Abstract
Mutations in the Membrane-type frizzled related protein (Mfrp) gene results in an early-onset retinal degeneration associated with retinitis pigmentosa, microphthalmia, optic disc drusen and foveal schisis. In the current study, a previously characterized mouse model of human retinal degeneration carrying homozygous c.498_499insC mutations in Mfrp (MfrpKI/KI) was used. Patients carrying this mutation have retinal degeneration at an early age. The model demonstrates subretinal deposits and develops early-onset photoreceptor degeneration. We observed large subretinal deposits in MfrpKI/KI mice which were strongly CD68 positive and co-localized with autofluorescent spots. Single cell RNA sequencing of MfrpKI/KI mice retinal microglia showed a significantly higher number of pan-macrophage marker Iba-1 and F4/80 positive cells with increased expression of activation marker (CD68) and lowered microglial homeostatic markers (TMEM119, P2ry13, P2ry13, Siglech) compared with wild type mice confirming microglial activation as observed in retinal immunostaining showing microglia activation in subretinal region. Trajectory analysis identified a small cluster of microglial cells with activation transcriptomic signatures that could represent a subretinal microglia population in MfrpKI/KI mice expressing higher levels of APOE. We validated these findings using immunofluorescence staining of retinal cryosections and found a significantly higher number of subretinal Iba-1/ApoE positive microglia in MfrpKI/KI mice with some subretinal microglia also expressing lowered levels of microglial homeostatic marker TMEM119, confirming microglial origin. In summary, we confirm that MfrpKI/KI mice carrying the c.498_499insC mutation had a significantly higher population of activated microglia in their retina with distinct subsets of subretinal microglia. Further, studies are required to confirm whether the association of increased subretinal microglia in MfrpKI/KI mice are causal in degeneration.
Subject terms: Hereditary eye disease, Retinal diseases
Introduction
Microglia, are the primary immune cell of the retina and central nervous system (CNS)1. In healthy retina, microglia are primarily located in the inner plexiform layer (IPL) and the outer plexiform layer (OPL) as well as in ganglion cell layer (GCL) where they assist retinal homeostasis and immune surveillance. Changes in the retinal microenvironment can induce rapid microglial activation with release of pro-inflammatory cytokines, neurotoxic molecules, complement proteins and free radicals such as oxygen free radicals (ROS)2–4. Retinal degeneration, resulting from inherited retinal dystrophies (IRDs), has also been shown to trigger microglial activation5,6. IRDs are a heterogeneous group of diseases resulting from genetic changes causing retinal degeneration or dysfunction7. These diseases are among the most common cause of blind registrations in developed countries8. Recent studies from animal models of IRD have demonstrated that microglia play a key role in photoreceptor degeneration, either directly, by phagocytosing cells, or indirectly, by secreting cytokines which induce photoreceptor apoptosis4–6,9,10. Membrane-type frizzled related protein (MFRP) is a transmembrane receptor protein expressed primarily by the retinal pigment epithelial (RPE) and ciliary body in the eye and is expressed in the apical microvilli of RPE. In humans, mutations in the Mfrp gene result in an early-onset retinitis pigmentosa associated with microphthalmia, foveoschisis and optic disc drusen in patients. Animal models of Mfrp associated retinal degeneration (MARD) have been developed which demonstrate many features of human disease including progressive early-onset retinal degeneration, low electrophysiological responses and microphthalmia11–13.
Interestingly, these mice also develop early-onset autofluorescent retinal spots which are thought to be the result of subretinal deposits14. Studies have demonstrated subretinal immune cell infiltration showing positive staining for macrophage15,16. Recent reports suggest that activation of resident microglia and monocyte derived macrophages could both contribute to subretinal microglia. Resident microglia can migrate to the subretinal region as an adaptive mechanism in response to ongoing damage in retinal degeneration models4,17. Some studies have investigated the activation of subretinal microglia in acute-onset retinal degeneration models18. On the other hand microglial depletion agent colony-stimulating factor 1 receptor (CSF1R) inhibitor was used to deplete resident microglia, showing that monocyte derived macrophages are recruited to the retina, including the subretina to exacerbate cone death in the rd10 model19. However, there has been relatively little exploration of microglial activation and migration in genetic models of human retinal degeneration. In the current study, we used a recently described MARD mouse model carrying homozygous c.498_499insC mutations13, to better understand the changes in retinal microglia in these mice. We hypothesize that in this early-onset IRD, resident retinal microglia are activated and are recruited to the subretina. To test our hypothesis, we performed single cell RNA sequencing (scRNA-seq) and validated our findings using immunostaining, coupled with in vivo and ex vivo retinal imaging.
Results
Retinal imaging of patients and mice homozygous for c.498_499insC mutations demonstrated discrete white spots and localized to the subretina
Four siblings with homozygous c.498_499insC Mfrp mutations were previously found to have retinitis pigmentosa, foveoschisis, microphthalmos with hyperopia, and optic disc drusen consistent with MARD. The original imaging studies in this family had primarily focused on central color fundus and electrophysiological changes13. However, it was not clear whether the cases also developed a discrete spot phenotype similar to mouse models of MARD. As a result, we performed ultra-wide field color fundus imaging (Fig. 1A–C) which identified discrete creamy-white spots in the mid-periphery and far-periphery of patients (Supplementary Fig. 1). However, subretinal changes were also different from that seen in mice with spots coalesced into a more reticular pattern and not covering the whole retina but located in the far and mid-peripheral retina. Additionally, the spots did not autofluoresce. These studies highlighted a possible subretinal location of deposits in the few cases where occasional spots were found more centrally in the parafovea and peripheral macula and were therefore accessible to SD-OCT imaging.
Figure 1.
Imaging from a 53-year-old female patient with homozygous c.498_499insC mutations in MFRP. (A,B) Ultra-widefield pseudo-color imaging from (A) right, (B) left eyes demonstrate signs of retinal degeneration with symmetrical retinal pigmentation and atrophy with relative sparing of central macula. Additionally, the imaging demonstrates multiple creamy white spots in the far and mid-periphery bilaterally (arrowheads). (C) Spectral domain optical coherence tomography aligned with near infrared imaging isolates one of the white spots in the macula (white arrowhead) and demonstrates a subretinal location of this spot (black arrowhead). (D,E) SLO images showing multiple autofluorescent spots in the fundus of WT and MfrpKI/KI mice retina.
As this was a novel finding, in order to see that the spots in patients were not mutation specific, and limited to patients harboring the c.498_499insC Mfrp mutation, we similarly performed ultra-widefield imaging and SD-OCT in a 44-year-old male patient carrying c.523C > T, p.(Gln175*) and c.649G > A, p.(Gly217Arg) variants in Mfrp (Supplementary Table A) who also exhibited classical features of MARD including retinal degeneration, nanophthalmos and foveoschisis. Imaging identified discrete creamy white spots which appeared in a subretinal location. The c.523C > T, p.(Gln175*) mutation has been reported previously and identified as pathogenic. There are 10 individuals heterozygous for this variant in gnomAD. The Mfrp c.523C > T, p.(Gln175*) variant generates a premature stop codon in exon 5 (out of a total of 15 exons) and is predicted to lead to loss of normal protein function and is potentially novel. This variant is absent in gnomAD. MFRP c.649G > A, p.(Gly217Arg) replaces a moderately conserved amino acid glycine (Gly) to arginine (Arg) at protein position 217 (exon 6). All in silico tools utilized (PolyPhen, SIFT, Mutationtaster) predicted this variant to be damaging to protein structure and function, however as the variant has not been reported before it was classified as a variant of undetermined significance. Family member testing is under way to help further variant classification and phase analysis. Overall, the pattern of the dystrophy is very similar between the new patient and the previously described patients with creamy/white spots mainly seen in the mid to far periphery, suggesting that the subretinal white/creamy spot phenotype is not restricted to a particular mutation in MFRP (Supplementary Fig. 2). Fundus imaging in 4–5-month-old MfrpKI/KI mice had shown regular dispersed autofluorescent spots throughout the retina which were co-localized with a subretinal location on SD-OCT. The white spots in humans had a different distribution, as described above, in that there were fewer present in the central macula where fewer rods are known to be found. In the mouse the AF spots were found throughout the retina. The mouse retina is rod dominant with rods distributed throughout the retina with no macula. This suggests that the spots may be associated with a rod distribution13 (Fig. 1D,E).
