Abstract
DNA motifs at several informative loci in more than 500 strains of Helicobacter pylori from five continents were studied by PCR and sequencing to gain insights into the evolution of this gastric pathogen. Five types of deletion, insertion, and substitution motifs were found at the right end of the H. pylori cag pathogenicity island. Of the three most common motifs, type I predominated in Spaniards, native Peruvians, and Guatemalan Ladinos (mixed Amerindian-European ancestry) and also in native Africans and U.S. residents; type II predominated among Japanese and Chinese; and type III predominated in Indians from Calcutta. Sequences in the cagA gene and in vacAm1 type alleles of the vacuolating cytotoxin gene (vacA) of strains from native Peruvians were also more like those from Spaniards than those from Asians. These indications of relatedness of Latin American and Spanish strains, despite the closer genetic relatedness of Amerindian and Asian people themselves, lead us to suggest that H. pylori may have been brought to the New World by European conquerors and colonists about 500 years ago. This thinking, in turn, suggests that H. pylori infection might have become widespread in people quite recently in human evolution.
Helicobacter pylori is a microaerophilic bacterium with the extraordinary ability to establish infections in human stomachs that can last for years or decades, despite immune and inflammatory responses and normal turnover of the gastric epithelium and overlying mucin layer in which it resides. It is carried by more than half of all people worldwide and has attracted great attention as a major cause of peptic ulcer disease and an early risk factor for gastric cancer, one of the most frequently lethal of malignancies worldwide (for reviews see references 23, 48, and 60).
H. pylori is one of the most genetically diverse of bacterial species, with any given isolate easily distinguished from most others by DNA fingerprinting (3, 4, 55) or the sequencing of representative gene segments (typically some 3 to 5% DNA sequence divergence between isolates, even in essential genes) (1, 31). This mutational diversity has been enhanced by extensive interstrain gene transfer and recombination (1, 4, 40, 53). In contrast, much stronger clonality, with the predominance of relatively fewer clones, is seen in populations of several other much-studied bacterial species (see, e.g., references 29, 33, 52).
The great diversity among H. pylori strains implies a striking lack of selection for just one or a few genotypes that might be best adapted for all humans. Some of this may reflect preferential transmission of H. pylori within families and among people in close contact (see, e.g., references 10 and 21). In consequence, no given strain need compete simultaneously against many other strains, despite occasional cases of mixed infection by unrelated strains (12, 28, 40). There is also a sense that humans differ in traits that could be important to individual strains, such as specificity and strength of immune and inflammatory responses and availability and distribution of receptors to which H. pylori adheres (18, 19, 27, 28, 35). Such host heterogeneity would select for divergence among H. pylori strains.
Superimposed on the diversity among H. pylori strains in a given community are indications of differences at certain loci between strains from different parts of the world or human ethnic groups. In particular, the DNA sequence motifs predominating in two virulence-associated genes, vacA (vacuolating cytotoxin) and cagA (cytotoxin-associated gene), in strains from the United States and Europe were found to differ from those predominating in southern coastal China and Japan (1, 8, 36, 47, 57–59, 62), although less phylogenetic clustering was found in sequences of several housekeeping genes (1, 57). Even though the factors that underlie the geographic partitioning of vacA and cagA alleles are not known, it is attractive to imagine that further studies of H. pylori genotypes from different well-separated human populations may identify new factors that affect colonization or disease in peoples of particular ethnicities and may also help us better understand the origin and evolution of H. pylori itself.
Here we report on a set of insertion, deletion, and substitution motifs at the extreme right end of the cag pathogenicity island (PAI) that are well suited to population level surveys of H. pylori genotypes. We show that these motifs, and also DNA sequence motifs in the vacA and cagA genes, are nonrandomly distributed geographically. The patterns observed should help us understand how H. pylori arrived in the Americas and the possible evolutionary origins of this bacterium as a human pathogen.
MATERIALS AND METHODS
Bacterial strains.
All H. pylori strains used contained the cag PAI, as determined by PCR and/or hybridization tests, carried out as described in reference 2. They were cultured from gastric biopsies from adult patients who had been referred for upper gastrointestinal endoscopy to collaborating gastroenterologists at clinics in the various countries listed in Tables 1 to 3. All samples were obtained with informed consent under protocols approved by the local human studies committee at each institution.
TABLE 1.
Distribution of cag right-junction motifs among major types of H. pylori strains
| Geographic region | No. (%) of strains
|
||||
|---|---|---|---|---|---|
| Total | Type I | Type II | Type III | Other | |
| South Europe | 36 | 33 (92%) | 1 (3%) | 2 (6%) | 0 |
| Spain | 36 | 33 | 1 | 2 | 0 |
| Latin America | 96 | 89 (93%) | 0 | 6 (6%) | 1 (1%) |
| Peru | 68 | 62 | 0 | 6 | 0 |
| Guatemala | 28 | 27 | 0 | 0 | 1a |
| Africa | 40 | 40 (100%) | 0 | 0 | 0 |
| Gambia | 8 | 8 | 0 | 0 | 0 |
| South Africa | 32 | 32 | 0 | 0 | 0 |
| North America | 51 | 45 (88%) | 0 | 2 (4%) | 4 (8%) |
| Louisiana | 16 | 15 | 0 | 0 | 1a |
| Missouri | 7 | 7 | 0 | 0 | 0 |
| Ohio | 13 | 11 | 0 | 1 | 1a |
| Tennessee | 5 | 5 | 0 | 0 | 0 |
| West Virginia | 10 | 7 | 0 | 1 | 2b |
| East Asia | 204 | 1 (0.5%) | 194 (95%) | 8 (4%) | 1 (0.5%) |
| South China | 96 | 0 | 92 | 3 | 1a |
| Hong Kong | 48 | 1 | 44 | 3 | 0 |
| North China | 13 | 0 | 13 | 0 | 0 |
| Japan | 47 | 0 | 45 | 2 | 0 |
| South Asia | 75 | 7 (9%) | 1 (1%) | 64 (85%) | 3 (4%) |
| Calcutta, India | 75 | 7 | 1 | 64c | 3d |
| Northeast Europe | 24 | 10 (42%) | 10 (42%) | 4 (17%) | 0 |
| Sweden | 16 | 7 | 5 | 4 | 0 |
| Lithuania | 8 | 3 | 5 | 0 | 0 |
TABLE 3.
Distribution of subtypes of type III strains
| Geographic region | No. of strains of type:
|
||
|---|---|---|---|
| Total | IIIa | IIIba | |
| India | 61 | 52 | 9 |
| United States | 2 | 2 | 0 |
| Latin America | 7 | 7 | 0 |
| East Asia | 8 | 8 | 0 |
Carries miniIS605.
