Abstract
Loss-of-function mutations at the retinoblastoma (RB1) gene are associated with increased mortality, metastasis, and poor therapeutic outcome in several cancers, including osteosarcoma. However, the mechanism(s) through which RB1 loss worsens clinical outcome remains understudied. Ubiquitin-like with PHD and Ring Finger domains 1 (UHRF1) has been identified as a critical downstream effector of the RB/E2F signaling pathway that is overexpressed in various cancers. Here, we determined the role and regulatory mechanisms of UHRF1 in rendering osteosarcoma cells more aggressive. Higher UHRF1 expression correlated with malignancy in osteosarcoma cell lines, clinical samples, and genetically engineered mouse models. Gain- and loss-of-function assays revealed that UHRF1 has cell-intrinsic and extrinsic functions promoting cell proliferation, migration, invasion, angiogenesis, and metastasis. UHRF1 overexpression induced angiogenesis by suppressing AMPK activation and Semaphorin 3E (SEMA3E) expression. Further, UHRF1-mediated migration and metastasis resulted, at least in part, through altered expression of extracellular vesicles and their cargo, including urokinase-type plasminogen activator (uPA). Novel osteosarcoma genetically engineered mouse models confirmed that knocking out Uhrf1 considerably decreased metastasis and reversed the poorer survival associated with Rb1 loss. This presents a new mechanistic insight into RB1 loss-associated poor prognosis and novel oncogenic roles of UHRF1 in the regulation of angiogenesis and exosome secretion, both critical for osteosarcoma metastasis. This provides substantial support for targeting UHRF1 or its downstream effectors as novel therapeutic options to improve current treatment for osteosarcoma.
Subject terms: Bone cancer, Tumour angiogenesis, Metastasis, Oncogenes
Introduction
Osteosarcoma is the most common primary bone cancer in children and young adults. As a highly metastatic cancer, ~20% of osteosarcoma patients are diagnosed after cancer has already metastasized (typically to the lungs), which translates to 5-year survival rates of <40% [1]. Unfortunately, pulmonary metastases occur in nearly half of osteosarcoma patients [2]. Thus, there is a pressing clinical need to determine the molecular mechanisms responsible for metastasis in osteosarcoma to facilitate the development of new therapeutic strategies.
The recurrent genetic mutations found in osteosarcoma are in tumor suppressors TP53 and RB1 [3, 4]. Loss-of-function mutations in RB1 are associated with poor therapeutic outcome: increased mortality, metastasis, and poor response to chemotherapy [5–9]. However, the mechanism(s) through which RB1 loss leads to poor prognosis remain unclear. We previously identified Ubiquitin-like, containing PHD and RING finger domains 1 (UHRF1) as a target of interest downstream of the RB/E2F signaling pathway [10]. UHRF1 is a multidomain protein that exerts various functions that include reading and writing of epigenetic codes. UHRF1 is most known for its role in maintaining DNA methylation [11, 12]. UHRF1 is overexpressed in different cancers that frequently present RB1 mutations, including lung, breast, bladder, colorectal cancer, and retinoblastoma [10, 13–16]. This suggests that UHRF1 upregulation may contribute to tumor progression following RB1 loss in osteosarcoma.
Here, we observed that UHRF1 is critical in osteosarcoma tumorigenesis, promoting proliferation, migration, angiogenesis, and metastasis. Mechanistically, we show that high UHRF1 decreases Semaphorin 3E (SEMA3E) expression through the suppression of AMPK activation, resulting in increased induction of angiogenesis. We also identified a novel role of UHRF1 in promoting exosome secretion which is associated, at least in part, with migration and invasion through the secretion of proteins, including urokinase-type plasminogen activator (uPA). Targeting uPA with small-molecule inhibitors resulted in a robust decrease in osteosarcoma migration and invasion. Uhrf1 loss drastically delayed tumor onset, decreased pulmonary metastasis, and increased the lifespan of developmental osteosarcoma mouse models carrying Rb1 mutation. These findings highlight a critical role of UHRF1 as a driver of the poor prognosis associated with RB1 loss and present UHRF1, and its downstream targets, as novel osteosarcoma therapeutic targets.
Results
UHRF1 expression correlates with osteosarcoma malignancy and metastasis
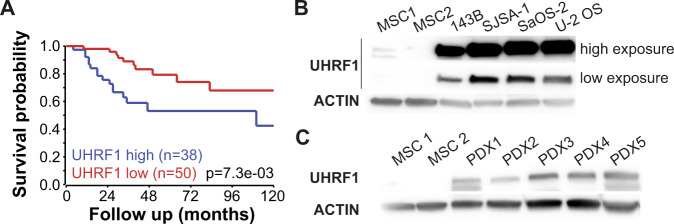
The Cancer Genome Atlas (TCGA) data suggest UHRF1 is significantly overexpressed in malignancies that frequently present RB pathway inactivation including sarcomas, breast invasive carcinomas, and lung adenocarcinomas, but not in cancers where RB1 loss is infrequent, like prostate adenocarcinomas (Supplementary Fig. 1A) [17]. In sarcomas, high UHRF1 expression is associated with poorer survival (Supplementary Fig. 1B). UHRF1 expression analysis using published RNA-Seq datasets from pretreatment biopsies from 88 osteosarcoma patients [18] associate high UHRF1 expression with poorer overall survival (Fig. 1A). UHRF1 mRNA in situ hybridization on an osteosarcoma tissue array confirmed a correlation between stage of malignancy and UHRF1 expression (Supplementary Fig. 1C, D).
Fig. 1. UHRF1 expression directly correlates with malignancy and metastasis in osteosarcoma.
A Overall survival probability of osteosarcoma patients comparing tumors with high UHRF1 (blue, n = 38) versus low UHRF1 (red, n = 50) expression at the time of diagnosis. B, C Western blot analysis of UHRF1 in (B) human osteosarcoma cell lines compared to MSC as control and (C) PDXs tumors. High exposure was used for UHRF1 detection in MSCs. β-actin was used as a loading control.
The clinical data led to examine UHRF1 mRNA and protein levels in 4 human osteosarcoma cell lines: 143B, SJSA-1, SaOS-2 (RB1-null cell line), and U-2 OS, and 5 patient-derived orthotopic xenografts (PDX1-5). qPCR analysis showed significant upregulation of UHRF1 mRNA levels in all samples except PDX2 when compared to MSC, independent of RB1 expression (Supplementary Fig. 1E–H). At the protein level, UHRF1 was highly expressed across all samples compared to MSC (Fig. 1B, C).
UHRF1 is a direct target of the RB/E2F signaling pathway in the osteogenic lineage
UHRF1 is reported as a direct E2F1 transcriptional target [19, 20]. Chromatin immunoprecipitation (ChIP) analysis using mesenchymal stem cells (MSCs) confirmed E2F1 enrichment at consensus binding motifs within the UHRF1 promoter (Supplementary Fig. 2A). However, knocking down E2F1 alone was insufficient to decrease UHRF1 expression in osteosarcoma cells (Supplementary Fig. 2B). We tested compensation by other activator E2Fs and found that knocking down E2F1 alongside E2F3, but not E2F2, reduced UHRF1 expression in osteosarcoma (Supplementary Fig. 2C, D). E2F3 knockdown alone was also insufficient to decrease UHRF1 expression (Supplementary Fig. 2E). Treatment with palbociclib, a CDK4/6 inhibitor, decreased UHRF1 mRNA and protein levels in all cell lines except SaOS-2, which is RB1-null and served as negative control (Supplementary Fig. 2F, G). Increased UHRF1 expression in RB1-wild-type cells correlated with upregulation of CDK4 and/or CDK6 transcripts and/or INK4A downregulation (Supplementary Fig. 2H, I). CDK4 amplification occurs in SJSA-1 and INK4A deletion in U2-OS [21, 22]. Thus, the RB/E2F pathway regulates UHRF1 through direct transcriptional activation by activator E2Fs, E2F1, and E2F3.
UHRF1 overexpression promotes osteosarcoma cell proliferation in vitro and in vivo
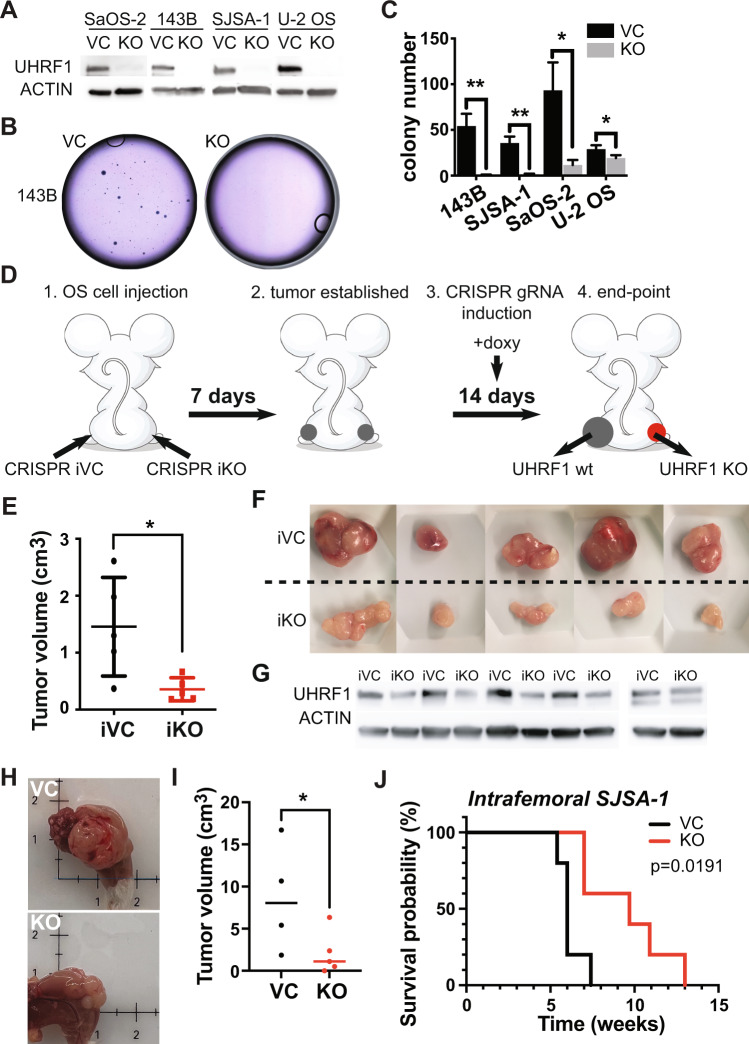
Emerging reports link UHRF1 overexpression in cancer with proliferation, migration/invasion, or both [10, 23–29]. To study the role of UHRF1 in osteosarcoma, we generated syngeneic UHRF1 CRISPR knockout clones (KO) and non-targeting vector controls (VC). Successful UHRF1 KO was achieved in all four osteosarcoma cell lines tested (Fig. 2A). Proliferation and EdU-incorporation analyses indicated UHRF1 KO cells have longer doubling times and lower proliferation rates compared to VC (Supplementary Fig. 3). This decrease in proliferation was confirmed by clonogenic assays showing significant reductions in both the number and the size of colonies in UHRF1 KO compared to VC cells (Fig. 2B, C).
