Abstract
Vascular endothelial cells have a critical role in the maintenance of cardiovascular function. Evidence suggests that endothelial function may be compromised under conditions of magnesium deficiency, which increases vulnerability to inflammation. Whole genome transcription analysis was used to explore the acute (24 h) effects of magnesium on human umbilical vascular endothelial cells (HUVEC) cultured in low (0.1 mM) or high (5 mM) concentrations. With low magnesium 2728 transcripts were differentially expressed compared to the 1 mM control cultures and 3030 were differentially expressed with high magnesium. 615 transcripts were differentially expressed under both conditions, of which only 34 showed a concentration-dependent response. Analysis indicated that cellular organisation and biogenesis and key cellular processes such as apoptosis were impacted by both low and high conditions. High magnesium also influenced protein binding functions, intracellular signal transduction, metabolic and catalytic processes. Both conditions impacted on stress-related processes, in particular the inflammatory response. Key mediators of calcium-dependent regulation of gene expression were responsive to both high and low magnesium conditions. The HUVEC transcriptome is highly sensitive to acute changes in the concentration of magnesium in culture medium. The findings of this study support the view that whilst inflammation is an important process that is responsive to magnesium, the function of the endothelium may be impacted by other magnesium-induced changes including maintenance of cellular integrity, receptor expression and metabolic functions. The high proportion of transcripts that did not show a concentration-dependent response suggests variation in magnesium may elicit indirect changes, possibly mediated by other ions.
Keywords: magnesium, vascular endothelium, microarray, transcriptome
1. Introduction
Vascular endothelial cells play a key role in controlling vascular function through regulation of blood flow [1], cellular adhesion, vascular inflammation, vessel tone and smooth muscle proliferation [2]. Key early steps in atherosclerosis and thrombosis involve injury or dysfunction of the endothelium. Vascular endothelial cell inflammation is the most important driver of atherosclerosis [3]. Cultured endothelial cells have been used extensively as an in vitro model in pathological and physiological experiments relating to vascular disease [4]. Gene expression in cultured endothelial cells has been shown to be responsive to a variety of nutritional stressors and challenges, including micronutrients and plant derived polyphenols [5,6,7]. In human populations the range of Mg intakes from food and in water is broad. Whilst hypermagnesemia is relatively uncommon, disorders related to poor magnesium status are a common problem worldwide [8]. The National Diet and Nutrition Survey in the United Kingdom reported that approximately 10% of men and 9% of women, aged 19 to 64, consumed less than the lower reference nutrient intake (LRNI), which is often an indicator of widespread deficiency in the population [9]. There are other causes of magnesium deficiency, such as malabsorption, gastrointestinal and renal loss, liver or pancreatic disease, vomiting, diarrhoea and chronic alcoholism. Moreover, hypomagnesaemia may be induced by diabetes due to excess excretion associated with diuresis [10]. Magnesium (Mg) is one micronutrient of potential interest in terms of vascular endothelial cell function, as epidemiological and experimental animal studies suggest that poor intakes of magnesium are associated with a greater risk of coronary heart disease [11]. In vitro, endothelial growth is impacted by magnesium deficiency through an increase in cytokine release in conditions of low magnesium concentration [12]. Magnesium status may impact on cardiovascular health through a number of mechanisms [13]. Our previous work suggests that low extracellular magnesium concentrations have a negative influence on endothelial cell proliferation, increase monocyte adhesion [14], inhibit cell migration [15], and exacerbate the response to inflammatory challenges [16]. These effects appear to be due to the activation of many types of cytokines, which induce overexpression of the inflammatory phenotype in endothelial cells [17] and are observable even after 24 h of exposure to variation in magnesium concentration. The cytokines orchestrate the inflammatory response to infection and injury, and play an important role in vascular endothelial cell biology. IL-8 is a critical cytokine that attaches to neutrophils and moves them to the inflamed area. IL-8 expression depends on the activation of NF-κB [18,19,20], and inflamed endothelial cells are the main producers of IL-8 [21]. In addition, GRO and GROα work with IL-8 in attracting neutrophils [22]. MCP-1 cytokines also bind to mononuclear leukocytes [23]. Moreover, IL-6 directs the generation of thrombocytes and the differentiation of B cells [24].
Such mechanisms have been identified through a candidate-led approach which may miss other changes that are associated with low or high magnesium concentrations. To date, there has been no systematic approach to understanding the impact of magnesium on the vascular endothelial cell transcriptome, an important first step towards fully determining the mechanisms which underpin the functional effects of magnesium. Therefore, this study aimed to perform an unbiased analysis of the acute effect of varying magnesium concentrations on the full transcriptome of human umbilical vascular endothelial cells (HUVECs).
2. Materials and Methods
2.1. Cell Culture
Primary HUVECs (C2519A; Lonza Basel, Basel, Switzerland) were cultured in endothelial cell growth medium (EGM-2, Lonza) with 2% fetal bovine serum (FBS), following the manufacturers instructions. The HUVEC cultures were incubated in six-well plates at seed density (7500/cm2), at 37 °C (5% CO2), with the medium changed every other day until the cells were grown to 80–90% confluence. At this point, the HUVECs were cultured for 24 hrs in a human endothelial Mg-free medium (Invitrogen, Waltham, MA, USA) supplemented with 10% FBS, 1% penicillin 100× 1%, 5% endothelial cell growth supplement (Sigma-Aldrich, Poole, UK), and MgSO4 concentrations of 0.1 mM or 5 mM following the method of Ferre et al., and Maier et al., [25,26]. The optimal concentration of FBS to use with the experimental medium, which lacked the growth factors present in the basal EGM-2, and the incubation period required to alter the cellular response to challenges, was determined through preliminary experiments. The samples were compared with cells cultured with 1 mM MgSO4, which is the physiological circulating concentration of Mg2+.
2.2. RNA Extraction
RNA was extracted from the HUVECs using a High Pure RNA Isolation Kit (Roche, Mannheim, Germany) according to the manufacturer’s protocol. A Thermo Scientific NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Wilmington, NC, USA) was used to quantify and determine RNA concentrations. RNA integrity was estimated using an Agilent RNA 6000 Nano Kit, 2100 Bioanalyzer, and Agilent 2100 Expert Software following the manufacturer’s instructions (Agilent Technologies, Waldbronn, Germany). The RNA integrity number (RIN) was measured using Agilent 2100 Bioanalyzer. All RNA preps showed RIN ≥ 9.2, where >7 is the accepted value.
2.3. Microarray Analysis
Whole-genome transcriptome analysis was conducted by hybridizing three biological samples of total RNA per treatment to the GeneChip® Human Genome U133 Plus 2.0 Array (#900470, Affymetrix, High Wycombe, Bucks, UK). All steps of sense cDNA synthesis, fragmentation, and hybridization were performed according to the manufacturer’s protocol (GeneChip® 3000 System, Affymetrix, Cleveland, OH, USA).
2.4. Bioinformatics and Statistical Analysis
Gene expression profile data was generated as CEL files and initially subjected to analysis by the Partek Genomics Suite 6.6 software. Quality Control (QC) metrics were checked by examining surface defects, hybridization, labeling, and a ratio of the 3′ probe set to the 5′ probe set (3′/5′ ratio) to provide the quality of the microarray data. The values were log2 transformed and quantile normalization using the Robust Multi-array Average (RMA). The list of genes of interest comprised genes up-regulated or down-regulated by at least one-fold (log2 fold-change) with an unadjusted p-value < 0.05. To analyse the effects of magnesium treatment, the high-Mg2+ and low Mg2+ groups were compared to the physiological concentration, 1 mM. Data in this paper are reported as Log2 fold-change. Transcript expression data was used to identify biological and molecular functions, which showed significant enrichment by accessing the Gene Ontology (GO) Consortium bioinformatics resource using appropriate portals (Ingenuity Pathway Analysis, Partek Genomics Suite, and PANTHER) [27].
