a 96-well black plate (Thermo Fisher Scientific) and incubated in BEGM for 24 hr. Then, the cells were stimulated with 10 ng/mL TGF-β (PeproTech) and incubated for 3 days. For FMT assay, HFL-1 cells (6 × 103) were seeded in a 96-well black plate (Thermo Fisher Scientific) and incubated in F‐12K medium with 10% FBS for 24 hr. The cells were washed 3 times with PBS buffer and starved in F‐12K medium with 0.1% FBS for 24 hr. Then, the cells were replaced with F‐12K medium with 0.5% FBS containing 10 ng/mL TGF-β (PeproTech) and incubated for 3 days. The BEAS-2B and HFL-1 cells were fixed with cold methanol (−20 ℃) for 30 min at room temperature. Following fixation, permeabilization, and blocking, the BEAS-2B and HFL-1 cells were incubated with a fibronectin antibody (FN1, 1:800 dilution, Cell Signaling) or with alpha smooth muscle actin antibody (α-SMA, 1:800 dilution, Cell Signaling) overnight at 4 ℃, respectively. After washing, the cells were incubated with Alexa Fluor 488 anti-rabbit IgG (1:1000 dilution, Cell Signaling). The cells were incubated with DAPI (5 μg/mL, Thermo Fisher Scientific) for nuclear staining. Images were captured with a Cytation 5.
2.15. Statistical analysis
GraphPad Prism (version 9.0; GraphPad Software, San Diego, CA) were used for data analysis. The results are presented as the mean ± SEM. Statistical analysis involved one-way ANOVA followed by the S-N-K t-test. Differences were considered statistically significant at p < 0.05.
2.16. Study approval
The studies involving human donors were reviewed and approved by the internal review board of Taipei Hospital, Ministry of Health and Welfare (IRB protocol #TH-IRB-0021–0008). The laboratory procedures involving animals were conducted in accordance with the Guide for the Care and Use of Laboratory Animals (National Research Council, 2011) and were approved by the Animal Research Committee of the National Research Institute of Chinese Medicine (ref no: NRICM-IACUC-110–355–1, NRICM-IACUC-111-912-2).
3. Results
3.1. Anti-SARS-CoV-2 activities of NRICM102
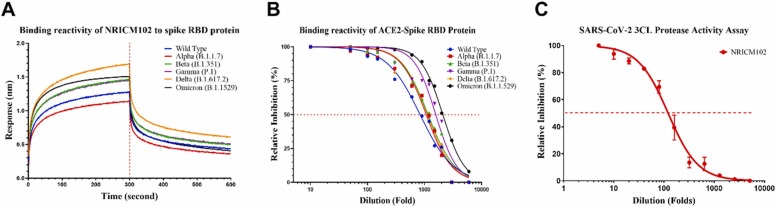
We investigated the antiviral effects of NRICM102 in inhibiting the spike protein/ACE2 interaction and 3CL protease activity. We tested the effect of NRICM102 on interrupting the binding of spike protein/ACE2 for strains Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and Omicron (B.1.1529) as well as the original wild type strain. As shown in Fig. 1A, BLI analysis revealed that NRICM102 (5-fold dilution) could bind the spike RBD protein of several SARS-CoV-2 variants of concern. The highest binding activity was observed with the Delta variant, followed by the Omicron, Beta, Gamma, wild type, and Alpha variants. The ELISA also showed the ability of NRICM102 to disrupt the binding of different variants to the human ACE2. The EC50 values of NRICM102 for the tested variants were a 2090-fold dilution (Omicron), a 1571-fold dilution (Gamma), a 1151-fold dilution (Beta), a 1117-fold dilution (Delta), a 1068-fold dilution (Alpha), and an 833.9-fold dilution (wild type) (Fig. 1B). In the 3CL protease activity assay, NRICM102 displayed a prominent inhibitory effect on 3CL protease with an IC50 of a 119-fold dilution (Fig. 1C). The results indicated that the antiviral properties of NRICM102 remained potent despite the variants of SARS-CoV-2.
Fig. 1.
Effects of NRICM102 on anti-SARS-CoV-2 activities. (A) The binding reactivity of NRICM102 to the spike RBD proteins of several SARS-CoV-2 variants, including the Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2) and Omicron (B.1.1529) variants and the original wild type strain, were determined by BLI. (B) Interaction of the spike RBDs of several SARS-CoV-2 variants with ACE2 was inhibited by serially diluted NRICM102 in the ACE2–spike RBD protein inhibition ELISA. The inhibition percentage was determined according to the binding signal normalized to the interaction of the spike RBD with ACE2 without NRICM102 treatment. (C) NRICM102 inhibited SARS-CoV-2 3CL protease activity. Serial dilutions of the decoction were used to investigate NRICM102’s inhibitory activity against 3CL protease.
3.2. NRICM102 reduced S1/thrombin-induced pulmonary embolism
To examine whether NRICM102 treatment could improve pulmonary embolism, hACE2 transgenic mice were treated with spike protein (S1, 400 μg/kg, intratracheal instillation (i.t.)) while ICR mice were treated with thrombin (50 U/kg, i.v.) to induce pulmonary embolism. Our results showed that S1 induced a 52% drop in the lung tissue perfusion ( Fig. 2A, S1+saline [white color] and Fig. 2C, OD: from 3.59 ± 0.13 [saline] to 1.72 ± 0.14 [S1+saline]) accompanied by a dramatic reduction in arterial oxygen saturation (SO2%) from 96.5 ± 0.6∼83.6 ± 3.7% (Fig. 2C). NRICM102 treatment after S1 administration for 3 consecutive days significantly improved pulmonary embolism (OD returned to 2.50 ± 0.18 for S1 +1.5 g/kg NRICM102 and 2.78 ± 0.16 for S1 +3.0 g/kg NRICM102) as well as arterial oxygen saturation (SO2%, >93%; Fig. 2C). Thrombin induced relatively severe pulmonary embolism (Fig. 2B) that killed 40% of the mice within 5 days and caused a 60% drop in pulmonary perfusion (OD from 3.54 ± 0.04 [saline] to 1.38 ± 0.18 [thrombin+saline]). NRICM102 (3.0 g/kg) treatment after pulmonary embolism induction for consecutive 5 days significantly improved survival rate to 100% (OD, p < 0.05). However, there were no significant differences among these groups in terms of arterial oxygen saturation (SO2%) and animal moving activity on day 5 (Fig. 2D).
Fig. 2.
Effects of NRICM102 on changes in S1 protein- and thrombin-induced pulmonary embolism assayed by Evans blue perfusion. (A) Experiments were performed on K18-hACE2 mice with S1 protein (400 μg/kg, intratracheal (i.t.))-induced pulmonary embolism that were divided into 4 subgroups, including a saline control (saline) group, an S1- and vehicle-treated (S1+saline) group and an S1- and NRICM102 (1.5 or 3.0 g/kg, p.o.)-treated (S1+NRICM102 [1.5 or 3.0 g]) group. (B) Experiments were performed on ICR mice with thrombin (50 U/kg, i.v.)-induced pulmonary embolism that were divided into 3 subgroups, including a saline control (saline) group, a thrombin- and vehicle-treated (thrombin+saline) group and a thrombin- and NRICM102 (3.0 g/kg, p.o.)-treated (thrombin+NRICM102 [3.0 g]) group. (C) S1 (400 μg/kg) was administered i.t. once on day 1 for pulmonary embolism induction. NRICM102 (1.5 g or 3.0 g/kg/day) was administered for 3 consecutive days after pulmonary embolism induction. A high OD indicates that the lungs were perfused well. §,* p < 0.05 compared to the saline or S1(400) group by one-way ANOVA followed by Dunnett test. (D) Thrombin (Tb, 50 U/kg) was administered i.v. once on day 1 for pulmonary embolism induction. NRICM102 (3.0 g/kg/day) was administered for 5 consecutive days at 1 hr after pulmonary embolism induction. A high OD indicates that the lungs perfused well. * , p < 0.05 compared to the Tb (50 U/kg) group by one-way ANOVA followed by Dunnett test. The survival rate was analyzed by log-rank test.
