Abstract
Background
Phospholipase A/acyltransferase (PLAAT) family exhibits O- and N-acyltransferase activity and biosynthesize N-acylated ethanolamine phospholipids. Previously, PLAAT4 was seen as a tumor suppressor, but the exact function of PLAAT4 in pancreatic cancer was still unknown. In this study, we investigated the relationship of PLAAT4 and pancreatic cancer.
Methods
Using the data from the cancer genome atlas (TCGA), Genotype-Tissue Expression (GTEx) database and Gene Expression Omnibus (GEO) datasets we compared the expression of PLAAT4 in normal and tumor tissues and analyzed the connections between PLAAT4 and several clinicopathological factors. Further, we conducted Gene ontology (GO) analysis, Gene set enrichment analysis (GSEA), single sample gene set enrichment analysis (ssGSEA) and estimate analysis to explore the association between PLAAT4 and biological function and immune infiltration. In addition, Kaplan-Meier (KM) analysis, univariate and multivariate Cox analysis were used to explore the association between PLAAT4 and prognosis. In addition, we plotted a nomogram according to the multivariate cox analysis visualizing the predictive ability of PLAAT4 on prognosis. In addition, we explore the influence of PLAAT4 on malignant behaviors of the pancreatic cancer cells in vitro.
Results
After comparing the expression of PLAAT4 in normal and tumor tissues, we found that the expression of PLAAT4 was significantly high in pancreatic ductal adenocarcinoma (PDAC) samples. In addition, the results of GO and GSEA found that the expression of PLAAT4 was related to cell cycle checkpoints, M phase, regulation by p53, cell cycle mitotic and etc. Further, ssGSEA has shown that PLAAT4 was positively related to the abundance of aDC, Th1 cells, Th2 cells and negatively related to the Th17 cells. Subsequently, KM analysis, univariate and multivariate Cox analysis were used to analyze the correlation between PLAAT4 and prognosis. Additionally, we found that higher expression of PLAAT4 was related to T stage, N stage, histologic grade, etc (P < 0.05) and has a significant correlation with poor Overall Survival (OS), Disease-Specific Survival (DSS) and Progression-Free Interval (PFI). At last, we proved that PLAAT4 contributed to the malignant behaviors of the pancreatic cancer cells.
Conclusion
This study indicated PLAAT4 as a novel prognostic biomarker and an important molecular that mediated immune response in pancreatic cancer.
Keywords: PLAAT4, Pancreatic cancer, Immune infiltration, Nomogram, Biomarker, The cancer genome atlas (TCGA)
PLAAT4; Pancreatic cancer; Immune infiltration; Nomogram; Biomarker; The Cancer Genome Atlas (TCGA).
1. Introduction
In the United States and China, pancreatic cancer is still the most lethal disease among all malignant tumors and the survival rate of patients with pancreatic cancer is about 11% [1, 2]. Moreover, the death rate of pancreatic cancer is still increasing and pancreatic cancer is considered to become the second leading reason of cancer-related death in the United States by 2030 [3, 4]. Most patients have few obvious symptoms and already show metastases at diagnosis, missing the opportunity of surgical resection, which is the only chance to cure the disease [5, 6]. Additionally, the lack of early diagnostic markers also makes it difficult to detect pancreatic cancer at an early stage. Currently, CA19-9 is a commonly used serum biomarker to detect the development of pancreatic cancer. But the sensitivity of 79–81% and specificity of 82–90% for diagnosis in patients with symptoms still limit the use of it [7]. Therefore, finding out more sensitive and specific biomarkers for early detection is urgently needed.
Phospholipase A/acyltransferase (PLAAT) family code PLAAT protein that exhibits O- and N-acyltransferase bioactivity, which synthetizes N-acylated ethanolamine phospholipids [8, 9]. The PLAAT family were named the H-Ras-like suppressor (HRASLS) family, due to its negative regulation of the oncogene H-Ras gene [10]. While all HRASLS members can to metabolize phospholipids, presenting PLA1/A2, O-acyltransferase (OAT) and N-acyltransferase (NAT) bioactivities, the member of typical phospholipid-metabolizing enzymes, thus were renamed Phospholipase A/acyltransferase [11, 12]. Previously, the PLAAT family members with low expression in tumor cells suppressed the proliferation ability of tumor cells, thus were classified as type II tumor suppressor genes [13]. However, the relationship between the PLAAT family and cancer is largely unknown.
Phospholipase A/acyltransferase 4 (PLAAT4), also known as tazarotene-induced gene 3 (TIG3), retinoid-inducible gene 1 (RIG1) and retinoic acid receptor responder 3 (RARRES3), is a growth regulatory gene induced by retinoid and the synthetic retinoid tazarotene could upregulate the expression of it [14]. As reported, the expression of PLAAT4 was reduced in a variety of primary human tumors, including uterine, kidney, rectal, ureter, compared with comparable adjacent normal tissues [15]. While the function of PLAAT4 in pancreatic cancer remains unclear.
In this paper, we concentrated on the significance of elevated PLAAT4 expression in pancreatic cancer. We obtained RNA expression information of pancreatic cancer patients and comparable clinical characteristics from TCGA datasets, therefore performing bioinformatics analysis, exemplified as Kaplan-Meier (KM) survival analysis, Cox & Logistic regression analysis, differentially expressed genes (DEGs) analysis, nomogram, Gene Ontology (GO) analysis, Gene Set Enrichment Analysis (GSEA), single-sample Gene Set Enrichment Analysis (ssGSEA) and ESTIMATE analysis. Additionally, we modulated the expression of PLAAT4 in vitro and investigated the influence on the cell proliferation and migration of pancreatic cancer cells.
2. Material and methods
2.1. Collecting and preprocessing data of PLAAT4
The mRNA expression information of PLAAT4 and clinical information of patients with pancreatic cancer (178 tumoral and 4 normal tissues, Data Type: HTSeq-FPKM) were obtained from TCGA project (https://portal.gdc.cancer.gov/) (https://tcga-data.nci.nih.gov/tcga/) [16]. We also obtained mRNA expression information of PLAAT4 of normal pancreatic tissues from Genotype-Tissue Expression (GTEx) database. Besides, we obtained mRNA expression data of pancreatic cancer and normal pancreatic tissues from the Gene Expression Omnibus (GEO) datasets (GSE15471, GSE46234). The exclusion criteria for this study were Pancreatic adenocarcinoma (PAAD) samples with an overall survival of less than 30 days. Level 3 HTSeq-FPKM data of 178 PAAD patients were then adapted into transcripts per million (TPM) reads to further analyze. Unknown or unattainable clinical characteristics in 178 samples were seen as missing data. These data were shown in Supplemental Table 1.
2.2. Differential expression of PLAAT4 in PAAD samples in TCGA and GEO database
Scatter plots as well as box plots were conducted to estimate the diverse expression of PLAAT4, using disease state (normal or tumor) as variables. Define the statistical ranking of PLAAT4 expression below or above the median value as PLAAT4 high or PLAAT4 low, respectively.
2.3. Identification of DEGs between lower and higher expression of PLAAT4 in PAAD groups
DEGs between PLAAT4-Low and PLAAT4-High expression PAAD groups were analyzed with the DESeq2 (4.0) package by the Student’s t-test. The genes with absolute logarithm foldchange (FC) higher than 1.5 and adjusted P-value < 0.05 were considered to be statistically significant. Heatmap and volcano plots were produced to demonstrate all DEGs.
2.4. Functional enrichment of DEGs and immune-related cell infiltration
The enrichment of PLAAT4 involved DEGs by pathway and process was explored by Metascape (http://metasape.org). To obtain obvious statistical differences, the criteria were set the enrichment factor >1.5, a minimum count of 3, and P < 0.01. GSEA was performed with PLAAT4 differentially expression matrix to investigate the differences between PLAAT4 -high and -low patients and to analyze the PLAAT4 related pathways and phenotypes. The changes in signaling pathways were analyzed by 1000 permutation tests. FDR <0.25 and adjusted p-value < 0.01 were recognized as significant differences. R package clusterProfiler (4.0) was used to perform Graphical plots and statistical analysis [17]. The protein-protein interaction (PPI) network was constructed by the results of DEGs, using the STRING database [18] and the PPI pairs with interaction score >0.95 were chosen. Using ssGSEA, the interrogating expression data of genes in published gene tables [19] to measure 24 types of immune cell relative infiltration levels. The signatures contained a series of innate and adaptive immune cell types, identified by 509 genes. Spearman correlation as well as Wilcoxon rank-sum test were conducted to evaluate the relations of PLAAT4 and immune cell infiltration as well as the different groups of PLAAT4 expression with immune cell infiltration. In addition, R package estimate (1.0.13) was applied to measure the tumour purity and stromal and immune cell admixture. Stromal Score, Immune Score and ESTIMATE Score were calculated to verify our results.
2.5. Clinical statistics of prognostic factors, model construction and estimation
By logistic regression and Wilcoxon rank-sum test, we analyzed the relation between PLAAT4 expression and clinicopathological characteristics, using the R package (Version 3.6.2). We counted the clinicopathologic characteristics associated with 10-year overall OS, DSS and PFI in TCGA samples, by the Kaplan-Meier and the Cox regression method. To analysis the PLAAT4 expression impact on survival rates together with other clinical pathologic features (TNM stage, Age, Gender, Histologic grade, Albumin, Tumor status, TP53 status, Vascular invasion, Residual tumor, Race, Adjacent pancreatic tissue inflammation), we used Multivariate Cox analysis. P-value < 0.05 was considered statistically significant and we choose the mean value of the PLAAT4 expression as the cut-off value. In PLAAT4 -low and -high group, we analyzed the difference of OS, DSS, and PFI separately, using two-sided log-rank test KM method. We constructed nomograms obtained from multivariate Cox analysis, using the independent prognostic factors to evaluate the survival rates for 1-, 3-, and 5-year, respectively, by RMS package (https://cran.r-project.org/web/packages/rms/index.html), including calibration plots and important clinical factors. The calibration curve was evaluated graphically by drawing the nomogram of speculated probabilities of these observed events, and the 45-degree line represented the most accurate prediction. A consistency index (c-index) was calculated by bootstrapping method with 1000 samples and applied to the discrimination of estimating nominal graphs, evaluating the prognostic factors and prediction accuracy of the nomogram. In this study, we used two-tailed statistical tests and set a significance level at 0.05.