Fig. 2. UHRF1 promotes osteosarcoma tumor growth in vitro and in vivo.
A Western blot analysis of UHRF1 in CRISPR/Cas9-mediated UHRF1 knockout of osteosarcoma cell lines (KO) in comparison to non-targeting vector control (VC). β-actin was used as a loading control. B Representative images from clonogenic assay plates with 143B VC and UHRF1 KO. C Histogram of colony counts from clonogenic assay. *P < 0.05, **P < 0.01 by unpaired two-tailed t test. D Cartoon representation for doxycycline-inducible in vivo knockout for flank-injected osteosarcoma cell lines carrying a non-targeting sgRNA (iCRISPR VC) or UHRF1 sgRNA (iCRISPR KO). E Quantification of the tumor volume for each of the replicates. *P < 0.05 by paired two-tailed t test. F Images from tumors collected from subcutaneous injection of iCRISPR VC (iVC, n = 5) and iCRISPR KO (iKO, n = 5) SJSA-1. G Western blot verification of UHRF1 knockout in iKO compared to iVC for tumors shown in (F). β-actin was used as a loading control. H Representative images from tumors collected from intrafemoral injection of SJSA-1 VC and UHRF1 KO. I Quantification of the tumor volume for each of the replicates, 5 weeks after intrafemoral injection. A solid linerepresents median from n = 4 for VC and n = 5 for KO. *P < 0.05 by paired two-tailed t test. J Kaplan–Meier curves showing the survival of mice with orthotopic SJSA-1 VC (black, n = 5) and UHRF1 KO (red, n = 5) xenografts.
To assess proliferative capacity in vivo, SJSA-1 UHRF1 KO and VC cells were injected subcutaneously at opposite flank regions. UHRF1 KO cells gave rise to significantly smaller tumors (average size 0.49 ± 0.37 cm3) when compared to VC (average volume 1.23 ± 0.75 cm3; P = 0.02; n = 10). The therapeutic potential of targeting UHRF1 in established tumors was tested using a doxycycline-inducible system to drive the expression of CRISPR/Cas9 gRNA against UHRF1. Inducible UHRF1 KO (iKO) and inducible VC (iVC) osteosarcoma cells were subcutaneously injected, and tumors allowed to establish for one week before inducing Cas9 expression via oral doxycycline delivery (Fig. 2D). Tumor growth was significantly reduced in UHRF1 iKO tumors (average tumor volume 0.356 ± 0.201 cm3; n = 5) compared to iVC tumors (1.455 ± 0.865 cm3; P = 0.02; n = 5) (Fig. 2E, F). Western blot analyses confirmed reduced UHRF1 protein at varying levels in UHRF1 iKO tumors (Fig. 2G). Thus, it is possible that residual tumors may be formed from cells not exposed to doxycycline or with incomplete UHRF1 KO. UHRF1 iKO tumors presented phenotypic features of less aggressive tumors, with lessened vasculature and ulceration compared to iVC tumors (Fig. 2F). Induction of a different UHRF1 gRNA in SJSA-1 resulted in similar tumor size reduction, as well as with other osteosarcoma cell lines (Supplementary Fig. 4A–D).
An orthotopic model using intrafemorally injected SJSA-1 confirmed UHRF1 KO tumors grow slower and have significantly higher survival than VC (Fig. 2H–J and Supplementary Fig. 4E). Together, our data suggest UHRF1 is critical for osteosarcoma growth.
UHRF1 promotes osteosarcoma metastasis
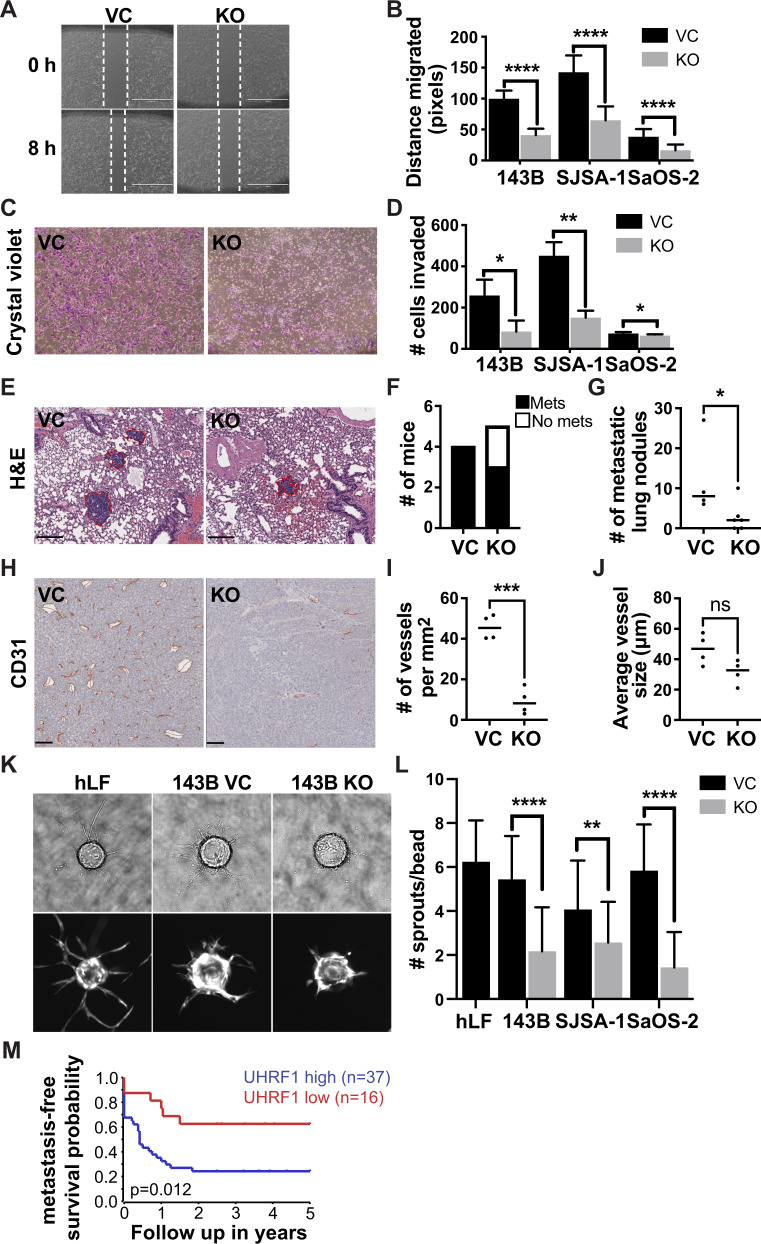
The high metastatic potential of osteosarcoma is the main cause of mortality in patients. UHRF1 was reported to promote cell invasion following exogenous UHRF1 overexpression in osteosarcoma [29]. Here, we evaluated whether the endogenously high UHRF1 protein levels present in osteosarcoma promote cell migration, invasion, angiogenesis, and metastasis. Scratch-wound assays were used to assess migration (Fig. 3A). UHRF1 KO resulted in a significant decrease in cell migration, with an average 56.8 ± 2.3% reduction in migration across three cell lines (Fig. 3B). Invasion assays also revealed a significant reduction in the number of UHRF1 KO cells capable of mobilizing across Matrigel-coated inserts (Fig. 3C, D).
Fig. 3. UHRF1 promotes osteosarcoma cell migration and invasion in vitro and in vivo.
