Abstract
In vivo expression technology was used to identify Salmonella enterica serovar Typhimurium genes that are transcriptionally induced when the bacteria colonize the small intestines of mice. These genes were subsequently screened for those that are transcriptionally inactive during the systemic stages of disease. This procedure identified gipA, a gene that is specifically induced in the small intestine of the animal. The gipA gene is carried on the lambdoid phage Gifsy-1. Consistent with the expression profile, the sole defect conferred by a gipA null mutation is in growth or survival in a Peyer's patch. The gipA strain is wild type in its ability to initially colonize the small intestine and invade the intestinal epithelium. The mutant also survives and propagates at wild-type levels during the systemic stages of disease. The gipA open reading frame is homologous to a family of putative insertion sequence elements, although our evidence shows that transposition is not required for gipA function in the Peyer's patch. These results suggest that the bacteria sense and respond to the particular environment of the Peyer's patch, a critical site for the replication of Salmonella serovar Typhimurium.
Salmonellae are invasive pathogens that cause a range of human diseases. Nontyphoid salmonellae usually cause gastroenteritis. Although this is often a self-limiting disease marked by diarrhea and abdominal cramps, the infection can be more severe, resulting in bacteremia, fever, or even death (36). After oral infection in mice, Salmonella spp. initially colonize the small intestine. The bacteria preferentially attach to and invade the M cells of the Peyer's patches (9, 26), mucosal lymphoid follicles that constitute the major site of antigen sampling and immune response in the small intestine (28). The bacteria gain access to the underlying lymphoid tissue and survive and multiply within the follicles. From this site, salmonellae eventually disseminate throughout the host, resulting in systemic infection (7, 26).
Analysis of the early steps of colonization and invasion by Salmonella enterica serovar Typhimurium using the BALB/c mouse model of infection and in vitro tissue culture models has significantly increased understanding of these processes (11). Three fimbriae have been implicated in the initial attachment of Salmonella serovar Typhimurium to the intestinal mucosa or Peyer's patch M cells: plasmid-encoded fimbriae (pef) (17), long polar fimbriae (lpf) (3, 4), and thin aggregative fimbriae (curli; agf) (47). Mutations that block the production of either Agf (48) or Lpf (4) increase the oral 50% lethal dose (LD50) three- to fivefold. These modest effects are apparently due to functional redundancy of the fimbriae; the oral LD50 of strains containing mutations in pef, lpf, agf, and fim, encoding type I fimbriae, is increased approximately 26-fold (48). After initial attachment, serovar Typhimurium uses the type III secretion system encoded on Salmonella pathogenicity island 1 (SPI1) to facilitate uptake by M cells (11, 19). The expression of the secretion system is controlled in response to a specific combination of environmental signals that presumably acts as a cue that the bacteria are in the appropriate anatomic location (2, 41, 44).
Although much is known about the mechanism by which Salmonella gains entry through M cells to the Peyer's patches (19), very little is understood about the growth of Salmonella in Peyer's patch tissue. The SipB protein, injected into the host cell via the SPI1 type III secretion system, can induce apoptosis in macrophages (22, 30, 37), presumably in the macrophages that initially engulf serovar Typhimurium in the Peyer's patches. Whether the SPI1 system is important throughout the growth of serovar Typhimurium in the Peyer's patches is not clear. Recent work suggests that diarrhea caused by Salmonella is an inflammatory response mediated by invasion of intestinal epithelial cells and subsequent infiltration of neutrophils (11). However, even in, for example, calves, where diarrhea is the major symptom of Salmonella infection, Peyer's patches are a primary site of invasion that may lead to systemic disease (18). Therefore, understanding the growth of the bacteria in this particular environment is vital to understanding Salmonella pathogenesis.
In this study, we used in vivo expression technology (IVET) to identify operons that are transcriptionally induced in the animal during the infection process (32, 45). IVET is based on the premise that many genes whose products are specifically required for the infection process will be regulated such that they are expressed only at the proper time and place in the host. Thus, this system was designed to identify promoters that are transcriptionally active in the mouse (in vivo) but not significantly transcribed on normal laboratory media (in vitro). In the pIVET1 system, random bacterial promoters are fused to a promoterless copy of the purA gene in a ΔpurA background. Serovar Typhimurium strains containing purA mutations are extremely attenuated at all stages of infection (13, 42). Therefore, only bacteria containing a purA fusion to a sufficiently active promoter can survive within host tissues. These bacteria are recovered from various host organs and screened for those that have low transcriptional activity in vitro by monitoring the expression of the lac operon located downstream of purA. Using this system, we have identified a locus that is specifically active during growth in the small intestine but is not significantly transcribed during the later, systemic stages of infection. We show here that this locus is carried on the Gifsy-1 bacteriophage and is important for the survival of serovar Typhimurium in the Peyer's patches of BALB/c mice.
MATERIALS AND METHODS
Bacterial strains and genetic manipulations.
The bacterial strains and plasmids used in this study are listed in Table 1. Unless otherwise indicated, bacteria were cultured at 37°C in Luria-Bertani (LB) medium (43) containing, when appropriate, ampicillin at 50 μg ml−1, kanamycin at 50 μg ml−1, chloramphenicol at 20 μg ml−1, or tetracycline at 25 μg ml−1. Strains containing ΔpurA were grown in medium supplemented with 27 μg of adenine ml−1 and 16 μg of thiamine ml−1 (AdB1). Lactose MacConkey (43) agar containing kanamycin, ampicillin, and AdB1 (lactose MacConkey/A/K/AdB1) was used to monitor lac expression.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype | Source or referencea |
|---|---|---|
| Strains | ||
| Salmonella serovar Typhimurium | ||
| 14028 | Wild type | ATCCb |
| JS120 | 14028 ΔpurA3141::kan | 25 |
| JS130 | 14028 zjg8103::pir | 25 |
| JS131 | JS120 constitutive pIVET1 fusion | |
| JS132 | JS120 pIVET1::gipA′ | |
| JS133 | 14028 zjg8101::kan pIVET1::gipA′ | |
| JS134 | 14028 gipA1::MudCm | |
| JS135 | 14028 zii8104::Tn10dTc | |
| JS136 | JS135 gipA2::MudJ | |
| JS137 | 14028 hilA339::kan | |
| JS200 | JS135 ΔGifsy-1::kan | |
| JS201 | JS135 gfoJ::MudJ | |
| JS202 | JS135 ΔgfoA::kan | |
| E. coli BW19851 | RP4-2-Tc::Mu-1 kan::Tn7 integrant/creB510 hsdR17 endA1 zbf-5 uid(ΔMluI)::pir recA1 thi | 35 |
| Plasmids | ||
| pWKS30 | pSC101 Apr | 50 |
| pJS302 | pWKS30 gipA | |
| pCtsPara-GBE | pSC101 ts ori gam bet exo | 12 |
| pKD4 | R6K ori FRT-kan-FRT | 12 |
This study, unless otherwise noted.
