Abstract
Longitudinal changes in gene expression during islet autoimmunity (IA) may provide insight into biological processes that explain progression to type 1 diabetes (T1D). We identified individuals from Diabetes Autoimmunity Study in the Young (DAISY) who developed IA, autoantibodies present on two or more visits. Illumina’s NovaSeq 6000 was used to quantify gene expression in whole blood. With linear mixed models we tested for changes in expression after IA that differed across individuals who progressed to T1D (progressors) (n = 25), reverted to an autoantibody-negative stage (reverters) (n = 47), or maintained IA positivity but did not develop T1D (maintainers) (n = 66). Weighted gene coexpression network analysis was used to identify coexpression modules. Gene Ontology pathway analysis of the top 150 differentially expressed genes (nominal P < 0.01) identified significantly enriched pathways including leukocyte activation involved in immune response, innate immune response, and regulation of immune response. We identified a module of 14 coexpressed genes with roles in the innate immunity. The hub gene, LTF, is known to have immunomodulatory properties. Another gene within the module, CAMP, is potentially relevant based on its role in promoting β-cell survival in a murine model. Overall, results provide evidence of alterations in expression of innate immune genes prior to onset of T1D.
Introduction
Type 1 diabetes (T1D) is an autoimmune disease that requires lifelong insulin treatment as a result of immune-mediated destruction of pancreatic β-cells (1). Development of preventative interventions has been hampered by an incomplete understanding of T1D etiology, which is believed to involve a complex interaction of genetic susceptibility, immune dysfunction, and environmental perturbation (2). There is a strong need for better understanding of the heterogenous preclinical, islet autoimmunity (IA) phase of the disease process. Improved knowledge of the mechanisms that explain progression or resolution of IA has important implications for the development of preventative interventions.
Gene expression has been used to study T1D. Only a handful of studies have included examination of gene expression prior to T1D (3–5). Microarray analyses of blood samples identified significant associations between T1D and genes related to type 1 interferon signaling (3,5,6). Among individuals with IA, the expression of genes related to lymphocyte activation and function were associated with the hazard of T1D (4). However, most studies of pre-T1D expression have been focused on average differences prior to T1D rather than testing for differences in the change in gene expression during the IA phase. In contrast, Xhonneux et al. (7) performed a transcriptional network analysis based on longitudinal measures of gene expression in whole blood samples from The Environmental Determinants of Diabetes in the Young (TEDDY). This novel work resulted in identification of an insulin autoantibody (IAA)-specific coexpression signature that differentiated between case and control subjects, was enriched for NK cell–specific transcripts, and was potentially responsive to pharmaceutical interventions targeting the G-protein–coupled receptor (7).
The IA phase of the T1D disease process is challenging to study because it is asymptomatic (8). The presence of autoantibodies confers an increase in risk that is commensurate with antibody level, type, and number of distinct autoantibodies that are present (9). IA progression is variable in terms of timing of progression to clinical T1D or reversion of autoantibodies (10). The study of biomarkers within disease subgroups defined based on changes in the presence or absence of these autoantibodies may provide more specific insight into the IA disease process.
The purpose of this study was to build on prior work by studying longitudinal changes in gene expression within individuals who developed IA. We tested for longitudinal changes in gene expression during IA among individuals grouped together based on autoantibody progression patterns: individuals who developed T1D (progressors), continued to produce autoantibodies but did not develop T1D (maintainers), or reverted to an autoantibody-negative state (reverters) during Diabetes Autoimmunity Study in the Young (DAISY). Our study is unique in that we included the reversion phenotype.
Research Design and Methods
Study Population
We identified individuals who developed IA between February 1994 and February 2019 in DAISY and underwent autoantibody testing at two or more visits (n = 214) (Supplementary Material). IA was defined as the presence of at least one autoantibody (IAA, GAA or glutamic acid decarboxylase, IA-2, or ZnT8) above the 99th percentile (based on healthy control subjects) on two or more consecutive visits. The DAISY testing protocol includes rigorous duplicate testing and confirmation of all positive results as well as a subset of negative results. The study design for DAISY has previously been described (11,12). The Colorado Multiple Institutional Review Board approved all DAISY protocols (COMIRB 92-080). Informed consent or assent was obtained from the parents/legal guardians of all children.
We divided individuals into three IA progression phenotypes. The reverter group was defined as individuals who reverted for all autoantibodies (two or more consecutive visits in which no autoantibodies were detected), did not develop T1D, and were autoantibody negative for all autoantibodies at their last DAISY visit (n = 47). The maintainer group was defined as individuals who continued to test positive for autoantibodies and did not develop T1D during follow-up (n = 66). The progressor group was defined as individuals who developed T1D (n = 25).
RNA Collection
In DAISY, venous blood samples are collected at 9, 15, and 24 months and yearly thereafter (12). Beginning in November of 2007, RNA samples were collected from all DAISY participants with Tempus tubes (Applied Biosystems). We selected samples from two visits (termed IA-1 and IA-2) that occurred after the onset of IA for mRNA sequencing. The IA-1 visit was selected as the first available visit after the onset of IA for all groups. In the progressor group, the visit that preceded clinical onset of T1D was selected as the IA-2 visit. In the maintainer and reverter groups, the IA-2 visit was selected as the visit where age was closest to median age in the progressor group at the IA-2 visit.
The study of Jin et al. (4) and the current study include blood samples from DAISY. The two studies do not include any overlapping samples (same study visit). Approximately 56% (80 of 138) of subjects in the current study are also included in the study of Jin et al. However, the samples in the current study were obtained later during IA (range of difference in time between samples used in the two studies: 3 months–7.6 years).
