Abstract
Aggression is an ethologically important social behavior, but excessive aggression can be detrimental to fitness. Social experiences among conspecific individuals reduce aggression in many species, the mechanism of which is largely unknown. We found that loss-of-function mutation of nervy (nvy), a Drosophila homolog of vertebrate myeloid translocation genes (MTGs), increased aggressiveness only in socially experienced flies and that this could be reversed by neuronal expression of human MTGs. A subpopulation of octopaminergic/tyraminergic neurons labeled by nvy was specifically required for such social experience–dependent suppression of aggression, in both males and females. Cell type–specific transcriptomic analysis of these neurons revealed aggression-controlling genes that are likely downstream of nvy. Our results illustrate both genetic and neuronal mechanisms by which the nervous system suppresses aggression in a social experience–dependent manner, a poorly understood process that is considered important for maintaining the fitness of animals.
A transcriptional regulator links the genes and neurons that mediate reduced aggression caused by social experience.
INTRODUCTION
Animals adjust their aggressiveness toward conspecifics according to the perceived costs or benefits of the interaction (1). Although innate variability in the level of aggression has been observed across animal species (2–5), an animal’s prior social experiences play an important role in either promoting or suppressing the intensity of aggression (5, 6). Even in laboratory animals with relatively homogeneous genetic backgrounds (rodents and the fruit fly), interactions with conspecific males (7–12) and females (13, 14) are known to profoundly alter the level of aggression. Most notably, across species, individuals reared as a group have markedly reduced aggressiveness compared with individuals reared in isolation (7, 10, 12). This phenotype has been linked to social isolation–induced stress, with substantial implications for human psychological and cognitive health (15). The experience of group rearing can have diverse consequences depending on the genetic and behavioral variabilities within a group (16–18), although several theoretical (19, 20) and experimental (17, 20) works argue that reduction of aggressive behavior under high population density is generally a favorable trait. Here, we focused on understanding at both the genetic and neuronal levels how the brain of the fruit fly Drosophila melanogaster, a genetically tractable model, decreases aggression after group rearing.
Recent studies have focused primarily on neuromodulatory factors and circuits that promote aggression (21–24) and dominance (25–28). Reported genetic and neural substrates that are associated with suppression of aggression (29–32) have not been directly linked to social experience. In the fruit fly, specific chemosensory inputs are implicated in the suppression of aggression after rearing with males (7, 33) and females (14), while the visual system is mostly dispensable (34). However, the precise mechanism by which the central brain converts the social experience to lower levels of aggression remains poorly understood despite recent progress in uncovering neuronal and molecular mechanisms that link social experience and behavioral changes (35–39).
In this study, we found that the neuronal transcription regulator nervy (nvy) plays a key role in the social experience–dependent suppression of aggression. Null mutation and neuronal knockdown of nvy result in an unusually high level of aggression specifically in group-reared flies. Detailed behavioral analysis revealed that functional abrogation of nvy increases the probability of attacks when the fly is in a striking position. This high level of aggression causes a mating disadvantage in a competitive environment. nvy is specifically required in a subset of octopaminergic/tyraminergic (OA/TA) neurons to influence aggression. In contrast to the nvy-negative OA/TA subpopulation [which promotes aggression (40–44)], neural activity in the nvy-expressing OA/TA subpopulation is necessary and sufficient to suppress aggression. Transcriptomic analysis of OA/TA neurons identified several genes that likely act downstream of nvy in this neuronal subpopulation. Our findings illustrate a mechanism by which a dedicated group of neuromodulatory cells converts social experience into the appropriate level of aggression. The function of these neurons is supported by an evolutionarily conserved transcriptional regulator, which orchestrates the molecular machinery that is required for the proper control of aggression. These findings provide insight into a common neural mechanism underlying experience-dependent suppression of aggressive behavior, a phenomenon widely observed from invertebrates to vertebrates.
RESULTS
nvy is necessary for social experience–dependent suppression of aggression
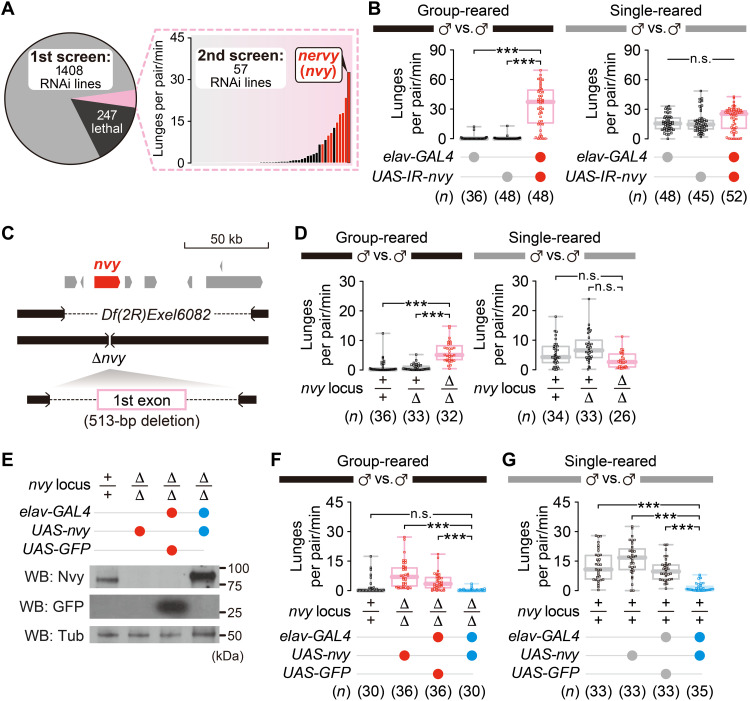
We used the fruit fly D. melanogaster to identify the genes necessary for suppressing aggression under group-rearing conditions, which plastically reduce aggression (7). We performed a systematic behavioral screen using group-reared adult male flies in which candidate genes were knocked down in neurons via RNA interference (RNAi). Fly aggressiveness was quantified by automated counting of lunges (45, 46), a male-type aggressive behavior (40, 47). We began our screen with 1408 RNAi effector lines, each controlled by the pan-neuronal elav-GAL4 driver, and found that 57 lines passed the initial threshold of increased aggression after group rearing (Fig. 1A, left, and table S1: see Materials and Methods for details and Source Data file for the complete dataset). Among those, flies from 11 lines showed a significant increase in lunges compared with both driver-only and effector-only genetic controls in the secondary round of assays (Fig. 1A, right, and table S2; see Table 1 for the list of all 11 genes and Source Data file for the complete dataset). The strongest phenotype was produced by neuronal knockdown of the gene nvy (Fig. 1A; Fig. 1B, left; and fig. S1A), which we focus on herein. The heads of the nvy RNAi flies showed reduced levels of nvy mRNA (less than 40% of controls) and Nvy protein expression (fig. S1B).
Fig. 1. The nvy gene suppresses aggression specifically in socially experienced flies.
(A) Pan-neuronal RNAi screen for Drosophila genes that suppress aggression after group rearing. The pie chart (left) summarizes the result of the first screening. Bars on the right represent median lunge numbers in the second screening of the 57 lines that passed the first screening. The 11 RNAi lines that showed a significant increase in lunges compared with two genetic controls are indicated in red. All flies used in the screenings were group-reared. (B) Pan-neuronal RNAi of the nvy gene increased the lunges performed by group-reared (left), but not single-reared (right), males. (C) Genome schematics of two deletion alleles for nvy. (D) Deletion of nvy increased the lunges in group-reared (left), but not in single-reared (right), flies, mirroring RNAi of nvy. (E) Pan-neuronal expression of Nvy in ∆nvy males verified by Western blot (WB). (F) Reversal of the hyperaggressive phenotype in group-reared ∆nvy males by pan-neuronal nvy expression. (G) Reduced aggression in single-reared flies by pan-neuronal nvy overexpression. ***P < 0.0005 and not significant (n.s.) P ≥ 0.05 [(B, D, F, and G) Kruskal-Wallis one-way analysis of variance (ANOVA) and post hoc Mann-Whitney U test with Bonferroni correction].
Table 1. Details of 11 “hit” genes in the secondary RNAi screening.
cAMP, adenosine 3′,5′-monophosphate.
| Annotation symbol | Gene name | Function | Median lunge number per pair/min |
| CG3385 | nvy | Transcription repressor, A kinase anchoring protein (AKAP) |
32.7 |
| CG15862 | PKA-R2 | Regulatory subunit of the cAMP- dependent protein kinases |
23.6 |
| CG7986 | Atg18a | Autophagy regulation | 14.7 |
| CG9436 | CG9436 |
d-threo-aldose 1-dehydrogenase, aldose reductase |
11.0 |
| CG2346 | FMRFa | Neuropeptide signaling | 10.0 |
| CG13948 | Gr21a | Gustatory receptor, response to carbon dioxide |
6.9 |
| CG8930 | rk | G protein–coupled receptor, neuropeptide signaling |
2.7 |
| CG32742 | Cdc7 | Cell cycle regulation | 0.6 |
| CG7529 | Est-Q | Carboxylesterase | 0.4 |
| CG7331 | Atg13 | Autophagy regulation | 0.3 |
| CG5954 | l(3)mbt | Cell proliferation regulation | 0.3 |
We generated a CRISPR-Cas9–mediated null mutation of the nvy gene (∆nvy; Fig. 1C) and confirmed that nvy is indeed necessary to dampen aggressiveness after group rearing. The homozygous ∆nvy mutation, as well as trans-heterozygosity of ∆nvy and a small chromosomal deficiency, led to increased aggressiveness relative to genetic controls after group rearing (Fig. 1D, left; fig. S1C, left; and movies S1 and S2). When ∆nvy homozygous mutants were reared in isolation, a condition known to elevate aggression (7), they showed levels of aggression similar to those of wild-type flies (Fig. 1D, right). This result was replicated with the trans-heterozygous flies with the deficiency (fig. S1C, right). Likewise, pan-neuronal RNAi knockdown of nvy did not increase aggression among socially isolated flies (Fig. 1B, right). Pan-neuronal nvy expression via the UAS-nvy transgene (Fig. 1E) reversed the hyperaggressive phenotype of the group-reared ∆nvy mutants (Fig. 1F). In addition, nvy overexpression in the wild-type background (fig. S1D) reduced high aggressiveness in single-reared flies (Fig. 1G). These results suggest that nvy is required specifically for social experience–dependent suppression of aggression.
While pairs of group-reared ∆nvy males were more active (traveled a longer distance) than group-reared wild-type flies (fig. S1E, left), levels of aggression and activity are tightly correlated (40), presumably because aggressive interactions involve frequent chasing (48, 49). The locomotor activity of flies introduced into the arena solitarily was comparable between wild-type and ∆nvy males (fig. S1F). Moreover, the distance traveled by pairs of socially isolated wild-type and ∆nvy males (which show similar levels of aggression) was statistically indistinguishable (fig. S1E, right). Thus, it is unlikely that ∆nvy causes general hyperactivity. Overexpression of nvy in the nervous system did not reduce the locomotion of solitary flies compared with a control that expresses green fluorescent protein (GFP) (fig. S1G), suggesting that reduction of aggressiveness in this genotype is not due to general inactivity. We also normalized the number of lunges by the distance traveled (40) to ask whether the observed changes in lunges were simply proportional to the changes in activity level. In all cases, statistical results using a normalized lunge number were consistent with the results using raw lunge numbers described above (fig. S1, H to K), arguing that activity level alone cannot account for nvy’s effect on aggression.
nvy controls a behavioral transition leading to lunges
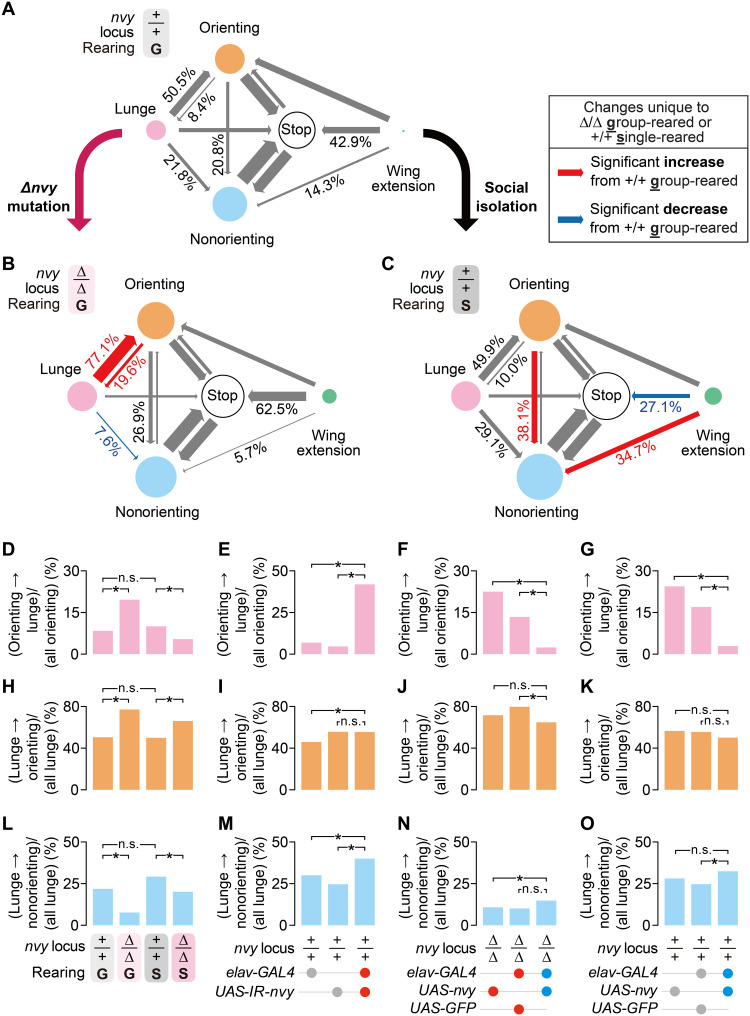
Our results thus far indicate that a reduction of nvy in the nervous system elevates aggression only in socially experienced (group-reared) flies. One possible function of nvy may be to gate the process that leads to the group-reared state. The mutation in nvy may force the flies to maintain a more aggressive, socially isolated state even after group rearing. Alternatively, nvy mutation may alter the behavioral dynamics during aggressive interactions, but the number of lunges alone does not illuminate such a transformation. To understand how nvy mutation affects fly interactions, we analyzed the behavioral transition patterns. We annotated all frames into five mutually exclusive behaviors: (i) stopping (the state where a fly moves very little), (ii) orienting (the state where a fly moves toward the other fly within a short distance with above-threshold speed), (iii) nonorienting (the state where a fly moves above-threshold speed without orienting toward the other fly), (iv) lunging, and (v) performing a wing extension (see Materials and Methods for the precise definitions of each behavior). “Orienting” can be largely equated with “chasing.” We then constructed ethograms to summarize the first-order transition probabilities from each behavior, for each experimental group. Fly courtship behavior, which also consists of complex motor subprograms (50, 51), is largely characterized by three relatively simple latent states (52), suggesting that this level of analysis is sufficient to reveal important aspects of behavioral dynamics during social interactions.
We noticed that the group-reared ∆nvy pairs generally had more behavioral events (including lunges) than group-reared wild-type pairs (fig. S2, A to F). In other words, the ∆nvy mutants switched behaviors more often than the group-reared wild-type flies, which tended to dwell at “stopping” events for a longer duration (fig. S2B2; see also movies S1 and S2). Theoretically, a uniform reduction in all behaviors would reduce the number of lunges as well while preserving the overall structure of behavioral transitions. We identified several transitions that were significantly different between group-reared ∆nvy mutants and wild-type flies (Fig. 2, A and B). These differences should also be present between single-reared and group-reared wild-type flies if ∆nvy mutation simply prevents social experience–dependent behavioral transformations. However, ethograms revealed that the ∆nvy mutation altered transition probabilities in a distinctly different manner from single rearing, relative to group-reared wild-type flies. The transition probabilities to and from lunges were comparable between group-reared and single-reared wild-type flies (Fig. 2, A and C), suggesting that socially isolated flies lunge more than group-reared flies by scaling up the number of events that lead to lunges. By contrast, ∆nvy mutants transitioned from “orienting” to “lunges” and from “lunges” to “orienting” more frequently (Fig. 2B), forming a recurrent loop between these two behaviors. This loop was further enhanced by a reduction in the “lunge”-to-“nonorienting” transition probability relative to group-reared wild-type flies. As a result, the group-reared ∆nvy mutants generally showed shorter intervals between lunges than single-reared wild-type flies (fig. S2G). These observations suggest that ∆nvy mutation elevates aggression by altering behavioral dynamics in response to group rearing differently from wild-type flies rather than maintaining the behavioral characteristics of single-reared wild-type flies. In support of this hypothesis, group-reared ∆nvy mutants performed fewer orienting behaviors (the major source behavior leading to lunges; see Source Data for details) than single-reared wild-type flies (fig. S2C), although the number of lunges was comparable between these two groups (fig. S2E1: see data S1 for statistical results).
Fig. 2. nvy suppresses aggression by modulating probability to lunge while orienting.