ATCC, American Type Culture Collection.
Kanamycin cassette insertion/deletions were constructed using a method developed by Datsenka and Wanner (12). PCR primers 60 nucleotides (nt) long were synthesized with 40 nt on the 5′ ends corresponding to the ends of the desired deletions. The 3′ 20 nt of each primer was annealed to the 5′ or 3′ end of an FRT-kan-FRT cassette in plasmid pKD4, where FRT is the site of Flp-mediated recombination. PCRs were carried out according to the Taq manufacturer's instructions (Life Technologies, Inc.). Plasmid pCtsPara-GBE was introduced into serovar Typhimurium strain 14028. This plasmid synthesizes the lambda recombination proteins Gam, Bet, and Exo when induced with arabinose. The plasmid-bearing strain was grown at 30°C in LB medium containing chloramphenicol and 1 mM arabinose and made electrocompetent (33). Approximately 800 ng of PCR product was transformed, and the cells were plated on LB medium containing kanamycin at 37°C. This procedure generally resulted in 30 to 50 Knr colonies. Each insertion/deletion was characterized by genetic mapping and PCR. The insertion/deletions were transduced via P22 to construct isogenic strains for subsequent analysis.
IVET library.
Chromosomal DNA was isolated from JS120 (ΔpurA) as described previously (43) and partially digested with Sau3AI to give 4- to 6-kb fragments, which were subsequently separated by size using a 10 to 40% sucrose density gradient (1, 43). These fragments were cloned into a BglII site directly 5′ of the promoterless purA-lacZY operon of pIVET1 (32, 45) and electroporated into Escherichia coli BW19851 (Pi+ Tra+). The Pi protein is required for replication of pIVET1 derivatives. Over 42,000 independent clones were obtained, sufficient to represent, with 99% confidence, a fusion to every kilobase of the chromosome in both orientations (10). This library was then mated with JS120 (ΔpurA), where the plasmid must integrate into the chromosome via homologous recombination with the cloned fragment in order to be stably maintained. The resulting exconjugants were pooled, and this library was used for the initial selection.
IVET selection.
The integrated pIVET1 library in JS120 was grown overnight in LB medium with ampicillin, kanamycin, and AdB1 (LB/A/K/AdB1). The culture was washed and resuspended in an equal volume of sterile 0.1 M sodium phosphate buffer (pH 8.0). Four 6-week-old female BALB/c mice were anesthetized with CO2 and inoculated orally with 0.2 ml of the culture (approximately 109 CFU). After 5 days, the mice were sacrificed by CO2 asphyxiation; their small intestines were harvested and homogenized in 2.0 ml of sterile 150 mM NaCl. The homogenate was used to inoculate a separate, fresh LB/A/K/AdB1 culture, which was grown overnight. This culture was used to orally inoculate naive mice for a second passage (four mice total) as described above. Five days after inoculation, the mice were sacrificed; their small intestines were harvested and homogenized as before. Serial dilutions were plated on lactose MacConkey/A/K/AdB1 and incubated overnight. Ninety colonies that displayed a Lac− phenotype were subsequently chosen for future analysis.
Cloning and sequencing of gipA.
The integrated pIVET1::gipA′ plasmid was recovered by P22 transduction (31) into the Pi-producing strain, JS130. Sequencing of the chromosomal region at the fusion joint was carried out at the W. M. Keck Center for Comparative and Functional Genomics at the University of Illinois using a primer for the 5′ end of purA. Further sequence data were obtained by cloning, via partial Sau3AI digestion, a Cmr insertional mutation located near the gipA open reading frame (ORF) into pWKS30. Restriction mapping was performed, and a suitable ClaI subclone of 1.8 kb that contained the entire gipA ORF (pJS302) was constructed. This clone was sequenced with primers specific for the plasmid and by primer walking. Sequence analyses were performed using the Wisconsin Package (Genetics Computer Group, Inc.).
Virulence studies and competition assays.
Six- to 8-week-old female BALB/c mice were inoculated either orally or intraperitoneally (i.p.) with 0.2 ml of a bacterial suspension diluted in sterile 0.1 M sodium phosphate buffer (oral) or sterile 150 mM NaCl (i.p.). After 4 to 7 days, the mice were sacrificed by CO2 asphyxiation; the ceca, small intestines, and/or spleens were recovered. These organs were homogenized in 2.0 ml of sterile 150 mM NaCl. Serial dilutions of the homogenates were plated on appropriate media to determine CFU per organ. Subsequent replica plating allowed us to determine the percentages of wild-type and mutant cells.
For Peyer's patch quantitation, mice were inoculated orally and sacrificed after 6 days. The intestines were removed, and the lumen was washed twice with 10 ml of sterile 150 mM NaCl using a syringe. The Peyer's patches were individually dissected and homogenized in 2.0 ml of 150 mM NaCl. Appropriate dilutions were plated on selective medium. Subsequent replica plating allowed us to determine the percentages of wild-type and mutant cells.
The LD50 was calculated according to the protocol of Reed and Muench (40) using groups of six mice for each dose. The competitive index (CI) was defined as the output ratio (mutant/wild type) divided by the input ratio (mutant/wild type). Student's t test was used for statistical analysis.
IOC.
Intestinal organ culturing (IOC) was performed essentially as described previously (4). Briefly, the small intestine was removed from a mouse and incubated in tissue culture medium in a petri dish. The intestine was washed with 10 ml of 150 mM NaCl, the ends were tied, and the intestine was filled with 107 CFU of an approximately 1:1 mixture of mutant and wild-type bacteria. The intestine was incubated for 1.0 h at 37°C in 5% CO2 to allow for attachment and invasion. The intestine was then washed extensively, and the most distal Peyer's patch was dissected. This Peyer's patch and the remaining small intestinal tissue were separately homogenized in 2.0 ml 150 mM NaCl, and dilutions were plated for CFU determinations. Subsequent replica plating allowed us to determine the ratio of wild-type to mutant recovered bacteria. The mean and standard deviation of the CI for five assays are reported.
Invasion of Henle-407 cultured epithelial cells.