3.3. NRICM102 reduced S1-induced infiltration of CD11b/Ly6G positive neutrophil and inflammation in the lungs
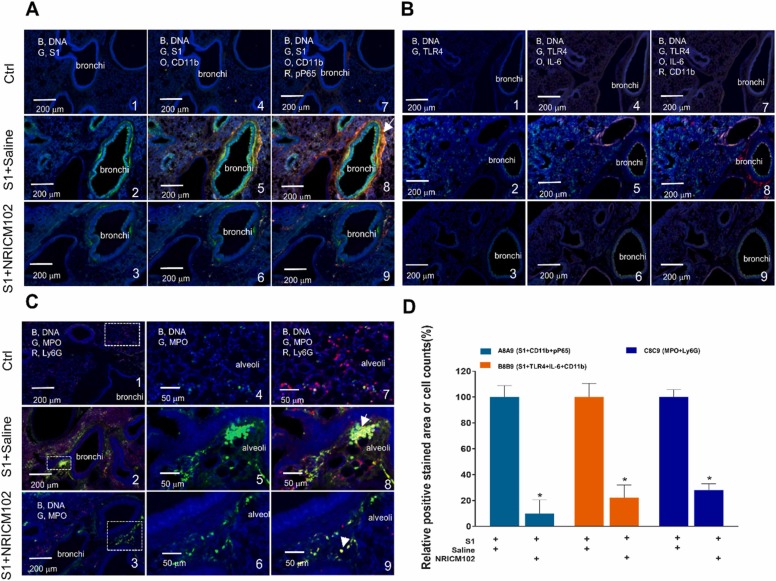
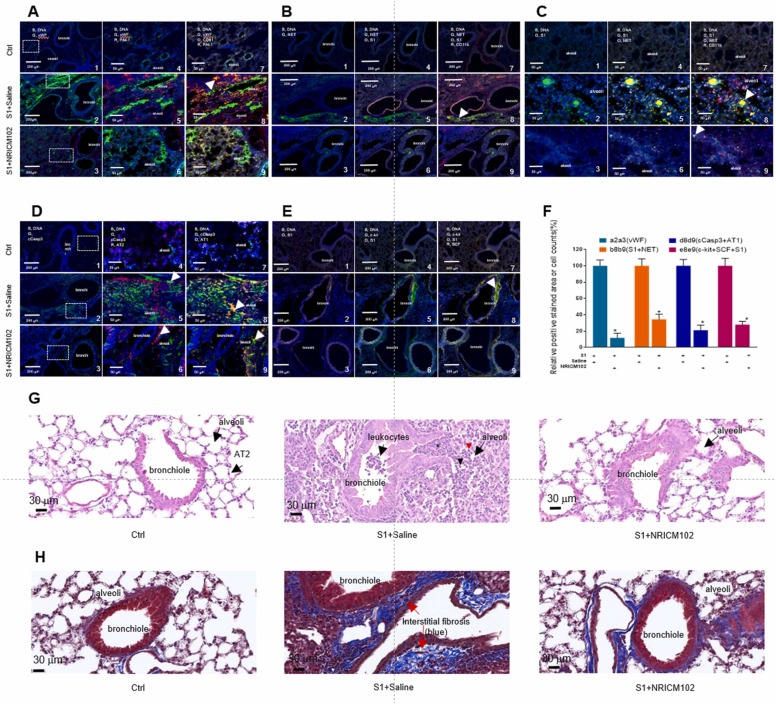
To examine the molecular mechanisms underlying the ameliorative effects of NRICM102 treatment on pulmonary embolism in the S1-induced animal model, several experiments were performed. First, we examined whether NRICM102 treatment could reduce S1’s effects in lung tissues and consequently suppress neutrophil infiltration and inflammatory responses. Our results showed that S1 accumulated to high levels in bronchi and bronchioles and that the accumulation was accompanied by strong CD11b and/or Ly6G positive neutrophil and strong inflammation responses including the expression of p-NFκB P65, MPO, TLR4, and IL-6 ( Fig. 3A-C). In the NRICM102 treatment groups, the accumulation of S1 and the levels of all these inflammatory markers (S1, CD11b, Ly6G, p-NFκB P65, MPO, TLR4, and IL-6) were significantly reduced. Statistical summary of fluorescence staining (relative stained area or cell no. [%]) for selected markers is shown in Fig. 3D.
Fig. 3.
Effects of NRICM102 on changes in S1 protein expression and lung inflammation (at 72 hr) in S1 protein-induced lung injury in K18-hACE2 mice. Typical confocal images from lung tissue showing (A) S1 expression (G, green) and CD11b positive leukocytes infiltration (O, orange)-associated activation of the inflammatory transcription factor p-NFκB (P65) (R, red, pP65). The arrow indicates strong inflammation with extensive colocalization of CD11b and pP65 staining. (B) TLR4 expression (G, green and IL-6 (O, orange)-associated CD11b positive leukocytes infiltration (R, red). (C) MPO expression (G, green) and Ly6G positive leukocytes infiltration (R, red). The arrow indicates strong inflammation with extensive colocalization of MPO and Ly6G staining. (D) Statistical summary of fluorescence staining (relative stained area or cell no. [%]) for some selected markers. The experimental animals were divided into 3 subgroups as described in the legend for Fig. 2A. The data are the mean±SEM (n = 3 for each group). * p < 0.05 compared with the corresponding S1+saline group by one-way ANOVA followed by the S-N-K t-test.
3.4. NRICM102 reduced S1-induced the expression of prothrombotic and fibrotic factors and apoptosis in the lungs
We examined whether NRICM102 treatment could reduce prothrombotic factor (vWF and PAI-1) expression and NET (CitH3) formation in S1-treated lung tissue and therefore suppress pulmonary embolism. Our results showed that vWF and PAI-1 protein were highly expressed within lung tissue treated with S1 ( Fig. 4A) and that strong NET (CitH3) formation and neutrophil infiltration occurred in association with S1 treatment (Fig. 4B and 4C). However, the levels of all these prothrombotic factors (vWF, PAI-1, NETs) were significantly reduced by NRICM102 treatment. Next, we examined whether AT1 and AT2 alveolar cells were lost in S1-treated animals through induction of apoptosis. Our results showed that S1 triggered strong apoptosis (indicated by the expression of cleaved caspase 3, cCasp3) all around lung tissues, especially within bronchi and alveoli composed of AT1 and AT2 cells (Fig. 4D). NRICM102 treatment significantly reduced apoptosis (cCasp3) and loss of AT1 and AT2 in S1-treated lung tissues. Finally, we examined whether the levels of fibrotic factors (c-Kit and stem cell factor (SCF)) were enhanced in S1-treated lung tissues. Our results showed that the fibrotic factors (c-Kit and SCF) were strongly expressed around bronchi in tissues treated with S1 and that their levels were significantly reduced by NRICM102 treatment (Fig. 4E). Statistical summary of positive fluorescence staining (relative stained area or cell no. [%]) for selected markers is shown in Fig. 4F. H&E staining showed that typical leukocyte infiltration into bronchioles, neutrophil (arrowhead, possess distinctive multi-lobulated nuclei) and monocyte (red arrow head, with large round or oval cell with an eccentrically placed nucleus) infiltration, and alveolar wall thickening with increased interstitial edema were clearly observed in the S1+saline group but not in the control (Ctrl) and S1+NRICM102 groups (Fig. 4G). Moreover, Masson's trichrome staining for tissue fibrosis detection also showed that the typical tissue fibrosis (blue) was clearly observed in the S1+saline group but not in the control group; however, the effect was attenuated in the S1+NRICM102 group (Fig. 4H).
Fig. 4.
Effects of NRICM102 on changes in levels of prothrombotic factor (vWF, PAI-1), NET, apoptosis and fibrotic factor (c-Kit and SCF) (at 72 hr) in S1 protein-induced lung injury in K18-hACE2 mice. Typical confocal images of lung tissue showing (A) vWF (G, green) and PAI-1 (R, red) colocalized with endothelial cell or platelet (CD31, PECAM-1) (O, orange). The arrow indicates extensive colocalization of CD31 and PAI-1. (B) S1 protein expression (O, orange) closely associated with NET formation (G, green) and leukocyte (CD11b) infiltration (R, red); the arrow indicates strong colocalization (yellow) of CD11b and NET staining (30X). (C) S1 expression (G, green) closely associated with NET formation (O, orange) and neutrophil (CD11b) infiltration (R, red); the arrow indicates strong colocalization of S1 and NET staining (100X). (D) Cleaved caspase 3 (cCasp3) (G, green) colocalized with AT1 cells (O, orange) or AT2 cells (R, red); the arrow indicates strong colocalization (yellow) of cCasp3 and AT2 or AT1. (E) c-Kit expression (G, green) colocalized with S1 protein accumulation (O, orange) and SCF (R, red); the arrow indicates strong colocalization (yellow) of S1 and c-Kit. (F) Statistical summary of positive fluorescence staining (relative stained area or cell no. counted [%]) for some selected markers. The experimental animals were divided into 3 subgroups as described in the legend for Fig. 2A. The data are the mean±SEM (n = 3 for each group). * p < 0.05 compared with the corresponding S1 +saline group by one-way ANOVA followed by the S-N-K t-test. (G) H&E staining of lung tissue. In the S1+saline group, typical leukocyte infiltration into bronchioles including neutrophils (black arrow head, possess distinctive multi-lobulated nuclei) and monocytes (red arrow head, with large round or oval cell with an eccentrically placed nucleus) infiltration, and thickened alveolar walls with interstitial edema (*) clearly observed, which less observed in the control (Ctrl) and S1+NRICM102 groups. (H) Masson's trichrome staining for typical tissue fibrosis (blue) clearly observed in the S1+saline and less observed in S1 +NRICM102.
