Summary
Background
Point-Of-Care (POC) diagnosis of life-threatening community-acquired meningitis currently relies on multiplexed RT-PCR assays, that lack genotyping and antibiotic susceptibility profiling. We assessed the usefulness of real-time metagenomics (RTM) directly applied to the cerebrospinal fluid (CSF) for the identification, typing and susceptibility profiling of pathogens responsible for community-acquired meningitis.
Methods
A series of 52 CSF samples from patients suspected of having community-acquired meningitis, were investigated at POC by direct RTM in parallel to routine real-time multiplex PCR (RT-PCR) and bacterial culture, for the detection of pathogens. RTM-generated sequences were blasted in real-time against an in-house database incorporating the panel of 12 most prevalent pathogens and against NCBI using EPI2ME online software, for pathogen identification. In-silico antibiogram and genotype prediction were determined using the ResFinder bio-tool and MLST online software.
Findings
Over eight months, routine multiplex RT-PCR yielded 49/52 positive CSFs, including 21 Streptococcus pneumoniae, nine Neisseria meningitidis, eight Haemophilus influenzae, three Streptococcus agalactiae, three Herpesvirus-1, two Listeria monocytogenes, and one each of Escherichia coli, Staphylococcus aureus and Varicella-Zoster Virus. Parallel RTM agreed with the results of 47/52 CSFs and revealed two discordant multiplex RT-PCR false positives, one H. influenzae and one S. pneumoniae. Both multiplex RT-PCR and RTM agreed on the negativity of three CSFs. While multiplex RT-PCR routinely took 90 min, RTM took 120 min, although the pipeline analysis detected the pathogen genome after 20 min of sequencing in 33 CSF samples; and after two hours in 14 additional CSFs; yielding > 50% genome coverage in 19 CSFs. RTM identified 14 pathogen genotypes, including a majority of H. influenzae b, N. meningitidis B and S. pneumoniae 11A and 3A. In all 16 susceptible cultured bacteria, the in-silico antibiogram agreed with the in-vitro antibiogram in 10 cases, available within 48 h in routine bacteriology.
Interpretation
In addition to pathogen detection, RTM applied to CSF samples offered supplementary information on bacterial profiling and genotyping. These data provide the proof-of-concept that RTM could be implemented in a POC laboratory for one-shot diagnostic and genomic surveillance of pathogens responsible for life-threatening meningitis.
Funding
This work was supported by the French Government under the Investments in the Future programme managed by the National Agency for Research reference: Méditerranée Infection 10-IAHU-03.
Keywords: Community-acquired meningitis, Real-time metagenomics, Point-of-care (POC) laboratory diagnosis, Antibiotic resistance, Genotyping
Research in context.
Evidence before this study
Community-acquired bacterial meningitis is a life-threatening infection that can progress to mortality within 48 h. The emergency diagnosis of infectious meningitis is currently based on multiplex real-time amplification using a syndromic panel limited by the most frequent microorganisms which lead to a central nervous system prognosis. Bacterial characterisation and drug resistance profiling require additional in-vitro investigations which take over 48 h, delaying pathogen-targeted treatment. Genomic surveillance and antibiotic resistance testing are based only on bacteria isolated from cerebrospinal fluid, failing in 60% of cases. We searched PubMed up to 30 November 2021 for research articles published in English, using the following search terms “real-time metagenomics sequencing”, “meningitis”, and “direct diagnosis”. Several articles had been published testing the Oxford Nanopore technologies sequencing on CSF, but no investigations were found into the direct diagnosis of CSF series. When the three terms were used together, only two articles previously published by us were found. As previously reported, we implemented real-time metagenomic sequencing (RTM) at the POC laboratory for the diagnosis of life-threatening infectious meningitis, in addition to the BioFire FilmArray® investigation. In light of its simplicity, rapidity and additional information collected, we propose RTM as a powerful diagnostic tool for the investigation of prospective series of CSF samples collected from patients with meningitis.
Added value of this study
In this study, we diagnosed a series of community-acquired meningitis cases by RTM directly from CSF samples. Over eight months, 52 CSFs were investigated directly by RTM using a four-hour workflow. Thirty-three CSF samples (63.5%) were diagnosed as positive after a 20-min sequencing run and an additional 14 were diagnosed as positive after two hours. The pipeline analysis of antibiotic resistance and bacteria genotyping was provided in-silico at the same time as sequencing, in contrast to conventional diagnostics. In addition, uncultured bacteria were successfully profiled in-silico, basing on pathogen genome analysis, independently of genome coverage.
Implications of all the available evidence
Despite the limited sample size in this study, using a four-hours workflow, RTM proved successful in diagnosing, genotyping, and profiling bacteria directly from CSF samples. At two discordances with conventional multiplex RT-PCR, RTM is a suitable method for the diagnosis of life-threatening meningitis at the POC laboratory.
Alt-text: Unlabelled box
Introduction
The rapid diagnosis of life-threatening, community-acquired meningitis (CAM) remains challenging in point-of-care (POC) laboratories.1 Bacterial meningitis charged with a 24-h mortality of 8–15%,2 results in an estimated 290,000 deaths every year, causing more than 50% deaths annually from all meningitis causes, and leaving one in five people who recover with chronic handicap.3 Community-acquired bacterial meningitis around the world is mainly due to Streptococcus pneumoniae (S. pneumoniae), Neisseria meningitidis (N. meningitidis), Haemophilus influenzae (H. influenzae) and Streptococcus agalactiae (S. agalactiae).4 In Europe, S. pneumoniae and N. meningitidis are the most common causes of bacterial meningitis,2 usually affecting children ≤ 5 years in 22.5% and 47% of cases, respectively.2 Current POC diagnosis of bacterial meningitis is based on real-time multiplex PCR (RT-PCR) assays incorporating a syndromic meningitis and encephalitis panel,5, 6, 7, 8, 9, 10 targeting small specific pieces of the pathogen genome.5,6 These approaches overlook serotype/genotype diversity, a major limitation for the microbiological diagnosis of bacterial meningitis, do not provide sufficient information for pathogen genotyping, and require bacterial culture to characterise different serotypes and antimicrobial resistance.5,6 Accordingly, additional specific PCRs have to be performed for genotyping N. meningitidis B and C serotypes,11, 12, 13 and H. influenzae b serotype associated to invasive diseases.2,14
Real-time metagenomics sequencing (RTM) could, theoretically, overcome this limitation, identifying the causative agent of meningitis,15 as well as its genotype/serotype directly from the cerebrospinal fluid (CSF) based on pathogen genome sequence.14,16 Indeed, we and others have already published evidence that RTM could be implemented in a POC laboratory, for one-shot diagnostic, genotyping as well as in-silico antibiotic resistance prediction, which is competitive in time and cost with commercial multiplex RT-PCR.14, 15, 16, 17, 18
In this study, we prospectively diagnosed a series of cases of community-acquired meningitis, directly using RTM on left-over CSF samples in a POC laboratory.
Methods
Ethics
As per French legislation, no specific patient consent was required. The analysis of biological samples obtained in the medical care context was considered as non-interventional research (article L1221-1.1 of the French Public Health Code), requiring only the non-opposition of the patient during sampling (article L1211-2 of the French Public Health Code). All data were generated as part of routine laboratory work at the Assistance Publique-Hôpitaux de Marseille and Nîmes university hospital, in the context of the routine clinical management of patients suspected of having community-acquired meningitis. No specific clinical sampling was performed for this study and RTM was applied to anonymised left-over CSF samples for which the age and sex of patient were anonymously collected, following a standard routine laboratory protocol including multiplex real-time PCR, which was carried out in full respect of the French law regarding clinical research. Accordingly, this study was approved by IHU Méditerranée Infection Ethics Committees under number: 2021-004 before the study began in Marseille; and further approval was granted by “Interface Recherche Bioéthique Institutional Review Board” Ethics Committee of Nimes CHU under the following number: 21.0016 before the study began in Nîmes.