(A to C) Ethograms between five classified behaviors for group-reared wild-type (A), group-reared ∆nvy (B), and single-reared wild-type (C) males. Numbers represent transition probabilities from the source of arrows. (D to O) Comparisons of transition probabilities from orienting to lunge (D to G), from lunge to orienting (H to K), and from lunge to nonorienting (L to O) among wild-type and ∆nvy males [(D, H, and L) dataset from Fig. 1D], group-reared flies with pan-neuronal RNAi knockdown of nvy [(E, I, and M) dataset from Fig. 1B, left], group-reared ∆nvy males in which nvy was pan-neuronally expressed [(F, J, and N) dataset from Fig. 1F], and single-reared males in which nvy was overexpressed pan-neuronally [(G, K, and O) dataset from Fig. 1G]. Only the orienting-to-lunge transition correlates with the changes in the number of lunges. *P < 0.01 and n.s. P ≥ 0.01 [(B to O) permutation test with Bonferroni-like correction].
To further investigate which of the three transitions that are distinctly altered in group-reared ∆nvy mutants are relevant for the increased aggression in this genotype, we compared the probabilities of these transitions in flies subjected to the genetic manipulations performed in Fig. 1. “Orienting-to-lunge” transitions were more frequent in ∆nvy mutants than wild-type flies specifically under group-reared (and not single-reared) conditions (Fig. 2D), suggesting a unique gene-environment interaction in this genotype. We found that wild-type flies and ∆nvy mutants altered several behavioral transitions differently (fig. S2, H and I) after group rearing. The frequency of “orienting-to-lunge” transitions was correlated with the number of lunges in flies with neuron-specific RNAi knockdown of nvy (Fig. 2E), neuron-specific rescue of the ∆nvy mutation (Fig. 2F), and neuron-specific overexpression of nvy (Fig. 2G). “Lunge-to-orienting” transitions were more frequent in ∆nvy mutants regardless of social experience (Fig. 2H) but were not consistently different from genetic controls in any of the RNAi, rescue, or overexpression experiments (Fig. 2, I to K). Last, “lunge-to-nonorienting” transitions were reduced in both single- and group-reared ∆nvy mutants (Fig. 2L) but were not consistently coupled to the amount of nvy in the RNAi, rescue, or overexpression experiments (Fig. 2, M to O). These results suggest that ∆nvy mutation promotes aggression after group rearing by increasing the probability of lunging while the mutant fly chases or confronts an opponent.
The dense behavioral annotations prompted us to analyze other behaviors in detail. We first asked whether male-to-male courtship was also affected by nvy. Male-to-male wing extensions are observed infrequently in wild-type flies (25, 48, 53, 54). Consistent with a previous report (55), single-reared wild-type flies performed more wing extensions than group-reared wild-type flies (fig. S2F), although even in single-reared wild-type flies, wing extensions accounted for only 0.3% of the total behavioral events (fig. S2A). The duration of wing extensions in both single-reared and group-reared ∆nvy mutant flies was comparable to that in single-reared wild-type flies (fig. S2F). While RNAi knockdown of nvy in neurons increased male-to-male wing extensions (fig. S3A), genetic rescue of ∆nvy (fig. S3B) and nvy overexpression (fig. S3C) did not alter wing extensions relative to genetic controls. Together, nvy does not influence male-to-male courtship as consistently as it does aggression. The small effect size of male-to-male wing extensions relative to male-to-female wing extensions (see Fig. 3 for the wing extension index toward females) may be secondary to increased opportunities to interact (55, 56).
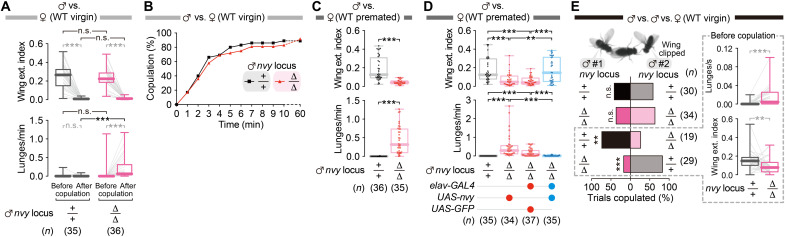
Fig. 3. Behavior phenotypes of Δnvy males under various social contexts.
(A) ∆nvy males performed wing extensions as vigorously as wild-type (WT) males before copulation (top), but, unlike wild-type males, they lunged toward females after copulation (bottom). (B) ∆nvy males copulated with virgin females at a rate comparable to wild-type males. (C) Virgin ∆nvy males had lower scores on the wing extension index (top) and performed more lunges (bottom) toward premated females (i.e., females mated before the experiment: see Materials and Methods for details) than wild-type males. (D) Reversal of the male-to-female wing extensions (top) and lunges (bottom) in ∆nvy by pan-neuronal expression of nvy. Genotypes of male testers are indicated below the plots. (E) Competitive copulation assay using two males and one virgin female. Left: Copulation success of either male #1 or #2 (wing clipped). Right: Lunge bouts (top) or wing extension indices (bottom) quantified in wild-type versus ∆nvy groups during the precopulation period. ***P < 0.0005, **P < 0.01, and n.s. P ≥ 0.05 [(A) (in black) and (C): Mann-Whitney U test; (A) (in gray) and (E) right: Wilcoxon signed-rank test; (D): Kruskal-Wallis one-way ANOVA and post hoc Mann-Whitney U test with Bonferroni correction; (E) left: Fisher’s exact test].
Chasing is observed in both courtship (50, 57–59) and aggressive interactions (48, 49, 60). Considering the low level of wing extensions among males, as discussed above, the higher level of orienting events observed among group-reared ∆nvy mutants than among wild types (fig. S2C) likely reflects a difference in the level of aggression rather than courtship. We also analyzed whether the locomotor patterns showed any obvious temporal structure. The binned distance traveled by pairs of flies was overall uniform across the 30-min assay duration in wild types and ∆nvy mutants (fig. S3, D and E) and in the RNAi against nvy (fig. S3, F and G), genetic rescue of ∆nvy (fig. S3, H and I), and nvy overexpression (fig. S3, J and K) experiments. Although analysis at higher temporal resolution may reveal differences among genotypes, we conclude that nvy manipulations do not have a gross impact on the temporal structure of fly locomotion. Lunges were also distributed relatively uniformly throughout the duration of the recordings (fig. S3L).
nvy prevents inappropriate aggression in the presence of a female
We next addressed whether nvy is also necessary for another type of social interaction, i.e., male-to-female interactions. Wild-type males vigorously courted a virgin female (Fig. 3A) (50), and more than 80% of males successfully copulated within 10 min (Fig. 3B). ∆nvy males performed wing extensions toward a virgin female (Fig. 3A) and copulated (Fig. 3B) at rates comparable to the wild-type males, indicating that ∆nvy mutation does not affect male courtship capability. However, ∆nvy males lunged toward a female after copulation, which was rarely observed in wild-type males (Fig. 3A). Virgin ∆nvy males also lunged toward premated females and had lower scores on the wing extension index relative to wild-type males (Fig. 3C and movies S3 and S4), suggesting that mating is not required for the ∆nvy males to attack females. The restoration of nvy expression in neurons suppressed male-to-female aggression in ∆nvy males (Fig. 3D). ∆nvy males formed courtship memories in the same way as wild-type males (fig. S4A), suggesting that ∆nvy males do not misidentify a mated female as a male. It is possible that ∆nvy males are sensitized to 11-cis-vaccenyl acetate, an aggression-promoting male pheromone (61) that is transferred to the female during mating (62). However, decapitated male (fig. S4B) or premated female (fig. S4C) opponents provoked no lunges from ∆nvy males, indicating that male chemical cues alone are insufficient to promote aggression in ∆nvy mutants. These data argue that nvy is necessary for suppressing aggression toward both male and female targets, without affecting sex recognition per se.
Contrary to a popular view, experimental results have remained inconclusive about whether high levels of aggression are advantageous to fitness (9, 20, 47), possibly because the context can change the payoff of aggression (1, 17). We next wished to address whether highly aggressive ∆nvy males gain a benefit in a competitive environment. We created a situation in which one wild-type male and one ∆nvy male competed over one virgin female and observed which of the two genotypes was more successful in copulating with the female (63). The ∆nvy males had less copulation success than wild-type rivals (Fig. 3E, left). We found that the ∆nvy males performed more lunges (all of which were directed toward the wild-type rivals) and had lower scores on the wing extension index (Fig. 3E, right), suggesting that the reduced mating success of the ∆nvy males was due to their failure to appropriately suppress aggression and commit to courtship toward the female target. These results collectively indicate that nvy is necessary for flies to prevent unnecessary aggression in multiple social contexts, which, in turn, can help increase fitness.
nvy-expressing QA neurons suppress aggression
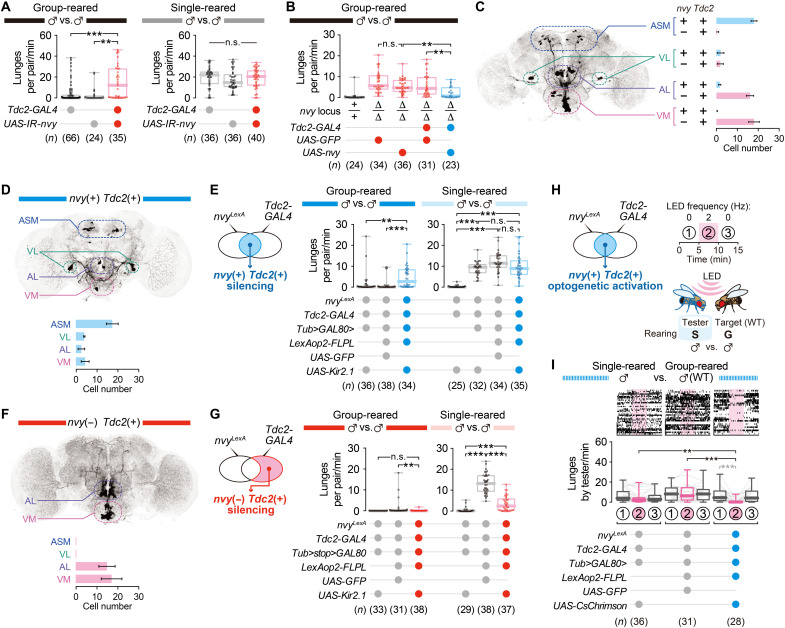
To identify the neuronal mechanisms by which nvy suppresses aggression, we screened selected GAL4 lines to restrict nvy RNAi to relatively small neuronal populations. Among those tested, loss of nvy in neurons labeled by Tyrosine decarboxylase 2 (Tdc2)–GAL4 increased aggression most markedly in group-reared flies (Fig. 4A, left, and table S3). Lunges by the same genotype under social isolation were not different from those of genetic controls (Fig. 4A, right), consistent with the results from pan-neuronal RNAi manipulation (Fig. 1B). Moreover, nvy expression driven by Tdc2-GAL4 suppressed the hyperaggressive phenotype of ∆nvy (Fig. 4B).
Fig. 4. Selective manipulation of nvy-expressing Tdc2 neurons suppresses aggression.
(A) Increase in lunges by group-reared (left), but not single-reared (right), males in which nvy was knocked down by RNAi in Tdc2 neurons. (B) Reversal of the hyperaggressive phenotype in group-reared ∆nvy males by nvy expression in Tdc2 neurons. (C) Number of neurons colabeled with nvyLexA and Tdc2-GAL4 in four neuronal subtypes, according to a previously described nomenclature (65). (D to G) Selective silencing of nvy-positive or nvy-negative Tdc2 neurons in males. Expression of GFP in nvy-positive (D) or nvy-negative (F) Tdc2 neurons is visualized by immunohistochemistry. Altered aggression after Kir2.1-mediated silencing of either nvy-positive (E) or nvy-negative (G) Tdc2 neurons is shown in box plots. Silencing of nvy-positive Tdc2 neurons increased aggression specifically in group-reared flies (E) (left), whereas silencing of nvy-negative Tdc2 neurons decreased aggression in single-reared flies (G) (right). (H) Optogenetic stimulation paradigm. (I) Reduced aggression in socially isolated males by optogenetic stimulation of the nvy-positive Tdc2 neurons. ***P < 0.0005, **P < 0.005, and n.s. P ≥ 0.05 [(A, B, E, G, and I) in black: Kruskal-Wallis one-way ANOVA and post hoc Mann-Whitney U test with Bonferroni correction; (I) in gray: Kruskal-Wallis one-way ANOVA and post hoc Wilcoxon signed-rank test; (C, D, and F) error bars indicate means ± SD of 8 to 10 brains].
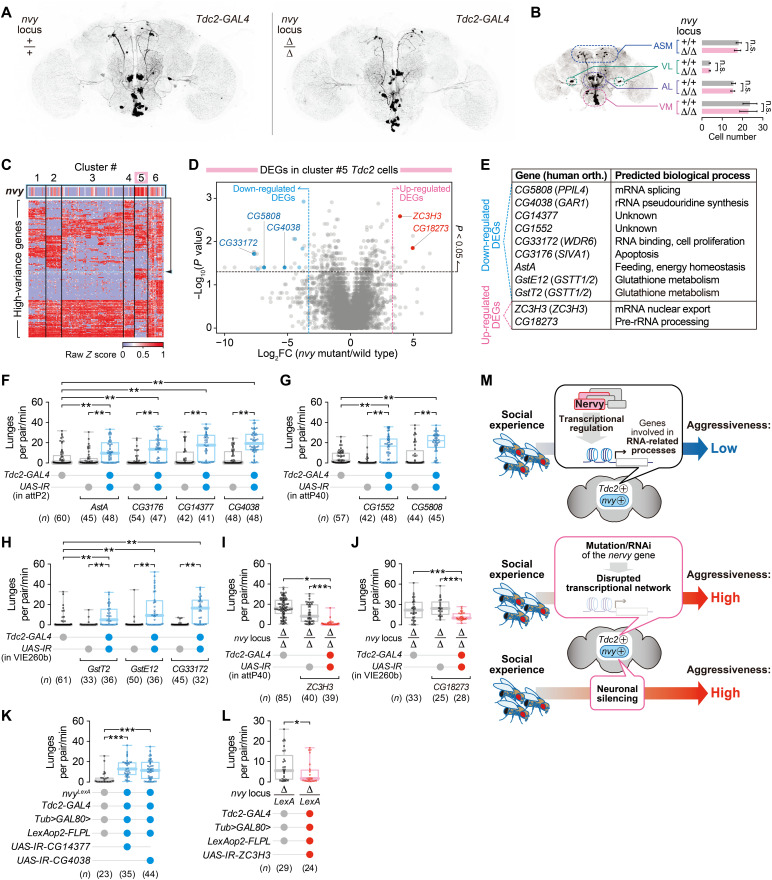
Tdc2 encodes the biosynthetic enzyme for octopamine/tyramine (OA/TA) (64), the invertebrate counterparts of norepinephrine/epinephrine. Since OA itself and Tdc2 neurons have been reported to promote fly aggression (40–43), identification of Tdc2 neurons as the site of nvy-dependent suppression of aggression intrigued us. We gained genetic access to the nvy-expressing cells by creating a CRISPR-Cas9–mediated knock-in allele of the nvy locus that expresses the bacterial transcription factor LexA in place of nvy (nvyLexA; fig. S5, A and B). This knock-in allele is null for nvy, as the heteroallelic combination of nvyLexA and ∆nvy reduced Nvy protein expression to an undetectable level, and resulted in hyperaggressiveness, similarly to homozygous ∆nvy (fig. S5, C and D). Although Nvy protein was expressed in a large population of neurons in the central brain (fig. S5E), we found that only a subset of Tdc2-GAL4 neurons was colabeled by nvyLexA in the central brain (Fig. 4C). From the locations of cell bodies and previously published nomenclature (65), we identified most nvyLexA-labeled Tdc2 neurons as being in the anterior superior medial (ASM) and ventrolateral (VL) clusters. Both clusters showed immunoreactivity to Nvy (fig. S5E) (66). nvyLexA-negative Tdc2 neurons were mostly in the VPM, VUMa, VUMd (collectively known as the VM cluster), and AL2 clusters (Fig. 4C).
Selective expression of Kir2.1, an inwardly rectifying potassium channel that induces neuronal hyperpolarization, in this nvyLexA-positive Tdc2 population (Fig. 4D and movie S5) markedly increased aggression in group-reared males that normally have low basal aggressiveness (Fig. 4E). By contrast, the high intensity of basal aggression in single-reared males was not affected by the same manipulation (Fig. 4E), similar to the ∆nvy mutant phenotype (Fig. 1E). Hyperpolarization of the nvyLexA-negative Tdc2 population (Fig. 4F and movie S6) had opposing effects; group-reared males remained nonaggressive, whereas single-reared males performed fewer lunges (Fig. 4G). This result is consistent with the previously reported aggression-promoting function of Tdc2 neurons (40–44).
