Abstract
Aim
The aim was to evaluate the impact of intermittent fasting (IF) on human body mass index (BMI) and serum lipid profile thorough constructive rectification of gut microbiota.
Methods and results
Fourteen healthy women and thirty-one men were included in the study. Their blood and fecal samples were collected before and at the end of the study. Blood parameters, anthropometric values, and gut microbiology were noted to investigate the impact of intermittent fasting (IF) on human gut microbiota and physiology. Our data revealed that IF reduces the body weight and improves blood lipid profile, such as increasing high-density lipoprotein (HDL) and decreasing total cholesterol, triglycerides, and low- and very low-density lipoprotein levels. IF also decreases culturable aerobic bacterial count and increased fungal count. It was also found that the gut metagenome is altered considerably after IF. The human fecal bacterial diversity exhibited significant changes in decreased overall bacterial population, increased bacterial diversity (alpha diversity), and promoted evenness within the bacterial population at the species level. Anti-inflammatory bacteria Lactobacillus and Bifidobacterium were favorably increased, while pathogenic bacteria were decreased.
Conclusion
Collectively, these results indicated that IF could improve lipid profile and body weight in humans, and the potential mechanisms might be via regulating gut microbiota.
Significance and impact of the study
We demonstrated for the first time that IF improved body weight and blood lipid profile, indicating that IF could mitigate gut microbiota in humans.
Keywords: human Gut microbiota, intermittent fasting, serum lipid profile, BMI, obesity
Introduction
Metabolic disorders and associated diseases like diabetes, cardiovascular diseases, inflammatory bowel disease, and various cancers including breast and colorectal cancer are the leading causes of death worldwide (Gibson and Macfarlane, 1995; Berg, 1996; Arumugam et al., 2011; Faulds and Dahlman-Wright, 2012; Rajilić-Stojanović et al., 2015; Blake and Suchodolski, 2016). The diversity and composition of gut microbiota have been identified as an important contributor to metabolic associated disorders, as healthy individuals contain highly diverse gut microbiota, while diseased individuals harbor aberrant and less diverse microbiota in the gut (Friedman, 2009; Le Chatelier et al., 2013; Harmsen and de Goffau, 2016). Moreover, the use of antibiotics in infants predisposes them to unusual weight gain and deposition of adipose tissues later in life, reaffirming that disruptions in gut microbiota composition can lead to the onset of obesity and then metabolic diseases (Cox and Blaser, 2015; Turta and Rautava, 2016). Fecal transplantation from healthy individuals has been proposed to restore normal microbiota. Several studies have demonstrated that fecal transplantation from healthy individuals improved metabolic disorders (cardiovascular disease, type diabetes, and obesity) in diseased individuals (Elinav et al., 2019). Although effective, this approach can lead to a reshuffling of gut microbiome dysbiosis and the emergence of pathobionts (Glauser, 2011; Weil and Hohmann, 2015). Similarly, other available weight loss strategies and modulation of gut microbiota by using antibiotics, prebiotics, and probiotics have many limitations. Like antibiotics kill good microbiota along with pathogens, similarly, it is challenging for probiotic strains to persist in the gut for a longer period. The impact of prebiotic products is also not specific (Druart et al., 2014; Erejuwa et al., 2014). Gonzalez’s group has recently demonstrated in mice that intermittent calorie restriction activates brown adipose tissue and ameliorates obesity by most likely modulating the composition of gut microbiota (Li et al., 2017). In humans, fasting reduces LDL, Vldl, and triglyceride levels, as well as increases HDL levels, as observed in type 2 diabetes patients (Shehab et al., 2012). Intermittent fasting also enhances cardiovascular health, regulates blood pressure by reducing brain natriuretic peptide levels, enhances renal activity (Masoro, 2005; Fontana et al., 2010; Abargouei et al., 2012; Trabelsi et al., 2012; Tiboura et al., 2015), and therefore has become a popular practice around the globe to achieve effective weight loss and improve metabolism. It is still not known how intermittent fasting influences the gut microbiota in humans and whether the regime can support the growth of microbial communities that increase metabolism associated with increased thermogenesis. Also, continuous energy restriction requires a well-defined diet plan that has met with non-compliance issues and adaption. Intermitted fasting or complete abstention from all or selected foods and fluids aiming to restrict food intake for a defined period of time is an easy alternate, is simple to follow, and has shown equal effectiveness in insulin sensitivity, cardiovascular functions, weight loss, and other health biomarkers (Rizza et al., 2014; Mattson et al., 2017).
Here, before and after controlled experimental study in humans was designed to test the effect of intermittent fasting on weight loss, body mass index (BMI), blood lipid profile, C-reactive protein (CRP), and gut microorganisms of obese, normal, and lean but otherwise healthy adult volunteers. We demonstrate that IF improves BMI and reduces triglycerides and low-density lipoprotein levels. Intriguingly, our results also demonstrate that the IF not only decreases gut microbe numbers but also increases their diversity and promotes the growth of microbial species that increase bodily energy metabolism, as identified by the metagenomic analyses.
Materials and methods
This study was carried out at the Department of Microbiology, Faculty of Biological Sciences, Quaid-i-Azam University, Islamabad, Pakistan.
Study population and setup
The Intermittent Fasting 91 (16/8 IF) or lean gain method was employed in this study as previously described (Berkhan, 2010). According to this method, participants abstain from foods and liquids for at least 16 h for 26 days. The remaining 8 h of the day serve as the feeding window. During this time, users may eat as many (or few) meals as desired, with the most frequent iteration being two and three meals.
Approval of the study
The study was approved by the Bioethical Committee (BEC) of Quaid-i-Azam University, Islamabad, Pakistan, under protocol number BEC-FBS-QAU2018-109, and all the methods were performed in accordance with the relevant guidelines and regulations advised by the Bioethical Committee (BEC) of Quaid-i-Azam University, Islamabad, Pakistan.
A campaign for the person-to-person meeting was conducted to meet the students (both male and female). The protocol and expected outcomes of the study were orally explained to everyone during the meetings. Students who agreed to participate were enrolled in the study. The age of the volunteers ranged from 18 to 35 years.
Informed consent for study: An informed consent was obtained from all the participants for participation in the study, handling of the samples, and data anonymously, and confidentially was also ensured by the research team through this consent (sample of the consent is given in Supplementary material in Supplementary Figure 1). Information about diet and living habits was recorded for all volunteers (given in the Supplementary material in Supplementary Figure 2). Participants were asked to follow the protocol. They were asked to continue their ad libitum eating habits during eating widows and live as per routine.
The volunteers who followed the protocol completed the study and provided the required samples (two blood samples and two fecal samples), and their data were included in the current manuscript for the conclusion of the results. The volunteers who were unable to follow the protocol, or complete the study, or were not able to provide the required samples were excluded from the conclusion of the results. Statistical analysis (mean, standard deviation, standard error, paired t-test, and agglomerative hierarchical clustering) was performed using Microsoft Excel 2016 and XLSTAT 2019.1.2.
Initially, an equal number of male and female volunteers were selected for the study, but there was a dropout in the samples, and finally, 31 male volunteers and 14 female volunteers were able to complete the study and provide the samples. Furthermore, volunteers were placed in any group of lean, underweight, normal, and overweight or obese where applicable.
Selection criteria included healthy (physically and mentally fit upon looking and feeling), non-smokers, no antimicrobial use (in last 4 weeks), no laxatives use (in last 2 months), no weight reducer use, no diarrhea (in last 1 week), no gastrointestinal tract disease, no exercise more than 10 h/week, and no supplement consumption; those participants provided the samples (two blood samples and two fecal samples), followed the protocol, and completed the study.
Blood collection and analysis
In total, 10 ml of blood (at once) was collected from the volunteers two times in specified blood collection tubes. One sample was collected 2 days earlier to the study, and the second was collected on the 25th or 26th day of IF (upon convivence). The samples were processed soon after collection, and serum was separated and kept at −20°C until further analysis. Different parameters were recorded from blood analysis. These factors include C-reactive proteins, serum total cholesterol level, serum triglycerides level (TG), high-density lipoproteins (HDL) levels, low-density lipoproteins (LDL) levels, and very low-density lipoproteins (vLDL) levels.
Fecal sample collection and analysis
The stool samples were collected twice from volunteers. The samples were collected in sterile stool collection containers following the protocols (Wu et al., 2010; Escobar et al., 2014). Soon after collection, each fecal sample was separated into two portions. One portion was stored at −80°C for later use and DNA extraction. In total, 1 g of the fecal sample was taken, diluted in sterile phosphate buffer saline (PBS), taken this as initial dilution, and diluted serially. In total, 100 μl from each dilution was poured and spread on different culture media for the isolation of fecal microorganisms (Castillo et al., 2006). For total microbial aerobic count (TMAC), BHIA (Brain Heart Infusion Agar; Oxoid United Kingdom) media were used. The de Man–Rogosa–Sharpe Agar (MRSA, Oxoid United Kingdom) with 5.4 pH was used to selectively grow the Lactobacillus spp. The M17 (Oxoid United Kingdom) was used to grow Enterococcus, Streptococcus, and Lactococcus spp. MacConkey agar was used to grow the family of Enterobacteriaceae, and OGA (Oxytetracycline Glucose Agar; Oxoid United Kingdom) was used for fungal growth. The inoculated plates were incubated at 37°C for 48–72 h (Lim et al., 2004). After incubation, plates were examined for microbial growth. The color, size, and morphology of the grown colonies were noted. The total number of colonies on each plate was counted using a colony counter. An average of 30–300 colonies was used for the actual colony count.
