Abstract
The bacterial enhancer-binding protein NtrC is a well-studied response regulator in a two-component regulatory system. The amino (N)-terminal receiver domain of NtrC modulates the function of its adjacent output domain, which activates transcription by the ς54 holoenzyme. When a specific aspartate residue in the receiver domain of NtrC is phosphorylated, the dimeric protein forms an oligomer that is capable of ATP hydrolysis and transcriptional activation. A chemical protein cleavage method was used to investigate signal propagation from the phosphorylated receiver domain of NtrC, which acts positively, to its central output domain. The iron chelate reagent Fe-BABE was conjugated onto unique cysteines introduced into the N-terminal domain of NtrC, and the conjugated proteins were subjected to Fe-dependent cleavage with or without prior phosphorylation. Phosphorylation-dependent cleavage, which requires proximity and an appropriate orientation of the peptide backbone to the tethered Fe-EDTA, was particularly prominent with conjugated NtrCD86C, in which the unique cysteine lies near the top of α-helix 4. Cleavage occurred outside the receiver domain itself and on the partner subunit of the derivatized monomer in an NtrC dimer. The results are commensurate with the hypothesis that α-helix 4 of the phosphorylated receiver domain of NtrC interacts with the beginning of the central domain for signal propagation. They imply that the phosphorylation-dependent interdomain and intermolecular interactions between the receiver domain of one subunit and the output domain of its partner subunit in an NtrC dimer precede—and may give rise to—the oligomerization needed for transcriptional activation.
The bacterial enhancer-binding protein nitrogen regulatory protein C (NtrC) is a response regulator of a two-component regulatory system (6, 21, 27, 30). Such systems, which dominate signal transduction in the bacteria, are composed minimally of a sensor-autokinase that serves as phosphodonor and a response regulator that is phosphorylated on a specific aspartate residue in its receiver domain. NtrC is composed of three domains (23, 27): the N-terminal receiver domain (∼124 residues), which contains the site of phosphorylation, aspartate 54; a central output domain (∼240 residues), which is directly responsible for activation of transcription by the ς54 holoenzyme form of RNA polymerase; and a C-terminal DNA-binding domain (∼90 residues), which carries the major dimerization determinants for the protein and mediates binding to transcriptional enhancers. The receiver domain of NtrC is connected to its central output domain, the only portion of the molecule for which no structural information is available, via a flexible and protease-sensitive linker (∼16 residues) (6, 15).
The unphosphorylated form of NtrC is a dimer in solution and is capable of binding both an enhancer and the ligand ATP (37). Upon phosphorylation, NtrC forms oligomers (likely to be octamers) that can hydrolyze ATP and couple the energy available from ATP hydrolysis to the formation of open complexes by ς54 holoenzyme (1, 31, 36, 42, 43). Oligomerization, ATP hydrolysis, and interaction with ς54 holoenzyme all appear to be functions of the central domain, which has been predicted to adopt a mononucleotide-binding fold (25). It is known that the phosphorylated receiver domain of NtrC must act positively on its central domain, because removing the receiver domain does not substitute for phosphorylation (10, 44). However, the mechanism of signal propagation has not been determined.
The isolated receiver domain of NtrC appears to remain monomeric even after phosphorylation (16, 40). To control the activity of the central domain positively, regions of the phosphorylated receiver domain are likely to interact with regions of the central domain to bring about the oligomerization required for ATP hydrolysis (10, 16, 40). Whether phosphorylation also affects contact with and/or energy coupling to ς54 holoenzyme is not known. It has been proposed that the interaction of the phosphorylated N-terminal domain of NtrC with the central domain may be with a long α-helix that precedes the mononucleotide-binding fold (25).
To identify interdomain interface regions involved in signal propagation in the NtrC protein from Salmonella enterica serovar Typhimurium, we have constructed mutant proteins carrying single cysteines, which can be derivatized with different sulfhydryl-specific probes to study conformational changes (12). Unique cysteines were placed at three positions in the N-terminal domain, positions 86, 89, and 115, and one position at the beginning of the central domain, position 160. These were positions at which other amino acid substitutions had resulted in activity in the absence of phosphorylation (“constitutive” substitutions) (10). The single-cysteine-containing proteins retained good ability to activate transcription when phosphorylated but were not constitutively active (12). Both positions 86 and 89 lie in α-helix 4 (α4) of the receiver domain (40). Nuclear magnetic resonance (NMR) spectroscopy indicated that α4 underwent structural changes in mutant forms of the isolated N-terminal domain carrying constitutive substitutions at positions 86 and 89 (22). Hence, α4 was proposed to constitute at least part of the interdomain interface between the receiver and central domains.
We previously studied phosphorylation-dependent conformational changes in α4 of the receiver domain of NtrC by derivatizing proteins carrying single cysteine substitutions in this helix with a cysteine-specific nitroxide spin label and monitoring the mobility of the nitroxide side chain so introduced by electron paramagnetic resonance (EPR) spectroscopy (12). These studies indicated that there was a dramatic phosphorylation-dependent change which resulted in a large decrease in mobility of the nitroxide side chain at position 86 (protein designated NtrCD86C) and a similar lesser decrease at position 89. Decreases in the mobility of the nitroxide side chain at these positions did not occur in the isolated, derivatized N-terminal receiver domains and apparently did not occur within a monomer of NtrC. However, EPR analysis could not give us any information on where the interdomain interaction between α4 of the N-terminal domain and the remainder of the protein was occurring, and we were unable to determine whether the interaction occurred within a phosphorylated NtrC dimer or required the formation of larger oligomers.
To obtain information additional to that provided by EPR spectroscopy, we derivatized the unique cysteines at positions 86, 89, 115, and 160 of NtrC with the sulfhydryl-specific iron chelate derivative (S)-1-[p-(bromoacetamido)benzyl]-EDTA-Fe (Fe-BABE) (4, 11) so that we could assess Fe-mediated cleavage of the peptide backbone (32–34). We found phosphorylation-dependent changes in the cleavage pattern for derivatized NtrCD86C, referred to as NtrCD86CFe, that gave clear evidence of a contact outside the receiver domain of the protein. Additional studies employing a cysteine-specific biotin derivative of NtrCD86C (called NtrCD86CBio), which was used to form heterodimers with NtrCD86CFe, allowed us to show that the phosphorylation-dependent cleavage was an intermolecular event. Finally, cleavage of NtrCD86CFe at various concentrations allowed us to show that the phosphorylation-dependent cleavage appeared to occur within a dimer of NtrC rather than depending on the formation of larger oligomers.
MATERIALS AND METHODS
Purification and conjugation of proteins.
