Abstract
HetR is a serine-type protease required for heterocyst differentiation in heterocystous cyanobacteria under conditions of nitrogen deprivation. We have identified the active Ser residue of HetR from Anabaena sp. strain PCC 7120 by site-specific mutagenesis. By changing the S152 residue to an Ala residue, the mutant protein cannot be labeled by Dansyl fluoride, a specific serine-type protein inhibitor. The mutant protein showed no autodegradation in vitro. The mutant hetR gene was introduced into Anabaena strain 884a, a hetR mutant. The resultant strain, Anabaena strain S152A, could not form heterocysts under conditions of nitrogen deprivation even though the up-regulation of the mutant hetR gene was induced upon removal of combined nitrogen. The Anabaena strain 216, which carries a mutant hetR gene encoding S179N HetR and could not form heterocysts, also produced HetR protein upon induction. Sequence comparison shows that Ser152 is conserved in all cyanobacterial HetR. Immunoblotting was used to study HetR induction in both the wild-type and mutant strains. The amount of mutant HetR in strain S152A and in strain 216 increased continuously for 24 h after nitrogen step-down, while the amount of HetR in wild-type cells reached a maximum level within 6 h after nitrogen step-down. Our results show the Ser152 is the active site of HetR. The protease activity is required for heterocyst differentiation and might be needed for repression of HetR overproduction under conditions of nitrogen deprivation.
Some filamentous cyanobacteria form specialized cells called heterocysts for providing a micro-oxygen environment for nitrogen fixation under nitrogen-limiting conditions. Mature heterocysts differ from vegetative cells metabolically and morphologically in several ways (3, 6, 10, 25): they lack photosystem II and oxygen-generating ability and have little phycobiliprotein, they lack the ability to fix CO2, they have a high rate of respiratory electron transfer, and they have a thick envelope consisting of an inner glycolipid layer and an outer polysaccharide layer. Another important feature of heterocysts is that they are usually spaced regularly along the filaments so that they form a pattern (23–25).
The process of differentiation from a vegetative cell to a heterocyst is complex, and many genes are involved (3, 25). Some gene products control early differentiation, while the others are essential for later stages of heterocyst differentiation. Among the gene products which are required for initiation of heterocyst differentiation are NtcA and HetR. NtcA is a global nitrogen regulator present in both unicellular and filamentous cyanobacteria (5, 22). HetR is an autoregulatory serine-type protease present in heterocystous and some nonheterocystous cyanobacteria (1, 2, 12, 28). The expression of the hetR gene in Anabaena sp. strain PCC 7120, a heterocystous cyanobacterium, is induced upon removal of combined nitrogen from the growth medium (2, 28). The hetR gene product is accumulated in the cells that are destined to become heterocysts (2) and in mature heterocysts (28). It has been shown that a normal hetR gene is required for its own induction in Anabaena sp. strain PCC 7120 (1) and a functional hetR gene is absolutely required for heterocyst differentiation (1, 2). HetR also controls pattern formation in heterocystous cyanobacteria (2, 25).
The hetR gene was initially discovered in Anabaena strain 216, which was unable to form heterocysts (2). Strain 216 carried a mutant hetR gene with a single nucleotide mutation which converted Ser179 to an Asn residue. The recombinant wild-type HetR showed auto-proteolysis while the mutant HetR protein (S179N HetR) has no auto-degrading activity in vitro (29). However, as in the case of wild-type HetR, S179N HetR could be labeled with Dansyl fluoride (DnsF), a specific serine-type protease inhibitor (21, 29). This result indicates that Ser179 may not be the active serine of HetR even though it is required for heterocyst differentiation and its protease activity. Peptide mapping of DnsF-labeled HetR showed that the active Ser of HetR could be located in the region between residues 140 and 164 (29), the region which contains two Ser residues.
The mechanism by which HetR controls heterocyst differentiation has not been fully elucidated. A database search revealed no similarity of HetR to any other proteins. The substrates of HetR remain unknown except for the fact that HetR undergoes auto-degradation (28, 29). The active Ser residue has not been identified even though it is known that Ser179 is required for its functions. In this communication, we report identification of the active site of HetR from Anabaena sp. strain PCC 7120. The roles of the active Ser residue in heterocyst differentiation and hetR gene expression are also reported.