The OA/TA system (especially in the ventral nerve cord (VNC)] has been implicated in locomotion (67). Normalizing the number of lunges by the distance traveled by the pair of flies showed that silencing of the nvyLexA-positive Tdc2 population (fig. S5G) or the nvyLexA-negative Tdc2 population (fig. S5I) did not change the level of aggression proportional to overall locomotor activity. We also quantified the speed of pairs of flies during nonorienting phases (in which flies were walking without interacting) when either the nvyLexA-positive or nvyLexA-negative Tdc2 populations were silenced. Both populations were present in the VNC, although nvyLexA-negative Tdc2 cells were more numerous (fig. S5F). Silencing of the nvyLexA-positive Tdc2 population in group-reared flies did not alter speed during nonorienting compared with one of the two negative controls (fig. S5H, left). In single-reared flies, speed was mildly decreased compared with genetic controls (fig. S5H, right), although the number of lunges was comparable across experimental and control flies (Fig. 4E). These findings argue that a difference in activity levels cannot account for the difference in aggression among genotypes. Silencing of the complementary, nvyLexA-negative Tdc2 population decreased speed compared with flies in which GFP was expressed instead of Kir2.1, although not to the extent observed when all Tdc2 neurons were silenced (fig. S5J). While the nvyLexA-negative Tdc2 population may contain neurons that are necessary for normal locomotion (67), it has been previously shown that aggression-promoting Tdc2 neurons reside in the central brain (41, 43). We therefore conclude that Tdc2 neurons contain both an nvy-expressing subpopulation that suppresses aggression and a non–nvy-expressing subpopulation that promotes aggression. This functional segregation within aminergic neurons can provide a complementary “push-and-pull” mechanism for the control of aggression according to social experience. Both populations are necessary for flies to adjust their level of aggression appropriately. Our findings parallel recent studies showing that the lateral habenula subpopulations respond in opposite directions during aggressive encounters (26, 68).
We addressed whether OA/TA themselves are necessary for the function of both the nvyLexA-positive and the nvyLexA-negative Tdc2 populations by using RNAi to knock down Tyramine β hydroxylase (Tbh: another key enzyme in the OA/TA biosynthetic pathway) and Tdc2. Consistent with the phenotypes of null mutants (40, 41, 44), knockdown of both enzymes in the entire population of Tdc2-GAL4 neurons reduced aggression among single-reared flies (fig. S6A). RNAi against these two genes in only the nvyLexA-positive population did not increase aggression in group-reared flies (fig. S6B), in contrast to the flies in which this neuronal population was silenced (Fig. 4E). This result implies that glutamate, a neurotransmitter often coexpressed in Tdc2-expressing neurons (43), but not OA/TA, may be responsible for the function of the nvyLexA-positive Tdc2 population. However, we cannot rule out the possibility that RNAi-mediated knockdown was not effective in this neuronal population. Also, it remains possible that Tdc2-GAL4 neurons contain non-OA aggression-suppressing nvyLexA-positive neurons [but see our single-cell RNA sequencing (scRNA-seq) data below].
The above chronic silencing results encouraged us to probe the aggression-suppressing role of Tdc2 neurons in socially isolated animals using optogenetics, which allows for greater temporal control of neural activity. Single-reared testers that express the channelrhodopsin CsChrimson specifically in the nvyLexA-positive Tdc2 population were photostimulated by a red light-emitting diode (LED) (Fig. 4H). We found that tester males performed significantly fewer lunges during the stimulation period compared with both the prestimulation period and with genetic controls during stimulation (Fig. 4I; fig. S7, A and B; and movies S7 and S8). General locomotion (fig. S7, C and D), speed during the nonorienting phase (fig. S7, E and F), and male-to-female courtship (fig. S7, G and H) were largely unaffected by stimulation. Thus, activation of the nvyLexA-positive Tdc2 population specifically blocks the execution of lunges without affecting other social behaviors. Our data collectively indicate that the previously uncharacterized nvy-expressing Tdc2 subpopulation serves as a neuronal switch that controls social experience–dependent changes in aggressiveness.
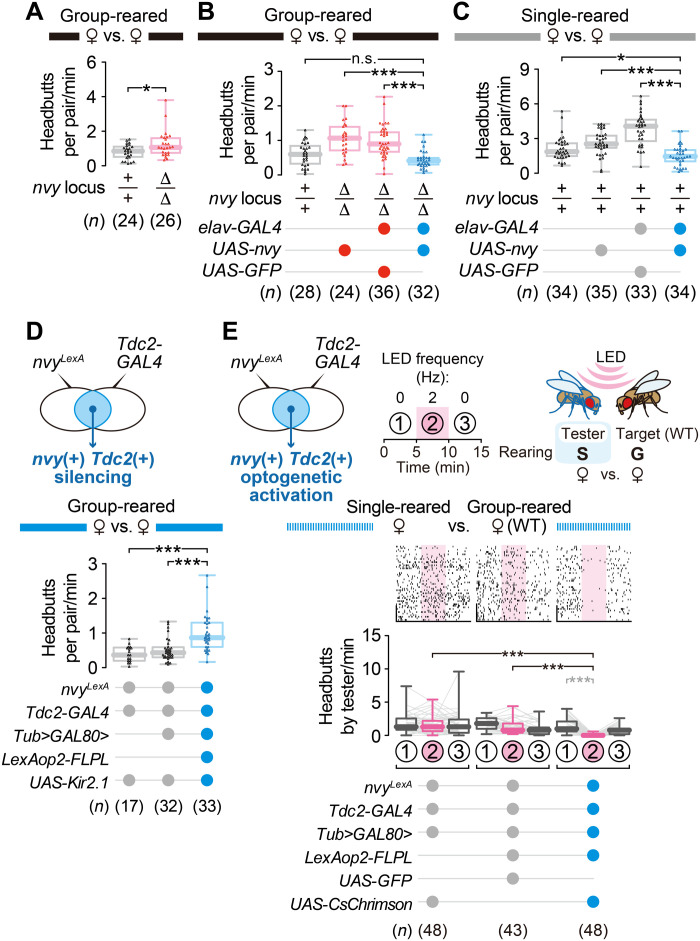
Sex-invariant function of nvy and nvy-expressing octopaminergic neurons
Similar to males, female flies also alter their aggressiveness according to their social experience (69). Although a few Tdc2 neurons in the VM cluster express the sex-determining gene fruitless (70), the gross population-level morphology in the central brain (fig. S8A) and the number of cell bodies in each of the four Tdc2 neuronal clusters (fig. S8B) were comparable between the two sexes, suggesting that the female brain contains the aggression-suppressing nvyLexA-positive Tdc2 population. To investigate whether our findings on the role of nvy are common to both sexes, we applied the above genetic and neuronal manipulations to female flies. Female aggressiveness was assessed by quantifying headbutts, a female-type aggressive action (60). The ∆nvy mutation increased headbutts in group-reared females (Fig. 5A), and this increase was suppressed by transgenic nvy expression (Fig. 5B and fig. S8C). As with males, the elevated basal aggressiveness of single-reared wild-type females was reduced by overexpression of nvy (Fig. 5C). Moreover, both silencing (Fig. 5D) and optogenetic activation (Fig. 5E) of the nvy-expressing Tdc2 population in females had similar effects on aggression as in males. Although it is relatively rare that sex-invariant neurons (or neurons specified by the same GAL4 line) affect fly aggression in both sexes (49, 71), recent studies in mice demonstrated that the subpopulation of neurons in the ventromedial hypothalamus (72, 73) and medial amygdala (74) controls aggression in both males and females. Our work provides an entry point to elucidate underexplored sex-invariant mechanisms underlying sexually dimorphic behaviors.
Fig. 5. Both the nvy gene and nvy-expressing Tdc2 neurons suppress female aggression.
(A) The ∆nvy mutation increased aggression in group-reared virgin females, as measured by the number of headbutts. (B) Rescue of the hyperaggressive phenotype in ∆nvy females by pan-neuronal nvy expression. (C) Reduced aggression in socially isolated females by pan-neuronal nvy overexpression. (D) Increased aggression in socially experienced females after Kir2.1-mediated silencing of nvy-positive Tdc2 neurons. (E) Reduced aggression in socially isolated females by optogenetic stimulation of nvy-positive Tdc2 neurons. Single-reared tester females with the indicated genotypes were paired with group-reared wild-type target females. The optogenetic stimulation paradigm was the same as in Fig. 4H. Raster plots (top) and box plots (bottom) of headbutts performed by the tester females are shown. ***P < 0.0005, *P < 0.05, and n.s. P ≥ 0.05 [(A): Mann-Whitney U test, (B to E) in black: Kruskal-Wallis one-way ANOVA and post hoc Mann-Whitney U test with Bonferroni correction; (E) in gray: Kruskal-Wallis one-way ANOVA and post hoc Wilcoxon signed-rank test].
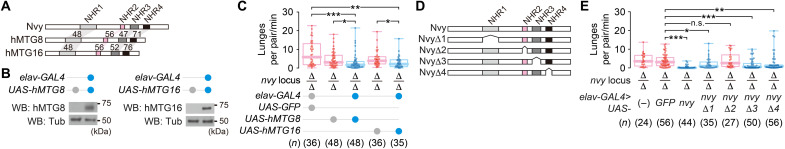
Functional conservation between nvy and its human homologs
The results described above prompted us to explore the molecular mechanism through which nvy acts to modulate aggression. nvy has been identified as a Drosophila homolog of vertebrate myeloid translocation genes (MTGs), proto-oncogenes encoding nuclear scaffold proteins that form transcription repressor complexes (75–78). In line with the high sequence similarities, pan-neuronal transgenic expression of human MTG8 and MTG16 significantly reduced the number of lunges (Fig. 6, A to C) as well as lunges normalized to the distance traveled (fig. S9A) in ∆nvy mutants, suggesting that nvy is a functional ortholog of these human genes. The expression of human MTG proteins was unlikely to cause a major locomotor deficit, as speed during the nonorienting state was comparable across genotypes (fig. S9B). We also created transgenes that express Nvy proteins lacking each of the four highly conserved Nvy homology regions (NHRs) (79). Of the truncated nvy constructs, only the variant that lacked the NHR2 domain (Fig. 6D and fig. S9C) failed to reverse the ∆nvy phenotype, measured both by the number of lunges (Fig. 6E) and by lunges normalized to the distance traveled (fig. S9D). Expression of the truncated constructs did not alter speed during the nonorienting state (except for a moderate decline in speed in flies expressing the variant lacking NHR3) (fig. S9E). NHR2 is required for the formation of homomultimers of Nvy proteins (fig. S9F), consistent with its scaffold role in mammalian MTGs (80, 81). These results collectively point to NHR2 as the key functional Nvy domain for controlling aggression and suggest that ∆nvy affects aggression by altering the gene expression patterns in functionally relevant populations of neurons.
Fig. 6. Human MTGs reverse the hyperaggressive phenotype of Δnvy males.
(A) Gene structures of human MTGs with amino acid sequence identities (%) against nvy within each NHR domain. (B) Pan-neuronal expression of human MTG genes by elav-GAL4, verified in fly head extracts by Western blot. (C) Rescue of the ∆nvy phenotype by pan-neuronal expression of human MTGs under the control of UAS. (D) Schematic of the truncated UAS-nvy constructs lacking each NHR domain. (E) Pan-neuronal expression of UAS-nvy lacking the NHR2 domain failed to rescue the ∆nvy phenotype. ***P < 0.001, **P < 0.01, *P < 0.05, and n.s. P ≥ 0.05 [(C and E): Kruskal-Wallis one-way ANOVA and post hoc Mann-Whitney U test with Bonferroni correction].
We therefore investigated the transcriptional profiles of Tdc2 neurons. As shown in Fig. 1, ∆nvy mutation elevates aggression relative to wild type in group-reared, but not single-reared, flies. We therefore reasoned that genes differentially expressed in wild-type and ∆nvy Tdc2 neurons are expected to be important for the control of aggressive behavior. To obtain cell type–specific transcriptional information from the Tdc2 neurons [which consist of several anatomically and functionally distinct subtypes (65)], we used scRNA-seq. The number of cells labeled by Tdc2-GAL4 was comparable in wild-type and ∆nvy brains (Fig. 7, A and B, and movies S9 and S10). Clonal labeling of single neurons revealed that several specific subtypes of Tdc2 neurons retain their overall neuroanatomy in ∆nvy brains as well (fig. S10). Although there is a possibility that the fine morphology, synapse distribution, or other fine structures of some Tdc2 neuronal subtypes may be altered in ∆nvy brains, we conclude that the ∆nvy mutation largely preserves the anatomically defined Tdc2 neuronal subtypes. Consistent with the anatomical data, fluorescence-activated cell sorting captured GFP-labeled Tdc2 neurons from wild-type and ∆nvy brains at similar rates (0.028 and 0.024% of the input, respectively) (fig. S11A). Hierarchical clustering analysis (82) of the data collected from 171 Tdc2 cells (fig. S11B) revealed six clusters with distinct gene expression patterns (Fig. 7C and fig. S11, C to E). Virtually all cells expressed Tdc2 mRNA (fig. S11F), suggesting that all Tdc2-GAL4–positive cells are indeed OA/TA neurons. Among them, cluster #5 was enriched in cells expressing nvy at relatively high levels (Fig. 7C and fig. S11, G to I; note that nvy mRNA is detectable in homozygous ∆nvy as the mutation removes only the first exon). We then performed differentially expressed gene (DEG) analysis on this cluster. Comparison of wild-type and ∆nvy cells within cluster #5 identified 25 down-regulated and 12 up-regulated genes with fold changes greater than 10 (Fig. 7D), whereas no such DEGs were detected in an analysis of all Tdc2 cells as a whole (fig. S11J). These data suggest that nvy affects gene expression in a cell type–specific manner.
Fig. 7. nvy functions in Tdc2 neurons to control aggression via transcriptional modulation.
(A) Neuronal morphology of Tdc2 neurons in the ∆nvy brain. GFP expressed under the control of Tdc2-GAL4 was visualized by immunohistochemistry in brains from the nvy locus of wild-type (left) or ∆nvy (right) males. (B) Cell counts of Tdc2 neurons in the ∆nvy brain. Subtypes of Tdc2-GAL4 neurons were classified according to a previous anatomical study (65). Note that the values for the wild-type nvy locus are replotted from fig. S8B. (C) Hierarchical iterative clustering analysis of Tdc2 cells. (D) Volcano plot of DEGs in cluster #5 cells. Pale colored dots represent genes that pass the Benjamini-Hochberg FDR test; dark colored dots correspond to genes that showed behavioral phenotypes in the following RNAi experiments. (E) Names and functions of DEGs in cluster #5 that exhibited aggression phenotypes following the Tdc2-GAL4–driven RNAi knockdown. (F to H) Increased aggression by Tdc2-GAL4–driven RNAi of nine down-regulated DEGs. (I and J) Reduced aggression in the ∆nvy mutants following the Tdc2-GAL4–driven RNAi of two up-regulated DEGs. (F) to (J) were organized according to the landing sites of UAS-IR insertions. (K) Increased aggression by RNAi of two down-regulated DEGs specifically in nvy-positive Tdc2 neurons. (L) Reduced aggression in the ∆nvy mutants by RNAi of ZC3H3, an up-regulated gene, specifically in nvy-positive Tdc2 neurons. (M) Schematic summary of the modulation of social experience–dependent aggression through manipulation of nvy and nvy-expressing Tdc2 neurons. ***P < 0.0005, **P < 0.01, and *P < 0.05 [(F to K): Kruskal-Wallis one-way ANOVA and post hoc Mann-Whitney U test with Bonferroni correction; (L): Mann-Whitney U test].
If nvy controls aggression through transcriptional regulation, DEGs found in the nvy-expressing Tdc2 neurons may contain effector genes necessary for appropriate modulation of aggression. Supporting this idea, Tdc2-GAL4–driven RNAi targeting nine down-regulated protein-coding genes found in cluster #5 significantly increased lunge numbers in group-reared males (Fig. 7, E to H, and fig. S12, A to C), phenocopying the nvy RNAi. Knocking down two up-regulated genes reduced the aggressiveness of ∆nvy mutants (Fig. 7, E, I, and J, and fig. S12, D to F), suggesting that these genes normally act downstream of nvy. The behavioral phenotypes shown above remained the same when lunges normalized by the distance traveled were analyzed (fig. S12, G to J), except for RNAi knockdown of CG18273 (fig. S12K), one of the two up-regulated DEGs in cluster #5. Speed during the nonorienting phase was elevated by RNAi knockdown of two DEGs (both among nine down-regulated genes in cluster #5; fig. S12, L to P), compared with two genetic controls. We conclude that changes in aggressiveness by RNAi against most, if not all, DEGs are not secondary to the changes in activity levels. We found that RNAi knockdown of three DEGs specifically in the nvyLexA-expressing subset of Tdc2 neurons was sufficient to modulate aggression, demonstrating that this subpopulation is where the behaviorally relevant DEGs function (Fig. 7, K and L). It is noteworthy that five DEGs that showed behavioral phenotypes have proposed roles in RNA-related processes (Fig. 7E). The fact that genes controlled by nvy can both promote and suppress aggression suggests that nvy serves as a molecular hub that coordinates gene expression changes within a specific subpopulation of Tdc2 neurons to control the level of aggression.