A Representative image from scratch-wound assay comparing wound closure of non-targeting vector control (VC) and UHRF1 KO (KO) cells over 8 h in SJSA-1 cells. White dashed lines represent the wound edge. Scale bar, 1000 μm. B Quantification of distance (pixels) migrated in scratch-wound assays for each of the osteosarcoma cell lines. Each data point is mean ± s.d. of ten measurements in triplicate samples. C Representative images from Transwell assays with cells stained with crystal violet (purple) comparing levels of invasion between VC and UHRF1 KO in SJSA-1 cells. D Quantification of the number of cells invaded across the Transwell membrane for each of the osteosarcoma cell lines. Each data point is mean ± s.d. of triplicate samples. E Representative histological images of H&E-stained lung section from intrafemoral-injected mice with VC and UHRF1 KO SJSA-1 cells, 5 weeks after injection. Scale bar, 200 μm. Quantification of the burden of lung metastases was quantified as (F) the number of mice with or without lung metastases and (G) the number of metastatic lung nodules per mouse. A solid line represents median from n = 4 for VC and n = 5 for KO. *P = 0.03 by unpaired t test. H Representative histological images of CD31 expression in intrafemoral tumors derived from VC and UHRF1 KO SJSA-1 cells, 5 weeks after injection and (I) quantification of the average number of vessels per mm2 and (J) quantification of the mean vessel diameter. Solid line represents median from n = 4. ns not significant; ***P = 0.002 by unpaired t test. Scale bars, 100 μm. K Representative brightfield (top) and fluorescent (bottom) images of late phase sprouting angiogenesis assay (day 6) using human lung fibroblasts as assay control and 143B VC and KO conditioned media in the fibrin gel bead assay. This is quantified as (L) the number of sprouts per bead. n = 30. M 5-year metastasis-free survival probability of osteosarcoma patients comparing tumors with high UHRF1 (blue, n = 37) versus low UHRF1 (red, n = 16) expression at the time of diagnosis. For all graphs: *P < 0.05, **P < 0.01, ****P < 0.0001 by unpaired two-tailed t test.
To determine if UHRF1 overexpression is sufficient to induce cell migration, we performed gain-of-function studies in MSCs. Using a doxycycline-inducible system to drive UHRF1 overexpression (pCW57-UHRF1; Supplementary Fig. 6A, B), we observed a significant increase in MSC migration and invasion in cells induced to overexpress UHRF1 compared to controls (Supplementary Fig. 5C–F). We also utilized our pCW57-UHRF1 vector to rescue UHRF1 expression in UHRF1 KO cells. Doxycycline treatment for 24 h was able to restore UHRF1 protein expression back to levels comparable to wild-type and significantly increase migration in all osteosarcoma cell lines (Supplementary Fig. 5G, H).
The reduction in migration and invasion observed in vitro translated into a lessened metastatic potential in vivo. We assessed the rate of spontaneous lung metastasis in the NSG mice bearing intrafemoral SJSA-1 UHRF1 KO and VC tumors (Fig. 2H, I). At 5 weeks after injection, 100% of VC mice (4/4 mice) had metastatic lung nodules compared to only 60% of UHRF1 KO mice (3/5 mice) and the number of metastatic lung nodules was significantly lower in UHRF1 KO compared to VC (Fig. 3E–G). To test if UHRF1 contributes to tumor intravasation or extravasation, SJSA-1 UHRF1 KO and VC cells were injected into the tail vein of NSG mice to assess the rate of lung colonization. At 3-weeks post injection, UHRF1 KO mice had fewer lung metastatic nodules, but only the nodule size was significantly smaller than VC (Supplementary Fig. 6A–F).
Following the observation that UHRF1 loss appears to reduce tumor vascularization (Fig. 2F, H), we probed UHRF1 VC and KO orthotopic xenografts for the vasculature marker CD31. UHRF1 KO tumors exhibited reduced number of tumor vessels without reducing the overall tumor vessel caliber compared to VC (Fig. 3H–J). Thus, we evaluated a novel role of UHRF1 in cancer cell induction of endothelial cell migration (sprouting) using a fibrin gel bead assay (Fig. 3K). UHRF1 KO osteosarcoma cells displayed a significant lower ability to induce endothelial cell sprouting compared to VC (Fig. 3L). These data suggest a novel role of UHRF1 in osteosarcoma cell’s ability to affect early stages of angiogenesis.
UHRF1 expression analysis using pretreatment biopsies from 53 osteosarcoma patients [18] associate low UHRF1 expression with increased rates of 5-year metastasis-free survival (Fig. 3M). Together, these results support the role of UHRF1 in the early stages of osteosarcoma metastasis, including tumor migration, invasion, and angiogenesis, and promotes tumor growth at secondary sites.
High PLAU/uPA expression is associated with enhanced osteosarcoma cell migration and invasion
Given UHRF1 role in heterochromatin maintenance, we analyzed the effect of UHRF1 loss on DNA methylation, chromatin structure, and transcription to determine how UHRF1 serves as an oncogene in osteosarcoma. Decreased genomic DNA methylation was detected in UHRF1 KO compared to VC in most osteosarcoma cells examined (Supplementary Fig. 6A). Reduced representation bisulfite sequencing (RRBS) analysis of differentially methylated regions across the genome of VC and UHRF1 KO cells confirmed a reduction in DNA methylation, with most changes occurring at non-coding regions of the genome (Supplementary Fig. 8B). We also assessed chromatin landscape changes upon UHRF1 loss by performing ATAC-seq on UHRF1 KO compared to VC (Supplementary Fig. 8C). Despite the role of UHRF1 in heterochromatin maintenance and decreased genomic DNA methylation levels, we identified only 16 chromatin regions with significant changes in chromatin accessibility across three biological replicates (Supplementary Table 1). Within these changes, the majority were consistent with the role of UHRF1 in chromatin repression, with ~69% (11/16) resulting in the opening of chromatin upon UHRF1 loss.
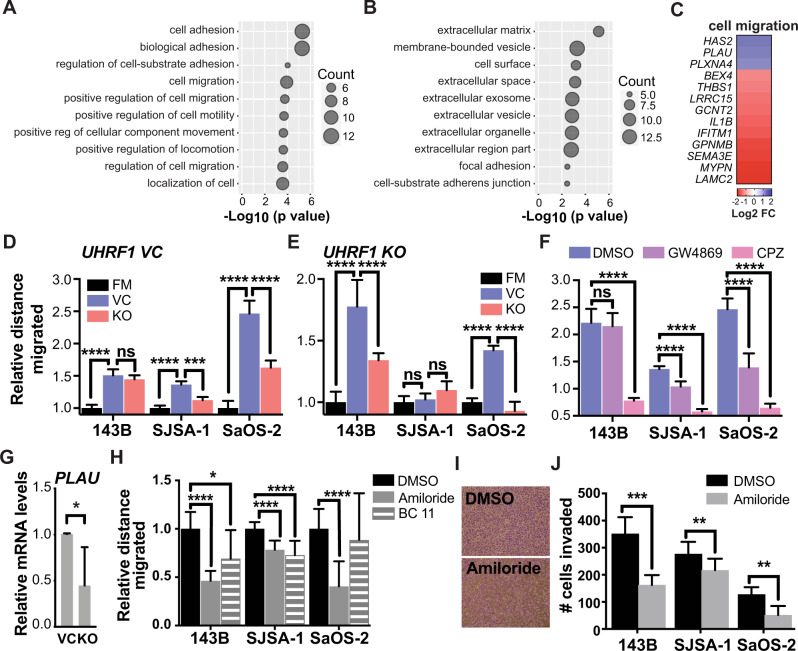
In line with modest changes in DNA methylation and chromatin accessibility, transcriptome analysis using RNA-seq revealed that the gene expression profile from UHRF1 KO cell lines present minor variability from VC cells (Supplementary Fig. 8D). We identified 272 differentially expressed genes (DEGs; 191 upregulated and 81 downregulated) in UHRF1 KO compared to VC cells. Gene ontology (GO) analysis for biological processes of the DEGs revealed an enrichment of genes involved in the regulation of cell adhesion and migration (Fig. 4A). GO analysis of cellular components of the DEGs from the RNA-seq data also exposed an enrichment of genes involved in extracellular vesicle formation and cell adhesion (Fig. 4B). We confirmed the changes in gene expression observed among top the 13 genes involved in the regulation of cell migration (Fig. 4C) through qPCR (Supplementary Fig. 6E).
Fig. 4. UHRF1 controls the expression of genes involved in exosomes and urokinase plasminogen activator production to drive osteosarcoma migration.
A, B Gene ontology (GO) analysis for (A) biological processes and (B) cellular components of the downregulated differentially expressed genes (DEGs) identified through RNA-seq. C Heatmap of top 13 genes involved in the regulation of cell migration, decreased (blue) or increased (red) in expression level upon UHRF1 loss. D, F Quantification of relative distance migrated in scratch-wound assays for (D) osteosarcoma VC cells assayed with fresh media (FM) or conditioned media (24 h) from VC cells or UHRF1 KO cells, normalized to FM; E osteosarcoma UHRF1 KO cells assayed with FM or conditioned media from VC cells or UHRF1 KO cells, normalized to FM; F each of the osteosarcoma cell lines treated with DMSO, 10 µM GW4869 or 20 µM CPZ, normalized to DMSO control. Each data point is mean ± s.d. of ten measurements in triplicate samples. G qPCR analysis of PLAU mRNA levels in flank-implanted tumors from control (VC) and UHRF1 KO (KO) SJSA-1 cell lines. *P < 0.05 by paired two-tailed t test. H Quantification of relative distance migrated in scratch-wound assays for each of the osteosarcoma cell lines treated with DMSO, 150 µM amiloride, or 16.4 µM BC11 hydrobromide, normalized to DMSO control. Each data point is mean ± s.d. of ten measurements in triplicate samples. I Representative images from Transwell assays with cells stained with crystal violet comparing levels of invasion between cells treated with DMSO or 150 µM amiloride in SJSA-1. J Quantification of the number of cells invaded in Transwell invasion assay for each of the osteosarcoma cell lines treated with DMSO or 150 µM amiloride. Each data point is mean ± s.d. of triplicate samples. For all graphs: ns not significant, **P < 0.01, ***P < 0.001, ****P < 0.0001 by unpaired two-tailed t test.
