Abstract
An xps gene cluster composed of 11 open reading frames is required for the type II protein secretion in Xanthomonas campestris pv. campestris. Immediately upstream of the xpsD gene, which encodes an outer membrane protein that serves as the secretion channel by forming multimers, there exists an open reading frame (previously designated ORF2) that could encode a protein of 261 amino acid residues. Its N-terminal hydrophobic region is a likely membrane-anchoring sequence. Antibody raised against this protein could detect in the wild-type strain of X. campestris pv. campestris a protein band with an apparent molecular mass of 36 kDa by Western blotting. Its aberrant slow migration in sodium dodecyl sulfate-polyacrylamide gels might be due to its high proline content. We designated this protein XpsN. By constructing a mutant strain with an in-frame deletion of the chromosomal xpsN gene, we demonstrated that it is required for the secretion of extracellular enzyme by X. campestris pv. campestris. Subcellular fractionation studies indicated that the XpsN protein was tightly associated with the membrane. Sucrose gradient sedimentation followed by immunoblot analysis revealed that it primarily appeared in the cytoplasmic membrane fractions. Immune precipitation experiments indicated that the XpsN protein was coprecipitated with the XpsD protein. In addition, the XpsN protein was co-eluted with the (His)6-tagged XpsD protein from the metal affinity chromatography column. All observations suggested that the XpsN protein forms a stable complex with the XpsD protein. In addition, immune precipitation analysis of the XpsN protein with various truncated XpsD proteins revealed that the C-terminal region of the XpsD protein between residues 650 and 759 was likely to be involved in complex formation between the two.
The type II secretion pathway is widespread among gram-negative bacteria (48, 49). It is also known as the main terminal branch of the general secretory pathway (38). In all cases, the secreted proteins possess a typical N-terminal signal sequence, which is processed while being exported across the inner membrane. Between 11 and 14 genes are required for the second step of the secretion pathway. Each encodes a protein that has amino acid sequence homology to different degrees with other proteins of the same family. An outer membrane protein belonging to the GspD protein family has been demonstrated to form multimeric complex (2, 7, 25, 31) and suggested to serve as a gated secretion channel (33). A second outer membrane-associated protein, GspS (PulS in the case of Klebsiella oxytoca), was recently demonstrated to be part of the GspD secretion channel (36). Four pseudopilin proteins belonging to the GspG, -H, -I, -J families were located mainly in the inner membrane (37, 43). Bleves et al. (5) have identified XcpX, the GspK homologue of Pseudomonas aeruginosa, as a fifth pseudopilin. The GspG proteins have been demonstrated to form either homodimers or heterodimers with the GspH-, -I, -J proteins, and the pilin subunit PilA protein in the case of P. aeruginosa, from cross-linking studies (32, 39). Five other inner membrane proteins include those belonging to the GspC, GspF, GspL, GspM, and GspN families. Secondary structure prediction indicated that these proteins possess either one (GspC, -L, -M, -N) or three (GspF) membrane-spanning sequences. Topological analysis of either PhoA or BlaM fusions confirmed such predictions (4, 42, 54). The GspE protein family, with a characteristic nucleotide-binding motif, was demonstrated to have autokinase activity (50). Various protein-protein interactions among different Gsp proteins have been demonstrated in several cases (32, 34, 50, 52). A multicomponent model was proposed for the type II secretory machinery of P. aeruginosa by Filloux et al. (15).
The two out gene clusters of Erwinia chrysanthemi and Erwinia carotovora share strong sequence homology and gene organization. However, the outN gene is absent in the former. Neither is the gene encoding the GspN homologue present in the P. aeruginosa xcp gene cluster, while it is encoded by most other type II secretion gene clusters (19, 40, 41, 51). Pairwise amino acid sequence alignments among the GspN proteins revealed the highest sequence identities of 34.7% between the PulN protein of K. oxytoca (accession no. P15753) and the OutN protein of E. carotovora (accession no. P31710) and 39% between the ExeN protein of Aeromonas hydrophila (accession no. P41852) and the EpsN protein of Vibrio cholerae (accession no. P45784). In contrast, with the same comparison parameters (blosum 62; gap creation of 8 and gap extension of 2 [18]), the sequence identity between PulD of K. oxytoca (accession no. P15644) and OutD of E. carotovora (accession no. P31701) is 72.5% and that between ExeD of A. hydrophila (accession no. P31780) and EpsD of V. cholerae (accession no. P45779) is 55.8%. These analyses suggest that the amino acid sequence of the GspN protein is probably not as well conserved as that of the GspD protein. Upstream of the xpsD gene in Xanthomonas campestris pv. campestris, we have previously identified two open reading frames (ORFs) (ORF1 and ORF2 [20]). ORF2 could encode a protein of 261 or 257 amino acid residues. We tentatively assigned the first in-frame ATG as the initiation codon (Fig. 1), because we located a putative Shine-Dalgarno sequence—GGAGG—6 nucleotides upstream of it. Searching in the GenBank database with filters to exclude input sequences with low complexity or repeat regions (1) retrieved no other protein. Its N-terminal amino acid sequence resembles a likely membrane-anchoring sequence (Fig. 1). Such analysis raised our interest to determine the significance of ORF2 in extracellular protein secretion in X. campestris pv. campestris. We designate it the xpsN gene for the following reasons. Its location in the xps gene cluster and the predicted molecular weight of its putative protein product are similar to those of all gspN genes of the type II protein secretion pathway. Upstream of ORF2, we have identified nine ORFs (unpublished results; accession no. L02630 and M81648), the deduced amino acid sequences of which have sequence homologies to various extents, as well as the same gene order, with the pulEFGHIJKLM genes of K. oxytoca. The first five have been designated the xpsEFGHI genes by Dums et al. (12). Furthermore, as discussed previously, the amino acid sequence homology among all known GspN proteins is not as high as other Gsp protein families.
FIG. 1.
Deduced amino acid sequence of the XpsN protein. The underlined portion is a likely membrane-anchoring sequence.
In this study, we constructed an in-frame deletion mutant of the xpsN gene on the chromosome of X. campestris pv. campestris. Subcellular fractionation of α-amylase revealed that it is accumulated in the periplasm of the xpsN mutant strain. Sucrose gradient sedimentation followed by immunoblot analysis of the parental strain showed that the XpsN protein is a cytoplasmic membrane protein. However, coimmune precipitation and affinity chromatography indicated that it may be associated with the outer membrane protein XpsD. The results suggested that the XpsN protein is probably involved in the type II secretion process by its association with the secretion channel located in the outer membrane.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Escherichia coli BL21(DE3) was a kind gift from F. W. Studier. X. campestris pv. campestris XC1701 was originally isolated as a spontaneous Rifr mutant from a wild isolate, XC17. XC1708 and XC17433 are both secretion mutants generated from Tn5 mutagenesis (20). Plasmids used are summarized in Table 1. Plasmid pNC2, expressing the xpsN gene, was obtained by cloning a 1.1-kb SacII-BclI fragment from pKC107 into the broad-host-range vector pCPP30 digested with HindIII and BamHI. A HindIII linker was ligated to the SacII-cleaved end that had been treated with ExoIII/S1 nuclease. DNA sequencing results revealed that 14 bp were removed from the SacII-cleaved end. To construct plasmid pSYP9, for expressing the XpsD(His)6 protein, we made use of the C-terminal (His)6 codon in the expression plasmid pET21b (Novagen), into which the xpsD coding sequence was introduced in three steps. We first generated an XhoI site upstream of the stop codon of the xpsD gene by performing PCR of the 3′-end fragment of the xpsD gene using the 5′ primer pSUDA6 (5′-GATGTTTCGGGTAGCGT-3′) and the 3′ primer pXhoI (5′-CTAGCGAATTACTCGAGTTTATGAACATCC-3′). By cloning the PCR product digested with BamHI and XhoI into pET21b, we obtained plasmid pX. In a similar way, an NdeI site was created upstream of the initiation codon of the xpsD gene. The 5′-end fragment of the xpsD gene was reproduced upon PCR using the 5′ primer pNdeI (5′-CAGACCCATATGAGTGA-3′) and the 3′ primer pR21 (5′-GTTCGGGCGCGCGTACGG-3′). The PCR fragment digested with NdeI and BamHI was then cloned into pX, giving rise to plasmid pXN. In the third step, a central BamHI fragment of the xpsD gene was introduced into pXN, producing plasmid pXNB. To introduce the xpsD(His)6 gene into a broad-host-range vector, we generated plasmid pCXNB15 by inserting XbaI-linearized pXNB into pCPP30. The pET21b moiety was subsequently removed from pCXNB15 by digestion with EcoRI and BlpI, followed by a filling-in reaction and self-ligation, producing plasmid pCXNB9. The XpsD(His)6 protein produced from pCXNB9 was low in abundance. For an unknown reason, when we replaced the 3′-end fragment of the xpsD gene in pCXNB9 with an MluI-EcoRI fragment generated upon PCR using the 5′ primer pSUDA6 and the 3′ primer pETEH (5′-GGGAATTCAGCAGCCAACTCAGCTTC-3′) to create plasmid pSYP9, the abundance of the XpsD(His)6 protein produced in X. campestris pv. campestris was increased. All PCR fragments were confirmed to be unaltered with DNA sequencing data.
