Abstract
The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated (Cas) system is widely used as a genome-editing tool in various organisms, including plants, to elucidate the fundamental understanding of gene function, disease diagnostics, and crop improvement. Among the CRISPR/Cas systems, Cas9 is one of the widely used nucleases for DNA modifications, but manipulation of RNA at the post-transcriptional level is limited. The recently identified type VI CRISPR/Cas systems provide a platform for precise RNA manipulation without permanent changes to the genome. Several studies reported efficient application of Cas13 in RNA studies, such as viral interference, RNA knockdown, and RNA detection in various organisms. Cas13 was also used to produce virus resistance in plants, as most plant viruses are RNA viruses. However, the application of CRISPR/Cas13 to studies of plant RNA biology is still in its infancy. This review discusses the current and prospective applications of CRISPR/Cas13-based RNA editing technologies in plants.
Keywords: type VI CRISPR systems, CRISPR/Cas13, RNA targeting, RNA editing, RNA interference
1. CRISPR/Cas Systems
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) systems are found in prokaryotes such as archaea (~80%) and bacteria (~40%) [1]. CRISPR consists of identical direct repetitive DNA sequences (direct repeats), interspaced by highly variable sequence (spacer), and are often associated with CRISPR-associated (Cas) proteins [1,2]. CRISPR, along with the Cas protein, provides an adaptive immune system in prokaryotes targeting the genetic material of the invading pathogen [2]. The discovery of this mechanism in prokaryotes has driven scientists to adopt the CRISPR/Cas system as an advanced tool to modify DNA and RNA in various organisms. CRISPR/Cas systems are classified mainly into class 1, where a multi-protein effector complex is present, and class 2, which utilizes a single protein to edit the target [3]. These classes are further divided into types and sub-types based on the variations and functions of Cas proteins [4,5].
Class 1 Cas systems comprise types I, III, and IV with diverse variants and effector complexes, whereas class 2 Cas systems consist of types II, V, and VI with an effector module containing a single multifunctional protein [5]. A typical CRISPR/Cas genome editing tool consists of a guide RNA (gRNA or CRISPR RNA (crRNA)) complementary to the gene target site and a Cas effector protein, which acts as an endonuclease to cleave the target site [6]. Because of their more straightforward organization, class 2 Cas systems are most widely employed as genome editing tools in which type II and V systems use Cas9 and Cas12 enzymes to edit DNA. Notably, the class 2 CRISPR/Cas9 protein of type II is one of the first Cas proteins studied in detail, which led to its widespread application for DNA editing in animals, plants, and bacteria [7,8,9,10,11]. However, there are certain limitations in DNA editing using CRISPR, such as the requirement of a Protospacer Adjacent Motif (PAM) site, off-target mutations, and low efficiency against viruses [12,13]. Some of these limitations can be circumvented by the recently identified type VI CRISPR/Cas systems that use the Cas13 protein to enable sequence-specific cleavage of ssRNA molecules [14]. RNA manipulation is advantageous over DNA editing as it prevents unwanted pleiotropic effects, and RNA products can be precisely and spatiotemporally regulated [15]. Here, we discuss the recent advances in CRISPR/Cas13 systems for RNA editing, focusing on their applications in plants and future directions.
2. Overview of CRISPR/Cas13 Systems
A single multifunctional Cas13 effector protein contains two higher eukaryotes and prokaryotes nucleotide-binding domains (HEPN) that provide RNase activity. The Cas13 protein, when associated with crRNA, forms an RNA-guided RNA targeting complex to recognize and cleave ssRNA targets. Based on Cas13 phylogeny, features, and functional characterization, this system is further classified into six subtypes: VI-A (Cas13a, C2c2), VI-B (Cas13b, C2c4), VI-C (Cas13c, C2c7), VI-D (Cas13d), and recently, VI-X (Cas13X) and VI-Y (Cas13Y) [16,17,18,19,20] (Table 1). All Cas13 proteins possess two enzymatically distinct RNase activities, which include processing pre-crRNA into mature functional crRNA and the degradation of target RNA by the HEPN domains [17,21,22,23]. The location of these HEPN domains differs based on the type of Cas13 proteins. In Cas13a, 13c, and 13d, the HEPN domains are present at the center and C terminus [24], whereas in Cas13b, Cas13X, and Y, they are located at the N-terminus and C-terminus of the proteins [17,20]. The HEPN domains of Cas13 proteins can cleave not only the desired target, but also exhibit a non-specific collateral cleavage activity resulting in the degradation of the RNA near the Cas13 complex [17,18,24]. The length of the crRNA or the spacer sequence varies (24–30 nt) with the type of Cas13. Of all the Cas13 enzymes, Cas13c is the least functionally characterized. This section discusses different subtypes of the type VI CRISPR/Cas system.
CRISPR/Cas13 Classification
Cas13a, or type VI-A, is the first subtype identified among the type VI systems and was characterized in Leptotrichia shahii (Lsh) [16,24]. In addition, Cas13a has many orthologs such as Listeria seeligeri (Lse) and Leptotrichia wadei (Lwa) [25], Leptotrichia buccalis (Lbu) [21], and Lachnospiraceae bacterium (Lba) [26] (Table 1). The Cas13a CRISPR array typically consists of a 5′ 28 nt direct repeat (DR) unique to each ortholog and a 28-30 nt spacer sequence (complementary to the target sequence). Some orthologs such as LshCas13a have a 3′ H (non-G), a single base protospacer flanking site (PFS) preference, whereas LwaCas13a and LbuCas13a do not show any PFS preference.
Cas13b has its DR on the 3′ end of crRNA compared to the 5′ DR present in Cas13a, Cas13c, and Cas13d. Cas13b orthologs such as Bergeyella zoohelcum (BzCas13b) and Porphyromonas gulae (PguCas13b) prefer 5′ PFS of D (A, U, or G) and 3′ PFS of NAN or NNA (Table 1) [17]. However, Cas13b from Prevotella sp. (PspCas13b) has no PFS requirement [27]. Cas13b is further differentiated into two types based on the presence of regulatory accessory proteins csx27 and csx28 that can repress or enhance the RNA interference activity of Cas13b, respectively [17].