The potential role of UHRF1 in migration and invasion through the control of exosome-mediated pathways was examined by testing the effect of conditioned media collected from VC and UHRF1 KO cell cultures on osteosarcoma cell migration. Conditioned media from osteosarcoma VC cells significantly increased autologous osteosarcoma cell migration compared to fresh media (Fig. 4D, blue bars). While conditioned media derived from osteosarcoma UHRF1 KO cells also increased cell migration compared to fresh media, VC cell migration was lower when exposed to UHRF1 KO conditioned media than autologous VC conditioned media (Fig. 4D, red bars). Interestingly, UHRF1 KO cells also displayed a robust increase in cell migration when exposed to VC conditioned media, but not when exposed to autologous UHRF1 KO conditioned media (Fig. 4E). Thus, UHRF1 loss decreases the secretion of pro-migratory factors to the extracellular matrix that can serve as an autologous signal to stimulate osteosarcoma cell migration but does not affect the ability of the cells to respond to these extracellular factors. Inhibition of exosome biogenesis/release using GW4869 and inhibition of endocytosis using chlorpromazine (CPZ) significantly reduced the cell migration induced by osteosarcoma conditioned media (Fig. 4F). Together, this indicates that UHRF1 overexpression alters the production and/or secretion of exosomes and/or its cargo which contribute, at least in part, to increased osteosarcoma cell migration.
Plasminogen activator, urokinase (PLAU) was one of the DEGs identified through transcriptome analysis as potentially involved in UHRF1-mediated cell migration. Encoded by PLAU, uPA is associated with cell migration and is a known cargo protein in osteosarcoma-secreted exosomes [30]. PLAU transcript levels were significantly decreased in UHRF1 KO osteosarcoma cells (Supplementary Fig. 6E) and subcutaneous xenografts, with an average 2.2-fold decrease compared to VC cell-derived tumors (P = 0.038, Fig. 4G). Thus, we tested whether uPA inhibition could decrease migratory potential and invasiveness of osteosarcoma cells. Amiloride treatment resulted in significantly reduced migration, with an average 40.3 ± 11.9% decrease across all osteosarcoma cells examined (Fig. 4H). BC11 hydrobromide treatment, a selective uPA inhibitor, resulted in a 23.2 ± 10.2% decrease in migration across all lines (Fig. 4H). Reduced invasiveness was also observed upon uPA inhibition (Fig. 4I, J). Further, amiloride treatment also inhibited cell migration in MSCs with induced UHRF1 expression (Supplementary Fig. 7). Thus, uPA is an important contributor of UHRF1-induced migration in osteosarcoma.
UHRF1 repression of AMPK activation and SEMA3E expression induces angiogenesis
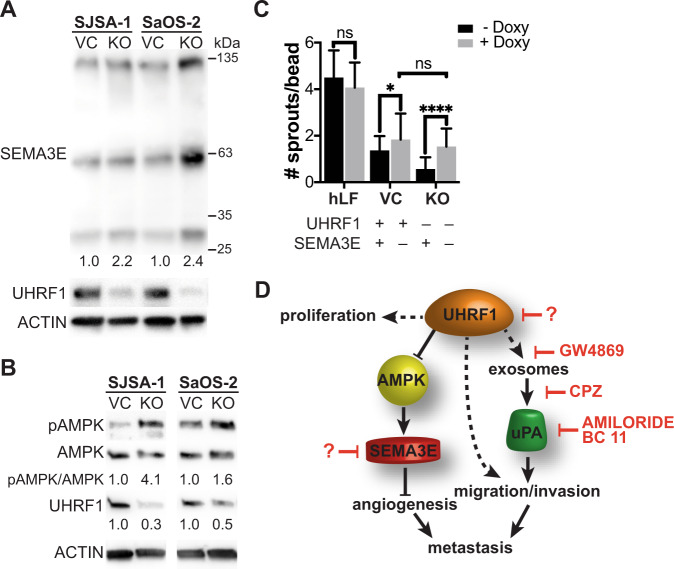
Another DEG identified in our RNA-seq analysis was SEMA3E (Fig. 4C). Sema3E acts as a repulsive factor for plexin-D1-expressing endothelial cells, leading to decreased neoangiogenesis and reduced tumor growth [31]. We confirmed that the increased SEMA3E transcript expression correlated with increased SEMA3E protein levels in the KO cells compared to VC (Fig. 5A). A recent study identified that UHRF1 suppresses AMPK activity [32], and AMPK activation was shown to induce SEMA3E expression [33]. Thus, we tested whether increased SEMA3E expression is correlated with increased phosphorylated AMPK (pAMPK) levels in UHRF1 KO cells. We found that while the total AMPK is unaltered by UHRF1 loss, pAMPK is elevated in UHRF1 KO cells, confirming a direct correlation between AMPK activity and SEMA3E expression (Fig. 5B). To determine if elevated SEMA3E is, at least in part, associated with the lower ability of UHRF1 KO cells to induce angiogenesis (Fig. 3N), we next tested whether knocking out SEMA3E could reverse the decreased angiogenesis observed in UHRF1 KO cells. Inducing SEMA3E KO resulted in a significant increase in the number of endothelial cells sprouting on the surface of fibrin gel beads both in UHRF1 VC and KO cells (Fig. 5C). Interestingly, upon SEMA3E KO, the extent of angiogenesis induced by UHRF1 VC and KO cells is similar (Fig. 5C). This suggests decreased SEMA3E secretion is associated with increased angiogenesis upon UHRF1 overexpression.
Fig. 5. UHRF1 decreases SEMA3E expression through suppression of AMPK activation to induce angiogenesis.
A, B Western blot analysis of (A) SEMA3E in SJSA-1 and SaOS-2 VC and KO osteosarcoma cell lines quantification of KO normalized to VC; and (B) activated AMPK (pAMPK) and total AMPK with the quantification of the ratio. UHRF1 used to confirm VC and KO status. β-actin was used as a loading control. C Early phase sprouting angiogenesis (day 3) using human lung fibroblasts as assay control and SJSA-1 VC and KO cells with inducible sgRNA to knockout SEMA3E upon doxycycline addition in the fibrin gel bead assay. Quantified as the number of sprouts per bead. n = 30. ns not significant, *P < 0.05, ****P < 0.0001, by unpaired t test. D Model of UHRF1 oncogenic function in osteosarcoma. UHRF1 overexpression stimulates proliferation, exosome and uPA production that stimulates migration, invasion, and metastasis. UHRF1 also suppresses AMPK activation to inhibit SEMA3E and induce angiogenesis. Downstream of UHRF1, inhibitors of exosome secretion (e.g., GW4869), exosome endocytosis (CPZ), or uPA inhibitors (e.g., amiloride, BC11 hydrobromide) are attractive therapeutic options to decrease migration and metastasis. The development of UHRF1-targeted therapeutics might result in a beneficial decrease in both tumor growth and pulmonary metastases.
UHRF1 overexpression is a critical driver of metastasis and the poor survival observed in Rb1-null osteosarcoma
Finally, we evaluated UHRF1’s pro-oncogenic function in osteosarcoma tumorigenesis using developmental mouse models. Loss of Tp53 in conditional knockout mice driven by osterix-cre recombinase (Tp53 cKO; Osx-cre Tp53lox/lox), a transgene expressed in preosteoblasts, results in osteosarcoma formation with complete penetrance [34, 35]. This model is potentiated by Rb1 loss (Tp53/Rb1 DKO; Osx-cre Tp53lox/lox Rb1lox/lox), mimicking the poor clinical outcome of RB1 loss in human osteosarcoma [34, 35]. Analysis of osteosarcoma tumors arising from Tp53 cKO and Tp53/Rb1 DKO mice revealed that UHRF1 is highly expressed at the mRNA (Fig. 6A) and protein level (Supplementary Fig. 8B) in these mouse models.
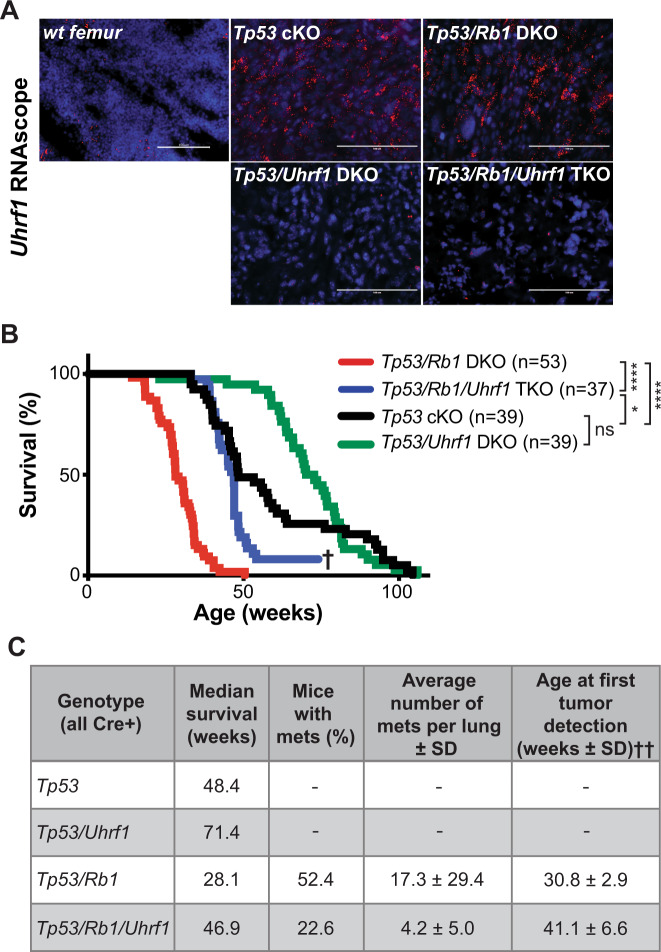
Fig. 6. UHRF1 is a critical driver of the increased malignancy observed in RB-null osteosarcoma.
