Abstract
The genome sequence of the extremely thermophilic bacterium Aquifex aeolicus encodes alternative sigma factor ςN (ς54, RpoN) and five potential ςN-dependent transcriptional activators. Although A. aeolicus possesses no recognizable nitrogenase genes, two of the activators have a high degree of sequence similarity to NifA proteins from nitrogen-fixing proteobacteria. We identified five putative ςN-dependent promoters upstream of operons implicated in functions including sulfur respiration, nitrogen assimilation, nitrate reductase, and nitrite reductase activity. We cloned, overexpressed (in Escherichia coli), and purified A. aeolicus ςN and the NifA homologue, AQ_218. Purified A. aeolicus ςN bound to E. coli core RNA polymerase and bound specifically to a DNA fragment containing E. coli promoter glnHp2 and to several A. aeolicus DNA fragments containing putative ςN-dependent promoters. When combined with E. coli core RNA polymerase, A. aeolicus ςN supported A. aeolicus NifA-dependent transcription from the glnHp2 promoter. The E. coli activator PspFΔHTH did not stimulate transcription. The NifA homologue, AQ_218, bound specifically to a DNA sequence centered about 100 bp upstream of the A. aeolicus glnBA operon and so is likely to be involved in the regulation of nitrogen assimilation in this organism. These results argue that the ςN enhancer-dependent transcription system operates in at least one extreme environment, and that the activator and ςN have coevolved.
The special form of bacterial RNA polymerase (RNAP) containing alternative sigma factor ςN or ς54 (ςN-RNAP), initiates transcription by a mechanism quite distinct from RNAP containing the major ς70 sigma factor (35, 39, 48). Transcription initiation by ςN-RNAP requires the hydrolysis of nucleoside triphosphate, catalyzed by activator proteins bound to upstream activator sequences (enhancer elements). The mechanisms of regulation involving this form of RNAP are among the most sophisticated in bacteria (52).
Since this novel form of RNAP was first recognized in enteric bacteria (25, 29, 47), ςN has been discovered in many other bacteria, including several proteobacteria, as well as in the gram-positive Bacillus subtilis (14) and in Planctomyces limnophila (36). Furthermore, ςN appears to be encoded by the genomes of the hyperthermophile Aquifex aeolicus (15), the spirochete Borrelia burgdorferi (18), and the obligate intracellular pathogens Chlamydia trachomatis (51) and Chlamydia pneumoniae (31).
The ςN proteins from Klebsiella pneumoniae and Escherichia coli have been the subject of much genetic and biochemical analysis (e.g., see references 8, 20, and 23). Although considerable progress has been made towards understanding the mechanisms by which ςN performs its function, to date no high-resolution structural data are available for this system. Thermophilic homologues of mesophilic proteins have often been shown to be tractable for structural studies. For example, the crystal structures of the histidine kinase domain of CheA from Thermotoga maritima and the core RNAP from Thermus aquaticus have recently been elucidated (4, 57). The publication of the complete genome sequence (15) of A. aeolicus, a hyperthermophilic bacterium capable of growth at temperatures as high as 95°C, provides a potentially valuable resource for the study of thermostable proteins.
A tRNA-binding protein has previously been purified from A. aeolicus (41), and Klenk et al. (33) have demonstrated activity and thermostability in the RNAP holoenzyme from its relative Aquifex pyrophilus. Characteristics of the hyperthermophile A. aeolicus pose important questions about the evolutionary origins and relationships of the bacteria. According to 16S ribosomal DNA (rDNA)-based phylogenies, the order Aquifecales (of which A. aeolicus is a member) represents the deepest branch of the bacterial evolutionary tree (7). Thus, it has been argued that by studying these organisms we can gain insights about the original bacterial ancestor. In particular, the hyperthermophilic nature of the deepest-branching organisms has been cited to support the hypothesis of a hyperthermophilic origin for the bacteria (1). However, several protein-based phylogenetic analyses, including that of the RNAP β and β′ subunits, have thrown into question the status of the Aquifecales as the deepest-branching group (e.g., see reference 33). Based on sequences of ς70 sigma factors, Aquifecales appears to be an early-branching member of the proteobacteria (22).
In this paper we describe the heterologous overexpression and purification of the ςN and show its core RNAP-binding activity and sequence-specific DNA binding. We also purified a NifA-like protein from A. aeolicus and demonstrate activator-dependent transcription activity of a holoenzyme containing A. aeolicus ςN in vitro. We predict possible functions of the ςN-RNAP mode of transcription in A. aeolicus.
MATERIALS AND METHODS
Computer-based analysis.
DNA sequences were examined for potential ςN-dependent promoters using SEQSCAN (B. T. Nixon: http://www.bmb.psu.edu/seqscan/seqform1.htm). Similarity searches were performed using the PSI-BLAST server (2).
Strains, plasmids, and DNA.
Plasmids are listed in Table 1. Samples of genomic DNA from A. aeolicus and expression strain E. coli C41(DE3) were kind gifts from R. Huber and J. Walker, respectively (15, 40).
TABLE 1.
Plasmids used in this study
| Plasmid | Relevant feature | Source | Reference |
|---|---|---|---|
| pFC60 | E. coli glnHp2 wild-type promoter | B. Magasanik | 10 |
| pFC60-m11 | E. coli glnHp2-m11 mutant promoter | B. Magasanik | 10 |
| pDJS42.12 | pET29b+::A. aeolicus rpoN | Our collection | This work |
| pDJS48.8 | pET29b+::A. aeolicus AQ_218 | Our collection | This work |
| pMTHςN | pET28b+::K. pneumoniae rpoN | Our collection | 19 |
Oligonucleotides, enzymes, and PCR.
PCR was carried out using standard protocols with BioTaq (Bioline) or, where proofreading activity was required, Bio-X-Act thermostable DNA polymerase (Bioline). E. coli core RNAP was purchased from Epicentre Technologies.
Cloning.
The sequences of rpoN, encoding ςN, and nifA, encoding AQ_218, were amplified from A. aeolicus genomic DNA by PCR using primers (Table 2, sets A and B) which introduced NdeI sites immediately upstream of the start codons and BamHI sites immediately downstream of the stop codons. These restriction sites were exploited to clone the PCR-amplified fragments into pET29b+ (Novagen). The inserts in the resulting plasmids (pDJS42.12 and pDJS48.8) were verified by DNA sequencing.