2.6. Cell culture and transfection
Pancreatic cancer cell lines (HPNE, CFPAC-1, AsPC-1, BxPC-3, Panc-1, PATU8988, MIA PaCa-2) were bought from the American Type Culture Collection (ATCC, Manassas, VA, USA) and were separately cultured in Dulbecco’s Modified Eagle Medium (DMEM, Hyclone, Logan, UT, USA) or Roswell Park Memorial Institute Medium (RPMI, Corning, NY, USA) added 10% fetal bovine serum (Gibco, Carlsbad, CA, USA), incubating under a temperature of 37 °C with 5% CO2. We used Lipofectamine 3000 and OPTI-MEM (Invitrogen, Thermo Fisher Scientific, Waltham, MA) to transfect cells with siRNA, according to the instructions. The siNC and siPLAAT4 (CCCGCUGUAAACAGGUGGAAA) were bought from RiboBio (Guangzhou, China). The final concentration of siRNA was 50nM when introduced. The over-expression plasmid was bought from Tsingke Biotechnology (Beijing, China). 2.5μg plasmid was introduced to cell in a 6-well plate following the instructions. After transfection, the cells were incubating for 48 h to perform further experiments.
2.7. Western blotting
Western blot experiment was conducted as previously described [20]. Briefly, we performed Western blot with 10% gel and 20μg sample protein in each experiment, following the protocol of antibodies. Anti-PLAAT4 (Catalog number: 12065-1-AP) and anti-GAPDH (Catalog number: 10494-1-AP) antibodies, bought from Proteintech Group (Rosemont, USA).
2.8. Cell proliferation, colony formation assays
We used colony formation and CCK-8 assay to evaluate the ability of cell proliferation. 3000 cells were plated into each well in the 96-well plate. We added 10 μL of CCK-8 solution (Beyotime biotechnology Co. Ltd., Shanghai, China) in each well of 96-well plate and after incubating for 2 h, the optical density of the mixture at 450 nm was further measured by a microplate spectrophotometer. For colony formation experiment, 1000 cells were seed in each cell of the 6-well plate and the culture medium was changed every 3 days. We used 4% paraformaldehyde to fix the cells and crystal violet to dye the colonies.
2.9. Wound healing and transwell assays
Wound healing assays were used to assess the cell migration ability. In brief, 5 × 105 cells in 70 μl medium containing 10% FBS were planted into each hole of ibidi Culture-Insert (ibidi, Germany). After the cells were found well adhering to the wall, ibidi Culture-Inserts were thus removed. Cells were washed by PBS and photographed under a microscope (0 h). Then the cells were cultured in serum-free medium. Images of the scratch were taken at different time points. Transwell system (Jet Bio-Filtration Co. Ltd., Guangzhou, China) was purchased to perform migration experiment. After transfected with siPLAAT4 for 24 h, 4 × 105 cells were digested and seeded into the upper layer of transwell chamber within 100 μl serum-free medium. 600 μl medium containing 10% FBS were put in the lower layer. After incubating for 24 h, cells on the upper side of the chamber were removed with cotton swab scratching and migrated cells on the lower side of the chamber were fixed by 4% paraformaldehyde and dyed by crystal violet. Pictures were taken under the microscope after drying. The counts of metastatic cells were calculated in five microscope fields randomly selected in each group.
2.10. Cell cycle experiment
Cell cycle and apoptosis analysis kit (C1052, Beyotime, China) was utilized to perform cell cycle experiment. In brief, transfected cells were digested and washed by precooled PBS and fixed by 70% ethanol water solution for 12 h at 4 °C. Then the cells were collected by centrifugation and washed by precooled PBS. Propidium iodide staining solution was used to dyed the double-strand DNA. The excitation wavelength at 488 nm was detected by flow cytometry. The results were analyzed by FlowJo software.
2.11. Statistical analysis
Statistical analyses were performed with the R software (V 3.8.0) and the RStudio software. Two-tailed Student’s t-test and one-way analysis of variance (ANOVA) were conducted to perform analyses of the initial data. Statistical findings are identified by P-value < 0.05.
3. Results
3.1. PLAAT4 expression in pan-cancer and pancreatic cancer
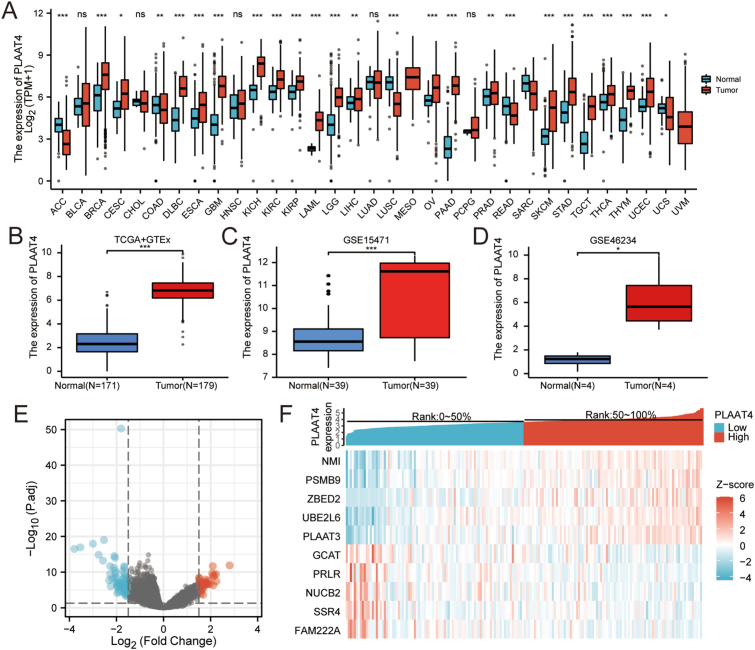
We firstly evaluated PLAAT4 expression in pan-cancer tissues versus corresponding normal tissues from TCGA and Genotype-Tissue Expression (GTEx) database. Among 33 cancer types, PLAAT4 expression was elevated in 22 types of cancer and downregulated in 6 types of cancer, indicating that PLAAT4 is widely expressed in cancer tissues. In PAAD, Wilcoxon rank-sum test demonstrates that PLAAT4 is significantly higher expressed in breast infiltrating carcinoma (BRCA), bladder urothelial carcinoma (BLCA), cervical squamous cell carcinoma and adenocarcinoma (CESC), colon adenocarcinoma (COAD), diffuse large B cell lymphoma (DLBCL), esophageal carcinoma (ESCA), pleomorphic glioma (GBM), renal chromophobe cell carcinoma (KICH), renal clear cell carcinoma (KIRC), renal papillary cell carcinoma (KIRP), acute myeloid leukeMIA PaCa-2 (LAML), Brain Lower Grade Glioma (LGG), liver hepatocellular carcinoma (LIHC), ovarian serous cystadenocarcinoma (OV), pancreatic cancer (PAAD), prostate cancer (PRAD), skin melanoma (SKCM), gastric cancer (STAD), testicular germ cell tumors (TGCT), thyroid cancer (THCA), thymic cancer (THYM), endometrial cancer (UCEC) (P < 0.05) (Figure 1A). Besides, we analyzed the PLAAT4 expression in pancreatic cancer, which demonstrated that PLAAT4 expression was higher in the tumor tissues than in normal tissues (P < 0.05; Figure 1B-D).
Figure 1.
The PLAAT4 expression in different malignant tumors and PLAAT4-associated differentially expressed genes (DEGs). (A) Differential expression of PLAAT4 in different cancers compared with comparable normal tissues in the TCGA and GTEx database. (B) The expression level of PLAAT4 in PAAD. (C,D) The expression level of PLAAT4 in GEO databases. (E) Volcano plots of the DEGs. (F) Top 10 DEGs are demonstrated in the heatmap.
3.2. Identification and analysis of DEGs in PAAD
89 PLAAT4-high samples with 90 PLAAT4-low samples in PAAD were compared. The results demonstrated that totally 105 DEGs were found statistically significant (absolute Log2-fold change >1.5 and adjusted p-value < 0.05) (Figure 1E; Supplemental Table 2), including 35 upregulated genes and 70 downregulated genes. The top 10 DEGs ranked by relative expression were shown in Figure 1F.
3.3. Functional enrichment of PLAAT4-Related genes in PAAD
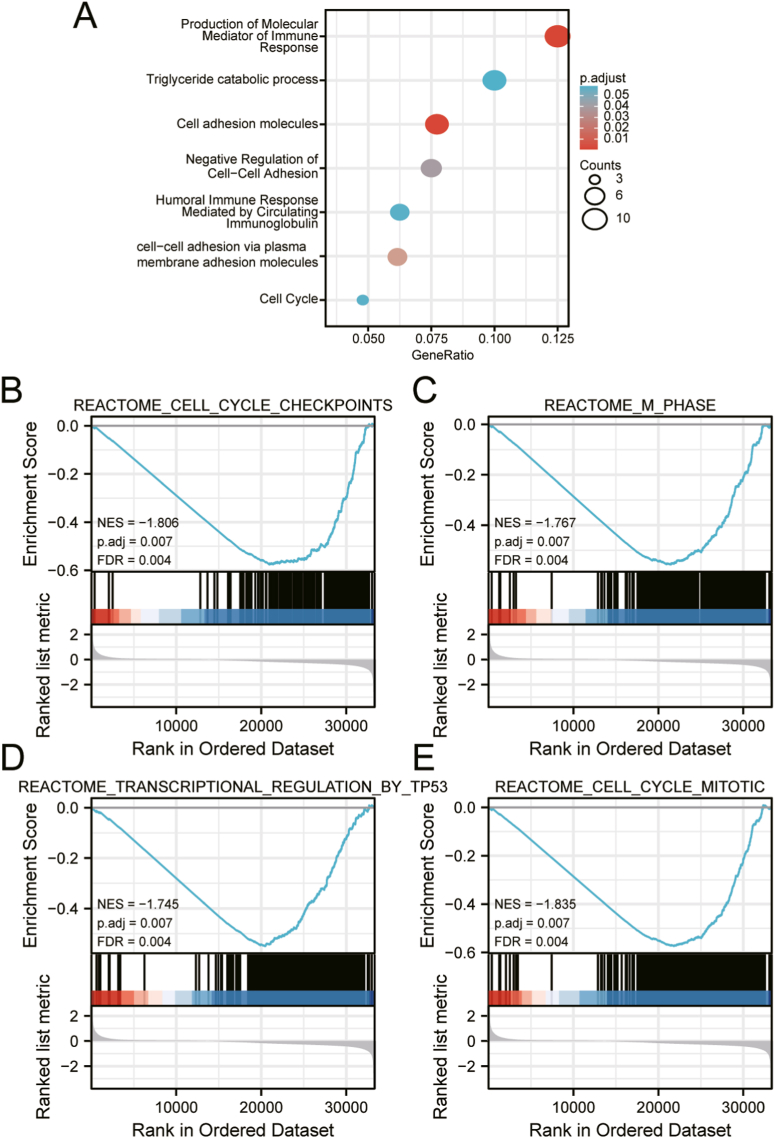
We analyzed the PLAAT4-related genes and conducted GO enrichment in Metascape to better understand the function of PLAAT4. The results revealed that the function of PLAAT4-related genes was related to several molecular functions, including production of molecular mediator of the immune response, triglyceride catabolic process, cell adhesion molecules, negative regulation of cell-cell adhesion, humoral immune response mediated by circulating immunoglobulin, cell-cell adhesion via plasma membrane adhesion molecules and cell cycle (Figure 2A; Supplemental Table 3).
