2021 has been a productive year for fungal research. Key studies focused on intestinal inflammation and inflammatory bowel disease highlight antibody-mediated immunity in control of fungal commensalism, commensal and dietary fungi in intestinal inflammation and wound healing, and the therapeutic potential of transgenic yeast engineered to sense and target factors during intestinal inflammation.
Fungi are an indivisible component of the environmental, animal and plant microbiomes, and have a key role in multiple physiological and ecological processes, yet their role in the intestinal ‘ecosystem’, intestinal immune homeostasis and pathophysiology of inflammatory bowel disease (IBD) was only recently acknowledged. Studies published in 2021 uncover additional mechanisms of host–mycobiota interactions in the gut during health and IBD.
Commensal Candida species such as Candida albicans play a part in intestinal homeostasis and inflammation in a context-dependent manner. New studies demonstrate that dietary and environmental C. famata (teleomorph: Debaryomyces hansenii) 1 and C. krusei (teleomorph: Pichia kudraverzii)2 are also in the gut and influence local and gut-distal host physiology. Jian at al.1 demonstrate that D. hansenii used widely by the food industry can be detrimental in a specific context. Tissue repair processes are important in intestinal homeostasis and are impaired in several chronic diseases including IBD. Broad spectrum antibiotic treatment in mice led to the expansion of D. hansenii specifically inside intestinal wounds to interfere with the wound healing process (Figure 1a) 1. Whilst normally important for intestinal antifungal immunity and fungal clearance3, upon encountering D. hansenii in the wound bed, intestinal macrophages express CCL5 and impair wound healing. Wound healing was restored after treatment with the antifungal drug amphotericin B, as well as in mice lacking CCL5 or the IFNα receptor (necessary to potentiate CCL5 production by macrophages) (Figure 1a). Although D. hansenii is rarely present in samples from healthy individuals, it was enriched in inflamed intestinal tissue of patients with Crohn’s disease. The study suggests that food-derived fungi can take an unusual residence in inflamed tissue and that specific dietary restrictions for patients with IBD might be an important avenue to pursue in future clinical studies.
Figure 1.
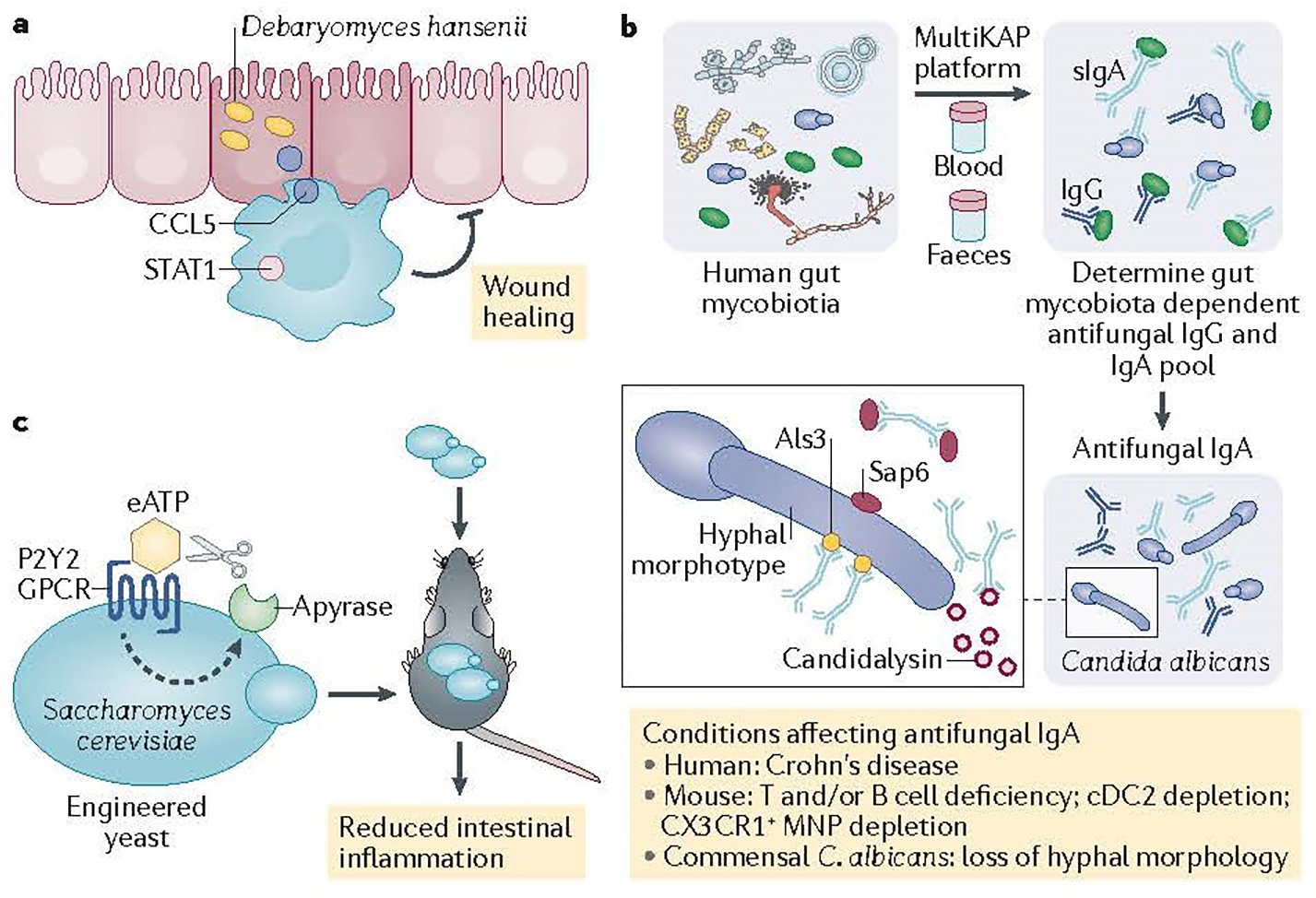
Intestinal inflammation and wound healing in with inflammatory bowel disease (IBD) patients are affected by intestinal fungi (the mycobiota). a) Debaryomyces hansenii takes unusual residence in intestinal wounds and perturbs wound healing by activation of CCL5-producton by macrophages. b) Gut mycobiota induces systemic IgG5 and secretory IgA antibodies 5, 6, 7 to tune host antifungal immunity5 and fungal commensalism in the gut by targeting C. albicans adhesins6 (Als3) and secreted virulence factors7 (candidalysin and SAP6) that are dysregulated in Crohn’s disease patients7. c) Saccharomyces cerevisiae engineered to express P2Y2 receptor senses and responds to intestinal eATP by the production of apyrase, and suppress intestinal inflammation in mouse models9.