TABLE 2.
Oligonucleotide PCR primers
| Primer set | Forward primera | Reverse primerb | Description |
|---|---|---|---|
| A | NNNCATATGTTAAATCAGAGATTAGAAGTAAGG | NNNGGATCCTTAAATCCTTCTTTCCCTTGAGGAGGG | A. aeolicus rpoN coding sequence |
| B | NNNCATATGGATTTAAAGGTAGAGATCGAAACT | GTGCGACAGAACGGATCCTTACTC | A. aeolicus nifA coding sequence |
| C | CCACATCATCACACAATCG | CAGACTTCATAGCATTTCC | E. coli glnHp2 and glnHp2-m11 promoters |
| D | GGCGTAAACTATTTTGTAATC | ACCCATACTTCTTACCTCCATT | A. aeolicus dhsU upstream region |
| E | CTTCCTCGCCGTGTTAGAA | TCATCTTCCTTCCTCCTTGTAAGTT | A. aeolicus glnB upstream region |
| F | CCCGTGGGAACACCTTCGAGT | CATTCTTTCACTCCTCCTGAG | A. aeolicus nirB upstream region |
| G | AAACACGAAAAATTTCCATACCTT | GAGGGCGTCCTTCACTTCGTCG | A. aeolicus glnB region 147-bp fragment |
| H | GGAAATTAAGCTGATTTAGTACC | GAGGGCGTCCTTCACTTCGTCG | A. aeolicus glnB region 203-bp fragment |
NdeI site is underlined.
BamHI site is underlined.
Proteins, overexpression, and purification.
Overexpression and purification of K. pneumoniae ςN from pMTHςN has been described previously (19). For A. aeolicus ςN and AQ_218, cultures of C41(DE3)(pDJS42.12) and C41(DE3)(pDJS48.8) were grown at 37°C until mid-log phase, when they were induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 12 to 16 h at 25°C. Cells were harvested by centrifugation and resuspended in 50 mM Tris-HCl (pH 8.0), 50 mM NaCl, and 5% glycerol containing a protease inhibitor cocktail (Boehringer). Cells were disrupted by two passages through a French press. Following centrifugation at 20,000 × g for 40 min, the overexpressed protein was found predominantly in the soluble fraction, which was then heated to 80°C for 10 min and again centrifuged at 20,000 × g for 40 min. The A. aeolicus protein remained in the soluble fraction and was purified (to more than 95% homogeneity as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis by affinity chromatography on a heparin-Sepharose column and eluted with a NaCl gradient, using fast-protein liquid chromatography (Pharmacia). The purified proteins were dialyzed into 50 mM Tris-HCl (pH 8.0)–50 mM NaCl–50% glycerol–1 mM dithiothreitol (DTT) for storage at −20°C.
Core-binding assays.
Core-binding assays were carried out as described previously (20) but using a Tris–3-(cyclohexylamino)-1-propanesulfonic acid (Tris-CAPS) buffer system (38) at pH 9.5.
Promoter DNA fragments.
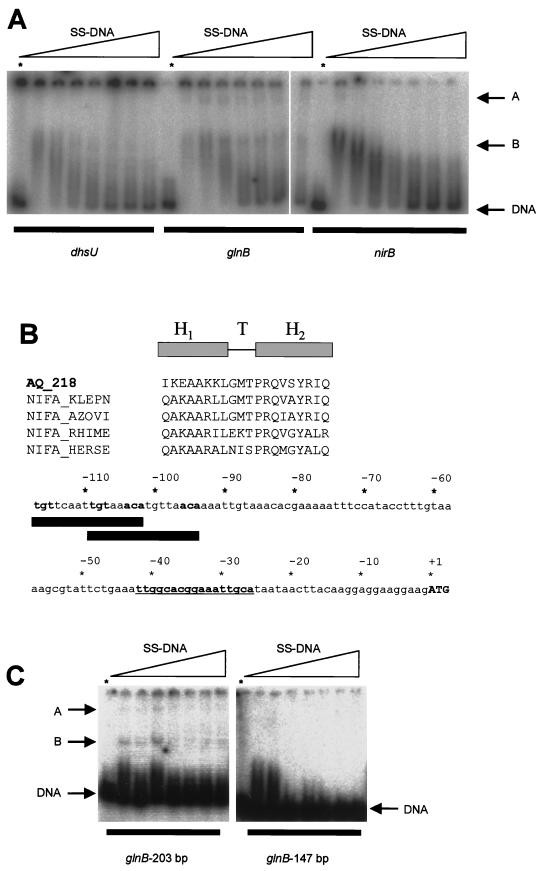
The wild-type E. coli glnHp2 and mutant glnHp2-m11 fragments (approximately 250 bp) were amplified from pFC60 and pFC60-m11, respectively, using the primers listed in Table 2 (set C). Three putative A. aeolicus promoter DNA fragments, dhsU, glnB, and nirB, were amplified from genomic DNA by PCR using primers in Table 2 (sets D to F). Each of these fragments contained the ATG start codon plus about 240 bp of upstream sequence. All promoter DNA fragments were radioactively end labeled using T4 polynucleotide kinase as described in reference 8. Two additional fragments were amplified from the A. aeolicus glnB region (Table 2, sets G and H). These two fragments differed from each other in that the shorter fragment lacked a 56-bp sequence from the upstream end (see Fig. 4).
FIG. 4.
DNA-binding activities of A. aeolicus NifA-like protein (AQ_218). (A) Binding to radioactively labeled 240-bp DNA fragments containing the dhsU, glnB, and nirB upstream regions. Assays were carried out in the presence of various concentrations of competitor DNA (17, 34, 68, 102, 136, 170, and 204 μg ml−1). Lanes marked with an asterisk contained DNA only (no protein). (B) Comparison of the amino acid sequences of the putative HTH motifs of AQ_218 and those of K. pneumoniae (NIFA_KLEPN), Azotobacter vinelandii (NIFA_AZOVI), Sinorhizobium meliloti (NIFA_RHIME), and Herbaspirillum seropedicae (NIFA_HERSE) and the DNA sequence upstream of the A. aeolicus glnB gene. Two matches to the NifA-binding motif TGTN10ACA are marked with thick solid lines. The ςN-binding site is highlighted in boldface, as is the ATG start codon. (C) Binding to radioactively labeled A. aeolicus glnB-region DNA fragments. These two fragments were 147 and 203 bp, the shorter fragment lacking the −141 to −86 (with respect to the start codon) sequence GGAAATTAAGCTGATTTAGTACCTTTGTTCAATTGTAAACATGTTAACAAAATTGT (with the TGTN10ACA motifs underlined) at the upstream end. Assays were carried out in the presence of varying concentrations of competitor DNA (17, 34, 68, 102, 136, 170, and 204 μg of salmon sperm DNA [SS-DNA] ml−1). Lanes marked with an asterisk contained DNA only (no protein).