Figure 2.
Significantly enriched GO annotations of PLAAT4 related genes in PAAD. (A) The most significant biological processes enriched in PLAAT4-related genes with bar graph. (B–E) GSEA enrichment plots revealed that cell cycle checkpoints, M phase, regulation by p53 and cell cycle mitotic were significantly enriched in patients with high-PLAAT4. NES, normalized enrichment score; p.adj, adjusted P value; FDR, false discovery rate.
3.4. Protein-protein interaction (PPI) network analysis
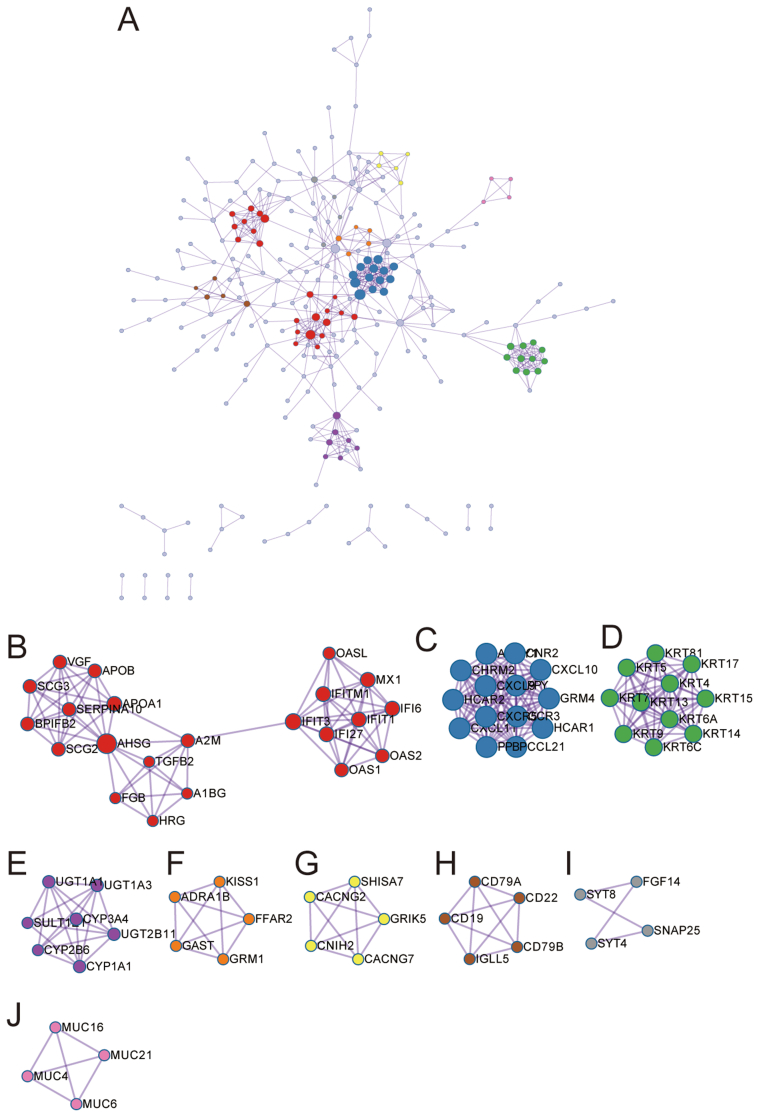
We performed a protein-protein interaction network by the STRING database to find out the interactions between DEGs in PAAD and we set the interaction score at high confidence (0.70). 645 edges 1 and 42 proteins were screened out and 9 hub clusters were selected in the PPI network (Supplymental Figure 1A-J; Supplemental Table 4).
3.5. Potential mechanism of PLAAT4 controlling PAAD progression
We performed GSEA analysis between PLAAT4-high and -low expression sample to investigate PLAAT4-related signaling pathways and significant differences (false discovery rate, FDR q-value < 0.25; nominal, NOM p-value < 0.05) were enriched in the Molecular Signatures Database (MSigDB) Collection (c2.cp.reactome/biocarta/kegg.v6.2.symbols.gmt). We screened out signaling pathways enriched with the most significance, according to the normalized enrichment score (NES). GSEA enrichment plots revealed that cell cycle checkpoints, M phase, regulation by p53 and cell cycle mitotic were significantly enriched in patients with high-PLAAT4 (Figure 2B-E; Supplemental Table 5).
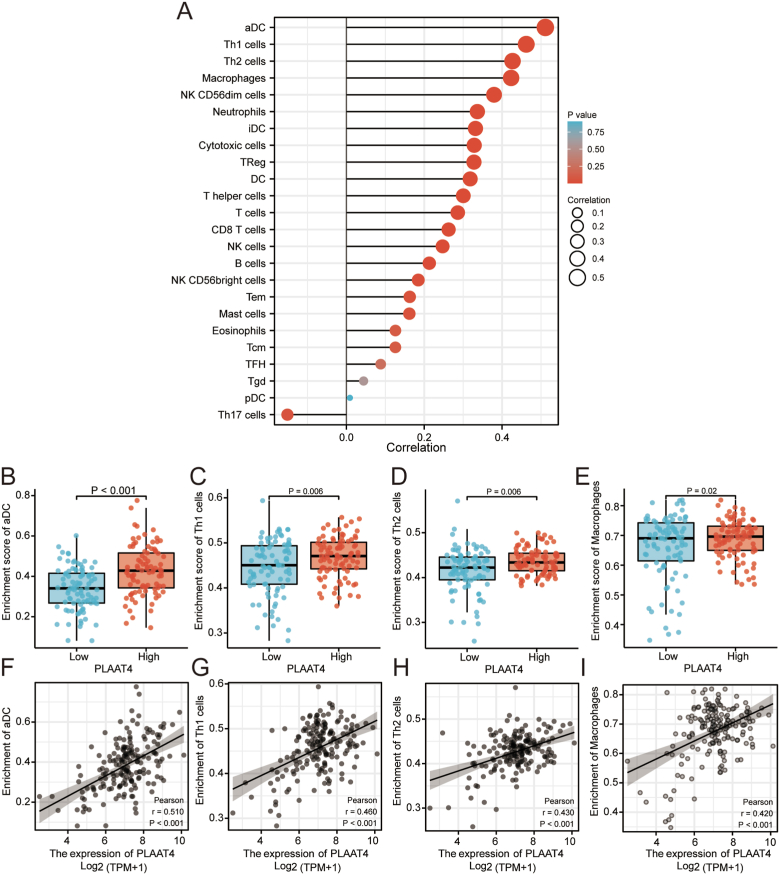
3.6. Connections between immune infiltration and PLAAT4 expression level
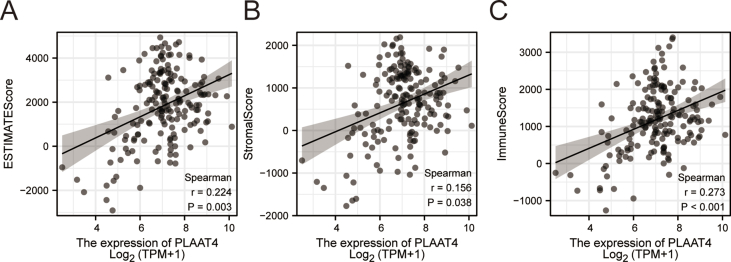
We performed ssGSEA analysis in PAAD samples and used Spearman correlation to determine the significant association between immune infiltration and PLAAT4 expression level. The expression of PLAAT4 was positively related to the abundance of several immune cells, such as aDC (R = 0.510, P < 0.001), Th1 cells (R = 0.460, P = 0.006), Th2 cells (R = 0.430, P = 0.006), Macrophages (R = 0.420, P < 0.001), NK CD56dim cells, Neutrophils, iDC, Cytotoxic cells and TReg, DC, T helper cells, T cells, CD8T cells, NK cells, B cells, NK CD56 bright cells, Tem, Mast cells, Eosinophils, Tcm, TFH, Tgd, pDC. The expression of PLAAT4 was negatively related to the abundance of Th17 cells (Figure 3A-I). In addition, we conducted the estimate analysis to explore the tumor purity and stromal and immune cell admixture from expression data. The StromalScore (R = 0.335, P < 0.001), the ImmuneScore (R = 0.374, P < 0.001) and the ESTIMATEScore (R = 0.374, P < 0.001) were calculated to demonstrate the expression of PLAAT4 was significantly associated with immune infiltration (Supplymental Figure 2A–C).
Figure 3.
The correlation of PLAAT4 expression with immune infiltration in tumor microenvironment. (A) The correlation between the expression level PLAAT4 and the relative abundances of the immune infiltrated cells. The size of the dots represents the results of Spearman R. (B–I) The differential infiltration level of Th1, Th2, aDC, macrophages between the high and low PLAAT4 groups in the Correlation diagrams and scatter plots.