Following are notes on ethnicities of patients and clinical disease associations that may be useful in evaluating the data presented here, where this information was available. Twenty-two of the 34 Spanish strains for which records were available were from patients with peptic ulcers, and the other 12 were from patients with gastritis only. The 68 Peruvian strains studied here were from Amerindian residents of the shanty towns of Las Pampas and San Juan de Miraflores, in Lima. Most patients were born in agricultural communities in the countryside; they had immigrated to Lima during the last few decades but have had rather little contact with Peruvians of other ethnicities. Each of the 35 strains for which records were available (including 3 containing type III motifs) were from patients with gastritis only, not peptic ulcers. The 28 Guatemalan strains were from patients at the Gastroenterology Clinic of the Social Security System Hospital in Guatemala City. Most patients were of Ladino (mixed Amerindian and European) ancestry and were generally of lower socioeconomic class. Of the 24 strains for which records were available, 15 were from patients with duodenal ulcers and 9 were from patients with chronic active gastritis. Each of the 32 South African strains were from native black African residents of Soweto. The ancestry of the patients (purely native African) is distinct from that of the “Cape colored” (mixed native African, Indian, and European), whose strains had been studied earlier (1, 53). Two of the Soweto patients had gastric ulcers, and 6 had duodenal ulcer disease; the other 24 patients had more-benign infections (generally gastritis only). The eight Gambian strains were also from native black Africans. Of the 51 U.S. strains studied, the 16 from Louisiana and the 13 from Ohio came from patients of African-American ethnicity (54; S. Czinn, unpublished data). The ethnicities of patients from whom the other U.S. strains were recovered are not known. The 48 strains from Hong Kong were from ethnic Chinese patients: 6 with gastric ulcers, 11 with duodenal ulcers, and 31 with gastritis only. The 96 strains from south China were from patients living in Shanghai and Guangzhou and had been studied previously in terms of vacA and cagA sequence motifs (47, 57). Among 81 Shanghai strains, 46 were from patients with peptic ulcers and 35 were from patients with gastritis only. The 13 north Chinese strains were from patients in Shenyang with benign infections (gastritis only). Of the 47 Japanese strains, half were from Ube in western Honshu and half were from Hokkaido. These strains were from patients with gastric cancer (13 strains), gastric ulcers (20 strains), or gastritis only (14 strains). The 75 Indian strains were from middle or lower middle class residents of Calcutta with peptic ulcers (53 strains) or with gastritis only (22 strains) and are described more fully in the accompanying paper (45). Also included in our study were the two strains whose genome sequences have been determined completely: 26695 (56) from a British patient and J99 (6) from a Caucasian resident of Pulaski, Tenn. (T. L. Cover, personal communication).
Bacterial growth.
Standard methods (12) were used for H. pylori growth in a microaerobic atmosphere on Difco brain heart infusion agar supplemented with 10% horse blood.
DNA methods.
Chromosomal DNA was isolated from confluent plate cultures using the QIAamp tissue kit (Qiagen, Chatsworth, Calif.) or by the hexadecyltrimethylammonium bromide method (9). Specific PCR was generally carried out in 20-μl volumes containing 5 ng of genomic DNA, 0.5 U of Taq polymerase (Promega Corp., Madison, Wis.), or KlenTaq (Clontech Corp, Palo Alto, Calif.), 5 pmol of each primer, and 0.25 mmol of each deoxynucleoside triphosphate in a standard buffer. Cycling conditions were usually 30 cycles at 94°C for 30 s, 52°C for 30 s, and 72°C for a time dependent on the expected product size (1 min per kb). The PCR primers used in this study are listed in Table 4; their use is validated in Fig. 1 by using strains that were also characterized by DNA sequencing of relevant regions, and the positions of these primers in the cag right junction region are diagrammed in Fig. 2. The relatively low stringency (52°C for the annealing step) used for most PCRs was chosen to increase the chance of getting informative PCR products, even from strains with point mutation differences in sequences used as primer binding sites.
TABLE 4.
Primers and strategies used for strain characterization
| Primer (no.) | Sequence | Remarks |
|---|---|---|
| cagF4584 (1) | 5′-GTTAATACAAAAGGTGGTTTCCAAAAATC | 5′ end 29 bp before 3′ end of cagA |
| cagF4580 (2) | 5′-GGAATTATCACACCTTATAATGCCC | 5′ end 309 bp before 3′ end of hel |
| cagR5280 (3) | 5′-GGTTGCACGCATTTTCCCTTAATC | 5′ end in glr, 198 bp after right end of cag PAI |
| Diagnostic primers for types I and IV | ||
| cagF4856 (4) | 5′-GCGATGAGAAGAATATCTTTAGCG | 5′ end 47 bp before 3′ end of hel |
| cagR5280 (3) | Same as above | |
| Diagnostic primers for type II | ||
| IS606-1692 (5) | 5′-CTAACAATTTGCCATTATGCTGT | 5′ end 43 bp before right end of IS606* fragment |
| cagR5280 (3) | Same as above | |
| Diagnostic primers for type III | ||
| fcn unk | ||
| (6) | 5′-TGGATTAAATCTTAATGAATTATCG | 5′ end 109 bp before right end of unk1 |
| (6a) | 5′-ACTCT(A/G)TTTTGCTTGC(A/G)TGCTTTTGG | 5′ end 138 bp before right end of unk1 |
| cagR5280 (3) | Same as above | |
| Diagnostic primers for type IV | ||
| cagF4584 (1) | Same as above | |
| Xins.R (7) | 5′-CGCTCTCTAATTGTTCTAGGA | 5′ end 226 bp after beginning of hel (type IV DNAs also yielded PCR product with type I primers; product size is similar to that obtained from subtype Ib) |
| Diagnostic primers for subtypes Ib and IIIb | ||
| cagF4584 (1) | Same as above | |
| miniIS605.R (8) | 5′-CCGCTAAAGACGATTGGGCTT | 5′ end 261 bp from left end, 13 bp from right end of miniIS605 |
| Diagnostic primers for subtype Ic | ||
| IS606orfB-F (9) | 5′-GCCGTAGGGTGTTAAAGTCTAA | 5′ end 563 bp before right end of IS606 |
| cagR5280 (3) | Same as above | |
| Diagnostic primers for cagA allele sequencing | ||
| cagA2 | 5′-GGAAATCTTTAATCTCAGTTCGG | |
| cagA5 | 5′-GGCAATGGTGGTCCTGGAGCTAGGC | |
| cagA5P | 5′-GGCAATGGTGGTTTTGGAGCCA | |
| Diagnostic primers for vacA allele typing | ||
| m1a | ||
| VA3-F | 5′-GGTCAAAATGCGGTCATGG | |
| VA3-R | 5′-CCATTGGTACCTGTAGAAAC | |
| m1b | ||
| VAm-F3 | 5′-GGCCCCAATGCAGTCATGGAT | |
| VAm-R3 | 5′-GCTGTTAGTGCCTAAAGAAGCAT | |
| m2 | ||
| VA4-F | 5′-GGAGCCCCAGGAAACATTG | |
| VA4-R | 5′-CATAACTAGCGCCTTGCAC | |
| Primers for miniIS605 detection | ||
| miniIS605L | 5′-TCTTTAGTATTTGGGATGAATGCT | |
| miniIS605R (8) | Same as above |
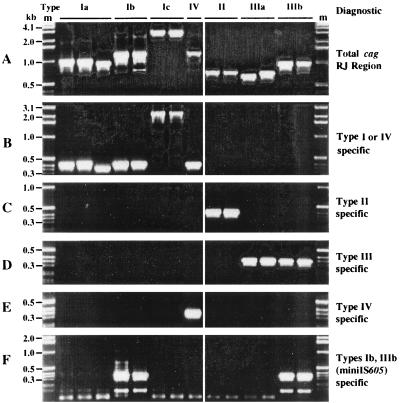
FIG. 1.
