Abstract
Conserved motif C, identified within members of the major facilitator superfamily (MFS) of transport proteins that mediate drug export, was examined in the tetracycline resistance efflux protein TetA(K) from Staphylococcus aureus; motif C is contained within transmembrane segment 5. Using site-directed mutagenesis, the importance of the conserved glycine (G151, G155, G159, and G160) and proline (P156) residues within this motif was investigated. Over 40 individual amino acid replacements were introduced; however, only alanine and serine substitutions for glycine at G151, G155, and G160 were found to retain significant levels of tetracycline resistance and transport activity in cells expressing mutant proteins. Notably, P156 and G159 appear to be crucial, as amino acid replacements at these positions either significantly reduced or abolished tetracycline/H+ activity. The highly conserved nature of motif C and its distribution throughout drug exporters imply that the residues of motif C play a similar role in all MFS proteins that function as antiporters.
The major facilitator superfamily (MFS), also known as the uniporter-symporter-antiporter family, comprises over 500 membrane-bound transport proteins which are present in all classes of living organisms (21, 26). These proteins perform a wide variety of cellular processes, including the uptake of essential ions and nutrients and the removal of toxic compounds, and typically utilize the proton motive force to drive the transport process (10). Seventeen families can be identified within the MFS based on sequence homology, where a correlation exists between each phylogenetic family and the class of compound transported (26). Despite conveying substrates vastly different in structure, MFS proteins are alike in their predicted membrane topologies. Fourteen out of the 17 families comprise proteins which have been shown or are predicted to possess 12 transmembrane-spanning segments (TMS), and the remaining three MFS families contain proteins with 14 TMS (26). The two families consisting of drug efflux systems, DHA12 and DHA14, possess 12 and 14 TMS, respectively. These families include the well-characterized Staphylococcus aureus multidrug efflux protein QacA (24, 25, 28) and the tetracycline export proteins TetA(B) (22) and TetA(K) (9, 12), from Escherichia coli and S. aureus, respectively, which in comparison to QacA display more limited substrate specificities.
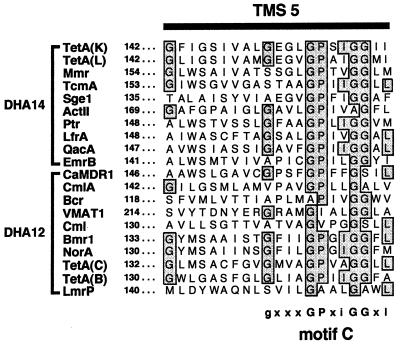
Transport proteins of the MFS contain a number of conserved amino acid sequence motifs which are either ubiquitous within the MFS or family specific (21, 28). Conservation of such motifs among proteins responsible for the transport of a wide variety of structurally disparate compounds implies that they play some vital structural or functional role rather than directly interacting with their substrate(s) (21). Of these, motif C, also known as the antiporter motif (33), is common to all members of the DHA12 and DHA14 families (29). This motif is positioned in TMS 5 of drug export proteins and is typified by the amino acid sequence gxxxGPxiGGxl (28, 30), where x represents any amino acid, residues in uppercase are conserved in more than 90% of export proteins, and residues in lowercase occur at a frequency of at least 40% (see Fig. 1 below).
FIG. 1.
Multiple-amino-acid-sequence alignment of TMS 5 from representative members of the MFS DHA14 and DHA12 drug efflux families (sequence names are shown on the left). GenBank (GB), Swiss-Prot (SP), or EMBL (EM) accession numbers for each protein are as follows: TetA(K), EM M16217; TetA(L), SP P07561; Mmr, GB M18263; TcmA, GB M80674; Sge1, SP P33335; ActII, GB M64683; Ptr, GB X84072; LfrA, GB U40487; QacA, EM X56628; EmrB, SP P27304; CaMDR1, SP P28873; CmlA, SP 32482; Bcr, SP P28246; VMAT1, GB M97380; Cml, SP P31141; Bmr1, SP P33449; NorA, SP P21191; TetA(C), EM J01749; TetA(B), EM J01830; LmrP, GB X89779. Numbers on the left refer to the position of the leftmost amino acid residue for each protein. Shaded and boxed residues represent amino acids that are conserved in at least 40% of proteins. The highly conserved motif C consensus sequence is displayed below the alignment, where x represents any amino acid, residues in uppercase are conserved in more than 90% of export proteins, and residues in lowercase occur at a frequency of at least 40%.
The S. aureus tetracycline resistance efflux protein TetA(K) is comprised of 14 TMS (9) and has been shown to function as a metal-tetracycline/H+ antiporter in both everted membrane vesicles (38) and proteoliposomes (J. Cheng, A. A. Guffanti, and T. A. Krulwich, Abstr. 1997 Meet. Microb. Pathog. Host Response, p. 144, 1997). Additionally, TetA(K) can mediate net potassium (K+) ion uptake (14, 15) in a manner analogous to that of TetA(C), encoded by the E. coli plasmid pBR322 (7). However, the highly related TetA(B) protein, encoded by the transposon Tn10, is unable to transport K+, thereby demonstrating that the function is not common to tetracycline resistance determinants (7).
The present study examined the role of the proline residue and four glycine residues of motif C that are conserved among members of the DHA12 and DHA14 families of the MFS (see Fig. 1). Using site-directed mutagenesis, these residues were altered to a number of different amino acids, including conservative and nonconservative amino acid replacements, and the ability of mutant proteins to transport tetracycline was determined in everted membrane vesicles. Hybrid TetA(K) proteins in which TMS 5 was replaced by the corresponding helix from the tetracycline efflux protein TetA(B) or the multidrug exporter QacA, members of the DHA12 and DHA14 MFS families, respectively, were constructed and analyzed. Taken together, these studies indicate that all conserved residues of motif C are essential for TetA(K)-mediated tetracycline transport.
MATERIALS AND METHODS
Bacterial strains, bacteriophages, plasmids, and growth media.
