Abstract
Chronic lower respiratory tract infections are a leading contributor to morbidity and mortality in persons with cystic fibrosis (pwCF). Traditional respiratory tract surveillance culturing has focused on a limited range of classic pathogens; however, comprehensive culture and culture-independent molecular approaches have demonstrated complex communities highly unique to each individual. Microbial community structure evolves through the lifetime of pwCF and is associated with baseline disease state and rates of disease progression including occurrence of pulmonary exacerbations. While molecular analysis of the airway microbiome has provided insight into these dynamics, challenges remain including discerning not only “who is there” but “what they are doing” in relation to disease progression. Moreover, the microbiome can be leveraged as a multi-modal biomarker for both disease activity and prognostication. In this article, we review our evolving understanding of the role these communities play in pwCF and identify challenges in translating microbiome data to clinical practice.
Keywords: biomarker, bronchiectasis, lung, microbiota, Pseudomonas aeruginosa, review
In healthy airways, until recently considered sterile, microaspirated microbiota shapes early colonizing communities through the balance of microbial immigration, colonization, and subsequent elimination (Table 1). In cystic fibrosis (CF), microbial elimination, as a function of mucociliary clearance and host defense, is critically impaired. Consequently, respiratory infections and associated inflammation are the primary contributors of morbidity and mortality for persons with CF (pwCF) [1]. With advancements in molecular technologies, it has become increasingly apparent that CF airways are colonized by a community much larger than merely those identified through routine clinical laboratory protocols [2–4]. In this review, we explore the complexity of these polymicrobial communities and their potential for impact on CF pathogenesis.
Table 1.
Terminology and Nomenclature of the CF Microbiotaa
| Term | Definition |
|---|---|
| Ecological | |
| Microbiota | The entire collection of microbial organisms at a particular site. |
| Microbiome | Defined as a characteristic microbial community occupying a reasonably well-defined habitat that has distinct physicochemical properties. The term thus not only refers to the microorganisms involved but also encompasses their theater of activity. |
| Mycome | Collective genomes and gene products of fungi within and on humans. |
| Virome | Collective genomes and gene products of viruses within and on humans. |
| Diversity | General term used to describe the number of different species of microbes present and their distribution within an ecosystem. |
| Dysbiosis | Imbalance of ecological homeostasis and loss of diversity in microbial communities often associated with disease states or acute antibacterial therapies. Characterized by altered bacterial landscapes, pathogen domination, and colonization resistance. |
| Metagenome | Collection of genomes within members of the microbiota (ie, what functional genes are present—but cannot determine who or what is active). |
| Transcriptome | High-throughput process to identify and quantify microbial genes expressed by the microbiota (ie, who is active and expressing genes). |
| Metabolome | Analysis of the complete set of metabolites present in a population (ie, what end-products, such as short-chain fatty acids, are present). |
| Resistome | Collection of all genes from pathogenic and commensal organisms associated with antibiotic resistance. |
| Multi-omics | Assimilation of data from various “omics” technologies, such as microbiomic, metagenomic, transcriptomic, and metabolomic. |
| Factors shaping the microbiome | |
| Immigration | The movement of microbes into a new environment. For example, in CF, this may be seen in the context of lower airways that includes aspiration, subclinical microaspiration, and inhalation of microbes leading to direct dispersal across airway mucosa. |
| Elimination | The movement of microbes out of an environment. For example, in CF, this may be seen in the context of lower airways done through adjunctive airways clearance measures, antimicrobial therapies, cough, and host immune defenses. |
| Relative reproduction | Bacterial growth influenced by regional growth conditions, including (i) environmental (ie, nutrient availability, temperature, pH, and oxygen tension), (ii) host (ie, concentration and activation of inflammatory cells), and (iii) bacterial (ie, local microbial composition/competition). |
| Methodology | |
| 16S Ribosomal RNA (rRNA/rDNA) gene | Amplification and sequencing of part of the 16SrRNA gene (SSU rRNA gene), typically including ≥1 hypervariable region(s) that can provide taxonomic resolution of the community structure. |
| Shotgun sequencing | Direct sequencing and analysis of total DNA extracted from a sample. This approach provides information on all genes present and can provide genome scale information on the more abundant community members. |
| Culture-independent | Analysis of the microbiome based on nucleic acid extracted directly from a sample (eg, 16S rRNA gene profiling, metagenomics, metatranscriptomics). |
| Culture-enriched metagenomics | Coupling culture enrichment methods with shotgun metagenomic approaches to improve the resolution of community analysis. |
| Operational taxonomic unit (OTU) | Clusters of similar sequence variants of the 16S rRNA gene used to identify taxa. 97% similarity is commonly used as a species-specific cutoff. |
| Amplicon sequence variant (ASV) | Alternative to OTUs. Infers the biological sequences prior to the introduction of amplification and sequencing errors. ASVs offer higher sensitivity to biological variation, as a change in one nucleotide in the 16S rRNA gene of a bacterial strain can indicate large variations within the rest of the genome relative to OTU. |
| Analysis | |
| Abundance | Total number of bacteria within a sample. |
| Relative abundance | Proportion of the microbiome made up of specific bacteria (ie, more dominant bacteria have higher relative abundances). Often denoted as a percentage or proportion (0-1). |
| Absolute abundance | Actual abundance of a taxon in a unit volume of an ecosystem (ie, a measure of bioburden). |
| Alpha-diversity | A measure of the composition of microbial community (single sample) based on richness (number of species) and may include measures of evenness (different abundances of community members). Common alpha-diversity measurements include observed species, Chao 1, Shannon Diversity Index, and/or Simpson’s Index. |
| Beta-diversity | A measure of the differences in community composition inclusive of taxonomy between samples (eg, longitudinal within a subject, or between subjects). The measures can be based on the presence/absence (unweighted) or different abundances of community members(weighted) and some measures incorporate phylogenetic relatedness within a community. Common beta-diversity metrics include Weighted- and Unweighted-Unifrac, Aitchison Distance, and Bray Curtis Dissimilarity). |
| Core microbiome | Group that contains species that affect a large proportion of individuals with high relative abundance. |
| Satellite microbiome | Group that contains species that are present in low relative abundance and at limited locations. Often detected infrequently, may be transient. |
| Pulmotype | Partitioning of airway bacterial communities into distinct types across patients. |
Adapted from references [5–10].
EVALUATION OF THE CF MICROBIOME: EVOLVING SAMPLING STRATEGIES
To unravel the microbiome’s role in CF pathogenesis, a comprehensive understanding of the microbial milieu and sampling modalities is necessary. The challenge of adequate sample collection for infection surveillance has long been recognized as it pertains to children and those with mild lung disease; however, this is further convoluted with molecular approaches to microbiome studies. Culture-independent microbiome studies are complicated by the lower respiratory tract microbiome being a low-microbial biomass system with high host DNA (>90% DNA is from neutrophils), resulting in very low depth of sequencing of microbial DNA, unlike other sites including the gastrointestinal tract [11]. Moreover, the respiratory environment is changing for many pwCF with the widespread use of cystic fibrosis transmembrane conductance (CFTR) modulators (Figure 1).