Extraction of metagenomic DNA from stool
Favor prep Stool DNA isolation mini kit (Favorgen) was used for DNA extraction. Following their designed protocol, 100mg stool was taken in a microtube to which 200mg glass beads were added; frequently, 300 μl of SDE1 buffer and 20 μl of Proteinase K were transferred to the same microtube for disruption of microbial cells. Then, this mixture was vortexed at high speed for 5–7 min and was incubated at 70°C first for 10 min and for an additional 5 min to lyse gram-positive cells. To enhance homogenization, the stool sample was vortexed three times during the period of incubation. The sample was let to cool and added 100 μl of SDE2 buffer. The sample was kept on an ice pack for 5 min and then centrifuged for 5 min at 14,000 rpm. The supernatant was taken and shifted to another microtube with the addition of SDE3, 200 μl buffer. The mixture after mixing was incubated for 2 min at room temperature. The mixture again was centrifuged at said speed for 2 min, and the supernatant was transferred to another sterile microtube. In total, 1 μl of RNase of concentration 100 mg/ml was transferred to the supernatant for the degradation of RNA. After removing the drops from the lid via spinning, it was supplemented with 250 μl SDE4 buffer and 250 μl cooled ethanol and mixed by pulse overtaxing. Collection columns were positioned in the collection tube, and the sample was shifted to it. The sample was centrifuged at maximum speed; the column was shifted to another collection tube, and flow-through was discarded; columns were transferred to the next clean collection tube, and flow-through was wasted. Wash buffer (750 μl) was used for washing the impurities by adding them to the column and then centrifuging at maximum speed for 2 min. This step was repeated. For drying the column and avoiding residual contamination, the collection column was centrifuged for 3 min.
The elution tube was taken, and the column was shifted to it. The 70 μl elution buffer was transferred right to the center of the column for elution of the DNA. For complete absorbance of elution buffer into the column, it was centrifuged later to the addition of elution buffer. DNA was eluted after centrifugation and was stored at −20°C for further processing.
Processing of extracted metagenomic DNA for sequencing
Amplification and purification of extracted DNA
PCR was carried out for the amplification of the V4 region of the 16S rRNA gene using forward primer 515F (GTGCCAGCMGCCGCGGTAA) and reverse primer 806R (GGATCACNVGGGTWTCTAAT). The amplification was carried out in triplicates using a volume reaction of 20 μl (Caporaso et al., 2011). HotStar Taq Plus Master Mix (Qiagen, Valencia, CA, United States) and samples were used for the preparation of reaction volume. PCR was performed at these settings: early denaturing, 94°C for 3 min, then tailed by 30 cycles of denaturation at 94°C for 30 sec, annealing at 53°C for 40 s, and finally amplification at 72°C for 1 min. After the entire process, confirmation of the amplification and quality of the gene fragment were tested in 2% agarose gel. Then, as per molecular weight and concentration of DNA, all the samples were joined to each other and were purified by using calibrated Ampure XP beads (Bioscience Corporation, MA, United States).
Library preparation
Following the Illumina TruSeq DNA library preparation protocol, libraries were prepared from the input of 1 ug of total DNA along with the aforementioned primers via the Illumina TruSeq PCR-Free Library Preparation Kit.
Sequencing
Sequencing was performed at MRDNA sequencing laboratory (Shallowater, TX, United States) using Illumina MiSeq next-generation sequencer. The MiSeq platform performs clonal amplification, genomic DNA sequencing, and data analysis (from assembly to functional classification) with base calling, alignment, variant calling, and reporting in a single run. It performs de novo assembly and generates contigs (FASTA) without using a reference genome. It also generates FASTQ files that can be used by third-party tools for analysis. It further aligns the reads against the reference genome and generates a sample report (bam format). Finally, it classifies bacteria according to their 16SrDNA.
Analysis of the data
All physiochemical data was analysed for statistical significances by using R software (version 3.3.3). One way and two-way ANOVA was used, and results are presented as highly significant (p < 0.01) and significant (p < 0.05). The post hoc test; Student Newman Keuls used to differentiate means after ANOVA significance. In addition to this the two-sample Wilcoxon signed-rank test was used to compare serum lipid profile before and after IF.
Raw sequencing data analysis was done by using MRDNA pipelines (MR DNA, Shallowater, TX, United States). Errors were minimized, and noises were removed. Finally, operational taxonomic units (OTUs) were generated, and chimeras were extruded. OTUs were further defined by clustering at 3% divergence (similarity 97%). Final OTUs were classified taxonomically using BLASTn against a database derived from GreenGenes, RDPII, and RDPI (Desantis et al., 2006). Diversity analysis was done by using QIME2 Microbiome.
Results
Impact of intermittent fasting on BMI
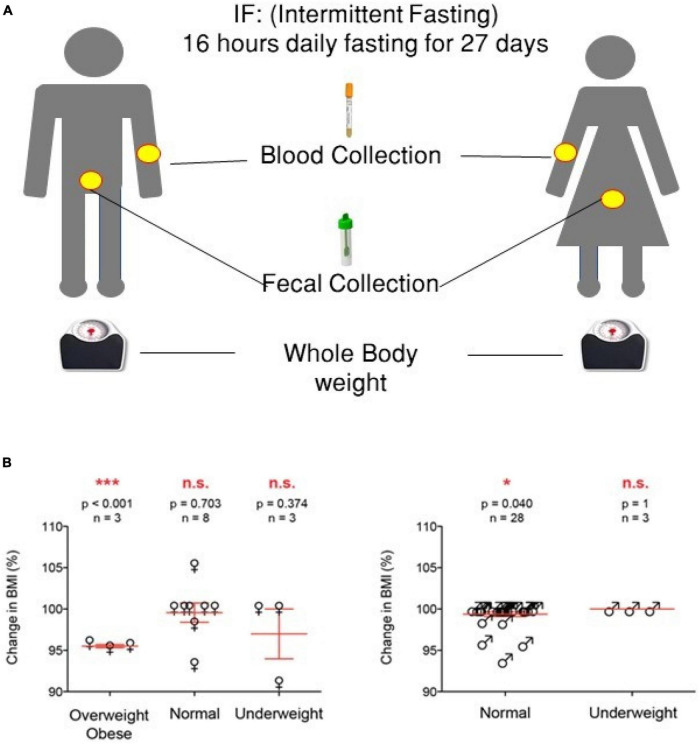
The impact of IF on weight was personalized, the weight in the majority of normal-weighted male volunteers (25/28; p = 0.040) did not change, and only three volunteers reduced their weight, but their BMI remained in the normal range (18.5–24.9; p = 1). The weight of underweighted male volunteers (2/3) increased (1–2 kg) (p = 0.040). The weight of normal female volunteers (5/16) increased significantly (p = 0.001) within the normal BMI range (18.5–24.9), while underweighted female volunteers (2/2) body weight increased and the weight of obese/overweight (3/3 out of total 14) decreased significantly (p < 0.001) (Figure 1 and Tables 1, 2).
FIGURE 1.
Experimental design of the study and summary of the anthropometric results. (A) Experimental design. (B) Summary of the anthropometric values [one-way ANOVA was used, highly significant (p < 0.01) and significant (p < 0.05) values are presented with *** and *, respectively, while non-significant values are presented by N.S].
TABLE 1.
Detail of the anthropometric values of the male volunteers.
| Sample ID | Age (years) | Height (inches) | Weight before study (Kg) | Weight at the end of study (Kg) | BMI before study | BMI at end of study | Fold change BMI | BMI status Before/After study |
| M1 | 24 | 66 | 65 | 65 | 23.13 | 23.13 | NIL | Normal |
| M2 | 22 | 66 | 60 | 60 | 21.35 | 21.35 | NIL | Normal |
| M3 | 22 | 64 | 65 | 65 | 24.59 | 24.59 | NIL | Normal |
| M4 | 23 | 71 | 57 | 57 | 17.52 | 17.52 | NIL | under weight |
| M5 | 22 | 67 | 57 | 57 | 19.68 | 19.68 | NIL | Normal |
| M6 | 23 | 69 | 64 | 62 | 20.83 | 20.18 | NIL | Normal |
| M7 | 22 | 69 | 65 | 64 | 21.16 | 20.83 | 0.95 | Normal |
| M8 | 23 | 63 | 54 | 54 | 21.08 | 21.08 | NIL | Normal |
| M9 | 22 | 68 | 70 | 70 | 23.46 | 23.46 | NIL | Normal |
| M10 | 22 | 71 | 68 | 68 | 20.90 | 20.90 | NIL | Normal |
| M11 | 23 | 63 | 56 | 56 | 21.86 | 21.86 | NIL | Normal |
| M12 | 22 | 65 | 54 | 54 | 19.80 | 19.80 | NIL | Normal |
| M13 | 23 | 64 | 53 | 53 | 20.05 | 20.05 | NIL | Normal |
| M14 | 23 | 67 | 58 | 58 | 20.02 | 20.02 | NIL | Normal |
| M15 | 24 | 68 | 66 | 63 | 22.12 | 21.11 | 0.95 | Normal |
| M16 | 24 | 60 | 75 | 72 | 32.28 | 30.99 | 0.96 | Obese |
| M17 | 23 | 63 | 50 | 50 | 19.52 | 19.52 | NIL | under weight |
| M18 | 22 | 68 | 63 | 61 | 21.11 | 20.44 | 0.97 | Normal |
| M19 | 23 | 65 | 54 | 54 | 19.80 | 19.80 | NIL | Normal |
| M20 | 24 | 60 | 53 | 53 | 22.81 | 22.81 | NIL | Normal |
| M21 | 23 | 64 | 57 | 57 | 21.56 | 21.56 | NIL | Normal |
| M22 | 22 | 77 | 57 | 58 | 14.89 | 15.16 | 1.0 | under weight |
| M23 | 24 | 64 | 63 | 60 | 23.83 | 22.70 | 0.95 | Normal |
| M24 | 24 | 68 | 68 | 66 | 22.79 | 22.12 | 0.97 | Normal |
| M25 | 24 | 68 | 67 | 65 | 22.45 | 21.78 | 0.97 | Normal |
| M26 | 24 | 65 | 58 | 58 | 21.27 | 21.27 | NIL | Normal |
| M27 | 24 | 72 | 68 | 68 | 20.32 | 20.32 | NIL | Normal |
| M28 | 25 | 69 | 70 | 68 | 22.78 | 22.13 | 0.97 | Normal |
| M29 | 23 | 62 | 53 | 53 | 21.36 | 21.36 | NIL | Normal |
| M30 | 24 | 72 | 65 | 64 | 19.43 | 23.12 | 1.2 | Normal |
| M31 | 23 | 66 | 56 | 56 | 21.34 | 21.34 | NIL | Normal |
TABLE 2.