The Nvy protein was detected in the ASM and VL clusters of both group-reared (fig. S13A) and socially isolated (fig. S13B) flies, suggesting that nvy expression does not markedly change with social experience. However, it remains possible that nvy expression is modulated specifically in a subtype of OA/TA neurons in response to social experience.
DISCUSSION
Our current work identified the genetic and neuronal nodes that are necessary for both male and female flies to adequately suppress aggression after group rearing, preventing excessive and maladaptive aggression (Fig. 7M). Although several mutations are known to make mice unusually aggressive (83), the neurogenetic mechanisms necessary for animals to reduce their levels of aggression in a social experience–dependent manner have not been well understood. The neuropeptide Drosulfakinin (Dsk) was reported to be necessary for suppressing aggression specifically in socially isolated flies, from the observation that deletion of the Dsk gene enhanced aggression in socially isolated, but not group-reared, flies (37). However, another group reported that Dsk mutation, as well as silencing of Dsk-expressing neurons, reduced aggression in socially isolated flies (84). Further studies will be necessary to clarify the function of Dsk on social experience–dependent modulation of aggressive behavior. Recently, mutation of another gene, hts, has been reported to increase aggression specifically among socially isolated flies (36). Last, RNAi-mediated knockdown of the transcription factor Tailless in pars intercerebralis neurons increased aggression in flies that were socially isolated for 2.5 days (85). Isolation for 3 days was sufficient to increase aggressiveness to a level comparable to that of flies isolated since the time of eclosion (6 days) (7), suggesting that Tailless knockdown is effective in enhancing social isolation–induced aggression. It is unclear whether Tailless knockdown can elevate aggression among group-reared flies. In summary, a function of these genes is to enhance aggression elevated by social isolation but not to suppress aggression in a social experience–dependent manner. In contrast to these works, our results clearly demonstrate that nvy gene products and nvyLexA-positive Tdc2 neurons are necessary for the suppression of aggression specifically among group-reared flies, providing a crucial link between social experience and the neurogenetic machinery in the central brain that controls the intensity of aggression. Functional loss of nvy appears to transform behavioral predispositions in response to group rearing rather than simply maintaining the socially isolated behavioral status. It has been proposed that different genotypes of Drosophila may respond to social experiences differently, in part by altering the very experience that each fly undergoes (16, 17). It will be interesting to address whether nvy underlies the variability in aggressive behaviors that has been observed among naturally isolated populations of Drosophila (3).
Nvy and its vertebrate homologs (MTGs) belong to an evolutionarily conserved group of transcription regulator proteins (79). Two of the human MTGs (MTG8 and MTG16) are targeted by chromosomal translocations that create a fusion protein with the MTGs and the DNA binding domain of the transcription factor RUNX1 (also known as AML1), which is associated with acute myeloid leukemia (79). Biochemical and genetic data suggest that MTGs are involved in cellular differentiation and organ development by serving as a scaffold protein for a transcription regulatory complex that mainly represses gene expression (75, 76, 79, 80, 86, 87). This is consistent with our finding that more genes were up-regulated (disinhibited) in nvy mutants, although whether the homologous genes are regulated in vertebrates remains currently unknown. In Drosophila, Nvy protein is proposed to bind to A-kinase anchoring proteins (88) [but see (89, 90)] via the NHR3 domain. Our result indicates that NHR2, and not NHR3, is the important domain for suppression of aggression by Nvy, which is consistent with the finding in MTGs that multimer formation via NHR2 is critical for their function as transcriptional regulators (80, 81).
How genetic manipulations of nvy affect the function of the Tdc2 neuronal subset is an intriguing question. One possibility is that nvy, as well as the candidates downstream of nvy identified in this study, is necessary for the maintenance of the connectivity and physiology of the nvyLexA-positive Tdc2 neurons. The underlying molecular mechanism remains unclear, although enrichment of genes related to RNA processing among candidate downstream genes implies that nvy-controlled genes may regulate actuators of neural functions at pre- and posttranscriptional stages, including trafficking of mRNAs and ribosomes to proper locations (91). Alternatively, considering the role of nvy and its vertebrate homologs in early development and cell differentiation (66, 92), nvy may be required for the proper establishment of nvyLexA-positive Tdc2 neurons. These two possibilities are not mutually exclusive. The role of Nvy/MTG proteins as a scaffold for histone deacetylases and other chromatin modifiers (75, 76, 79) suggests that functional abrogation of nvy can affect gene expression patterns across developmental stages rather than gating a specific neuronal differentiation process. Developmental stage–specific manipulations of nvy expression may clarify the function of nvy in aggressive behavior at each developmental stage. The fact that Nvy protein is detectable at the adult stage and that DEGs between nvy mutant and wild-type Tdc2 neurons at the adult stage modulate aggressive behavior strongly suggest that nvy plays a role in the developed nervous system. Genes involved in early development can be “repurposed” for neural functions at later stages (93, 94). In summary, further functional characterizations will be required to elucidate the molecular mechanism by which nvy influences the neuronal function of Tdc2 neurons and whether the behavioral impact of DEGs can be indeed attributed to specific subtypes of the Tdc2 neurons.
Inactivation of both the nvy gene and nvyLexA-positive Tdc2 neurons had the same behavioral effect (increased aggression), consistent with the idea that nvy is necessary for the proper function of nvyLexA-positive Tdc2 neurons. It is interesting that activation of the same neurons and overexpression of nvy both suppressed aggression. As a transcriptional repressor, an increased amount of Nvy protein complex may change the neuronal properties of nvyLexA-positive Tdc2 neurons so as to make them more sensitive to excitatory inputs (for instance, by altering the postsynaptic strength of synaptic inputs or by elevating membrane potential). Another important challenge is to understand how the amount of nvy influences the function of nvyLexA-positive Tdc2 neurons. Because the gross morphology of several subtypes of Tdc2 neurons remained unaltered in nvy mutants, behaviorally relevant changes within nvyLexA-positive Tdc2 neurons likely reside in the fine structures or in the physiological properties. The neural connectivity and physiology of Tdc2 neurons that belong to the so-called “VM” classes (65), which include aggression-promoting subtypes (41, 43), have been relatively well characterized particularly in the context of olfactory modulation (95–97), thanks in part to the complete description of the connectome and genetic drivers for several VM Tdc2 neurons (98, 99). By contrast, the ASM and VL clusters of Tdc2 neurons, which constitute most of the nvyLexA-positive Tdc2 neurons, have been less studied. The ASM cluster is reported to promote wakefulness by modulating pars intercerebralis neurons (100). The VL cluster, on the other hand, mediates starvation-induced desensitization of bitter-tasting gustatory receptor neurons (101). Both the ASM and VL clusters consist of multiple subtypes (65). Functional manipulation of specific subtypes will be necessary to elucidate which subtype suppresses aggression, whether they modulate the other behaviors described above as well, and where in the putative aggression-controlling circuit they are positioned. Although olfactory cues are implicated in social experience–dependent suppression of aggression in the fly (7, 33), none of the ASM or VL neurons innervate the major olfactory centers (98, 99, 102). These neurons may modulate olfactory perception indirectly or may act upon sensory inputs associated with social experiences after these are integrated.
Social experience—or the lack of it—influences subsequent interactions with conspecific individuals in both animals and humans. Our findings will serve as an entry point for understanding the circuit and molecular mechanisms that mediate a behavioral transformation associated with social experience in the fly. Comparative studies across animal species will help identify evolutionarily conserved genetic and neuronal motifs that are necessary for adaptive behavioral changes according to different levels of social experiences.
MATERIALS AND METHODS
Fly strains
Origins of fly lines
Full genotypes of flies used in experiments are listed in table S4. Canton-S originally from the laboratory of M. Heisenberg (University of Wurzburg) was used as the wild-type strain. UAS-IR lines used in the pan-neuronal RNAi screen (tables S1 and S2) were selected from the KK collection in the Vienna Drosophila Resource Center (VDRC), including UAS-IR-nvy (KK107374; VDRC, #100273, RRID:FlyBase_FBst0472147) used in Fig. 1B and fig. S1B. Other UAS-IR lines were obtained from the TRiP collection in the Bloomington Drosophila Stock Center (BDSC; University of Indiana), including another UAS-IR-nvy (JF03349; RRID:BDSC_29413) used in fig. S1A, UAS-IR-Tbh (JF02746; RRID:BDRC_27667), and UAS-IR-Tdc2 (JF01910; RRID:BDRC_25871) used in fig. S6. The Exelixis deficiency Df(2R)Exel6082 was obtained from BDSC (RRID:BDSC_7561). The following GAL4 lines were obtained from BDSC: Akh (RRID:BDSC_25684), AstA1 (RRID:BDSC_51978), AstA2 (RRID:BDSC_51977), AstC (RRID:BDSC_52017), Burs (RRID:BDSC_51980), Capa (RRID:BDSC_51969), Crz (RRID:BDSC_51975), Dh31 (RRID:BDSC_51988), Dh44 (RRID:BDSC_51987), Dsk (RRID:BDSC_51981), ETH (RRID:BDSC_51982), FMRFa (RRID:BDSC_51990), Mip (RRID:BDSC_51983), NPF (III) (RRID:BDSC_25682), Pdf (II) (RRID:BDSC_6900), Proc (RRID:BDSC_51971), amon (RRID:BDSC_30554), ato10 (RRID:BDSC_9494), ato14a (RRID:BDSC_6480), ey3–8 (RRID:BDSC_5534), ey4–8 (RRID:BDSC_5535), GH146 (RRID:BDSC_30026), Orco11.17 (RRID:BDSC_26818), Poxn1–7 (RRID:BDSC_66685), Ddc4.3D (RRID:BDSC_7010), Ddc4.36 (RRID:BDSC_7009), Tdc2 (RRID:BDSC_9313), Trh (RRID:BDSC_38389), 5-HT1B (II) (RRID:BDSC_27636), 5-HT1B (III) (RRID:BDSC_27637), R11H09 (RRID:BDSC_48478), R15F02 (RRID:BDSC_48698), R16F12 (RRID:BDSC_48739), R17C11 (RRID:BDSC_48763), R20G01 (RRID:BDSC_48611), R27G01 (RRID:BDSC_49233), R38G08 (RRID:BDSC_50020), R70B01 (RRID:BDSC_39511), R84H09 (RRID:BDSC_47803), and R93G12 (RRID:BDSC_40667). 8XLexAop2-FLPL (in attP40) (RRID:BDSC_55820) and the maternal stock for the MultiColor FlpOut (MCFO) (RRID:BDSC_64085) were obtained from BDSC. UAS-Dicer2 (X) from BDSC (RRID:BDSC_24644) was used in the w+ background. fruGAL4, elav-GAL4 (III), and ppk23-GAL4 were gifts from B. Dickson [Howard Hughes Medical Institute (HHMI) Janelia Research Campus]. dsxGAL4 was a gift from S. Goodwin (University of Oxford). NP2631 was a gift from D. Yamamoto (Tohoku University). ppk25-GAL4 and 10XUAS-IVS-Kir2.1eGFP (in attP2) were shared by D. Anderson (California Institute of Technology). 10XUAS-IVS-mCD8::GFP (in VK00005), 20XUAS-IVS-Syn21-GFP-p10 (in attP2), and 20XUAS-IVS-Syn21-CsChrimson::tdTomato3.1 (in attP2) were created by B. Pfeiffer in the laboratory of G. Rubin (HHMI Janelia Research Campus) and shared by D. Anderson. Tub-FRT-GAL80-FRT was a gift from K. Scott (University of California, Berkeley). Tub-FRT-stop-FRT-GAL80 was a gift from B. Zhang (University of Missouri). hs-Cre (X) was a gift from K. Basler (University of Zürich).
Genetic intersection labeling nvy-positive or nvy-negative Tdc2 neurons
Genetic access to each subpopulation shown in Fig. 4 (D to I) was achieved by selective expression of GAL80 by the following genotypes: (i) nvy(+) Tdc2(+) neurons were labeled by the genotype: (w; Tdc2-GAL4, nvyLexA/Tub-FRT-GAL80-FRT, 8XLexAop2-FLPL; 10XUAS-IVS-XX/+). In these flies, GAL80 is ubiquitously expressed under the tubulin promoter. Cells labeled by nvyLexA express flippase, which excises the GAL80 coding sequence flanked by flippase recognition targets (FRTs). This allows Tdc2-GAL4 to express effectors (XX: GFP or Kir2.1eGFP) selectively in nvy-positive cells; (ii) nvy(−) Tdc2(+)neurons were labeled by the genotype: (w; Tdc2-GAL4, nvyLexA/Tub-FRT-stop-FRT-GAL80, 8XLexAop2-FLPL; 10XUAS-IVS-XX/+).
As a transcriptional stop cassette flanked by FRTs precedes the GAL80 coding sequence, nvy-positive Tdc2 cells flip out the stop cassette, leading to GAL80-dependent suppression of GAL4. The remaining Tdc2 cells (the nvy-negative Tdc2 subpopulation) can express the effector.
Generation of transgenic lines
UAS-nvy, NHR domain–deleted versions of UAS-nvy, LexAop2-nvy, UAS-hMTG8, and UAS-hMTG16 lines were generated by ΦC31 integrase–mediated transgenesis as previously described (103). Primer sequences are provided in table S5.
The nvy coding sequence (CDS) [2232 base pairs (bp)] was amplified from the complementary DNA (cDNA) of the Canton-S strain. The CDS confirmed by sequencing is shown in data S1. Either three-tandem c-Myc (3xMyc; EQKLISEEDLEQKLISEEDLEQKLISEEDL) or HA (3xHA; YPYDVPDYAGYPYDVPDYAGSYPYDVPDYA) epitope tag was attached to the 5′ end of the nvy CDS. As for the domain-deletion mutants of nvy, primers were designed to skip each NHR region (NHR1, 631st to 924th; NHR2, 1360th to 1440th; NHR3, 1540th to 1686th; NHR4, 1777th to 1890th nucleotides within the nvy CDS). The original CDSs of the human MTG8b (1815 bp; GenBank: D14821.1) and MTG16b (1704 bp; GenBank: AB010420.1) genes were codon-optimized for expression in Drosophila (nucleotide sequences shown in data S1) by GenScript USA Inc. (Piscataway, NJ).
To make the 10XUAS constructs, the backbone plasmid pJFRC-MUH (RRID:Addgene_26213) was inserted with the intervening sequence (IVS) (104) downstream of the hsp70 promoter, between Bgl II and Not I sites. As for the 13XLexAop2 constructs, pJFRC48-13XLexAop2-myr::tdTomato [a derivative of pJFRC19-13XLexAop2-myr::GFP (RRID:Addgene_26224) originally described in (104)] was used as the backbone plasmid. Linker sequences containing the Not I site and the Kozak sequence (CAAA) were added right upstream to each CDS. Fragments and vectors were double-digested with Not I (NEB, #R3189) and Xba I (NEB, #R0145), followed by ligation using T4 DNA ligase (NEB, #M0202). Integrities of the resulting plasmids were confirmed by DNA sequencing. Plasmids were targeted to the attP site at VK00005 (RRID:BDSC_9725) using ΦC31 integrase–mediated transgenesis by BestGene Inc. (Chino Hills, CA). Transformants were selected by the eye color marker, and the presence of inserted CDSs was confirmed by polymerase chain reaction (PCR) genotyping. All transgenic lines were backcrossed to the wild-type Canton-S for six generations before experiments.
CRISPR-Cas9–mediated generation of nvy mutants
∆nvy and nvyLexA lines were created on the basis of the CRISPR-Cas9–mediated genome editing (105) as follows. Primer sequences are provided in table S5.
Target sites for guide RNAs (gRNAs) were searched by using the online CRISPR Target Finder available at the flyCRISPR website (https://flycrispr.org/) with default settings. Within the genome region surrounding the first exon of the nervy gene (Dmel\CG3385), the following sites with no detectable off-targets were selected [protospacer adjacent motif (PAM) sequence underlined]: gRNA target #1: 5′-TGATGTTTTCGTCTATCGCCCGG-3′; gRNA target #2: 5′-TCATTGTTTGGAACTATAATAGG-3′.
Primers containing linkers attached to each target without the PAM sequence were used for PCR with pCFD4 (RRID:Addgene_49411) as a template. The amplified 598-bp fragment was ligated with Bbs I–digested pCFD4, and the resulting pCFD4-nvy-gRNA-1 plasmid was injected into embryos of the vas-Cas9 (X) strain (RRID:BDSC_51323). The F0 adults (17 males and 17 females) were crossed individually with a balancer line, and F1 flies (5 to 12 males and 5 females from each F0 cross) were screened by PCR genotyping. Among 354 F1 individuals, two sibling lines with an identical 513-bp deletion (from −132 bp to +134 bp of the first exon) were found, designated herein as ∆nvy. The ∆nvy line was backcrossed to the wild-type Canton-S for 11 generations before experiments.
