Abstract
Gastrointestinal stromal tumor (GIST) is commonly characterized by activating mutations in the receptor tyrosine kinase KIT. Tyrosine kinase inhibitors are the only approved therapy for GIST, and complementary treatment strategies are urgently needed. As GIST lacks oncogene amplification and relies upon an established network of transcription factors, we hypothesized that unique chromatin modifying enzymes are essential in orchestrating the GIST epigenome. We identified through genome-scale CRISPR screening that MOZ and Menin-MLL chromatin regulatory complexes are cooperative and unique dependencies in GIST. These complexes were enriched at GIST-relevant genes and regulated their transcription. Inhibition of MOZ and Menin-MLL complexes decreased GIST cell proliferation by disrupting interactions with transcriptional/chromatin regulators, such as DOT1L. MOZ and Menin inhibition caused significant reductions in tumor burden in vivo, with superior effects observed with combined Menin and KIT inhibition. These results define unique chromatin regulatory dependencies in GIST and identify potential therapeutic strategies for clinical application.
INTRODUCTION
Gastrointestinal stromal tumor (GIST) is among the most common sarcomas and arises from the interstitial cells of Cajal (ICC) or their progenitors (1,2). The majority of GIST cases are characterized by activating mutations in the receptor tyrosine kinase KIT, and inhibition of KIT with tyrosine kinase inhibitors (TKIs) is the only established therapeutic strategy for this disease (3,4). Though TKIs are efficacious for the majority GIST patients, evolution of TKI resistance mutations is inexorable in the metastatic setting (5,6), leading to poor patient outcomes. Novel targeted therapeutic strategies are urgently needed for GIST clinical care, either as monotherapy or in combination with TKIs, though translational efforts to date have not yet met with success.
In contrast to many other malignancies, GIST does not amplify the mutant KIT gene, but instead relies upon epigenetic mechanisms of gene regulation for oncogene expression (7). We and others have previously characterized the enhancer and transcriptional landscapes of GIST, which emphasized a conserved core network of transcription factors (TFs) which are essential for GIST viability, regulate the expression of GIST-associated genes, including KIT, and are predictive of patient outcomes (8–11). Therapeutic targeting of these key transcriptional dependencies represents an attractive treatment strategy for this disease. In addition, understanding how these TFs establish co-dependency with essential chromatin regulators would provide significant insight into gene regulation and epigenetic mechanisms of disease (12,13). Recent advances in the targeting of chromatin modifying enzymes using inhibitory small molecules has led to new discoveries on the function of these epigenetic regulators and opened opportunities to therapeutically disrupt their activity; several of these drugs have been approved or are under active clinical investigation for cancer treatment.
Here, using genome-scale CRISPR/Cas9 dependency screening, we further determine the paramount importance of epigenetic regulation in GIST oncogenesis, defining essential chromatin regulators that maintain the GIST epigenome and providing a map of experimentally determined therapeutic targets. Among chromatin modifying enzymes enriched in GIST, KAT6A/MOZ and KMT2A/MLL1 were established as previously unknown co-dependencies, and, more broadly, were found to exhibit similar co-regulation across select cancer subtypes. MOZ is a histone acetyltransferase and, together with the assembly and chromatin reader proteins BRPF1, MEAF6 and ING5, constitutes the MOZ complex which mediates activating histone acetylation including locus-specific H3K9ac (14,15). KMT2A/MLL1 is a member of the Menin-MLL complex and responsible for H3K4 methylation and transcriptional activation (16,17). MOZ and Menin-MLL complexes were found to co-localize to GIST-associated genes genome-wide and regulate oncogenic transcription and cell cycle progression by regulation of transcription factor gene expression programs. Catalytic inactivation of MOZ or disruption of Menin-MLL protein interaction using small molecule inhibitors led to changes in the chromatin association and protein-protein interactions of key mediators of transcriptional regulation, such as disruptor of telomeric silencing 1-like (DOT1L). Inhibition of MOZ and Menin significantly reduced GIST tumor growth in vivo, with the combination of Menin and KIT inhibition showing combinatorial toxicity in cell line and patient derived xenograft (PDX) models of GIST. Our findings identify novel complementary roles for MOZ and Menin-MLL complexes in maintaining the cancer-associated transcriptional program in GIST and nominate novel therapeutic strategies either alone or in combination with TKIs.
RESULTS
Genome-scale CRISPR screening in GIST identifies unique epigenetic dependencies.
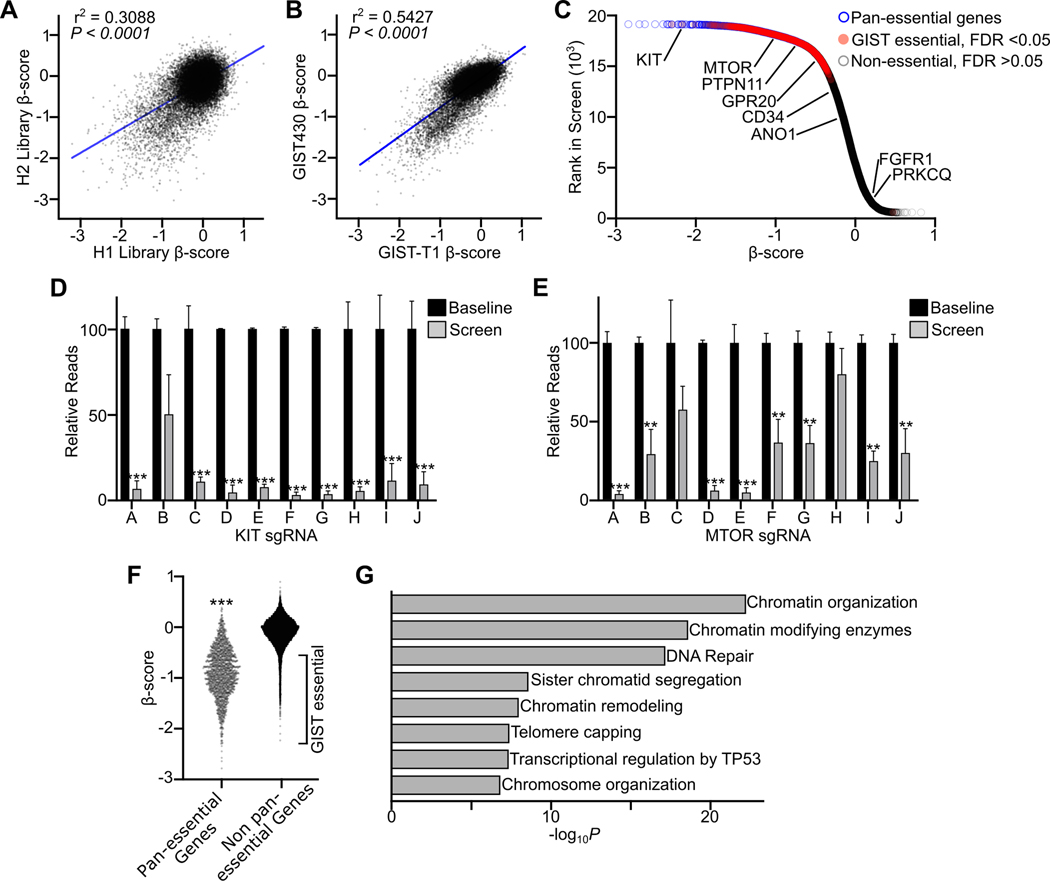
Previously, we and others have characterized the global transcriptional and enhancer landscape of GIST using RNA-seq, chromatin immunoprecipitation with sequencing (ChIP-seq), and assay for transposase-accessible chromatin using sequencing (ATAC-seq). These investigations focused on TFs relevant to GIST biology, including core TFs such as ETV1, FOXF1, HIC1 and OSR1 present across GIST samples, and accessory TFs BARX1 and HAND1 expressed in disease-state specific patterns (11). However, how these TFs integrate with other epigenetic regulators to establish the GIST-associated gene expression program is unknown. To establish genes essential to GIST biology, we performed a genome-wide CRISPR/Cas9-based dropout screen in two KIT mutant GIST cell lines, GIST-T1 and GIST430. A split-library approach utilizing paired human whole-genome sgRNA libraries (denoted H1 and H2) was used, with approximately 5 sgRNAs per gene in each library targeting 18,436 genes with a total of 185,634 sgRNAs in the screen. Significant correlation in dependency (β) scores was observed between the H1 and H2 libraries (Fig. 1A) as well as between the two GIST cell lines (Fig. 1B); data sets were then merged for subsequent analysis to improve statistical power. Genes were stratified as ‘pan-essential’, having been previously determined to be universally essential for cellular viability (18,19), ‘GIST essential’ with an FDR <0.05 in the screen but absent from the pan-essential list, or ‘Non-essential’ (Fig. 1C, Table S1).
Figure 1. Identification of GIST epigenetic dependencies through genome-scale CRISPR dependency screens.
A, Correlation of β-score between H1 and H2 sgRNA libraries, each targeting 18,436 genes with 5 sgRNAs per library. Data derived from replicate screens in GIST-T1 and GIST430 cell lines were merged (per library, n=4). B, Correlation of β-score between GIST430 and GIST-T1 cell lines (per cell line, n=4 libraries). C, Plot of rank in screen and β-score merging H1 and H2 libraries and GIST cell lines. Pan-essential genes (18) are indicated in blue, genes significantly depleted in GIST but not pan-essential are in red, and non-essential genes lacking significant depletion are in gray. Select GIST-associated genes are labeled. D-E, Relative reads for individual sgRNAs targeting KIT or MTOR comparing baseline plasmid library sequencing (n=2 per library) to results at the end of the screen (n=4). F, Plot comparing β-scores of pan-essential (n=1,702) and non-pan-essential (n=16,757) genes, with GIST essential genes labeled on the plot. G, Plot showing 8 of the top 18 significantly enriched gene ontology terms among genes uniquely essential in GIST. Conditions in D, E and F were compared by t-test (compared to non-essential genes or baseline sgRNA; **, P<0.01; ***, P<0.001). Pearson correlation was performed in A and B with P value and r2 shown.
As anticipated, KIT was among the strongest detected dependencies, with sgRNA-level data showing near complete dropout of most (9/10) sgRNAs during the screen (Fig. 1D). Other canonical downstream signaling mediators of the KIT pathway, such as mTOR, showed significant dropout in the screen, but less so compared to KIT (Fig. 1E). To identify biological processes which may be specifically enriched in GIST, we focused on the ‘GIST essential’ subset of genes (Fig. 1F). These unique dependencies in GIST were evaluated by gene ontology enrichment analysis, which revealed that 8 of the top 18 terms were associated with epigenetic regulatory mechanisms including chromatin and chromosomal organization (Fig. 1G, Fig. S1A). Taken together, data from these unbiased dependency screens characterize GIST as having remarkable reliance upon epigenetic mechanisms to maintain its oncogenic program.
MOZ and Menin-MLL complexes are unique and complementary dependencies.