Innate and type 17 antifungal immunity have a clear role in the maintenance of intact antifungal immunity and homeostasis in the gut, but humoral immunity to gut mycobiota remains poorly understood. The development of systemic antibodies against fungal mannan (ASCA antibodies) in patients with Crohn’s disease links antifungal humoral immunity and intestinal inflammation4. Doron et al.5 explored the human antibody repertoires against gut mycobiota and determined a large population of fungi bound by systemic IgG1 and secretory IgA (sIgA) antibodies in the lumen, with the human gut commensal C. albicans being the main target (Figure 1b).
In contrast to bacteria, C. albicans cells in the gut exist in a mixed yeast and hyphal morphotypes that differ by both cell size and function. Using mouse models and two genetic approaches to target hyphal formation in C. albicans, two 2021 studies6,7 determined that sIgA antibodies bind preferentially to hyphal morphotypes (Figure 1b). Antifungal sIgA production and germinal centre IgA+ B cell activation by C. albicans was also dependent on the ability to produce hyphae. Doron et al.3 determined that human sIgA limited the frequency of C. albicans hyphal morphologies7 whilst both studies found higher frequency of hyphae in B-cell-deficient mice 6, 7, implicating sIgA in the control of hyphal morphotypes in the gut. Ost et al6 demonstrated that binding of IgA to C. albicans was substantially reduced in T-cell-deficient mice, suggesting T-cell-dependent regulation of antifungal IgA. However, Doron et al.7 explored the innate immune requirements for this process and determined that cDC2 and CX3CR1+ mononuclear phagocytes are necessary, but play an interdependent part, in antifungal sIgA induction by affecting IgA+ B cells in the Peyer’s patches and the lamina propria, respectively. Loss of these populations of phagocytes led to increased granular hyphal C. albicans morphologies in the gut, suggesting that, in addition to T cells, innate immunity plays a part in this process (Figure 1b).
The capacity of C. albicans to reversibly change its morphology is crucial for its virulence; the formation of hyphae correlates with the upregulation of adhesins and effector molecule genes (Als3 and Ece1), involved in adhesion, iron acquisition, or host cell damage8. To identify C. albicans genes that are required for IgA binding Ost et al6 applied a screening approach of homozygous deletion mutant libraries and determined adhesion and hyphal formation regulator ahr1 as necessary for IgA binding. Ahr1 controls the transcriptional regulation of C. albicans adhesins such as Als3, Als1 and Hwp18. Using S. cerevisiae expression system, they showed that Als3 is sufficient to promote IgA binding (Figure 1b). Intestinal conditioning of Als3-expressing C. albicans in a wild-type mice, but not in Rag1−/− mice, reduced competitive advantage over Als3-non-expressing strain, confirming the role of adaptive immunity. Finally, induction of hyphal program or overexpression of Als1 in C. albicans aggravated the severity of chemically induced colitis in mice6, whereas vaccination against Als3 improved the disease outcome, suggesting that immune targeting of ALS3 in C. albicans might reduce colitis severity.