Routine microbial diagnosis
In the POC laboratory, all CSF samples were routinely examined to count white and red blood cells directly using NucleoView NC-3000 equipment and NucleoView/ChemoMetec software (ChemoMetec NucleoCounter, Allerod, Denmark). In parallel, 200 µL samples of CSF were used for multiplex RT-PCR diagnosis (BioFire FilmArray®, bioMérieux, Marcy-l’Étoile, France) as previously described.5 Further in the core laboratory, 200 µL of CSF was inoculated on chocolate agar PolyViteX (bioMérieux) and Columbia agar enriched with a 5% sheep's blood (bioMérieux) medium incubated at 37 °C under 5% CO2 for five days, and on Columbia agar enriched with 5% sheep blood (bioMérieux) under anaerobic conditions for ten days at 37 °C for bacterial culture and in-vitro antibiogram in the standard bacteriology laboratories. For any isolate, antibiograms were validated according to the antibiotic panel approved by the French Antibiogram Comity of Microbiology Society (CA-SFM, version V1 2020) (Appendix 1).
Installation of the RTM platform in the POC
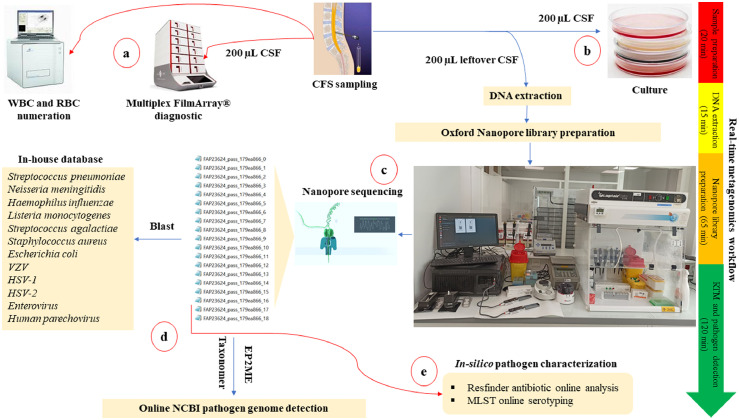
We updated the equipment available in our POC laboratories by setting up MinION sequencers (Oxford Nanopore Technologies, Oxford, UK) (Figure 1, Appendix 2). As an example, in the Marseille POC laboratory, an RTM bench was set up on a surface area of 210 × 70 cm2, the atmosphere was stabilised at one atm and the temperature was controlled by a central air conditioning at 20 °C ± 2 °C. The RTM bench was equipped with a Biocap® hood (Erlab, Val-de-Reuil, France), a clean area for DNA preparation, Qubit® for DNA quantification, a thermal cycler (ThermoFisher, Illkirch, France), an incubator at 20 °C with agitation for the different incubation steps, a vortex for mixing reagents and buffers, magnetic rack, a tube ice rack for enzyme storage during the manipulation, a mini centrifuge at 12,000 g, micro-pipettes with different volumes and a biological waste container (Figure 1, Appendix 2). Metagenomic handling was performed in an 1800 cm2 workspace. For library sequencing, four MinION instruments were attached in parallel to a powerful computer equipped with minimum Windows 10 or a Linux version (16.04 LTS) operating system, an i7 processor, RAM ≥ 8Gb, USB 3 port, with and enough disk space to store the data (∼ 1 Tb) (Lenovo, China), and an internet connection. MinION-Sequencer reading, and data storage were performed using Minknow Oxford Nanopore software version (8.3.1). In addition, Oxford Nanopore EPI2ME software was installed for real-time data analysis.
Figure 1.
CSF workflow for the diagnosis of community-acquired meningitis in the POC laboratory for the 52 CSF series prospective investigation. When the CSF sample was received at the POC laboratory, several tests were performed to detect the meningitis causative agent. a) Systematically, the emergency multiplex BioFire FilmArray® assay performed using 200 µL CSF when the sample was received, followed by quantification of the blood cells. b) As per routine diagnosis, all CSF samples received at the POC were routinely cultured. c) The RTM diagnosis was performed in a total workflow that did not exceed four hours. Sample preparation and DNA extraction from left over CSF samples took 35 min, Oxford Nanopore library preparation took 65 min, and the MinION library sequencing took two hours. d) Real-time genome identification was performed directly by blast of the MinION generated data against an in-house database, then against the NCBI database using EPI2ME and Taxonomer online software. e) Antibiotic resistance and pathogen genotyping were in-silico predicted using the ResFinder online database (https://cge.cbs.dtu.dk/services/ResFinder-4.1/), and MLST online database (https://pubmlst.org/organisms).
Abbreviations: CSF: cerebrospinal fluid. WBC: white blood cell. RBC: red blood cell. RTM: real-time metagenomics. MLST: Multi-Locus Sequence Typing.
RTM procedure
Total DNA was extracted from 200 µL of left-over CSF samples using an EZ1 DNA Tissue Kit (Qiagen, Courtaboeuf, France), after 15 min of incubation at 56 °C with 20 µL proteinase K (Qiagen), then eluted in a 50-µL volume. For the real-time next-generation sequencing, the Oxford Nanopore library preparation was performed in a 75-µL final volume as previously described.14 Briefly, 48 µL DNA was prepared and end-repaired in 60 µL containing 3.5 µL of NEBNext FFPE DNA Repair buffer and 3.5 µL of Ultra II End-prep reaction buffer, 2 µL of NEBNext FFPE DNA Repair mix (New England BioLabs, Evry-Courcouronnes, France) and 3 µL of Ultra II End-prep enzyme mix (New England BioLabs). The repair reaction Master Mix was incubated for five minutes at 20 °C followed by a five-minute incubation at 65 °C on a GeneAmp PCR System Thermal Cycler (Applied Biosystems, Foster City, CA, USA). Repaired DNA was purified using equal volumes of Agencourt Ampure XP beads (Beckman Coulter, Villepinte, France), and eluted in 25 µL of sterile water after incubation for five minutes at room temperature and two washes with 70% ethanol. A barcoding step was added to the standard Oxford Nanopore protocol to avoid any cross-contamination and to reduce the cost of the tests. A 22.5 µL volume of repaired DNA was barcoded in 50 µL containing 2.5 µL of native barcoding and 25 µL of Blunt/TA Ligase Master Mix (BioLabs), incubated for ten minutes at room temperature. Barcoded DNA was purified using 50 µL of Agencourt Ampure XP and eluted in 65 µL sterile water, after incubation for five minutes at room temperature and two washes with 70% ethanol. A 65-µL volume of the barcoded DNA was indexed in 100 µL containing 20 µL NEBNext Quick Ligation Reaction Buffer (5X) buffer, 5 µL of Adapter Mix II (AMII) and 10 µL of T4 DNA Ligase and incubated for ten minutes at room temperature. 60 µL of Agencourt Ampure XP beads were then added to the ligation master mix and incubated for five minutes at room temperature. Two washes were performed using an LFB buffer, then eluted in a 15-µL volume and incubated for ten minutes at room temperature. Finally, 12 µL of the eluted library were added to 37.5 µL sequencing buffer and 25.5 µL loading beads and sequenced for up to two hours on a MinION sequencer (Oxford Nanopore, Oxford Science Park, UK) (Figure 1). For rapid pathogen genome identification, the output fastq_pass was generated every 1500 reads per file to favour real time analysis, using Minknow specific parameters before starting sequencing run.