RNA Processing
Total RNA was isolated and purified from frozen (−80°C), peripheral whole blood samples of n = 138 subjects meeting the inclusion criteria (Supplementary Material). We also included 12 control samples and 12 technical replicates. NuGEN library preparation kits were used to construct the strand-specific total RNA libraries. The Globin AnyDeplete kit was used to remove globin transcripts. RNA integrity number (RIN) scores for all samples were quantitated with use of the TapeStation (Agilent). Paired-end 150-cycle 2 × 150 sequencing was performed with the Illumina NovaSeq 6000 system. Samples were pooled together in a single flow cell and were sequenced twice to obtain the desired read depth, resulting in a median read depth of 90 million paired end reads across the samples (range 54–128 million).
FASTQ files for the two runs for each sample were concatenated. Reads were trimmed to remove adapters and low-quality base calls with Cutadapt (13). Reads were aligned to the reference human genome (hg19) with use of TopHat2 (14). Individual samples were quantified against the most up-to-date Ensembl reference transcriptome with the RSEM (RNA-Seq by Expectation-Maximization) algorithm (15). The expected counts were used for further processing and normalization. For removal of any technical variation, the data first were quantile normalized with DESeq2 (16) and then normalized with removal of unwanted variation (17). Based on scree plots, we adjusted for one factor, as it explained the most variability in the data. The data were then transformed with the regularized log function (16). The transformed data were used in all subsequent analyses. We filtered out genes if ≥20% of the samples did not have a read mapped to the gene (transcripts per million value of 0) or the average transcripts per million value across all samples was <1. A total of 12,477 genes passed the filtering criteria.
Statistical Methods
Linear mixed models were used to test for differences in the change in gene expression levels between the IA-1 and IA-2 visits across the three groups (group * visit interaction). A random effect was used to account for within-subject correlation. Separate models were also used to test for differences in average gene expression across groups (group effect model). The group effect models also included the first two principal components representative of genetic ancestry in the study population. Two individuals were excluded from this analysis because of missing ancestry data. In a separate study, ancestry principal components were estimated at the University of Virginia School of Medicine Center for Public Health Genomics based on exome sequencing (Illumina HumanCoreExome-24 BeadChip) (N = 283) or whole genome sequencing (N = 162) from the larger DAISY population. The interaction models are expected to be robust to confounding from time-invariant variables such as population ancestry, and thus, to maximize power, we did not adjust for ancestry in the interaction models. The Benjamini-Hochberg false discovery rate (FDR) was used to correct for multiple comparisons (18). Individual genes differentially expressed at an FDR-adjusted P < 0.10 were considered statistically significant.
Gene Ontology (GO) term and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment analysis was performed with the top differentially expressed genes (nominal P < 0.01). The biomaRt (19) R package (v2.44.4) was used to convert the Ensembl gene identifiers into Entrez Gene identifiers. GO and KEGG enrichment analyses were performed with the goseq (20) (v1.4.0) and clusterProfiler (21) (v3.16.1) R packages, respectively. FDR-adjusted P values were calculated for all terms. We manually reviewed FDR significant terms to remove parent terms. REVIGO (22) was used to remove redundant terms.
Weighted Gene Co-Expression Network Analysis (WGCNA) (23) (v1.70.3) was used to identify modules of ≥10 genes. As the use of longitudinal gene expression changes was a nonstandard application of WGCNA, linear mixed models were used to regress out age and sex from the rlog gene expression values. The within-individual differences in the residual gene expression values at each visit (IA-2 − IA-1) were used to represent changes in gene expression between the visits (WGCNA difference model). The residuals from IA-2 and IA-1 were also averaged for each individual to represent overall gene expression (WGCNA average model). All genes that passed the filtering criteria were used in the WGCNA models. Only modules where the eigengene explained >60% of variance across all genes in the module were considered as possible candidates. The eigengene represents the first principal component from a principal components analysis of change in gene expression for all genes in the module. Among candidate modules, linear models were used to test for differences in the eigengenes across the groups. The Benjamini-Liu (24) step-down procedure was used to adjust for multiple testing. Among differentially coexpressed modules (FDR-adjusted P < 0.10), bootstrapping was used to evaluate the stability of modules. Modules were considered highly preserved if the z summary score statistic was >9 (25).
Methylation Measurements
To identify genomic features that regulate the coexpression module and are potentially responsive to environmental perturbation, we tested the association between the LTF module eigengene and DNA methylation (DNAm). We identified a subset of individuals (n = 55) with overlapping DNAm and gene expression data (see Appendix 2 overlapping samples vs. complete population). We tested the association between methylation M values and the WGCNA eigengenes (198,025 overlapping probes from the Infinium HumanMethylation450K Beadchip [450K] [Illumina, San Diego, CA] and the Infinium HumanMethylation EPIC Beadchip [EPIC] platforms). DNAm data are described by Vanderlinden et al. (26). Multiple variable linear regression models, with adjustment for cell proportions (estimated with the minfi [v1.12.0] package [27]), age, sex, and platform (450K vs. EPIC), were used to test the association between the eigengene and DNAm. The Benjamini-Hochberg FDR was used to correct for multiple comparisons (18). DNAm sites with an FDR-adjusted P < 0.10 were considered statistically significant.
Data and Resource Availability
The data sets generated during or analyzed during the current study are accessible through Gene Expression Omnibus (GEO) series accession no. GSE142512. The RNA-sequencing data will be added to the GEO series after manuscript publication.
Results
Study Population
There was no difference in sex, family history, prevalence of high risk DR3/4 genotype, ethnicity, age at the IA-1 study visit, or age at the IA-2 study visit across groups (Table 1). The mean ± SD duration of time between the two expression study visits was similar in the progressor (3.0 ± 2.0 years), maintainer (2.8 ± 2.7 years), and reverter (3.0 ± 2.7 years) groups (P = 0.8959). As expected, age at onset of IA and prevalence of multiple autoantibodies at onset of IA significantly differed in the progressor group relative to the other groups (Table 1). HbA1c levels were available in a subset of individuals (Appendix 3). Consistent with the findings of Stene et al. (28), HbA1c levels increased to a greater extent between IA-1 and IA-2 visits in the progressor group relative to the reverters and maintainers.