Immunostaining of retinal cross sections and flow cytometry confirms significant immune infiltration in MfrpKI/KI mice retina
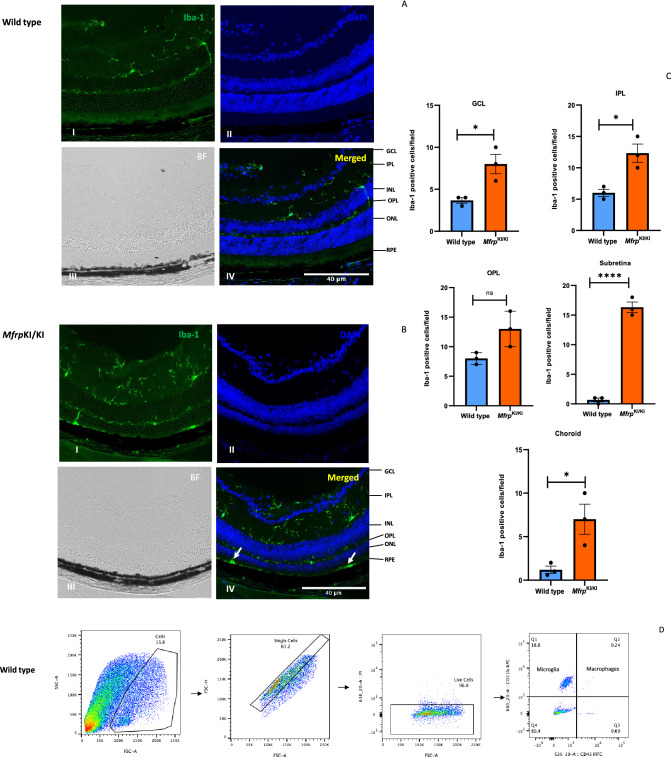
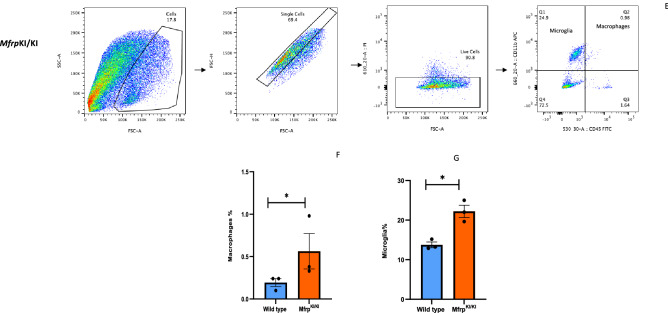
Retinal cryosections, which were immunostained for Iba-1, demonstrated large cells strongly positive for Iba-1 in the subretina. Iba-1 positive cells were also present in other layers in the MfrpKI/KI mice (Fig. 2A,B). We compared the microglial numbers between WT and MfrpKI/KI and observed significantly greater numbers of microglia in almost all the retinal layers in MfrpKI/KI mice, particularly in the subretina (p < 0.0001), GCL (p < 0.05), IPL (p < 0.05), choroid (p < 0.05) (Fig. 2C). Immune cells were also analyzed in retinal cell suspension. Using flow cytometry, microglia were counted as CD11bhighCD45low and monocyte-derived macrophages as CD11bhighCD45high, as described previously20–22 (Fig. 2D,E). We observed a significant increase in microglia (p < 0.05) and monocyte-derived macrophages, in MfrpKI/KI mice retina. (p < 0.05) (Fig. 2F,G).
Figure 2.
Immunostaining and flow cytometry for microglial and macrophage markers. (A,B) Retinal cryosections stained with Iba-1 antibody showed large numbers of positive cells in MfrpKI/KI mice retina (I-Iba-1, II-DAPI, III-Brightfield (BF), IV-Merged) (B) particularly in the subretinal region (white arrows) compared with WT mice retinal cryosections (I-Iba-1, II-DAPI, III-Brightfield (BF), IV-Merged) (A). (C) Significantly higher numbers of microglia were present in MfrpKI/KI mice retina when compared to WT mice retina in the subretina (****p < 0.0001), GCL (*p < 0.05), IPL (*p < 0.05) and choroid (*p < 0.05) region (n = 3). (D,E) Flow cytometry of immune cells from mouse retina was performed. First, single cells were gated, followed by live cells. Microglia were gated as CD11bhigh CD45low cells (Q1) and macrophages as CD11bhighCD45high (Q2). (F,G) There were significantly increased numbers of CD11bhigh CD45low microglial cells (*p < 0.05) (n = 6) and monocyte-derived CD11bhighCD45high macrophages in MfrpKI/KI retina compared with WT mouse retina (*p < 0.05) (n = 6) (GCL ganglion cell layer, IPL inner plexiform layer, INL inner nuclear layer, OPL outer plexiform layer, ONL outer plexiform layer, RPE retinal pigment epithelial cell layer).
Subretinal microglia accumulate in MfrpKI/KI mouse retina demonstrating strong activation marker expression
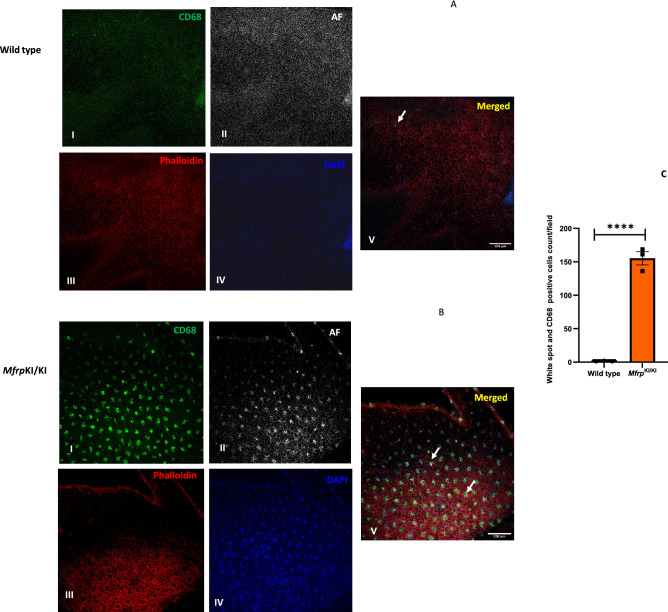
Microglia in WT retina had a typical ramified appearance, which has been described for quiescent resident microglial phenotypes, in both the retina (Fig. 2A–C) and the brain23,24. However, a few ramified microglia were still present in MfrpKI/KI retina, mostly in the inner retina, which was consistent with our scRNA-seq cluster analysis which identified a transcriptionally quiescent resident-type microglia cluster in MfrpKI/KI mice. Immunostaining also demonstrated significantly greater numbers of CD68 and Iba-1 positive cells in MfrpKI/KI mice retina (Iba-1, p < 0.01, CD68, p < 0.01) compared with WT mouse retina (Fig. 3A,B). There were significantly greater large, round and strongly CD68 positive cells localized to the subretinal region (p < 0.0001) in MfrpKI/KI mice retina compared with WT mice (Fig. 3C–E). In WT mice, microglia were mostly present in the IPL and OPL, whereas in MfrpKI/KI microglia were present in the GCL, IPL, OPL, choroid layers and subretina (Figs. 2A, 3A,B). To further investigate the autofluorescent spot phenotype in the MfrpKI/KI mice model, and to confirm that microglia were responsible for this in vivo imaging finding, retinal wholemounts of both WT type and MfrpKI/KI mice were prepared. In the retinal wholemounts we sought to understand whether the autofluorescent spots co-localized with subretinal deposits which were observed during in vivo imaging. Autofluorescent spots on retinal wholemounts were co-localized with CD68 positive activated microglia in MfrpKI/KI mice retina. The findings confirm an immune component to the autofluorescent spots, which were significantly (p < 0.0001) greater in number in MfrpKI/KI mice retina when compared with WT type retina (Fig. 4A–C). Taken together, the findings in these set of studies identified activated subretinal microglia which correspond with autofluorescent spots, similar to the ones seen on fundus autofluorescence.
Figure 3.
Immunohistochemistry studies for CD68 and Iba-1 positive cells in retinal cryosections (A,B) Iba-1(green) and CD68 (red) positive cells were prominent and strongly positive subretinal region (white arrows) and with occasional cells with co-localized immunostaining in the OPL and IPL in MfrpKI/KI retina, whereas in WT mice, microglia with Iba-1 cells were confined to the OPL and IPL and completely absent from the subretinal region (I-Iba-1, II-CD68, III-DAPI, IV-Brightfield (BF), V-Merged) and had few cells with CD68 immunostaining. (C,D) Iba-1 and CD68 count in both groups (**p < 0.01 and **p < 0.01) (n = 3). (E) The number of subretinal microglia were significantly higher in MfrpKI/KI retina compared to WT retina (****p < 0.0001) (n = 3) (GCL ganglion cell layer, IPL inner plexiform layer, INL inner nuclear layer, OPL outer plexiform layer, ONL outer plexiform layer, RPE retinal pigment epithelial cell layer).
Figure 4.
Confirmation of the microglial origin of autofluorescent spots in Mfrp KI/KI mice retina. (A,B) Retinal wholemount immunostaining for CD68 positive cells and autofluorescent spots using confocal microscopy with × 20 lens (Right panel), enlarged view of individual spots (I-CD68, II-AF (Autofluorescence), III-phalloidin (for actin, staining) IV-DAPI, V-Merged), (Left panel) show that (C) a significantly higher number of autofluorescent spots were present in MfrpKI/KI mice retina (****p < 0.0001) (n = 3).
Microglia in MfrpKI/KI mice retina demonstrate increased numbers of cells with an active expression profile
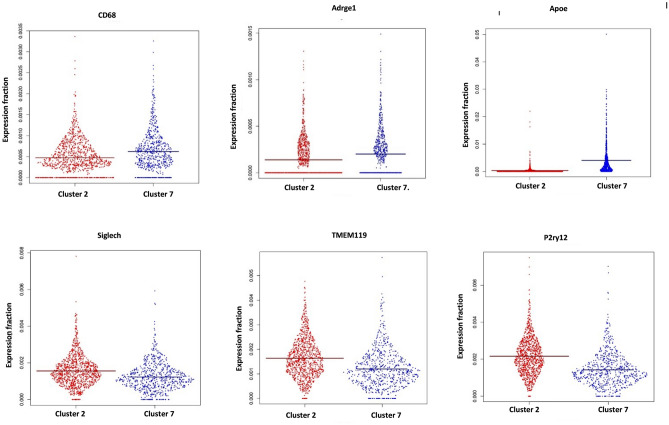
In order to better understand the expression profile of retinal microglia in MfrpKI/KI mice, we performed scRNA-seq of CD11bhighCD45low FACS sorted cells from age-matched MfrpKI/KI and WT type mice retina. Clustering analysis of this data from both MfrpKI/KI and WT type mice retina resulted in the separation of 20 microglial clusters, demonstrating a heterogeneous population numbered from 1 to 20 with cell numbers ranging from 20 to 1000 cells. We first compared MfrpKI/KI and WT type gene expression at a local false discovery rate (lfdr) < 0.1 (Fig. 5A–E) and identified significantly increased numbers of microglia expressing microglial activation markers including CD68 and an increase in other microglial markers such as F4/80, Iba-1 and decreased numbers of microglia expressing microglial homeostatic markers, associated with quiescent microglia, including TMEM119, Siglech, P2ry13 and P2ry12 in MfrpKI/KI retina (Fig. 5F) (Supplementary Table B). There was also increased expression of inflammatory markers in MfrpKI/KI as compared to WT (Supplementary Table C). Two clusters contained the highest number of cells; clusters 2 and 7. To identify the phenotype of cells in these clusters we looked at their composition more closely. Cluster 2 was composed of 80.3% WT microglia while cluster 7 was composed of 88.2% MfrpKI/KI microglia. We compared cluster 2 and 7 gene expression and found that cluster 2 had a higher number of cells expressing homeostatic microglial markers including TMEM119 (fold change = − 1.36, lfdr = 1.25E−08), Siglech (fold change = − 1.27, lfdr = 7.97E−05) and P2ry12 (fold change = − 1.51, lfdr = 1.07E−18) and a lower number of cells expressing microglial activation markers including CD68 (fold change = 1.31, lfdr = 0.000526) and microglia marker F4/80 (fold change = 1.44, lfdr = 0.013281). Hence, cluster 2 appeared to be composed of quiescent retinal microglia, which is likely to explain the greater proportion of cells in cluster 2 from WT mice retina. In contrast, expression characteristics for cluster 7 suggested activated microglia, which is likely to explain why cells in cluster 7 were predominantly derived from MfrpKI/KI mice retina (Fig. 5G–I) (Supplementary Tables D, E).