Detail of the anthropometric values of the female volunteers.
| Sample ID | Age (years) | Height (inches) | Weight (Kg) before study | Weight (Kg) at the end of study | BMI before study | BMI at end of study | Fold change BMI | BMI status |
| F1 | 25 | 64 | 52 | 52 | 19.67 | 19.67 | NIL | Normal |
| F2 | 23 | 62 | 62 | 62 | 24.99 | 24.99 | NIL | Underweight |
| F3 | 35 | 64 | 94 | 90 | 35.56 | 34.05 | 0.96 | Obese II |
| F4 | 30 | 68 | 115 | 110 | 38.54 | 36.86 | 0.96 | Obese II |
| F6 | 24 | 63 | 53 | 63 | 20.69 | 24.60 | 1.19 | Normal |
| F7 | 23 | 62 | 62 | 59 | 24.99 | 23.78 | 0.95 | Overweight/normal |
| F9 | 24 | 63 | 52 | 63 | 20.30 | 24.60 | 1.21 | |
| F11 | 25 | 64 | 55 | 62 | 20.81 | 23.45 | 1.13 | Normal |
| F12 | 25 | 65 | 62 | 57 | 22.74 | 20.90 | 0.92 | Normal |
| F14 | 25 | 62 | 58 | 56 | 23.38 | 22.57 | 0.97 | Normal |
| F20 | 24 | 64 | 50 | 64 | 18.91 | 24.21 | 1.28 | Normal |
| F23 | 23 | 63 | 55 | 58 | 21.48 | 22.64 | 1.05 | Normal |
| F25 | 22 | 62 | 45 | 62 | 18.14 | 24.99 | 1.38 | Normal |
| F29 | 34 | 63 | 63 | 63 | 24.60 | 24.60 | NIL | Underweight Normal |
Effect of intermittent fasting on acute inflammatory biomarker blood C-reactive protein
Blood C-reactive protein (CRP) was negative for all subject samples (data not shown).
Effect of Intermittent fasting on serum lipid profile
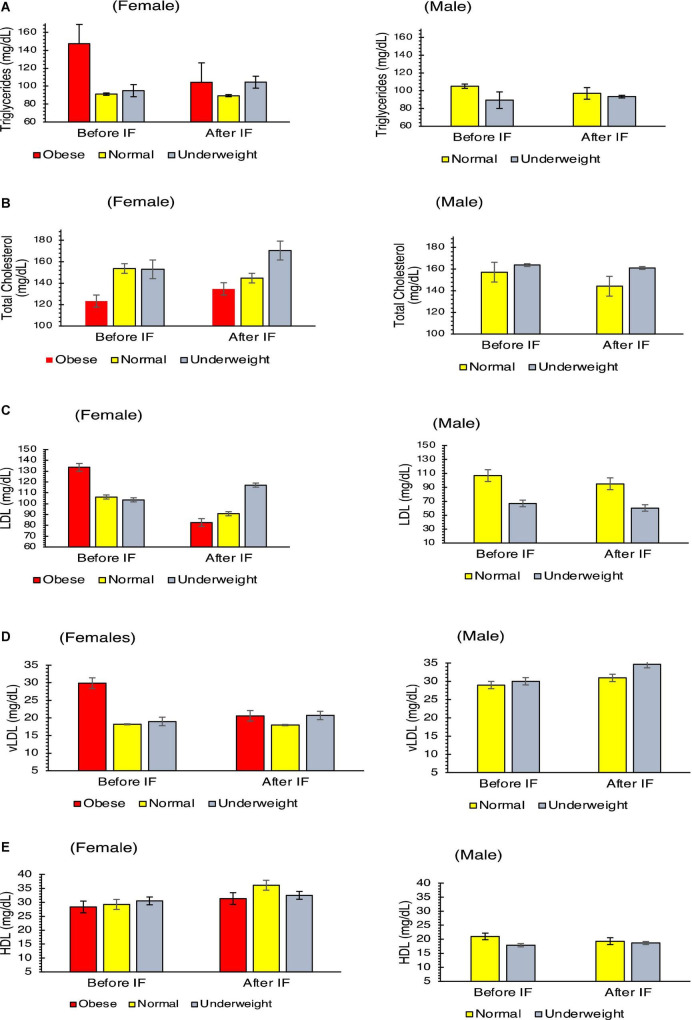
By using two-sample Wilcoxon signed-rank test, significant alternation was noticed in total triglycerides (p = 0.032), total cholesterol (p = 0.008), and low-density lipoprotein (p = 0.004) except in very low-density lipoprotein (p = 0.117). Total triglycerides decreased from 104 to 100 gm/dl and from 89 to 95 mg/dl (Figure 2A and Supplementary Tables 1, 2), total cholesterol decreased from 152 to 141 mg/dl and from 155 to 133 mg/dl (Figure 2B and Supplementary Tables 1, 2), low-density lipoprotein decreased from 104 to 93 mg/dl and from 102 to 87 mg/dl (Figure 2C and Supplementary Tables 1, 2), and very low-density lipoprotein decreased from 21 to 19 mg/dl and from 19 to 18 mg/dl (Figure 2D and Supplementary Tables 1, 2), while total high-density lipoprotein (HDL) increased from 27 to 28 mg/dl and from 28 to 31 mg/dl (Figure 2E and Supplementary Tables 1, 2) for both the female and male samples, respectively.
FIGURE 2.
Physiological parameters of blood serum lipid profile of the volunteers before and after intermittent fasting: (A) triglycerides, (B) total cholesterol, (C) low-density lipoproteins, (D) very low-density lipoproteins, and (E) high-density lipoproteins. Two-sample Wilcoxon signed-rank test was used, all parameters shown a Significant (p < 0.05) alteration except vLDL (p = 0.117).
Intermittent fasting alters the diversity and composition of the fecal microbiota
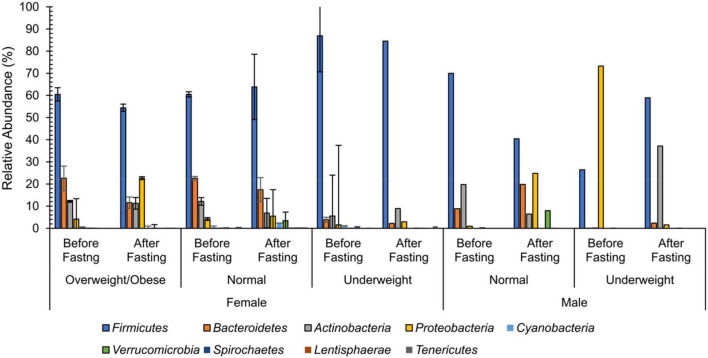
No differences were observed in cultured aerobic bacteria and fungi (Supplementary Figures 1A–E). The impact of intermittent fasting on gut microbiota by culture-independent microbiota was observed. In this assay, ten representative stool samples from individual groups of the human cohort were profiled through 16S DNA sequencing. Reads obtained from sequencing were filtered, low-quality reads were removed, and the remaining reads were classified on phylum, genera, and species level. Similarity tagging was 97% which confirmed the identification of all the species. Richness, α diversity, beta diversity, and dominance of the bacterial population are designed by using the number of reads, OTUs, Shannon, Simpson index, and PCoA (Figure 3 and Table 3). Generally, the total number of OTUs was high in the samples collected before IF while decreasing in samples collected at the end of the study.
FIGURE 3.
Weighted mean relative abundance of bacteria by 16S rDNA region at the phylum level for female and male volunteers before and after intermittent fasting.
TABLE 3.
Detail of the bacterial diversity of all the samples.
| Metagenomic data | Female |
Male |
||||||||
| Obese |
Normal |
Underweight |
Normal |
Underweight |
||||||
| Before fasting | After fasting | Before fasting | After fasting | Before fasting | After fasting | Before fasting | After fasting | Before fasting | After fasting | |
| No. of Reads | 389328 | 250457 | 370675 | 390956 | 263153 | 261845 | 320605 | 370675 | 336297 | 407028 |
| OTUs (Genera) | 197 | 223 | 212 | 215 | 205 | 200 | 194 | 212 | 193 | 206 |
| Genera with ≥ 0.1% | 82 | 57 | 64 | 83 | 64 | 61 | 21 | 64 | 19 | 52 |
*All the numbers are given in mean.
At the phylum level, 99% of the intestinal microbiota of this study subject consisted of nine phyla namely Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, Cyanobacteria, Verrucomicrobia, Spirochaetes, Lentisphaerae, and Tenericutesirmicutes. Except for one group (male underweight before fasting), Firmicutes were the dominant phylum among all the groups. In the female overweight/obese group, in response to intermittent fasting, Firmicutes decreased from the mean percentage of 60.47 to 54.43, Proteobacteria increased from 4.17 to 22.61%, Bacteroidetes decreased from 22.57 to 11.57%, and Actinobacteria decreased from 12.12 to 11.26%. In the female normal group, Firmicutes increased from the mean percentage of 60.47 to 63.81, Proteobacteria increased from 4.17 to 5.5%, Bacteroidetes decreased from 22.57 to 17.41%, and Actinobacteria decreased from 12.12 to 6.90%. Similarly, in the female underweight group, Firmicutes decreased from the mean of 86.91 to 84.54%, Proteobacteria increased from 1.57 to 2.95%, Bacteroidetes decreased from 3.86 to 2.18%, and Actinobacteria increased from 5.53 to 8.92%.