Table 1.
Demographics and family history across the IA progression phenotypes
| Maintainer (n = 66) | Progressor (n = 25) | Reverter (n = 47) | P | |
|---|---|---|---|---|
| Multiple autoantibodies at onset of IA* | 11 (16.7) | 11 (44.0) | 1 (2.1) | <0.0001 |
| Non-Hispanic White ethnicity | 47 (71.2) | 23 (92.0) | 33 (70.2) | 0.0873 |
| Female sex | 35 (53.0) | 11 (44.0) | 21 (44.7) | 0.6007 |
| DR3/4 genotype | 17 (25.8) | 11 (44.0) | 12 (25.5) | 0.1878 |
| Family history/affected first-degree relative | 42 (63.6) | 16 (64.0) | 23 (48.9) | 0.2465 |
| Age at onset of IA (years)* | 8.6 ± 4.5 | 5.4 ± 4.1 | 6.9± 4.4 | 0.0068 |
| Age at IA-1 study visit (years) | 10.9 ± 4.5 | 9.5 ± 3.5 | 10.6 ± 4.3 | 0.3626 |
| Age at IA-2 study visit (years) | 13.8 ± 2.6 | 12.5 ± 3.8 | 13.6 ± 2.2 | 0.1572 |
Data are n (%) or mean ± SD.
Onset of IA defined as the first study visit among the first two consecutive study visits where the individual tested positive for one or more autoantibodies. χ2 or Fisher exact test was used to test for differences in categorical variables across groups, and ANOVA was used to test for differences in continuous variables across groups.
Gene Expression During IA
We did not identify any genes for which the average of the expression levels at the IA-1 and IA-2 visits differed across groups (group effect model) at FDR-adjusted P < 0.10. The 57 differentially expressed genes (nominal P < 0.01) were included in the enrichment analysis (Supplementary Material). We did not identify any significantly enriched GO or KEGG terms based on this differentially expressed gene list.
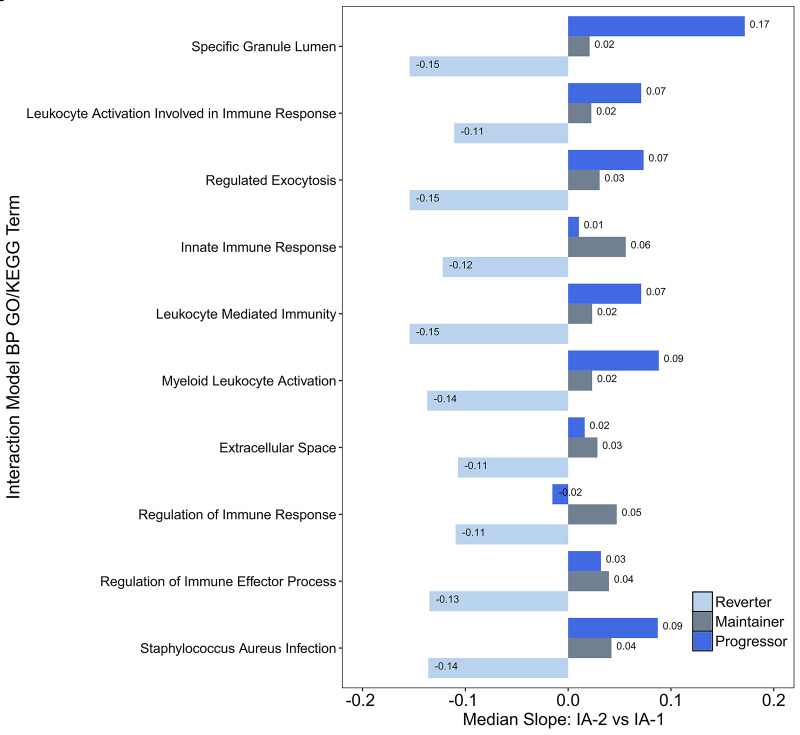
We did not identify any individual genes for which the change in gene expression between the IA-1 and IA-2 visits differed across groups (interaction model) at FDR-adjusted P < 0.10. The 150 differentially changing genes (nominal P < 0.01) were included in the enrichment analysis (Supplementary Material). We identified nine significantly enriched GO terms (Table 2). We also identified a single KEGG term (Table 3). The median group-specific slopes across all differentially changing genes included in each of the significantly enriched GO or KEGG terms are described in Fig. 1. The reverter group was associated with decreased expression during IA (negative slope) relative to a trend toward increasing expression in the progressor and maintainer groups (positive slope).
Table 2.