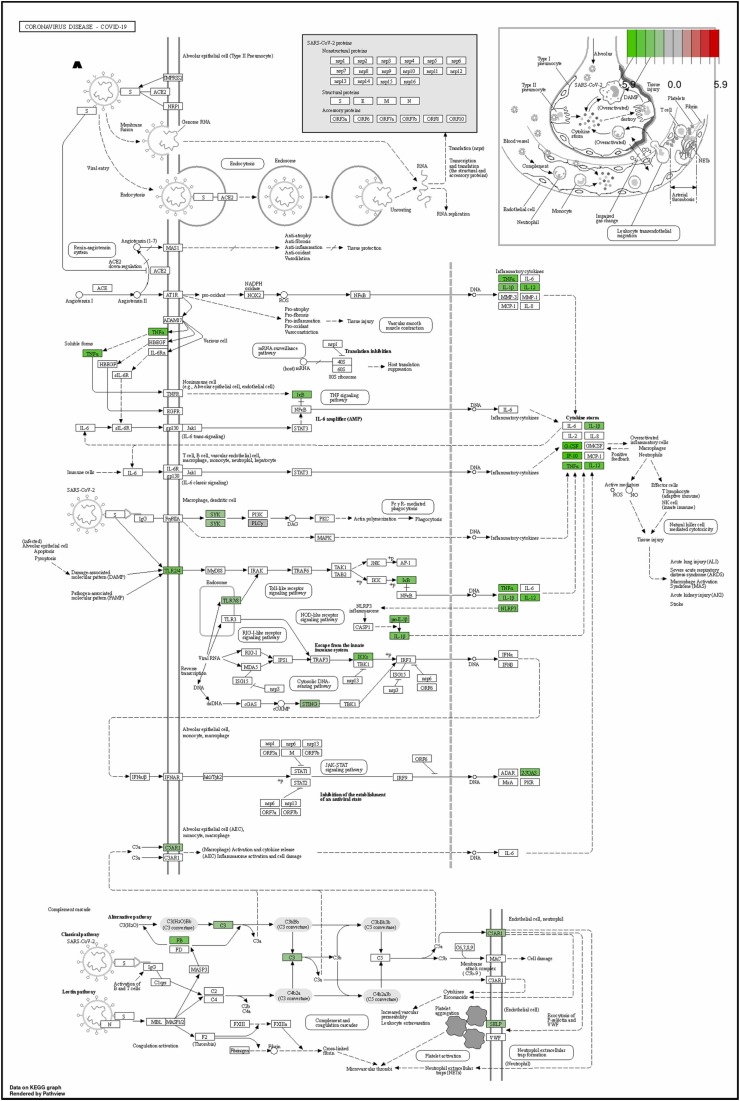
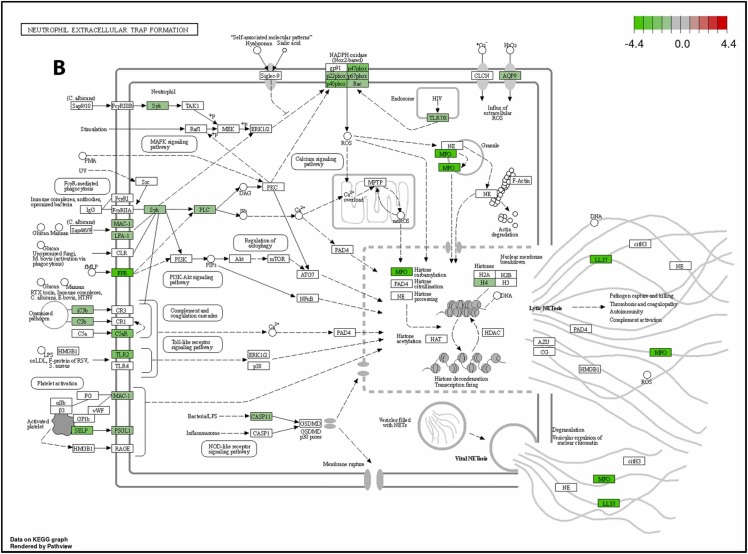
To further illustrate how TLR (Toll-like receptor) signaling mediated NET (neutrophil extracellular trap) formation and exacerbated lung inflammation and fibrosis in K18-hACE2 mice, the RNA-Seq profiles of mRNA expression in S1-stimulated K18-hACE2 mice was performed. We conducted Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of differentially expressed genes (DEGs) between the S1-stimulated group (S1+saline), S1+NRICM102, and sham group. KEGG analysis reveal that S1-stimulated group (S1+saline) displayed high levels of inflammatory markers associate with COVID-19 disease including TLR2/4, NOX2, TNFα, IL-1β, IL-12, G-CSF, IP-10, and NLRP3 ( Fig. 5A), and NET formation including TLR2, TLR7/8, MAC-1, LFA-1, FPR, FcγRIIIB, CASP11, PSGL1, MPO, and NOX2 (Fig. 5B). However, most of these COVID-19- and NET-associated inflammatory makers (genes) were significantly reduced in S1+NRICM102 group (Fig. 5). This result suggests that TLR signaling mediated NET formation and exacerbated lung inflammation and fibrosis through production of cytokine storm and activation of oxidative enzymes (NOX2 and MPO), and this TLR/NET signaling axis was targeted by NRICM102 to ameliorate pulmonary embolism and fibrosis.
Fig. 5.
The KEGG pathway analysis of NRICM102 on the RNA-Seq profiles of mRNA expression in S1-stimulated K18-hACE2 mice. (A) The inhibitory effect of NRICM102 on the KEGG Toll-like receptor signaling pathway mediating COVID-19 disorder. (B) The inhibitory effect of NRICM102 on the KEGG signaling pathway mediating NET formation. Green boxes indicate significantly downregulated expression in the transcriptomic profile.
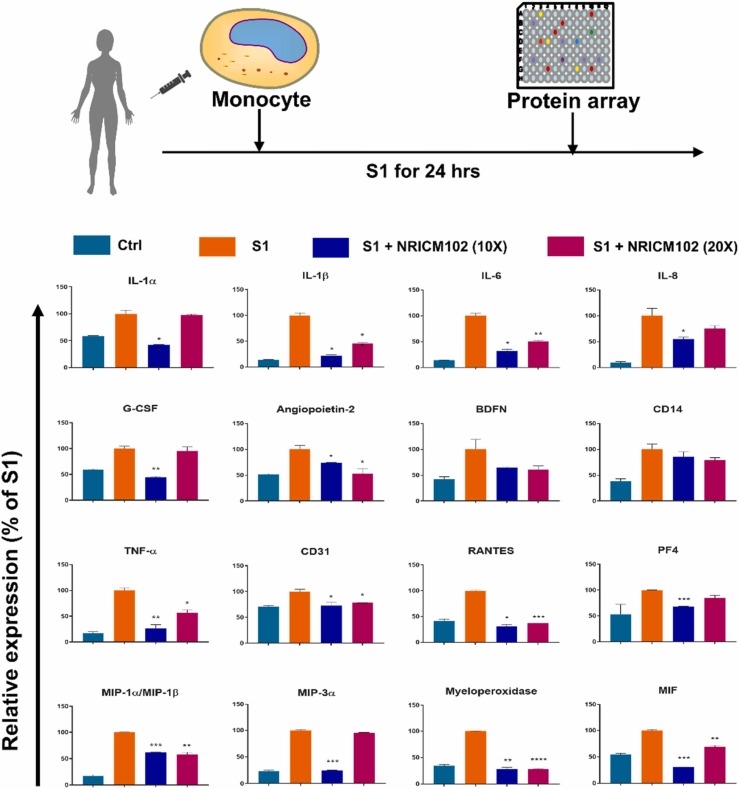
3.5. NRICM102 suppressed the S1-induced expression of multiple chemokines and cytokines in human monocytes
The results from the S1-induced pulmonary embolism animal model showed that NRICM102 suppressed the infiltration of neutrophils and monocytes in the bronchi and bronchioles. In addition, it suppressed strong inflammatory responses including IL-6 cytokine expression. It has been demonstrated that monocyte-driven cytokine storm and aberrant neutrophil activation are key mediators of COVID-19 disease severity [14]. In order to determine the inhibitory effect of NRICM102 on cytokine expression in monocytes, monocytes from human peripheral blood were stimulated with S1 and co-treated with NRICM102. Our results showed NRICM102 significantly suppressed S1-induced expression of various chemokines and cytokines, including TNF-α, CD31, RANTES, platelet factor 4 (PF4), IL-1α, IL-1β, IL-6, IL-8, macrophage inflammatory protein-1 alpha/beta (MIP-1α/β), MIP-3α, myeloperoxidase (MPO), macrophage migration inhibitory factor (MIF), granulocyte colony-stimulating factor (G-CSF), and angiopoietin-2, which are known factors related to the occurrence of cytokine storm ( Fig. 6).
Fig. 6.
Effects of NRICM102 on S1-induced expression of cytokines and chemokines in human monocytes. Ten- and twenty-fold dilutions of the decoction were used to investigate NRICM102’s inhibitory activity against cytokine and chemokines. The data are the mean±SEM. * p < 0.05, * * p < 0.01, * ** p < 0.005 compared with the corresponding S1 group by one-way ANOVA followed by the S-N-K t-test.
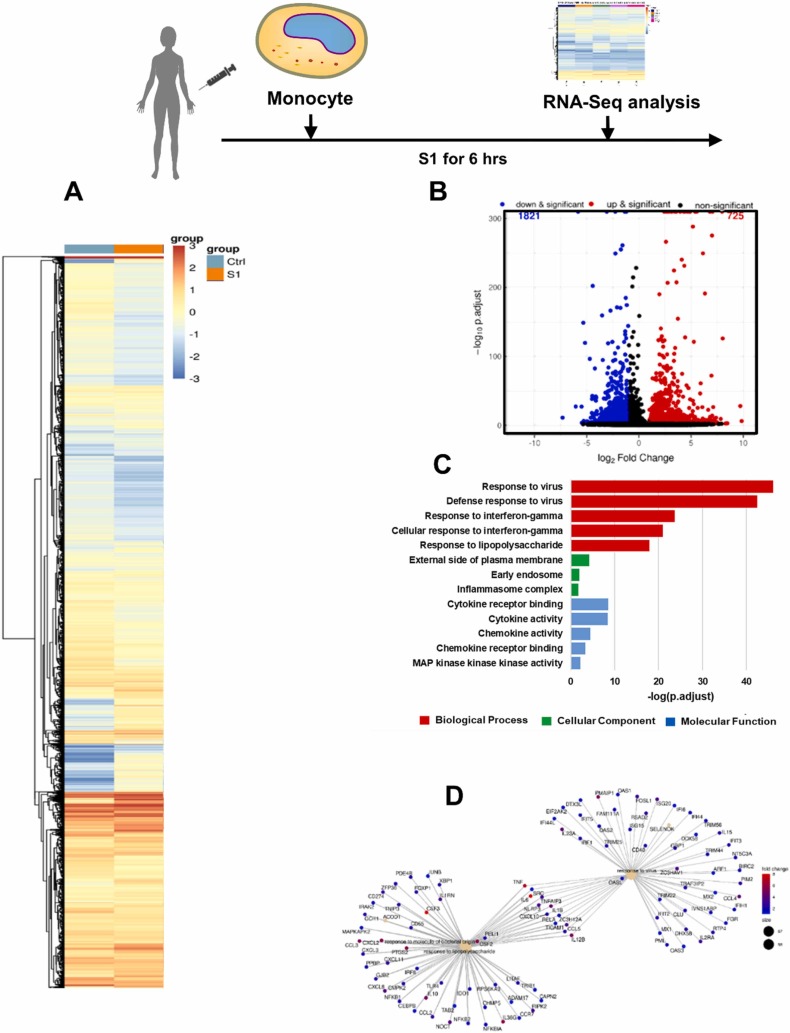
3.6. Activated phenotype of S1-stimulated monocytes determined by RNA-Seq
To examine transcriptional changes in monocytes in the context of SARS-CoV-2 pathogenesis, the RNA-Seq profiles of mRNA expression in S1-activated monocytes were analyzed. First, we identified differentially expressed genes (DEGs) in S1-stimulated monocytes compared to untreated control monocytes. Genes with an adjusted p value of less than 0.005 were considered to be differentially expressed. A total of 2546 genes exhibited altered expression in the S1-treated versus control cells. Among these DEGs, 725 (28%) genes were upregulated, and 1821 genes (72%) were downregulated in the S1-treated cells. The gene expression heatmap shows the up-and downregulated genes after S1 stimulation and indicates the distinct differences in gene expression between control and S1-treated cells ( Fig. 7A). The DEGs in S1-treated vs. control cells are also illustrated with a volcano plot (Fig. 7B). The 13 selected enriched upregulated GO terms are shown in Fig. 7C. The adjusted p values of the three upregulated GO terms were less than 0.005; thus, these GO terms were considered significantly enriched GO terms. The terms included the responses to viruses, defense response to virus, response to IFN-γ, cellular response to IFN-γ and response to lipopolysaccharide (LPS). These significantly upregulated GO terms were heavily associated with immune-related GO terms, indicating there was an increase in the immune response in the S1-treated cells. The category netplot (cnetplot) depicts the linkages of genes and biological concepts as a network. Our results showed that the genes upregulated by S1 stimulation were involved in signaling pathways associated with responses to molecules of bacterial origin, the response to LPS and the response to viruses (Fig. 7D).