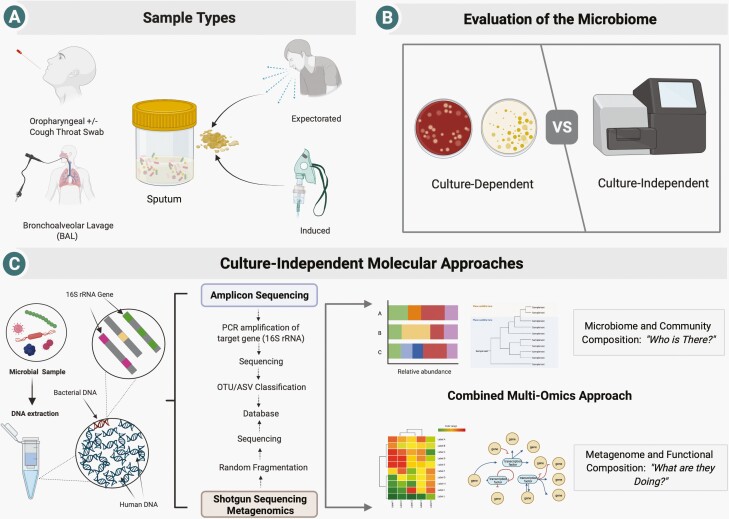
Figure 1.
Establishing the structure and composition of the CF microbiome. (A) A range of respiratory sample types can be assessed, each of which differs with respect to its ease of collection, sensitivity, specificity, and relevance to the lower airways. (B) Samples can be assessed using routine and augmented culture protocols to identify specifically targeted organisms (which allow for pathogen characterization) or an agnostic approach in which next-generation sequencing is used to define the entirety of community constituents. (C) After DNA extraction, microbial communities can be defined based on establishing their gene content either using amplicon (16S ribosomal RNA) amplification or shotgun sequencing (±host DNA depletion strategies) enabling downstream analysis. Figure created with Biorender. Abbreviation: CF, cystic fibrosis.
Bronchoalveolar lavage (BAL) is the gold standard for lower respiratory tract surveillance [12] but limited by its invasive nature, requirement for sedation, and cost—all of which preclude serial assessment. In CF, bronchoscopy is primarily utilized in children who often cannot spontaneously produce sputum [13]. However, even in this highest needs population, it has not been associated with improved outcomes [14].
Sputum, composed of lower airway-derived mucus plugs, is the mainstay of microbiome analysis and offers several advantages including its non-invasive nature and ease of collection in those who expectorate, thereby lending itself to serial collection, particularly important in longitudinal studies [15]. Given that sputum inherently represents a mixture of upper and lower airway microbiota, critiques include traveling through the upper respiratory tract with inevitable contamination by oropharyngeal microbiota. A study of end-stage pwCF identified microbial communities from explanted lungs as less diverse than expectorated sputum on the day of transplantation [16]. However, others have demonstrated sputum as representative of the lower airway microbiota established from BAL [17]. Indeed, paired samples of sputum and saliva found only modest community overlap, where expectorated sputum contained higher bacterial loads, lower richness, and less diversity [18].
Oropharyngeal swabs have been used as a surrogate for lower airway bacteria identification; however, the utility of routine use has been complicated by insensitivity and disproportionate recovery of oral commensal microbiota [19]. Cough swabs are easy to collect and have long been used as a pediatric surveillance tool [20, 21]. Recently, paired cough swabs and sputum samples from a cohort of pediatric and adult pwCF were assessed for microbiome composition and validity between testing modalities [22]. Despite similar diversity measures, poor concordance between swabs and sputum to discern CF pathogens was observed. Inducing sputum production with hypertonic saline is an alternative technique with good bacteriologic correlation to BAL in both children [21, 23] and adults [24], and offers superior detection of pathogens compared to cough swabs [21]. Moreover, induced sputum microbiome composition closely resembles that of expectorated sputum [19]. Taken together, sputum (expectorated or induced) may be considered as an acceptable, safe, and minimally invasive respiratory microbiome sampling strategy.
WHO ARE THE MICROBIAL PLAYERS?
For much of the 80 years since the initial description of CF, microbiologists have focused on a narrow range of canonical pathogens including Pseudomonas aeruginosa, Staphylococcus aureus, Haemophilus influenzae, and the Burkholderia cepacia complex. Work by Rogers et al first identified bacterial species not previously associated with CF airways, setting the stage for rapid scientific advancement [2, 25]. With more comprehensive culture methods and newer culture-independent-based molecular approaches, we now appreciate that CF respiratory tract samples reflect complex and dynamic microbial communities (Figure 2).
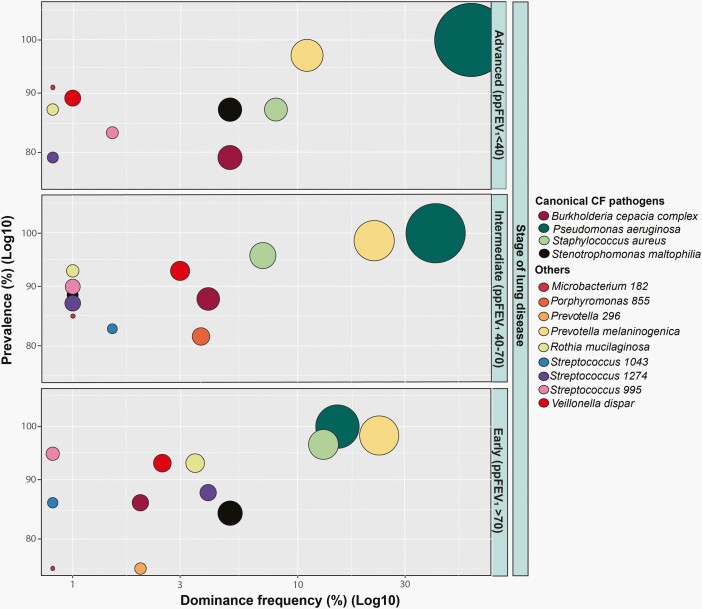
Figure 2.
The core constituents of the CF microbiome by lung disease stage. Data presented correspond to the prevalence (%) and dominance frequency (%) of canonical CF pathogens and other members of the CF microbiota found in a multicenter cohort of 297 pwCF respiratory samples from the study described by Cuthbertson et al [26]. pwCF and their microbiota are stratified by stage of lung disease: early (percentage predicted (ppFEV1) > 70) (n = 57), intermediate (ppFEV1 40-70) (n = 139), and advanced (ppFEV1 < 40) (n = 101). Prevalence for each taxon was defined as the proportion of patients in which a given taxon was detected for each stage of lung disease. Dominance frequency was defined as the percentage of samples that had a particular taxon as the most abundant. Size of the different taxa shown represents the median relative abundance (RA) across the samples for each stage with the lowest value corresponding to RA = 0 and maximum of RA = 40. Both prevalence and dominance frequency are on a log10 axis. Abbreviations: CF, cystic fibrosis; pwCF persons with cystic fibrosis.
Using a range of media and growth conditions, Sibley et al demonstrated the majority of bacteria present can be cultured (43 of the 48 families, with those recovered solely by culture-independent approaches present at very low abundance) [27]. Whelan et al utilized culture-enriched molecular profiling to culture ~80% of operational taxonomic units (OTUs) identified by molecular sequencing in sputum samples, representing >99% of the relative abundance (RA) identified from sequencing [28]. Moreover, culture enrichment identified over 60% more OTUs than identified by direct sequencing—highlighting the utility of integrated approaches (Table 1).