Significantly enriched GO terms based on genes for which change in expression between IA-1 and IA-2 visits differed (nominal P < 0.01) across the three groups
| Term | Description | N in category | N differentially expressed | Nominal P | FDR Adj. P | Top differentially expressed gene |
|---|---|---|---|---|---|---|
| Biological Process (BP) | ||||||
| GO:0002366 | Leukocyte activation involved in immune response | 389 | 17 | 7.82E−06 | 1.49E−02 | ARSB (P = 7.91E−5) |
| GO:0045055 | Regulated exocytosis | 403 | 17 | 1.19E−05 | 1.55E−02 | PDGFA (P = 9.34E−5) |
| GO:0045087 | Innate immune response | 429 | 17 | 2.75E−05 | 2.44E−02 | RSAD2 (P = 2.02E−3) |
| GO:0002443 | Leukocyte-mediated immunity | 439 | 17 | 4.19E−05 | 2.87E−02 | ARSB (P = 7.91E−5) |
| GO:0002274 | Myeloid leukocyte activation | 354 | 15 | 4.27E−05 | 2.87E−02 | ARSB (P = 7.91E−5) |
| GO:0050776 | Regulation of immune response | 492 | 17 | 1.51E−04 | 8.93E−02 | MAPK9 (P = 1.06E−6) |
| GO:0002697 | Regulation of immune effector process | 236 | 11 | 1.79E−04 | 9.99E−02 | CD86 (P = 1.62E−3) |
| Cellular Component (CC) | ||||||
| GO:0035580 | Specific granule lumen | 46 | 7 | 2.11E−06 | 6.86E−03 | HP (P = 1.54E−3) |
| GO:0005615 | Extracellular space | 1,359 | 34 | 6.06E−05 | 3.94E−02 | PDGFA (P = 9.34E−5) |
The three groups include progressors, reverters, and maintainers. N in category = number of genes within each GO term that were tested in current study (background-adjusted gene set). N differentially expressed = number of genes within each GO term included in differentially expressed gene set (Supplementary Material). Top differentially expressed gene = within each GO term, representing the differentially expressed gene with the smallest P value. The P value was obtained from linear mixed models testing group * visit hypothesis. Adj., adjusted.
Table 3.
Significantly enriched KEGG term based on genes for which change in expression between IA-1 and IA-2 visits differed (nominal P < 0.01) across the three groups
| Description (KEGG term) | Nominal P | FDR Adj. P | Differentially expressed genes (P)* |
|---|---|---|---|
| S. aureus infection (hsa05150) | 2.40E−04 | 3.80E−02 | C1QB (2.18E−3), C3AR1 (3.33E−3), CAMP (4.84E−3), DEFA4 (5.47E−3) |
The three groups include progressors, reverters, and maintainers. Adj., adjusted.
Nominal P value from linear mixed model testing group * visit hypothesis.
Figure 1.
Median within-group changes (IA-2 vs. IA-1) in gene expression among all differentially expressed genes included in each of the significantly enriched GO or KEGG categories. The y-axis represents each of the significantly enriched GO terms (nine terms ranked based on test statistics) as well as the significantly enriched KEGG term (S. aureus infection). We identified all differentially expressed genes included within each GO or KEGG category (P < 0.01) (Supplementary Material) and calculated the median slope for each group. The slope represents change in expression between the IA-2 and IA-1 visit. Positive slopes indicate an average increase in expression during IA. BP, Biological Process.
Coexpressed Modules
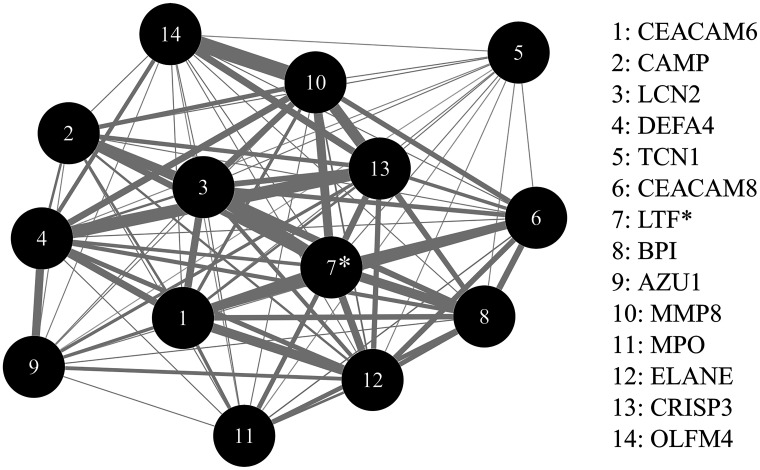
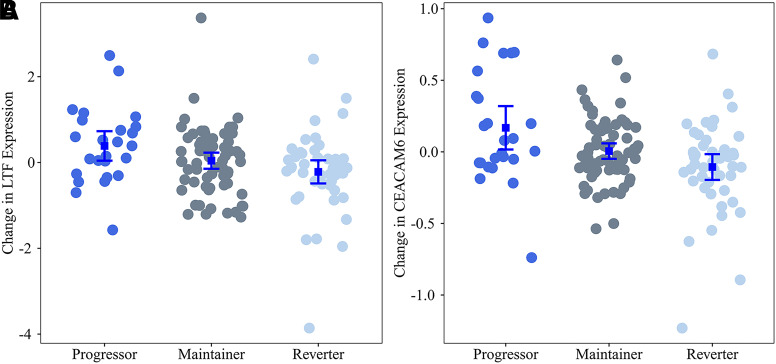
We identified four candidate modules where changes in gene expression during IA (WGCNA difference model) were similar across all genes (Appendix 6). We identified a single module, LTF module, where change in coexpression of 14 genes within the module significantly differed across the groups (FDR-adjusted P value = 0.0223) (Fig. 2 and Appendix 7). The module was named after its hub gene, LTF (Fig. 2). All 14 genes in the LTF module demonstrated a consistent pattern of increasing expression during IA in progressors in contrast to a consistent pattern of decreasing expression in all genes in reverters and decreasing expression in 8 of 14 genes in maintainers (Table 4 and Fig. 3).
Figure 2.
LTF network module. Each node represents a gene within the LTF module. Genes are ranked (1, most significant to 14, least significant) based on P values representing the difference in change in gene expression during IA across the three groups, reverters, progressors, and maintainers. (Group * visit interaction P values for each gene are displayed in Table 4.) The edges are weighted based on the correlation between the pairs of genes. Only correlation coefficients >0.65 are weighted. *Represents the hub gene for the network.
Table 4.