Doron at al.7 took another approach exploring sIgA binding to factors released by C. albicans and observed sIgA binding to the C. albicans secreted toxin candidalysin (Ece-III; encoded by Ece1) and to the secreted aspartyl proteinase protease (SAP6) (Figure 1b). Ahr1 and Efg1 are involved in transcriptional regulation of both Als3 and Ece1 in C. albicans 8. However, whilst Als3 is expressed by multiple Candida species orthologs of C. albicans candidalysin are so far described in C. dubliniensis and C. tropicalis. Doron et al.7 found sIgA against candidalysin and SAP6 in mice colonized with C. albicans. In patients with Crohn’s disease, decreased sIgA to candidalysin and SAP6 was associated with increased granular hyphal morphologies in mucosal washings. Altogether, these studies5, 7 demonstrate an involvement of sIgA in the regulation of fungal commensalism in the gut. Whether C. albicans secreted factors such as candidalysin and SAP6 that have strong ability to cause host damage are involved in the processes of intestinal inflammation and IBD remains to be elucidated but their exploration opens an attractive therapeutic avenue to pursue given their production specifically by pathogenic Candida species and not by other known gut commensal fungi.
In addition to its use as a powerful model organism, Saccharomyces cerevisiae (and specifically var. boulardii) have been long used as a probiotic for the prevention of travelers’ diarrhea and other gastrointestinal symptoms. Scott et al.9 took advantage of available tools in combination with synthetic biology approaches to engineer S. cerevisiae to sense luminal extracellular (eATP) through a high affinity eATP receptor P2Y2 and ‘respond’ with the release of the eATP-degrading enzyme apyrase (Figure 1c). Intestinal delivery of this strain as a probiotic ameliorated intestinal inflammation in several mouse models when supplemented daily. Although the use of transgenic yeast (and transgenic microorganisms in general) has not yet gained approval for a commercial use, the study provides a powerful example of how fungal genetics and biology can be harnessed for the development of mycobiota-centered future therapeutics.
Inside and outside the gut, 2021 has been a productive year for fungal research. In the upper gastrointestinal tract, Break at al10, demonstrate that T cell-dependent type 1 immunity inflammation during AIRE immunodeficiency promotes STAT1-dependent epithelial barrier defects that become a critical driver of susceptibility to chronic fungal infection in the oral mucosa. Altogether, these new studies highlight the interdependent relationship between fungi and the host and reveal multiple facets of host mycobiota interactions. Because strain-dependent features play a key role in phenotypes, exploration of the gut fungi and mycobiota mediated immunity with a strain-deep resolution would be likely the next frontier that will propel the field of the mycobiota, immunity and inflammation to further heights.
Key advances.
Mycobiota-induced antibodies influence systemic antifungal immunity5 and fungal commensalism in the gut by targeting Candida albicans adhesins6 (Als3) and secreted virulence factors7 (candidalysin and Sap6). Targeting Als3 influenced the severity of colitis in mice6, whereas secretory IgA antibodies to candidalysin and Sap6 are dysregulated in Crohn’s disease7.
Food-associated Candida species such as Debaryomyces hansenii can influence intestinal physiology1. D. hansenii found in mouse and human intestinal wounds prevent healing by turning protective macrophages into CCL5-producing perpetuators of tissue recovery1.
Transgenic Saccharomyces cerevisiae were engineered to sense and respond to intestinal extracellular ATP and suppress intestinal inflammation in mouse models9, providing a powerful example of mycobiota-centered future therapeutics.
Acknowledgements
Owing to reference limitations, several seminal and new works could not be cited.
References
- 1.Jain U et al. Debaryomyces is enriched in Crohn’s disease intestinal tissue and impairs healing in mice. Science 371, 1154–1159 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boutin RC et al. Bacterial-fungal interactions in the neonatal gut influence asthma outcomes later in life. Elife 10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leonardi I et al. CX3CR1(+) mononuclear phagocytes control immunity to intestinal fungi. Science 359, 232–236 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quinton JF et al. Anti-Saccharomyces cerevisiae mannan antibodies combined with antineutrophil cytoplasmic autoantibodies in inflammatory bowel disease: prevalence and diagnostic role. Gut 42, 788–791 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doron I et al. Human gut mycobiota tune immunity via CARD9-dependent induction of anti-fungal IgG antibodies. Cell (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ost KS et al. Adaptive immunity induces mutualism between commensal eukaryotes. Nature 596, 114–118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doron I.e.a. Mycobiota-induced IgA antibodies regulate fungal commensalism in the gut and are dysregulated in Crohn’s Disease. Nature microbiology In press (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruben S et al. Ahr1 and Tup1 Contribute to the Transcriptional Control of Virulence-Associated Genes in Candida albicans. mBio 11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott Benjamin M., C.G.-V., Sanmarco Liliana M., da Silva Pereira Jessica A., Li Zhaorong, Plasencia Agustín, Hewson Patrick, Cox Laura M., O’Brien Madelynn, Chen Steven K., Moraes-Vieira Pedro M., Chang Belinda S. W., Peisajovich Sergio G. & Quintana Francisco J. Self-tunable engineered yeast probiotics for the treatment of inflammatory bowel disease. Nature medicine (2021). [DOI] [PubMed] [Google Scholar]
- 10.Break TJ et al. Aberrant type 1 immunity drives susceptibility to mucosal fungal infections. Science 371 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]