Fig. 7.
Activated phenotype of S1-stimulated monocytes based on RNA-Seq. (A) Heatmap of DEGs between S1-stimulated and untreated monocytes. (B) Volcano plot for DEGs between S1-stimulated and untreated monocytes. (C) The 13 selected GO terms of DEGs upregulated in S1-stimulated monocytes vs. untreated monocytes. (D) Cnetplot of DEGs upregulated in S1-stimulated monocytes vs. untreated monocytes.
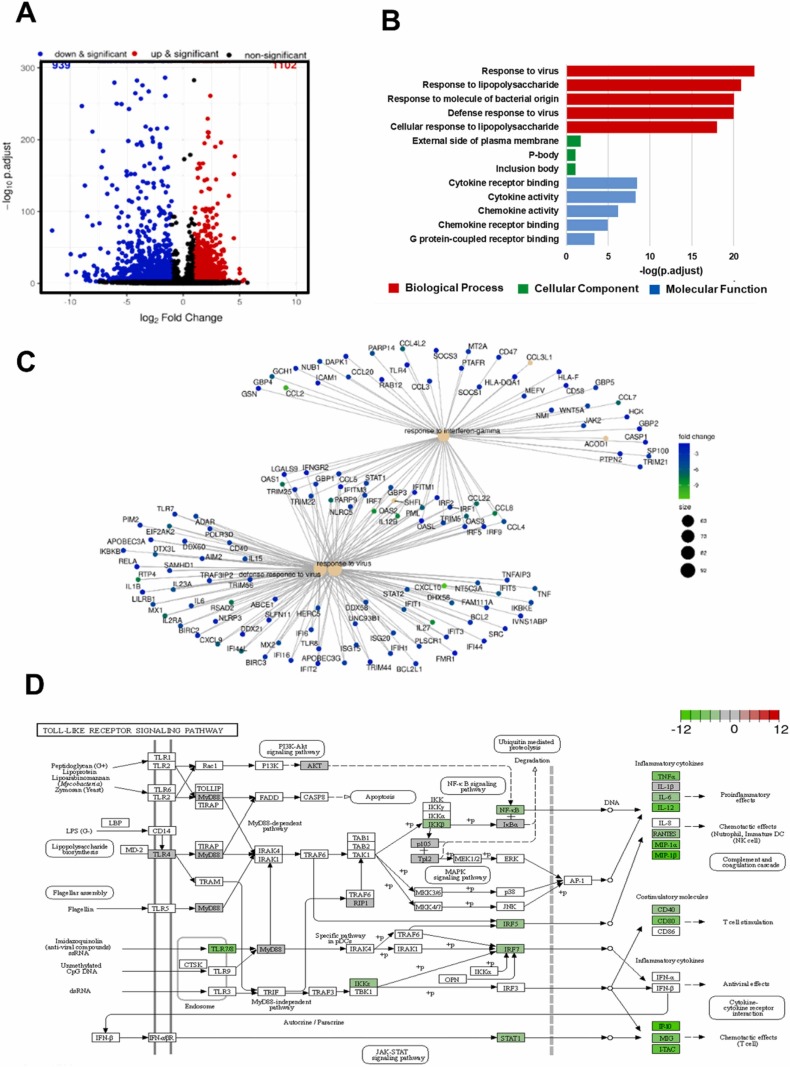
3.7. NRICM102 inhibits S1-stimulated effects on monocytes
We identified DEGs in S1 +NRICM102-treated monocytes compared to S1-treated monocytes. Genes with an adjusted p value of less than 0.005 were considered to be differentially expressed. A total of 2041 genes exhibited altered expression in the spike-treated versus control cells. Among these DEGs, 1102 (54%) genes were upregulated, and 939 genes (46%) were downregulated in the S1-treated cells. The volcano plot shows the upregulated and downregulated DEGs of the S1+NRICM102-treated and the spike-only cells ( Fig. 8A). The 13 selected enriched downregulated GO terms are shown in Fig. 8B. The adjusted p values of the three downregulated GO terms were less than 0.005; these GO terms were considered significantly enriched GO terms and included the responses to viruses and LPS, the response to molecule of bacterial origin, defense response to virus and cellular response to LPS (Fig. 8B). These significantly downregulated GO terms were heavily associated with immune-related GO terms, indicating a decrease in the immune response in the S1+NRICM102-treated cells. The cnetplot network of the DEGs showed that the genes downregulated by NRICM102 were involved in signaling pathways associated with the response to interferon-γ, the defense response to viruses and the response to viruses (Fig. 8C). The current research shows that the Toll-like receptor (TLRs) signaling pathway is one of the inflammatory signaling pathways that is dysregulated in patients with COVID-19. In addition, it has been reported that the SARS-CoV-2 spike protein induces inflammation via the TLR2 and TLR4 signaling pathways [42], [43]. To illustrate the role of NRICM102 in the Toll-like receptor signaling pathway in S1-stimulated monocytes, we conducted Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of DEGs in the S1+NRICM102 group compared with the S1-only group. The KEGG relationships of the Toll-like receptor signaling pathway is shown in Fig. 8D. NRICM102 suppressed the S1-activated expression of inflammatory cytokines, including TNF-α, IL-1β, IL-6, IL-12, RANTES, MIP-1α, MIP-1β, IP-10, MIG and interferon-inducible T-cell alpha chemoattractant (I-TAC), and costimulatory molecules, including CD40 and CD80, via regulation of PI3K/AKT, myeloid differentiation primary response 88 (MyD88)-dependent, MyD88-independent and JAK/STAT signaling pathways.
Fig. 8.
Effect of NRICM102 on the S1-activated phenotype of monocytes. (A) Volcano plot for DEGs between S1- and S1+NRICM102‐treated monocytes. (B) The 13 selected GO terms of DEGs downregulated in S1- and S1+NRICM102‐treated monocytes. (C) Cnetplot of DEGs downregulated in S1- and S1+NRICM102‐treated monocytes. (D) The inhibitory effect of NRICM102 on the KEGG Toll-like receptor signaling pathway. Green boxes indicate significantly downregulated expression in the transcriptomic profile.
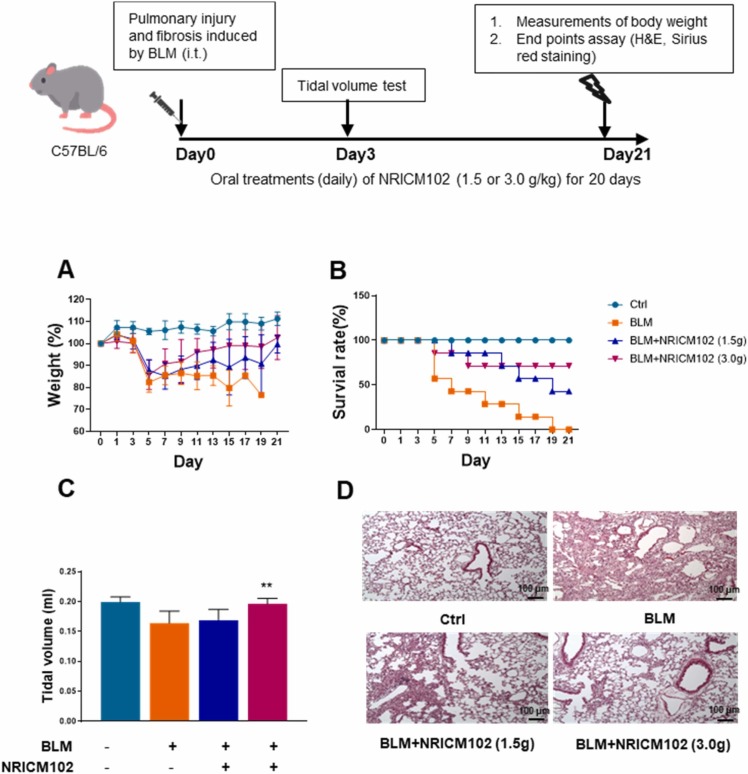
3.8. NRICM102 administration ameliorated bleomycin (BLM)-induced lung injury and respiratory function
To investigate the effect of NRICM102 on BLM-induced lung injury, mice were treated with NRICM102 for 20 days after exposure to BLM. A body weight reduction of 20% in mice was selected as the endpoint for humane sacrifice. The results showed that the weight loss occurred at day 5 after BLM treatment in the treated group compared to the control group. NRICM102 administration significantly ameliorated BLM-induced weight loss ( Fig. 9A). The survival rate of BLM-treated mice significantly declined from day 7 and reached zero at day 21 (Fig. 9B). Administration of 1.5 g and 3.0 g NRICM102 after BLM treatment for 20 consecutive days significantly improved the survival rate to 42.8% and 71.4%, respectively (Fig. 9B). The tidal volumes (TVs) of mice were measured 3 days after BLM treatment. BLM treatment reduced the tidal volumes of mice, and administration of NRICM102 (3.0 g) significantly attenuated the BLM-induced reduction in TV (Fig. 9C). Data from H&E staining confirmed that administration significantly ameliorated BLM-induced lung injury (highly inflamed and edema with collapse of alveoli) (Fig. 9D).