Figure 5.
Single cell RNA sequencing analysis. tSNE plots and clustering of microglia from WT (1345 Cells) and MfrpKI/KI (2059 cells) retina. (A) tSNE plot of microglia from WT and MfrpKI/KI mouse retina. (B) tSNE plot dividing microglia from WT and MfrpKI/KI mouse retina into clusters. (C) Histogram representing microglia divided into clusters from both groups, (Black-WT cells, Gray-MfrpKI/KI. (D) WT mouse retina microglia tSNE plot with clusters. (E) MfrpKI/KI mouse retina microglia tSNE plot with clusters. Heat maps of generated by “pheatmap” package in R program. (F) Differentially expressed microglial markers in WT and MfrpKI/KI mice. (G) Microglial activation, (H) homeostatic markers after comparing cluster 2 and 7 (Blue = Downregulation, Red = upregulation). (I) Violin plots showing expression of CD68, F4/80, ApoE (microglial activation markers) Siglech, Tmem119, P2ry12 (homeostatic markers) lfdr < 0.1.
MfrpKI/KI retinal microglia had a small cluster of cells with a unique gene signature resembling subretinal phenotypes
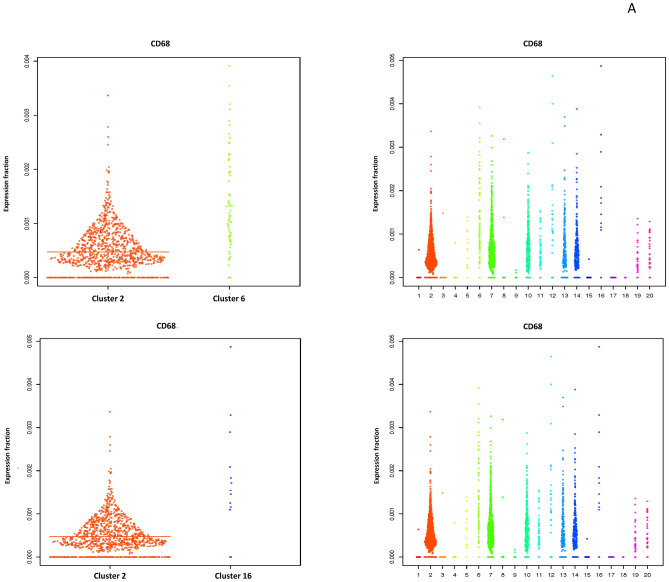
We next tried to isolate a cluster of cells which represented the activated subretinal macrophages that we identified in our retinal immunohistochemistry studies. Since we found that cluster 7 had greater numbers of activated microglia in the combined MfrpKI/KI and WT analysis (Fig. 5C), we hypothesized that subretinal cells would generate a smaller activated cell population, from our retinal wholemount studies. We first confirmed the characteristics of the two largest clusters, cluster 2 and 7. Cluster 7 showed an active microglial signature present in exclusively in Mfrp KIKI (cluster 7 CD68 (fold change = 1.31, lfdr = 0.000526) and cluster 2, mostly inactive microglia present in WT (Fig. 5) (Supplementary Tables D, E). We next compared cluster 2, the quiescent microglial cluster, with all other 19 clusters with a cut off lfdr < 0.1. We looked initially for clusters with a high number of cells expressing CD68 and found that cluster 6 and 16 had high CD68 expression (cluster 6 (CD68, fold change = 2.78, lfdr = 3.32E−10), and 16 (CD68, fold change = 3.26, lfdr = 0.0015) along with other activation markers and homeostatic static markers downregulation within lfdr cut off (Fig. 6A) (Supplementary Tables F, G). Among other clusters which were noted to have a CD68 expression, cluster 5 had lfdr value of 0.1 (CD68, fold change = 1.30 lfdr = 0.10), cluster 10 (CD68, fold change = 1.2 lfdr = 0.4) and 12 (CD68, fold change = 1.5 lfdr = 1) had increased CD68 expression but non-significant lfdr values and lastly cluster 11,13,14 had baseline CD68 expression (fold change = 1), clusters 1, 3, 4, 9,15 had low expression of CD68 with lfdr < 0.1 representing inactive clusters and cluster 8, 19, 20 had low expression of CD68 with a non-significant lfdr > 0.1.
Figure 6.
Violin plots: (A) comparing CD68 expression cluster 2 with cluster 6, cluster 2 with cluster 16. (B) Comparing key genes expressed in clusters 6 and 7.
As we aimed to identify activated microglial clusters in Mfrp KI/KI mice which may represent subretinal microglia we focused on clusters 6 and 16 and investigated microglial activation and homeostatic markers. We observed that cluster 6 and 16 had cells with increased expression of many myeloid markers. We also analyzed cluster 10, 13 and 14 in detail looking closely with respect to other marker expression (Iba-1 and F4/80) (Supplementary Table H). We found that cluster 10 and 14 were are also active clusters, with a large proportion of cells from Mfrp KIKI retina. We further tried to analyze small candidate in detail and compared cluster 6 with 40 cells and cluster 7 representing largest activated retinal microglia, we compared cluster 7 with cluster 6, one of the active small clusters. We found that cluster 7 had cells having higher expression of resident markers and lowered activation markers compared to cluster 6 TMEM119 (fold change = 1.95, lfdr = 0.061), Siglech (fold change = 2.94, lfdr = 8.16E−08) and P2ry12 (fold change = 1.99, lfdr = 0.005), CD68 (fold change = − 2.12 lfdr = 1.23E−05) and ApoE (fold change = − 8.27, lfdr = 2.20E−17) (Fig. 6B) Supplementary Table I). These results suggest cluster 6 was one of clusters which could represent subretinal microglia, however other clusters cluster 7, 10, 14 and 16 could also share these active populations and there was a possibility that the subretinal population was not represented by a single cluster but a combination of these clusters representing an active subretinal microglial population with transient states and heterogeneity.
Subretinal microglia in MfrpKI/KI mice have a well-defined activation trajectory
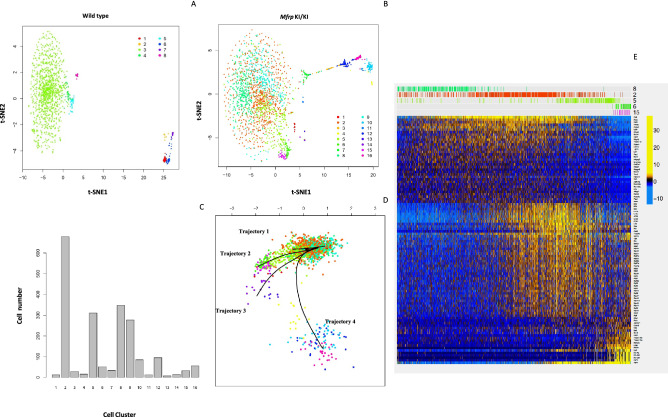
After identifying a candidate cluster of subretinal microglia, we next aimed to better understand activation stages of retinal microglia in MfrpKI/KI mice. We performed clustering of WT and MfrpKI/K retinal microglia populations separately. A total of 8 and 16 clusters were identified in WT and MfrpKI/KI mice respectively demonstrating increased heterogeneity in microglia in the MfrpKI/KI mice (Fig. 7A–C).Microglia from both groups were analyzed in a multidimensional coordinate space defined by relative expression of 18,727 genes describing the state of each cell. We found that MfrpKI/KI microglial cells were less localized, broader and probability density whereas WT microglial cells had a localized probability density (Supplementary Fig. 3A). Further, entropy of distribution function analysis of microglial cells from MfrpKI/KI mice was significantly greater than that from WT mice calculated by creating a random bootstrap ensemble of WT and MfrpKI/KI samples of equal size (Supplementary Fig. 3B).
Figure 7.
Separate analysis of Microglia from MfrpKI/KI and WT mouse retina and trajectory analysis. Microglia from MfrpKI/KI and WT mouse retina were clustered separately (A) tSNE plot of microglia from WT mouse retina, (B) tSNE plot of microglia from MfrpKI/KI mouse retina. (C) Histogram of clusters generated from MfrpKI/KI retinal microglia. (D) Trajectory analysis of MfrpKI/KI microglia demonstrating four trajectories 1 to 4, Trajectory 1: 8 2 5 4 10 16 12, Trajectory 2: 8 2 9 1 14 13, Trajectory 3: 8 2 5 6 15, Trajectory 4: 8 2 7. (E) Heat map, generated by “pheatmap” package in R program, shows gradual changes in gene expression beginning with cluster 8 and ending in cluster 15 in Trajectory 3.