Diversity is often maintained in patients with stable respiratory function and decreased in patients with deteriorating lung function over time [29]. Not surprisingly, the microbial community in advanced CF disease is particularly skewed with multiple studies having established a pattern of decreasing microbial diversity [30–35] and increasing RA in dominant taxa by traditional pathogens with increasing age and severity of lung disease [26, 29, 36, 37]. For instance, as P. aeruginosa colonization becomes chronic (often in late adolescence and early adulthood), community richness and diversity are lost and these changes are associated with disease progression [29, 38]. The emergence of canonical pathogens as dominant community members is a harbinger of advanced disease and postulated to be driven in part by frequent/recurrent antibiotic exposure in response to pulmonary exacerbations (PEx) [1, 29, 39].
The lungs are an oxygen-rich environment; however, in chronic inflammatory lung disease regions of hypoxia develop within infected airways, further compounded in CF due to thickened respiratory secretions and mucus plugs [40]. Consequently, anaerobic bacteria, once attributed to simply oropharyngeal contamination [41] are prevalent in the lungs of pwCF, including Prevotella, Veillonella, Streptococcus, Fusobacterium, Atopobium, Peptostreptococcus, and Porphyromonas (Figure 2) [42]. While ample studies have established that these organisms can colonize the airways of patients at densities comparable to canonical pathogens [27, 43–45], how they might influence pathogenesis remains controversial [46]. Muhlebach et al found that both the presence and RA of anaerobes were associated with milder disease, including improved lung function [47]. In contrast, harmful associations have been observed including increased anaerobe abundance correlating to PEx occurrence [3, 30–33, 48]. Anaerobes have also been identified as carriers of antibiotic resistance genes relevant to therapeutics commonly employed in CF [49, 50] including genes encoding several β-lactamases which may impart protection to neighboring organisms [44, 50, 51] (Figure 3). Finally, anaerobic metabolism facilitates the production of several pro-inflammatory short-chain fatty acids (SCFAs) that may further amplify host immune responses [52].
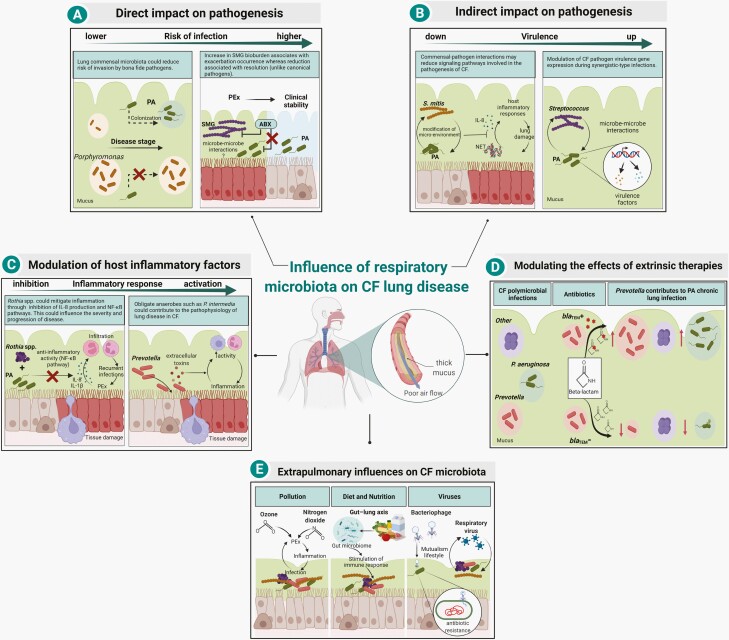
Figure 3.
Mechanisms by which respiratory microbiota influence CF lung disease. (A) Members of the CF microbiota have been associated with risk of infection in the airways. Colonization of commensal microbiota, such as Porphyromonas catoniae was found as a biomarker associated with a lower risk of P. aeruginosa (PA) early infection in CF [53]. In contrast, infection of Streptococcus milleri/anginosus group (SMG) at the onset of pulmonary exacerbations (PEx) is associated with symptomatic deterioration in clinical status, whereas its relative reduction is associated with symptom resolution (unlike canonical pathogens, such as P. aeruginosa) [3, 54]. (B) Commensal bacteria may negatively or positively influence the virulence of CF pathogens. Co-infection models in human epithelial cell lines with P. aeruginosa and commensal CF microbiota have shown that different strains of Streptococcus mitis reduce P. aeruginosa-induced inflammation through reduction of interleukin 8 (IL-8) production and neutrophil extracellular trap (NET) formation. The mechanism of action is still unknown but thought to be through modification of the micro-environment by metabolism adjustment by the commensal bacteria [55]. In contrast, some oral commensal streptococci enhance P. aeruginosa pathogenicity by increasing its virulence factor expression (eg, pyocyanin and elastase) [4, 56, 57]. (C) The CF microbiota contain bacteria with immunomodulatory activity that may alter host inflammatory response, which in turn could influence the progression of lung disease. Rothia mucilaginosa potentially mitigates host inflammation through the inhibition of the IL-8 production and NF-κB pathway activation in a human lung epithelial cell line [58]. Conversely, Prevotella intermedia was reported to be able to contribute to disease progression by secretion of cytotoxic extracellular toxins that induce the influx of macrophages and neutrophils in the airway lumen [59]. (D) CF microbiota influence disease through the modulation of extrinsic therapies. Extended-spectrum β-lactamases (ESBLs)-producing Prevotella isolates were reported to influence pathogenesis in vitro by shielding pathogens, such as P. aeruginosa from the action of β-lactam antibiotics [51]. (E) CF microbiota may be affected by a range of external factors, including pollution, diet, and viruses. Pollution may play a role in triggering PEx, leading to microbiota changes and further airway irritation and injury, which consequently could affect the extent of respiratory infections [60]. In the gut-lung axis, diet plays an important role in shaping the composition of the gut microbiota. Metabolites produced by the gut microbiota not only modulate gastrointestinal immunity but also impact immune responses in the lung [61, 62]. Bacteriophages may impact the fitness of members of the CF microbiota through horizontal gene transfer (HGT) of antimicrobial resistance genes [63]. Additionally, the progression of lung disease is influenced by infection with respiratory viruses which could indirectly promote community changes and host response [64]. Figure created with Biorender. Abbreviations: CF, cystic fibrosis; NF-κB, nuclear factor kappa B.
While beyond the scope of this review, both viruses and fungi exist within the microbiome of CF. Respiratory viruses are common and detected in 13%-60% of CF sputum samples, predominantly in children [65, 66] and are frequently identified as potential triggers of PEx. Viral infections are associated with poorer response to treatment, greater deterioration in lung function, and reduced time to next PEx [67]. Fungi, particularly Aspergillus and Candida spp, are frequently recovered from sputum in pwCF but their role in pathogenesis remains unclear [68].
USING MOLECULAR METHODS TO UNRAVEL THE COMPLEXITY OF THE MICROBIOME
Complex ecosystems are more than simply the “sum of their parts” and require evaluation beyond the presence or absence of individual species. Long-term decreases in community microbial diversity are clearly associated with worse lung function [69]; however, short-term dynamics are less clear. Identifying differences at the transition point between clinical stability and PEx has been a sought-after microbiologic mechanism to explain disease progression. While many groups report community structure transiently disrupted during antimicrobial treatment and/or PEx with a return to baseline after discontinuation [69–71], others suggest microbial community shifts can occur at the onset of PEx even preceding antimicrobial therapy [30, 32]. Moreover, even among canonical pathogens, such as P. aeruginosa, there is little evidence that bacterial density changes during PEx [31, 72–74] and the degree of reduction in bacterial load following antibacterial therapy does not correlate with clinical outcomes [75, 76].