3. Results
3.1. Microarray Approach: Effect of Magnesium Concentration on the HUVEC Transcriptome
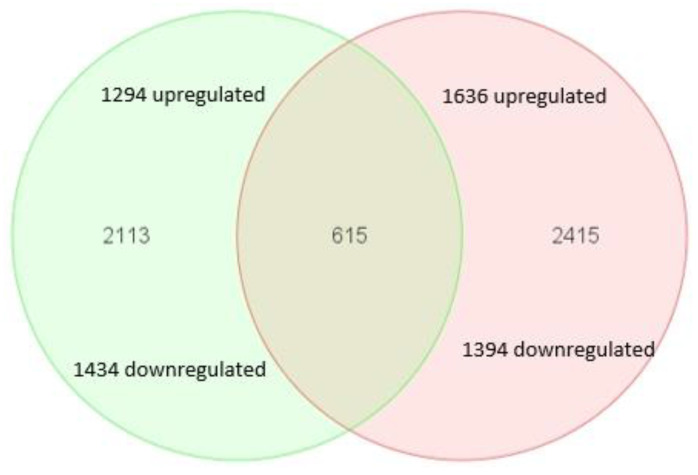
The microarray data were initially examined using principal component analysis, which showed a clear separation between mRNA expression across the three groups (data not shown). Low variance was observed, indicating well-conserved mRNA expression within each group. 5758 out of approximately 47,000 transcripts present on the U133 Plus 2.0 Array chip were differentially expressed in the high- and low-magnesium treatments as compared to the control. Figure 1 shows that 2728 transcripts were differentially expressed after the HUVECs were cultured in 0.1 mM MgSO4, of which 1434 were down-regulated, and 1294 were up-regulated. HUVECs cultured in 5 mM MgSO4 expressed 3030 transcripts differently to the control, with 1394 being down-regulated and 1636 up-regulated (Supplementary Table S1). Six hundred and fifteen transcripts were differentially expressed in both the high- and low-magnesium treatments relative to the control (307 up-regulated, 308 down-regulated; (Supplementary Table S2). Table 1 and Table 2 list the top 20 genes that were differentially expressed in HUVECs cultured in 0.1 and 5 mM MgSO4 based on Log2 fold change.
Figure 1.
Venn diagram of HUVECs genes that were significantly expressed after culturing for 24 h in 0.1 mM MgSO4, indicated by green, and 5 mM MgSO4, indicated by red, as compared to 1 mM MgSO4. 615 genes were differentially expressed relative to control in both conditions.
Table 1.
List of top 20 genes based on fold change that were differentially expressed in HUVECs culture in 0.1 mM MgSO4.
| Prob Set Name | Entrez Gene | Symbol | Gene Title | p-Value | Log 2 Fold-Change |
|---|---|---|---|---|---|
| 225160_x_at | 4193 | MDM2 | Mdm2 p53 binding protein homolog (mouse) | 0.046 | 2.1 |
| 231108_at | 2521 | FUS | Fused in sarcoma | 0.031 | 1.7 |
| 232423_at | 414 | ARSD | Arylsulfatase D | 0.038 | 1.7 |
| 215123_at | 23,117 | NPIPL3 | Nuclear pore complex interacting protein-like 3 | 0.031 | 1.7 |
| 244872_at | 5928 | RBBP4 | retinoblastoma binding protein 4 | 0.023 | 1.6 |
| 213742_at | 9295 | SRSF11 | Serine/arginine-rich splicing factor 11 | 0.046 | 1.6 |
| 209936_at | 10,181 | RBM5 | RNA binding motif protein 5 | 0.021 | 1.5 |
| 232898_at | 1601 | DAB2 | Disabled homolog 2, mitogen-responsive phosphoprotein | 0.003 | 1.5 |
| 243599_at | 100,507,226 | LOC100507226 | Hypothetical LOC100507226 | 0.04 | 1.5 |
| 216983_s_at | 7767 | ZNF224 | Zinc finger protein 224 | 0.008 | 1.4 |
| 224576_at | 57,222 | ERGIC1 | Endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 | 0.045 | −1.7 |
| 211611_s_at | 1388///7148 | ATF6B | Activating transcription factor 6 beta | 0.045 | −1.7 |
| 1555561_a_at | 55,757 | UGGT2 | UDP-glucose glycoprotein glucosyltransferase 2 | 0.001 | −1.6 |
| 210932_s_at | 6049 | RNF6 | Ring finger protein (C3H2C3 type) 6 | 0.023 | −1.5 |
| 229667_s_at | 3218 | HOXB8 | Homeobox B8 | 0.024 | −1.5 |
| 242157_at | 80,205 | CHD9 | Chromodomain helicase DNA binding protein 9 | 0.019 | −1.5 |
| 1555106_a_at | 51,496 | CTDSPL2 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like | 0.042 | −1.5 |
| 221440_s_at | 10,741 | RBBP9 | Retinoblastoma binding protein 9 | 0.001 | −1.4 |
| 213606_s_at | 396 | ARHGDIA | Rho GDP dissociation inhibitor (GDI) alpha | 0.042 | −1.4 |
| 206288_at | 5229 | PGGT1B | Protein geranylgeranyltransferase type I, beta subunit | 0.001 | −1.4 |
Table 2.
List of top 20 genes based on fold change that were differentially expressed in HUVECs culture in 5 mM MgSO4.
| Prob Set Name | Entrez Gene | Gene Symbol | Gene Title | p-Value | Log2 Fold-Change |
|---|---|---|---|---|---|
| 228656_at | 5629 | PROX1 | Prospero homeobox 1 | 0.001 | 1.6 |
| 205410_s_at | 493 | ATP2B4 | ATPase, Ca++ transporting, plasma membrane 4 | 0.01 | 1.5 |
| 209401_s_at | 6560 | SLC12A4 | Solute carrier family 12 (potassium/chloride transporters), member 4 | 0.034 | 1.5 |
| 241985_at | 133,746 | JMY | Junction mediating and regulatory protein, p53 cofactor | 0.04 | 1.4 |
| 243323_s_at | 463 | ZFHX3 | Zinc finger homeobox 3 | 0.002 | 1.4 |
| 221085_at | 9966 | TNFSF15 | Tumor necrosis factor (ligand) superfamily, member 15 | 0.018 | 1.4 |
| 207332_s_at | 7037 | TFRC | Transferrin receptor (p90, CD71) | p < 0.001 | 1.4 |
| 219772_s_at | 23,676 | SMPX | Small muscle protein, X-linked | 0.004 | 1.4 |
| 216950_s_at | 100,132,417///2209 | FCGR1A | Fc fragment of IgG, high affinity Ia, receptor (CD64) | 0.018 | 1.4 |
| 210954_s_at | 9819 | TSC22D2 | TSC22 domain family, member 2 | 0.038 | 1.3 |
| 236361_at | 117,248 | GALNTL2 | UDP-N-acetyl-alpha-D-galactosamine | p < 0.001 | −2.0 |
| 1556325_at | 27,145 | FILIP1 | Filamin A interacting protein 1 | p < 0.001 | −1.9 |
| 206336_at | 6372 | CXCL6 | Chemokine (C-X-C motif) ligand 6 | p < 0.001 | −1.8 |
| 230543_at | 8239 | USP9X | Ubiquitin specific peptidase 9, X-linked | 0.039 | −1.7 |
| 240757_at | 23,332 | CLASP1 | Cytoplasmic linker associated protein 1 | 0.007 | −1.6 |
| 203889_at | 6447 | SCG5 | Secretogranin V | 0.004 | −1.6 |
| 1570515_a_at | 27,145 | FILIP1 | Filamin A interacting protein 1 | p < 0.001 | −1.6 |
| 219825_at | 56,603 | CYP26B1 | Cytochrome P450, family 26, subfamily B, polypeptide 1 | 0.004 | −1.5 |
| 207542_s_at | 358 | AQP1 | Aquaporin 1 (Colton blood group) | p < 0.001 | −1.5 |
| 211506_s_at | 3576 | IL8 | Interleukin 8 | 0.011 | −1.5 |
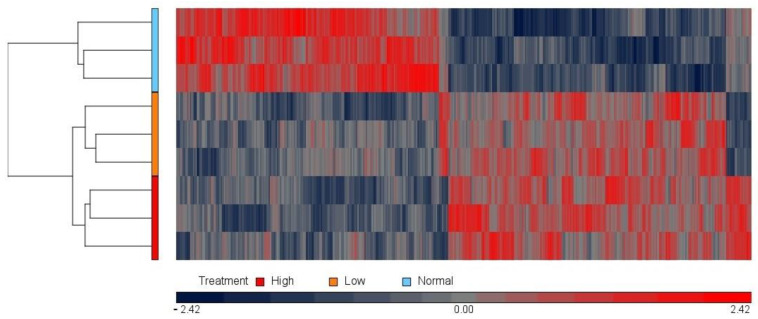
Before examining in detail which genes, processes, and pathways were differentially expressed in low- or high-magnesium conditions, a sub-analysis was conducted to assess the 615 transcripts that were differentially expressed in both conditions, Figure 2 shows the hierarchical cluster analysis (heat map) of these 615 transcripts, which revealed distinctly different expression patterns between the control (1 mM MgSO4) and the high- and low-magnesium treatment groups. As is clear from the heat map, most of these transcripts (581 transcripts) were up-or down-regulated in both low- and high-magnesium conditions, rather than the expected opposing effects of the two treatments. The Gene Ontology (GO) Consortium bioinformatics resource (GOC, http://www.geneontology.org/ 1 July 2022) was used to classify and describe functions of the transcripts that were differentially expressed in both high- and low-magnesium conditions. This analysis showed that the two conditions had effects consistent with changes to cell component organisation (p = 2.49 × 10−5), anion binding (p = 1.15 × 10−4), and protein binding (p = 7.9 × 10−10), and nucleic acid binding (p = 8.7 × 10−3).