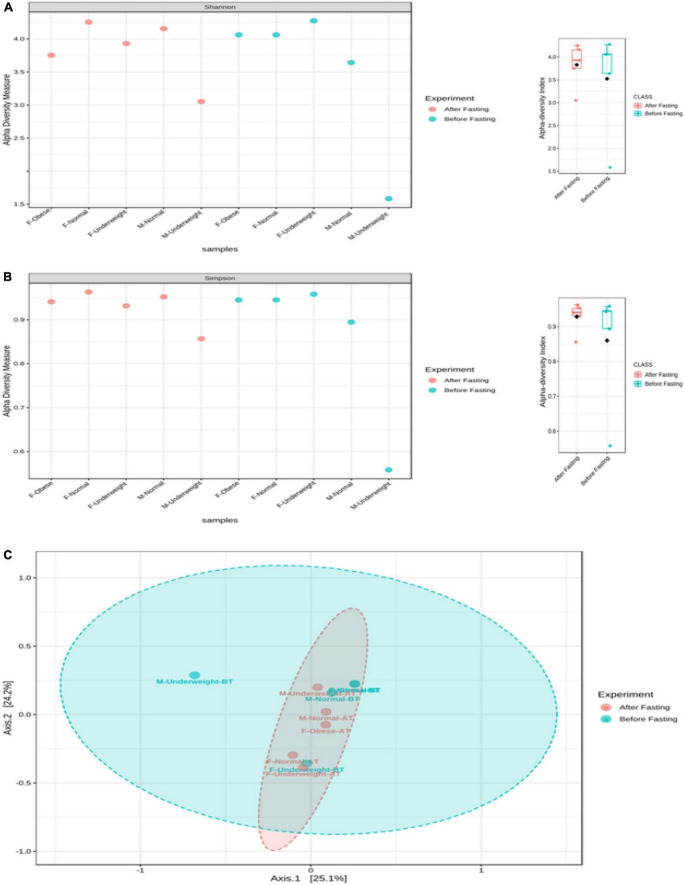
The effect of intermittent fasting on male volunteers is unique for both groups. In the male normal group, Firmicutes decreased from 69.95 to 40.42% at the end of intermittent fasting, Proteobacteria increased from 0.95 to 24.78%, Bacteroidetes increased from 8.87 to 19.81%, and Actinobacteria decreased from 19.72 to 6.43%. In the male underweight group in response to intermittent fasting, Firmicutes increased from the mean percentage of 26.37 to 58.91, Proteobacteria decreased from 73.28 to 1.56%, Bacteroidetes increased from 0.11 to 2.31%, and Actinobacteria increased from 0.19 to 37.14%. Alpha diversity was not significantly altered, while beta diversity was shown significantly altered in obese (Figure 4). The results for the remaining phyla are given in Supplementary Table 3.
FIGURE 4.
Alpha and beta diversity in volunteer’s intestinal microbiota in response to intermittent fasting. Diversity is shown for each sample and comparative for treatment. (A) Alpha diversity (Shannon index), (B) alpha diversity (Simpson index), and (C) beta diversity (PCoA). The box plot shows diversity for each sample and for experimental group.
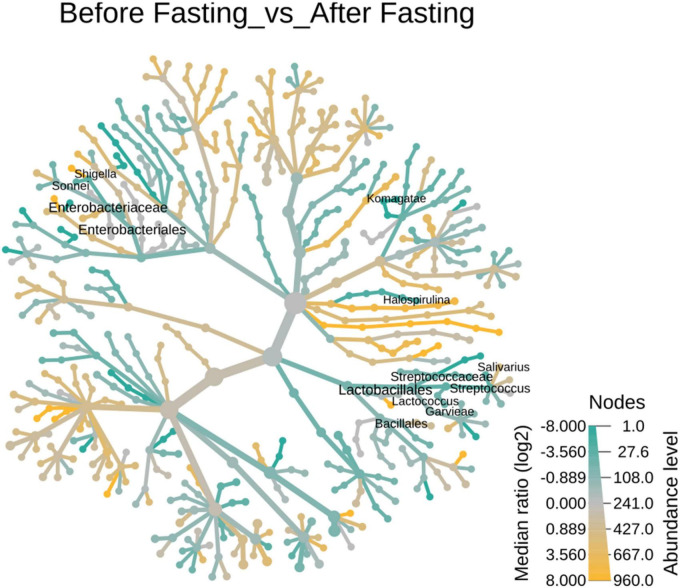
The number and type of bacterial genera altered in response to intermittent fasting. Comparative abundance analysis of bacterial diversity involved 365 genera. In total, 82 genera above 0.1% were identified in the fecal microbiota of the female overweight/obese group before intermittent fasting and 57 genera after fasting. In contrast, we found that women of normal BMI had 64 genera before fasting and 83 genera after intermittent fasting. It was also observed that underweight women had 63 genera before fasting and 61 after intermittent fasting. Normal male volunteers had initially 21 genera that changed to 64. Underweight male volunteer’s genera increase from 19 to 52 (Figure 5).
FIGURE 5.
Heat tree of the abundance of taxa at different ranks of the 23 tree hollows in New South Wales, Australia. The size and color of nodes and edges are correlated with the abundance of taxa. The central node is the total of all the other nodes in the tree for each phylum.
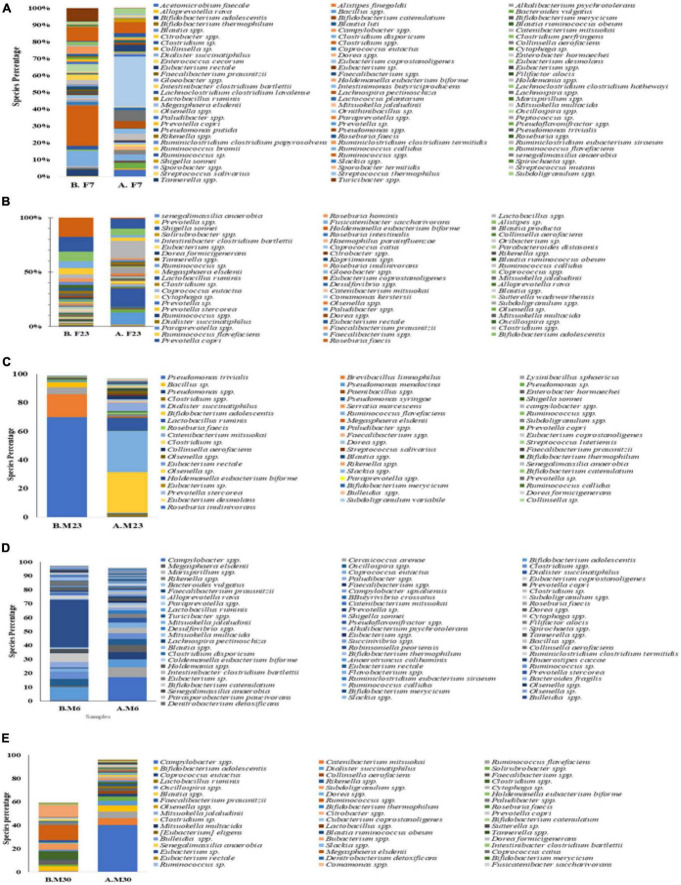
In the female overweight/obese group, Roseburia faecis was the dominant genera making up 15.34% of all the bacterial population that decreased to 0.97% at the end of the trial. The Prevotella copri was the second most dominant group with a percentage of 11.57 that decreased to 7.01% during intermittent fasting. The third most populous genera, Bifidobacterium adolescentis, also decreased from 8.41 to 5.80% at the end of the study. The Faecalibacterium also decreased from 5.76 to 2.91% at the end of the study. The Ruminococcaceae, before fasting, had 4.82% of the overall bacterial population that decreased to 2.91%. Some genera also increased in number when observed at the end of the study trial, for example, Shigella sonnei to 16.35% and Clostridium spp increased to 14.4% at the end of the trial. The results for other genera are depicted in Figure 6A, and detail is given in Supplementary Table 4.
FIGURE 6.
Mean weighted distribution of dominant intestinal bacteria at genus level in response to intermittent fasting: (A) obese female volunteers, (B) normal female volunteers, (C) underweight female volunteers, (D) male normal volunteers, and (E) male normal volunteers.
In the female normal volunteers’ cohort, the Campylobacter spp. were the dominant one making up 16.94% of the total fecal microbiota population that decreased to 0.12% at the end of the study trial. The Clostridiales also changed from 7.28 to 1.3% at the end of the trial. The Puniceicoccales initially was the third major group with 7.23% of the total, but at the end of the study, it was not detected in the countable form. The Bifidobacterium adolescentis also changed in response to intermittent fasting from 5.61 to 0.2%. The Sphingobacteriales initially was 5.28% that later on changed to neglectable count. Dominant genera at the end of the intermittent fasting in female normal volunteers cohort were Ruminococcus, Eubacteriaceae, Barnesiella, Clostridium, and Subdoligranulum with respective percentages of 12.60, 10.91, 5.44, 5.34, and 4.82. For more results, see Figure 6B and Supplementary Table 5.
Before intermittent fasting, the fecal microbiota of the female underweighted volunteer’s cohort contained Clostridium spp. Being the most prominent genera with 18.01% that the end of the trial increased to 24.23%, with Clostridium disporicum being the second most prominent genera with 8.63% of total fecal microbiota. The Ruminococcus was the third highest in number (7.23%), and the Erysipelotrichaceae was the fourth one (6.79%). At the end of the intermittent fasting trial along with clostridium, the Eubacteriaceae with 6.97% and the Bifidobacterium adolescentis with 3.63% were also among the dominant genera. Detail of the other genera is given in Figure 6C and Supplementary Table 6.