Protein-DNA-binding assays.
Binding of ςN and holoenzyme to radioactively labeled promoter DNA fragments was detected by a gel mobility shift assay (8, 9). Holoenzyme was prepared by mixing E. coli core RNAP and ςN subunit (from K. pneumoniae or A. aeolicus) in a 1:2 molar ratio. Assay reactions included concentrations up to 1 μM of the ςN protein or 100 nM holoenzyme (see figure legends for details), 25 nM labeled DNA, and 680 μg of salmon sperm DNA ml−1 in buffer (40 mM Tris-HCl [pH 8.0], 0.1 mM EDTA, 10% [vol/vol] glycerol, 100 nM NaCl, 250 mM KCl, and 1 mM DTT). For AQ_218 binding to DNA, except where stated otherwise, assays contained 100 nM AQ_218 protein (with respect to monomer), 25 nM labeled DNA, and up to 204 μg of salmon sperm DNA ml−1 in TAPS buffer (50 mM Tris-acetate [pH 8.0], 100 mM potassium acetate, 8 mM magnesium acetate, 27 mM ammonium acetate, 1 mM DTT, 3.5% [wt/vol] polyethylene glycol 8000).
In vitro ςN transcription activity assays.
Transcription assays were performed as described previously (20) except that supercoiled pFC60 and pFC60-m11 plasmid DNAs, containing the wild-type E. coli glnHp2 and mutant glnHp2-m11 promoters, were used as templates. The ςN-RNAP activator PspFΔHTH (30) is a useful test activator, since it is constitutively active without a requirement for covalent modification and is stable. It lacks DNA-binding activity and exerts its activity while in solution. The assays were performed at 30, 37, and 48°C and contained 50 nM RNAP holoenzyme assembled from the E. coli core and a sixfold excess of ςN. Template DNA (10 nM) was preincubated with holoenzyme for 15 min in STA buffer (25 mM Tris-acetate) [pH 8.0], 8 mM Mg acetate, 10 mM KCl, 1 mM DTT, 3.5% [wt/vol] polyethylene glycol 6000) with 4 mM GTP, 100 nM activator protein (A. aeolicus NifA-like or E. coli PspFΔHTH [30]), and 0.1 mM UTP before heparin, ATP, CTP, and radioactively labeled UTP (1.5 μCi) were added. Since the initial three nucleotides of the transcript are TGT, this preincubation would favor the formation of stable initiated complexes.
RESULTS AND DISCUSSION
The genome of A. aeolicus contains an open reading frame (ORF) apparently encoding a ςN protein and five ORFs predicted to encode NtrC/NifA-family transcriptional activators (15), designated AQ_218, AQ_164, AQ_230, AQ_1117, and AQ_1792. ORFs AQ_164, AQ_230, and AQ_1117 resemble the NtrC/AtoC subfamily of activators, members of which contain the receiver component of a two-component signal transduction system, whereas AQ_218 and AQ_1792 closely resemble the VnfA/NifA subfamily (unpublished data), whose members lack the conserved aspartate residue which undergoes phosphorylation in the two-component signal transduction system (43).
Purification of A. aeolicus ςN and NifA proteins.
ORFs AQ_599 and AQ_218, encoding A. aeolicus ςN and NifA proteins, respectively, were cloned, and the proteins were overproduced and purified using the T7 system (see Materials and Methods). The purified ςN was recognized by anti-E. coli ςN antiserum in Western blots (data not shown).
A. aeolicus ςN binding to core RNAP.
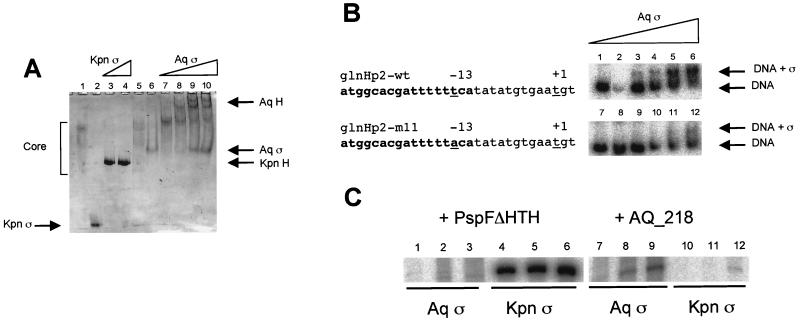
The purified ςN from A. aeolicus bound E. coli core RNAP, as demonstrated by a gel mobility shift assay (Fig. 1A). The band comprising free core RNAP was depleted, and new species formed upon addition of the A. aeolicus ςN. The lower degree of mobility of the A. aeolicus ςN-E. coli core complex compared to that of the K. pneumoniae ςN-E. coli core complex may be explained by the relatively high pI of the A. aeolicus protein (predicted to be 8.15, versus 4.60 for K. pneumoniae ςN; also compare Fig. 1, lanes 2 and 6). Also, whereas the K. pneumoniae ςN-E. coli core complex ran as a single band on the gel, the A. aeolicus ςN-E. coli core complex ran as at least two discrete bands, suggesting more than one conformation, a phenomenon that has previously been reported for several mutant K. pneumoniae ςN-E. coli core complexes (45, 54).
FIG. 1.