To date, the majority of CF respiratory microbiota studies utilize sputum collected at clinically relevant time points (ie, during regular quarterly clinical visits, or acute need, such as that at the outset of PEx) [31, 36]. Unlike longitudinal studies, cross-sectional and observational studies cannot determine microbiome predictors of PEx, nor can they capture dynamic changes during periods of PEx. Carmody et al collected daily sputum samples from 4 pwCF over a 25-day timeframe, including at initiation of PEx, and identified clear changes in the microbiome at PEx onset in a subset of participants [77]. Cuthbertson et al evaluated 10 pwCF across multiple time points longitudinally, including pre- and post-exacerbation, observing a relatively resilient core microbiota resistant to PEx and associated antimicrobial treatments regardless of clinical status [38].
METAGENOMIC ANALYSIS OF THE RESPIRATORY MICROBIOME
Much of the CF microbiome analysis to date has been carried out by 16S rDNA profiling, providing taxonomic composition of the community, but lacking information on species/strain diversity or important clinical features including virulence and antibiotic resistance. Culture-enriched metagenomics allows for greater depth of sequencing of the CF microbiome and provides greater sensitivity than culture-independent methods alone [28]. Shotgun metagenomics could address the limitations of 16S rDNA profiling; however, it is confounded by two factors: high concentration of host DNA in sputum and a significant burden of bacterial DNA from dead cells. Practical sequencing depths necessary to get comprehensive data requires methods to reduce host DNA, which may also help deplete extracellular microbial DNA. Lysing host cells followed by DNAse treatment [71] or depletion of human DNA using host methylation-specific-binding proteins [78] are the most common approaches. These methods reduce human DNA reads in the metagenomic data although there remains significant room for improvement. As a result, there are currently only a small number of metagenomic studies, most of which still have human DNA accounting for ≥80% of reads. These studies have largely focused on taxonomic profiling with the improved resolution possible by metagenomics and largely agreeing with the 16S rDNA profiling [71, 79–81]. These studies are starting to provide high-resolution mapping of sequence variants in the most abundant organisms with metagenomic assembled genomes [11, 71, 78]. Further reductions in sequencing costs (for both short and long-read sequencing), improved methods for enrichment of microbial DNA, and improvements in bioinformatics tools should manifest in a significant increase in metagenome-focused CF studies.
POLYMICROBIAL INTERACTIONS AND VIRULENCE
The lower airways can be evaluated from a polymicrobial perspective given abundant microbe-microbe and microbe-host-pathogen interactions (Figure 3) [82]. Several animal and in vitro studies have demonstrated microbial interactions contributing to pathogenesis potential. For example, increased virulence activity of P. aeruginosa in the presence of what are generally considered benign commensal microbiota is partially mediated by both the general bacterial signaling molecule AI-2 [4, 56] and 2,3-butanediol [83] metabolic cross-feeding of P. aeruginosa by Rothia [84]. Crosstalk may be observed between pathogens, such as where P. aeruginosa senses bacterial components of the S. aureus cell wall to upregulate virulence [85]. Moreover, it is also likely that we underestimate the virulence potential of some upper respiratory tract microbiota that are present in the lungs. Understanding and being able to discern these interactions would allow targeting of the partnering organisms where conventional antibiotic therapy is not effective.
Microbial metabolites produced locally in the airways or from the gut, such as SCFAs, can affect host responses, although there are conflicting data as to whether these are net beneficial vs harmful [86]. Although most studies to date have identified interactions that increase virulence of CF pathogens, it is expected that antagonistic interactions abound. Recently, in vitro data have demonstrated that several commensal isolates of Streptococcus mitis and Streptococcus oralis from sputum can reduce pro-inflammatory responses of patient-derived airway epithelial cells to P. aeruginosa [55]. Importantly, these findings were strain, not species, specific further highlighting the need for detailed profiling beyond 16S community level in the CF microbiome. No clear mechanism was identified, but active strains of S. mitis contained distinct genes absent in strains failing to suppress inflammation. At other mucosal sites, colonization resistance mediated by direct commensal-pathogen inhibition, largely mediated by bacteriocins, is prevalent [87] but has not been explored thoroughly in the CF microbiome.
THE MICROBIOME AS A BIOMARKER: FORECAST OF OUTCOMES AND TREATMENT RESPONSE
Recently, more groups have established and begun to interrogate CF-specific biobanks, enabling longitudinal studies to better understand host outcomes as a function of their CF microbiota. One particularly important goal of microbiome research is the identification of biomarkers to predict short (ie, PEx) and long-term outcomes (ie, lung function decline), and treatment response. This is particularly relevant as existing CF microbiology protocols used to guide clinical interventions (ie, routine culture and susceptibility testing) poorly correlate with clinical outcomes, creating a strong appetite for novel infection-based biomarkers that better correlate with clinical outcomes [88, 89].
Acosta et al assessed sputum from 104 pwCF to understand how features of the microbiome correlated with future clinical outcomes [90]. Whereas traditional microbiological endpoints, including the presence of canonical pathogens, failed to correlate with clinical outcomes, several measures of the microbiota (reduced alpha-diversity, enrichment of Pseudomonas, and depletion of Streptococcus) were associated with progression to end-stage lung disease and disproportionate FEV1 decline. Notably, the RA of Pseudomonas and Stenotrophomonas (as opposed to their mere presence or absence in aerobic culture) were associated with decline, suggesting that culture alone may lack the ability to discern the role of those agents. Efforts to incorporate machine learning to augment predictive models are underway but already demonstrate promise [91].
The CF Microbiome-determined Antibiotic Therapy Trial in Exacerbations Study (CFMATTERS) was the first prospective multicenter randomized controlled study intended to assess if antibacterial therapy prescribed on the basis of microbiome composition (collected months earlier) would improve outcomes of PEx [92]. While not fully reported, the empiric addition of a “microbiota-targeting agent” (predicted to have activity against the top four most abundant organisms) to standard of care (tobramycin and either ceftazidime or aztreonam) did not result in improved FEV1 recovery after PEx [5, 93]. While subject to the same bias as other randomized studies assessing novel CF microbiology-directed treatment algorithms [94, 95] (ie, decisions that are based on sputum collected potentially months preceding PEx and therefore not necessarily reflective of community composition at PEx), this study demonstrated for the first time in a large multicenter cohort that large-scale prospective microbiome-based intervention studies are indeed possible, with hopefully more to follow.