Henle-407 epithelial cells were obtained from the American Type Culture Collection and propagated according to supplier instructions. Invasion assays were performed as described previously (39). Bacterial cultures were grown under invasion conditions, and a 1:1 mixture of wild-type serovar Typhimurium and either the gipA mutant or a noninvasive hilA mutant was used as an inoculum. After lysis of the eukaryotic cells, dilutions were plated on the appropriate selective media. Subsequent replica plating allowed us to determine the percentages of wild-type and mutant cells. The mean and standard deviation of the CI for three assays are reported.
Isolation of chromosomal DNA and Southern blot analysis.
Chromosomal DNA from colonies of 14028 isolated from various tissues of an infected animal was isolated using the Qiagen chromosomal preparation protocol (Qiagen Inc.). Samples were digested with ClaI and run on a 1% agarose gel. The DNA was subsequently transferred to a nitrocellulose membrane as previously described (1). The probe, plasmid pJS302, was labeled (along with the parent plasmid containing no chromosomal insert), and hybridization was performed using a nonradioactive labeling and detection kit from DuPont NEN. Hybrids were detected using SuperSignal detection reagents (Pierce).
In vitro analysis of the regulation of gipA.
Expression of the IVET fusion in vitro was determined following overnight growth under various environmental conditions. Previous experiments had shown that growth phase has no effect on the expression of gipA. Iron starvation assays were performed with LB medium in the presence of 100 mM diethylenetriamine-pentaacetic acid, an extracellular iron chelator. This concentration was determined empirically to cause a reasonable amount of growth inhibition in strain 14028. Microtiter β-galactosidase assays were performed as previously described (46).
Nucleotide sequence accession number.
The gipA sequence has been entered into GenBank under accession number AF246666.
RESULTS
Rationale and identification of gipA.
We have used the IVET system (32) to identify serovar Typhimurium genes that are induced while the bacterium is in the mouse (45; data not shown). For IVET selection, a strain must contain a fusion of purA to a gene that is transcriptionally active in the animal. For the purposes of this study, we concentrated on genes that were identified as being transcriptionally induced in the small intestine. We reasoned that a subset of these genes would include those whose products are specifically required for some aspect of the early infection process. This subset of genes would be expressed during the early stages but not during the systemic stages of disease. Accordingly, we screened for fusions that were induced only in the small intestine and not in the spleen. To accomplish this, we performed a competition assay with a bacterial strain containing an IVET fusion to an in vivo-induced gene of interest and JS131, a strain containing a pIVET1 fusion to a promoter that is sufficiently active to allow for the survival of the strain in any host tissue (data not shown). Mice were inoculated either orally or i.p. with an equal mixture of the two strains. After 3 to 5 days, the mice were sacrificed; the small intestines were removed from the orally infected animals, while the spleens were removed from the i.p. infected animals. We screened for fusion strains that were recovered from the small intestines after oral infection in numbers equal to or larger than those of the constitutive fusion strain but that were recovered in relatively small numbers from the spleen after i.p. infection. Of the 10 orally induced fusions that we tested, one clearly met the screening requirements. We named the corresponding gene gipA (for growth in Peyer's patches).
In order to confirm the induction of gipA in the small intestine, we repeated the competition assay. After oral inoculation, the gipA fusion strain invariably outcompeted JS131 in the small intestine, with a median CI of 3.27 (Table 2). In contrast, after i.p. inoculation, the gipA fusion strain was significantly outcompeted by JS131 (median CI, 0.07) (Table 2). These results confirm that the gipA gene is regulated such that it is specifically induced only in the early stages of infection.
TABLE 2.
Competition assay with pIVET1 fusion-containing strains JS131 (constitutive) and JS132 (gipA)
| Inoculum | Mouse | Organ | Total CFU recovered | CIa |
|---|---|---|---|---|
| Oral | A | Small intestine | 3.7 × 103 | 1.44 |
| B | 2.7 × 103 | 4.51 | ||
| C | 2.4 × 103 | 1.66 | ||
| D | 2.2 × 103 | 3.27 | ||
| E | 8.8 × 102 | 4.15 | ||
| i.p. | F | Spleen | 4.8 × 106 | 0.09 |
| G | 3.8 × 107 | 0.03 | ||
| H | 9.6 × 106 | 0.07 | ||
| I | 6.2 × 106 | 0.07 | ||
| J | 1.8 × 106 | 0.13 |
Defined as the output ratio (JS131/JS132) divided by the input ratio (JS131/JS132). Results for oral and i.p. inocula are statistically different (P < 0.0005).
Molecular characterization of gipA.
The pIVET1::gipA′ plasmid was recovered from the fusion strain via P22 transduction into a Pi-producing strain of serovar Typhimurium, where the plasmid can recircularize and replicate autonomously (31). The portion of the chromosomal insert closest to the fusion joint was sequenced using a primer that hybridized to the 5′ end of the purA gene in pIVET1. The sequence data indicated that the fusion joint was 143 bp from the 5′ end of an ORF. In order to clone the entire gipA locus, an MudCm insertion mutation was isolated in the chromosomal insert of the pIVET1::gipA′ plasmid and recombined into the chromosome by P22 transduction (34) to yield JS134. Chromosomal DNA fragments containing the Cmr marker were cloned into the low-copy-number pSC101 derivative pWKS30 (50). Restriction analysis and subcloning allowed the construction of pJS302, which contains a ClaI fragment with approximately 400 bp 5′ and 200 bp 3′ of the gipA ORF.
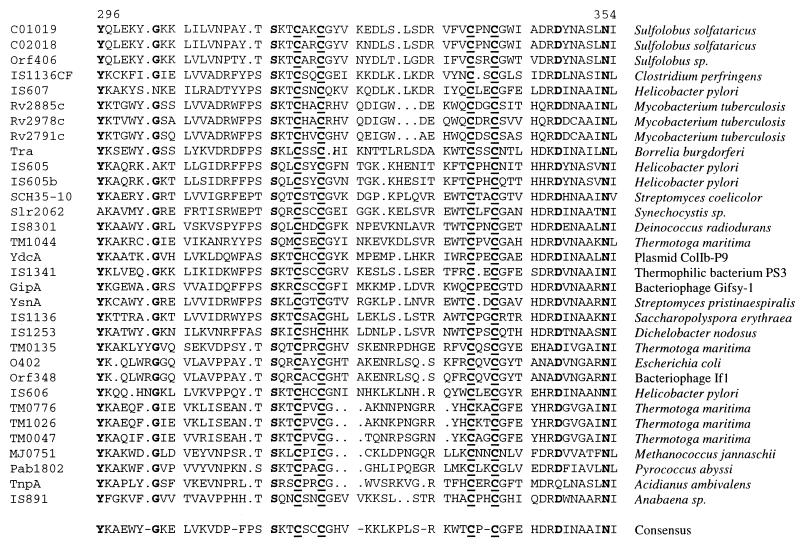
The complete DNA sequence of the gipA ORF and flanking regions was obtained. Analysis of this sequence suggested that gipA consists of a single ORF that shares significant homology with a group of IS891-like insertion elements found in a diverse group of organisms. Near the carboxy terminus of the predicted 411-amino-acid protein lies a cluster of highly conserved amino acids that include four completely conserved Cys residues (Fig. 1). The gipA ORF is located in the late operon of the Gifsy-1 bacteriophage between gfoZ (named gfo for Gifsy one) and gfoU, the orthologs of the phage lambda Z and U genes. However, the gipA ORF is in the opposite orientation with respect to these potential phage genes, both of which are intact.