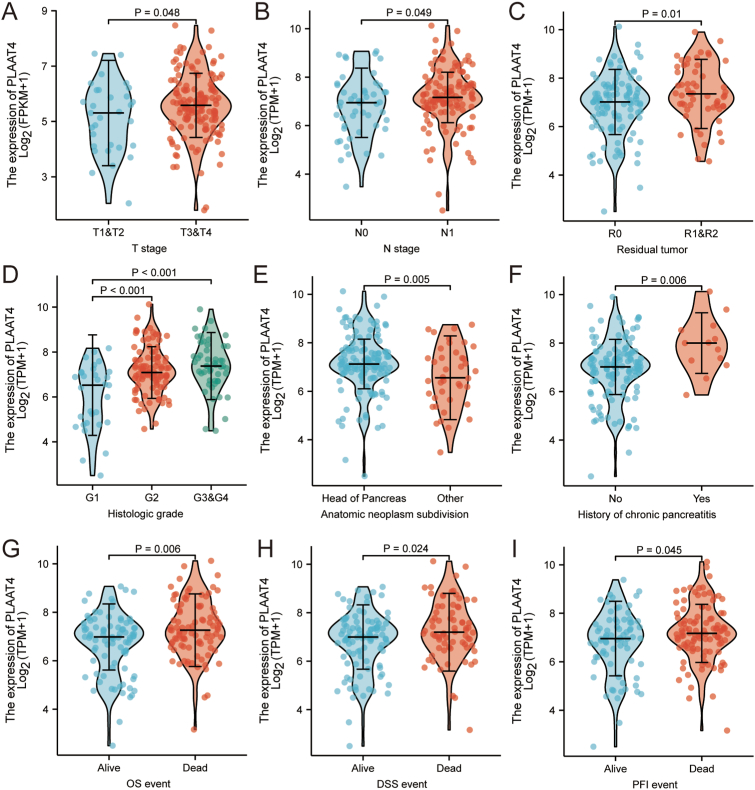
3.7. Association of PLAAT4 expression with clinicopathological variables
To better explore the function of PLAAT4 in PAAD, we analyzed the expression of PLAAT4 and clinical features in 178 PAAD patients from TCGA (Table 1). Increased PLAAT4 expression in PAAD robustly associated with clinical T stage (T3 & T4 vs. T1 & T2, P = 0.048), clinical N stage (N1 vs. N0, P = 0.049), residual tumor (R1 & R2 vs. R0, p = 0.01), histologic grade (G3/G4 vs. G1, P < 0.001; G2 vs. G1, P < 0.001), anatomic neoplasm subdivision (Head of Pancreas vs. Other, P = 0.005) (Figure 4A-I), history of chronic pancreatitis (Yes vs. No, P = 0.006), OS event (Dead vs. Alive, P = 0.006), DSS event (Dead vs. Alive, P = 0.024) and PFI event (Dead vs. Alive, P = 0.045). Univariate analysis via logistic regression showed that PLAAT4 expression was associated with poor prognostic clinicopathological characteristics (Table 2). High PLAAT4 expression in PAAD was associated with T stage (T3 & T4 vs. T1 & T2, OR = 1.373, P = 0.044), Pathologic stage (Stage II & III & IV vs. Stage I, OR = 1.473, P = 0.031), Residual tumor (R1 & R2 vs. R0, OR = 1.423, P = 0.015), Histologic grade (G3 & G4 vs. G1 & G2, OR = 1.411, P = 0.021), Anatomic neoplasm subdivision (Other vs. Head of Pancreas, OR = 0.079, P = 0.017) and History of chronic pancreatitis (Yes vs. No, OR = 2.199, P = 0.006). These results suggest that PAAD patients with higher expression of PLAAT4 are possible to experience a higher possibility of tumor progression.
Table 1.
The correlation between clinicopathological variables and PLAAT4 expression in PAAD patients.
| Characteristic | Low expression of PLAAT4 | High expression of PLAAT4 | p | method |
|---|---|---|---|---|
| n | 89 | 89 | ||
| T stage, n (%) | 0.493 | Fisher.test | ||
| T1 | 3 (1.7%) | 4 (2.3%) | ||
| T2 | 15 (8.5%) | 9 (5.1%) | ||
| T3 | 67 (38.1%) | 75 (42.6%) | ||
| T4 | 2 (1.1%) | 1 (0.6%) | ||
| N stage, n (%) | 0.098 | Chisq.test | ||
| N0 | 30 (17.3%) | 20 (11.6%) | ||
| N1 | 55 (31.8%) | 68 (39.3%) | ||
| M stage, n (%) | 0.187 | Fisher.test | ||
| M0 | 36 (42.9%) | 43 (51.2%) | ||
| M1 | 4 (4.8%) | 1 (1.2%) | ||
| Pathologic stage, n (%) | 0.245 | Fisher.test | ||
| Stage I | 13 (7.4%) | 8 (4.6%) | ||
| Stage II | 67 (38.3%) | 79 (45.1%) | ||
| Stage III | 2 (1.1%) | 1 (0.6%) | ||
| Stage IV | 4 (2.3%) | 1 (0.6%) | ||
| Primary therapy outcome, n (%) | 0.167 | Fisher.test | ||
| PD | 20 (14.4%) | 29 (20.9%) | ||
| SD | 4 (2.9%) | 5 (3.6%) | ||
| PR | 3 (2.2%) | 7 (5%) | ||
| CR | 41 (29.5%) | 30 (21.6%) | ||
| Radiation therapy, n (%) | 1 | Chisq.test | ||
| No | 60 (36.8%) | 58 (35.6%) | ||
| Yes | 23 (14.1%) | 22 (13.5%) | ||
| Residual tumor, n (%) | 0.213 | Fisher.test | ||
| R0 | 58 (35.4%) | 49 (29.9%) | ||
| R1 | 21 (12.8%) | 31 (18.9%) | ||
| R2 | 2 (1.2%) | 3 (1.8%) | ||
| Age, n (%) | 1 | Chisq.test | ||
| ≤65 | 47 (26.4%) | 46 (25.8%) | ||
| >65 | 42 (23.6%) | 43 (24.2%) | ||
| Race, n (%) | 0.057 | Fisher.test | ||
| Asian | 9 (5.2%) | 2 (1.1%) | ||
| Black or African American | 4 (2.3%) | 2 (1.1%) | ||
| White | 74 (42.5%) | 83 (47.7%) | ||
| Gender, n (%) | 0.88 | Chisq.test | ||
| Female | 41 (23%) | 39 (21.9%) | ||
| Male | 48 (27%) | 50 (28.1%) | ||
| Histologic grade, n (%) | <0.001 | Fisher.test | ||
| G1 | 23 (13.1%) | 8 (4.5%) | ||
| G2 | 48 (27.3%) | 47 (26.7%) | ||
| G3 | 15 (8.5%) | 33 (18.8%) | ||
| G4 | 2 (1.1%) | 0 (0%) | ||
| Anatomic neoplasm subdivision, n (%) | 0.209 | Chisq.test | ||
| Head of Pancreas | 65 (36.5%) | 73 (41%) | ||
| Other | 24 (13.5%) | 16 (9%) | ||
Figure 4.
The correlation of PLAAT4 expression with clinicopathological characteristics, including (A) T stage, (B) N stage, (C) Residual tumor, (D) Histologic stage, (E) Anatomic neoplasm subdivision, (F) History of chronic pancreatitis, (G) OS event, (H) DSS event, and (I) PFI event in PAAD patients in the TCGA cohort. TCGA, The Cancer Genome Atlas; PAAD, pancreatic adenocarcinoma.
Table 2.
PLAAT4 expression associated with clinicopathological characteristics (logistic regression).
| Characteristics | Total(N) | Odds Ratio (OR) | P value |
|---|---|---|---|
| T stage (T3 & T4 vs. T1 & T2) | 176 | 1.373 (1.009–1.882) | 0.044 |
| N stage (N1 vs. N0) | 173 | 1.292 (0.989–1.706) | 0.063 |
| M stage (M1 vs. M0) | 84 | 0.581 (0.290–1.148) | 0.104 |
| Pathologic stage (Stage II & Stage III & Stage IV vs. Stage I) | 175 | 1.473 (1.034–2.111) | 0.031 |
| Radiation therapy (Yes vs. No) | 163 | 0.916 (0.698–1.206) | 0.529 |
| Primary therapy outcome (PR & CR vs. PD & SD) | 139 | 0.802 (0.591–1.068) | 0.141 |
| Gender (Male vs. Female) | 178 | 0.977 (0.769–1.239) | 0.847 |
| Race (White vs. Asian & Black or African American) | 174 | 1.218 (0.822–1.776) | 0.312 |
| Age (>65 vs. ≤65) | 178 | 0.917 (0.721–1.161) | 0.473 |
| Residual tumor (R1 & R2 vs. R0) | 164 | 1.423 (1.082–1.911) | 0.015 |
| Histologic grade (G3 & G4 vs. G1 & G2) | 176 | 1.411 (1.065–1.912) | 0.021 |
| Anatomic neoplasm subdivision (Other vs. Head of Pancreas) | 178 | 0.709 (0.529–0.937) | 0.017 |
| Smoker (Yes vs. No) | 144 | 1.182 (0.911–1.550) | 0.212 |
| Alcohol history (Yes vs. No) | 166 | 1.118 (0.872–1.439) | 0.378 |
| History of diabetes (Yes vs. No) | 146 | 0.992 (0.737–1.345) | 0.956 |
| History of chronic pancreatitis (Yes vs. No) | 141 | 2.199 (1.292–3.994) | 0.006 |
3.8. Connection of PLAAT4 expression with prognosis of patients with PAAD
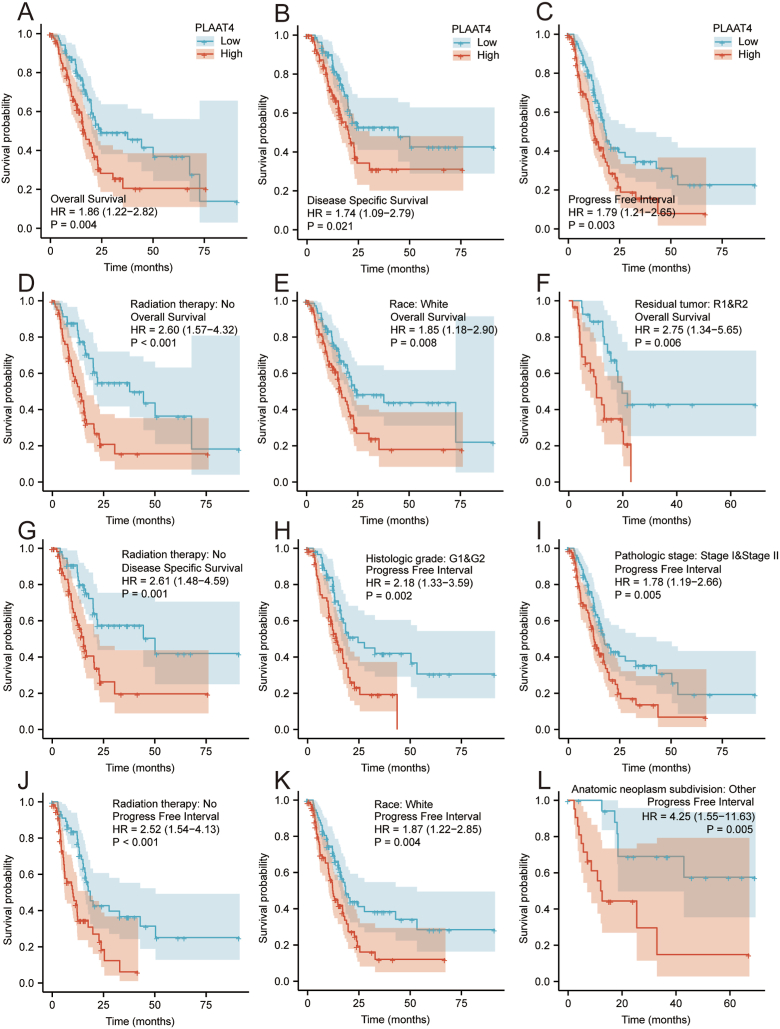