The E. coli strains used were BL21(DE3)/pLysS [F− ompT hsdSB(rB− mB−) dcm gal λ(DE3) pLysS (Cmr)] (32), CJ236 [F′ cat (=pCJ105; M13S) dut-1 ung-1 thi-1 relA1 pJC105 (Cmr)] (18), DH5α [supE44 ΔlacU169 (φ80dlacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1] (16), TG1 [F′ (traD36 proAB+ lacIq lacZΔM15) supE hsdΔ5 thi Δ(lac-proAB)] (31), and TK2205 (F− thi lacZ rha nagA kdpABC5 trkA405 trkD1) (7). Plasmids used were pBluescript II SK (Stratagene), pBR322 (22), pNK81 (36), pSK4236 (27), and pSK4646 (this study). The bacteriophage M13K07 (34) was employed in the creation of site-directed mutants. E. coli strains were grown in Luria-Bertani medium unless otherwise stated. Antibiotics for plasmid selection were used at the following concentrations: ampicillin, 100 μg/ml; tetracycline, 10 μg/ml; and chloramphenicol, 25 μg/ml.
Measurement of bacterial growth in the presence of tetracyclines.
Inhibitory concentrations (IC) of antibiotics were determined by an agar plate method. Duplicate Luria-Bertani agar plates containing the desired concentrations of tetracycline or the tetracycline analogs chlortetracycline, doxycycline, or oxytetracycline were inoculated with approximately 104 CFU of a stationary-phase E. coli strain TG1 culture, harboring the plasmid to be tested, using a multipoint replicator. Plates were incubated at 37°C for 24 h. Each test was performed in triplicate to ensure reproducibility of the data. The IC was defined as the lowest concentration of antibiotic (in micrograms per milliliter) which inhibited bacterial growth.
Potassium uptake complementation by TetA(K) and derivatives.
To test the ability of TetA(K) and various TetA(K) mutants to phenotypically complement the K+ uptake defect of E. coli strain TK2205, plasmids carrying a wild-type or mutant tetA(K) gene were transformed into this strain. Minimal medium plates (37) supplemented with K+ at various concentrations (0.5, 1, 1.5, 2, 2.5, 3, 4, 5, and 10 mM) were inoculated with the strains as described above for the measurement of bacterial resistance to the tetracyclines, incubated, and scored for growth. Bacterial growth on plates containing at least 1.5 mM K+ was taken as an ability of the tetA(K) gene to overcome the K+ transport defect of E. coli strain TK2205.
DNA manipulations.
Single-stranded DNA was prepared as described by Sambrook et al. (31). Plasmid DNA was isolated from E. coli by using either an alkaline lysis method (31) or the Quantum Prep plasmid miniprep kit (Bio-Rad). Restriction endonucleases were used in accordance with the manufacturers' instructions. Primers for mutagenesis and nucleotide sequencing were made using a Beckman Oligo 1000 synthesizer. Nucleotide sequencing was performed by using the SequiTherm cycle sequencing kit (Epicentre Technologies) as recommended by the manufacturer or by the Sydney University and Prince Alfred Macromolecular Analysis Centre, using the ABI ready reaction kit. Sequences were assembled and stored using the program SEQUENCHER (Gene Codes Corp.). The multiple-amino-acid-sequence alignment of motif C was prepared by using PILEUP (5) and SEQVU (Garvan Institute of Medical Research, Sydney, Australia).
Mutagenesis of tetA(K).
Site-directed mutants were constructed by using oligonucleotide-directed mutagenesis according to the method of Kunkel et al. (18). The following oligonucleotide primers were employed in the mutagenesis procedure: G151X, 5′-GCCTTTGGTTTTATAGGATCGATTGTAGCTTTA(ACG)(ACGT)(CG)GAAGGGTTAGGTCCTTCAATAGG-3′; G155X, 5′-GTAGCTTTAGGTGAAGGGTTA(ACG)(CG)(CT)CCTTCGATCGGGGGAATAATAGCACATTATATTC-3′; P156X, 5′-GCTTTAGGTGAAGGGTTAGGT( ACG )( ACGT )( CG )TCGATCGGGGGAATAATAGCACATTATATTC-3′; G159X, 5′-GCTTTAGGTGAAGGGTTAGGTCCATCGATA(ACG)(ACGT)(CG)GGAATAATAGCACATTATATTC-3′; and G160X, 5′-GCTTTAGGTGAAGGGTTAGGTCCATCGATAGGG(ACG)(ACGT)(CG)ATAATAGCACATTATATTCATTGG-3′. Underlining indicates mismatching of base pairs with the wild-type tetA(K) sequence so as to introduce a restriction endonuclease site without altering the coding sequence, and parentheses indicate degeneracy introduced into each oligonucleotide, resulting in the alteration of the amino acid sequence at that position. As a first step in the creation of TetA(K) hybrid proteins, StuI and NsiI sites were introduced into tetA(K), flanking the region encoding TMS 5 at amino acid positions 138 and 166, by using the QuickChange site-directed mutagenesis kit (Stratagene) and the oligonucleotides (i) 5′-CAAGAAAAAAACAAGGCAAGGCCTTTGGTTTTATAGGATC-3′ and its complement and (ii) 5′-GGGGGAATAATAGCACATTATATGCATTGGTCTTACCTACTTATAC-3′ and its complement, respectively. Oligonucleotide primers used for the amplification of TMS 5 of QacA (nucleotides 1254 to 1358, GenBank accession no. X56628; 5′-CGCAAGGCCTTAGCTGTATGGTCAATCGC-3′ and 5′-CGCATGCATTTGCTCAAGTAAAGCTCCTC-3′) and TMS 5 of TetA(B) (nucleotides 1998 to 2072, GenBank accession no. J01830; 5′-CGCAAGGCCTTAGCTGTATGGTCAATCGC-3′ and 5′-CGCATGCATTTGCTCAAGTAAAGCTCCTC-3′) incorporated StuI and NsiI sites at the 5′ and 3′ ends, respectively. Chemical modification of tetA(K) was achieved utilizing hydroxylamine as described by McNicholas et al. (23). Second-site suppressor mutations of E. coli DH5α cells harboring the tetA(K) mutant, pSK4541, were obtained by using the method of Kimura et al. (17). The DNA sequence of the tetA(K) gene for each mutant constructed was confirmed and analyzed to ensure that no spurious mutations had been introduced.