TABLE 1.
Plasmids
| Plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| pCPP30 | Tra− Mob+ Tetr IncP replicon, with multiple cloning sites downstream of Plac | D. W. Bauer |
| pET-3d | Ampr, PT7-containing derivative of pBR322 | F. W. Studier |
| pET21b | Ampr, PT7-containing expression vector for expressing C-terminal (His)6 tag fusion | Novagen |
| pUCD4121 | Camr Sucs, derivative of pTZ18R | C. I. Kado |
| pMH7 | XpsD(Δ414–759) expressed from Plac on pCPP30 | 7 |
| pKdPs | XpsD(Δ448–650) expressed from Plac on pCPP30 | 7 |
| pKDT | XpsD(Δ553–759) expressed from Plac on pCPP30 | 7 |
| pCD105 | XpsD(Δ29–428) expressed from Plac on pCPP30 | 7 |
| pYL4 | xpsD(Δ74–303) derivative of pKC107 | 7 |
| pKC107 | xps′JKLMND expressed from Plac on pCPP30 | Present study |
| pKC108 | xpsN(R34L) derivative of pKC107 | Present study |
| pKC111 | xpsN(Δ34–131) derivative of pKC107 | Present study |
| pKC114 | xpsN(G132L) derivative of pKC107 | Present study |
| pKC118 | XpsD expressed from Plac on pCPP30 | Present study |
| pYL2 | XpsN(Δ1–21) expressed from PT7 on pET-3d | Present study |
| pNC2 | XpsN expressed from Plac on pCPP30 | Present study |
| pSYP9 | XpsD(His)6 expressed from Plac on pCPP30 | Present study |
| pUKM | XbaI-SalI fragment of 5.5 kb from pKC111 subcloned in pUCD4121 | Present study |
Production of antibody against the XpsN protein.
An N-terminally truncated XpsN protein was overexpressed from the T7 promoter located on the T7 expression vector pET-3d (44). The overexpression plasmid pYL2 was constructed by cloning the xpsN gene from the 22nd amino acid of the XpsN protein in-frame into the filled-in NcoI site of pET-3d. E. coli BL21(DE3)(pYL2), grown in 2YT medium (tryptone, 1.6 g per liter; yeast extract, 1 g per liter; NaCl, 1 g per liter) with 0.4% glucose to an optical density at 600 nm (OD600) of 0.6 to 0.8, was induced with the addition of 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and incubated at 37°C for 50 min. Rifampin was then added at a final concentration of 100 μg/ml before incubation at 37°C for two more hours. Total cell lysates were prepared by treatment with lysozyme followed by sonication, as described by Hu et al. (22). The major protein band was collected from a sodium dodecyl sulfate (SDS)-polyacrylamide gel for immunization of a rabbit. Sera were prepared by the procedures of Harlow and Lane (16). Immunoblot analysis indicated that the antiserum could detect in X. campestris pv. campestris XC1701 a protein with an apparent molecular mass of 36 kDa that was absent in XC17433, in which the entire xps gene cluster was deleted (20).
Construction of the xpsN mutant strain.
An in-frame deletion mutant xpsN gene that produced a truncated protein, XpsN(Δ34–131), was constructed by first introducing a single HindIII site at two respective sites via site-directed mutagenesis. Plasmids pKC108 and pKC114 contain an upstream and a downstream HindIII site, respectively. Recombination of the upstream fragment from pKC108 with the downstream fragment from pKC114 generated plasmid pKC111, which produced a truncated XpsN protein migrating in SDS-polyacrylamide gel electrophoresis (PAGE) with an apparent molecular mass of approximately 25 kDa.
Subsequently, the deleted xpsN gene was introduced into the X. campestris pv. campestris genome by the procedures of Kamoun et al. (23) with slight modifications. An XbaI-SalI fragment of 5.5 kb containing the deleted xpsN gene was subcloned from pKC111 into a narrow-host-range plasmid, pUCD4121, that contains a cat gene for selecting the primary recombination event that integrates the plasmid into the genome and a sacB gene for selecting the secondary recombination event that excises the wild-type xpsN gene from the genome. The recombinants from the primary recombination event appeared at a much lower frequency than that of spontaneously arising chloramphenicol-resistant colonies. In order to overcome this problem, a kanamycin resistance gene cassette was introduced at the SalI site for primary selection. Despite the low frequency, we were able to obtain a recombinant that was resistant to both kanamycin and chloramphenicol but sensitive to sucrose, following electroporation of the recombinant plasmid pUKM, which contained the deleted xpsN gene, into XC1701. Electroporation procedures of Chen et al. (7) were followed. By incubating the recombinant X. campestris pv. campestris in LB with shaking at 30°C for 4 h, cells were plated on LA plus 5% sucrose. Colonies that were able to grow on sucrose were streaked on plates containing 50 μg of kanamycin per ml. Only those that became sensitive to kanamycin were picked and analyzed for α-amylase secretion on a starch plate. Approximately half of them were defective in secretion. PCR analysis using the upstream primer pMX (5′-GCAATGCGCCTTGAAATGATCGGCCTGCGC-3′) and the downstream primer pXA-1 (5′-TCGGCGCACGTCCGGTGGCGGAGTGGTGG-3′) confirmed that indeed only the deleted xpsN gene was present in the genome. We designated this strain XC1709.
Preparation of total membrane and its washing with NaCl or urea.
Total membrane was prepared following the procedures of Hu et al. (22). For every 100-ml culture grown to an OD600 of 1, the membrane from ultracentrifugation was suspended in 6 ml of 10 mM HEPES (pH 7.5). One-milliliter aliquots were mixed with NaCl or urea stocks made in the same buffer to respective final concentrations and incubated on ice for 20 min before centrifugation at 65,000 × g for 30 min. Both the supernatant concentrated upon trichloroacetic acid (TCA) precipitation and the pellet were analyzed by SDS-PAGE, followed by immunoblot analysis.
Sucrose gradient sedimentation analysis.
Total membrane was analyzed on a step sucrose gradient (25 to 61% [wt/wt]) by the procedures of Hu et al. (22). Each fraction was analyzed for density (determined from the refractive index) and for distribution of the XpsN and OprF (OmpA homologue) proteins by immunoblotting of TCA-precipitated samples. Upon dilution, each fraction was also tested for succinate dehydrogenase activity by the procedures of Kasahara and Anraku (24). One unit of succinate dehydrogenase was defined as the amount of enzyme catalyzing reduction of 1 nmol of DCPIP (2,6-dichlorophenol-indophenol) in 1 min at room temperature.