All plasmids were constructed as described previously (12), and maltose-binding protein (MBP)-NtrC fusion proteins with single cysteine residues were purified as described previously (12). (MBP is fused to the N terminus of NtrC). The NtrC protein concentration was determined by measuring the UV absorbance at 280 nm in the presence of 6 M guanidine hydrochloride, using an extinction coefficient of 1.11 × 105 M−1 cm−1 for the MBP-NtrC fusion protein (7). Fe-BABE was synthesized as described previously (11). Conjugation with Fe-BABE was initiated by adding a stock solution of Fe-BABE in dimethyl sulfoxide to NtrC mutant proteins in conjugation buffer [50 mM 3-(N-morpholino)propanesulfonic acid (MOPS) (pH 8), 50 mM KCl, 5% (vol/vol) glycerol, 5 mM EDTA] to yield a final Fe-BABE concentration of 0.5 or 1 mM. The reaction was allowed to proceed for 90 min at room temperature. For biotinylation, a 12-fold molar excess of the cysteine-specific biotinylation reagent N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio)propionamide (biotin-HPDP; Pierce, Rockford, Ill.) in dimethyl formamide was added to NtrC in PBS buffer (20 mM sodium phosphate [pH 7.4], 150 mM NaCl, 1 mM EDTA). The reaction was allowed to proceed for 90 min at room temperature. Excess, unreacted Fe-BABE or biotin-HPDP was removed by dialysis at 4°C against cleavage buffer (50 mM MOPS [pH 8], 50 mM KCl, 5% [vol/vol] glycerol, 1 mM EDTA). The percent conjugation of Fe-EDTA or biotin-HPDP reagent was assessed by the Ellman assay (8) or the microfluorometric assay of Parvari et al. (28), which measure free thiol. Conjugation was around 50%, and derivatized NtrC proteins retained some ability to activate transcription. Although it is likely that conjugation of NtrCD86C resulted in significant loss of activity, the effects of derivatization were hard to assess due to partial conjugation. Subunit exchange between NtrCD86CFe and NtrCD86CBio was performed by mixing the proteins in a 2:1 ratio and incubating the mixture for 30 min at 37°C (17). The protein was used immediately after subunit exchange.
Fe-mediated cleavage of NtrC.
Protein cleavage reactions (4) using NtrC proteins derivatized with Fe-BABE were performed in cleavage buffer by the sequential addition of freshly prepared stock solutions of sodium ascorbate (pH 7.5) (Fluka, New York, N.Y.) and hydrogen peroxide (Ultrex grade; J. T. Baker, Phillipsburg, N.J.) to final concentrations of 10 mM each. EDTA (1 mM final concentration) was added to all reagents and buffers (11). After addition of the cleavage reagents, the samples were gently mixed by tapping. After a 10-s reaction, cleavage was quenched by the addition of 4× sodium dodecyl sulfate (SDS) protein sample buffer (18) to a final concentration of 1× and the samples were immediately frozen in liquid nitrogen and stored at −70°C until used for SDS-polyacrylamide gel electrophoresis analysis. Cleavage buffer was used in place of sodium ascorbate and/or hydrogen peroxide for control reactions. Where indicated, protein was phosphorylated by adding MgCl2 and carbamoyl phosphate to final concentrations of 8 and 10 mM, respectively, and incubating the mixture at room temperature for 10 min before initiating cleavage. Under these conditions, NtrC was 50 to 100% phosphorylated as judged by mass spectrometry (I. Hwang and D. Yan, unpublished data).
Chemical cleavage of NtrC at cysteine residues.
To produce marker fragments, the method of Jacobson et al. (13) was modified as follows to cleave NtrCD54C at cysteine residues. The protein buffer was exchanged for 0.1 M MOPS (pH 8.5)–8 M urea, and the protein was then incubated for 2 h at 37°C. A fivefold molar excess of 2-nitro-5-thiocyanatobenzoic acid (NTCB; Aldrich, Milwaukee, Wis.) was added to the protein, and it was incubated at 37°C overnight. Cleavage was quenched with 1% SDS and 1% β-mercaptoethanol.
Separation and visualization of protein cleavage fragments.
Immediately upon being thawed, approximately 10 μg of cleaved protein was loaded on Laemmli (18) SDS–8% polyacrylamide gels without a heating step. (The use of 6 or 12% polyacrylamide gels or longer gels did not yield significantly improved separation of cleavage fragments.) Protein bands were visualized using Fast Stain (Zoion, Newton, Mass.). Electroblotting (4 μg of protein per lane) was performed using polyvinylidene difluoride membranes and transfer buffer [10 mM 3-(Cyclohexylamino)-1-propanesulfonic acid (CAPS) (pH 11), 10% (vol/vol) methanol] at 70 V for 45 min in a cold room (4°C). The molecular weight lanes were then cut from the blot membrane and stained with Fast Stain. The blots were incubated with primary antibodies raised against MBP (New England Biolabs, Beverly, Mass.), NtrC, the N-terminal domain of NtrC (124 residues; Zymed, South San Francisco, Calif.), the C-terminal domain of NtrC (90 residues; Zymed), or the metal chelate (CHA255 [35]; a kind gift from David Goodwin, Stanford University). Alkaline phosphatase (AP)-conjugated anti-rabbit or anti-mouse antibody (Zymed) was used as the secondary antibody. Streptavidin-AP conjugate (Pierce) was used for direct detection of biotinylated NtrC fragments on Western blots. All bands were visualized using AP conjugate substrate kits from Bio-Rad (Hercules, Calif.).
Determination of the N-terminal sequence.
Amino-terminal sequencing was performed by automated Edman degradation using an Applied Biosystems 470A gas phase sequencer at the Protein Structure Laboratory, University of California, Davis, Calif., and was attempted on bands 2/2′, 4, and 5 (see Results). If cleavage of a polypeptide backbone by a tethered iron chelate is hydrolytic (33), the free N terminus that is generated can be identified by Edman sequencing. By contrast, if cleavage is generated by radicals, the predominant products do not have a free primary amino group (29) and cannot be sequenced by automated Edman degradation.
Quantification of cleavage fragments and activity on dilution of NtrCD86C.
The same amount (approximately 13 μg) of NtrCD86CFe was cleaved at four different concentrations as follows: 10 μM (7.5 μl), 2 μM (37.5 μl), 1 μM (75 μl), and 0.2 μM (375 μl). After phosphorylation of the protein, 80 mM stock solutions of hydrogen peroxide and sodium ascorbate were added to final peroxide and ascorbate concentrations of 10 mM each. At 10 s after the sequential addition of cleavage reagents, cleavage buffer was added to the samples at 10, 2, and 1 μM, to increase their volumes to 500 μl, the same as that of the most diluted sample (0.2 μM). All samples were then concentrated from 500 to ∼5 μl in Microcon 30 concentrators (Amicon, Beverly, Mass.) by centrifuging for 25 to 30 min. After the addition of 15 μl of 1.5× SDS sample buffer, concentrated proteins were collected by centrifuging for 3 min. The samples were then immediately loaded onto an SDS-polyacrylamide gel along with a control sample that was thawed just prior to loading. The control sample (10 μM) was prepared by phosphorylating and cleaving as above and freezing immediately after cleavage without the dilution and concentration steps. The intensities of the bands on the stained and dried gels were quantified with a Molecular Dynamics Personal Densitometer.