The results illustrated that the OS and DSS of patients with high expression of PLAAT4 were lower than those of patients with low expression of PLAAT4 (Figure 5A-B, P = 0.004, P = 0.021). Besides, the PFI was lower in the PLAAT4-high cohort of PAAD patients (Figure 5C, P = 0.003). Subgroup analysis of OS, DSS, PFI illustrated that high PLAAT4 expression of PAAD patients in Radiation therapy: NO, Race: White and Residual Tumor: R1 & R2 subgroups have a lower OS (Figure 5D-F, P < 0.05). PAAD patients in Radiation therapy: NO subgroup also has a lower DSS (Figure 5G, P < 0.05). PAAD patients in Histologic grade: G1 & G2, Pathologic stage: Stage I & Stage II, Radiation therapy: NO, Race: White and Anatomic neoplasm subdivision: Other subgroups have worse PFI (Figure 5H-L, P < 0.05). By univariate Cox analysis, we found that patients with high PLAAT4 expression had poor OS (HR = 1.943 95% CI: 1.279–2.953; P = 0.002), DSS (HR = 1.832 95% CI: 1.145–2.932; P = 0.012) and PFI (HR = 1.898 95% CI: 1.282–2.810; P = 0.001) (Table 3, Supplemental Tables 6 and Supplemental Tables 7). We further performed multivariate Cox analysis with PLAAT4 expression, T stage, N stage, Radiation therapy, Primary therapy outcome, Residual tumor and Anatomic neoplasm subdivision to identify features related to prognosis. It is important that patients with high PLAAT4 expression have poor OS (HR = 1.806 95% CI: 1.080–3.020; P = 0.024) in multivariate Cox analysis. But no significant difference was found in DSS (HR = 1.394 95% CI: 0.791–2.457; P = 0.251) and PFI (HR = 1.319 95% CI: 0.804–2.165; P = 0.273) in patients with high PLAAT4 expression (Table 3, Supplemental Tables 6 and Supplemental Tables 7). We further analyzed the prognostic value of OS, DSS, PFI of PLAAT4 expression in PAAD patients grouped according to independent prognostic features screened by multivariate Cox analysis (Table 4, Supplemental Table 8 and Supplemental Table 9). The PLAAT4-high PAAD patients had low OS in the subgroups of T3 & 4 stage (HR: 1.69; CI: 1.08–2.62; P = 0.02), M0 stage (HR: 1.94; CI: 1.02–3.70; P = 0.045), pathologic stage I & II (HR: 1.17; CI: 1.12–2.62; P = 0.013) and histologic grade G1 & G2 (HR: 1.79; CI: 1.05–3.07; P = 0.033) (Table 4). However, the PLAAT4-high PAAD patients had low DSS only in the subgroups of histologic grade G1 & G2 (HR: 1.83; CI: 1.01–3.34; P = 0.047, Supplemental Table 8). The PLAAT4-high PAAD patients had low PFI in the subgroups of T3 & 4 stage (HR: 1.60; CI: 1.06–2.41; P = 0.026), N0 stage (HR: 2.74; CI: 1.19–6.32; P = 0.18), M0 stage (HR: 1.83; CI: 1.04–3.23; P = 0.035), pathologic stage I & II (HR: 2.74; CI: 1.19–6.32; P = 0.18), histologic grade G1 & G2 (HR: 2.18; CI: 1.33–3.59; P = 0.002) (Supplemental Table 9).
Figure 5.
Kaplan Meier survival plots comparing with high and low expression of PLAAT4 in patients with PAAD. (A–C) Survival curves of OS, DSS, and PFI between high and low expression of PLAAT4 in patients with PAAD. (D–F) OS survival curves of Radiation therapy: No, Race: White and Residual tumor: R1 & R2 subgroups between PLAAT4-high and -low patients with PAAD. (G) DSS survival curves of Radiation therapy: No subgroup between PLAAT4-high and -low patients with PAAD. (H–L) PFI survival curves of Histologic grade: G1 & G2, Pathologic stage: Stage I & Stage II, Radiation therapy: No, Race: White subgroups between PLAAT4-high and -low patients with PAAD. PAAD, pancreatic adenocarcinoma; OS, overall survival; DSS, disease specific survival; PFI, progression free interval.
Table 3.
Univariate and multivariate Cox analysis (Overall Survival) of prognostic covariates in patients with pancreatic cancer.
| Characteristics | Total(N) | Univariate analysis |
Multivariate analysis |
||
|---|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | ||
| T stage (T3 & T4 vs. T1 & T2) | 176 | 2.023 (1.072–3.816) | 0.030 | 1.057 (0.516–2.166) | 0.879 |
| N stage (N1 vs. N0) | 173 | 2.154 (1.282–3.618) | 0.004 | 1.524 (0.818–2.838) | 0.184 |
| M stage (M1 vs. M0) | 84 | 0.756 (0.181–3.157) | 0.701 | ||
| Radiation therapy (Yes vs. No) | 163 | 0.508 (0.298–0.866) | 0.013 | 0.430 (0.227–0.813) | 0.009 |
| Primary therapy outcome (PR & CR vs. PD&SD) | 139 | 0.425 (0.267–0.677) | <0.001 | 0.524 (0.315–0.871) | 0.013 |
| Gender (Male vs. Female) | 178 | 0.809 (0.537–1.219) | 0.311 | ||
| Race (White vs. Asian & Black or African American) | 174 | 1.161 (0.582–2.318) | 0.672 | ||
| Age (>65 vs. ≤65) | 178 | 1.290 (0.854–1.948) | 0.227 | ||
| Residual tumor (R1 & R2 vs. R0) | 164 | 1.645 (1.056–2.561) | 0.028 | 1.503 (0.887–2.546) | 0.130 |
| Histologic grade (G3 & G4 vs. G1 & G2) | 176 | 1.538 (0.996–2.376) | 0.052 | ||
| Anatomic neoplasm subdivision (Other vs. Head of Pancreas) | 178 | 0.417 (0.231–0.754) | 0.004 | 0.559 (0.265–1.178) | 0.126 |
| PLAAT4 (High vs. Low) | 178 | 1.943 (1.279–2.953) | 0.002 | 1.806 (1.080–3.020) | 0.024 |
Table 4.
The prognostic value of PLAAT4 (Overall Survival) in subgroups of patients with pancreatic cancer.
| Characteristics | N (%) | HR (95% CI) | P value |
|---|---|---|---|
| T stage | |||
| T1 & T2 | 31 (17) | 1.95 (0.55–6.92) | 0.299 |
| T3 & T4 | 145 (83) | 1.69 (1.08–2.62) | 0.02 |
| N stage | |||
| N0 | 50 (28.9) | 1.58 (0.59–4.20) | 0.364 |
| N1 | 123 (71.1) | 1.33 (0.83–2.11) | 0.231 |
| M stage | |||
| M0 | 79 (94) | 1.94 (1.02–3.70) | 0.045 |
| M1 | 5 (6) | – | – |
| Pathologic stage | |||
| Stage I & Stage II | 167 (96) | 1.71 (1.12–2.62) | 0.013 |
| Stage III & Stage IV | 8 (4) | – | – |
| Histologic grade | |||
| G1 & G2 | 126 (71) | 1.79 (1.05–3.07) | 0.033 |
| G4 & G3 | 50 (29) | 1.35 (0.66–2.74) | 0.409 |
3.9. Construction a nomogram plot to evaluate the predictive ability of PLAAT4
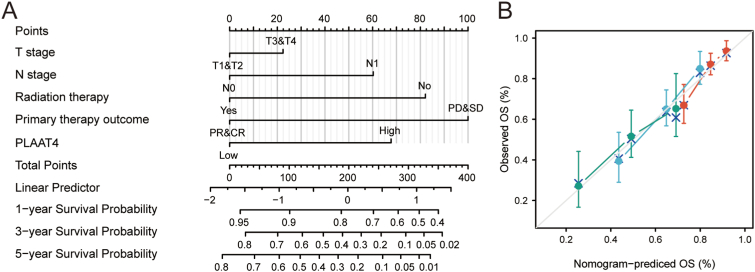
We plotted a nomogram according to PLAAT4 expression and other independent clinicopathologic features to predict the survival of PAAD patients (Figure 6A). According to multivariate Cox analysis, we plotted the nomogram with a point scale of 0–100 that was assigned to these variables. The total scores of these variables were recorded and seen as measurement units. The prognostic values of each variable were exhibited by a vertical line drawing from the total point axis to the outcome axis. For example, a PAAD patient with high PLAAT4 risk (67.5 points), T3 stage (22.5 points), N0 stage (0 points), Radiation therapy YES (0 points), PD (100 points) has a total point of 190 and the 1-, 3-, 5- year predictive survival rates of this patients are 80%, 40% and 28% respectively. Further, the prognostic accuracy of the nomogram was assessed by the Hosmer test of the calibration curve. In the TCGA-PAAD cohort, the calibration curve of this nomogram was 0.669, which suggested moderate accuracy of this prognostic model (Figure 6B).
Figure 6.
A quantitative scoring system to predict the probability of OS at the 1, 3, and 5 year in PAAD patients. (A) The nomogram for estimating the probability of OS at the 1, 3, and 5 year in PAAD patients. (B) The calibration plots of the nomogram for estimating the probability of OS at the 1, 3, and 5 year in PAAD patients. PAAD, pancreatic adenocarcinoma; OS, overall survival.