PCR amplification to detect and identify different insertion, deletion, and substitution motifs at the extreme right end of the cag PAI. The primers used in these amplifications are listed in Table 4, and their positions are diagrammed in Fig. 2. The same DNAs were used for each of the six PCR tests shown here, and in most cases their sequences in this region are known. The strains used (Genbank accession numbers or references) are as follows (from left to right): OhioM6 (AF190992), Gambia94/24 (AF190658), Peru002B (AF190994) (all type Ia); India120A (AF191014) and NTCT11638 (20) (type Ib); 84-183 (AF190660) and OhioP46 (AF190995 and AF190996) (type Ic); 26695 (56) (type IV); JapanHU38 (AF191000); Peruvian Hp1 (from an ethnic Japanese in Peru) (AF190661) (type II); NCTC11637 (AF191010); India17A (AF191016) (type IIIa); and India75A (AF190663) and India68A (not sequenced) (type IIIb). (A) Amplification of entire segment between cagA and glr using primers 1 and 3 (Table 4; Fig. 2). The difference in PCR product size between the two type IIIa strains is due to a 29-bp deletion in NCTC11637, relative to India17A. (B) Type I- and IV-specific amplification using primers 3 and 4 (Table 4; Fig. 2). (C) Type II-specific amplification using primers 3 and 5 (Table 4; Fig. 2). (D) Type III-specific amplification using primers 3 and 6 (Table 4; Fig. 2). (E) Type IV-specific amplification using primers 1 and 7 (Table 4; Fig. 2). (F) MiniIS605-specific amplification using primers 1 and 8 (Table 4; Fig. 2) (type Ib or IIIb).
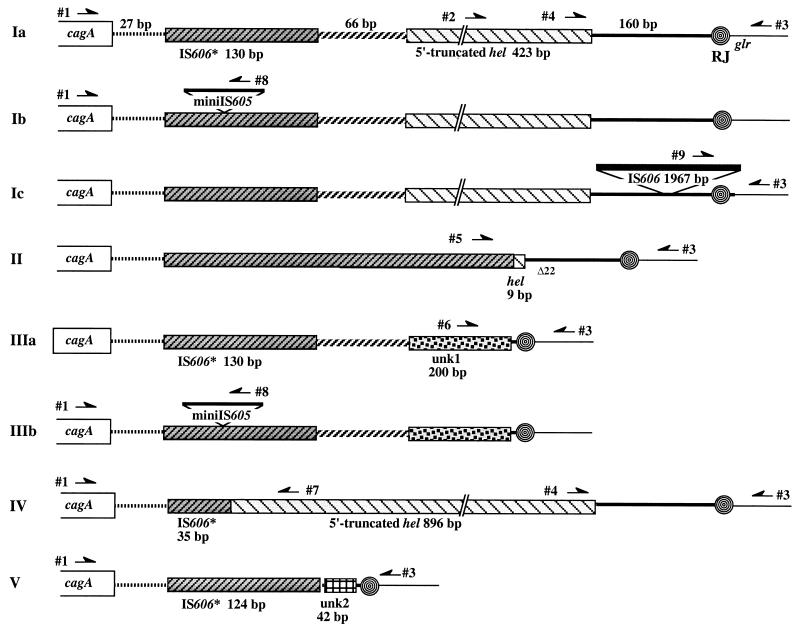
FIG. 2.
Diagram of DNA sequence motif types at the right end of the cag PAI of H. pylori and positions of primers that were most useful for distinguishing different motif types and subtypes. RJ, right junction, the 31 bp that contains the 3′ end of the glutamate racemase gene (glr) and that is also repeated directly at the left end of the cag PAI (2, 20, 56). The cagA gene is relatively near the right end of the cag PAI. Closely related sequences in different motif types are indicated by common symbols. The sequences that led to the interpretations diagrammed here have been deposited under GenBank accession no. AF190658, AF190992 to AF190994, and AF191013 (type Ia); AF190659 and AF191014 (type Ib); AF190660, AF190995, and AF190996 (type Ic); AF190661 and AF190997 to AF1901008 (type II); AF190662, AF191009 to AF191012, AF191015, and AF191016 (type IIIa); AF190663 (type IIIb); and AF200689, AF201074, and AF201075 (type V). The type IV sequence was determined in reference 56 as part of the strain 26695 genome sequencing effort. In terms of other published sequences, NCTC11638 (2, 20) and J99 (6) are of types Ib and Ia, respectively. The ancestral IS606* element is inferred to have been about 2 kb long, based on its homologs (e.g., canonical IS606 and IS605) (39, 56), and the IS606* remnants shown here are closely related to corresponding regions at one end of canonical IS606, with sequence matches of 82 to 85%. The ancestral form of the DNA hel gene found in this cag PAI right junction region may have been more than 1 kb long, based on the sizes of its homologs in the database (see, e.g., MJ0104 of M. janaschii) (17). It is inferred to have undergone various deletion or substitution events that removed much of its 5′ end and upstream sequences in different lineages. The two remnant forms of this gene were designated omega (in NCTC11638, a type Ia strain) (20) and HP0548 (in 26695, a type IV strain) (56). The ∼200-bp unk1 (function unknown) sequence that replaces nearly all of hel in type III strains is not obviously related to other sequences found to date in GenBank and seems not to be a fragment of any complete open reading frame. When present, the ∼268-bp miniIS605 element diagrammed here was always found inserted at the same site in the IS606* remnant, just downstream of TTTAA (Fig. 1F and 5). Five of the nine type III strains analyzed by sequencing contained a 39-bp deletion in the 66-bp region between the remnants of IS606* and the helicase (hel) gene, starting 3 bp after the IS606* deletion breakpoint: ChinaF30A (accession no. AF191009), JapanHU54 (accession no. AF191011), India17A (accession no. AF191016), and NCTC11637 (AF191010; also has an additional 29-bp deletion). Type III strains without this 39-bp deletion include India75A (accession no. 190663), India7A (accession no. AF191015), Sweden53 (accession no. 190662), and Peru466 (accession no. AF191012). One type III strain (India47A; accession no. AF200690) also contained a 158-bp deletion within the unk1 sequence. The three type V strains, all from India, were very similar in sequence.
PCR fragments for sequencing were purified by a QIAquick PCR purification kit (Qiagen). Sequencing was done automatically with Big Dye Terminator cycle sequencing kit (PE Applied Biosystems, Foster City, Calif.). DNA sequence editing and analysis were performed with programs in the GCG package (Genetics Computer Group, Madison, Wis.) and programs and data in H. pylori genome sequence databases (6, 56) (http://www.tigr.org/tdb/mdb/hpdb.html; http://scriabin.astrazeneca-boston.com/hpylori/).
The lengths of given deleted or rearranged segments in divergent H. pylori strains can sometimes be determined only approximately, even if in isogenic wild-type and mutant strains such segments could be described more exactly. Two factors contribute to this: insertion and deletion polymorphisms (one or a few base pairs) within the segment, which can lead to real variation among strains in segment length, and base substitutions and/or other point mutations that can lead to uncertainty as to the exact end point of the deletion or other rearrangement, since this corresponds to a site of fusion to new DNA sequences, not the end of the DNA per se.