Cas13c is not as functionally characterized compared to other types of Cas13. However, the known characteristics, such as the presence of DR on the 5′ end of crRNA and the spacer length (28–30 nt), are similar to that of Cas13a and Cas13d (Table 1) [19,28]. Cas13c is less efficient at RNA targeting when compared to the Cas13a, b, and d efficiencies [28]. Therefore, most current Cas13 studies rarely employ Cas13c orthologs such as Fusobacterium perfoetens (FpeCas13c).
Cas13d is the smallest of Cas13a-d and is one of the most efficient type VI systems for RNA targeting [18,22] (Table 1). The Cas13d from the Ruminococcus flavefaciens XPD3002 (CasRx/RfxCas13d) is a prominent homolog for multiple organisms [29,30,31]. In addition to the Cas13d effector protein, type VI-D contains a WYL domain consisting of accessory proteins, one of which can positively modulate target and collateral RNase activity [18]. Type VI-D is known for its versatility as it has no PFS constraints like the other Cas13 enzymes.
Table 1.
Summary table of CRISPR/Cas13 classification.
| Type of Cas13 | Orthologues | Structural Composition | Functional Region | Application Scope | Reference |
|---|---|---|---|---|---|
| Cas13a (1250 aa) |
LshCas13a | HEPN Domains (center and C terminus), pre-CrRNA processing, 3′ Non-G PFS preference (except Lwa and LbuCas13a), DR present on the 5′ end |
ssRNA (spacer length 28–30 nt) |
Virus resistance, RNA knockdown, disease diagnostics | [21,24,26,32] |
| LseCas13a | |||||
| LwaCas13a | |||||
| LbuCas13a | |||||
| LbaCas13a | |||||
| Cas13b (1150 aa) |
BzCas13b | HEPN Domains (N and C terminus), pre-CrRNA processing, 5′ PFS preference of D, 3′ PFS NAN/NNA (Except PspCas13b), DR present on 3′ end | ssRNA (spacer length 30 nt) |
Virus resistance, RNA base editing, RNA knockdown | [17,33] |
| PguCas13b | |||||
| PspCas13b | |||||
| Cas13c (1120 aa) |
FpeCas13c | HEPN domains (center and C terminus), pre-CrRNA processing, No PFS preference, DR present on 5′ end | ssRNA (spacer length 28–30 nt) |
RNA knockdown | [19,28] |
| Cas13d (930 aa) |
RfxCas13d | HEPN domains (center and C terminus), pre-CrRNA processing, No PFS preference, DR present on 5′ end | ssRNA (spacer length 23–30 nt) |
Virus resistance, RNA knockdown, alternative splicing modulation | [34,35] |
3. Applications of CRISPR/Cas13 Systems in Plants
Type VI CRISPR/Cas systems provide various applications in various organisms through different RNA technologies such as RNA interference, RNA detection, RNA editing, and RNA targeting [36,37,38]. Cas13 has been used in plants to target RNA viruses, with very few studies on endogenous plant RNA. However, the recent applications of Cas13 in transcriptome studies of humans, animals, and pathogens are opening possibilities for its application in plants. Here, we discuss different CRISPR/Cas13-based RNA technologies and their current plant applications.
3.1. RNA Interference against Viruses
RNA interference (RNAi) is an innate antiviral immunity mechanism that has been successfully used to combat various plant viruses. Nevertheless, the availability of such antiviral strategies is still limited to specific virus groups. Many viruses readily mutate and have developed different counter-defense mechanisms, leading to the rapid emergence of new antiviral approaches. The recently developed CRISPR/Cas13 is a promising tool for engineering plant immunity against a broad range of RNA viruses that constitute a majority of plant viruses (Figure 1a). Although similar to RNAi technology, Cas13 can be highly specific, resulting in fewer off-targets and high knockdown efficiency [28,39]. Aman et al. have successfully demonstrated viral RNA interference using LshCas13a against the Turnip Mosaic Virus (TuMV) RNA genome, targeting its helper component proteinase (HC-Pro) and coat protein (CP) sequences in Nicotiana benthamiana and Arabidopsis thaliana, respectively [40,41] (Table 2). Subsequently, CRISPR/Cas13 has been applied to target RNA viruses such as the potato virus Y (PVY), tobacco mosaic virus (TMV), southern rice black-streaked dwarf virus (SRBSDV), and rice stripe mosaic virus (RSMV) in various plants [42,43] (Table 2). Mahas et al. [30] have demonstrated that CasRx/Cas13d mediates high interference compared to Cas13a and Cas13b simultaneously against one or two different RNA viruses. A recent study on Sweet Potato Virus Disease (SPVD) resistance using the CRISPR/Cas13 system demonstrated that LwaCas13a and RfxCas13d exhibited efficient targeting activity against multiple RNA viruses simultaneously [44].
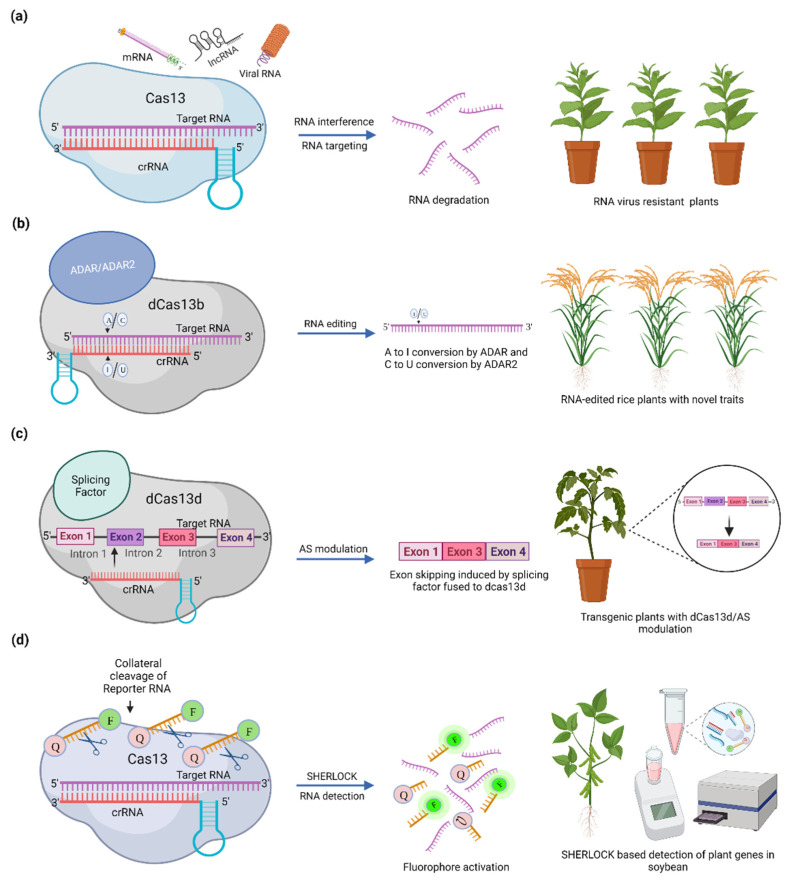
Figure 1.