In-silico data analysis
Pathogen genome identification
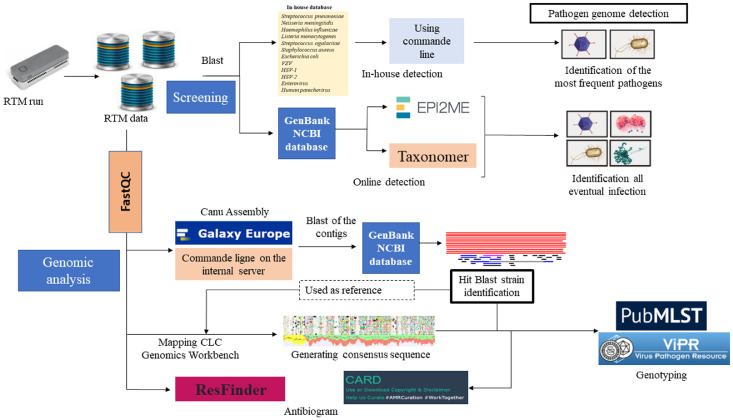
Pathogen genome sequences were detected in real-time using an in-house pipeline. First, total MinION data were aligned against an in-house database constructed in reference to Biofire FilmArray® panel (Appendix 6), including complete genome sequences for each one of N. meningitidis, S. pneumoniae, H. influenzae, S. agalactiae, Escherichia coli, Staphylococcus aureus and Listeria monocytogenes (100 sequences per pathogen), VZV (148 sequences), HSV-1 (51 sequences), HSV-2 (33 sequences), Parechovirus (200 sequences) and Enterovirus (300 sequences) using a blast nucleotide command line with default parameters on the IHU server. This analysis was interpreted as positive when the number of sequences for any specific pathogen was ≥ 2. Further, detection was queried against NCBI GenBank database to increase probability pathogen genome detection using Taxonomer (https://www.taxonomer.com) and Oxford Nanopore EPI2ME online software (Figures 1, 2). To confirm real-time identification, reads quality control was performed using FastQC online platform and assembled by “Canu assembler” tool (Version 2.1.1) on Galaxy Europe online software (https://usegalaxy.eu/) and generated contigs were then aligned by Blastn against NCBI GenBank database. The identified hit-blast strain, defined by maximum sequence similarity, was used as a reference sequence for mapping of the total MinION reads using CLC Genomics Workbench software version 21.0.3 (Qiagen) with default parameters. Consensus sequences were extracted in fasta files for further analysis (Figure 2). The in-silico prediction of antibiotic resistance-encoding genes was carried out using ResFinder (https://cge.cbs.dtu.dk/services/ResFinder/) and Resistance Gene Identification (https://card.mcmaster.ca/analyze/rgi) online software using total pathogen reads and hit-blast strains identified by Blastn against NCBI GenBank database after assembly of the MinION reads, in reference to antibiotics routinely assayed in-vitro (Appendix 1). In-silico prediction of antibiotic resistance was performed only in the case genome coverage was ≥ 1%.
Figure 2.
Bioinformatic pipeline. Rapid pathogen genome identification performed by direct alignment of MinION reads with an in-house database using blastn command line on the IHU server, further against NCBI GenBank by Taxonomer and EPI2ME online software. The analysis was interpreted as positive when ≥ 2 pathogen-specific reads were identified. To confirm pathogen identification, the quality of MinION reads was controlled by FastQC before assembly by “Canu assembler” on Galaxy Europe online software (https://usegalaxy.eu/). Hit-blast strains were identified by Blastn of the generated contigs against GenBank database, then used as reference genome for mapping the total MinION reads by CLC Genomics Workbench software version 21.0.3 (Qiagen). The consensus genomes were extracted in fasta files for pathogens genotyping on MLST on PubMLST (https://pubmlst.org/organisms) for bacteria genotyping and on ViPR online database (https://www.viprbrc.org/) for virus genotyping. The in-silico antibiogram was predicted on ResFinder (https://cge.cbs.dtu.dk/services/ResFinder/) and Resistance Gene Identification (https://card.mcmaster.ca/analyze/rgi) platforms using the total MinION reads and the generated fasta sequences with default settings.
*In-silico antibiogram and genotype were derived from genome sequence only in the case of > 1% genome coverage.
Pathogen genotyping
Microbial genotypes were predicted using multi-locus sequence typing tools (MLST) on PubMLST (https://pubmlst.org/organisms), based on total specific sequences data and hit-blast stains identified by blast against NCBI GenBank database. Virus genotyping was performed directly on ViPR online database (https://www.viprbrc.org/) (Figure 2). Genotyping was in-silico predicted only in the case genome coverage was ≥ 1%.
Graphic representation of data
Graphical representation was performed using R software (version 4.0.3). Pie and donut charts were created using the PieDonut function in the webr package (https://www.R-project.org/ and https://CRAN.R-project.org/package=webr).
Cost analysis
We compared estimations of global cost for Biofire FilmArray®-based diagnosis of CAM at the POC laboratory with that of RTM-based diagnosis. Noteworthy, these estimations incorporated mean cost by sample for RTM (144€) and FilmArray® (114€), based on values calculated with reference prices for materials and reagents, in our laboratories. We estimated that 30 RTM assays costed 4320 € including pathogen identification, genotyping, and in-silico antibiogram; while we estimated that 30 FilmArray® assays costed 3411€ for pathogen identification only; eventually increased by cost of additional pathogen-specific PCRs, in case of negative FilmArray® assay; and that of additional in-vitro investigations for pathogen characterization. Moreover, relative cost obviously decreased in case of series of CSF samples tested at the same time: as an example, RTM cost decreased to less than 70 € / CSF sample for series of 12 samples tested with the same MinION flow-cell, while FilmArray® cost was independent of the number of tested CSF samples, being series-insensitive (Table 3, Appendix 4).
Table 3.
Cost analysis and comparison of RTM and routine FilmArray® test.
| Approach | Blunt/TA Ligase Master Mix | NEBNext® FFPE DNA Repair Mix | NEBNext® FFPE DNA Repair Mix | NEBNext® Quick Ligation™* | Flow Cell (R9) | Ligation Kit | Flow Cell Priming Kit* | Flow Cell Wash Kit | Native Barcoding (PCR-free) * | Biofire FilmArray cardridge | Veritable cost by test |
|---|---|---|---|---|---|---|---|---|---|---|---|
| RTM | 1,44 | 6 € | 9 € | 12.72 | 54 | 45 € | 5 € | 13 € | 11 € | 0 € | 143,3€ |
| Biofire FilmArray | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 114 | 114€ |
Role of funding source
The funders did not have any role in the study design, data collection, data analyses, interpretation, or writing of the report.
Results
General data
CSF samples collected from 52 patients prospectively investigated in this study included 24 CSF samples at the IHU Méditerranée Infection POC laboratory in Marseille and 28 CSF samples at the bacteriology and hygiene laboratory at Nîmes University Hospital. These 52 CSF samples were collected from 24 female patients and 28 male patients, aged between 0 and 90 years (median, 38 years old), investigated between December 2020 and July 2021 (Table 1, Appendix 3).
Table 1.
Concordance and discordance of routine diagnosis with RTM, in-vitro and in-silico antibiogram. Antibiotics not used in the routine are in green characters. The final diagnosis is the diagnosis validated in routine by clinicians.
