Abstract
The splicing of transfer RNA (tRNA) introns is a critical step of tRNA maturation, for intron containing tRNAs. In eukaryotes, tRNA splicing is a multi-step process that relies on several RNA processing enzymes to facilitate intron removal and exon ligation. Splicing is initiated by the tRNA splicing endonuclease (TSEN) complex which catalyzes the excision of the intron through its two nuclease subunits. Mutations in all four subunits of the TSEN complex are linked to a family of neurodegenerative and neurodevelopmental diseases known as pontocerebellar hypoplasia (PCH). Recent studies provide molecular insights into the structure, function, and regulation of the eukaryotic TSEN complex and are beginning to illuminate how mutations in the TSEN complex lead to neurodegenerative disease. Using new advancements in the prediction of protein structure, we created a 3D model of the human TSEN complex. We review functions of the TSEN complex beyond tRNA splicing by highlighting recently identified substrates of the eukaryotic TSEN complex and discuss mechanisms for the regulation of tRNA splicing, by enzymes that modify cleaved tRNA exons and introns. Finally, we review recent biochemical and animal models that have worked to address the mechanisms that drive PCH and synthesize these studies with previous studies to try to better understand PCH pathogenesis.
Graphical Abstract
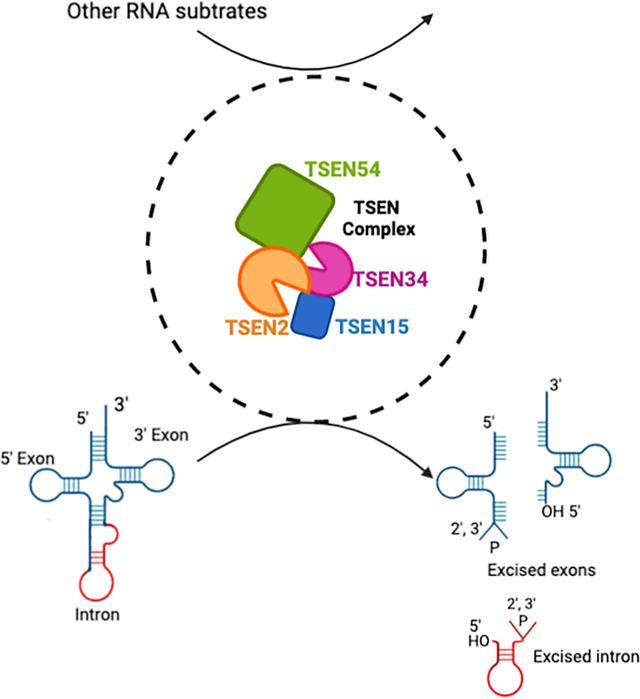
An overview graphic of the TSEN complex’s role in RNA processing. The eukaryotic TSEN complex is comprised of two endonucleases, TSEN2 (orange) and TSEN34 (pink), and two structural proteins TSEN15 (blue) and TSEN54 (green). The figure highlights the TSEN complexes’ known role in cleaving introns from intron containing pre-tRNAs and also that is has a role in targeting other RNA substrates, which is discussed more within this review. This figure was created with BioRender.com.
1. OVERVIEW OF TRNA SPLICING IN EUKARYOTES
Transfer RNAs (tRNAs) are critical players in the central dogma of molecular biology (Crick, 1970). They function as molecular decoders by converting the genetic code stored within our genes into the amino acid building blocks of proteins (O’Donoghue et al., 2018). While historically tRNAs were thought to be static players in translation, more recent work has revealed many additional functions of tRNAs such as regulating the proteome in response to stress and modulating rates of translation (Berg & Brandl, 2021; Hinnebusch, 2005; Phizicky & Hopper, 2010; Wilusz, 2015). In eukaryotes, tRNA biogenesis begins with transcription of the tRNA genes by RNA Pol III, which is highly regulated in response to nutrient availability and environmental stress (Willis & Moir, 2018). The initial pre-tRNA transcript then undergoes several processing and modification steps to generate the mature tRNA with the classic cloverleaf architecture including the acceptor stem, D-loop, Tψc-loop, variable arm, and anti-codon stem loop (Hopper & Nostramo, 2019; Huang & Hopper, 2016; Kimura et al., 2020; Phizicky & Hopper, 2010). Across all walks of life, a sub-set of pre-tRNAs contain introns that must be removed to form the anti-codon stem loop. While tRNA introns themselves do not appear to be essential for life (Cherry & White, 2018; Hayashi et al., 2019), they can control cellular growth and responses to physiological changes (Hayashi et al., 2019). The high inclusion of introns within tRNA genes of the same isoacceptors means that failure to remove tRNA introns would result in a loss of important tRNAs for translation. In humans, approximately 8% of tRNAs contain introns (Chan & Lowe, 2016) and the majority of those introns are clustered into similar isoacceptors such at Tyr-GTA, Leu-CAA, and Ile-TAT. The splicing of introns from tRNAs is a well conserved process from archaea to eukaryotes, yet there are notable differences in the RNA processing machinery required for splicing. Bacteria also contain intron containing pre-tRNAs, but they are self-spliced and do not require RNA processing machinery (reviewed in (Fujishima & Kanai, 2014).)
Splicing of tRNA introns in eukaryotes is a multi-step process that requires several RNA processing enzymes. The first step of the splicing reaction is excision of the intron, which is carried out by the heterotetrameric tRNA splicing endonuclease complex (TSEN) (Abelson et al., 1998; Hayne et al., 2020; Paushkin et al., 2004). The TSEN complex is comprised of the endonucleases TSEN2 and TSEN34 and two structural proteins, TSEN15 and TSEN54, the latter of which is believed to be responsible for orienting the complex through a ruler-like mechanism (Reyes & Abelson, 1988). Recent reconstitution studies with the recombinant human TSEN complex have shown that this complex in humans, as with other eukaryotes, is sufficient for the cleavage step of splicing (Figure 1) (Hayne et al., 2020; Sekulovski et al., 2021). The TSEN2 subunit is responsible for cleaving the 5′ splice site while the TSEN34 subunit is responsible for cleaving the 3′ splice site. The cleavage reaction generates three products, the 5′-exon containing a 2′3′-cyclic phosphate, the 3′-exon containing a 5′-hydroxyl and the excised intron containing 5′-hydroxyl and 2′3′-cyclic phosphate ends.
Figure 1: tRNA splicing pathway. A cartoon of the tRNA splicing pathway.
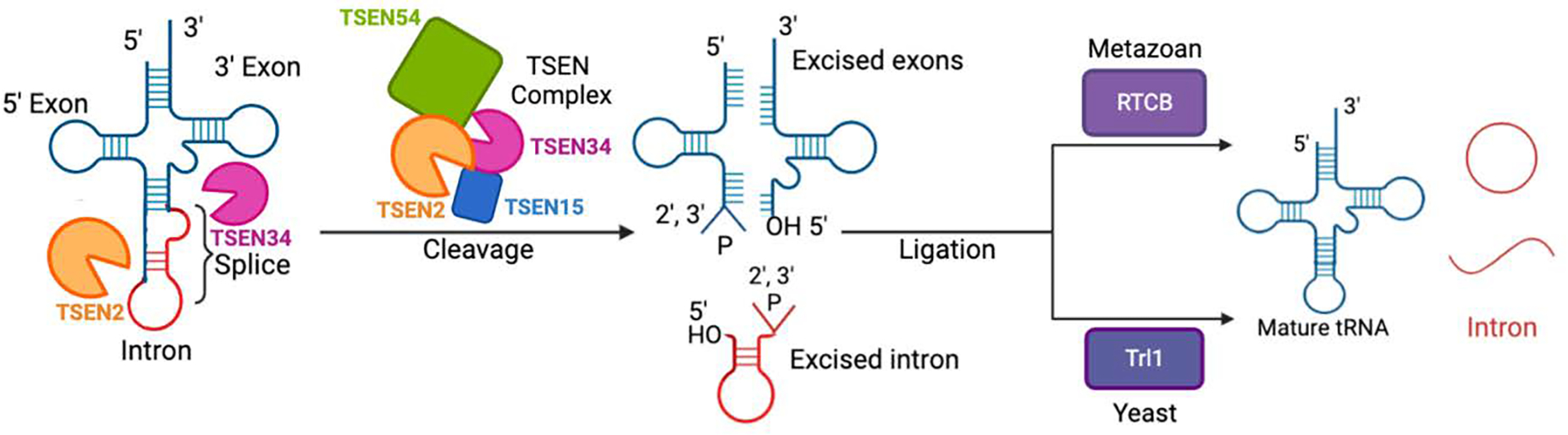
An intron-containing pre-tRNA is labelled showing the exons (teal) and intron (red). The endonuclease TSEN2 (orange) cuts at the 5′ splice site junction with the intron while the endonuclease TSEN34 (pink) cuts the 3′ exon/intron junction. The cleavage is conducted by the TSEN complex TSEN54 (green), TSEN2 (orange), TSEN34 (pink), TSEN15 (blue), resulting in an excised intron (red) and the exons (teal). The newly cleaved ends are a 5′-exon containing a 2′3′-cyclic phosphate, the 3′-exon containing a 5′-hydroxyl and the excised intron containing 5′-hydroxyl and 2′3′-cyclic phosphate ends. Finally, ligation proceeds the cleavage. In metazoans, the tRNA Ligase RTCB is the ligase while in yeast, Trl1 performs the ligation. Both ligation pathways lead to mature tRNA and linear and circular intron products. This figure was created with BioRender.com.
The next step of tRNA splicing is the ligation of the exon halves together to form the mature tRNA with an anti-codon loop. While the excision of the intron appears to be conserved across eukaryotes, the ligation pathways diverge from yeast and metazoans (Figure 1). In humans, the cleaved exons are directly re-sealed by the RTCB tRNA ligase complex which is a large multi-protein complex containing the ligase RTCB (sometimes referred to as HSPC117) along with several additional cofactors including archease, the DEAD-box helicase DDX1, ASW(C2orf49), FAM98B, and CGI-99 (Kroupova et al., 2021; Popow et al., 2011; Popow et al., 2014). In addition to sealing the exons halves together, RTCB can also drive the circularization of tRNA introns (Schmidt et al., 2019). This direct ligation is referred to as the “direct ligation pathway” because RTCB directly ligates RNAs containing 5′-hydroxyl and 2′3′-cyclic phosphate ends. In contrast, yeast utilize a different three-step ligation process called the “heal and seal pathway”, which is driven by the trifunctional enzyme Trl1 (Figure 1) (Greer et al., 1983; Phizicky et al., 1986). Trl1 contains three enzymatic domains including an RNA ligase, RNA kinase, and cyclic phosphodiesterase domain. During the healing stage, the cyclic phosphodiesterase opens the 2′3′-cyclic phosphate while the RNA kinase phosphorylates the 5′-hydroxyl. The exon halves are then ligated together during the sealing step, by Trl1’s RNA ligase domain. Beyond tRNA splicing, RTCB and Trl1 have been shown to function in other RNA processing pathways such as mRNA splicing during the unfolded protein response (Cherry et al., 2019; Kosmaczewski et al., 2014; Peschek & Walter, 2019). While RTCB and Trl1 have been shown to function in other RNA processing pathways, little is known about what role the TSEN complex plays in processing RNAs beyond intron containing pre-tRNAs. In addition to diverging ligation pathways, the spatial regulation of the TSEN complex differs from yeast and metazoans. The yeast TSEN complex localizes to the mitochondria while the human complex is found in the nucleus and this difference likely has a significant influence on substrates (Paushkin et al., 2004; Yoshihisa et al., 2007; Yoshihisa et al., 2003).
Mutations within all four subunits of the TSEN complex are associated with pontocerebellar hypoplasia (PCH), which is a family of rare neurodevelopmental and neurodegenerative diseases characterized by genotype and clinical phenotypes which include microcephaly, seizures, cognitive impairment, among others (van Dijk et al., 2018). Beyond the TSEN complex, PCH is also associated with mutations in several other RNA processing enzymes suggesting a strong link between defects in RNA processing and PCH. The underlying pathogenesis for PCH remains poorly understood but recent studies are beginning to illuminate the connections between altered RNA metabolism and PCH. In this review, we feature recent findings into the structure, function, and regulation of the eukaryotic TSEN complex. Progress in 3D structure prediction (Jumper et al., 2021; Tunyasuvunakool et al., 2021) coupled with homology to the archaeal TSEN complex allowed us to build a 3D model for the human TSEN complex. We also explore the connections between the TSEN complex and PCH and highlight outstanding questions in tRNA splicing.
2. ARCHITECTURE OF THE EUKARYOTIC TSEN COMPLEX
Identification and Purification of the Eukaryotic TSEN Complex
TSEN2 was the first subunit of the TSEN complex to be identified in S. cerevisiae in 1988 from genetic screens looking for genes defective in tRNA splicing (Winey & Culbertson, 1988). The endogenous complex was initially isolated from S. cerevisiae and shown to contain at least three subunits with apparent molecular weights of 31, 42, and 51 kDa (Rauhut et al., 1990), but the identities of these proteins remained unknown at the time. Using FLAG-tagged TSEN2 as bait Trotta et al. were able to purify and identify the TSEN15, TSEN34, and TSEN54 subunits, who were named based on their molecular weights (Trotta et al., 1997). Comparison of the sequences of the TSEN2 and TSEN34 subunits revealed that both genes share a region of ~130 amino acids with homology to previously identified archaeal tRNA splicing endonucleases (Trotta et al., 1997). This ground-breaking work revealed that eukaryotic TSEN complexes are heterotetramers composed of two nuclease subunits (TSEN2 and TSEN34) and two additional subunits (TSEN15 and TSEN54) (Figure 1). This subunit composition is very different from the archaeal TSEN complexes which are composed of homotetramers, homodimers, or heterodimers (for comprehensive reviews on the archaeal enzymes please see (Calvin & Li, 2008; Hirata, 2019; Yoshihisa, 2014)), however one unifying feature is that both eukaryotic and archaeal TSENs have at least two individual endonuclease subunits to facilitate cleavage of the 5′ and 3′ splice sites and two structural domains or subunits to support substrate recognition and complex stability.
The human TSEN subunits were identified based upon a combination of sequence similarity to the yeast subunits and affinity purification with individually tagged TSEN subunits transfected into HEK293 cells (Paushkin et al., 2004). SDS-PAGE analysis of the coimmunoprecipitations from HEK293 cells also revealed the presence of additional proteins that co-purify with the TSEN complex, one of which was identified as the polynucleotide kinase CLP1 (Paushkin et al., 2004). CLP1 participates in multiple RNA processing pathways and several putative roles for CLP1 in tRNA splicing have been proposed (reviewed in (Weitzer et al., 2015)). Recent reconstitution experiments from both E. coli and insect cell expression systems have shown that CLP1 is not essential to facilitate splicing (Hayne et al., 2020; Sekulovski et al., 2021) and likely plays an important regulatory role discussed later in this review. The four subunits of the TSEN complex have also been recently identified in Drosophila and knockdown of any individual subunit blocks tRNA splicing (Schmidt et al., 2019). Collectively this work established that the heterotetrameric architecture of the TSEN complex is well conserved across eukaryotes, the core TSEN subunits are sufficient for intron excision, and that the mammalian TSEN complex has additional binding partners that may be important for regulation of tRNA splicing in vivo and/or supporting the processing of other RNA substrates (Paushkin et al., 2004).