Pictorial representation of CRISPR/Cas13-based RNA technologies. (a) RNA interference of virus and RNA targeting using Cas13 to generate virus-resistant plants. (b) RNA editing illustration of A to I and C to U conversions when dCas13b is fused with ADAR or ADAR2 enzymes to manipulate a gene’s function and generate transgenic plants with novel traits. (c) Alternative splicing (AS) modulation with splicing factor fused to dCas13d to elucidate the function of different alternatively spliced transcripts. (d) RNA detection using Cas13-based SHERLOCK method for rapid detection of plant genes and pathogens.
Table 2.
Summary table of CRISPR/Cas13 applications in plants.
| Application Scope | Cas13 Type | Plant Species | Target | Reference |
|---|---|---|---|---|
| Viral RNA interference | LshCas13a | Nicotiana benthamiana | Turnip Mosaic Virus (TuMV) | [40] |
| Arabidopsis thaliana | [41] | |||
| Solanum tuberosum | Potato Virus Y (PVY) | [42] | ||
| Nicotiana benthamiana, Oryza sativa | Southern Rice Black-Streaked Dwarf Virus (SRBSDV), Rice Stripe Mosaic Virus (RSMV) | [43] | ||
| LshCas13a, LwaCas13a, PspCas13b, BzCas13b, RfxCas13d | Nicotiana benthamiana |
Turnip Mosaic Virus (TuMV), Tobacco Mosaic Virus (TMV), Potato Virus X (PVX) |
[30] | |
| LshCas13a, LwaCas13a, PspCas13b, RfxCas13d | Nicotiana benthamiana, Ipomoea batatas | Turnip Mosaic Virus (TuMV), Cucumber Mosaic Virus (CMV), Sweet Potato Chlorotic Stunt Virus (SPCSV)-RNase3 | [44] | |
| mRNA knockdown | LwaCas13a |
Oryza sativa ssp. Japonica var. Nipponbare (Protoplasts) |
5-enolpyruvylshikimate-3-phosphate synthase (EPSPS), hydroxycinnamoyl transferase (HCT), and phytoene desaturase (PDS) | [25] |
| LbaCas13a, LbuCas13a | Nicotiana benthamiana, Arabidopsis thaliana, Solanum lycopersicum | PDS transcript | [15] | |
| RNA detection | LwaCas13a, PsmCas13b | (Glyphosate resistant) Glycine max | EPSPS from Agrobacterium sp. strain CP4 (CP4 EPSPS) | [32] |
3.2. RNA Targeting/Knockdown
The CRISPR/Cas13 system can target any single-stranded RNA for degradation without altering DNA (Figure 1a). Accordingly, in addition to RNA interference against viruses, several Cas13 variants have been successfully applied to target endogenous RNA transcripts of various organisms, including LwaCas13a with efficient mRNA knockdown in human cancer cells, RfxCas13d in mRNA knockdown of zebrafish embryos [45], porcine cell, and parthenogenetic embryos [31]. In plants, mRNA knockdown using LwaCas13a was successfully performed in rice protoplasts targeting three different endogenous transcripts, such as 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS), hydroxycinnamoyl transferase (HCT), and phytoene desaturase (PDS), resulting in more than 50% knockdown efficiency in 48 h [25] (Table 2). Recently, Sharma et al. demonstrated the knockdown of PDS transcript in N. benthamiana, A. thaliana, and Solanum lycopersicum using LbaCas13a and LbuCas13a via Agrobacterium infiltration. This study also found that the crRNA can induce gene silencing even in the absence of Cas by utilizing the Argonaute proteins and the plant RNAi machinery [15]. Apart from mRNA, functional studies targeting different non-coding RNA such as circular RNA (circRNA) [46,47] and long non-coding RNA (lncRNA) [48,49] were demonstrated in several human cancer studies. In plants, non-coding RNA knockdown using CRISPR/Cas13 systems has not yet been performed and could be a potential application to characterize the roles and relations of lncRNA with protein-coding mRNA in plants.
3.3. RNA Editing
RNA editing involves the post-transcriptional editing of RNA in a site-specific manner. Recently, Cox et al. engineered a deactivated Cas13b (dCas13b) that binds to target RNA but lacks endonuclease activity [27], allowing the authors to associate the dCas13b enzyme with the ADAR (Adenosine Deaminase Acting on RNA) family enzyme that provides Adenosine to Inosine (A to I) editing of double-stranded RNA (dsRNA). This system can be further adapted to base edit the endogenous transcripts, increase the diversity of the transcriptome, and determine its significance. Based on this concept, RNA editing tools such as RNA Editing for Programmable Adenosine to Inosine (A to I) Replacement (REPAIR) [27] and RNA Editing for Specific Cytosine to Uracil (C to U) Exchange (RESCUE) [50] have been engineered and successfully applied in insects and mammals [20,27,28,50,51]. The ADAR2 deaminase domain provides the base editing from A to I (REPAIR) and C to U (with RNA cytosine deaminase from evolved ADAR2) (RESCUE) without cleaving the RNA transcript (Figure 1b). The A to I and C to U editing of RNA can provide mRNA manipulation, causing alterations in splicing and translation without making permanent changes at the genomic level. Base editing of C to T and A to G of the nuclear genome was previously demonstrated in plants using cytidine base editors (CBEs) and adenine base editors (ABEs), respectively, using a Cas9 nickase [52,53,54,55,56]. These techniques open doors for CRISPR/Cas13 base editors for potential editing in plants at the RNA level.