Nucleotide sequence accession numbers. The nucleotide sequences of the right-end junction regions of cag PAIs were deposited in GenBank under the following accession numbers: AF190658, AF190992 to AF190994, and AF191013 (type Ia); AF190659 and AF191014 (type (Ib); AF190660, AF190995, and AF190996 (type Ic); AF190661, AF190997 to AF191008 (type II); AF190662, AF19009 to AF19012, AF191015, AF191016, and AF200690 (type IIIa); AF190663 (type IIIb); and AF200689, AF201074, and AF201075 (type V). cagA gene sequences were deposited under the following accession numbers: AF198468 to AF198482 (European type) and AF198483 to AF198486 (East Asian type).
RESULTS
DNA deletion, insertion, and substitution diversity at the right end of the cag PAI.
PCR tests were carried out on H. pylori strains from various parts of the world using primers specific for sequences near and just past the right end of the H. pylori cag PAI (within cagA and glr, respectively). The products generated ranged from about 0.5 to 3 kb (Fig. 1A), indicating considerable DNA length diversity in this region. PCR-amplified DNAs from 34 representative strains were sequenced. This identified a series of deletions, insertions, and substitutions involving primarily remnants of a transposable (insertion sequence [IS]) element designated IS606*, a second small [IS] element designated miniIS605, a putative helicase (hel) gene, and small DNA segments with no significant homology to other entries in the database (designated unk) (Fig. 2).
Five types and several subtypes of DNA motifs were distinguished based on (i) lengths of the IS606* remnant (about 130 bp in types I and III, 312 bp in type II, 35 bp in type IV, and 124 bp in type V (i.e., 6 bp shorter in type V than in types I and III)); (ii) lengths of the hel gene fragment (about 423 bp in type I, just 9 bp in type II, and 896 bp in type IV); (iii) various other sequences, either between the IS606* and hel gene remnants (66 bp) or apparently replacing hel (unk1 or unk2) in certain strain types; (iv) the presence or absence of a miniIS605 element in the IS606* remnant; and (v) in a few cases, the presence of full-length canonical IS606 (Fig. 2). The type II strains were also distinguished by a specific 22-bp deletion just downstream of the hel sequences (Fig. 2 and 3) that was not found in other strains. Many additional base substitution and small insertion or deletion differences were also found by sequencing in each of the motif types (as expected) (1). Some of these were sufficiently large to cause perceptible differences in sizes of PCR products, as illustrated by the products in Fig. 1A from the two strains with type IIIa motifs (reflecting a 29-bp length difference). These and smaller DNA size differences did not affect the overall classification of motif types summarized in Fig. 2 and thus were not considered in detail in the present analyses.
FIG. 3.
DNA sequences at 3′ end of helicase (hel) gene. GenBank accession numbers or references for sequences presented here are as follows: type II strains (A) JapanC7, AF190999; ChinaR30, AF190997; Lith5-1, AF190998; Hp1, AF190661; type I and IV strains (B) NCTC11638, 20; 26695, 56. The data in panel A suggest that just the last three codons at the 3′ end of the hel gene (boldface) are retained in type II strains. Also shown is the 22-bp deletion that was found in all type II strains but not in any type I or IV strains (see also Fig. 2). Corresponding sequences from representative type I and IV strains, which have a much longer hel gene fragment, are shown in panel B.
The left end of IS606* (Fig. 2) was present in all strains. Based on comparisons with its complete (2-kb) homologs, e.g., IS606 (GenBank accession no. U95957 and AE001512 [39]), this left-end sequence seemed likely to correspond to the natural end of an ancestral full-length IS606* element. Similarly, the right end of the putative helicase (hel) gene fragments (Fig. 2), when present, seemed to correspond to the natural 3′ end of this gene (Fig. 3). Even though cloned parts of hel from 26695 can affect gene expression in Escherichia coli (43), its closest homologs in other organisms are more than 1 kb long (GenBank accession no. Q57568 [Methanococcus jannaschii], AE000770 [Aquifex aeolicus], and AAD35099 [Thermotoga maritima]), which in turn suggests that the hel alleles found here are truncated, even in strains with type IV motifs such as 26695 (inferred here to contain 896 bp of hel, although annotated as 825 bp [http://www.tigr.org/tdb/mdb/hpdb.html]). The site of deletion in IS606*, adjacent sequences, and the right end point of this 66-bp sequence were closely related in type I and type III motifs (Fig. 2), implying that these two motifs derive from the same ancient deletion or substitution event, or possibly from recurrent events at an insertion hotspot. The inferred junction between the 66-bp sequence and a putative remnant of helicase in the type III motif seemed to match that in type I strains, although this inference is tenuous since it depends on a correct guess that one codon (Fig. 4) derives from the hel gene. If correct, however, this would indicate a common deletion end point in the hel gene in the type I and III lineages, implying that they might be derived from one ancestral event.
FIG. 4.
DNA sequences that identify the location and termini of the unk1 (function unknown) sequence. GenBank accession numbers or references for these sequences are as follows: AF19009 (ChinaF30A), AF191011 (JapanHU54), AF191015 (India74), AF191012 (Peru466), AF191014 (India20A), AF190994 (Peru2B). The unk1 sequence is in lowercase. It seems to be inserted just after the first codon of the remnant of the hel gene found in type I strains (boldface) and to replace hel gene and downstream sequences almost to the 31-bp direct repeat that marks the right end of the cag PAI. Corresponding sequences from two representative type I strains (which have a much longer hel gene fragment; boldface) are also shown.
A subset of strains with type I and type III motifs contained a ∼268-bp miniIS605 element in the cag right-junction region. This element was at the same site in all cases analyzed by PCR (as in Fig. 1 and 2) or sequencing: just downstream of the sequence TTTAA (Fig. 5). It was striking that no type II motif contained miniIS605, even though PCR tests had indicated that most strains with type II motifs contained miniIS605 somewhere in their genomes (54 of 61 tested), and DNA sequencing indicated that about half of them contained the TTTAA motif at this site (data not shown). We also note that the leftmost 8 bp of miniIS605 itself, identified here by comparing inserted elements and empty sites, are variable and usually do not perfectly match the left end of the canonical full-length IS605 element of strain NCTC11638 (Fig. 5). These leftmost 8 bp had not been included in the initial description of miniIS605 (20).
FIG. 5.
Representative sequences at sites of insertion of miniIS605 elements. Representative sequences at termini of miniIS605 or full-length canonical IS605 are in capitals and boldface. Flanking sequences are in lowercase. (A) Examples of miniIS605 insertions at a site in the remnant of IS606* in the right junction region of the cag PAI. (B) Corresponding empty sites in strains that lack miniIS605 at this site. (C) The ends of canonical IS605 and its TTTAA target site sequence (39). GenBank accession numbers or references from which present sequences were extracted are as follows: India75A (a type IIIb strain), AF190663; India120A (a type Ib strain), AF191014; JapanHU54 (a type III strain), AF191011; NCTC11638 (a type Ib strain), 20; Gambia9424 (a type Ia strain), AF190658; Lith5-1 (a type II strain), AF190998. PCR tests also indicated that miniIS605 was inserted at the same site in all strains of type Ib and IIIb. Note that the leftmost 8 bp of miniIS605 are variable in sequence and were initially (20) not recognized as being part of this element.
Geographic differences in predominant H. pylori genotypes.