Summary of all genes included in the LTF network module as well as the group-specific changes in expression for each gene between the IA-1 and IA-2 visits
| Ensembl identifier | Gene name | Chr | Interaction P | Progressors | Reverters | Maintainers | |||
|---|---|---|---|---|---|---|---|---|---|
| β | P | β | P | β | P | ||||
| ENSG00000086548† | CEACAM6‡ | 19 | 0.0010 | 0.16 | 0.008 | −0.11 | 0.0163 | <0.01 | 0.9859 |
| ENSG00000164047† | CAMP | 3 | 0.0048 | 0.34 | 0.0028 | −0.1 | 0.2139 | 0.02 | 0.7698 |
| ENSG00000148346† | LCN2 | 9 | 0.0049 | 0.4 | 0.0129 | −0.23 | 0.0543 | 0.02 | 0.8257 |
| ENSG00000164821† | DEFA4 | 8 | 0.0055 | 0.05 | 0.0427 | −0.05 | 0.0111 | −0.02 | 0.2974 |
| ENSG00000134827† | TCN1 | 11 | 0.0056 | 0.28 | 0.0122 | −0.15 | 0.06 | −0.03 | 0.684 |
| ENSG00000124469† | CEACAM8 | 19 | 0.0087 | 0.43 | 0.0197 | −0.24 | 0.0758 | 0.05 | 0.6588 |
| ENSG00000012223 | LTF* ‡ | 3 | 0.0184 | 0.38 | 0.0346 | −0.22 | 0.1029 | 0.04 | 0.7461 |
| ENSG00000101425 | BPI | 20 | 0.0355 | 0.15 | 0.1955 | −0.21 | 0.0181 | −0.07 | 0.3658 |
| ENSG00000172232 | AZU1 | 19 | 0.0480 | 0.06 | 0.0728 | −0.03 | 0.1679 | −0.02 | 0.3197 |
| ENSG00000118113 | MMP8 | 11 | 0.0531 | 0.18 | 0.2574 | −0.28 | 0.0189 | −0.05 | 0.6067 |
| ENSG00000005381 | MPO | 17 | 0.0761 | 0.28 | 0.0435 | −0.06 | 0.5496 | −0.05 | 0.5694 |
| ENSG00000197561 | ELANE | 19 | 0.1068 | 0.1 | 0.1586 | −0.08 | 0.1373 | −0.02 | 0.6358 |
| ENSG00000096006 | CRISP3 | 6 | 0.1125 | 0.03 | 0.2295 | −0.04 | 0.0769 | −0.02 | 0.3907 |
| ENSG00000102837 | OLFM4 | 13 | 0.321 | <0.1 | 0.9876 | −0.03 | 0.0706 | <0.01 | 0.9854 |
Interaction P = nominal P value from linear mixed model in testing null hypothesis that the difference in expression between visits does not differ by group (group * visit). β = group-specific slopes, with adjustment for age and sex, representing change in gene expression between IA-2 and IA-1 visits; positive values indicate higher level of expression at the IA-2 visit relative to the IA-1 visit. P = nominal P value for the group-specific slopes in testing null hypothesis β = 0. Chr, chromosome.
Hub gene (boldface) was identified based on WGCNA analysis.
Genes also included in the Supplementary Material in the list of differentially changing genes (nominal P < 0.01).
Longitudinal changes in expression illustrated in Fig. 3.
Figure 3.
Change in expression of LTF and CEACAM6 by IA progression phenotype. Shown is the change in expression of LTF (A), the hub gene for the LTF module, as well as of CEACAM6 (B), representative of the largest differences between groups. Each dot represents the difference in expression between the IA-2 and IA-1 visits for each individual. The error bars represent mean ± SE change in expression. Positive values represent increase in expression during IA.
We also identified four candidate modules where average gene expression during IA (WGCNA average module) was similar across genes in the module. However, none of eigengenes representing coexpression were significantly different across groups (Appendix 6).
Based on our relatively small sample, we reviewed the stability of the candidate modules. The LTF module was highly preserved across the bootstrap iterations (mean z score 11.70, 95% CI 9.30–13.40), supporting the stability and reliability of this module.
Heterogeneity in the WGCNA model results due to the first appearing autoantibody was explored in Appendix 8.
Eigengene Expression Quantitative Trait Methylation (eQTM) Analysis
To identify genomic features that regulate the coexpression modules and are potentially responsive to environmental perturbation, we tested the association between the LTF module eigengene and DNAm. We reviewed both cis and trans effects, given that the LTF module included multiple genes across multiple chromosomes. We identified three DNAm sites that were significantly associated with the eigengene at FDR-adjusted P < 0.10 (Table 5).
Table 5.
Methylation probes significantly associated with the LTF module eigengene
| Name | Chr | Position | β | Nominal P | FDR Adj. P | Nearest gene | Relation to island | Regulatory annotation |
|---|---|---|---|---|---|---|---|---|
| cg00848392 | 6 | 31734401 | 2.05 | 1.44E−06 | 9.47E−02 | VWA7 | Open sea | Gene body, DNase I hypersensitivity site* |
| cg04171425 | 16 | 4654740 | 1.75 | 9.50E−07 | 9.41E−02 | C16orf96 | Open sea | |
| cg20233727 | 11 | 1316693 | −1.99 | 4.59E−07 | 9.09E−02 | TOLLIP | Island | Gene body |
β = slope from linear model testing association between eigengene and methylation, with adjustment for age, sex, and cell proportions. Gene body = between the ATG and stop codon. Adj., adjusted; Chr, chromosome.
Experimentally determined by the ENCODE project.
Discussion
The IA phase of the T1D disease process varies widely. To identify potential mechanisms that underlie this heterogenous phase, we grouped individuals together based on autoimmunity patterns. We observed gene expression changes during IA that differed between groups defined by progression to T1D (progressors), reversion to an autoantibody-negative state (reverters), or maintenance of IA positivity (maintainers). In enrichment and coexpression analyses we identified relevant biological pathways related to immune system function, response, regulation, and activation. Our results support similar findings in previous studies of gene expression in blood (Supplementary Material) with identification of differentially expressed genes related to pathways including lymphocyte activation and function (4), immune response (6), and signaling in immune system (Reactome 6900) (5).