Radioactive labeling of mutant proteins.
A modified method of Maneewannakul et al. (20) was used to detect mutant proteins. E. coli BL21(DE3)/pLysS cells harboring the desired plasmid were grown to an optical density at 600 nm of 0.7, harvested, and resuspended in 1 volume of assay medium (5% 18-amino-acid mixture, 0.2% glucose, 2 mM MgSO4, 1× M9 salts). Cells were incubated at 37°C for 1 h before the addition of 0.25 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and incubation was continued for a further 1 h at 37°C. Rifampin was added to a final concentration of 200 μg/ml, and the incubation temperature was switched to 42°C for 10 min and then returned to 37°C for 40 min. One milliliter of cells was transferred to an Eppendorf tube, and proteins were radioactively labeled for 10 min at 37°C by the addition of 10 μCi of Tran35S-label (ICN) per ml of cells. The cells were then collected by centrifugation and resuspended in sodium dodecyl sulfate gel-loading buffer, and the proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (19). Following electrophoresis, the gel was incubated in 1 M sodium salicylate, and proteins were identified by fluorography (2); fluorographs were scanned with a Nikon ScanTouch Controller AX-1200. Quantitation of TetA(K) proteins was performed by using the software package Quantity One (Bio-Rad). A host-encoded protein, present in all samples, was used as an internal standard and assigned a value of 100; this was subsequently used to normalize the level of TetA(K) expression. To evaluate the reproducibility of the technique and to ensure that any deviations observed in protein level expression were not due to variations caused either by the labeling procedure or by plasmid loss, experiments were performed twice, and plasmids from all samples were isolated and analyzed.
Preparation of membrane vesicles.
Everted membrane vesicles were prepared from E. coli TG1 cells harboring wild-type or mutant plasmids essentially as described by Yamaguchi et al. (38). Everted vesicles were prepared by disruption of cells by a single passage at 4,500 lb/in2 through an Aminco French pressure cell, and unlysed cells were removed by centrifugation. Vesicles were sedimented by using ultracentrifugation and gently resuspended by using a glass rod in 50 mM MOPS (morpholinepropanesulfonic acid)-KOH (pH 7.0) containing 100 mM KCl. The vesicles were then frozen in a dry ice-ethanol bath and stored at −70°C. The protein concentration in membrane vesicles was determined by using the DC protein assay (Bio-Rad) as described by the manufacturer.
Tetracycline transport assays.
A suspension of membrane vesicles in 50 mM MOPS-KOH buffer (pH 7.0) containing 100 mM KCl and 500 μM CoCl2 was preincubated at 31°C for 10 min with shaking in the presence of 5 mM NADH as an energy source. The uptake of tetracycline was initiated by the addition of [3H]tetracycline (DuPont New England Nuclear) to a final concentration of 10 μM. Aliquots were removed at the required time intervals and immediately mixed with 5 ml of 50 mM MOPS-KOH (pH 7.0) buffer containing 150 mM LiCl and filtered through a 0.45-μm-pore-size Gelman Metricel filter, followed by a 2-ml wash with the LiCl-containing buffer. The radioactivity on the filter was determined by using a Tri-Carb liquid scintillation analyzer (Packard), and tetracycline accumulation (nanomoles per milligram of protein) was calculated by subtracting the radioactivity retained by the filter alone.
RESULTS
Mutagenesis of motif C.
To identify functionally important amino acids within motif C of TetA(K) (151-GEGLGPSIGGII-162) (Fig. 1), random and site-directed mutageneses were performed on the cloned tetA(K) gene carried by pSK4646. This construct, a derivative of pSK4607 (9), incorporated a C-terminal tag which encodes six consecutive histidine residues prior to the termination codons. The residues targeted for mutagenesis were the glycines G151, G155, G159, and G160 and the proline P156 of the TetA(K) protein. The majority of these represent the highly conserved residues of motif C. However, more variation among DHA12 and DHA14 MFS members exists at G151; this residue is conserved in at least 40% of drug exporters (28) (lowercase in Fig. 1). By utilizing the mutagenic oligonucleotides listed in Materials and Methods, a number of substitutions were obtained at each of these positions (Table 1). Additionally, a random mutant created by hydroxylamine treatment, which expressed a G155D substitution (pSK4400), was included in this analysis.
TABLE 1.
Tetracycline resistance levels conferred by TetA(K) motif C mutantsa
| Plasmid | Mutation | IC (μg/ml) |
|---|---|---|
| pBluescript II SK | 5 | |
| pBR322 | 96 | |
| pSK4646 | 48 | |
| G151 mutations | ||
| pSK4591 | G151R | 5 |
| pSK4592 | G151V | 10 |
| pSK4594 | G151E | 5 |
| pSK4595 | G151D | 5 |
| pSK4596 | G151L | 5 |
| pSK4597 | G151K | 5 |
| pSK4598 | G151A | 48 |
| pSK4599 | G151S | 48 |
| G155 mutations | ||
| pSK4400 | G155D | 5 |
| pSK4538 | G155A | 48 |
| pSK4539 | G155R | 5 |
| pSK4541 | G155P | 5 |
| pSK4543 | G155S | 48 |
| pSK4544 | G155T | 5 |
| pSK4549 | P155S-P156S | 32 |
| P156 mutations | ||
| pSK4551 | P156A | 5 |
| pSK4552 | P156R | 5 |
| pSK4555 | P156L | 5 |
| pSK4556 | P156T | 5 |
| pSK4559 | P156V | 5 |
| pSK4560 | P156N | 5 |
| pSK4561 | P156Q | 5 |
| pSK4562 | P156G | 12 |
| pSK4563 | P156H | 5 |
| pSK4564 | P156I | 5 |
| pSK4565 | P156K | 5 |
| pSK4566 | P156S | 5 |
| G159 mutations | ||
| pSK4568 | G159A | 8 |
| pSK4570 | G159R | 5 |
| pSK4571 | G159D | 5 |
| pSK4572 | G159N | 5 |
| pSK4573 | G159E | 5 |
| pSK4574 | G159P | 5 |
| pSK4575 | G159S | 8 |
| pSK4576 | G159T | 5 |
| pSK4577 | G159V | 8 |
| G160 mutations | ||
| pSK4579 | G160V | 10 |
| pSK4580 | G160T | 5 |
| pSK4581 | G160L | 5 |
| pSK4582 | G160I | 5 |
| pSK4583 | G160E | 5 |
| pSK4584 | G160Q | 5 |
| pSK4585 | G160R | 5 |
| pSK4586 | G160A | 48 |
| pSK4588 | G160S | 32 |
All mutants were constructed by site-directed mutagenesis procedures, with the exception of the hydroxylamine-generated G155 mutation of pSK4400 and the second-site suppressor mutation of pSK4541, pSK4549 (see Materials and Methods for details).
