Abstract
Purpose:
Dynamic changes in circulating tumor DNA (ctDNA) are under investigation as an early indicator of treatment outcome.
Experimental Design:
Serial plasma ctDNA (baseline, 8 weeks and at progression) was prospectively incorporated into the SWOG S1403 clinical trial of afatinib +/− cetuximab in TKI-naïve, EGFR-mutation tissue-positive NSCLC.
Results:
EGFR mutations were detected in baseline ctDNA in 77% (82/106) of patients, associated with the presence of brain and/or liver metastases and M1B stage. Complete clearance of EGFR-mutations in ctDNA by 8 weeks was associated with a significantly decreased risk of progression, compared to those with persistent ctDNA at C3D1 (HR (95% CI): 0.23 (0.12-0.45), p<0.0001); with a median PFS of 15.1 (95% CI: 10.6-17.5) months in the group with clearance of ctDNA versus 4.6 (1.7-7.5) months in the group with persistent ctDNA. Clearance was also associated with a decreased risk of death (HR (95% CI): 0.44 (0.21-0.90), p=0.02; mOS: 32.6 versus 15.6 (4.9-28.3) months.
Conclusions:
Plasma clearance of mutant EGFR ctDNA at 8-weeks was highly and significantly predictive of progression-free and overall survival, outperforming RECIST response for predicting long-term benefit.
Statement of Translational Relevance
An algorithm measuring treatment-induced EGFR-mutant ctDNA clearance, defined as positive at baseline and negative at 8-weeks, is associated with extended progression-free and overall survival in patients treated on an afatinib-based regimen. ctDNA clearance significantly outperformed RECIST response in predicting long-term progression-free and overall survival. Residual ctDNA can identify patients at significantly enhanced risk of rapid progression. These studies support a plasma ctDNA-based paradigm for early identification of patients with innate resistance to therapy.
Introduction
Circulating tumor DNA (ctDNA) represents the fraction of cell free DNA in plasma that originated specifically in the tumor, exclusive of other sources. A key indicator of ctDNA is the presence of a cancer-specific mutation. In advanced non-small cell lung cancer (NSCLC), circulating tumor DNA (ctDNA) has demonstrated clinical utility as a qualitative marker for the presence of actionable driver mutations to guide therapeutic decision-making when tumor tissue is unavailable or insufficient for comprehensive molecular analysis, or when results are needed urgently [1, 2]. Similarly, ctDNA can be used to identify emergent resistance mechanisms at clinical progression, and further is being explored for response monitoring and as a marker of residual disease in early stage following definitive therapy. Modern analytical techniques, including next-generation sequencing, can provide a quantitative calculation in the form of mutant allele frequency (MAF), which is a measure of the percent of DNA fragments at a particular locus that contain the mutation versus the wild-type sequence. MAF is expressed as a percentage of the mutant form within the total population, thus a 1% MAF would indicate 1 mutant species for every 99 WT species. Because the half-life of ctDNA fragments is short, estimated at 1-2 hours, MAF is considered to be representative of the current state of tumor DNA shedding, under the assumption that, if unperturbed, both the tumor-specific DNA fragments and the background wild-type fragments are in a steady state of generation.
A growing body of evidence suggests that MAF, if consistently measured over time, can be effective for tracking treatment response in advanced NSCLC [3-9]. Recent reports have demonstrated the relationship between treatment-associated decreases in MAF and favorable patient outcomes (progression-free and overall survival). For immunotherapy, algorithms have been used to calculate changes in MAF using a consensus of multiple gene mutations to generate a Plasma Molecular Response (PMR) score [8-10]. Critical elements to these approaches are assay sensitivity in patients with low volume disease and exclusion of cell-free DNA variant alleles that are not of tumor origin, particularly those arising from clonal hematopoiesis of indeterminate potential (CHIP). Additionally, algorithms need to account for MAFs that fall at or near the limit of detection, where observation of changes may be subject to stochastic effects. Additionally, gene-specific MAFs are influenced by gene copy number abnormalities, which are frequent to some oncogenes, including EGFR.
Previous studies have demonstrated activity of the pan-HER tyrosine kinase inhibitor (TKI) afatinib plus the EGFR-targeted monoclonal antibody cetuximab in patients with EGFR-mutated NSCLC and acquired resistance to EGFR TKIs [11, 12]. We hypothesized that moving this combination into first-line therapy would delay or abrogate the onset of acquired resistance. S1403 investigated the activity of afatinib plus cetuximab in newly diagnosed patients with advanced NSCLC harboring an EGFR L858R or exon 19 deletion (E19del) mutation [13]. Patients were randomized to single-agent afatinib or the combination of afatinib plus cetuximab. Collection and analysis of ctDNA was a prospective, integrated component of this study, with serial plasma collection at baseline, after completing 8 weeks of treatment, and at progression [14-16]. Here, we analyze the relationship between patient outcome and changes in ctDNA MAFs after 8 weeks of therapy, demonstrating that clearance of EGFR mutations in plasma was a strong and significant predictor of long-term therapeutic benefit.
Methods
Patients and Plasma Collection Time Points
Patients were enrolled on S1403, a first-line phase 2 trial of afatinib with or without cetuximab in EGFR-mutant NSCLC. Details of the design and results of this trial have been published previously [13]. Written informed consent was obtained from all patients, and these institutional review board-approved studies were conducted in accordance with the Declaration of Helsinki, the Belmont Report, and the U.S. Common Rule. All patients had stage IV disease with an E19del or L858R EGFR activating mutation determined through local tissue testing. Asymptomatic brain metastases were allowed. Plasma collection was prospectively planned in S1403 on all patients at three timepoints: baseline (BL, prior to initiation of treatment), after 8-weeks on treatment coinciding with Cycle 3 Day 1 (C3D1) and at progression (PRG). BL plasma was available from 106 of 168 (63%) eligible patients enrolled on the S1403 clinical trial. Characteristics between those with and without baseline plasma did not appear different with the exception that newly diagnosed patients were more likely to have ctDNA results (see Supplementary Table 1). Not all timepoints were available for each patient; the collection rate is detailed in Table 1. Of the 106 with BL, 81 (76%) provided C3D1 ctDNA samples. Forty-six (44%) provided both C3D1 and progression samples, 47 (44%) had both BL and progression samples and 36 (34%) provided samples for all three time points.