3.4. Modulation of Alternative Splicing
Alternative splicing is a process regulating gene expression in which exons of a gene are spliced to form multiple alternative transcripts. The CRISPR/Cas13 system can regulate RNA splicing pathways by introducing point mutations into the 5′ and 3′ ends of a splice donor site (GU) and a splice acceptor site (AG), respectively, resulting in mis-splicing and losing targeted splice variants. Konerrman et al. [22] demonstrated a proof of concept for the manipulation of alternative splicing by fusing a catalytically inactive CasRx (dCasRx) with a Gly-rich C-terminal domain of a heterogeneous nuclear ribonucleoprotein (hnRNPa1) splice factor to induce exon skipping in Microtubule Associated Protein Tau (MAPT) pre-mRNA that encodes tau proteins in human neurodegeneration. Recently, Cas13-based splicing modulation tools such as CRISPR Artificial splicing factors (CASFx) have been engineered to demonstrate simultaneous exon inclusion and exclusion induced in different RNA targets based on the positioning of CASFx [35] (Figure 1c). In plants, the potential use of CRISPR/Cas13 to modulate alternative splicing in serine/arginine-rich (SR) family proteins has been described to analyze the role of splice isoforms in stress responses [57]. Nevertheless, experimental studies on CRISPR/Cas13 to manipulate alternative splicing in plants are yet to be demonstrated. The studies mentioned above provide feasibility for potentially modulating alternative splicing in plant disease-susceptible genes to achieve crop improvement.
3.5. RNA Tracking and Nucleic Acid Detection
The target RNA recognition and binding abilities of CRISPR/Cas13 provide means for its application in various functional and diagnostic studies. Previously, Abudayyeh et al. [25] harnessed the catalytically inactive LwaCas13a (dLwaCas13a) to bind targeted RNA transcripts and track stress granule formation in mammalian cells. Later, Yang et al. [58] performed dynamic RNA imaging in live cells using different CRISPR/Cas13 systems. RNA tracking using Cas13 has not been implemented in plants yet. However, in addition to RNA tracking, Cas13 is becoming a prominent tool in diagnostics for nucleic acid detection focusing on viral diseases [33,59]. The targeted cleavage of ssRNA by Cas13 is followed by a non-specific collateral cleavage of proximal RNA. East-Seletsky et al. [21] leveraged this collateral activity of Cas13 to develop an RNA detection tool that can be utilized in vitro to collaterally cleave a fluorophore quencher-labeled reporter RNA that can result in increased fluorescence upon target-RNA-triggered RNase activation (Figure 1d). The Cas13-based RNA detection tool Specific High sensitivity Enzyme Reporter unlocking (SHERLOCK) [60] was further developed based on this concept (Figure 1d). Recently, Ackerman et al. [61] developed a high throughput tool that uses Combinatorial Arrayed Reactions for Multiplexed Evaluation of Nucleic acids (CARMEN). CARMEN, combined with Cas13, can simultaneously detect 4500 crRNA target pairs on a single array, thereby providing a gateway for multiplexed pathogen detection using CRISPR/Cas13. Nucleic acid detection of plant genes using CRISPR/Cas13 was demonstrated by Abudayyeh et al. [32] in soybeans to detect multiple genes in a single reaction using SHERLOCK and to quantify levels of a glyphosate resistance gene, EPSPS from Agrobacterium sp. strain CP4 (CP4 EPSPS), in a mixture of soybeans (Table 2). This platform could be used in other agricultural contexts, such as detecting and quantifying genes, and can also be leveraged for the early detection of plant pathogens or pests, enabling rapid responses by farmers to reduce the use of pesticides or herbicides.
4. Prospective Directions of CRISPR/Cas13 RNA Targeting Systems in Plants
CRISPR/Cas technology has become a powerful genome-editing tool due to its applications in various fields for genetic alterations, disease diagnostics, and crop improvement. For the past decade, the DNA targeting CRISPR/Cas9 system has evolved into a remarkable tool for its implementation in various DNA technologies such as gene knockout, gene activation, gene editing, gene therapy, and DNA detection in multiple organisms [62,63,64,65,66,67]. The recent discovery of CRISPR/Cas13 RNA targeting systems aids in the advancement of existing RNA technologies and has become a promising tool applicable to various organisms [28,45,48]. However, the recent studies on CRISPR/Cas13 systems in plants have mainly focused on viral RNA interference. Cas13-mediated knockdown of endogenous mRNA has been demonstrated in a few studies [15,25], targeting known genes as a proof of concept that can be applied to target the mRNA of disease-susceptible plant genes or study the function of unknown genes through transient knockdown without disrupting the DNA sequence.
In addition, targeting non-coding RNA such as long non-coding RNA, microRNA, and circular RNA in plants using CRISPR/Cas13 can become a promising tool to study their role in plants. Previously, targeting non-coding RNA in plants was performed using CRISPR/Cas9 at the DNA level to find their roles in plant growth, development, and stress responses [52,68,69,70,71,72]. Similarly, CRISPR/Cas13 can also be employed to target non-coding RNA in plants at the RNA level, thereby promoting plant resistance without altering the genome. In the recent past, base editing in plants was performed using an impaired Cas9 (nCas9 or dCas9) to make single base alterations (C to T or T to A) in the genome [52,71,72]. Similarly, RNA base editing can be performed through REPAIR and RESCUE tools that use dCas13 fused with deaminase enzymes causing A to G and C to U conversions [27,73]. However, questions were raised addressing the challenges in the expression stability of CRISPR base editors and in identifying the phenotypic variations caused by base changes in RNA transcript in recent reviews on base editing using CRISPR/Cas13 in plants [74,75]. Therefore, more research on RNA editing in plants using CRISPR/Cas13 is needed for its application in future crop improvement. [74,75].