A Representative fluorescent images of in situ hybridizations using RNAscope on wild-type femurs or tumors from genetically engineered osteosarcoma mice using a probe against Uhrf1 (red). Nuclei were counterstained with DAPI (blue). Uhrf1 expression is detected in Tp53 cKO and Tp53/Rb1 DKO mouse tumors but is low or not detected in wt, Tp53/Uhrf1 DKO, and Tp53/Rb1/Uhrf1 TKO tumors. B Kaplan–Meier curves showing the survival of osteosarcoma mouse models. Mice bearing Rb1 mutations Tp53/Rb1 DKO: Osx-Cre; p53lox/lox; Rb1lox/lox (red; n = 53) have significantly shorter lifespan compared to Tp53 cKO: Osx-Cre; p53lox/lox; Rb1lox/lox (black; n = 39). This survival time was significantly increased in Tp53/Rb1/Uhrf1 TKO: Osx-Cre; p53lox/lox; Rb1lox/lox; Uhrf1lox/lox (blue; n = 37) mice. †Three surviving mice were removed from the study to confirm tumor absence. Tp53/Uhrf1 DKO: Osx-Cre; p53lox/lox; Uhrf1lox/lox (green; n = 39) showed an overall survival comparable to Tp53 cKO (black; n = 39). Mantel–Cox test were used for curve comparisons. ns not significant, *P < 0.05, ****P < 0.0001. C Summary table for each osteosarcoma mouse model. ††Age of mice (weeks) at earliest tumor detection via microCT and PET scans comparison: P < 0.05 by unpaired two-tailed t test; Tp53/Rb1 DKO (n = 4) and Tp53/Rb1/Uhrf1 TKO (n = 3).
Uhrf1 KO mice result in embryonic lethality [36]; thus, we generated Uhrf1 cKO mice (Osx-cre Uhrf1lox/lox), Tp53/Uhrf1 DKO (Osx-cre Tp53lox/lox Uhrf1lox/lox) and Tp53/Rb1/Uhrf1 triple knockout (Tp53/Rb1/Uhrf1 TKO; Osx-cre Tp53lox/lox Rb1lox/lox Uhrf1lox/lox) to study the role of UHRF1 in osteosarcoma development. Uhrf1 cKO mice display normal bone development (Supplementary Fig. 8C, D). For Tp53/Uhrf1 DKO and Tp53/Rb1/Uhrf1 TKO mice, we tracked overall survival and tumor formation compared to the corresponding littermate controls (Tp53 cKO and Tp53/Rb1 DKO, respectively). In situ hybridization confirmed reduced Uhrf1 expression in Tp53/Uhrf1 DKO and Tp53/Rb1/Uhrf1 TKO osteosarcoma tumors (Fig. 6A). All strains presented osteosarcoma with 100% penetrance but with distinct survival rates and disease presentation (Fig. 6B, C). Consistent with previous reports, Rb1 loss in Tp53/Rb1 DKO mice resulted in shorter median survival compared to Tp53 cKO mice (Fig. 6B). Strikingly, Uhrf1 genetic ablation in Tp53/Rb1/Uhrf1 TKO mice resulted in significantly increased survival compared to Tp53/Rb1 DKO mice. Further, the median survival of Tp53/Rb1/Uhrf1 TKO was comparable to Tp53 cKO (P = 0.0577, Gehan–Breslow–Wilcoxon test), suggesting that Uhrf1 overexpression is critical for the poor prognosis associated with Rb1 loss (Fig. 1B). However, the overall survival of Tp53/Rb1/Uhrf1 TKO and Tp53 cKO mice is distinct (P = 0.0126, Mantel–Cox test), suggesting that early UHRF1 overexpression is not the only factor contributing to Rb1 loss-associated outcomes. Three Tp53/Rb1/Uhrf1 TKO mice did not acquire tumors well beyond the average of the rest of their study group (>69 weeks; Fig. 6B†). FDG-PET/microCT scans in these three mice revealed no detectable tumors (Supplementary Fig. 8E), which was confirmed upon autopsy.
Since UHRF1 is also overexpressed in Tp53 cKO tumors (Fig. 6A and Supplementary Fig. 8B), we examined the effect of Uhrf1 loss in Tp53/Uhrf1 DKO compared to Tp53 cKO mice. We observed a significant increase in median survival of Tp53/Uhrf1 DKO mice, compared to Tp53 cKO (P = 0.0007, Gehan–Breslow–Wilcoxon test), but not in overall survival (P = 0.1189, Mantel–Cox test; Fig. 6B). Interestingly, we found that all Tp53 cKO mice with early morbidity (<50 weeks) presented tumors with increased Uhrf1 mRNA levels, suggesting spontaneous RB pathway inactivation (Supplementary Fig. 8F). High Uhrf1 mRNA expression correlated with increased Cdk4 expression (Supplementary Fig. 8G). No correlation was found with the expression of other RB regulators, including p16 and Cdk6 (Supplementary Fig. 8H, I). No significant changes on Rb1 expression were observed between these groups (Supplementary Fig. 8J). Histopathology analysis of tumors from the four mouse models analyzed linked Uhrf1 loss with lower mitotic index and decreased anaplasia (Supplementary Fig. 9 and Supplementary Table 2).
Most remarkably, in line with our in vitro observations, Uhrf1 loss resulted in a significant reduction of pulmonary metastases, with only 22.6% of Tp53/Rb1/Uhrf1 TKO mice presenting lung metastases compared to 52.4% of Tp53/Rb1 DKO mice. A smaller number of metastatic nodules was also observed in the Tp53/Rb1/Uhrf1 TKO mice that developed lung metastases (Fig. 6C).
To determine whether Uhrf1 contributes to tumor promotion or tumor progression, we performed periodic FDG-PET/microCT scans on Tp53/Rb1 DKO and Tp53/Rb1/Uhrf1 TKO mice to determine the age at which tumors are detectable (Supplementary Fig. 8A). In Tp53/Rb1/Uhrf1 TKO mice, tumors were detected significantly later than in Tp53/Rb1 DKO mice (P = 0.0362; Fig. 6C). All test subjects reached humane end-of-study within 3 weeks of tumor detection. These data suggest that Uhrf1 plays a pivotal role in the early developmental process of osteosarcoma promotion following the loss of Rb1.
Discussion
RB1 genetic alterations are associated with increased mortality, metastasis, and poor response to chemotherapy in osteosarcoma [5–9, 37, 38]. However, the precise mechanism through which this occurs remains to be elucidated. Studies from retinoblastoma, a cancer initiated by biallelic inactivation of the RB1 gene, defined a central role of RB in epigenetic control and a key role of RB/E2F-regulated chromatin remodelers in tumorigenesis [10, 39, 40]. Among others, UHRF1 was identified as a potential critical regulator of tumor initiation and progression following RB pathway inactivation [10, 13–16]. Here, we identified UHRF1 overexpression as directly correlated with increased osteosarcoma malignancy and metastatic disease.
UHRF1 expression is positively correlated with cell proliferation and cell mobility in both normal and malignant cells in vitro [10, 25–28]. Here, we show that UHRF1 KO decreases clonogenicity and tumor growth both in vitro and in vivo. UHRF1 also has a clear effect on migration. UHRF1 overexpression induced migration and invasion of normal MSCs and targeting UHRF1 drastically reduced migration and invasion of osteosarcoma cell lines. It is important to note that although RB1-null status has been clinically associated with poor disease outlook, the RB1-null cell line, SaOS-2, does not exhibit higher aggressiveness in comparison to other cell lines. This could be explained by the limitation of utilizing cell lines, in which loss of clinical representation is commonly seen.
This study also presents novel roles of UHRF1 in controlling exosome formation and angiogenesis. Exosomes and their cargo are implicated in osteosarcoma metastasis [41]. Our study shows that UHRF1 alters the expression of exosomal components and their protein cargo composition. We identified uPA as a critical factor in UHRF1-mediated migration, in line with a previous study describing the uPA/uPAR axis as a metastatic driver in osteosarcoma [42]. uPA is a serine protease associated with cell migration and a known cargo protein in osteosarcoma-secreted exosomes [30]. Upon plasminogen conversion into plasmin, uPA triggers a proteolytic cascade leading to degradation of extracellular matrix, an event necessary for angiogenesis and metastasis. While our results suggest that targeting UHRF1 might result in a more robust therapeutic outcome, the observation that cell migration can be reversed by treatment with uPA inhibitors like amiloride [43] and BC11 hydrobromide, provides therapeutic alternatives that can be further explored (Fig. 5D).
Arising from the observation that UHRF1 KO xenografts presented with reduced tumor vascularization, we identified a new role of UHRF1 in the induction of angiogenesis. Mechanistically, we propose a UHRF1-AMPK-SEMA3E axis where SEMA3E repels endothelial sprouting leading to decreased angiogenesis (Fig. 5D). This is supported by the recent report showing that UHRF1 suppresses AMPK activity in the nucleus by recruiting PP2A to dephosphorylate AMPK [32] and previous findings showing that AMPK activation induces SEMA3E expression [33]. SEMA3E is known to act as a repulsive factor for endothelial cells that express plexin-D1, resulting in decreased neoangiogenesis and reduced tumor growth [31].