Transcriptional signature analysis found that retinal microglia in cluster 8 from MfrpKI/KI retina contained mostly resident quiescent microglia like cluster 2 in the original combined WT and MfrpKI/KI analysis. Trajectory analysis was performed, setting cluster 8 as the baseline, to understand potential microglial activation. Analysis resulted in the identification of 4 distinct trajectories labelled as Trajectory 1, Trajectory 2, Trajectory 3 and Trajectory 4. As cluster 8 represented the resident microglia, each trajectory diverged from cluster 8 to the four different branches (Fig. 7D). We analyzed changes in gene expression along each trajectory by serially comparing clusters in each trajectory. Of the four trajectories, only trajectory 3 demonstrated reduced numbers of cells expressing microglial homeostatic markers from cluster 8 and ending at cluster 15, TMEM119 (fold change = − 35.77, lfdr = 2.42E−19), Siglech (fold change = − 1.74, lfdr = 1.35E−07), P2ry13 (fold change = − 5.65, lfdr = 0.0008) and P2ry12 (fold change = − 10.16, lfdr = 4.12E-21). Additionally, the serial analysis showed increasing numbers of cells expressing activation markers, previously found in subretinal cells4, along the trajectory. Lyz2 (fold change = 19.07, lfdr = 8.06E−54, Lgals1(fold change = 1770.32 lfdr = 4.24E−07), CD74 (fold change = 167.25 lfdr = 9.40E−09) and F4/80 (fold change = 3.13, lfdr = 5.98E−07) increased (Fig. 7E, Supplementary Tables J, K). Particularly, there was a marked increase in the number of cells expressing ApoE (fold change = 38.45 lfdr = 2.77E−45). We concluded that cluster 15 in the MfrpKI/KI only analysis and cluster 6 in the combined comparison, each with approximately 40 cells, could represent cells which played a part of the subretinal microglial population, consistent with earlier reports of subretinal cells in RD25,26. These results suggest that activated subretinal microglia may result from a stepwise activation of resident retinal microglia in MfrpKI/KI mice retina. These microglial clusters may represent terminally active subretinal microglia with a loss of homeostatic microglial markers but retaining activation marker expression, as has been reported previously4.
Subretinal microglia in MfrpKI/KI retina are activated resident cells strongly express APOE
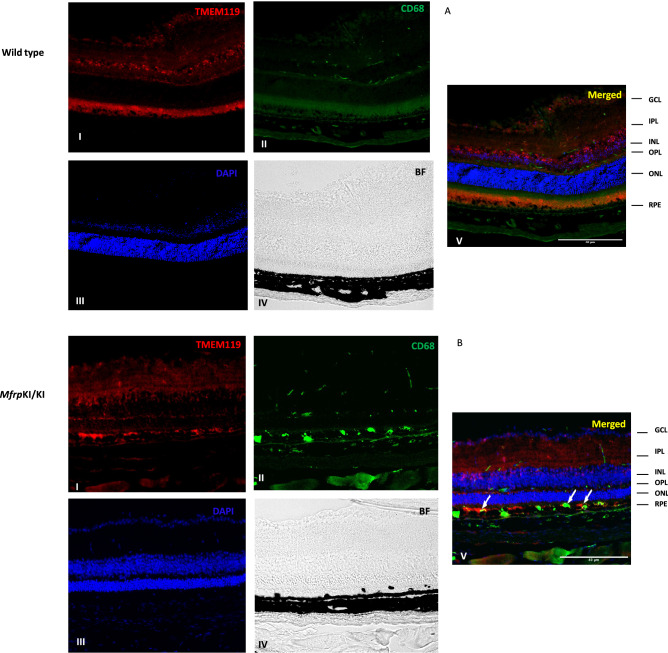
We observed a marked increase in the number of cells expressing microglial activation markers in the scRNA-seq analysis of microglia derived from MfrpKI/KI mice retina, we aimed to validate these transcriptional findings using immunofluorescence in retinal cryosections. Since the trajectory analysis supported a microglial origin of our subretinal candidate cluster, we next performed a validation. The scRNA-seq data had shown that APOE was highly expressed in the microglia, in our candidate subretinal cluster 6. We found significantly higher number of Iba-1/ApoE positive cells in the MfrpKI/KI mice retina compared to WT mice retinal cryosections, when analyzed in all layers (p < 0.05), and we also found APOE and Iba-1 co-expressing microglia were present mostly in the subretina (p < 0.001) (Fig. 8A–D) however, not all microglia in the subretina expressed ApoE suggesting heterogeneity in the sub retina. We also found that a significantly greater number of F4/80 positive cells were present in the GCL, IPL, OPL and subretina in MfrpKI/KI retinal cryosections when compared to WT retinal cryosections (p < 0.05) (Supplementary Figs. 5, 6) validating our scRNA-seq data. In order to try to confirm that the subretinal cells include cells of microglial origin, we performed immunostaining for resident microglia marker TMEM119 and activation marker CD68. We observed lowered expression of TMEM119 in MfrpKI/KI mice in the GCL, IPL and OPL layers but also found subretinal microglial staining for TMEM119 and CD68 (Fig. 9A,B) suggesting a resident microglial source for some of the subretinal cells.
Figure 8.
Iba-1 and APOE immunostaining (A,B) (I-Iba-1, II-APOE, III-DAPI, IV-Brightfield (BF),V-Merged) was performed, microglial expressing APOE were higher in MfrpKI/KI mice retina compared with WT retina in all layers (C) (*p < 0.01) (D) in the subretina APOE positive microglia were significantly greater in MfrpKI/KI (***p < 0.001) (n = 3) observed under fluorescence microscope using a × 20 lens and particularly found in the GCL, IPL, INL, OPL and in the subretina (white arrows) (GCL ganglion cell layer, IPL inner plexiform layer, INL inner nuclear layer, OPL outer plexiform layer, ONL outer plexiform layer, RPE retinal pigment epithelial cell layer).
Figure 9.
TMEM119 and CD68 immunostaining (A&B) (I-TMEM119-1, II-CD68, III-DAPI, IV-Brightfield (BF), V-Merged) was performed, microglial expressing TMEM119 were higher in WT mice retina compared with MfrpKI/KI. In subretinal region TMEM119 and CD68 positive microglia were particularly present in MfrpKI/KI which completely absent from WT (white arrows) (GCL ganglion cell layer, IPL inner plexiform layer, INL inner nuclear layer, OPL outer plexiform layer, ONL outer plexiform layer, RPE retinal pigment epithelial cell layer).
Discussion
In the present study, we used a mouse model with homozygous c.498_499insC mutations in Mfrp. This pathogenic variant has previously been reported in human cases of MARD13. The model shares features of clinical pathology seen in human subjects with homozygous c.498_499InsC. These features include nanophthalmos and rod/cone degeneration. Another key features of Mfrp mouse models is the presence of white/creamy spots in retina (Fig. 1A–C)27,28. We showed that human cases carrying these mutations also had a distribution of autofluorescent spots in the fundus, using ultra-widefield imaging (Supplementary Fig. 1). Other mutations in MFRP have been reported to cause multiple retinal alterations29. In the present study, we also described the findings in a 44-year-old male with nanophthalmos associated with high hypermetropia and angle closure glaucoma, who had two different variants in MFRP, c.523C > T, p. (Gln175*) and c.649G > A, p. (Gly217Arg), but also demonstrated similar subretinal white/creamy spots suggesting that this finding was not limited to the c.498_499InsC mutation (Supplementary Fig. 2, Supplementary Table A). To our knowledge this is the first report of this white spot phenotype in MARD. This is likely due the relatively recent availability of ultra-widefield imaging.
We also found autofluorescent spots in the fundus of our MfrpKI/KI mice at 4 months of age. The autofluorescent spots had a subretinal localization on SD-OCT imaging (Fig. 1D,E) and were also seen on retinal wholemounts co-localizing with CD68 microglial activation marker (Fig. 4A–C). These findings were consistent with another Mfrp mouse model, homozygous for c.174delG mutations11 which also have autofluorescent spots in retinal whole mounts, although immune cells in the RPE whole mounts were not co-localized with autofluorescent spots. Another Mfrp mouse model, the Rd6 mouse, has been reported to have accumulated phagocytic cells present in the subretina at an early disease onset, as identified by transmission electron microscopy12. Similar reports of F4/80 positive microglia contributing to autofluorescent spots have been reported in retinal rosettes of mice with other gene mutations30. The rd7 mouse model carries homozygous R311Q mutations in the NR2E3 gene and has been shown to have subretinal microglia31. It is unclear why the immune cells are found in the subretinal region in the present c.498_499insC MfrpKI/KI model. Immune cells may have been initially recruited to the subretinal region, because of an innate immune response to photoreceptor degeneration. Resident microglia are the major immune cell present in the retina responsible for crucial immune responses to any changes or alterations in the retinal microenvironment. It has been reported that resident microglia or monocyte-derived macrophages can move towards degenerating photoreceptors, the subretina or RPE32. Recruitment of microglia to the outer retinal layers, as part of an inflammatory response, has been reported in several retinal pathologies33. Complement dependent microglial clearance in the rd10 model leads to neurotoxicity and extensive damage whereas complement deficiency, with lowered microglial phagocytosis resulted in less photoreceptor loss. Thus, microglial activation was thought to be an adaptive mechanism to photoreceptor degeneration9. Microglial activation has also been reported to be beneficial, and an adaptive response to protect retina from ongoing damage. One group reported that subretinal microglial protects RPE in a light damage model and ensures proper functioning of photoreceptors4. In the MfrpKI/KI mouse model we found a significant increase in overall microglial activation, including subretinal activation, however the contribution of activated microglia to disease progression has yet to be established. To evaluate the role of immune cells, in the current model, we performed immunostaining of retinal cryosections with Iba-1 and observed that there were significantly higher numbers of microglia in MfrpKI/KI mice compared to WT mice retina. Iba-1 positive cells were also present in the subretina (Fig. 2B, panel IV), but were absent in WT mouse retinal cryosections (Fig. 2A, panel IV), suggesting a strong immune response in MfrpKI/KI mice retina. A similar observation was found when microglial numbers were compared in GCL, IPL and the subretina of WT and MfrpKI/KI mice (Fig. 2C). Flow cytometry analysis revealed that microglia numbers were doubled in MfrpKI/KI mice suggesting a strong immune response present shown. There were also increased numbers of monocyte-derived macrophages in MfrpKI/KI retina (Fig. 2D–G). It is not completely clear why there is such a profound cellular inflammatory response in the present model.