Activity of purified A. aeolicus ςN protein. (A) Core RNAP binding. Reaction mixtures contained 1 μM E. coli core RNAP and 0.5, 1, 2, and 3 μM A. aeolicus ςN (lanes 7 to 10, respectively). Lane 6, 3 μM A. aeolicus ςN and no core; lanes 1 and 5, 1 μM E. coli core RNAP and no ςN; lanes 3 and 4, 1 μM E. coli core RNAP plus K. pneumoniae ςN (0.5 and 1 μM, respectively); lane 2, 3 μM K. pneumoniae ςN and no core. The positions of core, K. pneumoniae ςN-RNAP holoenzyme (Kpn H), K. pneumoniae ςN (Kpn ς), A. aeolicus ςN-RNAP holoenzyme (Aq H), and A. aeolicus ςN (Aq ς) are indicated. (B) Sequence-specific binding of purified A. aeolicus ςN protein to wild-type E. coli glnHp2 (glnHp2-wt) and mutant glnHp2-m11 promoter DNA. The nucleotide sequences of the promoters are shown with the transcription start site (+1) and site of the mutation (−13) underlined. DNA-binding reaction mixtures contained 0 to 1 μM ςN. Positions of free DNA (DNA) and bound DNA (DNA + ς) are indicated. Assays contained 0 (lanes 1 and 7), 0.5 (lanes 2 and 8), 1 (lanes 3 and 9), 2 (lanes 4 and 10), 4 (lanes 5 and 11), and 5 (lanes 6 and 12) μM A. aeolicus ςN. (C) Activator-dependent transcription activity of ςN-RNAP holoenzymes. Transcription from the E. coli wild-type glnHp2 promoter was assayed using holoenzymes containing either K. pneumoniae ςN (lanes 4 to 6 and 10 to 12) or A. aeolicus ςN (lanes 1 to 3 and 7 to 9) in the presence of either E. coli PspFΔHTH activator protein (lanes 1 to 6) or A. aeolicus protein AQ_218 (lanes 7 to 12). Reactions were carried out at 30°C (lanes 1, 4, 7, and 10), 37°C (lanes 2, 5, 8, and 11), and 48°C (lanes 3, 6, 9, and 12).
Sequence-specific DNA-binding activity of A. aeolicus ςN.
The purified A. aeolicus ςN bound to the E. coli glnHp2 wild-type promoter with significantly greater affinity than the mutant glnHp2-m11, which has a single base substitution at position −13 (Fig. 1B). This DNA-binding activity was resistant to high levels of nonspecific competitor DNA (680 ng ml−1), further supporting the sequence specificity of the interaction. In contrast to the ς70 family of sigma factors, the A. aeolicus ςN, like those of the enteric bacteria (5), clearly has its DNA-binding determinants available without requiring core binding.
In vitro transcription activity.
The heterologous RNAP holoenzyme, containing A. aeolicus ςN and E. coli core subunits, supported activator-dependent transcription from the wild-type E. coli glnHp2 promoter in vitro, giving a transcript of the same length as that from the holoenzyme containing K. pneumoniae ςN (see Fig. 1C). When the E. coli activator protein PspFΔHTH was added to the transcription assays in place of A. aeolicus AQ_218, transcription levels were very low (PspFΔHTH is a good activator of RNAP containing K. pneumoniae ςN). Furthermore, there was only a very low level of transcription activation by AQ_218 with a holoenzyme containing K. pneumoniae ςN. This requirement for both A. aeolicus ςN and A. aeolicus activator for appreciable levels of transcription but tolerance of heterologous core subunits suggests that the A. aeolicus ςN and activators have coevolved and might directly interact with each other during activation of transcription initiation. Transcription could not be detected from the glnHp2-m11 promoter, demonstrating promoter-specific interaction (data not shown).
The transcriptional activity of the RNAP holoenzyme containing A. aeolicus ςN was lower than that of the holoenzyme containing K. pneumoniae ςN. It is possible that at the relatively low temperatures at which the assays were performed (30, 37, and 48°C, while the optimal growth temperature for A. aeolicus is about 85°C) the thermophile proteins are in a “frozen” state that is kinetically unfavorable to the conformational changes required for transcription initiation. Indeed, activity increased with temperature over the range tested. Higher temperatures were not used, since 48°C is close to the upper temperature limit for E. coli RNAP activity (55).
Many ςN-dependent promoters have integration host factor binding sites (26). The A. aeolicus genome encodes no obvious homologues of himA or himB, and so involvement, if any, of a sequence-specific DNA bending protein in A. aeolicus ςN-dependent transcription is presently unclear. Also, the fact that purified A. aeolicus NifA did not require additional factors for the activation of transcription suggests its activity might be negatively regulated in vivo.
We were unable to rigorously demonstrate that nucleoside hydrolysis by A. aeolicus NifA is required for transcription initiation due to the apparent instability of open complexes using the heterologous RNAP holoenzyme (data not shown). However, based on similarity of central domain sequences in the A. aeolicus NtrC/NifA family activators to those of well-studied members of this family (48), we believe that the mechanism of activation is likely to be similar.
Identification of potential ςN-dependent promoters in the A. aeolicus genome.
RNAP containing ςN transcribes from promoters with a consensus sequence of YTGGCACGRNNNTTGCW with the highly conserved GG and GC motifs at −24 and −12, respectively, relative to the +1 transcription start site (3). The A. aeolicus genome sequence was examined for potential ςN-dependent promoters. Six good matches to the consensus were found in predicted noncoding regions immediately upstream of predicted coding regions (Fig. 2). Four of the six predicted coding regions showed significant similarity to genes of known function and are designated glnB, fhp, dhsU, and nirB. The remaining two predicted coding regions encode the hypothetical proteins AQ_087 and AQ_1119.
FIG. 2.
Identification of putative A. aeolicus ςN-dependent promoters. Close matches to the consensus ςN-binding motif were identified immediately upstream of dhsU, glnB, nirB, AQ_087, and AQ_1119 ORFs in the A. aeolicus genome sequence. Putative ςN-binding motifs are shown in boldface, as are the ATG start codons. Possible ribosome binding sites are underlined.
In A. aeolicus, it appears that glnB, which encodes regulatory protein PII, is the first gene in an operon with glnA. This glnBA operon structure is found in several other bacteria, including Azospirillum brasilense (16) and Rhodobacter sphaeroides (58). It is well documented that gln genes are transcribed by ςN-RNAP in several proteobacteria (37), where their transcription is activated by the nitrogen-regulatory protein C (NtrC). However, in the gram-positive B. subtilis, transcriptional regulation is not dependent on the ςN homologue, ςL (28). It should also be noted that glnA, encoding glutamine synthetase, is found in the genomes of Thermotoga maritima (44) and Mycobacterium spp. (13), which have been completely sequenced, and yet no homologues of ςN have been identified.
