Abstract
Global identification of differentially regulated genes in prokaryotes is constrained because the mRNA does not have a 3′ polyadenylation extension; this precludes specific separation of mRNA from rRNA and tRNA and synthesis of cDNAs from the entire mRNA population. Knowledge of the entire genome sequence of Synechocystis sp. strain PCC 6803 has enabled us to develop a differential display procedure that takes advantage of a short palindromic sequence that is dispersed throughout the Synechocystis sp. strain PCC 6803 genome. This sequence, designated the HIP (highly iterated palindrome) element, occurs in approximately half of the Synechocystis sp. strain PCC 6803 genes but is absent in rRNA and tRNA genes. To determine the feasibility of exploiting the HIP element, alone or in combination with specific primer subsets, for analyzing differential gene expression, we used HIP-based primers to identify light intensity-regulated genes. Several gene fragments, including those encoding ribosomal proteins and phycobiliprotein subunits, were differentially amplified from RNA templates derived from cells grown in low light or exposed to high light for 3 h. One novel finding was that expression of certain genes of the pho regulon, which are under the control of environmental phosphate levels, were markedly elevated in high light. High-light activation of pho regulon genes correlated with elevated growth rates that occur when the cells are transferred from low to high light. These results suggest that in high light, the rate of growth of Synechocystis sp. strain PCC 6803 exceeds its capacity to assimilate phosphate, which, in turn, may trigger a phosphate starvation response and activation of the pho regulon.
Several techniques have been used to define differential gene expression in both prokaryotic and eukaryotic organisms; these include high-density microarrays (5–7, 15, 18, 24, 27, 34), subtractive libraries (22), and differential display (20). Generally, the use of these techniques to examine prokaryotic gene expression is more problematic than for the study of eukaryotic gene expression because prokaryotic mRNA is not polyadenylated and it is difficult to remove rRNA from prokaryotic RNA preparations.
Differential display, a powerful technique based on reverse transcriptase (RT)-mediated PCR (RT-PCR), permits rapid screening for genes that are expressed under specific conditions. It has been used extensively for the analysis of eukaryotic gene expression but to only a limited extent for examining gene expression in prokaryotes (2, 9–11, 17). The complete sequence of the Synechocystis sp. strain PCC 6803 genome has been elucidated; it predicts 3,168 potential open reading frames (ORFs) (16; Cyanobase [http://www.kazusa.or.jp /cyano]). A highly repeated, decameric palindromic sequence, 5′GGCGATCGCC, designated HIP1D (highly iterated palindrome), is dispersed throughout the Synechocystis sp. strain PCC 6803 genome with an average spacing of about 1.2 kbp (16). We will refer to HIP1D as the HIP element in this communication. Since the HIP element is absent in rRNA and tRNA genes, it serves as a feature of the genome that can be exploited for developing inexpensive strategies for analyzing global gene expression in Synechocystis sp. strain PCC 6803. In 1995, Robinson et al. showed that an octameric palindromic sequence (5′GCGATCGC) designated HIP1 occurred abundantly in several cyanobacterial genomes, including that of Synechococcus sp. strain PCC 6301, and suggested its use as a possible diagnostic tool (25).
In this study, we synthesized primers based on the decameric HIP element to identify genes specifically regulated when cells are transferred to high light (HL) from low light (LL). The primers included HIP elements with 3′ extensions to add specificity to the RT reactions and PCR amplifications. Furthermore, at two positions in the sequence we replaced G and C, which have the potential for strong pairing, with the weaker pairing A-T; this reduces the probability of duplex formation between the palindromic sequences. Even using a limited set of different primers, several differentially expressed genes were identified. The expression patterns of all genes identified by this procedure were confirmed by Northern blot hybridizations, RT-PCR, or RNase protection assays (RPAs). The results suggest that HIP element-based primers can be used alone or in combination with other synthetic primers to identify differentially regulated genes in Synechocystis sp. strain PCC 6803.
Some of the differentially expressed genes identified using HIP element-based differential display could have been predicted from previous work with cyanobacteria, while others were novel. For example, HL caused a decrease in the accumulation of the cpcBAHCD transcript and an increase in the level of the rpl11 or rpl1 transcript. Both of these cases corroborate what is already known about light responses of cyanobacteria and serve as proof-of-concept examples. However, unexpectedly, expression of the phoA gene, which encodes an alkaline phosphatase that was previously shown to be controlled by the level of phosphate in the environment (1, 21, 23, 36), was markedly elevated when the cells were exposed to HL. These results are discussed in the context of the acclimation of cells to both HL and nutrient limitation.
MATERIALS AND METHODS
Culture and growth conditions.
The nonmotile strain of Synechocystis sp. strain PCC 6803 (originally from John Williams) was obtained from Teruo Ogawa, Bioscience Centre, Nagoya University, Japan. Cells were grown in BG-11 medium in moderate light (70 μmol of photons m−2 s−1) at 30°C. The cultures were bubbled with 3% CO2 in air and harvested when they reached mid-logarithmic phase of growth (between 107 and 108 cells/ml). To elicit a light intensity-dependent change in the pattern of gene expression, Synechocystis sp. strain PCC 6803 cells were transferred from growth in 30 μmol of photons m−2 s−1 (i.e., LL conditions) to 500 μmol of photons m−2 s−1 for 3 h (or for periods of time as indicated in figure legends).
DNA and RNA isolation.
Molecular techniques were performed according to standard procedures (26). DNA was isolated from Synechocystis sp. strain PCC 6803 according to the method of Tandeau de Marsac et al. (32). RNA was isolated from pelleted cells frozen at −80°C, using a slight modification of the method of De Saizieu (8) as described by Bhaya et al. (3). Briefly, 500 μl of acidified phenol and 500 μl of NAES (50 mM sodium acetate [pH 5.1], 10 mM EDTA, 1% sodium dodecyl sulfate) were added to cell pellets from a 50-ml culture; after the addition of 100 mg of glass beads (0.1-μm, average diameter), the suspension was agitated in a Mini-Bead Beater (Biospec Products, Bartlesville, Okla.) three times for 20 s each at 5,000 rpm. This was followed by two phenol-chloroform (1:1) and one chloroform extraction. The RNA preparations were treated for 30 min at room temperature with RNase-free DNase I (20 U; Roche Molecular Biochemicals, Indianapolis, Ind.) followed by phenol-chloroform extraction (1:1) and precipitation in 2 volumes of ethanol. The final pellet was dissolved in 50 μl of 10 mM Tris (pH 8.0)–1 mM EDTA. RNA yields ranged from 150 to 250 μg from a 50-ml culture (approximately 5 × 108 cells).