An abundance of clinical trial and real-world evidence have established nebulized antibiotics as cornerstones of CF maintenance, improving the health and well-being of pwCF [96, 97]. While cycled therapies (licensed in 28 days on/off increments) induce transient reductions of P. aeruginosa [98], patient improvements do not generally correlate with changes in bioburden, suggesting additional “off-target” effects may exist. As inhaled antibiotics achieve exceedingly high concentrations within the CF airways, it was postulated that a range of microbial constituents are affected beyond P. aeruginosa. Accordingly, investigators have sought to understand the relationship between CF microbiome and inhaled anti-Pseudomonal agents. In a cohort of pwCF treated with nebulized aztreonam, Heirali et al did not observe changes in microbiome community structure or composition during treatment with aztreonam but did observe improvements in lung function [97] and quality of life indices [96] in persons with communities deplete for Staphylococcus. In contrast, a retrospective analysis of 41 pwCF naive to inhaled tobramycin found individuals demonstrating FEV1 improvements had communities that clustered together and were disproportionately enriched with Staphylococcus [99]. Whereas both aztreonam and tobramycin have broad aerobic gram-negative antibacterial activity, only the latter also has potent anti-S. aureus activity—suggesting that chronic suppression of Staphylococcus may be important, and strategies focusing inhaled antibiotics on individuals chronically infected with P. aeruginosa may be inappropriately exclusionary. Nelson et al similarly observed in a prospective cohort using a combination of qPCR and metagenomic sequencing that tobramycin primarily induced changes in “off-target” non-dominant community members and not P. aeruginosa [71]. These observations yield hope that the microbiome may be used to identify previously unrecognized organisms as contributors to disease pathogenesis, potentially serving as a biomarker that can be adapted to personalize chronic suppressive antibacterial therapies.
Several barriers limit the potential of adapting microbiome-based learnings to the clinic setting. Samples collected in the context of new systemic illness and/or new acute antibiotics may confound interpretation and should be avoided. Day-to-day variation in sputum composition has been observed in those few individuals followed for protracted periods of time, suggesting caution about inferring too much from any individual sample [100]. Relative to culture-based identification, molecular analysis is costly, slow, and requires considerable technical and bioinformatics expertise. Most importantly, the highly individualized nature of the CF microbiome means that a one-solution-for-all approach is unlikely, and that adequately powered studies will be required to discern complex relationships.
CFTR MODULATORS AND THE MICROBIOME
The use of highly effective CFTR modulators, designed to target the underlying genetic defect and improve protein function, has dramatically improved the well-being of many pwCF. Lung function can increase 3%-14% within 4 weeks of initiation [101]. However, even with these improvements in lung function, structural lung damage remains. Persistent infections have been identified in pwCF after modulator therapy [102–104] despite observations of restructured microbiomes [105]. In a study of 31 pwCF (with ≥1 G551D mutation) pre- and post-ivacaftor therapy, no significant changes in diversity, specific bacterial pathogens, or markers of inflammation were observed [102]. Similarly, neither total bacterial load nor the presence of Pseudomonas changed significantly. In contrast, one group used quantitative culture to demonstrate ivacaftor reduced both P. aeruginosa density and associated lung inflammation [103]. In recent work by Sosinski et al sputum microbiome diversity and evenness were increased in 24 pwCF (with ≥1 F508del mutation) pre- and post-elexacaftor-tezacaftor-ivacaftor (ETI) therapy but with no specific microbial taxa changes other than the log-ratio of canonical CF pathogens to anaerobes [105]. Furthermore, and consistent with almost all other longitudinal studies, microbiome structure is more similar within an individual pre- and post-treatment than between-subject after-modulator initiation.
The long-term sequelae of modulators on the composition of the microbiome are unknown—in fact, rebounding of P. aeruginosa density has been observed during the second year of treatment with re-emergence of strains that were transiently not cultured immediately after the initiation of ivacaftor in a small study [103]. The PROMISE study (NCT04038047), a large US multidisciplinary prospective study on the broad impacts of long-term ETI therapy in pwCF aged 6 years and older aims to clarify some of these questions raised around durability of modulator-related effects by evaluating sputum microbiology and quantitative measures of targeted pathogens serially over 24 months [106]. Management of chronic airway infections is critical to the care of pwCF; thus, understanding the effects of modulators on the microbiome is of utmost importance and an area for further exploration.
FUTURE OF THE MICROBIOME IN CF MANAGEMENT
Taken together over the course of multiple molecular taxonomic [25, 28, 29, 69, 100, 107, 108] and metabolomic [109] studies, a clear message is apparent: microbial composition of the CF respiratory tract is highly personalized and may variably contribute to patient outcomes by a range of mechanisms. The expansion of multi-omic approaches, including microbiomic, metabolomic, and transcriptomic analysis has provided insight into both taxonomic and functional processes. Individual microbiome fluctuations could potentially be used as a prognostic tool to delineate not only onset of PEx, but also severity and future risk. The use of culture-independent data, such as RA and measures of diversity, may in the future add another dimension to the care of pwCF to enable stratification of those at highest risk of future negative outcomes and allow targeted clinical interventions. Furthermore, a greater understanding of the microbiome’s role in CF pathogenesis may enable strategies to manipulate community structure and thereby impart benefits to pwCF.
Note
Supplement sponsorship. This supplement was sponsored by the Cystic Fibrosis Foundation.
Potential conflicts of interest. All authors: No potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Contributor Information
Christina S Thornton, Department of Pediatrics, University of Michigan, Ann Arbor, Michigan, USA; Department of Medicine, Cumming School of Medicine, University of Calgary, Calgary, Alberta, Canada.
Nicole Acosta, Department of Microbiology, Immunology and Infectious Diseases, University of Calgary, Calgary, Alberta, Canada.
Michael G Surette, Department of Biochemistry and Biomedical Sciences, McMaster University, Hamilton, Ontario, Canada; Department of Medicine, McMaster University, Hamilton, Ontario, Canada.
Michael D Parkins, Department of Medicine, Cumming School of Medicine, University of Calgary, Calgary, Alberta, Canada; Department of Microbiology, Immunology and Infectious Diseases, University of Calgary, Calgary, Alberta, Canada.