The geographic distribution of the various DNA motifs diagrammed in Fig. 2 was determined by PCR using more than 500 H. pylori strains from five continents. Representative data are shown in Fig. 1B to F, and the full set of results is summarized in Tables 1 to 3. These motifs were nonuniformly distributed geographically (Tables 1 to 3). In particular, type I DNA motifs predominated in Spanish strains (33 of 36 tested) but were less common in our north European collection. Type I motifs also predominated in the largely Amerindian population in a shanty town in Lima, Peru (62 of 68 strains tested), in a Ladino (Amerindian-European mixed ancestry) population in Guatemala City (27 of 28 strains tested), and in native Africans (each of 32 strains from Soweto and 8 strains from The Gambia). In contrast, type II DNA motifs predominated in Chinese and Japanese strains (194 of 204 tested) and type III motifs predominated in strains from Calcutta, India (64 of 75 tested). Type I, II, and III motifs were each common in the north European (Swedish and Lithuanian) strains studied to date. The type IV motif, although rare among the isolates screened to date, was found in our one English strain (26695), whose genome was sequenced in full (55), and in two strains from West Virginia; the type V motif was found in a few strains from Calcutta. Further study will be needed to learn if these two motifs are more abundant in certain as yet unstudied human populations.
Some of the strains used in these studies were from patients with peptic ulcer disease, although most were from patients with gastritis only (see Materials and Methods). No correlation of motif type with patterns of disease versus benign infection in a given region was found. That is, the motif types seem to be representative of the cag+ strains of a region, not a reflection of the criteria used at various centers to choose strains for further study. It was particularly striking to us that type I motifs predominated in the largely Amerindian Peruvian population and in Guatemalans, given the Asian ancestry of the native peoples of the Americas, as discussed below.
cagA and vacA DNA sequences.
Two major types of cagA alleles have been found, one in most U.S. and European strains and the other in most east Asian strains (1, 57, 62). These ethnic European and east Asian allelic types were also identified in 19 of our representative strains by sequencing an ∼250-bp segment that had been chosen earlier to distinguish Dutch from Chinese strains (57). In particular, ethnic European type cagA sequences were found in each of 10 Latin American strains with type I motifs (9 Peruvian strains [GenBank accession no. AF198473 to AF198481] and one Guatemalan strain [GenBank accession no. AF198472]) and also in each of four sub-Saharan Africa strains (GenBank accession no. AF198468 to AF198471) that we tested. East Asian type cagA alleles were found in two Hong Kong Chinese strains with type II motifs (GenBank accession no. AF198485 and AF198486), as expected based on prior studies of Shanghai and Guangzhou strains (1, 57). The east Asian type cagA allele was also found in a rare Japanese strain with a type III motif (GenBank accession no. AF198484), and one east Asian type cagA allele and one European type cagA allele were found in two unusual type III motif-containing native Peruvian strains (GenBank accession no. AF198483 and AF198482, respectively).
Separate PCR tests indicated that vacAm1 alleles of H. pylori strains from Peruvian natives contained “ethnic European” vacAm1a motifs, not east Asian vacAm1b motifs (each of 27 strains tested), as did each of 11 Spanish control strains (Fig. 6). In addition, each of 34 vacAm1 alleles from H. pylori from native Africans from The Gambia and South Africa were also of the vacAm1a type. None of these contained the easily distinguished east Asian vacAm1b motifs defined in earlier studies (36, 47). An independent study of the separate vacAs1 region had similarly concluded that vacA alleles of Latin American strains match those of strains from the Iberian Peninsula, not those of strains from east Asia (58). Because vacA and the cag PAI are far apart in H. pylori chromosomes (6, 56), these outcomes strengthen the sense of genetic relatedness between the H. pylori strains of Spain and of Amerindians in Latin America and also between those of south Europe and Africa.
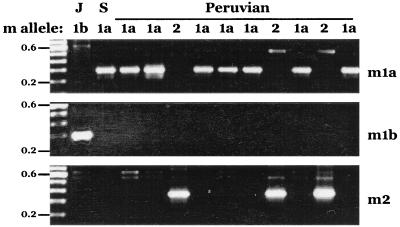
FIG. 6.
PCR analysis of middle-region alleles of vacA (vacuolating cytotoxin gene). Three allelic types were recognized (vacAm1a, -m1b, and -m2) (8, 47). Sizes (0.6 and 0.2 kb) of marker DNAs in the 100-bp ladder are shown. J and S, representative Japanese and Spanish strains, respectively. Other strains for which PCR profiles are shown are from native Peruvians.
DISCUSSION
A highly polymorphic DNA region at the right end of the cag PAI (Fig. 1 and 2) and sequences in the cagA and vacA genes of more than 500 strains of H. pylori from five continents were studied in order to gain insights into the evolution of this gastric pathogen. Five main types of DNA sequence motifs were found at the end of the cag PAI. Each showed evidence of DNA deletion and/or substitution events that had removed a segment extending from within IS606* on one side through part or all of a putative helicase gene on the other. Several end points were evident, implying several different ancient events. The cag PAI, like other such PAIs (34), is thought to have been transferred as a discrete unit by conjugation among bacterial species. Dedicated DNA helicases can be important for such transfer (30), suggesting that a larger, functional version of the hel gene may have contributed to the acquisition of the cag PAI by H. pylori. In this scenario, deletions in the IS606*-hel gene segment might have (i) been selected directly if, e.g., any part of this segment including hel itself were deleterious in H. pylori or (ii) accumulated by attrition, if functions encoded in this segment did not contribute to bacterial fitness in the H. pylori context.
Many type I and III motifs contained miniIS605, in each case inserted at the same site. This distribution could reflect (i) one ancient insertion event, followed by recombinational scrambling among type I and III lineages or (ii) independent insertions of miniIS605 into the same hotspot in each lineage. In either case, the lack of miniIS605 at this site in type II strains is in accord with an idea that the type II motif is distinct phylogenetically from type I and III motifs. This is reinforced by the finding of a 22-bp deletion just downstream of hel sequences in all strains with type II motifs that was not found in any strains with other motif types.
The distributions of informative markers (types of cag PAI right-end sequences and also cagA and vacA alleles) in H. pylori strains in different human populations were studied. It was remarkable that strains from Peruvians of primarily Amerindian ancestry were most similar to those of Spaniards, not those of east Asians, in tests of each marker used (cag right-junction motifs and alleles of cagA and of vacA). The H. pylori strains of Spain were distinct from those of north Europe (Sweden and Lithuania) in their greater abundance of type I motifs. This is in accord with a recent study of vacAs1 alleles and findings that different types predominate in north and south European strains and that vacAs1 alleles of south Europe match those from Colombia and Costa Rica (58). We have also found that motifs in the iceA1 gene of Peruvian strains resemble those of Spanish strains, not those of east Asian strains (Y. Ito, T. Azuma, and D. E. Berg, unpublished data), and have identified two mobile DNA elements that are common in Latin American and European strains but rare in east Asian strains (A. K. Mukhopadhyay, Z. J. Pan, and D. E. Berg, unpublished data).