Structure and Function of the TSEN Subunits
We currently lack high-resolution structures of the eukaryotic TSEN complex bound to a pre-tRNA which has hindered our understanding of how the TSEN complex recognizes and processes intron containing pre-tRNAs. Our current knowledge of the structure and function of each individual TSEN subunit comes from sequence and structural similarity to archaeal homologues, combined with biochemical and genetics experiments, and a partial structure of TSEN15 and the TSEN15:TSEN34 sub-complex. While structures for the rest of the TSEN complex and the TSEN complex bound to pre-tRNA are eagerly awaited, advances in protein structure prediction have begun to shed light on the architecture of the human complex (Table 1).
Table 1:
Available TSEN Structures
| PDB_ID(s) | Reference(s) | |
|---|---|---|
| α4 | 1A79 (Methanocaldococcus jannashii) | (Li et al., 1998) |
| αβ2 | 1RLV (Archaeoglobus fulgidus) 2GJW (RNA bound, Archaeoglobus fulgidus) 1R11, 1R0V (truncation, Archaeoglobus fulgidus) 2OHC, 2OHE (Thermoplasma acidophilum) |
(Li & Abelson, 2000) (Xue et al., 2006) (Zhang & Li, 2004) (Kim et al., 2007) |
| (αβ)2 | 3P1Z, 3P1Y, 3AJV (Aeropyrum pernix) 3IEY, 3IF0 (Nanoarchaeum equitans) 2ZYZ (Pyrobaculum aerophilum) 5X89 (Methanopyrus kandleri) |
(Hirata et al., 2011);(Okuda et al., 2011) (Mitchell et al., 2009) (Yoshinari et al., 2009) (Kaneta et al., 2018) |
| ε2 | 4FZ2 (Candidatus Micrarchaeum acidiphilum ARMAN-2) | (Hirata et al., 2012) |
| TSEN15 TSEN34 TSEN2 TSEN54 |
2GW6 (homo sapiens, TSEN15) 6Z9U (truncation, homo sapiens TSEN15/TSEN34) N/A N/A |
(Song & Markley, 2007) (Sekulovski et al., 2021) |
Archaeal building block:
The four individual subunits of the eukaryotic complex are thought to have evolved from a common archaeal ancestor (reviewed in (Calvin & Li, 2008; Hirata, 2019)). To date, four classes of archaeal TSEN complexes (also referred to as EndA) have been discovered and classified based upon the subunit composition (Table 1). The α4 class is a homotetramer composed of four identical α-subunits. Each α-subunit is made up of two subdomains: an αN-terminal subdomain composed of 4 β-strands and 3 α-helices followed by the αC-terminal subdomain composed of 5 β-strands and 2 α-helices (Figure 2A) (Li et al., 1998). The αC-subdomain also contains the well conserved tyrosine, histidine, and lysine residues that compose the catalytic triad in the active site that is responsible for facilitating the RNAse A-like transesterification reaction. Oligomerization of the α-subunit is supported by two key interaction interfaces including a β-β dimer interface formed by the final β-strand of two αC subdomains and a negatively charged loop (L10) from αC that fits into a positively charged pocket between the αN and αC subdomains of another α-subunit (Figure 2B).
Figure 2: The archaeal conserved α-subunit as a building block for a complex.
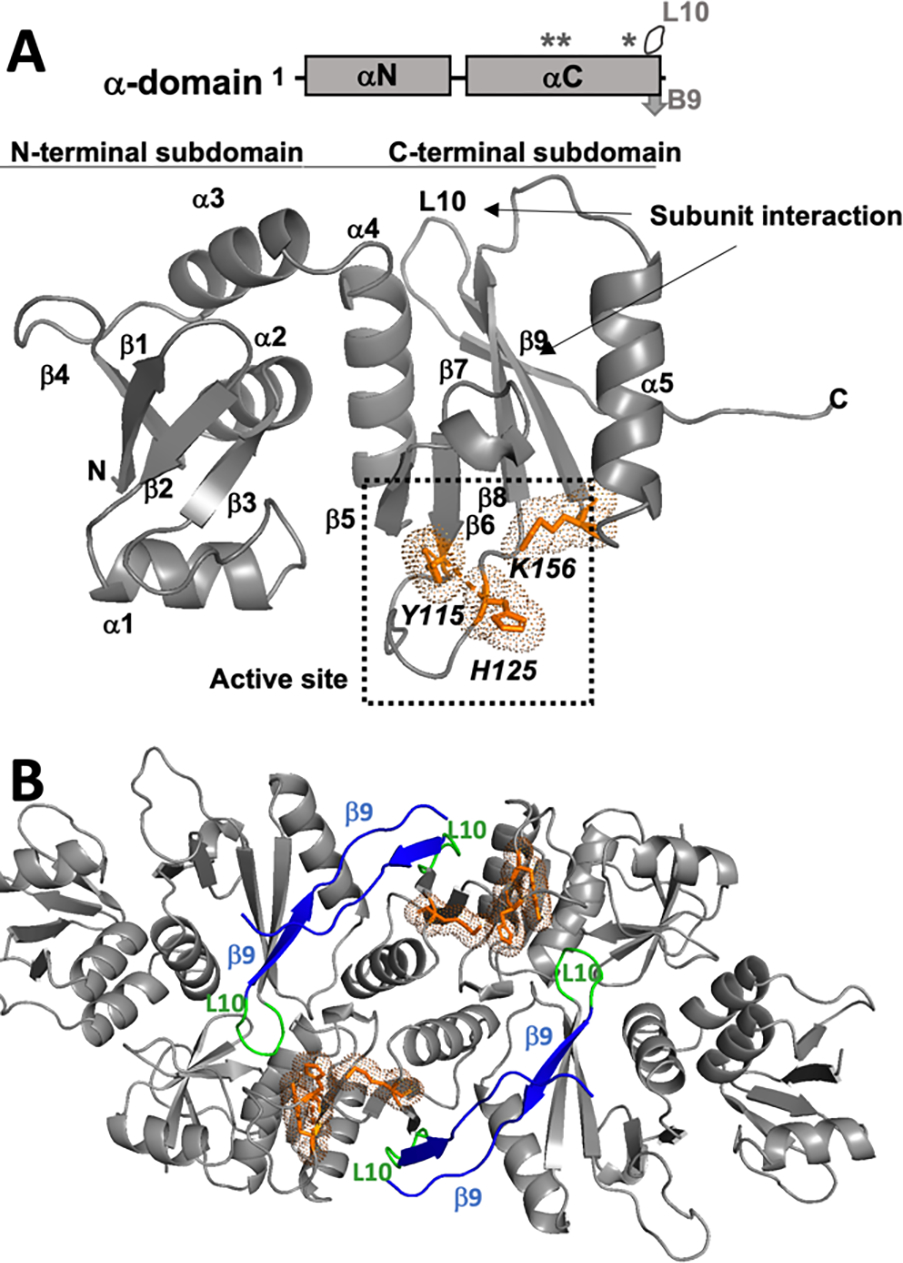
A) A stick cartoon showing the archaeal α-domains, labelled with the L10 loop, B9 β-sheet, and the active site residues (designated by*). The archaeal building block is a subunit comprised of an αN-terminal subdomain composed of 4 β-strands and 3 α-helices, followed by the αC-terminal subdomain composed of 5 β-strands and 2 α-helices. The active site is within the αC-terminal subdomain and is made up of Y115, H125, and K156. (PDB: 1A79). B) An α4 (PDB: 1A79) archaeal tRNA splicing complex, which is comprised of 4 α-subunits. The 4 β9-strands (blue), each from conserved inter-subunit interactions, supported by the two interior L10 loops (green). Two other L10 loops (also green) are exterior. Finally, the two interior active sites that would engage an RNA substrate are shown in orange with sticks and dots.
The other three classes of archaeal TSENs have modifications to the α-subunit building block but the overall structures and the interfaces between subunits are similar to one another (Hirata et al., 2012; Hirata et al., 2011; Kaneta et al., 2018; Kato-Murayama, 2005; Kim et al., 2007; Li & Abelson, 2000; Li et al., 1998; Mitchell et al., 2009; Okuda et al., 2011; Xue et al., 2006; Yoshinari et al., 2009; Zhang & Li, 2004). For example, in the α′2 class, two α-subunits are linked together to form a single chain (α′) that dimerizes. The (αβ)2 class is made-up of two α-subunits which function as the active endonucleases and two β-subunits, that are structurally very similar to the α-subunits but play a supportive structural role in the complex. The final class of archaeal TSEN is the ε2 class which is a homodimer. Each ε subunit contains a linked α- and β-subunit on a single chain that is a result of domain shuffling, with the nuclease α-subunit inserted between the βN and βC-subdomains of the structural β-subunit and connected by two linkers.
TSEN15:
TSEN15, is the smallest subunit of the complex (Figure 3A) and the first eukaryotic TSEN subunit to have its structure determined. The NMR solution structure of TSEN15 revealed that TSEN15 has a disordered N-terminus followed by a small domain composed of a 6-stranded β-sheet and three α-helices, that is structurally similar to the αC-subdomain from the archaeal Methanocaldococcus jannashii (MJ) TSEN (Song & Markley, 2007), however TSEN15 lacks the αN-subdomain of the α-subunit building block. In solution, purified TSEN15 is a homodimer with the dimer interface supported by a β-strand swap (Song & Markley, 2007). Reconstitution experiments with the full TSEN complex suggest that the stoichiometry of TSEN54:TSEN2:TSEN34:TSEN15 is 1:1:1:1 and contains only a single TSEN15 subunit (Hayne et al., 2020). This is further supported by a recent crystal structure of TSEN15 bound to a portion of TSEN34, which has a 1:1 stoichiometry in solution (Sekulovski et al., 2021). The TSEN15-TSEN34 structure revealed an extensive hydrophobic interface between TSEN15 and TSEN34 that is mediated by the final C-terminal β-strand of each subunit (Sekulovski et al., 2021). This β-β interface is reminiscent to the interfaces between protomers of the archaeal TSENs (Hirata et al., 2012; Hirata et al., 2011; Kaneta et al., 2018; Kato-Murayama, 2005; Kim et al., 2007; Li & Abelson, 2000; Li et al., 1998; Mitchell et al., 2009; Okuda et al., 2011; Xue et al., 2006; Yoshinari et al., 2009; Zhang & Li, 2004) and supports the hypothesis that TSENs evolved from a common ancestor (Hirata, 2019).
Figure 3. Structures and predicted structures of the TSEN proteins.
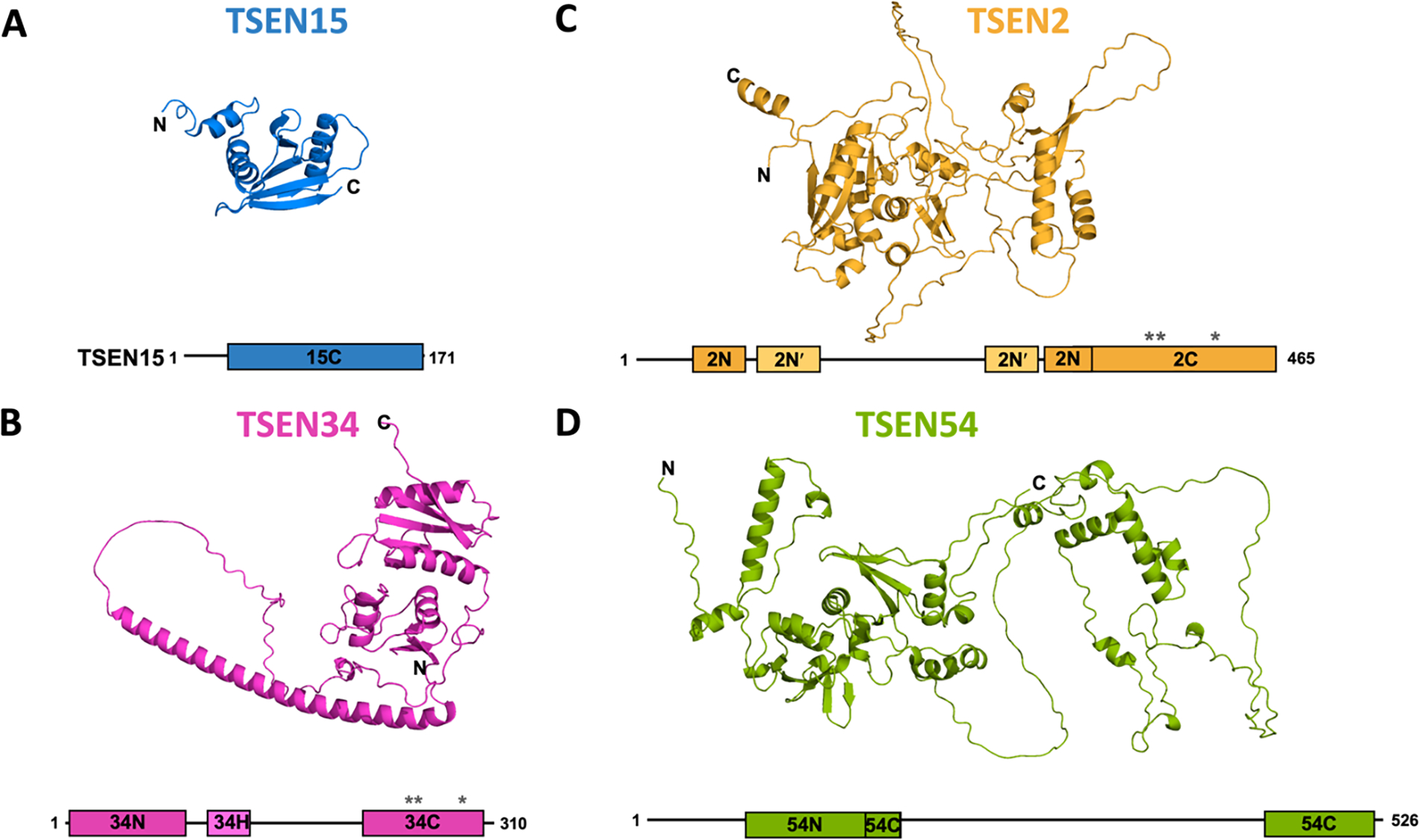
Depiction of each of the TSEN proteins showing known or predicted structures for each protein, paired with a bar cartoon of the protein. The cartoons show the conserved archaeal building block αN and αC - terminal subdomains and active site residues of the endonucleases are designated by “*”.
A) NMR structure of the TSEN15 (PDB:2GW6). B) Alphafold predicted structure for TSEN34. C) Alphafold predicted structure for TSEN2. D) Alphafold predicted structure for TSEN54.
TSEN34:
TSEN34 is one of the two endoribonuclease subunits and is responsible for catalyzing the excision of the 3′ splice site. Human TSEN34 is composed of 310 residues (Figure 3B). Structural prediction suggests that TSEN34 has ordered domains at its N(34N)- and C(34C)-termini that are separated by a large disordered region that is predicted to contain a long α-helix (Tunyasuvunakool et al., 2021). Limited experimental structural information for TSEN34 comes from a crystal structure of a TSEN34 fragment (residues 208–310) bound to N-terminal truncated TSEN15 (residues 23–170), described above. The fragment was selected by limited proteolysis of the full human TSEN complex (Sekulovski et al., 2021). This stable subcomplex is a dimer in solution but crystalized as a heterotetramer as the result of a domain swap within TSEN34, presumably because of the absence of the 34N domain.