We observed that there were more CD68, and Iba-1 positive cells present in GCL, ONL, OPL and IPL region. This typical topological distribution of microglia was not observed in WT retina. Also, WT microglia had a more ramified appearance, which has been reported as a resident quiescent phenotype. Additionally, these quiescent microglia were mainly localized in the IPL and OPL. The number of subretinal microglia were significantly higher in MfrpKI/KI when compared with WT retina (p < 0.001) (Fig. 3A–E)34,35.
To better understand retinal microglial characteristics and heterogeneity in the MfrpKI/KI mice, we analyzed microglial gene expression by scRNA-seq compared with age-matched WT control. We found MfrpKI/KI microglial cells were highly heterogenous, as analyzed by their distribution in multidimensional coordinate space and entropy distribution (Supplementary Fig. 3A,B). We observed a significant increase in microglial activation markers CD68 including F4/80 and Iba-1 which represent microglia or monocyte-derived macrophages and a decrease in resident homeostatic microglial markers, including TMEM119, Siglech, P2ry12 and P2ry13, in MfrpKI/KI mice retina (lfdr > 0.1) (Fig. 5A–I, Supplementary Table B). Retinal degenerative diseases associated with microglial activation have been reported to have a similar gene signature with lowered homeostatic and increased activation markers in the central nervous system and retina26,36,37. TMEM119, a transmembrane protein found in microglia, is specifically expressed on the ramified microglial forms and has been used to distinguish homeostatic microglia from monocyte-derived macrophages in the human brain38,39 whereas, Siglech, (Sialic acid-binding immunoglobulin type lectins) another well-known marker of resident microglia, exhibits its inhibitory effects by immunoreceptor tyrosine based inhibition motif, downregulating any unnecessary pro-inflammatory immune responses in the retina as well as in brain40,41. A change in the local microenvironment can stimulate the loss of these resident microglial markers and increasing expression of CD68 and Iba-1 active forms42. Validating our scRNA data, we observed that TMEM119 expression was lowered in MfrpKI/KI retinal cryosections as compared to WT but that CD68 and TMEM119 co-expressing microglia were present in the subretina. This suggests a resident microglial origin for some of the subretinal microglia. These subretinal cells were completely absent from WT retina (Fig. 9A,B).
CD68 has been extensively identified as a microglia activation markers in retinal degeneration studies43. Activated microglia express CD68 or macrosialin which is the only known member of the class D scavenger receptors in the retina44. We also found a decrease in expression of fractalkine/CX3CR1 in MfrpKI/KI mice, which may explain the increase in microglia and inflammatory marker expression in this model. Another study, using the rd10 mouse model of retinitis pigmentosa, suggested that lowered or altered CX3CR1 expression was associated with increased expression of inflammatory cytokines and microglial activation markers45. The role of fractalkine/CX3CR1 signaling has been suggested to play a crucial role in mediating microglia recruitment to apoptotic photoreceptors in the light induced retinal degeneration model46. Apart from CX3CR1, other pro-inflammatory cytokines include interleukin (IL)-1 beta, tumor necrosis factor (TNF)-alpha and CCL2, are crucial for microglial induced photoreceptor apoptosis47. Previously, Chekuri et al., had reported photoreceptor degeneration because of Mfrp mutations, in the MfrpKI/KI model, which may trigger this pro-inflammatory response. To our knowledge microglial induced apoptosis of photoreceptors has not been confirmed in Mfrp mouse models, however we observed increased expression of apoptotic markers (PARP9, ANXA5, TSPO and phagocytic markers (CTSB, CTSS, CSTL) in MfrpKI/KI mice when compared with WT. The presence or absence of apoptosis would be interesting to investigate further in future studies (Supplementary Fig. 4, Supplementary Table L). Next, we aimed to analyze major microglial sub-types in MfrpKI/KI mice retina. We aimed to find gene signatures associated with activated subretinal microglia, described in previous RD models with autofluorescent spots31. After confirming that cluster 2 was mostly composed of cells expressing homeostatic microglial markers, and that cluster 7 was composed of relatively more active microglia (Fig. 5G,I) (Supplementary Tables D, E), we looked in more detail at each cluster and found that cluster 6 and 16 (Supplementary Tables F, G), had several upregulated markers previously identified in active subretinal deposits (Lyz2, Ms4a7 and Lgals3bp). Lyz2, is a lysosomal marker, was consistently high in active microglia clusters in all comparisons. It has been reported to be involved in the early stages of microglial activation, in neurodegenerative diseases26. Lgals3bp, a cell–cell or cell–matrix interaction modulating protein, along with Lyz2, was high in subretinal active microglia in another study of photoreceptor degeneration model4. A recent scRNA-seq study in brain reported a distinct population of cerebral associated macrophages (CAMs) positive for Ms4a7 (membrane-spanning 4-domains subfamily A member 7) present throughout the course of neuroinflammation25.
Each of these genes are crucial for processes such as phagocytic clearance of degenerating photoreceptors, as has been reported in the current model. Cluster 6 and 16 could potentially represent subretinal microglia. As cluster 10, 13 and 14 share a good proportion of cells from MfrpKI/KI mice we revisited cluster analysis to find out their characteristics. As previously explained, we first considered CD68 high expressing clusters, but we also tried to investigate other clusters with a high expression of other markers expressed by microglia validated in current study such as Iba-1 or Aif-1, adgre-1 or F4/80 through immunostaining. Coincidentally, we found that cluster 10 had non-significant CD68. In addition, cluster 10 also had Aif-1 or Iba-1 expression, but had significant adgre1 or F4/80 (lfdr < 0.1). Cluster 10 also had cells with a low expression of homeostatic markers (P2ry13,P2ry12 and TMEM119). Other markers known to be high in the activation state (Lgals3bp, Ms4a7, Ms4a6c), were highly expressed (lfdr < 0.1). This suggests that cluster 10 could be made of active microglia with a different expression profile present in MfrpKI/KI mice compared to WT as we also found adgre-1 or F4/80 positive cells present in the subretina of MfrpKI/KI mice (Supplementary Figs. 5, 6). Cluster 13 also had a relatively good population of cells, but with a low or insignificant expression of activation markers Iba-1, Aif-1and adgre-1 or F4/80 which suggests these could be microglia in an intermediate state. Similarly, cluster 14 had a higher expression of Aif-1/iba-1, with a large number of cells expressing activation markers (Lyz2 and Lgals3bp) suggesting that a further subset of active microglia exist in MfrpKI/KI mice retina (Supplementary Table H). In summary, we conclude that microglia in MfrpKI/KI mice are present in several activation states and our studies suggest increased heterogeneity in the MfrpKI/KI mice retina as discussed earlier (Supplementary Fig. 3A,B).
We next compared cluster 7, which we define as most active large size cluster, with cluster 6 which was one of small candidate active clusters. We observed that, apart from a significantly higher number of cells expressing known activation markers, there were also other cells expressing myeloid markers (Lyz2, Ms4a7, Lgals3bp) (Fig. 6B, Supplementary Table I). We next performed a trajectory analysis to understand activation stages of microglia in MfrpKI/KI mice retina (Fig. 7D). Trajectory 3 had shown increased activation marker expression beginning from putative resident microglial cluster 8 then 2, 5 and 6, finally ending in cluster 15. The progression along the trajectory resulted in a stepwise loss of homeostatic markers, along with higher expression of subretinal markers Lyz2, Ms4a7, Lgals1 and ApoE (Fig. 7E, Supplementary Tables J, K). Microglial activation results in their migration into an injury site, where they phagocytize injured photoreceptors. These findings provide an insight into the microglial response and function during RD in the MfrpKI/KI model. We hypothesize that activated subretinal microglia remove potentially damaging cell debris15,48. We also observed that there was a significant increase in cells expressing genes associated with phagocytosis and apoptosis pathways in MfrpKI/KI retinal microglia (PARP9, ANXA5, TSPO) (Supplementary Fig. 4, Supplementary Table L). Poly-ADP ribose polymerase (PARP) is known to be activated as a result of DNA damage, in another model retinal degeneration model, the rd10 model49. High cGMP levels in dying photoreceptors were found to correlate with increased activity of PARP50. We also observed high PARP14 and PARP9 in MfrpKI/KI microglia along with ANXA5, which contributes to αvβ5 integrin-dependent photoreceptor phagocytosis clearance at the apical phagocytic surface of the RPE51. Similarly, translocator protein (TSPO), was also high in the Mfrp KI/KI microglia, which promotes ROS production, by increasing calcium levels and hence NADPH oxidase 1 (NOX1) activation. TSPO knock out has resulted in retinal protection from angiogenesis and inflammation35.
We validated scRNA-seq findings using immunofluorescence of retinal cryosections. We observed that greater numbers of F4/80 positive cells were distributed in the GCL, IPL and OPL, in MfrpKI/KI (Supplementary Fig. 5). We observed prominent F4/80 positive cells present in MfrpKI/KI subretina, whereas no cells were detected in WT retina (Supplementary Fig. 6). In addition, we had noted the higher number of cells expressing APOE, which plays important role in lipid metabolism and it has been reported in subretinal microglia in other retinal degeneration models37. Co-staining of Iba-1/APOE in retinal cryosections was performed (Fig. 8A,B). We observed Iba-1/APOE positive cells, which were prominently present in MfrpKI/KI subretina, which were completely absent from WT mice, which appears to validate findings of some of our candidate clusters in our scRNA seq data (Fig. 8C,D). Not all of the subretinal cells were positive for APOE, suggesting a heterogenous population. This warrants further study, and investigation as to the potential source of these other cells. Of note, other cells in the retina, including glial cells such as astrocytes also express APOE52. APOE positive cells were also found in the subretina, IPL and OPL in MfrpKI/KI mice retina. We observed a higher number of microglia expressing APOE in Mfrp KI/KI retina in scRNA-seq. APOE is a cholesterol transporter associated with retinal degeneration and hence its accumulation in the subretinal microglia has been reported to exacerbate RD53,54.