| Patient | Age | Gender | Final diagnostic | Biofire FilmArray | MinION | Culture | Specific RT-PCR | In-vitro antibiogram | In-silico antibiogram |
|---|---|---|---|---|---|---|---|---|---|
| Sample 1 | 90 | M | Streptococcus pneumoniae | Positive | Positive | Positive | Not realized | Macrolides and related groups (erythromycin, clindamycin, streptogramin b), tetracycline, trimethoprim | Macrolides and related groups (erythromycin, clindamycin, lincomycin, quinupristin, pristinamycine, virginiamycin), tetracycline, rifamycin, fluoroquinolone, aminoglycoside, phenicol |
| Sample 2 | 70 | M | Streptococcus pneumoniae | Positive | Positive | Positive | Not realized | Macrolides and related groups (erythromycin, clindamycin, streptogramin b), tetracycline, trimethoprim | Macrolides and related groups (erythromycin, clindamycin, lincomycin), tetracycline, phenicol |
| Sample 3 | 25 | M | Neisseria meningitidis | Positive | Positive | Positive | Not realized | Susceptible | Susceptible |
| Sample 4 | 80 | M | Streptococcus pneumoniae | Positive | Positive | Negative | Not realized | Not realized | Not realized |
| Sample 5 | 84 | M | Listeria monocytogenes | Positive | Positive | Positive | Not realized | Clindamycin, Trimethoprim | Not realized |
| Sample 6 | 66 | F | Streptococcus pneumoniae | Positive | Positive | Positive | Not realized | Macrolides and related groups (erythromycin, clindamycin, streptogramin b), tetracycline | Macrolides and related groups (erythromycin, clindamycin, streptogramin b, quinupristin, pristinamycine, virginiamycin), tetracycline |
| Sample 7 | 65 | F | Haemophilus influenzae | Positive | Positive | Positive | Not realized | Susceptible | Amoxicillin/ampicillin, piperacillin, ticarcillin, cephalothin |
| Sample 8 | 64 | F | Neisseria meningitidis | Positive | Positive | Negative | Not realized | Not realized | Not realized |
| Sample 9 | 30 | F | Streptococcus pneumoniae | Positive | Positive | Positive | Not realized | Susceptible | Not realized |
| Sample 10 | 57 | F | Streptococcus pneumoniae | Positive | Positive | Negative | Not realized | Not realized | Susceptible |
| Sample 11 | 2 | M | Streptococcus pneumoniae | Positive | Positive | Positive | Not realized | Susceptible | Susceptible |
| Sample 12 | 45 | M | Streptococcus pneumoniae | Positive | Positive | Negative | Not realized | Not realized | Susceptible |
| Sample 13 | 0 | F | Streptococcus agalactiae | Positive | Positive | Negative | Not realized | Not realized | Macrolides and related groups (erythromycin, spiramycin, azithromycin) |
| Sample 14 | 7 | F | Neisseria meningitidis | Positive | Positive | Negative | Not realized | Not realized | Susceptible |
| Sample 15 | 58 | M | Neisseria meningitidis | Positive | Positive | Negative | Not realized | Not realized | Susceptible |
| Sample 16 | 0 | F | Streptococcus agalactiae | Positive | Positive | Negative | Not realized | Not realized | Susceptible |
| Sample 17 | 33 | M | Streptococcus pneumoniae | Positive | Positive | Positive | Not realized | Susceptible | Macrolides and related groups (erythromycin, clindamycin, streptogramin b |
| quinupristin, pristinamycine, virginiamycin) | |||||||||
| Sample 18 | 0 | M | Haemophilus influenzae | Positive | Positive | Negative | Not realized | Not realized | Amoxicillin/ampicillin, piperacillin, ticarcillin, cephalothin |
| Sample 19 | 32 | M | Negative | Negative | Negative | Negative | Not realized | Not realized | Not realized |
| Sample 20 | 28 | F | Negative | Haemophilus influenzae | Negative | Negative | Negative | Not realized | Not realized |
| Sample 21 | 68 | M | Herpes Simplex Virus 1 | Positive | Positive | Negative | Positive | Not realized | Not realized |
| Sample 22 | 14 | F | Herpes Simplex Virus 1 | Positive | Positive | Negative | Positive | Not realized | Not realized |
| Sample 23 | 77 | F | Herpes Simplex Virus 1 | Positive | Positive | Negative | Positive | Not realized | Not realized |
| Sample 24 | 29 | F | Varicella Zoster Virus | Positive | Positive | Negative | Positive | Not realized | Not realized |
| Sample 25 | 89 | F | Haemophilus influenzae | Positive | Positive | Negative | Not realized | Not realized | Susceptible |
| Sample 26 | 74 | F | Streptococcus pneumoniae | Positive | Positive | Negative | Not realized | Not realized | Not realized |
| Sample 27 | 49 | F | Haemophilus influenzae | Positive | Positive | Negative | Not realized | Not realized | Susceptible |
| Sample 28 | 22 | M | Listeria monocytogenese | Positive | Positive | Positive | Not realized | Susceptible | Not realized |
| Sample 29 | 0 | M | Streptococcus pneumoniae | Positive | Positive | Positive | Positive | Susceptible | Susceptible |
| Sample 30 | 78 | F | Streptococcus pneumoniae | Positive | Positive | Negative | Positive | Not realized | Susceptible |
| Sample 31 | 11 | M | Streptococcus pneumoniae | Positive | Negative | Negative | Positive>35Ct | Negative | Not realized |
| Sample 32 | 22 | M | Neisseria meningitidis | Positive | Positive | Negative | Positive | Not realized | Penicillin A |
| Sample 33 | 18 | M | Neisseria meningitidis | Positive | Positive | Negative | Positive | Not realized | Susceptible |
| Sample 34 | 42 | M | Neisseria meningitidis | Positive | Positive | Negative | Positive | Not realized | Susceptible |
| Sample 35 | 0 | M | Haemophilus influenzae | Positive | Positive | Positive | Positive | Susceptible | Susceptible |
| Sample 36 | 0 | M | Haemophilus influenzae | Positive | Positive | Negative | Positive | Not realized | Not realized |
| Sample 37 | 40 | M | Haemophilus influenzae | Positive | Positive | Negative | Positive | Not realized | Susceptible |
| Sample 38 | 57 | M | Staphylococcus aureus | Positive | Positive | Positive | Positive | Susceptible | Cefoxitin (mecA) |
| Sample 39 | 40 | M | Negative | Negative | Negative | Negative | Negative | Not realized | Not realized |
| Sample 40 | 59 | F | Streptococcus pneumoniae | Positive | Positive | Negative | Positive | Not realized | Susceptible |
| Sample 41 | 33 | F | Neisseria meningitidis | Positive | Positive | Positive | Positive | Susceptible | Susceptible |
| Sample 42 | 18 | F | Streptococcus agalactiae | Positive | Positive | Negative | Positive | Not realized | Susceptible |
| Sample 43 | 0 | M | Streptococcus pneumoniae | Positive | Positive | Positive | Positive | Susceptible | Susceptible |
| Sample 44 | 20 | F | Neisseria meningitidis | Positive | Positive | Positive | Positive | Susceptible | Penicillin A |
| Sample 45 | 23 | F | Negative | Negative | Negative | Negative | Negative | Not realized | Not realized |
| Sample 46 | 51 | M | Streptococcus pneumoniae | Positive | Positive | Positive | Positive | Susceptible | Susceptible |
| Sample 47 | 55 | F | Streptococcus pneumoniae | Positive | Positive | Negative | Positive | Not realized | Susceptible |
| Sample 48 | 64 | F | Streptococcus pneumoniae | Positive | Positive | Positive | Positive | Susceptible | Susceptible |
| Sample 49 | 0 | M | Escherichia coli | Positive | Positive | Negative | Positive | Not realized | Not realized |
| Sample 50 | 6 | M | Streptococcus pneumoniae | Positive | Positive | Negative | Positive | Not realized | Not realized |
| Sample 51 | 85 | F | Streptococcus pneumoniae | Positive | Positive | Positive | Positive | Susceptible | Susceptible |
Routine investigations
Routine FilmArray® assays detected a microorganism in 49 CSF samples. Bacterial pathogens found in 45 CSF samples included 21 S. pneumoniae, nine N. meningitidis, eight H. influenzae, three S. agalactiae, two L. monocytogenes and one each of E. coli and S. aureus. In addition, three Herpesvirus-1 and one Varicella-zoster virus (VZV) were detected. All viral cases resulted from PCR, while 20 bacterial meningitis cases were confirmed by culture and RT-PCR, 12 by RT-PCR only, including one case (sample 31) of S. pneumoniae > 35 Ct which was interpreted as negative in routine POC diagnostic, 12 cases with FilmArray® only, and PCR and culture failed to identify one case of H. influenzae (sample 20). The 20 culture positive CSFs grew 12 S. pneumoniae, three N. meningitidis, two H. influenzae, two L. monocytogenes and one S. aureus, while the other 26 CSFs detected positive for bacteria by RT-PCR were culture-negative. In-vitro antibiotic investigation yielded 16 susceptible bacteria (nine S. pneumoniae, three N. meningitidis, two H. influenzae, and one each of L. monocytogenes and S. aureus), three S. pneumoniae which were resistant to erythromycin, clindamycin, pristinamycine, doxycycline, one N. meningitidis which was resistant to amoxicillin and rifampicin, and one L. monocytogenes which was resistant to trimethoprim and clindamycin (Table 1).