MATERIALS AND METHODS
Strains and culture conditions.
Anabaena sp. strain PCC 7120 and its mutant strains were grown in BG11 medium (17) containing appropriate antibiotics either in liquid culture or on plates. The BG11 medium was supplemented with 10 mM HEPES at pH 7.5. The cultures were grown under a moderate light intensity (50 to 100 μmol · m−2 · s−1) at 28°C. The illumination was provided with cool-white fluorescent lamps. NaNO3 at a concentration of 1 g · liter−1 was used when needed. Escherichia coli strains were grown in LB medium containing appropriate antibiotics at 38°C. E. coli strain DH5α was used for all cloning purposes, and BL21(DE3) was used to express hetR genes with pET3a (18) as the expression vector as reported previously (28, 29).
Site-specific mutagenesis and other DNA methods.
Plasmid isolation and analysis were performed according to standard procedures. Restriction endonucleases (Promega, Beijing, China) were used according to the manufacturer's recommendations. To generate site-specific mutant hetR genes, the pALTER-ex1 system from Promega was used according to the manufacturer's recommendation. The hetR gene was first cloned into the pALTER-ex1 plasmid, and the resultant plasmid was isolated and denatured under alkaline conditions. The in vitro synthesis of the second strand was then carried out. An oligonucleotide (5′ GAGCATAAGTTACCCGCAAATCTTCCCG 3′) was used to generate the S142A hetR gene. Another oligonucleotide (5′ CAGTTAGTTACTGCTTTTGAGTTT 3′) was used to generate the S152A hetR gene. Oligonucleotides (5′ CAGTTAGTTACTNNNTTTGAGTTT 3′) were used to generate other possible mutations at the Ser152 position. The mutant hetR genes were digested with XbaI and BamHI and cloned into a pBluescript KS+ vector (Stratagene). The inserts were sequenced with an automatic DNA sequencer (ABI377; PE-Applied Biosystems) to determine their exact mutations. The desired mutant hetR genes were then cloned into a pET3a vector as described previously (28) for overproduction of HetR proteins.
The expression vectors pET3a/HetRs were used to generate suicidal plasmids (19) containing different hetR genes to transform the Anabaena sp. strain PCC 7120 mutant derivative strain 884a (1). The 1.2-kb fragment containing hetR promoters was first amplified by PCR. The oligonucleotides for the PCR were 5′ TTAGATCTGATCAATAAATTAGGT 3′ and 5′ AACATATGACAAATAGTTGAATAGC 3′. The amplification conditions were as described by Zhou et al. (28). The amplified fragment was digested with BglII and NdeI and gel purified before ligation into pET3a/HetRs, which were also digested with BglII and NdeI. The Ω fragment containing the gene for streptomycin resistance (7) was then inserted into the HindIII site of the plasmids mentioned above to generate the suicidal plasmids, pHetRΩ.
Total DNA extraction, enzyme digestion, gel electrophoresis, and Southern blotting were performed according to the method of Zhao et al. (27). The isolated total DNA was digested with HindIII and DraI and separated with a 0.8% agarose gel. The DNA fragments in gel were denatured with NaOH and transferred to a nitrocellulose membrane. The membrane was baked and used for hybridization. Probes used for hybridization were prepared by using two different DNA templates. The first DNA template was the 0.9-kb DNA fragment containing the hetR gene encoding HetR in its entirety from Anabaena sp. strain PCC 7120 as described previously (28). The second DNA template was obtained by PCR amplification of an internal fragment of the hetR gene in Anabaena sp. strain PCC 7120. The primers used for the PCR were 5′ GGCATGGAGCATTCTTAG 3′ and 5′ AgATCCTCTTgCgATCgC 3′. The amplified region of the hetR gene was deleted in construction of Anabaena strain 884a (1). Radioisotope-labeled probes for Southern hybridization were generated with [α-32P]dATP (100 TBq mmol−1) by using a random-labeling kit from Amersham.