Although our study compared findings in a mouse model of MARD with those of age-matched mice on the same background, the present study only looked at the mice at one age point. This age-point was chosen as this was the approximate age at which degeneration was shown to be at a maximum, from previous longitudinal studies. However, retinal degeneration is not a single event and may change with the stage of degeneration. It would be important to look at earlier age points to confirm our trajectory analysis and to identify triggers for microglial activation. In addition, these studies show an association of immune cells with RD. They do not confirm causation, although expression markers for phagocytosis do suggest that they are involved in photoreceptor phagocytosis. The present study also identified an increased number of monocyte-derived macrophages in the MfrpKI/KI retina using flow cytometry. Further, study will be required to understand the role of monocyte-derived macrophages in MARD. The presence of a heterogenous population of microglia likely represents a changing microenvironment in the retina leading to dynamic changes in the immune response with ongoing RD. In future studies, validation of these markers will help better understanding of the immune response. Taken together, the validation studies confirm the findings from flow cytometry and scRNA-seq data demonstrating an increased number of activated microglia in MfrpKI/KI mice retina compared to age-matched WT mouse retina. In addition, the immunostaining also appears to confirm the scRNA-seq findings that cells in cluster 6 are likely to make up part of the subretinal microglial population seen in the MfrpKI/KI mice, as the subretinal microglia in MfrpKI/KI mice demonstrate strong CD68, F4/80 and APOE expression. However, the subretinal microglial population is heterogeneous. Further microglial knockout, targeted depletion or suppression studies will likely be required to confirm the role of retinal microglia in degeneration in MfrpKI/KI mice.
Materials and methods
Experimental animals
All animal procedures were performed with approval by the University of California, San Diego, Institutional Animal Care and Use Committee (IACUC). All methods were conducted in accordance with relevant guidelines and regulations, including the National Institute of Health (NIH), the Association for Research in Vision and Ophthalmology (ARVO) and Animal Research: Reporting of In Vivo Experiments (ARRIVE). MfrpKI/KI mice (4–5 months old) were generated on C57BL/6J background and have been previously characterized13. In summary, the c.498_499insC mutation in exon 5 of Mfrp gene was generated by targeting exons 3–9, using site directed mutagenesis. The targeting vector (15.25 kb) was generated by sub-cloning a 11.15 kb region from a C57BL/6 BAC clone (RP23:270P20) having exons 1–13 exons Mfrp with a LoxP site upstream (exon 3) and a LoxP/FRT‐flanked Neo cassette downstream (exon 9). Age matched WT mice were used as controls. All mice were housed in standard condition at standard temperature (25 °C) and with a 12-h light: dark cycle on a standard diet. All mice were genotyped for the above-mentioned mutation using tail tip samples for DNA before study.
Clinical evaluation of human patients with Mfrp mutation
Institutional Review Board (IRB) approval was acquired from the University of California, San Diego for the review of patient’s data. Informed consent from patients or their legal gradians were obtained as per institutional protocol for participation and publication of information including images. All data and images were anonymized for patient’s safety. All patient studies were conducted in accordance with the protocol approved by the University of California, San Diego and as per the Declaration of Helsinki (https://www.wma.net/policies-post/wma-declaration-of-helsinki-ethical-principles-for-medical-research-involving-human-subjects/). Human subjects with c.498_499insC mutations were examined and had fundus imaging as described earlier13. Four siblings of a consanguineous Mexican family with confirmed homozygous c.498_499insC mutations in MFRP were recruited at the Institute of Ophthalmology, “Conde de Valenciana” Faculty of Medicine, UNAM, Mexico City for the study. Spectral domain-optical coherence tomography (SD-OCT) imaging of the fundus was performed using SD-OCT Spectralis® HRA + OCT (Heidelberg Engineering, Heidelberg, Germany) at 850 nm wavelength in all patients. Patients also underwent, ultra-widefield pseudo color and autofluorescence imaging (Optos, Dunfermline, United Kingdom). A further 44-year-old male patient was examined at the Shiley Eye Institute, University of California using the same imaging techniques and was confirmed to have the carrying c.523C > T, p.(Gln175*) and c.649G > A, p.(Gly217Arg) mutations in MFRP using whole exome sequencing.
Autofluorescence and optical coherence tomography (OCT) imaging of MfrpKI/KI mice
Fundus imaging of MfrpKI/KI and WT (4 months old, n = 3) mice was performed as described earlier13. In brief, mice were anesthetized with intraperitoneal injection of ketamine (93 mg/kg) and xylazine (8 mg/kg), followed by1% atropine and 0.5% tropicamide application to eyes for dilating the pupil. Autofluorescence and OCT imaging of both the eyes were performed at 488 nm excitation SLO and 850 nm wavelength using Spectralis™ HRA + OCT ((Heidelberg Engineering, Heidelberg, Germany) and analyzed by using Heidelberg eye explorer software V2.
Fluorescence activated cell sorting (FACS)
Microglia were sorted from the retina of each group (n = 6) separately as described with some modifications55,56. In brief, mice were euthanized by carbon dioxide inhalation and death was confirmed by cervical dislocation. Eyecups were collected in ice cold 1xphosphate buffer saline (PBS). Eyes were dissected under microscope removing lens and cornea, and retinae were dissociated mechanically in 5 ml cold PBS. The resulting cell suspension was passed through wide bore pipette several times before passing through 70 µm cell strainers to remove large cell clumps. Cells were pellet down at 400 rcf at 4 °C for 5 min and resuspended in 1 ml FACS buffer containing 3% fetal bovine serum in 1× PBS. 1 × 106 cells from each group were stained with rat anti-mouse CD16/CD32 (Mouse BD Fc Block™, BD Pharmingen™, 553141, 1:500) for 20 min at 4 °C followed by staining with anti-CD45 (Biolegend 103108, 0.25 µg per/ul), anti-CD11b (Biolegend, 101212, 0.25 µg per/µl) for 30 min at 4 °C in the dark. Cells were washed in FACS buffer and resuspended in 500 ul 1× PBS containing 0.1% Bovine serum albumin (BSA). Propidium iodide was used to exclude the dead cells during gating, and compensation beads (UltraComp eBeads, Invitrogen, 01-2222) were used to set the experimental compensation per manufacturer instructions. Microglia were sorted as a CD11bhighCD45low population in 500 µl 1× PBS containing 40 U/ml RNAase (New England Biolabs, M0314S) and 0.04% BSA in BDFACS Aria II using BDFACS Diva™ softwareV9. Experiments were repeated three times (n = 3) and analyzed using FlowJo™ softwareV10.8 to. Doublets were excluded first, and single live cells were gated followed by CD11bhigh CD45high as monocyte-derived macrophages and CD11bhigh CD45low cells as microglia population. Percentage of microglia and macrophages were calculated out of live cells taken as parent population in each group.
Single cell RNA sequencing
FACS sorted retinal microglia (7000 cells) from MfrpKI/KI and WT mice (4-month-old) were processed for ScRNA-seq using 10× Genomics chromium controller and chromium single cell 3ʹ v3 reagent kit as per manufacturer’s instructions (10× Genomics) and libraries were sequenced using an Illumina NovaSeq 6000 sequencer. Initial data was analyzed by Cell Ranger Single-Cell Software Suite (10× Genomics). A total of 1345 cells from WT and 2059 cells from Mfrp KI/KI mice had passed the quality control criteria based on Cell Ranger software. There were three final output files each for gene count, barcodes, and feathers. A pre-built mouse transcriptome reference file refdata-cellranger-mm10-3.0.0 provided by 10× (https://support.10xgenomics.com/single-cell-gene-xpression/software/downloads/latest) was used for mapping of reads to transcripts. Pairwise distances between cells were calculated as the Jensen-Shannon distances based on per cell gene count frequencies as in Ref.57. 20 communities (clusters) were found among the cells using the full distance matrix using affinity propagation algorithm. Communities were visualized using t-SNE dimensional reduction58. Heterogeneity in each group was analyzed by calculating probability density in multi-dimensional coordinate space and entropy measurement by creating a random bootstrap ensemble of WT and Mfrp MfrpKI/KI samples of equal size 1345 cells. Differential expression between clusters was assessed with the bootstrap method of Pollard and van der Laan59, with lfdr calculated using the empirical Bayes method of Efron60. A cutoff of lfdr = 0.1 was chosen to define significant differential gene expression in Mfrp KI/KI and WT mice. Additionally, cells from MfrpKI/KI and WT mice each were also clustered separately to perform trajectory analysis using the Slingshot method61. Heat maps were generated by “pheatmap” package in R program (https://www.R-project.org) showing differential expression of genes, red color for upregulated and blue color for downregulated genes respectively.