The ATPase activity of unconjugated NtrCD86C at 1 and 0.2 μM was assayed as described previously (10, 43), with the following exceptions: cleavage buffer was used instead of the standard ATPase buffer and the assay was performed at room temperature instead of 37°C so that conditions for assessing activity would be similar to those used for protein cleavage.
RESULTS
Phosphorylation-dependent chemical cleavage of mutant NtrC proteins by Fe-EDTA chelates tethered to single cysteines in α4 of the N-terminal domain.
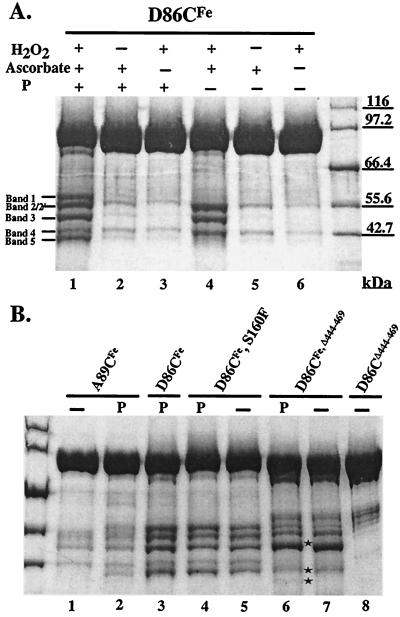
To identify regions of NtrC that undergo a phosphorylation-dependent conformational change, protein cleavage experiments were performed with Fe-EDTA derivatives of mutant NtrC proteins (MBP fusion versions; 94 kDa) carrying single cysteine residues in positions described in the Introduction (NtrCD86CFe, NtrCA89CFe, NtrCV115CFe, and NtrCS160FFe). MBP-NtrC fusion proteins have properties very similar to those of NtrC itself (17). In the absence of phosphorylation, NtrCD86CFe, which carries its single cysteine in α4 of the N-terminal domain, yielded four fragments of approximately 54.5, 54.5, 49, and 43 kDa (referred to as bands 2, 2′, 3, and 4, respectively) (Fig. 1A, lane 4). Bands 2 and 2′ were not separated in full-length NtrC. Upon phosphorylation, NtrCD86CFe yielded two additional bands of approximately 58 and 39.5 kDa (referred to as bands 1 and 5, respectively) (Fig. 1A, lane 1, and Fig. 1B, lane 3). Bands 1 and 5, whose combined size is close to that of full-length NtrC, provided evidence for a phosphorylation-dependent conformational change in NtrC that affected α4 of its N-terminal domain. To confirm that phosphorylating conditions were not sufficient to yield bands 1 and 5 but that phosphorylation of NtrCD86CFe was actually required, we showed that NtrCD54A,D86CFe, which cannot be phosphorylated, did not yield these bands (data not shown). Rather, it yielded only bands 2, 2′, 3, and 4 under both nonphosphorylating and phosphorylating conditions. Control experiments in which NtrCD86CFe was treated with only hydrogen peroxide or ascorbate yielded only traces of cleavage products (Fig. 1A, lanes 2, 3, 5, and 6), as reported by Rana and Meares (32, 33). Under phosphorylating conditions, several percent of the total protein was cleaved and the cleavage fragments were easily detected on Coomassie blue-stained gels.
FIG. 1.
Cleavage products of various NtrC proteins. (A) NtrCD86CFe was cleaved with (lanes 1 to 3) or without (lanes 4 to 6) phosphorylation. (B) NtrCA89CFe (lanes 2 and 1), NtrCD86CFe,S160F (lanes 4 and 5), and NtrCD86CFe,Δ444–469 (lanes 6 and 7) were cleaved with or without phosphorylation, respectively. Cleaved, phosphorylated NtrCD86CFe (lane 3) was used as a reference, and unconjugated NtrCD86C,Δ444–469 (lane 8) was used as control for background impurities of truncated monomeric protein. Stars indicate bands 2′, 4, and 5 from the truncated monomeric protein. Band 2′ from the truncated monomeric protein had a slightly lower mobility than band 3 and was only slightly separated from it. The designations “P” and “−” above the lanes indicate phosphorylation and lack of phosphorylation, respectively. Cleavage products were separated by electrophoresis on SDS–8% polyacrylamide gels, which were stained with Coomassie blue.
NtrCA89CFe, which also contains a single cysteine in α4 of the N-terminal domain, showed similar cleavage patterns to NtrCD86CFe in both the absence and presence of phosphorylation (Fig. 1B, lanes 1 and 2, respectively), but the cleaved products were present in smaller amounts, indicating that cleavage was less efficient. Neither NtrCV115CFe, which contains a single cysteine in α5 of the N-terminal domain, nor NtrCS160CFe, which contains a single cysteine at the beginning of the central domain of NtrC, yielded more than small amounts of cleavage products and these were unchanged by phosphorylation (data not shown).
When D86C was combined with the S160F constitutive substitution, the derivatized protein, NtrCD86CFe,S160F, yielded all five cleavage bands characteristic of phosphorylated NtrCD86CFe, whether or not it was phosphorylated (Fig. 1B, lanes 4 and 5, respectively). When phosphorylated, it yielded slightly more of bands 1 and 5 (lane 4 versus lane 5), which depend on phosphorylation of NtrCD86CFe. These results differed from those obtained by EPR spectroscopy of NtrCD86C,S160F derivatized with a nitroxide spin label (12). In that case, a decrease in the mobility of the label observed upon phosphorylation of derivatized NtrCD86C also required phosphorylation of derivatized NtrCD86C,S160F. The earlier result was interpreted to mean that formation of active oligomers by a bypass mechanism which did not entail phosphorylation was not sufficient to yield a phosphorylation-dependent change in conformation. We speculate that the discrepancy between the two results may be due to a greater sensitivity of detecting Fe-mediated cleavage products than was possible by measuring a population-dependent change in the EPR spectrum.
Assignment of cleavage products.
To determine the positions of the single phosphorylation-dependent cleavage and the two phosphorylation-independent cleavages in NtrCD86CFe, the six cleavage products yielded by NtrCD86CFe and by a C-terminally truncated form of the protein, NtrCD86CFe,Δ444–469 (24), were characterized immunologically (see below; summarized in Table 1). Hereafter, we refer to a pair of cleavage products as the N- and C-terminal cleavage fragments, respectively. The NtrCD86CFe,Δ444–469 protein, which is largely monomeric (∼20% dimer at the concentrations used for cleavage [12, 17, 24, 37]), yielded only a small amount of band 5 upon phosphorylation (Fig. 1B, lane 6), commensurate with the view that the phosphorylation-dependent cleavage which yields bands 1 and 5 occurs between monomers of NtrC rather than within a single monomer (see below) (band 1 in the truncated protein was obscured by a contaminant). Because the truncated form of NtrC lacks the last 25 residues of the protein, C-terminal cleavage fragments should migrate faster on SDS-polyacrylamide gels, and this was the case for bands 4 and 5 (Fig. 1B, lane 6, indicated by stars) and also for band 2′ (indicated by a star), which was more difficult to see.