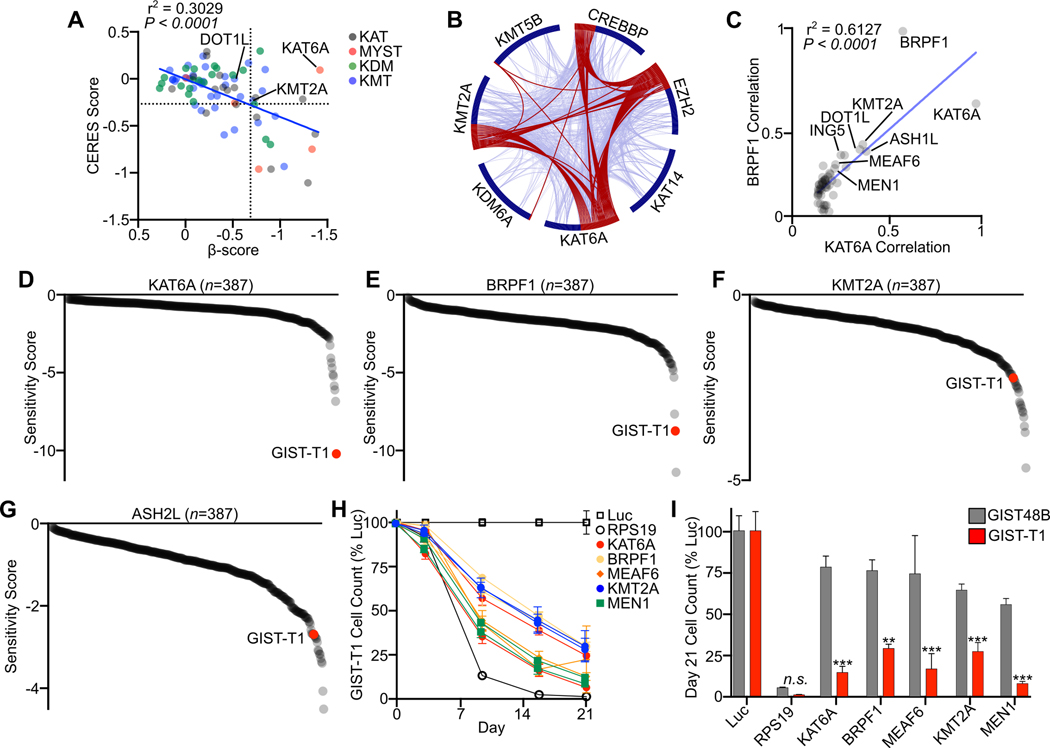
To better define which chromatin regulatory complexes may be most relevant and unique to GIST biology, we compared β-scores for all chromatin modifying enzymes in GIST cell lines to analogous CERES dependency scores averaged across all cell lines in the DepMap project (20). Only 7 of the 77 assessed chromatin modifying enzymes were unique and essential to both GIST cell lines, with β-score <−0.7 and CERES score >−0.25 (Fig. 2A, S1B), with dependency score cutoffs chosen to select for chromatin regulators likely to be unique dependencies. Enriched enzymes included members of the lysine acetyltransferase (KAT), MYST, lysine demethylase (KDM), and lysine methyltransferase (KMT) families. KAT6A was a significant outlier in this comparison, with the highest β-score among all chromatin modifying enzymes and average CERES score near zero. To establish which of these seven chromatin modifying enzymes may function collaboratively to maintain the epigenome, we evaluated gene-level co-dependency data within DepMap. Comparative analysis of the top 50 co-dependencies of each chromatin modifying enzyme showed the highest interaction at gene and ontology term levels between KAT6A, KMT2A and EZH2 (Fig. 2B), suggesting their genetic co-dependency. KAT6A, the catalytic member of MOZ, and complex members MEAF6 and BRPF1 were all significant dependencies in GIST (Fig. S1C). KMT2A, the catalytic member of the Menin-MLL complex, also had multiple recognized complex members with significant dependency scores including MEN1/Menin and ASH2L (Fig. S1D). Plotting the top DepMap co-dependencies between KAT6A and BRPF1, multiple members of the Menin-MLL complex are highly correlated co-dependencies, indicating genetic interaction between the MOZ and Menin-MLL complexes (Fig. 2C). As DepMap CRISPR dependency screening efforts have not yet profiled GIST, we leveraged available comparative screening results from Project DRIVE (21), which included GIST-T1 among nearly 400 cell lines profiled by RNAi. Among all cell lines profiled, GIST-T1 had the highest sensitivity score for KAT6A, second highest for BRPF1, and was in the top 5% for KMT2A and ASH2L (Fig. 2D–G), further indicating through an independent and comparative screening approach the essential and co-dependent nature of the MOZ and Menin-MLL complexes in GIST. Several other chromatin regulatory complexes were found to have multiple members with significant dependencies in our screen and also showed enrichment for GIST-T1 in Project Drive, including members of the INO80 complex, NuA4 histone acetyltransferase complex, FACT complex, and PAF1 complex (Fig. S1E–O). EZH2, SUZ12 and EED, the core members of the PRC2 complex (22), were all dependencies in our screen, and GIST-T1 had among the highest sensitivity scores for EZH2 and EED in Project DRIVE (Fig. S2A–C). Though few cell lines in DepMap had significant dependencies on core PRC2 complex members, the top correlated co-dependencies of EZH2, including DOT1L, EP300 and MEN1, showed overlap with MOZ and Menin-MLL complex co-dependencies (Fig. S2D–E), indicating the complementary function of the transcriptionally repressive PRC2 complex.
Figure 2. MOZ and Menin-MLL Complexes are Unique Co-Dependencies:
A, Plot of merged β-score in GIST-T1 and GIST430 and average CERES score of all cell lines in DepMap (n=796) for chromatin modifying enzymes (n=77) including lysine acetyltransferases (KAT), MYST type domain containing lysine acetyltransferases (MYST), lysine demethylases (KDM) and lysine methyltransferases (KMT). The dotted lines divide the plot into quadrants, with the right upper quadrant containing 7 genes that were dependencies in GIST but not common dependencies across DepMap cell lines. B, Circos plot showing overlap of the top 50 DepMap correlated dependencies of the seven chromatin modifying enzymes with enriched dependencies in GIST. Red lines connect genes shared on multiple co-dependency lists. Blue lines connect genes that fall into the same ontology term. C, Top 50 gene dependency correlations between KAT6A and BRPF1 in DepMap. Co-dependent chromatin modifying enzymes and complex members are labeled. D-G, Ranked Sensitivity Score from Project Drive cell lines (n=387) for MOZ complex members KAT6A and BRPF1 and Menin-MLL complex members KMT2A and ASH2L, with GIST-T1 highlighted in red. H, Growth over time assay following transduction of the indicated sgRNAs targeting Menin-MLL and MOZ complex members in GIST-T1, with two independent sgRNAs per gene. sgRNAs targeting Luc and RPS19 are shown in open boxes and circles, respectively (n=3 per sgRNA). I, Day 21 cell count in a growth over time assay comparing GIST-T1 to GIST48B (n=6 per gene from two sgRNAs). Data were analyzed by two-way ANOVA with Tukey’s multiple comparisons test, compared to GIST48B in the same treatment condition; ***,P<0.001; **,P<0.01. The Pearson correlation coefficient is shown in A and C.
To validate dependency upon MOZ and Menin-MLL complex members, we performed a growth-over time assay utilizing unique sgRNAs targeting MOZ complex members KAT6A, BRPF1 and MEAF6, and Menin-MLL complex members KMT2A and MEN1. For each gene, and with two independent sgRNAs, sgRNA treatment significantly reduced cell proliferation (Fig. 2H). To compare the relative toxicity of these sgRNAs to a control cell line, we utilized GIST48B, which has a similar growth rate as GIST-T1 but through in vitro selection has lost KIT expression and the GIST-associated epigenetic and transcriptional program (10,23). While all sgRNAs targeting MOZ and Menin-MLL complexes significantly reduce GIST-T1 cell proliferation, GIST48B showed little or no change in cell proliferation (Fig. 2I). Taken together, these data show a complementary co-dependency in GIST, and across select cell lines in DepMap, between MOZ and Menin-MLL complexes and suggests their collaborative role in maintaining the GIST epigenome.
MOZ and Menin-MLL complexes localize to TSSs genome-wide and are enriched at GIST-associated genes.
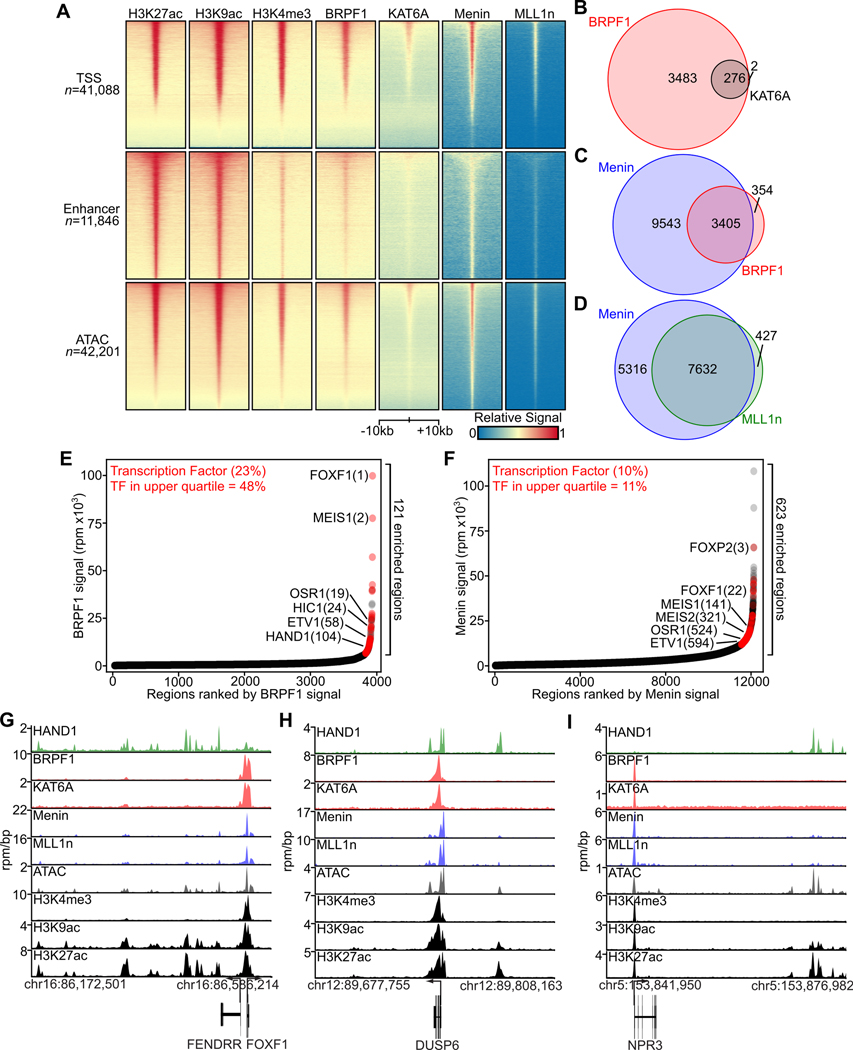
To define where in the GIST genome MOZ and Menin-MLL complexes bind and modify histones, we performed ChIP-seq for histone marks H3K27ac, H3K9ac (deposited by MOZ and other acetyltransferases), H3K4me3 (deposited by the Menin-MLL complexes), BRPF1 and KAT6A. While we were unable to successfully perform ChIP-seq for the Menin-MLL complex in GIST cell lines, we were able to identify genomic regions of binding of Menin and MLL1 using the analogous method CUT&Tag (24). Both MOZ and Menin-MLL complex members were found to be localized at the TSSs of active genes, as determined by their co-occupancy with H3K27ac and H3K9ac (Fig. 3A, top row). In contrast, we observed very little occupancy of these chromatin complex members at H3K27ac-defined enhancers (Fig. 3A, middle row). ATAC peaks, which include accessible DNA sites at both TSSs and enhancers, showed an intermediate level of MOZ and Menin-MLL complex binding (Fig. 3A, bottom row). As suggested by heatmaps, there was considerable overlap in peaks within and between MOZ and Menin-MLL complexes (Fig. 3B–D), indicating that the genomic binding of these complexes converges at TSSs and further suggesting their co-localization.
Figure 3. Genomic Localization of MOZ and Menin-MLL Complexes in GIST:
A, Heat maps demonstrating genomic localization in GIST-T1 of H3K27ac, H3K9ac, H3K4me3, BRPF1, and KAT6A by ChIP-seq, and Menin and MLL1n by CUT&Tag. Scaled read densities ± 10 kb from the TSS, H3K27ac-defined enhancers or ATAC-defined peaks are shown in rows. B, Overlap in MACS-defined peaks for BRPF1 and KAT6A. C, Overlap in MACS-defined peaks for Menin and BRPF1. D, Overlap in MACS-defined peaks for Menin and MLL1n. E-F, Enriched genomic regions of BRPF1 or Menin binding, with TFs indicated in red; the percent of all enriched regions associated with TF genes and those in the upper quartile are indicated. G-I, Tracks showing regions of genomic occupancy of the TF HAND1, MOZ complex members BRPF1 and KAT6A, Menin-MLL complex members Menin and MLL1n and histone marks H3K4me3, H3K9ac and H3K27ac at the FOXF1, DUSP6 and NPR3 loci.
We next evaluated genomic regions that displayed strong enrichment for BRPF1 and Menin in our ChIP-seq and CUT&Tag datasets, reasoning that these factors are representative of MOZ and Menin-MLL complexes. While both proteins bind to thousands of sites genome-wide, disproportionate enrichment was seen in 3–5% of these genomic regions, many of which have clear relevance to GIST (Fig. 3E–F, Fig. S3A). TFs, particularly those among the group of core and accessory GIST TFs (10,11), were included in these enriched regions, as were negative regulators of KIT signaling from DUSP and sprouty families and genes used as biomarkers for GIST (e.g. GPR20, CD34 (25)). ChIP-seq and CUT&Tag tracks show binding of MOZ and Menin-MLL complex members at the TSS and gene body of these enriched genes, analogous to H3K4me3, such as at core TF members, DUSP6 and NPR3 (Fig. 3G–I, Fig. S3B); by contrast, H3K27ac and H3K9ac are enriched at both enhancer regions and gene bodies, and the GIST accessory TF HAND1 binds exclusively to enhancers. MOZ and Menin-MLL complex members were notably not enriched at the KIT locus, though both these regulators bind to the TSS and Menin and MLL1 to a region downstream of the gene body (Fig. S3C). Other regions of enrichment show maximal binding of MOZ and Menin-MLL complex members within and immediately downstream of the TSS, with detectable signal evident at some enhancers of these highly regulated genes (Fig. S3D–F). These data indicate that MOZ and Menin-MLL complexes are globally present at active genes, with enrichment at a subset of genes relevant to the GIST transcriptional program.
MOZ and Menin inhibition disrupts GIST cell proliferation without causing apoptosis.