Because the native peoples of the Americas are of Asian ancestry (albeit most related to people now living in central Asia) (16, 38, 51), a priori we would have expected H. pylori strains of native Peruvians and of at least some Ladinos in Guatemala to be more related to those of east Asia than to those of any other region studied here. To explain the remarkable match of Latin American and Spanish strains, we suggest that H. pylori was brought to the New World by European conquerors some 500 years ago, as had happened with several other microbial pathogens (13, 22, 25, 44). Others had also suggested that current Latin American and European H. pylori strains may be related (58). If this is correct, then H. pylori might not have been endemic in pre-Columbian native Americans, or even in their central Asian ancestors, although there are at least two alternative interpretations for our results: (i) unexpected similarity between the putative ancestral H. pylori strains of central Asia and those of Spain, and (ii) inability of resident Amerindian strains to compete with newly introduced European strains. Possible support for these alternative explanations is found in an abstract on H. pylori-like antigens in 2 of 15 pre-Columbian mummy fecal samples (P. Correa, D. Willis, M. J. Allison, and E. Gerstzon, Abstr. Dig. Dis. Week Meet., abstr. 3155, 1998; also cited in reference 5). Such a result is not definitive, however, because it can be interpreted in several ways, including spurious cross-reaction of the antibodies used with antigens from other microbes in these mummified feces (see, e.g., references 7 and 37).
Any proposal that pre-Columbian Amerindians and their central Asian ancestors were H. pylori free disagrees with an assumption (14, 15) that H. pylori had been nearly universal in humans from our earliest days until the 20th century, when advances in hygiene and medicine began interrupting long-established patterns of transmission and persistent infection. We wish to consider the following alternative model. As with several other human pathogens, H. pylori might have become established in human populations quite recently, possibly in early agricultural societies (less than 10,000 years ago), as the result of close contact between H. pylori-infected domesticated animals or rodent pests and the people who lived closely with them. This model depends on H. pylori being able to jump from animals to humans. A few human H. pylori strains can infect mice (32, 41, 42), and recent studies have indicated that about half of the strains can infect Mongolian gerbils (61; F. Hirayama, personal communication), an apparently much more permissive host. Natural or experimental H. pylori infections of dogs, cats, rats, pigs, and sheep have also been reported (26, 41, 46, 49, 50), and many H. pylori strains can infect at least certain monkey species (11, 27, 28). Thus, although H. pylori is typically considered human specific, the species actually has a broader host range. We propose that the first Amerindians might have been H. pylori free because they were hunter-gatherers not agriculturists, and had crossed to the Americas well before animal-based agriculture began in their ancestral Asian homelands (25, 44).
The uniformity of strains studied to date from native sub-Saharan Africans and the similarity of the strains to those of Spain suggested that African strains might also be of European origin. This would be reminiscent of introductions of several other human pathogens into Africa through European and Asian contacts in recent centuries (24, 25). More analysis of H. pylori genotypes across the vast African continent is needed to better evaluate this possibility.
The great genetic diversity of H. pylori suggests to us that this species may have jumped from animals to people more than once. If this is correct, the geographic differences in H. pylori genotypes found to date (variously in cag PAI right-end motifs and cagA and vacA alleles) might reflect selection and genetic drift in ancient animal hosts and transmission from different animal sources to people in various early societies.
Most of our understanding of H. pylori genome organization and the bacterial traits that are important in colonization and disease is based on studies of strains from ethnic Europeans. Independent of how H. pylori became associated with humans, the dramatic differences found to date between various Asian and European H. pylori populations in at least a few loci should encourage further analyses of strains from relatively understudied geographic regions and human ethnic groups. Such “geographic genomics” may uncover new genes that affect human infection, increase our understanding of bacterium-host interactions in colonization and disease, and provide new insights into the evolution of this diverse and globally distributed human pathogen.
TABLE 2.
Distribution of subtypes of type I strains
| Geographic region | No. (%) of strains of subtype:
|
|||
|---|---|---|---|---|
| Total | Ia | Iba | Icb | |
| Latin America | 89 | 64 (82%) | 25 (28%) | 0 |
| Guatemala | 27 | 21 | 6 | 0 |
| Peru | 62 | 43 | 19 | 0 |
| Europe | 43 | 28 (65%) | 15 (35%) | 0 |
| Spain | 33 | 23 | 10 | 0 |
| Sweden | 7 | 2 | 5 | 0 |
| Lithuania | 3 | 3 | 0 | 0 |
| Africa | 40 | 32 (80%) | 8 (20%) | 0 |
| Gambia | 8 | 8 | 0 | 0 |
| South Africa | 32 | 24 | 8 | 0 |
| United States | 45 | 36 (80%) | 3 (7%) | 6 (13%) |
| Tennessee | 5 | 4 | 1 | 0 |
| Ohio | 11 | 5 | 1 | 5 |
| West Virginia | 7 | 6 | 1 | 0 |
| Missouri | 7 | 7 | 0 | 0 |
| Louisiana | 15 | 14 | 0 | 1 |
| East Asia (Hong Kong) | 1 | 0 | 1 | 0 |
| South Asia (India) | 7 | 2 | 5 | 0 |
Carries miniIS605.
Carries IS606.
ACKNOWLEDGMENTS
We are grateful to M. Asaka and T. Sugiyama (Hokkaido University, Sapporo, Japan) for the gift of some of the Japanese H. pylori strains used here.
This work was supported in part by NIH grants AI38166, DK53727, and TW00611 to D.E.B., P30 DK52574 to Washington University, JSPS-RFTF97L00101 to T.N., and by grants from the Swedish Cancer Society and The Swedish Medical Research Council (11218) to T.B.