TABLE 1.
Summary of the immunological reactivities of cleavage bands produced from NtrCD86CFe and NtrCD86CFe,Δ444–469a
| Band | Band detected with:
|
|||
|---|---|---|---|---|
| Anti-MBP | Anti-N-ter | Anti-C-ter | Anti-metal chelate | |
| 1b | + | + | − | + |
| 2c | + | + | − | + |
| 2′c | − | + | + | + |
| 3 | + | + | − | − |
| 4 | − | + | + | − |
| 5b | − | − | + | − |
Analysis was performed by Western blotting using antisera directed against MBP, the N-terminal domain of NtrC (N-ter), the C-terminal domain of NtrC (C-ter), intact NtrC, or the metal chelate (CHA255) (see Materials and Methods). Antibodies directed against full-length NtrC detected all bands detected by other antibodies and no additional bands. The latter confirmed that there were no cleavage fragments that lacked both termini of NtrC.
Produced only when proteins were phosphorylated.
Separated only in NtrCD86CFe,Δ444–469.
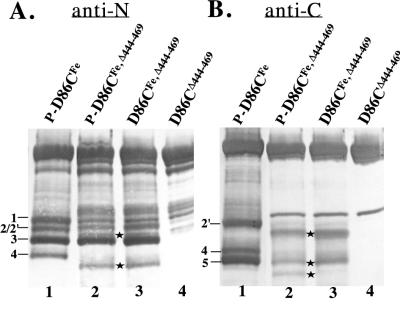
When cleavage products were probed by Western blotting with antibody directed against the C terminus of NtrC (last 90 residues) (Fig. 2B), bands 4 and 5 were detected from both full-length (lane 1) and C-terminally truncated NtrC proteins (lanes 2 and 3), as expected. In addition, one or both of the overlapping bands 2 and 2′ from full-length NtrC were detected whereas only band 2′ from the truncated protein was detected. Thus, bands 2′ and 4 appear to be C-terminal cleavage fragments from unphosphorylated NtrC (also seen when the protein is phosphorylated), whereas band 5 is a C-terminal fragment obtained only when the protein is phosphorylated. We were able to obtain an amino-terminal sequence of fragment 4 but not fragment 5 (see Materials and Methods). The sequence from band 4, QQGAFDY, corresponded to residues 95 to 101, indicating that the cleavage which gave rise to band 4 occurred between residues 94 and 95 of the N-terminal domain (Fig. 3). Because band 5 has a higher mobility than band 4, the cleavage which gave rise to it must have occurred downstream of residue 94.
FIG. 2.
Western analysis of cleavage products from the truncated monomeric protein NtrCD86CFe,Δ444–469 using antibodies directed against the N and C termini of NtrC. In both panels, phosphorylated NtrCD86CFe (lane 1) was used as reference and unconjugated monomeric protein NtrCD86C,Δ444–469 (lane 4) was used as a background control for contaminants. Phosphorylation is designated by “P-.” Cleavage products were separated by electrophoresis on SDS–8% polyacrylamide gels. (A) Detection with N-terminal antibodies. Stars indicate bands 2′ and 4 from the truncated protein. (B) Detection with C-terminal antibodies. Stars indicate bands 2′, 4, and 5 from the truncated protein.
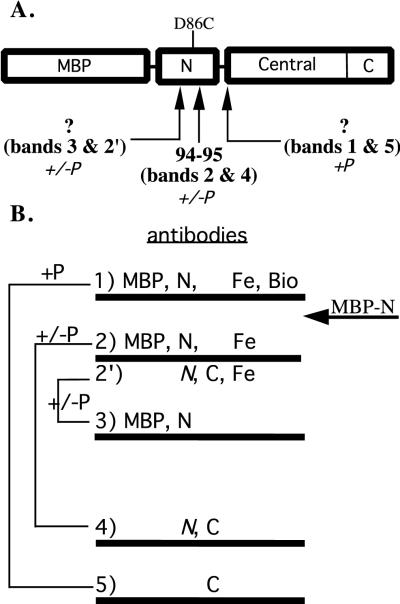
FIG. 3.
Summary of cleavage sites and Western analysis. (A) Diagram of cleavage sites, indicated by arrows. The D86C substitution in the N-terminal domain of NtrC, which was derivatized with Fe-BABE, is indicated. For purposes of illustration, all cleavage sites are indicated within a monomer. The site of the phosphorylation-independent cleavage that yielded bands 3 and 2′ lies after position 54 and before position 86. The site of the phosphorylation-dependent cleavage that yielded bands 1 and 5 lies outside the N-terminal domain and is predicted to lie at the end of the linker or the beginning of the central domain. This cleavage occurs between the monomers of a dimer. (B) Partner bands resulting from phosphorylation-dependent (+P) or -independent (+/−P) cleavages are indicated by their mobilities on SDS–8% polyacrylamide gels. For each band, the results of Western analysis using antibodies directed against MBP, the N-terminal domain of NtrC (N), the C-terminal domain (C), or the iron chelate (Fe) are indicated, as is the reaction of streptavidin with biotin (Bio). The mobility of the MBP–N-terminal protein (a protein containing only MBP and the N-terminal domain of NtrC) is indicated by an arrow.
Western blotting with antibodies directed against the entire N terminus of NtrC (residues 1 to 124) (Fig. 2A) detected bands 1, 2/2′, 3, and 4 from full-length NtrCD86CFe (lane 1) and bands 2, 2′, 3, and 4 from the truncated protein NtrCD86CFe,Δ444–469 (lanes 2 and 3) (band 1 was obscured by a contaminant in the truncated protein). Band 1, which was detected only with antibodies directed against the N terminus of NtrC, is an N-terminal fragment. As expected, it was also detected with antibodies directed against MBP, which was fused to the N terminus, and with antibodies directed against the iron chelate (CHA255) (reference 35 and data not shown [summarized in Table 1 and Fig. 3]). Band 5, which was generated by the same cleavage that gave rise to band 1, was not detected by any of the N-terminally directed antibodies. Together with the size of band 1 (see below), immunological evidence was commensurate with the view that the cleavage that gave rise to bands 1 and 5 occurred outside the N-terminal domain of NtrC. Thus, the cleavage that occurred uniquely in phosphorylated NtrC apparently occurred outside its N-terminal domain.