RTM investigations
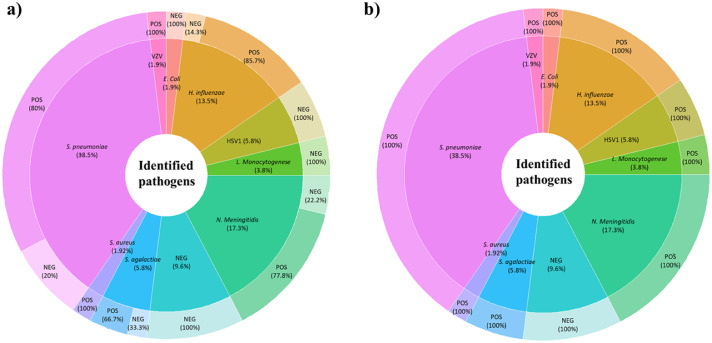
In total, RTM detected pathogen genomes in 47 leftover CSF samples. Bacteria detected in 43 CSF samples included 20 S. pneumoniae, nine N. meningitidis, seven H. influenzae, three S. agalactiae, two L. monocytogenes, and one each of E. coli and Staphylococcus aureus. Viral pathogens detected in four CSF samples included three Herpesvirus-1 and one Varicella-Zoster Virus. In addition, RTM yielded five negative CSF samples. In 63.5% CSF samples, pathogen genome detection in a 20-min sequencing run (Figure 3, Table 2) yielded 16 S. pneumoniae, seven N. meningitidis, six H. influenzae, one S. agalactiae and one VZV (median number of reads = 23), resulting from a blast analysis of MinION data against the in-house database and EPI2ME online analysis. A total of 47/52 CSF samples were detected as positive after two hours (median number of reads of 456.5). Genomic data analysis showed 19/47 (40.4%) of positive cases with >50% genome coverage including eight S. pneumoniae, five N. meningitidis, four H. influenzae, one each of S. agalactiae and VZV (Figure 1, Table 2). Viral cases were identified directly by blast against the in-house database and EPI2ME online software and confirmed by specific RT-PCR. False negative S. pneumoniae (sample 31) was confirmed by negative Illumina pair-end metagenomics, as previously described,16 may be due to failed DNA extraction and/or the limitation of RTM to detect low pathogen levels in the CSF (>35 Ct). The in-silico antibiogram analysis yielded 24/43 susceptible bacteria, 10 resistant bacteria and not realized in nine bacteria due to the low quantity of pathogens sequences (< 1% genome coverage) generated by MinION (Table 1).
Figure 3.
Real-time metagenomics data analysis and pathogen characterisation. a) Real-time data analysis and pathogen identification at 20 min sequencing. Thirty-one CSF samples were diagnosed as positive after a 20-min sequencing run, including 16 S. pneumoniae, seven N. meningitidis, six H. influenzae, one S. agalactiae, and one VZV. b) Total generated data and pathogen identification after a two-hour sequencing run. A total of 47/52 CSFs diagnosed positive after two hours RTM, included 20 S. pneumoniae, nine N. meningitidis, seven H. influenzae, three S. agalactiae, three Herpes Simplex Virus, two L. monocytogenes, one case each of E. coli and S. aureus, and one VZV.
Table 2.
partial and total genomic data, RTM analysis and bacteria genotyping.
| Samples | Run time | Identied pathogen | Specific reads at 20 min | Total reads at 2 h | Specific reads at 2 h | Specific reads (%) | Number of nucleotide | Genome coverage | Genotyping |
|---|---|---|---|---|---|---|---|---|---|
| Sample 1 | 2H | Streptococcus pneumoniae | 5897 | 174,156 | 33,465 | 19.22 | 1,940,109 | 95.17 | Serotype 16F |
| Sample 2 | 2H | Streptococcus pneumoniae | 7457 | 125,520 | 35,055 | 27.93 | 1,959,603 | 87.48 | Serotype 6B |
| Sample 3 | 2H | Neisseria meningitidis | 45 | 116 ,752 | 905 | 0.78 | 762,285 | 34.01 | Serotype B |
| Sample 4 | 2H | Streptococcus pneumoniae | 0 | 9137 | 6 | 0.065667068 | 4183 | 0.02 | Not realized |
| Sample 5 | 2H | Listeria monocytogenes | 0 | 480,000 | 21 | 0.00004375 | 14,597 | 0.5 | Not realized |
| Sample 6 | 2H | Streptococcus pneumoniae | 7 | 151,187 | 135 | 0.089293392 | 73,893 | 3.36 | Serotype 11A |
| Sample 7 | 2H | Haemophilus influenzae | 3471 | 180,169 | 26,014 | 14.43866592 | 1,751,169 | 88.37 | Serotype b |
| Sample 8 | 2H | Neisseria meningitidis | 0 | 3830 | 3 | 0.078328982 | 2687 | 0.1 | Not realized |
| Sample 9 | 2H | Streptococcus pneumoniae | 0 | 34,860 | 12 | 0.034423408 | 4578 | 0.22 | Not realized |
| Sample 10 | 2H | Streptococcus pneumoniae | 5 | 11,260 | 132 | 1.172291297 | 65,775 | 3 | Serotype A5 |
| Sample 11 | 2H | Streptococcus pneumoniae | 3451 | 30,015 | 17,074 | 56.88489089 | 1,994,683 | 93.65 | Serotype 3A |
| Sample 12 | 2H | Streptococcus pneumoniae | 245 | 131,943 | 1652 | 1.252055812 | 1,346,254 | 63.2 | Serotype 3A |
| Sample 13 | 2H | Streptococcus agalactiae | 123 | 206,130 | 1017 | 0.493377965 | 390,297 | 19.38 | Serotype V |
| Sample 14 | 2H | Neisseria meningitidis | 4513 | 60,134 | 18,965 | 31.53789869 | 2,023,530 | 93.96 | Serotype C |
| Sample 15 | 2H | Neisseria meningitidis | 411 | 57,227 | 3713 | 6.488196131 | 1,644,843 | 73.43 | Serotype A |
| Sample 16 | 2H | Streptococcus agalactiae | 341 | 125,815 | 2058 | 1.635735008 | 1,504,297 | 71.3 | Serotype V |
| Sample 17 | 2H | Streptococcus pneumoniae | 239 | 6501 | 1175 | 18.07414244 | 517,575 | 24.3 | Serotype 11A |
| Sample 18 | 2H | Haemophilus influenzae | 314 | 9283 | 1406 | 15.14596574 | 1,318,171 | 66.52 | Serotype b |
| Sample 19 | 2H | Negative | 0 | 39,810 | 0 | 0 | 0 | 0 | Not realised |
| Sample 20 | 2H | Negative | 0 | 8110 | 0 | 0 | 0 | 0 | Not realised |
| Sample 21 | 2H | Herpes Simplex Virus 1 | 0 | 4760 | 5 | 0.105042017 | 501 | 0.3 | Not realized |
| Sample 22 | 2H | Herpes Simplex Virus 1 | 0 | 14,110 | 2 | 0.014174344 | 271 | 0.2 | Not realized |
| Sample 23 | 2H | Herpes Simplex Virus 1 | 0 | 7082 | 9 | 0.127082745 | 1519 | 1 | Unkown |
| Sample 24 | 2H | Varicella Zoster Virus | 41 | 251,411 | 608 | 0.241835083 | 86,736 | 69.38 | Unkown |
| Sample 25 | 2H | Haemophilus influenzae | 967 | 775,438 | 4352 | 0.005612312 | 1,555,463 | 83.8 | Serotype f |
| Sample 26 | 2H | Streptococcus pneumoniae | 2 | 74,278 | 31 | 0.000417351 | 18,086 | 0.8 | Not realized |
| Sample 27 | 2H | Haemophilus influenzae | 8 | 14,994 | 63 | 0.004201681 | 2,8751 | 1.45 | Serotype f |
| Sample 28 | 2H | Listeria monocytogenese | 0 | 511,274 | 6 | 1.17354E−05 | 9104 | 0.3 | Not realized |
| Sample 29 | 2H | Streptococcus pneumoniae | 29 | 305,696 | 2566 | 0.00839396 | 1,138,209 | 51.8 | Serotype 6B |
| Sample 30 | 2H | Streptococcus pneumoniae | 46 | 52,353 | 136 | 0.00259775 | 44,612 | 3 | Serotype 3A |
| Sample 31 | 2H | Negative | 0 | 52,000 | 0 | 0 | 0 | 0 | Not realised |