Based on IC data, it appears that at positions 151, 155, and 160 the only substitutions that preserved wild-type tetracycline resistance levels were alanine and serine (Table 1). In contrast, no substitutions for G159 maintained any significant resistance to tetracycline. This indicated that there is an absolute requirement for the glycine residue at this position in TetA(K); G159A and G159S mutations conferred only marginal tetracycline resistance (8 μg/ml) (Table 1). Although this seems to be true for TetA(K), it is not the case for all MFS drug export proteins, because other amino acids naturally occur at this position; e.g., the actinhordin efflux protein from Streptomyces coelicolor, ActII, has an alanine residue at the equivalent position (Fig. 1 and see reference 28). The requirement for P156 also appears to be vital for TetA(K)-mediated tetracycline resistance; despite the numerous replacements introduced at P156, only the P156G mutation retained tetracycline resistance, albeit at a low level (12 μg/ml) (Table 1).
Expression of TetA(K) mutants.
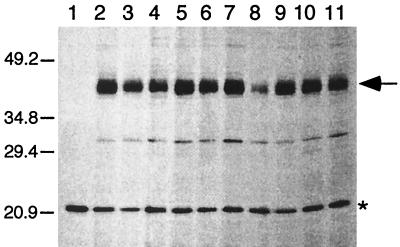
To determine whether any of the introduced mutations affected the expression level of TetA(K), proteins were radioactively labeled and visualized as described in Materials and Methods. With the wild type and all TetA(K) mutants, a band corresponding to a size of approximately 43 kDa was detected (Fig. 2 and data not shown). This is considerably smaller than the predicted size of 50.7 kDa for TetA(K) (12). However, this phenomenon has also been observed for the highly related Bacillus subtilis TetA(L) protein, which, although having a predicted size of 49.9 kDa, was determined experimentally to be 37.5 kDa (3); the difference between the observed and predicted molecular masses might be due to the high hydrophobicity of these membrane proteins. Radioactive labeling of mutant TetA(K) proteins indicated that, in general, the level of protein detected did not deviate greatly from that expressed by cells carrying the wild-type TetA(K) (Fig. 2 and data not shown). However, one exception was the introduction of a proline residue at G159 (pSK4574), which did not confer tetracycline resistance (Table 1) and resulted in a less intense TetA(K) band (24.7%) (Fig. 2, lane 8), reflecting either reduced protein expression or reduced stability.
FIG. 2.
Fluorograph of radioactively labeled TetA(K) proteins prepared from E. coli BL21(DE3)/pLysS cell extracts containing plasmids encoding mutations at G159 in TetA(K). Lanes [extracts from cells harboring the following plasmids, with the normalized value for detected TetA(K) protein denoted as a percentage of the wild-type value]: 1, pBluescript II SK; 2, pSK4646 (wild type; 100%); 3, pSK4568 (G159A; 101.0%); 4, pSK4570, (G159R; 63.6%); 5, pSK4571 (G159D; 102.6%); 6, pSK4572 (G159N; 96.1%); 7, pSK4573 (G159E; 97.9%); 8, pSK4574 (G159P; 24.7%); 9, pSK4575 (G159S; 87.7%); 10, pSK4576 (G159T; 78.5%); and 11, pSK4577 (G159V; 64.8%). The positions of migration and sizes (in kilodaltons) of coelectrophoresed standard proteins are shown on the left. Products corresponding to the TetA(K) protein (43 kDa) are indicated on the right by the arrow, and a host-encoded protein, used for normalization of TetA(K) expression levels, is indicated by the asterisk. Only the relevant portion of the fluorograph is shown. Similar studies were performed for all TetA(K) mutant proteins; only one representative data set is presented here.
It has been demonstrated that the TetA(K) polypeptide can complement E. coli cells defective in K+ import (14, 15). This collateral phenotype has been exploited in the present study as an indication of whether mutant TetA(K) proteins were inserted into the membrane in a functional conformation. It was found that the presence of either wild-type TetA(K) or any of its mutant derivatives enabled the growth of the K+ transport-defective E. coli strain TK2205 on medium supplemented with as little as 1.5 mM K+. As a positive control, E. coli TK2205 cells harboring the plasmid pBR322, which encodes the TetA(C) protein, were able to grow on medium containing low K+ concentrations, as previously reported (11); E. coli TK2205 cells either with or without the vector pBluescript II SK were unable to grow at a K+ concentration of 3 mM. The finding that all mutant TetA(K) proteins, including cells expressing the G159P mutation, which resulted in the production of a less-intense TetA(K) band (Fig. 2, lane 8), appeared to be able to complement the K+ uptake defect at a level reminiscent of the wild type suggested that the introduced changes did not significantly affect the folding of mutant proteins into the cytoplasmic membrane.
Tetracycline transport assays in everted membrane vesicles.