Ni-NTA affinity chromatography.
Membrane protein extract was prepared by mixing total membrane with 2% deoxycholate at 4°C overnight, followed by ultracentrifugation at 50,000 rpm (Beckman TLA 100.3) for 1 h. It was subsequently mixed with 1 ml of Ni-nitrilotriacetic acid (NTA) resin (Qiagen), which had been preequilibrated with a 15× volume of balance buffer (10 mM Tris-HCl [pH 8.0], 0.1% deoxycholate, 100 mM NaCl), in the presence of 10 mM imidazole and 1 mM phenylmethylsulfonyl fluoride at room temperature for 2 h. The resin mixture was then loaded on a 10- by 1-cm-diameter column (Bio-Rad) with the flow rate set at 0.5 ml/min. Following sequential washings with a 15× volume of washing buffer A (10 mM Tris-HCl [pH 8.0], 0.1% deoxycholate, 500 mM NaCl) and washing buffer B (washing buffer A plus 50 mM imidazole), the bound protein was eluted with elution buffer (washing buffer A plus 250 mM imidazole) and collected at 1.5 ml/fraction. Proteins in each fraction collected as flowthrough, washings, and eluates were precipitated with cold 10% TCA and analyzed by SDS-PAGE, followed by immunoblotting for the XpsN and XpsD(His)6 or XpsD proteins.
Coimmune precipitation.
Total membrane extracted with 2% Triton X-100 made in 10 mM Tris-HCl (pH 8.0)–10 mM EDTA at 4°C overnight was centrifuged at 11,000 × g for 30 min. The collected supernatant was mixed with antibody against XpsN (at a 1:40 dilution) or XpsD (at a 1:80 dilution) in Triton buffer (2% Triton X-100, 10 mM Tris-HCl [pH 8.0], 200 mM NaCl, 10 mM EDTA) and incubated overnight at 4°C, followed by incubation with protein A-Sepharose CL-4B (10% [wt/vol] in Triton buffer) at room temperature for 1 h. After centrifugation at 700 × g for 5 min, the pellet was washed two or three times with Triton buffer containing 200 mM NaCl and once with Triton buffer alone before being resuspended in electrophoresis sample buffer. Two types of antibody against the XpsD protein were used in this study. One (designated anti-XpsDN antibody), against a LacZ-XpsD (residues 37 to 589) fusion protein (22), was used in detecting the full-length and C-terminally truncated XpsD proteins. The other (designated anti-XpsDC antibody) was prepared by immunizing rabbits with a truncated XpsD protein containing residues 429 to 759 (7) and was used for detecting N-terminally truncated XpsD proteins.
Immunoblot analysis.
Procedures of Hu et al. (22) were followed for immunoblot analysis. Antibody against the XpsN protein was added at a 1:250 dilution, that against XpsDN at a 1:1,000 dilution, and that against α-amylase (21) at a 1:1,000 dilution. Antibody against the E. coli OmpA protein, a kind gift from U. Henning, was included at a 1:10,000 dilution.
RESULTS
XpsN is required for α-amylase secretion.
We constructed an in-frame deletion mutant xpsN gene on a narrow-host-range vector, pUCD4121, that can not replicate in X. campestris, introduced it into the parental strain XC1701, and obtained an xpsN mutant strain, XC1709. PCR analysis indicated that the wild-type xpsN gene on the chromosome was replaced with the deleted gene (data not shown). Introduction of plasmid pNC2, which encodes a wild-type xpsN gene alone, could complement the secretion defect in XC1709. This result suggested that the deletion introduced into the xpsN gene did not have a polar effect on downstream xpsD gene expression.
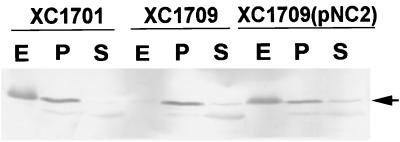
By analyzing α-amylase secretion on starch plates, we found that the clear zone produced by the xpsN mutant strain XC1709 was much smaller than that produced by the parental strain (data not shown). We further conducted subcellular fractionation and examined α-amylase distribution by immunoblot analysis. The α-amylase was no longer detectable in the extracellular fraction of the mutant strain XC1709. Instead, it appeared in the periplasm (Fig. 2). When the wild-type xpsN gene encoded by plasmid pNC2 was reintroduced into XC1709, α-amylase reappeared in the extracellular fraction. These results suggested that the XpsN protein is indeed required for α-amylase secretion across the outer membrane. In the periplasmic fraction of XC1701 and XC1709(pNC2), α-amylase was detected in significant amounts. It may represent extracellular α-amylase that remained cell-associated. We have detected cell-bound α-amylase by immunofluorescence labeling of intact cells with antibody against α-amylase in a wild-type strain, but not in a nonsecretive mutant (22). In support of this suggestion, proteinase K treatment of intact cells followed by immunodetection of α-amylase revealed that the amount of cell-associated α-amylase of the wild-type strain XC1701 and the complemented mutant strain XC1709(pNC2) was reduced with increasing concentrations of proteinase K and remained unchanged in the mutant strain XC1709 (data not shown).
FIG. 2.
Immunoblot analysis of subcellular distribution of α-amylase in XC1701 (xpsN+), XC1709 (xpsN), and XC1709(pNC2). Plasmid pNC2 contains the wild-type xpsN gene cloned in the broad-host-range vector pCPP30. Equivalent amounts of material were loaded in each lane. E, extracellular fraction; P, periplasmic fraction; S, spheroplast fraction. Antibody against α-amylase was included at a 1:1,000 dilution.
XpsN is tightly membrane associated.
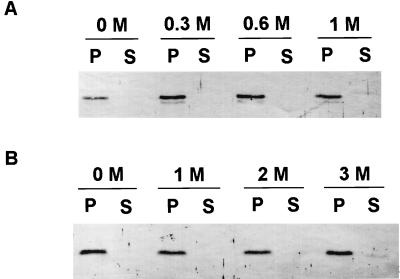
Examination of the amino acid sequence of the XpsN protein revealed a likely membrane-anchoring sequence (Fig. 1). However, immediately downstream of the hydrophobic amino acid residues there appears the amino acid sequence AVA, similar to an N-terminal signal peptide cleaved by signal peptidase I (58). When we examined the distribution of the XpsN protein in different subcellular fractions, we observed that it cofractionated with the membrane fraction. Furthermore, the XpsN protein remained membrane-bound after washings with up to 1 M NaCl or 2 M urea (Fig. 3). Only a minor portion appeared in the supernatant after washing with 3 M urea (Fig. 3B).
FIG. 3.
Effects of washing with NaCl (A) or urea (B) on membrane association of the XpsN protein in XC1701. S, supernatants from ultracentrifugation at 65,000 × g for 30 min; P, pellets from ultracentrifugation. Antibody against the XpsN protein was included at a 1:250 dilution. Molar concentrations of NaCl and urea are indicated above the respective panels.
XpsN localizes mainly in the cytoplasmic membrane.