References
- 1. Lipuma JJ. The changing microbial epidemiology in cystic fibrosis. Clin Microbiol Rev 2010; 23:299–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rogers GB, Hart CA, Mason JR, Hughes M, Walshaw MJ, Bruce KD.. Bacterial diversity in cases of lung infection in cystic fibrosis patients: 16S ribosomal DNA (rDNA) length heterogeneity PCR and 16S rDNA terminal restriction fragment length polymorphism profiling. J Clin Microbiol 2003; 41:3548–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sibley CD, Parkins MD, Rabin HR, Duan K, Norgaard JC, Surette MG.. A polymicrobial perspective of pulmonary infections exposes an enigmatic pathogen in cystic fibrosis patients. Proc Natl Acad Sci USA 2008; 105:15070–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Duan K, Dammel C, Stein J, Rabin H, Surette MG.. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol Microbiol 2003; 50:1477–91. [DOI] [PubMed] [Google Scholar]
- 5. Lee AJ, Einarsson GG, Gilpin DF, Tunney MM.. Multi-omics approaches: the key to improving respiratory health in people with cystic fibrosis? Front Pharmacol 2020; 11:569821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cox MJ, Cookson WO, Moffatt MF.. Sequencing the human microbiome in health and disease. Hum Mol Genet 2013; 22(R1):R88–94. [DOI] [PubMed] [Google Scholar]
- 7. Johnson CL, Versalovic J.. The human microbiome and its potential importance to pediatrics. Pediatrics 2012; 129:950–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lanaspa M, Bassat Q, Medeiros MM, Munoz-Almagro C.. Respiratory microbiota and lower respiratory tract disease. Expert Rev Anti Infect Ther 2017; 15:703–11. [DOI] [PubMed] [Google Scholar]
- 9. Dickson RP, Huffnagle GB.. The lung microbiome: new principles for respiratory bacteriology in health and disease. PLoS Pathog 2015; 11:e1004923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Widder S, Zhao J, Carmody LA, et al. Association of bacterial community types, functional microbial processes and lung disease in cystic fibrosis airways. ISME J 2022; 16:905–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Feigelman R, Kahlert CR, Baty F, et al. Sputum DNA sequencing in cystic fibrosis: non-invasive access to the lung microbiome and to pathogen details. Microbiome 2017; 5:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brennan S, Gangell C, Wainwright C, Sly PD.. Disease surveillance using bronchoalveolar lavage. Paediatr Respir Rev 2008; 9:151–9. [DOI] [PubMed] [Google Scholar]
- 13. Stafler P, Davies JC, Balfour-Lynn IM, Rosenthal M, Bush A.. Bronchoscopy in cystic fibrosis infants diagnosed by newborn screening. Pediatr Pulmonol 2011; 46:696–700. [DOI] [PubMed] [Google Scholar]
- 14. Jain K, Wainwright C, Smyth AR.. Bronchoscopy-guided antimicrobial therapy for cystic fibrosis. Cochrane Database Syst Rev 2018; 9:CD009530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dickson RP, Erb-Downward JR, Martinez FJ, Huffnagle GB.. The microbiome and the respiratory tract. Annu Rev Physiol 2016; 78:481–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goddard AF, Staudinger BJ, Dowd SE, et al. Direct sampling of cystic fibrosis lungs indicates that DNA-based analyses of upper-airway specimens can misrepresent lung microbiota. Proc Natl Acad Sci USA 2012; 109:13769–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hogan DA, Willger SD, Dolben EL, et al. Analysis of lung microbiota in bronchoalveolar lavage, protected brush and sputum samples from subjects with mild-to-moderate cystic fibrosis lung disease. PLoS One 2016; 11:e0149998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lu J, Carmody LA, Opron K, et al. Parallel analysis of cystic fibrosis sputum and saliva reveals overlapping communities and an opportunity for sample decontamination. mSystems 2020; 5:e00296–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zemanick ET, Wagner BD, Robertson CE, et al. Assessment of airway microbiota and inflammation in cystic fibrosis using multiple sampling methods. Ann Am Thorac Soc 2015; 12:221–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Breuer O, Caudri D, Akesson L, et al. The clinical significance of oropharyngeal cultures in young children with cystic fibrosis. Eur Respir J 2018; 51:1800238. [DOI] [PubMed] [Google Scholar]
- 21. Ronchetti K, Tame JD, Paisey C, et al. The CF-Sputum Induction Trial (CF-SpIT) to assess lower airway bacterial sampling in young children with cystic fibrosis: a prospective internally controlled interventional trial. Lancet Respir Med 2018; 6:461–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fenn D, Abdel-Aziz MI, Brinkman P, et al. Comparison of microbial composition of cough swabs and sputum for pathogen detection in patients with cystic fibrosis. J Cyst Fibros 2022; 21:52–60. [DOI] [PubMed] [Google Scholar]
- 23. Blau H, Linnane B, Carzino R, et al. Induced sputum compared to bronchoalveolar lavage in young, non-expectorating cystic fibrosis children. J Cyst Fibros 2014; 13:106–10. [DOI] [PubMed] [Google Scholar]
- 24. Ordonez CL, Henig NR, Mayer-Hamblett N, et al. Inflammatory and microbiologic markers in induced sputum after intravenous antibiotics in cystic fibrosis. Am J Respir Crit Care Med 2003; 168:1471–5. [DOI] [PubMed] [Google Scholar]
- 25. Rogers GB, Carroll MP, Serisier DJ, Hockey PM, Jones G, Bruce KD.. Characterization of bacterial community diversity in cystic fibrosis lung infections by use of 16S ribosomal DNA terminal restriction fragment length polymorphism profiling. J Clin Microbiol 2004; 42:5176–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cuthbertson L, Walker AW, Oliver AE, et al. Lung function and microbiota diversity in cystic fibrosis. Microbiome 2020; 8:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sibley CD, Grinwis ME, Field TR, et al. Culture enriched molecular profiling of the cystic fibrosis airway microbiome. PLoS One 2011; 6:e22702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Whelan FJ, Waddell B, Syed SA, et al. Culture-enriched metagenomic sequencing enables in-depth profiling of the cystic fibrosis lung microbiota. Nat Microbiol 2020; 5:379–90. [DOI] [PubMed] [Google Scholar]
- 29. Zhao J, Schloss PD, Kalikin LM, et al. Decade-long bacterial community dynamics in cystic fibrosis airways. Proc Natl Acad Sci USA 2012; 109:5809–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Carmody LA, Caverly LJ, Foster BK, et al. Fluctuations in airway bacterial communities associated with clinical states and disease stages in cystic fibrosis. PLoS One 2018; 13:e0194060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Carmody LA, Zhao J, Schloss PD, et al. Changes in cystic fibrosis airway microbiota at pulmonary exacerbation. Ann Am Thorac Soc 2013; 10:179–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Quinn RA, Whiteson K, Lim YW, et al. A Winogradsky-based culture system shows an association between microbial fermentation and cystic fibrosis exacerbation. ISME J 2015; 9:1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Layeghifard M, Li H, Wang PW, et al. Microbiome networks and change-point analysis reveal key community changes associated with cystic fibrosis pulmonary exacerbations. NPJ Biofilms Microbiomes 2019; 5:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Twomey KB, Alston M, An SQ, et al. Microbiota and metabolite profiling reveal specific alterations in bacterial community structure and environment in the cystic fibrosis airway during exacerbation. PLoS One 2013; 8:e82432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smith DJ, Badrick AC, Zakrzewski M, et al. Pyrosequencing reveals transient cystic fibrosis lung microbiome changes with intravenous antibiotics. Eur Respir J 2014; 44:922–30. [DOI] [PubMed] [Google Scholar]
- 36. Coburn B, Wang PW, Diaz Caballero J, et al. Lung microbiota across age and disease stage in cystic fibrosis. Sci Rep 2015; 5:10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zemanick ET, Wagner BD, Robertson CE, et al. Airway microbiota across age and disease spectrum in cystic fibrosis. Eur Respir J 2017; 50:1700832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cuthbertson L, Rogers GB, Walker AW, et al. Respiratory microbiota resistance and resilience to pulmonary exacerbation and subsequent antimicrobial intervention. ISME J 2016; 10:1081–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Millar FA, Simmonds NJ, Hodson ME.. Trends in pathogens colonising the respiratory tract of adult patients with cystic fibrosis, 1985-2005. J Cyst Fibros 2009; 8:386–91. [DOI] [PubMed] [Google Scholar]