TSEN2:
TSEN2 is responsible for catalyzing the excision of the 5′ splice site. Yeast two hybrid experiments revealed that TSEN2 forms a homodimer with TSEN54 (Trotta et al., 1997). Currently there are no structures available for any eukaryotic TSEN2. Secondary structure prediction along with the Alphafold model suggests that TSEN2 has three structured domains (Tunyasuvunakool et al., 2021). The first domain (2N) resembles the NTD of the archaeal α-building block with 4 β-strands and 3 α-helices, but it has a very large insertion between residues 70 and 300. Part of this insertion is predicted to fold into another small domain, which we have named the 2N′ domain (Figure 3C). Following the 2N domain is the predicted 2C domain containing the catalytic residues. Alphafold predicts the 2C domain to be structurally similar to the C-terminal subdomain of the alpha building block.
TSEN54:
TSEN54, is the largest subunit of the complex and earlier work suggests that it functions as a molecular ruler to help orient the pre-tRNA for cleavage (Reyes & Abelson, 1988). Similar to TSEN34 and TSEN2, TSEN54 is predicted to have two structured domains (54N and 54C) (Figure 3D). The predicted 54N domain, which shares some structural homology to the αN building block, is preceded by a large (~80 amino acids) unstructured region. While the 54C domain is predicted to be structurally similar to the C-terminal subdomain of the α-building block, it contains a very large, disordered insertion following the first β-strand.
TSEN Tetramer Model:
Using the archaeal structure from MJ-TSEN (PDB: 1A79) as a starting point we created a 3D model of the eukaryotic TSEN complex (Figure 4A). We docked the crystal structure of the TSEN15-TSEN34 subcomplex (PDB: 6Z9U) onto two α-subunits from MJ-TSEN. Next, we used the Alphafold model of TSEN34 – to model in the 34N domain missing from the crystal structure. Then, we took the Alphafold models of TSEN2 and TSEN54 and docked them onto the remaining two α-subunits from MJ-TSEN (Figure 4B). The TSEN2 and TSEN54 interface is predicted to be formed by the final C-terminal x-strand from each C-terminal subdomain. TSEN34, TSEN2, and TSEN54 have large regions predicted to be unstructured, therefore for clarity we removed the predicted disordered insertions from TSEN34, TSEN2, and TSEN54 to produce the final 3D model (Figure 4C). This model is structurally reminiscent of the MJ-TSEN complex, with the exception of the absence of the αN subdomain from TSEN15. Intriguingly, when the full-length TSEN2 alpha-fold model was superimposed on the MJ-TSEN complex, the extra 2N′ is roughly positioned next to the 15C domain. This rough docking suggests that the additional domain from TSEN2 could have arisen from a domain shuffling as observed with the ε2 class from archaea. Taken together, our human TSEN 3D model suggests that the core architecture of the mammalian complex is very similar to the archaeal complexes, but this model awaits structural validation. Moreover, the structure and significance of the large insertions within TSEN2, TSEN34, and TSEN54 remains unknown. These regions likely play critical roles in eukaryotic specific features of the TSEN complex and could facilitate RNA substrate recruitment/binding or interaction with co-factors/binding partners.
Figure 4: Alphafold modeled human TSEN tetramer.
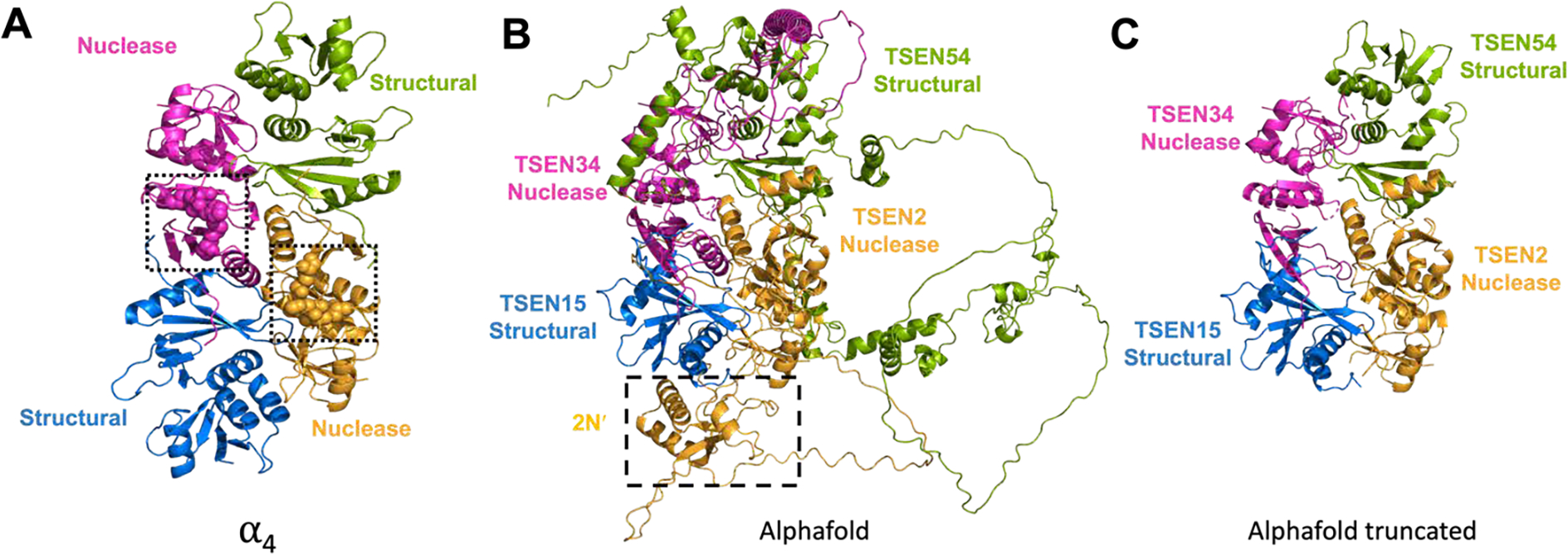
A) The archaeal tRNA splicing complex (PDB:1A79) that was used as the docking interface of the model, with two domains highlighted as nuclease domains (pink and orange), with the active sites shown as spheres and boxed, and the structural domains (green and blue). B) Alphafold models of the TSEN proteins docked onto the previous archaeal tRNA splicing complex. Boxed and labelled is a 2N′ region of the TSEN2 prediction that appears to supplement for part of TSEN15. C) The Alphafold model from part B, with the disordered regions and 2N′ truncated.
3. SUBSTRATES OF THE EUKARYOTIC TSEN COMPLEX
Universal Substrate and Mechanism of Cleavage
The universal substrate of all TSEN complexes is thought to be the bulge-helix-bulge (BHB) motif. This motif is composed of two three-nucleotide bulges separated by a helix containing four base pairs. The first base pair in the helix, known as the proximal base pair (previously referred to as the A-I base pair for anticodon-intron) is critical for cleavage (Baldi et al., 1992; Fabbri et al., 1998; Schmidt et al., 2019; Schmidt & Matera, 2019) Cleavage occurs 1 nucleotide from the 3′ and 5′ ends of the bulges. In most tRNAs, the intron containing the BHB motif is inserted in the same position within the pre-tRNA in the anti-codon stem loop between bases 37 and 38, however exceptions to this have been identified across archaea and in red algae (Kanai, 2015; Maruyama et al., 2010; Randau & Söll, 2008). Alterations to the x-subunit building block and composition of the TSEN subunits impacts substrate specificity (reviewed in (Hirata, 2019)). The α4 class has the strictest substrate specificity, while the human TSEN complex is predicted to be broader (reviewed in (Hirata, 2019)). A crystal structure of the α′2 class from Archaeoglobus fulgidus (AF) bound to a BHB motif revealed the molecular basis for recognition of the BHB motif (PDB:2GJW) (Figure 5A) (Xue et al., 2006). The three residues from each bulge are flipped out from the typical stacking arrangement and positioned within each nuclease active site. The interactions between the α′2 AF-TSEN and the BHB motif are primarily through the RNA backbone and are not nucleotide specific. The orientation of the BHB motif in the crystal structure suggests that there is cooperativity between the two nuclease active sites because of the presence of cross-subunit stabilization of each bulge through a cation-π sandwich. However, experiments in S. cereviase showed that this interaction is only required for cleavage at the 5′ splice site in eukaryotic TSEN complexes (Trotta et al., 2006).
Figure 5. Substrate Recognition by tRNA splicing complexes.
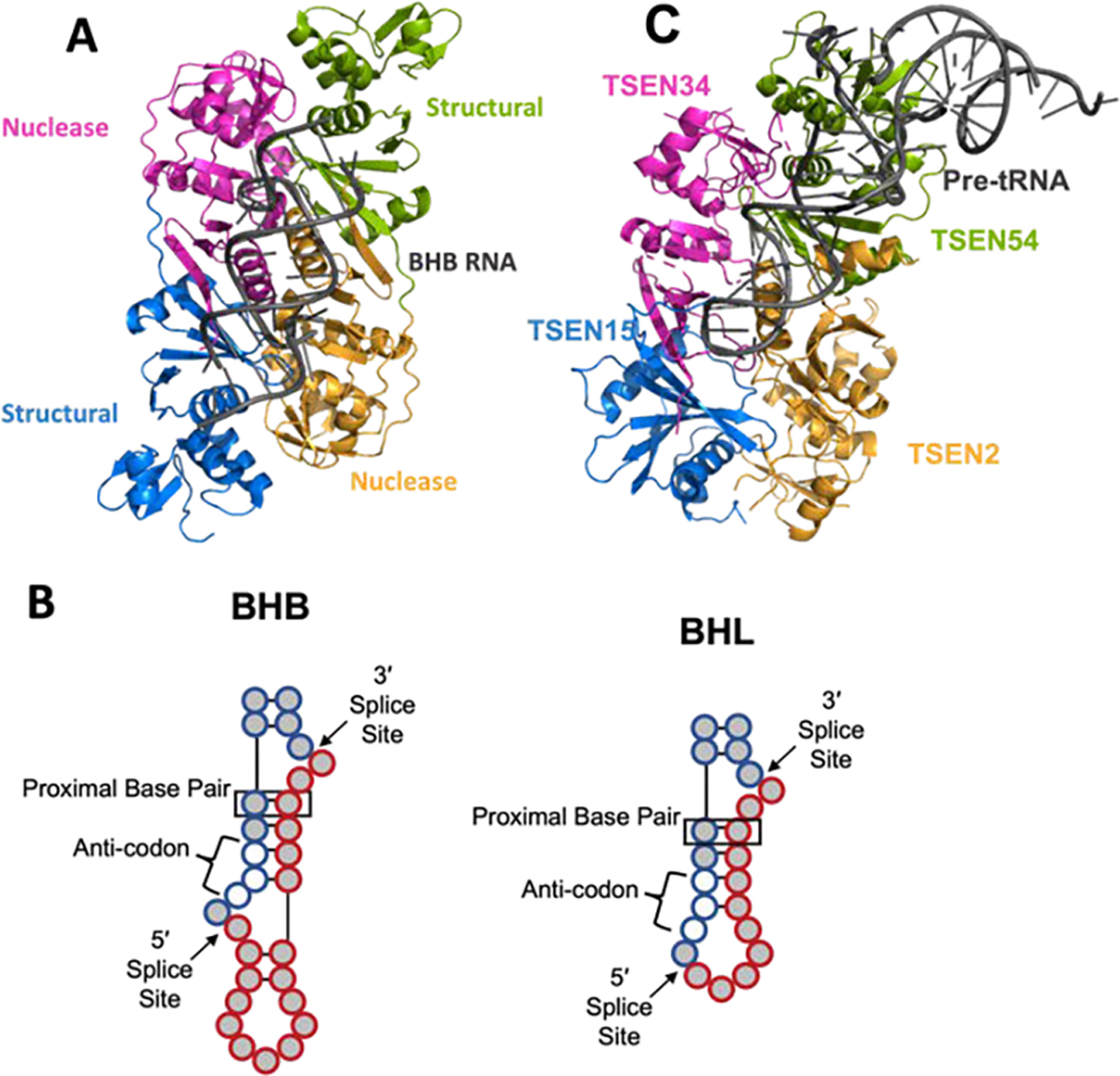
A) The archaeal tRNA splicing complex (PDB:2GJW) bound to a BHB RNA. B) A depiction of the bulge-helix bulge (BHB-type) and bulge-helix-loop (BHL-type) tRNA intron substrate for splicing with the intron (red filled circles) and exon (blue filled circles) shown as well as the anticodon (unfilled circles) and the proximal base pair (boxed). C) The Alphafold model with a tRNA (PDB:4tra) docked onto the complex similar to the BHB with the tRNA arms oriented in the region of TSEN54.
The structure of the AF-TSEN complex bound to the BHB motif also revealed critical information about the mechanism of RNA cleavage. The active sites of archaeal and eukaryotic TSEN endonuclease subunits contain three well conserved catalytic residues including a histidine, tyrosine, and lysine. This active site arrangement is similar to RNase A and other endoribonucleases such as the viral uridine specific endoribonuclease which support a mechanism of transesterification (Cuchillo et al., 2011; Pillon et al., 2021), however one of the conserved histidine residues from RNase A is replaced by a tyrosine in each TSEN active site. The tyrosine in the AF-TSEN structure is positioned for in-line attack on the 2′-hydroxyl, which activates the hydroxyl for nucleophilic attack on the phosphate and generates the 2′3′-cyclic phosphate. The histidine residue functions as a general-acid to donate a proton to the leaving group (the 5′-hydroxyl) while the lysine residue is thought to be important for stabilizing the transition state. The universal conservation of these residues supports the model that all TSEN nucleases cleave RNA through this conserved mechanism (reviewed in (Calvin & Li, 2008)). While the cleavage mechanism is thought to be conserved, alterations to TSEN subunit architecture give rise to altered substrate specificity and the cleavage of substrates with relaxed BHB motifs, such as the Bulge-Helix-Loop (BHL) (Figure 5B). This has been well-studied in the archaeal (αβ)2 and ε2 classes which have specific loop insertions near the active sites that give rise to broad specificity (reviewed in (Hirata, 2019)).
Eukaryotic pre-tRNAs
The well-established substrates of the eukaryotic TSEN complex are intron containing pre-tRNA. The number of intron containing tRNA genes varies widely across eukaryotes (reviewed in (Schmidt & Matera, 2019)). There is also variability in the size of the intron as they can range in size from around 14 nucleotides to over 100. There are 415 high-confidence human tRNA genes, of which 28 are predicted to contain an intron (Chan & Lowe, 2016). Recent developments in high-throughput sequencing technologies have led to an explosion in our knowledge of mature and pre-tRNA transcript levels. A hydrolysis-based (Hydo)-tRNA sequencing method coupled with PAR-CLIP led to the validation of the expression of 26 of the predicted intron containing pre-tRNAs in HEK293 cells (Table 2) (Gogakos et al., 2017). This method further highlighted the significance of the human TSEN complex as no intron-less isoleucine-TAT or leucine-CAA isodecoders were detected. Therefore, the human TSEN complex is essential for the maturation of isoleucine-TAT and leucine-CAA tRNAs. All of the 26 confirmed human intron containing pre-tRNAs have the intron inserted in the canonical position and RNA structure prediction suggest that all the introns fold into either a BHB or relaxed BHL motif (Table 2)(Figure 5B). Additional tRNA sequencing methods have also begun to reveal heterogeneity in the levels of tRNA isodecoders within different cells lines and tissues (Behrens et al., 2021; Pinkard et al., 2020). Continued tRNA sequencing analysis will no doubt begin to reveal how expression levels of intron containing tRNA genes vary across different cells types and tissues.