FIG. 1.
Alignment of the carboxy-terminal region of GipA and homologs. Amino acids 296 to 354 of the 411-amino-acid GipA protein are shown. This region corresponds to the most highly conserved region of the protein. Amino acids that are identical in at least 30 of 32 proteins are shown in bold. Underlining indicates four completely conserved Cys residues. The designation of each putative protein is shown on the left, and the source is shown on the right. The accession numbers for the protein sequences, in order of appearance, are as follows: C01019, CAA69469; C02018, CAA69450; Orf406, CAA09118; IS1136CF, CAA60219; IS607, AF189015; Rv2885c, Q10809; Rv2978c, CAB05434; Rv2791c, CAA15586; Tra, AAC97568; IS605, AAC44690; IS605b, AAD08043; SCH35-10, CAB44417; Slr2062, BAA17870; IS8301, BAA32390; TM1044, AAD36121; YdcA, BAA75131; IS1341, JC4292; GipA, AF246666; YsnA, P54992; IS1136, JN0563; IS1253, AAB16749; TM0135, AE001699; O402, AAC74514; Orf348, AAC62159; IS606, AAD06403; TM0776, AAD35858; TM1026, AAD36103; TM0047, AAD35141; MJ0751, Q58161; Pab1802, CAB49750; TnpA, CAB56750; and IS891, AAA90935.
Phenotypes of a gipA null mutation.
The data presented above suggest that the gipA locus is specifically induced in the small intestine. Accordingly, we predicted that a strain containing a null mutation in gipA would be attenuated for survival after oral inoculation but would be unaffected for survival during a strictly systemic infection. To test this hypothesis, an MudJ insertion mutation was isolated within the ORF of gipA. The location of the insertion in the gipA ORF was confirmed by PCR.
To test the role of gipA in pathogenesis, we performed oral and i.p. competition assays between the gipA2::MudJ insertion mutant and an isogenic wild-type strain. The results, shown in Table 3, indicate that the gipA mutant was outcompeted more than twofold in the small intestine. Although this phenotype was subtle, it was very reproducible and statistically significant. In contrast, no defect was conferred by the gipA insertion mutation when the bacteria were injected i.p. Indeed, the gipA mutant outcompeted the wild-type strain, although this result was not statistically significant. Thus, consistent with the expression profile, the gipA insertion mutation confers a virulence defect in an oral but not an i.p. infection.
TABLE 3.
In vivo competition assays with wild-type serovar Typhimurium and isogenic mutants
| Mutant | Route of inoculationa | Median CIb | No. of mice | Pc |
|---|---|---|---|---|
| gipA2::MudJ | Oral | 0.45 | 16 | <0.05 |
| i.p. | 2.32 | 9 | NS | |
| Oral (Peyer's patch)d | 0.10 | 5 | <0.0005 | |
| gipA2::MudJ/pJS203 | Oral (Peyer's patch)d | 0.59 | 5 | NS |
| gfoJ::MudJ | Oral | 1.5 | 5 | NS |
| ΔgfoA::kan | Oral | 1.3 | 5 | NS |
| ΔGifsy-1::kan | Oral | 0.28 | 12 | <0.00005 |
All inocula contained a 1:1 ratio of mutant to isogenic wild-type serovar Typhimurium (JS135). Doses given to BALB/c mice were 102 CFU i.p. and 108 CFU orally. Bacteria were recovered from the small intestine after oral infection or the spleen after i.p. infection unless otherwise indicated.
Defined as output (mutant/wild type) divided by input (mutant/wild type).
Student's t test was used to compare output versus inoculum. NS, not significant.
Bacteria were recovered from individual dissected Peyer's patches after oral infection.
Mutations in Gifsy-1 structural genes do not confer a virulence phenotype.
To ensure that the defect was not due to some effect on Gifsy-1 phage production, we isolated a MudJ insertion in the putative tail fiber gene gfoJ. An isogenic strain containing the gfoJ::MudJ insertion competed equally with the wild type after oral inoculation (Table 3). In addition, we constructed a deletion/kanamycin insertion in gfoA. Again, the isogenic mutant competed equally well with the wild type after oral inoculation (Table 3). Thus, insertions or deletions in the late tail operon of Gifsy-1 do not confer a virulence defect. Thus, the insertion in gipA confers a specific defect.
Finally, we constructed a deletion/kanamycin insertion of the entire Gifsy-1 phage. In oral competition experiments, the Gifsy-1 deletion strain showed a sevenfold-decreased ability to grow or survive in the small intestine (Table 3). As has previously been reported (14), we found no defect conferred by the loss of Gifsy-1 after i.p. infection (data not shown). Thus, the loss of Gifsy-1 confers a phenotype essentially identical to that of the gipA2::MudJ insertion.
The gipA null mutation decreases growth or survival in the Peyer's patches.
Previous studies (23) and observations from our own work indicated that, several days after an oral infection, most of the serovar Typhimurium found in the small intestine of mice is associated with the Peyer's patches. We reasoned that IVET selection would target genes that were transcriptionally active in the Peyer's patches because significant growth is required for selection. In order to compare the survival of the mutant and the wild type in the Peyer's patches, an oral competition assay was performed by inoculating a 1:1 mixture of the gipA2::MudJ mutant (JS136) and an isogenic wild-type strain (JS135). At the dose used for this analysis (∼107 CFU), any given Peyer's patch is likely to be colonized by only a single bacterium, resulting in a largely clonal population within a single patch. This phenomenon can be exploited to compare the growth of different bacterial strains within the same mouse. Three to five days after infection, individual Peyer's patches were excised, homogenized, and plated for total CFU determinations. Subsequent replica plating allowed us to determine the percentages of wild-type and gipA2::MudJ strains. The data indicated that the wild-type and mutant strains invaded and colonized Peyer's patches equally well (Table 4). However, the total number of CFU was consistently lower in Peyer's patches colonized by the mutant than in patches predominantly colonized by the wild type (Table 4). On average, the CFU of the mutant in a single colonized Peyer's patch was 5- to 10-fold lower than that in a Peyer's patch colonized by the wild type. These data suggested that the gipA product participates in some function required for survival and/or replication in the Peyer's patch.