Triparental mating was used to introduce the hetR genes into Anabaena strain 884a, a hetR mutant derivative of Anabaena sp. strain PCC 7120 (1), according to the method of Elhai and Wolk (4). The exconjugants were selected on BG11 plates containing neomycin (25 μg · ml−1) and streptomycin (5 μg · ml−1).
HetR protein characterization.
Overproduction, refolding, and purification of both wild-type and mutant HetR proteins were performed as described previously (28). Dansyl fluoride (DnsF) labeling of the HetR proteins was performed according to the method of Zhou et al. (29). The DnsF-labeled proteins were separated from free DnsF by acetone precipitation. Electroblotting was carried out after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (15) of the proteins, and the fluorescence images of the DnsF-HetR adducts were visualized under UV light and recorded with a digital camera.
The nitrogen step-down of Anabaena sp. strain PCC 7120 culture and immunoblotting for detection of HetR proteins were performed as described previously (9, 28). The protein concentration was determined with dye-binding assays by using the kit from Bio-Rad. The amount of HetR was determined by immunoblotting as an average of three blots.
Protein sequence alignment was carried out with the computer program CLUSTAL W (20). The relevant HetR sequences were obtained from GenBank.
Other methods.
The chlorophyll concentration was determined with 80% acetone (16). Observation and photography of Anabaena filaments were performed with a Leica light microscope equipped with a digital camera. Light intensity was measured with a Li-cor light intensity meter.
RESULTS AND DISCUSSION
Identification of the active Ser of the HetR from Anabaena sp. strain PCC 7120.
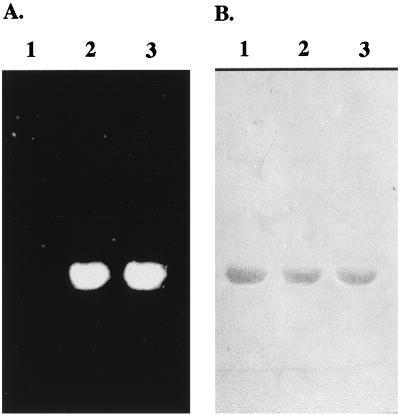
It has been shown that Dansyl fluoride (DnsF), a specific serine-type protease inhibitor, could specifically label both wild-type HetR and S179N HetR, a mutant HetR carried in strain 216 that cannot form heterocysts (2, 29). Tryptic mapping and sequencing have revealed that one of the two Ser residues (Ser142 and Ser152) on the tryptic peptide between residues 140 and 164 of the HetR protein from Anabaena sp. strain PCC 7120 could be the site for DnsF labeling. To determine which Ser residue is the site labeled by DnsF, site-specific mutagenesis was performed. Ser142 and Ser152 were changed into an Ala residue separately. The mutant genes as well as wild-type hetR were overexpressed as described previously (28), and the recombinant HetR proteins were purified. The purified HetR proteins were used for labeling with DnsF; the results are shown in Fig. 1. DnsF could label both the wild-type HetR (lane 3) and S142A HetR (lane 2), while S152A HetR (lane 1) could not be labeled. We also performed other mutagenesis at the site of Ser152 and found that no residues could replace Ser at this position for DnsF labeling (data not shown). These results strongly suggest that Ser152 is the active site of HetR.
FIG. 1.
DnsF labeling of HetR proteins. S152A HetR (lanes 1), S142A HetR (lanes 2), and wild-type HetR (lanes 3) were overproduced in E. coli and purified to homogeneity. DnsF was then reacted with these proteins, followed by 80% acetone precipitation. The proteins were separated from free DnsF by SDS-PAGE, followed by electroblotting. (A) Fluorescence image of DnsF-labeled HetR proteins on a polyvinylidene difluoride membrane recorded with a digital camera. (B) The same membrane shown in panel A, but stained with Coomassie blue.
HetR undergoes auto-degradation in the absence of a protease inhibitor (29). The fact that S152A HetR could not be labeled with DnsF predicted that the protein would not undergo auto-proteolysis. To test this prediction, recombinant HetR proteins were incubated at 37°C for auto-degradation in vitro (Fig. 2). While both the wild-type HetR and S142A HetR showed auto-degradation, S152A HetR showed no auto-degradation. On the basis of these results, we came to the conclusion that Ser152 is the active Ser of HetR in Anabaena sp. strain PCC 7120.