Figure 2.
Hierarchical clustering of the 615 genes that were commonly expressed in high- and low-magnesium treatments, with blue blocks representing down-regulated expression and red blocks representing upregulated expression. Each row represents one sample, and each column represents one gene. The red bar on the side of the heat map represents the 5 mM MgSO4 group, n = 3. The blue bar represents the 1 mM MgSO4 group (control), n = 3. The orange bar represents the 0.1 mM MgSO4 group. The scale at the bottom of the diagram represents Log2 fold-change values.
3.2. Gene Ontology Analysis
Table 3 and Table 4 show the enrichment scores and numbers of genes associated with significantly enriched GO processes for the full datasets. All transcript expression data is provided in Supplementary Table S1 which shows all of the differentially expressed transcripts, with the fold-change and is searchable. In terms of biological processes, low magnesium conditions markedly impacted RNA splicing (590 transcripts), cellular processes (819 transcripts), and cellular component organisation and biogenesis (243 transcripts), whilst high magnesium led to significant enrichment of cellular processes (1052 transcripts), metabolic processes (748 transcripts), cellular component organisation and biogenesis (296 transcripts), primary metabolic processes (604 transcripts), cell adhesion (68 transcripts), phosphate-containing compound metabolic processes (228 transcripts), intracellular signal transduction (160 transcripts), cell death (65 transcripts), apoptosis (62 transcripts), G coupled receptor signalling (24 transcripts), nucleobase-containing compound metabolic processes (364 transcripts) and perception of chemical signals (6 transcripts).
Table 3.
GO enrichment analysis of transcripts regulated by high magnesium concentration. Analysis applied FDR p < 0.005 for enrichment p values.
| GO Processes | Enrichment Score | Enrichment p Value | Number of Transcripts Differentially Regulated |
|---|---|---|---|
| Biological processes | |||
| Cellular process | 1.19 | 3.71 × 10−9 | 1052 |
| Sensory perception of chemical signals | 0.20 | 2.21 × 10−5 | 6 |
| Metabolic process | 1.19 | 2.64 × 10−5 | 748 |
| Cellular component organisation or biogenesis | 1.31 | 2.87 × 10−4 | 296 |
| Primary metabolic process | 1.18 | 4.31 × 10−4 | 604 |
| Cell adhesion | 1.78 | 4.94 × 10−4 | 68 |
| Phosphate containing compound metabolic process | 1.33 | 9.64 × 10−4 | 228 |
| Intracellular signal transduction | 1.39 | 2.22 × 10−3 | 160 |
| Cell death | 1.70 | 2.90 × 10−3 | 65 |
| Apoptotic process | 1.72 | 3.46 × 10−3 | 62 |
| G-protein coupled receptor signalling pathway | 0.49 | 3.46 × 10−3 | 24 |
| Nucleobase-containing compound metabolic processes | 1.21 | 3.69 × 10−3 | 364 |
| Molecular functions | |||
| Catalytic activity | 1.21 | 4.30 × 10−4 | 547 |
| Protein binding | 1.23 | 6.15 × 10−3 | 365 |
| Binding | 1.15 | 8.45 × 10−3 | 608 |
| Hydrolase activity | 1.27 | 3.14 × 10−4 | 251 |
| Cellular components | |||
| Intracellular | 1.23 | 1.91 × 10−7 | 694 |
| Cell part | 1.21 | 4.49 × 10−7 | 720 |
| Cytoplasm | 1.25 | 4.99 × 10−5 | 427 |
| Organelle | 1.21 | 1.71 × 10−4 | 507 |
| Nucleus | 1.27 | 1.91 × 10−3 | 265 |
Table 4.
GO enrichment analysis of transcripts regulated by low magnesium concentration. The analysis applied FDR p < 0.005 for enrichment p values.
| GO Processes | Enrichment Score | Enrichment p Value | Number of Transcripts Differentially Regulated |
|---|---|---|---|
| Biological processes | |||
| Metabolic process | 1.15 | 4.75 × 10−3 | 590 |
| Cellular component organisation or biogenesis | 1.33 | 1.02 × 10−3 | 243 |
| Cellular process | 1.14 | 3.73 × 10−4 | 819 |
| Cellular components | |||
| Ribosome macromolecular complex | 1.32 | 3.24 x10−4 | 247 |
| Cytoplasm | 1.29 | 3.08 × 10−5 | 355 |
| Intracellular | 1.27 | 2.39 × 10−8 | 583 |
| Nucleus | 1.32 | 6.89 × 10−4 | 223 |
| Organelle | 1.24 | 3.94 × 10−5 | 424 |
No molecular functions were enriched in conditions in low magnesium at an FDR of p < 0.005 (Table 4). With high magnesium, significant enrichment was noted for catalytic activity, binding activity, protein binding, and hydrolase activity (Table 3). Binding was heavily related to metal iron binding (522 transcripts) and ATP binding (226 transcripts). Conditions of high magnesium significantly influenced the expression of 547 transcripts associated with catalytic activity. The majority of these were hydrolases (251 transcripts; e.g., Ggh, Abdh, Atic, Abhd1, Hibch, Sgsh, Ddah2), and the range of enzymes responding to magnesium mainly included those involved in protein and peptide metabolism (e.g., Sppl3, Sppl13, Usp34, Usp36, Tpp1), and carboxylic acid metabolism (e.g., Csad, Mccc2, Paics). Cofactor binding was also influenced by high magnesium (e.g., Hpgd, Scp2, Gale, Mutyh1, Csad, Lancl1, Acly, Phyh), although there was no evidence that these effects were limited to enzymes with magnesium at their catalytic centers. All parts of the cell (intracellular, cytoplasm, nucleus, organelle, ribosome-associated GO processes) were impacted by the variation in magnesium concentrations).