TABLE 4.
Competition between wild type and the gipA mutant in individual Peyer's patches at an inoculum of 107 CFUa
| Peyer's patch | Total CFU recovered | % gipA mutant recovered |
|---|---|---|
| A | 8.4 × 104 | 0.1 |
| B | 1.4 × 104 | 0.8 |
| C | 3.4 × 104 | 5 |
| D | 1.5 × 103 | 72 |
| E | 2.2 × 103 | 84 |
| F | 3.2 × 103 | 88 |
| G | 8.1 × 103 | 100 |
The strains used were JS135 and JS136.
To test the above hypothesis further, we inoculated a second group of mice using a 10-fold-larger inoculum, thus increasing the probability that each Peyer's patch would be colonized by both strains. In this manner, we could more easily compare the growth rates of the strains within a single Peyer's patch. We explicitly isolated the most distal Peyer's patch (7) in each of five mice 6 days postinfection and determined that the wild-type strain consistently out-competed the gipA mutant strain by approximately 10-fold (Table 3). Thus, the gipA mutation confers a defect in growth or survival in the Peyer's patches.
We performed an identical experiment with a gipA2::MudJ strain containing the gipA+ plasmid pJS302. The results (Table 3) show that the plasmid did complement the Peyer's patch defect, but perhaps not to wild-type virulence. However, the remaining defect in the plasmid-bearing strain was not statistically significant. These data suggest that the Peyer's patch defect conferred by the gipA2::MudJ mutation is due to a functional loss of GipA. This conclusion is further supported by several data presented above. First, gipA is a single ORF that is transcribed in the orientation opposite that of all the surrounding genes. Hence, a polar effect of the MudJ insertion is unlikely. Second, other mutations in the late operon, including an MudJ insertion, do not confer a virulence defect. Third, the Gifsy-1 deletion confers the same phenotype as the insertion mutation. Our data suggest that gipA is tightly regulated such that it is expressed only at the appropriate time. The fact that the plasmid-bearing strain did not behave exactly like the wild-type strain is perhaps due to inappropriate expression of gipA from the plasmid.
Mutations in gipA do not affect attachment to or invasion of the small intestine or growth in the cecum.
The data presented in Table 4 show that equal numbers of randomly chosen Peyer's patches were colonized with the gipA mutant and the wild type, suggesting that mutations in gipA do not confer an attachment or invasion defect. To further examine this possibility, we quantified attachment and invasion of the gipA mutant with respect to the wild type using the IOC technique developed by Bäumler et al. (4). Ex vivo small intestines were inoculated with an equal mixture of the wild type (JS135) and the gipA2::MudJ mutant (JS136). After a 1-h incubation period, the gipA mutant and the wild type were recovered equally from both the distal Peyer's patch (CI = 1.02 ± 0.09; n = 5) and the small intestine (CI = 1.11 ± 0.08; n = 5). This result suggests that the gipA mutant is unaffected in its ability to initially attach to and invade the Peyer's patches and small intestine.
To confirm that gipA does not play a role in attachment to or invasion of intestinal epithelial cells, we performed in vitro invasion assays with Henle-407 cultured epithelial cells (39) using a hilA mutant as a negative control (2). After incubation of eukaryotic cells with a 1:1 mixture of wild-type and mutant bacteria, the gipA2::MudJ strain was equal to the wild-type strain in its ability to invade the eukaryotic cells (CI = 0.88 ± 0.26; n = 3). In contrast, the hilA strain was significantly outcompeted in control experiments (CI = 0.08 ± 0.07; n = 3). Thus, the gipA mutation does not affect attachment or invasion of intestinal epithelial cells in tissue cultures.
Finally, we tested if the gipA insertion mutation had any effect on the ability of serovar Typhimurium to colonize or propagate in the cecum, which is devoid of Peyer's patches. A 1:1 mixture of the gipA2::MudJ mutant strain (JS136) and an isogenic wild-type strain (JS135) was used to orally infect a group of six mice. The mutant was recovered from the ceca in numbers equal to those of the wild type (CI = 0.95 ± 0.70; n = 6). These same mice did have a significantly lower number of mutant bacteria in the small intestine, and these data are included in those for the group of orally infected mice in Table 3. Taken together, the above results suggest that the gipA gene product is specifically involved in growth or survival in Peyer's patches and does not play a role in attachment to or invasion of the small intestine or growth in other areas of the intestine.
The gipA mutation does not confer a general growth defect.
In order to test if the Peyer's patch phenotype could be accounted for by a simple growth defect, we performed competition assays with the gipA2::MudJ mutant (JS136) and the wild type (JS135) during growth in both rich and minimal media starting with an inoculum of approximately 200 cells and growing to saturation. The gipA strain showed no growth defect under these conditions (CI = 1.06 ± 0.19 for rich and 1.08 ± 0.08 for minimal; n = 10), suggesting that the defect in the Peyer's patch is not the result of a simple growth defect in the mutant.
A gipA mutation increases the oral LD50.
To further quantify the effects of the gipA insertion mutation, we determined the oral and i.p. LD50s of our wild-type strain and the isogenic mutant strain. The oral LD50 of the wild type in this study was similar to previously reported values (1.3 × 104 organisms). The oral LD50 of the gipA2::MudJ mutant was 6.6 × 104 organisms. Thus, the gipA insertion resulted in an approximately fivefold attenuation after oral inoculation. In contrast, the LD50 did not differ between the mutant and wild-type strains following i.p. inoculation. All mice inoculated with either the gipA mutant or the wild type died following i.p. inoculation of fewer than five organisms. These results are consistent with the expression profile and the previous data suggesting that gipA is specifically involved in an early stage of infection.
Transposition is not required for gipA function in the Peyer's patches.
The sequence of the gipA ORF shows homology to those of a family of putative transposases. To determine whether this locus actively transposes during an infection, we isolated wild-type serovar Typhimurium from various sites of infection 6 days after oral inoculation. Bacteria were recovered from three independent Peyer's patches, the remaining small intestine, and the spleen by homogenizing the organs and plating for single colonies. Two or three individual colonies were chosen to represent each tissue, and chromosomal DNA was extracted from the isolates. This chromosomal DNA, as well as chromosomal DNA from the original inoculum, was digested with the restriction enzyme ClaI, which cuts outside of the gipA ORF. Southern blotting was performed using the intact gipA ClaI fragment as a probe. All samples produced an identical pattern representing the wild-type conformation of this locus (data not shown). The results indicated that a single copy of the gipA locus was present in the strains before and after growth in the Peyer's patches. The results suggest that a simple transposition event is not required for gipA function in the Peyer's patches.