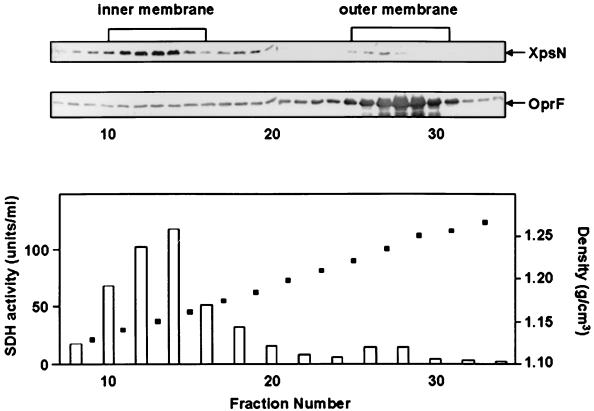
When we analyzed the XpsN protein by sucrose gradient sedimentation, we found that the major peak of the XpsN protein appeared in fractions 10 to 15, cofractionating with the peak of the cytoplasmic membrane marker succinate dehydrogenase (Fig. 4). Some weak but reproducible XpsN signals were also detectable in fractions 25 to 27, which overlapped the major peak of the outer membrane protein OprF appearing in fractions 25 to 31. Likewise, a very small but discernible peak of succinate dehydrogenase activity was observed in fractions 26 to 28. The proportion of the XpsN protein detected in the outer membrane fractions is no greater than that of succinate dehydrogenase in the same fractions. Moreover, similar distributions of the XpsN protein and succinate dehydrogenase activity were observed in the xpsD mutant strain XC1708 (data not shown), suggesting that appearance of the XpsN protein in the outer membrane fractions is probably unrelated to the XpsD protein.
FIG. 4.
Sucrose gradient sedimentation analysis of the XpsN protein in XC1701 on a 25 to 61% (wt/wt) sucrose gradient. The top two panels are immunoblots of fractions 7 to 34 collected from the gradient (top left) detected with antibody against the XpsN protein and with antibody against the E. coli OmpA protein (OprF). In the bottom panel, open bars represent succinate dehydrogenase (SDH) activity and solid squares represent density.
XpsN coimmunoprecipitates with XpsD.
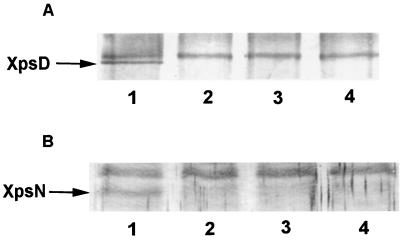
We conducted immune precipitation experiments by preparing Triton X-100–EDTA extracts of total membrane from various strains. When we conducted immune precipitation with anti-XpsN antibody and probed the blot with antibody against the XpsD protein, a protein band with an apparent molecular mass of 80 kDa was detected in XC1701 (Fig. 5A, lane 1) but not in XC1708 (xpsD mutant) or XC1709 (xpsN mutant) (Fig. 5A, lanes 2 and 3). Nor was it detectable in the negative control (Fig. 5A, lane 4), where membrane extract was omitted from the entire immune precipitation process. When we reversed the immune precipitation process by precipitating with antibody against the XpsD protein and probed with antibody against the XpsN protein, we observed a protein band migrating at a distance where the XpsN protein was expected in XC1701 but not in XC1708, XC1709, or the negative control (Fig. 5B). These results suggested that the XpsN protein may form a stable complex with the outer membrane protein XpsD. The bands migrating behind the XpsD or XpsN protein probably represent cross-reactive materials, since they appeared in all samples.
FIG. 5.
Coimmune precipitation of XpsN with XpsD in the parental strain XC1701. (A) Triton X-100 membrane extracts precipitated with antibody against the XpsN protein and detected on a Western blot with anti-XpsDN antibody (1:1,000 dilution). (B) Triton X-100 membrane extracts precipitated with anti-XpsDN antibody and detected on a Western blot with antibody against the XpsN protein (1:250 dilution). Samples were loaded in the following order: lane 1, XC1701 (xpsN+ xpsD+); lane 2, XC1708 (xpsN+ xpsD); lane 3, XC1709 (xpsN xpsD+); lane 4, no membrane extract.
XpsN cofractionates with XpsD(His)6 upon affinity chromatography.
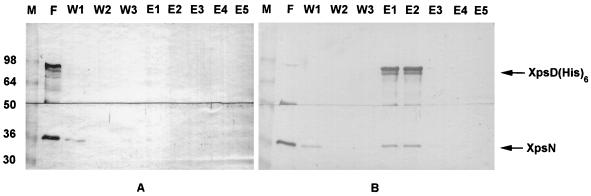
In order to confirm the interactive relationship between the XpsN and XpsD proteins, we made use of the specific binding property of the (His)6-tagged XpsD protein with a Ni-NTA column and examined the eluted protein profile with antibodies. Our unpublished results indicated that the C-terminally (His)6-tagged XpsD protein could complement the xpsD mutant XC1708 as well as the wild-type XpsD protein. As observed in Fig. 6, both the XpsD(His)6 and the XpsN proteins bound to the Ni-NTA column were eluted simultaneously in the first and second fractions. In these fractions, two protein bands detected with antibody against the XpsD protein were observed. The faster-migrating band may be a degradation product of the XpsD(His)6 protein, since it is not detected in any other fractions. Neither did it appear in other experiments. The control experiment indicated that without the (His)6 tag, neither XpsD nor XpsN was bound to the column or eluted with 250 mM imidazole. The XpsN protein that was coeluted with the XpsD(His)6 protein constitutes approximately 2.8% of the total XpsN protein recovered from the column in all fractions. Considering that approximately 30% of the XpsD(His)6 protein was detected in the flowthrough, we estimate the proportion of the XpsN protein cofractionating with the XpsD(His)6 protein to be approximately 4%.
FIG. 6.
Elution profiles of the XpsN and XpsD(His)6 proteins from the Ni-NTA affinity chromatography column. Deoxycholate membrane extracts were prepared from XC1708(pKC118) (A) and XC1708(pSYP9) (B). F, flowthrough; W1 to W3, fractions collected from washings with washing buffer A (W1) or washing buffer B (W2, W3); E1 to E5, fractions eluted with elution buffer; M, molecular size markers (sizes, in kilodaltons, are shown at left); top panels, immunodetection with anti-XpsDN antibody; bottom panels, immunodetection with antibody against the XpsN protein. Semiquantitative estimation of the relative amounts of proteins detected by immunoblotting was carried out by densitometric analysis using Bio Image Intelligent Quantifier software on a Sun workstation. The amounts of protein samples loaded in E fractions are approximately 20 times those loaded in F fractions and 10 times those loaded in W fractions.
The C-terminal domain of XpsD is involved in its association with XpsN.
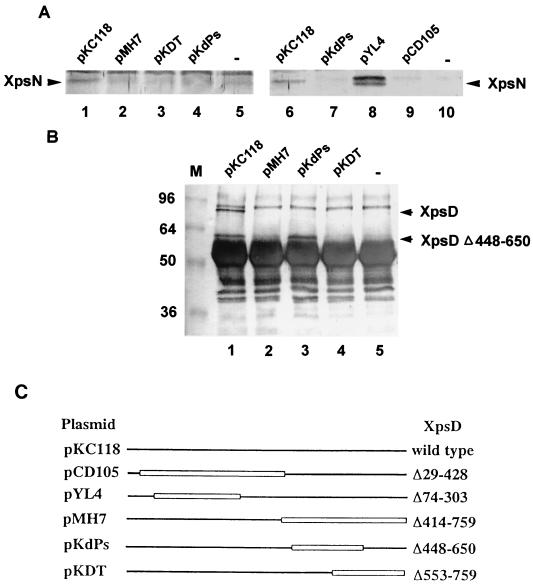
In order to locate the region of the XpsD protein that is involved in its interaction with the XpsN protein, we performed immune precipitation experiments using various XC1708 (xpsD mutant) strains carrying plasmids that express different truncated XpsD proteins. By performing immune precipitation with antibody against the XpsD protein, we observed that the XpsN protein coprecipitated with the full-length XpsD protein encoded by pKC118 (Fig. 7A, lanes 1 and 6). In contrast, precipitation with antibody against the XpsD protein did not give rise to the XpsN signal in any of the three strains containing plasmid pMH7, pKdPs, or pKDT (Fig. 7A, lanes 2 to 4 and 7). As shown in Fig. 7C, each of them encodes a C-terminally truncated XpsD protein with expected molecular sizes of 43, 59, and 58 kDa, respectively. On the other hand, the strain containing pYL4 clearly gave XpsN signals (Fig. 7A, lane 8), one of which may be a degradation product. The plasmid encodes an N-terminally truncated XpsD protein with an expected molecular size of 56 kDa (Fig. 7C). The XpsN signal in the strain containing pCD105, which encodes another N-terminally truncated XpsD protein, with an expected molecular size of 39 kDa, was barely visible (Fig. 7A, lane 9). It was probably the result of the protein level of the latter which was lower than that of the former (data not shown).