Table 1. Serial Blood Collection.
Summary of specimen collections categorized by timepoints, indicating EGFR ctDNA positivity and median VAF (with range and IQR). Blue section: individual totals for each timepoint; green section: all cases with both a BL and C3D1 timepoint available; orange section: all cases with both a BL and PRG timepoint available: purple section: all cases with all three timepoints collected.
| Baseline | C3D1 | PRG | BL + C3D1 | C3D1 + PRG | All 3 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Number | 106 | 98 | 77 | 81 | 46 | 36 | ||||
| BL | C3D1 | C3D1 | PRG | BL | C3D1 | PRG | ||||
| - EGFR neg | 24 | 80 | 23 | 19 | 65 | 33 | 14 | 5 | 25 | 10 |
| - EGFR pos | 82 | 18 | 54 | 62 | 16 | 13 | 32 | 31 | 11 | 26 |
| % Positive | 77.3% | 18.4% | 70.1% | 76.5% | 19.8% | 28.2% | 69.6% | 86.1% | 30.6 | 72.2% |
| Med VAF | 1.26 | 0 | 0.43 | 0.65 | 0 | 0 | 0.43 | 2.13 | 0 | 0.59 |
| - Range | 0-83 | 0-21 | 0-50 | 0-55 | 0-3 | 0-21 | 0-38 | 0-55 | 0-3 | 0-38 |
| IQR | 0.10-10.14 | 0 | 0-6.61 | 0.08-7.0 | 0 | 0-0.02 | 0-5.44 | 0.47-11.91 | 0-0.07 | 0-6.03 |
Tumor Tissue EGFR Sequencing
Pre-treatment tumor specimens were obtained from participating sites and enriched for malignant cells by manual microdissection of paraffin sections. EGFR E19del or L858R mutations were confirmed in these samples by pyrosequencing.
Plasma Collection and ctDNA Analysis
Plasma was collected in Streck tubes, frozen, and banked. ctDNA was then isolated from banked plasma and was analyzed using the Guardant360 assay. Plasma collection used the G360 kits, following the manufacture’s guidelines with the exception that after processing, DNA was banked for batch analysis. Analysis of ctDNA was conducted using the Guardant360 next generation sequencing platform. The reported lower limits of detection for each class of abnormality are: for SNVs, indels, fusions and CNAs > 0.04%, > 0.02%, > 0.04%, and > 2.12 copies, respectively, with a >99.9999% per-position analytic specificity [17].
Plasma kinetics were evaluated using two methodologies. First, the MAF of the activating EGFR mutation (Exon19del or L858R) from each case as provided in the G360 report was used to quantitate changes over time. Cases with a positive EGFR MAF detectable at BL were followed longitudinally for the main analyses if timepoints were available. Plasma kinetics were also evaluated using a Plasma Molecular Response (PMR) score conducted using the Guardant 360 assay[8, 9] for assessing ctDNA molecular response score in a manner largely analogous to the methodology used by Zhang et al. and Thompson et al. [8,9]. To calculate the PMR score, single nucleotide variants (SNVs), insertions and deletions (indels) and fusion events with a mutant molecular count above proprietary thresholds were averaged in baseline and C3D1 samples to generate a ratio of baseline mean MAF values and C3D1 mean MAF values. Samples lacking detectable somatic variants with sufficient mutant molecular counts at both baseline and C3D1 or that did not have any somatic alterations were considered to have low ctDNA level and were not evaluable for PMR. To ensure that changes in PMR score are attributable to alterations of tumor origin and not by germline alterations or clonal hematopoiesis, a proprietary bioinformatics filter was used to remove alterations calculated as being of likely hematologic origin. The PMR algorithm was applied to this data set in a two-category analysis comparing complete PMR (ctDNA clearance) to non-complete PMR (residual ctDNA).
Copy number gain (CNG) calling via Guardant360 has been previously described in detail [17]. In brief, probe-level unique molecular coverage was normalized for overall unique molecule throughput, probe efficiency, GC content, and signal saturation and robustly summarized at the gene level. CNG determinations were based on training set-established decision thresholds for both absolute and copy number deviation from per-sample diploid baseline and deviation from the baseline variation of probe-level normalized signal in the context of background variation within each sample’s own diploid baseline.
Statistical methodology
A survival landmark analysis at 8 weeks was used to evaluate the association between ctDNA clearance at C3D1 and clinical outcomes (OS, PFS). Survival curves were estimated using the method of Kaplan-Meier, 95% confidence intervals for median (m) OS/PFS were calculated using the method of Brookmeyer-Crowley and were compared using log rank test. A Cox regression model was used to estimate the hazard ratio (HR) and 95% CIs. A natural log transform was applied on continuous variable MAF when to build Cox regression model to evaluate the association between MAF and survival outcomes. Chi-square testing was performed to find the proportional differences among groups. When the sample size was small, exact fisher test was applied instead of chi-square test. T-test was performed to find the difference between continuous variables. All tests performed were two-sided. PFS was defined as the date of randomization to the date of first documentation of progression, symptomatic deterioration, or death due to any cause. PFS for patients last known to be alive, progression free, and free of symptomatic deterioration was censored at the date of last contact. Overall survival is defined as the date of randomization to the date of death due to any cause. Patients last known to be alive are censored at date of last contact. Overall Response Rate is defined as confirmed and unconfirmed complete and partial responses per RECIST 1.1 among patients with measurable disease at baseline. Only eligible and analyzable patients from the main clinical trial were included in this analysis.
Data availability of the secondary analysis presented in this manuscript is subject to policies found at the following links: https://nctn-data-archive.nci.nih.gov/about-us and https://www.swog.org/sites/default/files/docs/2019-12/Policy43_0.pdf
Data requests can be made to the SWOG Network Group Operations Center (https://www.swog.org/).
Results Section
EGFR mutations in ctDNA
Matched tissue was available in 89 of the 106 cases with BL blood draws, and centralized testing confirmed the reported mutation in 100% of cases (data not shown). Of the 106 cases with BL draws, 82 (77%) had detectable mEGFR in plasma ctDNA; at C3D1, 18/98 (18%) of samples had detectable ctDNA, and at progression, 54/77 (70%) had detectable ctDNA. The median, interquartile range, and range of MAFs among those with detectable ctDNA at these time points are shown in Table 1.