Our initial attempt for knock-in line generation using the above gRNA plasmid failed (none of 478 F1 individuals from 16 F0 crosses were DsRed positive) presumably due to the low efficacy of genome editing. To overcome this issue, we prepared a secondary plasmid (pCFD4-nvy-gRNA-2) for additional supply of gRNAs that target the following sites: gRNA target #3: 5′-GTTTCCAAGTTCCCAGGTTCCGG-3′; gRNA target #4: 5′-CACCAACAACACAACATCGGCGG-3′.
To construct the nvyLexA knock-in plasmid, pHD-DsRed (RRID:Addgene_51434) was used as the backbone. Left (from −1688 to −1 bp of the first exon) and right (from +140 to +1859 bp of the first exon) homologous arms were amplified from the genome DNA of the vas-Cas9 (X) strain. Point mutations were introduced within the PAM sequences of gRNA target sites to avoid plasmid cleaving by Cas9. The nls::LexA::p65 CDS was amplified from pBPnlsLexA::p65Uw (RRID:Addgene_26230). The first exon of nvy, of which the start codon was mutated from ATG to TAG, followed by the 139-bp downstream region was amplified from the genome DNA. The knock-in plasmid was constructed by using an In-Fusion HD Cloning kit (Takara Bio USA, #639650) or a NEBuilder HiFi DNA Assembly kit (New England Biolabs, #M5520) in two steps: The left homologous arm, nls::LexA::p65 CDS, and the first exon of nvy were first fused with the Xho I/Spe I–digested pHD-DsRed vector; then, the resulting plasmid was digested with Not I/Eco RI followed by insertion of the homologous right arm to generate pHD-DsRed-nvy-LexA.
Three plasmids (pCFD4-nvy-gRNA-1, pCFD4-nvy-gRNA-2, and pHD-DsRed-nvy-LexA) were coinjected to embryos of the vas-Cas9 (X) strain. The F0 adults (24 males) were crossed each with the balancer line, and F1 offspring were screened for DsRed expression in compound eyes under a fluorescent microscope. From 382 F1 males collected from seven F0 crosses, 15 individuals were found positive for DsRed. Insertion of LexA was confirmed by genotyping PCR with the primers used for the pHD-DsRed-nvy-LexA plasmid construction. Three candidate lines were backcrossed to the wild-type Canton-S for six generations, and the DsRed sequence flanked by two loxP sites was excised by crossing with hs-Cre (X). Genomic regions surrounding the first exon of nvy were analyzed by Southern blot as described below. One of the validated alleles was used as nvyLexA for further experiments. Plasmid injections to fly embryos were performed by BestGene Inc.
Southern blot
Two hundred adult flies per genotype were ground in 800 μl of TE buffer [10 mM tris-HCl (pH 9) and 1 mM EDTA] supplemented with 1% SDS, followed by incubation at 65°C for 30 min. Samples were added with 300 μl of 3 M potassium acetate and placed on ice for 30 min. After centrifugation at 13,000 rpm for 20 min at 4°C, the supernatant (600 μl) was collected and mixed with a half volume of isopropanol. Samples were centrifuged at 13,000 rpm for 10 min, and the pellet was washed with 70% ethanol. Precipitates were dried and dissolved in 500 μl of TE buffer. Samples were then treated with ribonuclease (RNase) A (diluted to 0.4 to 0.8 mg/ml; Sigma-Aldrich, #R4642) at 37°C for 15 min. For purification, each sample was mixed vigorously with the same volume of PCI [phenol:chloroform:isoamyl alcohol = 25:24:1 (v/v); Thermo Fisher Scientific, #15593031]. After centrifugation at 13,000 rpm at 5 min, the aqueous upper layer was collected and mixed vigorously with the same volume of chloroform, followed by another centrifugation. The upper layer (400 μl) was further subjected to ethanol precipitation. The final precipitates obtained were dried and dissolved in 100 μl of TE buffer. The typical yield of genomic DNA extracted from 200 flies was 0.2 to 0.5 mg.
Genomic DNA (10 to 20 μg) for each genotype was digested with Hind III at 37°C overnight. Electrophoresis was performed using a 0.7% agarose gel. Digoxigenin (DIG)–labeled DNA molecular weight maker III (Roche, #11218603910) was loaded as a marker. The gel placed on a shaker was sequentially subjected to depurination (in 0.25 N HCl for 10 min), denaturation (in 0.5 M NaOH and 1.5 M NaCl for 15 min × 2), neutralization [in 0.5 M tris-HCl (pH 7.5) and 1.5 M NaCl for 15 min × 2], and equilibration (in 20× SSC for 10 min). DNA was transferred to a nylon membrane (Roche, #1120929901) overnight, by sandwiching between paper towels soaked in 20× SSC under a weight of 1.5 kg. DNA was immobilized onto the membrane by using UV Stratalinker 2400 (Stratagene).
DIG-labeled DNA probes were synthesized using the PCR DIG Probe Synthesis Kit (Roche, #11636090910). Primers were designed to target either the external (676 bp; the genomic region from −1986 to −2661 bp upstream of the nvy exon 1) or internal (621 bp; 660th to 1280th nucleotides within the nls::LexA::p65 CDS) regions of the LexA knock-in construct, as shown in table S5. The DIG-labeled probes were hybridized to the membrane in DIG Easy Hyb hybridization buffer (Roche, #11603558001) at 49°C overnight. The membrane was sequentially washed with a low-stringency buffer (2× SSC and 0.1% SDS) at room temperature for 5 min × 2 and with a prewarmed high-stringency buffer (5× SSC and 0.1% SDS) at 68°C for 15 min × 2. After another brief wash with a buffer (from the DIG Easy Hyb kit), the membrane was soaked in a blocking buffer (from the DIG Easy Hyb kit) at 4°C overnight. Alkaline phosphatase–conjugated anti-DIG Fab fragments (Roche, #11093274910, RRID:AB_514497) were freshly added to the blocking buffer at 1:10,000, and the membrane was incubated at room temperature for 30 min. The membrane was washed with the wash buffer for 15 min × 2, followed by a brief equilibration in a detection buffer (from the DIG Easy Hyb kit). As a chemiluminescence substrate, CDP-Star (Roche, #11759051001) was freshly diluted to 1:200 in the same buffer. Signals were developed on autoradiography films (Genesee Scientific, #30-507).
Western blot
Sixty to 90 adult flies (5 to 7 days after eclosion) were snap-frozen in liquid nitrogen. The fly heads were separated from other body parts in liquid nitrogen–chilled metal sieves. Collected heads were ground in 60 to 90 μl of ice-cold extraction buffer [20 mM Hepes (pH 7.5), 100 mM KCl, 10 mM EDTA, 0.1% Triton X-100, 1 mM dithiothreitol, and 5% glycerol; according to (106)] with disposable pestles, followed by centrifugation at 1600g for 20 min at 4°C. The supernatant was mixed with 4× Laemmli Sample Buffer (Bio-Rad, #1610747), and samples were heated in boiling water for 5 min.
Proteins were separated in 4 to 20% Mini-PROTEAN TGX Precast Protein Gels (Bio-Rad, #4561096) and transferred to 0.45-μm–pore size nitrocellulose membranes (Bio-Rad, #1620215). Membranes were shaken in TBST [20 mM tris-HCl (pH 7.6), 150 mM NaCl, and 0.1% Tween 20] supplemented with 5% blotting-grade blocker (Bio-Rad, #1706404) at room temperature for 2 to 3 hours. After washing in TBST for 10 min × 3, membranes were incubated with primary antibodies (1:1000 to 1:10,000 dilution in 2 to 5% skim milk/TBST or Can Get Signal solution 1; Toyobo, #NKB-201) at room temperature for 1 to 2 hours. Membranes were washed in TBST for 10 min × 3, followed by reaction with horseradish peroxidase–conjugated secondary antibodies (1:10,000 dilution in 2 to 5% skim milk/TBST or Can Get Signal solution 2; Toyobo, #NKB-301) at room temperature for 1 to 2 hours. After the final wash in TBST for 10 min × 3, membranes were treated with Clarity Western ECL Substrate (Bio-Rad, #1705061). Signals were developed on autoradiography films (Genesee Scientific, #30-507). Detailed information for antibodies and incubation conditions are provided in table S6.
Immunoprecipitation
Immunoprecipitation of Myc- and HA-tagged Nvy proteins was preformed essentially as described previously (107). Tagged Nvy proteins were pan-neuronally expressed under the control of elav-GAL4. Heads from 100 to 120 flies were isolated using liquid nitrogen–chilled metal sieves as described above, followed by homogenization in 700 μl of buffer B {20 mM tris-HCl (pH 7.6), 150 mM NaCl, 5 mM MgCl2, 10% sucrose, 1% glycerol, 1 mM EDTA, and protease inhibitors [one tablet of cOmplete Protease Inhibitor Cocktail (Roche, #11697498001) dissolved in 50 ml]} supplemented with 1% CHAPS. Homogenates were first centrifuged at 16,000g for 30 min at 4°C, and the supernatants were centrifuged again at 16,000g for 20 min at 4°C. Cleared lysates (650 μl) were collected carefully using capillary pipet tips. Lysates were separated into three groups of 200 μl each and added with 800 μl of buffer A [20 mM tris-HCl (pH 7.6), 150 mM NaCl, 1 mM dithiothreitol, 3 mM MgCl2, and 1 mM EGTA]. The remaining lysates were stored at −20°C to be used as “inputs.”
Protein G PLUS-Agarose (Santa Cruz Biotechnology, #sc-2002, RRID:AB_10200697) was washed with buffer A, and 10 μl of 50% bead slurry was added to each sample. As a precleaning step, samples were gently rotated for 1 hour at 4°C. Samples were then centrifuged at 1000g for 30 s at 4°C, and collected supernatants were centrifuged again at 3000g for 30 s at 4°C. For each genotype, one sample was kept as a negative control without antibody, another sample was added with 2.5 μl of normal rat immunoglobulin G (IgG; 0.4 mg/ml; Santa Cruz Biotechnology, #sc-2026, RRID:AB_737202), and the last sample was added with 1 μl of anti–c-Myc rat IgG1 (1 mg/ml; clone JAC6, Abcam, #ab10910, RRID: AB_297569). The antibody binding was performed for 2 to 3 hours at 4°C on a rotator. To prepare the beads for precipitation, Protein G PLUS-Agarose was washed and suspended in buffer B supplemented with 0.2% CHAPS and 1% bovine serum albumin and incubated for 30 min at 4°C on a rotator. Beads were washed twice in buffer B and then suspended to make 50% slurry. For immunoprecipitation, each sample was added with 40 μl of bead slurry and incubated overnight at 4°C on a rotator. After centrifugation at 1000g for 30 s at 4°C, precipitated samples were washed twice with 0.5 ml of buffer A supplemented with 0.2% CHAPS. The final precipitates were suspended in 20 μl of 2× Laemmli buffer and heated in boiling water for 10 min. Western blot was performed as described above.
Immunohistochemistry
Standard immunostaining
Immunohistochemistry of fly brains essentially followed the method described in (108). Fly brains were dissected in phosphate-buffered saline (PBS) and then incubated in the fixing solution (2% formaldehyde and 75 mM l-lysine in PBS) at room temperature for 1 to 1.5 hours. All reactions from fixation to clearing were carried out in a well of 6 × 10 microwell minitray (Thermo Fisher Scientific, #439225). Brains were washed in PBST (0.3% Triton X-100 in PBS) for 5 min × 3, followed by incubation in a blocking solution (5% heat-inactivated normal goat serum and 0.3% Triton X-100 in PBS) for 30 min. Primary antibodies diluted with the blocking solution [1:10 (supernatant) or 1:100 (concentrated) for mouse anti-bruchpilot (BRP) (Developmental Studies Hybridoma Bank nc82, RRID: AB_2314866), 1:1000 for chicken anti-GFP (Abcam, #ab13970, RRID: AB_300798), 1:1000 for rabbit anti-DsRed (Takara Bio USA, #632496, RRID: AB_10013483), and 1:1000 for rabbit anti-Nvy (a gift from R. Mann)] were applied to the samples at 4°C for 2 days. The brains were washed in PBST for 10 min × 3 and then incubated in secondary antibodies diluted with the blocking solution [1:100 for goat anti-mouse Alexa 633 (Thermo Fisher Scientific, #A-21052, RRID: AB_2535719), 1:100 for goat anti-chicken Alexa 488 (Thermo Fisher Scientific, #A-11039, RRID: AB_2534096), and 1:100 for goat anti-rabbit Alexa 568 (Thermo Fisher Scientific, #A-11036, RRID: AB_10563566)] at 4°C overnight. Brains were washed in PBST for 10 min × 3 and then incubated in the clearing solution (50% glycerol/PBS) at room temperature for 2 hours. Samples were mounted in VECTASHIELD (Vector Laboratories, #H-1000) onto a glass slide. Images were acquired by FV-3000 confocal microscopy (Olympus America; shared by S. Pfaff at Salk Institute). Stacked images of maximum z-projections were generated on Fiji software (RRID: SCR_002285; https://fiji.sc/). FluoRender (RRID: SCR_014303; www.sci.utah.edu/software/fluorender.html) was used to create a three-dimensional rendering of a stacked confocal image.
Single-cell stochastic labeling
Single-cell stochastic labeling was performed by the MCFO approach described in (109). For wild-type (for nvy) flies, virgin females of the MCFO maternal strain (RRID:BDSC_64085) were crossed with Tdc2-GAL4 males. The F1 male offspring with the following genotype were used for the MCFO experiment: w,hs-FLPG5-PEST in attP3/Y; Tdc2-GAL4/+; 10XUAS-FRT-stop-FRT-myr::smGdP-HA in VK00005,10XUAS-FRT-stop-FRT-myr::smGdP-V5-THS-10XUAS-FRT-stop-FRT-myr::smGdP-FLAG in Su(Hw)attP1/+.
For nvy mutant flies, the ∆nvy allele was first combined with the MCFO maternal strain. Virgin females from this stock (that carries ∆nvy on the second chromosome) were subsequently crossed with the males that carried both the ∆nvy allele and the Tdc2-GAL4 insertion. The F1 male offspring with the following genotype were used for the MCFO experiment: w,hs-FLPG5-PEST in attP3/Y; ∆nvy,Tdc2-GAL4/∆nvy; 10XUAS-FRT-stop-FRT-myr::smGdP-HA in VK00005,10XUAS-FRT-stop-FRT-myr::smGdP-V5-THS-10XUAS-FRT-stop-FRT-myr::smGdP-FLAG in Su(Hw)attP1/+. With the use of Tdc2-GAL4 driver, preliminary investigation of heat shock conditions revealed that optimally sparse labeling was achieved when flies were treated as follows: reared at 25°C from embryo to adult 4 to 7 days after eclosion, warmed at 37°C for 10 to 11 min, and then kept at 25°C for another 1 to 2 weeks before dissection.
Subsequent immunostaining steps were performed according to (109) with minor modifications. Brains were dissected in ice-cold Schneider’s medium and fixed in 2% paraformaldehyde (PFA)/Schneider’s medium at room temperature for 55 min, followed by PBT (0.5% Triton X-100/PBS) washes for 10 min × 4. Samples were incubated in a blocking solution (5% goat serum/PBT) at room temperature for 90 min. Primary antibodies {1:300 dilution of anti-HA rabbit monoclonal antibody (mAb) (C29F4, Cell Signaling Technologies, #3724S, RRID:AB_1549585), 1:200 dilution of anti-FLAG rat mAb [DYKDDDDK Epitope Tag Antibody (L5), Novus Biologicals, #NBP1-06712, RRID:AB_1625981], and 1:10 dilution of supernatant anti-BRP mouse mAb (Developmental Studies Hybridoma Bank nc82, RRID: AB_2314866), all diluted in 5% goat serum/PBT} were applied at 4°C for 2 days. The brains were washed in PBT for 30 min × 3 and then incubated with secondary antibodies [1:500 dilution of anti-rabbit Alexa Fluor 594 (Jackson ImmunoResearch, #711-585-152, RRID:AB_2340621), 1:150 dilution of anti-rat Alexa Fluor 647 (Jackson Immuno Research, #712-605-153, RRID:AB_2340694), and 1:150 dilution of anti-mouse Alexa Fluor 488 (Jackson ImmunoResearch, #715-545-151, RRID:AB_2341099), all diluted in 5% goat serum/PBT] for 4 hours at room temperature followed by one to two overnights at 4°C. Brains were washed in PBT for 30 min × 3 and then incubated with DyLight 550–conjugated mouse anti-V5 (Bio-Rad, #MCA1360D550GA, RRID:AB_2687576) (1:500 dilution in 5% goat serum/PBT) at 4°C overnight.