Table 2: High confidence human intron containing pre-tRNAs.
Human tRNA genes with confirmed expression. The intron sequence is shown in red lowercase (Gogakos et al., 2017).
| tRNA Gene | Sequence |
|---|---|
| tRNA-Arg-TCT-2-1 | GGCTCTGTGGCGCAATGGAtAGCGCATTGGACTTCTAgtgacgaatagagcaATTCAAAGGtTGTGGGTTCGAATCCCACCAGAGTCG |
| tRNA-Arg-TCT-1-1 | GGCTCCGTGGCGCAATGGAtAGCGCATTGGACTTCTAgaggctgaaggcATTCAAAGGtTCCGGGTTCGAGTCCCGGCGGAGTCG |
| tRNA-Arg-TCT-3-2 | GGCTCTGTGGCGCAATGGAtAGCGCATTGGACTTCTAgatagttagagaaATTCAAAGGtTGTGGGTTCGAGTCCCACCAGAGTCG |
| tRNA-Arg-TCT-3-1 | GCTCTGTGGCGCAATGGAtAGCGCATTGGACTTCTAgctgagcctagtgtggtcATTCAAAGGtTGTGGGTTCGAGTCCCACCAGAGTCG |
| tRNA-Ile-TAT-1-1 | GCTCCAGTGGCGCAATCGGTtAGCGCGCGGTACTTATAtgacagtgcgagcggagcaATGCCGAGGtTGTGAGTTCGATCCTCACCTGGAGCA |
| tRNA-Ile-TAT-2-3 | GCTCCAGTGGCGCAATCGGTtAGCGCGCGGTACTTATAcaacagtatatgtgcgggtgATGCCGAGGtTGTGAGTTCGAGCCTCACCTGGAGCA |
| tRNA-Ile-TAT-2-1 | GCTCCAGTGGCGCAATCGGTtAGCGCGCGGTACTTATAcagcagtacatgcagagcaATGCCGAGGtTGTGAGTTCGAGCCTCACCTGGAGCA |
| tRNA-Ile-TET-2-2 | GCTCCAGTGGCGCAATCGGTtAGCGCGCGGTACTTATAtggcagtatgtgtgcgagtgATGCCGAGGtTGTGAGTTCGAGCCTCACCTGGAGCA |
| tRNA-Ile-TAT-3-1 | GCTCCAGTGGCGCAATCGGTtAGCGCGCGGTACTTATAagacagtgcacctgtgagcaATGCCGAGGtTGTGAGTTCAAGCCTCACCTGGAGCA |
| tRNA-Leu-CAA-1-2 | GTCAGGATGGCCGAGTGGTctAAGGCGCCAGACTCAAGcttggcttcctcgtgttgaggaTTCTGGTCTCCAATGGAGGCGTGGGTTCGAATCCCACTTCTGACA |
| tRNA-Leu-CAA-2-1 | GTCAGGATGGCCGAGTGGTctAAGGCGCCAGACTCAAGcttactgcttcctgtgttcgggtcTTCTGGTCTCCGTATGGAGCGTGGGTTCGAATCCCACTTCTGACA |
| tRNA-Leu-CAA-1-1 | GTCAGGATGGCCGAGTGGTctAAGGCGCCAGACTCAAGctaagcttcctccgcggtggggaTTCTGGTCTCCAATGGAGGCGTGGGTTCGAATCCCACTTCTGACA |
| tRNA-Leu-CAA-3-1 | GTCAGGATGGCCGAGTGGTctAAGGCGCCAGACTCAAGttgctacttcccaggtttggggcTTCTGGTCTCCGCATGGAGGCGTGGGTTCGAATCCCACTTCTGACA |
| tRNA-Leu-CAA-4-1 | GTCAGGATGGCCGAGTGGTctAAGGCGCCAGACTCAAGgtaagcaccttgcctgcgggctTTCTGGTCTCCGGATGGAGGCGTGGGTTCGAATCCCACTTCTGACA |
| tRNA-Tyr-GTA-3-1 | CCTTCGATAGCTCAGTTGGTAGAGCGGAGGACTGTAGgctcattaagcaaggtATCCTTAGGtCGCTGGTTCGAATCCGGCTCGGAGGA |
| tRNA-Tyr-GTA-1-1 | CTTCGATAGCTCAGTTGGTAGAGCGGAGGACTGTAGttggctgtgtccttagacATCCTTAGGtCGCTGGTTCGAATCCGGCTCGAAGGA |
| tRNA-Tyr-GTA-2-1 | CCTTCGATAGCTCAGTTGGTAGAGCGGAGGACTGTAGtggatagggcgtggcaATCCTTAGGtCGCTGGTTCGATTCCGGCTCGAAGGA |
| tRNA-Tyr-GTA-4-1 | CCTTCGATAGCTCAGCTGGTAGAGCGGAGGACTGTAGattgtatagacatttgcggacATCCTTAGGTCGCTGGTTCGATTCCAGCTCGAAGGA |
| tRNA-Tyr-GTA-5-5 | CCTTCGATAGCTCAGCTGGTAGAGCGGAGGACTGTAGtacttaatgtgtggtcATCCTTAGGTCGCTGGTTCGATTCCGGCTCGAAGGA |
| tRNA-Tyr-GTA-5-3 | CCTTCGATAGCTCAGCTGGTAGAGCGGAGGACTGTAGcctgtagaaacatttgtggacATCCTTAGGTCGCTGGTTCGATTCCGGCTCGAAGGA |
| tRNA-Tyr-GTA-5-4 | CCTTCGATAGCTCAGCTGGTAGAGCGGAGGACTGTAGattgtacagacatttgcggacATCCTTAGGTCGCTGGTTCGATTCCGGCTCGAAGGA |
| tRNA-Tyr-GTA-6-1 | CCTTCGATAGCTCAGCTGGTAGAGCGGAGGACTGTAGgggtttgaatgtggtcATCCTTAGGTCGCTGGTTCGAATCCGGCTCGGAGGA |
| tRNA-Tyr-GTA-5-1 | CTTCGATAGCTCAGCTGGTAGAGCGGAGGACTGTAGctacttcctcagcaggagacATCCTTAGGTCGCTGGTTCGATTCCGGCTCGAAGGA |
| tRNA-Tyr-GTA-8-1 | CTTTCGATAGCTCAGTTGGTAGAGCGGAGGACTGTAGgttcattaaactaaggcATCCTTAGGTCGCTGGTTCGAATCCGGCTCGAAGGA |
| tRNA-Tyr-GTA-7-1 | CCTTCGATAGCTCAGCTGGTAGAGCGGAGGACTGTAGactgcggaaacgtttgtggacATCCTTAGGtCGCTGGTTCAATTCCGGCTCGAAGGA |
| tRNA-Tyr-GTA-5-2 | CCTTCGATAGCTCAGCTGGTAGAGCGGAGGACTGTAGgcgcgcgcccgtggccATCCTTAGGtCGCTGGTTCGATTCCGGCTCGAAGGA |
How the TSEN complex recognizes intron containing pre-tRNA is still poorly understood. Lack of a structure of any TSEN complex bound to an intron containing pre-tRNA has hindered our understanding of how the TSEN complex engages the pre-tRNA, outside of the universal BHB region. Biochemical experiments have established several general principles concerning recognition of the pre-tRNA (Culbertson & Winey, 1989; Schmidt & Matera, 2019; Trotta, 1999; Yoshihisa, 2014). The first is that, with the exception of the proximal base-pair, the sequence and size of the intron does not matter. Eukaryotic TSEN complexes do not have the strict specificity requirement of base-pairing interactions within the anti-codon-intron helix of the BHB motif (Reyes & Abelson, 1988; Schmidt et al., 2019). The second is that mutations to distinct structural features of the mature tRNA lead to alterations in splicing. For example, mutation of bases that mediate tertiary interactions within the mature tRNA disrupt splicing (Greer et al., 1987; Reyes & Abelson, 1988). The third principle is that the length of the anticodon stem loop dictates the cleavage site. Insertion or deletion of bases within the anticodon stem loop alters the position of the cleavage site (Reyes & Abelson, 1988). Collectively these key observations led to establishment of the “Ruler Mechanism” whereby the TSEN complex uses structural features of the mature tRNA to position the intron within the active sites for cleavage. Modelling of a tRNA onto the AF- TSEN BHB co-crystal structure roughly positions the D-arm and acceptor stem of the tRNA near one of the structural subunits of AF-TSEN, suggesting that TSEN54 is likely mediating the ruler mechanism (Xue et al., 2006). We updated this tRNA docking model using the Alphafold model of the human TSEN complex. In this updated model the majority of the interfaces between the mature tRNA and the TSEN complex are mediated by TSEN54, however TSEN34 also lies near the predicted tRNA binding site, suggesting that both of these subunits contribute to recognition and binding of the pre-tRNA (Figure 5C).
Another outstanding question about eukaryotic intron containing tRNA is, what is the biological significance of the intron (for a recent review on this topic please refer to (Schmidt & Matera, 2019)). Recent work in S. cerevisiae has shown that removal of all the introns is not lethal, however removal of specific introns does impact cell growth under certain conditions such as cold temperatures (Hayashi et al., 2019). This observation hints toward a potential role of introns in optimizing cell fitness following physiological stress.
Beyond pre-tRNAs: Additional Substrates of the Eukaryotic TSEN Complex
While tRNA introns are not essential in yeast, all four members of the TSEN complex are essential. A genetic bypass system, in which all the intron containing tRNAs were removed revealed that each TSEN subunit is essential even in the absence of intron containing tRNAs (Cherry & White, 2018). This bypass system elegantly suggests that the TSEN complex must have additional essential substrates beyond the intron containing pre-tRNAs. The first non-tRNA TSEN substrate to be identified was the CBP1 mRNA (Tsuboi et al., 2015). Cbp1 (cytochrome b mRNA processing) is a non-essential protein that is imported into the mitochondria and suggested to play a role in regulating post-transcriptional processing and translation of cytochrome b. The yeast TSEN complex was shown to cleave a stem loop structure within CBP1 mRNA on the mitochondrial outer membrane both in vivo and in vitro. A recent bioinformatic approach termed comPARE (comparative parallel analysis of RNA ends) led to the identification of several additional yeast TSEN mRNA substrates (Hurtig et al., 2021). Interestingly all these new mRNA substrates encode for mitochondrial proteins, suggesting that the spatial regulation of the TSEN complex influences its substrates. The role of the TSEN complex in cleaving these additional mRNAs appears to be in mediating their decay via TED (tRNA endonuclease-initiated mRNA decay), whereby cleavage initiates subsequent exonuclease decay. Beyond mRNAs, connections have also been made between the yeast TSEN complex and pre-rRNA processing. Mutations in both TSEN34 and TSEN2 have been shown to lead to accumulation of pre-rRNAs (Dhungel & Hopper, 2012; Volta et al., 2005).
The spatial localization of the TSEN complex is not conserved across eukaryotes. The yeast complex localizes to the outer membrane of the mitochondria (Yoshihisa et al., 2003) while the human complex is localized to the nucleus (Paushkin et al., 2004). To date, no additional substrates have been identified for the human complex but given the difference in cellular localization between yeast and human, the non-tRNA substrates are not likely to be conserved. Adaptation of the comPARE methodology to mammalian cell lines along with 2′3′-cyclic phosphate and 5′-hydroxyl sequencing approaches will hopefully aid in the future determination of non-tRNA substrates of the human complex (Hurtig et al., 2021; Peach et al., 2015; Shigematsu et al., 2019).
4. REGULATORS OF TRNA SPLICING
The timely and accurate processing of intron containing tRNAs is important for maintaining cellular homeostasis. Several recent studies have provided new insights into ways that tRNA splicing may be regulated by factors that target the newly liberated ends of cleaved exons and introns: the 5′-hydroxyl polynucleotide kinase CLP1 and the recently discovered 2′3′-cyclic phosphatase ANGEL2 (Pinto et al., 2020). The tRNA ligase RTCB ligates the 2′3′-cyclic phosphate and 5′-hydroxyl ends left by the TSEN complex (Figure 1) (Popow et al., 2011); however, CLP1 and ANGEL2 both have enzymatic activities that could target the cleavage ends left by the TSEN complex, this results in impaired ligation by RTCB and suggests multiple levels of cellular regulation of tRNA splicing.
We recently discovered that CLP1 regulates tRNA maturation, likely through its catalytic activity (Hayne et al., 2020). CLP1 was previously known to interact with the TSEN complex (Paushkin et al., 2004), but its role in tRNA splicing, whether supportive or catalytic, remained unclear. While CLP1 has also been known to play a non-catalytic role in the mRNA pre-adenylation complex (de Vries et al., 2000), mouse models of kinase-dead CLP1 exhibit PCH (Hanada et al., 2013; Szoták-Ajtay et al., 2020), this suggests that CLP1’s kinase activity is essential for its proper function. CLP1 is a 5′-hydroxyl polynucleotide kinase, that phosphorylates 5′-hydroxyls on ss- and ds-RNA substrates with broad specificity (Dikfidan et al., 2014). It has also been demonstrated that Human CLP1 can phosphorylate the available 5′-hydroxyls on both the cleaved 3′-exon and intron (Weitzer & Martinez, 2007) (Figure 6A). CLP1 phosphorylation of the TSEN cleavage products presumably triggers kinase mediated decay by a 5′-3′-exonuclease (Cherry et al., 2019), such as XRN1 and XRN2, and also blocks RTCB ligation, preventing mature tRNA formation and blocking formation of tRNA intron circles (tricRNAs) (Figure 6A). The idea that CLP1 is required to help promote degradation of introns is supported by a recent CLP1 R140H PCH mouse model that revealed CLP1 R140H mice had an increase in both linear and circularized tRNA introns (Monaghan et al., 2021). This study further supports that CLP1 may drive kinase mediated exonuclease degradation, similar to intron degradation in yeast (Wu & Hopper, 2014). Thus, through its dual role in blocking ligation and targeting substrates for degradation, CLP1 may be a powerful negative regulator of tRNA splicing.
Figure 6: Regulation of tRNA splicing.