3.10. Effect of PLAAT4 0n the Proliferation Ability of Pancreatic Cancer Cells
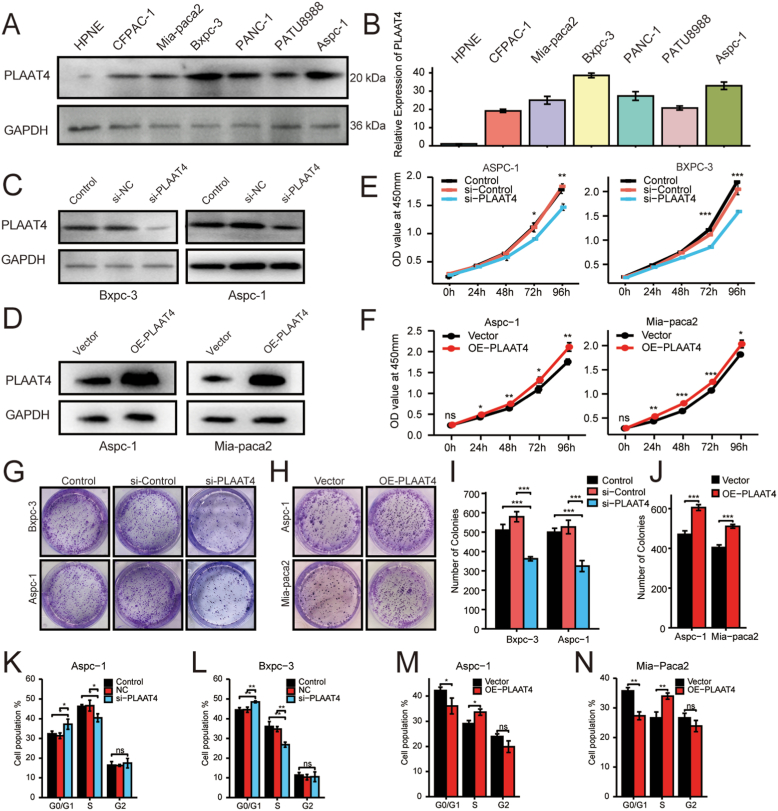
Firstly, we analyzed the expression level of PLAAT4 in pancreatic cancer cell lines (HPNE, CFPAC-1, AsPC-1, BxPC-3, PANC-1, PATU8988, MIA PaCa-2) by Western blot and qPCR assays. The expression of PLAAT4 was higher in AsPC-1 and BxPC-3 than in other cell lines (Figure 7A-B), thus selected for knockdown of PLAAT4. The efficiency of si-PLATT4 was verified by Western blot (Figure 7C). CCK8 experiments and colony formation showed that the si-PLAAT4 group had lower proliferation ability and colony formation rate in AsPC-1 and BxPC-3 (Figure 7E, G, I). Besides, we performed the PLAAT4-overexpression in moderately PLAAT4-expression AsPC-1 and MIA PaCa-2 cell lines for further verification. The efficiency was verified by Western blot (Figure 7D). CCK8 experiments and colony formation showed that the PLAAT4-overexpression group had a higher proliferation ability and colony formation rate in AsPC-1 and MIA PaCa-2 (Figure 7F, H, J). These results demonstrated the promoting effect of PLAAT4 on the proliferation ability.
Figure 7.
PLAAT4 Knockdown attenuated the Proliferation Ability of Pancreatic Cancer Cells. (A) The expression of PLAAT4 in pancreatic cancer cell lines (HPNE, CFPAC-1, AsPC-1, BxPC-3, Panc-1, PATU8988, MIA PaCa-2) were separately measured by western blotting. (B) The expression of PLAAT4 in pancreatic cancer cell lines (HPNE, CFPAC-1, AsPC-1, BxPC-3, Panc-1, PATU8988, MIA PaCa-2) were measured by qPCR. (C) The efficiency of si-PLAAT4 in AsPC-1 and BxPC-3 cell lines were tested by western blotting. (D) The efficiency of PLAAT4-overexpression in AsPC-1 and MIA PaCa-2 cell lines were verified by western blotting. (E) The CCK-8 assay was used to detect the proliferation ability after si-PLAAT4 in AsPC-1 and BxPC-3 cell lines. (F) The CCK-8 assay was used to detect the proliferation ability after PLAAT4-overexpression in AsPC-1 and MIA PaCa-2 cell lines. Representational images of the colony formation assay after si-PLAAT4 in AsPC-1 and BxPC-3 cell lines. (G–J) Digitization of the colony formation assay after si-PLAAT4 in AsPC-1 and BxPC-3 cell lines and after PLAAT4-overexpression in AsPC-1 and MIA PaCa-2 cell lines. (K–N) Digitization of the results of cell cycle assay after si-PLAAT4 in AsPC-1 and BxPC-3 cell lines and after PLAAT4-overexpression in AsPC-1 and MIA PaCa-2 cell lines. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
3.11. Effect of PLAAT4 in the cell cycle of pancreatic cancer cells
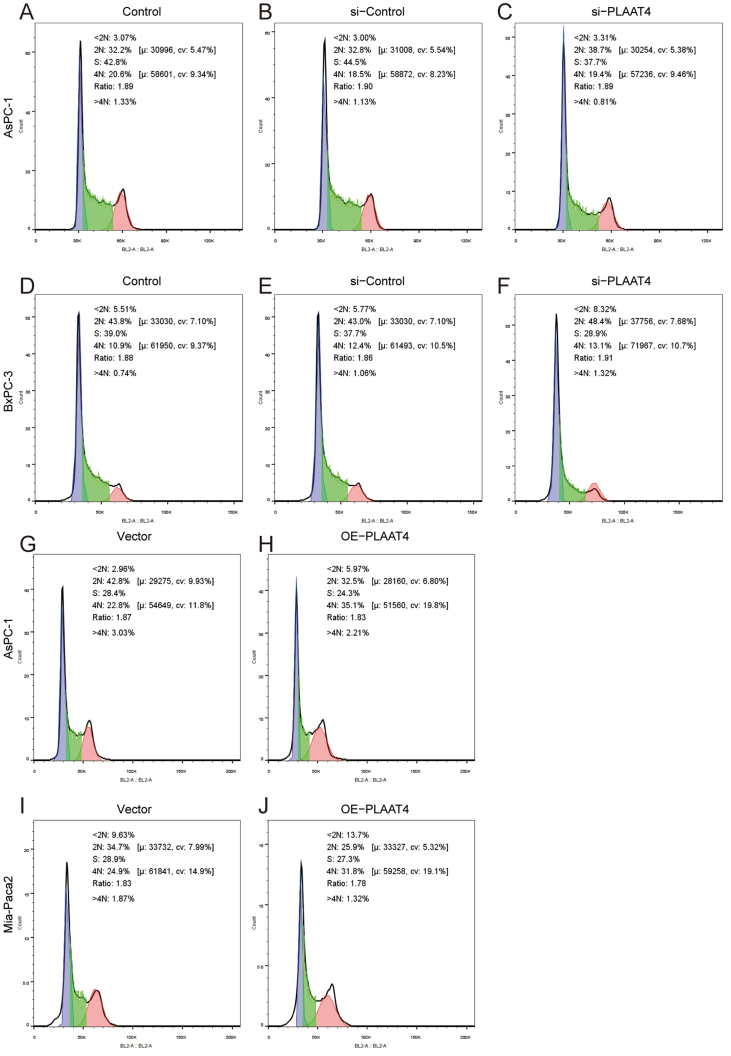
In order to further verify the inner mechanism of cell proliferation, we performed the cell cycle experiment. The results illustrated that in AsPC-1, there were a higher percentage of G0/G1 phase fraction of cells and lower percentage of S phase fraction of cells after knockdown of PLAAT4 (Figure 7K, Supplymental Figure 3A–C). The cells were increased in G0/G1 phase and decreased in the S phase after the knockdown of PLAAT4 in BxPC-3 (Figure 7L, Supplymental Figure 3D–F). Similarly, in AsPC-1 and MIA PaCa-2, there were a lower percentage of G0/G1 phase fraction of cells and a higher percentage of S phase fraction of cells after PLAAT4-overexpression (Figure 7M, N, Supplymental Figure 3G–J). These results indicated the significant effect of PLAAT4 on the cell cycle.
3.12. Effect of PLAAT4 in the Migration Ability of Pancreatic Cancer Cells
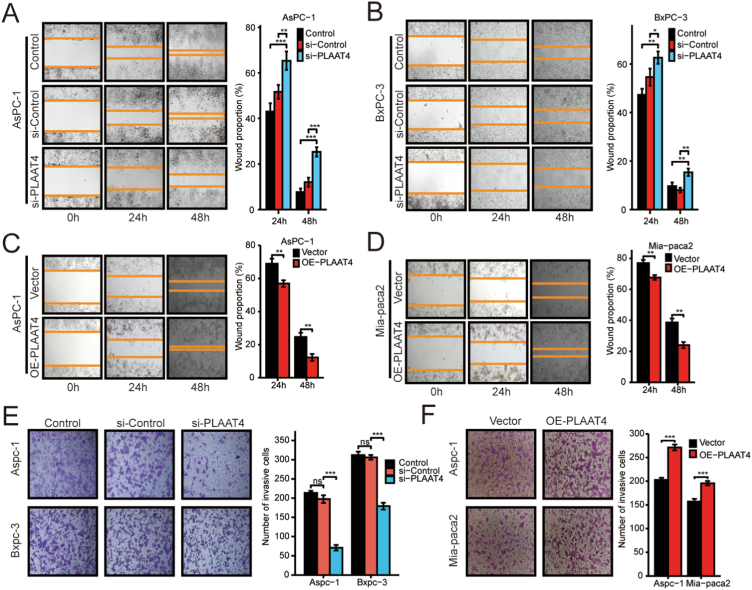
Further, we conducted the wound healing assay, which indicated that the si-PLAAT4 group had lower migration ability in AsPC-1 and BxPC-3 (Figure 8A, B). Meanwhile, we performed the transwell experiment, which suggested lower migration ability after PLAAT4 knockdown in AsPC-1 and BxPC-3 (Figure 8E). Similarly, wound healing assay and transwell experiment were performed with PLAAT4-overexpression cells in AsPC-1 and MIA PaCa-2, which indicated higher migration ability after PLAAT4-overexpression (Figure 8C-D, F). These results indicated that PLAAT4 mediated the proliferation and migration phenotype of pancreatic cancer cells. The potential molecular mechanism needs further studies.
Figure 8.