FIG. 2.
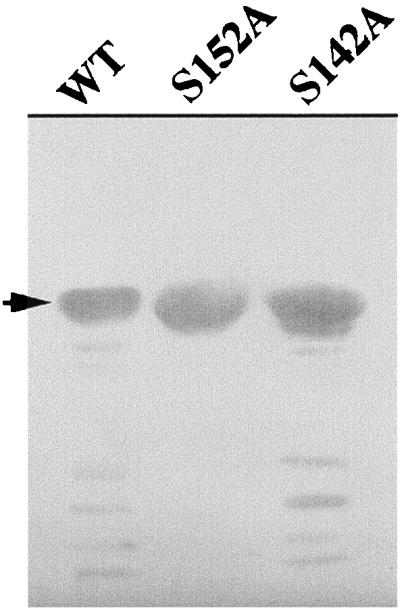
Degradation of HetR protein in vitro. Purified HetR protein at a concentration of 2 mg · ml−1 was incubated at 37°C for 1 h before sample loading buffer was added to stop the reaction. SDS-PAGE separation of the proteins was performed, and the gel was stained with Coomassie blue. The arrow indicates the position of intact HetR in the gel.
Ser152 is required for heterocyst differentiation.
To investigate the roles of the mutant HetR in heterocyst differentiation and pattern formation, suicidal plasmids containing either the wild-type hetR gene or mutant hetR genes were constructed and introduced into Anabaena strain 884a, a hetR mutant strain (1). Strain 884a lacks the middle part of the hetR gene, encoding a region from residue 35 through residue 168. The backbone of the suicidal plasmids is the expression vector for hetR genes, pET3a/HetRs. The 1.2-kb upstream fragment, which contains several promoters for the hetR gene (2, 11), was inserted upstream of the hetR genes, and the Ω fragment containing the Strr gene was inserted into the HindIII site of pET3a/HetR. The suicidal plasmids were introduced into strain 884a by triparental mating, and the exconjugants were selected with streptomycin and neomycin. The resultant strains, containing the wild-type hetR gene, the S142A hetR gene, and the S152A hetR gene, are named strain WT211, S142A, and S152A, respectively.
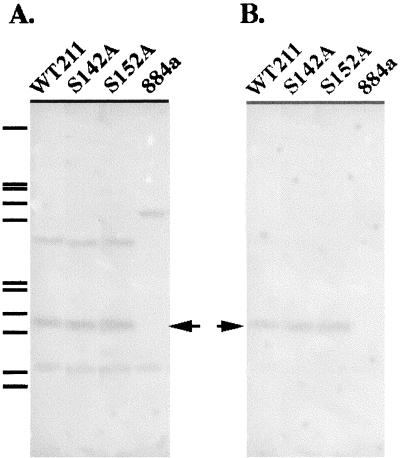
Southern hybridization was performed to confirm that the hetR genes were integrated into chromosomal DNA of strain 884a (Fig. 3). When hybridized with a probe prepared by using the entire hetR gene fragment as a template, two bands, with sizes of 3.8 and 1.0 kb, were observed in strain 884a DNA digested with DraI and HindIII. Three bands were observed in DraI- and HindIII-digested DNA of WT211, S142A, and S152A when the DNA was hybridized with the same probe. The sizes of the hybridized bands were 3.2, 1.5, and 1.0 kb. Because strain 884a retains about approximately 100 bp of the 5′ coding region and 400 bp of the 3′ coding region of the hetR gene, the two bands that were observed in strain 884a DNA (Fig. 3A) were likely the fragments containing these regions. The 1.5-kb bands in DraI- and HindIII-digested DNA of WT211, S142A, and S152A were expected on the basis of restriction sites of the suicidal plasmids. To confirm that the 1.5-kb bands were indeed from the hetR genes introduced into strain 884a through single recombination, a probe was prepared using a 0.4-kb DNA fragment as a template. This fragment was deleted in construction of strain 884a (1). The hybridization results (Fig. 3B) show that the region was missing in strain 884a, as was expected, and present in strains WT211, S142A, and S152A. Thus, it is evident that the hetR genes have been successfully introduced into strain 884a and integrated into the chromosome. Figure 3B also shows that the recombination between chromosomal DNA and the suicidal plasmids likely occurred in the region of hetR promoters.