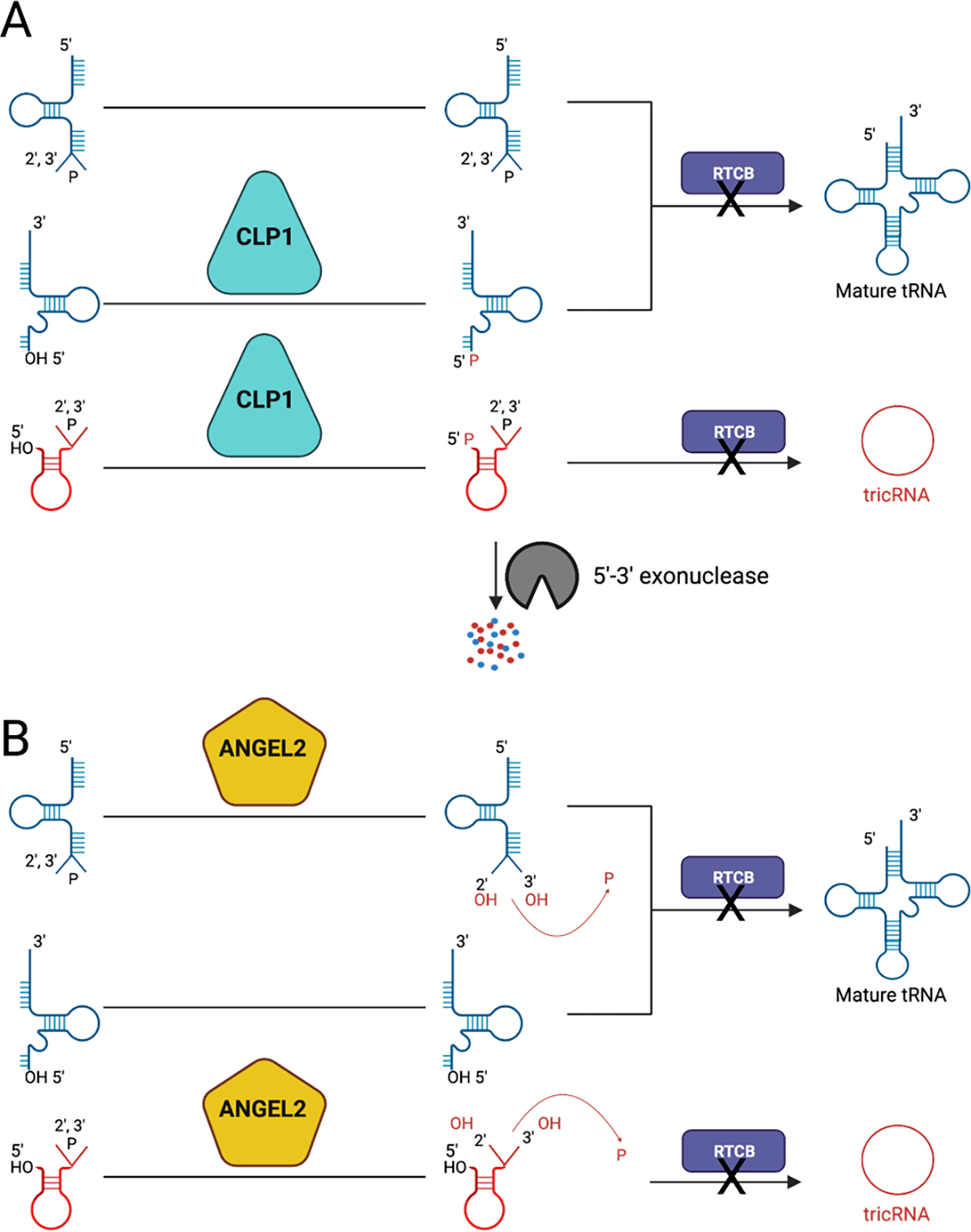
A) A cartoon of how CLP1 may modify splicing intermediates in which CLP1, a 5′-hydroxyl polynucleotide kinase, results in phosphorylation of the 5′-hydroxyl on the 3′-exon and the intron. The phosphorylated products are then unable to be ligated by the tRNA ligase RTCB, preventing the formation of mature tRNA and tricRNAs. At the bottom, it is shown that the phosphorylated intron may be targeted for kinase mediate intron degradation by a 5′-3′-exonuclease. B) A cartoon of how ANGEL2 may modify splicing intermediates. ANGEL2, as a 2′,3′-cyclic phosphatase, is believed to open the 2′,3′-cyclic phosphate on the 5′-exon and the intron, resulting in 2′ and 3′-hydroxyls and liberating a phosphate. These hydroxyl ends prevent the ligation of the splicing intermediates by the tRNA ligase RTCB, and thus preventing the formation of mature tRNA and tricRNAs. This figure was created with BioRender.com.
Similar to CLP1, a recent study by Pinto et al showed that ANGEL2 may also function as a negative regulator of tRNA maturation through a mechanism that targets the 2′3′-cyclic phosphate ends left by the TSEN complex (Pinto et al., 2020). Through a two-step mechanism, ANGEL2 opens the cyclic phosphate, breaks the 3′-bond to the phosphate before removing the phosphate altogether leaving hydroxyls in the 2′ and 3′ positions (Pinto et al., 2020) (Figure 6B). The authors discovered that ANGEL2 cell lysates accumulated cleaved tRNA splicing intermediates. Like CLP1, ANGEL2 may serve to regulate the re-ligation of splicing intermediates, while targeting the opposite ends from CLP1. In support of this hypothesis, it was found that in cellular extracts in which an N-terminal truncated version of ANGEL2 was overexpressed, catalytically active, but not catalytic deficient ANGEL2, resulted in accumulation of unligated introns and dephosphorylated 5′-exons, while having no impact on pre-tRNA cleavage (Pinto et al., 2020). The chemistry of ANGEL2 would result in hydroxyls on both the cleaved 3′ and 5′ exons, preventing their ligation. The fate of these unligated tRNA fragments in vivo remains unknown, although it was previously shown that the unphosphorylated product, in addition to oxidative stress, results in cellular death (Schaffer et al., 2014). Furthermore, the authors found that ANGEL2 also potentially serves a role in tRNA maturation through mediating CCA addition on the 3′-termini of pre-tRNAs (Pinto et al., 2020), signifying that ANGEL2 could play a dual role in regulating tRNA maturation.
While CLP1 and ANGEL2 seem to be able to regulate tRNA splicing and maturation, a major outstanding question remains: is there a rescue mechanism to rescue the intermediate RNA species? As mentioned above, the tRNA ligation pathway differs in yeast and humans but we cannot rule out the possibility of the existence of an alternative yeast like ligation pathway in metazoans. If CLP1 and ANGEL2 both acted on the same tRNA intermediates, they would result in products that could be ligated through a yeast-like ligation pathway. Previous studies suggest that CLP1 (Ramirez et al., 2008) and CNPase (Schwer et al., 2008) could account for the first two steps of a yeast-like ligation pathways in humans. Yeast like ligase activity has been detected in HeLa cells (Zillmann et al., 1991), but the identification of this ligase remains elusive. Furthermore, the role these regulators play in the splicing of other RNA species in vivo needs to be explored further. ANGEL2 has been implicated in processing of the unfolded protein response mRNA XBP1, which is cleaved by inositol-requiring enzyme 1 (IRE1), an endonuclease that also leaves 2′3′-cyclic phosphate and 5′-hydroxyl ends (Pinto et al., 2020). Moreover, given that the specificity of ANGEL2 and CLP1 is broad, these two RNA processing enzymes could play regulatory roles in many additional RNA processing pathways.
One important aspect related to the function and regulation of all of these enzymes appears to be their spatial regulation; from the substrate specificity of the TSEN complex on the mitochondria, to the direct interaction between CLP1 and TSEN that’s disrupted by the known CLP1 PCH mutation R140H, it’s clear that spatial regulation is critical for proper tRNA splicing and regulation by these enzymes. At this time, it is not clear if ANGEL2 and TSEN directly interact, which may impact the role of ANGEL2’s in splicing in vivo.
5. TRNA SPLICING DEFECTS AND HUMAN DISEASE
Mutations in the TSEN complex and CLP1 are the most common proteins with mutations linked to PCH, an autosomal recessive disease (van Dijk et al., 2018). All five of these proteins are essential for life (Cherry & White, 2018; Hanada et al., 2013; Schaffer et al., 2014) and it remains unclear if these proteins cause disease through a shared or divergent mechanism. It may be that PCH mutations cause rather weak defects in complex assembly, function, or regulation; therefore, allowing the complex to still perform some of its essential function, but in a reduced capacity that still permits life.
CLP1
A single mutation in CLP1, R140H, is linked to PCH and causes the unique PCH10 subtype (Figure 7A, B) (Karaca et al., 2014; Schaffer et al., 2014). CLP1 is composed of three conserved domains including a central kinase domain flanked by N- and C-terminal domains (Saito et al., 2019). R140 lies within the kinase domain and is thought to coordinate intra-domain stability. Mutation of R140 to a histidine (R140H) disrupts a bond to the NTD, leading to impairment in both CLP1’s kinase activity and protein:protein interactions. This mutation does not impact CLP1’s overall protein stability (Karaca et al., 2014; Schaffer et al., 2014). In vitro, R140H, has impaired kinase function and association with the TSEN complex, in addition, R140H reduces CLP1’s nuclear localization in cells (Hanada et al., 2013; Karaca et al., 2014; Schaffer et al., 2014). CLP1 is a multifunctional enzyme. In addition to regulating tRNA splicing, CLP1 is also linked to pre-mRNA 3′ end processing as a structural subunit of cleavage factor II that bridges the cleavage factor complex to rest of the cleavage and polyadenylation complex (de Vries et al., 2000). It remains difficult to assess if PCH is linked to CLP1’s mRNA or tRNA processing function as a kinase dead version of CLP1 (K127A) and the R140H mutation both impact the protein’s ability to associate with the TSEN complex and both have impaired kinase function. While CLP1’s role in mRNA processing is not believed to be related to its kinase activity, it remains unclear if the R140H mutation interferes with CLP1’s ability to act as a bridge for the mRNA processing machinery. If this mutation does impact mRNA maturation, it may be that PCH10 is caused by defects in both mRNA and tRNA processing. In support of this, a recent R140H mouse model found defects in both mRNA and tRNA processing and suggests that PCH disease pathogenesis arises because of defects in 3’ mRNA processing (Monaghan et al., 2021).
Figure 7. PCH mutations in CLP1 and the TSEN complex.
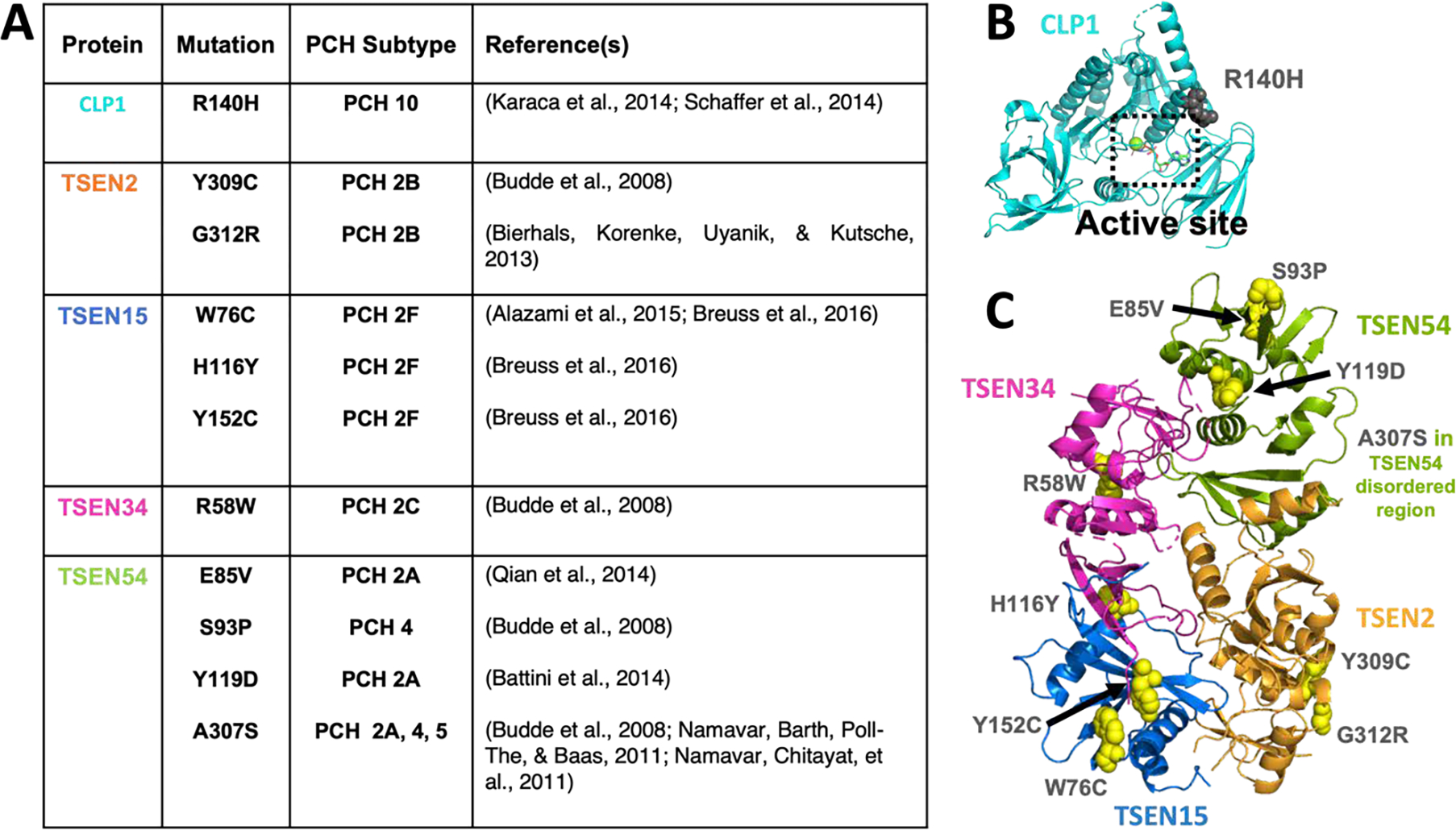
A) A table of PCH mutations in CLP1 and the TSEN proteins that have been characterized in the literature. B) CLP1 structure (PDB:4OHV) with the R140H PCH mutant in grey spheres and the active site boxed. C) The Alphafold TSEN complex model with disordered regions removed and with PCH mutations shown as yellow spheres and labelled. TSEN54’s mutation A307S was not in the regions shown.
Several studies have sought to uncover the mechanism by which CLP1 causes disease by evaluating changes in RNA processing in PCH patient fibroblasts, K127A and R140H mouse models, and in vitro and cell-based assays. These studies mostly focused on evaluating levels of pre-tRNAs, tRNA fragments (including introns) and mature tRNAs. In a Drosophila cellular system, knockdown of the CLP1 homolog, cbc, showed that CLP1 may be able to regulate mature tRNA levels, but in vivo models generally have been unable to detect significant differences in mature tRNA levels in either PCH mouse models or patient samples, suggesting that PCH10 is not caused by a deficiency in available tRNAs (Hanada et al., 2013; Karaca et al., 2014; Monaghan et al., 2021). Results from studies looking at tRNA products appear to vary with regard to tissue specificity, further complicating interpretations.
One school of thought is that tRNA fragments may play a role in neuron death through p53-mediated cell death. Early studies showed an accumulation of a tRNA 5′-Tyr exon fragment (5′-tRFs) with an intact 5′-leader in K127A MEFs and these fragments were recently shown to play a role in p53-mediated cell death (Inoue et al., 2020). These 5′-tRFs were also found to accumulate in two recent R140H mouse models (Monaghan et al., 2021; Morisaki et al., 2021), along with Ile-introns. One study concluded that the 5′ fragments accumulated more in the spinal cord than the cerebellum, questioning if these fragments are relevant to the development of PCH (Monaghan et al., 2021). Along similar lines to the 5′-tRFs, a previous study also observed Ile–intron accumulation and found that unphosphorylated 3′-exons (a presumable CLP1 substate) were cytotoxic, especially when paired with oxidative stress (H2O2). The same study also found that knockdown of p53 rescued the embryonic lethality of K127A on a mouse background for which it was previously lethal, further supporting a role for p53 in PCH (Schaffer et al., 2014).