We also used a network-based analysis to identify coexpression networks (Fig. 2). We identified a single reproducible module where coexpression was significantly different across groups. All 14 genes are included within the innate immune system pathway according to the Reactome database (29). This overlaps with our enrichment analysis of differentially changing genes that identified innate immune response as a significantly enriched GO term (Table 2). Staphylococcus aureus (S. aureus) infection (hsa05150) was also significantly enriched based on the differentially expressed gene set, which included C1QB, C3AR1, CAMP, and DEFA4 (Table 3), all of which are included within Reactome innate immune system pathways.
The overlap between the LTF module genes and innate immunological pathways is consistent with previous work implicating innate system function in T1D etiology (3,5,6). Ferreira et al. (3) observed increased expression of IFN-inducible genes among individuals who did versus did not develop islet autoantibodies. The increased type 1 interferon (IFN) gene signature was observed during and prior to IA but was not consistently observed after T1D onset (3), indicating that a disproportionate activation of the IFN-mediated immune response may play a role in the onset of T1D. Kallionpää et al. (5) also identified differentially expressed genes and pathways related to innate immune function including IFN signaling that were altered before and during IA. Overall, it has been hypothesized that a hyperactivated innate immune inflammatory state in combination with age-dependent failure of immunoregulatory pathways may play a key role in the etiology of T1D (30).
Several individual genes within the LTF module have potential relevance to T1D. The hub gene, LTF, encodes a glycoprotein, lactoferrin, with immunoregulatory functions (31). LTF is able to ameliorate damage during periods of increased inflammation and, furthermore, contributes to tissue repair (31). In a cross-sectional study (32), lactoferrin levels were positively correlated with insulin sensitivity and were inversely correlated with HbA1c levels and measures of obesity. Another gene within the module, CAMP, is potentially relevant based on its role in innate immunity through the generation of the antimicrobial peptide cathelicidin (propeptide)/LL-37(processed peptide), as well its role in β-cell function (33–35). In mouse and rat β-cells, increased expression of CRAMP (CAMP homolog) has been shown to promote β-cell survival, modulate β-cell apoptosis in inflammatory conditions, and promote insulin secretion (35). Furthermore, CAMP/LL-37 treatment in BBdp rats enhanced β-cell neogenesis and induced a beneficial change in the gut microbiome environment (34). In humans, the CAMP promoter region includes a vitamin D response element that induces CAMP expression in the presence of elevated levels of calcitriol, the active form or vitamin D (33,36). Although calcitriol supplementation following recent onset of T1D failed to show a beneficial effect on residual β function (37,38), increased levels of 25-hydroxyvitamin D [25(OH)D] (precursor to calcitriol) were protective against IA in TEDDY (39) as well as Trial to Reduce IDDM in the Genetically at Risk (TRIGR) (40). Evidence of a protective effect of 25(OH)D suggests that early supplementation before or during IA may be more promising than supplementation following T1D onset. Furthermore, as demonstrated by the interaction between 25(OH)D levels and VDR (rs7975232) regarding the risk of IA in the work of Norris et al. (39), the potential therapeutic potential for vitamin D may also depend on genetic variation.
Although the individual genes within the LTF module support its relevance to the T1D disease process, the LTF module should be interpreted as a coexpression signature rather than on the basis of individual genes within the module. To better understand relevance of the LTF module, we tested the association between DNAm and the LTF eigengene. We identified three FDR significant CpGs, including cg00848392 on chromosome 6 within the MHC region. DNAm at this site is known to be correlated with expression of complement pathway genes C4A and C4B (41). Complement genes (C1QB and C3AR1) were also among the differentially expressed genes within the S. aureus KEGG pathway term. The complement system plays an important role in innate immunity and may have both causative and protective roles in autoimmune disease (42). The complement coagulation cascade pathway was significantly enriched in a proteomics analysis of cadaveric pancreatic tissue samples from subjects with T1D and age-matched control subjects (43). Interestingly, C1QB was upregulated in T1D pancreas tissue (43), which supports our study results of an increase in C1QB expression during IA among progressors (Supplementary Material). However, additional work is needed to better understand the potential protective versus causative role of complement activation in T1D. Furthermore, the methylation sites identified in the eigengene expression quantitative trait methylation (eQTM) analysis were >1 MB from the transcription start sites for all genes included in the module, indicating trans effects. Additional work is needed to understand the role of methylation in the LTF module and, more importantly, whether methylation at these sites is influenced by modifiable environmental risk factors.
A major strength of our study is the inclusion of longitudinal measurements of gene expression that preceded T1D. Few studies have included longitudinal measurements prior to T1D (3,5,7). Ferreira et al. (3) tested for longitudinal changes in an IFN inducible gene signature. Among the 225 unique IFN inducible genes identified in the work by Ferreira et al. (3), 15 overlapped with our differential gene set (Supplementary Material). Similar to the comparison of the progressors and maintainers in the current study, Kallionpää et al. (5) tested for longitudinal differences in gene expression during IA among individuals who seroconverted and progressed to T1D versus individuals who seroconverted but did not develop T1D. Among the 54 differentially expressed genes (5), 4 (IFI35, IFI44, OAS3, and RSAD2) were related to innate immunity and/or interferon γ signaling, and all 4 overlapped with our differentially expressed gene set (Supplementary Material). Notably, RSAD2, a protein with antiviral properties, has been shown to be overexpressed in virus-infected murine islet cells (44) and in recent work in DAISY (45) investigators identified a novel single nucleotide polymorphism (rs55900661) associated with progression from IA to T1D and expression of RSAD2. Xhonneux et al. (7) used WGCNA to identify transcriptional networks based on longitudinal changes in gene expression before and after IA. Similar to the comparison of progressors and maintainers in the current study, Xhonneux et al. (7) studied longitudinal coexpression changes in T1D case subjects relative to “control” subjects who seroconverted. This work identified an IAA-specific signature enriched for NK cell–specific transcripts that, relative to those of control subjects who did not seroconvert, was elevated at a young age in control subjects who seroconverted as well as T1D case subjects. The signature was also elevated prior to onset of T1D. Among genes included in the IAA signature from Xhonneux et al. (7), two genes (MATK and PTGDR) were differentially expressed, on average, between progressors, maintainers, and reverters in the current study, supporting potential relevance of these genes to the latter stages of IA.