| Sample 32 | 2H | Neisseria meningitidis | 63 | 64,276 | 1107 | 0.017222603 | 1,918,857 | 85.6 | Serotype B |
| Sample 33 | 2H | Neisseria meningitidis | 51 | 92,000 | 591 | 0.006423913 | 796,395 | 37.12 | Serotype C |
| Sample 34 | 2H | Neisseria meningitidis | 4 | 228,118 | 238 | 0.00104332 | 359,034 | 16.73 | Serotype B |
| Sample 35 | 2H | Haemophilus influenzae | 192 | 84,000 | 2751 | 0.03275 | 1,869,540 | 99.06 | Non Typable |
| Sample 36 | 2H | Haemophilus influenzae | 23 | 287,552 | 561 | 0.001950951 | 378,275 | 20.66 | Serotype b |
| Sample 37 | 2H | Haemophilus influenzae | 0 | 69,418 | 5 | 7.20274E−05 | 1282 | 0.06 | Non Typable |
| Sample 38 | 2H | Staphylococcus aureus | 24 | 288,211 | 477 | 0.001655037 | 2,19,011 | 7.81 | Not realised |
| Sample 39 | 2H | Negative | 0 | 60,069 | 0 | 0 | 0 | 0 | Not realised |
| Sample 40 | 2H | Streptococcus pneumoniae | 97 | 88,376 | 1125 | 0.0127297 | 6,20,988 | 29.15 | Serotype 3A |
| Sample 41 | 2H | Neisseria meningitidis | 356 | 91,198 | 8726 | 0.095681923 | 2,054,566 | 91.29 | Serotype B |
| Sample 42 | 2H | Streptococcus agalactiae | 0 | 7409 | 5 | 0.000674855 | 573 | 0 | Serotype V |
| Sample 43 | 2H | Streptococcus pneumoniae | 0 | 74,260 | 8 | 0.00010773 | 27,083 | 1.3 | Serotype 12F |
| Sample 44 | 2H | Neisseria meningitidis | 0 | 7160 | 1638 | 0.22877095 | 1,161,716 | 51.16 | Serotype B |
| Sample 45 | 2H | Negative | Negative | 82,070 | 0 | 0 | 0 | 0 | Not realised |
| Sample 46 | 2H | Streptococcus pneumoniae | 562 | 199,270 | 6047 | 0.030345762 | 2,064,033 | 96.9 | Serotype 11A |
| Sample 47 | 2H | Streptococcus pneumoniae | 12 | 4238 | 456 | 0.107597924 | 1,011,157 | 47.5 | Serotype 3A |
| Sample 48 | 2H | Streptococcus pneumoniae | 4358 | 163,052 | 82,665 | 0.506985502 | 2,081,073 | 97.7 | Serotype 11A |
| Sample 49 | 2H | Escherichia coli | 0 | 2950 | 2 | 0.000677966 | / | / | Not realised |
| Sample 50 | 2H | Streptococcus pneumoniae | 2 | 12,370 | 14 | 0.00113177 | 4009 | 0.2 | Not realized |
| Sample 51 | 2H | Streptococcus pneumoniae | 0 | 3221 | 2,094 | 0.650108662 | 783,321 | 36.8 | Serotype 11A |
| Sample 52 | 2H | Streptococcus pneumoniae | 435 | 82,070 | 7632 | 0.092993786 | 2,028,482 | 95.23 | Serotype 3A |
Comparison between RTM and routine investigations
Two discordances in pathogen detection were noted by comparing RTM and routine investigation data. One case of H. influenzae (sample 20) detected by FilmArray® but not by RTM was eventually interpreted as a false-positive of multiplex RT-PCR,5 in agreement with a negative specific RT-PCR and culture. One case of S. pneumoniae (sample 31) was detected by multiplex RT-PCR but not by RTM. Further control by routine specific RT-PCR yielded a >35 Ct and culture remained negative. A total of 23/43 (53.5%) positive cases of bacterial meningitis diagnosed by BioFire FilmArray® failed in culture, and it was not possible to carry out an in-vitro antibiogram (Table 1). From the 16 susceptible bacteria which were identified in routine bacteriology, antibiotic susceptibility testing of cultured bacteria yielded 10/16 concordant in-vitro and in-silico antibiograms, while four bacteria (H. influenzae, S. pneumoniae, N. meningitidis and S. aureus) were in-silico resistant for beta-lactamins, macrolides and related antibiotics, penicillin A, and cefoxitin respectively (Table 1), probably due to the absence of expression of resistance despite the possession of the antibiotic resistance encoding genes. In two cultured S. pneumoniae and L. monocytogenes the in-silico antibiogram had been not realised faced to the low quantity of data <1% (Table 1). Furthermore, partial discordance was observed between in silico and in vitro antibiogram for CSF sample “1” and “2”. For both CSFs, in vitro antibiogram identified a resistant S. pneumoniae for trimethoprim and streptogramin b. However, in silico antibiogram yielded a resistant S. pneumoniae to lincomycin, quinupristin, pristinamycine, rifamycin, fluoroquinolone, aminoglycoside and phenicol for CSF sample “1”, and a resistant S. pneumoniae to lincomycin and phenicol in CSF sample “2” (Table 1). In addition, an in-silico antibiogram was successfully performed in 22 uncultured bacteria, including 14 susceptible bacteria (five S. pneumoniae, four N. meningitidis, three H. influenzae, and two S. agalactiae), one S. agalactiae was in-silico predicted resistant to erythromycin, azithromycin and spiramycin; one H. influenzae in-silico predicted resistant for amoxicillin, ampicillin, cephalothin, piperacillin, ticarcillin and one S. pneumoniae in-silico predicted resistant for erythromycin, streptogramin B, chloramphenicol and lincomycin. In addition, in-silico antibiogram could not be derived from genome sequencing following low quantity of genomic data, in five additional uncultured bacteria.
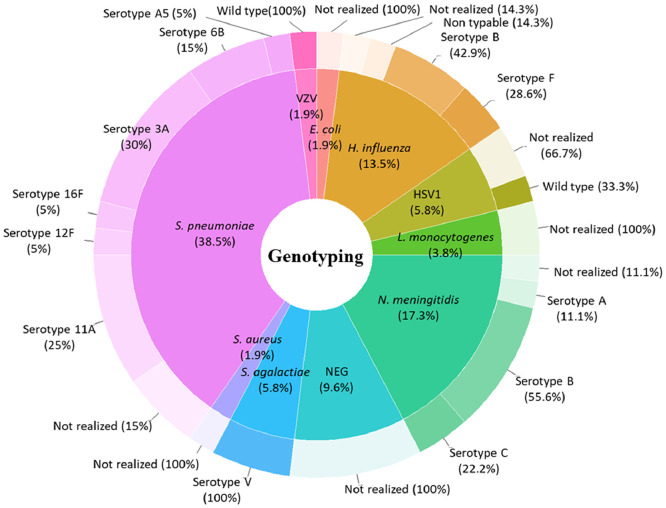
Bacteria genotyping
Genome-sequence-derived-MLST analysis yielded 13 bacterial serotypes derived from 34 MLST profiles. Sixteen genotyped S. pneumoniae yielded six serotypes (six 3A serotype, five 11A, two 6B, one 16F, one 5A and one 12F serotype), three N. meningitidis serotypes (five B serotype, two C serotype and one A serotype), three H. influenzae serotypes (three B serotype, two F serotype and one non-typable), three cases of S. agalactiae V serotype (Figure 4, Table 2). In addition, 50% of patients positive for S. pneumoniae have more than 55 years-old, with a dominance of 3A and 6B serotypes. Six out of nine N. meningitidis cases were aged 30 or under, with a dominance of the B-serotype. Four out of seven H. influenzae cases were aged 25 years or under and three were over the age of 45 years. The b-serotype was identified in patients aged 0, 45 and 65 years old, the F-serotype was identified in two patients aged 25 and 90 years old. One non-typable H. influenzae was identified in a 42-year-old patient. There was no association between bacteria serotypes, age, and gender of the patients. According to the low number of viral cases diagnosed in this series, virus genotyping was not performed.