Our group is the first to model the critical role of UHRF1 in osteosarcomagenesis, with Uhrf1 KO resulting in a dramatic increase in overall survival and a decrease in metastases, particularly in Rb1-null osteosarcoma. The survival and rate of metastasis of Tp53/Rb1/Uhrf1 TKO closely resembled that of Tp53 cKO mice, indicating that UHRF1 is a major driver of Rb1 loss-associated malignancy. Interestingly, our data also indicate that the increased survival in Tp53/Rb1/Uhrf1 TKO mice is largely a result of delayed tumor promotion, with negligible differences in tumor growth rates once the tumors have been detected. However, decreased growth rates were observed when UHRF1 KO was induced in established tumors. This distinct observation suggests that there might be compensatory pathways arising in the developmental tumor models. Further analyses on tumors arising from these developmental osteosarcoma models might provide insight on potential compensatory mechanisms of Uhrf1 loss which may be relevant for predicting mechanisms of resistance for future UHRF1-targeted therapeutics.
While targeting Uhrf1 in developmental mouse models showed drastic improvement in survival in Rb1-null tumors over Rb1-wild-type, such distinctions were not observed in UHRF1 overexpressing human osteosarcoma cell lines. Both RB1-null and RB1-wild-type osteosarcoma cell lines appeared to equally benefit from targeting UHRF1. It is possible that the timing of UHRF1 overexpression is crucial for the resulting poor prognosis. Early Rb1 loss would result in immediate UHRF1 overexpression which quickly promotes tumorigenesis compared to tumors that eventually inactivate the RB/E2F pathway through alternative mechanisms. Supporting this theory, Tp53/Uhrf1 DKO mice showed improvement in survival compared to a subpopulation of Tp53 cKO mice that exhibited characteristics of RB/E2F pathway inactivation and therefore presented early morbidity. Alternatively, the RB/E2F complex is known not only for its role in direct transcriptional repression, but also as epigenetic modifiers through the recruitment of chromatin remodelers [39, 44–47]. Two UHRF1 domains, PHD and RING, contain LXCXE RB-binding motifs [48]. Unpublished data from our group show UHRF1 preferentially binds to hyperphosphorylated RB. Thus, while the transcriptional regulation of UHRF1 is aberrant by means of RB/E2F pathway inactivation, RB1-wild-type osteosarcoma cells may preserve protein–protein interactions between RB and UHRF1. As a complex, RB and UHRF1 may exert regulation on downstream targets, including UHRF1 itself. The possible loss of this layer of regulation in RB1-null tumors could account for the drastic difference in clinical prognosis, supporting the view that the inactivation of RB by phosphorylation is not functionally equivalent to the mutation of the RB1 gene [49].
This study supports targeting UHRF1 as a novel therapeutic strategy and provides alternative therapeutic interventions along the RB-UHRF1 axis for osteosarcoma treatment. Targeting UHRF1 holds great potential in overcoming the highly metastatic characteristic of osteosarcoma, the main reason why the survival rate has remained stagnant in past decades. We are the first to model the effect of UHRF1 throughout development and showed minimal on-target toxicity, as opposed to various chemotherapeutic agents known to affect bone homeostasis [50]. Moreover, the degree to which UHRF1 loss reverted the increased malignancy upon Rb1 loss suggests that the benefit of UHRF1 targeting could go beyond the scope of osteosarcoma by improving current treatment paradigms of other cancers harboring RB/E2F pathway inactivation.
Materials and methods
Xenografts and mouse models
The orthotopic xenografts used in this study (all TP53 mutant): SJSO010930 (PDX1; RB1 mutant), SJOS001112 (PDX2; RB1 mutant), SJOS001107 (PDX3; RB1 wt), SJOS001105 (PDX4; RB1 wt), and SJOS001121 (PDX5; RB1 mutant) were obtained from the Childhood Solid Tumor Network [51]. Athymic nude (NU/J) and NOD scid gamma (NSG) mice were obtained from The Jackson Laboratories. p53lox and Rb1lox mice were obtained from the Mouse Models of Human Cancer Consortium at the National Cancer Institute; Osx-cre mice from The Jackson Laboratory. Uhrf1 cKO mice obtained from EMMA were backcrossed to flipasse (Flp) mice to remove the neo-cassette (tm1c conversion) and backcrossed to C57BL/6N mice for Flp removal. Mice were monitored weekly for signs of osteosarcoma. Moribund status defined as the point when tumors had reached 10% body weight or induced mouse paralysis. Experiments include animals from both sexes. The University of California Irvine Institutional Animal Care and Use Committee approved all animal procedures.
Cell culture
Osteosarcoma cell lines 143B, SJSA-1, SaOS-2, and U-2 OS, as well as bone marrow-derived MSCs and HEK293T cells were acquired from ATCC and maintained according to their specifications. Human umbilical vein endothelial cells (HUVEC) expressing blue fluorescent protein were cultured in EGM-2 (Lonza CC-3162). Normal human lung fibroblasts (hLF) were routinely cultured in DMEM + 10% FBS. All cells were routinely checked for mycoplasma contamination, and stocks were STR profiled at the time of freezing.
Cell lines were analyzed as clones (n = 3) or as pools examined within 2–3 weeks after being generated.
Western blotting
Western blots were performed as described [52] using the following antibodies and dilutions: 1:1000 Uhrf1 (sc-373750, Santa Cruz Biotechnology), 1:1000 Rb (9313T, Cell Signaling), actin (A1978, Sigma), E2F1 (3742S, Cell Signaling), E2F2 (sc-9967, Santa Cruz Biotechnology), E2F3 (MA5-11319, Nalgene Nunc); 1:1000 SEMA3E (PA547469, Thermo Scientific); 1:1000 AMPK (5831S, Cell Signaling); 1:1000 pAMPK (ab133448, Abcam). Secondary antibodies were diluted 1:1000 (PI-1000, PI-2000 or PI-9500, Vector Laboratories). Bands were visualized using chemiluminescence (SuperSignal West Pico Chemiluminescent Substrate, Thermo Scientific). Band intensities were analyzed using ImageJ software.
In situ hybridization
Formalin-fixed, paraffin-embedded (FFPE) slides were incubated at 60 °C for 1 h for deparaffinization, washed twice in xylene and twice in 100% ethanol, 5 min each at RT before air-drying. RNAscope was performed following the manufacturer’s protocol. Human and mouse UHRF1-specific probes were customized by Bio-Techne. RNAscope® Positive Control Probe- Hs-PPIB/Mm-PP1B and RNAscope® Negative Control Probe- DapB were used. OpalTM 570 fluorophore was used at a 1:1000 dilution. Slides were mounted with ProLong Gold Antifade Mountant.
Real-time RT-PCR (qPCR)
We performed qPCR as described [52]. Primers were designed using IDT Real-Time PCR tool (Integrated DNA Technologies). Reaction was carried out using 7500 Real-Time PCR system (Applied Biosystems). Data were normalized to those obtained with endogenous control 18 S mRNA and analyzed using ΔΔCt method. Primer sequence for PCR amplification are as follows: UHRF1 (5’-GCTGTTGATGTTTCTGGTGTC-3’; 5’-TGCTGTCAGGAAGATGCTTG-3’), PLAU (5’-GAGCAGAGACACTAACGACTTC-3’; 5’-CTCACACTTACACTCACAGCC-3’), SEMA3E (5’-CTGGCTCGAGACCCTTACTG-3’; 5’- CAAAGCATCCCCAACAAACT-3’), HAS2 (5’-TCCATGTTTTGACGTTTGCAG-3’; 5’-AGCAGTGATATGTCTCCTTTGG-3’), LAMC2 (5’-CACCATACTCCTTGCTTCCTG-3’; 5’-GTGCAGTTTGTCTTTCCATCC-3’), 18 S (5’-GTAACCCGTTGAACCCCATT-3’; 5’-CCATCCAATCGGTAGTAGCG-3’).
Lentivirus production and transduction
Lentiviral particles were produced as previously described [52]. plentiCRISPRv2 (GenScript) with UHRF1 gRNA (gRNA sequence: TCAATGAGTACGTCGATGCT), inducible CRISPR/Cas9 plasmid TLCV2 (Addgene plasmid #87360) with UHRF1 gRNA (gRNA1 sequence: CGCCGACACCATGTGGATCC and gRNA2 sequence: ACACCATGTGGATCCAGGTT) were used.
Clonogenic assay
Colony formation assay was performed as previously described [52].
Subcutaneous injection
In total, 2 × 106 cells were resuspended in 100 μl PBS and injected subcutaneously into the flank region of mice. Tumors were caliper-measured and collected 3 weeks post injection. For doxycycline induction, 2 mg/ml doxycycline hyclate supplemented water was administered ad libitum. 10 mg/ml sucrose was added to doxycycline-supplemented water to increase consumption.
Intrafemoral injection
Cell-PBS mixture (4 × 106 cells/ml) was mixed with an equal volume of 7.7 mg/ml Matrigel (Corning, 356230) to achieve a final concentration of 2 × 106 cells/ml. Cell mixture, needle, and 50-µl syringe (Hamilton, 80920) were left in ice until ready for injection. NSG mice were laid down in dorsal recumbency and the injection area was shaved and prepared. The right femur was stabilized and positioned perpendicular to the hood. A 25 G needle was inserted into the femur through the kneecap with a twist motion to create the space for injection later. A new 25 G needle loaded with 10 µL cell–Matrigel mixture was inserted back to the femur. The cell mixture was slowly injected to prevent leakage. Mice were monitored weekly after injection. Primary tumors, lungs, and the injected femurs were collected and used for analysis.
RNA sequencing
RNA sequencing was performed as described [53]. All sequencing data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus [54] and are accessible through GEO Series accession number GSE144418.