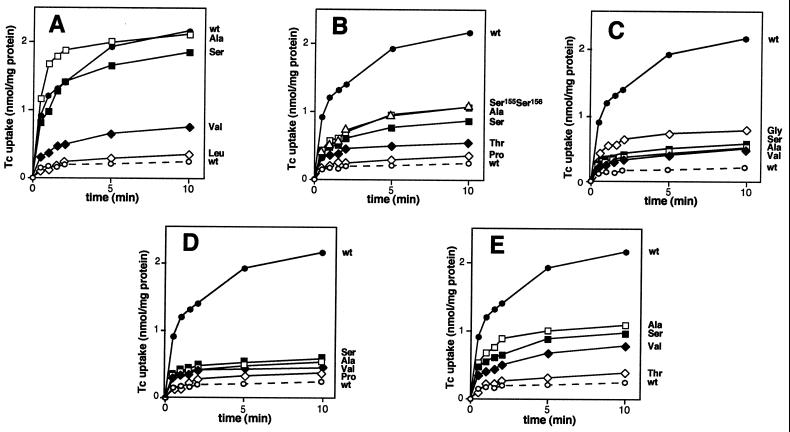
IC analyses revealed that most E. coli cells expressing mutant TetA(K) proteins were unable to sustain growth in the presence of as little as 5 μg of tetracycline per ml (Table 1), suggesting that these mutant derivatives are incapable of extruding tetracycline from the cell. This was confirmed for selected mutants, carrying representative amino acid substitutions, by direct measurements of tetracycline accumulation in E. coli TG1 everted membrane vesicles (Fig. 3). Relative tetracycline transport activities of the mutant TetA(K) proteins, as compared to that of the wild type and corrected for protein expression levels as determined by radioactive labeling, are shown in Table 2. Usually, a high tetracycline IC was reflected by significant TetA(K)-mediated transport activity (Fig. 3 and Table 1). For example, replacements at position 151, where glycine was replaced by either alanine or serine, maintained essentially wild-type tetracycline transport levels (Fig. 3A and Table 2), with some activity also being observed when valine was introduced at this position, consistent with the IC data (Table 1). In cases where no tetracycline resistance was observed, it was demonstrated that the active uptake of tetracycline was negligible (Fig. 3 and Table 1). These findings, together with the potassium uptake complementation data and the radioactive labeling studies, which indicate normal expression, support the notion that the inability to transport tetracycline as seen with a number of mutations (Fig. 3) is likely to be due solely to the introduced amino acid substitutions within motif C.
FIG. 3.
Tetracycline (Tc) accumulation by everted membrane vesicles prepared from E. coli TG1 cells harboring plasmids encoding mutations at the following amino acid positions in TetA(K): A, G151; B, G155; C, P156; D, G159; and E, G160. Data for cells carrying plasmids encoding the wild-type (wt) or mutant TetA(K) proteins are labeled with the three-letter code corresponding to the incorporated amino acid. Solid and dashed lines indicate the uptake of tetracycline by vesicles in the presence and absence, respectively, of NADH as an energy source.
TABLE 2.
Relative protein expression levels and transport activities of representative TetA(K) motif C mutants
| Mutation | TetA(K) protein expression (%)a | Relative tetracycline transport activity (%)b |
|---|---|---|
| G151 mutations | ||
| G151V | 92.0 | 28.0 |
| G151L | 117.4 | 4.3 |
| G151A | 110.1 | 88.3 |
| G151S | 80.6 | 103.3 |
| G155 mutations | ||
| G155A | 141.4 | 30.0 |
| G155P | 82.8 | 6.3 |
| G155S | 143.4 | 22.2 |
| G155T | 112.1 | 13.1 |
| P155S-P156S | 134.6 | 31.6 |
| P156 mutations | ||
| P156A | 91.1 | 16.1 |
| P156V | 104.0 | 13.8 |
| P156G | 82.7 | 27.2 |
| P156S | 93.0 | 12.5 |
| G159 mutations | ||
| G159A | 101.1 | 14.4 |
| G159P | 24.7 | 24.3 |
| G159S | 87.7 | 20.1 |
| G159V | 64.9 | 15.9 |
| G160 mutations | ||
| G160V | 142.4 | 19.5 |
| G160L | 110.7 | 5.0 |
| G160A | 103.6 | 42.9 |
| G160S | 120.1 | 31.1 |
TetA(K) expression levels were normalized using a host-encoded protein (see Materials and Methods for details) and are expressed as a percentage of the level detected with the wild-type TetA(K) encoded by pSK4646 (Table 1).
Tetracycline transport activity, corrected for the TetA(K) protein expression levels shown, is represented as a percentage of wild-type TetA(K) transport activity.
All amino acid substitutions of G159, including those with similar chemical properties, resulted in the complete loss of tetracycline transport activity (Fig. 3D), and, with the exception of glycine, no changes at P156 were tolerated (Fig. 3C). However, replacements at positions 155 and 160 with alanine or serine, or also valine in the case of G160, while conferring near-wild-type levels of tetracycline resistance (Table 1), displayed only 19.5 to 42.9% of wild-type TetA(K) transport function, suggesting that even minor structural changes affect the tetracycline/H+ antiport activity (Fig. 3B and E and Table 2). Therefore, although a good correlation exists between the levels of tetracycline resistance and transport activity of cells expressing mutant TetA(K) proteins in general, the data obtained for replacements at G155 and G160 demonstrated that the transport assay system was more sensitive than IC analysis for determining the effect of amino acid replacements in TetA(K).
Isolation of a suppressor mutation to TetA(K) G155P.
Replacement of G155 with proline (pSK4541) caused a complete loss of both tetracycline resistance (Table 1) and transport function compared to wild-type TetA(K) (Fig. 3B and Table 2). A spontaneous revertant (pSK4549) exhibiting near-wild-type levels of tetracycline resistance (32 μg/ml) (Table 1) was isolated from E. coli DH5α cells carrying the tetA(K) gene with this mutation. Sequence analysis of pSK4549 revealed that two amino acid replacements had occurred within motif C of TetA(K). The first changed the original G155P mutation to serine, and the second substitution occurred at the adjacent residue, P156, also replacing this amino acid with serine. Analysis of this double serine (S155-S156) mutant in everted membrane vesicles revealed that it possessed 31.6% of wild-type tetracycline transport function (Fig. 3B and Table 2). Not only is this level greater than that obtained for cells carrying the single G155S mutation (Table 2), it is significantly greater than that observed for cells with the P156S mutation, for which no tetracycline resistance or significant tetracycline transport activity was observed (Fig. 3C and Tables 1 and 2).