Band 4, which is a C-terminal cleavage product (see the sequence above), was detected by antibodies directed against both termini of NtrC. Presumably, detection by antibodies directed against the N terminus was due to the fact that the cleavage which gave rise to band 4 occurred within the N-terminal domain (Fig. 3) and residues 95 to 124 of the N-terminal domain were present in this band. As expected if this interpretation is correct, band 4 was not detected by antibodies directed against MBP or by antibodies directed against the iron chelate (data not shown; summarized in Table 1). Based on size, band 2 appears to be the N-terminal partner of band 4. As expected if this is the case, band 2 from the C-terminally truncated protein NtrCD86C,Δ444–469 was detected by antibodies directed against the N terminus of NtrC, by anti-MBP antibodies, and by antibodies directed against the iron chelate. Because band 2 overlapped with band 2′ in cleavage products generated from full-length NtrC, meaningful information about these bands could not be derived. Antibodies directed against both termini of NtrC, against MBP, and against the iron chelate reacted with this overlapped band.
Finally, bands 3 and 2′ appear to be N- and C-terminal partners, respectively, of a cleavage that occurs within the N-terminal domain of unphosphorylated NtrC (Fig. 2 and 3; Table 1). Band 3 is detected by N-terminal antibodies and anti-MBP antibodies. It migrates faster than band 2, indicating that the cleavage which gave rise to it is likely to occur before residue 94, and it is not detected by antibodies directed against the iron chelate, indicating that the cleavage occurs before position 86. Band 2′ from truncated NtrCD86CFe,Δ444–469 was detected by both anti-N-terminal and anti-C-terminal antibodies to NtrC and was also detected by antibodies to the iron chelate, indicating that it contains residue 86. When NtrCD54C was chemically cleaved at cysteine residues (see Materials and Methods) and the fragments so generated were used as molecular weight standards, bands 2/2′ migrated slightly faster than the C-terminal fragment carrying the sequence from residue 54 to the end of NtrC. Thus, the cleavage that resulted in bands 2′ and 3 is predicted to occur after residue 54 but before residue 86. As expected if it was a C-terminal cleavage product, band 2′ was not detected by anti-MBP antibodies. Given the predicted position of cleavage that gave rise to band 2′, i.e., that it occurred prior to position 86 in the N-terminal domain of NtrC, detection by anti-N-terminal antibodies is expected based on the results obtained for band 4. As was the case for band 2, meaningful information about band 2′ could not be derived from cleavage products of full-length NtrC because bands 2 and 2′ overlapped on the SDS-polyacrylamide gels. The overlap of bands 2 and 2′ is probably due to the aberrant mobility of band 2′, because the mobility of the C-terminal fragment beginning with residue 54 was lower than predicted (data not shown).
Evidence that the phosphorylation-dependent cleavage occurs outside the N-terminal domain of NtrC.
To provide further evidence that the phosphorylation-dependent cleavage of NtrCD86CFe occurred outside the N-terminal domain of the protein, we compared the size of band 1, the N-terminal cleavage product, to that of the intact N-terminal domain (residues 1 to 124) of NtrC (MBP fusion proteins in both cases) (12). The mobility of band 1 (∼58 kDa) was lower than that of MBP–N-terminus (calculated to be 55.3 kDa) (data not shown) and was close to the predicted mobility for a fragment that carries MBP–N-terminus and the entire linker region (residues 1 to 140; calculated to be 57.1 kDa). Based on the size of band 1, the cleavage that gave rise to it is likely to occur at the beginning of the central domain or within the exposed, protease-sensitive linker that connects the receiver and central domains (Fig. 3). We favor the latter because a small amount of cleavage occurs at this position even in NtrCD86C that has not been derivatized with Fe-BABE (data not shown).
As expected based on the sequence data for band 4, which indicated that bands 2 and 4 were produced by cleavage between residues 94 and 95, band 2 migrated faster than MBP–N-terminus did. The mobility of MBP–N-terminus was intermediate between those of bands 1 and 2 (data not shown).
Evidence that the phosphorylation-dependent cleavage occurs between monomers whereas phosphorylation-independent cleavages occur within a monomer.
To determine whether the three cleavages giving rise to bands 1 and 5, bands 2 and 4, and bands 3 and 2′ occurred within a monomer or between monomers, we first assessed whether monomerization of NtrC by means of a C-terminal truncation [Δ(444–469)] affected the efficiencies with which these cleavages occurred. Whereas the C-terminal truncation had little effect on the intensities of bands 2, 3, and 4, it greatly decreased the intensity of band 5 (e.g., note the relative intensities of bands 5 and 4 generated from truncated and from intact NtrC in Fig. 1B, lanes 6 and 3, respectively). (The intensity of band 1 generated from the truncated protein could not be assessed due to the presence of contaminants with similar mobility [Fig. 1B, lane 8].) Thus, the phosphorylation-independent cleavages occurred efficiently within an NtrC monomer whereas the phosphorylation-dependent cleavage did not. Residual phosphorylation-dependent cleavage of NtrCD86CFe,Δ444–469 is probably accounted for by residual dimerization.
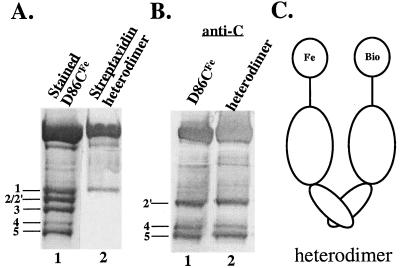
To demonstrate directly that phosphorylation-dependent scission occurred between the monomers of an NtrC dimer, we formed heterodimers between NtrCD86CFe and the cysteine-specific biotin derivative NtrCD86CBio (see Materials and Methods). A 2:1 ratio of the two proteins was used in the subunit exchange process (17) to produce heterodimers that contain one NtrCD86CFe subunit and one NtrCD86CBio subunit (Fig. 4C), in addition to the two types of homodimers. After allowing phosphorylation-dependent cleavage and separating products on a gel, blotted cleavage products were analyzed with AP-streptavidin to detect fragments containing biotin, which can be generated only by intermolecular scission. Band 1 was detected, whereas bands 2 and 2′, which appear to be intramonomer scission products, were not (Fig. 4A, lane 2). Thus, the phosphorylation-dependent cleavage yielding band 1 is an intermolecular event. To demonstrate that the same cleavages occurred in populations containing heterodimers as in populations of NtrCD86CFe homodimers, we demonstrated that antibodies directed against the C terminus of NtrC detected bands 2′, 4, and 5 (Fig. 4B, lane 2).
FIG. 4.