Figure 4.
Pathogen genotyping and distribution according to the causative bacteria. Genotype investigation was performed on PubMLST database (https://pubmlst.org/) for bacteria pathogens and ViPR database (https://www.viprbrc.org/) for virus genotyping. Total of 13 bacterial serotypes identified from 34 MLST profiles: Six S. pneumoniae serotype (six 3A serotype, five 11A, four 6B, one 16F, one 5A and one 12F serotype), (six 3A serotype, five 11A, two 6B, one 16F, one 5A and one 12F serotype), three N. meningitidis serotypes (five B serotype, two C serotype and one A serotype), three H. influenzae serotypes (three B serotype, two F serotype and one non-typable), three cases of S. agalactiae V serotype. Whereas all viruses detected in this series were for wild-type serotype.
Virology data
Four viral infections were diagnosed in immunocompetent patients in this series (Table 1). Only DNA viruses were detected here, including three reactivated HSV-1 (samples 21, 22, 23), diagnosed in women aged 14 and 77 years and one 68-year-old man, all diagnosed with meningoencephalitis. In addition, wild-type VZV (https://www.viprbrc.org/) was detected in only one CSF (sample 24) collected in a seemingly immunocompetent 29-year-old woman with a past medical history of childhood VZV infection, no further recent contact with the virus including no vaccination, and no clinical zona; all data suggestive of VZV-reactivation (Table 2, Figure 4).
Discussion
We investigated RTM directly applied to left over CSF samples for the diagnosis of community-acquired meningitis at the POC laboratories in two university hospitals in southern France. Multiplex RT-PCR assays currently used routinely in the POC laboratory for this purpose only detect pieces of pathogen genome, providing detection and identification.5 This study indicated that, in addition to detection and identification, an RTM diagnostic strategy using Oxford Nanopore sequencing performed well on the diagnosis of known and non-routinely detectable pathogens in CSF samples, the antibiotic susceptibility profile, as well as their genotype.14,16,19 Moreover, cross-contamination, shown to limit the interpretation of positive multiplex assay results in several original studies and resulting meta-analysis,5,20,21 was removed by the addition of a barcoding step; as illustrated here in one case of false-positive BioFire filmArray® H. influenzae.
In contrast to multiplex RT-PCR approaches which require specific conditions and equipment,5,6,22 RTM can be implemented in a surface area of less than two square metres in a POC laboratory using simple materials and with no requirement for advanced bioinformatics knowledge. This makes RTM a useful POC diagnostic tool, based on the simplicity, rapidity and cost-effectiveness of the process.15,23,24 Additional pieces of information were added concerning bacterial genotype/serotype, which are not applicable using conventional methods limited by the pathogen genome detection and culture, failing in 22/43 CSF, which required a specific PCR target for all pathogens.5,6 In-silico antibiogram investigation allowed to detect the presence of genes encoding for antibiotic resistance, which was concordant with the in-vitro investigation in 10/16 (62.5%) of cultured CSFs. In addition, RTM detected the presence of genes encoding for antibiotic resistance further phenotypically detected in three culture-positive CSFs and four additional failed bacteria cultures, despite the low level of pathogen in the CSF. In addition in-silico analysis of RTM data relieved supplementary information about antibiotic resistance mechanism detected in one N. meningitidis by the presence of farB gene encoding for efflux pumps, which was in-vitro resistant to rifampicin.25 In addition, we found significant pathogen genotype diversity mostly represented by S. pneumoniae 3A, 11A and 6B serotypes, followed by N. meningitidis B and C-serotypes. The H. influenzae b-serotype was identified most often in this study, followed by the non-typable and H. influenzae A serotype. This enabled the real-time genomic fine and accurate surveillance of bacteria genotypes and variants circulating in southern France, based on pathogen genome sequences (Figure 4), for the definition of a new strategy of infectious disease control including vaccination, as previously described in a case of non-typable H. influenzae meningitis identified in a patient vaccinated with the b-serotype.14 In addition, this strategy successfully detected four wild type viral DNA samples in agreement with routine multiplex RT-PCR,26 validating its application for the direct investigation of DNA pathogens.
The limits we encountered reflect ways in which the method can be improved. The failure of RTM in one CSF (sample 31) which was detected positive in routine multiplex-RT-PCR indicated the need to increase RTM sensitivity for RT-PCR-detected pathogens with Ct > 35, given the higher sensitivity of the BioFire FilmArray® assay based on nested multiplex-RT-PCR.5,7 Increased sensitivity could be achieved by improving DNA extraction through an adapted automatic library preparation protocol for low pathogen levels, including microbial genome enrichment and human genome depletion.7,23,27 Also, the enlargement of the microbial panel included in the in-house database and its combination into one protocol DNA and RNA RTM is needed. This enlargement would allow for the one-shot detection of most pathogens responsible for community-acquired meningitis and meningoencephalitis, especially RNA viruses, the most frequently encountered causative agents of meningitis 28 and non-routinely diagnosed bacterial meningitis at the POC laboratory.19
Conclusion
This study goes beyond a few previous reports14,16,19 which all indicated that RTM has the potential to complement current multiplex RT-PCR assays for the rapid detection, genotyping and in silico antibiotic resistance profiling of pathogens responsible for community-acquired meningitis. This technique is already competitive in terms of time with the routine multiplex-based diagnostics in POC laboratories. Implementation of RTM as a POC diagnostic tool for life-threatening meningitis may provide real-time genomic surveillance of meningitis causative pathogen variants circulating in the study area, to define a new strategy of epidemiological control and vaccination. The authors are in the way to implement RTM in routine POC in selected situations including potential multiplex-PCR failures, based on herein reported diagnosis results along with a preliminary cost analysis and preliminary formation course for residents in medical biology (Table 3, Appendix 4). Further developments may include the application of RTM on cases of undocumented meningitis and RNA virus cases to enrich the repertoire of meningitis causative pathogens, non-routinely diagnosed in CNS diseases.
Contributors
MM experimental design, implementation of RTM workbench at the POC, software, creation of database, bioinformatics data analysis, interpretation and writing the original draft paper. QK ensured sample collection, helped with implementation of the RTM workbench at the POC, and data analysis and writing. AB, RS sample collection and clinical data. FS statistical analysis. CR, LH, and PEF conceptualisation, reagents, implementation of RTM workbench at the POC and validation. JPL and MD writing of the original draft paper, critical reviewing of the paper, validation, management of the work and funding.
All authors read and approved the final version of the manuscript
Data sharing statement
All the extracted scaffold and contigs Fasta sequences corresponding to different pathogens were submitted to GenBank NCBI, available through BioProject No. PRJEB49201.
Declaration of interests
The authors declare no conflicts of interest. In particular, the authors did not receive any contribution from any of the suppliers mentioned in this report.
Acknowledgements
Madjid Morsli is a PhD student supported by the Fondation Méditerranée Infection. This work (lab material and reagents) was supported by the French Government under the “Investissements d'Avenir “(Investments in the Future) programme managed by the Agence Nationale de la Recherche (ANR, fr: National Agency for Research) [reference: Méditerranée Infection 10-IAHU-03]. This work was also supported by the Fondation Méditerranée Infection at the IHU Méditerranée Infection and bacteriology and hygiene laboratory at Nîmes University Hospital.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104247.