A second operon that appears to have a ςN-RNAP-dependent promoter contains genes designated fhp, cynS, glnBi, nasA, and narB. Each of the ORFs designated fhp (flavohemoprotein), cynS (cyanate hydrolase), nasA (nitrate transporter), and narB (nitrate reductase) shows similarity to genes associated with nitrate reduction (reviewed in reference 42). The glnBi ORF appears to encode a protein similar to the PII and GlnK family of nitrogen regulatory proteins.
A potential ςN-dependent promoter is found in the A. aeolicus genome upstream of dhsU, predicted to encode a flavocytochrome C sulfide dehydrogenase (10). It appears that dhsU is part of an operon with soxF and fccB′, which are predicted to encode a Rieske-I iron-sulfur protein and a sulfide dehydrogenase flavoprotein, respectively (49, 50). Therefore, this operon is almost certainly involved in sulfur respiration.
In A. aeolicus we have identified a potential ςN-dependent promoter upstream of nirB, predicted to encode the large subunit of cytoplasmic NADH-dependent nitrite reductase, whose function is detoxification of nitrite formed as a result of nitrate respiration (12). It appears that cobA and trpD2 may also be in the nirB operon, and they are predicted, respectively, to encode products involved in cobalamin and tryptophan biosynthesis (17, 27). Although nitrate respiration has not been demonstrated in A. aeolicus, it has been observed in the close relative A. pyrophilus (7). Previously, ςN-RNAP had been implicated in the regulation of nitrite reductase in Pseudomonas stutzeri (24). ORFs AQ_087 and AQ_1119 have no significant similarity to sequences of known function. A PSI-BLAST search using AQ_1119 as a probe revealed significant similarity to an ORF of unknown function, AF0913, in the genome of the archaeon Archaeglobus fulgidus.
To date, ςN-RNAP has been implicated in the transcriptional regulation of such diverse functions as degradation of xylene and toluene, transport of dicarboxylic acids, pilin synthesis, nitrogen fixation, hydrogen uptake, flagellar assembly, arginine catabolism, alginate production, rhamnolipid production, acetoin catabolism, mannose uptake, proline iminopeptidase activity, nitrogen assimilation, nitrate respiration (reviewed in references 3, 35, and 39), pathogenesis (34), development (32), and RNA modification (21). It is also predicted to be involved in the transcriptional regulation of other ς factors (46, 53). Here, we have extended the range of potential functions further to include sulfur respiration and have provided evidence that ςN-RNAP is probably involved in nitrogen assimilation and nitrate respiration in a hyperthermophile.
ςN- and ςN-RNAP holoenzyme binding to putative A. aeolicus ςN-dependent promoters.
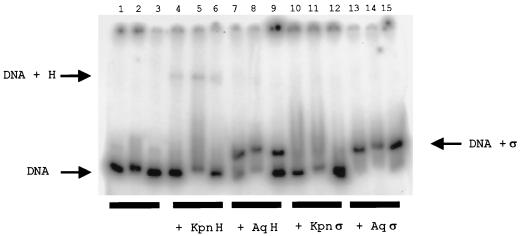
We tested ςN and ςN-RNAP holoenzyme binding to 240-bp fragments containing noncoding sequences upstream of A. aeolicus dhsU, glnB, and nirB (Fig. 3) in the presence of high levels of competitor DNA. A. aeolicus ςN bound to all three fragments, reducing the electrophoretic mobility of the labeled DNA (Fig. 3, lanes 13 to 15). K. pneumoniae ςN clearly also bound the dhsU and glnB upstream fragments (Fig. 3, lanes 10 and 11), albeit less stably than did A. aeolicus ςN, and only very weakly bound the nirB upstream fragment (lane 12). The RNAP holoenzyme containing K. pneumoniae ςN bound to all three fragments to produce species with much lower degrees of electrophoretic mobility (Fig. 3, lanes 4 to 6). However, the binding of the RNAP holoenzyme containing A. aeolicus ςN appeared to give rise to complexes with mobility similar to that of those arising from ςN and DNA alone (lanes 4 to 6). The amount of stable holoenzyme-DNA complex formed was below the limit of detection by this assay. The very low levels of DNA-holoenzyme complex formation, perhaps partly due to the heterologous nature of the system, might explain the relatively low levels of the transcription (Fig. 1C) on the glnHp2 promoter by RNAP containing A. aeolicus ςN.
FIG. 3.
Binding of RNAP holoenzyme and ςN to radioactively labeled 240-bp DNA fragments containing the dhsU (lanes 1, 4, 7, 10, and 13), glnB (lanes 2, 5, 8, 11, and 14), and nirB (lanes 3, 6, 9, 12, and 15) upstream regions was carried out in the presence of high concentrations of competitor DNA (680 μg of salmon sperm DNA ml−1). Lanes 1 to 3, DNA only (no protein); lanes 4 to 6, holoenzyme with K. pneumoniae ςN; lanes 7 to 9, holoenzyme with A. aeolicus ςN; lanes 10 to 12, K. pneumoniae ςN; lanes 13 to 15, A. aeolicus ςN. The positions of free DNA (DNA), holoenzyme-DNA complex (DNA + H), and ςN-DNA complex (DNA + ς) are indicated.
It is noteworthy that A. aeolicus ςN appeared to bind the A. aeolicus DNA fragments more tightly than did K. pneumoniae ςN (compare lanes 10 to 12 with lanes 13 to 15 in Fig. 3). This was also the case for binding on the E. coli glnHp2 promoter DNA fragment (data not shown). We speculate that a stronger DNA-binding activity may be advantageous to the thermophile in overcoming the kinetic effects of high temperature.
DNA-binding activity of A. aeolicus NifA-like protein (AQ_218).
The NtrC/NifA family ςN-dependent transcriptional activators bind to enhancerlike elements, usually located up to 200 bp upstream of the transcription start site (35, 48). Therefore, we tested whether purified protein AQ_218 bound to 240-bp fragments containing noncoding sequences upstream of A. aeolicus dhsU, glnB, and nirB. In a gel mobility shift assay, AQ_218 protein bound to all three DNA fragments to give complexes with reduced mobility (band B in Fig. 4A). The stability of these complexes was very sensitive to the concentration of competitor DNA (salmon sperm DNA). Binding to the glnB fragment appeared to be significantly more resistant to the presence of competitor DNA than was binding to the dhsU and nirB fragments. Furthermore, binding to the glnB upstream sequence gave rise to an additional species that was not observed in assays containing dhsU and nirB. Therefore, we examined the glnB upstream DNA sequence and its interaction with AQ_218 in more detail.