Presumably, PCH may not just be linked to the accumulation of one tRNA fragment. Another possibility is that introns accumulate and cause cellular death through a similar mechanism to the 5′-tRFs. The role of mammalian tRNA introns in cellular regulation remains a mystery, but it seems very plausible that they may play a role in cellular regulation just as yeast introns have been shown to do (Hayashi et al., 2019). Cell based assays revealed an accumulation of circular introns upon CLP1 depletion, and a recent R140H mouse model found accumulation of two intron species, assumed to correspond to linear and circular introns (Hayne et al., 2020; Monaghan et al., 2021). A major challenge to detecting introns, particularly in patient samples, is the small size of human tRNA introns (Table 2). These recent studies support early work that found intron accumulations, and highlight the dual role CLP1 may play, both in driving kinase-mediated cell death and in turn preventing the production and accumulation of the stable circular tRNA introns. Furthermore, while a recent study by Inoue et al. did not find that linear ILE-introns sensitizes cells to death, it remains unclear if other introns may impede cellular growth and homeostasis, and further if circular tRNA introns are cytotoxic (Inoue et al., 2020).
Finally, differences in pre-tRNA levels vary between studies and model systems. Patient fibroblasts revealed no difference in pre-tRNA levels while accumulations of pre-tRNAs were observed following the loss of wild-type CLP1 in fibroblasts reverted to induced neurons, the Drosophila cell system, and a new R140H mouse model of PCH all finding to an accumulation pre-tRNAs (Hayne et al., 2020; Karaca et al., 2014; Monaghan et al., 2021; Schaffer et al., 2014). Nevertheless, the accumulated pre-tRNAs are not believed to be toxic (Schaffer et al., 2014). These findings, in light of additional work revealing CLP1 does not impact tRNA cleavage by the TSEN complex, suggest that the increase in pre-tRNA levels could be linked to CLP1’s role in mRNA processing, which is further supported by the finding that R140H mice have changes in the polyA site selection (Monaghan et al., 2021).
TSEN
TSEN mutations are linked to unique PCH subtypes depending on the individual TSEN subunits (Figure 7B). TSEN54 causes a spectrum of disease that spans PCH 2A, PCH 4, and PCH 5, with PCH 4 and 5 being the most severe (van Dijk et al., 2018). Similar to CLP1, it is not fully understood why each of the mutations in the individual TSEN subunits cause disease or how that dysfunction leads to PCH. A recent crystal structure of stable fragments of TSEN34 bound to TSEN15 revealed that the TSEN15 H116Y PCH mutation lies at the interface of the TSEN34 and TSEN15 subunits (Sekulovski et al., 2021); however, the structural basis for the remaining PCH mutants remains unclear. To begin to understand how the additional PCH mutations impact the TSEN complex, we mapped the known PCH mutations onto our 3D Alphafold model (Figure 7C) and found that similar to CLP1, the mutations do not appear near the active sites. Instead, we observed that the mutations are generally found on the exterior interfaces of each of the subunits. In some cases, the mutations may interfere with intra and inter-subunit architecture and binding. In the case of the most common disease mutant, TSEN54 A307S, the mutation appears in a disordered region, which may be important for binding regulatory factors or recruiting substrates. Mutations on the exterior regions of the TSEN complex proteins suggest that mutations are unlikely to just impact the folding and stability of the individual proteins, but may also impact TSEN complex assembly or protein:protein interactions, such as CLP1, in vivo. Additional interaction partners of the TSEN complex have yet to be discovered for metazons, but interacting partners that impact TSEN spatial or enzymatic regulation could also lead to PCH in a similar manner to how CLP1 R140H seems to disrupt CLP1’s spatial localization.
In support of the hypothesis that mutations hinder complex assembly, previous studies have shown that when only the TSEN subunit of interest is overexpressed, the mutants are typically deficient in forming an entire complex. We and others have found that overexpression of all four subunits concurrently is required for efficient reconstitution of the TSEN complex in vitro (Hayne et al., 2020; Sekulovski et al., 2021). Sekulovski et al. recently showed that immunoprecipitations of the full TSEN complex does not result in obvious differences in complex formation or in vitro cleavage activity for several of the PCH mutations (Sekulovski et al., 2021). However, they found that the PCH mutations did impact the thermal stability of the complex in vitro (Sekulovski et al., 2021). Interestingly, while the authors observed no change in the catalytic activity of the purified complex, they did observe splicing defects when assaying cell extracts from patient fibroblasts. Furthermore, unlike CLP1 patient samples, TSEN54 PCH patient fibroblasts did not accumulate Ile-introns (as excepted for a cleavage defect) and had an expected increase in those pre-tRNAs, yet interestingly there was no detectable difference in mature tRNAs. Much is yet to be understood about how mutations alter TSEN complex function as it relates to pre-tRNAs and other yet to be discovered substrates. Even more of a mystery, how those dysregulated RNAs may impact cell survival. In light of previous data suggesting that pre-tRNAs were not cytotoxic and that the TSEN patient fibroblasts had no lack of mature tRNAs, it may be, that similar to CLP1, the TSEN complex dysregulates other RNA substrates that underlie disease progression.
PCH: a rare disease caused by a shared mechanism or a family of unrelated, yet similar diseases
PCH is a rare autosomal recessive disease primarily caused by mutations in RNA processing factors. In addition to the many fly model organisms mentioned above, several new fly PCH models have recently been developed (Morton et al., 2020; Schmidt et al., 2021; Slavotinek et al., 2020), which may be useful tools to better characterize and understand PCH. PCH appears to primarily be caused by dysfunction in RNA processing machinery, with a couple of exceptions. There appears to be two pathways that PCH-linked factors target: several proteins have direct roles in tRNA maturation factors (such as TSEN, CLP1, RARS2, and SEPSECS) and other factors are linked to mRNA processing, such as two Exosome subunits and CLP1 (van Dijk et al., 2018). While no non-tRNA substrates have been identified for the TSEN complex in humans, several newly identified substrates of the yeast complex implicate TSEN in eukaryotic mRNA processing, further suggesting a possible overlap in substrates. It may be that there is an underlying shared substrate and that non-RNA processing factors linked to PCH disrupt the target of the substate at another point, or perhaps all the proteins share a similar mechanism by which they cause disease. A major theme seems to be p53-mediated cell death. Indeed, in addition to the links to p53-mediated death through CLP1, there appear to be links between p53 and mutations in two other PCH-linked proteins Target of Egr1 1 (TOE1) and the Exosome (Hanada et al., 2013; Inoue et al., 2020; Müller et al., 2020; Sperandio et al., 2009). In contrast to these previous reports, a recent study by Monaghan et al 2021, suggests p53 may not be responsible for CLP1-linked PCH (Monaghan et al., 2021). There is still much to be discovered about the cellular changes behind PCH.
In addition, the full role these proteins play in the development of PCH and other diseases is an outstanding question. ANGEL2 is not yet known to have any links to disease, and is not essential for life, but it may play a role in stress responses and play a role in disease progression when dysregulated. Along these lines, mutations in TSEN54 have also been linked to late-onset ataxia (Qian et al., 2014) and leukodystrophy in canines (Störk et al., 2019), further supporting that these proteins may have uncharacterized roles in other neurological diseases. More recently, a spliced variant of TSEN2 was recently identified to cause a PCH like phenotype with additional vascular and renal issues, which has been termed TSEN2 Related Atypical hemolytic uremic syndrome, Craniofacial malformations, Kidney failure (TRACK)(Canpolat et al., 2021). RNA sequencing revealed these patients had abnormal tRNA transcripts, and the phenotype was found to be similar in zebrafish model of the TSEN2 defect (Canpolat et al., 2021).
The role of the TSEN proteins in these diseases may be directly linked to splicing defects, or they may be linked to other shared roles for tRNA fragments in other diseases, such as Alzheimer’s, diabetes, and aging (Li et al., 2021; Wu et al., 2021; Z. Zhou et al., 2021). It remains unclear why these diseases are tissue specific, but this draws parallels to other tissue-specific diseases associated with RNA processing factors such as ribosomopathies, which arise from mutations or altered expression of proteins involved in ribosome maturation (reviewed in (Farley-Barnes et al., 2019; McCann & Baserga, 2013)). One hypothesis is that neurons are particularly sensitive to the accumulation of RNAs that results from disruption of these pathways. It remains to be understood if the proteins linked to PCH share a common substrate, cellular pathway targeting neuronal death, or are linked in some other manner.
6. CONCLUSION & OPEN QUESTIONS IN TRNA SPLICING
Major discoveries in the last decade have reinvigorated the characterization of human tRNA splicing with studies linking new mutations to PCH and refined characterization of the TSEN complex in vitro. However, there remain several major questions about the role of these proteins in both healthy and disease states. The recent discoveries that CLP1 and ANGEL2 can regulate tRNA splicing raises the question of if other factors may also be involved in splicing and if a yeast-like ligation pathway may exist in humans, serving as a rescue mechanism. Furthermore, it remains a mystery what other substrates the TSEN complex and CLP1 may target and if these unknown targets underlie PCH. Recent work in yeast has identified several mitochondrial mRNA targets of the TSEN complex and discovery of non-tRNA substrates of the mammalian TSEN complex are eagerly awaited. Beyond facilitating tRNA splicing the TSEN complex has also been shown to be a useful tool for the generation of circular RNAs (Litke & Jaffrey, 2019; Noto et al., 2017; Schmidt et al., 2016), suggesting that the TSEN complex could be repurposed to process other RNAs.
Removal of the intron is just one of many tRNA processing events and it remains unclear if tRNA splicing is coordinated with tRNA transcription, processing, and/or modification (reviewed in (Jarrous et al., 2021). Following transcription by RNA Pol III nascent intron containing pre-tRNAs are processed by three nucleases including the TSEN complex, RNase P and RNase Z and there is growing evidence that suggests co-transcriptional tRNA processing is occurring (reviewed in (Jarrous et al., 2021)). tRNAs are the most heavily post-transcriptional modified RNAs inside the cell and dysregulation of these modifications has been linked to neurological disease (reviewed in (Das et al., 2021; Suzuki, 2021)). tRNA modifications impact the fidelity of translation and many of these modifications surround the anti-codon stem loop (Manickam et al., 2015; J. B. Zhou et al., 2021). Recent work revealed that a pseudoU modification in yeast tRNA-Ile is intron dependent, supporting a role for introns in mediating RNA modifications (Hayashi et al., 2019).The cellular function of eukaryotic introns remains poorly understood but new links to RNA modification, cell fitness, and stress response coupled with their presence across eukaryotes suggests that they have been retained to fulfil important but still undiscovered cellular roles. Finally, while excision of the intron by the TSEN complex is well conserved across archaea and eukaryotes, we still do not fully understand how this complex recognizes pre-tRNAs substrates or how CLP1 and ANGEL2 are regulated to ensure cellular proper regulation of tRNA splicing products.
Acknowledgments
The authors thank Oya Bermek and Meghan Warden for their review of this work.
Funding Information
This work was supported by the US National Institute of Health Intramural Research Program; US National Institute of Environmental Health Sciences (ZIA ES103247 to R.E.S.).
Additional support was provided to C.K.H. by the US National Institute of Health Extramural Research Program; US National Institute of General Medical Sciences through the NIH Maximizing Opportunities for Scientific and Academic Independent Careers K99/R00 Pathway to Independence Award (1K99-GM143534). T.A.L. was supported by the 2019–2020 NIEHS Scholars Connect Program (NSCP). Funding for open access charge: NIEHS.
Footnotes
Conflict of Interest
The authors have no conflicts to disclose.
Contributor Information
Cassandra K. Hayne, Signal Transduction Laboratory, National Institute of Environmental Health Sciences, National Institutes of Health, Department of Health and Human Services, 111 T. W. Alexander Drive, Research Triangle Park, NC 27709, USA.
Tanae A. Lewis, Department of Chemistry, North Carolina Agricultural and Technical State University, Greensboro, NC 27411, USA.
Robin E. Stanley, Signal Transduction Laboratory, National Institute of Environmental Health Sciences, National Institutes of Health, Department of Health and Human Services, 111 T. W. Alexander Drive, Research Triangle Park, NC 27709, USA.