The GO process Cellular Processes was significantly enriched under both low and high magnesium conditions (Table 3 and Table 4). This term covers a broad range of processes, including cell growth and maintenance, cell cycle, cell death, cellular communication, signal transduction, and immune system processes, many of which were separately identified as significantly enriched with high magnesium. As magnesium concentration is known to impact on the proliferation and viability of HUVECs in culture analysis principally focused on cell death and apoptosis. Of 65 transcripts identified as differentially expressed with high magnesium, 19 were also differentially expressed with low magnesium. Interestingly 8 were up-regulated in both conditions and were 7 down-regulated. Four transcripts were of particular interest as they showed different expression changes with low and high magnesium (Pcbp2, Pak4, MAP2K6 up-regulated in high magnesium, down-regulated in low; Cflar down-regulated with high magnesium, and up-regulated with low). MAP2K6, Pak4, and Pcbp2 are all associated with cell proliferation, whilst Cflar is an inhibitor of apoptosis.
Of 160 transcripts associated with intracellular signal transduction differentially regulated in high magnesium conditions, 31 were also differentially regulated with low magnesium (17 up-regulated in both states and 8 down-regulated in both states). Two (Ddr2 and Cflar) were down-regulated with high magnesium and down-regulated with low magnesium, whilst 4 were up-regulated with high and down-regulated with low (Smad2, Arhgap18, Pak4, and MAP2K6). Among transcripts that were differentially regulated in both conditions were NFATC1 (up-regulated) and NFATC3 (down-regulated) which encode transcription factors that play a critical role in responses to calcium. Several of the intracellular signaling transcripts were found to be calcium-sensitive or functionally related to calcium (e.g., Myo1c, Myh7B, Dab2, Grm1, Smad3, Ccl4) and a key steps in the calcium-sensing and gene regulation pathways were also differentially regulated by magnesium (Ryr1 down-regulated in both states; Ryr2 down-regulated with low magnesium; calmodulin 1 up-regulated in both states; Cabin1 (calcineurin binding protein) up-regulated with high magnesium). cAMP response element binding proteins also showed a response to magnesium (CREB1 down-regulated by low magnesium; CREB3 up-regulated with high magnesium).
High magnesium conditions resulted in differential expression of 296 transcripts associated with cellular organisation and biogenesis (Table 3 and Table 4). Of these 54 were also differentially expressed with magnesium deficiency, with all but 3 responding in the same manner (up- or down-regulation) in both states. The exceptions were Tfb1m, Col4A2 and Arhgap18. In magnesium-depleted cells the changes were mostly related to cell component biogenesis (assembly of macromolecules and cell components), whilst magnesium supplementation had a larger impact on organelles and their organisation (including cytoskeletal effects).
3.3. Effects of Magnesium Concentration on Genes Involved in Inflammation, Mediated by the Chemokine and Cytokine Signalling Pathway
As previous studies had demonstrated that variation in magnesium concentration altered the response of HUVECs to inflammatory challenges we focused analysis on pathways involved in inflammation. Interleukin-8 (IL-8) was one of the transcripts shown to be responsive to low and high magnesium concentrations in culture. Given this and the fact that inflammation plays a key role in endothelial dysfunction, we examined the effects of magnesium on IL-8 associated inflammatory processes using the Gene Ontology (GO) Consortium bioinformatics resource. High-magnesium treatment significantly affected 444 transcripts that respond to stress, 33 of which were involved in inflammation mediated by the chemokine and cytokine signaling pathway (Table 5). Ten of these, including IL-8, were involved in the interleukin signaling pathway (Table 6). In contrast, the low-magnesium treatment significantly altered mRNA expression of 376 genes that respond to stress, 28 of which belonged to the inflammation mediated by chemokine and cytokine signaling pathway (Table 7). Five of these genes were a part of the interleukin signaling pathway (Table 8).
Table 5.
Differential expression of 33 transcripts based on fold change annotated to genes that are involved in inflammation mediated by chemokine and cytokine signaling pathway (High magnesium condition).
| Prob Set Name | Entrez Gene | Gene Symbol | Gene Title | Gene Ontology Biological Process | p-Value | Log2 Fold-Change |
|---|---|---|---|---|---|---|
| 207445_s_at | 10,803 | CCR9 | Chemokine (C-C motif) receptor 9 | Chemotaxis/inferred from electronic annotation/chemotaxis | 0.042 | 1.3 |
| 211230_s_at | 5293 | PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | B cell homeostasis/inferred from electronic annotation/ | 0.004 | 1.3 |
| 216834_at | 5996 | RGS1 | Regulator of G-protein signaling 1 | Immune response/traceable author statement/signal transduce | 0.047 | 1.2 |
| 230202_at | 5970 | RELA | Transcription factor p65 | Liver development/inferred from electronic annotation | 0.047 | 1.2 |
| 221244_s_at | 5170 | PDPK1 | 3-phosphoinositide dependent protein kinase-1 | Protein phosphorylation/inferred from electronic annotation/ | 0.022 | 1.2 |
| 224909_s_at | 57,580 | PREX1 | Phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 | Superoxide metabolic process/traceable author statement | 0.019 | 1.1 |
| 210162_s_at | 4772 | NFATC1 | Nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 | G1/S transition of mitotic cell cycle/inferred from electronic annotation | 0.036 | 1.1 |
| 222912_at | 408 | ARRB1 | Arrestin, beta 1 | G-protein coupled receptor internalization/inferred from mutant phenotype | 0.015 | 1.1 |
| 203175_at | 391 | RHOG | Rho-related GTP-binding protein, member G (rho G) | Small GTPase mediated signal transduction/inferred from electronic annota | 0.035 | 1.1 |
| 212777_at | 6654 | SOS1 | Son of sevenless homolog 1 | Apoptosis/not recorded signal transduction | 0.017 | 1.1 |
| 211543_s_at | 2870 | GRK6 | G protein-coupled receptor kinase 6 | Protein phosphorylation/inferred from electronic annotation | 0.006 | 1.1 |
| 205884_at | 3676 | ITGA4 | Integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) | Blood vessel remodeling inferred from electronic annotation | 0.029 | 1.1 |
| 205127_at | 5742 | PTGS1 | Prostaglandin-endoperoxide synthase 1 | Prostaglandin biosynthetic process/inferred from sequence or structural s | 0.017 | 1.1 |
| 33814_at | 10,298 | PAK4 | P21 protein (Cdc42/Rac)-activated kinase 4 | Protein phosphorylation/inferred from electronic annotation | 0.019 | 1.1 |
| 228388_at | 4793 | NFKBIB | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta | Transcription/traceable author statement/signal transducti | 0.01 | 1.1 |
| 200885_at | 389 | RHOC | Rho-related GTP binding protein, member C | Small GTPase mediated signal transduction/inferred from electronic annota | 0.018 | 1.1 |
| 208075_s_at | 6354 | CCL7 | Chemokine (C-C motif) ligand 7 | Cellular calcium ion homeostasis/traceable author statement | 0.042 | 1.1 |
| 201188_s_at | 3710 | ITPR3 | Inositol 1,4,5-triphosphate receptor, type 3 | Transport/inferred from electronic annotation/ion transport | 0.038 | 1.1 |
| 225363_at | 5728 | PTEN | Phosphatase and tensin homolog | Regulation of cyclin-dependent protein kinase activity/traceable authors | 0.03 | 1.1 |
| 205125_at | 5333 | PLCD1 | Phospholipase C, delta 1 | Lipid metabolic process/inferred from electronic annotation/ | 0.03 | 1.1 |
| 201895_at | 369 | ARAF | Serine/threonine protein kinase A raf | Protein modification process/traceable author statement | p < 0.001 | 1.1 |
| 212647_at | 6237 | RRAS | Related RAS viral (r-ras) oncogene homolog | Signal transduction/inferred from electronic annotation | 0.041 | 1.1 |
| 224994_at | 817 | CAMK2D | Calcium/calmodulin-dependent protein kinase II delta | G1/S transition of mitotic cell cycle//inferred from electronic annotation | 0.038 | 1.1 |
| 206336_at | 6372 | CXCL6 | Chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) | Chemotaxis/inferred from electronic annotation/chemotaxis | p < 0.001 | −1.8 |
| 211506_s_at | 3576 | IL8 | Interleukin 8 | Angiogenesis/traceable author statement/cellular component | 0.011 | −1.5 |
| 204470_at | 2919 | CXCL1 | Chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) | Chemotaxis/traceable author statement inflammatory response | 0.027 | −1.2 |
| 225139_at | 4775 | NFATC3 | Nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 | Transcription/inferred from electronic annotation | 0.033 | −1.0 |
| 204010_s_at | 3845 | KRAS | GTPase KRas | Apoptosis/inferred from electronic annotation/signal trans | 0.029 | −1.1 |
| 38290_at | 10,636 | RGS14 | Regulator of G-protein signaling 14 | Mitosis/inferred from electronic annotation/signal transduce | 0.001 | −1.1 |
| 204103_at | 6351 | CCL4 | Chemokine (C-C motif) ligand 4 | Cellular component movement/traceable author statement | 0.008 | −1.1 |
| 207952_at | 3567 | IL5 | Interleukin 5 (colony-stimulating factor, eosinophil) | Inflammatory response/inferred from electronic annotation | 0.024 | −1.1 |
| 213044_at | 6093 | ROCK1 | Rho-associated, coiled-coil containing protein kinase 1 | Cytokinesis/inferred from electronic annotation/protein ph | 0.002 | −1.1 |
| 209774_x_at | 2920 | CXCL2 | Chemokine (C-X-C motif) ligand 2 | Chemotaxis/inferred from electronic annotation/chemotaxis | 0.045 | −1.1 |
Table 6.