Appendix. Supplementary materials
References
- 1.Beaman MH. Encephalitis: a narrative review. 2018;Box 1:1–6. doi: 10.5694/mja17.01073. [DOI] [PubMed] [Google Scholar]
- 2.Oordt-Speets AM, Bolijn R, Van Hoorn RC, Bhavsar A, Kyaw MH. Global etiology of bacterial meningitis: a systematic review and meta-analysis. PLoS One. 2018;13(6):1–16. doi: 10.1371/journal.pone.0198772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Https://apps.who.int/gb/ebwha/pdf_files/WHA73/A73_6-en.pdf. Global vaccine action plan. Seventy-third world heal assem provisional agenda item 113. 2020;31(May):B5–B31. 10.1016/j.vaccine.2013.02.015 [DOI]
- 4.Tubiana S, Varon E, Biron C, et al. Community-acquired bacterial meningitis in adults: in-hospital prognosis, long-term disability and determinants of outcome in a multicentre prospective cohort. Clin Microbiol Infect. 2020;26(9):1192–1200. doi: 10.1016/j.cmi.2019.12.020. [DOI] [PubMed] [Google Scholar]
- 5.Vincent JJ, Zandotti C, Baron S, et al. Point-of-care multiplexed diagnosis of meningitis using the FilmArray® ME panel technology. Eur J Clin Microbiol Infect Dis. 2020;39(8):1573–1580. doi: 10.1007/s10096-020-03859-y. [DOI] [PubMed] [Google Scholar]
- 6.Boudet A, Pantel A, Carles MJ, et al. A review of a 13-month period of filmarray meningitis/encephalitis panel implementation as a first-line diagnosis tool at a university hospital. PLoS One. 2019;14(10):1–14. doi: 10.1371/journal.pone.0223887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Launes C, Casas-Alba D, Fortuny C, Valero-Rello A, Cabrerizo M, Muñoz-Almagro C. Utility of filmArray meningitis/encephalitis panel during outbreak of brainstem encephalitis caused by enterovirus in Catalonia in 2016. J Clin Microbiol. 2017;55(1):336–338. doi: 10.1128/JCM.01931-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soucek DK, Dumkow LE, VanLangen KM, Jameson AP. Cost justification of the biofire filmarray meningitis/encephalitis panel versus standard of care for diagnosing meningitis in a community hospital. J Pharm Pract. 2019;32(1):36–40. doi: 10.1177/0897190017737697. [DOI] [PubMed] [Google Scholar]
- 9.Chang D, Okulicz JF, Nielsen LE, White BK. A tertiary care center's experience with novel molecular meningitis/encephalitis diagnostics and implementation with antimicrobial stewardship. Mil Med. 2018;183(1-2):e24–e27. doi: 10.1093/milmed/usx025. [DOI] [PubMed] [Google Scholar]
- 10.Moffa MA, Bremmer DN, Carr D, et al. Impact of a multiplex polymerase chain reaction assay on the clinical management of adults undergoing a lumbar puncture for suspected community-onset central nervous system infections. Antibiotics. 2020;9(6):1–8. doi: 10.3390/antibiotics9060282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tin Tin Htar M, Christopoulou D, Schmitt HJ. Pneumococcal serotype evolution in Western Europe. BMC Infect Dis. 2015;15:419. doi: 10.1186/s12879-015-1147-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van de Beek D, Brouwer M, Hasbun R, Koedel U, Whitney CG, Wijdicks E. Community-acquired bacterial meningitis. Nat Publ Gr. 2016;2 doi: 10.1038/nrdp.2016.74. [DOI] [PubMed] [Google Scholar]
- 13.Massenet D, Birguel J, Azowé F, et al. Epidemiologic pattern of meningococcal meningitis in northern Cameroon in 2007–2010:contribution of PCR-enhanced surveillance. Pathog Glob Health. 2013;107:15–20. doi: 10.1179/2047773212Y.0000000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morsli M, Kerharo Q, Delerce J, Roche P, Troude L, Drancourt M. Haemophilus influenzae meningitis direct diagnosis by metagenomic next-generation sequencing: a case report. Published online 2021:3–7. [DOI] [PMC free article] [PubMed]
- 15.Gu W, Deng X, Lee M, et al. Rapid pathogen detection by metagenomic next-generation sequencing of infected body fluids. Nat Med. 2021;27(1):115–124. doi: 10.1038/s41591-020-1105-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morsli M, Kerharo Q, Amrane S, Parola P, Fournier PE, Drancourt M. Real-time whole genome sequencing direct diagnosis of Streptococcus pneumoniae meningitis: a case report. J Infect. 2021;10(01634453):14–16. doi: 10.1016/j.jinf.2021.10.002. [DOI] [PubMed] [Google Scholar]
- 17.Wilson MR, Sample HA, Zorn KC, et al. Clinical metagenomic sequencing for diagnosis of meningitis and encephalitis. N Engl J Med. 2019;380(24):2327–2340. doi: 10.1056/NEJMoa1803396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leggett RM, Alcon-Giner C, Heavens D, et al. Rapid MinION profiling of preterm microbiota and antimicrobial-resistant pathogens. Nat Microbiol. 2020;5(3):430–442. doi: 10.1038/s41564-019-0626-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morsli M, Bechah Y, Coulibaly O, et al. Direct diagnosis of Pastuerella multocida meningitis using next-generation sequencing. Lancet Microbe. 2021;5247(21):5247. doi: 10.1016/s2666-5247(21)00277-9. [DOI] [PubMed] [Google Scholar]
- 20.Tansarli GS, Chapin KC. Diagnostic test accuracy of the BioFire® FilmArray® meningitis/encephalitis panel: a systematic review and meta-analysis. Clin Microbiol Infect. 2020;26(3):281–290. doi: 10.1016/j.cmi.2019.11.016. [DOI] [PubMed] [Google Scholar]
- 21.Leber AL, Everhart K, Balada-Llasat JM, et al. Multicenter evaluation of biofire filmarray meningitis/encephalitis panel for detection of bacteria, viruses, and yeast in cerebrospinal fluid specimens. J Clin Microbiol. 2016;54(9):2251–2261. doi: 10.1128/JCM.00730-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drancourt M, Michel-Lepage A, Boyer S, Raoult D. The point-of-care laboratory in clinical microbiology. Clin Microbiol Rev. 2016;29(3):429–447. doi: 10.1128/CMR.00090-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng X, Achari A, Federman S, et al. Metagenomic sequencing with spiked primer enrichment for viral diagnostics and genomic surveillance. Nat Microbiol. 2020;5(3):443–454. doi: 10.1038/s41564-019-0637-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grumaz C, Hoffmann A, Vainshtein Y, et al. Rapid next-generation sequencing–based diagnostics of bacteremia in septic patients. J Mol Diagn. 2020;22(3):405–418. doi: 10.1016/j.jmoldx.2019.12.006. [DOI] [PubMed] [Google Scholar]
- 25.Shafer WM, Veal WL, Lee EH, Zarantonelli L, Balthazar JT, Rouquette C. Genetic organization and regulation of antimicrobial efflux systems possessed by Neisseria gonorrhoeae and Neisseria meningitidis. J Mol Microbiol Biotechnol. 2001;3(2):219–224. [PubMed] [Google Scholar]
- 26.Ramachandran PS, Wilson MR, Catho G, et al. Meningitis caused by the live varicella vaccine virus: metagenomic next generation sequencing, immunology exome sequencing and cytokine multiplex profiling. Viruses. 2021;13(11):2286. doi: 10.3390/v13112286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oechslin CP, Lenz N, Liechti N, et al. Limited correlation of shotgun metagenomics following host depletion and routine diagnostics for viruses and bacteria in low concentrated surrogate and clinical samples. Front Cell Infect Microbiol. 2018;8:375. doi: 10.3389/fcimb.2018.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Broberg EK, Simone B, Jansa J, et al. The Eu/Eea Member StateContributors. Upsurge in echovirus 30 detections in five EU/EEA countries, April to September, 2018. Euro Surveill. 2018;23(44):1800537. doi: 10.2807/1560-7917.ES.2018.23.44.1800537. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.