Mounting of stained brains was also performed according to (109) and the “DPX Mounting” protocol shared at the FlyLight website (www.janelia.org/project-team/flylight/protocols). Coverslips (22 mm by 22 mm square no.1, Thermo Fisher Scientific, #12-542B) were dipped in a coating solution [0.08% (w/v) poly-l-lysine (PLL) hydrobromide (Sigma-Aldrich, #P1524) and 0.2% Kodak Photo Flo 200 (Electron Microscopy Sciences, #74257) in water, stored at 4°C] and air-dried beforehand. Brains were washed with PBT for 30 min × 3 and fixed once again with 4% PFA/PBS (without Triton X-100) at room temperature for 4 hours followed by PBT washes for 15 min × 4. Slides with spacers were made as described in the above website. Before mounting, brains were washed in PBS (without Triton X-100) for 5 min × 2 to remove detergents. Several drops of PBS were made on the PLL-coated coverslip, and brains were transferred into the drops and gently placed on the glass surface. With the brains stuck to one side, the coverslips were briefly washed with water to remove PBS and then subjected to dehydration series by 10-min soaks in successive baths of ethanol solutions (30, 50, 75, 95, 100, 100, and 100%), followed by 5-min soaks in three successive baths of xylene for clearing. The coverslips taken out of xylene were held horizontally and applied immediately with seven drops of DPX mountant (Electron Microscopy Sciences, #13512). The coverslips were flipped over the slides and placed between the spacers and gently pressed down. DPX was further applied near the edges of coverslips. The DPX-mounted slides were air-dried in the dark for 2 to 3 days before microscopic observations. Images were acquired by a Zeiss LSM 880 (at the Waitt Advanced Biophotonics Core, Salk Institute). Maximum z-projections were generated by Fiji as described above.
Social behavior experiments
Behavioral apparatus
Twelve-well acrylic chambers were designed as previously described (110). Each arena had a diameter of 16 mm and a height of 10 mm. The entire floor was covered with apple juice gel [Minute Maid 100% apple juice, 2.25% agarose, and 2.5% sucrose (w/v)] as food source. The inner wall and ceiling were coated with Insect-A-Slip (BioQuip Products #2871C) and Surfasil Siliconizing Fluid (Thermo Fisher Scientific, #TS-42800), respectively.
The chambers were lit from underneath by LED backlights. For optogenetic experiments, 850-nm infrared backlights (Sobel Imaging Systems, #SOBL-150x100-850) were used instead. Movies were taken using the Point Grey Flea3 USB3.0 digital cameras (FLIR, #FL3-U3-13Y3M-C) controlled by the BIAS acquisition software (IORodeo; http://stuff.iorodeo.com/notes/bias/). The camera was mounted with a machine vision lens (Fujinon, #HF35HA-1B). For optogenetic experiments with the infrared backlights, an infrared long-pass filter (Midwest Optical Systems, #LP780-25.5) was attached to the camera. Recording was performed either at 30 fps for the first round of RNAi screen (Fig. 1A, left) or at 60 fps for the rest of all experiments. The optogenetic stimulation was performed using 655-nm red light LEDs controlled by the Arduino Uno board (Arduino) with a custom program as described previously (111).
Fly preparation and behavioral assays
Parental flies (no more than 20 females and 10 males per bottle) were reared on 50 ml of standard cornmeal-based food and were transferred to fresh food every 2 to 3 days. Offspring flies were collected on the day of eclosion into vials with standard fly food medium. Adult males and females were kept separately to avoid mating, except when premated females were prepared for targets in some experiments. The premated females (Fig. 3, C and D, and fig. S7H) were created by cohousing 10 to 15 virgin females and five males (both 3 to 4 days after eclosion) in the same vials 2 to 3 days before the day of the experiment. For optogenetic experiments, adult testers were reared on food supplied with 0.2 mM all-trans retinal (Sigma-Aldrich, #R2500, 20 mM stock solution prepared in 95% ethanol), and the vials were covered with aluminum foil to avoid light exposure. Flies were kept either as a group of up to 15 (“group reared”) or 1 (“single reared”) per vial at 25°C with 60% relative humidity, in a 12-hour light/dark cycle (light phase, 9:00 a.m. to 9:00 p.m.). Flies were transferred to new vials with fresh food after every 3 days. To keep track of each fly’s identity within a pair of males with different genotypes, the tip of either one of the wings was clipped by a razor under brief CO2 anesthesia. This marking treatment itself does not reduce the level of lunge or wing extension by males under our experimental conditions (112).
Behavior experiments were performed in the late afternoon (4:00 to 9:00 p.m.) at 22° to 25°C. When pairing group-reared flies of the same genotypes (including RNAi screening), two flies were always taken from different vials to make a pair that has never met each other during their adulthood. For male-male and female-female pairs tested in the 12-well chamber, flies were introduced by gentle aspiration and acclimated for 5 min before the 30-min recording. In the case of male-female pairs (Fig. 3, A to D, and fig. S7H), either virgin or premated females were first introduced into the arenas, and males were trapped between two small plastic tips set upon the lid to prevent contact with females. After 5 min of acclimation, all males were simultaneously introduced to the arenas by sliding the lid, and the 1-hour recording was immediately started. In competitive copulation assays, group-reared male pairs were first loaded into the 12-well chamber, and one group-reared virgin female was trapped upon each well as described above. After acclimating the male pairs for 15 min, the females were simultaneously introduced to all arenas, and recording was immediately started. For experiments using the large chamber where we aimed to capture the flies’ behaviors from the first encounter, the recording was started before the introduction of flies to the arenas, and the movie was taken for 35 min. The 30-min time window after the entrance of the second fly to the arena was used for behavior analysis.
Courtship memory assay was performed essentially as described previously (113). In brief, a sexually naïve male was introduced into a 1.5-ml tube with food and either kept alone as the sham-trained group or paired with an unreceptive mated female as the trained group. Females were removed after 5 hours of training, and the males were left in the same tubes for 1 hour. Males were then transferred into the 12-well chamber, and a new set of wild-type mated females was loaded as above. The 30-min recording was started immediately after the females were introduced into the arenas.
Behavioral quantification
The 30-fps movies recorded in the first round of RNAi screen (Fig. 1A, left, and table S1) were analyzed by CADABRA software (45) on MATLAB (RRID: SCR_001622; The MathWorks). The program was slightly modified to make it compatible with later MATLAB versions (2014b and 2019a) without affecting its functionality. Flies were tracked using the “qtrak” function. Lunges were detected using the analysis program that accompanies CADABRA, by applying the parameters originally described in (45). The radius of the circular region of interest was set to 6.5 mm, which is approximately one fly body length smaller than the actual well size (8-mm radius), to exclude the movements of flies staying close to or climbing on the wall that may lead to false-positive detections.
The 60-fps movies recorded for the rest of all experiments were processed by the FlyTracker program (56) (version 1.0.5) on MATLAB2014b. For pairs of flies with different conditions (sexes, genotypes, and/or rearing conditions), the identities of tester and target flies (marked by the clipped wing) were manually corrected throughout the movie. Behaviors were quantified using automated classifiers based on the machine learning system JAABA (RRID: SCR_021597). The details of the classifiers for lunges, headbutts, and wing extensions are described elsewhere (46). As a postprocessing step to remove false positives, extremely short bouts (<50 ms for lunges and headbutts and <100 ms for wing extensions) were omitted from quantification.
In general, our lunge classifier has been trained to detect lunge bouts with reasonably high precision (89%) and recall (88%) (46). When testing male-female pairs, however, even a few cases of false positives due to tracking errors might significantly affect the statistical analyses of rarely observed male-to-female lunges. For this reason, we manually confirmed all male-to-female lunges detected in Fig. 3 (A and C to E).
Behavioral transition analysis
A fly in each frame was labeled with one of five mutually exclusive behaviors: stopping, orienting, nonorienting, lunge, and wing extension. Lunge and wing extension were quantified as described above. Stopping, orienting, and nonorienting were defined on the basis of the fly’s frame-wise kinematic features. First, a fly was classified as “stopping” when its speed was below the first threshold of 1 mm/s. To avoid rapid switching of behavior labels around this speed threshold, a Schmitt trigger similar to that used in (114) was adopted with the second (high) threshold of 2.5 mm/s. The fly was classified as moving (i.e., orienting or nonorienting) at above 2.5 mm/s; the frames in between the two thresholds maintained their previous classification until the second threshold was crossed. Among all the moving frames, the fly was classified as “orienting” toward the other fly when it had a facing angle magnitude smaller than 45° and at a distance no larger than 5.0 mm. Schmitt triggers on facing angle (high threshold: 60°) and on distance (high threshold: 7.5 mm) were also introduced. All the remaining moving frames were classified as “nonorienting.” As discussed in the Results section titled “nvy controls a behavioral transition leading to lunges”, “orienting” corresponds to chasing observed in both courtship and aggressive interactions (48–50, 57, 60), although the term “orienting” was chosen because it was unclear whether the orienting was evoked by the presence of the other fly in our assays, as opposed to an assay in which a programmed visual stimulus was presented to trigger an active chasing from the fly (57, 58, 115, 116). Last, “lunge” and “wing extension” labels were added by overriding stop, orienting, and nonorienting labels. An “event” of a behavior was defined as a segment of consecutive frames with the same behavior label. Events with the same behavior labels that were no more than 50 ms apart were merged by overriding the labels of intervening frames. Events that were shorter than 100 ms were eliminated from the subsequent analysis. This postprocessing was deemed appropriate to prevent excessive fragmentation [or “splitting” as described in (117)] of behavioral events, which can hinder transition analysis.
A behavioral transition matrix (48) was generated in the following procedure. First, all first-order transitions from one event to the following event were tallied for each fly. The result was represented as a matrix, in which the source event specifies the row, and the following event specifies the column. The number in each cell corresponds to the total number of transitions from the source event to the following event performed by each fly. Our definition of behavior events precludes self-transition. Matrices across all flies of the same genotype and rearing condition were then summed to have a sufficient number of instances per transition. This population-level tally was normalized for each row (the total number of events for the given behavior) to generate the transition probability matrix. Each probability in the transition matrix provides an estimate of how likely a certain behavior was to occur after another behavior for a given genotype under the single- or group-reared condition. Abundance, duration, and interval of the behaviors were separately analyzed (see fig. S2, A to F) because the transition probability matrix does not provide information about these.
An ethogram is a simplified version of a transition matrix. For clarity, transitions with a probability of less than 10% were eliminated except when comparing it to the probability in other groups. A diameter of a circle (behavior) corresponds to a logarithm of the average number of events per fly.
Single-cell RNA sequencing
Preparation of single-cell suspensions
Virgin male and female flies expressing mCD8::GFP under the control of Tdc2-GAL4 in either wild-type nvy locus or homozygous ∆nvy mutant background were used. Adults were collected upon eclosion and kept as a group of 15 per vial for 5 to 7 days at 25°C, as done for social behavior experiments (see above).
Single-cell suspensions were prepared according to the protocol detailed in (118). Fly brains were dissected and stored in ice-cold Schneider’s insect medium (Sigma-Aldrich, #21720024) for up to 2 hours. The brains were rinsed in cold RNase-free PBS and transferred to freshly made dissociation buffer [300 μl of heat-activated papain (100 U/ml; Worthington Biochemical Corporation, #LK003178) added with 6 μl of Liberase (2.5 mg/ml; Sigma-Aldrich, #5401119001)], followed by an incubation for 20 min at 25°C under continuous shaking at 1000 rpm. During this 20-min incubation, the suspension was pipetted 30 times at the 5- and 10-min time points and then forced through a 25-gauge 5/8 needle 7 times at the 15-min mark. One milliliter of ice-cold Schneider’s insect medium was added to terminate the enzymatic digestion. The suspension was then filtered through a cell strainer with a mesh size of 35 μm (BD Biosciences, #352235) and centrifuged at 600g for 7 min at 4°C. The pellet was resuspended in cold Schneider’s insect medium supplemented with DAPI (4′,6-diamidino-2-phenylindole; 1 μg/ml; Thermo Fischer Scientific, #D-1306). Samples were sorted using the BD Vantage DiVa Cell Sorter (BD Biosciences). Gates were set to collect viable (DAPI-negative) GFP-positive cells as shown in fig. S11A. Single cells were collected into individual wells of 96-well PCR plates containing 9.5 μl per well of freshly made lysis buffer (provided in a SMART-Seq v4 Ultra Low Input kit for Sequencing; Takara Bio USA, #634893). After sorting, samples were immediately placed on dried ice and stored at −80°C until use. To prevent nonphysiological transcriptional activities triggered during the single-cell preparation process, all solutions were supplemented with actinomycin D (Sigma-Aldrich, #A1410) at the final concentration of 5 μg/ml.
In total, brains were dissected from 359 nvy wild-type (213 males and 146 females) and 381 ∆nvy (242 males and 139 females) flies in 10 experimental days. Lysates of 197 nvy wild-type (93 male- and 104 female-derived) and 216 ∆nvy (112 male- and 104 female-derived) cells were processed as below.
Single-cell sequencing
mRNA in the cell lysate was reverse-transcribed and amplified for 25 cycles using the SMART-Seq v4 Ultra Low Input kit for Sequencing (Takara Bio USA, #634893) according to the manufacturer’s instructions. To confirm the presence of GFP transcripts, each cDNA was subjected to PCR genotyping using Emerald AMP HS PCR Master Mix (Takara Bio USA, #RR330B) and primers shown in table S5. As a result, 104 nvy wild-type (55 male- and 49 female-derived) and 114 ∆nvy (58 male- and 56 female-derived) GFP-positive samples were selected for sequencing. The amplified cDNAs were quantified by the Qubit 3.0 Fluorometer (Thermo Fischer Scientific, #Q33216) and normalized to a concentration of 0.22 ng/μl. Sequencing libraries were prepared using the Nextera XT kit (Illumina, #FC-131-1096) and mixed into 24 pools (12 samples per pool). After purification using the Agencourt AMPure XT beads (Beckman Coulter, #A63881), the sample quality was checked with both the Qubit 3 Fluorometer and the High Sensitivity D1000 ScreenTape assay (Agilent Technologies, #5067-5584). The libraries were equimolarly pooled, and the final concentration was estimated by qPCR using primers shown in table S5 and the KAPA Library Quantification Kit Illumina Platforms KK4828 (Roche, #07960166001) according to the manufacturer’s instructions. Sequencing of 75-bp paired-end reads was performed with the Illumina NextSeq 500 sequencer.
Bioinformatics analysis
In total, 218 cells were sequenced. Reads were quality-tested using FASTQC (RRID: SCR_014583; www.bioinformatics.babraham.ac.uk/projects/fastqc) and aligned to the D. melanogaster genome dm6 (from The FlyBase Consortium/Berkeley Drosophila Genome Project/Celera Genomics) using the alignment algorithm STAR version 2.5.3a (RRID: SCR_004463). Mapping was carried out using default parameters (up to 10 mismatches per read and up to nine multimapping locations) with additional code to filter out alignments that contain noncanonical junctions (--outFilterIntronMotifs RemoveNoncanonical). Raw gene expression was quantified using the software HOMER (RRID: SCR_010881) across exons, and the top isoform value was used to represent gene expression. Raw counts were processed using the supplied R script. In brief, cells containing the bottom 10% of raw sequence counts were filtered (22 cells), and TMM (trimmed mean of M values) normalization/size-factor correction was applied using the edgeR package version 3.24.3 (RRID: SCR_012802). Then, the bottom 10% of cells with genes having normalized counts of >32 per cell were removed as cells with low gene expression (19 cells). As a summary, a sequence depth of 1 × 106 reads per cell with 6 × 103 average genes per cell (4157 genes after the bottom 10% cutoff) was achieved for 171 cells (fig. S11B).
Expression values were log2-transformed, and t-distributed stochastic neighbor embedding (tSNE) (119) was performed to generate the plots. The scrattch.hicat R package version 1.0.0 (RRID: SCR_018099) was used to perform hierarchical iterative clustering (https://CRAN.R-project.org/package=gplots) (82) on the normalized expression dataset (table S7). Default parameters were adjusted (see the supplied script), and a stochastic sampling and consensus clustering approach (run_consensus_clust) was used to assign cell cluster identity, as recommended by the authors of the original code. Cell cluster co-occurrence was plotted with heatmap.2 from gplots in R. For differential expression analysis between the wild type and ∆nvy mutant, genes with expression values of 0 in 75% or more of the cells were filtered out. For DEGs that showed behavioral phenotypes in the RNAi experiments, predicted biological processes and human orthologs were taken from the “Gene Ontology” and “Human Orthologs (via DIOPT v7.1)” sections in FlyBase (http://flybase.org/), respectively. Custom R codes used in the analysis are shown in data S2.
Statistical analysis
Behavioral data were analyzed with nonparametric tests. For the RNAi screen, P values were calculated by Mann-Whitney U test using the MATLAB2014b function (“ranksum”). RNAi mutants that (i) passed the Benjamini-Hochberg false discovery rate (FDR) test of 0.05 and (ii) showed the median lunge numbers (per pair in 30 min) more than 3 were selected as hits. Statistical analyses for the rest of all experiments were carried out using Prism 6 (GraphPad Software), except for Fig. 2 and fig. S2 where MATLAB built-in functions including “kruskalwallis,” “multcompare,” and “prctile” were used. Multiple comparisons among different genotypes were performed using the Kruskal-Wallis test followed by the post hoc Mann-Whitney U test. When comparing paired datasets among different genotypes within a pair of flies or optogenetic stimulation periods within the same fly groups, the post hoc Wilcoxon matched-pairs signed-rank test was used. Bonferroni correction was applied to adjust the P values.