RT-PCR.
The primers used for RT-PCRs were variations of HIP elements, as shown in Tables 1 and 2. For the RT reaction between 10 and 100 ng of RNA was incubated with 4 pmol of primer for 10 min at 70°C prior to adding 100 U of Superscript II RT (GIBCO BRL, Grand Island, N.Y.). The reaction was allowed to proceed for 50 min at 42°C and terminated by incubation at 72°C for 15 min; 1 μl of the RT reaction, which contained 0.5 to 2.5 ng of DNA, was used for PCR. Approximately 5 pmol of primer and 2 U of Platinum Taq DNA polymerase (GIBCO BRL) were used in a 25-μl reaction volume. PCR was initiated with a hot start step at 94°C for 120 s, followed by 30 cycles of 94°C for 30 s, 40°C for 30 s, 72°C for 150 s, and termination after the last cycle at 72°C for an additional 5 min. The samples were maintained at 4°C until they were analyzed on a 2% agarose gel in TAE (40 mM Tris-acetate [pH 8.0, 1 mM EDTA]) buffer; the amplified fragments were visualized by staining with ethidium bromide. For confirmation of differential expression of the cpcBACHD genes, we used the specific primer pair 5′AATTGCTTTCGGTCGTCTA-5′GCGTAATCGAGGTAGGA for RT-PCR. The conditions for RT-PCR were the same as described above.
TABLE 1.
Primers based on HIP
| Primer | No. of possibilities | Example(s) |
|---|---|---|
| 10-mer (HIP) | 1 | HIP |
| 11-mer (HIPX) | 4 | HIPA, HIPC, HIPG, HIPT |
| 12-mer (HIPXY) | 16 | HIPAA, HIPAC, HIPAG, etc. |
| 13-mer (HIPXYZ) | 64 | HIPAAA, HIPAAC, HIPAAG, etc. |
TABLE 2.
Modified HIP primers
| Modification | Sequencea (5′→3′) |
|---|---|
| HIP | GGCGATCGCC |
| W1-HIP | AGAGATCGCC |
| W2-HIP | WGWGATCGCC |
| W3-HIP | GWCWATCGCC |
| W4-HIP | GGWGATCGCC |
W = A or T.
Cloning and sequencing.
PCR fragments were extracted from the agarose gel using a gel extraction kit (Qiagen, Chatsworth, Calif.) and ligated into pGEM-T or pGEM-T Easy vectors (Promega, Madison, Wis.). The cloned fragments were sequenced from both directions with either the T7 or SP6 primer using the recommended Big Dye protocol (PE Biosystems, Foster City, Calif.).
Northern blot hybridizations.
Northern blot hybridizations and preparation of radiolabeled DNA probes were performed as previously described (3).
RPA.
RPAs were performed according to the protocol supplied with the Hyperspeed RPA kit (Ambion, Tex.). To amplify the rpl1 and rpl11 fragments from genomic DNA, the primer pairs 5′AGGTAGATGACAGCAAAC-5′GTGGCCTCCTTGACCTTT and 5′AAAGTCGTCGCTCTGATT-5′TGCCATGATATTAACCCC, respectively, were used. The PCR products were ligated into the pGEM-T Easy vector, and the labeled RNA (antisense) probe was synthesized with a STRIP-EZ RNA kit (Ambion), using the T7 promoter for rpl11 and the SP6 promoter for rpl1. Labeled antisense RNA was incubated with 5 μg of total RNA (DNase treated), and the assay along with the appropriate controls were performed according to recommendations of the manufacturer. A portion of each RNA sample was subjected to electrophoresis on a formaldehyde agarose gel to confirm the RNA concentration, which was also determined spectrophotometrically.
RESULTS
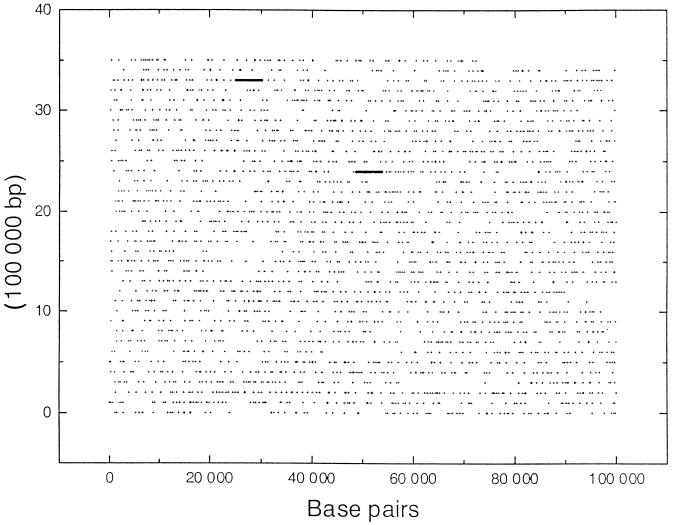
The palindromic, decameric HIP element, 5′GGCGATCGCC, occurs a total of 2,823 times throughout the Synechocystis sp. strain PCC 6803 genome, of which 2,562 (or 90.7%) are located in ORFs (14a, 16). The genome wide distribution of HIP elements is shown in Fig. 1 (positions of the HIP elements are available at http://www.sb-roscoff.fr/Phyto/Syn6803_HIP_sequences.html). Of the 3,168 ORFs identified in the genome, 1,653 (52.2%) contain HIP elements; 1,008 contain a single HIP element, while 645 contain multiple HIP elements. Within a coding region, the position of a HIP element appears to be random. Notably, it is absent in all rRNA and tRNA genes as well as in genes encoding transposases. It is significantly underrepresented among genes encoding chaperones, ribosomal proteins, proteins involved in photosynthetic function, and certain hypothetical ORFs (D. Vaulot, A. R. Grossman, D. Bhaya, J. Mrázek, and S. Karlin, unpublished data).