- 40. Page LK, Staples KJ, Spalluto CM, Watson A, Wilkinson TMA.. Influence of hypoxia on the epithelial-pathogen interactions in the lung: implications for respiratory disease. Front Immunol 2021; 12:653969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Harris JK, De Groote MA, Sagel SD, et al. Molecular identification of bacteria in bronchoalveolar lavage fluid from children with cystic fibrosis. Proc Natl Acad Sci USA 2007; 104:20529–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Thornton CS, Surette MG.. Potential contributions of anaerobes in cystic fibrosis airways. J Clin Microbiol 2021; 59:e01813–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bittar F, Richet H, Dubus JC, et al. Molecular detection of multiple emerging pathogens in sputa from cystic fibrosis patients. PLoS One 2008; 3:e2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Field TR, Sibley CD, Parkins MD, Rabin HR, Surette MG.. The genus Prevotella in cystic fibrosis airways. Anaerobe 2010; 16:337–44. [DOI] [PubMed] [Google Scholar]
- 45. Tunney MM, Field TR, Moriarty TF, et al. Detection of anaerobic bacteria in high numbers in sputum from patients with cystic fibrosis. Am J Respir Crit Care Med 2008; 177:995–1001. [DOI] [PubMed] [Google Scholar]
- 46. Thornton CS, Caverly LJ, LiPuma JJ.. Coming up for air: the role of anaerobes in cystic fibrosis. Ann Am Thorac Soc 2022; 19(5):713–716. [DOI] [PubMed] [Google Scholar]
- 47. Muhlebach MS, Hatch JE, Einarsson GG, et al. Anaerobic bacteria cultured from cystic fibrosis airways correlate to milder disease: a multisite study. Eur Respir J 2018; 52:1800242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Caverly LJ, LiPuma JJ.. Good cop, bad cop: anaerobes in cystic fibrosis airways. Eur Respir J 2018; 52:1801146. [DOI] [PubMed] [Google Scholar]
- 49. Sherrard LJ, Tunney MM, Elborn JS.. Antimicrobial resistance in the respiratory microbiota of people with cystic fibrosis. Lancet 2014; 384:703–13. [DOI] [PubMed] [Google Scholar]
- 50. Lamoureux C, Guilloux CA, Courteboeuf E, Gouriou S, Beauruelle C, Hery-Arnaud G.. Prevotella melaninogenica, a sentinel species of antibiotic resistance in cystic fibrosis respiratory niche? Microorganisms 2021; 9:1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sherrard LJ, McGrath SJ, McIlreavey L, et al. Production of extended-spectrum beta-lactamases and the potential indirect pathogenic role of Prevotella isolates from the cystic fibrosis respiratory microbiota. Int J Antimicrob Agents 2016; 47:140–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mirkovic B, Murray MA, Lavelle GM, et al. The role of short-chain fatty acids, produced by anaerobic bacteria, in the cystic fibrosis airway. Am J Respir Crit Care Med 2015; 192:1314–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Keravec M, Mounier J, Guilloux CA, et al. Porphyromonas, a potential predictive biomarker of Pseudomonas aeruginosa pulmonary infection in cystic fibrosis. BMJ Open Respir Res 2019; 6:e000374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sibley CD, Grinwis ME, Field TR, et al. McKay agar enables routine quantification of the “Streptococcus milleri” group in cystic fibrosis patients. J Med Microbiol 2010; 59:534–40. [DOI] [PubMed] [Google Scholar]
- 55. Tony-Odigie A, Wilke L, Boutin S, Dalpke AH, Yi B.. Commensal bacteria in the cystic fibrosis airway microbiome reduce P. aeruginosa induced inflammation. Front Cell Infect Microbiol 2022; 12:824101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sibley CD, Duan K, Fischer C, et al. Discerning the complexity of community interactions using a Drosophila model of polymicrobial infections. PLoS Pathog 2008; 4:e1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Whiley RA, Sheikh NP, Mushtaq N, et al. Differential potentiation of the virulence of the Pseudomonas aeruginosa cystic fibrosis Liverpool epidemic strain by oral commensal Streptococci. J Infect Dis 2014; 209:769–80. [DOI] [PubMed] [Google Scholar]
- 58. Rigauts C, Aizawa J, Taylor S, et al. Rothia mucilaginosa is an anti-inflammatory bacterium in the respiratory tract of patients with chronic lung disease. Eur Respir J 2022; 59:2101293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ulrich M, Beer I, Braitmaier P, et al. Relative contribution of Prevotella intermedia and Pseudomonas aeruginosa to lung pathology in airways of patients with cystic fibrosis. Thorax 2010; 65:978–84. [DOI] [PubMed] [Google Scholar]
- 60. Goss CH, Newsom SA, Schildcrout JS, Sheppard L, Kaufman JD.. Effect of ambient air pollution on pulmonary exacerbations and lung function in cystic fibrosis. Am J Respir Crit Care Med 2004; 169:816–21. [DOI] [PubMed] [Google Scholar]
- 61. Anand S, Mande SS.. Diet, microbiota and gut-lung connection. Front Microbiol 2018; 9:2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Enaud R, Prevel R, Ciarlo E, et al. The gut-lung axis in health and respiratory diseases: a place for inter-organ and inter-kingdom crosstalks. Front Cell Infect Microbiol 2020; 10:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rolain JM, Fancello L, Desnues C, Raoult D.. Bacteriophages as vehicles of the resistome in cystic fibrosis. J Antimicrob Chemother 2011; 66:2444–7. [DOI] [PubMed] [Google Scholar]
- 64. Kiedrowski MR, Bomberger JM.. Viral-bacterial co-infections in the cystic fibrosis respiratory tract. Front Immunol 2018; 9:3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. van Ewijk BE, van der Zalm MM, Wolfs TF, van der Ent CK.. Viral respiratory infections in cystic fibrosis. J Cyst Fibros 2005; 4:31–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Asner S, Waters V, Solomon M, et al. Role of respiratory viruses in pulmonary exacerbations in children with cystic fibrosis. J Cyst Fibros 2012; 11:433–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Flight W, Jones A.. The diagnosis and management of respiratory viral infections in cystic fibrosis. Expert Rev Respir Med 2017; 11:221–7. [DOI] [PubMed] [Google Scholar]
- 68. Poore TS, Hong G, Zemanick ET.. Fungal infection and inflammation in cystic fibrosis. Pathogens 2021; 10:618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Price KE, Hampton TH, Gifford AH, et al. Unique microbial communities persist in individual cystic fibrosis patients throughout a clinical exacerbation. Microbiome 2013; 1:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fodor AA, Klem ER, Gilpin DF, et al. The adult cystic fibrosis airway microbiota is stable over time and infection type, and highly resilient to antibiotic treatment of exacerbations. PLoS One 2012; 7:e45001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Nelson MT, Wolter DJ, Eng A, et al. Maintenance tobramycin primarily affects untargeted bacteria in the CF sputum microbiome. Thorax 2020; 75:780–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Stressmann FA, Rogers GB, Marsh P, et al. Does bacterial density in cystic fibrosis sputum increase prior to pulmonary exacerbation? J Cyst Fibros 2011; 10:357–65. [DOI] [PubMed] [Google Scholar]
- 73. Chin M, De Zoysa M, Slinger R, et al. Acute effects of viral respiratory tract infections on sputum bacterial density during CF pulmonary exacerbations. J Cyst Fibros 2015; 14:482–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fothergill JL, Ledson MJ, Walshaw MJ, McNamara PS, Southern KW, Winstanley C.. Comparison of real time diagnostic chemistries to detect Pseudomonas aeruginosa in respiratory samples from cystic fibrosis patients. J Cyst Fibros 2013; 12:675–81. [DOI] [PubMed] [Google Scholar]
- 75. Regelmann WE, Elliott GR, Warwick WJ, et al. Reduction of sputum Pseudomonas aeruginosa density by antibiotics improves lung function in cystic fibrosis more than do bronchodilators and chest physiotherapy alone. Am Rev Respir Dis 1990; 141(4 Pt 1):914–21. [DOI] [PubMed] [Google Scholar]
- 76. Lam JC, Somayaji R, Surette MG, Rabin HR, Parkins MD.. Reduction in Pseudomonas aeruginosa sputum density during a cystic fibrosis pulmonary exacerbation does not predict clinical response. BMC Infect Dis 2015; 15:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Carmody LA, Zhao J, Kalikin LM, et al. The daily dynamics of cystic fibrosis airway microbiota during clinical stability and at exacerbation. Microbiome 2015; 3:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Dmitrijeva M, Kahlert CR, Feigelman R, et al. Strain-resolved dynamics of the lung microbiome in patients with cystic fibrosis. mBio 2021; 12:e02863–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kirst ME, Baker D, Li E, Abu-Hasan M, Wang GP.. Upper versus lower airway microbiome and metagenome in children with cystic fibrosis and their correlation with lung inflammation. PLoS One 2019; 14:e0222323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Pust MM, Wiehlmann L, Davenport C, Rudolf I, Dittrich AM, Tummler B.. The human respiratory tract microbial community structures in healthy and cystic fibrosis infants. NPJ Biofilms Microbiomes 2020; 6:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Neerincx AH, Whiteson K, Phan JL, et al. Lumacaftor/ivacaftor changes the lung microbiome and metabolome in cystic fibrosis patients. ERJ Open Res 2021; 7:00731-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. LiPuma JJ. Placeholder for article included as part of review series. 2022.