5. Potential Limitations of CRISPR/Cas13
Though CRISPR/Cas13 can be used to target ssRNA in many organisms, the RNA targeting and RNA binding in Cas13 have constraints, some of which might limit its use. One major limitation is the collateral activity of Cas13 that causes non-specific RNA cleavage [76,77,78]. To overcome this limitation, Tong et al. [79] recently developed high-fidelity Cas13 variants with a dual-fluorescent-reporter system to detect and screen Cas13 variants in mammalian cells, providing efficient RNA targeting and minimizing collateral effects. However, no such collateral activity has been observed in plants yet. Apart from this, few studies in insects and plants report the knockdown of target RNA by crRNA alone in the absence of Cas13 [15,80], causing an RNAi phenomenon. Therefore, individual crRNA and Cas13 controls are necessary to recognize such activity, and further study is required to determine if this can occur in the presence of both Cas13 and crRNA. Another constraint that limits the application of Cas13 is the neurotoxic and embryotoxic effects of some Cas13 enzymes observed in mammalian cells [45,81]. Recently Charles et al. [82] repurposed the inactive Cas13 enzymes for targeted translational repression in bacteria and mitigating their toxicity. Nonetheless, alternative studies are needed to determine the potential of this new system to reduce the toxic effects of Cas13 in different organisms.
6. Conclusions
RNA targeting and editing in plants using CRISPR/Cas13 systems can provide great insights into plant transcriptomic studies. CRISPR/Cas13 application in plants has mainly been to impart tolerance against plant viruses. However, it is also emerging as a prominent tool to target mRNA, circRNA, and other non-coding RNAs. In addition, nucleic acid detection using CRISPR/Cas13 is gaining attention in many recent studies for its high specificity, especially in viral diagnostics. Though Cas13 has a few limitations such as a PFS requirement and non-specific collateral activity affecting its editing efficiency, orthologs such as LwaCas13a, PspCas13b, RfxCas13d can be used to avoid the PFS preference, and collateral RNA cleavage of Cas13 has not been observed in plants yet. Therefore, based on current applications and future perspectives, CRISPR/Cas13 systems can potentially emerge into a robust RNA targeting platform in plants, providing novel opportunities in modern agricultural applications.
Acknowledgments
We thank Renesh Bedre, Sonia Irigoyen, and Shreya Udawant (Texas A&M AgriLife Research) for helpful suggestions during the manuscript preparation.
Author Contributions
N.R.K. prepared the first draft of the manuscript. M.R., Y.Q. and K.M. supervised the work, and reviewed and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This study was supported in part by funds from USDA-NIFA (2021-70029-36056), FFAR, Texas A&M AgriLife Research Insect-vectored Disease Seed Grants (124185-96210), and the Texas A&M AgriLife Institute for Advancing Health Through Agriculture to K.M. Y.Q. is supported by NSF (IOS-1758745, IOS-2029889, IOS-2132693, and MCB-2141560), USDA-NIFA (2018-33522-28789, 2020-33522-32274, 2020-70029-33161, and 2021-67013-34554), FFAR (21010111), and USAID (22010332).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jansen R., Embden J.D.V., Gaastra W., Schouls L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 2.Horvath P., Barrangou R. CRISPR/Cas, the immune system of bacteria and Archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 3.Makarova K.S., Wolf Y.I., Alkhnbashi O.S., Costa F., Shah S.A., Saunders S.J., Barrangou R., Brouns S.J., Charpentier E., Haft D.H. An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koonin E.V., Makarova K.S., Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017;37:67–78. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Makarova K.S., Wolf Y.I., Koonin E.V. Classification and nomenclature of CRISPR-Cas systems: Where from here? CRISPR J. 2018;1:325–336. doi: 10.1089/crispr.2018.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li D., Qiu Z., Shao Y., Chen Y., Guan Y., Liu M., Li Y., Gao N., Wang L., Lu X. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat. Biotechnol. 2013;31:681–683. doi: 10.1038/nbt.2661. [DOI] [PubMed] [Google Scholar]
- 8.Xie K., Yang Y. RNA-guided genome editing in plants using a CRISPR–Cas system. Mol. Plant. 2013;6:1975–1983. doi: 10.1093/mp/sst119. [DOI] [PubMed] [Google Scholar]
- 9.Zhang H., Cheng Q.-X., Liu A.-M., Zhao G.-P., Wang J. A novel and efficient method for bacteria genome editing employing both CRISPR/Cas9 and an antibiotic resistance cassette. Front. Microbiol. 2017;8:812. doi: 10.3389/fmicb.2017.00812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sretenovic S., Qi Y. Plant prime editing goes prime. Nat. Plants. 2022;8:20–22. doi: 10.1038/s41477-021-01047-0. [DOI] [PubMed] [Google Scholar]
- 11.Zhou C., Zhou H., Ma X., Yang H., Wang P., Wang G., Zheng L., Zhang Y., Liu X. Genome-wide identification and characterization of main histone modifications in Sorghum decipher regulatory mechanisms involved by mRNA and long noncoding RNA genes. J. Agric. Food Chem. 2021;69:2337–2347. doi: 10.1021/acs.jafc.0c07035. [DOI] [PubMed] [Google Scholar]
- 12.Uddin F., Rudin C.M., Sen T. CRISPR gene therapy: Applications, limitations, and implications for the future. Front. Oncol. 2020;10:1387. doi: 10.3389/fonc.2020.01387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmad S., Wei X., Sheng Z., Hu P., Tang S. CRISPR/Cas9 for development of disease resistance in plants: Recent progress, limitations and future prospects. Brief. Funct. Genom. 2020;19:26–39. doi: 10.1093/bfgp/elz041. [DOI] [PubMed] [Google Scholar]