Differential expression of 10 transcripts based on fold change annotated to genes that are involved the interleukin signalling pathway (High magnesium condition).
| Prob Set Name | Entrez Gene | Gene Symbol | Gene Title | Gene Ontology Biological Process | p-Value | Log2 Fold-Change |
|---|---|---|---|---|---|---|
| 221244_s_at | 5170 | PDPK1 | 3-phosphoinositide dependent protein kinase-1 | Protein phosphorylation/inferred from electronic annotation | 0.022 | 1.2 |
| 228388_at | 4793 | NF-κB IB | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta | Transcription/traceable author statement/signal transducti | 0.01 | 1.1 |
| 202284_s_at | 1026 | CDKN1A | Cyclin-dependent kinase inhibitor 1A (p21, Cip1) | Regulation of cyclin-dependent protein kinase activity/traceable authors | 0.006 | 1.1 |
| 205170_at | 6773 | STAT2 | Signal transducer and activator of transcription 2 | Transcription/inferred from electronic annotation | 0.04 | 1.0 |
| 211506_s_at | 3576 | IL8 | Interleukin 8 | Angiogenesis/traceable author statement//cellular component | 0.011 | −1.5 |
| 234066_at | 9173 | IL1RL1 | Interleukin 1 receptor-like 1 | Immune response/non-traceable author statement/signal tran | p < 0.001 | −1.3 |
| 243541_at | 133,396 | IL31RA | Interleukin 31 receptor A | MAPKKK cascade/non-traceable author statement anti-apoptos | 0.028 | −1.2 |
| 207952_at | 3567 | IL5 | Interleukin 5 (colony-stimulating factor, eosinophil) | Inflammatory response/inferred from electronic annotation | 0.024 | −1.2 |
| 209821_at | 90,865 | IL33 | Interleukin 33 | Positive regulation of macrophage activation/inferred from direct assay | 0.014 | −1.2 |
| 226333_at | 3570 | IL6R | Interleukin 6 receptor | Hepatic immune response/traceable author statement | 0.044 | −1.1 |
Table 7.
Differential expression of 28 transcripts based on fold change annotated to genes that are involved in inflammation mediated by chemokine and cytokine signalling pathway (Low magnesium condition).
| Prob Set Name | Entrez Gene | Gene Symbol | Gene Title | Gene Ontology Biological Process | p-Value | Log2 Fold-Change |
|---|---|---|---|---|---|---|
| 211230_s_at | 5293 | PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | B cell homeostasis/inferred from electronic annotation | 0.006 | 1.3 |
| 205127_at | 5742 | PTGS1 | Prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | Prostaglandin biosynthetic process/inferred from sequence or structural | 0.008 | 1.2 |
| 221244_s_at | 5170 | PDPK1 | 3-phosphoinositide dependent protein kinase-1 | Protein phosphorylation/inferred from electronic annotation | 0.01 | 1.2 |
| 201188_s_at | 3710 | ITPR3 | Inositol 1,4,5-triphosphate receptor, type 3 | Transport/inferred from electronic annotation ion transpor | 0.03 | 1.2 |
| 233254_x_at | 5728 | PTEN | Phosphatase and tensin homolog | Regulation of cyclin-dependent protein kinase activity/traceable authors | 0.006 | 1.2 |
| 232043_at | 2788 | GNG7 | Guanine nucleotide binding protein (G protein), gamma 7 | Behavioral fear response/inferred from electronic annotation | 0.03 | 1.1 |
| 1568926_x_at | 91,807 | MYLK3 | Myosin light chain kinase 3 | Protein phosphorylation/inferred from electronic annotation | 0.018 | 1.1 |
| 216190_x_at | 3688 | ITGB1 | Integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, M | G1/S transition of mitotic cell cycle/inferred from electronic annotation | 0.014 | 1.1 |
| 224994_at | 817 | CAMK2D | Calcium/calmodulin-dependent protein kinase II delta | G1/S transition of mitotic cell cycle//inferred from electronic annotation | 0.01 | 1.1 |
| 205962_at | 5062 | PAK2 | P21 protein (Cdc42/Rac)-activated kinase 2 | Protein phosphorylation/inferred from direct assay/protein | 0.017 | 1.1 |
| 243829_at | 673 | BRAF | V-raf murine sarcoma viral oncogene homolog B1 | MAPKKK cascade/inferred from electronic annotation/protein | 0.003 | 1.1 |
| 210162_s_at | 4772 | NFATC1 | Nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 | G1/S transition of mitotic cell cycle/inferred from electronic annotation | 0.04 | 1.1 |
| 209201_x_at | 7852 | CXCR4 | Chemokine (C-X-C motif) receptor 4 | Activation of MAPK activity traceable author statement | 0.01 | −1.3 |
| 1560524_at | 400,581 | GRAP | GRB2-related adaptor protein | Ras protein signal transduction/traceable author statement | 0.03 | −1.2 |
| 224965_at | 54,331 | GNG2 | Guanine nucleotide binding protein (G protein), gamma 2 | Signal transduction/inferred from electronic annotation | 0.01 | −1.2 |
| 208641_s_at | 5879 | RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | Endocytosis/inferred from electronic annotation/apoptosis | 0.01 | −1.1 |
| 204174_at | 241 | ALOX5AP | Arachidonate 5-lipoxygenase-activating protein | Leukotriene production involved in inflammatory response/inferred from el | 0.04 | −1.1 |
| 222912_at | 408 | ARRB1 | Arrestin, beta 1 | G-protein coupled receptor internalization/inferred from mutant phenotype | 0.04 | −1.1 |
| 201179_s_at | 2773 | GNAI3 | Guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 | Transport/non-traceable author statement/vesicle fusion/ | 0.007 | −1.1 |
| 212590_at | 22,800 | RRAS2 | Related RAS viral (r-ras) oncogene homolog 2 | Signal transduction/inferred from electronic annotation | 0.013432 | −1.1 |
| 204103_at | 6351 | CCL4 | Chemokine (C-C motif) ligand 4 | Cellular component movement/traceable author statement | 0.01 | −1.1 |
| 225141_at | 4775 | NFATC3 | Nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 | Transcription/inferred from electronic annotation | 0.036 | −1.1 |
| 202647_s_at | 4893 | NRAS | Neuroblastoma RAS viral (v-ras) oncogene homolog | Apoptosis/inferred from electronic annotation/signal trans | 0.005 | −1.1 |
| 211434_s_at | 9034 | CCRL2 | Chemokine (C-C motif) receptor-like 2 | Chemotaxis/traceable author statement/signal transduction | 0.03 | −1.1 |
| 39402_at | 3553 | IL1B | Interleukin 1, beta | Activation of MAPK activity/inferred from direct assay | 0.02 | −1.1 |
| 203154_s_at | 10,298 | PAK4 | P21 protein (Cdc42/Rac)-activated kinase 4 | Protein phosphorylation/inferred from electronic annotation | 0.03 | −1.05 |
| 38290_at | 10,636 | RGS14 | Regulator of G-protein signaling 14 | Mitosis/inferred from electronic annotation/signal transdu | 0.004 | −1.0 |
| 207157_s_at | 2787 | GNG5 | Guanine nucleotide binding protein (G protein), gamma 5 | Signal transduction/non-traceable author statement/signal | 0.03 | −1.0 |
Table 8.