- 83. Nguyen M, Sharma A, Wu W, et al. The fermentation product 2,3-butanediol alters P. aeruginosa clearance, cytokine response and the lung microbiome. ISME J 2016; 10(12):2978–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Gao B, Gallagher T, Zhang Y, et al. Tracking polymicrobial metabolism in cystic fibrosis airways: Pseudomonas aeruginosa metabolism and physiology are influenced by Rothia mucilaginosa-derived metabolites. mSphere 2018; 3:e00151–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Korgaonkar A, Trivedi U, Rumbaugh KP, Whiteley M.. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc Natl Acad Sci USA 2013; 110:1059–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Price CE, O’Toole GA.. The gut-lung axis in cystic fibrosis. J Bacteriol 2021; 203:e0031121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Sorbara MT, Pamer EG.. Interbacterial mechanisms of colonization resistance and the strategies pathogens use to overcome them. Mucosal Immunol 2019; 12:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Waters VJ, Kidd TJ, Canton R, et al. Reconciling antimicrobial susceptibility testing and clinical response in antimicrobial treatment of chronic cystic fibrosis lung infections. Clin Infect Dis 2019; 69:1812–6. [DOI] [PubMed] [Google Scholar]
- 89. Somayaji R, Parkins MD, Shah A, et al. Antimicrobial susceptibility testing (AST) and associated clinical outcomes in individuals with cystic fibrosis: a systematic review. J Cyst Fibros 2019; 18:236–43. [DOI] [PubMed] [Google Scholar]
- 90. Acosta N, Heirali A, Somayaji R, et al. Sputum microbiota is predictive of long-term clinical outcomes in young adults with cystic fibrosis. Thorax 2018; 73:1016–25. [DOI] [PubMed] [Google Scholar]
- 91. Zhao CY, Hao Y, Wang Y, et al. Microbiome data enhances predictive models of lung function in people with cystic fibrosis. J Infect Dis 2021; 223(12 Suppl 2):S246–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Einarsson G, Flanagan E, Lee A, et al. Longitudinal airway microbiota profiling in cystic fibrosis patients enrolled in the CFMATTERS clinical trial. J Cyst Fibros 2017; 16:S4. [Google Scholar]
- 93. Einarsson G, Flanagan E, Lee A, et al. Microbiota profiling during 1-year of clinical stability in people with cystic fibrosis—CFMATTERS Consortium. J Cyst Fibros 2019; 18:S35. [Google Scholar]
- 94. Aaron SD, Vandemheen KL, Ferris W, et al. Combination antibiotic susceptibility testing to treat exacerbations of cystic fibrosis associated with multiresistant bacteria: a randomised, double-blind, controlled clinical trial. Lancet 2005; 366:463–71. [DOI] [PubMed] [Google Scholar]
- 95. Yau YC, Ratjen F, Tullis E, et al. Randomized controlled trial of biofilm antimicrobial susceptibility testing in cystic fibrosis patients. J Cyst Fibros 2015; 14:262–6. [DOI] [PubMed] [Google Scholar]
- 96. Heirali AA, Acosta N, Storey DG, et al. The effects of cycled inhaled aztreonam on the cystic fibrosis (CF) lung microbiome. J Cyst Fibros 2019; 18:829–37. [DOI] [PubMed] [Google Scholar]
- 97. Heirali AA, Workentine ML, Acosta N, et al. The effects of inhaled aztreonam on the cystic fibrosis lung microbiome. Microbiome 2017; 5:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Ratjen F, Munck A, Kho P, Angyalosi G, Group ES.. Treatment of early Pseudomonas aeruginosa infection in patients with cystic fibrosis: the ELITE trial. Thorax 2010; 65:286–91. [DOI] [PubMed] [Google Scholar]
- 99. Heirali A, Thornton C, Acosta N, et al. Sputum microbiota in adults with CF associates with response to inhaled tobramycin. Thorax 2020; 75:1058–64. [DOI] [PubMed] [Google Scholar]
- 100. Whelan FJ, Heirali AA, Rossi L, Rabin HR, Parkins MD, Surette MG.. Longitudinal sampling of the lung microbiota in individuals with cystic fibrosis. PLoS One 2017; 12:e0172811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Cuevas-Ocana S, Laselva O, Avolio J, Nenna R.. The era of CFTR modulators: improvements made and remaining challenges. Breathe 2020; 16:200016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Harris JK, Wagner BD, Zemanick ET, et al. Changes in airway microbiome and inflammation with ivacaftor treatment in patients with cystic fibrosis and the G551D mutation. Ann Am Thorac Soc 2020; 17:212–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Hisert KB, Heltshe SL, Pope C, et al. Restoring cystic fibrosis transmembrane conductance regulator function reduces airway bacteria and inflammation in people with cystic fibrosis and chronic lung infections. Am J Respir Crit Care Med 2017; 195:1617–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Bessonova L, Volkova N, Higgins M, et al. Data from the US and UK cystic fibrosis registries support disease modification by CFTR modulation with ivacaftor. Thorax 2018; 73:731–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Sosinski LM, H CM, Neugebauer KA, et al. A restructuring of microbiome niche space is associated with Elexacaftor-Tezacaftor-Ivacaftor therapy in the cystic fibrosis lung. J Cyst Fibros 2021:S1569-1993(21)02131-7. doi: 10.1016/j.jcf.2021.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Nichols DP, Donaldson SH, Frederick CA, et al. PROMISE: working with the CF community to understand emerging clinical and research needs for those treated with highly effective CFTR modulator therapy. J Cyst Fibros 2021; 20:205–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Filkins LM, Hampton TH, Gifford AH, et al. Prevalence of streptococci and increased polymicrobial diversity associated with cystic fibrosis patient stability. J Bacteriol 2012; 194:4709–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Hampton TH, Green DM, Cutting GR, et al. The microbiome in pediatric cystic fibrosis patients: the role of shared environment suggests a window of intervention. Microbiome 2014; 2:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Lim YW, Schmieder R, Haynes M, et al. Metagenomics and metatranscriptomics: windows on CF-associated viral and microbial communities. J Cyst Fibros 2013; 12:154–64. [DOI] [PMC free article] [PubMed] [Google Scholar]