A specific transition probability across different genotypes or housing conditions was statistically analyzed using a permutation test. First, for two groups of flies, differences in their behavior transitions were calculated by subtracting the transition probability matrix of one group from that of the other (“differential transition matrix”). Next, data from all flies in both groups were pooled, and each fly was “reassigned” randomly to one of the two groups (note that this procedure maintained the entire transition sequence of each fly). A transition matrix from the reassigned flies was generated. This procedure was repeated 10,000 times, which produced a resampled distribution of the differential transition matrix. Last, the observed differential transition probability for the transition of interest was compared against its resampled distribution. An observed difference in the top 0.05% of the distribution is considered a significant increase in transition probability; an observed difference in the bottom 0.05% of the distribution is considered a significant decrease. To maintain the level of stringency when simultaneously comparing 20 possible transitions, we applied “Bonferroni-like” correction to set the critical value of 0.05% (0.05% × 20 = 1%).
To determine DEGs in cluster #5 of the Tdc2 neurons (Fig. 7D), the following criteria were considered: (i) P values by Mann-Whitney U tests lower than 0.05, (ii) passed the Benjamini-Hochberg FDR test of 0.2, and (iii) fold change greater than 10 (|log2FC| > 3.321928095). Data summary, statistical methods, and P values for all panels are shown in data S1.
Acknowledgments
We thank D. Anderson, G. Rubin, and B. Pfeiffer for sharing unpublished transgenic Drosophila strains with us; S. Goodwin, D. Yamamoto, K. Scott, and B. Zhang for other Drosophila strains; S. Pfaff for sharing the Olympus FV-3000 confocal microscopy with us; T. G. Partin, C. Ambrosius, S. Saxena, and A. Samulak for technical assistance with mutant construction and fly maintenance; and C. Kintner, G. Lemke, E. Azim, and members of the Asahina laboratory for critical comments on the manuscript. The rabbit anti-Nvy antibody was provided by R. Mann (Columbia University). The antiserum nc82 (anti-BRP), developed by E. Buchner, was obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. Technical assistance for confocal imaging was provided by U. Manor and T. Zhang at the Waitt Advanced Biophotonics Core Facility of the Salk Institute (supported by NCI CCSG: P30 014195). The RNAi screen was partly conducted with support from D. J. Anderson’s research group at the California Institute of Technology.
Funding: This work was supported by NIH grant R35GM119844 (to K.A.), The Naito Foundation Grant for Studying Overseas (to K.I.), and JSPS Postdoctoral Fellowship for Research Abroad (28-869) (to K.I.).
Author contributions: K.I. performed the behavior experiments, immunohistochemistry, and statistical analyses; generated the ethograms; and designed the figures. M.C. carried out the scRNA-seq experiments, performed the bioinformatic analyses, and generated the tSNE plots and volcano plots. X.L. wrote the MATLAB code, conducted behavioral transition analyses from recorded movies, and performed statistical analyses on them. M.N.S. wrote the R code, assisted with bioinformatic analyses for scRNA-seq, and generated the cell cluster co-occurrence matrix plot. K.A. performed a substantial portion of the primary RNAi screen, supervised the project, and provided intellectual support. K.I. and K.A. wrote the manuscript with input from other authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data necessary to reproduce the figure panels and statistical analyses are available as Source Data file. The raw count table and the FASTQ files have been deposited to the Gene Expression Omnibus (GEO) database under the accession code GSE148630 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE148630). Code necessary to reproduce the behavioral analyses is available via Zenodo (https://doi.org/10.5281/zenodo.6672695). R scripts necessary to reproduce the statistical analyses on sequencing data are provided in data S2. Software information is listed in table S8.
Supplementary Materials
This PDF file includes:
Figs. S1 to S13
Table S3
References
Other Supplementary Material for this manuscript includes the following:
Tables S1 and S2, S4 to S8
Movies S1 to S10
Data S1 and S2
Source data
REFERENCES AND NOTES
- 1.J. M. Smith, Evolution and the Theory of Games (Cambridge Univ. Press, 1982). [Google Scholar]
- 2.Dierick H. A., Greenspan R. J., Molecular analysis of flies selected for aggressive behavior. Nat. Genet. 38, 1023–1031 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Shorter J., Couch C., Huang W., Carbone M. A., Peiffer J., Anholt R. R. H., Mackay T. F. C., Genetic architecture of natural variation in Drosophila melanogaster aggressive behavior. Proc. Natl. Acad. Sci. U.S.A. 112, E3555–E3563 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Golden S. A., Heshmati M., Flanigan M., Christoffel D. J., Guise K., Pfau M. L., Aleyasin H., Menard C., Zhang H., Hodes G. E., Bregman D., Khibnik L., Tai J., Rebusi N., Krawitz B., Chaudhury D., Walsh J. J., Han M.-H., Shapiro M. L., Russo S. J., Basal forebrain projections to the lateral habenula modulate aggression reward. Nature 534, 688–692 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briffa M., Sneddon L. U., Wilson A. J., Animal personality as a cause and consequence of contest behaviour. Biol. Lett. 11, 20141007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsu Y., Earley R. L., Wolf L. L., Modulation of aggressive behaviour by fighting experience: Mechanisms and contest outcomes. Biol. Rev. Camb. Philos. Soc. 81, 33–74 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Wang L., Dankert H., Perona P., Anderson D. J., A common genetic target for environmental and heritable influences on aggressiveness in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 105, 5657–5663 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Penn J. K., Zito M. F., Kravitz E. A., A single social defeat reduces aggression in a highly aggressive strain of Drosophila. Proc. Natl. Acad. Sci. U.S.A. 107, 12682–12686 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann A. A., Cacoyianni Z., Territoriality in Drosophila melanogaster as a conditional strategy. Anim. Behav. 40, 526–537 (1990). [Google Scholar]
- 10.Scott J. P., Fredericson E., The causes of fighting in mice and rats. Physiol. Zool. 24, 273–309 (1951). [Google Scholar]
- 11.Diaz V., Lin D., Neural circuits for coping with social defeat. Curr. Opin. Neurobiol. 60, 99–107 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.An D., Chen W., Yu D.-Q., Wang S.-W., Yu W.-Z., Xu H., Wang D.-M., Zhao D., Sun Y.-P., Wu J.-C., Tang Y.-Y., Yin S.-M., Effects of social isolation, re-socialization and age on cognitive and aggressive behaviors of Kunming mice and BALB/c mice. Anim. Sci. J. 88, 798–806 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Flannelly K. J., Blanchard R. J., Muraoka M. Y., Flannelly L., Copulation increases offensive attack in male rats. Physiol. Behav. 29, 381–385 (1982). [DOI] [PubMed] [Google Scholar]
- 14.Yuan Q., Song Y., Yang C.-H., Jan L. Y., Jan Y. N., Female contact modulates male aggression via a sexually dimorphic GABAergic circuit in Drosophila. Nat. Neurosci. 17, 81–88 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cacioppo J. T., Hawkley L. C., Perceived social isolation and cognition. Trends Cogn. Sci. 13, 447–454 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saltz J. B., Genetic variation in social environment construction influences the development of aggressive behavior in Drosophila melanogaster. Heredity (Edinb) 118, 340–347 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kilgour R. J., McAdam A. G., Betini G. S., Norris D. R., Experimental evidence that density mediates negative frequency-dependent selection on aggression. J. Anim. Ecol. 87, 1091–1101 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Ramdya P., Schneider J., Levine J. D., The neurogenetics of group behavior in Drosophila melanogaster. J. Exp. Biol. 220, 35–41 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Smith J. M., Price G. R., The logic of animal conflict. Nature 246, 15–18 (1973). [Google Scholar]
- 20.Knell R. J., Population density and the evolution of male aggression. J. Zool. 278, 83–90 (2009). [Google Scholar]
- 21.Asahina K., Neuromodulation and strategic action choice in Drosophila aggression. Annu. Rev. Neurosci. 40, 51–75 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Hoopfer E. D., Neural control of aggression in Drosophila. Curr. Opin. Neurobiol. 38, 109–118 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Lischinsky J. E., Lin D., Neural mechanisms of aggression across species. Nat. Neurosci. 23, 1317–1328 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Chen P., Hong W., Neural circuit mechanisms of social behavior. Neuron 98, 16–30 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang F., Zhu J., Zhu H., Zhang Q., Lin Z., Hu H., Bidirectional control of social hierarchy by synaptic efficacy in medial prefrontal cortex. Science 334, 693–697 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Chou M.-Y., Amo R., Kinoshita M., Cherng B.-W., Shimazaki H., Agetsuma M., Shiraki T., Aoki T., Takahoko M., Yamazaki M., Higashijima S., Okamoto H., Social conflict resolution regulated by two dorsal habenular subregions in zebrafish. Science 352, 87–90 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Rillich J., Stevenson P. A., A fighter’s comeback: Dopamine is necessary for recovery of aggression after social defeat in crickets. Horm. Behav. 66, 696–704 (2014). [DOI] [PubMed] [Google Scholar]
- 28.A. C. Nelson, V. Kapoor, E. Vaughn, J. A. Gnanasegaram, N. D. Rubinstein, V. N. Murthy, C. Dulac, Molecular and circuit architecture of social hierarchy. bioRxiv 838664 (2019).
- 29.Dierick H. A., Greenspan R. J., Serotonin and neuropeptide F have opposite modulatory effects on fly aggression. Nat. Genet. 39, 678–682 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Cases O., Seif I., Grimsby J., Gaspar P., Chen K., Pournin S., Muller U., Aguet M., Babinet C., Shih J. C., De Maeyer E., Aggressive behavior and altered amounts of brain serotonin and norepinephrine in mice lacking MAOA. Science 268, 1763–1766 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Z., Autry A. E., Bergan J. F., Watabe-Uchida M., Dulac C. G., Galanin neurons in the medial preoptic area govern parental behaviour. Nature 509, 325–330 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kayser M. S., Mainwaring B., Yue Z., Sehgal A., Sleep deprivation suppresses aggression in Drosophila. eLife 4, e07643 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu W., Liang X., Gong J., Yang Z., Zhang Y.-H., Zhang J.-X., Rao Y., Social regulation of aggression by pheromonal activation of Or65a olfactory neurons in Drosophila. Nat. Neurosci. 14, 896–902 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Ramin M., Domocos C., Slawaska-Eng D., Rao Y., Aggression and social experience: Genetic analysis of visual circuit activity in the control of aggressiveness in Drosophila. Mol. Brain 7, 55 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Donlea J. M., Ramanan N., Shaw P. J., Use-dependent plasticity in clock neurons regulates sleep need in Drosophila. Science 324, 105–108 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eddison M., A genetic screen for Drosophila social isolation mutants and analysis of sex pistol. Sci. Rep. 11, 17395 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agrawal P., Kao D., Chung P., Looger L. L., The neuropeptide Drosulfakinin regulates social isolation-induced aggression in Drosophila. J. Exp. Biol. 223, jeb207407 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li W., Wang Z., Syed S., Lyu C., Lincoln S., O’Neil J., Nguyen A. D., Feng I., Young M. W., Chronic social isolation signals starvation and reduces sleep in Drosophila. Nature 597, 239–244 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eddison M., Guarnieri D. J., Cheng L., Liu C.-H., Moffat K. G., Davis G., Heberlein U., arouser reveals a role for synapse number in the regulation of ethanol sensitivity. Neuron 70, 979–990 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Hoyer S. C., Eckart A., Herrel A., Zars T., Fischer S. A., Hardie S. L., Heisenberg M., Octopamine in male aggression of Drosophila. Curr. Biol. 18, 159–167 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Zhou C., Rao Y., Rao Y., A subset of octopaminergic neurons are important for Drosophila aggression. Nat. Neurosci. 11, 1059–1067 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Watanabe K., Chiu H., Pfeiffer B. D., Wong A. M., Hoopfer E. D., Rubin G. M., Anderson D. J., A circuit node that integrates convergent input from neuromodulatory and social behavior-promoting neurons to control aggression in Drosophila. Neuron 95, 1112, 1128.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sherer L. M., Catudio Garrett E., Morgan H. R., Brewer E. D., Sirrs L. A., Shearin H. K., Williams J. L., McCabe B. D., Stowers R. S., Certel S. J., Octopamine neuron dependent aggression requires dVGLUT from dual-transmitting neurons. PLOS Genet. 16, e1008609 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andrews J. C., Fernandez M. P., Yu Q., Leary G. P., Leung A. K., Kavanaugh M. P., Kravitz E. A., Certel S. J., Octopamine neuromodulation regulates Gr32a-linked aggression and courtship pathways in Drosophila males. PLOS Genet. 10, e1004356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dankert H., Wang L., Hoopfer E. D., Anderson D. J., Perona P., Automated monitoring and analysis of social behavior in Drosophila. Nat. Methods 6, 297–303 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leng X., Wohl M., Ishii K., Nayak P., Asahina K., Quantifying influence of human choice on the automated detection of Drosophila behavior by a supervised machine learning algorithm. PLOS ONE 15, e0241696 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dow M. A., von Schilcher F., Aggression and mating success in Drosophila melanogaster. Nature 254, 511–512 (1975). [DOI] [PubMed] [Google Scholar]
- 48.Chen S., Lee A. Y., Bowens N. M., Huber R., Kravitz E. A., Fighting fruit flies: A model system for the study of aggression. Proc. Natl. Acad. Sci. U.S.A. 99, 5664–5668 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chiu H., Hoopfer E. D., Coughlan M. L., Pavlou H. J., Goodwin S. F., Anderson D. J., A circuit logic for sexually shared and dimorphic aggressive behaviors in Drosophila. Cell 184, 507–520.e16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hall J. C., The mating of a fly. Science 264, 1702–1714 (1994). [DOI] [PubMed] [Google Scholar]
- 51.Yamamoto D., Koganezawa M., Genes and circuits of courtship behaviour in Drosophila males. Nat. Rev. Neurosci. 14, 681–692 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Calhoun A. J., Pillow J. W., Murthy M., Unsupervised identification of the internal states that shape natural behavior. Nat. Neurosci. 22, 2040–2049 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hall J. C., Courtship among males due to a male-sterile mutation in Drosophila melanogaster. Behav. Genet. 8, 125–141 (1978). [DOI] [PubMed] [Google Scholar]
- 54.Thistle R., Cameron P., Ghorayshi A., Dennison L., Scott K., Contact chemoreceptors mediate male-male repulsion and male-female attraction during Drosophila courtship. Cell 149, 1140–1151 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Svetec N., Ferveur J.-F., Social experience and pheromonal perception can change male-male interactions in Drosophila melanogaster. J. Exp. Biol. 208, 891–898 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Eyjolfsdottir E., Branson S., Burgos-Artizzu X. P., Hoopfer E. D., Schor J., Anderson D. J., Perona P., Detecting social actions of fruit flies. Computer Vision – ECCV 8690, 772–787 (2014). [Google Scholar]
- 57.Cook R., The courtship tracking of Drosophila melanogaster. Biol. Cybern. 34, 91–106 (1979). [Google Scholar]
- 58.Ribeiro I. M. A., Drews M., Bahl A., Machacek C., Borst A., Dickson B. J., Visual projection neurons mediating directed courtship in Drosophila. Cell 174, 607–621.e18 (2018). [DOI] [PubMed] [Google Scholar]
- 59.Kohatsu S., Yamamoto D., Visually induced initiation of Drosophila innate courtship-like following pursuit is mediated by central excitatory state. Nat. Commun. 6, 6457 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Nilsen S. P., Chan Y.-B., Huber R., Kravitz E. A., Gender-selective patterns of aggressive behavior in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 101, 12342–12347 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang L., Anderson D. J., Identification of an aggression-promoting pheromone and its receptor neurons in Drosophila. Nature 463, 227–231 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kurtovic A., Widmer A., Dickson B. J., A single class of olfactory neurons mediates behavioural responses to a Drosophila sex pheromone. Nature 446, 542–546 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Lin H.-H., Cao D.-S., Sethi S., Zeng Z., Chin J. S. R., Chakraborty T. S., Shepherd A. K., Nguyen C. A., Yew J. Y., Su C.-Y., Wang J. W., Hormonal modulation of pheromone detection enhances male courtship success. Neuron 90, 1272–1285 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cole S. H., Carney G. E., McClung C. A., Willard S. S., Taylor B. J., Hirsh J., Two functional but noncomplementing Drosophila tyrosine decarboxylase genes: Distinct roles for neural tyramine and octopamine in female fertility. J. Biol. Chem. 280, 14948–14955 (2005). [DOI] [PubMed] [Google Scholar]