References
- Abelson J, Trotta CR, & Li H (1998, May 22). tRNA splicing. J Biol Chem, 273(21), 12685–12688. 10.1074/jbc.273.21.12685 [DOI] [PubMed] [Google Scholar]
- Baldi MI, Mattoccia E, Bufardeci E, Fabbri S, & Tocchini-Valentini GP (1992, Mar 13). Participation of the intron in the reaction catalyzed by the Xenopus tRNA splicing endonuclease. Science, 255(5050), 1404–1408. [DOI] [PubMed] [Google Scholar]
- Behrens A, Rodschinka G, & Nedialkova DD (2021, 2021/04/15/). High-resolution quantitative profiling of tRNA abundance and modification status in eukaryotes by mim-tRNAseq. Mol Cell, 81(8), 1802–1815.e1807. 10.1016/j.molcel.2021.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg MD, & Brandl CJ (2021, Mar). Transfer RNAs: diversity in form and function. RNA Biol, 18(3), 316–339. 10.1080/15476286.2020.1809197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvin K, & Li H (2008, Apr). RNA-splicing endonuclease structure and function. Cell Mol Life Sci, 65(7–8), 1176–1185. 10.1007/s00018-008-7393-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canpolat N, Liu D, Atayar E, Saygili S, Kara NS, Westfall TA, Ding Q, Brown BJ, Braun TA, Slusarski D, Karli Oguz K, Ozluk Y, Tuysuz B, Tastemel Ozturk T, Sever L, Sezerman OU, Topaloglu R, Caliskan S, Attanasio M, & Ozaltin F (2021, Dec 28). A splice site mutation in the TSEN2 causes a new syndrome with craniofacial and central nervous system malformations, and atypical hemolytic uremic syndrome. Clin Genet. 10.1111/cge.14105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PP, & Lowe TM (2016, Jan 4). GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res, 44(D1), D184–189. 10.1093/nar/gkv1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry PD, Peach SE, & Hesselberth JR (2019, 2019/03/15). Multiple decay events target HAC1 mRNA during splicing to regulate the unfolded protein response. eLife, 8, e42262. 10.7554/eLife.42262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry PD, & White LK (2018, Mar). Genetic bypass of essential RNA repair enzymes in budding yeast. RNA, 24(3), 313–323. 10.1261/rna.061788.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F (1970, 1970/08/01). Central Dogma of Molecular Biology. Nature, 227(5258), 561–563. 10.1038/227561a0 [DOI] [PubMed] [Google Scholar]
- Cuchillo CM, Nogués MV, & Raines RT (2011, 2011/09/20). Bovine Pancreatic Ribonuclease: Fifty Years of the First Enzymatic Reaction Mechanism. Biochemistry, 50(37), 7835–7841. 10.1021/bi201075b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson MR, & Winey M (1989). Split tRNA genes and their products: A paradigm for the study of cell function and evolution. Yeast, 5(6), 405–427. 10.1002/yea.320050602 [DOI] [PubMed] [Google Scholar]
- Das AS, Alfonzo JD, & Accornero F (2021). The importance of RNA modifications: From cells to muscle physiology. WIREs RNA, n/a(n/a), e1700. 10.1002/wrna.1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries H, Rüegsegger U, Hübner W, Friedlein A, Langen H, & Keller W (2000). Human pre-mRNA cleavage factor IIm contains homologs of yeast proteins and bridges two other cleavage factors. Embo j, 19(21), 5895–5904. 10.1093/emboj/19.21.5895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhungel N, & Hopper AK (2012, Mar 1). Beyond tRNA cleavage: novel essential function for yeast tRNA splicing endonuclease unrelated to tRNA processing. Genes Dev, 26(5), 503–514. 10.1101/gad.183004.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikfidan A, Loll B, Zeymer C, Magler I, Clausen T, & Meinhart A (2014). RNA Specificity and Regulation of Catalysis in the Eukaryotic Polynucleotide Kinase Clp1. Mol Cell, 54(6), 975–986. 10.1016/j.molcel.2014.04.005 [DOI] [PubMed] [Google Scholar]
- Fabbri S, Fruscoloni P, Bufardeci E, Di Nicola Negri E, Baldi MI, Attardi DG, Mattoccia E, & Tocchini-Valentini GP (1998, Apr 10). Conservation of substrate recognition mechanisms by tRNA splicing endonucleases. Science, 280(5361), 284–286. [DOI] [PubMed] [Google Scholar]
- Farley-Barnes KI, Ogawa LM, & Baserga SJ (2019, Oct). Ribosomopathies: Old Concepts, New Controversies. Trends Genet, 35(10), 754–767. 10.1016/j.tig.2019.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujishima K, & Kanai A (2014, 2014-May-26). tRNA gene diversity in the three domains of life [Review]. Front Genet, 5(142). 10.3389/fgene.2014.00142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogakos T, Brown M, Garzia A, Meyer C, Hafner M, & Tuschl T (2017, Aug 8). Characterizing Expression and Processing of Precursor and Mature Human tRNAs by Hydro-tRNAseq and PAR-CLIP. Cell Rep, 20(6), 1463–1475. 10.1016/j.celrep.2017.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer CL, Peebles CL, Gegenheimer P, & Abelson J (1983, Feb). Mechanism of action of a yeast RNA ligase in tRNA splicing. Cell, 32(2), 537–546. 10.1016/0092-8674(83)90473-7 [DOI] [PubMed] [Google Scholar]
- Greer CL, Söll D, & Willis I (1987, Jan). Substrate recognition and identification of splice sites by the tRNA-splicing endonuclease and ligase from Saccharomyces cerevisiae. Mol Cell Biol, 7(1), 76–84. 10.1128/mcb.7.1.76-84.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada T, Weitzer S, Mair B, Bernreuther C, Wainger BJ, Ichida J, Hanada R, Orthofer M, Cronin SJ, Komnenovic V, Minis A, Sato F, Mimata H, Yoshimura A, Tamir I, Rainer J, Kofler R, Yaron A, Eggan KC, Woolf CJ, Glatzel M, Herbst R, Martinez J, & Penninger JM (2013, Mar 28). CLP1 links tRNA metabolism to progressive motor-neuron loss. Nature, 495(7442), 474–480. 10.1038/nature11923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Mori S, Suzuki T, Suzuki T, & Yoshihisa T (2019). Impact of intron removal from tRNA genes on Saccharomyces cerevisiae. Nucleic Acids Res, 47(11), 5936–5949. 10.1093/nar/gkz270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayne CK, Schmidt CA, Haque MI, Matera AG, & Stanley RE (2020). Reconstitution of the human tRNA splicing endonuclease complex: insight into the regulation of pre-tRNA cleavage. Nucleic Acids Res. 10.1093/nar/gkaa438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG (2005). Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol, 59, 407–450. 10.1146/annurev.micro.59.031805.133833 [DOI] [PubMed] [Google Scholar]
- Hirata A (2019). Recent Insights Into the Structure, Function, and Evolution of the RNA-Splicing Endonucleases. Front Genet, 10, 103. 10.3389/fgene.2019.00103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata A, Fujishima K, Yamagami R, Kawamura T, Banfield JF, Kanai A, & Hori H (2012, Nov 1). X-ray structure of the fourth type of archaeal tRNA splicing endonuclease: insights into the evolution of a novel three-unit composition and a unique loop involved in broad substrate specificity. Nucleic Acids Res, 40(20), 10554–10566. 10.1093/nar/gks826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata A, Kitajima T, & Hori H (2011, Nov). Cleavage of intron from the standard or non-standard position of the precursor tRNA by the splicing endonuclease of Aeropyrum pernix, a hyper-thermophilic Crenarchaeon, involves a novel RNA recognition site in the Crenarchaea specific loop. Nucleic Acids Res, 39(21), 9376–9389. 10.1093/nar/gkr615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper AK, & Nostramo RT (2019). tRNA Processing and Subcellular Trafficking Proteins Multitask in Pathways for Other RNAs. Front Genet, 10, 96. 10.3389/fgene.2019.00096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HY, & Hopper AK (2016, Mar 23). Multiple Layers of Stress-Induced Regulation in tRNA Biology. Life (Basel), 6(2). 10.3390/life6020016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtig JE, Steiger MA, Nagarajan VK, Li T, Chao TC, Tsai KL, & van Hoof A (2021, Mar 9). Comparative parallel analysis of RNA ends identifies mRNA substrates of a tRNA splicing endonuclease-initiated mRNA decay pathway. Proc Natl Acad Sci U S A, 118(10). 10.1073/pnas.2020429118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue M, Hada K, Shiraishi H, Yatsuka H, Fujinami H, Morisaki I, Nishida Y, Matsubara E, Ishitani T, Hanada R, Matsumoto M, Penninger JM, Ihara K, & Hanada T (2020, May 7). Tyrosine pre-transfer RNA fragments are linked to p53-dependent neuronal cell death via PKM2. Biochem Biophys Res Commun, 525(3), 726–732. 10.1016/j.bbrc.2020.02.157 [DOI] [PubMed] [Google Scholar]
- Jarrous N, Mani D, & Ramanathan A (2021, April 30, 2021). Coordination of transcription and processing of tRNA. The FEBS Journal, n/a(n/a). 10.1111/febs.15904 [DOI] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, & Hassabis D (2021, 2021/08/01). Highly accurate protein structure prediction with AlphaFold. Nature, 596(7873), 583–589. 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai A (2015). Disrupted tRNA Genes and tRNA Fragments: A Perspective on tRNA Gene Evolution. Life, 5(1), 321–331. https://www.mdpi.com/2075-1729/5/1/321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneta A, Fujishima K, Morikazu W, Hori H, & Hirata A (2018). The RNA-splicing endonuclease from the euryarchaeaon Methanopyrus kandleri is a heterotetramer with constrained substrate specificity. Nucleic Acids Res, 46(4), 1958–1972. 10.1093/nar/gky003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaca E, Weitzer S, Pehlivan D, Shiraishi H, Gogakos T, Hanada T, Jhangiani SN, Wiszniewski W, Withers M, Campbell IM, Erdin S, Isikay S, Franco LM, Gonzaga-Jauregui C, Gambin T, Gelowani V, Hunter JV, Yesil G, Koparir E, Yilmaz S, Brown M, Briskin D, Hafner M, Morozov P, Farazi TA, Bernreuther C, Glatzel M, Trattnig S, Friske J, Kronnerwetter C, Bainbridge MN, Gezdirici A, Seven M, Muzny DM, Boerwinkle E, Ozen M, Clausen T, Tuschl T, Yuksel A, Hess A, Gibbs RA, Martinez J, Penninger JM, & Lupski JR (2014, Apr 24). Human CLP1 mutations alter tRNA biogenesis, affecting both peripheral and central nervous system function. Cell, 157(3), 636–650. 10.1016/j.cell.2014.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato-Murayama M, Bessho Y, Shirouzu M, Yokoyama S,. (2005). Crystal structure of tRNA-intron endonuclease from Sulfolobus tokodaii. RIKEN Structural Genomics/Proteomics Initiative (RSGI). 10.2210/pdb2cv8/pdb [DOI] [Google Scholar]
- Kim YK, Mizutani K, Rhee KH, Nam KH, Lee WH, Lee EH, Kim EE, Park SY, & Hwang KY (2007, Nov). Structural and mutational analysis of tRNA intron-splicing endonuclease from Thermoplasma acidophilum DSM 1728: catalytic mechanism of tRNA intron-splicing endonucleases. J Bacteriol, 189(22), 8339–8346. 10.1128/jb.00713-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Srisuknimit V, & Waldor MK (2020, Oct). Probing the diversity and regulation of tRNA modifications. Curr Opin Microbiol, 57, 41–48. 10.1016/j.mib.2020.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmaczewski SG, Edwards TJ, Han SM, Eckwahl MJ, Meyer BI, Peach S, Hesselberth JR, Wolin SL, & Hammarlund M (2014, Dec). The RtcB RNA ligase is an essential component of the metazoan unfolded protein response. EMBO Rep, 15(12), 1278–1285. 10.15252/embr.201439531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroupova A, Ackle F, Asanović I, Weitzer S, Boneberg FM, Faini M, Leitner A, Chui A, Aebersold R, Martinez J, & Jinek M (2021, Dec 2). Molecular architecture of the human tRNA ligase complex. eLife, 10. 10.7554/eLife.71656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, & Abelson J (2000, Sep 22). Crystal structure of a dimeric archaeal splicing endonuclease. J Mol Biol, 302(3), 639–648. 10.1006/jmbi.2000.3941 [DOI] [PubMed] [Google Scholar]
- Li H, Trotta CR, & Abelson J (1998, Apr 10). Crystal structure and evolution of a transfer RNA splicing enzyme. Science, 280(5361), 279–284. 10.1126/science.280.5361.279 [DOI] [PubMed] [Google Scholar]
- Li X, Liu X, Zhao D, Cui W, Wu Y, Zhang C, & Duan C (2021, Sep 18). tRNA-derived small RNAs: novel regulators of cancer hallmarks and targets of clinical application. Cell Death Discov, 7(1), 249. 10.1038/s41420-021-00647-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litke JL, & Jaffrey SR (2019, 2019/06/01). Highly efficient expression of circular RNA aptamers in cells using autocatalytic transcripts. Nature Biotechnology, 37(6), 667–675. 10.1038/s41587-019-0090-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manickam N, Joshi K, Bhatt MJ, & Farabaugh PJ (2015). Effects of tRNA modification on translational accuracy depend on intrinsic codon–anticodon strength. Nucleic Acids Res, 44(4), 1871–1881. 10.1093/nar/gkv1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama S, Sugahara J, Kanai A, & Nozaki H (2010, May). Permuted tRNA genes in the nuclear and nucleomorph genomes of photosynthetic eukaryotes. Mol Biol Evol, 27(5), 1070–1076. 10.1093/molbev/msp313 [DOI] [PubMed] [Google Scholar]
- McCann KL, & Baserga SJ (2013). Mysterious Ribosomopathies. Science, 341(6148), 849–850. 10.1126/science.1244156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell M, Xue S, Erdman R, Randau L, Söll D, & Li H (2009, Sep). Crystal structure and assembly of the functional Nanoarchaeum equitans tRNA splicing endonuclease. Nucleic Acids Res, 37(17), 5793–5802. 10.1093/nar/gkp537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan CE, Adamson SI, Kapur M, Chuang JH, & Ackerman SL (2021, Sep 28). The Clp1 R140H mutation alters tRNA metabolism and mRNA 3’ processing in mouse models of pontocerebellar hypoplasia. Proc Natl Acad Sci U S A, 118(39). 10.1073/pnas.2110730118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisaki I, Shiraishi H, Fujinami H, Shimizu N, Hikida T, Arai Y, Kobayashi T, Hanada R, Penninger JM, Fujiki M, & Hanada T (2021, Sep 17). Modeling a human CLP1 mutation in mouse identifies an accumulation of tyrosine pre-tRNA fragments causing pontocerebellar hypoplasia type 10. Biochem Biophys Res Commun, 570, 60–66. 10.1016/j.bbrc.2021.07.036 [DOI] [PubMed] [Google Scholar]
- Morton DJ, Jalloh B, Kim L, Kremsky I, Nair RJ, Nguyen KB, Rounds JC, Sterrett MC, Brown B, Le T, Karkare MC, McGaughey KD, Sheng S, Leung SW, Fasken MB, Moberg KH, & Corbett AH (2020, Jul). A Drosophila model of Pontocerebellar Hypoplasia reveals a critical role for the RNA exosome in neurons. PLoS Genet, 16(7), e1008901. 10.1371/journal.pgen.1008901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller JS, Burns DT, Griffin H, Wells GR, Zendah RA, Munro B, Schneider C, & Horvath R (2020, Aug). RNA exosome mutations in pontocerebellar hypoplasia alter ribosome biogenesis and p53 levels. Life Sci Alliance, 3(8). 10.26508/lsa.202000678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noto JJ, Schmidt CA, & Matera AG (2017, Aug 3). Engineering and expressing circular RNAs via tRNA splicing. RNA Biol, 14(8), 978–984. 10.1080/15476286.2017.1317911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donoghue P, Ling J, & Söll D (2018). Transfer RNA function and evolution. RNA Biol, 15(4–5), 423–426. 10.1080/15476286.2018.1478942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda M, Shiba T, Inaoka D-K, Kita K, Kurisu G, Mineki S, Harada S, Watanabe Y. i., & Yoshinari S (2011, 2011/01/07/). A Conserved Lysine Residue in the Crenarchaea-Specific Loop is Important for the Crenarchaeal Splicing Endonuclease Activity. J Mol Biol, 405(1), 92–104. 10.1016/j.jmb.2010.10.050 [DOI] [PubMed] [Google Scholar]