NifA and VnfA are involved in the transcriptional regulation of the nitrogenase complex in azotrophic proteobacteria. Since A. aeolicus lacks recognizable nifHDK genes, the two NifA-like proteins (AQ_218 and AQ_1792) presumably perform a different function in this organism. Two canonical NifA binding sites (6), TGTN10ACA, are found centered around 105 bp upstream of the glnBA operon of A. aeolicus (Fig. 4B), so it is possible that one (or both) of the NifA homologues is involved in the regulation of nitrogen assimilation rather than of nitrogen fixation. The second helix (H2 in Fig. 4B) of the NifA helix-turn-helix (HTH) motif is believed to be responsible for recognition of specific DNA sequences. Although at present we cannot predict DNA-protein-binding affinities from sequence alone, the high degree of similarity between the putative HTH motifs of AQ_218 and known NifA sequences (Fig. 4B) suggested that they might recognize similar DNA sequences. Therefore, we tested the binding of AQ_218 to two A. aeolicus DNA sequences which differed in that one (of 203 bp) contained the two TGTN10ACA motifs and the other (of 147 bp) was truncated so that it lacked a 56-bp sequence containing these motifs (see Materials and Methods; Fig. 4C). Under the conditions of this binding assay, the A. aeolicus protein AQ_218 bound to the 203-bp fragment to form species A and B (Fig. 4C) but failed to form these complexes with the 147-bp fragment lacking the putative NifA binding motifs. Therefore, we conclude that the A. aeolicus glnB upstream region contains determinants for sequence-specific DNA binding by AQ_218 and that this protein is likely to be involved in positive regulation of the glnBA operon in A. aeolicus.
The occurrence of several potential ςN-dependent promoters and five ORFs potentially encoding ςN-dependent activators supports the proposition that the enhancer-dependent ςN-RNAP mode of transcription is functional in A. aeolicus. Direct binding of A. aeolicus promoter specificity factor ςN to these sequences supports this conclusion. We present indirect evidence that the ςN-RNAP holoenzyme is involved in sulfur respiration, nitrogen assimilation, reduction of nitrate, and nitrite reductase activity, as well as having at least two unknown functions, in A. aeolicus. Furthermore, based on a demonstrated specific DNA-binding activity, we propose that AQ_218, a NifA-like protein from A. aeolicus, is involved in the regulation of the glnBA operon. Clearly, the NifA subfamily of proteins is unlikely to have arisen twice independently, so there are two possible explanations for these proteins' occurrence both in A. aeolicus and in the proteobacteria: (i) horizontal gene transfer between these bacteria occurred, or (ii) the lineage containing A. aeolicus split from the proteobacteria after the evolution of the NifA subfamily. Since NifA has not been found in any bacteria other than A. aeolicus and azotrophic proteobacteria, this would suggest that the split was much more recent than is implied by the 16S rDNA data and is more consistent with protein-based models of bacterial evolution (e.g., see references 22 and 33). The RNAP holoenzyme containing purified A. aeolicus ςN appears to be silent for transcription, and the A. aeolicus NifA-like protein (AQ_218) overcomes this inhibition to allow transcription, as is the case for all homologous mesophile systems so far studied. It seems that the interactions that silence the polymerase, and others that allow enhancer dependence and which are known to critically involve ςN region I sequences (20), are at least partly intact when A. aeolicus ςN combines with E. coli core RNAP. To the best of our knowledge, this is the first report of transcriptional activation using components from a hyperthermophilic bacterium.
ACKNOWLEDGMENTS
This work was supported by CEC contract Bio4 CT 97-2143 to M.B. M.-T.G. was supported by a CEC Marie Curie fellowship. S.R.W. was supported by the Local Education Authority of Karlsruhe, Germany.
We are grateful to R. Huber for the A. aeolicus genomic DNA, to J. Walker for C41(DE3), to B. Magasanik for plasmids, and to A. Ishihama for the anti-ςN antiserum. We thank B. T. Nixon for making available his SEQSCAN program, R. Wassem for helpful discussion about NifA, and J. Bartley for automated DNA sequencing service.
REFERENCES
- 1.Achenbach-Richter L, Gupta R, Stetter K O, Woese C R. Were the original eubacteria thermophiles? Syst Appl Microbiol. 1987;9:34–39. doi: 10.1016/s0723-2020(87)80053-x. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Koonin E V. Iterated profile searches with PSI-BLAST—a tool for discovery in protein databases. Trends Biochem Sci. 1998;23:444–447. doi: 10.1016/s0968-0004(98)01298-5. [DOI] [PubMed] [Google Scholar]
- 3.Barrios H, Valderrama B, Morett E. Compilation and analysis of ς54-dependent promoter sequences. Nucleic Acids Res. 1999;27:4305–4313. doi: 10.1093/nar/27.22.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bilwes A M, Alex L A, Crane B R, Simon M I. Structure of CheA, a signal-transducing histidine kinase. Cell. 1999;96:131–141. doi: 10.1016/s0092-8674(00)80966-6. [DOI] [PubMed] [Google Scholar]
- 5.Buck M, Cannon W. Specific binding of the transcription factor sigma-54 to promoter DNA. Nature. 1992;358:422–424. doi: 10.1038/358422a0. [DOI] [PubMed] [Google Scholar]
- 6.Buck M, Miller S, Drummond M, Dixon R. Upstream activator sequences are present in the promoters of nitrogen fixation genes. Nature. 1986;320:374–378. [Google Scholar]
- 7.Burggraf S, Olsen G J, Stetter K O. A phylogenetic analysis of Aquifex pyrophilus. Syst Appl Microbiol. 1992;15:352–356. doi: 10.1016/S0723-2020(11)80207-9. [DOI] [PubMed] [Google Scholar]