Construction of hybrid TetA(K) proteins.
In general, individual replacement of the conserved residues that comprise motif C within TetA(K) resulted in the loss of tetracycline resistance and tetracycline/H+ antiport activity. Topological studies performed on TetA(K) demonstrated that all 14 membrane segments of this protein were required to confer tetracycline resistance (9). Therefore, studies were undertaken to determine whether TMS 5 of the TetA(K) polypeptide could be replaced with the corresponding membrane segments from other drug export proteins also containing motif C, a strategy we term TMS replacement mutagenesis. To achieve this, unique restriction endonuclease recognition sites were introduced into the cloned tetA(K) gene carried by pSK4646, as described in Materials and Methods, such that these sites flanked the region encoding TMS 5. Although the introduction of the StuI recognition site did not alter the coding sequence of the TetA(K) protein, the incorporation of the NsiI recognition sequence resulted in an I166M mutation within TetA(K). Cells that carried this construct, pSK4410, had wild-type tetracycline resistance (48 μg/ml) and, in addition to complementation of E. coli cells defective in K+ import, radioactive labeling demonstrated that the level of TetA(K) protein detected was not affected (98.0% of the wild-type level).
By using PCR, the regions encoding TMS 5 from the tetracycline exporter TetA(B) and the multidrug resistance efflux protein QacA were amplified from pNK81 (36) and pSK4236 (27), respectively, and cloned into pSK4410 after removal of its cognate TMS 5. The resulting plasmids, pSK4411 and pSK4412, encode TetA(K)-QacA and TetA(K)-TetA(B) chimeras, respectively. IC analysis revealed that E. coli cells expressing these hybrid proteins conferred only background levels of resistance to tetracycline and the tetracycline analogs chlortetracycline, doxycycline, and oxytetracycline, which are also substrates of TetA(K) and TetA(B) (13). Additionally, cells expressing the TetA(K)-QacA hybrid were unable to grow on plates containing ethidium bromide, a substrate of the QacA polypeptide (30). Radioactive labeling of these chimeras, as described for Fig. 2, revealed that, in comparison to cells expressing wild-type TetA(K), the TetA(K)-QacA and TetA(K)-TetA(B) hybrid proteins were present at reduced levels (14.5 and 9.6%, respectively).
DISCUSSION
Motif C, a motif in TMS 5 of the TetA(K) polypeptide, is conserved throughout members of the DHA12 and DHA14 drug export families of the MFS. Conservation of this motif within these two families and its absence from proteins that function as importers suggest that it plays an important role in linking proton translocation to the antiport, but not symport, of the substrate or in influencing the polarity of substrate transport (29, 33). Previously it has been postulated that the G155-P156 dipeptide of motif C influences the conformation of the TMS 5 helix and, in conjunction with the other glycine residues, forms a pocket on one face of TMS 5 (33). Indeed, in proteins where glycine residues do not naturally occur in motif C, alanine and serine residues are prevalent (Fig. 1); like glycine, alanine and serine possess small side chains. By using site-directed mutagenesis combined with IC analysis and the more sensitive tetracycline transport assay system, we have demonstrated experimentally that G155, along with the glycine residues G151 and G160, is important for tetracycline/H+ antiport activity, although alanine and serine substitutions in lieu of glycine are well tolerated at these positions (Fig. 3A, B, and E and Tables 1 and 2). In contrast to these glycine residues, glycine residue G159 and proline residue P156 appear to be crucial, since all single-amino-acid replacements at these positions either significantly reduced or abolished TetA(K)-mediated tetracycline transport activity (Fig. 3C and D and Tables 1 and 2). The ability of the double serine mutant with mutations at positions 155 and 156 to mediate significant levels of tetracycline resistance (Table 1) suggests that the tetracycline transport defect conferred by the P156S single mutant can be partially compensated for by also replacing the adjacent residue with serine.
Proline residues are frequently embedded in the membrane-spanning portions of ion channels and transporters but are absent from the transmembrane segments of proteins with no transport function. This has led to the proposition that membrane-bound proline residues play an important functional role in transport proteins (1). However, proline residues are unfavorable in α-helical structures due to the steric hindrance caused by their ring structure; their backbone nitrogen is unavailable for normal hydrogen bonding (4). Consequently, proline residues introduce kinks into α-helices, with the convex side of the kink packed away from the lipid, potentially forming a binding site or channel vestibule (35). The ability of the proline-to-glycine substitution at position 156 to retain partial tetracycline resistance (12 μg/ml) (Table 1) may be explained by the hypothesis that both proline and glycine act as helix-breaking residues (4). Thus, the introduced glycine residue may preserve the natural secondary structure of TMS 5. Indeed, this role has been suggested for the proline residue within motif C, where this residue may insert a bend in the α-helix (33). It is worth noting that in drug export proteins lacking a proline residue at this position, such as the chloramphenicol export protein Cml from Streptomyces lividans (6), one is often located at the adjacent position (Fig. 1).
Based on the results of TMS replacement mutagenesis, TMS 5 of the TetA(K) polypeptide was unable to be replaced by the corresponding helix from the drug export protein TetA(B) or QacA. Radioactive-labeling experiments demonstrated that these hybrid proteins were present at reduced levels in comparison to the wild-type TetA(K) polypeptide. Fujihira and coworkers (8) have shown that the glutamic acid residue E152 in TMS 5 of TetA(K) is important for tetracycline transport function; only a charge-conserved replacement to aspartic acid retained transport activity. This raises the possibility that the inability of TetA(K) chimeras to confer tetracycline resistance may be due to the fact that TetA(B) and QacA possess the neutral residues leucine and alanine, respectively, at the corresponding position (Fig. 1). Thus, if E152 plays an important structural role within the TetA(K) polypeptide (for example, if it was involved in an ionic interaction), a reduction in the amount of TetA(K) hybrid proteins detected would not be unexpected.