FIG. 1.
Positions of HIP elements in the Synechocystis sp. strain PCC 6803 genome. The position of each HIP element is marked with a cross (+). The sequence begins at the lower left (nucleotide 1 of the sequence) and ends at the top right (nucleotide 3573470). Positions of the rRNA genes which lack HIP elements are shown by thick lines.
Initial experiments were performed to test the use of HIP primers for the implementation of differential display in Synechocystis sp. strain PCC 6803. Two types of modifications of HIP primers were used in these experiments. First, the primers were extended at their 3′ ends by one, two, and three nucleotides (HIPX, HIPXY, and HIPXYZ, respectively) to add specificity to the RT reactions and/or PCR (Table 1). Second, because the primers are palindromes and tend to anneal to each other, we modified positions by changing strongly pairing bases (G and C) to the weakly pairing bases (A and T); four different kinds of modifications (designated W1-HIP, W2-HIP, W3-HIP, and W4-HIP; [Table 2]) were incorporated into the primers.
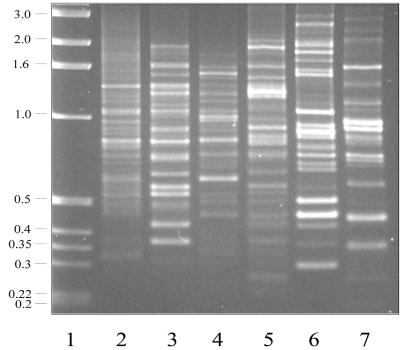
Figure 2 shows PCR of total genomic DNA using the primers HIPA, HIPC, W2-HIPA, W2-HIPC (11-mers), W2-HIPAC (12-mer), and HIPACC (13-mer). Each of the primers, when used separately, amplified several specific genomic DNA fragments. The inclusion of modified primers (compare lanes 2 and 4 or lanes 3 and 5) in the reaction mixture tends to bias the amplification of specific fragments. Furthermore, results obtained with the modified primers were more reproducible than those generated using nonmodified primers; thus, in most experiments only the former were used. Under the conditions used, we generally obtained PCR products ranging in size from 0.2 to 3.0 kbp.
FIG. 2.
PCR from genomic DNA using different HIP primers. HIP primers were used for PCR with 5 ng of genomic DNA template. Primers used were HIPA (lane 2), HIPC (lane 3), W2-HIPA (lane 4), W2-HIPC (lane 5), W2-HIPAC (lane 6), and HIPACC (lane 7). Molecular weight markers, in kilobase pairs, are shown in lane 1. PCR conditions are described in Materials and Methods.
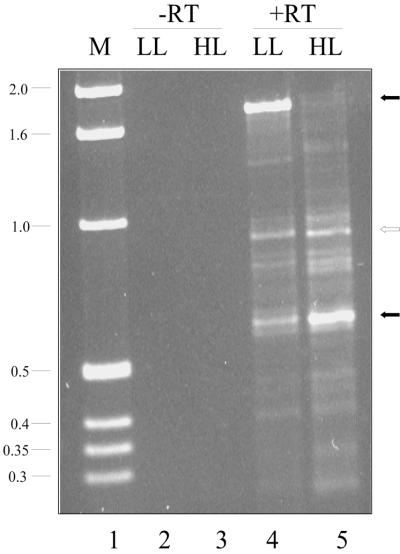
To determine if the HIP elements could be used for generating and amplifying cDNAs, specific HIP primers were used to reverse transcribe total Synechocystis sp. strain PCC 6803 mRNA and then to amplify the cDNA product. In the initial experiments, the same primer was used for both the RT reaction and PCR. To identify genes that are differentially expressed with respect to light intensity, RNA was isolated from cells grown in LL (30 μmol of photons m−2 s−1) or from cells exposed to HL (500 μmol of photons m−2 s−1) for 3 h after growth in LL. Figure 3 shows the amplified products when the primer W2-HIPG was used for both the RT reaction and PCR. No products are observed in the no-RT control (lanes 2 and 3). Some of the products generated from the RNA isolated from cells grown in LL or exposed to HL were the same in size and intensity, suggesting that they did not originate from differentially expressed genes. However, there were also products that exhibited differential accumulation between the two conditions. For instance, there was a strong band at 1.75 kbp in LL but not HL samples (lanes 4 and 5). Conversely, a fragment of approximately 0.6 kbp was present at a higher level in HL samples than in LL samples.
FIG. 3.
Differential display of W2-HIPG-primed RNA from cells grown in LL and exposed to HL. RT-PCR was performed as described in Materials and Methods with 100 ng of RNA from cells either grown in LL (lane 4) or exposed to HL for 3 h (lane 5). In control reactions (in which the RT step was omitted [-RT]), RNA from LL-grown (lane 2) and HL-exposed (3 h; lane 3) cells was used. Filled arrows mark products that differentially accumulate; the open arrow marks a product that does not differentially accumulate. Molecular weight markers, in kilobase pairs, are shown in lane 1.