Fig. 9.
Effect of NRICM102 on BLM-induced lung injury in C57BL/6 mice. Body weights (A) and survival rates (B) of mice in the control (Ctrl), bleomycin (BLM), BLM+NRICM102 (1.5 g) and BLM+NRICM102 (3.0 g). (C) Tidal volumes of mice in the control (Ctrl), bleomycin (BLM), BLM+NRICM102 (1.5 g) and BLM+NRICM102 (3.0 g) groups (n = 7, ** p < 0.01 by one-way ANOVA followed by the S-N-K t-test.). (D) Hematoxylin and eosin (H&E)-stained images (×40 magnification) of the lungs.
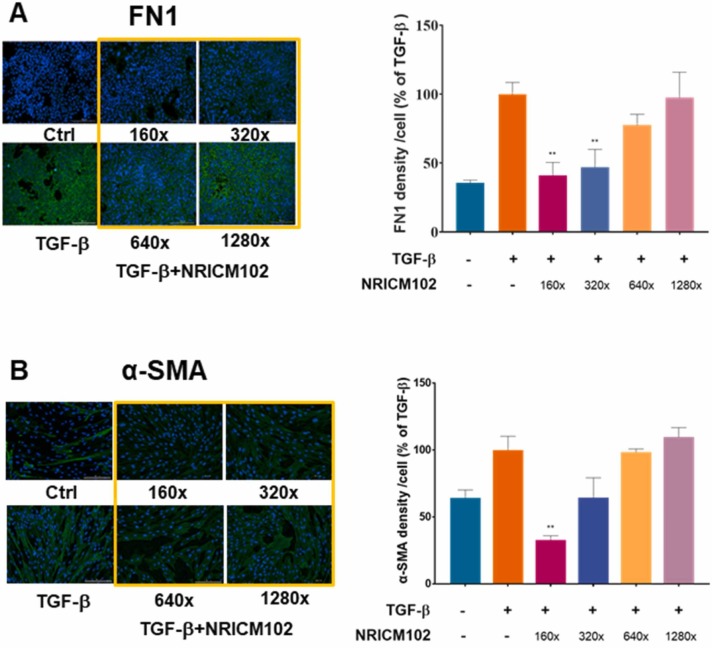
3.9. NRICM102 reduced TGF-β-induced epithelial–mesenchymal transition (EMT) and fibroblast to myofibroblast transformation (FMT)
EMT of bronchial epithelial cells and fibroblast to myofibroblast transformation (FMT) are the key process in the development of pulmonary fibrosis [44]. Epithelial cells which have undergone EMT which subsequently promotes the generation of FMT and fibrogenesis [44], [45], [46]. TGF-β has been reported to induce EMT and FMT which were characterized by the expression of fibronectin (FN1) and alpha smooth muscle actin (α-SMA), respectively [47], [48]. In order to evaluate the effect of NRICM102 on the EMT of human bronchial epithelial cells (BEAS-2B cells), the cells were treated with TGF-β or co-treated with TGF-β and NRICM102 for 3 days. In order to evaluate the effect of NRICM102 on the FMT of human fibroblast cells (HFL-1 cells), the cells were treated with TGF-β or co-treated with TGF-β and NRICM102 for 3 days. The results showed that TGF-β treatment significantly increased the expression of FN1 and α-SMA compared with that in the untreated cells. NRICM102 treatment significantly decreased the expression of FN1 and α-SMA in a dose-dependent manner compared with the expression in the TGF-β treated cells ( Fig. 10A and 10B). These results suggest that NRICM102 exhibited the capability to inhibit the TGF-β induced transition of BEAS-2B cells from bronchial epithelial cells to a mesenchymal-like phenotype and formation of HFL-1 cells from fibroblast to myofibroblast.
Fig. 10.
Effect of NRICM102 on TGF-β-induced EMT and FMT. (A) Representative immunofluorescence images of EMT of human bronchial epithelial cells (BEAS-2B cells). Green indicates the expression of fibronectin (FN1), and blue indicates DAPI staining. Ratio of the average FN1 fluorescence intensity per cell obtained after stimulation with TGF-β or TGF-β + NRICM102 at different concentrations. (B) Representative immunofluorescence images of FMT of human fibroblast cells (HFL-1 cells). Green indicates the expression of α-SMA, and blue indicates DAPI staining. Ratio of the average α-SMA fluorescence intensity per cell obtained after stimulation with TGF-β or TGF-β + NRICM102 at different concentrations. (** p < 0.01 by one-way ANOVA followed by the S-N-K t-test).
3.10. Identification of the isolated compounds and HPLC profiling
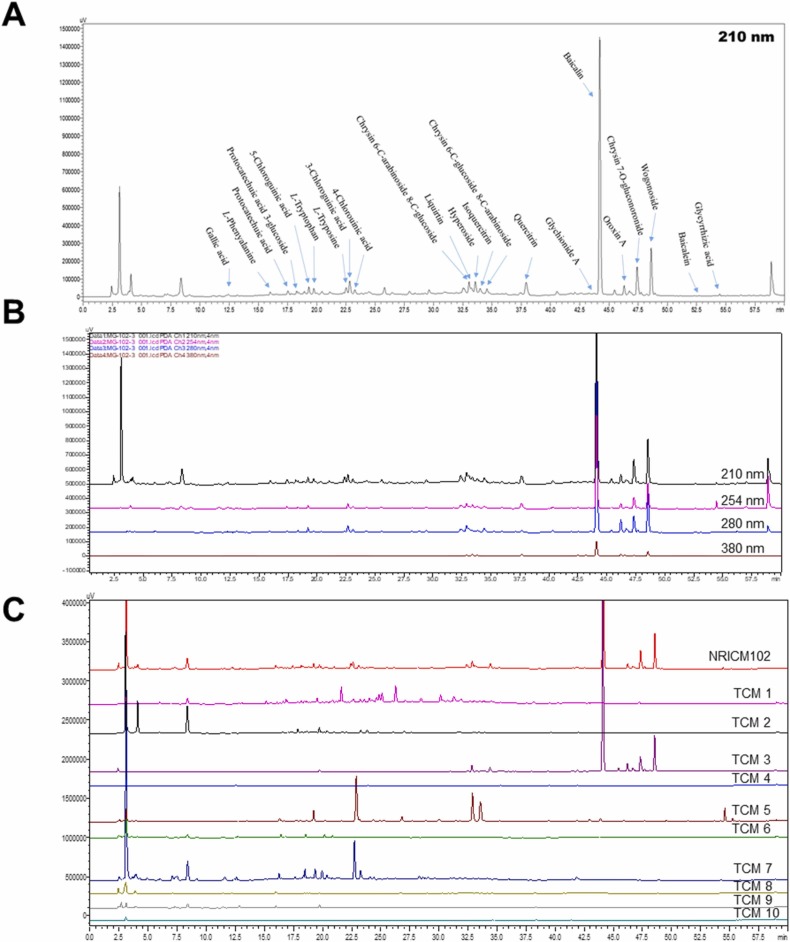
To identify the chemical compounds in the NRICM102 decoction, the HPLC fingerprint profiles of NRICM102 and its constituent TCMs were established by reverse-phase HPLC with diode-array detector (DAD) ( Fig. 11A-C). These samples were successfully analyzed. Briefly, the mobile phase consisted of water containing 0.3% phosphoric acid and ACN and was used with the following gradient program: 0.01–5.00 min, 0% ACN; 10.00 − 15.00 min, 3–10% ACN; 15.00 − 25.00 min, 10–15% ACN; 25 − 30 min, 15–20% ACN; 30 − 35 min, 20% ACN, 35 − 50 min, 20–40% ACN, 50–60 min, and 40–100% ACN. Separation was achieved on a Cosmosil 5 C18-AR-II column (5 µm, 250 mm × 4.6 mm i.d.) with a flow rate of 1.0 mL/min at 40 °C. As shown in Fig. 11A, twenty-two compounds were isolated from the NRICM102 decoction, including gallic acid (Rt: 12.45 min), L-phenylalanine (Rt: 16.11 min), protocatechuic acid (Rt: 17.59 min), protocatechuic acid 3-glucoside (Rt: 18.29 min), 5-caffeoylquinic acid (Rt: 19.34 min), L-tryptophan (Rt: 19.76 min), L-tyrosine (Rt: 22.53 min), 3-caffeoylquinic acid (Rt: 22.84 min), 4-caffeoylquinic acid (Rt: 23.27 min), chrysin 6-C-arabinoside-8-C-glucoside (Rt: 33.04 min), liquiritin (Rt: 33.23 min), quercetin 3-galactoside (Rt: 33.58 min), quercetin 3-glucoside (Rt: 33.98 min), chrysin 6-C-glucoside-8-C-arabinoside (Rt: 34.57 min), quercetin 3-rhamnoside (Rt: 37.93 min), oroxin A (Rt: 43.78 min), baicalin (Rt: 44.23 min), norwogonin 7-glucuronide (Rt: 46.31 min), oroxyloside (Rt: 47.42 min), wogonoside (Rt: 48.63 min), baicalein (Rt: 52.71 min), and glycyrrhizic acid (Rt: 54.48 min). These compounds were also recognized in the HPLC fingerprint profile of the NRICM102 decoction.
Fig. 11.
HPLC fingerprint profiles of the NRICM102 decoction and its single TCM extracts. (A) HPLC profiles of the NRICM102 decoction with the isolated compounds at a UV wavelength of 210 nm. (B) HPLC profiles of the NRICM102 decoction at 210, 254, 280, and 380 nm. (C) HPLC profiles of the 10 single TCMs at 210 nm. TCM1: Magnolia Bark (Magnolia officinalis), TCM2: Heartleaf Houttuynia Herb (Houttuynia cordata), TCM3: Scutellaria Root (Scutellaria baicalensis), TCM4: Prepared Monkshood Daughter Root (Aconitum carmichaelii), TCM5: Baked Licorice Root and Rhizome (Glycyrrhiza glabra), TCM6: Mongolian Snakegourd Fruit (Trichosanthes kirilowii), TCM7: Oriental Wormwood Herb (Artemisia scoparia), TCM8: Pinellia tuber (Pinellia ternata), TCM9: Fragrant Solomonseal Rhizome (Polygonatum odoratum), and TCM10: Indian Bread (Poria cocos).