Immunofluorescence
Eyes were removed after mice were euthanized by carbon dioxide inhalation and death confirmed by cervical dislocation. Eyecups were fixed immediately in 4% paraformaldehyde for 24 h at 4 °C. After washing in PBS, Eye cups were cryoprotected in sucrose gradient (from 10, 20 to 30%) for 2 h each followed by embedding in Optimal cutting temperature (OCT) compound and cut into 12 µm thin cryosections. For immunodetection of Iba-1, F4/80and co-staining of Iba-1and CD68 sections were fixed in cold acetone for 10 min and washed with 1xPBS followed by blocking in 5% normal goat or donkey serum prepared in PBST (0.1% triton in 1× PBS) for 45 min at room temperature (RT). Further, sections were incubated with primary antibodies: CD68 (rat-anti mouse, 1:500;MCA1957), Iba-1 (rabbit anti-mouse, 1:1000; ab153696), F4/80 rat anti-mouse (1:200; AB664) overnight at 4 °C. Further, for APOE (rabbit anti-mouse, 1:100, ab183597) and Iba-1 (goat anti-mouse, 1:500, 011-27991,Wako chemicals) co-staining and for TMEM119 rabbit anti-mouse, 1:500 ab209064), and CD68 (rat-anti mouse, 1:500;MCA1957) co-staining slightly modified protocol was used. In brief, cryosections were first washed with 1× PBS then incubated for 1 h with 1% sodium borohydride in PBS for antigen retrieval Blocking was done in 5% BSA and 5% donkey serum in PBST (0.3% triton in 1 × PBS) for 1 h at RT. Further, sections were incubated with primary antibodies: Iba-1 (goat anti-mouse, 1:200; AB5076), and APO-E (rabbit anti mouse, 1:200; ab183597) at 4 °C overnight. Antibodies were diluted in 1% BSA in PBST v.
In a separate experiment another F4/80 primary antibody (rabbit anti- mouse 1:500, 70076 Cell signaling) was used for detection with different antigen retrieval system. In brief, sections were incubated with for 1 h (Iba-1/ApoE) or 10 min (TMEM119/CD68) in 1% sodium borohydride in PBS at RT for antigen retrieval and blocked with 3% donkey serum, 0.01% azide and 0.3 M Glycine for 1 h followed by incubation with primary antibody overnight at 4 °C. Compatible secondary antibodies (1:750) were used for detection at RT for 2 h diluted in 1% BSA in PBST. Sections were mounted in vectashield mounting medium with DAPI and all images were acquired under × 20 magnification using Keyence Z-BX800 microscope (Keyence, Itasca, IL). For analyzing single immunodetection three retinal cryosections from each mouse (n = 3) in each group were counted in individual retinal layers. For CD68/Iba-1 and all APOE/Iba-1 positive cells in the subretina we counted 3 complete eye cups each mouse (n = 3) from both group by a person blinded to study conditions.
Retinal wholemount immunostaining
Retinal wholemount staining was performed as described earlier62. Briefly, both the eyes were collected after mice were euthanized by carbon dioxide inhalation and death was confirmed by cervical dislocation. Eyecups were immediately fixed for an hour in 4% PFA at 4 °C, followed by fixation in 70% alcohol for another hour. Eyes were then dissected in cold 1× PBS under dissection microscopes, cornea, lens and vitreous were removed carefully. Retinae were blocked with 3% milk and 3% BSA in 1× PBS for an hour. Further, incubated in rabbit anti mouse CD68 antibody (rat-anti mouse, 1:500; MCA1957) in PBS 1%BSA solution overnight at RT. Retina were rinsed three times in 1xPBS (20 min each) and incubated with secondary antibody (Alexa fluor 488,1:500; A32790) and Rhodamine-Phalloidin (Thermo scientific, R415:1:10) for an hour at RT. After three final washings with PBS (20 min each), each retina was flattened using four radial incisions under light microscope and mounted with vectashield antifade mounting medium with DAPI. Autofluorescent spots were detected at 670 nm excitation and 450 nm emission range. Autofluorescent and Iba-1 positive cells colocalization images were acquired under × 20 magnification using a Nikon AIR confocal microscope by Z-stacking and the closest images to the RPE was used for analysis. Three retinal wholemount images from each group were analyzed for quantitative comparison (n = 3).
Statistical analysis
All data were compared using unpaired student t-test and presented as mean ± SEM. Data was analyzed using Graph Pad Prism 9.0 (La Jolla, CA, USA) and statistical significance was considered at p < 0.05.
Ethical declarations
All methods and experimental procedures involving animal were performed in accordance with the protocols approved by the University of California, San Diego, USA, Institutional Animal Care and Use committee (IACUC), relevant guidelines and regulations including the National Institute of Health (NIH), the Association for Research in Vision and Ophthalmology (ARVO) and Animal Research: Reporting of In Vivo Experiments (ARRIVE). All patient studies were conducted in accordance with the protocol approved by the University of California, San Diego and as per the Declaration of Helsinki (https://www.wma.net/policies-post/wma-declaration-of-helsinki-ethical-principles-for-medical-research-involving-human-subjects/).
Supplementary Information
Acknowledgements
Dr. Shyamanga Borooah is supported by a Foundation Fighting Blindness Career Development award and a Nixon Visions Foundation grant. ScRNA-seq was conducted at the Institute for Genomic Medicine, Genomics Center, UCSD, San Diego, LA Jolla, CA National Institutes of Health SIG Grant (#S10 OD026929). ScRNA-seq data was analyzed with support from the center for Computational biology and Bioinformatics (CCBB) services at UCSD, San Diego, LA Jolla. They thank Jacinta Lucero, BS, Senior research associate at UCSD, San Diego, LA Jolla, for performing TMEM119/CD68 immunofluorescence co-staining and performing F4/80 detection with modified antigen retrieval system. We thank the Department of Ophthalmology, UCSD Histology and UCSD microscopy imaging core facilities which was supported by a National Institute of Health core grant (P30EY022589). They would also like to thank Stephanie Landeros and Megan Bliss for their support with histology and mice colony maintenance.
Author contributions
S.B. designed the study, analyzed the data and helped write the manuscript, R.A.* reviewed and edited the manuscript and generated the mouse used in the studies. A.K. designed the study, performed experiments, analyzed the data and wrote the majority of the manuscript. K.H. performed IF (Figs. 7A,B, 8) and reviewed the manuscript. R.A. and J.C. collected human case fundus history and imaging, S.R. analyzed the sc-RNA Seq data and helped write the manuscript. The manuscript was edited and approved by all the authors for final submission.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Radha Ayyagari, Email: rayyagari@health.ucsd.edu.
Shyamanga Borooah, Email: sborooah@health.ucsd.edu.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-19351-w.
References
- 1.Hanisch U-K, Kettenmann H. Microglia: Active sensor and versatile effector cells in the normal and pathologic brain. Nat. Neurosci. 2007;10:1387–1394. doi: 10.1038/nn1997. [DOI] [PubMed] [Google Scholar]
- 2.Santos AM, et al. Embryonic and postnatal development of microglial cells in the mouse retina. J. Comp. Neurol. 2008;506:224–239. doi: 10.1002/cne.21538. [DOI] [PubMed] [Google Scholar]
- 3.Rashid K, Akhtar-Schaefer I, Langmann T. Microglia in retinal degeneration. Front. Immunol. 2019;10:1975. doi: 10.3389/fimmu.2019.01975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Koren EG, et al. Microglial function is distinct in different anatomical locations during retinal homeostasis and degeneration. Immunity. 2019;50:723–737. doi: 10.1016/j.immuni.2019.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou T, et al. Microglia polarization with M1/M2 phenotype changes in rd1 mouse model of retinal degeneration. Front. Neuroanat. 2017 doi: 10.3389/fnana.2017.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao L, et al. Microglial phagocytosis of living photoreceptors contributes to inherited retinal degeneration. EMBO Mol. Med. 2015;7:1179–1197. doi: 10.15252/emmm.201505298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang B, et al. Retinal degeneration mutants in the mouse. Vis. Res. 2002;42:517–525. doi: 10.1016/s0042-6989(01)00146-8. [DOI] [PubMed] [Google Scholar]
- 8.Liew G, Michaelides M, Bunce C. A comparison of the causes of blindness certifications in England and Wales in working age adults (16–64 years), 1999–2000 with 2009–2010. BMJ Open. 2014;4:e004015. doi: 10.1136/bmjopen-2013-004015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silverman SM, Ma W, Wang X, Zhao L, Wong WT. C3- and CR3-dependent microglial clearance protects photoreceptors in retinitis pigmentosa. J. Exp. Med. 2019;216:1925–1943. doi: 10.1084/jem.20190009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noailles A, Fernández-Sánchez L, Lax P, Cuenca N. Microglia activation in a model of retinal degeneration and TUDCA neuroprotective effects. J. Neuroinflamm. 2014;11:186. doi: 10.1186/s12974-014-0186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fogerty J, Besharse JC. 174delG mutation in mouse MFRP causes photoreceptor degeneration and RPE atrophy. Investig. Ophthalmol. Vis. Sci. 2011;52:7256–7266. doi: 10.1167/iovs.11-8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kameya S, et al. Mfrp, a gene encoding a frizzled related protein, is mutated in the mouse retinal degeneration 6. Hum. Mol. Genet. 2002;11:1879–1886. doi: 10.1093/hmg/11.16.1879. [DOI] [PubMed] [Google Scholar]