- 14.Wolter F., Puchta H. The CRISPR/Cas revolution reaches the RNA world: Cas13, a new Swiss Army knife for plant biologists. Plant J. 2018;94:767–775. doi: 10.1111/tpj.13899. [DOI] [PubMed] [Google Scholar]
- 15.Sharma V.K., Marla S., Zheng W., Mishra D., Huang J., Zhang W., Morris G.P., Cook D.E. CRISPR guides induce gene silencing in plants in the absence of Cas. Genome Biol. 2022;23:6. doi: 10.1186/s13059-021-02586-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shmakov S., Abudayyeh O.O., Makarova K.S., Wolf Y.I., Gootenberg J.S., Semenova E., Minakhin L., Joung J., Konermann S., Severinov K. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol. Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smargon A.A., Cox D.B., Pyzocha N.K., Zheng K., Slaymaker I.M., Gootenberg J.S., Abudayyeh O.A., Essletzbichler P., Shmakov S., Makarova K.S. Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol. Cell. 2017;65:618–630. doi: 10.1016/j.molcel.2016.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan W.X., Chong S., Zhang H., Makarova K.S., Koonin E.V., Cheng D.R., Scott D.A. Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol. Cell. 2018;70:327–339. doi: 10.1016/j.molcel.2018.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shmakov S., Smargon A., Scott D., Cox D., Pyzocha N., Yan W., Abudayyeh O.O., Gootenberg J.S., Makarova K.S., Wolf Y.I. Diversity and evolution of class 2 CRISPR–Cas systems. Nat. Rev. Microbiol. 2017;15:169–182. doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu C., Zhou Y., Xiao Q., He B., Geng G., Wang Z., Cao B., Dong X., Bai W., Wang Y. Programmable RNA editing with compact CRISPR–Cas13 systems from uncultivated microbes. Nat. Methods. 2021;18:499–506. doi: 10.1038/s41592-021-01124-4. [DOI] [PubMed] [Google Scholar]
- 21.East-Seletsky A., O’Connell M.R., Knight S.C., Burstein D., Cate J.H., Tjian R., Doudna J.A. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. 2016;538:270–273. doi: 10.1038/nature19802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Konermann S., Lotfy P., Brideau N.J., Oki J., Shokhirev M.N., Hsu P.D. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell. 2018;173:665–676. doi: 10.1016/j.cell.2018.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Connell M.R. Molecular mechanisms of RNA targeting by Cas13-containing type VI CRISPR–Cas systems. J. Mol. Biol. 2019;431:66–87. doi: 10.1016/j.jmb.2018.06.029. [DOI] [PubMed] [Google Scholar]
- 24.Abudayyeh O.O., Gootenberg J.S., Konermann S., Joung J., Slaymaker I.M., Cox D.B., Shmakov S., Makarova K.S., Semenova E., Minakhin L. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abudayyeh O.O., Gootenberg J.S., Essletzbichler P., Han S., Joung J., Belanto J.J., Verdine V., Cox D.B., Kellner M.J., Regev A. RNA targeting with CRISPR–Cas13. Nature. 2017;550:280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knott G.J., East-Seletsky A., Cofsky J.C., Holton J.M., Charles E., O’Connell M.R., Doudna J.A. Guide-bound structures of an RNA-targeting A-cleaving CRISPR–Cas13a enzyme. Nat. Struct. Mol. Biol. 2017;24:825–833. doi: 10.1038/nsmb.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cox D.B., Gootenberg J.S., Abudayyeh O.O., Franklin B., Kellner M.J., Joung J., Zhang F. RNA editing with CRISPR-Cas13. Science. 2017;358:1019–1027. doi: 10.1126/science.aaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huynh N., Depner N., Larson R., King-Jones K. A versatile toolkit for CRISPR-Cas13-based RNA manipulation in Drosophila. Genome Biol. 2020;21:279. doi: 10.1186/s13059-020-02193-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He B., Peng W., Huang J., Zhang H., Zhou Y., Yang X., Liu J., Li Z., Xu C., Xue M. Modulation of metabolic functions through Cas13d-mediated gene knockdown in liver. Protein Cell. 2020;11:518. doi: 10.1007/s13238-020-00700-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahas A., Aman R., Mahfouz M. CRISPR-Cas13d mediates robust RNA virus interference in plants. Genome Biol. 2019;20:263. doi: 10.1186/s13059-019-1881-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bi D., Yao J., Wang Y., Qin G., Zhang Y., Wang Y., Zhao J. CRISPR/Cas13d mediated efficient KDM5B mRNA knockdown in porcine cells and parthenogenetic embryos. Reproduction. 2021;162:149–160. doi: 10.1530/REP-21-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abudayyeh O.O., Gootenberg J.S., Kellner M.J., Zhang F. Nucleic acid detection of plant genes using CRISPR-Cas13. CRISPR J. 2019;2:165–171. doi: 10.1089/crispr.2019.0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Freije C.A., Myhrvold C., Boehm C.K., Lin A.E., Welch N.L., Carter A., Metsky H.C., Luo C.Y., Abudayyeh O.O., Gootenberg J.S. Programmable inhibition and detection of RNA viruses using Cas13. Mol. Cell. 2019;76:826–837. doi: 10.1016/j.molcel.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wessels H.-H., Méndez-Mancilla A., Guo X., Legut M., Daniloski Z., Sanjana N.E. Massively parallel Cas13 screens reveal principles for guide RNA design. Nat. Biotechnol. 2020;38:722–727. doi: 10.1038/s41587-020-0456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du M., Jillette N., Zhu J.J., Li S., Cheng A.W. CRISPR artificial splicing factors. Nat. Commun. 2020;11:2973. doi: 10.1038/s41467-020-16806-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kordyś M., Sen R., Warkocki Z. Applications of the versatile CRISPR-Cas13 RNA targeting system. Wiley Interdiscip. Rev. RNA. 2021;13:e1694. doi: 10.1002/wrna.1694. [DOI] [PubMed] [Google Scholar]
- 37.Wang F., Wang L., Zou X., Duan S., Li Z., Deng Z., Luo J., Lee S.Y., Chen S. Advances in CRISPR-Cas systems for RNA targeting, tracking and editing. Biotechnol. Adv. 2019;37:708–729. doi: 10.1016/j.biotechadv.2019.03.016. [DOI] [PubMed] [Google Scholar]