Detection of biotin-containing cleavage products of heterodimers. Subunit exchange between NtrCD86CFe and NtrCD86CBio (2:1) yielded the heterodimers represented in panel C. All samples were phosphorylated and cleaved. (A) Biotin-containing cleavage fragments from heterodimers (4.5 μg of total protein) were detected with AP-streptavidin (lane 2). Cleavage fragments from NtrCD86CFe (10 μg) were stained with Coomassie blue to serve as markers (lane 1). (B) Antibodies directed against the C terminus of NtrC were used to detect fragments from heterodimers (4.5 μg of total protein; lane 2) and from the control NtrCD86CFe (3 μg; lane 1). (C) Heterodimers (see above).
Evidence that the phosphorylation-dependent cleavage occurs within a dimer.
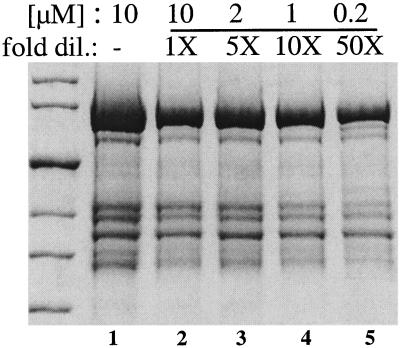
Because dimers of NtrC oligomerize upon phosphorylation, the intermolecular cleavage unique to the phosphorylated protein could have occurred within a dimer or alternatively could have required oligomer formation. To distinguish between these two possibilities, we assessed this cleavage as a function of the concentration of NtrCD86CFe (Fig. 5). Equal amounts of the protein were cleaved at different concentrations between 10 and 0.2 μM (lanes 2 to 5). Each sample was then diluted to the lowest concentration, 0.2 μM, and reconcentrated. An additional sample at 10 μM, the usual concentration for the reaction, was analyzed directly as a control (lane 1). Recovery after concentration (lanes 2 to 5) was 40 to 65% (compared to the control in lane 1) and was least good for the smaller fragments, bands 4 and 5, whose sizes were close to those for exclusion by the Microcon filter (30 kDa). The lowest recovery (40%) was obtained for the sample that was least concentrated at the time of cleavage (0.2 μM). However, the ratio of cleavage products to uncleaved protein was similar at all four concentrations tested. To assess the efficiency of the phosphorylation-dependent cleavage at different NtrC concentrations, we quantified the intensities of the large N-terminal cleavage products, bands 1 and 3, by densitometry and calculated the ratios of band 1 to band 3. For samples at 10 and 2 μM, the ratio was similar to that of the control (Table 2). At lower concentrations (1 and 0.2 μM), this ratio decreased slightly, and at 0.2 μM it was 60% of that at 10 μM. The amounts of NtrC oligomer at the different concentrations tested for cleavage were assessed by gel filtration chromatography (10 and 1 μM) or by assay of the ATPase activity of the protein, which is a function of the amount of oligomer. The amount of oligomer decreased 4-fold between 10 and 1 μM protein (D. Yan, personal communication) and a further 14-fold between 1 and 0.2 μM (data not shown). Thus, the amount of oligomer decreased approximately 50-fold over the 50-fold range of concentrations tested whereas the ratio of phosphorylation-dependent to phosphorylation-independent cleavage decreased by less than 2-fold. Therefore, we infer that the phosphorylation-dependent cleavage is occurring within a dimer.
FIG. 5.
Effect of dilution on the phosphorylation-dependent cleavage of NtrCD86CFe. NtrCD86CFe was phosphorylated and cleaved at four different concentrations between 10 and 0.2 μM (lanes 2 to 5), diluted, and then concentrated before being loaded on the gel (see Results and Materials and Methods). The control sample in lane 1 was not diluted and reconcentrated.
TABLE 2.
Quantification of band 1 as a function of dilution of NtrCD86CFea
| Sample concn (μM) | Ratio of band 1 to band 3b (%) | Intensity normalized to sample at 10 μM |
|---|---|---|
| 10 (control)c | 87 | 1.06 |
| 10 | 82 | 1.00 |
| 2 | 71 | 0.87 |
| 1 | 57 | 0.70 |
| 0.2 | 49 | 0.60 |
Samples were phosphorylated and were analyzed as described in Results and Materials and Methods (Fig. 5).
Calculated ratio of the intensity of band 1 to the intensity of band 3 on a Coomassie-blue stained gel, measured densitometrically.
10 μM, frozen immediately after cleavage.
DISCUSSION
In the absence of structural information on either the phosphorylated receiver domain of NtrC or its central output domain, we derivatized single cysteine residues at strategic locations in the receiver domain with an Fe-EDTA chelate to explore the pathway of signal transduction between domains. We obtained evidence for a phosphorylation-dependent conformational change that brought the beginning of α4 (positions 86 and 89) of the receiver domain in one monomer into contact with (i.e., within ca. 12 Å of) (26) the end of the flexible linker region and the beginning of the central domain in the opposite monomer of a dimer (Fig. 6) (40). By analogy to the case for muscle and yeast glycogen phosphorylases (14, 19, 20), this intermolecular interaction may rearrange the dimer interface to promote phosphorylation-dependent oligomerization of NtrC. To explore the site of intermonomer interaction in more detail, we have placed single cysteine residues at 10 locations within the first long α-helix in the central domain of NtrC, which has been postulated to be responsible for communication between the receiver domain and the remainder of the central domain (25). Interestingly, five of the cysteine substitutions appear to result in loss of transcriptional activation by NtrC in vivo (J. Lee, unpublished results), commensurate with the postulated role of this helix in signal propagation.
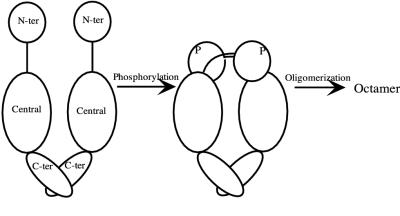
FIG. 6.
Proposed model for phosphorylation-induced signal propagation in NtrC. A phosphorylation-dependent conformational change in α4 of the N-terminal (N-ter) receiver domain of NtrC (positions 86 and 89 at the beginning of the helix) results in a demonstrable contact with the beginning of the central domain and the end of the flexible interdomain linker in the opposite monomer of a dimer. This change in the dimer interface leads to the formation of active oligomers (probably octamers). The diagram, which is schematic, is not meant to imply a large interdomain movement of the entire N-terminal domain with respect to the central domain, nor is it meant to imply a complete absence of contact between the two domains in unphosphorylated NtrC.
NMR spectra of the phosphorylated receiver domain of wild-type NtrC and of constitutive mutant forms with amino acid substitutions in α4 indicated that conformational changes occurred in the so-called 3445 face of the molecule (22). This face extends from α3 through β-strand 5 (β5). The recently obtained structure of the phosphorylated domain (16) indicates that there is a profound rearrangement of α4 that results in the formation of a solvent-exposed hydrophobic surface constituted by the side chains of L87, A90, and V91. In intact NtrC, this surface, which is the one we have derivatized, presumably contacts the remainder of the protein (Fig. 6).