Baseline ctDNA: Demographics and Clinical Outcomes
Younger age, presence of brain and/or liver metastases, and M1B stage were associated with detectable mutant EGFR ctDNA at baseline (p<0.05 for all). No difference was seen in baseline detectability by EGFR mutation type (L858R versus E19del). Demographic and baseline characteristics for patients with detectable BL EGFR ctDNA versus those without detectable ctDNA are described in Table 2. The rate of BL positivity for EGFR ctDNA differed by Stage 4 subtype (based on 7th edition of lung cancer staging): patients classified as M0 (recurrent)/M1A (limited metastases) had ctDNA detectable in 22/37 cases (60%) compared to M1B (at least 2 distal sites) with 60/69 detectable (87%)(p=0.001)(Table 2). Patients with liver metastases were universally detectable at BL in this study (23/23; 100%), significantly higher compared to those without known liver metastases (59/83; 71%)(p=0.002). The presence of brain metastases was also independently and significantly associated with BL EGFR MAF detectability at 26/28 (93%) compared to those with no known brain metastases 56/78 (72%)(p=0.033).
Table 2. Patient demographics and Clinical Features.
Tabulation of patient demographics, smoking history, histology and clinical variables, broken out by EGFR ctDNA VAF positivity at baseline.
| ctDna Results | No ctDNA Results | Total | |||||
|---|---|---|---|---|---|---|---|
| (N = 106) | (N = 62) | (N = 168) | P-value | ||||
| Age | |||||||
| median (range) | 65.8 | (39, 91) | 66.9 | (28, 93) | 66.2 | (28, 93) | 0.66 |
| Sex | |||||||
| Females | 69 | (65%) | 43 | (69%) | 112 | (67%) | 0.572 |
| Males | 37 | (35%) | 19 | (31%) | 56 | (33%) | . |
| Race | |||||||
| Black | 5 | (5%) | 6 | (10%) | 11 | (7%) | 0.252 |
| Multi-Racial | 1 | (1%) | 0 | (0%) | 1 | (1%) | . |
| Native American | 0 | (0%) | 2 | (3%) | 2 | (1%) | . |
| Pacific Islander | 0 | (0%) | 1 | (2%) | 1 | (1%) | . |
| Asian | 13 | (12%) | 8 | (13%) | 21 | (13%) | . |
| Unknown | 7 | (7%) | 3 | (5%) | 10 | (6%) | . |
| White | 80 | (75%) | 42 | (68%) | 122 | (73%) | . |
| Hispanic | |||||||
| Not Hispanic | 99 | (93%) | 52 | (84%) | 151 | (90%) | 0.048 |
| Hispanic | 7 | (7%) | 10 | (16%) | 17 | (10%) | . |
| Performance Status | |||||||
| PS = 0 | 41 | (39%) | 29 | (47%) | 70 | (42%) | 0.590 |
| PS = 1 | 55 | (52%) | 28 | (45%) | 83 | (49%) | . |
| PS = 2 | 10 | (9%) | 5 | (8%) | 15 | (9%) | . |
| Smoking History | |||||||
| Current | 12 | (11%) | 2 | (3%) | 14 | (8%) | 0.072 |
| Former | 35 | (33%) | 29 | (47%) | 64 | (38%) | . |
| Never | 59 | (56%) | 31 | (50%) | 90 | (54%) | . |
| Weight Loss within 6 months prior to randomization | |||||||
| < 5% | 71 | (67%) | 43 | (69%) | 114 | (68%) | 0.67 |
| 5 - < 10% | 17 | (16%) | 11 | (18%) | 28 | (17%) | . |
| 10 - < 20% | 13 | (12%) | 8 | (13%) | 21 | (13%) | . |
| > = 20% | 1 | (1%) | 0 | (0%) | 1 | (1%) | . |
| Not reported | 4 | (4%) | 0 | (0%) | 4 | (2%) | . |
| Disease Type | |||||||
| Newly Diagnosed | 93 | (88%) | 48 | (77%) | 141 | (84%) | 0.044 |
| Recurrent | 11 | (10%) | 14 | (23%) | 25 | (15%) | . |
| Not reported | 2 | (2%) | 0 | (0%) | 2 | (1%) | . |
| Histology | |||||||
| Adenocarcinoma | 99 | (93%) | 62 | (100%) | 161 | (96%) | 0.326 |
| Squamous | 3 | (3%) | 0 | (0%) | 3 | (2%) | . |
| Mixed adeno-squamous | 2 | (2%) | 0 | (0%) | 2 | (1%) | . |
| Other NSCLC | 2 | (2%) | 0 | (0%) | 2 | (1%) | . |
| Asymptomatic Brain Metastases | |||||||
| No Brain Mets | 78 | (74%) | 42 | (68%) | 120 | (71%) | 0.42 |
| Brain Mets | 28 | (26%) | 20 | (32%) | 48 | (29%) | . |
| Liver Metastases | |||||||
| No Liver Mets | 83 | (78%) | 53 | (85%) | 136 | (81%) | 0.253 |
| Liver Mets | 23 | (22%) | 9 | (15%) | 32 | (19%) | . |
| M staging | |||||||
| M0 | 6 | (6%) | 4 | (6%) | 10 | (6%) | 0.832 |
| M1A | 31 | (29%) | 20 | (32%) | 51 | (30%) | . |
| M1B | 69 | (65%) | 38 | (61%) | 107 | (64%) | . |
| EGFR Mutation Type from Study# | |||||||
| Exon 19 | 69 | 65% | 38 | 61% | 107 | 64% | 0.620 |
| Exon 21 | 37 | 35% | 24 | 39% | 61 | 36% | . |
When patients were dichotomized by base mEGFR detectability, PFS was not significant but OS was significantly different (PFS HR = 1.46, 95% CI: 0.90 – 2.38, p = 0.12; OS HR = 2.16, 95% CI: 1.02 – 4.58, p = 0.04) (Supplementary Figure 1). After adjusting for other baseline factors: Age, M stage, Liver or Brain metastases, the adjusted HR of mEGFR detectability for PFS is 1.30, 95%CI: 0.73-2.31, p = 0.38; OS HR = 2.23, 95%CI: 0.97 - 5.15), p = 0.06. When evaluating MAF levels as a continuous variable, higher baseline EGFR MAF was associated with an increased risk of progression and death; specifically, a natural log-fold difference in MAF was associated with a 59% increased risk of progression and 53% increased risk of death (PFS HR = 1.59, 95% CI: 1.21- 2.07, p = 0.0007; OS HR = 1.53, 95%CI: 1.10 – 2.13, p = 0.0108), after adjusting for CNV (Supplementary Table 2).