4. Discussion
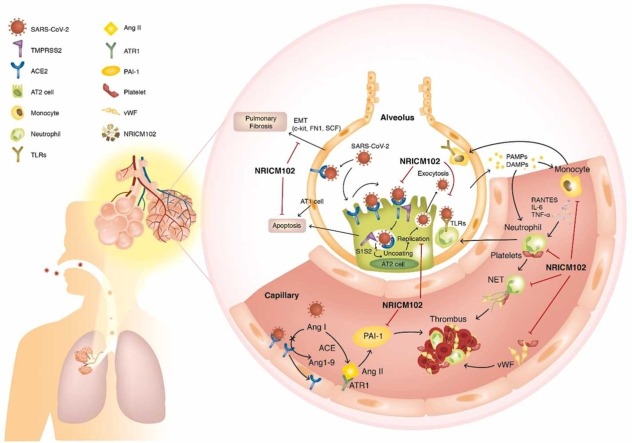
This research demonstrates promising effects of NRICM102 in ameliorating pulmonary embolism and injury. It provides scientific evidence to support the convincing clinical observations that this TCM formula improves pulmonary function and mortality among severe COVID-19 patients. The presented in vitro and in vivo data provide sound evidence of a compromised spike protein/ACE2 interaction and 3CL protease activity as well as reduced activation of monocytes and neutrophils and reduced expression of cytokines, chemokines and prothrombotic and fibrotic factors. These effects impede viral spread and progression to embolic and fibrotic pulmonary injury through specific signaling pathways ( Fig. 12).
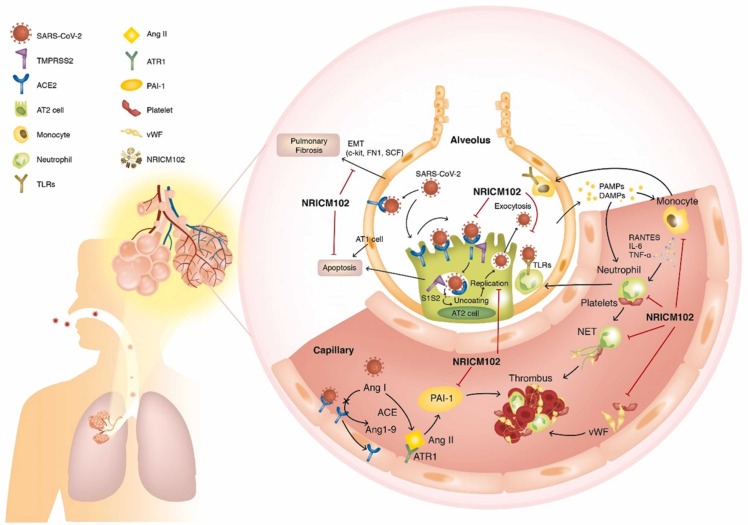
Fig. 12.
SARS-CoV-2 (S1) infection-induced alveolar type II (AT2) progenitor cell dysfunction and loss (apoptosis or necrosis) are deleterious to injured lungs for the following reasons: 1) decreases in surfactant lipids increase alveolar collapse and atelectasis; 2) decreases in AT2 cells cause impaired AT1 cell replacement, affecting alveolar repair and promoting fibrosis (EMT); 3) ACE2 downregulation by S1 drives overactivity of the ACE/Angiotensin II (Ang II)/AT1 receptor (ATR1) axis, exacerbating the tissue-destroying effects of the inflammatory responses and increasing production of plasminogen activator inhibitor − 1 (PAI-1), reducing plasmin activation and fibrinolysis; and 4) SARS-CoV-2 (S1) infection triggers AT1/AT2 and endothelial cells to release cytokines, which cause increased capillary permeability, enabling adhesion and extravasation of neutrophils and monocytes into the alveolar interstitial space. When stimulated by pathogen-associated and damage-associated molecular patterns (PAMPs and DAMPs), neutrophils and macrophages secrete massive amounts of cytokines, procoagulants, and complement, which induces further vascular injury, enhancing the risk for thrombosis. Therefore, several key factors contribute to the promotion of intravascular thrombus formation. 1) Neutrophil-mediated secretion of NETs (neutrophil extracellular traps) and vWF upregulation on cytokine-/virus-activated endothelial cells or macrophages, and lung residential megakaryocytes produce locally available platelets, which enhances platelet aggregation. 2) Cytokine-triggered secretion of tissue factor (TF) by endothelial cells and macrophages stimulates the coagulation cascade and increases fibrin clot formation. 3) Overactivation of the ACE (angiotensin-converting enzyme)/Ang II (angiotensin II)/AT1 receptor (ATR1) axis increases production of plasminogen activator inhibitor-1 (PAI-1), reducing plasmin activation and fibrinolysis. Here, NRICM102 displays multitargeted effects by preventing SARS-CoV-2 S1 entry; reducing AT1/AT2 cell apoptosis; downregulating inflammatory responses involving activation of neutrophils and monocytes, release of cytokines (IL-6, MCP-1) and expression of inflammatory receptors (TLR4); and reducing the production of prothrombotic factors (vWF and PAI-1) and fibrotic factors (c-Kit, FN1 and SCF) to ameliorate pulmonary embolism and fibrosis in hACE2 mice.
Oral Paxlovid (nirmatrelvir plus ritonavir) has been shown to result in a risk of progression to severe COVID-19 that was 89% lower than the risk with placebo among high-risk, non-hospitalized adults, without evident safety concerns [49]. Early administration of molnupiravir was found to reduce the risk of hospitalization or death in at-risk, unvaccinated adults with COVID-19 by 30% [19]. However, drug-drug interactions, side effects and various restrictions strongly limit the clinical application of these antiviral agents. A dire need exists for an effective treatment for hospitalized patients suffering from severe diseases (requiring oxygen supply) who are likely to present multiple organ dysfunction in this stage. NRICM102 is designed to treat severe-to-critical COVID-19 patients who suffer from acute respiratory distress syndrome, cardiac dysfunction or an exaggerated inflammatory response. This formula maintained the antiviral properties of NRICM101 while adding new components for the management of excessive immune responses (such as cytokine storm) and improvement of the cardiovascular function as a rescue therapy for patients at risk of experiencing organ failure. Lung injury can be ameliorated when excessive release of inflammatory mediators is suppressed by compounds in Scutellaria baicalensis, Houttuynia cordata and Poria cocos [50], [51], [52]. H. cordata and G. glabra might also contribute to this outcome by attenuating oxidative stress [51], [53]. Furthermore, G. glabra and Aconitum carmichaelii are known to stabilize cardiac function [54].
According to TCM’s principle of treating critically ill patients who have weak body constitution, special supportive therapy is strongly recommended. Compared with its predecessor NRICM101, which was used for mild cases, NRICM102 contains new ingredients (P. odoratum, P. cocos, and A. carmichaelii) that are particularly beneficial for severe and critical cases. In a previous study on NRICM101 [54], patients receiving intensive care were additionally given A. carmichaelii to warm the Yang and strengthen the heart, P. cocos to invigorate the spleen and remove dampness, and P. odoratum to nourish the Yin of the lungs in order to prevent cardiopulmonary failure. Further research is warranted to investigate the role of these herbs.
The mechanisms leading to the inflammatory and prothrombotic environment and finally to development of embolic and fibrotic pulmonary injury in patients with COVID-19 are very complicated (Fig. 12). In one mechanism, SARS-CoV-2 infection causes downregulation of ACE2, in turn causing upregulation of platelet activator inhibitor-1 (PAI-1) to elicit microvascular thrombosis. In another, SARS-CoV-2 infection of endothelial cells triggers the release of cytokines (e.g., IL-6) that cause increased capillary permeability, enabling adhesion and extravasation of neutrophils and monocytes, and promotes neutrophil extracellular trap (NET) formation and cytokine production. The NETs, in turn, increase coagulation by enhancing thrombin generation and platelet adhesion in the alveolar interstitial space. Therefore, it is very challenging to treat COVID-19 patients with pulmonary embolism using preferred clinical agents such as non-vitamin K antagonist oral anticoagulants (NOACs). Here, NRICM102 displayed multi-targeted effects by preventing SARS-CoV-2 S1-induced damage (AT1/AT2 cell loss); reducing inflammatory responses including neutrophil and monocyte activation, cytokine (TNF-α, IL-6, MCP-1) release and inflammatory receptors (TLR4/TLR2) expression; and reducing prothrombotic factor (vWF and PAI-1) levels to ameliorate embolic and fibrotic pulmonary injury in hACE2 mice. Based on our KEGG pathway analysis of DEGs in S1-induced human PBMC model, and S1-stimulated K18-hACE2 mice model, NRICM102 significantly suppressed cytokine storm via regulation of PI3K/AKT, MyD88-dependent, MyD88-independent and JAK/STAT signaling pathways, and mitigated the TLR mediated NET formation and exacerbated lung inflammation and fibrosis in K18-hACE2 mice, most possibly by downregulating COVID-19- and NET-associated inflammatory genes.
5. Conclusion
This study evaluated the contribution of the NRICM102 formula in response to the global pandemic. While the recent WHO expert meeting acknowledged the benefit of TCMs as add-on interventions to conventional treatment, it also called for scientific evaluation on the TCM for severe COVID-19 [20], [55]. Thus, this pharmacology study on NRICM102 is important in demonstrating significant therapeutic properties to reverse disease progress in patients experiencing embolic and fibrotic pulmonary injury, and also in clarifying why the formula works. Despite inherent limitations, the results from the current study justifies serious attention in the management of severe COVID-19.