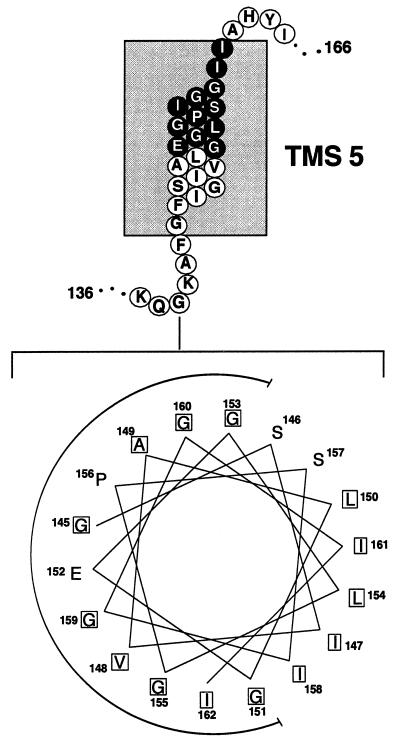
Examination of a helical-wheel projection of TMS 5 from the TetA(K) polypeptide revealed that the conserved residues of motif C are all located on the same face of the helix (Fig. 4), furthering the proposition that these residues are close to each other and form a pocket in drug export proteins (33). Interestingly, the glutamic acid residue E152, shown previously to be crucial for TetA(K)-mediated tetracycline resistance (8), appears to be embedded within the center of the recess formed by these glycine residues, between P156 and G159 (Fig. 4), residues which are both vital for high-level transport activity. In addition to the conserved glycines that comprise motif C, TetA(K) contains another three glycine residues on the same helical face, located at positions 142, 145, and 153 (Fig. 4). Thus, in TetA(K) the glycine residues comprise one-third of the amino acids of TMS 5, seven glycine residues in total (Fig. 1); other TMS of TetA(K) contain between zero and four glycine residues. Significantly, the high glycine content of TMS 5 is not unique to TetA(K); the drug efflux proteins TcmA, ActII, Bmr1, and NorA all contain six glycine residues within this TMS (Fig. 1). Conformational transitions, which occur within or adjacent to membrane domains of membrane transport proteins during substrate flux, are likely to require regions of the protein structurally responsive to the immediate environment. The abundance of glycine residues in TMS 5 may confer such conformational plasticity to the MFS drug export proteins.
FIG. 4.
Schematic diagram of the proposed topology of TMS 5 from the TetA(K) polypeptide (9); residues comprising motif C are depicted in reverse type. Shown at the bottom is a helical-wheel projection of 18 amino acid residues from TetA(K) TMS 5 created using the Helicalwheel program of the Genetics Computer Group package (5). Each residue within the helical wheel is offset from the preceding one by 100°; hydrophobic amino acids are boxed. The glycine-rich face of TMS 5 is indicated by the semicircle.
ACKNOWLEDGMENTS
We thank A. M. George for many useful discussions regarding transport assays, N. Firth for critical reading of the manuscript, and J. K. Griffith for providing the E. coli strain TK2205.
This work was supported in part by a grant from the Australian Research Council. S.L.G. is the recipient of an Australian Postgraduate Award.
REFERENCES
- 1.Brandl C J, Deber C M. Hypothesis about the function of membrane-buried proline residues in transport proteins. Proc Natl Acad Sci USA. 1986;83:917–921. doi: 10.1073/pnas.83.4.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chamberlain J P. Fluorographic detection of radioactivity in polyacrylamide gels with the water-soluble fluor, sodium salicylate. Anal Biochem. 1979;98:132–135. doi: 10.1016/0003-2697(79)90716-4. [DOI] [PubMed] [Google Scholar]
- 3.Cheng J B, Hicks D B, Krulwich T A. The purified Bacillus subtilis tetracycline efflux protein TetA(L) reconstitutes both tetracycline-cobalt/H+ and Na+ (K+)/H+ exchange. Proc Natl Acad Sci USA. 1996;93:14446–14451. doi: 10.1073/pnas.93.25.14446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chou P Y, Fasman G D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- 5.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dittrich W, Betzler M, Schrempf H. An amplifiable and deletable chloramphenicol-resistance determinant of Streptomyces lividans 1326 encodes a putative transmembrane protein. Mol Microbiol. 1991;5:2789–2797. doi: 10.1111/j.1365-2958.1991.tb01987.x. [DOI] [PubMed] [Google Scholar]
- 7.Dosch D C, Salvacion F F, Epstein W. Tetracycline resistance element of pBR322 mediates potassium transport. J Bacteriol. 1984;160:1188–1190. doi: 10.1128/jb.160.3.1188-1190.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujihira E, Kimura T, Shiina Y, Yamaguchi A. Transmembrane glutamic acid residues play essential roles in the metal-tetracycline/H+ antiporter of Staphylococcus aureus. FEBS Lett. 1996;391:243–246. doi: 10.1016/0014-5793(96)00743-0. [DOI] [PubMed] [Google Scholar]
- 9.Ginn S L, Brown M H, Skurray R A. Membrane topology of the metal-tetracycline/H+ antiporter TetA(K) from Staphylococcus aureus. J Bacteriol. 1997;179:3786–3789. doi: 10.1128/jb.179.11.3786-3789.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffith J K, Baker M E, Rouch D A, Page M G P, Skurray R A, Paulsen I T, Chater K F, Baldwin S A, Henderson P J F. Membrane transport proteins: implications of sequence comparisons. Curr Opin Cell Biol. 1992;4:684–695. doi: 10.1016/0955-0674(92)90090-y. [DOI] [PubMed] [Google Scholar]
- 11.Griffith J K, Kogoma T, Corvo D L, Anderson W L, Kazim A L. An N-terminal domain of the tetracycline resistance protein increases susceptibility to aminoglycosides and complements potassium uptake defects in Escherichia coli. J Bacteriol. 1988;170:598–604. doi: 10.1128/jb.170.2.598-604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guay G G, Khan S A, Rothstein D M. The tet(K) gene of plasmid pT181 of Staphylococcus aureus encodes an efflux protein that contains 14 transmembrane helices. Plasmid. 1993;30:163–166. doi: 10.1006/plas.1993.1045. [DOI] [PubMed] [Google Scholar]