In the absence of phosphorylation, the iron chelate tethered at positions 86 and 89 of NtrC cleaved at two positions within the receiver domain of the protein (Fig. 3). One cleavage, which gave rise to bands 2 and 4 on SDS-polyacrylamide gels, lay at the end of α4 between residues 94 and 95 and was well defined because it was possible to obtain the amino-terminal sequence of band 4. Based on the NMR structure of the receiver domain of NtrC (40), the position of cleavage is some 12 to 15 Å distant from the site at which the chelator was placed, commensurate with the length of its spacer arm (26). The second cleavage, which gave rise to bands 3 and 2′ occurred prior to position 86, apparently somewhere between residues 54 and 86 (Fig. 3).
There is considerable structural information on unphosphorylated receiver domains, all of which adopt the same general β5-α5 fold (2, 3, 5, 9, 38–41). However, there is virtually no structural information on their phosphorylated counterparts, and only two structures of unphosphorylated multidomain response regulators have been solved (2, 3, 5), the transcriptional regulator NarL and the methylesterase CheB, which regulates the adaptation phase of bacterial chemotaxis. In both cases, the structures revealed that the receiver domains block active sites in the output domains, and hence it is inferred that phosphorylation-induced conformational changes in the receiver domains disrupt inhibitory interdomain interactions (2, 3, 5). For CheB, it has been proposed that α4 and the C-terminal ends of β5 and α5 of the receiver domain constitute the interdomain interface that blocks the catalytic site (5), whereas for NarL, the loops connecting α2 and β3, α3 and β4, and α4 and β5, i.e., a different surface, are implicated in blocking the helix-turn-helix DNA-binding motif (2, 3). It has recently been recognized that the phosphorylated receiver domain of CheB must play a positive role in stimulating methylesterase activity, in addition to its role in relieving inhibition caused by the unphosphorylated domain (5). Thus, as is true for NtrC, the phosphorylated receiver domain of CheB apparently forms a new contact with the remainder of the protein.
ACKNOWLEDGMENTS
We thank Eric Soupene and Dalai Yan for criticism of the manuscript and O. Carmi for help in its preparation.
This work was supported by NIH grants GM25909 and GM38361 to C.M. and S.K., respectively.
REFERENCES
- 1.Austin S, Dixon R. The prokaryotic enhancer binding protein NTRC has an ATPase activity which is phosphorylation and DNA dependent. EMBO J. 1992;11:2219–2228. doi: 10.1002/j.1460-2075.1992.tb05281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baikalov I, Schröder I, Kaczor-Grzeskowiak M, Cascio D, Gunsalus R P, Dickerson R E. NarL dimerization? Suggestive evidence from a new crystal form. Biochemistry. 1998;37:3665–3676. doi: 10.1021/bi972365a. [DOI] [PubMed] [Google Scholar]
- 3.Baikalov I, Schröder I, Kaczor-Grzeskowiak M, Grzeskowiak K, Gunsalus R P, Dickerson R E. Structure of the Escherichia coli response regulator NarL. Biochemistry. 1996;35:11053–11061. doi: 10.1021/bi960919o. [DOI] [PubMed] [Google Scholar]
- 4.Datwyler, A., and C. F. Meares. Probing of proteins by metal ions and their low-molecular-weight complexes. Metal Ions Biol. Syst., in press.
- 5.Djordjevic S, Goudreau P N, Xu Q, Stock A M, West A H. Structural basis for methylesterase CheB regulation by a phosphorylation-activated domain. Proc Natl Acad Sci USA. 1998;95:1381–1386. doi: 10.1073/pnas.95.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drummond M, Whitty P, Wootton J. Sequence and domain relationships of ntrC and nifA from Klebsiella pneumoniae: homologies to other regulatory proteins. EMBO J. 1986;5:441–447. doi: 10.1002/j.1460-2075.1986.tb04230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edelhoch H. Spectroscopic determination of tryptophan and tryrosine in proteins. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 8.Ellman G. Tissue sulfhydryl groups. Arch Biochem Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 9.Feher V A, Zapf J W, Hoch J A, Dahlquist F W, Whiteley J M, Cavanagh J. 1H, 15N, and 13C backbone chemical shift assignments, secondary structure, and magnesium-binding characteristics of the Bacillus subtilis response regulator, Spo0F, determined by heteronuclear high-resolution NMR. Protein Sci. 1995;4:1801–1814. doi: 10.1002/pro.5560040915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flashner Y, Weiss D S, Keener J, Kustu S. Constitutive forms of the enhancer-binding protein NtrC: evidence that essential oligomerization determinants lie in the central activation domain. J Mol Biol. 1995;249:700–713. doi: 10.1006/jmbi.1995.0330. [DOI] [PubMed] [Google Scholar]
- 11.Greiner D P, Miyake R, Moran J K, Jones A D, Negishi T, Ishihama A, Meares C F. Synthesis of the protein cutting reagent iron (S)-1-(p-bromoacetamidobenzyl)ethylenediaminetetraacetate and conjugation to cysteine side chains. Bioconjugate Chem. 1997;8:44–48. doi: 10.1021/bc9600731. [DOI] [PubMed] [Google Scholar]
- 12.Hwang I, Thorgeirsson T, Lee J, Kustu S, Shin Y K. Physical evidence for a phosphorylation-dependent conformational change in the enhancer-binding protein NtrC. Proc Natl Acad Sci USA. 1999;96:4880–4885. doi: 10.1073/pnas.96.9.4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacobson G R, Schaffer M H, Stark G R, Vanaman T C. Specific chemical cleavage in high yield at the amino peptide bonds of cysteine and cystine residues. J Biol Chem. 1973;248:6583–6591. [PubMed] [Google Scholar]
- 14.Johnson L N, O'Reilly M. Control by phosphorylation. Curr Opin Struct Biol. 1996;6:762–769. doi: 10.1016/s0959-440x(96)80005-4. [DOI] [PubMed] [Google Scholar]
- 15.Keener J, Kustu S. Protein kinase and phosphoprotein phosphatase activities of nitrogen regulatory proteins NTRB and NTRC of enteric bacteria: roles of the conserved amino-terminal domain of NTRC. Proc Natl Acad Sci USA. 1988;85:4976–4980. doi: 10.1073/pnas.85.14.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kern D, Volkman B F, Luginbühl P, Nohaile M J, Kustu S, Wemmer D E. Structure of a transiently phosphorylated switch in bacterial signal transduction. Nature. 1999;402:894–898. doi: 10.1038/47273. [DOI] [PubMed] [Google Scholar]