- Paushkin SV, Patel M, Furia BS, Peltz SW, & Trotta CR (2004, Apr 30). Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3’ end formation. Cell, 117(3), 311–321. [DOI] [PubMed] [Google Scholar]
- Peach SE, York K, & Hesselberth JR (2015, Sep 30). Global analysis of RNA cleavage by 5’-hydroxyl RNA sequencing. Nucleic Acids Res, 43(17), e108. 10.1093/nar/gkv536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschek J, & Walter P (2019, Jun 25). tRNA ligase structure reveals kinetic competition between non-conventional mRNA splicing and mRNA decay. eLife, 8. 10.7554/eLife.44199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky EM, & Hopper AK (2010, Sep 1). tRNA biology charges to the front. Genes Dev, 24(17), 1832–1860. 10.1101/gad.1956510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky EM, Schwartz RC, & Abelson J (1986, Feb 25). Saccharomyces cerevisiae tRNA ligase. Purification of the protein and isolation of the structural gene. J Biol Chem, 261(6), 2978–2986. [PubMed] [Google Scholar]
- Pillon MC, Frazier MN, Dillard LB, Williams JG, Kocaman S, Krahn JM, Perera L, Hayne CK, Gordon J, Stewart ZD, Sobhany M, Deterding LJ, Hsu AL, Dandey VP, Borgnia MJ, & Stanley RE (2021, 2021/01/27). Cryo-EM structures of the SARS-CoV-2 endoribonuclease Nsp15 reveal insight into nuclease specificity and dynamics. Nat Commun, 12(1), 636. 10.1038/s41467-020-20608-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkard O, McFarland S, Sweet T, & Coller J (2020, 2020/08/14). Quantitative tRNA-sequencing uncovers metazoan tissue-specific tRNA regulation. Nat Commun, 11(1), 4104. 10.1038/s41467-020-17879-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto PH, Kroupova A, Schleiffer A, Mechtler K, Jinek M, Weitzer S, & Martinez J (2020, Jul 31). ANGEL2 is a member of the CCR4 family of deadenylases with 2’,3’-cyclic phosphatase activity. Science, 369(6503), 524–530. 10.1126/science.aba9763 [DOI] [PubMed] [Google Scholar]
- Popow J, Englert M, Weitzer S, Schleiffer A, Mierzwa B, Mechtler K, Trowitzsch S, Will CL, Luhrmann R, Soll D, & Martinez J (2011, Feb 11). HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science, 331(6018), 760–764. 10.1126/science.1197847 [DOI] [PubMed] [Google Scholar]
- Popow J, Jurkin J, Schleiffer A, & Martinez J (2014, Jul 3). Analysis of orthologous groups reveals archease and DDX1 as tRNA splicing factors. Nature, 511(7507), 104–107. 10.1038/nature13284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y, Wang H, Jin T, Wang Y, Fang L, Chen Y, & Chen L (2014, 2014/09/01). A familial late-onset hereditary ataxia mimicking pontocerebellar hypoplasia caused by a novel TSEN54 mutation. Mol Med Rep, 10(3), 1423–1425. 10.3892/mmr.2014.2342 [DOI] [PubMed] [Google Scholar]
- Ramirez A, Shuman S, & Schwer B (2008, Sep). Human RNA 5’-kinase (hClp1) can function as a tRNA splicing enzyme in vivo. RNA, 14(9), 1737–1745. 10.1261/rna.1142908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randau L, & Söll D (2008). Transfer RNA genes in pieces. EMBO Rep, 9(7), 623–628. 10.1038/embor.2008.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauhut R, Green PR, & Abelson J (1990, October 25, 1990). Yeast tRNA-splicing endonuclease is a heterotrimeric enzyme. Journal of Biological Chemistry, 265(30), 18180–18184. http://www.jbc.org/content/265/30/18180.abstract [PubMed] [Google Scholar]
- Reyes VM, & Abelson J (1988, 1988/11/18/). Substrate recognition and splice site determination in yeast tRNA splicing. Cell, 55(4), 719–730. 10.1016/0092-8674(88)90230-9 [DOI] [PubMed] [Google Scholar]
- Saito M, Sato A, Nagata S, Tamaki S, Tomita M, Suzuki H, & Kanai A (2019, Oct 1). Large-Scale Molecular Evolutionary Analysis Uncovers a Variety of Polynucleotide Kinase Clp1 Family Proteins in the Three Domains of Life. Genome Biol Evol, 11(10), 2713–2726. 10.1093/gbe/evz195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer Ashleigh E., Eggens Veerle R. C., Caglayan Ahmet O., Reuter Miriam S., Scott E, Coufal Nicole G., Silhavy Jennifer L., Xue Y, Kayserili H, Yasuno K, Rosti Rasim O., Abdellateef M, Caglar C, Kasher Paul R., Cazemier JL, Weterman Marian A., Cantagrel V, Cai N, Zweier C, Altunoglu U, Satkin NB, Aktar F, Tuysuz B, Yalcinkaya C, Caksen H, Bilguvar K, Fu X-D, Trotta CR, Gabriel S, Reis A, Gunel M, Baas F, & Gleeson Joseph G. (2014, 2014/04/24/). CLP1 Founder Mutation Links tRNA Splicing and Maturation to Cerebellar Development and Neurodegeneration. Cell, 157(3), 651–663. 10.1016/j.cell.2014.03.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt CA, Giusto JD, Bao A, Hopper AK, & Matera AG (2019, Jul 9). Molecular determinants of metazoan tricRNA biogenesis. Nucleic Acids Res, 47(12), 6452–6465. 10.1093/nar/gkz311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt CA, & Matera AG (2019, Dec 28). tRNA introns: Presence, processing, and purpose. WIREs RNA, e1583. 10.1002/wrna.1583 [DOI] [PubMed] [Google Scholar]
- Schmidt CA, Min LY, McVay MH, Giusto JD, Brown JC, Salzler HR, & Matera AG (2021). Mutations in Drosophila tRNA processing factors cause phenotypes similar to Pontocerebellar Hypoplasia. bioRxiv, 2021.2007.2009.451847. 10.1101/2021.07.09.451847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt CA, Noto JJ, Filonov GS, & Matera AG (2016). A Method for Expressing and Imaging Abundant, Stable, Circular RNAs In Vivo Using tRNA Splicing. Methods Enzymol, 572, 215–236. 10.1016/bs.mie.2016.02.018 [DOI] [PubMed] [Google Scholar]
- Schwer B, Aronova A, Ramirez A, Braun P, & Shuman S (2008, Feb). Mammalian 2’,3’ cyclic nucleotide phosphodiesterase (CNP) can function as a tRNA splicing enzyme in vivo. RNA, 14(2), 204–210. 10.1261/rna.858108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekulovski S, Devant P, Panizza S, Gogakos T, Pitiriciu A, Heitmeier K, Ramsay EP, Barth M, Schmidt C, Tuschl T, Baas F, Weitzer S, Martinez J, & Trowitzsch S (2021, Sep 28). Assembly defects of human tRNA splicing endonuclease contribute to impaired pre-tRNA processing in pontocerebellar hypoplasia. Nat Commun, 12(1), 5610. 10.1038/s41467-021-25870-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigematsu M, Morichika K, Kawamura T, Honda S, & Kirino Y (2019). Genome-wide identification of short 2′,3′-cyclic phosphate-containing RNAs and their regulation in aging. PLoS Genet, 15(11), e1008469. 10.1371/journal.pgen.1008469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavotinek A, Misceo D, Htun S, Mathisen L, Frengen E, Foreman M, Hurtig JE, Enyenihi L, Sterrett MC, Leung SW, Schneidman-Duhovny D, Estrada-Veras J, Duncan JL, Haaxma CA, Kamsteeg EJ, Xia V, Beleford D, Si Y, Douglas G, Treidene HE, van Hoof A, Fasken MB, & Corbett AH (2020, Aug 3). Biallelic variants in the RNA exosome gene EXOSC5 are associated with developmental delays, short stature, cerebellar hypoplasia and motor weakness. Hum Mol Genet, 29(13), 2218–2239. 10.1093/hmg/ddaa108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, & Markley JL (2007, Feb 9). Three-dimensional structure determined for a subunit of human tRNA splicing endonuclease (Sen15) reveals a novel dimeric fold. J Mol Biol, 366(1), 155–164. 10.1016/j.jmb.2006.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandio S, Tardito S, Surzycki A, Latterich M, & de Belle I (2009, 2009/07/07/). TOE1 interacts with p53 to modulate its transactivation potential. FEBS Letters, 583(13), 2165–2170. 10.1016/j.febslet.2009.06.004 [DOI] [PubMed] [Google Scholar]
- Störk T, Nessler J, Anderegg L, Hünerfauth E, Schmutz I, Jagannathan V, Kyöstilä K, Lohi H, Baumgärtner W, Tipold A, & Leeb T (2019, Oct). TSEN54 missense variant in Standard Schnauzers with leukodystrophy. PLoS Genet, 15(10), e1008411. 10.1371/journal.pgen.1008411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T (2021, 2021/06/01). The expanding world of tRNA modifications and their disease relevance. Nature Reviews Molecular Cell Biology, 22(6), 375–392. 10.1038/s41580-021-00342-0 [DOI] [PubMed] [Google Scholar]
- Szoták-Ajtay K, Szõke D, Kovács G, Andréka J, Brenner GB, Giricz Z, Penninger J, Kahn ML, & Jakus Z (2020). Reduced Prenatal Pulmonary Lymphatic Function Is Observed in Clp1 (K/K) Embryos With Impaired Motor Functions Including Fetal Breathing Movements in Preparation of the Developing Lung for Inflation at Birth. Front Bioeng Biotechnol, 8, 136. 10.3389/fbioe.2020.00136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta CR, Abelson John. (1999). 21 tRNA Splicing: An RNA World Add-on or an Ancient Reaction? In The RNA World, 2nd Ed.: The Nature of Modern RNA Suggests a Prebiotic RNA World (Vol. 37). Cold Spring Harbor. 10.1101/0.561-584 [DOI] [Google Scholar]
- Trotta CR, Miao F, Arn EA, Stevens SW, Ho CK, Rauhut R, & Abelson JN (1997, 1997/06/13/). The Yeast tRNA Splicing Endonuclease: A Tetrameric Enzyme with Two Active Site Subunits Homologous to the Archaeal tRNA Endonucleases. Cell, 89(6), 849–858. 10.1016/S0092-8674(00)80270-6 [DOI] [PubMed] [Google Scholar]
- Trotta CR, Paushkin SV, Patel M, Li H, & Peltz SW (2006, May 18). Cleavage of pre-tRNAs by the splicing endonuclease requires a composite active site. Nature, 441(7091), 375–377. 10.1038/nature04741 [DOI] [PubMed] [Google Scholar]
- Tsuboi T, Yamazaki R, Nobuta R, Ikeuchi K, Makino S, Ohtaki A, Suzuki Y, Yoshihisa T, Trotta C, & Inada T (2015, Jun 26). The tRNA Splicing Endonuclease Complex Cleaves the Mitochondria-localized CBP1 mRNA. J Biol Chem, 290(26), 16021–16030. 10.1074/jbc.M114.634592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunyasuvunakool K, Adler J, Wu Z, Green T, Zielinski M, Žídek A, Bridgland A, Cowie A, Meyer C, Laydon A, Velankar S, Kleywegt GJ, Bateman A, Evans R, Pritzel A, Figurnov M, Ronneberger O, Bates R, Kohl SAA, Potapenko A, Ballard AJ, Romera-Paredes B, Nikolov S, Jain R, Clancy E, Reiman D, Petersen S, Senior AW, Kavukcuoglu K, Birney E, Kohli P, Jumper J, & Hassabis D (2021, Aug). Highly accurate protein structure prediction for the human proteome. Nature, 596(7873), 590–596. 10.1038/s41586-021-03828-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk T, Baas F, Barth PG, & Poll-The BT (2018, June 15). What’s new in pontocerebellar hypoplasia? An update on genes and subtypes [journal article]. Orphanet Journal of Rare Diseases, 13(1), 92. 10.1186/s13023-018-0826-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volta V, Ceci M, Emery B, Bachi A, Petfalski E, Tollervey D, Linder P, Marchisio PC, Piatti S, & Biffo S (2005, 2005/11/11/). Sen34p depletion blocks tRNA splicing in vivo and delays rRNA processing. Biochemical and Biophysical Research Communications, 337(1), 89–94. 10.1016/j.bbrc.2005.09.012 [DOI] [PubMed] [Google Scholar]
- Weitzer S, Hanada T, Penninger JM, & Martinez J (2015, Jan-Feb). CLP1 as a novel player in linking tRNA splicing to neurodegenerative disorders. Wiley Interdiscip Rev RNA, 6(1), 47–63. 10.1002/wrna.1255 [DOI] [PubMed] [Google Scholar]
- Weitzer S, & Martinez J (2007, 2007/05/01). The human RNA kinase hClp1 is active on 3′ transfer RNA exons and short interfering RNAs. Nature, 447(7141), 222–226. 10.1038/nature05777 [DOI] [PubMed] [Google Scholar]
- Willis IM, & Moir RD (2018, Jun 20). Signaling to and from the RNA Polymerase III Transcription and Processing Machinery. Annu Rev Biochem, 87, 75–100. 10.1146/annurev-biochem-062917-012624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE (2015). Controlling translation via modulation of tRNA levels. WIREs RNA, 6(4), 453–470. 10.1002/wrna.1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, & Culbertson MR (1988, Apr). Mutations affecting the tRNA-splicing endonuclease activity of Saccharomyces cerevisiae. Genetics, 118(4), 609–617. 10.1093/genetics/118.4.609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, & Hopper AK (2014, Jul 15). Healing for destruction: tRNA intron degradation in yeast is a two-step cytoplasmic process catalyzed by tRNA ligase Rlg1 and 5’-to-3’ exonuclease Xrn1. Genes Dev, 28(14), 1556–1561. 10.1101/gad.244673.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Lee I, Spratt H, Fang X, & Bao X (2021). tRNA-Derived Fragments in Alzheimer’s Disease: Implications for New Disease Biomarkers and Neuropathological Mechanisms. J Alzheimers Dis, 79(2), 793–806. 10.3233/jad-200917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, Calvin K, & Li H (2006, May 12). RNA recognition and cleavage by a splicing endonuclease. Science, 312(5775), 906–910. 10.1126/science.1126629 [DOI] [PubMed] [Google Scholar]
- Yoshihisa T (2014). Handling tRNA introns, archaeal way and eukaryotic way. Front Genet, 5, 213. 10.3389/fgene.2014.00213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihisa T, Ohshima C, Yunoki-Esaki K, & Endo T (2007, Mar). Cytoplasmic splicing of tRNA in Saccharomyces cerevisiae. Genes Cells, 12(3), 285–297. 10.1111/j.1365-2443.2007.01056.x [DOI] [PubMed] [Google Scholar]
- Yoshihisa T, Yunoki-Esaki K, Ohshima C, Tanaka N, & Endo T (2003). Possibility of Cytoplasmic pre-tRNA Splicing: the Yeast tRNA Splicing Endonuclease Mainly Localizes on the Mitochondria. Molecular Biology of the Cell, 14(8), 3266–3279. 10.1091/mbc.e02-11-0757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinari S, Shiba T, Inaoka D-K, Itoh T, Kurisu G, Harada S, Kita K, & Watanabe Y. i. (2009). Functional importance of Crenarchaea-specific extra-loop revealed by an X-ray structure of a heterotetrameric crenarchaeal splicing endonuclease. Nucleic Acids Res, 37(14), 4787–4798. 10.1093/nar/gkp506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, & Li H (2004, Mar). Structure determination of a truncated dimeric splicing endonuclease in pseudo-face-centered space group P2(1)2(1)2. Acta Crystallogr D Biol Crystallogr, 60(Pt 3), 447–452. 10.1107/s0907444903029482 [DOI] [PubMed] [Google Scholar]
- Zhou JB, Wang ED, & Zhou XL (2021, Oct 4). Modifications of the human tRNA anticodon loop and their associations with genetic diseases. Cell Mol Life Sci. 10.1007/s00018-021-03948-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Sun B, Yu D, & Bian M (2021, May 26). Roles of tRNA metabolism in aging and lifespan. Cell Death Dis, 12(6), 548. 10.1038/s41419-021-03838-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillmann M, Gorovsky MA, & Phizicky EM (1991). Conserved mechanism of tRNA splicing in eukaryotes. Molecular and Cellular Biology, 11(11), 5410–5416. 10.1128/mcb.11.11.5410-5416.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]