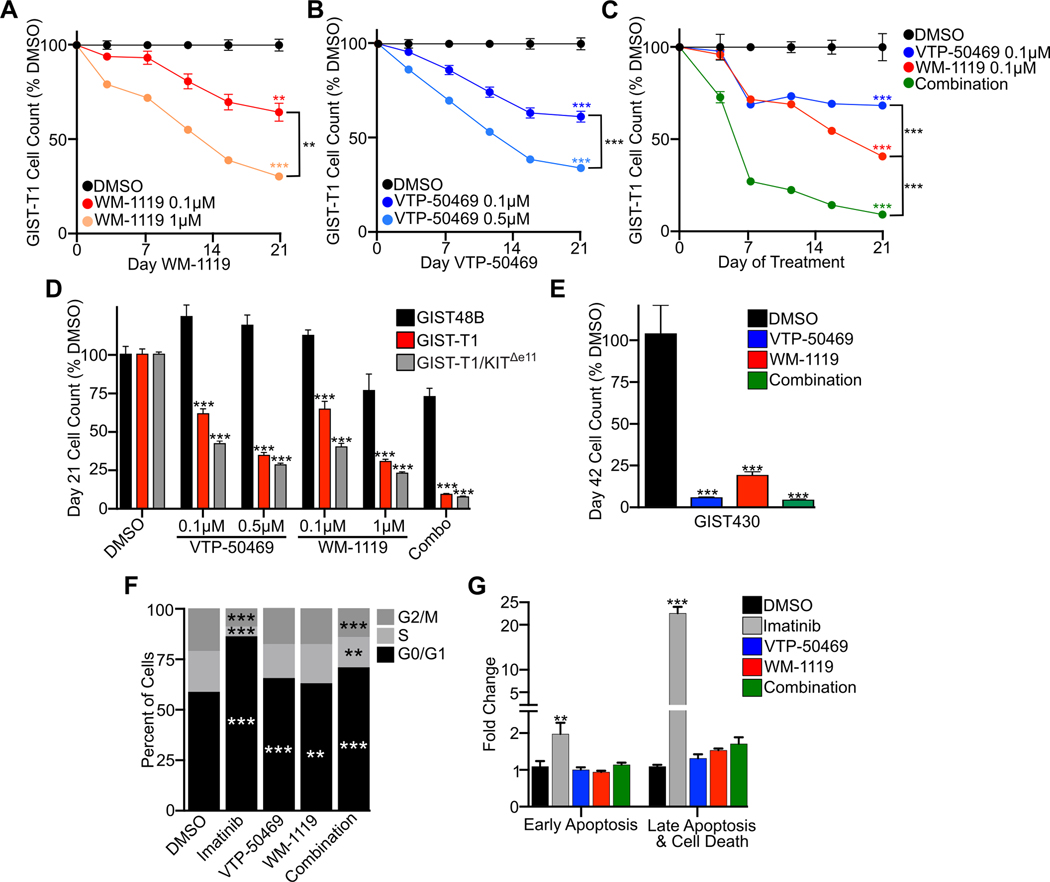
Based on the genetic data and genomic colocalization of MOZ and Menin-MLL complexes we reasoned that small molecule inhibitors targeting these complexes would be a viable therapeutic approach. To explore the functional consequences of MOZ and Menin-MLL disruption, we treated GIST-T1 with the selective KAT6A inhibitor WM-1119 (26) alone or in combination with the Menin inhibitor VTP-50469 (27). At sub-micromolar concentrations, both WM-1119 and VTP-50469 decreased GIST cell proliferation in a growth over time assay (Fig. 4A–B), with a greater effect seen with combination of the two drugs (Fig. 4C). Unlike imatinib (Fig. S4A), antiproliferative effects of MOZ and Menin inhibitors are seen with prolonged drug exposure, in line with other studies utilizing these compounds in leukemia (26,27). Demonstrating selective toxicity of these inhibitors in KIT-dependent GIST cell lines, the KIT-independent GIST48B cell line showed modest or no alterations in proliferation after a 21-day drug treatment (Fig. 4D), in agreement with genetic data from CRISPR experiments (Fig. 2I). In previous studies of the KIT enhancer, we used an sgRNA directed at the KIT TSS to ablate endogenous KIT expression while simultaneously rescuing cell viability by expressing a viral promoter-driven KIT construct bearing the same activating mutation (23). This KIT-dependent KIT rescue cell line was similarly susceptible to WM-1119 and VTP-50469 alone or in combination (Fig. 4D), indicating that regulation of the endogenous KIT locus is not the principal mechanism of toxicity of these compounds. To confirm the proliferative effects of these inhibitors in additional GIST cell lines, we treated the slower growing KIT mutant cell lines GIST430, GIST882 and GIST48 (23) with WM-1119 and VTP-50469 alone or in combination, observing analogous anti-proliferative effects arising from drug treatment (Fig. 4E, Fig. S4B).
Figure 4. Inhibition of MOZ and Menin-MLL Complexes leads to cell cycle arrest.
A, Growth over time assay in GIST-T1 with the indicated concentrations of KAT6A inhibitor WM-1119. B, Growth over time assay in GIST-T1 with the indicated concentrations of Menin inhibitor VTP-50469. C, Growth over time assay in GIST-T1 treated with WM-1119, VTP-50469 or combination treatment at 0.1 μM of each inhibitor. D, Day 21 cell count normalized to DMSO following treatment of GIST48B, GIST-T1 or KIT enhancer independent cell line GIST-T1/KITΔe11 (with endogenous KIT knocked out with rescue of CMV promoter driven mutant KIT) treated with the indicated inhibitors. Statistical comparisons reference GIST48B under the identical treatment. E, Growth over time assay in GIST430, with relative cell count shown at day 42 following treatment with VTP-50469 at 0.5 μM, WM-1119 at 1 μM or the combination with each drug at 0.1 μM. F, Cell cycle analysis showing the percentage of cells in G0/G1, S or G2/M comparing DMSO to 72 hours imatinib or 8 days of VTP-50469 at 0.5 μM, WM-1119 at 1 μM or the combination with each drug at 0.1 μM. G, Fold change compared to DMSO control of cells in early apoptosis or late apoptosis and cell death following 72 hours treatment with imatinib at 0.5 μM or 8 days of VTP-50469 at 0.5 μM, WM-1119 at 1 μM or the combination with each drug at 0.1 μM (n=3–5 per condition). Data were analyzed by two-way or one-way ANOVA, where appropriate, with Tukey’s post-hoc test, compared to DMSO or the indicated condition; ***,P<0.001; **,P<0.01.
To evaluate the cellular phenotypic consequences of WM-1119 and VTP-50469 treatment, we performed cell cycle and apoptosis assays utilizing these drugs and the TKI imatinib as comparator. While imatinib acutely and potently caused G0/G1 cell cycle arrest within 72 hours, eight days of treatment with either WM-1119 or VTP-50469 alone lead to a modest increase in the fraction of cells in G0/G1; the combination of WM-1119 and VTP-50469 led to more marked disruption of the cell cycle after an 8 day treatment (Fig. 4F). While 72 hour imatinib treatment produced a significant increase in early and late-phase apoptosis and cell death, an 8 day treatment of WM-1119 and VTP-50469 alone or in combination did not significantly increase apoptosis or cell death compared to DMSO control (Fig. 4G). We next evaluated molecular disruptors of the MOZ complex that bind outside the KAT6A active site. BRPF1, considered a scaffolding protein for its multiple DNA and chromatin binding domains, bears a bromodomain that can be selectively and potently targeted by inhibitors PFI-4 and GSK6853, among others (28,29). Despite the genetic dependency upon BRPF1 in GIST, neither PFI-4 nor GSK6853 disrupted GIST cell proliferation (Fig. S4C). A similar absence of toxicity arising from these compounds has been observed in other cell types (28,29), though concomitant genetic dependency upon BRPF1 has not previously been well established; these findings suggest that BRPF1 bromodomain targeting compounds may represent ideal candidates for the development of proteolysis targeting chimera (PROTAC)-based molecular degraders. Taken together, these data show that MOZ and Menin-MLL complexes are targetable and unique vulnerabilities in GIST, and in keeping with their distribution across the genome at TSSs of active genes, that they may play essential roles in gene regulation greater than just KIT gene expression. Disruption of these complexes alone or in combination causes alterations in the cell cycle but not in programmed cell death.
MOZ and Menin-MLL disruption alters core transcriptional programs in GIST.
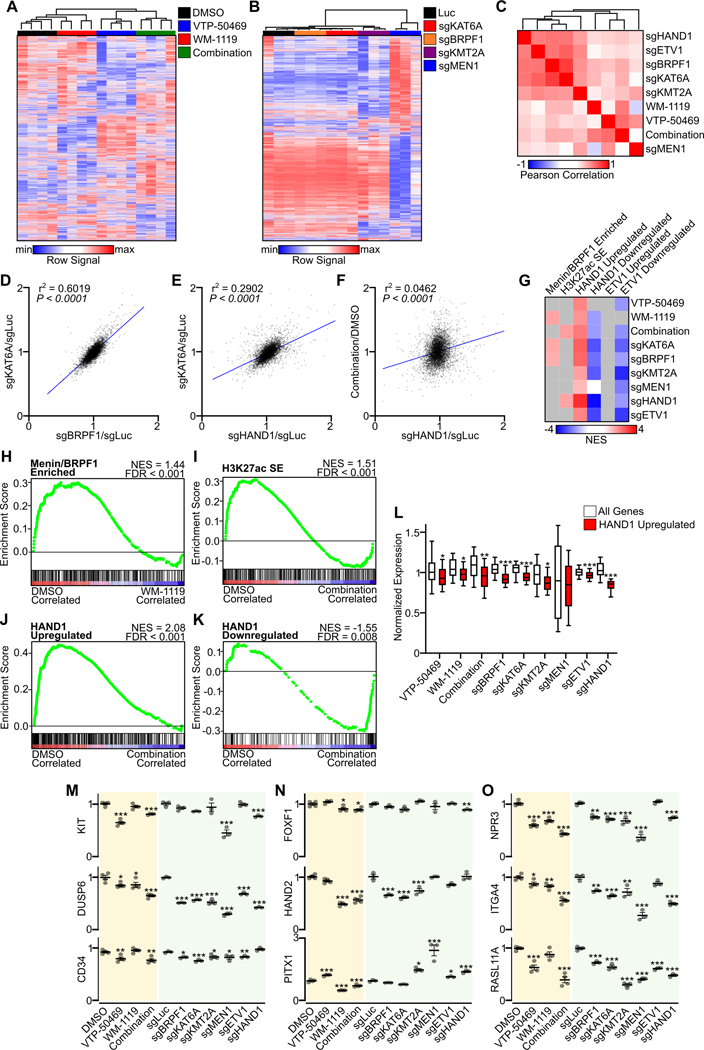
Observing the genome-wide occupancy of MOZ and Menin-MLL complexes, with limited regions of enrichment, we next probed for selective changes in gene expression arising from pharmacologic or genetic disruption of these complexes. We performed RNA-seq on GIST-T1 cells treated for five days with WM-1119, VTP-50469 or the combination of these two drugs. Compared to DMSO control, all treatment groups showed changes in global gene expression, with VTP-50469 and combination groups showing the greatest deviation from control (Fig. 5A). We similarly evaluated gene expression changes following genetic disruption of MOZ and Menin-MLL complexes using sgRNAs targeting two complex members each. Using this genetic system, sgRNAs targeting MEN1 led to global changes in gene expression, with disruption of other MOZ and Menin-MLL1 complex members showing less dramatic changes (Fig. 5B). To integrate and compare transcriptional changes arising from either pharmacologic or genetic disruption, we evaluated the correlation of gene expression changes from control across the transcriptome. For comparison, we also evaluated transcriptional changes from sgRNA mediated disruption of GIST TFs HAND1 and ETV1 (11). sgRNAs targeting MOZ complex members KAT6A and BRPF1 showed the highest degree of correlation, and moreover induced similar global transcriptional changes as did disruption of HAND1, ETV1 or KMT2A (Fig. 5C–E). Pharmacologic inhibition of MOZ and/or Menin-MLL complexes had a comparatively weak but positive correlation with sgRNAs targeting GIST TFs, MOZ complex members and KMT2A, whereas genetic disruption of MEN1 showed the least correlation with other conditions (Fig. 5C,F).
Figure 5. Genetic or pharmacologic MOZ and/or Menin disruption alters GIST TF-driven programs.
A, Unsupervised hierarchical clustering of RNA-seq data comparing 5-day treatment of GIST-T1 with VTP-50469 at 0.5 μM, WM-1119 at 1 μM or the combination with each drug at 0.1 μM (n=4 per condition) for all expressed genes (>10 FPKM, n=7,106). B, Unsupervised hierarchical clustering of all expressed genes from RNA-seq data comparing GIST-T1/Cas9 cells transduced with sgRNAs targeting KAT6A, BRPF1, KMT2A, MEN1 or luciferase as control (n=3 per condition) and collected at day 5. C, Heatmap showing Pearson correlation of control normalized RNA-seq data from A and B, with inclusion of control-normlized RNA-seq data from GIST-T1/Cas9 cells transduced with sgRNAs targeting HAND1 and ETV1 (11). D-F, Correlation of gene expression changes in the top 5,000 expressed transcripts comparing control-normalized sgRNAs or drug treatments for sgKAT6A and sgBRPF1, sgKAT6A and sgHAND1, and combination drug treatment and sgHAND1. Pearson correlation was performed with P value and r2 shown. G, Heatmap of NES from GSEA gene sets including genes showing enrichment for Menin and BRPF1 (n=385), GIST-T1 SE-associated genes defined by H3K27ac (n=366), genes upregulated (n=421) or downregulated (n=165) by HAND1 and genes upregulated (n=438) or downregulated (n=31) by ETV1. Only gene sets with significant FDRs are displayed using the color scale, with those bearing non-significant FDRs indicated in gray. H-K, GSEA plots showing changes in Menin/BRPF1 enriched genes, SE-associated genes and HAND1 regulated genes. L, Control-normalized expression of genes upregulated by HAND1 (i.e. downregulated following sgHAND1 treatment) in each sgRNA and drug treatment condition. Data were analyzed by Welch’s t test, compared to all expressed genes; ***,P<0.001; **,P<0.01; *,P<0.05. M-O, Expression of select genes associated with GIST lineage, transcription factors, or HAND1-regulation across drug and sgRNA treatment conditions. Data were analyzed by one-way ANOVA with Dunnett’s multiple comparisons test, compared to DMSO or sgLuc control; ***,P<0.001; **,P<0.01; *,P<0.05.