- 17.Klose K E, North A K, Stedman K M, Kustu S. The major dimerization determinants of the nitrogen regulatory protein NTRC from enteric bacteria lie in its carboxy-terminal domain. J Mol Biol. 1994;241:233–245. doi: 10.1006/jmbi.1994.1492. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Lin K, Hwang P K, Fletterick R J. Mechanism of regulation in yeast glycogen phosphorylase. J Biol Chem. 1995;270:26833–26839. doi: 10.1074/jbc.270.45.26833. [DOI] [PubMed] [Google Scholar]
- 20.Lin K, Rath V L, Dai S C, Fletterick R J, Hwang P K. A protein phosphorylation switch at the conserved allosteric site in GP. Science. 1996;273:1539–1542. doi: 10.1126/science.273.5281.1539. [DOI] [PubMed] [Google Scholar]
- 21.Morett E, Segovia L. The ς54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J Bacteriol. 1993;175:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nohaile M, Kern D, Wemmer D, Stedman K, Kustu S. Structural and functional analyses of activating amino acid substitutions in the receiver domain of NtrC: evidence for an activating surface. J Mol Biol. 1997;273:299–316. doi: 10.1006/jmbi.1997.1296. [DOI] [PubMed] [Google Scholar]
- 23.North A K, Klose K E, Stedman K M, Kustu S. Prokaryotic enhancer-binding proteins reflect eukaryote-like modularity: the puzzle of nitrogen regulatory protein C. J Bacteriol. 1993;175:4267–4273. doi: 10.1128/jb.175.14.4267-4273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.North A K, Kustu S. Mutant forms of the enhancer-binding protein NtrC can activate transcription from solution. J Mol Biol. 1997;267:17–36. doi: 10.1006/jmbi.1996.0838. [DOI] [PubMed] [Google Scholar]
- 25.Osuna J, Soberón X, Morett E. A proposed architecture for the central domain of the bacterial enhancer-binding proteins based on secondary structure prediction and fold recognition. Protein Sci. 1997;6:543–555. doi: 10.1002/pro.5560060304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Owens J T, Miyake R, Murakami K, Chmura A J, Fujita N, Ishihama A, Meares C F. Mapping the sigma70 subunit contact sites on Escherichia coli RNA polymerase with a sigma70-conjugated chemical protease. Proc Natl Acad Sci USA. 1998;95:6021–6026. doi: 10.1073/pnas.95.11.6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parkinson J S. Signal transduction schemes of bacteria. Cell. 1993;73:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- 28.Parvari R, Pecht I, Spreq H. A microfluorometric assay for cholinesterases, suitable for multiple kinetic determinations of picomoles of released thiocholine. Anal Biochem. 1983;133:450–456. doi: 10.1016/0003-2697(83)90107-0. [DOI] [PubMed] [Google Scholar]
- 29.Platis I E, Ermácora M R, Fox R O. Oxidative polypeptide cleavage mediated by EDTA-Fe covalently linked to cysteine residues. Biochemistry. 1993;32:12761–12767. doi: 10.1021/bi00210a027. [DOI] [PubMed] [Google Scholar]
- 30.Porter S C, North A K, Kustu S. Mechanism of transcriptional activation by NtrC. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: American Society for Microbiology; 1995. pp. 147–158. [Google Scholar]
- 31.Porter S C, North A K, Wedel A B, Kustu S. Oligomerization of NTRC at the glnA enhancer is required for transcriptional activation. Genes Dev. 1993;7:2258–2273. doi: 10.1101/gad.7.11.2258. [DOI] [PubMed] [Google Scholar]
- 32.Rana T M, Meares C F. Iron chelate mediated proteolysis: protein structure dependence. J Am Chem Soc. 1991;113:1859–1861. [Google Scholar]
- 33.Rana T M, Meares C F. Specific cleavage of a protein by an attached iron chelate. J Am Chem Soc. 1990;112:2457–2458. [Google Scholar]
- 34.Rana T M, Meares C F. Transfer of oxygen from an artificial protease to peptide carbon during proteolysis. Proc Natl Acad Sci USA. 1991;88:10578–10582. doi: 10.1073/pnas.88.23.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reardan D T, Meares C F, Goodwin D A, McTigue M, David G S, Stone M R, Leung J P, Bartholomew R M, Frincke J M. Antibodies against metal chelates. Nature. 1985;316:265–268. doi: 10.1038/316265a0. [DOI] [PubMed] [Google Scholar]
- 36.Rombel I, North A, Hwang I, Wyman C, Kustu S. The bacterial enhancer-binding protein NtrC as a molecular machine. Cold Spring Harbor Symp Quant Biol. 1998;63:157–166. doi: 10.1101/sqb.1998.63.157. [DOI] [PubMed] [Google Scholar]
- 37.Rombel I, Peters-Wendisch P, Mesecar A, Thorgeirsson T, Shin Y K, Kustu S. MgATP binding and hydrolysis determinants of NtrC, a bacterial enhancer-binding protein. J Bacteriol. 1999;181:4628–4638. doi: 10.1128/jb.181.15.4628-4638.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Solá M, Gomis-Rüth F X, Serrano L, González A, Coll M. Three-dimensional crystal structure of the transcription factor PhoB receiver domain. J Mol Biol. 1999;285:675–687. doi: 10.1006/jmbi.1998.2326. [DOI] [PubMed] [Google Scholar]
- 39.Stock A M, Mottonen J M, Stock J B, Schutt C E. Three-dimensional structure of CheY, the response regulator of bacterial chemotaxis. Nature. 1989;337:745–749. doi: 10.1038/337745a0. [DOI] [PubMed] [Google Scholar]
- 40.Volkman B F, Nohaile M J, Amy N K, Kustu S, Wemmer D E. Three-dimensional solution structure of the N-terminal receiver domain of NTRC. Biochemistry. 1995;34:1413–1424. doi: 10.1021/bi00004a036. [DOI] [PubMed] [Google Scholar]
- 41.Volz K, Matsumura P. Crystal structure of Escherichia coli CheY refined at 1.7-A resolution. J Biol Chem. 1991;266:15511–15519. doi: 10.2210/pdb3chy/pdb. [DOI] [PubMed] [Google Scholar]
- 42.Wedel A, Kustu S. The bacterial enhancer-binding protein NTRC is a molecular machine: ATP hydrolysis is coupled to transcriptional activation. Genes Dev. 1995;9:2042–2052. doi: 10.1101/gad.9.16.2042. [DOI] [PubMed] [Google Scholar]
- 43.Weiss D S, Batut J, Klose K E, Keener J, Kustu S. The phosphorylated form of the enhancer-binding protein NTRC has an ATPase activity that is essential for activation of transcription. Cell. 1991;67:155–167. doi: 10.1016/0092-8674(91)90579-n. [DOI] [PubMed] [Google Scholar]
- 44.Weiss D S, Klose K E, Hoover T R, North A, Porter S, Wedel A B, Kustu S. Prokaryotic transcriptional enhancers. In: McKnight S L, Yamamoto K R, editors. Transcriptional regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 667–694. [Google Scholar]