- 13.Guay G G, Rothstein D M. Expression of the tetK gene from Staphylococcus aureus in Escherichia coli: comparison of substrate specificities of TetA(B), TetA(C), and TetK efflux proteins. Antimicrob Agents Chemother. 1993;37:191–198. doi: 10.1128/aac.37.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guay G G, Tuckman M, McNicholas P, Rothstein D M. The tet(K) gene from Staphylococcus aureus mediates the transport of potassium in Escherichia coli. J Bacteriol. 1993;175:4927–4929. doi: 10.1128/jb.175.15.4927-4929.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guffanti A A, Cheng J, Krulwich T A. Electrogenic antiport activities of the Gram-positive Tet proteins include a Na+(K+)/K+ mode that mediates net K+ uptake. J Biol Chem. 1998;273:26447–26454. doi: 10.1074/jbc.273.41.26447. [DOI] [PubMed] [Google Scholar]
- 16.Hanahan D. Studies on transformation in Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 17.Kimura T, Sawai T, Yamaguchi A. Remote conformational effects of the gly-62→leu mutation of the Tn10-encoded metal-tetracycline/H+ antiporter of Escherichia coli and its second-site suppressor mutation. Biochemistry. 1997;36:6941–6946. doi: 10.1021/bi9631879. [DOI] [PubMed] [Google Scholar]
- 18.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Maneewannakul K, Maneewannakul S, Ippen-Ihler K. Sequence alterations affecting F plasmid transfer gene expression: a conjugation system dependent on transcription by the RNA polymerase of phage T7. Mol Microbiol. 1992;6:2961–2973. doi: 10.1111/j.1365-2958.1992.tb01755.x. [DOI] [PubMed] [Google Scholar]
- 21.Marger M D, Saier M H., Jr A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends Biochem Sci. 1993;18:13–20. doi: 10.1016/0968-0004(93)90081-w. [DOI] [PubMed] [Google Scholar]
- 22.McMurry L M, Petrucci R E, Jr, Levy S B. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc Natl Acad Sci USA. 1980;77:3974–3977. doi: 10.1073/pnas.77.7.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McNicholas P, Chopra I, Rothstein D M. Genetic analysis of the tetA(C) gene on plasmid pBR322. J Bacteriol. 1992;174:7926–7933. doi: 10.1128/jb.174.24.7926-7933.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitchell B A, Brown M H, Skurray R A. QacA multidrug efflux pump from Staphylococcus aureus: comparative analysis of resistance to diamidines, biguanidines, and guanylhydrazones. Antimicrob Agents Chemother. 1998;42:475–477. doi: 10.1128/aac.42.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitchell B A, Paulsen I T, Brown M H, Skurray R A. Bioenergetics of the staphylococcal multidrug export protein QacA: identification of distinct binding sites for monovalent and divalent cations. J Biol Chem. 1999;274:3541–3548. doi: 10.1074/jbc.274.6.3541. [DOI] [PubMed] [Google Scholar]
- 26.Pao S S, Paulsen I T, Saier M H., Jr The major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62:1–32. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paulsen I T, Brown M H, Littlejohn T G, Mitchell B A, Skurray R A. Multidrug resistance proteins QacA and QacB from Staphylococcus aureus: membrane topology and identification of residues involved in substrate specificity. Proc Natl Acad Sci USA. 1996;93:3630–3635. doi: 10.1073/pnas.93.8.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paulsen I T, Brown M H, Skurray R A. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paulsen I T, Skurray R A. Topology, structure and evolution of two families of proteins involved in antibiotic and antiseptic resistance in eukaryotes and prokaryotes—an analysis. Gene. 1993;124:1–11. doi: 10.1016/0378-1119(93)90755-r. [DOI] [PubMed] [Google Scholar]
- 30.Rouch D A, Cram D S, DiBerardino D, Littlejohn T G, Skurray R A. Efflux-mediated antiseptic resistance gene qacA from Staphylococcus aureus: common ancestry with tetracycline- and sugar-transport proteins. Mol Microbiol. 1990;4:2051–2062. doi: 10.1111/j.1365-2958.1990.tb00565.x. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 32.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 33.Varela M F, Sansom C E, Griffith J K. Mutational analysis and molecular modelling of an amino acid sequence motif conserved in antiporters but not symporters in a transporter superfamily. Mol Membr Biol. 1995;12:313–319. doi: 10.3109/09687689509072433. [DOI] [PubMed] [Google Scholar]
- 34.Vieira J, Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- 35.von Heijne G. Proline kinks in transmembrane α-helices. J Mol Biol. 1991;218:499–503. doi: 10.1016/0022-2836(91)90695-3. [DOI] [PubMed] [Google Scholar]
- 36.Way J C, Davis M A, Morisato D, Roberts D E, Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984;32:369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- 37.Willetts N S, Finnegan D J. Characteristics of E. coli K12 strains carrying both an F prime and an R factor. Genet Res. 1970;16:113–122. doi: 10.1017/s0016672300002329. [DOI] [PubMed] [Google Scholar]
- 38.Yamaguchi A, Shiina Y, Fujihira E, Sawai T, Noguchi N, Sasatsu M. The tetracycline efflux protein encoded by the tet(K) gene from Staphylococcus aureus is a metal-tetracycline/H+ antiporter. FEBS Lett. 1995;365:193–197. doi: 10.1016/0014-5793(95)00455-i. [DOI] [PubMed] [Google Scholar]