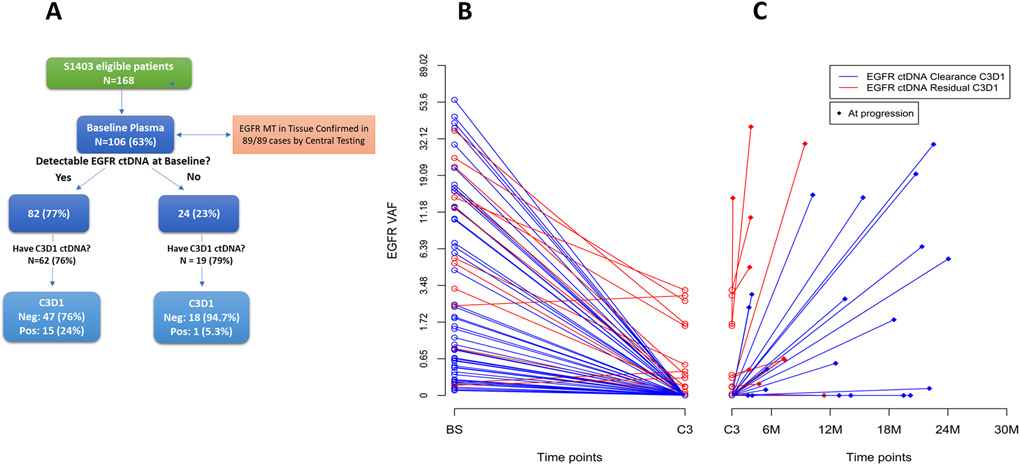
Treatment-induced changes in MAFs
The BL detectability of EGFR mutant ctDNA is summarized in Figure 1A, along with the subsets which remained detectable at C3D1. For patients with a detectable mEGFR ctDNA at BL, tracking the EGFR MAF in individual patients across the three timepoints (BL, C3D1 and PRG) typically produced a “V” shaped curve, where EGFR MAFs initially decreased on treatment and then rebounded at progression. Figure 1B-C shows treatment-induced changes in EGFR MAF over time, color coded by ctDNA clearance (blue) or residual ctDNA (red) at C3D1. Of 81 cases where both BL and C3D1 plasma were available, 62 (77%) had detectable levels at BL. Of these, 47 of 62 (76%) dropped to below the level of detection (ctDNA negative) at C3D1 (Figure 1B Blue lines), whereas 15 (19%) did not clear at C3D1 (Figure 1B Red lines). Figure 1C shows the change in EGFR MAF for all patients with both C3D1 and PRG timepoints (n = 46). Of these, 13 (28%) had detectable ctDNA at C3D1 and 32 (70%) had detectable ctDNA at PRG, 10 (22%) had detectable ctDNA at both time points and 11 (24%) had no ctDNA detectable at either time point. The median and range of mutant allele frequencies (MAFs) among those with detectable ctDNA at these time points were: baseline median: 4.52 range: 0.07 – 55.29; C3D1 median: 0.33 range: 0.02 – 3.21; PD median: 2.53 range: 0.05 – 38.00 The 36 cases with all three timepoints available are graphed in Supplementary Figure 2.
Figure 1.
A) Flow diagram of patient plasma availability and EGFR mutation positivity in ctDNA. Time course showing treatment-induced changes in EGFR MAF for B) all patients with a positive BL EGFR MAF and a C3D1 timepoint (n = 62), and C) for all patients with both C3D1 and PRG timepoints (right side, n = 46). Red lines indicate cases with residual ctDNA at C3D1; blue lines indicate cases with complete clearance at C3D1. The graph is log-transformed to highlight differences at low MAFs.
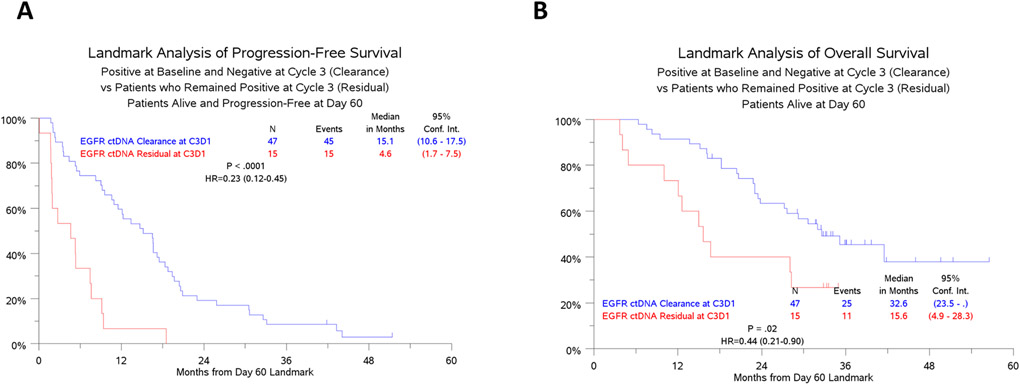
Mutant EGFR Plasma Clearance and Clinical Outcomes
Among patients with detectable EGFR ctDNA at baseline, clearance of ctDNA by C3D1 was associated with a decreased risk of progression, compared to those with persistent ctDNA at C3D1 (HR (95% CI): 0.23 (0.12-0.45), p<0.0001); with a median PFS of 15.1 (95% CI: 10.6-17.5) months in the group with clearance of ctDNA and 4.6 (1.7-7.5) months in the group with persistent ctDNA (Figure 2A). Clearance by C3D1 was also associated with a decreased risk of death (HR (95% CI): 0.44 (0.21-0.90), p=0.02); median OS was 32.6 (23.5-not estimable) months in the cleared group and 15.6 (4.9-28.3) months in the persistence group (Figure 2B) This association appears to not vary by treatment arm or EGFR mutation type (Supplementary Figure 3A-B).
Figure 2.
Kaplan-Meier analysis of A) PFS and B) OS for patients with EGFR MAF clearance (blue lines) or residual (red lines) ctDNA at C3D1 (8 weeks). All patients here were BL positive for mutant EGFR ctDNA.