As these results disclosed disparate global transcriptional consequences arising from genetic or chemical disruption of these epigenetic regulators, we sought to identify core GIST transcriptional programs exhibiting common alterations that could account for central functions of MOZ and Menin-MLL complexes. Using GSEA, we evaluated for changes in the expression of genes enriched for MOZ and Menin-MLL complexes, H3K27ac-defined super-enhancer (SE) associated genes, and genes regulated by TFs HAND1 and ETV1. Among these GIST-associated gene lists, genes regulated by TFs were the most markedly affected, with drug or sgRNA treatment causing reduced expression of genes upregulated by HAND1 and increased expression of genes normally downregulated by HAND1 or ETV1 function (Fig. 5G). Genetic or pharmacologic MOZ disruption exhibited the greatest effect on genes bound by Menin or BRPF1 (Fig. 5H), while only targeting HAND1 or the combination of WM-1119 and VTP-50469 decreased the expression of SE-associated genes (Fig. 5I). However, common to all conditions was the disruption of HAND1-associated gene expression, with expression values of MOZ and Menin-MLL complex disruption using either inhibitors or sgRNAs phenocopying direct HAND1 knockout (Fig. 5J–L).
KIT gene expression was most markedly affected by pharmacologic or genetic disruption of Menin, while there was common loss of DUSP6, a negative regulator of KIT signaling, and the GIST biomarker CD34 (Fig. 5M). Expression of several core GIST TFs was altered by genetic or pharmacologic MOZ or Menin-MLL complex disruption, most notably FOXF1, HAND2 and PITX1, with WM-1119 exerting the greatest global reduction in TF expression (Fig. 5N, Fig. S5A–B). Several other genes highly regulated by HAND1 expression, including NPR3, ITGA4 and RASL11A, showed similar loss of expression with disruption of MOZ and Menin-MLL complexes (Fig. 5O). Comparable reductions in gene expression of DUSP6 and NPR3 were seen in the KIT-dependent GIST cell lines GIST430, GIST882 and GIST48 by qRT-PCR (Fig. S5C–F). Among all Reactome gene sets, processes related to protein translation were the most recurrently altered gene sets among treatment conditions, with most drug and sgRNA data sets showing a decrease in gene expression (Fig. S5G–H). Taken together, these results show that both genetic and pharmacologic means of MOZ and Menin-MLL complex disruption leads to selective alteration of transcriptional programs associated with GIST TFs, and most remarkably HAND1. Further, dual inhibition of Menin and MOZ with small molecules induces complementary effects on global gene expression and reduces expression of GIST SE-associated genes.
Disruption of chromatin and transcriptional regulatory protein interactions with MOZ or Menin inhibition.
As MOZ and Menin-MLL complexes function in a coordinated fashion at highly regulated genomic regions together with other chromatin regulators, we sought to evaluate effects on local protein interactions in the presence or absence of WM-1119 and/or VTP-50469. We utilized the BioID system (30) to append the biotin ligase BirA* to the N-terminus of the MOZ complex member MEAF6, which enabled the covalent labeling of proteins localized within 10 nm with a biotin moiety. To ensure appropriate incorporation of the BirA*-tagged MEAF6 into the MOZ complex, we used CRISPR/Cas9 and a MEAF6-targeted sgRNA to disrupt the endogenous MEAF6, which would otherwise be lethal if not functionally replaced by the stably expressed MEAF6-BirA construct (Fig. 2H). Stable expression of the N-terminally tagged MEAF6-BirA in GIST-T1 to led to high levels of protein production, with evidence of N-terminal degradation products observed on Western blot (Fig. 6A). As a BioID control, we used a construct that fused BirA* to the DNA binding domain of IKZF1, which retained its nuclear localization signal and served as a nuclear background control. Following labeling of cells for 24 hours with biotin, we performed a streptavidin pulldown followed by mass spectrometry, identifying 243 proteins labeled by MEAF6-BirA and enriched above control (Fig. 6B, Table S2). Labeled interactors included chromatin regulatory proteins such as KMT2A/MLL1, KMT2B/MLL2, JADE3 and RUVBL1 in addition to MOZ complex members, enhancer-associated proteins such as BRD4, and the core GIST TF HIC1. Ordering these MOZ-proximal proteins by gene ontology, there was enrichment for cellular processes including DNA repair, mRNA processing and chromatin complex regulation (Fig. 6C). These data demonstrate the integrated cellular function of these transcriptional regulatory proteins and their complex interactions between splicing factors, enhancers and chromatin complexes.
Figure 6. Alteration in protein interactions following MOZ or Menin inhibition.
A, Western blot of parental GIST-T1 cells or those following sgRNA deletion and rescue with a codon optimized MEAF6 construct fused to BirA* (R118G). Western blots include MEAF6 and HA, indicating the endogenous and full-length rescue construct, or actin as loading control. B, Plot of PSM and log2 signal intensity of proximal proteins identified by MEAF6 BioID. MEAF6-enriched proteins, indicated in blue, show >2-fold intensity enrichment compared to background control (n=243). Select interactors are labeled. C, GO term enrichment for MEAF6 proximal proteins. D, Log2 ratio of VTP-50469/DMSO or WM-1119/DMSO signal intensity for MEAF6-enriched proteins following 3 days pre-treatment with inhibitors with an additional 24 hour treatment during biotin labeling. The Pearson correlation is shown. E, Heatmap showing unsupervised hierarchical clustering of DMSO-normalized signal intensity of 67 proteins significantly changing in at least one condition in response to VTP-50469, WM-1119 or the combination treatment. F-H, Plots of DMSO-normalized signal intensity for protein interactors enriched with WM-1119 or combination treatment (F), interactors lost with VTP-50469 or combination treatment (G), or showing up- or down-regulation in response to either or both inhibitors (H). I, Heat maps demonstrating spike-in normalized signal of DOT1L at MACS-defined peaks (n=67,769) in GIST-T1 cells treated with DMSO, VTP-50469 or WM-1119 for 3 days. Scaled read densities ± 1.25 kb from the peak center are shown in rows. J-K, Box plots showing spike-in normalized DOT1L (J) or MEAF6 (K) signal at MACS-defined peaks (DOT1L n=67,769, MEAF6 n=22,581). Data were analyzed by one-way ANOVA with Dunnett’s multiple comparisons test; compared to DMSO control ***,P<0.001; **, P<0.01; *,P<0.05. L, Tracks showing regions of genomic occupancy of spike-in normalized DOT1L under the indicated treatments, H3K79me2, MEAF6 and H3K27ac at the HAND1 locus. M, Day 21 cell count normalized to DMSO following treatment of GIST-T1 or GIST48B with the indicated concentrations of EPZ-5676 (n=5 per condition). Growth over time experiments were analyzed by two-way ANOVA with Tukey’s post-hoc test, compared to GIST48B; ***,P<0.001; *,P<0.05. N, Heatmap showing Pearson correlation of control normalized RNA-seq data from cells treated for 5 days with the indicated inhibitors. O, Correlation of gene expression changes in expressed transcripts (n=5,000) comparing control-normalized drug treatments with EPZ-5676 and VTP-50469. Pearson correlation was performed with P value and r2 shown. P, GSEA plots showing changes in HAND1 regulated genes arising from EPZ-5676 treatment. Q-R, Expression of select genes associated with GIST lineage and transcription factors (n=4 per condition). Data were analyzed by two-tailed t test, compared to DMSO; ***,P<0.001; **,P<0.01; *,P<0.05.
To evaluate for changes in the MEAF6 proximal proteome as a consequence of MOZ and Menin inhibition, we pre-treated MEAF6-BirA expressing GIST-T1 cells for 3 days with drug prior to labeling with biotin and subsequent label-free quantification using mass spectrometry. While the majority of MEAF6-proximal proteins remained the same, a subset of proteins showed significant alterations in abundance with drug treatment, with significant correlation in changes seen with WM-1119 and/or VTP-50469 treatment (Fig. 6D–E). A subset of proximal proteins increased only with WM-1119 or combination treatment, including MOZ complex members KAT6A and BRPF1 and splicing factor XAB2 (Fig. 6F); that BRPF1 association increased only with the combination treatment may suggest differences in MOZ complex interactions following dual inhibitor treatment as compared to initial complex formation (31). In contrast, MLL family members KMT2A/MLL1, KMT2B/MLL2 and DNA and RNA binding anti-apoptotic protein GPATCH4 (32) interactions were decreased only with VTP-50469 treatment (Fig. 6G). Proximity of select chromatin regulators, splicing factors and polymerase regulatory proteins were altered in a similar fashion with either drug, most notably DOT1L which was significantly decreased in all treatment conditions (Fig. 6H). Leveraging DepMap data, DOT1L dependency was found to be highly correlated with co-dependency upon Menin-MLL complex, MOZ complex and PRC2 complex members (Fig. S6A), suggesting the importance of this protein-protein interaction. To determine how MOZ or Menin inhibition altered chromatin association of DOT1L, we performed spike-in normalized ChIP-seq in GIST-T1 cells treated with WM-1119 or VTP-50469. Both drugs significantly decreased DOT1L association with chromatin at all DOT1L binding sites without changing DOT1L expression (Fig. 6I–J, S6B–E).
We similarly performed spike-in normalized ChIP-seq for MEAF6 using the HA tagged MEAF6-BirA construct, which showed no loss of signal with VTP-50469 treatment, but modest reduction with WM-1119 treatment (Fig. 6K, S6F). Together with BioID data, which showed increased association of other MOZ complex members with MEAF6-BirA following WM-1119 treatment, these data suggest that inhibition of KAT6A leads to loss of the intact MOZ complex and associated factors from chromatin; this accounts for both the reduction in ChIP-seq signal and increased MOZ complex association in BioID data following WM-1119 treatment. In an analogous fashion, VTP-50469 treatment was found to decrease labeling of MLL proteins by chromatin-bound MEAF6-BirA, suggesting loss of Menin-MLL complexes from chromatin analogous to what has been shown in MLL-rearranged leukemia (27).
Like other MOZ and Menin-MLL complex members, DOT1L, H3K79me2, the histone mark deposited by DOT1L, and MEAF6 all showed enrichment genome wide at the TSS and gene body of active genes, with enrichment at loci relevant to GIST biology and reduction in DOT1L signal with WM-1119 or VTP-50469 treatment (Fig. 6L, S6B, G–J). To further explore whether loss of DOT1L function could constitute a mechanism of cellular toxicity downstream of MOZ and Menin inhibition, we treated GIST-T1 cells, or GIST48B as control, in a growth over time assay at various doses of the selective DOT1L inhibitor EPZ-5676 (33). At all doses tested, GIST-T1 exhibited significantly reduced cellular proliferation compared to DMSO control or GIST48B, indicating selective toxicity of DOT1L inhibition in a manner similar to MOZ and Menin inhibition (Fig. 6M). DOT1L-targeting sgRNAs led to significant reductions in GIST-T1 cell proliferation, though more modest than that seen with Menin-MLL and MOZ complex targeting sgRNAs, consistent with findings of the genome-scale CRISPR screen (Fig. S6K, 2A). To better characterize the transcriptional consequences of DOT1L inhibition in GIST, we treated GIST-T1 cells with EPZ-5676 for 5 days followed by performing RNA-seq. Globally, transcriptional changes associated with EPZ-5676 treatment were highly correlated with those arising from Menin inhibition with VTP-50469 (Fig. 6N–O), a phenomenon previously observed in MLL-rearranged leukemia (27). Like Menin-MLL and MOZ inhibition, DOT1L inhibition led to significant disruption in HAND1-regulated transcriptional programs (Fig. 6P), with alterations in the expression of KIT, CD34, NPR3 and GIST TFs (Fig. 6Q–R). Together, these data demonstrate the complexity of proximal protein interactions between these epigenetic complexes, alterations in the protein and chromatin associations of multiple transcriptional regulators with MOZ or Menin inhibition, and that DOT1L function is a dependency in KIT-dependent GIST with loss of DOT1L chromatin association serving as a downstream consequence of MOZ or Menin inhibition.
Therapeutic effects of MOZ and Menin inhibition in vivo.