- 13.Chekuri A, et al. Long-term effects of gene therapy in a novel mouse model of human MFRP-associated retinopathy. Hum. Gene Ther. 2019;30:632–650. doi: 10.1089/hum.2018.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fogerty J, Besharse JC. Subretinal infiltration of monocyte derived cells and complement misregulation in mice with AMD-like pathology. Adv. Exp. Med. Biol. 2014;801:355–363. doi: 10.1007/978-1-4614-3209-8_45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makabe K, Sugita S, Mandai M, Futatsugi Y, Takahashi M. Microglia dynamics in retinitis pigmentosa model: Formation of fundus whitening and autofluorescence as an indicator of activity of retinal degeneration. Sci. Rep. 2020;10:14700. doi: 10.1038/s41598-020-71626-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Veremeyko T, Starossom SC, Weiner HL, Ponomarev ED. Detection of microRNAs in microglia by real-time PCR in normal CNS and during neuroinflammation. J. Vis. Exp. 2012 doi: 10.3791/4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu C, Roubeix C, Sennlaub F, Saban DR. Microglia versus monocytes: Distinct roles in degenerative diseases of the retina. Trends Neurosci. 2020;43:433–449. doi: 10.1016/j.tins.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lavalette S, et al. CD36 deficiency inhibits retinal inflammation and retinal degeneration in Cx3cr1 knockout mice. Front. Immunol. 2019;10:3032. doi: 10.3389/fimmu.2019.03032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Funatsu J, et al. Circulating inflammatory monocytes oppose microglia and contribute to cone cell death in retinitis pigmentosa. PNAS Nexus. 2022 doi: 10.1093/pnasnexus/pgac003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou L, Li Q. Isolation of region-specific microglia from one adult mouse brain hemisphere for deep single-cell RNA sequencing. J. Vis. Exp. 2019 doi: 10.3791/60347. [DOI] [PubMed] [Google Scholar]
- 21.Bell OH, et al. Single eye mRNA-Seq reveals normalisation of the retinal microglial transcriptome following acute inflammation. Front. Immunol. 2020;10:3033. doi: 10.3389/fimmu.2019.03033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DePaula-Silva AB, et al. Differential transcriptional profiles identify microglial- and macrophage-specific gene markers expressed during virus-induced neuroinflammation. J. Neuroinflamm. 2019;16:152. doi: 10.1186/s12974-019-1545-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis E, Foster T, Thomas W. Cellular forms and functions of brain microglia. Brain Res. Bull. 1994;34:73–78. doi: 10.1016/0361-9230(94)90189-9. [DOI] [PubMed] [Google Scholar]
- 24.Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314–1318. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- 25.Jordão MJC, et al. Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science. 2019;363:7554. doi: 10.1126/science.aat7554. [DOI] [PubMed] [Google Scholar]
- 26.Keren-Shaul H, et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell. 2017;169:1276–1290. doi: 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 27.Tsujikawa A, et al. Punctate hyperfluorescent spots associated with central serous chorioretinopathy as seen on indocyanine green angiography. Retina. 2010;30:801–809. doi: 10.1097/IAE.0b013e3181c72068. [DOI] [PubMed] [Google Scholar]
- 28.Secondi R, Kong J, Blonska AM, Staurenghi G, Sparrow JR. Fundus autofluorescence findings in a mouse model of retinal detachment. Investig. Ophthalmol. Vis. Sci. 2012;53:5190–5197. doi: 10.1167/iovs.12-9672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sundin OH, et al. Extreme hyperopia is the result of null mutations in MFRP, which encodes a Frizzled-related protein. Proc. Natl. Acad. Sci. U.S.A. 2005;102:9553–9558. doi: 10.1073/pnas.0501451102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang NK, et al. Cellular origin of fundus autofluorescence in patients and mice with a defective NR2E3 gene. Br. J. Ophthalmol. 2009;93:1234–1240. doi: 10.1136/bjo.2008.153577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang N-K, et al. Origin of fundus hyperautofluorescent spots and their role in retinal degeneration in a mouse model of Goldmann-Favre syndrome. Dis. Model. Mech. 2013;6:1113–1122. doi: 10.1242/dmm.012112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karlstetter M, et al. Retinal microglia: Just bystander or target for therapy? Prog. Retin. Eye Res. 2015;45:30–57. doi: 10.1016/j.preteyeres.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 33.Xu H, Chen M, Forrester JV. Para-inflammation in the aging retina. Prog. Retin. Eye Res. 2009;28:348–368. doi: 10.1016/j.preteyeres.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 34.Scholz R, et al. Targeting translocator protein (18 kDa)(TSPO) dampens pro-inflammatory microglia reactivity in the retina and protects from degeneration. J. Neuroinflamm. 2015;12:1–12. doi: 10.1186/s12974-015-0422-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolf A, Herb M, Schramm M, Langmann T. The TSPO-NOX1 axis controls phagocyte-triggered pathological angiogenesis in the eye. Nat. Commun. 2020;11:2709. doi: 10.1038/s41467-020-16400-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krasemann S, et al. The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity. 2017;47:566–581. doi: 10.1016/j.immuni.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu C, Saban DR. Identification of a unique subretinal microglia type in retinal degeneration using single cell RNA-seq. Adv. Exp. Med. Biol. 2019;1185:181–186. doi: 10.1007/978-3-030-27378-1_30. [DOI] [PubMed] [Google Scholar]
- 38.Satoh J-I, et al. TMEM119 marks a subset of microglia in the human brain. Neuropathology. 2016;36:39–49. doi: 10.1111/neup.12235. [DOI] [PubMed] [Google Scholar]
- 39.Bennett ML, et al. New tools for studying microglia in the mouse and human CNS. Proc. Natl. Acad. Sci. 2016;113:E1738. doi: 10.1073/pnas.1525528113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Linnartz-Gerlach B, Kopatz J, Neumann H. Siglec functions of microglia. Glycobiology. 2014;24:794–799. doi: 10.1093/glycob/cwu044. [DOI] [PubMed] [Google Scholar]
- 41.Böttcher C, et al. Human microglia regional heterogeneity and phenotypes determined by multiplexed single-cell mass cytometry. Nat. Neurosci. 2019;22:78–90. doi: 10.1038/s41593-018-0290-2. [DOI] [PubMed] [Google Scholar]
- 42.Kurushima H, et al. Surface expression and rapid internalization of macrosialin (mouse CD68) on elicited mouse peritoneal macrophages. J. Leukoc. Biol. 2000;67:104–108. doi: 10.1002/jlb.67.1.104. [DOI] [PubMed] [Google Scholar]
- 43.Wohl SG, Schmeer CW, Witte OW, Isenmann S. Proliferative response of microglia and macrophages in the adult mouse eye after optic nerve lesion. Investig. Ophthalmol. Vis. Sci. 2010;51:2686–2696. doi: 10.1167/iovs.09-4537. [DOI] [PubMed] [Google Scholar]
- 44.Wang M, et al. Macroglia-microglia interactions via TSPO signaling regulates microglial activation in the mouse retina. J. Neurosci. 2014;34:3793–3806. doi: 10.1523/JNEUROSCI.3153-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zabel MK, et al. Microglial phagocytosis and activation underlying photoreceptor degeneration is regulated by CX3CL1-CX3CR1 signaling in a mouse model of retinitis pigmentosa. Glia. 2016;64:1479–1491. doi: 10.1002/glia.23016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang M, Xu G, Liu W, Ni Y, Zhou W. Role of fractalkine/CX3CR1 interaction in light-induced photoreceptor degeneration through regulating retinal microglial activation and migration. PLoS ONE. 2012;7:e35446. doi: 10.1371/journal.pone.0035446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feng C, et al. Expression of CCL2 and its receptor in activation and migration of microglia and monocytes induced by photoreceptor apoptosis. Mol. Vis. 2017;23:765–777. [PMC free article] [PubMed] [Google Scholar]
- 48.Okunuki Y, et al. Microglia inhibit photoreceptor cell death and regulate immune cell infiltration in response to retinal detachment. Proc. Natl. Acad. Sci. 2018;115:E6264–E6273. doi: 10.1073/pnas.1719601115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arango-Gonzalez B, et al. Identification of a common non-apoptotic cell death mechanism in hereditary retinal degeneration. PLoS ONE. 2014;9:e112142. doi: 10.1371/journal.pone.0112142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vidal-Gil L, Sancho-Pelluz J, Zrenner E, Oltra M, Sahaboglu A. Poly ADP ribosylation and extracellular vesicle activity in rod photoreceptor degeneration. Sci. Rep. 2019;9:3758. doi: 10.1038/s41598-019-40215-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu C, Muñoz LE, Mallavarapu M, Herrmann M, Finnemann SC. Annexin A5 regulates surface αvβ5 integrin for retinal clearance phagocytosis. J. Cell Sci. 2019;132:232439. doi: 10.1242/jcs.232439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chung W-S, et al. Novel allele-dependent role for APOE in controlling the rate of synapse pruning by astrocytes. Proc. Natl. Acad. Sci. 2016;113:10186–10191. doi: 10.1073/pnas.1609896113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Levy O, et al. APOE isoforms control pathogenic subretinal inflammation in age-related macular degeneration. J. Neurosci. 2015;35:13568–13576. doi: 10.1523/JNEUROSCI.2468-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rao SR, Fliesler SJ. Cholesterol homeostasis in the vertebrate retina: Biology and pathobiology. J. Lipid Res. 2020;62:100057. doi: 10.1194/jlr.TR120000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rajan WD, et al. Dissecting functional phenotypes of microglia and macrophages in the rat brain after transient cerebral ischemia. Glia. 2019;67:232–245. doi: 10.1002/glia.23536. [DOI] [PubMed] [Google Scholar]
- 56.O'Koren EG, Mathew R, Saban DR. Fate mapping reveals that microglia and recruited monocyte-derived macrophages are definitively distinguishable by phenotype in the retina. Sci. Rep. 2016;6:20636. doi: 10.1038/srep20636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ntranos V, Kamath GM, Zhang JM, Pachter L, Tse DN. Fast and accurate single-cell RNA-seq analysis by clustering of transcript-compatibility counts. Genome Biol. 2016;17:112. doi: 10.1186/s13059-016-0970-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van der Maaten L, Hinton G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008;9:1. [Google Scholar]
- 59.Pollard KS, van der Laan MJ. Resampling-Based Multiple Testing: Asymptotic Control of Type I Error and Applications to Gene Expression Data. University of California; 2003. [Google Scholar]
- 60.Efron B. Microarrays, empirical Bayes and the two-groups model. Stat. Sci. 2008;23:1–22. [Google Scholar]
- 61.Perraudeau F, Risso D, Street K, Purdom E, Dudoit S. Bioconductor workflow for single-cell RNA sequencing: Normalization, dimensionality reduction, clustering, and lineage inference. F1000Research. 2017;6:1158. doi: 10.12688/f1000research.12122.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lipski DA, et al. MHC class II expression and potential antigen-presenting cells in the retina during experimental autoimmune uveitis. J. Neuroinflamm. 2017;14:136. doi: 10.1186/s12974-017-0915-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.