FIG. 7.
Coimmune precipitation of XpsN with various truncated XpsD proteins produced from plasmid-encoded genes in XC1708. (A) Triton X-100 membrane extracts precipitated with anti-XpsDN antibody (lanes 1 to 5) or with anti-XpsDC antibody (lanes 6 to 10) and detected on the blot with antibody against the XpsN protein (1:250 dilution). (B) Triton X-100 membrane extracts precipitated with antibody against the XpsN protein and detected on the blot with anti-XpsDN antibody (1:1,000 dilution). M, molecular size standards (sizes, in kilodaltons, are shown at left). The thick band migrating between 48 and 60 kDa in all samples is the heavy chain of IgG. (C) XpsD proteins produced from each plasmid represented by straight lines. Open boxes represent truncated regions.
To confirm these results, we performed reciprocal experiments by precipitating the membrane extract with antibody against the XpsN protein and probing the blot with antibody against the XpsD protein. In agreement with previous observations, we observed that the full-length XpsD protein encoded by pKC118 coprecipitated with the XpsN protein (Fig. 7B, lanes 1 and 6). Moreover, we did not detect in the strain containing pMH7 or pKDT (Fig. 7B, lanes 2 and 4) any XpsD signal, besides those present in the negative control, with the expected molecular size of the truncated XpsD protein. To our surprise, an XpsD protein signal migrating with an apparent molecular size of approximately 60 kDa was clearly detected in the strain containing pKdPs (Fig. 7B, lane 3). A similar signal, although slightly weaker, was present in the strain containing pKC118 (Fig. 7B, lane 1), probably a degradation product from the full-length protein. The signal was absent in the negative control (Fig. 7B, lane 5), where no membrane extract from any strain was included in the precipitation mixture. Nor was it present in the strain containing pMH7 or pKDT (Fig. 7B, lanes 2 and 4). We did not anticipate detecting the truncated XpsD proteins encoded by pYL4 (56 kDa) or by pCD105 (39 kDa) in this experiment, since each was expected to comigrate with the heavy chain of immunoglobulin G (IgG) or cross-reactive materials, respectively.
Both positive signals observed (Fig. 7A, lane 8; Fig. 7B, lane 3) suggested that the C-terminal region, between amino acid residues 650 and 759, of the XpsD protein is probably involved in its interaction with the XpsN protein. The reason for our not detecting the XpsN signal in the strain containing pKdPs when precipitated with either anti-XpsDN (Fig. 7A, lane 4) or anti-XpsDC antibody (Fig. 7A, lane 7) was probably the close proximity between the XpsN-binding domain and the XpsD antibody-binding domain of the XpsD(Δ448–650) protein encoded by pKdPs. Binding of either XpsD antibody with the XpsD(Δ448–650) protein may have prevented its association with the XpsN protein. The possibility that the XpsD(Δ448–650) protein may lack the epitopes necessary for it to be precipitated by antibodies against the XpsD protein was excluded by our control experiments (data not shown).
DISCUSSION
By constructing a nonpolar chromosomal xpsN gene mutant strain, XC1709, we demonstrated in this study that the xpsN gene is clearly required for extracellular protein secretion by X. campestris pv. campestris. α-Amylase, which was secreted extracellularly in the normal strain, accumulated in the periplasm of the mutant strain. This conclusion agrees with the observation that nonpolar transposon insertion in the pulN gene of K. oxytoca abolished pullulanase secretion (40). On the other hand, the gspN gene was not found in the out gene cluster of E. chrysanthemi (29), the xcp gene cluster of P. aeruginosa (14), or the gsp gene cluster of E. coli (58). Interestingly, expression in E. coli of the out gene cluster of E. chrysanthemi was sufficient for extracellular enzyme secretion (17). Likewise, d'Enfert et al. (9) demonstrated that expression of the pul gene cluster of K. oxytoca enabled E. coli to secrete pullulanase extracellularly. It appears that the gspN gene may be required for some type II protein secretions but not for others. Alternatively, the outN gene may be located outside the out gene cluster in E. chrysanthemi, and an E. coli gene, outside its gsp gene cluster, can substitute for it.
When analyzed on a sucrose gradient, majority of the XpsN protein cofractionated with succinate dehydrogenase, which was demonstrated to be a cytoplasmic membrane protein in X. campestris pv. campestris (11). The PulN protein of K. oxytoca was also observed in the cytoplasmic membrane (40). A stretch of hydrophobic amino acids between residues 10 and 32 could serve to anchor the XpsN protein in the cytoplasmic membrane. Disruption of the hydrophobic region either by changing amino acid residues 21 and 22 (L21R, L22P) or by truncating a major portion of the hydrophobic region (Δ6–25), as well as its replacement with a cleavable N-terminal signal peptide, rendered the XpsN protein unstable (unpublished results). Hence we could not determine the subcellular locations of these mutated XpsN proteins. Nevertheless, our observations implied that the hydrophobic region might be significant in maintaining the stability of the XpsN protein, probably by anchoring it in the cytoplasmic membrane. The prediction was supported by our observation that both XpsN protein stability and its membrane localization were restored by replacing the hydrophobic region (residues 6 to 25) of the XpsN protein with a hydrophobic segment of the TetA protein (residues 2 to 26; accession no. M10786) (data not shown). Moreover, the TetA-XpsN hybrid is not functional, suggesting that either the specific sequence of the XpsN protein, the length of the hydrophobic region, or both are significant in its function. An N-terminal hydrophobic region is also predicted in all known GspN proteins.
Almost the entire amino acid sequence of the XpsN protein is C-terminal to the membrane-anchoring sequence. According to the positive-inside rule proposed by von Heijne (57), the major part of the XpsN protein is predicted to face the periplasm. In agreement with this prediction, we were able to detect alkaline phosphatase activity in an E. coli strain that expressed an XpsN-PhoA fusion protein (unpublished results). Similar topology was deduced for the PulN protein of K. oxytoca and the OutN protein of E. carotovora, as suggested by alkaline phosphatase-positive PulN-PhoA fusions (40) and β-lactamase fusion studies (42), respectively. To examine the interactive relationship between the periplasmically facing cytoplasmic membrane protein XpsN and the channel-forming outer membrane protein XpsD, we conducted two types of experiments. Both suggested stable complex formation between the two. The XpsN protein extracted from the membrane with either Triton X-100 or deoxycholate, which are known to form micelles of different sizes (55), was found to cofractionate with the XpsD protein. Furthermore, only those truncated XpsD proteins containing amino acid residues 650 to 759 were coprecipitable with the XpsN protein. All of these results argue for the conclusion that cofractionation of the XpsN protein with the full-length XpsD protein probably represents authentic association between the two. In intact cells, they, or portions of them, may transiently maintain stable association with each other. The proportion of the XpsN protein that cofractionated with the XpsD(His)6 protein was estimated to be approximately 4%. In close agreement, the amount of XpsN protein that was coprecipitated with the XpsD protein was estimated to be approximately 4.1% of the XpsN protein precipitated with anti-XpsN antibody (data not shown). On the other hand, the amount of XpsD protein that was coprecipitated with the XpsN protein was estimated to be approximately 1.6% of the XpsD protein precipitated with anti-XpsD antibody (data not shown). Since less total XpsD protein cofractionated with the XpsN protein, an XpsN excess cannot account for the small percentage of XpsN protein that cofractionated with the XpsD protein. It is possible that the XpsN-XpsD complex is not entirely stable to the detergents necessary to solubilize it. Alternatively, the association between XpsN and XpsD may not be a permanent assembly. It would be interesting to find out if the abundance of the XpsN-XpsD complex varies with secretion activity.