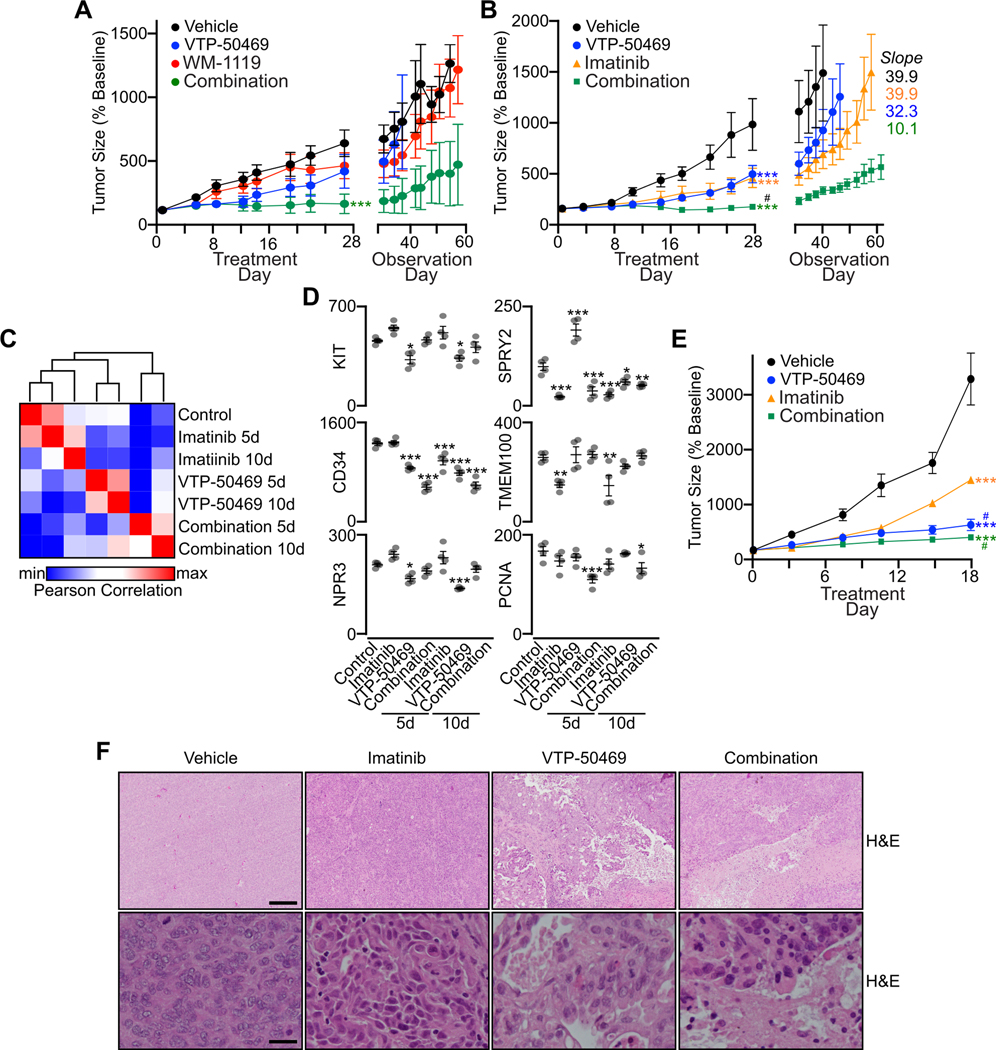
To evaluate the effects of genetic loss of KAT6A, Menin or DOT1L on tumor growth in vivo, we expressed and selected for cells expressing an sgRNA directed against each of these targets or luciferase control in GIST-T1 cells co-expressing Cas9. Following implantation of an equal number of modified cells, mice were monitored for tumor formation and growth. While all implants generated tumors, those derived from cells treated with sgKAT6A or sgMEN1 had significantly reduced growth compared to sgLuc control, whereas expression of sgDOT1L led to a non-significant trend towards reduced growth (Fig. S7A). Though growth restriction was more modest than analogous in vitro experiments (Fig. 2H, S6K), sgKAT6A and sgMEN1 conditions required selection and propagation for two weeks in vitro to produce enough cells for implantation, likely selecting for cells with less deleterious gene alterations. As these genetic results supported a more prominent role for MOZ and Menin-MLL complexes in tumor growth, we next evaluated the effects of MOZ and Menin pharmacologic inhibition in vivo. We treated GIST-T1 engrafted mice with either WM-1119, dosed three times daily 7 days/week, or VTP-50469 given continuously in chow. WM-1119 has been shown to be rapidly cleared after administration (26), necessitating frequent daily administration to achieve therapeutic effects. Whereas 28 day administration of WM-1119 or VTP-50469 alone led to non-significant reductions in tumor size (72 and 65% of vehicle control, respectively), the combination treatment completely halted tumor growth over the treatment period (Fig. 7A). We continued monitoring tumor recovery without further drug treatment, and all conditions showed comparable tumor growth rates similar to vehicle control. Mice tolerated treatment with WM-1119, VTP-50469 or the combination without evidence of overt toxicity or weight loss (Fig. S7B).
Figure 7. Menin and MOZ disruption regulates GIST growth in vivo.
A, GIST-T1 cell line xenografts were treated for 28 days with WM-1119 (50 mg/kg gavage three times daily, 7d/week; n=6), VTP-50469 (0.1% in chow; n=5), combination WM-1119 and VTP-50469 (n=6) or vehicle control (n=5). After the 28 day treatment period, tumors were continuously measured to monitor for recovery of growth. Data were analyzed by two-way ANOVA, compared to vehicle; ***,P<0.001. B, GIST-T1 cell line xenografts were treated for 28 days with imatinib (50 mg/kg gavage 5d/week; n=5), VTP-50469 (0.1% in chow; n=4), combination imatinib and VTP-50469 (n=5) or vehicle control (n=5). After the 28 day treatment period, tumors were continuously measured to monitor for recovery of growth. Data were analyzed by two-way ANOVA, compared to vehicle; ***,P<0.001; compared to monotherapy; #,P<0.01. C, Data from RNA-seq performed on GIST-T1 cell line xenografts treated for 5 or 10 days with imatinib (50 mg/kg gavage 5d/week; n=4), VTP-50469 (0.1% in chow; n=4), combination imatinib and VTP-50469 (n=4) or vehicle control (n=4) with heatmap showing the Pearson correlation of group-averaged FPKM of all expressed genes (FPKM >10, n=7,434). D, Expression in FPKM of select genes associated with GIST lineage, imatinib regulation or cell proliferation. Data were analyzed by one-way ANOVA with Dunnett’s multiple comparisons test, compared to Control; ***,P<0.001; **,P<0.01; *,P<0.05. E, PG27 PDX were treated for 18 days with imatinib (50 mg/kg gavage 5d/week; n=5), VTP-50469 (0.1% in chow; n=5), combination imatinib and VTP-50469 (n=5) or vehicle control (n=5). Data were analyzed by two-way ANOVA, compared to vehicle; ***,P<0.001; compared to imatinib; #,P<0.01. F, PG27 tumors were harvested at the end of the treatment period and fixed tissues sectioned and stained with H&E. Representative images are shown from treatment groups at 4x (upper panels, scale bar = 250 μm) and 40x (lower panels, scale bar = 25 μm).
Multiple Menin inhibitors have advanced to early phase clinical trials for the treatment of leukemia (NCT04067336, NCT04811560, NCT04065399) and may represent an optimal strategy for targeting GIST’s chromatin regulatory dependencies in combination with TKIs. To assess the effects of Menin inhibition alone or in combination with imatinib in vivo, we treated mice engrafted with GIST-T1 cells with imatinib, VTP-50469, the combination of imatinib and VTP-50469 or vehicle as control. At the end of a 28 day treatment period, both monotherapy treatment groups showed similar significant reductions in tumor growth compared to vehicle, while the combination group showed complete cessation of tumor growth. In a subsequent post-treatment observation period, while the tumors from imatinib and VTP-50469 monotherapy groups regained a tumor growth trajectory similar to the vehicle group, the combination of imatinib and VTP-50469 sustained a 3–4-fold reduced slope of tumor recovery (Fig. 7B). All treatment arms tolerated drug administration without overt toxicity or weight loss (Fig. S7C).
To evaluate for changes in the GIST transcriptional program arising from Menin and/or KIT inhibition in vivo, we performed RNA-seq on GIST-T1 xenografts after 5 and 10 days of imatinib and/or VTP-50469 treatment. While all treatment conditions led to global transcriptional changes compared to vehicle control, greater changes were seen following treatment with VTP-50469 and the combination of imatinib and VTP-50469 at both timepoints, with the gene expression profile of imatinib treatment more closely correlating with vehicle-treated tumors (Fig. 7C). Imatinib, VTP-50469 and combination treatments all led to a decrease in expression of genes regulated by HAND1 (Fig. S7D). While GIST-associated transcripts including KIT, CD34 and NPR3 were preferentially reduced by VTP-50469 treatment, other KIT signaling-dependent transcripts including TMEM100 and SPRY2 (34) were preferentially reduced with imatinib treatment; PCNA, a marker of cellular proliferation, was only reduced with the combination of imatinib and VTP-50469 at both 5 and 10 day timepoints (Fig. 7D), consistent with the greater effect of the combination treatment on tumor growth.
We next evaluated the effects of imatinib and VTP-50469 treatment on PG27, a KIT mutant patient derived xenograft (PDX) model of GIST (23). While imatinib administration below the maximal tolerated dose had a significant but modest growth inhibitory effect compared to the GIST-T1 cell line xenograft, treatment with VTP-50469 alone or in combination with imatinib resulted in a significant reduction in tumor growth (Fig. 7E). At the end of the treatment period, PG27 tumors were harvested, fixed and sectioned to evaluate tumor histology. While vehicle and imatinib treated tumors had monomorphic sheets of tumor cells, xenografts treated with VTP-50469 or drug combination exhibited areas of tumor necrosis (Fig. 7F). Whereas mice treated with VTP-50469 in chow showed no weight loss in the GIST-T1 xenograft experiments, PG27-engrafted mice treated with VTP-50469 at the same concentration exhibited modest weight loss (Fig. S7E), possibly related to systemic effects of the observed tumor necrosis. Despite restriction of tumor growth with VTP-50469 treatment, viable areas of tumor showed similar levels of Ki-67 and cleaved caspase-3 across conditions (Fig. S7F). These data demonstrate the therapeutic activity of VTP-50469 alone or in combination with imatinib, with Menin inhibition decreasing GIST xenograft growth, producing tumor necrosis and, when combined with imatinib, generating durable tumor responses after cessation of treatment.
DISCUSSION
Organization and remodeling of chromatin is essential to cellular lineage, identity and function. Post-translational modifications of histones serve as a nexus of epigenetic regulation that controls binding of TFs and chromatin regulators, ultimately administrating gene expression and chromosomal structure. As chromatin modifications are dynamic and reversible, they require active maintenance by cell type and state specific chromatin modifying enzymes. Cancer exploits or appropriates the chromatin state of its precursor cells to sustain a malignant phenotype, through maintenance of an environment permissive of oncogene activation or by gain-of-function alterations in chromatin regulators such as MLL gene fusions (17). Here, we show that specific chromatin regulators are essential to sustain the GIST epigenome, with the MOZ and Menin-MLL complexes binding to actively expressed genes genome-wide, regulating GIST-associated gene expression programs, coordinating protein-protein interactions between multiple regulators of gene expression, and ultimately regulating cellular proliferation and tumor growth.
MOZ is essential for embryonic development and locus-specific gene regulation (14,35). Through the development of selective KAT6A/B inhibitors, this complex has been found to induce cell cycle arrest and senescence by maintaining H3K9ac at cell type-specific genes (26). Menin, encoded by the multiple endocrine neoplasia 1 (MEN1) gene, has classically been described as a tumor suppressor, with mutations in MEN1 promoting endocrine tumor formation. However, in other tissues multiple functions have been ascribed to Menin arising from the protein’s ability to positively or negatively regulate gene expression, associate with different chromatin complexes, integrate inputs from upstream signaling pathways and modulate DNA replication and repair (36). Menin has been best studied as an oncogenic dependency in the context of MLL-rearranged leukemia, where it binds to the MLL fusion protein and, together with recruitment of DOT1L, executes the leukemogenic gene expression program (27,37,38). In GIST, both MOZ and Menin-MLL complex members are essential for global chromatin regulation and, ultimately, tumor cell proliferation. Compared to hundreds of other cell types profiled in Project DRIVE and DepMap, GIST has exceptional sensitivity to targeted disruption of MOZ and Menin-MLL complex members. In agreement with the conservation of TFs and transcriptional and chromatin landscapes in KIT-dependent GIST (10,37), sensitivity to genetic or pharmacologic MOZ and Menin-MLL complex disruption was lost in a KIT-independent GIST cell line. These data indicate that, unlike the oncogenic hijacking seen in MLL-rearranged leukemias, GIST depends upon the native function of the MOZ and Menin-MLL complexes and their associated dependencies to maintain a chromatin landscape that provides a foundation for a malignant gene expression program.