- 8.Cannon W, Gallegos M T, Casaz P, Buck M. Amino-terminal sequences of ςN (ς54) inhibit RNA polymerase isomerization. Genes Dev. 1999;13:357–370. doi: 10.1101/gad.13.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casaz P, Buck M. Probing the assembly of transcription initiation complexes through changes in ςN protease sensitivity. Proc Natl Acad Sci USA. 1997;94:12145–12150. doi: 10.1073/pnas.94.22.12145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Z W, Koh M, Van Driessche G, Van Beeumen J J, Bartsch R G, Meyer T E, Cusanovich M A, Mathews F S. The structure of flavocytochrome c sulfide dehydrogenase from a purple phototrophic bacterium. Science. 1994;266:430–432. doi: 10.1126/science.7939681. [DOI] [PubMed] [Google Scholar]
- 11.Claverie-Martin F, Magasanik B. Positive and negative effects of DNA bending on activation of transcription from a distant site. J Mol Biol. 1992;227:996–1008. doi: 10.1016/0022-2836(92)90516-m. [DOI] [PubMed] [Google Scholar]
- 12.Cole J. Nitrate reduction to ammonia by enteric bacteria: redundancy, or a strategy for survival during oxygen starvation? FEMS Microbiol Lett. 1996;136:1–11. doi: 10.1111/j.1574-6968.1996.tb08017.x. [DOI] [PubMed] [Google Scholar]
- 13.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell B G, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 14.Debarbouille M, Martin-Verstraete I, Kunst F, Rapoport G. The Bacillus subtilis sigL gene encodes an equivalent of sigma 54 from gram-negative bacteria. Proc Natl Acad Sci USA. 1991;88:9092–9096. doi: 10.1073/pnas.88.20.9092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, Huber R, Feldman R A, Short J M, Olsen G J, Swanson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 16.de Zamaroczy M, Paquelin A, Elmerich C. Functional organization of the glnB-glnA cluster of Azospirillum brasilense. J Bacteriol. 1993;175:2507–2515. doi: 10.1128/jb.175.9.2507-2515.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Escalante-Semerena J C, Suh S-J, Roth J R. cobA function is required for both de novo cobalamin biosynthesis and assimilation of exogenous corrinoids in Salmonella typhimurium. J Bacteriol. 1990;172:273–280. doi: 10.1128/jb.172.1.273-280.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, Gwinn M, Dougherty B, Tomb J F, Fleischmann R D, Richardson D, Peterson J, Kerlavage A R, Quackenbush J, Salzberg S, Hanson M, van Vug R, Palmer N, Adams M D, Gocayne J, Venter J C, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 19.Gallegos M T, Buck M. Sequences in ςN determining holoenzyme formation and properties. J Mol Biol. 1999;288:539–553. doi: 10.1006/jmbi.1999.2704. [DOI] [PubMed] [Google Scholar]
- 20.Gallegos M T, Cannon W V, Buck M. Functions of the ς54 Region I in trans and implications for transcription activation. J Biol Chem. 1999;274:25285–25290. doi: 10.1074/jbc.274.36.25285. [DOI] [PubMed] [Google Scholar]
- 21.Genschik P, Drabikowski K, Filipowicz W. Characterization of the Escherichia coli RNA 3′-terminal phosphate cyclase and its ς54-regulated operon. J Biol Chem. 1998;273:25516–25526. doi: 10.1074/jbc.273.39.25516. [DOI] [PubMed] [Google Scholar]
- 22.Gruber T M, Bryant D A. Characterization of the group 1 and group 2 sigma factors of the green sulfur bacterium Chlorobium tepidum and the green non-sulfur bacterium Chloroflexus aurantiacus. Arch Microbiol. 1998;170:285–296. doi: 10.1007/s002030050644. [DOI] [PubMed] [Google Scholar]
- 23.Guo Y, Wang L, Gralla J D. A fork junction DNA-protein switch that controls promoter melting by the bacterial enhancer-dependent sigma factor. EMBO J. 1999;18:3736–3745. doi: 10.1093/emboj/18.13.3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartig E, Zumft W G. The requirement of RpoN (sigma factor ς54) in denitrification by Pseudomonas stutzeri is indirect and restricted to the reduction of nitrite and nitric oxide. Appl Environ Microbiol. 1998;64:3092–3095. doi: 10.1128/aem.64.8.3092-3095.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirschman J, Wong P K, Sei K, Keener J, Kustu S. Products of nitrogen regulatory genes ntrA and ntrC of enteric bacteria activate glnA transcription in vitro: evidence that the ntrA product is a sigma factor. Proc Natl Acad Sci USA. 1985;82:7525–7529. doi: 10.1073/pnas.82.22.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoover T R, Santero E, Porter S, Kustu S. The integration host factor stimulates interaction of RNA polymerase with NifA, the transcriptional activator of nitrogen fixation operons. Cell. 1990;63:11–22. doi: 10.1016/0092-8674(90)90284-l. [DOI] [PubMed] [Google Scholar]
- 27.Horowitz H, Christie G E, Platt T. Nucleotide sequence of the trpD gene, encoding anthranilate synthetase component II of Escherichia coli. J Mol Biol. 1982;156:245–256. doi: 10.1016/0022-2836(82)90326-6. [DOI] [PubMed] [Google Scholar]
- 28.Hu P, Leighton T, Ishkhanova G, Kustu S. Sensing of nitrogen limitation by Bacillus subtilis: comparison to enteric bacteria. J Bacteriol. 1999;181:5042–5050. doi: 10.1128/jb.181.16.5042-5050.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunt T P, Magasanik B. Transcription of glnA by purified Escherichia coli components: core RNA polymerase and the products of glnF, glnG, and glnL. Proc Natl Acad Sci USA. 1985;82:8453–8457. doi: 10.1073/pnas.82.24.8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jovanovic G, Rakonjac J, Model P. In vivo and in vitro activities of the Escherichia coli ς54 transcription activator, PspF, and its DNA-binding mutant, PspFΔHTH. J Mol Biol. 1999;285:469–483. doi: 10.1006/jmbi.1998.2263. [DOI] [PubMed] [Google Scholar]
- 31.Kalman S, Mitchell W, Marathe R, Lammel C, Fan J, Hyman R W, Olinger L, Grimwood J, Davis R W, Stephens R S. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat Genet. 1999;21:385–389. doi: 10.1038/7716. [DOI] [PubMed] [Google Scholar]