PLAAT4 Knockdown attenuated the Migration Ability of Pancreatic Cancer Cells. (A) and (B) Representational images of the wound healing assay after si-PLAAT4 in AsPC-1 and BxPC-3 cell lines and the digitization of the results. (C) and (D) Representational images of the wound healing assay after PLAAT4-overexpression in AsPC-1 and MIA PaCa-2 cell lines. (E) Representational images of the transwell assay after si-PLAAT4 in AsPC-1 and BxPC-3 cell lines. (F) Digitization of the transwell assay after PLAAT4-overexpression in AsPC-1 and MIA PaCa-2 cell lines. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
4. Discussion
The PLAAT family possess O- and N-acyltransferase activity, serve as phospholipid metabolizing enzymes and are widely existed in eukaryotes, bacteria and viruses. There are five PLAAT members in Homo sapiens, named PLAAT1-5 separately [9]. PLAAT2-5 depletion in human were found related to the occurrence of Poland Syndrome [21]. PLAAT members were seen as tumor suppressors, reducing H-RAS signaling and expressed little in a variety of cancer cells [13]. But the exact function in pancreatic cancer was not clear.
PLAAT4 exerts low N-acyltransferase activity in contrast to its PLA activity [22]. Previously, PLAAT4 has been confirmed to suppress cancer progression in multiple cancer types. PLAAT4 suppressed epithelial-mesenchymal transition and metastasis in colorectal cancer [23], and attenuated tumor adhesion and differentiation, suppressing lung metastasis in breast cancer [24]. PLAAT4 also showed anti-tumor effect in cervical cancer [25], skin cancer [26], testis cancer [27], and liver cancer [28, 29], etc. However, PLAAT4 was highly expressed in glioblastoma and predicted poor survival in patients with primary glioblastoma [30]. Moreover, a previous study has shown that PLAAT4 was correlated with poor prognosis in patients with pancreatic cancer, as a putative downstream of the oncogene EPS8 [31]. PLAAT4 was also implicated as a vital regulator of the oncogenetic RAS signaling binding with the hyper-variable regions of RAS proteins [32]. The exact function of PLAAT4 in pancreatic cancer is still unclear.
In our study, we found that PLAAT4 was differently expressed in tumor tissues in contrast to parallel normal tissues and that PLAAT4 was significantly elevated in a variety of cancer types, including PAAD. And in PAAD, we found the prognostic value of PLAAT4 was rather high with an estimated AUC of 0.983 in TCGA. The diagnostic value of PLAAT4 needs further study.
In PLAAT4-high samples in PAAD, we conducted GO analysis and GSEA analysis to fully understand the function of PLAAT4. We found that PLAAT4 was connected with the production of molecular mediator of immune response, negative regulation of cell-cell adhesion, humoral immune response mediated by circulating immunoglobulin, cell adhesion molecules and cell-cell adhesion via plasma membrane adhesion molecules, triglyceride catabolic process and cell cycle. Studies have proved that cancer-derived immunoglobulin exerts a prompting effect on cancer cells, mediating various pathways that contributed to cell malignancy [33]. But the specific role of PLAAT4 to affect the immune response and immunoglobulin production is unclear. Multiple studies have proved that cell-cell adhesion contributed to tumor invasion, migration and metastasis, and simultaneously connected with tumor metabolism and immune suppression [34, 35, 36], suggesting that PLAAT4 has a variety of connections with tumor malignant phenotype. As a member of the phospholipase A/acyltransferase (PLAAT) family and typical phospholipid-metabolizing enzymes, it is easy to comprehend the function of the PLAAT4 mediating triglyceride catabolic process.
We also found that PLAAT4 related to cell cycle checkpoints, M phase, regulation by p53 and cell cycle mitotic using GSEA analysis. These results uncovered the essential role of PLAAT4 in mediating mitosis of tumor cells and that PLAAT4 may sustain tumor growth by regulating cell cycle. Additionally, the PLAAT4 was possibly regulated by p53. As a recent study declared, targeting cell cycle checkpoints could be a promising strategy to enhance chemosensitivity [37].
Tumor-infiltrating immune cells (TIICs) were seen as prognostic markers in a variety of cancers [38, 39], mediating cancer progression and development [40]. According to our study, the PLAAT4 expression level was positively correlated with the abundance of several TIICs, such as aDC, Th1 and Th2 cells, and negatively correlated with the abundance of Th17 cells, which was in accordance with the GO and GSEA analysis. DCs served as antigen-presenting cells, regulating innate immunity and adaptive immunity [41]. The activated form of DCs, the aDCs, loaded antigens initiate antigen-specific immunity and thus promoted the proliferation and differentiation of T cells into helper cells and effector cells [42]. A recent study has shown that neoantigen expression could activate specific immunity, cause an aggravated microenvironment and increase tumor progression in pancreatic cancer. While restoring conventional DCs rather than activating DCs could increase CD8+ T cells viability and attenuate tumor progression in pancreatic cancer [43]. Combined with our results, it can be inferred that the abundance of aDC in PLAAT4-high samples might activate antigen-specific immunity, but mediate malignant phenotypes and lead to tumor progression in PAAD. Th1, Th2 cells and Th17 cells are all differentiated from the primitive CD4 T cells, stimulated by T-cell receptor (TCR) signaling [44]. A previous study has shown that the ratio of Th2 to Th1 cells intrastromal is an independent predictor of reduced postoperative survival in patients with PDAC, implying Th2 cells as a tumor-promoting factor [45]. In patients with colorectal cancer, high infiltration of Th1 was related to a prolonged survival, while Th2 expression had no relation with survival and high infiltration of Th17 predicted poor prognosis [46]. In pancreatic cancer, Th17 cells and their associated cytokines were found significantly higher in patients with high tumor stages and served as prognostic biomarkers of pancreatic cancer patients. Our results only pointed out the relevance of PLAAT4 expression with aDC, Th1, Th2 and Th 17 cells, etc. but not reflected the function of them in pancreatic cancer. Further studies are needed to reveal the inner mechanism.
In this study, high PLAAT4 expression in PAAD patients was found connected with several progressive clinicopathological characteristics, such as higher clinical T stage, Pathologic stage II & III & IV, R1 & R2 residual tumor, G3/G4 histologic grade, pancreatic head carcinoma, pancreatitis history. Moreover, high PLAAT4 expression was connected with poor prognosis and served as an independent biomarker of OS and DSS in PAAD. As our nomogram demonstrated, the expression level of PLAAT4 with other characteristics, exemplified as tumor T stage, radiation therapy, primary therapy outcome, etc. can be quantified to predict OS and DSS. The C-index of this prediction is 0.669 and presented high prediction efficiency.
We further tested the proliferation and migration capabilities of PLAAT4 in pancreatic cancer cells. The malignant phenotype of pancreatic cancer cells was restrained after PLAAT4 knockdown and promoted after PLAAT4-overexpression. Targeting PLAAT4 could be a promising strategy for pancreatic cancer therapy. However, the underlying molecular mechanism is still unclear and needs further investigation.
There are still some limitations in this study. Firstly, the clinical characteristics to predict the prognosis of PAAD patients are not limited to the index presented in our study. Some clinical factors were missing because of the heterogeneity in different treatment centers and were unable to access. Secondly, we preferred to test the prognosis value of PLAAT4 in a large cohort with more samples. Unfortunately, the verification could not be realized because of the limited condition and we only performed the analysis in the public dataset. And the last, we didn’t verify the putative biological function or molecular mechanism of PLAAT4 in pancreatic cancer. We would carry out more studies on PLAAT4 in deep soon afterward.
5. Conclusion
In this study, we found PLAAT4 as a prognostic biomarker in PAAD patients, which is highly expressed in pancreatic cancer, and has connections with specific molecular functions, such as negative regulation of cell-cell adhesion, cell adhesion molecules and cell cycle, etc. This research revealed the connection between PLAAT4 and clinicopathological significance in PAAD patients. PLAAT4 is expected to be a therapeutic target for pancreatic cancer, therefore, further research is urgently needed to elucidate its intrinsic molecular mechanism.
Declarations
Author contribution statement
Wang Yuan-Yang: Performed the experiments; Wrote the paper.
Zhao Bang-Bo: Analyzed and interpreted the data.
Qin Cheng; Li Ze-Ru: Contributed reagents, materials, analysis tools or data.
Wang Wei-Bin; Li Tian-Hao: Conceived and designed the experiments.
Funding statement
Dr. Weibin Wang was supported by the Natural Science Foundation of China [No. 81773215].
Dr. Weibin Wang was supported by the CAMS Innovation Fund for Medical Sciences [No. 2021-I2M-1-002].
Data availability statement
Data included in article/supp. material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
TCGA PAAD patient characteristics.
PLAAT4-related differential expressed Genes.
GO enrichment analysis of PLAAT4.
Protein to protein interactions of PLAAT4.
PLAAT4-Gene sets enriched in phenotype with high adjusted P-value.
Univariate and multivariate survival results (DSS) of prognostic covariates in PAAD patients.
Univariate and multivariate survival results (PFI) of prognostic covariates in PAAD patients.
The prognostic value of PLAAT4 (DSS) in various PAAD subgroups.
Supplymental Figure 1-PLAAT4 PPI.
The interactions network of protein to protein built based on interactions pairs of protein to protein by the STRING dataset.
Supplymental Figure 2-PLAAT4 estimate score.
The Estimate related score built based on TCGA database.
Supplymental Figure 3-PLAAT4 cell cycle.