Differential expression based on fold change of 5 transcripts annotated to genes that are involved the interleukin signalling pathway (Low magnesium condition).
| Prob Set Name | Entrez Gene | Gene Symbol | Gene Title | Gene Ontology Biological Process | p-Value | Log2 Fold-Change |
|---|---|---|---|---|---|---|
| 204906_at | 6196 | RPS6KA2 | Ribosomal protein S6 kinase, 90 kDa, polypeptide 2 | Mitotic metaphase/inferred from electronic annotation | 0.01 | 1.2 |
| 221244_s_at | 5170 | PDPK1 | 3-phosphoinositide dependent protein kinase-1 | Protein phosphorylation/inferred from electronic annotation | 0.01 | 1.2 |
| 243829_at | 673 | BRAF | V-raf murine sarcoma viral oncogene homolog B1 | MAPKKK cascade/inferred from electronic annotation/protein | 0.003 | 1.1 |
| 202647_s_at | 4893 | NRAS | Neuroblastoma RAS viral (v-ras) oncogene homolog | Apoptosis/inferred from electronic annotation/signal trans | 0.005 | −1.1 |
| 207538_at | 3565 | IL4 | Interleukin 4 | Chemotaxis/traceable author statement/immune response | 0.04 | −1.1 |
4. Discussion
Magnesium deficiency has been reported to be associated with a greater risk of cardiovascular disease in humans, and animal studies have shown that it promotes changes to arterial architecture and exacerbates atherosclerosis in transgenic strains [12,17,28]. Vascular endothelial cells play a central role in the development of atherosclerosis, and there has been interest in whether magnesium status is associated with cardiovascular risk due to the development of endothelial dysfunction in magnesium deficient states. We and others have previously shown that growing HUVECs under acute conditions of magnesium deficiency results in an exaggerated inflammatory response [16,29]. Our previous work reported greater expression of cytokines and adhesion molecules both before and after endotoxin challenge, and that high magnesium concentrations greatly blunted the inflammatory response [30]. The aim of the current work was to evaluate the extent of the HUVEC transcriptome response to variation in magnesium concentration. This provides a greater mechanistic understanding of the association between magnesium and endothelial cell dysfunction and identifies priorities for future study. The work is novel as few studies have evaluated the impact of magnesium on the whole transcriptome in human cells. Martin et al., reported that the impact of lifelong magnesium deficiency or supplementation had only modest effects on the rat liver transcriptome, but this study did not consider acute effects of magnesium, the response to further physiological challenges and looked at a whole tissue rather than specific cell type [31].
Findings presented in this paper show that 12.3% of the transcripts on the microarray were responsive to magnesium treatment, with 6.5% changing due to high magnesium treatment and the remaining 5.8% changing due to low magnesium treatment. In addition, 54% of the transcripts that were affected by high magnesium were up-regulated, and 46% were down-regulated, while 47% of the low magnesium group transcripts were up-regulated, and 52% were down-regulated. These data suggest that magnesium has a powerful effect on the HUVEC transcriptome. Importantly, the study found that magnesium can alter a high percentage of HUVECs genes involved in essential physiological pathways and highlighted the critical role of magnesium in inflammation. In comparison, Nicholson, Tucker and Brameld [5] found that around 6% of the genome was differentially expressed when HUVECs were treated with three polyphenolic compounds, ferulic acid, quercetin and resveratrol. In contrast, Chen, et al. [32] found that only 1.5% of the HUVECs genome was impacted by E coli derived LPS. Bal, et al. [33] stimulated HUVECs with different interleukins and found that stimulation of HUVECs with IL-1β changed 1% of the gene profile, while IL-3 affected 1.4% and IL-6 changed around 1.5% of the HUVECs genome.
The analysis showed that varying the magnesium concentration in culture medium had effects across all cellular components, including the cytosol, nucleus and other organelles. Given the large numbers of transcripts responding in the experiment and the fundamental role of magnesium in processes such as binding to ATP in enzyme catalysed reactions, synthesis of nucleic acids and stabilizing membranes, this finding was perhaps unsurprising. The impact upon such basic processes will have fed into other aspects of the response to magnesium as categorised as ‘cellular processes’ and ‘cellular organisation’ by the GO analysis.
Our previous work [30] and a robust body of literature [15,17,34] demonstrate that magnesium deficiency is detrimental to the function of HUVECs in culture, in particular reducing cell proliferation, increasing cell death, and exacerbating the response to inflammatory insults. The findings of the current study are consistent with such observations as they suggest perturbation of inflammatory pathways and apoptosis. The phenotype that we have previously shown to be associated with high magnesium under inflammatory conditions includes a dampened down inflammatory response, reduced cell death, and suppression of adhesion to monocytes [30]. It was apparent from the current experiment that modulation of intracellular signaling, apoptosis, binding processes, cell adhesion, and metabolic activities was widespread even in the absence of inflammatory challenge. These responses can all be envisaged as contributing to such a phenotype. The clear lack of dose-responses to magnesium was an interesting aspect of the study. Only 34 transcripts that responded to magnesium showed evidence of differential responses with magnesium deficiency and excess. These did not cluster in any process or pathway. With almost 20% of the transciptomic response to high magnesium being the same as with magnesium deficiency, it is clear that excess magnesium could be detrimental to HUVEC function.
The key starting point for this study was that magnesium regulates endothelial cell function and modulates the response to an inflammatory challenge [4,28,35]. Analysis of the array results demonstrated that mRNA expression for many of the interleukin genes, such as IL8, IL5, IL33, IL6R, IL1RL1, and IL31RA, and chemokine genes, such as CXCL6 (GCP-2), CXCL1 (GROα) and CXCL2 (GROβ), was down-regulated by high magnesium treatment, consistent with the hypothesis that maintaining magnesium concentrations suppresses the inflammatory response in HUVECs. In contrast, and at variance with our previous finding that expression of cytokine proteins such as IL-8, MCP-1, GRO, GROα, IL-2 and IL-3 was enhanced with magnesium deficiency [14], the array analysis found little impact of low magnesium on interleukins and their associated signaling pathway. This inconsistency may be because some effects of magnesium deficiency are mediated at the level of translation rather than transcription. The microarray data and associated GO analysis has clearly indicated that magnesium is a potent modulator of gene expression in HUVECs and that inflammatory processes are among those which are responsive to variation in magnesium concentration.
The heat map for the 615 transcripts that were commonly differentially expressed in both high and low magnesium treatment showed that the physiological magnesium concentration group matrix clustered differently between the high and low groups and that, surprisingly, the majority of these transcripts responded in the same manner in both conditions. Further analysis of these transcripts found that 13% of these genes were related to the cytoskeleton, and 17% and 52% were involved in anion and protein binding, respectively. Magnesium plays a role in cytoskeleton formation during the mitotic spindle and cytokinesis [36], and the present data suggest that deviation away from optimal concentration (either deficiency or excess) can disrupt this process. Moreover, any variation in the magnesium concentration can affect the other cell ions, such as Ca2+, Na+, and K+ [36,37]. The efflux of Ca2+ can be influenced by the concentration of Mg2+ on the membrane binding sites because the magnesium ion is a Ca2+ antagonist. In addition, K+ transport is controlled by Mg2+ through the Na+-K+ ATPase pump. Further, extracellular magnesium can impact the ionized and protein bonds [38].