FIG. 3.
Southern hybridization analysis of integration of the hetR genes into chromosomal DNA of Anabaena strain 884a. Total DNA was isolated from strains 884a, WT211, S142A, and S152A and digested with DraI and HindIII. The digested DNA was separated with a 0.8% agarose gel and NaOH denatured, followed by blotting onto nitrocellulose membranes. The membranes were hybridized either with a probe of full-length hetR (A) or with a probe consisting of a 0.4-kb internal fragment of hetR (B) as described in Materials and Methods. The membrane was then washed three times and dried before exposure to an X-ray film. The arrows indicate the positions of DNA fragments containing hetR genes (1.5 kb). The λ phage DNA fragments generated by EcoRI and HindIII digestions were used as molecular size markers (shown on the left). They were (from the top down): 21.2, 5.1, 4.9, 4.3, 3.5, 2.0, 1.9, 1.6, 1.4, 0.95, and 0.83 kb.
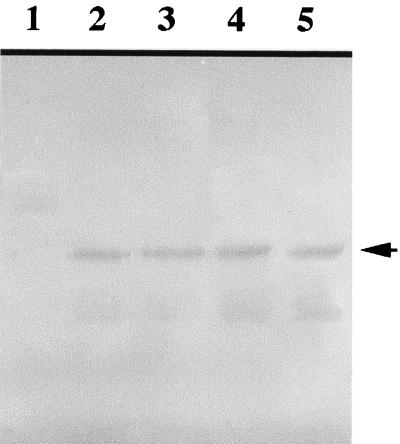
The expression of the hetR genes in these strains was studied by immunoblotting using anti-HetR antibodies. Cells were first grown in nitrogen-replete media and then subjected to nitrogen step-down for 3 h before immunoblotting. The results of immunoblotting are shown in Fig. 4. Both the wild-type hetR gene and the mutant hetR genes were expressed, and similar amount of HetR proteins were accumulated in these strains 3 h after nitrogen step-down. Strain 216, which carries a S179N hetR gene and is unable to differentiate heterocysts (2), is also studied for its ability to produce HetR. The result shows that a similar amount of HetR protein was produced in strain 216 3 h after nitrogen step-down.
FIG. 4.
Immunoblotting analysis of hetR gene expression in Anabaena strains 884a (lane 1), WT211 (lane 2), S142A (lane 3), S152A (lane 4), and 216 (lane 5). The Anabaena cultures at a chlorophyll concentration of 3 μg · ml−1 under nitrogen-replete conditions were washed three times with nitrogen-depleted medium and resuspended at a chlorophyll concentration of 3 μg · ml−1. The cultures were incubated in light for 3 h at 28°C. Five milliliters of culture was collected and lysed with SDS-gel loading buffer at 100°C before SDS-PAGE and immunoblotting. The arrow indicates the position of intact HetR in the gel.
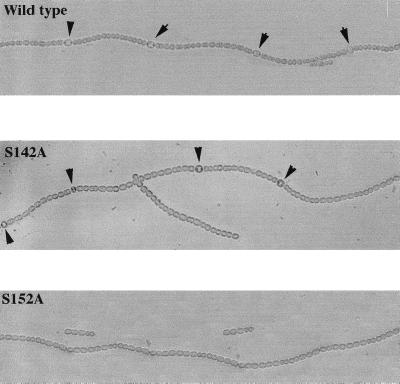
To determine whether strain S152A is capable of differentiating heterocysts under nitrogen-deprived conditions, filaments of the wild type, S142A, and S152A were transferred to BG11 medium containing no combined nitrogen and incubated for 36 h. While it is obvious that both the wild type and S142A strains form heterocysts, strain S152A could not form heterocysts (Fig. 5). Longer incubation of strain S152A under nitrogen deprivation resulted in filament fragmentation and cell lysis (data not shown). The pattern formation in strain S142A is normal, showing that S142A HetR did not change heterocyst distribution along the filaments of Anabaena sp. strain PCC 7120.
FIG. 5.