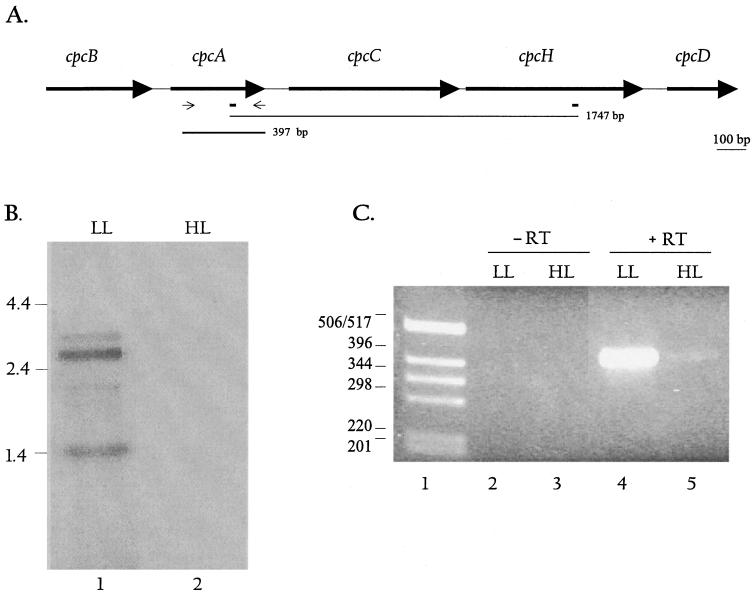
The differential display product of 1.75 kbp was cloned into the pGEM T-Easy vector, and the sequences at both ends of the cloned fragment were determined. This identified the fragment as part of the cpcBACHD operon, which contains genes encoding phycocyanin, and the phycocyanin linker polypeptides. We could locate the two W2-HIPG-like priming sites (5′CTTGATCGCCG and 5′CGGCGATCGTG, where underlined letters represent deviations from the W2-HIPG sequence) on the genomic DNA; the distance between them is 1,747 nucleotides, which corresponds well with the size of the differential display product. Figure 4A shows the cpcBACHD operon (sll1577 to sll1580 and ssl3093) and the locations of W2HIP-G sequence. The amplified sequence starts within cpcA, spans cpcC, and ends within cpcH, strongly suggesting that these genes are transcribed together and are part of an operon. Previous work has demonstrated that cpc genes are clustered into operons in a number of different organisms (13). Furthermore, accumulation of transcripts encoding phycobiliproteins has been shown to be regulated by both light intensity and in some cases light quality (4, 12, 14, 19, 31, 33). In general, cells grown in HL have significantly lower levels of the transcripts encoded by the cpcBA operon than cells grown in LL.
FIG. 4.
Map and expression of the cpcBACHD operon. (A) The map of the cpcBACHD operon shows the relative positions and sizes of the five cpc genes. The positions of the W2-HIPG-like elements are shown as rectangles below the map. The thin line below the map depicts the position and size of the differentially displayed product, while the thick line shows the size and position of the cpcA gene-specific probe. Thin arrows indicate positions of primers used to generate the cpcA-specific probe. (B) Northern blot hybridization using the cpcA-specific probe. Each lane contained 5 μg of RNA. Lanes 1 and 2 show signals from RNA isolated from LL- and HL-grown cells, respectively, hybridized to the cpcA probe. The major transcripts in lane 1 are 3.5, 2.6, 2.0, and 1.4 kb. RNA size markers are given on the left. (C) RT-PCR was performed with RNA from LL- and HL-grown cells using cpcA-specific primers; 100 ng of RNA was used for the reaction, as described in Materials and Methods. Lanes 2 and 3 show the no-RT control; Lanes 4 and 5 show the RT-PCR product (397 bp) generated from LL- and HL-grown cells, respectively.
To confirm that the cpcBACHD transcripts decline upon exposure of cells to HL conditions, we performed Northern blot hybridizations (Fig. 4B). The RNA was hybridized with a 397-bp gene fragment specific for the cpcA gene which overlaps with the 1.75-kbp fragment that was identified by differential display (the position of the fragment is shown in Fig. 4A). The gene-specific probe hybridized to a number of transcripts (with sizes of approximately 3.5, 2.6, 2.0, and 1.4 kb) that are synthesized from the cpcBACHD operon (lane 1) in LL. The 3.5-kb transcript may be just long enough to encode all of the genes of the operon (cpcBACHD). The shorter transcripts may cover a subset of these genes; for instance, the strong transcript of 1.4 kb may cover cpcBA. Barely detectable transcript levels are present in cells exposed to HL for 3 h (lane 2). RT-PCR using primers specific for the cpcA gene (positions of the primers are shown in Fig. 4A) also confirmed that expression declined dramatically upon exposure of cells to HL (Fig. 4C). While there were no bands in lanes 2 and 3 (no-RT controls), a strong band of 397 bp was present in lane 4 (LL) and a faint band was present in lane 5 (HL). In fact, a 30-min exposure to HL caused a marked reduction in transcript levels (data not shown).
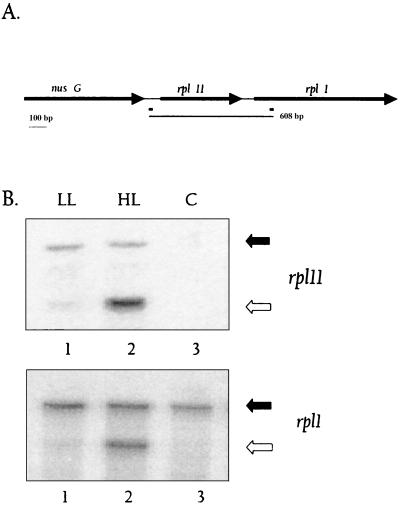
Results presented in Fig. 3 also show a product of approximately 0.6 kbp that was significantly elevated when RNA from cells exposed to HL was used for differential display (compare lanes 4 and 5). This fragment was cloned, sequenced, and determined to have originated from the tandemly arranged genes encoding ribosomal protein subunits Rpl1 and Rpl11. A map showing the arrangement of the rpl1 and rpl11 genes and positions of the W2-HIPG sequences is shown in Fig. 5A. The amplified fragment encompassed the entire rpl11 gene and the 5′ end of the rpl1 gene. These results strongly suggest that these ribosomal protein genes increase in expression upon exposure of Synechocystis sp. strain PCC 6803 to HL and that rpl11 and rpl1 are cotranscribed. RPAs were performed to confirm these results and to quantify levels of the RNAs encoding Rpl11 and Rpl1. As shown in Fig. 5B, the protected fragments specific for rpl1 and rpl11 were three- and sixfold higher, respectively, when RNA from cells exposed to HL (lane 2) was used in the assay relative to RNA from LL-grown cells (compare lanes 1 and 2 in the top and bottom panels).
FIG. 5.