In the male normal volunteers’ cohort, the number and type of genera also changed. It was observed that before fasting, the five dominant genera were Ruminococcus (35.46%), Bifidobacterium (12.31%), Eubacterium (7.42%), Clostridium (6.33%), and Oscillospira (5.04%). Later after the intermittent fasting trial, the five prominent genera were Campylobacter spp. (16.94%), Clostridiales (728%), Puniceicoccales (7.23%), Bifidobacterium adolescentis (5.61%), and Sphingobacteriales (5.28%). Detail for the remaining genera is given in Figure 7D and Supplementary Table 7.
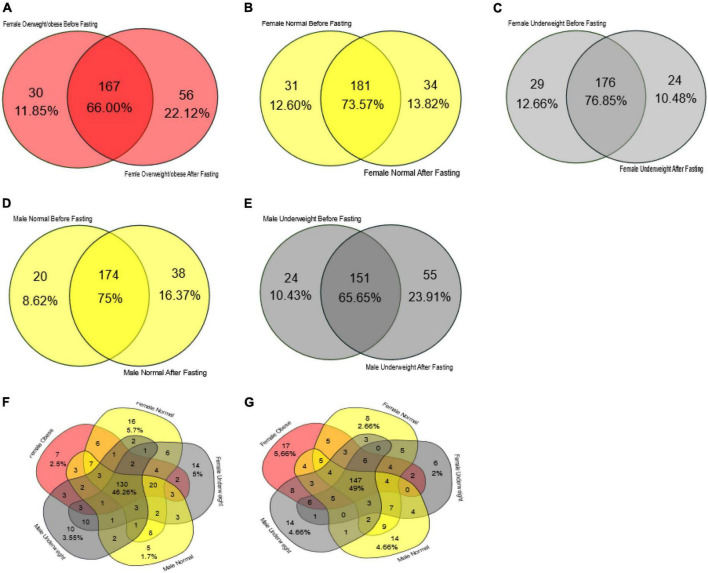
FIGURE 7.
Venn diagram representing core unique and shared intestinal microbiota of female and male volunteers in response to intermittent fasting. (A) Obese/overweight female volunteers, (B) normal female volunteers, (C) underweight female volunteers, (D) male normal volunteers, (E) male underweight volunteers, (F) all volunteers before intermittent fasting, and (G) all volunteers after fasting.
Similarly, it was observed that the fecal microbiota of underweighted male volunteers has changed in response to intermittent fasting. Before the trial, Pseudomonas trivialis (69.47%), Brevibacillus limnophilus (16.23%), Lysinibacillus sphaericus (4.63), and Bacillus (3.64) were dominating while at end of the trial Bifidobacterium adolescentis (28.12%), Ruminococcaceae (25.68), Lactobacillus ruminis (9.15%), Catenibacterium mitsuokai (5.55%), and Clostridiales (3.58%). Detail for the remaining genera is given in Figure 6E and Supplementary Table 8.
In the female samples, the overweight/obese group had 167 bacterial genera as core, and it makes 66.0% of the total bacterial count. At the start of the study, this group had 30 genera unique while at the end of the study it had 56 genera unique (Figure 7A). The female normal group had 181 bacterial genera as the core that makes up 73.57% of the total bacteria present in the fecal sample. Initially, it had 31 bacterial genera which increased to 34 at the end of the study (Figure 7B). The female underweight group had 176 (76.85%) bacterial genera as the core; at the start of the trial, 29 unique genera were associated with this group later on reduced to 24 (Figure 7C). In the male samples, the normal group had 174 bacterial genera as a core that makes up almost 75% of the total bacterial flora. In the remaining 25%, initially, it had eight unique genera, while at the end of the study, there was an increase in unique species and it had 38 genera “Figure 7D.” The male underweight group had 151 species as core, making 65.65% of the total bacterial flora. Initially, there were 24 bacterial genera unique, which increased to 55 later on at the end of the study (Figure 7E). The number of core bacterial genera in all the groups before intermittent fasting was 130 (46.26%) (Figure 7F) and after intermittent fasting was 147 (49%) (Figure 7G).
Discussion
Human physiology is significantly impacted by trillions of gut microbiota having nine million non-redundant genes (Hooper and Gordon, 2001; Yang et al., 2009). Dysbiosis of the gut microbiome causes a variety of metabolic disorders that leads to chronic inflammations which are depicted by overweight and obesity as phenotypic outcomes (Vuksan et al., 2016). Body weight is a physical indicator of metabolic diseases (Swanton et al., 2007). Different strategies like the use of probiotics (Fooks and Gibson, 2002), prebiotics, fecal transplantation (Delzenne et al., 2013), and the use of antibiotics are being tried to restore the gut microbiota to normal. In this context, fasting is considered an ancient therapy (Handschin, 2016). Numerous research studies find out that IF mostly results in weight loss and improvement of blood profile (Aloui et al., 2016).
In this study, it was found that IF has an impact on body mass indexes, blood lipid profiles, and gut microbial profiles. It was found that observing IF reduced the BMI of obese/overweight individuals, and there was a slight reduction of BMI in normal men, while no change was found in normal women. Surprisingly, weight increase was observed in underweight women. These results show that this IF regimen tends to normalize the total body mass without its unwarranted depletion in healthy individuals. Reduction of body weight by IF has been reported (Adlouni et al., 1997; Ley et al., 2006; Kul et al., 2014); however, normalization of body weight is reported for the first time. The number of Bifidobacteria and Lactobacillus increased in all five groups, and it is well established that reduced body weight is linked to an increasing trend in gut microbial diversity, and overweight or obesity is linked to reduced gut bacterial diversity (Ley et al., 2006). In our study, the weight reduction was found linked to an increasing trend in bacterial diversity, while a decreasing trend in bacterial diversity was observed especially in women those gained weights. In an earlier report, the impact of IF on a female subject showed an increase in weight was due to the use of energy-dense diets and less physical activities during the fasting days (Sadeghirad et al., 2014; Savitri et al., 2016). However, the impact of IF is not gender-specific, when tested in the mice model (Piotrowska et al., 2016). Abdelgadir in 2015 conducted a study on three different nations and reported that people using normal diets with physical activities have improved lipid profiles after IF while people eating more or energy-rich diets or doing no or limited physical activities have no change in their lipid profile at the end of study (Abdelgadir et al., 2015). Yeoh et al. (2015) performed a study in 2015 on 29 volunteers in Singapore, and at the end of the study, he concluded that weight was reduced, and lipid profile was also improved in volunteers who were taking routine diets and doing physical activities (Yeoh et al., 2015).
It was also observed that the IF trial lowered the level of total cholesterol but retained it in the normal recommended range (<200 mg/dl). The level of triglycerides decreased in 24/31 male volunteers while increased in 5/31 male volunteers. In the same way, the level of triglycerides is retained in the normal range (< 150 mg/dl). The level of low-density lipoprotein (LDL) decreased in 20/31 male volunteers and increased in 6/31 male volunteers in the same recommended ranges of <100 mg/dl. The level of high-density lipoprotein (HDL) increased in 24/31 and decreased in 3/31 male volunteers but in the normal range (>40 mg/dl), and the level of very low-density lipoproteins decreased in 22/31 male volunteers and increased in 6/31 male volunteers. The total cholesterol level decreased in all female volunteers. The total triglycerides decreased in 11/14 female volunteers and increased only in three volunteers, but within the normal recommended range, low-density lipoproteins decreased in 12/14 female volunteers, and HDL increased in 13/14 female volunteers. IF lowered total aerobic bacterial count in all the male and female volunteers when checked through conventional culturing methods. The number of total Lactococcus and total Lactobacillus, when assessed through culturing method, seen increased at the end of the IF trial in both male and female volunteers.
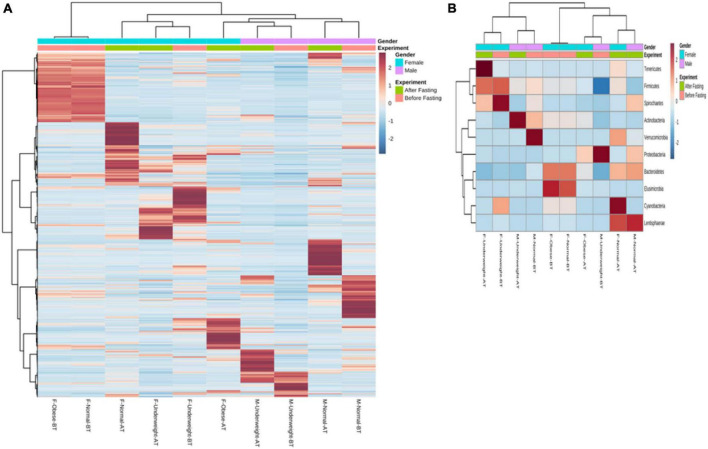
Recently, a study established the link between gut bacterial diversity and lipid blood profile, which showed that fasting sugar level, total triglycerides, total cholesterol, and low-density lipoproteins are associated with reduced gut bacterial diversity as compared to the control cohort (Rebolledo et al., 2017). Another study conducted on Finnish-men also showed that elevated blood lipids are associated with increased gut bacterial diversity (Org et al., 2017). Yang et al. (2017), in 2017, conducted a study on pigs, which showed that increased blood lipid levels are associated with altered bacterial diversity while Lactobacillus is negatively correlated with the blood lipid profile (Yang et al., 2017; Liu et al., 2018). The in vitro cholesterol assimilation activity of gut microbiota isolated from healthy individuals has been confirmed by different researchers recently, and the presence of bile salt hydrolase bsh genes is reported in Lactobacillus and Bifidobacteria, mostly used for cholesterol assimilation activities (Pereira and Gibson, 2002; Begley et al., 2006; Kim et al., 2009, 2013). These bacteria are known for anti-inflammatory activities, and inulin is the key component that favorably increases the number of Bifidobacteria spp (de Souza et al., 2017). These findings favored our results where all the samples carried Lactobacillus spp while their lipid profile was also positively improved. It has been demonstrated that overweight individuals carry a high microbial load in their intestines when compared with normal ones (Mokkala et al., 2016). Another study showed that increased bacterial richness is associated with parasite-positive guts (Andersen et al., 2016). It was explained by Maine Margo that increased diversity in the intestine is considered a healthy flora. They have more potential to degrade and metabolize diverse nutrient components (Maine and Kelly, 2016). Therefore, the reduction in bacterial richness and increase in diversity confirm a positive alteration in the intestinal bacteria. Changes were seen in the percentages of bacterial phylum at the end of the study (Figure 8). Normal weight male group had a higher percentage of Proteobacteria 73.28% which decreased to 1.56% by intermittent fasting. Firmicutes were initially 26% while increased to 58% making the highest portion of all the phylum at the end of the study. Increase in Actinobacteria from 0.19 to 37.14% was observed at the end of the study. Male underweighted group before IF had the highest Firmicutes of 69.95% and later decreased to 40.42%. The Actinobacteria also reduced from 19.72 to 6.43%, while Proteobacteria increased from 0.95 to 24.78%, Bacteroides from 8.87 to 19.81%, and Verrucomirobia from 0.21 to 7.98% at the end of the study. In female underweighted group, Firmicutes and Bacteroides decreased from 63.18 to 38.55% and 17.41 to 5.43%, respectively, while Proteobacteria increased from 5.52 to 41.96% and Actinobacteria from 6.90 to 13.97% at the end of the study. Female overweight/obese group before IF carried high Firmicutes than Bacteroides and low Proteobacteria. At the end of the study, Firmicutes and Bacteriodes were decreased and Proteobacteria was increased up to 11%. In female normal group, no significant changes at phylum level were observed. It has been concluded that too high and too low Proteobacteria are signs of dysbiosis. A minor percentage of Proteobacteria (<12%) is known healthy one at phylum level (Shin et al., 2015).