Scratch-wound-healing assay
The scratch-wound assay was performed as previously described [52]. Two hours prior scratch, cells were treated with 5 µg/ml mitomycin C (S8146, Sigma) to control for proliferation differences. Amiloride was used at 150 µM, BC11 hydrobromide at 16.5 µM, GW4869 at 10 µM, which were supplemented in growth medium and given to the cells at the 0 h time point. Chlorpromazine (CPZ) was used at 20 µM for 30 min prior to scratch. For experiments using conditioned media, serum-free media was used and exposed to cells for 24 h.
Transwell invasion assay
In all, 8-µm pore PET membranes (353097, Corning) were coated with Matrigel (25 µg/insert). In all, 2 × 104 cells were seeded onto the membrane in 100 µl of appropriate cell culture medium and placed into the incubator for 2 h at 37 °C with 5% CO2 to allow cell attachment. After cells have attached, 100 µl of media were added to the inner chamber, amiloride (150 µM final concentration) or BC11 hydrobromide (16.4 µM final concentration) or doxycycline (1 µg/ml final concentration) were supplemented at this time. In total, 600 µl of appropriate cell culture medium supplemented with 100 ng/ml fibroblast growth factor (FGF) was added to the outer chamber. Cells were incubated for 16 h at 37 °C with 5% CO2. The cell culture medium was aspirated, and cotton swabs were used to remove the remaining cells inside the inner chamber. Cells at the bottom side of the insert were then fixed and stained with 0.5% crystal violet for 2 h. Excess dye was washed out with tap water. Images of five different fields within each insert were taken for analysis by cell count.
Tail vein injection
In total, 2 × 106 cells were resuspended in 200 µl PBS and injected intravenously through the tail vein. Lungs were collected 3 weeks after injection and fixed in 4% formaldehyde for histological analysis.
3D fibrin gel angiogenesis assay
Fibrin gel angiogenesis assays were performed as previously described [55]. Briefly, HUVECs were coated onto Cytodex 3 microcarrier beads (GE 17048501) at a concentration of 150 cells/bead for 4 h and allowed to adhere overnight. HUVEC-coated beads were then resuspended in a 2.5 mg/ml fibrinogen solution (MP Biomedicals) at a concentration of 250 beads/ml. Gels were formed by adding 500 µl of the fibrinogen/bead suspension to each well of a 24-well plate containing 0.5 U of thrombin (Sigma-Aldrich). Once gels were clotted, 1 ml of EMG2 conditioned media from osteosarcoma VC or KO cells or containing 20,000–50,000 hLFs (as assay control) was added to each well. Assays were quantified between days 6 by live-culture imaging using bright-field and fluorescence microscopy. Thirty beads per condition were quantified per experiment.
Statistical analysis
All graphing and statistical analyses were performed using GraphPad Prism (version 9 for Mac, GraphPad Software, San Diego, CA, USA). All microscopy images were randomly taken from different areas. Details of sample size (n), statistical test, and P value applied for each experiment were indicated in the figure legends. Results are presented as median ± SD. The statistical significance of differences between the two groups was assessed by Student’s t test. P values < 0.05 were considered statistically significant. Survival curves were analyzed using log-rank (Mantel–Cox) and Gehan–Breslow–Wilcoxon tests. Animal numbers were calculated from pilot studies and biostatistical calculation of a minimum of 75% power at 0.05 significance. Animals euthanized for non-study-related pre-established criteria were excluded from the analysis. Phenotypic measurements were scored blinded.
Supplementary information
Acknowledgements
We thank Dr. Argyrios Ziogas for power and statistical analysis of animal studies. Erin Yamanaka, Josselyn Peña, Sara Akhlaghi, and Annie Jeon for help with mouse genotyping and cell culture; Christopher Liang for help with FDG-PET/microCT scans. This work was supported in part by grants to CAB from the NIH (CA178207 and CA229696), the American Cancer Society (129801-IRG-16-187-13 and 133403-RSG-19-031-01-DMC) and the 2018 AACR-Aflac, Inc. Career Development Award for Pediatric Cancer Research (18-20-10-BENA). JL, KS, and SS were supported by the NIH-MARC U-STAR training grant (T34GM136498). This work was also made possible through access to the Genomics High Throughput Facility Shared Resource of the Cancer Center Support Grant (P30CA-062203) at the University of California, Irvine and NIH shared instrumentation grants 1S10RR025496-01, 1S10OD010794-01, and 1S10OD021718-01.
Author contributions
SCW and CAB designed research; SCW, AK, YG, DIM, JL, KS, and SS performed experiments and data collection. CCC and JW performed bioinformatics analyses. AN performed pathology analysis. SCW and CAB wrote the paper.
Data availability
All data supporting the findings of this study are available within the article and its supplementary information files and from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41389-022-00430-6.
References
- 1.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 2.Huang YM, Hou CH, Hou SM, Yang RS. The metastasectomy and timing of pulmonary metastases on the outcome of osteosarcoma patients. Clin Med Oncol. 2009;3:99–105. doi: 10.4137/cmo.s531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin JW, Squire JA, Zielenska M. The genetics of osteosarcoma. Sarcoma. 2012;2012:627254. doi: 10.1155/2012/627254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen X, Bahrami A, Pappo A, Easton J, Dalton J, Hedlund E, et al. Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep. 2014;7:104–12. doi: 10.1016/j.celrep.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feugeas O, Guriec N, Babin-Boilletot A, Marcellin L, Simon P, Babin S, et al. Loss of heterozygosity of the RB gene is a poor prognostic factor in patients with osteosarcoma. J Clin Oncol. 1996;14:467–72. doi: 10.1200/JCO.1996.14.2.467. [DOI] [PubMed] [Google Scholar]
- 6.Benassi MS, Molendini L, Gamberi G, Sollazzo MR, Ragazzini P, Merli M, et al. Altered G1 phase regulation in osteosarcoma. Int J Cancer. 1997;74:518–22. doi: 10.1002/(SICI)1097-0215(19971021)74:5<518::AID-IJC7>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 7.Goto A, Kanda H, Ishikawa Y, Matsumoto S, Kawaguchi N, Machinami R, et al. Association of loss of heterozygosity at the p53 locus with chemoresistance in osteosarcomas. Jpn J Cancer Res. 1998;89:539–47. doi: 10.1111/j.1349-7006.1998.tb03295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wadayama B, Toguchida J, Shimizu T, Ishizaki K, Sasaki MS, Kotoura Y, et al. Mutation spectrum of the retinoblastoma gene in osteosarcomas. Cancer Res. 1994;54:3042–8. [PubMed] [Google Scholar]
- 9.Ren W, Gu G. Prognostic implications of RB1 tumour suppressor gene alterations in the clinical outcome of human osteosarcoma: a meta-analysis. Eur J Cancer Care. 2017;26:e12401. [DOI] [PubMed]
- 10.Benavente CA, Finkelstein D, Johnson DA, Marine J-C, Ashery-Padan R, Dyer MA. Chromatin remodelers HELLS and UHRF1 mediate the epigenetic deregulation of genes that drive retinoblastoma tumor progression. Oncotarget. 2014;5:9594–608. doi: 10.18632/oncotarget.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–4. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 12.Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–12. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 13.Mousli M, Hopfner R, Abbady AQ, Monte D, Jeanblanc M, Oudet P, et al. ICBP90 belongs to a new family of proteins with an expression that is deregulated in cancer cells. Br J Cancer. 2003;89:120–7. doi: 10.1038/sj.bjc.6601068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Unoki M, Kelly JD, Neal DE, Ponder BA, Nakamura Y, Hamamoto R. UHRF1 is a novel molecular marker for diagnosis and the prognosis of bladder cancer. Br J Cancer. 2009;101:98–105. doi: 10.1038/sj.bjc.6605123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daskalos A, Oleksiewicz U, Filia A, Nikolaidis G, Xinarianos G, Gosney JR, et al. UHRF1-mediated tumor suppressor gene inactivation in nonsmall cell lung cancer. Cancer. 2011;117:1027–37. doi: 10.1002/cncr.25531. [DOI] [PubMed] [Google Scholar]
- 16.Kofunato Y, Kumamoto K, Saitou K, Hayase S, Okayama H, Miyamoto K, et al. UHRF1 expression is upregulated and associated with cellular proliferation in colorectal cancer. Oncol Rep. 2012;28:1997–2002. doi: 10.3892/or.2012.2064. [DOI] [PubMed] [Google Scholar]
- 17.Aparicio A, Den RB, Knudsen KE. Time to stratify? The retinoblastoma protein in castrate-resistant prostate cancer. Nat Rev Urol. 2011;8:562–8. doi: 10.1038/nrurol.2011.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuijjer ML, Rydbeck H, Kresse SH, Buddingh EP, Lid AB, Roelofs H, et al. Identification of osteosarcoma driver genes by integrative analysis of copy number and gene expression data. Genes Chromosomes Cancer. 2012;51:696–706. doi: 10.1002/gcc.21956. [DOI] [PubMed] [Google Scholar]