What is the biological significance of the association between the XpsN and XpsD proteins in the type II protein secretion process? An interactive relationship between a cytoplasmic membrane protein and an outer membrane protein has been demonstrated in the export of the assembling filamentous phage, as well as in TonB-dependent outer membrane transport systems. In the former case, interaction between the cytoplasmic membrane protein pI and the outer membrane pretein pIV, which has been demonstrated to form a gated channel for the passage of the assembling phage (33), was supported by strong genetic evidence (45, 46). However, attempts to demonstrate pI-pIV association with cross-linking or immune precipitation experiments were not successful (25) until recently (13). A model proposed by Russel (47) suggested that the pI protein may stimulate channel opening by transiently interacting with the channel-forming protein pIV. In contrast, positive cross-linking between the cytoplasmic membrane protein TonB and the outer membrane protein FepA receptor of ferric enterobactin and colicins B and D (53) was supportive of their close contact with each other, or even stable complex formation between the two. Further support for stable complex formation between the TonB protein and the outer membrane protein was suggested by the appearance of 30% of the TonB protein in the outer membrane fractions upon sucrose gradient sedimentation analysis (27, 28). TonB is proposed to be the energy transducer that couples the proton motive force of the cytoplasmic membrane to drive active transport across the outer membrane (6).
Similarities between the XpsN and TonB proteins are fourfold. Both are proline-rich, periplasmically facing, cytoplasmic membrane proteins that associate with an outer membrane protein. However, the proline-rich region of the TonB protein could be tandemly duplicated (56) or deleted (26) without affecting TonB activity. Furthermore, although we detected XpsN-XpsD association in immune precipitation or XpsN-XpsD(His)6 cofractionation in affinity chromatography experiments, the proportion of the XpsN protein appearing in the outer membrane fractions upon sucrose gradient sedimentation was much lower than that of the TonB protein in the same fractions. It is possible that we have not found the conditions for detecting significant amounts of the XpsN protein in the outer membrane. Alternatively, the XpsN protein could serve as an energy transducer for the type II protein secretion apparatus by interacting with the channel-forming protein XpsD without being detectable in significant amounts in the outer membrane. Nevertheless, we cannot yet rule out the possibility that the XpsN protein may serve to stimulate opening of the protein-translocating channel by interacting with the XpsD protein, similar to the role suggested for the pI protein in export of the assembling phage particle. Alternatively, the GspN protein might be a major component in determining species specificity. Primary sequence identity between individual components of the two Out systems of E. carotovora and E. chrysanthemi ranges from 50 to 83%. However, He et al. (17) demonstrated clearly that the pectate lyase secreted by the former cannot be secreted by the latter. Complementation of deletion mutations in the out gene of E. chrysanthemi with E. carotovora out homologues reveals the OutC and OutD proteins as possible determinants for species specificity (30). The OutN protein was not included in the test because the outN gene was absent from the out gene cluster of E. chrysanthemi. Thus, the possibility for the GspN protein to be the candidate as species-specific determinant cannot be excluded entirely.
Pairwise global alignments of seven complete GspN proteins that could be retrieved from the GenBank database (Table 2) reveal that the overall conservation of primary sequences among GspN proteins is poor. In particular, the XpsN protein (XANCP) is barely similar to the other GspN proteins. Nevertheless, in addition to its molecular size and gene location in the xps gene cluster, we observed in this study that the XpsN protein is similar to the PulN protein of K. oxytoca in cytoplasmic membrane location as well as topology in the membrane (40). Primary sequence conservation may not be essential for proteins to have the same function. A conditional lethal E. coli DNA topoisomerase I mutant was complemented by human topoisomerase I despite the lack of sequence similarity between them (3). Moreover, we noted that, among all GspD proteins, the XpsD protein is the most remotely related, as are the other Xps proteins. Until another one with a more similar primary sequence is identified, we assume the XpsN protein characterized in this study to be a GspN homologue. In that case, regardless of role the GspN protein may be assigned in the type II secretion pathway, the primary sequence divergence should be taken into consideration. Perhaps the GspN protein plays a role in the secretion pathway such that it does not have a strict requirement for a particular tertiary structure, or the specific structure required for the GspN protein could be attained by all homologues despite significant differences in their primary sequences. Poor conservation in primary sequences among the GspN proteins is also supportive of a role as a determinant of species specificity.
TABLE 2.
Pairwise alignments of GspN proteinsa
| Proteinb | % Sequence identity (alignment quality)c
|
||||||
|---|---|---|---|---|---|---|---|
| AERHY | VIBCH | KLEPN | ERWCA | PSEPU | BURPS | XANCP | |
| AERHY (252) | 100 (1334) | 39.0 (383) | 33.6 (269) | 33.2 (322) | 31.5 (83) | 25.5 (52) | 29.5 (7) |
| VIBCH (251) | 100 (1297) | 31.3 (228) | 23.4 (203) | 34.7 (82) | 22.3 (37) | 23.0 (6) | |
| KLEPN (252) | 100 (1314) | 34.7 (358) | 29.0 (56) | 26.1 (14) | 0 (0) | ||
| ERWCA (248) | 100 (1281) | 24.3 (6) | 24.9 (40) | 15.4 (5) | |||
| PSEPU (219) | 100 (1188) | 31.1 (32) | 24.4 (10) | ||||
| BURPS (263) | 100 (1357) | 23.1 (6) | |||||
| XANCP (261) | 100 (1338) | ||||||
Global alignment was performed using SeqWeb, version 1.1.
AERHY, ExeN of A. hydrophila (accession no. P41852); VIBCH, EpsN of V. cholerae (accession no. P45784); KLEPN, PulN of K. oxytoca (accession no. P15753); ERWCA, OutN of E. carotovora (accession no. P31710); PSEPU, GspN of P. putida (accession no. X81085) (8); BURPS, GspN of B. pseudomallei (accession no. AF110185); (10); XANCP, XpsN of X. campestris pv. campestris (accession no. M81648) (20). Values in parentheses are predicted numbers of amino acid residues.
Determined based on the alignment method of Needleman and Wunsch (35).
ACKNOWLEDGMENTS
We thank S.-W. Tyan for performing experiments to determine the subcellular distribution of α-amylase. We are also grateful to the reviewers for providing many helpful comments and suggestions.