- 38.Burmistrz M., Krakowski K., Krawczyk-Balska A. RNA-targeting CRISPR–Cas systems and their applications. Int. J. Mol. Sci. 2020;21:1122. doi: 10.3390/ijms21031122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Granados-Riveron J.T., Aquino-Jarquin G. CRISPR–Cas13 precision transcriptome engineering in cancer. Cancer Res. 2018;78:4107–4113. doi: 10.1158/0008-5472.CAN-18-0785. [DOI] [PubMed] [Google Scholar]
- 40.Aman R., Ali Z., Butt H., Mahas A., Aljedaani F., Khan M.Z., Ding S., Mahfouz M. RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol. 2018;19:1. doi: 10.1186/s13059-017-1381-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aman R., Mahas A., Butt H., Ali Z., Aljedaani F., Mahfouz M. Engineering RNA virus interference via the CRISPR/Cas13 machinery in Arabidopsis. Viruses. 2018;10:732. doi: 10.3390/v10120732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhan X., Zhang F., Zhong Z., Chen R., Wang Y., Chang L., Bock R., Nie B., Zhang J. Generation of virus-resistant potato plants by RNA genome targeting. Plant Biotechnol. 2019;17:1814–1822. doi: 10.1111/pbi.13102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang T., Zhao Y., Ye J., Cao X., Xu C., Chen B., An H., Jiao Y., Zhang F., Yang X. Establishing CRISPR/Cas13a immune system conferring RNA virus resistance in both dicot and monocot plants. Plant Biotechnol. J. 2019;17:1185. doi: 10.1111/pbi.13095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu Y., Pan Z., Wang X., Bian X., Wang W., Liang Q., Kou M., Ji H., Li Y., Ma D. Targeting of SPCSV-RNase3 via CRISPR-Cas13 confers resistance against sweet potato virus disease. Mol. Plant Pathol. 2022;23:104–117. doi: 10.1111/mpp.13146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kushawah G., Hernandez-Huertas L., Del Prado J.A.-N., Martinez-Morales J.R., DeVore M.L., Hassan H., Moreno-Sanchez I., Tomas-Gallardo L., Diaz-Moscoso A., Monges D.E. CRISPR-Cas13d induces efficient mRNA knockdown in animal embryos. Dev. Cell. 2020;54:805–817. doi: 10.1016/j.devcel.2020.07.013. [DOI] [PubMed] [Google Scholar]
- 46.Li S., Li X., Xue W., Zhang L., Yang L.-Z., Cao S.-M., Lei Y.-N., Liu C.-X., Guo S.-K., Shan L. Screening for functional circular RNAs using the CRISPR–Cas13 system. Nat. Methods. 2021;18:51–59. doi: 10.1038/s41592-020-01011-4. [DOI] [PubMed] [Google Scholar]
- 47.Ishola A.A., Chien C.-S., Yang Y.-P., Chien Y., Yarmishyn A.A., Tsai P.-H., Chen J.C.-Y., Hsu P.-K., Luo Y.-H., Chen Y.-M. Oncogenic circRNA hsa_circ_0000190 modulates EGFR/ERK pathway in promoting NSCLC. Cancer Res. 2021;82:75–89. doi: 10.1158/0008-5472.CAN-21-1473. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Z., Chen J., Zhu Z., Zhu Z., Liao X., Wu J., Cheng J., Zhang X., Mei H., Yang G. CRISPR-Cas13-mediated knockdown of lncRNA-GACAT3 inhibited cell proliferation and motility, and induced apoptosis by increasing p21, Bax, and E-cadherin expression in bladder cancer. Front. Mol. Biosci. 2021;7:433. doi: 10.3389/fmolb.2020.627774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu D., Cai Y., Tang L., Han X., Gao F., Cao H., Qi F., Kapranov P. A CRISPR/Cas13-based approach demonstrates biological relevance of vlinc class of long non-coding RNAs in anticancer drug response. Sci. Rep. 2020;10:1794. doi: 10.1038/s41598-020-58104-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abudayyeh O.O., Gootenberg J.S., Franklin B., Koob J., Kellner M.J., Ladha A., Joung J., Kirchgatterer P., Cox D.B., Zhang F. A cytosine deaminase for programmable single-base RNA editing. Science. 2019;365:382–386. doi: 10.1126/science.aax7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kannan S., Altae-Tran H., Jin X., Madigan V.J., Oshiro R., Makarova K.S., Koonin E.V., Zhang F. Compact RNA editors with small Cas13 proteins. Nat. Biotechnol. 2022;40:194–197. doi: 10.1038/s41587-021-01030-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zong Y., Wang Y., Li C., Zhang R., Chen K., Ran Y., Qiu J.-L., Wang D., Gao C. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 2017;35:438–440. doi: 10.1038/nbt.3811. [DOI] [PubMed] [Google Scholar]
- 53.Kang B.-C., Yun J.-Y., Kim S.-T., Shin Y., Ryu J., Choi M., Woo J.W., Kim J.-S. Precision genome engineering through adenine base editing in plants. Nat. Plants. 2018;4:427–431. doi: 10.1038/s41477-018-0178-x. [DOI] [PubMed] [Google Scholar]
- 54.Hua K., Tao X., Yuan F., Wang D., Zhu J.-K. Precise A· T to G· C base editing in the rice genome. Mol. Plant. 2018;11:627–630. doi: 10.1016/j.molp.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 55.Li J., Sun Y., Du J., Zhao Y., Xia L. Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Mol. Plant. 2017;10:526–529. doi: 10.1016/j.molp.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 56.Molla K.A., Sretenovic S., Bansal K.C., Qi Y. Precise plant genome editing using base editors and prime editors. Nat. Plants. 2021;7:1166–1187. doi: 10.1038/s41477-021-00991-1. [DOI] [PubMed] [Google Scholar]
- 57.Morton M., AlTamimi N., Butt H., Reddy A.S., Mahfouz M. Serine/Arginine-rich protein family of splicing regulators: New approaches to study splice isoform functions. Plant Sci. 2019;283:127–134. doi: 10.1016/j.plantsci.2019.02.017. [DOI] [PubMed] [Google Scholar]
- 58.Yang L.-Z., Wang Y., Li S.-Q., Yao R.-W., Luan P.-F., Wu H., Carmichael G.G., Chen L.-L. Dynamic imaging of RNA in living cells by CRISPR-Cas13 systems. Mol. Cell. 2019;76:981–997. doi: 10.1016/j.molcel.2019.10.024. [DOI] [PubMed] [Google Scholar]