FIGURE 8.
Hierarchically clustered heatmap of the bacterial distribution of different communities. Double hierarchical dendrogram shows the bacterial distribution. The bacterial phylogenetic tree was calculated using the neighbor-joining method, and the relationship among samples was determined by Bray–Curtis distance and the complete clustering method. The heatmap plot depicts the relative percentage of each bacterial phylum (variables clustering on the vertical axis) within each sample group (horizon-axis clustering). The relative values for bacterial phylum are indicated by the color intensity with the legend indicated at the bottom of the figure.
At the genera level, before IF, it was seen that the normal-weighted male group had a high percentage of Pseudomonas triavilis, that is, 69%. This bacterium is isolated from grasses and is kept in risk group one, and it may cause infection in humans (Söhngen et al., 2013). Later, at the end of the study, it decreased to 0.04%. The Brevibacillus limnophilus was in the second number (4%) followed by Bacillus spp. At the end of the study, the number of Ruminococcus flavefaciens increased to 28%, Bifidobacterium adolescentis to 28%, and Lactobacillus rumins to 9%. A study conducted by Cucchi et al. (2013) showed that the fecal material of vegetarians has a high number of Ruminococcus, Lactobacillus, and Bifidobacteria sp. Participants consumed a vegetable-rich diet during the study period. The Ruminococcus flavefaciens are fiber digesting bacteria mainly involved in interactive digestion and acquisition of the nutrients in the intestine (Cucchi et al., 2013). Ejtahed et al. (2012), investigated and found that increase in the number of Bifidobacterium is responsible for blood cholesterol lowering. In another study the effect of energy restriction on body weight and gut microbiota was evaluated. It was concluded that an increase in the number of Bifidobacterium sp and lactobacillus spp is associated with weight loss (Santacruz et al., 2009).
A normal-weighted male group at the end of the IF study had 5% Catenibacterium mitsoukai of the total bacterial population. This species is heterofermentative and has a broad range of substrate utilization, thus providing more nutrients to their host (Kageyama and Benno, 2000). Before IF, there were no detectable species of Collinsella aerofaciens, but later on, it was observed that they make up 2% of all the total bacterial population present in the gut. The Collinsella aerofaciens was associated with the prevention and cure of irritable bowel diseases (Kassinen et al., 2007). This group carried 2% Subdoligranulum spp. These species have been used for the treatment of type 2 diabetes (Wang and Jia, 2016). Less number of Dorea spp was observed, and it is known that an increased number of Dorea spp can contribute to the development of irritable bowel disease (Rajilić-Stojanović et al., 2015). These findings present an improvement in the health status of the intestine and the whole host. In the male underweighted group, evenness and bacterial diversity were increased after the trial. It carried more percentage of Campylobacter spp, that is, 16% of the total bacterial population. Studies have shown that Campylobacter spp are normally present in animals and poultry, and their count goes high in warmer months. Not all Campylobacter spp are pathogenic to humans (Keener et al., 2004). The Campylobacter spp. have shown the colonization of the large intestine (Saint-Cyr et al., 2016). This study was performed in warmer months, while the volunteers were consuming poultry meat as a part of their routine diet. Hence, the high concentration of Campylobacter sp in the fecal samples of the volunteer’s cohort might be the competitive exclusion of these undesired microorganisms by the intestinal flora. The Megasphaera elsdenii were 5% of the total bacterial population. This bacterium is responsible for the treatment of metabolic acidosis (Marx et al., 2011). The Oscillospira spp. were decreased in the groups. According to a study, a decrease in Oscillospira spp is considered anti-obesity signature bacteria (Walters et al., 2014). Before IF, overweight/obese female group carried a high number of Roseburia spp (17%) and Prevotell copri (16%). At the end of IF, they were decreased to 1 and 8%, respectively. Increased Roseburia spp. are associated with gall stone formation (Tamanai-Shacoori et al., 2017), while increased Prevotella copri is associated with autoimmune diseases (Campbell, 2014).
Strength of the study: Blood lipid profile was improved and modulated gut microbiota even though the volunteers were taking their ad libitum diets during the eating windows of the experiment and were living their routine life. No dietary or living habits were advised to be changed or restricted, but, in many gut modulation studies, experimental subjects are passed through dietary restriction or changing living styles. Recently, a study conducted on a mice model for the improvement of blood lipid profile and modulation of gut microbiota kept the experimental mice restricted to a diet having the main portion of isoquercetin and inulin (Tan et al., 2018). In another study, antibiotics were administered in the diet for the improvement of blood lipid profile and modulation of gut microbiota (Rune et al., 2016). We have conducted our experimental trial on the human population, and it reflects the actual outcome and can be used as a reference for further studies, while in many cases, such studies are performed on mice model. There is a difference in the gut microbiota of humans and mice which affects the outcome of any experimental trial. Recently, the gut microbiota of mice and humans was compared using large datasets, and it was found that both the gut microbiota are similar at phylum level but have large variations at the species level (Xiao et al., 2015). The core microbiota of mice is small, while the core of human gut microbiota is large (Kostic et al., 2013). It was found that 85% of genera that are present in mice are not present in humans (Nguyen et al., 2015), while, when compared at the gene level, only 4% of genes were found in mice microbiota (Xiao et al., 2015). The gut microbiota of mice can be changed by the diet within 1 week, while human gut microbiota does not change so quickly (Wang and Jia, 2016). The difference between the results of women and men may be due to the fasting days, and women were exempted to fast during their menstrual cycle. The number of fasting days is not defined yet.
Limitation of the study: The number of male and female samples was not equal and thus may have an effect on the results. This may have influenced the results.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA477600, PRJNA477600.
Ethics statement
The studies involving human participants were reviewed and approved by BEC-FBS-QAU2018-109. The patients/participants provided their written informed consent to participate in this study.
Author contributions
MI and MIR conceived the study and experimental designing. MI supervised the study. AA and MI analyzed the data and critically revised the manuscript. MYK contributed to sample analysis. MNK and SIK conducted experimental work and drafted the manuscript. All authors contributed to the preparation of the final manuscript.
Acknowledgments
We would like to acknowledge Hafiz Abrab Sikandar for his guidance and all the volunteers of the study who participated willingly.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.922727/full#supplementary-material
Informed consent form for participants.
Diet and living habits of Participants.
Impact of intermittent fasting on culturable aerobic bacteria and fungi from fecal material of participants.
Impact of intermittent fasting of serum lipid profile of male participants.
Impact of intermittent fasting of serum lipid profile of female participants.
Impact of intermittent fasting of gut microbiota at phylum level of participants.
Impact of intermittent fasting of gut microbiota at genera level of obese female participants.
Impact of intermittent fasting of gut microbiota at genera level of normal weight female participants.
Impact of intermittent fasting of gut microbiota at genera level of under-weight female participants.
Impact of intermittent fasting of gut microbiota at genera level of normal weight male participants.
Impact of intermittent fasting of gut microbiota at genera level of under-weight male participants.