Map and expression of the rpl11-rpl1 operon. (A) The map of the rpl11-rpl1 operon shows the relative positions and sizes of nusG, rpl11, and rpl1. The positions of the W2-HIPG elements are shown as black rectangles below the map. The thin line below the map represents the size and position of the differentially displayed product. (B) Probes specific for rpl11 and rpl1 were made as described in Materials and Methods and used for RPA. In the RPA for rpl11 (top), the undigested probe migrates at 162 bp and is indicated with a filled arrow. A strongly protected fragment of 102 bp, indicated by an open arrow, is seen in lane 2 (HL), with a much fainter protected fragment in lane 1 (LL). Similar results are observed in the lower panel, in which a rpl1-specific probe was used (sizes of the unprotected and protected fragments are 304 and 223 bp, respectively). Lanes 3 in both panels show the control (C) (total RNA replaced with yeast RNA), in which no protected fragment is seen.
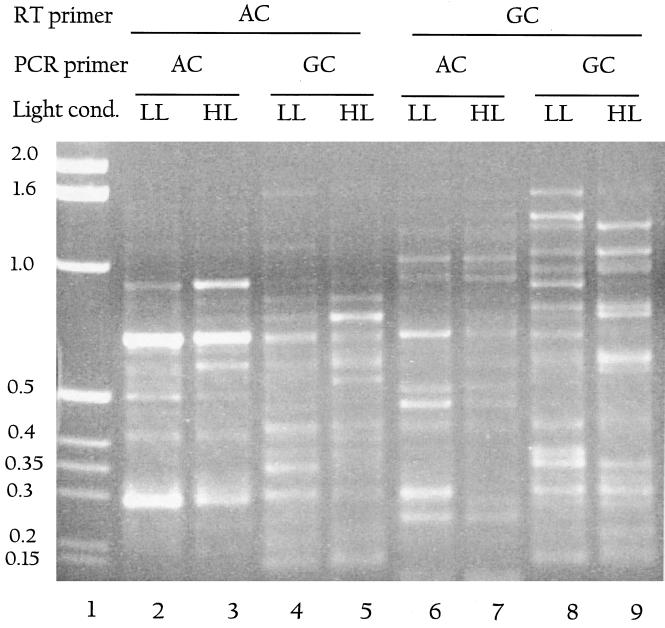
To increase the range of differential display products generated, we used combinations of primers for RT reactions and PCR on RNA from LL- and HL-grown cells (Fig. 6). W2-HIPAC (lanes 2 to 5) or W2-HIPGC (lanes 6 to 9) was used for the RT reaction followed by PCR in which either W2-HIPAC or W2-HIPGC was used. As a control, PCR was performed directly on RNA that was not reverse transcribed (data not shown). We found that depending on the types and combinations of primers used, we either get no products or a few products in our no-RT control. But even in cases where there are products in the control, the pattern changes significantly when the RT step is included. While some of the amplified products show similar levels of accumulation in LL and HL, others either change in intensity or are specific to LL or HL. Several of these products were sequenced, including fragments from the genes listed as slr2123/2124, sll1069/1070, slr1968, slr0872, slr1841 sll0328, and sll1452 (these are reduced in HL) and slr1277, sll0721, slr1923, slr0724, and sll0533 (increased in HL). We have not yet confirmed these results with secondary tests. However, these and other data were used to ascertain the percentage of mismatches between the HIP primers used and the genomic sequences to which they hybridized. In most cases we found that the mismatches were at weakly pairing bases, implying that the use of weakly pairing bases increased the number of genes identified by this method. Almost no mismatches were found at other positions (data not shown). Of the more than 20 differentially displayed products we have sequenced, only two turned out to be within rRNA.
FIG. 6.
Use of HIP primer combinations for differential display. RT-PCR conditions as described in Materials and Methods were used. W2-HIPAC was used as the RT primer in lanes 2 to 5, while W2-HIPGC was used as the RT primer in lanes 6 to 9. This was followed by PCR using either W2-HIPAC (lanes 2, 3, 6, and 7) or W2-HIPGC (lanes 4, 5, 8, and 9). Alternate rows are from LL (lanes 2, 4, 6, and 8) or HL (lanes 3, 5, 7, and 9). Molecular weight markers are shown in kilobase pairs in lane 1. Several putative differentially displayed products are visible in all lanes.
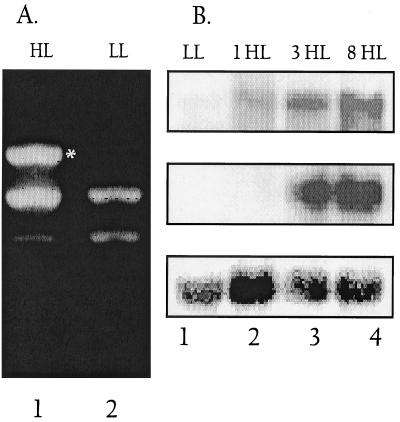
Another example of a product that preferentially accumulated when RNA from HL-grown cells was used as a template for RT-PCR is shown in Fig. 7A. In this case, HIPAAA was used for both the RT reaction and PCR. A band of approximately 1 kbp, absent from reactions performed with RNA from LL-grown cells, was a dominant product when RNA from HL-grown cells was used for RT-PCR (Fig. 7A, compare lanes HL and LL). The fragment was cloned, sequenced, and found to encode part of a putative alkaline phosphatase (sll0654). The sll0654 gene contains two HIPAAA-like sequences separated by 1,075 bp (5′GGCGATCGCCAAA at position 2,793 of the ORF and 5′CTTGGCGATCGCC at position 3868). It was surprising to find that light has a major influence on the level of the alkaline phosphatase transcript; however, the RT-PCR results were confirmed by Northern blot hybridizations, as shown in Fig. 7B. The alkaline phosphatase transcript (the size of the transcript is ∼4.3 kb, which is close to the size of the gene, 4.23 kbp) accumulated after 1 h of exposure to HL and was still abundant after 8 h in HL (Fig. 7B, top panel). Transcripts from the gene encoding PstS (sll0680), known to be part of the phosphate transport system, also accumulated when cells were exposed to HL (middle panel). The kinetics of accumulation were somewhat different from those of the alkaline phosphatase mRNA; the mRNA was low after 1 h of HL but continued to increase for 8 h. Both of these genes also become active when cells are starved for phosphorus (data not shown). These results clearly show that genes of the pho regulon are activated when Synechocystis sp. strain PCC 6803 is exposed to HL.