This work was supported by the following grants from the National Science Council of the Republic of China: NSC86-2311-B005-009, NSC87-2311-B005-031, and NSC86-2313-B005-032.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bitter W, Koster M, Latijnhouwers M, de Cock H, Tommassen J. Formation of oligomeric rings by XcpQ and PilQ, which are involved in protein transport across the outer membrane of Pseudomonas aeruginosa. Mol Microbiol. 1998;27:209–219. doi: 10.1046/j.1365-2958.1998.00677.x. [DOI] [PubMed] [Google Scholar]
- 3.Bjornsti M-A, Benedetti P, Viglianti G A, Wang J C. Expression of human topoisomerase I in yeast cells lacking yeast DNA topoisomerase I: restoration of sensitivity of the cells to the antitumor drug camptothecin. Cancer Res. 1989;49:6318–6323. [PubMed] [Google Scholar]
- 4.Bleves S, Lazdunski A, Filloux A. Membrane topology of three Xcp proteins involved in exoprotein transport by Pseudomonas aeruginosa. J Bacteriol. 1996;178:4297–4300. doi: 10.1128/jb.178.14.4297-4300.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bleves S, Voulhoux R, Michel G, Lazdunski A, Tommassen J, Filloux A. The secretion apparatus of Pseudomonas aeruginosa: identification of a fifth pseudopilin, XcpX (GspK family) Mol Microbiol. 1998;27:31–40. doi: 10.1046/j.1365-2958.1998.00653.x. [DOI] [PubMed] [Google Scholar]
- 6.Braun V. Energy-coupled transport and signal transduction through the Gram-negative outer membrane via TonB-ExbB-ExbD-dependent receptor proteins. FEMS Microbiol Rev. 1995;16:295–307. doi: 10.1111/j.1574-6976.1995.tb00177.x. [DOI] [PubMed] [Google Scholar]
- 7.Chen L-Y, Chen D-Y, Miaw J, Hu N-T. XpsD, an outer membrane protein required for protein secretion by Xanthomonas campestris pv. campestris, forms a multimer. J Biol Chem. 1996;271:2703–2708. doi: 10.1074/jbc.271.5.2703. [DOI] [PubMed] [Google Scholar]
- 8.de Groot A, Krijger J J, Filloux A, Tommassen J. Characterization of type II protein secretion (xcp) genes in the plant growth-stimulating Pseudomonas putida, strain WCS358. Mol Gen Genet. 1996;250:491–504. doi: 10.1007/BF02174038. [DOI] [PubMed] [Google Scholar]
- 9.d'Enfert, Antoinette C R, Pugsley A P. Cloning and expression in Escherichia coli of the Klebsiella pneumoniae genes for production, surface localization and secretion of the lipoprotein pullulanase. EMBO J. 1987;6:3531–3538. doi: 10.1002/j.1460-2075.1987.tb02679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeShazer D, Brett P J, Burtnick M N, Woods D E. Molecular characterization of genetic loci required for secretion of exoproducts in Burkholderia pseudomallei. J Bacteriol. 1999;181:4661–4664. doi: 10.1128/jb.181.15.4661-4664.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dianese J C, Schaad N W. Isolation and characterization of inner and outer membrane of Xanthomonas campestris pv. campestris. Phytopathology. 1982;72:1284–1289. [Google Scholar]
- 12.Dums F, Dow J M, Daniels M J. Structural characterization of protein secretion genes of the bacterial phytopathogen Xanthomonas campestris pathovar campestris: relatedness to secretion systems of other gram-negative bacteria. Mol Gen Genet. 1991;229:357–364. doi: 10.1007/BF00267456. [DOI] [PubMed] [Google Scholar]
- 13.Feng J, Model P, Russel M. A trans-envelope protein complex needed for filamentous phage assembly and export. Mol Microbiol. 1999;34:745–755. doi: 10.1046/j.1365-2958.1999.01636.x. [DOI] [PubMed] [Google Scholar]
- 14.Filloux A, Bally M, Ball G, Akrim M, Tommassen J, Lazdunski A. Protein secretion in Gram-negative bacteria: transport across the outer membrane involves common mechanisms in different bacteria. EMBO J. 1990;9:4323–4329. doi: 10.1002/j.1460-2075.1990.tb07881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filloux A, Gerard M, Bally M. GSP-dependent protein secretion in Gram-negative bacteria: the Xcp system of Pseudomonas aeruginosa. FEMS Microbiol Rev. 1998;22:177–198. doi: 10.1111/j.1574-6976.1998.tb00366.x. [DOI] [PubMed] [Google Scholar]
- 16.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. p. 119. [Google Scholar]
- 17.He S Y, Lindeberg M, Chatterjee A K, Collmer A. Cloned Erwinia chrysanthemi out genes enable Escherichia coli to selectively secrete a diverse family of heterologous proteins to its milieu. Proc Natl Acad Sci USA. 1991;88:1079–1083. doi: 10.1073/pnas.88.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henikoff S, Henikoff J G. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howard S P, Critch J, Bedi A. Isolation and analysis of eight exe genes and their involvement in extracellular protein secretion and outer membrane assembly in Aeromonas hydrophila. J Bacteriol. 1993;175:6695–6703. doi: 10.1128/jb.175.20.6695-6703.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu N-T, Hung M-N, Chiou S-J, Tang F, Chiang D-C, Huang H-Y, Wu C-Y. Cloning and characterization of a gene required for the secretion of extracellular enzymes across the outer membrane by Xanthomonas campestris pv. campestris. J Bacteriol. 1992;174:2679–2687. doi: 10.1128/jb.174.8.2679-2687.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu N-T, Hung M-N, Huang A-M, Tsai H-F, Yang B-Y, Chow T-Y, Tseng Y-H. Molecular cloning, characterization and nucleotide sequence of the gene for secreted α-amylase from Xanthomonas campestris pv. campestris. J Gen Microbiol. 1992;138:1647–1655. doi: 10.1099/00221287-138-8-1647. [DOI] [PubMed] [Google Scholar]
- 22.Hu N-T, Hung M-N, Liao C-T, Lin M-H. Subcellular location of XpsD, a protein required for extracellular protein secretion by Xanthomonas campestris pv. campestris. Microbiology. 1995;141:1395–1406. doi: 10.1099/13500872-141-6-1395. [DOI] [PubMed] [Google Scholar]
- 23.Kamoun S, Tola E, Kamdar H, Kado C I. Rapid generation of directed and unmarked deletions in Xanthomonas. Mol Microbiol. 1992;6:809–816. doi: 10.1111/j.1365-2958.1992.tb01531.x. [DOI] [PubMed] [Google Scholar]
- 24.Kasahara M, Anraku Y. Succinate dehydrogenase of Escherichia coli membrane vesicles. J Biochem. 1974;76:959–966. [PubMed] [Google Scholar]
- 25.Kazmierczak B I, Mielke D L, Russel M, Model P. pIV, a filamentous phage protein that mediates phage export across the bacterial cell envelope, forms a multimer. J Mol Biol. 1994;238:187–198. doi: 10.1006/jmbi.1994.1280. [DOI] [PubMed] [Google Scholar]
- 26.Larsen R A, Wood G E, Postle K. The conserved proline-rich motif is not essential for energy transduction by Escherichia coli TonB protein. Mol Microbiol. 1993;10:943–953. doi: 10.1111/j.1365-2958.1993.tb00966.x. [DOI] [PubMed] [Google Scholar]
- 27.Larsen R A, Thomas M G, Postle K. Protonmotive force, ExbB and ligand-bound FepA drive conformational changes in TonB. Mol Microbiol. 1999;31:1809–1824. doi: 10.1046/j.1365-2958.1999.01317.x. [DOI] [PubMed] [Google Scholar]
- 28.Letain T E, Postle K. TonB protein appears to transduce energy by shuttling between the cytoplasmic membrane and the outer membrane in Escherichia coli. Mol Microbiol. 1997;24:271–283. doi: 10.1046/j.1365-2958.1997.3331703.x. [DOI] [PubMed] [Google Scholar]