Funding
Ministry of Health and Welfare of Republic of China grant (NRICM-110T88, MOHW110-NRICM-B-325-000400, MOHW111-NRICM-M-325-122400); Ministry of Science and Technology of Republic of China grant (MOST 108-2320-B-077-003-MY3, MOST 110-2320-B-077-003).
CRediT authorship contribution statement
Wen-Chi Wei: Conceptualization, Methodology, Investigation, Visualization, Project administration, Writing – original draft. Chia-Ching Liaw: Methodology, Investigation, Visualization, Writing – original draft. Keng-Chang Tsai: Methodology, Investigation, Visualization, Project administration, Writing – original draft. Chun-Tang Chiou: Methodology, Investigation, Visualization, Writing – original draft. Yu-Hwei Tseng: Investigation, Visualization, Writing – original draft. Wen-Fei Chiou: Investigation, Writing – review & editing. Yu-Chi Lin: Investigation. Chia-I Tsai: Investigation. Chen-Shien Lin: Investigation. Chen-Sung Lin: Investigation. Kuo-Tong Liou: Investigation, Writing – review & editing. I-Shing Yu: Investigation. Yuh-Chiang Shen: Conceptualization, Methodology, Investigation, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing. Yi-Chang Su: Conceptualization, Funding acquisition, Supervision, Visualization.
Competing interests
The authors have declared that no conflict of interest exists.
Acknowledgments
The authors thank the members of the authors’ laboratory in the NRICM, who assisted in and supported this study.
Data Availability
Data will be made available on request.
References
- 1.Del Valle D.M., Kim-Schulze S., Huang H.H., Beckmann N.D., Nirenberg S., Wang B., Lavin Y., Swartz T.H., Madduri D., Stock A., Marron T.U., Xie H., Patel M., Tuballes K., Van Oekelen O., Rahman A., Kovatch P., Aberg J.A., Schadt E., Jagannath S., Mazumdar M., Charney A.W., Firpo-Betancourt A., Mendu D.R., Jhang J., Reich D., Sigel K., Cordon-Cardo C., Feldmann M., Parekh S., Merad M., Gnjatic S. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020;26(10):1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poissy J., Goutay J., Caplan M., Parmentier E., Duburcq T., Lassalle F., Jeanpierre E., Rauch A., Labreuche J., Susen S. Lille ICUHC-G. pulmonary embolism in patients with COVID-19: awareness of an increased prevalence. Circulation. 2020;142(2):184–186. doi: 10.1161/CIRCULATIONAHA.120.047430. [DOI] [PubMed] [Google Scholar]
- 3.Xiong M., Liang X., Wei Y.D. Changes in blood coagulation in patients with severe coronavirus disease 2019 (COVID-19): a meta-analysis. Br. J. Haematol. 2020;189(6):1050–1052. doi: 10.1111/bjh.16725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wichmann D., Sperhake J.P., Lutgehetmann M., Steurer S., Edler C., Heinemann A., Heinrich F., Mushumba H., Kniep I., Schroder A.S., Burdelski C., de Heer G., Nierhaus A., Frings D., Pfefferle S., Becker H., Bredereke-Wiedling H., de Weerth A., Paschen H.R., Sheikhzadeh-Eggers S., Stang A., Schmiedel S., Bokemeyer C., Addo M.M., Aepfelbacher M., Puschel K., Kluge S. Autopsy findings and venous thromboembolism in patients with COVID-19: a prospective cohort study. Ann. Intern Med. 2020;173(4):268–277. doi: 10.7326/M20-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Middeldorp S., Coppens M., van Haaps T.F., Foppen M., Vlaar A.P., Muller M.C.A., Bouman C.C.S., Beenen L.F.M., Kootte R.S., Heijmans J., Smits L.P., Bonta P.I., van Es N. Incidence of venous thromboembolism in hospitalized patients with COVID-19. J. Thromb. Haemost. 2020;18(8):1995–2002. doi: 10.1111/jth.14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klok F.A., Kruip M., van der Meer N.J.M., Arbous M.S., Gommers D., Kant K.M., Kaptein F.H.J., van Paassen J., Stals M.A.M., Huisman M.V., Endeman H. Confirmation of the high cumulative incidence of thrombotic complications in critically ill ICU patients with COVID-19: An updated analysis. Thromb. Res. 2020;191:148–150. doi: 10.1016/j.thromres.2020.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berlin D.A., Gulick R.M., Martinez F.J. Severe Covid-19. New Engl. J. Med. 2020;383(25):2451–2460. doi: 10.1056/NEJMcp2009575. [DOI] [PubMed] [Google Scholar]
- 8.Wu Z., McGoogan J.M. Characteristics of and important lessons from the Coronavirus Disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the chinese center for disease control and prevention. JAMA. 2020;323(13):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 9.Brosnahan S.B., Jonkman A.H., Kugler M.C., Munger J.S., Kaufman D.A. COVID-19 and respiratory system disorders: current knowledge, future clinical and translational research questions. Arterioscler. Thromb. Vasc. Biol. 2020;40(11):2586–2597. doi: 10.1161/ATVBAHA.120.314515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barnes B.J., Adrover J.M., Baxter-Stoltzfus A., Borczuk A., Cools-Lartigue J., Crawford J.M., Dassler-Plenker J., Guerci P., Huynh C., Knight J.S., Loda M., Looney M.R., McAllister F., Rayes R., Renaud S., Rousseau S., Salvatore S., Schwartz R.E., Spicer J.D., Yost C.C., Weber A., Zuo Y., Egeblad M. Targeting potential drivers of COVID-19: neutrophil extracellular traps. J. Exp. Med. 2020;217:6. doi: 10.1084/jem.20200652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zuo Y., Yalavarthi S., Shi H., Gockman K., Zuo M., Madison J.A., Blair C., Weber A., Barnes B.J., Egeblad M., Woods R.J., Kanthi Y., Knight J.S. Neutrophil extracellular traps in COVID-19. JCI Insight. 2020;5:11. doi: 10.1172/jci.insight.138999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hui K.P.Y., Cheung M.C., Perera R., Ng K.C., Bui C.H.T., Ho J.C.W., Ng M.M.T., Kuok D.I.T., Shih K.C., Tsao S.W., Poon L.L.M., Peiris M., Nicholls J.M., Chan M.C.W. Tropism, replication competence, and innate immune responses of the coronavirus SARS-CoV-2 in human respiratory tract and conjunctiva: an analysis in ex-vivo and in-vitro cultures. Lancet Respir. Med. 2020;8(7):687–695. doi: 10.1016/S2213-2600(20)30193-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong M., Liang X., Wei Y.D. Changes in blood coagulation in patients with severe coronavirus disease 2019 (COVID‐19): a meta‐analysis. Br. J. Haematol. 2020 doi: 10.1111/bjh.16725. 0-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vanderbeke L., Van Mol P., Van Herck Y., De Smet F., Humblet-Baron S., Martinod K., Antoranz A., Arijs I., Boeckx B., Bosisio F.M., Casaer M., Dauwe D., De Wever W., Dooms C., Dreesen E., Emmaneel A., Filtjens J., Gouwy M., Gunst J., Hermans G., Jansen S., Lagrou K., Liston A., Lorent N., Meersseman P., Mercier T., Neyts J., Odent J., Panovska D., Penttila P.A., Pollet E., Proost P., Qian J., Quintelier K., Raes J., Rex S., Saeys Y., Sprooten J., Tejpar S., Testelmans D., Thevissen K., Van Buyten T., Vandenhaute J., Van Gassen S., Velasquez Pereira L.C., Vos R., Weynand B., Wilmer A., Yserbyt J., Garg A.D., Matthys P., Wouters C., Lambrechts D., Wauters E., Wauters J. Monocyte-driven atypical cytokine storm and aberrant neutrophil activation as key mediators of COVID-19 disease severity. Nat. Commun. 2021;12(1):4117. doi: 10.1038/s41467-021-24360-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang G., Yang M.L., Duan Z.L., Liu F.L., Jin L., Long C.B., Zhang M., Tang X.P., Xu L., Li Y.C., Kamau P.M., Yang L., Liu H.Q., Xu J.W., Chen J.K., Zheng Y.T., Peng X.Z., Lai R. Dalbavancin binds ACE2 to block its interaction with SARS-CoV-2 spike protein and is effective in inhibiting SARS-CoV-2 infection in animal models. Cell Res. 2021;31(1):17–24. doi: 10.1038/s41422-020-00450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin C., Li Y., Zhang Y., Liu Z., Mu X., Gu C., Liu J., Li Y., Li G., Chen J. Ceftazidime is a potential drug to inhibit SARS-CoV-2 infection in vitro by blocking spike protein-ACE2 interaction. Signal Transduct. Target Ther. 2021;6(1):198. doi: 10.1038/s41392-021-00619-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamb Y.N. Nirmatrelvir plus ritonavir: first approval. Drugs. 2022:1–7. doi: 10.1007/s40265-022-01692-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., Lopez de Castilla D., Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R., Touloumi G., Lye D.C., Ohmagari N., Oh M.D., Ruiz-Palacios G.M., Benfield T., Fatkenheuer G., Kortepeter M.G., Atmar R.L., Creech C.B., Lundgren J., Babiker A.G., Pett S., Neaton J.D., Burgess T.H., Bonnett T., Green M., Makowski M., Osinusi A., Nayak S., Lane H.C. Members A-SG. Remdesivir for the treatment of Covid-19 - final report. N. Engl. J. Med. 2020;383(19):1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jayk Bernal A., Gomes da Silva M.M., Musungaie D.B., Kovalchuk E., Gonzalez A., Delos Reyes V., Martin-Quiros A., Caraco Y., Williams-Diaz A., Brown M.L., Du J., Pedley A., Assaid C., Strizki J., Grobler J.A., Shamsuddin H.H., Tipping R., Wan H., Paschke A., Butterton J.R., Johnson M.G., De Anda C. MO-OS. Molnupiravir for oral treatment of Covid-19 in nonhospitalized patients. New Engl. J. Med. 2022;386(6):509–520. doi: 10.1056/NEJMoa2116044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.WHO. WHO Expert Meeting on Evaluation of Traditional Chinese Medicine in the Treatment of COVID-19. In.; 2022. p. 17.