Transcriptional regulation of gipA.
The expression profile for gipA suggests that gipA is induced only in the small intestine. The constitutive fusion strain (JS131) used in the competition assay (Table 2) produces approximately 120 U of β-galactosidase in vitro. The pIVET1::gipA′ fusion strain produces approximately 10 U of β-galactosidase activity. However, in animals, the gipA fusion outcompetes JS131 in the small intestine (Table 2). Since growth is dependent on expression of the fusion, this result suggests that gipA is induced at least 12-fold in the early stages of disease. We attempted to find laboratory conditions that would induce the gipA fusion in vitro. The pIVET1::gipA′ fusion was moved into a purA+ background to ensure that there was no selection for expression of the fusion (JS133). We then measured the β-galactosidase activity produced by the fusion in response to a variety of in vitro conditions, including low iron, changes in pH, anaerobiosis, and PhoPQ-activating conditions (20). We also tested conditions known to induce other genes in the small intestine, such as the serovar Typhimurium invasion locus (2), and cholera toxin in both classical (24) and El Tor (21) Vibrio cholerae strains. None of the in vitro conditions that we used caused a significant induction of gipA, giving β-galactosidase activities ranging from 7.8 to 19 U, with a mean of 12.4 U. It is possible that gipA is induced in response to a signal that is specific to the small intestine. However, we cannot rule out the possibility that this gene is regulated in response to some environmental condition or combination of conditions that we have not tested.
DISCUSSION
We have used the IVET system (32, 45) to identify a serovar Typhimurium gene, gipA, that is specifically induced when the bacteria colonize the small intestine. By identifying genes that are expressed in a tissue-specific fashion, we expect to focus on gene products that are required for particular aspects of the infection process. We anticipated that oral selection would target genes induced in a Peyer's patch, since this is the site of significant bacterial growth in the small intestine (23) and growth is required for IVET selection. Indeed, we believe that gipA is specifically induced in the Peyer's patches and are performing experiments to explicitly address this issue. If true, then the gipA promoter is ideal for use in vaccine constructs, where the synthesis of antigens could be targeted to the Peyer's patches to induce mucosal immunity (16).
Our evidence suggests that a gipA null mutation confers a growth or survival defect only in the Peyer's patches. The gipA mutant is unaffected in its ability to initially colonize the small intestine and invade Peyer's patches. Several lines of evidence support this conclusion. First, equal numbers of Peyer's patches were infected with primarily mutant or wild-type bacteria in a mixed inoculation. Second, the mutant showed no defect in colonization in an IOC model. Third, the mutant showed no defect in invasion of epithelial cells in tissue cultures, an assay that is well correlated with defects in entry into M cells of Peyer's patches (19). Moreover, no defect was conferred when the bacteria were grown systemically in animals or in laboratory cultures. Thus, the gipA defect is specific to the Peyer's patches, consistent with the expression profile. These data suggest that the bacteria sense the particular environment of the Peyer's patch and respond by producing products required at this site of replication.
The results presented here help to elucidate the early steps in serovar Typhimurium pathogenesis. In mice, it appears that a limited number of bacteria enter a Peyer's patch and propagate in this site (7, 23). The number of organisms in a Peyer's patch normally reaches approximately 104 (Table 4). However, the numbers of bacteria need only increase to 102 to 103 before the bacteria are able to initiate a systemic infection. Our mutant had an approximately 10-fold-decreased ability to propagate in the Peyer's patches, but because it could still grow to some extent, there was only a fivefold increase in the LD50. Once past this bottleneck, the mutant is fully capable of causing disease.
The fact that the gipA mutant shows a defect in the Peyer's patches and not in the spleen emphasizes the inherent differences between these two lymphoid tissues. Distinct B-cell and T-cell subsets comprise a Peyer's patch (6). Indeed, the particular B cells apparently initiate differentiation of the lymphoid follicle (27). Although it is presumed that growth in macrophages is important for serovar Typhimurium after M-cell invasion, it is not clear in what cell types, if any, the bacteria grow in Peyer's patch tissue. For example, serovar Typhimurium is capable of invading both B cells and T cells in tissue cultures (49). There is apparently something unique about growth in the Peyer's patches that requires a specific set of bacterial gene products. Our results suggest that serovar Typhimurium senses this particular environment and responds accordingly.
The gipA ORF shows homology at the protein level to a family of putative insertion sequence element proteins from a wide array of prokaryotic species (Fig. 1). The elements present in Dichelobacter nodosus, a pathogen responsible for ovine foot rot (5), and Helicobacter pylori, the causative agent of duodenal and gastric ulcers (8), are both associated with clinically relevant pathogenicity determinants. Our evidence suggests that transposition is not required for gipA function in the Peyer's patches. Indeed, it is difficult to imagine how a transposition event would specifically enhance growth in a Peyer's patch. Alternatively, GipA could be a regulatory protein that is involved in the control of one or more virulence factors. An example is the Piv protein in Moraxella lacunata, which is homologous to a family of insertion sequence element “transposases” but acts via site-specific recombination to control pilin phase variation (29).
The putative protein sequences from the various GipA homologs are approximately 50% identical over their entire lengths. There is a particularly well-conserved motif near the C terminus of the protein that contains a number of invariant residues, including the amino acid sequence CXXC-15X-CXXC (Fig. 1). This motif is similar to motifs involved in metal ion binding in several other protein families. It most closely resembles putative zinc finger motifs found in certain ribosomal proteins (51) but could also be involved in binding other metals (38). Since it is reasonable to believe that this protein may be involved in DNA binding, this highly conserved sequence could function as a zinc finger DNA binding domain.