With 20% of all of the transcripts that responded to magnesium showing the same response in both low and high magnesium conditions, it appears that some of the effects of magnesium may be indirect and related to other factors. Magnesium shares key transporters with calcium, particularly TRPM6 and TRMP7 [39]. The latter is also a zinc transporter. The effects of low and high magnesium concentrations on the overall intracellular ion balance of the cell was not evaluated in this study, but would certainly be of interest. We noted that key components of the apparatus which enables calcium to regulate gene expression were perturbed by both low and high magnesium. Calcium binding to calmodulin (expressed in our study) and calcineurin activates the nuclear factor of activated T-cells family of transcription factors (NFATs-differentially regulated in the current study) leading to transcription. The ryanodine receptors (Ryr1 and Ryr2 responded to magnesium concentration in this study) regulate the release of calcium from intracellular stores enabling calmodulin to activate CREB sites on DNA.
It is important to acknowledge that this was an exploratory study that has been useful in generating hypotheses for further studies and demonstrating the complexity of the cellular response to variation in magnesium concentrations. We acknowledge that changes at the transcriptomic level do not necessarily reflect changes at the protein or functional level. We have shown elsewhere that the differential mRNA expression described in the inflammatory pathways reported in this paper are matched by differential expression (to a similar degree) in the protein expression of interleukins (IL-2, IL-3, IL-8, IL-15) and MCP-1 [14,30]. This confirms observations in the current work. The diverse range of pathways and processes that were responsive to magnesium was surprising, particularly given the acute challenge, although the magnitude of the expression changes was relatively small in most cases. This is not unusual in studies where nutrients are the stimulus or insults applied to cells and tissues. It is unclear whether the same differences in expression associated with low and/or high magnesium in culture would be present in the presence of an inflammatory insult and this requires further investigation either using a similar whole genome or a more targeted approach. Certainly the finding that IL-1β for example was down-regulated in conditions of low magnesium, differs from previous reports that low magnesium alone results in up-regulation [40,41]. Our data is consistent with the view that acute variation in magnesium concentration establishes the conditions for an altered response to inflammatory challenge [30,32] and we have previously reported that the combined insult may differ from the effects of varying magnesium concentration alone [30].
5. Conclusions
This study has shown that the expression of transcripts in HUVECs is extremely sensitive to the concentration of magnesium in culture. Considered in the context of observations that magnesium deficiency is associated with an exaggerated inflammatory response in this cell type, whilst high magnesium has a protective effect, it can be inferred that fluctuations in magnesium concentration are sensed within cells and modulate gene expression in diverse pathways. The data suggests that there are a number of responses to magnesium that may be indirectly mediated through variation in intracellular concentrations of other ions. However, magnesium should be recognised as an important factor in determining inflammatory responses in endothelial cells, and this may explain reports of associations between poor magnesium status and cardiovascular disease in humans and animals.
Acknowledgments
The authors extend thanks to Princess Nourah Bint Abdulrahman University Researchers Supporting Project number (PNURSP2022R129), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia for funding this work.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/nu14173586/s1, Supplementary Table S1: Transcripts showing differential expression in response to high magnesium concentration; Supplementary Table S2: 615 transcripts showing differential expression to both low- and high-magnesium concentrations.
Author Contributions
Conceptualization, L.A.A., A.M.S. and S.C.L.-E.; Data curation, L.A.A.; Formal analysis, M.C. and S.T.M.; Funding acquisition, L.A.A.; Project administration, A.M.S. and S.C.L.-E.; Software, M.C. and S.T.M.; Supervision, A.M.S. and S.C.L.-E.; Validation, L.A.A., A.M.S. and S.C.L.-E.; Visualization, A.M.S. and S.C.L.-E.; Writing—original draft, L.A.A.; Writing—review & editing, S.C.L.-E. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by Princess Nourah Bint Abdulrahman University Researchers Supporting Project number (PNURSP2022R129), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bouïs D., Hospers G., Meijer C., Molema G., Mulder N. Endothelium In Vitro: A review of human vascular endothelial cell lines for blood vessel-related research. Angiogenesis. 2001;4:91–102. doi: 10.1023/A:1012259529167. [DOI] [PubMed] [Google Scholar]
- 2.Deanfield J.E., Halcox J.P., Rabelink T.J. Endothelial function and dysfunction: Testing and clinical relevance. Circulation. 2007;115:1285–1295. doi: 10.1161/CIRCULATIONAHA.106.652859. [DOI] [PubMed] [Google Scholar]
- 3.Zhang J., DeFelice A., Hanig J., Colatsky T. Biomarkers of endothelial cell activation serve as potential surrogate markers for drug-induced vascular injury. Toxicol. Pathol. 2010;38:856–871. doi: 10.1177/0192623310378866. [DOI] [PubMed] [Google Scholar]
- 4.Bernardini D., Nasulewic A., Mazur A., Maier J.A. Magnesium and microvascular endothelial cells: A role in inflammation and angiogenesis. Front. Biosci. J. Virtual Libr. 2005;10:1177–1182. doi: 10.2741/1610. [DOI] [PubMed] [Google Scholar]
- 5.Nicholson S., Tucker G., Brameld J. Effects of dietary polyphenols on gene expression in human vascular endothelial cells. Proc. Nutr. Soc. 2008;67:42–47. doi: 10.1017/S0029665108006009. [DOI] [PubMed] [Google Scholar]
- 6.Koltsova S., Trushina Y., Haloui M., Akimova O.A., Tremblay J., Hamet P., Orlov S.N. Ubiquitous [Na+] i/[K+] i-sensitive transcriptome in mammalian cells: Evidence for Ca2+ i-independent excitation-transcription coupling. PLoS ONE. 2012;7:e38032. doi: 10.1371/journal.pone.0038032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ambra R., Manca S., Palumbo M., Leoni G., Natarelli L., De Marco A., Consoli A., Pandolfi A., Virgili F. Transcriptome analysis of human primary endothelial cells (HUVEC) from umbilical cords of gestational diabetic mothers reveals candidate sites for an epigenetic modulation of specific gene expression. Genomics. 2014;103:337–348. doi: 10.1016/j.ygeno.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 8.Chakraborti S., Chakraborti T., Mandal M., Mandal A., Das S., Ghosh S. Protective role of magnesium in cardiovascular diseases: A review. Mol. Cell. Biochem. 2002;238:163–179. doi: 10.1023/A:1019998702946. [DOI] [PubMed] [Google Scholar]