The relationship between plasma clearance and response by RECIST, was evaluated in two ways: 1) at the first disease assessment coinciding with the 8-week plasma draw at C3D1, and 2) for best objective response at any time during the study. In either category, tumor response by imaging was not associated with detection or clearance of ctDNA at C3D1 (Table 3). Overall, 89 patients had baseline measurable disease on imaging; of these, 73% (65/89) had a RECIST response on study, with 53% (47/89) having the response by C3D1 (8-week assessment). The RECIST response rate by C3D1 was 64% in cases with persistent ctDNA and 53% in cases with clearance. The overall response rate by imaging any time on study was 79% in cases with persistent ctDNA and 78% in cases with clearance (Table 3).
Table 3. Relationship between RECIST response by imaging and EGFR ctDNA clearance.
Clinical Response by C3D1 (8 weeks) did not correlate with plasma clearance (p = 0.47). Clinical Response any time on study did not correlate with plasma clearance (p = 1). * The total also includes the 15 patients with no baseline ctDNA samples
| Best response by RECIST |
EGFR ctDNA at Baseline (N=74) | Overall* (N=89) | ||||
|---|---|---|---|---|---|---|
| Positive (N=59) | Negative (N=15) | |||||
| Cycle 3 Day 1: | Positive (N=14) |
Negative (N=45) |
Positive (N=1) |
Negative (N=14) |
Positive (N=17) |
Negative (N=72) |
| Response by C3D1 | 9 (64%) | 24 (53%) | 0 | 6 (43%) | 9 (53%) | 38 (53%) |
| Response on study | 11 (79%) | 35 (78%) | 0 | 9 (64%) | 11 (65%) | 54 (75%) |
In contrast to the predictive nature of ctDNA clearance, radiographic response did not appear to predict PFS. To compare the predictive utility of RECIST response rate versus plasma ctDNA clearance, RECIST response by imaging and patient survival were analyzed using the Kaplan-Meier curves. In the subgroup of patients with detectable ctDNA at baseline, imaging response by C3D1 was not associated with subsequent PFS (HR (95% CI): 0.97 (0.60-1.55), p=0.90). Overall survival was statistically different between the groups (HR (95% CI): 0.52 (0.29-0.94), p=0.03). See Supplemental Figure 4.
Plasma Molecular Response (PMR) score and Clinical Outcomes.
We also evaluated a Plasma Molecular Response (PMR) score conducted using the Guardant360 assay, and a molecular response calculation analogous to the methodology reported by Zhang et al [9] and Thompson et al [8] for assessing ctDNA molecular response score. Using the PMR score, complete molecular response was associated with a decreased risk of progression, but not significantly (HR (95% CI) = 0.67 (0.38-1.17), p = 0.16) (Supplemental Figure 5A). Complete molecular response was not associated with risk of death (HR (95% CI) =0.83 (0.42-1.65), p = 0.60) (Supplemental Figure 5B).
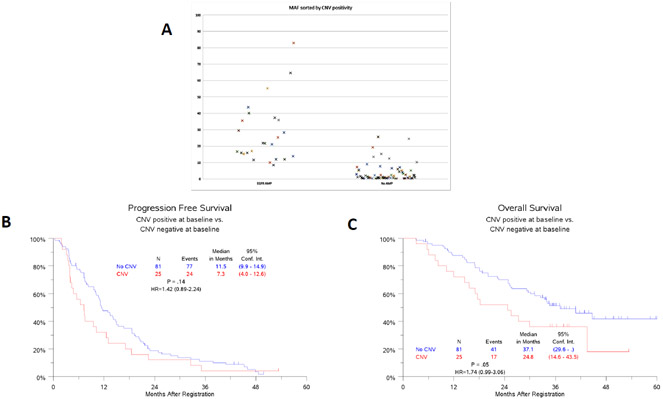
EGFR Copy Number Gain (CNG) is associated with EGFR MAF
There was a statistically significant association between cases classified as positive for EGFR CNG and elevated EGFR MAF (Figure 3A). EGFR CNG allocation is determined independently from EGFR MAF in the G360 assay. At BL, 24% (25/106) of patients were determined to have CNG specifically for the EGFR gene. Median MAF was 27.3% (8.5 - 82.9%) for CNG-positive cases and 2.1% (0 – 24.5%) for those with no detectable CNG (p < 0.0001). As shown in Figure 3B and C, patients who were positive for EGFR CNG as measured in ctDNA performed worse than those where it was absent [PFS: HR (95% CI) =1.42 (0.89-2.24), p = 0.14; OS: HR (95% CI) =1.74 (0.99-3.06), p = 0.05].
Figure 3.
EGFR Copy number Gain. A) Dot plot showing EGFR MAF (Y-axis) categories by positivity or negativity for EGFR copy number gain. B) PFS and C) OS for patients dichotomized by CNG.
Mutant TP53 ctDNA kinetics
We hypothesized that co-mutations would generally follow the same ctDNA trajectory as the primary driver mutation in EGFR following treatment. An oncoplot showing somatic mutations derived from ctDNA for all patients at baseline is shown in Supplemental Figure 6. TP53 was the most common co-mutation detected; of the 106 patients with baseline plasma, 64 (60%) were ctDNA-positive for TP53 mutations. TP53 ctDNA detection was associated with EGFR ctDNA positivity, with EGFR detected in 94% (60/64) of samples that were positive for TP53 versus 52% (22/42) of samples without TP53 detected (p <0.0001). No attempt was made in this analysis to discriminate tumor versus CHIP provenance for TP53 alterations.
Of the 81 cases with BL and C3D1 samples, TP53 mutations were detected in 46 (57%) at BL and 17 (21%) at C3D1, with 16/46 with persistent TP53 and 30/46 with TP53 clearance by C3D1 (Supplemental Figure 7A). One case was BL negative and C3D1 positive (rapid progression). Treatment-associated clearance of TP53 MAFs significantly correlated with improved PFS, (HR (95% CI) =0.44 (0.22-0.87), p = 0.01) (Supplemental Figure 7B). Patients negative for TP53 at C3D1 had significantly higher EGFR clearance rate at C3D1 (43/48, 89%) than patients with C3D1 TP53 positivity (4/14, 29%), p < 0.0001.