REFERENCES
- 1.Achtman M, Azuma T, Berg D E, Ito Y, Morelli G, Pan Z J, Suerbaum S, Thompson S A, van der Ende A, van Doorn L J. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol Microbiol. 1999;32:459–470. doi: 10.1046/j.1365-2958.1999.01382.x. [DOI] [PubMed] [Google Scholar]
- 2.Akopyants N S, Clifton S W, Kersulyte D, Crabtree J E, Youree B E, Reece C A, Bukanov N O, Drazek E S, Roe B A, Berg D E. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol Microbiol. 1998;28:37–54. doi: 10.1046/j.1365-2958.1998.00770.x. [DOI] [PubMed] [Google Scholar]
- 3.Akopyants N S, Fradkov A, Diatchenko L, Siebert P D, Lukyanov S, Sverdlov E D, Berg D E. PCR-based subtractive hybridization and differences in gene content among strains of Helicobacter pylori. Proc Natl Acad Sci USA. 1998;95:13108–13113. doi: 10.1073/pnas.95.22.13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akopyanz N, Bukanov N O, Westblom T U, Kresovich S, Berg D E. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 1992;20:5137–5142. doi: 10.1093/nar/20.19.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allison M J, Bergman T, Gerszten E. Further studies on fecal parasites in antiquity. Am J Clin Pathol. 1999;112:605–609. doi: 10.1093/ajcp/112.5.605. [DOI] [PubMed] [Google Scholar]
- 6.Alm R A, Ling L S, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 7.Andersen L P, Espersen F. Immunoglobulin G antibodies to Helicobacter pylori in patients with dyspeptic symptoms investigated by the Western immunoblot technique. J Clin Microbiol. 1992;30:1743–1751. doi: 10.1128/jcm.30.7.1743-1751.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atherton J C, Sharp P M, Cover T L, Gonzalez-Valencia G, Peek R M, Thompson S A, Hawkey C J, Blaser M J. Vacuolating cytotoxin (vacA) alleles of Helicobacter pylori comprise two geographically widespread types, m1 and m2, and have evolved through limited recombination. Curr Microbiol. 1999;39:211–218. doi: 10.1007/s002849900447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing and Wiley-Interscience; 1993. [Google Scholar]
- 10.Bamford K B, Bickley J, Collins J S, Johnston B T, Potts S, Boston V, Owen R J, Sloan J M. Helicobacter pylori: comparison of DNA fingerprints provides evidence for intrafamilial infection. Gut. 1993;34:1348–1350. doi: 10.1136/gut.34.10.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baskerville A, Newell D G. Naturally occurring chronic gastritis and C. pylori infection in the rhesus monkey: a potential model for gastritis in man. Gut. 1988;29:465–472. doi: 10.1136/gut.29.4.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berg D E, Gilman R H, Lelwala-Guruge J, Valdez Y, Watanabe J, Miyagi J, Akopyants N S, Srivastava K, Ramirez-Ramos A, Yoshiwara T H, Recavarren S, Leon Barua R. Helicobacter pylori populations in individual peruvian patients. Clin Infect Dis. 1997;25:996–1002. doi: 10.1086/516081. [DOI] [PubMed] [Google Scholar]
- 13.Bianchine P J, Russo T A. The role of epidemic infectious diseases in the discovery of America. Allergy Proc. 1992;13:225–232. doi: 10.2500/108854192778817040. [DOI] [PubMed] [Google Scholar]
- 14.Blaser M J. Helicobacters are indigenous to the human stomach: duodenal ulceration is due to changes in gastric microecology in the modern era. Gut. 1998;43:721–727. doi: 10.1136/gut.43.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blaser M J. Hypothesis: the changing relationships of Helicobacter pylori and humans: implications for health and disease. J Infect Dis. 1999;179:1523–1530. doi: 10.1086/314785. [DOI] [PubMed] [Google Scholar]
- 16.Bonatto S L, Salzano F M. A single and early migration for the peopling of the Americas supported by mitochondrial DNA sequence data. Proc Natl Acad Sci USA. 1997;94:1866–1871. doi: 10.1073/pnas.94.5.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 18.Cann H M. Human genome diversity. C R Acad Sci III. 1998;321:443–446. doi: 10.1016/s0764-4469(98)80774-9. [DOI] [PubMed] [Google Scholar]
- 19.Cavalli-Sforza L L, Menozzi P, Piazza A. The history and geography of human genes. Princeton, N.J: Princeton University Press; 1994. [Google Scholar]
- 20.Censini S, Lange C, Xiang Z, Crabtree J E, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. cag, a pathogenicity island of Helicobacter pylori encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chalkauskas H, Kersulyte D, Cepuliene I, Urbonas V, Ruzeviciene D, Barakauskiene A, Raudonikiene A, Berg D E. Genotypes of Helicobacter pylori in Lithuanian families. Helicobacter. 1998;3:296–302. [PubMed] [Google Scholar]
- 22.Cook N D. Born to die: disease and New World conquest, 1492–1650. Cambridge, England: Cambridge University Press; 1998. [Google Scholar]
- 23.Cvacci A, Telford J L, Del Giudice G, Parsonnet J, Rappuoli R. Helicobacter pylori virulence and genetic geography. Science. 1999;284:1328–1333. doi: 10.1126/science.284.5418.1328. [DOI] [PubMed] [Google Scholar]
- 24.Curtin P D. Disease exchange across the tropical Atlantic. Pubbl Stn Zool Napoli II. 1993;15:329–356. [PubMed] [Google Scholar]
- 25.Diamond J. Guns, germs, and steel: the fates of human societies. W. W. New York, N.Y: Norton and Co.; 1997. [Google Scholar]
- 26.Dore M P, Sepulveda A R, Osato M S, Realdi G, Graham D Y. Helicobacter pylori in sheep milk. Lancet. 1999;354:132. doi: 10.1016/S0140-6736(99)01724-9. [DOI] [PubMed] [Google Scholar]
- 27.Dubois A, Berg D E, Incecik E T, Fiala N, Heman-Ackah L M, Perez-Perez G I, Blaser M J. Transient and persistent infection of nonhuman primates with Helicobacter pylori: implications for human disease. Infect Immun. 1996;64:2885–2891. doi: 10.1128/iai.64.8.2885-2891.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dubois A, Berg D E, Incecik E T, Fiala N, Heman-Ackah L M, Del Valle J, Yang M, Wirth H-P, Perez-Perez G I, Blaser M J. Individual host specificity of Helicobacter pylori strains and host responses in experimentally challenged non-human primates. Gastroenterology. 1999;116:90–96. doi: 10.1016/s0016-5085(99)70232-5. [DOI] [PubMed] [Google Scholar]
- 29.Feil E J, Maiden M C, Achtman M, Spratt B G. The relative contributions of recombination and mutation to the divergence of clones of Neisseria meningitidis. Mol Biol Evol. 1999;16:1496–1502. doi: 10.1093/oxfordjournals.molbev.a026061. [DOI] [PubMed] [Google Scholar]
- 30.Firth N, Ippen-Ihler K, Skurray R A. Structure and function of the F factor and mechanism of conjugation. In: Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 2377–2401. [Google Scholar]
- 31.Garner J A, Cover T L. Analysis of genetic diversity in cytotoxin-producing and non-cytotoxin-producing Helicobacter pylori strains. J Infect Dis. 1995;172:290–293. doi: 10.1093/infdis/172.1.290. [DOI] [PubMed] [Google Scholar]
- 32.Guruge J L, Falk P G, Lorenz R G, Dans M, Wirth H-P, Blaser M J, Berg D E, Gordon J I. Epithelial attachment alters the outcome of Helicobacter pylori infection. Proc Natl Acad Sci USA. 1998;95:3925–3930. doi: 10.1073/pnas.95.7.3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guttman D S, Dykhuizen D E. Detecting selective sweeps in naturally occurring Escherichia coli. Genetics. 1994;138:993–1003. doi: 10.1093/genetics/138.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hacker J, Blum-Oehler G, Muhldorfer I, Tschape H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 35.Ilver D, Arnqvist A, Ogren J, Frick I-M, Kersulyte D, Incecik E T, Berg D E, Covacci A, Engstrand L, Boren T. The Helicobacter pylori Lewis b blood group antigen binding adhesin revealed by retagging. Science. 1998;279:373–377. doi: 10.1126/science.279.5349.373. [DOI] [PubMed] [Google Scholar]