- 9.Whitton C., Nicholson S.K., Roberts C., Prynne C.J., Pot G., Olson A., Fitt E., Cole D., Teucher B., Bates B. National Diet and Nutrition Survey: UK food consumption and nutrient intakes from the first year of the rolling programme and comparisons with previous surveys. Br. J. Nutr. 2011;106:1899. doi: 10.1017/S0007114511002340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swaminathan R. Magnesium metabolism and its disorders. Clin. Biochem. Rev. 2003;24:47. [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang L., He P., Chen J., Liu Y., Liu D., Qin G., Tan N. Magnesium levels in drinking water and coronary heart disease mortality risk: A meta-analysis. Nutrients. 2016;8:5. doi: 10.3390/nu8010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maier J. Low magnesium and atherosclerosis: An evidence-based link. Mol. Asp. Med. 2003;24:137–146. doi: 10.1016/S0098-2997(02)00095-X. [DOI] [PubMed] [Google Scholar]
- 13.De Baaij J., Hoenderop J., Bindels R. Magnesium in man: Implications for health and disease. Physiol. Rev. 2015;95:1–46. doi: 10.1152/physrev.00012.2014. [DOI] [PubMed] [Google Scholar]
- 14.Almousa L., Salter A., Langley-Evans S. Magnesium deficiency heightens lipopolysaccharide-induced inflammation and enhances monocyte adhesion in human umbilical vein endothelial cells. Magnes. Res. 2018;31:39–48. doi: 10.1684/mrh.2018.0436. [DOI] [PubMed] [Google Scholar]
- 15.Banai S., Haggroth L., Epstein S., Casscells W. Influence of extracellular magnesium on capillary endothelial cell proliferation and migration. Circ. Res. 1990;67:645–650. doi: 10.1161/01.RES.67.3.645. [DOI] [PubMed] [Google Scholar]
- 16.Ferrè S., Baldoli E., Leidi M., Maier J. Magnesium deficiency promotes a pro-atherogenic phenotype in cultured human endothelial cells via activation of NFkB. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2010;1802:952–958. doi: 10.1016/j.bbadis.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 17.Maier J. Endothelial cells and magnesium: Implications in atherosclerosis. Clin. Sci. 2012;122:397–407. doi: 10.1042/CS20110506. [DOI] [PubMed] [Google Scholar]
- 18.Guo F., Tang J., Zhou Z., Dou Y., Van Lonkhuyzen D., Gao C., Huan J. GEF-H1-RhoA signaling pathway mediates LPS-induced NF-κB transactivation and IL-8 synthesis in endothelial cells. Mol. Immunol. 2012;50:98–107. doi: 10.1016/j.molimm.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 19.Hoffmann E., Dittrich-Breiholz O., Holtmann H., Kracht M. Multiple control of interleukin-8 gene expression. J. Leukoc. Biol. 2002;72:847–855. [PubMed] [Google Scholar]
- 20.Berg A., Baldwin A. The Ikb proteins: Multifunctional regulators of rel/NFk-B transcription. Genes Dev. 1993;7:2064–2070. doi: 10.1101/gad.7.11.2064. [DOI] [PubMed] [Google Scholar]
- 21.Li A., Varney M.L., Valasek J., Godfrey M., Dave B.J., Singh R.K. Autocrine role of interleukin-8 in induction of endothelial cell proliferation, survival, migration and MMP-2 production and angiogenesis. Angiogenesis. 2005;8:63–71. doi: 10.1007/s10456-005-5208-4. [DOI] [PubMed] [Google Scholar]
- 22.Baggiolini M., Loetscher P., Moser B. Interleukin-8 and the chemokine family. Int. J. Immunopharmacol. 1995;17:103–108. doi: 10.1016/0192-0561(94)00088-6. [DOI] [PubMed] [Google Scholar]
- 23.Lukacs N.W., Strieter R., Elner V., Evanoff H., Burdick M., Kunkel S. Production of chemokines, interleukin-8 and monocyte chemoattractant protein-1, during monocyte: Endothelial cell interactions. Blood. 1995;86:2767–2773. doi: 10.1182/blood.V86.7.2767.2767. [DOI] [PubMed] [Google Scholar]
- 24.Romano M., Sironi M., Toniatti C., Polentarutti N., Fruscella P., Ghezzi P., Faggioni R., Luini W., Van Hinsbergh V., Sozzani S., et al. Role of IL-6 and its soluble receptor in induction of chemokines and leukocyte recruitment. Immunity. 1997;6:315–325. doi: 10.1016/S1074-7613(00)80334-9. [DOI] [PubMed] [Google Scholar]
- 25.Ferrè S., Mazur A., Maier J. Low-magnesium induces senescent features in cultured human endothelial cells. Magnes. Res. 2007;20:66–71. [PubMed] [Google Scholar]
- 26.Maier J., MalpuechBrugère C., Zimowska W., Rayssiguier Y., Mazur A. Low magnesium promotes endothelial cell dysfunction: Implications for atherosclerosis, inflammation and thrombosis. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2004;1689:13–21. doi: 10.1016/j.bbadis.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 27.Mi H., Muruganujan A., Casagrande J., Thomas P. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013;8:1551. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mazur A., Maier J., Rock E., Gueux E., Nowacki W., Rayssiguier Y. Magnesium and the inflammatory response: Potential physiopathological implications. Arch. Biochem. Biophys. 2007;458:48–56. doi: 10.1016/j.abb.2006.03.031. [DOI] [PubMed] [Google Scholar]
- 29.Shechter M., Sherer Y. Endothelial dysfunction: A crystal ball prediction for enhanced cardiovascular risk? IMAJ. 2003;5:736–738. [PubMed] [Google Scholar]
- 30.Almousa L., Salter A., Langley-Evans S. The effect of varying concentrations of magnesium on expression of human endothelia adhesion molecules. J. Nutr. Food Sci. 2016;6:53. [Google Scholar]
- 31.Martin H., Staedtler F., Lamboley C., Adrian M., Schumacher M., Chibout S.-D., Laurant P., Richert L., Berthelot A. Effects of long-term dietary intake of magnesium on rat liver transcriptome. Magnes. Res. 2007;20:259–265. [PubMed] [Google Scholar]
- 32.Chen C., Coats S., Bumgarner R., Darveau R. Hierarchical gene expression profiles of HUVEC stimulated by different lipid A structures obtained from Porphyromonas gingivalis and Escherichia coli. Cell. Microbiol. 2007;9:1028–1038. doi: 10.1111/j.1462-5822.2006.00849.x. [DOI] [PubMed] [Google Scholar]
- 33.Bal G., Kamhieh-Milz J., Futschik M., Häupl T., Salama A., Moldenhauer A. Transcriptional profiling of the hematopoietic support of interleukin-stimulated human umbilical vein endothelial cells (HUVECs) Cell Transplant. 2012;21:251–267. doi: 10.3727/096368911X580581. [DOI] [PubMed] [Google Scholar]
- 34.Wolf F., Trapani V., Cittadini A. Magnesium and the control of cell proliferation: Looking for a needle in a haystack. Magnes. Res. 2008;21:83–91. [PubMed] [Google Scholar]
- 35.Sugimoto J., Romani A., Valentin-Torres A., Luciano A., Kitchen C., Funderburg N., Mesiano S., Bernstein H.B. Magnesium decreases inflammatory cytokine production: A novel innate immunomodulatory mechanism. J. Immunol. 2012;188:6338–6346. doi: 10.4049/jimmunol.1101765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolf F., Trapani V. Cell (patho) physiology of magnesium. Clin. Sci. 2008;114:27–35. doi: 10.1042/CS20070129. [DOI] [PubMed] [Google Scholar]
- 37.Saris N.-E., Mervaala E., Karppanen H., Khawaja J., Lewenstam A. Magnesium: An update on physiological, clinical and analytical aspects. Clin. Chim. Acta. 2000;294:1–26. doi: 10.1016/S0009-8981(99)00258-2. [DOI] [PubMed] [Google Scholar]
- 38.Noronha L., Matuschak G. Magnesium in critical illness: Metabolism, assessment, and treatment. Intensive Care Med. 2002;28:667–679. doi: 10.1007/s00134-002-1281-y. [DOI] [PubMed] [Google Scholar]
- 39.Yee N., Kazi A., Yee R. Cellular and developmental biology of TRPM7 channel-kinase: Implicated roles in cancer. Cells. 2014;3:751–777. doi: 10.3390/cells3030751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nielsen F. Magnesium deficiency and increased inflammation: Current perspectives. J. Inflamm. Res. 2018;18:25–34. doi: 10.2147/JIR.S136742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Almousa L.A., Salter A.M., Langley-Evans S.C. Varying magnesium concentration elicits changes in inflammatory response in human umbilical vein endothelial cells (HUVECs) Magnes. Res. 2018;31:99–109. doi: 10.1684/mrh.2018.0439. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author upon reasonable request.