References
- Abargouei A. S., Janghorbani M., Salehi-Marzijarani M., Esmaillzadeh A. (2012). Effect of dairy consumption on weight and body composition in adults: a systematic review and meta-analysis of randomized controlled clinical trials. Int. J. Obes. 36 1485–1493. 10.1038/ijo.2011.269 [DOI] [PubMed] [Google Scholar]
- Abdelgadir E., Rashid F., Bashier A., Makeen S., Alawadi F. F., Hassanein M. (2015). “Effect of ramadan fasting on lipid profile, HBA1C, fasting blood glucose and body Mass Index in healthy muslim adults from three different nationalities in Dubai Hospital, United Arab Emirates,” in Proceedings of the Endocrine Society 9th Annual Meeting Health Outcomes/Quality Improvement Research. Endocrine Society, San Diego, CA. [Google Scholar]
- Adlouni A., Ghalim N., Benslimane A., Lecerf J. M., Saïle R. (1997). Fasting during Ramadan induces a marked increase in high-density lipoprotein cholesterol and decrease in low-density lipoprotein cholesterol. Ann. Nutr. Metab. 41 242–249. 10.1159/000177999 [DOI] [PubMed] [Google Scholar]
- Aloui A., Chtourou H., Briki W., Tabben M., Chaouachi A., Souissi N., et al. (2016). Rapid weight loss in the context of Ramadan observance: recommendations for judokas. Biol. Sport 33 407–413. 10.5604/20831862.1224098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen L. B., Karim A., Roager H. M., Vigsnæs L. K., Krogfelt K., Licht T. R., et al. (2016). Associations between common intestinal parasites and bacteria in humans as revealed by qPCR. Eur. J. Clin. Microbiol. Infect. Dis. 35 1427–1431. 10.1007/s10096-016-2680-2 [DOI] [PubMed] [Google Scholar]
- Arumugam M., Raes J., Pelletier E., Le Paslier D., Yamada T., Mende D. R., et al. (2011). Enterotypes of the human gut microbiome. Nature 473 174–180. 10.1038/nature09944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley M., Hill C., Gahan C. G. (2006). Bile salt hydrolase activity in probiotics. Appl. Environ. Microbiol. 72 1729–1738. 10.1128/AEM.72.3.1729-1738.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg R. D. (1996). The indigenous gastrointestinal microflora. Trends Microbiol. 4 430–435. 10.1016/0966-842X(96)10057-3 [DOI] [PubMed] [Google Scholar]
- Berkhan M. (2010). The Leangains Guide. Intermittent Fasting Diet for Fat Loss, Muscle Gain and Health. London: Leangains, 14. [Google Scholar]
- Blake A. B., Suchodolski J. S. (2016). Importance of gut microbiota for the health and disease of dogs and cats. Anim. Front. 6 37–42. 10.2527/af.2016-0032 32704858 [DOI] [Google Scholar]
- Campbell A. W. (2014). Autoimmunity and the gut. Autoimmune Dis. 2014:152428. 10.1155/2014/152428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Lozupone C. A., Turnbaugh P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108 4516–4522. 10.1073/pnas.1000080107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo M., Martín-Orúe S. M., Manzanilla E. G., Badiola I., Martín M., Gasa J. (2006). Quantification of total bacteria, enterobacteria and lactobacilli populations in pig digesta by real-time PCR. Vet. Microbiol. 114 165–170. 10.1016/j.vetmic.2005.11.055 [DOI] [PubMed] [Google Scholar]
- Cox L. M., Blaser M. J. (2015). Antibiotics in early life and obesity. Nat. Rev. Endocrinol. 11:182. 10.1038/nrendo.2014.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucchi T., Kovács Z. E., Berthon R., Orth A., Bonhomme F., Evin A., et al. (2013). On the trial of Neolithic mice and men towards Transcaucasia: zooarchaeological clues from Nakhchivan (Azerbaijan). Biol. J. Linn. Soc. 108 917–928. 10.1111/bij.12004 [DOI] [Google Scholar]
- de Souza C. H. B., Gioielli L. A., Saad S. M. I. (2017). Inulin increases Bifidobacterium animalis Bb-12 in vitro gastrointestinal resistance in margarine. LWT Food Sci. Technol. 79 205–212. 10.1016/j.lwt.2017.01.032 [DOI] [Google Scholar]
- Delzenne N. M., Neyrinck A. M., Cani P. D. (2013). Gut microbiota and metabolic disorders: how prebiotic can work? Br. J. Nutr. 109 S81–S85. 10.1017/S0007114512004047 [DOI] [PubMed] [Google Scholar]
- Desantis T. Z., Hugenholtz P., Larsen N., Rojas M., Brodie E. L., Keller K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72 5069–5072. 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druart C., Alligier M., Salazar N., Neyrinck A. M., Delzenne N. M. (2014). Modulation of the gut microbiota by nutrients with prebiotic and probiotic properties. Adv. Nutr. Int. Rev. J. 5 624S–633S. 10.3945/an.114.005835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejtahed H. S., Mohtadi-Nia J., Homayouni-Rad A., Niafar M., Asghari-Jafarabadi M., Mofid V. (2012). Probiotic yogurt improves antioxidant status in type 2 diabetic patients. Nutrition 28 539–543. 10.1016/j.nut.2011.08.013 [DOI] [PubMed] [Google Scholar]
- Elinav E., Leshem A., Horesh N. (2019). Fecal microbial transplantation and its potential application in cardiometabolic syndrome. Front. Immunol. 10:1341. 10.3389/fimmu.2019.01341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erejuwa O. O., Sulaiman S. A., Wahab M. S. A. (2014). Modulation of gut microbiota in the management of metabolic disorders: the prospects and challenges. Int. J. Mol. Sci. 15 4158–4188. 10.3390/ijms15034158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar J. S., Klotz B., Valdes B. E., Agudelo G. M. (2014). The gut microbiota of Colombians differs from that of Americans, Europeans and Asians. BMC microbiol. 14:311. 10.1186/s12866-014-0311-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulds M. H., Dahlman-Wright K. (2012). Metabolic diseases and cancer risk. Curr. Opin. Oncol. 24 58–61. 10.1097/CCO.0b013e32834e0582 [DOI] [PubMed] [Google Scholar]
- Fontana L., Klein S., Holloszy J. O. (2010). Effects of long-term calorie restriction and endurance exercise on glucose tolerance, insulin action, and adipokine production. Age 32 97–108. 10.1007/s11357-009-9118-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fooks L., Gibson G. R. (2002). Probiotics as modulators of the gut flora. Br. J. Nutr. 88 s39–s49. 10.1079/BJN2002628 [DOI] [PubMed] [Google Scholar]
- Friedman J. M. (2009). Obesity: causes and control of excess body fat. Nature 459 340–342. 10.1038/459340a [DOI] [PubMed] [Google Scholar]
- Gibson G. R., Macfarlane G. T. (1995). Human Colonic Bacteria: Role in Nutrition, Physiology and Pathology. Boca Raton, FL: CRC Press. [Google Scholar]
- Glauser W. (2011). Risk and rewards of fecal transplants. Can. Med. Assoc. 183 541–542. 10.1503/cmaj.109-3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschin C. (2016). Caloric restriction and exercise “mimetics”: ready for prime time? Pharmacol. Res. 103 158–166. 10.1016/j.phrs.2015.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmsen H. J., de Goffau M. C. (2016). The human gut microbiota. Adv. Exp. Med. Biol. 902 95–108. 10.1007/978-3-319-31248-4_7 [DOI] [PubMed] [Google Scholar]
- Hooper L. V., Gordon J. I. (2001). Commensal host-bacterial relationships in the gut. Science 292 1115–1118. 10.1126/science.1058709 [DOI] [PubMed] [Google Scholar]
- Kageyama A., Benno Y. (2000). Catenibacterium mitsuokai gen. nov., sp. nov., a gram-positive anaerobic bacterium isolated from human faeces. Int. J. Syst. Evol. Microbiol. 50 1595–1599. 10.1099/00207713-50-4-1595 [DOI] [PubMed] [Google Scholar]
- Kassinen A., Krogius-Kurikka L., Mäkivuokko H., Rinttilä T., Paulin L., Corander J., et al. (2007). The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology 133 24–33. 10.1053/j.gastro.2007.04.005 [DOI] [PubMed] [Google Scholar]
- Keener K., Bashor M., Curtis P., Sheldon B., Kathariou S. (2004). Comprehensive review of Campylobacter and poultry processing. Compr. Rev. Food Sci. Food Saf. 3 105–116. 10.1111/j.1541-4337.2004.tb00060.x [DOI] [PubMed] [Google Scholar]
- Kim E., Coelho D., Blachier F. (2013). Review of the association between meat consumption and risk of colorectal cancer. Nutr. Res. 33 983–994. 10.1016/j.nutres.2013.07.018 [DOI] [PubMed] [Google Scholar]
- Kim J. Y., Kim D. H., Choi J., Park J.-K., Jeong K.-S., Leeuwenburgh C., et al. (2009). Changes in lipid distribution during aging and its modulation by calorie restriction. Age 31 127–142. 10.1007/s11357-009-9089-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic A. D., Howitt M. R., Garrett W. S. (2013). Exploring host–microbiota interactions in animal models and humans. Genes Dev. 27 701–718. 10.1101/gad.212522.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kul S., Savaş E., Öztürk Z. A., Karadağ G. (2014). Does Ramadan fasting alter body weight and blood lipids and fasting blood glucose in a healthy population? A meta-analysis. J. Religion Health 53 929–942. 10.1007/s10943-013-9687-0 [DOI] [PubMed] [Google Scholar]
- Le Chatelier E., Nielsen T., Qin J., Prifti E., Hildebrand F., Falony G., et al. (2013). Richness of human gut microbiome correlates with metabolic markers. Nature 500 541–546. 10.1038/nature12506 [DOI] [PubMed] [Google Scholar]
- Ley R. E., Turnbaugh P. J., Klein S., Gordon J. I. (2006). Microbial ecology: human gut microbes associated with obesity. Nature 444:1022. 10.1038/4441022a [DOI] [PubMed] [Google Scholar]
- Li G., Xie C., Lu S., Nichols R. G., Tian Y., Li L., et al. (2017). Intermittent fasting promotes white adipose browning and decreases obesity by shaping the gut microbiota. Cell Metab. 26 672.e4–685.e4. 10.1016/j.cmet.2017.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H.-J., Kim S.-Y., Lee W.-K. (2004). Isolation of cholesterol-lowering lactic acid bacteria from human intestine for probiotic use. J. Vet. Sci. 5 391–395. 10.4142/jvs.2004.5.4.391 [DOI] [PubMed] [Google Scholar]
- Liu B., Zhang Y., Wang R., An Y., Gao W., Bai L., et al. (2018). Western diet feeding influences gut microbiota profiles in apoE knockout mice. Lipids Health Dis. 17:159. 10.1186/s12944-018-0811-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maine M., Kelly J. (2016). Pursuing Perfection: Eating Disorders, Body Myths, and Women at Midlife and Beyond. Abingdon: Routledge. 10.4324/9781315710099 [DOI] [Google Scholar]