The gipA ORF is carried in the late operon of the lambdoid phage Gifsy-1. This phage is fully functional and is capable of moving from wild-type serovar Typhimurium and lysogenizing cured strains (14). It is found in all serovar Typhimurium strains that have been examined (14). Our results (unpublished) suggest that Gifsy-1 is not present in other serovars of Salmonella, including enteritidis, typhi, dublin, and paratyphi. Although we cannot rule out the possibilities that the bacteriophage moves throughout the salmonellae and we just happened to examine serovar Typhimurium strains that were lysogenic and other isolates that were not, the simplest explanation for the results is that serovar Typhimurium strains particularly benefit from maintaining Gifsy-1. There is a complicated and poorly understood relationship between Gifsy-1 and Gifsy-2, another lambdoid phage that is lysogenic in strains of serovar Typhimurium (14, 15). This relationship pertains to phage production as well as virulence. Although, as in our studies, these authors showed that Gifsy-1-cured strains were fully virulent in an i.p. infection, they noted that Gifsy-1 can apparently complement certain virulence defects conferred by the loss of Gifsy-2 (14). Whether any genes carried on Gifsy-2 can specifically contribute to growth in the Peyer's patches is difficult to answer given the other virulence defects conferred by the loss of the Gifsy-2 phage (14).
Virulence in serovar Typhimurium is the sum action of a large number of virulence factors, each of which may contribute only slightly to overall pathogenesis. In order to completely understand the molecular mechanisms of pathogenesis, we must investigate all of these factors. By using tissue-specific gene expression as a guide, we have identified a gene whose product is involved in survival in the Peyer's patches. Although the defect conferred by a gipA null mutation is subtle and the molecular explanation for the defect is not immediately obvious, this information gives us a starting point for understanding the genes and gene products required for this critical aspect of serovar Typhimurium pathogenesis.
ACKNOWLEDGMENTS
We thank Catherine Lee for generously providing us with the hilA339::Kn allele. We thank members of the Slauch laboratory for valuable discussions.
This study was supported by NIH grant AI37530 and ACS Junior Faculty research award JFRA-633.
REFERENCES
- 1.Ausubel F M. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 2.Bajaj V, Lucas R L, Hwang C, Lee C A. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol Microbiol. 1996;22:703–714. doi: 10.1046/j.1365-2958.1996.d01-1718.x. [DOI] [PubMed] [Google Scholar]
- 3.Bäumler A J, Heffron F. Identification and sequence analysis of lpfABCDE, a putative fimbrial operon of Salmonella typhimurium. J Bacteriol. 1995;177:2087–2097. doi: 10.1128/jb.177.8.2087-2097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bäumler A J, Tsolis R M, Heffron F. The lpf fimbrial operon mediates adhesion of Salmonella typhimurium to murine Peyer's patches. Proc Natl Acad Sci USA. 1996;93:279–283. doi: 10.1073/pnas.93.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Billington S J, Sinistaj M, Cheetham B F, Ayres A, Moses E K, Katz M E, Rood J I. Identification of a native Dichelobacter nodosus plasmid and implications for the evolution of the vap regions. Gene. 1996;172:111–116. doi: 10.1016/0378-1119(96)00032-7. [DOI] [PubMed] [Google Scholar]
- 6.Butcher E C. Lymphocyte homing and intestinal immunity. In: Ogra P L, Mestecky J, Lamm M E, Strober W, Bienenstock J, McGhee J R, editors. Mucosal immunology. San Diego, Calif: Academic Press, Inc.; 1999. pp. 507–522. [Google Scholar]
- 7.Carter P B, Collins F M. The route of enteric infection in normal mice. J Exp Med. 1974;139:1189–1203. doi: 10.1084/jem.139.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Censini S, Lange C, Xiang Z, Crabtree J E, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark M A, Jepson M A, Simmons N L, Hirst B H. Preferential interaction of Salmonella typhimurium with mouse Peyer's patch M cells. Res Microbiol. 1994;145:543–552. doi: 10.1016/0923-2508(94)90031-0. [DOI] [PubMed] [Google Scholar]
- 10.Clarke L, Carbon J. A colony bank containing synthetic Col E1 hybrid plasmids representative of the entire E. coli genome. Cell. 1976;9:91–99. doi: 10.1016/0092-8674(76)90055-6. [DOI] [PubMed] [Google Scholar]
- 11.Darwin K H, Miller V L. Molecular basis of the interaction of Salmonella with the intestinal mucosa. Clin Microbiol Rev. 1999;12:405–428. doi: 10.1128/cmr.12.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datsenka K A, Wanner B L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fields P I, Swanson R V, Haidaris C G, Heffron F. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci USA. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Figueroa-Bossi N, Bossi L. Inducible prophages contribute to Salmonella virulence in mice. Mol Microbiol. 1999;33:167–176. doi: 10.1046/j.1365-2958.1999.01461.x. [DOI] [PubMed] [Google Scholar]
- 15.Figueroa-Bossi N, Coissac E, Netter P, Bossi L. Unsuspected prophage-like elements in Salmonella typhimurium. Mol Microbiol. 1997;25:161–173. doi: 10.1046/j.1365-2958.1997.4451807.x. [DOI] [PubMed] [Google Scholar]
- 16.Frey A, Neutra M R. Targeting of mucosal vaccines to Peyer's patch M cells. Behring Inst Mitt. 1997;98:376–389. [PubMed] [Google Scholar]
- 17.Friedrich M J, Kinsey N E, Vila J, Kadner R J. Nucleotide sequence of a 13.9 kb segment of the 90 kb virulence plasmid of Salmonella typhimurium: the presence of fimbrial biosynthetic genes. Mol Microbiol. 1993;8:543–558. doi: 10.1111/j.1365-2958.1993.tb01599.x. [DOI] [PubMed] [Google Scholar]
- 18.Frost A J, Bland A P, Wallis T S. The early dynamic response of the calf ileal epithelium to Salmonella typhimurium. Vet Pathol. 1997;34:369–386. doi: 10.1177/030098589703400501. [DOI] [PubMed] [Google Scholar]
- 19.Galan J E. Interaction of Salmonella with host cells through the centisome 63 type III secretion system. Curr Opin Microbiol. 1999;2:46–50. doi: 10.1016/s1369-5274(99)80008-3. [DOI] [PubMed] [Google Scholar]
- 20.Garcia V E, Soncini F C, Groisman E A. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell. 1996;84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 21.Gardel C L, Mekalanos J J. Regulation of cholera toxin by temperature, pH, and osmolarity. Methods Enzymol. 1994;235:517–526. doi: 10.1016/0076-6879(94)35167-8. [DOI] [PubMed] [Google Scholar]