Multiple lines of evidence suggest collaboration between MOZ and Menin-MLL complexes in transcriptional regulation. Here, we show genome-wide co-localization of MOZ and Menin-MLL complex members at the TSS of actively expressed genes, changes in gene expression arising from inhibition of either complex, proximal protein interactions between these two complexes, coordinated regulation of DOT1L and other transcription-associated proteins, and that effects on cell cycle and proliferation were more marked when inhibiting both complexes rather than one at a time. In agreement with our findings, interactions between MOZ and MLL complexes promoting gene expression have been previously described at the HOXA locus in hematopoietic progenitor cells (39), as well as their coordinated co-localization genome-wide (40). Leveraging DepMap data, we highlight a previously underappreciated and complementary genetic co-dependency of these chromatin regulatory complexes in a minority of cancer cell lines. Dependency upon the PRC2 complex was also seen in GIST, with similar co-dependency observed across DepMap data, suggesting the contrasting but complementary role of PRC2 in chromatin silencing balancing the activating functions of MOZ and Menin-MLL complexes. These data suggest that MOZ, Menin-MLL and PRC2 complexes cooperatively function genome-wide to control chromatin state and transcriptional output. We found superior activity of simultaneous inhibition of MOZ and Menin-MLL complexes with WM-1119 and VTP-50469 on cell cycle, cellular proliferation assays, transcriptional output, and tumor growth. Whereas the MOZ complex was enriched at core GIST TF genes and WM-1119 treatment decreased TF expression/function, the Menin-MLL complex showed greater binding at the KIT locus and VTP-50469 decreased KIT expression, indicating at least some non-overlapping functions of these complexes. While combination drug treatment led to greater alterations in global and SE-associated transcription, the expression of some GIST-associated genes and disruption in protein-protein interactions showed many similarities between monotherapy and combination treatments. While the mechanism of combinatorial toxicity requires further investigation, these results suggest that disruption of one complex may maximally deregulate both at specific target loci, and that non-overlapping functions of MOZ and Menin-MLL complexes may also exist.
In keeping with their association with the TSSs of active genes genome-wide, genetic or pharmacologic disruption of MOZ and Menin-MLL complexes led to alterations in transcription that were enriched in specific pathways. We found these transcriptional changes to closely phenocopy those observed with acute loss of GIST TFs, and most notably HAND1. Selective disruption of HAND1 and other GIST TF-mediated gene expression may represent a primary anti-neoplastic mechanism of MOZ and Menin-MLL complex inhibition in GIST. This toxic mechanism may explain the delayed and progressive effects on GIST cell proliferation, as has been observed in leukemias (27,38,41), and is contrasted by the acutely toxic effects of direct KIT inhibition. In agreement with our findings, KAT6A was recently identified to support oncogenic MYC-associated gene expression and block differentiation in acute myeloid leukemia (AML) through H3K9 acetylation and ENL recruitment (42). ENL (MLLT1) was also found to be a dependency in our GIST genome-scale CRISPR screen (Table S1) and was identified in MOZ complex proximity proteomics experiments (Table S2), suggesting similar chromatin regulation strategies between GIST and AML. These results suggest a common utilization of epigenetic regulators supporting oncogenic transcriptional programs in subsets of these mesoderm-derived cancers.
Using BioID to assess the proximal proteome of MOZ complex member MEAF6, we identified multiple potential regulatory interactions between proteins involved in chromatin modification, transcriptional elongation, mRNA processing and DNA repair. Several interactions with enhancer-associated proteins were also observed, including BRD4, MED1 and the core GIST TF HIC1 which, in light of the principal localization of MOZ and Menin-MLL complexes at the TSS of target genes, suggests possible interaction with looping enhancers. In agreement with these findings, Menin has previously been shown to interact with looping enhancers and cancer-specific TFs to promote oncogenic gene expression (43). That HAND1 was not detected in proximity proteomics experiments, yet HAND1’s gene expression program was highly sensitive to the disruption of MOZ and Menin-MLL complexes, suggests these complexes may control TF programs by both direct association as well as indirectly through the modulation of downstream transcriptional regulatory processes. Our findings suggest that MOZ and Menin-MLL complexes serve as central integrators of the GIST gene expression program, regulating multiple steps in transcription through interactions with enhancer elements and chromatin and transcriptional regulators, depositing histone marks supportive of productive gene transcription. Downstream consequences of MOZ or Menin inhibition include disruption of proximal interactions between transcriptional regulators, including loss of DOT1L from chromatin. DOT1L methylates H3K79 to support an active transcriptional state, and has been investigated in leukemia where its recruitment by the MLL fusion protein is essential for leukemogenesis (44). In solid tumors, DOT1L has been found to cooperate with oncogenic transcription factors (45,46), though DOT1L inhibitors have thus far not been evaluated in clinical trials for solid tumors. With prior studies demonstrating TF dependencies in GIST, and current work showing the vulnerability of GIST cells to both DOT1L genetic and pharmacologic disruption, DOT1L may function as a downstream integrator of TF, MOZ and Menin-MLL complex activity in establishing a transcriptionally active state of select cancer-associated genes. Unexpectedly, using BioID we identified proximal protein interactions with regulators of ribonucleoprotein and ribosomal biogenesis, and associated loss of expression of genes involved in protein translation upon disruption of MOZ and/or Menin-MLL complexes. While the relative contribution of changes in translation to the observed anti-neoplastic activity of these inhibitors is uncertain, this may represent an understudied downstream consequence of such chromatin complex alterations. Taken together, these data demonstrate the essential function of MOZ and Menin-MLL complexes in GIST, which serve as integral components of chromatin regulation and the oncogenic GIST gene expression program.
Through in vivo studies, we found that genetic or pharmacologic disruption of MOZ and Menin-MLL complexes led to reductions in GIST tumor burden, with the combination of WM-1119 and VTP-50469 fully halting tumor growth. Multiple Menin inhibitors have been developed that disrupt the association between Menin and MLL and are now under clinical investigation (27,47,48), and represent ideal strategies to combine with TKIs in GIST. To assess the in vivo effect of Menin inhibition on xenograft models of GIST, we treated cell line and patient-derived xenografts with imatinib, VTP-50469 or the combination treatment, which demonstrated activity of VTP-50469 as a monotherapy and superior activity with the combination of KIT and Menin inhibition. After the treatment period, tumors in both monotherapy arms regained their growth trajectory, while tumors treated with the combination of imatinib and VTP-50469 sustained prolonged tumor suppressive effects observed weeks after treatment withdrawal. Both imatinib and VTP-50469 treatment led to distinctive transcriptional changes in GIST xenografts, with the combination of KIT and Menin inhibition resulting in greater deviation from the baseline tumor transcriptional state. In a PDX model of GIST, we saw potent anti-tumor activity of VTP-50469, with histology showing areas of necrosis interspersed with viable tumor. These results support the clinical development of Menin inhibitors for GIST patients, either alone or ideally in combination with TKIs.
As TKIs are the only clinically available therapeutic strategy in GIST, which has native resistance to cytotoxic chemotherapy (49), targeting Menin and other essential components of the GIST epigenome may prove therapeutically advantageous. The conserved transcriptional and enhancer landscape seen in GIST tumors and cell lines, together with oncogenic KIT gene expression being regulated by disease-specific TFs and enhancer elements, harbingers this disease’s dependency upon epigenetic mechanisms. We describe here the collaborating chromatin regulators responsible for maintaining the GIST epigenome, and how their disruption at multiple disparate nodes with small molecule inhibitors (WM-1119, VTP-50469, EPZ-5676) displays promising and selective anti-cancer activity; members of each of these inhibitor classes have reached clinical trial (e.g. NCT04606446, NCT02141828). As compared with leukemias bearing oncogenic alterations in chromatin regulators, GIST may be an outlier among solid tumors in its dependency upon these pathways and susceptibility to their disruption.
MATERIALS AND METHODS
Cell Culture and Virus Production.
All cell lines tested negative for mycoplasma infection on routine surveillance (MilliporeSigma Cat# MP0025–1KT). Human embryonic kidney (HEK) 293FT (Thermo Fisher Scientific Cat# R70007, RRID: CVCL_6911) and the GIST cell lines GIST-T1 (Cosmo Bio Cat# PMC-GIST01-COS, RRID:CVCL_4976; KIT mutation in exon 11 Δ560–578), GIST430 (RRID: CVCL_7040; KIT mutation in exon 11 Δ560–576), GIST48 (RRID: CVCL_7041; KIT mutations in exon 11 V560D and exon 17 D820A), GIST48B (RRID: CVCL_M441; KIT-independent), and GIST882 (RRID: CVCL_7044; KIT mutation in exon 13 K642E) were cultured in Dulbecco’s modified Eagle’s medium containing 10% FBS, 2 mM L-glutamine, 100 mg/ml penicillin, and 100 mg/ml streptomycin. KIT rescue cell lines, which are independent of the KIT enhancer, were generated as previously described (23). Non-commercial cell lines were obtained from the laboratory of Jonathan Fletcher between 2014 and 2016. KIT exons were sequenced to confirm the expected coding mutations and cell identity of GIST cell lines, and cells were thawed from original or derived stocks and used in the described experiments within approximately 3 months. Transfections were performed with X-tremeGene (Roche, Cat# 6365809001). Lentiviral production was performed as previously described (50). Briefly, 293FT cells were cotransfected with pMD2.G (Addgene #12259), psPAX2 (Addgene #12260) and the lentiviral expression plasmid. Viral supernatant was collected at approximately 72 h and debris removed by centrifugation at 1,000g for 5 min. Cells were transduced with viral supernatant and polybrene at 8 μg/mL by spinoculation at 680g for 60 min. Drugs were used at the indicated concentrations and included imatinib (LC Laboratories Cat# I-5508), WM-1119 (Selleck Chemicals Cat# S8776 or MedChemExpress Cat# HY-102058), VTP-50469 (gift of Syndax Pharmaceuticals), and EPZ-5676 (Selleck Chemicals Cat# S7062). For growth over time assays, 15 × 103 cells were dispensed per well in a 96 well plate, transduced with virus or treated with drug, and cell count performed approximately twice per week on a Guava easyCyte Flow Cytometer (Luminex Corporation), with normalization of cell count to the control condition.
Genome-scale CRISPR Screen.
The Liu Human CRISPR Knockout Library (Addgene #1000000132 (51)) targeting 18,436 genes with 185,634 sgRNAs is divided into two pooled libraries, H1 and H2, containing approximately 5 sgRNAs per gene in each library. Each virion contained an sgRNA, Cas9 and a puromycin resistance gene derived from lentiCRISPRv2. The cell lines GIST-T1 and GIST430 were transduced in duplicate with each library (n=8 total). For each library transduction, 44.64 × 106 cells were transduced at a target MOI of 0.3, with an estimated library coverage of 134x. Puromycin was applied at 72 h for selection. Cells were passaged at confluency to maintain a library coverage >134x for approximately 30 days. At termination of the experiment, genomic DNA was extracted from 30 × 106 cells per library. The region of the sgRNA between U6 and EF-1α was amplified from 200 μg of genomic DNA from each experimental replicate in 32 separate 100 μL reactions. The product was pooled and a second PCR reaction was performed to incorporate Illumina adaptors and a 6bp barcode. A third PCR reaction was performed to enrich for full-length amplicons (primers detailed in Table S3). Final amplicon libraries were purified by agarose gel electrophoresis and extraction with a QIAquick Gel Extraction Kit (Qiagen Cat# 28704). Next generation sequencing was performed on a NovaSeq 6000 (Illumina). MAGeCK software (version 0.5.8) was used to analyze screen data (52). The “count” command was used to generate read counts of all libraries (n=8) with the initial plasmid library (n=2) used as baseline control. Total counts were normalized between samples to minimize effects of sequencing depth. The maximum likelihood estimate command was used to generate β-scores for each screen, with data normalized to control AAVS1 sgRNAs contained within H1 and H2 libraries. Metascape was used for gene ontology enrichment analysis (53).
Cloning and CRISPR.
Cell lines stably expressing a human codon-optimized Streptococcus pyogenes Cas9 (Addgene #73310) were generated by viral transduction. CRISPR single-guide RNAs (sgRNAs) were designed using CHOPCHOP (54) (chopchop.cbu.uib.no), cloned into Lenti-sgRNA-EFS-GFP (LRG, Addgene #65656) modified with GFP replaced by copGFP linked to a puromycin resistance gene by a 2A peptide, and detailed in Table S3. The BioID expression vectors were synthesized with codon optimization to alter sgRNA binding sequences (Twist Bioscience). Dependency Map (DepMap) portal data was accessed through depmap.org (20), utilizing the CRISPR (Avana) Public 20Q3 through 20Q4 releases.
Cell cycle and apoptosis.
Cell cycle analysis was performed following drug treatment for 72 h (imatinib) or 8 days (VTP-50469, WM-1119). Cells were trypsinized, washed in PBS and fixed in 70% ethanol. Propidium iodide at 25 μg/mL (Life Technologies Cat# P1304MP) and RNAse A at 0.2 mg/mL (Thermo Fischer Scientific Cat# EN0531) were used to stain nuclear DNA. Analysis was performed on a Guava easyCyte Flow Cytometer (Luminex Corporation), and single cells were assessed for nuclear content using Guava InCyte software. Apoptosis and cell death were measured following 72 h or 8 days of drug treatment using Guava Nexin Reagent (Luminex Corporation Cat# 4500–0450) per manufacturer’s recommendations. Non-apoptotic cells stain negative for Annexin V and 7-AAD, early apoptotic cells stain positive for Annexin V but negative for 7-AAD and late apoptotic and dead cells stain positive for both Annexin V and 7-AAD. Staining was assayed on a Guava easyCyte Flow Cytometer and data analyzed using Guava InCyte software.
Quantitative RT-PCR.
Cells were trypsinized and washed in PBS for RNA extraction using the RNeasy Mini Kit (Qiagen Cat# 74106). Libraries of cDNA were made using SuperScript IV VILO cDNA Synthesis Kit (Invitrogen Cat# 11766050). RT-PCR was performed using Power SYBR Green PCR Master Mix (Life Technologies Cat# 4367659) on a QuantStudio 6 Flex Real-Time PCR System (Thermo Fischer Scientific). Relative mRNA levels were calculated by the ΔΔCt method using GAPDH expression as reference. Primers are listed in Table S3.