FIG. 7.
Differential expression of genes in the pho regulon with respect to light intensity. (A) Differential display shows in the induction of the phoA gene (encoding alkaline phosphatase) in HL. LL- or 18-h HL-grown cells were used for RNA isolation. RT-PCR was performed using HIPAAA and conditions described in Materials and Methods. Lanes 1 and 2 show products from LL- HL-grown cells, respectively. A band at 1 kbp is visible only in lane 2 (marked with an asterisk), while bands at 800 and 750 bp appear in both lanes (including in the no-RT control lanes [data not shown]). (B) Gene-specific probes were used for hybridizations to phoA (top panel) and pstS (middle panel) transcripts. RNA was isolated from LL-grown cells (lane 1) or cells transferred to HL for 1 h (lane 2), 3 h (lane 3), or 8 h (lane 4). Blots were also probed with rDNA-specific probes as a loading control (lower panel).
DISCUSSION
There are obstacles to the use of differential display to study gene expression in bacteria. Most of these difficulties stem from the fact that bacterial mRNAs do not have a poly(A) tail, making it impossible to use oligo(dT) for the RT reaction (as done for differential display of eukaryotic mRNA) that generates the template for the subsequent PCR. Thus, either random primers or specific sets of pooled primers must be used for the RT reactions. Random primers are likely to anneal to the rRNA, which could constitute up to 98% of the RNA in the reaction mixture; this could strongly skew the population of mRNAs that is effectively converted into cDNAs.
The use of the HIP element to generate primers helps eliminate problems discussed in the preceding paragraph since this element does not readily anneal to either the 16S or 23S rRNA. However, while using HIP elements as primers for differential display with Synechocystis sp. strain PCC 6803 mRNA has advantages, it also has distinct shortcomings. First, the HIP element is present in only 52% of the ORFs, and thus several genes and some gene categories will not be readily amplified by HIP elements. The Synechocystis sp. strain PCC 6803 genes encoding photosynthetic proteins, ribosomal proteins, and transposases are nearly devoid of HIP elements. It is not known why certain classes of genes lack HIP elements. Furthermore, the functional significance of the HIP element has not been explored, although it has been suggested that it may play a role in recombination (16). There are small repeat elements in several bacterial genomes, and in at least one case, they have been assigned a function. In Haemophilus influenzae, the 9-bp uptake sequence (5′AAGTGCGGT) is known to be involved in natural competence for the recognition and efficient uptake of homospecific DNA (29, 30). However, the characteristics of uptake sequence repeats and their genomic organization are very different from those of HIP elements.
Our major concern in using HIP elements for differential display was that not all of the genes would be amenable to amplification. However, our initial data suggested that the number of genes that can potentially be identified as differentially regulated may significantly exceed the number predicted based simply on the number of genes that contain two or more HIP elements. This stems from modifications that we have made to the initial methodology. Of these, the most significant is the substitution of G-C base pairs at specific positions in the HIP element with the weakly pairing A and T bases. Although the substitutions were primarily included to reduce the chances of self-annealing of the palindromic HIP primers, the modified primers are able to anneal to an increased number of sites within the mRNA population. These primers anneal to several mRNAs that contain HIP-like elements (sequences that do not precisely match the HIP palindrome); a certain degree of mismatch appears to be tolerated in both the RT reaction and subsequent PCR. The practical outcome of this result is that we observe a greater number of amplified products following differential display.
Another important observation is that in some instances we observe amplified products derived from polycistronic mRNAs. The fragment of the gene amplified from mRNA derived from the cpcBACDH operon (down-regulated in HL) spanned cpcA and cpcB. Additionally, up-regulation of the ribosomal protein genes in HL was reflected in an amplified fragment that spanned rpl1 and rpl11. In fact, it has been shown that a cluster of genes in this region may be transcribed as a 9.5-kbp transcript which includes rpl11, rpl1, rpl10, rpl12, and aroC (28). Once it is confirmed that a fragment isolated by differential display that spans more than one gene is not a false positive resulting from amplification of contaminating genomic DNA (as was done for the two cases mentioned above), the data will help elucidate operonic structure of the genome. Furthermore, the fact that many cyanobacterial genes are cotranscribed will increase the theoretical number of amplified fragments using single primers since products will be generated if the primers anneal to sites within clusters of genes that are part of a single transcript.
Identification of differentially expressed genes and operons that contain either no HIP (or HIP-like) elements, a single HIP element, or nonidentical HIP elements will require strategies that amplify a broader set of mRNAs. Different HIP primers can be used in combination with each other, and individual HIP primers can be used in combination with sets of random primers for both the RT reaction and PCR. Furthermore, other methods of differential display have recently been developed in which random primers are used to synthesize cDNAs, followed by restriction endonuclease treatment of the cDNAs. The restriction fragments generated are ligated to specific adapter sequences, which are then PCR amplified (Display Systems Biotech, Vista, Calif.). By combining the different approaches, we should be able to assay a large subset of Synechocystis sp. strain PCC 6803 genes for differential gene expression. Another possibility is to use currently available techniques that might allow for the polyadenylation of the bacterial mRNAs (2). If this approach were specific, it would increase the spectrum of mRNAs that can be easily screened for expression.
Finally, high-density DNA microarrays can also be used to evaluate global gene expression in Synechocystis sp. strain PCC 6803. Although such an approach has the greatest potential for the analysis of genomewide gene expression, it is very costly and there can be difficulties in obtaining strong signals for many of the genes. These difficulties arise because the mRNAs are not readily separated from rRNA, and many of the primers used to synthesize the fluorescently labeled cDNAs anneal to rRNA, which can create a strong bias in the hybridization signals. Furthermore, signals for many genes that are not highly transcribed may not be intense enough relative to background fluorescence emitted from the coating on the chip to evaluate expression levels.