Discussion
In this report, we demonstrate that early treatment-associated changes in mutant EGFR ctDNA levels were highly predictive of long-term outcome for patients treated on the S1403 study in which all patients had EGFR E19del or L858R mutations in tumor tissue, were EGFR TKI-naive and received the second-generation EGFR TKI afatinib (with or without cetuximab)[13]. At baseline (prior to initiation of therapy), 77% of patients had EGFR mutations detectable in plasma ctDNA. For patients where mutant EGFR ctDNA “cleared” after 8 weeks (e.g. went from positive to undetectable), the median PFS was 15.1 months. In contrast, those who had “residual” ctDNA (e.g. started positive and stayed positive) had a median PFS of only 4.6 months, presenting a HR of 0.23 (p<0.0001). This latter group included cases where ctDNA stayed level, increased, or decreased but not to undetectable levels. The association between ctDNA clearance and PFS held true for patients receiving either afatinib alone or the combination of afatinib-plus-cetuximab, as well as for cases with EGFR E19del or L858R. ctDNA clearance proved to be far more predictive of long-term PFS than RECIST response criteria at the first imaging assessment. This predictive relationship was also observed for overall survival: those with plasma ctDNA clearance had an OS of 33 months compared to 16 months for those with residual ctDNA after 8 weeks of treatment (HR: 0.44, p=0.02).
Exploratory analysis of the FLAURA study of osimertinib found that ctDNA progression preceded or co-occurred with clinical progression, using a ddPCR approach [18]. Also using ddPCR, similar results were reported in patients treated with osimertinib in second line for T790M-positive disease, where EGFR activating mutations detectable in plasma within 8 weeks after first dose of osimertinib had significantly shorter PFS times [19].Plasma ctDNA clearance can only be assessed in patients where ctDNA MAFs are detectable at baseline. The assay used here, Guardant360, has been reported to detect ctDNA in approximately 85% of patients with advanced NSCLC, at a median VAF of 0.41 (range 0.03-97.6), with lower limits of detection at 0.04 and 0.02% MAF for SNV and deletions, respectively [15, 17]. Factors that contribute to increased shedding of ctDNA include stage, tumor burden, genomic instability, proliferation rate, extent of metastases and previous lines of therapy [20-23]. In S1403, 77% of patients had detectable EGFR mutations in plasma ctDNA, each matching the mutation detected in available tumor tissue. Previous reports have suggested that an absence of detectable ctDNA at baseline is a positive prognostic indicator [24]. In our study, all patients had EGFR mutant-positive tumor by tissue testing prior to enrollment, indicating that cases with undetectable baseline EGFR MAFs harbor low-shedding tumors. Collectively, patients with undetectable baseline EGFR MAFs had a PFS of 11.2M (95%CI: 8.2 – 15.0M). Statistically significant differences were observed between shedders and non-shedders: patients classified as M0/M1A (recurrence or limited metastases often completely localized to the lung) had significantly less frequent detection of baseline ctDNA compared to those classified as M1B (more disseminated disease with at least two distant sites) (60% vs 87%, p= 0.001). Similar results were reported by Aggarwal et al [25]. Furthermore, patients with liver and/or brain metastases had significantly higher ctDNA detection rates than those without these features. In this study, all but one patient who had undetectable EGFR MAF at baseline remained undetectable at the 8-week assessment. The one case where levels changed from undetectable to detectable at this timepoint developed progressive disease at 6.3M, died at 18.1M, with stable disease as best response.
Another factor that can affect ctDNA MAFs is gene amplification. In EGFR mutant lung adenocarcinoma, EGFR gene amplification is reported at a frequency of over 30% [26]. In liquid biopsies, EGFR CNGs are often reported in tandem with EGFR activating mutations [14]. The interpretation and clinical significance of CNG in liquid biopsies remains controversial. In the G360 assay, determination of CNG is derived from the comparison of probe-level molecular coverage based on deviation from the per-sample diploid baseline. In our study, the G360 assay reported EGFR copy number gain (CNG) in plasma in 24% of cases that were ctDNA-positive for EGFR mutations at baseline. There was a highly significant correlation between the presence of EGFR CNG and elevated EGFR MAF: those reported to have CNG had a median MAF of 27% compared to 2% for CNG-negative cases (p<0.0001). We further observed that BL EGFR CNG was associated with a significantly worse OS compared to those where it was not detected. In this study where all patients had tumors harboring an EGFR mutation and were all treated with an EGFR TKI, it is difficult to determine whether this a prognostic effect related to baseline ctDNA detection. Nevertheless, this observation provides credence to the veracity of CNG measurements in plasma ctDNA, and further has important implications to the growing field of ctDNA kinetics. For instance, a number of previously explored algorithms utilize the highest gene MAF as a basis for determining subclonality (i.e., the inference that lower MAF genes may not be truncal mutations). Such calculations need to include an assessment of CNG in order to appropriately deduce subclonality. If not accounted for, CNG may also affect algorithms that utilize multiple gene MAFs to estimate plasma response.
With regard to plasma response measurements, several recent studies have also explored the use of on-treatment changes in ctDNA levels as a predictor of patient outcome. Goldberg et al reported significant correlations between patient outcomes and ctDNA plasma response in 28 patients with metastatic NSCLC receiving immune checkpoint inhibitor therapy, concluding that decreasing ctDNA levels were predictive of prolonged patient survival. [5]. Mok et al reported significant associations between outcomes and ctDNA levels at baseline and cycle 3, suggesting a role for dynamic changes in ctDNA as a monitoring tool [4]. For metastatic NSCLC patients treated with immune checkpoint inhibitors, a series of recent studies have presented quantitative algorithms to refine the predictive utility of treatment-induced changes in ctDNA MAFs [8, 9]. These Plasma Molecular Response (PMR) algorithms use a consensus approach of all baseline and on-treatment gene variants with in silico methodologies to identify and rule out variant cell-free DNA originating from clonal hematopoiesis of indeterminate potential (CHIP) or other non-tumor sources. Both studies demonstrate significant associations between PMR and long-term patient outcomes, with further improved outcomes for patients who clear ctDNA versus those who decrease by more than 50%.