RNA-seq.
Total RNA was isolated using an RNeasy Plus Kit (Qiagen Cat# 74136), and concentration measured by Nanodrop (Thermo Fisher Scientific) and quality by TapeStation 4200 (Agilent). Library preparation was performed using the NEBNext Ultra II DNA Library Prep Kit (New England Biolabs Cat# E7645S). Paired-end 150 bp sequencing was performed on a NovaSeq 6000 (Illumina). RNA-seq data were aligned to hg19 using STAR (55) with expression quantification using Cufflinks (56) to generate gene expression values in fragments per kilobase of transcript per million mapped reads (FPKM) units. Gene set enrichment analysis (GSEA, RRID:SCR_003199 (57)) was performed using custom gene lists or those in the Molecular Signatures Database (software.broadinstitute.org/gsea/).
ChIP-seq and Cut&Tag.
For ChIP-seq, approximately 20 × 106 cells were incubated in 1% formaldehyde for 10 min. Following fixation, excess formaldehyde was quenched with glycine at 0.125 M for 5 min. Samples were washed with PBS, and intact nuclei suspended in SDS Buffer (0.5% SDS, 50 mM Tris, 100 mM NaCl, 5 mM EDTA with protease inhibitor cocktail (Roche Cat# 11873580001)) and sonicated in a E220 Focused-ultrasonicator (Covaris, Inc.). Sonicated samples were spun 20,000g for clarification and supernatant diluted to <0.1% SDS then incubated with Dynabeads Protein A (Life Technologies Cat# 10002D) pre-bound with antibody (H3K9ac, Active Motif Cat# 39137, RRID:AB_2561017; H3K4me3, Abcam Cat# ab8580, RRID:AB_306649; BRPF1, Thermo Fisher Scientific Cat# PA5–27783, RRID:AB_2545259; KAT6A, Cell Signaling Technology Cat# 78462; HA, Cell Signaling Technology Cat# 3724, RRID:AB_1549585; DOT1L, Cell Signaling Technology Cat# 77087, RRID:AB_2799889; H3K79me2, Cell Signaling Technology Cat# 5427, RRID:AB_10693787) overnight. Samples were washed serially with Buffer A (150 mM NaCl, 5 mM EDTA, 5% sucrose, 1% Triton X-100, 0.2% SDS, 20 mM Tris), Buffer B (5 mM EDTA, 1% Triton X-100, 0.1% Deoxycholate, 20 mM Tris), Buffer C (250 mM LiCl, 1 mM EDTA, 0.5% NP40, 0.5% Deoxycholate, 10 mM Tris) and TE following resuspension of beads in Elution Buffer (200 mM NaCl, 100 mM NaHCO3, 1% SDS) and incubation at 65°C to reverse crosslinks for 12–15 h. DNA was purified using AMPureXP beads (Beckman Coulter Cat# A63881) per manufacturer recommendation, and quality assessed by Qubit dsDNA HS Assay Kit (Life Technologies Cat# Q32854) and TapeStation 4200 (Agilent). Sequencing libraries were prepared using a ThruPLEX DNA-seq Kit (Takara Bio Cat# R400675) and sequenced on a NextSeq 500 or 550 System (Illumina). ChIP-seq spike-in normalization was performed by pre-binding spike-in antibody (Active Motif Cat# 61686) together with the IP antibody of interest to Dynabeads. Equal amounts of Drosophila melanogaster chromatin (Active Motif Cat# 53083) was added to prepared GIST cell chromatin per manufacturers recommendations. Resultant sequenced samples were aligned to the Drosophila genome, with total Drosophila read counts used to normalize Homo sapiens read counts across samples.
Cut&TAG was performed as previously described (24) using a protein A and Tn5 Transposase fusion protein (Addgene #124601). In brief, 100,000 GIST-T1 cells were washed in Wash Buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM Spermidine, protease inhibitor cocktail) and bound to Concanavalin A beads (Bangs Laboratories Cat# BP531) for 15 min at room temperature. Bound cells were resuspended in 50 μL Dig Wash Buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM Spermidine, protease inhibitor cocktail, 2 mM EDTA, 0.05% Digitonin) and incubated with antibody diluted 1:100 overnight at 4°C (Menin, Bethyl Cat# A300–105A, RRID:AB_2143306; MLL1n, Bethyl Cat# A300–086A, RRID:AB_242510). A magnet was used to collect beads, and cells were resuspended in 100 μL Dig-Wash buffer with a secondary antibody diluted 1:100 and incubated at room temperature for 30 min. Cells were washed three times in Dig-Wash buffer and resuspended in Dig-Med Buffer (0.05% Digitonin, 20 mM HEPES, pH 7.5, 300 mM NaCl, 0.5 mM Spermidine, protease inhibitor cocktail) containing 1:250 pA-Tn5 transposase and incubated at room temperature for 1 h. Cells were washed three times in Dig-Med Buffer and resuspended in 300 μL Tagmentation Buffer (10 mM MgCl2 in Dig-Med Buffer) and incubated at 37°C for 1 h. Tagmentation was stopped by adding 10 μL of 0.5 M EDTA, 3 μL of 10% SDS and 2.5 μL of 20 mg/mL Proteinase K (Invitrogen Cat# 25530049) and samples incubated 50°C for 1 h. Tagmented DNA was purified by phenol:cholorphorm:isoamyl alcohol extraction, and aqueous layer subjected to ethanol precipitation, and DNA was resuspended in 30 μL TE. For each sample, 21 μL DNA was mixed with a universal i5 and uniquely barcoded i7 primer and amplified using NEBNext High Fidelity 2x PCR Master Mix (New England Biolabs Cat# M0541S) in a thermocycler using the following conditions: 98°C for 30 sec; 14 cycles of 98°C for 10 sec, 63°C for 10 sec; 72°C for 2 min. DNA was purified with AMPureXP beads per manufacturer recommendation, and quality assessed by Qubit dsDNA HS Assay Kit and TapeStation 4200. Samples were sequenced on a NextSeq 550 System (Illumina).
All sequencing data were aligned to the human reference genome assembly hg19 using Bowtie2 (58). Normalized read density was calculated using Bamliquidator (version 1.0) read density calculator. Aligned reads were extended by 200 bp and the density of reads per base pair was calculated. In each region, the density of reads was normalized to the total number of million mapped reads, generating read density in units of reads per million mapped reads per bp (rpm/bp). Peak finding was performed by Model-based Analysis for ChIP-seq (MACS, version 1.4.2, (59)), and ROSE2 (60) was used to identify regions of signal enrichment. Individual ChIP-seq track displays were generated using bamplot (github.com/linlabbcm). Heat map visualizations of ChIP-seq data were generated using ChAsE (61).
Immunoblotting.
Cells were lysed in RIPA buffer containing protease inhibitor cocktail (Roche Cat# 11873580001) and centrifuged at 14,000g for 10 min to remove genomic DNA and debris. Protein concentrations were determined using a bicinchoninic acid-based assay (Pierce Biotechnology Cat# 23225). Protein samples were subjected to SDS-PAGE and Western blotting with the following antibodies: HA (1:1,000, Cell Signaling Technology Cat# 2367, RRID:AB_10691311), MEAF6 (1:500, Proteintech Cat# 26465–1-AP, RRID:AB_2880524), Actin (1:1,000, Cell Signaling Technology Cat# 4967, RRID:AB_330288), Menin (1:10,000, Bethyl Cat# A300–105A, RRID:AB_2143306), DOT1L (1:1,000; Cell Signaling Technology Cat# 77087, RRID:AB_2799889), or streptavidin-HRP (1:40,000, Abcam Cat# ab7403). Western blots were probed with anti-mouse or anti-rabbit secondary antibodies and detected using the Odyssey CLx infrared imaging system (LI-COR Biosciences), or streptavidin-HRP by chemiluminescence (MilliporeSigma Cat# WBKLS0500). Immunoblots shown are representative of at least three independent experiments.
Mass Spectrometry and BioID.
GIST-T1 cell lines were generated which stably expressed control or experimental mutant biotin ligase (BirA* R118G)-tagged fusion proteins. 24 h biotin-labeled whole cell lysate was subject to affinity pulldown overnight at 4°C using streptavidin-sepharose beads (GE Healthcare Cat# 17–5113-01). Beads were washed three times in 2% SDS in 50 mM Tris, twice in BioID buffer (50 mM Tris, 500 mM NaCl, 0.4% SDS), six times in 50 mM Tris and resuspended in 100 μL of ammonium bicarbonate. Samples were subject to tryptic digestion, and beads and salts removed in a reverse-phase cleanup step. Extracts were dried on a speed-vac, and later reconstituted in 5–10 μl of 2.5% acetonitrile and 0.1% formic acid. A nano-scale reverse-phase HPLC capillary column was created by packing 2.6 μm C18 spherical silica beads into a fused silica capillary (100 μm inner diameter x ~30 cm length) with a flame-drawn tip. After equilibrating the column each sample was loaded via a Famos Auto Sampler (LC Packings). A gradient was formed and peptides were eluted with increasing concentrations of 97.5% acetonitrile and 0.1% formic acid. As peptides eluted they were subjected to electrospray ionization and then entered into an LTQ Orbitrap Velos Pro ion-trap mass spectrometer (Thermo Fisher Scientific). Peptides were detected, isolated, and fragmented to produce a tandem mass spectrum of specific fragment ions for each peptide. Peptide sequences (and hence protein identity) were determined by matching protein databases with the acquired fragmentation pattern by Sequest (Thermo Fisher Scientific). All databases include a reversed version of all the sequences and the data was filtered to between a one and two percent peptide false discovery rate. Label-free quantification of signal intensity was used in replicate samples for quantitative comparisons. Heat maps of log2 fold change in signal compared to DMSO were generated using Morpheus (software.broadinstitute.org/morpheus).
Xenograft Models.
The PG27 patient derived xenograft was obtained from a patient undergoing clinically indicated surgery and following written informed consent to a Dana-Farber Cancer Institute IRB-approved research protocol. Cryopreserved tumor or the GIST-T1 cell line mixed 1:1 with matrigel were implanted subcutaneously into 6 week old female nude mice (NU/NU; Charles River Laboratories). GIST-T1 tested negative for mycoplasma and rodent pathogens (Charles River Laboratories). For in vivo assessment of CRISPR/Cas9 modified cell line growth, GIST-T1/Cas9 cells were treated with the indicated sgRNAs and selected with puromycin in vitro for 14 days prior to bilateral flank implantation. For drug treatment studies, singly engrafted mice were enrolled into treatment groups when tumors reached approximately 100–200 mm3 in size, as measured by calipers and determined by the tumor volume equation: volume = long diameter2 × short diameter × 0.5. Mice were randomly assigned to treatment groups administered imatinib (50 mg/kg gavage daily, 5 days per week), WM-1119 (50 mg/kg gavage three times daily, 7 days per week), VTP-50469 (0.1% in chow) or combination treatments. Imatinib was administered below the maximal tolerated dose to facilitate testing of combination therapy. No statistical methods were used to predetermine sample size, and no animals died from drug treatment. Two GIST-T1 cell line xenograft mice in the control groups were excluded from analysis as the subcutaneous implants failed to grow. One outlier tumor-bearing mouse in the VTP-50469 arm in Fig. 7A was excluded due to early termination from rapid tumor growth (full data available in Table S4). Tumors were dissected and fixed in 10% formalin for corollary studies including H&E staining and immunohistochemistry of sectioned tumors. 4 μm sections were cut from fixed and embedded tumors and stained with Ki-67 (1:400, Cell Signaling Technology Cat# 9027, RRID:AB_2636984) and cleaved caspase-3 (1:250, Cell Signaling Technology Cat# 9579, RRID:AB_10897512). Reactions were developed using DAB (Cell Signaling Technology Cat# 8059) or NovaRed (Vector Laboratories Cat# SK-4800) substrate kits per manufacturer recommendations. All procedures were conducted under protocols approved by the Institutional Animal Care and Use Committee at Dana-Farber Cancer Institute.
Statistical analysis.
Center values, error bars, P-value cutoffs, number of replicates and statistical tests are identified in the corresponding figure legends. For box plots, the box extends from the 25th to 75th percentiles, with the center line indicating the median and whiskers drawn to the 10th and 90th percentile. Samples sizes were not predetermined.
Data and Materials Availability.
Novel sequencing data is available through the GEO Publication Reference ID GSE172154. Additional RNA-seq, ATAC-seq and ChIP-seq data sets analyzed in this study include GSE95864 (10), GSE113207 and GSE113217 (23).
Supplementary Material
STATEMENT OF SIGNIFICANCE.
While many malignancies rely on oncogene amplification, GIST instead depends upon epigenetic regulation of KIT and other essential genes. Utilizing genome-scale CRISPR dependency screens, we identified complementary chromatin modifying complexes essential to GIST and characterize consequences of their disruption, elucidating a novel therapeutic approach to this disease.
ACKNOWLEDGEMENTS
We thank the members of the Armstrong lab, the Experimental Therapeutics Core at DFCI (Prafulla Ghokhale, Benjamin Springer) and the Taplin Mass Spectrometry Facility (Ross Tomaino) for experimental guidance and support. Funding for this study was provided by NIH award R01 CA176745 (S.A. Armstrong), the Harvard Catalyst Medical Research Investigator Training Program, NIH Award UL 1TR002541 (M.L. Hemming), NIH Award K08 CA245235 (M.L. Hemming), and the DFCI Medical Oncology Grant Program (M.L. Hemming).