- 59.Myhrvold C., Freije C.A., Gootenberg J.S., Abudayyeh O.O., Metsky H.C., Durbin A.F., Kellner M.J., Tan A.L., Paul L.M., Parham L.A. Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018;360:444–448. doi: 10.1126/science.aas8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., Verdine V., Donghia N., Daringer N.M., Freije C.A. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ackerman C.M., Myhrvold C., Thakku S.G., Freije C.A., Metsky H.C., Yang D.K., Simon H.Y., Boehm C.K., Kosoko-Thoroddsen T.-S.F., Kehe J. Massively multiplexed nucleic acid detection with Cas13. Nature. 2020;582:277–282. doi: 10.1038/s41586-020-2279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma X., Zhang Q., Zhu Q., Liu W., Chen Y., Qiu R., Wang B., Yang Z., Li H., Lin Y. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant. 2015;8:1274–1284. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 63.Chen Y., Cao J., Xiong M., Petersen A.J., Dong Y., Tao Y., Huang C.T.-L., Du Z., Zhang S.-C. Engineering human stem cell lines with inducible gene knockout using CRISPR/Cas9. Cell Stem Cell. 2015;17:233–244. doi: 10.1016/j.stem.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jia Y., Xu R.-G., Ren X., Ewen-Campen B., Rajakumar R., Zirin J., Yang-Zhou D., Zhu R., Wang F., Mao D. Next-generation CRISPR/Cas9 transcriptional activation in Drosophila using flySAM. Proc. Natl. Acad. Sci. USA. 2018;115:4719–4724. doi: 10.1073/pnas.1800677115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y., Qin W., Lu X., Xu J., Huang H., Bai H., Li S., Lin S. Programmable base editing of zebrafish genome using a modified CRISPR-Cas9 system. Nat. Commun. 2017;8:118. doi: 10.1038/s41467-017-00175-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rosenblum D., Gutkin A., Kedmi R., Ramishetti S., Veiga N., Jacobi A.M., Schubert M.S., Friedmann-Morvinski D., Cohen Z.R., Behlke M.A. CRISPR-Cas9 genome editing using targeted lipid nanoparticles for cancer therapy. Sci. Adv. 2020;6:eabc9450. doi: 10.1126/sciadv.abc9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou W., Hu L., Ying L., Zhao Z., Chu P.K., Yu X.-F. A CRISPR–Cas9-triggered strand displacement amplification method for ultrasensitive DNA detection. Nat. Commun. 2018;9:5012. doi: 10.1038/s41467-018-07324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li R., Fu D., Zhu B., Luo Y., Zhu H. CRISPR/Cas9-mediated mutagenesis of lncRNA1459 alters tomato fruit ripening. Plant J. 2018;94:513–524. doi: 10.1111/tpj.13872. [DOI] [PubMed] [Google Scholar]
- 69.Zhou J., Deng K., Cheng Y., Zhong Z., Tian L., Tang X., Tang A., Zheng X., Zhang T., Qi Y. CRISPR-Cas9 based genome editing reveals new insights into microRNA function and regulation in rice. Front. Plant Sci. 2017;8:1598. doi: 10.3389/fpls.2017.01598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou J., Yuan M., Zhao Y., Quan Q., Yu D., Yang H., Tang X., Xin X., Cai G., Qian Q. Efficient deletion of multiple circle RNA loci by CRISPR-Cas9 reveals Os06circ02797 as a putative sponge for OsMIR408 in rice. Plant Biotechnol. 2021;19:1240. doi: 10.1111/pbi.13544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Veillet F., Perrot L., Chauvin L., Kermarrec M.-P., Guyon-Debast A., Chauvin J.-E., Nogué F., Mazier M. Transgene-free genome editing in tomato and potato plants using agrobacterium-mediated delivery of a CRISPR/Cas9 cytidine base editor. Int. J. Mol. Sci. 2019;20:402. doi: 10.3390/ijms20020402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qin L., Li J., Wang Q., Xu Z., Sun L., Alariqi M., Manghwar H., Wang G., Li B., Ding X. High-efficient and precise base editing of C• G to T• A in the allotetraploid cotton (Gossypium hirsutum) genome using a modified CRISPR/Cas9 system. Plant Biotechnol. 2020;18:45–56. doi: 10.1111/pbi.13168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chester A., Scott J., Anant S., Navaratnam N. RNA editing: Cytidine to uridine conversion in apolipoprotein B mRNA. Biochim. Biophys. Acta Gene Struct. Expr. 2000;1494:1–13. doi: 10.1016/S0167-4781(00)00219-0. [DOI] [PubMed] [Google Scholar]
- 74.Bharat S.S., Li S., Li J., Yan L., Xia L. Base editing in plants: Current status and challenges. Crop J. 2020;8:384–395. doi: 10.1016/j.cj.2019.10.002. [DOI] [Google Scholar]
- 75.Mishra R., Joshi R.K., Zhao K. Base editing in crops: Current advances, limitations and future implications. Plant Biotechnol. 2020;18:20–31. doi: 10.1111/pbi.13225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang Q., Liu X., Zhou J., Yang C., Wang G., Tan Y., Wu Y., Zhang S., Yi K., Kang C. The CRISPR-cas13a gene-editing system induces collateral cleavage of RNA in glioma cells. Adv. Sci. 2019;6:1901299. doi: 10.1002/advs.201901299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Buchman A.B., Brogan D.J., Sun R., Yang T., Hsu P.D., Akbari O.S. Programmable RNA targeting using CasRx in flies. CRISPR J. 2020;3:164–176. doi: 10.1089/crispr.2020.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li Y., Xu J., Guo X., Li Z., Cao L., Liu S., Guo Y., Wang G., Luo Y., Zhang Z. Collateral cleavage of 28s rRNA by RfxCas13d causes death of mice. bioRxiv. 2022 doi: 10.1101/2022.01.17.476700. [DOI] [Google Scholar]
- 79.Tong H., Huang J., Xiao Q., He B., Dong X., Liu Y., Yang X., Han D., Wang Z., Wang X. High-fidelity Cas13 variants for targeted RNA degradation with minimal collateral effects. Nat. Biotechnol. 2022 doi: 10.1038/s41587-022-01419-7. [DOI] [PubMed] [Google Scholar]
- 80.Tng P.Y.L., Carabajal Paladino L., Verkuijl S.A.N., Purcell J., Merits A., Leftwich P.T., Fragkoudis R., Noad R., Alphey L. Cas13b-dependent and Cas13b-independent RNA knockdown of viral sequences in mosquito cells following guide RNA expression. Commun. Biol. 2020;3:413. doi: 10.1038/s42003-020-01142-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu Q.-W., Kapfhammer J.P. The bacterial enzyme Cas13 interferes with neurite outgrowth from cultured cortical neurons. Toxins. 2021;13:262. doi: 10.3390/toxins13040262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Charles E.J., Kim S.E., Knott G.J., Smock D., Doudna J., Savage D.F. Engineering improved Cas13 effectors for targeted post-transcriptional regulation of gene expression. bioRxiv. 2021 doi: 10.1101/2021.05.26.445687. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.