- 21.Tsai K.C., Huang Y.C., Liaw C.C., Tsai C.I., Chiou C.T., Lin C.J., Wei W.C., Lin S.J., Tseng Y.H., Yeh K.M., Lin Y.L., Jan J.T., Liang J.J., Liao C.C., Chiou W.F., Kuo Y.H., Lee S.M., Lee M.Y., Su Y.C. A traditional Chinese medicine formula NRICM101 to target COVID-19 through multiple pathways: a bedside-to-bench study. Biomed. Pharm. 2021;133 doi: 10.1016/j.biopha.2020.111037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tseng Y.H., Lin S.H.S., Hou S.M., Wang C.H., Cheng S.P., Tseng K.Y., et al. Curbing COVID-19 progression and mortality with traditional Chinese medicine among hospitalized patients with COVID-19: a propensity score-matched analysis. Pharmacol. Res. 2022;184 doi: 10.1016/j.phrs.2022.106412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colunga Biancatelli R.M.L., Solopov P.A., Sharlow E.R., Lazo J.S., Marik P.E., Catravas J.D. The SARS-CoV-2 spike protein subunit S1 induces COVID-19-like acute lung injury in Kappa18-hACE2 transgenic mice and barrier dysfunction in human endothelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 2021;321(2):L477–L484. doi: 10.1152/ajplung.00223.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banno F., Kita T., Fernandez J.A., Yanamoto H., Tashima Y., Kokame K., Griffin J.H., Miyata T. Exacerbated venous thromboembolism in mice carrying a protein S K196E mutation. Blood. 2015;126(19):2247–2253. doi: 10.1182/blood-2015-06-653162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brandt M., Giokoglu E., Garlapati V., Bochenek M.L., Molitor M., Hobohm L., Schonfelder T., Munzel T., Kossmann S., Karbach S.H., Schafer K., Wenzel P. Pulmonary arterial hypertension and endothelial dysfunction is linked to NADPH oxidase-derived superoxide formation in venous thrombosis and pulmonary embolism in mice. Oxid. Med. Cell Longev. 2018;2018 doi: 10.1155/2018/1860513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen H., Liu D., Ge L., Wang T., Ma Z., Han Y., Duan Y., Xu X., Liu W., Yuan J., Liu J., Li R., Du R. Catestatin prevents endothelial inflammation and promotes thrombus resolution in acute pulmonary embolism in mice. Biosci. Rep. 2019;39:11. doi: 10.1042/BSR20192236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williamson J.D., Sadofsky L.R., Hart S.P. The pathogenesis of bleomycin-induced lung injury in animals and its applicability to human idiopathic pulmonary fibrosis. Exp. Lung Res. 2015;41(2):57–73. doi: 10.3109/01902148.2014.979516. [DOI] [PubMed] [Google Scholar]
- 28.Izbicki G., Segel M.J., Christensen T.G., Conner M.W., Breuer R. Time course of bleomycin-induced lung fibrosis. Int J. Exp. Pathol. 2002;83(3):111–119. doi: 10.1046/j.1365-2613.2002.00220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milton P.L., Dickinson H., Jenkin G., Lim R. Assessment of respiratory physiology of C57BL/6 mice following bleomycin administration using barometric plethysmography. Respiration. 2012;83(3):253–266. doi: 10.1159/000330586. [DOI] [PubMed] [Google Scholar]
- 30.Van De Vlekkert D., Machado E., d'Azzo A. Analysis of generalized fibrosis in mouse tissue sections with Masson's trichrome staining. Bio Protoc. 2020;10(10) doi: 10.21769/BioProtoc.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei W.C., Liu C.P., Yang W.C., Shyur L.F., Sheu J.H., Chen S.S., Yang N.S. Mammalian target of rapamycin complex 2 (mTORC2) regulates LPS-induced expression of IL-12 and IL-23 in human dendritic cells. J. Leukoc. Biol. 2015;97(6):1071–1080. doi: 10.1189/jlb.2A0414-206RR. [DOI] [PubMed] [Google Scholar]
- 32.Ewels P., Magnusson M., Lundin S., Kaller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32(19):3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sahraeian S.M.E., Mohiyuddin M., Sebra R., Tilgner H., Afshar P.T., Au K.F., Bani Asadi N., Gerstein M.B., Wong W.H., Snyder M.P., Schadt E., Lam H.Y.K. Gaining comprehensive biological insight into the transcriptome by performing a broad-spectrum RNA-seq analysis. Nat. Commun. 2017;8(1):59. doi: 10.1038/s41467-017-00050-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang L., Feng Z., Wang X., Wang X., Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26(1):136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 37.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schurch N.J., Schofield P., Gierlinski M., Cole C., Sherstnev A., Singh V., Wrobel N., Gharbi K., Simpson G.G., Owen-Hughes T., Blaxter M., Barton G.J. How many biological replicates are needed in an RNA-seq experiment and which differential expression tool should you use? RNA. 2016;22(6):839–851. doi: 10.1261/rna.053959.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., Katayama T., Kawashima S., Okuda S., Tokimatsu T., Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36(Database Issue) 2008:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanehisa M., Sato Y., Furumichi M., Morishima K., Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019;47(D1):D590–D595. doi: 10.1093/nar/gky962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao Y., Kuang M., Li J., Zhu L., Jia Z., Guo X., Hu Y., Kong J., Yin H., Wang X., You F. Publisher correction: SARS-CoV-2 spike protein interacts with and activates TLR4. Cell Res. 2021;31(7):825. doi: 10.1038/s41422-021-00501-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khan S., Shafiei M.S., Longoria C., Schoggins J.W., Savani R.C., Zaki H. SARS-CoV-2 spike protein induces inflammation via TLR2-dependent activation of the NF-kappaB pathway. Elife. 2021:10. doi: 10.7554/eLife.68563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weigle S., Martin E., Voegtle A., Wahl B., Schuler M. Primary cell-based phenotypic assays to pharmacologically and genetically study fibrotic diseases in vitro. J. Biol. Methods. 2019;6(2) doi: 10.14440/jbm.2019.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pakshir P., Hinz B. The big five in fibrosis: macrophages, myofibroblasts, matrix, mechanics, and miscommunication. Matrix Biol. 2018;68–69:81–93. doi: 10.1016/j.matbio.2018.01.019. [DOI] [PubMed] [Google Scholar]
- 46.Grande M.T., Sanchez-Laorden B., Lopez-Blau C., De Frutos C.A., Boutet A., Arevalo M., Rowe R.G., Weiss S.J., Lopez-Novoa J.M., Nieto M.A. Snail1-induced partial epithelial-to-mesenchymal transition drives renal fibrosis in mice and can be targeted to reverse established disease. Nat. Med. 2015;21(9):989–997. doi: 10.1038/nm.3901. [DOI] [PubMed] [Google Scholar]
- 47.Mattey D.L., Dawes P.T., Nixon N.B., Slater H. Transforming growth factor beta 1 and interleukin 4 induced alpha smooth muscle actin expression and myofibroblast-like differentiation in human synovial fibroblasts in vitro: modulation by basic fibroblast growth factor. Ann. Rheum. Dis. 1997;56(7):426–431. doi: 10.1136/ard.56.7.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanjore H., Xu X.C., Polosukhin V.V., Degryse A.L., Li B., Han W., Sherrill T.P., Plieth D., Neilson E.G., Blackwell T.S., Lawson W.E. Contribution of epithelial-derived fibroblasts to bleomycin-induced lung fibrosis. Am. J. Respir. Crit. Care Med. 2009;180(7):657–665. doi: 10.1164/rccm.200903-0322OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hammond J., Leister-Tebbe H., Gardner A., Abreu P., Bao W., Wisemandle W., Baniecki M., Hendrick V.M., Damle B., Simon-Campos A., Pypstra R., Rusnak J.M., Investigators E.-H. Oral nirmatrelvir for high-risk, nonhospitalized adults with Covid-19. New Engl. J. Med. 2022;386(15):1397–1408. doi: 10.1056/NEJMoa2118542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liao H., Ye J., Gao L., Liu Y. The main bioactive compounds of Scutellaria baicalensis Georgi. for alleviation of inflammatory cytokines: a comprehensive review. Biomed. Pharmacother. 2021;133 doi: 10.1016/j.biopha.2020.110917. [DOI] [PubMed] [Google Scholar]
- 51.Shingnaisui K., Dey T., Manna P., Kalita J. Therapeutic potentials of Houttuynia cordata Thunb. against inflammation and oxidative stress: a review. J. Ethnopharmacol. 2018;220:35–43. doi: 10.1016/j.jep.2018.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tian H., Liu Z., Pu Y., Bao Y. Immunomodulatory effects exerted by Poria Cocos polysaccharides via TLR4/TRAF6/NF-κB signaling in vitro and in vivo. Biomed. Pharmacother. 2019;112 doi: 10.1016/j.biopha.2019.108709. [DOI] [PubMed] [Google Scholar]
- 53.El-Saber Batiha G., Magdy Beshbishy A., El-Mleeh A., Abdel-Daim M.M., Prasad Devkota H. Traditional uses, bioactive chemical constituents, and pharmacological and toxicological activities of glycyrrhiza glabra L. (Fabaceae) Biomolecules. 2020;10(3):352. doi: 10.3390/biom10030352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsai K.-C., Huang Y.-C., Liaw C.-C., Tsai C.-I., Chiou C.-T., Lin C.-J., Wei W.-C., Lin S.J.-S., Tseng Y.-H., Yeh K.-M. A traditional Chinese medicine formula NRICM101 to target COVID-19 through multiple pathways: a bedside-to-bench study. Biomed. Pharmacother. 2021;133 doi: 10.1016/j.biopha.2020.111037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tseng Y.H., Chang F.R. Bringing scientific methods to traditional medicine. J. Formos. Med. Assoc. 2019;118(12):1574–1575. doi: 10.1016/j.jfma.2019.08.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.