The original figure of cell cycle experiment.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA A Cancer J. Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F., et al. Cancer statistics in China, 2015. CA A Cancer J. Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Rahib L., Smith B.D., Aizenberg R., Rosenzweig A.B., Fleshman J.M., Matrisian L.M. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 4.Zhang T., Cao Z., Chen R., Du Y., Hong D., Jiang K., et al. A Chinese consensus statement on the diagnosis and treatment of pancreatic exocrine insufficiency after pancreatic surgery (2018) Journal of Pancreatology. 2018;1:30–34. [Google Scholar]
- 5.Mizrahi J.D., Surana R., Valle J.W., Shroff R.T. Pancreatic cancer. Lancet. 2020;395:2008–2020. doi: 10.1016/S0140-6736(20)30974-0. [DOI] [PubMed] [Google Scholar]
- 6.Zhang T., Wu W., Yang Y., Zhao Y. The Chinese guidelines for neoadjuvant therapy of pancreatic cancer (2020) Journal of Pancreatology. 2021;4:135–145. [Google Scholar]
- 7.Ballehaninna U.K., Chamberlain R.S. The clinical utility of serum CA 19-9 in the diagnosis, prognosis and management of pancreatic adenocarcinoma: an evidence based appraisal. J. Gastrointest. Oncol. 2012;3:105–119. doi: 10.3978/j.issn.2078-6891.2011.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uyama T., Tsuboi K., Ueda N. An involvement of phospholipase A/acyltransferase family proteins in peroxisome regulation and plasmalogen metabolism. FEBS Lett. 2017;591:2745–2760. doi: 10.1002/1873-3468.12787. [DOI] [PubMed] [Google Scholar]
- 9.Mardian E.B., Bradley R.M., Duncan R.E. The HRASLS (PLA/AT) subfamily of enzymes. J. Biomed. Sci. 2015;22:99. doi: 10.1186/s12929-015-0210-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hajnal A., Klemenz R., Schäfer R. Subtraction cloning of H-rev107, a gene specifically expressed in H-ras resistant fibroblasts. Oncogene. 1994;9:479–490. [PubMed] [Google Scholar]
- 11.Shinohara N., Uyama T., Jin X.H., Tsuboi K., Tonai T., Houchi H., et al. Enzymological analysis of the tumor suppressor A-C1 reveals a novel group of phospholipid-metabolizing enzymes. J. Lipid Res. 2011;52:1927–1935. doi: 10.1194/jlr.M015081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ueda N., Tsuboi K., Uyama T. Metabolism of endocannabinoids and related N-acylethanolamines: canonical and alternative pathways. Febs j. 2013;280:1874–1894. doi: 10.1111/febs.12152. [DOI] [PubMed] [Google Scholar]
- 13.Sers C., Emmenegger U., Husmann K., Bucher K., Andres A.C., Schäfer R. Growth-inhibitory activity and downregulation of the class II tumor-suppressor gene H-rev107 in tumor cell lines and experimental tumors. J. Cell Biol. 1997;136:935–944. doi: 10.1083/jcb.136.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang S.-L., Shyu R.-Y., Yeh M.-Y., Jiang S.-Y. Cloning and characterization of a novel retinoid-inducible gene 1(RIG1) deriving from human gastric cancer cells. Mol. Cell. Endocrinol. 2000;159:15–24. doi: 10.1016/s0303-7207(99)00207-5. [DOI] [PubMed] [Google Scholar]
- 15.DiSepio D., Ghosn C., Eckert R.L., Deucher A., Robinson N., Duvic M., et al. Identification and characterization of a retinoid-induced class II tumor suppressor/growth regulatory gene. Proc. Natl. Acad. Sci. U. S. A. 1998;95:14811–14815. doi: 10.1073/pnas.95.25.14811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z., Jensen M.A., Zenklusen J.C. A practical guide to the cancer genome atlas (TCGA) Methods Mol. Biol. 2016;1418:111–141. doi: 10.1007/978-1-4939-3578-9_6. [DOI] [PubMed] [Google Scholar]
- 17.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47 doi: 10.1093/nar/gky1131. D607-d13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bindea G., Mlecnik B., Tosolini M., Kirilovsky A., Waldner M., Obenauf A.C., et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Wang W., Xu L., Yang Y., Dong L., Zhao B., Lu J., et al. A novel prognostic marker and immunogenic membrane antigen: prohibitin (PHB) in pancreatic cancer. Clin. Transl. Gastroenterol. 2018;9:178. doi: 10.1038/s41424-018-0044-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaccari C.M., Romanini M.V., Musante I., Tassano E., Gimelli S., Divizia M.T., et al. De novo deletion of chromosome 11q12.3 in monozygotic twins affected by Poland Syndrome. BMC Med. Genet. 2014;15:63. doi: 10.1186/1471-2350-15-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hussain Z., Uyama T., Tsuboi K., Ueda N. Mammalian enzymes responsible for the biosynthesis of N-acylethanolamines. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2017;1862:1546–1561. doi: 10.1016/j.bbalip.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 23.Wang Z., Wang L., Hu J., Fan R., Zhou J., Wang L., et al. RARRES3 suppressed metastasis through suppression of MTDH to regulate epithelial-mesenchymal transition in colorectal cancer. Am J Cancer Res. 2015;5:1988–1999. [PMC free article] [PubMed] [Google Scholar]
- 24.Morales M., Arenas E.J., Urosevic J., Guiu M., Fernández E., Planet E., et al. RARRES3 suppresses breast cancer lung metastasis by regulating adhesion and differentiation. EMBO Mol. Med. 2014;6:865–881. doi: 10.15252/emmm.201303675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai F.M., Shyu R.Y., Jiang S.Y. RIG1 suppresses Ras activation and induces cellular apoptosis at the Golgi apparatus. Cell. Signal. 2007;19:989–999. doi: 10.1016/j.cellsig.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 26.Duvic M., Helekar B., Schulz C., Cho M., DiSepio D., Hager C., et al. Expression of a retinoid-inducible tumor suppressor, Tazarotene-inducible gene-3, is decreased in psoriasis and skin cancer. Clin. Cancer Res. 2000;6:3249–3259. [PubMed] [Google Scholar]
- 27.Wu C.C., Shyu R.Y., Wang C.H., Tsai T.C., Wang L.K., Chen M.L., et al. Involvement of the prostaglandin D2 signal pathway in retinoid-inducible gene 1 (RIG1)-mediated suppression of cell invasion in testis cancer cells. Biochim. Biophys. Acta. 2012;1823:2227–2236. doi: 10.1016/j.bbamcr.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 28.Wei L., Chiu D.K., Tsang F.H., Law C.T., Cheng C.L., Au S.L., et al. Histone methyltransferase G9a promotes liver cancer development by epigenetic silencing of tumor suppressor gene RARRES3. J. Hepatol. 2017;67:758–769. doi: 10.1016/j.jhep.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 29.Xu Y., Chen T., Liao D., Wu X., Zhong Y., Liu S., et al. The antitumor effect of TIG3 in liver cancer cells is involved in ERK1/2 inhibition. Tumour Biol. 2016;37:11311–11320. doi: 10.1007/s13277-016-4998-x. [DOI] [PubMed] [Google Scholar]
- 30.Wang H., Xu H., Xu T., Tan C., Jiang M., Chen Y., et al. High expression of TIG3 predicts poor survival in patients with primary glioblastoma. Tumour Biol. 2017;39 doi: 10.1177/1010428317712135. [DOI] [PubMed] [Google Scholar]
- 31.Fukuhisa H., Seki N., Idichi T., Kurahara H., Yamada Y., Toda H., et al. Gene regulation by antitumor miR-130b-5p in pancreatic ductal adenocarcinoma: the clinical significance of oncogenic EPS8. J. Hum. Genet. 2019;64:521–534. doi: 10.1038/s10038-019-0584-6. [DOI] [PubMed] [Google Scholar]
- 32.Engin H.B., Carlin D., Pratt D., Carter H. Modeling of RAS complexes supports roles in cancer for less studied partners. BMC Biophys. 2017;10:5. doi: 10.1186/s13628-017-0037-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu D., Zheng H., Liu H., Li M., Ren W., Liao W., et al. Immunoglobulin expression and its biological significance in cancer cells. Cell. Mol. Immunol. 2008;5:319–324. doi: 10.1038/cmi.2008.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janiszewska M., Primi M.C., Izard T. Cell adhesion in cancer: beyond the migration of single cells. J. Biol. Chem. 2020;295:2495–2505. doi: 10.1074/jbc.REV119.007759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sousa B., Pereira J., Paredes J. The crosstalk between cell adhesion and cancer metabolism. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20081933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Läubli H., Borsig L. Altered cell adhesion and glycosylation promote cancer immune suppression and metastasis. Front. Immunol. 2019;10:2120. doi: 10.3389/fimmu.2019.02120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kong Y.W., Dreaden E.C., Morandell S., Zhou W., Dhara S.S., Sriram G., et al. Enhancing chemotherapy response through augmented synthetic lethality by co-targeting nucleotide excision repair and cell-cycle checkpoints. Nat. Commun. 2020;11:4124. doi: 10.1038/s41467-020-17958-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li L., Ouyang Y., Wang W., Hou D., Zhu Y. The landscape and prognostic value of tumor-infiltrating immune cells in gastric cancer. PeerJ. 2019;7 doi: 10.7717/peerj.7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ye L., Zhang T., Kang Z., Guo G., Sun Y., Lin K., et al. Tumor-infiltrating immune cells act as a marker for prognosis in colorectal cancer. Front. Immunol. 2019;10:2368. doi: 10.3389/fimmu.2019.02368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Domingues P., González-Tablas M., Otero Á., Pascual D., Miranda D., Ruiz L., et al. Tumor infiltrating immune cells in gliomas and meningiomas. Brain Behav. Immun. 2016;53:1–15. doi: 10.1016/j.bbi.2015.07.019. [DOI] [PubMed] [Google Scholar]
- 41.Wculek S.K., Cueto F.J., Mujal A.M., Melero I., Krummel M.F., Sancho D. Dendritic cells in cancer immunology and immunotherapy. Nat. Rev. Immunol. 2020;20:7–24. doi: 10.1038/s41577-019-0210-z. [DOI] [PubMed] [Google Scholar]
- 42.Martinek J., Wu T.C., Cadena D., Banchereau J., Palucka K. Interplay between dendritic cells and cancer cells. Int Rev Cell Mol Biol. 2019;348:179–215. doi: 10.1016/bs.ircmb.2019.07.008. [DOI] [PubMed] [Google Scholar]
- 43.Hegde S., Krisnawan V.E., Herzog B.H., Zuo C., Breden M.A., Knolhoff B.L., et al. Dendritic cell paucity leads to dysfunctional immune surveillance in pancreatic cancer. Cancer Cell. 2020;37:289–307. doi: 10.1016/j.ccell.2020.02.008. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu J.T. Helper cell differentiation, heterogeneity, and plasticity. Cold Spring Harbor Perspect. Biol. 2018;10:a030338. doi: 10.1101/cshperspect.a030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Monte L., Reni M., Tassi E., Clavenna D., Papa I., Recalde H., et al. Intratumor T helper type 2 cell infiltrate correlates with cancer-associated fibroblast thymic stromal lymphopoietin production and reduced survival in pancreatic cancer. J. Exp. Med. 2011;208:469–478. doi: 10.1084/jem.20101876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tosolini M., Kirilovsky A., Mlecnik B., Fredriksen T., Mauger S., Bindea G., et al. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res. 2011;71:1263–1271. doi: 10.1158/0008-5472.CAN-10-2907. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TCGA PAAD patient characteristics.
PLAAT4-related differential expressed Genes.
GO enrichment analysis of PLAAT4.
Protein to protein interactions of PLAAT4.
PLAAT4-Gene sets enriched in phenotype with high adjusted P-value.
Univariate and multivariate survival results (DSS) of prognostic covariates in PAAD patients.
Univariate and multivariate survival results (PFI) of prognostic covariates in PAAD patients.
The prognostic value of PLAAT4 (DSS) in various PAAD subgroups.
Data Availability Statement
Data included in article/supp. material/referenced in article.