Photographic images of Anabaena sp. strain PCC 7120 (wild type), strain S142A, and strain S152A after incubation under nitrogen-depleted conditions for 36 h in light at 28°C. The nitrogen step-down procedure was the same as that described in the legend to Fig. 4. The images of the filaments were recorded with a digital camera attached to a Leica light microscope. Arrows indicate the positions of heterocysts.
Sequence comparison has often been used to determine whether an amino acid is important at a certain position on a protein. A database search revealed that Ser152 was conserved in all HetR proteins deduced from hetR genes in other cyanobacteria (data not shown), suggesting that Ser152 is a critical residue for HetR.
Immunoblotting study of the expression of the mutant hetR genes.
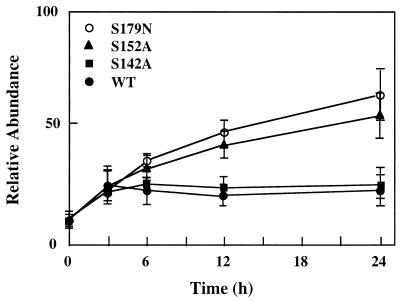
The expression of the wild-type hetR gene from Anabaena sp. strain PCC 7120 has been shown to be auto-regulatory (1). To study the expression of the mutant hetR genes, cells from the wild type and from strains S142A and S152A at 3, 6, 12, and 24 h after nitrogen step-down were collected and the amount of HetR protein was determined by immunoblotting (Fig. 6). Figure 6 also includes the immunoblotting results for strain 216, which contains S179N hetR. In both the wild type and strain S142A, the up-regulation of HetR production occurred within the first 6 h after nitrogen step-down, as reported previously (28). The overall increase in the HetR level was approximately two- to threefold. In strains 216 and S152A, the up-regulation of HetR continued for at least 24 h after nitrogen step-down. The overall increase in HetR in both strains upon induction was approximately five- to sixfold. Longer incubation of the strain 216 and S152A resulted in smearing in immunoblotting, making it difficult to determine the amount of HetR (data not shown).
FIG. 6.
Induction of HetR proteins in Anabaena strains 216, WT211, S142A, and S152A in the absence of combined nitrogen from the growth medium. The nitrogen step-down procedure was the same as that described in the legend to Fig. 5. Cells at times indicated after nitrogen step-down were collected and analyzed by immunoblotting as described in the legend to Fig. 5. The amount of HetR proteins in each culture was determined by band intensity after staining. Each set of data from different strains was normalized against the point at zero time nitrogen deprivation. Each point shown in the figure was an average from three blots.
The integration of the hetR genes introduced by suicidal plasmids will result in two copies of the hetR promoter region in Anabaena. The induction results of S142A shown in Fig. 6 suggest that the duplication of the hetR promoter region did not change the pattern of hetR gene expression as compared with that of the wild-type strain (28). The spatial pattern of heterocysts was not altered either in strain S142A (Fig. 5).
It has been reported that hetR gene is auto-regulatory in its expression. By using luxAB genes as reporter genes, Black et al. (1) showed that a functional hetR gene product was required for its own induction in expression. The strain used in the study by Black et al. was the strain 884a, which produced no HetR due to a partial deletion of the hetR gene. In the present report, we show that both strain 216, which carries the S179N hetR gene, and strain S152A, which carries the S152A hetR gene, exhibit induction of hetR gene expression upon shifting to nitrogen deprivation conditions (Fig. 4 and Fig. 6). These results suggest that the protease activity of HetR is not required for hetR gene induction, although this activity is required for heterocyst formation. Figure 6 also shows that the induction of expression of S152A hetR and S179N hetR is quite different from that of the wild-type hetR gene. The production of wild-type hetR gene product reached its maximum level within 6 h after nitrogen step-down (28) (Fig. 6) and was maintained at this level thereafter. The level of HetR in strains 216 and S152A continued to increase for at least 24 h after nitrogen step-down (Fig. 6). The expression of hetR genes upon induction with nitrogen-limiting conditions in these two mutant strains is apparently derepressed, suggesting that the protease activity of HetR is required not only for heterocyst differentiation but also to prevent overaccumulation of HetR protein in cells. A similar conclusion has been reached on the basis of results using the green fluorescent protein gene as a reporter gene under control of the hetR promoter (11). The prevention of overaccumulation of HetR in Anabaena sp. strain PCC 7120 could be achieved by HetR's auto-proteolytic activity. It is also possible that a proteolytic product generated by HetR could be functioning as a repressor for blocking hetR gene induction in neighboring vegetative cells. The pentapeptide of PatS (26) could be generated in such a way, as was suggested by Haselkorn (11).