- 22.Hersh D, Monack D M, Smith M R, Ghori N, Falkow S, Zychlinsky A. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc Natl Acad Sci USA. 1999;96:2396–2401. doi: 10.1073/pnas.96.5.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hohmann A W, Schmidt G, Rowley D. Intestinal colonization and virulence of Salmonella in mice. Infect Immun. 1978;22:763–770. doi: 10.1128/iai.22.3.763-770.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwanaga M, Yamamoto K, Higa N, Ichinose Y, Nakasone N, Tanabe M. Culture conditions for stimulating cholera toxin production by Vibrio cholerae O1 El Tor. Microbiol Immunol. 1986;30:1075–1083. doi: 10.1111/j.1348-0421.1986.tb03037.x. [DOI] [PubMed] [Google Scholar]
- 25.Janakiraman A, Slauch J M. The putative iron transport system SitABCD encoded on SPI1 is required for full virulence of Salmonella typhimurium. Mol Microbiol. 2000;35:1146–1155. doi: 10.1046/j.1365-2958.2000.01783.x. [DOI] [PubMed] [Google Scholar]
- 26.Jones B D, Ghori N, Falkow S. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer's patches. J Exp Med. 1994;180:15–23. doi: 10.1084/jem.180.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerneis S, Bogdanova A, Kraehenbuhl J P, Pringault E. Conversion by Peyer's patch lymphocytes of human enterocytes into M cells that transport bacteria. Science. 1997;277:949–952. doi: 10.1126/science.277.5328.949. [DOI] [PubMed] [Google Scholar]
- 28.Kraehenbuhl J P, Neutra M R. Molecular and cellular basis of immune protection of mucosal surfaces. Physiol Rev. 1992;72:853–879. doi: 10.1152/physrev.1992.72.4.853. [DOI] [PubMed] [Google Scholar]
- 29.Lenich A G, Glasgow A C. Amino acid sequence homology between Piv, an essential protein in site-specific DNA inversion in Moraxella lacunata, and transposases of an unusual family of insertion elements. J Bacteriol. 1994;176:4160–4164. doi: 10.1128/jb.176.13.4160-4164.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lundberg U, Vinatzer U, Berdnik D, von Gabain A, Baccarini M. Growth phase-regulated induction of Salmonella-induced macrophage apoptosis correlates with transient expression of SPI-1 genes. J Bacteriol. 1999;181:3433–3437. doi: 10.1128/jb.181.11.3433-3437.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahan M J, Slauch J M, Mekalanos J J. Bacteriophage P22 transduction of integrated plasmids: single-step cloning of Salmonella typhimurium gene fusions. J Bacteriol. 1993;175:7086–7091. doi: 10.1128/jb.175.21.7086-7091.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahan M J, Slauch J M, Mekalanos J J. Selection of bacterial virulence genes that are specifically induced in host tissues. Science. 1993;259:686–688. doi: 10.1126/science.8430319. [DOI] [PubMed] [Google Scholar]
- 33.Maloy S R, Stewart V J, Taylor R K. Genetic analysis of pathogenic bacteria: a laboratory manual. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 34.Mann B A, Slauch J M. Transduction of low-copy number plasmids by bacteriophage P22. Genetics. 1997;146:447–456. doi: 10.1093/genetics/146.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Metcalf W W, Jiang W, Wanner B L. Use of the rep technique for allele replacement to construct new Escherichia coli hosts for maintenance of R6K gamma origin plasmids at different copy numbers. Gene. 1994;138:1–7. doi: 10.1016/0378-1119(94)90776-5. [DOI] [PubMed] [Google Scholar]
- 36.Miller S I, Pegues D A. Salmonella species, including Salmonella typhi. In: Mandell G L, Bennett J E, Dolin R, editors. Principles and practice of infectious diseases. Philadelphia, Pa: Churchill Livingstone; 2000. pp. 2344–2363. [Google Scholar]
- 37.Monack D M, Raupach B, Hromockyj A E, Falkow S. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc Natl Acad Sci USA. 1996;93:9833–9838. doi: 10.1073/pnas.93.18.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Halloran T V. Transition metals in control of gene expression. Science. 1993;261:715–725. doi: 10.1126/science.8342038. [DOI] [PubMed] [Google Scholar]
- 39.Rakeman J L, Bonifield H R, Miller S I. A HilA-independent pathway to Salmonella typhimurium invasion gene transcription. J Bacteriol. 1999;181:3096–3104. doi: 10.1128/jb.181.10.3096-3104.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reed L J, Muench H. A simple method for estimating fifty percent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 41.Schechter L M, Damrauer S M, Lee C A. Two AraC/XylS family members can independently counteract the effect of repressing sequences upstream of the hilA promoter. Mol Microbiol. 1999;32:629–642. doi: 10.1046/j.1365-2958.1999.01381.x. [DOI] [PubMed] [Google Scholar]
- 42.Sigwart D F, Stocker B A, Clements J D. Effect of a purA mutation on efficacy of Salmonella live-vaccine vectors. Infect Immun. 1989;57:1858–1861. doi: 10.1128/iai.57.6.1858-1861.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 44.Slauch J M. Regulation of virulence gene expression in vivo. In: Brouckaert P, editor. Virulence mechanisms of bacterial pathogens. Washington, D.C.: ASM Press; 2000. pp. 241–249. [Google Scholar]
- 45.Slauch J M, Mahan M J, Mekalanos J J. In vivo expression technology for selection of bacterial virulence genes specifically induced in host tissues. In: Clark V L, Bavoil P M, editors. Bacterial pathogenesis. San Diego, Calif: Academic Press, Inc.; 1997. pp. 309–320. [Google Scholar]
- 46.Slauch J M, Silhavy T J. cis-Acting ompF mutations that result in OmpR-dependent constitutive expression. J Bacteriol. 1991;173:4039–4048. doi: 10.1128/jb.173.13.4039-4048.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sukupolvi S, Lorenz R G, Gordon J I, Bian Z, Pfeifer J D, Normark S J, Rhen M. Expression of thin aggregative fimbriae promotes interaction of Salmonella typhimurium SR-11 with mouse small intestinal epithelial cells. Infect Immun. 1997;65:5320–5325. doi: 10.1128/iai.65.12.5320-5325.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Velden A W, Baumler A J, Tsolis R M, Heffron F. Multiple fimbrial adhesins are required for full virulence of Salmonella typhimurium in mice. Infect Immun. 1998;66:2803–2808. doi: 10.1128/iai.66.6.2803-2808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verjans G M, Ringrose J H, van Alphen L, Feltkamp T E, Kusters J G. Entrance and survival of Salmonella typhimurium and Yersinia enterocolitica within human B- and T-cell lines. Infect Immun. 1994;62:2229–2235. doi: 10.1128/iai.62.6.2229-2235.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang R F, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 51.Wool I R. The bifunctional nature of ribosomal proteins and speculations on their origins. In: Nierhaus K H, Franceschi F, Subramanian A R, Erdmann V A, Wittmann-Liebold B, editors. The translational apparatus. New York, N.Y: Plenum Press; 1993. pp. 727–737. [Google Scholar]