Footnotes
COMPETING INTERESTS
S.A.A. has been a consultant and/or shareholder for Neomorph Inc, Imago Biosciences, Vitae/Allergan Pharma, Cyteir Therapeutics, C4 Therapeutics, Accent Therapeutics, and Mana Therapeutics. S.A.A. has received research support from Janssen, Novartis, Syndax, and AstraZeneca. B.J.A. is a former employee of the Walter and Eliza Hall Institute of Medical Research, Melbourne, Australia, and has received proceeds from Royalties and Milestone payments related to the BCL2-inhibitor, ABT-199/venetoclax, and the aryl sulfonohydrazide MOZ inhibitors. None of these relationships constitute a conflict of interest for the present work. The remaining authors declare no conflict of interest.
REFERENCES
- 1.Corless CL, Heinrich MC. Molecular Pathobiology of Gastrointestinal Stromal Sarcomas. Annu Rev Pathol Mech Dis. 2008;3:557–86. [DOI] [PubMed] [Google Scholar]
- 2.Hemming ML, Heinrich MC, Bauer S, George S. Translational insights into gastrointestinal stromal tumor and current clinical advances. Annals of Oncology. 2018;3:557–9. [DOI] [PubMed] [Google Scholar]
- 3.Demetri GD, Mehren von M, Blanke CD, Van den Abbeele AD, Eisenberg B, Roberts PJ, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002;347:472–80. [DOI] [PubMed] [Google Scholar]
- 4.Blay J-Y, Serrano C, Heinrich MC, Zalcberg J, Bauer S, Gelderblom H, et al. Ripretinib in patients with advanced gastrointestinal stromal tumours (INVICTUS): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2020;21:923–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antonescu CR, Besmer P, Guo T, Arkun K, Hom G, Koryotowski B, et al. Acquired resistance to imatinib in gastrointestinal stromal tumor occurs through secondary gene mutation. Clin Cancer Res. 2005;11:4182–90. [DOI] [PubMed] [Google Scholar]
- 6.Demetri GD, van Oosterom AT, Garrett CR, Blackstein ME, Shah MH, Verweij J, et al. Efficacy and safety of sunitinib in patients with advanced gastrointestinal stromal tumour after failure of imatinib: a randomised controlled trial. The Lancet. 2006;368:1329–38. [DOI] [PubMed] [Google Scholar]
- 7.Tabone S, Théou N, Wozniak A, Saffroy R, Deville L, Julié C, et al. KIT overexpression and amplification in gastrointestinal stromal tumors (GISTs). Biochimica et Biophysica Acta. 2005;1741:165–72. [DOI] [PubMed] [Google Scholar]
- 8.Chi P, Chen Y, Zhang L, Guo X, Wongvipat J, Shamu T, et al. ETV1 is a lineage survival factor that cooperates with KIT in gastrointestinal stromal tumours. Nature. 2010;467:849–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ran L, Chen Y, Sher J, Wong EWP, Murphy D, Zhang JQ, et al. FOXF1 Defines the Core-Regulatory Circuitry in Gastrointestinal Stromal Tumor. Cancer Discov. 2018;8:234–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hemming ML, Lawlor MA, Zeid R, Lesluyes T, Fletcher JA, Raut CP, et al. Gastrointestinal stromal tumor enhancers support a transcription factor network predictive of clinical outcome. Proc Natl Acad Sci USA. 2018;115:E5746–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hemming ML, coy S, Lin J-R, Andersen JL, Przybyl J, Mazzola E, et al. HAND1 and BARX1 Act as Transcriptional and Anatomic Determinants of Malignancy in Gastrointestinal Stromal Tumor. Clin Cancer Res. 2021;27:1706–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voss TC, Hager GL. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat Rev Genet. 2014. Feb;15(2):69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen T, Dent SYR. Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat Rev Genet. 2014. Feb;15(2):93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voss AK, Collin C, Dixon MP, Thomas T. Moz and Retinoic Acid Coordinately Regulate H3K9 Acetylation, Hox Gene Expression, and Segment Identity. Developmental Cell. 2009;17:674–86. [DOI] [PubMed] [Google Scholar]
- 15.Yang X-J, Ullah M.MOZ and MORF, two large MYSTic HATs in normal and cancer stem cells. Oncogene. 2007;26:5408–19. [DOI] [PubMed] [Google Scholar]
- 16.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Molecular Cell. 2007;25:15–30. [DOI] [PubMed] [Google Scholar]
- 17.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–33. [DOI] [PubMed] [Google Scholar]
- 18.Blomen VA, Majek P, Jae LT, Bigenzahn JW, Nieuwenhuis J, Staring J, et al. Gene essentiality and synthetic lethality in haploid human cells. Science. 2015;350:1092–6. [DOI] [PubMed] [Google Scholar]
- 19.Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, et al. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDonald ER, de Weck A, Schlabach MR, Billy E, Mavrakis KJ, Hoffman GR, et al. Project DRIVE: A Compendium of Cancer Dependencies and Synthetic Lethal Relationships Uncovered by Large-Scale, Deep RNAi Screening. Cell. 2017;170:577–592. [DOI] [PubMed] [Google Scholar]
- 22.Laugesen A, Højfeldt JW, Helin K. Role of the Polycomb Repressive Complex 2 (PRC2) in Transcriptional Regulation and Cancer. Cold Spring Harbor Perspectives in Medicine. 2016;6:a026575–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hemming ML, Lawlor MA, Andersen JL, Hagan T, Chipashvili O, Scott TG, et al. Enhancer Domains in Gastrointestinal Stromal Tumor Regulate KIT Expression and Are Targetable by BET Bromodomain Inhibition. Cancer Research. 2019;79:994–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaya-Okur HS, Wu SJ, Codomo CA, Pledger ES, Bryson TD, Henikoff JG, et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Comms. 2019:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corless CL, Fletcher JA, Heinrich MC. Biology of gastrointestinal stromal tumors. Journal of Clinical Oncology. 2004;22:3813–25. [DOI] [PubMed] [Google Scholar]
- 26.Baell JB, Leaver DJ, Hermans SJ, Kelly GL, Brennan MS, Downer NL, et al. Inhibitors of histone acetyltransferases KAT6A/B induce senescence and arrest tumour growth. Nature. 2018:1–21. [DOI] [PubMed] [Google Scholar]
- 27.Krivtsov AV, Evans K, Gadrey JY, Eschle BK, Hatton C, Uckelmann HJ, et al. A Menin-MLL Inhibitor Induces Specific Chromatin Changes and Eradicates Disease in Models of MLL- Rearranged Leukemia. Cancer Cell. Elsevier Inc; 2019;36:660–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bamborough P, Barnett HA, Becher I, Bird MJ, Chung C-W, Craggs PD, et al. GSK6853, a Chemical Probe for Inhibition of the BRPF1 Bromodomain. ACS Med Chem Lett. 2016;7:552–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meier JC, Tallant C, Fedorov O, Witwicka H, Hwang S-Y, van Stiphout RG, et al. Selective Targeting of Bromodomains of the Bromodomain-PHD Fingers Family Impairs Osteoclast Differentiation. ACS Chem Biol. 2017;12:2619–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lambert J-P, Tucholska M, Go C, Knight JDR, Gingras A-C. Proximity biotinylation and affinity purification are complementary approaches for the interactome mapping of chromatin-associated protein complexes. Journal of Proteomics. 2015;118:81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ullah M, Pelletier N, Xiao L, Zhao SP, Wang K, Degerny C, et al. Molecular Architecture of Quartet MOZ/MORF Histone Acetyltransferase Complexes. Mol Cell Biol. 2008;28:6828–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blotta S, Tassone P, Prabhala RH, Tagliaferri P, Cervi D, Amin S, et al. Identification of novel antigens with induced immune response in monoclonal gammopathy of undetermined significance. Blood. 2009;114:3276–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, et al. Selective Killing of Mixed Lineage Leukemia Cells by a Potent Small-Molecule DOT1 L Inhibitor. Cancer Cell. 2011;20:53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li F, Huynh H, Li X, Ruddy DA, Wang Y, Ong R, et al. FGFR-Mediated Reactivation of MAPK Signaling Attenuates Antitumor Effects of Imatinib in Gastrointestinal Stromal Tumors. Cancer Discov. 2015;5:438–51. [DOI] [PubMed] [Google Scholar]
- 35.Voss AK, Vanyai HK, Collin C, Dixon MP, McLennan TJ, Sheikh BN, et al. MOZ Regulates the Tbx1 Locus, and Moz Mutation Partially Phenocopies DiGeorge Syndrome. Developmental Cell. 2012;23:652–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matkar S, Thiel A, Hua X. Menin: a scaffold protein that controls gene expression and cell signaling. Trends in Biochemical Sciences. 2013;38:394–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yokoyama A, Somervaille TCP, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123:207–18. [DOI] [PubMed] [Google Scholar]
- 38.Dafflon C, Craig VJ, Méreau H, Gräsel J, Schacher Engstler B, Hoffman G, et al. Complementary activities of DOT1L and Menin inhibitors in MLL-rearranged leukemia. Leukemia. 2017;31:1269–77. [DOI] [PubMed] [Google Scholar]
- 39.Paggetti J, Largeot A, Aucagne R, Jacquel A, Lagrange B, Yang X-J, et al. Crosstalk between leukemia-associated proteins MOZ and MLL regulates HOX gene expression in human cord blood CD34+ cells. Oncogene. 2010;29:5019–31. [DOI] [PubMed] [Google Scholar]
- 40.Miyamoto R, Okuda H, Kanai A, Takahashi S, Kawamura T, Matsui H, et al. Activation of CpG-Rich Promoters Mediated by MLL Drives MOZ-Rearranged Leukemia. CellReports. 2020;32:108200. [DOI] [PubMed] [Google Scholar]
- 41.Dzama MM, Steiner M, Rausch J, Sasca D, Schönfeld J, Kunz K, et al. Synergistic targeting of FLT3 mutations in AML via combined menin-MLL and FLT3 inhibition. Blood. 2020;136:2442–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan F, Li J, Milosevic J, Petroni R, Liu S, Shi Z, et al. KAT6A and ENL Form an Epigenetic Transcriptional Control Module to Drive Critical Leukemogenic Gene-Expression Programs. Cancer Discov. 2022. Mar 1;12(3):792–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dreijerink KMA, Groner AC, Vos ESM, Font-Tello A, Gu L, Chi D, et al. Enhancer-Mediated Oncogenic Function of the Menin Tumor Suppressor in Breast Cancer. CellReports. 2017;18:2359–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, et al. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–78. [DOI] [PubMed] [Google Scholar]
- 45.Wong M, Tee AEL, Milazzo G, Bell JL, Poulos RC, Atmadibrata B, et al. The Histone Methyltransferase DOT1L Promotes Neuroblastoma by Regulating Gene Transcription. Cancer Research. 2017;77:2522–33. [DOI] [PubMed] [Google Scholar]
- 46.Vatapalli R, Sagar V, Rodriguez Y, Zhao JC, Unno K, Pamarthy S, et al. Histone methyltransferase DOT1L coordinates AR and MYC stability in prostate cancer. Nat Comms. 2020:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klossowski S, Miao H, Kempinska K, Wu T, Purohit T, Kim E, et al. Menin inhibitor MI-3454 induces remission in MLL1-rearranged and NPM1-mutated models of leukemia. J Clin Invest. 2020;130:981–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu S, Aguilar A, Huang L, Xu T, Zheng K, McEachern D, et al. Discovery of M-808 as a Highly Potent, Covalent, Small-Molecule Inhibitor of the Menin-MLL Interaction with Strong In Vivo Antitumor Activity. J Med Chem. 2020;63:4997–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maki RG, Blay J-Y, Demetri GD, Fletcher JA, Joensuu H, Martin-Broto J, et al. Key Issues in the Clinical Management of Gastrointestinal Stromal Tumors: An Expert Discussion. The Oncologist. 2015;20:823–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hemming ML, Elias JE, Gygi SP, Selkoe DJ. Proteomic profiling of gamma-secretase substrates and mapping of substrate requirements. PLoS Biol. 2008;6:e2571–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fei T, Li W, Peng J, Xiao T, Chen C-H, Wu A, et al. Deciphering essential cistromes using genome-wide CRISPR screens. Proc Natl Acad Sci USA. 2019;116:25186–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang B, Wang M, Zhang W, Xiao T, Chen C-H, Wu A, et al. Integrative analysis of pooled CRISPR genetic screens using MAGeCKFlute. Nature Protocols. 2019:1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou Y, Zhou Bin, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Comms. 2019:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Labun K, Montague TG, Gagnon JA, Thyme SB, Valen E. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Research. 2016;44:W272–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2012;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25.1–R25.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nature Protocols. 2012;7:1728–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, et al. Selective Inhibition of Tumor Oncogenes by Disruption of Super-Enhancers. Cell. 2013;153:320–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Younesy H, Nielsen CB, Lorincz MC, Jones SJM, Karimi MM, Möller T. ChAsE: chromatin analysis and exploration tool. Bioinformatics. 2016;32:3324–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.