Limitations
We did not detect any FDR significant changes in individual transcripts. Our experience highlights challenges associated with conducting discovery analyses using RNA-sequencing data. Given heterogeneity in the phenotype (autoantibody endotype, age at IA, etc.) as well as tissue-related heterogeneity (whole blood), large sample sizes are required to detect significant differences in individual transcripts. Collection of RNA samples in DAISY began after 2007. T1D case subjects who seroconverted and developed T1D early during the DAISY enrollment period are not represented in our study. We focused on gene changes following the onset of IA. Whether the differences are relevant prior to onset of IA is unclear. Due to sample availability, the current analysis included blood samples obtained later during the IA process. RNA samples were available at the onset of IA (seroconversion visit) for ∼25% of the population. The median time from IA onset was 24.5 months (interquartile range 0–56.9) for IA-1 and 80.4 months (59.8–125.3) for IA-2. As a result, we may not have detected changes that occur very early in the IA process. Furthermore, the median age at T1D diagnosis was 12.6 years—older than the median age at T1D across the full DAISY population, 9.8 years. As a result, the gene expression changes within the progressor group are generalizable to the T1D subpopulation in DAISY who tend to be older at T1D diagnosis. We used whole blood samples rather than sorted cells, and it is possible that the changes in gene expression that we observed may reflect changes in cell populations. We did not adjust for cell type composition due to absence of gold standard method for cell type adjustment in bulk RNA sequencing. Furthermore, changes in the blood subpopulations may be an important mechanism in the pathogenesis of T1D, and thus adjustment for cell type could adjust out important biological mechanisms. Therefore, enrichment results and changes in gene expression within the LTF module may be due to changes in individual genes, changes in cell populations within whole blood, or both. Additional work is needed to better understand the specific mechanisms connecting LTF module coexpression to progression versus reversion of IA.
Conclusion
We identified changes in gene expression prior to T1D that distinguished progression from reversion and maintenance of IA. Enrichment analysis identified pathways related to immune regulation, immune activation, and innate immune response. We identified a set of coexpressed genes, also related to innate immune system pathways. Coexpression of these genes during IA were higher among progressors, indicating potential activation of innate immunological system pathways prior to onset of T1D. Additional work is needed to understand whether changes in gene expression were initiated prior to IA, whether the observed changes were causal versus compensatory responses to progression of the IA disease process, and whether the changes were driven by modifiable environmental factors.
Article Information
Funding. This work was funded by National Institutes of Health (NIH) grants R01-DK104351 and R01-DK32493 and National Institute of Allergy and Infectious Diseases, NIH, grant R21AI142483.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. P.M.C. designed the study, performed the data analysis, interpreted data, and drafted the manuscript. L.A.V., A.K.S., I.Y., T.E.F., K.K., and J.M.N. designed the study, interpreted data, and edited the manuscript. K.W., R.K.J., T.B., and M.R. reviewed and edited the manuscript. J.M.N. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains supplementary material online at https://doi.org/10.2337/figshare.20060480.
References
- 1. Tree TI, Peakman M. Autoreactive T cells in human type 1 diabetes. Endocrinol Metab Clin North Am 2004;33:113–133, ix–x [DOI] [PubMed] [Google Scholar]
- 2. Rewers M, Ludvigsson J. Environmental risk factors for type 1 diabetes. Lancet 2016;387:2340–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ferreira RC, Guo H, Coulson RM, et al. A type I interferon transcriptional signature precedes autoimmunity in children genetically at risk for type 1 diabetes. Diabetes 2014;63:2538–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jin Y, Sharma A, Bai S, et al. Risk of type 1 diabetes progression in islet autoantibody-positive children can be further stratified using expression patterns of multiple genes implicated in peripheral blood lymphocyte activation and function. Diabetes 2014;63:2506–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kallionpää H, Elo LL, Laajala E, et al. Innate immune activity is detected prior to seroconversion in children with HLA-conferred type 1 diabetes susceptibility. Diabetes 2014;63:2402–2414 [DOI] [PubMed] [Google Scholar]
- 6. Reynier F, Pachot A, Paye M, et al. Specific gene expression signature associated with development of autoimmune type-I diabetes using whole-blood microarray analysis. Genes Immun 2010;11:269–278 [DOI] [PubMed] [Google Scholar]
- 7. Xhonneux LP, Knight O, Lernmark Å, et al. Transcriptional networks in at-risk individuals identify signatures of type 1 diabetes progression. Sci Transl Med 2021;13:eabd5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bonifacio E, Mathieu C, Nepom GT, et al. Rebranding asymptomatic type 1 diabetes: the case for autoimmune beta cell disorder as a pathological and diagnostic entity. Diabetologia 2017;60:35–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ziegler AG, Rewers M, Simell O, et al. Seroconversion to multiple islet autoantibodies and risk of progression to diabetes in children. JAMA 2013;309:2473–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Steck AK, Vehik K, Bonifacio E, et al.; TEDDY Study Group . Predictors of progression from the appearance of islet autoantibodies to early childhood diabetes: The Environmental Determinants of Diabetes in the Young (TEDDY). Diabetes Care 2015;38:808–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rewers M, Bugawan TL, Norris JM, et al. Newborn screening for HLA markers associated with IDDM: diabetes autoimmunity study in the young (DAISY). Diabetologia 1996;39:807–812 [DOI] [PubMed] [Google Scholar]