- 19.Magri L, Swiss VA, Jablonska B, Lei L, Pedre X, Walsh M, et al. E2F1 coregulates cell cycle genes and chromatin components during the transition of oligodendrocyte progenitors from proliferation to differentiation. J Neurosci. 2014;34:1481–93. doi: 10.1523/JNEUROSCI.2840-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Unoki M, Nishidate T, Nakamura Y. ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene. 2004;23:7601–10. doi: 10.1038/sj.onc.1208053. [DOI] [PubMed] [Google Scholar]
- 21.Gump J, Stokoe D, McCormick F. Phosphorylation of p16INK4A correlates with Cdk4 association. J Biol Chem. 2003;278:6619–22. doi: 10.1074/jbc.C200622200. [DOI] [PubMed] [Google Scholar]
- 22.Al-Assar O, Rees-Unwin KS, Menasce LP, Hough RE, Goepel JR, Hammond DW, et al. Transformed diffuse large B-cell lymphomas with gains of the discontinuous 12q12-14 amplicon display concurrent deregulation of CDK2, CDK4 and GADD153 genes. Br J Haematol. 2006;133:612–21. doi: 10.1111/j.1365-2141.2006.06093.x. [DOI] [PubMed] [Google Scholar]
- 23.Guan D, Factor D, Liu Y, Wang Z, Kao HY. The epigenetic regulator UHRF1 promotes ubiquitination-mediated degradation of the tumor-suppressor protein promyelocytic leukemia protein. Oncogene. 2013;32:3819–28. doi: 10.1038/onc.2012.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Huang Z, Zhu Z, Zheng X, Liu J, Han Z, et al. Upregulated UHRF1 promotes bladder cancer cell invasion by epigenetic silencing of KiSS1. PLoS ONE. 2014;9:e104252. [DOI] [PMC free article] [PubMed]
- 25.Zhang Z-Y, Cai J-J, Hong J, Li KK-W, Ping Z, Wang Y, et al. Clinicopathological analysis of UHRF1 expression in medulloblastoma tissues and its regulation on tumor cell proliferation. Medical Oncol. 2016;33:99. [DOI] [PubMed]
- 26.Zhu M, Xu Y, Ge M, Gui Z, Yan F. Regulation of UHRF1 by microRNA-9 modulates colorectal cancer cell proliferation and apoptosis. Cancer Sci. 2015;106:833–9. doi: 10.1111/cas.12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui L, Chen J, Zhang Q, Wang X, Qu J, Zhang J, et al. Up-regulation of UHRF1 by oncogenic Ras promoted the growth, migration, and metastasis of pancreatic cancer cells. Mol Cell Biochem. 2015;400:223–32. doi: 10.1007/s11010-014-2279-9. [DOI] [PubMed] [Google Scholar]
- 28.Li X-L, Xu J-H, Nie J-H, Fan S-J. Exogenous expression of UHRF1 promotes proliferation and metastasis of breast cancer cells. Oncol Rep. 2012;28:375–83. doi: 10.3892/or.2012.1792. [DOI] [PubMed] [Google Scholar]
- 29.Liu W, Qiao RH, Wang DM, Huang XW, Li B, Wang D. UHRF1 promotes human osteosarcoma cell invasion by downregulating the expression of Ecadherin in an Rb1dependent manner. Mol Med Rep. 2016;13:315–20. doi: 10.3892/mmr.2015.4515. [DOI] [PubMed] [Google Scholar]
- 30.Endo-Munoz L, Cai N, Cumming A, Macklin R, de Long LM, Topkas E, et al. Progression of osteosarcoma from a non-metastatic to a metastatic phenotype is causally associated with activation of an autocrine and paracrine uPA axis. PLoS ONE. 2015;10:e0133592. [DOI] [PMC free article] [PubMed]
- 31.Sakurai A, Gavard J, Annas-Linhares Y, Basile JR, Amornphimoltham P, Palmby TR, et al. Semaphorin 3E initiates antiangiogenic signaling through plexin D1 by regulating Arf6 and R-Ras. Mol Cell Biol. 2010;30:3086–98. doi: 10.1128/MCB.01652-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu X, Ding G, Liu C, Ding Y, Chen X, Huang X, et al. Nuclear UHRF1 is a gate-keeper of cellular AMPK activity and function. Cell Res. 2022;32:54–71. doi: 10.1038/s41422-021-00565-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kemmerer M, Finkernagel F, Cavalcante MF, Abdalla DS, Muller R, Brune B, et al. AMP-activated protein kinase interacts with the peroxisome proliferator-activated receptor delta to induce genes affecting fatty acid oxidation in human macrophages. PLoS ONE. 2015;10:e0130893. doi: 10.1371/journal.pone.0130893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walkley CR, Qudsi R, Sankaran VG, Perry JA, Gostissa M, Roth SI, et al. Conditional mouse osteosarcoma, dependent on p53 loss and potentiated by loss of Rb, mimics the human disease. Genes Dev. 2008;22:1662–76. doi: 10.1101/gad.1656808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berman SD, Calo E, Landman AS, Danielian PS, Miller ES, West JC, et al. Metastatic osteosarcoma induced by inactivation of Rb and p53 in the osteoblast lineage. Proc Natl Acad Sci USA. 2008;105:11851–6. doi: 10.1073/pnas.0805462105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muto M, Kanari Y, Kubo E, Takabe T, Kurihara T, Fujimori A, et al. Targeted disruption of Np95 gene renders murine embryonic stem cells hypersensitive to DNA damaging agents and DNA replication blocks. J Biol Chem. 2002;277:34549–55. doi: 10.1074/jbc.M205189200. [DOI] [PubMed] [Google Scholar]
- 37.Benassi MS, Molendini L, Gamberi G, Ragazzini P, Sollazzo MR, Merli M, et al. Alteration of pRb/p16/cdk4 regulation in human osteosarcoma. Int J Cancer. 1999;84:489–93. doi: 10.1002/(SICI)1097-0215(19991022)84:5<489::AID-IJC7>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 38.Maitra A, Roberts H, Weinberg AG, Geradts J. Loss of p16(INK4a) expression correlates with decreased survival in pediatric osteosarcomas. Int J Cancer. 2001;95:34–38. doi: 10.1002/1097-0215(20010120)95:1<34::AID-IJC1006>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 39.Zhang J, Benavente CA, McEvoy J, Flores-Otero J, Ding L, Chen X, et al. A novel retinoblastoma therapy from genomic and epigenetic analyses. Nature. 2012;481:329–34. doi: 10.1038/nature10733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benavente CA, Dyer MA. Genetics and epigenetics of human retinoblastoma. Annu Rev Pathol. 2015;10:547–62. doi: 10.1146/annurev-pathol-012414-040259. [DOI] [PubMed] [Google Scholar]
- 41.Jerez S, Araya H, Hevia D, Irarrazaval CE, Thaler R, van Wijnen AJ, et al. Extracellular vesicles from osteosarcoma cell lines contain miRNAs associated with cell adhesion and apoptosis. Gene. 2019;710:246–57. doi: 10.1016/j.gene.2019.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Endo-Munoz L, Cai N, Cumming A, Macklin R, Merida de Long L, Topkas E, et al. Progression of osteosarcoma from a non-metastatic to a metastatic phenotype is causally associated with activation of an autocrine and paracrine uPA axis. PLoS ONE. 2015;10:e0133592. doi: 10.1371/journal.pone.0133592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vassalli JD, Belin D. Amiloride selectively inhibits the urokinase-type plasminogen activator. FEBS Lett. 1987;214:187–91. doi: 10.1016/0014-5793(87)80039-X. [DOI] [PubMed] [Google Scholar]
- 44.Chi P, Allis CD, Wang GG. Covalent histone modifications-miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–69. [DOI] [PMC free article] [PubMed]
- 45.Lu J, Ruhf ML, Perrimon N, Leder P. A genome-wide RNA interference screen identifies putative chromatin regulators essential for E2F repression. Proc Natl Acad Sci USA. 2007;104:9381–6. doi: 10.1073/pnas.0610279104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benetti R, Gonzalo S, Jaco I, Munoz P, Gonzalez S, Schoeftner S, et al. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol. 2008;15:998. doi: 10.1038/nsmb0908-998b. [DOI] [PubMed] [Google Scholar]
- 47.Wen H, Andrejka L, Ashton J, Karess R, Lipsick JS. Epigenetic regulation of gene expression by Drosophila Myb and E2F2-RBF via the Myb-MuvB/dREAM complex. Genes Dev. 2008;22:601–14. doi: 10.1101/gad.1626308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jeanblanc M, Mousli M, Hopfner R, Bathami K, Martinet N, Abbady A-Q, et al. The retinoblastoma gene and its product are targeted by ICBP90: a key mechanism in the G1/S transition during the cell cycle. Oncogene. 2005;24:7337–45. doi: 10.1038/sj.onc.1208878. [DOI] [PubMed] [Google Scholar]
- 49.Dyson NJ. RB1: a prototype tumor suppressor and an enigma. Genes Dev. 2016;30:1492–502. doi: 10.1101/gad.282145.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Madan B, McDonald MJ, Foxa GE, Diegel CR, Williams BO, Virshup DM. Bone loss from Wnt inhibition mitigated by concurrent alendronate therapy. Bone Res. 2018;6:17. doi: 10.1038/s41413-018-0017-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stewart E, Federico S, Karlstrom A, Shelat A, Sablauer A, Pappo A, et al. The childhood solid tumor network: a new resource for the developmental biology and oncology research communities. Dev Biol. 2016;411:287–93. doi: 10.1016/j.ydbio.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu SC, Benavente CA. Chromatin remodeling protein HELLS is upregulated by inactivation of the RB-E2F pathway and is nonessential for osteosarcoma tumorigenesis. Oncotarget. 2018;9:32580–92. doi: 10.18632/oncotarget.25953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zocchi L, Mehta A, Wu SC, Wu J, Gu Y, Wang J, et al. Chromatin remodeling protein HELLS is critical for retinoblastoma tumor initiation and progression. Oncogenesis. 2020;9:25. doi: 10.1038/s41389-020-0210-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakatsu MN, Hughes CC. An optimized three-dimensional in vitro model for the analysis of angiogenesis. Methods Enzymol. 2008;443:65–82. doi: 10.1016/S0076-6879(08)02004-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the article and its supplementary information files and from the corresponding author upon reasonable request.