- Marx H., Graf A. B., Tatto N. E., Thallinger G. G., Mattanovich D., Sauer M. (2011). Genome sequence of the ruminal bacterium Megasphaera elsdenii. J. Bacteriol. 193 5578–5579. 10.1128/JB.05861-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoro E. J. (2005). “Are age-associated diseases an integral part of aging?,” in Handbook of the Biology of Aging, Sixth Edn, eds Musi N., Hornsby P. (Amsterdam: Elsevier; ). 10.1016/B978-012088387-5/50005-4 [DOI] [Google Scholar]
- Mattson M. P., Longo V. D., Harvie M. (2017). Impact of intermittent fasting on health and disease processes. Ageing Res. Rev. 39 46–58. 10.1016/j.arr.2016.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokkala K., Röytiö H., Munukka E., Pietilä S., Ekblad U., Rönnemaa T., et al. (2016). Gut microbiota richness and composition and dietary intake of overweight pregnant women are related to serum zonulin concentration, a marker for intestinal permeability. J. Nutr. 146 1694–1700. 10.3945/jn.116.235358 [DOI] [PubMed] [Google Scholar]
- Nguyen T. L. A., Vieira-Silva S., Liston A., Raes J. (2015). How informative is the mouse for human gut microbiota research? Dis. Models Mech. 8 1–16. 10.1242/dmm.017400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Org E., Blum Y., Kasela S., Mehrabian M., Kuusisto J., Kangas A. J., et al. (2017). Relationships between gut microbiota, plasma metabolites, and metabolic syndrome traits in the METSIM cohort. Genome Biol. 18:70. 10.1186/s13059-017-1194-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira D. I., Gibson G. R. (2002). Cholesterol assimilation by lactic acid bacteria and bifidobacteria isolated from the human gut. Appl. Environ. Microbiol. 68 4689–4693. 10.1128/AEM.68.9.4689-4693.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowska K., Tarnowski M., Zgutka K., Pawlik A. (2016). Gender differences in response to prolonged every-other-day feeding on the proliferation and apoptosis of hepatocytes in mice. Nutrients 8:176. 10.3390/nu8030176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajilić-Stojanović M., Jonkers D. M., Salonen A., Hanevik K., Raes J., Jalanka J., et al. (2015). Intestinal microbiota and diet in IBS: causes, consequences, or epiphenomena? Am. J. Gastroenterol. 110 278–287. 10.1038/ajg.2014.427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebolledo C., Cuevas A., Zambrano T., Acuña J. J., Jorquera M. A., Saavedra K., et al. (2017). Bacterial community profile of the gut microbiota differs between hypercholesterolemic subjects and controls. BioMed. Res. Int. 2017:8127814. 10.1155/2017/8127814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizza W., Veronese N., Fontana L. (2014). What are the roles of calorie restriction and diet quality in promoting healthy longevity? Ageing Res. Rev. 13 38–45. 10.1016/j.arr.2013.11.002 [DOI] [PubMed] [Google Scholar]
- Rune I., Rolin B., Larsen C., Nielsen D. S., Kanter J. E., Bornfeldt K. E., et al. (2016). Modulating the gut microbiota improves glucose tolerance, lipoprotein profile and atherosclerotic plaque development in ApoE-deficient mice. PLoS One 11:e0146439. 10.1371/journal.pone.0146439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghirad B., Motaghipisheh S., Kolahdooz F., Zahedi M. J., Haghdoost A. A. (2014). Islamic fasting and weight loss: a systematic review and meta-analysis. Public Health Nutr. 17 396–406. 10.1017/S1368980012005046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Cyr M. J., Guyard-Nicodème M., Messaoudi S., Chemaly M., Cappelier J.-M., Dousset X., et al. (2016). Recent advances in screening of anti-campylobacter activity in probiotics for use in poultry. Front. Microbiol. 7:553. 10.3389/fmicb.2016.00553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santacruz A., Marcos A., Wärnberg J., Martí A., Martin-Matillas M., Campoy C., et al. (2009). Interplay between weight loss and gut microbiota composition in overweight adolescents. Obesity 17 1906–1915. 10.1038/oby.2009.112 [DOI] [PubMed] [Google Scholar]
- Savitri A. I., Amelia D., Painter R. C., Baharuddin M., Roseboom T. J., Grobbee D. E., et al. (2016). Ramadan during pregnancy and newborn’s birth weight. Matern. Nutr. Newborn Health Outcome 27:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehab A., Abdulle A., El Issa A., Al Suwaidi J., Nagelkerke N. (2012). Favorable changes in lipid profile: the effects of fasting after Ramadan. PLoS One 7:e47615. 10.1371/journal.pone.0047615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin N.-R., Whon T. W., Bae J.-W. (2015). Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 33 496–503. 10.1016/j.tibtech.2015.06.011 [DOI] [PubMed] [Google Scholar]
- Söhngen C., Bunk B., Podstawka A., Gleim D., Overmann J. (2013). BacDive—the bacterial diversity metadatabase. Nucleic Acids Res. 42 D592–D599. 10.1093/nar/gkt1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanton K., Frost M., Maryon-Davis A. (2007). Lightening the Load: Tackling Overweight and Obesity: A Toolkit for Developing Local Strategies to Tackle Overweight and Obesity in Children and Adults. Washington, DC: Wordworks. [Google Scholar]
- Tamanai-Shacoori Z., Smida I., Bousarghin L., Loreal O., Meuric V., Fong S. B., et al. (2017). Roseburia spp.: a marker of health? Future Microbiol. 12 157–170. 10.2217/fmb-2016-0130 [DOI] [PubMed] [Google Scholar]
- Tan S., Caparros-Martin J. A., Matthews V. B., Koch H., O’Gara F., Croft K. D., et al. (2018). Isoquercetin and inulin synergistically modulate the gut microbiome to prevent development of the metabolic syndrome in mice fed a high fat diet. Sci. Rep. 8:10100. 10.1038/s41598-018-28521-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiboura G., Khaled B. M., Diaf M., Semeria S., Bouanani G. (2015). Effect of Ramadan fasting on serum glucose and lipid profile among Algerian type 2 diabetes patients. Roman. J. Diabetes Nutr. Metab. Dis. 22 385–392. 10.1515/rjdnmd-2015-0045 [DOI] [Google Scholar]
- Trabelsi K., Stannard S. R., Maughan R. J., Jammoussi K., Zeghal K., Hakim A. (2012). Effect of resistance training during Ramadan on body composition and markers of renal function, metabolism, inflammation, and immunity in recreational bodybuilders. Int. J. Sport Nutr. Exerc. Metab. 22 267–275. 10.1123/ijsnem.22.4.267 [DOI] [PubMed] [Google Scholar]
- Turta O., Rautava S. (2016). Antibiotics, obesity and the link to microbes-what are we doing to our children? BMC Med. 14:57. 10.1186/s12916-016-0605-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuksan V., Jenkins A., Brissette C., Choleva L., Jovanovski E., Gibbs A., et al. (2016). Salba-chia (Salvia hispanica l.) in the treatment of overweight and obese patients with type 2 diabetes: a double-blind randomized controlled trial. Nutr. Metab. Cardiovasc. Dis. 27 138–146. 10.1016/j.numecd.2016.11.124 [DOI] [PubMed] [Google Scholar]
- Walters W. A., Xu Z., Knight R. (2014). Meta−analyses of human gut microbes associated with obesity and IBD. FEBS Lett. 588 4223–4233. 10.1016/j.febslet.2014.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Jia H. (2016). Metagenome-wide association studies: fine-mining the microbiome. Nat. Rev. Microbiol. 14 508–522. 10.1038/nrmicro.2016.83 [DOI] [PubMed] [Google Scholar]
- Weil A. A., Hohmann E. L. (2015). Fecal Microbiota Transplant: Benefits and Risks. Open Forum Infectious Diseases, 2015. Oxford: Oxford University Press, ofv005. 10.1093/ofid/ofv005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G. D., Lewis J. D., Hoffmann C., Chen Y.-Y., Knight R., Bittinger K., et al. (2010). Sampling and pyrosequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags. BMC Microbiol. 10:206. 10.1186/1471-2180-10-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L., Feng Q., Liang S., Sonne S. B., Xia Z., Qiu X., et al. (2015). A catalog of the mouse gut metagenome. Nat. Biotechnol. 33:1103. 10.1038/nbt.3353 [DOI] [PubMed] [Google Scholar]
- Yang H., Huang X., Fang S., He M., Zhao Y., Wu Z., et al. (2017). Unraveling the fecal microbiota and metagenomic functional capacity associated with feed efficiency in pigs. Front. Microbiol. 8:1555. 10.3389/fmicb.2017.01555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Xie L., Li Y., Wei C. (2009). More than 9,000,000 unique genes in human gut bacterial community: estimating gene numbers inside a human body. PLoS One 4:e6074. 10.1371/journal.pone.0006074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeoh E., Zainudin S. B., Loh W. N., Chua C. L., Fun S., Subramaniam T., et al. (2015). Fasting during ramadan and associated changes in glycaemia, caloric intake and body composition with gender differences in Singapore. Ann. Acad. Med. Singapore 44 202–206. 10.47102/annals-acadmedsg.V44N6p202 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Informed consent form for participants.
Diet and living habits of Participants.
Impact of intermittent fasting on culturable aerobic bacteria and fungi from fecal material of participants.
Impact of intermittent fasting of serum lipid profile of male participants.
Impact of intermittent fasting of serum lipid profile of female participants.
Impact of intermittent fasting of gut microbiota at phylum level of participants.
Impact of intermittent fasting of gut microbiota at genera level of obese female participants.
Impact of intermittent fasting of gut microbiota at genera level of normal weight female participants.
Impact of intermittent fasting of gut microbiota at genera level of under-weight female participants.
Impact of intermittent fasting of gut microbiota at genera level of normal weight male participants.
Impact of intermittent fasting of gut microbiota at genera level of under-weight male participants.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA477600, PRJNA477600.