We compared a PMR scoring system from Guardant Health to the approach used here of tracking EGFR MAFs. The predictive capacity of PMR trended non-significantly with PFS (p = 0.16, HR = 0.67) when using complete clearance versus all others. In contrast to previous studies focused on the use of PMR to predict response to immune therapy, our study is limited exclusively to cases with an EGFR driver abnormality – a truncal mutation expected to be present in all tumor cell lineages. Since the majority of our patients derived some degree of benefit from treatment, a corresponding drop in tumor-associated ctDNA variants was generally observed. Furthermore, it is notable that 23% of patients had no EGFR driver mutation detected in baseline ctDNA, and EGFR-negative cases are more subject to misinterpretation due to the presence of CHIP (non-tumor) MAFs. Known CHIP-associated variants such as GNAS remained at consistent MAFs or slowly increased over time in S1403 patients. Over half of PMR “non-responders” had no mutant EGFR detectable at baseline and some did very well on study. In the EGFR-based scoring criteria, EGFR MAF-negative cases were excluded from analysis. Though several suspected CHIP-associated variants were successfully filtered out of the PMR algorithm, it is very likely that unconfirmed CHIP-associated variants were confounding elements in the PMR score of certain samples. Overall, the PMR algorithm did not perform as well in this study as has been shown previously; However, given the small number of molecular non-responders in this cohort, and the confounding prognostic implications of a lack of EGFR mutation at baseline and co-occurring CHIP, it seems possible that the lack of EGFR baseline detection limited the utility of the PMR calls in a number of patients. Additional analysis of potential CHIP-based DNA in circulation is planned for this study cohort.
Limitations of this study include the moderate sample size, missing blood draws on a subset of patients, the study of afatinib instead of the first-line standard osimertinib, and any confounding effects from the use of cetuximab administered to half of the patients. We observed no meaningful difference between those who had plasma drawn versus the entire S1403 cohort, or any effect from the addition of cetuximab, but these still represent additional variables that could dilute the study. Strengths include the verification of EGFR mutations in tumor tissue, the fact that this was a first-line study and the prospective, hypothesis-driven inclusion of serial plasma collection for the purpose of investigating plasma kinetics using a highly validated, commercially available ctDNA assay (Guardant 360).
In summary, these data, generated prospectively in a clinical trial setting, demonstrate a clear and convincing correlation between rapid clearance of detectable ctDNA and progression-free and overall survival in patients with EGFR-driven NSCLC. In this cohort, tracking the primary driver (in this case EGFR mutations) was able to identify a subset of patients where additional or alternative therapeutic options should be studied as an early intervention to improve long-term outcomes. Exploration of the value of examining co-occurring alterations when tracking response to targeted therapy needs to be further examined.
Supplementary Material
Acknowledgements:
This work was supported by National Institutes of Health/National Cancer Institute grants: U10CA180888, U10CA180819; and in part by Boehringer-Ingelheim Pharmaceuticals, Inc. and Eli Lilly and Company. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, Boehringer-Ingelheim Pharmaceuticals, Inc. (BIPI) or Eli Lilly and Company.
Footnotes
Conflict of Interest Disclosures
PCM: Honorarium Guardant, Amgen
JielingM MWR, JamesM: No reported disclosures
SBG: Research funds and honorarium from AstraZeneca and Boehringer Ingelheim, and honorarium from Bristol-Myers Squibb, Genentech, Amgen, Blueprint Medicine, Sanofi Genzyme, Daiichi-Sankyo, Regeneron, Takeda, and Janssen.
RSH: see attachment
MAM: No reported disclosures
ZW: No reported disclosures
FRH: Scientific advisory boards (compensated): AstraZeneca/Daiichi, Genentech, Sanofi/Regeneron, Merck, Bristol-Myers Squibb, Novartis, Amgen, OncoCyte, Nectin Therapeutics. Institutional Research grants (University of Colorado): Amgen, Biodesix, Rain Therapeutics
KP: co-inventor on a patent licensed to Molecular MD for EGFR(T790M) mutation testing (through MSKCC). K.P. has received Honoraria/Consulting fees from Takeda, NCCN, Novartis, Merck, AstraZeneca, Tocagen, Maverick Therapeutics, Dynamo Therapeutics, Halda and research support from AstraZeneca, Kolltan, Roche, Boehringer Ingelheim and Symphogen.
KK: Consultant or advisory role: AstraZeneca, Regeneron, Novartis, Takeda, Lilly, Amgen, EMD Serono,Genmab, Targeted Oncology, Genentech, Debiopharm Group, Abbvie, Daiichi Sanko, Janssen, Eisai, Sanofi; institutional research grants: Company: EMD Serono, Genentech, Abbvie, Regeneron, Astellas Pharma, Tizona Therapeutics, Inc., Lilly, Novartis, Amgen, Bristol-Meyers Squibb, Five Prime Therapeutics, Jounce Therapeutics, Seattle Genetics; Author Royalties for UpToDate, an evidence based, peer reviewed information resource, available via the web, desktop, and PDA. Travel or accommodations: Lilly, EMD Serono, Novartis, Takeda
DRG: Institutional Research Grants: Amgen, Astex, Genentech; Consultant/Advisory Board: Adagene, AstraZeneca (institutional), Boehringer Ingelheim, Guardant Health (institutional), IO Biotech (institutional), Merck, Novartis, Oncocyte (institutional), Oceans Genomics (institutional), Regeneron, Roche-Genentech, Sanofi
RSH: Board of Directors: Immunocore, Junshi Pharmaceuticals, Consulting: AstraZeneca, Bolt Biotherapeutics , Bristol-Myers Squibb, Candel Therapeutics, Inc., Checkpoint Therapeutics, Cybrexa Therapeutics, DynamiCure Biotechnology LLC, eFFECTOR Therapeutics Inc., Eli Lilly and Company, EMD Serono, Genentech, Gilead, HiberCell Inc., I-Mab Biopharma, Immune-Onc Therapeutics Inc., Immunocore, Janssen, Johnson and Johnson, Loxo Oncology, Mirati Therapeutics, NextCure, Novartis, Ocean Biomedical Inc., Oncocyte Corp, Oncternal Therapeutics, Pfizer, Regeneron Pharmaceuticals, Revelar Biotherapeutics Inc, Ribbon Therapeutics, Roche, Sanofi, WindMIL Therapeutics, Xencor Inc; Research Support: AstraZeneca, Eli Lilly and Company Genentech/Roche Merck and Company. Leadership roles: American Association for Cancer Research (Board Member/ Committee Chair), International Association for the Study of Lung Cancer (Board Member/ Committee Chair), Society for Immunotherapy of Cancer (Committee Chair), Southwest Oncology Group (Committee Chair/ Principal Investigator)