- 29.Lindeberg M, Collmer A. Analysis of eight out genes in a cluster required for pectic enzyme secretion by Erwinia chrysanthemi: sequence comparison with secretion genes from other gram-negative bacteria. J Bacteriol. 1992;174:7385–7397. doi: 10.1128/jb.174.22.7385-7397.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindeberg M, Salmond G P C, Collmer A. Complementation of deletion mutations in a cloned functional cluster of Erwinia chrysanthemi out genes with Erwinia carotovora out homologues reveals OutC and OutD as candidate gatekeepers of species-specific secretion of proteins via the type II pathway. Mol Microbiol. 1996;20:175–190. doi: 10.1111/j.1365-2958.1996.tb02499.x. [DOI] [PubMed] [Google Scholar]
- 31.Linderoth N A, Simon M N, Russel M. The filamentous phage pIV multimer visualized by scanning transmission electron microscopy. Science. 1997;278:1635–1638. doi: 10.1126/science.278.5343.1635. [DOI] [PubMed] [Google Scholar]
- 32.Lu H M, Motley S T, Lory S. Interactions of the components of the general secretion pathway—role of Pseudomonas aeruginosa type IV pilin subunits in complex formation and extracellular protein secretion. Mol Microbiol. 1997;25:247–259. doi: 10.1046/j.1365-2958.1997.4561818.x. [DOI] [PubMed] [Google Scholar]
- 33.Marciano D K, Russel M, Simon S M. An aqueous channel for filamentous phage export. Science. 1999;284:1516–1519. doi: 10.1126/science.284.5419.1516. [DOI] [PubMed] [Google Scholar]
- 34.Michel G, Bleves S, Ball G, Lazdunski A, Filloux A. Mutual stabilization of the XcpZ and XcpY components of the secretory apparatus in Pseudomonas aeruginosa. Microbiology. 1998;144:3379–3386. doi: 10.1099/00221287-144-12-3379. [DOI] [PubMed] [Google Scholar]
- 35.Needleman S B, Wunsch C D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 36.Nouwen N, Ranson N, Saibil H, Wolpensinger B, Engel A, Ghazi A, Pugsley A P. Secretin PulD: association with pilot PulS, structure, and ion-conducting channel formation. Proc Natl Acad Sci USA. 1999;96:8173–8177. doi: 10.1073/pnas.96.14.8173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nunn D N, Lory S. Cleavage, methylation, and localization of the Pseudomonas aeruginosa export proteins XcpT, -U, -V, and -W. J Bacteriol. 1993;175:4375–4382. doi: 10.1128/jb.175.14.4375-4382.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pugsley A P. Multimers of the precursor of a type IV pilin-like component of the general secretory pathway are unrelated to pili. Mol Microbiol. 1996;20:1235–1245. doi: 10.1111/j.1365-2958.1996.tb02643.x. [DOI] [PubMed] [Google Scholar]
- 40.Pugsley A P, Reyss I. Five genes at the 3′ end of the Klebsiella pneumoniae pulC operon are required for pullulanase secretion. Mol Microbiol. 1990;4:365–379. doi: 10.1111/j.1365-2958.1990.tb00604.x. [DOI] [PubMed] [Google Scholar]
- 41.Reeves P J, Whitcombe D, Wharam S, Gibson M, Allison G, Bunce N, Barallon R, Douglas P, Mulholland V, Stevens S, Walker D, Salmond G P C. Molecular cloning and characterization of 13 out genes from Erwinia carotovora subspecies carotovora: genes encoding members of a general secretion pathway (GSP) widespread in Gram-negative bacteria. Mol Microbiol. 1993;8:443–456. doi: 10.1111/j.1365-2958.1993.tb01589.x. [DOI] [PubMed] [Google Scholar]
- 42.Reeves P J, Douglas P, Salmond G P C. Beta-lactamase topology probe analysis of the OutO NMePhe peptidase, and six other Out protein components of the Erwinia carotovora general secretion pathway apparatus. Mol Microbiol. 1994;12:445–457. doi: 10.1111/j.1365-2958.1994.tb01033.x. [DOI] [PubMed] [Google Scholar]
- 43.Reyss I, Pugsley A P. Five additional genes in the pulC-O operon of the gram-negative bacterium Klebsiella oxytoca UNF5023 which are required for pullulanase secretion. Mol Gen Genet. 1990;222:176–184. doi: 10.1007/BF00633815. [DOI] [PubMed] [Google Scholar]
- 44.Rosenberg A H, Lade B N, Chui D-S, Lin S-W, Dunn J J, Studier F W. Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene. 1987;56:125–135. doi: 10.1016/0378-1119(87)90165-x. [DOI] [PubMed] [Google Scholar]
- 45.Russel M. Interchangeability of related proteins and autonomy of function: the morphogenetic proteins of filamentous phage f1 and IKe cannot replace one another. J Mol Biol. 1992;227:453–462. doi: 10.1016/0022-2836(92)90900-5. [DOI] [PubMed] [Google Scholar]
- 46.Russel M. Protein-protein interactions during filamentous phage assembly. J Mol Biol. 1993;231:689–697. doi: 10.1006/jmbi.1993.1320. [DOI] [PubMed] [Google Scholar]
- 47.Russel M. Phage-assembly: a paradigm for bacterial virulence factor export? Science. 1994;265:612–614. doi: 10.1126/science.8036510. [DOI] [PubMed] [Google Scholar]
- 48.Russel M. Macromolecular assembly and secretion across the bacterial cell envelope: type II protein secretion systems. J Mol Biol. 1998;279:485–499. doi: 10.1006/jmbi.1998.1791. [DOI] [PubMed] [Google Scholar]
- 49.Salmond G P C, Reeves P J. Membrane traffic wardens and protein secretion in gram-negative bacteria. Trends Biochem Sci. 1993;18:7–12. doi: 10.1016/0968-0004(93)90080-7. [DOI] [PubMed] [Google Scholar]
- 50.Sandkvist M, Bagdasarian M, Howard S P, DiRita V J. Interaction between the autokinase EpsE and EpsL in the cytoplasmic membrane is required for extracellular secretion in Vibrio cholerae. EMBO J. 1995;14:1664–1673. doi: 10.1002/j.1460-2075.1995.tb07155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sandkvist M, Michel L O, Hough L P, Morales V M, Bagdasarian M, Kommey M, DiRita V J, Bagdasarian M. General secretion pathway (eps) genes required for toxin secretion and outer membrane biogenesis in Vibrio cholerae. J Bacteriol. 1997;179:6994–7003. doi: 10.1128/jb.179.22.6994-7003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sandkvist M, Hough L P, Bagdasarian M M, Bagdasarian M. Direct interaction of the EpsL and EpsM proteins of the general secretion apparatus in Vibrio cholerae. J Bacteriol. 1999;181:3129–3135. doi: 10.1128/jb.181.10.3129-3135.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skare J T, Ahmer B M M, Seachord C L, Darveau R P, Postle K. Energy transduction between membranes: TonB, a cytoplasmic membrane protein, can be chemically cross-linked in vivo to the outer membrane receptor FepA. J Biol Chem. 1993;268:16302–16308. [PubMed] [Google Scholar]
- 54.Thomas S R, Reeves P J, Salmond G P. The general secretion pathway of Erwinia carotovora subsp. carotovora: analysis of the membrane topology of OutC and OutF. Microbiology. 1997;143:713–720. doi: 10.1099/00221287-143-3-713. [DOI] [PubMed] [Google Scholar]
- 55.Thomas T C, McNamee M G. Purification of membrane proteins. Methods Enzymol. 1990;182:499–520. doi: 10.1016/0076-6879(90)82040-9. [DOI] [PubMed] [Google Scholar]
- 56.Traub I, Gaisser S, Braun V. Activity domains of the TonB protein. Mol Microbiol. 1993;8:409–423. doi: 10.1111/j.1365-2958.1993.tb01584.x. [DOI] [PubMed] [Google Scholar]
- 57.von Heijne G. Control of topology and mode of assembly of a polytopic membrane protein by positively charged residues. Nature. 1989;341:456–458. doi: 10.1038/341456a0. [DOI] [PubMed] [Google Scholar]
- 58.von Heijne G. The signal peptide. J Membr Biol. 1990;115:195–201. doi: 10.1007/BF01868635. [DOI] [PubMed] [Google Scholar]
- 59.Whitchurch C B, Mattick J S. Escherichia coli contains a set of genes homologous to those involved in protein secretion, DNA uptake and the assembly of type-4 fimbriae in other bacteria. Gene. 1994;150:9–15. doi: 10.1016/0378-1119(94)90851-6. [DOI] [PubMed] [Google Scholar]