- 36.Ito Y, Azuma T, Ito S, Suto H, Miyaji H, Yamazaki Y, Kohli Y, Kuriyama M. Full-length sequence analysis of the vacA gene from cytotoxic and noncytotoxic Helicobacter pylori. J Infect Dis. 1998;178:1391–1398. doi: 10.1086/314435. [DOI] [PubMed] [Google Scholar]
- 37.Johansen H K, Norgaard A, Andersen L P, Jensen P, Nielsen H, Hoiby N. Cross-reactive antigens shared by Pseudomonas aeruginosa, Helicobacter pylori, Campylobacter jejuni, and Haemophilus influenzae may cause false-positive titers of antibody to H. pylori. Clin Diagn Lab Immunol. 1995;2:149–155. doi: 10.1128/cdli.2.2.149-155.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karafet T M, Zegura S L, Posukh O, Osipova L, Bergen A, Long J, Goldman D, Klitz W, Harihara S, de Knijff P, Wiebe V, Griffiths R C, Templeton A R, Hammer M F. Ancestral Asian source(s) of new world Y-chromosome founder haplotypes. Am J Hum Genet. 1999;64:817–831. doi: 10.1086/302282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kersulyte D, Akopyants N S, Clifton S W, Roe B A, Berg D E. Novel sequence organization and insertion specificity of IS605 and IS606: chimaeric transposable elements of Helicobacter pylori. Gene. 1998;223:175–186. doi: 10.1016/s0378-1119(98)00164-4. [DOI] [PubMed] [Google Scholar]
- 40.Kersulyte D, Chalkauskas H, Berg D E. Emergence of recombinant strains of Helicobacter pylori during human infection. Mol Microbiol. 1999;31:31–43. doi: 10.1046/j.1365-2958.1999.01140.x. [DOI] [PubMed] [Google Scholar]
- 41.Lee A. Animal models for host-pathogen interaction studies. Br Med Bull. 1998;54:163–173. doi: 10.1093/oxfordjournals.bmb.a011666. [DOI] [PubMed] [Google Scholar]
- 42.Marchetti M, Arico B, Burroni D, Figura N, Rappuoli R, Ghiara P. Development of a mouse model of Helicobacter pylori infection that mimics human disease. Science. 1995;267:1655–1658. doi: 10.1126/science.7886456. [DOI] [PubMed] [Google Scholar]
- 43.McGee D J, May C A, Garner R M, Himpsl J M, Mobley H L T. Isolation of Helicobacter pylori genes that modulate urease activity. J Bacteriol. 1999;181:2477–2484. doi: 10.1128/jb.181.8.2477-2484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McNeil W H. Plagues and peoples. Garden City, N.Y: Anchor Press/Doubleday; 1976. [Google Scholar]
- 45.Mukhopadhyay A K, Kersulyte D, Jeong J-Y, Datta S, Ito Y, Chowdhury A, Chowdhury S, Santra A, Bhattacharya S K, Azuma T, Nair G B, Berg D E. Distinctiveness of genotypes of Helicobacter pylori in Calcutta, India. J Bacteriol. 2000;182:3219–3227. doi: 10.1128/jb.182.11.3219-3227.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nedrud J G. Animal models for gastric Helicobacter immunology and vaccine studies. FEMS Immunol Med Microbiol. 1999;24:243–250. doi: 10.1111/j.1574-695X.1999.tb01290.x. [DOI] [PubMed] [Google Scholar]
- 47.Pan Z-J, Berg D E, van der Hulst R W M, Su W-W, Raudonikiene A, Xia S-D, Dankert J, Tytgat G N J, van der Ende A. High prevalence of vacuolating cytotoxin production and unusual distribution of vacA alleles in Helicobacter pylori from China. J Infect Dis. 1998;178:220–226. doi: 10.1086/515601. [DOI] [PubMed] [Google Scholar]
- 48.Parsonnet J. Helicobacter and gastric adenocarcinoma. In: Parsonnet J, editor. Microbes and malignancy: infection as a cause of human cancers. New York, N.Y: Oxford University Press; 1999. pp. 372–408. [Google Scholar]
- 49.Perkins S E, Fox J G, Marini R P, Shen Z, Dangler C A, Ge Z. Experimental infection in cats with a cagA+ human isolate of Helicobacter pylori. Helicobacter. 1998;3:225–235. doi: 10.1046/j.1523-5378.1998.08037.x. [DOI] [PubMed] [Google Scholar]
- 50.Rossi G M, Rossi C G, Vitali D, Fortuna D, Burroni L, Pancotto S, Capecchi S, Renzoni G, Braca G, Del Giudice G, Rappuoli R, Ghiara P, Taccini E. A conventional beagle dog model for acute and chronic infection with Helicobacter pylori. Infect Immun. 1999;67:3112–3120. doi: 10.1128/iai.67.6.3112-3120.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Santos F R, Pandya A, Tyler-Smith C, Pena S D, Schanfield M, Leonard W R, Osipova L, Crawford M H, Mitchell R J. The central Siberian origin for native American Y chromosomes. Am J Hum Genet. 1999;64:619–628. doi: 10.1086/302242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith J M, Smith N H, O'Rourke M, Spratt B G. How clonal are bacteria? Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suerbaum S, Maynard Smith J, Bapumia K, Morelli G, Smith N H, Kuntsmann E, Dyrek I, Achtman M. Free recombination within Helicobacter pylori. Proc Natl Acad Sci USA. 1998;95:12619–12624. doi: 10.1073/pnas.95.21.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taylor N S, Fox J G, Akopyants N S, Berg D E, Thompson N, Shames B, Yan L, Fontham E, Janney F, Hunter F M, Correa P. Long term colonization with single and multiple strains of Helicobacter pylori assessed by DNA fingerprinting. J Clin Microbiol. 1995;33:918–923. doi: 10.1128/jcm.33.4.918-923.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tee W, Lambert J, Smallwood R, Schembri M, Ross B C, Dwyer B. Ribotyping of Helicobacter pylori from clinical specimens. J Clin Microbiol. 1992;30:1562–1567. doi: 10.1128/jcm.30.6.1562-1567.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzegerald L M, Lee N, Adams M D, Hickey E K, Berg D E, Gocayne J D, Utterback T R, Peterson J D, Kelley J M, Cotton M D, Weidman J M, Fujii C, Bowman C, Watthey L, Wallin E, Hayes W S, Borodovsky M, Karp P D, Smith H O, Fraser C M, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 57.van der Ende A, Pan Z J, Bart A, van der Hulst R W, Feller M, Xiao S D, Tytgat G N, Dankert J. cagA-positive Helicobacter pylori populations in China and The Netherlands are distinct. Infect Immun. 1998;66:1822–1826. doi: 10.1128/iai.66.5.1822-1826.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Doorn L J, Figueiredo C, Megraud F, Pena S, Midolo P, Queiroz D M, Carneiro F, Vanderborght B, Pegado M D, Sanna R, De Boer W, Schneeberger P M, Correa P, Ng E K, Atherton J, Blaser M J, Quint W G. Geographic distribution of vacA allelic types of Helicobacter pylori. Gastroenterology. 1999;116:823–830. doi: 10.1016/s0016-5085(99)70065-x. [DOI] [PubMed] [Google Scholar]
- 59.van Doorn L J, Figueiredo C, Sanna R, Blaser M J, Quint W G. Distinct variants of Helicobacter pylori cagA are associated with vacA subtypes. J Clin Microbiol. 1999;37:2306–2311. doi: 10.1128/jcm.37.7.2306-2311.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Westblom T U, Czinn S J, Nedrud J G. Gastroduodenal disease and Helicobacter pylori: pathophysiology, diagnosis and treatment. Curr Top Microbiol Immunol. 1999;241:1–313. [Google Scholar]
- 61.Wirth H P, Beins M H, Yang M, Tham K T, Blaser M J. Experimental infection of Mongolian gerbils with wild-type and mutant Helicobacter pylori strains. Infect Immun. 1998;66:4856–4866. doi: 10.1128/iai.66.10.4856-4866.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamaoka Y, Kodama T, Kashima K, Graham D Y, Sepulveda A R. Variants of the 3′ region of the cagA gene in Helicobacter pylori isolates from patients with different H. pylori-associated diseases. J Clin Microbiol. 1998;36:2258–2263. doi: 10.1128/jcm.36.8.2258-2263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]