HetR plays a very critical role in heterocyst differentiation and pattern formation (3, 6, 24). Experimental evidence shows that HetR is a serine-type protease (29). The evidence presented here clearly demonstrates that Ser152 of HetR in Anabaena sp. strain PCC 7120 is the active site and is required for heterocyst differentiation. The Ser179 residue is also critical to the function of HetR in Anabaena sp. strain PCC 7120. Strain 216, which carries a Ser-to-Asn mutation of HetR, could not form heterocysts under conditions of nitrogen deprivation (2). The recombinant S179N HetR protein lacks auto-degradation, while wild-type HetR undergoes auto-proteolysis (29). On the basis of results that all revertants in the S179N hetR gene exhibit an Asn-to-Ser codon change (11), it is likely that Ser179 plays an active role in HetR. Ser179 could participate in substrate binding. The lack of Ser179 in HetR in Anabaena sp. strain PCC 7120 could then lead to a total loss of HetR activity.
Proteases play important roles in cell differentiation and development in many organisms (14). Cell cycles are also influenced strongly by protease activity (13). Proteases are also required for bacterial development, such as sporulation in Bacillus subtilis (8). HetR is another example of proteases involved in cell differentiation. One possible role of HetR in heterocyst differentiation is to specifically cleave a repressor(s) which blocks transcription of heterocyst-specific genes. More study is required to understand the mechanism of HetR in cyanobacterial cell differentiation. The identification of the active Ser of HetR would certainly help in the understanding of its biochemical functions.
ACKNOWLEDGMENTS
We thank C. P. Wolk (Michigan State University) for strain 884a and R. Haselkorn (University of Chicago) for strain 216. We also thank C. Dong for her skillful technical assistance.
This work was supported by the National Natural Science Foundation of China to J. Z. (grants 39535002 and 39570067).
REFERENCES
- 1.Black T A, Cai Y, Wolk C P. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol Microbiol. 1993;9:77–84. doi: 10.1111/j.1365-2958.1993.tb01670.x. [DOI] [PubMed] [Google Scholar]
- 2.Buikema W J, Haselkorn R. Characterization of a gene controlling heterocyst differentiation in the cyanobacterium Anabaena 7120. Genes Dev. 1991;5:321–330. doi: 10.1101/gad.5.2.321. [DOI] [PubMed] [Google Scholar]
- 3.Buikema W J, Haselkorn R. Molecular genetics of cyanobacterial development. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:33–52. [Google Scholar]
- 4.Elhai J, Wolk C P. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 1988;167:747–754. doi: 10.1016/0076-6879(88)67086-8. [DOI] [PubMed] [Google Scholar]
- 5.Frias J E, Flores E, Herrero A. Requirement of the regulatory protein NtcA for the expression of nitrogen assimilation and heterocyst development genes in the cyanobacterium Anabaena sp. PCC 7120. Mol Microbiol. 1994;14:823–832. doi: 10.1111/j.1365-2958.1994.tb01318.x. [DOI] [PubMed] [Google Scholar]
- 6.Golden J W, Yoon H S. Heterocyst formation in Anabaena. Curr Opin Microbiol. 1998;1:623–629. doi: 10.1016/s1369-5274(98)80106-9. [DOI] [PubMed] [Google Scholar]
- 7.Golden J W, Wiest D R. Genome rearrangement and nitrogen fixation in Anabaena blocked by inactivation of xisA gene. Science. 1988;242:1421–1423. doi: 10.1126/science.3144039. [DOI] [PubMed] [Google Scholar]
- 8.Gottesman S. Regulation by proteolysis: developmental switches. Curr Opin Microbiol. 1999;2:142–47. doi: 10.1016/S1369-5274(99)80025-3. [DOI] [PubMed] [Google Scholar]