Even using single HIP elements for both the RT reaction and PCR enabled us to identify genes controlled by light levels, providing proof-of-concept examples. Genes encoding phycocyanin subunits were shown to be strongly down-regulated in HL. In HL, the cells do not require much light harvesting capacity since absorption of the light by reaction center and core antenna chlorophyll would be enough to saturate photosynthetic electron flow. Cells grown in HL exhibit a marked reduction in the levels of the phycobiliproteins. As shown here, this is reflected in greatly reduced mRNA levels, although reduced synthesis of the phycobiliproteins immediately following exposure of cells to HL (when phycobiliprotein mRNA is still present) suggests that the synthesis of the phycobiliproteins is also regulated at the level of translation (N. Dolganov and A. R. Grossman, unpublished data). Furthermore, genes encoding the ribosomal proteins Rpl1 and Rpl11 were shown to be strongly up-regulated in HL. When LL-grown Synechocystis sp. strain PCC 6803 was transferred to HL, its doubling time decreased from approximately 10 h to approximately 4 h. Increased synthesis of ribosomes and mRNAs encoding ribosomal polypeptides was also shown to accompany an elevated rate of Escherichia coli growth (34).
A number of other genes that we have isolated by the differential display procedure also appear to be modulated by light levels. For example, some transcripts encoding specific enzymes of key metabolic pathways, such as a transketolase (sll1070) or components of the secretory pathway (gspD or slr1277), appear strongly modulated by light, but we have not yet investigated the significance of this result. The most surprising finding of this study is that genes that consititute the pho regulon exhibit increased expression when Synechocystis sp. strain PCC 6803 is exposed to HL. How do we explain the HL activation of genes present in the pho regulon? In HL in liquid medium, during rapid cell proliferation (doubling time of 4 to 5 h), the acquisition of phosphate may not be able to keep pace with the growth potential of the cells; initially, cells use the phosphate more rapidly than it can be acquired. Therefore, although there are high levels of phosphate in the medium (1 mM), the cells are starved for phosphorus, which could lead to activation of the pho regulon. An alternative possibility is that the pho regulon is directly activated by high light, perhaps via a SphR/SphS-like regulator/sensor pair (1, 21). Thus, it is interesting that SphS and its homolog in Synechocystis sp. strain PCC 6803, sll0337, may contain a somewhat diverged PAS domain (Igor Zhulin, personal communication). PAS domains are cytosolic sensor modules that monitor changes in light, redox potential, or oxygen (35). These unexpected results serve to illustrate two points: (i) differential display may allow us novel insights into the interactions between various stress conditions, although (ii) such interactions may be a consequence of indirect effects.
ACKNOWLEDGMENTS
We thank Samuel Karlin and Jan Mrázek, Department of Mathematics, Stanford University, for helping with analysis of the HIP elements. We had several helpful discussions with Jeffrey Shrager, Chung-Soon Im, Chao-Jung Tu, and Hideki Takahashi concerning both the procedure and the data that were generated.
This work was supported by National Science Foundation grant MCB 9727836 and USDA grant 98-35301-6445 to A.R.G. as well as a bilateral NSF-CNRS grant to A.R.G. and D.V. The sabbatical stay of D.V. was supported by a NATO fellowship.
Footnotes
Carnegie Institution of Washington publication no. 1436.
REFERENCES
- 1.Aiba H, Nagaya M, Mizuno T. Sensor and regulator proteins from the cyanobacterium Synechococcus species PCC7942 that belong to the bacterial signal-transduction protein families: implication in the adapative response to phosphate limitation. Mol Microbiol. 1993;8:81–91. doi: 10.1111/j.1365-2958.1993.tb01205.x. [DOI] [PubMed] [Google Scholar]
- 2.Amara R R, Vijaya S. Specific polyadenylation and purification of total messenger RNA from Escherichia coli. Nucleic Acids Res. 1997;25:3465–3470. doi: 10.1093/nar/25.17.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhaya D, Watanabe N, Ogawa T, Grossman A R. The role of an alternate sigma factor in motility and pili formation in the cyanobacterium Synechocystis sp. strain PCC6803. Proc Natl Acad Sci USA. 1999;96:3188–3193. doi: 10.1073/pnas.96.6.3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell D, Eriksson M J, Oquist G, Gustafsson P, Clarke A K. The cyanobacterium Synechococcus resists UV-B by exchanging photosystem II reaction-center D1 proteins. Proc Natl Acad Sci USA. 1998;95:364–369. doi: 10.1073/pnas.95.1.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castellino A M. When the chips are down. Genome Res. 1997;7:943–946. doi: 10.1101/gr.7.10.943. [DOI] [PubMed] [Google Scholar]
- 6.Cho R J, Campbell M J, Winzeler E A, Steinmetz L, Conway A, Wodicka L, Wolfsberg T G, Gabrielian A E, Landsman D, Lockhart D J, Davis R W. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell. 1998;2:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- 7.DeRisi J L, Iyer V R, Brown P O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 8.De Saizieu A, Certa U, Warrington J, Gray C, Keck W, Mous J. Bacterial transcript imaging by hybridization of total RNA to oligonucleotide arrays. Nat Biotechnol. 1998;16:45–48. doi: 10.1038/nbt0198-45. [DOI] [PubMed] [Google Scholar]
- 9.Fislage R. Differential display approach to quantitation of environmental stimuli on bacterial gene expression. Electrophoresis. 1998;19:613–616. doi: 10.1002/elps.1150190426. [DOI] [PubMed] [Google Scholar]