- 65.Busch S., Selcho M., Ito K., Tanimoto H., A map of octopaminergic neurons in the Drosophila brain. J. Comp. Neurol. 513, 643–667 (2009). [DOI] [PubMed] [Google Scholar]
- 66.Wildonger J., Mann R. S., Evidence that nervy, the Drosophila homolog of ETO/MTG8, promotes mechanosensory organ development by enhancing Notch signaling. Dev. Biol. 286, 507–520 (2005). [DOI] [PubMed] [Google Scholar]
- 67.Selcho M., Pauls D., El Jundi B., Stocker R. F., Thum A. S., The role of octopamine and tyramine in Drosophila larval locomotion. J. Comp. Neurol. 520, 3764–3785 (2012). [DOI] [PubMed] [Google Scholar]
- 68.Flanigan M. E., Aleyasin H., Li L., Burnett C. J., Chan K. L., LeClair K. B., Lucas E. K., Matikainen-Ankney B., Durand-de Cuttoli R., Takahashi A., Menard C., Pfau M. L., Golden S. A., Bouchard S., Calipari E. S., Nestler E. J., DiLeone R. J., Yamanaka A., Huntley G. W., Clem R. L., Russo S. J., Orexin signaling in GABAergic lateral habenula neurons modulates aggressive behavior in male mice. Nat. Neurosci. 23, 638–650 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ueda A., Kidokoro Y., Aggressive behaviours of female Drosophila melanogaster are influenced by their social experience and food resources. Physiol. Entomol. 27, 21–28 (2002). [Google Scholar]
- 70.Certel S. J., Leung A., Lin C.-Y., Perez P., Chiang A.-S., Kravitz E. A., Octopamine neuromodulatory effects on a social behavior decision-making network in Drosophila males. PLOS ONE 5, e13248 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Koganezawa M., Kimura K., Yamamoto D., The neural circuitry that functions as a switch for courtship versus aggression in Drosophila males. Curr. Biol. 26, 1395–1403 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Lee H., Kim D.-W., Remedios R., Anthony T. E., Chang A., Madisen L., Zeng H., Anderson D. J., Scalable control of mounting and attack by Esr1+ neurons in the ventromedial hypothalamus. Nature 509, 627–632 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hashikawa K., Hashikawa Y., Tremblay R., Zhang J., Feng J. E., Sabol A., Piper W. T., Lee H., Rudy B., Lin D., Esr1+ cells in the ventromedial hypothalamus control female aggression. Nat. Neurosci. 20, 1580–1590 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Unger E. K., Burke K. J. Jr., Yang C. F., Bender K. J., Fuller P. M., Shah N. M., Medial amygdalar aromatase neurons regulate aggression in both sexes. Cell Rep. 10, 453–462 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang J., Hoshino T., Redner R. L., Kajigaya S., Liu J. M., ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc. Natl. Acad. Sci. U.S.A. 95, 10860–10865 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lutterbach B., Westendorf J. J., Linggi B., Patten A., Moniwa M., Davie J. R., Huynh K. D., Bardwell V. J., Lavinsky R. M., Rosenfeld M. G., Glass C., Seto E., Hiebert S. W., ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol. Cell. Biol. 18, 7176–7184 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Feinstein P. G., Kornfeld K., Hogness D. S., Mann R. S., Identification of homeotic target genes in Drosophila melanogaster including nervy, a proto-oncogene homologue. Genetics 140, 573–586 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wildonger J., Mann R. S., The t(8;21) translocation converts AML1 into a constitutive transcriptional repressor. Development 132, 2263–2272 (2005). [DOI] [PubMed] [Google Scholar]
- 79.Davis J. N., McGhee L., Meyers S., The ETO (MTG8) gene family. Gene 303, 1–10 (2003). [DOI] [PubMed] [Google Scholar]
- 80.Zhang J., Hug B. A., Huang E. Y., Chen C. W., Gelmetti V., Maccarana M., Minucci S., Pelicci P. G., Lazar M. A., Oligomerization of ETO is obligatory for corepressor interaction. Mol. Cell. Biol. 21, 156–163 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu Y., Cheney M. D., Gaudet J. J., Chruszcz M., Lukasik S. M., Sugiyama D., Lary J., Cole J., Dauter Z., Minor W., Speck N. A., Bushweller J. H., The tetramer structure of the Nervy homology two domain, NHR2, is critical for AML1/ETO’s activity. Cancer Cell 9, 249–260 (2006). [DOI] [PubMed] [Google Scholar]
- 82.Tasic B., Yao Z., Graybuck L. T., Smith K. A., Nguyen T. N., Bertagnolli D., Goldy J., Garren E., Economo M. N., Viswanathan S., Penn O., Bakken T., Menon V., Miller J., Fong O., Hirokawa K. E., Lathia K., Rimorin C., Tieu M., Larsen R., Casper T., Barkan E., Kroll M., Parry S., Shapovalova N. V., Hirschstein D., Pendergraft J., Sullivan H. A., Kim T. K., Szafer A., Dee N., Groblewski P., Wickersham I., Cetin A., Harris J. A., Levi B. P., Sunkin S. M., Madisen L., Daigle T. L., Looger L., Bernard A., Phillips J., Lein E., Hawrylycz M., Svoboda K., Jones A. R., Koch C., Zeng H., Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nelson R. J., Chiavegatto S., Aggression in knockout mice. ILAR J. 41, 153–162 (2000). [DOI] [PubMed] [Google Scholar]
- 84.Wu F., Deng B., Xiao N., Wang T., Li Y., Wang R., Shi K., Luo D.-G., Rao Y., Zhou C., A neuropeptide regulates fighting behavior in Drosophila melanogaster. eLife 9, e54229 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Davis S. M., Thomas A. L., Nomie K. J., Huang L., Dierick H. A., Tailless and atrophin control Drosophila aggression by regulating neuropeptide signalling in the pars intercerebralis. Nat. Commun. 5, 3177 (2014). [DOI] [PubMed] [Google Scholar]
- 86.Hug B. A., Lazar M. A., ETO interacting proteins. Oncogene 23, 4270–4274 (2004). [DOI] [PubMed] [Google Scholar]
- 87.Fischer M. A., Moreno-Miralles I., Hunt A., Chyla B. J., Hiebert S. W., Myeloid translocation gene 16 is required for maintenance of haematopoietic stem cell quiescence. EMBO J. 31, 1494–1505 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Terman J. R., Kolodkin A. L., Nervy links protein kinase a to plexin-mediated semaphorin repulsion. Science 303, 1204–1207 (2004). [DOI] [PubMed] [Google Scholar]
- 89.Ice R. J., Wildonger J., Mann R. S., Hiebert S. W., Comment on “Nervy links protein kinase a to plexin-mediated semaphorin repulsion”. Science 309, 558 (2005). [DOI] [PubMed] [Google Scholar]
- 90.Terman J. R., Kolodkin A. L., Response to comment on “Nervy links protein kinase a to plexin-mediated semaphorin repulsion”. Science 309, 558 (2005). [DOI] [PubMed] [Google Scholar]
- 91.Holt C. E., Martin K. C., Schuman E. M., Local translation in neurons: Visualization and function. Nat. Struct. Mol. Biol. 26, 557–566 (2019). [DOI] [PubMed] [Google Scholar]
- 92.Koyano-Nakagawa N., Kintner C., The expression and function of MTG/ETO family proteins during neurogenesis. Dev. Biol. 278, 22–34 (2005). [DOI] [PubMed] [Google Scholar]
- 93.Yoon K., Gaiano N., Notch signaling in the mammalian central nervous system: Insights from mouse mutants. Nat. Neurosci. 8, 709–715 (2005). [DOI] [PubMed] [Google Scholar]
- 94.Inestrosa N. C., Arenas E., Emerging roles of Wnts in the adult nervous system. Nat. Rev. Neurosci. 11, 77–86 (2010). [DOI] [PubMed] [Google Scholar]
- 95.Burke C. J., Huetteroth W., Owald D., Perisse E., Krashes M. J., Das G., Gohl D., Silies M., Certel S., Waddell S., Layered reward signalling through octopamine and dopamine in Drosophila. Nature 492, 433–437 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Youn H., Kirkhart C., Chia J., Scott K., A subset of octopaminergic neurons that promotes feeding initiation in Drosophila melanogaster. PLOS ONE 13, e0198362 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sayin S., De Backer J.-F., Siju K. P., Wosniack M. E., Lewis L. P., Frisch L.-M., Gansen B., Schlegel P., Edmondson-Stait A., Sharifi N., Fisher C. B., Calle-Schuler S. A., Lauritzen J. S., Bock D. D., Costa M., Jefferis G. S. X. E., Gjorgjieva J., Grunwald Kadow I. C., A neural circuit arbitrates between persistence and withdrawal in hungry Drosophila. Neuron 104, 544–558.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Aso Y., Hattori D., Yu Y., Johnston R. M., Iyer N. A., Ngo T.-T., Dionne H., Abbott L. F., Axel R., Tanimoto H., Rubin G. M., The neuronal architecture of the mushroom body provides a logic for associative learning. eLife 3, e04577 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li F., Lindsey J. W., Marin E. C., Otto N., Dreher M., Dempsey G., Stark I., Bates A. S., Pleijzier M. W., Schlegel P., Nern A., Takemura S., Eckstein N., Yang T., Francis A., Braun A., Parekh R., Costa M., Scheffer L. K., Aso Y., Jefferis G. S., Abbott L. F., Litwin-Kumar A., Waddell S., Rubin G. M., The connectome of the adult Drosophila mushroom body provides insights into function. eLife 9, e62576 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Crocker A., Shahidullah M., Levitan I. B., Sehgal A., Identification of a neural circuit that underlies the effects of octopamine on sleep:wake behavior. Neuron 65, 670–681 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.LeDue E. E., Mann K., Koch E., Chu B., Dakin R., Gordon M. D., Starvation-induced depotentiation of bitter taste in Drosophila. Curr. Biol. 26, 2854–2861 (2016). [DOI] [PubMed] [Google Scholar]
- 102.Schlegel P., Bates A. S., Sturner T., Jagannathan S. R., Drummond N., Hsu J., Serratosa Capdevila L., Javier A., Marin E. C., Barth-Maron A., Tamimi I. F., Li F., Rubin G. M., Plaza S. M., Costa M., Jefferis G. S. X. E., Information flow, cell types and stereotypy in a full olfactory connectome. eLife 10, e66018 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Groth A. C., Fish M., Nusse R., Calos M. P., Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166, 1775–1782 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pfeiffer B. D., Ngo T.-T., Hibbard K. L., Murphy C., Jenett A., Truman J. W., Rubin G. M., Refinement of tools for targeted gene expression in Drosophila. Genetics 186, 735–755 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gratz S. J., Ukken F. P., Rubinstein C. D., Thiede G., Donohue L. K., Cummings A. M., O’Connor-Giles K. M., Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196, 961–971 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Thomas A., Lee P.-J., Dalton J. E., Nomie K. J., Stoica L., Costa-Mattioli M., Chang P., Nuzhdin S., Arbeitman M. N., Dierick H. A., A versatile method for cell-specific profiling of translated mRNAs in Drosophila. PLOS ONE 7, e40276 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lee S.-J., Xu H., Montell C., Rhodopsin kinase activity modulates the amplitude of the visual response in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 101, 11874–11879 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hummel T., Vasconcelos M. L., Clemens J. C., Fishilevich Y., Vosshall L. B., Zipursky S. L., Axonal targeting of olfactory receptor neurons in Drosophila is controlled by Dscam. Neuron 37, 221–231 (2003). [DOI] [PubMed] [Google Scholar]
- 109.Nern A., Pfeiffer B. D., Rubin G. M., Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proc. Natl. Acad. Sci. U.S.A. 112, E2967–E2976 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Asahina K., Watanabe K., Duistermars B. J., Hoopfer E., Gonzalez C. R., Eyjolfsdottir E. A., Perona P., Anderson D. J., Tachykinin-expressing neurons control male-specific aggressive arousal in Drosophila. Cell 156, 221–235 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Inagaki H. K., Jung Y., Hoopfer E. D., Wong A. M., Mishra N., Lin J. Y., Tsien R. Y., Anderson D. J., Optogenetic control of Drosophila using a red-shifted channelrhodopsin reveals experience-dependent influences on courtship. Nat. Methods 11, 325–332 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ishii K., Wohl M., DeSouza A., Asahina K., Sex-determining genes distinctly regulate courtship capability and target preference via sexually dimorphic neurons. eLife 9, e52701 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Keleman K., Kruttner S., Alenius M., Dickson B. J., Function of the Drosophila CPEB protein Orb2 in long-term courtship memory. Nat. Neurosci. 10, 1587–1593 (2007). [DOI] [PubMed] [Google Scholar]
- 114.Robie A. A., Straw A. D., Dickinson M. H., Object preference by walking fruit flies, Drosophila melanogaster, is mediated by vision and graviperception. J. Exp. Biol. 213, 2494–2506 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Agrawal S., Dickinson M. H., The effects of target contrast on Drosophila courtship. J. Exp. Biol. 222, jeb203414 (2019). [DOI] [PubMed] [Google Scholar]
- 116.Pan Y., Meissner G. W., Baker B. S., Joint control of Drosophila male courtship behavior by motion cues and activation of male-specific P1 neurons. Proc. Natl. Acad. Sci. U.S.A. 109, 10065–10070 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Datta S. R., Anderson D. J., Branson K., Perona P., Leifer A., Computational neuroethology: A call to action. Neuron 104, 11–24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li H., Horns F., Wu B., Xie Q., Li J., Li T., Luginbuhl D. J., Quake S. R., Luo L., Classifying Drosophila olfactory projection neuron subtypes by single-cell RNA sequencing. Cell 171, 1206–1220.e22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.van der Maaten L., Hinton G., Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008). [Google Scholar]
- 120.Luo L., Liao Y. J., Jan L. Y., Jan Y. N., Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 8, 1787–1802 (1994). [DOI] [PubMed] [Google Scholar]
- 121.Yapici N., Kim Y.-J., Ribeiro C., Dickson B. J., A receptor that mediates the post-mating switch in Drosophila reproductive behaviour. Nature 451, 33–37 (2008). [DOI] [PubMed] [Google Scholar]
- 122.Gordon M. D., Scott K., Motor control in a Drosophila taste circuit. Neuron 61, 373–384 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Baines R. A., Seugnet L., Thompson A., Salvaterra P. M., Bate M., Regulation of synaptic connectivity: Levels of fasciclin II influence synaptic growth in the Drosophila CNS. J. Neurosci. 22, 6587–6595 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.von Reyn C. R., Breads P., Peek M. Y., Zheng G. Z., Williamson W. R., Yee A. L., Leonardo A., Card G. M., A spike-timing mechanism for action selection. Nat. Neurosci. 17, 962–970 (2014). [DOI] [PubMed] [Google Scholar]
- 125.Pfeiffer B. D., Truman J. W., Rubin G. M., Using translational enhancers to increase transgene expression in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 109, 6626–6631 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bohm R. A., Welch W. P., Goodnight L. K., Cox L. W., Henry L. G., Gunter T. C., Bao H., Zhang B., A genetic mosaic approach for neural circuit mapping in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 107, 16378–16383 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Klapoetke N. C., Murata Y., Kim S. S., Pulver S. R., Birdsey-Benson A., Cho Y. K., Morimoto T. K., Chuong A. S., Carpenter E. J., Tian Z., Wang J., Xie Y., Yan Z., Zhang Y., Chow B. Y., Surek B., Melkonian M., Jayaraman V., Constantine-Paton M., Wong G. K.-S., Boyden E. S., Independent optical excitation of distinct neural populations. Nat. Methods 11, 338–346 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Stockinger P., Kvitsiani D., Rotkopf S., Tirián L., Dickson B. J., Neural circuitry that governs Drosophila male courtship behavior. Cell 121, 795–807 (2005). [DOI] [PubMed] [Google Scholar]
- 129.Rideout E. J., Dornan A. J., Neville M. C., Eadie S., Goodwin S. F., Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nat. Neurosci. 13, 458–466 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yu J. Y., Kanai M. I., Demir E., Jefferis G. S. X. E., Dickson B. J., Cellular organization of the neural circuit that drives Drosophila courtship behavior. Curr. Biol. 20, 1602–1614 (2010). [DOI] [PubMed] [Google Scholar]
- 131.Toda H., Zhao X., Dickson B. J., The Drosophila female aphrodisiac pheromone activates ppk23(+) sensory neurons to elicit male courtship behavior. Cell Rep. 1, 599–607 (2012). [DOI] [PubMed] [Google Scholar]
- 132.Starostina E., Liu T., Vijayan V., Zheng Z., Siwicki K. K., Pikielny C. W., A Drosophila DEG/EnaC subunit functions specifically in gustatory neurons required for male courtship behavior. J. Neurosci. 32, 4665–4674 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wan Y., Otsuna H., Holman H. A., Bagley B., Ito M., Lewis A. K., Colasanto M., Kardon G., Ito K., Hansen C., FluoRender: Joint freehand segmentation and visualization for many-channel fluorescence data analysis. BMC Bioinformatics 18, 280 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kabra M., Robie A. A., Rivera-Alba M., Branson S., Branson K., JAABA: Interactive machine learning for automatic annotation of animal behavior. Nat. Methods 10, 64–67 (2013). [DOI] [PubMed] [Google Scholar]
- 136.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H., Glass C. K., Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S13
Table S3
References
Tables S1 and S2, S4 to S8
Movies S1 to S10
Data S1 and S2
Source data