- 9.Harlow E, Lane D. Antibodies, a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 10.Haselkorn R. Heterocysts. Annu Rev Plant Physiol. 1978;29:319–344. [Google Scholar]
- 11.Haselkorn R. How cyanobacteria count to 10. Science. 1998;30:891–892. doi: 10.1126/science.282.5390.891. [DOI] [PubMed] [Google Scholar]
- 12.Janson S, Matveyev A, Bergman B. The presence and expression of hetR in the non-heterocystous cyanobacterium Symploca PCC 8002. FEMS Microbiol Lett. 1998;15:173–179. doi: 10.1111/j.1574-6968.1998.tb13270.x. [DOI] [PubMed] [Google Scholar]
- 13.King R W, Deshaies R J, Peters J-M, Kirschner M W. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 14.Kumar S. ICE-like proteases in apoptosis. Trends Biochem Sci. 1995;20:198–202. doi: 10.1016/s0968-0004(00)89007-6. [DOI] [PubMed] [Google Scholar]
- 15.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.MacKinney G. Absorption of light by chlorophyll solutions. J Biol Chem. 1941;140:315–322. [Google Scholar]
- 17.Rippka R, Deruelles J, Waterbury J B, Herdman M, Stanier R Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–61. [Google Scholar]
- 18.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1991;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 19.Thiel T. Genetic analysis of cyanobacteria. In: Bryant D A, editor. The molecular biology of cyanobacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 581–611. [Google Scholar]
- 20.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaz W L, Schoellmann G. Specific fluorescent derivatives of macromolecules. A fluorescence study of some specifically modified derivatives of chymotrypsin, trypsin and subtilisin. Biochim Biophys Acta. 1976;439:206–218. doi: 10.1016/0005-2795(76)90176-8. [DOI] [PubMed] [Google Scholar]
- 22.Wei T-F, Ramasubramanian T S, Golden J W. Anabaena sp. strain PCC 7120 ntcA gene required for growth on nitrate and heterocyst development. J Bacteriol. 1994;176:4473–4482. doi: 10.1128/jb.176.15.4473-4482.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilcox M, Mitchison G J, Smith R J. Pattern formation in the blue-green alga, Anabaena. I. Basic mechanisms. J Cell Sci. 1973;12:707–725. doi: 10.1242/jcs.12.3.707. [DOI] [PubMed] [Google Scholar]
- 24.Wolk C P, Quine M P. Formation of one-dimensional patterns by stochastic processes and by filamentous blue-green algae. Dev Biol. 1975;46:370–382. doi: 10.1016/0012-1606(75)90113-x. [DOI] [PubMed] [Google Scholar]
- 25.Wolk C P, Ernst A, Elhai J. Heterocyst metabolism and development. In: Bryant D A, editor. The molecular biology of cyanobacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 769–823. [Google Scholar]
- 26.Yoon H S, Golden J W. Heterocyst pattern formation controlled by a diffusible peptide. Science. 1998;282:935–938. doi: 10.1126/science.282.5390.935. [DOI] [PubMed] [Google Scholar]
- 27.Zhao J, Snyder W B, Muhlenhoff U, Rhiel E, Warren P V, Golbeck J H, Bryant D A. Cloning and characterization of the psaE gene of the cyanobacterium Synechococcus sp. PCC 7002: characterization of a psaE mutant and overproduction of the protein in Escherichia coli. Mol Microbiol. 1993;9:183–194. doi: 10.1111/j.1365-2958.1993.tb01680.x. [DOI] [PubMed] [Google Scholar]
- 28.Zhou R, Cao Z, Zhao J. Characterization of HetR protein turnover in Anabaena sp. PCC 7120. Arch Microbiol. 1998;169:417–423. doi: 10.1007/s002030050592. [DOI] [PubMed] [Google Scholar]
- 29.Zhou R, Wei X, Jiang N, Li H, Dong Y, Hsi K L, Zhao J. Evidence that HetR protein is an unusual serine-type protease. Proc Natl Acad Sci USA. 1998;95:4959–4963. doi: 10.1073/pnas.95.9.4959. [DOI] [PMC free article] [PubMed] [Google Scholar]