- 10.Fislage R, Berceanu M, Humboldt Y, Wendt M, Oberender H. Primer design for a prokaryotic differential display RT-PCR. Nucleic Acids Res. 1997;25:1830–1835. doi: 10.1093/nar/25.9.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fleming J T, Yao W-H, Sayler G S. Optimization of differential display of prokaryotic mRNA: application to pure culture and soil microcosms. Appl Environ Microbiol. 1998;64:3698–3706. doi: 10.1128/aem.64.10.3698-3706.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujita Y, Murakami A, Aizawa K, Ohki K. Short-term and long-term adaptation of the photosynthetic apparatus: homeostatic properties of thylakoids. In: Bryant D A, editor. The molecular biology of cyanobacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 677–692. [Google Scholar]
- 13.Grossman A R, Bhaya D, Apt K E, Kehoe D M. Light-harvesting complexes in oxygenic photosynthesis: Diversity, control and evolution. Annu Rev Genet. 1995;29:231–287. doi: 10.1146/annurev.ge.29.120195.001311. [DOI] [PubMed] [Google Scholar]
- 14.Grossman A R, Kehoe D M. Phosphorelay control of phycobilisome biogenesis during complementary chromatic adamptation. Photosynth Res. 1997;53:95–108. [Google Scholar]
- 14a.Karlin S, Mrazek J, Campbell A M. Frequent oligonucleotides and peptides of the Hemophilus influenzae genome. Nucleic Acids Res. 1996;24:4263–4272. doi: 10.1093/nar/24.21.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kehoe D M, Villand P, Somerville S. DNA microarrays for studies of higher plants and other photosynthetic organisms. Trends Plant Sci. 1999;4:38–41. doi: 10.1016/s1360-1385(98)01354-5. [DOI] [PubMed] [Google Scholar]
- 16.Kotani H, Tabata S. Lessons from sequencing of the genome of a unicellular cyanobacterium, Synechocystis sp. PCC6803. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:151–171. doi: 10.1146/annurev.arplant.49.1.151. [DOI] [PubMed] [Google Scholar]
- 17.Kwaik Y, Pederson L. The use of differential display-PCR to isolate and characterize a Legionella pneumophila locus induced during the intracellular infection of macrophages. Mol Microbiol. 1996;21:543–556. doi: 10.1111/j.1365-2958.1996.tb02563.x. [DOI] [PubMed] [Google Scholar]
- 18.Lashkari D A, DeRisi J L, McCusker J H, Namath A F, Gentile C, Hwang S Y, Brown P O, Davis R W. Yeast microarrays, for genome wide parallel genetic and gene expression analysis. Proc Natl Acad Sci USA. 1997;94:13057–13062. doi: 10.1073/pnas.94.24.13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemaux P G, Grossman A R. Major light-harvesting polypeptides encoded in polycistronic transcript in eukaryotic algae. EMBO J. 1985;4:1911–1919. doi: 10.1002/j.1460-2075.1985.tb03870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang P, Pardee A B. Differential display methods and protocols. Totowa, N.J: Humana Press; 1997. [Google Scholar]
- 21.Nagaya M, Aiba H, Mizuno T. The sphR product, a two-component system response regulator protein, regulates phosphate assimilation in Synechococcus sp. strain PCC 7942 by binding to two sites upstream from the phoA promoter. J Bacteriol. 1994;176:2210–2215. doi: 10.1128/jb.176.8.2210-2215.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plum G, Clark Curtiss J E. Induction of Mycobacterium avium gene expression following phagocytosis by human macrophages. Infect Immun. 1994;62:476–483. doi: 10.1128/iai.62.2.476-483.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ray J M, Bhaya D, Block M A, Grossman A R. Isolation, transcription, and inactivation of the gene for an atypical alkaline phosphatase of Synechococcus sp. strain PCC 7942. J Bacteriol. 1991;173:4297–4309. doi: 10.1128/jb.173.14.4297-4309.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richmond C S, Glasner J D, Mau R, Jin H F, Blattner F R. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 1999;27:3821–3835. doi: 10.1093/nar/27.19.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson N J, Robinson P J, Gupta A, Bleasby A J, Whitton B A, Morby A P. Singular over-representation of an octameric palindrome, HIPI, in DNA from many cyanobacteria. Nucleic Acids Res. 1995;23:729–735. doi: 10.1093/nar/23.5.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Schena M, Shalon D, Heller R, Chai A, Brown P O, Davis R W. Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc Natl Acad Sci USA. 1996;93:10516–10619. doi: 10.1073/pnas.93.20.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt J, Bubenenko M, Subramanian A R. A novel operon organization involving the genes for chorismate synthase (aromatic biosynthesis pathway) and ribosomal GTPase center proteins (L11, L1, L10, L12: rplKAJL) in cyanobacterium Synechocystis PCC 6803. J Biol Chem. 1993;268:27447–27457. [PubMed] [Google Scholar]
- 29.Smith H O, Gwinn M S, Salzberg S L. DNA uptake signal sequences in naturally transformable bacteria. Res Microbiol. 1999;150:603–616. doi: 10.1016/s0923-2508(99)00130-8. [DOI] [PubMed] [Google Scholar]
- 30.Smith H O, Tomb J-F, Dougherty B A, Fleischmann R D, Venter J C. Frequency and distribution of DNA uptake signal sequences in the Haemophilus influenzae Rd genome. Science. 1995;269:538–540. doi: 10.1126/science.7542802. [DOI] [PubMed] [Google Scholar]
- 31.Sobczyk A, Schyns G, Tandeau de Marsac N, Houmard J. Transduction of the light signal during complementary chromatic adaptation in the cyanobacterium Calothrix sp. PCC7601: DNA-binding proteins and modulation by phosphorylation. EMBO J. 1993;12:997–1004. doi: 10.1002/j.1460-2075.1993.tb05740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tandeau de Marsac N, Borrias W E, Kuhlemeier C J, Castets A M, van Arkel G A, van den Hondel C A M J J. A new approach for molecular cloning in cyanobacteria: cloning of an Anacystis nidulans met gene using a Tn901-induced mutant. Gene. 1982;20:111–119. doi: 10.1016/0378-1119(82)90092-0. [DOI] [PubMed] [Google Scholar]
- 33.Tandeau de Marsac N, Houmard J. Adaptation of cyanobacteria to environmental stimuli: new steps towards molecular mechanisms. FEMS Microbiol Rev. 1993;104:119–190. [Google Scholar]
- 34.Tao H, Bausch C, Richmond C, Blattner F R, Conway T. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J Bacteriol. 1999;181:6425–6440. doi: 10.1128/jb.181.20.6425-6440.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor B B, Zhulin I B. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wanner B L. Gene regulation by phosphate in enteric bacteria. J Cell Biochem. 1993;51:47–54. doi: 10.1002/jcb.240510110. [DOI] [PubMed] [Google Scholar]