References
- 1.Rolfo C, et al. , Liquid Biopsy for Advanced Non-Small Cell Lung Cancer: A Consensus Statement from The International Association for the Study of Lung Cancer (IASLC). J Thorac Oncol, 2021. [DOI] [PubMed] [Google Scholar]
- 2.Leighl NB, et al. , Clinical Utility of Comprehensive Cell-free DNA Analysis to Identify Genomic Biomarkers in Patients with Newly Diagnosed Metastatic Non-small Cell Lung Cancer. Clin Cancer Res, 2019. 25(15): p. 4691–4700. [DOI] [PubMed] [Google Scholar]
- 3.Diehl F, et al. , Circulating mutant DNA to assess tumor dynamics. Nat Med, 2008. 14(9): p. 985–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mok T, et al. , Detection and Dynamic Changes of EGFR Mutations from Circulating Tumor DNA as a Predictor of Survival Outcomes in NSCLC Patients Treated with First-line Intercalated Erlotinib and Chemotherapy. Clin Cancer Res, 2015. 21(14): p. 3196–203. [DOI] [PubMed] [Google Scholar]
- 5.Goldberg SB, et al. , Early Assessment of Lung Cancer Immunotherapy Response via Circulating Tumor DNA. Clin Cancer Res, 2018. 24(8): p. 1872–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hellmann MD, et al. , Circulating Tumor DNA Analysis to Assess Risk of Progression after Long-term Response to PD-(L)1 Blockade in NSCLC. Clin Cancer Res, 2020. 26(12): p. 2849–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anagnostou V, et al. , Dynamics of Tumor and Immune Responses during Immune Checkpoint Blockade in Non-Small Cell Lung Cancer. Cancer Res, 2019. 79(6): p. 1214–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson JC, et al. , Serial Monitoring of Circulating Tumor DNA by Next-Generation Gene Sequencing as a Biomarker of Response and Survival in Patients With Advanced NSCLC Receiving Pembrolizumab-Based Therapy. JCO Precis Oncol, 2021. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Q, et al. , Prognostic and Predictive Impact of Circulating Tumor DNA in Patients with Advanced Cancers Treated with Immune Checkpoint Blockade. Cancer Discov, 2020. 10(12): p. 1842–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou W, et al. , ctDNA Predicts Overall Survival in Patients With NSCLC Treated With PD-L1 Blockade or With Chemotherapy. JCO Precis Oncol, 2021. 5: p. 827–838. [DOI] [PubMed] [Google Scholar]
- 11.Janjigian YY, et al. , Dual inhibition of EGFR with afatinib and cetuximab in kinase inhibitor-resistant EGFR-mutant lung cancer with and without T790M mutations. Cancer Discov, 2014. 4(9): p. 1036–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pirazzoli V, et al. , Afatinib plus Cetuximab Delays Resistance Compared to Single-Agent Erlotinib or Afatinib in Mouse Models of TKI-Naive EGFR L858R-Induced Lung Adenocarcinoma. Clin Cancer Res, 2016. 22(2): p. 426–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldberg SB, et al. , Randomized Trial of Afatinib Plus Cetuximab Versus Afatinib Alone for First-Line Treatment of EGFR-Mutant Non-Small-Cell Lung Cancer: Final Results From SWOG S1403. J Clin Oncol, 2020. 38(34): p. 4076–4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mack PC, et al. , Spectrum of driver mutations and clinical impact of circulating tumor DNA analysis in non-small cell lung cancer: Analysis of over 8000 cases. Cancer, 2020. 126(14): p. 3219–3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lanman RB, et al. , Analytical and Clinical Validation of a Digital Sequencing Panel for Quantitative, Highly Accurate Evaluation of Cell-Free Circulating Tumor DNA. PLoS One, 2015. 10(10): p. e0140712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zill OA, et al. , The Landscape of Actionable Genomic Alterations in Cell-Free Circulating Tumor DNA from 21,807 Advanced Cancer Patients. Clin Cancer Res, 2018. 24(15): p. 3528–3538. [DOI] [PubMed] [Google Scholar]
- 17.Odegaard JI, et al. , Validation of a Plasma-Based Comprehensive Cancer Genotyping Assay Utilizing Orthogonal Tissue- and Plasma-Based Methodologies. Clin Cancer Res, 2018. 24(15): p. 3539–3549. [DOI] [PubMed] [Google Scholar]
- 18.Reungwetwattana T, et al. , Longitudinal circulating tumour DNA (ctDNA) monitoring for early detection of disease progression and resistance in advanced NSCLC in FLAURA. Annals of Oncology, 2019. 30. [Google Scholar]
- 19.Buder A, et al. , EGFR mutation tracking predicts survival in advanced EGFR- mutated non-small cell lung cancer patients treated with osimertinib. Translational Lung Cancer Research, 2020. 9(2): p. 239–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho MS, et al. , Clinicopathological parameters for circulating tumor DNA shedding in surgically resected non-small cell lung cancer with EGFR or KRAS mutation. PLoS One, 2020. 15(3): p. e0230622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chabon JJ, et al. , Integrating genomic features for non-invasive early lung cancer detection. Nature, 2020. 580(7802): p. 245–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lam VK, et al. , Genotype-Specific Differences in Circulating Tumor DNA Levels in Advanced NSCLC. J Thorac Oncol, 2021. 16(4): p. 601–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abbosh C, et al. , Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature, 2017. 545(7655): p. 446–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gray JE, et al. , Tissue and Plasma EGFR Mutation Analysis in the FLAURA Trial: Osimertinib versus Comparator EGFR Tyrosine Kinase Inhibitor as First-Line Treatment in Patients with EGFR-Mutated Advanced Non-Small Cell Lung Cancer. Clin Cancer Res, 2019. 25(22): p. 6644–6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aggarwal C, et al. , Clinical Implications of Plasma-Based Genotyping With the Delivery of Personalized Therapy in Metastatic Non-Small Cell Lung Cancer. JAMA Oncol, 2019. 5(2): p. 173–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruiz-Patino A, et al. , EGFR Amplification and Sensitizing Mutations Correlate with Survival in Lung Adenocarcinoma Patients Treated with Erlotinib (MutP-CLICaP). Target Oncol, 2018. 13(5): p. 621–629. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.