- 12. Rewers M, Norris JM, Eisenbarth GS, et al. Beta-cell autoantibodies in infants and toddlers without IDDM relatives: diabetes autoimmunity study in the young (DAISY). J Autoimmun 1996;9:405–410 [DOI] [PubMed] [Google Scholar]
- 13. Martin M: Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 2011;17:3. Available from ttps://journal.embnet.org/index.php/embnetjournal/article/view/200/479
- 14. Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009;10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Risso D, Ngai J, Speed TP, Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol 2014;32:896–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 1995;57:289–300 [Google Scholar]
- 19. Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc 2009;4:1184–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 2010;11:R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 2011;6:e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Benjamini Y, Liu W. A step-down multiple hypotheses testing procedure that controls the false discovery rate under independence. J Stat Plan Inference 1999;82:163–170 [Google Scholar]
- 25. Langfelder P, Luo R, Oldham MC, Horvath S. Is my network module preserved and reproducible? PLOS Comput Biol 2011;7:e1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vanderlinden LA, Johnson RK, Carry PM, et al. An effective processing pipeline for harmonizing DNA methylation data from Illumina’s 450K and EPIC platforms for epidemiological studies. BMC Res Notes 2021;14:352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014;30:1363–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stene LC, Barriga K, Hoffman M, et al. Normal but increasing hemoglobin A1c levels predict progression from islet autoimmunity to overt type 1 diabetes: Diabetes Autoimmunity Study in the Young (DAISY). Pediatr Diabetes 2006;7:247–253 [DOI] [PubMed] [Google Scholar]
- 29. Fabregat A, Sidiropoulos K, Viteri G, et al. Reactome diagram viewer: data structures and strategies to boost performance. Bioinformatics 2018;34:1208–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cabrera SM, Henschel AM, Hessner MJ. Innate inflammation in type 1 diabetes. Transl Res 2016;167:214–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kruzel ML, Zimecki M, Actor JK. Lactoferrin in a context of inflammation-induced pathology. Front Immunol 2017;8:1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Moreno-Navarrete JM, Ortega FJ, Bassols J, Ricart W, Fernández-Real JM. Decreased circulating lactoferrin in insulin resistance and altered glucose tolerance as a possible marker of neutrophil dysfunction in type 2 diabetes. J Clin Endocrinol Metab 2009;94:4036–4044 [DOI] [PubMed] [Google Scholar]
- 33. Gombart AF, Borregaard N, Koeffler HP. Human cathelicidin antimicrobial peptide (CAMP) gene is a direct target of the vitamin D receptor and is strongly up-regulated in myeloid cells by 1,25-dihydroxyvitamin D3. FASEB J 2005;19:1067–1077 [DOI] [PubMed] [Google Scholar]
- 34. Pound LD, Patrick C, Eberhard CE, et al. Cathelicidin antimicrobial peptide: a novel regulator of islet function, islet regeneration, and selected gut bacteria. Diabetes 2015;64:4135–4147 [DOI] [PubMed] [Google Scholar]
- 35. Sun J, Xu M, Ortsäter H, et al. Cathelicidins positively regulate pancreatic β-cell functions. FASEB J 2016;30:884–894 [DOI] [PubMed] [Google Scholar]
- 36. Gombart AF. The vitamin D-antimicrobial peptide pathway and its role in protection against infection. Future Microbiol 2009;4:1151–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Walter M, Kaupper T, Adler K, Foersch J, Bonifacio E, Ziegler AG. No effect of the 1alpha,25-dihydroxyvitamin D3 on beta-cell residual function and insulin requirement in adults with new-onset type 1 diabetes. Diabetes Care 2010;33:1443–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bizzarri C, Pitocco D, Napoli N, et al.; IMDIAB Group . No protective effect of calcitriol on beta-cell function in recent-onset type 1 diabetes: the IMDIAB XIII trial. Diabetes Care 2010;33:1962–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Norris JM, Lee HS, Frederiksen B, et al.; TEDDY Study Group . Plasma 25-hydroxyvitamin D concentration and risk of islet autoimmunity. Diabetes 2018;67:146–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Miettinen ME, Niinistö S, Erlund I, et al.; TRIGR Investigators . Serum 25-hydroxyvitamin D concentration in childhood and risk of islet autoimmunity and type 1 diabetes: the TRIGR nested case-control ancillary study. Diabetologia 2020;63:780–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bonder MJ, Luijk R, Zhernakova DV, et al.; BIOS Consortium . Disease variants alter transcription factor levels and methylation of their binding sites. Nat Genet 2017;49:131–138 [DOI] [PubMed] [Google Scholar]
- 42. Conigliaro P, Triggianese P, Ballanti E, Perricone C, Perricone R, Chimenti MS. Complement, infection, and autoimmunity. Curr Opin Rheumatol 2019;31:532–541 [DOI] [PubMed] [Google Scholar]
- 43. Woo J, Sudhir PR, Zhang Q. Pancreatic tissue proteomics unveils key proteins, pathways, and networks associated with type 1 diabetes. Proteomics Clin Appl 2020;14:e2000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Marroqui L, Lopes M, dos Santos RS, et al. Differential cell autonomous responses determine the outcome of coxsackievirus infections in murine pancreatic α and β cells. eLife 2015;4:e06990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Onengut-Gumuscu S, Paila U, Chen WM, et al. Novel genetic risk factors influence progression of islet autoimmunity to type 1 diabetes. Sci Rep 2020;10:19193. [DOI] [PMC free article] [PubMed] [Google Scholar]