- 32.Keseler I M, Kaiser D. ςN, a vital protein for Myxococcus xanthus. Proc Natl Acad Sci USA. 1997;94:1979–1984. doi: 10.1073/pnas.94.5.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klenk H P, Meier T D, Durovic P, Schwass V, Lottspeich F, Dennis P P, Zillig W. RNA polymerase of Aquifex pyrophilus: implications for the evolution of the bacterial rpoBC operon and extremely thermophilic bacteria. J Mol Evol. 1999;48:528–541. doi: 10.1007/pl00006496. [DOI] [PubMed] [Google Scholar]
- 34.Klose K E, Mekalanos J J. Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol Microbiol. 1998;28:501–520. doi: 10.1046/j.1365-2958.1998.00809.x. [DOI] [PubMed] [Google Scholar]
- 35.Kustu S, Santero E, Keener J, Popham D, Weiss D. Expression of ς54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol Rev. 1989;53:367–376. doi: 10.1128/mr.53.3.367-376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leary B A, Ward-Rainey N, Hoover T R. Cloning and characterization of Planctomyces limnophilus rpoN: complementation of a Salmonella typhimurium rpoN mutant strain. Gene. 1998;221:151–157. doi: 10.1016/s0378-1119(98)00423-5. [DOI] [PubMed] [Google Scholar]
- 37.Magasanik B. Regulation of transcription of the glnALG operon of Escherichia coli by protein phosphorylation. Biochimie. 1989;71:1005–1012. doi: 10.1016/0300-9084(89)90104-1. [DOI] [PubMed] [Google Scholar]
- 38.McLellan T. Electrophoresis buffers for polyacrylamide gels at various pH. Anal Biochem. 1982;126:94–99. doi: 10.1016/0003-2697(82)90113-0. [DOI] [PubMed] [Google Scholar]
- 39.Merrick M J. In a class of its own—the RNA polymerase sigma factor ςN (ς54) Mol Microbiol. 1993;10:903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 40.Miroux B, Walker J E. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 41.Morales A J, Swairjo M A, Schimmel P. Structure-specific tRNA-binding protein from the extreme thermophile Aquifex aeolicus. EMBO J. 1999;18:3475–3483. doi: 10.1093/emboj/18.12.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moreno-Vivián C, Cabello P, Martínez-Luque M, Blasco R, Castillo F. Prokaryotic nitrate reduction: molecular properties and functional distinction among bacterial nitrate reductases. J Bacteriol. 1999;181:6573–6584. doi: 10.1128/jb.181.21.6573-6584.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morett E, Segovia L. The ς54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J Bacteriol. 1993;175:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nelson K E, Clayton R A, Gill S R, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Nelson W C, Ketchum K A, McDonald L, Utterback T R, Malek J A, Linher K D, Garrett M M, Stewart A M, Cotton M D, Pratt M S, Phillips C A, Richardson D, Heidelberg J, Sutton G G, Fleischmann R D, Eisen J A, Fraser C M, et al. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 45.Oguiza J A, Gallegos M T, Chaney M K, Cannon W V, Buck M. Involvement of the ςN DNA-binding domain in open complex formation. Mol Microbiol. 1999;33:873–885. doi: 10.1046/j.1365-2958.1999.01542.x. [DOI] [PubMed] [Google Scholar]
- 46.Pallen M. RpoN-dependent transcription of rpoH? Mol Microbiol. 1999;31:393. doi: 10.1046/j.1365-2958.1999.01148.x. [DOI] [PubMed] [Google Scholar]
- 47.Reitzer L J, Magasanik B. Transcription of glnA in E. coli is stimulated by activator bound to sites far from the promoter. Cell. 1986;45:785–792. doi: 10.1016/0092-8674(86)90553-2. [DOI] [PubMed] [Google Scholar]
- 48.Rombel I, North A, Hwang I, Wyman C, Kustu S. The bacterial enhancer-binding protein NtrC as a molecular machine. Cold Spring Harbor Symp Quant Biol. 1998;63:157–166. doi: 10.1101/sqb.1998.63.157. [DOI] [PubMed] [Google Scholar]
- 49.Schmidt C L, Anemuller S, Schafer G. Two different respiratory Rieske proteins are expressed in the extreme thermoacidophilic crenarchaeon Sulfolobus acidocaldarius: cloning and sequencing of their genes. FEBS Lett. 1996;388:43–46. doi: 10.1016/0014-5793(96)00511-x. [DOI] [PubMed] [Google Scholar]
- 50.Simon J, Gross R, Klimmek O, Ringel M, Kroger A. A periplasmic flavoprotein in Wolinella succinogenes that resembles the fumarate reductase of Shewanella putrefaciens. Arch Microbiol. 1998;169:424–433. doi: 10.1007/s002030050593. [DOI] [PubMed] [Google Scholar]
- 51.Stephens R S, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov R L, Zhao Q, Koonin E V, Davis R W. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 52.Stock J B, Stock A M, Mottonen J M. Signal transduction in bacteria. Nature. 1990;344:395–400. doi: 10.1038/344395a0. [DOI] [PubMed] [Google Scholar]
- 53.Studholme D J, Buck M. Novel roles of ςN in small genomes. Microbiology. 2000;146:4–5. doi: 10.1099/00221287-146-1-4. [DOI] [PubMed] [Google Scholar]
- 54.Studholme D J, Finn R D, Chaney M K, Buck M. The C-terminal 13 amino acids of ςN are required for structure and function. Arch Biochem Biophys. 1999;371:234–240. doi: 10.1006/abbi.1999.1426. [DOI] [PubMed] [Google Scholar]
- 55.Wang J T, Syed A, Gralla J D. Multiple pathways to bypass the enhancer requirement of sigma 54 RNA polymerase: roles for DNA and protein determinants. Proc Natl Acad Sci USA. 1997;94:9538–9543. doi: 10.1073/pnas.94.18.9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang L, Gralla J D. Multiple in vivo roles for the −12-region elements of sigma 54 promoters. J Bacteriol. 1998;180:5626–5631. doi: 10.1128/jb.180.21.5626-5631.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang G, Campbell E A, Minakhin L, Richter C, Severinov K, Darst S A. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- 58.Zinchenko V, Churin Y, Shestopalov V, Shestakov S. Nucleotide sequence and characterization of the Rhodobacter sphaeroides glnB and glnA genes. Microbiology. 1994;140:2143–2151. doi: 10.1099/13500872-140-8-2143. [DOI] [PubMed] [Google Scholar]